Abstract
Since the first diacylglycerol acyltransferase (DGAT) gene was characterized in plants, a number of studies have focused on understanding the role of DGAT activity in plant triacylglycerol (TAG) biosynthesis. DGAT enzyme is essential in controlling TAGs synthesis and is encoded by different genes. DGAT1 and DGAT2 are the two major types of DGATs and have been well characterized in many plants. On the other hand, the DGAT3 and WS/DGAT have received less attention. In this study, we present the first general view of the presence of putative DGAT3 and WS/DGAT in several plant species and report on the diversity and evolution of these genes and its relationships with the two main DGAT genes (DGAT1 and DGAT2). According to our analyses DGAT1, DGAT2, DGAT3 and WS/DGAT are very divergent genes and may have distinct origin in plants. They also present divergent expression patterns in different organs and tissues. The maintenance of several types of genes encoding DGAT enzymes in plants demonstrates the importance of DGAT activity for TAG biosynthesis. Evolutionary history studies of DGATs coupled with their expression patterns help us to decipher their functional role in plants, helping to drive future biotechnological studies.
Keywords: Triacylglycerol biosynthesis, DGAT, phylogeny, gene structure
Introduction
Triacylglycerols (TAGs) are the major seed storage lipids, providing carbon and energy reserves to support the growth of the seedling during germination (Lisa et al., 2009). TAGs are also important for pollen development and sexual reproduction in many plant species (Wolters-Arts et al., 1998; Zheng et al., 2003; Zhang et al., 2009). These storage lipids have been intensely explored as a source of edible oils for human consumption and have also been increasingly used for non-food applications, such as fuel and industrial feedstocks (Lung and Weselake,, 2006; Durrett et al., 2008; Dyer et al., 2008). Oilseeds primarily accumulate five common fatty acids, namely saturated palmitic acid (C16:0), stearic acid (C18:0), unsaturated oleic acid (C18:1), and the polyunsaturated linoleic (C18:2) and α-linolenic acid (C18:3) (Millar et al., 2000; Cagliari et al., 2011). In addition, some plant species are able to accumulate high amounts of unusual fatty acids, such as hydroxy (Ricinus communis) (Li et al., 2010), epoxy (Vernonia galamensis) (Yu et al., 2008) or acetylenic fatty acids (Euonymus alatus) (Durrett et al., 2010).
Although multiple pathways for TAG biosynthesis have been described in different organisms and tissues (Liu et al., 2012), the Kennedy or sn-glycerol-3-phosphate (G3P) (Kennedy and Weiss, 1956) is the canonical pathway leading to TAG synthesis. In this pathway, the biosynthesis of TAG occurs through successive acylation reactions, which begins with the trans esterification of acyl-CoA to glycerol-3-phosphate to form phosphatidic acid (PA) through the action of glycerol-3-phosphate acyltransferase (G3PAT; EC 2.3.1.15) and lysophosphatidic acid acyltransferase (LPAAT; EC 2.3.1.51) enzymes. Subsequently, PA is dephosphorylated to generate diacylglycerol (DAG), which is converted to TAG through the action of acylCoA:diacylglycerol acyltransferase (DGAT; EC 3.2.1.20) (Kenedy and Weiss, 1956; Ohlrogge and Browse, 1995). Some studies about evolutionary history of Kennedy pathway enzymes were performed in the last years (Turchetto-Zolet et al., 2011; Smart et al., 2014, Korbes et al., 2016). DGAT is considered a key enzyme in the conversion of DAG to TAG and therefore has been proposed as the rate-limiting enzyme in plant storage lipid accumulation (Ichihara et al., 1988; Perry and Harwood, 1993). DGAT activity was first reported by Weiss et al. (1960), and in the last decade, genes encoding DGAT enzymes have been identified and studied in a variety of plant species (Hobbs et al., 1999; Hobbs and Hills, 2000; He et al., 2004a,b, 2006; Kroon et al., 2006; Chen et al., 2007; Cagliari et al., 2010; Durrett et al., 2010; Banilas et al., 2011, Liu et al., 2012). Several studies have demonstrated that DGAT plays an essential role in controlling both the quantitative and qualitative flow of fatty acids into storage TAGs (He et al., 2004a; Sorensen et al., 2005; Lung and Weselake, 2006). A recent study in Brassica napus demonstrated that suppression of the DGAT1 gene results in a reduction in seed oil content and germination rates, in addition to severe developmental abnormalities (Lock et al., 2009). The study demonstrated that some DGAT genes might also have additional functions, as verified in vitro for DGAT1, which showed wax ester synthase and acyl-CoA-retinyl acyltransferase activities (Yen et al., 2005). Thereby, genes encoding proteins with DGAT activity have become targets for biotechnological approaches to improve the oil content and fatty acid composition in oleaginous crops (Settlage et al., 1998; Slabas et al., 2001; Lung and Weselake, 2006, Lardizabal et al., 2008; Xu et al., 2008, Andrianov et al., 2010). For example, an increase in seed oil content has been reported in Arabidopsis thaliana (Jako et al., 2001) and Brassica napus (Zheng et al., 2003) after DGAT1 overexpression. The heterologous expression of a fungal DGAT2 in soybean (Glycine max) resulted in an increase in the seed oil content (Lardizabal et al., 2008). Finally, the results of forward and reverse genetic studies have also revealed that mutations in DGAT1 directly affect oil content in some plant species (Zou et al., 1999).
DGAT enzymatic activity is encoded by different genes, which reinforces its importance in the synthesis of TAG in plants, and their distinct roles in determining the quality and quantity of acyl-CoA flux into TAG synthesis. The two major types of DGATs, designated as DGAT1 and DGAT2 genes, have been broadly studied in most eukaryote organisms, including fungi, animals, algae and plants. Phylogenetic and evolutionary analyses of these genes demonstrated that DGAT1 and DGAT2 evolved separately with functional convergence during eukaryotic evolution (Turchetto-Zolet et al., 2011). In addition to the ubiquitous occurrence of DGAT1 and DGAT2 genes in plants, other DGAT-related genes have also been identified. A soluble DGAT (DGAT3) that participates on the cytosolic pathway of TAG synthesis was first identified in peanuts (Arachis hypogaea) (Saha et al., 2006), and more recently in A. thaliana (Hernández et al., 2012) and yeast (Rani et al., 2013). In addition, a bifunctional DGAT/wax ester synthase (WS/DGAT), homologous to Acinetobacter calcoaceticus WS/DGAT (Kalscheuer and Steinbuchel, 2003), was characterized in A. thaliana (WSD1) (Li et al., 2008). The A. thaliana WS/DGAT predominantly catalyzes the synthesis of wax esters, but it is also responsible for the synthesis of minor amounts of TAGs. While DGAT1 and DGAT2 have been well characterized in most plant species, DGAT3 and WS/DGAT were studied in very few species. Until now, little is known about the roles of DGAT3 and WS/DGAT genes in most plant species. Hence, some issues such as (i) the presence of the homologous to DGAT3 and WS/DGAT genes in other plant species, (ii) the origin of these genes, and (iii) its relationships with DGAT1 and DGAT2 genes, remain unsolved. Therefore, the identification of putative DGAT3 and WS/DGAT genes and the understanding of their evolutionary history in plant species represent an important step to fully explore the DGAT potential in oilseed metabolic engineering and biotechnology.
Here, using homology searches in several plant genomes available we identified putative DGAT3 and WS/DGAT genes and used a phylogenetic approach and gene structure comparison to report on the diversity and evolution of these putative DGAT genes. The relationship of DGAT3 and WS/DGAT with the two main DGAT genes (DGAT1 and DGAT2) was also discussed. In addition, aiming to the understanding of the role of these genes in an oleaginous plant during the accumulation of lipid reserves in developing seeds we evaluated the expression profile of the putative DGAT3 and WS/DGAT genes in soybean (Glycine max). This oilseed species is one of the most economically important oilseed crops worldwide (Dornbos Jr and Muller, 1992; Saski et al., 2005; Vijav et al., 2009), which makes this species a potential biofuel feedstock (Hausman 2012; Hou et al., 2011). The combination of experimental and in silico analyses allowed us to describe the molecular evolution of these DGAT genes and to infer about their possible functions. We found that like DGAT1 and DGAT2 genes, DGAT3 and WS/DGAT also have experienced a distinct evolutionary history with different origins. Combined, our findings improve the current understanding about plant TAG biosynthesis, and will guide future functional and biotechnological studies.
Materials and Methods
Data sources and sequence retrieving
DGAT3 and WS/DGAT genes and proteins sequences were obtained through BLAST searches (TBLASTX, BLASTX and BLASTP) of the protein and genome databases with the default parameters and an e-value threshold of 1.0 e-20 at the NCBI (National Center for Biotechnology Information), and the completed genome projects at the Phytozome database. The DGAT3 and WSD1 sequences from Arabidopsis thaliana were used as queries in the BLAST searches. Supplementary Table S1 (491.8KB, pdf) provides a detailed description of the sequences used in this study and their corresponding accession numbers. Taxa terminologies are abbreviated using the first letter of the genus and two letters of the species name (e.g., Gma corresponds to Glycine max).
Sequence alignment and phylogenetic analysis
The nucleotide and protein sequences were aligned using MUSCLE (Edgar 2004) implemented in Molecular Evolutionary Genetics Analysis (MEGA version 5.0; Tamura et al., 2011). The multiple alignments were manually inspected and edited and only unambiguously aligned positions were included in the final analysis. The phylogenetic analysis was constructed after protein sequence alignments using Bayesian method, carried out in BEAST1.7 software (Drummond and Rambaut, 2007). The model of protein evolution used in this analysis was the JTT model for protein matrix substitution. The Yule tree was selected as a tree prior to Bayesian analysis and 20,000,000 generations were performed with Markov chain Monte Carlo (MCMC) algorithms. The trees were visualized and edited using FigTree v1.3.1 software.
Gene and protein structure analyses
The structural organization of the putative DGAT3 and WS/DGAT genes was determined by analyzing the genomic and coding sequences. We use the GSDraw web server, an interface for gene structure annotation available in PIECE database (Wang et al., 2012). Basically, we submitted a query sequence set (in multi-FASTA format) consisting of genomic and CDS to GSDraw and retrieved the gene structures with conserved protein motifs and phylogenetic trees. In addition, we searched for predicted transmembrane structures using the transmembrane prediction server TMHMM-2.0 and SMART database with the complete putative protein sequences.
Plant material, RNA extraction and cDNA preparation
Soybean leaf tissue (Glycine max cv. Conquista) and four seed developmental stages, representing R-stages (Supplementary Figure S1 (533KB, pdf) ) (R5: beginning seed; R6: full seed; R7: beginning maturity and R8: full maturity) were collected (Egli, 1994; Egli and Bruening, 2000). Total RNA was extracted using Trizol (Invitrogen), and the RNA quality was evaluated by electrophoresis on a 1.0% agarose gel. The reverse transcription of first-strand cDNA was performed with 2 μg of purified mRNA, T25V primer (1 μg/μL) and 200 units of M-MLV reverse transcriptase (Promega) in a final volume of 50 μL. The reverse transcription reaction included a denaturation step at 70 °C for 5 min, followed by a rapid thaw on ice, and an elongation step at 42 °C for 1 h. The cDNA products were diluted 1:10 and stored at -80 °C.
RT-qPCR expression analysis of putative soybean DGAT3 and WS/DGAT genes
To analyze expression pattern of the putative DGAT3 and WS/DGAT genes in soybean tissues, comparing with DGAT1 and DGAT2 expression, quantitative real time PCR (RT-qPCR) was performed using the CFX384 Real Time PCR system (BioRad) with SYBR-Green according to the manufacturer's protocol. Briefly, 10 μL of 1:100 diluted cDNA was mixed with primer pairs (0.2 μM), dNTPs (25 μM), 1X reaction buffer, MgCl2 (3 mM), 0.1X SYBR-Green Platinum Taq polymerase (0.25 U/μL) and DNase-free water to a final reaction volume of 20 μL. The RT-qPCR conditions were: an initial hot-start step at 94 °C for 5 min followed by 40 cycles of denaturation at 94 °C for 15 s, annealing at 60 °C for 10 s, extension at 72 °C for 15 s and an additional data recording step at 60 °C for 35 s. After cycling, an additional melting curve step was performed.
The four protein-coding genes, ELF1B, CYP2, ACT and TUA were selected based on previous reports as reference genes for soybean (Jian et al., 2008; Hu et al., 2009; Kulcheski et al., 2010). The primers used in these experiments are listed in Table S2 (452.2KB, pdf) . The experiments were performed using biological and technical quadruplicates. The relative expression of the DGAT genes was calculated using the 2-ΔΔCt method (Livak and Schmittgen, 2001). The statistical analyses were performed with SPSS v.20. One-way ANOVA was applied, with the Tukey's test (p ≤ 0.05) to compare pairwise differences in the expression for all genes.
In silico expression analysis
Tissue specificity and intensity of expression of DGAT genes were examined using microarray data at the GENEVESTIGATOR web site (Hruz et al., 2008). The available Hierarchical Clustering tool was used to perform this analysis. The highest expression values were considered for genes with more than one probe set. The expression data were gene-wise normalized and hierarchically clustered based on Pearson's coefficients.
Results
Homology search for putative DGAT3 and WS/DGAT genes in plant genomes
Putative homologs of the DGAT3 and WS/DGAT genes were searched in fully sequenced genomes from 20 plant and two algae from the Phytozome database using TBLASTX, BLASTX and BLASTP (see Material and Methods). Using DGAT3 and WSD1 (WS/DGAT) from A. thaliana as queries in blast searches, we were able to identify putative DGAT3 and WS/DGAT homologous sequences in all genomes. The exception was the green algae species Volvox carteri, that present putative DGAT3 gene but no match to WS/DGAT gene. The complete list of genes and species studied are summarized in Table S1 (491.8KB, pdf) . In total, we identified 25 putative DGAT3 and 80 putative WS/DGAT genes in plant and algae genomes (Table 1). While one or two putative DGAT3 genes were identified in all species, a larger number of putative WS/DGAT genes were found in the majority of plant species. The species included in phylogenetic and exon-intron comparisons analyses are indicated in Table 1.
Table 1. Number of putative DGAT3 and WS/DGAT sequences retrieved in this study and identification of species used for the phylogenetic and exon-intron structure comparative analyses.
| Species name | Taxa terminologies | N of putative DGAT3 genes | N of putative WS/DGAT genes |
|---|---|---|---|
| Arabidopsis thaliana | Ath* # Δ | 1 | 11 |
| Arabidopsis lyrata | Aly* | 1 | 7 |
| Brassica rapa | Bra* # Δ | 1 | 12 |
| Gossypium raimondii | Gra* | 1 | 5 |
| Theobroma cacao | Tca | 1 | 2 |
| Ricinus communis | Rco* # Δ | 1 | 3 |
| Manihot esculenta | Mes* Δ | 2 | 4 |
| Populus trichocarpa | Ptr* Δ | 1 | 4 |
| Medicago truncatula | Mtr* # Δ | 1 | 1 |
| Glycine max | Gma* # Δ | 2 | 1 |
| Solanum tuberosum | Stu* | 1 | 4 |
| Solanum lycopersicum | Sly* Δ | 1 | 4 |
| Aquilegia coerulea | Aco* Δ | 1 | 1 |
| Sorghum bicolor | Sbi* Δ | 1 | 3 |
| Oryza sativa | Osa* # Δ | 1 | 3 |
| Setaria italica | Sit* | 1 | 5 |
| Zea mays | Zma* # Δ | 1 | 1 |
| Brachypodium distachyon | Bdi* | 1 | 5 |
| Selaginella moellendorfii | Smo* # Δ | 1 | 2 |
| Physcomitrella patens | Ppa* # Δ | 2 | 1 |
| Volvox carteri | Vca* # Δ | 1 | – |
| Ostreococus lucimarinus | Olu* | 1 | 1 |
Phylogenetic relationship of DGAT genes in plants
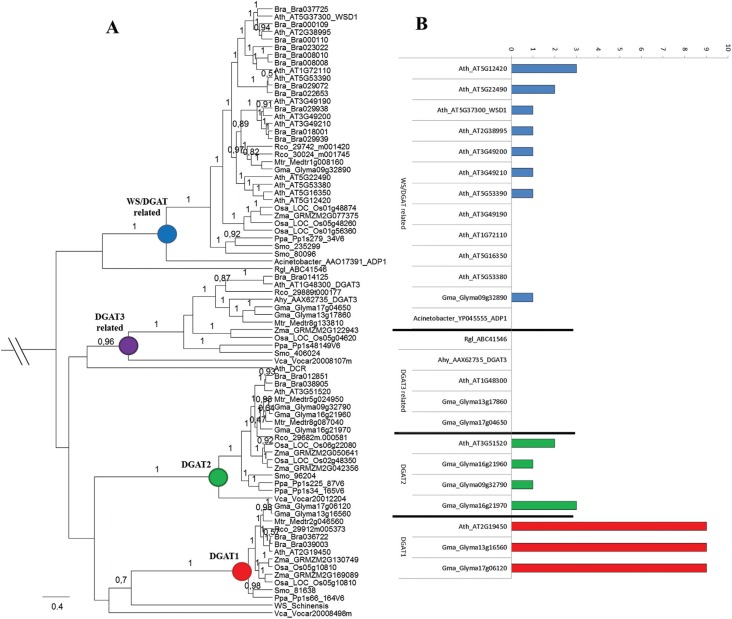
To understand the evolutionary relationships of the four different DGAT types in plant and algae species, we conducted a phylogenetic analysis using the protein sequence of putative DGAT3 and WS/DGAT identified by homology search and the DGAT1 and DGAT2 protein sequences reported in Turchetto-Zolet et al. (2011) (Figure 1A). For this analysis, we used DGATs sequences from nine plant and one algae species. We also included DGAT3 sequence from Arachis hypogaea, DCR sequence from A. thaliana, Wax ester synthase (WS) from Simmondsia chinensis, DGAT3 from Rhodotorula glutinis and WS/DGAT from Acinetobacter sp. A total of 80 sequences and 253 positions were included in the final dataset. We also performed a phylogenetic analysis of DGAT3 and WS/DGAT including a larger number of plants (Figure S2 (1.4MB, pdf) ). For this analysis, we used DGAT3 and WS/DGAT sequences from 21 plant and one algae species. A total of 105 sequences and 247 positions were included in the final dataset. The phylogenetic analysis of the DGATs amino acid sequences resulted in a well-resolved tree, revealing the formation of four well-supported clades separating the different DGAT types (Figure 1A and Figure S2 (1.4MB, pdf) ). Within each clade, we also observed that monocots and eudicots form distinct clusters, as was previously observed for DGAT1 and DGAT2 genes (Turchetto-Zolet et al., 2011). The ADP1 (WS/DGAT) sequence from Acinetobacter sp. grouped within the WS/DGAT clade, together with A. thaliana WS/DGAT and putative WS/DGAT from other plant and algae species, with high support, suggesting that diversification of this DGAT type occurred before the origin of plants. The DCR (Defective Cuticle Ridge) from A. thaliana, which is a soluble protein that belongs to the BAHD family of acyltransferases, was related to soluble DGAT3 clade. The Wax synthase (WS) sequence, which catalyzes the final step in the synthesis of linear esters (waxes) in Simmondsia chinensis, is closely related with DGAT1 sequences, suggesting a common origin for DGAT1 and WS/DGAT (Figure 1A). Within DGAT3 clade, the two putative soybean DGAT3 genes grouped closest to the DGAT3 from peanut. Another interesting result observed in Figure 1A and Figure S2 (1.4MB, pdf) was the gene duplication during DGATs gene family evolution. The pattern of gene duplication was distinct within each DGAT1, DGAT2, DGAT3 and WS/DGAT. While WS/DGAT was the most diversified gene with all plants presenting more than two WS/DGATs, DGAT3 genes was maintained as single copy in plants, except for G. max that has suffered gene duplication (Figure 1A and Figure S2 (1.4MB, pdf) ). In DGAT1 and DGAT2 more than one gene was observed in most analyzed plants (Figure 1A). All duplication events seemed to have occurred after plant diversification, since one gene of each DGAT1, DGAT2, DGAT3 and WS/DGAT was identified in algae.
Figure 1. Phylogenetic relationship among plant DGAT1, DGAT2, DGAT3 and WS/DGAT protein sequences. (A) The phylogenetic analysis was performed with DGAT protein sequences from Glycine max (Gma), Arabidopsis thaliana (Ath), Brassica rapa (Bra), Ricinus communis (Rco), Medicago truncatula (Mtr), Arachis hypogaea (Ahy), Oryza sativa (Osa), Zea mays (Zma), Selaginella moellendorffii (Smo), Physcomitrella patens and Volvox carteri (Vca). The DCR (AT5G23940) from A. thaliana, WS (AAD38041) from Simmondsia chinensis, the DGAT3 (ABC41546) from Rhodotorula glutinis and WS/DGAT (YP045555) from Acinetobacter sp. were also included in the analysis. The posteriori probabilities are labeled above the branches. Only values higher than 0.5 are presented. (B) Predicted transmembrane domain for DGAT1, DGAT2, DGAT3 and WS/DGAT from A. thaliana (Ath) and G. max (Gma). The WS/DGAT from Acinetobacter sp. and DGAT3 from R. glutinis and A. hypogaea were also analyzed. TMHMM web tools of the Center for Biological Sequence Analysis, Technical University of Denmark TMHMM Server plots showing the probability of the ALDH sequence forming a transmembrane helix (0-1.0 on the y-axis) (shown in red for the relevant amino acid sequences).
The transmembrane (TrM) domains were predicted and compared among the four DGAT types in soybean and A. thaliana species (Figure 1B). This analysis demonstrated distinct structure pattern among the different DGAT types within these two species. A. thaliana and soybean DGAT1 proteins contain nine putative transmembrane domains (TrM) (Figure 1B). DGAT2 from soybean and A. thaliana have two to three TrM and the WSD1 from A. thaliana and the putative soybean homologous contained one TrM. In contrast, no TrM regions were detected in DGAT3, supporting their status of soluble enzymes (Figure 1B). Interestingly, some A. thaliana WSD1 homologous presented two and three TrM regions, while others presented no TrM regions.
Structural organization of DGAT3 and WS/DGAT genes in plants
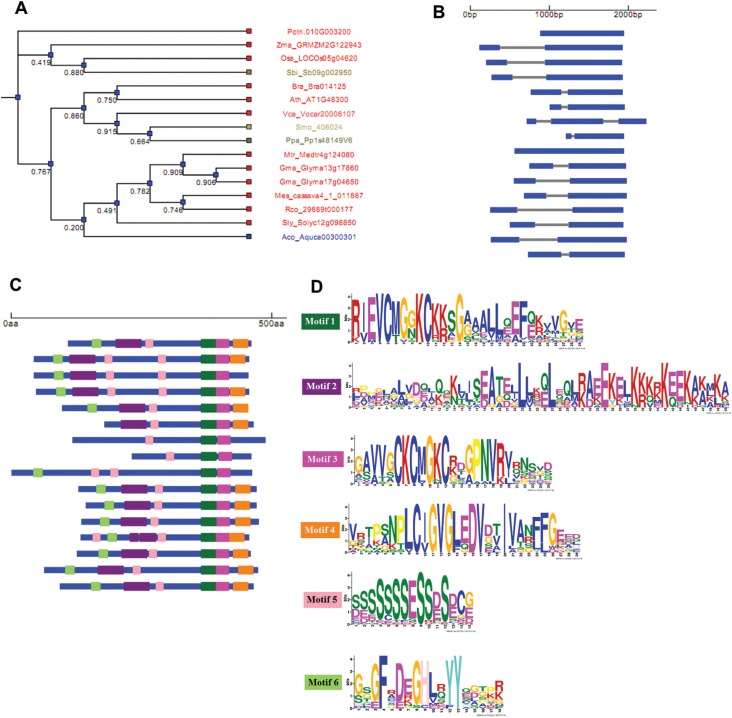
We performed a comparative analysis of the exon-intron organization of DGAT3 and WS/DGAT genes in plants and algae genomes to unveil their structural organization and to infer about their molecular evolution. For this analysis, genes from 14 species of plant and one species of algae were used (Table 1). The gene structure and conserved protein motif pattern diagram linked to a bootstrapped similarity dendrogram was obtained (Figure 2A-D and Figure 3A-D). The putative DGAT3 genes present in most species of this study contain two exons (Figure 2B). Exceptions were the green algae V. carteri that presents three exons, and the moss Physcomitrella patens and the tree species Populus trichocarpa that lack introns, suggesting the occurrence of gain and loss of introns during plant evolution. This analysis revealed a high degree of conservation among species regarding their gene structure, as shown in the cladogram of Figure 2B. The well characterized DGAT3 from A. thaliana presents two exons. Figure 2C shows the conserved motifs identified in the protein sequences of all putative DGAT3 analyzed. We observed that these protein motifs are present in most species with a high degree of conservation. The sequences of the six domains identified are showed in Figure 2D and in the alignment of Figure S3 (1.7MB, pdf) .
Figure 2. DGAT3 gene structure and organization in plant genomes. Dendrogram of sequences clustered according to the presence and similarity of identified protein motifs (A). Diagram displaying information of the gene structure for each sequence (B). Conserved motifs identified on protein sequences (C) and sequence logo of the conserved motif (D). Exon sequences are represented as blue boxes and the gray bars represent introns. The species included in this analysis are listed in the Table 1. The bootstrap values are given below the branches of the tree.
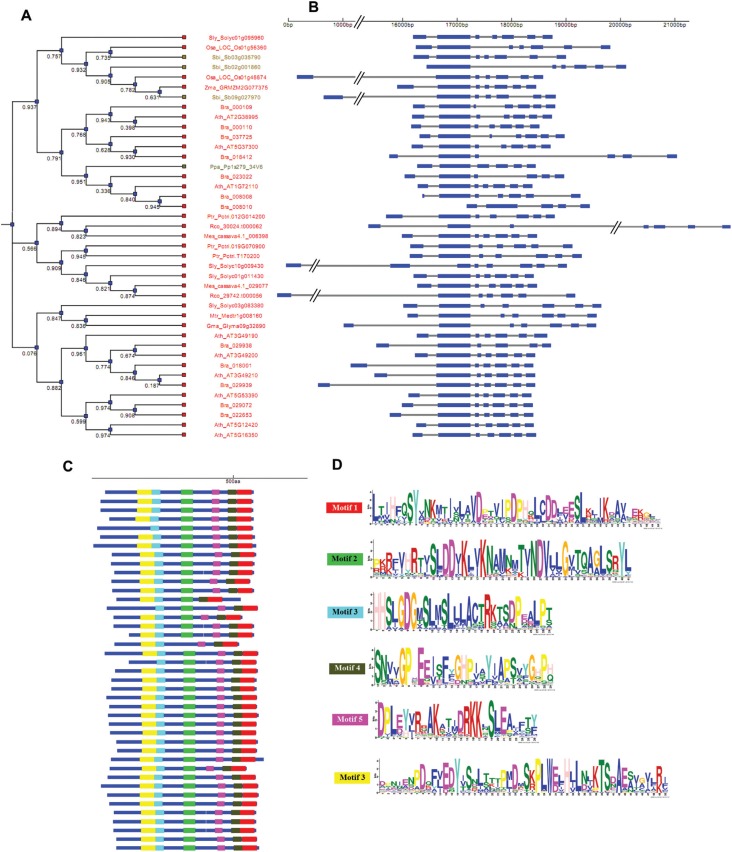
Figure 3. WS/DGAT gene structure and organization and conserved motifs identified in plant genomes. Dendrogram of sequences clustered according to the presence and similarity of identified protein motifs (A). Diagram displaying information of the gene structure for each sequence (B). Conserved motifs identified on protein sequences (C) and sequence logo of the conserved motif (D). The species included in this analysis are listed in the Table 1. Exon sequences are represented as blue boxes and the gray bars represent introns. The bootstrap values are given below the branches of the tree.
The putative WS/DGAT genes from most species present seven exons, which were also observed in the well characterized Arabidopsis WDS1 gene. The WDS1 and putative WS/DGAT genes identified in our study are highly conserved in terms of their structural organization in all species (Figure 3 A-D). This conservation is observed even among those genes that present six and eight exons (Figure 3B), which may be related to exon loss or gain during evolution. Concerning the distribution of protein motifs in the WS/DGAT protein sequences, we observed that the six identified domains are highly conserved in most species (Figure 3C). Likewise the A. calcoaceticus WS/DGAT and the A. thaliana WSD1 protein sequences, we observed the presence of the proposed active-site motif (228HHXXXDG234) in the N-terminal region in all putative WS/DGAT identified (Figure 3C, D and Figure S4 (2.2MB, pdf) ).
Comparison of the structural organization among the four different types of DGAT (DGAT1, DGAT2, DGAT3 and WS/DGAT) genes from soybean and Arabidopsis revealed distinct degrees of conservation in gene structure among these genes (Figure S5 (846.5KB, pdf) ). However, the comparisons clearly demonstrate a high degree of conservation within each type of DGAT gene between soybean and A. thaliana species. The A. thaliana and soybean DGAT1 genes contained 16 exons, the DGAT2 genes contained 5 to 9 exons, DGAT3 contained 2 and WS/DGAT contained 7 exons. This demonstrates that the four types of DGAT genes have experienced different evolutionary history.
Expression profiles of soybean DGAT3 and WS/DGAT
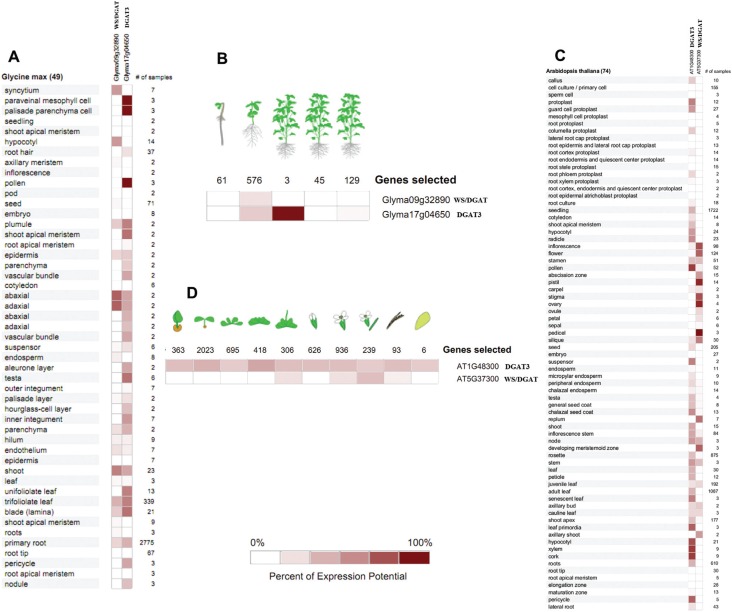
For inference on a role of the DGAT3 and WS/DGAT genes in lipid accumulation during seed development, we performed an expression analyses of the putative soybean DGAT3 and WS/DGAT genes and compared the results with the microarray expression data publicly available for A. thaliana. First, we performed an in silico comparative gene expression analysis with soybean and Arabidopsis DGAT3 and WS/DGAT genes using the GENEVESTIGATOR web-based software (Figure 4A). The probe sets used for in silico expression analysis are shown in Table S3 (461.4KB, pdf) . The analysis of the microarray expression data showed that the soybean and A. thaliana DGAT3 and WS/DGAT genes present different expression patterns across different tissues and plant developmental stages within each species (Figure 4). DGAT3 transcripts of soybean were detected in 30 of 49 analyzed tissues and in three of five plant development stages, while WS/DGAT transcripts were detected in 20 of 49 tissues and in one of five analyzed plant development stages (Figure 4A,B). The same pattern was observed for A. thaliana, where DGAT3 transcripts were detected in 47 of 74 tissues and in all analyzed plant development stages, while WS/DGAT transcripts were detected in 23 of 74 tissues and four of 10 plant development stages (Figure 4C, D). The putative soybean DGAT3 gene was highly expressed in paraveinal mesophyll cells, palisade parenchyma cells, pollen, plumule of the seed, shoot apical meristem, testa, unifoliolate and trifoliolate leaves, while the putative soybean WS/DGAT was highly detected in syncytium, hypocotyl, adaxial and abaxial cotyledon (Figure 4A,B). In A. thaliana, DGAT3 was more expressed in radicle, pollen, senescent leaf, leaf primordia, xylem and cork, while WS/DGAT was higher in inflorescence, flower, pistil, stigma, ovary and pedicel (Figure 4C,D).
Figure 4. In silico expression analysis of the selected DGAT genes in soybean and Arabidopsis. The soybean and Arabidopsis DGAT genes were analyzed for organ-specific and developmental expression patterns using the GENEVESTIGATOR program. Expression profile of organ-specific (A) and plant developmental expression profiles (B) of soybean DGAT3 and WS/DGAT genes. Expression profile of organ-specific (C) and plant developmental expression profiles (D) of Arabidopsis DGAT3 and WS/DGAT genes.
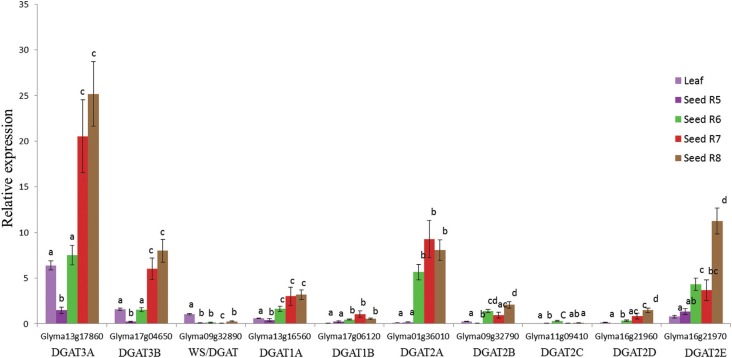
Subsequently, we checked the expression profile of putative soybean DGAT3 and WS/DGAT genes, as well as the expression of DGAT1 and DGAT2 genes throughout four seed development stages and the leaf tissue by RT-qPCR (Figure 5). The expression analysis of DGA1 and DGAT2 genes was performed to compare the expression levels among the four different DGAT types in soybean. The expression levels of the putative DGAT3 gene was higher in the seeds than in the leaves, with higher expression from mid to late stages of soybean seed development (R7 and R8) compared with leaf tissue and initial seed development stages. This result was similar to that found for DGAT1 and DGAT2 genes. In contrast, the expression levels of the putative soybean WS/DGAT genes were higher in leaf than in seed (Figure 5). The two putative soybean DGAT3 genes had similar expression patterns with significantly higher expression levels observed at the full maturity stage (Figure 5). DGAT1A and DGAT1B were both highly expressed from stages R6 (Full seed) to R8 (seed maturation phase). DGAT1B and DGAT1A did not show any significant differences among the R6, R7 and R8 stages. Except for DGAT2C, all five DGAT2 genes presented similar expression profiles (highly expressed at R6 to R8); the genes diverged, however, in their expression amplitude throughout soybean seed development. Comparing the expression pattern of soybean DGAT1, DGAT2, DGAT3 and WS/DGAT, we found DGAT3 as the highest expressed gene among the DGAT members. Also, one putative DGAT3 gene (Glyma13g17860) had the highest transcript levels detected, suggesting that this gene is probably involved in TAG synthesis in seed tissue.
Figure 5. Expression profiles of the DGAT1, DGAT2, DGAT3 and WS/DGAT genes during soybean seed development using RT-qPCR analysis. Expression profiles of each DGAT3 (Glyma13g17860, Glyma17g04650), WS/DGAT (Glyma09g32890), DGAT1 (Glyma13g16560, Glyma17g06120) and DGAT2 (Glyma01g36010, Glyma09g32790, Glyma11g09410, Glyma16g21960, Glyma16g21970) genes. The comparison of expression profiles between DGAT3 and WS/DGAT genes was done using an endogenous normalization with the leaf tissue of WS/DGAT gene. Standard error bars are based on four biological replicates. One-way ANOVA followed by the Tukey's test was used in the statistical analysis of the RT-qPCR data. mRNA input normalization was performed with four constitutive genes (Actin, ELF, CPY and TUA).
Discussion
The characterization of plant DGAT genes is highy relevant in studies directed towards the control of oilseed storage. However, more information is still required regarding the genes of TAG synthesis in plants, since different DGAT genes have been identified in some plant species, but are not yet well characterized. The comprehension of the evolutionary history of these genes and the presence of so many different genes encoding DGATs in plants is crucial to better understand their role in TAG biosynthesis. In the present study, taking advantage of the genome data available for several species and the large amount of current analytical methods, we identified putative DGAT3 and WS/DGAT genes in several plant species and present a view about their evolution. The search for DGAT3 and WS/DGAT genes in plant and algae genomes revealed that putative homologous of these genes are present in most analyzed species, suggesting that these genes could have essential function(s) in the survival of these plants, as has been shown with DGAT1 and DGAT2 genes. The function of the WS/DGAT gene was first described in Acinetobacter calcoaceticus (Kalscheuer and Steinbuchel, 2003) and was associated to the synthesis of both wax ester and TAG. In some bacteria, TAG formation is catalyzed by this bifunctional membrane-associated enzyme. A homologous of this gene was lately identified in A. thaliana (Li et al., 2008). It catalyzes, predominantly, the synthesis of wax esters, but also has a DGAT activity. We identified the highly conserved condensing domain with a proposed active-site motif (228HHXXXDG234) in the N-terminal region of all putative WS/DGAT, suggesting that all studied plants present an ortologous of this gene. This motif was suggested to be essential for catalytic activity in the acylCoA acyltransferase reactions involved in wax ester and TAG formation (Kalscheuer and Steinbuchel, 2003, Li et al., 2008). DGAT3 function was discovered and first characterized in peanuts (Arachis hypogaea) (Saha et al., 2006) and was recently identified in arabidopsis to be involved in active recycling of 18:2 and 18:3 fatty acids (FAs) into TAG when seed oil breakdown is blocked (Hernández et al., 2012). DGAT3 is proposed to be part of an alternative pathway for TAG synthesis (Saha et al., 2006; Hernández et al., 2012). This pathway occurs in the cytosol and involves the acylation of monoacylglycerol to DAG and the further acylation of DAG to TAG by the action of DGAT3. A soluble protein with DGAT activity has also been identified in oleaginous yeast (Rani et al., 2013). In addition, another soluble acyltransferase involved in TAG syntheses (LPAAT: lysophosphatidic acid acyltransferases) has been identified in Arabidopsis (Ghosh et al., 2009). Although the transmembrane domains are present in DGAT2 and in some WS/DGAT sequences, DGAT1 is the only gene that belongs to the superfamily of membrane-bound O-acyltransferases (MBOAT), which have transmembrane domains and histidine within a long hydrophobic invariant region (Hofmann, 2000). All members of the MBOAT superfamily are biochemically characterized by the transfer of organic acids, typically fatty acids, onto hydroxyl groups of membrane-embedded targets (Hofmann, 2000).
The phylogenetic analysis of putative DGAT3, WS/DGAT, DGAT1 and DGAT2 genes in plants revealed that they form monophyletic groups, suggesting that they probably have diverged early during plant evolution, or may have independent origins, as previously shown for DGAT1 and DGAT2 in eukaryotes (Turchetto-Zolet et al., 2011). The independent origin hypothesis is the most likely, since WS/DGAT genes from plants grouped together Acinetobacter calcoaceticus WS/DGAT, indicating that this type of DGAT has originated before plant diversification, and the DGAT activity was maintained due the importance of TAGs in all organisms. Recently we demonstrated a distinct origin for lysophosphatidic acid acyltransferases (LPAAT) genes, a group of genes involved in TAG synthesis (Korbes et al., 2016). Another interesting result from the phylogenetic analysis was the identification of different isoforms within each DGAT groups (DGAT1, DGAT2, DGAT3 and WS/DGAT) in some plant species. This indicates that they may have originated from gene duplication during plant evolution. This demonstrates that duplication events were important for the evolution and diversification of these genes. Gene duplication has also driven the evolution and diversification of LPAAT members during plant evolution (Korbes et al., 2016). This gene encodes a soluble protein that belongs to the BAHD family (Rani et al., 2010). Even though the soluble DGAT identified in the oleaginous yeast R. glutinis was considered a member of DGAT3 (Rani et al., 2013), this sequence has an uncertain position on our phylogenetic tree and more studies including a higher number of yeast species will be necessary to clarify the phylogenetic relationship between DGATs from yeast and plant species.
Comparative analyses of exon-intron organization are very important to understand rules of gene structure and organization, protein functionality and evolutionary changes among species (Wang et al., 2012). Our analysis demonstrated that the putative DGAT3 and WS/DGAT genes of most analyzed species present a high degree of conservation with the well-characterized A. thaliana DGAT3 and WS/DGAT genes, respectively. Nonetheless, comparison analysis of the four DGAT genes (DGAT1, DGAT2, DGAT3 and WS/DGAT) showed that they differ in their gene (exon/intron) organization, suggesting a distinct evolutionary history for these DGAT genes, unveiling the diversity of DGATs in plant species. We also observed that the loss/gain of introns is an evolutionary pattern for DGAT genes evolution. The loss/gain of introns may be caused by different processes, such as insertions of transposable elements, nucleotide substitutions or indels (Roy and Penny, 2006). We identified two DGAT1 homologous sequences that were actually part of a single gene corresponding to DGAT1 that was interrupted by the insertion of two transposons of the Copia family in the soybean genome (Glyma09g07510, Glyma09g07520) (data not shown). Glyma09g07510 and Glyma09g07520 lacked a DAG-binding signature motif and have probably lost their DGAT function. This suggests that in DGATs, transposable element insertions could have an important role also in tintron loss and gain.
There has been increasing evidence that DGAT enzymes play a key role in TAG biosynthesis, emphasizing the importance of understanding their roles, since TAGs are fundamental to all plant species. Distinct roles of two main DGATs (DGAT1 and DGAT2) enzymes in TAG metabolism have been demonstrated by molecular and functional characterization of these genes (Liu et al., 2012). The hypothesis received supported from gene expression studies, where in some plant species, DGAT1 and DGAT2 were shown to have different expression profiles, acting differently in some plant species, and presenting non-redundant functions in plants (Shockey et al., 2006; Chen et al., 2007). Examining when and where a gene is expressed in a cell or in the whole organism can provide clues to gene function. Here, we analyzed the expression profile of putative DGAT3 and WS/DGAT genes in soybean and found the same diversified pattern of transcript levels in both genes. The in silico and RT-qPCR analyses showed distinct expression patterns for these two DGAT3 and WS/DGAT genes in both soybean and A. thaliana species. The soybean and A. thaliana DGAT3 transcripts are more ubiquitously expressed, as they are detected in several tissues, than soybean and A. thaliana WS/DGAT transcripts, which are restricted to fewer tissues. In soybean, the transcript levels for DGAT3 were more abundant in the final stages of seed maturation, whereas WS/DGAT mRNA was higher in the leaf tissue samples, indicating different gene expression and distinct regulatory mechanisms. In A. thaliana, Li et al. (2008) demonstrated that the WS/DGAT (WSD1) gene is transcribed in flowers, top parts of stems, and leaves.
Interestingly, when comparing the expression patterns among four DGAT genes in soybean, we found that the putative DGAT3 (Glyma13g17860) sequence was the most abundant one in developing soybean seeds compared to the other DGAT genes. The putative DGAT3 (Glyma17g04650) gene was also highly expressed, suggesting a possible involvement of these sequences in TAG synthesis in this species, as was demonstrated for Arachis hypogaea (Saha et al., 2006), A. thaliana (Hernández et al., 2012) and oleaginous yeast (Rani et al., 2013). A transcriptome analysis during Arabidopsis seed development showed that the expression pattern of DGAT1 was similar to DGAT3 (Peng and Weselake 2011), but the expression of DGAT3 was higher during late seed maturation.
Many studies have demonstrated differences in the expression levels between DGAT1 and DGAT2 genes in a number of plant species. A study comparing gene expression across seed development in four different oilseeds (Brassica napus, Ricinus comunis, Euonimus alatus and Tropaeolum majus) using transcriptome analysis showed that in B. napus DGAT1 was more expressed than DGAT2, but contrasting results were observed in R. communis, where DGAT1 expression is essentially absent and DGAT2 is expressed at high levels (Troncoso-Ponce et al., 2011). Another study with Ricinus communis showed DGAT2 is higher expressed than DGAT1 during seed development (Cagliari et al., 2010). DGAT2 has been associated with the accumulation of unusual TAGs in R. comunis and in the tung tree (Shockey et al., 2006; Chen et al., 2007; Burgal et al., 2008). DGAT2 transcripts are also found with relatively high abundance in olive (Alagna et al., 2009) and palm (Bourgis et al., 2011; Tranbarger et al., 2011), which typically undergo TAG accumulation.
The phylogenetic relationship among DGAT1, DGAT2, DGAT3 and WS/DGAT and the characteristics of exon-/ntron organization, as well as of protein sequence motifs suggest that they have evolved in an independent way in plants. It is interesting to note that although these four types of DGATs present many structural differences, the DGAT activity encoded by them has been demonstrated in several plant species. Hence, the maintenance of all these different genes encoding DGAT enzymes appears to be closely associated with the increased genomic and metabolic complexity of plants, and may be explained by the essential importance of DGAT activity in triglyceride synthesis through an evolutionarily conserved process (Ichihara et al., 1988; Perry and Harwood 1993). We have demonstrated that purifying selection seems to have driven the evolution of DGAT1 and DGAT2 genes (Turchetto Zolet et a. 2011), suggesting a functional constraint. Thus, the observed distinct expression patterns of these genes may play a pivotal role in the development of such complex organisms, highlighting the importance of gene regulation for gene function during evolution.
In summary, the approaches used in this study allowed us to present a first general view about the presence of two DGAT genes (DGAT3 and WS/DGAT) in several plant species and showed a picture about their diversity and evolution in plants. We also observed that although the DGAT1, DGAT2, DGAT3 and WS/DGAT genes encode enzymes with a common function in TAG formation, they may have divergent expression patterns in different species and in different organs and tissues within a species. The diversity of genes encoding DGAT enzymes and their involvement in the control of TAG biosynthesis reinforces the need of functional studies of all DGAT genes in plants. Thereby, further comparative studies of these genes in oilseed species will be essential to identify new potential target genes for the manipulation of TAG fatty acid content through biotechnology techniques.
Acknowledgments
This work was financially supported through the CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) grant numbers 559636/2009-1 and 472574/2011-6; CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior), Grant Agroestruturante do RS: FAPERGS (Fundação de Amparo a Pesquisa do Estado do Rio Grande do Sul, FINEP (Financiadora de Projetos) and MCT (Ministério de Ciência e Tecnologia). FRK has DOCFIX (CAPES/FAPERGS) fellowships. MMP and RM are recipients of CNPq research fellowships (307868/2011-7 and 303967/2008-0, respectively). We also thank Joseane Biso de Carvalho for sample collection and Luiz Felipe Valter de Oliveira for helping with figures.
Supplementary material
The following online material is available for this article:
Footnotes
Associate Editor: Carlos F. M. Menck
References
- Alagna F, D'Agostino N, Torchia L, Servili M, Rao R, Pietrella M, Giuliano G, Chiusano ML, Baldoni L, Perrotta G. Comparative 454 pyrosequencing of transcripts from two olive genotypes during fruit development. BMC Genomics. 2009;10:e399–e399. doi: 10.1186/1471-2164-10-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrianov V, Borisjuk N, Pogrebnyak N, Brinker A, Dixon J, Spitsin S, Flynn J, Matyszczuk P, Andryszak K, Laurelli M, et al. Tobacco as a production platform for biofuel: Overexpression of Arabidopsis DGAT and LEC2 genes increases accumulation and shifts the composition of lipids in green biomass. Plant Biotechnol J. 2010;8:277–287. doi: 10.1111/j.1467-7652.2009.00458.x. [DOI] [PubMed] [Google Scholar]
- Banilas G, Karampelias M, Makariti I, Kourti A, Hatzopoulos P. The olive DGAT2 gene is developmentally regulated and shares overlapping but distinct expression patterns with DGAT1. J Exp Bot. 2011;62:521–532. doi: 10.1093/jxb/erq286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgis F, Kilaru A, Cao X, Ngando-Ebongue G-F, Drira N, Ohlrogged JB, Arondel V. Comparative transcriptome and metabolite analysis of oil palm and date palm mesocarp that differ dramatically in carbon partitioning. Proc Natl Acad Sci U S A. 2011;108:12527–12532. doi: 10.1073/pnas.1106502108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgal J, Shockey J, Lu CF, Dyer J, Larson T, Graham I, Browse J. Metabolic engineering of hydroxy fatty acid production in plants: RcDGAT2 drives dramatic increases in ricinoleate levels in seed oil. Plant Biotechnol J. 2008;6:819–831. doi: 10.1111/j.1467-7652.2008.00361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagliari A, Margis-Pinheiro M, Loss G, Mastroberti AA, Mariath JEA, Margis R. Identification and expression analysis of castor bean (Ricinus communis) genes encoding enzymes from the triacylglycerol biosynthesis pathway. Plant Sci. 2010;179:499–509. doi: 10.1016/j.plantsci.2010.07.015. [DOI] [PubMed] [Google Scholar]
- Cagliari A, Margis M, Maraschin FS, Turchetto-Zolet AC, Loss G, Margis-Pinheiro M. Biosynthesis of triacylglycerols (TAGs) in plants and algae. Int J Plant Sci. 2011;2:e10–e10. [Google Scholar]
- Chen GQ, Turner C, He X, Nguyen T, McKeon TA, Laudencia-Chingcuanco D. Expression profiles of genes involved in fatty acid and triacylglycerol synthesis in castor bean (Ricinus communis L.) Lipids. 2007;42:263–274. doi: 10.1007/s11745-007-3022-z. [DOI] [PubMed] [Google Scholar]
- Dornbos DL, Jr, Mullen RE. Soybean seed protein and oil contents and fatty acid composition adjustments by drought and temperature. J Am Oil Chem Soc. 1992;69:228–231. [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:e214–e214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrett TP, Benning C, Ohlrogge J. Plant triacylglycerols as feedstocks for the production of biofuels. Plant J. 2008;54:593–607. doi: 10.1111/j.1365-313X.2008.03442.x. [DOI] [PubMed] [Google Scholar]
- Durrett TP, McClosky DD, Tumaney AW, Elzinga DA, Ohlrogge J, Pollard M. A distinct DGAT with sn-3 acetyltransferase activity that synthesizes unusual, reduced-viscosity oils in Euonymus and transgenic seeds. Proc Natl Acad Sci U S A. 2010;107:9464–9469. doi: 10.1073/pnas.1001707107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer JM, Stymne S, Green AG, Carlsson AS. High-value oils from plants. Plant J. 2008;54:640–655. doi: 10.1111/j.1365-313X.2008.03430.x. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli DB. Cultivar maturity and reproductive growth duration in soybean. J Agron Crop Sci - Z Acker Pflanzenbau. 1994;173:249–254. [Google Scholar]
- Egli DB, Bruening WP. Potential of early-maturing soybean cultivars in late plantings. Agronomy J. 2000;92:532–537. [Google Scholar]
- Ghosh AK, Chauhan N, Rajakumari S, Daum G, Rajasekharan R. At4g24160, a soluble acyl-coenzyme A-dependent lysophosphatidic acid acyltransferase 1[W][OA] Plant Physiol. 2009;151:869–881. doi: 10.1104/pp.109.144261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausman C. Biofuels and land use change: Sugarcane and soybean acreage response in Brazil. Environ Resource Econ. 2012;51:163–163. [Google Scholar]
- He XH, Turner C, Chen GQ, Lin JT, McKeon TA. Cloning and characterization of a cDNA encoding diacylglycerol acyltransferase from castor bean. Lipids. 2004a;39:311–318. doi: 10.1007/s11745-004-1234-2. [DOI] [PubMed] [Google Scholar]
- He XH, Chen GQ, Lin JT, McKeon TA. Regulation of diacylglycerol acyltransferase in developing seeds of castor. Lipids. 2004b;39:865–871. doi: 10.1007/s11745-004-1308-1. [DOI] [PubMed] [Google Scholar]
- He XH, Chen GQ, Lin JT, McKeon TA. Diacylglycerol acyltransferase activity and triacylglycerol synthesis in germinating castor seed cotyledons. Lipids. 2006;41:281–285. doi: 10.1007/s11745-006-5098-2. [DOI] [PubMed] [Google Scholar]
- Hernández ML, Whitehead L, He Z, Gazda V, Gilday A, Kozhevnikova E, Vaistij FE, Larson TR, Graham IA. A cytosolic acyltransferase contributes to triacylglycerol synthesis in sucrose-rescued Arabidopsis seed oil catabolism mutants1[W][OA] Mol Genet Genomics. 2012;160:215–225. doi: 10.1104/pp.112.201541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs DH, Hills MJ. Expression and characterization of diacylglycerol acyltransferase from Arabidopsis thaliana in insect cell cultures. Biochem Soc Trans. 2000;28:687–689. [PubMed] [Google Scholar]
- Hobbs DH, Lu CF, Hills MJ. Cloning of a cDNA encoding diacylglycerol acyltransferase from Arabidopsis thaliana and its functional expression. FEBS Lett. 1999;452:145–149. doi: 10.1016/s0014-5793(99)00646-8. [DOI] [PubMed] [Google Scholar]
- Hofmann K. A superfamily of membrane-bound O-acyltransferases with implications for Wnt signaling. Trends Biochem Sci. 2000;25:111–112. doi: 10.1016/s0968-0004(99)01539-x. [DOI] [PubMed] [Google Scholar]
- Hou J, Wang C, Hong X, Zhao J, Xue C, Guo N, Gai J, Xing H. Association analysis of vegetable soybean quality traits with SSR markers. Plant Breed. 2011;130:444–449. [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv Bioinform 2008. 2008:e420747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R, Fan C, Li H, Zhang Q, Fu Y-F. Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol Biol. 2009;10:e93. doi: 10.1186/1471-2199-10-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihara K, Takahashi T, Fujii S. Diacylglycerol acyltransferase in maturing safflower seeds - its influences on the fatty acid composition of triacylglycerol and on the rate of triacylglycerol synthesis. Biochim Biophys Acta. 1988;958:125–129. doi: 10.1016/0005-2760(88)90253-6. [DOI] [PubMed] [Google Scholar]
- Jako C, Kumar A, Wei YD, Zou JT, Barton DL, Giblin EM, Covello PS, Taylor DC. Seed-specific over-expression of an Arabidopsis cDNA encoding a diacylglycerol acyltransferase enhances seed oil content and seed weight. Plant Physiol. 2001;126:861–874. doi: 10.1104/pp.126.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian B, Liu B, Bi Y, Hou W, Wu C, Han T. Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol Biol. 2008;9:e59. doi: 10.1186/1471-2199-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalscheuer R, Steinbuchel A. A novel bifunctional wax ester synthase/acyl-CoA:diacylglycerol acyltransferase mediates wax ester and triacylglycerol biosynthesis in Acinetobacter calcoaceticus ADP1. J Biol Chem. 2003;278:8075–8082. doi: 10.1074/jbc.M210533200. [DOI] [PubMed] [Google Scholar]
- Kennedy EP, Weiss SB. Function of cytidine coenzymes in the biosynthesis of phospholipides. J Biol Chem. 1956;222:193–213. [PubMed] [Google Scholar]
- Korbes AP, Kulcheski FR, Margis R, Margis-Pinheiro M. Molecular evolution of the lysophosphatidic acid acyltransferase (LPAAT) gene family. Mol Phylogenet Evol. 2016;96:55–69. doi: 10.1016/j.ympev.2015.12.001. [DOI] [PubMed] [Google Scholar]
- Kroon JTM, Wei WX, Simon WJ, Slabas AR. Identification and functional expression of a type 2 acyl-CoA: Diacylglycerol acyltransferase (DGAT2) in developing castor bean seeds which has high homology to the major triglyceride biosynthetic enzyme of fungi and animals. Phytochemistry. 2006;67:2541–2549. doi: 10.1016/j.phytochem.2006.09.020. [DOI] [PubMed] [Google Scholar]
- Kulcheski FR, Marcelino-Guimaraes FC, Nepomuceno AL, Abdelnoor RV, Margis R. The use of microRNAs as reference genes for quantitative polymerase chain reaction in soybean. Anal Biochem. 2010;406:185–192. doi: 10.1016/j.ab.2010.07.020. [DOI] [PubMed] [Google Scholar]
- Lardizabal K, Effertz R, Levering C, Mai J, Pedroso MC, Jury T, Aasen E, Gruys K, Bennett K. Expression of Umbelopsis ramanniana DGAT2A in seed increases oil in soybean. Plant Physiol. 2008;148:89–96. doi: 10.1104/pp.108.123042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Wu X, Lam P, Bird D, Zheng H, Samuels L, Jetter R, Kunst L. Identification of the wax ester synthase/acyl-coenzyme A: Diacylglycerol acyltransferase WSD1 required for stem wax ester biosynthesis in Arabidopsis. Plant Physiol. 2008;148:97–107. doi: 10.1104/pp.108.123471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Yu K, Hildebrand DF. DGAT1, DGAT2 and PDAT expression in seeds and other tissues of epoxy and hydroxy fatty acid accumulating plants. Lipids. 2010;45:145–157. doi: 10.1007/s11745-010-3385-4. [DOI] [PubMed] [Google Scholar]
- Lisa M, Holcapek M, Bohac M. Statistical evaluation of triacylglycerol composition in plant oils based on high-performance liquid chromatography-atmospheric pressure chemical ionization mass spectrometry data. J Agricult Food Chem. 2009;57:6888–6898. doi: 10.1021/jf901189u. [DOI] [PubMed] [Google Scholar]
- Liu Q, Siloto RMP, Lehner R, Stone S, Weselake RJ. Acyl-CoA:diacylglycerol acyltransferase: Molecular biology, biochemistry and biotechnology. Prog Lipid Res. 2012;51:350–377. doi: 10.1016/j.plipres.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lock Y-Y, Snyder CL, Zhu W, Siloto RMP, Weselake RJ, Shah S. Antisense suppression of type 1 diacylglycerol acyltransferase adversely affects plant development in Brassica napus . Physiol Plant. 2009;137:61–71. doi: 10.1111/j.1399-3054.2009.01258.x. [DOI] [PubMed] [Google Scholar]
- Lung SC, Weselake RJ. Diacylglycerol acyltransferase: A key mediator of plant triacylglycerol synthesis. Lipids. 2006;41:1073–1088. doi: 10.1007/s11745-006-5057-y. [DOI] [PubMed] [Google Scholar]
- Millar AA, Smith MA, Kuns L. All fatty acids are not equal: Discrimination in plant membrane lipids. Trends Plant Sci. 2000;5:7–7. doi: 10.1016/s1360-1385(00)01566-1. [DOI] [PubMed] [Google Scholar]
- Ohlrogge J, Browse J. Lipid biosynthesis. Plant Cell. 1995;7:957–970. doi: 10.1105/tpc.7.7.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng FY, Weselake RJ. Gene coexpression clusters and putative regulatory elements underlying seed storage reserve accumulation in Arabidopsis . BMC Genomics. 2011;12:e286. doi: 10.1186/1471-2164-12-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry HJ, Harwood JL. Changes in the lipid content of developing seeds of Brassica napus . Phytochemistry. 1993;32:1411–1415. [Google Scholar]
- Rani SH, Krishna TH, Saha S, Negi AS, Rajasekharan R. Defective in cuticular ridges (DCR) of Arabidopsis thaliana, a gene associated with surface cutin formation, encodes a soluble diacylglycerol acyltransferase. J Biol Chem. 2010;285:38337–38347. doi: 10.1074/jbc.M110.133116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani HR, Saha S, Rajasekharan R. A soluble diacylglycerol acyltransferase is involved in triacylglycerol biosynthesis in the oleaginous yeast Rhodotorula glutinis . Microbiology. 2013;159:155–166. doi: 10.1099/mic.0.063156-0. [DOI] [PubMed] [Google Scholar]
- Roy SW, Penny D. Patterns of intron loss and gain in plants: Intron loss-dominated evolution and genome-wide comparison of O. sativa and A. thaliana . Mol Biol Evol. 2006;24:171–181. doi: 10.1093/molbev/msl159. [DOI] [PubMed] [Google Scholar]
- Saha S, Enugutti B, Rajakumari S, Rajasekharan R. Cytosolic triacylglycerol biosynthetic pathway in oilseeds. molecular cloning and expression of peanut cytosolic diacylglycerol acyltransferase. Plant Physiol. 2006;141:1533–1543. doi: 10.1104/pp.106.082198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saski L, Lee SB, Daniel H, Wood TC, Tomkins J, Kim HG, Jansem PK. Complete chloroplast genome sequence of Glycine max and comparative analysis with other legume genomes. Plant Mol Biol. 2005;59:309–322. doi: 10.1007/s11103-005-8882-0. [DOI] [PubMed] [Google Scholar]
- Settlage SB, Kwanyuen P, Wilson RF. Relation between diacylglycerol acyltransferase activity and oil concentration in soybean. J Am Oil Chem Soc. 1998;75:775–781. [Google Scholar]
- Shockey JM, Gidda SK, Chapital DC, Kuan JC, Dhanoa PK, Bland JM, Rothstein SJ, Mullen RT, Dyer JM. Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell. 2006;18:2294–2313. doi: 10.1105/tpc.106.043695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slabas AR, Simon JW, Brown AP. Biosynthesis and regulation of fatty acids and triglycerides in oil seed rape. Current status and future trends. Eur J Lipid Sci Technol. 2001;103:455–466. [Google Scholar]
- Smart HC, Mast FD, Chilije MFJ, Tavassoli M, Dacks JB, Zaremberg V. Phylogenetic analysis of glycerol 3-phosphate acyltransferases in opisthokonts reveals unexpected ancestral complexity and novel modern biosynthetic components. PLoS One. 2014;9:e110684. doi: 10.1371/journal.pone.0110684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen BM, Furukawa-Stoffer TL, Marshall KS, Page EK, Mir Z, Forster RJ, Weselake RJ. Storage lipid accumulation and acyltransferase action in developing flaxseed. Lipids. 2005;40:1043–1049. doi: 10.1007/s11745-005-1467-0. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranbarger TJ, Dussert S, Joet T, Argout X, Summo M, Champion A, Cros D, Omore A, Nouy B, Morcillo F. Regulatory mechanisms underlying oil palm fruit mesocarp maturation, ripening, and functional specialization in lipid and carotenoid metabolism. Plant Physiol. 2011;156:564–584. doi: 10.1104/pp.111.175141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troncoso-Ponce MA, Kilaru A, Cao X, Durrett TP, Fan J, Jensen JK, Thrower NA, Pauly M, Wilkerson C, Ohlrogge JB. Comparative deep transcriptional profiling of four developing oilseeds. Plant J. 2011;68:1014–1027. doi: 10.1111/j.1365-313X.2011.04751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchetto-Zolet AC, Maraschin FS, de Morais GL, Cagliari A, Andrade CMB, Margis-Pinheiro M, Margis R. Evolutionary view of acyl-CoA diacylglycerol acyltransferase (DGAT), a key enzyme in neutral lipid biosynthesis. BMC Evol Biol. 2011;11:e263. doi: 10.1186/1471-2148-11-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, You FM, Lazo GR, Luo M-C, Thilmony R, Gordon S, Kianian SF, Gu YQ. PIECE: A database for plant gene structure comparison and evolution. J Lipid Res. 2012;41:D1159–D1166. doi: 10.1093/nar/gks1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay D, Dadlani M. Seed longevity and water absorption patterns in maize, soybean and safflower. Ind J Plant Physiol. 2003;8(Special issue):244–248. [Google Scholar]
- Weiss SB, Kennedy EP, Kiyasu JY. The enzymatic synthesis of triglycerides. J Biol Chem. 1960;235:40–44. [PubMed] [Google Scholar]
- Wolters-Arts M, Lush WM, Mariani C. Lipids are required for directional pollen-tube growth. Nature. 1998;392:818–821. doi: 10.1038/33929. [DOI] [PubMed] [Google Scholar]
- Xu JY, Francis T, Mietkiewska E, Giblin EM, Barton DL, Zhang Y, Zhang M, Taylor DC. Cloning and characterization of an acyl-CoA-dependent diacylglycerol acyltransferase 1 (DGAT1) gene from Tropaeolum majus, and a study of the functional motifs of the DGAT protein using site-directed mutagenesis to modify enzyme activity and oil content. Plant Biotechnol J. 2008;6:799–818. doi: 10.1111/j.1467-7652.2008.00358.x. [DOI] [PubMed] [Google Scholar]
- Yen CLE, Monetti M, Burri BJ, Farese RV. The triacylglycerol synthesis enzyme DGAT1 also catalyzes the synthesis of diacylglycerols, waxes, and retinyl esters. J Lipid Res. 2005;46:1502–1511. doi: 10.1194/jlr.M500036-JLR200. [DOI] [PubMed] [Google Scholar]
- Yu K, Li R, Hatanaka T, Hildebrand D. Cloning and functional analysis of two type 1 diacylglycerol acyltransferases from Vernonia galamensis . Phytochemistry. 2008;69:1119–1127. doi: 10.1016/j.phytochem.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Zhang M, Fan J, Taylor DC, Ohlrogge JB. DGAT1 and PDAT1 Acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell. 2009;21:3885–3901. doi: 10.1105/tpc.109.071795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng ZF, Xia Q, Dauk M, Shen WY, Selvaraj G, Zou J. Arabidopsis AtGPAT1, a member of the membrane-bound glycerol-3-phosphate acyltransferase gene family, is essential for tapetum differentiation and male fertility. Plant Cell. 2003;15:1872–1887. doi: 10.1105/tpc.012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou JT, Wei YD, Jako C, Kumar A, Selvaraj G, Taylor DC. The Arabidopsis thaliana TAG1 mutant has a mutation in a diacylglycerol acyltransferase gene. Plant J. 1999;19:645–653. doi: 10.1046/j.1365-313x.1999.00555.x. [DOI] [PubMed] [Google Scholar]
Internet resources
- GENEVESTIGATOR Tools. Apr 01, 2015. https://www.genevestigator.com.
- FigTree Software. 2015 Mar 01; http://tree.bio.ed.ac.uk/software/figtree/
- The Plant Genomics Resouces (Phytozome) Jan 01, 2015. https://phytozome.jgi.doe.gov/pz/portal.html.
- National Center for Biotechnology Information (NCBI) Jan 01, 2015. http://www.ncbi.nlm.nih.gov/
- PIECE Database. Apr 01, 2015. http://wheat.pw.usda.gov/piece/GSDraw.php.
- TMHMM-2.0 Software. 2015 Mar 01; http://www.cbs.dtu.dk/services/
- SMART Software. 2015 Mar 01; http://smart.embl-heidelberg.de/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- GENEVESTIGATOR Tools. Apr 01, 2015. https://www.genevestigator.com.
- The Plant Genomics Resouces (Phytozome) Jan 01, 2015. https://phytozome.jgi.doe.gov/pz/portal.html.
- National Center for Biotechnology Information (NCBI) Jan 01, 2015. http://www.ncbi.nlm.nih.gov/
- PIECE Database. Apr 01, 2015. http://wheat.pw.usda.gov/piece/GSDraw.php.