Changes in cytology and gene expression are reversible during Cunninghamia lanceolata seed dormancy cycling.
Abstract
Seeds with physiological dormancy usually experience primary and secondary dormancy in the nature; however, little is known about the differential regulation of primary and secondary dormancy. We combined multiple approaches to investigate cytological changes, hormonal levels, and gene expression dynamics in Cunninghamia lanceolata seeds during primary dormancy release and secondary dormancy induction. Light microscopy and transmission electron microscopy revealed that protein bodies in the embryo cells coalesced during primary dormancy release and then separated during secondary dormancy induction. Transcriptomic profiling demonstrated that expression of genes negatively regulating gibberellic acid (GA) sensitivity reduced specifically during primary dormancy release, whereas the expression of genes positively regulating abscisic acid (ABA) biosynthesis increased during secondary dormancy induction. Parallel analysis of RNA ends revealed uncapped transcripts for ∼55% of all unigenes. A negative correlation between fold changes in expression levels of uncapped versus capped mRNAs was observed during primary dormancy release. However, this correlation was loose during secondary dormancy induction. Our analyses suggest that the reversible changes in cytology and gene expression during dormancy release and induction are related to ABA/GA balance. Moreover, mRNA degradation functions as a critical posttranscriptional regulator during primary dormancy release. These findings provide a mechanistic framework for understanding physiological dormancy in seeds.
Seed dormancy mediates the crucial interaction between the environment and the plant life cycle transition from durable seeds to vulnerable seedlings (Willis et al., 2014). Different plant species have adopted various classes of dormancy to regulate the timing of seed germination, help seedlings emerge under favorable conditions, and maximize the growing season (Baskin and Baskin, 2004; Donohue et al., 2010). Physiological dormancy, the most widely occurring type, is caused by inadequate growth potential of the embryo, which prevents it from overcoming the resistance of its surrounding tissues (Baskin and Baskin, 2014). Seeds with physiological dormancy can change their degree of dormancy, a process known as dormancy cycling: Primary dormant seeds enter nondormancy during seasons favorable for germination, and nondormant seeds may reenter dormancy (secondary dormancy) under unfavorable conditions if they have not germinated (Footitt et al., 2011; Cao et al., 2014). Due to the importance of seed dormancy in the plant life cycle, particularly in agriculture and vegetative restoration, many studies have focused on the regulatory mechanisms underlying seed dormancy.
Genetic studies have revealed some genes that regulate seed dormancy and germination, especially genes involved in abscisic acid (ABA) and gibberellic acid (GA) pathways (Shu et al., 2013; Vaistij et al., 2013; Wang et al., 2013). Also exogenous application of these phytohormones modifies seed germination (Ogawa et al., 2003; Oh et al., 2007), suggesting that they play a central role in the physiological dormancy of seeds. Recently, emerging “-omics” technologies have increased our understanding of the regulation of physiological dormancy at the whole-genome level. Indeed, species of stored mRNAs account for more than half of the genome mRNAs in dry Arabidopsis (Arabidopsis thaliana) seeds (Nakabayashi et al., 2005), and numerous genes with various functions show differential expression during dormancy release, indicating that a highly complex network regulates seed dormancy, in addition to the crucial roles of ABA and GA (Cadman et al., 2006; Carrera et al., 2008).
Despite the distinct gene expression programs between dormant and nondormant seeds (Cadman et al., 2006; Carrera et al., 2008), little is known about the regulation of gene expression during dormancy release. mRNA degradation is involved in rapid changes and precise adjustments in gene expression in eukaryotic cells (Goeres et al., 2007; Belasco, 2010; Haimovich et al., 2013), and mRNA decapping mediated by decapping enzymes (DCPs) represents a crucial step in bulk mRNA degradation (Coller and Parker, 2004; Franks and Lykke-Andersen, 2008), after which decapped mRNA intermediates are digested by the 5′-3′ exonuclease XRN1 or degraded in 3′-5′ orientation by the exosome complex (Houseley and Tollervey, 2009; Haimovich et al., 2013). Alternatively, mRNA degradation can be initiated by internal cleavage by endonucleases or small RNAs, including microRNAs (miRNAs) and small interfering RNAs (Houseley and Tollervey, 2009; Belasco, 2010). Both the activity of decapping enzymes and internal cleavage by endonucleases and small RNAs produce uncapped mRNAs, which are characterized by a 5′ monophosphate and a shortened poly(A) tail (Jiao et al., 2008). In addition, nonsense-mediated mRNA decay also produces uncapped mRNAs (Schoenberg and Maquat, 2012). Parallel analysis of RNA ends (PARE), which was first developed to identify miRNA cleavage on target mRNAs (German et al., 2008), has been proven a powerful tool for profiling uncapped mRNAs in eukaryotic cells (Harigaya and Parker, 2012; Zhang et al., 2013). This method takes advantage of a modified 5′-RACE technique to selectively capture the mRNA degradation intermediates, poly(A) RNAs with 5′ monophosphate (German et al., 2009). To date, much of our understanding of mRNA degradation is based on studies in yeast and mammalian models; however, the process of mRNA degradation and its possible roles remain largely unknown in plants, particularly during seed dormancy.
Previous studies on regulation of seed dormancy have focused on herbaceous species (e.g. Arabidopsis, rice [Oryza sativa], and barley [Hordeum vulgare]); by contrast, information on trees remains limited. Cunninghamia lanceolata (Cupressaceae) is an evergreen conifer that is widely distributed in China, North Vietnam, Laos, and Cambodia (Fu et al., 1999; Kusumi et al., 2000). This commercially important tree (with a forestland area of >11 million hm2) produces a large amount of soft and highly durable scented wood (with a total standing stock volume of 734 million m3; State Forest Administration P.R. China, 2014). Due to the rapid growth and commercial importance, much effort has focused on exploring the regulatory mechanisms underlying wood production and reproduction of this species, and high-throughput sequencing technologies have been used extensively in recent years to identify functional genes in this species (Shi et al., 2010; Wan et al., 2012; Qiu et al., 2013).
To increase our understanding of the regulatory mechanisms underlying seed dormancy in trees, we applied light microscopy and transmission electron microscopy to investigate the cellular and subcellular changes that occur during dormancy release and induction in C. lanceolata seeds. We also profiled global mRNA expression in C. lanceolata seeds with various dormancy statuses (primary dormancy, secondary dormancy, and nondormancy) to explore the regulatory network of seed dormancy. Furthermore, we used PARE to investigate the role of mRNA degradation in gene expression of C. lanceolata seeds. The long-term objective of our research is to provide reference maps of the regulatory mechanisms underlying seed dormancy, with an emphasis on the regulation of orchestrated gene expression during dormancy release and induction.
RESULTS
Obtaining C. lanceolata Seeds with Various Dormancy Statuses
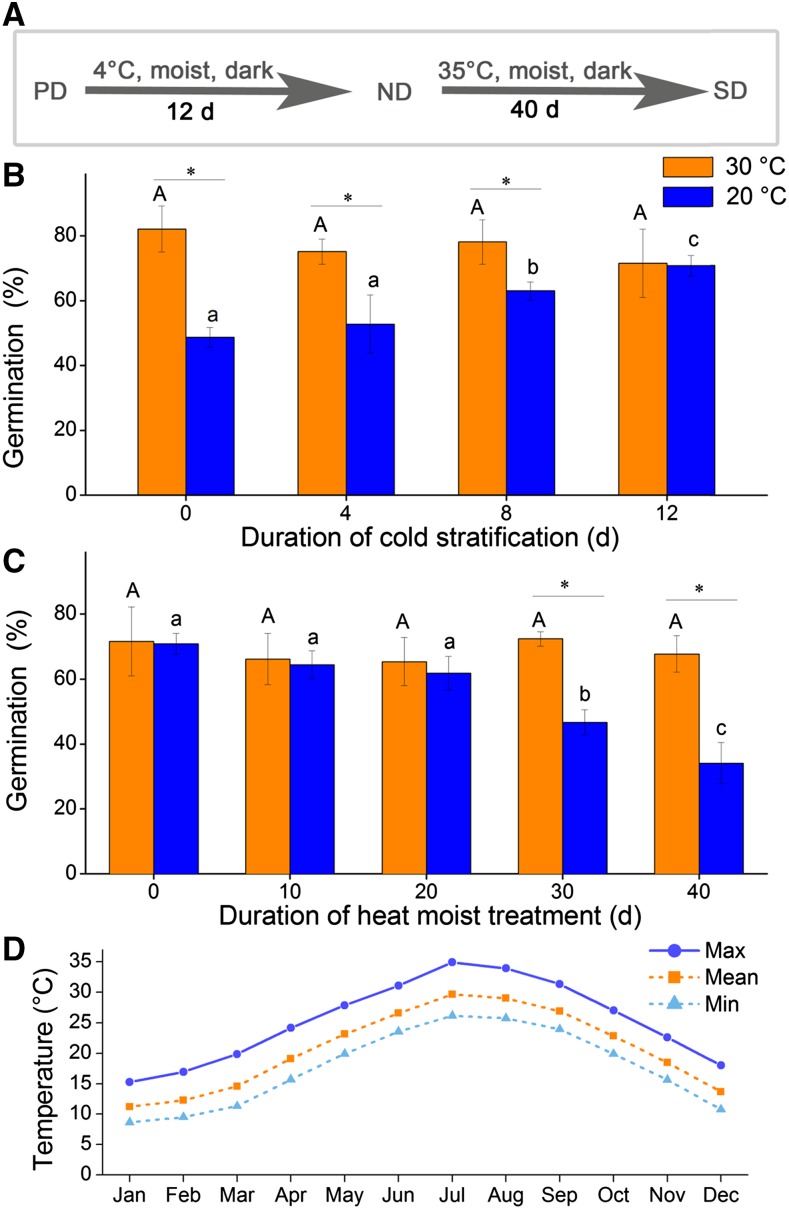
To obtain seeds with various dormancy statuses, we first tested the germination percentages of seeds that had been subjected to different environmental conditions (Fig. 1A). C. lanceolata seeds maintained relatively high germination percentages at 30°C throughout the experiment (Fig. 1, B and C). The freshly matured seeds germinated to 48.7% ± 3.0% at 20°C, and germination gradually increased in response to cold stratification, increasing significantly (P < 0.05) to 70.8% ± 3.2% after 12 d of cold stratification (Fig. 1B). After eight additional days of cold stratification, the germination percentage did not significantly change (P > 0.05; Supplemental Fig. S1). These results suggested that the seeds matured under primary dormancy, and dormancy release occurred during the 12-d cold stratification. When cold-stratified (for 12 d) seeds were subjected to high temperature (35°C), the subsequent germination percentage at 20°C strongly decreased, reaching 34.1% ± 6.4% after 40 d, indicating that secondary dormancy was induced (Fig. 1C). For simplicity, freshly matured, 12-d-cold-stratified and 40-d-stressed seeds are referred to as primary dormant (PD), nondormant (ND), and secondary dormant (SD) seeds, respectively.
Figure 1.
Germination of seeds with various dormancy statuses. A, Schematic depiction of pretreatments used to obtain seeds with various dormancy statuses. The fresh seeds were cold stratified (4°C) for 12 d, and seed germination was tested at 4-d intervals; the 12-d-cold-stratified seeds were transferred to 35°C moist conditions and tested for germination at 10-d intervals. B and C, Germination of seeds after pretreatment: (B) 4°C cold stratification and (C) 35°C heat stress. Vertical bars in B and C indicate ses. The same uppercase letters indicate no significant difference (P > 0.05) among germination percentages at 30°C. The same lowercase letters indicate no significant differences (P > 0.05) among germination percentages at 20°C. The asterisks indicate a significant difference (P < 0.05) between germination percentages of the same seed lot at 20°C and 30°C. D, Average monthly mean, maximum, and minimum temperatures in the natural habitat of the C. lanceolata trees used in this study. Data are from the 2000 to 2010 record from the Portal of Chinese Science and Technology Resource (http://www.escience.gov.cn).
Cytological Changes in C. lanceolata Seeds during Release and Induction of Dormancy
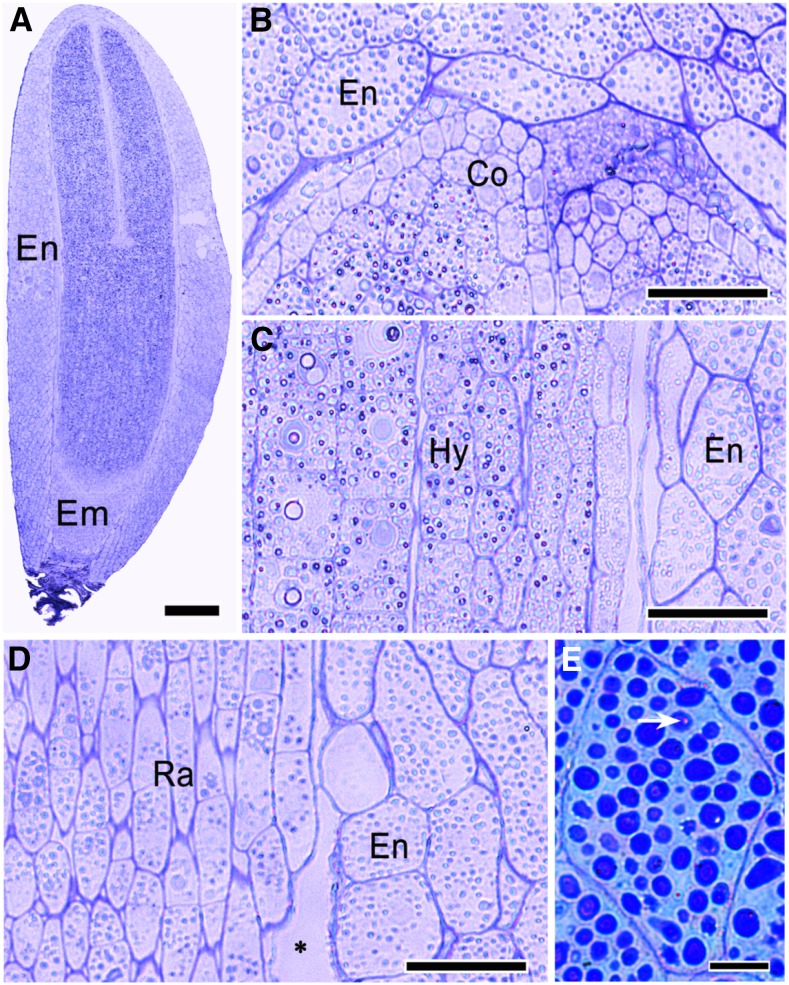
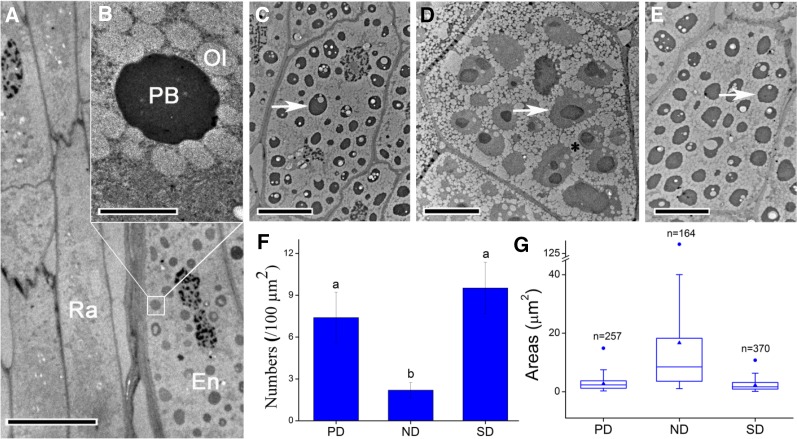
In freshly matured C. lanceolata seeds, the embryos were fully developed and surrounded by several layers of endosperm cells (Fig. 2A). Cells of C. lanceolata seeds contained large amounts of reserve substances (Fig. 2, A–D). The reserve substances were stained blue by Coomassie Brilliant Blue (Fig. 2E), indicating that they were composed of proteins. The cells also contained some polysaccharides, as revealed by periodic acid-Schiff’s reagent staining (Fig. 2E). Given that interactions between radicle cells and the surrounding endosperm determine physiological dormancy in endospermic seeds (Morris et al., 2011; Zhang et al., 2014), we applied transmission electron microscopy to investigate subcellular changes in these cells during dormancy release and induction (Fig. 3, A–E). From Figure 3, C to E, it is evident that considerable changes occurred in the endosperm cells. The protein bodies in the endosperm cells of PD seeds were small and present at high concentrations (Fig. 3, F and G). After a 12-d cold stratification, we observed larger but fewer protein bodies in ND seeds (Fig. 3, F and G). During secondary dormancy induction, the number of protein bodies increased but the size decreased (Fig. 3E). The protein bodies in endosperm cells were surrounded by oleosomes (Fig. 3B), which showed similar dynamics as the protein bodies did (Fig. 3D).
Figure 2.
Light micrographs of C. lanceolata seed sections. A, Semithin section stained with toluidine blue O indicates that the embryo is fully developed in a freshly matured seed. B to D, There is considerable variation in the morphology of cells in different parts of the embryo: (B) cotyledon, (C) hypocotyl, and (D) radicle. E, Protein bodies of an endosperm cell stained blue by Coomassie Brilliant Blue R. The asterisk in D indicates that there is a visible gap between the embryo and the endosperm in the radicle region. The arrow in E indicates the presence of polysaccharide in the cells stained by periodic acid-Schiff’s reagent. Co, Cotyledon; Em, embryo; En, endosperm; Hy, hypocotyl; Ra, radicle. Bars = 200 μm in A, 25 µm in B to D, and 10 μm in E.
Figure 3.
Electron micrographs of ultra-thin sections of the radicle region. A, Radicle cells and neighboring endosperm of a freshly matured seed. The endosperm cells contain many protein bodies, but the radicle cells do not. B, A higher magnification image of the region of interest in A, showing that protein bodies are surrounded by small oleosomes. C to E, Endosperm cells close to radicle cells in seeds with various dormancy statuses: (C) primary dormant, (D) nondormant, and (E) secondary dormant seeds. F and G, There is a notable change in number and size of protein bodies in the endosperm cells during dormancy release and induction: (F) number and (G) size. Numbers and sizes of protein bodies were calculated in 10 cells each of PD, ND, and SD seeds. There were 257, 164, and 370 protein bodies in the 10 cells of PD, ND, and SD seeds, respectively. The same lowercase letters in F and G indicate no significant difference (P > 0.05) between the values. Values in F are means ± sd; solid triangles indicate mean values, and solid circles indicate the maximum values in G. En, Endosperm; Ol, oleosome; PB, protein body; Ra, radicle. Arrows in C to E indicate PBs. The asterisk in D indicates that there was a coalescence of smaller oleosomes into a larger one. Bars = 10 μm in A and C to E and 2 μm in B.
De Novo Assembly and Gene Annotation of the Global Transcriptome
We conducted de novo transcriptome assembly according to the Trinity pipeline (Grabherr et al., 2011), using all clean reads from the three libraries (PD, ND, and SD seeds), as well as our previously published mixed library from C. lanceolata leaves, stems, roots, and seedlings (Qiu et al., 2013). After stringent data cleanup and quality checks, we obtained 95,630,372, 102,209,268, and 100,202,696 clean reads from the PD, ND, and SD seed libraries, respectively. The assembly generated 81,997 transcripts with a total length of 100,852,690 bp and an average length of 1,229 bp. Homologous transcripts with >95% similarity were clustered, producing 64,189 unigenes. The length of the unigenes ranged from 201 to 17,728 bp, with an average of 1,132 bp (Supplemental Table S1).
All unigenes were annotated using BLASTX searches against the NCBI nonredundant protein sequences (NR), Swiss-Prot, and Pfam databases. This identified 36,398 unigenes matching known genes in the NR database with E-value of <10−5, accounting for 56.70% of total unigenes (Supplemental Table S2). Similarly, 25,880 (40.32%) unigenes were identified in the Swiss-Prot and 31,740 (49.45%) in Pfam databases. To complete the annotation of unigenes in our global transcriptome, we used GO (Gene Ontology), KOG (Eukaryotic Ortholog Groups), and KEGG (Kyoto Encyclopedia of Genes and Genomes) assignments to classify their functions. Based on sequence similarity, 23,173 unigenes (36.10% of total unigenes) were annotated in the GO database (http://geneontology.org/). A total of 22,327 unigenes (34.78% of total unigenes) were categorized into 25 KOG functional groups, among which “general function prediction only” represented the largest group, followed by “signal transduction mechanisms,” “posttranslational modification, protein turnover, chaperones,” and “translation, ribosomal structure, and biogenesis” (Supplemental Fig. S2). There were 18,338 (28.57%) unigenes assigned to 276 KEGG pathways; “ribosome,” “oxidative phosphorylation,” “cell cycle,” and “purine metabolism” were the most highly represented pathways.
Transcriptomic Alterations in Seeds during Dormancy Release/Induction
To assess alterations in gene expression during release and induction of dormancy in C. lanceolata seeds, we globally profiled mRNA expression in PD, ND, and SD seeds. We detected 38,294, 41,521, and 35,473 unigenes (>1 read per kilobase per million clean reads [RPKM]) in PD, ND, and SD seeds, respectively (Supplemental Table S3). By further analyzing differential gene expression between ND and dormant (PD and SD) seeds, we found 5,911 differentially (|log2 fold change| > 2, P < 0.05) expressed genes (DEGs) between ND and PD seeds, 3,464 DEGs between ND and SD seeds, and 2,424 DEGs between PD and SD seeds (Supplemental Table S4). Hierarchical cluster analysis indicated that these DEGs can be grouped into five clusters (Supplemental Fig. S3). Genes in Cluster 1 showed increased expression during primary dormancy release and decreased expression during secondary dormancy induction. By contrast, genes in Cluster 3 showed decreased expression followed by increased expression during these two processes. The opposite change patterns of gene expression during dormancy release and induction suggested a tight linkage of these genes with dormancy of C. lanceolata seeds.
We further performed GO enrichment analysis to investigate biological functions of these crucial DEGs. It was revealed that Cluster 1 genes were enriched in GO terms (biological process subcategory) associated with respiration, translation, cell cycle, and stress responses (Supplemental Fig. S4). Cluster 3 genes were enriched in GO terms related to ABA responses, cell wall modification, and responses to stimulus. For the rest three gene clusters (Clusters 2, 4, and 5), enriched GO terms were mostly associated with regulation of gene expression, development maturation, and secondary metabolism. It was of interest to note that Cluster 5 genes were also enriched in GO terms related to respiration and stress responses. Using quantitative real-time PCR (qRT-PCR) to validate the DEGs identified by high-throughput sequencing, we found similar expression patterns, which supported the reliability of the sequencing data (Supplemental Fig. S5). Pearson correlation analysis also indicated a close correlation between RNA-seq and qRT-PCR data (r = 0.651, P < 0.0001).
Genes Regulating Respiration Play a Role in Dormancy Release
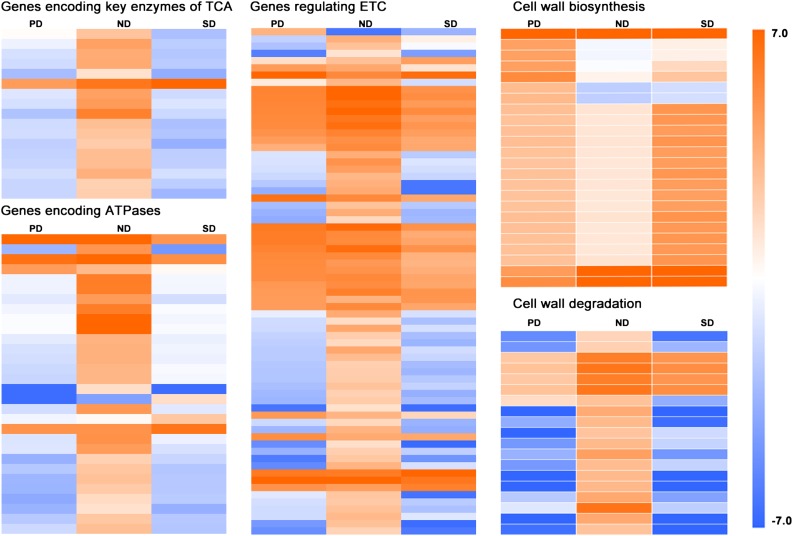
Since GO enrichment analysis showed that genes in Clusters 1 and 5 were enriched in GO terms related to respiration, we paid special attention to genes involved in respiration. We totally identified 73 unigenes encoding critical enzymes of the tricarboxylic acid cycle, 231 unigenes involved in the electron transport chain, and 71 ATPase unigenes in the three C. lanceolata seed libraries (Supplemental Table S5). Among these respiration- and energy-related genes, 117 were differentially expressed between dormant (PD or SD) and ND seeds. Notably, all DEGs encoding critical tricarboxylic acid cycle enzymes, including aconitase, citrate synthase, fumarase, malate dehydrogenase, pyruvate dehydrogenase, succinate dehydrogenase, succinyl-CoA synthetase, and dihydrolipoylsuccinyl transferase, were more highly expressed in ND seeds. Moreover, 90% of the electron transport chain-related and ATPase DEGs were upregulated in ND seeds (Fig. 4).
Figure 4.
Expression profiles of DEGs that regulate respiration and cell wall turnover in PD, ND, and SD seeds. The expression levels of genes (RPKM) based on RNA-seq were log2 transformed and color-coded (blue, low expression levels; orange, high expression levels). TCA, Tricarboxylic acid cycle; ETC, electron transport chain.
Cell Wall Turnover during the Release and Induction of Dormancy
Based on key genes in the regulation of cell wall biosynthesis and degradation discussed in recent reviews (Cosgrove, 2005; Mohnen, 2008; Harholt et al., 2010; Vanholme et al., 2010), we detected 223 genes regulating cell wall turnover in C. lanceolata seeds (Supplemental Table S6). Furthermore, 43 genes exhibited differential expression between dormant (PD or SD) and ND seeds (Fig. 4). Intriguingly, genes regulating cell wall biosynthesis and degradation exhibited distinct expression patterns. The expression levels of genes regulating cell wall biosynthesis were significantly higher (log2 fold change > 2, and P < 0.05) in dormant (PD, SD, or both) seeds than in ND seeds, including CSLH1 (cellulose synthesis-like H1), CESA8 (cellulose synthesis 8), KOR (KORRIGAN, a membrane-bound endoglucanase), 4CLL10 (4-coumarate-CoA ligase-like 10), GAUT1 (galacturonosyltransferase 1), and GAUT14 (Fig. 4). By contrast, the expression levels of genes regulating cell wall degradation were higher in ND seeds than in dormant (PD and SD) seeds, including genes encoding β-glucosidase, β-glucosidase-like protein, endo-1,3(4)- β-glucanase, xylanase, and pectaselyases (Fig. 4).
Hormonal Regulation of C. lanceolata Seeds
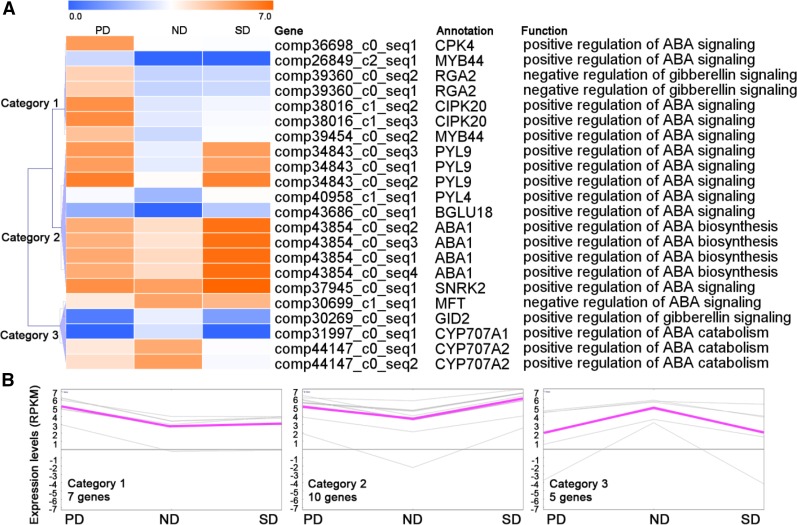
To investigate the hormonal regulation of C. lanceolata seed dormancy, we analyzed the expression of the GA and ABA related genes. We detected 109 genes (>1 RPKM) regulating GA and ABA biosynthesis, catabolism, and signaling in C. lanceolata seeds (Supplemental Table S7), among which 22 exhibited significantly differential expression between dormant (PD or SD) and ND seeds (Fig. 5). Using hierarchical cluster analyses, we grouped the DEGs into three categories (Fig. 5). Categories 1 and 2 genes were more highly expressed in dormant (PD or SD) seeds than in ND seeds (Fig. 5). By contrast, category 3 genes, including GID2, CYP707A1, and CYP707A2, were expressed at higher levels in ND seeds than in dormant (PD and SD) seeds (Fig. 5). Nevertheless, expression patterns of genes in Categories 1 and 2 exhibited some variations. Specifically, genes in Category 1, including MYB44, RGA2, CPK4, and CIPK20, were expressed at higher levels in PD seeds than in ND seeds, and genes in Category 2, including ABA1, SNRK2, PYL9, and BGLU19, were expressed at higher levels in SD seeds than in ND seeds (Fig. 5). Using qRT-PCR to validate the expression patterns of some GA and ABA associated genes, we found that expression patterns of these validated genes in qRT-PCR analysis were in agreement with those in RNA-seq (Supplemental Fig. S6).
Figure 5.
Expression profiles of genes involved in the regulation of biosynthesis, catabolism, and sensitivity to GA and ABA. A, Hierarchical cluster analysis grouped the DEGs associated with GA and ABA into three categories. B, Gene expression patterns in each category. The expression levels of genes (RPKM), based on RNA-seq, were log2 transformed and color-coded (blue, low expression levels; orange, high expression levels).
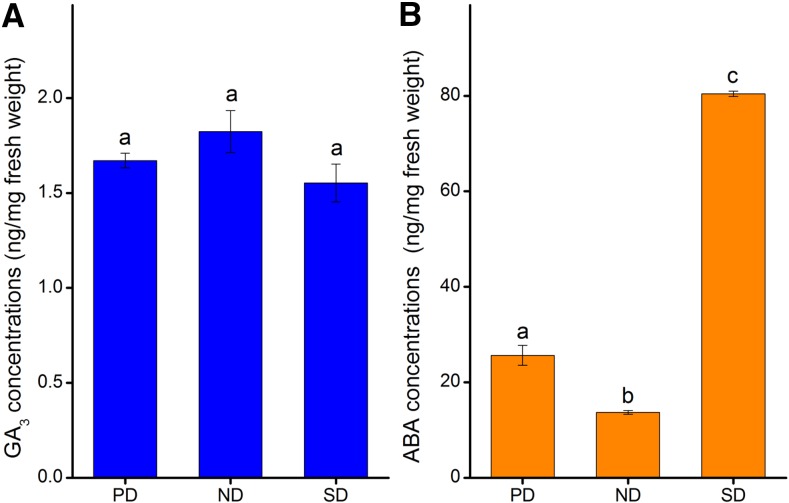
Exogenous GA and ABA was also applied to seeds to verify their influences on seed germination of C. lanceolata. We found that GA3 and GA4+7 increased seed germination slightly from 28% ± 3.65% (in water) to 36% ± 3.65% and 32% ± 6.53%, respectively (Supplemental Fig. S7), while ABA inhibited germination dramatically, with no germination in the 100 µm ABA solution (Supplemental Fig. S7). During the analyses of endogenous GA and ABA levels in C. lanceolata seeds, we found that freshly matured seeds (PD seeds) had an average GA3 concentration of 1.67 ± 0.04 ng mg−1 and an average ABA concentration of 25.65 ± 2.09 ng mg−1 fresh weight (FW; Fig. 6). Endogenous GA3 concentrations remained constant in C. lanceolata seeds during dormancy release and induction, while ABA concentrations varied significantly (P < 0.05) in PD, ND, and SD seeds (Fig. 6). During dormancy release, ABA concentration decreased to 13.71 ± 0.40 ng mg−1 FW in ND seeds, and it increased during the induction of secondary dormancy, reaching 80.43 ± 0.55 ng mg−1 FW in SD seeds (Fig. 6).
Figure 6.
Hormonal concentrations in C. lanceolata seeds. A, GA contents in PD, ND, and SD seeds. B, ABA contents in the seeds. Values with different letters are significantly different (P < 0.05) from each other. Error bars represent ses.
Transcriptional and Translational Regulation of Seed Dormancy
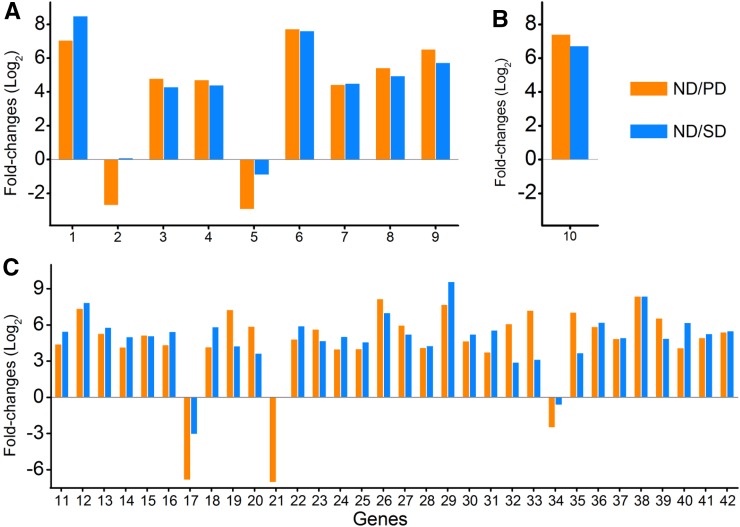
The significant alteration in gene expression programs between dormant and ND seeds prompted us to investigate the regulation of gene expression during C. lanceolata seed dormancy. We measured the expression levels of genes encoding transcriptional initiation factors (TFIIs), transcription elongation factors, and translation initiation factors (eIF1-5) in C. lanceolata seeds. We detected nine TFII genes, one transcription elongation factor gene (SPT6), and 32 eIF genes showed more than 5-fold differences (P < 0.05) in expression levels between dormant (PD or SD) and ND seeds (Fig. 7). Moreover, seven out of nine TFII genes and 19 out of 22 eIF genes were more highly expressed in ND seeds than in dormant (PD and SD) seeds (Fig. 7). These genes exhibited strong variation in expression between dormant and ND seeds, with an average fold change of 83.9 and a maximum of 363 (for TFII).
Figure 7.
Differential expression of genes regulating transcription and translation. A, Transcription initiation factors; B, transcription elongation factors; C, translation initiation factors. The numbers 1 to 42 of the x axis represent unigenes comp27016_c2_seq2, comp33593_c0_seq1, comp35471_c0_seq1, comp35471_c0_seq2, comp42802_c0_seq1, comp55492_c0_seq1, comp72530_c0_seq1, comp98783_c0_seq1, comp98928_c0_seq1, comp13373_c0_seq1, comp12692_c0_seq1, comp14777_c1_seq1, comp14818_c0_seq1, comp15208_c0_seq1, comp15932_c0_seq1, comp15977_c0_seq1, comp17459_c0_seq1, comp17480_c0_seq1, comp19806_c0_seq1, comp20250_c0_seq1, comp20598_c0_seq1, comp20650_c0_seq1, comp20784_c0_seq1, comp20807_c0_seq1, comp22810_c0_seq1, comp24591_c0_seq2, comp25333_c0_seq1, comp26384_c0_seq2, comp26433_c0_seq3, comp26701_c1_seq1, comp31465_c0_seq1, comp32700_c1_seq1, comp32700_c1_seq2, comp34970_c4_seq2, comp35211_c0_seq1, comp46043_c0_seq1, comp70931_c0_seq1, comp75085_c0_seq1, comp8129_c0_seq1, comp81330_c0_seq1, comp8710_c0_seq1, and comp9914_c0_seq1, respectively.
mRNA Degradation in C. lanceolata Seeds
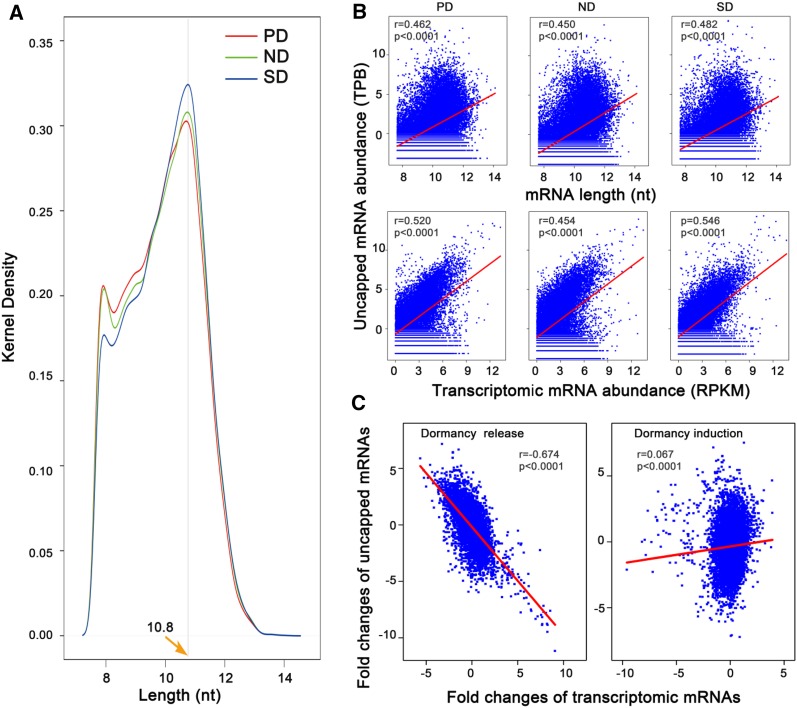
To globally analyze mRNA degradation in C. lanceolata seeds, we performed high-throughput sequencing of PARE libraries from PD, ND, and SD seeds. In total, we identified 33,438 uncapped mRNA species in C. lanceolata seeds, among which 26,263, 25,568, and 22,611 were detected in the PD, ND, and SD libraries, respectively (Supplemental Fig. S8). We found that 28,325 unigenes were detected in both capped (transcriptome libraries) and uncapped forms (PARE libraries), accounting for 54.89% of transcriptome unigenes (Supplemental Fig. S8). In the three libraries, mRNA decay products were detected for mRNAs 201 to ∼17,728 nucleotides in length, with ∼1,783-nucleotide mRNAs having the most abundant uncapped decay products (Fig. 8A). Correlation analysis revealed a positive correlation (Pearson correlation = 0.462, P < 0.0001) between uncapped mRNA abundance and transcriptomic mRNA length in PD seeds (Fig. 8B). Transcriptomic mRNA abundance was also positively correlated to uncapped mRNA levels in PD seeds (Pearson correlation = 0.520, P < 0.0001; Fig. 8B). Analysis of the ND and SD libraries confirmed these positive correlations (Fig. 8B).
Figure 8.
Characterization of uncapped mRNA expression in C. lanceolata seeds. A, Length distribution of mRNAs with uncapped decay products. The y axis shows the relative frequency of mRNA species detected in the uncapped form. B, Correlations between uncapped mRNA abundance and transcriptomic mRNA length and abundance. The three upper panels show correlations between expression levels of uncapped mRNAs and transcriptomic mRNA length, and the lower three show correlations between expression levels of uncapped mRNAs and transcriptomic mRNA abundance in PD, ND, and SD seeds. C, Correlation between fold changes of uncapped mRNAs and those of transcriptomic mRNAs. A close negative correlation was found between fold changes of uncapped mRNAs and that of transcriptomic mRNAs during the release of primary dormancy (left), but a loose correlation was detected during secondary dormancy induction (right). Data for gene expression levels and mRNA length were log2 transformed.
To evaluate the contribution of mRNA decay to the regulation of mRNA levels, we analyzed the relationship between fold changes of uncapped mRNA levels and those of transcriptomic mRNAs. During primary dormancy release of C. lanceolata seeds, a significantly negative correlation (Pearson correlation = −0.674, P < 0.0001) was observed between the fold change (ND/PD) of uncapped mRNA levels and that of transcriptomic mRNAs (Fig. 8C). A loose correlation (Pearson correlation = 0.067, P < 0.0001) was observed between the fold change (SD/ND) of uncapped mRNA levels and that of transcriptomic mRNAs during secondary dormancy induction (Fig. 8C). Furthermore, we detected 44 genes encoding mRNA decay factors (including CCR4-NOT, DCP, EXOSC10, PARN, XRN, UPF, and ZC3H12A) in C. lanceolata seeds. The expression levels of these genes were similar among PD, ND, and SD seeds, except for five genes which were significantly more highly expressed (|log2 fold change| > 2, P < 0.05) in PD seeds than in ND and SD seeds (Supplemental Table S8).
When analyzing the DEGs detected in RNA-Seq and PARE libraries, we found that 269 DEGs showed opposite expression patterns in the capped and uncapped forms (Supplemental Table S9), with 124 DEGs found during the process of primary dormancy release and 150 DEGs found during the process of secondary dormancy induction (there was an overlap of five genes between the 124 and 150 DEGs). The 269 genes covered 19 KOG categories, indicating that mRNA degradation regulated expression levels of genes in a variety of functional groups (Supplemental Fig. S9). Especially, seven out of the 269 genes encoded proteins involved in GA and ABA metabolism and signaling, suggesting a critical role of mRNA degradation in seed dormancy regulation of C. lanceolata seeds (Supplemental Table S10).
Roles of miRNAs in Seed Dormancy in C. lanceolata
PARE analysis revealed 1,978 miRNA-target mRNA pairs in the three libraries (PD, ND, and SD) of C. lanceolata seeds (Supplemental Table S11). To further confirm the miRNAs, we identified the precursors of these candidate miRNAs. Only candidate miRNAs having precursors that could form stem-loop structures were identified as authentic miRNAs. We identified19 authentic miRNAs in C. lanceolata seeds (Supplemental Table S12). Most targets of the 19 miRNAs act in hormone signal sensing (including PLT1, TIR1, 14-3-3, and Scarecrow-like) and gene expression regulation (including NFYA, AGO1, DCL1, UBQL1, and genes encoding homeobox-Leu zipper protein 15, WD repeat-containing proteins, and ribosomal protein S3Ae; Supplemental Table S13). In addition, SPL7, which regulates cell wall biogenesis, was identified as a target of cln-miR156a.
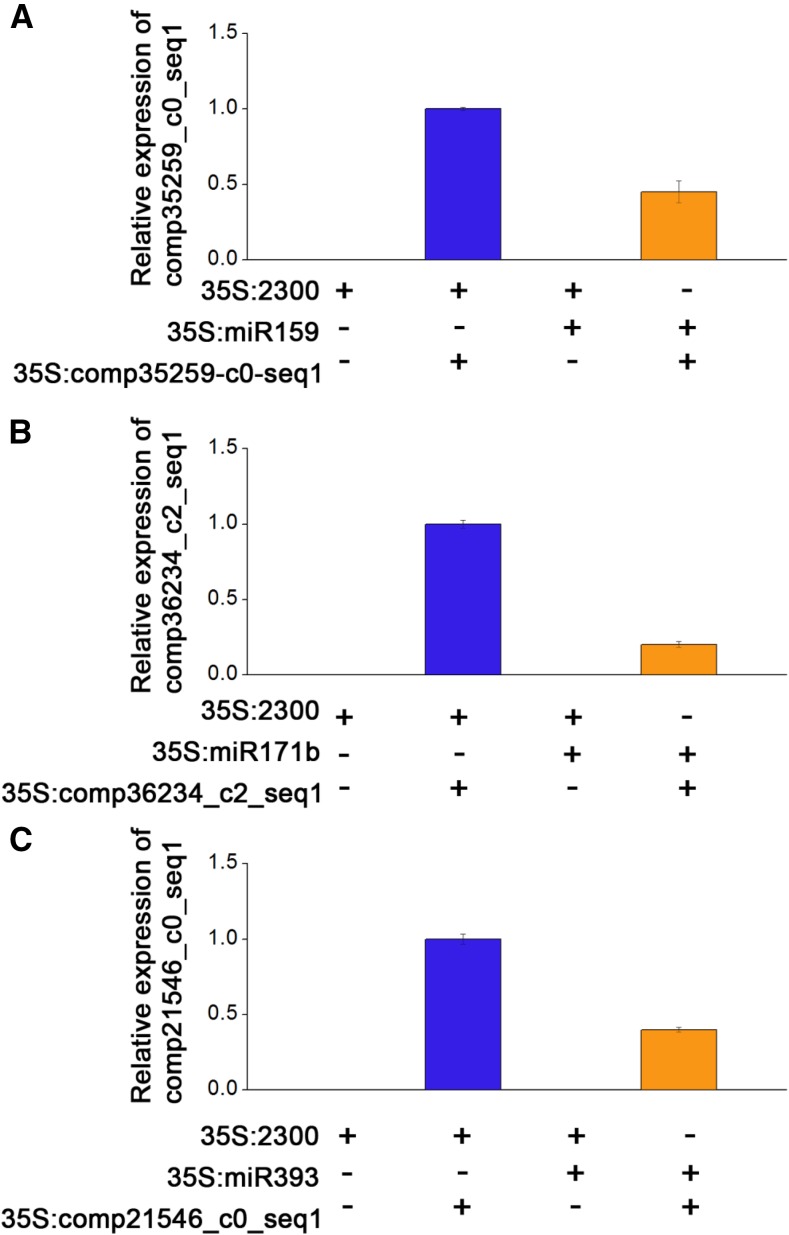
To validate the miRNA/target pairs predicated via degradome sequencing, we undertook transient coexpression assays in Nicotiana benthamiana leaves. We chose three miRNA/target pairs, including miR159/comp35259_c0_seq1, miR171b/comp36234_c2_seq1, and miR393/comp21546_c0_seq1, to conduct the coexpression experiments. After 2 d of coexpression in N. benthamiana leaves, RNA was extracted and expression levels of the target genes analyzed via qRT-PCR. As expected, the target genes expression was decreased considerably by the miRNAs (Fig. 9), demonstrating that the miRNAs exhibited differential efficiency in regulating expression levels of their target genes.
Figure 9.
Validation of the predicted miRNA/target pairs using the transient coexpression assay. Coexpression of various combinations of miRNA and the target gene expression constructs in N. benthamiana: A, the validation of miR159/comp35259_c0_seq1; B, the validation of miR171b/comp36234_c2_seq1; and C, the validation of miR393/comp21546_c0_seq1. Expression levels were quantified using qRT-PCR. Values are means ± se.
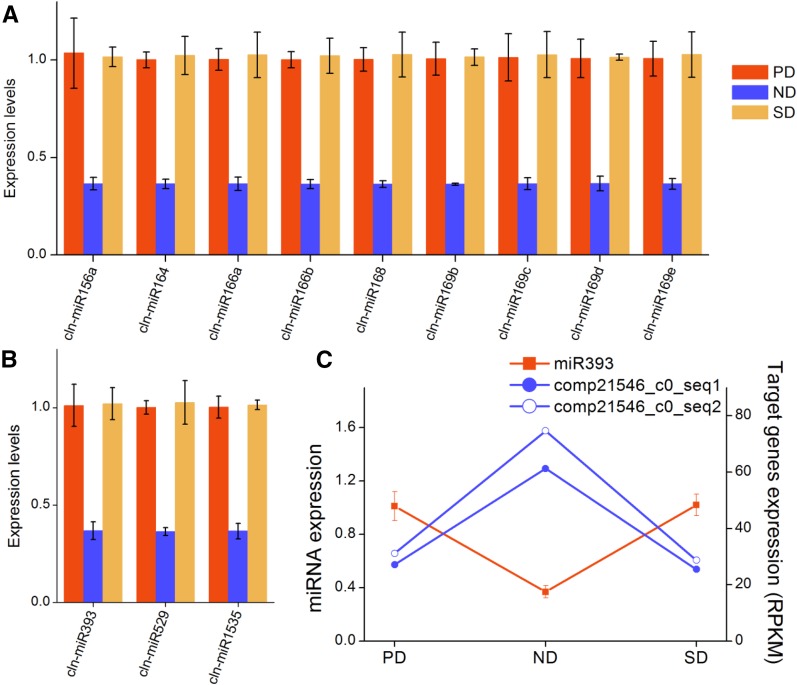
We also performed qRT-PCR of the mature miRNAs using sequence-specific primers to characterize the expression patterns of these authentic miRNAs. In total, we detected the expression of 12 miRNAs via qRT-PCR (Fig. 10, A and B). The expression of all 12 miRNAs decreased during the release of primary dormancy and increased during the induction of secondary dormancy. It is interesting to note that the two targets of miR393, which were comp21546_c0_seq1 and comp21546_c0_seq2, showed increased expression during primary dormancy release and decreased expression during secondary dormancy induction (Fig. 10C). The two target genes of miR393 were homologous to TIR1 in Arabidopsis and rice, which encodes auxin receptor F-box proteins.
Figure 10.
Expression of miRNAs in C. lanceolata seeds. A and B, Twelve miRNAs were expressed at detectable levels using qRT-PCR. C, The expression pattern of miR393 was opposite to that of its target transcripts. Unigenes comp21546_c0_seq1 and comp21546_c0_seq2 are homologous to auxin receptor F-box gene TIR1 in rice and Arabidopsis and are predicted cleavage targets of miR393 by PARE analysis.
DISCUSSION
Many plants have adopted physiological dormancy to cope with harsh environments during the seed stage (Baskin and Baskin, 2014). This type of dormancy is caused by low growth potential of the embryo and can be disrupted by a period of cold or warm stratification (Baskin and Baskin, 2003, 2014). In this study, freshly matured C. lanceolata seeds had fully developed embryos, and the germination increased in response to 12-d cold stratification, indicating dormancy release. Furthermore, the seeds exhibited reduced germination after exposure to 35°C, indicating that secondary dormancy was induced. Compared with changes observed in herbaceous species such as Arabidopsis (Wang et al., 2013; Xiang et al., 2014), the changes in dormancy status of C. lanceolata seeds were mild; however, these changes appeared quite robust and were supported by changes in ABA levels. Overall, these results suggest that C. lanceolata seeds undergo physiological dormancy.
The cytological changes during dormancy release have been examined in seeds with physiological dormancy. For example, in apple (Pyrus malus), only minor changes in the endoplasmic reticulum occur in embryo cells during dormancy release (Bouvier-Durand et al., 1981). In other species, such as barley, cytological changes may be absent during dormancy release (Barrero et al., 2009). Similar to Arabidopsis seeds (Bethke et al., 2007), we found an increase of size but a decrease of number of protein bodies and oleosomes in endosperm cells of C. lanceolata seeds during dormancy release, which might be as a result of a coalescence process. Nevertheless, we revealed that the cytological changes were reversible (coalescence versus separation of protein bodies and oleosomes) in C. lanceolata seeds during dormancy release and induction. Compared with the significant and rapid changes during seed germination in Brachypodium distachyon and maize (Zea mays; Barrero et al., 2012; Liu et al., 2013a), relative mild cytological changes occur during dormancy release in C. lanceolata seeds, suggesting that the cytological and morphological basis of germination is well established in mature seeds with physiological dormancy, while germination is blocked by a physiological barrier.
The dynamic balance in ABA/GA concentration and sensitivity plays a central role in physiological dormancy in various seeds (Barrero et al., 2012; Cantoro et al., 2013; Vaistij et al., 2013). For example, increased GA biosynthesis and ABA catabolism lead to dormancy release in Arabidopsis (Footitt et al., 2011), and ABA sensitivity is closely related to dormancy level in barley seeds (Benech-Arnold et al., 2000). It was reported that ABA synthesis, GA degradation, and ABA signaling were enhanced during dormancy induction and that GA synthesis, ABA degradation, and GA signaling were enhanced during dormancy release of Arabidopsis seeds (Cadman et al., 2006; Finch-Savage et al., 2007; Footitt et al., 2011). However, this set of GA/ABA balance was not proved the case in C. lanceolata seeds. Here, we found that negative regulation of GA signaling plays a specific role in primary dormancy release, while positive regulation of ABA biosynthesis specifically acts on secondary dormancy induction in C. lanceolata seeds. In addition, we found that during dormancy release and induction, endogenous GA concentrations maintained relatively constant, whereas ABA concentrations varied, thereby modifying the ABA/GA balance. Furthermore, we detected reduced expression of RGA2 (encoding a negative regulator of GA signaling) during primary dormancy release and increased expression of ABA1 (encoding a positive regulator of ABA biosynthesis) during secondary dormancy induction, indicating that there are distinct modes of hormonal regulation during primary and secondary dormancy in C. lanceolata seeds. These findings suggest that seeds with physiological dormancy utilize distinct strategies to fine-tune ABA/GA balance during primary and secondary dormancy.
Differential gene expression in dormant and nondormant seeds may explain the variation of germination capacity. Previous investigations reported up-regulation of genes regulating translation, cell division, and energy production during dormancy release in Arabidopsis seeds (Cadman et al., 2006). In this study, we also observed increased expression of genes regulating translation, cell cycles, and respiration in nondormant C. lanceolata seeds, indicating key roles of these functional groups of genes in seed dormancy and germination. More strikingly, GO enrichment analysis revealed differential expression of genes associated with cell wall turnover during dormancy release and induction of C. lanceolata seeds. We further found that genes regulating cell wall biosynthesis were down-regulated during dormancy release and up-regulated during dormancy induction. By contrast, genes regulating cell wall degradation were up-regulated during dormancy release and down-regulated during dormancy induction. Although genes involved in cell wall turnover were not among the most differentially expressed gene groups, several genes encoding expansin, α-xylosidase, β-galactosidase/lactase, endo-xyloglucan transferase, and pectinesterase were found to be more highly expressed in after-ripened seeds than dormant Arabidopsis seeds (Cadman et al., 2006). Since cell wall degradation was critical for radicle protrusion via weakening the surrounding endosperm (Barrero et al., 2009; Zhang et al., 2014), we suggest that intensive cell wall modification may be necessary for dormancy release and induction to change germination capacity of seeds during dormancy cycles.
Besides the reversible changes of gene expression relating to cell wall modification, energy production, translation, and hormone responses during dormancy release and induction, we also found some differences in gene expression between PD and SD seeds. Intriguingly, some genes involved in secondary metabolism showed higher expression levels in SD seeds than in PD seeds, while genes regulating stress responses, responses to virus, as well as responses to ozone showed lower expression in SD than in PD seeds. In addition, PD and SD seeds showed differential expression of some other genes, e.g. genes regulating ion (including potassium, nitrogen, and hydrogen) transport. These findings suggested that some unknown regulation underlying seed dormancy remained to be illustrated in future studies.
PARE analysis revealed extensive posttranscriptional regulation of physiological dormancy in C. lanceolata seeds. We detected uncapped transcripts for more than half of the expressed genes in C. lanceolata seeds. In Arabidopsis flowers, >90% of all expressed genes produce uncapped transcripts (Jiao et al., 2008). In seedlings of another herbaceous plant, B. distachyon, uncapped transcripts were detected for ∼67% of all genes (Zhang et al., 2013). These results suggest that mRNA degradation is broadly involved in regulating gene expression in plants. Furthermore, we demonstrated that increased mRNA degradation activity considerably reduce mRNA expression levels during primary dormancy release. Especially, our study revealed that expression levels of several key genes regulating ABA and GA metabolism and signaling could significantly be reduced by mRNA degradation. However, we detected only a loose positive correlation between mRNA degradation and reduced mRNA levels during secondary dormancy induction. Therefore, mRNA degradation appears to function as an efficient posttranscriptional regulator of gene expression during primary dormancy in C. lanceolata seeds and affects RNA homeostasis in cells during secondary dormancy induction.
Many miRNAs play important roles in plant development and stress responses (Giacomelli et al., 2012; Zhan et al., 2012; Lu et al., 2013; Muraro et al., 2014). Some miRNAs have been identified in seeds via high-throughput sequencing, suggesting that miRNAs are broadly involved in regulating various biological processes in seeds (Song et al., 2011; Shamimuzzaman and Vodkin, 2012). Moreover, miR393 is involved in root development and stress resistance via modulating the expression of auxin F-box genes (Vidal et al., 2010; Sunkar et al., 2012; Yan et al., 2015). Here, we detected a close association between miR393/TIR1 abundance and dormancy depth. TIR1 is an F-box protein acting as an auxin receptor (Vanneste and Friml, 2009), and accumulating evidence suggests a potential involvement of auxin in seed dormancy (Liu et al., 2007, 2013b). Specifically, miR393 was less abundant in ND seeds than in dormant seeds, whereas TIR1 transcripts were more abundant, suggesting that the miR393/TIR1 module also regulates seed dormancy. In addition, PARE analysis revealed miR162/DCL1 and miR168/AGO1 interactions in posttranscriptional regulation during seed dormancy of C. lanceolata. Furthermore, we found five miRNAs, including miR159, miR166a, miR166b, miR171b, and miR396a, to target genes that were involved in GA and ABA signaling, supporting the critical roles of miRNAs in fine-tuning hormonal balance of C. lanceolata seeds. Considering that expression levels of these miRNAs were closely related to seed dormancy depth, which was regulated by the GA/ABA balance, these findings suggested a feedback regulation of GA and ABA pathways in seed dormancy.
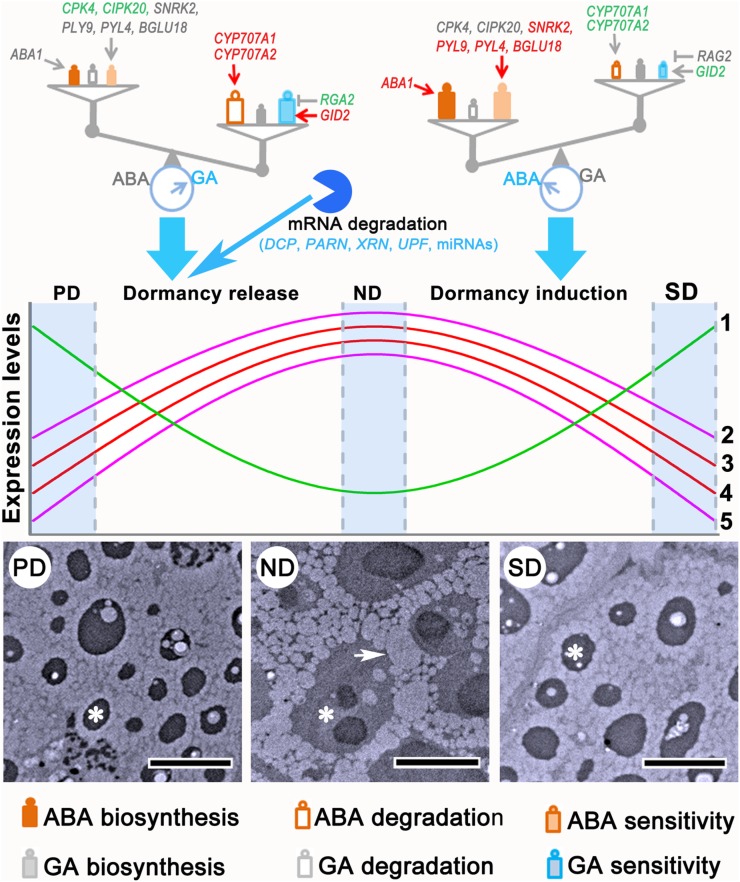
Based on previously published findings and these results, we propose a conceptual model for the regulatory network of physiological dormancy in the tree species C. lanceolata (Fig. 11). When a seed experiences certain environmental conditions, the existing ABA/GA balance is disturbed; this induces distinct gene expression programs, depending on the environmental conditions. Cool and wet conditions may lead to the dominance of GA, leading to dormancy release. In such cases, expression of genes positively regulating transcription initiation, translation initiation, respiration and energy production, cell division, and cell wall degradation would be induced, and while genes regulating cell wall biogenesis would be down-regulated. This gene expression program would prepare the seed for germination, resulting in increased ability to germinate during dormancy release. When the seed experiences unfavorable conditions, e.g. high temperature stress, ABA may play a dominant role in the ABA/GA balance, resulting in dormancy induction. As a result, an opposite pattern of gene expression would be induced, which would decrease the ability of the seed to germinate, thus promoting dormancy. Furthermore, distinct strategies may be adopted to fine-tune the ABA/GA balance during primary and secondary dormancy. Negative regulation of GA signaling plays a specific role in primary dormancy release, while positive regulation of ABA biosynthesis specifically acts on secondary dormancy induction. In addition to the central roles played by the ABA/GA balance, mRNA degradation is also required to reduce RNA levels during primary dormancy release.
Figure 11.
Diagram of the universal regulatory mechanisms of physiological dormancy and differences between primary and secondary dormancy. Environmental cues are sensed by the seeds and may disturb the existing ABA/GA balance, which opens the door for dormancy release or induction. The reversible changes in gene expression and cytology provide a basis for dormancy cycling. mRNA degradation mediated by DCPs, PARN, XRN, UPFs, and miRNAs also contributes to transcriptional regulation of gene expression during primary dormancy release. Colors of gene names in the ABA/GA balance diagram indicate different gene expression patterns. Red, green, and gray indicate increased, reduced, and stable expression, respectively. Group 1 genes are those regulating cell wall biogenesis, including CSL, CES, KOR, 4CLL10, and GAUT; Group 2, genes regulating transcription initiation and elongation and translation initiation, including TFIIs, eIFs, and SPT; Group 3, genes regulating respiration, including ACO, CSY, DLST, and FUM; Group 4, genes regulating cell cycle and division, including CYC, CDC, and CUL; Group 5, genes regulating cell wall degradation, including XYN and genes encoding β-glucosidase and pectase lyases. The transmission electron microscopy images are fitted from Figure 3, C to E. The asterisks in the transmission electron microscopy images indicate protein bodies, and the arrow indicates an oleosome. Bars in the transmission electron microscopy images represent 5 μm.
Our sequential transcriptome and degradome analyses have provided important insights into primary and secondary dormancy in C. lanceolata seeds. Three novel findings are as follows: (1) the changes in cytology and mRNA expression during dormancy release and induction, which are related to alteration in the ABA/GA balance, are reversible; (2) mRNA degradation is involved in regulating gene expression during primary dormancy; and (3) some miRNAs are involved in seed physiological dormancy via targeting crucial genes in hormonal signaling pathways. Taken together, these findings substantially increase our understanding of the complex regulatory framework underlying seed physiological dormancy.
MATERIALS AND METHODS
Plant Material and Germination Assays
Freshly matured seeds were collected from 20 Cunninghamia lanceolata (Cupressaceae) trees in Minhou County (N 26°35′, E 119°24′, a.s.l. 1020 m), Fujian Province, China on December 20, 2013. The trees had an average height of approximately 25 m and a mean diameter at breast height of 19.4 cm. The collected seeds were air-dried at room temperature for 10 d before initiation of germination tests. For all germination experiments, seeds were incubated at 20°C and 30°C in the light for 20 d. The testing temperatures were chosen based on the seasonal air temperature of the natural habitat for seed production (Fig. 1D). Four replicates each of 25 seeds were used, and radicle protrusion was the criterion for germination. Following germination tests, the viability of nongerminated seeds was assessed based on tetrazolium tests. Only viable seeds were taken into account when calculating germination percentages.
Seeds with various dormancy statuses were obtained by sequential treatments of cold stratification and warm incubation (Fig. 1A). Briefly, 40 petri dishes, each containing 400 seeds were cold stratified at 4°C in the dark. During cold stratification, four petri dishes were arbitrarily selected for germination tests every 4 d, with 50 seeds arbitrarily selected from each of the four dishes (25 seeds tested at 20°C and 25 at 30°C). After 12 d of cold stratification, all remaining dishes were transferred to 35°C in the dark, except for eight dishes maintained in cold stratification. The transferred seeds were stored for 40 d and tested for germination every 10 d. During the 40 d, ∼1 mL of water was sprayed onto each petri dish every 10 d to simulate occasional rainfall events in the wild. The dormancy status of the seeds was determined based on germination tests, allowing PD, ND, and SD seeds to be selected: PD seeds were fresh seeds; ND seeds were cold stratified for 12 d; and SD seeds were stored at 35°C for 40 d (Fig. 1, B and C).For each germination test, the remaining seeds in the selected dishes were stored at −80°C for RNA extraction and hormonal profiling.
Light Microscopy and Transmission Electron Microscopy
Semithin sections were stained with toluidine blue O, Coomassie Brilliant Blue (R 250), and periodic acid-Schiff’s reagent. Seed sections were observed under a Leica DM 2500 microscope equipped with a digital camera. Ultrathin sections (70 nm thick) of PD, ND, and SD seeds were cut with an ultramicrotome (Leica Ultracut R; Leica) and observed under a transmission electron microscope (JEM-1230; JEOL). Images were processed (sharpened, brightness, and contrast adjusted) and assembled using Photoshop CS5 (Adobe).
Analysis of Plant Hormones
Plant hormones were analyzed according to the standard protocol described by Pan et al. (2010). Triplicate samples of ∼50 mg of PD, ND, and SD seeds were used, and liquid chromatography-mass spectrometry analyses of GA and ABA were performed by the Testing and Analysis Center of Beijing Forestry University, Beijing, China.
RNA Isolation and RNA-Sequencing Analysis
Total RNAs was isolated separately from PD, ND, and SD seeds using RNAiso Plus and RNAiso-Mate for Plant Tissue (Takara) following the manufacturer’s protocol. The integrity and quantity of total RNA were assessed using a BioAnalyzer (Agilent Technologies). Three cDNA libraries (PD, ND, and SD) were prepared and sequencing was performed by LC Sciences using the Illumina Hi-Seq 2500 platform. The raw reads were filtered for adapter contamination, ambiguous residues (Ns), and low-quality reads prior to transcriptome assembly. The clean reads (after filtering) of the three libraries were pooled for de novo assembly of the global transcriptome using Trinity (Grabherr et al., 2011). BLAST analysis of unigene sequences of the assembled transcriptome was performed against the NCBI nonredundant protein database (NR), Swiss-Prot, Pfam, KOG, and KEGG for gene annotation. After assembly and gene annotation of the global transcriptome, clean reads of the PD, ND, and SD libraries were mapped to the transcriptome using Bowtie 2 (Langmead and Salzberg, 2012), and the number of mapped clean reads per unigene was calculated and normalized to the number of RPKMs (Mortazavi et al., 2008). GO enrichment was performed using the agriGO analysis tools (http://bioinfo.cau.edu.cn/agriGO/; Du et al., 2010), and the enriched GO terms were visualized via REVIGO (Supek et al., 2011) and Cytoscape (Cline et al., 2007) applications.
PARE Analysis of Uncapped Transcripts
Total RNA was extracted from PD, ND, and SD seeds using RNAiso Plus and RNAiso-Mate for Plant Tissue (Takara) in the same manner as for transcriptome sequencing. PARE libraries from PD, ND, and SD seeds were separately constructed following the protocol of German et al. (2009). Briefly, a modified RLM 5′-RACE technique was used to globally isolate RNAs with a 5′ monophosphate and a 3′ poly(A) tail. An RNA adapter was ligated to the 5′ ends of single-stranded degraded RNAs with a 5′ monophosphate, after which reverse transcription was performed to generate first strand cDNA using an oligo(dT) with a 3′ adapter. The cDNA was amplified using a short PCR and then digested with the restriction enzyme MmeI cleaving 20 nucleotides 3′ of the recognition site. After gel purification, the products were subjected to high-throughput sequencing by LC Sciences using an Illumina Hi-Seq 2000 instrument.
Analysis of uncapped mRNAs was conducted as described by Zhang et al. (2013). Briefly, raw sequencing reads were filtered by removing those corresponding to known rRNAs, tRNAs, small nuclear RNAs, small nucleolar RNAs, repeats, and transposons. The remaining clean reads were then mapped to the assembled transcriptome using Bowtie 2. When a PARE signature was mapped to multiple genes, reads were divided among genes. The total number of reads for specific transcripts was identified via custom Perl scripts and normalized to the number of tags per billion reads.
Identification of miRNAs and Their Target Genes
The miRNAs of C. lanceolata seeds were identified using PAREsnip (Folkes et al., 2012) and LC small RNA analysis pipeline ACGT101-miR (V4.2, LC Sciences). Since the goal was to examine miRNAs during seed dormancy, which is a general phenomenon in various plant species, conserved rather than novel miRNAs were investigated in C. lanceolata. We used previously identified conserved miRNA sequences from C. lanceolata (Wan et al., 2012) and all registered miRNAs in the latest miRBase (V21, June 2014, www.mirbase.org) as sRNAome input for PAREsnip analysis. The assembled transcriptome obtained in this study was used as transcriptome input.
The miRNA/target interactions identified via PAREsnip were further processed using ACGT101-miR (V4.2; LC Sciences) to explore precursors of the candidate miRNAs. The ACGT101-miR workflow is shown in detail in Supplemental Figure S10. Briefly, the miRNA candidates were multialigned to the latest miRBase and the reference genome (the assembled transcriptome in this study, EST sequences of C. lanceolata in the NCBI database) and the forest tree genome database (http://dendrome.ucdavis.edu/). The miRNA candidates were identified as true miRNAs when (1) the candidate was mapped to the reference genome and (2) its registered premiRNA was mapped to the reference genome or to the extended genome sequences from the mapping loci with the potential to form hairpins.
Validation of miRNA/Target Interactions
To validate the miRNA/target interactions predicted via degradome sequencing, we experimentally analyzed their transient coexpression in Nicotiana benthamiana leaves as described by Li et al. (2008). Three miRNA/target pairs were chosen, including miR159/comp35259_c0_seq1, miR171b/comp36234_c2_seq1, and miR393/comp21546_c0_seq1. Briefly, genomic fragments surrounding the miRNAs including the fold-back structure were amplified from genomic DNA from the seeds with sequence-specific primers (Supplemental Table S14). Similarly, gene products of the target genes harboring the miRNA complementary sites were also amplified from the seeds (primers were listed in Supplemental Table S14). The amplified fragments were initially introduced into TA-vector and then cloned into pCAMBIA2300 under the control of a 35S promoter. The constructs harboring the cloned genes were transformed into Agrobacterium tumefaciens strain GV1301. Overnight cultures were infiltrated into N. benthamiana leaves as described by English et al. (1997). For coexpression analysis, an equal amount of A. tumefaciens cultures containing the miRNAs and their corresponding targets was mixed before infiltration as described by Zheng et al. (2012). Leaves were harvested 2 d after the infiltration for qRT-PCR analyses. Quantification of the qRT-PCR analyses was normalized to the expression level of tobacco tubulin gene. Gene-specific primers for qRT-PCR were listed in Supplemental Table S14.
qRT-PCR Analysis
The expression levels of the transcriptome sequences and identified miRNAs were validated using qRT-PCR. Total RNA was isolated from PD, ND, and SD seeds as described above. First-strand cDNA was synthesized using Transcript One-Step gDNA Removal and cDNA Synthesis SuperMix (TianGen Biotech) following the manufacturer’s instructions. Gene-specific primers for qRT-PCR of mRNAs and all miRNAs are listed in Supplemental Table S14. PCR reactions were conducted in triplicate in an ABI 7500 real-time PCR detection system (Applied Biosystems) using SYBR Green qRT-PCR Mix (Toyobo). Relative gene expression levels were calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001), with GAPDH used as an internal reference for mRNAs and 5.8s rRNA gene for miRNAs.
Accession Numbers
RNA-seq data generated in this work has been submitted to the Sequence Read Archive database in NCBI under the accession number SRP063368.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Germination of seeds in eight additional days of cold stratification.
Supplemental Figure S2. Eukaryotic Ortholog Groups (KOG) categories of the transcriptome.
Supplemental Figure S3. Hierarchical cluster analysis of the DEGs and expression patterns for the gene clusters.
Supplemental Figure S4. Enriched GO (BP) terms for the five gene clusters.
Supplemental Figure S5. Comparison of gene expression levels detected by RNA-seq versus qRT-PCR.
Supplemental Figure S6. qRT-PCR for GA and ABA associated genes.
Supplemental Figure S7. Germination of C. lanceolata seeds in GA3, GA4+7, and ABA solutions.
Supplemental Figure S8. Number of uncapped mRNA species in primary dormant (PD), nondormant (ND), and secondary dormant (SD) seeds.
Supplemental Figure S9. Eukaryotic Ortholog Groups (KOG) categories of the 269 DEGs showing opposite expression patterns in capped and uncapped forms.
Supplemental Figure S10. The workflow of the ACGT101-miR pipeline.
Supplemental Table S1. Summary of the C. lanceolata transcriptome.
Supplemental Table S2. Summary of gene annotation for the transcriptome.
Supplemental Table S3. Unigenes detected at >1 RPKM level in the PD, ND, and SD seeds.
Supplemental Table S4. DEGs revealed by RNA-seq.
Supplemental Table S5. Expression of genes regulating respiration.
Supplemental Table S6. Expression of genes regulating cell wall turnover.
Supplemental Table S7. Expression of genes related to GA and ABA metabolism and signaling.
Supplemental Table S8. Differentially expressed genes regulating mRNA degradation.
Supplemental Table S9. DEGs showing opposite expression patterns in capped and uncapped forms.
Supplemental Table S10. Seven DEGs involved in GA and ABA signaling and metabolism showed opposite expression patterns in capped and uncapped forms.
Supplemental Table S11. miRNA/target interactions identified by PARE analysis in C. lanceolata seeds.
Supplemental Table S12. Authentic miRNAs identified in C. lanceolata seeds.
Supplemental Table S13. Targets of authentic miRNAs identified using PAREsnip.
Supplemental Table S14. Sequence-specific primers used for qRT-PCR and the transient coexpression assay.
Supplementary Material
Acknowledgments
We thank Ms. Fengqin Dong of the Institute of Botany, Chinese Academy of Sciences for help in preparing the semithin and ultrathin sections. We also thank Ms. Wenhong Deng of the Analytical and Testing Center of Beijing Forestry University for help in performing the hormone profiling experiment and Ms. Xiaoyu Zhao of Beijing Forestry University for kind assistance with the seed germination tests. We thank Dr. Yanwei Wang of Beijing Forestry University for advice on the first draft.
Glossary
- ABA
abscisic acid
- GA
gibberellic acid
- PARE
parallel analysis of RNA ends
- miRNA
microRNA
- PD
primary dormant
- ND
nondormant
- SD
secondary dormant
- GO
Gene Ontology
- KOG
Eukaryotic Ortholog Groups
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- DEG
differentially expressed gene
- qRT-PCR
quantitative real-time PCR
- FW
fresh weight
- RPKM
read per kilobase per million clean reads
References
- Barrero JM, Jacobsen JV, Talbot MJ, White RG, Swain SM, Garvin DF, Gubler F (2012) Grain dormancy and light quality effects on germination in the model grass Brachypodium distachyon. New Phytol 193: 376–386 [DOI] [PubMed] [Google Scholar]
- Barrero JM, Talbot MJ, White RG, Jacobsen JV, Gubler F (2009) Anatomical and transcriptomic studies of the coleorhiza reveal the importance of this tissue in regulating dormancy in barley. Plant Physiol 150: 1006–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin CC, Baskin JM (2014) Seeds: Ecology, Biogeography, and Evolution of Dormancy and Germination, Ed 2. Academic Press, San Diego, CA [Google Scholar]
- Baskin JM, Baskin CC (2003) Classification, biogeography, and phylogenetic relationships of seed dormancy. In Smith RD, ed, Seed Conservation: Turning Science into Practice. The Royal Botanic Gardens, London, pp 517-544 [Google Scholar]
- Baskin JM, Baskin CC (2004) A classification system for seed dormancy. Seed Sci Res 14: 1–16 [Google Scholar]
- Belasco JG. (2010) All things must pass: contrasts and commonalities in eukaryotic and bacterial mRNA decay. Nat Rev Mol Cell Biol 11: 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benech-Arnold RL, Enciso S, Sánchez RA (2000) Involvement of ABA and GAs in the regulation of dormancy in developing sorghum seeds. In Black M, Bradford KJ, Vazquez-Ramos J, eds, Seed Biology: Advances and Applications. CAB International, Wallingford, UK, pp 101–111 [Google Scholar]
- Bethke PC, Libourel IGL, Aoyama N, Chung YY, Still DW, Jones RL (2007) The Arabidopsis aleurone layer responds to nitric oxide, gibberellin, and abscisic acid and is sufficient and necessary for seed dormancy. Plant Physiol 143: 1173–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier-Durand M, Dereuddre J, Côme D (1981) Ultrastructural changes in the endoplasmic reticulum during dormancy release of apple embryos (Pyrus malus L.). Planta 151: 6–14 [DOI] [PubMed] [Google Scholar]
- Cadman CSC, Toorop PE, Hilhorst HWM, Finch-Savage WE (2006) Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. Plant J 46: 805–822 [DOI] [PubMed] [Google Scholar]
- Cantoro R, Crocco CD, Benech-Arnold RL, Rodríguez MV (2013) In vitro binding of Sorghum bicolor transcription factors ABI4 and ABI5 to a conserved region of a GA 2-OXIDASE promoter: possible role of this interaction in the expression of seed dormancy. J Exp Bot 64: 5721–5735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Baskin CC, Baskin JM, Yang F, Huang Z (2014) Dormancy cycling and persistence of seeds in soil of a cold desert halophyte shrub. Ann Bot (Lond) 113: 171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera E, Holman T, Medhurst A, Dietrich D, Footitt S, Theodoulou FL, Holdsworth MJ (2008) Seed after-ripening is a discrete developmental pathway associated with specific gene networks in Arabidopsis. Plant J 53: 214–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, et al. (2007) Integration of biological networks and gene expression data using Cytoscape. Nat Protoc 2: 2366–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J, Parker R (2004) Eukaryotic mRNA decapping. Annu Rev Biochem 73: 861–890 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861 [DOI] [PubMed] [Google Scholar]
- Donohue K, de Casas RR, Burghardt L, Kovach K, Willis CG (2010) Germination, postgermination adaptation, and species ecological ranges. Annu Rev Ecol Evol Syst 41: 293–319 [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English JJ, Davenport GF, Elmayan T, Vaucheret H, Baulcombe DC (1997) Requirement of sense transcription for homology-dependent virus resistance and trans-inactivation. Plant J 12: 597–603 [Google Scholar]
- Finch-Savage WE, Cadman CSC, Toorop PE, Lynn JR, Hilhorst HWM (2007) Seed dormancy release in Arabidopsis Cvi by dry after-ripening, low temperature, nitrate and light shows common quantitative patterns of gene expression directed by environmentally specific sensing. Plant J 51: 60–78 [DOI] [PubMed] [Google Scholar]
- Folkes L, Moxon S, Woolfenden HC, Stocks MB, Szittya G, Dalmay T, Moulton V (2012) PAREsnip: a tool for rapid genome-wide discovery of small RNA/target interactions evidenced through degradome sequencing. Nucleic Acids Res 40: e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Footitt S, Douterelo-Soler I, Clay H, Finch-Savage WE (2011) Dormancy cycling in Arabidopsis seeds is controlled by seasonally distinct hormone-signaling pathways. Proc Natl Acad Sci USA 108: 20236–20241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J (2008) The control of mRNA decapping and P-body formation. Mol Cell 32: 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu LG, Yu YF, Robert RM (1999) Taxodiaceae. In Flora of China Editorial Committee, ed, Flora of China, Vol 4. Science Press and Missouri Botanical Garden Press, Beijing, China and St. Louis, MO, pp 54–61 [Google Scholar]
- German MA, Luo S, Schroth G, Meyers BC, Green PJ (2009) Construction of Parallel Analysis of RNA Ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome. Nat Protoc 4: 356–362 [DOI] [PubMed] [Google Scholar]
- German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, et al. (2008) Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol 26: 941–946 [DOI] [PubMed] [Google Scholar]
- Giacomelli JI, Weigel D, Chan RL, Manavella PA (2012) Role of recently evolved miRNA regulation of sunflower HaWRKY6 in response to temperature damage. New Phytol 195: 766–773 [DOI] [PubMed] [Google Scholar]
- Goeres DC, Van Norman JM, Zhang W, Fauver NA, Spencer ML, Sieburth LE (2007) Components of the Arabidopsis mRNA decapping complex are required for early seedling development. Plant Cell 19: 1549–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimovich G, Medina DA, Causse SZ, Garber M, Millán-Zambrano G, Barkai O, Chávez S, Pérez-Ortín JE, Darzacq X, Choder M (2013) Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell 153: 1000–1011 [DOI] [PubMed] [Google Scholar]
- Harholt J, Suttangkakul A, Vibe Scheller H (2010) Biosynthesis of pectin. Plant Physiol 153: 384–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harigaya Y, Parker R (2012) Global analysis of mRNA decay intermediates in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 109: 11764–11769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Tollervey D (2009) The many pathways of RNA degradation. Cell 136: 763–776 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Riechmann JL, Meyerowitz EM (2008) Transcriptome-wide analysis of uncapped mRNAs in Arabidopsis reveals regulation of mRNA degradation. Plant Cell 20: 2571–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi J, Tsumura Y, Yoshimaru H, Tachida H (2000) Phylogenetic relationships in Taxodiaceae and Cupressaceae sensu stricto based on matK gene, chlL gene, trnL-trnF IGS region, and trnL intron sequences. Am J Bot 87: 1480–1488 [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WX, Oono Y, Zhu J, He XJ, Wu JM, Iida K, Lu XY, Cui X, Jin H, Zhu JK (2008) The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 20: 2238–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PP, Montgomery TA, Fahlgren N, Kasschau KD, Nonogaki H, Carrington JC (2007) Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J 52: 133–146 [DOI] [PubMed] [Google Scholar]
- Liu WY, Chang YM, Chen SCC, Lu CH, Wu YH, Lu MYJ, Chen DR, Shih AC-C, Sheue C-R, Huang HC, et al. (2013a) Anatomical and transcriptional dynamics of maize embryonic leaves during seed germination. Proc Natl Acad Sci USA 110: 3979–3984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhang H, Zhao Y, Feng Z, Li Q, Yang HQ, Luan S, Li J, He ZH (2013b) Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc Natl Acad Sci USA 110: 15485–15490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lu S, Li Q, Wei H, Chang MJ, Tunlaya-Anukit S, Kim H, Liu J, Song J, Sun YH, Yuan L, et al. (2013) Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proc Natl Acad Sci USA 110: 10848–10853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohnen D. (2008) Pectin structure and biosynthesis. Curr Opin Plant Biol 11: 266–277 [DOI] [PubMed] [Google Scholar]
- Morris K, Linkies A, Muller K, Oracz K, Wang XF, Lynn JR, Leubner-Metzger G, Finch-Savage WE (2011) Regulation of seed germination in the close Arabidopsis relative Lepidium sativum: a global tissue-specific transcript analysis. Plant Physiol 155: 1851–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Muraro D, Mellor N, Pound MP, Help H, Lucas M, Chopard J, Byrne HM, Godin C, Hodgman TC, King JR, et al. (2014) Integration of hormonal signaling networks and mobile microRNAs is required for vascular patterning in Arabidopsis roots. Proc Natl Acad Sci USA 111: 857–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E (2005) Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J 41: 697–709 [DOI] [PubMed] [Google Scholar]
- Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y, Yamaguchi S (2003) Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15: 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Yamaguchi S, Hu J, Yusuke J, Jung B, Paik I, Lee HS, Sun TP, Kamiya Y, Choi G (2007) PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19: 1192–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Welti R, Wang X (2010) Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat Protoc 5: 986–992 [DOI] [PubMed] [Google Scholar]
- Qiu Z, Wan L, Chen T, Wan Y, He X, Lu S, Wang Y, Lin J (2013) The regulation of cambial activity in Chinese fir (Cunninghamia lanceolata) involves extensive transcriptome remodeling. New Phytol 199: 708–719 [DOI] [PubMed] [Google Scholar]
- Schoenberg DR, Maquat LE (2012) Regulation of cytoplasmic mRNA decay. Nat Rev Genet 13: 246–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamimuzzaman M, Vodkin L (2012) Identification of soybean seed developmental stage-specific and tissue-specific miRNA targets by degradome sequencing. BMC Genomics 13: 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Zhen Y, Zheng RH (2010) Proteome profiling of early seed development in Cunninghamia lanceolata (Lamb.) Hook. J Exp Bot 61: 2367–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu K, Zhang H, Wang S, Chen M, Wu Y, Tang S, Liu C, Feng Y, Cao X, Xie Q (2013) ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis. PLoS Genet 9: e1003577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song QX, Liu YF, Hu XY, Zhang WK, Ma B, Chen SY, Zhang JS (2011) Identification of miRNAs and their target genes in developing soybean seeds by deep sequencing. BMC Plant Biol 11: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- State Forest Administration P.R. China (2014) Forest resources in China (2009–2013). China Forestry Publishing House, Beijing, China [Google Scholar]
- Sunkar R, Li Y-F, Jagadeeswaran G (2012) Functions of microRNAs in plant stress responses. Trends Plant Sci 17: 196–203 [DOI] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T (2011) REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6: e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaistij FE, Gan Y, Penfield S, Gilday AD, Dave A, He Z, Josse E-M, Choi G, Halliday KJ, Graham IA (2013) Differential control of seed primary dormancy in Arabidopsis ecotypes by the transcription factor SPATULA. Proc Natl Acad Sci USA 110: 10866–10871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W (2010) Lignin biosynthesis and structure. Plant Physiol 153: 895–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S, Friml J (2009) Auxin: a trigger for change in plant development. Cell 136: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Vidal EA, Araus V, Lu C, Parry G, Green PJ, Coruzzi GM, Gutiérrez RA (2010) Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 4477–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan LC, Wang F, Guo X, Lu S, Qiu Z, Zhao Y, Zhang H, Lin J (2012) Identification and characterization of small non-coding RNAs from Chinese fir by high throughput sequencing. BMC Plant Biol 12: 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Cao H, Sun Y, Li X, Chen F, Carles A, Li Y, Ding M, Zhang C, Deng X, Soppe WJ, Liu YX (2013) Arabidopsis paired amphipathic helix proteins SNL1 and SNL2 redundantly regulate primary seed dormancy via abscisic acid-ethylene antagonism mediated by histone deacetylation. Plant Cell 25: 149–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis CG, Baskin CC, Baskin JM, Auld JR, Venable DL, Cavender-Bares J, Donohue K, Rubio de Casas R; NESCent Germination Working Group (2014) The evolution of seed dormancy: environmental cues, evolutionary hubs, and diversification of the seed plants. New Phytol 203: 300–309 [DOI] [PubMed] [Google Scholar]
- Xiang Y, Nakabayashi K, Ding J, He F, Bentsink L, Soppe WJJ (2014) Reduced Dormancy5 encodes a protein phosphatase 2C that is required for seed dormancy in Arabidopsis. Plant Cell 26: 4362–4375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Hossain MS, Arikit S, Valdés-López O, Zhai J, Wang J, Libault M, Ji T, Qiu L, Meyers BC, Stacey G (2015) Identification of microRNAs and their mRNA targets during soybean nodule development: functional analysis of the role of miR393j-3p in soybean nodulation. New Phytol 207: 748–759 [DOI] [PubMed] [Google Scholar]
- Zhan X, Wang B, Li H, Liu R, Kalia RK, Zhu JK, Chinnusamy V (2012) Arabidopsis proline-rich protein important for development and abiotic stress tolerance is involved in microRNA biogenesis. Proc Natl Acad Sci USA 109: 18198–18203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen B, Xu Z, Shi Z, Chen S, Huang X, Chen J, Wang X (2014) Involvement of reactive oxygen species in endosperm cap weakening and embryo elongation growth during lettuce seed germination. J Exp Bot 65: 3189–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Mao Z, Chong K (2013) A global profiling of uncapped mRNAs under cold stress reveals specific decay patterns and endonucleolytic cleavages in Brachypodium distachyon. Genome Biol 14: R92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Li YF, Sunkar R, Zhang W (2012) SeqTar: an effective method for identifying microRNA guided cleavage sites from degradome of polyadenylated transcripts in plants. Nucleic Acids Res 40: e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.