Abstract
To specify the C-terminal peroxisome targeting signal type 1 (PTS1) and the N-terminal PTS2 for higher plants, a maximum number of plant cDNAs and expressed sequence tags that are homologous to PTS1- and PTS2-targeted plant proteins was retrieved from the public databases and the primary structure of their targeting domains was analyzed for conserved properties. According to their high overall frequency in the homologs and their widespread occurence in different orthologous groups, nine major PTS1 tripeptides ([SA][RK][LM]> without AKM> plus SRI> and PRL>) and two major PTS2 nonapeptides (R[LI]x5HL) were defined that are considered good indicators for peroxisomal localization if present in unknown proteins. A lower but significant number of homologs contained 1 of 11 minor PTS1 tripeptides or of 9 minor PTS2 nonapeptides, many of which have not been identified before in plant peroxisomal proteins. The region adjacent to the PTS peptides was characterized by specific conserved properties as well, such as a pronounced incidence of basic and Pro residues and a high positive net charge, which probably play an auxiliary role in peroxisomal targeting. By contrast, several peptides with assumed peroxisomal targeting properties were not found in any of the 550 homologs and hence play—if at all—only a minor role in peroxisomal targeting. Based on the definition of these major and minor PTS and on the recognition of additional conserved properties, the accuracy of predicting peroxisomal proteins can be raised and plant genomes can be screened for novel proteins of peroxisomes more successfully.
Peroxisomes are ubiquitous small cell organelles that are involved in a variety of oxidative metabolic processes. Plant peroxisomes participate in recycling of P-glycolate produced by the oxygenase activity of Rubisco during photorespiration and they are the site of fatty acid β-oxidation. Some enzymes of the main metabolic pathways of peroxisomes have been cloned only recently, such as the photorespiratory enzymes Ala-glyoxylate aminotransferase (AGT) and Glu-glyoxylate aminotransferase (GGT; Liepman and Olsen, 2001, 2003; Igarashi et al., 2003), two isoforms of peroxisomal acyl-CoA synthetase (LACS; Fulda et al., 2002; Hayashi et al., 2002; Shockey et al., 2002) as well as four isoforms of acyl-CoA oxidase with distinct substrate specificity (ACX; Hayashi et al., 1998, 1999; Hooks et al., 1999; Froman et al., 2000). Cloning of several other genes in the past few years and localization of the encoded proteins to plant peroxisomes support the idea that the metabolic function of plant peroxisomes is more diverse than expected and far from being understood in its complexity. For instance, plant peroxisomes do also play an important role in metabolism of reactive oxygen species (Kliebenstein et al., 1998; Lopez-Huertas et al., 2000; del Rio et al., 2002), in the biosynthesis of the hormone jasmonic acid (Sanders et al., 2000; Stintzi and Browse, 2000; Strassner et al., 2002) and the osmoprotectant Glybetaine (Nakamura et al., 1997), in Val catabolism (Zolman et al., 2001), and possibly sulfur metabolism (Eilers et al., 2001; Nakamura et al., 2002). In line with the functional diversity of plant peroxisomes, the proteome of plant peroxisomes seems to be larger than in mammals and fungi (Emanuelsson et al., 2003), which most likely is a result of the large number of metabolically specialized plant microbodies, i.e. leaf peroxisomes, glyoxysomes, and senescent- and nodule-specific peroxisomes as well as hardly characterized unspecialized peroxisomes.
Peroxisomal proteins are encoded in the nucleus and synthesized in the cytoplasm on free ribosomes with targeting signals that specify their delivery to peroxisomes (Lazarow and Fujiki, 1985; de Hoop and Ab, 1992; Subramani 1993). Peroxisome targeting signals (PTS) as well as the targeting pathways are conserved to large extent throughout the eukaryotic kingdom. Most known matrix proteins are targeted to peroxisomes by a noncleaved C-terminal tripeptide of the prototype SKL>, the PTS type 1 (PTS1), or conservative variations thereof (Gould et al., 1987, 1989). Proteins with a PTS1 are recognized in the cytosol by the soluble receptor Pex5 (Van der Leij et al., 1993; Kragler et al., 1998; Wimmer et al., 1998) and are guided to and translocated across the peroxisomal membrane still complexed with Pex5 (Dammai and Subramani, 2001). A smaller group of matrix proteins contain a conserved nonapeptide that acts as a PTS2 and is embedded in a longer N-terminal presequence (e.g. RLx5HL; Swinkels et al., 1991; Glover et al., 1994). These proteins are recognized by the cytoplasmic receptor Pex7 (Marzioch et al., 1994; Rehling et al., 1996), and both targeting pathways merge subsequently at the peroxisomal surface at a single protein import complex. In plants and mammals, but not in fungi or trypanosomes, the N-terminal presequence of PTS2-targeted proteins is proteolytically processed upon import into the peroxisomal matrix. Some peroxisomal matrix proteins do not contain either of the two PTS (Karpichev and Small, 2000) and are thought to be targeted to the organelle by internal sequences to be specified. Regarding membrane proteins, the targeting signals have so far only been identified for a small subset of proteins, and general algorithms remain to be deduced (Dyer et al., 1996; Pause et al., 2000; Sacksteder et al., 2000; Jones et al., 2001; Murphy et al., 2003).
Experimental studies revealed a functional degeneracy of the PTS1 motif: a small uncharged residue at position −3, a basic residue at position −2, and a nonpolar residue at position −1 ([SAC][KRH]L>; Gould et al., 1989; Swinkels et al., 1992). Despite its general conservation between fungi, mammals, and plants, the PTS1 motif seems to display a certain diversity and to differ in some details between the kingdoms (Hansen et al., 1992; Sommer et al., 1992; Elgersma et al., 1996; Amery et al., 1998). In addition, upstream sequence elements modulate targeting efficiency and have partly been characterized (Lametschwandtner et al., 1998; Neuberger et al., 2003a). Plant-specific variations of the PTS1 motif were studied in detail by Hayashi et al. (1996a, 1997) by comparing the targeting efficiency of various tripeptides fused to the C terminus of β-glucuronidase to peroxisomes in transgenic Arabidopsis. The plant-specific PTS1 motif was specified accordingly to the conservative pattern [CASP][KR][ILM]> (Hayashi et al., 1997). Mullen et al. (1997a) used a slightly different experimental system consisting of the reporter protein chloramphenicol acetyltransferase and transient expression in tobacco (Nicotiana tabacum) BY-2 suspension-cultured cells. Because some noncanonical tripeptides, in which one amino acid deviated from the Hayashi motif, caused peroxisomal targeting as well, a more permissive PTS1 motif was deduced ([ACGST][HKLNR][ILMY]>; Mullen et al., 1997a). Kragler et al. (1998) studied the interaction of tobacco Pex5 with a wide range of C-terminal tripeptides in the yeast two-hybrid system and provided further support for the permissive PTS1 motif, including additionally P at position −3 ([ACGPST][HKLNR][ILMY]>).
The PTS2 of mammalian and yeast proteins has been defined as a nonapeptide with four conserved amino acid residues separated by five variable amino acid residues ([RK][LVI]x5[HQ][LA]; Swinkels et al., 1991; de Hoop and Ab, 1992; Osumi et al., 1992). In plants, enzymes such as malate dehydrogenase, citrate synthase (CS), and thiolase contain a PTS2 that is necessary and sufficient for peroxisomal targeting (Gietl et al., 1994; Kato et al., 1996, 1998). Kato et al. (1996, 1998) deduced from mutational analysis of the presequences of CS and malate dehydrogenase and from reported sequences of plant PTS2-targeted proteins the consensus motif R[ILQ]x5HL for plants. Based on targeting experiments with the presequence of rat thiolase modified by site-directed mutagenesis, Flynn et al. (1998) suggested a more relaxed PTS2 motif for eukaryotes with negligible restriction regarding the residues at position 2 ([RK]x6[HQ][ALF]).
Prediction of the subcellular localization of unknown proteins is a challenge of the postgenomic era but requires targeting motifs of high specificity to maximize the number of true positives and minimize that of false positives. Major difficulties in predicting PTS1-targeted peroxisomal proteins are (1) the small size of the signal, (2) the missing cleavage site that would provide additional diagnostic information, (3) the lower hierarchy of the PTS1 as compared to N-terminal signals (Neuberger et al., 2003b), and (4) the fact that PTS1-like C-terminal tripeptides are found in many nonperoxisomal proteins in which the tripeptide is not properly exposed on the surface of the folded protein, preventing recognition by Pex5 (Emanuelsson et al., 2003). In contrast to the prediction of transit and signal peptides as well as mitochondrial presequences, most prediction programs lack algorithms for predicting peroxisomal proteins or are underdeveloped in that noncanonical PTS with experimentally demonstrated peroxisomal targeting properties as well as plant-specific variations are not considered yet. Only recently, new approaches have been undertaken to optimize the prediction of PTS1-targeted proteins, but also in these projects plant-specific algorithms have not been developed (PeroxiP, Emanuelsson et al., 2003; PTS1 predictor, http://mendel.imp.univie.ac.at/mendeljsp/sat/pts1/PTS1predictor.jsp; Neuberger et al., 2003a, 2003b). Even though experimental studies have provided important information to define plant PTSs, it needs to be considered that even in the case of the most conservative variants, a large number of tri- and nonapeptides, formally included in the motifs, were not investigated nor had been found in any homologs. Therefore, the conclusion that residues of functional PTS can be combined freely to form further functional PTS may be premature. In addition, the possible role of accessory elements was neglected but can have accounted for partially contradictory results (Hayashi et al., 1997; Mullen et al., 1997a; Kragler et al., 1998).
We aimed to identify novel genes encoding putative peroxisomal proteins in the Arabidopsis genome (Arabidopsis Genome Initiative, 2000) and to predict their intracellular localization with high accuracy. To specify the PTS1 and PTS2 motifs, a bioinformatics approach was applied comprising the identification of a maximum number of homologous cDNAs and expressed sequence tags (ESTs) in the database and a thorough analysis of the primary and secondary structure of the PTS as well as adjacent regions for yet unrecognized conserved properties. The different PTS1 and PTS2 peptides were found to differ largely in the frequency at which they are widespread in nature. Major and minor PTS have been defined accordingly and novel structural properties of the targeting domain have been identified. These results are expected to provide valuable information to increase the accuracy of predicting peroxisomal proteins.
RESULTS
Identification of Sequences Homologous to PTS1- and PTS2-Targeted Proteins
Thanks to large-scale sequencing projects of whole plant genomes (Arabidopsis, Oryza sativa) and ESTs (as of August 2003, 27 plant species with >20,000 ESTs each) the large number of DNA sequences in the public databases provides an enormous amount of biological information that can possibly be used to specify the signals that are indicative of targeting proteins to plant peroxisomes. According to our current knowledge, the targeting mechanism of specific peroxisomal proteins is conserved within the plant kingdom. No peroxisomal protein has been reported that contains a PTS1 in one plant species while carrying a PTS2 or another peroxisomal targeting signal in an ortholog in another plant species or vice versa. Thus, if a specific plant protein is targeted to peroxisomes by a PTS1 (or PTS2), all plant orthologs of this protein are expected to possess a PTS1 (or PTS2). About two-thirds of the currently known matrix proteins from plant peroxisomes contain a C-terminal canonical tripeptide of the PTS1 motif defined by Hayashi et al. (1997) and about one-third a PTS2 (Supplemental Table SI, which can be viewed at www.plantphysiol.org). For many of these proteins, experimental evidence has been provided that the peptide is necessary and sufficient for protein targeting to peroxisomes. With respect to hydroxyisobutyryl-CoA hydrolase (HIBCH, AKL>; Zolman et al., 2001), evidence for the function of the C-terminal tripeptide as a PTS1 is still indirect but was considered sufficient to include the protein in the set of PTS1-targeted proteins (see “Materials and Methods”). Because of their differing C-terminal targeting motifs and the recent discrepancy regarding the localization of the targeting peptide within the C-terminal domain (Mullen et al., 1997b; Kamigaki et al., 2003), the homologs of catalase were excluded from this study.
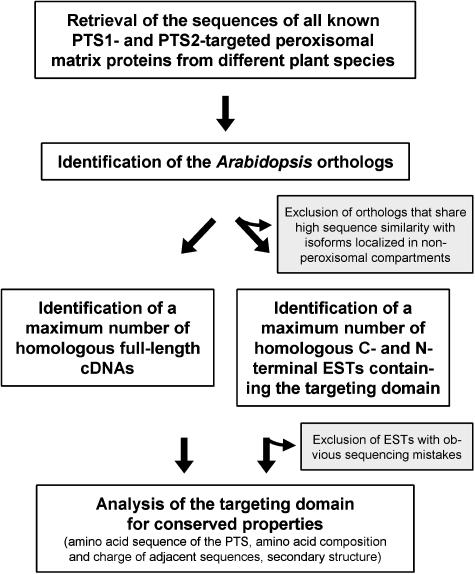
The starting point of the database search for homologs of PTS1/2-targeted peroxisomal proteins was a retrieval of the Arabidopsis orthologs of all PTS1/2-targeted plant proteins from the protein database (Fig. 1). The genes that were cloned from Arabidopsis were supplemented by the Arabidopsis orthologs of known PTS1/2-targeted matrix proteins, which were identified by sequence similarity. In case of unusually large gene families in Arabidopsis, such as that of the glycolate oxidase (GOX)-related proteins comprising five genes (Reumann, 2002), the orthologs corresponding to the reported proteins were identified according to maximum sequence similarity and similar tissue-specific expression pattern deduced from a “digital northern” (Mekhedov et al., 2000; data not shown). In a digital northern a maximum number of ESTs corresponding to the gene of interest are retrieved from all publicly accessible EST collections of the same plant species and analyzed semiquantitatively for the source from which the RNA was isolated (e.g. plant tissue, abiotic stress conditions). The remaining members of these gene families were not considered due to a current lack of experimental evidence for their localization in peroxisomes. Next, using these Arabidopsis proteins as query against the nonredundant database it was investigated if the homologs from other plant species corresponding to the peroxisomal isoforms could be identified unambiguously based on maximum sequence similarity. Two PTS1-targeted proteins, namely LACS7 and superoxide dismutase, as well as the PTS2-targeted peroxisomal Hsp70, had to be excluded because they shared high sequence similarity with the PTS2-targeted isoform LACS6 or isoforms localized in different cellular compartments (Supplemental Table SI). To enlarge the data set, the search was extended to the database of ESTs, and a maximum number of homologous C-terminal (for PTS1 proteins) and N-terminal sequences (for PTS2 proteins) of sufficient quality were retrieved irrespective of the amino acid sequence of the putative targeting peptide.
Figure 1.
Strategy of the specification of peroxisome targeting signals (PTS) for higher plants by bioinformatics analyses. The genes of peroxisomal matrix proteins that were cloned from Arabidopsis were supplemented by the Arabidopsis orthologs of known PTS1- and PTS2-targeted matrix proteins from other plant species identified by sequence similarity. These proteins were used as query for the identification of a maximum number of homologous full-length cDNA sequences as well as EST sequences containing the targeting domain in the databases by bioinformatics analyses. The peroxisomal targeting domains of all homologs were then analyzed for conserved properties, such as amino acid sequence of the PTS, amino acid composition and charge of adjacent sequences, and secondary structure.
Bioinformatics Specification of the PTS1 Motif for Higher Plants
In total, 391 sequences were retrieved that are homologous to PTS1-targeted peroxisomal matrix proteins and derive from various higher plant species. Of these sequences, 73 represented full-length homologs and 318 homologous ESTs (81%). About one-fourth of the sequences were derived from monocotyledons and three-quarters from dicotyledons and in total from about 80 different plant species. The total number of different C-terminal tripeptides was 39, one-half of which were found several times and represented 95% of the sequences. The remaining 19 tripeptides were not found in any other homologous sequence. Considering the large number of possible tripeptides, these 19 sequences with unique C-terminal tripeptides (5%) are estimated to represent the maximum number of false positives, i.e. homologs corresponding to nonperoxisomal isoforms, incorrectly annotated genes, or incorrectly sequenced ESTs. It can be concluded that the criteria to select the ESTs were reasonably chosen and that the statistical analysis was not considerably disturbed by sequencing errors or an unwanted extraction of sequences derived from nonperoxisomal isoforms.
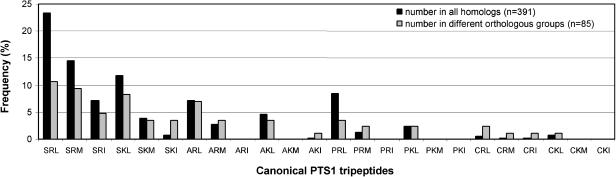
Within each orthologous group, on average 30.1 ± 14.5 homologs from different plant species were found, reflecting the high sensitivity of the homology analysis as well as the fact that most known proteins of plant peroxisomes are abundant enzymes of primary metabolism, the cDNAs of which are represented in many EST collections. The homologs of each enzyme contained on average 6.5 ± 2.5 different C-terminal tripeptides (all sequences included) and at least 5.2 ± 1.9 different true PTS1 tripeptides (sequences with unique tripeptides excluded). This number indicates that the sequence of the PTS1 is medium conserved within an orthologous group and that a considerable number of different PTS1 tripeptides are produced if summarized over all 13 orthologous groups. The large majority of PTS1 sequences of an orthologous group contained a canonical C-terminal tripeptide included in the plant-specific PTS1 motif defined by Hayashi (93% ± 4%, SOX homologs excluded, [SACP][KR][LMI]>; Hayashi et al., 1997). These data show that the 6 additional amino acids and the more than 90 additional tripeptides of the permissive motif (i.e. G, T, H, L, N, and Y; Mullen et al., 1997a; Kragler et al., 1998) play only a minor role in targeting the homologs of currently known proteins to peroxisomes. The highest number of different canonical PTS1 tripeptides was detected for GOX, malate synthase (MS), and multi-functional protein with 7 to 8 (data not shown). The other extreme is AGT, the entire C-terminal domain of which is highly conserved with the result that the C-terminal tripeptide SRI> is identical in 24 out of 27 homologs. Because a particular PTS1 tripeptide is considered a stronger indicator for peroxisomal localization due to not only the higher the number of homologs containing this tripeptide in total but also the higher its frequency in different orthologous groups, the C-terminal tripeptides of the homologs were analyzed in two different ways. First, all homologs including those of different higher plant species with identical C-terminal tripeptides were included in the statistical analysis (Fig. 2). Alternatively and to avoid an overestimation of the frequency of some tripeptides that are predominantly found in a single orthologous group, only one homolog with a particular tripeptide per enzyme was included. In this case, for instance, only a single sequence out of the 24 AGT homologs terminating with SRI> and a single GOX homolog out of 31 with PRL> were considered. This restriction reduced the number of included PTS1 sequences from 391 to 85 (Fig. 2).
Figure 2.
Frequency of canonical PTS1 tripeptides in plant homologs of PTS1-targeted proteins. The Arabidopsis orthologs of PTS1-targeted proteins were blasted against the nonredundant protein and against organism-specific EST databases, and a maximum number of homologous sequences were identified from various of higher plants species by sequence similarity (73 full-length cDNAs and 318 C-terminal ESTs). Canonical tripeptides are defined as included in the conservative variant of the plant-specific PTS1 motif ([SAPC][KR][LMI]>; Hayashi et al., 1997). All homologs irrespective of the nature of their C-terminal tripeptide were analyzed (n = 391) or only a single sequence with a specific tripeptide was included per orthologous group (n = 85) to avoid an overestimation of the abundance of specific tripeptides, such as SRI> and PRL>, which are predominantly found in specific orthologous groups (AGT and GOX, respectively). The data for the noncanonical tripeptides are not shown (SRV>, 1.5% in all homologs, 4.7% in different orthologous groups; SNL>, 1.5%/1.2%; SNM>, 0.8%/2.4%; SML>, 0.8%/1.2%; ANL>, 0.8%/1.2%; SSM>, 0.5%/1.2%; unique sequences, AKS>, ALL>, FRL>, FRV>, FYL>, ISR>, IYI>, KAL>, LKR>, LRL>, LWQ>, QFL>, SAM>, SRF>, SSL>, and TEP>). Nine major PTS1 tripeptides ([SA][RK][LM]> without AKM> plus SRI> and PRL>) are considered strong indicators for peroxisomal localization.
The possible 24 tripeptides of the conservative Hayashi motif ([SACP][KR][LMI]>) turned out to differ largely in the frequency at which they occur in the homologs of PTS1-targeted proteins. Nine major PTS1 tripeptides can be defined, each of them comprising at least 10 ESTs and occurring in at least 3 different orthologous groups (Fig. 3). They are headed by far by SRL> (91 sequences, 23.3%), which accordingly represents the prototypical plant PTS1, followed by SRM> and SKL>, PRL>, ARL>, and SRI>, and finally AKL>, SKM>, and ARM>. These major PTS1 tripeptides ([SA][RK][LM]> without AKM> plus SRI> and PRL>) are thought to represent the most reliable indicators for peroxisomal localization and covered altogether 84% of the homologous sequences of PTS1-targeted proteins (Fig. 3).
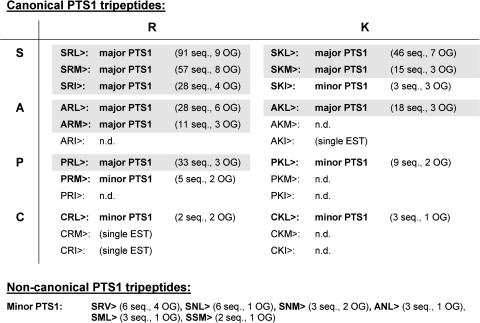
Figure 3.
Definition of major and minor PTS1 tripeptides of peroxisomal proteins from higher plants. Major PTS1 tripeptides are defined as C-terminal peptides present in at least 10 sequences and 3 different orthologous groups. Minor PTS1 tripeptides are present in at least two sequences. Major PTS1 tripeptides are given in bold print and shaded in gray. Minor PTS1 tripeptides are given in bold print. Canonical PTS1 tripeptides are included in the restrictive plant-specific motif determined by Hayashi et al. (1997). OG, orthologous groups; n.d., not detected.
A smaller number of sequences contained any of the tripeptides PKL>, PRM>, SKI>, CKL>, or CRL>, most of which were found in ESTs and have not been reported in plant peroxisomal matrix proteins before. These tripeptides are defined as minor PTS1 tripeptides and considered functional but low-abundant PTS1. If only a single sequence with a particular C-terminal tripeptide per orthologous group is considered, the overall result is similar even though the frequency of some tripeptides is lower (e.g. SRL>, SRM>, SRI>, SKL>, and PRL>), indicating their dominance in particular orthologous groups (Fig. 2), whereas that of others is higher (e.g. SKI>, PRM>, and CRL>), reflecting the fact that these tripeptides occur in a number of orthologous groups above average. The tripeptides AKI>, CRM>, and CRI> were found in single sequences and probably also represent low-abundant variations of the plant PTS1. By contrast, no homologs of PTS1-targeted proteins were detected that contained any of the remaining tripeptides included in the most conservative pattern of the plant-specific PTS1 (ARI>, AKM>, PRI>, PKM>, PKI>, CKM>, and CKI>; Hayashi et al., 1997). The data imply that these undetectable tripeptides do not indicate accurately targeting to plant peroxisomes. The enzyme sulfite oxidase (SOX), which contains a noncanonical PTS1 tripeptide in Arabidopsis itself (SNL>), is unusual because the majority of its homologs (14 out of 19) contain a noncanonical PTS1 as well, such as SNM>, SSM>, SAM>, or ANL>. Because SNL> is found in several PTS1 proteins from yeast (e.g. d-Asp oxidase; Amery et al., 1998) and because a few SOX homologs contained a canonical PTS1 (e.g. SKL> and SKM>; Supplemental Table SI), the targeting pathway of SOX is not expected to differ substantially from the well-characterized PTS1 pathway. In total, 6 noncanonical tripeptides were found in several ESTs and considered low-abundant minor PTS1 (Fig. 3). Of these the tripeptide SRV> was most widespread, being found in 6 ESTs and 4 different orthologous groups.
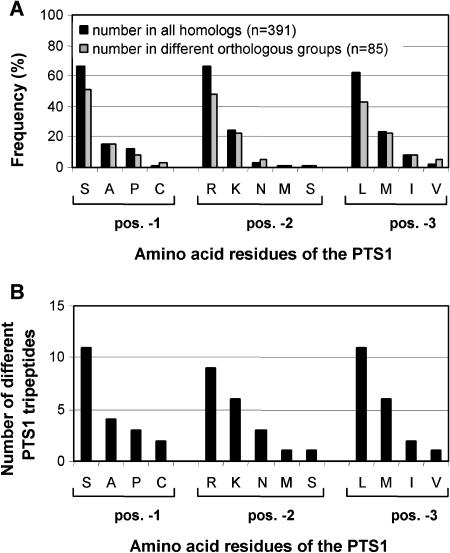
If particular amino acid combinations are by far more widespread as PTS1 tripeptides than other closely related combinations, the question arises whether rules can be deduced about which combinations yield a functional PTS1. The frequency of each amino acid residue at its specific position within the tripeptide was calculated and the amino acids classified accordingly (Fig. 4A). Highly abundant amino acids in PTS1 are S, R, and L of the tripeptide SRL>, medium abundant K and M, and the remaining low abundant (Fig. 4A). The amino acids differed also in the number of different PTS1 tripeptides in which they occurred. The residues S, R, and L were found in a large number of different tripeptides (9–11) and seem to reveal a broad tolerance with respect to the residues at the remaining positions, whereas other amino acids, such as P, C, N, and I, were restricted to a small number of very specific tripeptides and mostly to combinations with two residues of the motif S[RK]L> (Fig. 4B). Combinations exclusively of amino acids none of which belongs to the prototypical plant PTS1 SRL> were not found in any plant protein (e.g. A, P, and C at position −3; K, N, M, and S at position −2; and M, I, and V at position −1).
Figure 4.
Position-specific frequency of amino acids in PTS1 tripeptides (A) and number of different PTS1 tripeptides in which these amino acids are present (B). Highly abundant amino acids of PTS1 tripeptides that are present in about 60% of the sequences are S (position −3), R (position −2), and L (position −1), medium abundant (>20%) K (position −2) and M (position −1), and the remaining low abundant (≤15%; A, P, and C at position −3; N, M, and S at position −2, and I and V at position −1). A, The position-specific frequency of amino acids roughly correlates with the number of different PTS1 peptides in which they are present (B). Sequences with unique C-terminal tripeptides were not considered.
Conserved Properties of the PTS1 Domain
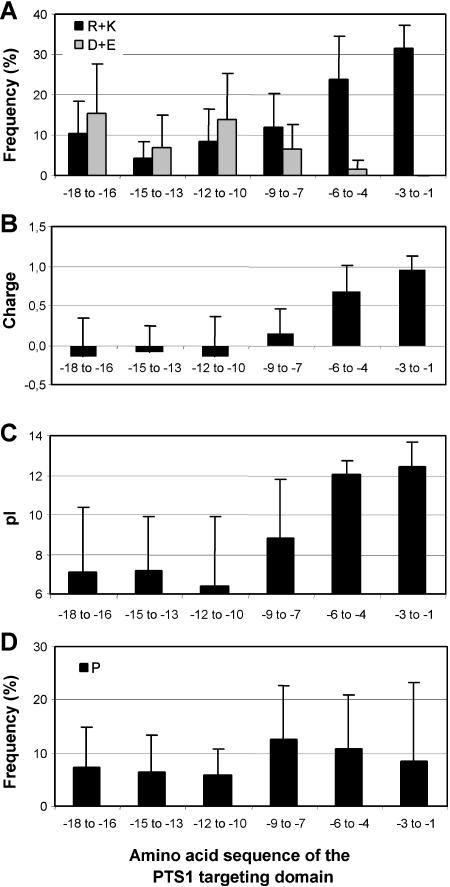
Sequences upstream of the PTS1 may have an auxiliary function as accessory elements in targeting proteins to peroxisomes and can provide additional indications for peroxisomal localization of unknown proteins. To investigate whether PTS1-targeted proteins contain conserved sequences upstream of the PTS1 tripeptide, the C-terminal 18 amino acids of the PTS1 targeting domains were analyzed successively in groups of tripeptides both per orthologous group (data not shown) and per group of identical PTS1 tripeptides (Fig. 5). The content of basic residues, R and K, was found to increase significantly from an average value in the core protein (about 10%, position −18 to −7) to 24% in the tripeptide in front of the PTS1 and to 32% in the PTS1 tripeptide itself (Fig. 5A). Thus, most PTS1-targeted proteins carry a second basic residue closely in front of the PTS1 tripeptide. The rise in the content of basic residues is accompanied by a decrease in the content of acidic residues toward the C terminus (Fig. 5A). The uneven distribution of charged residues leads to an increasing positive net charge toward the C-terminal end with a total positive charge of the C-terminal 6-mer of 1.6 and results in a significant increase of the pI to 12 (Fig. 5, B and C). Because of the low sd, the pI in particular is a useful additional criterion to confirm the postulated peroxisomal localization of unknown proteins. In addition the PTS1 domain is characterized by a high probability of P occurring in front of the PTS1 (Fig. 5D), showing that plant PTS1 proteins contain on average 0.70 ± 0.61 P residues within the 6-mer preceeding the PTS1 tripeptide.
Figure 5.
Conserved properties of the PTS1 targeting domain. A, Relative content of basic (R + K) and acidic amino acids (D + E). B, Net charge, determined as the number of basic (R + K) minus acidic residues (D + E). C, pI. D, Relative content of P. The homologs of PTS1-targeted proteins were grouped according to their PTS1 tripeptide. Sequences of groups containing less than five sequences were analyzed together but sequences with unique C-terminal tripeptides were excluded. The amino acid composition of the C-terminal 18-mer was analyzed in groups of three amino acids. Apart from an enrichment in basic and P residues, the PTS1 domain is characterized by a low content of S-containing (C, M) and aromatic residues and a high content of hydrophobic residues, especially A and L, as well as hydroxylated amino acids (A + L, 19% in the C-terminal 18 amino acids; S + T, 15% between position −7 and −15; data not shown).
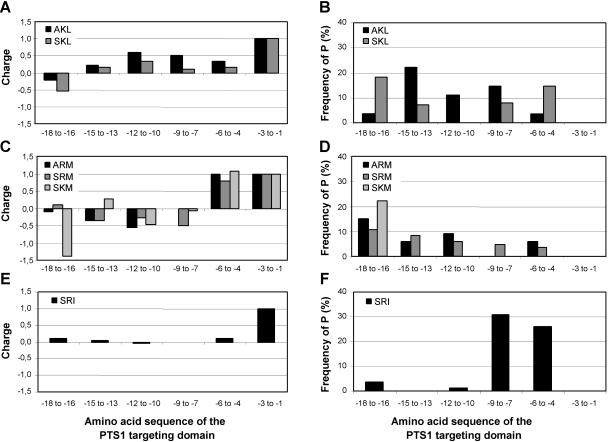
For proteins with an unusually low positive charge directly in front of the PTS1 peptide, such as those carrying AKL> and SKL> (Fig. 6, A and B), the positive charge seems to be spread over a larger region of 9 to 12 residues upstream of the PTS1 tripeptide, leading to similar values for the entire domain compared with the other PTS1 groups. In parallel, the position of P also is shifted further upstream (Fig. 6B). Closer inspection of sequence variations in the targeting domain within an orthologous group revealed that changes of the PTS from the prototypical targeting peptide SRL> or SKL> to low-abundant peptides of presumably weaker targeting efficiency (see “Discussion”) are often accompanied by a simultaneous addition of further basic and/or P residues in close proximity to the PTS1 peptide (Fig. 7, A–C). Overall, the second basic residue was mostly located at position −4 or at position −6. Only for proteins with M at position −1 of the PTS1 tripeptide, such as ARM>, PRM>, SKM>, and SRM>, which all contain a second basic residue in close proximity to the PTS1 tripeptide except for one-half of the GGT homologs, a preferential localization of the second basic residue at position −4 was found (Fig. 7D).
Figure 6.
Net charge and Pro content of the PTS1 targeting domain of proteins with SKL> or AKL> (A and B), with M-containing PTS1 tripeptides (C and D), and of proteins with SRI> (E and F). The most C-terminal 18 amino acids were analyzed in groups of three amino acids and the charge (A, C, and E) and the relative content of P (B, D, and F) were calculated. In proteins with the tripeptides AKL> and SKL> the positive net charge is less pronounced in the 3-mer preceeding the PTS1 but is spread over a longer peptide from position −4 to −12 (A and B). Proteins with M-containing PTS1 have an unusually low P content directly in front of the PTS1 tripeptide (position −4 to −9) but possess a high P content further upstream of the PTS1 tripeptide (position −10 to −18) and a pronounced positive net charge in front of the PTS1 (C and D). The proteins with the tripeptide SRI>, which are predominantly AGT homologs, do not carry a positive net charge in the targeting domain except for the R of the PTS1 tripeptide itself but possess about two P residues directly in front of the PTS1 (E and F).
Figure 7.
Sequence comparison of the PTS1 targeting domain of homologs from uricase (uric; A), 12-oxophytodienoate reductase (OPR3; B), and malate synthase (MS; C), and proteins with M at position −1 of the PTS1 tripeptide (D). Basic residues are shaded in gray and P residues in black. Most uric homologs with the PTS1 tripeptide SKL> (8 out of 9; 3 homologs shown) lack another basic residue at position −4, whereas 4 homologs (out of 11; 2 homologs shown) with SKM> and the 2 homologs with weaker PTS1 tripeptides (AKI> and SNM>) contain an additional basic residue at position −4 (A). Similarly, none of the 7 homologs of ORP3 with the PTS1 SRL> carry a second basic residue at position −4 (only one at position 5; 3 homologs shown), whereas all 6 homologs with SRM> or ARM> and the homolog with ARL> possess a second basic residue at position −4 (B). In addition to the conserved P at position −7/−8, all 10 MS homologs with SRL> (3 homologs shown), most homologs with SKL> (5 out of 8; 4 homologs shown), and the homolog with PRL> lack a second P at position −4. By contrast, all 3 homologs with CKL> as well as the 2 homologs with SKI> and FRL> contain an additional P at position −4 or −5 (C). Regarding those proteins with M at position −1 of the PTS1 tripeptide (S[RK]M>, [AP]RM>), the large majority contains a second basic residue in front of the PTS1 (position −4 to −6) with a strong preference for position −4, even though the proteins are derived from 9 different orthologous groups in many of which the second basic residue is not conserved (ARM>, all 11 with R/K at position −4; PRM>, all 5 with R/K at position −4 to −6; SKM>, 13 out of 15 with R/K at position −4 to −6; SRM>, 38 out of 57 with R/K at position −4) (D). The homologs of GGT (e.g. 15 GGT homologs with SRM>) represent an exception in that most of them do not carry a second basic residue or P in front of the PTS1. Per group of PTS1 tripeptide (S[RK]M>, [AP]RM>) 2 sequences are shown at maximum for each orthologous group. Ao, Asparagus officinalis; As, Aegilops speltoides; At, Arabidopsis; Ca, Capsicum annuum; Cr, Ceratopteris richardi; Cs, Cucumis sativus; Gm, Glycine max; Gh, Gossypium hirsutum; Ha, Helianthus annuus; Hv, Hordeum vulgare; Ib, Ipomoea batatas; Jr, Juglans regia; Lc, Lotus corniculatus var japonicus; Le, Lycopersicon esculentum; Lj, Lotus japonicus; Ls, Lactuca sativa; Ma, Musa acuminate; Mt, Medicago truncatula; Os, Oryza sativa; Pt, Pinus taeda; Pv, Phaseolus vulgaris; Rc, Ricinus communis; Sb, Sorghum bicolor; Sc, Secale cereale; Ta, Triticum aestivum; Vv, Vitis vinifera; Zm, Zea mays.
In light of targeting prediction an important question is whether different conserved properties function independently of each other or can compensate for each other. In case of an auxiliary function of specific amino acids in front of the PTS1 tripeptide in facilitating recognition by the cytosolic receptor Pex5, basic and P residues seem to be able to complement the accessory targeting function of each other. Most proteins with M at position −1 of the PTS1 peptide lack a P residue in front of the PTS1 tripeptide but are characterized by a high positive charge above average in the 3-mer in front of the PTS1 tripeptide (Fig. 6, C and D). Vice versa, proteins terminating with SRI>, most of which are AGT homologs, do not carry a pronounced positive net charge outside of the PTS1 tripeptide but contain about 2 P residues in the 6-mer adjacent to the PTS1 (Fig. 6, E and F). In summary, the PTS1 targeting domain comprises about 12 to 15 residues with characteristic properties, and the 3-mer directly in front of the PTS1 tripeptide is characterized by a positive net charge and a high P content. At least one of these properties seems to be required in plant PTS1 proteins to enhance targeting to peroxisomes.
Specification of the PTS2 Motif of Plants
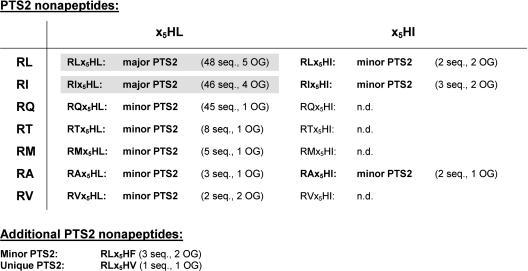
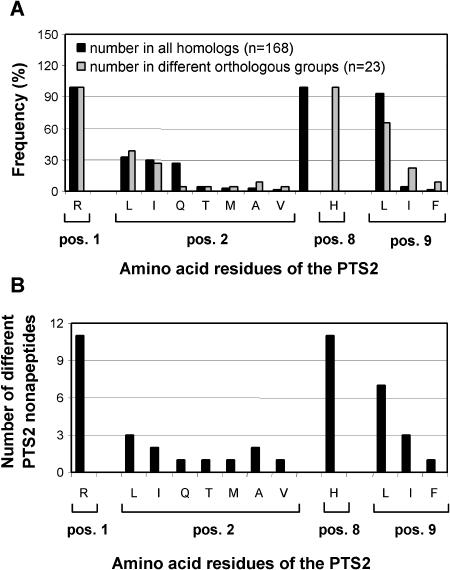
About one-third of the proteins from plant peroxisomes contain an N-terminal PTS2 (Supplemental Table SI). Except for the peroxisomal Hsp70 homolog (Wimmer et al., 1997), all PTS2-targeted plant proteins turned out to be suitable for analysis. In total, 168 homologous sequences of PTS2-targeted peroxisomal matrix proteins from higher plants were retrieved (31 full-length genes, 137 homologous ESTs). The total number of different N-terminal nonapeptides was only 12 including 1 unique nonapeptide (RLx5HV). The value of 23.9 ± 13.3 PTS2 homologs/orthologous group is lower as compared to PTS1 proteins. The homologs of each enzyme contained on average 3.1 ± 1.3 different nonapeptides (sequence variation in the x5 region excluded), indicating a higher conservation of the 4 residues of the nonapeptide as compared to the PTS1. The region outside of the nonapeptide showed in general a lower sequence conservation as compared to PTS1-targeted proteins, except for thiolase the entire N-terminal domain of which is highly conserved. Almost all homologs of PTS2-targeted proteins (94%) contained a nonapeptide of the motif R[LIQTMAV]x5HL, showing that three amino acids of the prototypical PTS2 nonapeptide, RLx5HL, are highly conserved and that most exchanges are tolerated at position 2 (Supplemental Fig. S2). The peptides RLx5HL and RIx5HL are defined as major PTS2 nonapeptides (present in ≥ 10 sequences and ≥ 3 orthologous groups; Fig. 8). By contrast, the nonapeptide RQx5HL was restricted to thiolase and is therefore defined as a minor PTS peptide due to its lower indicative targeting properties. Further minor PTS2 nonapeptides contain either other amino acid exchanges at position 2 (R[TMAV]x5HL) or an exchange at position 9 (RLx5H[IF]). Isoleucine is the second most frequent residue after L at both position 2 and 9 (Fig. 9A), and, apart from the totally conserved residues R and H, the residues L and I are present in the largest number of different nonapeptides as well (Fig. 9B).
Figure 8.
Definition of major and minor PTS2 nonapeptides of peroxisomal proteins from higher plants. Major PTS2 nonapeptides are defined as N-terminal peptides present in at least 10 sequences and 3 different orthologous groups. Minor PTS2 nonapeptides are present in at least two sequences. Major PTS2 nonapeptides are printed in bold and shaded in gray. Minor PTS2 nonapeptides are printed in bold. OG, orthologous groups; n.d., not detected.
Figure 9.
Position-specific frequency of amino acids in PTS2 nonapeptides (A) and number of different PTS2 nonapeptides in which these amino acids are present (B). The position-specific frequency of amino acids roughly correlates with the number of different PTS2 peptides in which they are present. One sequence with a unique nonapeptide was not considered.
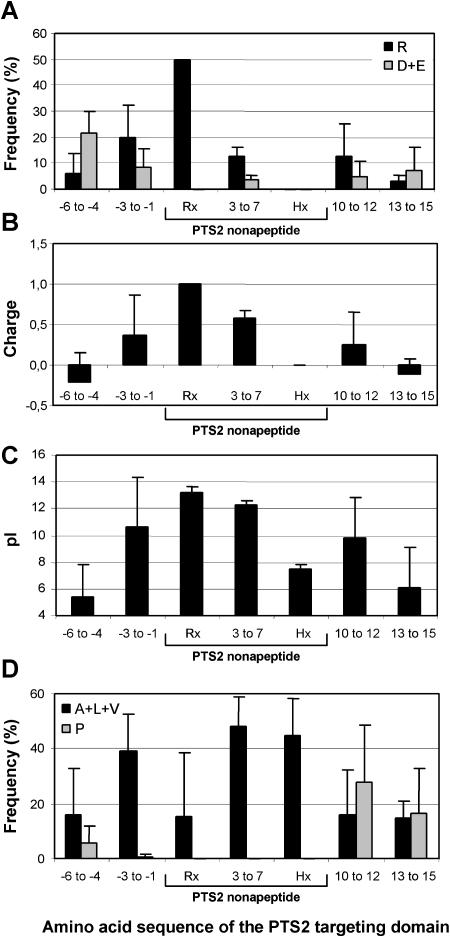
Upon analysis of the neighboring regions of the PTS2 nonapeptide for conserved properties, the size of the PTS2 targeting domain was defined as a region of approximately 15 residues surrounding roughly symmetrically the PTS2 nonapeptide (position −3 to 12). Common features of the PTS2 domain are the following (Fig. 10): (1) a high incidence of R, (2) a low content of acidic residues (Fig. 10A), (3) a high positive charge of the targeting domain of on average 2.2 ± 0.7 (Fig. 10B), (4) a pronounced increase in pI, of which the low sd within the x5 region is most noteworthy (Fig. 10C), (5) a strict absence of P in front of the nonapeptide and within the x5 region, contrasting with a frequent presence immediately downstream of the PTS2, and (6) a high incidence of A, L, and V in front of the nonapeptide and within the x5 region (Fig. 10D). In addition, the sequences imply a conserved secondary structure of the PTS2 domain because in the large majority of sequences examined, the nonapeptide seemed to be located in the end of a short α-helix (data not shown).
Figure 10.
Conserved properties of the PTS2 targeting domain. A, Relative content of R and acidic residues (D + E). B, Net charge, determined as the number of basic minus acidic residues. C, pI. D, Relative content of hydrophobic residues (L + A + V) and P residues. The homologs of PTS2-targeted proteins were grouped according to their PTS2 nonapeptide and analyzed. Sequences of groups containing less than five sequences were analyzed together but the sequence with a unique nonapeptide was excluded. The amino acid composition of PTS2 proteins was analyzed in groups of two to five amino acids. The PTS2 targeting domain was defined as a region of approximately 15 residues surrounding roughly symmetrically the PTS2 nonapeptide (position −3 to 12). The content of K as well as G and I were significantly lower as compared to that of R and L, A, and V, respectively (data not shown). Apart from these characteristics, the PTS2 domain was characterized by a low content of S-containing (C, M) and aromatic residues (data not shown).
DISCUSSION
The large amount of biological information in the public databases opens the possibility to apply bioinformatics tools to build up novel hypotheses and to answer biological questions. The complete genome sequences of Arabidopsis and Oryza allow now the extraction of genes encoding unknown proteins with a putative PTS1 or PTS2. To increase the prediction accuracy of peroxisomal localization, an exact definition of the PTS is crucial. Experimental studies have provided valuable information on plant-specific PTS motifs but suffer from three important drawbacks. First, the number of experimentally analyzed PTS peptides is limited, and solid support for the assumption that the amino acids of different functional PTS can freely be combined is currently lacking. For instance, inclusion of G and T (position −3) in the permissive PTS1 motif is solely based on positive targeting results of three tripeptides (GRL>, GKL>, and TKL>; Mullen et al., 1997a; Kragler et al., 1998), and data are lacking on whether the remaining 28 tripeptides constitute functional PTS1 ([GT][HNRK][IMY]> and [GT][HN]L>). Second, targeting efficiency is difficult to quantify experimentally, resulting in mostly qualitative results. The yeast-two-hybrid system can partially overcome these limits but restricts the multi-step targeting pathway to the binding affinity between the PTS1 tripeptide and Pex5 and thus relies for plant proteins on a heterologous system (Kragler et al., 1998). Third, the use of different expression systems (constitutive or transient expression) and a negligence of accessory sequences may have accounted for some discrepant experimental targeting results, for instance, those of the tripeptides SHL>, SLL>, and GRL> (Hayashi et al., 1997; Mullen et al., 1997a; Kragler et al., 1998).
Advantages and Pitfalls of PTS Specification by Bioinformatics Analysis
Bioinformatics analyses can possibly provide additional information to specify targeting motifs. Complete plant genome sequences provide novel essential information on the size of gene families, the subcellular localization of different isoforms, and on sequence similarity shared between orthologs and paralogs, all of which are prerequisite for an unambiguous identification of homologous sequences that correspond to specific isoforms in species lacking a sequenced genome. Especially kingdom-specific variations of targeting signals can be deduced from such bioinformatics analyses. The frequency at which PTS1 and PTS2 peptides occur in plant proteins reflects a close to final stage of the ongoing evolutionary optimization of targeting signals and reflects semiquantitatively the targeting efficiency of these peptides (see below). The large data set of PTS-targeted sequences also allows a study of the targeting signals within their native context and facilitates the detection of yet unrecognized accessory sequences with an auxiliary targeting function by sequence conservation.
Critical to the specificity of targeting motifs deduced from such analyses is a reasonable compromise between the identification of a maximum number of homologous sequences and a minimum number of false positives. A major factor to increase the size of the data set to about 550 sequences (catalase homologs excluded) and thereby more than 10-fold as compared to the number of cloned genes was the use of EST databases, which contributed about 80% of the sequences. To prevent that the higher rate of sequencing errors of ESTs led to an erroneous identification of PTS1 tripeptides particularly because of their localization next to a stop codon, reasonable requirements for the selection of ESTs were chosen and all sequences manually inspected. In cases of a problematic differentiation between homologs corresponding to peroxisomal or nonperoxisomal isoforms due to the small length of ESTs, further bioinformatics tools were applied to identify the homologs of interest but three peroxisomal proteins had to be excluded beforehand.
The number of false PTS peptides determines the minimum number of sequences required to judge a particular tri- or nonapeptide a functional PTS and was estimated. Considering that the number of different C-terminal tripeptides is 8,000 and that the number of noncanonical tripeptides that can by created by single amino acid substitutions from canonical PTS1 is 448, it is not expected that 2 identical tripeptides can derive from canonical PTS1 by random point mutations within a total number of 39 different detected tripeptides and 23 sequences that contained a tripeptide found only once or twice. Therefore, those sequences containing unique targeting peptides are considered to represent the maximum number of false positives, which is relatively low (PTS1, 5%; PTS2, 0.6%). Because many of these unique peptides are either included in the conservative PTS1 motif defined by Hayashi et al. (1997) or closely related peptides, the number of false positives is probably largely overestimated. Experimental support for peroxisomal targeting has even been provided for some of these unique tripeptides (e.g. CRM>, SSL>; Kragler et al., 1998; Cutler et al., 2000). Vice versa, all tripeptides that were found in more than one sequence were considered functional PTS1 in plants, including a possibly pronounced auxiliary targeting function of accessory elements.
A certain degree of sequence variation of the PTS within an orthologous group is required to allow the identification of PTS peptides rather independently of accessory elements specific to particular orthologous groups and to reach high coverage of all PTS1/2 peptides present in higher plants despite the relatively low number of input proteins as compared to the large size of the estimated peroxisomal proteome (Fukao et al., 2002, 2003; Emanuelsson et al., 2003). Apart from AGT and thiolase, the PTS was medium conserved within an orthologous group, and on average 5 functional PTS1 and 3 PTS2 peptides were found per orthologous group and 20 functional PTS1 and 11 PTS2 peptides were identified in total. The higher conservation of the PTS2 may indicate that further nonapeptides target proteins to plant peroxisomes.
Specification of the Plant PTS1
The high number of homologous sequences identified, the low number of false positives, and the medium degree of sequence variability of the PTS within orthologous groups allowed a statistically significant analysis of the frequency of particular PTS tripeptides in the plant kingdom. Regarding first the PTS1 proteins, not expected and most important is the result that only a small specific subset of PTS1 tripeptides is widespread in plant proteins and seems to constitute functional PTS peptides. Nine PTS1 tripeptides ([SA][RK][LM]> without AKM> plus SRI> and PRL>) have been defined as major PTS, of which PRL> and SRI> had shown only weak targeting efficiencies in experimental studies (Hayashi et al., 1997). Eleven PTS1 tripeptides were defined as minor PTS1 and are regarded low-abundant but functional plant PTS1 tripeptides, most of which have not been reported in plant peroxisomal matrix proteins before nor have been described experimentally as functional PTS1. Experimental support for the targeting function of some of these PTS1 tripeptides has been provided (e.g. CRL>, CKL>, SML>; Kragler et al., 1998), whereas others failed to target passenger proteins to peroxisomes or to interact with the plant PTS1 receptor (PKL>; Mullen et al., 1997a; SKI>; Kragler et al., 1998). The noncanonical peptide SRV> also did not target a fusion protein to Arabidopsis peroxisomes (Hayashi et al., 1997) but is clearly recognized as a functional PTS1 due to its presence in four different orthologous groups (AGT, ICL, and ACX1/4). By contrast, several peptides with assumed peroxisomal targeting properties and included in the most restricted PTS1 motif were completely absent in all sequences identified, even though several of them could easily be created from widespread PTS peptides by single amino acid substitutions.
From a position-specific quantitative analysis of the amino acids, it can be concluded which amino acid residues are tolerated in PTS1 peptides. Overall, the eleven amino acids of the restrictive PTS1 motif were indeed present in functional PTS ([CASP][KR][ILM]>; Hayashi et al., 1997), whereas the seven additional amino acids of more permissive motifs ([GST][HLN][Y]>) were rare and can mostly be neglected. Regarding position −3, apart from the well-known residues S and A, the residue P is clearly an alternative option in plant PTS1, even though it revealed negative targeting properties in one experimental study (PKL>; Mullen et al., 1997a). The residue C was found in a significant but lower number of proteins, even though the efficiency of Cys-containing PTS1 tripeptides in peroxisomal targeting or in Pex5-interaction was comparable to that of the prototypical tripeptides SRL> and SKL> (Hayashi et al., 1997; Mullen et al., 1997a; Kragler et al., 1998). No sequence carried G or T (position −3), even though the peptides GKL>, GRL>, and TKL> conferred peroxisomal targeting or Pex5 interaction in some studies (Hayashi et al., 1997; Mullen et al., 1997a; Kragler et al., 1998). Regarding position −2, R occurred more frequently than K, and the remaining amino acids, such as N, S, and M were only found in a small number of sequences and mostly SOX homologs. The surprisingly strong interaction of the tripeptide SML> with Pex5 may have been enhanced by the high content of basic residues of the linker region (RRLRSML>; Kragler et al., 1998). Unexpectedly, none of the 400 homologous sequences carried H at position −2, even though the tripeptide SHL> is a common PTS1 in yeast and mammals, targeted a passenger protein to plant peroxisomes and interacted with Pex5 (Mullen et al., 1997a; Kragler et al., 1998; Emanuelsson et al., 2003). Whether a weak interaction between SHL> and Pex5 was significantly enhanced by the presence of auxiliary residues upstream of the PTS1 (PLRSHL>; Kragler et al., 1998) requires further investigations. Regarding position −1, PTS1 tripeptides carrying L and M were most widespread in plant proteins, but I was also present in one major (SRI>) and one minor PTS1 (SKI>). The residue V is so far restricted to the tripeptide SRV>, and none of the minor PTS1 contained F or Y (position −1), even though good targeting properties have been demonstrated for peptides containing these residues (SK[FY]>; Mullen et al., 1997a). In summary and in contrast to mammalian and yeast PTS1 tripeptides, the plant PTS1 is characterized by good targeting properties of PTS1 tripeptides carrying P at position −3, a preference of R over K as well as a lack of H at position −2, and a tolerance of I at position −1. With these features the plant PTS1 is more similar to the mammalian PTS1, both of which differ fundamentally from fungi, as stated earlier (Kragler et al., 1998; Lametschwandtner et al., 1998).
The strikingly different abundance of PTS1 peptides in plant proteins is interpreted from an evolutionary point of view in the following way: (1) a strong selection pressure has optimized the primary structure of PTS1 tripeptides with the result that the abundance of specific peptides in nature differs drastically nowadays, and (2) the current abundance of specific peptides in plant proteins correlates roughly with their targeting efficiency. In line with experimental results, highly abundant peptides, defined as major PTS peptides in this study, can be regarded as peptides with strong peroxisomal targeting properties (Kragler et al., 1998), whereas low-abundant minor peptides seem to reveal weaker targeting properties. Similarly, single amino acids of the PTS1 seem to differ considerably in their targeting efficiency and obviously cannot be freely combined to form functional PTS. For instance, the three highly abundant amino acids of the PTS1 tripeptide SRL> show a broad tolerance with respect to the choice of amino acids at the remaining positions for maintenance of a functional PTS1, suggesting strong targeting properties. By contrast, low-abundant amino acids (e.g. A, P, C, N, and I) are restricted to specific tripeptides and seem to reveal weaker targeting properties that need to be compensated by the presence of one or two strong targeting amino acids at the remaining positions to maintain peroxisomal targeting (e.g. PR[LM]> and PKL> are found but not PRI> or PK[MI]>). This conclusion holds also for noncanonical tripeptides. Thus, the combination of amino acids to yield functional PTS1 tripeptides follows strict rules that are determined by the apparent targeting strength of the single residues, and the targeting strength of the entire tripeptide seems to be an additive function of the targeting strength of the three amino acid residues. The reason why low-abundant minor PTS1 tripeptides can target some but not all proteins to peroxisomes is probably the dependence of its targeting function on cryptic auxiliary elements in the targeting domain that would need to be acquired by mutation simultaneously to the PTS mutation.
The region upstream of the PTS1 tripeptide was also characterized by conserved properties. According to our analysis, the PTS1 targeting domain with characteristic and diagnostic properties comprises about 12 to 15 amino acids, similar to recent results for fungi and mammals (Neuberger et al. 2003a). About 4 to 5 residues upstream of the PTS1 tripeptide are thought to interact with the surface of Pex5 (Gatto et al., 2003). According to this study, the PTS1 targeting domain of plant proteins is characterized by a frequent presence of P in front of the PTS1 tripeptide and a high content of basic residues accompanied by a lack of acidic residues, which provides the targeting domain with a high pI and a pronounced positive charge of about 2. The second basic residue is located in general in a 4-mer preceeding the PTS1 (position −7 to −4), but in M-containing PTS1 mostly directly in front of the tripeptide at position −4. Enhanced targeting properties have been reported earlier for tripeptides with a basic residue at position −4 (KANL>; Mullen et al., 1997b). A similar distribution of charged residues has recently been observed for metazoan sequences in the stretch between position −4 and −9 with a predominant location of positively charged residues at position −4 (called position −3 in Neuberger et al., 2003a). A frequent presence of P residues in a region of about 6 amino acids upstream of the PTS1 tripeptide, however, has not been noticed previously. The importance of P in mediating protein-protein interactions has been recognized in many cases. The P residue in front of the plant PTS1 and behind the PTS2 may guarantee PTS exposure and accessibility for the cytosolic receptors Pex5 and Pex7 and prevent further extension of the postulated α-helix.
Taken together, conserved properties upstream of the PTS1 tripeptide represent important information to further increase the prediction accuracy and shed new light on previous experimental results. The negative charge present upstream of the PTS1 in β-glucuronidase fusion proteins may have reduced the targeting efficiency of PRL> and SRI> and prevented the detection of peroxisomal targeting for some weaker tripeptides, such as SRV> and SSL> (ELxxx> in Hayashi et al., 1997; compare with GAxxx> in Mullen et al., 1997a). In another study, the region adjacent to the PTS1 differed substantially with respect to the content of basic and P residues at position −4 for the tripeptides investigated, constraining the comparability of Pex5 affinities determined for these peptides (e.g. KSQL>, PCRL>, CSSL>; Kragler et al., 1998).
Specification of the Plant PTS2
As for the PTS1, only a specific subset of PTS2 nonapeptides seems to be widespread in the plant kingdom and to constitute functional PTS. Two major (R[LI]x5HL) and nine minor PTS2 nonapeptides were defined. In support of the conservative variant of the plant PTS2 motif (R[LIQ]x5HL, Kato et al., 1998), the two amino acid residues R and H were absolutely and L (position 9) highly conserved, and position 2 was the most flexible position with L and I being the most frequent residues, followed by Q, T, M, A, and V. Analogously to the PTS1, the possible exchanges of L at position 2 and 9 from the prototypical nonapeptide RLx5HL were not present in random combinations but occurred preferentially only at one position at a time. If two amino acids deviated from L at position 2 and 9, then at least one of the exchanges was I, the second most abundant amino acid at both positions.
The PTS2 RQx5HL is unusual in several aspects. The nonapeptide was present in all plant thiolase homologs and exclusively present in this orthologous group. Moreover, to the best of our knowledge, Q (position 2) is absent in all known PTS2-targeted proteins from mammals and fungi including thiolase homologs. The fact that no stable single point mutation [LITMAV]→Q has occurred in any of the 110 PTS2 sequences of nonthiolase proteins strongly suggests that yet unrecognized features of the PTS2 domain of thiolase need to act synergistically with this nonapeptide to evoke targeting to peroxisomes. This hypothesis is further supported by the result that the mutation R(L→Q)x5HL abolished peroxisomal targeting in plants (Flynn et al., 1998). In contrast to experimentally determined motifs, none of the 168 plant sequences of PTS2-targeted proteins contained K at position 1 or Q at position 8 even though these residues are present in PTS2-targeted proteins from fungi and trypanosomes. Thus, the two basic residues, R and K (position 1), apparently are not interchangeable in plants and H (position 8) is absolutely conserved. To the best of our knowledge, peroxisomal Hsp70 from Citrullus lanatus with dual targeting to chloroplasts and peroxisomes by an alternative use of two translation starts is the only reported plant PTS2 protein with K at position 8 (RTx5KL; Wimmer et al., 1997). Unfortunately, homologs corresponding to peroxisomal Hsp70 from other plant species including Arabidopsis could not be identified unambiguously. However, the mutation H→K (position 8) introduced into the presequence of rat thiolase did not target a fusion protein to plant peroxisomes, and neither did the entire nonapeptide of peroxisomal Hsp70 embedded in the same presequence (Flynn et al., 1998). Taken together, the PTS2 nonapeptide of peroxisomal Hsp70 possesses unusual and probably context-dependent targeting properties and does not currently indicate peroxisomal targeting of unknown proteins.
Strikingly similar to PTS1-targeted proteins, the PTS2 targeting domain was characterized by a high content of basic residues accompanied by a lack of acidic residues, resulting in a basic pI and a high positive charge. In this case, the positive net charge was concentrated upstream of the nonapeptide and within the x5 region, whereas P was predominantly present downstream of the PTS2. In addition, the PTS2 domain was significantly enriched in three hydrophobic residues, namely A, L, and V, and mostly forms a short α-helix terminating in the end of the nonapeptide.
Prediction of Peroxisomal Proteins
Application of the major PTS peptides to genome screens are expected to lead to the identification of a large number of interesting novel proteins from plant peroxisomes. By contrast, those peptides that have been included in experimentally determined plant PTS motifs but not been defined as functional PTS in this study are not recommended for any genome screens. For minor PTS peptides, true positives are expected, but an elevated number of nonperoxisomal proteins needs to be anticipated as well. It needs to be pointed out that the indicative value of peroxisomal targeting differs significantly within the group of minor PTS peptides. The tripeptide PKL>, for instance, is relatively close to the empirical threshold for peptides defined as major PTS peptides and close to the frequency of the major PTS1 ARM>, whereas other minor PTS1 tripeptides were rare and only found in single orthologous groups (e.g. GOX, SML>; SOX, SNL>, ANL>, SNM>, SSM>). In this regard, unknown structural properties of SOX are suspected to be responsible for the unusual tolerance of these noncanonical PTS1 peptides as compared to other PTS1-targeted proteins and need to be investigated experimentally. Taken together, targeting to peroxisomes should only be predicted if a minor PTS is detected in at least two to three different orthologous groups and present in combination with other conserved properties of peroxisomal targeting domains (Table I). As a general rule of thumb, peroxisomal targeting of atypical putative PTS1 can be predicted in the following way: First, the higher the abundance of each single amino acid of the tripeptide of interest in all PTS1 proteins examined in this study, the higher the probability that this tripeptide targets a protein to plant peroxisomes. Second, a low-abundant amino acid is only found in a functional tripeptide in the presence of one to two high-abundant amino acids with presumably high targeting properties. To improve targeting prediction, the amino acids of the plant-specific motif are now given in order of abundance and deduced targeting strength (PTS1, [SAPC][RKNMS][LMIV]>). For PTS2 proteins, R (position 1), L (position 2, 9), H (position 8), and, to a lesser degree, I (position 2, 9) represent residues of presumably strong targeting properties; at least three of these residues need to be present to constitute a functional PTS2 nonapeptide (R[LIQTMAV]x5H[LIF]).
Table I.
Conserved properties of the targeting domains of PTS1- and PTS2-targeted plant peroxisomal proteins
| PTS1 Proteins | PTS2 Proteins | |
|---|---|---|
| Major PTS | [SA][RK][LM]> without AKM> plus SRI> and PRL> | R[LI]x5HL |
| Minor PTS | Canonical PTS1 tripeptides: SKI>, PRM>, PKL>, C[RK]L> | R[QTMAV]x5HL, RLx5H[IF], R[AI]x5HI |
| Noncanonical PTS1 tripeptides: S[MN]L>, SNM>, SRV>, SSM>, ANL> | ||
| Amino acids (in order of abundance) | [SAPC][RKNMS][LMIV]> | R[LIQTMAV]x5H[LIF] |
| Targeting domain | ||
| Size | C-terminal 12 to 15 residues | Approximately 15 residues, thus about three residues on each side of the PTS2 nonapeptide |
| Basic residues | High content at position −4 to −6 | High content in the targeting domain (mostly R) |
| Acidic residues | Low content at position −4 to −6 | Low content at position 1 to 10 |
| Charge and pI | Positive charge and basic pI at position −4 to −6 (or spread over the entire targeting domain) | Positive charge (2.2 ± 0.7) and basic pI |
| Proline | High content at position −4 to −9 | Absence in front of the PTS2 nonapeptide (position −1 to −3) and within the x5 region but high content immediately downstream of the PTS2 (position 10 to 15) |
| Hydrophobic residues | High content in the C-terminal 18 residues (A and L) | High content in front of the PTS2 nonapeptide (position −3 to −1) and within the x5 region (A, L, and V) |
| Secondary structure | n.d. | Short α-helix terminating in the end of the nonapeptide |
n.d., not detected.
Before starting time-consuming experiments, confirmation of the postulated peroxisomal localization of unknown proteins by further bioinformatics analyses is recommended. Postulated targeting to plant peroxisomes can sometimes be supported by results provided by new subcellular prediction software for peroxisomes despite their lack of plant-specific algorithms (PeroxiP; Emanuelsson et al., 2003; PTS1 predictor, http://mendel.imp.univie.ac.at/mendeljsp/sat/pts1/PTS1predictor.jsp; Neuberger et al., 2003a, 2003b). Dual subcellular localization of peroxisomal proteins has been reported (Hayashi et al., 1996b; Wimmer et al., 1997; Kunze et al., 2002) but is expected to represent an exception from the general rule of an exclusive localization of peroxisomal proteins in microbodies. Thus, the presence of accurately predicted plastidic transit peptides, mitochondrial presequences, or ER signal peptides can mostly be considered as evidence against a localization of the same protein in peroxisomes. The peroxisomal predictor PeroxiP even excludes proteins with secretory signals as potential peroxisomal matrix proteins (Emanuelsson et al., 2003). Because the prediction of a protein being peroxisomal is strengthened if its orthologs in other organisms possess a PTS1/2 as well, strongest support for the predicted peroxisomal localization can be provided by identification of homologous proteins with a PTS in other plant species similar to the strategy of this study. Finally, the prediction accuracy of PTS1/2-targeted proteins can be further raised by analyzing the neighboring region for accessory elements found to be conserved in PTS1/2-targeted proteins. These properties are more pronounced in proteins carrying a minor PTS and should be considered as positive complimentary indications for peroxisomal localization because we are only in the beginning of understanding the structure of these accessory elements and because not all properties are strictly conserved in the orthologous groups.
The number of plant genomes, the number and size of EST collections, and—most important—the number of known proteins of plant peroxisomes will increase in the near future and affect the results provided in this study. Therefore, this definition of functional PTS peptides represents an intermediate result. Whereas the frequency and the strong indicative properties of major PTS are not expected to change significantly, new database entries will affect the frequency of minor and currently undetected PTS. The tripeptide CRM> found in a single sequence, for instance, was currently excluded but is expected to represent a rare but functional PTS1 (Kragler et al., 1998). Most importantly, the example of SOX demonstrates that localization of novel proteins with unusual plant PTS peptides to peroxisomes (e.g. SHL> or AHL>) can lead to the identification of a large number of new noncanonical PTS1 peptides. The hypothesis that peptides other than those defined as major and minor PTS in this study represent functional PTS in plant proteins is supported by proteome studies of leaf peroxisomes and glyoxysomes from Arabidopsis (Fukao et al., 2002, 2003). Provided that the purity of the organelles was sufficient to prevent analysis of contaminating proteins, the fact that many novel proteins contained an unusual noncanonical C-terminal tripeptide (e.g. SKD>, SNI>, IRL>, SKP>, and IIL>) or even lacked any putative PTS, suggest a large number of additional PTS in higher plants. Interestingly, recent evidence shows that even proteins without a PTS1 motif may also be imported into plant peroxisomes via the PTS1 pathway. Acyl-CoA oxidase from Saccharomyces cerevisiae lacking a PTS1 tripeptide was shown to bind to Pex5 via an internal sequence and to be imported into peroxisomes by the PTS1 pathway as well (Klein et al., 2002).
CONCLUSIONS
In summary, the major and minor PTS deduced from this study are recommended for genome screens for novel peroxisomal matrix protein. In the Arabidopsis genome, about 170 proteins with a major PTS and 110 with a minor PTS are detected (S. Reumann, C. Ma, S. Lemke, and L. Babujee, unpublished data). We are currently setting up a database of Arabidopsis proteins with a putative PTS1 or PTS2 that will be presented in a forthcoming article and publicly available in the near future.
MATERIALS AND METHODS
Selection of the Arabidopsis Proteins Suitable for the Bioinformatics Analysis
All peroxisomal sequences that were cloned from Arabidopsis and encode PTS1/2-targeted proteins were retrieved. The Arabidopsis orthologs of PTS1/2-targeted proteins that were only cloned from other plant species were identified based on sequence similarity using blastp (http://www.ncbi.nlm.nih.gov/BLAST/; nonredundant database; E value threshold: 10; matrix BLOSUM62, gap costs: existence: 11, extension: 1; Supplemental Table SI). Proteins were used for subsequent bioinformatics analyses if they fulfilled the following criteria: (1) localization of one protein of an orthologous group to plant peroxisomes by biochemical studies using native proteins or by in vivo localization studies using fusion proteins; (2) identification of the PTS by redirection of the protein with deleted PTS to the cytosol as compared to the full-length protein; and (3) possible differentiation between homologs corresponding to peroxisomal isoforms or to isoforms from other cell compartments by sequence similarity. With respect to the enzyme CHY1 (Zolman et al., 2001), complementation of the phenotype of the chy1 mutant by the mammalian homolog redirected from mitochondria to peroxisomes together with the nature of its canonical C-terminal tripeptide AKL> was considered as sufficient support for the subcellular localization of the CHY1 protein in plant peroxisomes. Because the peroxisomal enzymes superoxide dismutase (Kliebenstein et al., 1998) and LACS7 share high sequence similarity with cytosolic isoforms and the PTS2-targeted LACS6, respectively (Fulda et al., 2002; Hayashi et al., 2002), the corresponding peroxisomal homologs could not clearly be identified by sequence similarity or multiple sequence alignment analysis and had to be excluded. Regarding the peroxisomal Hsp70 homolog from Citrullus lanatus with a dual subcellular localization that can either be translated from a first M followed by a transit peptide or from a second M followed by a PTS2 (Wimmer et al., 1997), this protein had to be excluded as well due to inconsistent findings of the second conserved M lacking in several homologous proteins and the presence of divergent putative PTS2 nonapeptides in other sequences. The isoforms of betaine aldehyde dehydrogenase from monocots that were experimentally localized to peroxisomes (Nakamura et al., 1997) were also excluded because of accumulating evidence for two closely related peroxisomal and nonperoxisomal homologs in monocots as well as in dicots (S. Lemke and S. Reumann, unpublished data). The splice variant HPR2 (LGI>; Cucurbita cv Kurokawa Amakuri; accession no. S68165) that encodes a polypeptide without an SKL motif and is localized in the cytosol (Hayashi et al., 1996b) was excluded as well. Similarily, HPR from Cucumis sativus (accession no. P13443; GNA>; Schwartz et al., 1991) was excluded. In case of unusually large novel gene families in Arabidopsis, such as that of the GOX homologs comprising five genes (Reumann 2002), the Arabidopsis homolog(s) corresponding to the reported proteins were identified according to maximum sequence similarity and similar expression pattern deduced from a digital northern (Mekhedov et al., 2000; e.g. GOX1/2, At3g14415/20). The remaining members of the gene family were not considered due to a current lack of experimental evidence for their localization in peroxisomes (e.g. GOX3, At4g18360 and HAOX1/2, At3g14130/50; Reumann, 2002).
Identification and Selection of Homologous cDNAs and ESTs from Various Higher Plant Species
To identify the plant homologs of known PTS1-targeted proteins from various plant species, the entire amino acid sequence of the Arabidopsis orthologs was blasted against the nonredundant database and the database of ESTs of GenBank at the National Center for Biotechnology Information using blastp and tblastn, respectively (http://www.ncbi.nlm.nih.gov/; E value threshold: 10; matrix BLOSUM62, gap costs: existence: 11, extension: 1; Supplemental Table SI). All blast searches were updated in August and September 2003, when the nonredundant database contained about 1,500,000 sequences and the database dbEst at NCBI the following number of sequences (for different species of the same genus, only the species with the largest EST collection is listed): 200,000 ≤ x < 500,000 ESTs: Triticum aestivum, Zea mays, Hordeum vulgare, Glycine max, and Oryza sativa; 100,000 ≤ x < 200,000 ESTs: Arabidopsis, Medicago truncatula, Lycopersicon esculentum, Sorghum bicolor, and Vitis vinifera; 50,000 ≤ x < 100,000 ESTs: Solanum tuberosum, Pinus taeda, Lactuca sativa, Helianthus annuus, and Populus tremula × Populus tremuloides; 20,000 ≤ x < 50,000 ESTs: Gossypium arboreum, Brassica napus, Lotus corniculatus, Ipomoea nil, Mesembryanthemum crystallinum, Capsicum annuum, and Phaseolus coccineus; 5,000 ≤ x < 20,000 ESTs: Beta vulgaris, Prunus persica, Nicotiana tabacum, Zinnia elegans, Secale cereale, Citrus sinensis, and Stevia rebaudiana. To increase the sensitivity of the database searches and the number of homologous sequences containing the targeting domain of interest, organism-specific databases were chosen and the query sequence of the full-length protein was shortened to the terminal 100 amino acids. To identify homologous sequences that correspond to the peroxisomal isoform based on a shortened protein fragment, the targeting domain of the peroxisomal proteins was first blasted against the nonredundant database to analyze the sequence similarity shared with peroxisomal isoforms on the one hand and nonperoxisomal isoforms on the other hand. If the sequence similarity shared with peroxisomal sequences differed significantly from that shared with nonperoxisomal sequences, the targeting domain was judged suitable for searches of homologous ESTs and appropriate thresholds were deduced. Homologs of Clamydomonas reinhardtii and Physcomitrella patens were omitted due to the restriction to higher plants.
EST clones were selected for further analysis if they met the following requirements: (1) sufficient size (mostly longer than 150 bp), (2) sufficient sequence quality (no undefined nucleotides “x” in the sequence) and no stop codon within the C-terminal 20 amino acids (PTS1 homologs) or the N-terminal 50 amino acids (PTS2), (3) homology throughout the entire domain, and (4) identification as the homolog corresponding to the peroxisomal isoforms based on sequence similarity or analysis by multiple sequence alignments. Some ESTs were excluded because an abrupt reduction of sequence conservation in the C-terminal region suggested an upstream sequencing error leading to a frame shift. For the selected ESTs, the nucleotide sequence was then translated (http://www.expasy.org/tools/dna.html). Regarding the homologs of PTS1-targeted proteins, EST sequences were only included if the peptide that aligned with the C terminus of the open reading frame was followed by a stop codon (about 5 amino acids up- or downstream of the PTS1 of the query). Regarding the homologs of PTS2-targeted proteins, EST sequences were only included if the peptide that aligned with the N terminus of the peroxisomal protein was preceded by an M (about 20 amino acids upstream of the PTS2 of the query). The amino acid sequence of the C-terminal 20 amino acids (PTS1 proteins) or that of the N-terminal 50 amino acids (PTS2 proteins) was saved for later analysis of the targeting domain for conserved properties.
For plants with large EST collections, several homologous cDNAs or ESTs from the same plant species or the same genus were present in the database that differed only slightly (e.g. Sorghum bicolor and S. propinquum, Triticum aestivum and T. monococcum, Oryza sativa and O. minuta). In an attempt to distinguish sequences with sequencing errors from sequences of closely related genes, an empirical threshold was chosen. Regarding homologs of PTS1-targeted proteins, additional sequences were excluded unless the C-terminal 20 amino acids differed by at least three amino acids (<90% sequence identity) compared to another homologous sequence from the same plant species or genus. The N-terminal 20 amino acids of PTS2-targeted proteins were handled analogously. To facilitate detection of these ESTs, homologous ESTs of the plant species or genus of interest were often used as query. For peroxisomal proteins that are encoded by several genes in Arabidopsis (e.g. ACX1, HIBCH, thiolase, citrate synthase, multi-functional protein, etc.), the application of this empirical rule resulted in a number of homologous sequences in large EST collections that was close to the size of the gene family in Arabidopsis.
Analysis of Amino Acid Composition and pI
For analysis of amino acid composition, charge, and pI of the targeting domain, the sequences were grouped according to their PTS peptide. Groups of canonical PTS1 tripeptides containing less than five sequences (CKL>, CRL>, SKI>) and the noncanonical tripeptides (ANL>, SML>, SNL>, SNM>, SRV>, and SSM>) were combined and analyzed together. Similarly, groups of PTS2 nonapeptides containing less than five sequences (RLx5HF, RLx5HI, RIx5HI, RAx5HL, RAx5HI, and RVx5HL) were analyzed together. For PTS1 sequences, the C-terminal 18 amino acids were analyzed in groups of three amino acids and provided with position-specific numbers relative to the PTS1 tripeptide (PTS1, position −1 to −3; 3-mer preceeding the PTS1, position −4 to −6). For PTS2 sequences, the residues of the nonapeptide were numbered according to the traditional system (PTS2, position 1 to 9) and residues in front and behind the nonapeptide were provided with negative and positive numbers, respectively. The amino acid composition of PTS2 proteins was analyzed in two groups of three amino acids in front of the nonapeptide (position −6 to −4 and position −3 to −1), the first two conserved residues of the nonapeptide (Rx, position 1, 2), the five residues of the unspecific x5 region (position 3 to 7), the other two conserved residues of the nonapeptide (Hx, position 8, 9), as well as two groups of three amino acids behind the nonapeptide (position 10 to 12 and position 13 to 15). For these groups, the amino acid composition, the absolute content of basic and acidic residues, the charge (difference of basic and acidic residues), and the pI were calculated (http://us.expasy.org/tools/protparam.html) and mean values and sd determined. Secondary structure was analyzed using PredictProtein (http://www.embl-heidelberg.de/predictprotein/predictprotein.html).
Supplementary Material
Acknowledgments
I thank Martin Fulda, Ivo Feussner, Hans-Walter Heldt, and the students of my group for stimulating discussions and critical comments on the manuscript with special thanks to Katharina Pawlowski.
This work was supported by the Deutsche Forschungsgemeinschaft (Re1304/2–1).
The online version of this article contains Web-only data.
References
- Amery L, Brees C, Baes M, Setoyama C, Miura R, Mannaerts GP, Van Veldhoven PP (1998) C-terminal tripeptide Ser-Asn-Leu (SNL) of human D-aspartate oxidase is a functional peroxisome-targeting signal. Biochem J 336: 367–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR (2000) Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA 97: 3718–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammai V, Subramani S (2001) The human peroxisomal targeting signal receptor, Pex5p, is translocated into the peroxisomal matrix and recycled to the cytosol. Cell 105: 187–196 [DOI] [PubMed] [Google Scholar]
- de Hoop MJ, Ab G (1992) Import of proteins into peroxisomes and other microbodies. Biochem J 286: 657–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Rio LA, Corpas FJ, Sandalio LM, Palma JM, Gomez M, Barroso JB (2002) Reactive oxygen species, antioxidant systems and nitric oxide in peroxisomes. J Exp Bot 53: 1255–1272 [PubMed] [Google Scholar]
- Dyer JM, McNew JA, Goodman JM (1996) The sorting sequence of the peroxisomal integral membrane protein PMP47 is contained within a short hydrophilic loop. J Cell Biol 133: 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers T, Schwarz G, Brinkmann H, Witt C, Richter T, Nieder J, Koch B, Hille R, Hansch R, Mendel RR (2001) Identification and biochemical characterization of Arabidopsis thaliana sulfite oxidase. A new player in plant sulfur metabolism. J Biol Chem 276: 46989–46994 [DOI] [PubMed] [Google Scholar]
- Elgersma Y, Vos A, van den Berg M, van Roermund CW, van der Sluijs P, Distel B, Tabak HF (1996) Analysis of the carboxyl-terminal peroxisomal targeting signal 1 in a homologous context in Saccharomyces cerevisiae. J Biol Chem 271: 26375–26382 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Elofsson A, von Heijne G, Cristobal S (2003) In silico prediction of the peroxisomal proteome in fungi, plants and animals. J Mol Biol 330: 443–456 [DOI] [PubMed] [Google Scholar]
- Flynn CR, Mullen RT, Trelease RN (1998) Mutational analyses of a type 2 peroxisomal targeting signal that is capable of directing oligomeric protein import into tobacco BY-2 glyoxysomes. Plant J 16: 709–720 [DOI] [PubMed] [Google Scholar]
- Froman BE, Edwards PC, Bursch AG, Dehesh K (2000) ACX3, a novel medium-chain acyl-coenzyme A oxidase from Arabidopsis. Plant Physiol 123: 733–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao Y, Hayashi M, Hara-Nishimura I, Nishimura M (2003) Novel glyoxysomal protein kinase, GPK1, identified by proteomic analysis of glyoxysomes in etiolated cotyledons of Arabidopsis thaliana. Plant Cell Physiol 44: 1002–1012 [DOI] [PubMed] [Google Scholar]
- Fukao Y, Hayashi M, Nishimura M (2002) Proteomic analysis of leaf peroxisomal proteins in greening cotyledons of Arabidopsis thaliana. Plant Cell Physiol 43: 689–696 [DOI] [PubMed] [Google Scholar]
- Fulda M, Shockey J, Werber M, Wolter FP, Heinz E (2002) Two long-chain acyl-CoA synthetases from Arabidopsis thaliana involved in peroxisomal fatty acid beta-oxidation. Plant J 32: 93–103 [DOI] [PubMed] [Google Scholar]
- Gatto GJ Jr, Maynard EL, Guerrerio AL, Geisbrecht BV, Gould SJ, Berg JM (2003) Correlating structure and affinity for PEX5:PTS1 complexes. Biochemistry 42: 1660–1666 [DOI] [PubMed] [Google Scholar]
- Gietl C, Faber KN, van der Klei IJ, Veenhuis M (1994) Mutational analysis of the N-terminal topogenic signal of watermelon glyoxysomal malate dehydrogenase using the heterologous host Hansenula polymorpha. Proc Natl Acad Sci USA 91: 3151–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover JR, Andrews DW, Subramani S, Rachubinski RA (1994) Mutagenesis of the amino targeting signal of Saccharomyces cerevisiae 3-ketoacyl-CoA thiolase reveals conserved amino acids required for import into peroxisomes in vivo. J Biol Chem 269: 7558–7563 [PubMed] [Google Scholar]
- Gould SJ, Keller GA, Hosken N, Wilkinson J, Subramani S (1989) A conserved tripeptide sorts proteins to peroxisomes. J Cell Biol 108: 1657–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SG, Keller GA, Subramani S (1987) Identification of a peroxisomal targeting signal at the carboxy terminus of firefly luciferase. J Cell Biol 105: 2923–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen H, Didion T, Thiemann A, Veenhuis M, Roggenkamp R (1992) Targeting sequences of the two major peroxisomal proteins in the methylotrophic yeast Hansenula polymorpha. Mol Gen Genet 235: 269–278 [DOI] [PubMed] [Google Scholar]
- Hayashi M, Aoki M, Kato A, Kondo M, Nishimura M (1996. a) Transport of chimeric proteins that contain a carboxy-terminal targeting signal into plant microbodies. Plant J 10: 225–234 [DOI] [PubMed] [Google Scholar]
- Hayashi M, Aoki M, Kato A, Nishimura M (1997) Changes in targeting efficiencies of proteins to plant microbodies caused by amino acid substitutions in the carboxyl-terminal tripeptide. Plant Cell Physiol 38: 759–768 [DOI] [PubMed] [Google Scholar]
- Hayashi H, De Bellis L, Ciurli A, Kondo M, Hayashi M, Nishimura M (1999) A novel acyl-CoA oxidase that can oxidize short-chain acyl-CoA in plant peroxisomes. J Biol Chem 274: 12715–12721 [DOI] [PubMed] [Google Scholar]
- Hayashi H, De Bellis L, Hayashi Y, Nito K, Kato A, Hayashi M, Hara-Nishimura I, Nishimura M (2002) Molecular characterization of an Arabidopsis acyl-coenzyme A synthetase localized on glyoxysomal membranes. Plant Physiol 130: 2019–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Toriyama K, Kondo M, Nishimura M (1998) 2,4-Dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid beta-oxidation. Plant Cell 10: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Tsugeki R, Kondo M, Mori H, Nishimura M (1996. b) Pumpkin hydroxypyruvate reductases with and without a putative C-terminal signal for targeting to microbodies may be produced by alternative splicing. Plant Mol Biol 30: 183–189 [DOI] [PubMed] [Google Scholar]
- Hooks MA, Kellas F, Graham IA (1999) Long-chain acyl-CoA oxidases of Arabidopsis. Plant J 20: 1–13 [DOI] [PubMed] [Google Scholar]
- Igarashi D, Miwa T, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Ohsumi C (2003) Identification of photorespiratory glutamate:glyoxylate aminotransferase (GGAT) gene in Arabidopsis. Plant J 33: 975–987 [DOI] [PubMed] [Google Scholar]
- Jones JM, Morrel JC, Gould SJ (2001) Multiple distinct targeting signals in integral peroxisomal membrane proteins. J Cell Biol 153: 1141–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamigaki A, Mano S, Terauchi K, Nishi Y, Tachibe-Kinoshita Y, Nito K, Kondo M, Hayashi M, Nishimura M, Esaka M (2003) Identification of peroxisomal targeting signal of pumpkin catalase and the binding analysis with PTS1 receptor. Plant J 33: 161–175 [DOI] [PubMed] [Google Scholar]
- Karpichev IV, Small GM (2000) Evidence for a novel pathway for the targeting of a Saccharomyces cerevisiae peroxisomal protein belonging to the isomerase/hydratase family. J Cell Sci 113: 533–544 [DOI] [PubMed] [Google Scholar]
- Kato A, Hayashi M, Kondo M, Nishimura M (1996) Targeting and processing of a chimeric protein with the N-terminal presequence of the precursor to glyoxysomal citrate synthase. Plant Cell 8: 1601–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, Takeda-Yoshikawa Y, Hayashi M, Kondo M, Hara-Nishimura I, Nishimura M (1998) Glyoxysomal malate dehydrogenase in pumpkin: cloning of a cDNA and functional analysis of its presequence. Plant Cell Physiol 39: 186–195 [DOI] [PubMed] [Google Scholar]
- Klein AT, van den Berg M, Bottger G, Tabak HF, Distel B (2002) Saccharomyces cerevisiae acyl-CoA oxidase follows a novel, non-PTS1, import pathway into peroxisomes that is dependent on Pex5p. J Biol Chem 277: 25011–25019 [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Monde RA, Last RL (1998) Superoxide dismutase in Arabidopsis: an eclectic enzyme family with disparate regulation and protein localization. Plant Physiol 118: 637–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragler F, Lametschwandtner G, Christmann J, Hartig A, Harada JJ (1998) Identification and analysis of the plant peroxisomal targeting signal 1 receptor NtPEX5. Proc Natl Acad Sci USA 95: 13336–13341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze M, Kragler F, Binder M, Hartig A, Gurvitz A (2002) Targeting of malate synthase 1 to the peroxisomes of Saccharomyces cerevisiae cells depends on growth on oleic acid medium. Eur J Biochem 269: 915–922 [DOI] [PubMed] [Google Scholar]
- Lametschwandtner G, Brocard C, Fransen M, Van Veldhoven P, Berger J, Hartig A (1998) The difference in recognition of terminal tripeptides as peroxisomal targeting signal 1 between yeast and human is due to different affinities of their receptor Pex5p to the cognate signal and to residues adjacent to it. J Biol Chem 273: 33635–33643 [DOI] [PubMed] [Google Scholar]
- Lazarow PB, Fujiki Y (1985) Biogenesis of peroxisomes. Annu Rev Cell Biol 1: 489–530 [DOI] [PubMed] [Google Scholar]
- Liepman AH, Olsen LJ (2001) Peroxisomal alanine:glyoxylate aminotransferase (AGT1) is a photorespiratory enzyme with multiple substrates in Arabidopsis thaliana. Plant J 25: 487–498 [DOI] [PubMed] [Google Scholar]
- Liepman AH, Olsen LJ (2003) Alanine aminotransferase homologs catalyze the glutamate:glyoxylate aminotransferase reaction in peroxisomes of Arabidopsis. Plant Physiol 131: 215–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Huertas E, Charlton WL, Johnson B, Graham IA, Baker A (2000) Stress induces peroxisome biogenesis genes. EMBO J 19: 6770–6777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzioch M, Erdmann R, Veenhuis M, Kunau WH (1994) PAS7 encodes a novel yeast member of the WD-40 protein family essential for import of 3-oxoacyl-CoA thiolase, a PTS2-containing protein, into peroxisomes. EMBO J 13: 4908–4918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhedov S, de Ilarduya OM, Ohlrogge J (2000) Toward a functional catalog of the plant genome. A survey of genes for lipid biosynthesis. Plant Physiol 122: 389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen RT, Lee MS, Flynn CR, Trelease RN (1997. a) Diverse amino acid residues function within the type 1 peroxisomal targeting signal. Implications for the role of accessory residues upstream of the type 1 peroxisomal targeting signal. Plant Physiol 115: 881–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen RT, Lee MS, Trelease RN (1997. b) Identification of the peroxisomal targeting signal for cottonseed catalase. Plant J 12: 313–322 [DOI] [PubMed] [Google Scholar]
- Murphy MA, Phillipson BA, Baker A, Mullen RT (2003) Characterization of the targeting signal of the Arabidopsis 22-kD integral peroxisomal membrane protein. Plant Physiol 133: 813–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Meyer C, Sano H (2002) Molecular cloning and characterization of plant genes encoding novel peroxisomal molybdoenzymes of the sulphite oxidase family. J Exp Bot 53: 1833–1836 [DOI] [PubMed] [Google Scholar]
- Nakamura T, Yokota S, Muramoto Y, Tsutsui K, Oguri Y, Fukui K, Takabe T (1997) Expression of a betaine aldehyde dehydrogenase gene in rice, a glycinebetaine nonaccumulator, and possible localization of its protein in peroxisomes. Plant J 11: 1115–1120 [DOI] [PubMed] [Google Scholar]
- Neuberger G, Maurer-Stroh S, Eisenhaber B, Hartig A, Eisenhaber F (2003. a) Motif refinement of the peroxisomal targeting signal 1 and evaluation of taxon-specific differences. J Mol Biol 328: 567–579 [DOI] [PubMed] [Google Scholar]
- Neuberger G, Maurer-Stroh S, Eisenhaber B, Hartig A, Eisenhaber F (2003. b) Prediction of peroxisomal targeting signal 1 containing proteins from amino acid sequence. J Mol Biol 328: 581–592 [DOI] [PubMed] [Google Scholar]
- Osumi T, Tsukamoto T, Hata S (1992) Signal peptide for peroxisomal targeting: replacement of an essential histidine residue by certain amino acids converts the amino-terminal presequence of peroxisomal 3-ketoacyl-CoA thiolase to a mitochondrial signal peptide. Biochem Biophys Res Commun 186: 811–818 [DOI] [PubMed] [Google Scholar]
- Osumi T, Tsukamoto T, Hata S, Yokota S, Miura S, Fujiki Y, Hijikata M, Miyazawa S, Hashimoto T (1991) Amino-terminal presequence of the precursor of peroxisomal 3-ketoacyl-CoA thiolase is a cleavable signal peptide for peroxisomal targeting. Biochem Biophys Res Commun 181: 947–954 [DOI] [PubMed] [Google Scholar]
- Pause B, Saffrich R, Hunziker A, Ansorge W, Just WW (2000) Targeting of the 22 kDa integral peroxisomal membrane protein. FEBS Lett 471: 23–28 [DOI] [PubMed] [Google Scholar]
- Rehling P, Marzioch M, Niesen F, Wittke E, Veenhuis M, Kunau WH (1996) The import receptor for the peroxisomal targeting signal 2 (PTS2) in Saccharomyces cerevisiae is encoded by the PAS7 gene. EMBO J 15: 2901–2913 [PMC free article] [PubMed] [Google Scholar]
- Reumann S (2002) The photorespiratory pathway of leaf peroxisomes. In A Baker, IA Graham, eds, Plant Peroxisomes: Biochemistry, Cell Biology and Biotechnological Applications, Ed 1. Kluwer Academic Publishers, Dordrecht, the Netherlands, pp 141–189
- Sacksteder KA, Jones JM, South ST, Li X, Liu Y, Gould SJ (2000) PEX19 binds multiple peroxisomal membrane proteins, is predominantly cytoplasmic, and is required for peroxisome membrane synthesis. J Cell Biol 148: 931–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders PM, Lee PY, Biesgen C, Boone JD, Beals TP, Weiler EW, Goldberg RB (2000) The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12: 1041–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz BW, Sloan JS, Becker WM (1991) Characterization of genes encoding hydroxypyruvate reductase in cucumber. Plant Mol Biol 17: 941–947 [DOI] [PubMed] [Google Scholar]
- Shockey JM, Fulda MS, Browse JA (2002) Arabidopsis contains nine long-chain acyl-coenzyme A synthetase genes that participate in fatty acid and glycerolipid metabolism. Plant Physiol 129: 1710–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer JM, Cheng QL, Keller GA, Wang CC (1992) In vivo import of firefly luciferase into the glycosomes of Trypanosoma brucei and mutational analysis of the C-terminal targeting signal. Mol Biol Cell 3: 749–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A, Browse J (2000) The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci USA 97: 10625–10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strassner J, Schaller F, Frick UB, Howe GA, Weiler EW, Amrhein N, Macheroux P, Schaller A (2002) Characterization and cDNA-microarray expression analysis of 12-oxophytodienoate reductases reveals differential roles for octadecanoid biosynthesis in the local versus the systemic wound response. Plant J 32: 585–601 [DOI] [PubMed] [Google Scholar]
- Subramani S (1993) Protein import into peroxisomes and biogenesis of the organelle. Annu Rev Cell Biol 9: 445–478 [DOI] [PubMed] [Google Scholar]
- Swinkels BW, Gould SJ, Bodnar AG, Rachubinski RA, Subramani S (1991) A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J 10: 3255–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinkels BW, Gould SJ, Subramani S (1992) Targeting efficiencies of various permutations of the consensus C-terminal tripeptide peroxisomal targeting signal. FEBS Lett 305: 133–136 [DOI] [PubMed] [Google Scholar]
- Van der Leij I, Franse MM, Elgersma Y, Distel B, Tabak HF (1993) PAS10 is a tetratricopeptide-repeat protein that is essential for the import of most matrix proteins into peroxisomes of Saccharomyces cerevisiae. Proc Natl Acad Sci USA 90: 11782–11786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer B, Lottspeich F, van der Klei I, Veenhuis M, Gietl C (1997) The glyoxysomal and plastid molecular chaperones (70-kDa heat shock protein) of watermelon cotyledons are encoded by a single gene. Proc Natl Acad Sci USA 94: 13624–13629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer C, Schmid M, Veenhuis M, Gietl C (1998) The plant PTS1 receptor: similarities and differences to its human and yeast counterparts. Plant J 16: 453–464 [DOI] [PubMed] [Google Scholar]
- Zolman BK, Monroe-Augustus M, Thompson B, Hawes JW, Krukenberg KA, Matsuda SPT, Bartel B (2001) chy1, an Arabidopsis mutant with impaired β-oxidation, is defective in a peroxisomal β-hydroxyisobutyryl-CoA hydrolase. J Biol Chem 276: 31037–31046 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.