Abstract
The structural determinants involved in the targeting of multitransmembrane-span proteins to the plasma membrane (PM) remain poorly understood. The plasma membrane H+-ATPase (PMA) from Nicotiana plumbaginifolia, a well-characterized 10 transmembrane–span enzyme, was used as a model to identify structural elements essential for targeting to the PM. When PMA2 and PMA4, representatives of the two main PMA subfamilies, were fused to green fluorescent protein (GFP), the chimeras were shown to be still functional and to be correctly and rapidly targeted to the PM in transgenic tobacco. By contrast, chimeric proteins containing various combinations of PMA transmembrane spanning domains accumulated in the Golgi apparatus and not in the PM and displayed slow traffic properties through the secretory pathway. Individual deletion of three of the four cytosolic domains did not prevent PM targeting, but deletion of the large loop or of its nucleotide binding domain resulted in GFP fluorescence accumulating exclusively in the endoplasmic reticulum. The results show that, at least for this polytopic protein, the PM is not the default pathway and that, in contrast with single-pass membrane proteins, cytosolic structural determinants are required for correct targeting.
INTRODUCTION
Eukaryotic cells are characterized by an endomembrane system consisting of organelles with a distinct protein composition, which is partly because of the presence of specific targeting information on the resident polypeptides. This information is quite well characterized for soluble proteins but not for membrane proteins, especially polytopic proteins, such as the plasma membrane (PM) proteins of plant cells.
PM integral proteins are thought to be first integrated into the endoplasmic reticulum (ER), then transported to the cell surface through the secretory pathway. The organization and functioning of the plant secretory pathway is attracting much interest (reviewed in Jiang and Rogers, 1999; Vitale and Denecke, 1999; Nebenfuhr, 2002; Ueda and Nakano, 2002), and some recent studies have identified morphological and functional aspects specific to plant cells (Boevink et al., 1998; Nebenfuhr et al., 1999; Batoko et al., 2000; Brandizzi et al., 2002a; Ritzenthaler et al., 2002; Saint-Jore et al., 2002). Segregation between the vacuolar and PM routes mainly takes place in the Golgi apparatus, which functions as a sorting gateway. In yeast cells, the default compartment for polytopic proteins is thought to be the vacuole (Roberts et al., 1992), whereas in mammalian cells, it is still generally thought to be the PM because of the identification of lysosomal targeting determinants (Peters et al., 1990). However, recent data have shown that, in mammalian cells, mutated or truncated PM polytopic proteins accumulate in the Golgi, suggesting that the PM might not be the default pathway (Stockklausner and Klocker, 2003).
In plants, the tonoplast (Hofte and Chrispeels, 1992) or the PM (Vitale and Raikhel, 1999) for several years have been suggested to be the default destination for polytopic proteins, although no strong evidence was provided. However, at least two types of vacuoles are now known to exist in some plant cells (Paris et al., 1996; Di Sansebastiano et al., 1998), and a cytosolic determinant segregates between them (Jiang and Rogers, 1998).
Although no targeting information has been clearly identified for vacuolar or PM polytopic proteins, in the case of single-span proteins, it has been proposed that the length of the transmembrane span (TM) might define the final localization of the protein because of differences in the thickness and composition of the lipid bilayer of the different subcellular compartments. Increasing or decreasing the length of TMs of yeast, mammalian, or plant proteins or the use of synthetic sequences of different lengths fused to soluble reporters showed that the length could be correlated with the final localization (Masibay et al., 1993; Munro, 1995; Pedrazzini et al., 1996; Rayner and Pelham, 1997; Watson and Pessin, 2001; Brandizzi et al., 2002b). The requirement for a long TM in PM proteins may be related to the high sphingolipid and sterol content of the PM, especially in the microdomains, known as rafts, and characterized by their resistance to some detergents, which are found in animal and yeast cells (Harder and Simons, 1997; Bagnat et al., 2000). However, traffic occurs slowly and is not fully efficient, suggesting that other signals might be involved in efficient targeting of a native protein. Proteins are transported through the secretory pathway by the exchange of membrane-bound intermediates between compartments. The loading of nonresident proteins into vesicles originating from the ER was described for many years as passive, but recent data in yeast showed that specific concentration of proteins occurs in budding vesicles (Muniz et al., 2000; Malkus et al., 2002) and that there are several routes from the ER to the Golgi (Muniz et al., 2001; Morsomme et al., 2003), suggesting a sorting mechanism. However, passive, slow traffic and active, selective, fast traffic might coexist in a eukaryotic cell (Muniz et al., 2000).
We are interested in determining the structural motifs involved in the targeting of plant PM polytopic proteins. As a model, we have chosen to use the well-characterized PM H+-ATPase, which is composed of 10 TMs and four cytosolic regions, the N- and C-terminal regions, the so-called small loop delimited by TM2 and TM3, and the large loop between TM4 and TM5. The PM H+-ATPase is a P-type ATPase found in plants and fungi that pumps protons out of the cell and maintains the electrical and pH gradients across the PM, which provide the energy used by channels and secondary transporters for nutrient and ion transport across the membrane (reviewed in Morsomme and Boutry, 2000a; Palmgren, 2001; Lefebvre et al., 2003).
Although little is known about PM H+-ATPase targeting information, the sorting and trafficking of Saccharomyces cerevisae PM H+-ATPase1 (PMA1) have been studied to some extent (reviewed in Ferreira et al., 2001; Lefebvre et al., 2003). After translation and insertion into the ER membrane, PMA1 is packaged into COPII vesicles (Shimoni et al., 2000), then transported to the PM via the Golgi, because in various secretory mutants, it remains in the Golgi apparatus (sec7) or in the secretory vesicles trafficking between the Golgi and the PM (sec6 and sec1) (Holcomb et al., 1988; Chang and Slayman, 1991). Moreover, data obtained in yeast suggest a relationship between (1) protein oligomerization and/or association with detergent-resistant membranes and (2) the PM targeting and/or stability of the protein (Patton et al., 1992; Bagnat et al., 2000, 2001; Gong and Chang, 2001; Lee et al., 2002; Luo et al., 2002; Wang and Chang, 2002).
In this report, we used green fluorescent protein (GFP) as a reporter linked to truncated forms of PMA4, a PMA from Nicotiana plumbaginifolia, to determine whether PM targeting was the default pathway and, if not, whether the TMs alone were sufficient for PM targeting.
RESULTS
GFP Tagging Does Not Interfere with PM Targeting of Plant PM H+-ATPase
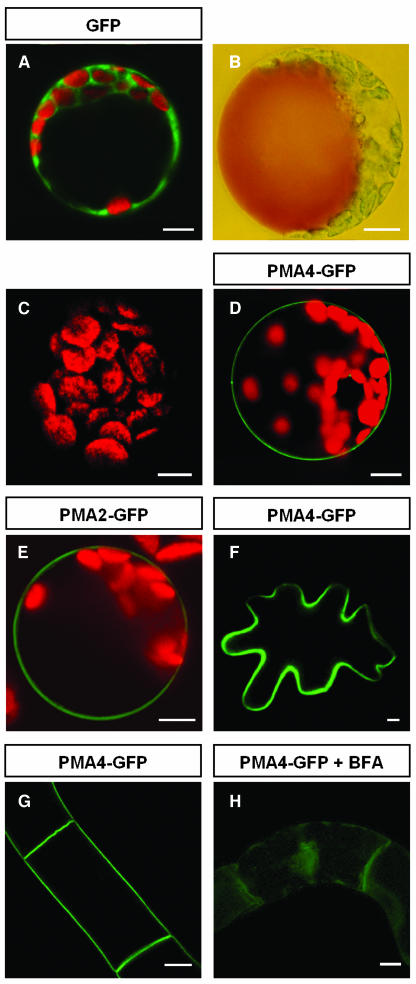
To study the subcellular localization of the plant PM H+-ATPase, three N. plumbaginifolia isogenes, PMA2 and PMA3 (subfamily I) and PMA4 (subfamily II) (Arango et al., 2003), were translationally fused at their 3′-coding end to the GFP gene and put under the control of the strong transcriptional promoter, PMA4-En50 (Zhao et al., 1999), then the resulting plasmids were used to transiently transform N. tabacum protoplasts by electroporation. As expected, free GFP (Figure 1A) was found in the cytosol surrounding the large vacuole visualized by neutral red staining (shown for another protoplast in Figure 1B). Using the same imaging settings, expression of PMA4-GFP (Figure 1D), PMA2-GFP (Figure 1E), or PMA3-GFP (data not shown) resulted in GFP fluorescence located exclusively in the PM, whereas nontransformed protoplasts showed no detectable green fluorescence (Figure 1C). Although most observations were performed at 24 h after transformation, PM localization of PMA4-GFP was seen within 8 h, depending on the protoplast batch (data not shown). No detectable GFP was associated with internal membranes, suggesting that PM targeting was rapid and specific. Microscopy or protein gel blotting analysis showed similar levels of expression of the three isoforms (data not shown). The fusion proteins were detected at the expected size, ∼125 kD, by protein gel blotting (see PMA4-GFP in Figure 6I).
Figure 1.
Validation of the PMA4-GFP Fusion System.
(A) to (E) N. tabacum protoplasts were transformed by electroporation with pTZ19U-gfp (A), pma4-gfp (D), pma2-gfp (E), or no DNA ([B] and [C]). A cell was stained with neutral red (B).
(F) A leaf epidermal cell transiently expressing PMA4-GFP 48 h after A. tumefaciens infiltration.
(G) BY2 cells stably expressing PMA4-GFP.
(H) BY2 cells stably expressing PMA4-GFP treated for 8 h with 10 μg/mL of brefeldin A (BFA).
The cells were analyzed either by a Bio-Rad MRC-1024 confocal microscope using emission band-pass filters of 506 to 538 nm and 664 to 696 nm ([A], [C] to [E], and [G]) or by a Leica DRM microscope under visible light (B) or with a GFP filter ([F] and [H]). The red fluorescence corresponds to chloroplast autofluorescence. Bars = 10 μm.
Figure 6.
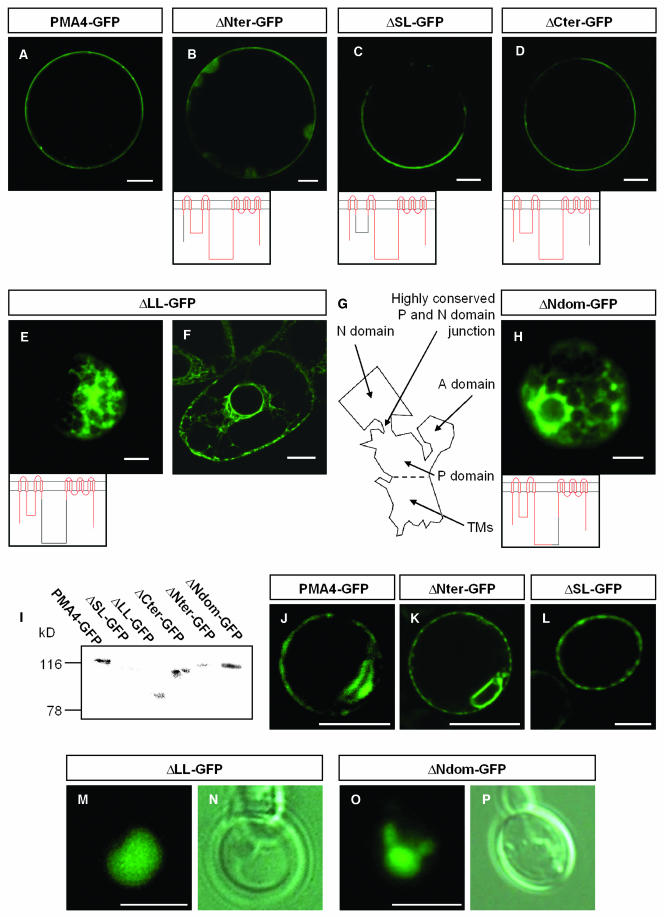
In Vivo and in Vitro Analysis of PM H+-ATPase-GFP Chimeras Lacking Different Cytosolic Regions.
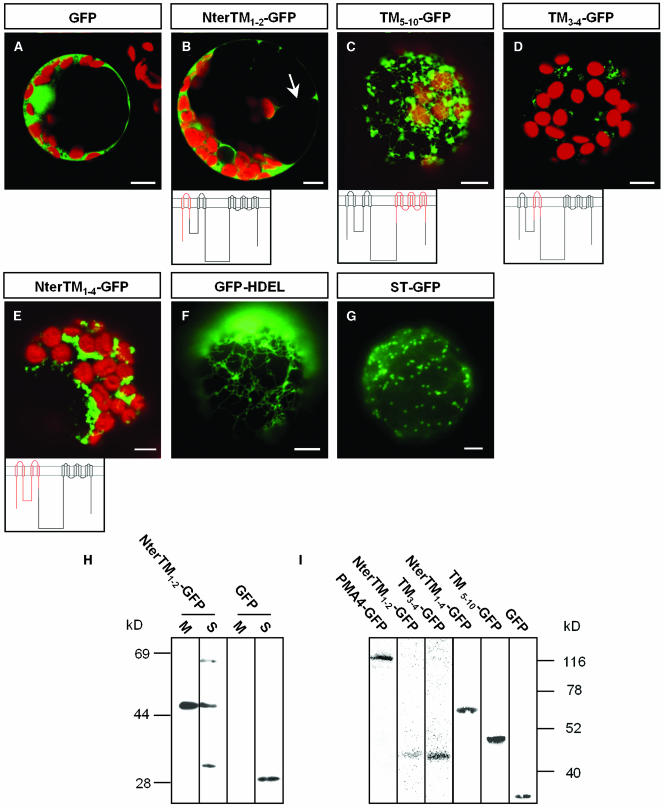
(A) to (E) and (H) N. tabacum protoplasts were transformed by electroporation with the indicated constructs. In (B) to (D), because of the low GFP signal, the microscope settings were adjusted to detect GFP, with the result that the chloroplast green autofluorescence is also seen in (B). Below the images, the PM H+-ATPase is schematized with the region fused to GFP in red.
(F) BY2 cells stably expressing ΔLL-GFP.
(G) Schematic representation of the P-type ATPase structure based on the 3D structure of rabbit sarcoplasmic Ca2+-ATPase (Toyoshima et al., 2000; Toyoshima and Nomura, 2002). The large loop is composed of the phosphorylation (P) domain and the nucleotide binding (N) domain. The A domain puts together the N-terminal region and the small loop.
(I) Protein gel blot analysis of protoplast extracts as described in Figure 4 using anti-GFP antibodies.
(J) to (P) S. cerevisae cells (YAK2) expressing the indicated constructs. Note that in some cases ([J] and [K]), GFP was localized in karmellae in addition to the PM. (N) and (P), corresponding to the same cells as in (M) and (O), respectively, were observed using a Leica DRM microscope with differential interference contrast. Fluorescence was imaged with either a Leica DRM microscope using a GFP filter ([E], [H], [M], and [O]) or with a Bio-Rad MRC-1024 confocal microscope using an emission band-pass filter of 506 to 538 nm ([A] to [D], [F], and [J] to [L]). Bars in (A) to (H) = 10 μm; bars in (J) to (P) = 5 μm.
To confirm these data, we genetically transformed tobacco cells using Agrobacterium tumefaciens. In a leaf transient expression system, PMA4-GFP was exclusively detected in the PM as soon as 2 d after infiltration of the A. tumefaciens strain into the tobacco leaf, with no intermediate accumulation in the secretory pathway (Figure 1F). A similar time course was seen for the appearance of free GFP in the cytosol (data not shown), indicating that the longer time required to observe PMA-GFP fluorescence in the PM of tobacco leaf epidermal cells as compared with the protoplast electroporation system was attributable to the biological transformation process, rather than to inefficient PM targeting. Epifluorescence analysis of stably transformed tobacco Bright Yellow (BY2) cell lines or various organs sampled from independent transgenic plants also showed that the GFP fusion proteins localized to the PM (Figure 1G), corroborating the observations made after transient expression.
In the presence of brefeldin A, a fungal drug known to affect protein traffic from the ER to the Golgi (Ritzenthaler et al., 2002; Saint-Jore et al., 2002), PMA4-GFP accumulated in the ER in protoplasts, leaf epidermal cells (data not shown), or BY2 cells (Figure 1H). In addition, in cells expressing transiently the dominant negative mutant AtRAB1b (N121I), known to inhibit the pathway from the ER to the Golgi (Batoko et al., 2000), PMA4-GFP also accumulated in the ER of tobacco leaf epidermal cells (data not shown), confirming that the protein was transported to the PM via the ER.
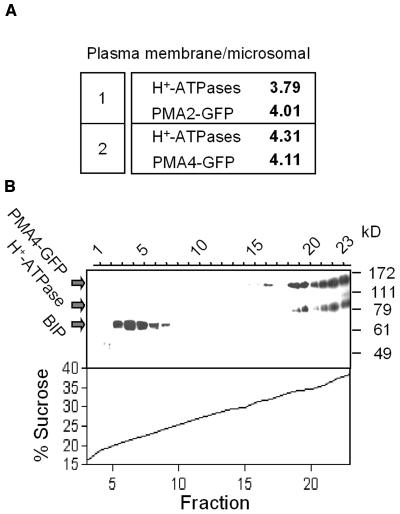
These microscopic data were further corroborated by analysis of PM-enriched subcellular fractions. Protein gel blotting showed that the levels of both PMA4-GFP and PMA2-GFP in the PM fraction were fourfold higher than those in the microsomal fraction, a similar result to that seen for the endogenous PM H+-ATPases (Figure 2A). Moreover, sucrose gradient centrifugation of a cellular homogenate, followed by protein gel blotting, revealed that the endogenous PM H+-ATPases and PMA4-GFP cofractionated in the heaviest fractions (Figure 2B).
Figure 2.
In Vitro Subcellular Localization of PMA4-GFP.
(A) A sample (30 μg of protein) of the microsomal fraction or the PM-enriched fraction from plants expressing PMA2-GFP (1) or PMA4-GFP (2) was electrophoresed, then subjected to protein gel blotting using anti-GFP or anti-PM H+-ATPase antibodies, followed by 125I protein A. The PM/microsomal fraction distribution ratio of the PMA4-GFP and PM H+-ATPase signals is indicated.
(B) A homogenate from plants expressing PMA4-GFP was centrifuged on a discontinuous 15 to 40% sucrose gradient (see Methods) and the fractions analyzed by protein gel blotting using successive antibodies against GFP, PM H+-ATPase, and BIP (used as an ER marker).
Taken together, these results demonstrate that the C-terminal tagging of different plant PM H+-ATPase isoforms with GFP did not affect the targeting of the protein to the PM and that these fusion proteins can be used to mark the PM.
PM H+-ATPase-GFP Fusion Proteins Are Functional
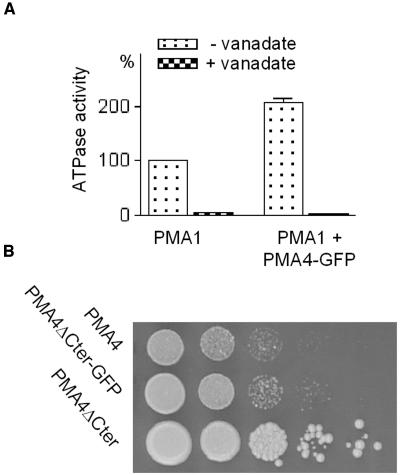
We then examined whether the GFP tag affected the folding of PM H+-ATPase by checking whether the chimeras were active. We employed a yeast expression system previously used to characterize plant PM H+-ATPases (de Kerchove d'Exaerde et al., 1995). PMA4 was chosen for this and subsequent characterization studies because it is the most widely expressed isoform in tobacco (Arango et al., 2003). PMA4-GFP was cloned into a multicopy yeast expression vector and introduced into a yeast strain (YAK2, de Kerchove d'Exaerde et al., 1995) in which the host PM H+-ATPase genes had been deleted, but which had been rendered viable by transformation with a plasmid bearing the main yeast PM H+-ATPase gene PMA1 under the control of a galactose-induced transcription promoter and the URA3 auxotrophic marker. When the ability of the plant PMA4-GFP to complement the absence of the yeast PM H+-ATPases was checked by counterselecting the yeast PMA1-bearing plasmid on 5-fluoroorotic acid–containing medium (conditions under which URA3 is toxic), the results showed that PMA4-GFP alone was unable to sustain yeast growth (data not shown). However, when the cells were shifted to glucose medium to prevent yeast PMA1 expression, the ATPase-specific activity of the microsomal fraction was twice as high in the presence of PMA4-GFP (Figure 3A), indicating that, although PMA4-GFP was unable to complement the yeast PM H+-ATPase deletion, it was still active, suggesting that GFP tagging reduced, but did not prevent, ATPase activity. To confirm this, we expressed in yeast a variant of the same fusion (PMA4ΔCter-GFP), in which the last 98 codons, corresponding to the ATPase autoinhibitory domain, were deleted; this is predicted to generate an activated form of PM H+-ATPase (Zhao et al., 2000). As shown in Figure 3B, PMA4ΔCter-GFP was able to complement the yeast PM H+-ATPase deletion, although to a lesser extent than the untagged form (PMA4ΔCter). We therefore concluded from the yeast expression data that GFP tagging resulted in substantial retention of ATPase activity and thus did not markedly interfere with PM H+-ATPase folding.
Figure 3.
Functional Expression of PMA4-GFP and PMA4ΔCter-GFP in Saccharomyces cerevisae.
(A) A null mutant strain of S. cerevisae relying on conditional expression of S. cerevisae PM H+-ATPase PMA1 was transformed with a plasmid bearing PMA4-GFP. After preculture in galactose medium, transformed and untransformed cells were transferred to glucose medium for 16 h to switch off PMA1 expression, then the microsomal fraction was prepared and an ATPase assay performed in the presence or absence of vanadate, a PM H+-ATPase inhibitor. The specific activity of membranes containing only residual PMA1 (0.043 μmol Pi × min−1 × mg−1 protein) was taken as 100%.
(B) Serial 10-fold dilutions of strains expressing the indicated PMA4-derived constructs after 5-fluoroorotic acid selection.
Subsets of PM H+-ATPase TM Spans Are Not Sufficient for PM Targeting
To determine which structural elements in PMA4 were involved in PM targeting, we first tested whether different combinations of TMs were able to direct GFP to the PM. We expressed the TMs as one or more pairs because several reports have suggested that the TMs of polytopic proteins, including the Neurospora crassa PM H+-ATPase, are integrated in pairs into the lipid bilayer of biological membranes (Skach and Lingappa, 1993, 1994; Lin and Addison, 1995). The following PMA4 regions were fused to GFP: the N-terminal region (Nter) together with TM1 and TM2 (NterTM1-2-GFP), Nter and TM1 to TM4 (including the cytosolic small loop) (NterTM1-4-GFP), TM3 and TM4 (TM3-4-GFP), and TM5 to TM10 (TM5-10-GFP) (Figure 4). In all constructs, the 30 amino acids on either side of the TM pair(s) were retained to try to avoid destabilizing the membrane insertion of the corresponding TM. The resulting fusions were transiently expressed in tobacco leaf protoplasts by electroporation or in tobacco leaf epidermal cells using A. tumefaciens infiltration.
Figure 4.
In Vivo and in Vitro Analysis of PM H+-ATPase TM-GFP Fusion Proteins Transiently Expressed in Protoplasts.
(A) to (G) N. tabacum protoplasts were transformed by electroporation with the indicated constructs. Below the images, the PM H+-ATPase is schematized, with the region fused to GFP in red. Cells were imaged with a Bio-Rad MRC-1024 confocal microscope using emission band-pass filters of 506 to 538 nm and 664 to 696 nm ([A] to [E]); the red fluorescence corresponds to chloroplasts. In (B), the arrow indicates a transvacuolar strand. Fluorescence was imaged with a Leica DRM microscope using a GFP filter ([F] and [G]). Bars = 10 μm.
(H) Microsomal (M) and soluble (S) fractions were prepared from protoplasts expressing NterTM1-2-GFP or GFP, then samples (50 μL) were electrophoresed and analyzed by protein gel blotting using anti-GFP antibodies.
(I) A homogenate prepared from protoplasts transiently expressing the indicated PM H+-ATPase-GFP fusion protein was electrophoresed and analyzed by protein gel blotting using anti-GFP antibodies.
In protoplasts transformed with NterTM1-2-GFP, confocal microscopic analysis performed 4 h (data not shown) or 24 h (Figure 4B) after electroporation showed GFP fluorescence in the cytoplasm, including transvacuolar strands (Figure 4B, arrow). Analysis of optical sections showed that no GFP fluorescence was seen in the nucleus, in contrast with in protoplasts expressing GFP alone (Figure 4A). This observation prompted us to analyze the membrane partitioning of NterTM1-2-GFP by protein gel blotting using anti-GFP antibodies. As shown in Figure 4H, in contrast with free GFP, which was found only in the soluble (S) fraction, the chimera was detected in both the S and microsomal (M) fractions, with a large part of the chimera being membrane bound. However, two bands other than the expected 45-kD band were detected in the soluble fraction, the smallest band being slightly larger than free GFP and half the size of the largest band. These bands could be, respectively, monomers and dimers of a degradation product of the chimera not integrated into membranes, and the fluorescence observed in the cytosol might therefore correspond to a fraction of the chimeras and to a GFP-containing degradation product.
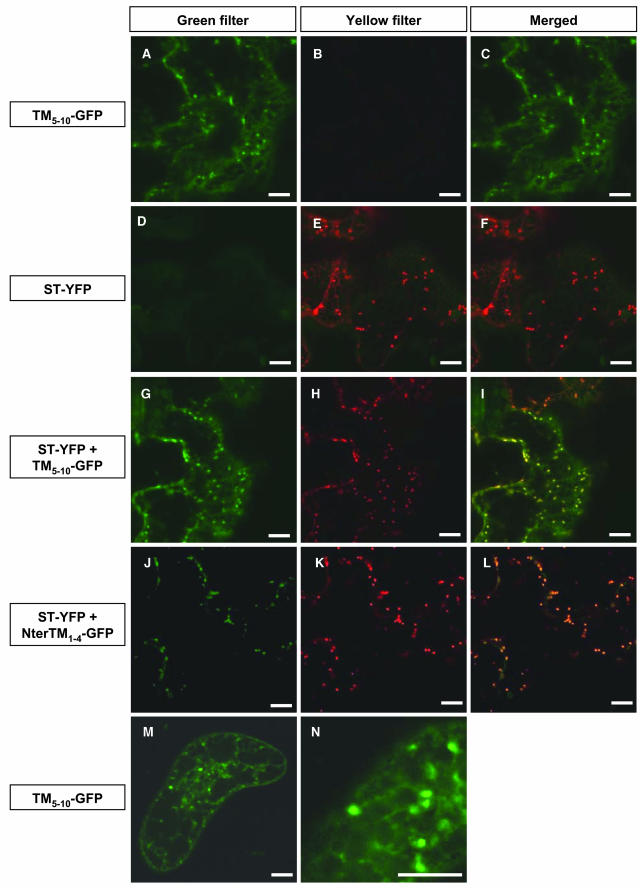
Confocal microscopic analysis of protoplasts expressing TM5-10-GFP showed GFP to be localized in a network and in mobile structures superimposed on this network (Figure 4C). The network was similar to that seen in protoplasts transformed with GFP-HDEL (Figure 4F), a GFP sequence fused to a signal peptide and the ER retrieval sequence HDEL, which can be used as an ER marker (Haseloff et al., 1997). By contrast, the mobile structures labeled by TM5-10-GFP resembled those labeled by GFP fused to truncated rat sialyl transferase (ST), commonly used as a Golgi marker in plant cells (Boevink et al., 1998; Batoko et al., 2000; Saint-Jore et al., 2002) (Figure 4G).
Using a leaf transient expression system, during the early stage of expression, generally 2 d after infiltration of the A. tumefaciens strain into the tobacco leaves, TM5-10-GFP fluorescence in epidermal cells also showed a dual distribution in a tubular network and mobile structures (Figure 5A). One day later, in some cells, only the mobile structures were labeled (data not shown). Leaf epidermal cells were also transformed with a plasmid encoding the Golgi marker, ST, fused to the yellow fluorescent protein (YFP), a spectral variant of GFP. During the early stage of expression, <48 h after infiltration, in addition to the mobile Golgi stacks, ST-YFP fluorescence was detected in most of the epidermal cells in a cytoplasmic network (data not shown), identified as the ER and representing fusion proteins not yet translocated to the Golgi (Boevink et al., 1998). One day later, the distribution of ST-YFP mainly shifted to mobile spots (Figure 5E), although both ER and Golgi labeling still could be observed in some cells (see the cell on the left in Figure 5E). In cells in which ST-YFP and TM5-10-GFP were coexpressed, the two fusion proteins showed a similar distribution at either 2 or 3 d after infiltration, as demonstrated by the merging of the green and yellow channels (Figure 5I). This showed that TM5-10-GFP was located in the same membrane compartments as ST-YFP, which corresponded mainly to the ER at 2 d after infiltration and mainly to the Golgi at 3 d after infiltration and that, in contrast with PMA4-GFP, traffic of TM5-10-GFP and ST-YFP from the ER to the Golgi was slow. To confirm this, we generated transgenic BY2 cell lines stably expressing TM5-10-GFP and observed labeling of a tubular network and a punctuate structure but not the PM (Figures 5M and 5N), suggesting that the accumulation in the endomembrane system seen with the transient expression system was not an intermediate step in a slow PM targeting process.
Figure 5.
Colocalization of PM H+-ATPase TM-GFP Chimeras and ST-YFP.
(A) to (L) Tobacco leaves were transiently transformed by A. tumefaciens containing the indicated constructs, then the lower epidermis was observed using a Zeiss LSM 510 confocal microscope. In the left panels (green filter), GFP was excited with a 458 laser from the argon line and the emission detected using a 475/525 band-pass filter; in the center panels (yellow filter), YFP was simultaneously excited with a 514 laser from the same argon line, and the resulting emission detected using a 535/590 band-pass filter; and in the right panels (merged), the left and central panels are superimposed. The microscope settings and postcapture processing were identical for all pictures, except for (J) to (L), in which the brightness was increased. Images were taken at 2 d ([A] to [I]) or 3 d ([J] to [L]) after A. tumefaciens infiltration of leaves.
(M) and (N) A BY2 cell stably expressing TM5-10-GFP was analyzed by a Bio-Rad MRC-1024 confocal microscope using an emission band-pass filter of 506 to 538 nm. Bars = 10 μm.
In protoplasts transformed with TM3-4-GFP or NterTM1-4-GFP, the fluorescence distribution (Figures 4D and 4E) could not be unambiguously assigned to membrane structures that have been morphologically described in plant cells. It is possible that these unidentified membrane compartments labeled by TM3-4-GFP or NterTM1-4-GFP represent remnant or modified well-characterized compartments that are disturbed by the expression of TM3-4-GFP or NterTM1-4-GFP. Protoplasts recovering from wall lysis treatment might have a less efficient cellular metabolism and might thus be more susceptible to subcellular perturbation. Indeed, analysis of epidermal cells coexpressing ST-YFP and either TM3-4-GFP (data not shown) or NterTM1-4-GFP (Figures 5J to 5L) showed that both PMA4-derived fusion proteins were located first in the ER, then in the Golgi, similarly to TM5-10-GFP.
Protein gel blotting analysis of the expression of the various TM pair-GFP chimeras in protoplasts using anti-GFP antibodies revealed antigens of the expected sizes (Figure 4I). Although the data shown in Figure 4I cannot be quantitatively compared because they were obtained using different protoplast preparations, the amounts of TM3-4-GFP and NterTM1-4-GFP expressed did not seem to be lower than for the other chimeras, whereas the fluorescent emission from these chimeras was much lower than that seen with TM5-10-GFP, suggesting an effect of these fusions on the formation or maturation of the GFP chromophore.
Finally, we also attempted to express, in protoplasts and leaves, a GFP fusion protein containing all 10 TMs but none of the cytosolic domains. However, no GFP was detectable either by microscopy or protein gel blotting, suggesting that this fusion protein was unstable and rapidly degraded.
The N-Terminal Region, Small Loop, and C-Terminal Region Are Not Required for PM Targeting
Because the TMs pair(s) allowed traffic to the Golgi, but not to the PM, we suspected that cytosolic domains of the full-length protein might be necessary. We addressed this possibility by searching for essential, rather than sufficient, determinants by individually deleting the N- or C-terminal region or replacing each of the internal cytosolic regions of PMA4 with a 5-Gly bridge, while retaining all 10 PMA4 membrane spans. Ten flanking amino acids were retained between the deleted domain and the adjacent TM.
Confocal microscopic analysis of protoplasts expressing PMA4-GFP in which the N-terminal region (ΔNter-GFP), small loop (ΔSL-GFP), or C-terminal region (ΔCter-GFP) was deleted showed that, like full-length PMA4-GFP (Figure 6A), these chimeras were localized in the PM (Figures 6B to 6D). However, the fluorescence signal from these three truncated PMA4-GFPs was reduced compared with that from PMA4-GFP. Protein gel blotting analysis of protoplast homogenates (Figure 6I) showed that the molecular masses of the chimeras were those expected but that the amount expressed was reduced compared with that seen using PMA4-GFP, suggesting that the decreased fluorescence seen using identical microscope settings was because of a lower amount of protein.
Chimeras resulting from double deletion of the small loop and either the N-terminal region (ΔNterΔSL-GFP) or the C-terminal region (ΔSLΔCter-GFP) were also localized to the PM (data not shown). However, no expression of the N-terminal/C-terminal double deletion chimera (ΔNterΔCter-GFP) or the triple deletion chimera (ΔNterΔSLΔCter-GFP) was detectable by microscopy or protein gel blotting.
The Nucleotide Binding Domain from the Large Loop Is Essential for PM Targeting
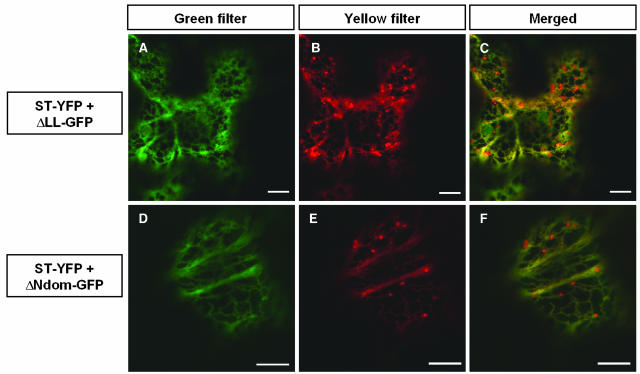
In contrast with the above deletions, removal of the large loop (ΔLL-GFP) prevented PM localization. Using transient expression in either protoplasts (Figure 6E) or epidermal cells (Figure 7A), ΔLL-GFP fluorescence was seen in protoplasts 24 h after electroporation and in leaves 2 or 3 d after infiltration in both cases in a cytoplasmic network morphologically similar to the ER labeled in GFP-HDEL–expressing cells. To show that this ER accumulation was not transient localization because of slow traffic, we produced BY2 cell lines stably expressing ΔLL-GFP and found that fluorescence was again restricted to the ER (Figure 6F). The absence of ΔLL-GFP in the PM seen in vivo in transgenic BY2 cells was supported by protein gel blotting results, which showed that the amounts of both ΔLL-GFP and binding protein (BIP), an ER resident chaperone, in the PM-enriched fractions were fivefold lower than those in the microsomal fraction, whereas in a cell line expressing PMA4-GFP, the amount of PMA4-GFP in the PM was fivefold higher than that in the microsomal fraction, a ratio similar to that seen for the endogenous PM H+-ATPases (data not shown).
Figure 7.
Colocalization of ST-YFP and Either ΔLL-GFP or ΔNdom-GFP Transiently Coexpressed in Leaves.
Tobacco leaves were transiently transformed by A. tumefaciens containing the indicated constructs; abaxial epidermal cells were imaged 2 d after infection using a Zeiss LSM 510 confocal microscope under the same settings as in Figure 5. In the left (green filter) and central (yellow filter) panels, GFP and YFP imaging parameters were as described in Figure 5; in the right panel (merged), the left and center panels are superimposed. The microscope and image settings were identical for all images. Bars = 10 μm.
Because these data suggested that the large loop was required for PM targeting, we wondered whether it was also sufficient. We therefore tested its ability to target different TM combinations to the PM. However, no GFP was detected in protoplasts bearing constructs containing the large loop fused to TM3 plus TM4, TM5-TM10, TM3-TM10, or all 10 TMs both in vivo or by protein gel blotting using anti-GFP or anti-PM H+-ATPase antibodies.
To analyze the large loop in more detail, we relied on the three-dimensional (3D) structure of another P-type ATPase, a rabbit Ca2+-ATPase (Toyoshima et al., 2000). In this structure, the large loop is divided into two domains, one of which, the phosphorylation (P) domain, lies close to the membrane, whereas the second, the nucleotide binding (N) domain, is more or less isolated from the rest of the enzyme and is connected to it by a narrow linking region, which is conserved in all P-type ATPases (Figure 6G). A similar structure has been modeled for the N. crassa PM H+-ATPase (Kuhlbrandt et al., 2002). We therefore replaced the N domain (N340-P491 of PMA4) with a bridge consisting of a single Gly and Asn residue, a combination of amino acids frequently found in loops (Chou and Fasman, 1978), resulting in ΔNdom-GFP. The Gly and Asn residues used to bridge Asn340 to Pro491 were predicted from the Ca2+-ATPase structure to maintain the distance seen between Asn340 and Pro491 in the native protein. In leaf epidermal cells (Figure 7D) and in protoplasts (Figure 6H) expressing ΔNdom-GFP, the fluorescence was localized in a cytoplasmic network, suggesting an ER localization. To clarify the localization of ΔLL-GFP and ΔNdom-GFP, these constructs were coexpressed with ST-YFP in epidermal cells, and both ΔLL-GFP and ΔNdom-GFP were found to colocalize with ST-YFP present in the tubular network but not with that in the mobile Golgi stacks (Figures 7A to 7F), suggesting that both ΔLL-GFP and ΔNdom-GFP were localized to the ER.
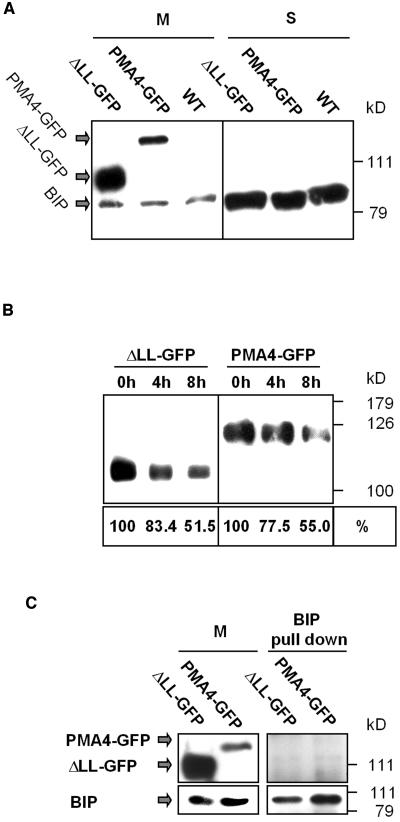
The hypothesis that ΔLL-GFP might be retained in the ER by the quality control system is unlikely for several reasons. First, in plant protoplasts or epidermal cells, ΔLL-GFP was stable, and this was confirmed in transgenic BY2 cell lines. In fact, in some transgenic BY2 lines, the amount of ΔLL-GFP detected by protein gel blotting was even higher than the amount of PMA4-GFP (Figure 8A, M). Stability of ΔLL-GFP could possibly be further addressed by pulse-chase experiments. However, we did not succeed in immunoprecipitating the GFP chimeras using our antibodies. A possible reason is that the latter did not recognize the native form of the protein. We therefore addressed the question by another approach. When stably transformed BY2 cells were treated with cycloheximide, a protein translation inhibitor, and the amounts of protein measured by protein gel blotting 4 or 8 h later, the rate of decrease in the amounts of ΔLL-GFP was no greater than that of PMA4-GFP (Figure 8B). Secondly, the ER chaperone BIP was shown to be overexpressed when a modified GFP was retained by the quality control system (Brandizzi et al., 2003). However, no increase in amounts of BIP (Figure 8A) or of another chaperone, calreticulin (data not shown), was detected by protein gel blotting in M or S fractions of BY2 cell lines expressing ΔLL-GFP or PMA4-GFP compared with the nontransformed cells. Finally, we checked whether BIP was associated with ΔLL-GFP. Microsomal membrane proteins from BY2 cells expressing either ΔLL-GFP or PMA4-GFP were solubilized with dodecyl maltoside. This detergent was previously shown to keep the native conformation of N. plumbaginifolia PM H+-ATPases and their interaction with regulatory 14-3-3 proteins (Maudoux et al., 2000). Using anti-BIP antibodies, we pulled down BIP from the solubilized fraction, but no GFP fusion was found to copurify with BIP even after overexposition (Figure 8C). We can therefore conclude that ΔLL-GFP does not seem to be retained in the ER by the quality control system.
Figure 8.
BIP Expression and Chimera Stability Analysis.
(A) Microsomal (M) and soluble (S) fractions prepared from BY2 cells stably expressing PMA4-GFP or ΔLL-GFP or nontransformed BY2 cells (WT) were electrophoresed and analyzed by protein gel blotting using successive antibodies against BIP and GFP.
(B) BY2 cell lines stably expressing PMA4-GFP or ΔLL-GFP were treated with 500 μM cycloheximide for 0, 4, or 8 h, and then microsomal fractions were prepared, electrophoresed, and analyzed by protein gel blotting using anti-GFP antibodies. Below is indicated the protein amount (0 h taken as 100%) obtained in a parallel experiment using anti-GFP antibodies followed by 125I protein A.
(C) Proteins from a microsomal fraction (M) of BY2 cells expressing ΔLL-GFP and PMA4-GFP were solubilized in dodecyl maltoside and recovered in the supernatant after centrifugation. Anti-BIP antibodies and then agarose-protein A were added (see Methods) to supernatant. Twenty micrograms of M and the totality of pull-down proteins were electrophoresed and analyzed by protein gel blotting using successive antibodies against BIP and GFP. The top right panel was overexposed compared with the other panels to show the background level.
LL and Ndom, but Not Nter, SL, and Cter, Are Essential for PM Targeting of Plant H+-ATPase in Yeast
Because the yeast PM H+-ATPase has been shown to oligomerize (reviewed in Lefebvre et al., 2003), it could be argued that the truncated PMA4 chimeras ΔNter-GFP, ΔSL-GFP, and ΔCter-GFP that localized to the PM actually bound to the endogenous PM H+-ATPase and were carried by it into the PM, thus masking the absence of targeting motifs in the three cytosolic domains. To rule out this hypothesis, we expressed these chimeras in yeast. The plant PM H+-ATPases were indeed shown to be targeted to the yeast PM (Figure 6J), although a fraction of the protein also accumulated in karmellae, internal membranes around the nucleus (as already shown by de Kerchove d'Exaerde et al., 1995). This yeast expression system is convenient because, given the low sequence identity between plant and yeast PM H+-ATPases (29%), it is unlikely that they will interact. This is supported by the observation that 6-His tagged Arabidopsis thaliana PM H+-ATPase expressed in yeast can be purified uncontaminated by the yeast PM H+-ATPases (Jahn et al., 2001). In yeast, like in plants, ΔLL-GFP (Figures 6M and 6N) and ΔNdom-GFP (Figures 6O and 6P) failed to reach the PM and were found in the cytoplasm. However, these GFP fusions seem to localize to the vacuole, whereas they were found in the ER in plants. By contrast, similar amounts of ΔNter-GFP (Figure 6K), ΔSL-GFP (Figure 6L), ΔCter-GFP (data not shown), and PMA4-GFP (Figure 6J) were detected in the PM, showing that the truncated chimeras were targeted to the PM without any assistance from the wild-type plant PM H+-ATPase and confirming that the Nter, SL, and Cter regions were not necessary for targeting to the PM.
DISCUSSION
Localization of H+-ATPases to the PM
We used GFP as a reporter to follow the subcellular localization of PM H+-ATPases and the deletion mutants. Expression of the full-length chimera in yeast showed that the fusion protein was still active, although the presence of GFP reduced the activity to some extent. The reporter system was therefore validated.
GFP had previously been successfully used to define the localization of AHA2, an Arabidopsis PM H+-ATPase closely related to the N. plumbaginifolia PMA4 used in this work (Jin et al., 2001; Kim et al., 2001). In this study, the PM localization of PMA4 was confirmed by in vitro subcellular fractionation. We also extended our observations to PMA2 and PMA3, members of PM H+-ATPase subfamily I, which diverged early in plant evolution from subfamily II containing PMA4 and AHA2 (Arango et al., 2003). PMA2-GFP and PMA3-GFP, like PMA4-GFP, were found only in the PM. The exclusive PM localization of PM H+-ATPases has been questioned in the past (Hager et al., 1991; Wada et al., 1994; DeWitt et al., 1996). This question is not trivial, because within the related plant Ca2+-ATPase family that is classified as a PM-type according to the animal classification, some isoforms localize to the PM, whereas others are found in endomembranes (Baxter et al., 2003). Therefore, we now have experimental data supporting the exclusive localization in the PM of the two major plant PM H+-ATPase families. This does not exclude the possibility that under some circumstances these enzymes remain in, or are internalized in, endomembranes.
TM Spans Alone Are Not Sufficient for PM Targeting
It has always been accepted, but never demonstrated, that the plant PM H+-ATPase follows the secretory pathway to reach the PM. In this study, we showed that, in the presence of brefeldin A or the dominant negative mutant AtRab1 (N121I), the enzyme was localized in the ER membrane. This was not the case for the direct traffic of αTIP from the ER to the storage vacuole, bypassing the Golgi (Park et al., 2004). In addition, the yeast PM H+-ATPase has been shown to traffic through the Golgi. We can therefore conclude that the plant PM H+-ATPase is also most likely transported to the PM through the Golgi.
Expression in N. tabacum protoplasts and leaf epidermal cells of various N. plumbaginifolia PMA4 TMs fused to GFP led to the conclusion that sets of TM pairs (TM3-4, NterTM1-4, or TM5-10) were not able to direct the fusion proteins to the PM, leaving it associated with internal membranes. These in vivo results confirm earlier observations in vitro (Skach and Lingappa, 1993, 1994; Lin and Addison, 1995) that pairs of TM spans of a polytopic protein can be inserted independently of other TMs in the ER lipid bilayer, probably through the signal recognition particle mechanism. The NterTM1-2 pair was an exception, in that a fraction of the protein was cytosolic. Similar results were obtained using the fusion protein containing the NterTM1-2 region from PMA2 (data not shown). These results are in agreement with the observation that a truncated form of Arabidopsis AHA2 containing TM1 and TM2 fused to the reporter β-glucuronidase also failed to reach the PM and was found partly in the soluble fraction and partly associated with membranes (DeWitt et al., 1996). Analysis of the first two TMs predicts a shorter length and lower hydrophobicity than for TM3 and TM4. This may explain why the former pair did not completely integrate into membranes and suggests that membrane integration of the native protein might be achieved only after synthesis of the third and fourth TMs, as shown for the chimera NterTM1-4-GFP. Alternatively, NterTM1-2-GFP could be partly retrotranslocated to the cytosol by the quality control system.
In both a transient and a stable expression system, TM3-4-GFP, NterTM1-4-GFP, and TM5-10-GFP were detected in the ER and/or the Golgi. The Golgi seemed to be the final destination for these TM combinations, with no GFP fluorescence being detected in downstream compartments, such as the prevacuole, vacuole, or PM. Retention of Golgi resident proteins could be triggered either by a targeting sequence usually located in the cytoplasmic tail and involved in recycling (between Golgi and vacuoles or between the different Golgi cisternae) or by their TM (reviewed in Saint-Jore-Dupas et al., 2004). In the latter case, the TM length might be the determinant for Golgi retention or exit. Predicted lengths of the PMA-GFP chimera TMs are compatible with trafficking to the PM. However, this prediction has to be considered with caution because several TM might be tilted compared with the lipid bilayer plane. This is the case for the majority of Ca2+-ATPase TMs (Toyoshima et al., 2000; Toyoshima and Nomura, 2002). Cys and His residues in TMs were shown to play a role in the retention of an animal Golgi resident protein (Aoki et al., 1992). Only PMA4 TM3 possesses two Cys, but no His is found in the chimeras TMs. In another study, a stretch of four Phe in the single TM was shown to make a small contribution to Golgi retention (Munro, 1995). Although there are several Phe in TM5-10-GFP, there is only one in TM3-4-GFP. Thus, these various features characterizing Golgi retention, based only on the analysis of single-span proteins that represent the large majority of the Golgi proteins, does not seem to explain the localization of PMA4-GFP chimeras, suggesting that Golgi retention of polytopic protein possibly follows a different model. In conclusion, these results suggest that at least all of the 10 TMs are required for efficient Golgi exit of the PM H+-ATPase and its further targeting to the PM.
Cytosolic Domain Deletions
In plant protoplasts, the single deletions, ΔNter, ΔSL, and ΔCter, and the double deletions, ΔNterΔSL and ΔSLΔCter, did not affect PM H+-ATPase targeting to the PM, except that the fluorescent and protein gel blotting signals were weaker compared with the full-length chimera. These results could be explained by lower stability and higher turnover of these chimeras. PM localization was also obtained when these constructs were expressed in yeast. We can therefore conclude that the PM targeting of these truncated PM H+-ATPases in plants is unlikely to be mediated in trans by an associated wild-type PM H+-ATPase.
Like other PM H+-ATPases, the Cter region of PMA4 contains a regulatory domain (Dambly and Boutry, 2001). Its sequence is poorly conserved between plant and yeast PM H+-ATPases, suggesting that this region was acquired recently during evolution and, therefore, that it does not play a role in targeting to the PM. This was confirmed by the stable PM expression in transgenic plants of PMA4 lacking the Cter region (Zhao et al., 2000). The Nter region and the small loop make up the A domain, which is well separated from the rest of the protein in the 3D structure of the closely related Ca2+-ATPase (Figure 6G). The small loop is essential for catalysis in yeast PM H+-ATPase (Wang et al., 1996) and human Ca2+-ATPase (Clarke et al., 1990), but its role is still debated. Our results show that it does not play a major role in targeting and, therefore, that PM H+-ATPase does not have to be functional to be targeted to the PM. In the 3D structure, the A domain is quite separate from the rest of the molecule, and its deletion is therefore not expected to markedly modify the structure of the rest of the enzyme.
In contrast with the other three cytosolic regions, deletion of the large loop resulted in retention of PMA4-GFP in the ER. Deletion of the entire large loop might be expected to drastically affect the whole structure because it constitutes a central region with connections to the other PM H+-ATPase domains. It is therefore difficult to separate two possible effects, namely the loss of a specific sequence involved in targeting to the PM (e.g., by interacting with the trafficking machinery) or a change in the overall structure, resulting in the modification of targeting sequences outside the large loop. For instance, in yeast, several point mutations in various positions in the PM H+-ATPase large loop result in misfolding, thus preventing targeting to the PM (Morsomme et al., 2000b), suggesting that the 3D structure plays a role in targeting.
To more precisely define regions possibly involved in PM targeting, we deleted the nucleotide binding domain (N domain in Figure 6G), which is joined to the P domain by a narrow bridge consisting of two extended sequences, and found that the resulting GFP fusion protein was also localized to the ER. Given the remote position of the N domain, its deletion is less likely to have a marked structural effect on the rest of the enzyme than deletion of the entire large loop, raising the question of how to interpret the failure of PM targeting. Considering the absence of ΔLL-GFP and ΔNdom-GFP in downstream membrane compartments of the secretory pathway and the highly dynamic nature of the Golgi, an active retention mechanism may be responsible for their ER-specific localization. Some yeast PM H+-ATPase mutants (Wang and Chang, 2002) or mutants of other polytopic proteins, such as the yeast PDR5 (Plemper et al., 1998) or the human CFTR (Jensen et al., 1995), are retained in the ER and degraded via the proteasome. Such a degradation process might occur in the case of the construct containing only the 10 TMs, the double and triple deletions (ΔNterCter-GFP and ΔNterSLCter-GFP), and those in which the large loop was fused to TM3 plus TM4, TM5 to TM10, TM3 to TM10, or all 10 TMs, as no protein could be detected in transformed cells. By contrast, ΔLL-GFP or ΔNdom-GFP were stable because their protein levels, determined by fluorescence or protein gel blotting, and their turnover were not different from those of full-length PMA4-GFP, arguing against intervention of the quality control system. Moreover, no overexpression of two ER chaperones was detected in a BY2 cell line stably expressing ΔLL-GFP. Finally, no ΔLL-GFP copurified when BIP was pulled down with anti-BIP antibodies, although we cannot exclude the possibility that the detergent used for solubilization modifies the protein–protein interactions. Another hypothesis that a hidden ER or Golgi retention signal became fortuitously uncovered is highly unlikely because this would have had to occur independently in several of the chimeras used (TM3-4, NterTM1-4, TM5-10, ΔLL, or ΔNdom), some of which have nothing in common except the GFP tag. Alternatively, ER retention of these chimeras may result from the deletion or modification of an ER export signal, together with inefficient bulk flow (if this actually exists) to the Golgi (Sevier et al., 2000; Ma et al., 2001; Votsmeier and Gallwitz, 2001; Epping and Moye-Rowley, 2002; Malkus et al., 2002). Based on the ER retention of ΔLL-GFP and ΔNdom-GFP, we can propose that the cytosolic N domain contains an ER export motif. In this case, ER export of the chimeras TM3-4, NterTM1-4, and TM5-10 might be because of slow leakage of these chimeras to the Golgi.
One more point must be taken into account. The yeast PM H+-ATPase oligomerizes during the early step of the secretory pathway (Bagnat et al., 2001; Lee et al., 2002; Wang and Chang, 2002), and this seems to be essential for PM targeting. If this also holds for plant PM H+-ATPases, one hypothesis could be that deletion of the N domain affects oligomerization and therefore prevents exit from the ER. Whatever the reason(s) for the ER retention, the PM H+-ATPase-GFP chimeras are an interesting tool for studying the poorly characterized mechanism of stable ER accumulation.
While in the plant, ΔLL-GFP and ΔNdom-GFP were found in the ER; in yeast, these chimeras were found in the vacuole. Several yeast PMA1 mutants were found to be misdirected to the vacuole (Bagnat et al., 2001; Gong and Chang, 2001; Wang and Chang, 2002). This observation indicates that, although the plant and yeast trafficking machineries share common properties, they also differ in some aspects.
The Default Compartment for Polytopic Proteins in the Secretory Pathway
The default final destination of polytopic proteins is far from clear, although in plant cells, it has been proposed to be the PM (Vitale and Raikhel, 1999). The observation that several membrane-bound, truncated PMA4 forms fused to GFP failed to reach the PM and were found in endomembrane compartments, but never in the vacuolar membrane, strongly suggests that neither the PM nor the vacuolar membrane is the default compartment, at least for this polytopic protein, and that one or several signals are required for sorting through the secretory pathway. This situation would be similar to that in animal cells in which ER and Golgi export motifs have been found for a K+ channel (Ma et al., 2001; Stockklausner and Klocker, 2003), which led to the hypothesis that a default pathway might not exist for membrane proteins (Cereijido et al., 2003).
Efficiency of Traffic through the Secretory Pathway
All the full-length PMA-GFP fusions were detected in the PM after a short time interval. This occurred only a few hours after electroporation in protoplasts. A longer interval of 2 d was needed using A. tumefaciens–mediated transformation of tobacco leaves, but as suggested using the soluble GFP control, this discrepancy in detection timing is likely because of the biological transformation process. We have never detected transient accumulation of PMA-GFP in the secretory pathway. By contrast, it appears that the traffic of the chimeras TM3-4-GFP, NterTM1-4-GFP, and TM5-10-GFP to the Golgi was slow, allowing them to be transiently detected in the ER on the way to the Golgi, as seen with the Golgi marker ST-GFP (Figure 5). These contrasting results might reflect the coexistence of two types of traffic from the ER to the Golgi, one being a slow traffic process, which could be passive (either through vesicles or by direct connections between organelles), and the other a fast traffic process, which could be active and selective. Fast traffic is expected for enzymes that are required quickly in their final destination for physiological needs or that can be toxic if allowed to accumulate in intermediate compartments during their traffic. Most of the evidence for slow traffic through the secretory pathway was obtained using heterologous or modified proteins, such as ST (a truncated rat protein) or modified TM domains (Brandizzi et al., 2002b), all of which were fused to GFP, and their slow mobility could result from the absence of a signal necessary for efficient traffic. The fast moving PM H+-ATPase is not unique because an ABC transporter has shown by in situ immunodecoration to be present in the PM as early as 18 h after induction of its expression (Jasinski et al., 2001).
In conclusion, the data reported here support the idea that the PM or vacuolar membrane is not the default compartment for a plant polytopic protein, such as PM H+-ATPase. The Nter, small loop, and Cter regions play no part in targeting to the PM because they can be deleted without mistargeting and with no loss of traffic rate. The membrane spans are not sufficient for targeting to the PM, so their length and/or hydrophobicity cannot be the sole targeting determinant, in contrast with the situation for single-span membrane proteins (Masibay et al., 1993; Munro, 1995; Pedrazzini et al., 1996; Rayner and Pelham, 1997; Watson and Pessin, 2001; Brandizzi et al., 2002b). Finally, the nucleotide binding domain within the large loop seems to be essential for correct targeting and should be the focus of further studies.
METHODS
Plasmids and DNA Constructs
All constructs for transient protoplast transformation were prepared in the cloning vector pTZ19U (Stratagene, La Jolla, CA). The expression vector, pTZ19U-gfp (Duby et al., 2001), contains the PMA4-En50 transcriptional promoter (Zhao et al., 1999), the PMA4 5′ untranslated region, BglII, BamHI, and KpnI restriction sites, mGFP4 S65T (Heim et al., 1994; Haseloff et al., 1997), and the nopaline synthase transcription terminator. cDNAs coding for the Nicotiana plumbaginifolia PM H+-ATPase isoforms PMA2 (Boutry et al., 1989), PMA3 (Boutry et al., 1989), and PMA4 (Moriau et al., 1993) were amplified by PCR to generate a fragment containing appropriate cloning sites at each end. For PMA2, the primer 5′-GGGGTACCGGATCCAACAGTGTATGATTGCTG-3′ was used to remove the stop codon and add a BamHI restriction site (underlined) at the end of the coding sequence from a PMA2 already preceded by a BamHI site (de Kerchove d'Exaerde et al., 1995). Modified PMA2 cDNA was digested with BamHI and cloned into pTZ19U-gfp opened with BglII and BamHI to generate plasmid pma2-gfp. For PMA4, the primers 5′-CGGGATCCAGATCTGAGATGGCAAAAGCTATCAGCC-3′ and 5′-AGCGGAAGCTTGGTACCGGATCCAACTGTATAATGCTGCTGG-3′ (pma4-3′) were used, and the recovered PCR product cloned into pTZ19U-gfp was opened with BglII and BamHI (underlined) to generate plasmid pma4-gfp. For PMA3, primers 5′-GGACTCAGATTAGCGTGAACAGACAAACC-3′ and 5′-CGGGATCCAACGGTGTACGCCTGCTG-3′ were used, and the PCR product was digested with XbaI/BamHI (underlined) and cloned into pTZ19U-gfp digested with BglII/BamHI after filling in of the incompatible XbaI and BglII ends, resulting in plasmid pma3-gfp.
For pma4-gfp and each derived construct, the translation ATG context was optimized (Lukaszewicz et al., 2000). Primers 5′-GGAAGATCTAAAATGGCAAAAGCTATCAGC-3′ (1) and 5′-AGCGGGGTACCCCCTGGAACCAGAATAGC-3′ were used to amplify PMA4 from nucleotide 1 to 462, and the PCR product was cloned into pTZ19U-gfp opened with BglII and KpnI (underlined) to generate plasmid NterTM1-2-gfp. Primers 5′-GGAAGATCTAAAATGGCTGTAGTTATTGCTACTGG-3′ and 5′-AAGCGGGGTACCGTCACTACAAAGCACATC-3′ (2) were used to amplify PMA4 from nucleotide 634 to 996, and the PCR product was cloned into pTZ19U-gfp opened with BglII and KpnI (underlined) to generate plasmid TM3-4-gfp. Primers 5′-CGCGGATCCAAAATGGCTGACATCGTGCTCACTGAACC-3′ and 5′-CGCGGATCCTTCTTTGCCATAGTCTTTCTT-3′ were used to amplify PMA4 from nucleotide 1858 to 2702, and the PCR product was cloned into pTZ19U-gfp opened with BglII and BamHI (underlined) to generate plasmid TM5-10-gfp. Primers (1) and (2) were used to amplify PMA4 from nucleotide 1 to 996, and the PCR product was cloned into pTZ19U-gfp opened with BglII and KpnI to generate plasmid NterTM1-4-gfp. For deletion of the small loop, the two PCR products obtained using primers (1) plus 5′-ACCTCCGCCTCCACCAGCTGCAGCAGCATT-3′ or 5′-GGTGGAGGCGGAGGTGACAGCACCACCAAT-3′ plus (pma4-3′) (overlapping regions between the two primers are underlined) were combined and amplified, and the final product used to replace the region corresponding to nucleotides 379 to 690 of PMA4 by five Gly codons. The final PCR product was cloned into pTZ19U-gfp opened with BglII and KpnI to generate plasmid ΔSL-gfp. Similarly, to obtain plasmid ΔLL-gfp, primers 5′-GCCTCCACCACCGCCGATGGCACCTTGCTG-3′ and 5′-ATCGGCGGTGGTGGAGGCAGTAGAGCTATTTTCCAGAGG-3′ (overlapping regions between the two primers are underlined) were used to replace the region corresponding to nucleotides 739 to 1911 of PMA4 by five Gly codons. For deletion of the N domain, primers 5′-GCGGGGTACCCTTACATTGGGAAACCCCAGGCATGATAGTGCT-3′ (the KpnI restriction site is underlined) and pma4-3′ were used to replace the region corresponding to nucleotides 1018 and 1473 of PMA4 by a sequence coding for Gly-Asn (bold nucleotides). The PCR product was cloned into pma4-gfp opened with KpnI, resulting in plasmid ΔNdom-gfp. To obtain plasmid ΔNter-gfp, primer 5′-GGAAGATCTAAAAATGGCTAAAATACTCAAGTTCCTTGGG-3′ (BglII restriction site underlined) was used to amplify PMA4 from nucleotide 178. To obtain plasmid ΔCter-gfp, primer 5′-TTCGCCGGATCCATTCCAAGCCTTTCC-3′ (BamHI restriction site underlined) was used to amplify PMA4 up to nucleotide 2562. All plasmids were verified by sequencing. Double or triple deletions were obtained by combining simple deletions by restriction and ligation.
Protoplast Isolation and Transient and Stable Expression Methods
Protoplasts were prepared from N. tabacum leaves and transiently transformed by electroporation as described by Lukaszewicz et al. (1998) using 6.45 pmol of each of the appropriate plasmids and incubated for 24 h in the dark at 25°C before analysis.
Vectors for stable transformation or transient expression in leaf epidermal cells were obtained by replacing the gusA gene in pBi101 (Clontech, Palo Alto, CA) with the various GFP expression cassettes cut from pTz19U (HindIII/SacI). The resulting plasmids were mobilized in Agrobacterium tumefaciens (strain C58 GV301; Koncz and Schell, 1986) by electroporation. Stable transformation of N. tabacum SR1 (Maliga et al., 1973) was performed as described previously (Horsch et al., 1986). Stable transformation of N. tabacum BY2 cells (Nagata et al., 1992) was performed as described previously (Grec et al., 2003).
Transient expression in tobacco leaf epidermal cells was achieved by infiltrating greenhouse-maintained N. tabacum leaves at the abaxial surface with a suspension (OD600 = 0.3) of A. tumefaciens (Batoko et al., 2000).
Subcellular Fractionation and Protein Analysis
Transformed protoplasts (1 to 1.5 × 106) were collected by centrifugation at 100g for 5 min. For whole-cell protein gel blotting, they were resuspended in 150 μL of solubilization buffer (80 mM Tris-HCl, pH 6.8, 2% SDS, and 10% glycerol). To prepare the microsomal fraction, the protoplasts were resuspended in 800 μL of 20 mM KH2PO4, pH 7.8, 5 mM MgCl2, 330 mM sucrose, 5 mM β-mercaptoethanol, and protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 μg/mL of each of leupeptin, aprotinin, antipain, chymostatin, and pepstatin; Sigma; St. Louis, MO); all subsequent steps were at 4°C. The suspension was ground with 500 mg of glass beads (0.8 mm diameter) in an MSK cell homogenizer (Braun Biotech, Melsungen, Germany). The homogenate was centrifuged at 5000g for 5 min, and the resulting supernatant centrifuged again at 20,800g for 45 min. The pellet (microsomal fraction) was resuspended in 100 μL of solubilization buffer, and the supernatant (soluble fraction) was made to 10% with trichloroacetic acid and kept overnight on ice and then centrifuged at 20,800g for 20 min. The pellet was washed twice in ether-ethanol (1:1) and once in ether, vacuum dried, and resuspended in 100 μL of solubilization buffer.
PM fractions from transgenic plant leaves or BY2 cell lines were prepared as described by Zhao et al. (2000) and Jasinski et al. (2001).
For sucrose gradient subcellular fractionation, a leaf disc (∼6 cm2) from a transgenic plant was homogenized for 3 × 30 s at 4°C with 150 μL of glass beads (0.8 mm diameter) and 300 μL of 100 mM Tris-HCl, pH 8.5, 250 mM sucrose, 20 mM β-mercaptoethanol, 2 mM EDTA, and protease inhibitors. All subsequent steps were at 4°C. The homogenate plus a 150-μL wash (same buffer) of the beads was centrifuged at 10,000g for 5 min, and the supernatant loaded on top of a discontinuous sucrose gradient (16 layers of 215 μL each, ranging from 15 to 40% sucrose in 10 mM Tris-HCl, pH 7.5, and 2 mM EDTA). After centrifugation at 100,000g for 18 h, 25 fractions of 150 μL were collected, and their protein content analyzed by protein gel blotting.
For cycloheximide treatment, 4-d cultures of stably transformed and nontranformed BY2 cells were washed, resuspended in fresh medium, and incubated overnight. Cycloheximide (500 μM final concentration; Aldrich, St. Louis, MO) was added, and then samples of 15 mL were collected after 0, 4, and 8 h and frozen immediately in liquid nitrogen. Microsomal fractions were then prepared, resuspended in 50 mM Tris-HCl, pH 8.8, 2% SDS, 30% glycerol, and 6 M urea, and boiled for 5 min before further analysis.
For pull-down experiments, microsomal fractions from BY2 cells were diluted to 4 mg/mL in 10 mM imidazole, 20% (w/v) glycerol, 150 mM KCl, 1 mM MgCl2, and protease inhibitors. The same volume of 20 mg/mL dodecyl maltoside (Alexis, Lausen, Switzerland) in the same buffer was added drop by drop. After a 30-min incubation at 4°C, the supernatant from a 20-min centrifugation at 100,000g was kept, and 3 μL of anti-BIP serum (Hofte and Chrispeels, 1992) were added in 100 μL of supernatant and incubated for 1 h at room temperature. Then, 15 μL of agarose-protein A beads (Fluka, Buchs, Switzerland) were added and incubated overnight at 4°C. The beads were centrifuged for 10 s at 20,000g, and the pellet was washed twice in 500 μL of the above buffer with 1 mg/mL of dodecyl maltoside, then directly resuspended in 30 μL of solubilization buffer. All samples were boiled for 5 min before gel loading to dissociate the antibody chains.
For protein gel blotting, proteins were resolved by SDS-PAGE and transferred to a nitrocellulose membrane according to standard procedures. The blot was probed with polyclonal antibodies raised against either PMA2 that was expressed in, and purified from, yeast (Maudoux et al., 2000), BIP, or GFP (Duby et al., 2001) followed by either 125I-protein A [100 μCi/L] (Amersham Bioscience, Buckinghamshire, UK) or peroxidase-coupled anti-rabbit IgG antibody (Chemicon International, Temecula, CA). Bound protein A was quantified using a phosphoimager GS-525 molecular imager system (Bio-Rad, Hercules, CA), and peroxidase activity was detected by a chemiluminescence reaction (Roche, Basel, Switzerland).
Yeast Transformation and Enzyme Assays
The yeast plasmid 2μp(PMA1)pma4 (Luo et al., 1999) was used as a backbone to generate Saccharomyces cerevisiae strains expressing the various PMA-GFP chimeras. DNA fragments corresponding to PMA4, PMA4ΔNter, PMA4ΔSL, PMA4ΔLL, PMA4ΔNdom, and PMA4ΔCter fused to GFP were isolated from pTZ19U by XbaI and PvuI (trimmed by T4 DNA polymerase) and introduced into 2μp(PMA1)pma4 between XbaI and HindIII (filled in by the Klenow fragment). The resulting plasmids were used to transform strain YAK2 (de Kerchove d'Exaerde et al., 1995). Transformation, selection, and maintenance of the resulting strains were performed as described previously (Luo et al., 1999). The construct coding for PMA4ΔCter (Q882ochre) has been described previously (Luo et al., 1999).
Yeast microsomal fractions were obtained as described previously (de Kerchove d'Exaerde et al., 1995) from a 300-mL culture grown for 16 h in glucose-rich medium after preculture for 24 h in 50 mL of galactose minimal medium. ATPase activity was measured as described previously (de Kerchove d'Exaerde et al., 1995), except that the reaction medium was adjusted to pH 6.9.
Microscopic Analysis
Epifluorescence microscopy was performed using a Leica DRM microscope coupled to a Leica DC200 camera (Leica Microsystems, Wetzlar, Germany). A GFP filter (excitation 470 ± 20 nm and emission 525 ± 25 nm) was used to detect GFP. For vacuolar staining, protoplasts were resuspended in electroporation buffer (Lukaszewicz et al., 1998) containing 1 mg/mL of Neutral Red (Molecular Probes, Eugene, OR) and incubated for 30 min at room temperature, and then stained protoplasts were observed under visible light. Confocal microscopy was performed using a Bio-Rad MRC-1024 laser scanning system as described previously (Duby et al., 2001). Simultaneous detection of GFP and YFP fluorescence was performed on a Zeiss LSM 510 as described previously (Geelen et al., 2002).
Acknowledgments
We thank C. Hawes (Oxford Brookes University, UK), M.J. Chrispeels (University of California, San Diego), and I. Moore (University of Oxford, UK) for the markers. H.B. is a research associate of the Belgian Fund for Scientific Research. This work was supported by grants from the Belgian Fund for Scientific Research, the Human Frontier Science Program, the European Union RTN program, and the Interuniversity Attraction Poles Program-Belgian Science Policy.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Marc Boutry (boutry@fysa.ucl.ac.be).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.022277.
References
- Aoki, D., Lee, N., Yamaguchi, N., Dubois, C., and Fukuda, M.N. (1992). Golgi retention of a trans-Golgi membrane protein, galactosyltransferase, requires cysteine and histidine residues within the membrane-anchoring domain. Proc. Natl. Acad. Sci. USA 89, 4319–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arango, M., Gévaudant, F., Oufattole, M., and Boutry, M. (2003). The plasma membrane proton-ATPase: The significance of gene subfamilies. Planta 216, 355–365. [DOI] [PubMed] [Google Scholar]
- Bagnat, M., Chang, A., and Simons, K. (2001). Plasma membrane proton ATPase Pma1p requires raft association for surface delivery in yeast. Mol. Biol. Cell 12, 4129–4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnat, M., Keranen, S., Shevchenko, A., and Simons, K. (2000). Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc. Natl. Acad. Sci. USA 97, 3254–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batoko, H., Zheng, H.Q., Hawes, C., and Moore, I. (2000). A Rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell 12, 2201–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter, I., Tchieu, J., Sussman, M.R., Boutry, M., Palmgren, M.G., Gribskov, M., Harper, J.F., and Axelsen, K.B. (2003). Genomic comparison of P-type ATPase ion pumps in Arabidopsis and rice. Plant Physiol. 132, 618–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boevink, P., Oparka, K., Cruz, S.S., Martin, B., Betteridge, A., and Hawes, C. (1998). Stacks on tracks: The plant Golgi apparatus traffics on an actin/ER network. Plant J. 15, 441–447. [DOI] [PubMed] [Google Scholar]
- Boutry, M., Michelet, B., and Goffeau, A. (1989). Molecular cloning of a family of plant genes encoding a protein homologous to plasma membrane H+-translocating ATPases. Biochem. Biophys. Res. Commun. 162, 567–574. [DOI] [PubMed] [Google Scholar]
- Brandizzi, F., daSilva, L.L.P., Boevink, P., Evans, D., Oparka, K., Denecke, J., and Hawes, C. (2003). ER quality control can lead to retrograde transport from the ER lumen to the cytosol and the nucleoplasm in plants. Plant J. 34, 269–281. [DOI] [PubMed] [Google Scholar]
- Brandizzi, F., Frangne, N., Marc-Martin, S., Hawes, C., Neuhaus, J.M., and Paris, N. (2002. b). The destination for single-pass membrane proteins is influenced markedly by the length of the hydrophobic domain. Plant Cell 14, 1077–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandizzi, F., Snapp, E.L., Roberts, A.G., Lippincott-Schwartz, J., and Hawes, C. (2002. a). Membrane protein transport between the endoplasmic reticulum and the Golgi in tobacco leaves is energy dependent but cytoskeleton independent: Evidence from selective photobleaching. Plant Cell 14, 1293–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereijido, M., Contreras, R.G., Shoshani, L., and Garcia-Villegas, M.R. (2003). Membrane targeting. Prog. Biophys. Mol. Biol. 81, 81–115. [DOI] [PubMed] [Google Scholar]
- Chang, A., and Slayman, C.W. (1991). Maturation of the yeast plasma membrane H+-ATPase involves phosphorylation during intracellular transport. J. Cell Biol. 115, 289–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, P.Y., and Fasman, G.D. (1978). Empirical predictions of protein conformation. Annu. Rev. Biochem. 47, 251–276. [DOI] [PubMed] [Google Scholar]
- Clarke, D.M., Loo, T.W., and MacLennan, D.H. (1990). Functional consequences of alterations to polar amino acids located in the transmembrane domain of the Ca2+-ATPase of sarcoplasmic reticulum. J. Biol. Chem. 265, 6262–6267. [PubMed] [Google Scholar]
- Dambly, S., and Boutry, M. (2001). The two major plant plasma membrane H+-ATPases display different regulatory properties. J. Biol. Chem. 276, 7017–7022. [DOI] [PubMed] [Google Scholar]
- de Kerchove d'Exaerde, A., Supply, P., Dufour, J.P., Bogaerts, P., Thines, D., Goffeau, A., and Boutry, M. (1995). Functional complementation of a null mutation of the yeast Saccharomyces cerevisiae plasma membrane H+-ATPase by a plant H+-ATPase gene. J. Biol. Chem. 270, 23828–23837. [DOI] [PubMed] [Google Scholar]
- DeWitt, N.D., Hong, B.M., Sussman, M.R., and Harper, J.F. (1996). Targeting of two Arabidopsis H+-ATPase isoforms to the plasma membrane. Plant Physiol. 112, 833–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Sansebastiano, G.P., Paris, N., Marc-Martin, S., and Neuhaus, J.M. (1998). Specific accumulation of GFP in a non-acidic vacuolar compartment via a C-terminal propeptide-mediated sorting pathway. Plant J. 15, 449–457. [DOI] [PubMed] [Google Scholar]
- Duby, G., Oufattole, M., and Boutry, M. (2001). Hydrophobic residues within the predicted N-terminal amphiphilic alpha-helix of a plant mitochondrial targeting presequence play a major role in in vivo import. Plant J. 27, 539–549. [DOI] [PubMed] [Google Scholar]
- Epping, E.A., and Moye-Rowley, W.S. (2002). Identification of interdependent signals required for anterograde traffic of ATP-binding cassette transporter protein Yor1p. J. Biol. Chem. 277, 34860–34869. [DOI] [PubMed] [Google Scholar]
- Ferreira, T., Mason, A.B., and Slayman, C.W. (2001). The yeast Pma1 proton pump: A model for understanding the biogenesis of plasma membrane proteins. J. Biol. Chem. 276, 29613–29616. [DOI] [PubMed] [Google Scholar]
- Geelen, D., Leyman, B., Batoko, H., Di Sansebastiano, G.P., Moore, I., and Blatt, M.R. (2002). The abscisic acid-related SNARE homolog NtSyr1 contributes to secretion and growth: Evidence from competition with its cytosolic domain. Plant Cell 14, 387–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, X., and Chang, A. (2001). A mutant plasma membrane ATPase, Pma1-10, is defective in stability at the yeast cell surface. Proc. Natl. Acad. Sci. USA 98, 9104–9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grec, S., Vanham, D., de Ribaucourt, J.C., Purnelle, B., and Boutry, M. (2003). Identification of regulatory sequence elements within the transcription promoter region of NpABC1, a gene encoding a plant ABC transporter induced by diterpenes. Plant J. 35, 237–250. [DOI] [PubMed] [Google Scholar]
- Hager, A., Debus, H., Edel, H.G., Stransky, H., and Serrano, R. (1991). Auxin induces exocytosis and the rapid synthesis of a high-turnover pool of plasma-membrane H+-ATPase. Planta 185, 527–537. [DOI] [PubMed] [Google Scholar]
- Harder, T., and Simons, K. (1997). Caveolae, DIGs, and the dynamics of sphingolipid-cholesterol microdomains. Curr. Opin. Cell Biol. 9, 534–542. [DOI] [PubMed] [Google Scholar]
- Haseloff, J., Siemering, K.R., Prasher, D.C., and Hodge, S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94, 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim, R., Prasher, D.C., and Tsien, R.Y. (1994). Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc. Natl. Acad. Sci. USA 91, 12501–12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofte, H., and Chrispeels, M.J. (1992). Protein sorting to the vacuolar membrane. Plant Cell 4, 995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcomb, C.L., Hansen, W.J., Etcheverry, T., and Schekman, R. (1988). Secretory vesicles externalize the major plasma membrane ATPase in yeast. J. Cell Biol. 106, 641–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch, R.B., Klee, H.J., Stachel, S., Winans, S.C., Nester, E.W., Rogers, S.G., and Fraley, R.T. (1986). Analysis of Agrobacterium tumefaciens virulence mutants in leaf discs. Proc. Natl. Acad. Sci. USA 83, 2571–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn, T., Dietrich, J., Andersen, B., Leidvik, B., Otter, C., Briving, C., Kuhlbrandt, W., and Palmgren, M.G. (2001). Large scale expression, purification and 2D crystallization of recombinant plant plasma membrane H+-ATPase. J. Mol. Biol. 309, 465–476. [DOI] [PubMed] [Google Scholar]
- Jasinski, M., Stukkens, Y., Degand, H., Purnelle, B., Marchand-Brynaert, J., and Boutry, M. (2001). A plant plasma membrane ATP binding cassette-type transporter is involved in antifungal terpenoid secretion. Plant Cell 13, 1095–1107. [PMC free article] [PubMed] [Google Scholar]
- Jensen, T.J., Loo, M.A., Pind, S., Williams, D.B., Goldberg, A.L., and Riordan, J.R. (1995). Multiple proteolytic systems, including the proteasome, contribute to Cftr processing. Cell 83, 129–135. [DOI] [PubMed] [Google Scholar]
- Jiang, L.W., and Rogers, J.C. (1998). Integral membrane protein sorting to vacuoles in plant cells: Evidence for two pathways. J. Cell Biol. 143, 1183–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, L.W., and Rogers, J.C. (1999). Sorting of membrane proteins to vacuoles in plant cells. Plant Sci. 146, 55–67. [Google Scholar]
- Jin, J.B., Kim, Y.A., Kim, S.J., Lee, S.H., Kim, D.H., Cheong, G.W., and Hwang, I. (2001). A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 13, 1511–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D.H., Eu, Y.J., Yoo, C.M., Kim, Y.W., Pih, K.T., Jin, J.B., Kim, S.J., Stenmark, H., and Hwang, I. (2001). Trafficking of phosphatidylinositol 3-phosphate from the trans-Golgi network to the lumen of the central vacuole in plant cells. Plant Cell 13, 287–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz, C., and Schell, J. (1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204, 383–396. [Google Scholar]
- Kuhlbrandt, W., Zeelen, J., and Dietrich, J. (2002). Structure, mechanism, and regulation of the Neurospora plasma membrane H+-ATPase. Science 297, 1692–1696. [DOI] [PubMed] [Google Scholar]
- Lee, M.C., Hamamoto, S., and Schekman, R. (2002). Ceramide biosynthesis is required for the formation of the oligomeric H+-ATPase Pma1p in the yeast endoplasmic reticulum. J. Biol. Chem. 277, 22395–22401. [DOI] [PubMed] [Google Scholar]
- Lefebvre, B., Boutry, M., and Morsomme, P. (2003). The yeast and plant plasma membrane H+ pump ATPase: Divergent regulation for the same function. Prog. Nucleic Acid Res. Mol. Biol. 74, 203–237. [DOI] [PubMed] [Google Scholar]
- Lin, J.L., and Addison, R. (1995). A novel integration signal that is composed of 2 transmembrane segments is required to integrate the Neurospora plasma membrane H+-ATPase into microsomes. J. Biol. Chem. 270, 6935–6941. [DOI] [PubMed] [Google Scholar]
- Lukaszewicz, M., Feuermann, M., Jerouville, B., Stas, A., and Boutry, M. (2000). In vivo evaluation of the context sequence of the translation initiation codon in plants. Plant Sci. 154, 89–98. [DOI] [PubMed] [Google Scholar]
- Lukaszewicz, M., Jerouville, B., and Boutry, M. (1998). Signs of translational regulation within the transcript leader of a plant plasma membrane H+-ATPase gene. Plant J. 14, 413–423. [DOI] [PubMed] [Google Scholar]
- Luo, H., Morsomme, P., and Boutry, M. (1999). The two major types of plant plasma membrane H+-ATPases show different enzymatic properties and confer differential pH sensitivity of yeast growth. Plant Physiol. 119, 627–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, W.J., Gong, X.H., and Chang, A. (2002). An ER membrane protein, Sop4, facilitates ER export of the yeast plasma membrane H+-ATPase, Pma1. Traffic 3, 730–739. [DOI] [PubMed] [Google Scholar]
- Ma, D.K., Zerangue, N., Lin, Y.F., Collins, A., Yu, M., Jan, Y.N., and Jan, L.Y. (2001). Role of ER export signals in controlling surface potassium channel numbers. Science 291, 316–319. [DOI] [PubMed] [Google Scholar]
- Maliga, P., Sz-Breznovits, A., and Marton, L. (1973). Streptomycin-resistant plants from callus culture of haploid tobacco. Nat. New Biol. 244, 29–30. [DOI] [PubMed] [Google Scholar]
- Malkus, P., Jiang, F., and Schekman, R. (2002). Concentrative sorting of secretory cargo proteins into COPII- coated vesicles. J. Cell Biol. 159, 915–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masibay, A.S., Balaji, P.V., Boeggeman, E.E., and Qasba, P.K. (1993). Mutational analysis of the Golgi retention signal of bovine beta-1,4-galactosyltransferase. J. Biol. Chem. 268, 9908–9916. [PubMed] [Google Scholar]
- Maudoux, O., Batoko, H., Oecking, C., Gevaert, K., Vandekerckhove, J., Boutry, M., and Morsomme, P. (2000). A plant plasma membrane H+-ATPase expressed in yeast is activated by phosphorylation at its penultimate residue and binding of 14-3-3 regulatory proteins in the absence of fusicoccin. J. Biol. Chem. 275, 17762–17770. [DOI] [PubMed] [Google Scholar]
- Moriau, L., Bogaerts, P., Jonniaux, J.L., and Boutry, M. (1993). Identification and characterization of a second plasma-membrane H+-ATPase gene subfamily in Nicotiana plumbaginifolia. Plant Mol. Biol. 21, 955–963. [DOI] [PubMed] [Google Scholar]
- Morsomme, P., and Boutry, M. (2000. a). The plant plasma membrane H+-ATPase: Structure, function and regulation. Biochim. Biophys. Acta 1465, 1–16. [DOI] [PubMed] [Google Scholar]
- Morsomme, P., Prescianotto-Baschong, C., and Riezman, H. (2003). The ER v-SNAREs are required for GPI-anchored protein sorting from other secretory proteins upon exit from the ER. J. Cell Biol. 162, 403–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morsomme, P., Slayman, C.W., and Goffeau, A. (2000. b). Mutagenic study of the structure, function and biogenesis of the yeast plasma membrane H+-ATPase. Biochim. Biophys. Acta 1469, 133–157. [DOI] [PubMed] [Google Scholar]
- Muniz, M., Morsomme, P., and Riezman, H. (2001). Protein sorting upon exit from the endoplasmic reticulum. Cell 104, 313–320. [DOI] [PubMed] [Google Scholar]
- Muniz, M., Nuoffer, C., Hauri, H.P., and Riezman, H. (2000). The Emp24 complex recruits a specific cargo molecule into endoplasmic reticulum-derived vesicles. J. Cell Biol. 148, 925–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro, S. (1995). An investigation of the role of transmembrane domains in Golgi protein retention. EMBO J. 14, 4695–4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata, T., Nemoto, Y., and Hasezawa, S. (1992). Tobacco by-2 cell-line as the Hela-cell in the cell biology of higher-plants. Int. Rev. Cytol. 132, 1–30. [Google Scholar]
- Nebenfuhr, A. (2002). Vesicle traffic in the endomembrane system: A tale of COPs, Rabs and SNAREs. Curr. Opin. Plant Biol. 5, 507–512. [DOI] [PubMed] [Google Scholar]
- Nebenfuhr, A., Gallagher, L.A., Dunahay, T.G., Frohlick, J.A., Mazurkiewicz, A.M., Meehl, J.B., and Staehelin, L.A. (1999). Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiol. 121, 1127–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmgren, M.G. (2001). Plant plasma membrane H+-ATPases: Powerhouses for nutrient uptake. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 817–845. [DOI] [PubMed] [Google Scholar]
- Paris, N., Stanley, C.M., Jones, R.L., and Rogers, J.C. (1996). Plant cells contain two functionally distinct vacuolar compartments. Cell 85, 563–572. [DOI] [PubMed] [Google Scholar]
- Park, M., Kim, S.J., Vitale, A., and Hwang, I. (2004). Identification of the protein storage vacuole and protein targeting to the vacuole in leaf cells of three plant species. Plant Physiol. 134, 625–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton, J.L., Srinivasan, B., Dickson, R.C., and Lester, R.L. (1992). Phenotypes of sphingolipid-dependent strains of Saccharomyces cerevisiae. J. Bacteriol. 174, 7180–7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrazzini, E., Villa, A., and Borgese, N. (1996). A mutant cytochrome b(5) with a lengthened membrane anchor escapes from the endoplasmic reticulum and reaches the plasma membrane. Proc. Natl. Acad. Sci. USA 93, 4207–4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, C., Braun, M., Weber, B., Wendland, M., Schmidt, B., Pohlmann, R., Waheed, A., and von Figura, K. (1990). Targeting of a lysosomal membrane protein: A tyrosine-containing endocytosis signal in the cytoplasmic tail of lysosomal acid phosphatase is necessary and sufficient for targeting to lysosomes. EMBO J. 9, 3497–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plemper, R.K., Egner, R., Kuchler, K., and Wolf, D.H. (1998). Endoplasmic reticulum degradation of a mutated ATP-binding cassette transporter Pdr5 proceeds in a concerted action of Sec61 and the proteasome. J. Biol. Chem. 273, 32848–32856. [DOI] [PubMed] [Google Scholar]
- Rayner, J.C., and Pelham, H.R.B. (1997). Transmembrane domain-dependent sorting of proteins to the ER and plasma membrane in yeast. EMBO J. 16, 1832–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritzenthaler, C., Nebenfuhr, A., Movafeghi, A., Stussi-Garaud, C., Behnia, L., Pimpl, P., Staehelin, L.A., and Robinson, D.G. (2002). Reevaluation of the effects of brefeldin A on plant cells using tobacco bright yellow 2 cells expressing Golgi-targeted green fluorescent protein and COPI antisera. Plant Cell 14, 237–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, C.J., Nothwehr, S.F., and Stevens, T.H. (1992). Membrane-protein sorting in the yeast secretory pathway: Evidence that the vacuole may be the default compartment. J. Cell Biol. 119, 69–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Jore, C.M., Evins, J., Batoko, H., Brandizzi, F., Moore, I., and Hawes, C. (2002). Redistribution of membrane proteins between the Golgi apparatus and endoplasmic reticulum in plants is reversible and not dependent on cytoskeletal networks. Plant J. 29, 661–678. [DOI] [PubMed] [Google Scholar]
- Saint-Jore-Dupas, C., Gomord, V., and Paris, N. (2004). Protein localization in the plant Golgi apparatus and the trans-Golgi network. Cell. Mol. Life Sci. 61, 159–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevier, C.S., Weisz, O.A., Davis, M., and Machamer, C.E. (2000). Efficient export of the vesicular stomatitis virus G protein from the endoplasmic reticulum requires a signal in the cytoplasmic tail that includes both tyrosine-based and di- acidic motifs. Mol. Biol. Cell 11, 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoni, Y., Kurihara, T., Ravazzola, M., Amherdt, M., Orci, L., and Schekman, R. (2000). Lst1p and Sec24p cooperate in sorting of the plasma membrane ATPase into COPII vesicles in Saccharomyces cerevisiae. J. Cell Biol. 151, 973–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skach, W.R., and Lingappa, V.R. (1993). Amino-terminal assembly of human P-glycoprotein at the endoplasmic reticulum is directed by cooperative actions of two internal sequences. J. Biol. Chem. 268, 23552–23561. [PubMed] [Google Scholar]
- Skach, W.R., and Lingappa, V.R. (1994). Transmembrane orientation and topogenesis of the third and fourth membrane-spanning regions of human P-glycoprotein (MDR1). Cancer Res. 54, 3202–3209. [PubMed] [Google Scholar]
- Stockklausner, C., and Klocker, N. (2003). Surface expression of inward rectifier potassium channels is controlled by selective Golgi export. J. Biol. Chem. 278, 17000–17005. [DOI] [PubMed] [Google Scholar]
- Toyoshima, C., Nakasako, M., Nomura, H., and Ogawa, H. (2000). Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 A resolution. Nature 405, 647–655. [DOI] [PubMed] [Google Scholar]
- Toyoshima, C., and Nomura, H. (2002). Structural changes in the calcium pump accompanying the dissociation of calcium. Nature 418, 605–611. [DOI] [PubMed] [Google Scholar]
- Ueda, T., and Nakano, A. (2002). Vesicular traffic: An integral part of plant life. Curr. Opin. Plant Biol. 5, 513–517. [DOI] [PubMed] [Google Scholar]
- Vitale, A., and Denecke, J. (1999). The endoplasmic reticulum—Gateway of the secretory pathway. Plant Cell 11, 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale, A., and Raikhel, N.V. (1999). What do proteins need to reach different vacuoles? Trends Plant Sci. 4, 149–155. [DOI] [PubMed] [Google Scholar]
- Votsmeier, C., and Gallwitz, D. (2001). An acidic sequence of a putative yeast Golgi membrane protein binds COPII and facilitates ER export. EMBO J. 20, 6742–6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada, M., Shono, M., Urayama, O., Satoh, S., Hara, Y., Ikawa, Y., and Fujii, T. (1994). Molecular cloning of P-type ATPases on intracellular membranes of the marine alga Heterosigma akashiwo. Plant Mol. Biol. 26, 699–708. [DOI] [PubMed] [Google Scholar]
- Wang, G.F., Tamas, M.J., Hall, M.J., Pascual-Ahuir, A., and Perlin, D.S. (1996). Probing conserved regions of the cytoplasmic LOOP1 segment linking transmembrane segments 2 and 3 of the Saccharomyces cerevisiae plasma membrane H+-ATPase. J. Biol. Chem. 271, 25438–25445. [DOI] [PubMed] [Google Scholar]
- Wang, Q., and Chang, A. (2002). Sphingoid base synthesis is required for oligomerization and cell surface stability of the yeast plasma membrane ATPase, Pma1. Proc. Natl. Acad. Sci. USA 99, 12853–12858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson, R.T., and Pessin, J.E. (2001). Transmembrane domain length determines intracellular membrane compartment localization of syntaxins 3, 4, and 5. Am. J. Physiol. Cell Physiol. 281, 215–223. [DOI] [PubMed] [Google Scholar]
- Zhao, R.M., Dielen, V., Kinet, J.M., and Boutry, M. (2000). Cosuppression of a plasma membrane H+-ATPase isoform impairs sucrose translocation, stomatal opening, plant growth, and male fertility. Plant Cell 12, 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, R.M., Moriau, L., and Boutry, M. (1999). Expression analysis of the plasma membrane H+-ATPase pma4 transcription promoter from Nicotiana plumbaginifolia activated by the CaMV 35S promoter enhancer. Plant Sci. 149, 157–165. [Google Scholar]