Abstract
Previous studies have shown that Listeria monocytogenes flagellar motility genes, including flaA, encoding flagellin, are transcriptionally down-regulated at 37°C. For some L. monocytogenes strains, temperature-dependent motility gene expression is less stringent. By using flaA-lacZ transcriptional fusions, we identified regions upstream of the –35/–10 promoter elements that are necessary for temperature-dependent expression of flaA in L. monocytogenes strain EGDe. Whereas the sequence of the flaA promoter region was identical in L. monocytogenes strain 10403S, transcriptional activity was only partially down-regulated at 37°C in 10403S. This finding suggested that a transacting regulatory protein with differential expression or activity in EGDe might be involved in temperature-dependent transcription of flaA. Indeed, a protein factor capable of specifically binding to the flaA promoter region was identified in cytoplasmic extracts of EGDe by using affinity purification and MS. Deletion of the factor-encoding gene (lmo0674) resulted in loss of temperature-dependent flaA expression and an increase in flaA promoter activity. Expression of other motility genes was also deregulated in the lmo0674 deletion. We have designated lmo0674 as mogR, indicating its role as a motility gene repressor. In tissue culture models, MogR repression of flaA during intracellular infection was independent of temperature and a deletion of mogR reduced the capacity for cell-to-cell spread. During in vivo infection, a deletion of mogR resulted in a 250-fold decrease in virulence. These studies indicate that regulation of flagellar motility gene expression and/or other genes controlled by MogR is required for full virulence of L. monocytogenes.
Listeria monocytogenes is a food-borne bacterial pathogen of humans and is best known for its intricate intracellular lifestyle (1). The majority of genes encoding virulence factors required for intracellular infection, such as ActA, which is necessary for actin-based motility and cell-to-cell spread (2, 3), are coordinately regulated by the transcriptional activator protein PrfA (4). L. monocytogenes can also swim by means of flagella-based motility in extracellular environments. Previous studies have shown that flagellar motility gene expression in L. monocytogenes is regulated by temperature. L. monocytogenes strains are highly flagellated and motile at low temperatures, 30°C and below, and are typically not motile at temperatures of 37°C or above (5, 6). Furthermore, bacterial flagellins serve as pattern recognition molecules for Toll-like receptor 5-mediated signaling, leading to activation of innate immune responses to infection (7, 8). Because previous studies (9, 10) have shown that transcription of L. monocytogenes flaA, encoding flagellin, is down-regulated at physiological temperature (37°C), it has been proposed that down regulation of flaA expression during in vivo infection by L. monocytogenes may serve as an adaptive mechanism to avoid host recognition and mobilization of host innate immune responses (6, 11).
Several global regulatory factors have been implicated in the control of motility gene expression in L. monocytogenes, including PrfA, the major transcriptional activator of virulence gene expression (12); FlaR, a histone-like protein (13); and CtsR, a negative regulator of class III heat shock genes (14). However, these proteins influence motility gene expression indirectly. To date, no regulatory protein that controls flagellar motility gene expression by binding to promoter regions of these genes has been identified. In this report, we show that temperature-dependent expression of motility genes in Listeria spp. is independent of PrfA. In addition, we have identified a regulator protein that directly binds to the flaA promoter region and provide evidence that this protein, designated as motility gene repressor (MogR), functions as a repressor of motility gene expression and is required for full virulence of L. monocytogenes.
Materials and Methods
Determination of flaA Promoter and lmo0674 Sequences. The flaA promoter region sequences of L. monocytogenes strains EGDe (obtained from M. Loessner, Institute of Food Science and Nutrition, Zurich) and 10403S (15) were found to be identical to the published sequence of L. monocytogenes strain EGD-e (16). The promoter and coding sequence of lmo0674 from strain EGDe were also identical to that published for EGD-e; however, base changes were found in the coding sequence of lmo0674 from strain 10403S (see Fig. 6, which is published as supporting information on the PNAS web site). The lmo0674 sequence from 10403S was submitted to the GenBank database, accession no. AY590468.
Bacterial and Eukaryotic Cell Growth Conditions. Listeria and Escherichia coli strains used in this study are listed in Table 1, which is published as supporting information on the PNAS web site. Listeria strains were grown at temperatures of 30°C, 37°C, or at room temperature (RT; 18–25°C) in brain heart infusion medium (BHI; Difco). E. coli strains were grown in LB medium at 37°C with shaking. The mouse cell lines J774 and L2 were maintained at 37°C in a 5% CO2-air atmosphere (17).
Strain and Plasmid Construction. The construction of the bacterial strains and plasmids and the antibiotic concentrations used in this work are described in Supporting Methods, which is published as supporting information on the PNAS web site. Primer sequences are in Table 2, which is published as supporting information on the PNAS web site.
Analysis of Surface-Extracted and Cytoplasmic Proteins by SDS/PAGE. Eight-milliliter-aliquots of BHI medium were inoculated with single colonies of Listeria strains and incubated without shaking for ≈24 h at RT or at 37°C. Optical density (OD600) readings were taken and a culture volume equivalent to 4 ml of an OD600 = 1.2 was centrifuged to collect bacteria. Samples were washed once with PBS, pH 7.1, resuspended in 100 μl of 2× protein sample buffer, boiled for 5 min to extract surface proteins, and cells were collected by centrifuging for 5 min at 16,000 × g. The supernatant (containing surface-extracted proteins) was stored on ice. The remaining pellet was washed once with PBS, pH 7.1, and resuspended in 500 μl of PBS, pH 7.1. Samples were placed in FastProtein Blue tubes (Q-BIOgene, Carlsbad, CA), and bacteria were lysed in a FastPrep apparatus FP120 (Q-BIOgene) for 30 sec on setting 6.0. Tubes were centrifuged at 16,000 × g for 8 min at 4°C, and supernatants (containing cytoplasmic protein fractions) were recovered. Five microliters of surface-extracted samples, and 25 μl of cytoplasmic samples were mixed with 25 μl of 2× protein sample buffer, and proteins were separated on SDS/10% PAGE gels. Proteins were visualized by Western blot by using a rabbit α-Listeria FlaA antibody (Denka Seike, Tokyo) and a goat α-rabbit alkaline phosphatase-conjugated secondary antibody (Kirkegaard & Perry Laboratories).
Plaque Formation Assay in L2 Fibroblasts and Determination of LD50 Values. Plaquing assays were performed as described (18). A gentamicin concentration of 40 μg/ml was used in the agarose-medium overlays, and plaquing assays were scanned to digital images 5 days after infection. Assays were performed with four independent cultures, and plaque sizes are given as the percentage of wild-type EGDe plaque size. LD50 values were determined as described (19).
Additional Methods. See Supporting Methods for a description of additional methods used for strain and plasmid construction, detection of flagella, primer extension analysis, β-galactosidase assays, and affinity purification of Lmo0674 from bacterial cytoplasmic extracts.
Results
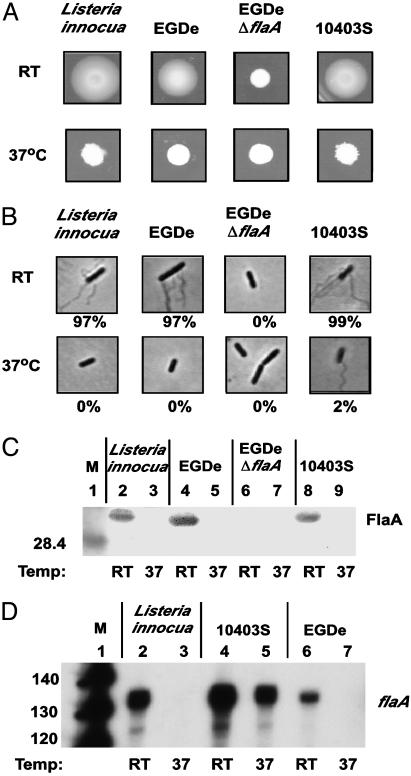
Temperature-Dependent Regulation of Motility Gene Expression in Listeria Is PrfA-Independent. We analyzed motility, flagella production, and flaA gene expression in nonpathogenic Listeria innocua (which lacks the prfA containing virulence gene cluster) in comparison with the virulent L. monocytogenes strains EGDe and 10403S. We found that when grown at RT, all wild-type Listeria strains examined were motile on soft agar plates (Fig. 1A), highly flagellated (Fig. 1B), and expressed high levels of FlaA protein on the bacterial surface (Fig. 1C). Consistent with temperature-dependent FlaA expression, flagella could not be detected on the surface of L. innocua or L. monocytogenes EGDe when grown at 37°C. Of interest, ≈2% of L. monocytogenes 10403S had a single detectable flagellum on the bacterial surface when grown at 37°C (Fig. 1B). This single flagellum was randomly distributed on the bacterial surface and not necessarily polar located as depicted in Fig. 1B. This finding is in agreement with a published observation (6) that strain 10403S remains motile at higher temperatures. However, the number of flagella and the amount of FlaA protein on the bacterial surface of strain 10403S is drastically reduced at higher temperatures (Fig. 1 B and C). Differences in flaA expression between L. innocua and EGDe compared with 10403S were more pronounced when transcript levels were examined (Fig. 1D). Whereas flaA specific transcripts were only detected at RT in L. innocua and L. monocytogenes EGDe, significant levels of flaA transcripts were detected in 10403S, when grown at 37°C (Fig. 1D, lane 5). Because L. monocytogenes strains EGDe and 10403S have identical flaA promoter region sequences, this finding suggested that the observed temperature-dependent difference in gene expression is not due to differences in promoter sequences, but might be due to a transacting factor(s). Because L. innocua lacks PrfA, and the results in Fig. 1 demonstrate that temperature-dependent motility gene expression is observed in nonpathogenic L. innocua, temperature-dependent motility gene expression in Listeria spp. can be PrfA-independent. Furthermore, results depicted in Fig. 1 indicated that temperature-dependent motility and flagellation in Listeria is not only regulated on a transcriptional level but also on a posttranscriptional level. Despite the presence of a significant amount of flaA transcripts in 10403S when grown at 37°C (Fig. 1D, lane 5), there was no FlaA protein detectable on the bacterial surface by Western blot (Fig. 1C, lane 9), and only a few (2%) 10403S bacteria contained a single flagellum on the surface (Fig. 1B).
Fig. 1.
Analysis of temperature-dependent motility and flagellar expression in Listeria stains. L. innocua and L. monocytogenes strains were grown ≈24 h at RT or at 37°C and motility and flagellar expression was analyzed. (A) Two microliters of culture were spotted on low-agar (0.375%) BHI plates and were incubated at 37°C or at RT. (B) Bacterial cultures were stained for flagella and were analyzed microscopically. The percentage (%) of bacteria within a population that possessed at least one visible flagellum is given. (C) Detection of FlaA protein by Western blot. Surface-extracted proteins were separated on an SDS/10% PAGE gel and analyzed by Western blot using a FlaA-specific antibody (lanes 2–9). Lane 1, molecular mass marker (M) with size given in kilodaltons. (D) RNA was isolated from bacteria and analyzed by primer extension for detection of flaA-specific transcripts (lanes 2–7). Lane 1, radiolabeled DNA standard with sizes given in nucleotides.
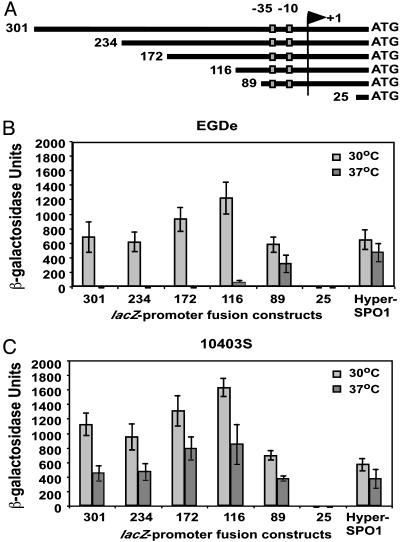
DNA Sequences Upstream of the flaA Promoter Are Required for Temperature-Dependent Gene Expression. Given the suggestion from data in Fig. 1 that a transacting factor may be required for temperature-dependent motility gene expression, we sought to determine what regions of the flaA promoter are required for temperature-dependent regulation. We used chromosomal DNA from strain EGDe flaA::Tn containing a Tn917-lacZ insertion within the flaA gene to generate flaA promoter-lacZ reporter fusions ranging from 25 bp (lacking the promoter) to 301 bp upstream of the flaA translational start site (Fig. 2A). Each fusion was cloned into the site-specific integration vector pPL3e. The different-length flaA promoter-lacZ fusions and a control lacZ fusion under the constitutive HyperSPO1 promoter were integrated as single copies into the chromosome of L. monocytogenes strains EGDe and 10403S.
Fig. 2.
Determination of flaA promoter activity by using lacZ fusions. (A) Schematic representation of flaA promoter-lacZ fusions. Numbers at the beginning of each line indicate the number of base pairs upstream of the flaA translational start site included in each construct. The –35 and –10 promoter elements and the flaA transcriptional start site (10) are indicated as boxes or an arrowhead, respectively. (B) lacZ fusions were introduced in single copy onto the chromosome of EGDe and β-galactosidase activities were determined from cultures grown at 30°C (light shaded bars) or 37°C (dark shaded bars). (C) Same as in B with lacZ fusions integrated into strain 10403S. β-galactosidase activities represent the means plus SD of four individual experiments.
We determined that expression of β-galactosidase depended on temperature in strain EGDe flaA::Tn. We detected high β-galactosidase activity at 30°C (2,172 ± 704 units) and low activity at 37°C (34 ± 26 units), indicating that this fusion was regulated in a manner similar to the flaA gene. β-galactosidase activities were determined for the different-length flaA promoter-lacZ fusions and the HyperSPO1-lacZ control fusion from cultures grown at low (30°C) or high (37°C) temperature. No significant β-galactosidase activity was detected from the promoterless 25-bp flaA-lacZ construct, whereas β-galactosidase activity from the 89-bp minimal flaA promoter-lacZ fusion was similar to the activity measured from the HyperSPO1-lacZ control construct (Fig. 2 B and C). Temperature-dependent gene expression was essentially absent in the minimal promoter construct and was only regained when additional upstream sequences were included (Fig. 2B, 116 bp or longer constructs). Consistent with primer extension results shown in Fig. 1D, the flaA promoter-lacZ fusions showed less stringent temperature regulation of β-galactosidase activity when the same flaA promoter-lacZ constructs were integrated into the chromosome of strain 10403S (Fig. 2C). These results showed that DNA sequences upstream of the minimal flaA promoter are required for temperature-dependent flaA regulation and support the idea that a factor (or multiple factors) may differentially bind to the flaA upstream DNA region within EGDe compared with 10403S, resulting in differential repression of gene expression at elevated temperature (37°C).
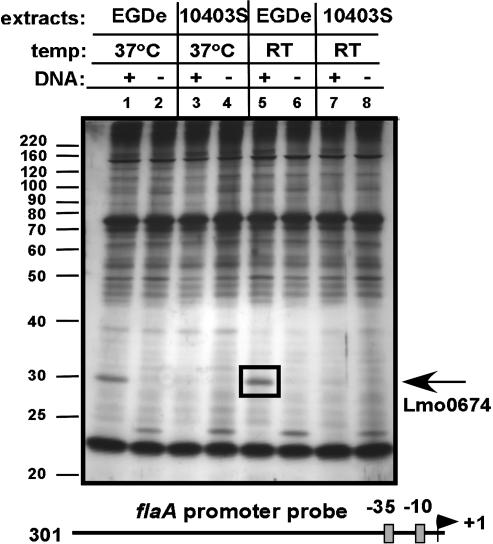
Lmo0674 Protein in EGDe Cytoplasmic Extracts Binds flaA Promoter DNA. To determine whether a factor(s) exists that binds to the flaA promoter region, we coupled biotinylated flaA promoter region DNA to streptavidin-coated magnetic beads (Dynabeads M280) and incubated DNA/magnetic bead complexes with EGDe or 10403S cytoplasmic extracts obtained from cultures grown at RT or at 37°C. Uncoupled beads were also incubated with cytoplasmic extracts as a control. After washing of the magnetic beads to minimize nonspecific binding, proteins that remained bound to the flaA promoter DNA or uncoupled beads were eluted with SDS/PAGE sample buffer, separated on a denaturing polyacrylamide gel, and visualized by silver staining. As shown in Fig. 3, the majority of recovered proteins was found in both EGDe and 10403S extracts, and was eluted from both control and DNA-coupled magnetic beads. However, one prominent protein band with a molecular mass of ≈30 kDa was found only in samples containing flaA promoter DNA-coupled beads and EGDe extracts. Comparable amounts of this protein were recovered from EGDe extracts isolated from bacteria grown at RT or at 37°C. This protein band was excised from the gel and was identified by MS as the hypothetical L. monocytogenes protein Lmo0674. Strikingly, the gene encoding Lmo0674 is located immediately upstream of the L. monocytogenes motility gene cluster (16).
Fig. 3.
Affinity purification of Lmo0674 from EGDe cytoplasmic extracts. Cytoplasmic extracts of L. monocytogenes strains EGDe or 10403S grown at RT or at 37°C were incubated with magnetic Dynabeads coupled or not coupled to a flaA-specific promoter fragment (see illustration at the bottom). Proteins associated with the Dynabeads or DNA/bead complexes were separated by SDS/PAGE and were visualized by silver staining. Bacterial extracts incubated with the Dynabeads and the temperature at which bacterial cultures were grown is indicated at the top of each lane. The presence or absence of flaA promoter DNA in the sample is indicated by a + or –, respectively. The electrophoretic mobility of protein standards in kilodaltons is indicated on the left. The boxed band indicates an abundant protein of ≈30 kDa observed only by using EGDe extracts and DNA coupled Dynabeads. The protein band was excised and identified by MS as L. monocytogenes hypothetical protein Lmo0674.
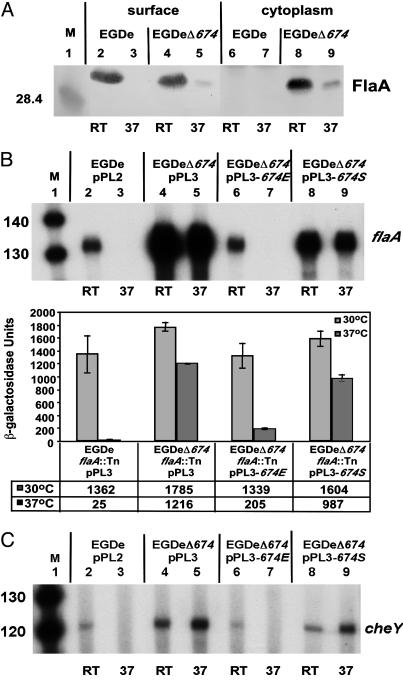
Deletion of lmo0674 Results in Increased Motility Gene Expression and Loss of Temperature Dependency. The data depicted in Fig. 3, indicating that Lmo0674 binds to flaA promoter region DNA and the presence of lmo0674 adjacent to the motility gene cluster, suggested that Lmo0674 might play a role in regulation of flagellar gene expression. To gain insight into the function of Lmo0674, we constructed strain EGDeΔ674 containing an in-frame deletion in the lmo0674 gene. A role for Lmo0674 in regulation of flagellar gene expression was first shown by staining of EGDe and EGDeΔ674 for flagella and microscopic examination of bacteria. Although no flagella could be detected on the surface of EGDe when grown at 37°C (Fig. 1B), we found that ≈1% of EGDeΔ674 bacteria contained a single flagellum when grown at 37°C (data not shown). This result correlated with the detection of small amounts of FlaA protein on the surface of strain EGDeΔ674, but not EGDe when cultures were grown at 37°C (Fig. 4A, compare lanes 3 and 5). Furthermore, significant levels of FlaA protein were detected in the cytoplasmic fraction of strain EGDeΔ674 at RT and at 37°C (Fig. 4A, lanes 6–9). When grown at RT, similar amounts of FlaA protein were detected on the bacterial surface of strains EGDe and EGDeΔ674 (Fig. 4A, lanes 2 and 4).
Fig. 4.
Comparison of motility gene expression in EGDe- and EGDeΔ674-derived strains. Cultures of EGDe- or EGDeΔ674-derived strains (indicated in each image) were grown ≈24 h at RT or at 37°C, and motility gene expression was analyzed. (A) Western blot analysis. Surface-localized (lanes 2–5) or cytoplasmic (lanes 6–9) FlaA protein was detected by Western blot using a FlaA-specific antibody. Lane 1: Molecular mass marker (M) with size given in kilodaltons. (B) Analysis of flaA transcript levels and flaA promoter activity by using primer extension (Upper) or β-galactosidase activity assays by using flaA::Tn transposon insertion strains (Lower). β-galactosidase activities represent the means plus SD of four individual experiments. (C) Primer extension analysis of cheY transcripts. Lane 1 (M) in B and C show mobility of a radiolabeled DNA standard with sizes indicated in nucleotides.
We next examined flaA promoter activity in strain EGDeΔ674 by using primer extension analysis and flaA-lacZ promoter fusions. The flaA::Tn transposon insertion of strain EGDe flaA::Tn was transduced into strain EGDeΔ674, resulting in strain EGDeΔ674 flaA::Tn. Both primer extension analysis and β-galactosidase assays revealed that flaA promoter activity was significantly altered in the strain lacking Lmo0674. Deletion of lmo0674 abolished temperature regulation of the flaA promoter and a significant increase in flaA transcript levels compared with wild-type EGDe was observed (Fig. 4B, lanes 4 and 5). The increase in flaA promoter activity in the EGDeΔ674 strain indicated that the presence of Lmo0674 led to repression of flaA transcription, even at RT, and is consistent with the affinity purification of Lmo0674 protein from both RT- and 37°C-grown EGDe extracts (Fig. 3). As confirmation for a role of Lmo0674 in repression of flaA transcription, integration of a single copy of the lmo0674 gene from strain EGDe into the chromosome of strain EGDeΔ674 by using plasmid pPL3-674E resulted in restoration of temperature-dependent transcription of flaA, similar to that observed in EGDe (Fig. 4B, lanes 6 and 7). Interestingly, when we introduced a similar construct into EGDeΔ674 containing lmo0674 cloned from strain 10403S (pPL3-674S), only partial complementation of temperature-dependent flaA expression was observed (Fig. 4B, lanes 8 and 9). Sequence analysis of the lmo0674 genes of 10403S and EGDe revealed seven nucleotide changes within the coding region (see Fig. 6). All of the nucleotide changes within the coding region were at the third position of codons and would not lead to an altered Lmo0674 amino acid sequence. However, 10403S contained a G-to-A nucleotide substitution 12 nt upstream of the translational start site, presumably within the ribosome binding site (Fig. 6). This change could cause a decrease in translation efficiency of Lmo0674, resulting in partial complementation and less stringent temperature-dependent flaA expression in strain EGDeΔ674 pPL3-674S (Fig. 4B, lanes 8 and 9) and strain 10403S (Fig. 1D, lanes 4 and 5).
Lmo0674 Modulates Expression of Other Motility Genes. We next determined whether Lmo0674 affects expression of other genes involved in motility of L. monocytogenes. Transcript levels for cheY (Fig. 4C), a gene encoding a chemotaxis protein and lmo0675, a gene showing homology to the Bacillus cereus fliN flagellar switch protein (data not shown), were both dramatically increased and were no longer temperature-dependent in an lmo0674 deletion strain. Similar to complementation of flaA transcripts (Fig. 4B), integration of plasmids pPL3-674E or pPL3-674S resulted in full or partial complementation of temperature-dependent cheY transcription, respectively (Fig. 4C). Collectively, these data demonstrated that the L. monocytogenes Lmo0674 protein is a negative regulator of motility gene expression and is essential for temperature-dependent transcription of motility genes. We have thus designated Lmo0674 as MogR, indicating its role as a motility gene repressor.
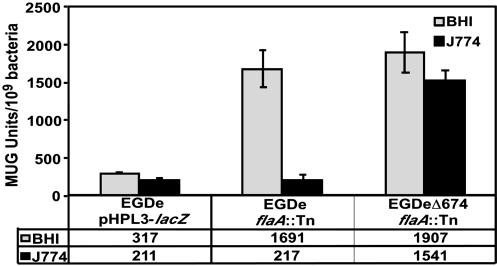
MogR Is Required for Down-Regulation of Motility Gene Expression Within Host Cells and Contributes to Virulence of L. monocytogenes. To determine whether MogR influenced regulation of flaA expression within host cells, we infected murine J774 cells at a low temperature (27°C) with strains EGDe flaA::Tn or EGDeΔ674 flaA::Tn and compared β-galactosidase activity of bacteria isolated from infected host cells with that of bacteria grown in BHI at the same temperature. Strain EGDe pHPL3-lacZ was used as a control to confirm that lacZ expression from a constitutive promoter (HyperSPO1 promoter) was not altered within host cells (Fig. 5, EGDe pHPL3-lacZ). Low β-galactosidase activity (217 units) was measured from EGDe flaA::Tn bacteria that were isolated from infected host cells, indicating that flaA promoter activity was down-regulated during intracellular infection, even at a temperature permissive for high flaA promoter activity (1,691 units) outside of host cells (Fig. 5, EGDe flaA::Tn). This down-regulation of flaA promoter activity within host cells depended on MogR because high β-galactosidase activities were detected during extracellular and intracellular growth of the EGDeΔ674 flaA::Tn strain (1,907 and 1,541 units, respectively).
Fig. 5.
Comparison of flaA promoter activity during intracellular and extracellular growth in the presence or absence of MogR. Murine J774 cells were infected at 27°C with strain EGDe flaA::Tn or EGDeΔ674 flaA::Tn, containing a flaA promoter-lacZ fusion, or strain EGDe pHPL3-lacZ, containing a constitutive HyperSPO1-lacZ promoter fusion. β-galactosidase activity from bacteria grown within host cells (dark shaded bars) was determined and compared with β-galactosidase activity from bacteria grown in BHI broth (light shaded bars). β-galactosidase activities depicted represent the means plus SD of three individual experiments. The numerical value of the mean for each condition is also given below the bar graphs.
Furthermore, deletion of mogR resulted in a decrease in the ability of L. monocytogenes to spread from cell to cell. By using in vitro plaquing assays to assess the efficiency of cell-to-cell spread (18), plaque sizes decreased by 20% after infection with EGDeΔ674 compared with infection with strain EGDe. Finally, given that deletion of MogR has an effect on cell-to-cell spread, it would be predicted that EGDeΔ674 would exhibit reduced virulence. To assess whether MogR contributes to virulence of L. monocytogenes, we determined the LD50 values for the EGDe, EGDeΔ674, and the complemented EGDeΔ674 pPL3-674E strains after infection of BALB/c mice. We found that the LD50 for EGDe was 3–5 × 103, whereas the LD50 for EGDeΔ674 was ≈1 × 106, which equates to a 250-fold decrease in virulence for EGDeΔ674. Confirmation that the virulence defect was attributed to the lack of MogR expression was obtained, because the LD50 for the EGDeΔ674 pPL3-674E strain was ≈5 × 103. These results indicate that MogR is required for full virulence of L. monocytogenes.
Discussion
In this report, we identified a regulatory protein (MogR, formerly Lmo0674) that represses expression of flagellar motility genes in L. monocytogenes. MogR was identified by affinity binding purification using flaA promoter region DNA and MS (Fig. 3). Deletion of the gene encoding MogR in L. monocytogenes strain EGDeΔ674 dramatically altered flaA gene expression such that transcription of flaA was no longer regulated by temperature and increased in comparison with wild-type EGDe. Transcription of at least two other motility genes, cheY and lmo0675, was also deregulated in a similar manner in the mogR deletion strain (Fig. 4). The biochemical and genetic data presented in this report demonstrate that MogR binds to the flaA promoter region and represses gene expression of flaA and other motility genes. MogR is not only required for temperature-dependent motility gene repression during extracellular growth but also appears to function during intracellular infection to down-regulate motility gene expression independent of temperature (Fig. 5). Misregulation of gene expression upon deletion of mogR resulted in a decreased ability of bacteria to spread from cell to cell as well as a 250-fold increase in LD50 during infection of BALB/c mice. Therefore, coordinate regulation of motility gene expression or other genes regulated by MogR contributes to full virulence of L. monocytogenes.
To gain further insight into the mechanism of MogR regulation, we used the profilescan analysis program (20) to search for known motifs within the amino acid sequence of MogR. A weak helix–turn–helix motif similarity, indicative of a DNA-binding protein, was found within MogR, suggesting that MogR might bind directly to DNA without the need of an adaptor protein. However, MogR does not show striking homology to any previously characterized protein in known databases. Nonetheless, uncharacterized ORFs that show homology to MogR are found in L. innocua, Bacillus cereus, and Bacillus anthracis. All genes encoding for MogR homologs are located adjacent to a cluster of genes displaying homology to flagellar-based motility genes. Indeed, in this report, we have shown that motility gene expression in nonpathogenic L. innocua is regulated by temperature in a similar manner as in L. monocytogenes EGDe (Fig. 1). Therefore, we believe that the MogR homolog in L. innocua has a similar function as in L. monocytogenes in regulating motility gene expression. Moreover, the finding that temperature-dependent motility is observed in L. innocua negates a direct role for PrfA, a transcriptional regulator of virulence gene expression in L. monocytogenes, in regulating temperature-dependent motility gene expression.
Data from our group and others (6, 21) have shown that temperature-dependent motility gene expression is less stringent in L. monocytogenes strain 10403S. It is very plausible that the molecular basis for this difference is on the level of MogR expression. Indeed, a MogR deletion strain could be complemented when mogR, under the control of its native promoter, was cloned from strain EGDe, but was only partially complemented when mogR was cloned from strain 10403S (Fig. 4). Sequence analysis revealed several nucleotide changes in mogR of strain 10403S compared with EGDe. One of these changes is predicted to be within the ribosome-binding site, presumably resulting in decreased translation of MogR in strain 10403S. Preliminary results from our laboratory show that MogR from strain 10403S, when expressed from a constitutive promoter with an altered ribosome binding-site sequence, can effectively repress flaA promoter activity (data not shown).
A deletion of mogR decreases the virulence of L. monocytogenes in a mouse model of infection, as well as its ability to spread from cell to cell during in vitro infection of mouse L2 fibroblasts. We envision multiple possibilities for the requirement of MogR for full virulence of L. monocytogenes. Flagellin proteins from several bacteria have been shown to possess potent proinflammatory activity and to activate host inflammatory responses through Toll-like receptor 5 (7, 8). Indeed, the L. monocytogenes FlaA protein has been show to activate host cells by means of Toll-like receptor 5 signaling (6, 8). However, no differences were observed in virulence, bacterial clearance, or induction of L. monocytogenes-specific T lymphocyte responses between strain 10403S and an isogenic flaA deletion strain in the mouse infection model (6). This study concluded that L. monocytogenes flagellin is not essential for pathogenesis or an essential trigger for innate or adaptive immunity to L. monocytogenes infection. However, data presented here indicated that only a small percentage of bacteria in a 10403S population actually possessed flagella at elevated temperature (Fig. 1B). We further showed that despite high flaA transcript levels generated at elevated temperature, no FlaA protein could be detected on the bacterial surface of strain 10403S by Western blot (Fig. 1C), yet small amounts of FlaA protein could be detected on the surface of EGDeΔ674 (Fig. 4A). This result indicates that both transcriptional and posttranscriptional levels of regulation play a role in temperature-dependent flagellation in L. monocytogenes. Furthermore, we detected FlaA protein within the cytoplasmic fraction of strain EGDeΔ674 (Fig. 4A), which is in contrast to strain 10403S (data not shown). Thus, an increase in the total amount of FlaA produced in the mogR deletion strain EGDeΔ674 might be sufficient to elicit a more robust innate immune response, resulting in faster clearance of infection, in contrast to what was observed for 10403S (6). Alternatively, increased expression of one or more MogR-regulated motility genes might compromise the integrity of L. monocytogenes, leading to the observed increase in LD50 and reduced plaque size in the in vitro cell-to-cell spreading assay. Finally, MogR might be necessary for the regulation of genes other than those required for flagellar motility. This latter possibility would be of great interest, in light of the fact that a mogR homolog is found in the genome of the nonmotile bacterial pathogen Bacillus anthracis.
Several questions regarding MogR remain to be answered and include: how are temperature and other environmental signals integrated through MogR to affect gene expression in L. monocytogenes? What is the full complement of genes regulated by MogR? How does MogR repress gene expression on a molecular level? Regions upstream of the minimal promoter sequence are required for repression of flaA promoter activity, as determined by β-galactosidase activity using different-length flaA promoter-lacZ fusions (Fig. 2). This finding could indicate that MogR binds to a region upstream of the RNA polymerase-binding site, potentially interacting with components of RNA polymerase. Indeed, by using gel shift analysis, we observed weak binding of a protein or proteins from cytoplasmic extracts of EGDe, but not 10403S, to a flaA-specific DNA fragment containing sequences upstream of the RNA polymerase-binding site (data not shown). However, significantly stronger binding was observed to a flaA-specific DNA fragment, including both upstream sequences and the flaA promoter region sequence (data not shown), indicating that MogR might require RNA polymerase or part of the RNA polymerase-binding site for its action. Future studies may clarify how MogR functions in regulation of gene expression and virulence and will further our understanding of how gene expression is coordinately regulated in bacterial pathogens.
Supplementary Material
Acknowledgments
We thank Aimee Shen and Erin Troy for construction of plasmids pPL3, pHPL3, and pPL3e; Hélène Marquis (Cornell University, Ithaca, NY) and Nancy Freitag (Seattle Biomedical Research Institute, Seattle) for sending the α-Listeria FlaA-antibody and the Listeria innocua strain, respectively; and Christiaan van Ooij and Hélène Marquis for helpful review of the manuscript. This work was supported by U.S. Public Health Service Grant AI-53669 (to D.E.H.), National Institutes of Health Grant AI-44376 (to H.G.A.B.), and Austrian Science Foundation FWF Erwin Schrödinger Postdoctoral Fellowship J2183 (to A.G.). L.S.B. is a Howard Hughes Medical Institute Predoctoral Fellow.
Abbreviations: RT, room temperature; BHI, brain heart infusion.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY590468).
References
- 1.Vazquez-Boland, J. A., Kuhn, M., Berche, P., Chakraborty, T., Dominguez-Bernal, G., Goebel, W., Gonzalez-Zorn, B., Wehland, J. & Kreft, J. (2001) Clin. Microbiol. Rev. 14, 584–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tilney, L. G. & Portnoy, D. A. (1989) J. Cell Biol. 109, 1597–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cossart, P. & Lecuit, M. (1998) EMBO J. 17, 3797–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Portnoy, D. A., Chakraborty, T., Goebel, W. & Cossart, P. (1992) Infect. Immun. 60, 1263–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peel, M., Donachie, W. & Shaw, A. (1988) J. Gen. Microbiol. 134, 2171–2178. [DOI] [PubMed] [Google Scholar]
- 6.Way, S. S., Thompson, L. J., Lopes, J. E., Hajjar, A. M., Kollmann, T. R., Freitag, N. E. & Wilson, C. B. (2004) Cell. Microbiol. 6, 235–242. [DOI] [PubMed] [Google Scholar]
- 7.Gewirtz, A. T., Navas, T. A., Lyons, S., Godowski, P. J. & Madara, J. L. (2001) J. Immunol. 167, 1882–1885. [DOI] [PubMed] [Google Scholar]
- 8.Hayashi, F., Smith, K. D., Ozinsky, A., Hawn, T. R., Yi, E. C., Goodlett, D. R., Eng, J. K., Akira, S., Underhill, D. M. & Aderem, A. (2001) Nature 410, 1099–1103. [DOI] [PubMed] [Google Scholar]
- 9.Dons, L., Rasmussen, O. F. & Olsen, J. E. (1992) Mol. Microbiol. 6, 2919–2929. [DOI] [PubMed] [Google Scholar]
- 10.Dons, L., Olsen, J. E. & Rasmussen, O. F. (1994) DNA Seq. 4, 301–311. [DOI] [PubMed] [Google Scholar]
- 11.Dons, L., Eriksson, E., Jin, Y., Rottenberg, M. E., Kristensson, K., Larsen, C. N., Bresciani, J. & Olsen, J. E. (2004) Infect. Immun. 72, 3237–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michel, E., Mengaud, J., Galsworthy, S. & Cossart, P. (1998) FEMS Microbiol. Lett. 169, 341–347. [DOI] [PubMed] [Google Scholar]
- 13.Sanchez-Campillo, M., Dramsi, S., Gomez-Gomez, J. M., Michel, E., Dehoux, P., Cossart, P., Baquero, F. & Perez-Diaz, J. C. (1995) Mol. Microbiol. 18, 801–811. [DOI] [PubMed] [Google Scholar]
- 14.Karatzas, K. A., Wouters, J. A., Gahan, C. G., Hill, C., Abee, T. & Bennik, M. H. (2003) Mol. Microbiol. 49, 1227–1238. [DOI] [PubMed] [Google Scholar]
- 15.Bishop, D. K. & Hinrichs, D. J. (1987) J. Immunol. 139, 2005–2009. [PubMed] [Google Scholar]
- 16.Glaser, P., Frangeul, L., Buchrieser, C., Rusniok, C., Amend, A., Baquero, F., Berche, P., Bloecker, H., Brandt, P., Chakraborty, T., et al. (2001) Science 294, 849–852. [DOI] [PubMed] [Google Scholar]
- 17.Dancz, C. E., Haraga, A., Portnoy, D. A. & Higgins, D. E. (2002) J. Bacteriol. 184, 5935–5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun, A. N., Camilli, A. & Portnoy, D. A. (1990) Infect. Immun. 58, 3770–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barry, R. A., Bouwer, H. G., Portnoy, D. A. & Hinrichs, D. J. (1992) Infect. Immun. 60, 1625–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gribskov, M., Homyak, M., Edenfield, J. & Eisenberg, D. (1988) Comput. Appl. Biosci. 4, 61–66. [DOI] [PubMed] [Google Scholar]
- 21.Gorski, L., Palumbo, J. D. & Mandrell, R. E. (2003) Appl. Environ. Microbiol. 69, 258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.