Abstract
miRNAs are essential factors of an extensively conserved post-transcriptional process controlling gene expression at mRNA level. Varoius biological processes such as growth and differentiation are regulated by miRNAs. Web of Science and PubMed databases were searched using the Endnote software for the publications about the role miRNA-183 family in inner ear: hair cell development and deafness published from 2000 to 2016. A triplet of these miRNAs particularly the miR-183 family is highly expressed in vertebrate hair cells, as with some of the peripheral neurosensory cells. Point mutations in one member of this family, miR-96, underlie DFNA50 autosomal deafness in humans and lead to abnormal hair cell development and survival in mice. In zebrafish, overexpression of the miR-183 family induces extra and ectopic hair cells, while knockdown decreases the number of hair cell. The miR-183 family (miR-183, miR-96 and miR-182) is expressed abundantly in some types of sensory cell in the eye, nose and inner ear. In the inner ear, mechanosensory hair cells have a robust expression level. Despite much similarity of these miRs sequences, small differences lead to distinct targeting of messenger RNAs targets. In the near future, miRNAs are likely to be explored as potential therapeutic agents to repair or regenerate hair cells, cell reprogramming and regenerative medicine applications in animal models because they can simultaneously down-regulate dozens or even hundreds of transcripts.
Keywords: miRNA, miR-183 family, Hair cell, Hearing loss
Introduction
The present review article was aimed to discuss distinct and common functions of the three members of miR-183 in cell fate determination in the inner ear. In this regards, Web of Science and PubMed databases were searched for the publications about the role of miRNA-183 family in inner ear: hair cell development and deafness between 2000 and 2016 using the EndNote software. The used search terms were miRNA-183 and deafness or hearing loss and miRNA-183 family or miRNA or microRNA in Title/Keywords/Abstract. Each database was searched independently. The articles retrieved from both databases were analyzed once. Abstracts were reviewed based on predefined inclusion and exclusion criteria. When necessary, full texts were retrieved to assess study eligibility. The articles without English abstract and English available full texts were excluded. Only the articles directly addressing the effect of miRNA-183 family in inner ear: Hair cell development and deafness were selected and analyzed. From the Web of Science, 5 articles were retrieved and from the PubMed, 8 articles retrieved. Overal, 13 articles were retrieved from both databases and 90 were included in the final analysis. After reviewing the abstracts, we excluded 17 articles from the analysis as they did not meet the inclusion criteria. 73 articles investigated the role of miRNA-183 family in inner ear: hair cell development and deafness.
History of miRNA and Biogenesis
The first miRNA (lin-4) was discovered in 1993 [1]. Generally, miRNAs are small RNAs originated from a stem-loop shaped precursor RNA that mainly intervenes in the regulation of gene expression. This regulation occurs after targeting and leading particular mRNAs to degradation [2]. The seed region of the 5' mature miRNA molecule including seven nucleotides contribute to binding to the 3'–UTR of complementary mRNA [3]. The miRNA molecule can completely bind to the mRNA target leading to full degradation of the target mRNA or partially bind to the target mRNA and cause the inhibition of mRNA translation. A wide variety of miRNAs exist in different cells and tissues which play an important role in many biological processes such as cell cycle, apoptosis, tumorigenesis and neurogenesis [4,5]. In addition based on bioinformatic predictions, almost one third of protein-coding genes in the human genome are regulated by miRNA [6]. To date, more than 100 gene loci have been demonstrated to be involved in deafness, 44 of which have been reported to possess at least on mutation; however, there are still a large number of unexplained cases. Approximately 98% of RNA in mammalian cells do not code protein [7] and non-coding RNA such as miRNAs may be involved in the development of the inner ear and thus have a role in hearing loss [8]. In 2005 miRNAs were identified in the vestibular system for the first time. Since the discovery of a tissue-specific miR-183 family and its correlation with deafness in mice and humans, miRNAs have been shown to be involved in some other diseases such as cholesteatoma, schwannomas and otitis media.
The miR-183 Family
miR-96, detected in human cancer cell lines, was the first discovered member of human miRNA of this family [9]. Bioinformatics tools allowed the discovery of miR-182 and miR-183 through comparison between human or mouse RNAs with Fugu rubripes genome (a type of Japanese puffer-fish) [10]. Considering the same expression pattern in retina and very close chromosomal loci, both miR-182 and miR-183 are categorized in the family [11]. In addition, miR-96 was added to this family due to its sequence homology with miR-182 and miR-183, and also its chromosomal locus that is between miR-182 and miR-183 [12,13]. miR-183 family consists of three homologous miRNA, miR-183, miR-182, and miR-96 (Fig. 1) which are expressed in sensory neurons and hair cells in vertebrates and sensory cells of all species of animals [14]. Among vertebrates, members of miR-183 family are expressed in the olfactory epithelium, eye, neuromast and ear. In the inner ear of mice, miRNAs are expressed in sensory neurons and hair cells of the corti organ and end organs of vestibule (Table 1) [15]. Minor differences in seed sequence among this family members causes them to have independent target RNAs (Fig. 1). This miRNAs is expressed in a completely synchronous manner throughout development and is needed for maturation of sensory organs [16]. miR-96, mir-182, and miR-183 comprise a fully conserved family of miRNAs and localize in intergenic loci of chromosome 7 long arm (Fig. 1) [17]. In vertebrates, the expression of miR-183 family seems to be limited to ciliated sensory epithelial cells and certain cranial and spiral ganglia. In zebrafish, these miRNAs have been detected in eye, nasal epithelium, sensory hair cells in ear, and neuromasts [18,19]. miR-182, alongside miR-183 and miR-96, are from a polycistronic miRNA cluster which localize in a 4 kb region on the long arm of chromosome 6. They are expressed particularly in certain organs such as eye, nose, and inner ear [12].
Fig. 1. A: Family chromosomal loci of miR-183, 182, and 96 in human and mouse. B: Seed sequence of miRNAs. Adapted from Wei, et al. Thorac Cancer 2015;6:2-9. [20].
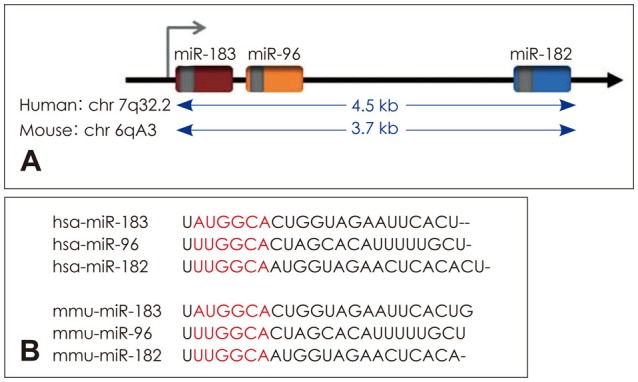
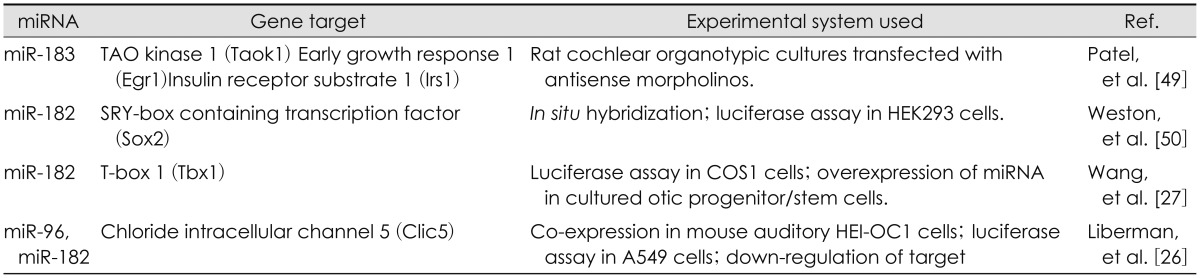
Table 1. Validation of miRNA-gene targets found in the inner ear.
miR-96
miR-96 could indirectly cause increase in the expression of significant developmental genes such as Gfi1, Ptprq, and Tmc1 [21]. In olfactory system, this miRNA family is vital for differentiation of neural precursor into mature olfactory neurons, but has no contribution to onset of specific differentiation or maturation steps [22]. miR-96 is likely to play a similar role in auditory system. Understanding miR-96 mechanism may help to explain the complicated expression of the genes required for cochlear activity and contribute to advances in non syndromic progressive deafness. Studies of diminuendo mice (a mice model to investigate the causes of hearing loss and its progression) have demonstrated that a mutation in miR-96 gene causes a mutation at locus 7 of miR-96 seed sequence [23]. Functional activity of miR-96 as an miRNA-183 family member was confirmed in a mice model with a mutation at seed regions loci. The mutation was caused by N-ethyl-Nitrosurea and led to partial hearing loss in heterozygous mice and absolute hearing loss in homozygous mice. The mechanism through which miRNAs affect inner ear development has not been yet explained fully. In a study to determine the miR-96 target genes, 12 genes were predicted by miRanda software with strong filtering, five of which (Aqp5, Celsr2, Myrip, Odf2, and Ryk) were confirmed as miR-96 target by analysis of luciferase. In a study to compare the genes expression in inner ear between wild and mutant diminuendo mice using microarray, 96 transcripts were affected significantly and the expression of five genes (Pitpnm1, oncomodulin, Preston, Ptprq, and Gfi1) was found to decline. None of these genes have a binding site for miR-96. They are likely to affect downstream targets indirectly and the changes in expression could phenotype diminuendo mice. Studies of diminuendo mice have indicated that miR-96 is responsible for maturation of stereocilia bundle in inner and external ear [24]. In addition, synaptic morphology of mutant mice remained immature and miR-96 is speculated to contribute to developing cochlear nerve. C1ic5 is an inner chloride cell canal alongside stereocilia in inner ear and has been known as a target for both miR-96 and miR-182 [25]. Mutant mice for C1ic5 have a similar stereocilial morphology to ENU mice which has been already described. C1ic5 has a binding site for miR-96, which has been confirmed by luciferase. Transfecting miR-96-contained liposomes into HEI-OC1-derived hearing cell leads to decline of C1ic5 at surfaces of both mRNA and protein [26].
miR-182
miR-182 is expressed as hair cell-like at differentiation of fundamental precursor cell of inner ear. Differentiation of fundamental precursor cell of inner ear into hair cell-like is accelerated. miR-182 activity could be associated with targeting Tbx1, which is a transcription factor playing role in development and differentiation of hair cell [27]. Increased expression of miR-182 causes increase in the number of ectopic hair cells in the inner ear. Ectopic expression of miR-182 in supporting cell could suppress and convert their gene into hair cell. Therefore if deafness is due to losing hair cells, direct transfection of miR-182 into SC could facilitate their transforming into hair cells (hair cell regeneration). Previous studies have shown that miR-183 family members are likely to suppress prosensory or SC genes to help to differentiate hair cell specifically. SOX2 as a marker prosensory cells which is also targeted by miR-182 [28,29]. In several genome wide studies on autism, the susceptibility gene to this disease, which is near 7q32/2, has been detected close to miR-182 chromosomal locus. Defective activity of miR-182 family could cause defect in sensory development steps and contribute to autism incidence [30,31,32,33,34,35]. C>A mutation has been detected at a binding site of miR-182 on radixin (RDX) 3'UTR in a human family with recessive autosomal non syndromic deafness [36]. Moreover in addition to the mutation at protein-coding loci, mutation at 3'UTR non coding loci, where miRNAs usually bind, could cause disorder in gene activity. Although in vitro investigation of luciferase did not confirm the mutual effect between RDX and miR-182 [29]. In vitro studies have indicated that Microphthalmia-associated transcription factor is a transcription factor required for retinal development and maintenance. A gene is directly targeted by miR-182. miR-182 is likely to contribute to regulating visual circadian rhythm, which is probably done by changing ADCY6 adenylyl cyclase expression [12].
Expression and Activity of the miR-183 Family in the Developing Hearing Organ of Chicken
Basillar papilla is similar to mammals auditory organ (corti organ) in various dimensions [37,38,39]. In studies, chicken embryo was used to investigate whether this family was expressed. Basillar papilla, an auditory organ, exhibited a mild increase in expression (mostly on peripheral neuron) in prosensory cells at seventh embryonic day and an increase in expression (mostly in apical hair cells) nine years later for this family. However transfection had no effect on phenotype. No detectable change in hair cell or bundle morphology was seen due to increasd expression by miR-183 family vector [40].
Expression and Activity of the miR-183 Family in Hearing Organ of Mouse
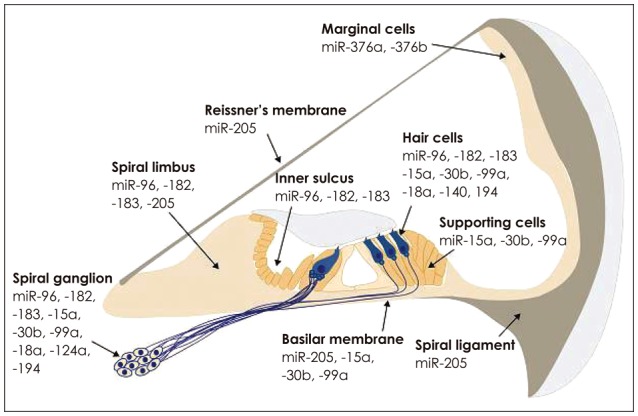
Throughout development of mice embryos, miR-183 family is initially expressed across the entire otic and spiral vesicle and atrial ganglionic neurons, and then only in hair cells and neurons accompanying them. miR-183 family has been also seen in greater epithelial ridge at 15.5–17.5 embryonic days, and is expressed in inner sulcus and spiral lumbus 4–11 days after birth [41]. Although miR-183 family in the inner ear of nature mice become evident by microarray data, in situ hybridization technique demonstrated that they disappeared in hari cell and spiral ganglion at the second and third weeks after birth (Fig. 2)[14,41].
Fig. 2. miRNAs expression pattern in mice cochlea [8,14,41,42,43,44,45,46,47,48]. Adapted from Friedman et al. Proc Natl Acad Sci U S A 2009; 106:7915-20. [8]. Adapted from Kent WJ, et al. Genome Res 2002;12:996-1006. [14]. Adapted from Sacheli et al. Gene Expr Patterns 2009;9:364-70, with permission of Elsevier. [41]. Adapted from Soukup et al. Dev Biol 2009;328:328-41, with permission of Elsevier. [42]. Adapted from Wang et al. Neuroreport 2010;21:611-7, with permission of Lippincott Williams & Wilkins. [43]. Adapted from Wang et al. Brain Res 2010;1346:14-25, with permission of Elsevier. [44]. Adapted from Elkan-Miller, et al. PLoS One 2011;6: e18195. [45]. Adapted from Hertzano, et al. PLoS Genet 2011;7: e1002309. [46]. Adapted from Yan, et al. Int J Exp Pathol 2012;93: 450-7. [47]. Adapted from Ushakov, et al. Front Mol Neurosci 2013;6:52. [48].
miR-183 Family Activity in Hair Cell
The significance of miR-183 family for hair cell development has been recently confirmed in human, mice, and zebrafish (Fig. 3). In genetic investigation of deafness locus of human autosomal the two families had a common point mutation in seed sequence which was transferred to mature miRNA and cause loss of miRNA function. Knocking down miR-183 family members cause defect in semicircular canals and inner ear [51].
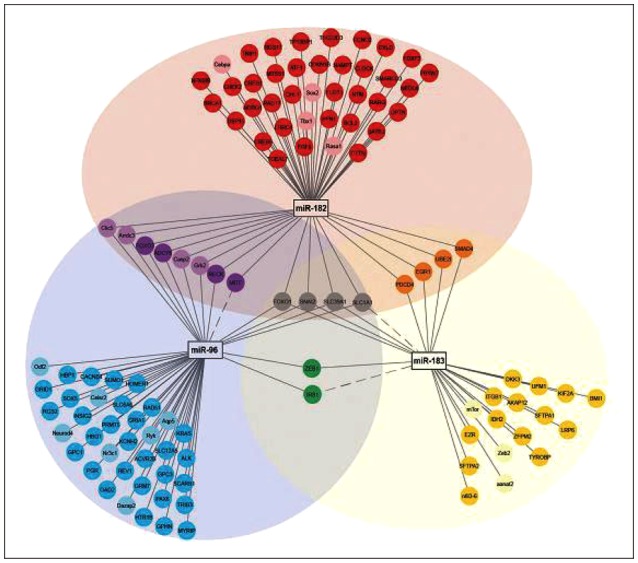
Fig. 3. Illustrates the network and diagram of mRNAs targeted by miR-183 family, and overlapping and differentiated mRNAs. The network diagram was developed by cyto scape 3.1 [53]. Target mRNA-miR-182 is shown in red color, miR-183 in yellow, miR-96 in blue, miR-182 and mir-183 in orange, miR-96 and miR-183 in green, miR-96 and miR-182 in purple, miR-182 and miR-183 in grey [16]. Adapted from Smoot, et al. Bioinformatics 2011;27:431-2. [53]. Adapted from Dambal, et al. Nucleic Acids Res 2015;43:7173-88. [16].
The Functional Role of miR-183 Family in the Development of the Inner Ear
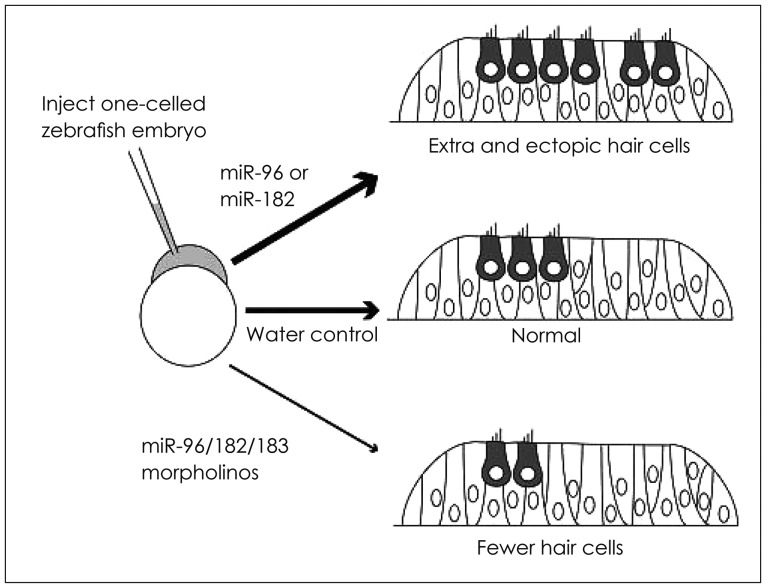
The functional role of miR-183 family members has been explored in inner ear development in zebrafish embryo [52]. Synthetic injection of miR-182 and miR-96 into embryos accelerates hair cell ectopic growth while mir-183 leads to hair cell normal development (Fig. 4). Although this family are a scended from similar initial transcript, their role varies probably due to difference in their downstream targets. miR-183 family inhibition using morpholino antisense oligonucleotides affects ear normal development at frontier and posterior macula. The severity of hair cell loss depends on the number of inhibited miRNA. All miR-183 family members are involved in controlling sensory cell. Since both the inhibition of this family proliferation and increase in its expression cause disorder in development, the balance in expression of this family could be critical to normal development of hair cell.
Fig. 4. miR-183 regulates the number of hair cells. In developing zebrafish, injecting miR-183 family reagents into an embryonic cell and then analyzing the inner ear were done 1-2 days after fertilization. Injecting miR-96 or miR-182 at monocellular embryonic step causes production of a large number of hair cells after one day. Declined level of any members of miR-183 family after morpholino antisense injection leads to producing few hair cells two days after injection [29,52]. Adapted from Li, et al. Curr Opin Otolaryngol Head Neck Surg 2010;18:459-65. [29]. Adapted from Li, et al. J Neurosci 2010;30:3254-63. [52].
miRNAs and Regeneration of Hair Cells
Treatment of hair cell regeneration is still a big challenge. Due to hair cell inability to regenerate, the stem cells with hair cell-like phenotype have been genetically manipulated [54]. A challenge of this approach is that how cells proliferation and differentiation should be regulated in reprogramming and differentiating. miRNAs could contribute greatly to initiating cell differentiation, maintaining differentiation steps, and terminating differentiation. To date, hundreds of miRNAs have been identified in cultured cochlear precursor cells. Expression patterns are different in differentiated and undifferentiated cells [55]. Therefore, the role of any involved miRNAs in cochlear (Fig. 2) cells differentiation has been yet explained fully. Understanding miRNAs regulatory roles could elucidate the hair cell regeneration in mammals inner ear.
Family miR-183 and Age-Related Hearing Loss
The above-mentioned miR-183 expression in hair cells could help to differentiate and maintain the apical structures of these cells. At older ages, the hair cells gradually lose their apical structure and hearing threshold increases. By increasing miR-183 family level in auditory cell, the apical structures and hearing ability may be recovered.
Regulatory Factor Atoh1 and miR-183 Family
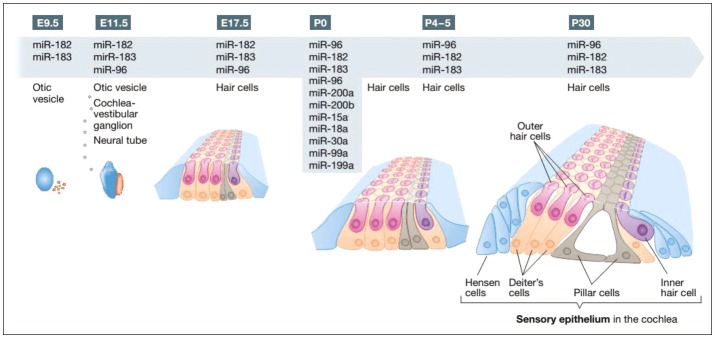
Atoh1 is an important transcription factor which can initiate hair cell differentiation and begins to express approximately at E12.5-E14.5 in hair cells [50]. Studies have indicated that miR-183 in developing cochlea did not express at E12.5 and expressed partially at E14.5. At E16.5, miR-183 family is expressed in all sensory ganglia and 17.5 entire inner ear epithelium. These findings confirm no evidence of miR-183 family in vivo (Fig. 5) expression prior to Atoh1. Therefore, this family has no contribution to onset of development although it may play a significant role in development. The expression level of miR-183 family ranges within the initial steps of inner ear development. However miR-183 family expression is specific to hair cells in the subsequent steps of development [55].
Fig. 5. Shows the time table of mi-RNAs expression at development and early steps after birth. miR-183 family is a subgroup of miRNAs that has been already investigated more frequently. miR-183 are expressed initially in otic vesicle at E9.5, which continues till reaching into cochlear-vestibular ganglion and neural tube at E11.5 and E17.5. The above expression has been reported in cochlear hair cells till p30 in some studies. Other miRNAs were identified by ISH. *represents the miRNAs that were tested only at this step [56]. Adapted from Rudnicki, et al. EMBO Mol Med 2012;4:849-59. [56].
Conclusions
Because of the limitations of studying miRNAs and their targets in human inner ear, animal models are very important in this regard. Therefore miRNAs role in development and activity of inner ear has been investigated in zebrafish and mice. Transcriptomic and proteomic investigations help to figure out miRNAs role and targets. miRNAs are thought to be used for regeneration of inner ear hair cells and finally contribute to treatment [56]. Normal expression of miR-183 family is specific to sensory organs and essential for their development. Knocking down miR-183 causes increase in the expression of the genes that are through the signaling axis PI3K/AKT/mTOR as they undergo apoptosis and dysregulation. Migration and invasion are upregulated by miR-183 family and mainly mediated by miR-182. miR-183 expression has been confirmed in various cancers [57,58,59,60,61,62,63,64,65,66,67,68,69]. The association of miR-183 family with development and diseases could be also explained by beta-catenin being able to regulate this family expression [70,71]. Beta-catenin is activated in downstream Wnt signaling pathway and is a survived transcription factor which plays a role in embryonic period and contributes to regulation of organ development. Under normal biological conditions, miR-183 expression takes place only in ciliated sensory cells, where the length and durability of survival activities are important for the entire organism. These findings make the assumption that the miR-183 family is a constituent of developmental mechanism for maintenance and longevity of sensory neuron. Obviously when miRNAs are removed in rodents, neurodegeneration takes place, confirming the hypothesis stating that this family plays a critical role in maintaining neurons health. These findings have been obtained by combined in vitro/bioinformatic studies [72]. Studies have speculated that miR-183 family is likely to suppress prosensory cells genes or to suppress SC genes to assist hair cells in specific differentiation, and SOX2 as a prosensory marker has been predicted to be a target for miR-182 [28]. Recent studies of nervous system in several organism models have demonstrated the critical role of miRNAs in development and function of nervous system. miRNAs have a great potential to help to develop a new generation of drugs but potential problems continue to raise about them, including high dose-dependent side effects and toxicity (if used in vivo) [73]. However researchers argue that miRNAs-based therapeutic interventions will be definitely helpful for the patients in the near future thanks to rapid developments in science and technology.
Footnotes
Conflicts of interest: The authors have no financial conflicts of interest.
References
- 1.Appasani K. MicroRNAs: from basic science to disease biology. Cambridge: Cambridge University Press; 2008. [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Shi M, Liu D, Duan H, Shen B, Guo N. Metastasis-related miRNAs, active players in breast cancer invasion, and metastasis. Cancer Metastasis Rev. 2010;29:785–799. doi: 10.1007/s10555-010-9265-9. [DOI] [PubMed] [Google Scholar]
- 4.Gao FB. Context-dependent functions of specific microRNAs in neuronal development. Neural Dev. 2010 Oct 01; doi: 10.1186/1749-8104-5-25. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shivdasani RA. MicroRNAs: regulators of gene expression and cell differentiation. Blood. 2006;108:3646–3653. doi: 10.1182/blood-2006-01-030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuss AW, Chen W. MicroRNAs in brain function and disease. Curr Neurol Neurosci Rep. 2008;8:190–197. doi: 10.1007/s11910-008-0031-0. [DOI] [PubMed] [Google Scholar]
- 7.Provenzano MJ, Domann FE. A role for epigenetics in hearing: establishment and maintenance of auditory specific gene expression patterns. Hear Res. 2007;233:1–13. doi: 10.1016/j.heares.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedman LM, Dror AA, Mor E, Tenne T, Toren G, Satoh T, et al. MicroRNAs are essential for development and function of inner ear hair cells in vertebrates. Proc Natl Acad Sci U S A. 2009;106:7915–7920. doi: 10.1073/pnas.0812446106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, et al. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16:720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 11.Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. 2003;9:175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J Biol Chem. 2007;282:25053–25066. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- 13.Weston MD, Pierce ML, Rocha-Sanchez S, Beisel KW, Soukup GA. MicroRNA gene expression in the mouse inner ear. Brain Res. 2006;1111:95–104. doi: 10.1016/j.brainres.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 14.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dambal S, Shah M, Mihelich B, Nonn L. The microRNA-183 cluster: the family that plays together stays together. Nucleic Acids Res. 2015;43:7173–7188. doi: 10.1093/nar/gkv703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho YJ, Tsherniak A, Tamayo P, Santagata S, Ligon A, Greulich H, et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol. 2011;29:1424–1430. doi: 10.1200/JCO.2010.28.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 19.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 20.Wei Q, Lei R, Hu G. Roles of miR-182 in sensory organ development and cancer. Thorac Cancer. 2015;6:2–9. doi: 10.1111/1759-7714.12164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 22.Choi PS, Zakhary L, Choi WY, Caron S, Alvarez-Saavedra E, Miska EA, et al. Members of the miRNA-200 family regulate olfactory neurogenesis. Neuron. 2008;57:41–55. doi: 10.1016/j.neuron.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mencía A, Modamio-Høybjør S, Redshaw N, Morín M, Mayo-Merino F, Olavarrieta L, et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat Genet. 2009;41:609–613. doi: 10.1038/ng.355. [DOI] [PubMed] [Google Scholar]
- 24.Kuhn S, Johnson SL, Furness DN, Chen J, Ingham N, Hilton JM, et al. miR-96 regulates the progression of differentiation in mammalian cochlear inner and outer hair cells. Proc Natl Acad Sci U S A. 2011;108:2355–2360. doi: 10.1073/pnas.1016646108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu C, Li X, Tan Q, Wang Z, Chen L, Liu Y. MiR-183 family regulates chloride intracellular channel 5 expression in inner ear hair cells. Toxicol In Vitro. 2013;27:486–491. doi: 10.1016/j.tiv.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 26.Liberman MC, Tartaglini E, Fleming JC, Neufeld EJ. Deletion of SLC19A2, the high affinity thiamine transporter, causes selective inner hair cell loss and an auditory neuropathy phenotype. J Assoc Res Otolaryngol. 2006;7:211–217. doi: 10.1007/s10162-006-0035-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang XR, Zhang XM, Du J, Jiang H. MicroRNA-182 regulates otocyst-derived cell differentiation and targets T-box1 gene. Hear Res. 2012;286:55–63. doi: 10.1016/j.heares.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 28.Soukup GA. Little but loud: small RNAs have a resounding affect on ear development. Brain Res. 2009;1277:104–114. doi: 10.1016/j.brainres.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Fekete DM. MicroRNAs in hair cell development and deafness. Curr Opin Otolaryngol Head Neck Surg. 2010;18:459–465. doi: 10.1097/MOO.0b013e32833e0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barrett S, Beck JC, Bernier R, Bisson E, Braun TA, Casavant TL, et al. An autosomal genomic screen for autism. Collaborative linkage study of autism. Am J Med Genet. 1999;88:609–615. doi: 10.1002/(sici)1096-8628(19991215)88:6<609::aid-ajmg7>3.3.co;2-c. [DOI] [PubMed] [Google Scholar]
- 31.Schellenberg GD, Dawson G, Sung YJ, Estes A, Munson J, Rosenthal E, et al. Evidence for multiple loci from a genome scan of autism kindreds. Mol Psychiatry. 2006;11:1049, 1060–1979. doi: 10.1038/sj.mp.4001874. [DOI] [PubMed] [Google Scholar]
- 32.Lamb JA, Barnby G, Bonora E, Sykes N, Bacchelli E, Blasi F, et al. Analysis of IMGSAC autism susceptibility loci: evidence for sex limited and parent of origin specific effects. J Med Genet. 2005;42:132–137. doi: 10.1136/jmg.2004.025668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Philippe A, Martinez M, Guilloud-Bataille M, Gillberg C, Råstam M, Sponheim E, et al. Genome-wide scan for autism susceptibility genes. Paris Autism Research International Sibpair Study. Hum Mol Genet. 1999;8:805–812. doi: 10.1093/hmg/8.5.805. [DOI] [PubMed] [Google Scholar]
- 34.Risch N, Spiker D, Lotspeich L, Nouri N, Hinds D, Hallmayer J, et al. A genomic screen of autism: evidence for a multilocus etiology. Am J Hum Genet. 1999;65:493–507. doi: 10.1086/302497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shao Y, Wolpert CM, Raiford KL, Menold MM, Donnelly SL, Ravan SA, et al. Genomic screen and follow-up analysis for autistic disorder. Am J Med Genet. 2002;114:99–105. doi: 10.1002/ajmg.10153. [DOI] [PubMed] [Google Scholar]
- 36.Hildebrand MS, Witmer PD, Xu S, Newton SS, Kahrizi K, Najmabadi H, et al. miRNA mutations are not a common cause of deafness. Am J Med Genet A. 2010;152A:646–652. doi: 10.1002/ajmg.a.33299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fuchs PA, Nagai T, Evans MG. Electrical tuning in hair cells isolated from the chick cochlea. J Neurosci. 1988;8:2460–2467. doi: 10.1523/JNEUROSCI.08-07-02460.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tilney LG, Tilney MS, DeRosier DJ. Actin filaments, stereocilia, and hair cells: how cells count and measure. Annu Rev Cell Biol. 1992;8:257–274. doi: 10.1146/annurev.cb.08.110192.001353. [DOI] [PubMed] [Google Scholar]
- 39.Lim DJ. Cochlear anatomy related to cochlear micromechanics. A review. J Acoust Soc Am. 1980;67:1686–1695. doi: 10.1121/1.384295. [DOI] [PubMed] [Google Scholar]
- 40.Zhang KD, Stoller ML, Fekete DM. Expression and Misexpression of the miR-183 Family in the Developing Hearing Organ of the Chicken. PLoS One. 2015 Jul 15; doi: 10.1371/journal.pone.0132796. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sacheli R, Nguyen L, Borgs L, Vandenbosch R, Bodson M, Lefebvre P, et al. Expression patterns of miR-96, miR-182 and miR-183 in the development inner ear. Gene Expr Patterns. 2009;9:364–370. doi: 10.1016/j.gep.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 42.Soukup GA, Fritzsch B, Pierce ML, Weston MD, Jahan I, McManus MT, et al. Residual microRNA expression dictates the extent of inner ear development in conditional Dicer knockout mice. Dev Biol. 2009;328:328–341. doi: 10.1016/j.ydbio.2009.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang XR, Zhang XM, Zhen J, Zhang PX, Xu G, Jiang H. MicroRNA expression in the embryonic mouse inner ear. Neuroreport. 2010;21:611–617. doi: 10.1097/WNR.0b013e328338864b. [DOI] [PubMed] [Google Scholar]
- 44.Wang Z, Liu Y, Han N, Chen X, Yu W, Zhang W, et al. Profiles of oxidative stress-related microRNA and mRNA expression in auditory cells. Brain Res. 2010;1346:14–25. doi: 10.1016/j.brainres.2010.05.059. [DOI] [PubMed] [Google Scholar]
- 45.Elkan-Miller T, Ulitsky I, Hertzano R, Rudnicki A, Dror AA, Lenz DR, et al. Integration of transcriptomics, proteomics, and microRNA analyses reveals novel microRNA regulation of targets in the mammalian inner ear. PLoS One. 2011 Apr 05; doi: 10.1371/journal.pone.0018195. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hertzano R, Elkon R, Kurima K, Morrisson A, Chan SL, Sallin M, et al. Cell type-specific transcriptome analysis reveals a major role for Zeb1 and miR-200b in mouse inner ear morphogenesis. PLoS Genet. 2012 Sep 29; doi: 10.1371/journal.pone.0018195. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan D, Xing Y, Ouyang X, Zhu J, Chen ZY, Lang H, et al. Analysis of miR-376 RNA cluster members in the mouse inner ear. Int J Exp Pathol. 2012;93:450–457. doi: 10.1111/j.1365-2613.2012.00840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ushakov K, Rudnicki A, Avraham KB. MicroRNAs in sensorineural diseases of the ear. Front Mol Neurosci. 2013 Dec 23; doi: 10.3389/fnmol.2013.00052. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patel M, Cai Q, Ding D, Salvi R, Hu Z, Hu BH. The miR-183/Taok1 target pair is implicated in cochlear responses to acoustic trauma. PLoS One. 2013;8:e58471. doi: 10.1371/journal.pone.0058471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weston MD, Pierce ML, Jensen-Smith HC, Fritzsch B, Rocha-Sanchez S, Beisel KW, et al. MicroRNA-183 family expression in hair cell development and requirement of microRNAs for hair cell maintenance and survival. Dev Dyn. 2011;240:808–819. doi: 10.1002/dvdy.22591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shibata SB, Ranum PT, Moteki H, Pan B, Goodwin AT, Goodman SS, et al. RNA Interference Prevents Autosomal-Dominant Hearing Loss. Am J Hum Genet. 2016;98:1101–1113. doi: 10.1016/j.ajhg.2016.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H, Kloosterman W, Fekete DM. MicroRNA-183 family members regulate sensorineural fates in the inner ear. J Neurosci. 2010;30:3254–3263. doi: 10.1523/JNEUROSCI.4948-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qin H, Zhao LD, Sun JH, Ren LL, Guo WW, Liu HZ, et al. The differentiation of mesenchymal stem cells into inner ear hair cell-like cells in vitro. Acta Otolaryngol. 2011;131:1136–1141. doi: 10.3109/00016489.2011.603135. [DOI] [PubMed] [Google Scholar]
- 55.Hei R, Chen J, Qiao L, Li X, Mao X, Qiu J, et al. Dynamic changes in microRNA expression during differentiation of rat cochlear progenitor cells in vitro. Int J Pediatr Otorhinolaryngol. 2011;75:1010–1014. doi: 10.1016/j.ijporl.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 56.Rudnicki A, Avraham KB. microRNAs: the art of silencing in the ear. EMBO Mol Med. 2012;4:849–859. doi: 10.1002/emmm.201100922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abraham D, Jackson N, Gundara JS, Zhao J, Gill AJ, Delbridge L, et al. MicroRNA profiling of sporadic and hereditary medullary thyroid cancer identifies predictors of nodal metastasis, prognosis, and potential therapeutic targets. Clin Cancer Res. 2011;17:4772–4781. doi: 10.1158/1078-0432.CCR-11-0242. [DOI] [PubMed] [Google Scholar]
- 58.Han Y, Chen J, Zhao X, Liang C, Wang Y, Sun L, et al. MicroRNA expression signatures of bladder cancer revealed by deep sequencing. PLoS One. 2011;6:e18286. doi: 10.1371/journal.pone.0018286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hannafon BN, Sebastiani P, de las Morenas A, Lu J, Rosenberg CL. Expression of microRNA and their gene targets are dysregulated in preinvasive breast cancer. Breast Cancer Res. 2011;13:R24. doi: 10.1186/bcr2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lehmann U, Streichert T, Otto B, Albat C, Hasemeier B, Christgen H, et al. Identification of differentially expressed microRNAs in human male breast cancer. BMC Cancer. 2010;10:109. doi: 10.1186/1471-2407-10-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma Q, Yang L, Wang C, Yu YY, Zhou B, Zhou ZG. [Differential expression of colon cancer microRNA in microarry study] Sichuan Da Xue Xue Bao Yi Xue Ban. 2011;42:344–348. [PubMed] [Google Scholar]
- 62.Miko E, Czimmerer Z, Csánky E, Boros G, Buslig J, Dezso B, et al. Differentially expressed microRNAs in small cell lung cancer. Exp Lung Res. 2009;35:646–664. doi: 10.3109/01902140902822312. [DOI] [PubMed] [Google Scholar]
- 63.Motoyama K, Inoue H, Takatsuno Y, Tanaka F, Mimori K, Uetake H, et al. Over- and under-expressed microRNAs in human colorectal cancer. Int J Oncol. 2009;34:1069–1075. doi: 10.3892/ijo_00000233. [DOI] [PubMed] [Google Scholar]
- 64.Schaefer A, Jung M, Mollenkopf HJ, Wagner I, Stephan C, Jentzmik F, et al. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int J Cancer. 2010;126:1166–1176. doi: 10.1002/ijc.24827. [DOI] [PubMed] [Google Scholar]
- 65.Vaksman O, Stavnes HT, Kaern J, Trope CG, Davidson B, Reich R. miRNA profiling along tumour progression in ovarian carcinoma. J Cell Mol Med. 2011;15:1593–1602. doi: 10.1111/j.1582-4934.2010.01148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamada Y, Enokida H, Kojima S, Kawakami K, Chiyomaru T, Tatarano S, et al. MiR-96 and miR-183 detection in urine serve as potential tumor markers of urothelial carcinoma: correlation with stage and grade, and comparison with urinary cytology. Cancer Sci. 2011;102:522–529. doi: 10.1111/j.1349-7006.2010.01816.x. [DOI] [PubMed] [Google Scholar]
- 67.Yin Y, Li M, Li H, Jiang Y, Cao LY, Zhang HF, et al. [Expressions of 6 microRNAs in prostate cancer] Zhonghua Nan Ke Xue. 2010;16:599–605. [PubMed] [Google Scholar]
- 68.Jacobs DK, Nakanishi N, Yuan D, Camara A, Nichols SA, Hartenstein V. Evolution of sensory structures in basal metazoa. Integr Comp Biol. 2007;47:712–723. doi: 10.1093/icb/icm094. [DOI] [PubMed] [Google Scholar]
- 69.Dvornyk V, Vinogradova O, Nevo E. Origin and evolution of circadian clock genes in prokaryotes. Proc Natl Acad Sci U S A. 2003;100:2495–2500. doi: 10.1073/pnas.0130099100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tang X, Zheng D, Hu P, Zeng Z, Li M, Tucker L, et al. Glycogen synthase kinase 3 beta inhibits microRNA-183-96-182 cluster via the β-Catenin/TCF/LEF-1 pathway in gastric cancer cells. Nucleic Acids Res. 2014;42:2988–2998. doi: 10.1093/nar/gkt1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chiang CH, Hou MF, Hung WC. Up-regulation of miR-182 by β-catenin in breast cancer increases tumorigenicity and invasiveness by targeting the matrix metalloproteinase inhibitor RECK. Biochim Biophys Acta. 2013;1830:3067–3076. doi: 10.1016/j.bbagen.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 72.Weeraratne SD, Amani V, Teider N, Pierre-Francois J, Winter D, Kye MJ, et al. Pleiotropic effects of miR-183~96~182 converge to regulate cell survival, proliferation and migration in medulloblastoma. Acta Neuropathol. 2012;123:539–552. doi: 10.1007/s00401-012-0969-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu G, Detloff MR, Miller KN, Santi L, Houlé JD. Exercise modulates microRNAs that affect the PTEN/mTOR pathway in rats after spinal cord injury. Exp Neurol. 2012;233:447–456. doi: 10.1016/j.expneurol.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]