Abstract
Weedy rice is a conspecific form of cultivated rice (Oryza sativa L.) that infests rice fields and results in severe crop losses. Weed strains in different world regions appear to have originated multiple times from different domesticated and/or wild rice progenitors. In the case of Malaysian weedy rice, a multiple-origin model has been proposed based on neutral markers and analyses of domestication genes for hull color and seed shattering. Here, we examined variation in pericarp (bran) color and its molecular basis to address how this trait evolved in Malaysian weeds and its possible role in weed adaptation. Functional alleles of the Rc gene confer proanthocyanidin pigmentation of the pericarp, a trait found in most wild and weedy Oryzas and associated with seed dormancy; nonfunctional rc alleles were strongly favored during rice domestication, and most cultivated varieties have nonpigmented pericarps. Phenotypic characterizations of 52 Malaysian weeds revealed that most strains are characterized by the pigmented pericarp; however, some weeds have white pericarps, suggesting close relationships to cultivated rice. Phylogenetic analyses indicate that the Rc haplotypes present in Malaysian weeds likely have at least three distinct origins: wild O. rufipogon, white-pericarp cultivated rice, and red-pericarp cultivated rice. These diverse origins contribute to high Rc nucleotide diversity in the Malaysian weeds. Comparison of Rc allelic distributions with other rice domestication genes suggests that functional Rc alleles may confer particular fitness benefits in weedy rice populations, for example, by conferring seed dormancy. This may promote functional Rc introgression from local wild Oryza populations.
Keywords: adaptive introgression, seed dormancy, targeted genome sequencing, Rc gene, weedy crop relatives
Rice (Oryza sativa L.) is one of the world’s most important staple foods, providing the primary calorie source to over one-third of the world’s population (Cheng et al. 2003; Khush 1997). One of the major challenges faced by rice farmers worldwide is weedy rice, a conspecific relative of the crop that infests rice fields and aggressively outcompetes desirable cultivars. Weedy rice seedlings are often morphologically nearly identical to cultivated rice plants before they reach the reproductive stage, hindering detection and selective weeding. With a rapid growth rate, highly shattering seed, persistent seed dormancy, and dark, undesirable grains that contaminate harvests, weedy rice is considered one of the primary constraints on rice production and marketability in both temperate and tropical regions (Estorninos et al. 2005; Gealy et al. 2012; Merotto et al. 2016). In addition, the potential for crop–weed gene flow threatens the long-term sustainability of weed control strategies that rely on herbicide-resistant rice cultivars (Pusadee et al. 2013; Burgos et al. 2014; Merotto et al. 2016).
The genetic diversity and origins of weedy rice have been studied extensively, and research to date suggests that weed strains in different world regions have evolved independently from different cultivated rice varieties, including indica, aus, and japonica rice (Reagon et al. 2010; Song et al. 2014). In Southeast Asia, where rice is grown in close proximity to populations of its wild ancestor, O. rufipogon, hybridization with wild populations also appears to have also contributed to the genetic composition of weed populations (Pusadee et al. 2013; Song et al. 2014). Although weedy rice is specifically adapted to agricultural fields, there are some traits in wild Oryzas that would likely be adaptive for the weedy life history strategy if introgressed into nearby weed populations. These include strong seed dormancy, allowing seeds to persist in the soil seed bank over multiple years, as well as increased seed shattering. Both of these traits were selected against during rice domestication and are absent or greatly reduced in domesticated rice varieties. The potential for both wild and domesticated rice to shape the genetic composition of Southeast Asian weed strains makes this region a particularly interesting focus for studying weedy rice evolution (Pusadee et al. 2013; Song et al. 2014).
In Malaysia, rice fields occupy ∼14% of agricultural lands (Ahmed 2012; Karim et al. 2004). Weedy rice was first reported in Malaysia in 1998 (Wahab and Suhaimi 1991), and it has become a major problem over the last two decades (Azmi and Baki 2002), with crop losses in Peninsular Malaysia exceeding $20 million by 2004 (Anuar et al. 2014). The proliferation of weedy rice in this region appears to be a direct consequence of shifts to industrialized rice production. Mechanized planting of rice paddies reduces opportunities for detection and hand-weeding in the field; in addition, large-scale commercial rice farming has led to the widespread introduction of elite (high-yielding) modern indica cultivars that appear to have given rise to some Malaysian weed strains (Song et al. 2014). In an analysis based on 24 nuclear SSRs and sequence variation at two domestication genes (sh4, controlling shattering; and Bh4, controlling hull color), Song et al. (2014) found that both wild and cultivated rice have likely contributed to the composition of contemporary Malaysian weedy rice populations.
Pericarp pigmentation is, together with seed shattering, one of the most defining features of weedy rice (Gross et al. 2010a). Most weed strains worldwide possess proanthocyanidin-pigmented (red) pericarps, a phenotype that is characteristic of wild Oryzas and associated with persistent seed dormancy (Warwick and Stewart 2005; Gu et al. 2011). Pericarp color variation in rice is primarily controlled by the regulatory gene Rc, which encodes a bHLH transcription factor that was a genomic target of selection during domestication (Sweeney et al. 2007, 2006). The functional (wild-type) Rc allele produces a red pericarp; in contrast, most cultivated rice varieties have nonpigmented (white) pericarps, and ∼97% of these carry a loss-of-function rc allele characterized by a 14-bp frame shift deletion in exon 7 (Sweeney et al. 2007). Besides this predominant rc allele, which originated in japonica rice and was selectively introgressed into indica varieties, an independently evolved domestication allele (Rc-s, characterized by a C-to-A nonsense substitution in exon 7) is found specifically in aus rice varieties (Sweeney et al. 2007, 2006; see also Wang et al. 2016). Other Rc loss-of-function mutations were selected in the independently domesticated African rice species, O. glaberrima (Gross et al. 2010b).
The Rc-encoded transcription factor pleiotropically regulates both the proanthocyanidin pigment synthesis pathway and abscisic acid-mediated seed dormancy (Gu et al. 2011). QTL mapping has indicated that pericarp color variation is correlated with seed dormancy, with Rc alleles accounting for 30% of the phenotypic variance in dormancy; germination tests have shown red-pericarp seeds to have 16% lower average germination rates at 7 d (Gu et al. 2004, 2011). Thus, the strong selection for Rc loss-of-function mutations during rice domestication is likely to reflect both human preferences for nonpigmented grains and selection for crop seeds that readily germinate upon planting (Sweeney et al. 2007; Gu et al. 2011).
Given its association with seed dormancy, Rc is of particular interest in weedy rice evolution (Gross et al. 2010a; Li et al. 2014; Sweeney et al. 2006). For Malaysian weeds, information on the genetic basis of pericarp color variation can provide a useful complement to neutral genetic markers for understanding the relative roles of wild Oryzas, recently introduced elite cultivars, and traditional crop landraces in the weed’s evolution. In this study, we examined pericarp color variation and corresponding sequence variation at the Rc locus in a set of 52 Malaysian weedy rice accessions. We compared these data to newly generated and previously published Rc sequences from wild, cultivated, and United States weedy rice accessions (N = 309 Rc sequences in total). The aims of our study were to: (1) determine how phenotypic variation in pericarp color in Malaysian weedy rice corresponds to Rc sequence variation; (2) assess phylogenetic relationships at Rc to draw inferences on evolutionary origins of Malaysian weed haplotypes; and (3) compare patterns observed at Rc with three other rice domestication genes, sh4 (controlling seed shattering), Bh4 (controlling hull color), and An-1 (controlling awn length), to consider the broader adaptive significance of pericarp color variation within Malaysian weedy rice.
Materials and Methods
Plant materials and DNA extraction
Data used in this study include newly generated Rc sequences and phenotype data, as well as previously published DNA sequences and phenotypes (Gross et al. 2010a) (Table 1). The newly generated data are from 156 Oryza accessions, including domesticated rice varieties (28 indica, 18 aus, 12 japonica, three aromatic), 52 Malaysian weedy rice accessions, 41 O. rufipogon accessions (including eight accessions of the annual form, O. nivara), and two accessions of the African wild rice species O. barthii for use as an outgroup (Supplemental Material, Table S1). The 52 Malaysian weed samples, representing 17 populations distributed across three major rice planting areas of Peninsular Malaysia (northwestern, northeastern, and central-western; Table S1), were collected in 2011 and 2012. Data for pericarp color of wild and cultivated rice were obtained from the online databases of the United States Department of Agriculture (https://npgsweb.ars-grin.gov) and the International Rice Research Institute (IRRI) (http://www.irgcis.irri.org:81/grc/SearchData.htm). Besides the common designations of red or white pericarp, a few cultivated rice accessions are categorized in online databases as has having “brown” or “light brown” pericarps; for those accessions, the alternative designations were included as such. Pericarp color of Malaysian weeds (scored as red or white) was determined by examination of five or more seeds per accession. All 156 rice accessions were grown in the greenhouse at Washington University in St. Louis. Fresh leaf tissue was collected and frozen in liquid nitrogen; DNA was extracted by a modified CTAB procedure (Gross et al. 2009).
Table 1. Pericarp and Rc variations for O. sativa and O. rufipogon accessions used in this study.
| Group | Pericarp Color | |||||
|---|---|---|---|---|---|---|
| Red (Rc) | White | Light Brown (rc) | Brown (Rc-s) | Mixed (White and Red) | ||
| (rc) | (Rc-s) | |||||
| Domesticated rice (129) | ||||||
| indica (46) | 11 | 29 | 6 | |||
| aus (27) | 12 | 4 | 4 | 7 | ||
| tropical japonica | ||||||
| Asian varieties (24) | 10 | 14 | ||||
| United States varieties (13) | 0 | 13 | ||||
| temperate japonica (11) | 1 | 8 | 2 | |||
| aromatic (8) | 0 | 7 | 1 | |||
| Malaysian weedy rice (52) | 43 | 9 | ||||
| United States weedy rice (57) | ||||||
| Black hull awned (24) | 24 | |||||
| Straw hull awnless (24) | 24 | |||||
| Brown hull (5) | 5 | |||||
| Crop–weed hybrids (4) | 3 | 1 | ||||
| O. rufipogon (67) | 60 | 4 | 3 | |||
Previously published data included 153 Oryza accessions consisting of domesticated rice (18 indica, nine aus, 29 tropical japonica, seven temperate japonica, five aromatic); 26 O. rufipogon accessions; 57 United States weedy rice strains (comprising 24 straw hull awnless, 24 black hull awned, five brown hull, and four strains of putative crop–weed hybrid origin); and two O. glumaepatula outgroup accessions (Table S2).
DNA sequencing of the Rc genomic region
DNA sequences for the Rc genomic region were obtained through targeted genome sequencing using SureSelect (Agilent) technology and Illumina Hi-Seq 2500 sequencing performed at the Whitehead Institute (Massachusetts Institute of Technology). Probes for the Rc region were designed based on the rice reference genome (MSU 6.0 assembly). Because the targeted genome sequencing approach failed to reliably detect the rc 14-bp deletion (or other indel variations), we designed PCR primers to amplify and direct-sequence the exon 7 region of Rc to definitively determine the presence or absence of this functional variation in all Malaysian weedy accessions. Primers were designed using Primer3 (Rozen and Skaletsky 1999), and PCR amplifications were carried out in 20 μl reactions containing the following: 4 μl 5× Promega GoTaq green Flexi Buffer, 2 μl 25 mM MgCl2 (2.5 mM in each reaction), 0.4 μl 10 mM dNTPs (200 μM in reaction), 1.0 μl forward and reverse primers (20 μM) respectively, 0.1 μl GoTaq polymerase, and 2 μl genomic DNA template (50 ng/μl). The following PCR profile was used: 2 min at 94° for initial denaturation; followed by 35 cycles of 30 sec at 94°, 30 sec at 55°, and 1 min at 72°; and lastly, 7 min at 72°. DNA sequencing was performed by standard methods on an ABI 3130 capillary sequencer in the Washington University Biology core facility.
Data analysis
Raw reads from Illumina sequencing were assessed for quality using FastQC software (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and the low quality reads were filtered using NGStoolkit (Patel and Jain 2012). Clean reads were mapped onto the reference genome (MSU 6.0 assembly) using BWA (Li and Durbin 2009). Single nucleotide polymorphisms (SNPs) and indels were recorded using SAMtools (Li et al. 2009). We then employed a series of Perl scripts to convert the polymorphisms from variant call format (VCF) into FASTA format for sequence alignment. Previously published Rc sequences were downloaded from GenBank and combined with our newly obtained data. Sequence alignment was performed using ClustalX (Larkin et al. 2007), with editing by hand in Bioedit (Hall 1999) to produce the final alignment.
A maximum likelihood (ML) tree was generated for the Rc locus based on high quality SNPs (excluding indels) using the Galaxy website GTR+gamma model (https://usegalaxy.org/root) and the complete dataset consisting of 309 Oryza accessions. Indels in the sequence alignment were excluded as they cannot be incorporated in the mutation model employed in this analysis. Statistical support for branches was assessed by bootstrap analysis via 1000 replicated simulations of the dataset. In addition to the ML analysis, we performed a haplotype network analysis with the same SNP dataset using TCS v 1.21 (Clement et al. 2000).
Estimates of nucleotide diversity (π and θW) were calculated for all accessions and subgroups using DnaSP version 5 (Librado Sanz and Rozas Liras 2009). Tajima's D (Tajima 1989) and Fu and Li's F and D statistics (Fu and Li 1993) were calculated to test for signatures of selection in each group, with significance assessed using 10,000 coalescent simulations and the recombination parameter set to 0. Diversity analyses examined the 6.5 kb Rc coding region, as well as a larger 9.7 kb region that included sequences spanning 2.07 kb upstream to 1.17 kb downstream of the Rc coding region.
To assess the potential adaptive significance of observed Rc allelic distributions within Malaysian weeds, patterns at this gene were compared to functional nucleotide polymorphism (FNP) distributions at three other domestication genes that also control phenotypic variation in cultivated and weedy rice: sh4 (controlling shattering), Bh4 (controlling hull color), and An-1 (controlling awn length). Genotype data for sh4 and Bh4 were extracted from Song et al. (2014) (N = 197 and 185 genotypes for sh4 and Bh4, respectively). For the An-1 gene, 17 Malaysian weed accessions were selected to represent plants that vary in awn length. The following An-1 PCR primers were designed to amplify and sequence a 775-bp FNP region previously identified by Luo et al. (2013), spanning exons 1 and 2: forward primer An-01F, 5′-AGCGCCAACAACTCCTGCTAC-3′; reverse primer An-01R, 5′-GCTTCATCCTCTCGCTTATCCTC-3′. Amplified products were sequenced directly using Sanger sequencing (ABI PRISM BigDye Terminator Cycle Sequencing Reaction Kit, Perkin Elmer) at the First BASE Laboratories Sdn. Bhd. (Malaysia). Accessions were scored for three FNPs identified by Luo et al. (2013): a GCC/– – – deletion, a C/G substitution, and a G/– deletion.
Data availability
Newly generated DNA sequences are available in GenBank (accession nos. KX549104–KX549259). Table S1 contains IDs, phenotypes, and genotypes of newly characterized accessions, including Rc genotypes confirmed by direct sequencing of exon 7 and An-1 genotype data. Table S2 contains previously published data included in analyses. The Rc alignment used in phylogeny construction is available on Dryad (doi: 10.5061/dryad.631kf).
Results
Distribution of Rc alleles and pericarp color variation
Malaysian weedy rice accessions were variable with respect to pericarp color, with 43 red-pericarp and 9 white-pericarp accessions in the sample set (Table 1 and Table S1). This stands in contrast to United States weeds, where the red pericarp is nearly universally present (Gross et al. 2010a) and the weed is commonly referred to as “red rice.” Most domesticated rice varieties in the sample set possessed white pericarps (although sample selection was intentionally enriched to represent red-pericarp varieties), and most wild rice (O. rufipogon) had red pericarps; this pattern was consistent with previous studies (Sweeney et al. 2006, 2007; Gross et al. 2010a) (Table S2).
The length of the aligned Rc dataset generated by targeted genome sequencing is 9773 bp. This spans 2068 bp upstream of the start codon, the entire 6528 bp coding region, and 1177 bp downstream of the stop codon. Consistent with previous findings, all white-pericarp (and light brown pericarp) accessions of cultivated rice were found to carry one of the two previously reported O. sativa loss-of-function alleles (rc and Rc-s), of which the rare Rc-s allele was only observed in four aus and one aromatic accession (Table 1). In addition, as previously reported, all United States weedy rice strains carried putatively functional Rc alleles (Gross et al. 2010a). Malaysian weeds carried both functional Rc sequences and the rc loss-of-function allele, consistent with the phenotypic variation observed in this group.
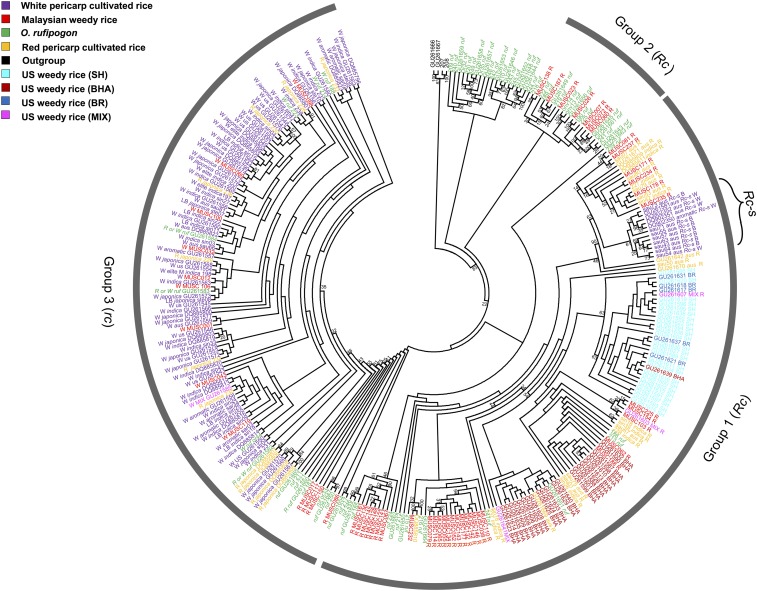
To examine the phylogenetic origin of pericarp color variation in Malaysian weedy rice, we performed ML tree construction and TCS haplotype network analyses using a combined dataset of the newly generated and previously published Rc sequences; the aligned dataset contained 721 SNPs with indels excluded (Dryad doi: 10.5061/dryad.631kf). On the resulting ML tree, Malaysian weedy rice accessions fall into three phylogenetically distinct groups (Figure 1); this pattern is also evident in the TCS haplotype network (Figure S1). The largest group of Malaysian weeds (32 of 52 accessions) occur in a large clade (labeled group 1) where they are clustered with United States weeds, red-pericarp domesticated rice (12 aus and 11 indica varieties), white- or brown-pericarp domesticated rice varieties that carry the Rc-s nonsense mutation (11 aus and one aromatic variety), and a few O. rufipogon accessions. A second clade (group 2) contains seven Malaysian weeds that are grouped exclusively with O. rufipogon accessions. The third group of Malaysian weeds (group 3) is characterized by haplotypes that either carry the rc 14-bp deletion or have functional Rc sequences closely related to rc haplotypes. All white-pericarp Malaysian weeds occur in this group.
Figure 1.
Maximum likelihood (ML) tree of Rc haplotypes. Different colors indicate identity of accessions, as indicated in the key. Numbers on branches indicate bootstrap support. BHA, black hull awned; BR, brown hull; MIX, crop–weed hybrid; SH, straw hull awnless.
Direct Sanger sequencing of the Rc exon 7 region confirmed the presence of the rc 14-bp frame shift deletion in eight of the nine white-pericarp Malaysian weeds. The one exception (MUSC032) showed no other obvious evidence of an independent loss-of-function mutation in the Rc gene. This genotype–phenotype discrepancy may reflect an instance of heterozygosity at the Rc locus in this weed strain, such that the individual plant grown for DNA sequencing (potentially a red-pericarp Rc/rc heterozygote) differed genetically from the seed material used in assessing pericarp color (potentially a white-pericarp rc homozygote). Recent hybridization between plants carrying Rc and rc alleles could account for such heterogeneity. Consistent with this hypothesis, all four red-pericarp Malaysian weeds that are clustered in group 3 were revealed by Sanger sequencing to be heterozygotes for the rc mutation (Table S1), which further suggests recent hybridization between plants with functional and nonfunctional Rc alleles.
Taken together, the haplotype groupings in the ML tree and TCS network suggest that the Rc sequences present in Malaysian weed strains are derived from at least three distinct sources: wild O. rufipogon populations (most evident for accessions in group 2); white-pericarp domesticated rice (group 3); and red-pericarp indica and aus rice varieties that did not undergo selection for the white-pericarp phenotype found in most contemporary rice (group 1). Thus, it appears that reproductively compatible O. rufipogon populations, together with phenotypically diverse domesticated rice varieties, have contributed to the genetic and phenotypic complexity of Malaysian weeds.
In order to determine which SNPs contribute to the Rc haplotype differentiation between the three groups, we examined the nucleotide variation in the genomic region spanning 2068 bp upstream of the start codon to 1177 bp downstream of the stop codon in the Malaysian weed accessions. For white-pericarp Malaysian weeds (group 3), there are 13 SNPs that are diagnostic of this group, in comparison to two unique SNPs for group 1 (containing most weed accessions) and no unique SNPs for the O. rufipogon-like group 2 (Table S3). The greater differentiation of group 3 haplotypes from the other two groups is consistent with the known origin of the rc allele from a japonica background (Sweeney et al. 2007), which would be expected to show high differentiation from the indica- and aus-related accessions occurring in groups 1 and 2 (Figure 1). Multiple sequence alignment of the Rc coding region and predicted amino acid sequences confirmed that the rc 14-bp deletion occurs specifically in white-pericarp cultivated rice and Malaysian weedy rice (genomic sequence positions 1429–1442 bp; Table S4). Among the 24 SNPs within exon regions, two are nonsynonymous, one of which is predicted to produce a Q-to-H gain-of-charge amino acid replacement (position 21; Table S4). This replacement occurs in Malaysian weeds and indica varieties in group 3, as well as one Malaysian weed in group 1. Both of the nonsynonymous substitutions occur outside the functionally critical DNA-binding HLH domain (Sweeney et al. 2006), however, and may have no phenotypic effect.
Genetic diversity analysis
Artificial selection during domestication is expected to decrease the nucleotide diversity at the genomic target of selection, as the favored domestication allele rises to high frequency and displaces neutral variation. To examine changes in nucleotide variation at the Rc locus, we calculated the nucleotide diversity of wild, cultivated, and weedy rice. As expected, white-pericarp cultivated rice (carrying the rc allele) shows reduced nucleotide diversity (silent-site π = 0.04/kb) compared to O. sativa overall (silent-site π = 0.95/kb) and the wild progenitor O. rufipogon (silent-site π = 1.30/kb) (Table 2; see also Table S5). Correspondingly, the Tajima’s D value of white-pericarp cultivated rice shows a statistically significant deviation from neutrality in a direction suggesting strong positive selection for the domestication allele (D = −2.283; P < 0.01). Red-pericarp crop varieties, which were not subject to such selection, show no significant deviation from neutrality at Rc. These results are consistent with previous findings of selection signatures at Rc for the white-pericarp domestication phenotype (Gross et al. 2010a; Sweeney et al. 2007, 2006). Interestingly, the wild progenitor, O. rufipogon, also shows a significantly negative Tajima’s D value (D = −2.570; P < 0.001), despite most of the wild accessions possessing red pericarps and functional Rc alleles; in this case, however, the deviation reflects the low-frequency presence of rc alleles in wild rice, which generate a statistical excess of rare SNPs in the sample set (Table S4).
Table 2. Nucleotide diversity and Tajima’s D at the Rc locus in wild, cultivated, and Malaysian weedy rice.
| O. rufipogon | O. sativa | Malaysian Weedy Rice | |||||||
|---|---|---|---|---|---|---|---|---|---|
| (n = 67) | All Cultivated Rice (n = 129) | White Pericarp Cultivated Rice (rc)a (n = 83) | Red Pericarp Cultivated Rice (n = 34) | All Malaysian Weeds (n = 52) | White Pericarp Malaysian Weeds (n = 9) | Malaysian Weeds in Group 1 (n = 32) | Malaysian Weeds in Group 2 (n = 7) | Malaysian Weeds in Group 3 (n = 13) | |
| π per kb (silent sites) | 1.30 | 0.95 | 0.04 | 2.57 | 1.44 | 0.31 | 1.50 | 4.01 | 0.12 |
| π per kb (all sites) | 1.13 | 0.87 | 0.04 | 2.26 | 1.24 | 0.26 | 1.28 | 3.47 | 0.10 |
| θw per kb (silent sites) | 5.06 | 1.44 | 0.30 | 2.21 | 2.66 | 0.51 | 1.51 | 4.30 | 0.08 |
| θw per kb (all sites) | 4.34 | 1.33 | 0.30 | 1.81 | 2.35 | 0.44 | 1.34 | 3.72 | 0.07 |
| Tajima’s D | −2.5700*** | −1.0633 | –2.2830** | −0.91202 | −1.6765 | –1.8613* | −0.1526 | 0.3886 | −0.1526 |
***, **, and * for Tajima’s D values indicates statistical significance at P < 0.001, P < 0.01, and P < 0.05, respectively.
Includes varieties scored as “light brown” in online databases.
Like domesticated rice, white-pericarp Malaysian weeds also show reductions in Rc nucleotide diversity and a statistically significant deviation from neutrality in a direction consistent with selection for the rc allele (silent-site π = 0.31/kb; Tajima’s D = −1.861; P < 0.05) (Table 2 and Table S5). However, most of the weed strains do not have this domestication phenotype, and nucleotide diversity for Malaysian weeds overall (silent site π = 1.44/kb) exceeds that of all cultivated O. sativa, and is even marginally higher than the wild species O. rufipogon (silent site π = 1.30/kb). The high Rc nucleotide diversity present in these weeds supports our conclusion based on phylogenetic analyses that Malaysian weed haplotypes have diverse origins from both cultivated and wild rice.
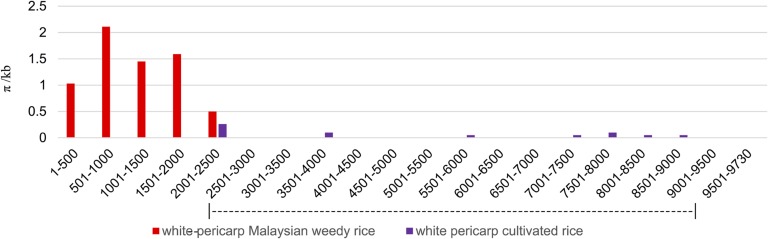
To further explore the distribution of genetic diversity at the Rc locus, we compared nucleotide diversity across the larger 9.7 kb region (spanning upstream and downstream sequences) for white-pericarp weeds with cultivated rice accessions carrying the rc allele. Despite the shared presence of the rc allele within the coding region, the white-pericarp weeds show higher genetic diversity in the immediate 5′ flanking sequence (Figure 2). This elevated diversity is attributable to a single weed accession, MUSC098, which carries an upstream sequence that differs from the other white-pericarp weed accessions by 15 SNPs, all of which are shared with red-pericarp weed accessions (Table S3). This pattern suggests that MUSC098 carries a recombinant haplotype, with the rc allele in the coding region and a sequence characteristic of red-pericarp weeds in its upstream sequence.
Figure 2.
Comparison of nucleotide diversity across the Rc genomic region for white-pericarp Malaysian weedy rice vs. white-pericarp cultivated rice. Diversity is shown for accessions that carry the rc allele, using 500-bp windows across the 5′ upstream region (0–2 kb), the Rc coding region (2–8.5 kb), and the 3′ downstream region (8.5–9.7 kb). The last window is 230 bp. The dashed line corresponds to the approximate location of the Rc coding region.
Disproportionate representation of wild Rc alleles in Malaysian weeds
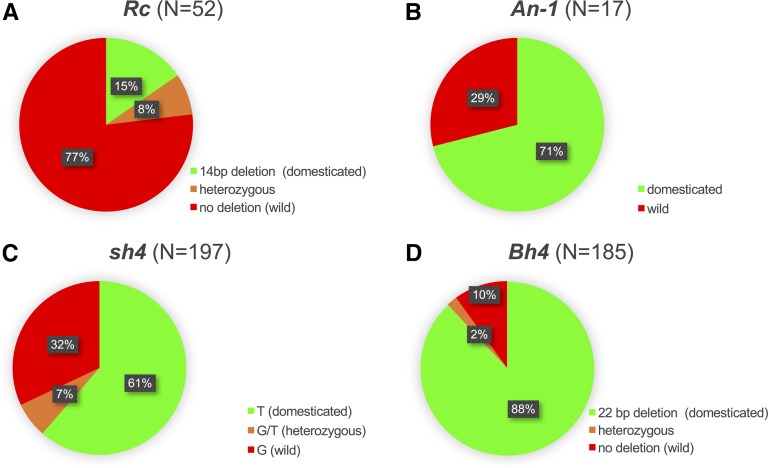
Rc differs markedly from the other domestication genes examined in the proportions of wild vs. domestication alleles represented in Malaysian weeds. For An-1 (controlling awn length), Bh4 (controlling hull color), and sh4 (controlling seed shattering), the majority of Malaysian weeds are homozygous for domestication alleles (ranging from 61% of accessions for sh4 to 88% for Bh4; Figure 3 and Table S1). This preponderance of domestication alleles is consistent with previous inferences that domesticated rice has played a major role in the evolution of Malaysian weeds (Song et al. 2014). In contrast, for Rc the opposite pattern is observed: the majority of Malaysian weeds (77%) are homozygous for the wild-type functional allele that is characteristic of wild Oryzas and absent in the vast majority of modern cultivated rice varieties (Figure 3A). This Rc-specific pattern suggests that selection may be playing a role in elevating Rc allele frequencies in Malaysian weedy rice. Given the known functional importance of Rc in maintaining rice seed dormancy (Gu et al. 2011; Subudhi et al. 2012), this disproportionate occurrence of wild-type Rc alleles could potentially reflect selection to maintain seed dormancy alleles introgressed from wild O. rufipogon into Malaysian weed populations.
Figure 3.
Percentage of Malaysian weedy rice accessions with wild-type vs. domestication alleles for four domestication genes: (A) Rc, (B) An-1, (C) sh4, and (D) Bh4. N indicates numbers of accessions genotyped per gene. Genotyping details for each sampled accession are listed in Table S1. Data for sh4 and Bh4 were extracted from Song et al. (2014).
Discussion
As one of the primary genetic determinants of pericarp color, Rc was a target of strong selection during rice domestication because humans selected for nonpigmented grains (Sweeney et al. 2007, 2006). Given the pleiotropic role of the Rc-encoded transcription factor in regulating seed dormancy (Gu et al. 2011), this selective event may also reflect human selection for crop seeds that readily germinate upon sowing. In contrast to cultivated rice, seed dormancy is highly adaptive in agricultural weeds, as it allows seeds to persist in crop fields over multiple seasons (Delouche and Labrada 2007). Here, we find evidence that in Malaysian weedy rice, Rc allelic diversity has likely been enhanced by both wild O. rufipogon and cultivated rice (Figure 1, Table 2, and Figure S1), and that selection may specifically favor elevated frequencies of functional wild-type Rc alleles in weed populations (Figure 3), which would be expected to confer seed dormancy. Below we discuss these findings and their potential implications for weedy rice evolution and adaptation.
Contributors to Rc diversity in Malaysian weedy rice
Oryza rufipogon:
The Rc locus has been studied extensively in cultivated rice (Gulick et al. 2009; Sweeney et al. 2007, 2006; Wang et al. 2016) and to a lesser extent in United States weedy rice (Gross et al. 2010a); it has not, however, previously been examined in weedy rice populations that grow in regions where they can be influenced by gene flow from local wild Oryzas. For Malaysian weeds, which often occur in proximity to wild O. rufipogon, the likely influence of wild populations is evident in the Rc phylogenetic analysis, where we detect Rc haplotypes that are characteristic of the wild species in the weeds (e.g., group 2, Figure 1; see also Figure S1), and in our comparison across domestication genes, where we find disproportionate representation of wild-type alleles compared to other domestication genes (Figure 3). Notably, more than three-quarters (77%) of weed accessions in our sample set carry functional Rc alleles, a marked contrast from allelic distributions at the domestication genes sh4, Bh4, and An-1. In a recent morphology based study of a larger sampling of Malaysian weedy rice (N = 193 accessions), Sudinato et al. (2016) observed red or brown pericarp color in a comparable proportion of the samples (68%). This suggests that Rc allelic distribution observed here may be generally representative of Malaysian weed populations.
Red-pericarp rice:
Unlike domestication genes such as the shattering locus sh4, where cultivated rice worldwide is nearly 100% fixed for the domestication allele, domestication alleles of Rc are not completely fixed in cultivated rice, and while rare nowadays, some red-pericarp Malaysian landraces are still grown that carry functional Rc haplotypes. A proportion of the functional Rc alleles observed in Malaysian weeds may therefore be derived from red-pericarp crop landraces rather than O. rufipogon. Weed haplotypes in Rc group 1 are the most likely candidates to be of crop origin, as most red-pericarp rice varieties cluster within this group (see Figure 1 and Figure S1). Cultivation of red-pericarp landraces in Malaysia has declined to very low levels in recent decades, particularly following the industrialization of rice production. No red-pericarp landraces have been grown in any of the locations sampled for the present study for at least 30 yr (B. K. Song, unpublished data), and aus rice varieties (which make up the majority of red-pericarp rice landraces) do not occur in Malaysia. This suggests that red-pericarp landraces probably play a limited role, if any, in the contemporary evolution of Malaysian weed populations, although they may have played a more important role in the past (see Song et al. 2014).
White-pericarp rice:
Whereas red-pericarp cultivated rice may have a limited role in ongoing Malaysian weed evolution, the white-pericarp varieties are clearly influencing contemporary phenotypic and genetic diversity of the weed. Nine of the 52 sampled accessions (17%) have white pericarps, eight of which were confirmed to carry the 14-bp deletion rc allele (Figure 1 and Table 1). Moreover, direct Sanger sequencing revealed that four additional red-pericarp weeds carry the rc allele in a heterozygous state (Table S1). Like cultivated rice, weedy rice is predominantly self-fertilizing, and heterozygosity would be expected to decline to negligible levels within just a few generations of a hybridization event. The observation here of four Rc/rc heterozygotes, along with the substantial proportion of white-pericarp weeds (Table S1), strongly suggests that crop–weed hybridization is contributing to the genetic diversity of Malaysian weedy rice, and that this is an ongoing process.
While Rc sequences do not provide insight on which specific crop varieties are most likely to be contributing rc alleles to Malaysian weedy rice, a complementary analysis based on neutral SSR markers has implicated recently introduced elite cultivars in the weed’s evolution (Song et al. 2014). The study revealed some Malaysian weed strains to be genetically highly similar to elite indica varieties; these modern cultivars were originally developed at the IRRI (Dalrymple 1986) and were later mass-distributed in Malaysia with the advent of industrialized rice production.
It should be noted that because the Rc alignment excluded indels (which could be scored less reliably from genome sequence data; see Materials and Methods), the rc 14-bp deletion was not included as a phylogenetic character in generating the ML tree, and the presence or absence of this mutation was treated as unknown for previously published Rc sequences (Table S1). The rc allele is known to have a single mutational origin from a japonica rice background (Sweeney et al. 2007), and as such, the haplotypes that carry this mutation (characterizing group 3 sequences; Figure 1 and Figure S1) differ by multiple SNPs from the indica, aus, and O. rufipogon sequences that characterize groups 1 and 2. All of the accessions in groups 1 and 2 for which we have phenotypic data are characterized by red pericarps (consistent with carrying functional Rc alleles), or they have white pericarps resulting from the previously described Rc-s nonsense mutation (Table S1). Thus, it appears unlikely that any of the previously published accessions within groups 1 and 2 would carry the 14-bp rc deletion.
Consequences for genetic and phenotypic diversity:
The cumulative effects of these multiple contributors to Malaysian weedy rice are evident not only in the Rc phylogeny (Figure 1 and Figure S1) but also in levels of nucleotide diversity at this locus. For both silent sites and all sites across the Rc gene, the average pairwise nucleotide diversity (π) of the Malaysian weeds exceeds that of the cultivated rice samples, and even exceeds that of a geographically wide sample of wild rice (O. rufipogon) (Table 2 and Table S5). This high diversity provides a particularly striking contrast to Rc variation previously observed in United States weedy rice (Gross et al. 2010a), where the reported nucleotide diversity (π/kb = 0.34 and 0.41 for total and silent sites, respectively) is <30% of the values observed here. This large difference in Rc diversity between United States and Malaysian weeds is likely to reflect not only the more dynamic crop–weed gene flow interactions that can occur in Southeast Asia, but also the sharp demographic bottleneck that occurred with the introduction of weedy rice into North America from Asia (Reagon et al. 2010).
Rc and weedy rice adaptation
The disproportionate representation of functional (wild-type) Rc alleles in Malaysian weedy rice compared to other domestication genes (Figure 3) is consistent with Rc serving an adaptive function in weed populations. Pericarp pigmentation could conceivably provide camouflage for weed seeds in crop fields (Vigueira et al. 2013 a,b), or protection from abiotic and biotic stress (Ithal and Reddy 2004; Martínez-Castillo et al. 2008). However, the adaptive value of functional Rc alleles may be more likely to come from the gene’s effects in maintaining seed dormancy. The ability of agricultural weed seeds to remain dormant in the soil over multiple planting seasons plays a key role in their persistence and proliferation (Delouche and Labrada 2007; Vigueira et al. 2013b), and the Rc locus has been identified as one of the two major effect QTL for seed dormancy in United States weedy rice (Subudhi et al. 2012). While the functional effects of Rc sequence variation on seed dormancy have not been explicitly examined, it is reasonable to expect that wild-type sequences that can successfully regulate proanthocyanidin synthesis would also be functional in establishing seed dormancy. Phenotypic assays of seed dormancy in the Malaysian weeds would be useful for testing this prediction.
If functional Rc alleles are, as proposed, adaptive for seed dormancy in Malaysian weed populations, the question then arises as to why 17% of the sampled accessions were homozygous for the loss-of-function rc allele. One possibility is that these accessions represent recently derived, maladapted descendants of crop–weed hybridization that are unlikely to persist over multiple generations. The observation of four Rc/rc heterozygotes in these highly selfing weeds supports the hypothesis that the crop-derived rc allele is of recent origin in the weeds, and may be only transiently present. Studies that explicitly examine the relationships between Rc sequence variation, gene expression, phenotypic variation (both seed dormancy and pericarp color), and fitness in field plantings would be valuable for definitively establishing the adaptive role of Rc alleles in weedy rice.
Malaysian weedy rice: a Trojan horse for contamination of crop and wild germplasm?
Our inferences for the Rc locus, together with previous neutral-marker analyses (Song et al. 2014), strongly suggest that both cultivated and wild rice populations are contributing to the genetic composition of Malaysian weedy rice through hybridization, and that this may be contributing to its adaptation and rapid proliferation. If Malaysian weeds are, in fact, a crucible where crop and wild alleles can recombine, the weeds could also potentially serve as a bridge for the introduction of crop alleles into wild populations (Snow et al. 2010; Campbell et al. 2016) or vice versa (Félix et al. 2014). Of these two gene flow possibilities, wild-to-crop gene flow may be the less serious problem. Although in principle, the proximity of weedy rice to crops within rice fields could allow them to serve as a Trojan horse for the introgression of undesirable wild traits, crop seed-stock certification can be expected to minimize the contamination of rice germplasm. More problematic is the possibility of genetic swamping of the wild progenitor populations (e.g., Martínez-Castillo et al. 2008; Cornille et al. 2015; Fuchs et al. 2016), which, in the case of Malaysian O. rufipogon, are increasingly rare and have no in situ conservation status. Introgression from cultivated rice into weedy rice is not uncommon (Xia et al. 2011; Ellstrand et al. 2013; Jiang et al. 2012), and the recent widespread adoption of herbicide-resistant rice production will likely increase hybridization due to selection for introgression of resistance alleles into weed populations (Burgos et al. 2014). Combined with the recent demographic explosion in weedy rice populations across Southeast Asia (Azmi et al. 2013; Pusadee et al. 2013), weedy rice-mediated genetic erosion of wild O. rufipogon may become an increasing concern in this region.
Conclusions
Our results show that weedy rice in Peninsular Malaysia appears to have diverse origins, resulting in high Rc genetic diversity in the weeds. The multiple-origin model of Rc alleles in Malaysian weedy rice is in line with co-occurrence of cultivated and wild rice at the edges of rice fields in this region. This is also consistent with the previous analyses of domestication genes of Malaysian weedy populations, where multiple origins were demonstrated for alleles of the sh4 and Bh4 domestication genes (Song et al. 2014). The disproportionate representation of functional Rc alleles in Malaysian weeds compared to other domestication genes suggests that crop–weed gene flow may differentially promote high frequencies of functional Rc alleles. With the potential disadvantageous features incurred by the domestication rc allele (e.g., lack of seed dormancy), we predict that the nonfunctional rc alleles may be purged from the weedy populations unless continued crop–weed hybridization maintains their presence. Future work is needed, however, to assess the relationship between Rc expression and its full range of phenotypic and fitness effects. A follow-up study should also consider whether and how crop–weed introgression may differentially shape allele frequencies at other rice domestication genes. The evidence that red-pericarp cultivated rice, possibly the traditional landraces planted before the cultivation of modern elite varieties, may be one of the potential origins of weedy rice should be an alarming signal to the rice industry. A practical implication of our work is that more stringent seed-stock certification guidelines should be in place in local rice breeding programs, in order to avoid severe and long-term impacts that could compromise crop productivity.
Supplementary Material
Acknowledgments
We thank Linda Small for technical assistance and the Olsen laboratory group for helpful comments on the manuscript. We acknowledge Washington University greenhouse staff for plant care support. Y.C. thanks the China Scholarship Council for financial support and Qianming Huang for guidance in scientific writing. Having worked on this study during a research sojourn in the United States, B.K.S. is grateful to Washington University in St. Louis and Monash University Malaysia for hospitality and support. Funding for this project was provided by grants from the United States National Science Foundation (IOS-1032023) to K.M.O., A.L.C., and Y.J. and the Malaysian Ministry of Education (FRGS/1/2015/ST03/MUSM/02/1) to B.K.S. The United States Department of Agriculture is an equal opportunity provider and employer.
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.035881/-/DC1.
Communicating editor: J. Ross-Ibarra
Literature Cited
- Ahmed Q. N., 2012. Vegetative and reproductive growth of weedy rice in Selangor, Malaysia: a comparative study with commercial rice varieties. Malaysian Applied Biology 41: 29–35. [Google Scholar]
- Anuar N. H. S., Mazlan N., Ariff E. A. K. E., Juraimi A. S., Yusop M. R., 2014. A comparative study of vegetative and reproductive growth of local weedy and Clearfield® rice varieties in Malaysia. J. ISSAAS. 20: 41–51. [Google Scholar]
- Azmi, M., and B. Baki, 2002 Impact of continuous direct seeding rice culture on weed species diversity in the Malaysian rice ecosystem. Proceedings of Regional Symposium on Environment and Natural Resources (Kuala Lumpur, Malaysia April 10–11, 2002), Universiti Kebangsaan Malaysia, Bangi, Vol. 1, pp. 61–67. [Google Scholar]
- Burgos N. R., Singh V., Tseng T. M., Black H., Young N. D., et al. , 2014. The impact of herbicide-resistant rice technology on phenotypic diversity and population structure of United States weedy rice. Plant Physiol. 166: 1208–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell L. G., Lee D., Shukla K., Waite T. A., Bartsch D., 2016. An ecological approach to measuring the evolutionary consequences of gene flow from crops to wild or weedy relatives. Appl. Plant Sci. 4: 1500114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C., Motohashi R., Tsuchimoto S., Fukuta Y., Ohtsubo H., et al. , 2003. Polyphyletic origin of cultivated rice: based on the interspersion pattern of SINEs. Mol. Biol. Evol. 20: 67–75. [DOI] [PubMed] [Google Scholar]
- Clement M., Posada D., Crandall K. A., 2000. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9: 1657–1659. [DOI] [PubMed] [Google Scholar]
- Cornille A., Feurtey A., Gélin U., Ropars J., Misvanderbrugge K., et al. , 2015. Anthropogenic and natural drivers of gene flow in a temperate wild fruit tree: a basis for conservation and breeding programs in apples. Evol. Appl. 8: 373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalrymple D. G., 1986. Development and Spread of High-Yielding Rice Varieties in Developing Countries. Agency for International Development, Washington, DC. [Google Scholar]
- Delouche J. C., Labrada R., 2007. Weedy rices: origin, biology, ecology and control. Food Agr. Org. 188: 45–62. [Google Scholar]
- Ellstrand N. C., Meirmans P., Rong J., Bartsch D., Ghosh A., et al. , 2013. Introgression of crop alleles into wild or weedy populations. Annu. Rev. Ecol. Evol. Syst. 44: 325–345. [Google Scholar]
- Estorninos L. E., Gealy D. R., Talbert R. E., Gbur E. E., 2005. Rice and red rice interference. I. Response of red rice (Oryza sativa) to sowing rates of tropical japonica and indica rice cultivars. Weed Sci. 53: 676–682. [Google Scholar]
- Félix D. T., Coello-Coello J., Martínez-Castillo J., 2014. Wild to crop introgression and genetic diversity in lima bean (Phaseolus lunatus L.) in traditional Mayan milpas from Mexico. Conserv. Genet. 15: 1315–1328. [Google Scholar]
- Fu Y.-X., Li W.-H. 1993. Statistical tests of neutrality of mutations. Genetics. 133: 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E. J., Martínez A. M., Calvo A., Muñoz M., Arrieta Espinoza G., 2016. Genetic diversity in Oryza glumaepatula wild rice populations in Costa Rica and possible gene flow from O. sativa. PeerJ 4: e1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gealy D. R., Agrama H., Jia M. H., 2012. Genetic analysis of atypical US red rice phenotypes: indications of prior gene flow in rice fields? Weed Sci. 60: 451–461. [Google Scholar]
- Gross B. L., Skare K. J., Olsen K. M., 2009. Novel Phr1 mutations and the evolution of phenol reaction variation in US weedy rice (Oryza sativa L.). New Phytol. 184: 842–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross B. L., Reagon M., Hsu S. C., Caicedo A. L., Jia Y., et al. , 2010a Seeing red: the origin of grain pigmentation in US weedy rice. Mol. Ecol. 19: 3380–3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross B. L., Steffen F. T., Olsen K. M., 2010b Molecular basis of white pericarps in African domesticated rice: novel mutations at the Rc gene. J. Evol. Biol. 23: 2747–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X. Y., Kianian S. F., Foley M. E., 2004. Multiple loci and epistases control genetic variation for seed dormancy in weedy rice (Oryza sativa). Genetics 166: 1503–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X. Y., Foley M. E., Horvath D. P., Anderson J. V., Feng J., et al. , 2011. Association between seed dormancy and pericarp color is controlled by a pleiotropic gene that regulates abscisic acid and flavonoid synthesis in weedy red rice. Genetics 189: 1515–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulick P., Lee D., Lupotto E., Powell W., 2009. G-string slippage turns white rice red. Genome 52: 490–493. [DOI] [PubMed] [Google Scholar]
- Hall T. A., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41: 95–98. [Google Scholar]
- Ithal N., Reddy A. R., 2004. Rice flavonoid pathway genes, OsDfr and OsAns, are induced by dehydration, high salt and ABA, and contain stress responsive promoter elements that interact with the transcription activator, OsC1-MYB. Plant Sci. 166: 1505–1513. [Google Scholar]
- Jiang Z., Xia H., Basso B., Lu B. R., 2012. Introgression from cultivated rice influences genetic differentiation of weedy rice populations at a local spatial scale. Theor. Appl. Genet. 124: 309–322. [DOI] [PubMed] [Google Scholar]
- Karim R. S., Man A. B., Sahid I. B., 2004. Weed problems and their management in rice fields of Malaysia: an overview. Weed Biol. Manage. 4: 177–186. [Google Scholar]
- Khush G. S., 1997. Origin, dispersal, cultivation and variation of rice. Plant Mol. Biol. 35: 25–34. [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., et al. , 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Qiang S., Song X., Cai K., Sun Y., et al. , 2014. Allele types of Rc gene of weedy rice from Jiangsu Province, China. Rice Sci. 21: 252–261. [Google Scholar]
- Librado Sanz P., Rozas Liras J. A., 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25(11): 1451–1452. [DOI] [PubMed] [Google Scholar]
- Luo J., Liu H., Zhou T., Gu B., Huang X., et al. , 2013. An-1 encodes a basic helix-loop-helix protein that regulates awn development, grain size, and grain number in rice. Plant Cell 25: 3360–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Castillo J., Colunga-GarcíaMarín P., Zizumbo-Villarreal D., 2008. Genetic erosion and in situ conservation of lima bean (Phaseolus lunatus L.) landraces in its Mesoamerican diversity center. Genet. Resour. Crop Evol. 55: 1065–1077. [Google Scholar]
- Merotto A., Goulart I., Nunes A., Kalsing A., Markus C., et al. , 2016. Evolutionary and social consequences of introgression of non‐transgenic herbicide resistance from rice to weedy rice in Brazil. Evol. Appl. 9: 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R. K., Jain M., 2012. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7: e30619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusadee T., Schaal B. A., Rerkasem B., Jamjod S., 2013. Population structure of the primary gene pool of Oryza sativa in Thailand. Genet. Resour. Crop Evol. 60: 335–353. [Google Scholar]
- Reagon M., Thurber C. S., Gross B. L., Olsen K. M., Jia Y., et al. , 2010. Genomic patterns of nucleotide diversity in divergent populations of US weedy rice. BMC Evol. Biol. 10: 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H., 1999. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132: 365–386. [DOI] [PubMed] [Google Scholar]
- Snow A., Culley T., Campbell L., Sweeney P. M., Hegde S. G., et al. , 2010. Long‐term persistence of crop alleles in weedy populations of wild radish (Raphanus raphanistrum). New Phytol. 186: 537–548. [DOI] [PubMed] [Google Scholar]
- Song B. K., Chuah T. S., Tam S. M., Olsen K. M., 2014. Malaysian weedy rice shows its true stripes: wild Oryza and elite rice cultivars shape agricultural weed evolution in Southeast Asia. Mol. Ecol. 23: 5003–5017. [DOI] [PubMed] [Google Scholar]
- Subudhi P. K., Parco A., Singh P. K., DeLeon T., Karan R., et al. , 2012. Genetic architecture of seed dormancy in U.S. weedy rice in different genetic backgrounds. Crop Sci. 52: 2564–2575. [Google Scholar]
- Sudinato E., Neik T. X., Tam S. M., Chuah T., Idris A. A., et al. , 2016. Morphology of Malaysian weedy rice (Oryza sativa): diversity, origin and implications for weed management. Weed Sci. 64: 501–512. [Google Scholar]
- Sweeney M. T., Thomson M. J., Pfeil B. E., McCouch S., 2006. Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell 18: 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney M. T., Thomson M. J., Cho Y. G., Park Y. J., Williamson S. H., et al. , 2007. Global dissemination of a single mutation conferring white pericarp in rice. PLoS Genet. 3: e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigueira C., Li W., Olsen K. M., 2013a The role of Bh4 in parallel evolution of hull colour in domesticated and weedy rice. J. Evol. Biol. 26: 1738–1749. [DOI] [PubMed] [Google Scholar]
- Vigueira C. C., Olsen K. M., Caicedo A. L., 2013b The red queen in the corn: agricultural weeds as models of rapid adaptive evolution. Heredity 110: 303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahab A., Suhaimi O., 1991. Padi angin characteristics, adverse effects and methods of its eradication. Teknologi Padi. 7: 21–31. [Google Scholar]
- Wang H., Xu X., Vieira F. G., Xiao Y., Li Z., et al. , 2016. The power of inbreeding: NGS based GWAS of rice reveals convergent evolution during rice domestication. Mol. Plant 9: 975–985. [DOI] [PubMed] [Google Scholar]
- Warwick S. I., Stewart C, 2005. Crops come from wild plants: how domestication, transgenes, and linkage together shape ferality, pp. 9–30 in Crop Ferality and Volunteerism, edited by Gressel J. CRC Press, Boca Raton, FL. [Google Scholar]
- Xia H. B., Wang W., Xia H., Zhao W., Lu B. R., 2011. Conspecific crop-weed introgression influences evolution of weedy rice (Oryza sativa f. spontanea) across a geographical range. PLoS One 6: e16189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Newly generated DNA sequences are available in GenBank (accession nos. KX549104–KX549259). Table S1 contains IDs, phenotypes, and genotypes of newly characterized accessions, including Rc genotypes confirmed by direct sequencing of exon 7 and An-1 genotype data. Table S2 contains previously published data included in analyses. The Rc alignment used in phylogeny construction is available on Dryad (doi: 10.5061/dryad.631kf).