Abstract
Dendritic cells (DCs) play critical roles in activating innate immune cells and initiating adaptive immune responses. The functions of DCs were originally obscured by their overlap with other mononuclear phagocytes, but new mouse models have allowed for the selective ablation of subsets of DCs and have helped to identify their non-redundant roles in the immune system. These tools have elucidated the functions of DCs in host defense against pathogens, autoimmunity, and cancer. This review will describe the mouse models generated to interrogate the role of DCs, and will discuss how their use has progressively clarified our understanding of the unique functions of DC subsets.
eTOC
Durai et al review the progress made in developing new mouse models for the analysis of the functions of dendritic cell subsets and what these models have revealed about the roles of these cells in immune responses
Introduction
The vertebrate immune system has evolved the remarkable capacity to robustly and precisely eliminate the wide variety of potential threats it encounters, from single cell bacteria to multicellular parasites to even transformed oncogenic versions of its own cellular components. To achieve this goal, many diverse lineages of effector cells must act together in different capacities throughout the course of the immune response. As with any system possessing such complexity, the careful control and coordination of the numerous components of the immune system is critical for its proper functioning. As our understanding of each cell type acting within this system has grown, it has become increasingly apparent that dendritic cells (DCs) act as the central regulators of the entire immune response, responsible both for sensing the nature of the threats faced and for activating the precise combination of effectors required to eradicate them.
First isolated by Ralph Steinman and Zanvil Cohn, DCs were identified by their stellate morphology and capacity to stimulate naïve T cells (Steinman and Cohn, 1973; Steinman and Witmer, 1978; Nussenzweig et al., 1980). DCs comprise two major branches, the classical DCs (cDCs) identified by Steinman and the lymphocyte-like plasmacytoid DCs (pDCs) that produce Type 1 interferon in response to pathogens (Perussia et al., 1985; Heath and Carbone, 2009; Cella et al., 1999; Siegal et al., 1999). cDCs can be further divided into two major subsets recently renamed cDC1s and cDC2s (Guilliams et al., 2014). All DCs originate from bone marrow (BM) progenitors arising from hematopoietic stem cells, starting with the macrophage/dendritic cell progenitor (MDP) (Fogg et al., 2006; Auffray et al., 2009), which gives rise to the common dendritic cell progenitor (CDP) (Naik et al., 2007; Onai et al., 2007), which finally gives rise to committed progenitors for each branch of DC such as the pre-cDC1 and the pre-cDC2 (Grajales-Reyes et al., 2015; Schlitzer et al., 2015).
cDCs express the integrin CD11c and MHC class II (Steinman et al., 1979; Metlay et al., 1990), and each subset can be distinguished by additional markers. Resident cDC1s in the spleen and lymph nodes (LNs) express CD8α, CD24, and XCR1, while cDC2s express CD4 and Sirpα (Mildner and Jung, 2014; Murphy et al., 2016). In nonlymphoid tissues, all cDCs express CD24, which distinguishes them from macrophages that instead express CD64 (Schlitzer et al., 2013; Plantinga et al., 2013; Langlet et al., 2012). Nonlymphoid tissue cDC1s also express XCR1 and CD103, while cDC2s express CD11b and Sirpα. Migratory cDCs that traffic from nonlymphoid tissues to LNs express these same markers they expressed in the periphery. There are several exceptions to these rules, however, such as CD11b+ cDC2s in the small intestine that comprise both CD103+ and CD103− fractions (Bogunovic et al., 2009; Satpathy et al., 2013). While these varied markers have historically been used to identify cDC subsets, a recent analysis suggests that a more simple and consistent identification of these cells across most tissues is possible by gating cDCs as CD11c+MHCII+CD26+CD64−F4/80−, and within this population cDC1s as XCR1+ and cDC2s as Sirpα+ (Guilliams et al., 2016). pDCs also express CD11c and MHCII, but can be segregated by their additional expression of B220, Siglec-H, and Bst2 (Blasius et al., 2006; Zhang et al., 2006).
While cDCs were discovered for their ability to serve as potent antigen-presenting cells (APCs), it is now clear that they also have non-redundant roles in innate immune responses (Mashayekhi et al., 2011; Satpathy et al., 2013). Their early recognition of pathogens and rapid cytokine production activates innate immune cells such as innate lymphoid cells (ILCs) and natural killer (NK) cells to limit pathogen spread until adaptive immunity can be initiated. Indeed, the heterogeneity of cDCs can itself be viewed as an evolutionary adaptation for the coordinated activation of the specific innate and adaptive effector responses best suited to control various forms of pathogens (Briseno et al., 2014). cDC1s, for example, recognize intracellular pathogens and initiate type 1 immune responses that require the early activation of ILC1s and NK cells as well as eventual Th1 polarization (Mashayekhi et al., 2011). Some cDC2s, on the other hand, govern type 2 immune responses against parasites in which they activate ILC2s and Th2 cells (Tussiwand et al., 2015). Other cDC2s sense extracellular bacteria and initiate type 3 immune responses by activating ILC3s and Th17 cells (Lewis et al., 2011; Satpathy et al., 2013). In this way, each cDC subset acts as the gatekeeper for a specific module of the immune response, recognizing a particular type of threat and activating both the innate and adaptive defenses best suited for overcoming it.
Since the time of their discovery, there has been considerable debate over whether DCs possess unique functions or whether they are redundant with macrophages and other mononuclear cells (Hume, 2008). Fortunately, increasingly sophisticated mouse models for the specific ablation of DCs or for the conditional deletion of genes in these cells have helped resolve some of these controversies, revealing how immune responses are compromised in the absence of DCs and thus the roles these cells play in the normal response. These mouse models have been critical for elucidating the unique functions of DCs and resolving previously ambiguous conclusions. This review will provide an overview of the mouse models that have been developed to interrogate DC function and how these models have progressively clarified the role of DCs in various types of immune responses. As recent reviews have discussed DC development (Murphy et al., 2016; Mildner and Jung, 2014), this review will focus primarily on how DCs function in immune responses to pathogens, autoimmunity, and cancer.
Mouse models for studying dendritic cells
Numerous models of constitutive and inducible DC depletion have been generated and used to identify the specific functions of DC subsets. In this section, we will first describe the different strains developed, their specificity, and their limitations. We will organize this by categories of DTR/DTA based models, Cre strains, and transcription factor knockout mice. In the next section we will discuss the discoveries made with these models regarding the functions of DCs in various immune responses.
Depletion of dendritic cells by DTR/DTA systems
The first models of genetic ablation of cell lineages were transgenic mice in which cell type-specific promoters drove expression of the diphtheria toxin (DT) A-chain (Palmiter et al., 1987; Breitman et al., 1987). This toxin disrupts protein translation by catalyzing the ADP-ribosylation of polypeptide chain elongation factor 2 and eventually leads to cell death (Honjo et al., 1968; Robinson et al., 1974; Van Ness et al., 1980). While DT efficiently ablates the cells in which it is expressed, it can be problematic as even low levels of off-target expression can lead to death of unintended cells or even to effects on embryogenesis or morphogenesis (Breitman et al., 1990). Later mouse models overcame this problem by expressing the human or simian DT receptor (DTR) under the control of a cell type-specific promoter, with subsequent administration of DT to these mice (Saito et al., 2001). As the mouse ortholog of the DTR is orders of magnitude less sensitive to DT (Mitamura et al., 1995; Pappenheimer, Jr. et al., 1982), this allows for the efficient depletion of only DTR-expressing cells and has the added benefit of allowing for inducible depletion of these target cells rather than constitutive ablation.
The first DTR based model used for DCs was the Itgax-DTR strain, a transgenic line expressing a DTR-GFP fusion protein under the control of the murine promoter for the gene Itgax, which codes for the integrin CD11c (Jung et al., 2002). DT administration completely depleted CD11c+ DCs within 24 hours, with no observable depletion of splenic B cells or F4/80+ macrophages. DCs began to reappear 3 days after DT treatment. While this strain was vital for early work confirming the functions of DCs in T cell priming and pathogen responses, several limitations have emerged. First, repeated administration of DT led to death in these mice, restricting the duration through which depletion could be studied (Jung et al., 2002). This lethality was likely due to off-target expression of the Itgax-DTR transgene in radioresistant or non-hematopoietic tissues, since BM chimeras of WT BM into Itgax-DTR recipients also died after repeated DT treatment. This limitation can be overcome by generating chimeras of Itgax-DTR BM into WT recipients, which tolerate repeated DT treatment (Zaft et al., 2005). Another caveat derives from CD11c expression by non-DCs and the depletion by DT of such cells, which include splenic metallophilic and marginal zone macrophages, LN sinusoidal macrophages (Probst et al., 2005), alveolar macrophages (van Rijt et al., 2005), activated CD8 T cells (Jung et al., 2002), and plasma cells (Hebel et al., 2006). Ablation of these cells complicates analysis with this strain, since phenotypes observed may result from their depletion rather than that of DCs. Some CD11c+ cells, notably pDCs, are not depleted in this model, indicating that the level of CD11c expressed by cells influences whether they are depleted (Sapoznikov et al., 2007).
A similar DTR strain, Itgax-DOG (Hochweller et al., 2008), is a BAC transgenic line in which the Itgax promoter drives expression of a fusion protein composed of DTR, a portion of ovalbumin, and GFP (DOG). Continuous DT treatment in these mice does not cause lethality, perhaps due to more faithful expression of the BAC compared with the promoter-based transgene. Depletion of splenic macrophages was also observed after DT treatment in these mice, but whether CD8 T cells and plasma cells were similarly affected has not been evaluated.
The recently developed Zbtb46-DTR mouse allows for more specific depletion of cDCs (Meredith et al., 2012). This strain has sequences for an internal ribosome entry site (IRES) and a DTR-mCherry fusion protein inserted into the 3′ untranslated region (UTR) of the endogenous Zbtb46 gene, which codes for the zinc finger transcription factor Zbtb46 (also known as zDC) whose expression in hematopoietic lineages is restricted to cDCs. However, a single administration of DT into Zbtb46-DTR mice or into chimeras of WT BM into Zbtb46-DTR recipients proved lethal, indicating zDC expression in vital radioresistant or non-hematopoietic cells. Further work indicated these vital cells may be endothelial cells (Satpathy et al., 2012). DT treatment of Zbtb46-DTR BM into WT recipient chimeras depleted cDCs alone without ablation of B cells, T cells, neutrophils, natural killer cells, pDCs, or monocytes. This strain therefore allows for the specific and complete deletion of cDCs, with the caveat that BM chimeras must be used. To circumvent this need, the Zbtb46-LSL-DTR mouse was generated (Loschko et al., 2016a), in which the IRES-mCherry-DTR is preceded by a loxP flanked transcriptional Stop cassette (LSL) that terminates transcription of the locus before the DTR. Transcription of the DTR proceeds only after Cre-mediated excision of the Stop cassette. After crossing the Zbtb46-LSL-DTR strain to a Csf1r-cre strain (Loschko et al., 2016a), DT administration specifically depleted cDCs and did not lead to lethality. This model allows for the continuous depletion of cDCs for at least 4 weeks without the need for BM chimeras.
Several DTR strains allow for depletion of specific DC subsets. The first were two strains that expressed DTR from the endogenous Cd207 gene that codes for langerin, a C-type lectin expressed on epidermal Langerhans cells and dermal cDC1s. One strain was generated by inserting an IRES DTR-egfp into the 3′ UTR of the Cd207 gene (Kissenpfennig et al., 2005), while in the second strain the first Cd207 coding exon was replaced with a DTR-egfp cassette (Bennett et al., 2005). In both strains, DT treatment depletes Langerhans cells as well as cDC1s in skin-draining LNs. Additionally, a BAC transgenic CD207-DTA mouse strain was generated using a BAC in which an IRES DTA was inserted into the 3′ UTR of the human CD207 gene that codes for langerin (Kaplan et al., 2005). This strain has constitutive ablation of Langerhans cells but not of cDC1s in skin-draining LNs, suggesting that the human CD207 promoter is controlled differently from its murine equivalent. It was later demonstrated that these mice also lack CD103+CD11b+ cDC2s in the intestinal lamina propria (Welty et al., 2013).
The Ly75-DTR strain was generated by inserting an IRES DTR-eGFP into the 3′ UTR of the Ly75 gene that codes for CD205, an endocytic type I C-type lectin-like receptor also known as DEC-205 (Fukaya et al., 2012). DT administration to this mouse also resulted in lethality, necessitating the use of Ly75-DTR into WT recipient BM chimeras. DT treatment of such chimeras depleted CD205+ cDCs, the majority of which were CD8α+ cDC1s. CD205 is also expressed on B cells at 10–50 fold lower levels than on BM derived DCs (Inaba et al., 1995) and accordingly a minor decrease in splenic B cells was observed upon DT treatment to Ly75-DTR mice. However, CD205 is also expressed at much higher levels in germinal center (GC) B cells (Victora et al., 2010) as well as all migratory cDCs and Langerhans cells (Idoyaga et al., 2013), but it was not determined whether DT treatment also depleted these cells.
The Clec9a-DTR BAC transgenic mouse, in which the first Clec9a coding exon in the BAC was replaced with DTR, also allows for depletion of cDC1s (Piva et al., 2012). DT treatment completely ablates cDC1s, but also causes a ~50% decrease in pDCs, which may be due to the lower expression of Clec9a in mature pDCs (Sancho et al., 2008; Caminschi et al., 2008). Clec9a is expressed in the common dendritic cell progenitor (CDP) (Schraml et al., 2013), so continuous DT treatment may deplete this progenitor and therefore all DCs over prolonged periods. Though this issue was not examined in the original study, at least one report indicated that cDC2s were unaffected after 15 days of DT treatment (Muzaki et al., 2016).
Two more recent DTR strains specifically deplete cDC1s. In the Xcr1-DTRvenus strain, the first Xcr1 coding exon was replaced by a cassette encoding a DTR-venus fusion protein (Yamazaki et al., 2013). XCR1 is a chemokine receptor uniquely expressed by cDC1s in humans, mice, and sheep (Crozat et al., 2010; Crozat et al., 2011). DT treatment to this strain completely depleted cDC1s with no decrease in T cells, B cells, NK cells, granulocytes, monocytes, cDC2s, or pDCs. Expression of the Venus fluorescent protein was likewise restricted to cDC1s. This subset was depleted by 24 hours after DT treatment and began to recover on day 4 after treatment. A similar mouse strain, referred to as Karma, has an IRES tdTomato-2A-DTR inserted into the 3′ UTR of the endogenous a530099j19rik gene (Alexandre et al., 2016), which like XCR1 is highly specific for cDC1s. Both tdTomato expression and cell ablation after DT treatment were specific to cDC1s.
The Clec4a4-DTR strain, a BAC transgenic line in which an IRES DTR was inserted into the 3′ UTR of the Clec4a4 gene on the BAC, allows for ablation of cDC2s (Muzaki et al., 2016). Clec4a4 (also known as DCIR2) is a C-type lectin expressed by cDC2s and recognized by the 33D1 antibody first used to define this subset (Nussenzweig et al., 1982; Dudziak et al., 2007). DT treatment in this strain severely reduces CD103+CD11b+ cDC2s in the colonic lamina propria and CD11b+ cDC2s in the mesenteric LNs, and partially depletes CD103−CD11b+ cDC2s and CD64+CX3CR1+ macrophages in the colonic lamina propria. Whether depletion of other cells occurs and whether cDC2s in other organs are similarly depleted remains to be determined. A second strain thought to deplete a subset of cDC2s is the Mgl2-DTR strain, in which the first coding exon of the endogenous Mgl2 gene was replaced by a DTR-gfp cassette (Kumamoto et al., 2013). DT treatment in this strain depletes the Mgl2+ population of dermal cDC2s, but whether DCs in other tissues are also depleted remains to be analyzed.
Several DTR strains have been generated for pDC depletion. The first was the CLEC4C-DTR BAC transgenic strain, which replaced the exons of the human gene CLEC4C in the BAC with the sequence for DTR (Swiecki et al., 2010). CLEC4C codes for BDCA-2, a C-type lectin uniquely expressed by human pDCs with no equivalent in mouse pDCs, but its human promoter remains active in mouse pDCs as DT treatment in CLEC4C-DTR mice specifically and completely depleted pDCs. No reduction was observed in B cells, cDCs, T cells, NK cells, macrophages, monocytes, or neutrophils. Second, a Siglech-DTR strain was generated by knocking an IRES DTR-egfp sequence into the 3′ UTR of the endogenous murine Siglech gene (Takagi et al., 2011). Homozygous Siglech-DTR mice lacked expression of Siglec-H on pDCs, implying that the IRES DTR cassette somehow altered native Siglec-H expression. In this strain, DT administration completely depletes pDCs in the spleen, mesenteric LNs, and BM, with no apparent effect on cDCs. However, Siglec-H deficiency has been associated with altered cytokine responses (Swiecki et al., 2014) and perhaps abnormal kidney and testes function (Orr et al., 2013), which may influence the phenotypes of this strain. Third, a Siglech-DTR BAC transgenic strain was generated on the Balb/c background by replacing the coding exons of Siglech in a BAC with the sequence for DTR (Piva et al., 2012). DT treatment in these mice completely depleted pDCs with no effect on cDCs. A separate BAC transgenic Siglech-DTR line on the C57BL/6 background was later generated, and analysis of both Siglech-DTR lines found that Siglec-H was also expressed by marginal zone macrophages and DT treatment ablated these cells as well as pDCs (Swiecki et al., 2014). This indicates that cell ablation in Siglech-DTR strains may not be limited to pDCs, complicating analysis with these lines.
Finally, several DTR based systems for the depletion of monocytes and macrophages (reviewed in (Lauvau et al., 2015)), have been generated that are driven by genes such as CCR2 and CX3CR1. These have helped segregate the independent functions of monocytes and cDCs. One useful model for this purpose is the Cx3cr1-LSL-DTR strain (Diehl et al., 2013), in which an LSL-DTR was inserted into the endogenous Cx3cr1 locus. This strain was crossed to Itgax-cre so that DT treatment specifically depletes CD11c+CX3CR1+ cells, which include CD103−CD11b+ cDC2s and monocyte-derived CD11b+ macrophages in the intestinal lamina propria (Bogunovic et al., 2009). Another useful model is the MM-DTR strain, which was a cross between the Csf1r-LSL-DTR and LysM-cre strains, and in which DTR expression is limited to monocytes and monocyte-derived macrophages (Schreiber et al., 2013). Table 1 provides a summary of DTR strains useful for the study of DC function.
Table 1.
Comparison of DTA/DTR based mouse models for dendritic cell depletion
| Strain | Cells Depleted | Caveats |
|---|---|---|
| Itgax-DTR (Jung et al., 2002) | cDCs, metallophilic, marginal zone, sinusoidal, and alveolar macrophages, activated CD8 T cells, plasma cells | Repeated DT treatment leads to death, requires CD11c-DTR into WT BM chimeras No depletion of pDCs |
| Itgax-DOG (Hochweller et al., 2008) | cDCs, splenic macrophages | Depletion of other cell types not evaluated |
| Zbtb46-DTR (Meredith et al., 2012) | cDCs | DT treatment leads to death, requires zDC-DTR into WT BM chimeras |
| Zbtb46-LSL-DTR (Loschko et al., 2016a) | cDCs | Requires crossing to Cre strain active in cDC lineage |
| Cd207-DTR (Knockin) (Kissenpfennig et al., 2005) | Langerhans cells, cDC1s | |
| Cd207-DTR (BAC) (Bennett et al., 2005) | Langerhans cells, cDC1s | |
| CD207-DTA (Kaplan et al., 2005) | Langerhans cells, CD103+CD11b+ cDC2s in small intestine | |
| Ly75-DTR (Fukaya et al., 2012) | cDC1s, ~15% splenic B cells | DT treatment leads to death, requires CD205-DTR into WT BM chimeras Depletion of germinal center B cells, migratory cDCs, and Langerhans cells untested but possible |
| Clec9a-DTR (Piva et al., 2012) | cDC1s, ~50% pDCs | |
| Xcr1-DTRvenus (Yamazaki et al., 2013) | cDC1s | |
| Karma (a530099j19rik-DTR) (Alexandre et al., 2016) | cDC1s | |
| Clec4a4-DTR (Muzaki et al., 2016) | cDC2s and CD64+CX3CR1+ macrophages in intestinal lamina propria and MLNs | DC depletion in other organs not analyzed |
| Mgl2-DTR (Kumamoto et al., 2013) | Mgl2+ dermal cDC2s | |
| CLEC4C-DTR (Swiecki et al., 2010) | pDCs | |
| Siglech-DTR (Knockin) (Takagi et al., 2011) | pDCs | Loss of SiglecH expression in homozygous knockin mice Depletion of marginal zone macrophages untested but possible |
| Siglech-DTR (BAC) (Piva et al., 2012) | pDCs, marginal zone macrophages | |
| Cx3cr1-LSL-DTR Itgax-cre (Diehl et al., 2013) | CD103−CD11b+CX3CR1+ cDC2s, CD11b+CX3CR1+ macrophages | Requires crossing to Cre strain active in cDCs |
| MM-DTR (Schreiber et al., 2013) | Monocytes and monocyte-derived macrophages |
An important caveat for some DTR strains is the reduced LN cellularity seen even without DT treatment (van Blijswijk et al., 2015). In a cross between the Clec9a-cre strain and a Rosa26-LSL-DTR strain (Buch et al., 2005), reduced cellularity was observed in skin-draining and mesenteric LNs even without DT treatment. There was also reduced cellularity in skin-draining LNs of Itgax-DTR mice, Cd207-DTR mice, and a cross between Itgax-cre and Rosa26-LSL-DTR mice. There was no decrease observed in Itgax-DOG mice, so this phenomenon must be evaluated in each individual strain. The basis for this effect is unclear.
Cre strains for conditional deletion of genes in dendritic cells
The Cre-loxP system allows for conditional deletion of genes in specific cell lineages (Sharma and Zhu, 2014). In this system, two 34-bp loxP sites are inserted on either side of a gene or exon, which is then said to be “floxed”. Cell type-specific promoters are then used to express the bacteriophage P1 cre gene, which encodes an integrase that mediates recombination between two adjacent loxP sites, leading to deletion of the intervening DNA and inactivation of the floxed gene in Cre+ cells.
The earliest Cre strains developed for cDCs were based on CD11c expression. First was an Itgax-cre BAC transgenic line in which the first coding exon of Itgax was replaced with cre (Caton et al., 2007). When crossed to the Rosa26-LSL-yfp strain (Srinivas et al., 2001), in which Cre-mediated excision of a STOP cassette allows YFP expression, splenic DCs were >95% labeled and pDCs were ~86% labeled. However, background deletion of 5–12% was seen in splenic T, B, and NK cells. A later study also found deletion in 20–40% of blood monocytes, nearly ~100% of alveolar macrophages, 70% of splenic red pulp macrophages, 35% of marginal zone macrophages, and 20% of peritoneal macrophages (Abram et al., 2014). Therefore, as with Itgax-DTR strains, Itgax-cre is active in non-DC populations. The second Itgax-cre strain is a transgenic line expressing cre-IRES-gfp under the control of the Itgax promoter (Stranges et al., 2007). GFP expression in this line was found to be uniformly high in cDC1s and cDC2s as well as in pDCs, but no expression was seen in T cells or B cells. Whether deletion of genes occurred in cell types other than DCs, especially other CD11c+ populations such as macrophages, was not evaluated and remains to be resolved.
The Clec9a-cre strain was generated by replacing the first two exons of the endogenous Clec9a gene with cre (Schraml et al., 2013). Clec9a is first expressed at the CDP stage and is maintained in cDC1s and pDCs, but not in cDC2s (Schraml et al., 2013; Sancho et al., 2008; Caminschi et al., 2008). The Clec9a-cre strain achieved deletion in ~100% of cDC1s, ~50% of cDC2s, and ~20% of pDCs, suggesting that the transient expression of Cre at the CDP stage is insufficient for full deletion in cDC2 precursors and that pDCs do not express high enough levels of Cre for complete deletion. Deletion was not observed in any other cell types.
Recently a Zbtb46-cre line was produced by inserting an IRES-cre cassette into the 3′ UTR of the endogenous Zbtb46 gene, allowing for more specific deletion of genes in cDCs (Loschko et al., 2016b). When crossed to Rosa26-LSL-yfp mice, the Zbtb46-cre achieved deletion in ~65% of cDCs, which was lower than the ~100% deletion with Itgax-cre, but also demonstrated <10% deletion in T cells, B cells, and monocytes, which was also consistently lower than the deletion seen with Itgax-cre. Also <10% deletion was seen in red pulp macrophages, pDCs, and small intestinal macrophages, in contrast to the Itgax-cre strain where ~70% of red pulp macrophages and pDCs and ~100% of small intestinal lamina propria macrophages showed deletion. Zbtb46-cre also induced ~95% deletion of a floxed MHCII allele in cDCs, indicating that deletion of some alleles may be more complete than others. It was not evaluated whether Zbtb46-cre is as active in endothelial cells as the Zbtb46-DTR strain is.
The first Cre strain for targeting specific DC subsets was the Cd207-cre, in which a cre cassette was inserted into the second exon of the endogenous Cd207 gene (Zahner et al., 2011). This strain achieved nearly complete deletion in Langerhans cells and langerin+ cDC1s in the dermis, skin-draining LNs, and lung. A second Cre line useful for targeting particular DC subsets is the Xcr1-cre strain, in which the first exon of the endogenous Xcr1 gene was replaced by cre (Ohta et al., 2016). This was crossed to Rosa26-lacZbpAfloxDTA mice (Brockschnieder et al., 2006), in which Cre excises a loxP flanked lacZ-polyadenylation (bpA) sequence and allows for DTA expression and cell ablation. This cross resulted in nearly complete ablation of cDC1s in the spleen, MLN, and intestinal lamina propria, but no reduction in cDC2s in these organs. There was no reduction in CD4 or CD8 T cells in the spleen or MLN, but T cells and intraepithelial lymphocytes in the intestinal lamina propria were reduced, which was attributed to the absence of cDC1s. Finally, in the BAC transgenic Siglech-cre strain, the first exon of Siglech was replaced with a cre IRES mCherry cassette (Puttur et al., 2013). However, this strain only achieved deletion in ~30% of pDCs and ~2% of all lymphoid cells, and so is better suited for lineage tracing than functional analysis.
Transcription factor based depletion of dendritic cells
Several transcription factors have been identified whose deletion selectively depletes specific subsets of DCs, and these knockout mice provide useful models for studying DC function. One of the earliest identified was the Irf8−/− strain, which lacks cDC1s (Schiavoni et al., 2002; Aliberti et al., 2003), but also has impairments in B cells (Wang et al., 2008), monocytes (Kurotaki et al., 2013), eosinophils (Milanovic et al., 2008), and basophils (Sasaki et al., 2015). Irf8−/− mice were also thought to lack pDCs (Schiavoni et al., 2002), but more recently it was determined that pDCs do not require IRF8 for their development but instead that loss of this factor affects their expression of cell-surface markers and their ability to produce interferon (Sichien et al., 2016). Irf8−/− mice therefore do not serve as a model of specific cDC1 depletion, but this can be overcome by crossing a floxed Irf8 allele to the Itgax-cre strain (Luda et al., 2016), which depletes cDC1s and CD64+CD11b+ macrophages in the intestinal lamina propria, or to the Zbtb46-cre strain (Esterhazy et al., 2016), which specifically depletes cDC1s.
Id2−/− mice lack cDC1s (Hacker et al., 2003), but also have defects in NK cells (Yokota et al., 1999) and ILCs (Serafini et al., 2015). Nfil3−/− mice similarly lack cDC1s (Kashiwada et al., 2011), NK cells (Gascoyne et al., 2009), and ILCs (Geiger et al., 2014; Seillet et al., 2014). The similarity of Id2−/− and Nfil3−/− mice may result from an epistatic action of Nfil3 on the expression of Id2 during DC development, as has been suggested for NK cell development (Male et al., 2014).
Batf3−/− mice are selectively deficient in cDC1s (Hildner et al., 2008). Batf3 and the AP-1 transcription factor Jun form a heterodimer that interacts with IRF8 to bind AP-1/IRF consensus elements (AICEs) (Tussiwand et al., 2012; Glasmacher et al., 2012). Batf3 acts to maintain the expression of IRF8 by autoactivation during development of cDC1s, without which cDC1s divert to cDC2 fate (Grajales-Reyes et al., 2015). Batf3−/− mice on the 129SvEv and Balb/C backgrounds have an almost complete lack of cDC1s in lymphoid and nonlymphoid tissues (Edelson et al., 2010), but interestingly in the C57BL/6 background they retain cells resembling cDC1s in skin-draining LNs (Tussiwand et al., 2012).
Few known transcription factors selectively control cDC2 development. Relb−/− mice have DCs with reduced immunogenicity (Burkly et al., 1995), and reportedly a partial cell-intrinsic defect in cDC2 development (Wu et al., 1998). RelB acts in the noncanonical NFκB signaling pathway activated by lymphotoxin-β receptor (LTβR) signaling, and Ltbr−/− mice also have a cell-intrinsic but partial reduction in CD4+ cDC2s (Kabashima et al., 2005). It has not been tested whether the impact of RelB deficiency on DCs is secondary to deficient LTβR signaling. As Relb−/− mice develop severe multiorgan inflammation and hematopoietic abnormalities (Weih et al., 1995), and Ltbr−/− mice lack lymph nodes (Futterer et al., 1998), neither strain is a convenient model for testing DC function. Conditional deletion of Ltbr with Itgax-cre has been used to overcome this limitation (Tumanov et al., 2011).
Other factors that influence cDC2 development include Notch2 and Klf4. Conditional deletion of the Notch cofactor Rbpj (Caton et al., 2007) or Notch2 (Lewis et al., 2011; Satpathy et al., 2013) using Itgax-cre demonstrated a specific depletion of CD11b+ESAM+ cDC2s in the spleen and CD103+CD11b+ cDC2s in the intestinal lamina propria and MLNs, without apparent loss of other cell types. Notch2f/f Itgax-cre mice did exhibit gene expression changes in remaining cDC1s and cDC2s, however, indicating possible effects of Notch2 in these lineages (Satpathy et al., 2013). Conditional deletion of the transcription factor Klf4 with Itgax-cre also led to the depletion of subsets of cDC2s (Tussiwand et al., 2015) similar to migratory CD24− CD11b− cDC2s in skin-draining LNs (Ochiai et al., 2014).
Irf4−/− mice do not lack cDC2s, but instead have partial impairments in the number or functioning of these cells. CD11b+ cDC2s are present in the skin of Irf4−/− mice, but fail to migrate to LNs (Bajana et al., 2012; Bajana et al., 2016). Similarly, conditional deletion of Irf4 with Itgax-cre resulted in a partial reduction in CD11b+ cDC2s in the lung but an almost complete absence of CD11b+ cDC2s in the lung draining LNs (Schlitzer et al., 2013), as well as a partial reduction in CD103+CD11b+ cDC2s in the intestinal lamina propria and a more complete reduction in mesenteric LNs (Schlitzer et al., 2013; Persson et al., 2013). Finally, conditional deletion of Irf4 with Itgax-cre depletes a PDL2+Mgl2+ population of cDC2s in skin-draining LNs but not the dermis (Gao et al., 2013). But although Irf4f/f Itgax-cre mice can serve as a useful model for cDC2 deficiency, IRF4 also plays a role in other CD11c+ cells such as M2 macrophages (Satoh et al., 2010) and GM-CSF activated monocytes (Briseno et al., 2016), and these may also be affected in this strain.
Several transcription factors have been identified that regulate pDC development. The first model of specific pDC deficiency was conditional deletion of Tcf4, which encodes the factor E2-2, with Itgax-cre (Cisse et al., 2008). Recently, conditional deletion of the transcription factor Zeb2 using Itgax-cre also demonstrated selective pDC deficiency (Scott et al., 2016). Table 3 provides a summary of the transcription factor knockout mice useful for the study of DCs.
Table 3.
Comparison of transcription factor knockout mice that affect dendritic cell development
| Strain | Cells Depleted | Caveats |
|---|---|---|
| Irf8−/− (Schiavoni et al., 2002; Aliberti et al., 2003) | cDC1s, B cells, monocytes, eosinophils, basophils | Myeloid neoplasia eventually results with age Conditional deletion with Itgax-cre or Zbtb46-cre can prevent depletion of other cell types seen in Irf8−/− mice |
| Id2−/− (Hacker et al., 2003) | cDC1s, NK cells, ILCs | |
| Nfil3−/− (Kashiwada et al., 2011) | cDC1s, NK cells, ILCs | |
| Batf3−/− (Hildner et al., 2008) | cDC1s | In certain genetic backgrounds residual cDC1-like cells remain in skin-draining LNs |
| Relb−/− (Burkly et al., 1995) | CD11b+ESAM+ cDC2s in spleen | Fatal multiorgan inflammation |
| Ltbr−/− (Weih et al., 1995) | CD11b+ESAM+ cDC2s in spleen | Loss of lymph nodes, can overcome with conditional deletion with Itgax-cre |
| Notch2f/f Itgax-cre (Lewis et al., 2011; Satpathy et al., 2013) | CD11b+ESAM+ cDC2s in spleen, CD103+CD11b+ cDC2s in small intestine lamina propria | Transcriptional changes also evident in cDC1s and CD11b+ cDC2s |
| Klf4f/f Itgax-cre (Tussiwand et al., 2015) | Subset of cDC2s similar to CD24−CD11b− cDC2s in skin-draining LNs | |
| Irf4f/f Itgax-cre (Bajana et al., 2012) (Schlitzer et al., 2013) (Persson et al., 2013) (Gao et al., 2013) | ~50% CD11b+ cDC2s in lung, ~50% CD103+CD11b+ cDC2s in small intestine lamina propria, complete absence of cDC2s in lymph nodes |
cDC2s cannot migrate, failure to migrate may explain many observed phenotypes May also affect M2 macrophages or GM-CSF activated monocytes |
| Tcf4f/f Itgax-cre (Cisse et al., 2008) | pDCs | |
| Zeb2f/f Itgax-cre (Scott et al., 2016) | pDCs |
Functions of Dendritic Cells
Since their discovery, cDCs were recognized as possessing a remarkable capacity to stimulate naïve T cells and to initiate adaptive immune responses (Steinman and Witmer, 1978; Nussenzweig et al., 1980). Subsequent work, based in large part on the models described above, has shown that DCs are also critical for - not just participants in - early innate immune responses, activating innate lymphoid cells (ILCs) and other cells involved in the immediate response to pathogens. Additionally, subsets of cDCs play varied roles in autoimmunity and responses to tumors. Below we discuss the studies that identified these functional aspects of DCs, what tools were used to arrive at these conclusions, and confounding factors that may require further resolution.
General Adaptive Immune Responses
The first study to use Itgax-DTR mice showed that the absence of CD11c+ cells led to the loss of CD8 T cell priming to cell-associated antigens and to the intracellular pathogens Listeria monocytogenes and malaria (Jung et al., 2002). This provided the first direct in vivo evidence for the role of CD11c+ cells in T cell priming. Subsequent use of the Itgax-DTR strain showed a requirement for CD11c+ cells in priming CD8 T cells against lymphocytic choriomeningitis virus (LCMV) (Probst and Van Den, 2005), herpes simplex virus (HSV) (Kassim et al., 2006), vesicular stomatitis virus (VSV) (Ciavarra et al., 2006), and influenza (GeurtsvanKessel et al., 2008; Kim et al., 2010), as well as in CD4 T cell priming against HSV (Kassim et al., 2006), Mycobacterium tuberculosis (Mtb) (Tian et al., 2005), and immune complexes (de Jong et al., 2006). Depletion of CD11c+ cells in Itgax-DTR mice also impaired the expansion of antigen-specific memory CD8 T cells during rechallenge with Listeria, VSV, or influenza (Zammit et al., 2005) or with LCMV (Lauterbach et al., 2006).
Later studies used additional models to validate the specific role of cDCs and not other CD11c+ cells in T cell priming. Specific ablation of cDCs with the Zbtb46-DTR strain resulted in a complete inability to prime CD8 or CD4 T cells against soluble antigen (Meredith et al., 2012), and also a failure to prime CD4 T cells against Mtb (Samstein et al., 2013). Finally, cDC specific deletion of MHCII using Zbtb46-cre led to a complete reduction in CD4 T cell priming to soluble antigen (Loschko et al., 2016b).
Targeting antigen to specific DC subsets has indicated that there may be a specialization of roles between subsets, with cDC1s preferentially priming CD8 T cells and cDC2s priming CD4 T cells (Dudziak et al., 2007). cDC1s have also been recognized as the cells most efficient in cross-presentation of exogenous antigens on MHC class I (den Haan et al., 2000). Studies using mouse models with selective depletion of one subset have provided evidence to support this dichotomy. Batf3−/− mice failed to prime CD8 T cells against cell-associated antigen or West Nile virus (WNV), but still had intact priming of CD4 T cells (Hildner et al., 2008). Batf3−/− mice also mounted reduced CD8 T cell responses to cytomegalovirus (CMV) (Torti et al., 2011), influenza (Helft et al., 2012; Waithman et al., 2013), cowpox virus (Gainey et al., 2012), HSV (Nopora et al., 2012; Zelenay et al., 2012), and malaria (Lau et al., 2014). Depleting cDC1s using the Xcr1-DTRvenus strain also abrogated CD8 T cell priming against soluble and cell-associated antigen as well as against Listeria infection (Yamazaki et al., 2013). CD4 T cells could still be primed against soluble antigen in these mice. A study using Karma mice similarly showed that depletion of cDC1s led to deficient CD8 T cell priming to soluble protein as well as failure to reactivate memory CD8 T cells in response to Listeria, vaccinia virus, and VSV (Alexandre et al., 2016). Taken together these studies suggest that cDCs alone are responsible for priming T cells, with a specific role for cDC1s in priming naïve CD8 T cells and activating memory CD8 T cells during recall responses. Future work with specific depletion of cDC2s is needed to confirm their unique role in priming CD4 T cells.
Recent work has also indicated that CD4 T cell help in CD8 T cell responses might be mediated through “licensing” of cDC1s. Analysis of immune responses suggests that CD4 T cells are primed by DCs earlier than CD8 T cells, and that later in the response clusters of these three cell types form during which CD4 T cells may “license” cDC1s in order to enhance priming of CD8 T cells (Hor et al., 2015; Eickhoff et al., 2015). In agreement with this model, one study found that mixed bone marrow chimeric mice made of Xcr1-DTR:H2-Ab1−/− BM, in which following DT treatment all remaining cDC1s lack MHCII expression, generated fewer antigen-specific CD8 T cells upon vaccinia virus infection (Eickhoff et al., 2015). MHCII expression in cDC1s may therefore serve as a mechanism by which they receive CD4 T cell help rather than for their direct priming of naïve CD4 T cells.
Several studies have implicated DCs in the induction of Tfh cells and the initiation of the germinal center (GC) response. Depletion of all CD11c+ cells by Itgax-DTR was found to diminish the number of Tfh cells induced in response to Toxoplasma infection (Goenka et al., 2011). Specific cDC depletion with Zbtb46-DTR mice also abolished GC responses and antibody production against allogeneic red blood cells (RBCs) (Calabro et al., 2016). This was attributed to the function of cDC2s, as Batf3−/− mice had intact antibody responses but Irf4f/f Itgax-cre mice did not (Calabro et al., 2016). Another study found that CD25 expression by cDCs was crucial for Tfh induction (Li et al., 2016). In this study, mixed BM chimera mice reconstituted with Zbtb46-DTR:Il2ra−/− BM, in which after DT treatment all remaining cDCs lack the IL-2 receptor alpha chain (CD25), failed to generate antibodies or GC B cells upon sheep RBC immunization. cDC2s in the spleen can induce expression of CD25, suggesting these may be the specific cells involved in this process (Li et al., 2016). Finally, IgA class switching in Peyer’s patches was also found to be regulated by DCs (Reboldi et al., 2016). This study utilized mixed BM chimeras made from Itgax-DTR:Ltbr−/− BM, in which following DT treatment all remaining DCs lack the LTβ receptor and thus certain cDC2s fail to develop. After DT treatment, these chimeras had fewer IgA class-switched GC B cells in Peyer’s patches. This appears to involve the activation of latent TGF-β by the integrin chains Itgβ8 and Itgαv on DCs and presentation of the active cytokine by DCs to B cells, as Itgax-cre mediated deletion of Itgb8 also resulted in reduced IgA+ GC B cells (Reboldi et al., 2016).
Type 1 Immune Responses
Type 1 immune responses are mounted against intracellular pathogens that require IFN-γ activated macrophages and cytotoxic CD8 T cells for their clearance. Early nonspecific sources of IFN-γ are NK cells and ILC1s, while antigen-specific Th1 and CD8 T cells are responsible for producing this cytokine later in the immune response. IL-12 is critical for the activation of type 1 responses, as it induces NK cells and ILC1s to secrete IFN-γ (Sonnenberg and Artis, 2015), and is also responsible for the polarization of naïve T cells to Th1 cells (Hsieh et al., 1993). Several studies have established that cDC1s are vital for mounting type 1 responses because of their non-redundant production of IL-12 and their ability to prime CD8 T cells.
The first indication that cDC1s are important in these responses was a study demonstrating that Irf8−/− mice had increased susceptibility to Toxoplamsa gondii infection and reduced serum IL-12 levels (Scharton-Kersten et al., 1997), though given the myriad defects in this mouse this could not be attributed specifically to cDC1s. Later, Itgax-DTR mice were used to show that CD11c+ cells were required for IL-12 and IFN-γ production and protection against Toxoplasma infection (Liu et al., 2006). Finally, Batf3−/− mice that specifically lack cDC1s were found to be susceptible to Toxoplasma, demonstrating that these cells were indeed critical for resistance (Mashayekhi et al., 2011). Mixed chimeras made from Batf3−/−:Il12a−/− BM, in which all cDC1s that develop are deficient in IL-12 production, were susceptible to Toxoplasma infection, indicating that cDC1s are the non-redundant source of IL-12 necessary for resistance to infection (Mashayekhi et al., 2011). Additionally, depletion of cDC1s using the Karma strain abolished IL-12 production in response to soluble Toxoplasma antigen (Alexandre et al., 2016). Batf3−/− mice also have reduced IFN-γ production from NK cells during Toxoplasma infection, suggesting that IL-12 from cDC1s is critical for NK cell activation in this response (Askenase et al., 2015).
The role of cDCs in type 1 responses to several other intracellular pathogens has also been documented. During infection by Listeria, Itgax-DTR (Neuenhahn et al., 2006), Batf3−/− (Edelson et al., 2011), Ly75-DTR (Fukaya et al., 2012), and Xcr1-DTRvenus (Yamazaki et al., 2013) mice all had reduced CD8 T cell responses. However, many of these strains also exhibited a reduced splenic Listeria burden and Batf3−/− mice actually demonstrated increased resistance to infection. This is because Listeria must infect cDC1s in the splenic marginal zone in order to spread to and proliferate in the lymphoid areas of the spleen (Neuenhahn et al., 2006), so Batf3−/− mice lacking these cells show reduced susceptibility to infection. Finally, constitutive deletion of CD11c+ cells, achieved by crossing Itgax-cre mice to Rosa26-LSL-DTA mice (Voehringer et al., 2008), diminished IFN-γ production from NK cells, NKT cells, and T cells during Listeria infection (Kang et al., 2008).
Leishmania major, another intracellular pathogen, similarly requires cDC1s for its clearance, as Batf3−/− mice exhibit greater disease burden from infection (Ashok et al., 2014) and generate fewer Th1 cells (Martinez-Lopez et al., 2015). Another study used Batf3−/− and Cd207-DTR mice to demonstrate that cDC1s are required for the priming of CD8 T cells and Th1 cells against skin infection by the fungus Candida albicans (Igyarto et al., 2011).
Several studies have implicated DCs in NK cell homeostasis. Depletion of CD11c+ cells in Itgax-DTR mice diminished steady-state numbers of NK cells (Guimond et al., 2010) and reduced their cytotoxicity and IFN-γ production in several infection models (Lucas et al., 2007; Schleicher et al., 2007; Kassim et al., 2006). NK cell survival and activation requires trans-presentation of IL-15, whereby a non-NK cell that expresses the non-signaling receptor chain IL-15Rα binds IL-15 and presents to the full IL-15αβγ receptor on NK cells (Burkett et al., 2004). One study implicated a CD11c+ cell as the trans-presenting cell by using mixed BM chimera mice made from Itgax-DTR:Il15−/− BM or from Itgax-DTR:Il15rα−/− BM (Mortier et al., 2008). In these chimeras, DT treatment causes all remaining CD11c+ cells to lack IL-15 or IL-15Rα, respectively, and after DT treatment these chimeras show severely reduced NK cell activation in vivo. Future studies are needed to clarify whether cDCs are involved in this activity, and if so which subsets.
Type 2 Immune Responses
Type 2 responses are carried out against multicellular parasites at barrier surfaces in order to aid in their expulsion. Many cytokines play key roles in this response. These include IL-25 and IL-33, which activate ILC2s to produce effector cytokines such as IL-4, IL-5, and IL-13 (Sonnenberg and Artis, 2015), and IL-4, which polarizes naïve T cells to Th2 cells (Le Gros et al., 1990). Much remains unknown about the initiation of type 2 immune responses, and it has been controversial whether DCs are even involved directly.
An early study found that depletion of CD11c+ cells in Itgax-DTR mice did not impair Th2 cell polarization in response to immunization with papain (Sokol et al., 2009), although another study using the same strain found that it did (Tang et al., 2010). A later study also found that depletion of CD11c+ cells in Itgax-DTR mice abrogated type 2 responses, this time to inhaled house dust mite (HDM) allergen (Hammad et al., 2010). This study implicated an FcεRI expressing CD11c+MHCII+ cell population as being the principal APC responsible. Subsequent studies also found that CD11c+ cell depletion by Itgax-DTR reduced numbers of Th2 cells after infections with the helminthes Schistosoma mansoni (Phythian-Adams et al., 2010), Heligmosomoides polygyrus (Smith et al., 2011), and Nippostrongylus brasiliensis (Smith et al., 2012).
Other models have suggested putative Th2 inducing DC subsets. Two groups demonstrated that depletion of an Mgl2+ subset of cDC2s, either with the Mgl2-DTR strain (Kumamoto et al., 2013) or by conditional deletion of Irf4 with Itgax-cre (Gao et al., 2013), diminished Th2 priming in response to papain and Nippostrongylus infection. Irf4f/f Itgax-cre mice also had reduced Th2 responses after HDM allergen challenge (Williams et al., 2013). Another study found that depletion of a subset of cDC2s in the Klf4f/f Itgax-cre strain increased susceptibility to Schistosoma mansoni infection and diminished allergic inflammation after intransal HDM challenge (Tussiwand et al., 2015). Also, a subset of CXCR5+ DCs, which was depleted by using Itgax-DTR:Cxcr5−/− BM chimeras, appeared to be required for Th2 responses to Heligmosomoides infection (Leon et al., 2012). Finally, Batf3−/− mice develop somewhat stronger Th2 responses to helminth infection, suggesting that the IL-12 produced by cDC1s regulates Th2 polarization carried out by other DCs (Everts et al., 2016).
Cytokine production by DCs has not been established in any of these models, so whether they control type 2 responses by this mechanism or some other remains to be determined. Recent studies have suggested that ILC2s may be the critical source of cytokines acting to induce type 2 responses (Halim et al., 2014; Halim et al., 2016). Conceivably, ILC2s and DCs may cooperate in Th2 priming, but this area awaits further studies.
Type 3 Immune Responses
Type 3 immune responses at barrier surfaces such as the lungs and intestines control infections by extracellular bacteria and fungi and require several cytokines, including IL-23 and IL-6. IL-6 and TGF-β initiate Th17 cell polarization (Bettelli et al., 2006), and IL-23 increases the survival and expansion of committed Th17 cells (Veldhoen et al., 2006). IL-23 is also crucial in innate responses for activating ILC3s to produce IL-22, which in turn promotes production of bactericidal lectins such as RegIIIγ from small intestinal epithelial cells (Sonnenberg and Artis, 2015; Kinnebrew et al., 2012).
Citrobacter rodentium, a mouse pathogen used to study type 3 immune responses, requires IL-23 and IL-22 for its clearance (Zheng et al., 2008; Basu et al., 2012; Mundy et al., 2005). An early study that implicated cDCs in response to oral Citrobacter infection found that conditional deletion of Ltbr by Itgax-cre, which depleted a subset of cDC2s, reduced colonic IL-22 production and increased pathogen burden (Tumanov et al., 2011). Later studies demonstrated that depletion of all cDCs in Zbtb46-DTR mice (Satpathy et al., 2013; Schreiber et al., 2013) led to severe susceptibility to Citrobacter infection, as did depletion of cDC2s with Notch2f/f Itgax-cre mice (Satpathy et al., 2013). Mixed BM chimeras reconstituted with Notch2f/f Itgax-cre:Il23a−/− BM were also severely susceptible to Citrobacter infection, indicating that Notch2-dependent cDC2s are the critical source of IL-23 in defense against this pathogen (Satpathy et al., 2013). NFκB signaling in CD11c+ cells is also critical, as mice with conditional deletion of Myd88 with Itgax-cre also showed susceptibility to Citrobacter (Longman et al., 2014). Importantly, mice depleted of all cDCs or Notch2-dependent cDC2s die at ~day 10 after Citrobacter infection, while Rag2−/− mice lacking B and T cells die at ~day 30 after infection (Zheng et al., 2008). This suggests that cDC2s are critical for activating innate defenses during Citrobacter infection and not just for initiating adaptive immunity.
Several recent studies have suggested contributions from multiple cDC2 subsets in defense against Citrobacter. First, CD207-DTA mice, which lack intestinal CD103+CD11b+ cDC2s, were not found to be susceptible to Citrobacter infection (Welty et al., 2013). Thus, the lethality seen with Notch2 deficiency may be due to an impact on the function of CD103-CD11b+ cDC2s, rather than the depletion of CD103+CD11b+ cDC2s. In accordance with this possibility, Cx3cr1-LSL-DTR mice crossed to Itgax-cre, in which DT treatment depletes CD103−CD11b+CX3CR1+ cDC2s and CD11b+ macrophages, also showed susceptibility to Citrobacter and reduced IL-22 production (Longman et al., 2014). Finally, MM-DTR mice, which lack monocytes and monocyte-derived macrophages, did not display susceptibility to Citrobacter, suggesting that CD103−CD11b+ cDC2s and not CD11b+ macrophages are the critical cells for defense against this pathogen (Schreiber et al., 2013). However, direct comparison of these different models may be required to eliminate confounding effects such as variable microbiota in the strains used in each study.
Studies have also implicated cDC2s in various other type 3 responses, but the reported mechanisms by which they act have varied. Several groups found that depletion of intestinal CD103+CD11b+ cDC2s, using Notch2f/f Itgax-cre, Irf4f/f Itgax-cre, or CD207-DTA mice, resulted in fewer small intestinal Th17 cells at steady state (Lewis et al., 2011; Schlitzer et al., 2013; Welty et al., 2013). Another study using the Irf4f/f Itgax-cre strain similarly found reduced numbers of Th17 cells at steady state in the small intestine lamina propria and mesenteric lymph nodes, and also reduced Th17 polarization after immunization with antigen plus αCD40 and LPS (Persson et al., 2013). This study observed reduced levels of IL-6 in Irf4−/− DCs, which might explain the reduced Th17 priming. A second group found deficient Th17 priming in Irf4f/f Itgax-cre mice after lung infection by the fungal pathogen Aspergillus fumigatus, which was attributed to the loss of CD11b+ cDC2s that normally express high levels of IL-23, IL-6, and TGF-β (Schlitzer et al., 2013).
In agreement, another study used BM chimeras to show that CD11c+ cells must produce IL-6 and TGF-β to induce Th17 responses to Streptococcus pyogenes (Linehan et al., 2015). They found that BM chimeras of Itgax-DTR:Il6−/− or Itgax-DTR:Tgfb1f/f BM, in which DT treatment causes all remaining DCs to be deficient in either IL-6 or TGF-β respectively, had reduced Th17 responses to this pathogen. This was confirmed in Tgfb1f/f Itgax-cre mice and specifically found to involve Mgl2+ cDC2s, as DT treated Mgl2-DTR:Il6−/− mixed BM chimeric mice also had reduced Th17 induction upon Streptococcus pyogenes infection (Linehan et al., 2015). A separate study used Mgl2-DTR:Il23a−/− mixed BM chimeras to determine that Mgl2+ cDC2s must produce IL-23 in order to activate IL-17 secretion from dermal γδ T cells during cutaneous Candida albicans infection (Kashem et al., 2015). Finally, a study using the Il23af/f Itgax-cre strain concluded that IL-23 from CD11c+ cells was important for Th17 and Th1 induction in response to Helicobacter hepaticus infection (Arnold et al., 2016), although the relative contributions of cDCs and macrophages in this infection remains to be defined with more specific methods. While depletion of cDC2s has revealed defects in type 3 responses to several pathogens, responses to many of these infections, specifically Citrobacter (Satpathy et al., 2013), Streptococcus (Linehan et al., 2015), and Candida (Kashem et al., 2015; Trautwein-Weidner et al., 2015), are intact in mice lacking cDC1s. This again underscores the functional specialization of different cDC subsets and the non-redundant roles they play in host defense.
Control of type 3 responses against segmented filamentous bacteria (SFB) may differ from those described above. SFB are a commensal organism present in some mice microbiota that induce an antigen-specific Th17 response in colonized mice (Ivanov et al., 2009). Conditional deletion of MHCII with Itgax-cre reduced Th17 induction after inoculation with SFB, which was interpreted to mean cDCs were responsible for priming against this organism. But Batf3−/−CD207-DTA mice, which lack cDC1s and CD103+CD11b+ cDC2s, and Flt3l−/− mice, which have substantially reduced numbers of all cDCs (McKenna et al., 2000), had no impairment in Th17 priming (Goto et al., 2014; Geem et al., 2014). Instead, depletion of monocytes with the Ccr2-DTR strain abrogated Th17 induction against SFB, and this could be restored by transfer of wildtype monocytes, suggesting that monocyte-derived intestinal macrophages prime this response (Panea et al., 2015). This example illustrates how some conclusions regarding cDC function drawn from CD11c-based systems may need re-interpretation with follow-up studies using more precise genetic models.
Dendritic cells in autoimmunity
DCs are important for the induction of tolerance to self-antigens and for the pathogenicity of several autoimmune syndromes. Early studies found a relationship between DC and Treg numbers, and that depletion of Tregs with a Foxp3-DTR strain greatly expanded cDCs (Kim et al., 2007). This expansion was due to increased numbers of cDC progenitors and was dependent on Flt3L, a crucial growth factor for cDCs (Liu et al., 2009). Conversely, depletion of DCs in Itgax-DTR mice or in Flt3l−/− mice reduced splenic Treg numbers, while expansion of DCs by exogenous Flt3L increased Treg numbers (Darrasse-Jeze et al., 2009). This phenomenon held for certain antigen-specific Treg clones as well (Leventhal et al., 2016).
DCs primarily control Treg numbers by regulating their proliferation rather than their induction, as Tregs transferred into Itgax-DTR mice treated with DT divided less than when transferred into WT mice (Darrasse-Jeze et al., 2009). This control of proliferation required MHCII expression by DCs, as both Itgax-DTR:H2-Ab1−/− mixed BM chimeric mice treated with DT and H2-Ab1f/− Itgax-cre mice also showed a decrease in Treg numbers and proliferation (Darrasse-Jeze et al., 2009). Later work also concluded that DCs do not play a primary role in Treg induction in the thymus, as neither constitutive depletion of DCs in mice with Itgax-cre crossed to Rosa26-LSL-DTA nor H2-Ab1 f/f Itgax-cre mice had differences in the absolute number of Foxp3+ cells that develop in the thymus (Birnberg et al., 2008; Liston et al., 2008).
However, several recent studies have demonstrated that at least some Treg clones are induced in the thymus by DCs, especially clones that recognize self-antigens expressed by the transcription factor AIRE. One group identified certain T cell clones that convert to Tregs in the thymus in response to AIRE-dependent antigens in wildtype mice, but not in mice that were depleted of CD11c+ cells by crossing the Itgax-cre to the Rosa26-LSL-DTA strain (Perry et al., 2014). These same T cells also did not convert in Batf3−/− mice, suggesting that cDC1s controlled their induction. A subsequent study reported that both polyclonal Tregs and a specific AIRE-dependent Treg were reduced in number in mice with conditional deletion of MHCII by Itgax-cre, but in contrast to the previous study found that this AIRE-dependent Treg still developed in Batf3−/− mice (Leventhal et al., 2016). This suggests that the specific Treg clone or the nature of the antigen it recognizes may determine which cDC subset is responsible for its induction.
DCs also play a role in several models of autoimmunity. Studies using Itgax-DTR mice show that CD11c+ cells are required for pathogenesis in ovalbumin-driven asthma (van Rijt et al., 2005) and in Th2-mediated peanut food allergy (Chu et al., 2014). Conditional deletion of Myd88 with Itgax-cre also diminished colitis in Il10−/− mice (Hoshi et al., 2012) and decreased lupus-like symptoms in Lyn−/− mice (Hua et al., 2014), suggesting that NFκB signaling in CD11c+ cells contributes to autoimmunity. Constitutively depleting CD11c+ cells in a model of lupus, achieved by crossing the Rosa26-LSL-DTA and Itgax-cre alleles onto the lupus-prone MRL.Faslpr background, reduced disease severity, in part by decreasing T and B cell activation (Teichmann et al., 2010). Disease incidence partly involved MyD88 signaling in CD11c+ cells, since Myd88f/f Itgax-cre mice on this background also had reduced dermatitis and T cell activation, but not reduced nephritis (Teichmann et al., 2013). In the nonobese diabetic (NOD) model of murine diabetes, cDC1s are required for disease initiation, as Batf3−/− NOD mice are completely free of lymphocyte infiltration into and destruction of pancreatic islets (Ferris et al., 2014). Conversely, increased activity of DCs can lead to autoimmunity, often from aberrant cytokine production or inappropriate activation of autoreactive T cells. This was seen when Itgax-cre was used to delete inhibitory signaling molecules such as Fas (Stranges et al., 2007) β-catenin (Manicassamy et al., 2010), FADD (Young et al., 2013), A20 (Hammer et al., 2011), TRAF6 (Han et al., 2013), Lyn (Lamagna et al., 2013), and Caspase-8 (Cuda et al., 2014).
Recent work has also implicated cDCs in establishing oral tolerance to ingested antigens. Using Zbtb46-DTR mice, one study demonstrated a loss of oral tolerance in mice lacking all cDCs (Esterhazy et al., 2016). These mice failed to induce Tregs against orally fed antigen and generated an immune response to subsequent peripheral antigen challenge. Oral tolerance was intact in Irf8f/f Zbtb46-cre mice that lack cDC1s, although antigen-specific Treg conversion was partially impaired. This suggests cDC1s are important in establishing oral tolerance but that other cDCs can compensate for their loss. A separate study found that for orally fed antigen to be tolerized it is phagocytosed by CX3CR1+ macrophages in the intestinal lamina propria and subsequently transferred to CD103+CD11b+ cDC2s through gap junctions made up of connexin-43 (Mazzini et al., 2014). Conditional deletion of Gja1, which encodes connexin-43, with Itgax-cre reduced the number of Tregs induced to orally fed antigen and also diminished oral tolerance established to this same antigen. Also, conditional deletion of Itgb8, which encodes the integrin Itgβ8 that activates latent TGF-β, with Itgax-cre resulted in autoimmune inflammatory bowel disease and reduced numbers of colonic Tregs, suggesting that CD11c+ cells are critical sources of active TGF-β for the induction of Tregs in response to gut antigens (Travis et al., 2007).
Dendritic cells in tumor immunity
Recent work has expanded our understanding of the roles of DCs in antitumor immune responses. Studies using Itgax-DTR mice demonstrated that depletion of CD11c+ cells reduced CD8 T cell responses against transferred tumor cells (Casares et al., 2005; Shimizu et al., 2007) and reduced survival in oncogene-driven tumor models (Scarlett et al., 2012). cDC1s in particular were critical for antitumor responses, as Batf3−/− mice failed to reject transplanted immunogenic fibrosarcomas (Hildner et al., 2008) and failed to induce T cell infiltration into autochthonous oncogene-driven melanoma (Spranger et al., 2015). cDC1s are also required for the antitumor effects of many cancer therapies, as Batf3−/− mice have reduced responses to therapeutic PD-L1 blockade, Flt3L injection, or polyI:C injection (Salmon et al., 2016) as well as to anti-CD137 or anti-PD1 therapy (Sanchez-Paulete et al., 2016). cDC1s must migrate to LNs to activate tumor-specific CD8 T cells, as mixed BM chimeras made with Xcr1-DTR:Ccr7−/− BM, in which DT treatment causes all remaining cDC1s to lack CCR7, are not able to reject tumors (Roberts et al., 2016). Finally, cDCs play a role in opposing the establishment of metastatic foci, as Zbtb46-DTR mice depleted of cDCs mice had increased numbers of melanoma metastases to the lung (Headley et al., 2016).
Two studies have demonstrated that interferon signaling in DCs is necessary for the initiation of antitumor immune responses. In the first, conditional deletion of the type 1 interferon receptor Ifnar1 with Itgax-cre prevented rejection of transplanted fibrosarcomas (Diamond et al., 2011). IFNAR deficient cDC1s also displayed deficient cross-presentation of antigens to CD8 T cells, suggesting that an inability to cross-present tumor antigens and activate CD8 T cells underlies the inability to reject tumors in this strain (Diamond et al., 2011). In the second, Ifnar1−/−:Batf3−/− mixed BM chimeras failed to prime CD8 T cells to tumor antigens, suggesting that interferon signaling is specifically necessary in cDC1s (Fuertes et al., 2011).
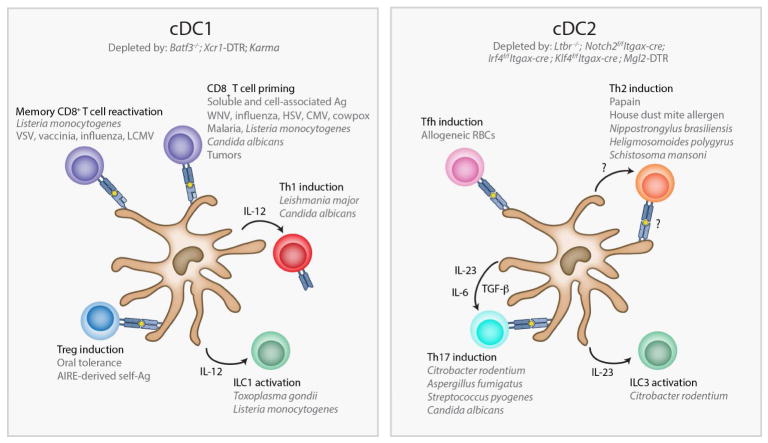
Comparison of Zbtb46-DTR and Itgax-DTR mice reveals a partial redundancy between cDCs and other CD11c+ cells in some aspects of the antitumor response. When immunized with tumor antigens and challenged with melanoma, DT treatment in either strain led to a failure to reject tumors (Meredith et al., 2012). But mice depleted of CD11c+ cells succumbed to tumors faster than mice depleted of cDCs, suggesting that CD11c+ non-cDCs can compensate in driving antitumor responses. Others studies have distinguished various macrophage and DC populations present within the tumor microenvironment (Franklin et al., 2014; Broz et al., 2014), and future studies may provide a better understanding of the roles of each in tumor immunity. Figure 1 summarizes the many known roles of cDCs in immune responses.
Fig. 1. Functions of dendritic cell subsets in the immune response.
Classical dendritic cells (cDCs) play critical roles in both innate and adaptive immunity. Furthermore, each subset of cDC appears to possess unique functions and to control the immune response against specific forms of pathogens. cDC1s control type 1 immune responses against viruses and intracellular pathogens (left panel). In these responses they prime naïve CD8 T cells, reactivate memory CD8 T cells, activate ILC1s, and induce Th1 cells. Their production of the cytokine IL-12 is vital for many of these functions. cDC1s also play a role in inducing Tregs against orally fed antigens and AIRE-dependent self-antigens expressed in the thymus, though it appears that in some cases cDC2s can also mediate Treg conversion to these antigens. Some cDC2s, on the other hand, regulate type 2 immune responses against parasites in which they induce Th2 cells (right panel). The exact mechanism by which they do this is unclear. Other cDC2s control type 3 immune responses against extracellular bacteria and fungi (right panel). In these responses, cDC2s produce IL-23 in order to activate ILC3s and to induce Th17 cells. Their production of IL-6 and TGF-β also contributes to the polarization of Th17 cells. Finally, cDC2s are also responsible for the induction of T follicular helper (Tfh) cells that regulate the germinal center response. DTR, diphtheria toxin receptor; VSV, vesicular stomatitis virus; LCMV, lymphocytic choriomeningitis virus; WNV, west nile virus; HSV, herpes simplex virus; CMV, cytomegalovirus; AIRE, autoimmune regulator; Ag, antigen.
Concluding Remarks
The increasing precision by which cDC subsets can be ablated in mouse models has greatly expanded our understanding of their functions. It is now clear that cDCs are a unique lineage comprised of distinct functional subsets critical for both innate and adaptive immunity. Further work is needed to resolve the apparent heterogeneity in cDC2s (Jaitin et al., 2014), and to test for their potential activities. In particular, how cDC2s control type 2 immune responses is unclear, including whether they provide instructive signals themselves, whether they are the primary APCs, and whether they, or other cells such as epithelial tuft cells, directly sense the pathogen. While these and other questions remain, it is now clear that DCs are the critical regulators of the entire immune response and that to understand the immune system we must fully analyze these remarkable cells.
Table 2.
Comparison of Cre strains for conditional deletion of genes in dendritic cells
| Strain | Cells Affected | Caveats |
|---|---|---|
| Itgax-cre (BAC) (Caton et al., 2007) | ~100% of cDCs, pDCs, alveolar macrophages, ~70% red pulp macrophages, 20–40% monocytes, marginal zone macrophages, peritoneal macrophages, ~5–12% T, B, NK cells | Germline deletion possible |
| Itgax-cre (Transgene) (Stranges et al., 2007) | cDCs, pDCs | Deletion in other cell types was not analyzed, most likely similar to other Itgax- cre strain |
| Clec9a-cre (Schraml et al., 2013) | ~100% of cDC1s, ~50% of cDC2s, ~20% of pDCs | Begins deletion at CDP stage, low or transient expression may affect deletion efficiency in cDC2s and pDCs |
| Zbtb46-cre (Loschko et al., 2016b) | 65–95% cDCs, <10% in all other cell types | Deletion efficiency variable depending on floxed allele |
| Cd207-cre (Zahner et al., 2011) | Langerhans cells, Langerin+ cDC1s in dermis, skin-draining LNs, and lung | |
| Xcr1-cre (Ohta et al., 2016) | cDC1s | |
| Siglech-cre (Puttur et al., 2013) | ~30% of pDCs, ~2% of other lymphoid cells | Incomplete deletion in pDCs Deletion in marginal zone macrophages untested but possible |
Acknowledgments
The authors acknowledge Carlos G. Briseño, Takeshi Egawa, Ansuman T. Satpathy, and Xiaodi Wu for their helpful suggestions. VD is supported by a Ruth L. Kirschstein National Research Service Award F30 DK108498. KMM is a Howard Hughes Medical Institute Investigator. The authors have no financial conflicts of interest to disclose.
Footnotes
Author Contributions
VD wrote the text and created the figures. KMM edited the text.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Abram CL, Roberge GL, Hu Y, Lowell CA. Comparative analysis of the efficiency and specificity of myeloid-Cre deleting strains using ROSA-EYFP reporter mice. J Immunol Methods. 2014;408:89–100. doi: 10.1016/j.jim.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandre YO, Ghilas S, Sanchez C, Le Bon A, Crozat K, Dalod M. XCR1+ dendritic cells promote memory CD8+ T cell recall upon secondary infections with Listeria monocytogenes or certain viruses. J Exp Med. 2016;213:75–92. doi: 10.1084/jem.20142350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliberti J, Schulz O, Pennington DJ, Tsujimura H, Sousa Reis E, Ozato K, Sher A. Essential role for ICSBP in the in vivo development of murine CD8alpha + dendritic cells. Blood. 2003;101:305–310. doi: 10.1182/blood-2002-04-1088. [DOI] [PubMed] [Google Scholar]
- Arnold IC, Mathisen S, Schulthess J, Danne C, Hegazy AN, Powrie F. CD11c(+) monocyte/macrophages promote chronic Helicobacter hepaticus-induced intestinal inflammation through the production of IL-23. Mucosal Immunol. 2016;9:352–363. doi: 10.1038/mi.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashok D, Schuster S, Ronet C, Rosa M, Mack V, Lavanchy C, Marraco SF, Fasel N, Murphy KM, Tacchini-Cottier F, Acha-Orbea H. Cross-presenting dendritic cells are required for control of Leishmania major infection. Eur J Immunol. 2014;44:1422–1432. doi: 10.1002/eji.201344242. [DOI] [PubMed] [Google Scholar]
- Askenase MH, Han SJ, Byrd AL, Morais dF, Bouladoux N, Wilhelm C, Konkel JE, Hand TW, Lacerda-Queiroz N, Su XZ, Trinchieri G, Grainger JR, Belkaid Y. Bone-Marrow-Resident NK Cells Prime Monocytes for Regulatory Function during Infection. Immunity. 2015;42:1130–1142. doi: 10.1016/j.immuni.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffray C, Fogg DK, Narni-Mancinelli E, Senechal B, Trouillet C, Saederup N, Leemput J, Bigot K, Campisi L, Abitbol M, Molina T, Charo I, Hume DA, Cumano A, Lauvau G, Geissmann F. CX3CR1+ CD115+ CD135+ common macrophage/DC precursors and the role of CX3CR1 in their response to inflammation. J Exp Med. 2009;206:595–606. doi: 10.1084/jem.20081385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajana S, Roach K, Turner S, Paul J, Kovats S. IRF4 promotes cutaneous dendritic cell migration to lymph nodes during homeostasis and inflammation. J Immunol. 2012;189:3368–3377. doi: 10.4049/jimmunol.1102613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajana S, Turner S, Paul J, Ainsua-Enrich E, Kovats S. IRF4 and IRF8 Act in CD11c+ Cells To Regulate Terminal Differentiation of Lung Tissue Dendritic Cells. J Immunol. 2016;196:1666–1677. doi: 10.4049/jimmunol.1501870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu R, O’Quinn DB, Silberger DJ, Schoeb TR, Fouser L, Ouyang W, Hatton RD, Weaver CT. Th22 cells are an important source of IL-22 for host protection against enteropathogenic bacteria. Immunity. 2012;37:1061–1075. doi: 10.1016/j.immuni.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CL, van Rijn E, Jung S, Inaba K, Steinman RM, Kapsenberg ML, Clausen BE. Inducible ablation of mouse Langerhans cells diminishes but fails to abrogate contact hypersensitivity. J Cell Biol. 2005;169:569–576. doi: 10.1083/jcb.200501071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, Kuchroo VK. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- Birnberg T, Bar-On L, Sapoznikov A, Caton ML, Cervantes-Barragan L, Makia D, Krauthgamer R, Brenner O, Ludewig B, Brockschnieder D, Riethmacher D, Reizis B, Jung S. Lack of conventional dendritic cells is compatible with normal development and T cell homeostasis, but causes myeloid proliferative syndrome 1. Immunity. 2008;29:986–997. doi: 10.1016/j.immuni.2008.10.012. [DOI] [PubMed] [Google Scholar]
- Blasius AL, Giurisato E, Cella M, Schreiber RD, Shaw AS, Colonna M. Bone marrow stromal cell antigen 2 is a specific marker of type I IFN-producing cells in the naive mouse, but a promiscuous cell surface antigen following IFN stimulation. J Immunol. 2006;177:3260–3265. doi: 10.4049/jimmunol.177.5.3260. [DOI] [PubMed] [Google Scholar]
- Bogunovic M, Ginhoux F, Helft J, Shang L, Hashimoto D, Greter M, Liu K, Jakubzick C, Ingersoll MA, Leboeuf M, Stanley ER, Nussenzweig M, Lira SA, Randolph GJ, Merad M. Origin of the lamina propria dendritic cell network. Immunity. 2009;31:513–525. doi: 10.1016/j.immuni.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitman ML, Clapoff S, Rossant J, Tsui LC, Glode LM, Maxwell IH, Bernstein A. Genetic ablation: targeted expression of a toxin gene causes microphthalmia in transgenic mice. Science. 1987;238:1563–1565. doi: 10.1126/science.3685993. [DOI] [PubMed] [Google Scholar]
- Breitman ML, Rombola H, Maxwell IH, Klintworth GK, Bernstein A. Genetic ablation in transgenic mice with an attenuated diphtheria toxin A gene. Mol Cell Biol. 1990;10:474–479. doi: 10.1128/mcb.10.2.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briseno CG, Haldar M, Kretzer NM, Wu X, Theisen DJ, KCW, Durai V, Grajales-Reyes GE, Iwata A, Bagadia P, Murphy TL, Murphy KM. Distinct Transcriptional Programs Control Cross-Priming in Classical and Monocyte-Derived Dendritic Cells. Cell Rep. 2016;15:2462–2474. doi: 10.1016/j.celrep.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briseno CG, Murphy TL, Murphy KM. Complementary diversification of dendritic cells and innate lymphoid cells. Curr Opin Immunol. 2014;29C:69–78. doi: 10.1016/j.coi.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockschnieder D, Pechmann Y, Sonnenberg-Riethmacher E, Riethmacher D. An improved mouse line for Cre-induced cell ablation due to diphtheria toxin A, expressed from the Rosa26 locus. Genesis. 2006;44:322–327. doi: 10.1002/dvg.20218. [DOI] [PubMed] [Google Scholar]
- Broz ML, Binnewies M, Boldajipour B, Nelson AE, Pollack JL, Erle DJ, Barczak A, Rosenblum MD, Daud A, Barber DL, Amigorena S, Van’t Veer LJ, Sperling AI, Wolf DM, Krummel MF. Dissecting the tumor myeloid compartment reveals rare activating antigen-presenting cells critical for T cell immunity. Cancer Cell. 2014;26:638–652. doi: 10.1016/j.ccell.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buch T, Heppner FL, Tertilt C, Heinen TJ, Kremer M, Wunderlich FT, Jung S, Waisman A. A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat Methods. 2005;2:419–426. doi: 10.1038/nmeth762. [DOI] [PubMed] [Google Scholar]
- Burkett PR, Koka R, Chien M, Chai S, Boone DL, Ma A. Coordinate expression and trans presentation of interleukin (IL)-15Ralpha and IL-15 supports natural killer cell and memory CD8+ T cell homeostasis. J Exp Med. 2004;200:825–834. doi: 10.1084/jem.20041389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkly L, Hession C, Ogata L, Reilly C, Marconi LA, Olson D, Tizard R, Cate R, Lo D. Expression of relB is required for the development of thymic medulla and dendritic cells. Nature. 1995;373:531–536. doi: 10.1038/373531a0. [DOI] [PubMed] [Google Scholar]
- Calabro S, Gallman A, Gowthaman U, Liu D, Chen P, Liu J, Krishnaswamy JK, Nascimento MS, Xu L, Patel SR, Williams A, Tormey CA, Hod EA, Spitalnik SL, Zimring JC, Hendrickson JE, Stowell SR, Eisenbarth SC. Bridging channel dendritic cells induce immunity to transfused red blood cells. J Exp Med. 2016;213:887–896. doi: 10.1084/jem.20151720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caminschi I, Proietto AI, Ahmet F, Kitsoulis S, Shin TJ, Lo JC, Rizzitelli A, Wu L, Vremec D, van Dommelen SL, Campbell IK, Maraskovsky E, Braley H, Davey GM, Mottram P, van d V, Jensen K, Lew AM, Wright MD, Heath WR, Shortman K, Lahoud MH. The dendritic cell subtype-restricted C-type lectin Clec9A is a target for vaccine enhancement. Blood. 2008;112:3264–3273. doi: 10.1182/blood-2008-05-155176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casares N, Pequignot MO, Tesniere A, Ghiringhelli F, Roux S, Chaput N, Schmitt E, Hamai A, Hervas-Stubbs S, Obeid M, Coutant F, Metivier D, Pichard E, Aucouturier P, Pierron G, Garrido C, Zitvogel L, Kroemer G. Caspase-dependent immunogenicity of doxorubicin-induced tumor cell death. J Exp Med. 2005;202:1691–1701. doi: 10.1084/jem.20050915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caton ML, Smith-Raska MR, Reizis B. Notch-RBP-J signaling controls the homeostasis of CD8− dendritic cells in the spleen. J Exp Med. 2007;204:1653–1664. doi: 10.1084/jem.20062648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella M, Jarrossay D, Facchetti F, Alebardi O, Nakajima H, Lanzavecchia A, Colonna M. Plasmacytoid monocytes migrate to inflamed lymph nodes and produce large amounts of type I interferon [see comments] Nature Medicine. 1999;5:919–923. doi: 10.1038/11360. [DOI] [PubMed] [Google Scholar]
- Chu DK, Jimenez-Saiz R, Verschoor CP, Walker TD, Goncharova S, Llop-Guevara A, Shen P, Gordon ME, Barra NG, Bassett JD, Kong J, Fattouh R, McCoy KD, Bowdish DM, Erjefalt JS, Pabst O, Humbles AA, Kolbeck R, Waserman S, Jordana M. Indigenous enteric eosinophils control DCs to initiate a primary Th2 immune response in vivo. J Exp Med. 2014;211:1657–1672. doi: 10.1084/jem.20131800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciavarra RP, Stephens A, Nagy S, Sekellick M, Steel C. Evaluation of immunological paradigms in a virus model: are dendritic cells critical for antiviral immunity and viral clearance? J Immunol. 2006;177:492–500. doi: 10.4049/jimmunol.177.1.492. [DOI] [PubMed] [Google Scholar]
- Cisse B, Caton ML, Lehner M, Maeda T, Scheu S, Locksley R, Holmberg D, Zweier C, den Hollander NS, Kant SG, Holter W, Rauch A, Zhuang Y, Reizis B. Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development 1. Cell. 2008;135:37–48. doi: 10.1016/j.cell.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozat K, Guiton R, Contreras V, Feuillet V, Dutertre CA, Ventre E, Vu Manh TP, Baranek T, Storset AK, Marvel J, Boudinot P, Hosmalin A, Schwartz-Cornil I, Dalod M. The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to mouse CD8alpha+ dendritic cells. J Exp Med. 2010;207:1283–1292. doi: 10.1084/jem.20100223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozat K, Tamoutounour S, Vu Manh TP, Fossum E, Luche H, Ardouin L, Guilliams M, Azukizawa H, Bogen B, Malissen B, Henri S, Dalod M. Cutting edge: expression of XCR1 defines mouse lymphoid-tissue resident and migratory dendritic cells of the CD8alpha+ type. J Immunol. 2011;187:4411–4415. doi: 10.4049/jimmunol.1101717. [DOI] [PubMed] [Google Scholar]
- Cuda CM, Misharin AV, Gierut AK, Saber R, Haines GK, III, Hutcheson J, Hedrick SM, Mohan C, Budinger GS, Stehlik C, Perlman H. Caspase-8 acts as a molecular rheostat to limit RIPK1- and MyD88-mediated dendritic cell activation. J Immunol. 2014;192:5548–5560. doi: 10.4049/jimmunol.1400122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrasse-Jeze G, Deroubaix S, Mouquet H, Victora GD, Eisenreich T, Yao KH, Masilamani RF, Dustin ML, Rudensky A, Liu K, Nussenzweig MC. Feedback control of regulatory T cell homeostasis by dendritic cells in vivo. J Exp Med. 2009;206:1853–1862. doi: 10.1084/jem.20090746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong JM, Schuurhuis DH, Ioan-Facsinay A, Welling MM, Camps MG, van der Voort EI, Huizinga TW, Ossendorp F, Verbeek JS, Toes RE. Dendritic cells, but not macrophages or B cells, activate major histocompatibility complex class II-restricted CD4+ T cells upon immune-complex uptake in vivo. Immunol. 2006;119:499–506. doi: 10.1111/j.1365-2567.2006.02464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Haan JM, Lehar SM, Bevan MJ. CD8(+) but not CD8(-) dendritic cells cross-prime cytotoxic T cells in vivo. J Exp Med. 2000;192:1685–1696. doi: 10.1084/jem.192.12.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond MS, Kinder M, Matsushita H, Mashayekhi M, Dunn GP, Archambault JM, Lee H, Arthur CD, White JM, Kalinke U, Murphy KM, Schreiber RD. Type I interferon is selectively required by dendritic cells for immune rejection of tumors. J Exp Med. 2011;208:1989–2003. doi: 10.1084/jem.20101158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl GE, Longman RS, Zhang JX, Breart B, Galan C, Cuesta A, Schwab SR, Littman DR. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX(3)CR1(hi) cells. Nature. 2013;494:116–120. doi: 10.1038/nature11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudziak D, Kamphorst AO, Heidkamp GF, Buchholz VR, Trumpfheller C, Yamazaki S, Cheong C, Liu K, Lee HW, Park CG, Steinman RM, Nussenzweig MC. Differential antigen processing by dendritic cell subsets in vivo. Science. 2007;315:107–111. doi: 10.1126/science.1136080. [DOI] [PubMed] [Google Scholar]
- Edelson BT, Bradstreet TR, Hildner K, Carrero JA, Frederick KE, KCW, Belizaire R, Aoshi T, Schreiber RD, Miller MJ, Murphy TL, Unanue ER, Murphy KM. CD8a+ Dendritic Cells Are an Obligate Cellular Entry Point for Productive Infection by Listeria monocytogenes. Immunity. 2011;35:236–248. doi: 10.1016/j.immuni.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelson BT, KCW, Juang R, Kohyama M, Benoit LA, Klekotka PA, Moon C, Albring JC, Ise W, Michael DG, Bhattacharya D, Stappenbeck TS, Holtzman MJ, Sung SS, Murphy TL, Hildner K, Murphy KM. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8alpha+ conventional dendritic cells. J Exp Med. 2010;207:823–836. doi: 10.1084/jem.20091627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickhoff S, Brewitz A, Gerner MY, Klauschen F, Komander K, Hemmi H, Garbi N, Kaisho T, Germain RN, Kastenmuller W. Robust Anti-viral Immunity Requires Multiple Distinct T Cell-Dendritic Cell Interactions. Cell. 2015;162:1322–1337. doi: 10.1016/j.cell.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esterhazy D, Loschko J, London M, Jove V, Oliveira TY, Mucida D. Classical dendritic cells are required for dietary antigen-mediated induction of peripheral T(reg) cells and tolerance. Nat Immunol. 2016;17:545–555. doi: 10.1038/ni.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everts B, Tussiwand R, Dreesen L, Fairfax KC, Huang SC, Smith AM, O’Neill CM, Lam WY, Edelson BT, Urban JF, Jr, Murphy KM, Pearce EJ. Migratory CD103+ dendritic cells suppress helminth-driven type 2 immunity through constitutive expression of IL-12. J Exp Med. 2016;213:35–51. doi: 10.1084/jem.20150235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris ST, Carrero JA, Mohan JF, Calderon B, Murphy KM, Unanue ER. A minor subset of Batf3-dependent antigen-presenting cells in islets of Langerhans is essential for the development of autoimmune diabetes. Immunity. 2014;41:657–669. doi: 10.1016/j.immuni.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogg DK, Sibon C, Miled C, Jung S, Aucouturier P, Littman DR, Cumano A, Geissmann F. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science. 2006;311:83–87. doi: 10.1126/science.1117729. [DOI] [PubMed] [Google Scholar]
- Franklin RA, Liao W, Sarkar A, Kim MV, Bivona MR, Liu K, Pamer EG, Li MO. The cellular and molecular origin of tumor-associated macrophages. Science. 2014;344:921–925. doi: 10.1126/science.1252510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuertes MB, Kacha AK, Kline J, Woo SR, Kranz DM, Murphy KM, Gajewski TF. Host type I IFN signals are required for antitumor CD8+ T cell responses through CD8{alpha}+ dendritic cells. J Exp Med. 2011;208:2005–2016. doi: 10.1084/jem.20101159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaya T, Murakami R, Takagi H, Sato K, Sato Y, Otsuka H, Ohno M, Hijikata A, Ohara O, Hikida M, Malissen B, Sato K. Conditional ablation of CD205+ conventional dendritic cells impacts the regulation of T-cell immunity and homeostasis in vivo. Proc Natl Acad Sci U S A. 2012;109:11288–11293. doi: 10.1073/pnas.1202208109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futterer A, Mink K, Luz A, Kosco-Vilbois MH, Pfeffer K. The lymphotoxin beta receptor controls organogenesis and affinity maturation in peripheral lymphoid tissues. Immunity. 1998;9:59–70. doi: 10.1016/s1074-7613(00)80588-9. [DOI] [PubMed] [Google Scholar]
- Gainey MD, Rivenbark JG, Cho H, Yang L, Yokoyama WM. Viral MHC class I inhibition evades CD8+ T-cell effector responses in vivo but not CD8+ T-cell priming. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1217111109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Nish SA, Jiang R, Hou L, Licona-Limon P, Weinstein JS, Zhao H, Medzhitov R. Control of T helper 2 responses by transcription factor IRF4-dependent dendritic cells. Immunity. 2013;39:722–732. doi: 10.1016/j.immuni.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoyne DM, Long E, Veiga-Fernandes H, de Boer J, Williams O, Seddon B, Coles M, Kioussis D, Brady HJ. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat Immunol. 2009;10:1118–1124. doi: 10.1038/ni.1787. [DOI] [PubMed] [Google Scholar]
- Geem D, Medina-Contreras O, McBride M, Newberry RD, Koni PA, Denning TL. Specific microbiota-induced intestinal Th17 differentiation requires MHC class II but not GALT and mesenteric lymph nodes. J Immunol. 2014;193:431–438. doi: 10.4049/jimmunol.1303167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger TL, Abt MC, Gasteiger G, Firth MA, O’Connor MH, Geary CD, O’Sullivan TE, van den Brink MR, Pamer EG, Hanash AM, Sun JC. Nfil3 is crucial for development of innate lymphoid cells and host protection against intestinal pathogens. J Exp Med. 2014;211:1723–1731. doi: 10.1084/jem.20140212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GeurtsvanKessel CH, Willart MAM, van Rijt LS, Muskens F, Kool M, Baas C, Thielemans K, Bennett C, Clasen BE, Hoogsteden HC, Osterhaus ADME, Rimmelzwaan GF, Lambrecht BN. Clearance of influenza virus from the lung depends on migratory langerin(+)CD11b(-) but not plasmacytoid dendritic cells. J Exp Med. 2008;205:1621–1634. doi: 10.1084/jem.20071365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasmacher E, Agrawal S, Chang AB, Murphy TL, Zeng W, Vander LB, Khan AA, Ciofani M, Spooner C, Rutz S, Hackney J, Nurieva R, Escalante CR, Ouyang W, Littman DR, Murphy KM, Singh H. A Genomic Regulatory Element That Directs Assembly and Function of Immune-Specific AP-1-IRF Complexes. Science. 2012;338:975–980. doi: 10.1126/science.1228309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goenka R, Barnett LG, Silver JS, O’Neill PJ, Hunter CA, Cancro MP, Laufer TM. Cutting edge: dendritic cell-restricted antigen presentation initiates the follicular helper T cell program but cannot complete ultimate effector differentiation. J Immunol. 2011;187:1091–1095. doi: 10.4049/jimmunol.1100853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Panea C, Nakato G, Cebula A, Lee C, Diez MG, Laufer TM, Ignatowicz L, Ivanov II. Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity. 2014;40:594–607. doi: 10.1016/j.immuni.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grajales-Reyes GE, Iwata A, Albring J, Wu X, Tussiwand R, KCW, Kretzer NM, Briseno CG, Durai V, Bagadia P, Haldar M, Schonheit J, Rosenbauer F, Murphy TL, Murphy KM. Batf3 maintains autoactivation of Irf8 for commitment of a CD8alpha(+) conventional DC clonogenic progenitor. Nat Immunol. 2015;16:708–717. doi: 10.1038/ni.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilliams M, Dutertre CA, Scott CL, McGovern N, Sichien D, Chakarov S, Van Gassen S, Chen J, Poidinger M, De Prijck S, Tavernier SJ, Low I, Irac SE, Mattar CN, Sumatoh HR, Low GH, Chung TJ, Chan DK, Tan KK, Hon TL, Fossum E, Bogen B, Choolani M, Chan JK, Larbi A, Luche H, Henri S, Saeys Y, Newell EW, Lambrecht BN, Malissen B, Ginhoux F. Unsupervised High-Dimensional Analysis Aligns Dendritic Cells across Tissues and Species. Immunity. 2016;45:669–684. doi: 10.1016/j.immuni.2016.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilliams M, Ginhoux F, Jakubzick C, Naik SH, Onai N, Schraml BU, Segura E, Tussiwand R, Yona S. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat Rev Immunol. 2014;14:571–578. doi: 10.1038/nri3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimond M, Freud AG, Mao HC, Yu J, Blaser BW, Leong JW, Vandeusen JB, Dorrance A, Zhang J, Mackall CL, Caligiuri MA. In vivo role of Flt3 ligand and dendritic cells in NK cell homeostasis. J Immunol. 2010;184:2769–2775. doi: 10.4049/jimmunol.0900685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker C, Kirsch RD, Ju XS, Hieronymus T, Gust TC, Kuhl C, Jorgas T, Kurz SM, Rose-John S, Yokota Y, Zenke M. Transcriptional profiling identifies Id2 function in dendritic cell development. Nat Immunol. 2003;4:380–386. doi: 10.1038/ni903. [DOI] [PubMed] [Google Scholar]
- Halim TY, Hwang YY, Scanlon ST, Zaghouani H, Garbi N, Fallon PG, McKenzie AN. Group 2 innate lymphoid cells license dendritic cells to potentiate memory TH2 cell responses. Nat Immunol. 2016;17:57–64. doi: 10.1038/ni.3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halim TY, Steer CA, Matha L, Gold MJ, Martinez-Gonzalez I, McNagny KM, McKenzie AN, Takei F. Group 2 innate lymphoid cells are critical for the initiation of adaptive T helper 2 cell-mediated allergic lung inflammation. Immunity. 2014;40:425–435. doi: 10.1016/j.immuni.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammad H, Plantinga M, Deswarte K, Pouliot P, Willart MA, Kool M, Muskens F, Lambrecht BN. Inflammatory dendritic cells--not basophils--are necessary and sufficient for induction of Th2 immunity to inhaled house dust mite allergen. J Exp Med. 2010;207:2097–2111. doi: 10.1084/jem.20101563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer GE, Turer EE, Taylor KE, Fang CJ, Advincula R, Oshima S, Barrera J, Huang EJ, Hou B, Malynn BA, Reizis B, DeFranco A, Criswell LA, Nakamura MC, Ma A. Expression of A20 by dendritic cells preserves immune homeostasis and prevents colitis and spondyloarthritis. Nat Immunol. 2011;12:1184–1193. doi: 10.1038/ni.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D, Walsh MC, Cejas PJ, Dang NN, Kim YF, Kim J, Charrier-Hisamuddin L, Chau L, Zhang Q, Bittinger K, Bushman FD, Turka LA, Shen H, Reizis B, DeFranco AL, Wu GD, Choi Y. Dendritic cell expression of the signaling molecule TRAF6 is critical for gut microbiota-dependent immune tolerance. Immunity. 2013;38:1211–1222. doi: 10.1016/j.immuni.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Headley MB, Bins A, Nip A, Roberts EW, Looney MR, Gerard A, Krummel MF. Visualization of immediate immune responses to pioneer metastatic cells in the lung. Nature. 2016;531:513–517. doi: 10.1038/nature16985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath WR, Carbone FR. Dendritic cell subsets in primary and secondary T cell responses at body surfaces. Nat Immunol. 2009;10:1237–1244. doi: 10.1038/ni.1822. [DOI] [PubMed] [Google Scholar]
- Hebel K, Griewank K, Inamine A, Chang HD, Muller-Hilke B, Fillatreau S, Manz RA, Radbruch A, Jung S. Plasma cell differentiation in T-independent type 2 immune responses is independent of CD11c(high) dendritic cells. Eur J Immunol. 2006;36:2912–2919. doi: 10.1002/eji.200636356. [DOI] [PubMed] [Google Scholar]
- Helft J, Manicassamy B, Guermonprez P, Hashimoto D, Silvin A, Agudo J, Brown BD, Schmolke M, Miller JC, Leboeuf M, Murphy KM, Garcia-Sastre A, Merad M. Cross-presenting CD103+ dendritic cells are protected from influenza virus infection. J Clin Invest. 2012;122:4037–4047. doi: 10.1172/JCI60659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, Kohyama M, Calderon B, Schraml BU, Unanue ER, Diamond MS, Schreiber RD, Murphy TL, Murphy KM. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochweller K, Striegler J, Hammerling GJ, Garbi N. A novel CD11c.DTR transgenic mouse for depletion of dendritic cells reveals their requirement for homeostatic proliferation of natural killer cells. Eur J Immunol. 2008;38:2776–2783. doi: 10.1002/eji.200838659. [DOI] [PubMed] [Google Scholar]
- Honjo T, Nishizuka Y, Hayaishi O. Diphtheria toxin-dependent adenosine diphosphate ribosylation of aminoacyl transferase II and inhibition of protein synthesis. J Biol Chem. 1968;243:3553–3555. [PubMed] [Google Scholar]
- Hor JL, Whitney PG, Zaid A, Brooks AG, Heath WR, Mueller SN. Spatiotemporally Distinct Interactions with Dendritic Cell Subsets Facilitates CD4+ and CD8+ T Cell Activation to Localized Viral Infection. Immunity. 2015;43:554–565. doi: 10.1016/j.immuni.2015.07.020. [DOI] [PubMed] [Google Scholar]
- Hoshi N, Schenten D, Nish SA, Walther Z, Gagliani N, Flavell RA, Reizis B, Shen Z, Fox JG, Iwasaki A, Medzhitov R. MyD88 signalling in colonic mononuclear phagocytes drives colitis in IL-10-deficient mice. Nat Commun. 2012;3:1120. doi: 10.1038/ncomms2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh CS, Macatonia SE, Tripp CS, Wolf SF, O’Garra A, Murphy KM. Development of TH1 CD4+ T cells through IL-12 produced by Listeria-induced macrophages. Science. 1993;260:547–549. doi: 10.1126/science.8097338. [DOI] [PubMed] [Google Scholar]
- Hua Z, Gross AJ, Lamagna C, Ramos-Hernandez N, Scapini P, Ji M, Shao H, Lowell CA, Hou B, DeFranco AL. Requirement for MyD88 signaling in B cells and dendritic cells for germinal center anti-nuclear antibody production in Lyn-deficient mice. J Immunol. 2014;192:875–885. doi: 10.4049/jimmunol.1300683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume DA. Macrophages as APC and the dendritic cell myth. J Immunol. 2008;181:5829–5835. doi: 10.4049/jimmunol.181.9.5829. [DOI] [PubMed] [Google Scholar]
- Idoyaga J, Fiorese C, Zbytnuik L, Lubkin A, Miller J, Malissen B, Mucida D, Merad M, Steinman RM. Specialized role of migratory dendritic cells in peripheral tolerance induction. J Clin Invest. 2013;123:844–854. doi: 10.1172/JCI65260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igyarto BZ, Haley K, Ortner D, Bobr A, Gerami-Nejad M, Edelson BT, Zurawski SM, Malissen B, Zurawski G, Berman J, Kaplan DH. Skin-Resident Murine Dendritic Cell Subsets Promote Distinct and Opposing Antigen-Specific T Helper Cell Responses. Immunity. 2011 doi: 10.1016/j.immuni.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba K, Swiggard WJ, Inaba M, Meltzer J, Mirza A, Sasagawa T, Nussenzweig MC, Steinman RM. Tissue distribution of the DEC-205 protein that is detected by the monoclonal antibody NLDC-145. I. Expression on dendritic cells and other subsets of mouse leukocytes. Cell Immunol. 1995;163:148–156. doi: 10.1006/cimm.1995.1109. [DOI] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A, Amit I. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S, Unutmaz D, Wong P, Sano G, De los SK, Sparwasser T, Wu S, Vuthoori S, Ko K, Zavala F, Pamer EG, Littman DR, Lang RA. In vivo depletion of CD11c(+) dendritic cells abrogates priming of CD8(+) T cells by exogenous cell-associated antigens. Immunity. 2002;17:211–220. doi: 10.1016/s1074-7613(02)00365-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashima K, Banks TA, Ansel KM, Lu TT, Ware CF, Cyster JG. Intrinsic lymphotoxin-beta receptor requirement for homeostasis of lymphoid tissue dendritic cells. Immunity. 2005;22:439–450. doi: 10.1016/j.immuni.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Kang SJ, Liang HE, Reizis B, Locksley RM. Regulation of Hierarchical Clustering and Activation of Innate Immune Cells by Dendritic Cells. Immunity. 2008;29:819–833. doi: 10.1016/j.immuni.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan DH, Jenison MC, Saeland S, Shlomchik WD, Shlomchik MJ. Epidermal langerhans cell-deficient mice develop enhanced contact hypersensitivity. Immunity. 2005;23:611–620. doi: 10.1016/j.immuni.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Kashem SW, Riedl MS, Yao C, Honda CN, Vulchanova L, Kaplan DH. Nociceptive Sensory Fibers Drive Interleukin-23 Production from CD301b+ Dermal Dendritic Cells and Drive Protective Cutaneous Immunity. Immunity. 2015;43:515–526. doi: 10.1016/j.immuni.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwada M, Pham NL, Pewe LL, Harty JT, Rothman PB. NFIL3/E4BP4 is a key transcription factor for CD8{alpha}+ dendritic cell development. Blood. 2011;117:6193–6197. doi: 10.1182/blood-2010-07-295873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassim SH, Rajasagi NK, Zhao X, Chervenak R, Jennings SR. In vivo ablation of CD11c-positive dendritic cells increases susceptibility to herpes simplex virus type 1 infection and diminishes NK and T-cell responses. J Virol. 2006;80:3985–3993. doi: 10.1128/JVI.80.8.3985-3993.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Rasmussen JP, Rudensky AY. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat Immunol. 2007;8:191–197. doi: 10.1038/ni1428. [DOI] [PubMed] [Google Scholar]
- Kim TS, Hufford MM, Sun J, Fu YX, Braciale TJ. Antigen persistence and the control of local T cell memory by migrant respiratory dendritic cells after acute virus infection. J Exp Med. 2010;207:1161–1172. doi: 10.1084/jem.20092017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnebrew MA, Buffie CG, Diehl GE, Zenewicz LA, Leiner I, Hohl TM, Flavell RA, Littman DR, Pamer EG. Interleukin 23 production by intestinal CD103(+)CD11b(+) dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity. 2012;36:276–287. doi: 10.1016/j.immuni.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissenpfennig A, Henri S, Dubois B, Laplace-Builhe C, Perrin P, Romani N, Tripp CH, Douillard P, Leserman L, Kaiserlian D, Saeland S, Davoust J, Malissen B. Dynamics and function of Langerhans cells in vivo: dermal dendritic cells colonize lymph node areas distinct from slower migrating Langerhans cells. Immunity. 2005;22:643–654. doi: 10.1016/j.immuni.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Kumamoto Y, Linehan M, Weinstein JS, Laidlaw BJ, Craft JE, Iwasaki A. CD301b(+) dermal dendritic cells drive T helper 2 cell-mediated immunity. Immunity. 2013;39:733–743. doi: 10.1016/j.immuni.2013.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurotaki D, Osato N, Nishiyama A, Yamamoto M, Ban T, Sato H, Nakabayashi J, Umehara M, Miyake N, Matsumoto N, Nakazawa M, Ozato K, Tamura T. Essential role of the IRF8-KLF4 transcription factor cascade in murine monocyte differentiation. Blood. 2013;121:1839–1849. doi: 10.1182/blood-2012-06-437863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamagna C, Scapini P, van Ziffle JA, DeFranco AL, Lowell CA. Hyperactivated MyD88 signaling in dendritic cells, through specific deletion of Lyn kinase, causes severe autoimmunity and inflammation. Proc Natl Acad Sci U S A. 2013;110:E3311–E3320. doi: 10.1073/pnas.1300617110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlet C, Tamoutounour S, Henri S, Luche H, Ardouin L, Gregoire C, Malissen B, Guilliams M. CD64 expression distinguishes monocyte-derived and conventional dendritic cells and reveals their distinct role during intramuscular immunization. J Immunol. 2012;188:1751–1760. doi: 10.4049/jimmunol.1102744. [DOI] [PubMed] [Google Scholar]
- Lau LS, Fernandez-Ruiz D, Mollard V, Sturm A, Neller MA, Cozijnsen A, Gregory JL, Davey GM, Jones CM, Lin YH, Haque A, Engwerda CR, Nie CQ, Hansen DS, Murphy KM, Papenfuss AT, Miles JJ, Burrows SR, Koning-Ward T, McFadden GI, Carbone FR, Crabb BS, Heath WR. CD8+ T cells from a novel T cell receptor transgenic mouse induce liver-stage immunity that can be boosted by blood-stage infection in rodent malaria. PLoS Pathog. 2014;10:e1004135. doi: 10.1371/journal.ppat.1004135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauterbach H, Zuniga EI, Truong P, Oldstone MB, McGavern DB. Adoptive immunotherapy induces CNS dendritic cell recruitment and antigen presentation during clearance of a persistent viral infection. J Exp Med. 2006;203:1963–1975. doi: 10.1084/jem.20060039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauvau G, Loke P, Hohl TM. Monocyte-mediated defense against bacteria, fungi, and parasites. Semin Immunol. 2015;27:397–409. doi: 10.1016/j.smim.2016.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gros G, Ben-Sasson SZ, Seder R, Finkelman FD, Paul WE. Generation of interleukin 4 (IL-4)-producing cells in vivo and in vitro: IL-2 and IL-4 are required for in vitro generation of IL-4-producing cells. J Exp Med. 1990;172:921–929. doi: 10.1084/jem.172.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon B, Ballesteros-Tato A, Browning JL, Dunn R, Randall TD, Lund FE. Regulation of T(H)2 development by CXCR5+ dendritic cells and lymphotoxin-expressing B cells. Nat Immunol. 2012;13:681–690. doi: 10.1038/ni.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leventhal DS, Gilmore DC, Berger JM, Nishi S, Lee V, Malchow S, Kline DE, Kline J, Vander Griend DJ, Huang H, Socci ND, Savage PA. Dendritic Cells Coordinate the Development and Homeostasis of Organ-Specific Regulatory T Cells. Immunity. 2016;44:847–859. doi: 10.1016/j.immuni.2016.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis KL, Caton ML, Bogunovic M, Greter M, Grajkowska LT, Ng D, Klinakis A, Charo IF, Jung S, Gommerman JL, Ivanov II, Liu K, Merad M, Reizis B. Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity. 2011;35:780–791. doi: 10.1016/j.immuni.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lu E, Yi T, Cyster JG. EBI2 augments Tfh cell fate by promoting interaction with IL-2-quenching dendritic cells. Nature. 2016;533:110–114. doi: 10.1038/nature17947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linehan JL, Dileepan T, Kashem SW, Kaplan DH, Cleary P, Jenkins MK. Generation of Th17 cells in response to intranasal infection requires TGF-beta1 from dendritic cells and IL-6 from CD301b+ dendritic cells. Proc Natl Acad Sci U S A. 2015;112:12782–12787. doi: 10.1073/pnas.1513532112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston A, Nutsch KM, Farr AG, Lund JM, Rasmussen JP, Koni PA, Rudensky AY. Differentiation of regulatory Foxp3+ T cells in the thymic cortex. Proc Natl Acad Sci U S A. 2008;105:11903–11908. doi: 10.1073/pnas.0801506105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CH, Fan YT, Dias A, Esper L, Corn RA, Bafica A, Machado FS, Aliberti J. Cutting edge: dendritic cells are essential for in vivo IL-12 production and development of resistance against Toxoplasma gondii infection in mice 1. J Immunol. 2006;177:31–35. doi: 10.4049/jimmunol.177.1.31. [DOI] [PubMed] [Google Scholar]
- Liu K, Victora GD, Schwickert TA, Guermonprez P, Meredith MM, Yao K, Chu FF, Randolph GJ, Rudensky AY, Nussenzweig M. In vivo analysis of dendritic cell development and homeostasis. Science. 2009;324:392–397. doi: 10.1126/science.1170540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman RS, Diehl GE, Victorio DA, Huh JR, Galan C, Miraldi ER, Swaminath A, Bonneau R, Scherl EJ, Littman DR. CX(3)CR1(+) mononuclear phagocytes support colitis-associated innate lymphoid cell production of IL-22. J Exp Med. 2014;211:1571–1583. doi: 10.1084/jem.20140678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loschko J, Rieke GJ, Schreiber HA, Meredith MM, Yao KH, Guermonprez P, Nussenzweig MC. Inducible targeting of cDCs and their subsets in vivo. J Immunol Methods. 2016a doi: 10.1016/j.jim.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loschko J, Schreiber HA, Rieke GJ, Esterhazy D, Meredith MM, Pedicord VA, Yao KH, Caballero S, Pamer EG, Mucida D, Nussenzweig MC. Absence of MHC class II on cDCs results in microbial-dependent intestinal inflammation. J Exp Med. 2016b;213:517–534. doi: 10.1084/jem.20160062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M, Schachterle W, Oberle K, Aichele P, Diefenbach A. Dendritic cells prime natural killer cells by trans-presenting interleukin 15. Immunity. 2007;26:503–517. doi: 10.1016/j.immuni.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luda KM, Joeris T, Persson EK, Rivollier A, Demiri M, Sitnik KM, Pool L, Holm JB, Melo-Gonzalez F, Richter L, Lambrecht BN, Kristiansen K, Travis MA, Svensson-Frej M, Kotarsky K, Agace WW. IRF8 Transcription-Factor-Dependent Classical Dendritic Cells Are Essential for Intestinal T Cell Homeostasis. Immunity. 2016;44:860–874. doi: 10.1016/j.immuni.2016.02.008. [DOI] [PubMed] [Google Scholar]
- Male V, Nisoli I, Kostrzewski T, Allan DS, Carlyle JR, Lord GM, Wack A, Brady HJ. The transcription factor E4bp4/Nfil3 controls commitment to the NK lineage and directly regulates Eomes and Id2 expression. J Exp Med. 2014;211:635–642. doi: 10.1084/jem.20132398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manicassamy S, Reizis B, Ravindran R, Nakaya H, Salazar-Gonzalez RM, Wang YC, Pulendran B. Activation of beta-catenin in dendritic cells regulates immunity versus tolerance in the intestine. Science. 2010;329:849–853. doi: 10.1126/science.1188510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Lopez M, Iborra S, Conde-Garrosa R, Sancho D. Batf3-dependent CD103+ dendritic cells are major producers of IL-12 that drive local Th1 immunity against Leishmania major infection in mice. Eur J Immunol. 2015;45:119–129. doi: 10.1002/eji.201444651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashayekhi M, Sandau MM, Dunay IR, Frickel EM, Khan A, Goldszmid RS, Sher A, Ploegh HL, Murphy TL, Sibley LD, Murphy KM. CD8a+ Dendritic Cells Are the Critical Source of Interleukin-12 that Controls Acute Infection by Toxoplasma gondii Tachyzoites. Immunity. 2011;35:249–259. doi: 10.1016/j.immuni.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzini E, Massimiliano L, Penna G, Rescigno M. Oral tolerance can be established via gap junction transfer of fed antigens from CX3CR1(+) macrophages to CD103(+) dendritic cells. Immunity. 2014;40:248–261. doi: 10.1016/j.immuni.2013.12.012. [DOI] [PubMed] [Google Scholar]
- McKenna HJ, Stocking KL, Miller RE, Brasel K, De Smedt T, Maraskovsky E, Maliszewski CR, Lynch DH, Smith J, Pulendran B, Roux ER, Teepe M, Lyman SD, Peschon JJ. Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood. 2000;95:3489–3497. [PubMed] [Google Scholar]
- Meredith MM, Liu K, Darrasse-Jeze G, Kamphorst AO, Schreiber HA, Guermonprez P, Idoyaga J, Cheong C, Yao KH, Niec RE, Nussenzweig MC. Expression of the zinc finger transcription factor zDC (Zbtb46, Btbd4) defines the classical dendritic cell lineage. J Exp Med. 2012;209:1153–1165. doi: 10.1084/jem.20112675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metlay JP, Witmer-Pack MD, Agger R, Crowley MT, Lawless D, Steinman RM. The distinct leukocyte integrins of mouse spleen dendritic cells as identified with new hamster monoclonal antibodies. J Exp Med. 1990;171:1753–1771. doi: 10.1084/jem.171.5.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milanovic M, Terszowski G, Struck D, Liesenfeld O, Carstanjen D. IFN consensus sequence binding protein (Icsbp) is critical for eosinophil development. J Immunol. 2008;181:5045–5053. doi: 10.4049/jimmunol.181.7.5045. [DOI] [PubMed] [Google Scholar]
- Mildner A, Jung S. Development and function of dendritic cell subsets. Immunity. 2014;40:642–656. doi: 10.1016/j.immuni.2014.04.016. [DOI] [PubMed] [Google Scholar]
- Mitamura T, Higashiyama S, Taniguchi N, Klagsbrun M, Mekada E. Diphtheria toxin binds to the epidermal growth factor (EGF)-like domain of human heparin-binding EGF-like growth factor/diphtheria toxin receptor and inhibits specifically its mitogenic activity. J Biol Chem. 1995;270:1015–1019. doi: 10.1074/jbc.270.3.1015. [DOI] [PubMed] [Google Scholar]
- Mortier E, Woo T, Advincula R, Gozalo S, Ma A. IL-15Ralpha chaperones IL-15 to stable dendritic cell membrane complexes that activate NK cells via trans presentation. J Exp Med. 2008;205:1213–1225. doi: 10.1084/jem.20071913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy R, Macdonald TT, Dougan G, Frankel G, Wiles S. Citrobacter rodentium of mice and man. Cell Microbiol. 2005;7:1697–1706. doi: 10.1111/j.1462-5822.2005.00625.x. [DOI] [PubMed] [Google Scholar]
- Murphy TL, Grajales-Reyes GE, Wu X, Tussiwand R, Briseno CG, Iwata A, Kretzer NM, Durai V, Murphy KM. Transcriptional Control of Dendritic Cell Development. Annu Rev Immunol. 2016;34:93–119. doi: 10.1146/annurev-immunol-032713-120204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzaki AR, Tetlak P, Sheng J, Loh SC, Setiagani YA, Poidinger M, Zolezzi F, Karjalainen K, Ruedl C. Intestinal CD103(+)CD11b(-) dendritic cells restrain colitis via IFN-gamma-induced anti-inflammatory response in epithelial cells. Mucosal Immunol. 2016;9:336–351. doi: 10.1038/mi.2015.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik SH, Sathe P, Park HY, Metcalf D, Proietto AI, Dakic A, Carotta S, O’Keeffe M, Bahlo M, Papenfuss A, Kwak JY, Wu L, Shortman K. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat Immunol. 2007;8:1217–1226. doi: 10.1038/ni1522. [DOI] [PubMed] [Google Scholar]
- Neuenhahn M, Kerksiek KM, Nauerth M, Suhre MH, Schiemann M, Gebhardt FE, Stemberger C, Panthel K, Schroder S, Chakraborty T, Jung S, Hochrein H, Russmann H, Brocker T, Busch DH. CD8alpha+ dendritic cells are required for efficient entry of Listeria monocytogenes into the spleen. Immunity. 2006;25:619–630. doi: 10.1016/j.immuni.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Nopora K, Bernhard CA, Ried C, Castello AA, Murphy KM, Marconi P, Koszinowski U, Brocker T. MHC class I cross-presentation by dendritic cells counteracts viral immune evasion. Front Immunol. 2012;3:348. doi: 10.3389/fimmu.2012.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussenzweig MC, Steinman RM, Gutchinov B, Cohn ZA. Dendritic cells are accessory cells for the development of anti-trinitrophenyl cytotoxic T lymphocytes. J Exp Med. 1980;152:1070–1084. doi: 10.1084/jem.152.4.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussenzweig MC, Steinman RM, Witmer MD, Gutchinov B. A monoclonal antibody specific for mouse dendritic cells. Proc Natl Acad Sci U S A. 1982;79:161–165. doi: 10.1073/pnas.79.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai S, Roediger B, Abtin A, Shklovskaya E, Fazekas de St GB, Yamane H, Weninger W, Le Gros G, Ronchese F. CD326loCD103loCD11blo Dermal Dendritic Cells Are Activated by Thymic Stromal Lymphopoietin during Contact Sensitization in Mice. J Immunol. 2014;193:2504–2511. doi: 10.4049/jimmunol.1400536. [DOI] [PubMed] [Google Scholar]
- Ohta T, Sugiyama M, Hemmi H, Yamazaki C, Okura S, Sasaki I, Fukuda Y, Orimo T, Ishii KJ, Hoshino K, Ginhoux F, Kaisho T. Crucial roles of XCR1-expressing dendritic cells and the XCR1-XCL1 chemokine axis in intestinal immune homeostasis. Sci Rep. 2016;6:23505. doi: 10.1038/srep23505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onai N, Obata-Onai A, Schmid MA, Ohteki T, Jarrossay D, Manz MG. Identification of clonogenic common Flt3(+) M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat. 2007;8:1207–1216. doi: 10.1038/ni1518. [DOI] [PubMed] [Google Scholar]
- Orr SL, Le D, Long JM, Sobieszczuk P, Ma B, Tian H, Fang X, Paulson JC, Marth JD, Varki N. A phenotype survey of 36 mutant mouse strains with gene-targeted defects in glycosyltransferases or glycan-binding proteins. Glycobiology. 2013;23:363–380. doi: 10.1093/glycob/cws150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmiter RD, Behringer RR, Quaife CJ, Maxwell F, Maxwell IH, Brinster RL. Cell lineage ablation in transgenic mice by cell-specific expression of a toxin gene. Cell. 1987;50:435–443. doi: 10.1016/0092-8674(87)90497-1. [DOI] [PubMed] [Google Scholar]
- Panea C, Farkas AM, Goto Y, Abdollahi-Roodsaz S, Lee C, Koscso B, Gowda K, Hohl TM, Bogunovic M, Ivanov II. Intestinal Monocyte-Derived Macrophages Control Commensal-Specific Th17 Responses. Cell Rep. 2015;12:1314–1324. doi: 10.1016/j.celrep.2015.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappenheimer AM, Jr, Harper AA, Moynihan M, Brockes JP. Diphtheria toxin and related proteins: effect of route of injection on toxicity and the determination of cytotoxicity for various cultured cells. J Infect Dis. 1982;145:94–102. doi: 10.1093/infdis/145.1.94. [DOI] [PubMed] [Google Scholar]
- Perry JS, Lio CW, Kau AL, Nutsch K, Yang Z, Gordon JI, Murphy KM, Hsieh CS. Distinct contributions of Aire and antigen-presenting-cell subsets to the generation of self-tolerance in the thymus. Immunity. 2014;41:414–426. doi: 10.1016/j.immuni.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson EK, Uronen-Hansson H, Semmrich M, Rivollier A, Hagerbrand K, Marsal J, Gudjonsson S, Hakansson U, Reizis B, Kotarsky K, Agace WW. IRF4 Transcription-Factor-Dependent CD103(+)CD11b(+) Dendritic Cells Drive Mucosal T Helper 17 Cell Differentiation. Immunity. 2013;38:958–969. doi: 10.1016/j.immuni.2013.03.009. [DOI] [PubMed] [Google Scholar]
- Perussia B, Fanning V, Trinchieri G. A leukocyte subset bearing HLA-DR antigens is responsible for in vitro alpha interferon production in response to viruses. Nat Immun Cell Growth Regul. 1985;4:120–137. [PubMed] [Google Scholar]
- Phythian-Adams AT, Cook PC, Lundie RJ, Jones LH, Smith KA, Barr TA, Hochweller K, Anderton SM, Hammerling GJ, Maizels RM, MacDonald AS. CD11c depletion severely disrupts Th2 induction and development in vivo. J Exp Med. 2010;207:2089–2096. doi: 10.1084/jem.20100734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piva L, Tetlak P, Claser C, Karjalainen K, Renia L, Ruedl C. Cutting edge: Clec9A+ dendritic cells mediate the development of experimental cerebral malaria. J Immunol. 2012;189:1128–1132. doi: 10.4049/jimmunol.1201171. [DOI] [PubMed] [Google Scholar]
- Plantinga M, Guilliams M, Vanheerswynghels M, Deswarte K, Branco-Madeira F, Toussaint W, Vanhoutte L, Neyt K, Killeen N, Malissen B, Hammad H, Lambrecht BN. Conventional and Monocyte-Derived CD11b(+) Dendritic Cells Initiate and Maintain T Helper 2 Cell-Mediated Immunity to House Dust Mite Allergen. Immunity. 2013 doi: 10.1016/j.immuni.2012.10.016. [DOI] [PubMed] [Google Scholar]
- Probst HC, Tschannen K, Odermatt B, Schwendener R, Zinkernagel RM, Van Den BM. Histological analysis of CD11c-DTR/GFP mice after in vivo depletion of dendritic cells. Clin Exp Immunol. 2005;141:398–404. doi: 10.1111/j.1365-2249.2005.02868.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst HC, Van Den BM. Priming of CTLs by lymphocytic choriomeningitis virus depends on dendritic cells. J Immunol. 2005;174:3920–3924. doi: 10.4049/jimmunol.174.7.3920. [DOI] [PubMed] [Google Scholar]
- Puttur F, Arnold-Schrauf C, Lahl K, Solmaz G, Lindenberg M, Mayer CT, Gohmert M, Swallow M, van Helt C, Schmitt H, Nitschke L, Lambrecht BN, Lang R, Messerle M, Sparwasser T. Absence of Siglec-H in MCMV infection elevates interferon alpha production but does not enhance viral clearance. PLoS Pathog. 2013;9:e1003648. doi: 10.1371/journal.ppat.1003648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboldi A, Arnon TI, Rodda LB, Atakilit A, Sheppard D, Cyster JG. IgA production requires B cell interaction with subepithelial dendritic cells in Peyer’s patches. Science. 2016;352:aaf4822. doi: 10.1126/science.aaf4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts EW, Broz ML, Binnewies M, Headley MB, Nelson AE, Wolf DM, Kaisho T, Bogunovic D, Bhardwaj N, Krummel MF. Critical Role for CD103+/CD141+ Dendritic Cells Bearing CCR7 for Tumor Antigen Trafficking and Priming of T Cell Immunity in Melanoma. Cancer Cell. 2016 doi: 10.1016/j.ccell.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson EA, Henriksen O, Maxwell ES. Elongation factor 2. Amino acid sequence at the site of adenosine diphosphate ribosylation. J Biol Chem. 1974;249:5088–5093. [PubMed] [Google Scholar]
- Saito M, Iwawaki T, Taya C, Yonekawa H, Noda M, Inui Y, Mekada E, Kimata Y, Tsuru A, Kohno K. Diphtheria toxin receptor-mediated conditional and targeted cell ablation in transgenic mice. Nat Biotechnol. 2001;19:746–750. doi: 10.1038/90795. [DOI] [PubMed] [Google Scholar]
- Salmon H, Idoyaga J, Rahman A, Leboeuf M, Remark R, Jordan S, Casanova-Acebes M, Khudoynazarova M, Agudo J, Tung N, Chakarov S, Rivera C, Hogstad B, Bosenberg M, Hashimoto D, Gnjatic S, Bhardwaj N, Palucka AK, Brown BD, Brody J, Ginhoux F, Merad M. Expansion and Activation of CD103(+) Dendritic Cell Progenitors at the Tumor Site Enhances Tumor Responses to Therapeutic PD-L1 and BRAF Inhibition. Immunity. 2016;44:924–938. doi: 10.1016/j.immuni.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samstein M, Schreiber HA, Leiner IM, Susac B, Glickman MS, Pamer EG. Essential yet limited role for CCR2(+) inflammatory monocytes during Mycobacterium tuberculosis-specific T cell priming. Elife. 2013;2:e01086. doi: 10.7554/eLife.01086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Paulete AR, Cueto FJ, Martinez-Lopez M, Labiano S, Morales-Kastresana A, Rodriguez-Ruiz ME, Jure-Kunkel M, Azpilikueta A, Aznar MA, Quetglas JI, Sancho D, Melero I. Cancer Immunotherapy with Immunomodulatory Anti-CD137 and Anti-PD-1 Monoclonal Antibodies Requires BATF3-Dependent Dendritic Cells. Cancer Discov. 2016;6:71–79. doi: 10.1158/2159-8290.CD-15-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho D, Mourao-Sa D, Joffre OP, Schulz O, Rogers NC, Pennington DJ, Carlyle JR, Sousa Reis E. Tumor therapy in mice via antigen targeting to a novel, DC-restricted C-type lectin. J Clin Invest. 2008;118:2098–2110. doi: 10.1172/JCI34584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapoznikov A, Fischer JA, Zaft T, Krauthgamer R, Dzionek A, Jung S. Organ-dependent in vivo priming of naive CD4+, but not CD8+, T cells by plasmacytoid dendritic cells. J Exp Med. 2007;204:1923–1933. doi: 10.1084/jem.20062373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Kurotaki D, Osato N, Sato H, Sasaki I, Koizumi S, Wang H, Kaneda C, Nishiyama A, Kaisho T, Aburatani H, Morse HC, III, Ozato K, Tamura T. Transcription factor IRF8 plays a critical role in the development of murine basophils and mast cells. Blood. 2015;125:358–369. doi: 10.1182/blood-2014-02-557983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Takeuchi O, Vandenbon A, Yasuda K, Tanaka Y, Kumagai Y, Miyake T, Matsushita K, Okazaki T, Saitoh T, Honma K, Matsuyama T, Yui K, Tsujimura T, Standley DM, Nakanishi K, Nakai K, Akira S. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol. 2010;11:936–944. doi: 10.1038/ni.1920. [DOI] [PubMed] [Google Scholar]
- Satpathy AT, Briseno CG, Lee JS, Ng D, Manieri NA, KCW, Wu X, Thomas SR, Lee WL, Turkoz M, McDonald KG, Meredith MM, Song C, Guidos CJ, Newberry RD, Ouyang W, Murphy TL, Stappenbeck TS, Gommerman JL, Nussenzweig MC, Colonna M, Kopan R, Murphy KM. Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat Immunol. 2013;14:937–948. doi: 10.1038/ni.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpathy AT, KCW, Albring JC, Edelson BT, Kretzer NM, Bhattacharya D, Murphy TL, Murphy KM. Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J Exp Med. 2012;209:1135–1152. doi: 10.1084/jem.20120030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarlett UK, Rutkowski MR, Rauwerdink AM, Fields J, Escovar-Fadul X, Baird J, Cubillos-Ruiz JR, Jacobs AC, Gonzalez JL, Weaver J, Fiering S, Conejo-Garcia JR. Ovarian cancer progression is controlled by phenotypic changes in dendritic cells. J Exp Med. 2012;209:495–506. doi: 10.1084/jem.20111413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharton-Kersten T, Contursi C, Masumi A, Sher A, Ozato K. Interferon consensus sequence binding protein-deficient mice display impaired resistance to intracellular infection due to a primary defect in interleukin 12 p40 induction. J Exp Med. 1997;186:1523–1534. doi: 10.1084/jem.186.9.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavoni G, Mattei F, Sestili P, Borghi P, Venditti M, Morse HC, III, Belardelli F, Gabriele L. ICSBP is essential for the development of mouse type I interferon-producing cells and for the generation and activation of CD8alpha(+) dendritic cells. J Exp Med. 2002;196:1415–1425. doi: 10.1084/jem.20021263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleicher U, Liese J, Knippertz I, Kurzmann C, Hesse A, Heit A, Fischer JA, Weiss S, Kalinke U, Kunz S, Bogdan C. NK cell activation in visceral leishmaniasis requires TLR9, myeloid DCs, and IL-12, but is independent of plasmacytoid DCs. J Exp Med. 2007;204:893–906. doi: 10.1084/jem.20061293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlitzer A, McGovern N, Teo P, Zelante T, Atarashi K, Low D, Ho AW, See P, Shin A, Wasan PS, Hoeffel G, Malleret B, Heiseke A, Chew S, Jardine L, Purvis HA, Hilkens CM, Tam J, Poidinger M, Stanley ER, Krug AB, Renia L, Sivasankar B, Ng LG, Collin M, Ricciardi-Castagnoli P, Honda K, Haniffa M, Ginhoux F. IRF4 Transcription Factor-Dependent CD11b(+) Dendritic Cells in Human and Mouse Control Mucosal IL-17 Cytokine Responses. Immunity. 2013;38:970–983. doi: 10.1016/j.immuni.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlitzer A, Sivakamasundari V, Chen J, Sumatoh HR, Schreuder J, Lum J, Malleret B, Zhang S, Larbi A, Zolezzi F, Renia L, Poidinger M, Naik S, Newell EW, Robson P, Ginhoux F. Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat Immunol. 2015 doi: 10.1038/ni.3200. [DOI] [PubMed] [Google Scholar]
- Schraml BU, van Blijswijk J, Zelenay S, Whitney PG, Filby A, Acton SE, Rogers NC, Moncaut N, Carvajal JJ, Sousa Reis E. Genetic tracing via DNGR-1 expression history defines dendritic cells as a hematopoietic lineage. Cell. 2013;154:843–858. doi: 10.1016/j.cell.2013.07.014. [DOI] [PubMed] [Google Scholar]
- Schreiber HA, Loschko J, Karssemeijer RA, Escolano A, Meredith MM, Mucida D, Guermonprez P, Nussenzweig MC. Intestinal monocytes and macrophages are required for T cell polarization in response to Citrobacter rodentium. J Exp Med. 2013;210:2025–2039. doi: 10.1084/jem.20130903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott CL, Soen B, Martens L, Skrypek N, Saelens W, Taminau J, Blancke G, Van Isterdael G, Huylebroeck D, Haigh J, Saeys Y, Guilliams M, Lambrecht BN, Berx G. The transcription factor Zeb2 regulates development of conventional and plasmacytoid DCs by repressing Id2. J Exp Med. 2016;213:897–911. doi: 10.1084/jem.20151715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seillet C, Rankin LC, Groom JR, Mielke LA, Tellier J, Chopin M, Huntington ND, Belz GT, Carotta S. Nfil3 is required for the development of all innate lymphoid cell subsets. J Exp Med. 2014;211:1733–1740. doi: 10.1084/jem.20140145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafini N, Vosshenrich CA, Di Santo JP. Transcriptional regulation of innate lymphoid cell fate. Nat Rev Immunol. 2015;15:415–428. doi: 10.1038/nri3855. [DOI] [PubMed] [Google Scholar]
- Sharma S, Zhu J. Immunologic applications of conditional gene modification technology in the mouse. Curr Protoc Immunol. 2014;105:10–13. doi: 10.1002/0471142735.im1034s105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K, Kurosawa Y, Taniguchi M, Steinman RM, Fujii S. Cross-presentation of glycolipid from tumor cells loaded with alpha-galactosylceramide leads to potent and long-lived T cell mediated immunity via dendritic cells. J Exp Med. 2007;204:2641–2653. doi: 10.1084/jem.20070458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sichien D, Scott CL, Martens L, Vanderkerken M, Van Gassen S, Plantinga M, Joeris T, De Prijck S, Vanhoutte L, Vanheerswynghels M, Van Isterdael G, Toussaint W, Madeira FB, Vergote K, Agace WW, Clausen BE, Hammad H, Dalod M, Saeys Y, Lambrecht BN, Guilliams M. IRF8 Transcription Factor Controls Survival and Function of Terminally Differentiated Conventional and Plasmacytoid Dendritic Cells, Respectively. Immunity. 2016;45:626–640. doi: 10.1016/j.immuni.2016.08.013. [DOI] [PubMed] [Google Scholar]
- Siegal FP, Kadowaki N, Shodell M, Fitzgerald-Bocarsly PA, Shah K, Ho, Antonenko S, Liu YJ. The nature of the principal type 1 interferon-producing cells in human blood. Science. 1999;284:1835–1837. doi: 10.1126/science.284.5421.1835. [DOI] [PubMed] [Google Scholar]
- Smith KA, Harcus Y, Garbi N, Hammerling GJ, MacDonald AS, Maizels RM. Type 2 innate immunity in helminth infection is induced redundantly and acts autonomously following CD11c(+) cell depletion. Infect Immun. 2012;80:3481–3489. doi: 10.1128/IAI.00436-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KA, Hochweller K, Hammerling GJ, Boon L, MacDonald AS, Maizels RM. Chronic helminth infection promotes immune regulation in vivo through dominance of CD11cloC. J Immunol. 2011;186:7098–7109. doi: 10.4049/jimmunol.1003636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol CL, Chu NQ, Yu S, Nish SA, Laufer TM, Medzhitov R. Basophils function as antigen-presenting cells for an allergen-induced T helper type 2 response. Nat Immunol. 2009;10:713–720. doi: 10.1038/ni.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg GF, Artis D. Innate lymphoid cells in the initiation, regulation and resolution of inflammation. Nat Med. 2015;21:698–708. doi: 10.1038/nm.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature. 2015;523:231–235. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman RM, Cohn ZA. Identification of a novel cell type in peripheral lymphoid organs of mice. I. Morphology, quantitation, tissue distribution. J Exp Med. 1973;137:1142–1162. doi: 10.1084/jem.137.5.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman RM, Kaplan G, Witmer MD, Cohn ZA. Identification of a novel cell type in peripheral lymphoid organs of mice. V. Purification of spleen dendritic cells, new surface markers, and maintenance in vitro. J Exp Med. 1979;149:1–16. doi: 10.1084/jem.149.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman RM, Witmer MD. Lymphoid dendritic cells are potent stimulators of the primary mixed leukocyte reaction in mice. Proc Natl Acad Sci U S A. 1978;75:5132–5136. doi: 10.1073/pnas.75.10.5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranges PB, Watson J, Cooper CJ, Choisy-Rossi CM, Stonebraker AC, Beighton RA, Hartig H, Sundberg JP, Servick S, Kaufmann G, Fink PJ, Chervonsky AV. Elimination of antigen-presenting cells and autoreactive T cells by Fas contributes to prevention of autoimmunity. Immunity. 2007;26:629–641. doi: 10.1016/j.immuni.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiecki M, Gilfillan S, Vermi W, Wang Y, Colonna M. Plasmacytoid dendritic cell ablation impacts early interferon responses and antiviral NK and CD8(+) T cell accrual. Immunity. 2010;33:955–966. doi: 10.1016/j.immuni.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiecki M, Wang Y, Riboldi E, Kim AH, Dzutsev A, Gilfillan S, Vermi W, Ruedl C, Trinchieri G, Colonna M. Cell depletion in mice that express diphtheria toxin receptor under the control of SiglecH encompasses more than plasmacytoid dendritic cells. J Immunol. 2014;192:4409–4416. doi: 10.4049/jimmunol.1303135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi H, Fukaya T, Eizumi K, Sato Y, Sato K, Shibazaki A, Otsuka H, Hijikata A, Watanabe T, Ohara O, Kaisho T, Malissen B, Sato K. Plasmacytoid dendritic cells are crucial for the initiation of inflammation and T cell immunity in vivo. Immunity. 2011;35:958–971. doi: 10.1016/j.immuni.2011.10.014. [DOI] [PubMed] [Google Scholar]
- Tang H, Cao W, Kasturi SP, Ravindran R, Nakaya HI, Kundu K, Murthy N, Kepler TB, Malissen B, Pulendran B. The T helper type 2 response to cysteine proteases requires dendritic cell-basophil cooperation via ROS-mediated signaling. Nat Immunol. 2010;11:608–617. doi: 10.1038/ni.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichmann LL, Ols ML, Kashgarian M, Reizis B, Kaplan DH, Shlomchik MJ. Dendritic cells in lupus are not required for activation of T and B cells but promote their expansion, resulting in tissue damage. Immunity. 2010;33:967–978. doi: 10.1016/j.immuni.2010.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichmann LL, Schenten D, Medzhitov R, Kashgarian M, Shlomchik MJ. Signals via the adaptor MyD88 in B cells and DCs make distinct and synergistic contributions to immune activation and tissue damage in lupus. Immunity. 2013;38:528–540. doi: 10.1016/j.immuni.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Woodworth J, Skold M, Behar SM. In vivo depletion of CD11c+ cells delays the CD4+ T cell response to Mycobacterium tuberculosis and exacerbates the outcome of infection. J Immunol. 2005;175:3268–3272. doi: 10.4049/jimmunol.175.5.3268. [DOI] [PubMed] [Google Scholar]
- Torti N, Walton SM, Murphy KM, Oxenius A. Batf3 transcription factor-dependent DC subsets in murine CMV infection: differential impact on T-cell priming and memory inflation. Eur J Immunol. 2011;41:2612–2618. doi: 10.1002/eji.201041075. [DOI] [PubMed] [Google Scholar]
- Trautwein-Weidner K, Gladiator A, Kirchner FR, Becattini S, Rulicke T, Sallusto F, LeibundGut-Landmann S. Antigen-Specific Th17 Cells Are Primed by Distinct and Complementary Dendritic Cell Subsets in Oropharyngeal Candidiasis. PLoS Pathog. 2015;11:e1005164. doi: 10.1371/journal.ppat.1005164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis MA, Reizis B, Melton AC, Masteller E, Tang Q, Proctor JM, Wang Y, Bernstein X, Huang X, Reichardt LF, Bluestone JA, Sheppard D. Loss of integrin alpha(v)beta8 on dendritic cells causes autoimmunity and colitis in mice. Nature. 2007;449:361–365. doi: 10.1038/nature06110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumanov AV, Koroleva EP, Guo X, Wang Y, Kruglov A, Nedospasov S, Fu YX. Lymphotoxin controls the IL-22 protection pathway in gut innate lymphoid cells during mucosal pathogen challenge. Cell Host Microbe. 2011;10:44–53. doi: 10.1016/j.chom.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tussiwand R, Everts B, Grajales-Reyes GE, Kretzer NM, Iwata A, Bagaitkar J, Wu X, Wong R, Anderson DA, Murphy TL, Pearce EJ, Murphy KM. Klf4 expression in conventional dendritic cells is required for T helper 2 cell responses. Immunity. 2015;42:916–928. doi: 10.1016/j.immuni.2015.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tussiwand R, Lee WL, Murphy TL, Mashayekhi M, Wumesh KC, Albring JC, Satpathy AT, Rotondo JA, Edelson BT, Kretzer NM, Wu X, Weiss LA, Glasmacher E, Li P, Liao W, Behnke M, Lam SS, Aurthur CT, Leonard WJ, Singh H, Stallings CL, Sibley LD, Schreiber RD, Murphy KM. Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature. 2012;490:502–507. doi: 10.1038/nature11531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Blijswijk J, Schraml BU, Rogers NC, Whitney PG, Zelenay S, Acton SE, Sousa Reis E. Altered lymph node composition in diphtheria toxin receptor-based mouse models to ablate dendritic cells. J Immunol. 2015;194:307–315. doi: 10.4049/jimmunol.1401999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ness BG, Howard JB, Bodley JW. ADP-ribosylation of elongation factor 2 by diphtheria toxin. Isolation and properties of the novel ribosyl-amino acid and its hydrolysis products. J Biol Chem. 1980;255:10717–10720. [PubMed] [Google Scholar]
- van Rijt LS, Jung S, KleinJan A, Vos N, Willart M, Duez C, Hoogsteden HC, Lambrecht BN. In vivo depletion of lung CD11c+ dendritic cells during allergen challenge abrogates the characteristic features of asthma. J Exp Med. 2005;201:981–991. doi: 10.1084/jem.20042311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Victora GD, Schwickert TA, Fooksman DR, Kamphorst AO, Meyer-Hermann M, Dustin ML, Nussenzweig MC. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 2010;143:592–605. doi: 10.1016/j.cell.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voehringer D, Liang HE, Locksley RM. Homeostasis and effector function of lymphopenia-induced “memory-like” T cells in constitutively T cell-depleted mice. J Immunol. 2008;180:4742–4753. doi: 10.4049/jimmunol.180.7.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waithman J, Zanker D, Xiao K, Oveissi S, Wylie B, Ng R, Togel L, Chen W. Resident CD8(+) and migratory CD103(+) dendritic cells control CD8 T cell immunity during acute influenza infection. PLoS One. 2013;8:e66136. doi: 10.1371/journal.pone.0066136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Lee CH, Qi C, Tailor P, Feng J, Abbasi S, Atsumi T, Morse HC., III IRF8 regulates B-cell lineage specification, commitment, and differentiation. Blood. 2008;112:4028–4038. doi: 10.1182/blood-2008-01-129049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weih F, Carrasco D, Durham SK, Barton DS, Rizzo CA, Ryseck RP, Lira SA, Bravo R. Multiorgan inflammation and hematopoietic abnormalities in mice with a targeted disruption of RelB, a member of the NF-kappa B/Rel family. Cell. 1995;80:331–340. doi: 10.1016/0092-8674(95)90416-6. [DOI] [PubMed] [Google Scholar]
- Welty NE, Staley C, Ghilardi N, Sadowsky MJ, Igyarto BZ, Kaplan DH. Intestinal lamina propria dendritic cells maintain T cell homeostasis but do not affect commensalism. J Exp Med. 2013 doi: 10.1084/jem.20130728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JW, Tjota MY, Clay BS, Vander LB, Bandukwala HS, Hrusch CL, Decker DC, Blaine KM, Fixsen BR, Singh H, Sciammas R, Sperling AI. Transcription factor IRF4 drives dendritic cells to promote Th2 differentiation. Nat Commun. 2013;4:2990. doi: 10.1038/ncomms3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, D’Amico A, Winkel KD, Suter M, Lo D, Shortman K. RelB is essential for the development of myeloid-related CD8alpha- dendritic cells but not of lymphoid-related CD8alpha+ dendritic cells. Immunity. 1998;9:839–847. doi: 10.1016/s1074-7613(00)80649-4. [DOI] [PubMed] [Google Scholar]
- Yamazaki C, Sugiyama M, Ohta T, Hemmi H, Hamada E, Sasaki I, Fukuda Y, Yano T, Nobuoka M, Hirashima T, Iizuka A, Sato K, Tanaka T, Hoshino K, Kaisho T. Critical roles of a dendritic cell subset expressing a chemokine receptor, XCR1. J Immunol. 2013;190:6071–6082. doi: 10.4049/jimmunol.1202798. [DOI] [PubMed] [Google Scholar]
- Yokota Y, Mansouri A, Mori S, Sugawara S, Adachi S, Nishikawa S, Gruss P. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature. 1999;397:702–706. doi: 10.1038/17812. [DOI] [PubMed] [Google Scholar]
- Young JA, He TH, Reizis B, Winoto A. Commensal microbiota are required for systemic inflammation triggered by necrotic dendritic cells. Cell Rep. 2013;3:1932–1944. doi: 10.1016/j.celrep.2013.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaft T, Sapoznikov A, Krauthgamer R, Littman DR, Jung S. CD11chigh dendritic cell ablation impairs lymphopenia-driven proliferation of naive and memory CD8+ T cells. J Immunol. 2005;175:6428–6435. doi: 10.4049/jimmunol.175.10.6428. [DOI] [PubMed] [Google Scholar]
- Zahner SP, Kel JM, Martina CA, Brouwers-Haspels I, van Roon MA, Clausen BE. Conditional deletion of TGF-betaR1 using Langerin-Cre mice results in Langerhans cell deficiency and reduced contact hypersensitivity. J Immunol. 2011;187:5069–5076. doi: 10.4049/jimmunol.1101880. [DOI] [PubMed] [Google Scholar]
- Zammit DJ, Cauley LS, Pham QM, Lefrancois L. Dendritic cells maximize the memory CD8 T cell response to infection. Immunity. 2005;22:561–570. doi: 10.1016/j.immuni.2005.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelenay S, Keller AM, Whitney PG, Schraml BU, Deddouche S, Rogers NC, Schulz O, Sancho D, Sousa Reis E. The dendritic cell receptor DNGR-1 controls endocytic handling of necrotic cell antigens to favor cross-priming of CTLs in virus-infected mice. J Clin Invest. 2012;122:1615–1627. doi: 10.1172/JCI60644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Raper A, Sugita N, Hingorani R, Salio M, Palmowski MJ, Cerundolo V, Crocker PR. Characterization of Siglec-H as a novel endocytic receptor expressed on murine plasmacytoid dendritic cell precursors. Blood. 2006;107:3600–3608. doi: 10.1182/blood-2005-09-3842. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, Ouyang W. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]