Abstract
A typical genome-wide association study tests correlation between a single phenotype and each genotype one at a time. However, single-phenotype analysis might miss unmeasured aspects of complex biological networks. Analyzing many phenotypes simultaneously may increase the power to capture these unmeasured aspects and detect more variants. Several multivariate approaches aim to detect variants related to more than one phenotype, but these current approaches do not consider the effects of population structure. As a result, these approaches may result in a significant amount of false positive identifications. Here, we introduce a new methodology, referred to as GAMMA for generalized analysis of molecular variance for mixed-model analysis, which is capable of simultaneously analyzing many phenotypes and correcting for population structure. In a simulated study using data implanted with true genetic effects, GAMMA accurately identifies these true effects without producing false positives induced by population structure. In simulations with this data, GAMMA is an improvement over other methods which either fail to detect true effects or produce many false positive identifications. We further apply our method to genetic studies of yeast and gut microbiome from mice and show that GAMMA identifies several variants that are likely to have true biological mechanisms.
Keywords: multivariate analysis, population structure, mixed models
OVER the past few years, genome-wide association studies (GWAS) have been used to find genetic variants that are involved in disease and other traits by testing for correlations between these traits and genetic variants across the genome. A typical GWAS examines the correlation of a single phenotype and each genotype one at a time. Recently, large amounts of genomic data, including expression data, have been collected from GWAS cohorts. This data often contains thousands of phenotypes per individual. The standard approach to analyzing this type of data involves performing a single-phenotype analysis: a GWAS on each phenotype individually.
The genomic loci that are of the most interest are the loci that simultaneously affect many phenotypes. For example, researchers often seek genetic variants that affect the profile of gut microbiota, which encompass 10s of 1000s of species (Lockhart et al. 1996; Gygi et al. 1999). Another example is when researchers want to detect regulatory hotspots in expression quantitative trait loci (eQTL) studies. Many genes are known to be regulated by a small number of genomic regions called trans-regulatory hotspots (Wang et al. 2004; Cervino et al. 2005; Hillebrandt et al. 2005), which strongly indicate the presence of master regulators of transcription. Moreover, current strategies for analyzing phenotypes independently are underpowered. A more powerful approach could capture the unmeasured aspects of complex biological networks, such as protein mediators, together with many phenotypes that might otherwise be missed when using an approach that focuses on a single phenotype or a few phenotypes (O’Reilly et al. 2012).
Many multivariate methods have been proposed that are designed to jointly analyze large numbers of genomic phenotypes. Most of the methods perform some form of data reduction, such as cluster analysis and factor analysis (Alter et al. 2000; Quackenbush 2001). However, these data-reduction methods have many issues such as the difficulty of determining the number of principal components, doubts about the generalizability of principal components, etc. (Nievergelt et al. 2007). Aschard et al. (2014) discussed the performance of different principal component analysis-based strategies for multiple-phenotype analysis and showed that testing only the top principal components often have low power, whereas combining signals across all principal components can have greater power in the analysis. Alternatively, Zapala and Schork (2012) proposed a way of analyzing high dimensional data called multivariate distance matrix regression (MDMR) analysis. MDMR uses a distance matrix whose elements are tested for association with independent variables of interest. This method is simple and directly applicable to high dimensional multiple-phenotype analysis. In addition, users can flexibly choose appropriate distance matrices (Webb 2002; Wessel and Schork 2006).
Each of the previous methods is based on the assumption that the phenotypes of the individuals are independently and identically distributed (i.i.d.). Unfortunately, as has been shown in GWAS, this assumption is invalid due to a phenomenon referred to as population structure. Allele frequencies are known to vary widely from population to population, because each population carries its own unique genetic and social history. These differences in allele frequencies, along with the correlation of a phenotype with its populations, may cause spurious correlation between genotypes and phenotypes and induce spurious associations (Kittles et al. 2002; Freedman et al. 2004; Marchini et al. 2004; Campbell et al. 2005; Helgason et al. 2005; Reiner et al. 2005; Voight and Pritchard 2005; Berger et al. 2006; Foll and Gaggiotti 2006; Seldin et al. 2006; Flint and Eskin 2012). These errors potentially compound when analyzing multiple phenotypes because biases in test statistics accumulate from each phenotype, which is shown in our experiments. Unfortunately, none of the previously discussed multivariate methods are able to correct for population structure and may cause a significant number of false positive results. Recently, multiple-phenotypes analysis methods have been developed that consider population structure (Korte et al. 2012; Zhou and Stephens 2014). However, these methods are impractical for cases with large number of phenotypes (>100) since their computational time scales quadratically with the number of phenotypes considered.
In this article, we propose a method, called GAMMA (generalized analysis of molecular variance for mixed-model analysis), which efficiently analyzes large numbers of phenotypes while simultaneously considering population structure. Recently, the linear mixed model has become a popular approach for GWAS as it can correct for population structure (Kang et al. 2008, 2010; Lippert et al. 2011; Segura et al. 2012; Svishcheva et al. 2012; Zhou and Stephens 2012; Hormozdiari et al. 2015). The linear mixed model incorporates genetic similarities between all pairs of individuals, known as kinship, into their model and corrects for population structure. We take the key principles behind MDMR (Nievergelt et al. 2007; Zapala and Schork 2012), which performs multivariate regression using distance matrices to form a statistic for testing the effects of covariates on multiple phenotypes. To correct for population structure, we extend the statistical procedure of MDMR to incorporate the linear mixed model.
To demonstrate the utility of GAMMA, we use both simulated and real data sets and compared our method with representative previous approaches. These approaches include the standard t-test, one of the standard and the simplest method for GWAS; efficient mixed-model association (EMMA) (Kang et al. 2008), a representative single-phenotype analysis method that implements linear mixed model and corrects for population structure (Lippert et al. 2011; Zhou and Stephens 2012); and MDMR (Zapala and Schork 2012), a multiple-phenotypes analysis method. In a simulated study, GAMMA corrects for population structure and accurately identifies genetic variants associated with phenotypes. In comparison, the previous approaches we tested, which analyze each phenotype individually, do not have enough power to detect associations and are not able to detect variants. MDMR (Zapala and Schork 2012) predicts many spurious associations produced due to population structure. We further applied GAMMA to two real data sets. When applied to a yeast data set, GAMMA identified most of the regulatory hotspots identified as related to regulatory elements in a previous study (Joo et al. 2014); while the previous approaches we tested failed to detect those hotspots. When applied to a gut microbiome data set from mice, GAMMA corrected for population structure and identified regions of the genome that harbor variants responsible for taxa abundances. In comparison, the previous methods we tested either failed to identify any of the variants in the region or produced a significant number of false positives.
Materials and Methods
Linear mixed models
For analyzing the ith SNP, we assume the following linear mixed model as the generative model:
| (1) |
Let n be the number of individuals and m be the number of genes. Here, Y is an matrix, where each column vector contains the jth phenotype values; is a vector of length n with genotypes of the ith SNP; and β is a vector of length m, where each entry contains an effect of the ith SNP on the jth phenotype. U is an matrix, where each column vector contains the effect of population structure of the jth phenotype. E is an matrix, where each column vector contains i.i.d. residual errors of the jth phenotype. We assume the random effects, and follow multivariate normal distribution, and where K is a known genetic similarity matrix and I is an identity matrix with unknown magnitudes and respectively.
Multiple-phenotypes analysis
Let us say we are analyzing associations between the ith SNP and the jth phenotype. Traditional univariate analysis is based on the following linear model:
| (2) |
Here, is a vector of length n with the jth phenotype values, is a vector of length n with the ith SNP values, is a value contains an effect of the ith SNP on the jth phenotype, and is a vector of length n with i.i.d. residual errors of the jth phenotype. To test associations, we test the null hypothesis against the alternative hypothesis We can perform an F-test for the analysis by comparing two models, model 1: and model 2: The standard F-statistic is given as follows:
| (3) |
where and are the residual sum of squares (RSS) of model 1 and model 2, respectively; and and are the number of parameters in model 1 and model 2, respectively. Applying this statistic (Equation 3) to our case, we find the following:
| (4) |
where and Applying Equation 4 to Equation 3, we find the following F-statistic:
| (5) |
Using the fact that the statistics follow we could extend the univariate case into a multivariate case in the following:
| (6) |
where Y is an matrix, where each column vector contains the jth phenotype values; β is a vector of length m, where each entry contains an effect of the ith SNP on the jth phenotype; and E is an matrix, where each column vector contains i.i.d. residual errors of the jth phenotype. Here, we assume that the random effect follows multivariate normal distribution, where I is an identity matrix with unknown magnitude In the multivariate case, both and are matrices, where the diagonal element is for the jth phenotype as computed in the univariate case. Given this, if we take the trace of this matrix, we obtain a sum of statistics. Thus in the multivariate case (Equation 6), we can estimate a pseudo-F-statistic as follows:
| (7) |
where The reason why we call this a “pseudo” F-statistic is because it is not guaranteed that we are summing independent statistics, and when they are not independent we do not expect that the result is also
Here we note that the trace of an inner product matrix is the same as the trace of an outer product matrix: and The advantage of this duality is that we can estimate the trace of and from the outer product matrix by using the fact that and The outer product matrix could be obtained from any symmetric matrix of distances (Gower 1966; McArdle and Anderson 2001). Let us say we have a distance matrix D with each element Let A be a matrix where each element and we can center the matrix by taking Gower’s centered matrix G (Gower 1966; McArdle and Anderson 2001):
| (8) |
where 1 is a column of 1’s of length n. Then this matrix G is an outer-product matrix and we can generate a pseudo-F-statistic from a distance matrix as follows:
| (9) |
Correcting for population structure
In GWAS, it is widely known that genetic relatedness, referred to as population structure, complicates analysis by creating spurious associations. The linear model (Equation 6) does not account for population structure, and applying the model to the multiple-phenotypes analysis may induce false positive identifications. Recently, the linear mixed model has emerged as a powerful tool for GWAS as it could correct for the population structure. GAMMA incorporates the effect of population structure by assuming a linear mixed model (Equation 1), which has an extra term U accounting for the effects of population structure, instead of the conventional linear model (Equation 6). This is an extension of the following widely used linear mixed model for a univariate analysis:
Based on the linear mixed model (Equation 1), each phenotype follows a multivariate normal distribution with mean and variance as follows:
where is the variance of the jth phenotype. We compute a covariance matrix, as described in Implementation, and the alternate model is transformed by the inverse square root of this matrix as follows:
Thus, to incorporate population structure, we transform genotypes and phenotypes, and and apply them to Equation 9 to get an alternative pseudo-F-statistic as follows:
where and is a Gower’s centered matrix estimated from in turn estimated from where each column vector of is
Efficiency of GAMMA
There are several multiple-phenotypes analysis methods considering population structure (Korte et al. 2012; Zhou and Stephens 2014). These methods explicitly model the dependencies of phenotypes to accurately estimate associations between a SNP and phenotypes. However, their computational time is quadratic or cubic to the number of phenotypes; thus, they are only applicable for data sets with no more than 100 phenotypes. These methods are impractical for data sets with a large number of phenotypes such as eQTL studies, which often contain 1000s of gene expressions. On the other hand, the computational time for GAMMA increases linearly to the number of phenotypes, which is useful for analyzing high dimensional data. Let n be the number of samples, m be the number of phenotypes, and p be the number of SNPs. The time complexity of estimating a kinship matrix; variance components; and transforming genotypes and phenotypes with the inverse squared root of a covariance matrix, is However, this needs to be performed only once for the complete analysis for the data set. The most computationally expensive part of GAMMA is the permutation step, which we can get in for each SNP, where T is the number of permutations. To reduce the cost of permutations, GAMMA performs an adaptive permutation where we increase the number of permutations from 100, increasing by 10 times. As most of the SNPs are under the null, our adaptive permutation reduces time dramatically. In addition, we note that the time complexity of each step could be reduced using various special mathematical techniques (Kang et al. 2010; Lippert et al. 2011; Williams 2011; Davie and Stothers 2013; Gall 2014; Loh et al. 2015). On an Intel Xeon 2.5 GHz Linux machine, GAMMA takes 2.79 hr for the yeast data set, which has 6138 probes and 2956 genotyped loci in 112 segregants.
Distance matrix
GAMMA uses the Bray–Curtis measure (Bray and Curtis 1957; Gower 1966) to compute the distance matrix for MDMR and GAMMA. The Bray–Curtis measures a distance as the summation of absolute differences between abundances of elements divided by the sum of the abundances. Let us say n is the number of individuals and we have a phenotype matrix Y with each element Then, we derive an distance matrix D with each element as follows:
| (10) |
Permutation
The distribution of the pseudo-F-statistic is complex and does not follow distribution as described in Multiple-phenotypes analysis in the Materials and Methods section. Therefore, to assess statistical significance, we performed a permutation test. Permutation tests can be pursued by permuting the transformed genotypes or the transformed phenotypes or simultaneously permuting the rows and columns of the matrix. To reduce the cost of permutations, GAMMA performs an adaptive permutation where we increase the number of permutations from 100, increasing by 10 times. Up to permutations were performed for the simulated data set and permutations were performed for the yeast and the microbiome data sets.
Implementation
For running GAMMA, we need to compute the covariance matrix To do this, we need the estimates of and Let and be the two variance components of the jth phenotype, where We follow the approach taken in efficient mixed-model association expedited (EMMAX) (Kang et al. 2010) or factored spectrally transformed linear mixed models (FaST-LMM) (Lippert et al. 2011) and estimate and in the null model, with no SNP effect. As we take multiple phenotypes into account, a median value of is used for and a median value of is used for which practically worked well in both of our real data sets. R package vegan is used to perform permutational multivariate analysis and the C package of EMMA is used to perform mixed-model association test.
Simulated data set
We sampled data from a multivariate normal distribution based on our generative model to generate a simulated data set containing 1000 genes, 100 SNPs, and >96 samples (Equation 1). SNPs are extracted from a Hybrid Mouse Diversity Panel (HMDP) (Bennett et al. 2010), which is a mouse association study panel containing significant amounts of population structure. Five randomly selected trans-regulatory hotspots are simulated, and 20% of the genes in each hotspot have trans effects of size 1. Cis effect is simulated with the size of 2. = 0.8 and = 0.2 is used.
Real data sets
We evaluated our method using a yeast data set (Brem and Kruglyak 2005). The data set contains 6138 probes and 2956 genotyped loci in 112 segregants. In addition, we evaluated our method using a gut microbiome data set (Org et al. 2015) collected from 592 mice representing 110 HMDP strains. The study protocol has been described in detail by Parks et al. (2013). Bacterial 16S ribosomal RNA gene V4 region was sequenced using the Illumina MiSeq platform and data were analyzed using established guidelines (Bokulich et al. 2013). The relative abundance of each taxon was computed by dividing the sequences pertaining to a specific taxon by the total number of bacterial sequences for that sample. We focused on abundant microbes, operational taxonomic units with at least 0.01% relative abundance; and for GWAS we used 197,885 SNPs and 26 genus-level taxa. Because of the nature of meta-genomics data, the distributions of abundances of species are often highly aggregated or skewed (McArdle and Anderson 2001). Thus, we applied arcsine transformation on the phenotype values. Minor allele frequency <5% and missing values >10% are filtered out. We expect the data set contains a strong population structure effect, because the mouse genome is known to contain a significant amount of population structure.
Data availability
The HMDP data set (Bennett et al. 2010) is available at Gene Expression Omnibus (GEO) accession number GSE16780, yeast data set (Brem and Kruglyak 2005) is available at GEO accession number GSE9376, and microbiome data set (Parks et al. 2013) is available at Sequence Read Archive under accession number SRP059760. The software, source codes, installation package, and instructions are available at http://genetics.cs.ucla.edu/GAMMA/. GAMMA is offered under the GNU Affero general public license, version 3 (AGPL-3.0). For the details of the license please see https://www.gnu.org/licenses/why-affero-gpl.html.
Results
Correcting for population structure in multivariate analysis
Unlike traditional univariate analyses that test associations between each phenotype and each genotype, our goal is to identify SNPs that are simultaneously associated with multiple phenotypes. Let us say with n as the number of samples and m as the number of phenotypes, we are analyzing an association between the ith SNP and m phenotypes. The standard multivariate regression analysis assumes a linear model as follows:
where Y is an matrix, where each column vector contains the jth phenotype values; is a vector of length n containing genotypes of the ith SNP; β is a vector of length m, where each entry contains an effect of the ith SNP on the jth phenotype; and E is an matrix, where each column vector contains i.i.d. residual errors of the jth phenotype. Here, we assume that each column of the random effect E follows a multivariate normal distribution, where I is an identity matrix with unknown magnitude
To test an association between the ith SNP and m phenotypes, we test whether any of is 0 or not from the linear model. The standard least-squares solution for is However, this is problematic when which is often the case in genomics data as there could be many solutions when there are more unknown variables than observations. Alternatively, MDMR (Zapala and Schork 2012) forms a statistic to test the effect of a variable on multiple phenotypes by leveraging the sums of squares associated with the linear model. These sums can be directly computed from an distance matrix D estimated from Y, where each element reflects the distance between sample i and j. This is because the standard multivariate analysis proceeds through a partitioning of the total sum of squares and cross products (SSCP) matrix, and the relevant information contained in required inner product matrices could be achieved by an outer-product matrix YY′, which could be obtained from an distance matrix estimated from Y.
However, in GWAS, it has been widely known that genetic relatedness, referred to as population structure, complicates the analysis by creating spurious associations. The linear model does not account for population structure and may induce numerous false positive identifications. Moreover, these problems may compound in multiple-phenotypes analysis where biases accumulate from each phenotype as their test statistics are summed over the phenotypes (see details in Material and Methods.). Recently, the linear mixed model has emerged as a powerful tool for GWAS as it could correct for population structure. To incorporate effects of population structure, GAMMA assumes a linear mixed model instead of the linear model as follows:
which has an extra matrix term U, where each column vector contains effects of population structure of the jth phenotype. This is an extension of the following widely used linear mixed model for univariate analysis:
where and K is the kinship matrix that encodes the relatedness between individuals, and is the variance of the phenotype accounted for by the genetic variation in the sample. To estimate a test statistic for the multiple-phenotype analysis, we perform a multivariate regression analysis through partitioning of the total SSCP matrix based on the linear mixed model. Details of how we perform the inference including test statistics, distance matrix, and permutations are described in Materials and Methods.
GAMMA corrects for population structure and accurately identifies genetic variances in a simulated study
Our goal is to detect an association between a variant and multiple phenotypes. A trans-regulatory hotspot is a variant that regulates many genes, thus, detecting trans-regulatory hotspots is a good application for GAMMA. In testing the accuracy of GAMMA, we assessed the approach’s potential for eliminating effects of population structure and identifying true trans-regulatory hotspots. We created a simulated data set that has 96 samples with 100 SNPs and 1000 gene expression levels. To incorporate the effects of population structure, we took SNPs from a subset of an HMDP (Bennett et al. 2010) containing significant amounts of population structure. To incorporate the effects of trans-regulatory hotspots, we simulated five trans-regulatory hotspots on the gene expression. For each of the trans-regulatory hotspots, we added trans effects to 20% of the genes. In addition, we added cis effects (Michaelson et al. 2009), which are associations between SNPs and genes in close proximity, as they are well-known eQTLs that exist in real organisms.
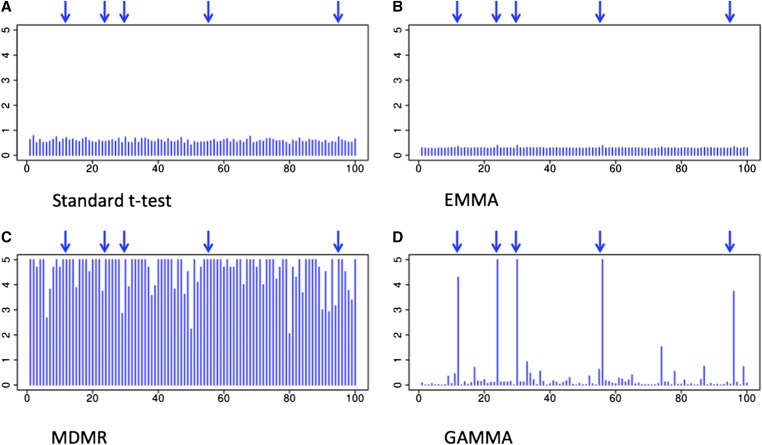
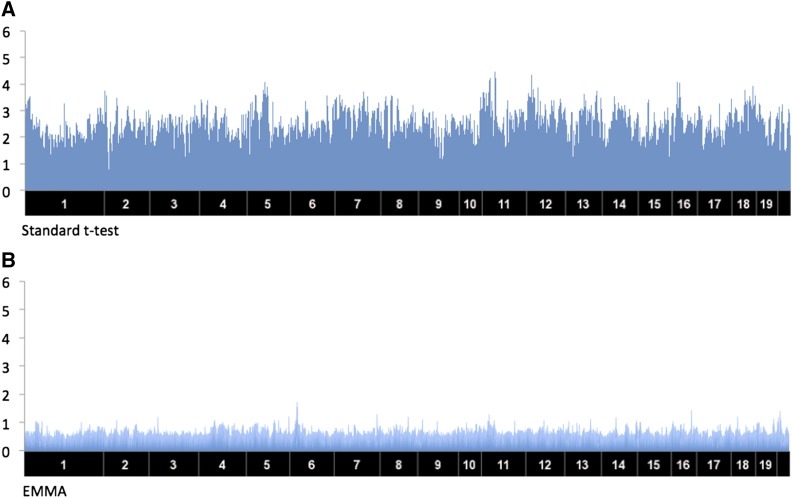
We applied the standard t-test, EMMA (Kang et al. 2008), MDMR (Zapala and Schork 2012), and GAMMA on the simulated data set. We visualized results in a plot (Figure 1), where the x-axis shows SNP locations and the y-axis shows −log10 P-values. For the t-test and EMMA, we averaged the P-values over all of the phenotypes for each SNP, because they give a P-value for each phenotype. In each plot, we marked the locations of true trans-regulatory hotspots with blue arrows. As a result, the plot clearly indicates that GAMMA successfully identifies the true trans-regulatory hotspots without producing false positive identifications induced by population structure (Figure 1D). However, the standard t-test and EMMA fail to identify the true trans-regulatory hotspots, because they lack sufficient power to detect the associations (Figure 1, A and B). As it does not account for population structure, MDMR results in many false positive identifications induced by spurious associations (Figure 1C).
Figure 1.
The results of different methods applied to a simulated data set. The x-axis shows SNP locations and the y-axis shows P-values of associations between each SNP and all the genes. Blue ↓ shows the location of the true trans-regulatory hotspots. (A) The result of the standard t-test. (B) The result of EMMA. For (A) and (B), we averaged the P-values over all of the genes for each SNP. (C) The result of MDMR. (D) The result of GAMMA.
GAMMA identifies regulatory hotspots related to regulatory elements of a yeast data set
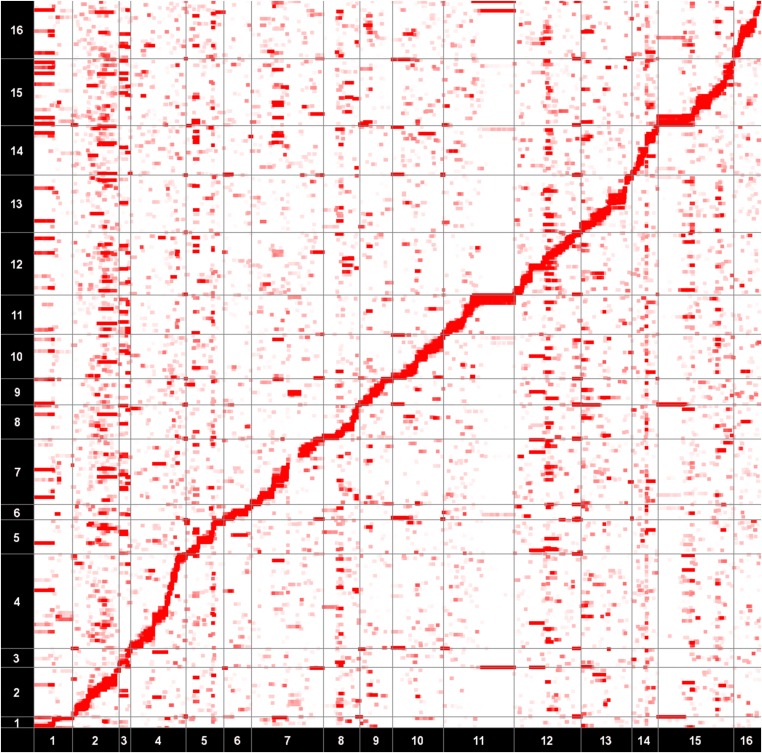
Yeast is one of the model organisms that are known to contain several trans-regulatory hotspots. For example, in a comprehensively characterized yeast data set, validation with additional lines of evidence, such as protein measurements, identified multiple hotspots as having true genetic effects (Foss et al. 2007; Perlstein et al. 2007). Unfortunately, expression data are known to contain significant amounts of confounding effects stemming from various technical artifacts, such as batch effects. To correct for these confounding effects, we applied NICE (Next generation Intersample Correlation Emended) (Joo et al. 2014), a recently developed method that corrects for the heterogeneity in expression data, to the yeast data set and drew an eQTL map (Figure 2). On the map, the x-axis corresponds to SNP locations, and the y-axis corresponds to gene locations. The intensity of each point on the map represents the significance of the association between a gene and a SNP. There are some vertical bands in the eQTL map that represent trans-regulatory hotspots. However, the eQTL map does not visually indicate which bands are the trans-regulatory hotspots as it only depicts associations between each SNP and a single gene.
Figure 2.
An eQTL map of a real yeast data set. P-values are estimated from NICE (Joo et al. 2014). The x-axis corresponds to SNP locations and the y-axis corresponds to the gene locations. The intensity of each point on the map represents the significance of the association. The diagonal band represents the cis effects and the vertical bands represent trans-regulatory hotspots.
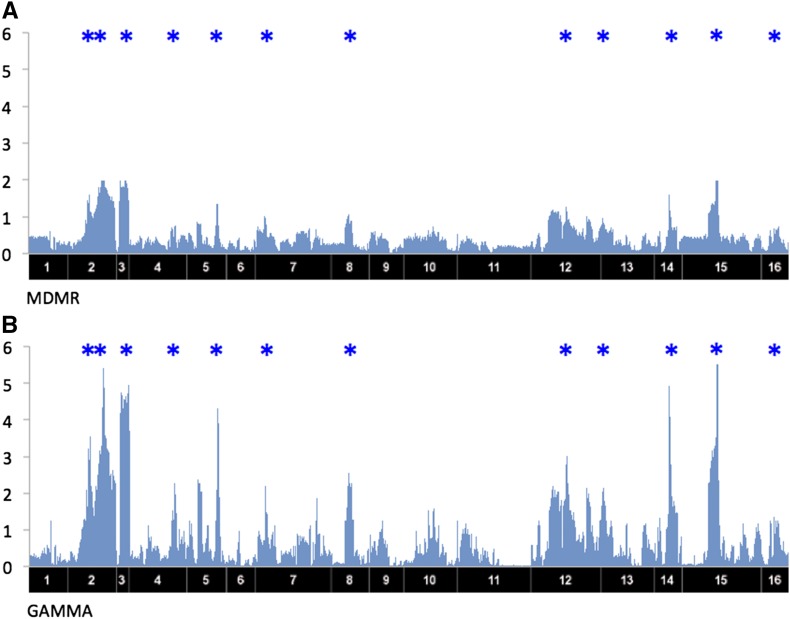
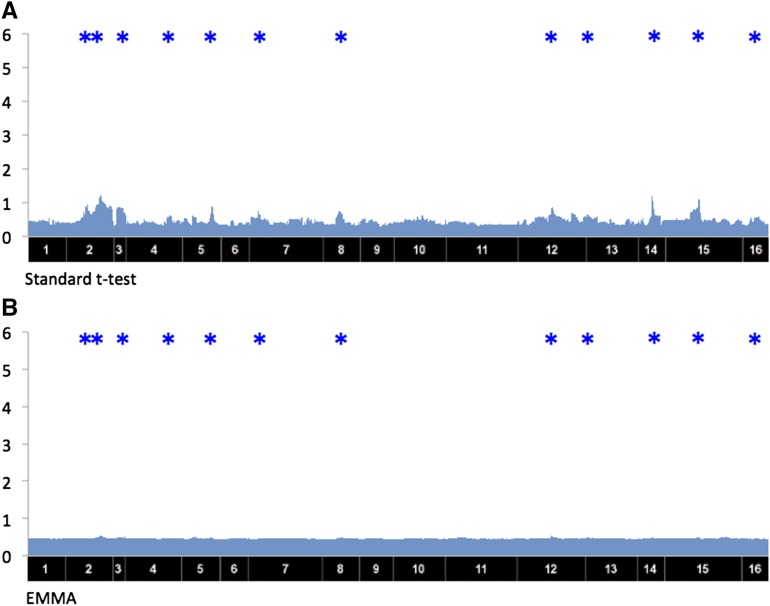
We applied the standard t-test, EMMA (Kang et al. 2008), MDMR (Zapala and Schork 2012), and GAMMA to the yeast data set to detect the trans-regulatory hotspots. To remove the confounding effects and other effects from various technical artifacts, we applied genomic control λ, which is a standard way of removing unknown plausible effects (Devlin et al. 2001). The inflation factor λ shows how much the statistics of obtained P-values are departed from a uniform distribution; indicates an inflation and indicates a deflation. The λ values are 1.20, 0.86, 3.64, and 0.98 for the t-test, EMMA, MDMR, and GAMMA, respectively. As the yeast data set does not contain a significant amount of population structure, the λ value is not very big even for the t-test. However, the λ value is very big for MDMR which shows that even a small amount of bias could cause significant problems in multiple-phenotypes analysis. GAMMA could successfully correct for the bias, and the λ value for GAMMA is close to 1. Figure 3, A and B, shows the results of MDMR and GAMMA, respectively. The x-axis shows locations of the SNPs and the y-axis shows −log10 P-values. The blue stars above each plot show hotspots that a previous study (Joo et al. 2014) identified as putative trans-regulatory hotspots for the yeast data. As a result, GAMMA (Figure 3B) shows significant signals on most of the putative hotspots. Details of the functions of the hotspots are described in Yvert et al. (2003). However, MDMR (Figure 3A) does not show significant signals on those sites. The t-test and EMMA fail to identify the trans-regulatory hotspots, because each phenotype is expected to have a genetic effect too small to detect with single-phenotype analysis (Figure 4).
Figure 3.
The results of MDMR and GAMMA applied to a yeast data set. The x-axis corresponds to SNP locations and the y-axis corresponds to gene locations. The y-axis corresponds to −log10 of P-values. Blue * above each plot shows putative hotspots that were reported in a previous study (Joo et al. 2014) for the yeast data. (A) The result of MDMR. (B) The result of GAMMA.
Figure 4.
The results of the standard t-test and EMMA applied to a yeast data set. The x-axis corresponds to SNP locations and the y-axis corresponds to gene locations. The y-axis corresponds to sum of −log10 of P-value over the genes. Blue * above each plot shows putative hotspots that were reported in a previous study (Joo et al. 2014) in the yeast data. (A) The result of the standard t-test. (B) The result of EMMA.
GAMMA identifies variants associated with a gut microbiome
An increasing body of evidence supports the idea that both diet and host genetics affect the composition of gut microbiota, and that shifts in microbial communities can lead to cardio-metabolic diseases such as obesity (Ley et al. 2005), diabetes (Ley et al. 2005), and other metabolic diseases (Karlsson et al. 2013). The ecosystem of gut bacteria is comprised of many complex interactions that remain largely unidentified. Accounting for the relationship between gut microbiota and disease mechanisms is a challenge, as some taxa could be coexpressed and there could be clinical overlap between the taxa. Our incomplete understanding of how the gut microbiota network poses a challenge to characterizing how a SNP simultaneously affects multiple gut microbiome taxa. Performing a multiple-phenotypes analysis with microbiome data may produce results that allow more complete reconstruction of these networks. We applied the standard t-test, EMMA (Kang et al. 2008), MDMR (Zapala and Schork 2012), and GAMMA on a gut microbiome data set (Org et al. 2015) from HMDP that contains 26 common genus-level taxa identified from 592 mice samples, including 197,885 SNPs.
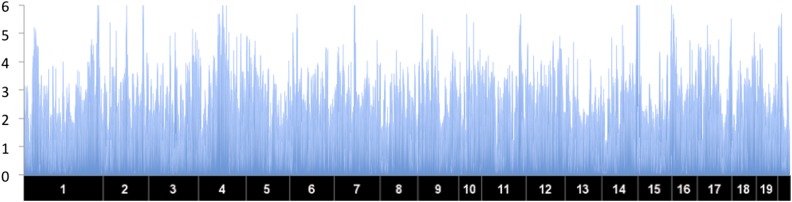
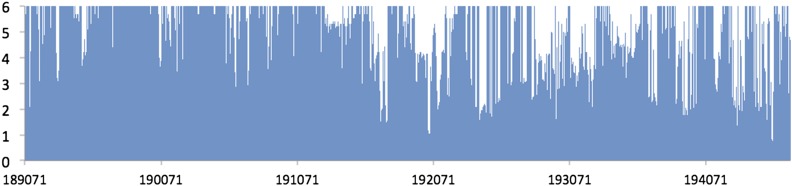
Meta-genomics data are highly heterogeneous, and studies frequently produce highly aggregated or skewed distributions of species abundance (McArdle and Anderson 2001). In addition, many of the individuals have no abundance for specific taxa, which further affects the distribution. Therefore, when we integrate all of the taxa together, the taxa with these distribution problems drive very high λ values (>5) in our combined statistic, except EMMA, which is known to have a deflation problem (Lippert et al. 2011; Joo et al. 2014). For this reason, we did not apply the genomic control on the data. Figure 5 shows the result of GAMMA applied on the data set. We defined the peaks with P-value as significant peaks, and in mouse genome we found nine loci that are likely to be associated with the genus-level taxa. Many of the identified loci contain a number of strong candidate genes based on the literature and overlap with signals of clinical traits and functional variations such as cis-eQTL (see Table 1 for a list of loci). For example, chromosome 1 and 2 loci are the same regions detected with obesity traits in our previous study using the same mice (Parks et al. 2013). In addition, global gene expression in epididymal adipose tissue and liver showed a significant cis-eQTL with genes residing in six out of nine detected loci. On the other hand, MDMR predicts many false positives as mouse data are known to contain significant amounts of population structure. We applied MDMR on one of the smallest chromosomes, chromosome 19. Even in this small region, MDMR produces 1989 significant peaks out of 5621 loci, which demonstrates that MDMR is not advantageous for data sets with population structure (Figure 6). The t-test and EMMA both fail to detect significant signals due to the low power (Figure 7).
Figure 5.
The result of GAMMA applied to a gut microbiome data set. The x-axis corresponds to SNP locations and the y-axis corresponds to gene locations. The y-axis corresponds to −log10 of P-value.
Table 1. The list of significant associations with a gut microbiome data set.
| Chr | Peak SNP | Position (Mb) | Associated region (Mb) | Number of genes | Clinical QTL | cis-eQTL | Overlapping with single genus GWAS |
|---|---|---|---|---|---|---|---|
| 1 | rs31797108 | 182,072,111 | 18.1–18.2 | 21 | Body fat percentage increase | ||
| 2 | rs27323290 | 157,697,578 | 11.4–15.8 | 7 | Food intake, weight | Ctnnbl1 | Akkermansia muciniphila |
| 4 | rs28319212 | 95,462,396 | 82.1–10.5 | 74 | Food intake | Caap1, Ift74 | Oscillospira spp. |
| 6 | rs50368681 | 38,026,365 | 37.5–38.0 | 16 | Atp6v0a4, Replin1, Zfp467 | Sarcina spp. | |
| 7 | rs33129247 | 68,944,648 | 68.5–71.4 | 3 | TG, Gonadal Fat | Nr2f2, Igf1r | Akkermansia muciniphila |
| 11 | rs3680824 | 104,011,091 | 10.2–10.4 | 47 | Ccdc85a, Efemp1 | ||
| 14 | rs30384023 | 120,051,254 | 11.9–12.1 | 5 | Dnajc3, Uggt2, Farp1 | ||
| 16 | rs4154709 | 6,236,151 | 62.3–75.0 | 1 | |||
| X | rs29064137 | 87,504,122 | 87.2–88.6 | 1 |
FaST-LMM (Lippert et al. 2011) is used for single genus GWAS. Chr, chromosome; Ctnnbl1, catenin, β-like 1; Capp1, caspase activity and apoptosis inhibitor; Ift74, intraflagellar transport 74; Atp6v0a4, ATPase, H+ transporting, lysosomal V0 subunit A4; Zfp467, zinc finger protein 467; TG, thyroglobulin; Nr2f2, nuclear receptor subfamily 2, group F, member 2; Igf1r, insulin-like growth factor 1 receptor; Ccdc85a, coiled-coil domain containing 85A; Efemp1, EGF containing fibulin-like extracellular matrix protein 1; Dnajc3, DnaJ (Hsp40) homolog, subfamily C, member 3; Uggt2, UDP-glucose glycoprotein glucosyltransferase 2; Farp1, FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1.
Figure 6.
The result of MDMR applied to chromosome 19 of a gut microbiome data set. The x-axis corresponds to SNP locations and the y-axis corresponds to gene locations. The y-axis corresponds to −log10 of P-value.
Figure 7.
The results of the standard t-test and EMMA applied to a gut microbiome data set. The x-axis corresponds to SNP locations and the y-axis corresponds to gene locations. The y-axis corresponds to sum of −log10 of P-value over the genus. (A) The result of the standard t-test. (B) The result of EMMA.
Discussion
In this article we present GAMMA, an accurate and efficient method for identifying genetic variants associated with multiple phenotypes while simultaneously considering population structure. Population structure is a widespread confounding factor that creates genetic relatedness between samples. This confounding factor makes both genotypes and phenotypes dependent on each other. In these cases, previous multivariate methods that assume i.i.d. between samples will produce erroneous results. Moreover, the bias accumulates for each phenotype, thus, even a small degree of population structure may produce significant errors in multiple-phenotypes analysis.
GAMMA successfully identifies the variants associated with multiple phenotypes in both simulated and real data sets, including yeast and gut microbiomes from mice. GAMMA is an improvement over other methods (Kang et al. 2008; Zapala and Schork 2012) that either fail to identify true signals or produce many false positives. We used a pseudo-F-statistic that Brian et al. (2001) introduced as a test statistic. This method quickly and efficiently estimates a test statistic and is especially useful in cases with a larger number of phenotypes than total number of samples, which is often the case in genomics data. However, other appropriate multivariate statistics could be applied to GAMMA as well.
We further tailored our method to address several potential problems. First, in the single-phenotype analysis, we use the average P-value of all the phenotypes for each SNP. This method could be a naive way of comparing the results of a single-phenotype analysis and multiple-phenotypes analysis. Second, we use a median value of variance components that are estimated from genes to compute a covariance matrix when transforming phenotypes and genotypes. Empirically, median values give good results in both of our experiments with real data sets. However, variance components could be widespread across genes and median values may not be suitable in some data sets. Finding an appropriate value could be an excellent direction for future work. Third, GAMMA does not provide information that allows us to assess whether individual phenotypes in a set are associated with the SNP; GAMMA results only suggest whether a set of phenotypes is or is not associated with a SNP. There are several methods for determining which individual phenotype the SNP is associated with, including the m-values of Han and Eskin (2012). Lastly, GAMMA uses the Bray–Curtis measure (Bray and Curtis 1957; Gower 1966) to compute the distance matrix, but other distance matrices could be used. There are various potential distance measures that could be used to construct the distance matrix (Webb 2002). Unfortunately, very little investigative work has been published that guides selection of a distance measure most appropriate for a given case. Zapala and Schork (2006) discussed the influences of a distance measure by comparing distance matrices derived by various distance measures. The choice of a distance matrix explains the proportion of variation in the distance matrix, but does not necessarily explain the significance of the relationship between the predictor variables and the distance matrix entries. A more exhaustive study may be needed to thoroughly understand the effects of the distance matrix.
Acknowledgments
J.W.J.J., E.Y.K., N.F., and E.E. are supported by National Science Foundation grants 0513612, 0731455, 0729049, 0916676, 1065276, 1302448, and 1320589; and National Institutes of Health grants K25 HL-080079, U01 DA-024417, P01 HL-30568, P01 HL-28481, R01 GM-083198, R01 MH-101782, and R01 ES-022282. We acknowledge the support of the National Institute of Neurological Disorders and Stroke Informatics Center for Neurogenetics and Neurogenomics (P30 NS-062691). E.O. is supported by FP7 grant number 330381. No competing financial interests exist.
Footnotes
Communicating editor: R. Nielsen
Literature Cited
- Alter O., Brown P. O., Botstein D., 2000. Singular value decomposition for genome-wide expression data processing and modeling. Proc. Natl. Acad. Sci. USA 97: 10101–10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschard H., Vilhjálmsson B. J., Greliche N., Morange P.-E. E., Trégouët D.-A. A., et al. , 2014. Maximizing the power of principal-component analysis of correlated phenotypes in genome-wide association studies. Am. J. Hum. Genet. 94: 662–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett B. J., Farber C. R., Orozco L., Kang H. M., Ghazalpour A., et al. , 2010. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res. 20: 281–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger M., Stassen H. H., Köhler K., Krane V., Mönks D., et al. , 2006. Hidden population substructures in an apparently homogeneous population bias association studies. Eur. J. Hum. Genet. 14: 236–244. [DOI] [PubMed] [Google Scholar]
- Bokulich N. A., Subramanian S., Faith J. J., Gevers D., Gordon J. I., et al. , 2013. Quality-filtering vastly improves diversity estimates from illumina amplicon sequencing. Nat. Methods 10: 57–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray J. R., Curtis J. T., 1957. An ordination of the upland forest communities of southern wisconsin. Ecol. Monogr. 27: 325–349. [Google Scholar]
- Brem R. B., Kruglyak L., 2005. The landscape of genetic complexity across 5,700 gene expression traits in yeast. Proc. Natl. Acad. Sci. USA 102: 1572–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell C. D., Ogburn E. L., Lunetta K. L., Lyon H. N., Freedman M. L., et al. , 2005. Demonstrating stratification in a European American population. Nat. Genet. 37: 868–872. [DOI] [PubMed] [Google Scholar]
- Cervino A. C., Li G., Edwards S., Zhu J., Laurie C., et al. , 2005. Integrating qtl and high-density snp analyses in mice to identify insig2 as a susceptibility gene for plasma cholesterol levels. Genomics 86: 505–517. [DOI] [PubMed] [Google Scholar]
- Davie A. M., Stothers A. J., 2013. Improved bound for complexity of matrix multiplication. P. Roy. Soc. Edinb. A Math. 143: 351–369. [Google Scholar]
- Devlin B., Roeder K., Wasserman L., 2001. Genomic control, a new approach to genetic-based association studies. Theor. Popul. Biol. 60: 155–166. [DOI] [PubMed] [Google Scholar]
- Flint J., Eskin E., 2012. Genome-wide association studies in mice. Nat. Rev. Genet. 13: 807–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foll M., Gaggiotti O., 2006. Identifying the environmental factors that determine the genetic structure of populations. Genetics 174: 875–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss E. J., Radulovic D., Shaffer S. A., Ruderfer D. M., Bedalov A., et al. , 2007. Genetic basis of proteome variation in yeast. Nat. Genet. 39: 1369–1375. [DOI] [PubMed] [Google Scholar]
- Freedman M. L., Reich D., Penney K. L., McDonald G. J., Mignault A. A., et al. , 2004. Assessing the impact of population stratification on genetic association studies. Nat. Genet. 36: 388–393. [DOI] [PubMed] [Google Scholar]
- Gall F. L., 2014. Powers of tensors and fast matrix multiplication. arXiv DOI: 1401.7714.
- Gower J. C., 1966. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 53: 325–338. [Google Scholar]
- Gygi S. P., Rist B., Gerber S. A., Turecek F., Gelb M. H., et al. , 1999. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 17: 994–999. [DOI] [PubMed] [Google Scholar]
- Han B., Eskin E., 2012. Interpreting meta-analyses of genome-wide association studies. PLoS Genet. 8: e1002555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgason A., Yngvadóttir B., Hrafnkelsson B., Gulcher J., Stefánsson K., 2005. An icelandic example of the impact of population structure on association studies. Nat. Genet. 37: 90–95. [DOI] [PubMed] [Google Scholar]
- Hillebrandt S., Wasmuth H. E., Weiskirchen R., Hellerbrand C., Keppeler H., et al. , 2005. Complement factor 5 is a quantitative trait gene that modifies liver fibrogenesis in mice and humans. Nat. Genet. 37: 835–843. [DOI] [PubMed] [Google Scholar]
- Hormozdiari F., Kichaev G., Yang W.-Y., Pasaniuc B., Eskin E., 2015. Identification of causal genes for complex traits. Bioinformatics 31: i206–i213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo J. W. J., Sul J. H., Han B., Ye C., Eskin E., 2014. Effectively identifying regulatory hotspots while capturing expression heterogeneity in gene expression studies. Genome Biol. 15: r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. M., Ye C., Eskin E., 2008. Accurate discovery of expression quantitative trait loci under confounding from spurious and genuine regulatory hotspots. Genetics 180: 1909–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. M., Sul J. H., Service S. K., Zaitlen N. A., Kong S.-Y. Y., et al. , 2010. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42: 348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson F. H., Tremaroli V., Nookaew I., Bergström G., Behre C. J., et al. , 2013. Gut metagenome in european women with normal, impaired and diabetic glucose control. Nature 498: 99–103. [DOI] [PubMed] [Google Scholar]
- Kittles R. A., Chen W., Panguluri R. K., Ahaghotu C., Jackson A., et al. , 2002. Cyp3a4-v and prostate cancer in african americans: causal or confounding association because of population stratification? Hum. Genet. 110: 553–560. [DOI] [PubMed] [Google Scholar]
- Korte A., Vilhjalmsson B. J., Segura V., Platt A., Long Q., et al. , 2012. A mixed-model approach for genome-wide association studies of correlated traits in structured populations. Nat. Genet. 44: 1066–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R. E., Bäckhed F., Turnbaugh P., Lozupone C. A., Knight R. D., et al. , 2005. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 102: 11070–11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippert C., Listgarten J., Liu Y., Kadie C. M., Davidson R. I., et al. , 2011. Fast linear mixed models for genome-wide association studies. Nat. Methods 8: 833–835. [DOI] [PubMed] [Google Scholar]
- Lockhart D. J., Dong H., Byrne M. C., Follettie M. T., Gallo M. V., et al. , 1996. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 14: 1675–1680. [DOI] [PubMed] [Google Scholar]
- Loh P.-R. R., Tucker G., Bulik-Sullivan B. K., Vilhjálmsson B. J., Finucane H. K., et al. , 2015. Efficient bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 47: 284–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J., Cardon L. R., Phillips M. S., Donnelly P., 2004. The effects of human population structure on large genetic association studies. Nat. Genet. 36: 512–517. [DOI] [PubMed] [Google Scholar]
- McArdle B. H., Anderson M. J., 2001. Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology 82: 290–297. [Google Scholar]
- Michaelson J. J., Loguercio S., Beyer A., 2009. Detection and interpretation of expression quantitative trait loci (eqtl). Methods 48: 265–276. [DOI] [PubMed] [Google Scholar]
- Nievergelt C. M., Libiger O., Schork N. J., 2007. Generalized analysis of molecular variance. PLoS Genet. 3: e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly P. F., Hoggart C. J., Pomyen Y., Calboli F. C. F., Elliott P., et al. , 2012. Multiphen: joint model of multiple phenotypes can increase discovery in gwas. PLoS One 7: e34861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Org E., Parks B. W. W., Joo J. W. J., Emert B., Schwartzman W., et al. , 2015. Genetic and environmental control of host-gut microbiota interactions. Genome Res. 25: 1558–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks B. W., Nam E., Org E., Kostem E., Norheim F., et al. , 2013. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab. 17: 141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlstein E. O., Ruderfer D. M., Roberts D. C., Schreiber S. L., Kruglyak L., 2007. Genetic basis of individual differences in the response to small-molecule drugs in yeast. Nat. Genet. 39: 496–502. [DOI] [PubMed] [Google Scholar]
- Quackenbush J., 2001. Computational analysis of microarray data. Nat. Rev. Genet. 2: 418–427. [DOI] [PubMed] [Google Scholar]
- Reiner A. P., Ziv E., Lind D. L., Nievergelt C. M., Schork N. J., et al. , 2005. Population structure, admixture, and aging-related phenotypes in African American adults: the cardiovascular health study. Am. J. Hum. Genet. 76: 463–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura V., Vilhjalmsson B. J., Platt A., Korte A., Seren U., et al. , 2012. An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat. Genet. 44: 825–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seldin M. F., Shigeta R., Villoslada P., Selmi C., Tuomilehto J., et al. , 2006. European population substructure: clustering of northern and southern populations. PLoS Genet. 2: e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svishcheva G. R., Axenovich T. I., Belonogova N. M., van Duijn C. M., Aulchenko Y. S., 2012. Rapid variance components-based method for whole-genome association analysis. Nat. Genet. 44: 1166–1170. [DOI] [PubMed] [Google Scholar]
- Voight B. F., Pritchard J. K., 2005. Confounding from cryptic relatedness in case-control association studies. PLoS Genet. 1: e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Korstanje R., Higgins D., Paigen B., 2004. Haplotype analysis in multiple crosses to identify a qtl gene. Genome Res. 14: 1767–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb A. R., 2002. Statistical Pattern Recognition, Ed. 2 John Wiley & Sons, Chichester, United Kingdom. [Google Scholar]
- Wessel J., Schork N. J., 2006. Generalized genomic distance-based regression methodology for multilocus association analysis. Am. J. Hum. Genet. 79: 792–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams V. V., 2012. Multiplyng Matrices Faster than Coppersmith-winograd. Proceedings of the fourty-fourth Annual Symposium on the Theory of Computing. ACM, pp. 887–98. [Google Scholar]
- Yvert G., Brem R. B., Whittle J., Akey J. M., Foss E., et al. , 2003. Trans-acting regulatory variation in saccharomyces cerevisiae and the role of transcription factors. Nat. Genet. 35: 57–64. [DOI] [PubMed] [Google Scholar]
- Zapala M. A., Schork N. J., 2006. Multivariate regression analysis of distance matrices for testing associations between gene expression patterns and related variables. Proc. Natl. Acad. Sci. USA 103: 19430–19435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapala M. A., Schork N. J., 2012. Statistical properties of multivariate distance matrix regression for high-dimensional data analysis. Front. Genet. 3: 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Stephens M., 2012. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 44: 821–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Stephens M., 2014. Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nat. Methods 11: 407–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The HMDP data set (Bennett et al. 2010) is available at Gene Expression Omnibus (GEO) accession number GSE16780, yeast data set (Brem and Kruglyak 2005) is available at GEO accession number GSE9376, and microbiome data set (Parks et al. 2013) is available at Sequence Read Archive under accession number SRP059760. The software, source codes, installation package, and instructions are available at http://genetics.cs.ucla.edu/GAMMA/. GAMMA is offered under the GNU Affero general public license, version 3 (AGPL-3.0). For the details of the license please see https://www.gnu.org/licenses/why-affero-gpl.html.