Abstract
Telomerase is required for the maintenance of telomere length and is an important determinant for cell immortalization. In human cells, telomerase activity is due to the expression of its enzymatic subunit, human telomerase reverse transcriptase (hTERT). The expression of hTERT is not typically detectable in healthy somatic human cells but is present in cancerous tissues and immortalized cells. We have previously shown that hTERT promoter activity is up-regulated by the Kaposi's sarcoma-associated herpesvirus (KSHV)-encoded latency-associated nuclear antigen (LANA). LANA is expressed in all forms of human malignancies associated with KSHV. The hTERT promoter sequence located at positions −130 to +5 contains several Sp1 binding motifs and was shown be important for up-regulation by LANA. In this report, we demonstrate that hTERT promoter activity is due to the direct interaction of LANA with Sp1. The interaction of LANA with Sp1 was demonstrated through in vitro binding experiments and coimmunoprecipitation and is supported by the colocalization of these two molecules in the nuclei of KSHV-infected cells. Moreover, LANA modulates Sp1-mediated transcription in transient GAL4 fusion reporter assays. Mapping of the regions involved in binding and transcriptional activation showed that the amino terminus of LANA is the major site for interaction and up-regulation but that it can cooperate with the carboxy terminus to enhance these functions. An analysis of Sp1 binding to its cognate sequence corroborated the binding data. Together, our results suggest that the interaction of LANA with Sp1 up-regulates the telomerase promoter, potentially contributing to the immortalization of KSHV-infected cells.
Latency-associated nuclear antigen (LANA), encoded by open reading frame (ORF) 73 of the Kaposi's sarcoma-associated herpesvirus (KSHV) genome, is one of the major latent proteins detected in all forms of KS-associated malignancies, including body-cavity-based lymphomas (BCBLs) and multicentric Castleman's disease, an aggressive lymphoproliferative disorder (6-8, 62, 75). LANA is a highly immunogenic nuclear protein that was initially detected in sera from KS patients by the use of immunofluorescence assays; it is composed of 1,162 amino acid (aa) residues and runs in sodium dodecyl sulfate (SDS)-polyacrylamide gels as a 222- to 234-kDa protein (24, 42). The secondary structure of the amino acid sequence reveals three major protein domains. The N-terminal domain contains a nuclear localization sequence, the chromosome binding sequence (aa 5 to 22), and a proline-rich region, which has been shown to be important for tethering the protein to the host chromatin (65, 68, 73). The central domain, which is rich in glutamine (Q) and glutamic acid (E) residues and is reported to have varying sizes in different isolates, may account for its slow electrophoretic migration. The C-terminal domain contains a leucine zipper motif (25). Leucine zipper motifs have been shown to be important for the interaction of LANA with various cellular proteins, including p53, pRb, histone H1, and CREB2/ATF (11, 20, 57, 67). The C-terminal domain of LANA has also been shown to have a nuclear localization signal sequence, which may be important for the characteristic nuclear speckling of LANA (71). Binding to the terminal repeat DNA of the KSHV genome, which is crucial for tethering the viral genome to the host chromosome, has also been mapped to the C terminus (12, 26).
Studies into the role of LANA as a transcription factor demonstrated its ability to function as a transcriptional modulator of various cellular and viral promoters, including its own promoter (1, 26, 31, 37, 46, 67, 69). Recent studies on the role of LANA in modulating cellular pathways strongly suggest that LANA may act as an oncoprotein, driving cell proliferation and tumor development. LANA physically interacts with the tumor suppressor p53 and down modulates its activity (20), and it also interacts with the hypophosphorylated form of the retinoblastoma protein (pRb), releasing the E2F transcription factor, which in turn activates E2F-responsive genes (67). Recently, LANA was shown to interact with serine-threonine kinase glycogen synthetase kinase 3β, a negative regulator of β-catenin activity, leading to the up-regulation of β-catenin-Tef/Lef-responsive genes, including the genes involved in cellular proliferative responses (21, 22).
KSHV was shown to transform human umbilical vein endothelial cells and had activated levels of telomerase in transformed cells (19). Telomerase is a multisubunit ribonucleoprotein holoenzyme that prevents chromosome degradation, end-to-end chromosome fusions, and chromosome loss, thus maintaining telomere length by the addition of new repeat sequences at the ends of the chromosomes (29, 30). Telomerase is active in dividing cells, such as lymphocytes, keratinocytes, hematopoietic progenitor cells, and uterine endometrial cells, and in tumor-derived cell lines as well as malignant tissues (44, 61, 72). Human telomerase reverse transcriptase (hTERT), the enzymatic component of the telomerase, has been shown to be important and activated during cell immortalization and down-regulated during cell differentiation (39, 48, 61, 79). Telomerase-positive cell lines and cancer cells have been shown to have an active hTERT promoter. However, in telomerase-negative cells, hTERT promoter activity is repressed (36, 78). Recently, our group showed that KSHV-encoded LANA can up-regulate the hTERT promoter, demonstrating for the first time that a gene product from an oncogenic herpesvirus can activate the promoter (46). Promoter truncation studies showed that the sequence at positions −130 to +5 was enough for the LANA-mediated promoter activity. This region is highly GC rich and contains five GC boxes, the consensus binding sites for the Sp1 transcription factor.
Sp1 belongs to the zinc finger family of transcription factors, which recognize GC-rich DNA sequences and have been shown to play an important role in early embryonic development (5, 50, 60). Sp1 is a ubiquitous transcription factor that is involved in the modulation of various cellular transcriptional pathways by physically interacting with various transcriptional activators, including NF-κB (33-35), TBP (18), dTAFII110/hTAFII130 (28), YY1 (10, 52), E2F (41, 59), and the retinoblastoma-related protein pRb/p107 (14, 80).
A previous report demonstrated that LANA transactivates the hTERT promoter; however, the mechanism of activation was not understood. Here we show by in vitro and in vivo binding that LANA physically interacts with Sp1, predominantly through its N-terminal domain, and synergistically activates Sp1-mediated transcription. We suggest that the ability of LANA to function as an oncoprotein is in part due to its ability to increase transactivation of the hTERT promoter by targeting the Sp1 transcription factor, thus contributing to cell immortalization.
MATERIALS AND METHODS
Cell lines and antibodies.
Human embryonic kidney (HEK) 293 and 293T cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, and penicillin-streptomycin (5 U/ml and 5 μg/ml, respectively). BJAB (KSHV negative), BC-3, and BCBL-1 (KSHV infected) cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, and penicillin-streptomycin (5 U/ml and 5 μg/ml, respectively). All cell lines were grown at 37°C in a humidified environment supplemented with 5% CO2.
Antibodies for the Sp1 transcription factor and the GAL4 DNA binding domain (DBD) were purchased from Santa Cruz Biotechnologies Inc. (Santa Cruz, Calif.).
Plasmids and expression constructs.
The Sp1 bacterial expression constructs pGEX-Sp1-(83-778), pGEX-Sp1 516C, pGEX-Sp1 N619, and pGEX-Δint349 were generous gifts from Dimitris Kardassis, University of Crete Medical School, and from R. Tijan, University of California, Berkeley (40). The GAL4 (DBD)-Sp1 fusion constructs GAL4Sp1, GAL4Sp1 A+B, and GAL4Sp1 B were also gifts from Dimitris Kardassis. The pFR Luc reporter plasmid, which contains five GAL4 binding sites, was purchased from Stratagene Inc. (La Jolla, Calif.). Deletion mutants of LANA were constructed by PCR amplification, with pA3 M LANA used as a template (2).
Purification of GST fusion protein.
Glutathione S-transferase (GST) fusion proteins were expressed in Escherichia coli strain DH5α. Overnight bacterial cultures were inoculated into fresh Luria broth at a 1:100 dilution and induced with 100 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 h at 37°C after reaching an A600 of 0.7. The bacteria were harvested and washed with 5.0 ml of STE buffer (100 mM NaCl, 10 mM Tris, and 1.0 mM EDTA, pH 8.0). Pellets were then resuspended in 1.5 ml of NETN (100 mM NaCl, 1 mM EDTA [pH 8.0], 20 mM Tris [pH 8.0], and 0.5% NP-40 [pH 8.0]) supplemented with protease inhibitors (100 mM phenylmethylsulfonyl fluoride [PMSF], 1 μg of pepstatin per ml, and 1 μg of aprotinin per ml). The cells were sonicated, followed by the addition of 75 μl of dithiothreitol (DTT) (1 M) and 900 μl of 10% Sarkosyl for 1 min on ice, resulting in a clear lysate. The cell debris was pelleted by centrifugation at 10,000 × g for 10 min at 4°C. The supernatant containing the GST fusion protein was incubated with glutathione 4B-Sepharose beads (Amersham Inc.) in the presence of Triton X-100 for 2 h with rotation at 4.0°C. The beads were washed three times with 10 volumes of NETN with protease inhibitors, and the GST fusion proteins were visualized by Coomassie blue staining after being resolved by SDS-polyacrylamide gel electrophoresis (SDS-PAGE).
Expression of proteins in vitro.
Proteins were expressed in vitro by the use of a coupled in vitro transcription-translation system (TNT) from Promega Inc. (Madison, Wis.) according to the manufacturer's instructions. Labeling of the in vitro-translated protein was done by the addition of [35S]methionine-cysteine (Perkin-Elmer Inc., Boston, Mass.) to the TNT reaction mixture.
GST protein interaction assay.
GST fusion proteins bound to glutathione-Sepharose beads were used for in vitro binding. The lysates were first precleared with glutathione-Sepharose beads in binding buffer (1× phosphate-buffered saline [PBS], 0.1% NP-40, 0.5 mM DTT, 10% glycerol, 1 mM PMSF, 2 mg of aprotinin per ml) for 30 min. 35S-labeled rabbit reticulocyte lysates were then incubated with purified GST fusion proteins in binding buffer in a final volume of 500 μl. The reaction mixture was incubated with rotation for 2 h at 4°C. The beads were then collected and washed three times with 1 ml of binding buffer. The bound proteins were eluted by denaturation in SDS-β-mercaptoethanol lysis buffer and were fractionated by SDS-PAGE. Bound proteins were analyzed after the gel was dried on a phosphorimager plate (Molecular Dynamics).
Transient transfection and reporter assay.
HEK293, HEK293T, and BJAB cells were transiently transfected with the pFR Luc, GAL4Sp1, and LANA expression constructs by electroporation under previously described standard conditions (77). Briefly, 10 million cells were harvested, washed with PBS, resuspended in Dulbecco's modified Eagle or RPMI 1640 medium, and transfected by electroporation at 210 V and 975 μF with a Gene Pulser II (Bio-Rad Laboratories). The total amount of DNA was balanced by use of the parental vector, and the transfection efficiency was normalized by use of the green fluorescent protein-encoding vector pEGFPC1 (Clontech Inc., Palo Alto, Calif.). At 24 h posttransfection, the cells were collected, washed with PBS, and lysed with 200 μl of lysis buffer (Promega, Inc.). Luciferase activity was measured with an Opticomp I luminometer (MGM Instruments, Inc.). All of the transfections were done in triplicate, and the results shown represent the means of data from multiple experiments.
Western blot analysis.
Cell lysates from reporter assays were used for Western blot detection of GAL4Sp1 and LANA expression, SDS lysis buffer was added to the lysed cells, and the lysates were heated at 95°C for 5 min and resolved by SDS-PAGE with an 8% polyacrylamide gel. The resolved proteins were transferred to a 0.45-μm-pore-size nitrocellulose membrane. The membrane was blocked with 5% nonfat dried milk and then incubated with a mouse anti-GAL4 DBD antibody (Santa Cruz Biotechnology Inc.) and anti-Myc ascites 9E10 to detect LANA expression. The membranes were washed and then incubated with a horseradish peroxidase-conjugated secondary antibody, and cross-reactivity was visualized by chemiluminescence in a Fuji imager.
Immunoprecipitation.
For immunoprecipitation, 8 × 107 BJAB, BC-3, and BCBL-1 cells were lysed on ice with 1 ml of RIPA buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 0.1% NP-40, 1 mM EDTA [pH 8.0]) supplemented with protease inhibitors (1 mM PMSF, 10 μg of pepstatin/ml, 10 μg of leupeptin/ml, and 10 μg of aprotinin/ml). The lysates were centrifuged at high speed to remove the cell debris. A control serum was used to preclear the lysate before it was incubated with specific antibodies. Precleared lysates were then incubated with anti-Sp1 antibodies (Santa Cruz Biotechnology Inc.) overnight at 4°C with rotation and further incubated with protein G-Sepharose beads at 4°C for 1 h with rotation. The resulting immunoprecipitates were collected by centrifugation at 2,000 × g for 3 min at 4°C, and the pellets were washed four times with 1 ml of ice-cold RIPA butter. The immunoprecipitated pellets were resuspended in 30 μl of 2× SDS protein sample buffer (62.5 mM Tris [pH 6.8], 40 mM DTT, 2% SDS, 0.025% bromophenol blue, and 10% glycerol) and then resolved by SDS-PAGE with an 8% polyacrylamide gel. The separated proteins were transferred to a nitrocellulose membrane, and Western blot analysis was performed for the detection of LANA protein by the use of a polyclonal human serum or an anti-rabbit polyclonal antibody. The membrane was stripped and probed for the Sp1 protein by the use of an anti-Sp1 mouse monoclonal antibody (Santa Cruz Biotechnology Inc.). Similarly, reverse immunoprecipitation with an anti-LANA polyclonal serum was performed for BJAB, BC-3, and BCBL-1 cells, which were probed for the detection of Sp1 coimmunoprecipitating with LANA.
Immunolocalization of LANA and Sp1.
BC-3 cells were spread on a microscope slide and fixed in acetone-methanol (1:1) for 30 min at −20°C. Air-dried slides were incubated with 20% normal goat serum in 1× PBS to block nonspecific binding sites. Slides were then incubated with a primary anti-LANA polyclonal serum at room temperature in a humidified chamber, followed by three washes for 5 min each in PBS. The slides were then incubated with an anti-Sp1 mouse antibody for 2 h, and cross-reactivity was localized by the use of fluorescently labeled antibodies. The presence of LANA was localized by the use of goat anti-human antibodies labeled with an Alexa fluor, and Sp1 was localized by the use of Texas Red-conjugated goat anti-mouse secondary antibodies. The slides were then washed four times with 1× PBS and mounted with antifade solution. The slides were examined under an Olympus BX60 fluorescence microscope, and photographs were captured with a PixelFly digital camera (Cooke Inc., Warren, Mich.).
EMSAs.
An Sp1 DNA probe was prepared with the complementary strand of the hTERT promoter sequence from positions −119 to −98 (GCGCGGACCCCGCCCCGTCCCG [the underlined sequence is the consensus binding site for the Sp1 transcription factor]), as described previously (46). The mutant probe used as a control in electrophoretic mobility shift assays (EMSAs) had the sequence GCGCGGACCCCGAACCGTCCCG. The probes were end labeled with [α-32P]dCTP by the use of terminal transferase (New England Biolabs Inc., Beverly, Mass.). The labeled probes were purified through a NucTrap probe purification column (Stratagene Inc.) according to the manufacturer's instructions. BJAB nuclear extracts were used as a source of the Sp1 transcription factor, which was prepared as described previously (70). In vitro translation of LANA and its deletion mutants was done with a TNT quick transcription-translation couple kit (Promega Inc.) used according to the manufacturer's instructions. EMSA-binding reactions were prepared with a BJAB nuclear extract (approximately 5 μg of protein) and a GC box probe in a 50-μl reaction volume with binding buffer (20 mM HEPES [pH 7.5], 0.01% NP-40, 5.0% glycerol, 10 mM MgCl2, 100 μg of bovine serum albumin, 2 mM DTT, 1 mM PMSF, 40 mM KCl) and were then incubated at room temperature for 5 min. Sp1 mouse monoclonal immunoglobulin G (IgG) was used to supershift the mobility of the probe. Cold competitors (200×) were added 5 min prior to the addition of the radiolabeled probe. TLBR4, an unrelated DNA probe, was used as a nonspecific cold competitor. Bound complexes were resolved in a 5.0% nondenaturing polyacrylamide gel in 0.5× Tris-borate-EDTA. The gel was dried, and the signals were detected by use of a phosphorimager plate (Molecular Dynamics).
RESULTS
LANA interacts with the Sp1 transcription factor in vitro.
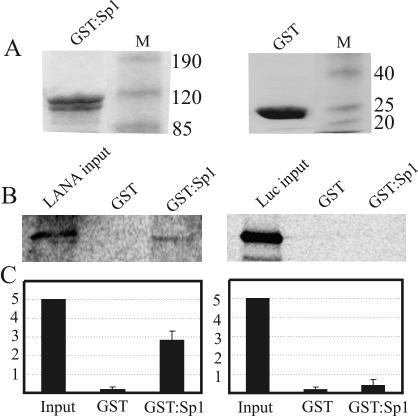
We have previously shown that the expression of LANA in eukaryotic cells can transactivate hTERT, and promoter truncation studies suggested that the minimum promoter sequence required for the transactivation activity lies from positions −130 to +5 of the promoter region (46). A nucleotide sequence analysis of this region showed that it contains five GC-rich boxes, which are the consensus binding sites for the transcription factor Sp1. Gel shifts in EMSAs with Sp1 in complex with the GC box were abolished when LANA was introduced into the complex (46). We hypothesized that LANA interacts physically with Sp1 to alter the mobility of the GC box probe. In this study, we show that LANA physically interacts with the Sp1 transcription factor. A GST-Sp1 fusion protein expressed in bacteria was purified by the use of glutathione-Sepharose beads and was then incubated with in vitro-translated [35S]methionine-labeled LANA and allowed to bind. Complexes of the GST-Sp1 fusion protein and LANA were resolved by SDS-PAGE. The results showed a significant amount of LANA binding to the complex, whereas little or no binding was seen with a GST control protein incubated with LANA, suggesting that the binding was specific for Sp1 (Fig. 1). The GST-Sp1 fusion protein was also incubated with an irrelevant protein, luciferase, as a negative control. The results indicated no specific binding, further supporting the observation that LANA bound specifically to Sp1.
FIG. 1.
In vitro binding of LANA and Sp1 transcription factor. (A) A GST-Sp1 fusion protein and GST control protein were expressed in E. coli, purified with glutathione-Sepharose beads, resolved by SDS-8.0% PAGE, and stained with Coomassie brilliant blue. (B) In vitro-translated LANA ([35S]methionine labeled) was incubated with the GST-Sp1 fusion protein or control GST protein in binding buffer and allowed to incubate, and the bound complex was resolved by SDS-PAGE. The GST-Sp1 lane shows the amount of LANA in complex with the Sp1 fusion protein compared to the LANA input lane (5%). Luciferase was used as an irrelevant control protein and did not bind to Sp1, suggesting the specificity of LANA binding to Sp1. (C) Intensities of LANA protein bands quantified with ImageQuant software (Molecular Dynamics).
LANA coimmunoprecipitates with Sp1 in KSHV-infected BCBL cells.
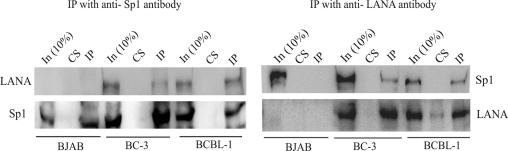
To confirm the association of LANA with the transcription factor Sp1 in vivo, we performed coimmunoprecipitation assays with LANA and Sp1, using anti-LANA and anti-Sp1 antibodies. Cell lysates prepared from BC-3 and BCBL-1 cells were incubated with anti-mouse monoclonal Sp1 antibodies, and the complexes were precipitated with protein A+G beads. A BJAB cell lysate was used as a negative control. The precipitated immune complex was resolved, Western blotted, and first incubated with an anti-LANA antibody, which showed the presence of the LANA protein immunoprecipitating with Sp1 (Fig. 2, left panel). A control serum did not precipitate LANA or Sp1 from the cell lysates of these two cell lines, supporting the specificity of the interaction with LANA. The same blot, when cross-reacted with an anti-Sp1 antibody, showed the presence of the Sp1 protein in BC-3, BCBL-1, and BJAB immunoprecipitation complexes. BJAB, which is a KSHV-negative cell line, did not show cross-reactivity with the anti-LANA serum. We also examined whether complexes with LANA also contained Sp1 by performing reverse immunoprecipitation assays using a rabbit polyclonal anti-LANA antibody. With these experiments, we demonstrated that Sp1 complexes contained LANA, since LANA coimmunoprecipitated with Sp1 complexes (Fig. 2, right panel). Fig. 2 shows the amount of LANA that immunoprecipitated with the anti-LANA antibody in BC-3 and BCBL-1 cells (right panel). Together, these data strongly suggest that LANA associates with Sp1 in KSHV-infected cell lines.
FIG. 2.
Coimmunoprecipitation assays with LANA and Sp1. (Left) BJAB (KSHV negative), BC-3, and BCBL-1 (KSHV infected) cells (8 × 107) were lysed in RIPA buffer. Ten percent of the total lysate was used as input; the rest of the lysate was incubated for 1 h with a control serum and precipitated with protein A+G beads. This precleared lysate was then incubated with a mouse monoclonal anti-Sp1 antibody. The protein complexed with the Sp1 antibody was precipitated with protein A and protein G beads, resolved by SDS-PAGE, and transferred to a nitrocellulose membrane. The membrane was cross-reacted with an anti-LANA serum followed by horseradish peroxidase-conjugated protein A and was visualized by a standard chemiluminescence assay. LANA was coimmunoprecipitated from KSHV-positive cell lysates but was not detected in BJAB cells (IP lane). The Sp1 protein was visualized with an anti-Sp1 antibody after stripping of the same blot (Sp1 panel). (Right) Reverse coimmunoprecipitation. Sp1 was coimmunoprecipitated with an anti-LANA antibody (Sp1 panel), and LANA was detected after stripping and reprobing of the blot with an anti-LANA antibody (LANA panel).
LANA colocalizes with Sp1 in the nuclei of KSHV-positive BCBL-1 cells.
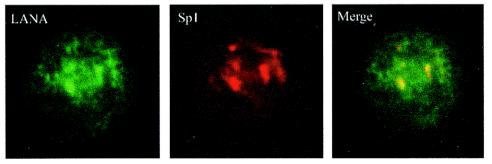
To further support the physical interaction of LANA with Sp1 in the BC-3 and BCBL-1 cell lines, we investigated the localization of these proteins by an immunofluorescence assay. Immunolocalization of LANA by use of an anti-LANA antibody detected the characteristic punctate expression of LANA in the nuclei of BC-3 and BCBL-1 cells, as reported previously, as well as signals in nuclear clusters. This was visualized with an Alexa fluor-conjugated donkey anti-rabbit secondary antibody (3, 11). Sp1, which is ubiquitously expressed in eukaryotic cells, also showed a distinct localization in the nucleus, seen as nuclear clusters, by use of an anti-mouse Sp1 antibody and a Texas red-conjugated goat anti-mouse secondary antibody (Fig. 3). Double staining for the detection of both Sp1 and LANA showed that LANA colocalized with Sp1 in similar nuclear compartments or clusters of BC-3 cells. However, some diffuse staining was also observed in the nucleus in addition to the punctate pattern. BJAB cells used as a control were incubated with the anti-LANA and anti-Sp1 antibodies. Sp1 was detected in a similar pattern to that seen in BC-3 cells, but no LANA signals were detected (data not shown).
FIG. 3.
Immunocolocalization of LANA and Sp1 from BC-3 cells. BC-3 cells were fixed in methanol-acetone (1:1) and incubated with an anti-LANA serum. LANA was localized by use of an Alexa fluor-conjugated secondary antibody. Sp1 was localized in the same cells by use of a mouse monoclonal anti-Sp1 antibody and a Texas red-conjugated secondary antibody. The merged image shows the colocalization of these two proteins in the nuclear clusters, where the predominant amount of the Sp1 signal was observed. LANA staining was observed in a somewhat speckled, fibrous pattern with additional signals in the nuclear clusters dominated by Sp1 staining.
The glutamine-rich and serine/threonine-rich regions of domain B and the amino-terminal region adjacent to the DNA binding domain of Sp1 interact with LANA.
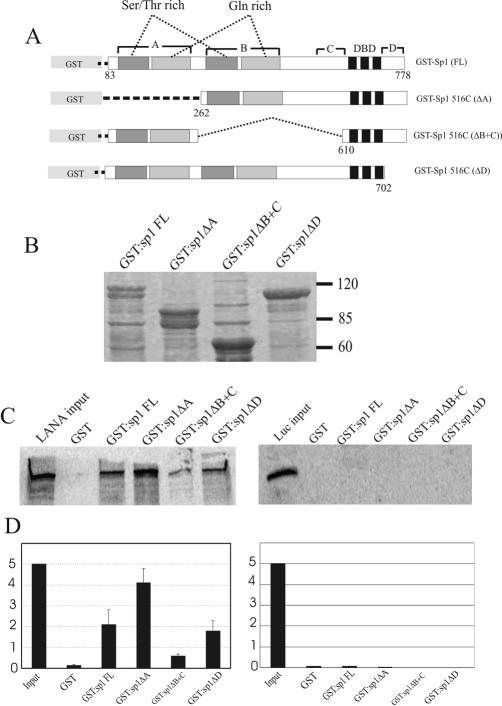
Sp1 belongs to the zinc finger family of transcription factors and has four distinct domains, namely, two identical glutamine-rich domains (A and B) that are required for transcriptional activation (13) and domains C and D, which include the DNA binding domain (40). The GST-Sp1 in vitro binding assay with LANA described above suggested that both proteins can physically interact. To further map the specific domains of Sp1 that are capable of mediating the physical interaction with LANA, we incubated Sp1 deletion mutant proteins with full-length LANA. Sp1516C (ΔA) lacks N-terminal aa 1 to 261 (domain A), Sp1516C (ΔB+C) contains an internal deletion of aa 263 to 609 (domains B and C), and Sp1N619 (ΔD) lacks C-terminal aa 703 to 778 (domain D); these mutants were expressed in E. coli and utilized as described above for in vitro binding assays as was done for full-length Sp1. This analysis showed that none of the Sp1 deletion mutants was able to completely eliminate the binding of LANA to Sp1 (Fig. 4C). However, there were two strikingly noticeable observations, as follows: (i) Sp1 with a deletion of domain A showed relatively stronger binding than full-length Sp1 and (ii) the Sp1 construct with a deletion of domains B and C showed almost 75% weaker binding than the full-length protein. There was little or no detectable interaction observed between LANA and a GST control protein. As expected, none of these Sp1 deletion mutants showed any significant binding to the luciferase protein control (Fig. 4C, right panel). These data strongly suggest that the LANA-Sp1 interaction requires the glutamic acid- and Ser/Thr-rich regions of domain B as well as the region adjacent to the DNA binding domain of Sp1.
FIG. 4.
Domain B of Sp1 is required for binding to LANA. (A) Schematic representation of wild-type [GST-Sp1 (FL)] and mutated Sp1 forms used in the GST interaction experiments shown in panel C. The Ser/Thr-rich regions of transactivation domains A and B are shown with dark gray boxes, and Gln-rich regions are shown with light gray boxes. (B) Expression profiles of wild-type (FL) and mutant GST-Sp1 forms in bacterial cells. GST-Sp1 fusion proteins were expressed in E. coli DH5α as described in Materials and Methods. Expressed fusion proteins were coupled to glutathione-Sepharose beads, and an aliquot of the bound protein was resolved by SDS-PAGE after extensive washing. Relative molecular masses of the fusion proteins are shown after Coomassie brilliant blue staining. (C) In vitro-transcribed and -translated LANA ([35S]methionine labeled) was incubated with GST- or GST-Sp1-coupled Sepharose beads as described in Materials and Methods. Sepharose bead fusion proteins were washed and separated by SDS-PAGE, and bound LANA was detected by autoradiography. Glutathione-Sepharose beads with Sp1 and mutant Sp1 were incubated with luciferase as a negative control. (D) The amount of LANA bound to different Sp1 mutants was quantified with ImageQuant software (Molecular Dynamics).
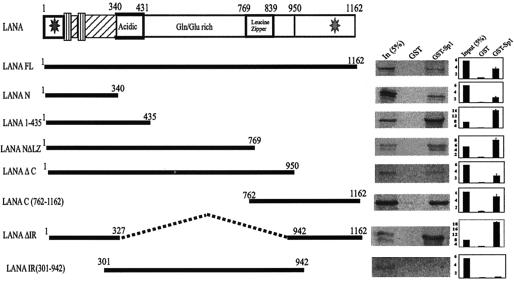
The amino and carboxy termini of LANA are required for interaction with Sp1.
LANA has three distinct domains, and the roles of this multifunctional protein have been mapped to different domains (12, 20, 26, 55, 57, 58, 66, 67, 73). To map the domains involved in binding to Sp1, we employed various deletion mutants of LANA for in vitro binding assays with the GST-Sp1 fusion protein. Figure 5 shows the various LANA mutants used in the binding assay. A fusion protein consisting of the entire Sp1 ORF fused to the C terminus of GST (GST-Sp1) or GST alone was incubated with [35S]methionine-labeled and in vitro-transcribed and -translated products of LANA mutant proteins. The bound complexes were resolved by SDS-PAGE with an acrylamide gel (with the concentration depending on the size of the protein) and were compared with the input (5% of each) after drying and autoradiography (Fig. 5). The results of the in vitro binding assay suggest that the amino terminus (aa 1 to 435) of the LANA protein binds very efficiently to Sp1. Other carboxy terminus deletion mutants of LANA (aa 1 to 769 and 1 to 950) containing the N terminus along with the central G/E-rich domain showed relatively less binding than LANA 1-435. However, the carboxy-terminal domain of LANA (aa 762 to 1162) also showed significant binding to GST-Sp1, albeit at a lower affinity than that observed with the amino-terminal region (Fig. 5). Strikingly, the LANA mutant with a deletion of the central G/E-rich domain showed the strongest binding (18% more than GST alone) to Sp1, which was significantly higher than that of full-length LANA. The central domain mutant LANA 301-942 (central G/E-rich domain) did not show significant binding to the GST-Sp1 fusion protein above that seen with the GST control (Fig. 5). These data suggest that the amino terminus of LANA is the primary site of interaction; however, its interaction with Sp1 also involves the carboxy-terminal 220 amino acids.
FIG. 5.
Mapping of LANA domains essential for binding to Sp1. Full-length LANA or various mutants of LANA were in vitro transcribed and translated (and [35S]methionine labeled) and then were incubated with glutathione-Sepharose-conjugated GST-Sp1 fusion proteins. Bound proteins were resolved by SDS-PAGE after extensive washing. Bound LANA was detected by autoradiography and was compared with the amount in the input lane (5%). Relative intensities of the bound LANA proteins were quantified with ImageQuant software (Molecular Dynamics) and are plotted in a bar diagram adjacent to each LANA mutant.
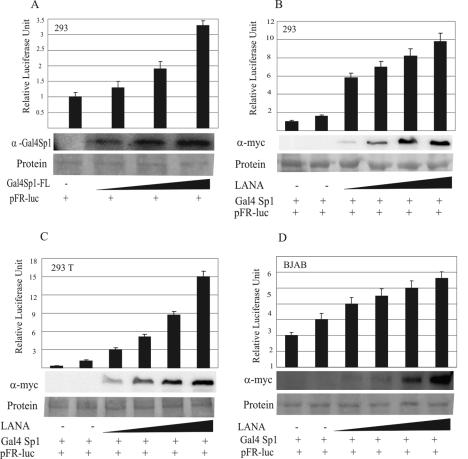
LANA upregulates Sp1-mediated transcriptional activation.
The proximal region of the hTERT promoter, extending from positions −130 to +5, is highly GC rich and contains five sequences that resemble the recognition sequences of the ubiquitous transcription factor Sp1 (5′-GGGCGG-3′). In previous studies, LANA was shown to upregulate the hTERT promoter in transient transfection assays (46). Here we have shown that LANA physically interacts with the Sp1 transcription factor. Therefore, we asked whether or not LANA can modulate Sp1-mediated transcription. We addressed this by expressing the GAL4Sp1 fusion protein (GAL4 DBD aa 1 to 147 fused in frame with the full-length Sp1 ORF) in the presence of a reiterated GAL4 response element (5XGAL4) cloned upstream of the luciferase gene (pFRLuc; Clontech) by using transient transfection reporter assays. Figure 6A shows, as expected, the dose-dependent response of GAL4Sp1 expression on the transactivation of the 5XGAL4 promoter in HEK293 cells, represented in relative luciferase units. The pFRLuc construct showed some basic luciferase activity, but when it was transfected with increasing amounts of GAL4Sp1, a more-than-threefold activation was observed (Fig. 6A). The coexpression of LANA in HEK293 cells with GAL4Sp1 increased the transactivation of the 5XGAL4 promoter 6- to 10-folds with increasing amounts of the LANA expression vector (Fig. 6B). This transactivation experiment was repeated in HEK293T and BJAB cells (Fig. 6C and D, respectively). HEK293T cells showed more (up to 15-fold) activation of the 5XGAL4 Sp1 promoter activity with increasing amounts of the LANA expression vector. BJAB cells also showed a dose-dependent effect on the promoter activity of 5XGAL4 by Sp1 (Fig. 6D). The expression of LANA was detected by Western blotting with an anti-Myc antibody of the same cell lysate as that used for luciferase activity assays, as described in Materials and Methods.
FIG. 6.
LANA synergistically activates Sp1-mediated transcription. (A) GAL4-Sp1 activates transcription. HEK293 cells were cotransfected with the GAL4 DBD fused in frame with Sp1 and the pFR reporter plasmid containing the 5XGAL4 response element. Approximately 107 cells were transfected, and at 24 h posttransfection, the cells were harvested and lysed for luciferase assays. Increasing amounts of GAL4-Sp1 showed proportional increases in luciferase activity. Fractions of the cell lysates were resolved by SDS-PAGE to demonstrate the increased expression of GAL4-Sp1 in samples with larger amounts of transfected DNA. (B) LANA modulates GAL4-Sp1-mediated transcription. HEK293 cells were transfected with increasing amounts of LANA to show the effect of LANA on GAL4-Sp1-mediated transcription. LANA showed a dose-dependent response of GAL4-Sp1-mediated luciferase activity, which was plotted in relative luciferase units. (C and D) LANA modulates GAL4-mediated transcription in HEK293T and BJAB cells. Approximately 107 cells were transfected in both cases; at 24 h posttransfection, the luciferase activity was measured as described above. All of these experiments were done independently three times in duplicate, and the average values are presented in the figure. The increased expression of LANA was detected by use of an anti-Myc monoclonal antibody because LANA has a Myc epitope at its C terminus.
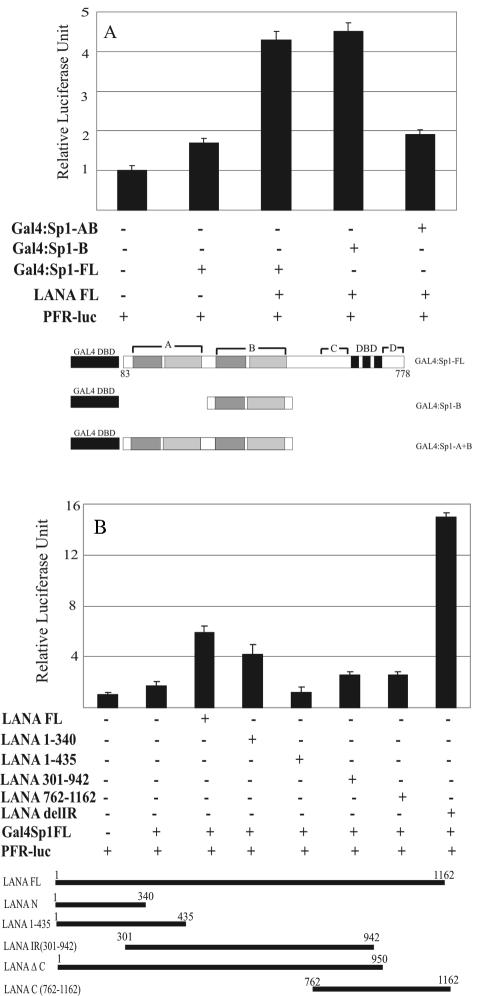
Domain B of Sp1, which is rich in glutamic acid and Ser/Thr, is sufficient to mediate an increased transcriptional activity of Sp1 in reporter assays.
The transcription factor Sp1 contains two homologous Gln and Ser/Thr domains (A and B) that are responsible for most of the associated transcriptional activities. These domains were assayed for their ability to transactivate the 5XGAL4 promoter-driven luciferase reporter in the presence of LANA. The Sp1 mutants employed were GAL4Sp1 B, which contains only domain B (aa 263 to 542), and GAL4Sp1 A+B, which contains both domains A and B (aa 1 to 542) along with full-length GAL4Sp1. These GAL4Sp1 mutants were tested by transient transfection reporter assays in HEK293 cells with the LANA expression constructs (Fig. 7A). The coexpression of LANA with GAL4Sp1 B increased the transactivation of the 5XGAL4 promoter 4.5-fold in BJAB cells, whereas LANA and GAL4Sp1 A+B did not show any significant activation, suggesting that domain A may have a negative regulatory function in the full-length molecule. The in vitro binding data also suggested that a mutant of Sp1 with a deletion of domain A bound more strongly than the full-length Sp1 molecule.
FIG. 7.
Mapping of functional domains of LANA and Sp1. (A) The B domain of Sp1 is enough for transcriptional activation by LANA. HEK293 cells were cotransfected with full-length (FL) GAL4-Sp1 and a vector containing either the A or the A and B domains in the presence of LANA. The number of relative luciferase units indicated that domain B is sufficient for the LANA-mediated transcription of luciferase. The GAL4 DBD-Sp1 fusion proteins used in the assay are shown below the bar diagram. (B) Different deletion mutants of LANA, shown below the bar diagram, were cotransfected into HEK293 cells (107) in the presence of GAL4-Sp1 and the pFR Luc reporter plasmid, and the luciferase activity was measured as described earlier after 24 h posttransfection. The N terminus of LANA (aa 1 to 340) showed an up-regulation in GAL4-mediated luciferase activity, whereas the C terminus itself did not show any effect and the C terminus fused to the N terminus had enhanced activation of the GAL4-Sp1-mediated luciferase activity, plotted in terms of relative luciferase activity. The data shown here are representative of three independent experiments done in triplicate.
The amino- and carboxy-terminal domains of LANA cooperate with Sp1 to enhance transcriptional activity.
The panel of LANA mutants used for the in vitro binding assay described above were used to map the domains of LANA which would suffice in mediating the activities of the GAL4 reiterated response element. The results from this assay, shown in Fig. 7B, indicate that both the amino- and carboxy-terminal regions cooperate to enhance the activity of Sp1. Coexpression of the amino terminus of LANA 1-340 with GAL4Sp1 caused a fourfold activation in luciferase activity in this assay. LANA 1-435 did not show transactivation of the promoter, whereas the in vitro binding of LANA 1-435 was stronger than that of LANA 1-340. Therefore, the acidic domain present in LANA 1-435 is essential for binding but has negative regulatory effects on Sp1-mediated transcriptional activities. The central domain of LANA (G/E-rich) did not show any effect on the 5XGAL4 promoter activity (Fig. 7B). The C-terminal domain of LANA (LANA 762-1162) also did not show any enhanced activation. However, the fusion protein which contained the N and C termini strongly enhanced (15-fold) the 5XGAL4 promoter 2.5 times more than the full-length LANA molecule (Fig. 7B).
These data indicate that the N-terminal domain of LANA is sufficient to mediate transcriptional activation; however, the critical C-terminal domain which is important for the binding of DNA can cooperate with the amino-terminal domain to synergize Sp1-mediated activation of the 5XGAL4-responsive promoter.
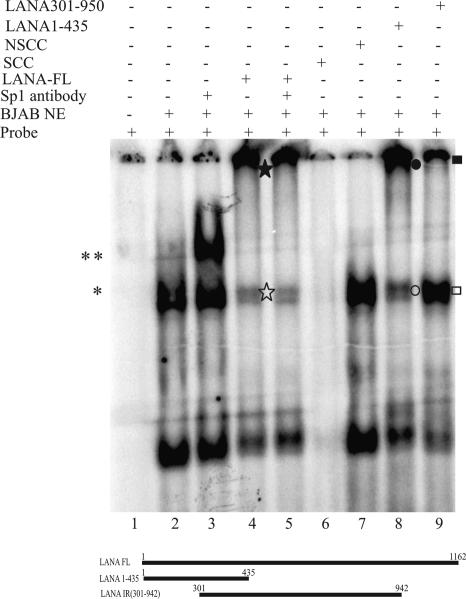
The amino terminal 1 to 435 aa of LANA target the GC box-Sp1 complex.
As shown previously, the hTERT promoter sequence from positions −130 to +5 contains five GC-rich boxes, which have been shown to be the binding sites for the Sp1 transcription factor by EMSAs (46). The interactions of Myc at the E box and Sp1 at the GC box were shown to be the major determinants of hTERT expression (49). Furthermore, LANA was shown to up-regulate the hTERT promoter and targets the Sp1-GC box complex shown by EMSAs (46). Here we showed that the various domains of LANA differed in their binding to Sp1. Therefore, we asked which region of LANA bound to Sp1 when it was complexed with its cognate sequence. The previously described double standard DNA probe of the hTERT promoter sequence between positions −119 and −98 (46) was labeled and tested for binding of the Sp1 complex compared with that in the presence of full-length LANA by EMSA. Nuclear extracts prepared from BJAB cells known to contain Sp1 complexes reduced the mobility of the GC box probe, as expected (Fig. 8). The mobility was further supershifted with an anti-Sp1 monoclonal antibody, suggesting that the specificity of the shift was due to the presence of Sp1. The addition of in vitro-translated LANA to the binding reaction resulted in the formation of an Sp1-LANA complex, thus abolishing or decreasing the observed intensity of the Sp1-specific band (Fig. 8, lane 4). This resulted in the formation of a large complex with limited mobility migrating at the top of the gel (Fig. 8, lane 4). Furthermore, the addition of an Sp1-specific antibody along with in vitro-translated LANA showed a similar pattern, indicating that the large LANA-Sp1-DNA complex is likely to be at the top of the gel, as seen by an increased intensity of the signal at that position that did not migrate into the gel. Additionally, the Sp1-specific shift was competed with the addition of a 200× cold specific competitor, but it was not affected by a similar amount of a nonspecific cold competitor (Fig. 8).
FIG. 8.
N terminus of LANA is sufficient for binding. A probe for the Sp1 binding sequence (GC box) spanning positions −119 to −98 of the hTERT promoter was labeled with [32P-α]dCTP and used for EMSAs in the presence of in vitro-transcribed and -translated LANA or different LANA mutants. Lanes 1 and 2, probe with and without BJAB nuclear extract (NE), respectively, showing activity due to Sp1 binding (*); lane 3, probe with BJAB nuclear extract and Sp1 mouse monoclonal IgG antibody, which supershifted the GC box probes (**); lane 4, in vitro-translated LANA, which abolished ([star]) the Sp1-specific shift, most likely due to the formation of a large complex that was unable to enter the gel ([starf]); lane 5, probe with Sp1 mouse monoclonal IgG along with in vitro-translated LANA and BJAB nuclear extract; lanes 6 and 7, probe with 200-fold molar excess of cold specific and nonspecific competitor, respectively. The LANA mutants were used for EMSAs, but only two mutants, LANA 1-435 (strong binding) and LANA 301-942 (very little or no binding), are shown in this figure. Lane 8, in vitro-translated N terminus of LANA (aa 1 to 435) along with the probe and BJAB nuclear extract, which abolished (○) the Sp1-specific band, forming a larger complex similar to that with full-length LANA (•); lane 9, LANA 301-942 did not show the elimination (□) of the Sp1-specific band and the accumulation of a larger complex (▪).
A panel of LANA mutants was tested for the ability to target the GC box-Sp1 complex. LANA aa 1 to 435 efficiently targeted the GC box-Sp1 complex, similar to the full-length LANA. However, LANA aa 301 to 942 did not efficiently target the complex compared to LANA 1-435 and full-length LANA (Fig. 8, lanes 4, 8, and 9). Additional LANA mutants showed some activity in terms of binding and elimination of the Sp1-specific band (data not shown). These results corroborate the above binding data, which indicate that the N terminus of LANA is essential for binding and that the C-terminal domain of LANA may contribute to binding and increased transcriptional activity.
DISCUSSION
LANA encoded by KSHV is expressed during all forms of KS-associated malignancies (17, 43, 68). LANA is critical for episomal maintenance of the viral DNA during latent infections through interactions with cellular proteins, including histone H1 (11, 73). Moreover, plasmids containing the KSHV terminal repeats are maintained when the N-terminal chromatin binding domain is deleted and LANA is expressed as a fusion with the chromatin-associated protein histone H1 (4, 32, 73). This suggests that the interaction between LANA and histone H1 may contribute to the chromosomal association. Recently, two separate cellular proteins (the methyl-CpG binding protein MeCP2 and the cellular DEK protein) associated with LANA were identified. These proteins may play a role in tethering the KSHV genome to chromosomes during mitosis, thus ensuring long-term viral persistence (47). Besides the maintenance of the episomal DNA, LANA also has transcriptional regulatory properties, including the down modulation of the activities of the tumor suppressors p53 and pRb (20, 67). LANA also may have transforming potential, as it is capable of transforming rat primary fibroblast cells in cooperation with the Hras oncogene (67).
Another role associated with LANA's oncogenicity is its transactivation of the hTERT promoter (46). LANA was shown for the first time by in vitro transcriptional reporter assays to be capable of activating the hTERT promoter (46). It was previously documented that KSHV-transformed human umbilical vein endothelial cells had active telomerase, as shown by a telomerase repeat amplification protocol (19). Promoter truncation studies revealed that the −130 to +5 sequence is sufficient for promoter activity and that this sequence contains five GC box sequences, which are known to be binding sites for the Sp1 transcription factor (46).
In vitro binding experiments suggested that LANA and Sp1 physically interact, and this was supported by in vivo immunoprecipitation studies of cell lysates of KSHV-positive cell lines (BC-3 and BCBL-1). Immunolocalization using anti-LANA and anti-Sp1 antibodies for LANA and Sp1, respectively, showed a speckled nuclear localization of these two proteins. Therefore, taken together, these studies strongly suggest that Sp1 and LANA can cooperate to functionally enhance or activate the hTERT promoter in KSHV-positive cells.
Furthermore, the physical interaction between these two molecules was assayed to detect changes in the functional activity of Sp1. Sp1 was fused in frame with the GAL4 DBD and was tested for the ability to upregulate luciferase expression from a reporter containing the 5XGAL4 response element upstream of the TATA box. As expected, LANA upregulated Sp1-mediated transcription and supported previous results which indicated that LANA transactivates the hTERT promoter (46). Therefore, these results demonstrated that the transactivation of the hTERT promoter is likely to be mediated through Sp1-activated transcription, which is enhanced through the direct association of Sp1 with LANA. Other viral proteins have also been shown to modulate Sp1-mediated transcriptional activation. The simian virus 40 small antigen has been shown to stimulate Sp1-dependent transcription mediated by protein kinase C and its upstream regulator phosphatidylinositol 3-kinase and dependent on a consensus TATA motif (27, 38, 74). The E1A protein encoded by adenovirus can also induce the p21 promoter activity mediated by Sp1 binding sites, most probably by interacting with the CR3 region of E1A, which has been shown to be the binding site for other transcriptional activators, including c-Jun, ATF-1, -2, and -3, CBF, TBP, TAF (II) 110, 135, and 250, and YY1 (23, 64). E1A has also been shown to upregulate the hTERT core promoter, which contains the binding site for Sp1 (45). Human papillomavirus-encoded E2 down modulates hTERT promoter activity, which agrees with data showing that E2 inhibits cell growth in human papillomavirus-infected cells and triggers apoptosis in HeLa cells (51). It can also activate the expression of p21(WAF1/CIP1) via its promoter-proximal 200 nucleotides, which contain several Sp1 binding sites and no E2 binding sites (76).
The binding of Sp1 to LANA and the supporting reported assays prompted us to map the domains of Sp1 that are involved in binding to LANA as well as the activation of the 5XGAL4 promoter. Our analysis suggests that the Gln-rich domain B of Sp1 is essential for binding LANA, and this domain was also previously shown to be the binding domain for the transcription activator c-Jun (40). Besides the Gln-rich domain of Sp1, the DNA binding domain has been shown to interact with various transcriptional activators, such as the p65/RelA subunit of NF-κB (33-35), the erythroid factor GATA-1 (53), the transcription factor YY1 (10, 52), and the cell cycle regulator E2F (41, 59). Since the activation domain of Sp1 maps as a DNA binding domain, our results indicate that enhanced transactivation of the promoter occurs through a direct protein-protein interaction. This is distinct from data reported for other transcriptional activators in that their cognate sequences lie adjacent to the binding sequence for Sp1 in the context of the activated promoter (9, 81).
In transient reporter assays, the Gln-rich homologous domains of Sp1 fused to the GAL4 DBD indicated that domain B is essential for transactivation, strongly suggesting that the binding region of Sp1 for LANA is sufficient for both binding and transcription activation. The Gln-rich region of the B domain of Sp1 is required for the transcriptional activation of Sp1 promoters by transforming growth factor beta (15, 16, 63). The Gln-rich region of domain B has also been shown to be important for an enhanced activation of Sp1-dependent transcription through an interaction with c-Jun and the Smad protein (40, 82). Our data also suggest that domain B is essential for the enhanced transactivation of Sp1-responsive promoters through its interaction with LANA, indicating that LANA may regulate Sp1-dependent transcription via a similar mechanism (40). Additionally, the A domain, which reduces the in vitro binding activity, may also have some negative regulatory effect on the Sp1-responsive hTERT promoter.
In this study, we further delineated the domains of LANA that are involved in binding to Sp1 and the domains that are essential for Sp1-mediated transcriptional activation. Different functions have been mapped to different domains of LANA in terms of its binding and transcription modulatory effects on many promoters (20, 54-58, 67, 69). The N terminus of LANA (aa 1 to 435) efficiently binds to Sp1. Additionally, the acidic domain in the N terminus of LANA is essential for the interaction, as LANA 1-435 binds somewhat less efficiently than a molecule with a deletion of the central region. LANA polypeptides with a deletion of the central Gln/Glu-rich region showed the highest affinity for Sp1, suggesting that the C-terminal domain may be a regulatory domain or may cooperate to increase the binding activity. The C-terminal domain itself also showed some level of binding to Sp1 independent of the N terminus. Therefore, the enhanced binding is most likely due to the cooperative effects of both the N- and C-terminal domains. The reduced binding of LANA 1-950 compared to the LANA 1-769 polypeptide was probably due to the leucine zipper region, as Sp1 also contains a leucine zipper (57). This may have affected the resulting binding activities of these two proteins in our assays. The results for the LANA 301-942 polypeptide clearly suggest that the central Gln/Glu-rich domain has little or no binding capacity for Sp1 and thus support the conclusion that binding to Sp1 is primarily due to the N- and C-terminal domains.
EMSAs with the GC box probe in the presence of BJAB nuclear extracts showed a binding activity of the probe which was shifted by the addition of an anti-Sp1 antibody. The addition of LANA to the reaction diminished the Sp1-specific complex, likely due to the formation of a larger heterogeneous complex with Sp1 that was incapable of entering the native PAGE gel (46). These data strongly suggest that LANA binds to Sp1, forming a heterogeneous LANA-Sp1-GC box complex. We showed here that the binding of Sp1 and LANA, independent of the Sp1 DNA binding sequence, was similar to the previously observed phenomena involving Rb-Sp1 and SREBP-1-Sp1 (9, 81). The binding of different domains of LANA to Sp1 also suggested that the domains identified by in vitro binding assays form complexes with Sp1, but the domain which had negligible binding to Sp1 in the in vitro binding assay also showed little or no affinity for Sp1 when it was complexed with its cognate binding sequence. Taken together, the in vitro binding and EMSA data strongly demonstrate that the N-terminal domain of LANA is critical for Sp1 binding.
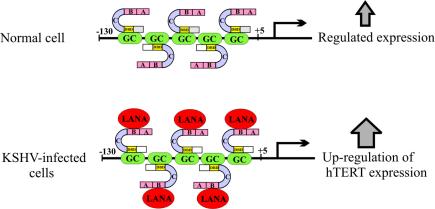
In the present study, we showed that LANA directly binds to Sp1 in vitro and associates in a complex in vivo. Therefore, Sp1 bound to its GC box is likely to form a complex with LANA that results in increased transcription activity. This was demonstrated in the context of the hTERT promoter with five GC boxes, which are known to be cognate binding sequences for Sp1. LANA expressed constitutively during latent infection enhances transactivation of the hTERT promoter by targeting the Sp1 transcription factor, thus contributing to the immortalization of KSHV-infected cells (Fig. 9).
FIG. 9.
Schematic model for Sp1-mediated transcriptional regulation of hTERT promoter enhanced by LANA. The hTERT promoter sequence contains five GC boxes from positions −130 to +5 which are binding sites for the Sp1 transcription factor. In KSHV-infected cells, LANA is expressed constitutively during latent infection and upregulates telomerase expression by physically interacting with the Sp1 transcription factor, resulting in increased telomerase activity and potentially contributing to immortalization of the infected cells.
Acknowledgments
This work was supported by grants from the Leukemia and Lymphoma Society of America and by public health service grants NCI CA072510 and CA091792 and NIDCR DE01436 (to E.S.R.). E.S.R. is a scholar of the Leukemia and Lymphoma Society of America.
REFERENCES
- 1.An, J., A. K. Lichtenstein, G. Brent, and M. B. Rettig. 2002. The Kaposi sarcoma-associated herpesvirus (KSHV) induces cellular interleukin 6 expression: role of the KSHV latency-associated nuclear antigen and the AP1 response element. Blood 99:649-654. [DOI] [PubMed] [Google Scholar]
- 2.Aster, J. C., E. S. Robertson, R. P. Hasserjian, J. R. Turner, E. Kieff, and J. Sklar. 1997. Oncogenic forms of NOTCH1 lacking either the primary binding site for RBP-Jkappa or nuclear localization sequences retain the ability to associate with RBP-Jkappa and activate transcription. J. Biol. Chem. 272:11336-11343. [DOI] [PubMed] [Google Scholar]
- 3.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 4.Ballestas, M. E., and K. M. Kaye. 2001. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 75:3250-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berg, J. M. 1992. Sp1 and the subfamily of zinc finger proteins with guanine-rich binding sites. Proc. Natl. Acad. Sci. USA 89:11109-11110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 7.Cesarman, E., P. S. Moore, P. H. Rao, G. Inghirami, D. M. Knowles, and Y. Chang. 1995. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood 86:2708-2714. [PubMed] [Google Scholar]
- 8.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 9.Chen, L. I., T. Nishinaka, K. Kwan, I. Kitabayashi, K. Yokoyama, Y. H. Fu, S. Grunwald, and R. Chiu. 1994. The retinoblastoma gene product RB stimulates Sp1-mediated transcription by liberating Sp1 from a negative regulator. Mol. Cell. Biol. 14:4380-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole, E. G., and K. Gaston. 1997. A functional YY1 binding site is necessary and sufficient to activate Surf-1 promoter activity in response to serum growth factors. Nucleic Acids Res. 25:3705-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cotter, M. A., II, and E. S. Robertson. 1999. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 264:254-264. [DOI] [PubMed] [Google Scholar]
- 12.Cotter, M. A., II, C. Subramanian, and E. S. Robertson. 2001. The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen binds to specific sequences at the left end of the viral genome through its carboxy-terminus. Virology 291:241-259. [DOI] [PubMed] [Google Scholar]
- 13.Courey, A. J., D. A. Holtzman, S. P. Jackson, and R. Tjian. 1989. Synergistic activation by the glutamine-rich domains of human transcription factor Sp1. Cell 59:827-836. [DOI] [PubMed] [Google Scholar]
- 14.Datta, P. K., P. Raychaudhuri, and S. Bagchi. 1995. Association of p107 with Sp1: genetically separable regions of p107 are involved in regulation of E2F- and Sp1-dependent transcription. Mol. Cell. Biol. 15:5444-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Datto, M. B., Y. Li, J. F. Panus, D. J. Howe, Y. Xiong, and X. F. Wang. 1995. Transforming growth factor beta induces the cyclin-dependent kinase inhibitor p21 through a p53-independent mechanism. Proc. Natl. Acad. Sci. USA 92:5545-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Datto, M. B., Y. Yu, and X. F. Wang. 1995. Functional analysis of the transforming growth factor beta responsive elements in the WAF1/Cip1/p21 promoter. J. Biol. Chem. 270:28623-28628. [DOI] [PubMed] [Google Scholar]
- 17.Dupin, N., C. Fisher, P. Kellam, S. Ariad, M. Tulliez, N. Franck, E. van Marck, D. Salmon, I. Gorin, J. P. Escande, R. A. Weiss, K. Alitalo, and C. Boshoff. 1999. Distribution of human herpesvirus-8 latently infected cells in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. Proc. Natl. Acad. Sci. USA 96:4546-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emili, A., J. Greenblatt, and C. J. Ingles. 1994. Species-specific interaction of the glutamine-rich activation domains of Sp1 with the TATA box-binding protein. Mol. Cell. Biol. 14:1582-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flore, O., S. Rafii, S. Ely, J. J. O'Leary, E. M. Hyjek, and E. Cesarman. 1998. Transformation of primary human endothelial cells by Kaposi's sarcoma-associated herpesvirus. Nature 394:588-592. [DOI] [PubMed] [Google Scholar]
- 20.Friborg, J., Jr., W. Kong, M. O. Hottiger, and G. J. Nabel. 1999. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402:889-894. [DOI] [PubMed] [Google Scholar]
- 21.Fujimuro, M., and S. D. Hayward. 2003. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus manipulates the activity of glycogen synthase kinase-3beta. J. Virol. 77:8019-8030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujimuro, M., F. Y. Wu, C. ApRhys, H. Kajumbula, D. B. Young, G. S. Hayward, and S. D. Hayward. 2003. A novel viral mechanism for dysregulation of beta-catenin in Kaposi's sarcoma-associated herpesvirus latency. Nat. Med. 9:300-306. [DOI] [PubMed] [Google Scholar]
- 23.Gallimore, P. H., and A. S. Turnell. 2001. Adenovirus E1A: remodelling the host cell, a life or death experience. Oncogene 20:7824-7835. [DOI] [PubMed] [Google Scholar]
- 24.Gao, S. J., L. Kingsley, M. Li, W. Zheng, C. Parravicini, J. Ziegler, R. Newton, C. R. Rinaldo, A. Saah, J. Phair, R. Detels, Y. Chang, and P. S. Moore. 1996. KSHV antibodies among Americans, Italians and Ugandans with and without Kaposi's sarcoma. Nat. Med. 2:925-928. [DOI] [PubMed] [Google Scholar]
- 25.Gao, S. J., Y. J. Zhang, J. H. Deng, C. S. Rabkin, O. Flore, and H. B. Jenson. 1999. Molecular polymorphism of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent nuclear antigen: evidence for a large repertoire of viral genotypes and dual infection with different viral genotypes. J. Infect. Dis. 180:1466-1476. [DOI] [PubMed] [Google Scholar]
- 26.Garber, A. C., M. A. Shu, J. Hu, and R. Renne. 2001. DNA binding and modulation of gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:7882-7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia, A., S. Cereghini, and E. Sontag. 2000. Protein phosphatase 2A and phosphatidylinositol 3-kinase regulate the activity of Sp1-responsive promoters. J. Biol. Chem. 275:9385-9389. [DOI] [PubMed] [Google Scholar]
- 28.Gill, G., E. Pascal, Z. H. Tseng, and R. Tjian. 1994. A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc. Natl. Acad. Sci. USA 91:192-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greider, C. W. 1991. Chromosome first aid. Cell 67:645-647. [DOI] [PubMed] [Google Scholar]
- 30.Greider, C. W. 1996. Telomere length regulation. Annu. Rev. Biochem. 65:337-365. [DOI] [PubMed] [Google Scholar]
- 31.Groves, A. K., M. A. Cotter, C. Subramanian, and E. S. Robertson. 2001. The latency-associated nuclear antigen encoded by Kaposi's sarcoma-associated herpesvirus activates two major essential Epstein-Barr virus latent promoters. J. Virol. 75:9446-9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grundhoff, A., and D. Ganem. 2003. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus permits replication of terminal repeat-containing plasmids. J. Virol. 77:2779-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirano, F., M. Chung, H. Tanaka, N. Maruyama, I. Makino, D. D. Moore, and C. Scheidereit. 1998. Alternative splicing variants of IkappaB beta establish differential NF-kappaB signal responsiveness in human cells. Mol. Cell. Biol. 18:2596-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirano, F., H. Tanaka, Y. Hirano, M. Hiramoto, H. Handa, I. Makino, and C. Scheidereit. 1998. Functional interference of Sp1 and NF-kappaB through the same DNA binding site. Mol. Cell. Biol. 18:1266-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirano, F., H. Tanaka, T. Miura, Y. Hirano, K. Okamoto, Y. Makino, and I. Makino. 1998. Inhibition of NF-kappaB-dependent transcription of human immunodeficiency virus 1 promoter by a phosphodiester compound of vitamin C and vitamin E, EPC-K1. Immunopharmacology 39:31-38. [DOI] [PubMed] [Google Scholar]
- 36.Horikawa, I., P. L. Cable, C. Afshari, and J. C. Barrett. 1999. Cloning and characterization of the promoter region of human telomerase reverse transcriptase gene. Cancer Res. 59:826-830. [PubMed] [Google Scholar]
- 37.Hyun, T. S., C. Subramanian, M. A. Cotter II, R. A. Thomas, and E. S. Robertson. 2001. Latency-associated nuclear antigen encoded by Kaposi's sarcoma-associated herpesvirus interacts with Tat and activates the long terminal repeat of human immunodeficiency virus type 1 in human cells. J. Virol. 75:8761-8771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johannessen, M., P. A. Olsen, R. Sorensen, B. Johansen, O. M. Seternes, and U. Moens. 2003. A role of the TATA box and the general co-activator hTAF(II)130/135 in promoter-specific trans-activation by simian virus 40 small t antigen. J. Gen. Virol. 84:1887-1897. [DOI] [PubMed] [Google Scholar]
- 39.Kao, W. Y., J. A. Briggs, M. C. Kinney, R. A. Jensen, and R. C. Briggs. 1997. Structure and function analysis of the human myeloid cell nuclear differentiation antigen promoter: evidence for the role of Sp1 and not of c-Myb or PU.1 in myelomonocytic lineage-specific expression. J. Cell Biochem. 65:231-244. [DOI] [PubMed] [Google Scholar]
- 40.Kardassis, D., P. Papakosta, K. Pardali, and A. Moustakas. 1999. c-Jun transactivates the promoter of the human p21 (WAF1/Cip1) gene by acting as a superactivator of the ubiquitous transcription factor Sp1. J. Biol. Chem. 274:29572-29581. [DOI] [PubMed] [Google Scholar]
- 41.Karlseder, J., H. Rotheneder, and E. Wintersberger. 1996. Interaction of Sp1 with the growth- and cell cycle-regulated transcription factor E2F. Mol. Cell. Biol. 16:1659-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kedes, D. H., E. Operskalski, M. Busch, R. Kohn, J. Flood, and D. Ganem. 1996. The seroepidemiology of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus): distribution of infection in KS risk groups and evidence for sexual transmission. Nat. Med. 2:918-924. [DOI] [PubMed] [Google Scholar]
- 43.Kellam, P., C. Boshoff, D. Whitby, S. Matthews, R. A. Weiss, and S. J. Talbot. 1997. Identification of a major latent nuclear antigen, LNA-1, in the human herpesvirus 8 genome. J. Hum. Virol. 1:19-29. [PubMed] [Google Scholar]
- 44.Kim, N. W., M. A. Piatyszek, K. R. Prowse, C. B. Harley, M. D. West, P. L. Ho, G. M. Coviello, W. E. Wright, S. L. Weinrich, and J. W. Shay. 1994. Specific association of human telomerase activity with immortal cells and cancer. Science 266:2011-2015. [DOI] [PubMed] [Google Scholar]
- 45.Kirch, H. C., S. Ruschen, D. Brockmann, H. Esche, I. Horikawa, J. C. Barrett, B. Opalka, and U. R. Hengge. 2002. Tumor-specific activation of hTERT-derived promoters by tumor suppressive E1A-mutants involves recruitment of p300/CBP/HAT and suppression of HDAC-1 and defines a combined tumor targeting and suppression system. Oncogene 21:7991-8000. [DOI] [PubMed] [Google Scholar]
- 46.Knight, J. S., M. A. Cotter II, and E. S. Robertson. 2001. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus transactivates the telomerase reverse transcriptase promoter. J. Biol. Chem. 276:22971-22978. [DOI] [PubMed] [Google Scholar]
- 47.Krithivas, A., M. Fujimuro, M. Weidner, D. B. Young, and S. D. Hayward. 2002. Protein interactions targeting the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus to cell chromosomes. J. Virol. 76:11596-11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kyo, S., T. Kanaya, M. Takakura, M. Tanaka, and M. Inoue. 1999. Human telomerase reverse transcriptase as a critical determinant of telomerase activity in normal and malignant endometrial tissues. Int. J. Cancer 80:60-63. [DOI] [PubMed] [Google Scholar]
- 49.Kyo, S., M. Takakura, T. Taira, T. Kanaya, H. Itoh, M. Yutsudo, H. Ariga, and M. Inoue. 2000. Sp1 cooperates with c-Myc to activate transcription of the human telomerase reverse transcriptase gene (hTERT). Nucleic Acids Res. 28:669-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lania, L., B. Majello, and P. De Luca. 1997. Transcriptional regulation by the Sp family proteins. Int. J. Biochem. Cell Biol. 29:1313-1323. [DOI] [PubMed] [Google Scholar]
- 51.Lee, D., H. Z. Kim, K. W. Jeong, Y. S. Shim, I. Horikawa, J. C. Barrett, and J. Choe. 2002. Human papillomavirus E2 down-regulates the human telomerase reverse transcriptase promoter. J. Biol. Chem. 277:27748-27756. [DOI] [PubMed] [Google Scholar]
- 52.Lee, J. S., K. M. Galvin, and Y. Shi. 1993. Evidence for physical interaction between the zinc-finger transcription factors YY1 and Sp1. Proc. Natl. Acad. Sci. USA 90:6145-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee, J. S., H. Ngo, D. Kim, and J. H. Chung. 2000. Erythroid Kruppel-like factor is recruited to the CACCC box in the beta-globin promoter but not to the CACCC box in the gamma-globin promoter: the role of the neighboring promoter elements. Proc. Natl. Acad. Sci. USA 97:2468-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liao, W., Y. Tang, S. F. Lin, H. J. Kung, and C. Z. Giam. 2003. K-bZIP of Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 (KSHV/HHV-8) binds KSHV/HHV-8 Rta and represses Rta-mediated transactivation. J. Virol. 77:3809-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lim, C., Y. Gwack, S. Hwang, S. Kim, and J. Choe. 2001. The transcriptional activity of cAMP response element-binding protein-binding protein is modulated by the latency associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Biol. Chem. 276:31016-31022. [DOI] [PubMed] [Google Scholar]
- 56.Lim, C., D. Lee, T. Seo, C. Choi, and J. Choe. 2003. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus functionally interacts with heterochromatin protein 1. J. Biol. Chem. 278:7397-7405. [DOI] [PubMed] [Google Scholar]
- 57.Lim, C., H. Sohn, Y. Gwack, and J. Choe. 2000. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) binds ATF4/CREB2 and inhibits its transcriptional activation activity. J. Gen. Virol. 81:2645-2652. [DOI] [PubMed] [Google Scholar]
- 58.Lim, C., H. Sohn, D. Lee, Y. Gwack, and J. Choe. 2002. Functional dissection of latency-associated nuclear antigen 1 of Kaposi's sarcoma-associated herpesvirus involved in latent DNA replication and transcription of terminal repeats of the viral genome. J. Virol. 76:10320-10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin, S. Y., A. R. Black, D. Kostic, S. Pajovic, C. N. Hoover, and J. C. Azizkhan. 1996. Cell cycle-regulated association of E2F1 and Sp1 is related to their functional interaction. Mol. Cell. Biol. 16:1668-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marin, M., A. Karis, P. Visser, F. Grosveld, and S. Philipsen. 1997. Transcription factor Sp1 is essential for early embryonic development but dispensable for cell growth and differentiation. Cell 89:619-628. [DOI] [PubMed] [Google Scholar]
- 61.Meyerson, M., C. M. Counter, E. N. Eaton, L. W. Ellisen, P. Steiner, S. D. Caddle, L. Ziaugra, R. L. Beijersbergen, M. J. Davidoff, Q. Liu, S. Bacchetti, D. A. Haber, and R. A. Weinberg. 1997. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell 90:785-795. [DOI] [PubMed] [Google Scholar]
- 62.Moore, P. S., and Y. Chang. 1995. Detection of herpesvirus-like DNA sequences in Kaposi's sarcoma in patients with and without HIV infection. N. Engl. J. Med. 332:1181-1185. [DOI] [PubMed] [Google Scholar]
- 63.Moustakas, A., and D. Kardassis. 1998. Regulation of the human p21/WAF1/Cip1 promoter in hepatic cells by functional interactions between Sp1 and Smad family members. Proc. Natl. Acad. Sci. USA 95:6733-6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Najafi, S. M., Z. Li, K. Makino, R. Shao, and M. C. Hung. 2003. The adenoviral E1A induces p21WAF1/CIP1 expression in cancer cells. Biochem. Biophys. Res. Commun. 305:1099-1104. [DOI] [PubMed] [Google Scholar]
- 65.Piolot, T., M. Tramier, M. Coppey, J. C. Nicolas, and V. Marechal. 2001. Close but distinct regions of human herpesvirus 8 latency-associated nuclear antigen 1 are responsible for nuclear targeting and binding to human mitotic chromosomes. J. Virol. 75:3948-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Platt, G. M., G. R. Simpson, S. Mittnacht, and T. F. Schulz. 1999. Latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts with RING3, a homolog of the Drosophila female sterile homeotic (fsh) gene. J. Virol. 73:9789-9795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Radkov, S. A., P. Kellam, and C. Boshoff. 2000. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat. Med. 6:1121-1127. [DOI] [PubMed] [Google Scholar]
- 68.Rainbow, L., G. M. Platt, G. R. Simpson, R. Sarid, S. J. Gao, H. Stoiber, C. S. Herrington, P. S. Moore, and T. F. Schulz. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J. Virol. 71:5915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Renne, R., C. Barry, D. Dittmer, N. Compitello, P. O. Brown, and D. Ganem. 2001. Modulation of cellular and viral gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:458-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Robertson, E. S., J. Lin, and E. Kieff. 1996. The amino-terminal domains of Epstein-Barr virus nuclear proteins 3A, 3B, and 3C interact with RBPJ(kappa). J. Virol. 70:3068-3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schwam, D. R., R. L. Luciano, S. S. Mahajan, L. Wong, and A. C. Wilson. 2000. Carboxy terminus of human herpesvirus 8 latency-associated nuclear antigen mediates dimerization, transcriptional repression, and targeting to nuclear bodies. J. Virol. 74:8532-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shay, J. W., and S. Bacchetti. 1997. A survey of telomerase activity in human cancer. Eur. J. Cancer 33:787-791. [DOI] [PubMed] [Google Scholar]
- 73.Shinohara, H., M. Fukushi, M. Higuchi, M. Oie, O. Hoshi, T. Ushiki, J. Hayashi, and M. Fujii. 2002. Chromosome binding site of latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus is essential for persistent episome maintenance and is functionally replaced by histone H1. J. Virol. 76:12917-12924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sontag, E., J. M. Sontag, and A. Garcia. 1997. Protein phosphatase 2A is a critical regulator of protein kinase C zeta signaling targeted by SV40 small t to promote cell growth and NF-kappaB activation. EMBO J. 16:5662-5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 76.Steger, G., C. Schnabel, and H. M. Schmidt. 2002. The hinge region of the human papillomavirus type 8 E2 protein activates the human p21 (WAF1/CIP1) promoter via interaction with Sp1. J. Gen. Virol. 83:503-510. [DOI] [PubMed] [Google Scholar]
- 77.Subramanian, C., and E. S. Robertson. 2002. The metastatic suppressor Nm23-H1 interacts with EBNA3C at sequences located between the glutamine- and proline-rich domains and can cooperate in activation of transcription. J. Virol. 76:8702-8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Takakura, M., S. Kyo, T. Kanaya, H. Hirano, J. Takeda, M. Yutsudo, and M. Inoue. 1999. Cloning of human telomerase catalytic subunit (hTERT) gene promoter and identification of proximal core promoter sequences essential for transcriptional activation in immortalized and cancer cells. Cancer Res. 59:551-557. [PubMed] [Google Scholar]
- 79.Takakura, M., S. Kyo, T. Kanaya, M. Tanaka, and M. Inoue. 1998. Expression of human telomerase subunits and correlation with telomerase activity in cervical cancer. Cancer Res. 58:1558-1561. [PubMed] [Google Scholar]
- 80.Udvadia, A. J., D. J. Templeton, and J. M. Horowitz. 1995. Functional interactions between the retinoblastoma (Rb) protein and Sp-family members: superactivation by Rb requires amino acids necessary for growth suppression. Proc. Natl. Acad. Sci. USA 92:3953-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yieh, L., H. B. Sanchez, and T. F. Osborne. 1995. Domains of transcription factor Sp1 required for synergistic activation with sterol regulatory element binding protein 1 of low density lipoprotein receptor promoter. Proc. Natl. Acad. Sci. USA 92:6102-6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang, Y., X. H. Feng, and R. Derynck. 1998. Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-beta-induced transcription. Nature 394:909-913. [DOI] [PubMed] [Google Scholar]