Abstract
A transient transfection assay using Drosophila S2 tissue culture cells and WT and mutant Drosophila vnd/NK-2 homeobox cDNAs was used to localize repression and activation domains of vnd/NK-2 homeodomain protein. A repression domain was identified near the N terminus of vnd/NK-2 homeodomain protein (amino acid residues 154–193), which contains many hydrophobic amino acid residues. The major determinants of the repression domain were shown to be amino acid residues F155, W158, I161, L162, L163, and W166. Truncated protein consisting of the N-terminal repression domain and the DNA-binding homeodomain repressed transcription as efficiently as WT vnd/NK-2 protein. An activation domain was identified between the tinman domain and the homeodomain (amino acid residues 277–543), which consists of a glutamine-rich subdomain and two acidic subdomains. No effect was detected of the tinman domain or the NK-2-specific domain on either activation or repression of a β-galactosidase reporter gene.
The ventral nerve cord of Drosophila melanogaster is derived from four anterior–posterior stripes of nuclei or cells. The neural pathway of gene expression arises independently and by different mechanisms in each of the four stripes of nuclei or cells that comprise the neurogenic analage. Expression of the ventral nervous system defective homeobox gene (vnd/NK-2) (1–3) initiates the neural pathway of gene expression in the ventrolateral column of nuclei (4); whereas, expression of the intermediate neuroblast defective gene (ind) initiates neural development in the adjacent intermediate column of neuroectodermal nuclei (5, 6). The mechanism of initiating the neural pathway of development in the dorsal column of neuroectodermal cells is unknown; however, the muscle segment homeobox gene (msh) is expressed in many neuroectodermal cells and neuroblasts that comprise the dorsal column of neural cells, and the msh homeodomain (HD) protein is known to be required for specification of some of the neuroblasts in the dorsal column (7, 8). Expression of a fourth gene that encodes a gene regulatory protein, singleminded, initiates neural development in the ventral midline column of mesoectodermal cells (9, 10). During embryonic development the vnd/NK-2 HD protein directly or indirectly activates the proneural achaete gene (4), which encodes a basic helix–loop–helix gene regulator, and represses ind (5) and msh (11, 12) gene expression. vnd/NK-2 HD protein also activates the expression of the NK-6 gene in brain precursors (13). In addition, vnd/NK-2 protein, directly or indirectly, promotes the continued expression of the vnd/NK-2 gene (11, 14).
We have used a transient assay employing Drosophila S2 tissue culture cells and WT and mutant vnd/NK-2 cDNAs to localize the repression and activation domains of vnd/NK-2 HD protein. Understanding the domain organization of the vnd/NK-2 HD protein is an important step in determining the mechanisms of regulating vnd/NK-2 target genes and in clarifying the problem of how different kinds of neuroblasts in the ventral nerve cord are generated along the dorsoventral axis of the embryo.
Regulation of transcription is mediated by proteins that interact with specific DNA sequences located in the promoter or enhancer regions of DNA at various distances from the start of transcription where the basal transcription complex is assembled (15–17). The basal transcription factors together with RNA polymerase II form a complex with DNA before initiation of transcription (preinitiation complex). Cis-acting transcription activators may stimulate transcription by increasing the rate of assembly or stability of the preinitiation complex.
In general, transcription activators and repressors are composed of at least two domains. The DNA binding domain is involved in site-specific DNA recognition, and the activation or repression domains interact directly with components of the general transcription machinery (18–20). There are at least three types of activation domains: acidic (21, 22), glutamine-rich (23, 24), and proline-rich (25). Transcription factors have modular structures in which distinct regions of the protein mediate particular functions such as DNA binding or interactions with other molecules to activate or repress transcription (26–28).
Vnd/NK-2 is a member of the NK-2 class of HD proteins. Most NK-2-class proteins contain three highly conserved regions: the HD, the EH-1 or tinman domain, and the NK-2 box, also termed the NK-2-specific domain (NK-2 SD). Other than these regions of protein, there is no homology between NK-2-class proteins in different species (2). The vnd/NK-2 HD is a highly conserved DNA binding domain that recognizes the consensus nucleotide sequence 5′-TCAAGTGG in DNA with the core sequence 5′-AAGT (29, 30), which is quite different from the 5′-TAAT core sequence that is recognized by antennapedia type HD proteins (31). The short tinman domain is located in the N-terminal region of most NK-2-class proteins. The tinman domains of engrailed (32) and NK-4 (20) have been shown to mediate repression. The NK-2-specific domain is located in the C-terminal part of vnd/NK-2 HD protein and is separated from the HD by a short linker. This domain is 17 aa in length and contains a hydrophobic central region, VAVPV-LVI, and flanking basic amino acid residues. The NK-2 SD of human NKX-2.2 was shown to function as an intramolecular inhibitor of a C-terminal activation domain (19). The C-terminal region of mouse Nkx-2.5 contains an inhibitory domain (33, 34), and a highly charged proline-rich activation domain is present in the N-terminal region of the protein. Apparently, different members of the NK-2 class of HD proteins have different activation or repression domains.
Whereas much is known about the interactions between vnd/NK-2 HD and DNA (29, 30, 35–37), little is known about activation or repression domains of this protein. In this report, we define the locations of repressor and activation domains of vnd/NK-2 HD protein.
Materials and Methods
Plasmid Construction and Site-Directed Mutagenesis. Expression plasmids were constructed by inserting the synthetic double-stranded oligonucleotide 5′-ACAACATGGGTCACCGATACCCCTACGATGTGCCCGATTACGCGGGCG 3′-CATGTGTTGTACCCAGTGGCTATGGGGATGCTACACGGGCTAATGCGCCCGCTTAA between the KpnI and EcoRI sites of pIB/V5-His plasmid (Invitrogen). This oligonucleotide contains a start codon and appropriate flanking nucleotides and encodes part (YPYDVPDYA) of the influenza hemagglutinin (HA) peptide (plasmid pIB/HA).
Expression vectors were constructed by the insertion of WT or mutant vnd/NK-2 cDNA into the EcoRI and XhoI sites of pIB/HA. A reporter gene construct was prepared by cloning a XhoI–HindIII DNA fragment containing an hsp40 minimal promoter (TATA box and start of transcription) and a β-galactosidase (β-gal) cDNA from the P-element, pCaSpeR4, into plasmid pUC18 with the polylinker NheI–SfiI–XhoI–HindIII (pUC/hsp40β-gal reporter gene). A double-stranded oligodeoxynucleotide containing five tandem consensus vnd/NK-2 binding sites separated by short linkers, or part of the vnd/NK-2 promoter region, kilobases –0.41 to –0.75, were cloned into pUC/hsp40β-gal plasmid upstream of the minimal hsp40 promoter (pUC/5NK-2hsp40β-gal or pUC/41-75hsp40β-gal, respectively) as shown in Fig. 1. vnd/NK-2 C- or N-terminal deletions were made by PCR with Drosophila vnd/NK-2 cDNA as the template. All constructs were confirmed by sequencing. SEAP cDNA sequence from plasmid pDNA-SEAP (Clontech) was cloned into plasmid pAC5 (Invitrogen) after the AC5 promoter, and the construct was used as an internal control for transfection efficiency.
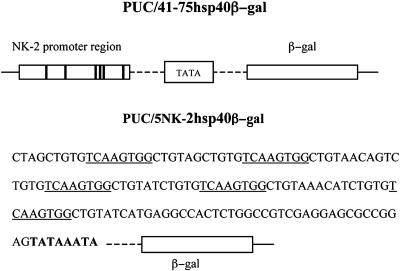
Fig. 1.
Schematic representation of plasmids with reporter genes. Plasmid pUC/41-75hsp40β-gal contains part of vnd/NK-2 genomic DNA between –0.41 and –0.75 kilobases from the start of transcription, a minimal hsp40 promoter with a TATA box, and a β-gal cDNA reporter gene. Positions of six vnd/NK-2 binding sites (two strong and four weak) in the vnd/NK-2 promoter region are shown as bold vertical lines. Plasmid pUC/5NK2hsp40β-gal contains a double-stranded oligonucleotide (sequence shown) with five, tandem vnd/NK-2 binding sites (underlined), a minimal hsp40 promoter with a TATA box, and a β-gal cDNA reporter gene.
Transient Transfection. For transient transfections, Drosophila S2 cells and Spodoptera Sf9 cells, which were cultured in Drosophila SFM or Sf-900 II SFM media with 10% FBS, respectively, were used. Reporter plasmids were transfected into cells alone or cotransfected with an expression vector. Forty-eight hours later, cells were removed from tissue culture dishes, pelleted, and lysed in buffer containing 50 mM Hepes (pH 7.5) and 5 mM CHAPS. Nuclei and cell debris were pelleted at 14,000 × g for 10 min, and the supernatant fraction was used to determine β-gal activity by colorimetric assay. The hydrolysis of the colorless substrate, o-nitrophenyl-β-d-galactopyranoside, is catalyzed by β-gal to produce o-nitrophenol, which adsorbs light at 420 nm with a molar extinction coefficient of 4,600 at basic pH. One unit of β-gal activity corresponds to the amount of β-gal that catalyzed the hydrolysis of 1 μmol of ONPG per min at 37°C. β-gal activity in the sample was calculated as follows: units/sample = (A420 × 0.8)/(4.6 × tmin), where 0.8 is the final reaction volume (ml) and tmin is minutes of incubation. All transcription experiments were repeated at least three times.
Nuclei Extract Preparation and Western Blot Analysis. The stabilities of WT mutant proteins in nuclei after cell transfection were assayed by Western blot. Nuclei were suspended in buffer containing 50 mM Hepes (pH 7.5), 100 mM NaCl, 0.5% Nonidet P-40, and protease and phosphatase inhibitors and sonicated, and equal amounts of total protein were applied to SDS gels. Proteins were subjected to electrophoresis and then transferred to nitrocellulose membranes and visualized by mouse anti-HA antibody/anti-mouse HRP antibody/ECL methods.
Results
Determination of vnd/NK-2 Repression Domains. To assess the ability of vnd/NK-2 to function as a transcription factor, we constructed a set of plasmids containing WT or mutant vnd/NK-2 cDNA, and these constructs were tested with reporter gene plasmids pUC/hsp40β-gal, pUC/5NK-2hsp40β-gal, or pUC/41–75-hsp40β-gal. In Fig. 2 are shown the results of cotransfection experiments of Drosophila S2 cells, using pUC/hsp40β-gal and pUC/41–75hsp40β-gal DNA. Previously, the vnd/NK-2 promoter region between kilobases –0.41 and –0.75 (from the start of transcription) was shown to strongly activate the transcription of a reporter gene in S2 cells (X. Shao and M.N., unpublished data). This region contains binding sites for different Drosophila transcription factors and two strong and four weak vnd/NK-2 binding sites. The control reporter construct, pUC/hsp40β-gal, does not contain vnd/NK-2 binding sites upstream of the minimal hsp40 promoter, and S2 cells transfected with this reporter have a relatively low basal level of β-gal activity. When the –0.41 to –0.75 vnd/NK-2 DNA region was inserted upstream of the hsp40 minimal promoter the activity of β-gal increased 46-fold in S2 cells (100% in Fig. 2) but only 1.6-fold in Sf9 cells. This finding suggests that S2 cells contain one or more transcription factors that activate β-gal transcription and that Sf9 cells do not express these factors and/or express factors that repress transcription of the reporter gene. Therefore, S2 cells were used for all further experiments. Cotransfection of S2 cells with pUC/41-75β-gal plus pIB/HAwtNK-2 resulted in a 26-fold decrease in β-gal activity, presumably because of binding of the WT vnd/NK-2 HD protein to its recognition sequences in the –0.41 to –0.75 vnd/NK-2 DNA fragment. The main DNA recognition element of the NK-2 family of proteins is the third helix of the HD, and a major determinant of DNA binding and recognition is Asn-595 of vnd/NK-2 protein (Asn-51 of the vnd/NK-2 HD) (37). Substitutions of Asn-51 have been shown to abolish the binding of some other HD proteins to DNA (38). Converting vnd/NK-2 Asn-595 to Ala resulted in the restoration of β-gal activity (Fig. 2, N595A mutant), indicating that binding of the vnd/NK-2 HD to its binding sites in DNA is required for repression of β-gal gene expression. Removal of the first 97 N-terminal amino acid residues, Δ(1–97), had no effect; however, deletion of the 194 N-terminal amino acids, Δ(1–194), resulted in a loss of ability to repress β-gal transcription and an increase in β-gal activity to 140%. Deletion of the 277 N-terminal amino acid residues resulted in a 2.2-fold increase in β-gal transcription compared to the control (transfection of cells only with the reporter gene plasmid, pUC/41-75hsp40β-gal). Deletion of the 392 N-terminal amino acid residues, Δ(1–392), decreased activation of β-gal transcription slightly by the mutant vnd/NK-2 protein. The vnd/NK-2 HD with some flanking amino acid residues had little or no effect on the expression of the reporter gene.
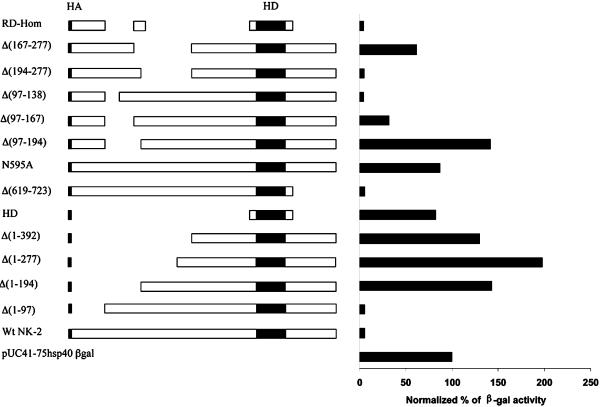
Fig. 2.
β-gal activity of Drosophila S2 cells transfected with pUC/41–75hsp40β-gal DNA alone or cotransfected with pIB/HANK-2 containing WT or deletion mutants of vnd/NK-2. AC5SEAP plasmid DNA was used as an internal control for transcription efficiency (see Materials and Methods). Transcription activity of the –0.41 to –0.75 vnd/NK-2 promoter (pUC/41-75hsp40β-gal vector) in transfected S2 cells corresponds to 100%. HD (black box) corresponds to the HD of vnd/NK-2. N591A represents a mutant with N591 replaced by A in the third helix of the HD. The small black box at the beginning of each construct, labeled HA, corresponds to the influenza hemagglutin peptide. The repression domain (RD)-Hom construct contains a small N-terminal part of vnd/NK-2 (amino acids 1–97), a repression domain (amino acids 154–193), and a HD.
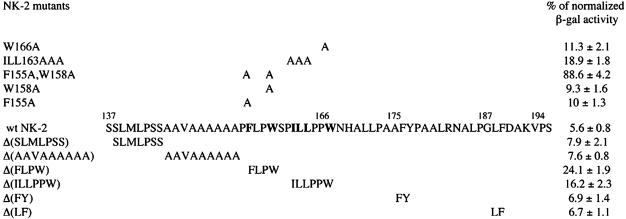
As shown in Fig. 2, deletion of the C-terminal part of vnd/NK-2 protein, Δ(619–723), had no effect on the ability of the vnd/NK-2 mutant protein to repress β-gal expression. More detailed mapping showed that the repression domain is located between amino acid residues 138 and 193 (Fig. 3). This region is very hydrophobic and contains alanine- and proline-rich sequences. One hundred percent corresponds to β-gal activity in cells transfected with the reporter gene construct only (pUC/41-75hsp40β-gal). β-gal activity in cells cotransfected with this reporter and pIB/HAwtNK-2 DNA was 5.6%. Deletion of the six amino acids S(138)LMLPSS(144) or hydrophobic amino acids A(145)AVAAAAAA(153) had no effect on vnd/NK-2 repression activity, whereas deletion of the four amino acids F(155)LPW(158) or six amino acids I(161)LLPPW(166) decreased NK-2 repressive activity 4.3- and 3.0-fold, respectively. Deletion of the hydrophobic amino acids F(175)Y(176) or amino acids L(187)F(188) had no effect on repression. Similar experiments with amino acids substitution in this region shows that amino acids F155 and W158, together with I(161)LL(163) and W166, are the major determinants of NK-2 repressive activity. W158A substitution leads only to a 1.7-fold decrease of NK-2 activity and 1.8-fold change for F155A substitution. The double mutant F155A, W158A has only 6.3% of wtNK-2 repressive activity. Nevertheless, these six amino acid residues do not completely determine the ability of vnd/NK-2 protein to repress transcription. As shown in Fig. 2, vnd/NK-2 mutants Δ(97–167) and Δ(167–277) retained some ability to repress the β-gal reporter gene. Thus, only two domains are necessary and sufficient for repression of β-gal expression: RD amino acid residues 154–193 and the HD. In our experiments, the repression domain was linked to the N-terminal region of NK-2 (RD-Hom mutant), because the region between amino acids 154 and 194 is extremely hydrophobic, and proteins containing homeo and repressive domains only are insoluble. As shown in Fig. 2, the first N-terminal 97 amino acids are not sufficient for NK-2 repressive activity.
Fig. 3.
β-gal activity of S2 cells cotransfected with pUC/41-75hsp40β-gal and pIB/HANK-2 DNAs encoding either WT or mutant vnd/NK-2 cDNAs. Activity of β-gal with the –0.41 to –0.75 vnd/NK-2 promoter in S2 cells corresponds to 100%. On the right is β-gal activity in cotransfection experiments with WT or mutant vnd/NK-2 cDNAs. Amino acid substitutions are shown in the top part of the figure, and positions of amino acid residues deleted are indicated in the bottom part of the figure.
Mapping the vnd/NK-2 Activation Domain. As shown in Fig. 2, vnd/NK-2 mutants Δ(1–194) and Δ(1–277) have completely lost the ability to repress β-gal expression but also have acquired the ability to activate transcription of β-gal. To identify potential transcription activation domains within the vnd/NK-2 protein, additional experiments were performed by using the pUC/5NK-2hsp40β-gal reporter construct because of the low basal β-gal activity found in S2 cells transfected with this construct (1.0% shown in Fig. 4).
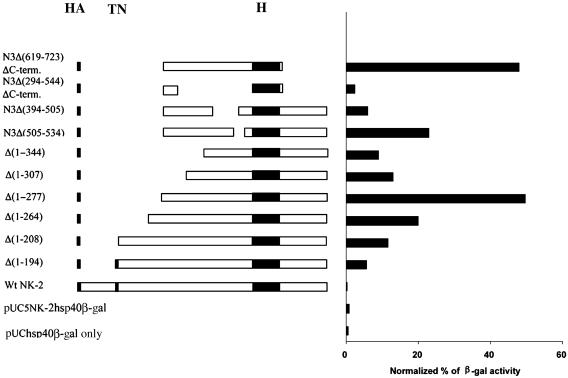
Fig. 4.
Analysis of the position of the vnd/NK-2 activation domain. S2 cells were transfected with pUC/5NK2hsp40β-gal DNA (which contains five tandem vnd/NK-2 binding sites) alone or cotransfected pIB/HANK-2 containing WT or mutant NK-2 cDNA. The activity of β-gal in cells transfected with pUC/5NK-2hsp40β-gal reporter gene corresponds to 1.0%. Activities of the same reporter gene cotransfected with mutant vnd/NK-2 constructs are shown in Right.N3 corresponds to the Δ(1–277) mutant that has maximal activation potential. This construct was used for mapping the position of the activation domain. PUC/hsp40β-gal is a reporter plasmid containing the hsp40 minimal promoter only. β-gal activity was normalized as described in Materials and Methods. The level of β-gal expression in cells transfected with pUC/hsp40βgal was the same as that found with pUC/5NK-2hsp40β-gal DNA.
As shown in Fig. 4, β-gal activity in S2 cells cotransfected with pIB/HAwtNK-2 and pUC/5NK-2hsp40β-gal DNA was reduced to 0.034%. Deletion of the first 194 amino acids from the N terminus, Δ(1–194), resulted in a 6-fold increase in transcription activity. Sequential removal of N-terminal amino acid residues up to 277 monotonically increased the transcription activity of the vnd/NK-2 mutants. The vnd/NK-2 Δ(1–277) mutant maximally activated β-gal expression 48-fold compared with the control value. Deletion of any amino acid residues tested in the region between amino acids 294 and 536 decreased the ability of vnd/NK-2 protein to activate β-gal expression. Deletion of the repression domain and the C-terminal region after the HD Δ(1–294), Δ(619–723) also resulted in a 48-fold activation of β-gal; hence, deletion of the C-terminal region of vnd/NK-2 protein had no effect on the activation of the β-gal gene. In addition, the results show that the entire 277–536 amino acid residue region is necessary for maximum transcription activation.
Discussion
vnd/NK-2 Inhibitory Domain. The inhibitory domains of a number of transcription factors inhibit either the assembly of the basal transcription complex or reduce the activity and/or stability after it has assembled (direct inhibition). For example, the inhibitory domain of the thyroid hormone receptor has been shown to interact directly with TFIIB (39). Drosophila transcription factor Kruppel binds to DNA as a monomer and activates transcription by interaction with TFIIB but, at a high concentration, Kruppel forms dimers and inhibits transcription by interaction with another component of the basal transcription complex, TFIIEβ. We studied the transcription activity of Drosophila vnd/NK-2 HD protein by assaying transient expression of the β-gal gene in transfected S2 cells. The results show that the vnd/NK-2 protein in its native state is a strong transcription repressor and that the repression domain is located in the N-terminal region of vnd/NK-2 protein between amino acid residues 154 and 193. As shown in Fig. 2, this region is necessary and sufficient for all of the repression activity of vnd/NK-2 protein. The N-terminal portion of the vnd/NK-2 repression domain is proline- and alanine-rich, 154-PFLPWSPILLPPWNHALLPAAFYPAAL-180. Several amino acid residues such as F155, W158, 161-ILL-163, and W158 were shown to be major determinants of the repression effect. Nevertheless, for full repression activity, the C-terminal part of the inhibitor y domain, 181-RNALPGLFDAKVP-193, is required. Inhibitory domains of several Drosophila transcription factors including eve (40) contain proline-rich domains. However, other inhibitory domains from Oct-2 (41) and E4BP4 (42) differ from each other and from proline-rich domains. Hence, there are several types of inhibitory domains. We assume that the inhibitory domain of vnd/NK-2 directly represses transcription by interaction with the basal transcription complex.
vnd/NK-2 Activation Domain. There are at least three classes of activation domains: acidic, glutamine-rich, and proline-rich. The activation domain of vnd/NK-2 consists of three regions. The first region, located between amino acid residues 293 and 334, contains repeats like QQQH or HQQ. The second region (amino acid residues 401–458) and the third region (amino acid residues 498–539) are rich in acidic amino acid residues (Fig. 5). Similar glutamine-rich regions have been found in the activation domains of Sp1 and Antp (43). The activation potential of a glutamine-rich domain is not defined by the primary amino acid sequence but rather by the high glutamine content of the domain. When this region was deleted, the activation potential of the Δ(1–344) vnd/NK-2 mutant protein decreased 5-fold compared with the Δ(1–277) mutant. An acidic region, another type of activation domain, is found in several transcription factors including the yeast factor GCN4, GAL4, glucocorticoid receptor, and VP16 (28). Although these proteins do not show any amino acid sequence homology to each other, each has a large proportion of acidic amino acids producing a strong negative charge. The 82-aa activation region of the glucocorticoid receptor contains 17 acidic residues. Increasing the negative charge of an acidic domain is correlated with its ability to activate transcription. Deletion of the second acidic domain Δ(505–534) together with Δ(1–294) resulted in a 2-fold decrease in activation of β-gal. Deletion of the first acidic region Δ(394–505) together with Δ(1–294) resulted in a 10-fold reduction in vnd/NK-2 activation of β-gal gene expression. Additional studies showed that the mutant protein N3Δ(294–544)Δ(C-term), lacking part of the glutamine-rich region (amino acid residues 294–330) and two acidic domains, had little ability to activate the β-gal gene (5% of the maximal value).
Fig. 5.
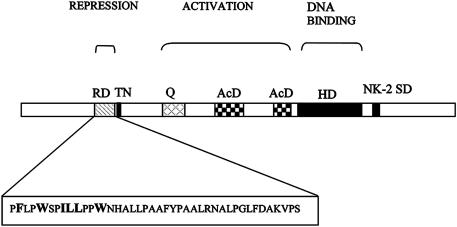
Schematic representation of the domains of vnd/NK-2 protein. Conserved vnd/NK-2 domains: tinman (TN) (amino acid residues 198–209); HD (545–604); and NK-2-specific domain (NK-2 SD) (631–647) are shown in black. The HD binds to DNA and recognizes nucleotide residues. The functions of TN and NK-2 SD are unknown. The vnd/NK-2 repression domain (154–193) is located just before the TN domain. The amino acid sequence of the repression domain is shown below. Amino acid residues F155, W158, I161 L162, L163, and W166 (bold) are major determinants required for repression. The vnd/NK-2 activation domain consists of a glutamine-rich (Q) subdomain (293–334) and two acidic subdomains (AcD) (401–458 and 509–528).
In the case of VP16, the negative charge of its acidic domain allows it to establish an electrostatic interaction with TAFII31, a component of TBP. Long-distance electrostatic interaction with TAFII31 induces a conformational change in the VP16 domain to an α-helical structure in which TAFII31, the residues aspartic acid (D472), phenylalanine (F479), and leucine (L481) are brought close to one another and bind to TAFII31 (22). There are clear differences in the abilities of the different activation domains to activate transcription. When the DNA binding site was placed close to the transcription initiation site, all three domains (proline-rich, glutamine-rich, and acidic) were able to activate transcription by enhancing the assembly of the basal transcription complex. In contrast, the glutamine-rich domain was unable to activate transcription when the binding site was distantly located (within the enhancer element) from the start of transcription. At increasing distances the activation domains are needed to recruit the other transcriptional factors. The acidic domain was strongly active from the enhancer position, whereas the proline-rich domain could also activate transcription from this position but only weakly (44, 45). Target factor(s) with which acidic domain interact are likely to be highly conserved in evolution.
In many cases, these factors are components of the basal transcription complex. We assume that inhibitory and activation domains of vnd/NK-2 HD protein directly repress or activate transcription by interaction with components of the basal transcription complex.
Acknowledgments
We thank Alessandra Rovescalli and James A. Ferretti for insightful discussions and Kathleen Blair for editorial assistance.
Abbreviations: β-gal, β-galactosidase; HA, hemagglutinin; HD, homeodomain; RD, repression domain.
References
- 1.Jimenez, F., Martin-Morris, L. E., Velaco, L., Chu, H., Sierra, J., Rosen, D. R. & White, K. (1995) EMBO J. 14, 3487–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim, Y. & Nirenberg, M. (1989) Proc. Natl. Acad. Sci. USA 86, 7716–7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nirenberg, M., Nakayama, K., Nakayama, N., Kim, Y., Mellerick, D., Wang, L.-H., Webber, K. O. & Lad, R. (1995) Ann. N.Y. Acad. Sci. 758, 224–242. [DOI] [PubMed] [Google Scholar]
- 4.Skeath, J. B., Panganiban, G. F. & Carrol, S. B. (1994) Development (Cambridge, U.K.) 120, 1517–1524. [DOI] [PubMed] [Google Scholar]
- 5.Weiss, J. B., Ohlen, T. V., Mellerick, D. M., Dressler, G., Doe, C. Q. & Scott, M. P. (1998) Genes Dev. 12, 3591–3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McDonald, J. A., Holbrook, S., Isshiki, T., Weiss, J., Doe, C. Q. & Mellerick, D. M. (1998) Genes Dev. 12, 3603–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D'Alessio, M. & Frasch, M. (1996) Mech. Dev. 58, 217–231. [DOI] [PubMed] [Google Scholar]
- 8.Isshiki, T., Takeichi, M. & Nose, A. (1997) Development (Cambridge, U.K.) 124, 3099–3109. [DOI] [PubMed] [Google Scholar]
- 9.Thomas, J. B., Crews, S. T. & Goodman, C. S. (1998) Cell 52, 133–141. [DOI] [PubMed] [Google Scholar]
- 10.Nambu, J. R., Franks, R. G., Hu, S. & Crews, S. T. (1990) Cell 63, 63–75. [DOI] [PubMed] [Google Scholar]
- 11.Chu, H., Parras, C., White, K. & Jimenez, F. (1998) Genes Dev. 12, 3613–3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Von Ohlen, T. & Doe, C. Q. (2000) Dev. Biol. 224, 362–372. [DOI] [PubMed] [Google Scholar]
- 13.Uhler, J., Carbern, J., Yang, L., Kamholz, J. & Mellerick, D. M. (2002) Mech. Dev. 116, 105–116. [DOI] [PubMed] [Google Scholar]
- 14.Saunders, H.-M. H., Koizumi, K., Odenwald, W. & Nirenberg, M. (1998) Proc. Natl. Acad. Sci. USA 95, 8316–8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitchell, P. J. & Tjian, R. (1989) Science 245, 371–378. [DOI] [PubMed] [Google Scholar]
- 16.Ptashne, M. & Gann, A. A. F. (1990) Nature 346, 329–331. [DOI] [PubMed] [Google Scholar]
- 17.Lin, Y. S. & Green, M. R. (1991) Cell 64, 971–981. [DOI] [PubMed] [Google Scholar]
- 18.DeFelice, M., Damante, G., Zannini, M., Francis-Lang, H. & DiLauro, R. (1995) J. Biol. Chem. 270, 26649–26656. [DOI] [PubMed] [Google Scholar]
- 19.Watada, H., Mirmira, R. G., Kalamaras, J. & German, M. S. (2000) Proc. Natl. Acad. Sci. USA 97, 9443–9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi, C. Y., Lee, Y. M., Kim, Y. H., Park, T., Jeon, B. H., Schultz, R. A. & Kim, Y. (1999) J. Biol. Chem. 274, 31543–31552. [DOI] [PubMed] [Google Scholar]
- 21.Ma, J. & Ptashne, M. (1987) Cell 48, 847–853. [DOI] [PubMed] [Google Scholar]
- 22.Cress, W. D. & Triezenberg, S. J. (1991) Science 251, 87–90. [DOI] [PubMed] [Google Scholar]
- 23.Courey, A. J. & Tjian, R. (1988) Cell 55, 887–898. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka, M. & Herr, W. (1990) Cell 60, 375–386. [DOI] [PubMed] [Google Scholar]
- 25.Mermod, N., O'Neill, E. A., Kelly, T. J. & Tjian, R. (1989) Cell 58, 741–753. [DOI] [PubMed] [Google Scholar]
- 26.Hope, I. A. & Struhl, K. (1986) Cell 46, 885–894. [DOI] [PubMed] [Google Scholar]
- 27.Hollenberg, S. M. & Evans, R. M. (1988) Cell 55, 899–906. [DOI] [PubMed] [Google Scholar]
- 28.Hans, S. (1993) Cell 72, 481–483.8440015 [Google Scholar]
- 29.Weiler, S., Gruschus, J. M., Tsao, D. H. H., Yu, L., Wang, L. H., Nirenberg, M. & Ferretti, J. A. (1998) J. Biol. Chem. 273, 10994–11000. [DOI] [PubMed] [Google Scholar]
- 30.Gruschus, J. M., Tsao, D. H., Wang, L. H., Nirenberg, M. & Ferretti, J. A. (1999) J. Mol. Biol. 289, 529–545. [DOI] [PubMed] [Google Scholar]
- 31.Wey, E. & Schafer, B. W. (1996) Biochem. Biophys. Res. Commun. 220, 274–279. [DOI] [PubMed] [Google Scholar]
- 32.Smith, S. T. & Jaynes, J. B. (1996) Development (Cambridge, U.K.) 122, 3141–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen, C. Y. & Schwartz, R. J. (1995) J. Biol. Chem. 270, 15628–15633. [DOI] [PubMed] [Google Scholar]
- 34.Sepulveda, J. L., Belaguli, N., Nigam, V., Chen, C. Y., Nemer, M. & Schwartz, R. J. (1998) Mol. Cell. Biol. 18, 3405–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiang, B., Weiler, S., Nirenberg, M. & Ferretti, J. A. (1998) Proc. Natl. Acad. Sci. USA 95, 7412–7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Desiree, H. H., Gruschus, J. M., Wang, L. H., Nirenberg, M. & Ferretti, J. A. (1994) Biochemistry 33, 15053–15060. [DOI] [PubMed] [Google Scholar]
- 37.Wang, L. H., Chmelik, R. & Nirenberg, M. (2002) Proc. Natl. Acad. Sci. USA 99, 12721–12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verrijzer, C. P. & Van der Vliet, P. C. (1993) Biochim. Biophys. Acta 1173, 1–21. [DOI] [PubMed] [Google Scholar]
- 39.Baniahmad, A., Ha, I., Reinberg, D., Tsai, M. J. & O'Malley, B. W. (1993) Proc. Natl. Acad. Sci. USA 90, 8832–8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han, K. & Manley, J. L. (1993) Genes Dev. 7, 491–503. [DOI] [PubMed] [Google Scholar]
- 41.Lillycrop, K. A., Dawson, S. J., Estridge, J. K., Gerster, T., Matthias, P. & Latchman, D. S. (1994) Mol. Cell. Biol. 14, 7633–7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cowell, I. G. & Hurst, H. C. (1994) Nucleic Acids Res. 22, 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lania, L., Majello, B. & de Luca, P. (1997) Int. J. Biochem. Cell Biol. 29, 1313–1323. [DOI] [PubMed] [Google Scholar]
- 44.Ranish, J. A. & Hahn, S. (1996) Curr. Opin. Genet. Dev. 6, 151–158. [DOI] [PubMed] [Google Scholar]
- 45.Stargell, L. A. & Struhl, K. (1996) Trends Genet. 12, 311–315. [DOI] [PubMed] [Google Scholar]