Abstract
The related protein kinases SPAK and OSR1 regulate ion homeostasis in part by phosphorylating cation cotransporter family members. The structure of the kinase domain of OSR1 was solved in the unphosphorylated inactive form, and like some other Ste20 kinases, exhibited a domain-swapped activation loop. To further probe the role of domain swapping in SPAK/OSR1, we have determined the crystal structures of SPAK 63–403 at 3.1 Å and SPAK 63–390 T243D at 2.5 Å resolutions. These structures encompass the kinase domain and different portions of the C-terminal tail, the longer without, and the shorter with an activating point mutation T243D. The structure of the T243D protein reveals significant conformational differences relative to unphosphorylated SPAK and OSR1, but also has some features of an inactive kinase. Both structures are domain-swapped dimers. Sequences involved in domain swapping were identified and mutated to create a SPAK monomeric mutant with kinase activity, indicating that monomeric forms are active. The monomeric mutant is activated by WNK1, but has reduced activity toward its substrate NKCC2, suggesting regulatory roles for domain swapping. The structure of the partially active SPAK T243D is consistent with a multi-stage activation process in which phosphorylation induces a SPAK conformation that requires further remodeling to build the active structure.
In animals, ion concentrations and cell volume are controlled to maintain blood pressure, hearing, neurotransmission, fluid secretion, and to preserve cell viability for all other physiological functions (1–3). The Ste20-related proline-alanine-rich kinase (SPAK; also called PASK and STK39) and its close relative oxidative stress-responsive kinase 1 (OSR1) directly contribute to regulation of ion balance through phosphorylation of the cytoplasmic tails of the SLC12 family of Na+-Cl−, Na+-K+-2Cl−, and K+-Cl− ion cotransporters (NCC, NKCCs 1 and 2, and KCCs 1–4) (4–8). Reflecting the close connection between salt flux and blood pressure, mutations in the gene encoding SPAK have been linked to increased susceptibility to hypertension (9). Additionally, kinase-dead SPAK knock-in mice are hypotensive, and show reduced activation and expression of NKCC2 and NCC in the kidney, whereas OSR1 knock-outs die during embryonic development with blood vessel defects (7;10–12). These results suggest that therapies specifically targeting SPAK or OSR1 have the potential to treat hypertension, which is estimated to cost over $130 billion annually worldwide (5;13;14).
SPAK and OSR1 are members of the Ste20 germinal center kinase (GCK)-VI subfamily of protein kinases (6;15). They share a common structural organization with the kinase domain near the N-terminus. The C-terminus contains two conserved domains only found in this subfamily, initially referred to as PF1 and PF2 (PF stands for PASK and Fray, the Drosophila homolog) (16). The PF1 domain is required for kinase activity and may fold with the kinase domain. The PF2 domain, also referred to as the conserved C-terminal (CCT) domain, is involved in protein-protein interactions, and binds a short consensus motif [RFX(V/I)] found in SCL12 family cotransporter substrates, as well as in upstream regulators, the WNK (with no lysine (K)) kinases (17–21). Structural studies illuminated the basis for this interaction (22).
Mutations in the upstream WNK1 and WNK4 kinases were shown by positional cloning to be responsible for single gene forms of inherited hypertension, pseudohypoaldosteronism type II (23). WNKs are activated by hypotonic, low Cl−, and hyperosmotic conditions, due at least in part to a direct Cl− sensing mechanism involving the WNK1 kinase domain (24). SPAK and OSR1 are activated by all four WNK kinases by phosphorylation at two conserved sites, T243 and S383 (mouse SPAK numbering) (18;20;25). Threonine 243 is located within the activation loop of the kinase domain and serine 383 is located in the PF1 domain and has been suggested to be part of an auto-inhibitory element, but its precise function remains unclear (26).
Previous crystal structures of the unphosphorylated OSR1 kinase domain have been solved (27;28). The structures revealed OSR1 activation loop domain-swapped dimers, where the active sites of both kinases in the dimer are formed with residues from both monomers (29). The structures of the inactive form of the OSR1 kinase domain left open many questions: What is the activation mechanism of the kinase? How does the PF1 domain, known to be important for kinase activity and regulation, fold with the kinase domain? What is the functional significance of dimerization?
To gain insight into these questions we have solved the structures of SPAK 63–403:ATP and the phosphomimetic mutant SPAK 63–390 T243D:AMP-PNP, which mimics the phosphorylation of the activation loop by WNKs. The structures encompass the kinase domain and part of the C-terminal regulatory region. Both structures reveal domain-swapped dimers with the N-terminal part of the PF1 domain folded onto the backside of the kinase domain. The new structural data offer insights into the function of domain-swapping and the function of the PF1 domain in stabilizing the kinase structure.
Experimental Procedures
Cloning
DNA encoding mouse SPAK was amplified from cDNA clone 6843981 (ATCC) and ligated into pHisParallel (30). Mutants were created using QuikChange (Stratagene) (20).
Protein expression and purification
SPAK 63–390 T243D was expressed in Rosetta (DE3) E. coli (Novagen). Cells were grown at 37 °C until OD600 reached 0.4–0.6 and then induced with 0.5 mM IPTG overnight at 16 °C. Cells were lysed in 1 mg/ml lysozyme in 50 mM HEPES, pH 8.0, 0.3 M NaCl, 10% glycerol, and protease inhibitors (1 mM phenylmethanesulfonyl fluoride, 10 mM benzamidine, 0.2 µg/ml leupeptin, 2 µg/ml aprotinin), on ice for 30 min followed by sonication. Soluble proteins were applied to nickel-nitrilotriacetic acid-agarose (Ni2+-NTA) and SPAK was eluted with 250 mM imidazole. Fractions with SPAK were identified by polyacrylamide gel electrophoresis in sodium dodecyl sulfate (SDS-PAGE), and dialyzed against 50 mM HEPES, pH 7.5, and 50 mM NaCl at 4°C overnight. The tag was cleaved with TEV protease at a ratio of 50:1 (protein: protease), and removed by re-chromatography on Ni2+-NTA agarose. SPAK was further purified on Mono S using a 0.05–1 M NaCl gradient in 50 mM HEPES, pH 7.5, 1 mM dithiothreitol (DTT), and 1 mM EDTA and on Superdex75 equilibrated in 50 mM HEPES, pH 7.0, 50 mM NaCl, 1 mM DTT, and 1 mM EDTA. Similar procedures were effective for all SPAK 63–390 mutants evaluated.
SPAK 63–403 was purified similarly. The lysis buffer was as above except that glycerol was increased to 15% and included 2.5 mM β-mercaptoethanol. The dialysis buffer contained 20 mM Hepes, pH 7.5, 0.1 M NaCl and 5% glycerol. Before gel filtration, the protein was further purified on Mono Q eluted with a gradient of 0.1–0.5 M NaCl in dialysis buffer also containing 1 mM DTT.
Crystallization
SPAK 63–390 T243D,~20 mg/ml, was pre-incubated with 5 mM AMP-PNP, 5 mM MgCl2 for 30 min on ice. Crystallization (from Hampton Screening Kit I) was carried out at 20°C in hanging drops by mixing protein with an equal volume of well solution, 0.2 M Mg (OAc)2, 0.1 M Tris, pH 8.5, 16% PEG3350 (Hampton), 0.01 M Na(CH3) 2AsO2 (Hampton), and 0.01 M CaCl2. Blimp-shaped rods 0.4 mm long with a maximum crossection of 0.05 mm were flash frozen in liquid nitrogen after sequentially soaking in well solution with an additional 5, 10, and 15% glycerol as cryoprotectant.
SPAK 63–403 at 15 mg/ml was pre-incubated with 5 mM ATP, 5 mM MgCl2 for 30 min on ice. Crystallization was carried out at 20° C in hanging drops by mixing protein with an equal volume of well solution, 100 mM MES, pH 5.5, 150 mM ammonium sulfate, and 18% PEG4000. Rectangular plates 0.2 × 0.1 mm long with a maximum cross-section of 10–15 µm were flash frozen in liquid nitrogen after soaking in cryoprotectant, 20% ethylene glycol, 22% PEG4000, 100 mM MES, pH 5.5, 150 mM ammonium sulfate, 100 mM NaCl, 5 mM MgCl2, 20 mM HEPES, pH 7.5, 5% glycerol.
Data collection and structure determination
Diffraction data were collected at beamline 19-ID at the Advanced Photon Source (Argonne National Laboratory, Argonne, IL). Data were indexed, integrated, and scaled in HKL2000 (31). The structure of SPAK 63–390 T243D was determined by molecular replacement in PHASER in the CCP4 suite (32;33). The p21-activated kinase PAK6 (pdb code 2c30) provided the best search model. The structure contains two molecules in the asymmetric unit (the solvent content was about 50% and VM=2.51 Å3/Da). Model building was carried out in Coot and refinement was performed with REFMAC5 in the CCP4 suite (34;35). As(CH3)2 was covalently linked to a subpopulation of SPAK T243D cysteine residues, as a result of the crystallization conditions. TLS (translation/libration/screw) refinement was carried out in TLSMD (36). The structure was checked on the MolProbity web server (37); figures were made in PyMOL (The PyMOL Molecular Graphics System, Schrödinger, LLC).
The structure of SPAK 63–403 was solved using molecular replacement in PHASER in Phenix (32;33;38). The refined structure of SPAK 63–390 T243D chain A N-terminal domain excluding helix αC and C-terminal domain excluding the activation loop were used as search models. The structure contains two SPAK molecules per asymmetric unit. Manual rigid body docking of the SPAK 63–390 T243D chain A activation loop and helix αC excluded from the molecular replacement search models was followed by iterative rounds of refinement (TLS, rigid body, and individual ADPs) in phenix.refine and model building in Coot (34;38;39). Secondary structure restraints were used until the final rounds of refinement. Model validation was carried out in phenix.refine, which uses analyses derived, in part, from the MolProbity web server (37–39). Dimer interface surface areas were calculated by directly inputting the structures into the PISA server (http://http://www.ebi.ac.uk/pdbe/pisa), and using the A to B ranges with symmetry operator x,y,z (40).
Kinase assays
0.5–1 µg of SPAK proteins were added to a 30 µl kinase reaction containing 20 mM Hepes, pH 7.6, 50 µM ATP (10 µCi [γ-32P]ATP), 10 mM MgCl2, 10 mM β-glycerophosphate, 1 mM DTT, and 1 mM benzamidine, with 5 µg of either GST-PAK1 1–230 or GST-NKCC2 1–175 as substrate for a 30-min reaction at 30°C. For the coupled assays using WNK1 pre-activated SPAK, SPAK was first phosphorylated by WNK1 kinase domain (residues 132–483) by incubation of 1 µg of SPAK with 2.5 µg of WNK1 in a 20 µl kinase reaction containing 1 mM DTT, 20 mM HEPES pH 7.6, 50 µM ATP, 75 mM NaCl, 5% w/v glycerol, 15 mM MgCl2 for 30 min at 25°C. Then 5 µl of a substrate mixture containing 2 µg GST-NKCC2 (residues 1–175), 250 µM ATP (10 µCi [γ-32P]ATP), and 75 mM MgCl2 was added to the reaction and incubated for 5 min at 25°C. Samples were analyzed by SDS-PAGE. Gels were stained with Coomassie blue, dried, and exposed to film.
In vitro binding assay
Equimolar amounts of GST, GST-SPAK 63–390 and GST-OSR 1–557 purified from bacteria were incubated with His6-SPAK 63–390 also purified from bacteria in buffer containing 50 mM Tris (pH 7.5), 100 mM NaCl, 5 mM MgCl2 with or without 1 mM ATP or ADP. Samples were washed extensively, separated by SDS-PAGE, and analyzed by immunoblotting using anti-His6 antibody.
Gel filtration chromatography
Gel filtration chromatography of 250 µM SPAK mutants was on a Superdex 75 10/300 GL column in 50 mM HEPES (pH 7.0), 50 mM NaCl, 1 mM DTT, and 1 mM EDTA.
Results
SPAK Crystallization and Structure Determination
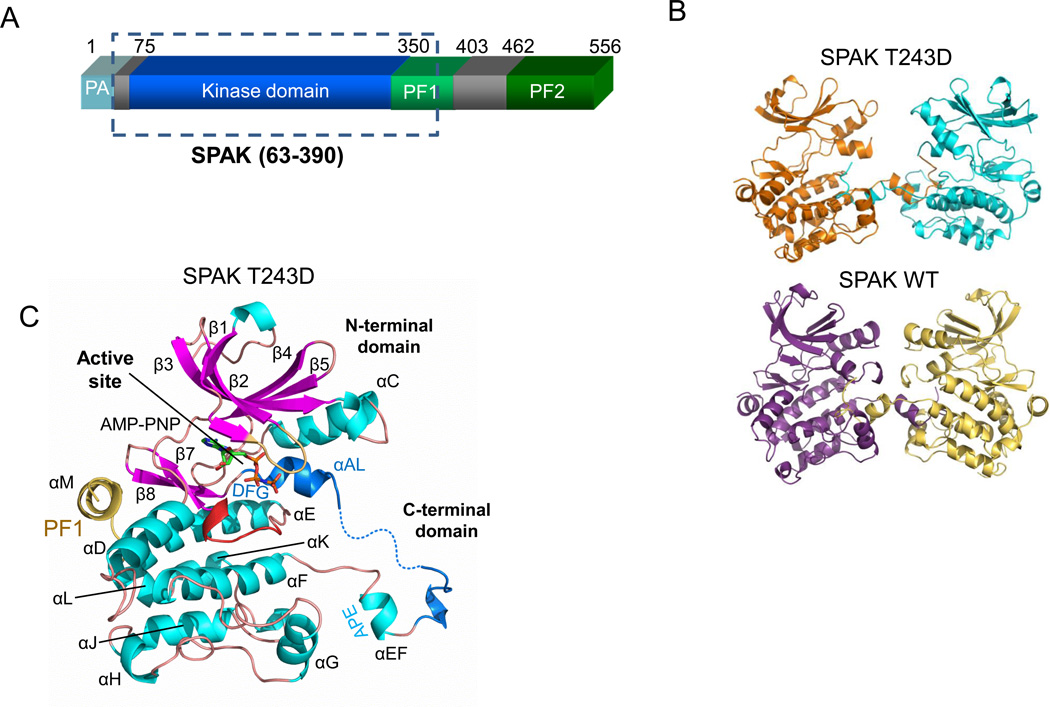
Extensive bacterial expression and crystallization screening of the SPAK kinase domain together with both N and C-terminal extensions led to crystals of SPAK 63–403 and SPAK 63–390 T243D. The constructs crystallized encompass the kinase domain (residues 75–350), an N-terminal extension (residues 63–74), and residues 351–390 or 351–403 from the PF1 homology box (residues 351–403) (Fig. 1A) (16;41). SPAK 63–403 will be referred to throughout as SPAK WT and SPAK 63–390 T243D will be referred to as SPAK T243D. The previously reported structure of wild type OSR1, residues 1–295, used for comparison in this study (PDB code 3DAK) will be referred to as OSR1 WT (28).
Figure 1. Domain organization and structure of SPAK WT and T243D.
(A) Domain organization of mouse SPAK. The kinase domain (residue 75–350) is shown in blue, the PF1 domain (residues 350–403) in green, and the PF2 domain (residue 462–556) in darker green. The SPAK T243D fragment crystallized is boxed. (B) Activation loop domain-swapped dimers of SPAK T243D and SPAK WT. The two SPAK T243D dimer subunits are colored orange and blue, and the two SPAK WT subunits are colored purple and yellow. (C) Ribbon diagram of a SPAK T243D subunit (or monomer) as observed in the domain-swapped dimer. α-helices-cyan; β-strands-magenta; glycine-rich loop- brown; catalytic loop- red; activation loop- blue; PF1 domain- yellow. AMP-PNP is shown in a stick representation.
SPAK WT and SPAK T243D crystallized in different conditions and space groups. SPAK WT crystallized in the presence of ATP and was solved at 3.1 Å resolution with a final Rwork = 23.7% and Rfree = 26.9%. SPAK T243D crystallized in the presence of AMP-PNP at 2.5Å resolution with a final Rwork = 20.6% and Rfree = 24.6%. The refined models display reasonable geometry and Ramachandran statistics (Table 1). Two SPAK molecules are in each asymmetric unit in both structures (Fig. 1B). The conformations of the two monomers in SPAK WT and SPAK T243D are similar, with average root mean square deviations (RMSD) in Cα positions of 0.31 Å and 0.45 Å, respectively. Complete information on crystallization, data collection, and refinement can be found in Experimental Procedures.
Table 1.
Crystal data and refinement statistics
| Data | SPAK 63–403 | SPAK 63–390 (T243D) |
|---|---|---|
| Spacegroup | P 21 | P 21 21 21 |
| Cell dimensions (Å) | a= 72.4, b= 56.1, c= 100.0 | a= 66.4, b= 101.7, c= 104.1 |
| α, β, γ | 90°, 108.3°, 90° | 90°, 90°, 90° |
| Resolution (Å) | 42.9–3.10 (3.20–3.10) | 50–2.45 (2.54–2.45) |
| Observed reflection | 38612 | 308327 |
| Unique reflection | 13658 | 27322 |
| Completeness | 85.6 (44.3) | 93 (80.7) |
| Rmerge (outer shell)1 | 0.069 (0.49) | 0.043 (0.41) |
| I/> (outer shell) | 15.1 (2.1) | 31.4 (1.98) |
| Refinement | ||
| Rwork2 (%) | 23.6 | 20.6 |
| Rfree3 (%) | 26.9 | 24.6 |
| R.m.s.d. bond length | 0.009 Å | 0.006 Å |
| R.m.s.d. bond angle | 1.18° | 1.03° |
| Ramachandran favored (%) | 92.8 | 96.6 |
| Ramachandran outliers (%) | 0.2 | 0.0 |
| Average B factor (Å2) | 76.9 | 70.6 |
SPAK Structural Overview
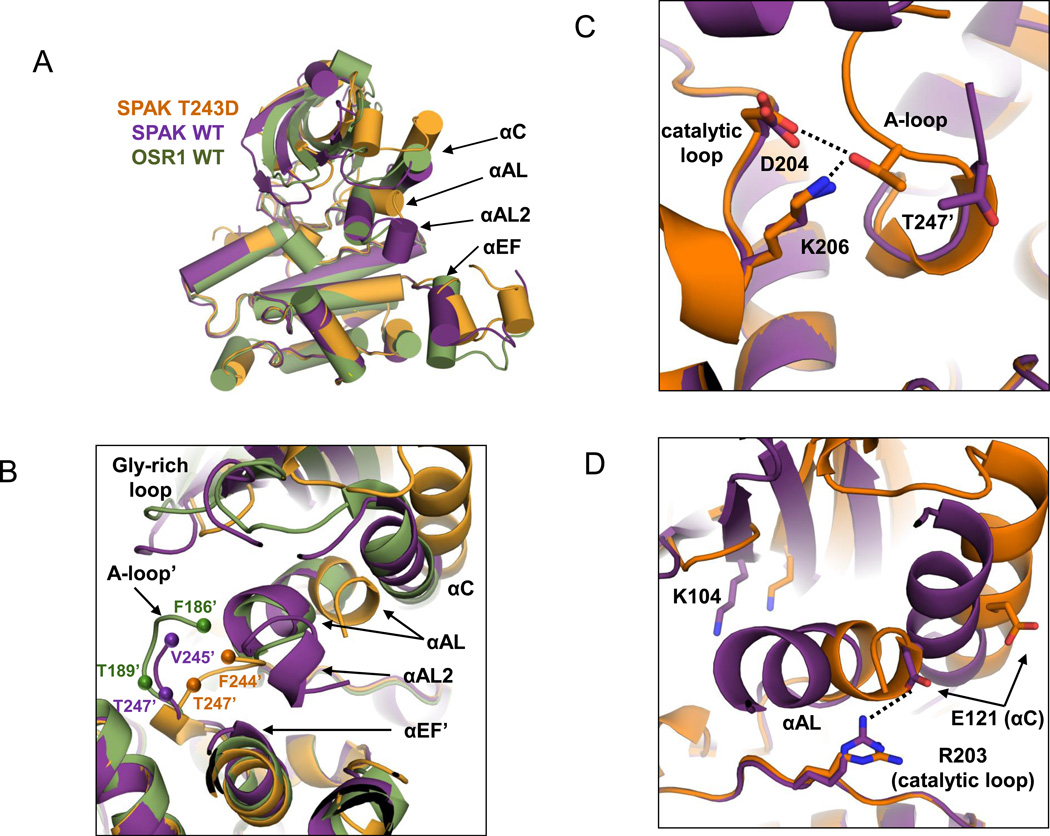
Both SPAK WT and SPAK T243D structures adopt a canonical two-domain kinase fold, with a small N-terminal domain (residues 63–155) connected to a larger C-terminal domain (residues 156–365) through a hinge (Fig. 1C). The C-terminal domain resembles that of OSR1, and has the canonical kinase helices D to I, as well as small, C-terminal helices J, K, and L. Both structures also possess an extra helix, αM (residues 355–363) that occupies a groove at the back of the kinase close to helix αD and strands β7 and β8. This helix is in a position similar to docking motif interactions found in MAPKs, which allosterically regulate sites distal to the docking site (42–45). The constructs used to solve the structures of OSR1 lacked the C-terminal extension, including residues in αM (28). Overlay of an individual subunit of both the SPAK WT and OSR1 WT structures reveals a RMSD of 0.99 Å. Apparently, neither the sequence variation between SPAK and OSR1, nor the presence of the PF1 domain in SPAK WT (of which helix αM is visible in the density) give rise to significant conformational differences between the molecules (Fig. 2A). However, conformational differences between the SPAK T243D and SPAK WT structures do exist. A slight (6°) domain rotation is present, with SPAK T243D in the more open configuration. αC and αAL (in the activation loop) show significant differences, again with αAL adopting the more open configuration in SPAK T243D (Fig 2A,B).
Figure 2. Conformational changes occurring in SPAK T243D compared to SPAK WT and OSR1 WT.
(A) Cartoon representation (α-helices represented as cylinders for clarity) of C-terminal domain alignments of SPAK WT (chain A; purple), SPAK T243D (chain A; orange), and OSR1 WT (chain A; green; PDB: 3DAK). Structural elements of particular interest are labeled. (B) Dimer interface near the domain-swapped activation loop and SPAK T243/OSR1 T185 WNK phosphorylation site. Labels marked with a prime (‘) correspond to the second monomer in the dimer. Spheres represent the first residue observed in the activation loop electron density and the SPAK T247'/OSR1 T189' catalytic residue. (C) The D-K-T catalytic triad interaction, formed by catalytic loop residues D204 and K206 and domain-swapped activation loop residue T247', is only present is SPAK T243D. (D) The essential K-E ion pair, between β3 K104 and αC E121, which is a hallmark of active kinases, is not present in SPAK WT or SPAK T243D. An interaction between catalytic loop residue R203 and E121 is present in SPAK WT but lost in SPAK T243D.
The nucleotide positions in SPAK T243D and SPAK WT primarily track with the N-terminal domains, and reflect the difference in domain rotation between the two structures (Fig. S1). In the open T243D structure, the phosphates are bound to the glycine-rich loop. In the more closed SPAK WT structure the γ-phosphate maintains contact with K206 in the catalytic loop in the C-terminal domain. Neither position is identical to that in PKA because PKA has a closed domain structure and ATP contacts both domains. The position of the ATP in the SPAK T243D structure is more similar to that in PKA.
Both SPAK WT and SPAK T243D have similar disordered regions. In the activation loop SPAK T243D only has helix αAL, while SPAK WT has two helices αAL and αAL2. Notably missing from the electron density is the C-terminal part of the PF1 domain (after residue R365 in SPAK T243D). To determine if this C-terminal region of the PF1 box affects activity, we compared wild-type SPAK 63–370 and 63–390, both phosphorylated by WNK1, and found that the longer construct had more activity (Fig. S2A). SPAK T243D has 10–20 fold more activity than wild-type SPAK 63–390, but only about 10% of the activity of SPAK 63–390 phosphorylated by WNK1 (Fig. S2B).
SPAK Activation Loop Domain Swapping
The most prominent feature of both the SPAK WT and SPAK T243D structures is that the activation loops are domain-swapped (Fig. 1B and Fig. S3A,B). To ensure that the dimers observed in the crystal structures were not artifacts of crystallization, we performed gel filtration experiments. Chromatography on Mono S revealed two peaks. Gel filtration showed that the peaks correspond to monomer and dimer (Fig. S3C). Rechromatography of proteins from each peak gave rise to both populations (data not shown) indicating SPAK exists in a monomer:dimer equilibrium in solution. In addition, ATP favors dimerization (Fig S3D).
Activation loop domain swapping encompasses residues G233-G261: G233-G246 of the activation loop and P+1 specificity pocket (binds P+1 site of substrate), T247 in the active site of the other monomer, helix αE/F (E254-V259), and the following loop leading to helix αF (Fig. S4A). In SPAK T243D, T247 is hydrogen-bonded to two invariant catalytic residues, D204 and K206, from the opposite monomer (Fig. 2C, S4B). The local structure in this area is similar to that of active canonical non-domain-swapped protein kinases, such as PKA (Fig. S4B).
The dimer interfaces in SPAK T243D and SPAK WT bury 1906 Å2 and 2003 Å2 of the dimer surface areas, respectively, or approximately 5% of the total surface areas of the dimers as calculated by the PISA server, and are almost entirely hydrophobic (40). Part of the activation loops in both SPAK WT and T243D are disordered; in SPAK T243D these residues encompass T231-D243 in the activation loop of one monomer, and D234-D243 in the activation loop of the other monomer (Fig. 2B). Similar residues are disordered in SPAK WT, with the exception of the presence of an additional helix αAL2 at the N-terminus of the activation loop. Notably, densities for T243 and the phosphomimetic mutation, T243D, are not present in either structure (Fig. 2B).
At the quaternary level, the dimeric arrangement of SPAK WT and SPAK T243D differ significantly. Both form two-fold symmetric dimers. However, these dimers are not equivalent due to differences in the orientation of the dimer axes (Fig. S5A). This difference in rotation correlates with tighter dimer packing in SPAK WT, including close contact between the αAL2 helices, which are only ordered in SPAK WT (Fig. S5C). In SPAK T243D, the two monomers are farther apart by a surprising 5 Å, although the function of this enhanced separation is unclear. Thus, these conformational differences are found not only in specific kinase substructures, but also in the overall organization of the two monomers. These differences might be responsible for alteration of the relative orientation and position of the N- and C-terminal domains, or activation loop accessibility, which are generally associated with kinase activation (46–50).
Comparison of the SPAK Structures Suggests a Multistage Activation Mechanism
SPAK WT and SPAK T243D monomers superimpose with a RMSD of ~1.7 Å. The superposition improves considerably using the C-terminal domain alone, with a RMSD of 0.31 Å. The large discrepancy in RMSD is primarily due to the relative positions of the N- and C-terminal domains, with SPAK T243D in the more open configuration, as discussed above (Fig 2A). Other specific differences occur in the glycine-rich loop, helix αC, and at both ends of the activation loop. The glycine-rich loop of SPAK T243D falls closer to the ATP binding pocket than it does in both SPAK WT and OSR1 WT, and adopts a closed conformation on the first two β strands. The C-termini of the activation loops adopt different conformations in the SPAK WT and T243D structures (Fig 2B). The SPAK T243D configuration is similar to active kinases, as discussed above, with a well-formed P+1 pocket, and T247 from the opposing monomer making standard active site contacts with D204 and K206 in the catalytic loop (Fig. 2C, Figs S4B). In contrast, SPAK WT and OSR1 WT have poorly formed P+1 pockets. In SPAK T243D, the N-terminus of helix αC is shifted about 5 Å relative to SPAK WT. As noted previously in the report on OSR1 WT, SPAK WT R203 from the catalytic loop makes an electrostatic interaction with E121 in αC (28). This interaction is lost in SPAK T243D, potentially as the result of the movement of helix αAL (Fig 2D). However, SPAK T243D still lacks the K104-E121 ion pair, which is a hallmark of active kinases (Fig. 2D, Figs. S7A,B). Thus, SPAK T243D appears more active near the P+1 pocket and T247, but is still in an inactive configuration, because it lacks the K-E ion pair.
Another difference between the SPAK WT and SPAK T243D structures lies in the orientation of the N-termini of the activation loops, and in particular the αAL helix. αAL helices occur in diverse classes of kinases, e.g., EGF receptor (51), and are known to be inhibitory by sterically blocking the inward movement of αC to form the K-E ion pair. Recent work has shown that deletion of the αAL helix in OSR1 leads to enhanced kinase activity (52). Interestingly, the positions of SPAK WT and OSR1 WT αAL helices are incompatible with the position of the SPAK T243D activation loop (Fig. 2B). We compared the position of αAL using both N and C-terminal domain structural alignments, and found that the movement of αAL was partially independent of the gross movements of the N- and C-terminal domains (Fig. S8A,B). Several hydrophobic interactions between αAL and αC, are lost in the SPAK T243D structure. In addition, αAL partially unfolds in SPAK T243D, and both αAL and αC exhibit higher B-factors (Fig. S8C–E).
Recent work has shown that the small activating protein MO25, also known as Cab39 (calcium binding protein 39) potently activates SPAK and OSR1 even in the presence of phosphomimetic mutation of the activation loop (46). Structures of MO25 in complex with the related MST kinases reveal that this occurs through stabilization of the K-E ion pair. Therefore, MO25 binding may be required for SPAK and OSR1 to attain fully active conformations (49;53).
Residues Permissive for Activation Loop Swapping
The exact N-terminal switch point for the swapping is within the disordered residues D234-T243. The C-terminus of the swapped region ends in the loop that connects helices αEF (short helix following activation loop) and αF (Fig. 1C, S4A). Several other protein kinases exhibit activation loop domain swapping. Ste20-like kinase (SLK), lymphocyte-originated kinase (LOK, also known as STK10), and OSR1 in the Ste group, and in the CaMK group, checkpoint kinase 2 (CHK2) and death-associated protein kinase 3 (DAPK3) show domain swapping (28;54–56). In each case the αE/F helices are intact but bound to the opposite subunit.
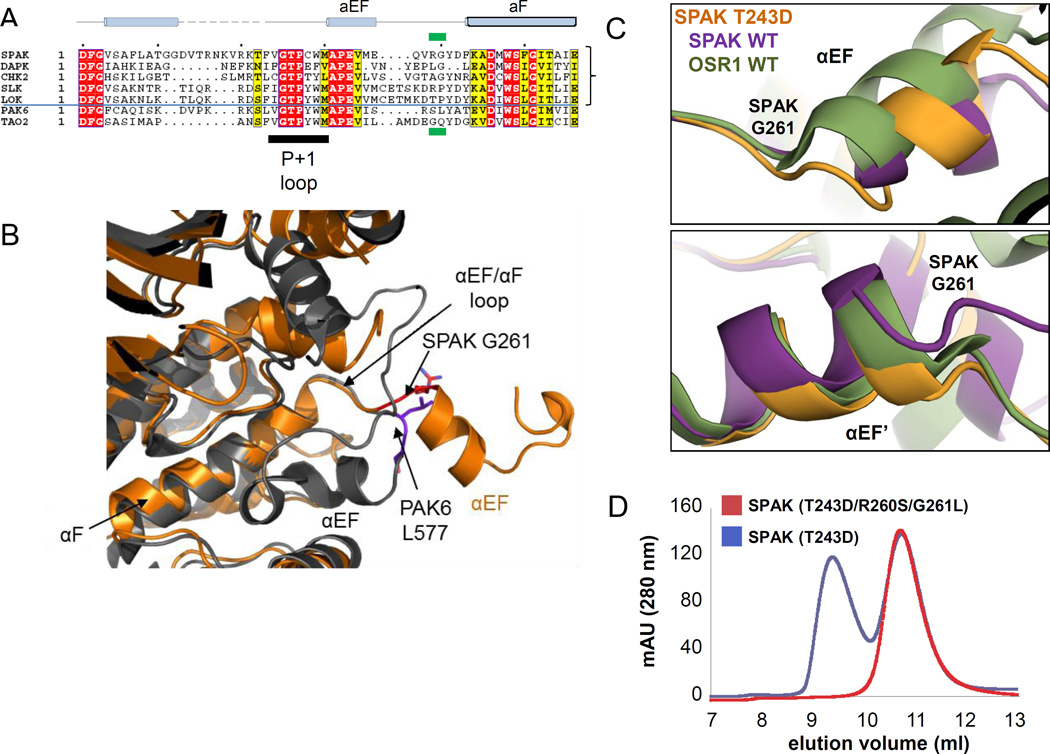
A structure-based multiple sequence alignment of SPAK and other kinases in the domain-swapped segment shows that all the domain-swapped kinases contain either a glycine or a proline at the position corresponding to G261 in SPAK, while the non-swapped kinases (for example, PAK6 and TAO2) have leucine and glutamine instead (Fig. 3A). Proline is frequently found in the hinge loops of domain-swapped proteins (29). Comparison of domain-swapped and non-swapped kinase structures reveals a major conformational difference in the αEF/αF loop in the backbone conformation between residues R260/G261 in SPAK and S576/L577 in PAK6 (Fig. 3B). In particular, G261, in SPAK T243D, adopts a left-handed conformation with φ= 83° and ψ= 175°. Additionally, by aligning chain A of the SPAK WT, OSR1 WT, and SPAK T243D dimer structures, the positional differences begin to occur around SPAK G261, or OSR1 G203 (Fig. 3C). Thus, we suspected that G261 is part of the domain-swap hinge-point (29).
Figure 3. Identification and mutation of the domain-swap hinge.
(A) Multiple sequence alignment of representative sequences of domain swapped kinases SPAK, DAPK3, CHK2, SLK, LOK, compared to the non-swapped kinases PAK6 and TAO2. The position corresponding to G261 in SPAK is boxed in green. (B) Superposition of SPAK T243D and the non-swapped kinase PAK6 (PDB 2C30) reveals the difference in the αEF/αF loop. SPAK is colored in orange and PAK6 is in gray. The switch point R260/G261 in SPAK and S576/L577 in PAK6 are represented in sticks and colored in red and purple, respectively. (C) View near αEF and αEF’ helices using chain A structural alignments of SPAK WT, SPAK T243D, and OSR1 WT dimers. Positional differences begin to occur near G261 and G261' (G203 in OSR1). (D) Gel filtration profile of SPAK T243D and SPAK (T243D/R260S/G261L).
To investigate the role of G261 in activation loop swapping, we mutated R260/G261 of SPAK to S/L as in PAK6. The Superdex75 gel filtration profile of SPAK 63–390 (T243D/R260S/G261L) suggests that it is primarily a monomer in solution, in contrast to SPAK T243D (Fig. 3D). Thus, apparently R260 and G261 are part of the domain-swap hinge.
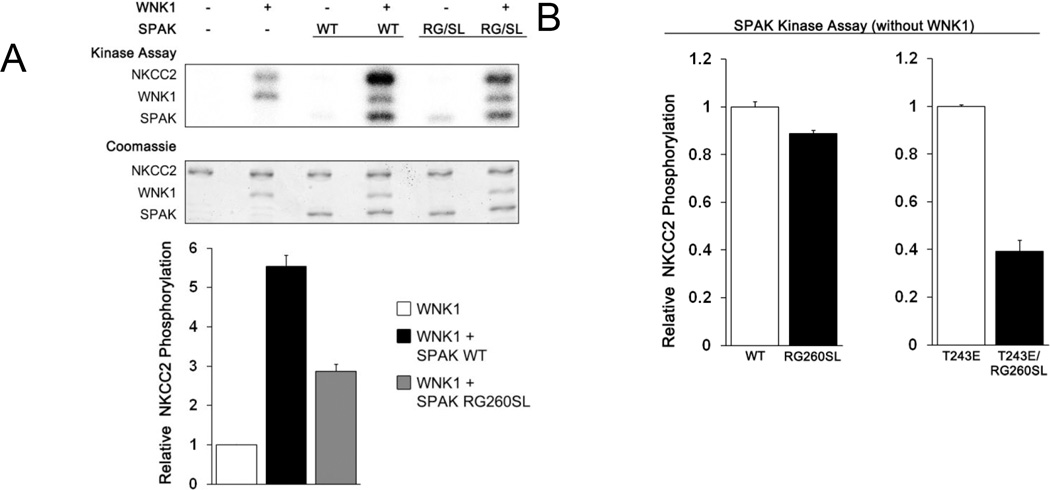
We then addressed the question of whether the monomer is active by comparing the activity of SPAK WT and dimerization-blocked mutant. The activity profile was studied in the context of constructs encompassing the entire PF1 domain (SPAK 63–403, and SPAK 63–403 R260S/G261L). In an in vitro kinase assay the SPAKs were first preactivated by phosphorylation by WNK1 for 30 min., and both were phosphorylated similarly (Fig. S9). The SPAK 63–403 R260S/G261L mutant had ~40% of the activity of SPAK WT towards the substrate GST-NKCC2 1–175 (Fig. 4A). Data is also presented that SPAK exhibits a small amount of autophosphorylation, and WNK1 exhibits some phosphorylation activity towards GST-NKCC2 1–175 in the absence of SPAK. Similarly, SPAK T243E/R260S/G261L had ~40% activity of SPAK T243E (Fig 4B). However, the dimerization blocking mutation has no effect on the basal activity of SPAK (Fig. 4B). Taken together we believe these results suggest that the monomeric form of SPAK can be activated by WNK1, and it has kinase activity.
Figure 4. Effect of the R260S/G261L mutation on SPAK activity.
(A) Coupled protein kinase assay. SPAK 63–403 WT and R260S/G261L mutant were preincubated with ATP-MgCl2 for 30 minutes with or without WNK1, after which [γ-32P]-ATP was added for radiolabeling, and GST-NKCC2 1–175 was added as substrate. (B) Kinase assays comparing activity of SPAK 63–403 with and without the R260S/G261L mutation. T243E is an activating phosphomimetic mutation in the SPAK activation loop. All results in (A) and (B) are reported as relative activities, and as the mean and standard deviation of triplicate measurements quantified by phosphoimaging.
Discussion
Domain swapping of activation loops is not uncommon among protein kinases. SLK, lymphocyte-originated kinase (LOK, also known as STK10) and OSR1, in the STE group, and checkpoint kinase 2 (CHK2) and death-associated protein kinase (DAPK3), in the CAMK group, are examples of domain-swapped enzymes (28;55;56). Several structures of domain-swapped kinases, some phosphorylated, have been reported. These structures vary considerably in conformation, with some having hallmarks of an active kinase, such as the essential K-E ion pair interaction between helix αC and strand β3, and others appearing inactive. These results show that domain swapping is not a reflection of the state of phosphorylation or activity of the kinase.
Here we report the domain-swapped structures of SPAK WT and SPAK T243D, which mimics the WNK activating phosphorylation site in the SPAK activation loop. SPAK WT adopts an inactive configuration very similar to the previously reported structure of OSR1 WT (28). SPAK T243D, in contrast, has some features reminiscent of active kinases. Thus, we hypothesize that the present structures reflect two different activity states in a multistage activation process: activation loop phosphorylation, mimicked here with the T243D mutation, induces a partially active conformation that can be further activated. Possible interactions that could lead to a fully active structure are with substrates or with the known activator protein MO25 (Cab39). In support of the multistage activation concept, SPAK and OSR1 harboring the phosphomimetic mutation in the activation loop are still activated by nearly 100-fold in the presence of MO25 (46). We also note that the SPAK T243D active site is much broader compared to SPAK WT and OSR1 WT due both to N-terminal domain rotation and outward movement of helix αAL. The more open SPAK T243D active site could potentially be more accessible to substrate, but the precise function is unclear from the present study.
By comparing structures and sequences of domain-swapped and non-swapped kinases, we identified a switch point for domain swapping. By mutating switch point residues to ones incompatible with domain swapping, we were able to generate a monomeric mutant form of SPAK, and show that it can be phosphorylated by WNK1 and retains activity, albeit lower than the wild-type kinase. The dimerization blocking mutation should prove valuable in future studies aimed at understanding the role of SPAK and OSR1 domain swapping.
Supplementary Material
Acknowledgments
We thank Diana Tomchick, Mischa Machius, and Zhe Chen for help in synchrotron data collection, Dominika Borek for suggestions about the structure, Radha Akella and Svetlana Earnest for technical assistance, Wen-Huang Ko for thoughts about interfaces, and Dionne Ware for administrative assistance. Results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by the University of Chicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357.
This work was funded by grants I1143 (MHC) and I1128 (EJG) from the Welch Foundation and R01 GM53032 (MHC) and R01 DK46993 (EJG) from the National Institutes of Health.
The work of CT, YJ, and SS was completed in partial fulfillment of the requirements for the Ph.D.
Abbreviations used
- SPAK
Ste20-related proline-alanine-rich kinase
- WNK
with no lysine (K)
- OSR1
oxidative stress-responsive kinase
- NKCC
Na+-K+-2Cl− cotransporter
- DTT
dithiothreitol
- SDS-PAGE
polyacrylamide gel electrophoresis in sodium dodecyl sulfate
- PKA
cyclic AMP-dependent protein kinase
- SLK
Ste20-like kinase
- LOK
lymphocyte-originated kinase
- CHK2
checkpoint kinase 2
- DAPK3
death-associated protein kinase
- PAK6
p21-activated protein kinase 6
- TAO2
thousand-and-one protein kinase 2
Footnotes
Supporting Information
Nine supporting figures.
This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.McCormick JA, Ellison DH. The WNKs: atypical protein kinases with pleiotropic actions. Physiol Rev. 2011;91:177–219. doi: 10.1152/physrev.00017.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Welling PA, Chang YP, Delpire E, Wade JB. Multigene kinase network, kidney transport, and salt in essential hypertension. Kidney Int. 2010;77:1063–1069. doi: 10.1038/ki.2010.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie J, Craig L, Cobb MH, Huang CL. Role of with-no-lysine [K] kinases in the pathogenesis of Gordon's syndrome. Pediatr. Nephrol. 2006;21:1231–1236. doi: 10.1007/s00467-006-0106-6. [DOI] [PubMed] [Google Scholar]
- 4.Darman RB, Forbush B. A regulatory locus of phosphorylation in the N terminus of the Na-K-Cl cotransporter, NKCC1. J. Biol. Chem. 2002;277:37542–37550. doi: 10.1074/jbc.M206293200. [DOI] [PubMed] [Google Scholar]
- 5.Alessi DR, Zhang J, Khanna A, Hochdorfer T, Shang Y, Kahle KT. The WNK-SPAK/OSR1 pathway: master regulator of cation-chloride cotransporters. Sci. Signal. 2014;7:re3. doi: 10.1126/scisignal.2005365. [DOI] [PubMed] [Google Scholar]
- 6.Gagnon KB, Delpire E. Molecular physiology of SPAK and OSR1: two Ste20-related protein kinases regulating ion transport. Physiol Rev. 2012;92:1577–1617. doi: 10.1152/physrev.00009.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Z, Xie J, Wu T, Truong T, Auchus RJ, Huang CL. Downregulation of NCC and NKCC2 cotransporters by kidney-specific WNK1 revealed by gene disruption and transgenic mouse models. Hum. Mol. Genet. 2011;20:855–866. doi: 10.1093/hmg/ddq525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Markadieu N, Delpire E. Physiology and pathophysiology of SLC12A1/2 transporters. Pflugers Arch. 2014;466:91–105. doi: 10.1007/s00424-013-1370-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, O'Connell JR, McArdle PF, Wade JB, Dorff SE, Shah SJ, Shi X, Pan L, Rampersaud E, Shen H, Kim JD, Subramanya AR, Steinle NI, Parsa A, Ober CC, Welling PA, Chakravarti A, Weder AB, Cooper RS, Mitchell BD, Shuldiner AR, Chang YP. From the Cover: Whole-genome association study identifies STK39 as a hypertension susceptibility gene. Proc. Natl. Acad. Sci. U. S. A. 2009;106:226–231. doi: 10.1073/pnas.0808358106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie J, Wu T, Xu K, Huang IK, Cleaver O, Huang CL. Endothelial-specific expression of WNK1 kinase is essential for angiogenesis and heart development in mice. Am. J Pathol. 2009;175:1315–1327. doi: 10.2353/ajpath.2009.090094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rafiqi FH, Zuber AM, Glover M, Richardson C, Fleming S, Jovanovic S, Jovanovic A, O'Shaughnessy KM, Alessi DR. Role of the WNK-activated SPAK kinase in regulating blood pressure. EMBO Mol. Med. 2010;2:63–75. doi: 10.1002/emmm.200900058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie J, Yoon J, Yang SS, Lin SH, Huang CL. WNK1 protein kinase regulates embryonic cardiovascular development through the OSR1 signaling cascade. J Biol. Chem. 2013;288:8566–8574. doi: 10.1074/jbc.M113.451575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castaneda-Bueno M, Gamba G. SPAKling insight into blood pressure regulation. EMBO Mol. Med. 2010;2:39–41. doi: 10.1002/emmm.200900059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heidenreich PA, Trogdon JG, Khavjou OA, Butler J, Dracup K, Ezekowitz MD, Finkelstein EA, Hong Y, Johnston SC, Khera A, Lloyd-Jones DM, Nelson SA, Nichol G, Orenstein D, Wilson PW, Woo YJ. Forecasting the future of cardiovascular disease in the United States: a policy statement from the American Heart Association. Circulation. 2011;123:933–944. doi: 10.1161/CIR.0b013e31820a55f5. [DOI] [PubMed] [Google Scholar]
- 15.Dan I, Watanabe NM, Kusumi A. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 2001;11:220–230. doi: 10.1016/s0962-8924(01)01980-8. [DOI] [PubMed] [Google Scholar]
- 16.Leiserson WM, Harkins EW, Keshishian H. Fray, a Drosophila serine/threonine kinase homologous to mammalian PASK, is required for axonal ensheathment. Neuron. 2000;28:793–806. doi: 10.1016/s0896-6273(00)00154-9. [DOI] [PubMed] [Google Scholar]
- 17.Delpire E, Gagnon KB. Genome-wide analysis of SPAK/OSR1 binding motifs. Physiol Genomics. 2007;28:223–231. doi: 10.1152/physiolgenomics.00173.2006. [DOI] [PubMed] [Google Scholar]
- 18.Vitari AC, Deak M, Morrice NA, Alessi DR. The WNK1 and WNK4 protein kinases that are mutated in Gordon's hypertension syndrome phosphorylate and activate SPAK and OSR1 protein kinases. Biochem. J. 2005;391:17–24. doi: 10.1042/BJ20051180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moriguchi T, Urushiyama S, Hisamoto N, Iemura S, Uchida S, Natsume T, Matsumoto K, Shibuya H. WNK1 regulates phosphorylation of cation-chloride-coupled cotransporters via the STE20-related kinases, SPAK and OSR1. J. Biol. Chem. 2005;280:42685–42693. doi: 10.1074/jbc.M510042200. [DOI] [PubMed] [Google Scholar]
- 20.Anselmo AN, Earnest S, Chen W, Juang YC, Kim SC, Zhao Y, Cobb MH. WNK1 and OSR1 regulate the Na+, K+, 2Cl− cotransporter in HeLa cells. Proc. Natl. Acad. Sci. U. S. A. 2006;103:10883–10888. doi: 10.1073/pnas.0604607103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gagnon KB, England R, Delpire E. Volume sensitivity of cation-chloride cotransporters is modulated by the interaction of two kinases: SPAK and WNK4. Am. J. Physiol Cell Physiol. 2006;290:C134–C142. doi: 10.1152/ajpcell.00037.2005. [DOI] [PubMed] [Google Scholar]
- 22.Villa F, Goebel J, Rafiqi FH, Deak M, Thastrup J, Alessi DR, van Aalten DM. Structural insights into the recognition of substrates and activators by the OSR1 kinase. EMBO Rep. 2007;8:839–845. doi: 10.1038/sj.embor.7401048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson FH, Disse-Nicodeme S, Choate KA, Ishikawa K, Nelson-Williams C, Desitter I, Gunel M, Milford DV, Lipkin GW, Achard JM, Feely MP, Dussol B, Berland Y, Unwin RJ, Mayan H, Simon DB, Farfel Z, Jeunemaitre X, Lifton RP. Human hypertension caused by mutations in WNK kinases. Science. 2001;293:1107–1112. doi: 10.1126/science.1062844. [DOI] [PubMed] [Google Scholar]
- 24.Piala AT, Moon TM, Akella R, He H, Cobb MH, Goldsmith EJ. Chloride sensing by WNK1 involves inhibition of autophosphorylation. Sci. Signal. 2014;7:ra41. doi: 10.1126/scisignal.2005050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gagnon KB, England R, Delpire E. Characterization of SPAK and OSR1, regulatory kinases of the Na-K-2Cl cotransporter. Mol. Cell Biol. 2006;26:689–698. doi: 10.1128/MCB.26.2.689-698.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gagnon KB, Delpire E. On the substrate recognition and negative regulation of SPAK, a kinase modulating Na+-K+-2Cl− cotransport activity. Am. J. Physiol Cell Physiol. 2010;299:C614–C620. doi: 10.1152/ajpcell.00074.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Villa F, Deak M, Alessi DR, van Aalten DM. Structure of the OSR1 kinase, a hypertension drug target. Proteins. 2008;73:1082–1087. doi: 10.1002/prot.22238. [DOI] [PubMed] [Google Scholar]
- 28.Lee SJ, Cobb MH, Goldsmith EJ. Crystal structure of domain-swapped STE20 OSR1 kinase domain. Protein Sci. 2009;18:304–313. doi: 10.1002/pro.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rousseau F, Schymkowitz JW, Itzhaki LS. The unfolding story of three-dimensional domain swapping. Structure. 2003;11:243–251. doi: 10.1016/s0969-2126(03)00029-7. [DOI] [PubMed] [Google Scholar]
- 30.Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of "parallel" expression vectors. Protein Expr. Purif. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- 31.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CW, Sweet RM, editors. Macromolecular Crystallography, part A. New York: Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 32.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr. D. Biol Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 34.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 36.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D. Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 37.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D. Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 41.Chen W, Yazicioglu M, Cobb MH. Characterization of OSR1, a member of the mammalian Ste20p/germinal center kinase subfamily. J Biol Chem. 2004;279:11129–11136. doi: 10.1074/jbc.M313562200. [DOI] [PubMed] [Google Scholar]
- 42.Chang CI, Xu B, Akella R, Cobb MH, Goldsmith EJ. Crystal structures of MAP kinase p38 complexed to the docking sites on its nuclear substrate MEF2A and activator MKK3b. Mol. Cell. 2002;9:1241–1249. doi: 10.1016/s1097-2765(02)00525-7. [DOI] [PubMed] [Google Scholar]
- 43.Goldsmith EJ, Akella R, Min X, Zhou T, Humphreys JM. Substrate and docking interactions in serine/threonine protein kinases. Chem Rev. 2007;107:5065–5081. doi: 10.1021/cr068221w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou T, Sun L, Humphreys J, Goldsmith EJ. Docking interactions induce exposure of activation loop in the MAP kinase ERK2. Structure. 2006;14:1011–1019. doi: 10.1016/j.str.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 45.Peti W, Page R. Molecular basis of MAP kinase regulation. Protein Sci. 2013;22:1698–1710. doi: 10.1002/pro.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Filippi BM, de Los HP, Mehellou Y, Navratilova I, Gourlay R, Deak M, Plater L, Toth R, Zeqiraj E, Alessi DR. MO25 is a master regulator of SPAK/OSR1 and MST3/MST4/YSK1 protein kinases. EMBO J. 2011;30:1730–1741. doi: 10.1038/emboj.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grimm PR, Taneja T, Liu J, Coleman R, Chen Y-Y, Delpire E, Wade JB, Welling PA. SPAK, OSR1 and Cab39/MO25 form an interdependent signaling system which regulates thiazide-sensitive salt transport, distal tubule mass and blood pressure. 2012:867–837. [Google Scholar]
- 48.Hao Q, Feng M, Shi Z, Li C, Chen M, Wang W, Zhang M, Jiao S, Zhou Z. Structural insights into regulatory mechanisms of MO25-mediated kinase activation. J. Struct. Biol. 2014;186:224–233. doi: 10.1016/j.jsb.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 49.Mehellou Y, Alessi DR, Macartney TJ, Szklarz M, Knapp S, Elkins JM. Structural insights into the activation of MST3 by MO25. Biochem. Biophys. Res. Commun. 2013;431:604–609. doi: 10.1016/j.bbrc.2012.12.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeqiraj E, Filippi BM, Deak M, Alessi DR, van Aalten DM. Structure of the LKB1-STRAD-MO25 complex reveals an allosteric mechanism of kinase activation. Science. 2009;326:1707–1711. doi: 10.1126/science.1178377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 52.Li C, Feng M, Shi Z, Hao Q, Song X, Wang W, Zhao Y, Jiao S, Zhou Z. Structural and biochemical insights into the activation mechanisms of germinal center kinase OSR1. J. Biol. Chem. 2014;289:35969–35978. doi: 10.1074/jbc.M114.592097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi Z, Jiao S, Zhang Z, Ma M, Zhang Z, Chen C, Wang K, Wang H, Wang W, Zhang L, Zhao Y, Zhou Z. Structure of the MST4 in complex with MO25 provides insights into its activation mechanism. Structure. 2013;21:449–461. doi: 10.1016/j.str.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 54.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 55.Pike AC, Rellos P, Niesen FH, Turnbull A, Oliver AW, Parker SA, Turk BE, Pearl LH, Knapp S. Activation segment dimerization: a mechanism for kinase autophosphorylation of non-consensus sites. EMBO J. 2008;27:704–714. doi: 10.1038/emboj.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oliver AW, Paul A, Boxall KJ, Barrie SE, Aherne GW, Garrett MD, Mittnacht S, Pearl LH. Trans-activation of the DNA-damage signalling protein kinase Chk2 by T-loop exchange. EMBO J. 2006;25:3179–3190. doi: 10.1038/sj.emboj.7601209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 58.Johnson LN, Noble ME, Owen DJ. Active and inactive protein kinases: structural basis for regulation. Cell. 1996;85:149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 59.Nolen B, Taylor S, Ghosh G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol. Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 60.Shi F, Telesco SE, Liu Y, Radhakrishnan R, Lemmon MA. ErbB3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation. Proc. Natl. Acad. Sci. U. S. A. 2010;107:7692–7697. doi: 10.1073/pnas.1002753107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.