Abstract
The huge variation in root system architecture (RSA) among different rice (Oryza sativa) cultivars is conferred by their genetic makeup and different growth or climatic conditions. Unlike model plant Arabidopsis, the molecular basis of such variation in RSA is very poorly understood in rice. Cultivars with stable variation are valuable resources for identification of genes involved in RSA and related physiological traits. We have screened for RSA and identified two such indica rice cultivars, IR-64 (OsAS83) and IET-16348 (OsAS84), with stable contrasting RSA. OsAS84 produces robust RSA with more crown roots, lateral roots and root hairs than OsAS83. Using comparative root transcriptome analysis of these cultivars, we identified genes related to root development and different physiological responses like abiotic stress responses, hormone signaling, and nutrient acquisition or transport. The two cultivars differ in their response to salinity/dehydration stresses, phosphate/nitrogen deficiency, and different phytohormones. Differential expression of genes involved in salinity or dehydration response, nitrogen (N) transport, phosphate (Pi) starvation signaling, hormone signaling and root development underlies more resistance of OsAS84 towards abiotic stresses, Pi or N deficiency and its robust RSA. Thus our study uncovers gene-network involved in root development and abiotic stress responses in rice.
A wide range of variations, in terms of yield, physiology and morphology including root system architecture (RSA) trait are observed among various cultivars of rice (Oryza sativa)1. O. sativa has two major varieties - the upland varieties, usually with small number of deeper and thicker roots and cultivated in dry fields of temperate East Asia, and lowland varieties, usually with higher number of shallow and thin roots with low root/shoot mass ratio for better adaptation to submergence and cultivated in submerged fields of tropical Asia2. RSA of different cultivars vary tremendously to meet the requirement of the plant to combat various growth conditions such as nutrient deficiency, drought or sanity stress. Some rice cultivars are tolerant to abiotic stresses; Nagina 22 (mutant NH219) is drought tolerant, Pokkali is salinity tolerant and Dular is low phosphate tolerant3,4. The variation in RSA is regulated by intrinsic factors, such as transcription factors, microRNAs and phytohormones, and influenced by several extrinsic factors (like light and water), and a cumulative effect of all these leads to development of a particular type of RSA5.
In monocotyledonous plants, RSA consists of primary root (PR), lateral roots (LRs), crown/seminal roots (CR) and root hairs (RHs)5,6. Root is developed from root stem cells, which are present in the root apical meristem (RAM)7. Different genes and transcription factors, such as PLETHORA (PLT), SHORT ROOT (SHR), SCARECROW (SCR) and WUSCHEL RELATED HOMEOBOX5 (WOX5) regulate the maintenance of root meristem and its pattern in Arabidopsis8. In rice, homologs of these genes have also been identified; OsWOX11 regulates CR emergence, OsWOX3A regulates LR development and RH formation, and OsSCR1 and OsSCR2 are involved in regulation of asymmetric division of cortex/endodermis progenitor cells and initiation of root development9,10,11.
Phytohormones such as auxin and cytokinin have been implicated in regulation of various RSA traits12. In rice, auxin and AUX/IAA (AUXIN/INDOLE ACETIC ACID) proteins regulate the expression of CROWN ROOTLESS1 (CRL1), which encodes for Arabidopsis homologs LBD16/LBD19, which are ASYMMETRIC LEAVES (AS2)/LATERAL ORGAN BOUNDARIES (LOB) family transcription factor13. crl1 mutant showed defects in CR formation and alteration in other auxin-related RSA traits such as reduced LR number, auxin insensitivity in LR formation and impaired root gravitropism13. The CRL1 promoter contains an auxin responsive element (ARE), which binds with the OsARF16, a ortholog of ARF7 and ARF19 of Arabidopsis14. ARF7 and ARF19 regulates the expression of LBD16 and LBD29 and control lateral organ development15. ARF7 and ARF19 also show close relationship to CRL1 in phylogenetic study16,17. Knockdown of OsARF16 resulted in reduced sensitivity of PR, LRs and RHs towards auxin and phosphate (Pi) deficiency18. Mutation in OsIAA23, another auxin responsive gene required for quiescent center (QC) maintenance, led to pleiotropic defects in root cap, LR and CR development19.
The major or minor nutrients also influence various aspects of root development, such as root length, diameter, LR branching, root angle, and other physiological changes20. Pi deficiency leads to reduced PR length and increased LR density in Arabidopsis with altered hormone sensitivity21,22,23. In rice, Pi deficiency leads to increased PR length24,25. Pi deficiency leads to redistribution of auxin content26,27. Low Pi condition for several days resulted in irreversible inhibition of root growth in Arabidopsis28. Availability of Nitrogen (N) has drastic and contrasting effects on development of RSA in different species or varieties. In Arabidopsis, both PR and LRs elongation is reduced on increased nitrate availability, whereas in rice and maize, PR gets elongated in N deprivation29,30.
Besides nutrient availability, soil composition and environmental stresses such as drought, high-salt and freezing factors also affect root development. Drought is one of the major constraints to many crop plants, especially to rice yields1. Under water deficiency, growth of leaves and stem is restricted, but root may continue to elongate31. Till date, many stress-related genes have been shown to play role in drought tolerance. Root specific overexpression of OsNAC10 and OsNAC5 resulted in enhanced root growth and increased drought tolerance32,33. In Arabidopsis, overexpression of a member of aspartic protease gene family, ASPARTIC PROTEASE IN GUARD CELL 1(ASPG1) leads to the reduction in water loss in ABA-dependent manner34.
Recent advances in various emerging phenomics tools, invasive or non-invasive imaging techniques and next generation sequencing (NGS) are helpful in correlating the genetic signature with RSA trait in rice35,36. Despite several efforts, our knowledge of regulation of monocot root development is still inadequate. Therefore dissecting the molecular and genetic regulatory mechanisms involved in root development is a prerequisite for the development of new rice varieties with improved RSA traits37. To uncover the molecular basis of RSA trait in rice, we have screened two dozen rice varieties and identified two indica rice cultivars, IR-64 and IET-16348 (OsAS83, lowland and OsAS84, upland, respectively) with stable and contrasting RSA trait as a model system. In this study, we have compared the root transcriptome of these cultivars and identified genes potentially involved in root development and related physiological responses. Our results indicate that differential expression of various transporters, transcription factors and genes involved in hormonal signaling could largely contribute to the phenotypic and physiological differences observed between two cultivars.
Results and Discussion
The OsAS83 and OsAS84 rice cultivars differ in their RSA
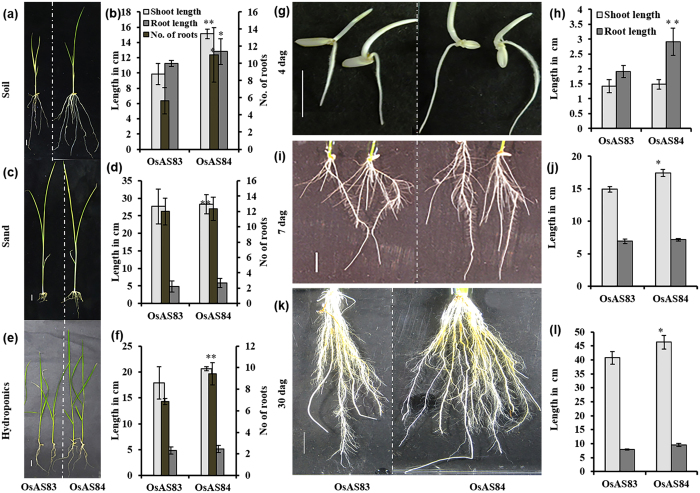
It has been reported earlier that root system is highly plastic and vary drastically in terms of RSA in different growth conditions5. We screened two dozen of selected indica rice cultivars in different growth conditions for detailed analysis of their RSA (Fig. S1). In different growth conditions (soil, sand and hydroponics), OsAS83 and OsAS84 cultivars maintained stable and contrasting root growth pattern and RSA at 14 days after germination (dag). In all the conditions tested, OsAS84 showed more robust RSA than OsAS83. In soil, OsAS84 showed 11% longer PR and two-times more CR number in comparison to OsAS83 (Fig. 1a and b). In sand, the PR length was found to be significantly affected in both the cultivars and difference between PR length and CR number between two cultivars was reduced in comparison to soil (Fig. 1c and d). In hydroponics system, OsAS84 showed 33% higher number of CRs in comparison to OsAS83 (Fig. 1e,f and Fig. S2). We performed growth assay at different developmental stages and found that OsAS84 has longer PR and more number of CRs than OsAS83 at 4 dag, 7 dag and 30 dag (Fig. 1g–l). Interestingly, shoot of OsAS84 was also found to be longer than OsAS83 at 4 dag, 7 dag and 30 dag.
Figure 1. OsAS84 has advanced root system architecture (RSA) than OsAS83.
(a–f) Phenotypic difference in RSA of OsAS83 and OsAS84 rice cultivars at 14 days after germination (dag). (a,c and e) Phenotypic difference between two cultivars in soil, sand and hydroponics, respectively. (b,d and f) Difference in primary root length, shoot length and number of crown roots in two cultivars in soil, sand and hydroponics, respectively. (g–l) Difference in RSA of OsAS83 and OsAS84 rice cultivars at different developmental stages in hydroponics. (g,i and k) Phenotypic difference of RSA between two cultivars at 4 dag, 7 dag and 30 dag, respectively. (h,j and l) Difference in primary root length and shoot length at 4 dag, 7 dag and 30 dag, respectively in two cultivars. Scale bar 1 cm. Error bars indicate standard error (n = 10). Asterisks indicate significant statistical differences, ***P < 0.001, **P < 0.01, *P < 0.05 (One-way ANOVA). Experiment was repeated three times with reproducible results.
To check the histological variation between these two cultivars, we analyzed the transverse sections of 2 dag PR tip (at 0–1 cm). The epidermal and cortical cell layer was more compact in OsAS84 than OsAS83. OsAS84 also showed higher number of cortical cell layers in comparison to OsAS83, although there is no difference in vascular patterning (Fig. S3a and 3b). RH density was also found to be more in OsAS84 (Fig. S3c). For survival in dry temperate regions, the upland rice varieties tends to develop thicker and bushy roots and plants exposed to salinity and drought stress developed deeper roots with more branches2,38. Our analysis showed that OsAS84, an upland cultivar, produces longer PR, more CR, LR and RH density in different growing mediums as well as in developmental stages in comparison to OsAS83, a lowland cultivar. These advanced RSA traits of OsAS84 is indicative of its better adaptability against stress.
OsAS83 and OsAS84 differ in shoot, flowering and seed traits
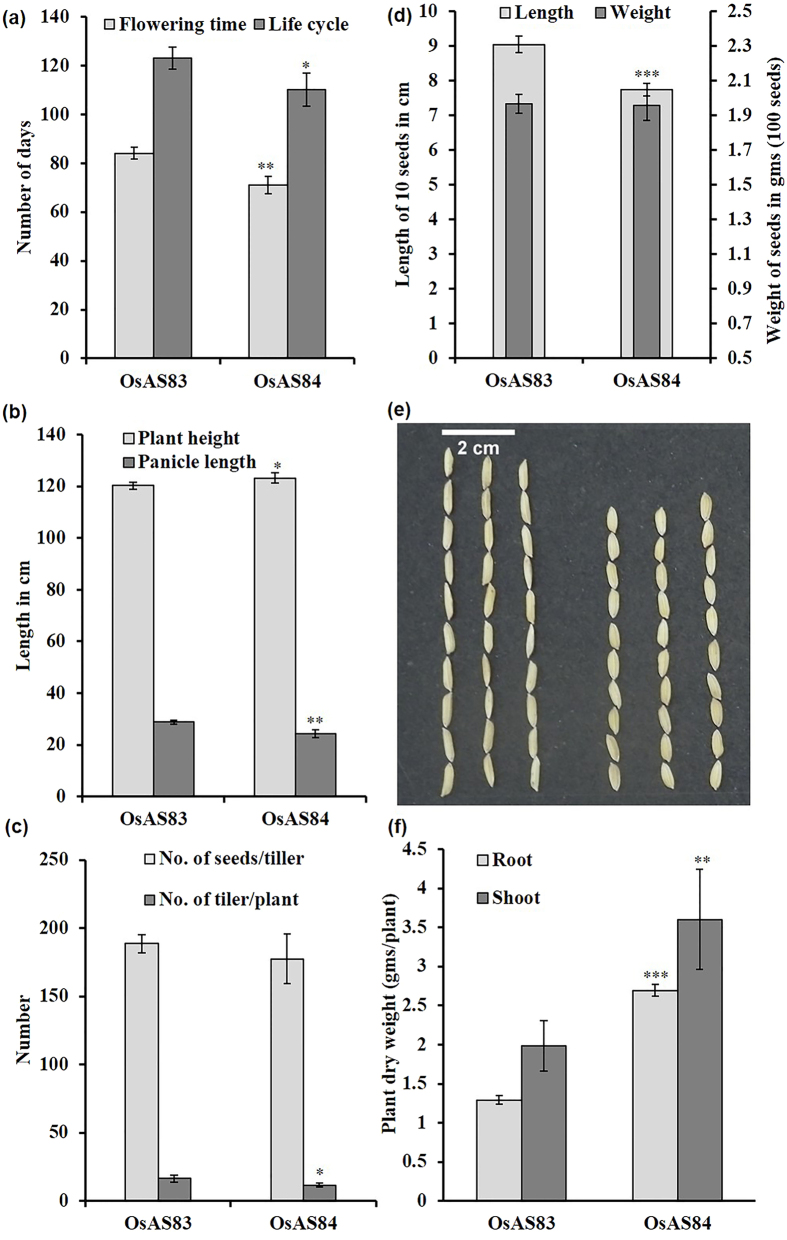
To understand the effect of differential RSA traits on overall plant growth various phenotypic traits were analyzed. OsAS84 has smaller life-cycle (10.5% less) as well as early flowering time (by 10 ± 2 days), in comparison to OsAS83 (Fig. 2a). Height of fully mature plant show slight variation (120 ± 5 cm and 123 ± 5 cm for OsAS83 and OsAS84, respectively). It is possible that advanced RSA trait helped OsAS84 to grow more efficiently in less time (Fig. 2b). Average tiller number/plant and seeds number/tiller were reduced in OsAS84 by ~6% and ~30%, respectively (Fig. 2c). Grain size of OsAS84 was found to be 1% shorter in length and thicker in diameter in relation to OsAS83 seeds. Weight of seeds (per 100 seeds) showed no difference between two cultivars (Fig. 2d and e). Dry weight of both root and shoot was found to be more in OsAS84 (Fig. 2f). Therefore, despite having advanced RSA, there was no yield penalty in OsAS84.
Figure 2. OsAS84 and OsAs83 differ in various agronomic traits.
(a) OsAS84 has shorter flowering time than OsAS83. Error bars indicate SD (n = 10). (b) OsAS84 have longer plant height, and lower panicle length than OsAS83. (c) Number of seeds/tiller, and number of tillers/plant was lower in OsAS84. Error bars indicate SD (n = 10). (d) and (e) OsAS84 has smaller seed length and equal seed weight as OsAS83. Scale bar is 1 cm. (f), OsAS84 have more root and shoot dry weight than OsAS83. Shorter life-span of OsAS84 does not hamper the seed-vigour and final productivity. Asterisks indicate significant statistical differences, ***P < 0.001, **P < 0.01, *P < 0.05 (One-way ANOVA). Each experiment was repeated three times with similar results. Error bars indicate SD (n = 10).
Differential root transcriptome signature underlies variation in RSA of OsAS83 and OsAS84
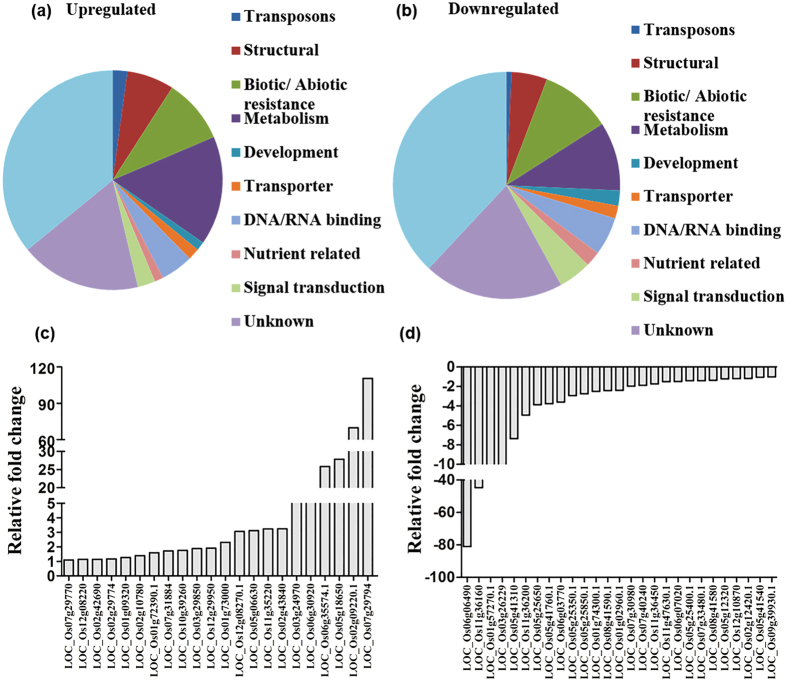
Analysis of variation in gene expression pattern between rice cultivars with stable contrasting RSA is likely to uncovers molecular factors responsible for RSA trait. To identify the differentially expressed genes between the root tissues (at 14 dag) of OsAS83 and OsAS84 cultivars, we performed gene expression microarray. After taking a cut-off of P ≤ 0.05 and fold change ≥2.0, a total of 231 genes were found to be upregulated and 276 genes were found to be downregulated in OsAS84 root in comparison to OsAS83. We performed Gene Ontology (GO) analysis to annotate the function of differentially expressed genes which were categorized in 11 classes, for e.g. transposon, structural, biotic/abiotic resistance, metabolism, developmental, transporter, DNA/RNA binding, nutrient related, signal transduction, unknown and unidentified (Fig. 3a and b). Microarray results were further validated for some of the selected genes by real time quantitative RT-PCR (qRT-PCR) analysis (Fig. 3c,d and Table S1).
Figure 3. Comparative root transcriptome of OsAS83 and OsAS84 identifies genes of developmental and physiological importance.
Microarray between 14 dag roots of two cultivars showed differential expression of several transporters, transcription factors and dehydration responsive genes. (a,b) Upregulated genes (231) and downregulated genes (276) in OsAS84 were classified according to their functional annotation. (c,d) Some of differentially expressed genes were further validated by real time qRT-PCR. All are statistically significant, P < 0.05 (n = 3).
Several transporters are differentially expressed in the roots of OsAS83 and OsAS84
We identified that several genes encoding for transporters are differentially expressed in roots of two cultivars. We observed that the metal cation transporter (LOC_Os03g29850.1), transmembrane amino acid transporter (LOC_Os05g14820.1), major facilitator transporter (LOC_Os02g02170.1), nitrate chloride transporter (LOC_Os12g29950.1), sugar transporter MtN3 (LOC_Os05g12320.1) and ABC transporter (LOC_Os06g03770.1) were differentially expressed in roots of these two cultivars (Table 1). Homologs of LOC_Os03g29850 (a metal cation transporter) and LOC_Os07g31884 (MATE efflux family protein) proteins in Arabidopsis are known to maintain Zinc homeostasis39. MATE transporter family is a large family with 58 orthologs in Arabidopsis and 40 members in rice40. In Arabidopsis, members of MATE transporters are known to be involved in pathogen infection, aluminum (Al) tolerance, sequestration of proanthocyanids, and glucose assimilation41,42,43,44. MATE transporter from sorghum, Hordeum, Triticum and maize have also been shown to participate in Al tolerance by forming non-toxic complexes of citrate with Al in soil solution45,46,47,48. Overexpression of OsMATE1 and OsMATE2 in Arabidopsis leads to altered development and pathogen susceptibility49. Overexpression of AtATM3, a homolog of ABC transporter (LOC_Os06g03770) showed enhanced Cd(II) and Pb(II) resistance in Arabidopsis50. Overexpression of AtATM3 in Brassica juncea also conferred increased tolerance to Cd(II) and Pb(II) stresses51. Higher expression of MATE and other transporters indicate possible ameliorate adaptation of OsAS84 towards metal toxicity, which need to be tested further, and also reflects the conservation of this mechanism in rice.
Table 1. Genes differentially expressed in OsAS84 in comparison to OsAS83 roots.
| Accession No. | Fold change in microarray | Arabidopsis ID | Function | References |
|---|---|---|---|---|
| Transporters | ||||
| LOC_Os03g29850.1 | 2.7012017 | AT1G55910 | Metal cation transporter | 39 |
| LOC_Os05g14820.1 | 2.8707578 | AT5G40780 | Transmembrane amino acid transporter protein | 96 |
| LOC_Os09g20490.1 | 2.8190773 | AT1G78130 | Transporter, putative | |
| LOC_Os03g40780.1 | 2.158206 | AT1G49870 | Transport protein-related, putative | |
| LOC_Os02g02170.1 | 16.11485 | AT1G08090 | Transporter, major facilitator family, putative | |
| LOC_Os12g29950.1 | 2.418649 | AT2G39210 | Nitrate chloride transporter | |
| LOC_Os07g31884.1 | 2.2769732 | AT3G23560 | MATE efflux family protein, putative | 97 |
| LOC_Os05g12320.1 | −2.959296 | AT5G53190 | Nodulin MtN3 family protein, putative | 53 |
| LOC_Os07g03960.1 | −3.7330253 | AT5G61520 | Transporter family protein, putative | |
| LOC_Os12g08090.1 | −4.244538 | AT1G77380 | Amino acid transporter, putative | 98 |
| LOC_Os06g03770.1 | −2.0081406 | AT5G58270 | ABC transporter, ATP-binding protein, putative | 99 |
| LOC_Os08g41590.1 | −6.2401733 | AT1G72125 | Peptide transporter PTR2, putative, expressed | 100 |
| LOC_Os01g17214.1 | −2.0424147 | AT3G43790 | Major facilitator superfamily antiporter, putative, expressed | 101 |
| Hormone signaling related | ||||
| LOC_Os09g27820.1 | 19.6028 | AT1G05010 | 1-aminocyclopropane-1-carboxylate oxidase protein, putative, expressed | 102 |
| LOC_Os02g43840.1 | 4.617959 | AT5G47310 | Ethylene-responsive element-binding protein, putative, expressed | 103 |
| LOC_Os11g13670.1 | 6.153131 | AT5G23530 | Gibberellin receptor GID1L2, putative, expressed | |
| LOC_Os07g40240.1 | −5.867474 | AT5G59845 | GASR9 - Gibberellin-regulated GASA/GAST/Snakin family protein precursor, expressed | 60 |
| LOC_Os06g07040.1 | −3.1850903 | AT2G33310 | OsIAA20 - Auxin-responsive Aux/IAA gene family member, expressed | 104 |
| LOC_Os11g29120.1 | −15.173324 | AT1G04910 | Growth regulator related protein, putative, expressed | |
| LOC_Os05g41760.1 | −2.0487518 | AT1G28360 | AP2 domain containing protein, expressed | 105 |
| Transcription factors | ||||
| LOC_Os05g41540.1 | −2.7472656 | AT2G16770 | bZIP transcription factor domain containing protein, expressed | 67 |
| LOC_Os05g14370.1 | −5.4798274 | AT3G56400 | WRKY82, expressed | 106 |
| LOC_Os06g33970.1 | −2.890836 | VQ domain containing protein | 91 | |
| LOC_Os02g42690.3 | 3.7169807 | AT4G03510 | Zinc finger, C3HC4 type domain containing protein, | 107 |
| LOC_Os03g24970.1 | 3.0067194 | N/A | SWIM zinc finger family protein | |
| LOC_Os07g29770.1 | 2.1135173 | AT1G70150 | Zinc finger protein, putative, expressed, ATROPGEF7/ROPGEF7 | |
| LOC_Os06g07020.1 | −2.8766186 | AT5G52010 | ZOS6-01 - C2H2 zinc finger protein, expressed | |
| LOC_Os11g47630.1 | −2.201833 | AT3G10470 | ZOS11-10 - C2H2 zinc finger protein, expressed | |
For adapting to various growth conditions like presence of heavy metals, availability of nutrients in the soil, and abiotic stresses, RSA has to undergo developmental changes which is regulated by various molecular regulators. These developmental changes may often involve changes in the distribution and transport of sugars in shoot and root. In our transcriptome study, the expression of a sugar transporter (MtN3; SWEET gene family member) was downregulated in OsAS84 root (Fig. 3d). SWEET proteins are uniporters, which facilitate diffusion of sugars across cell membranes, and loading of sucrose into phloem52. Mutant of a member of SWEET gene family in Arabidopsis, atsweet11/12 exhibited reduced root length upon germination on sugar-free media53. In rice, OsSWEET11 and OsSWEET14 have been reported to regulate rice reproductive development54,55. Expression of a nitrogen chloride transporter (OsNCT; LOC_Os12g29950.1) and OsSPX2 (LOC_Os02g10780) was higher in OsAS84. The homolog of a nitrate chloride transporter, major facilitator protein (AT2G39210.1), was found to be upregulated in salt tolerant halophyte salt cress in comparison to Arabidopsis, which suggests its involvement in salinity stress56. Differential expression of these transporters in OsAS83 and OsAS84 roots suggests that these two cultivars might differ in their ability for nutrition acquisition or heavy metal toxicity tolerance or stress response, which further may lead to developmental variation in RSA. Although a matter of further study, the expression level of these transporters possibly link the developmental adaptability of RSA and physiological responses of plants.
OsAS84 shows better ion absorption efficiency
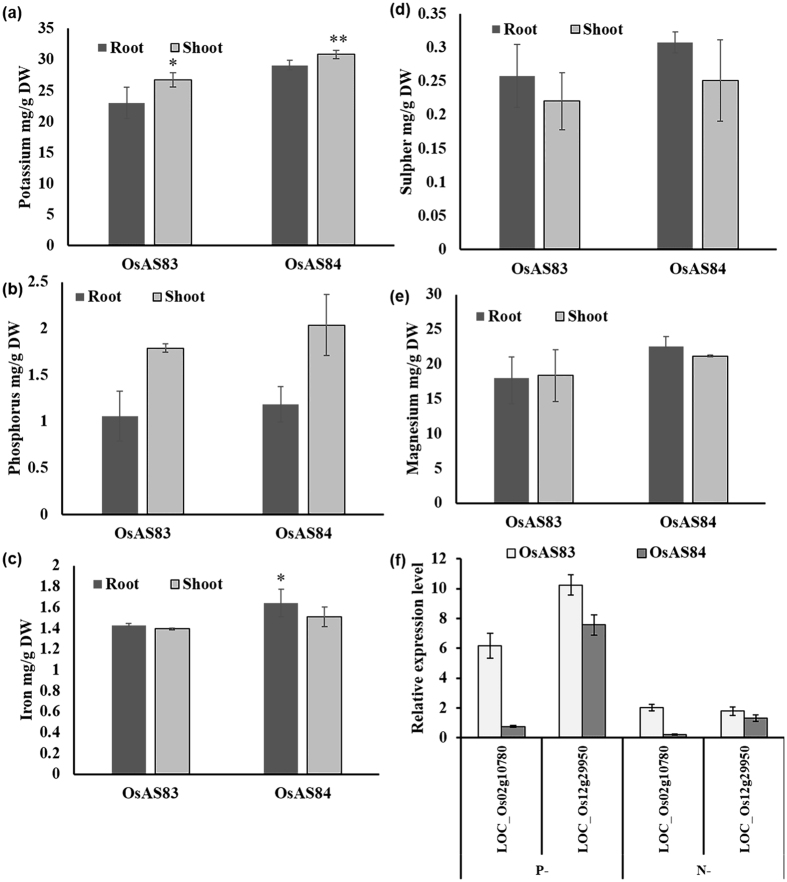
Differential expression of transporters may lead to differential acquisition of concerned micro/macro nutrients by roots. The concentration of various elements was measured in both roots and shoots of OsAS83 and OsAS84, which showed contrasting variation. We observed that the concentration of K, P, Fe, S, and Mg ions was more in both root and shoot tissue of OsAS84 than in OsAS83 (Fig. 4a–e). Higher concentration of various micro/macro elements in OsAS84 suggests that, this cultivar with advanced RSA has better ion absorption efficiency than OsAS83. More surface area of absorption (more root branches and RHs) and higher expression of several transporters (as indicated above) in OsAS84 roots could contribute to its enhanced ability of ion uptake. In same line, we have studied the expression of OsNCT and OsSPX2 in N and Pi deficit conditions, respectively. In N or Pi deprived conditions, the expression of these transporters were induced in both the cultivars in comparison to N or Pi sufficient conditions (Fig. 4f). Thus, some RSA traits and expression level of transporters may be associated with ion transport efficiency in rice cultivars.
Figure 4. OsAS84 shows better ion absorption efficiency.
(a–e) Concentration of K, P, Fe, S and Mg in root and shoot of OsAS83 and OsAS84. Error bars indicate SE (n = 3). (f) Expression of OsSPX2 (LOC_Os02g10780) and nitrate chloride transporter (LOC_Os12g29950) in Pi deficiency/N deficiency (Pi−/N−) conditions. For ΔΔCt study Pi+/N+ conditions were used as control. Error bars indicate SE (n = 3). Asterisks indicate significant statistical differences, ***P < 0.001, **P < 0.01, *P < 0.05 (One-way ANOVA).
Differential expression of hormone signaling genes is associated with RSA trait variation, in OsAS83 and OsAS84
Many genes involved in hormone signaling, are also involved in root development and in regulation of nutrient deficiency responses57. Hormone related genes like, ethylene responsive gene (LOC_Os02g43840), gibberellic acid stimulated transcript (GAST) (LOC_Os11g13670), and OsIAA20 (LOC_Os06g07040, an Auxin-responsive Aux/IAA gene family member) were differentially expressed in roots of two cultivars (Table 1). Similar to INDUCED BY PHOSPHATE STARVATION 1 (IPS1) of Arabidopsis, LOC_Os02g43840 of rice was also shown to have ability to modulate the activity of rice miR399, which is a key regulator of phosphate signaling58. Involvement of Ethylene Response Factors (ERFs) in Pi starvation was also supported by the observation that five members of ERFs showed upregulation in Pi deficient conditions59. Differential expression of LOC_Os02g43840 (a rice ERF having the potential to sequester to miR399) and LOC_Os02g10780 (OsSPX2, a key member of phosphate starvation-induced signaling), further strengthen the hypothesis that these two cultivar differs in their responses to Pi deficiency. Members of GA responsive, GAST gene family in rice and maize were shown to be expressed in RAM and in mutant defective in root branching, indicating their role in root development60. Role of Aux/IAA proteins in root development have been described previously13. In maize, a member of Aux/IAA domain containing protein, RUM1 (ROOTLESS WITH UNDETECTABLE MERISTEM 1) was shown to regulate crown and seminal root development61. Mutation in Arabidopsis IAA13 (At2g33310), a homolog of rice LOC_Os06g07040, resulted in seedlings with no root, due to failure of specification and abnormal cell division in the embryonic root meristem62. Differential accumulation of these genes in two cultivars indicates probable role of these genes in RSA regulation.
Recently it has been reported that overexpression of CYP71Z2 in rice leads to resistance to Xanthomonas oryzae and in reduction of OsIAA20 transcript level, indicating involvement of auxin signaling in disease resistance, besides its role in various aspects of plant development63. Majority of rice resistance (R) genes, which confer resistance to X. oryzae, belong to Nucleotide-Binding Site Leucine-Rich Repeat (NBS-LRR) or LRR Kinase super families64,65. In our microarray data, the expression of LOC_Os11g39190.1 and LOC_Os11g14110.1, two NBS-LRR domain containing proteins coding genes, were found to be upregulated in OsAS84 roots. Another member of cytochrome P450 family, CYP87A3 was found to be down regulated in OsAS84. This protein functions as a negative regulator for the auxin responsiveness of growth66. Accumulation of R genes, suppression of OsIAA20 and CYP genes, suggests that plant defense pathway and root developmental pathway share some common signaling mechanism, including hormone signaling.
Transcription factors involved in various signaling pathways are differentially expressed in OsAS83 and OsAS84 roots
Root transcriptome analysis of OsAS83 and OsAS84 revealed differential expression of many transcription factors involved in various signaling pathways, such as nutrient, pathogen, and stress responses, hormone signaling, development etc (Table 1). OsAS84 showed reduced expression of LOC_Os05g41540 (a bZIP transcription factor), LOC_Os05g14370.1 (WRKY82), LOC_Os02g42690 (a zinc finger, C3HC4 type domain containing protein) and LOC_Os06g33970.1 (a VQ domain containing protein) etc. Mutant of Arabidopsis homolog of LOC_Os05g41540, was significantly more sensitive to Zn depletion, and played role in uptake of Zn67. In plants, defense responsive pathways are regulated via several factors, and WRKY family proteins are one of the major regulators of this defense response68,69,70. Many of the WRKY genes are known to involved in jasmonic acid (JA) and salicyclic acid (SA) hormone mediated defense responsive pathways71. The expression of OsWRKY45, OsWRKY62, OsWRKY10, OsWRKY82, OsWRKY85, OsWRKY30, and OsWRKY83 was found to be responsive to SA and JA treatments71. Homolog of OsWRKY82 in Arabidopsis, WRKY70 was reported to be involved in osmotic stress, since double mutant of wrky54 wrky70 exhibited enhanced tolerance to osmotic stress72. Overexpression of ZmWRKY33 in Arabidopsis led to improved salt stress tolerance73. Overexpression of Arabidopsis homolog of LOC_Os02g42690, a E3 ubiquitin ligase RING membrane-anchor 1 (Rma1), conferred drought tolerance in Arabidopsis74. In silico analysis of few of the selected genes showed differential expression in various anatomical tissues of rice. Among the analyzed genes, expression of LOC_Os10g39260 (OsAPN), LOC_Os02g10780 (OsSPX2), LOC_Os02g42690 (OsZNC3HC4), LOC_Os06g03770 (OsABCT) and LOC_Os05g41540 (OsbZIP) were found to have higher in root tissues, which further suggests their probable role in root development (Fig. S4 and Table S1). Differential expression of these genes in contrasting roots of OsAS84 and OsAS83 indicate their involvement in rice root development and physiology. As discussed above, these transcription factors may also be recruited by plants in various other signaling pathways like nutrient transport, hormone regulated pathogen response, stress response, and root development, as a more economic strategy.
OsAS84 cultivar with advanced RSA shows less sensitivity to nutrient deficiency
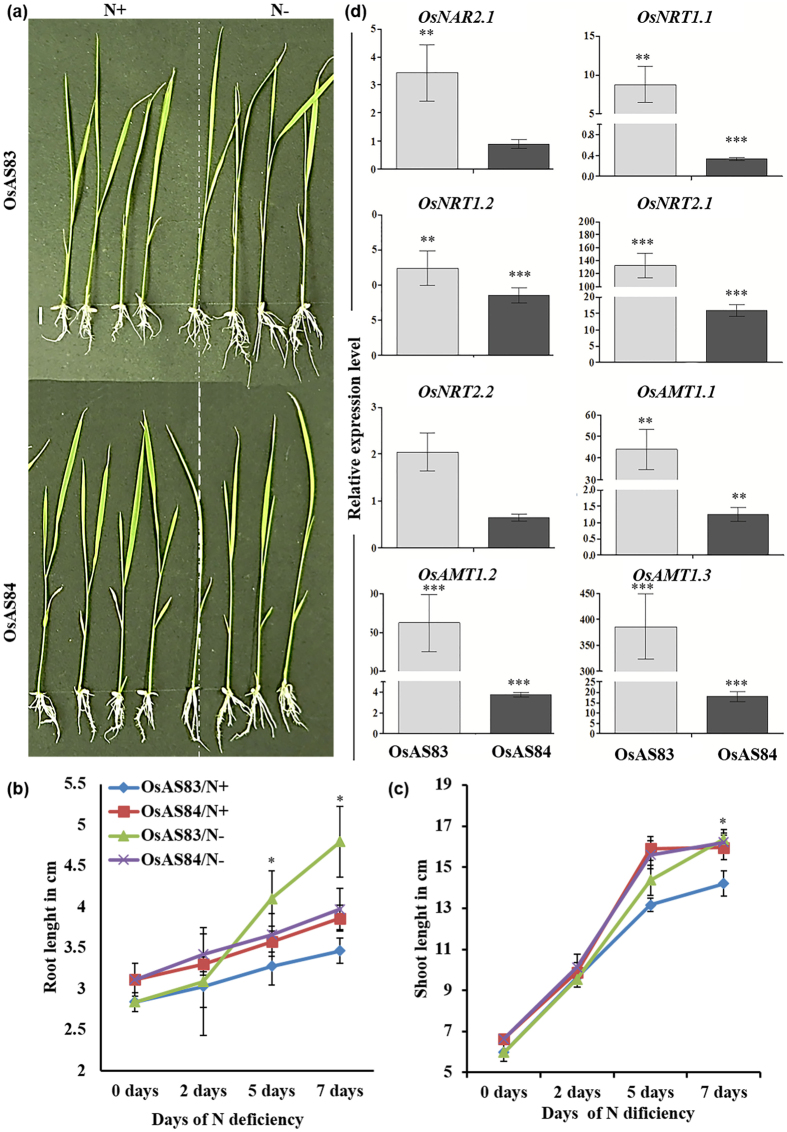
Increased RH density and stimulation of LR formation leads to enhanced nutrient uptake by increasing root surface area contact with soil. As OsAS84 RSA has higher number of CRs, high RH density in comparison to OsAS83, and differential expression of several transporters, we were interested to investigate the response of two varieties (with contrasting RSA) towards N and Pi nutrient deficit condition. Growth assay in N deficiency showed that RSA of OsAS84 was less sensitive in comparison to OsAS83, as indicated by increase in PR length by 2% and 38% increase, in case of OsAS84 and OsAS83, respectively (Fig. 5a and b). In case of shoot length, OsAS84 showed only 1.5% increase while OsAS83 showed 15% increase in comparison to nitrogen sufficient control condition (Fig. 5c).
Figure 5. OsAS84 showed less sensitivity towards nitrogen deficiency.
(a) Physiological responses of OsAS83 and OsAS84 to nitrogen deficiency. Scale bar 1 cm. (b,c) Primary root length and shoot length of OsAS83 and OsAS84 in N+ and N− conditions. Error bars represents standard error (n = 10). The experiment was repeated three times with similar results. (d) Expression pattern of OsNRTs and OsAMTs genes in root tissue harvested after 7th day, in both N deficit and normal conditions. Error bars indicate Standard error (n = 3). Asterisks indicate significant statistical differences, ***P < 0.001, **P < 0.01, *P < 0.05 (One-way ANOVA).
Differential expression of nitrogen (N) and ammonium transporter affects the ability of root to absorb nitrogen. Ammonium and nitrate ions are the basic source of N for the plants, which are absorbed by ammonium transporter (AMT family) and nitrogen transports (NRT family). To further investigate the molecular reason for differential response of these two cultivars to N deficiency, the expression of some of the ammonium/nitrogen transporters and their protein partner OsNAR2.1 was analysed. Rice seedlings of 3 dag were transferred to N sufficient and N deficit hydroponic media and RNA was isolated from the root tissues after 7th day of treatment. The member of AMT family in rice, OsAMT2.1, encodes an ammonium transporter which is expressed in root75. OsAMT1.1 and OsAMT1.3 are involved in influx of NH4+ 76. OsAMT1.1 and OsAMT2.3 showed varied expression in roots at different doses of nitrogen77,78. In rice, OsNRT2 (high-affinity) and a constitutively expressed OsNRT1 (low affinity) were identified as nitrogen transporters79,80. Both AMTs and NRTs along with OsNAR2.1 showed higher induction in expression level in OsAS83 in N deficiency (Figs 5d and S5). These results suggest that OsAS83 has to go through extensive transcriptional reprogramming to combat the N deficiency, which leads to drastic change in root growth pattern, while OsAS84 can withstand the N deficiency without any remarkable changes in root growth and transcriptional remodelling. This also suggests that root development and nitrogen transport mechanisms possess some common signalling crosstalk.
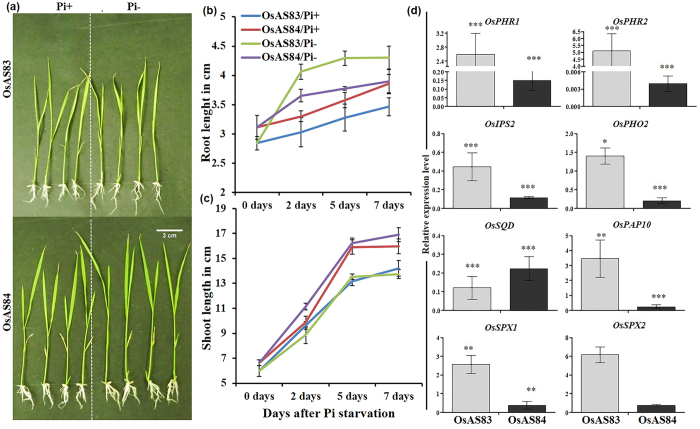
To enhance the capacity of plant to acquire more Pi from soil under Pi deficiency, the RSA gets altered showing formation of more LRs and increase in length and density of RH81,82. Growth assay under Pi deficiency showed that RSA of OsAS84 is less responsive in comparison to OsAS83, as PR length showed no difference and 24% increase, in case of OsAS84 and OsAS83, respectively (Fig. 6a and b). In OsAS84, presence of more root branches could be help to withstand the Pi starvation. In case of shoot length, OsAS84 showed only 5% increase while OsAS83 showed 4% decrease under Pi deficit condition (Fig. 6c). Plants adapt to low Pi by modulating the RSA for increasing topsoil foraging, through better Pi acquisition and efficient Pi utilization83. The adaptive strategies to cope up with Pi deficiency are tightly mediated by Pi signaling network and in rice, Pi starvation-induced signaling (PSI) is quite established. The central regulator of Pi signaling, OsPHR1 and OsPHR2 positively regulate OsIPS1, a non-protein coding gene which in turn sequesters miR39984. miR399 post-transcriptionally negatively regulates OsPHO2, which is involved in the enhancement of uptake and translocation of Pi. In root, OsSPX1 also negatively regulates OsIPS1 and other members of SPX family under Pi-supplied conditions85.
Figure 6. OsAS84 showed less sensitivity towards phosphate deficiency.
(a) Physiological responses of OsAS83 and OsAS84 to Pi deficiency. Scale bar 1 cm. (b,c) Primary root and shoot length of OsAS83 and OsAS84 grown under Pi + and Pi- conditions. Error bars represent standard error (n = 10). The experiment was repeated three times with similar results. (d) Expression pattern of PSI genes in root tissue harvested after 7th day, in both N deficit and normal conditions. Error bars indicates Standard error (n = 3). Asterisks indicate significant statistical differences, ***P < 0.001, **P < 0.01, *P < 0.05 (One-way ANOVA).
Since OsAS83 and OsAS84 cultivars showed significantly variability in terms of alteration of RSA in response to Pi deficiency, we also investigated the expression of PSI genes in roots of both the cultivars. Rice seedlings at 3 dag were transferred to Pi sufficient and Pi deficit hydroponic media and RNA was isolated from the root tissues after 7th day of treatment. OsSPX2 expression was highly induced in Pi sufficient conditions in OsAS84 roots, while in Pi deficiency, expression of OsSPX2 was found to be more induced in OsAS83 roots, which is also true in case of other PSI genes such as OsPHO2, OsPHR2, OsPAP10, and OsSQD (Fig. 6d and Fig. S6). This indicates that both varieties have differential regulation of PSI signaling pathway. OsSPX1 negatively regulates Pi accumulation in shoots and acts as a positive regulator of OsSPX2, OsSPX3 and OsSPX585. The expression of OsSPX1 was lower in Pi sufficient conditions and the level of Pi in shoot was higher in case of OsAS84 (Fig. S6), which indicates that OsSPX1 negatively regulates Pi accumulation in OsAS84 similar to previous published report85. Level of induction of OsSPX1 was also lower in OsAS84 roots, in comparison to OsAS83, under Pi starvation. The central regulator of PSI signaling, OsPHR1 and OsPHR2, showed higher level of expression in roots of OsAS84 under Pi sufficient conditions, however, under Pi starvation, the expression level of these two genes was downregulated (Fig. 6d). The level of OsIPS2 expression was more in OsAS84 roots under Pi sufficient condition while its expression level was reduced more in OsAS84 under Pi deficit conditions (Fig. 6d and Fig. S6). Target of rice miR399, OsPHO2 also followed a similar pattern of expression (Fig. 6d). These results suggest that OsAS84 is more resistant to Pi starvation, which is possibly due to naturally existing more robust RSA than OsAS83. This also suggests that plants recruit various common regulators for RSA trait and PSI pathway and that nutrient acquisition and RSA trait are strongly linked at molecular level.
OsAS84 with advanced RSA shows increased resistance to abiotic stresses
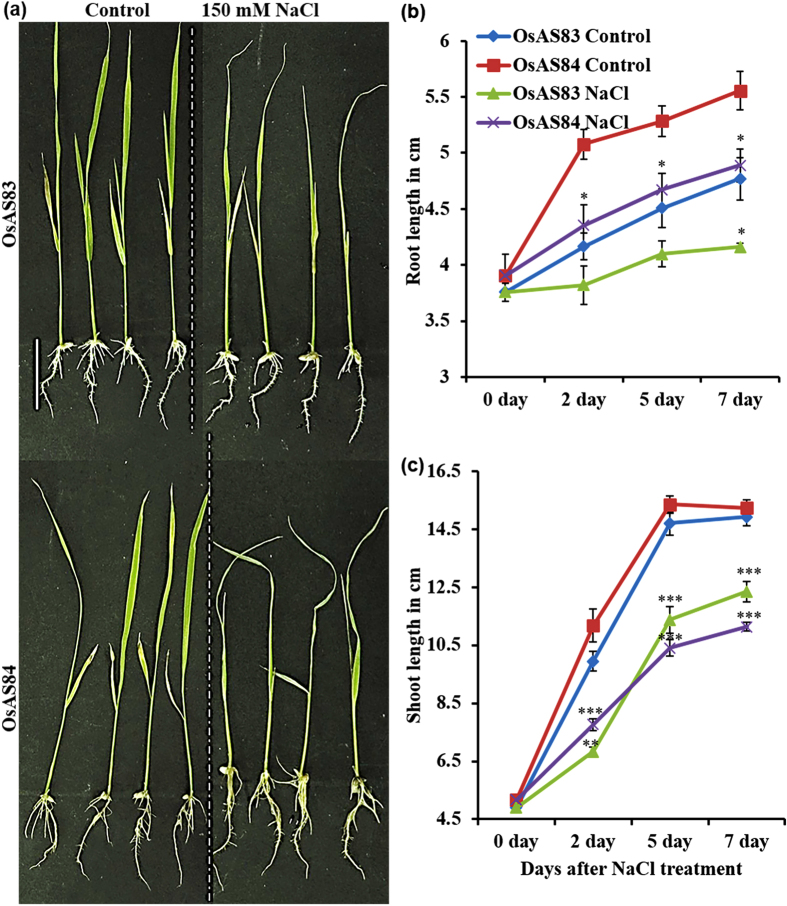
Several genes related to dehydration, salt or oxidative stress tolerance were found to be upregulated in OsAS84 roots, for e.g. genes involved in trehalose and ethylene biosynthesis, peroxidase, aspartic proteinase, ERE-binding protein, and NADH dehydrogenase. Abundant expression of these genes in advanced RSA of OsAS84 are indicative of its possible resistance to abiotic stresses. To test this hypothesis, we first analyzed the response of these two cultivars toward salinity stress. When 3 dag old seedlings of both cultivars were subjected to high salt conditions, OsAS84 showed less sensitivity to salinity stress, as evident by less wilting and yellowing in leaves in comparison to OsAS83 (Fig. 7a). Overall root growth was also found to be less affected in OsAS84. Development of seminal/CRs was less affected in OsAS84 than OsAS83 (Fig. S7). The PR length was reduced by 13% and 12%, while shoot length was reduced by 18% and 27% in OsAS83 and OsAS84, respectively (Fig. 7b and c). Under salinity stress, despite decrease in PR length, OsAS84 showed healthy shoots, which could be due to increased number and length of CR and LR. In OsAS84, the shoot length was reduced as a primary response to the salinity stress (osmotic phase), but the leaves showed less wilting and yellowing, and rate of dying was also slower (ion-specific, phase) in comparison to OsAS8386. These observations suggest that OsAS84 cultivar having robust RSA is more resistance to salinity stress than OsAS83, and indicate a positive correlation between RSA and stress response.
Figure 7. OsAS83 and OsAS4 shows variable physiological responses under salinity stress.
(a) Phenotypic variation in both the cultivars after treatment of 150 mM NaCl for 7 days. Scale bar 1 cm. (b,c) Primary root and shoot length of OsAS83 and OsAS84 at 0 days, 2 days, 5 days and 7 days after treatment. Error bars represents standard error (n = 10). The experiment was repeated three times with reproducible results. Asterisks indicate significant statistical differences, ***P < 0.001, **P < 0.01, *P < 0.05 (One-way ANOVA).
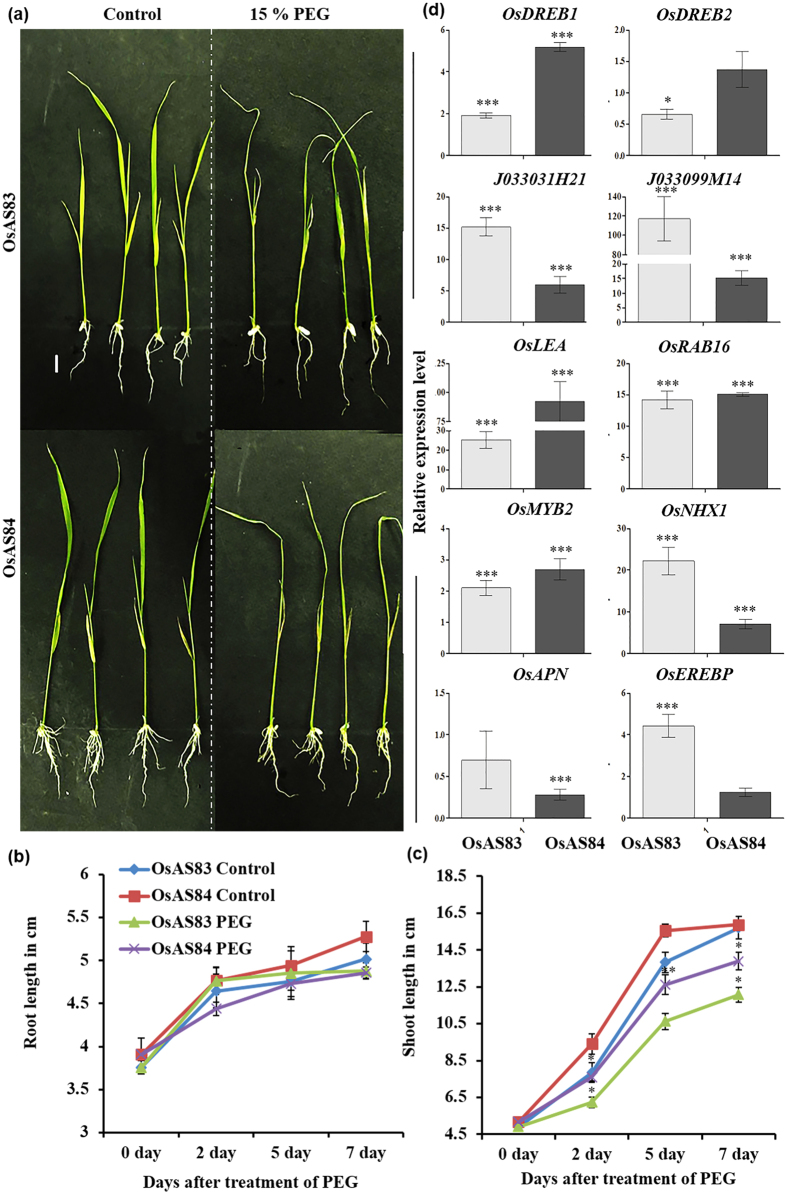
Then, we analyzed the response of these two cultivars to water deficit conditions. When 3 days old seedlings were subjected to dehydration stress (Fig. 8a and Fig. S8a), we observed 8% and 4% reduction in PR length and 12% and 24% reduction in shoot length in case of OsAS84 and OsAS83, respectively (Fig. 8b and c). Besides decrease in PR length, overall RSA of OsAS84 was found to be healthy (more branches) than OsAS83 under dehydration stress. Leaves of OsAS84 were found to be less affected by dehydration stress in comparison to OsAS83. These results indicate that OsAS84 with advance RSA is comparatively less sensitive to dehydration stress than OsAS83. We further analyzed if this dehydration response accompanied changes in the expression of relevant genes in roots of these cultivars. For this, 3 dag seedlings of OsAS83 and OsAS84 were transferred to control and 15% PEG containing hydroponic media and qRT-PCR was done using cDNA prepared from RNA isolated from the root tissues after 7th day of treatment. We observed that some of the dehydration responsive genes like OsMYB2, LATE EMBRYOGENESIS ABUNDANT(OsLEA), DEHYDRATION-RESPONSIVE ELEMENT (OsDREB1) and OsDREB2 were upregulated in OsAS84 roots in comparison to OsAS83, while OsNHX1 (maintain homeostasis in salt and water stress) and amino acid kinase genes, J033099M14 and J033031H21 (proline biosynthesis genes) were downregulated in OsAS84 under dehydration stress (Fig. 8d). Plants used several mechanisms to combat dehydration stress, such as ABA dependent/independent and DREB mediated signaling and accumulation of osmotolerant compounds like proline or glycine betaine. Upregulation of OsMYB2, OsLEA, OsDREB1 and OsDREB2 genes suggests that upon exposure to dehydration stress OsAS84 activates OsDREB2- dependent signaling while genes involved in proline accumulation were not induced in OsAS84 upon dehydration stress, indicating differential signaling pathways respond differently in OsAS83 and OsAS84 upon dehydration stress.
Figure 8. OsAS84 showed higher resistance to dehydration stress than OsAS83.
Physiological responses of OsAS83 and OsAS84 upon dehydration stress. (a) 3 dag seedlings of both the cultivars were treated with 15% Polyethylene glycol 6000 (PEG 6000) for 7 days in hydroponics. Scale bar 1 cm. (b,c) Primary root and shoot length of OsAS83 and OsAS84 in control and PEG conditions at 0, 2, 5, and 7 days. Error bars represents standard error (n = 10). The experiment was repeated three times with reproducible results. (d) Expression of dehydration responsive genes in root tissue harvested after 7 day of treatment. Error bars represent Standard error (n = 3). Asterisk indicates significant statistical differences, ***P < 0.001, **P < 0.01, *P < 0.05 (One-way ANOVA).
Under normal condition, dehydration responsive genes were upregulated in OsAS84 in comparison to OsAS83 (Fig. S8b). In our microarray analysis, dehydration responsive genes such as LOC_Os10g39260 (OsAPN) and LOC_Os02g43840 (OsEREBP) were differentially expressed in OsAS84 roots. Expression of both of these genes was higher in OsAS84 in normal conditions, but their expression was higher in OsAS83 (Fig. 8d and Fig. S8b) under dehydration stress. This indicates that OsAPN and OsEREBP, along with other dehydration responsive genes, probably act as molecular regulators for enhanced response towards dehydration stress in OsAS84. The expression pattern of dehydration responsive genes, under control and dehydration stress, indicate these genes play important role in conferring resistance to dehydration stress, and that sensitive OsAS83 with poor RSA had to undergo more rigorous changes at the relevant transcript level to combat dehydration stress through changes in RSA and physiology.
Phytohormones differentially regulates root growth in two cultivars
It is well known that phytohormones play very critical role in development of PR as well as CRs/LRs in rice or Arabidopsis. It has been shown previously with avr1 and crl1 mutants studies in rice that auxin promotes initiation of CRs13. Cytokinins, which act antagonistically to auxin, also play important role in controlling the initiation of LR primordia by affecting auxin distribution87,88,89. Cytokinin signaling works through WOX11 and ARABIDOPSIS RESPONSE REGULATORS) (ARRs) in rice and overexpression of OsRR6 (CK-inducible type-A response regulator) led to decreased LR outgrowth90. Abscisic acid (ABA) is known to regulate LR development differently under normal (low level of exogenous ABA) and stressed conditions91. Ethylene has also been reported to influence the LR development by stimulation of auxin biosynthesis92,93.
To understand the effect of phytohormones on RSA of two contrasting rice cultivars, we analyzed the difference in root and shoot growth pattern between two cultivars after treatment with various phytohormones (1 μM ABA, BAP, IAA and GA (Fig. 9S). Treatment of 1 μM ABA led to enhancement of root and shoot growth in OsAS83 by 33% and 7%, respectively. With same treatment in OsAS84, we observed 3% increase and 20% decrease in root and shoot length, respectively (Fig. S9b). ABA treatment also increased LR number and length in OsAS83 in comparison to control plant. In BAP (a CK) treatment, both the cultivars showed reduction in root and shoot growth, but OsAS84 show more reduction in both root (14%) and shoot growth (7%) in comparison to OsAS83, in which only 2.5% reduction was observed in root growth after 7 days of treatment (Fig. S9c). IAA showed stimulatory effect on root growth in both cultivars (2% increase in root length), since low dose of auxin is known to promote root growth (Fig. S9d). GA led to significant increase in shoot length by 118% and 96% in OsAS83 and OsAS84, respectively. However, treatment with 1 μM of GA led to 10% increase in root growth in OsAS83 and 18.5% reduction in root growth in OsAS84 (Fig. S9e). Although a matter of further study, the differential expression of several hormonal regulatory pathways related genes (such as, LOC_Os02g43840, LOC_Os11g13670, and LOC_Os06g07040) in roots of two cultivars (with contrasting RSA) may contribute to their variable responses to phytohormones. Similarly, hormonal regulatory pathway genes may also contribute largely to the variation in RSA trait observed among rice cultivars.
In the present study we have identified and analyzed two indica rice cultivars - OsAS83 and OsAS84, which differ significantly in their RSA traits and responses towards abiotic stresses and nutrient deficiency. Based on our gene expression microarray studies and marker analysis, we propose that the stable contrasting variation in RSA trait between these two cultivars is potentially conferred by the differential expression of various transcripts in their root. Both the cultivars (with robust or shallow RSA) behaved differently in nutrient deficiency and in abiotic stresses. We propose that longer PR, more number of CRs/LRs, and higher RH density makes OsAS84 cultivar more adaptive to nutrient deficiencies and other abiotic stresses (salinity/dehydration). Differential expression pattern of nitrogen transporters, PSI signaling genes and dehydration responsive genes in roots of rice cultivars indicates a strong correlation between molecular regulation of RSA trait and physiological responses in rice. Our results also indicate that plants may recruit its common molecular machinery, at least in part, for different regulatory pathways like root development, nutrient signaling, abiotic stresses or pathogen responses etc. Further elaborate studies of this common regulatory mechanism could be helpful to develop rice varieties with improved developmental and physiological traits of agronomic importance.
Methods
Plant materials, growth conditions and treatment
For rice study, IR-64 and IET-16348 seeds were germinated and grown in hydroponics media under controlled conditions (16 h:8 h::light:dark cycle, 28 °C ± 1 °C, light intensity 250 μmol m−2s−1)94. For Pi and N deficiency, seedlings were kept in hydroponics Yoshida medium lacking K2HPO4/KH2PO4 stocks and NH4NO3 stock, respectively94. For dehydration and salinity stress, hydroponics medium was supplemented with 15% of PEG 6000 and 150 mM NaCl, respectively. For hormone treatment, rice seedlings were grown in hydroponics media supplemented with various hormones (1 or 10 μM of ABA, BAP, GA, and IAA).
Detection of macro or micro elements
Plants were raised in laterite soil with farmyard manure (6:1 ratio) having a water holding capacity of 22%. Recommended dose of fertilizer was added to the soil in the form of urea, super phosphate and murate of Potash. Plants were harvested on 35th day for analysis. Samples were ground to a fine powder and acid digested using nitric acid and perchloric acid. Double distilled water was added to the digested samples and filtered using 0.45 μm filter paper and a vacuum pump. These samples were analyzed for metals using ICP-OES (Thermo Scientific iCAP 6000). In this, liquid samples are injected and atomic emission was recorded. Measurements were made using monochromator/photomultiplier combination for P and polychromator and an array detector combination for the Mg, K, Fe, Zn, and S.
RNA isolation and qRT-PCR
RNA was isolated from the different tissues of rice seedling using the TRI-reagent (Sigma) at different time intervals and was further treated with RNase-free DNase I (Thermo Scientific) as per manufacturer’s instructions. cDNA preparation and qRT-PCR was performed using the previously described method95. After checking the quality and spectrophotometric quantification, 2 μg RNA were converted into cDNA using M-MLV Reverse Transcriptase (Invitrogen, USA) following the manufacture’s protocol. Each sample was analyzed in triplicate using at least two cDNA preparations by qRT-PCR using 2X Brilliant II SYBR® Master Mix (Agilent Technologies, USA). PCR reaction was placed in ABI PRISM 7900 HT Fast Real Time PCR System (Applied Biosystems, USA). The specificity of the reactions was verified by carrying out melting (dissociation) curve analysis. GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (GAPDH) and ACTIN 11 were used as endogenous control. Primer sequences, which were used in study, are given in Table S2.
Gene chip Microarray
Total RNA was extracted from 14 days old root tissue of OsAS83 and OsAS84. Quality of total RNA was analyzed by Agilent 2100 Bioanalyser and RIN values were above 7. RNA (250 ng) was used from each sample in two biological replicates for setting up in-vitro transcription reaction; RNA purification, fragmentation and hybridization reaction was done using O. sativa gene chip arrays (Affymetrix). The array contains 51,279 independent probes corresponding two rice cultivars japonica and indica (http://www.affymetrix.com). Washing and scanning were followed as suggested in Affymetrix Gene Chip total RNA procedure. The processed raw signal intensities were subjected to normalization using the Gene spring GX software v11.5. Result obtained after scanning were analyzed using Gene Spring software. Expression of selected genes differentially expressed in microarray was further validated through real time qRT-PCR (as mentioned above).
Histological analysis
Primary root tips (0–1 cm) of 2 days old seedlings of OsAS83 and OsAS84 were serially sectioned by free hand. Sections were stained with 0.1% Safranin stain (Sigma) and were visualized under bright field microscope (Nikon 80i). Cell size and diameter was calculated using IMAGEJ software.
Additional Information
How to cite this article: Singh, A. et al. Root transcriptome of two contrasting indica rice cultivars uncovers regulators of root development and physiological responses. Sci. Rep. 6, 39266; doi: 10.1038/srep39266 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank Department of Biotechnology (DBT), India (grant no. BT/PR3292/AGR/2/811/2011) and National Institute of Plant Genome Research (NIPGR) for financial support and central instrument facility of NIPGR. Fellowship to AS was provided by DBT and fellowship to PK and VG was provided by Council of Scientific and Industrial Research (CSIR), India. BR was supported by above DBT grant and NIPGR fellowship. We also thank Suvakanta Barik and M. Karthikeyan for help in preliminary stage of the work.
Footnotes
Author Contributions A.S. and P.K. performed major experiments and wrote manuscript. V.G., M.U. and B.R. performed initial rice cultivars screening. M.U. laboratory performed I.C.P. analysis of elements. B.A. and M.U. provided some of the germplasms and provided scientific inputs on rice cultivars. A.K.S. conceptualized and designed the experiments, and revised the manuscript.
References
- Kumar A. et al. Breeding high-yielding drought-tolerant rice: genetic variations and conventional and molecular approaches. J Exp Bot. 65, 6265–78 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois B. et al. Genome-wide association mapping of root traits in a japonica rice panel. PLoS One. 8, e78037 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissuwa M. & Ae N. Genotypic variation for tolerance to phosphorus deficiency in rice and the potential for its exploitation in rice improvement. Plant Breeding. 120, 43–48 (2001). [Google Scholar]
- Kavitha P. G., Miller A. J., Mathew M. K. & Maathuis F. J. Rice cultivars with differing salt tolerance contain similar cation channels in their root cells. J Exp Bot. 63, 3289–96 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A. K., Mayandi K., Gautam V., Barik S. & Sarkar Das S. Improving the Plant Root System Architecture to Combat Abiotic Stresses Incurred as a Result of Global Climate Changes. In Climate Change and Plant Abiotic Stress Tolerance 305–324 (Wiley-VCH Verlag GmbH & Co. KGaA, 2013). [Google Scholar]
- Yu P., Gutjahr C., Li C. & Hochholdinger F. Genetic Control of Lateral Root Formation in Cereals. Trends Plant Sci. (2016). [DOI] [PubMed] [Google Scholar]
- Hardtke C. S. Root development–branching into novel spheres. Curr Opin Plant Biol. 9, 66–71 (2006). [DOI] [PubMed] [Google Scholar]
- Sarkar A. K. et al. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature. 446, 811–4 (2007). [DOI] [PubMed] [Google Scholar]
- Zhao Y., Hu Y., Dai M., Huang L. & Zhou D. X. The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. Plant Cell. 21, 736–48 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. H. et al. The rice narrow leaf2 and narrow leaf3 loci encode WUSCHEL-related homeobox 3A (OsWOX3A) and function in leaf, spikelet, tiller and lateral root development. New Phytol. 198, 1071–84 (2013). [DOI] [PubMed] [Google Scholar]
- Kamiya N., Itoh J., Morikami A., Nagato Y. & Matsuoka M. The SCARECROW gene’s role in asymmetric cell divisions in rice plants. Plant J. 36, 45–54 (2003). [DOI] [PubMed] [Google Scholar]
- Gao S. et al. CYTOKININ OXIDASE/DEHYDROGENASE4 Integrates Cytokinin and Auxin Signaling to Control Rice Crown Root Formation. Plant Physiol. 165, 1035–1046 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inukai Y. et al. Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell. 17, 1387–96 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D. et al. Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene. 394, 13–24 (2007). [DOI] [PubMed] [Google Scholar]
- Okushima Y., Fukaki H., Onoda M., Theologis A. & Tasaka M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell. 19, 118–30 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmoth J. C. et al. NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 43, 118–30 (2005). [DOI] [PubMed] [Google Scholar]
- Laplaze L. et al. GAL4-GFP enhancer trap lines for genetic manipulation of lateral root development in Arabidopsis thaliana. J Exp Bot. 56, 2433–42 (2005). [DOI] [PubMed] [Google Scholar]
- Shen C. et al. OsARF16, a transcription factor, is required for auxin and phosphate starvation response in rice (Oryza sativa L.). Plant Cell Environ. 36, 607–20 (2013). [DOI] [PubMed] [Google Scholar]
- Jun N. et al. OsIAA23-mediated auxin signaling defines postembryonic maintenance of QC in rice. Plant J. 68, 433–42 (2011). [DOI] [PubMed] [Google Scholar]
- Lopez-Bucio J., Cruz-Ramirez A. & Herrera-Estrella L. The role of nutrient availability in regulating root architecture. Curr Opin Plant Biol. 6, 280–7 (2003). [DOI] [PubMed] [Google Scholar]
- Williamson L. C., Ribrioux S. P., Fitter A. H. & Leyser H. M. Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiol. 126, 875–82 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bucio J. et al. Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol. 129, 244–56 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai V. et al. Iron Availability Affects Phosphate Deficiency-Mediated Responses, and Evidence of Cross-Talk with Auxin and Zinc in Arabidopsis. Plant Cell Physiol. 56, 1107–23 (2015). [DOI] [PubMed] [Google Scholar]
- Yi K. et al. OsPTF1, a novel transcription factor involved in tolerance to phosphate starvation in rice. Plant Physiol. 138, 2087–96 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y. F. et al. Responses of root architecture development to low phosphorus availability: a review. Ann Bot. 112, 391–408 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borch K., Bouma T. J., Lynch J. P. & Brown K. M. Ethylene: a regulator of root architectural responses to soil phosphorus availability. Plant, Cell & Environment. 22, 425–431 (1999). [Google Scholar]
- Nacry P. et al. A role for auxin redistribution in the responses of the root system architecture to phosphate starvation in Arabidopsis. Plant Physiol. 138, 2061–74 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Calderon L. et al. Phosphate starvation induces a determinate developmental program in the roots of Arabidopsis thaliana. Plant Cell Physiol. 46, 174–84 (2005). [DOI] [PubMed] [Google Scholar]
- Zhang J., Xu L., Wang F., Deng M. & Yi K. Modulating the root elongation by phosphate/nitrogen starvation in an OsGLU3 dependant way in rice. Plant Signal Behav. 7, 1144–5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun L., Mi G., Li J., Chen F. & Zhang F. Genetic Analysis of Maize Root Characteristics in Response to Low Nitrogen Stress. Plant and Soil. 276, 369–382 (2005). [Google Scholar]
- Parent B., Suard B., Serraj R. & Tardieu F. Rice leaf growth and water potential are resilient to evaporative demand and soil water deficit once the effects of root system are neutralized. Plant Cell Environ. 33, 1256–67 (2010). [DOI] [PubMed] [Google Scholar]
- Jeong J. S. et al. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 153, 185–97 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J. S. et al. OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol J. 11, 101–14 (2013). [DOI] [PubMed] [Google Scholar]
- Yao X., Xiong W., Ye T. & Wu Y. Overexpression of the aspartic protease ASPG1 gene confers drought avoidance in Arabidopsis. J Exp Bot. 63, 2579–93 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Ingram P. A., Benfey P. N. & Elich T. From lab to field, new approaches to phenotyping root system architecture. Curr Opin Plant Biol. 14, 310–7 (2011). [DOI] [PubMed] [Google Scholar]
- Gautam V., Singh A., Singh S. & Sarkar A. K. An Efficient LCM-Based Method for Tissue Specific Expression Analysis of Genes and miRNAs. Sci Rep. 6, 21577 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebouillat J. et al. Molecular Genetics of Rice Root Development. Rice. 2, 15–34 (2009). [Google Scholar]
- Uga Y. et al. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat Genet. 45, 1097–102 (2013). [DOI] [PubMed] [Google Scholar]
- van de Mortel J. E. et al. Large expression differences in genes for iron and zinc homeostasis, stress response, and lignin biosynthesis distinguish roots of Arabidopsis thaliana and the related metal hyperaccumulator Thlaspi caerulescens. Plant Physiol. 142, 1127–47 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazaki K. Transporters of secondary metabolites. Curr Opin Plant Biol. 8, 301–7 (2005). [DOI] [PubMed] [Google Scholar]
- Sun X. et al. ADS1 encodes a MATE-transporter that negatively regulates plant disease resistance. New Phytol. 192, 471–82 (2011). [DOI] [PubMed] [Google Scholar]
- Liu J., Magalhaes J. V., Shaff J. & Kochian L. V. Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J. 57, 389–99 (2009). [DOI] [PubMed] [Google Scholar]
- Debeaujon I., Peeters A. J., Leon-Kloosterziel K. M. & Koornneef M. The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell. 13, 853–71 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrett T. P., Gassmann W. & Rogers E. E. The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol. 144, 197–205 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes J. V. et al. A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat Genet. 39, 1156–61 (2007). [DOI] [PubMed] [Google Scholar]
- Furukawa J. et al. An aluminum-activated citrate transporter in barley. Plant Cell Physiol. 48, 1081–91 (2007). [DOI] [PubMed] [Google Scholar]
- Ryan P. R., Raman H., Gupta S., Horst W. J. & Delhaize E. A second mechanism for aluminum resistance in wheat relies on the constitutive efflux of citrate from roots. Plant Physiol. 149, 340–51 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maron L. G. et al. Two functionally distinct members of the MATE (multi-drug and toxic compound extrusion) family of transporters potentially underlie two major aluminum tolerance QTLs in maize. Plant J. 61, 728–40 (2010). [DOI] [PubMed] [Google Scholar]
- Tiwari M., Sharma D., Singh M., Tripathi R. D. & Trivedi P. K. Expression of OsMATE1 and OsMATE2 alters development, stress responses and pathogen susceptibility in Arabidopsis. Scientific Reports. 4, 3964 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. Y. et al. AtATM3 is involved in heavy metal resistance in Arabidopsis. Plant Physiol. 140, 922–32 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuiyan M. S. U. et al. Overexpression of AtATM3 in Brassica juncea confers enhanced heavy metal tolerance and accumulation. Plant Cell, Tissue and Organ Culture (PCTOC). 107, 69–77 (2011). [Google Scholar]
- Patil G. et al. Soybean (Glycine max) SWEET gene family: insights through comparative genomics, transcriptome profiling and whole genome re-sequence analysis. BMC Genomics. 16, 520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. Q. et al. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science. 335, 207–11 (2012). [DOI] [PubMed] [Google Scholar]
- Chu Z. et al. Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev. 20, 1250–5 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antony G. et al. Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os-11N3. Plant Cell. 22, 3864–76 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taji T. et al. Comparative genomics in salt tolerance between Arabidopsis and aRabidopsis-related halophyte salt cress using Arabidopsis microarray. Plant Physiol. 135, 1697–709 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T., Kudo T., Kojima M. & Sakakibara H. Hormonal control of nitrogen acquisition: roles of auxin, abscisic acid, and cytokinin. J Exp Bot. 62, 1399–409 (2011). [DOI] [PubMed] [Google Scholar]
- Meng Y., Shao C., Wang H. & Jin Y. Target mimics: an embedded layer of microRNA-involved gene regulatory networks in plants. BMC Genomics. 13, 197 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T., Suzuki K., Fujimura T. & Shinshi H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140, 411–32 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann R., Sakai H. & Hochholdinger F. The Gibberellic Acid Stimulated-Like gene family in maize and its role in lateral root development. Plant Physiol. 152, 356–65 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. et al. The Aux/IAA gene rum1 involved in seminal and lateral root formation controls vascular patterning in maize (Zea mays L.) primary roots. J Exp Bot. 65, 4919–30 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D. et al. Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. Embo j. 24, 1874–85 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. et al. Overexpressing CYP71Z2 enhances resistance to bacterial blight by suppressing auxin biosynthesis in rice. PLoS One. 10, e0119867 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwessinger B. & Ronald P. C. Plant innate immunity: perception of conserved microbial signatures. Annu Rev Plant Biol. 63, 451–82 (2012). [DOI] [PubMed] [Google Scholar]
- Sun X. et al. Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J. 37, 517–27 (2004). [DOI] [PubMed] [Google Scholar]
- Chaban C., Waller F., Furuya M. & Nick P. Auxin Responsiveness of a Novel Cytochrome P450 in Rice Coleoptiles. Plant Physiology. 133, 2000–2009 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba S. et al. Identification of putative target genes of bZIP19, a transcription factor essential for Arabidopsis adaptation to Zn deficiency in roots. Plant J. 84, 323–34 (2015). [DOI] [PubMed] [Google Scholar]
- Eulgem T., Rushton P. J., Schmelzer E., Hahlbrock K. & Somssich I. E. Early nuclear events in plant defence signalling: rapid gene activation by WRKY transcription factors. Embo j. 18, 4689–99 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulker B. & Somssich I. E. WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol. 7, 491–8 (2004). [DOI] [PubMed] [Google Scholar]
- Zhang Y. & Wang L. The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol Biol. 5, 1 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H. S. et al. A comprehensive expression analysis of the WRKY gene superfamily in rice plants during defense response. Plant Cell Rep. 25, 836–47 (2006). [DOI] [PubMed] [Google Scholar]
- Li J. et al. Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytol. 200, 457–72 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. ZmWRKY33, a WRKY maize transcription factor conferring enhanced salt stress tolerances in Arabidopsis. Plant Growth Regulation. 70, 207–216 (2013). [Google Scholar]
- Lee H. K. et al. Drought stress-induced Rma1H1, a RING membrane-anchor E3 ubiquitin ligase homolog, regulates aquaporin levels via ubiquitination in transgenic Arabidopsis plants. Plant Cell. 21, 622–41 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suenaga A. et al. Constitutive expression of a novel-type ammonium transporter OsAMT2 in rice plants. Plant Cell Physiol. 44, 206–11 (2003). [DOI] [PubMed] [Google Scholar]
- Kumar A., Silim S. N., Okamoto M., Siddiqi M. Y. & Glass A. D. Differential expression of three members of the AMT1 gene family encoding putative high-affinity NH4+ transporters in roots of Oryza sativa subspecies indica. Plant Cell Environ. 26, 907–914 (2003). [DOI] [PubMed] [Google Scholar]
- Gazzarrini S. et al. Three functional transporters for constitutive, diurnally regulated, and starvation-induced uptake of ammonium into Arabidopsis roots. Plant Cell. 11, 937–48 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur V., Singh U. S., Gupta A. & Kumar A. Influence of different nitrogen inputs on the members of ammonium transporter and glutamine synthetase genes in two rice genotypes having differential responsiveness to nitrogen. Molecular Biology Reports. 39, 8035–8044 (2012). [DOI] [PubMed] [Google Scholar]
- Li B., Xin W., Sun S., Shen Q. & Xu G. Physiological and Molecular Responses of Nitrogen-starved Rice Plants to Re-supply of Different Nitrogen Sources. Plant and Soil. 287, 145–159 (2006). [Google Scholar]
- Lin C. M. et al. Cloning and functional characterization of a constitutively expressed nitrate transporter gene, OsNRT1, from rice. Plant Physiol. 122, 379–88 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svistoonoff S. et al. Root tip contact with low-phosphate media reprograms plant root architecture. Nat Genet. 39, 792–6 (2007). [DOI] [PubMed] [Google Scholar]
- Perez-Torres C. A. et al. Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. Plant Cell. 20, 3258–72 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch J. P. Root phenes for enhanced soil exploration and phosphorus acquisition: tools for future crops. Plant Physiol. 156, 1041–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J. et al. OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol. 146, 1673–86 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. et al. Regulation of OsSPX1 and OsSPX3 on expression of OsSPX domain genes and Pi-starvation signaling in rice. J Integr Plant Biol. 51, 663–74 (2009). [DOI] [PubMed] [Google Scholar]
- Munns R. & Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 59, 651–81 (2008). [DOI] [PubMed] [Google Scholar]
- Sabatini S. et al. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell. 99, 463–72 (1999). [DOI] [PubMed] [Google Scholar]
- Dello Ioio R. et al. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol. 17, 678–82 (2007). [DOI] [PubMed] [Google Scholar]
- Moubayidin L. et al. The rate of cell differentiation controls the Arabidopsis root meristem growth phase. Curr Biol. 20, 1138–43 (2010). [DOI] [PubMed] [Google Scholar]
- Hirose N., Makita N., Kojima M., Kamada-Nobusada T. & Sakakibara H. Overexpression of a type-A response regulator alters rice morphology and cytokinin metabolism. Plant Cell Physiol. 48, 523–39 (2007). [DOI] [PubMed] [Google Scholar]
- Cheng W. H. et al. A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell. 14, 2723–43 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmont K. S., Sibout R. & Hardtke C. S. Hidden branches: developments in root system architecture. Annu Rev Plant Biol. 58, 93–113 (2007). [DOI] [PubMed] [Google Scholar]
- Stepanova A. N. & Alonso J. M. Ethylene signaling and response: where different regulatory modules meet. Curr Opin Plant Biol. 12, 548–55 (2009). [DOI] [PubMed] [Google Scholar]
- Yoshida Kaoru T. & Makoto Sakata S. F. Genkichi Takeda. Control of Organogenesis and Embryogenesis in Rice Calli. Japanese Journal of Breeding. 44, 355–360 (1994). [Google Scholar]
- Singh A., Singh S., Panigrahi K. C., Reski R. & Sarkar A. K. Balanced activity of microRNA166/165 and its target transcripts from the class III homeodomain-leucine zipper family regulates root growth in Arabidopsis thaliana. Plant Cell Rep. 33, 945–53 (2014). [DOI] [PubMed] [Google Scholar]
- Shin K. et al. Genetic identification of ACC-RESISTANT2 reveals involvement of LYSINE HISTIDINE TRANSPORTER1 in the uptake of 1-aminocyclopropane-1-carboxylic acid in Arabidopsis thaliana. Plant Cell Physiol. 56, 572–82 (2015). [DOI] [PubMed] [Google Scholar]
- Marinova K. et al. The Arabidopsis MATE transporter TT12 acts as a vacuolar flavonoid/H+ -antiporter active in proanthocyanidin-accumulating cells of the seed coat. Plant Cell. 19, 2023–38 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. H. & Tegeder M. Selective expression of a novel high-affinity transport system for acidic and neutral amino acids in the tapetum cells of Arabidopsis flowers. Plant J. 40, 60–74 (2004). [DOI] [PubMed] [Google Scholar]
- Teschner J. et al. A novel role for Arabidopsis mitochondrial ABC transporter ATM3 in molybdenum cofactor biosynthesis. Plant Cell. 22, 468–80 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Aliaga I. & Daniel H. Peptide transporters and their roles in physiological processes and drug disposition. Xenobiotica. 38, 1022–42 (2008). [DOI] [PubMed] [Google Scholar]
- Haydon M. J. & Cobbett C. S. A novel major facilitator superfamily protein at the tonoplast influences zinc tolerance and accumulation in Arabidopsis. Plant Physiol. 143, 1705–19 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter D. A., Yoo S. D., Butcher S. M. & McManus M. T. Expression of 1-aminocyclopropane-1-carboxylate oxidase during leaf ontogeny in white clover. Plant Physiol. 120, 131–42 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T. et al. Functional analysis of Arabidopsis ethylene-responsive element binding protein conferring resistance to Bax and abiotic stress-induced plant cell death. Plant Physiol. 138, 1436–45 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher K. A., Brown J., Saw R. E. & Callis J. The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell. 18, 699–714 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamuro J. K., Caster B., Villarroel R., Van Montagu M. & Jofuku K. D. The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc Natl Acad Sci USA. 94, 7076–81 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C. H. et al. Transcription factors WRKY70 and WRKY11 served as regulators in rhizobacterium Bacillus cereus AR156-induced systemic resistance to Pseudomonas syringae pv. tomato DC3000 in Arabidopsis. J Exp Bot. 67, 157–74 (2016). [DOI] [PubMed] [Google Scholar]
- Ma K., Xiao J., Li X., Zhang Q. & Lian X. Sequence and expression analysis of the C3HC4-type RING finger gene family in rice. Gene. 444, 33–45 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.