Abstract
DbpA is a DEAD-box RNA helicase implicated in RNA structural rearrangements in the peptidyl transferase center. DbpA contains an RNA binding domain, responsible for tight binding of DbpA to hairpin 92 of 23S ribosomal RNA, and a RecA-like catalytic core responsible for double-helix unwinding. It is not known if DbpA unwinds only the RNA helices that are part of a specific RNA structure, or if DbpA unwinds any RNA helices within the catalytic core’s grasp. In other words, it is not known if DbpA is a site-specific enzyme or region-specific enzyme. In this study, we used protein and RNA engineering to investigate if DbpA is a region-specific or a site-specific enzyme. Our data suggest that DbpA is a region-specific enzyme. This conclusion has an important implication for the physiological role of DbpA. It suggests that during ribosome assembly, DbpA could bind with its C-terminal RNA binding domain to hairpin 92, while its catalytic core may unwind any double-helices in its vicinity. The only requirement for a double-helix to serve as a DbpA substrate is for the double-helix to be positioned within the catalytic core’s grasp.
INTRODUCTION
Depending on their conserved amino acid sequence helicases are assigned to five super families (SF1–SF5)1. The DEAD-box family of RNA helicases are members of SF21. Similar to other members of SF2, DEAD-box proteins possess two RecA-like domains that perform double-helix unwinding2–5. However, the mechanism DEAD-box proteins employ for unwinding is different from that of other SF2 family members. While the other SF2 enzymes initiate their unwinding actions at the single-stranded regions 3’ or 5’ of the double-helix substrate and translocate toward the opposite end4,5, the DEAD-box RNA helicases directly attack the double-helix substrate and unwind it without translocation4–6. More precisely, the RecA-like domains of the DEAD-box proteins, which form their catalytic core, attack one strand of the RNA double-helix and bend it4–6. The bending process forces the release of the complementary RNA strand. The ATP-binding to the RecA-like domains provides the energy for the single-stranded RNA bending, while the ATP hydrolysis causes the release of the second strand of the double-helix from the catalytic core and the regeneration of the enzymes2–7.
Extensive structural and biochemical data have demonstrated that the interaction of the catalytic core with the 2’OH groups of the RNA strands that it loads upon is required for ATP hydrolysis and double-helix unwinding by DEAD-box proteins8–15. The absence of the 2’OH groups on the strand of the double-helix that does not interact with the catalytic core has no effect on DEAD-box proteins function8,10. Hence, the enzymes of this family are able to unwind RNA–RNA double-helices and DNA–RNA hybrid double-helices; however, they cannot unwind DNA double-helices8–11.
DbpA, which is the focus of this study, is an Escherichia coli (E. coli) DEAD-box RNA helicase. In addition to the two RecA-like domains, DbpA possesses a structured C-terminal domain, which is connected to the catalytic core via a peptide linker16–18. The C-terminal domain imparts DbpA’s specificity for hairpin 92 of 23S ribosomal RNA (rRNA)16–20, while, the RecA catalytic core houses DbpA’s ATPase and helicase activities. Hairpin 92 is located in the peptidyl transferase center. Hence, DbpA is believed to perform RNA structural isomerizations in a region of the ribosome that is evolutionarily conserved in all organisms and crucial for their survival.
The in vivo substrates of DbpA, like the substrates of the majority of DEAD-box proteins, remain unknown2,3; consequently, it is not known if DbpA, during in vivo ribosome assembly, unwinds specific double-helices, which belong to certain RNA structures, or if DbpA unwinds any double-helix that comes within its catalytic core’s reach. In other words, it is not known if DbpA is a site-specific RNA helicase, or a region-specific RNA helicase. Of course, if DbpA was a region-specific enzyme, its region of specificity would be the physical area around hairpin 92, where DbpA is anchored via its C-terminal domain. The knowledge concerning the DbpA protein’s site-specific or region-specific mode of action is important for a better understanding of DbpA’s physiological function during the ribosome assembly process.
Previous chemical footprinting experiments of a 172 nucleotide construct from the 23S rRNA in the presence of YxiN or DbpA, and the non-hydrolysable ATP analog, AMPPNP, show that the catalytic core interacts only with two distinct RNA sites in this RNA molecule16,21, implying that YxiN/DbpA are site-specific helicases, and the correct positioning of protein modules relative to each other is important for the helicase activity of YxiN/DbpA.
Other experimental work performed with short RNA constructs containing hairpin 92 demonstrated that DbpA unwinds sequence non-specific RNA helices located at various distances 3’ and 5’ of hairpin 92, suggesting that the exact fit of the protein modules to the RNA structure is not required for DbpA catalytic activity13, suggesting that DbpA is a region-specific enzyme. However, the RNA used in this study could fold into a three dimensional structure, and the folded structure could bring the double-helix substrates, that are far in sequence from hairpin 92, near the catalytic core and correctly position them to support DbpA activity. Therefore, it remains unknown if DbpA is a site-specific or a region-specific enzyme.
Since previous experiments have shown that the RNA binding domain of YxiN and its catalytic core do not directly interact with each other16, the only way that YixN/DbpA could achieve site-specificity is if the interdomain linker region correctly positioned the catalytic core to attack certain double-helix substrates but not others. To investigate the importance of the role of the peptide linker in positioning the catalytic core relative to the RNA structure and the C-terminal RNA binding domain, we increased the length of the interdomain peptide linker from 12 amino acids to 35 amino acids and measured the new DbpA construct’s catalytic activity in presence of various RNA substrates. This peptide linker insertion changes the position and distance of the catalytic core relative to the helix 92 and double-helix substrate, while keeping the local concentration of catalytic domains near hairpin 92 and the double-helix substrate unchanged. If the structural position of the catalytic core relative to the RNA binding domain and the RNA structure is important for the catalytic activity of DbpA—in other words, if DbpA is a site-specific enzyme—then modifying the linker connecting the DbpA RNA binding domain with its catalytic core would decrease the DbpA protein’s catalytic activity.
RESULTS
Extended DbpA Protein Design
The goal of these experiments was to place a polypeptide segment into the interdomain linker region connecting the DbpA catalytic core to the RNA binding domain and investigate if the insertion affects DbpA’s function. The interdomain region is a short polypeptide of 12 residues with an amino acid sequence of PANSSIATLEAE16. Neither the length nor the sequence of this region is conserved among different members of the DbpA family16. The structure of the interdomain linker is unknown, but is predicted to be flexible and unstructured16. In order to increase the peptide linker region length, a 23 amino acid residue polypeptide with a composition of NASSGSSASSPSASNSPGANGSS was inserted between the native interdomain region’s Ala and Thr residues. The sequence of the new extended interdomain linker is PANSSIANASSGSSASSPSASNSPGANGSSTLEAE. This peptide sequence was chosen because it has similar structural and dynamic properties to the native interdomain region; both the designed and the native interdomain linker are predicted to form a flexible and unstructured region22. In addition, since DbpA is purified as a native protein, small and polar amino acids, which promote peptide solubility, were placed into the polypeptide insert to discourage the aggregation of extended DbpA and its partition into inclusion bodies. The new interdomain linker was not digested by the E. coli proteolytic enzymes and the extended DbpA, as judged by SDS–PAGE and gel filtration chromatography (data not shown), was expressed as an intact and soluble protein in E. coli cells.
Insertion of the Peptide into the Interdomian Linker Region of DbpA Has a Minimal Affect in Its Function
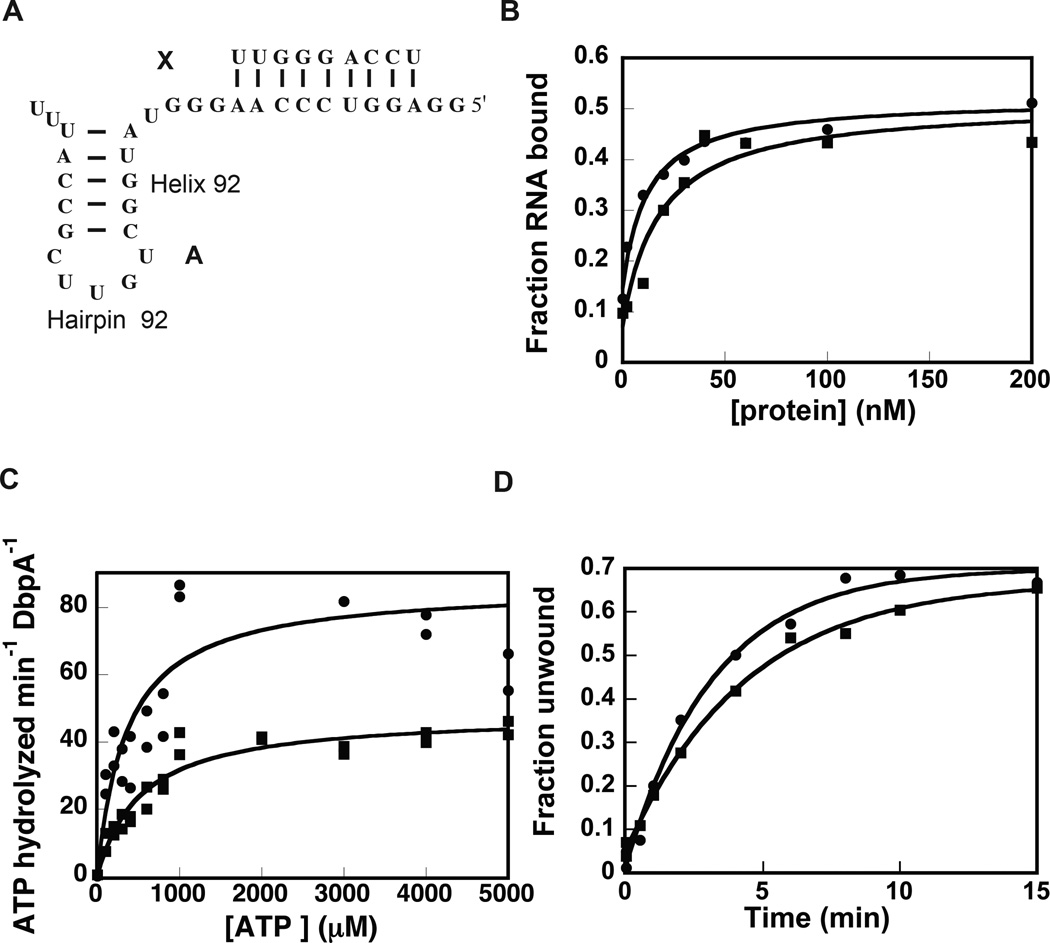
Once the extended DbpA construct was expressed and purified, its ability to bind RNA, hydrolyze ATP, and unwind RNA were measured. The RNA binding affinity was measured by electrophoretic mobility shift assay. The cognate RNA used for this experiment was a 32-mer RNA (Figure 1A, construct A), which contains hairpin 92 from 23S rRNA and has been previously used by our laboratory and others to investigate the DbpA protein’s functional properties10,19,23,24. Analysis of the extended and wild-type DbpA electrophoretic mobility shift data shows that the non-cooperative binding equation fits these data accurately (Figure 1B). The apparent equilibrium dissociation constants for both the wild-type and the extended proteins obtained from the non-cooperative binding equation are: 10.5 ± 2.3 nM for the wild-type DbpA and 33.7 ± 9.0 nM for the extended DbpA. Hence, the inserted peptide minimally affects the ability of DbpA to bind RNA. Previous biochemical and structural experiments have shown that neither the interdomain linker nor the catalytic core influence the affinity of the RNA binding domain of YxiN/DbpA for RNA16,21,25. Therefore, breaking the sequence of the interdomain peptide linker and inserting the 23 amino acids peptide segment was expected to have no effect on the binding affinity of DbpA for its cognate RNA. The fact that we observe a decrease in binding affinity is likely a consequence of formation of non-native interaction between the insert peptide and the RNA molecule or other regions of the protein and not a consequence of disrupting native interactions between the DbpA RNA binding domain and the interdomain linker.
Figure 1.
Effect of the extended peptide linker on the functional properties of DbpA. (A) The RNA construct used for these studies. (B) Binding of wild-type and extended DbpA to 32-mer RNA (construct A) as measured by gel shift assay. (C) ATPase activity of wild-type (circles) and extended DbpA (squares) as measured by coupled NADH/phosphoenolpyruvate assay. (D) Helicase activity of wild-type (circles) and extended DbpA (square). The data are representative of one experiment. The averages of multiple experiments and the standard deviations are shown in Table 1 and Table 2. Representative gel for the binding and the helicase assays are shown in the Supplemental Materials.
Subsequently, the ATP hydrolysis activity of the extended and wild-type DbpA were measured by the pyruvate kinase/lactate dehydrogenase coupled assay26. Figure 1C shows the dependence of ATP hydrolysis rate for the extended and wild-type DbpA as function of ATP concentration. The Michaelis-Menten equation was used to fit these data. Within experimental error, the Michaelis-Menten ATP binding constant is the same for extended and wild-type DbpA. Hence, the peptide extension is not effecting the formation of the proper ATP pocket.
The ATP turnover rate is affected by the peptide extension. The kcat of ATP hydrolysis by extended DbpA is 0.72 ± 0.08 s−1 and the kcat of ATP hydrolysis by wild-type DbpA is 1.28 ± 0.14 s−1. Although the ATP turnover of the extended DbpA is reduced when compared to wild-type DbpA, extended DbpA is a much more efficient enzyme than many members of DEAD-box family of proteins27. We believe the reduction on the ATP turnover of the extended DbpA is a consequence of its decrease in binding affinity for RNA.
The RNA substrate used in the initial helicase assay was the 32 nucleotide long RNA segment, used for the binding and ATPase experiments, annealed to a 5’-32P labeled 9 nucleotide long RNA (Figure 1A, molecule A:X). The fraction of double-helix unwound was calculated by the ratio of 32P counts on the displaced 9-mer band over the total 32P counts on the lane10,19,24. Within the experimental errors, the observed microscopic rate and the extent of RNA unwinding by the extended DbpA and the wild-type DbpA are the same (Table 2).
Table 2.
Helicase Parameters of wild-type and extended DbpA interacting with various RNA molecules.
| DbpA constructs | RNA | kobsa(min−1) |
|---|---|---|
| Wild-type | A:X | 0.34 ± 0.12 |
| F:Y | 0.35 ± 0.07 | |
| G:Y | ~ 0 | |
| Extended | A:X | 0.20 ± 0.03 |
| B:X | 2.0 ± 1.0 | |
| C:X | 0.27 ± 0.11 | |
| D:X | 0.44 ± 0.08 | |
| E:X | ~ 0 | |
| F:Y | 0.30 ± 0.04 | |
| G:Y | ~ 0 |
The observed rate of unwinding was obtained by fitting the single-exponential equation to the native gel helicase data as described in the Materials and Method section of the paper. The errors are the standard deviations from the means of at least two independent data sets.
The observation that a difference is detected in the ATPase activity of the two constructs, but no difference is observed in their helicase activity could be a consequence of the different protein and RNA concentrations used in each assay. The helicase assay was performed at 600 nM and 1 nM annealed RNA; instead, the ATPase assay was performed at 30 nM protein and 2000 nM RNA. At the very high protein to RNA ratio, the conditions under which the helicase assay was performed, the slight differences in binding affinity between wild-type and extended DbpA become non-significant and all the RNA molecules are saturated with protein.
The fact that molecule A:X supports the helicase activity of wild-type DbpA and extended DbpA with a similar observed microscopic rate and extent could be a fortuitous observation. To ensure that the peptide linker insert had no effect on the helicase activity of DbpA, the helicase activity of the extended DbpA was also measured in presence of a number of RNA–DNA chimeras. These RNA–DNA chimeras have been shown to support the helicase activity of wild-type DbpA at different extents and with different rates10.
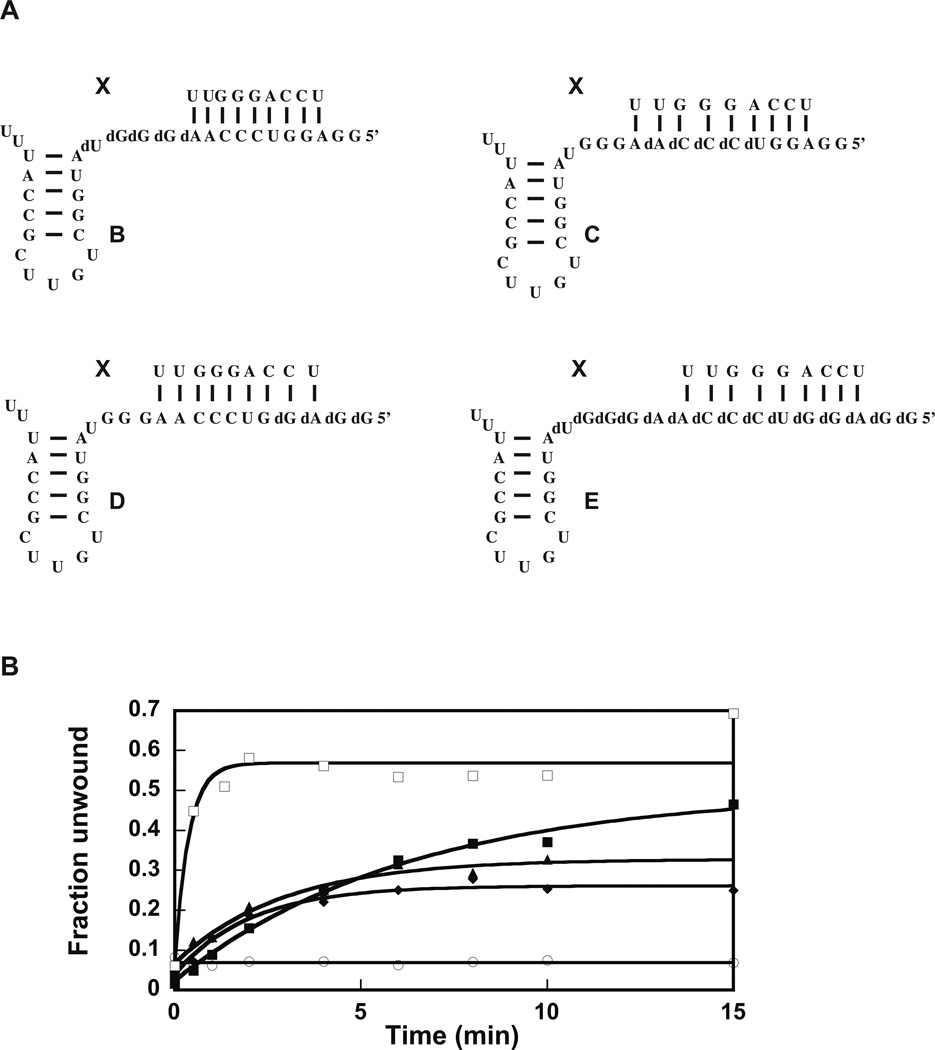
The chemical structures of the model molecules are shown in Figure 2A. Molecule B:X has five bases 5’ of helix 92 changed to DNA, molecule C:X has five bases 5’ of helix 92 and five bases removed from helix 92 change to DNA, molecule D:X has five bases 5’ of helix 92 and ten bases removed from helix 92 changed to DNA, and molecule E:X has all the fifteen bases 5’ of helix 92 changed to DNA.
Figure 2.
Helicase activity of the extended DbpA in presence of various RNA–DNA chimeras. (A) The RNA–DNA molecules used in the helicase experiments in (B). (B) Helicase activity of the 23 amino acid extended DbpA was measured in presence of molecule A:X (filled square), B:X (empty square), C:X (triangles), D:X (diamond), E:X (empty circle) plus 5 mM ATP. The data are representative of one experiment. The averages from multiple experiments and the standard deviations are shown in Table 2.
Molecule B:X supports the helicase activity of extended DbpA to a similar extent as the molecule A:X and with a higher observed microscopic rate constant (Figure 2B, Table 2). Within experimental errors, molecule B:X stimulates the helicase activity of extended and wild-type DbpA to the same extent and with a similar observed microscopic rate constant10.
Why molecule B:X stimulates the unwinding activity of the wild-type DbpA with a higher observed rate constant than molecule A:X is not well understood. Our previous results show that construct B binds wild-type DbpA with a similar affinity as construct A10. On the other hand, the rate of ATP hydrolysis in the presence of construct B is higher than that in the presence of construct A10. Based in these observations, our hypothesis is that the 5 DNA bases 5’ of hairpin 92 in construct B, interacts with the same affinity but in a different mode with the DbpA RNA binding domain than the 5 RNA bases 5’ of hairpin 92 in construct A. This different mode of interaction may increase the productive encounters between the DbpA catalytic core and the 10 RNA bases in construct B when compared with the same 10 RNA bases in construct A10.
The molecules C:X and D:X support the helicase activity of extended DbpA with a reduced extent of unwinding (Figure 2B, Table 2). The observed microscopic rate and the extent of unwinding in the presence of these molecules is the same within experimental error for the extended and wild-type DbpA (Table 2)10. Similar to the observation with wild-type DbpA, molecule E:X, in which all the RNA residues 5’ of helix 92 have been changed to DNA, does not support the helicase activity of extended DbpA.
The reduced extent of unwinding in presence of constructs C:X, D:X, and E:X was suggested to be a consequence of the interaction of the RNA binding domain of DbpA with the top strand of the double-helix substrate. As a result of this interaction, only the bottom strand of the double-helix substrate is accessible for the DbpA catalytic core to act upon. When the residues in the bottom strand of the double-helix are DNA, they inhibit both the ATPase and the helicase activity of DbpA10.
Combined, the helicase data in Figure 1D and Figure 2B suggest the extension of the interdomain linker region has no effect on the ability of DbpA to perform its helicase function. Thus, the physical connection of DbpA RNA binding domain to the catalytic core is unimportant for the helicase activity of DbpA, suggesting the DbpA protein is a region-specific enzyme, which would unwind any double-helix substrate near hairpin 92.
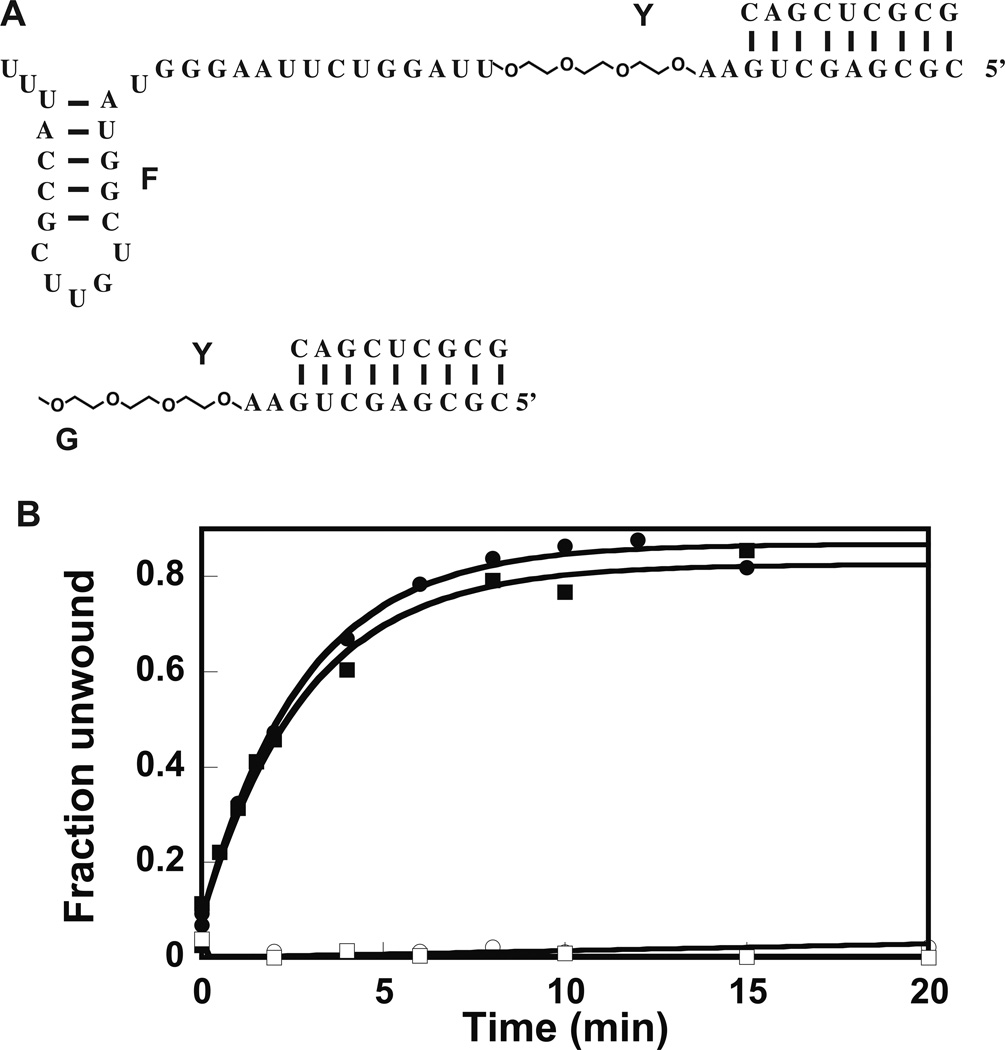
RNA–PEG Chimera Containing Hairpin 92 Supports the Helicase Activity of Extended and Wild-type DbpA
To further investigate if DbpA is a region-specific or a site-specific enzyme, two RNA–PEG chimeras were employed and the ability of the chimeras to support the helicase activity of wild-type and extended DbpA was investigated. In both chimeras, three residues of ethylene glycol were placed 3’of double-helix substrate and two RNA bases removed from it (Figure 3A). The ethylene glycol residues span a distance of 3Å; this distance is equivalent to the contour length of three single-stranded RNA bases28. The difference in structure between the two PEG chimeras is that one of them contains hairpin 92, while the other does not (Figure 3A). The RNA–PEG chimeras were annealed to a 32P-labeled 9-mer RNA (Figure 3A, construct Y). The ability of the extended and wild-type DbpA to unwind the 32P-labeled RNA was investigated via native gel electrophoresis. Figure 3B shows the results of native gel electrophoresis experiments. While molecule F:Y, which contains hairpin 92, was able to support the helicase activity of both wild-type and extended DbpA, molecule G:Y, which is missing hairpin 92, is unable to support the helicase activity of wild-type or extended DbpA (Table 2). Again, both the extended and wild-type DbpA show the same helicase catalytic activity in the presence of molecules F:Y and G:Y. More importantly, these experiments strongly suggest that the DbpA catalytic core will unwind any double-helix substrate in its vicinity, and how the catalytic core domain is connected to the RNA binding domain of DbpA, or how the RNA double-helix is connected to the hairpin 92 is mechanistically unimportant for the helicase activity of DbpA. On the other hand, when the local concentration of the catalytic core near the double-helix substrate is low because hairpin 92—where DbpA is anchored to the model RNA molecule via its RNA binding domain—is missing, the DbpA catalytic core is less likely to reach the double-helix substrate and perform unwinding.
Figure 3.
Effect of PEG linker on the functional properties of DbpA. (A) The RNA–PEG chimera constructs used for these experiments. The 9-atom ethylene glycol linker spans 3 RNA bases. (B) Helicase activity of 23 amino acid extended and wild-type DbpA in presence of RNA–PEG chimeras. Legend: Filled square stimulation of the unwinding activity of wild-type DbpA by molecule F:Y; empty circle stimulation of helicase activity of wild-type DbpA by molecule G:Y; filled square stimulation of the unwinding activity of 23 amino acid extended DbpA by molecule F:Y; empty square stimulation of the unwinding activity of 23 amino acid extended DbpA by molecule G:Y. All the experiments shown here were performed at 600 nM protein. The helicase assay in presence of molecule G:Y was also performed at 2000 nM of protein (data not shown). Even at this much higher protein concentration, the construct G:Y could not support the helicase activity of wild-type or 23 amino acid extended DbpA. The data are representative of one experiment. The means of multiple experiments and the averages from the means are shown in Table 2.
DISCUSSION
Helicase assays performed in the presence of a DbpA construct, in which the RNA binding domain and the catalytic core are separated by a non-native peptide linker, and RNA-PEG chimera, in which the double-helix substrate is separated from hairpin 92 by a PEG linker, suggest that the only requirement for the DbpA catalytic core to perform double-helix unwinding is for the catalytic core and the double-helix substrate to be physically near each other in space. How the catalytic core is physically linked to the RNA binding domain and how the double-helix substrate is physically linked to the hairpin 92 are mechanistically unimportant for DbpA helicase activity. Thus, these experiments suggest that DbpA’s double-helix substrates are not required to be unique and part of specific RNA structure; they are only required to be within the DbpA catalytic core’s reach, making DbpA a region-specific enzyme.
Exceptions to the above mechanism are the double-helix substrates located near the RNA binding domain of DbpA. Our previous data have suggested that when the double-helix substrate is placed near the RNA binding domain of DbpA, this domain could interact with one strand of the double-helix substrate, preventing the access of DbpA’s catalytic core to that stand of the double-helix substrate10. Nevertheless, the DbpA catalytic core could interact with the free RNA strand and unwind the double-helix.
A protein construct containing only the YxiN catalytic core showed ATP hydrolysis activity16. However, the ATP hydrolysis activity of the separated YxiN catalytic core was considerably diminished in comparison to that of whole wild-type YxiN16. The separated catalytic core of YxiN in presence of construct A has an estimated ATP turnover rate of 0.05 s−1 instead the whole YxiN protein in presence of the construct A has an ATP turnover rate of 0.95 s−116. The addition of the YxiN C-terminal domain in trans did not increase the ATP hydrolysis activity of the separated catalytic core construct, which demonstrates that the direct interaction of the C-terminal domain with the catalytic core is not important for the YxiN ATP hydrolysis activity16. The diminished ATP hydrolysis activity of the separated YxiN separated catalytic core could be a consequence of diminished local concentration of YxiN catalytic core near the double-helix substrate when the C-terminal RNA binding domain is not anchoring them to hairpin 92. Alternatively, the interaction of the C-terminal RNA binding domain with the hairpin 92 could trigger a signal that is transmitted through the interdomain peptide linker to the catalytic core, but when this signal is missing, the catalytic core is less active. Our data show that the peptide insertion in the interdomain linker region produced a minimal decrease in DbpA’s binding affinity for its cognate RNA, which consequently produced a decrease in the ATPase rate of DbpA measured at subsaturation concentration of RNA. On the other hand, at saturating protein concentration, where the differences in binding affinity between the wild-type DbpA and the extended DbpA become unimportant, extended and wild-type DbpA unwind a number of model substrates with a similar rate and extent of unwinding. Thus, the helicase experiments suggests that the interdomain linker region is not serving as a bridge transmitting information between the DbpA C-terminal RNA binding domain and its catalytic core. Taken together, our results and the previous data16 suggest that the DbpA RNA binding domain and its catalytic core are not interacting with each other directly or through the peptide linker region, and their distinct functions are independent.
Neither the amino acid composition nor the length of the interdomain linker are conserved among the members of the DbpA family. The linker length in different organisms varies from 5 to 35 amino acids16. Therefore, the question arises: why have bacteria developed different requirements for DbpA linker length and composition while keeping the sequence and structure of ribosomal RNA unchanged29? An attractive hypothesis is that during the ribosome assembly process, organisms form intermediate ribosomal particles of different misfolded structures in which the DbpA substrates are positions at different distances from hairpin 92. In order for DbpA catalytic core to unwind the double-helices present in different organisms, the peptide linker between the DbpA catalytic core and the RNA binding domain must change in each organism and match the distance of the DbpA substrates from hairpin 92. The knowledge of bacterial large subunit intermediate particles’ structures remains very limited30,31. Moreover, the knowledge of DbpA in particular and DEAD-box proteins in general in vivo substrates also is very limited2,3. Future experiments determining the ribosomal RNA intermediate structures and DbpA action sites in different organisms could shed light if indeed the length of the interdomain region has evolved to fulfill cells need for unwinding RNA double-helices position within a certain distance from hairpin 92.
An alternative hypothesis is that there may be no evolutionary pressure to preserve the DbpA interdomain linker size and its composition as long as this linker is flexible and allows DbpA catalytic domains movement around the intermediate structures formed during the ribosome assembly process. This hypothesis is supported by the fact that for many proteins that contain peptide linkers, which connect domains with distinct functions, neither the length nor the composition of the interdomain linker are conserved32–34. On the other hand, the flexibility of these linker regions has been shown to be important for these proteins’ functions, and it is believed that the role of the peptide linker regions is to facilitate the domains’ movement relative to each other or other macromolecular structures32–35.
In conclusion, our results suggest that during the ribosome assembly process, DbpA binds with its C-terminal domain to hairpin 92 and its catalytic core unwinds any RNA double-helices that come within its grasp. This property makes DbpA a region-specific, instead of a site-specific, enzyme. The region that DbpA is specific for is the spatial area in the vicinity of hairpin 92 that the DbpA catalytic core could sample while anchored to this hairpin via the DbpA RNA binding domain. The unwinding activity of DbpA in the spatial area near hairpin 92 could provide the 23S rRNA with the opportunity to anneal into the correct structure, or could give the ribosomal proteins or the maturation factors the opportunity to interact with the newly formed single-stranded RNA regions. Thus, during the ribosome assembly process, DbpA may act as an ATP dependent RNA chaperon in the peptidyl transferase center.
MATERIALS AND METHODS
RNA Substrates
All the RNA constructs and RNA–PEG chimera constructs were purchased HPLC purified from IDT. Poly(A) and pyruvate kinase/lactate dehydrogenase enzyme mixture was purchased from Sigma-Aldrich.
Extended DbpA Cloning
The DNA sequence of wild-type DbpA was cloned into pET-3a vector24. The DNA sequence for the extended DbpA was created by inserting a double-stranded DNA segment into the unique XcmI site of the wild-type DbpA sequence. The map of pET-3a vector bearing the wild-type or extended DbpA coding sequence is shown in Supplemental Materials part of the paper (Figure S2 and S3).
Protein Purification
Both the wild-type DbpA and the extended DbpA constructs were purified as previously described10,24. In brief, the DbpA constructs bearing an N-terminal His-tag were overexpressed in E. coli cells and purified via His-tag affinity chromatography followed by gel filtration chromatography10,24. The His-tag was not removed from the protein constructs.
RNA Binding Affinity Assay
The wild-type DbpA and the extended DbpA affinities for RNA were measured by gel mobility shift assay as previously described36. For these studies, 1 nM of 5’-32P-RNA was mixed with a series of protein concentrations in presence of 50 mM HEPES (pH 7.5), 50 mM KCl, 10 mM MgCl2, 1 mM DTT, 70 mM poly(A), 5% (v/v) glycerol. The reaction mixture was incubated at 22°C for 10 minutes to reach equilibrium and then applied to a cold 10% native acrylamide gel (29:1 acrylamide to bis-acrylamide ratio). The gel was run for two hours at 200 V and then dried and exposed on a phosphor screen. The fraction of protein bound RNA at each protein concentration was calculated from the ratio of the counts in the protein shifted RNA band over the sum of the counts in the protein free and shifted RNA bands. The fraction of protein bound RNA versus the protein concentration was fit to the equation:
where fB is the fraction of protein bound RNA, fB(0) and fB(max) are the lower and upper baselines of the binding curve, and Kd is the dissociation constant.
ATPase Activity
The rate of ATP hydrolysis was measured by pyruvate kinase/lactate dehydrogenase coupled assay as previously described26. In this assay, ATP hydrolysis by DbpA protein constructs is coupled to NADH oxidation, which produces a decrease in absorbance at 338 nm26. These measurements were performed at 22°C in the presence of 2 µM RNA, 30 nM protein, 50 mM HEPES (pH 7.5), 50 mM KCl, 10 mM MgCl2, 0.1% (v/v) Tween-20, 1 mM DTT, 1 mM phosphoenolpyruvate, 250 µM NADH, 10 units/mL pyruvate kinase, 15 units/mL lactate dehydrogenase and a series of ATP·Mg concentrations. The rate of ATP hydrolysis versus the ATP concentration was fit to the Michaelis-Menten equation and the Km (the Michaelis constant) and kcat (the turnover rate) were obtained from this fit.
Helicase Assay
The helicase activity of wild-type DbpA and the extended DbpA was investigated by measuring the unwinding of the 5’-32P labeled 9-mer annealed to the unlabeled 32-mer RNA, 32-mer RNA-DNA or the RNA–PEG chimera. The RNA was annealed by heating the reaction mixture containing 0.6 µM of 5’-32P labeled 9-mer RNA and 1.2 µM of unlabeled long RNA, RNA-DNA chimera or RNA-PEG chimera in presence of 50mM HEPES (pH 7.5), 50 mM KCl at 95°C for one minute, cooling the reaction down and holding it at 65°C for three minutes, cooling the reaction mixture to 22°C and adding 10 mM MgCl2 final, and incubating the reaction mixture at 22°C for 15 minutes. For the helicase assay, 1 nM of annealed RNA was incubated for 10 minutes with HEPES (pH 7.5), 50 mM KCl, 10 mM MgCl2, 1 mM DTT, 70 mM poly(A), 5% (v/v) glycerol and 600 nM or 2000 nM of DbpA. The helicase reaction was started by the addition of 5 mM ATP·Mg final. At different time points after the addition of ATP, aliquots of the reaction mixture were taken and the helicase reaction was quenched by adding a final concentration of 7.5 mM EDTA (pH 8.0), 0.15% SDS, 1.25% glycerol, 0.0025% (m/v) xylene cyanol. The quenched reaction was immediately loaded in 20% native polyacrylamide gel (29:1 acrylamide to bis-acrylamide ratio). The gel running buffer was 1/3 TBE plus 5 mM MgCl2. The gels were run for four hours at 200V and 4°C. Subsequently, the gels were dried and exposed to phosphor imager screens.
The fraction of unwound 9-mer was calculated from the ratio of 32P counts on the 9-mer band over the total 32P counts on the 9-mer annealed to the 32-mer RNA plus the counts on the separated 9-mer band. The data was fit to the equation:
fu is the fraction of 9-mer unwound, fu(0) is the fraction of 9-mer unwound before the addition of ATP, A is the amplitude of unwinding transition, and k is the observed rate constant of unwinding10,19,24,37.
With the exception of molecule G:Y, the double-helix was stable and did not unwind spontaneously during the course of the reaction. For molecule G:Y the extent of spontaneous and the DbpA construct assisted unwinding were very similar. The molecule G:Y data in Figure 3B were obtained by subtracting the fraction of spontaneously unwound RNA at different time points during the reaction progression from the fraction of the DbpA construct assisted unwinding.
Supplementary Material
Table 1.
Kinetics and Equilibrium Parameters of Wild-Type DbpA and Extended DbpA Interacting with 32-mer RNA.
| Protein | kcata (s−1) | Km (ATP)b (M) | kcat/Km (ATP) (s−1 M−1) |
KD (RNA)c (nM) |
|---|---|---|---|---|
| Wild-type DbpA | 1.27 ± 0.14 | (3.3 ± 0.5) × 10−4 | (3.9 ± 0.8) × 103 | 10.5 ± 2.3 |
| Extended DbpA | 0.72 ± 0.08 | (4.9 ± 0.2) × 10−4 | (1.5 ± 0.2) × 103 | 33.7 ± 9.0 |
The turnover number was calculated from the fit of Michaelis-Menten equation to the data similar as those shown in Figure 1C. The values represent the means from at least three independent data sets and the errors are the standard deviations from these means.
The Michaelis constant was obtained from the fit of the Michaelis-Menten equation to the data similar to those shown in Figure 1C. The values represent the mean from at least three independent experiments and the errors are the standard deviations from the means.
The apparent disassociation constant was measured by gel shift assay as described in the Materials and Methods section of this paper. The values represent the means from at least two independent experiments and the errors are the standard deviations from the means.
Acknowledgments
EK thanks O. Uhlenbeck and M. Saks for many helpful discussions. This work was supported by Start-up funds from the University of Central Florida Department of Chemistry, NIH (5R21CA175625-02 to E.K) and UCF (In-house grant to E.K).
REFERENCES
- 1.Caruthers JM, McKay DB. Curr. Opin. Struct. Biol. 2002;12:123. doi: 10.1016/s0959-440x(02)00298-1. [DOI] [PubMed] [Google Scholar]
- 2.Linder P, Jankowsky E. Nat. Rev. Mol. Cell Biol. 2011;12:505. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- 3.Putnam AA, Jankowsky E. Biochim. Biophys. Acta. 2013;1829:884. doi: 10.1016/j.bbagrm.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pyle AM. Annu. Rev. Biophys. 2008;37:317. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 5.Fairman-Williams ME, Guenther UP, Jankowsky E. Curr. Opin. Struct. Biol. 2010;20:313. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan C, Russell R. RNA Biol. 2010;7:28. doi: 10.4161/rna.7.6.13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henn A, Bradley MJ, De La Cruz EM. Annu. Rev. Biophys. 2012;41:247. doi: 10.1146/annurev-biophys-050511-102243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Q, Del Campo M, Lambowitz AM, Jankowsky E. Mol. Cell. 2007;28:253. doi: 10.1016/j.molcel.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 9.Rogers GW, Jr, Lima WF, Merrick WC. J. Biol. Chem. 2001;276:12598. doi: 10.1074/jbc.M007560200. [DOI] [PubMed] [Google Scholar]
- 10.Childs JJ, Gentry RC, Moore AF, Koculi E. RNA. 2016;22:408. doi: 10.1261/rna.052928.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jarmoskaite I, Russell R. Annu. Rev. Biochem. 2014;83:697. doi: 10.1146/annurev-biochem-060713-035546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Cell. 2006;125:287. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 13.Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CL, Pedersen JS, Seraphin B, Le Hir H, Andersen GR. Science. 2006;313:1968. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 14.Bono F, Ebert J, Lorentzen E, Conti E. Cell. 2006;126:713. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 15.Del Campo M, Lambowitz AM. Mol. Cell. 2009;35:598. doi: 10.1016/j.molcel.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karginov FV, Caruthers JM, Hu Y, McKay DB, Uhlenbeck OC. J. Biol. Chem. 2005;280:35499. doi: 10.1074/jbc.M506815200. [DOI] [PubMed] [Google Scholar]
- 17.Wang S, Hu Y, Overgaard MT, Karginov FV, Uhlenbeck OC, McKay DB. RNA. 2006;12:959. doi: 10.1261/rna.5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardin JW, Hu YX, McKay DB. J. Mol. Biol. 2010;402:412. doi: 10.1016/j.jmb.2010.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diges CM, Uhlenbeck OC. EMBO J. 2001;20:5503. doi: 10.1093/emboj/20.19.5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuller-Pace FV, Nicol SM, Reid AD, Lane DP. EMBO J. 1993;12:3619. doi: 10.1002/j.1460-2075.1993.tb06035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karginov FV, Uhlenbeck OC. Nucleic Acids Res. 2004;32:3028. doi: 10.1093/nar/gkh640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buchan DWA, Minneci F, Nugent TCO, Bryson K, Jones DT. Nucleic Acids Res. 2013;41:W349. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diges CM, Uhlenbeck OC. Biochemistry. 2005;44:7903. doi: 10.1021/bi050033x. [DOI] [PubMed] [Google Scholar]
- 24.Elles LM, Uhlenbeck OC. Nucleic Acids Res. 2008;36:41. doi: 10.1093/nar/gkm926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kossen K, Karginov FV, Uhlenbeck OC. J. Mol Biol. 2002;324:625. doi: 10.1016/s0022-2836(02)01140-3. [DOI] [PubMed] [Google Scholar]
- 26.Tsu CA, Uhlenbeck OC. Biochemistry. 1998;37:16989. doi: 10.1021/bi981837y. [DOI] [PubMed] [Google Scholar]
- 27.Garcia I, Albring MJ, Uhlenbeck OC. Biochemistry. 2012;51:10109. doi: 10.1021/bi301172s. [DOI] [PubMed] [Google Scholar]
- 28.Saenger W. Principles of Nucleic Acid Structure. New York: Springer-Verlag Inc.; 1988. [Google Scholar]
- 29.Doris SM, Smith DR, Beamesderfer JN, Raphael BJ, Nathanson JA, Gerbi SA. RNA. 2015;21:1719. doi: 10.1261/rna.051144.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shajani Z, Sykes MT, Williamson JR. Annu. Rev. Biochem. 2011;80:501. doi: 10.1146/annurev-biochem-062608-160432. [DOI] [PubMed] [Google Scholar]
- 31.Gamalinda M, Woolford JL., Jr Translation. 2015;3:e975018. doi: 10.4161/21690731.2014.975018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hrycyna CA, Airan LE, Germann UA, Ambudkar SV, Pastan I, Gottesman MM. Biochemistry. 1998;37:13660. doi: 10.1021/bi9808823. [DOI] [PubMed] [Google Scholar]
- 33.Joshi AK, Witkowski A, Berman HA, Zhang L, Smith S. Biochemistry. 2005;44:4100. doi: 10.1021/bi047856r. [DOI] [PubMed] [Google Scholar]
- 34.de Cock E, Springer M, Dardel F. Mol. Microbiol. 1999;32:193. doi: 10.1046/j.1365-2958.1999.01350.x. [DOI] [PubMed] [Google Scholar]
- 35.Perham RN. Biochemistry. 1991;30:8501. doi: 10.1021/bi00099a001. [DOI] [PubMed] [Google Scholar]
- 36.Polach KJ, Uhlenbeck OC. Biochemistry. 2002;41:3693. doi: 10.1021/bi012062n. [DOI] [PubMed] [Google Scholar]
- 37.Chen Y, Potratz JP, Tijerina P, Del Campo M, Lambowitz AM, Russell R. Proc. Natl. Acad. Sci. U.S.A. 2008;105:20203. doi: 10.1073/pnas.0811075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.