Abstract
Sterol C24-methyltransferases (SMTs) constitute a group of sequence-related proteins that catalyze the pattern of sterol diversity across eukaryotic kingdoms. The only gene for sterol alkylation in green algae was identified and the corresponding catalyst from Chlamydomonas reinhardtii (Cr) was characterized kinetically and for product distributions. The properties of CrSMT were similar to those predicted for an ancient SMT expected to possess broad C3-anchoring requirements for substrate binding and formation of 24β-methyl/ethyl Δ25(27)-olefin products typical of primitive organisms. Unnatural Δ24(25)-sterol substrates, missing a C4β-angular methyl group involved with binding orientation, convert to product ratios in favor of Δ24(28)-products. Remodeling the active site to alter the electronics of Try110 (to Leu) results in delayed timing of the hydride migration from methyl attack of the Δ24-bond, that thereby produces metabolic switching of product ratios in favor of Δ25(27)-olefins or impairs the second C1-transfer activity. Incubation of [27-13C]lanosterol or [methyl-2H3]SAM as co-substrates established the CrSMT catalyzes a sterol methylation pathway by the “algal” Δ25(27)-olefin route, where methylation proceeds by a conserved SN2 reaction and de-protonation proceeds from the pro-Z methyl group on lanosterol corresponding to C27. This previously unrecognized catalytic competence for an enzyme of sterol biosynthesis, together with phylogenomic analyses, suggest that mutational divergence of a promiscuous SMT produced substrate- and phyla-specific SMT1 (catalyzes first biomethylation) and SMT2 (catalyzes second biomethylation) isoforms in red and green algae, respectively, and in the case of SMT2 selection afforded modification in reaction channeling necessary for the switch in ergosterol (24β-methyl) biosynthesis to stigmasterol (24α-ethyl) biosynthesis during the course of land plant evolution.
Keywords: Chlamydmonas reinhardtii green algae, Sterol evolution, Sterol C24-methyltransferase, Ergosterol, Cholesterol, SMT2, SMT1
1. Introduction
Membrane-bound C28-ergosta- and C29-stigmasta-type sterols of different C24-stereochemistry have contributed to the evolution of primary metabolism in eukaryotes through biosynthetic pathways that can differ significantly (Nes, 2011; Volkman, 2005; Weete et al., 2010). The uneven distribution of fossil C28- and C29-steranes that date to the Precambrian (Brocks et al., 1999; Love et al., 2009; Summons et al., 2006) and phylogenomic analysis of genes of sterol biosynthesis (Desmond and Gribaldo, 2009) suggest that all the necessary enzymes for the formation of phytosterols (24-alkyl sterols), as well as cholesterol, may have existed in the last eukaryotic common ancestor. In the biosynthesis of ergosterol 3 from glucose via the acetate–mevalonate or mevalonate-independent pathways, it is now evident that quite separate, independent, molecular evolution occurred in fungi and green algae (Lichtenthaler, 1999; Lombard and Moreira, 2011). However, evidence also is available showing land plants can generate stigmasterol 4 by the same acetate–mevalonate pathways as used by fungi (Miller et al., 2012; Opitz et al., 2014) (Fig. 1). Based on the divergent structures of a 24β-methyl group and 24α-ethyl group in the final products, one might expect that functional differences in sterol methylating enzymes, sterol C24 methyltransferases (SMTs) responsible for biomethylation along the sterol side chain, would be phylogenetically significant, perhaps even crucial to generation of the panoply of sterol patterns observed throughout nature.
Fig. 1.
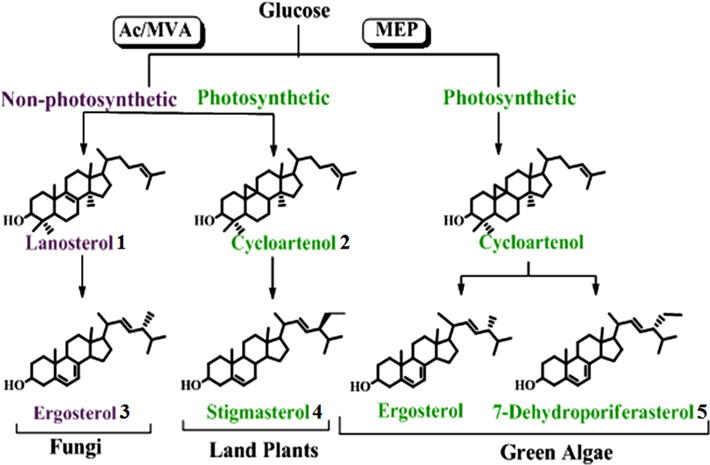
Sterol biosynthesis pathways of phylogenetic significance; AC–MVA is the acetate–mevalonate pathway to Δ3-IPP, and MEP is the methyl erythritol-D-phosphate pathway (or MVA-independent pathway) to Δ3-IPP.
The family of SMTs is considered to be a group of homologous enzymes derived from a common ancestor and are therefore structurally related (Nes, 2000). These slow-acting catalysts show a high degree of sequence similarity, possess tetrameric subunit organization and use comparable mechanistic features to carry out the C24-methylation reactions (Nes et al., 1998, 2003; Zhou et al., 2006). Testing variant acceptor molecules of heterologously expressed enzymes across kingdoms indicates that substrate specificities evolved differently in the two major classes of SMTs identified in the GenBank as SMT1 and SMT2. All SMT enzymes from fungi and plants accept Δ24(25)-substrates; the fungal SMT1 that prefers zymosterol 9 is given the designation EC 2.1.1.41, whereas the land plant SMT1 and SMT2 that prefer cycloartenol 2 and 24(28)-methylene lophenol 17 are given the designation EC 2.1.1.142 and EC 2.1.1.143, respectively (Benveniste, 2004; Zhou and Nes, 2003).
Fungi and green algae SMTs can convert protosterol intermediates to 24β-methyl sterols by convergent C24-methylation pathways commencing with the transfer of the electrophilic S-methyl group of SAM to C24 of a Δ24-sterol precursor (Nes, 2011). The cations produced can be eliminated through a phyla-specific de-protonation at C28 or C27 affording Δ24(28)-olefins in fungi or Δ25(27)-olefins d in green algae, respectively (Fig. 2 and Supplementary Figure S1). On the other hand, the Δ24(28)-route is expressed in the biosynthesis of 24α-ethyl sterols through the successive action of SMT1 and SMT2 (Bouvier et al., 2005; Neelakandan et al., 2009), suggesting a recapitulation of the fungal C24-methylation pathway through SMT1 that diverged to produce SMT2. Equally intriguing is the possibility based on bioinformatics analyses of amino acid sequences of SMTs annotated in the GenBank that the genome of green algae may be unique among primitive organisms and contain a single SMT2 gene bifunctional in substrate recognition (Supplementary Figure S2 and Table S1). This gene may have originated from a promiscuous SMT of the last eukaryotic common ancestor very early in the evolution of plants, and then diverged to yield SMT1 and SMT2 of land plants.
Fig. 2.
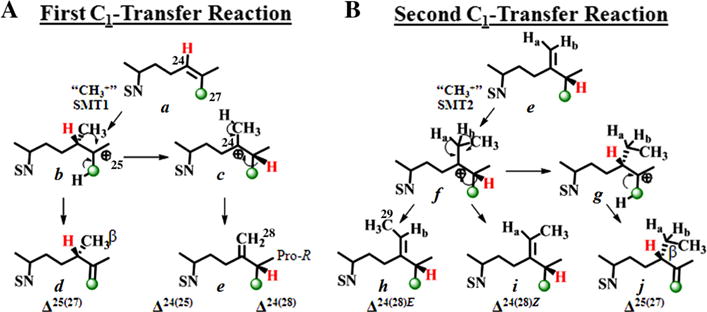
Alternative C24-alkylation pathways catalyzed by sterol C24-methyltransferase enzymes to Δ25(27)- or Δ24(28)-olefin products. 13C-labeled carbon is labeled in green. Stereospecific deprotonation at C28 of Ha-atom or Hb-atom yield the C24(28)E-ethylidene sterol or C24(28)Z-ethylidene sterol side chain geometry, respectively, as reported in Nes et al. (2003); SN, sterol nucleus. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
These opposing views for how SMTs evolved in distantly related groups led us to investigate the catalytic strategy of an early stage SMT at the root of the green lineage. Completion of the Chlamydomonas reinhardtii (Cr) genome project in green algae revealed that a SMT in the ergosterol biosynthesis (basal trait) pathway with homology to land plant SMT2 implicated in stigmasterol biosynthesis (derived trait) might be present in this primitive organism. Here, reported are functional characterization and mechanistic analysis of cloned CrSMT and product analysis of a remodeled active site from site-directed mutagenesis of a catalytically relevant Tyr110 (= Tyr81 of Erg6p) residue (Nes et al., 2002). By defining the overall rate enhancements and product specificities that distinguish catalytic behavior of an algal SMT from those of more-advanced SMT forms may provide insights into the evolutionary forces that determine the different side-chain constructions of phytosterols in plants and fungi and why SMT2s, which often mediate the balance of C24-methyl/ethyl sterols in plants (Schaller et al., 2000, 2001), appear to be restricted to the photosynthetic lineage.
2. Results
2.1. Functional expression of CrSMT
Using protocols previously successful to provide functional confirmation of the identity of a SMT gene in bacterial expression studies (Nes et al., 1998), CrSMT was incubated first with cycloartenol because this compound is considered the initial protosterol intermediate for ergosterol biosynthesis in C. reinhardtii (Nes, 2011; Summons et al., 2006). Activity assays performed under initial velocity conditions afforded approximate Km and Vmax values for cycloartenol of 35 μM ± 3 and 100 pmol/min/mg protein, respectively (Supplementary Figure S3), similar to the catalytic competency of other SMTs incubated with the corresponding natural substrate. Other properties of the CrSMT, such as pH optima (between 7.0 and 8.0) and temperature optima (between 30 and 35 °C), were similar to those reported previously for 24-SMTs (Nes et al., 1998, 2003; Zhou et al., 2006; Zhou and Nes, 2003). The conversion rate was linear with increasing time up to 90 min in a total lysate protein concentration of 1–2 mg/mL.
Product distribution by GC–MS analysis of the cycloartenol incubation with CrSMT indicated a mixture of three 24-alkyl (idene) sterols, formed in 70% overall yield in a ratio of 55:43:2 (Fig. 3A). From a preparative incubation with cycloartenol, individual 24-alkylated olefins were isolated by HPLC and the identity of each product verified from comparison with authentic standards as 24β-methyl cycloart-25(27)-enol (cyclolaudenol) 7 (product of the first biomethylation); ((EI-MS m/z [rel. int. (relative intensity)]): 440 (8) [M+ (molecular ion)], 425 (17) [M+-methyl], 422 (20) [M+-H2O], 407 (37) [M+-methyl-H2O], 379 (21) [M+-propyl-H2O], 353 (10) [M+-part of A ring-H], 315 (7) [M+-SC], 300 (32), 95 (100), 83 (31), 69 (92) [C-24–C-27 allylic cation+], 55 (70); δH: 4.667 (H2-27, brd), 1.640 (H3-26, s), 0.999 (H3-28, d, J 7.0), 0.957 (H6-18, s), 0.859 (H3-21, d, J 6.3), 0.550 (H1-19 endo, d, J 4.1), 0.330 (H1-19 exo, d, J 4.1)), 24(28)-methylene cycloartanol 14 (second product of the first biomethylation) (440 (7) [M+ (molecular ion)], 425 (17) [M+-methyl], 422 (23) [M+-H2O], 407 (40) [M+-methyl-H2O], 379 (21) [M+-propyl-H2O], 353 (7) [M+-part of A ring-H], 315 (5) [M+-SC], 300 (27), 95 (100), 83 (40) [C-23–C-27 allylic cation+], 69 (94), 55 (87); δH: 4.720 (H1-28, brs), 4.670 (H1-28, brs), 1.031 (H3-27, d, J 6.9), 1.026 (H3-26, d, J 6.8), 0.968 (H6-18, 30, s), 0.896 (H3-21, d, J 6.5), 0.555 (H1-19 endo, d, J 4.2), 0.334 (H1-19 exo, d, J 4.2)), and 24β-ethyl cycloart-25(27)-enol (product of the second biomethylation) (454 (8) [M+ (molecular ion)], 439 (16) [M+-methyl], 436 (18) [M+-H2O], 421 (31) [M+-methyl-H2O], 393 (17) [M+-propyl-H2O], 367 (8) [M+-part of A ring-H], 314 (26) [M+-SC-H], 95 (88), 83 (42) [C-23–C-27 allylic cation+], 69 (69), 55 (100); δH: 4.730 (H1-27, brd), 4.646 (H1-27, brd), 1.612 (H3-26, t, J 7.0), 0.965 (H3-18, s), 0.849 (H3-21, d, J 6.5), 0.808 (H1-29, t, J 7.5), 0.549 (H1-19 endo, d, J 4.0), 0.327 (H1-19 exo, d, J 4.0)), respectively. The chromatographic and spectroscopic analysis of the enzyme-generated sterol mixture clearly shows CrSMT can produce multiple 24-methyl/ethyl products from the successive methylation of a Δ24-substrate. These results, together with in vivo labeling studies (Miller et al., 2012), demonstrate methylation of the side-chains of ergosterol (24β-methyl) and 7-dehydroporiferasterol 5 (24β-ethyl) of green algae of the class Chlorococcales can be carried out by a single enzyme. In contrast, land plants utilize separate SMT1 and SMT2 enzymes in the methylation of the side-chains of campesterol (24α-methyl) and stigmasterol (24α-ethyl), respectively.
Fig. 3.
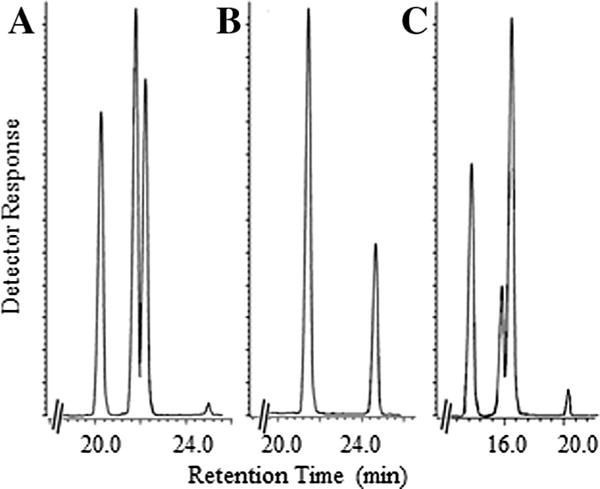
Total ion current chromatogram of the total sterol fraction recovered from wild-type CrSMT preparations incubated with natural and non-preferred substrates. Panel (A) is incubation of cycloartenol (first peak) followed by enzyme-generated products cyclolaudenol, 24(28)-methylene cycloartanol and 24β-ethyl cycloart-25(27)-enol; Panel (B) is incubation of 24(28)-methylene cycloartanol (first peak) followed by enzyme-generated product 24β-ethyl cycloart-25(27)-enol; and Panel (C) is incubation of zymosterol (first peak) followed by enzyme-generated products ergosta-8,25(27)-dienol, ergosta-8,24(28)-dienol and 24β-ethyl ergosta-8,25(27)-dienol, respectively.
2.2. Sequence of CrSMT
The CrSMT cDNA expressed in Escherichia coli generated a polypeptide with an open reading frame of 1161 bp that encodes for an apparent protein of 387 amino acids, which included Tyr110 (= Tyr81, ScSMT) and 62 other amino acids (16% of total residues) conserved in the primary structure. The four substrate binding segments typical of SMTs were arranged in the same order and separated by comparable intervals along the polypeptide side-chain (Fig. 4). Additionally, the residues found in the catalytic C-domain forming an α/β-Rossmann fold in the core of nucleotide binding proteins and SAM-dependent methyltransferases were also present in this class of SMT (Liscombe et al., 2012). The deduced amino acid sequence of CrSMT deposited in the GenBank matched our experimentally determined sequence and shows greater homology to known land plant SMT2s (48% similarity) than to known SMT1 (34% similarity) from fungi or protozoa (Supplementary Figure S4). As noted before, phylogenetic relationships of over 70 SMTs using alignments based on the CLUSTAL X program with standard settings for protein alignment show the CrSMT to cluster in the SMT2 protein family typically assigned to land plants (Supplementary Figure S2 and Table S1) (Zhou et al., 2008).
Fig. 4.

Deduced sterol C24-methyltransferase (SMT) amino acid sequence for Chlamydomonas reinhardtii (GenBank™ accession number, gi|158279461). Identical residues conserved in the primary structure of SMTs studied in this work are blocked in black. Substrate binding segments (boxed) are indicated for sterol-Regions I, III and IV and SAM-Region II.
2.3. Substrate acceptance for CrSMT catalysis
The influence of substrate structure on CrSMT sterol binding has been determined by using a series of substrates that differ in a single molecular feature or a combination of similar features. To evaluate the optimal substrate of CrSMT, it was incubated first with potential acceptor molecules synthesized by C. reinhardtii, cycloartenol, obtusifoliol 16, 24(28)-methylene cycloartanol, cyclolaudenol and ergosta-5,7,25(27)-trienol 13, then incubated with unnatural substrates considered preferred substrates of SMT1 and SMT2 of fungi and land plants, including zymosterol or lanosterol 1 and 24(28)-methylene lophenol, respectively. Initial steady-state kinetic analyses indicated the algal SMT can differentially recognize the test substrates within a relatively narrow range of Km values from 18 μM for 24(28)-methylene lophenol to 25 μM for cycloartenol, or there is no productive binding of the Δ25(27)-substrates ergosta-5,7,25(27)-trienol and cyclolaudenol (Table 1). A noticeable correlation between production rate (Vmax) and loss of substrate-active site complementarity is observed by modifications to the Δ24-substrate double bond undergoing alkylation. Thus, optimal binding interactions occurred with Δ24(25)-sterols that mimicked the structure of cycloartenol (Vmax/Km of 5.5 = 100% activity) and approximately 50–70% less activity for Δ24(28)-sterol substrates that mimicked the structure of obtusifoliol.
Table 1.
Kinetic parameters of sterol substrates assayed with CrSMT.
| Substratea | Km (μM) |
Vmax (pmol/min/mg) |
Vmax/Km |
|---|---|---|---|
| Cycloartenol | 25 | 137 | 5.48 (100) |
| Lanosterol | 24 | 109 | 4.54 (80) |
| Zymosterol | 22 | 120 | 5.45 (99) |
| 24(28)-Methylene lophenol | 20 | 52 | 2.89 (53) |
| Obtusifoliol | 19 | 42 | 2.21 (40) |
| 24(28)-Methylene cycloartanol | 18 | 29 | 1.61 (29) |
| Ergosta-5,7,25(27)-trienol | 0 | 0 | 0 |
| Cyclolaudenol | 0 | 0 | 0 |
Catalytic competence of cycloartenol conversion to product is normalized to 100% (in parenthesis). See Fig. 5 for a key to structures.
These results for the isolated enzyme are consistent with in vivo studies (Miller et al., 2012) showing the natural substrates of the first and second C1-transfer reactions are cycloartenol and obtusifoliol, respectively (cf. biosynthesis pathway in Supplementary Figure S1). The inability of CrSMT to metabolize 24β-methyl Δ25(27)-sterols, which is necessary to further alkylate the sterol side-chain of many uncommon triply alkylated sterols reported in marine organisms (Kerr and Baker, 1991), shows the progression of sterol methylation is limited in the green lineage. Alternatively, three acceptor molecules not synthesized by C. reinhardtii (lanosterol, zymosterol and 24(28)-methylene lophenol) are recognized with equal efficacy to the endogenous substrates cycloartenol or obtusifoliol which suggests a certain degree of substrate promiscuity in green algae which is not found in fungi. These unusual observations prompted further study of the product specificities of CrSMT by GC–MS analysis.
2.4. Variation in product specificity
The product outcomes of 17 acceptor molecules, incubated as before, can separate into two groups based on the side-chain olefin structure as Group I-Δ24(25) or Group II-Δ24(28) (Fig. 5). In the absence of competing Δ24(25)-substrate, substrates of Group II convert in relatively high yield to a single 24-ethyl product, typified by the conversion of 24(28)-methylene cycloartanol to 24-ethyl cycloart-25(27)-enol (Fig. 3, Panel B). A single product is formed whether the Δ24(28)-substrate contains zero, 1 or 2 methyl groups at C4 in the nucleus. To establish the generality of the 24β-ethyl stereochemistry in the C2-transfer product, obtusifoliol was incubated with CrSMT and the product purified by HPLC. The isolated product afforded: HPLC retention time of αc 1.2; EI-MS (m/z [rel. int. (relative intensity)]) 440 (42) [M+ (molecular ion)], 425 (100) [M+-methyl], 422 (2) [M+-H2O], 407 (18) [M+-methyl-H2O], 341 (7) [M+-part of A ring+H], 313 (4) [M+-A ring], 300 (4) [M+-SC-H], 95 (43), 83 (57) [C-23–C-27 allylic cation+], 69 (42), 55 (66); δH: 4.739 (H1-27, dd, J 2.0, 2.5), 4.653 (H1-27, d, J 2.5), 1.573 (H3-26, s), 0.974 (H3-19, s), 0.890 (H3-21, d, J 6.5) 0.707 (H3-18, s), 0.810 (H3-29, t, J 7.2) (Supplementary Figure S5). The NMR assignments of the isolated sterol are consistent with the product side-chain bearing the 24β-ethyl stereochemistry and a Δ25(27)-double bond (Goad and Akihisa, 1997).
Fig. 5.
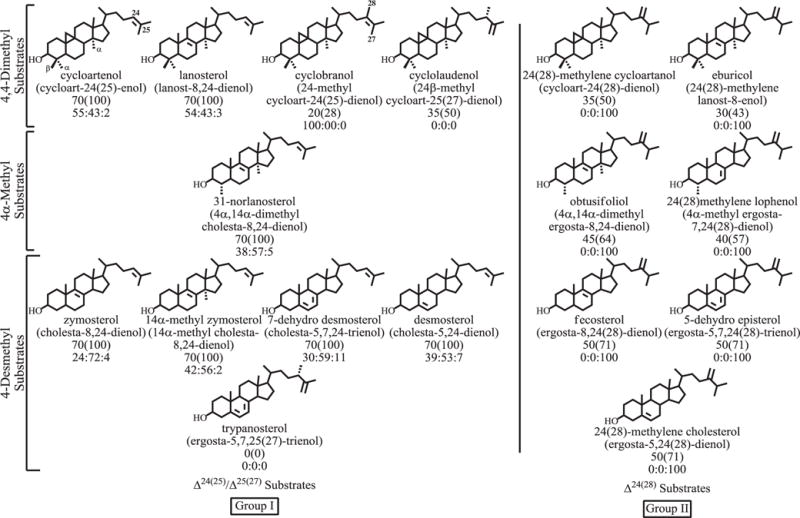
Product distributions of substrates incubated with the CrSMT. Figure is outlined as follows: name of substrate, in parenthesis the systematic name of sterol, percent conversion of substrate to product, in parenthesis catalytic competence normalized to 100% against cycloartenol, and the enzyme-generated product sets of C24-methyl C25(27) olefin to C24(28)-methylene olefin to C24-ethyl C25(27)-olefin.
Retention of the Δ25(27)-double bond was confirmed in the product of the second alkylation reaction of the substrate 24-methyl cycloart-24(25)-enol (cyclobranol) 6 by a combination of chromatographic and spectroscopic analysis (Supplementary Figure S6): HPLC retention time of αc 1.9; EI-MS, 454 (4) [M+ (molecular ion)], 439 (7) [M+-methyl], 436 (9) [M+-H2O], 421 (16) [M+-methyl-H2O], 393 (8) [M+-propyl-H2O], 367 (4) [M+-part of A ring-H], 314 (16) [M+-SC-H], 95 (100), 83 (91) [C-23–C-27 allylic cation+], 69 (73), 55 (98); δH: 4.722 (H2-27, brd), 4.661 (H2-27, brd), 1.607 (H3-26, t, J 11.0), 0.966 (H6-18,30, s), 0.861 (H3-21, s), 0.808 (H6-28,28′, s), 0.549 (H1-19 endo, d, J 4.5), 0.325 (H1-19 exo, d, J 4.5). This unnatural CrSMT-generated product cycloneolitsol contains an uncommon dimethyl group at C24 and is occasionally detected in marine organisms and protozoa (Goad and Akihisa, 1997). Alternatively, incubation of cyclobranol with land plant SMT1 from soybean yields the 24(28)-methylene 25-methyl side chain (Wang and Nes, 2008).
However, unexpected and unprecedented product ratios were observed from incubation of Δ24(25)-substrates that possess a different C4-atom substitution from cycloartenol that contains a 4,4-dimethyl group. Thus, Δ24(25)-substrates that contain a geminal C4-methyl group convert to Δ25(27)- and Δ24(28)-olefin product ratios of approximately 5:4, whereas substrates with no C4β-angular methyl group, as in 31-nor-lanosterol 8, convert to a product ratio of 4:6 and substrates with no C4-methyl groups, as in zymosterol, convert to a product ratio of 4:14 (Fig. 3, Panel C). This remarkable metabolic switching in product ratios likely result from changes in C4-steric effects on the polarity of the C3-anchor group, that therefore can posit the terminal methyl groups of the bound intermediate somewhat further away (as for C27) or closer (as for C28) to the deprotonating base (Supplementary Figure S7). Although the catalytic proficiencies for alternative substrates tested with CrSMT could be similar for natural and unnatural substrates, no substantial rate enhancements for Δ24(28)-substrates over the Δ24(25)-substrates are achieved. This contrasts with the situation of SMT2 from land plants where rate enhancements are detected for select Δ24(28)-substrates (e.g., 24(28)-methylene lophenol) over Δ24(25)-substrates (e.g., cycloartenol).
2.5. Mechanistic analysis
Early studies have established two parallel C24-alkylation pathways for the synthesis of 24β- and 24α-isomers in phytosterol biosynthesis across kingdoms (Goad and Akihisa, 1997; Kerr and Baker, 1991). The key to this chemodiversity originates in the stereochemical path and regiospecificity of the coupled-methylation-deprotonation reaction. It is well established that the 24-methyl and 24-ethyl groups (C28 and C29) in ergosterol and 7-dehydropo-riferasterol produced by green algae when grown in presence of [methyl-2H3 = CD3] methionine, should contain up to three and five deuterium atoms in the molecules, respectively (Miller et al., 2012; Nes, 1977). In contrast, campesterol and stigmasterol incorporate 2 and 4 deuterium atoms in the 24-alkyl group, respectively (Nes, 1977). Therefore, it was not surprising to observe that cloned CrSMT, incubated with [methyl-2H3]SAM and lanosterol, afforded 24-methyl(ene)-products that incorporated three and two deuterium atoms (M+ 443 and M+ 442, respectively; Table 2). GC–MS analysis of the sterol mixture showed a notable change in the proportion of olefin products in the direction of 24β-methyl lanosta-8,25(27)-dienol. This demonstrates the operation of a pronounced kinetic isotope effect (KIE) during the stabilization of C25 cation b and indicates that proton loss from C28 c is the rate-limiting step in Δ24(28)-sterol formation (Fig. 2). These data further support our recent conclusion using cloned soybean SMT1 that the coupled methylation-de-protonation pathway affording multiple products has a fast kinetic step as the alkylation of the Δ24-double bond to give initially a C25-carbonium ion and a slow de-protonation step that establishes the product specificity (Patkar et al., 2013).
Table 2.
Percentage composition of sterol mixtures generated by CrSMT incubated with 2H- or 13C-labeled substrates.
| Substrate pair | Product ratioa (M+)
|
||
|---|---|---|---|
| 1 | 2 | 3 | |
| Lanosterol/SAM | 55 (440) | 38 (440) | 7 (454) |
| [27-13C]Lanosterol/SAM | 54 (441) | 38 (441) | 8 (455) |
| [24-2H]Lanosterol/SAM | 54 (441) | 38 (441) | 8 (455) |
| Lanosterol/[2H3-methyl]SAM | 88 (443) | 12 (442) | 0 (459) |
CrSMT products determined by GC–MS analysis are: 1, 24β-methyl lanosta-8,25(27)-enol; 2, 24(28)-methylene lanosterol (eburicol), and 3, 24β-ethyl lanosta-8,25(27)-enol.
For the side-chain formation of ergosterol and 7-dehydroporif-erasterol in green algae, a C24-methylation reaction mechanism that involves β-face attack of the “methyl cation” on the Δ24-substrate bond has been proposed (Fig. 2) (Nes, 2011). Based on the product specificity of CrSMT reported above, an SN2-type SAM bio-alkylation is demonstrated through 1HNMR identification of the 24β-alkyl group stereochemistry for enzyme-generated products 24β-methyl cycloart-25(27)-enol and 24β-ethyl cycloart-25(27)-enol, respectively. Mass spectral data of the three enzyme-generated products show a one mass unit increase over the corresponding non-labeled product (Table 2). The molecular ion clusters for 2H[D]-labeled 24(28)-methylene product (M+ 441) show increased peak intensities in the high mass end region from loss of isopropyl group at M+-44 (m/z 397) and scission between C22 and C23 of m/z 84 at m/z 357, thus establishing the presence of deuterium label at C25. Alternatively, the enhanced fragment ion for allylic cleavage through the C22–23 bond at M+-70 (m/z 371) and fragment ion of m/z 84 in the deuterium labeled 24-methyl/ethyl Δ25(27)-products (M+ 441 and M+ 455, respectively) show the 2H-atom is retained at C24 in these molecules (Supplementary Figures S8–S10).
13CNMR spectroscopic analysis of the two diastereotopic methyl groups at C25 in the 13C-labeled product show the signal for pro-R methyl group (C27) (Dennis and Nes, 2002; Guo et al., 1996) is markedly enhanced at 21.86 ppm (Fig. 6). The general picture which emerges from these studies is one in which the pro-Z methyl group on cycloartenol, corresponding to C27, is transformed into the pro-R methyl group at C25 on 24(28)-methylenecycloartanol by migration of the hydrogen atom at C24 to C25 from the Re-face of the substrate double bond (Fig. 2). 13CNMR spectroscopic analysis of the other enzyme-generated products from incubation of [27-13C]lanosterol ([27-13C]1) or [27-13C]eburicol ([27-13C]15) (recovered from the sterol mixture of the [27-13C]lanosterol) show a single enhanced peak resonating at 109 ppm (Fig. 6). These observations indicate the biosynthetic inequality of chemically equivalent C26 and C27 and for regiospecific proton elimination (CH3 → CH2) at the cis terminal methyl group of the substrate double bond to become C27. The absence of scrambling of methyl groups during the biosynthesis of 24-methyl or 24-ethyl sterols as can occur in the biosynthesis of some pentacyclic triterpenoid (Kushiro et al., 1999) indicates the isopropyl group of the 24β-alkyl cation must be held tightly by CrSMT.
Fig. 6.
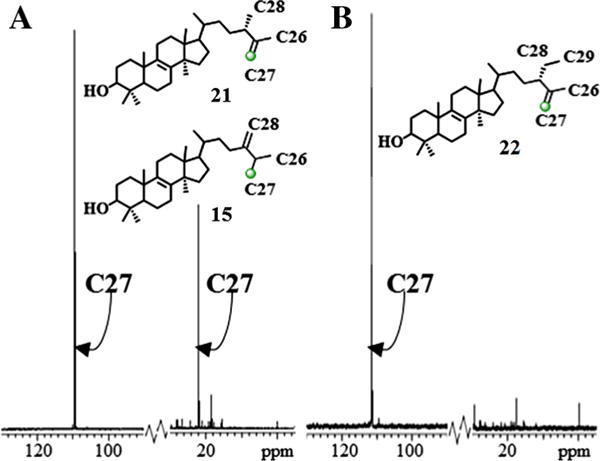
Partial 13C NMR spectra of 13C-labeled products from incubation of CrSMT with [27-13C]lanosterol (Panel A) or [27-13C]eburicol (Panel B).
2.6. Remodeling the CrSMT active site at position-110
A combination of sequence alignments, molecular modeling and site-directed mutagenesis experiments were used previously to identify residues that contribute to catalytic specificity within the substrate recognition segment of Region I, considered the signature motif for this class of catalyst (Ganapathy et al., 2008). Recent studies that focused on modification of the enzyme active site by site-directed mutagenesis indicated the aromatic amino acid at position-81(tyrosine) in Erg6p is essential for the successive methylation of the Δ24-bond (Ganapathy et al., 2008). Also discovered was that this residue functions mechanistically in the Michaelis complex to stabilize the bound intermediate through cation–π interactions (Nes et al., 2008). Additionally, the data herein on Erg6p inhibition show that ground-modeled inhibitors have little or no effect in binding due to Tyr81 mutation, whereas transition-state inhibitor binding is greatly affected because of the substitution on Tyr81 (Nes et al., 2008). This observation is consistent with the role of the Tyr81 residue during transition-state stabilization, while it may not be required for substrate binding. Taking into account these catalysis, two site-specific mutations targeted at the Tyr110 residue in CrSMT (= Tyr81 in Erg6p; ScSMT) were constructed to inspect the function of the polar residue. Residue numbers described hereafter correspond to the consecutive numbering of the full-length amino acid sequence of a given gene beginning with the start codon. For example, using Erg6p as reference the amino acid numbers in CrSMT are frame shifted up by 29 amino acids, whereas for TbSMT the residues are shifted down by 15 amino acids (Supplementary Figure S4).
GC–MS analysis of enzyme-generated products from the Phe mutant versus wild-type enzyme showed that removal of the OH group from tyrosine had little effect on catalysis (Table 3), suggesting a change in bulk at position-81 can be tolerated. These results were disappointing since in related studies of fungal and plant SMT1, the Tyr → Phe mutation afforded a gain-in-function such that the mutant Erg6p proceeds to carry out a plant-like second biomethylation of fecosterol 18 (Kushiro et al., 1999), or the mutation can in soybean enzyme produce unusual product ratios from biomethylation of 24(28)-methylene cycloartanol (Nes et al., 2006). On the other hand, substitution of Tyr81 by leucine, an aliphatic residue that can alter the polarity of the contact amino acid involved with stabilization of the bound intermediate cation, yielded a mutant enzyme that converts cycloartenol and zymosterol to product ratios that favor the formation of Δ25(27)-olefins (Fig. 3, Panel C), or as shown by incubation of 24(28)-methylene cycloartanol the second C1-transfer activity is inhibited. In most cases where the C25 cation is formed by attack of “ ” on the Δ24-bond, several alternatives exist for stabilization (Fig. 2). The one that is perhaps most expected, if the process is concerted, is removal of a proton from one of the gem-dimethyl groups, that thereby eliminates the tertiary cation (i.e., to form cyclolaudenol). However, for this outcome to take place at the expense of Δ24(28)-formation, as can occur during the second C1-transfer reaction, there must be proper positioning of a stabilizing residue to control the timing of the 1,2-hydride shift involved with elimination of the C24-cation. In particular, the secondary carbocations are significantly farther away to the deprotonating base than the preceding tertiary carbocations (Fig. 2 and Supplementary Fig. 7, Panel A). Accordingly, it is hypothesized that the introduced aliphatic residue in CrSMT at position-110, which is inert, fails to adequately stabilize the C24-cation such that the ensuing reaction channeling during the course of biomethylation is interrupted.
Table 3.
Product ratios of substrates incubated with wild-type and mutant CrSMT.
| Strain | Substrate | Δ25(27) to Δ24(28)-producta (% conversion) |
|---|---|---|
| Wild-type | Cycloartenol 24(28)-Methylene cycloartanol Zymosterol |
57/43 (70) 100/0 (35) 28/72 (70) |
| Tyr110Phe | Cycloartenol 24(28)-Methylene cycloartanol Zymosterol |
65/35 (45) 100/0 (60) 30/70 (43) |
| Tyr100Leu | Cycloartenol 24(28)-Methylene cycloartanol Zymosterol |
65/35 (10) NCb 55/45 (55) |
Product mixtures, analyzed by GC–MS, contain trace 24-ethyl sterol. In parenthesis is the percent conversion of substrate to product.
NC, no conversion.
3. Discussion
The current studies were undertaken, in part, to define further enzyme action responsible for the expansion of chemodiversity in phytosterol biosynthesis, and furthermore to probe in depth the fundamental properties of SMT enzymes that led to the switch in C24-methylation routes from producing algal ergosterol and 7-dehydroporiferasterol (24β-ethyl group) to producing land plant stigmasterol (24α-ethyl group). A simplified overview of phytosterol biosynthesis with SMT as a biomarker of the phyla-specific pathways is shown as Supplementary Figure S1. The class of Chlorophyceae within the Kingdom of Viridplantae has been placed at the bottom of the eukaryotic tree (Cavalier-Smith, 2004). In the present report, green algae is shown to encode a bifunctional SMT2 that catalyzes 24β-methyl/ethyl sterol products that strongly supports this theory. Our phylogenetic analysis shows that SMT2 and SMT1 lineages evolved repeatedly from a common ancestral gene, likely concurrent with the origin of the eukaryote domain, and then lost variably in some lineages because no SMT homologs from prokaryotes or advanced animals (above the sponges) were identified (Fig. 7). It is interesting that plant-derived SMT genes acquired different catalytic strategies upon mutational divergence into SMT1 (red algae) and SMT2 (green algae). The tendency for preferential binding of cycloartenol to plant SMT1 and zymosterol to fungal SMT1 compared to the preferential binding of 24(28)-methylene lophenol to land plant SMT2 support the notion these enzymes evolved their requirements for C4-substituted substrates independently. Given that land plant SMT1 possesses a vestigial Δ25(27)-olefin pathway (Patkar et al., 2013) and that green algal SMT2 possess a binding pocket for 24(28)-methylene lophenol suggest an algal-like SMT2 underwent mutational divergence affording separate land plant SMT1 and SMT2 isoforms; these enzymes acquired the Δ24(28)-route to form 24Z-ethylidene sterols (Benveniste, 2004; Bouvier et al., 2005; Neelakandan et al., 2009) required in the synthesis of 24α-alkyl sterols. In this way, SMT2 evolution has remained confined to the green lineage.
Fig. 7.
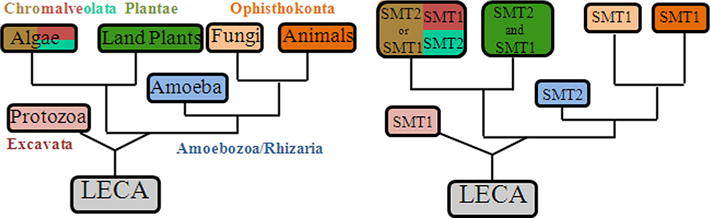
Phylogenetic arrangement of life forms (based on Cavalier-Smith, 2004) correlated to the distribution of SMT types reported in the GenBank. Color schemes for Chromalveolata indicate brown, green and red algae. LECA, last eukaryotic common ancestor. SMTs arranged according to sequence relatedness is reported in Supplementary Figure S2 and Supplementary Table 1. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In pursuing the discussion of SMT evolution, an empirical approach was taken which emphasizes a major determinant of molecular diversity in sterol side chain-structure, and which is the precise anchoring system of ring-A and the conformation of the bound intermediate at the active site (Howard et al., 2012). Comparative analysis with the genomes of plants, fungi, and protozoa indicates that the SMT genes involved with phytosterol biosynthesis are orthologous among these organisms. Accordingly, these gene products should display functional equivalence with increases in catalytic efficiency and improvements in product ratios resulting from changes in differential binding of substrates associated with the sterol shape and functional groups of the anchoring system (Albery and Knowles, 1977). In this evolutionary scenario, the charge-stabilizing mechanism producing the Δ24(28)Z-olefin pathway for land plant phytosterol biosynthesis depends on the proximity of the cationic center with the nearest substrate double bond during sterol methylation (Supplementary Figure S7). As shown here, the presence of Try110 in CrSMT is relevant to cation-π interactions by stabilizing the ensuing C25- and C24-cations in the intermediate in the active site. The loss of these interactions can lead to directed channeling and loss of the second biomethylation activity; as such this residue functions in algal SMT to control the expansion of the sterol side-chain.
SMTs have strongly conserved amino acid sequences and therefore the corresponding common ancestral gene can be envisioned to have possessed a finite number of introns, but then lost several of them during evolution giving rise to different sets of conserved intron–exon organization for the extant enzymes. A similar proposal has been presented for the evolution of intron–exon organization in RNA sequences of terpene synthases (Lange et al., 2000). Recently, Neelakandan et al. examined the intron–exon structures for 11 SMTs representing green algae, fungi, protozoa and land plants (Neelakandan et al., 2009). Most striking from the genomic organization of SMT from green algae to vascular plants is that these isoforms can be placed into a monophyletic clade and classed by the number of introns-5 (green algae) to 12 (embryophytes)-dispersed between exons in the SMT structure. On the other hand, land plant SMT2, protozoan SMT1 and some fungal SMT1 are marked by a single conserved intron suggesting that algal SMT2 might have diverged from an ancestral SMT early in plant evolution. Indeed, continued evolution of the green algal SMT2 into the Charophyta could provide the antecedent of land plant SMT1 and SMT2 (Patterson et al., 1991); notably, the Chara synthesize 24(28)Z-ethylidene cholesterol (isofucosterol) which is an intermediate to stigmasterol biosynthesis (Patterson et al., 1991). Interestingly, red algae typically synthesize cholesterol as their dominant sterol (Beastall et al., 1974; Kodner et al., 2008), and cholesterol is often present in variable amounts in other organisms as diverse as the oomycetous fungi and humans (Nes, 2011; Weete et al., 2010), suggesting a down-regulation or loss of SMT activity from these lineages. Therefore, it is evident that retrograde evolution of SMT could be class-specific, and may have arisen in select lineages due to loss of promoters in the cis-sequence motifs located upstream of the SMT coding sequence (Neelakandan et al., 2010). Thus, directed mutations of regulatory and transcribed regions associated with the SMT gene may be responsible for shifts in sterol biosynthesis pathways which in the lineage generating vertebrate animals led to coupled loss in phytosterols with a gain in cholesterol production.
4. Concluding remarks
The results described here provide important clues about the conservation of molecular mechanism versus diversity-creating channel switching in the C24-methylation reaction, especially with regard to the face of the substrate Δ24-bond that is methylated as well the stabilization of the resulting carbocationic intermediate(s) through cation–π interactions. The CrSMT relaxed specificity for unnatural substrates and remodeling of the active site through mutagenesis of a catalytically relevant aromatic residue appears not only to provide the framework for the continued expansion and variations in sterol side chain constructions but may also allow further modifications of active site residues by mutagenesis that produce novel sterol profiles affording value-added traits in plants and microorganisms.
5. Experimental
5.1. Chemicals and instrumental analysis
The source of 17 sterol substrates included in this study is described in earlier papers (Liu et al., 2011; Miller et al., 2012; Xu et al., 1988). All sterols were purified by HPLC to <95% by GC analysis. SAM iodide salt was purchased from Sigma, [methyl-3H3]-SAM (specific activity 10–15 Ci/mMol and diluted to 10 μCi/μmol, 1 Ci = 37 GBq) Tetraosylate [methyl-2H3]SAM (SAM is S-adenosyl-L-methionine = AdoMet; 99% atom enrichment) was purchased from C/D/N Isotopes (Pointe-Claire, QC), and [24-2H]lanosterol ([24-2H]1) and [27-13C]lanosterol (98% atom enrichment) were prepared as previously described (Schaller et al., 2001). The Bradford protein assay kit was purchased from Bio-Rad and isopropyl-1-thio-β-D-galactoside (IPTG) was from Research Products International Corp. The QuikChange site-directed mutagenesis kit-was purchased from Strategene. All other reagents and chemicals were from Sigma or Fisher unless otherwise noted.
Instrumental methods have been reported previously (Liu et al., 2011; Xu et al., 1988). Briefly, proton and carbon NMR spectra were recorded in CDCl3 at the indicated frequencies using a Varian Unity Inova 500 MHz spectrometer or JEOL ESC 400 MHz spectrometer. Chemical shifts (δ, ppm) are referenced to TMS (δ, 0). Mass spectra were obtained on a Hewlett-Packard 6890 GC-HP 5973 MSD instrument (electron impact, 70 eV, scan range 50–550 amu). HPLC was carried out using Phenomenex Luna C18-column (250 mm × 4.6 mm × 5 μm) connected to a diode array multiple wavelength detector (Liu et al., 2011). Capillary GC (0.25 mm i.d. by 30 m fused silica column coated with Zebron ZB-5 from Phenomenex) was operated as previously described (Miller et al., 2012; Liu et al., 2011). GC and HPLC of sterols are reported as retention times relative to the elution time of cholesterol as RRTc and αc, respectively. Product distributions are determined by approximate integration of chromatographic peaks.
5.2. Cloning of full length cDNA of CrSMT
By use of conventional, PCR based methods, a full length 1136 bp cDNA was assembled from CrSMT cDNA clones (1030123H02.y1, 102407H12.y1 and 963068B05.×1) available from the Chlamydomonas Resource Center (http://chlamycollection.org) and was identical to the published transcript sequence of CrSMT (NCBI: XM_001690723.1 and Merchant et al. (2007)). The cDNA was cloned into a pet30a+ (Novagen) between EcoR I and Xho I sites and transformed into E. coli BL21 strain affording a wild-type recombinant CrSMT protein of 384 amino acids, three short from the predicted sequence of 387 amino acids. The DNASTAR Lasergene 8 sequence analysis programs (DNA STAR Inc.,) was used to identify the largest open reading frame of the cDNA sequence of CrSMT. The protein sequence of CrSMT translated from cDNA was determined in SeqBuilder.
5.3. CrSMT assay and product analysis
For bacterial expression of cloned CrSMT protein, a 50 mL culture of E. coli BL21(DE3) strain harboring the CrSMT cDNA was supplemented with of kanamycin (50 μL, 50 mg/mL) and grown at 30 °C to an optical density (OD) of 0.7. A 20 mL aliquot plus 50 μL of kanamycin was transferred to 1 L of LB media and incubated at 30 °C for 3 h to an OD of 0.5–0.6 at 225 rpm. Cultures were then induced by the addition of 0.4 mM IPTG and grown for another 2 h to an OD of 1.1–1.3. Cells were harvested by centrifugation (7500×g) and re-suspended in 5 mL of phosphate buffer 20 mM containing 5% (v/v) glycerol at pH 7.5. The cell pellet was broken by French Press disruption and the resulting lysate preparations, maintained in phosphate buffer throughout, were assayed under initial velocity conditions (1–2 mg total protein) of 45 min using the appropriate sterol acceptor tested over the concentration range 5–150 μM and paired with saturating levels of [methyl-3H3] SAM at 100 μM (0.6 μCi) following standard protocols (Nes et al., 2008). Incubation mixtures were saponified with methanolic KOH and extracted with hexane (3×1 mL) to provide the non-saponifiable lipid fraction. This assay involves scintillation counting of the non-saponifiable lipid fraction to determine conversion rate which did not differ by more than 10%. Incubation of [methyl-3H3]SAM, in the absence of sterol, with enzyme preparation generated background levels of radioactivity between 500 to 1000 dpm. Data analysis was determined according to the Lineweaver–Burk equation using the computer program Sigmaplot 2001 plus the enzyme kinetics module software package (Nes et al., 2008). In control enzyme preparations of E. coli, there is no sterol contaminant detected by GC–MS as would be expected since these compounds are not synthesized by bacteria. Product distributions generated by wild-type and mutant CrSMT were determined by incubating saturating amounts of the co-substrates with sufficiently large preparations of protein (2–3 mg/mL) for 8–12 h to allow completion of the reaction; products were analyzed by GC–MS or in special cases the products were purified by HPLC and analyzed further by NMR spectroscopy. Product yields determined by capillary GC were accurate to approximately 1% (50 ng). Protein amounts were quantified by using the Bradford method (Bio-Rad) and bovine albumin as the protein standard.
5.4. Site-directed mutagenesis
Point mutants, constructed from the wild-type CrSMT gene, were made with a pet30a+ vector by using the standard Quik-Change mutagenesis method (Stratagene) and confirmed by DNA sequencing. Specific mutagenic primers (sense direction with mutagenic bases shown in boldface) are as follows: Y100FCTAGTCACT GACATTTTCGAGTGGGGCTGGGGC, Y110LCTAGTCACTGACATTCTCGAGTGGGGCTGGGGC. Plasmid DNA constructs were transformed into BL21 (DE3) cells, and expression of the CrSMT genes were induced by IPTG addition to the cultures as described for wild-type CrSMT. The product distribution generated by mutant SMTs were determined by GC–MS following overnight incubation of saturating amounts of co-substrates (100 μM) with lysate protein (3 mg).
Supplementary Material
Acknowledgments
This investigation was supported by a National Science Foundation Grant [Grant number MCB 0929212, to W.D. Nes] and National Institutes of Health Grant [Grant number GM-25661, to W.J.S.]. Its contents are solely the responsibilities of the authors and do not necessarily represent the official views of the federal agencies. The support of the National Science Foundation for NMR spectroscopy instrumentation [Grant number CHE-1048553] is also gratefully acknowledged.
Abbreviations
- SMT
sterol C24-methyltransferase
- SAM
S-adenosyl-L-methionine
- GC–MS
gas chromatography–mass spectroscopy
- SC
side chain
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.phytochem.2014.07.019.
Footnotes
This contribution is in honor of Professor Vincenzo de Luca’s 60th birthday
References
- Albery WJ, Knowles JR. Efficiency and evolution of enzyme catalysis. Angew Chem Int Ed. 1977;16:285–293. doi: 10.1002/anie.197702851. [DOI] [PubMed] [Google Scholar]
- Beastall GH, et al. Sterols in Porphyridium species. Eur J Biochem. 1974;41:301–309. doi: 10.1111/j.1432-1033.1974.tb03270.x. [DOI] [PubMed] [Google Scholar]
- Benveniste P. Biosynthesis and accumulation of sterols. Annu Rev Plant Biol. 2004;55:429–457. doi: 10.1146/annurev.arplant.55.031903.141616. [DOI] [PubMed] [Google Scholar]
- Bouvier FA, et al. Biogenesis, molecular recognition and function of plant isoprenoids. Prog Lipid Res. 2005;44:357–429. doi: 10.1016/j.plipres.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Brocks JJ, et al. Archean molecular fossils and the early rise of eukaryotes. Science. 1999;285:1033–1036. doi: 10.1126/science.285.5430.1033. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Only six kingdoms of life. Proc R Soc Lond B. 2004;271:1251–1262. doi: 10.1098/rspb.2004.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis AL, Nes WD. Sterol methyl transferase. Evidence for successive C-methyl transfer reactions generating Δ24(28)- and Δ25(27)-olefins by a single plant enzyme. Tetrahedron Lett. 2002;43:7017–7021. [Google Scholar]
- Desmond E, Gribaldo S. Phylogenomics of sterol synthesis: insights into the origin, evolution and diversity of a key eukaryotic feature. Genome Biol Evol. 2009;2009:364–381. doi: 10.1093/gbe/evp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganapathy K, et al. Molecular probing of the Saccharomyces cerevisiae sterol 24-C-methyltransferase (SMT) reveals multiple amino acids involved in C2-transfer activity. Biochim Biophys Acta. 2008;1781:344–351. doi: 10.1016/j.bbalip.2008.04.015. [DOI] [PubMed] [Google Scholar]
- Goad LJ, Akihisa T. Analysis of Sterols. Blackie Academic; London: 1997. [Google Scholar]
- Guo D, et al. Stereochemistry of hydrogen migration from C24 to C25 during phytosterol biomethylation. J Am Chem Soc. 1996;118:8507–8508. [Google Scholar]
- Howard AL, et al. Sterol C24-methyltransferase: physio- and stereo-chemical features of the sterol C3-group required for catalytic competence. Arch Biochem Biophys. 2012;512:43–50. doi: 10.1016/j.abb.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Kerr RG, Baker BJ. Marine sterols. Nat Prod Rep. 1991;14:465–497. [Google Scholar]
- Kodner RB, et al. Sterols in red and green algae: quantification, phylogeny, and relevance for the interpretation of geologic stearanes. Geobiology. 2008;6:411–420. doi: 10.1111/j.1472-4669.2008.00167.x. [DOI] [PubMed] [Google Scholar]
- Kushiro T, et al. Chimeric triterpene synthase. A possible model for multifunctional synthase. J Am Chem Soc. 1999;121:1208–1216. [Google Scholar]
- Lange BM, et al. Isoprenoid biosynthesis: the evolution of two ancient and distinct pathways across genomes. Proc Natl Acad Sci USA. 2000;97:13172–13177. doi: 10.1073/pnas.240454797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK. Evolution of carotenoid and isoprenoid biosynthesis in photosynthetic and nonphotosynthetic organisms. Annu Rev Plant Biol. 1999;50:47–65. doi: 10.1146/annurev.arplant.50.1.47. [DOI] [PubMed] [Google Scholar]
- Liscombe DK, et al. Architectures, mechanisms, molecular evolution of natural product methyltransferases. Nat Prod Rep. 2012;29:1238–1250. doi: 10.1039/c2np20029e. [DOI] [PubMed] [Google Scholar]
- Liu J, et al. Effect of substrate features and mutagenesis of active site tyrosine residues on the reaction course catalyzed by Trypanosoma brucei sterol C24-methyltransferase. Biochem J. 2011;439:413–422. doi: 10.1042/BJ20110865. [DOI] [PubMed] [Google Scholar]
- Lombard J, Moreira D. Origins and early evolution of the mevalonate pathway of isoprenoid biosynthesis in the three domains of life. Mol Biol Evol. 2011;28:87–99. doi: 10.1093/molbev/msq177. [DOI] [PubMed] [Google Scholar]
- Love GD, et al. Fossil steroids record the appearance of demospongiae during the Cryogenian period. Nature. 2009;5:718–721. doi: 10.1038/nature07673. [DOI] [PubMed] [Google Scholar]
- Merchant SS, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MB, et al. Evolutionary conserved Δ25(27)-olefin ergosterol biosynthesis pathway in the alga Chlamydomonas reinhardtii. J Lipid Res. 2012;53:1636–1645. doi: 10.1194/jlr.M027482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelakandan AK, et al. Cloning, functional expression, and phylogenetic analysis of plant 24C-methyltransferases involved in sitosterol biosynthesis. Phytochemistry. 2009;70:1982–1998. doi: 10.1016/j.phytochem.2009.09.003. [DOI] [PubMed] [Google Scholar]
- Neelakandan AK, et al. Molecular characterization and functional analysis of Glycine max sterol methyltransferase 2 genes involved in plant membrane sterol biosynthesis. Plant Mol Biol. 2010;74:503–518. doi: 10.1007/s11103-010-9692-6. [DOI] [PubMed] [Google Scholar]
- Nes WR. Biochemistry of plant sterols. Adv Lipid Res. 1977;15:315–324. [Google Scholar]
- Nes WD. Sterol methyltransferase: enzymology and inhibition. Biochim Biophys Acta. 2000;1529:63–88. doi: 10.1016/s1388-1981(00)00138-4. [DOI] [PubMed] [Google Scholar]
- Nes WD. Biosynthesis of cholesterol and other sterols. Chem Rev. 2011;111:6423–6451. doi: 10.1021/cr200021m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nes WD, et al. Overexpression, purification, and stereochemical studies of the recombinant (S)-adenosyl-L-methionine: Δ24(25)- to Δ24(28)-sterol methyl transferase enzyme from Saccharomyces cerevisiae. Arch Biochem Biophys. 1998;353:297–311. doi: 10.1006/abbi.1998.0665. [DOI] [PubMed] [Google Scholar]
- Nes WD, et al. Active site mapping and substrate channeling in the sterol methyltransferase pathway. J Biol Chem. 2002;277:42459–42556. doi: 10.1074/jbc.M204223200. [DOI] [PubMed] [Google Scholar]
- Nes WD, et al. Biosynthesis of phytosterols: kinetic mechanism for the enzymatic C-methylation of sterols. J Biol Chem. 2003;278:34505–34516. doi: 10.1074/jbc.M303359200. [DOI] [PubMed] [Google Scholar]
- Nes WD, et al. Probing the sterol binding site of soybean sterol methyltransferase by site-directed mutagenesis: functional analysis of conserved aromatic amino acids in region 1. Arch Biochem Biophys. 2006;448:23–30. doi: 10.1016/j.abb.2005.08.022. [DOI] [PubMed] [Google Scholar]
- Nes WD, et al. Yeast sterol C24-methyltransferase: role of highly conserved tyrosine in catalytic competence studied by site-directed mutagenesis and thermodynamic analysis. Arch Biochem Biophys. 2008;477:316–323. doi: 10.1016/j.abb.2008.05.016. [DOI] [PubMed] [Google Scholar]
- Opitz S, et al. Both methylerythritol phosphate and mevalonate pathways contribute to biosynthesis of each of the major isoprenoid classes in young cotton seedlings. Phytochemistry. 2014;98:110–119. doi: 10.1016/j.phytochem.2013.11.010. [DOI] [PubMed] [Google Scholar]
- Patkar P, et al. C24-Methylation of 26-fluorocyclartenols by recombinant sterol C24-methyltransferase from soybean: evidence for channel switching and its phylogenetic implications. Biochem J. 2013;456:253–262. doi: 10.1042/BJ20121818. [DOI] [PubMed] [Google Scholar]
- Patterson GW, et al. Sterols of the Charophycease. J Nat Prod. 1991;54:1141–1143. [Google Scholar]
- Schaller H, et al. Plant sterol-C24-methyltransferase: different profiles of tobacco transformed with SMT1 and SMT2. Lipids. 2000;35:263–269. doi: 10.1007/s11745-000-0522-1. [DOI] [PubMed] [Google Scholar]
- Schaller H, et al. The ratio campesterol to sitosterol which modulates growth in Arabidopsis is controlled by STEROL METHYLTRANSFERASE 2:1. Plant J. 2001;25:605–615. doi: 10.1046/j.1365-313x.2001.00994.x. [DOI] [PubMed] [Google Scholar]
- Summons RE, et al. Steroids, triterpenoids and molecular oxygen. Philos Trans R Soc. 2006;361:951–968. doi: 10.1098/rstb.2006.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman JK. Sterols and other triterpenoids: source, specificity and evolution of biosynthetic pathways. Org Geochem. 2005;6:139–159. [Google Scholar]
- Wang J, Nes WD. Cyclobranol: a substrate for C25 methyl sterol side chains and potent mechanistic based inactivator of plant sterol methyltransferase. Bioorg Med Chem Lett. 2008;18:3878–3881. doi: 10.1016/j.bmcl.2008.06.044. [DOI] [PubMed] [Google Scholar]
- Weete JD, et al. Phylogenetic distribution of fungal sterols. PLoS One. 2010;5(5):e10899. doi: 10.1371/journal.pone.0010899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, et al. Comparison of the chromatographic properties of sterols, select additional steroids and triterpenoids: gravity-flow liquid chromatography, thin-layer chromatography, gas–liquid chromatography, and high performance liquid chromatography. J Chromatogr. 1988;452:377–398. doi: 10.1016/s0021-9673(01)81462-x. [DOI] [PubMed] [Google Scholar]
- Zhou W, Nes WD. Sterol methyltransferase2: purification, properties and inhibition. Arch Biochem Biophys. 2003;420:18–34. doi: 10.1016/j.abb.2003.08.029. [DOI] [PubMed] [Google Scholar]
- Zhou W, et al. Mechanistic analysis of a multiple product sterol methyltransferase implicated in ergosterol biosynthesis in Trypanosoma brucei. J Biol Chem. 2006;281:6290–6296. doi: 10.1074/jbc.M511749200. [DOI] [PubMed] [Google Scholar]
- Zhou W, et al. Plant sterol methyltransferases: phytosterolomic analysis, enzymology, and bioengineering strategies. In: Lewis NG, editor. Methods in Plant Biochemistry and Molecular Biology. Vol. 1. Elsevier Science; Amsterdam: 2008. pp. 241–281. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


