Abstract
Animal models are essential for elucidating the molecular mechanisms of carcinogenesis. Hodgkin's and many diverse non-Hodgkin's lymphomas overexpress the Hodgkin's disease antigen CD30 (CD30hi), a tumor necrosis factor receptor II family member. Here we show that chicken Marek's disease (MD) lymphoma cells are also CD30hi and are a unique natural model for CD30hi lymphoma. Chicken CD30 resembles an ancestral form, and we identify a previously undescribed potential cytoplasmic signaling domain conserved in chicken, human, and mouse CD30. Our phylogeneic analysis defines a relationship between the structures of human and mouse CD30 and confirms that mouse CD30 represents the ancestral mammalian gene structure. CD30 expression by MD virus (MDV)-transformed lymphocytes correlates with expression of the MDV Meq putative oncogene (a c-Jun homologue) in vivo. The chicken CD30 promoter has 15 predicted high-stringency Meq-binding transcription factor recognition motifs, and Meq enhances transcription from the CD30 promoter in vitro. Plasma proteomics identified a soluble form of CD30. CD30 overexpression is evolutionarily conserved and defines one class of neoplastic transformation events, regardless of etiology. We propose that CD30 is a component of a critical intracellular signaling pathway perturbed in neoplastic transformation. Specific anti-CD30 Igs occurred after infection of genetically MD-resistant chickens with oncogenic MDV, suggesting immunity to CD30 could play a role in MD lymphoma regression.
Lymphomas, the sixth-leading cause of human cancer death and fourth greatest in economic impact, are increasing in incidence (1). Controlling lymphoma requires early diagnosis, accurate categorization, and understanding pathogenesis (2). Lymphomas were first categorized by gross morphology, then microscopic morphology, and now gene expression. An example of molecular definition for lymphomas is overexpression of the Hodgkin's disease antigen, CD30, a tumor necrosis factor receptor (TNFR) II family member. A diverse range of Hodgkin's and many non-Hodgkin's lymphomas are CD30hi, suggesting that common pathways are involved in lymphomagenesis. CD30 signaling in normal lymphocytes promotes not only survival and proliferation but also cell death. In contrast, CD30 expression in neoplastic cells promotes survival and proliferation (3, 4). Mechanistic understanding of carcinogenesis and cancer regression requires animal models. However, no natural CD30hi lymphoma animal models are known.
Marek's disease (MD) has contributed greatly to understanding herpesvirus oncogenicity and has many biological parallels with Epstein–Barr γ-herpesvirus (EBV) (5). Despite having different genome structures, EBV and MD α-herpesvirus (MDV) evolved the similar life strategies of lymphotrophy and lymphomagenesis. MDV-transformed T lymphocytes overexpress a host-encoded antigen recognized by the mAb AV37 (6).
The AV37 antigen is Marek's-associated tumor surface antigen (MATSA) (7). MATSAs were central to demonstrating that host-encoded antigens may be aberrantly expressed after herpesvirus transformation, yet no MATSA has ever been definitively identified. We show that AV37 recognizes chicken CD30 and lymphomagenic MDV of poultry is a naturally occurring model for CD30hi lymphoma. EBV-induced lymphomas are also CD30hi (8). We propose that CD30 overexpression is an evolutionarily conserved response after, or to, lymphoid neoplastic-transformation regardless of etiology and that EBV and MDV evolved convergent strategies to survive in lymphocytes by disrupting homologous molecular pathways.
Materials and Methods
Chickens and MDV. Specified pathogen-free, MDV maternal-antibody-negative inbred White Leghorn chickens [susceptible (line 72) or resistant (line 61) to MD] and commercial Ross × Ross broiler chickens and oncogenic MDV strain HPRS-16 were used (9).
Identification of Chicken CD30. The CD30 antigen was detected from an MDV-transformed cell line HP9 lysate by SDS/PAGE (native and reduced), Western blotting (Hybond-C membrane; Amersham Pharmacia Biosciences), and immunostaining using AV37 followed by goat anti-mouse Ig-horseradish peroxidase (10) and enhanced chemiluminescence (Amersham Pharmacia Biosciences). CD30 cDNA was cloned and sequenced after magnetic enrichment of COS7 cells transfected with a cDNA library derived from HP9 (in the eukaryote vector pCINx maintained in DH5α Escherichia coli), using AV37 as described (11) (GenBank accession nos. NM_204444, AJ276964.1, and GI:28950398). CD30 was isolated from 1010 HP9 cells by immunoaffinity chromatography (12) using protein A-purified AV37 coupled to CNBr-activated Sepharose (Amersham Pharmacia Biosciences), blotted to poly(vinylidene difluoride) membrane (Sequi-blot, Bio-Rad) from SDS/PAGE, stained (naphthol blue-black, Sigma), excised, and N-terminal-sequenced with a Procise 492 protein sequencer (Applied Biosystems).
The TNFR domains of chicken CD30 were aligned (clustalw; ref. 13) with the core set of domains defined in the Pfam database and other known chicken TNFR domains. protdist and fitch (phylip package; ref. 14) were used to generate a tree relating the domains. Rat sequences and branches most distal to the chicken CD30 domains were removed; the resultant set was analyzed in the same way, including bootstrap calculation (consense programs) with 100 data sets.
Chicken genome sequence data homologous to the 5′ UTR of our CD30 cDNA sequence was first identified by using traceblast (National Center for Biotechnology Information). A composite CD30 promoter sequence was then generated. Potential transcription factor binding sites were identified by using matinspector (15), core and matrix similarity values of 1.0 and ≥ 0.9, respectively, and alibaba-2 (16).
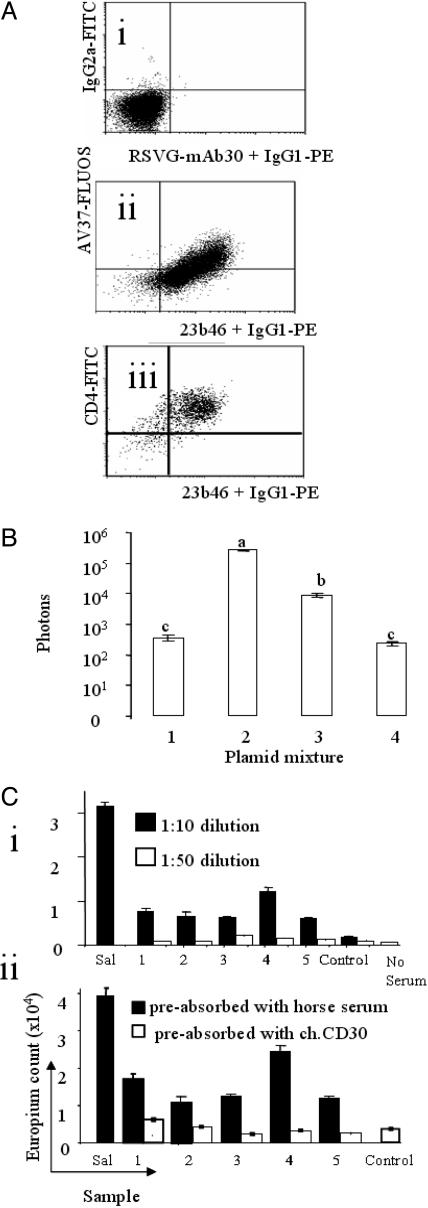
Association of CD30 and MDV Meq Putative Oncogene Expression in Vivo. The anti-Meq mAb (23b46) (17) and a mAb recognizing CD4 were used together with AV37 in double-staining flow cytometry experiments on permeabilized MD-lymphoma cells, isolated directly from line 72 lymphomas (four nerve, four ovary, two heart, and two liver) as described (6) (see Supporting Text, which is published as supporting information on the PNAS web site).
Chicken Cell Lines. Non-MDV-transformed chicken cell lines (Table 1), maintained as described (18), were examined by flow cytometry for CD30 expression (6).
Table 1. CD30hi expression in chicken transformed cell lines.
| Cell line | Transforming agent | Derivation cell type | CD30hi expression |
|---|---|---|---|
| OU2 | MNNG* | Embryo fibroblast | - |
| 1104 | ALV | Bursa lymphoma | + |
| DT40 | ALV | Bursa lymphoma | - |
| DT95 | ALV | Bursa lymphoma | - |
| HP46 | ALV | Bursa lymphoma | + |
| RP9 | ALV | Transplantable lymphoma | + |
| IAH16 | Reticuloendotheliousis virus T | Bursa lymphoma | + |
N-methyl-N′-nitro-N-nitrosoguanidine.
Luciferase Assay for Effect of Meq on CD30 Promoter. A 1,738-bp region of chicken DNA, 5′ to the first CD30 ATG, was amplified by PCR (Supporting Text). A reporter plasmid was constructed by replacing the cytomegalovirus (CMV) promoter in a pBK-CMV plasmid (Stratagene) containing the firefly luciferase-coding region (LUC) with this amplicon. SogE cells (4 × 105 per well; ref. 19) were transfected by using Lipofectin following the manufacturer's protocol (Invitrogen), with either this plasmid alone, together with a plasmid expressing the Meq protein, or a positive or negative control plasmid. All wells were cotransfected with a plasmid expressing destabilized GFP (pd2EGFP; BD Biosciences) to normalize for transfection efficiencies. The luciferase assay was done with standard methods (see Supporting Text). Statistical significance was tested by ANOVA.
Serum Proteomics to Identify Soluble CD30. We used 2D liquid chromatography electrospray ionization tandem MS to identify CD30 protein in pooled plasma taken from 14 Ross × Ross 6-week-old broiler chickens (see Supporting Text).
CD30 Serum Ig After MDV Infection. Ten line 61 and 10 line 72 chickens were infected with HPRS-16 at 1, 7, and 14 days of age with HPRS-16. Sera were taken at 28 days of age and tested in triplicate for anti-chicken CD30 Ig by dissociation enhanced lanthanide fluorescence immunoassay (DELFIA) with a Victor 1420 plate reader (Wallac) as described (20). Salmonella antigen plus anti-Salmonella Ig+ chicken serum was the positive control. Optimal antigen concentration was determined by using purified chicken CD30 and serially diluted AV37. At 1/10 dilution, 5 of 10 line 61 samples were positive, and these samples were then specificity-tested by preabsorption. DELFIA plates were incubated (16 h; 4°C) with either chicken CD30 or horse serum then washed. Matched sera samples (1:10 dilution) were incubated (16 h; 4°C) in each plate. After incubation, sera were removed, and the plates were washed and then analyzed for anti-chicken CD30 Ig by DELFIA.
Results
CD30 Is Overexpressed on Neoplastically Transformed MD Lymphoma Cells. The antigen recognized by AV37 has an apparent native and reduced molecular mass of ≈70 kDa (Fig. 1). blast searches showed our cDNA sequence (encoding 467 aa) was most similar to human and mouse CD30 but contained four extracellular TNFR repeats, rather than the three in human and mouse (Figs. 2 and 3).
Fig. 1.
Western blot of native (A) and reduced (B) antigen (≈70 kDa) expressed by HP9 cell line and identified by the mAb AV37.
Fig. 2.
Amino acid sequence alignment of chicken, human, and mouse CD30. Equals sign (=) indicates identity between all three sequences; – indicates biochemical similarity. TNFRSF repeats are marked above the sequence. Duplicated TNFRSF domains in human CD30 are labeled Hu′, and the position of their insertion is indicated by {DUP}. TM, transmembrane region. Dotted rectangle surrounds a TTRAP/TRAF6 region in mammalian CD30s. Solid rectangle surrounds a unique motif highly conserved between avian and mammalian CD30s (Table 2). TRAF 1, 2, and 3 binding motif is shown in bold. TRAF 1 and 2 motif is underlined.
Fig. 3.
Graphic representation of relationship between chicken, human, and mouse CD30. (A) Tree representing the genetic distance between selected TNFR superfamily repeats generated by using protdist and fitch programs from the phylip package (14). Leaves are labeled with TNFR number, species (C, chicken; H, human; M, mouse), and a domain number representing the order of domains in the sequences. Chicken domains are surrounded by rectangles, solid for CD30 (TNR8), dotted for others. Larger dotted rectangles contain nearest neighbors to the chicken CD30 repeats. Numbers in parentheses show the percentage of trees in the bootstrap analysis in which the repeats in these rectangles were in the same clade. The circle covers part of the tree where bootstrap values were all <50%. (B) Graphic representation of chicken, mouse, and human CD30 protein structures. Similar repeats are similarly shaded. Repeats are numbered from the N terminus with the proposed source of duplicated segments in parentheses.
The chicken TNFR repeats 1–3 are most similar to their mammalian equivalents, but the fourth was not clearly related to any other CD30 TNFR repeat (Figs. 2 and 3A).
Overall, the chicken CD30 cytoplasmic region has low similarity to its mammalian counterparts; it has binding motifs for signal transducing TNFR-associated factor (TRAF) 1, 2, and 3 molecules, but not the TRAF6/TNFR-associated protein (TTRAP) motif present in human and mouse CD30 (21) (Fig. 2 and Table 2). In the chicken either (i) TTRAP does not exist, (ii) CD30 does not bind TTRAP, or (iii) TTRAP has extremely low homology to human and mouse TTRAP. Notably, a unique 22-aa sequence in the cytoplasmic domain is highly conserved between chicken, human, and mouse (Fig. 2 and Table 2). This exceptional conservation, within an otherwise divergent region, suggests this sequence is important. Principles defined in multispecies comparative genomic analyses support this assertion (22). Possibly it is a novel signal transduction region. A potential phosphotyrosine, although not part of a typical Src homlogy 2 binding motif, is present. We found no existing reference to function, or potential function, for this or any similar sequence. blast searches reveal the sequence is unique to CD30.
Table 2. Amino acid identity/similarity (%) in cytoplasmic regions of chicken (ch.), human (hu.), and mouse (mo.) CD30.
| Domain | Species | Identical | Strongly similar | Weakly similar | Different |
|---|---|---|---|---|---|
| Entire cytoplasmic | hu. vs mo. | 65 | 12 | 8 | 15 |
| ch. vs hu. | 31 | 20 | 8 | 41 | |
| ch. vs mo. | 35 | 15 | 12 | 38 | |
| TTRAP binding | hu. vs mo. | 65 | 6 | 6 | 24 |
| ch. vs hu. | 24 | 24 | 18 | 35 | |
| ch. vs mo. | 35 | 12 | 12 | 31 | |
| Region with unknown function | hu. vs mo. | 91 | 9 | 0 | 0 |
| ch. vs hu. | 74 | 17 | 0 | 9 | |
| ch. vs mo. | 78 | 13 | 0 | 9 | |
| TRAF 1, 2, and 3 binding | hu. vs mo. | 85 | 11 | 0 | 4 |
| ch. vs hu. | 70 | 4 | 7 | 19 | |
| ch. vs mo. | 63 | 4 | 11 | 22 |
The chicken CD30 promoter includes sequences defined in human and mouse CD30 as transcription factor binding sites (23). Specifically, chicken CD30 has 15 predicted high-stringency AP-1 transcription factor binding sites (Supporting Text and Fig. 6, which is published as supporting information on the PNAS web site), suggesting the possibility of a direct functional relationship between the MDV AP-1 transcription factor homologue Meq and CD30 expression in MD.
CD30 and MDV Meq Oncogene Expression Are Associated in Vivo. MDV Meq is homologous to the host AP-1 trancription factor c-Jun, is unique to oncogenic MDV, and has oncogenic characteristics (24). Meq mRNA expression positively correlates with intensity of AV37 immunostaining in MD lymphoma cells (17). We suggest a functional link between Meq and CD30. However, Meq protein has never been associated with neoplastic transformation in vivo. We found Meq protein expression directly positively associates with CD30 expression, but not CD4 expression, on cells taken from MD lymphomas ex vivo (Fig. 4A), i.e., the CD30hi cells, which are the neoplastically transformed cells in MD lymphomas (6), contain most Meq protein in vivo. As Meq is expressed in both latent and productive MDV life cycles (24), the amount of Meq protein, rather than just its presence, appears important to its proposed oncogenicity in vivo.
Fig. 4.
CD30-Meq relationships and CD30 immunogeneicity. (A) Flow cytometry dot plots of nerve MD lymphoma cells. Negative control mAbs, IgG2a vs. RSVG-mAb30 (i). Chicken CD30 (mAb AV37) expression is proportional to MDV Meq expression (mAb 23b46) (ii), but not CD4 (iii). (B) Luciferase assay (mean of three replicates ± SEM) showing that MDV Meq protein can promote luciferase expression from a plasmid containing the CD30 promoter sequence 5′ to the luciferase (LUC) ORF (pCD30pLUC). SogE cells were transfected with plasmid mixtures containing pCD30pLUC (100 ng) (lane 1); the CMV promoter 5′ to LUC (100 ng; pBK-LUC, positive control) (lane 2); pCD30pLUC (100 ng) plus a plasmid encoding the Meq ORF 3′ to the CMV promoter (25 ng) (lane 3); and negative control plasmid pBK-CMV only (100 ng) (lane 4). A GFP-expressing plasmid (pd2EGFP, BD Biosciences; 50 ng) was used to normalize for differences in transfection efficiency. The plasmid pBK-CMV was added to all four transfection mixtures to give a total amount of plasmid DNA of 600 ng per well. P values are: a vs. b, <5 × 10–6; b vs. c, <5 × 10–7. (C) Chicken CD30-specific serum Ig after MDV HPRS-16 superinfection of genetically MD-resistant inbred chickens detected by dissociation enhanced lanthanide fluorescence immunoassay; chicken CD30-specific serum Ig titers could be diluted (i) and could be ablated by incubation with affinity-purified chicken CD30 but not horse serum (ii). Sal, Salmonella (positive control).
Meq Protein Activates the CD30 Promoter in Vitro. The relationship between CD30 and Meq protein expression in lymphoma cells, and the presence of 15 predicted high-stringency AP-1 transcription factor binding sites in the putative CD30 promoter, suggests that there may be a direct functional relationship between CD30 expression and the amount of Meq protein in the cell. We tested this suggestion in vitro by using a luciferase assay. Cotransfection of the plasmid expressing Meq with the plasmid encoding the CD30 promoter 5′ to the luciferase gene (pCD30pLUC) increased luciferase expression compared with transfection of pCD30pLUC alone (P < 5 × 10–7; Fig. 4B).
CD30 Overexpression in Avian Lymphomas Caused by Other Oncogenic Viruses. Human lymphomas of different etiologies are CD30hi. If CD30 is an evolutionarily conserved component of a neoplastic transformation pathway then CD30 overexpression may also be conserved between chicken lymphomas of different etiologies. We analyzed different chicken cell lines neoplastically transformed by different agents for CD30 overexpression (Table 1). Lymphocytes neoplastically transformed by three of the five avian leukosis viruses (ALVs) and reticuloendotheliosis virus (REV)-T overexpressed CD30. Two ALV-transformed lymphoid cell lines and the fibroblast cell line were CD30–. MDV, ALV, and REV-T all neoplastically transform lymphocytes by different mechanisms, yet their lymphomas overexpress CD30. Each virus may subvert the CD30 signaling pathway, and/or CD30 overexpression is a conserved response by chicken lymphocytes to some types of neoplastic transformation. The differences between the ALV-transformed cell lines are intriguing. These differences could simply be caused by loss of surface antigen expression after repeated culture; for example, one MDV-transformed cell line is unique among those we have tested in that it does not express CD30 (17). The mechanisms involved in CD30 overexpression in ALV-transformed cells remain to be elucidated.
Soluble CD30 Is Present in Chicken Plasma. In mammals, two forms of CD30 exist: a membrane-bound form and a soluble form found in blood. Soluble CD30 is a diagnostic maker for T helper 2-biased immune responses in infections and allergy (25) and lymphoproliferative disorders (26). The peptides IRGTSETDVSCEECPPGTFSDQSSSTDVCK and SCPMDPDEDCMRCGPEQYLNQSPK (charge, X-correlation and delta correlation value, respectively: +2, 2.72, and 0.25; +2, 2.60, and 0.10) were present in the plasma of uninfected outbred chickens. These peptides are present in the extracellular region of chicken CD30. blast searches of the chicken genome sequence using these peptides show that they are unique to CD30.
CD30-Specific Immunity. Five tumor antigen families are defined: differentiation antigens, tumor-specific shared antigens, tumor-specific mutated host antigens, virus antigens, and overexpressed normal/ubiquitous antigens (20). Intuitively, antigens in the first four families contain foreign immunogenic epitopes. Less intuitively, overexpressed normal/ubiquitous antigens are also immunogenic and, like autoantigens, overcome peripheral immune tolerance to activate low-affinity lymphocytes after antigen expression exceeds defined thresholds (27–29). Most identified tumor antigens to which weak T cell responses occur are nonmutated self-antigens (20).
We investigated whether anti-CD30-specific immunity exists after MDV infection. In the chicken, low-affinity cytotoxic lymphocyte (CTL) responses have never been measured. However, antigen-specific serum Ig directly correlates with antigen-specific CTL regardless of affinity (30). We detected chicken CD30-specific serum Ig only after HPRS-16 infection of genetically MD-resistant inbred line 61 chickens but not after infection of MD-susceptible line 72 chickens. This Ig could be titered and was eliminated after incubation with affinity-purified chicken CD30 but not horse serum (Fig. 4B).
Discussion
The chicken CD30 mRNA sequence and the chicken genome sequence data together allow inferences about the evolution of the CD30 molecule in vertebrates (Fig. 5, Supporting Text, and Fig. 7 and Tables 3 and 4, which are published as supporting information on the PNAS web site). The segment of human CD30 from the middle of repeat 1 to the middle of repeat 3 has exceptionally high similarity to the TNFR repeat regions further toward the C terminus (84% amino acid identity), suggesting that these two segments of human CD30 are the result of a duplication event. Duplication of a genomic region including exons 3 and 4 of a mouse-like ancestral gene, and its insertion within the intron between exons 6 and 7 would produce the exact structure of the modern human gene, including the preservation of all relevant splice sites and their phases. The same result could have been achieved by a direct tandem duplication of exons 3–6, followed by the deletion of the duplicated versions of exons 5 and 6. The effective result is the insertion of 2½ TNFR repeats within the stem portion of the ancestral molecule. The absence of this duplication in the even older chicken gene confirms that mouse CD30 represents the ancestral mammalian gene structure.
Fig. 5.
Comparison of chicken, mouse, and human CD30 genes. Intron/exon boundaries of the chicken CD30 gene were determined by comparison of cDNA sequence with unassembled shotgun genomic sequence (www.sanger.ac.uk). All introns started with GT and ended with AG. One ambiguity would allow lengths of 29 and 211 for exons 8 and 9, respectively. The exon structure of mouse and human genes was obtained from the ensembl database (www.ebi.ac.uk). Exons, shown by boxes, are numbered starting from the signal peptide encoding exon. Lengths of internal exons are shown below each box. The phase of each splice junction is shown by the number at the start of each intron. Braces indicate exons encoding transmembrane regions. Bars and arrow below the human gene show a duplication that occurred after the divergence of human and mouse from a common ancestor (see Supporting Text). Exons encoding TNFR repeats are indicated by dotted lines above each sequence. Human and mouse genes are variously annotated, sometimes with all or part of the fourth repeat that is clearly evident in the chicken sequence.
We suggest human CD30 evolved from a common precursor by duplication of a segment whose ends are within introns 2 and 4 and insertion of the duplicated segment into another intron. This could have been the result of a simpler mechanism in which a tandem duplication including exons 3–6, followed by a two-exon deletion. Such a route to the evolution of human CD30 is consistent with the gene structures of the human and mouse genes, including the phases of the intron/exon boundaries at all relevant locations (see Supporting Text). Our suggestion that the gene has evolved by this duplication, including one full and two half repeats, defines a relationship between the structures of human and mouse CD30 (reflected in Figs. 2 and 5). The human molecule is longer, potentially having two ligand binding sites. These changes may have significant effects on biological function.
In a cancer biology context, our work demonstrates that MD of poultry is a naturally occurring model for CD30hi lymphoma. The overexpression of CD30 by the neoplastically transformed cells in chicken MD lymphomas, and those induced by other oncogenic avian viruses, contributes to the axiom of comparative biology that a finite number of molecular pathways are available for viral manipulation in vertebrates. CD30 is overexpressed by the neoplastically transformed cells in human lymphomas of different etiologies (3). For example, EBV causes Hodgkin's lymphoma but it does not cause all CD30hi human lymphomas. CD30hi non-Hodgkin's lymphomas may, or may not, have other viral etiologies (8). CD30 overexpression after neoplastic transformation by chicken viruses suggests CD30 is a component of an evolutionary-conserved pathway defining one lymphoma category in vertebrates.
Many human viruses perturb CD30 expression (31–38). Disruption of the CD30 pathway must be fundamental, or at least beneficial, to virus survival. Notably, CD30 signaling promotes T helper 2 cytokine secretion (3, 4), and disruption of the normal CD30 signaling system in CD30-ligand deficient mice results in diminished primary CD8+ T lymphocyte clonal expansion, clonal contraction after primary expansion, and aberrant anamnestic CD8+ T lymphocyte expansion (39). We hypothesize that herpes-viruses convergently evolved mechanisms to increase CD30 expression and thus CD30 signaling to promote a T helper 2 environment, which would be inconsistent with cell-mediated immunity.
In addition, the longest-term persistence for any lymphotrophic virus would be achieved in memory lymphocytes. A logical explanation for herpesvirus-induced neoplastic transformation of lymphocytes is that it is an incidental consequence of increasing the lifespan of latently infected memory lymphocytes. Memory lymphocytes have increased proliferation and decreased death; deregulated cell proliferation and protection from cell death (40) are essential for survival of neoplastically transformed cells in vivo.Itis a small conceptual step to either further increase proliferation and/or decrease death of memory lymphocytes such that accumulation of these cells (i.e., lymphoma) results. CD30 up-regulation often coincides with memory lymphocyte marker expression (3, 4). We propose that EBV and MDV convergently evolved mechanisms to disrupt pathways involved in memory lymphocyte maintenance and that CD30 may be involved.
The EBV gene LMP-1 is critical for EBV neoplastic transformation (41, 42). LMP-1 disrupts CD30 signaling by associating with TRAF 1, 2, and 3 (43) at the cell membrane, which then activate NF-κB (44, 45), promote T helper 2 cytokine production and cell proliferation, and up-regulate costimulatory molecules (including CD30). We propose that MDV Meq also disrupts the CD30 signaling pathway, but does so in the nucleus at the level of CD30 transcription. Avian and human oncogenic herpesviruses apparently have convergently evolved mechanisms to perturb the CD30 signaling pathway. Meq is an AP-1 transcription factor homologue. AP-1 transcription factors are essential for cell proliferation; they activate oncogene, growth factor, and cytokine transcription (28). Meq/Meq and Meq/c-Jun dimers bind DNA with high affinity, and Meq must disrupt normal lymphocyte physiology (24). Our data suggest this disruption is proportional to the amount of Meq protein. We propose the following model for Meq function in a MD neoplastic transformation cycle. (i) Meq binds sequences in the chicken CD30 promoter and increases CD30 transcription. (ii) We identified (by using matinspector) an NF-κB transcription factor binding site in the Meq promoter (at –508 to –522). In normal physiology, after ligation with CD153, CD30 activates NF-κB (via TRAF 1, 2, and 3) (46). Activated T lymphocytes and monocytes, but not B cells or CD30hi lymphoma cells, express CD153 (47). Thus, ligation of CD30 by CD153+ CD30– cells within the heterogeneous MD lymphomas could further promote Meq expression by activating NF-κB. A cycle of cell proliferation and protection from death could be established. However, even if Meq is not involved, CD30 signaling in lymphoma environments is self-growth promoting (48).
High Meq protein expression raises an important immunological question. How do MD lymphoma cells, expressing high levels of MHC class I (6), avoid cell-mediated immunity? Both MD-susceptible and -resistant chickens have Meq-specific cytotoxic lymphocytes (49). The existence of tumor-infiltrating lymphocytes, specific to all classes of tumor antigens, is well established even in progressing tumors (20). Immune-escape mechanisms must allow lymphomagenesis in MD-susceptible chickens despite high amounts of Meq. A T helper 2 environment, as postulated above, may be critical in such immune escape.
Last, evidence for natural tumor immunity in MD exists, but immunogenic tumor antigens per se have never been definitively identified (50). We detected CD30-specific Ig after MDV infection of MD-resistant chickens. It could be that CD30 is only immunogenic in MD-resistant chickens, autoantigen immune tolerance in the susceptible chickens is greater than in resistant chickens, or B lymphocytolysis in susceptible chickens is so great that few specific B lymphocytes survive to produce CD30-specific Ig. Regardless, the presence of specific Ig to CD30 supports previous suggestions that antitumor immunity, especially that directed toward host-encoded antigens (50), may exist in MD.
Supplementary Material
Acknowledgments
We thank J. Kaufman, H.-J. Kung, and H. A. Wood for valuable advice; T. Pechan for running the mass spectrometer LCQ; A. N. Musselwhite for help with the IVIS Imaging System 100 Series; A. B. Chromiak for help with the Illumatool LT-9900; and Intervet UK LTD for financial support (S.C.B.). This work was partly supported by National Institutes of Health Grant P20 RR 017661.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ALV, avian leukosis virus; CMV, cytomegalovirus; EBV, Epstein–Barr γ-herpesvirus; LUC, luciferase-coding region; MD, Marek's disease; MDV, MD α-herpesvirus; TNFR, tumor necrosis factor receptor; TRAF, TNFR-associated factor; TTRAP, TRAF6/TNFR-associated protein.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. NM_204444, AJ276964.1, and GI:28950398).
References
- 1.Titcomb, C. P. J. (2001) J. Insur. Med. 33, 329–338. [PubMed] [Google Scholar]
- 2.National Cancer Institute Progress Review Group (2002) Strategic Plan for Addressing the Recommendations of the Leukemia, Lymphoma, and Myeloma Progress Review Group (National Institutes of Health, Bethesda).
- 3.Chiarle, R., Podda, A., Prolla, G., Gong, J., Thorbecke, G. J. & Inghirami, G. (1999) Clin. Immunol. 90, 157–164. [DOI] [PubMed] [Google Scholar]
- 4.Tarkowski, M. (1999) Arch. Immunol. Ther. Exp. (Warsz) 47, 217–221. [PubMed] [Google Scholar]
- 5.Epstein, M. A. (2001) Philos. Trans. R. Soc. London B 356, 413–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burgess, S. C. & Davison, T. F. (2002) J. Virol. 76, 7276–7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Witter, R. L., Stephens, E. A., Sharma, J. M. & Nazerian, K. (1975) J. Immunol. 115, 177–183. [PubMed] [Google Scholar]
- 8.Braylan, R. C., Orfao, A., Borowitz, M. J. & Davis, B. H. (2001) Cytometry 46, 23–27. [DOI] [PubMed] [Google Scholar]
- 9.Burgess, S. C., Basaran, B. H. & Davison, T. F. (2001) Vet. Pathol. 38, 129–142. [DOI] [PubMed] [Google Scholar]
- 10.Gallagher, S. R. & Smith, J. A. (2002) in Current Protocols in Immunology, eds. Coligan, J. E., Kruisbeek, A. M., Margulies, D. H., Shevach, E. M. & Strober, W. (Wiley, New York), Vol. 1, pp. 8.4.1–8.2.21. [Google Scholar]
- 11.O'Regan, M. N., Parsons, K. R., Tregaskes, C. A. & Young, J. R. (1999) Immunogenetics 49, 68–71. [DOI] [PubMed] [Google Scholar]
- 12.Springer, T. A. (2002) in Current Protocols in Immunology, eds. Coligan, J. E., Kruisbeek, A. M., Margulies, D. H., Shevach, E. M. & Strober, W. (Wiley, New York), Vol. 1, pp. 8.2.1–8.2.4. [Google Scholar]
- 13.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felsenstein, J. (1989) Cladistics 5, 164–166. [Google Scholar]
- 15.Quandt, K., Frech, K., Karas, H., Wingender, E. & Werner, T. (1995) Nucleic Acids Res. 23, 4878–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grabe, N. (2002) In Silico Biol. 2, S1–S15. [PubMed] [Google Scholar]
- 17.Ross, N., O'Sullivan, G., Rothwell, C., Smith, G., Burgess, S. C., Rennie, M., Lee, L. F. & Davison, T. F. (1997) J. Gen. Virol. 78, 2191–2198. [DOI] [PubMed] [Google Scholar]
- 18.Nazerian, K. (1987) Avian Pathol. 16, 527–544. [DOI] [PubMed] [Google Scholar]
- 19.Schumacher, D., Tischer, B. K., Teifke, J. P., Wink, K. & Osterrieder, N. (2002) J. Gen. Virol. 83, 1987–1992. [DOI] [PubMed] [Google Scholar]
- 20.Houghton, A. N., Gold, J. S. & Blachere, N. E. (2001) Curr. Opin. Immunol. 13, 134–140. [DOI] [PubMed] [Google Scholar]
- 21.Pype, S., Declercq, W., Ibrahimi, A., Michiels, C., Van Rietschoten, J. G., Dewulf, N., de Boer, M., Vandenabeele, P., Huylebroeck, D. & Remacle, J. E. (2000) J. Biol. Chem. 275, 18586–18593. [DOI] [PubMed] [Google Scholar]
- 22.Thomas, J. W., Touchman, J. W., Blakesley, R. W., Bouffard, G. G., Beckstrom-Sternberg, S. M., Margulies, E. H., Blanchette, M., Siepel, A. C., Thomas, P. J., McDowell, J. C., et al. (2003) Nature 424, 788–793. [DOI] [PubMed] [Google Scholar]
- 23.Durkop, H., Oberbarnscheidt, M., Latza, U., Bulfone-Paus, S., Krause, H., Pohl, T. & Stein, H. (2001) Biochim. Biophys. Acta 1519, 185–191. [DOI] [PubMed] [Google Scholar]
- 24.Kung, H. J., Xia, L., Brunovskis, P., Li, D., Liu, J.-L. & Lee, L. F. (2001) in Marek's Disease, ed. Hirai, K. (Springer, Tokyo), pp. 91–120.
- 25.Romagnani, S. (1995) J. Clin. Immunol. 15, 121–129. [DOI] [PubMed] [Google Scholar]
- 26.Vinante, F., Morosato, L., De Sabata, D. & Pizzolo, G. (1991) Leukemia Suppl. 5, 1, 18–21. [PubMed] [Google Scholar]
- 27.Lethe, B., van der Bruggen, P., Brasseur, F. & Boon, T. (1997) Melanoma Res. 7, S83–S88. [PubMed] [Google Scholar]
- 28.Hesketh, R., ed. (1997) in The Oncogene and Tumor Suppressor Gene Facts Book (Academic, London), pp. 65–66.
- 29.Ropke, M., Hald, J., Guldberg, P., Zeuthen, J., Norgaard, L., Fugger, L., Svejgaard, A., Van der Burg, S., Nijman. H, W., Melie, C. J. & Claesson, M. H. (1996) Proc. Natl. Acad. Sci. USA 93, 14704–14707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen, Y. T. (2000) Cancer J. 6, S208–S217. [PubMed] [Google Scholar]
- 31.Lemons-Estes, F. M., Capt, H. P., Skelton, H. & Smith, K. J. (2000) Int. J. Dermatol. 39, 521–527. [DOI] [PubMed] [Google Scholar]
- 32.Panus, J. F., Smith, C. A., Ray, C. A., Smith, T. D., Patel, D. D. & Pickup, D. J. (2002) Proc. Natl. Acad. Sci. USA 99, 8348–8353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seko, Y., Takahashi, N., Oshima, H., Shimozato, O., Akiba, H., Takeda, K., Kobata, T., Yagita, H., Okumura, K., Azuma, M. & Nagai, R. (2001) J. Pathol. 195, 593–603. [DOI] [PubMed] [Google Scholar]
- 34.Hong, S. & Krafft, A. E. (2001) AIDS Reader 11, 418–422. [PubMed] [Google Scholar]
- 35.Takahara, T., Masutani, K., Kajiwara, E., Sadoshima, S., Misago, M., Sasaguri, Y. & Onoyama, K. (1999) Intern. Med. 38, 824–828. [DOI] [PubMed] [Google Scholar]
- 36.Takeshita, M. (1999) Intern. Med. 38, 757–758. [DOI] [PubMed] [Google Scholar]
- 37.Vinante, F., Krampera, M., Morosato, L., Rigo, A., Romagnani, S. & Pizzolo, G. (1999) Haematologica 84, 683–689. [PubMed] [Google Scholar]
- 38.Funkhouser, A. W., Katzman, P. J., Sickel, J. Z. & Lambert, J. S. (1998) J. Pediatr. Hematol. Oncol. 20, 556–559. [PubMed] [Google Scholar]
- 39.Podack, E. R., Strbo, N., Sotosec, V. & Muta, H. (2002) Ann. N.Y. Acad. Sci. 975, 101–113. [DOI] [PubMed] [Google Scholar]
- 40.Evan, G. (1997) Int. J. Cancer 71, 709–711. [DOI] [PubMed] [Google Scholar]
- 41.Kaye, K. M., Izumi, K. M. & Kieff, E. (1993) Proc. Natl. Acad. Sci. USA 90, 9150–9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noguchi, T., Ikeda, K., Yamamoto, K., Yoshida, I., Ashiba, A., Tsuchiyama, J., Shinagawa, K., Yoshino, T., Takata, M. & Harada, M. (2001) Br. J. Haematol. 114, 84–92. [DOI] [PubMed] [Google Scholar]
- 43.Sandberg, M., Hammerschmidt, W. & Sugden, B. (1997) J. Virol. 71, 4649–4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Devergne, O., Hatzivassiliou, E., Izumi, K. M., Kaye, K. M., Kleijnen, M. F., Kieff, E. & Mosialos, G. (1996) Mol. Cell. Biol. 16, 7098–7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaye, K. M., Devergne, O., Harada, J. N., Izumi, K. M., Yalamanchili, R., Kieff, E. & Mosialos, G. (1996) Proc. Natl. Acad. Sci. USA 93, 11085–11090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horie, R. & Watanabe, T. (1998) Semin. Immunol. 10, 457–470. [DOI] [PubMed] [Google Scholar]
- 47.Barclay, A. N., Brown, M. H., Law, S. K. A., McKnight, A. J., Tomlinson, M. G. & van der Merwe, P. A. (1997) in The Leukocyte Antigen Facts Book, ed. Barclay, A. N. (Academic, London), pp. 417–418.
- 48.Horie, R., Higashihara, M. & Watanabe, T. (2003) Int. J. Hematol. 77, 37–47. [DOI] [PubMed] [Google Scholar]
- 49.Omar, A. R. & Schat, K. A. (1996) Virology 222, 87–99. [DOI] [PubMed] [Google Scholar]
- 50.Burgess, S. C. & Nair, V. K. (2002) in Modern Concept of Immunology in Veterinary Medicine: Poultry Immunology, ed. Mathew, T. (Thajema, New York), pp. 236–291.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.