Abstract
High pressure high temperature (HPHT) nanodiamonds (NDs) represent extremely promising materials for construction of fluorescent nanoprobes and nanosensors. However, some properties of bare NDs limit their direct use in these applications: they precipitate in biological solutions, only a limited set of bio-orthogonal conjugation techniques is available and the accessible material is greatly polydisperse in shape. In this work, we encapsulate bright 30-nm fluorescent nanodiamonds (FNDs) in 10–20-nm thick translucent (i.e., not altering FND fluorescence) silica shells, yielding monodisperse near-spherical particles of mean diameter 66 nm. High yield modification of the shells with PEG chains stabilizes the particles in ionic solutions, making them applicable in biological environments. We further modify the opposite ends of PEG chains with fluorescent dyes or vectoring peptide using click chemistry. High conversion of this bio-orthogonal coupling yielded circa 2000 dye or peptide molecules on a single FND. We demonstrate the superior properties of these particles by in vitro interaction with human prostate cancer cells: while bare nanodiamonds strongly aggregate in the buffer and adsorb onto the cell membrane, the shell encapsulated NDs do not adsorb nonspecifically and they penetrate inside the cells.
1. Introduction
Fluorescent nanodiamonds (FNDs) are a recently introduced class of biocompatible [1–6] luminescent probes. [7–10] Their exceptional optical properties for bioimaging are facilitated by the presence of nitrogen-vacancy centers, NV, which are localized defects of the diamond crystalline lattice. [11,12] The NV center is extremely photostable, showing no photobleaching or photoblinking. Upon excitation by green light (typically 532 nm wavelength laser), NV center emits in red to near infrared part of spectrum (emission maximum ∼700 nm for the negatively charged NV− center [12]) with high quantum efficiency (up to 90% in nanodiamond). [13] Its fluorescence therefore falls into the so-called tissue absorption window and is well-separated from cell or tissue autofluorescence. [14] FNDs were successfully used as fluorescent probes [15–18] and sensors [19,20] in vitro, allowing for single particle tracking inside cell. [21] They were also successfully visualized and tracked in small mammals. [2] Unique electron structure of NV center allows for selective turn-off of its emission by external electromagnetic field. [19,22,23] This property was successfully used for background-free imaging of nanodiamonds. [24,25]
Fluorescence lifetime of NV center (>10 ns) is substantionally longer than lifetime of autofluorescence (<4 ns). Therefore use of time-gated techniques like fluorescence lifetime imaging microscopy (FLIM) is also a way to suppress background signal, as shown for cells in culture [18] and recently demonstrated in vivo .[26]
In vivo long term cell tracking is an essential method in cell-based therapies such as adoptive immunotherapy and stem-cell therapy. [27,28] The biocompatibility of FNDs, their absolute optical stability and ability to observe them in vivo makes them an ideal candidate for in vivo long term tracking of stem cells, as was recently demonstrated. [29]
Although unmodified FNDs have been demonstrated as biocompatible nanoparticles, [1,3,4,17,30] a better control of nanodiamond properties is crucial for development of highly sophisticated probes enabling, for example, construction of multimodal nanoprobes or stealth [31] FNDs, which are able to target selectively cancer cells and tumors in vivo. Among the key issues of FND are still limited options for their bioorthogonal modifications by biomolecules, their colloidal behavior in buffers and biological media as well as a control of their size and shape. Overcoming these issues is essential for construction of advanced bioprobes.
We focused on optimizing properties of the brightest available FNDs that can be prepared from high-pressure high-temperature (HPHT) type nanodiamonds (NDs). [18,32–34] Although chemical modifications on the surface of detonation NDs is well described and many approaches for introduction of various functionalities were described, [35–38] the situation with HPHT NDs is more complicated due to their lower reactivity and surface/volume ratio. Direct covalent attachment of (bio)molecules to HPHT NDs surface is performed typically via amide bonds, [39] adsorption of proteins [2,40–42] or functionalization of artificially created surface graphene structures. [43] Because these conjugation reactions are rather nonspecific, the attention is currently paid onto sophisticated organic systems enabling selective, bioorthogonal bond formation. [44]
As typical colloidal dispersions, unmodified NDs in solution lose their colloidal stability at higher ionic strength and aggregate. [44,45] This behavior presents a serious disadvantage for direct biological application, as all biological liquids or cultivation media contain high concentrations of salts. Salt-caused aggregation could be overcome by chemical modification of the ND surface [46] or by attachment of hydrophilic polymers [47–50] or proteins [2,40–42] to the ND surface.
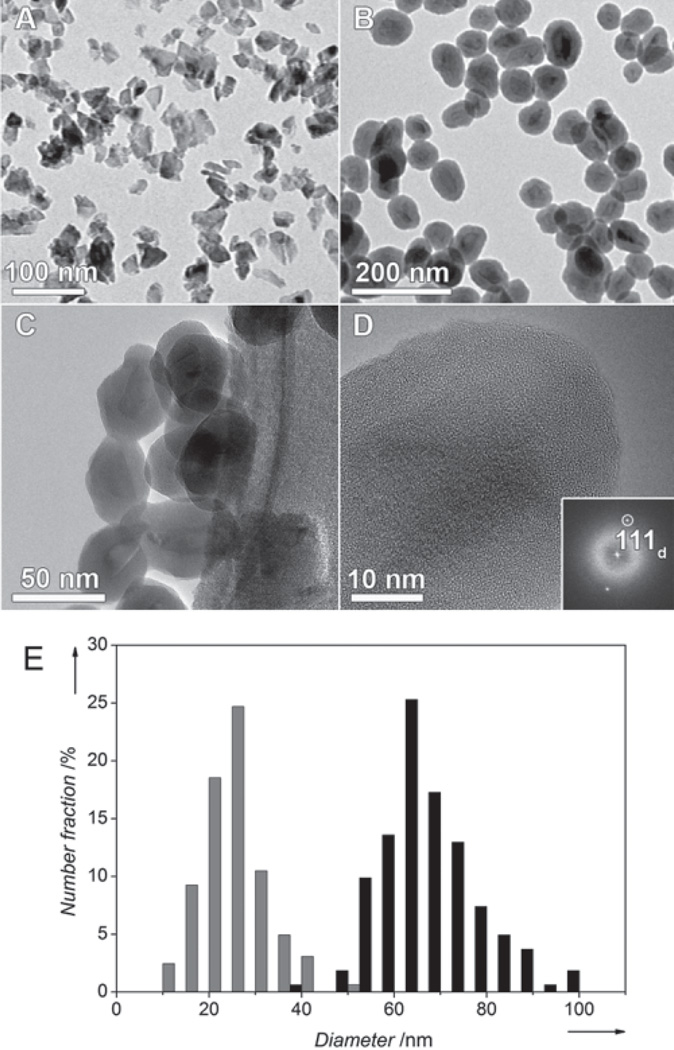
Strikingly, in contrast to many kinds of nanoparticles that can now be controllably engineered with well-defined geometric and chemical properties, [51,52] the control of HPHT ND shape is still in beginning. The available materials consist of heterogeneous nanoparticles of very irregular shape [21] bearing sharp edges and spiky vertexes (Figure 1A).
Figure 1.
BF-TEM images of non-coated ND1 (A) and aminosilica-coated ND3 (B) particles. Several coated ND3 particles are shown at higher magnification in (C). (D) High resolution TEM image showing the uniform silane surface coating. The crystallinity of the diamond core is evidenced by the presence of 111 diamond lattice planes, indexed in the Fourier Transform inset. (E) Histograms from image analysis of TEM micrographs of non-coated ND1 (gray) and aminosilica-coated ND3 (black) particles.
In a majority of publications the NDs are considered biocompatible and no toxicity was observed in cell studies [1–5,17,18,53] nor in animal models experiments [2,54–56] (for a review see ref. [4]). However, several recent works document toxic effects of NDs in vitro [57,58] as well as in vivo, [59] indicating that their biocompatibility should not be overgeneralized. Generally, a nanoparticle’s biological response strongly depends on its size and shape (for a review see refs. [31,60]) The shift from spherical to non-spherical shapes can significantly affect the behavior of a nanoparticle in biological environment [61] During finishing of this article three pioneering works dealing with the spiky ND shape appeared. [62–64] Among others it has been demonstrated that coating by silica shell can transform NDs to pseudo-spherical particles of improved shape homogeneity. [63,64]
Here we introduce a composite surface architecture on bright HPHT FNDs enabling selective bioorthogonal attachment of various (bio)molecules by click chemistry. The architecture comprises silica shell, which normalizes the spiky ND shape into pseudo-spherical. The silica surface allows simple modification with polyethyleneglycol (PEG) chains, responsible for colloidal stability of particles in buffers. The PEG chains can be further modified with a molecule of choice using click reaction. Obtained particles are fairly monodisperse in shape and colloidaly stable in ionic buffers. Noteworthy, the surface architecture has no adverse effects on the unique fluorescence of NV centers. Comparing to non-coated NDs, we demonstrate superior behavior of coated NDs in in vitro tissue culture; their application for prostate cancer cell labeling and imaging is shown.
2. Results and Discussion
2.1. Preparation and Structure of the Shell-Coated NDs
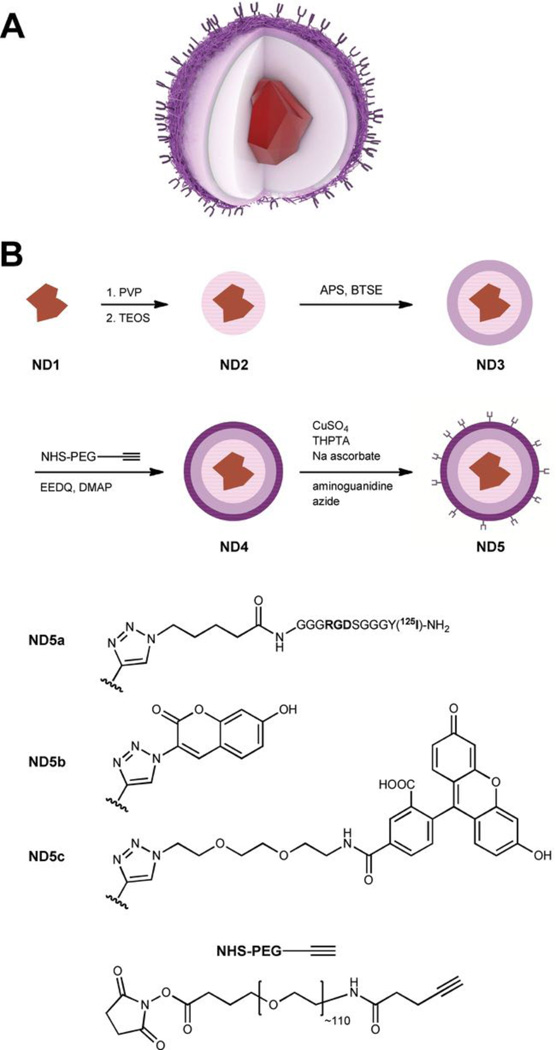
The architecture is designed from two covalently connected layers surrounding the ND particle: i) a solid crosslinked silica shell [65] bearing ii) a flexible polyethylene glycol (PEG) layer (Scheme 1A). The particle is therefore exposed to solution by a polymeric interface that protects it from electrolyte-induced precipitation. At the same time we took advantage of PEG heterofunctionality and we decorated the opposite ends of polymer chains by alkyne moieties enabling the use of click chemistry. [66]
Scheme 1.
The schematic structure (A) and preparation (B) of the particles. The fluorescent diamond nanocrystal is consecutively coated by silica shell (pink), thin crosslinked aminopropyl-silica shell (violet), and PEG-alkyne layer (dark violet). Azide-labeled 125 I-RGDS peptide (ND5a), coumarin (ND5b), and fluorescein (ND5c) moieties are attached. TEOS – tetraethoxysilane, BTSE – 1,2-bis(triethoxysilyl)ethane, APS – (3-aminopropyl)triethoxysilane, PVP – polyvinypyrrolidone, EEDQ – 2-ethoxy-1-ethoxycarbonyl-1,2-dihydroquinoline, DMAP – dimethylaminopyridine.
The silica shell can be grown by controlled hydrolysis of tetraalkoxysilylesters or substituted trialkoxyalkylsilylesters, [67] in which the alkyl group allows further surface modification. The presence of the alkyl group, on the other hand, weakens the compactness of the shell, making it hydrolytically labile and also colloidally unstable in aqueous media. [68] To avoid hydrolytic lability, we used a crosslinking agent, bis(triethoxysilyl)ethane (BTSE), that improves the shell’s resistance to hydrolysis [69] (Scheme 1B). First, the TEOS-based shell was formed (ND2), followed by growth of a crosslinked amino-functionalized layer from a mixture of APS and BTSE (ND3). Notably, this procedure enabled direct functionalization of nanodiamond by silane-based moieties without any reductive pretreatment that is an essential step in anhydrous silylation procedure introduced by Krueger and collaborators. [36] Amino-modified silica surface was used for attachment of the heterobifunctional PEG chain bearing N -hydroxysuccinimidyl group at one end of the chain and alkyne moiety at the other. N -hydroxysuccinimidyl group served for formation of an amidic anchor, whereas the alkyne moiety was used for attachment of various molecules to the particle by click chemistry in the next reaction step.
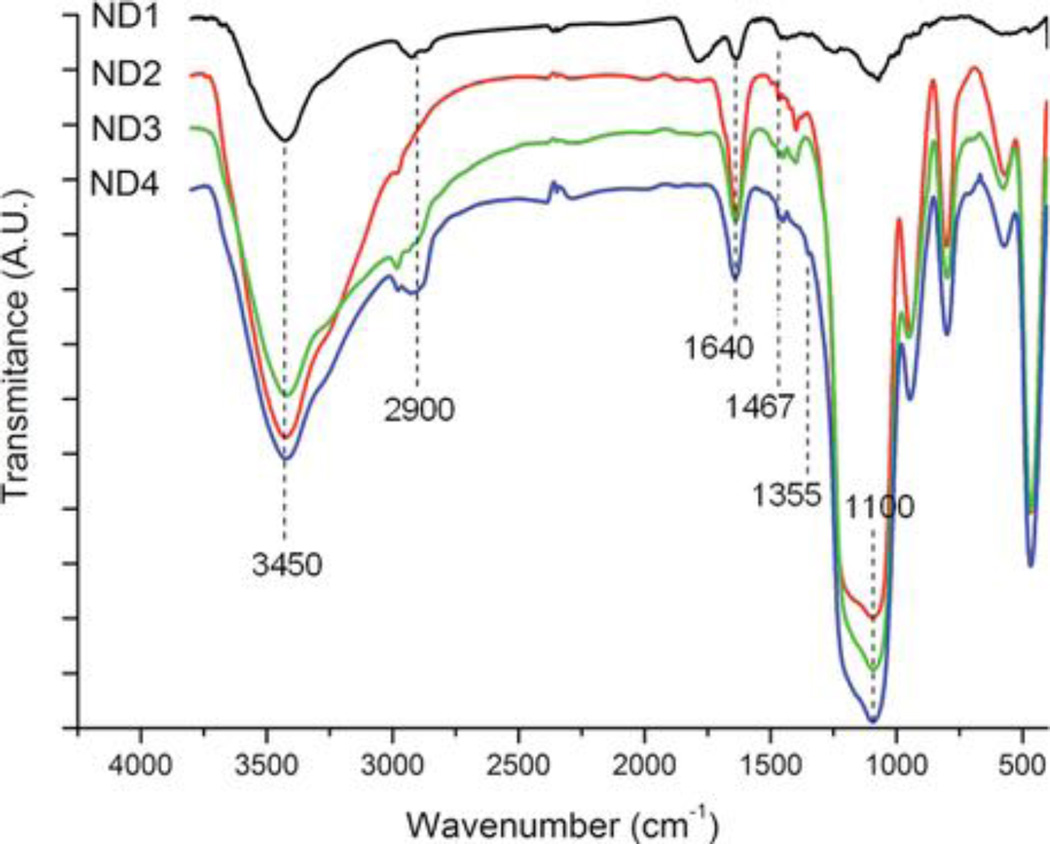
The composition of the shell was confirmed by IR spectroscopy indicating the presence of both Si-O bands and characteristic diamond bands (Figure 2). The characteristic peaks present in all silica-coated samples at about 1110 cm −1 correspond to the asymmetric stretching vibrations of the Si–O–Si from silica shells. The peaks at about 1640 cm −1 and 3450 cm −1 also indicate the formation of silica coating on the surface, corresponding to silanol groups (Si–OH) as well as to OH groups from nanodiamond and residual water. Bands at 2900, 1467, and 1355 cm −1 are characteristic for PEG.
Figure 2.
Normalized IR spectra of noncoated (ND1), silica-coated (ND2), aminosilica-coated (ND3), and PEGylated aminosilica-coated (ND4) fluorescent nanodiamonds. For clarity, spectra are shifted along the y-axis.
The bright field transmission electron microscopy (BF-TEM) images in Figure 1 show NDs before and after encapsulation. The diamond particles themselves (ND1, Figure 1A) are of irregular shape (circularity ∼0.67), with sharp edges and often elongated in one dimension (needle–like). Their diameter (expressed as average circular equivalent diameter – see Supporting information) is 27 ± 7 nm. After coating (Figure 1B–D), the particles become more spherical (circularity ∼0.87), and their diameter rises to 66 ± 10 nm (Figure 1E). Notably, spherical shape of nanoparticles is generally considered as the proper, biocompatible geometry for bio-applications. [61,70]
The optimal size of nanoparticular biolabels is considered to be between 10 and 100 nm, depending on specific application. [31] Both original and coated particles fall into this size range. The total mass of the sample rises approximately 4-fold upon encapsulation, which corresponds to the data from elemental analysis (Table 1). The thickness of the shell can be varied by changing the amounts of silylesters during encapsulation. Indeed, the thinner shell leads to smaller particles, however, their shape is more distant from spherical (Figure S1 in Supporting information).
Table 1.
Elemental composition of FND bearing different surface architectures.
| % C | % H | % N | |
|---|---|---|---|
| ND1 | 93.7 | 0.38 | 0.06 |
| ND2 | 19.6 | 1.28 | 0.29 |
| ND3 | 19.8 | 1.90 | 1.10 |
| ND4 | 21.5 | 2.07 | 0.66 |
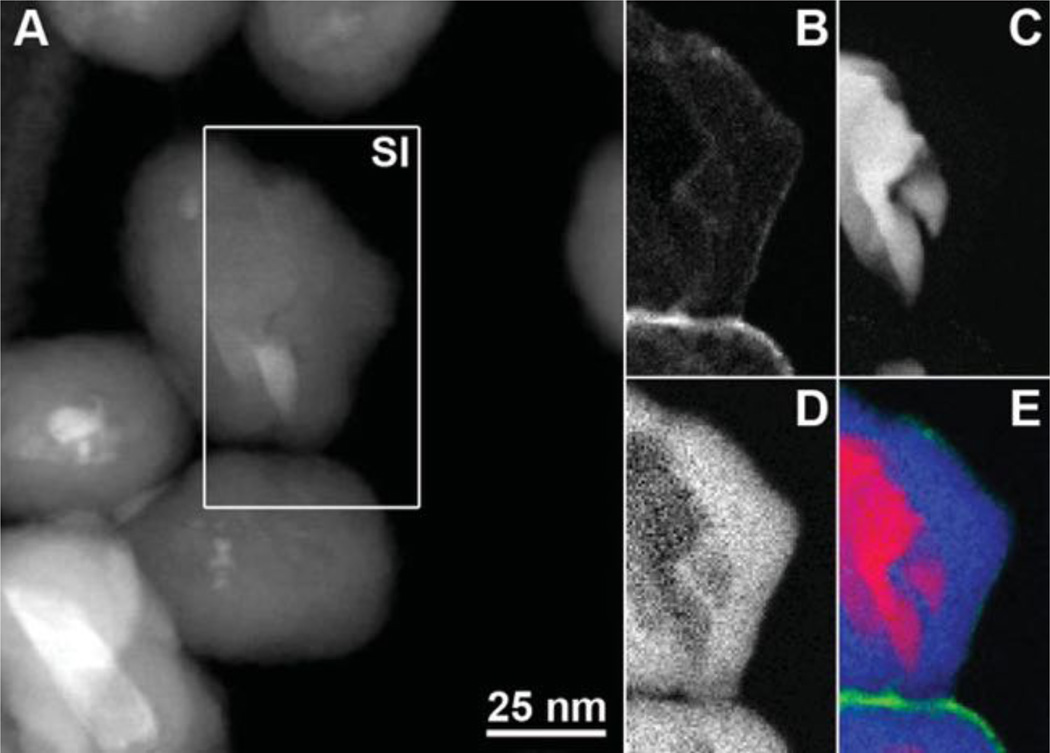
In order to confirm the presence of the various coatings on ND4 directly, we performed spatially resolved electron energy-loss spectroscopy (EELS) measurements using the spectrum imaging technique in STEM mode [71] (Figure 3). The acquired EELS data were used to generate an element-specific map for silicon and element- and bond-specific maps for amorphous carbon and diamond. The diamond map in Figure 3C confirms the size and irregular shape of the diamond core. The Si map shows the ∼10–20 nm thick silica shell surrounding the diamond core. Some amorphous carbon is measured at the surface of the diamond core in the amorphous carbon map, which is to be expected in most ND samples. [72] The predominant amorphous carbon signal, however, arises from the thin layer of PEG at the surface of the silica coating.
Figure 3.
STEM-EELS analysis of PEGylated aminosilica-coated ND4 nanoparticles. (A) Annular dark field STEM image with the 72*129 pixel spectrum image (SI) region indicated by the white rectangle. (B–E) STEM-EELS maps for (B) amorphous carbon, (C) diamond, (D) silicon, and (E) a color map showing diamond (red), silicon (blue), and amorphous carbon (green).
2.2. Stability Studies and Chemical Modifications
Because low colloidal stability of nanoparticles in buffers and biological liquids is a typical limitation for their applications in biodisciplines, we tested the stabilization effects of this PEG layer on the particles. The difference between ND1 and ND4 particles after dissolving in PBS buffer is clearly observable with the naked eye (Figure 4). Aggregation of colloidal aqueous solutions of ND4 at various pH and ionic strength were further monitored using dynamic light scattering (Figure S2 in Supporting Information). The particles were colloidally stable (i.e., their hydrodynamic diameter remained unchanged within the experimental error) across a wide working pH range (2–10) and extreme ionic strength (1 M NaCl). After one week a precipitation was observable in the alkaline solution (pH = 10) only, while the other alternatives were stable at least for two weeks. Furthermore, particles exhibited long term (at least 1 month) stability in PBS and tissue culture media, which is essential for their use in bioapplications.
Figure 4.
Photography of ND1 and ND4 dispersions (0.5 mg mL−1) in PBS, 30 min after mixing. ND1 (left) precipitates and finally gives sediment at the bottom of the tube, while ND4 (right) remains in the form of stable colloid scattering the laser beam (red laser pointer).
The function of the mid-size (5000 Da) heterobifunctional PEG in the surface architecture is not only a colloidal protection: its terminal alkyne group exposed to the solution is available for selective attachment of various (bio) molecules via click chemistry. In order to demonstrate the applicability of particles as a modular platform for construction of different probes, three selected azide-modified molecules were attached to the ND4 particles: 125I-labeled RGDS peptide[73–74] (ND5a), coumarin[75] (ND5b), and fluorescein[76] (ND5c) (for structures see Scheme 1B).
The RGDS peptide exhibits targeting properties towards tumor cells and here served as a model structure for our future studies. Its radiolabeling[73] enabled highly sensitive monitoring of conjugation yield by radioactivity measurements. The consecutive post-reaction washing steps (Figure S3 in Supporting information) revealed that only a small amount of peptide can be released after the excess from reaction has been washed away. The total load of peptide estimated from radioactive measurements was 8 µmol g−1. To confirm that the peptide is bound selectively by click chemistry and not only adsorbed to the particle surface, a control reaction without CuI catalyst was performed. The total amount of peptide attached in control was only ∼4% of the load reached by reaction in the presence of CuI catalyst. The vast majority of RGDS attached by reaction in presence of catalyst was therefore covalently bound to the PEG chains, and only a very small fraction of physisorbed peptide remained on the particles after post-reaction washings. Because presence of hydrophobic RGDS peptide on a nanoparticle can lead to its aggregation, we tested also the long-term colloidal stability of conjugate ND5a in PBS buffer using DLS. The colloidal behavior introduced already by PEG layer was preserved even after attachment of the peptide: the particles remained stable for at least one month.
To directly confirm the covalent grafting mechanism, the fluorogenic probe coumarin azide[75] was attached to the particles, providing ND5b. The dye itself is non-fluorescent, but a highly fluorescent structure is formed upon cycloaddition to an alkyne. Compared with the control sample reacted in the absence of CuI, the reaction of coumarin azide with the particles in the presence of CuI catalyst produced a characteristic bright fluorescent product (Figure S4 in Supporting information).
2.3. Spectroscopic Characterization
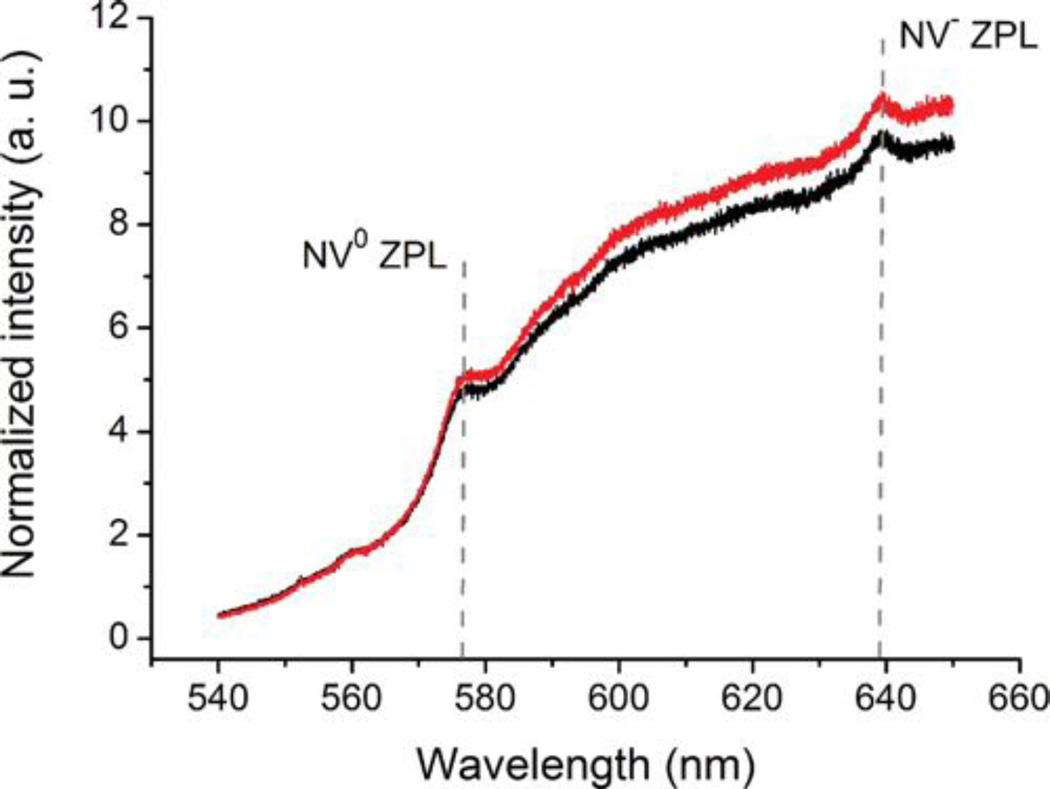
Modification of FND surface chemistry has been shown to produce changes in NV centers charge state which is strongly reflected in fluorescence spectra[77] To evaluate the possible influence of the silica shell on FND fluorescence, we measured the photoluminescence spectra for ND1 and ND4 (Figure 5). The normalization to the diamond Raman signal enabled us to compare the relative change in fluorescence caused by formation of the shell. The spectra show that the shell behaves as an inert translucent layer: its presence has no significant influence on the fluorescence and the spectrum retains its intensity and characteristic shape.
Figure 5.
Normalized photoluminescence spectra of non-coated (ND1, black) and PEGylated silica-coated (ND4, red) FNDs. For comparison of relative change in fluorescence caused by formation of the shell the spectra were normalized to the diamond Raman signal. The relative difference in photoluminescence intensity is within the standard deviation of the measured data. Each spectrum shows an average of 50 normalized measurements. The standard deviation was 8.9% for ND1and 9.4% for ND4. ZPL - zero phonon line.
For detailed understanding of the conjugation process and quality of the resulting conjugate we aimed to measure the fraction of FNDs that are not properly modified by either silica shell or PEG-alkyne and therefore cannot be further modified by click reaction. Using total internal reflection fluorescence microscopy (TIRF) we characterized conjugate ND5c bearing a secondary fluorescent probe, fluorescein,[76] at the single particle level. From 5 measurements, we counted 400 red spots that colocalize with green ones, and only 20 that do not colocalize (Figure S6 in Supporting information). This result confirmed us that the vast majority (>95%) of the FNDs are properly modified and the reaction procedure, although performed in very diluted solutions (240 µM fluorescein-azide), is robust enough to provide high yield conjugation (for experimental details and further discussion see Figures S5–S7 in Supporting information). It should be noted, that the total load of fluorescein on particles was 9 umol g−1 of NDs (estimated by UV-Vis spectroscopy) which is in agreement with conjugation yields of RGDS peptide (8 µmol g−1, see above). Because the loads of both molecules obtained by different methods (scintigraphy and UV-Vis spectroscopy, respectively) are in excellent agreement despite the different character of the molecules, this load represents apparently an upper limit for covalent attachment of molecules to the particles. The quantities can be recalculated, using simple spherical model, giving a number of approximately 2000 molecules attached to a single nanoparticle. This quantity is sufficient not only for targeting of a probe, but also for applications more demanding on a cargo payload, like MR imaging contrast agents or drug delivery systems.
2.4. Imaging of Coated NDs in Cells
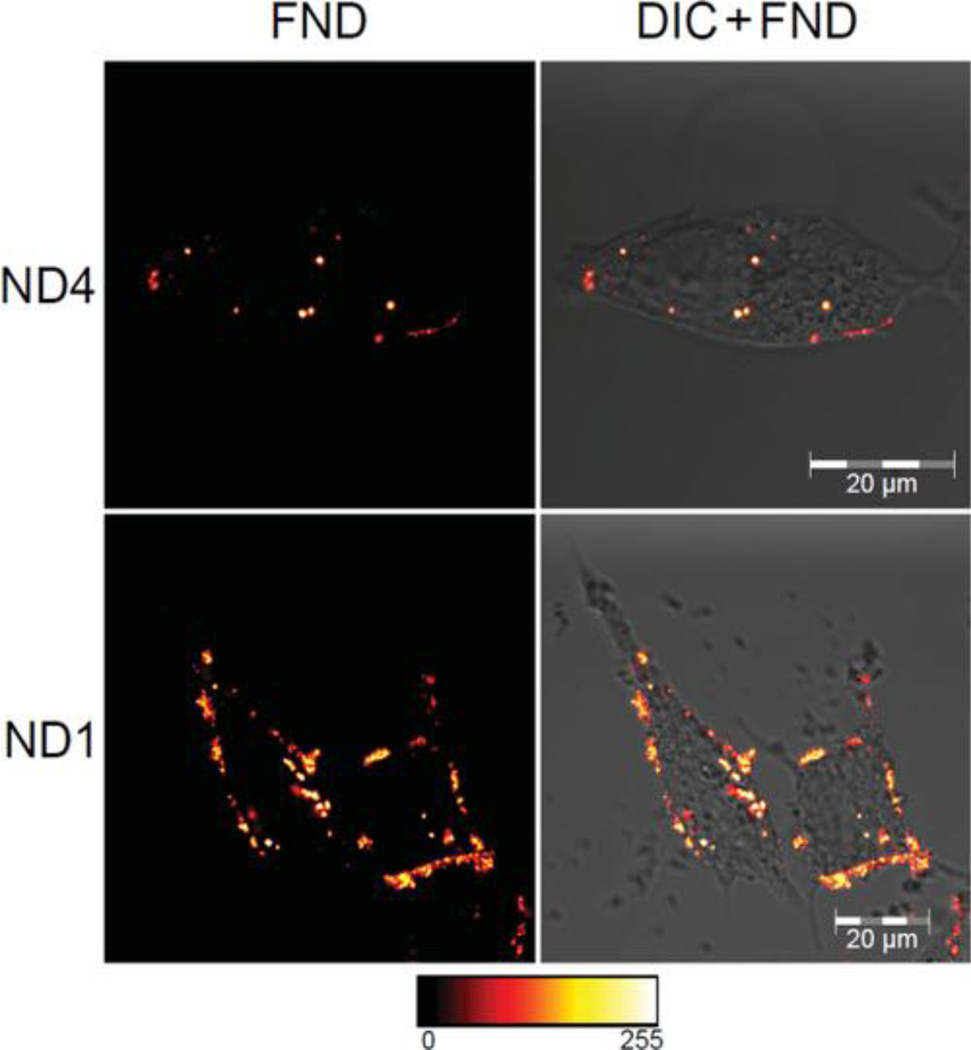
We examined the differences between ND1 and ND4 particles in interaction with human prostate cancer cells. The unmodified, “naked” nanoparticles have due to high free energy to the environment a much greater nonspecific affinity for the cell surface than particles modified by proteins[78] or sterically protecting polymers. Using in vitro tissue culture experiments, we examined whether the shell structure on ND4 is able to compensate this effect compared to “naked” ND1. For this experiment we choose serum-free PBS buffer, a well-defined medium enabling control and analysis of binding events on cells.[79] We dispersed both ND1 and ND4 particles in PBS buffer and exposed cells for one hour to this solution. After washing the cells with ND-free PBS buffer we observed the cellular localization of the particles using confocal microscopy (Figure 6). While the ND4 were found internalized inside prostate cancer cells (LNCaP cell line), the ND1 were mostly adhered on the cell membrane. This difference may be explained by the observation that ND1 are strongly aggregating and subsequently precipitating in PBS buffer (Figure 4), while the ND4 are colloidaly stable under these conditions (see above). We hypothesized that the presence of large aggregates of ND1 formed in the buffer promotes cell binding but prevents internalization, possibly because the larger aggregates are energetically not favored to be endocytosed.
Figure 6.
Confocal microscopy images of PEGylated aminosilica-coated (ND4) and noncoated (ND1) particles preincubated in PBS and internalized by LNCaP cells. The displayed images are fluorescence from NDs (FND) in false colours (left) and merged differential interference contrast (DIC) and FND fluorescence (right). FNDs were dipersed in PBS before addingto cells in medium (final concentration 200 µgml−1). Cells were washed by PBS after one hour incubation, incubated for subsequent 23 hours and observed after fixation. Fluorescence of FND was collected by Zeiss LSM 780 at 639–758 nm upon excitation at 532 nm. For full set of images see Figure S8 in the Supporting Information.
To prevent the aggregation as well as decrease the free energy of particles we preincubated ND1 in cell growth medium containing serum. Noncovalent adsorption of serum albumin has been already described as an option for stabilizing NDs in aqueous solutions[21] We tested the aggregation state of ND1 in cell media using DLS, which showed rapid formation of still ≈600 nm large aggregates. Due to serum proteins stabilization the ND-serum complex did not further agglomerate or precipitate for several days (in contrast to ND1 which precipitates in PBS in less than 30 minutes). In cell experiment, we indeed observed very similar behavior for both serum media treated ND1 and ND4 samples: the particles were internalized into cells without nonspecific cell membrane adhesion (Figure S8 in Supporting information). However, in case of ND1 the cells were interacting with very different, 600 nm particles resulting from association of ND1 nanoparticles in presence of serum proteins and ions from medium. In contrast, colloidal behavior of ND4 was not influenced by any biologically relevant liquid used in study (see Figure S2) and particles we taken up in non-aggregated form.
From these studies, it is clear that the shell architecture, or more precisely its bionanointerface formed by hydrophillic PEG, prevents the nonspecific adsorption of particles to the cell membrane. This property is a key starting point for any study on cell recognition or targeting.
3. Conclusion
In summary, this study introduces hybrid near-spherical nanoparticles combining the advantages of fluorescence of NV color centers in NDs with a bio-orthogonally reactive translucent shell. We have demonstrated in bulk solution as well as at the single particle level that the surface architecture is not quenching or otherwise modifying the unique fluorescent properties of NV centers. The attachment of molecules or biomolecules can be performed selectively at high conversion efficiencies using click chemistry in aqueous buffers or biological media while keeping the excellent colloidal stability of the particles. Morever, the bionanointerface of particles prevents their nonspecific adhesion on cell membrane, as proven on human prostate cancer cells. Our design based on unique photophysical properties of non-photobleachable NV center in combination with silica coating can therefore serve as a versatile biocompatible platform, providing new directions for construction of various sophisticated imaging probes or targeted systems[80]
4. Experimental Section
Chemicals and Solvents
Tetraethoxysilane (TEOS), l,2-bis(triethoxysilyl)ethane (BTSE), (3-aminopropyl)triethoxysilane (APS), polyvinypyrrolidone MW = 10 000 g mol−1 (PVP), 2-ethoxy-l-ethoxycarbonyl-l,2-dihydroquinoline (EEDQ), and dimethylaminopyridine (DMAP) were purchased from Sigma-Aldrich. NHS-PEG(5000)-alkyne (Scheme 1B) was purchased from Iris Biotech. 125I-labeled RGDS peptide azidopentanoyl-GGGRGDSGGGY(125I)-NH2,[73] coumarinazide,[75] fluoresceinazide[76] (Scheme 1B), and THPTA ligand[66] were synthesized according to published procedures.
Infrared (IR) spectroscopy
IR spectra were recorded from 4000–400 cm−1 on a Bruker Equinox spectrometer using KBr pellets. The sample weight was 1.0 mg/pellet for all samples.
Raman Spectroscopy
Photoluminescence spectra were measured with a Renishaw InVia Raman Microscope at an excitation wavelength of 514 nm with 25 mW laser power. Spectra were recorded at room temperature and normalized to the diamond Raman peak (excitation at 488 nm). The measurements were performed on aqueous solutions (as received, 1:1,1:2,1:5, and 1:10 dilutions) in a Hellma fluorescence cuvette (type no. 105.252-QS). A set of 10 measurements was performed on each sample. All spectra were normalized to the diamond Raman peak.
Elemental Analysis
Elemental analysis was performed with a CHN PE 2400 Series automatic analyzer. Each sample was measured three times, and the results were averaged.
UV-Vis Spectroscopy
UV-Vis spectra were recorded with a Specord 210 (Analytik Jena) spectrometer in the 300–700 nm range at room temperature with an optical path of 1 cm.
Luminescence Spectroscopy
Luminescence measurements were performed on an Edinburgh Instruments FS900 spectrofluorimeter, equipped with a 450 W xenon arc lamp, a microsecond flash lamp, and a red-sensitive photomultiplier (300–850 nm).
Electron Microscopy
To prepare the samples, a drop of diluted colloidal solution was placed on a carbon-coated copper grid and left to dry. Bright field transmission electron microscopy (TEM) experiments presented in Figure 1A and SI were performed using a JEOL JEM-1200EX electron microscope operated at 60 kV. All other TEM, high resolution TEM, STEM, and spatially-resolved EELS experiments were carried out on a FEI Titan 80–300 “cubed” microscope fitted with an aberration-corrector for the imaging lens and the probe forming lens and a GIF Quantum energy filter for spectroscopy, operated at 80 kV to minimize knock-on damage to the sample. STEM-EELS experiments were performed using a convergence semi-angle α of ∼21 mrad and a collection semi-angle β of ∼200 mrad, using a beam current of approximately 80 pA. A fine electron probe (diameter∼1.5 Å) was scanned over a region of the sample, acquiring an EELS spectrum at each point. All spectra were acquired at an energy dispersion of 0.4 eV per pixel and an energy resolution of approximately 1.2 eV. Chemical maps for the C signals (amorphous carbon and diamond) were generated by fitting the carbon K-edge to known references for diamond and amorphous carbon. The Si maps were generated by plotting the intensity under the background-subtracted Si L2,3-edge in each pixel using a 22 eV broad energy window.
The size distributions of coated and noncoated particles in TEM microphotographs were calculated with ImageJ software, using more than 150 particles for each sample. Average particle sizes are expressed in the form of average circular equivalent diameter. It is defined as the diameter of a spherical particle that has the same area as the observed particle.
Dynamic Light Scattering (DLS)
DLS was recorded with a Zetasizer Nano ZS system (Malvern Instruments) at room temperature. Sample concentrations were 0.05 mg/mL.
Radioactivity Measurements
Gamma emission of 125I was measured with an ionization-chamber Bqmetr 4 applied activity indicator (Empos, Prague, Czech Republic). The concentrations of RGDS in samples were calculated from a measured activity of a sample and known activity per 1 mg of labeled RGDS.[73]
Total Internal Reflection Fluorescence (TIRE) Microscopy
The TIRF microscope is a home-made setup relying on a Zeiss Axiovert 35 mount and a x100 and 1.49 numerical aperture microscope objective (Ref. UAPON 100XOTIRF, Olympus, Japan). We used continuous-wave diode-pumped solid-state lasers sources, one at a wavelength of 488 nm and 50 mW maximum power for fluorescein excitation (Sapphire 488–50 LP, Coherent Inc., USA) and the other at 561 nm for excitation of NV color centers in nanodiamonds, at 100 mW maximum power (SUM-561-100, Oxxius S.A., France). We used two different filter sets depending on the detection channel: the “green channel” dedicated to fluorescein and the “red channel” for FND fluorescence. The “green channel” filter set is composed of a dichroic beamsplitter (Ref. z488rdc Chroma, AHF Analysentechnik, Germany) and a 50 nm band-pass filter centered on 525 nm, which is close to the maximum of fluorescein fluorescence (ET525/50, AHF). The “red channel” filter set is composed of a dichroic beamsplitter with a sharp edge at 561 nm and a 97% flat transmission above this wavelength (Ref. ZT561rdc Chroma, AHF) and a 75 nm band-pass filter centered on 697 nm, which is the maximum of the NV color center fluorescence (HC 697/75, Ref. F39–697, AHF). The detection was performed with a cooled electron multiplied CCD array detector (iXon-DU885, Andor Technology, Ireland).
For colocalization statistics, five TIRF fields of view were analyzed (an example of one field is depicted in Figure S6). To obtain similar maximal counting rates of 160 kilo counts/s (kcts/s), excitation laser intensities of 32 kW cm−2 at a 561 nm excitation wavelength (Figure S6A) and 3 kW cm−2 at a 488 nm excitation wavelength (Figure S6B) were used.
Confocal Microscopy
Confocal microscope Zeiss LSM 780 was equipped with the In-Tune laser (with minimum power 1.5 mW per wavelength) tuned to 532 nm wavelength and an oil-immersion objective (Plan-Apochromat 63x/1.40 Oil DIC M27). The fluorescence images were collected in ZEN 2011 software at room temperature using InTune laser, intensity 100% at 532 nm, QUASAR PMT spectral channel #34 in range 639–758 nm with master gain set to 1200 V (or 850 V in case of ND1 pre-incubated in PBS), offset 0, digital gain 1, pinhole at diameter 67 µm (1.35 Airy unit) and with line averaging 2. For continuous maintenance of focus Definite focus was used. Images were processed using the GIMP2 Program where images were cropped to selection and input level of black points was set to 15. Fluorescence from nanodiamonds is displayed in false colors in red-to-white LUT.
Nanodiamond Pretreatment (ND1)
Nanodiamonds were supplied by Microdiamant Switzerland (MSY 0-0.05). The particles were oxidized by air in a Thermolyne 21100 tube furnace at 510 °C for 5 hours. The nanodiamonds were subsequently treated with a mixture of HNO3 and H2SO4 (85 °C, 3 days), washed with 0.1 M NaOH and 0.1 M HCI, washed five times with water, and freezedried.[41] Purified nanodiamond powder (160 mg), containing approximately 100–200 ppm of natural nitrogen impurities, was pressed in an aluminum target holder and irradiated with a 15.5 MeV proton beam extracted from the isochronous cyclotron U-120M for 70 min (fluence 6 × 1016 cnr2). The irradiated material was annealed at 900 °C for 1 h and subsequently oxidized for 6 h at 510 °C. The nanodiamonds were then treated with a mixture of HNO3 and H2SO4 (85 °C, 3 days), washed with 0.1 M NaOH and 0.1 M HCI, washed five times with water, and freezedried. Prior to use, the particles were dissolved in water (2 mg/mL) and sonicated with a probe (Cole-Parmer, 750 W) for 30 minutes. The resulting transparent colloid was filtered using a 0.2 µm PVDF microfilter to provide colloidal solution of ND1 particles.
Coating of Nanodiamond Particles with Amino-terminated Silica (ND3)
Polyvinylpyrrolidone (96 mg, 9.6 µmol) was dissolved in water (204 mL) and sonicated for 10 minutes in an ultrasonic bath. ND1 colloid (6 mL, 2 mg mL−1) was added, and the mixture was stirred for 24 hours. The colloid was then concentrated via centrifugation in two steps. In the first step (40,000 ref, 1 hour), the volume was reduced to approximately 12 mL. The second centrifugation step (30,000 ref, 30 min) was performed in microvials and reduced the solvent volume to approximately 0.4 mL. Sedimented nanodiamonds were resuspended in ethanol (12 mL) in a round bottom flask and sonicated in an ultrasonic bath for 2–4 min. TEOS (112 mg, 539 µmol) was added. After 2 minutes of vigorous stirring, ammonia solution (25%, 500 µL) was added, and the reaction mixture was stirred for 14 h, affording ND2. BTSE (12 mg, 34 µmol) and APS (12 mg, 54 µmol) were added to the solution. After approximately 2 h, the reaction mixture became turbid, a portion of particles slowly precipitated from solution and stuck to the walls of the reaction vessel. The reaction mixture was stirred for an additional 24 hours. The product was purified by centrifugation (14,000 ref, 5 min) with ethanol (12 mL, 4x) and MeCN (12 mL, 2x) and was dissolved in 6 mL of MeCN. The ND3 particles were stored in the freezer (−18 °C) as a stable colloid for several weeks without changes in particle characteristics (confirmed with TEM, and DLS) or reactivity.
Grafting of PEG Chains (ND4)
NHS-PEG-alkyne (42 mg, 8.4 µmol), EEDQ (8.4 mg, 34 µmol), and DMAP (4.1 mg, 34 µmol) were dissolved together in 6.4 mL MeCN. The resulting solution was placed into an ultrasonic bath cooled to 18 °C. Using a syringe pump, the ND3 colloid (6 mg ND, i.e., 24 mg of particles in 3 mL solution) was slowly added to the reaction mixture in 30 min. The mixture was sonicated at 18 °C for an additional 2.5 hours and then gently shaken for 16 hours. The product was purified by centrifugation (14 000 ref, 5 min) with MeCN (6 mL, 3x), ethanol (12 mL, 1x), and water (6 mL, 2x) and was dissolved in water or PBS (3 mL). For long term storage, the product (ND4) was kept in MeCN at −18 °C.
Functionalization of ND4 by 125I-Labeled RGDS, coumarin azide, and fluorescein azide (ND5a–c)
A solution of Cu-catalyst was prepared in a separate vial by mixing CuSO4 · 5H2O (20 µL of a 25 mM solution) and THPTA ligand (20 µL of a 50 mM solution).
The click reactions were performed similarly to those described in ref.[4] by mixing reactants in a 1.5 mL vial in the following order and quantities:
Water (87 pL)
Aqueous colloid of ND4 (250 pL containing 0.5 mg of ND, i.e., 2 mg of particles)
1251-labeled RGDS (222 µL of a 0.46 mM aqueous solution) or fluorescein azide (5.90 µL of a 17.3 mM DMSO solution) or coumarin azide (13.5 µL of a 7.6 mM DMSO solution)
Aminoguanidine hydrochloride (32 µL of a 100 mM solution)
Cu-catalyst solution (16.5 µL) (see above)
Sodium ascorbate (32 µL of a 100 mM solution)
The vials were well-sealed and kept for 3 h without stirring or shaking. Nanoparticles ND5a, ND5b, and ND5C were isolated in almost quantitative yield by centrifugation (14 000 ref, 5 min) and washed with water (1×, 1 mL) and PBS buffer (3×, 1 mL). The amount of molecules attached to the nanoparticles after every washing was quantified by UV-Vis spectrophotometry in the case of fluorescein azide or by radioactivity measurements in the case of 125I-labeled RGDS (see Figure S3).
Cell Culture and Cellular Uptake
LNCaP cells (human prostate cancer cell line, ATCC) were grown in RPMI medium (Sigma) with 10% FBS serum (Sigma) in Petri dish with No. 1.5 glass bottom D35C4–20–1.5-N (BioPort Europe, s.r.o.) at 37 °C in a humidified 5% CO 2 atmosphere. After 24 hours (the cells were 20–30% confluent) the culture medium was replaced with the fresh medium containing NDs (ND1 or ND4) at concentration 200 µg ml −1. NDs were pre-incubated either half an hour in RPMI medium or one hour in isotonic PBS solution (pH = 7.4) before adding to the cells. After one hour of incubation the cells were washed twice with RPMI medium for removing excess of nanodiamonds. After additional 23 hours of incubation cells were fixed by 2% formaldehyde solution in PBS at 4 °C. After fixation cells were washed twice with PBS and subjected to confocal microscopy.
Supplementary Material
Acknowledgments
The work was supported by GACR project P108/12/0640, MSMT CR grant No. LH11027. Part of this work was performed within OPPK project CZ.2.16/3.1.00/24016, OP VK project CZ.1.07/2.3.00/20.0306 (to VP), EU 7FP Program No. 262348 (European Soft Matter Infrastructure, ESMI), NIH/NIBIB T32 EB007509 (to A.M.W.), and Mt. Sinai Foundation (to N.F.S.). S.T. acknowledges support from the fund for scientific research flanders (FWO). P.S. has been supported by a grant of the Grant Agency of the Czech Republic P208/12/G016. The Titan microscope used for this work was partially funded by the Hercules foundation. We thank to Prof. M. Nesladek, Mr. J. Havlik, Dr. R. Hadravova and Mrs. C. Durieu for their very valuable help and to Dr. H. Hoffman for critical proofreading of the manuscript.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Ivan Rehor, Dr., Institute of Organic Chemistry and Biochemistry, AS CR, v.v.i. Flemingovo nam. 2, Prague 6, 166 10, Czech Republic
Jitka Slegerova, Institute of Organic Chemistry and Biochemistry, AS CR, v.v.i. Flemingovo nam. 2, Prague 6, 166 10, Czech Republic.
Jan Kucka, Dr., Institute of Macromolecular Chemistry AS CR, v.v.i. Heyrovského nám. 2, Prague 6, 162 06, Czech Republic Nuclear Physics Institute AS CR, v.v.i. Rez near Prague, 250 68, Czech Republic.
Vladimir Proks, Dr., Institute of Macromolecular Chemistry AS CR, v.v.i. Heyrovského nám. 2, Prague 6, 162 06, Czech Republic
Vladimira Petrakova, Dr., Faculty of Biomedical Engineering, Czech Technical University in Prague, Sítná sq. 3105, 272 01 Kladno (Czech Republic) and Institute of Physics AS CR, v.v.i, Prague 8, Czech Republic
Marie-Pierre Adam, Laboratoire de Photonique Quantique et Moléculaire, UMR 8537 CNRS and ENS Cachan, F-94235 Cachan (France), Laboratoire Aimé Cotton, CNRS, Université Paris Sud and ENS Cachan, F-91405, Orsay, France.
François Treussart, Prof., Laboratoire de Photonique Quantique et Moléculaire, UMR 8537 CNRS and ENS Cachan, F-94235 Cachan (France), Laboratoire Aimé Cotton, CNRS, Université Paris Sud and ENS Cachan, F-91405, Orsay, France
Stuart Turner, Dr., EMAT, University of Antwerp, Groenenborgerlaan 171, B-2020, Antwerp, Belgium
Sara Bals, Prof., EMAT, University of Antwerp, Groenenborgerlaan 171, B-2020, Antwerp, Belgium
Pavel Sacha, Dr., Institute of Organic Chemistry and Biochemistry, AS CR, v.v.i. Flemingovo nam. 2, Prague 6, 166 10, Czech Republic
Miroslav Ledvina, Dr., Institute of Organic Chemistry and Biochemistry, AS CR, v.v.i. Flemingovo nam. 2, Prague 6, 166 10, Czech Republic
Amy M. Wen, Department of Biomedical Engineering, Case Western Reserve University, School of Medicine and Engineering, 10990 Euclid Avenue, Cleveland, Ohio, USA
Nicole F. Steinmetz, Prof., Department of Biomedical Engineering, Case Western Reserve University, School of Medicine and Engineering, 10990 Euclid Avenue, Cleveland, Ohio, USA
Petr Cigler, Dr., Institute of Organic Chemistry and Biochemistry, AS CR, v.v.i. Flemingovo nam. 2, Prague 6, 166 10, Czech Republic.
References
- 1.Liu K-K, Cheng C-L, Chang C-C, Chao J-I. Nanotechnology. 2007;18:325102. [Google Scholar]
- 2.Vaijayanthimala V, Cheng P-Y, Yeh S-H, Liu K-K, Hsiao C-H, Chao J-I, Chang H-C. Biomaterials. 2012;33:7794–7802. doi: 10.1016/j.biomaterials.2012.06.084. [DOI] [PubMed] [Google Scholar]
- 3.Schrand AM, Dai L, Schlager JJ, Hussain SM, Osawa E. Diam. Relat. Mater. 2007;16:2118–2123. [Google Scholar]
- 4.Zhu Y. Theranostics. 2012:302. doi: 10.7150/thno.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schrand AM, Huang H, Carlson C, Schlager JJ, Ōsawa E, Hussain SM, Dai L. J. Phys. Chem. B. 2007;111:2–7. doi: 10.1021/jp066387v. [DOI] [PubMed] [Google Scholar]
- 6.Faklaris O, Joshi V, Irinopoulou T, Tauc P, Sennour M, Girard H, Gesset C, Arnault J, Thorel A, Boudou J-P, Curmi PA, Treussart F. ACS Nano. 2009;3:3955–3962. doi: 10.1021/nn901014j. [DOI] [PubMed] [Google Scholar]
- 7.Hui YY, Cheng CL, Chang HC. J. Phys. Appl. Phys. 2010;43:374021. [Google Scholar]
- 8.Badea I, Kaur R. Int. J. Nanomedicine. 2013:203–220. doi: 10.2147/IJN.S37348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mochalin VN, Shenderova O, Ho D, Gogotsi Y. Nat. Nanotechnol. 2011;7:11–23. doi: 10.1038/nnano.2011.209. [DOI] [PubMed] [Google Scholar]
- 10.Xing Y, Dai L. Nanomed. 2009;4:207–218. doi: 10.2217/17435889.4.2.207. [DOI] [PubMed] [Google Scholar]
- 11.Davies G, Lawson SC, Collins AT, Mainwood A, Sharp SJ. Phys. Rev. B. 1992;46:13157. doi: 10.1103/physrevb.46.13157. [DOI] [PubMed] [Google Scholar]
- 12.Jelezko F, Wrachtrup J. Phys. Status Solidi. 2006;203:3207–3225. [Google Scholar]
- 13.Mohtashami A, Femius Koenderink A. New J. Phys. 2013;15:043017. [Google Scholar]
- 14.Weissleder R, Ntziachristos V. Nat. Med. 2003;9:123–128. doi: 10.1038/nm0103-123. [DOI] [PubMed] [Google Scholar]
- 15.Fu CC, Lee HY, Chen K, Lim TS, Wu HY, Lin PK, Wei PK, Tsao PH, Chang HC, Fann W. Proc. Natl. Acad. Sci. USA. 2007;104:727–732. doi: 10.1073/pnas.0605409104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tzeng Y-K, Faklaris O, Chang B-M, Kuo Y, Hsu J-H, Chang H-C. Angew. Chem. Int. Ed. 2011;50:2262–2265. doi: 10.1002/anie.201007215. [DOI] [PubMed] [Google Scholar]
- 17.Vaijayanthimala V, Tzeng Y-K, Chang H-C, Li C-L. Nanotechnology. 2009;20:425103. doi: 10.1088/0957-4484/20/42/425103. [DOI] [PubMed] [Google Scholar]
- 18.Faklaris O, Garrot D, Joshi V, Druon F, Boudou JP, Sauvage T, Georges P, Curmi PA, Treussart F. Small. 2008;4:2236–2239. doi: 10.1002/smll.200800655. [DOI] [PubMed] [Google Scholar]
- 19.McGuinness LP, Yan Y, Stacey A, Simpson DA, Hall LT, Maclaurin D, Prawer S, Mulvaney P, Wrachtrup J, Caruso F, Scholten RE, Hollenberg LCL. Nat. Nanotechnol. 2011;6:358–363. doi: 10.1038/nnano.2011.64. [DOI] [PubMed] [Google Scholar]
- 20.Ermakova A, Pramanik G, Cai J-M, Algara-Siller G, Kaiser U, Weil T, Tzeng Y-K, Chang HC, McGuinness LP, Plenio MB, Naydenov B, Jelezko F. Nano Lett. 2013;13:3305–3309. doi: 10.1021/nl4015233. [DOI] [PubMed] [Google Scholar]
- 21.Chang Y-R, Lee H-Y, Chen K, Chang C-C, Tsai D-S, Fu C-C, Lim T-S, Tzeng Y-K, Fang C-Y, Han C-C, Chang H-C, Fann W. Nat. Nanotechnol. 2008;3:284–288. doi: 10.1038/nnano.2008.99. [DOI] [PubMed] [Google Scholar]
- 22.Balasubramanian G, Chan IY, Kolesov R, Al-Hmoud M, Tisler J, Shin C, Kim C, Wojcik A, Hemmer PR, Krueger A, Hanke T, Leitenstorfer A, Bratschitsch R, Jelezko F, Wrachtrup J. Nature. 2008;455:648–651. doi: 10.1038/nature07278. [DOI] [PubMed] [Google Scholar]
- 23.Maze JR, Stanwix PL, Hodges JS, Hong S, Taylor JM, Cappellaro P, Jiang L, Dutt MVG, Togan E, Zibrov AS, Yacoby A, Walsworth RL, Lukin MD. Nature. 2008;455:644–647. doi: 10.1038/nature07279. [DOI] [PubMed] [Google Scholar]
- 24.Hegyi A, Yablonovitch E. Nano Lett. 2013;13:1173–1178. doi: 10.1021/nl304570b. [DOI] [PubMed] [Google Scholar]
- 25.Igarashi R, Yoshinari Y, Yokota H, Sugi T, Sugihara F, Ikeda K, Sumiya H, Tsuji S, Mori I, Tochio H, Harada Y, Shirakawa M. Nano Lett. 2012;12:5726–5732. doi: 10.1021/nl302979d. [DOI] [PubMed] [Google Scholar]
- 26.Kuo Y, Hsu T-Y, Wu Y-C, Hsu J-H, Chang H-C. Proc. SPIE. 2013;8635:863503. [Google Scholar]
- 27.Kircher MF, Gambhir SS, Grimm J. Nat. Rev. Clin. Oncol. 2011;8:677–688. doi: 10.1038/nrclinonc.2011.141. [DOI] [PubMed] [Google Scholar]
- 28.Taylor A, Wilson KM, Murray P, Fernig DG, Lévy R. Chem. Soc. Rev. 2012;41:2707–2717. doi: 10.1039/c2cs35031a. [DOI] [PubMed] [Google Scholar]
- 29.Wu T-J, Tzeng Y-K, Chang W-W, Cheng C-A, Kuo Y, Chien C-H, Chang H-C, Yu J. Nat. Nanotechnol. 2013;8:682–689. doi: 10.1038/nnano.2013.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu S-J, Kang M-W, Chang H-C, Chen K-M, Yu Y-C. J. Am. Chem. Soc. 2005;127:17604–17605. doi: 10.1021/ja0567081. [DOI] [PubMed] [Google Scholar]
- 31.Prokop A, Davidson JM. J. Pharm. Sci. 2008;97:3518–3590. doi: 10.1002/jps.21270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaijayanthimala V, Chang H-C. Nanomed. 2009;4:47–55. doi: 10.2217/17435889.4.1.47. [DOI] [PubMed] [Google Scholar]
- 33.Smith BR, Inglis DW, Sandnes B, Rabeau JR, Zvyagin AV, Gruber D, Noble CJ, Vogel R, Ōsawa E, Plakhotnik T. Small. 2009;5:1649–1653. doi: 10.1002/smll.200801802. [DOI] [PubMed] [Google Scholar]
- 34.Havlik J, Petrakova V, Rehor I, Petrak V, Gulka M, Stursa J, Kucka J, Ralis J, Rendler T, Lee S-Y, Reuter R, Wrachtrup J, Ledvina M, Nesladek M, Cigler P. Nanoscale. 2013;5:3208–3211. doi: 10.1039/c2nr32778c. [DOI] [PubMed] [Google Scholar]
- 35.Krueger A, Lang D. Adv. Funct. Mater. 2012;22:890–906. [Google Scholar]
- 36.Krueger A. J. Mater. Chem. 2008;18:1485. [Google Scholar]
- 37.Schrand AM, Hens SAC, Shenderova OA. Crit. Rev. Solid State Mater. Sci. 2009;34:18. [Google Scholar]
- 38.Moore L, Chow EK-H, Osawa E, Bishop JM, Ho D. Adv. Mater. 2013;25:3532–3541. doi: 10.1002/adma.201300343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang B, Li Y, Fang C-Y, Chang C-C, Chen C-S, Chen Y-Y, Chang H-C. Small. 2009;5:2716–2721. doi: 10.1002/smll.200900725. [DOI] [PubMed] [Google Scholar]
- 40.Nguyen T, Chang HC, Wu VW. Diam. Relat. Mater. 2007;16:872–876. [Google Scholar]
- 41.Huang L-CL, Chang H-C. Langmuir. 2004;20:5879–5884. [PubMed] [Google Scholar]
- 42.Weng M-F, Chiang S-Y, Wang N-S, Niu H. Diam. Relat. Mater. 2009;18:587–591. [Google Scholar]
- 43.Chang IP, Hwang KC, Ho JA, Lin C-C, Hwu RJ-R, Horng J-C. Langmuir. 2010;26:3685–3689. doi: 10.1021/la903162v. [DOI] [PubMed] [Google Scholar]
- 44.Dahoumane SA, Nguyen MN, Thorel A, Boudou J-P, Chehimi MM, Mangeney C. Langmuir. 2009;25:9633–9638. doi: 10.1021/la9009509. [DOI] [PubMed] [Google Scholar]
- 45.Sreenivasan VKA, Ivukina EA, Deng W, Kelf TA, Zdobnova TA, Lukash SV, Veryugin BV, Stremovskiy OA, Zvyagin AV, Deyev SM. J. Mater. Chem. 2011;21:65–68. [Google Scholar]
- 46.Liang Y, Meinhardt T, Jarre G, Ozawa M, Vrdoljak P, Schöll A, Reinert F, Krueger A. J. Colloid Interface Sci. 2011;354:23–30. doi: 10.1016/j.jcis.2010.10.044. [DOI] [PubMed] [Google Scholar]
- 47.Rehor I, Mackova H, Filippov SK, Kucka J, Proks V, Slegerova J, Turner S, Van Tendeloo G, Ledvina M, Hruby M, Cigler P. ChemPlusChem. 2013 doi: 10.1002/cplu.201300339. [DOI] [PubMed] [Google Scholar]
- 48.Zhao L, Takimoto T, Ito M, Kitagawa N, Kimura T, Komatsu N. Angew. Chem. Int. Ed. 2011;50:1388–1392. doi: 10.1002/anie.201006310. [DOI] [PubMed] [Google Scholar]
- 49.Zhang X-Q, Chen M, Lam R, Xu X, Osawa E, Ho D. ACS Nano. 2009;3:2609–2616. doi: 10.1021/nn900865g. [DOI] [PubMed] [Google Scholar]
- 50.Marcon L, Kherrouche Z, Lyskawa J, Fournier D, Tulasne D, Woisel P, Boukherroub R. Chem. Commun. 2011;47:5178–5180. doi: 10.1039/c1cc10338e. [DOI] [PubMed] [Google Scholar]
- 51.Burda C, Chen X, Narayanan R, El-Sayed MA. Chem. Rev. 2005;105:1025–1102. doi: 10.1021/cr030063a. [DOI] [PubMed] [Google Scholar]
- 52.Tao AR, Habas S, Yang P. Small. 2008;4:310–325. [Google Scholar]
- 53.Vial S, Mansuy C, Sagan S, Irinopoulou T, Burlina F, Boudou J-P, Chassaing G, Lavielle S. ChemBioChem. 2008;9:2113–2119. doi: 10.1002/cbic.200800247. [DOI] [PubMed] [Google Scholar]
- 54.Chow EK, Zhang X-Q, Chen M, Lam R, Robinson E, Huang H, Schaffer D, Osawa E, Goga A, Ho D. Sci. Transl. Med. 2011;3:73ra21–73ra21. doi: 10.1126/scitranslmed.3001713. [DOI] [PubMed] [Google Scholar]
- 55.Yuan Y, Wang X, Jia G, Liu J-H, Wang T, Gu Y, Yang S-T, Zhen S, Wang H, Liu Y. Diam. Relat. Mater. 2010;19:291–299. [Google Scholar]
- 56.Puzyr AP, Baron AV, Purtov KV, Bortnikov EV, Skobelev NN, Mogilnaya OA, Bondar VS. Diam. Relat. Mater. 2007;16:2124–2128. [Google Scholar]
- 57.Xing Y, Xiong W, Zhu L, Osawa E, Hussin S, Dai L. ACS Nano. 2011;5:2376–2384. doi: 10.1021/nn200279k. [DOI] [PubMed] [Google Scholar]
- 58.Thomas V, Halloran BA, Ambalavanan N, Catledge SA, Vohra YK. Acta Biomater. 2012;8:1939–1947. doi: 10.1016/j.actbio.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang X, Yin J, Kang C, Li J, Zhu Y, Li W, Huang Q, Zhu Z. Toxicol. Lett. 2010;198:237–243. doi: 10.1016/j.toxlet.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 60.Daum N, Tscheka C, Neumeyer A, Schneider M. Wiley Inter-discip. Rev. Nanomed. Nanobiotechnol. 2012;4:52–65. doi: 10.1002/wnan.165. [DOI] [PubMed] [Google Scholar]
- 61.Hutter E, Boridy S, Labrecque S, Lalancette-Hébert M, Kriz J, Winnik FM, Maysinger D. ACS Nano. 2010;4:2595–2606. doi: 10.1021/nn901869f. [DOI] [PubMed] [Google Scholar]
- 62.Bumb A, Sarkar SK, Billington N, Brechbiel MW, Neuman KC. J. Am. Chem. Soc. 2013;135:7815–7818. doi: 10.1021/ja4016815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.von Haartman E, Jiang H, Khomich AA, Zhang J, Burikov SA, Dolenko TA, Ruokolainen J, Gu H, Shenderova OA, Vlasov II, Rosenholm JM. J. Mater. Chem. B. 2013;1:2358–2366. doi: 10.1039/c3tb20308e. [DOI] [PubMed] [Google Scholar]
- 64.Prabhakar N, Näreoja T, von Haartman E, Sen Karaman D, Jiang H, Koho S, Dolenko TA, Hänninen P, Vlasov DI, Ralchenko VG, Hosomi S, Vlasov I, Sahlgren C, Rosenholm JM. Nanoscale. 2013;5:3713–3722. doi: 10.1039/c3nr33926b. [DOI] [PubMed] [Google Scholar]
- 65.Selvan ST. Biointerphases. 2010;5:FA110. doi: 10.1116/1.3516492. [DOI] [PubMed] [Google Scholar]
- 66.Hong V, Presolski SI, Ma C, Finn MG. Angew. Chem. Int. Ed. 2009;48:9879–9883. doi: 10.1002/anie.200905087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Graf C, Vossen DLJ, Imhof A, van Blaaderen A. Langmuir. 2003;19:6693–6700. [Google Scholar]
- 68.Etienne M, Walcarius A. Talanta. 2003;59:1173–1188. doi: 10.1016/S0039-9140(03)00024-9. [DOI] [PubMed] [Google Scholar]
- 69.Zhang Z, Berns AE, Willbold S, Buitenhuis J. J. Colloid Interface Sci. 2007;310:446–455. doi: 10.1016/j.jcis.2007.02.024. [DOI] [PubMed] [Google Scholar]
- 70.Chithrani BD, Chan WCW. Nano Lett. 2007;7:1542–1550. doi: 10.1021/nl070363y. [DOI] [PubMed] [Google Scholar]
- 71.Egerton RF. Electron Energy-Loss Spectroscopy in the Electron Microscope. Springer; 2011. [Google Scholar]
- 72.Turner S, Lebedev OI, Shenderova O, Vlasov II, Verbeeck J, Van Tendeloo G. Adv. Funct. Mater. 2009;19:2116–2124. [Google Scholar]
- 73.Macková H, Proks V, Horák D, Kučka J, Trchová M. J. Polym. Sci. Part Polym. Chem. 2011;49:4820–4829. [Google Scholar]
- 74.Proks V, Jaroš J, Pop-Georgievski O, Kucˇka J, Popelka Š, Dvorˇák P, Hampl A, Rypáček F. Macromol. Biosci. 2012;12:1232–1242. doi: 10.1002/mabi.201200095. [DOI] [PubMed] [Google Scholar]
- 75.Sivakumar K, Xie F, Cash BM, Long S, Barnhill HN, Wang Q. Org Lett. 2004;6:4603–4606. doi: 10.1021/ol047955x. [DOI] [PubMed] [Google Scholar]
- 76.Berchel M, Haelters J-P, Couthon-Gourvès H, Deschamps L, Midoux P, Lehn P, Jaffrès P-A. Eur. J. Org. Chem. 2011;2011:6294–6303. [Google Scholar]
- 77.Petrakova V, Taylor A, Kratochvilova I, Fendrych F, Vacik Jan Kucka J, Stursa J, Cigler P, Ledvina M, Fiserova A, Kneppo P, Nesladek M. Adv. Funct. Mater. 2012;22:812–819. [Google Scholar]
- 78.Walczyk D, Bombelli FB, Monopoli MP, Lynch I, Dawson KA. J. Am. Chem. Soc. 2010;132:5761–5768. doi: 10.1021/ja910675v. [DOI] [PubMed] [Google Scholar]
- 79.Salvati A, Pitek AS, Monopoli MP, Prapainop K, Bombelli FB, Hristov DR, Kelly PM, Åberg C, Mahon E, a Dawson K. Nat. Nanotechnol. 2013;8:137–43. doi: 10.1038/nnano.2012.237. [DOI] [PubMed] [Google Scholar]
- 80.Ashley CE, Carnes EC, Phillips GK, Padilla D, Durfee PN, Brown PA, Hanna TN, Liu J, Phillips B, Carter MB, Carroll NJ, Jiang X, Dunphy DR, Willman CL, Petsev DN, Evans DG, Parikh AN, Chackerian B, Wharton W, Peabody DS, Brinker CJ. Nat. Mater. 2011;10:389–397. doi: 10.1038/nmat2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.