Abstract
The generation of multiciliated cells (MCCs) is required for the proper function of many tissues, including the respiratory tract, brain, and germline. Defects in MCC development have been demonstrated to cause a subclass of mucociliary clearance disorders termed reduced generation of multiple motile cilia (RGMC). To date, only two genes, Multicilin (MCIDAS) and cyclin O (CCNO) have been identified in this disorder in humans. Here, we describe mice lacking GEMC1 (GMNC), a protein with a similar domain organization as Multicilin that has been implicated in DNA replication control. We have found that GEMC1‐deficient mice are growth impaired, develop hydrocephaly with a high penetrance, and are infertile, due to defects in the formation of MCCs in the brain, respiratory tract, and germline. Our data demonstrate that GEMC1 is a critical regulator of MCC differentiation and a candidate gene for human RGMC or related disorders.
Keywords: cilia, ciliopathy, hydrocephaly, infertility, transcription
Subject Categories: Cell Adhesion, Polarity & Cytoskeleton; Development & Differentiation
Introduction
The generation of diverse types of cilia, microtubule‐based organelles that project from the cell surface, is required for many aspects of development and organismal function (Fliegauf et al, 2007; Choksi et al, 2014b). Most cells can form solitary, immotile cilia involved in sensory functions, while particular cell types exhibit motile cilia that are involved in sensory functions, motility or feeding. Multiciliated cells (MCCs), located in the brain, respiratory system, and reproductive tracts, contain dozens of motile cilia that act in a coordinated beating pattern to promote fluid flow or ovum transport along epithelia (Fliegauf et al, 2007; Brooks & Wallingford, 2014).
The events that trigger MCC differentiation are not completely understood and likely vary to some extent depending on the organism and tissue (Brooks & Wallingford, 2014; Choksi et al, 2014b). In many tissues, the inhibition of Notch/Delta signaling promotes the differentiation of progenitors into MCCs (Deblandre et al, 1999; Liu et al, 2007; Stubbs et al, 2008; Guseh et al, 2009; Tsao et al, 2009; Marcet et al, 2011; Wang et al, 2013; Song et al, 2014; Lafkas et al, 2015). Notch inhibition is mediated in part by the Mir34 and Mir449 microRNA (miRNA) families that directly control Notch1 and Delta‐like 1 (DLL1) (Marcet et al, 2011). Additionally, Notch inhibition leads to the activation of Multicilin (MCIDAS), considered one of the most upstream transcriptional activators of the MCC differentiation program. Multicilin promotes the transcription of key genes required for multiciliogenesis (Stubbs et al, 2012), including FOXJ1, MYB, RFX2, and RFX3 (Brooks & Wallingford, 2014; Choksi et al, 2014b) through interactions with the E2F4 and E2F5 transcription factors and their cofactor, DP1 (referred to collectively as the EDM complex) (Ma et al, 2014). This interaction occurs via a C‐terminal domain of Multicilin, dubbed the TIRT domain based on its amino acid sequence (Stubbs et al, 2012). Consistent with Multicilin being linked to the E2F4/5 transcription factors, loss of E2F4 in mice impairs the development of MCCs in the airway epithelium (Danielian et al, 2007) and E2F5‐deficient mice develop hydrocephaly, potentially due to defects in the formation of MCCs in the brain ependyma, although this has not been conclusively demonstrated (Davy & Robinson, 2003; Danielian et al, 2007).
As each cilium in MCCs is formed from a specialized type of centriole, called the basal body, the development of MCCs requires the massive expansion of centrioles to allow the generation of dozens of motile cilia per cell (Brooks & Wallingford, 2014). Centriole duplication is normally coupled to the cell cycle through the canonical mother‐centriole‐dependent pathway, but as MCCs are post‐mitotic, they rely on the de novo basal body formation pathway. This system of basal body expansion is dependent on deuterosomes, poorly described, electron dense ring structures that produce multiple centrioles simultaneously, enabling rapid formation of large number of centrioles (Sorokin, 1968). The genes required for deuterosome pathway centriole amplification in MCCs are transcriptionally activated by the EDM complex and include DEUP1, CCNO, and CCDC78 (Klos Dehring et al, 2013; Zhao et al, 2013; Boon et al, 2014; Ma et al, 2014; Funk et al, 2015).
Dysfunction of motile cilia causes mucociliary clearance disorders, such as primary ciliary dyskinesia (PCD), characterized by defects in the upper and lower respiratory system, heart malformations, polysplenia, hydrocephaly, subfertility, or infertility, and in some cases by defects in asymmetric organ patterning (known as situs inversus) (Zariwala et al, 2011). A distinct class of mucociliary clearance disorders, referred to as congenital mucociliary clearance disorder with reduced generation of multiple motile cilia (RGMC), was shown to result from mutations in the CCNO or MCIDAS genes that are required for the generation of a fully functional multiciliated epithelium (Boon et al, 2014; Wallmeier et al, 2014). Due to reduced numbers or function of MCCs, RGMC patients develop recurring respiratory infections and in some cases have exhibited hydrocephaly and reduced fertility (Boon et al, 2014).
The MCIDAS gene (encoding Multicilin) is located on chromosome 5q11.2 in humans (13 D2.2 in mice), in a locus that harbors other key regulators of MCC formation, including CCNO, CDC20B, and Mir‐449a/b/c (Marcet et al, 2011; Boon et al, 2014; Song et al, 2014; Wu et al, 2014). Multicilin is a member of the Geminin superfamily, sharing a similar central coiled‐coil (CC) domain with Geminin and Geminin‐like coiled coil containing 1 (GEMC1, encoded by GMNC, also referred to as Lynkeas) (Balestrini et al, 2010; Pefani et al, 2011). Multicilin can interact with Geminin through the CC domain and inhibit its function as a negative regulator of DNA replication (Caillat et al, 2013). Conversely, Geminin impairs the ability of Multicilin to activate genes required for centriole biogenesis in MCCs, suggesting opposing roles that favor proliferation or differentiation, respectively (Ma et al, 2014). Mutations identified in human RGMC patients are located in the TIRT domain of Multicilin (MCIDAS) and compromise interactions with E2Fs that are required for its transcriptional functions, but are unlikely to affect interactions with Geminin (Boon et al, 2014).
GEMC1 was originally identified due to its homology with the Geminin CC domain and was shown to function in DNA replication initiation through interactions with Cdc45 and CDK2/cyclin E that are important for replication initiation, and TOPBP1, a replication fork component critical for activating the ATR kinase that is mutated in Seckel syndrome (O'Driscoll et al, 2003; Mordes et al, 2008; Balestrini et al, 2010). Moreover, depletion of GEMC1 impaired DNA replication in both Xenopus extracts and mammalian cells (Balestrini et al, 2010). Like Multicilin, GEMC1 interacts with Geminin, but the functional relevance of this interaction has not yet been clearly established (Caillat et al, 2013, 2015). GEMC1 is structurally similar to Multicilin, and phylogenetic analysis strongly suggests that they are out‐paralogues (Fig EV1A). In addition to the central Geminin‐like CC domain, GEMC1 contains a C‐terminal TIRT domain with a high degree of homology to that of Multicilin (Fig EV1B).
Figure EV1. Evolutionary relationship and sequence homology of GEMC1 and Multicilin.
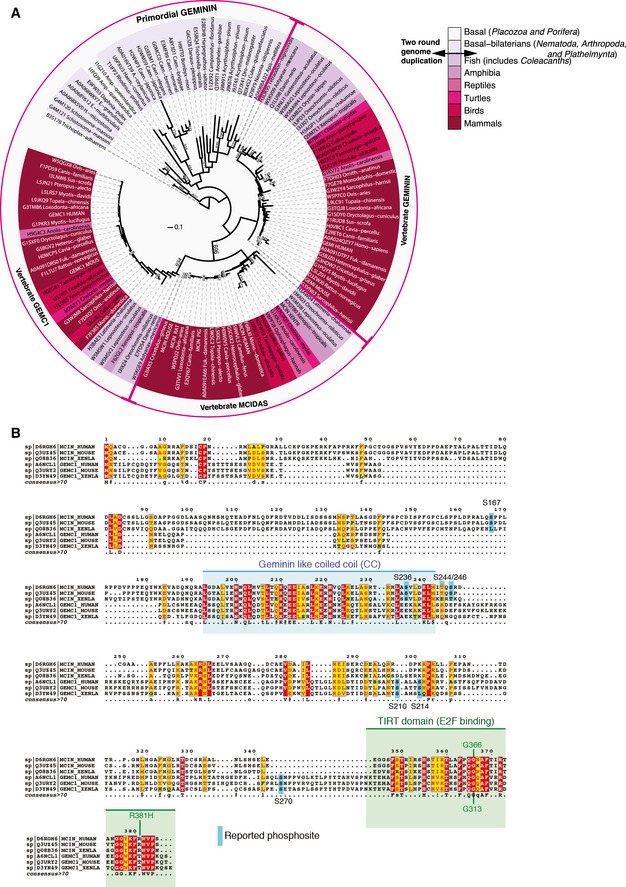
- Phylogenetic analysis strongly suggests that GEMC1 and MCIDAS are out‐paralogues that arose from primordial GEMININ, likely at the time of the two round genome duplication that shaped vertebrate genomes. This is supported by (i) the species distribution of proteins containing this domain, which resembles the species tree, (ii) the absence of identifiable similar proteins in the databases when excluding the common coiled‐coil sequence, (iii) the presence of a common additional domain at the C‐terminal region of both proteins, and (iv) by the bootstrap values separating the GEMININ‐MCIDAS group. A possible evolutionary scenario is that after duplication, the C‐terminal region was acquired by a domain‐shuffling event and GEMC1 began to diverge faster. A plausible explanation for this would be a relaxation of purifying selection in this region (the CC domain), which would yield a large divergence, although the acquisition of a novel function (positive selection) could also have played a role in such a divergence. Along these lines, we can observe large branches separating the GEMC1 group, while the branches separating the GEMC1 sequences are very short. This is indicative of strong purifying selection, although this should be explored in more depth in future analyses (see Appendix Supplementary Materials and Methods for further information).
- The amino acid sequences of GEMC1 and Multicilin (MCIDAS) from human, mouse, and Xenopus were aligned using T‐Coffee (Notredame et al, 2000) and alignments exported using ESPript (Robert & Gouet, 2014) and further annotated in Adobe Illustrator. The Geminin‐like coiled‐coil domain is indicated in blue and the TIRT domain in green. Phosphorylated residues reported in PhosphositePlus (Bateman et al, 2002; Li et al, 2002; Obenauer et al, 2003) are notated in blue and patient mutations in MCIDAS or corresponding mutations in GEMC1 analyzed in this study are indicated (Boon et al, 2014).
Here, we report that mice lacking GEMC1 are growth impaired, develop bilateral hydrocephaly, and are infertile. These defects are accompanied by a dramatic impairment in MCC development in the brain, respiratory system, and reproductive tract of embryonic and adult animals, as well as severe defects in spermatogenesis. The trachea and oviduct that normally have large numbers of MCCs showed highly reduced expression levels of many genes linked to deuterosome‐mediated centriole expansion and cilia development, including Mcidas, FoxJ1, Ccno, Ccdc78, and Deup1. We find that GEMC1 interacts with E2F4/5‐DP1 and Multicilin to control the transcriptional program of MCC cells and that these interactions are required for the activation of key regulators of MCC fate. Our results demonstrate that a primary function of GEMC1 in vivo is to promote the proper differentiation of progenitor cells into the MCC lineage in multiple tissues and suggest that GEMC1 should be considered as a candidate gene for human RGMC disorders.
Results
GEMC1‐deficient mice are runted and develop hydrocephaly
As previous work linked GEMC1 to the control of DNA replication, we sought to examine its functions in vivo (Balestrini et al, 2010). For this purpose, we generated mice with a targeted deletion of the Gemc1 gene (Fig 1A and Appendix Fig S1). Gemc1 −/− mice were notably smaller at birth, while no obvious phenotypes were observed in Gemc1 +/− animals (Fig 1B and Appendix Fig S2). Gemc1 −/− mice presented with symmetrical hydrocephaly with almost complete penetrance (Fig 1C and Appendix Fig S2), and for this reason, we were obligated to cull a number of animals in the first months of life (Fig 1D). However, those less severely affected lived up to 16 months (at which time the study was terminated) without incident. Postmortem analysis revealed hydrocephaly in all but a single Gemc1 −/− animal. Given the strong effect on brain development and growth, we examined the expression of Gemc1 in wild‐type animals. Although there was considerable variability between animals, Gemc1 was expressed at low levels in the kidney, spleen, heart, muscle, liver, and intestine, and at the highest levels in the brain, respiratory system and some reproductive tissues (Fig 1E and F). No Gemc1 mRNA expression was detected in any tissues examined from Gemc1 −/− mice (Fig 1F and Appendix Fig S1). These results established that Gemc1 was required for normal development and that its expression was variable between tissues.
Figure 1. Gemc1‐deficient animals are growth impaired and develop hydrocephaly.
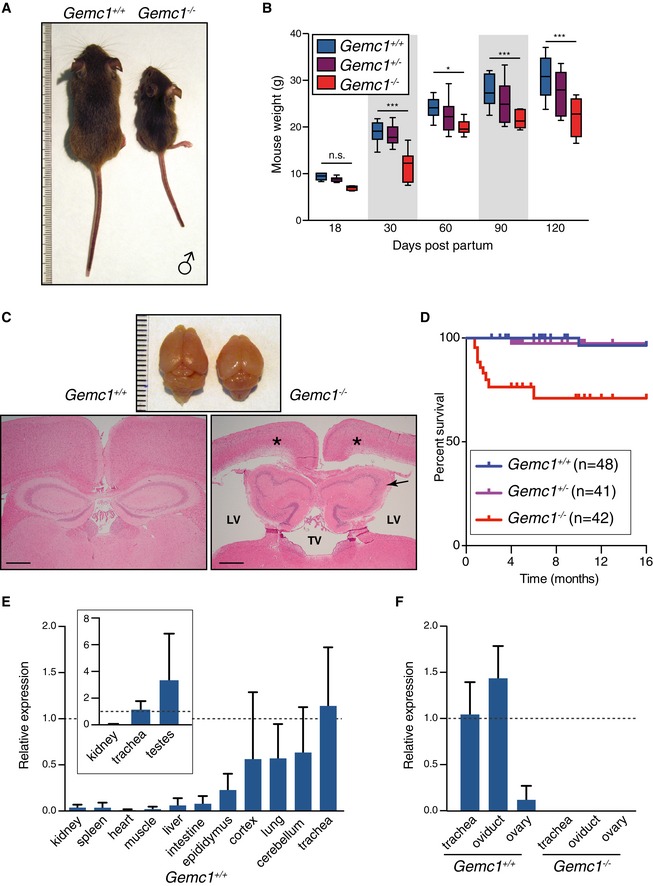
- Example of littermate males of the indicated genotypes 26 days postpartum. Additional examples are provided in Appendix Fig S2.
- Weights of mice of the indicated genotypes from 18 to 120 days postpartum. Box plots indicate 5th–95th percentile. A minimum of eight animals are plotted at each age. Asterisks denote statistical significance (n.s., not significant, *P‐value < 0.05 and ***P‐value < 0.001) determined by the unpaired two‐way Wilcoxon rank‐sum test.
- Example of dissected brains (top panel) and coronal sections (bottom left and right panels) stained with H&E of the indicated genotypes. Note the severe hydrocephaly and ventricular dilation of the lateral ventricle (LV) and third ventricle (TV) in a Gemc1 −/− mouse (bottom right panel) compared to a Gemc1 +/+ littermate animal (bottom left panel). Periventricular malacia (asterisks) and hippocampal atrophy (arrow) are also evident in the Gemc1 −/− sample. Scale bars = 500 μm.
- Kaplan–Meier survival curve of a cohort of mice of the indicated genotypes and numbers (n). The study was terminated at 16 months, and mice were examined for histology. About 97% (31/32) of the Gemc1 −/− mice examined to date exhibited hydrocephaly (additional histological examples in Appendix Fig S2).
- RT–qPCR analysis of Gemc1 expression in murine tissues from Gemc1 +/+ mice using a TaqMan probe. Data are compiled from duplicate samples of three male mice per tissue and mean and standard deviation are plotted.
- RT–qPCR analysis of Gemc1 in trachea, oviduct, and ovary tissues from Gemc1 +/+ and Gemc1 −/− mice shows that the knockout reduces Gemc1 mRNA to undetectable levels. Mean and standard deviation of duplicate samples from two female mice are plotted. Additional examples from other Gemc1 −/− tissues are shown in Appendix Fig S1.
GEMC1 is required for fertility and multiciliated cell generation
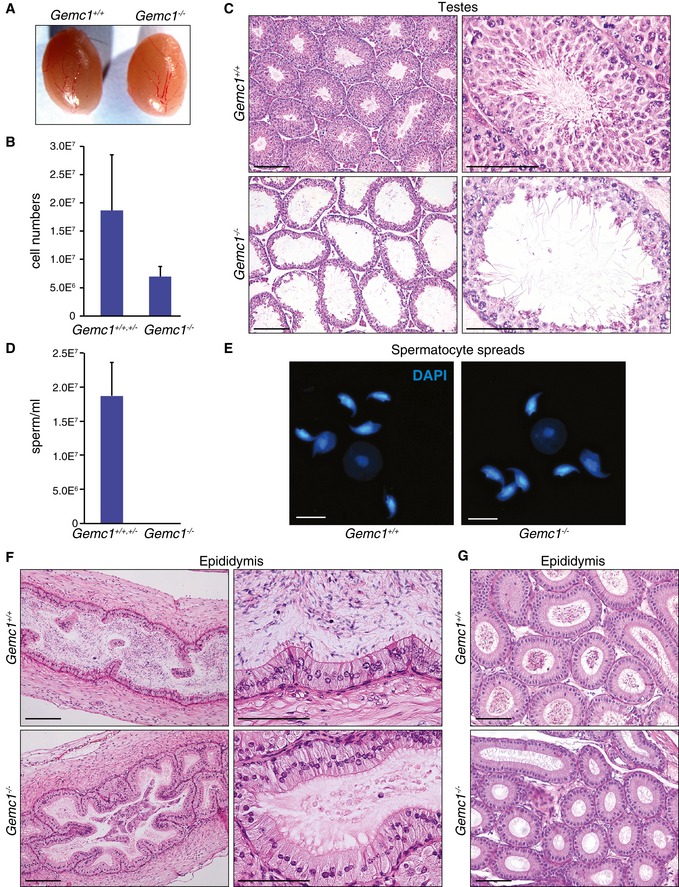
Despite numerous breedings and copulation with wild‐type mice, we did not obtain any offspring from Gemc1 −/− males or females. As Gemc1 was more highly expressed in the germline of wild‐type mice (Fig 1E and F), we histologically analyzed the reproductive tissues of Gemc1 −/− mice. Testes size was similar to that of wild type but cellularity was highly reduced in tubule sections and some cell death was evident (Fig 2C). Sperm counts from the cauda epididymis of Gemc1 −/− mice revealed no sperm, although some morphologically normal elongated spermatids were detected in seminiferous tubule sections (Fig 2C–E). Histological examination of the caput epididymis was consistent with sperm counts as no sperm were identified (Fig 2F). However, the epididymus appeared to be formed normally and the stereocilia lining its epithelium appeared to be intact (Fig 2G).
Figure 2. GEMC1 is required for spermatogenesis.
-
AExample of dissected testes from littermate mice of the indicated genotypes at 2 months of age.
-
BTestes from Gemc1 −/− mice have strongly reduced cellularity compared to Gemc1 +/+ or Gemc1 +/− animals, as measured by disaggregation of the tissue and cell counting with a Neubauer chamber (n = 3). Mean and standard deviation are indicated.
-
CHistological analysis of tubules from 6‐week‐old Gemc1 −/− mice revealed thinning of the spermatogenic cell layer and decreased numbers of elongated spermatids. Scale bars = 100 μm (left panels) and 50 μm (right panels).
-
DSperm counts from the cauda epididymis of wild‐type and Gemc1 −/− mice revealed no sperm in mice lacking GEMC1 (n = 3). Mean and standard deviation are indicated.
-
EExamples of sperm morphology from spermatocyte spreads from 2‐month‐old mice. Scale bars = 10 μm.
-
F, GHistological evaluation of the caput epididymis in sagittal (F) and coronal (G) orientation from 2‐ and 4‐month‐old animals, respectively. Note the absence of luminal spermatozoa in the Gemc1 −/− mice compared to wild type. Scale bars = 100 μm (F, left panels) and 50 μm (F, right panels and G).
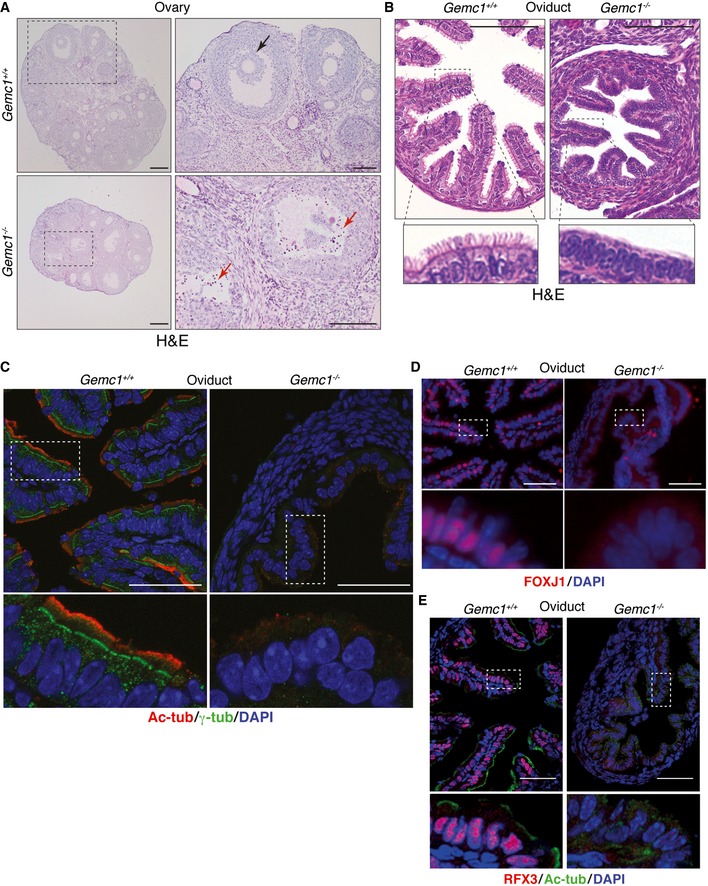
Similar to male mice, Gemc1 −/− females were infertile and presented with severe defects in germline development. The ovaries of the Gemc1 −/− mice examined were smaller, contained few primordial and secondary follicles, and were primarily composed of degenerated antral follicles (Fig 3A). Additionally, the oviduct was smaller in Gemc1 −/− mice and was completely devoid of MCCs, in comparison with the wild‐type epithelia where they were clearly abundant (Fig 3B). To address this phenotype in more detail, we performed immunofluorescence (IF) analysis with markers of cilia (γ‐tubulin (γ‐tub) for basal bodies and acetylated α‐tubulin (Ac‐tub) for cilia), as well as key transcription factors required for MCC differentiation, FOXJ1, and RFX3 (Choksi et al, 2014b). In wild‐type oviducts, an organized layer of basal bodies with cilia extending from them was clearly visible (Fig 3C, left panels). However, in Gemc1 −/− animals, only sporadic, punctate γ‐tubulin staining was observed, likely representing individual centrosomes, and there appeared to be a complete absence of detectable cilia, as no Ac‐tub staining was detected (Fig 3C, right panels). Consistent with this, staining for either FOXJ1 or RFX3 was clearly positive in wild‐type animals, but no signal was detected in the Gemc1 −/− oviducts. These results demonstrated that GEMC1 was required for fertility in both sexes and that in females, this was potentially due to its role in the differentiation of MCCs in the oviduct.
Figure 3. GEMC1 is required for female fertility and multiciliogenesis.
-
AExamples of ovaries from Gemc1 +/+ (top panels) and Gemc1 −/− mice (bottom panels) at 6 weeks of age. Ovaries were smaller in Gemc1 −/− mice and contained primarily degenerated antral follicles. A histologically normal antral follicle is indicated in the Gemc1 +/+ ovary (top right, black arrow) and degenerated antral follicles with high levels of cell death are indicated in the Gemc1 −/− ovaries (bottom right, red arrows). Scale bars = 200 μm (left panels) and 100 μm (right top panels and right bottom panel, respectively).
-
BMorphological abnormalities in the oviduct of 3‐week‐old Gemc1 −/− mice (right panels) compared to Gemc1 +/+ (left panels). A clear lack of cilia (bottom panels) in Gemc1 −/− oviducts is observed by H&E staining. Scale bars = 50 μm (top panels).
-
CStaining of frozen sections of oviduct with antibodies for Ac‐tubulin (cilia) and γ‐tubulin (basal bodies) reveals normal cilia organization in the wild‐type oviduct (left) while basal bodies and cilia are not detectable in Gemc1 −/− oviducts (right). Scale bars = 50 μm.
-
D, EFOXJ1 (D) and RFX3 (E) staining that is characteristic of MCCs was clearly evident in Gemc1 +/+ oviducts and absent in Gemc1 −/− mice. Scale bars = 50 μm.
GEMC1 is required for MCCs in the brain and respiratory system
Given the homology of GEMC1 to the Multicillin protein (Fig EV1) and the phenotypes we observed in the oviduct, we considered that GEMC1 could be important for the development of MCCs in other tissues, such as the brain and respiratory tract, where it was expressed (Fig 1E). To address this, we first examined cilia formation in the nasal and tracheal mucosa of wild‐type and Gemc1 −/− embryos at embryonic days 17.5 and 18.5 (E17.5 and E18.5). At this stage, wild‐type respiratory epithelia exhibit a ciliated, pseudostratified columnar morphology with large numbers of MCCs interspersed with secretory goblet cells (Figs 4A and EV2). In sharp contrast, Gemc1 −/− embryos presented with a flat columnar to squamous, non‐ciliated epithelium in both the nasal and tracheal mucosa (Figs 4A and EV2) and no cilia were identifiable by staining with Ac‐tubulin (Fig EV2). We next used periodic acid–Schiff (PAS) staining to detect secretory cells, as previous work demonstrated that the loss of E2F4 disrupted the development of the respiratory epithelium, leading to the loss of MCCs and increased numbers of PAS‐positive cells (Danielian et al, 2007). Gemc1 −/− mutants exhibited a similar phenotype as that reported for E2F4, as most of the epithelium in Gemc1 −/− mice had abundant PAS‐positive material (Figs 4B, right panels and EV2), in contrast to the normal epithelium where the distinct goblet cells were normally interspersed between columnar MCCs (Figs 4B, left panels and EV2).
Figure 4. GEMC1 is required for the generation of multiciliated tissues.
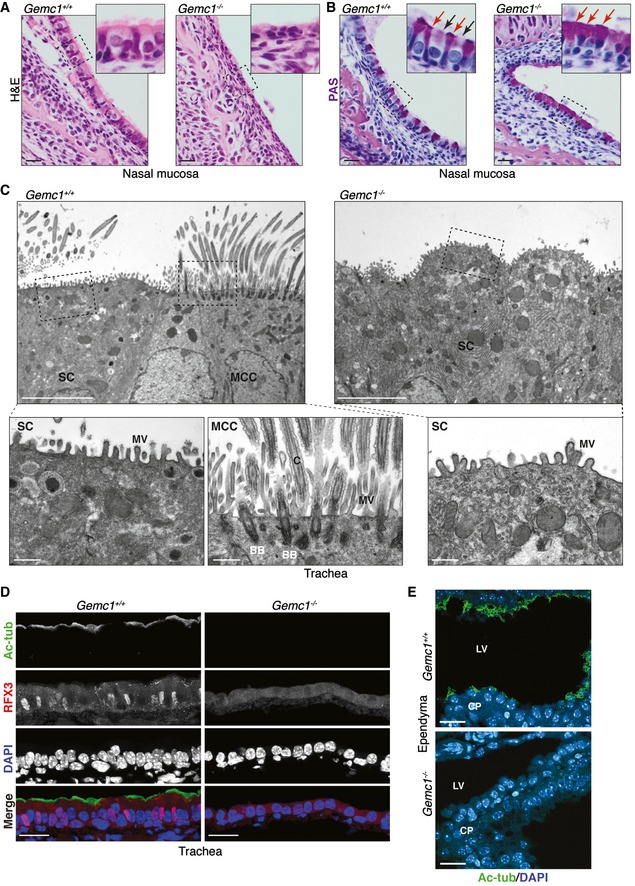
- H&E staining of the nasal mucosa of E17.5 embryos of the indicated genotype. Normal respiratory pseudostratified columnar ciliated epithelium is shown in Gemc1 +/+ embryos compared to the attenuated, flat columnar to squamous epithelium of Gemc1 −/− embryos. Scale bars = 20 μm.
- PAS staining (purple) of the nasal mucosa of E18.5 embryos of the indicated genotype. Normal PAS‐positive goblet cells (inset, red arrows) are interspersed between MCCs (inset, black arrows) with their characteristic goblet or cup‐like shape in Gemc1 +/+ embryos. The Gemc1 −/− mucosa has abundant PAS‐positive materials that accumulate in the apical borders of almost every columnar epithelial cell (inset, red arrows) and lacks typical goblet cells. Scale bars = 20 μm.
- Transmission electron micrographs of cross sections of adult trachea from Gemc1 +/+ animals revealed the presence of secretory cells (SC) and multiciliated cells (MCC). The apical membrane of MCCs accumulated basal bodies (BB) nucleating multiple cilia (C) and microvilli (MV). In the Gemc1 −/− airway epithelium, no MCCs are identifiable and only secretory‐like cells with MV at the apical surface are detectable. Scale bars = 5 μm. Higher magnifications of the boxed regions are shown below. Scale bars = 0.5 μm.
- Lack of Ac‐tubulin (top panel) and RFX3 (second panel from top) staining in the trachea of 1‐month‐old mice. Scale bars = 20 μm.
- Cilia (Ac‐tubulin staining) are not detectable in the ependymal cells of 2‐month‐old Gemc1 −/− mice (lower panel). Choroid plexus (CP) and lateral ventricles (LV) are indicated. Scale bars = 20 μm.
Figure EV2. Analysis of multiciliated epithelia in GEMC1‐deficient mice.
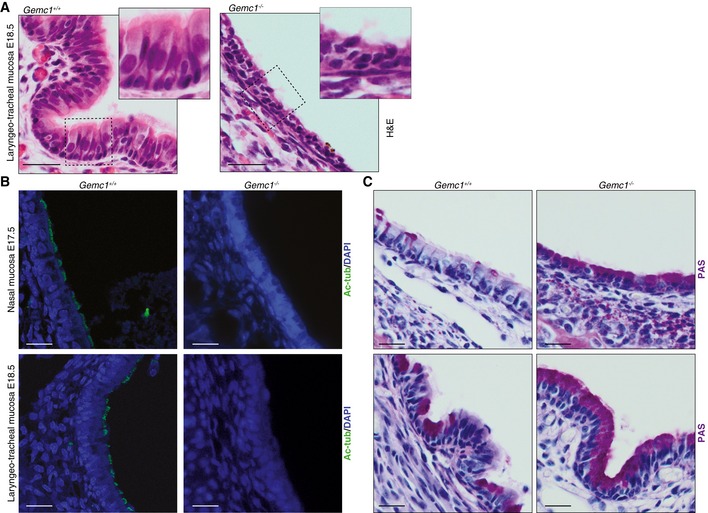
- H&E‐stained sections of the laryngeo‐tracheal mucosa of E18.5 embryos of the indicated genotype. In wild‐type mice, ciliated cells are clearly visible (left, inset) and in sections from Gemc1 −/− mice, no cilia were detectable (right, inset). Scale bars = 50 μm.
- IF staining of sections of the nasal mucosa of E17.5 and laryngeo‐tracheal mucosa of E18.5 embryos with Ac‐tubulin confirmed the lack of cilia in Gemc1 −/− embryos (right panels) compared to wild type where cilia are easily detectable (left panels). Scale bars = 50 μm.
- PAS staining of the nasal mucosa in E17.5 and laryngeo‐tracheal mucosa of E18.5 embryos shows the increased number of PAS‐positive cells in Gemc1 −/− respiratory epithelia. Scale bars = 20 μm.
To further characterize the identity of this abnormal population in the respiratory airways, we performed transmission electron microscopy (TEM) in adult tracheas. The normal MCC and goblet cells were readily observed in wild‐type mice (Fig 4C). In contrast, no ciliated cells were identifiable in the Gemc1 −/− respiratory epithelium and all cells appeared to have a secretory cell‐like morphology (Fig 4C). In addition, no evidence of basal body expansion was readily detectable (Fig 4C and Appendix Fig S3). These results were further confirmed by IF staining in the trachea, where no MCCs were detectable in Gemc1 −/− mice using antibodies for both RFX3 and Ac‐tubulin staining (Fig 4D). In addition, we observed no detectable cilia in the choroid plexus (CP) of Gemc1 −/− mice, compared to a robust Ac‐tubulin signal in wild‐type animals, consistent with the development of hydrocephaly in Gemc1 −/− mice (Figs 4E and 1C, and Appendix Fig S2). Thus, Gemc1 −/− mice exhibited an apparently complete lack of MCCs in multiple tissues.
GEMC1 controls the MCC transcriptional program
As we were unable to detect basal body expansion by IF or TEM, FOXJ1 or RFX3 expression, or MCCs in the oviduct, respiratory tract, or brain of Gemc1 −/− mice (Figs 3 and 4), we considered that GEMC1 likely played an early role in MCC differentiation. To address this, we performed transcriptional profiling of tissues enriched for MCCs (trachea and oviduct) using microarrays. Loss of Gemc1 led to a large number of gene expression differences in both tissues when compared to wild‐type littermate controls (Fig 5 and Dataset EV1). Consistent with our histopathological observations, gene ontology analysis of differentially expressed genes revealed that the category most enriched for downregulated genes in both tissues was cilium (Fig 5A). In addition, enrichment of downregulated genes related to the microtubule organizing center (MTOC) and protein folding was common to both tissues from Gemc1 −/− mice (Fig 5A). To further analyze the gene expression differences, we performed gene set enrichment analysis (GSEA) using the GOSlim dataset, several custom datasets collated from previously published work describing targets of E2F4, FOXJ1, and Myb, known ciliopathy genes, and other suspected genes in current human screening panels (Dataset EV2, Appendix Fig S4 and Appendix Table S1) (Geremek et al, 2011; Lee et al, 2011; Zhao et al, 2011; Choksi et al, 2014b; Ma et al, 2014; Treutlein et al, 2014; Li et al, 2015). We found that the genes downregulated in Gemc1 −/− mice were highly enriched for those implicated in human ciliopathies, FOXJ1, and E2F4 transcriptional targets and to a lesser extent Myb target genes (Fig 5B–D, Dataset EV2 and Appendix Fig S4 and Appendix Table S1).
Figure 5. GEMC1 controls the MCC transcriptional program in vivo .
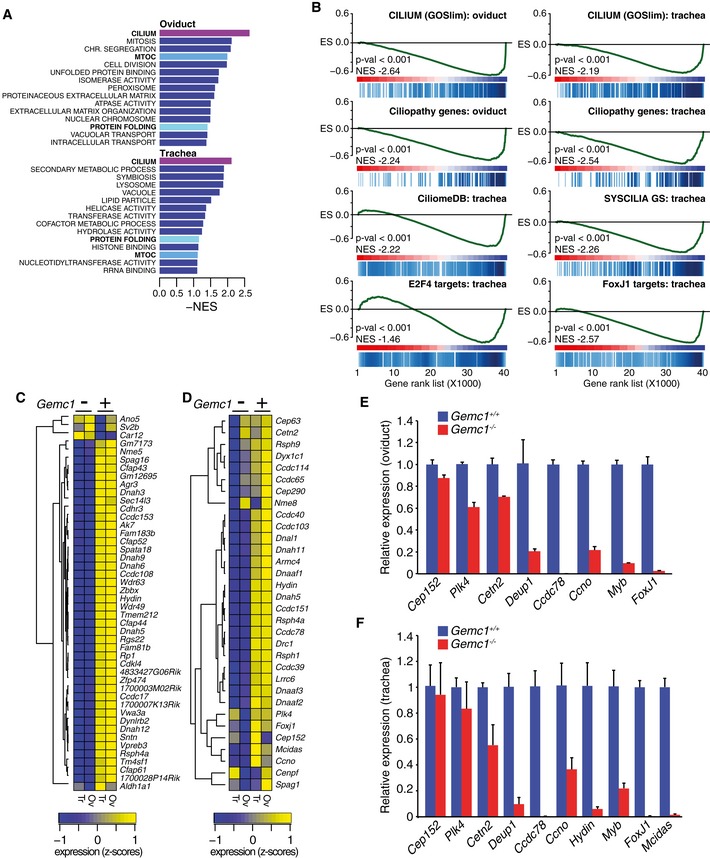
-
AGene ontology (GO) analysis of microarray data from the trachea and oviduct. GO scores (negative enrichment) of differentially expressed genes from the trachea and oviduct of Gemc1 −/− mice (additional information in Dataset EV2).
-
BGene set enrichment analysis of the microarray data using standard and custom gene sets (additional information in Appendix Fig S4 and Appendix Table S1). Gene rank lists from the individual tissues were analyzed against gene sets for gene ontology (GOSlim, top row), a custom gene set containing all known or suspected ciliopathy genes (second row), the Ciliome database (DB), the SYSCILIA consortium gold standard cilia gene list version 1 (SYSCILIA GS) (van Dam et al, 2013; Roosing et al, 2015), E24F target genes (Lee et al, 2011; Ma et al, 2014), and FoxJ1 target genes (Choksi et al, 2014a). Several additional gene sets generated from analysis of ciliopathy patient data (Geremek et al, 2011; Li et al, 2015) or multiciliated cells (Hoh et al, 2012; Treutlein et al, 2014) were also queried (detailed information in Appendix Table S1). Negative enrichment scores are shown for genes downregulated in Gemc1 −/− tissues.
-
CThe most differentially expressed genes related to cilia in the trachea (Tr) and oviduct (Ov) of Gemc1 −/− mice are plotted in a heatmap. Normalized, centered, and scaled Affymetrix probeset intensities are plotted.
-
DHeatmap of differential expression of known human ciliopathy genes in the trachea and oviduct of Gemc1 −/− mice compared to wild‐type littermates.
-
E, FRT–qPCR analysis of individual genes related to MCC formation in the oviduct (E) and trachea (F) of Gemc1 −/− mice compared to wild‐type littermates, using 18S rRNA as a reference (the mean and standard deviation of triplicate samples from two animals of each genotype is graphed). Genes examined generally fall into three classes: centriole duplication (Cep152, Plk4, Cetn2), deuterosome‐mediated centriole expansion (Deup1, Ccdc78, Ccno) and transcription of genes required for MCC differentiation (FoxJ1, Myb, Mcidas).
To validate these results and investigate the status of particular genes of interest in more detail, we performed real&hyphen ;time quantitative PCR (RT–qPCR) for a number of key genes normally induced in MCCs, including the transcription factors Mcidas, FoxJ1, and Myb, as well as genes involved in basal body expansion, such as Ccno, Deup1, and Ccdc78. In wild‐type mice, we observed robust expression of all of those genes in both the trachea and oviduct, but the expression levels were significantly reduced in both tissues of Gemc1 −/− mice (Fig 5E and F). In contrast, several other genes involved in centriole biogenesis including Cep152, Plk4, and Centrin‐2 (Cetn2) were only mildly affected or unaffected (Fig 5E and F). Thus, GEMC1 is required for the early induction of the MCC transcriptional program and affects a wide array of known targets of both E2F4 and FOXJ1 including genes required for the amplification of centrioles through the deuterosome‐mediated pathway.
GEMC1 interacts with E2F4/5‐DP1 and Multicilin via distinct domains
As GEMC1 loss had a pronounced effect on the expression of E2F4 targets, and E2F4 was reported to interact with Multicilin through its C‐terminal TIRT domain, which is highly conserved in GEMC1 (Fig EV1), we performed immunoprecipitation (IP) experiments to determine whether GEMC1 bound to E2F family members and/or Multicilin. The IP of FLAG‐tagged GEMC1 brought down co‐expressed HA‐tagged E2F4 or HA‐tagged DP1 (Fig 6A, lanes 5 and 6). However, the IP of both E2F4 and DP1 was greatly enriched when they were co‐expressed, similar to what has been reported for the Multicilin containing EDM complex (Fig 6A, lane 7) (Ma et al, 2014). GEMC1 could also pull down E2F5 that is the most similar E2F family member to E2F4 but not E2F1 (Fig 6B). Following the nomenclature established for the EDM complex, we termed this tripartite complex of E2F4/5‐DP1‐GEMC1, the EDG complex.
Figure 6. GEMC1 interacts with E2F4/5 and Multicilin through distinct domains.
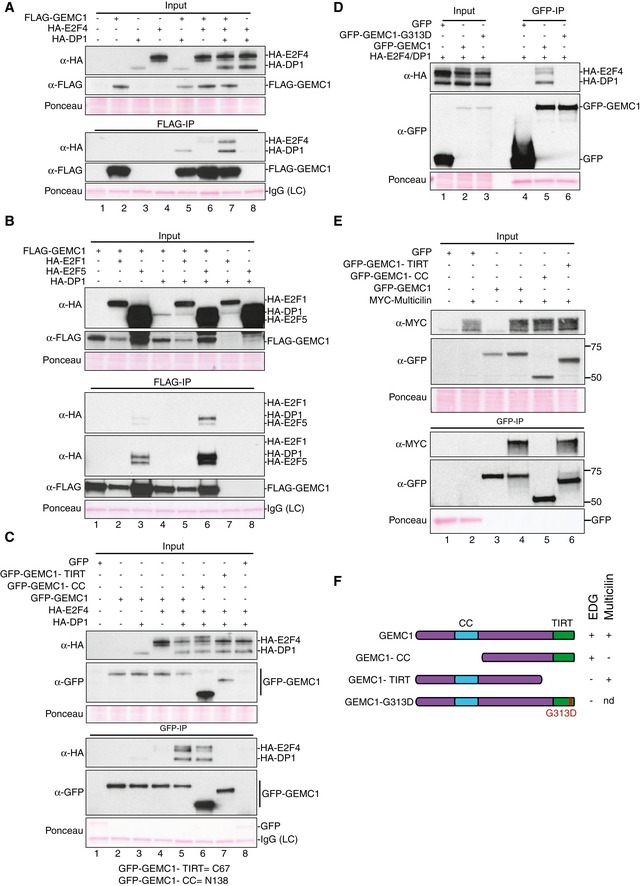
- GEMC1 interacts with E2F4 and DP1. Western blot of HA‐ or FLAG‐tagged protein expression in the input lysates are shown in the top panel. FLAG immunoprecipiation (IP) of GEMC1 co‐IPs DP1 (α‐HA, lane 5) and E2F4 (α‐HA, lane 6). The interaction is enhanced upon co‐expression of HA‐tagged E2F4 and DP1 (α‐HA, lane 7).
- GEMC1 interacts with E2F5 but not E2F1. Co‐IP of E2F5 (α‐HA, lane 3) and E2F5‐DP1 (α‐HA, lane 6) but not E2F1 or E2F1‐DP1 (α‐HA, lanes 2 and 5). In the FLAG‐IP panels, a short (top) and long (second from top) exposure of the Western are shown.
- Identification of the domain required for E2F4 interaction. GFP‐tagged GEMC1 and mutant forms were expressed, and IPs were performed against GFP. Wild‐type GEMC1 (lane 5) or GEMC1 lacking the CC domain (lane 6) pulled down E2F4‐DP1. A GEMC1 protein lacking the TIRT domain or GFP alone did not IP E2F4‐DP1 (lanes 7, 8).
- Mutation of the single amino acid analogous to that identified in MCIDAS in human RGMC patients (G313D) impairs the interaction with E2F4‐DP1 (compare land 5 and 6).
- GEMC1 interacts with Multicilin through its CC domain. Wild‐type GEMC1 or a mutant lacking the TIRT domain pull down Myc‐tagged Multicilin (anti‐Myc blot, lanes 4 and 6) while a CC domain mutant does not (anti‐Myc blot, lane 5).
- Schematic summary of the GEMC1 mutants used in (A–E) and their effect on EDG complex or Multicilin interactions.
Source data are available online for this figure.
We next sought to identify the domains of GEMC1 involved in EDG complex formation and found that the C‐terminal TIRT domain was required for EDG complex formation (Fig 6C, compare α‐HA lane 5 to lane 7 in the GFP‐IP panel), while the Geminin‐like CC domain was dispensable for this interaction (Fig 6C, lane 6 in the GFP‐IP panel). In addition, a point mutation in the conserved G313 to D residue, mimicking the Multicilin (MCIDAS) TIRT mutation identified in human RGMC patients, disrupted the E2F4‐DP1 interaction suggesting a similar mode of binding (Fig 6D) (Boon et al, 2014). In contrast to E2F4/5‐DP1, GEMC1 interacted with Multicilin through its CC domain and independently of its TIRT domain (Fig 6E, compare lanes 4–6 of the anti‐Myc blot). These data indicated that GEMC1 can interact with E2F4/5‐DP1 and Multicilin through distinct domains (Fig 6F).
GEMC1 activates the MCC transcriptional program
As a primary function of GEMC1 in vivo appeared to be the activation of transcriptional programs required for multiciliogenesis, similar to what has been reported for Multicilin, we asked whether the ectopic expression of GEMC1 was sufficient to activate endogenous FOXJ1 or MCIDAS. Transient overexpression of GEMC1 led to the robust activation of FOXJ1 and to a lesser extent MCIDAS (Figs 7A and EV3). Previous work established that the CC and TIRT domains of Multicilin contributed to the transcriptional activation of FOXJ1(Stubbs et al, 2012). To identify the domains of GEMC1 required for its transcriptional activity, we transfected a panel of mutant GEMC1 cDNAs. Both the CC and TIRT domains of GEMC1 were required for the efficient induction of FOXJ1 in both HEK293T and U2OS cell lines (Figs 7B and EV3).
Figure 7. GEMC1 transcriptionally activates the MCC program.
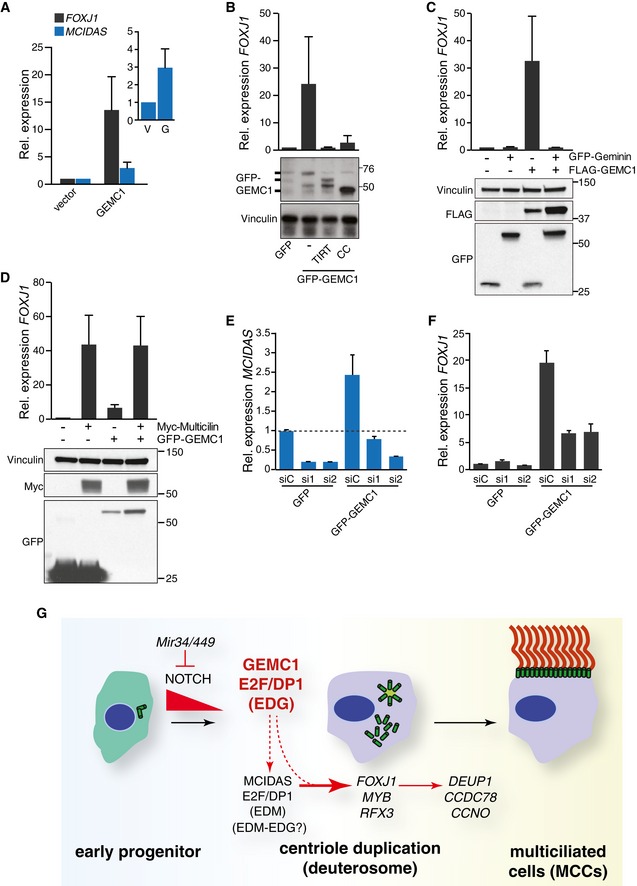
-
ATransient overexpression of GEMC1 leads to increased levels of FOXJ1 and MCIDAS expression (RT–qPCR) in HEK293T cells. Results for MCIDAS are graphed on a smaller scale in the right panel: V = vector and G = GEMC1. The mean and standard deviation of 3 individual experiments in HEK293T cells are graphed. Similar induction of FOXJ1 was observed in U2OS and T98G (Fig EV3).
-
BBoth the CC and the TIRT domain of GEMC1 are required to activate FOXJ1 (RT–qPCR). A corresponding Western blot showing relative expression levels of the mutant GFP‐tagged GEMC1 variants is shown from HEK293T cells. The mean and standard deviation of three individual experiments in HEK293T cells are graphed. Similar results were also observed in U2OS (Fig EV3).
-
CActivation of FOXJ1 by GEMC1 is inhibited by the co‐expression of Geminin (compare lanes 3 and 4). Example Western blot is shown below. Results graphed are the mean and standard deviation of three individual experiments in HEK293T cells.
-
DCo‐expression of GEMC1 and Multicilin results in similar levels of FOXJ1 expression (measured by RT–qPCR) as Multicilin alone in HEK293T cells. The mean and standard deviation of 4 individual experiments in HEK293T cells are graphed.
-
E, FThe activation of FOXJ1 by GEMC1 is partially dependent on Multicilin. HEK293T cells were transfected with a control siRNA (siC) or 2 different siRNAs against MCIDAS (si1 and si2) prior to being transfected with GFP or GFP‐GEMC1 expression vectors. Relative MCIDAS (E) and FOXJ1 (F) expression levels were assessed by RT–qPCR. The mean and standard deviation of a representative experiment performed in triplicate is graphed. An additional experiment is presented in Fig EV3.
-
GModel for the role of GEMC1 in the differentiation of MCCs. Inhibition of Notch signaling by Mir34/449 is required for MCC differentiation and the ability of GEMC1 and Multicilin to activate transcription (Liu et al, 2007; Tsao et al, 2009; Marcet et al, 2011; Stubbs et al, 2012; Kyrousi et al, 2015). GEMC1 acts early, in conjunction with E2F4/5‐DP1 (EDG), to activate genes including Mcidas and FoxJ1, likely through direct interaction with E2F4 at the promoters of FoxJ1 and some other E2F target genes (dotted line) (Figs 5, 6, 7). The activation of Multicilin leads to further amplification of the transcriptional program through E2Fs (EDM complex) (Ma et al, 2014) and GEMC1 and Multicilin may form heterocomplexes (Fig 6E) with E2F4/5‐DP1 (EDM‐EDG) (Caillat et al, 2013). This leads to the optimal activation of FOXJ1,MYB,RFX3, and other transcription factors required for the expression of CCDC78,DEUP1,CCNO, and other downstream components of the MCC differentiation pathway (Klos Dehring et al, 2013; Tan et al, 2013; Zhao et al, 2013; Ma et al, 2014; Wallmeier et al, 2014; Funk et al, 2015). See Discussion for further details.
Source data are available online for this figure.
Figure EV3. Transcriptional activation by GEMC1.
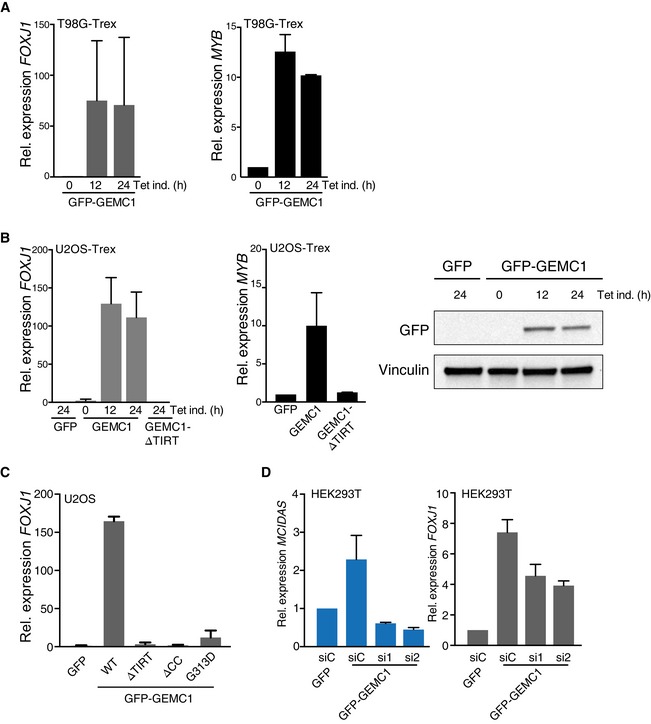
- Induction of FOXJ1 (left panel) and MYB (right panel) in T98G‐Trex cells measured by RT–qPCR following the induction of GFP‐GEMC1 with tetracycline (Tet) for the indicated time. The mean and standard deviation triplicate experiments is graphed.
- Induction of FOXJ1 (left panel) and MYB (middle panel) in U2OS‐Trex cells measured by RT–qPCR following the induction of GFP‐GEMC1 or GFP‐GEMC1‐∆TIRT with tetracycline (Tet). An example Western blot showing GFP‐GEMC1 levels after induction is shown (right panel). Induction times are 24 h unless indicated otherwise. The mean and standard deviation of triplicate experiments is graphed.
- Levels of FOXJ1 expression (RT–qPCR) following the transfection of U2OS cells with GFP or GFP‐GEMC1 constructs. The wild‐type (WT) or mutant form transfected is indicated. The mean and standard deviation of triplicate samples is shown.
- Depletion of Multicilin by siRNA reduces but does not eliminate GEMC1 activation of FOXJ1. HEK293T cells transfected with control siRNA (siC) or 2 different siRNAs against MCIDAS (si1 and si2) were transfected with GFP‐GEMC1 and MCIDAS and FOXJ1 levels were measured by RT–qPCR. Mean and standard deviation of triplicate samples are shown.
As available data indicated that GEMC1, Multicilin and Geminin could interact through their CC domains, we next examined their relative influence on FOXJ1 activation (Caillat et al, 2013, 2015). The co‐expression of Geminin with GEMC1 resulted in the near complete inhibition of FOXJ1 transcriptional activation (Fig 7C). In contrast, the co‐expression of Multicilin with GEMC1 did not lead to higher levels of FOXJ1 expression than Multicilin alone (Fig 7D), suggesting that GEMC1's influence on FOXJ1 could be partially indirect, through its ability to induce Multicilin. To determine the extent to which Multicilin contributed to the activation of FOXJ1 by GEMC1, we depleted MCIDAS mRNA levels using siRNA in cells transfected with GEMC1. The siRNA‐mediated depletion of MCIDAS reduced it to basal levels or below (Fig 7E), and FOXJ1 activation by GEMC1 was strongly attenuated, but not eliminated (Figs 7F and EV3). These data indicated that GEMC1 activation of MCIDAS contributed significantly, but was not absolutely required, for the activation of FOXJ1 observed upon GEMC1 overexpression.
Collectively, our results identify GEMC1 as a critical regulator of MCC differentiation that acts through its C‐terminal TIRT domain that facilitates the formation of the EDG complex (E2F4/5‐DP1‐GEMC1) and through its CC domain that mediates interactions with Geminin or Multicilin (Fig 7G). GEMC1 can activate MCIDAS and FOXJ1, as well as other genes involved in MCC differentiation (Fig 5 and Datasets EV1 and EV2), and this activation is strongly enhanced by the presence of Multicilin (Figs 7E and F, and EV3). Available data suggest that GEMC1 and Multicilin appear to initiate similar transcriptional programs required for MCC differentiation (Fig 5) and are similarly inhibited by Geminin (Fig 7E) (Stubbs et al, 2012; Ma et al, 2014). Although both GEMC1 and Multicilin can activate FOXJ1 (Fig 7E), and presumably other common targets, GEMC1 is required for Multicilin expression, the activation of FOXJ1, and the generation of MCCs in vivo.
Discussion
GEMC1 is one of the most upstream activators of the MCC transcriptional cascade that has been identified thus far, as it is required for the expression of key transcriptional regulators, including Mcidas, FoxJ1, Rfx3, and Myb, and components of the deuterosome‐mediated centriole amplification program, such as Ccno, Deup1, and Ccdc78 (Fig 5) (Klos Dehring et al, 2013; Zhao et al, 2013; Ma et al, 2014). Our results indicate that GEMC1 is a strong candidate gene for human RGMC disorders. Consistent with this possibility, Gemc1 −/− mice phenocopy many aspects of FoxJ1, E2F4, E2F5, Rfx2, Rfx3, and Ccno knockout mice and show similar pathologies to human patients with CCNO or MCIDAS mutations (Lindeman et al, 1998; Danielian et al, 2007; El Zein et al, 2009; Chung et al, 2012; Boon et al, 2014; Wallmeier et al, 2014; Funk et al, 2015). Notably, Gemc1 +/− mice did not present with obvious phenotypes, suggesting that it is not haploinsufficient, although we have observed a minor reduction in ciliary beat frequency in tracheal cultures from Gemc1 +/− mice (Appendix Fig S5). In addition, Gemc1 −/− mice may be a powerful tool for the identification of additional ciliopathy candidate genes, as we find a large number of common genes negatively affected in both the trachea and oviduct of these mice (Dataset EV1). Further work will be required to define their functional relevance and identify those related to ciliogenesis.
A previous GWAS study identified a number of single&hyphen ;nucleotide polymorphisms (SNPs) associated with Alzheimer's disease (AD) risk that mapped near the GMNC gene on chromosome 3q28 (Cruchaga et al, 2013). Our demonstration that GEMC1 plays a crucial role in brain development, particularly in the movement of cerebrospinal fluid, would be consistent with potential pathogenic effects caused by less severe impairments of GEMC1 regulation. However, whether these SNPs indeed have an impact on GEMC1 regulation remains to be determined. Several of the other SNPs identified in that, and another study, are also located near or in genes implicated in cilia formation, including NME8, GLIS3, and DNAH5 (Kang et al, 2009a,b; Lambert et al, 2013). While, to our knowledge, defects in MCCs or motile cilia have not been implicated in AD, it has been suggested that defects in primary cilia may play an important role in the pathology of AD, and defects in primary and motile cilia may influence Huntington's disease (Keryer et al, 2011; Armato et al, 2013). These links between neurodegenerative diseases and multiciliogenesis controlled by GEMC1 warrant further exploration given that GLIS3, NME8, and DNAH5 are downregulated in tracheal tissue lacking GEMC1 (Dataset EV1) and DNAH5 is also not detectable in multiciliated tissues in patients lacking functional Multicilin (MCIDAS) (Boon et al, 2014).
The expression of the Mir34 and Mir449 families of microRNAs represent one of the earliest events known to occur in MCC specification, as they have been implicated in the inhibition of Notch signaling (Marcet et al, 2011; Song et al, 2014). The Gemc1 mRNA contains a large 3′UTR with a highly conserved consensus binding sites for the Mir449/Mir34 microRNA families (Targetscan (Friedman et al, 2009; Garcia et al, 2011; Grimson et al, 2007; Lewis et al, 2005)) that are not present in the Mcidas or Ccno 3′UTR regions (Appendix Fig S6). In the testes, it appears that Gemc1 is a bona fide target of Mir449/Mir34, as its expression is upregulated in Mir449/Mir34a/Mir34bc triple knockout mice (Wu et al, 2014). Interestingly, the testicular phenotype of Gemc1 −/− mice, that is thus far the only pathology we have identified that is not clearly linked to MCC generation, is highly reminiscent of that reported in the Mir449/Mir34a/Mir34bc triple knockouts (Comazzetto et al, 2014; Wu et al, 2014; Yuan et al, 2015). At face value, this data would suggest that the expression of Mir449, which is highly expressed in MCCs, would paradoxically suppress Gemc1 expression that is required for MCC generation (Song et al, 2014). We propose that GEMC1 protein levels are likely to be strictly controlled by translational regulation. This could occur through RNA‐binding proteins, as consensus target sites for both Musashi and CPEBs (CPE elements) are present in the 3′UTR of Gemc1 (Appendix Fig S6). The predicted CPE element in the GEMC1 3′UTR overlaps with the Mir34/449 consensus target sequence, suggesting potential competition. A role for Musashi in the regulation of the MCC transcriptional program would be consistent with a previous report that the deletion of Musashi 1 and Musashi 2 (Msi1 and Msi2) leads to defects in the generation of ependymal cells and hydrocephaly in mice (Sakakibara et al, 2002). Further work will be needed to understand the complex mechanisms of GEMC1 regulation in different tissues.
We have shown that GEMC1 forms a ternary complex with E2F4/5 and DP1 through its C‐terminal TIRT domain that we have termed the EDG complex to differentiate it from the previously described EDM complex (Ma et al, 2014). GEMC1 was recently reported not to interact with E2F4/5‐DP1 using a similar approach (Zhou et al, 2015), and this discrepancy could be due to different experimental conditions or differences in the affinity tags used. Regardless, given the phenotypic similarity of Gemc1 and E2F4‐deficient mice or Xenopus (Fig 4), as well as the high negative enrichment in E2F4 targets in tissues of Gemc1 −/− mice (Fig 5 and Dataset EV2), GEMC1 is clearly required to activate E2F4 target genes in tissues that give rise to MCCs (Ma et al, 2014).
A major question remains as to the nature of the relationship between GEMC1 and Multicilin that appear to function in a mechanistically similar manner. GEMC1 is not located in the same genetic locus that contains Multicilin (Mcidas), Ccno, Mir449, and Cdc20B, and recent work has demonstrated that the expression of Gemc1 precedes that of Mcidas in developing mouse ependymal cells (Kyrousi et al, 2015). In addition, it was shown that Gemc1 is required for Mcidas expression in zebrafish and Xenopus (Zhou et al, 2015), similar to what we have observed here in mice, where we can detect little expression of Mcidas or Ccno in Gemc1 −/− tissues (Fig 5E and F). Although these data suggest that GEMC1 acts upstream of Multicilin, it does not exclude the possibility that GEMC1 and Multicilin function together to promote MCC differentiation and potentially regulate MCC homeostasis. Consistent with this, the expression of both genes was reported to be specific to MCCs from single‐cell analysis of lung tissues (Treutlein et al, 2014) and we (Fig 6E) and others have observed an interaction between the 2 proteins that may impact their function, for example, by allowing a transition between EDG and EDM transcriptional complexes that could have distinct targets, functions or modes of regulation (Caillat et al, 2015).
While the phenotypes of Gemc1 −/− mice indicate that a primary function of GEMC1 in vivo is to promote MCC differentiation, it thus far remains unclear whether its roles in the regulation of DNA replication impact the pathologies we observed or cause additional developmental defects that we have not yet identified. The smaller size and the defects in gametogenesis of Gemc1 −/− mice could result from reduced cellularity as a consequence of growth phenotypes secondary to defective DNA replication control. Alternatively, these phenotypes might be due to impaired endocrine control, for example, due to pituitary gland malfunction induced by hydrocephaly, or loss of specialized functions of GEMC1 unrelated to either DNA replication or transcriptional roles.
Given the limited expression of Gemc1 in adult tissues, we feel it is unlikely that GEMC1 is a general regulator of DNA replication in most somatic cell types. A non‐exclusive possibility is that GEMC1 and/or Multicilin could function to restrain Geminin activity in particular cell types where it is highly expressed, especially during embryogenesis. Compensatory mechanisms that attenuate Geminin mediated replication inhibition might mask the potential Geminin inhibitory role of GEMC1 in Gemc1 −/− cells. Increased Geminin expression has also been reported in many cancer cell types and the elevated expression of GEMC1 or Multicilin could provide an alternative means for facilitating DNA replication in this context (Kapoor, 2012). In addition, we have observed that the overexpression of Geminin that also has transcriptional roles, including the inhibition of Multicilin, inhibits GEMC1‐mediated activation of target genes (Fig 7D) (Yellajoshyula et al, 2012; Ma et al, 2014; Karamitros et al, 2015), suggesting the existence of reciprocal regulatory mechanisms between GEMC1 and Geminin.
It was proposed previously that Geminin mediated inhibition of Multicilin could act to prevent the activation of the MCC program prior to cell cycle exit, as the activation of deuterosome‐mediated centriole expansion could be deleterious to cycling cells (Stubbs et al, 2012). One possibility consistent with available data is that limited expression of Gemc1 precedes that of Mcidas and allows cells to be exquisitely sensitive to Geminin levels that act to dampen deleterious transcriptional events in cells that have not fully exited the cell cycle. The attenuation of Geminin levels during cell cycle exit, promoted by a shift in the stoichiometry between GEMC1 and Geminin, would then allow the activation of Mcidas, thus further committing cells to a MCC fate through the robust activation of transcriptional programs by the EDG and EDM (or hybrid/independent) complexes. Future work will be required to understand the complex regulatory control of GEMC1 expression and activity, its precise relationship with Geminin and Multicilin and the mechanisms by which it controls MCC development and other cellular processes.
Materials and Methods
Animal generation and husbandry
Mice lacking GEMC1 were generated in the IRB Barcelona mutant mouse core facility (Appendix Fig S1). Mouse embryonic stem (ES) cells were targeted with a Gemc1 targeting construct that was designed and constructed using standard recombineering methods (Copeland et al, 2001). The targeting vector was confirmed by sequencing and linearized vector was transfected into ES cells for targeting and blastocyst injections were performed. All animals were maintained in strict accordance with the European Community (86/609/EEC) guidelines in the specific‐pathogen‐free (SPF) animal facilities in the Barcelona Science Park (PCB). Protocols were approved by the Animal Care and Use Committee of the PCB (IACUC; CEEA‐PCB) in accordance with applicable legislation (Law 5/1995/GC; Order 214/1997/GC; Law 1201/2005/SG). All efforts were made to minimize use and suffering.
Total testicular cell numbers and sperm counts
Tunica albuginea was removed from testes and transferred to 10 ml of Hanks' balanced salt solution (HBSS, Lonza) with 0.5 mg/ml collagenase IV (Sigma) and 1 μg/ml DNase I (Sigma) for 15 min at 33°C. The suspension was incubated for 5 min at room temperature (RT) and washed with cold HBSS to remove the interstitial tissue. Cellular aggregates were disrupted using a Pasteur pipette and strained with a 40‐μm mesh to achieve a single‐cell suspension. To determine sperm counts, cauda epididymis was dissected from 8‐ to 10‐week‐old mice, cut with scissors in five places, and incubated in 1 ml of warm PBS at 37°C for 10 min to let the sperm exit the tissue. Then, total testicular extracts or sperm suspension were manually counted under the microscope on a Neubauer glass slide counter.
Immunofluorescence
Freshly dissected oviducts, tracheas, and brains of the indicated age were fixed in 4% PFA (Santa Cruz) overnight at 4°C, cryoprotected in 30% sucrose, and frozen in optimal cutting temperature (OCT) compound at −80°C. Cryosections were cut at 10 μm thickness and fixed to frosted glass slides by incubation for 20 min in ice‐cold acetone at −20°C. Slides were then washed several times in PBS followed with washing in PBS‐T (0.3% Triton X‐100 in PBS) and blocked with 5% goat serum in PBS‐T for 60 min. Slides were incubated overnight at 4°C with primary antibody, washed several times in PBS‐T, and stained with the Alexa Fluor‐conjugated complementary antibody for 60 min at RT. After final washing, DNA was counterstained with DAPI. Slides were imaged using a Leica TCS SP5 confocal microscope equipped with 63× NA 1.40 oil immersion objective and HyD detectors.
Histopathology and immunohistochemistry
Samples (tissues or embryos) were harvested and fixed in 4% PFA overnight at 4°C. Dissected testes, epididymis, ovaries, and oviducts of the indicated age were also fixed in Bouin's solution (Electron Microscopy Sciences) overnight at 4°C. Samples were embedded in paraffin using standard procedures. Sections were cut at 4–5 μm thickness and stained with hematoxylin and eosin or periodic acid–Schiff (PAS) reagent (Sigma‐Aldrich). Stained slides were examined microscopically, unbiased by 2 board‐certified pathologists (S.A.Y. and A.d.B). For immunohistochemistry, sections were deparaffinized, rehydrated, and rinsed in Tris‐buffered saline (TBS). For antigen retrieval, tissue sections were microwaved for 5 min in 10 mM citrate buffer at 40% power in an 1,100 W oven. For colorimetric visualization, sections were incubated with primary antibody overnight after quenching endogenous peroxidase activity using 0.5% H2O2 (vol/vol) in TBS for 10 min and 10% normal goat serum (NGS) for 1 h. Slides were washed and incubated with biotinylated anti‐rabbit secondary diluted in 1% BSA in TBS (Vector Labs BA‐1000 at 1:200) and 30 min with Vectastain Elite ABC reagent. Sections were incubated in DAB substrate kit for 4 min (Vector SK 4100) and counterstained in hematoxylin, dehydrated, and mounted in DPX (Fluka). For Ac‐tubulin staining, slides treated for antigen retrieval were washed in PBS with 0.1% Tween‐20 (PBST) and incubated with primary antibody overnight at 4°C. Slides were rinsed and incubated with secondary anti‐goat Alexa488 for 1 h and rinsed and mounted in Fluoroshield with DAPI (Sigma). Slides were imaged using Olympus DP25 camera mounted on Olympus BX45 microscope or using a E600 Nikon epifluorescence microscope equipped with 40x NA 0.75 objective and a charge‐coupled Olympus DP72 device camera.
Antibodies
| Antigen | Species | Source | Reference no. | Dilution |
|---|---|---|---|---|
| γ‐tubulin | rabbit | Sigma | T5192 | 1:500 |
| Acetylated‐α‐tubulin | mouse | Sigma | T6793 | 1:200‐1:300 |
| FoxJ1 (2A5) | mouse IgG1 | eBioscience | 14‐9965‐82 | 1:100 |
| RFX3 | rabbit | Sigma | HPA035689 | 1:100 |
Secondary antibodies: Alexa Fluor 488‐ and 568‐conjugated goat anti‐mouse, Alexa Fluor 488‐ and 568‐conjugated goat anti‐rabbit (Life Technologies).
Real&hyphen ;time quantitative PCR
Dissected tissues were collected on ice, washed in PBS, and frozen. Tissues were disrupted in Tri Reagent by zirconium beads in a mechanical tissue disruptor (Precellys 24, Bertin technologies). RNA was isolated according to manufacturer recommendations (PureLink RNA mini kit, Ambion). cDNA was generated using 1 μg of total RNA and a High Capacity RNA‐to‐cDNA Kit (ABI).
Real‐time quantitative PCR (RT–qPCR) was performed using the comparative CT method and a Step‐One‐Plus real‐time PCR Applied Biosystems Instrument. Amplification was performed using Power SYBR Green PCR Master Mix (ABI) or TaqMan Universal PCR Master Mix (ABI). All assays were performed in triplicate. For TaqMan assays, mActB probe was used as an endogenous control for normalization and a specific Taqman probe was used for mouse or human Gemc1, Multicilin, Ccno, and FoxJ1. Primers (Sigma) used for SYBR Green assays are provided in Appendix Supplementary Materials and Methods.
Cell culture, immunoprecipitation and Western blotting
HEK293 cells (ATCC) were cultured in DMEM (Gibco) with 10% FBS (Hyclone) and were routinely tested for mycoplasma and found negative. For pull‐down experiments, cells were transiently transfected with equal amounts of desired vectors using FuGENE 6 Transfection Reagent (Promega) according to the manufacturer's instructions. Cells were harvested 24 h after transfection, washed twice in cold PBS, and resuspended in FLAG‐IP buffer (50 mM Tris–HCl pH 7.4, 200 mM NaCl, 1 mM EDTA, 1% Triton X‐100) supplemented with fresh protease (Millipore) and phosphatase inhibitors (0.1 mM NaOVan, 10 mM NaF). Samples were incubated 10 min on ice, sonicated using a Bioruptor Standard sonication device (Diagenode) for 4 min with a 30‐s interval, and centrifuged for 15 min at 13,000 rpm. FLAG and GFP‐Flag immunoprecipitations were carried out using anti‐FLAG M2 affinity gel (Sigma A2220) or GFP‐Trap beads (ChromoTek). For a single reaction, 40 μl of gel suspension was washed twice with FLAG‐IP buffer and 1.5 mg of total protein was incubated with washed resin for 2 h on a rotating wheel at 4°C. Resin was then washed five times with FLAG‐IP buffer and once with TBS 1×. Proteins were eluted by boiling in Laemmli buffer, loaded on a SDS–PAGE gel, and transferred to PVDF membrane (Millipore) for Western blot analysis. Antibodies to the following epitope tags were used: Flag (Sigma), GFP‐tag (rabbit serum produced by Julian Gannon, London Research Institute), HA HRP conjugated (Sigma), and Myc (Santa Cruz). Primary antibodies were detected with appropriate secondary antibodies conjugated to HRP (DAKO) and visualized by ECL substrate (GE) or WesternBright ECL (Advansta) on Amersham Hyperfilm (GE). Ponceau staining of anti‐FLAG M2 IgG light chain was used as a loading control for immunoprecipitation.
For siRNA transfection, cells were co‐transfected with siRNAs and GFP or GFP‐GEMC1 using Lipofectamine 2000 (Invitrogen), and after 16 h, a second transfection with siRNAs was done using Lipofectamine RNAiMAX (Invitrogen). We used siGFP (GGCUACGUCCAGGAGCGCCGCACC) as a negative control for siRNA transfection unless indicated. siMCIDAS#1: CCACCAAACGGAAGCAGACUUCAAU; siMCIDAS#2: GAGACGCGCUUGU UGAGAAUAAUCA.
DNA constructs
Human GEMC1 cDNA corresponding to transcript NM_001146686.2 was amplified from total cDNA generated from human AG2603 cells and cloned into pCR‐Blunt II‐TOPO vector (Invitrogen) and then into the gateway‐compatible pDONR221 vector (Invitrogen) and pcDNA5FRT/TO (Invitrogen) vector, modified with an N‐terminal Flag‐tag (gift from Zuzana Horejsi, Clare Hall Laboratories). Full‐length GFP‐GEMC1 (1–334), GFP‐GEMC1‐∆TIRT (∆C67), and GFP‐GEMC1‐∆CC (∆N138) cDNAs were cloned into the pIRESpuro3 vector (Clontech), modified with an N‐terminal GFP‐Flag‐tag (gift from Tohru Takaki, Clare Hall Laboratories). GFP‐GEMC1‐G313D was generated by site‐directed mutagenesis (Quick‐Change site‐directed mutagenesis kit (Qiagen)) using pIRESpuro3‐GFP‐Flag‐GEMC1 as template and the primer 5′‐CATTCCTTCCACCAGGACCAAGCCTTTGTGCG‐3′. E2F4 and DP1 cDNAs (Addgene plasmids #10914 and #37968) were cloned into the mammalian expression vector pcDNA3.1‐HA (Addgene) at the EcoRI and BamHI restriction sites to produce proteins N‐terminally fused to HA under the control of the constitutive CMV promoter. pCMV‐HA‐E2F5 and pCMV‐HA‐E2F1 were a gift from Kristian Helin (Addgene plasmids #24213 and #24225). Human Geminin cDNA (OriGene SC319331) was cloned into pcDNA5FRT/TO (Invitrogen) vector, modified with an N‐terminal YFP‐tag. Human Multicilin Myc‐DDK‐tagged cDNA was obtained by OriGene (RC231199).
Microarray analysis and GSEA
Transcriptional profiling of trachea and oviduct tissues in wild‐type and Gemc1 −/− mice were performed by the Microarray Unit‐Cogentech at IFOM‐IEO campus. Biotin‐labeled cDNA targets were synthesized starting from 150 ng of total RNA. Double‐stranded cDNA synthesis and related cRNA were performed with GeneChip® WT Plus Kit (Affymetrix) that was also used to synthesize the sense strand cDNA before it was fragmented and labeled. All steps of the labeling protocol were performed as suggested by Affymetrix. Hybridization was performed using the GeneChip® Hybridization, Wash and Stain Kit. Targets were diluted in hybridization buffer at a concentration of 25 ng/μl, denatured at 99°C for 5 min incubated at 45°C for 5 min and centrifuged at maximum speed for 1 min prior to introduction into the GeneChip® cartridge. A single GeneChip® Mouse Transcriptome Array 1.0 was then hybridized with each biotin‐labeled sense target for 16 h at 45°C in a rotisserie oven. GeneChip® cartridges were washed and stained with GeneChip® Hybridization, Wash and Stain Kit in the Affymetrix Fluidics Station 450. GeneChip arrays were scanned using an Affymetrix GeneChip® Scanner 3000 7G using default parameters. Affymetrix GeneChip® Command Console software (AGCC) was used to acquire GeneChip® images and generate.CEL files for analysis. Arrays were imported to R applying RMA background correction and probe summarization. For each tissue, fold changes between conditions were computed after mean and variance normalization and centering using a generalized additive model (GAM). An empirical Bayesian partial density model was used to identify differentially expressed genes with a false discovery rate (FDR) of 5% and log2‐fold change larger (smaller) than 3 (−3). Gene set enrichment analysis (GSEA) was performed using all genes in the array sorted by fold change computed in each tissue (Subramanian et al, 2005). We tested the GOslim gene sets (version from January 2015) and a list of custom gene sets extracted from various publications or databases (Ashburner et al, 2000; Inglis et al, 2006; Geremek et al, 2011; Lee et al, 2011; Zhao et al, 2011; van Dam et al, 2013; Choksi et al, 2014a; Ma et al, 2014; Treutlein et al, 2014; Wheway et al, 2015). Additional information and references are provided in Appendix Table S1. The microarray data have been submitted to NCBI's Gene Expression Omnibus (GEO) and has been assigned the reference number GSE71746.
Transmission electron microscopy (TEM)
Adult trachea tissue was dissected, fixed in 4% PFA in PBS and cut into 200‐μm transverse sections using a vibratome (Leica VT1200S). After an overnight incubation with 0.25% glutaraldehyde in buffer, the tissue was post‐fixed with 1% osmium tetroxide, dehydrated, washed, and contrasted with 0.2% tannic acid in water for 10 min. After washing, it was incubated with 0.5% uranyl acetate in water for 1 h in dark and dehydrated through a graded series of ethanol at room temperature (15–30 min for each step), before infiltration with Epon replacement resin (Roth), and polymerization at 60°C. Ultrathin sections (70 nm) were cut on a UCT microtome (Leica Microsystems), stained with uranyl acetate and lead citrate, and viewed under an electron microscope (Morgagni; FEI Company). Micrographs were taken with a charge‐coupled device camera (Morada; EMSIS) and ITEM software (EMSIS).
Author contributions
THS, VC, BT, GP, SSB, AMR, and MAV wrote the manuscript; THS, VC, BT, GP, SSB, GGG, MWB, CJ, AMR, MM, PAK, SF, MAV, and AdB conceived or designed the experiments; BT, GP, SSB, GGG, CJ, MM, PAK, LP, TLR, SAY, SF, and AdB performed the experiments; THS, VC, BT, GP, SSB, GGG, SAY, CSOA, AMR, MWB, CJ, MM, PAK, LP, TLR, SF, WBH, MAV, and AdB analyzed the data; and AMR and CSOA performed bioinformatics analyses.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Source Data for Expanded View
Dataset EV1
Dataset EV2
Review Process File
Source Data for Figure 6
Source Data for Figure 7
Acknowledgements
We are grateful to the Stracker lab, Ignasi Roig, Raul Mendez, Jens Luders, Joan Roig, Angel Nebreda, and Julien Colombelli for helpful input, and Bill Keyes and Alba Mas for IHC advice. We thank the Ministerio de Economía y Competitividad (MINECO) for funding to TS (BFU2012‐39521) and MAV (SAF2012‐38140; Fondo de Investigación Sanitaria (RD12/0042/0014); FEDER Funds); G.G.‐G. is supported by ISCIII (PI13/00864); V.C. is funded by the Associazione Italiana per Ricerca sul Cancro (AIRC), the European Research Council (ERC) consolidator grant (614541), the Association for International Cancer Research (AICR) (13‐0026), the Giovanni Armenise Award to V.C., the Epigen Progetto Bandiera (4.7) and the Fondazione Telethon (GGP13071); G.P. was supported by AIRC Borsa: Fondazione Giovanna Ciani Rif. 16444; S.S.B. was supported by a fellowship from Fundació La Caixa; P.A.K. was supported by an Early Postdoc Mobility Fellowship from the Swiss National Science Foundation; and B.T. was supported by a Severo Ochoa FPI Fellowship (MINECO). IRB Barcelona is a Severo Ochoa Award of Excellence Recipient (MINECO).
The EMBO Journal (2016) 35: 942–960
References
- Armato U, Chakravarthy B, Pacchiana R, Whitfield JF (2013) Alzheimer's disease: an update of the roles of receptors, astrocytes and primary cilia (review). Int J Mol Med 31: 3–10 [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel‐Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestrini A, Cosentino C, Errico A, Garner E, Costanzo V (2010) GEMC1 is a TopBP1‐interacting protein required for chromosomal DNA replication. Nat Cell Biol 12: 484–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths‐Jones S, Howe KL, Marshall M, Sonnhammer EL (2002) The Pfam protein families database. Nucleic Acids Res 30: 276–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon M, Wallmeier J, Ma L, Loges NT, Jaspers M, Olbrich H, Dougherty GW, Raidt J, Werner C, Amirav I, Hevroni A, Abitbul R, Avital A, Soferman R, Wessels M, O'Callaghan C, Chung EM, Rutman A, Hirst RA, Moya E et al (2014) MCIDAS mutations result in a mucociliary clearance disorder with reduced generation of multiple motile cilia. Nat Commun 5: 4418 [DOI] [PubMed] [Google Scholar]
- Brooks ER, Wallingford JB (2014) Multiciliated cells. Curr Biol 24: R973–R982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillat C, Pefani DE, Gillespie PJ, Taraviras S, Blow JJ, Lygerou Z, Perrakis A (2013) The Geminin and Idas coiled coils preferentially form a heterodimer that inhibits Geminin function in DNA replication licensing. J Biol Chem 288: 31624–31634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillat C, Fish A, Pefani DE, Taraviras S, Lygerou Z, Perrakis A (2015) The structure of the GemC1 coiled coil and its interaction with the Geminin family of coiled‐coil proteins. Acta Crystallogr D Biol Crystallogr 71: 2278–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choksi SP, Babu D, Lau D, Yu X, Roy S (2014a) Systematic discovery of novel ciliary genes through functional genomics in the zebrafish. Development 141: 3410–3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choksi SP, Lauter G, Swoboda P, Roy S (2014b) Switching on cilia: transcriptional networks regulating ciliogenesis. Development 141: 1427–1441 [DOI] [PubMed] [Google Scholar]
- Chung MI, Peyrot SM, LeBoeuf S, Park TJ, McGary KL, Marcotte EM, Wallingford JB (2012) RFX2 is broadly required for ciliogenesis during vertebrate development. Dev Biol 363: 155–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comazzetto S, Di Giacomo M, Rasmussen KD, Much C, Azzi C, Perlas E, Morgan M, O'Carroll D (2014) Oligoasthenoteratozoospermia and infertility in mice deficient for miR‐34b/c and miR‐449 loci. PLoS Genet 10: e1004597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland NG, Jenkins NA, Court DL (2001) Recombineering: a powerful new tool for mouse functional genomics. Nat Rev Genet 2: 769–779 [DOI] [PubMed] [Google Scholar]
- Cruchaga C, Kauwe JS, Harari O, Jin SC, Cai Y, Karch CM, Benitez BA, Jeng AT, Skorupa T, Carrell D, Bertelsen S, Bailey M, McKean D, Shulman JM, De Jager PL, Chibnik L, Bennett DA, Arnold SE, Harold D, Sims R et al (2013) GWAS of cerebrospinal fluid tau levels identifies risk variants for Alzheimer's disease. Neuron 78: 256–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam TJ, Wheway G, Slaats GG, Huynen MA, Giles RH (2013) The SYSCILIA gold standard (SCGSv1) of known ciliary components and its applications within a systems biology consortium. Cilia 2: 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielian PS, Bender Kim CF, Caron AM, Vasile E, Bronson RT, Lees JA (2007) E2f4 is required for normal development of the airway epithelium. Dev Biol 305: 564–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davy BE, Robinson ML (2003) Congenital hydrocephalus in hy3 mice is caused by a frameshift mutation in Hydin, a large novel gene. Hum Mol Genet 12: 1163–1170 [DOI] [PubMed] [Google Scholar]
- Deblandre GA, Wettstein DA, Koyano‐Nakagawa N, Kintner C (1999) A two‐step mechanism generates the spacing pattern of the ciliated cells in the skin of Xenopus embryos. Development 126: 4715–4728 [DOI] [PubMed] [Google Scholar]
- El Zein L, Ait‐Lounis A, Morle L, Thomas J, Chhin B, Spassky N, Reith W, Durand B (2009) RFX3 governs growth and beating efficiency of motile cilia in mouse and controls the expression of genes involved in human ciliopathies. J Cell Sci 122: 3180–3189 [DOI] [PubMed] [Google Scholar]
- Fliegauf M, Benzing T, Omran H (2007) When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol 8: 880–893 [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk MC, Bera AN, Menchen T, Kuales G, Thriene K, Lienkamp SS, Dengjel J, Omran H, Frank M, Arnold SJ (2015) Cyclin O (Ccno) functions during deuterosome‐mediated centriole amplification of multiciliated cells. EMBO J 34: 1078–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP (2011) Weak seed‐pairing stability and high target‐site abundance decrease the proficiency of lsy‐6 and other microRNAs. Nat Struct Mol Biol 18: 1139–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geremek M, Bruinenberg M, Zietkiewicz E, Pogorzelski A, Witt M, Wijmenga C (2011) Gene expression studies in cells from primary ciliary dyskinesia patients identify 208 potential ciliary genes. Hum Genet 129: 283–293 [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett‐Engele P, Lim LP, Bartel DP (2007) MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 27: 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guseh JS, Bores SA, Stanger BZ, Zhou Q, Anderson WJ, Melton DA, Rajagopal J (2009) Notch signaling promotes airway mucous metaplasia and inhibits alveolar development. Development 136: 1751–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoh RA, Stowe TR, Turk E, Stearns T (2012) Transcriptional program of ciliated epithelial cells reveals new cilium and centrosome components and links to human disease. PLoS ONE 7: e52166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis PN, Boroevich KA, Leroux MR (2006) Piecing together a ciliome. Trends Genet 22: 491–500 [DOI] [PubMed] [Google Scholar]
- Kang HS, Beak JY, Kim YS, Herbert R, Jetten AM (2009a) Glis3 is associated with primary cilia and Wwtr1/TAZ and implicated in polycystic kidney disease. Mol Cell Biol 29: 2556–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HS, Kim YS, ZeRuth G, Beak JY, Gerrish K, Kilic G, Sosa‐Pineda B, Jensen J, Pierreux CE, Lemaigre FP, Foley J, Jetten AM (2009b) Transcription factor Glis3, a novel critical player in the regulation of pancreatic beta‐cell development and insulin gene expression. Mol Cell Biol 29: 6366–6379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor S (2012) Emerging role of geminin as a prognostic marker in systemic malignancies. Journal of Breast Cancer 15: 481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamitros D, Patmanidi AL, Kotantaki P, Potocnik AJ, Bahr‐Ivacevic T, Benes V, Lygerou Z, Kioussis D, Taraviras S (2015) Geminin deletion increases the number of fetal hematopoietic stem cells by affecting the expression of key transcription factors. Development 142: 70–81 [DOI] [PubMed] [Google Scholar]
- Keryer G, Pineda JR, Liot G, Kim J, Dietrich P, Benstaali C, Smith K, Cordelieres FP, Spassky N, Ferrante RJ, Dragatsis I, Saudou F (2011) Ciliogenesis is regulated by a huntingtin‐HAP1‐PCM1 pathway and is altered in Huntington disease. J Clin Invest 121: 4372–4382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klos Dehring DA, Vladar EK, Werner ME, Mitchell JW, Hwang P, Mitchell BJ (2013) Deuterosome‐mediated centriole biogenesis. Dev Cell 27: 103–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrousi C, Arbi M, Pilz GA, Pefani DE, Lalioti ME, Ninkovic J, Gotz M, Lygerou Z, Taraviras S (2015) Mcidas and GemC1/Lynkeas are key regulators for the generation of multiciliated ependymal cells in the adult neurogenic niche. Development 142: 3661–3674 [DOI] [PubMed] [Google Scholar]
- Lafkas D, Shelton A, Chiu C, de Leon Boenig G, Chen Y, Stawicki SS, Siltanen C, Reichelt M, Zhou M, Wu X, Eastham‐Anderson J, Moore H, Roose‐Girma M, Chinn Y, Hang JQ, Warming S, Egen J, Lee WP, Austin C, Wu Y et al (2015) Therapeutic antibodies reveal Notch control of transdifferentiation in the adult lung. Nature 528: 127–131 [DOI] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim‐Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier‐Boley B, Russo G, Thorton‐Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, Vardarajan BN, Kamatani Y et al (2013) Meta‐analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet 45: 1452–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BK, Bhinge AA, Iyer VR (2011) Wide‐ranging functions of E2F4 in transcriptional activation and repression revealed by genome‐wide analysis. Nucleic Acids Res 39: 3558–3573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20 [DOI] [PubMed] [Google Scholar]
- Li J, Ning Y, Hedley W, Saunders B, Chen Y, Tindill N, Hannay T, Subramaniam S (2002) The molecule pages database. Nature 420: 716–717 [DOI] [PubMed] [Google Scholar]
- Li Y, Klena NT, Gabriel GC, Liu X, Kim AJ, Lemke K, Chen Y, Chatterjee B, Devine W, Damerla RR, Chang C, Yagi H, San Agustin JT, Thahir M, Anderton S, Lawhead C, Vescovi A, Pratt H, Morgan J, Haynes L et al (2015) Global genetic analysis in mice unveils central role for cilia in congenital heart disease. Nature 521: 520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeman GJ, Dagnino L, Gaubatz S, Xu Y, Bronson RT, Warren HB, Livingston DM (1998) A specific, nonproliferative role for E2F‐5 in choroid plexus function revealed by gene targeting. Genes Dev 12: 1092–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Pathak N, Kramer‐Zucker A, Drummond IA (2007) Notch signaling controls the differentiation of transporting epithelia and multiciliated cells in the zebrafish pronephros. Development 134: 1111–1122 [DOI] [PubMed] [Google Scholar]
- Ma L, Quigley I, Omran H, Kintner C (2014) Multicilin drives centriole biogenesis via E2f proteins. Genes Dev 28: 1461–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcet B, Chevalier B, Luxardi G, Coraux C, Zaragosi LE, Cibois M, Robbe‐Sermesant K, Jolly T, Cardinaud B, Moreilhon C, Giovannini‐Chami L, Nawrocki‐Raby B, Birembaut P, Waldmann R, Kodjabachian L, Barbry P (2011) Control of vertebrate multiciliogenesis by miR‐449 through direct repression of the Delta/Notch pathway. Nat Cell Biol 13: 693–699 [DOI] [PubMed] [Google Scholar]
- Mordes DA, Glick GG, Zhao R, Cortez D (2008) TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev 22: 1478–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J (2000) T‐Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302: 205–217 [DOI] [PubMed] [Google Scholar]
- Obenauer JC, Cantley LC, Yaffe MB (2003) Scansite 2.0: proteome‐wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res 31: 3635–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Driscoll M, Ruiz‐Perez VL, Woods CG, Jeggo PA, Goodship JA (2003) A splicing mutation affecting expression of ataxia‐telangiectasia and Rad3‐related protein (ATR) results in Seckel syndrome. Nat Genet 33: 497–501 [DOI] [PubMed] [Google Scholar]
- Pefani DE, Dimaki M, Spella M, Karantzelis N, Mitsiki E, Kyrousi C, Symeonidou IE, Perrakis A, Taraviras S, Lygerou Z (2011) Idas, a novel phylogenetically conserved geminin‐related protein, binds to geminin and is required for cell cycle progression. J Biol Chem 286: 23234–23246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert X, Gouet P (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42: W320–W324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosing S, Hofree M, Kim S, Scott E, Copeland B, Romani M, Silhavy JL, Rosti RO, Schroth J, Mazza T, Miccinilli E, Zaki MS, Swoboda KJ, Milisa‐Drautz J, Dobyns WB, Mikati MA, Incecik F, Azam M, Borgatti R, Romaniello R (2015) Functional genome‐wide siRNA screen identifies KIAA0586 as mutated in Joubert syndrome. eLife 4: e06602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara S, Nakamura Y, Yoshida T, Shibata S, Koike M, Takano H, Ueda S, Uchiyama Y, Noda T, Okano H (2002) RNA‐binding protein Musashi family: roles for CNS stem cells and a subpopulation of ependymal cells revealed by targeted disruption and antisense ablation. Proc Natl Acad Sci USA 99: 15194–15199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song R, Walentek P, Sponer N, Klimke A, Lee JS, Dixon G, Harland R, Wan Y, Lishko P, Lize M, Kessel M, He L (2014) miR‐34/449 miRNAs are required for motile ciliogenesis by repressing cp110. Nature 510: 115–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin SP (1968) Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J Cell Sci 3: 207–230 [DOI] [PubMed] [Google Scholar]
- Stubbs JL, Oishi I, Izpisua Belmonte JC, Kintner C (2008) The forkhead protein Foxj1 specifies node‐like cilia in Xenopus and zebrafish embryos. Nat Genet 40: 1454–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs JL, Vladar EK, Axelrod JD, Kintner C (2012) Multicilin promotes centriole assembly and ciliogenesis during multiciliate cell differentiation. Nat Cell Biol 14: 140–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP (2005) Gene set enrichment analysis: a knowledge‐based approach for interpreting genome‐wide expression profiles. Proc Natl Acad Sci USA 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan FE, Vladar EK, Ma L, Fuentealba LC, Hoh R, Espinoza FH, Axelrod JD, Alvarez‐Buylla A, Stearns T, Kintner C, Krasnow MA (2013) Myb promotes centriole amplification and later steps of the multiciliogenesis program. Development 140: 4277–4286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein B, Brownfield DG, Wu AR, Neff NF, Mantalas GL, Espinoza FH, Desai TJ, Krasnow MA, Quake SR (2014) Reconstructing lineage hierarchies of the distal lung epithelium using single‐cell RNA‐seq. Nature 509: 371–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao PN, Vasconcelos M, Izvolsky KI, Qian J, Lu J, Cardoso WV (2009) Notch signaling controls the balance of ciliated and secretory cell fates in developing airways. Development 136: 2297–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallmeier J, Al‐Mutairi DA, Chen CT, Loges NT, Pennekamp P, Menchen T, Ma L, Shamseldin HE, Olbrich H, Dougherty GW, Werner C, Alsabah BH, Kohler G, Jaspers M, Boon M, Griese M, Schmitt‐Grohe S, Zimmermann T, Koerner‐Rettberg C, Horak E et al (2014) Mutations in CCNO result in congenital mucociliary clearance disorder with reduced generation of multiple motile cilia. Nat Genet 46: 646–651 [DOI] [PubMed] [Google Scholar]
- Wang L, Fu C, Fan H, Du T, Dong M, Chen Y, Jin Y, Zhou Y, Deng M, Gu A, Jing Q, Liu T, Zhou Y (2013) miR‐34b regulates multiciliogenesis during organ formation in zebrafish. Development 140: 2755–2764 [DOI] [PubMed] [Google Scholar]
- Wheway G, Schmidts M, Mans DA, Szymanska K, Nguyen TT, Racher H, Phelps IG, Toedt G, Kennedy J, Wunderlich KA, Sorusch N, Abdelhamed ZA, Natarajan S, Herridge W, van Reeuwijk J, Horn N, Boldt K, Parry DA, Letteboer SJ, Roosing S et al (2015) An siRNA‐based functional genomics screen for the identification of regulators of ciliogenesis and ciliopathy genes. Nat Cell Biol 17: 1074–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Bao J, Kim M, Yuan S, Tang C, Zheng H, Mastick GS, Xu C, Yan W (2014) Two miRNA clusters, miR‐34b/c and miR‐449, are essential for normal brain development, motile ciliogenesis, and spermatogenesis. Proc Natl Acad Sci USA 111: E2851–E2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yellajoshyula D, Lim JW, Thompson DM Jr, Witt JS, Patterson ES, Kroll KL (2012) Geminin regulates the transcriptional and epigenetic status of neuronal fate‐promoting genes during mammalian neurogenesis. Mol Cell Biol 32: 4549–4560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Tang C, Zhang Y, Wu J, Bao J, Zheng H, Xu C, Yan W (2015) mir‐34b/c and mir‐449a/b/c are required for spermatogenesis, but not for the first cleavage division in mice. Biol Open 4: 212–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zariwala MA, Omran H, Ferkol TW (2011) The emerging genetics of primary ciliary dyskinesia. Proc Am Thorac Soc 8: 430–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Glazov EA, Pattabiraman DR, Al‐Owaidi F, Zhang P, Brown MA, Leo PJ, Gonda TJ (2011) Integrated genome‐wide chromatin occupancy and expression analyses identify key myeloid pro‐differentiation transcription factors repressed by Myb. Nucleic Acids Res 39: 4664–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Zhu L, Zhu Y, Cao J, Li S, Huang Q, Xu T, Huang X, Yan X, Zhu X (2013) The Cep63 paralogue Deup1 enables massive de novo centriole biogenesis for vertebrate multiciliogenesis. Nat Cell Biol 15: 1434–1444 [DOI] [PubMed] [Google Scholar]
- Zhou F, Narasimhan V, Shboul M, Chong YL, Reversade B, Roy S (2015) Gmnc is a master regulator of the multiciliated cell differentiation program. Curr Biol 25: 3267–3273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Source Data for Expanded View
Dataset EV1
Dataset EV2
Review Process File
Source Data for Figure 6
Source Data for Figure 7


