Abstract
Species of several unrelated families within the angiosperms are able to constitutively produce pyrrolizidine alkaloids as a defense against herbivores. In pyrrolizidine alkaloid (PA) biosynthesis, homospermidine synthase (HSS) catalyzes the first specific step. HSS was recruited during angiosperm evolution from deoxyhypusine synthase (DHS), an enzyme involved in the posttranslational activation of eukaryotic initiation factor 5A. Phylogenetic analysis of 23 cDNA sequences coding for HSS and DHS of various angiosperm species revealed at least four independent recruitments of HSS from DHS: one within the Boraginaceae, one within the monocots, and two within the Asteraceae family. Furthermore, sequence analyses indicated elevated substitution rates within HSS-coding sequences after each gene duplication, with an increased level of nonsynonymous mutations. However, the contradiction between the polyphyletic origin of the first enzyme in PA biosynthesis and the structural identity of the final biosynthetic PA products needs clarification.
INTRODUCTION
Because of their sessile way of life, plants have developed a rich arsenal of chemicals, encompassing some 200,000 known compounds. These metabolites participate in all kinds of biotic and abiotic interactions with the environment, predominantly with regard to defense against herbivores and pathogens (Harborne, 1993), and are considered secondary metabolites because they are not involved in the primary processes of growth and development. One of the most conspicuous features of secondary metabolites is that they are often restricted to individual species or related groups of species, rather than being broadly distributed in the plant kingdom.
Numerous genes are required to catalyze the formation of this chemical arsenal and to control its storage and deployment. Based on the sequence of chromosome 4 of Arabidopsis thaliana, ∼10% of the predicted genes have been assigned to secondary metabolism and a further 14% to specific roles in plant disease resistance and other defensive functions (Bevan et al., 1998). The genes of secondary metabolism are also not usually generally distributed but are often restricted to the species synthesizing a particular class of secondary metabolites. They are believed to be recruited by gene duplication (Pichersky and Gang, 2000) and, as a consequence, are often found in gene families. The evolutionary signature of their gradual adjustment to novel functions by natural selection can be frequently found in these gene families (Ohta, 1991). Less frequently, one finds evidence indicating the integration of a gene copy into a completely different functional environment in which the gene is exposed to new selection pressures. One example of the latter is the recruitment of homospermidine synthase, the first specific enzyme of pyrrolizidine alkaloid biosynthesis.
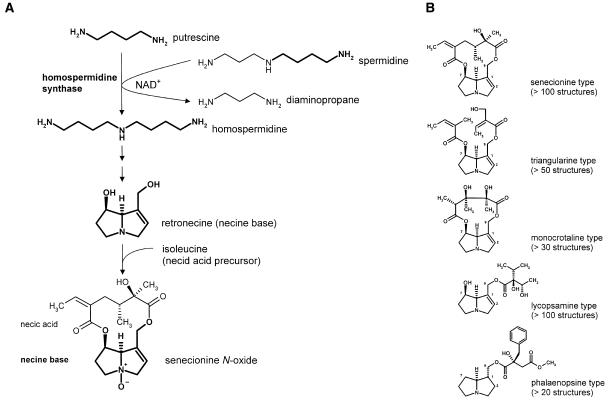
Pyrrolizidine alkaloids (PAs) are typical secondary compounds that are constitutively produced by the plant as a defense against herbivores (Hartmann, 1999; Hartmann and Ober, 2000; Ober, 2003). Their occurrence is restricted to the angiosperms in which they are found scattered in a few unrelated families. Approximately 95% of the more than 400 known structures are found within the tribes Senecioneae and Eupatorieae (Asteraceae), in several genera of the Boraginaceae, in the genus Crotalaria (Fabaceae), and in some genera of the Orchidaceae. Isolated occurrences have been described in single species of some additional families, such as the Apocynaceae, Celastraceae, Convolvulaceae, and Ranunculaceae (Hartmann and Witte, 1995). The characteristic skeleton of a PA molecule consists of a necine base moiety, which is esterified with necic acids. PAs can be classified into five structural types (Figure 1B). Homospermidine synthase (HSS) catalyzes the first specific step in necine base biosynthesis using the primary metabolites spermidine and putrescine as substrates (Figure 1A). The product, homospermidine, is exclusively incorporated into the necine base (Böttcher et al., 1993). Recently, we have been able to show that HSS was recruited from the ubiquitous enzyme deoxyhypusine synthase (DHS) (Ober and Hartmann, 1999a), which catalyzes the first of two reactions required for the posttranslational activation of the eukaryotic initiation factor 5A (eIF5A) (Park et al., 1997). Despite their completely different reaction products, the two enzymes share a common reaction mechanism. Both enzymes transfer the aminobutyl moiety of spermidine in a NAD+-dependent reaction to a second substrate (Ober et al., 2003a). In the case of DHS, two substrates may function as acceptors, namely, a specific Lys residue of its protein substrate eIF5A and the diamine putrescine, resulting in protein-bound deoxyhypusine and the triamine homospermidine, respectively (Ober and Hartmann, 1999b; Ober et al., 2003a). However, the latter activity appears to be silent under in vivo conditions. HSS shows identical kinetics with only putrescine as an acceptor but is unable to bind the protein substrate eIF5A (Ober et al., 2003a). Thus, during gene recruitment, the product of the duplicated dhs gene lost its protein-modifying activity and became exclusively an HSS (Figure 2). The ability to produce homospermidine allowed the biosynthesis of the carbon skeleton of PAs, whose abundance and further modification were presumably selected under herbivore pressure.
Figure 1.
Biosynthesis of PAs Exemplified with Senecionine N-Oxide and the Five Major Structural Types of PAs.
(A) HSS catalyzes the first specific step of the biosynthesis of the necine base moiety common to all PAs. Exemplified with senecionine N-oxide.
(B) Structural types of PAs according to Hartmann and Witte (1995).
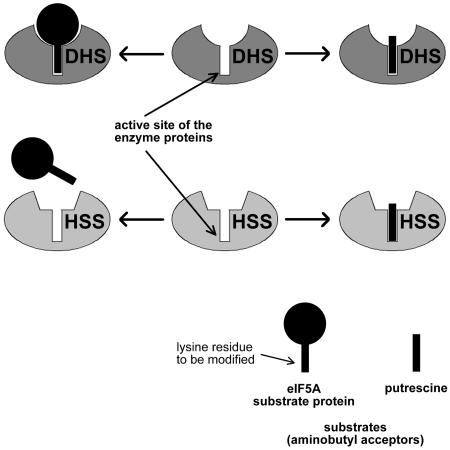
Figure 2.
Substrate Binding Properties of DHS and HSS.
For clarity, the figure shows only one of the four active sites present in the enzyme tetramer and only the potential acceptors of the aminobutyl moiety of spermidine. DHS binds the eIF5A substrate protein over an extensive area of the enzyme surface to ensure the proper orientation of the specific Lys residue in the active site, thereby allowing its aminobutylation. Putrescine, which mimics the Lys residue, is also accepted within the active site as a substrate. HSS, however, is unable to bind the eIF5A substrate protein because of a modified enzyme surface, whereas the binding of putrescine deep within the active site is not affected.
The isolated occurrence of PA-producing plants among unrelated angiosperm families provokes an interesting question about the origin of HSS. Was it recruited only once very early in angiosperm evolution, followed by independent losses in several lineages, or is the ability to produce PAs of polyphyletic origin? In the latter case, HSS would have had to be recruited several times independently, in each case followed by the establishment of the whole pathway leading to PAs of virtually identical structure. To answer this question, we have identified cDNAs coding for HSS and DHS in various PA-containing species from different angiosperm families and, after functional identification, have used them for phylogenetic analysis. Our results establish that HSS was recruited from DHS at least four times independently.
RESULTS
Identification and Characterization of cDNAs Coding for HSS and DHS
We identified 10 cDNAs encoding HSS and nine cDNAs encoding DHS from 12 species (Table 1). The sequence of a second HSS of Senecio vernalis (HSS2) was found incidentally using the full-length primers designed for the amplification of the cDNA coding for the previously published HSS of S. vernalis (HSS1; Ober and Hartmann, 1999a). To determine the enzymatic function of the proteins encoded by these cDNAs, the cDNAs were expressed in Escherichia coli (see Supplemental Figure 1 online), after which desalted crude extracts were used to assay the activities of HSS and DHS (Table 1). E. coli cultures containing the expression vectors pET3a or pET3a-mod showed neither HSS nor DHS activity (data not shown). All proteins with activity only in the HSS assay were designated as HSS, whereas proteins with HSS and DHS activity were designated as DHS. All DHS proteins showed unequivocal activity with the eIF5A substrate protein of S. vernalis (Ober and Hartmann, 1999a). All activities were in the same order of magnitude as those of the HSS and DHS of S. vernalis that were previously characterized in detail (Ober and Hartmann, 1999a, 1999b; Ober et al., 2003a). Differences in specific activity are mainly attributable to different concentrations of the tested enzymes in the desalted crude E. coli extracts. Supplemental Figure 1 online shows their expression level in the cultures used for the activity tests. In the case of Phalaenopsis, we were unable to identify a sequence coding for DHS. We identified, cloned, and expressed the eIF5A of Phalaenopsis to establish that the identified sequence indeed coded for an HSS and not for a DHS that was unable to bind the eIF5A substrate protein of S. vernalis. This orchid eIF5A substrate protein was well accepted by DHS of S. vernalis but not by the protein of Phalaenopsis (N. Nurhayati and D. Ober, unpublished results), which we consequently identified as HSS.
Table 1.
Specific Activities of DHS and HSS Encoded by cDNA Sequences Identified in Various Angiosperm Species
| cDNA | Size of ORF (bp) | DHS Activity (pkat/mg) | HSS Activity (pkat/mg) |
|---|---|---|---|
| HSS Heliotropium indicum | 1095 | 0.000 | 0.666 |
| DHS H. indicum | 1116 | 0.155 | 0.238 |
| HSS Cynoglossum officinale | 1110 | 0.000 | 0.902 |
| DHS C. officinale | 1119 | 0.036 | 0.113 |
| HSS Symphytum officinale | 1128 | 0.000 | 0.267 |
| DHS S. officinale | 1119 | 0.165 | 0.479 |
| HSS Eupatorium cannabinum | 1128 | 0.000 | 0.597 |
| DHS E. cannabinum | 1125 | 0.022 | 0.139 |
| HSS Senecio jacobaea | 1119 | 0.000 | 0.328 |
| HSS1 Senecio vernalis | 1113a | 0.000 | 1.124 |
| HSS2 S. vernalis | 1113 | 0.000 | 1.183 |
| DHS S. vernalis | 1116b | 0.147 | 0.280 |
| HSS Senecio vulgaris | 1113c | 0.000 | 0.293 |
| HSS Petasites hybridus | 1110 | 0.000 | 0.652 |
| DHS P. hybridus | 1098 | 0.018 | 0.048 |
| DHS Ipomoea hederifolia | 1149 | 0.147 | 0.631 |
| DHS Nicotianum tabacum | 1140d | 0.089 | 0.406 |
| DHS Crotolaria retusa | 1116 | 0.016 | 0.030 |
| HSS Phalaenopsis sp | 1116 | 0.000 | 0.504 |
The size of the ORF expressed heterologously in E. coli is given for each sequence, as are the specific activities according to the DHS and HSS assays of desalted crude extracts of E. coli expressing the respective cDNA. For SDS-PAGE analysis of the assayed E. coli cultures, see Supplemental Figure 1 online.
The identified sequences of HSS and DHS reveal a high degree of identity in comparison with the sequence similarities typically found among members of other gene families. Within the HSS/DHS family, even sequences as distantly related as the HSS of S. vernalis and the DHS of banana (Musa acuminate) show a sequence identity of 70% on the nucleic acid level and 71% on the amino acid level. Moreover, in comparison with human DHS, the DHS of S. vernalis shows a sequence identity of 66 and 63% on the amino acid and nucleic acid levels, respectively.
HSS Recruitment from DHS Occurred Several Times Independently
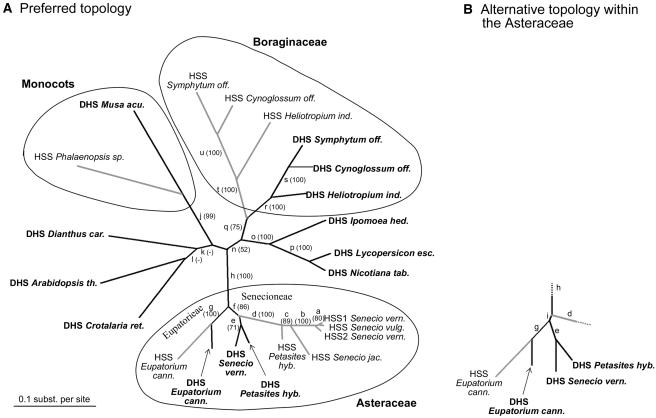
For phylogenetic analysis, we used the complete open reading frame (ORF) of all of the sequences of HSS and DHS listed in Table 1 and of the recently published sequences coding for DHS of tomato (Lycopersicon esculentum), Arabidopsis thaliana, banana (Musa acuminata), and carnation (Dianthus caryophyllus) (Wang et al., 2001). Figure 3A shows an unrooted maximum likelihood phylogram based on the nucleic acid sequences of these 23 cDNAs. The topology of the tree supports four independent origins of the HSS-coding genes from ancestors of the contemporary DHS genes by gene duplication. HSS was apparently recruited once early in the evolution of the Boraginaceae, once within the monocots, and twice within the Asteraceae. Only two tribes of the Asteraceae have species known to contain PAs, namely, the Senecioneae and the Eupatorieae. In each of the two lineages, we found an independent origin of HSS from DHS. Phylogenetic analyses of the Asteraceae confirmed that Senecioneae and Eupatorieae are not sister groups (Kim and Jansen, 1995; Jansen and Kim, 1996; Bayer and Starr, 1998). Phylograms obtained using different algorithms or based on amino acid sequences instead of nucleic acid sequences resulted in similar topologies that all supported the four independent origins of HSS (Table 2). The topologies differed mainly in the support of the branching pattern near the separation of the Eupatorieae and Senecioneae within the Asteraceae (Figure 3B). Whereas the preferred topology indicated a recruitment of HSS in the Senecioneae after the separation of the two lineages, the alternative topology suggests HSS recruitment before the separation. In this scenario, a common ancestor of the Eupatorieae and Senecioneae must have possessed an HSS. Because an independent recruitment of HSS within the Eupatorieae is well supported, one has to assume that this initial HSS was lost at the beginning of the Eupatorieae lineage. To avoid ambiguities that may have occurred because of the alignment used for the phylogenetic reconstruction (Brocchieri, 2001), we created a neighbor-joining tree based on a nucleic acid alignment generated with DIALIGN software. Although Clustal aligns sequences over their entire length, DIALIGN produces local multiple alignments by aligning whole segments rather than single residues (Morgenstern, 1999). The resulting phylogram confirmed the preferred topology (Figure 3A). The bootstrap proportions for all analyzed phylograms are given in Table 2.
Figure 3.
Unrooted Maximum Likelihood Tree Based on 23 cDNA Sequences Coding for HSS and DHS of Various Angiosperm Species.
(A) Preferred topology. HSS sequences are shown in gray, DHS sequences in black. Branch lengths were estimated by maximum likelihood based on the Jones-Taylor-Thornton model.
(B) Alternative topology that was found in some phylograms for the sequences within the Asteraceae. Bootstrap proportions to the branches are given in parentheses below the lower-case letters a to u, which refer to all calculated bootstrap proportions summarized in Table 2. Abbreviations are as follows: Symphytum off., Symphytum officinale; Cynoglossum off., Cynoglossum officinale; Heliotropium ind., Heliotropium indicum; Nicotiana tab., Nicotiana tabacum; Lycopersicon esc., Lycopersicon esculentum; Ipomoea hed., Ipomoea hederifolia; Senecio vern., Senecio vernalis; Senecio vulg., Senecio vulgaris; Senecio jac., Senecio jacobaea; Petasites hyb., Petasites hybridus; Eupatorium cann., Eupatorium cannabinum; Crotalaria ret., Crotalaria retusa; Arabidopsis th., Arabidopsis thaliana; Dianthus car., Dianthus caryophyllus; Musa acu., Musa acuminata.
Table 2.
Bootstrap Proportions of the Branches a to u in Figure 3
| Nucleic Acid Sequence
|
Amino Acid Sequence
|
|||||
|---|---|---|---|---|---|---|
| Branch | NJ | NJ-D | MP | ML | NJ | ML* |
| a | 99 | 100 | 91 | 80 | 54 | 60 |
| b | 100 | 100 | 100 | 100 | 100 | 100 |
| c | – | – | 94 | 89 | – | 77 |
| d | 100 | 100 | 100 | 100 | – | 100 |
| e | 100 | 100 | 67 | 71 | 100 | 91 |
| f | – | 52 | 53 | 86 | – | 71 |
| g | 100 | 100 | 100 | 100 | 56 | 98 |
| h | 100 | 100 | 100 | 100 | 83 | 99 |
| I | 56 | – | – | – | 75 | – |
| j | 100 | 93 | 100 | 99 | 65 | 95 |
| k | 68 | 76 | – | – | 89 | 79 |
| l | 52 | 69 | – | – | 83 | – |
| n | 73 | 60 | 75 | 52 | – | – |
| o | 100 | 100 | 100 | 100 | 92 | 97 |
| p | 100 | 100 | 100 | 100 | 100 | 100 |
| q | 89 | 90 | 93 | 75 | – | – |
| r | 100 | 100 | 100 | 100 | 100 | 100 |
| s | 100 | 100 | 100 | 100 | 94 | 81 |
| t | 100 | 100 | 100 | 100 | 100 | 100 |
| u | 100 | 100 | 100 | 100 | 100 | 100 |
The average bootstrap proportions resulting from 1000 replicates with the given method are shown. Average values <50 are indicated with a dash. For NJ-D, the calculation is based on an alignment generated with the DIALIGN software; all other calculations are based on ClustalX alignments. NJ, neighbor joining; NJ-D, neighbor joining based on DIALIGN alignment; MP, maximum parsimony; ML, maximum likelihood; ML*, log-likelihood (RELL) bootstrap proportions.
To analyze the phylogeny in more detail, we made pairwise comparisons of selected pairs of DHS- and HSS-coding cDNA sequences within the Boraginaceae family and the Asteraceae family (see Supplemental Table 1 online). The amino acid distance (daa) is defined as the number of amino acid substitutions per number of positions in the alignment corrected for multiple substitutions at the same site. The calculation of daa has revealed a relative low degree of sequence divergence between orthologous DHS sequences within the Boraginaceae and within the Asteraceae (daa between 5.9 and 13.5%). On the other hand, daa is noticeably increased when the paralogous DHS and HSS of the same species are compared (daa from 27.7 to 31.1% for Boraginaceae and from 17.1 to 23.5% for Asteraceae). A similar situation is found for the nucleotide distance (dna), which is defined as the number of nucleotide substitutions per length of the alignment corrected for multiple substitutions per site and for different substitution rates at different codon positions. Within the Boraginaceae, dna between orthologous DHS-coding sequences ranges from 10.5 to 21.1%, whereas between paralogous DHS- and HSS-coding sequences of the same species, dna ranges from 33.8 to 45.2%. For the Asteraceae, we calculated dna between orthologous DHS-coding sequences from 9.3 to 18.3% and between paralogous DHS- and HSS-coding sequences of the same species of 16.0 to 23.7%. Within the Boraginaceae, the distance between the functionally identical orthologous HSS sequences is not as high as that for paralogous HSS and DHS but is higher than that for orthologous DHS sequences (daa from 9.4 to 19.7%; dna from 16.0 to 35.9%), indicating reduced functional constraints on the HSS sequences, probably because of a reduced number of invariant sites within the amino acid sequence. Within the Asteraceae, the situation was the same when we compared only the HSS of the two species of the tribe Senecioneae (daa = 9.7%; dna = 11.6%). Instead, when we compared the two Senecioneae HSS with the HSS of Eupatorium cannabinum of the tribe Eupatorieae, these functionally identical orthologs were as different (daa = 24.2 and 22.7%; dna = 32.1 and 27.6%) as the paralogous DHS and HSS of the same species, supporting two independent origins of HSS within the tribes Eupatorieae and Senecioneae of the Asteraceae family.
Nonsynonymous Substitution Rates Seem to Be Elevated along HSS Lineages
To compare the synonymous and the nonsynonymous substitution rates, we analyzed the number of nonsynonymous (amino acid replacing) substitutions per nonsynonymous site (dN) and the number of synonymous (silent) substitutions per synonymous site (dS) to define the nonsynonymous/synonymous substitution rate ratio (ω = dN/dS) (Yang et al., 2000). The resulting data are summarized in Supplemental Table 1 online; ω is a measure of the selective pressure at the protein level because synonymous mutations are mostly invisible to natural selection, whereas nonsynonymous mutations may be under strong selective pressure (Yang and Nielsen, 2000). The higher the proportion of largely invariable amino acids within a protein attributable to strong functional constraints, the closer ω will approach to zero. Comparison of the estimates for ω between the paralogous DHS- and HSS-coding cDNAs within Cynoglossum and Heliotropium (0.118 and 0.147, respectively) and between the orthologous DHS-coding cDNAs (from 0.066 to 0.088) indeed suggests that DHS-coding cDNAs suffer a stronger selective pressure than the HSS-coding sequences. Only within Symphytum, whose paralogs are the most diverged sequence pair within the analyzed Boraginaceae cDNAs, is ω between these paralogs identical to the estimate for ω between the DHS of Symphytum and Cynoglossum (ω = 0.088). Within the Asteraceae, in which the analyzed sequences are less divergent than those of the Boraginaceae, this phenomenon becomes more obvious. The estimate for ω is 0.093 if the orthologous DHS-coding cDNAs of the Senecioneae (Senecio and Petasites) are compared, but a comparison of these DHS-coding sequences with their paralogous HSS-coding cDNA of the same species results in estimates for ω of 0.190 and 0.168, respectively. Moreover, a comparison of the orthologous DHS-coding sequences between Senecio and Eupatorium as members of the two analyzed tribes of the Asteraceae confirms the strong functional constraints on the DHS-coding sequences (ω = 0.111), whereas the estimate for ω is noticeably elevated when the paralogs of Eupatorium are compared (ω = 0.294).
DHS- and HSS-Coding Genes Were Not Equalized by Concerted Evolution
The genomic structures of selected DHS- and HSS-coding sequences of the Asteraceae and Boraginaceae and of tobacco (Nicotiana tabacum), Crotalaria, and Phalaenopsis were analyzed to identify the number, positions, and phases of potential introns. In comparison with the human dhs gene, which contains eight introns (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=DisplayandDB=gene), all analyzed dhs and hss genes and the dhs gene of Arabidopsis (http://www.arabidopsis.org: AT5G05920.1) possess six introns at identical positions and with the same phase. Only the genes coding for HSS1 and HSS2 of S. vernalis possess five introns missing the last intron (D. Ober, N. Nurhayati, and A. Reimann, unpublished results). We used the conserved intron–exon structure of the genomic sequences analyzed to test whether concerted evolution had produced a result different from our phylogeny. Nine potential exons were predicted for all cDNA sequences that were used for the phylogeny in Figure 3. In all these cDNAs, the sequences corresponding to the homologous exons were compared individually to construct neighbor-joining trees based on the two-parameter model of Kimura and maximum likelihood trees with global rearrangements, both with randomized input order. Although the branching pattern was not identical for all exons, no tree supported a common cluster of HSS-coding sequences (data not shown).
Catalytically Relevant Amino Acid Residues within the Active Site Are Identical between DHS and HSS
The binding of the eIF5A substrate protein to the surface of DHS requires precise interactions between the amino acid residues at the surfaces of the two interacting proteins to guarantee that the specific Lys residue of the eIF5A fits into the active center of DHS. Catalysis of homospermidine formation merely requires the binding of putrescine deep within the active site (Figure 2). Because the three-dimensional structure of human DHS (Liao et al., 1998) is closely related to other eukaryotic DHS, we know that DHS is a homotetramer comprising two tightly associated dimers with four active sites, two in each dimer interface. The cavity of the active site is formed by both contiguous subunits between which the NAD+ is sandwiched. Nine residues of one subunit and three residues of the other form hydrogen bonds to NAD+. Six residues (five of one and one of the other subunit) are involved in the binding and cleavage of spermidine and in the transfer of the aminobutyl moiety to the specific Lys residue of the eIF5A substrate protein. An alignment of human DHS with the 23 DHS- and HSS-coding cDNA sequences that we have used for the phylogenetic reconstruction shows that all these residues are identical in all plant sequences, with two exceptions: two amino acid residues in the DHS of Arabidopsis and one of these in the HSS of S. jacobaea differ from the consensus, both forming hydrogen bonds with the NAD+ molecule (data not shown).
DISCUSSION
We have identified, cloned, and functionally expressed 19 cDNAs coding for ubiquitous DHS and PA-specific HSS of various angiosperm species. Together with four functionally characterized DHS cDNAs available in sequence databases, we have used these 23 sequences for phylogenetic analysis. The branching patterns of trees generated by neighbor joining, maximum parsimony, and maximum likelihood methods support four independent recruitments of HSS from DHS during angiosperm evolution. Further such recruitments are possible because HSS sequences from other angiosperm families producing PAs have not as yet been studied.
The DHS-HSS System Is Conserved with Regard to Sequence, Gene Structure, and Biochemical Function
High sequence conservation related to strongly preserved biochemical function is a characteristic feature of the DHS system (Gordon et al., 1987; Bartig et al., 1992). DHS catalyzes the first step of the posttranslational activation of the eIF5A regulatory protein by modifying a specific protein-bound Lys residue (Park et al., 1997). The amino acid residues both within the active site and on the enzyme surface are essential to ensure the proper docking of the substrate protein with the enzyme. Thus, the proportion of amino acids within the DHS sequence with direct importance for enzymatic activity is high. These amino acids are largely invariant because of the strong purifying selection resulting in the highly conserved DHS-eIF5A system. Not only is the sequence within this system conserved but also the mode of action. Hence, the eIF5A substrate protein of a given eukaryotic species can be accepted as a substrate by the DHS proteins of many different eukaryotes. For example, the eIF5A substrate of the fungi Neurospora crassa is accepted by human DHS (Yan et al., 1996), and yeast DHS is active with the eIF5A substrate protein of human (Schwelberger et al., 1993; Magdolen et al., 1994; Kang et al., 1995), of CHO cells (Kang et al., 1995), and of Dictyostelium discoideum or Medicago sativa (Magdolen et al., 1994). Here, we have been able to show that, as expected, the eIF5A substrate protein of S. vernalis is modified by all DHS of plant origin so far analyzed. HSS, as the first pathway-specific enzyme of PA biosynthesis, shares a common ancestor with contemporary DHS, of which both originated by gene duplication (Ober and Hartmann, 1999a, 2000). Although both enzymes are involved in completely different metabolic processes of the plant's metabolism (i.e., protein modification in the case of DHS versus biosynthesis of plant defense compounds in the case of HSS), they share not only a high degree of sequence identity, but also many biochemical properties, such as pH dependency, molecular organization, substrate specificity, and kinetics, with one fundamental exception, namely, HSS is no longer able to bind the eIF5A protein substrate (Ober and Hartmann, 1999a; Ober et al., 2003a). The data that we present here confirm the high degree of sequence identity between DHS- and HSS-coding cDNAs.
There Is No Evidence for Concerted Evolution of HSS Sequences
The construction of a gene tree as a model for the evolutionary history of a gene or gene family of interest is problematic because in contrast with a species tree, the topology is based exclusively on the sequence data of the relevant gene. We have therefore thoroughly checked the topology that is shown in Figure 3. The topology showing clusters of both DHS and HSS sequences belonging to the monocots, the Boraginaceae, and the Asteraceae is most parsimoniously interpreted as supporting multiple origins of HSS. Alternatively, gene conversions and unequal crossing over could be responsible for the intrafamilial homogenization of paralogous sequences, a phenomenon termed concerted evolution. Concerted evolution has been hypothesized to obscure a hypothetical monophyletic origin of paralogous sequences (Zimmer et al., 1980). Homologous genes do not evolve independently but exchange sequence information with each other, thus maintaining a high degree of intrafamilial sequence similarity. Mechanisms of concerted evolution act mainly on tandem repeats of sequences belonging to multigene families that share a high degree of sequence identity. In typical cases the nucleotide differences do not exceed 1 to 3% (Li, 1997; Swanson and Vacquier, 1998), and only parts of the homologous genes are exchanged (Li, 1997).
Although nothing is known about the number of DHS- and HSS-coding genes or pseudogenes in PA-producing plants, several lines of evidence suggest that concerted evolution was not a major force in the evolution of DHS- and HSS-coding sequences. First, in all eukaryotic genomes completed so far, DHS is not encoded by a multigene family (see below); thus, we have no evidence for a tandem organization of the dhs and hss genes within PA-producing plants. Second, none of the tree topologies constructed independently of the nine potential exons separated DHS- and HSS-coding sequences into two distinct clusters. Third, the nucleotide differences among the paralogous genes from these species, of which the sequences are available, are considerable (>10%; see Supplemental Table 1 online). Fourth, the inability of HSS proteins to bind the eIF5A substrate protein shows that the dhs and hss genes diverged with respect to their biochemical properties, suggesting that they have diversified, rather than becoming homogenized. We therefore have no evidence for the hypothesis that the HSS-coding sequences share a common ancestor very early in angiosperm evolution, their antiquity being obscured by mechanisms of concerted evolution later in evolution.
The Single-Copy Gene dhs Was Duplicated Several Times
Gene duplications are relatively frequent events within genomes and have a high impact on the evolution of new biological functions (Wagner, 1998; Kondrashov et al., 2002). The most likely event after gene duplication is the production of pseudogenes from one of the gene copies by destructive mutations. However, in rare cases, one copy may acquire a completely new function as a result of beneficial mutations within its regulatory and structural components. According to a model proposed by Hughes (1994), the evolution of functionally distinct daughter genes should be preceded by a period in which the ancestral gene is bifunctional. This bifunctionality is realized by the DHS protein that originally possesses homospermidine synthesizing activity, which later will become the exclusive activity of the gene copy. The HSS-coding gene copy presumably lost the protein-modifying activity of DHS and escaped the strong selection pressure on this essential function of primary metabolism. Nevertheless, its remaining ability to synthesize homospermidine became the object of new selection pressure from herbivores, enabling some plants to recruit the gene copy to establish the first step in the biosynthesis of defense compounds.
There are several examples known in which genes have been independently recruited to a single function within a gene family. For instance, the resistance of insects to specific toxins seems to have been acquired through the independent recruitment of paralogous genes belonging to the cytochrome P450 superfamily (Scott and Wen, 2001; Wilson, 2001). Plant terpene synthases, such as limonene synthase, have been shown to be repeatedly recruited within their gene family from other terpene synthases (Bohlmann et al., 1998). For the repeated recruitment of HSS, the situation is different because DHS occurs as a single-copy gene in the eukaryotic genome projects finished to date. The larger a gene family within a genome, the larger the pool of genes from which new gene duplications can occur. Thus, it is surprising that DHS as a single-copy gene was the source of presumably several gene duplications, some of which were fixed independently in different plant lineages coding for the first pathway-specific enzyme in PA biosynthesis. Until now, no HSS has been identified from any plant unable to produce PAs. This may be attributable to the focus of our previous strategy of identifying primarily cDNAs of PA-producing plant species; however, the genome of Arabidopsis is also devoid of a HSS-coding gene. HSS might be found in other plant species, although the traces of homospermidine apparently occurring in all plants have been considered as a by-product of ubiquitous DHS (Ober et al., 2003b). This universal availability of homospermidine as the essential precursor of the necine base moiety of all PAs may have been a prerequisite for the repeated establishment of this pathway in angiosperm evolution. Elevated levels of homospermidine have been described for only a few plants, which is the reason for it being regarded as a rare polyamine (Kuehn et al., 1990; Hamana et al., 1992, 1994). Research is in progress to identify and characterize the enzymes responsible for the elevated homospermidine content in those plants.
All HSS Have Lost Their DHS Activity
It is remarkable that all the HSS that we have identified to date have lost the protein-modifying activity of DHS. There may be a metabolic need to shut off the DHS activity in HSS. Mechanistic and regulatory reasons may make the plant intolerant to two independent genes carrying the same essential function. As discussed by Hughes and Hughes (1993), a duplicated protein involved in interactions with other proteins may negatively interfere in these protein–protein interactions because of, for example, mutations altering its structure. If the protein–protein interaction is essential for the organism, as it is for DHS and eIF5A, a gene copy carrying the same function would be eliminated very rapidly. Probably only those gene copies that had lost their DHS activity had the chance to survive and to develop a novel function with regard to its residual HSS activity, but under a new selection pressure. This hypothesis is supported by the observation that, in all completed eukaryotic genome projects, DHS is a single-copy gene. Further support could be provided by the identification of those amino acid replacements that lie within the HSS protein and that are responsible for the loss of DHS activity. Positive Darwinian selection that is not detectable comparing the whole sequence may have acted on these residues (ω is well below 1 for all compared sequences; see Supplemental Table 1 online).
A comparison of the 23 cDNAs used for the phylogenetic analysis with the sequence for human DHS for which the three dimensional structure is available indicates that the loss of DHS activity during HSS recruitment is caused by the inability of the enzyme to bind the eIF5A substrate protein on the enzyme surface, but not by modifications within the active site. This observation is supported by comparative analyses of the kinetic properties of DHS and HSS of S. vernalis (Ober et al., 2003a). Both enzymes show the same unique substrate specificities and the same specific activities and exhibit almost identical Michaelis kinetics for the aminobutylation of putrescine. Furthermore, using a chromatography approach with immobilized eIF5A substrate protein, Ober et al. (2003a) have been able to establish the inability of HSS to bind this protein substrate.
The reduced number of invariant sites within HSS-coding cDNA in comparison with the paralogous DHS-coding cDNA should have two consequences: first, we expect a higher rate of sequence evolution within the HSS-coding lineages, as has been described as a consequence of other gene duplication events resulting in relaxed selective constraints on one of the gene copies (Li, 1985; Axelsen and Palmgren, 1998); second, we expect a higher rate of nonsynonymous mutations within the HSS-coding sequences. Longer branches for HSS sequences within the Boraginaceae and Asteraceae in Figure 3 indicate a higher rate of sequence evolution in comparison with their paralogous DHS sequences. Quantification of these differences by calculation of the amino acid distance (daa) and the nucleotide distance (dna) within these two families has shown longer distances between the paralogous DHS and HSS sequences than that for orthologous DHS. Moreover, the rate of nonsynonymous substitutions is increased in these lineages. Taken together, the increase of ω as a measure of the selective pressure on the protein level and the observation that modifications within the HSS-coding gene have resulted in the inability of HSS to bind the eIF5A substrate protein suggest decreased functional constraints on the HSS-coding gene.
Despite the Independent Origins of HSS, the Products of PA Biosynthesis Are Essentially Identical
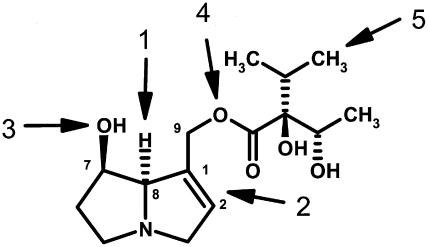
HSS is the first specific enzyme in the biosynthesis of the necine base moiety of PAs. Thus, at first glance, it appears that the recruitment of the hss gene had to be followed independently in each case by the recruitment of the rest of the pathway leading to the PAs. However, such an evolutionary scenario appears to be extremely unlikely. If the end products of the PA biosynthetic pathways in various families are compared, identical structures are often observed with a large number of specific features in common. One impressive example is the PAs of the lycopsamine type. Lycopsamine and its stereoisomers (Figure 4) are found independently in species of the Apocynaceae and in species of the Boraginaceae and Eupatorieae (Asteraceae) (Hartmann and Witte, 1995) in which we have demonstrated independent recruitments of HSS (Figure 3). The structure of lycopsamine possesses the necine base, retronecine, with a 1,2-double bond and two OH groups. One OH group is esterified with a unique branched-chain aliphatic C7-acid, which is, with the exception of two minor occurrences, only known as a structural element of PAs of the lycopsamine type. This acid is synthesized via a unique route related to Val biosynthesis (Weber et al., 1999). Two possibilities explain the co-occurrence of lycopsamine in these three nonrelated taxa: independent evolution (biochemical convergence) of all enzymes involved in PA biosynthesis downstream of HSS or monophyletic origin of this part of the biosynthetic pathway with homospermidine as a precursor, probably in a completely different metabolic context. To be able to decide between these possibilities, a more complete comparison of the entire biosynthetic pathway will be required at the genetic level.
Figure 4.
Structure of the PA Lycopsamine
PAs of this structural type are found in the unrelated families Boraginaceae, Apocynaceae, and Eupatorieae (Asteraceae). Common features of these PAs are (numbers refer to the arrows) retronecine with stereochemistry at carbon 8 of the necine base as given (1) and with a 1,2-double bond (2) and two OH groups, one at carbon 7 (3) and one esterified at carbon 9 (4) with a unique branched-chain aliphatic C7-acid (5).
A further unanswered question concerns the origin of the regulatory elements that allow the proper integration of these newly recruited enzymes into a single pathway. For HSS of S. vernalis (Asteraceae), we have been able to show that its expression has changed completely in comparison with DHS. Its localization in groups of specialized cells situated directly beside the phloem, namely, tissue involved in the root-to-shoot translocation of PAs, suggests a correlation of PA biosynthesis and transport, perhaps reflecting the way in which HSS expression has been adjusted to the specific needs of PA biosynthesis (Moll et al., 2002). Work is now in progress to elucidate further steps of PA biosynthesis at the molecular level to understand their evolutionary origin and role in regulating the function of PAs in plant defense.
METHODS
RNA Isolation and cDNA Synthesis
To extract total RNA from the plant tissues listed in Table 3, the RNeasy plant mini kit (Qiagen, Hilden, Germany) was used. Total RNA (2 μg) was used as template for reverse transcription as previously described (Ober and Hartmann, 1999b).
Table 3.
Identification and Cloning Strategy for HSS- and DHS-Coding cDNA Sequences of Various Angiosperm Species
| cDNA | Organ for Total RNA Preparation | Degenerate Primers | Primers for 3′-RACE and 5′-RACE | Full-Length Primers with Included Restriction Sites | Expression Vector with Used Restriction Sites |
|---|---|---|---|---|---|
| HSS H. indicum | Shoot | P9, P4 | P30, P31-33 | P80 (NdeI), P81 (BamHI) | pET3a (NdeI + BamHI) |
| DHS H. indicum | Root | P9, P4 | P34, P35-37 | P82 (XhoI), P83 (BamHI) | pET3a-mod (XhoI + BamHI) |
| HSS C. officinale | Root | I.P5, P3; II.P9, P8 | P49, P50+51 | P74 (NdeI), P75 (BamHI) | pET3a (NdeI + BamHI) |
| DHS C. officinale | Shoot | I.P5, P3; II.P9, P8 | P45, P46-48 | P76 (XhoI), P77 (BamHI) | pET3a-mod (XhoI + BamHI) |
| HSS S. officinale | Root | P9, P4 | P41, P42-44 | P72 (XhoI), P73 (BclI) | pET3a-mod (XhoI + BamHI) |
| DHS S. officinale | Shoot | I.P9, P3; II.P6, P8 | P78, P38-40 | P78 (XhoI), P79 (BamHI) | pET3a mod (XhoI+BamHI) |
| HSS E. cannabinum | Root | P1, P2 | P10, P11+12 | P64 (XhoI), P65 (BamHI) | pET3a-mod (XhoI + BamHI) |
| DHS E. cannabinum | Shoot | P7, P8 | P13, P11+14 | P66 (XhoI), P67 (BamHI) | pET3a-mod (XhoI + BamHI) |
| HSS S. jacobaea | Root | P1, P2 | P19, P20-22 | P70 (NdeI), P71 (BamHI) | pET3a (NdeI + BamHI) |
| HSS1 S. vernalis | Ober and Hartmann (1999a) | ||||
| HSS2 S. vernalis | Root | as for HSS1 of S. vernalis | pET3a (NdeI + BamHI) | ||
| DHS S. vernalis | Ober and Hartmann (1999a) | ||||
| HSS S. vulgaris | Ober et al. (2000) | ||||
| HSS P. hybridus | Root | P5, P4 | P52, P53-55 | P84 (NdeI), P85 (BamHI) | pET3a (NdeI + BamHI) |
| DHS P. hybridus | Shoot | P5, P4 | P56, P57-59 | P86 (XhoI), P87 (BamHI) | pET3a-mod (XhoI + BamHI) |
| DHS I. hederifolia | Shoot tip | P7, P4 | P15, P16-18 | P68 (NdeI), P69 (BamHI) | pET3a (NdeI + BamHI) |
| DHS N. tabacum | Ober and Hartmann (1999b) | ||||
| DHS C. retusa | Root | P7, P4 | P23, P24-26 | P60 (NdeI), P61 (BamHI) | pET3a (NdeI + BamHI) |
| HSS Phalaenopsis sp | Aerial root tip | P9, P8 | P27, P28+29 | P62 (NdeI), P63 (BamHI) | pET3a (NdeI + BamHI) |
Plant organs from which total RNA was extracted, the primers used for identification and cloning, the vectors for heterologuous protein expression in E. coli BL21(DE3), and the restriction enzymes used for cloning of the full-length cDNA into the expression vectors are listed. RACE, rapid amplification of cDNA ends.
Identification of cDNAs Coding for HSS and DHS
To identify cDNA sequences that code for HSS and DHS in the tissues of interest, a PCR approach with degenerate primers was used as described previously (Ober and Hartmann, 1999a, 1999b). The whole ORF of each cDNA was obtained by 3′-RACE and 5′-RACE techniques. Each sequence was analyzed to design gene-specific primers that allowed the amplification of the full-length cDNA with Pfu and Pfx DNA polymerase (Promega, Madison, WI and Invitrogen, Carlsbad, CA, respectively) for cloning in expression vectors pET3a (Novagen, Madison, WI) and pET3a-mod. pET3a-mod is identical to pET3a, except that the NdeI restriction site has been replaced by an XhoI restriction site. All primers are given in Table 4, as are the restriction sites used for cloning. The resulting constructs were sequenced and used for expression in Escherichia coli BL21(DE3) to identify unambiguously HSS and DHS from their substrate specificities in the enzyme assay. The expression and preparation of the crude cell extracts was performed as previously described for tobacco (Nicotiana tabacum) DHS (Ober and Hartmann, 1999b). Successful expression was confirmed by SDS-PAGE analysis of cell culture aliquots with Precision Protein Standard (Bio-Rad, Hercules, CA). HSS and DHS activity were assayed in the desalted crude extracts as previously described (Ober and Hartmann, 1999a) using the eIF5A precursor protein of Senecio vernalis as the aminobutyl acceptor in DHS assays (Ober and Hartmann, 1999a). Protein quantities were estimated according to Bradford (1976).
Table 4.
Sequences of Primers Used for the Identification and Cloning of the cDNAs Listed in Table 3
| No. | Primer Sequence | No. | Primer Sequence |
|---|---|---|---|
| P1 | 5′-GGNGGNRTNGARGARGAY-3′ | P52 | 5′-TTGCGATCTAAAGGATTAAATCGCATC-3′ |
| P2 | 5′-CCANSWNACNGCYTCRTC-3′ | P53 | 5′-ACAGCCTCATTATCCAT-3′ |
| P3 | 5′-GCYTCRTCNGGWCKNGMRCC-3′ | P54 | 5′-GACAACCAGACCAGGATTGGTAACT-3′ |
| P4 | 5′-CCCCANSWNACNGCYTCRTC-3′ | P55 | 5′-TGCCCAATATAGATATGAAGACTCGTT-3′ |
| P5 | 5′-AARATHTTYYTNGGNTWYAC-3′ | P56 | 5′-CTTGCGTTCAAAAGGACTAAATCGTATT-3′ |
| P6 | 5′-TNGTNCCANAYGAYAAYTAY-3′ | P57 | 5′-TGGAGCGTATAATATACT-3′ |
| P7 | 5′-AARTTYGARGAYTGGATHATHCC-3′ | P58 | 5′-GCAGTGTTAATGAAAACAGCGTAATCA-3′ |
| P8 | 5′-CCRTCRWAYTCYTGNGCNGTRTT-3′ | P59 | 5′-CTAAAATGATTATCCCGGTCTTCCTA-3′ |
| P9 | 5′-GARGARGAYYTNATHAARTGYYT-3′ | P60 | 5′-ATATATCATATGAGTGAAGAAGTAAAGGAAGC-3′ |
| P10 | 5′-GGGAAGGAGATTAATGACGAGAGTTCATA-3′ | P61 | 5′-ATGGATCCTATTGATGGCAAGGTTTTACTC-3′ |
| P11 | 5′-TATTTGCGTTGCATATGT-3′ | P62 | 5′-ATATATCATATGGGCTCGGCTGCGAGCGA-3′ |
| P12 | 5′-CTCATTGTCCATGGCCAGGACATCTT-3′ | P63 | 5′-TAAGGATCCTTAAGCTTTAATCCCATTGGCCTG-3′ |
| P13 | 5′-GCAAATATGATGCGCAATGGTGCAGATTAT-3′ | P64 | 5′-TATCTAGAATGGCGGCAGCAATTAAAGAAG-3′ |
| P14 | 5′-GCTGTTTCTTTTGTTCTTCCAACAGTTGAT-3′ | P65 | 5′-TAAGGATCCTTATGCGGAACATAGTTTCTTCT-3′ |
| P15 | 5′-GGAAAGAAATTAATGATGAAAGCTCGTATTTA-3′ | P66 | 5′-ATCTAGACTCGAGATGGGGGAACCCACTAAAGAAG-3′ |
| P16 | 5′-ATATTGGCATTGCAAATAT-3′ | P67 | 5′-TAAGGATCCCTATCCAGAACTTGGTTTTGAATC-3′ |
| P17 | 5′-GATTATTTGGATCTTCCTTTCTGAAAGAAT-3′ | P68 | 5′-ATATACATATGGGAGAAGATACCAGAGATC-3′ |
| P18 | 5′-AAACTAGGACAGAAGACAGGAATTTTGTT-3′ | P69 | 5′-ATGGATCCTATTGATTGCCTGTGGAATTTTT-3′ |
| P19 | 5′-TGGTGCAGATTACGCTGTTTTTATCAACA-3′ | P70 | 5′-ATATATCATATGGGAGAGACCAACAAATCAGC-3′ |
| P20 | 5′-CAACATTTGGTCAAAGAT-3′ | P71 | 5′-TAAGGATCCTTAAAACCCATTGAGTTTAGATGCTT-3′ |
| P21 | 5′-CGATTTAACCCTCTCGATCGCAAGTTA-3′ | P72 | 5′-TACTCGAGATGGGGGAAGTAGCCGCTGCT-3′ |
| P22 | 5′-TACCTCTAAATGTGCTTCCTAGACATTT-3′ | P73 | 5′-TATGATCATCACTTAGATAAATTATTTGCCTGTTT-3′ |
| P23 | 5′-CTGCACAAGAATTTGATGGGAGTGATTCT-3′ | P74 | 5′-ATATACATATGGGAGAAGTAGCAACCAGCAACAA-3′ |
| P24 | 5′-AATCGTGCAACTAACTTA-3′ | P75 | 5′-ATTGGATCCTCACTTAGATTTTTTATCTGCAATCTTC-3′ |
| P25 | 5′-GTCCATAAGACATTCTCTTCATTTTGTT-3′ | P76 | 5′-TACTCGAGATGGGGGAAGCCTTGAAAGAGCAA-3′ |
| P26 | 5′-CGAAGATAGGGATTATCCAATCTT-3′ | P77 | 5′-ATTGGATCCTCACAACTTGTTATCTGCAGATTTACT-3′ |
| P27 | 5′-CAATGGACGAGGAAGCTGTTCATGCAA-3′ | P78 | 5′-ATATACATATGGACGCCGTACGGGCTACAGTA-3′ |
| P28 | 5′-TCTCATTGTTTATCTCTTT-3′ | P79 | 5′-ATTGGATCCTCACAACTTGTTTTCTGCAGATTTAGT-3′ |
| P29 | 5′-GGCGTCCAGTGCGCTTTCTCATTAA-3′ | P80 | 5′-ATATACATATGGGAGAGGTAGCCACAAATGCT-3′ |
| P30 | 5′-ACATGCTCTACTTGCATGCTAACAACAATA-3′ | P81 | 5′-ATTGGATCCTTAACTTGATTTCTTCACCTTTGCAAA-3′ |
| P31 | 5′-CATTGCATATATGATGTTT-3′ | P82 | 5′-TACTCGAGATGGGAGATGTGAGTGAAGAGCAG-3′ |
| P32 | 5′-CCCCAAGAATTATTATTCCCGTCTTTCTT-3′ | P83 | 5′-ATTGGATCCCTAGCCAGATTTATTTACTTTTGTGG-3′ |
| P33 | 5′-TCATTGTCCATGGCC CTCACATCTTCA-3′ | P84 | 5′-ATATACATATGGGGTCTAACAAACAAGCAGAGA-3′ |
| P34 | 5′-CATGTTATACATACATGCTATCCGCAAAGA-3′ | P85 | 5′-ATTGGATCCTTAAAAGCCATTGACTTTAGATGCTTT-3′ |
| P35 | 5′-CCTGTGCTGTGTTAATA-3′ | P86 | 5′-TACTCGAGATGGGGGAATCTTTGAAACAAGCG-3′ |
| P36 | 5′-TTGGGCAACCCTCCCCCAAGAACAATA-3′ | P87 | 5′-ATTGGATCCTTATTTCTCTCGCTTTGCAGCAAATG-3′ |
| P37 | 5′-GTACAGCCTCACTGTTCATGGCCCTAAT-3′ | P88 | 5′-ATTGAGCAGCAGCAGCAGGCACAACAA-3′ |
| P38 | 5′-TTCATGGACCTAATATCT-3′ | P89 | 5′-TAAGGATCCTTAAAAACCATTGACTTTAGATGG-3′ |
| P39 | 5′-TAGACCAGGATCTTTGCGTATGGAATGTAT-3′ | P90 | 5′-CACTTAATAGAGAAATGGCCGAGTCAAA-3′ |
| P40 | 5′-AACCATCGGTCAAACCAGGGCAGTAGAT-3′ | P91 | 5′-ACAAGCCTCGGTGATGGAGAACTTGAGG-3′ |
| P41 | 5′-GATGTTAGAAGAACAAATAAACGAGAAAAAACT-3′ | P92 | 5′-TAAGGATCCTCAGGAACTCGGCTCAGC-3′ |
| P42 | 5′-CATCTTGTCCAAAATGTT-3′ | P93 | 5′-AGGTGTAAATTATGCGGAGCTTCTCAAAT-3′ |
| P43 | 5′-CCAGTCCTCAAACTTACAGTAATTA-3′ | P94 | 5′-TGACTTTGGCTGAGCCGTCAAGGTTTT-3′ |
| P44 | 5′-AGCATAGCTCCAGGTAAGGCAAAAT-3′ | P95 | 5′-GGCCCTCAACGTAATGGAGTCAGTAAGAT-3′ |
| P45 | 5′-AAATGATGGAGGAACAAAACACGAAGAAAATT-3′ | P96 | 5′-CGGAAAATCTAGAAGGTTCTGCCACAAAA-3′ |
| P46 | 5′-ACTGTTCATTGACCTAAT-3′ | P97 | 5′-TTGTTAATCAAATGGTCAGTTTTCACTTCTT-3′ |
| P47 | 5′-CAACTAGACCAGGGTCTTTGCGTATAGAA-3′ | P98 | 5′-TGAATGGCGTCACCAAGATTAGCAGCTT-3′ |
| P48 | 5′-CGTCAAACCAGGGCAGTAGACAGGTATA-3′ | P99 | 5′-TCTACATTAACACTGCACAAGAGTTTGAT-3′ |
| P49 | 5′-TTTGGACAAGATGTTAGAAGAGCAAATATCT-3′ | P100 | 5′-CAAAATCTGCACCATTGCGCATCATATT-3′ |
| P50 | 5′-CCTCCTCTCAGGATTA-3′ | P101 | 5′-GAATATGGTGCGCTGTTTCTTGTTGAAAT-3′ |
| P51 | 5′-CATTGACTGACTCATCGTCCATAGCACATA-3′ |
For degenerate primers P1 to P9, the following code is used: H = A + T + C, K = G + T, M = A + C, N = A + T + C + G, R = A + G, S = C + G, W = A + T, and Y = T + C.
Sequence Analyses
The MEGA program version 2.1 (Kumar et al., 2001) was used to calculate the following parameters in pairwise alignments of DHS- and HSS-coding cDNA sequences: dna as the number of nucleotide substitutions per number of positions in the alignment corrected according to the Jukes-Cantor model (Jukes and Cantor, 1969) and the γ distribution (Golding, 1983) for multiple substitutions per site and for different substitution rates at different codon positions, respectively; dN as the number of nonsynonymous substitutions per nonsynonymous site; dS as the number of synonymous substitutions per synonymous site with the Jukes-Cantor correction for multiple substitutions per site (Jukes and Cantor, 1969). In pairwise amino acid alignments of HSS and DHS sequences, daa was estimated as the number of amino acid substitutions per number of positions in the alignment using the Poisson correction in the MEGA program.
Phylogenetic Analysis
Twenty-three full-length cDNAs coding for HSS and DHS of different angiosperm species were aligned based on their nucleic acid sequence and their deduced amino acid sequence using ClustalX (Thompson et al., 1997) and, based on their nucleic acid sequence, using DIALIGN 2 (Morgenstern, 1999). The resulting alignments were improved by visual inspection and used to estimate phylogenies with the following software of the PHYLIP program package (Felsenstein, 2001): DNADIST with the two-parameter model of Kimura (Kimura, 1980) and PROTDIST, both in combination with NEIGHBOR as a neighbor-joining method (Saitou and Nei, 1987), DNAPARS for maximum parsimony analyses, and DNAML and PROML as maximum likelihood methods (Felsenstein, 1981). For PROTDIST and PROML, the Jones-Taylor-Thornton model (Jones et al., 1992) was applied. For all algorithms, the input order of the sequences was randomized. Bootstrap values were estimated with the programs SEQBOOT and CONSENSE of the PHYLIP package, but for the maximum likelihood analysis of the amino acid alignment, RELL bootstrap proportions were calculated with PROTML of the MOLPHY package (Adachi and Hasegawa, 1996). All bootstrap estimates are the result of 1000 replicates.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under the following accession numbers. Sequences taken from GenBank include the following: human DHS cDNA (L39068); Arabidopsis thaliana DHS (http://www.arabidopsis.org; AT5G05920.1) cDNA (NM_120674); Dianthus caryophyllus DHS cDNA (AF296080); Lycopersicon esculentum DHS cDNA (AF296077); Musa acuminata DHS cDNA (AF296079); Oryza sativa DHS genomic DNA (AC117988). Sequences analyzed in our lab include the following: Crotalaria retusa DHS cDNA (AJ704838); Cynoglossum officinale DHS cDNA (AJ704839); C. officinale HSS cDNA (AJ704840); Eupatorium cannabinum DHS cDNA (AJ704841); E. cannabinum HSS cDNA (AJ704842); Heliotropium indicum DHS cDNA (AJ704844); H. indicum HSS cDNA (AJ704843); Ipomoea hederifolia DHS cDNA (AJ704845); Nicotiana tabacum DHS cDNA (AJ242017); Petasites hybridus DHS cDNA (AJ704846); P. hybridus HSS cDNA (AJ704847); Phalaenopsis sp HSS cDNA (AJ704848); Senecio vernalis DHS cDNA (AJ238622); S. vernalis HSS1 cDNA (AJ238623); S. vernalis HSS2 cDNA (AJ704849); Senecio vulgaris HSS cDNA (AJ251500); Senecio jacobaea HSS cDNA (AJ704850); Symphytum officinale DHS cDNA (AJ704852); S. officinale HSS cDNA (AJ704851).
Supplementary Material
Acknowledgments
We thank T. Hartmann for relevant support of this work and W. Martin and C. Wheat for helpful discussions. We thank three anonymous reviewers and E. Pichersky and J. Gershenzon for critical and helpful comments on the manuscript. The work was supported by the Deutsche Forschungsgemeinschaft.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Dietrich Ober (d.ober@tu-bs.de).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.023176.
References
- Adachi, J., and Hasegawa, M. (1996). MolPhy Version 2.3: Programs for Molecular Phylogenetics Based on Maximum Likelihood. (Tokyo: Institute of Statistical Mathematics).
- Axelsen, K.B., and Palmgren, M.G. (1998). Evolution of substrate specificities in the P-type ATPase superfamily. J. Mol. Evol. 46, 84–101. [DOI] [PubMed] [Google Scholar]
- Bartig, D., Lemkemeier, K., Frank, J., Lottspeich, F., and Klink, F. (1992). The archaebacterial hypusine-containing protein. Structural features suggest common ancestry with eukaryotic translation initiation factor 5A. Eur. J. Biochem. 204, 751–758. [DOI] [PubMed] [Google Scholar]
- Bayer, R.J., and Starr, J.R. (1998). Tribal phylogeny of the Asteraceae based on two noncoding chloroplast sequences, the trnL intron and trnL/trnF intergenic spacer. Ann. Mo. Bot. Gard. 85, 242–256. [Google Scholar]
- Bevan, M., et al. (1998). Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature 391, 485–488. [DOI] [PubMed] [Google Scholar]
- Bohlmann, J., Meyer-Gauen, G., and Croteau, R. (1998). Plant terpenoid synthases: Molecular biology and phylogenetic analysis. Proc. Natl. Acad. Sci. USA 95, 4126–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher, F., Adolph, R.D., and Hartmann, T. (1993). Homospermidine synthase, the first pathway-specific enzyme in pyrrolizidine alkaloid biosynthesis. Phytochemistry 32, 679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Brocchieri, L. (2001). Phylogenetic inferences from molecular sequences: Review and critique. Theor. Popul. Biol. 59, 27–40. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. (1981). Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 17, 368–376. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. (2001). PHYLIP, Phylogeny Inference Package, Version 3.6 (alpha2). (Seattle: University of Washington).
- Golding, G.B. (1983). Estimates of DNA and protein sequence divergence: An examination of some assumptions. Mol. Biol. Evol. 1, 125–142. [DOI] [PubMed] [Google Scholar]
- Gordon, E.D., Mora, R., Meredith, S.C., Lee, C., and Lindquist, S.L. (1987). Eukaryotic initiation factor 4D, the hypusine-containing protein, is conserved among eukaryotes. J. Biol. Chem. 262, 16585–16589. [PubMed] [Google Scholar]
- Hamana, K., Matsuzaki, S., Niitsu, M., and Samejima, K. (1992). Distribution of unusual polyamines in leguminous seeds. Can. J. Bot. 70, 1984–1990. [Google Scholar]
- Hamana, K., Matsuzaki, S., Niitsu, M., and Samejima, K. (1994). Distribution of unusual polyamines in aquatic plants and gramineous seeds. Can. J. Bot. 72, 1114–1120. [Google Scholar]
- Harborne, J.B. (1993). Introduction to Ecological Biochemistry. (San Diego: Academic Press).
- Hartmann, T. (1999). Chemical ecology of pyrrolizidine alkaloids. Planta 207, 483–495. [Google Scholar]
- Hartmann, T., and Ober, D. (2000). Biosynthesis and metabolism of pyrrolizidine alkaloids in plants and specialized insect herbivores. In Topics in Current Chemistry, F.J. Leeper and J.C. Vederas, eds (Berlin: Springer-Verlag), pp. 207–244.
- Hartmann, T., and Witte, L. (1995). Pyrrolizidine alkaloids: Chemical, biological and chemoecological aspects. In Alkaloids: Chemical and Biological Perspectives, S.W. Pelletier, ed (Oxford: Pergamon Press), pp. 155–233.
- Hughes, A.L. (1994). The evolution of functionally novel proteins after gene duplication. Proc. R. Soc. Lond. B. Biol. Sci. 256, 119–124. [DOI] [PubMed] [Google Scholar]
- Hughes, M.K., and Hughes, A.L. (1993). Evolution of duplicate genes in a tetraploid animal, Xenopus laevis. Mol. Biol. Evol. 10, 1360–1369. [DOI] [PubMed] [Google Scholar]
- Jansen, R.K., and Kim, K.-J. (1996). Implications of chloroplast DNA data for the classification and phylogeny of the asteraceae. In Compositae: Systematics. Proceedings of the International Compositae Conference, Kew 1994, D.J.N. Hind and H. Beentje, eds (Kew: Royal Botanic Gardens), pp. 317–339.
- Jones, D.T., Taylor, W.R., and Thornton, J.M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282. [DOI] [PubMed] [Google Scholar]
- Jukes, T.H., and Cantor, C.R. (1969). Evolution of protein molecules. In Mammalian Protein Metabolism, H.N. Munro, ed (New York: Academic Press), pp. 21–132.
- Kang, K.R., Wolff, E.C., Park, M.H., Folk, J.E., and Chung, S.I. (1995). Identification of YHR068w in Saccharomyces cerevisiae chromosome VIII as a gene for deoxyhypusine synthase: Expression and characterization of the enzyme. J. Biol. Chem. 270, 18408–18412. [DOI] [PubMed] [Google Scholar]
- Kim, K.J., and Jansen, R.K. (1995). ndhF sequence evolution and the major clades in the sunflower family. Proc. Natl. Acad. Sci. USA 92, 10379–10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. [DOI] [PubMed] [Google Scholar]
- Kondrashov, F.A., Rogozin, I.B., Wolf, Y.I., and Koonin, E.V. (2002). Selection in the evolution of gene duplications. Genome Biol. 3, research0008.0001–0008.0009. [DOI] [PMC free article] [PubMed]
- Kuehn, G.D., Rodriguez-Garay, B., Bagga, S., and Phillips, G.C. (1990). Novel occurrence of uncommon polyamines in higher plants. Plant Physiol. 94, 855–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., Tamura, K., Jakobsen, I.B., and Nei, M. (2001). MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics 17, 1244–1245. [DOI] [PubMed] [Google Scholar]
- Li, W.-H. (1985). Accelerated evolution following gene duplication and its implication for the neutralist-selectionist controversy. In Population Genetics and Molecular Evolution, T. Ohta and K. Aoki, eds (Berlin: Springer-Verlag), pp. 333–352.
- Li, W.-H. (1997). Molecular Evolution. (Sunderland, MA: Sinauer Associates).
- Liao, D.I., Wolff, E.C., Park, M.H., and Davies, D.R. (1998). Crystal structure of the NAD complex of human deoxyhypusine synthase: An enzyme with a ball-and-chain mechanism from blocking the active site. Structure 6, 23–32. [DOI] [PubMed] [Google Scholar]
- Magdolen, V., Klier, H., Woehl, T., Klink, F., Hirt, H., Hauber, J., and Lottspeich, F. (1994). The function of the hypusine-containing proteins of yeast and other eukaryotes is well conserved. Mol. Gen. Genet. 244, 646–652. [DOI] [PubMed] [Google Scholar]
- Moll, S., Anke, S., Kahmann, U., Hänsch, R., Hartmann, T., and Ober, D. (2002). Cell-specific expression of homospermidine synthase, the entry enzyme of the pyrrolizidine alkaloid pathway in Senecio vernalis, in comparison with its ancestor, deoxyhypusine synthase. Plant Physiol. 130, 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern, B. (1999). DIALIGN 2: Improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics 15, 211–218. [DOI] [PubMed] [Google Scholar]
- Ober, D. (2003). Chemical ecology of alkaloids exemplified with the pyrrolizidines. In Integrative Phytochemistry: From Ethnobotany to Molecular Ecology, J.T. Romeo, ed (Amsterdam: Pergamon), pp. 203–230.
- Ober, D., Gibas, L., Witte, L., and Hartmann, T. (2003. b). Evidence for general occurrence of homospermidine in plants and its supposed origin as by-product of deoxyhypusine synthase. Phytochemistry 62, 339–344. [DOI] [PubMed] [Google Scholar]
- Ober, D., Harms, R., and Hartmann, T. (2000). Cloning and expression of homospermidine synthase from Senecio vulgaris: A revision. Phytochemistry 55, 305–309. [DOI] [PubMed] [Google Scholar]
- Ober, D., Harms, R., Witte, L., and Hartmann, T. (2003. a). Molecular evolution by change of function: Alkaloid-specific homospermidine synthase retained all properties of deoxyhypusine synthase except binding the eIF5A precursor protein. J. Biol. Chem. 278, 12805–12812. [DOI] [PubMed] [Google Scholar]
- Ober, D., and Hartmann, T. (1999. a). Homospermidine synthase, the first pathway-specific enzyme of pyrrolizidine alkaloid biosynthesis, evolved from deoxyhypusine synthase. Proc. Natl. Acad. Sci. USA 96, 14777–14782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober, D., and Hartmann, T. (1999. b). Deoxyhypusine synthase from tobacco: cDNA isolation, characterization, and bacterial expression of an enzyme with extended substrate specificity. J. Biol. Chem. 274, 32040–32047. [DOI] [PubMed] [Google Scholar]
- Ober, D., and Hartmann, T. (2000). Phylogenetic origin of a secondary pathway: The case of pyrrolizidine alkaloids. Plant Mol. Biol. 44, 445–450. [DOI] [PubMed] [Google Scholar]
- Ohta, T. (1991). Multigene families and the evolution of complexity. J. Mol. Evol. 33, 34–41. [DOI] [PubMed] [Google Scholar]
- Park, M.H., Lee, Y.B., and Joe, Y.A. (1997). Hypusine is essential for eukaryotic cell proliferation. Biol. Signals 6, 115–123. [DOI] [PubMed] [Google Scholar]
- Pichersky, E., and Gang, D.R. (2000). Genetics and biochemistry of secondary metabolites in plants: An evolutionary perspective. Trends Plant Sci. 5, 439–445. [DOI] [PubMed] [Google Scholar]
- Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Schwelberger, H.G., Kang, H.A., and Hershey, J.W.B. (1993). Translation initiation factor eIF-5A expressed from either of two yeast genes or from human cDNA: Functional identity under aerobic and anaerobic conditions. J. Biol. Chem. 268, 14018–14025. [PubMed] [Google Scholar]
- Scott, J.G., and Wen, Z. (2001). Cytochromes P450 of insects: The tip of the iceberg. Pest Manag. Sci. 57, 958–967. [DOI] [PubMed] [Google Scholar]
- Swanson, W.J., and Vacquier, V.D. (1998). Concerted evolution in an egg receptor for a rapidly evolving abalone sperm protein. Science 281, 710–712. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, A. (1998). The fate of duplicated genes: Loss or new function? Bioessays 20, 785–788. [DOI] [PubMed] [Google Scholar]
- Wang, T.W., Lu, L., Wang, D., and Thompson, J.E. (2001). Isolation and characterization of senescence-induced cDNAs encoding deoxyhypusine synthase and eucaryotic translation initiation factor 5A from tomato. J. Biol. Chem. 276, 17541–17549. [DOI] [PubMed] [Google Scholar]
- Weber, S., Eisenreich, W., Bacher, A., and Hartmann, T. (1999). Pyrrolizidine alkaloids of the lycopsamine type: Biosynthesis of trachelanthic acid. Phytochemistry 50, 1005–1014. [Google Scholar]
- Wilson, T.G. (2001). Resistance of Drosophila to toxins. Annu. Rev. Entomol. 46, 545–571. [DOI] [PubMed] [Google Scholar]
- Yan, Y.P., Tao, Y., and Chen, K.Y. (1996). Molecular cloning and functional expression of human deoxyhypusine synthase cDNA based on expressed sequence tag information. Biochem. J. 315, 429–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., and Nielsen, R. (2000). Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 17, 32–43. [DOI] [PubMed] [Google Scholar]
- Yang, Z., Nielsen, R., Goldman, N., and Pedersen, A.M.K. (2000). Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155, 431–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer, E.A., Martin, S.L., Beverley, S.M., Kan, Y.W., and Wilson, A.C. (1980). Rapid duplication and loss of genes coding for the alpha chains of hemoglobin. Proc. Natl. Acad. Sci. USA 77, 2158–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.