Standing genetic variation for flowering time in a nondomesticated grass encompasses known and novel regulators.
Abstract
The domestication of plants is underscored by the selection of agriculturally favorable developmental traits, including flowering time, which resulted in the creation of varieties with altered growth habits. Research into the pathways underlying these growth habits in cereals has highlighted the role of three main flowering regulators: VERNALIZATION1 (VRN1), VRN2, and FLOWERING LOCUS T (FT). Previous reverse genetic studies suggested that the roles of VRN1 and FT are conserved in Brachypodium distachyon yet identified considerable ambiguity surrounding the role of VRN2. To investigate the natural diversity governing flowering time pathways in a nondomesticated grass, the reference B. distachyon accession Bd21 was crossed with the vernalization-dependent accession ABR6. Resequencing of ABR6 allowed the creation of a single-nucleotide polymorphism-based genetic map at the F4 stage of the mapping population. Flowering time was evaluated in F4:5 families in five environmental conditions, and three major loci were found to govern flowering time. Interestingly, two of these loci colocalize with the B. distachyon homologs of the major flowering pathway genes VRN2 and FT, whereas no linkage was observed at VRN1. Characterization of these candidates identified sequence and expression variation between the two parental genotypes, which may explain the contrasting growth habits. However, the identification of additional quantitative trait loci suggests that greater complexity underlies flowering time in this nondomesticated system. Studying the interaction of these regulators in B. distachyon provides insights into the evolutionary context of flowering time regulation in the Poaceae as well as elucidates the way humans have utilized the natural variation present in grasses to create modern temperate cereals.
The coordination of flowering time with geographic location and seasonal weather patterns has a profound effect on flowering and reproductive success (Amasino, 2010). The mechanisms underpinning this coordination are of great interest for understanding plant behavior and distribution within natural ecosystems (Wilczek et al., 2010). Plants that fail to flower at the appropriate time are unlikely to be maximally fertile and, therefore, will be less competitive in the longer term. Likewise, optimal flowering time in crops is important for yield and quality: seed and fruit crops need to flower early enough to allow ripening or to utilize seasonal rains, while delayed flowering may be advantageous for leaf and forage crops (Distelfeld et al., 2009; Jung and Müller, 2009).
Although developmental progression toward flowering can be modulated in several ways, many plants have evolved means to detect seasonal episodes of cold weather and adjust their flowering time accordingly, a process known as vernalization (Ream et al., 2012). Despite the importance of flowering time, the molecular and genetic mechanisms underlying this dependency have been studied in only a few systems, notably the Brassicaceae, Poaceae, and Amaranthaceae (Andrés and Coupland, 2012; Ream et al., 2012). Three major VERNALIZATION (VRN) genes appear to act in a regulatory loop in temperate grasses. The wheat (Triticum aestivum) VRN1 gene is a MADS box transcription factor that is induced in the cold (Yan et al., 2003; Andrés and Coupland, 2012). This gene is related to the Arabidopsis (Arabidopsis thaliana) genes APETALA1 and FRUITFUL (Yan et al., 2003; Andrés and Coupland, 2012). VRN2 encodes a small CCT domain protein (Yan et al., 2004) that is repressed by VRN1 and, in turn, represses FLOWERING LOCUS T (FT), a strong universal promoter of flowering (Kardailsky et al., 1999; Yan et al., 2006; Andrés and Coupland, 2012; Ream et al., 2012). In cereals, active VRN2 alleles are necessary for a vernalization requirement. Spring barley (Hordeum vulgare) and spring wheat varieties, which do not require vernalization to flower, either lack VRN2 (Dubcovsky et al., 2005; Karsai et al., 2005; von Zitzewitz et al., 2005), have point mutations in the conserved CCT domain (Yan et al., 2004), or possess dominant constitutively active alleles of VRN1 (repressor of VRN2; Yan et al., 2003; Fu et al., 2005) or FT (repressed by VRN2; Yan et al., 2006).
Investigations of the regulation of flowering in the Poaceae have focused on rice (Oryza sativa), wheat, and barley, all domesticated species that have been heavily subjected to human selection over the past 10,000 years. Little information is available on wild species within this family that have not been subjected to human selection. Such a study could provide additional insights into the standing variation present within wild systems and its likely predomestication adaptive significance in the Poaceae (Schwartz et al., 2010). A favorable species for such a study is Brachypodium distachyon, a small, wild grass with a sequenced and annotated genome. B. distachyon was developed originally as a model system for the agronomically important temperate cereals (Draper et al., 2001; Opanowicz et al., 2008; International Brachypodium Initiative, 2010; Catalán et al., 2014). With the recent availability of geographically dispersed diversity collections, we can ask how wild grasses have adapted to different climatic zones.
Previous studies have begun to explore the molecular basis of vernalization in this system. Higgins et al. (2010) identified homologs of the various flowering pathway genes in B. distachyon, and several mainly reverse genetic studies have focused on characterizing these genes further (Schwartz et al., 2010; Lv et al., 2014; Ream et al., 2014; Woods et al., 2014, 2016b). Schwartz et al. (2010) did not find complete correlation between the expression of VRN1 and flowering and, therefore, hypothesized that VRN1 could have different activity or roles that are dependent on the genetic background. Yet, Ream et al. (2014) found low VRN1 and FT levels in B. distachyon accessions with delayed flowering, suggesting a conserved role of these homologs. Further support for a conserved role of VRN1 and FT comes from the observations that overexpression of these genes leads to extremely early flowering (Lv et al., 2014; Ream et al., 2014) and that RNA interference-based silencing of FT and artificial microRNA-based silencing of VRN1 prevent flowering (Lv et al., 2014; Woods et al., 2016b). The role of VRN2 in B. distachyon is less clear. Higgins et al. (2010) failed to identify a homolog of VRN2 in B. distachyon; however, other studies identified Bradi3g10010 as the best candidate for the B. distachyon VRN2 homolog (Schwartz et al., 2010; Ream et al., 2012). Recent research supports the functional conservation of VRN2 in its role as a flowering repressor but suggests that the regulatory interaction between VRN1 and VRN2 evolved after the diversification of the Brachypodieae and the core Pooideae (e.g. wheat and barley; Woods et al., 2016b).
To date, most studies on the regulation of flowering time of B. distachyon have used reverse genetic approaches to implicate the role of previously characterized genes from other species (Higgins et al., 2010; Lv et al., 2014; Ream et al., 2014; Woods et al., 2016b), while only a few studies have used the natural variation present among B. distachyon accessions to identify flowering loci (Tyler et al., 2016; Wilson et al., 2016). Currently lacking is the characterization of loci that control variation in flowering time in a biparental B. distachyon mapping population. The Iraqi reference accession Bd21 does not require vernalization (Vogel et al., 2006; Garvin et al., 2008); in addition, vernalization does not greatly reduce time to flowering in a 16- or 20-h photoperiod (Schwartz et al., 2010; Ream et al., 2014). In contrast, the Spanish accession ABR6 can be induced to flower following a 6-week vernalization period (Draper et al., 2001; Routledge et al., 2004).
In this article, we report on the genetic architecture underlying flowering time in a mapping population developed from ABR6 and Bd21. We observed the segregation of vernalization dependency during population advancement (Fig. 1) and characterized the genetic basis of this dependency in detail at the F4:5 stage in multiple environments. The ability to flower without vernalization was linked to three major loci, two of which colocalize with the B. distachyon homologs of VRN2 and FT. Notably, our results further support the role of the VRN2 locus as a conserved flowering time regulator in B. distachyon.
Figure 1.
Flowering behavior within the ABR6 × Bd21 mapping population. Three months after a 6-week vernalization period, ABR6 (left) is not flowering, whereas Bd21 (center) is flowering, and an individual in the ABR6 × Bd21 mapping population displays an intermediate flowering phenotype (right).
RESULTS
Development of a B. distachyon Mapping Population between Geographically and Phenotypically Distinct Accessions
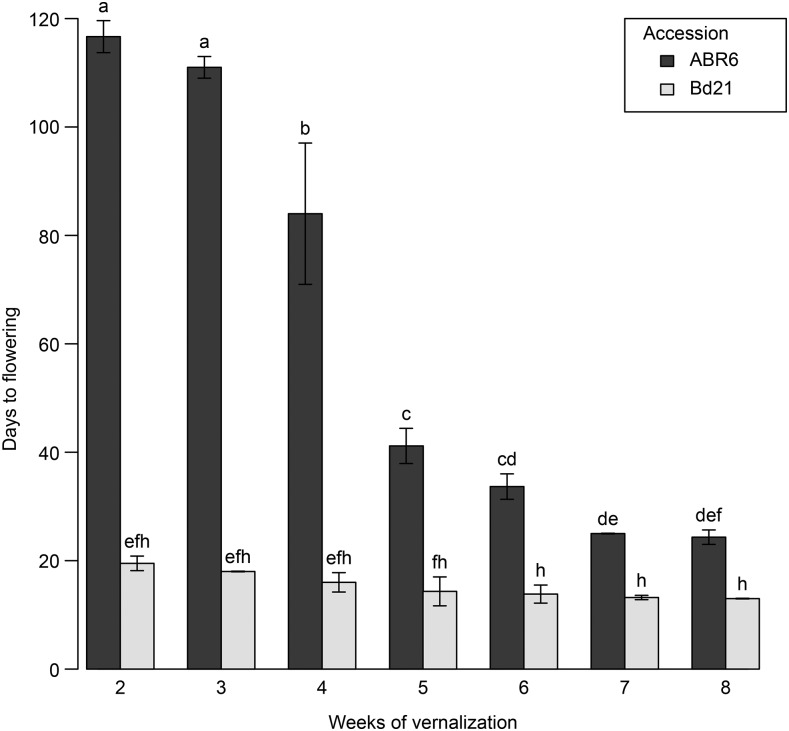
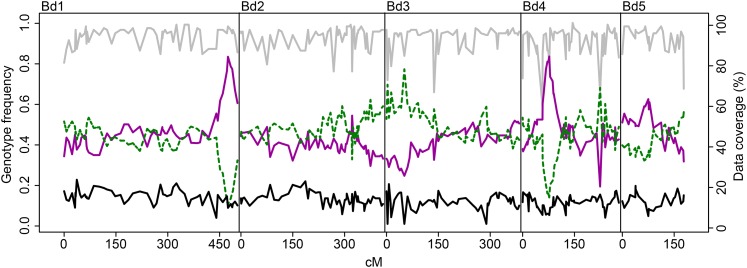
Initial investigations into the flowering time of ABR6 and Bd21 in response to different vernalization periods showed contrasting effects on the two accessions (Figs. 1 and 2). ABR6 responded strongly to increasing vernalization times with a reduction in flowering by 93 d, ranging from 117 d for a 2-week vernalization period to 24 d for an 8-week vernalization period. This reduction in flowering time for ABR6 was not linear, and the greatest drop of 43 d occurred between 4 and 5 weeks of vernalization (Fig. 2). In contrast, no statistically significant difference was found with respect to the vernalization response of Bd21, although a consistent trend toward a reduced flowering time was observed. A cross was generated from these phenotypically diverse accessions for the creation of a recombinant inbred line population. To develop a single-nucleotide polymorphism (SNP)-based genetic map, ABR6 was resequenced, and reads were aligned to the reference genome. A total of 1.36 million putative SNPs were identified between ABR6 and Bd21, of which 711,052 constituted nonambiguous polymorphisms based on a minimum coverage of 15× and a strict threshold for SNP calling (i.e. 100% of reads with an ABR6 allele, 0% of reads with a Bd21 allele). Following iterative cycles of marker selection, the final genetic map consists of 252 nonredundant markers and has a cumulative size of 1,753 centimorgan (cM; Supplemental Fig. S1). This size is comparable to that of the previously characterized Bd3-1 × Bd21 mapping population (Huo et al., 2011) and confirms that B. distachyon has a high rate of recombination compared with other grass species. The quality of the genetic map was verified by assessing the two-way recombination fractions for all 252 markers (Supplemental Fig. S2). All five chromosomes were scanned for segregation distortion by comparing observed and expected genotype frequencies for each marker. The expected heterozygosity at the F4 stage is 12.5%, and the expected allele frequency for each parental genotype is 43.75%. Although all five chromosomes contained regions of potential segregation distortion (Fig. 3), only two loci on chromosomes Bd1 (peak at 474.1 cM) and Bd4 (peak at 77 cM) deviated significantly from these expected frequencies.
Figure 2.
Effects of vernalization on flowering time in ABR6 and Bd21. Days to flowering was measured from the end of vernalization for seven different vernalization periods. After vernalization, plants were grown in a growth chamber (16-h photoperiod) for 35 d and then transferred to a greenhouse without light and temperature control (late April to mid July, 2013; Norwich, UK). Mean days to flowering and se are based on six biological replicates. Different letters represent statistically significant differences based on pairwise comparisons using Student’s t tests with pooled sd and Bonferroni correction for multiple comparisons.
Figure 3.
Segregation distortion in the ABR6 × Bd21 F4 population. For each marker of the genetic map, the frequencies of F4 individuals with homozygous ABR6 genotype (solid magenta line), homozygous Bd21 genotype (dashed green line), or heterozygous genotype (solid black line) were calculated (scale on left). Data coverage (percentage of F4 individuals with genotype calls per marker) is represented by the gray line (scale on right).
Multiple Quantitative Trait Loci Control Flowering in the ABR6 × Bd21 Mapping Population
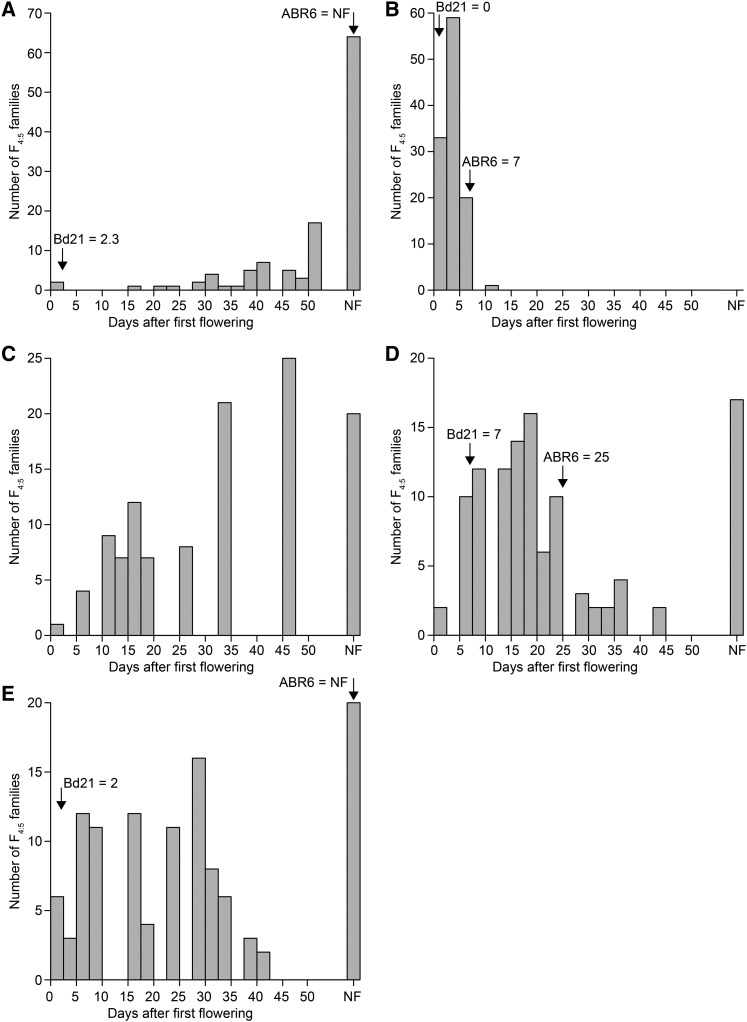
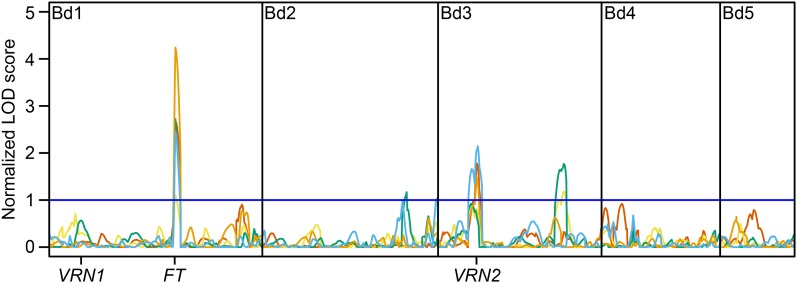
We evaluated the ABR6 × Bd21 F4:5 population in a number of environments to identify the genetic architecture underlying flowering time (Supplemental Table S1; Supplemental Data S1). Four sets of the population were grown without vernalization, whereas in one additional set, flowering was scored in response to 6 weeks of vernalization. In all experiments, the population was exposed to natural light, although in three experiments, supplemental light was used to ensure a minimum 16- or 20-h growth period. In addition, two experiments did not have any temperature control (i.e. plants were exposed to the natural temperature in the greenhouse), two experiments had the temperature controlled at 22°C/20°C during light/dark cycles, and one experiment had the temperature maintained at a minimum of 18°C/11.5°C during light/dark cycles. Analysis of the nonvernalized environments revealed a bimodal distribution between families that flowered and families that did not flower (Fig. 4). However, considerable residual variation in flowering time existed among the flowering families. For example, in environment 5, flowering occurred over a 42-d period from 63 to 105 d after germination (Fig. 4E). Flowering in the other nonvernalized environments occurred over a similar time period (Fig. 4). Interestingly, transgressive segregation for early- and late-flowering phenotypes was observed in environment 4 (Fig. 4D). Phenotypes in the vernalized environment were heavily skewed toward early flowering (Fig. 4B). Only limited residual variation existed among the vernalized F4:5 families, and all plants flowered within 11 d from the first observation of flowering in the population. The variation in flowering time for all five environments was found to be not normally distributed. Among these diverse environments, quantitative trait locus (QTL) analyses using binary and nonparametric models were conservative in detecting QTLs controlling flowering time (qFLT; Supplemental Tables S2 and S3), whereas transformation of flowering time consistently identified QTLs between environments (Tables I and II; Supplemental Tables S4 and S5). Three major QTLs were identified on chromosomes Bd1 and Bd3 that were robustly observed using parametric and nonparametric mapping approaches (Tables I and II; Fig. 5). The QTL on Bd1 (qFLT1; peak marker Bd1_47808182) appeared to be the major locus governing flowering time in this population, as it was the major QTL in all five environments, explaining the most phenotypic variation (PVE) compared with any other QTL (Table II). PVE values for this locus ranged from 15.9% to 37.5%. Another QTL on Bd3 (qFLT6; peak marker Bd3_8029207) also was detected in all five studies, although its contribution was significant in only three environments. PVE values for the statistically significant QTLs ranged from 11.8% to 18.7%. Bd21 alleles at these two loci promoted early flowering, whereas individuals with ABR6 alleles at both loci had maximal flowering time or did not flower within the time scale of the experiment (Fig. 6). Interestingly, in the two environments where this former locus did not have a significant contribution, two other QTLs were identified. A QTL on Bd3 (qFLT7; peak marker Bd3_44806296) explained 13.6% and 14% of the variation observed in these studies, and a QTL on Bd2 (qFLT3; peak marker Bd2_53097824) was identified through a combination of nonparametric and parametric analyses of environments 4 and 5. Additional QTLs on Bd1 (qFLT2), Bd2 (qFLT4), Bd3 (qFLT5), and Bd4 (qFLT8) were not significant in more than one of the environments tested (Table I).
Figure 4.
Frequency distribution of flowering time in the ABR6 × Bd21 population. Flowering time was measured from the first day that flowering was observed in the entire population. A, Environment 1 (April to July; natural light supplemented for 20 h, 22°C/20°C, no vernalization). B, Environment 2 (April to July; natural light supplemented for 20 h, 22°C/20°C, 6 weeks of vernalization). C, Environment 3 (May to July; natural light and temperatures, no vernalization). D, Environment 4 (September to November; natural light supplemented for 16 h, minimum 18°C/11.5°C, no vernalization). E, Environment 5 (March to May; natural light and temperatures, no vernalization). Flowering times for the parental lines are indicated by arrows (no data for environment 3). NF, Not flowering.
Table I. Significant flowering time QTLs (qFLT) in the different environments identified using several binary, nonparametric, and parametric approaches.
Dashes, Corresponding QTL was not detected within respective environment.
| Locus | Chra | cM | Alleleb | E1c | E2 | E3 | E4 | E5 |
|---|---|---|---|---|---|---|---|---|
| qFLT1 | Bd1 | 297.6 | Bd21 | B, T2, T3, NPd | T1, T3, NP | T2, T3, NP | T2, T3 | T1, T2, T3, NP |
| qFLT2 | Bd1 | 465.2 | Bd21 | T2 | – | – | – | – |
| qFLT3 | Bd2 | 338.3 | ABR6 | – | – | – | NP | T2, T3 |
| qFLT4 | Bd2 | 409.0 | Bd21 | – | T1, T3 | – | – | – |
| qFLT5 | Bd3 | 60.8 | Bd21 | – | – | – | T1 | – |
| qFLT6 | Bd3 | 91.2 | Bd21 | T2, T3 | T1, T3 | T2, T3 | – | – |
| qFLT7 | Bd3 | 294.6 | Bd21 | – | – | – | T2, T3, NP | B, T2, T3, NP |
| qFLT8 | Bd4 | 90.1 | Bd21 | – | – | – | NP | – |
Chromosome. bAllele that reduces flowering time. cE1 to E5, Environment (see Supplemental Table S1). dQTL analyses were performed with interval mapping using binary classification (B) and nonparametric analysis (NP) and composite interval mapping using transformed data (T1, T2, and T3).
Table II. Significant QTLs from composite interval mapping of transformed flowering time phenotypes (T3) in the ABR6 × Bd21 F4:5 families.
| ENVa | Locus | Chrb | cM | EWTc | LOD | AEEd | PVEe | 1 − LOD SIf |
|---|---|---|---|---|---|---|---|---|
| 1 | qFLT1 | Bd1 | 297.6 | 3.06 | 12.96 | 2.87 | 36.3% | 296.1–305.6 |
| 1 | qFLT6 | Bd3 | 91.2 | 3.06 | 4.51 | 1.64 | 11.8% | ND |
| 2 | qFLT1 | Bd1 | 297.6 | 3.09 | 7.59 | 0.82 | 20.0% | 296.1–305.6 |
| 2 | qFLT4 | Bd2 | 409.0 | 3.09 | 3.20 | 0.47 | 6.7% | 403.2–411.0 |
| 2 | qFLT6 | Bd3 | 93.2 | 3.09 | 6.64 | 0.79 | 18.2% | 72.9–97.0 |
| 3 | qFLT1 | Bd1 | 297.6 | 3.20 | 8.61 | 1.50 | 31.1% | 292.1–303.6 |
| 3 | qFLT6 | Bd3 | 91.2 | 3.20 | 5.69 | 1.20 | 18.7% | 74.9–97.0 |
| 4 | qFLT1 | Bd1 | 297.6 | 3.19 | 3.49 | 1.77 | 15.9% | 292.1–305.6 |
| 4 | qFLT7 | Bd3 | 294.6 | 3.19 | 3.79 | 1.59 | 14.0% | 273.9–300.7 |
| 5 | qFLT1 | Bd1 | 297.6 | 3.17 | 8.62 | 3.43 | 37.5% | 294.1–301.6 |
| 5 | qFLT3 | Bd2 | 338.3 | 3.17 | 3.70 | −1.75 | 9.9% | 323.7–348.0 |
| 5 | qFLT7 | Bd3 | 294.6 | 3.17 | 5.61 | 2.02 | 13.6% | 275.9–302.0 |
Environment (see Supplemental Table S1). bChromosome. cExperiment-wide permutation threshold. dAdditive effect estimate for transformed phenotypes. ePercentage of phenotypic variance explained. fThe 1 − LOD support interval (cM). ND denotes QTLs not detected using standard interval mapping.
Figure 5.
Linkage mapping of flowering time in the ABR6 × Bd21 population. Time to flowering for 114 F4:5 families of the population was transformed into ordered rank values, QTL analysis was performed using composite interval mapping under an additive model hypothesis test (H0:H1), and data were plotted based on normalized permutation thresholds. The blue horizontal line represents the threshold of statistical significance based on 1,000 permutations. Orange line = environment 1 (April to July; natural light supplemented for 20 h, 22°C/20°C, no vernalization), blue line = environment 2 (April to July; natural light supplemented for 20 h, 22°C/20°C, 6 weeks of vernalization), red line = environment 3 (May to July; natural light and temperatures, no vernalization), yellow line = environment 4 (September to November; natural light supplemented for 16 h, minimum 18°C/11.5°C, no vernalization), and green line = environment 5 (March to May; natural light and temperatures, no vernalization). For full environmental details, see Supplemental Table S1. The genetic positions of the previously identified homologs of VRN1, VRN2, and FT are indicated (compare Higgins et al., 2010, and Ream et al., 2012).
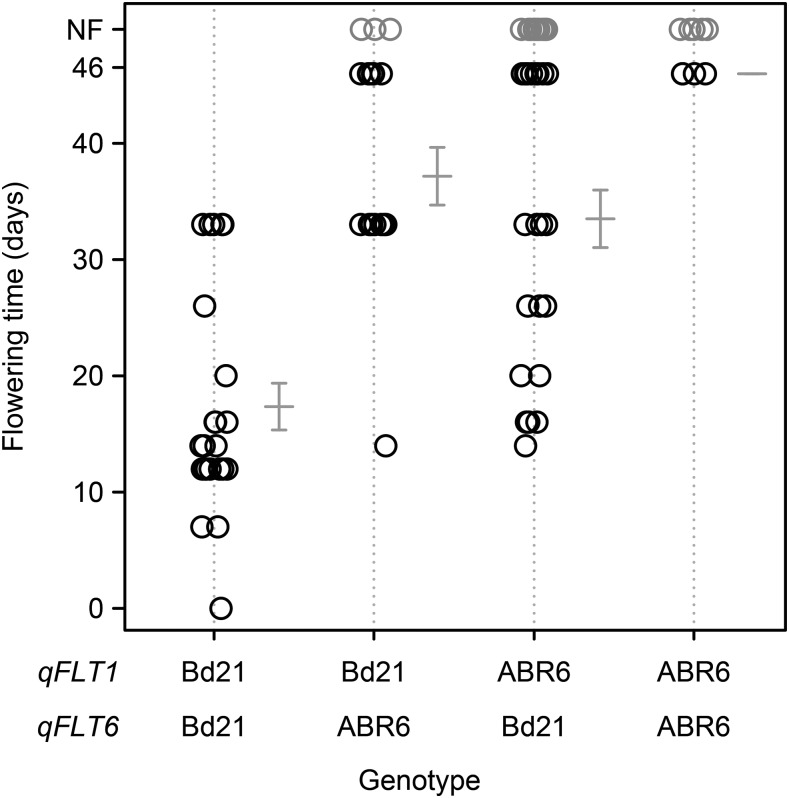
Figure 6.
Phenotype-by-genotype plot for the two major loci controlling flowering time in the ABR6 × Bd21 mapping population. Days to flowering in environment 3 for the ABR6 × Bd21 F4:5 families homozygous at qFLT1 and qFLT6 shows that the Bd21 alleles at these two loci promote early flowering. Error bars represent 1 se; NF, not flowering.
Previous studies identified the B. distachyon homologs of flowering regulators from Arabidopsis, wheat, barley, and rice (Higgins et al., 2010; Ream et al., 2012). The 1 − log of the odds (LOD) support intervals of all statistically significant QTLs were combined to identify the maximal 1 − LOD support interval for each QTL. Several of the previously identified B. distachyon homologs of flowering regulators are candidate genes underlying these QTLs (Table III). Although several homologs fall within the 1 − LOD support intervals of qFLT1 on Bd1 (292.1–305.6 cM) and qFLT6 on the short arm of Bd3 (72.9–97 cM), these loci also harbor the B. distachyon homologs of FT (Bradi1g48830) and VRN2 (Bradi3g10010), which have been implicated previously in flowering time regulation in B. distachyon through a series of mainly reverse genetic studies (Lv et al., 2014; Ream et al., 2014; Woods et al., 2014, 2016b).
Table III. Previously identified B. distachyon homologs of flowering regulators in Arabidopsis (At), hexaploid and diploid wheat (Ta and Tm), barley (Hv), and rice (Os) within the 1 − LOD support intervals of the statistically significant QTLs under transformation T3.
| Locus | Chra | 1 − LOD SIb | B. distachyon Gene | Homologous Genesc |
|---|---|---|---|---|
| qFLT1 | Bd1 | 292.1–305.6 | Bradi1g45810 | AtAGL24, TaVRT2, OsMADS55 |
| Bradi1g46060 | AtABF1 | |||
| Bradi1g48340 | AtCLF, OsCLF | |||
| Bradi1g48830 | AtTSF, HvFT1, OsHd3a/OsFTL2 | |||
| qFLT3 | Bd2 | 323.7–348.0 | Bradi2g53060 | AtFDP |
| Bradi2g54200 | AtNF-YB10 | |||
| Bradi2g55550 | AtbZIP67 | |||
| qFLT4 | Bd2 | 403.2–411.0 | Bradi2g60820 | AtFY, OsFY |
| Bradi2g62070 | AtLUX, OsLUX | |||
| qFLT6 | Bd3 | 72.9–97.0 | Bradi3g08890 | OsFTL13 |
| Bradi3g10010 | TaVRN2, TmCCT2, OsGhd7 | |||
| Bradi3g12900 | AtHUA2 | |||
| qFLT7 | Bd3 | 273.9–300.7 | Bradi3g41300 | OsMADS37 |
| Bradi3g42910 | AtSPY, OsSPY | |||
| Bradi3g44860 | OsRCN2 |
Chromosome. bCombined maximal 1 − LOD support interval (cM) from all significant QTLs. cIdentified by Higgins et al. (2010) and Ream et al. (2012).
Natural Variation in FT and VRN2
Analysis of the resequencing and RNA sequencing (RNAseq) data allowed an initial evaluation of candidate genes underlying these QTLs. A de novo assembly was created from the ABR6 resequencing reads, and the resulting contigs were probed with the Bd21 sequences of FT (Bradi1g48830) and VRN2 (Bradi3g10010), enabling the identification of structural variation between ABR6 and Bd21 (Fig. 7; Supplemental Table S6). Spliced alignment of RNAseq reads permits the further characterization of candidate genes underlying an identified QTL through the confirmation of polymorphisms between two parental genotypes, verification of annotated candidate gene models, qualitative assessment of the expression of candidate genes in the sampled tissue, and discovery of potential splice variants.
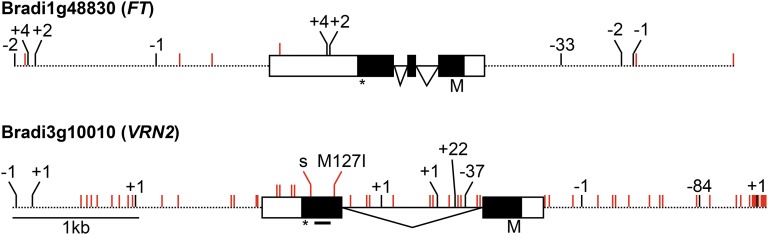
Figure 7.
Comparison of the flowering regulators FT and VRN2 between the B. distachyon accessions Bd21 and ABR6. Contigs of the ABR6 de novo assembly were aligned to the Bd21 reference sequence (version 3), and polymorphisms were identified in the genes of interest and 2-kb promoter and terminator sequences (1.9-kb promoter for VRN2). Red ticks represent SNPs, and black ticks represent insertions/deletions (indels). The length of indels (bp) is shown with + for insertion and – for deletion. The amino acid change of the nonsynonymous SNP in VRN2 is indicated. s = synonymous SNP; dashed line = promoter or terminator; white box = 5′ untranslated region (UTR) or 3′ UTR; black box = exon; black line = intron; M = Met/translation start; star = translation stop; black bar under VRN2 = CCT domain.
No polymorphisms were found in the coding sequence of Bradi1g48830, the B. distachyon homolog of FT. However, two indels (2 and 4 bp) and an SNP mapped to the 3′ UTR. Additionally, two SNPs and three indels (including a 33-bp indel 590 bp upstream of Bradi1g48830) were found in the promoter region (2 kb upstream). The terminator region (2 kb downstream) contained three SNPs and four indels. Bradi1g48830 was not expressed in ABR6 and was barely detectable in Bd21 (only two reads mapped to the gene). Owing to the low expression, it was not possible to confirm the published gene model with our RNAseq data.
Greater sequence variation was observed at Bradi3g10010, the B. distachyon homolog of VRN2, and its flanking regions. Only 1.9 kb of the promoter region is present on the Bradi3g10010 contig, but this region contains 29 SNPs and three indels (including an 84-bp indel 1.4 kb upstream of Bradi3g10010). The 2-kb terminator region contains 14 SNPs and three 1-bp indels. Additionally, 11 SNPs and four indels (including a 37-bp and a 22-bp indel) were localized in the intron, two SNPs in the coding sequence, and four SNPs in the 3′ UTR. Bradi3g10010 was expressed in leaves from both Bd21 and ABR6, and spliced alignment of RNAseq reads confirmed the published annotation of Bradi3g10010 for both ABR6 and Bd21. Moreover, the six SNPs predicted in the exons were supported by the RNAseq data, and these may contribute to the observed effect on flowering time in this mapping population. Two SNPs map to the annotated coding sequence and four SNPs map to the 3′ UTR. One of the two SNPs in the annotated coding sequence is predicted to cause a nonsynonymous mutation (Fig. 7).
Expression of VRN1, VRN2, and FT in Response to Vernalization
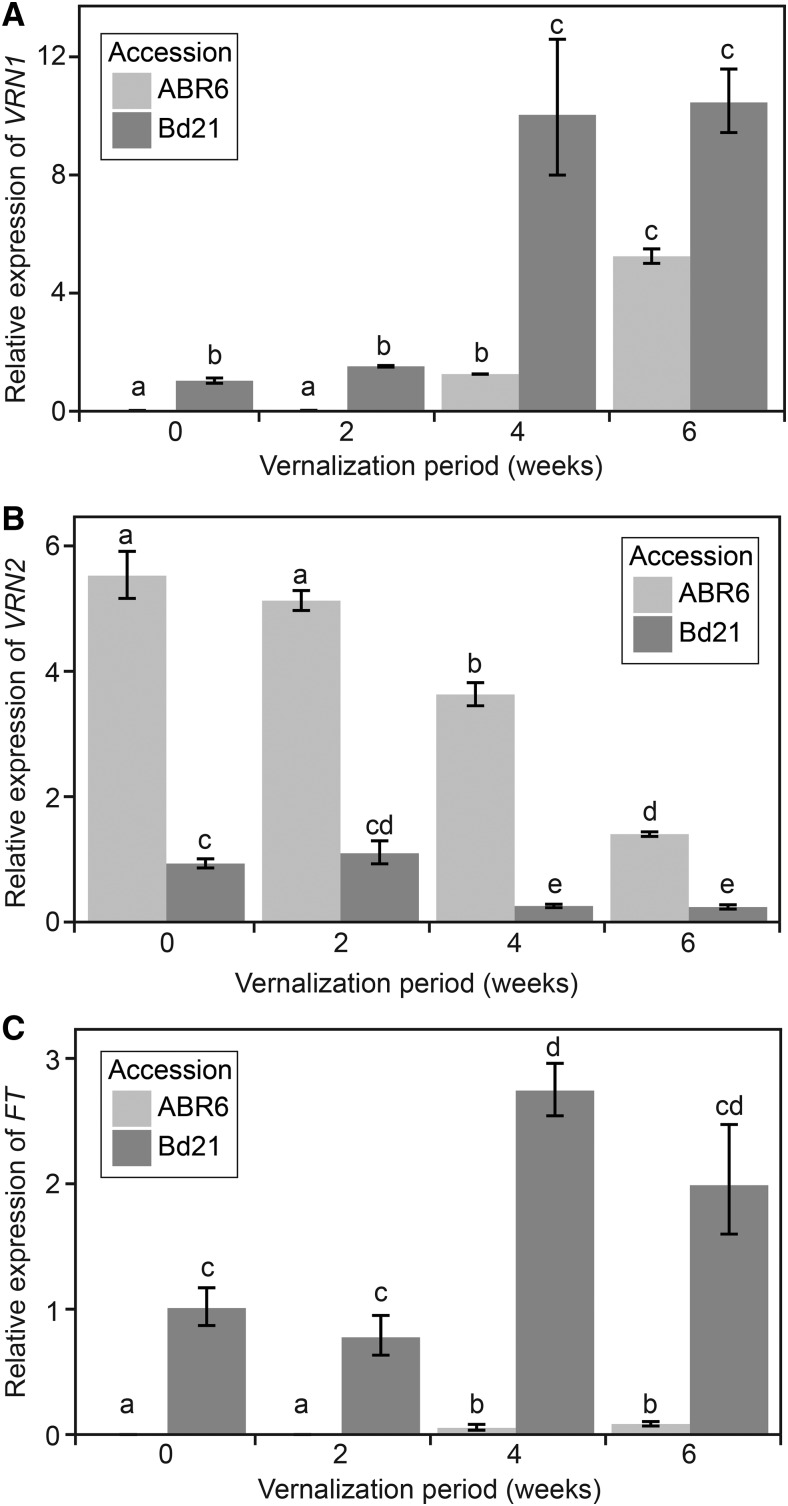
To understand the transcriptional dynamics of VRN1, VRN2, and FT in response to vernalization, we assessed steady-state levels of mRNA expression in plants at the fourth leaf stage after exposure to 2, 4, and 6 weeks of vernalization at 5°C or to no vernalization (Fig. 8). VRN1 and FT had a similar pattern in steady-state levels of gene expression in response to vernalization (Fig. 8, A and C). For both genes, very low levels of expression were observed in ABR6, whereas Bd21 had fairly high levels of transcript abundance. After experiencing 4 weeks of vernalization, ABR6 had similar levels of VRN1 transcript to Bd21 without vernalization treatment. In contrast, FT expression had a marginal increase after 4 and 6 weeks of vernalization in ABR6 relative to no vernalization or 2 weeks of vernalization. FT expression levels were significantly lower than in Bd21 across all periods of vernalization. Both VRN1 and FT expression increased significantly between Bd21 samples vernalized for 2 or 4 weeks. VRN2 expression in ABR6 was inversely correlated with the length of vernalization, with similar levels of expression after no vernalization and 2 weeks of vernalization and increasingly lower levels of expression after 4 and 6 weeks of vernalization (Fig. 8B). Bd21 exhibited a similar reduction in VRN2 expression, although lower levels of expression were observed without vernalization compared with ABR6 with 6 weeks vernalization. The trends of all three genes highlighted the importance of 4 weeks of vernalization as the inflection point in transcriptional abundance, which coincides with a significant reduction in days to flowering in ABR6 (Fig. 2).
Figure 8.
VRN1, VRN2, and FT expression in the fourth leaf of ABR6 and Bd21 after varying periods of cold treatment. Seeds were imbibed with water and not vernalized or vernalized for 2, 4, or 6 weeks and transferred to a growth chamber with parameters similar to environment 2. Fully expanded fourth leaves were harvested in the middle of the photoperiod. Relative gene expression of VRN1 (A), VRN2 (B), and FT (C) was determined using reverse transcription-quantitative PCR and analyzed using the 2−ΔΔCt method. All genes were normalized to 1 based on Bd21 expression with no cold treatment (0 weeks), and UBIQUITIN-CONJUGATING ENZYME18 was used as an internal control. Bars represent means of three biological replicates, with error bars showing 1 se. Different letters represent statistically significant differences based on pairwise Student’s t tests using a multiple hypothesis-corrected P value threshold of 0.05 with the Benjamini-Hochberg approach (Benjamini and Hochberg, 1995).
DISCUSSION
In our advancement of the ABR6 × Bd21 population, we observed substantial variation in flowering time. To define the genetic architecture of flowering time, we developed a comprehensive genetic map and assessed F4:5 families in multiple environments. We uncovered three major QTLs, with two QTLs coincident with the B. distachyon homologs of VRN2 and FT. Interestingly, VRN1 was not associated with flowering time and was found to have no mutations within the transcribed sequence (Supplemental Table S6). Further minor-effect QTLs were identified, suggesting that additional regulators play a role in controlling flowering time in B. distachyon.
Segregation Distortion in the ABR6 × Bd21 Population
Segregation distortion is a common observation in the development of mapping populations in plants, including grasses such as rice, Aegilops tauschii, maize (Zea mays), or barley (Xu et al., 1997; Faris et al., 1998; Lu et al., 2002; Muñoz-Amatriaín et al., 2011). In the ABR6 × Bd21 population, significant deviation from expected genotype frequencies was observed at two loci on chromosomes Bd1 and Bd4 (Fig. 3). Interestingly, heterozygosity was not affected at these loci, but the ABR6 allele was overrepresented. It is likely that these loci are linked to traits that were selected inadvertently during population advancement based on genetic and/or environmental factors. Several genetic mechanisms can contribute to segregation distortion in intraspecific crosses, including hybrid necrosis (Bomblies and Weigel, 2007), genes involved in vernalization requirement and flowering time (such as the vrn2 locus in the Haruna Nijo × OHU602 doubled-haploid barley population; Muñoz-Amatriaín et al., 2011), or preferential transmission of a specific parental genotype. While segregation distortion at these loci was not associated with the identified flowering time QTLs, canonical resistance genes encoding nucleotide-binding, Leu-rich repeat proteins are present at the Bd4 locus (Bomblies et al., 2007; Tan and Wu, 2012).
The Genetic Architecture of Flowering Time in B. distachyon
In Arabidopsis, natural variation has been used as a complementary forward genetics-based approach for investigating flowering time (Koornneef et al., 2004). In our work, we identified two major QTLs controlling flowering time (qFLT1 and qFLT6; Fig. 6) in both vernalized and nonvernalized environments that colocalized with the B. distachyon homologs of FT (Bradi1g48830) and VRN2 (Bradi3g10010). These observations are consistent with previous reverse genetic studies on the role of FT and VRN2 in controlling flowering time (Lv et al., 2014; Ream et al., 2014; Woods et al., 2014, 2016b). Two additional QTLs on chromosomes Bd2 (qFLT3) and Bd3 (qFLT7) were detected in two environments, whereas four minor-effect QTLs (qFLT2, qFLT4, qFLT5, and qFLT8) were found in individual environments only. Two recent genome-wide association studies used the natural variation found within B. distachyon germplasm to identify SNPs associated with flowering time (Tyler et al., 2016; Wilson et al., 2016). Tyler et al. (2016) identified nine significant marker-trait associations, none of which overlap with the QTLs identified in our study. In contrast, Wilson et al. (2016) identified a much simpler genetic architecture consisting of three significant marker-trait associations, one of which could be linked to FT. These additional QTLs and marker-trait associations identified in our study and the genome-wide association studies could either correspond to one of the identified homologs of flowering genes in B. distachyon (Table III; Higgins et al., 2010) or constitute novel loci as hypothesized by Schwartz et al. (2010). With the exception of the proximal QTL on Bd2 (qFLT3), all alleles that prolonged time to flowering in our study were contributed by ABR6 (Table I). Bd21 has been classified previously as a spring annual (Schwartz et al., 2010) or extremely rapid flowering (Ream et al., 2014). However, increased vernalization times still led to a modest reduction in flowering time (Fig. 2), which is explained by the detection of a QTL contributed by Bd21.
We hypothesized that structural variation between ABR6 and Bd21 would underlie the observed variation in flowering time. No structural variation in FT was observed between ABR6 and Bd21 in the coding sequence; however, several indels map to the promoter region (Fig. 7). These polymorphisms may explain the expression differences between these two accessions. As expected, no FT expression was found in ABR6 seedlings, and only two Bd21 RNAseq reads mapped to this gene. Steady-state expression levels of FT in the fourth leaf were significantly lower in ABR6 relative to Bd21 without vernalization (Fig. 8C). After 4 weeks of vernalization, FT expression levels increased in ABR6, although they were significantly lower than Bd21 steady-state levels after any level of vernalization. It was shown previously that in barley, wheat, and B. distachyon, FT expression is up-regulated after vernalization (Sasani et al., 2009; Chen and Dubcovsky, 2012; Ream et al., 2014). Our observations indicate that FT is expressed in Bd21 and increases less than VRN1 in response to vernalization. In contrast, FT in ABR6 increases only marginally after 4 weeks of vernalization and remains significantly below the levels observed in Bd21 after no vernalization.
Interestingly, an intact copy of the flowering repressor VRN2 also is present in Bd21 (Ream et al., 2012), which does not have a strong vernalization response (Vogel et al., 2006; Garvin et al., 2008). The lack of a vernalization requirement in some B. distachyon accessions, therefore, cannot be explained by an absence of VRN2 (Ream et al., 2012). Intriguingly, early-flowering mutants identified in genetic screens thus far have not mapped in the VRN2 region (Ream et al., 2014). Moreover, expression levels for VRN2 also did not vary among early- and late-flowering accessions, and VRN2 mRNA levels are likely not rate limiting (Ream et al., 2014). An earlier study by Schwartz et al. (2010) described a potential correlation between different VRN2 alleles and flowering time. The authors did not rule out the effects of population structure and proposed that elucidating the role of VRN2 in B. distachyon will require more in-depth genetic studies. A recent comprehensive analysis of population structure in B. distachyon collections revealed that flowering time, and not geographic origin, is indeed the major distinguishing factor between genotypically distinct clusters (Tyler et al., 2016). Our results confirm VRN2 as an important flowering regulator in the ABR6 × Bd21 mapping population and highlight structural and expression variation between parental accessions. However, none of the SNPs identified in the coding sequence map to the CCT domain. A point mutation in this domain results in a spring growth habit in cultivated Triticum monococcum accessions (Yan et al., 2004). It is unclear whether the structural variation surrounding VRN2 corresponds to the allelic variation observed by Schwartz et al. (2010). Woods and Amasino (2016) hypothesize that, even though VRN2 may not be involved in vernalization control in B. distachyon, it may still possess an ancestral role in flowering regulation. This is further supported by the observation that VRN2 expression is not controlled by VRN1 in B. distachyon, yet VRN2 was found to be a functional repressor of flowering in this species (Woods et al., 2016b). We observed a negative correlation between VRN2 transcript accumulation and vernalization period in ABR6 and Bd21 (Fig. 8B). Similar decreases were observed for ABR6 and Bd21, although transcript abundance in Bd21 was significantly lower than in ABR6 under any vernalization period. Therefore, our identification of natural variation in VRN2 among geographically diverse B. distachyon accessions further supports VRN2 as a core flowering regulator in this nondomesticated grass.
In our study of the natural variation between two morphologically and geographically diverse B. distachyon accessions, we failed to implicate VRN1 as a flowering regulator. However, VRN1 expression during and after cold treatment and the failure of VRN1-silenced lines to flower suggest a conserved role of VRN1 as a promoter of flowering (Woods and Amasino, 2016; Woods et al., 2016b). Interestingly, a QTL in the Bd21 × Bd1-1 B. distachyon mapping population colocalized with VRN1 and the light receptor PHYTOCHROME C (Woods et al., 2016a). Between ABR6 and Bd21, sequence variation was found in the promoter and terminator regions of VRN1, and a strong positive correlation was observed with extended periods of vernalization (Fig. 8A), particularly at 4 weeks of vernalization, which was a critical inflection point for flowering time in ABR6. Despite this sequence and expression variation, VRN1 was not found to contribute to flowering time in the ABR6 × Bd21 mapping population. Interestingly, an assessment of allelic variation in 53 B. distachyon accessions currently available in Phytozome (version 11.0.2; https://phytozome.jgi.doe.gov) found that none of these accessions possess structural variation in the VRN1 annotated coding sequence. These findings suggest that VRN1 is a crucial regulator of flowering in B. distachyon and under strong selection pressure.
CONCLUSION
Thanks to their economic and evolutionary importance, flowering time pathways are of particular interest in the cereals and related grasses. Our report adds to this body of research by using natural variation to map vernalization dependency in a B. distachyon mapping population. Since B. distachyon is partly sympatric with the wild relatives of wheat and barley, it seems likely that the species would have been subjected to similar selective pressure and, therefore, is a useful model for understanding predomestication or standing variation. We investigated this standing variation by assessing the segregation of flowering regulators in a mapping population derived from two geographically diverse accessions of B. distachyon. Notably, we found additional support for the roles of FT and VRN2 in controlling flowering in wild temperate grasses. Additionally, allelic variation may explain the ambiguity around the role of the VRN2 homolog observed in B. distachyon. Further fine-mapping will be required to confirm the roles of these genes in B. distachyon flowering time. However, we also detected novel components in the form of additional QTLs, which reflects the power of studying natural variation in mapping populations derived from phenotypically diverse parents. During population advancement, we observed a variety of additional morphological and pathological characteristics segregating in this population, and it will serve as a useful resource for other researchers investigating standing variation in nondomesticated grasses.
MATERIALS AND METHODS
Plant Growth for Assessing ABR6 and Bd21 Vernalization Responses
Six seeds for Brachypodium distachyon ABR6 and Bd21 were germinated on paper (in darkness at room temperature) and transferred to an equal mixture of the John Innes Cereal Mix and a peat and sand mix (Vain et al., 2008) 4 d after germination. Vernalization was initiated 14 d after germination for 2, 3, 4, 5, 6, 7, or 8 weeks (8-h daylength, 1.2 klux light intensity, and 5°C). The different sets were staggered to ensure that all sets left vernalization on the same date. After vernalization, plants were grown in a Sanyo Versatile Environmental Test Chamber (model MLR-351; 16-h photoperiod, 8 klux light intensity, and 22°C/20°C day/night temperatures) for 35 d and then transferred to a greenhouse without light and temperature control (late April to mid July, 2013; Norwich, UK). Days to flowering was measured from the end of vernalization until the emergence of the first spike and was averaged across all six biological replicates (only five replicates were available for Bd21 after 7 weeks of vernalization). Statistical significance was assessed by pairwise comparisons using Student’s t tests with pooled sd and Bonferroni correction for multiple comparisons.
Resequencing of ABR6
Seedlings were grown in a Sanyo Versatile Environmental Test Chamber (16-h photoperiod, 8 klux light intensity, and 22°C) in an equal mixture of the John Innes Cereal Mix and a peat and sand mix. Seven-week-old plants were placed in darkness for 3 d prior to collecting tissue. Genomic DNA was extracted using a standard cetyl-trimethyl-ammonium bromide protocol, and a library of 800-bp inserts was constructed and sequenced with 100-bp paired-end reads and an estimated coverage of 25.8× on an Illumina HiSeq 2500. Library preparation and sequencing were performed at The Genome Analysis Centre. The resulting reads were mapped to the Bd21 reference sequence (version 1; International Brachypodium Initiative, 2010) with the Galaxy wrapper, which used the BWA (version 0.5.9) aln and sampe options (Li and Durbin, 2009). Polymorphisms between ABR6 and Bd21 were identified with the mpileup2snp and mpileup2indel tools of VarScan (version 2.3.6) using default settings (Koboldt et al., 2009). A de novo assembly was created from the raw ABR6 reads using default settings of the CLC Assembly Cell (version 4.2.0) and default parameters. Potential structural variation between ABR6 and Bd21 was investigated by performing a BLAST search with the Bd21 regions of interest against the ABR6 de novo assembly and mapping contigs for hits with at least 95% identity and an E value under 1e−20 to the Bd21 reference sequence (version 3).
Development of the ABR6 × Bd21 F4 Population and Genetic Map
The B. distachyon accessions ABR6 and Bd21 were crossed, and three ABR6 × Bd21 F1 individuals, confirmed as hybrid by simple sequence repeat marker analysis (data not shown), were allowed to self-pollinate to generate a founder F2 population composed of 155 individuals. After single-seed descent, DNA was extracted from leaf tissue of 114 independent F4 lines using a cetyl-trimethyl-ammonium bromide genomic DNA extraction protocol modified for plate-based extraction (Dawson et al., 2016). SNPs for genetic map construction were selected based on a previously characterized Bd21 × Bd3-1 F2 genetic map to ensure an even distribution of markers relative to physical and genetic distances (Huo et al., 2011). SNPs without additional sequence variation in a 120-bp window were selected every 10 cM. The Agena Bioscience MassARRAY design suite was used to develop 17 assays that genotyped 449 putative SNPs using the iPLEX Gold assay at the Iowa State University Genomic Technologies Facility (Supplemental Data S2). Markers were excluded for being monomorphic (106), dominant (34), or for missing data for the parental controls (33). Heterozygous genotype calls for some markers were difficult to distinguish and classified as missing data. Additional SNPs between ABR6 and Bd21 in six markers developed for the Bd21 × Bd3-1 F2 genetic map (Barbieri et al., 2012) were converted into cleaved-amplified polymorphic sequence markers (Konieczny and Ausubel, 1993; Supplemental Table S7). The integrity of these 282 markers was evaluated using R/qtl (version 1.33-7) recombination fraction plots (Broman et al., 2003). Two markers were removed for not showing linkage, and one marker was moved to its correct position based on linkage. Genetic distances were calculated using the Kosambi function in MapManager QTX (version b20; Manly et al., 2001). Removal of unlinked and redundant markers produced a final ABR6 × Bd21 F4 genetic map consisting of 252 SNP-based markers (Supplemental Data S3). Segregation distortion was assessed using a χ2 test with Bonferroni correction for multiple comparisons.
Plant Growth and Phenotyping of Flowering Time in the ABR6 × Bd21 F4:5 Families
Three to five plants for each of the 114 ABR6 × Bd21 F4:5 families were grown under five different environmental conditions as detailed in Supplemental Table S1. For the phenotyping performed in Aberystwyth, individual seeds were sown in 6-cm pots with a mixture of 20% grit sand and 80% Levington F2 peat-based compost. Seeds were grown for 2 weeks in greenhouse conditions (22°C/20°C and natural light supplemented with 20 h of lighting) and then either maintained in the greenhouse or transferred to a vernalization room for 6 weeks (16-h daylength at 5°C). Plants were returned to the greenhouse following vernalization and grown to maturity. Flowering time was defined as the emergence of the first inflorescence and was measured from the first day that flowering was observed in the entire mapping population. Flowering time was averaged across the individuals of an F4:5 family. For the phenotyping performed in Norwich, plants were first subjected to growth conditions and pathogen assays as described by Dawson et al. (2015). Plants were germinated in a peat-based compost in 1-L pots and grown for 6 weeks in a controlled environment room (18°C/11°C and a 16-h light period). Six weeks post germination, the fourth or fifth leaf of each plant was cut off for pathological assays. The plants were transplanted into 9-cm pots with an equal mixture of the John Innes Cereal Mix and a peat and sand mix (Vain et al., 2008) and transferred to the respective growth environments for flowering assessment (Supplemental Table S1). Flowering time was defined as the emergence of the first inflorescence within an F4:5 family and was measured from the first day that flowering was observed in the entire mapping population. Families that did not flower 60 d after emergence of the first inflorescence in the mapping population were scored as not flowering.
QTL Analysis for Flowering Time
Flowering phenotypes were assessed for normality using the Shapiro-Wilk test (Royston, 1982). In an initial analysis, phenotypic values were converted into a binary classification based on whether families flowered or did not flower. Interval mapping was performed with the scanone function in R/qtl under a binary model with conditional genotype probabilities computed with default parameters and the Kosambi map function (Xu and Atchley, 1996; Broman et al., 2006). Simulation of genotypes was performed with a fixed step distance of 2 cM, 128 simulation replicates, and a genotyping error rate of 0.001. Statistical significance for QTLs was determined by performing 1,000 permutations and controlled at α = 0.05 (Doerge and Churchill, 1996). Nonparametric interval mapping was performed with similar parameters in R/qtl under an np model (Kruglyak and Lander, 1995). For parametric mapping, flowering time data were transformed using the following approaches: T1, the removal of all F4:5 families that did not flower within the time scale of the experiment; T2, transforming all nonflowering phenotypic scores to 1 d above the maximum observed; and T3, transforming by ranking families according to their flowering time. For the third transformation approach (T3), the earliest flowering family was given a rank score of 1, and subsequent ordered families were given incremental scores based on rank (2, 3, 4, etc.). When two or more families had a shared flowering time, they were given the same rank, and the next ranked family was given an incremental rank score based on the number of preceding shared rank families. Nonflowering families were given the next incremental rank after the last flowering rank. For all three transformations, composite interval mapping was performed under an additive model (H0:H1) using QTL Cartographer (version 1.17j) with the selection of five background markers, a walking speed of 2 cM, and a window size of 10 cM (Zeng, 1993, 1994; Basten et al., 2004). Statistical significance for QTLs was determined by performing 1,000 permutations with reselection of background markers and controlled at α = 0.05 (Doerge and Churchill, 1996; Lauter et al., 2008). The 1 − LOD support intervals were estimated based on interval mapping (Lander and Botstein, 1989).
RNAseq of ABR6 and Bd21
Plants were grown in a controlled environment room with 16 h of light at 22°C, and fourth and fifth leaves were harvested as soon as the fifth leaf was fully expanded (roughly 28 d after germination). RNA was extracted using TRI Reagent (Sigma-Aldrich) according to the manufacturer’s specifications. TruSeq libraries were generated from total RNA, and mean insert sizes were 251 and 254 bp for ABR6 and Bd21, respectively. Library preparation and sequencing were performed at The Genome Analysis Centre. Sequencing was carried out using 150-bp paired-end reads on an Illumina HiSeq 2500, and ABR6 and Bd21 yielded 38,867,987 and 37,566,711 raw reads, respectively. RNAseq data quality was assessed with FastQC, and reads were removed using Trimmomatic (version 0.32; Bolger et al., 2014) with parameters set at ILLUMINACLIP:TruSeq 3-PE.fa:2:30:10, LEADING:3, TRAILING:3, SLIDINGWINDOW:4:15, and MINLEN:100. These parameters will remove all reads with adapter sequence, ambiguous bases, or a substantial reduction in read quality. The sequenced reads were mapped to the Bd21 reference genome using the TopHat (version 2.0.9) spliced alignment pipeline (Trapnell et al., 2009).
Reverse Transcription-Quantitative PCR Analyses
ABR6 and Bd21 seeds were surface sterilized (70% ethanol for 30 s, washed in autoclaved deionized water, 1.3% sodium hypochlorite for 4 min, and washed in autoclaved water three times), transferred to moistened Whatman filter paper, left at room temperature in darkness overnight, and vernalized for 2, 4, or 6 weeks (in darkness at 5°C). A control set was surface sterilized and transferred to filter paper overnight but not vernalized. Following vernalization, plants were transferred to soil and grown in a Sanyo Versatile Environmental Test Chamber in conditions similar to environment 2 (20-h photoperiod, 4 klux light intensity, and 22°C/20°C). Once fully expanded, fourth leaves were collected in the middle of the photoperiod and flash frozen in liquid nitrogen.
Total RNA was extracted using TRI Reagent according to the manufacturer’s instructions (Sigma-Aldrich). RNA samples were treated with DNase I (Roche) prior to cDNA synthesis. The quality and quantity of RNA samples were assessed using a NanoDrop spectrophotometer followed by agarose electrophoresis. First-strand cDNA was synthesized according to the manufacturer’s instructions (Invitrogen). Briefly, 1 μg of total RNA, 1 μL of 0.5 μm poly-T primers, and 1 μL of 10 mm deoxyribonucleotide triphosphate were incubated at 65°C for 5 min and 4°C for 2 min, with subsequent reverse transcription reactions performed using 2 μL of 10× reverse transcription buffer, 4 μL of 25 mm MgCl2, 2 μL of 0.1 m dithiothreitol, 1 μL of RNaseOUT (40 units μL−1), and 1 μL of SuperScript III reverse transcriptase (200 units μL−1) at 50°C for 50 min. Reverse transcription was inactivated by incubating at 85°C for 5 min, and residual RNA was removed with the addition of 1 μL of RNase H (2 units μL−1) and incubation at 37°C for 20 min.
Quantitative real-time PCR was performed in 20-μL reaction volumes using 10 μL of SYBR Green mix (Sigma-Aldrich), 1 μL of 10 μm forward and reverse primers, 4 μL of water, and 4 μL of cDNA diluted 10-fold. The program for PCR amplification involved an initial denaturation at 95°C for 3 min and then 40 cycles of 94°C for 10 s, 60°C for 15 s, and 72°C for 15 s. Fluorescence data were collected at 72°C at the extension step and during the melting curve program on a CFX96 Real-Time system (Bio-Rad).
Relative gene expression was determined using the 2−ΔΔCT method described by Livak and Schmittgen (2001) using UBIQUITIN-CONJUGATING ENZYME18 (Hong et al., 2008; Schwartz et al., 2010) for normalization. All primers were used previously by Ream et al. (2014) and had PCR efficiency ranging from 95% to 110%. Statistical analysis of gene expression was performed using R (version 3.2.3). Comparisons between all genotype-by-treatment combinations were made with pairwise Student’s t tests using log-transformed relative expression levels, with P values corrected for multiple hypothesis testing based on the Benjamini-Hochberg approach (Benjamini and Hochberg, 1995).
Accession Numbers
Raw resequencing reads of ABR6 have been submitted to the National Center for Biotechnology Information Short Read Archive under the BioProject identifier PRJNA319372 and SRA accession number SRX1720894. The ABR6 de novo assembly has been deposited at the DNA Data Bank of Japan/European Nucleotide Archive/GenBank under accession number LXJM00000000. The version described in this article is version LXJM01000000. Raw RNAseq reads have been submitted to the National Center for Biotechnology Information Short Read Archive under the BioProject identifier PRJNA319373 and SRA accession numbers SRX1721358 (ABR6) and SRX1721359 (Bd21).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Linkage groups of the ABR6 × Bd21 genetic map.
Supplemental Figure S2. Two-way recombination fraction plot for the ABR6 × Bd21 F4 population.
Supplemental Table S1. Summary of the environmental conditions tested.
Supplemental Table S2. Significant QTLs from interval mapping of the binary classification of flowering time phenotypes in the ABR6 × Bd21 F4:5 families.
Supplemental Table S3. Significant QTLs from interval mapping using a nonparametric model for flowering time phenotypes in the ABR6 × Bd21 F4:5 families.
Supplemental Table S4. Significant QTLs from composite interval mapping of transformed flowering time phenotypes in the ABR6 × Bd21 F4:5 families (T1).
Supplemental Table S5. Significant QTLs from composite interval mapping of transformed flowering time phenotypes in the ABR6 × Bd21 F4:5 families (T2).
Supplemental Table S6. Summary of the structural variation between Bd21 and ABR6 for the flowering regulators Bradi1g48830 (FT), Bradi3g10010 (VRN2), and Bradi1g08340 (VRN1).
Supplemental Table S7. Five cleaved-amplified polymorphic sequence markers included in the ABR6 × Bd21 genetic map design.
Supplemental Data S1. Raw, binary, and transformed flowering time data for the ABR6 × Bd21 F4:5 families in the five environments tested.
Supplemental Data S2. Sequence information used to develop iPLEX assays for the 247 MassARRAY markers in the ABR6 × Bd21 genetic map design.
Supplemental Data S3. ABR6 × Bd21 genetic map.
Supplementary Material
Acknowledgments
We thank John Vogel for sharing preliminary sequencing data, Burkhard Steuernagel for assistance with the de novo assembly of the ABR6 genome, David Garvin and Luis Mur for providing seed, and Claire Collett, Ray Smith, Tom Thomas, and Aliyah Debbonaire for assistance with population progression. MassARRAY genotyping was performed at the Genomic Technologies Facility at Iowa State University.
Glossary
- SNP
single-nucleotide polymorphism
- cM
centimorgan
- QTL
quantitative trait locus
- LOD
log of the odds
- RNAseq
RNA sequencing
- UTR
untranslated region
- indel
insertion/deletion
- PVE
phenotypic variance explained
Footnotes
This work was supported by the Biotechnology and Biological Sciences Research Council Doctoral Training Programme (grant no. BB/F017294/1) and Institute Strategic Programme (grant no. BB/J004553/1), the Gatsby Charitable Foundation, the Leverhulme Trust (grant no. 10754), the 2Blades Foundation, and the Human Frontiers Science Program (grant no. LT000218/2011).
Articles can be viewed without a subscription.
References
- Amasino R. (2010) Seasonal and developmental timing of flowering. Plant J 61: 1001–1013 [DOI] [PubMed] [Google Scholar]
- Andrés F, Coupland G (2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13: 627–639 [DOI] [PubMed] [Google Scholar]
- Barbieri M, Marcel TC, Niks RE, Francia E, Pasquariello M, Mazzamurro V, Garvin DF, Pecchioni N (2012) QTLs for resistance to the false brome rust Puccinia brachypodii in the model grass Brachypodium distachyon L. Genome 55: 152–163 [DOI] [PubMed] [Google Scholar]
- Basten CJ, Weir BS, Zeng ZB (2004) QTL Cartographer, version 1.17. Department of Statistics, North Carolina State University, Raleigh, NC [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57: 289–300 [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K, Lempe J, Epple P, Warthmann N, Lanz C, Dangl JL, Weigel D (2007) Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol 5: e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K, Weigel D (2007) Hybrid necrosis: autoimmunity as a potential gene-flow barrier in plant species. Nat Rev Genet 8: 382–393 [DOI] [PubMed] [Google Scholar]
- Broman KW, Sen S, Owens SE, Manichaikul A, Southard-Smith EM, Churchill GA (2006) The X chromosome in quantitative trait locus mapping. Genetics 174: 2151–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA (2003) R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890 [DOI] [PubMed] [Google Scholar]
- Catalán P, Chalhoub B, Chochois V, Garvin DF, Hasterok R, Manzaneda AJ, Mur LA, Pecchioni N, Rasmussen SK, Vogel JP, et al. (2014) Update on the genomics and basic biology of Brachypodium: International Brachypodium Initiative (IBI). Trends Plant Sci 19: 414–418 [DOI] [PubMed] [Google Scholar]
- Chen A, Dubcovsky J (2012) Wheat TILLING mutants show that the vernalization gene VRN1 down-regulates the flowering repressor VRN2 in leaves but is not essential for flowering. PLoS Genet 8: e1003134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson AM, Bettgenhaeuser J, Gardiner M, Green P, Hernández-Pinzón I, Hubbard A, Moscou MJ (2015) The development of quick, robust, quantitative phenotypic assays for describing the host-nonhost landscape to stripe rust. Front Plant Sci 6: 876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson AM, Ferguson JN, Gardiner M, Green P, Hubbard A, Moscou MJ (2016) Isolation and fine mapping of Rps6: an intermediate host resistance gene in barley to wheat stripe rust. Theor Appl Genet 129: 831–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distelfeld A, Li C, Dubcovsky J (2009) Regulation of flowering in temperate cereals. Curr Opin Plant Biol 12: 178–184 [DOI] [PubMed] [Google Scholar]
- Doerge RW, Churchill GA (1996) Permutation tests for multiple loci affecting a quantitative character. Genetics 142: 285–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper J, Mur LA, Jenkins G, Ghosh-Biswas GC, Bablak P, Hasterok R, Routledge AP (2001) Brachypodium distachyon: a new model system for functional genomics in grasses. Plant Physiol 127: 1539–1555 [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky J, Chen C, Yan L (2005) Molecular characterization of the allelic variation at the VRN-H2 vernalization locus in barley. Mol Breed 15: 395–407 [Google Scholar]
- Faris JD, Laddomada B, Gill BS (1998) Molecular mapping of segregation distortion loci in Aegilops tauschii. Genetics 149: 319–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D, Szücs P, Yan L, Helguera M, Skinner JS, von Zitzewitz J, Hayes PM, Dubcovsky J (2005) Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol Genet Genomics 273: 54–65 [DOI] [PubMed] [Google Scholar]
- Garvin DF, Gu YQ, Hasterok R, Hazen SP, Jenkins G, Mockler TC, Mur LAJ, Vogel JP (2008) Development of genetic and genomic research resources for Brachypodium distachyon, a new model system for grass crop research. Crop Sci 48: S-69 [Google Scholar]
- Higgins JA, Bailey PC, Laurie DA (2010) Comparative genomics of flowering time pathways using Brachypodium distachyon as a model for the temperate grasses. PLoS ONE 5: e10065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SY, Seo PJ, Yang MS, Xiang F, Park CM (2008) Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biol 8: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo N, Garvin DF, You FM, McMahon S, Luo MC, Gu YQ, Lazo GR, Vogel JP (2011) Comparison of a high-density genetic linkage map to genome features in the model grass Brachypodium distachyon. Theor Appl Genet 123: 455–464 [DOI] [PubMed] [Google Scholar]
- International Brachypodium Initiative (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768 [DOI] [PubMed] [Google Scholar]
- Jung C, Müller AE (2009) Flowering time control and applications in plant breeding. Trends Plant Sci 14: 563–573 [DOI] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D (1999) Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Karsai I, Szücs P, Mészáros K, Filichkina T, Hayes PM, Skinner JS, Láng L, Bedö Z (2005) The Vrn-H2 locus is a major determinant of flowering time in a facultative × winter growth habit barley (Hordeum vulgare L.) mapping population. Theor Appl Genet 110: 1458–1466 [DOI] [PubMed] [Google Scholar]
- Koboldt DC, Chen K, Wylie T, Larson DE, McLellan MD, Mardis ER, Weinstock GM, Wilson RK, Ding L (2009) VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25: 2283–2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM (1993) A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J 4: 403–410 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Vreugdenhil D (2004) Naturally occurring genetic variation in Arabidopsis thaliana. Annu Rev Plant Biol 55: 141–172 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Lander ES (1995) A nonparametric approach for mapping quantitative trait loci. Genetics 139: 1421–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Botstein D (1989) Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauter N, Moscou MJ, Habiger J, Moose SP (2008) Quantitative genetic dissection of shoot architecture traits in maize: towards a functional genomics approach. Plant Genome 1: 99 [Google Scholar]
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lu H, Romero-Severson J, Bernardo R (2002) Chromosomal regions associated with segregation distortion in maize. Theor Appl Genet 105: 622–628 [DOI] [PubMed] [Google Scholar]
- Lv B, Nitcher R, Han X, Wang S, Ni F, Li K, Pearce S, Wu J, Dubcovsky J, Fu D (2014) Characterization of FLOWERING LOCUS T1 (FT1) gene in Brachypodium and wheat. PLoS ONE 9: e94171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manly KF, Cudmore RH Jr, Meer JM (2001) Map Manager QTX, cross-platform software for genetic mapping. Mamm Genome 12: 930–932 [DOI] [PubMed] [Google Scholar]
- Muñoz-Amatriaín M, Moscou MJ, Bhat PR, Svensson JT, Bartoš J, Suchánková P, Šimková H, Endo TR, Fenton RD, Lonardi S, et al. (2011) An improved consensus linkage map of barley based on flow-sorted chromosomes and single nucleotide polymorphism markers. Plant Genome 4: 238–249 [Google Scholar]
- Opanowicz M, Vain P, Draper J, Parker D, Doonan JH (2008) Brachypodium distachyon: making hay with a wild grass. Trends Plant Sci 13: 172–177 [DOI] [PubMed] [Google Scholar]
- Ream TS, Woods DP, Amasino RM (2012) The molecular basis of vernalization in different plant groups. Cold Spring Harb Symp Quant Biol 77: 105–115 [DOI] [PubMed] [Google Scholar]
- Ream TS, Woods DP, Schwartz CJ, Sanabria CP, Mahoy JA, Walters EM, Kaeppler HF, Amasino RM (2014) Interaction of photoperiod and vernalization determines flowering time of Brachypodium distachyon. Plant Physiol 164: 694–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routledge APM, Shelley G, Smith JV, Talbot NJ, Draper J, Mur LAJ (2004) Magnaporthe grisea interactions with the model grass Brachypodium distachyon closely resemble those with rice (Oryza sativa). Mol Plant Pathol 5: 253–265 [DOI] [PubMed] [Google Scholar]
- Royston JP. (1982) An extension of Shapiro and Wilk’s W test for normality to large samples. Appl Stat 31: 115 [Google Scholar]
- Sasani S, Hemming MN, Oliver SN, Greenup A, Tavakkol-Afshari R, Mahfoozi S, Poustini K, Sharifi HR, Dennis ES, Peacock WJ, et al. (2009) The influence of vernalization and daylength on expression of flowering-time genes in the shoot apex and leaves of barley (Hordeum vulgare). J Exp Bot 60: 2169–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz CJ, Doyle MR, Manzaneda AJ, Rey PJ, Mitchell-Olds T, Amasino RM (2010) Natural variation of flowering time and vernalization responsiveness in Brachypodium distachyon. BioEnergy Res 3: 38–46 [Google Scholar]
- Tan S, Wu S (2012) Genome wide analysis of nucleotide-binding site disease resistance genes in Brachypodium distachyon. Comp Funct Genomics 2012: 418208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler L, Lee SJ, Young ND, DeIulio GA, Benavente E, Reagon M, Sysopha J, Baldini RM, Troìa A, Hazen SP, et al. (2016) Population structure in the model grass Brachypodium distachyon is highly correlated with flowering differences across broad geographic areas. Plant Genome 9: 1–20 [DOI] [PubMed] [Google Scholar]
- Vain P, Worland B, Thole V, McKenzie N, Alves SC, Opanowicz M, Fish LJ, Bevan MW, Snape JW (2008) Agrobacterium-mediated transformation of the temperate grass Brachypodium distachyon (genotype Bd21) for T-DNA insertional mutagenesis. Plant Biotechnol J 6: 236–245 [DOI] [PubMed] [Google Scholar]
- Vogel JP, Garvin DF, Leong OM, Hayden DM (2006) Agrobacterium-mediated transformation and inbred line development in the model grass Brachypodium distachyon. Plant Cell Tissue Organ Cult 84: 199–211 [Google Scholar]
- von Zitzewitz J, Szücs P, Dubcovsky J, Yan L, Francia E, Pecchioni N, Casas A, Chen TH, Hayes PM, Skinner JS (2005) Molecular and structural characterization of barley vernalization genes. Plant Mol Biol 59: 449–467 [DOI] [PubMed] [Google Scholar]
- Wilczek AM, Burghardt LT, Cobb AR, Cooper MD, Welch SM, Schmitt J (2010) Genetic and physiological bases for phenological responses to current and predicted climates. Philos Trans R Soc Lond B Biol Sci 365: 3129–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson P, Streich J, Borevitz J (2016) Genomic diversity and climate adaptation in Brachypodium. In Vogel JP, ed, Genetics and Genomics of Brachypodium. Springer International Publishing, Cham, Switzerland, pp 107–127 [Google Scholar]
- Woods D, Amasino R (2016) Dissecting the control of flowering time in grasses using Brachypodium distachyon. In Vogel JP, ed, Genetics and Genomics of Brachypodium. Springer International Publishing, Cham, Switzerland, pp 259–273 [Google Scholar]
- Woods D, Bednarek R, Bouché F, Gordon SP, Vogel J, Garvin DF, Amasino R (2016a) Genetic architecture of flowering-time variation in Brachypodium distachyon. Plant Physiol 172: 269–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods DP, McKeown MA, Dong Y, Preston JC, Amasino RM (2016b) Evolution of VRN2/Ghd7-like genes in vernalization-mediated repression of grass flowering. Plant Physiol 170: 2124–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods DP, Ream TS, Minevich G, Hobert O, Amasino RM (2014) PHYTOCHROME C is an essential light receptor for photoperiodic flowering in the temperate grass, Brachypodium distachyon. Genetics 198: 397–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Atchley WR (1996) Mapping quantitative trait loci for complex binary diseases using line crosses. Genetics 143: 1417–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zhu L, Xiao J, Huang N, McCouch SR (1997) Chromosomal regions associated with segregation distortion of molecular markers in F2, backcross, doubled haploid, and recombinant inbred populations in rice (Oryza sativa L.). Mol Gen Genet 253: 535–545 [DOI] [PubMed] [Google Scholar]
- Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J (2006) The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci USA 103: 19581–19586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J (2004) The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303: 1640–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J (2003) Positional cloning of the wheat vernalization gene VRN1. Proc Natl Acad Sci USA 100: 6263–6268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng ZB. (1993) Theoretical basis for separation of multiple linked gene effects in mapping quantitative trait loci. Proc Natl Acad Sci USA 90: 10972–10976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng ZB. (1994) Precision mapping of quantitative trait loci. Genetics 136: 1457–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.