Genome-wide transcriptomic data in combination with H3K27me3 and protein localization data unveiled the roles of the AtBMI1s and their possible relationship with other PRC1 components.
Abstract
Polycomb Group regulation in Arabidopsis (Arabidopsis thaliana) is required to maintain cell differentiation and allow developmental phase transitions. This is achieved by the activity of three PcG repressive complex 2s (PRC2s) and the participation of a yet poorly defined PRC1. Previous results showed that apparent PRC1 components perform discrete roles during plant development, suggesting the existence of PRC1 variants; however, it is not clear in how many processes these components participate. We show that AtBMI1 proteins are required to promote all developmental phase transitions and to control cell proliferation during organ growth and development, expanding their proposed range of action. While AtBMI1 function during germination is closely linked to B3 domain transcription factors VAL1/2 possibly in combination with GT-box binding factors, other AtBMI1 regulatory networks require participation of different factor combinations. Conversely, EMF1 and LHP1 bind many H3K27me3 positive genes up-regulated in atbmi1a/b/c mutants; however, loss of their function affects expression of a different subset, suggesting that even if EMF1, LHP1, and AtBMI1 exist in a common PRC1 variant, their role in repression depends on the functional context.
The evolutionarily conserved Polycomb Group (PcG) machinery plays a crucial role in maintaining repression of genes that are not required during a specific cell fate (Ringrose and Paro, 2004). PcG proteins form multiprotein complexes with different histone modifying activities, including PcG repressive complex 2 (PRC2), which possesses histone H3 Lys 27 (H3K27) trimethyltransferase activity (Müller et al., 2002), and PRC1, which has histone H2A Lys 119 E3 ubiquitin ligase activity (Cao et al., 2005) as well as other nonenzymatic functions critical for chromatin compaction (Francis et al., 2004). The combined activity of both complexes is required for stable repression of target genes.
In Drosophila, single-copy genes encode the four core subunits of PRC2: Suppressor of Zeste 12 [Su(z)12], Extra sex combs (Esc), p55, and the catalytic subunit Enhancer of Zeste [E(z); Simon and Kingston, 2013]. Arabidopsis (Arabidopsis thaliana) has three E(z) homologs, CURLY LEAF (CLF), MEDEA (MEA), and SWINGER (SWN; Goodrich et al., 1997; Grossniklaus et al., 1998; Chanvivattana et al., 2004), and three Su(z)12 homologs, EMBRYONIC FLOWER2 (EMF2), VERNALIZATION2 (VRN2), and FERTILIZATION INDEPENDENT SEED2 (FIS2; Luo et al., 1999; Gendall et al., 2001; Yoshida et al., 2001), while MULTIPLE SUPPRESSOR OF IRA1 (MSI1), which is one of the five p55 homologs in Arabidopsis (Hennig et al., 2005), and the Esc homolog FERTILIZATION INDEPENDENT ENDOSPERM (FIE; Ohad et al., 1999) are common subunits to the different possible PRC2s (Mozgova et al., 2015).
Drosophila PRC1 contains Polycomb (Pc), Polyhomeotic (Ph), Posterior sex comb (Psc), and dRing1 (Shao et al., 1999; Peterson et al., 2004), each with multiple homologs in vertebrates (Schwartz and Pirrotta, 2013). Furthermore, vertebrate PRC1 complexes exist in canonical or noncanonical forms. Canonical variants harbor homologs to the four Drosophila core subunits (Schwartz and Pirrotta, 2013), while noncanonical PRC1 complexes contain RING1A or RING1B and one of the six different homologs of Drosophila Psc (PCGF) to form a H2A monoubiquitination module, along with additional subunits that further add specific biochemical properties and genomic localization to the different variants (Schwartz and Pirrotta, 2013). In Arabidopsis, several pieces of evidence suggest a similar high degree of complexity (Förderer et al., 2016). Two RING1 homologs, AtRING1A and AtRING1B, and three Psc/PCGF homologs, AtBMI1A, AtBMI1B, and AtBMI1C, have been characterized (Sanchez-Pulido et al., 2008; Xu and Shen, 2008; Bratzel et al., 2010, 2012; Chen et al., 2010; Yang et al., 2013; Calonje, 2014). Plants with mutations in these genes suggest a high degree of functional redundancy between AtRING1 or AtBMI1 proteins; thus, it is not clear whether each paralog can regulate a different subset of targets (Bratzel et al., 2010; Chen et al., 2010; Yang et al., 2013). The analysis is complicated by the observation that several mutant alleles are knock-downs rather than null alleles and that phenotypes show a wide range of stochastic variation among segregating siblings with “weak” and “strong” phenotypes (Bratzel et al., 2010; Chen et al., 2010).
Two other plant-specific proteins have been related to PRC1, EMBRYONIC FLOWER1 (EMF1) mediating chromatin compaction (Calonje et al., 2008; Beh et al., 2012) and LIKE-HETEROCHROMATIN PROTEIN1 (LHP1), which, as Drosophila Pc, binds H3K27me3 marks through its chromodomain (Turck et al., 2007). Although both proteins can interact with either AtRING1 or AtBMI1 (Bratzel et al., 2010; Chen et al., 2010), recent reports showed that they also copurify with PRC2 components (Derkacheva et al., 2013; Liang et al., 2015); thus, it is not clear in which context they carry out their functions. Additional proteins with chromatin-related functions have been shown to participate in PRC1-mediated repression of specific target genes, such as the VIVIPAROUS1 (VP1)/ABSCISIC ACID INSENSITIVE3 (ABI3)-Like1 and 2 proteins (VAL1/2; Yang et al., 2013), ALFIN1-like proteins (ALs; Molitor et al., 2014), and JMJ14 (Wang et al., 2014).
In plants, PcG repression maintains the differentiated state of the cells but also orchestrates developmental phase transitions by controlling the establishment of new cell identities. This likely requires different PRC1s, but little is known about their subunit composition. The repression of several seed maturation genes after germination requires the AtBMI1 and AtRING1 proteins (Bratzel et al., 2010; Chen et al., 2010; Yang et al., 2013), and a recent genome-wide study showed gene networks regulated by AtBMI1s and AtRING1s during the suppression of seed development in seedlings (Wang et al., 2016). As these results were derived from the analysis of atring1a/b and atbmi1a/b mutants developing a weak phenotype (Bratzel et al., 2010; Chen et al., 2010), their possible implication in other developmental processes or stages was not unveiled. Conversely, the repression of flower homeotic genes in seedlings requires EMF1 (Kim et al., 2012) and LHP1 (Gaudin et al., 2001), but their role in regulating other processes is not clear.
In this work, by analyzing the transcriptome of single, strong double, and triple atbmi1 mutants, we have identified a more comprehensive set of candidate genes regulated by AtBMI1 proteins. Our results indicate that in addition to switching off the seed maturation program after germination, AtBMI1s promote the transition from each developmental phase to the next throughout development and furthermore control cell proliferation during organ growth and development. By integrating transcriptomics datasets with previously published data, we show that AtBMI1 and VAL1/2 act together only in the regulation of seed maturation genes. Enrichment of cis-regulatory elements at VAL1/2-dependent and -independent genes suggests that AtBMI1-mediated gene repression requires different combinational modules always involving VAL-related B3 domain factors. Conversely, while EMF1 and LHP1 occupy a considerable number of genes up-regulated in atbmi1a/b/c mutants, loss of their function does not impact the expression of most but affects the expression of a different subset of genes. Together these results suggest that the different PRC1 variants may differ in subunit composition but also in the role that single components play all depending on the cis-regulatory context.
RESULTS
Genome-Wide Transcriptomic Data Analysis of atbmi1 Mutants
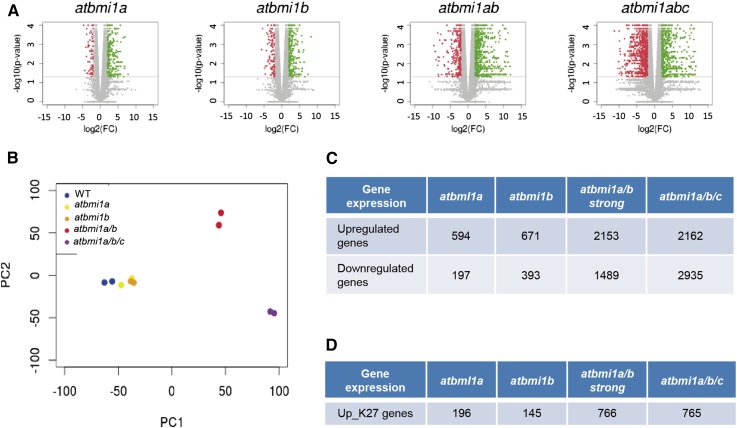
Previous data have suggested that AtBMI1A and AtBMI1B are ubiquitously expressed and act mostly redundantly throughout development (Bratzel et al., 2010), whereas AtBMI1C, which is expressed in roots, endosperm, and stamen, may have functionally diverged since it cannot fully rescue atbmi1a/b defects when overexpressed (Yang et al., 2013; Merini and Calonje, 2015); nevertheless, atbmi1a/c and atbmi1b/c do not show phenotypic alterations (Yang et al., 2013), suggesting that loss of AtBMI1C function is compensated by the other two AtBMI1s. Therefore, to gain insight into the regulatory roles of AtBMI1s, we performed genome-wide transcriptome analysis using RNA sequencing (RNA-seq) of wild-type Col-0, atbmi1a, atbmi1b, atbmi1a/b, and atbmi1a/b/c mutants at 10 d after germination (DAG). Since individual atbmi1a/b double mutants display a wide range of phenotypes (Bratzel et al., 2010), we chose to select the strong atbmi1a/b mutant phenotype for the analysis, which differs from the atbmi1a/b/c phenotype mainly in the root (Yang et al., 2013; Supplemental Fig. S1). The Tuxedo protocol (Trapnell et al., 2012) was used for transcript assembly and differential expression analysis. All sequencing samples were of high quality (Supplemental Fig. S2; Supplemental Table S1). Differentially expressed genes were determined using stringent criteria consisting of a combination of fold change >4 and a P value < 0.05. The number of genes scored as present in at least one of our samples was 24,503, representing 72.96% of the entire Arabidopsis transcriptome. We found <3% to 4% of the surveyed transcriptome affected in single mutants and around 15% and 20% differentially expressed in strong atbmi1a/b double and atbmi1a/b/c triple mutants, respectively (Fig. 1A; Supplemental Fig. S3). Principal components analysis showed that the transcriptomes of wild type, atbmi1a, and atbmi1b mutants clustered together, whereas the transcriptomes of atbmi1a/b and atbmi1a/b/c mutants constituted two distant and distinct clusters, indicating not only differences from the wild type and single mutant group but also in between (Fig. 1B). In any case, we found a considerable number of genes misregulated in the single mutants (Fig. 1C; Supplemental Table S2), of which a majority were a subset of those affected in double and triple mutants (Supplemental Fig. S4, A and B). The number of up-regulated genes for atbmi1a, atbmi1b, and atbmi1a/b was higher than down-regulated (Fig. 1C), which might confirm the role of AtBMI1 proteins in transcriptional repression. However, atbmi1a/b/c mutant showed higher number of down-regulated genes than up-regulated genes. This may be a consequence of the developmental stage of these mutants, in which all organs are stuck in a seed maturation phase. Up-regulation of some genes within this context may have a stronger negative impact on gene expression.
Figure 1.
Transcriptome analysis of wild type and selected atbmi1 mutants at 10 DAG. A, Volcano plots representing differentially expressed genes in atbmi1 mutants compared to wild type according to a 4-fold change and a P value of 0.05. Green color indicates significantly up-regulated genes and red color significantly down-regulated genes. B, Principal component analysis of the transcriptomes showing that wild type, atbmi1a, and atbmi1b cluster together, whereas atbmi1a/b and atbmi1a/b/c constitute two distinct clusters. C, Differentially expressed genes in the different genotypes, where the number of up- and down-regulated genes is indicated. D, Number of genes that were up-regulated in the different mutants and H3K27me3 marked in wild-type seedlings of the same age (up_K27).
Globally, the upregulated genes in the strong atbmi1a/b and atbmi1a/b/c mutants (Supplemental Figs. S5A and S6A) showed overrepresentation of Gene Ontology terms associated with response to different stimuli (e.g. water stress, temperature, hormones) and lipid metabolism (e.g. transport, biosynthesis, storage), whereas the down-regulated genes were enriched for Gene Ontology terms related to photosynthesis and metabolic processes (Supplemental Figs. S5B and S6B). This is consistent with the developmental fate of the mutants, which are trapped in the seed maturation phase (Yang et al., 2013). During this phase, seeds acquire desiccation tolerance and accumulate storage reserves, prevailing in the form of lipids (Vicente-Carbajosa and Carbonero, 2005), while chloroplast structure is disrupted (Delmas et al., 2013).
As PcG function is involved in the repression of master regulatory genes (Xiao and Wagner, 2015), misregulation in the different atbmi1 mutants may be an indirect or direct consequence of the loss of AtBMI1 function, or a mix of both. Conversely, a considerable number of AtBMI1 direct target genes may not display altered expression in the absence of their upstream transcriptional activators, as has been reported for other PcG loss of function mutants (Bouyer et al., 2011; Kim et al., 2012; Derkacheva et al., 2013). In any case, although the interrelationship between PRC1 and PRC2 is not clear yet, the activity of both complexes is required for stable PcG-mediated repression; therefore, selecting genes up-regulated in atbmi1 mutants and H3K27me3 marked in wild-type seedlings should enrich for a subset of candidate genes directly controlled by AtBMI1s. Accordingly, we intersected genes up-regulated in the different mutants with a set of 5360 H3K27me3 target genes previously identified in two independent analyses in seedlings (Supplemental Table S3; Bouyer et al., 2011; Kim et al., 2012) to selected up-regulated H3K27me3 positive (up_K27) genes (Fig. 1D). The analysis showed significant overlaps between H3K27me3 marked genes and up-regulated genes in the different mutants except for atbmi1b, probably because it is a knock-down mutant (Bratzel et al., 2010). The same analysis using down-regulated genes showed nonsignificant overlaps in all cases excluding atbmi1a/b/c due to the high number of down-regulated genes in this mutant (Supplemental Fig. S7; Supplemental Table S3).
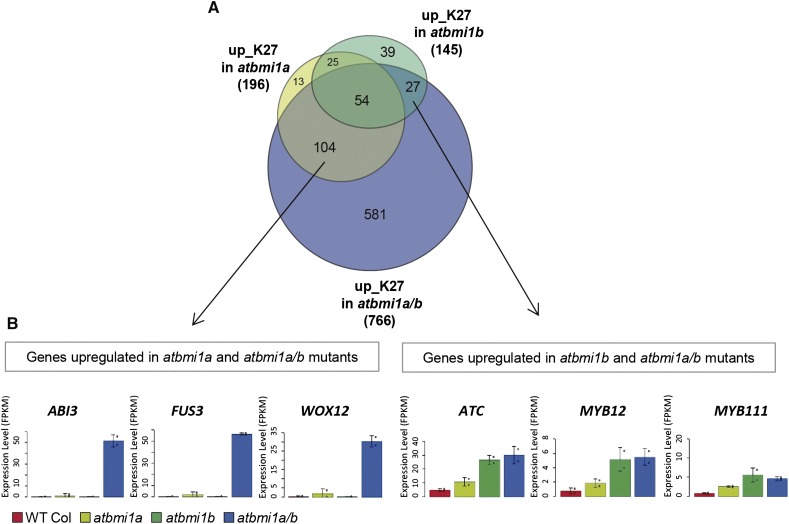
To determine whether there were AtBMI1A and AtBMI1B specific candidate targets, we compared up_K27 genes in the single and double mutants (Fig. 2A). Their number in the double mutant was considerably higher than in the single mutants, illustrating a high degree of functional redundancy. Also, most of the up_K27 genes in single mutants were included in the double mutants set of up_K27 genes; however, a group of genes seemed to be exclusively up-regulated in atbmi1a and atbmi1a/b or in atbmi1b and atbmi1a/b (104 and 27 genes, respectively). Up_K27 genes in atbmi1a and atbmi1a/b were expressed at very low levels in both single compared to the double mutants (Fig. 2B), indicating redundant regulation by AtBMI1A and B. The atbmi1b mutant shows some remnant expression of AtBMI1B, possibly explaining higher expression in atbmi1a versus atbmi1b and the greater number of affected genes in the atbmi1a single mutant (Bratzel et al., 2010). Nevertheless, some genes were indeed specifically sensitive to AtBMI1B being more affected in atbmi1b than atbmi1a and not further increased in double mutants (Fig. 2B).
Figure 2.
Genes regulated by AtBMI1A and AtBMI1B. A, Venn diagram showing the number of up_K27 genes that overlap among atbmi1a, atbmi1b, and atbmi1a/b mutants. All overlaps are significant with P < 2.2 × 10−16 and odds ratios >17 according to Fisher’s exact test. B, Expression of levels of genes that were apparently specifically up-regulated in atbmi1a or atbmi1b mutants in the different genotypes.
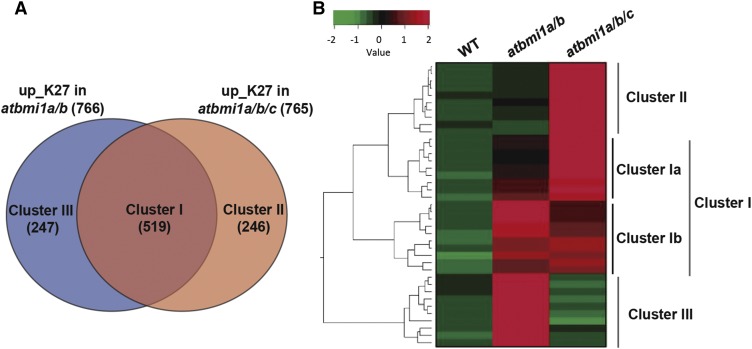
We next investigated the degree of redundancy between AtBMI1A/B and AtBMI1C by comparing the genes up_K27 in atbmi1a/b and atbmi1a/b/c (Fig. 3A). Clustering analysis showed that atbmi1a/b and atbmi1a/b/c shared two-thirds of the up_K27 genes (Cluster I, Supplemental Table S3) but the remaining one-third was genotype-specific (Cluster II, atbmi1a/b/c specific; and Cluster III, atbmi1a/b specific). The expression pattern of genes in Cluster I fell into two distinct subgroups. Cluster Ia included genes that displayed a gradual increase of expression in double and triple mutants, suggesting redundant regulation by AtBMI1A/B and AtBMI1C (Fig. 3B; Supplemental Fig. S8A). Cluster Ib contained genes whose regulation may depend exclusively on AtBMI1A/B, as the loss of AtBMI1C function did not affect significantly their overall expression levels (Fig. 3B; Supplemental Fig. SA8). Cluster II (Supplemental Table S3) included genes exclusively up-regulated in atbmi1a/b/c, indicating that these are AtBMI1C specific targets or, alternatively, that AtBMI1C fully compensates the loss of AtBMI1A/B function in regulating these genes (Fig. 3B; Supplemental Fig. S8A). To discern between these two possibilities, we measured the levels of a subset of cluster II genes in wild type, atbmi1c single, and atbmi1a/b/c mutants in whole seedlings and roots at 10 DAG by quantitative reverse transcription PCR (qRT-PCR). As they were not misexpressed in atbmi1c single mutants (Supplemental Fig. S8B), we concluded that AtBMI1C compensates for the loss of AtBMI1A/B function in the regulation of these genes. Finally, genes in Cluster III (Supplemental Table S3) were exclusively up-regulated in atbmi1a/b mutants, but not in atbmi1a/b/c (Fig. 3B; Supplemental Fig. S8A). Although a priori unexpected, the result can be explained if the activation of these genes requires a developmental stage that is not reached in atbmi1a/b/c.
Figure 3.
Functional redundancy between AtBMI1A/B and AtBMI1C. A, Clustering analysis of genes up_K27 in atbmi1a/b and atbmi1a/b/c mutants. This is a significant overlap with P < 2.2 × 10−16 and an odds ratio >21 according to Fisher’s exact test. B, Expression levels in wild type, atbmi1a/b, and atbmi1a/b/c of genes from the different clusters. The color code represents normalized expression values measured in FPKM.
All together, these data indicated that AtBMI1A and B regulate genes predominantly redundantly, whereas AtBMI1C affects only a subset of AtBMI1A/B possible targets.
Deregulated Developmental Programs in atbmi1 Mutants
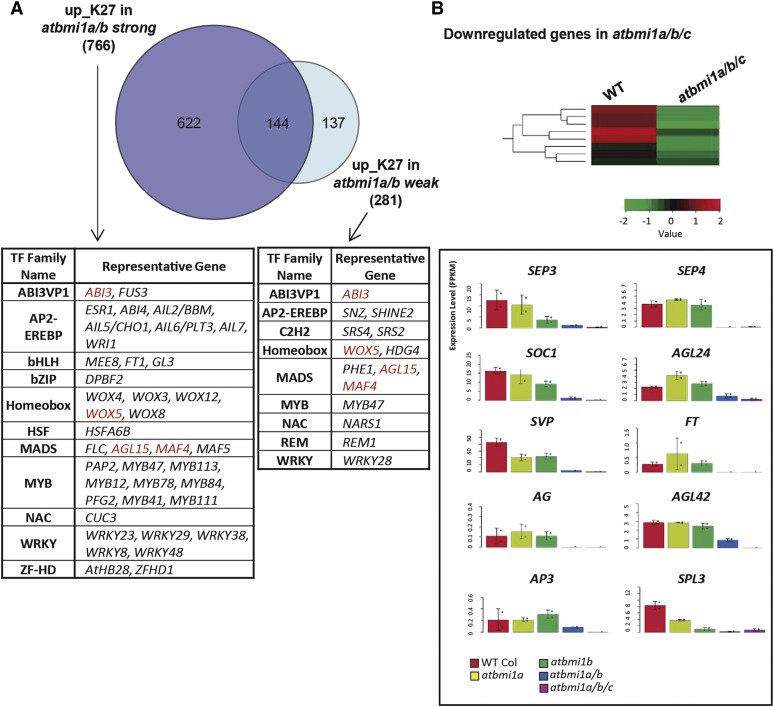
AtBMI1 proteins were previously shown to participate in the regulation of several seed maturation- (Bratzel et al., 2010; Chen et al., 2010; Yang et al., 2013) and germination-related genes (Molitor et al., 2014). In addition, a recent transcriptome analysis of atbmi1a/b weak phenotype confirmed the role of AtBMI1 function in regulating seed development (Wang et al., 2016). When we compared the H3K27me3 up-regulated genes in the atbmi1a/b weak (fold change ≥2, according to Wang et al., 2016; Supplemental Table S1) to those in atbmi1a/b strong phenotype mutants, we found significantly more genes in the stronger mutant (Fig. 4A). Among the genes up-regulated in both datasets were genes previously identified as AtBMI1 target genes, like ABI3, and DELAY OF GERMINATION1 (DOG1); however, other well-known AtBMI1 targets, such as FUSCA3 (FUS3) or BABYBOOM (BBM; Yang et al., 2013), were included only in the atbmi1a/b strong dataset. A similar picture was obtained comparing atbmi1a/b weak and atbmi1a/b/c datasets (Supplemental Fig. S9). Therefore, to obtain a more comprehensive picture of the developmental processes regulated by AtBMI1s, we examined the annotated developmental functions of up_K27 genes in atbmi1a/b/c mutants, as they displayed the strongest developmental alterations.
Figure 4.
Different gene expression patterns of atbmi1a/b weak and strong mutants. A, Venn diagram showing overlap between the genes up_K27 in atbmi1a/b weak and strong mutants. The overlap is significant with P < 2.2 × 10−16 and an odds ratio >15 according to Fisher’s exact test. Some representative TFs in each dataset are indicated. TFs found in the two data sets are highlighter in red. B, Key flowering genes are down-regulated in atbmi1a/b/c mutants. The color code in upper panel represents normalized expression values measured in FPKM.
Seed Maturation and Dormancy
Changes in the triple atbmi1a/b/c mutant uncovered additional genes involved in seed maturation and abscisic acid (ABA) response, such as FUS3 and ABI4, and in seed dormancy, like SOMNUS (SOM). Also, there were genes involved in regulating carbohydrate and lipid metabolism, like WRINKLED1 (Supplemental Fig. S9; Supplemental Table S3). Most of these genes are switched off after germination in wild type; however, the ABIs are required for plant responses to various biotic and abiotic stresses (Cutler et al., 2010), suggesting involvement of AtBMI1s in regulating responses to environmental conditions.
Endosperm-Specific Genes
Maturation genes were not the only seed genes up-regulated in atbmi1a/b/c mutants. We found up-regulation of genes that are predominantly expressed in endosperm but not in the seed coat and vegetative tissues (Wolff et al., 2011). Interestingly, among these were genes displaying a maternal (FLOWERING WAGENINGEN, HOMEODOMAIN GLABROUS8, and AtBMI1C) or paternal (PICKLE RELATED2 [PKR2], VARIANT IN METHYLATION5, AT2G21930, and AT3G49770) preferred expression in the endosperm (Supplemental Fig. S9; Supplemental Table S3).
Meristem Maintenance and Cell Proliferation-Related Genes
The atbmi1a/b/c mutant also up-regulated genes involved in meristem maintenance and cell proliferation throughout plant life. Remarkably, two gene families with crucial roles in these processes were up-regulated in the mutants. The first encompassed the PLETHORA (PLT) or AINTEGUMENTA-LIKE (AIL) genes. Six of eight members of this family were up_K27 in atbmi1a/b/c mutants (PLT1/2/3/5/7 and BBM; Supplemental Figs. S9 and S10; Supplemental Table S3). Some of these PLT genes have overlapping roles in regulating embryo patterning, shoot and root apical meristem maintenance, and organ primordia initiation (Horstman et al., 2014). The second was the WUS homeobox-containing (WOX) gene family, which comprises 14 members (van der Graaff et al., 2009), among which WUS and WOX2/3/4/5/8/9/11/12 were up-regulated in atbmi1a/b/c mutants (Supplemental Figs. S9 and S10; Supplemental Table S3). These factors promote cell division and prevent premature cell differentiation, which are crucial processes required for stem-cell maintenance and organ formation. In addition, we found up-regulation of other genes with related functions, for instance, CUP SHAPED COTYLEDON3, ENHANCER OF SHOOT REGENERATION1, and GROWTH REGULATING FACTOR5.
Root Development-Specific Genes
Apart from the genes involved in root meristem maintenance, we found in atbmi1a/b/c up-regulation of genes that play a crucial role in postembryonic root development, as CEGENDUO, MAGPIE, INDOLE-3-ACETIC ACID INDUCIBLE30, the ROOT MERISTEM GROWTH FACTOR2, and the Class IIB NAC transcription factor SOMBRERO (SMB), underpinning the importance of AtBMI1 function for root development (Supplemental Fig. S9; Supplemental Table S3).
Other Developmental Genes
Among the up_K27 genes in atbmi1a/b/c mutants were genes involved in regulating other developmental processes, such as gametophyte development, leaf development, and the flowering transition (e.g. KANADI2, KNUCKLES, DEVELOPMENT-RELATED PcG TARGET IN THE APEX4, SEPALLATA2, FLOWERING LOCUS C [FLC], MADS AFFECTING FLOWERING4 [MAF4], MAF5, FACTOR PROMOTING FLOWERING1; Supplemental Fig. S9; Supplemental Table S3).
Secondary Metabolic Processes
In addition, atbmi1a/b/c mutants up-regulated genes involved in secondary metabolic processes like those involved in phenylpropanoid metabolism. Up-regulated genes involved in this pathway were CHALCONE SYNTHASE (TRANSPARENT TESTA4 [TT4], CHALCONE ISOMERASE [TT5], FLAVONOID 3′-HYDROXYLASE [TT7], DIHYDROFLAVONOL 4-REDUCTASE, and transcription factors [TFs] such as AtMYB90 [PRODUCTION OF ANTHOCYANIN PIGMENT2]), AtMYB111, and AtMYB11 (Supplemental Fig. S9; Supplemental Table S3).
In summary, AtBMI1 function in Arabidopsis is required to regulate more developmental processes than previously thought.
Regulatory Cross Talk between Chromatin Complexes
RNA-seq data revealed up-regulation of several PcG or PcG-related genes in atbmi1a/b/c mutants, like AtRING1A, AtRING1B, VAL1, VAL2, and VIN3. Conversely, we did not find a significant change in the expression of CLF, SWN, MEA, EMF2, VRN2, FIS2, MSI1, FIE, EMF1, and LHP1 (Supplemental Fig. S10). On the other hand, the Trithorax Group genes ULTRAPETALA1 (ULT1), ULT2, and PKR2 that act antagonistically to PcG complexes were up-regulated in atbmi1a/b/c mutants (Supplemental Fig. S11). Misregulation of some of these chromatin factors could contribute to the strongly altered expression pattern of atbmi1a/b/c mutants.
Several Master Regulators of the Flowering Program Are Down-Regulated in atbmi1a/b/c Mutants
Several MADS-box transcription factors required to specify floral meristem identity or involved in floral organ development were down-regulated in atbmi1a/b/c mutants (Fig. 4B; Supplemental Table S2; e.g. AGL42, SUPPRESSOR OF CONSTANS1 [SOC1], SEP3, SEP4, AGL24, SHORT VEGETATIVE PHASE), but also other key regulatory flowering genes, such as TEMPRANILLO1 and several SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPLs; e.g. SPL2, 3, 4, 8, 12). In addition, we found that some flowering factors that have basal expression levels in wild-type seedlings at 10 DAG expressed at lower levels in atbmi1a/b/c (e.g. AGAMOUS [AG], APETALA3, FLOWERING LOCUS T [FT]; Fig. 4B) The fact that the flowering program seems to be more repressed in atbmi1a/b/c mutants than in wild-type seedlings points to a requirement of AtBMI1 function for proper regulation of flower development.
VAL1/2 and the AtBMI1s Coregulate a Subset of Potential AtBMI1 Targets
VAL1/2 and AtBMI1 proteins are required for the initial repression of several seed maturation genes after germination, such as FUS3, LEC1, and ABI3. Furthermore, we previously showed that the VAL1/2 recruit AtBMI1 proteins to these genes; accordingly, val1/2 and atbmi1a/b/c mutants display a very similar phenotype (Yang et al., 2013). However, WUS is an AtBMI1 but not a VAL1/2 regulated gene, indicating that there are also differences between those mutants (Yang et al., 2013). To determine to which extent the VAL1/2 and AtBMI1 proteins act together in regulating gene expression, we compared genes up-regulated in val1/2 (Suzuki et al., 2007; Supplemental Table S2) and H3K27me3 marked in wild type according to our dataset (Supplemental Table S3) with up_K27 genes in atbmi1a/b/c (Fig. 5A). We found that 70% of val1/2 up_K27 genes were included in the up_K27 atbmi1a/b/c dataset; these genes represented one-third of the genes up_K27 in atbmi1a/b/c, indicating that, despite the fact that they coregulate a considerable number of genes, AtBMI1 proteins clearly perform functions independently of VAL1/2.
Figure 5.
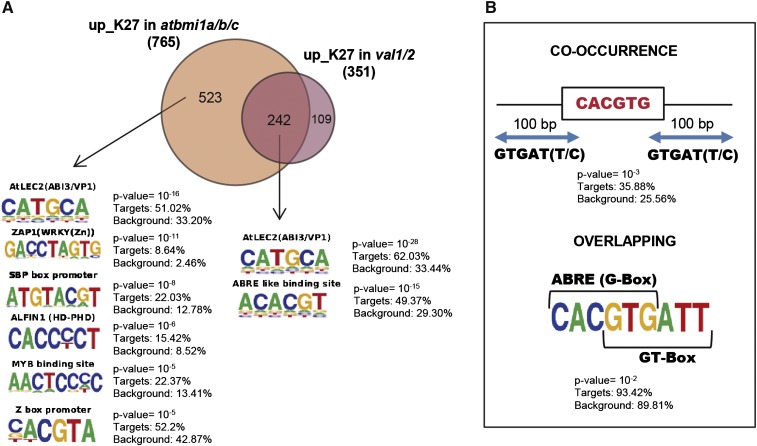
Interplay of AtBMI1 proteins with VAL1/2 proteins. A, Venn diagram showing overlap between the genes up_K27 in atbmi1a/b/c and val1/2 mutants. Sequence LOGOs of cis-regulatory elements enriched only in up_K27 atbmi1a/b/c and in atbmi1a/b/c and val1/2 overlapping genes. B, Co-occurrence and overlapping of ABRE/G-box and GT-box at the promoter of AtBMI1/VAL1/2 coregulated genes. P values and percentage in targets and background are indicated.
The VAL proteins (VAL1, 2, and 3) belong to a subfamily of plant-specific B3 domain-containing proteins (Swaminathan et al., 2008) that is predicted to bind to LEC2/ABI3/VP1 elements (also known as RY elements [CATGCA]; Suzuki et al., 2007); in fact, a recent report showed that a point mutation in a LEC2/ABI3/VP1 element located at the first intron of FLC prevents the epigenetic silencing of the gene during vernalization (Qüesta et al., 2016). FLC is up-regulated in val1/2 and atbmi1 mutants (Supplemental Tables S2 and S3). Therefore, we investigated whether this or other cis-regulatory motifs were enriched at the promoter of AtBMI1/VAL1/2 coregulated genes. Indeed, we found enrichment of LEC2/ABI3/VP1 motifs but also of ABA responsive elements (ABRE; ACGT or G-box; Choi et al., 2000; Fig. 5A). ABRE/G-box elements are recognized by bZIP transcription factors such as ABI5 (Carles et al., 2002). LEC2/ABI3/VP1 and ABRE elements are clustered in the 5′ upstream regions of genes regulated by ABI3/VP1 factors and ABA (Suzuki et al., 2005) and are required for the correct expression of seed maturation genes (Santos-Mendoza et al., 2008). On the other hand, the plant-specific trihelix DNA binding protein ARABIDOPSIS 6B-INTERACTING PROTEIN 1-LIKE1 (ASIL1) that is involved in the repression of seed maturation genes after germination binds GT-box elements (GTGATT and variations of this; Gao et al., 2009). These elements are closely associated with ABRE/G-box and LEC2/ABI3/VP1 elements at the promoter of several seed maturation genes. Furthermore, GT-box elements frequently overlap with ABRE/G-box elements, leading to the proposal that ASIL1 represses embryonic genes by competing with the binding of transcriptional activators (Gao et al., 2009). Therefore, we looked for co-occurrence of both elements at the promoter of AtBMI1/VAL1/2 coregulated genes. Co-occurrence was indeed significant (Fig. 5B); moreover, both elements significantly overlapped at the promoter of these genes (Fig. 5B). Therefore, the combination of LEC2/ABI3/VP1 and GT-box co-occurring with ABRE/G-box elements represents a landmark for the subset of AtBMI1/VAL1/2 coregulated genes.
Surprisingly, the LEC2/ABI3/VP1 elements were as highly overrepresented at promoter regions of genes exclusively up_K27 in atbmi1a/b/c, which suggests that their repression may be functionally connected to other B3 domain transcription factors. The specific combination of LEC2/ABI3/VP1 and ABRE/GT-box elements was not detected in this group. Conversely, other motifs were enriched in the VAL1/2-independent up_K27 subset, such as SQUAMOSA BINDING PROTEIN-, ZAP1-, ALFIN1-, and MYB-binding sites and a frequent Z-box promoter motif that is bound by a new class of transcription factors, the Z-box BINDING FACTORS, whose roles in regulating plant development have just started to be unraveled (Gangappa et al., 2013; Fig. 5A). ALFIN1 elements are bound by plant-specific ALFIN1-like proteins (AL1–7; Lee et al., 2009), which mediate gene repression (Wei et al., 2015) and interact with AtRING1 and AtBMI1 (Molitor et al., 2014), supporting the existence of other combinatorial modules involving B3 domain factors and diverse partners for AtBMI1-mediated gene repression.
Regulatory Networks of AtBMI1, EMF1, and LHP1
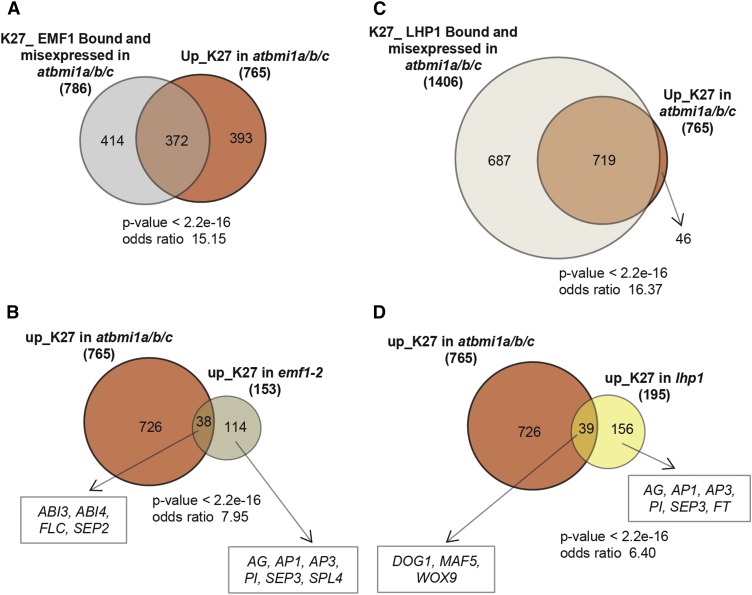
To investigate the functional relationship between AtBMI1 proteins and EMF1, we compared direct EMF1 targets as previously determined through genome-wide ChIP-chip analysis (Kim et al., 2012) with our WT_K27 gene dataset and with genes with altered expression (up- and down-regulated) in atbmi1a/b/c mutants (Supplemental Fig. S1A; Supplemental Table S4). Clustering analysis showed a subgroup of 786 overlapping genes, indicating that among the misexpressed genes in atbmi1a/b/c there is a significant amount of EMF1 targets. Then, we determined the number of up_K27 genes in atbmi1a/b/c that were included in this subgroup (Fig. 6A). We found that one-half of atbmi1a/b/c up_K27 genes were EMF1 targets, suggesting interplay of EMF1 and AtBMI1 proteins in the regulation of a considerable number of genes.
Figure 6.
AtBMI1, EMF1, and LHP1 regulatory networks. A, Comparison of genes H3K27me3 marked bound by EMF1 and misexpressed in atbmi1a/b/c and with genes up_K27 in atbmi1a/b/c. B, Venn diagram showing up_K27 genes in atbmi1a/b/c and emf1-2. C, Comparison of genes H3K27me3 marked bound by LHP1 and misexpressed in atbmi1a/b/c and with genes up_K27 in atbmi1a/b/c. D, Venn diagram showing up_K27 genes in atbmi1a/b/c and lhp1. Some overlapping and nonoverlapping representative genes are indicated. All these overlaps are significant (P values and Fisher’s exact test results are indicated).
There was little overlap between genes up_K27 in atbmi1a/b/c and emf1-2 (Fig. 6B; Supplemental Table S4; Kim et al., 2010); furthermore, the majority of EMF1 target genes up_K27 in atbmi1a/b/c was not up-regulated in emf1-2 mutants, which is consistent with the previous observation that expression of only a small percentage of EMF1 target genes is increased in emf1-2 mutants (Kim et al., 2012; Fig. 6C). LHP1 has been shown to colocalize with 85% to 90% of H3K27me3 marked sites in Arabidopsis (Turck et al., 2007; Zhang et al., 2007; Engelhorn et al., 2012); consistent with this, 92.3% of our list of H3K27me3 marked genes (4,949 of 5,360) were occupied by LHP1 according to a recently published data set of LHP1 targets (Veluchamy et al., 2016); of these genes, 1,406 significantly overlap with the genes misexpressed (up- and down-regulated) in atbmi1a/b/c mutants (Supplemental Table S4; Supplemental Fig. S1B). Furthermore, we found that 93.9% of atbmi1a/b/c up_K27 genes were LHP1 targets (Fig. 6C), suggesting that AtBMI1 and LHP1 coregulate a high number of genes. However, when we compared H3K27me3 marked genes up-regulated in lhp1 (fold change ≥2, according to Wang et al., 2016; Supplemental Table S3) with up_K27 atbmi1a/b/c genes (Fig. 6D), we found very little overlap, indicating that loss of LHP1 function has also little impact on the expression of AtBMI1 regulated genes. Loss of LHP1 function, as loss of EMF1 function, mostly impacts the expression of genes involved in reproductive development. These genes were not up-regulated in atbmi1a/b/c mutants and some were even repressed, suggesting that LHP1 and EMF1 play different roles in their regulation. In conclusion, regulation is not correlated to the codistribution of EMF1 and LHP1 and likely also AtBMI1 proteins at target genes.
DISCUSSION
PcG regulation in Arabidopsis requires the activity of three different PRC2s, which regulate different developmental stages and display partial target specificity, and PRC1, whose identity and function is not yet well defined. Although several putative subunits have been identified (Merini and Calonje, 2015) and some evidence suggested the existence of different functional PRC1 variants (Yang et al., 2013; Calonje, 2014; Wang et al., 2014; Merini and Calonje, 2015), little is known about their composition and function. In this work, we integrated genome-wide transcriptome data with H3K27me3 and protein localization data to shed some light on the role of different PRC1 components and their possible relationship throughout plant development.
Functional Redundancy among the AtBMI1s
The identification of three AtBMI1 paralogs in Arabidopsis raised the question of whether they display functional divergence (Sanchez-Pulido et al., 2008). We found that AtBMI1A and B display mainly redundant functions throughout development, although a small number of genes were specifically sensitive to AtBMI1B. A splice variant is annotated at the AtBMI1B locus (the Arabidopsis Information Resource), which encodes a variant isoform without the amino-terminal RING finger domain (Supplemental Fig. S12). It is possible that alternative roles of the variant protein explain the observed differences in gene expression between atbmi1a and atbmi1b mutants. Conversely, AtBMI1C regulates a subset of AtBMI1A/B targets. The fact that ectopic expression of AtBMI1C in double mutants (Yang et al., 2013; Supplemental Table S2) cannot rescue atbmi1a/b defects in the aerial part of the seedling points to a requirement of tissue-specific factors for AtBMI1C mediated repression. Accordingly, AtBMI1C acts redundantly to AtBMI1A/B in the regulation of a considerable number of genes involved in root development. Differences in protein sequence between AtBMI1C and AtBMI1A/B (Bratzel et al., 2010, 2012; Chen et al., 2010) may have restricted the possibilities of AtBMI1C to interact with some factors and/or favored interaction with others. Likewise, MEA cannot compensate the loss of CLF and SWN function despite its ectopic expression in clf/swn double mutants (Farrona et al., 2011). In any case, AtBMI1A, AtBMI1B, and in part AtBMI1C display functional redundancy, indicating how important it is to ensure AtBMI1 function throughout development.
Role of AtBMI1 Function in Plant Development
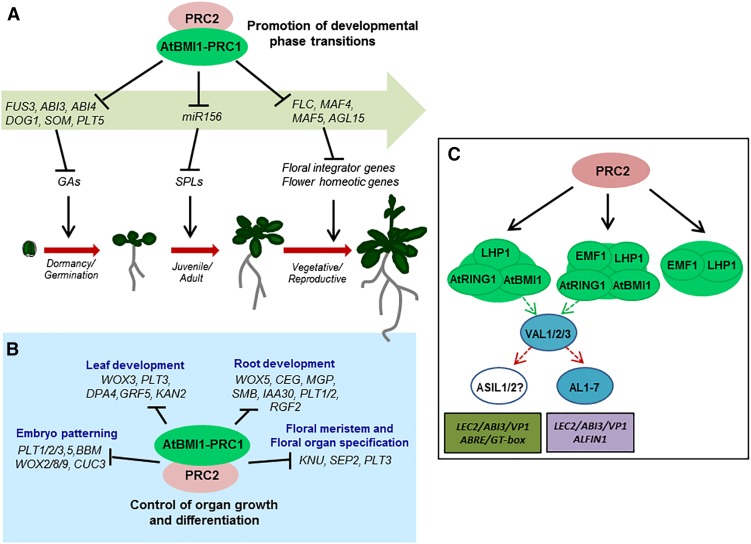
Transcriptome analysis revealed that 20% of the surveyed transcriptome was misregulated in atbmi1a/b/c mutants, a much higher percentage than the one reported for other PcG mutants, including clf/swn (Bouyer et al., 2011; Kim et al., 2012; Wang et al., 2016), thereby underlining the central role of AtBMI1s in gene regulation. To determine the AtBMI1 regulatory gene network, we focused on genes that were up-regulated in atbmi1 mutants and H3K27me3 marked in wild-type seedlings of the same age, even though these genes may represent a subset of candidate AtBMI1 targets. Our analysis supported a requirement of AtBMI1 function for the repression of the seed maturation/dormancy program after germination (Bratzel et al., 2010; Chen et al., 2010; Molitor et al., 2014; Wang et al., 2016); however, it also unveiled the crucial role of these proteins in promoting the transition from one developmental phase to the next throughout development (Fig. 7A). After embryogenesis, plants undergo the transition from seed dormancy to germination that is antagonistically regulated by two hormones, ABA and gibberellins (GA; Shu et al., 2016). During seed maturation, endogenous ABA accumulates in the seed, inducing and maintaining seed dormancy. In contrast, before the onset of germination, endogenous ABA levels in the seed are down-regulated, while the GA content is up-regulated. Among the up-regulated genes in atbmi1a/b/c mutants were genes involved in inducing ABA and/or inhibiting GA signaling (e.g. ABI3, ABI4, DOG1, PLT5, SOM; Fig. 6A), indicating that AtBMI1-mediated repression of these genes promotes this developmental transition. Following germination, plants pass through a phase of vegetative growth that can be further divided into a juvenile and an adult vegetative phase. The microRNA 156 (miR156) regulates a subset of SPL transcription factors that have been shown to promote the transition from juvenile to adult phase (Wu and Poethig, 2006); therefore, to allow phase transition, miR156 levels need to decrease. Although our transcriptome analysis could not detect mature miRNAs, it has been previously shown that pri-miR156 was up-regulated in atbmi1a/b mutants of all phenotypic severity (Picó et al., 2015); accordingly, we found down-regulation of several SPLs (e.g. SPL2/3/4/8/12; Fig. 6A), supporting that AtBMI1 function is required to allow this transition. Eventually, plants experience the transition from vegetative to reproductive development. This transition requires the repression of several flowering repressors such as FLC, MAF4/5 (Gu et al., 2013), and AGL15 (Fernandez et al., 2014), which are up-regulated in double and triple atbmi1 mutants (Fig. 7A). Consequently, flowering genes like FT, SOC1, and AGL24 were down-regulated in atbmi1 mutants; therefore, AtBMI1 activity is also required to switch from vegetative to reproductive development.
Figure 7.
Role of AtBMI1 proteins in regulating plant development. A, AtBMI1 proteins and PRC2 promote developmental phase transitions by the repression of key regulatory genes. B, AtBMI1 and PRC2 are required to control cell proliferation and differentiation during organ growth and development through the repression of master regulators. C, PRC1 variants differing in component composition and biochemical properties may collaborate with PRC2 activity in regulating phase transitions and different developmental processes throughout plant development. VAL and ASIL1/2 or AL1 to 7 proteins may recruit AtBMI1-containing complexes to target gene promoters by binding the appropriate combination of cis-regulatory elements.
Furthermore, our data revealed the key role of AtBMI1 activity in controlling stem cell niche specification and cell proliferation for proper organ growth and development via the repression of several master regulators (e.g. PLT and WOX genes; Fig. 7B), which is consistent with the widespread acquisition of proliferating capacity of atbmi1 strong mutants and the alterations in root, leaf, and flower development observed in different atbmi1 mutants (Bratzel et al., 2010; Yang et al., 2013).
Interplay of AtBMI1 with Other PcG-Related Factors
The function of AtBMI1 has been linked to the function of VAL1/2 proteins for the regulation of several seed maturation genes (Yang et al., 2013). Here, we show that VAL1/2 and AtBMI1s act together in the regulation of the seed maturation/dormancy program; however, they do not seem to collaborate in the regulation of other developmental processes. We found a specific enrichment of LEC2/ABI3/VP1 and ABRE/G-box overlapping with GT-box cis-regulatory elements at the promoters of genes coregulated by AtBMI1 and VAL1/2 proteins. An enrichment of LEC2/ABI3/VP1 and ABRE BINDING FACTOR1 elements has been previously reported at the promoter of genes up-regulated in atbmi1a/b weak phenotype (Wang et al., 2016). Genes coregulated by ABI3/VP1-like proteins and ABA contain these motifs at their promoters (Suzuki et al., 2005). Accordingly, ABI3 and ABI5 regulate gene expression synergistically. Moreover, ABI3 interacts physically with ABI5, thereby ABI3 is also recruited to the promoters of the target genes via protein-protein interaction (Nakamura et al., 2001). A similar mechanism could be assumed for repression in which the VAL1/2 proteins bind to LEC2/ABI3/VP1 and ASIL1 to the GT-box element, resulting in a direct competition with the transcriptional activators. The binding of VAL1/2 and possibly ASIL1 proteins could recruit the AtBMI1s and the other PcG proteins to establish chromatin modifications that maintain gene repression. Whether ASIL1-mediated repression involves in vivo interaction with VAL and/or PcG proteins remains to be investigated; however, in support of this, it has been shown that EMF1 interacts with ASIL1 (named EIP7) in yeast two hybrid experiments (Park et al., 2011).
We also found an enrichment of LEC2/ABI3/VP1 elements, but not ABRE or GT-box elements, at the promoter of genes exclusively up_K27 in atbmi1a/b/c mutants, suggesting an implication of B3 factors in the regulation of these genes as well. Interestingly, two VAL1 splice variants have been identified through RNA sequencing analysis: a full-length form and a truncated form lacking the plant homeodomain-like domain similar to VAL3, which also lacks the plant homeodomain-like domain (Schneider et al., 2016). It is possible that truncated VAL1 and VAL3 target this group of genes, explaining their lack of up-regulation in val1/2 mutants. Alternatively, since the B3 superfamily encompasses other subfamilies, such as the AUXIN RESPONSE FACTORS, the RELATED ABI3/VP1 and REPRODUCTIVE MERISTEM subfamilies (Swaminathan et al., 2008), some uncharacterized members of these might bind the LEC2/ABI3/VP1 element or a variation of it. In any case, the promoters of the VAL1/2-independent genes are also enriched in other cis-regulatory elements such as ALFIN1 motifs that are recognized in Arabidopsis by the ALs. Since the AL proteins interact with AtBMI1 proteins (Molitor et al., 2014), it is likely that a combination of B3 and AL factors participates in the regulation of a subset of these genes.
The relationship between AtBMI1 and EMF1 has been controversial. On one side, mutants in both display a very different phenotype and misexpress different subsets of PRC2 targets (Kim et al., 2010; Pu et al., 2013; Yang et al., 2013), which has led to propose the existence of PRC1 variants (Calonje, 2014; Merini and Calonje, 2015); however, they also coregulated a subset of targets (e.g. ABI3, ABI4, FLC) and in vitro they interact. Recent reports have shown that EMF1 copurifies with PRC2 components (Liang et al., 2015), questioning its exclusive association with PRC1. However, EMF1 colocalizes with only 45% of H3K27me3 marked genes showing a more narrow distribution at target genes than H3K27me3 marks (Kim et al., 2012). Another putative PRC1 component, LHP1, which broadly distributes across H3K27me3 marked sites (Turck et al., 2007; Zhang et al., 2007; Engelhorn et al., 2012), also copurifies with PRC2 (Derkacheva et al., 2013; Liang et al., 2015) and interacts with AtBMI1 and AtRING1 proteins in vitro (Xu and Shen, 2008; Bratzel et al., 2010). However, neither EMF1 nor LHP1 seem to be PRC2 core components, since they are required for H3K27me3 marking of only a subset of PRC2 targets (Kim et al., 2012; Wang et al., 2016).
Interestingly, when we compared the H3K27me3 marked genes that were up-regulated in atbmi1a/b/c with K27_EMF1 direct targets, we found that 50% of the up-regulated genes in atbmi1 mutants were also EMF1 targets, suggesting that AtBMI1 and EMF1 could be in a complex and potentially both impact the expression of these genes. Since LHP1 is at 93.9% of genes up_K27 in atbmi1a/b/c mutants, the same holds true also for this PRC1 component. However, the little overlap between the genes up-regulated in atbmi1a/b/c and emf1-2 or lhp1 suggests a decisive role of AtBMI1 function in maintaining their repression. There were also genes exclusively up-regulated in emf1-2 or lhp1, the majority of which are involved in flower development and these genes were not up-regulated in atbmi1a/b/c mutants. An interesting possibility could be that a PcG mechanism dependent on EMF1, LHP1, and PRC2 activities has evolved to specifically regulate the flower developmental program, which is consistent with the finding of these proteins copurifying with PRC2 (Liang et al., 2015).
CONCLUSION
In summary, our data point to different PRC1 functional networks in which genes may be regulated by AtBMI1 and/or EMF1 together with LHP1 and PRC2 and that additional proteins are required to regulate distinct subsets of genes. This is the case of VAL1/2 proteins in the seed development program, which built a network that apparently also includes ABRE/GT-box binding factors (Fig. 7C). Furthermore, it seems highly likely that other B3 domain transcription factors and ALs are part of AtBMI1-repressive circuits. In contrast, there seems little or no overlap in gene regulation by AtBMI1 on the one side and EMF1 and LHP1 on the other, although these factors may physically interact and be simultaneously present at target genes.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) atbmi1a (N645041 line), atbmi1b (CS855837 line), atbmi1a/b, and atbmi1a/b/c (atbmi1c is a GT21221.Ds5.09.01.2006.jz07.348 line) mutants were described previously (Bratzel et al., 2010; Yang et al., 2013). Segregation of “weak” and “strong” atbmi1a/b phenotypes has been previously shown (Bratzel et al., 2010; Picó et al., 2015). Plants were grown under long-day conditions at 21°C on Murashige and Skoog agar plates containing 1.5% Suc and 0.8% agar. Seedling samples were collected at zeitgeber time 2.
Transcriptomic Analysis by RNA Sequencing
The experimental design in our study consisted of two replicates for each genotype (wild-type Col-0, atbmi1a, atbmi1b, atbmi1a/b, and atbmi1a/b/c). RNA extraction was performed using Qiagen RNeasy mini kit, following the manufacturer’s instructions. RNA concentration and purity was tested using nanodrop-photometric quantification (Thermo Scientific). Library preparation was carried out following the manufacturer’s recommendations (TruSeq RNA Sample Prep Kit v2, Illumina). Sequencing of RNA libraries was performed with the Illumina HiSEquation 2000 sequencer, yielding an average of approximately 15 million 100 bp long paired-end reads for each sample. The software package FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used for quality control. All sequencing samples were of high quality, and no preprocessing of the reads was required to remove low-quality reads or read fragments (Supplemental Fig. S2). The Arabidopsis Col-0 reference genome and annotation were downloaded from the Phytozome database (TAIR10; Goodstein et al., 2012). Mapping of reads to the reference genome, transcript assembly, and differential expression were performed with the software tools Bowtie, TopHat, and Cufflinks (Trapnell et al., 2012) using default parameters producing a high percentage of concordant pair alignment rate (Supplemental Table S1). The R package from Bioconductor CummeRbund (http://www.bioconductor.org/) was used for subsequent analysis and graphical representation of the results. Differentially expressed genes were selected as those exhibiting an expression fold change greater than four when compared with the wild type and a P value < 0.05. Venn diagrams comparing the different sets of differentially expressed genes were generated with Venny 2.0.2 (http://bioinfogp.cnb.csic.es/tools/venny/index.html) and the significance of their intersections with H3K27me3 marked genes was performed using Fisher’s exact test. Gene ontology term enrichment was performed over the sets of differentially expressed genes with the web-based tools AgriGO and ReViGO (Supek et al., 2011; Yu et al., 2012) and the R bioconductor package ClusterProfiler (Du et al., 2010) using Singular Enrichment Analysis.
The clustering analysis was performed using the hierarchical algorithm implemented in the R package cluster over normalized expression levels measured using FPKM.
qRT-PCR
For qRT-PCR analysis, cDNAs were reverse-transcribed from total RNAs with QuantiTect reverse transcription kit (Qiagen). qRT-PCRs were performed using Sensi FAST SYBR & Fluorescein kit (Bioline) and an iQ5 Bio-Rad system. Expression was calculated relative to ACTIN. Primers used were as follows: WOX9-RT-Fw (5′ ACTGTCGGAGGGTTTGAAGGTATC 3′); WOX9-RT-Rev (5′ AGTGGTAGCGTAACAAATCTGAGTCT 3′); WOX2-RT-Fw (5′ GCTTACTTCAATCGCCTCCTCCACAA 3′); WOX2-RT-Rev (5′ GTCCGTTTCTCGTAGCCACCACTTG 3′); SMB-RT-Fw (5′ ACGAATATCGCTTGGACGATAG 3′); SMB-RT-Rev (5′ GCTCTTGTTCTTGGTGAAATCC 3′); ACT2-RT-Fw (5′ CACTTGCACCAAGCAGCATGAAGA 3′); ACT2-RT-Rev (5′ AATGGAACCACCGATCCAGACACT 3′).
Motif and Transcription Factor Binding Site Enrichment Analysis
Transcription Factor Binding Sites enrichment analysis was performed using HOMER (Heinz et al., 2010) and the known Transcription Factor Binding Sites sequences in plants from the databases AGRIS (Davuluri et al., 2003), JASPAR (Sandelin et al., 2004), and AthaMap (Steffens et al., 2004). The findMotifs.pl script was used with default parameters to perform known and de-novo motif overrepresentation analysis for DNA sequences of 6-, 7-, 8-, and 9-bp lengths. The target set consisted of all the gene promoters of interest. The background used for the overrepresentation analysis consisted of all the gene promoters annotated in the Arabidopsis TAIR10 genome. For the co-occurrence of the ABRE and GT-box motifs, we first identified the locations of the ABRE motif at the promoters and then extracted the DNA sequences 100 bp upstream and downstream from the center of the ABRE motif. We performed an enrichment analysis of the GT-box motif in these DNA sequences using the findMotifsGenome.pl HOMER script with default parameters. The significance of the overlapping between motifs was performed as an enrichment analysis of the DNA sequence resulting from the combination of both motifs. DNA sequences used in these analyses were downloaded using the BioMart functionality associated with Phytozome (Goodstein et al., 2012). Gene promoters were defined as the 1,000-bp DNA sequence upstream of the start codon of the corresponding gene.
Accession Numbers
The RNA-seq raw data generated in this study are publicly available from the GEO database identified with accession number GSE83568 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?&acc=GSE83568).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phenotypes of atbmi1a/b and atbmi1a/b/c mutants.
Supplemental Figure S2. Boxplots representing the read quality scores (Illumina 1.5 encoding) per base for the first replicate of all samples.
Supplemental Figure S3. Correlation among differentially expressed genes in wild type and the different genotypes.
Supplemental Figure S4. Altered gene expression in atbmi1 mutants.
Supplemental Figure S5. Gene ontology (GO) enrichment analysis of up- and down-regulated genes in atbmi1a/b mutants.
Supplemental Figure S6. Gene ontology (GO) enrichment analysis of up- and down-regulated genes in atbmi1a/b/c mutants.
Supplemental Figure S7. Putative AtBMI1direct target genes.
Supplemental Figure S8. Genes differentially expressed in atbmi1a/b and atbmi1a/b/c.
Supplemental Figure S9. Different gene expression patterns of atbmi1a/b weak and atbmi1a/b/c mutants.
Supplemental Figure S10. Expression levels of different important developmental genes in wild type and atbmi1a/b/c mutants.
Supplemental Figure S11. AtBMI1, EMF1, and LHP1 functional relationship.
Supplemental Figure S12. AtBMI1B (At1g06770) splice variants.
Supplemental Table S1. Number of reads and concurrent pair alignment rate per sequencing sample.
Supplemental Table S2. Up- and down-regulated genes in atbmi1 mutants.
Supplemental Table S3. Up-regulated genes in atbmi1 and val1/2 mutants that are marked with H3K27me3 marks in wild type, and genes in clusters I, II, and III after comparing genes up_K27 in atbmi1a/b and atbmi1a/b/c.
Supplemental Table S4. Up-regulated genes in emf1-2 and lhp1 mutants that are marked with H3K27me3 marks in wild type.
Supplementary Material
Glossary
- ABA
abscisic acid
- DAG
days after germination
- GA
gibberellin
- qRT-PCR
quantitative reverse transcription PCR
- RNA-seq
RNA sequencing
- TF
transcription factor
Footnotes
This work was supported by Marie Curie CIG Grant 333748 and BIO2013-44078-P Grant from Ministerio de Economía y Competitividad. F.T. and Y.Z. are supported by core funding from the Max Planck Gesellschaft.
Articles can be viewed without a subscription.
References
- Beh LY, Colwell LJ, Francis NJ (2012) A core subunit of Polycomb repressive complex 1 is broadly conserved in function but not primary sequence. Proc Natl Acad Sci USA 109: E1063–E1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouyer D, Roudier F, Heese M, Andersen ED, Gey D, Nowack MK, Goodrich J, Renou J-P, Grini PE, Colot V, et al. (2011) Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genet 7: e1002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratzel F, López-Torrejón G, Koch M, Del Pozo JC, Calonje M (2010) Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr Biol 20: 1853–1859 [DOI] [PubMed] [Google Scholar]
- Bratzel F, Yang C, Angelova A, López-Torrejón G, Koch M, del Pozo JC, Calonje M (2012) Regulation of the new Arabidopsis imprinted gene AtBMI1C requires the interplay of different epigenetic mechanisms. Mol Plant 5: 260–269 [DOI] [PubMed] [Google Scholar]
- Calonje M. (2014) PRC1 marks the difference in plant PcG repression. Mol Plant 7: 459–471 [DOI] [PubMed] [Google Scholar]
- Calonje M, Sanchez R, Chen L, Sung ZR (2008) EMBRYONIC FLOWER1 participates in polycomb group-mediated AG gene silencing in Arabidopsis. Plant Cell 20: 277–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Tsukada Y, Zhang Y (2005) Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell 20: 845–854 [DOI] [PubMed] [Google Scholar]
- Carles C, Bies-Etheve N, Aspart L, Léon-Kloosterziel KM, Koornneef M, Echeverria M, Delseny M (2002) Regulation of Arabidopsis thaliana Em genes: role of ABI5. Plant J 30: 373–383 [DOI] [PubMed] [Google Scholar]
- Chanvivattana Y, Bishopp A, Schubert D, Stock C, Moon Y-H, Sung ZR, Goodrich J (2004) Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131: 5263–5276 [DOI] [PubMed] [Google Scholar]
- Chen D, Molitor A, Liu C, Shen W-H (2010) The Arabidopsis PRC1-like ring-finger proteins are necessary for repression of embryonic traits during vegetative growth. Cell Res 20: 1332–1344 [DOI] [PubMed] [Google Scholar]
- Choi H, Hong J, Ha J, Kang J, Kim SY (2000) ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275: 1723–1730 [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Davuluri RV, Sun H, Palaniswamy SK, Matthews N, Molina C, Kurtz M, Grotewold E (2003) AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics 4: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas F, Sankaranarayanan S, Deb S, Widdup E, Bournonville C, Bollier N, Northey JGB, McCourt P, Samuel MA (2013) ABI3 controls embryo degreening through Mendel’s I locus. Proc Natl Acad Sci USA 110: E3888–E3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkacheva M, Steinbach Y, Wildhaber T, Mozgová I, Mahrez W, Nanni P, Bischof S, Gruissem W, Hennig L (2013) Arabidopsis MSI1 connects LHP1 to PRC2 complexes. EMBO J 32: 2073–2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64– 70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhorn J, Reimer JJ, Leuz I, Göbel U, Huettel B, Farrona S, Turck F (2012) Development-related PcG target in the apex 4 controls leaf margin architecture in Arabidopsis thaliana. Development 139: 2566–2575 [DOI] [PubMed] [Google Scholar]
- Farrona S, Thorpe FL, Engelhorn J, Adrian J, Dong X, Sarid-Krebs L, Goodrich J, Turck F (2011) Tissue-specific expression of FLOWERING LOCUS T in Arabidopsis is maintained independently of polycomb group protein repression. Plant Cell 23: 3204–3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez DE, Wang C-T, Zheng Y, Adamczyk BJ, Singhal R, Hall PK, Perry SE (2014) The MADS-domain factors AGAMOUS-LIKE15 and AGAMOUS-LIKE18, along with SHORT VEGETATIVE PHASE and AGAMOUS-LIKE24, are necessary to block floral gene expression during the vegetative phase. Plant Physiol 165: 1591–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förderer A, Zhou Y, Turck F (2016) The age of multiplexity: recruitment and interactions of Polycomb complexes in plants. Curr Opin Plant Biol 29: 169–178 [DOI] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE, Woodcock CL (2004) Chromatin compaction by a polycomb group protein complex. Science 306: 1574–1577 [DOI] [PubMed] [Google Scholar]
- Gangappa SN, Srivastava AK, Maurya JP, Ram H, Chattopadhyay S (2013) Z-box binding transcription factors (ZBFs): a new class of transcription factors in Arabidopsis seedling development. Mol Plant 6: 1758–1768 [DOI] [PubMed] [Google Scholar]
- Gao M-J, Lydiate DJ, Li X, Lui H, Gjetvaj B, Hegedus DD, Rozwadowski K (2009) Repression of seed maturation genes by a trihelix transcriptional repressor in Arabidopsis seedlings. Plant Cell 21: 54–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudin V, Libault M, Pouteau S, Juul T, Zhao G, Lefebvre D, Grandjean O (2001) Mutations in LIKE HETEROCHROMATIN PROTEIN 1 affect flowering time and plant architecture in Arabidopsis. Development 128: 4847–4858 [DOI] [PubMed] [Google Scholar]
- Gendall AR, Levy YY, Wilson A, Dean C (2001) The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107: 525–535 [DOI] [PubMed] [Google Scholar]
- Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz EM, Coupland G (1997) A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386: 44–51 [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus U, Vielle-Calzada JP, Hoeppner MA, Gagliano WB (1998) Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280: 446–450 [DOI] [PubMed] [Google Scholar]
- Gu X, Le C, Wang Y, Li Z, Jiang D, Wang Y, He Y (2013) Arabidopsis FLC clade members form flowering-repressor complexes coordinating responses to endogenous and environmental cues. Nat Commun 4: 1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L, Bouveret R, Gruissem W (2005) MSI1-like proteins: an escort service for chromatin assembly and remodeling complexes. Trends Cell Biol 15: 295–302 [DOI] [PubMed] [Google Scholar]
- Horstman A, Willemsen V, Boutilier K, Heidstra R (2014) AINTEGUMENTA-LIKE proteins: hubs in a plethora of networks. Trends Plant Sci 19: 146–157 [DOI] [PubMed] [Google Scholar]
- Kim SY, Lee J, Eshed-Williams L, Zilberman D, Sung ZR (2012) EMF1 and PRC2 cooperate to repress key regulators of Arabidopsis development. PLoS Genet 8: e1002512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Zhu T, Sung ZR (2010) Epigenetic regulation of gene programs by EMF1 and EMF2 in Arabidopsis. Plant Physiol 152: 516–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WY, Lee D, Chung W-I, Kwon CS (2009) Arabidopsis ING and Alfin1-like protein families localize to the nucleus and bind to H3K4me3/2 via plant homeodomain fingers. Plant J 58: 511–524 [DOI] [PubMed] [Google Scholar]
- Liang SC, Hartwig B, Perera P, Mora-García S, de Leau E, Thornton H, de Lima Alves F, Rappsilber J, Yang S, James GV, et al. (2015) Kicking against the PRCs - a domesticated transposase antagonises silencing mediated by polycomb group proteins and is an accessory component of polycomb repressive complex 2. PLoS Genet 11: e1005660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Koltunow A, Dennis ES, Peacock WJ, Chaudhury AM (1999) Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci USA 96: 296–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merini W, Calonje M (2015) PRC1 is taking the lead in PcG repression. Plant J 83: 110–120 [DOI] [PubMed] [Google Scholar]
- Molitor AM, Bu Z, Yu Y, Shen W-H (2014) Arabidopsis AL PHD-PRC1 complexes promote seed germination through H3K4me3-to-H3K27me3 chromatin state switch in repression of seed developmental genes. PLoS Genet 10: e1004091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozgova I, Köhler C, Hennig L (2015) Keeping the gate closed: functions of the polycomb repressive complex PRC2 in development. Plant J 83: 121–132 [DOI] [PubMed] [Google Scholar]
- Müller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA (2002) Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111: 197–208 [DOI] [PubMed] [Google Scholar]
- Nakamura S, Lynch TJ, Finkelstein RR (2001) Physical interactions between ABA response loci of Arabidopsis. Plant J 26: 627–635 [DOI] [PubMed] [Google Scholar]
- Ohad N, Yadegari R, Margossian L, Hannon M, Michaeli D, Harada JJ, Goldberg RB, Fischer RL (1999) Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11: 407–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H-Y, Lee S-Y, Seok H-Y, Kim S-H, Sung ZR, Moon Y-H (2011) EMF1 interacts with EIP1, EIP6 or EIP9 involved in the regulation of flowering time in Arabidopsis. Plant Cell Physiol 52: 1376–1388 [DOI] [PubMed] [Google Scholar]
- Peterson AJ, Mallin DR, Francis NJ, Ketel CS, Stamm J, Voeller RK, Kingston RE, Simon JA (2004) Requirement for sex comb on midleg protein interactions in Drosophila polycomb group repression. Genetics 167: 1225–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picó S, Ortiz-Marchena MI, Merini W, Calonje M (2015) Deciphering the role of Polycomb Repressive Complex 1 (PRC1) variants in regulating the acquisition of flowering competence in Arabidopsis. Plant Physiol 168: 1286–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu L, Liu M-S, Kim SY, Chen L-FO, Fletcher JC, Sung ZR (2013) EMBRYONIC FLOWER1 and ULTRAPETALA1 act antagonistically on Arabidopsis development and stress response. Plant Physiol 162: 812–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qüesta JI, Song J, Geraldo N, An H, Dean C (2016) Arabidopsis transcriptional repressor VAL1 triggers Polycomb silencing at FLC during vernalization. Science 353: 485–488 [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R (2004) Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet 38: 413–443 [DOI] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Devos D, Sung ZR, Calonje M (2008) RAWUL: a new ubiquitin-like domain in PRC1 ring finger proteins that unveils putative plant and worm PRC1 orthologs. BMC Genomics 9: 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A, Alkema W, Engström P, Wasserman WW, Lenhard B (2004) JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res 32: D91–D94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Mendoza M, Dubreucq B, Baud S, Parcy F, Caboche M, Lepiniec L (2008) Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J 54: 608–620 [DOI] [PubMed] [Google Scholar]
- Schneider A, Aghamirzaie D, Elmarakeby H, Poudel AN, Koo AJ, Heath LS, Grene R, Collakova E (2016) Potential targets of VIVIPAROUS1/ABI3-LIKE1 (VAL1) repression in developing Arabidopsis thaliana embryos. Plant J 85: 305–319 [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V (2013) A new world of Polycombs: unexpected partnerships and emerging functions. Nat Rev Genet 14: 853–864 [DOI] [PubMed] [Google Scholar]
- Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu CT, Bender W, Kingston RE (1999) Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98: 37–46 [DOI] [PubMed] [Google Scholar]
- Shu K, Liu XD, Xie Q, He ZH (2016) Two faces of one seed: hormonal regulation of dormancy and germination. Mol Plant 9: 34–45 [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE (2013) Occupying chromatin: polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell 49: 808–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens NO, Galuschka C, Schindler M, Bülow L, Hehl R (2004) AthaMap: an online resource for in silico transcription factor binding sites in the Arabidopsis thaliana genome. Nucleic Acids Res 32: D368–D372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T (2011) REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6: e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Ketterling MG, McCarty DR (2005) Quantitative statistical analysis of cis-regulatory sequences in ABA/VP1- and CBF/DREB1-regulated genes of Arabidopsis. Plant Physiol 139: 437–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Wang HH-Y, McCarty DR (2007) Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiol 143: 902–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan K, Peterson K, Jack T (2008) The plant B3 superfamily. Trends Plant Sci 13: 647–655 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Roudier F, Farrona S, Martin-Magniette M-L, Guillaume E, Buisine N, Gagnot S, Martienssen RA, Coupland G, Colot V (2007) Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet 3: e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Graaff E, Laux T, Rensing SA (2009) The WUS homeobox-containing (WOX) protein family. Genome Biol 10: 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veluchamy A, Jégu T, Ariel F, Latrasse D, Mariappan KG, Kim S-K, Crespi M, Hirt H, Bergounioux C, Raynaud C, et al. (2016) LHP1 regulates H3K27me3 spreading and shapes the three-dimensional conformation of the Arabidopsis genome. PLoS One 11: e0158936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Carbajosa J, Carbonero P (2005) Seed maturation: developing an intrusive phase to accomplish a quiescent state. Int J Dev Biol 49: 645–651 [DOI] [PubMed] [Google Scholar]
- Wang H, Liu C, Cheng J, Liu J, Zhang L, He C, Shen W-H, Jin H, Xu L, Zhang Y (2016) Arabidopsis flower and embryo developmental genes are repressed in seedlings by different combinations of polycomb group proteins in association with distinct sets of cis-regulatory elements. PLoS Genet 12: e1005771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Gu X, Yuan W, Schmitz RJ, He Y (2014) Photoperiodic control of the floral transition through a distinct polycomb repressive complex. Dev Cell 28: 727–736 [DOI] [PubMed] [Google Scholar]
- Wei W, Zhang Y-Q, Tao J-J, Chen H-W, Li Q-T, Zhang W-K, Ma B, Lin Q, Zhang J-S, Chen S-Y (2015) The Alfin-like homeodomain finger protein AL5 suppresses multiple negative factors to confer abiotic stress tolerance in Arabidopsis. Plant J 81: 871–883 [DOI] [PubMed] [Google Scholar]
- Wolff P, Weinhofer I, Seguin J, Roszak P, Beisel C, Donoghue MTA, Spillane C, Nordborg M, Rehmsmeier M, Köhler C (2011) High-resolution analysis of parent-of-origin allelic expression in the Arabidopsis Endosperm. PLoS Genet 7: e1002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Poethig RS (2006) Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133: 3539–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Wagner D (2015) Polycomb repression in the regulation of growth and development in Arabidopsis. Curr Opin Plant Biol 23: 15–24 [DOI] [PubMed] [Google Scholar]
- Xu L, Shen W-H (2008) Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Curr Biol 18: 1966–1971 [DOI] [PubMed] [Google Scholar]
- Yang C, Bratzel F, Hohmann N, Koch M, Turck F, Calonje M (2013) VAL- and AtBMI1-mediated H2Aub initiate the switch from embryonic to postgerminative growth in Arabidopsis. Curr Biol 23: 1324–1329 [DOI] [PubMed] [Google Scholar]
- Yoshida N, Yanai Y, Chen L, Kato Y, Hiratsuka J, Miwa T, Sung ZR, Takahashi S (2001) EMBRYONIC FLOWER2, a novel polycomb group protein homolog, mediates shoot development and flowering in Arabidopsis. Plant Cell 13: 2471–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang L-G, Han Y, He Q-Y (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16: 284–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Germann S, Blus BJ, Khorasanizadeh S, Gaudin V, Jacobsen SE (2007) The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat Struct Mol Biol 14: 869–871 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.