Summary
The MHC is a highly polymorphic genomic region that encodes the transplantation and immune regulatory molecules. It receives special attention for genetic investigation because of its important role in the regulation of innate and adaptive immune responses and its strong association with numerous infectious and/or autoimmune diseases. The MHC locus was first discovered in the mouse and for the past 50 years it has been studied most intensively in both mice and humans. However, in recent years the macaque species have emerged as some of the more important and advanced experimental animal models for biomedical research into MHC with important human immunodeficiency virus/simian immunodeficiency virus and transplantation studies undertaken in association with precise MHC genotyping and haplotyping methods using Sanger sequencing and next‐generation sequencing. Here, in this special issue on ‘Macaque Immunology’ we provide a short review of the genomic similarities and differences among the human, macaque and mouse MHC class I and class II regions, with an emphasis on the association of the macaque class I region with MHC polymorphism, haplotype structure and function.
Keywords: haplotype, macaque, major histocompatibility complex, polymorphism
Introduction
The genomic locus of the MHC encodes the polymorphic cell‐membrane‐bound glycoproteins known as MHC classical class I and class II molecules (antigens) that regulate the immune response by presenting peptides of fragmented proteins to circulating cytotoxic and helper T lymphocytes, respectively.1, 2, 3 The MHC classical class I antigens are produced by most tissues and they associate non‐covalently with β2‐microglobulin to present intracellularly processed peptide antigens (8–11 amino acids in length) to T‐cell receptors of specific CD8+ T cells in order to induce their activation and/or cytotoxicity.2 The processed peptides may arise from the cell's own proteome or from foreign intracellular pathogens.4 Mature dendritic cells use the MHC class I system to present peptides deriving from antigens captured by endocytosis.5 This process, called cross‐presentation, plays a crucial role in the initiation of responses of specific T CD8+ lymphocytes in peripheral lymphoid organs. In addition, the MHC classical class I proteins may act as ligands for killer‐cell immunoglobulin‐like receptors that regulate the cytotoxic activity of cytotoxic T cells and natural killer cell6, 7, 8, 9 and leucocyte immunoglobulin‐like receptors expressed on myelomonocytes and other leucocyte lineages.10 In contrast to the classical class I antigens, the classical class II antigens form heterodimeric structures specialized in the presentation of exogenous peptides (15–25 amino acids in length) on the surface of lymphoid cells to the CD4+ helper T lymphocytes of the immune system.2 The class II gene expression is predominantly restricted to the lymphoid cells, such as B cells, monocytes, macrophages, endothelial cells, dendritic cells and activated T cells. Both the classical class I and class II genes are often highly polymorphic, presumably to preserve the inter‐individual variability of the antigen‐presenting ability and help the species to defend against and survive the natural selection pressure from various infectious agents.11, 12 The non‐classical class I and class II antigens, although similar in structure to their classical class I or class II counterparts, are usually far less polymorphic, have variable or limited tissue expression and functions that are often distinctly different to those of the classical class I or class II antigens.13 Moreover, several non‐classical MHC class I genes are located outside the MHC.
The MHC was first discovered in mice more than 60 years ago and because it was a tumour‐resistant locus it soon became known as the histocompatibility locus H2.3 Its equivalent in humans was named the human leucocyte antigen (HLA) complex or human MHC after the pioneering description by Jean Dausset of the first alloantibodies against antigens expressed by human leucocytes of certain, but not all, individuals.14, 15, 16 It was later demonstrated that HLA donor/recipient incompatibility was critically involved in organ transplant rejection and graft‐versus‐host disease.3, 11 The MHC is now known to be a highly complex immune‐response genomic region composed of a large group of linked genes, some not necessarily directly associated with the class I and class II genes, but many of them involved functionally with the adaptive and innate immune response systems in all the jawed vertebrates studied so far.17 The MHC genomic regions have been completely sequenced in various representatives of the mammalian (chimpanzee, rhesus macaque, mouse, rat, pig, dog and opossum) and non‐mammalian (chicken, quail, shark and amphioxus) species and comparatively analysed for a better understanding of the evolutionary process responsible for the genetic diversity of the regions and their role in the immune system.2, 18 Although the human MHC has received considerable attention because of its role in immune regulation, transplantation and autoimmune diseases, experimental animal models have been developed to advance biomedical research on various aspects of the MHC such as gene expression and the mechanisms of peptide presentation in the mouse and rat, and diversity in the dog, pig and macaque.
Although the mouse still remains the premier animal model for MHC research,18 the macaque species such as the rhesus macaques (Macaca mulatta; Mamu), the cynomolgus macaques (Macaca fascicularis; Mafa) and southern pig‐tailed macaques (Macaca nemestrina; Mane) have emerged more recently as important MHC experimental models because of their relatively close phylogenetic relationship to humans (Fig. 1). In addition, the macaques have considerable immunological similarities with humans as demonstrated by frequent and high interspecies cross‐reactivity of antibodies raised against human antigens.19, 20 Although macaques are phylogenetically closer to humans than rodents, they differ significantly from humans by many blood group systems including their MHC.21 Also, murine anti‐HLA monoclonal antibodies to monomorphic and polymorphic epitopes have been compared for their reactivity in humans and M. nemestrina.22 One of these monoclonal antibodies, P77·1, which detects the human allo‐antigen HLA‐Bw6, was highly conserved and reacted with 87% of the macaque epitopes, a frequency very close to that observed in humans. Since the macaque species have immunological similarities with the human, they are often used for biomedical studies of infectious, neurological and reproductive diseases, transplantation and immunotherapy. Many of these studies would benefit from a better understanding of the MHC genetic background in these animals.
Figure 1.
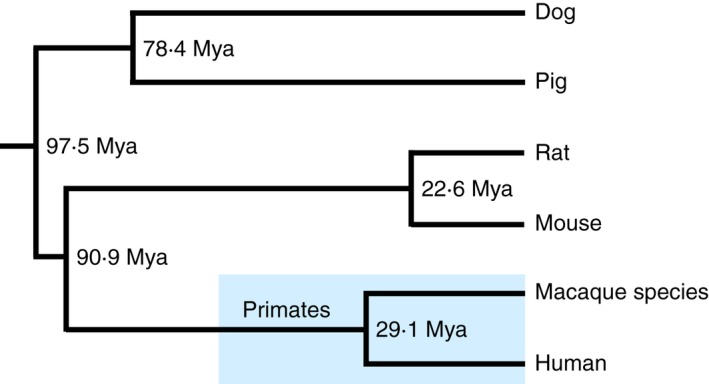
Phylogenetic relationships between the human and five representative experimental animals. The time of divergence is on the Time Tree website (http://www.timetree.org/).
Here, in this special issue on ‘Macaque Immunology’ we briefly review the genomic similarities and differences among the macaque, human and mouse MHC regions with particular emphasis on the comparative polymorphisms, haplotype structure and function of the class I region of humans and macaques.
Genomic characteristics of the HLA genomic region
The first fully sequenced and gene annotated human genomic MHC was published in 1999.23 This sequence was a ‘virtual MHC’ because it was composed of a mosaic of different human haplotypes rather than representing any one particular haplotype. Subsequently, the genomic sequences of at least eight different human ancestral MHC haplotypes11 were published for a more precise comparative genomic analysis of the similarities and differences.24 Figure 2 shows the current gene map of the HLA genomic region based on Genome Reference Consortium Human Build 38 patch release 2 (GRCh38.p2) in the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov/gene/). The HLA super locus is located on the short arm of chromosome 6, band p21.3, with the class I region located at the telomeric end and the class II region located at the centrometric end, both separated from each other by an extended class III region of 61 protein‐coding genes. Whereas the HLA class I and class II genomic regions encode the highly polymorphic gene complex of the HLA class I and HLA class II genes,25, 26 the class III region consists of many different genes that are involved in stress (HSPA1A, HSPA1B and HSPA1L), the complement cascade (C4A, C4B, C2, CFB), and inflammation (cytokine genes LTB, TNF, LTA and regulatory genes NFKBIL1 and DDX39B).2, 11 The class II region also contains some proteosome‐processing and peptide antigen transportation genes such as PSMB8, PSMB9, TAP1 and TAP2.4 The TAP‐binding protein, TAPBP, is in the extended class II region. The ‘Class I’ region ranges from HLA‐F to MICB, ‘Class III’ ranges from PPIAP9 to BTNL2, and ‘Class II’ ranges from HLA‐DRA to HLA‐DPA3. There are also sub‐regions from the telomeric side of the Class I sub‐regions and the centromeric side of the Class II sub‐regions that are called the ‘Extended class I’ (telomeric side of HCG4P11) and ‘Extended class II’ (centromeric side of COL11A2) sub‐regions, respectively.27 The class I region is additionally divided into three genomic blocks, α, β and κ, that include duplicated HLA genes and two framework gene blocks that include well‐conserved non‐MHC genes in mammalian species.18 HLA‐A, HLA‐G and HLA‐F are in the α block, HLA‐B and HLA‐C are in the β block, and HLA‐E is in the κ block.
Figure 2.
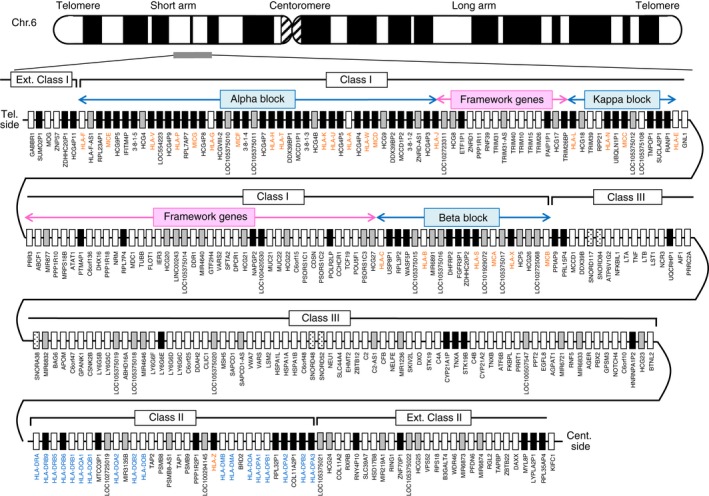
Gene map of the HLA genomic region. The MHC gene map corresponds to the genomic coordinates of 29 602 228 (GABBR1) to 33 410 226 (KIFC1) in the human genome GRCh38.p2 primary assembly of the NCBI map viewer. The regions separated by arrows show the HLA sub‐regions such as extended class I, class I, class III, classical class II and extended class II regions from telomere (left and top side) to centromere (right and bottom side). Blue and pink boxes show the spans of α, β and κ blocks and framework gene blocks, respectively. White, grey, dotted and black boxes show protein‐coding genes, non‐coding RNAs (ncRNAs), small nucleolar RNAs (snoRNAs) and pseudogenes, respectively. Red and blue letters indicate HLA class I / MIC and class II genes, respectively. The location of the α, β and κ blocks containing the cluster of duplicated HLA class I genes and framework gene blocks between them in the class I region are indicated by blue and pink arrows, respectively.
A total of 283 loci have now been identified and/or reclassified in the 3·78‐Mb HLA genomic region of the PGF haplotype28 from GABBR1 located on the Extended class I region to KIFC1 located on the Extended class II region (Fig. 2 and Table 1). When all the loci of the HLA genomic region are grouped into four categories of gene types, then 144 loci are classified as a protein‐coding gene, 53 loci are non‐coding RNA (ncRNA), five loci are small nucleolar RNA (snoRNA) and 81 loci are pseudogenes (Table 1, and see Supplementary material, Table S1). Of the 283 loci, 15·5% (44 loci) are occupied by HLA and HLA‐like genes (HLA class I, HLA class II and MHC class I polypeptide‐related sequence; MIC genes).
Table 1.
Gene numbers in the HLA genomic region
| Gene status | Protein coding | ncRNA | snoRNA | Pseudo | Total |
|---|---|---|---|---|---|
| Extended Class I (GABBR1–HCG4P11) | 3 | 0 | 0 | 3 | 6 |
| Class I | 47 | 30 | 0 | 55 | 132 |
| Class III | 61 | 12 | 5 | 8 | 86 |
| Class II | 18 | 4 | 0 | 10 | 32 |
| Extended Class I (COL11A2–KIFC1) | 15 | 7 | 0 | 5 | 27 |
| Total for all regions | 144 | 53 | 5 | 81 | 283 |
Of the HLA and HLA‐like genes, 18 HLA class I genes (six protein‐coding genes and 12 pseudogenes) and seven MIC genes (two protein‐coding genes and five pseudogenes) are located in the HLA class I region, and 18 HLA class II genes (13 protein‐coding genes and five pseudogenes) are located in the HLA class II region (Fig. 2 and Table 2). Interestingly, one HLA class I pseudogene (HLA‐Z) is located close to the HLA‐DMB gene in the HLA class II region. In addition, the classical HLA class I genes, HLA‐A, HLA‐B and HLA‐C, and the classical HLA class II genes, HLA‐DR, HLA‐DQ and HLA‐DP, are distinguished by their extraordinary polymorphisms, whereas the non‐classical HLA class I genes, HLA‐E, HLA‐F and HLA‐G, are distinguished by their tissue‐specific expression and limited polymorphism.25, 26
Table 2.
Numbers of HLA and MIC genes in the HLA genomic region
| Protein coding | Pseudo | Total | |
|---|---|---|---|
| HLA class I genes | 6 | 13 | 19 |
| HLA class II genes | 13 | 5 | 18 |
| MIC genes | 2 | 5 | 7 |
| Total for HLA‐like genes | 21 | 23 | 44 |
Genomic comparison among human, rhesus macaque and mouse
MHC regions
The MHC genomic regions of the mouse13, 18 and macaque29, 30 were still only partially sequenced 3–5 years after the human MHC genomic sequence was first published in 1999.23 Figure 3 shows a schematic, comparative MHC genomic map, considering the nucleotide length of each of the MHC class I, class II and class III regions in human, rhesus macaque and mouse. The entire Mamu region from the start of class I to the end of class II is 4·72 Mb in length based on the genomic map data29, 30 compared with the 3·41 Mb of the HLA (GRCh38.p2) and 2·91 Mb of the mouse MHC (H2) class I to class II regions [Genome Reference Consortium Mouse Build 38 patch release 3 (GRCm38.p3]. In comparing the length of each region, the HLA and Mamu class II regions have similar lengths (0·69–0·70 Mb), whereas the H2 orthologous region is much shorter at only 0·26 Mb. On the other hand, the length of the HLA, Mamu and H2 class I regions are 1·79 Mb, 3·07 Mb and 1·72 Mb, respectively, with significant differences observed in the lengths of the α, β and κ blocks where the MHC class I gene duplications are located. Namely, the α block of the Mamu is 690 kb in length in comparison to 286 kb in the HLA region and 15 kb in the H2 region. The κ block of the H2 is 791 kb in length, compared with 235 kb in the HLA region and 200 kb in the Mamu region. The β block of the Mamu is the longest at 1140 kb, in comparison to 242 kb in the HLA region and 203 kb in the H2 region (Fig. 3). These differences in the length of the blocks with the class I region of the three species were generated by segmental duplications and insertions and deletions involving the MHC class I genes and repeat sequences.11, 31 In contrast, the class III region is essentially the same length in the three species showing a high level of orthologous gene density.
Figure 3.
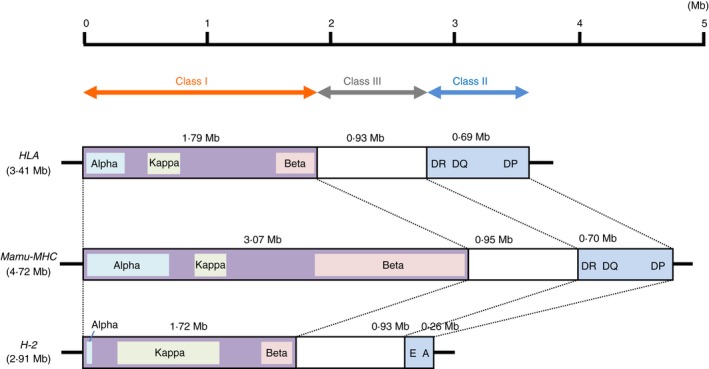
Comparison of genomic structures of the HLA, Mamu‐MHC and H2 regions. Genomic information of the HLA and H2 region was based on the current genome database at the NCBI site, and genomic information of the Mamu‐MHC region was referred from a previous report.25
MHC genes
Figure 4 shows a schematic, comparative gene map of the protein coding MHC class I and class II genes in the three species. The Mamu has 33 MHC (22 class I and 11 class II genes) and two MIC genes in the Mamu region compared with 19 HLA genes (six class I and 13 class II genes) and two MIC genes in the HLA region, and 39 H2 genes (30 class I and nine class II genes) in the H2 region. These numbers are increased by including the pseudogenes, 73 MHC (56 class I and 17 class II genes) and 11 MIC genes are located on the Mamu region compared with 44 (19 class I, 18 class II and seven MIC genes) and 47 (36 class I and 11 class II genes) in the HLA and H2 regions, respectively (see Supplementary material, Tables S2 and S3). Therefore, the overall structure of the Mamu class I region is more complex than the HLA and the H2 class I regions.
Figure 4.
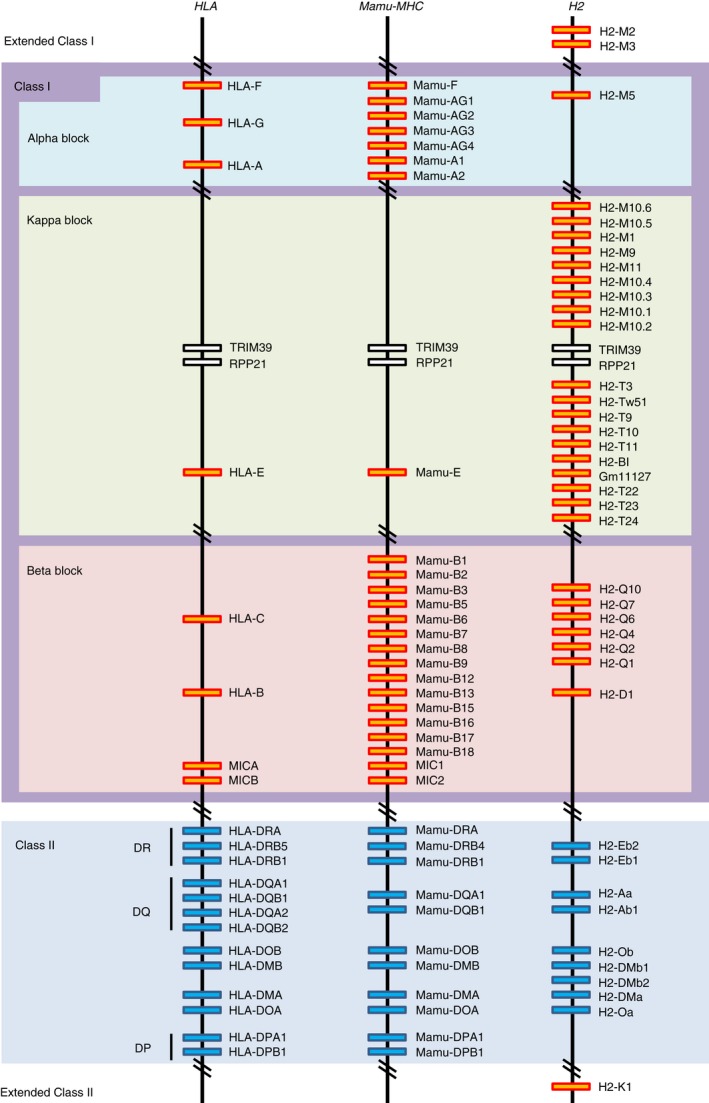
Comparative genomic map of the protein coding MHC loci among the HLA, Mamu and H2 regions. Orange and blue boxes indicate MHC class I and class II genes, respectively. The classification for protein‐coding genes and pseudogenes is shown in the Supplementary material (Table S3).
The rhesus macaque counterparts of the classical HLA‐A and HLA‐B genes and the non‐classical HLA‐E, HLA‐F and HLA‐G genes were identified in the α, β or κ block and designated Mamu‐A, Mamu‐B, Mamu‐E, Mamu‐F and Mamu‐G, respectively. Mamu‐G appears to be a pseudogene and its function may have been taken over by Mamu‐AG, which is expressed on the rhesus monkey placenta and shares unique features with HLA‐G.32, 33 There are two Mamu‐A and four Mamu‐AG in the α block, one Mamu‐E in the κ block and 14 Mamu‐B genes in the β block based on the Mamu genomic sequence used in Fig. 4. The orthologues of the HLA‐C gene have not been identified so far in rhesus macaques and in any other species of Old World monkeys, although some Mamu‐B genes (Mamu‐B5, ‐B9, ‐B18, ‐B6, ‐B2, ‐B8, ‐B1, ‐B7, ‐B3 and ‐B13) showed closer relationships with HLA‐C than with HLA‐B (Fig. 5). Also, there is a Mamu‐Z pseudogene l similar to the HLA‐Z pseudogene located in the class II orthologous region.
Figure 5.
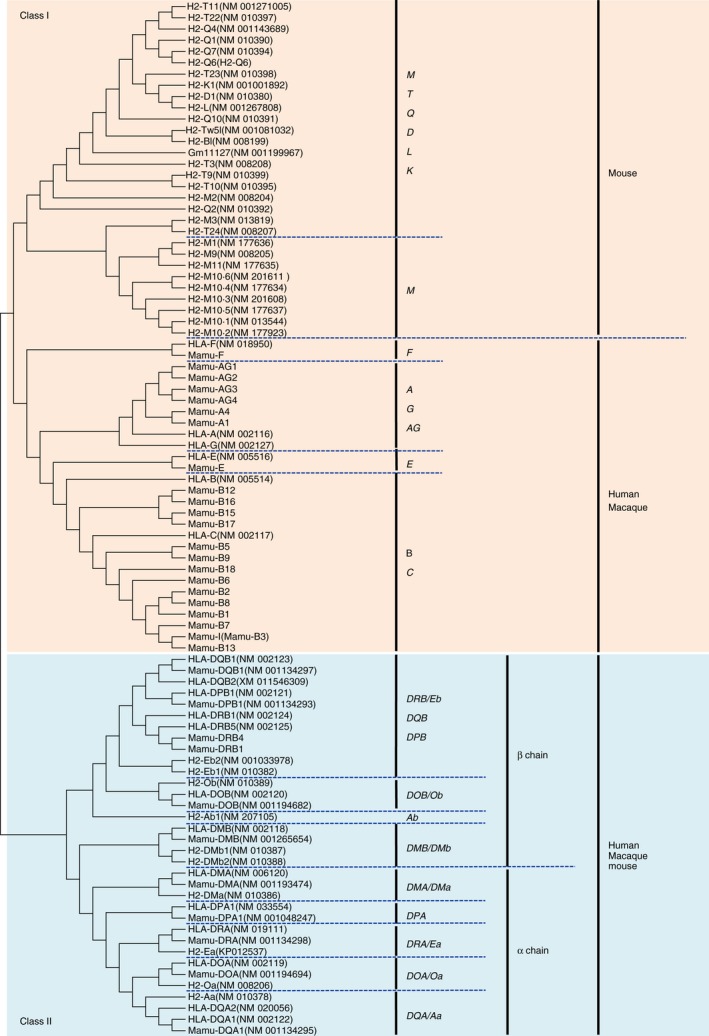
Nucleotide sequence‐based phylogenetic tree of MHC class I and class II genes. Multiple sequence alignment was created using the clustalW Sequence Alignment program of the Molecular Evolution Genetics Analysis software 5 (MEGA5: http://www.megasoftware.net/)107 Phylogenetic trees of the MHC genes were constructed by the neighbour‐joining method (MEGA5)108 with a Maximum Composite Likelihood model using exon 4 and exon 3 of the MHC class I and class II genes, respectively. Parentheses and bold letters indicate GenBank/EMBL/DDBJ accession numbers and human MHC genes, respectively.
In contrast to human and macaque, the mouse MHC genomic sequence determined for the C57BL/6J strain has classical class Ia genes, H2‐D1 and H2‐K1 in the β block and the extended class II region between RING1 and VPS52, respectively. Contrary to the mouse b haplotype, the mouse strain C57BL/6J and bc haplotype have an additional classical class Ia gene, H2‐L, located between the H2‐D1and H2‐Q cluster.34, 35 Of the protein coding non‐classical class Ib genes, 12 H2‐M genes are included in the extended class I region, and the α and κ blocks, and nine H2‐T, one H2‐BI and six H2‐Q genes are included in the κ and β blocks, respectively (Fig. 4). Interestingly, whereas the κ block of the Mamu and HLA each has only one MHC class I gene, Mamu‐E and HLA‐E, respectively, the mouse κ block has at least 19 MHC class I genes, with 10 H2‐T like genes and nine H2‐M like genes on either side of the TRIM39 and RPP21 orthologous framework genes. The reason for the large number of duplicated MHC class I genes in the κ block of the mouse H2 is not known, but many of them are considered to have non‐classical, non‐immune functions and are expressed differentially compared with the ubiquitous expression of the classical class Ia genes.13, 35, 36, 37, 38 For example, the H2‐M1 and H2‐M10 families of the class Ib genes interact specifically with the V2R class of pheromone receptors presented on the cell surfaces of the vomeronasal organ.18 The transcripts of some other H2‐M and H2‐T genes are expressed widely, including in the brains of adult and embryonic mice.13 Many reports are published concerning the gene function of the non‐classical MHC class Ib genes.13, 36, 37, 38, 39, 40, 41, 42, 43, 44
The protein‐coding MICA and MICB genes and MIC1 and MIC2 are identified in the β block of the HLA and Mamu class I regions, respectively. Two combinations of MIC1 and MIC2, and MIC3 and MIC2 are identified in the Mamu haplotypes of the β block. The MIC1 and MIC2 genes are the orthologues of the human MICA and MICB genes, respectively. The MIC3 gene is a hybrid of MICA and MICB generated by a crossing over event with one breakpoint in intron 3, and MIC3 is named as MICA/B.45 However, the MIC orthologous gene is not observed in the H2 region.
The MHC class II genes (MHC‐DRA, ‐DQA1, ‐DQB1, ‐DOB, ‐DMB, ‐DMA, ‐DOA, ‐DPA1 and ‐DPB1) are well conserved between the HLA and Mamu regions, excluding the MHC‐DRB, MHC‐DQA2 and MHC‐DQB2 genes that were generated by different evolutionary processes.46 The gene order within the HLA and Mamu class II orthologous gene regions is essentially the same (Fig. 4). In contrast, H2‐Ea, which is thought to be the HLA‐DRA orthologue, has not been observed in some H2 haplotypes including the C57BL/6J strain, and the MHC‐DPA1 and DPB1 genes are missing from all H2 haplotypes.
Non‐MHC genes
The 123 protein‐coding non‐MHC genes were identified in the HLA genomic framework gene region, the class III region and the extended class I and class II regions (Fig. 2 and Table 1). Of the 120 genes excluding three genes (C4A, C4B and CYP21A2) that have CYP21A‐C4 haplotype structure, 94·2% (113 genes) are commonly observed among the macaque MHC, HLA and H2 regions (see Supplementary material, Table S4). In contrast, two genes (LOC105375108 and PSORS1C1), four genes (MUC21, MCCD1, NCR3 and C6orf48) and one gene (BTNL2) are observed in only HLA, HLA and macaque species, and HLA and H2, respectively.
The paralogy and diversity of the MHC class I genes
To summarize the comparative data analysis, the basic organizational structure of the MHC regions is largely conserved among the mice, macaques and humans with the exception that the class I genes within the α, β and κ blocks are constantly remodelled between the species by ‘birth and death’ evolution in response to environmental pathogens.47 Although the framework genes within the MHC class I region essentially have remained conserved and orthologous, the number of MHC classical and non‐classical class I genes vary enormously between the three species. This suggests that despite the possibility of some trans‐species inheritance the majority of the reorganization of the MHC class genes has occurred during and/or after speciation. Human MHC gene sequences are closer to those of the macaque than the mouse and similar to the phylogenetic relationships shown in Fig. 1, but the MHC class I genes are also paralogous within each of the three species. In contrast, the MHC class II genes are essentially orthologous between the three species (Fig. 5). This paralogy within the MHC class I region highlights its uniqueness within these three mammalian genomes and easily distinguishes it from the other two major regions, class II and class III, within the MHC supra loci. Most of these paralogous genes have arisen from monogenic and multigenic duplication events involving retro‐elements, especially ancient endogenous retroviruses, as part of the duplication or inversion mechanisms during speciation.11, 13, 31, 48
MHC polymorphism and haplotype structure
The MHC class I and class II gene regions are among the most polymorphic nucleotide stretches within the genome of mammals.4, 11, 12 These polymorphisms include single nucleotide polymorphisms, deletions, substitutions, insertions and repeat elements including short tandem repeats or microsatellites. The reasons for this extensive and extreme polymorphism is not always readily clear, although the hypervariable exonic sites of the MHC class I and class II genes are generally interpreted as necessary for allowing the transport of a greater diversity of peptide sequences for MHC presentation to circulating immune cells. Another possibility is that many of the polymorphisms have evolved rapidly within and between species as a result of duplication events, indel activity, sequence mutations and associated hitchhiking diversity in response to adaptation of the MHC to the constantly evolving microbial antigenic attack.12
Of the total of 14 232 HLA allele sequences reported by the IMGT/HLA database release 3·23 in January 2016, 10 574 were in the class I region and 3658 were in the class II gene regions. In contrast to the HLA, a total of only 1704 Mafa, 1407 Mamu and 721 Mane MHC alleles were released by IPD‐MHC database on 2 April, 2016 (available from: http://www.ebi.ac.uk/ipd/mhc/; Table 3, and see Supplementary material, Table S5). Most of the macaque allele sequences were determined by RT‐PCR‐based Sanger sequencing and next generation sequencing and were probably free of contaminated PCR products originating from pseudogenes. Of the macaque MHC‐B alleles, 101 alleles were perfectly matched with at least two species (Fig. 1). These trans‐species polymorphisms were probably already generated before speciation of the macaques 2·4–4·2 million years ago.49 In addition, MICA, MICB and MICA/B genes in macaque species are polymorphic like the human MICA and MICB genes.45, 50
Table 3.
Allele numbers of MHC alleles in three macaque species
| Locus | Mafa | Mamu | Mane | |
|---|---|---|---|---|
| Class I | MHC‐F | 8 | 6 | 0 |
| MHC‐G | 10 | 4 | 0 | |
| MHC‐AG | 35 | 9 | 1 | |
| MHC‐A a | 380 | 336 | 150 | |
| MHC‐E | 5 | 22 | 8 | |
| MHC‐B b | 501 | 462 | 285 | |
| MHC‐I | 42 | 43 | 44 | |
| Class II | MHC‐DRA | 50 | 28 | 16 |
| MHC‐DRB c | 283 | 261 | 113 | |
| MHC‐DQA1 | 82 | 46 | 30 | |
| MHC‐DQB1 | 95 | 79 | 40 | |
| MHC‐DOA | 15 | 0 | 0 | |
| MHC‐DOB | 16 | 0 | 0 | |
| MHC‐DMA | 11 | 0 | 0 | |
| MHC‐DMB | 7 | 0 | 0 | |
| MHC‐DPA1 | 80 | 50 | 18 | |
| MHC‐DPB1 | 84 | 61 | 16 | |
| Total | 1704 | 1407 | 721 |
Total allele numbers of A1‐A8 loci.
Total allele numbers of B, B11L, B12, B16, B17, B20 and B21 loci.
Total allele numbers of DRB*W, DRB1, DRB3, DRB4, DRB5 and DRB6 loci. Detailed allele numbers are shown in the Supplementary material (Table S5).
It was observed in humans that HLA haplotypes are often ancestral in that they have been inherited largely intact over many generations because the polymorphisms within the Class I and Class II blocks have been frozen due to a suppression of meiotic recombination within and between these polymorphic regions.11, 24 Ancestral‐like MHC haplotypes also appear to exist in mice18 and macaques.45, 51, 52, 53, 54, 55 The macaque MHC class I, class II and/or entire MHC haplotypes were estimated using the MHC allele and/or pedigree information in each species such as Mafa,51, 52, 53, 54, 55, 56, 57, 58, 59, 60 Mamu 61, 62, 63 and Mane.64, 65 Copy number variations of the MHC‐A, MHC‐E, MHC‐B, MHC‐I and MHC‐DRB loci were observed in the different macaque species, similar to the HLA‐DR haplotype.66 For example, two MHC‐A, two MHC‐E, three MHC‐B, one MHC‐I and three MHC‐DRB loci were identified in the most frequent Mafa haplotype (tentatively named ‘HT1’) in the Filipino macaque population (Fig. 6).51 Hence, the HLA system has accumulated a large amount of allelic variation at only a few limited HLA loci, whereas the macaque MHC system has many regional configurations generated by birth and death evolution with little allelic variation at many different loci.9
Figure 6.
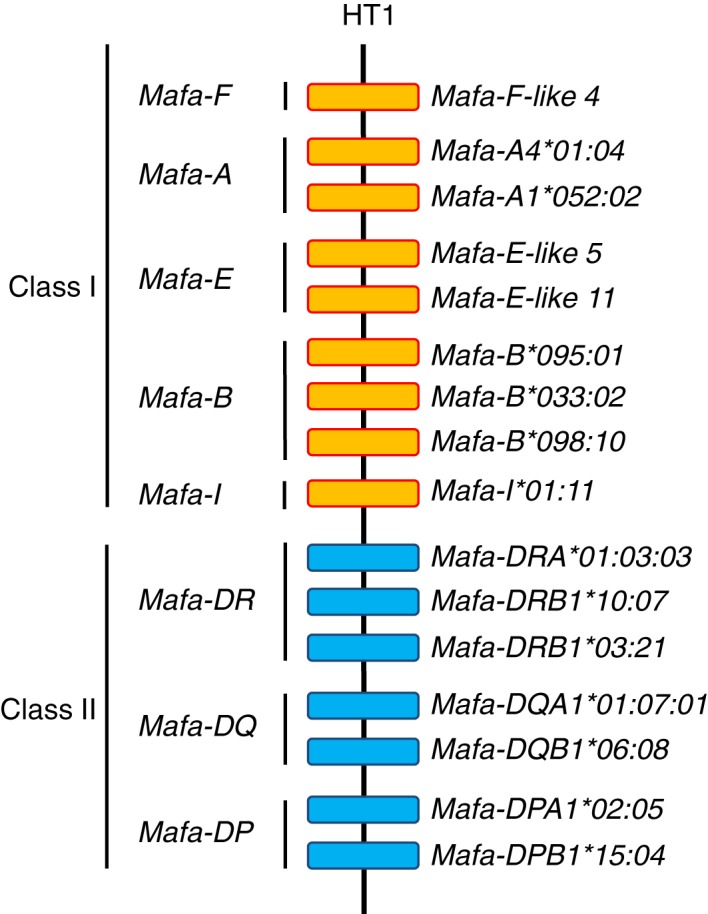
Gene organization of the most frequent Filipino Mafa‐MHC haplotype. Orange and blue boxes indicate MHC class I and MHC class II genes, respectively.
MHC class I gene functions and disease
Although the MHC was originally considered to be an important chromosomal region with a cluster of genes specifically involved in graft rejection,1, 3, 67 the modern view of the MHC is seen as a large, haplotypic genomic region of more than 250 genes with approximately 40% of them expressed in the immune system.23 It is beyond the scope of this review to elaborate on all the possible functions of these genes, suffice to say that many of them may have some ancillary role in regulating the innate and adaptive immune response. In this regard, we limit our attention here to a brief consideration of the class I genes, which are different to the class II genes, even though they both function to present peptides to T‐cell receptors. However, many MHC class I genes do not necessarily have the classical class I function of peptide presentation to T‐cell receptors. For example, in mice, some of the MHC class Ib molecules expressed by non‐classical class I genes such as H2‐M and H2‐T family members associate with V2R pheromone receptors and have vomeronasal sensory rather than immune functions.36, 39, 40 Hence, the number, type and function of classical and non‐classical class I genes in the MHC is often dependent on the species under investigation.
In humans, the MHC classical class I genes are involved critically in organ transplant rejection and graft‐versus‐host disease following haematopoietic stem cell transplants.1, 67 Various associations have been evidenced between HLA class I molecules and the numerous autoimmune diseases,68, 69, 70, 71 as well as infectious diseases72 and drug adverse reactions.73, 74 Apart from their essential role in the elaboration of adaptive immune responses, the role of MHC class I genes was demonstrated in various steps of reproduction such as pregnancy maintenance, mate selection and kin recognition.75 The MHC has also been considered to be a system primarily for sexual selection and avoidance of inbreeding with histocompatibility fulfilling a secondary role.76 The MHC class I gene products also have impact on central nervous system development and plasticity,13, 77, 78, 79, 80 neurological cell interactions,81, 82 synaptic function and behaviour,83, 84 cerebral hemispheric specialization,85 and neurological and psychiatric disorders.86, 87, 88 Hence, the human MHC class I region is one of the most biomedically diverse and important genomic regions that warrant special attention for genetic investigation.
The different macaque species have strong immunological similarities with the human and mouse,22 and therefore, they are often used for investigating the role of MHC class I genes in infectious diseases including human immunodeficiency virus/simian immunodeficiency virus,6 influenza,89 tuberculosis90, 91 and severe acute respiratory syndrome,92 neurological diseases including Alzheimer's disease93 and Parkinson's disease,94 reproduction,95 regenerative medicine using induced pluripotent stem cells and/or embryonic stem cells96, transplantation98, 99 and immunotherapy.100 In this regard, it has been important to know the MHC genetic background to understand how the MHC polymorphisms in the various populations affect the results of these various studies.
Recently, it was reported that one of the Mamu‐B genes, Mamu‐B*98, is capable of binding N‐myristoylated 5‐mer peptides and presenting them to specific cytotoxic T lymphocytes101 but an orthologue for this gene has not been identified in the human and mouse. The Mamu‐I gene was reported to be an oligomorphic MHC‐B‐like gene with classical and non‐classical characteristics.102 In addition, although the HLA‐E has very limited polymorphism and serves as the ligand for the inhibitory NKG2A receptor expressed by natural killer cells,103 recent studies have shown that the MHC‐E genes in macaque species are polymorphic.104 Because Mafa‐E and Mamu‐E have increased polymorphism and haplotype diversity, they may have a different function to HLA‐E gene. On the other hand, the Mafa‐F alleles are well conserved and limited in number like those of HLA‐F, which was recently identified to be one of the ligands for natural killer cell immunoglobulin‐like receptors (KIRs).105
Conclusion
Here, we have provided a short review of the genomic similarities and differences between the macaque, human and mouse MHC regions with a particular emphasis on the comparative polymorphisms, haplotype structure and function of the class I region of humans and macaques. As described above, the macaque species have had many more MHC class I genes generated by gene duplication events than those in humans, whereas the organization of MHC class II genes is well conserved between the two species. The MHC class I genes in macaque species have been divided into major expressed genes and minor expressed genes,106 but how most of these genes are regulated and what their functions are remain unknown. The gene clusters, polymorphisms and haplotypes of the KIR genomic region generally have been analysed independently of the MHC genomic region in the macaque species.6, 7, 8 However, the KIR gene cluster has a complex haplotype structure similar to the class I and class II gene clusters in the MHC genomic region, which suggests that the MHC and KIR gene polymorphisms and haplotypes might have co‐evolved in macaques by duplication, deletion and mutation to generate copy number variation.9 Much more genetic diversity and transcriptome data of MHC and KIR genes in macaque species will be necessary for biomedical studies, such as human immunodeficiency virus/simian immunodeficiency virus and transplantation studies, to progress towards a better understanding of the interrelated roles of the MHC and KIR. Hence, the detailed genomic comparison between three distinct mammalian species provided in this review suggests that more comprehensive genotyping and functional studies of the MHC, NK receptor and also MHC accessory genes relating to MHC expression are still required for the macaque species to provide a better insight into the roles of their MHC genes in immunity, neurology, transplantation and infectious diseases.
Disclosures
The authors declare that they have no conflict of interest.
Supporting information
Figure S1. Venn diagram of the number of Mamu, Mafa and Mane MHC‐B alleles shared among three macaque species.
Table S1. Locus information in the HLA genomic region (20 November 2015 to present).
Table S2. Genomic diversity of the MHC genes in the MHC region among the human, rhesus macaque and mouse (20 September 2015 to present).
Table S3. Summary for classification of MHC genes for each sub‐region.
Table S4. Conservation of non‐MHC loci in the MHC genomic region among human, macaque species and mouse (20 NJovember 2015 to present).
Table S5. Details of allele numbers of MHC alleles in representable macaque species.
Acknowledgements
TS, AB, HI and JKK wrote the paper. This work was supported by MEXT KAKENHI (No. 221S0002), JSPS KAKENHI (No. 21300155), and was partially supported by the programme ‘Research Centre Network for Realization of Regenerative Medicine’ in Japan Agency for Medical Research and Development (AMED).
References
- 1. Zinkernagel RM, Doherty PC. The discovery of MHC restriction. Immunol Today 1997; 18:14–17. [DOI] [PubMed] [Google Scholar]
- 2. Kulski JK, Inoko H. Major histocompatibility complex (MHC) Genes. Nat Encycl Hum Genom 2003; 3:778–85. [Google Scholar]
- 3. Thorsby E. A short history of HLA. Tissue Antigens 2009; 74:101–16. [DOI] [PubMed] [Google Scholar]
- 4. Danchin E, Vitiello V, Vienne A, Richard O, Gouret P, McDermott MF et al The major histocompatibility complex origin. Immunol Rev 2004; 198:216–32. [DOI] [PubMed] [Google Scholar]
- 5. Joffre OP, Segura E, Savina A, Amigorena S. Cross‐presentation by dendritic cells. Nat Rev Immunol 2012; 12:557–69. [DOI] [PubMed] [Google Scholar]
- 6. Walter L, Ansari AA. MHC and KIR Polymorphisms in Rhesus Macaque SIV Infection. Front Immunol 2015; 6:540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moreland AJ, Guethlein LA, Reeves RK, Broman KW, Johnson RP, Parham P et al Characterization of killer immunoglobulin‐like receptor genetics and comprehensive genotyping by pyrosequencing in rhesus macaques. BMC Genom 2011; 12:295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bimber BN, Moreland AJ, Wiseman RW, Hughes AL, O'Connor DH. Complete characterization of killer Ig‐like receptor (KIR) haplotypes in Mauritian cynomolgus macaques: novel insights into nonhuman primate KIR gene content and organization. J Immunol 2008; 181:6301–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. de Groot NG, Blokhuis JH, Otting N, Doxiadis GG, Bontrop RE. Co‐evolution of the MHC class I and KIR gene families in rhesus macaques: ancestry and plasticity. Immunol Rev 2015; 267:228–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Martin AM, Kulski JK, Witt C, Pontarotti P, Christiansen FT. Leukocyte Ig‐like receptor complex (LRC) in mice and men. Trends Immunol 2002; 23:81–8. [DOI] [PubMed] [Google Scholar]
- 11. Dawkins R, Leelayuwat C, Gaudieri S, Tay G, Hui J, Cattley S et al Genomics of the major histocompatibility complex: haplotypes, duplication, retroviruses and disease. Immunol Rev 1999; 167:275–304. [DOI] [PubMed] [Google Scholar]
- 12. Shiina T, Ota M, Shimizu S, Katsuyama Y, Hashimoto N, Takasu M et al Rapid evolution of major histocompatibility complex class I genes in primates generates new disease alleles in humans via hitchhiking diversity. Genetics 2006; 173:1555–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ohtsuka M, Inoko H, Kulski JK, Yoshimura S. Major histocompatibility complex (MHC) class Ib gene duplications, organization and expression patterns in mouse strain C57BL/6. BMC Genom 2008; 9:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dausset J. Iso‐leuko‐antibodies. Acta Haematol 1958; 20:156–66. [DOI] [PubMed] [Google Scholar]
- 15. Van Rood JJ, Eernisse JG, Van Leeuwen A. Leucocyte antibodies in sera from pregnant women. Nature 1958; 181:1735–6. [DOI] [PubMed] [Google Scholar]
- 16. Payne R, Rolfs MR. Fetomaternal leukocyte incompatibility. J Clin Invest 1958; 37:1756–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kulski JK, Shiina T, Anzai T, Kohara S, Inoko H. Comparative genomic analysis of the MHC: the evolution of class I duplication blocks, diversity and complexity from shark to man. Immunol Rev 2002; 190:95–122. [DOI] [PubMed] [Google Scholar]
- 18. Kumanovics A, Takada T, Lindahl KF. Genomic organization of the mammalian MHC. Annu Rev Immunol 2003; 21:629–57. [DOI] [PubMed] [Google Scholar]
- 19. Gaur LK, Heise ER, Hansen JA, Clark EA. Conservation of HLA class I private epitopes in macaques. Immunogenetics 1988; 27:356–62. [DOI] [PubMed] [Google Scholar]
- 20. Poignard P, Moldt B, Maloveste K, Campos N, Olson WC, Rakasz E et al Protection against high‐dose highly pathogenic mucosal SIV challenge at very low serum neutralizing titers of the antibody‐like molecule CD4‐IgG2. PLoS ONE 2012; 7:e42209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ahnini RT, Henry J, Pontarotti P. Geography and History of the Genes in the Human MHC: Can We Predict MHC Organization in Nonhuman Primates? In: Blancher A, Klein J, Socha WW, eds. Molecular Biology and Evolution of Blood Group and MHC Antigens in Primates. Berlin: Springer, 1997:325–39. [Google Scholar]
- 22. Gaur LK, Antonelli P, Clark EA, Hansen JA. Evolution of HLA class I epitopes defined by murine monoclonal antibodies: distribution in macaques. Hum Immunol 1986; 17:406–15. [DOI] [PubMed] [Google Scholar]
- 23. The MHC sequencing consortium . Complete sequence and gene map of a human major histocompatibility complex. Nature 1999; 401:921–3. [DOI] [PubMed] [Google Scholar]
- 24. Horton R, Gibson R, Coggill P, Miretti M, Allcock RJ, Almeida J et al Variation analysis and gene annotation of eight MHC haplotypes: the MHC Haplotype Project. Immunogenetics 2008; 60:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shiina T, Inoko H, Kulski JK. An update of the HLA genomic region, locus information and disease associations: 2004. Tissue Antigens 2004; 64:631–49. [DOI] [PubMed] [Google Scholar]
- 26. Shiina T, Hosomichi K, Inoko H, Kulski JK. The HLA genomic loci map: expression, interaction, diversity and disease. J Hum Genet 2009; 54:15–39. [DOI] [PubMed] [Google Scholar]
- 27. Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK et al Gene map of the extended human MHC. Nat Rev Genet 2004; 5:889–99. [DOI] [PubMed] [Google Scholar]
- 28. Stewart CA, Horton R, Allcock RJ, Ashurst JL, Atrazhev AM, Coggill P et al Complete MHC haplotype sequencing for common disease gene mapping. Genome Res 2004; 14:1176–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Watanabe A, Shiina T, Shimizu S, Hosomichi K, Yanagiya K, Kita YF et al A BAC‐based contig map of the cynomolgus macaque (Macaca fascicularis) major histocompatibility complex genomic region. Genomics 2007; 89:402–12. [DOI] [PubMed] [Google Scholar]
- 30. Daza‐Vamenta R, Glusman G, Rowen L, Guthrie B, Geraghty DE. Genetic divergence of the rhesus macaque major histocompatibility complex. Genome Res 2004; 14:1501–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kulski JK, Anzai T, Shiina T, Inoko H. Rhesus macaque class I duplicon structures, organization, and evolution within the α block of the major histocompatibility complex. Mol Biol Evol 2004; 21:2079–91. [DOI] [PubMed] [Google Scholar]
- 32. Boyson JE, Iwanaga KK, Golos TG, Watkins DI. Identification of a novel MHC class I gene, Mamu‐AG, expressed in the placenta of a primate with an inactivated G locus. J Immunol 1997; 159:3311–21. [PubMed] [Google Scholar]
- 33. Boyson JE, Iwanaga KK, Urvater JA, Hughes AL, Golos TG, Watkins DI. Evolution of a new nonclassical MHC class I locus in two Old World primate species. Immunogenetics 1999; 49:86–98. [DOI] [PubMed] [Google Scholar]
- 34. Flaherty L, Elliott E, Tine JA, Walsh AC, Waters JB. Immunogenetics of the Q and TL regions of the mouse. Crit Rev Immunol 1990; 10:131–75. [PubMed] [Google Scholar]
- 35. Lindahl KF, Byers DE, Dabhi VM, Hovik R, Jones EP, Smith GP et al H2‐M3, a full‐service class Ib histocompatibility antigen. Annu Rev Immunol 1997; 15:851–79. [DOI] [PubMed] [Google Scholar]
- 36. Loconto J, Papes F, Chang E, Stowers L, Jones EP, Takada T et al Functional expression of murine V2R pheromone receptors involves selective association with the M10 and M1 families of MHC class Ib molecules. Cell 2003; 112:607–18. [DOI] [PubMed] [Google Scholar]
- 37. Arepalli SR, Jones EP, Howcroft TK, Carlo I, Wang C, Lindahl KF et al Characterization of two class I genes from the H2‐M region: evidence for a new subfamily. Immunogenetics 1998; 47:264–71. [DOI] [PubMed] [Google Scholar]
- 38. Chen L, Reyes‐Vargas E, Dai H, Escobar H, Rudd B, Fairbanks J et al Expression of the mouse MHC class Ib H2‐T11 gene product, a paralog of H2‐T23 (Qa‐1) with shared peptide‐binding specificity. J Immunol 2014; 193:1427–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ishii T, Hirota J, Mombaerts P. Combinatorial coexpression of neural and immune multigene families in mouse vomeronasal sensory neurons. Curr Biol 2003; 13:394–400. [DOI] [PubMed] [Google Scholar]
- 40. Ishii T, Mombaerts P. Expression of nonclassical class I major histocompatibility genes defines a tripartite organization of the mouse vomeronasal system. J Neurosci 2008; 28:2332–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mashimo H, Chorney MJ, Pontarotti P, Fisher DA, Hood L, Nathenson SG. Nucleotide sequence of the BALB/c H‐2T region gene, T3d. Immunogenetics 1992; 36:326–32. [DOI] [PubMed] [Google Scholar]
- 42. Crowley MP, Fahrer AM, Baumgarth N, Hampl J, Gutgemann I, Teyton L et al A population of murine γδ T cells that recognize an inducible MHC class Ib molecule. Science 2000; 287:314–16. [DOI] [PubMed] [Google Scholar]
- 43. Guidry PA, Stroynowski I. The murine family of gut‐restricted class Ib MHC includes alternatively spliced isoforms of the proposed HLA‐G homolog, “blastocyst MHC”. J Immunol 2005; 175:5248–59. [DOI] [PubMed] [Google Scholar]
- 44. Lew AM, Maloy WL, Coligan JE. Characteristics of the expression of the murine soluble class I molecule (Q10). J Immunol 1986; 136:254–8. [PubMed] [Google Scholar]
- 45. Doxiadis GG, Heijmans CM, Otting N, Bontrop RE. MIC gene polymorphism and haplotype diversity in rhesus macaques. Tissue Antigens 2007; 69:212–19. [DOI] [PubMed] [Google Scholar]
- 46. Bontrop RE, Otting N, de Groot NG, Doxiadis GG. Major histocompatibility complex class II polymorphisms in primates. Immunol Rev 1999; 167:339–50. [DOI] [PubMed] [Google Scholar]
- 47. Nei M, Gu X, Sitnikova T. Evolution by the birth‐and‐death process in multigene families of the vertebrate immune system. Proc Natl Acad Sci USA 1997; 94:7799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shiina T, Tamiya G, Oka A, Takishima N, Inoko H. Genome sequencing analysis of the 1.8 Mb entire human MHC class I region. Immunol Rev 1999; 167:193–9. [DOI] [PubMed] [Google Scholar]
- 49. Hedges SB, Dudley J, Kumar S. TimeTree: a public knowledge‐base of divergence times among organisms. Bioinformatics 2006; 22:2971–2. [DOI] [PubMed] [Google Scholar]
- 50. Meyer A, Carapito R, Ott L, Radosavljevic M, Georgel P, Adams EJ et al High diversity of MIC genes in non‐human primates. Immunogenetics 2014; 66:581–7. [DOI] [PubMed] [Google Scholar]
- 51. Shiina T, Yamada Y, Aarnink A, Suzuki S, Masuya A, Ito S et al Discovery of novel MHC‐class I alleles and haplotypes in Filipino cynomolgus macaques (Macaca fascicularis) by pyrosequencing and Sanger sequencing: Mafa‐class I polymorphism. Immunogenetics 2015; 67:563–78. [DOI] [PubMed] [Google Scholar]
- 52. Saito Y, Naruse TK, Akari H, Matano T, Kimura A. Diversity of MHC class I haplotypes in cynomolgus macaques. Immunogenetics 2012; 64:131–41. [DOI] [PubMed] [Google Scholar]
- 53. Budde ML, Wiseman RW, Karl JA, Hanczaruk B, Simen BB, O'Connor DH. Characterization of Mauritian cynomolgus macaque major histocompatibility complex class I haplotypes by high‐resolution pyrosequencing. Immunogenetics 2010; 62:773–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Otting N, de Groot N, de Vos‐Rouweler AJ, Louwerse A, Doxiadis GG, Bontrop RE. Multilocus definition of MHC haplotypes in pedigreed cynomolgus macaques (Macaca fascicularis). Immunogenetics 2012; 64:755–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Otting N, Doxiadis GG, Bontrop RE. Definition of Mafa‐A and ‐B haplotypes in pedigreed cynomolgus macaques (Macaca fascicularis). Immunogenetics 2009; 61:745–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kita YF, Hosomichi K, Kohara S, Itoh Y, Ogasawara K, Tsuchiya H et al MHC class I A loci polymorphism and diversity in three Southeast Asian populations of cynomolgus macaque. Immunogenetics 2009; 61:635–48. [DOI] [PubMed] [Google Scholar]
- 57. Blancher A, Aarnink A, Yamada Y, Tanaka K, Yamanaka H, Shiina T. Study of MHC class II region polymorphism in the Filipino cynomolgus macaque population. Immunogenetics 2014; 66:219–30. [DOI] [PubMed] [Google Scholar]
- 58. Blancher A, Aarnink A, Tanaka K, Ota M, Inoko H, Yamanaka H et al Study of cynomolgus monkey (Macaca fascicularis) Mhc DRB gene polymorphism in four populations. Immunogenetics 2012; 64:605–14. [DOI] [PubMed] [Google Scholar]
- 59. O'Connor SL, Blasky AJ, Pendley CJ, Becker EA, Wiseman RW, Karl JA et al Comprehensive characterization of MHC class II haplotypes in Mauritian cynomolgus macaques. Immunogenetics 2007; 59:449–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Campbell KJ, Detmer AM, Karl JA, Wiseman RW, Blasky AJ, Hughes AL et al Characterization of 47 MHC class I sequences in Filipino cynomolgus macaques. Immunogenetics 2009; 61:177–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Naruse TK, Chen Z, Yanagida R, Yamashita T, Saito Y, Mori K et al Diversity of MHC class I genes in Burmese‐origin rhesus macaques. Immunogenetics 2010; 62:601–11. [DOI] [PubMed] [Google Scholar]
- 62. Karl JA, Bohn PS, Wiseman RW, Nimityongskul FA, Lank SM, Starrett GJ et al Major histocompatibility complex class I haplotype diversity in Chinese rhesus macaques. G3: Genes ‐ Genomes ‐ Genetics 2013; 3:1195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Doxiadis GG, Otting N, de Groot NG, de Groot N, Rouweler AJ, Noort R et al Evolutionary stability of MHC class II haplotypes in diverse rhesus macaque populations. Immunogenetics 2003; 55:540–51. [DOI] [PubMed] [Google Scholar]
- 64. Fernandez CS, Reece JC, Saepuloh U, De Rose R, Ishkandriati D, O'Connor DH et al Screening and confirmatory testing of MHC class I alleles in pig‐tailed macaques. Immunogenetics 2011; 63:511–21. [DOI] [PubMed] [Google Scholar]
- 65. Gooneratne SL, Alinejad‐Rokny H, Ebrahimi D, Bohn PS, Wiseman RW, O'Connor DH et al Linking pig‐tailed macaque major histocompatibility complex class I haplotypes and cytotoxic T lymphocyte escape mutations in simian immunodeficiency virus infection. J Virol 2014; 88:14310–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Andersson G. Evolution of the human HLA‐DR region. Front Biosci 1998; 3:d739–45. [DOI] [PubMed] [Google Scholar]
- 67. Sasazuki T, Juji T, Morishima Y, Kinukawa N, Kashiwabara H, Inoko H et al Effect of matching of class I HLA alleles on clinical outcome after transplantation of hematopoietic stem cells from an unrelated donor. Japan Marrow Donor Program. N Engl J Med 1998; 339:1177–85. [DOI] [PubMed] [Google Scholar]
- 68. Fernando MM, Stevens CR, Walsh EC, De Jager PL, Goyette P, Plenge RM et al Defining the role of the MHC in autoimmunity: a review and pooled analysis. PLoS Genet 2008; 4:e1000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Cotsapas C, Voight BF, Rossin E, Lage K, Neale BM, Wallace C et al Pervasive sharing of genetic effects in autoimmune disease. PLoS Genet 2011; 7:e1002254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pandit L, Ban M, Sawcer S, Singhal B, Nair S, Radhakrishnan K et al Evaluation of the established non‐MHC multiple sclerosis loci in an Indian population. Mult Scler 2011; 17:139–43. [DOI] [PubMed] [Google Scholar]
- 71. Raychaudhuri S, Sandor C, Stahl EA, Freudenberg J, Lee HS, Jia X et al Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet 2012; 44:291–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. International HIVCS , Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI et al The major genetic determinants of HIV‐1 control affect HLA class I peptide presentation. Science 2010; 330:1551–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. McCormack M, Alfirevic A, Bourgeois S, Farrell JJ, Kasperaviciute D, Carrington M et al HLA‐A*3101 and carbamazepine‐induced hypersensitivity reactions in Europeans. N Engl J Med 2011; 364:1134–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Illing PT, Vivian JP, Dudek NL, Kostenko L, Chen Z, Bharadwaj M et al Immune self‐reactivity triggered by drug‐modified HLA‐peptide repertoire. Nature 2012; 486:554–8. [DOI] [PubMed] [Google Scholar]
- 75. Ziegler A, Kentenich H, Uchanska‐Ziegler B. Female choice and the MHC. Trends Immunol 2005; 26:496–502. [DOI] [PubMed] [Google Scholar]
- 76. Jones JS, Partridge L. Tissue rejection: the price of sexual acceptance? Nature 1983; 304:484–5. [DOI] [PubMed] [Google Scholar]
- 77. Xiao BG, Link H. Immune regulation within the central nervous system. J Neurol Sci 1998; 157:1–12. [DOI] [PubMed] [Google Scholar]
- 78. Huh GS, Boulanger LM, Du H, Riquelme PA, Brotz TM, Shatz CJ. Functional requirement for class I MHC in CNS development and plasticity. Science 2000; 290:2155–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Boulanger LM, Shatz CJ. Immune signalling in neural development, synaptic plasticity and disease. Nat Rev Neurosci 2004; 5:521–31. [DOI] [PubMed] [Google Scholar]
- 80. Cullheim S, Thams S. The microglial networks of the brain and their role in neuronal network plasticity after lesion. Brain Res Rev 2007; 55:89–96. [DOI] [PubMed] [Google Scholar]
- 81. Matsuo R, Asada A, Fujitani K, Inokuchi K. LIRF, a gene induced during hippocampal long‐term potentiation as an immediate‐early gene, encodes a novel RING finger protein. Biochem Biophys Res Commun 2001; 289:479–84. [DOI] [PubMed] [Google Scholar]
- 82. Patino‐Lopez G, Hevezi P, Lee J, Willhite D, Verge GM, Lechner SM et al Human class‐I restricted T cell associated molecule is highly expressed in the cerebellum and is a marker for activated NKT and CD8+ T lymphocytes. J Neuroimmunol 2006; 171:145–55. [DOI] [PubMed] [Google Scholar]
- 83. Goddard CA, Butts DA, Shatz CJ. Regulation of CNS synapses by neuronal MHC class I. Proc Natl Acad Sci USA 2007; 104:6828–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tonelli LH, Postolache TT, Sternberg EM. Inflammatory genes and neural activity: involvement of immune genes in synaptic function and behavior. Front Biosci 2005; 10:675–80. [DOI] [PubMed] [Google Scholar]
- 85. Lengen C, Regard M, Joller H, Landis T, Lalive P. Anomalous brain dominance and the immune system: do left‐handers have specific immunological patterns? Brain Cogn 2009; 69:188–93. [DOI] [PubMed] [Google Scholar]
- 86. O'Keefe GM, Nguyen VT, Benveniste EN. Regulation and function of class II major histocompatibility complex, CD40, and B7 expression in macrophages and microglia: Implications in neurological diseases. J Neurovirol 2002; 8:496–512. [DOI] [PubMed] [Google Scholar]
- 87. Raha‐Chowdhury R, Andrews SR, Gruen JR. CAT 53: a protein phosphatase 1 nuclear targeting subunit encoded in the MHC Class I region strongly expressed in regions of the brain involved in memory, learning, and Alzheimer's disease. Brain Res Mol Brain Res 2005; 138:70–83. [DOI] [PubMed] [Google Scholar]
- 88. Bailey SL, Carpentier PA, McMahon EJ, Begolka WS, Miller SD. Innate and adaptive immune responses of the central nervous system. Crit Rev Immunol 2006; 26:149–88. [DOI] [PubMed] [Google Scholar]
- 89. Arikata M, Itoh Y, Okamatsu M, Maeda T, Shiina T, Tanaka K et al Memory immune responses against pandemic (H1N1) 2009 influenza virus induced by a whole particle vaccine in cynomolgus monkeys carrying Mafa‐A1*052:02. PLoS ONE 2012; 7:e37220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Capuano SV 3rd, Croix DA, Pawar S, Zinovik A, Myers A, Lin PL et al Experimental Mycobacterium tuberculosis infection of cynomolgus macaques closely resembles the various manifestations of human M. tuberculosis infection. Infect Immun 2003; 71:5831–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Kita Y, Tanaka T, Yoshida S, Ohara N, Kaneda Y, Kuwayama S et al Novel recombinant BCG and DNA‐vaccination against tuberculosis in a cynomolgus monkey model. Vaccine 2005; 23:2132–5. [DOI] [PubMed] [Google Scholar]
- 92. Lawler JV, Endy TP, Hensley LE, Garrison A, Fritz EA, Lesar M et al Cynomolgus macaque as an animal model for severe acute respiratory syndrome. PLoS Med 2006; 3:e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wang CY, Finstad CL, Walfield AM, Sia C, Sokoll KK, Chang TY et al Site‐specific UBITh amyloid‐β vaccine for immunotherapy of Alzheimer's disease. Vaccine 2007; 25:3041–52. [DOI] [PubMed] [Google Scholar]
- 94. Emborg ME. Nonhuman primate models of Parkinson's disease. ILAR J 2007; 48:339–55. [DOI] [PubMed] [Google Scholar]
- 95. Shively CA, Wood CE, Register TC, Willard SL, Lees CJ, Chen H et al Hormone therapy effects on social behavior and activity levels of surgically postmenopausal cynomolgus monkeys. Psychoneuroendocrinology 2007; 32:981–90. [DOI] [PubMed] [Google Scholar]
- 96. Morizane A, Doi D, Kikuchi T, Okita K, Hotta A, Kawasaki T et al Direct comparison of autologous and allogeneic transplantation of iPSC‐derived neural cells in the brain of a non‐human primate. Stem Cell Rep 2013; 1:283–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kawamura T, Miyagawa S, Fukushima S, Maeda A, Kashiyama N, Kawamura A, et al. Cardiomyocytes Derived from MHC‐Homozygous Induced Pluripotent Stem Cells Exhibit Reduced Allogeneic Immunogenicity in MHC‐Matched Non‐human Primates. Stem Cell Rep 2016; 6:312–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Larsen CP, Page A, Linzie KH, Russell M, Deane T, Stempora L et al An MHC‐defined primate model reveals significant rejection of bone marrow after mixed chimerism induction despite full MHC matching. Am J Transplant 2010; 10:2396–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Kisu I, Mihara M, Banno K, Hara H, Masugi Y, Araki J et al Uterus allotransplantation in cynomolgus macaque: a preliminary experience with non‐human primate models. J Obstet Gynaecol Res 2014; 40:907–18. [DOI] [PubMed] [Google Scholar]
- 100. Liu C, Noorchashm H, Sutter JA, Naji M, Prak EL, Boyer J et al B lymphocyte‐directed immunotherapy promotes long‐term islet allograft survival in nonhuman primates. Nat Med 2007; 13:1295–8. [DOI] [PubMed] [Google Scholar]
- 101. Morita D, Yamamoto Y, Mizutani T, Ishikawa T, Suzuki J, Igarashi T. Crystal structure of the N‐myristoylated lipopeptide‐bound MHC class I complex. Nat Commun. 2016; 7:10356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Urvater JA, Otting N, Loehrke JH, Rudersdorf R, Slukvin II, Piekarczyk MS et al Mamu‐I: a novel primate MHC class I B‐related locus with unusually low variability. J Immunol 2000; 164:1386–98. [DOI] [PubMed] [Google Scholar]
- 103. Lee N, Llano M, Carretero M, Ishitani A, Navarro F, Lopez‐Botet M et al HLA‐E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc Natl Acad Sci USA 1998; 95:5199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Boyson JE, McAdam SN, Gallimore A, Golos TG, Liu X, Gotch FM et al The MHC E locus in macaques is polymorphic and is conserved between macaques and humans. Immunogenetics 1995; 41:59–68. [DOI] [PubMed] [Google Scholar]
- 105. Goodridge JP, Burian A, Lee N, Geraghty DE. HLA‐F and MHC class I open conformers are ligands for NK cell Ig‐like receptors. J Immunol 2013; 191:3553–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Wiseman RW, Karl JA, Bimber BN, O'Leary CE, Lank SM, Tuscher JJ et al Major histocompatibility complex genotyping with massively parallel pyrosequencing. Nat Med 2009; 15:1322–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011; 28:2731–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Saitou N, Nei M. The neighbor‐joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 1987; 4:406–25. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Venn diagram of the number of Mamu, Mafa and Mane MHC‐B alleles shared among three macaque species.
Table S1. Locus information in the HLA genomic region (20 November 2015 to present).
Table S2. Genomic diversity of the MHC genes in the MHC region among the human, rhesus macaque and mouse (20 September 2015 to present).
Table S3. Summary for classification of MHC genes for each sub‐region.
Table S4. Conservation of non‐MHC loci in the MHC genomic region among human, macaque species and mouse (20 NJovember 2015 to present).
Table S5. Details of allele numbers of MHC alleles in representable macaque species.


