ABSTRACT
Glycosylphosphatidylinositol (GPI) anchoring of the prion protein (PrPC) influences PrPC misfolding into the disease-associated isoform, PrPres, as well as prion propagation and infectivity. GPI proteins are found in cholesterol- and sphingolipid-rich membrane regions called rafts. Exchanging the GPI anchor for a nonraft transmembrane sequence redirects PrPC away from rafts. Previous studies showed that nonraft transmembrane PrPC variants resist conversion to PrPres when transfected into scrapie-infected N2a neuroblastoma cells, likely due to segregation of transmembrane PrPC and GPI-anchored PrPres in distinct membrane environments. Thus, it remained unclear whether transmembrane PrPC might convert to PrPres if seeded by an exogenous source of PrPres not associated with host cell rafts and without the potential influence of endogenous expression of GPI-anchored PrPC. To further explore these questions, constructs containing either a C-terminal wild-type GPI anchor signal sequence or a nonraft transmembrane sequence containing a flexible linker were expressed in a cell line derived from PrP knockout hippocampal neurons, NpL2. NpL2 cells have physiological similarities to primary neurons, representing a novel and advantageous model for studying transmissible spongiform encephalopathy (TSE) infection. Cells were infected with inocula from multiple prion strains and in different biochemical states (i.e., membrane bound as in brain microsomes from wild-type mice or purified GPI-anchorless amyloid fibrils). Only GPI-anchored PrPC supported persistent PrPres propagation. Our data provide strong evidence that in cell culture GPI anchor-directed membrane association of PrPC is required for persistent PrPres propagation, implicating raft microdomains as a location for conversion.
IMPORTANCE Mechanisms of prion propagation, and what makes them transmissible, are poorly understood. Glycosylphosphatidylinositol (GPI) membrane anchoring of the prion protein (PrPC) directs it to specific regions of cell membranes called rafts. In order to test the importance of the raft environment on prion propagation, we developed a novel model for prion infection where cells expressing either GPI-anchored PrPC or transmembrane-anchored PrPC, which partitions it to a different location, were treated with infectious, misfolded forms of the prion protein, PrPres. We show that only GPI-anchored PrPC was able to convert to PrPres and able to serially propagate. The results strongly suggest that GPI anchoring and the localization of PrPC to rafts are crucial to the ability of PrPC to propagate as a prion.
KEYWORDS: GPI, NpL2, PrP, neuron, prion, raft, transmembrane, transmissible spongiform encephalopathy
INTRODUCTION
Misfolding and aggregation of the prion protein (PrPC) are associated with several infectious disorders affecting mammals, including Creutzfeldt-Jakob disease in humans, scrapie in sheep and goats, chronic wasting disease in cervids, and bovine spongiform encephalopathy in cattle, collectively known as transmissible spongiform encephalopathies (TSEs) (1). PrPC consists of a largely alpha-helical C-terminal folded domain, while the N-terminal half of the protein is unstructured (2–4). Misfolding of PrPC involves a conformational change in the protein combined with its self-assembly that usually imparts detergent insolubility and protease resistance onto the aggregated isoform, PrPres (5). In iatrogenic forms of prion disease, i.e., transmission through medical procedure, the conformational conversion process is initiated by PrPres in the inoculum inducing the conversion of host PrPC by a templating mechanism. The cell surface has been proposed as one site of conversion of PrPC to PrPres (6–12). Productive infection requires this process to become self-sustaining, with newly formed PrPres seeding further conversion of PrPC.
PrPres is typically identified using proteinase K (PK), which completely degrades PrPC but removes only the N-terminal ∼67 residues of PrPres to give characteristic TSE strain-associated banding patterns on Western blots of un-, mono-, and diglycosylated forms of PrPres (13). However, infectivity can be present in the absence of any detectable PrPres, possibly explained by the identification of strain-dependent infectious conformers that are either generally protease sensitive (14–19) or sensitive to PK specifically (20, 21). It is not clear whether these isoforms are generated on a different pathway from PrPres and are structurally unrelated or are simply smaller aggregates on the pathway to PrPres formation, although evidence points toward lower-molecular-weight assemblies being more infectious (22) and more PK sensitive (17, 19, 23). Thus, although protease resistance provides a useful tool to determine the presence of PrPres, it may not reveal all infectious material. These observations have led to the development of assays that do not involve any protease treatment, including the conformation-dependent immunoassay (18, 24), amyloid-seeding assay (25), protein-misfolding cyclic amplification (23), and quaking-induced conversion (26–28).
PrP is anchored to membranes by a glycosylphosphatidylinositol (GPI) moiety. Consequently, PrPres and PrPC are found associated with raft microdomains (29–35). PrPC and exogenous PrPres fibrils also undergo clathrin-dependent endocytosis after binding to lipoprotein receptor-related protein 1 (LRP1) via N-terminal basic amino acids in PrP (36, 37). Rafts have been defined as highly dynamic sterol- and sphingolipid-enriched membrane regions that range in size from 10 to 200 nm (38). Rafts are commonly extracted from cells as detergent-resistant membranes (DRMs) using nonionic detergents at low temperatures which solubilize nonraft domains only (39), although there is some controversy over whether these biochemically isolated fractions represent rafts as they exist within the cell (38, 40, 41).
PrP GPI anchoring and localization to raft regions may also be of importance in bona fide TSE pathology. Early evidence for the importance of PrPC raft association came from experiments showing that pharmacological disruption of rafts decreases levels of PrPres in scrapie-infected cells (42, 43). In vivo, transgenic mice expressing GPI-anchorless PrPC develop an amyloid disease upon infection that, although transmissible itself, differs from typical TSE with respect to pathology, clinical signs, and longer incubation periods (44, 45). In cell culture, anchorless PrPC is able to convert to PrPres acutely (46, 47) but requires coexpression of anchored PrPC to persistently infect cells, indicating that GPI anchoring of PrPC is important for stable infection in vitro (47). GPI anchor-dependent modulation of protein aggregation is not limited to PrP. Ectopic expression of the cytoplasmic amyloid-forming yeast prion protein Sup35NM as a GPI-anchored protein in mouse neuroblastoma cells has shown how GPI anchoring can change the behavior of other amyloidogenic proteins besides PrP. Addition of a GPI anchor to Sup35NM facilitated its prion-like propagation and intercellular spread in mammalian cells; aggregation was not observed in control cells expressing anchorless Sup35NM (48). Analogous to its effects on PrP aggregation, GPI anchoring also influenced the nature of the Sup35NM aggregates by directing the formation of nonfibrillar species that lack many defining characteristics of amyloid (49). Collectively, these data point toward GPI anchoring and raft localization as significant facets of prion propagation and TSE pathogenesis.
In order to test the hypothesis that raft localization promotes conversion of PrPC to PrPres, other groups have developed cell culture systems in which PrPC is anchored to membranes via a transmembrane (TM) domain instead of a GPI anchor (42, 50). In these studies, the constructs were expressed in persistently infected N2a cells already propagating PrPres; no exogenous inoculum was added, and in neither case were they found to convert to PrPres. An explanation for the lack of conversion could be that the PrPres in the cells resided in a different membrane environment (rafts) from the site of the PrPC substrate (nonraft); hence, the interaction required for templated conversion of transmembrane PrPC (TM PrP) was prohibited. This conclusion is supported by the observation that PrPC and PrPres must reside in a contiguous membrane for the former to undergo conversion, as the two must be sterically allowed to interact, likely in a specific orientation (7, 51). Other groups have examined PrPC glycosylation and trafficking using a construct containing a TM domain from CD4 or the C terminus of angiotensin-converting enzyme (ACE) (52–56). Although no infection studies were conducted, these experiments showed that TM PrP undergoes proper glycosylation and trafficking to the cell surface, suggesting that TM anchoring has no gross effect on PrP folding and, hence, TM PrP resistance to conversion to PrPres is likely due to the effects of TM anchoring on PrP localization.
To gain comprehensive insight into how membrane anchoring and raft association influence the propagation of PrPres, here we used a novel approach by stably expressing PrPC variants that traffic to different membrane subdomains, i.e., raft and nonraft, in a PrP knockout hippocampal cell line called NpL2, isolated from Zurich I Prnp−/− mice (57). Importantly, two different exogenous sources of PrPres were added to the cultures to attempt to initiate an infection, wild-type (WT) PrPres in the form of brain-derived membranes or amyloid fibrils purified from the brains of scrapie-infected mice expressing anchorless PrPC (44). Since the GPI anchor of WT PrPres might target this inoculum to raft membranes, the testing of both anchorless and anchored PrPres inocula provided assurance that cells expressing either PrPC variant were exposed to at least one type of PrPres seed with the potential to interact with and convert PrPC irrespective of where the PrPC is localized. Several methods were used to examine the persistent propagation of PrPres, including highly sensitive assays. WT or TM mouse PrPC constructs were stably expressed in NpL2 cells (57, 58), which have been used previously for studies on prion biology (59–61). Only WT, raft-associated PrPC supported persistent TSE infection, initiated by inocula containing GPI-anchored or anchorless PrPres. In addition to developing a novel model for studying TSE infection, these experiments provide a rigorous examination into the role of membrane subdomain location in the persistent propagation of PrPres and reveal that, in this model, raft association is a requirement for conversion.
RESULTS
NpL2 cells stably express wild-type and transmembrane forms of PrP which show a similar distribution.
In order to generate a cell line suitable for use in infection experiments, the parent cell line (NpL2) was transduced with viral particles containing the constructs shown in Fig. 1A. The WT PrP construct (top) contains the full-length sequence of 254 amino acids. The GPI anchor signal sequence begins at residue 230 (serine), and so the rest of the sequence was truncated in the TM construct. A flexible linker with sequence SAGAGS was modeled based on other, similar approaches (50, 62, 63) to approximate the flexibility and distance from the membrane imparted onto PrP by the GPI anchor (64). The C-terminal green fluorescent protein (GFP) domain contains an A206K substitution to avoid dimerization of the fluorophore (65), which could promote aggregation of PrP by bringing two molecules into close proximity. GFP was added for imaging purposes; any aggregation of the TM PrP construct would result in clusters of GFP fluorescence, which could be observed in real time as seen with GPI-anchored Sup35NM (48, 49). Furthermore, this domain serves to firmly anchor the TM PrP in the membrane such that it could not be removed without either proteolysis or severe compromise of the integrity of the plasma membrane.
FIG 1.
WT and TM PrPC are expressed, glycosylated, and trafficked to the cell surface in NpL2 cells. (A) Constructs of either WT or TM PrPC were stably expressed in PrP knockout NpL2 cells. The WT sequence was full-length mouse PrP (254 amino acids), whereas the TM sequence consisted of residues 1 to 230 of WT PrP followed by a flexible linker region (SAGAGS), a 22-amino-acid TM domain (Qa), a FLAG tag, and a GFP domain containing an A206K substitution to prevent dimerization of GFP. CMV, cytomegalovirus. (B) Representative Western blot of lysates from NpL2 cells either untransfected (NpL2) or expressing WT or TM PrPC. Brackets indicate full-length PrP bands. White vertical lines indicate removal of irrelevant lanes. (C) Western blot of lysates from untransfected or PrP-expressing cells that were untreated (U) or treated with PNGase F (P). Samples were analyzed on gels electrophoresed in MOPS running buffer to improve resolution of the TM PrPC glycoforms. (D) Immunolabeling of permeabilized (top row) and nonpermeabilized (middle row) cells using anti-PrP antibody 6D11 and imaging by wide-field fluorescence microscopy. Bright-field images of nonpermeabilized cells are shown in the bottom row. Bar, 20 μm. Numbers at left of blots in panels B and C are molecular masses in kilodaltons.
As expression levels in bulk cell populations were relatively low and it is known that some cell lines can express clone-dependent resistance to infection (66), we created multiple cloned cell lines for each construct. WT and TM PrP clones were matched for expression of total PrP (Fig. 1B). It should be noted that the expression levels were comparatively low, approximately 3-fold lower than those of PrPC expression in N2a cells (data not shown). This is not uncommon for NpL2 cell lines created using the pMSCV expression system (T. Onodera, personal communication) and reduces the possibility of spontaneous aggregation of PrPC into PrPres. WT lysates showed the typical banding pattern for PrP with a diglycosylated form running at ∼37 kDa, a monoglycosylated form at ∼30 kDa, and an unglycosylated form at ∼27 kDa. There was a predominant band for the TM PrP at ∼65 kDa; the greater molecular mass than that of WT PrP was consistent with the expected size increase due to the Qa TM domain, FLAG epitope, and 27-kDa GFP tag. Two weaker bands (which can be seen more clearly in Fig. 1C) were also present, corresponding to the mono- and unglycosylated forms of TM PrP as shown by peptide-N-glycosidase F (PNGase F) treatment. A faint, diffuse, glycosylated band at around 45 kDa was also present, possibly a cleavage product. Cloned cell lines were selected so that expression levels of PrPC both overall and on the cell surface were similar between clones (also Fig. 1D). Quantification of lysates from 3 independent cultures of the full-length forms of WT and TM PrP revealed that the two forms expressed similar levels of PrP (TM level was 82% ± 6.8% of WT level).
The addition of mature, complex N-linked sugars on PrPC is one indicator of proper folding and transport to the cell surface (52, 56, 67) and can influence conversion and TSE pathogenesis (68–74). In order to establish if WT and TM PrP were glycosylated similarly, lysates were treated with PNGase F, which cleaves N-linked oligosaccharides from glycoproteins (Fig. 1C). For both WT and TM cell lysates, the band in lysates digested with PNGase F comigrated with the presumptive unglycosylated PrP band in the untreated lane, indicating that the majority of the steady-state population of PrP detected was glycosylated correctly.
To further verify that the distributions of PrP were similar between TM and WT clones, cell surface and intracellular PrP was labeled with an anti-PrP antibody, 6D11 (Fig. 1D). Wide-field epifluorescence imaging of immunolabeled cells confirmed a similar subcellular localization of PrP to the cell surface and prominent perinuclear regions, akin to previously observed distributions of WT PrP with and without a GFP tag (75). Some background labeling was seen in the permeabilized nontransduced NpL2 negative-control cells. However, this was readily distinguished from the labeling in WT and TM cells. Labeling with other anti-PrP antibodies directed at different epitopes gave similar results, suggesting that WT and TM PrP have similar overall folds on a gross level (data not shown).
Collectively, these results show that TM PrP resembled WT PrP in terms of expression level, glycosylation pattern, and distribution, all of which are implicated in the ability of PrPC to convert to PrPres.
TM PrP does not localize to detergent-resistant membranes.
Cholesterol and sphingolipid-rich raft membranes contain GPI-anchored proteins, including PrP, and are insoluble in cold nonionic detergent (31–33, 39). To assay for raft localization of WT and TM PrP, we used a previously reported in situ method involving cell surface PrP immunofluorescence staining combined with detergent extraction (54, 76). Figure 2 shows that untreated cells stably expressing WT or TM PrP were labeled all around the plasma membrane (top row). The specificity of immunolabeling was shown by the absence of fluorescent labeling in untransduced NpL2 control cells (Fig. 2, left column). Only TM PrP was removed from the cell membrane following treatment with cold 1% Triton X-100 (TX-100) (Fig. 2F), suggesting that it is located in a different membrane subdomain from WT PrP, where a large fraction of WT PrP resisted cold TX-100 treatment (Fig. 2E). WT PrP displayed a more punctate distribution after TX-100 treatment, possibly due to raft coalescence following application of detergent or solubilization of some cell surface nonraft PrPC as described by Sunyach et al. (77). Methyl-beta cyclodextrin (MβCD) is a compound that disrupts rafts by binding to and depleting them of cholesterol. As expected, no discernible difference in WT or TM PrP distribution could be observed following MβCD treatment (Fig. 2H) because the spatial resolution limitations of our confocal imaging system would not allow the direct visualization of rafts, the upper limit of which has been defined as approximately 200 nm (38, 78). In order to demonstrate that MβCD had an effect, cells were exposed to MβCD and then extracted with cold TX-100. Following this treatment, WT PrP was removed completely from the membrane, presumably because rafts were no longer intact; thus, TX-100 was able to solubilize the entire membrane. These results showed that WT PrP was located in cholesterol-rich rafts in the plasma membrane, whereas TM PrP was located outside rafts.
FIG 2.
TM PrPC is not located in raft subdomains of the plasma membrane. Cells were immunolabeled with anti-PrP antibody 6D11 at 4°C and then either left untreated or exposed to 1% TX-100, 2 mM MβCD, or 2 mM MβCD and then 1% TX-100, followed by processing for fluorescent labeling as described in Materials and Methods. Cells were imaged by confocal microscopy. Images were deconvolved, and a representative Z slice from the middle of cells is shown. Bar, 20 μm.
WT but not TM PrP faithfully propagates PrPres upon infection independent of strain or inoculum type.
Having established that WT PrP resides in a different membrane subdomain from TM PrP in NpL2 cells, we wished to examine how this affected the ability of either construct to convert to PrPres following the application of inocula of different TSE strains and types to the cell lines in culture. Specifically, the persistent propagation, i.e., the generation of misfolded PrP that is able to transmit its fold to new PrPC substrate in daughter cells over several generations of PrPres, was examined. The blot shown in Fig. 3 corresponds to cells at passage 5, with serial 10-fold dilutions at each passage, by which stage the original inoculum has been diluted out, as shown by the control lanes (NpL2 group, lanes 2 to 4). WT cells infected with either 22L scrapie microsomes (lane 6) or GPI-anchorless fibrils extracted from mice infected with 22L (lane 7) or RML (lane 8) persistently propagated PK-resistant PrPres when infection was initiated with an inoculum containing 100 ng of PrPres. The banding pattern was typical of PrPres with the diglycosylated band at ∼30 kDa, monoglycosylated band at ∼25 kDa, and unglycosylated band at ∼20 kDa, all shifted to a ∼6- to 7-kDa-smaller apparent molecular mass than PrPC. Adding microsomes from uninfected mice to WT cultures did not produce any PrPres, showing that its induction required exposure to scrapie infectivity. Using these conditions (20 μg/ml PK and PrPres precipitation with 0.3% phosphotungstic acid [PTA]), no PrPres was detected in TM PrP-expressing cells (lanes 10 to 12).
FIG 3.
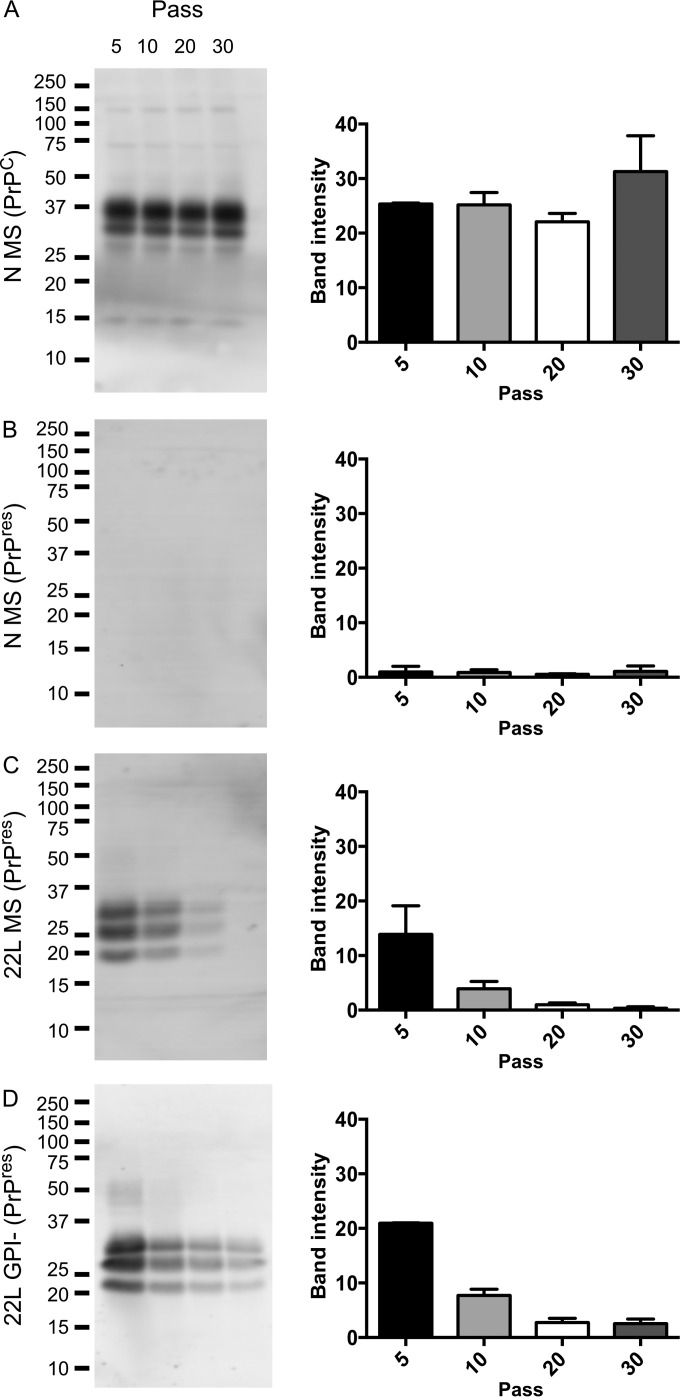
WT but not TM cells persistently propagate 22L and RML mouse scrapie. Western blot of cell lysates showing formation of PK-resistant PrP following infection with either 22L PrPres present in WT brain microsome fractions (MS) or as amyloid fibrils (GPI-) extracted from mice producing a GPI-anchorless form of 22L or RML PrPres. N MS, cells treated with control brain microsomes from uninfected mice. Cells are at passage 5 postinfection. White vertical lines indicate removal of irrelevant lanes. Numbers at left are molecular masses in kilodaltons.
WT cells were then examined at later passages to determine if PrPres propagation was stable. Figure 4A shows that expression of PrPC did not change significantly as cells were passaged. Control cells (Fig. 4B) showed that no spontaneous PrPres formation occurred when microsomes from uninfected mice were added to the cells, as observed in Fig. 3. Following treatment with either 22L microsomes (Fig. 4C) or 22L GPI-anchorless amyloid fibrils (Fig. 4D), PrPres was detected in cells using PK treatment of cell lysates followed by PTA precipitation to concentrate PrPres. In both cases, the level of PrPres decreased as cells were passaged following infection. However, at least for the cells infected with anchorless fibrils, PrPres propagated over extended serial passages (>30).
FIG 4.
PrPres propagation in infected WT cells over extended passages. Cells expressing WT PrP were treated with brain microsomes from uninfected mice (A and B) or 22L microsomes (C) or 22L GPI-anchorless fibrils (D). Lysates were either loaded without further treatment (A) or treated with PK and PTA (B to D). Quantification was carried out on lysates from two independent cultures run on separate gels. Error bars indicate standard deviations. Band intensity is shown in arbitrary units. Five hundred micrograms of total protein was loaded per lane. Numbers at left are molecular masses in kilodaltons.
TM PrP does not convert into a more protease-sensitive form of PrPres.
Using the conditions outlined above, i.e., digesting lysates with 20 μg/ml PK and precipitating aggregated PrPres with 0.3% PTA, no misfolded PrP was detected in infected TM cells after 5 serial passages. There have been reports in the literature of infectious, PK-sensitive forms of misfolded PrP (17, 18, 25, 79) that can be revealed by PTA precipitation alone or in combination with alternative proteases such as pronase E (21) or thermolysin (20). Initially, we characterized the PK resistance of WT PrPres in persistently infected WT cells by titrating in concentrations of PK from 0 to 200 μg/ml (Fig. 5A). Without PK, total PrP migrated as 4 to 5 bands, suggesting that both PrPC and PrPres had been precipitated by PTA (Fig. 5A and B). Background precipitation of PrPC with PTA was not unexpected and has been reported previously (80). The presence of TSE infection-specific, PTA-precipitated bands that comigrated with PK-truncated PrPres suggested that WT PrPres was truncated by endogenous proteases in the NpL2 cell model, as has been reported to occur in other cell lines (81, 82). With 1 μg/ml PK and more, all PrPC was degraded and only PrPres was visualized, at least some of which was resistant to very high PK concentrations of 200 μg/ml. In contrast, no infection-specific PTA-precipitated bands were detected in lysates from TM PrP cells without PK treatment, indicating that PTA precipitation alone did not detect any TM PrPres (Fig. 5B, upper gel). Knowing that all PrPC would be degraded with as little as 1 μg/ml PK, we applied these conditions to lysates from infected TM PrP cells (Fig. 5B, lower gel). As observed above, PrPres was detected only in control WT cells. PK concentrations as low as 0.01 μg/ml were tested; the lowest concentration that could successfully cleave most PrPC was 0.1 μg/ml, but this still did not reveal any PK-resistant TM PrP species, nor did treating the lysates with cold PK, which has also been used to detect nonnative abnormal PrP isomers (data not shown) (83–86).
FIG 5.
PK sensitivity of WT and TM PrP. (A) PK titration (0 to 200 μg/ml) followed by PTA precipitation of WT cell lysates 13 passages after infection with 22L scrapie microsomes. Quantification of PrPres bands on the gel presented is shown in the graph. Band intensity is shown in arbitrary units. (B) Cell lysates of untransfected NpL2, WT, or TM cells infected with normal brain microsomes (N MS), 22L microsomes (22L MS), or anchorless fibrils (22L GPI-) with either PTA precipitation alone (top) or 1 μg/ml PK followed by PTA precipitation (bottom). Numbers to the left of gels are molecular masses in kilodaltons.
In order to examine whether TM PrP had misfolded into an isoform sensitive to PK specifically, the alternative proteases chymotrypsin, pronase E, and thermolysin were used, all of which have previously revealed nonnative, misfolded PrP (20, 21, 87) (Fig. 6A). Once again, no PrPres was detected in TM lysates. Immunoblotting with alternative anti-PrP antibodies (6D11 and 31C6) was performed to ensure that TM PrPres was not detected as a result of adopting an unusual conformation that either resisted D13 detection after transfer to polyvinylidene difluoride (PVDF) or lacked the D13 epitope due to proteolytic cleavage at a more internal residue than that in WT PrPres. 6D11 has an epitope in a similar region as D13 (residues 93 to 109 compared to residues 96 to 104), and the epitope for 31C6 (residues 143 to 149) lies further toward the C terminus. While both of these antibodies detected WT PrPres, no PrPres bands were detected in the TM cell lysate, suggesting that TM PrP did not misfold into a nonnative, protease-sensitive conformation (Fig. 6B).
FIG 6.
Detecting abnormal PrP isoforms using alternative proteases and antibodies. (A) Proteases other than PK were used to digest cell lysates from infected cell cultures at passage 5. Indicated cell lines were infected with 22L or normal (N) microsomes as indicated. Samples were analyzed by Western blotting with D13 antibody. PE, pronase E; TL, thermolysin. The white vertical line indicates removal of irrelevant lanes. (B) Anti-PrP antibodies 6D11 and 31C6 were used to assay for misfolded PrP from infected cultures at passage 5. Samples were treated with PK and PTA prior to Western blot analysis with the antibodies indicated. The ∼30-kDa band present in the TM lanes corresponds to antibody cross-reactivity with PK in samples as shown by its presence in the corresponding NpL2 control lanes as well as other infection experiments initiated with normal microsomes (data not shown). This cross-reactivity may be more likely to occur in experiments when higher PK digest reaction equivalents are loaded on the gel. Numbers to the left of blots are molecular masses in kilodaltons.
Absence of RT-QuIC seeding activity in TM cells exposed to TSE infectivity.
The ability to seed conversion of new substrate is a requirement of any protein-based infectious agent able to spread and propagate its fold. To determine if exposure to TSE infectivity induced TM PrP conversion into a self-propagating misfolded conformation at a level below the detection limit of Western blot analysis, we employed real-time quaking-induced conversion (RT-QuIC), which can detect subnanomolar amounts of aggregates that can seed conversion of recombinant PrP (rPrP) to thioflavin T (ThT)-positive amyloid fibrils (28, 88). The minimal sample preparation (resuspension of a cell pellet into a low concentration of detergent followed by homogenization) eliminates any concerns that protease digestion and precipitation methods might remove any converted TM PrP. Lysates from WT cells persistently infected with anchorless 22L fibrils (open circles) or 22L microsomes (open triangles) were both able to seed rPrP conversion, shown in Fig. 7B. The culture infected with fibrils had particularly strong seeding activity, and an increase in fluorescence occurred within 10 h, comparable to the lag phase for the scrapie brain homogenate used as a positive control (gray solid line). Uninfected brain homogenate was used as a negative control; the rise seen in all three experiments at around 50 h was due to spontaneous aggregation of the recombinant protein that can occur at late time points in RT-QuIC (26, 28). Thus, any increase in fluorescence past this point would be excluded as nonspecific signal. NpL2 cells treated with TSE inocula followed by serial passage were all negative (Fig. 7A), showing that positive samples were due to newly induced self-propagating PrPres and not residual inoculum. RT-QuIC assays of corresponding TM samples showed that lysates of 22L microsome-treated cells induced an increase in ThT fluorescence beginning around 40 h (Fig. 7C, TM, 22L MS), a time point just prior to an increase in fluorescence from the negative controls [Fig. 7C, TM, N MS and BH (Sc-)]. Repeat analysis of the same lysate gave a similar result, but no positive samples were detected in subsequent independent attempts to infect TM PrP cells (data not shown). Given the modest separation between time points for the increase in ThT fluorescence of 22L microsome-treated TM PrP and the negative controls and the fact that only two of four replicate wells showed increased ThT fluorescence, this result could not be considered a robust positive signal. Nevertheless, it provided an indication that there may be extremely low levels of misfolded TM PrP, although, if present, this was in stark contrast to the highly PK-resistant, easily detected aggregated PrPres formed in WT cultures.
FIG 7.
Persistent propagation of prion seeding activity in 22L-infected WT PrP cells. Lysates from NpL2 (A), WT (B), or TM (C) cells that had been treated with either normal brain microsomes (crosses), 22L microsomes (open triangles), or anchorless 22L fibrils (open circles) and passaged eight times were tested for prion seeding activity by RT-QuIC. Brain homogenates (10−4-fold dilution) from mice either infected with scrapie strain RML (Sc+, gray solid line) or uninfected (Sc-, gray dashed line) were used as controls. At approximately 50 h, the uninfected brain homogenate control began to show an increase in fluorescence intensity, indicating that the recombinant substrate had begun to spontaneously aggregate. Traces represent the average values from 4 wells.
DISCUSSION
By adding a TM sequence to the C terminus of PrP, we aimed to explore how GPI anchoring and raft association can influence the persistent propagation of PrPres. In doing so, we have developed a new model for studying persistent TSE infection that we consider advantageous over others. The parental cell line used, NpL2, was derived from a PrP knockout mouse (57) and chosen because there would be no interference from endogenous PrP, which is expressed in many other models, including the commonly used N2a neuroblastoma cell line. Thus, we are able to categorically state that any new PrPres detected is formed exclusively by the expressed constructs without any contribution from WT PrPC. This is significant, as some phenotypes can be at least partially rescued when WT PrPC is coexpressed with mutant PrPC (89). Importantly, this approach eliminates the need to introduce changes to the PrP sequence that would be required to distinguish endogenous PrPres from exogenous PrPres, for example, by incorporating epitope tags, which under certain circumstances can influence PrP misfolding or the resulting conformation (90, 91). In addition, NpL2 cells are hippocampal in origin and unlike N2a cells do not require differentiation in order to enhance neuronal properties. It is well established that N2a cells are genetically unstable and display clone-dependent susceptibility to TSE infection (66, 92). Although we cannot discount the possibility of clonal effects in our model, all WT clones tested were able to support infection and all TM PrP clones resisted infection. Furthermore, NpL2 cells require highly enriched neurobasal medium supplemented with B27 formulated for culture of primary neurons, suggesting that they are physiologically closer to primary cells than other cell lines used to study TSE infection that are cultured in serum-supplemented rich medium such as Opti-MEM or Dulbecco's modified Eagle medium (DMEM). We have shown that NpL2 WT PrP cells can persistently propagate PrPres over more than 30 passages and that PrPres displays the typical properties of protease resistance and self-perpetuating formation following an initial infection, recapitulating important aspects of TSE disease. We did observe that the amount of PrPres propagated diminished as cells were passaged; the possible reasons for this, such as death of infected cells, selection for an infection-resistant population of cells during passage (S. Priola, personal communication), or culture medium, which has been shown to have lot-dependent efficiency in maintaining infection in N2a cells (G. Raymond, personal communication), are of interest but beyond the scope of this work. Nevertheless, at up to around 30 passages WT PrPres was still easily detectable by the methods used here, and it is possible that in clones with a higher expression level of PrPC this may be increased.
A careful characterization of WT- and TM PrPC-expressing cells was carried out and, consistent with prior reports from multiple labs (42, 50, 52, 54–56), they were similar in terms of expression level and distribution, but TM PrPC was located in a different plasma membrane subdomain from WT PrPC, as would be expected for a protein that was transmembrane rather than GPI anchored. The data presented here suggest that TM PrP is in theory a potential substrate for conversion for the following reasons: (i) the constructs have identical ectodomains and differ only in their membrane anchoring and the GFP addition at the C terminus of the TM PrP, (ii) Western blotting and deglycosylation showed that the majority of TM PrPC was the expected size and glycosylated with mature complex N-linked glycans, and (iii) immunolabeling revealed that there was a significant population of PrP on the cell surface where interaction with exogenous inoculum and conversion may take place (6, 11, 93). These data show very little difference between WT and TM PrPC other than a significant alteration in where each resides within the membrane due to the different membrane anchors.
The concept of raft domains in the plasma membrane of living cells has come under intense questioning, partly because DRMs may not be representative of fractions as they exist within the cell and may be formed artificially as a consequence of the biochemical isolation procedure. Despite the contention, generic heterogeneity within the membrane is accepted as being of functional importance (94). For example, it can serve to group together in close proximity the components required for a particular function, such as neurotransmitter signaling (95). Analogously, it is conceivable that raft-mediated clustering of PrPC substrate could improve efficiency of conversion to PrPres. The issue of whether rafts are representative of such distinct membrane compartmentalization will continue to be debated, but our results do suggest a degree of organization within the membrane where GPI-anchored PrP is located in a separate region from TM PrP, which is in a membrane environment that is easily solubilized by cold TX-100.
Despite extensive attempts using multiple approaches, no TM PrPres, which may adopt a misfolded conformation different from WT PrPres, was detected. Lysate from infected TM cells in multiple independent experiments showed possible trace seeding activity by RT-QuIC (Fig. 7C, TM, 22L MS). Although these data suggest that it is possible for TM PrPC to support persistent conversion to a nonnative isoform that contains seeding activity, the low incidence indicates that the efficiency of TM PrPC to convert is very poor. It might be that if expressed at higher levels, TM PrPC would support persistent conversion with greater efficiency. However, the WT control clone had similar PrPC expression levels and propagated PrPres at a readily detectable level, showing that susceptibility to persistent infection was not restricted by the amount of substrate available. Also, evidence from other cell culture models of TSE infection indicates that cells expressing low levels of PrPC support persistent infection (96, 97). Thus, our data argue that the membrane subdomain within which PrPC is localized has a strong influence on whether it can undergo conversion to PrPres.
These results are consistent with data from other laboratories that further suggest the importance of PrPC membrane anchoring and raft localization. Even for PrPC localized within rafts, there is evidence that posttranslational modification of PrPC may affect trafficking, raft composition, and other aspects of PrP biology. Sialylation of the GPI anchor has been proposed as influencing PrPC conversion to PrPres, as well as PrPres infectivity and toxicity (98–101). Using a GPI painting model with purified, N-linked glycan-deglycosylated (N-deglycosylated) PrPC, Bate and coworkers reported that asialo-GPI N-deglycosylated PrPC resisted conversion to PrPC and inhibited conversion of endogenously expressed PrPC (100). These effects were correlated with the N-deglycosylated asialo-GPI PrPC exhibiting a prolonged half-life and associating with rafts of altered lipid composition compared with the sialo-GPI PrPC control (100). In our system, unlike sialo-GPI, the SAGAGS linker of our TM anchor has a neutral charge. It is possible that this charge difference and/or other effects on the local membrane microenvironment may also contribute to the inability of TM PrPC to support PrPres propagation. However, the role of PrPC GPI anchor sialylation on conversion is somewhat controversial as asialo-GPI PrPres is generated both in vivo and in cell culture models of infection (98, 102). Since the asialo-GPI PrPC in these systems is endogenously expressed and contains N-linked glycans, these molecules are arguably more comparable to TM PrPC in the present study.
It should be noted that our data pertain to the analysis of PrPres propagation in cells. This is not to imply that generation of PrPres and TSE infectivity can occur only in association with membranes. There are now many reports of the protein misfolding cyclic amplification (PMCA) procedure that produce PrPres and infectivity under cell-free conditions with the assistance of detergents and sonication, circumstances under which one would expect PrPC and other molecules to be solubilized from membranes (101, 103–105). Several PMCA studies have also shown the importance of various cofactors in the conversion process (104, 106–110). In PMCA reactions, all of these cofactors, including any that associate with rafts, should be free to interact with PrPC, but all components might be expected to have a lower local concentration than that achieved for membrane-bound components in cells. Perhaps this in part accounts for the PrPC concentration dependence of PMCA reactions (111). Whatever the case, prion propagation in cells and PMCA are both complex and different but not mutually exclusive processes.
Our work extends the information provided by previous studies and introduces new facets that we believe to be important for determining how membrane anchoring influences PrP conversion. First, GPI anchoring of PrPC imparts a distancing from the membrane and consequentially a flexibility on it (64) that might be important in aiding conversion, possibly by allowing PrPC to interact with cofactors enriched in raft microdomains that either are required for or assist conversion, such as glycosaminoglycans (GAGs), the 37-kDa/67-kDa laminin receptor, and glypicans (50, 92, 112–115). This positioning may also be required for PrPres binding to PrPC substrate and subsequent templating of its fold (7, 116, 117). In designing the TM construct for this study, a spacer region was introduced to position PrPC away from the membrane in order to mimic WT PrPC, differentiating our construct from all published TM PrPC constructs. In addition, we attempted to infect the cells with an exogenous source of PrPres and assayed for TM PrPC conversion using sensitive assays for forms of TM-PrPres with reduced PK resistance, none of which has been attempted previously.
In the present study, two different types of PrPres inoculum were used: membrane-bound (in the form of microsomal brain membranes containing GPI-anchored PrPres) and purified GPI-anchorless PrPres fibrils. The PrPres aggregates in the two inocula have different ultrastructures, with fibrils adopting an amyloid conformation and microsome-bound aggregates adopting a nonfibrillar, apparently nonamyloid structure (7, 35, 44, 49, 118–121). Furthermore, PrPres in the two inocula might be expected to interact differently with distinct membrane regions. Cell-free studies have shown that raft-associated PrPC present in DRMs will convert only if the exogenously applied PrPres inserts into a contiguous membrane (7, 51). It is possible that during infection membrane-bound, raft-associated PrPres would likewise have to insert into the cell membrane and partition to host cell rafts (7, 8), perhaps by membrane fusion or GPI painting (122–125), in order to achieve the proximity and orientation required for templating WT PrPC substrate. If this membrane insertion and raft partitioning of GPI-anchored PrPres occur, TM PrPC, which is not in rafts, may not be expected to convert as it would not be proximal to PrPres. On the other hand, anchorless fibrils in solution have greater rotational freedom than GPI-anchored PrPres aggregates tethered to membranes and are not subject to GPI anchor-dependent raft targeting. It is possible that the PK-treated anchorless fibrils could indirectly associate with membranes via PrPres-binding membrane molecules such as GAGs (126, 127).
We observed that purified anchorless fibrils were also able to seed persistent conversion of raft-bound WT PrPC to PrPres, showing that exogenous PrPres does not have to be membrane bound or GPI anchored and that the conformation of the fibrils is sufficient to transmit their fold onto a new substrate. McNally and coworkers similarly observed efficient infection of WT PrPC-expressing cells using brain homogenate from 22L-infected transgenic mice expressing anchorless PrPC (47). However, anchorless fibrils were not able to induce self-propagating conversion of TM PrPC. We also showed that templating and propagation of PrPres were not strain dependent, as both RML and 22L scrapie strains were able to initiate stable infection of cells expressing WT PrPC but not TM PrPC (Fig. 3). Careful attempts were made to ensure that TM PrPC had the best possible opportunity to interact with the exogenous PrPres so that an infection could be initiated, including the use of inocula containing a very high dose of PrPres per cell. Despite this and using a variety of methods to detect protease-sensitive PrPres conformers, in this system TM anchoring of PrPC did not support aggregation that propagated persistently. It is possible that acute formation of TM PrPres occurred; however, here our interest was in persistent aggregation, as this is more relevant to intercellular transmission and establishment of a productive infection in vivo.
The resistance of TM PrPC-expressing cells to infection even with anchorless fibrils suggests that the ability to stably propagate a misfolded conformer is not due to conformational templating alone. There may be different mechanisms by which different forms of PrPres, such as fibrillar and membrane bound, are able to bind to and interact with PrPC, although membrane association might be important in both cases. It has been shown previously that PrPC does not solely interact with membranes via its GPI anchor, as GPI-anchorless forms of PrPC can bind to artificial raft membranes and other liposomes (51, 128, 129). Therefore, it might be expected that if membrane association were required, anchorless fibrils would have been able to induce misfolding of TM PrPC. This either did not happen at all or did so extremely inefficiently, pointing toward the possibility that the differential partitioning of WT and TM PrPC may be responsible for the inability of TM PrPC to misfold. These results corroborate the vast literature stating that a raft-associated cofactor may be required not only for PrPC conversion but also for its persistent propagation and infectivity in vivo. One example is glypican-1 (a GPI-anchored heparan sulfate proteoglycan), proposed as a scaffold that mediates the interaction of PrPres with PrPC within rafts (115, 130, 131).
MATERIALS AND METHODS
Generation of constructs, transfections, and cell culture.
The WT mouse PrP open reading frame (a kind gift from Ryuichiro Atarashi, Nagasaki University, Nagasaki, Japan) flanked by HindIII and XbaI sites on the 5′ and 3′ ends, respectively, and cloned into a pCR2.1 cloning vector (Gibco) was used as a source of WT mouse PrP. Cassettes of Qa PrP (containing 5′-mouse PrP 1–230-SAGAGS-Qa TM domain-FLAG epitope-3′) and A206K monomeric GFP (65, 132) were both custom synthesized by Blue Heron (Bothell, WA, USA). The Qa PrP cassette was engineered to contain flanking HindIII and XbaI restriction sites and a PacI site immediately 3′ to the FLAG epitope sequence. The A206K GFP cassette was inserted into the PacI site of the Qa PrP cassette using 5′- and 3′-flanking PacI restriction sites engineered into the A206K GFP cassette to create the final TM PrP construct. The WT PrP and TM PrP reading frames were excised as HindIII/XbaI fragments and subcloned into pcDNA3.1(+). Finally, the WT PrP and TM PrP inserts were then excised by PmeI digestion and ligated to HpaI-digested pMSCV (Clontech) for expression in mammalian cells using the murine stem cell virus retroviral expression system. The final constructs were verified by sequencing. The expression plasmids were first transfected using nucleofection (Lonza, Germany) into a packaging cell line, PT67 (3 to 5 μg/ml final DNA concentration). Stably transfected cells were selected for using 3 μg/ml puromycin. Viral particles containing the recombinant pMSCV constructs were collected in the supernatant from stably transfected cells and transduced into NpL2 cells. Stably transduced NpL2 cells were selected for using puromycin. The resulting bulk populations were screened for PrP expression by immunoblotting and immunolabeling (see below), and then cloned cell lines were isolated using limited-dilution cloning in 96-well plates. Several clones were screened again for PrP expression; those with similar expression levels were chosen for use in the remaining experiments. NpL2 cells were grown in neurobasal medium supplemented with B27, 1% penicillin-streptomycin, and 1% l-glutamine (all from Gibco) and maintained in a humidified environment with 5% CO2. Experiments were carried out on two WT PrP- and two TM PrP-expressing clones to verify trends (only one is shown in the results).
Ethics statement.
Animal experiments were conducted in an Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC)-accredited facility in accordance with animal welfare guidelines under animal study protocols (2010-30 and 2010-45) approved by the Animal Care and Use Committee of the Rocky Mountain Laboratories, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Infection of cells.
Medium was removed from NpL2 cells at 50 to 60% confluence in 96-well plates. One hundred nanograms of PrPres as contained in brain microsomes (small membrane vesicles representing the total membrane fraction derived from brain homogenates; for preparation, see reference 7) or purified PrPres fibrils (35) was diluted into 50 μl of neurobasal medium, all of which was added to each well (the low volume was intended to encourage contact between the inoculum and cell surface PrPC). Cells were incubated for 4 to 5 h, and then 150 μl fresh medium was added to each well and incubated for 2 days. After this time, cells were trypsinized and transferred to a 24-well plate. Cells were then passaged when they reached 80 to 90% confluence every 3 days. At passages where cells were to be assayed by Western blotting, a portion of the leftover cell suspension was transferred to a 6-well plate and allowed to reach confluence before transfer to a T25 flask, was again allowed to reach around 90% confluence, and then was lysed. Infection experiments were carried out 7 times (microsomes) or 5 times (anchorless fibrils) on two independent TM PrP clones with similar expression levels.
Raft assay.
Cells at 50% confluence were washed with wash buffer (25% Superblock [Gibco] in phosphate-buffered saline [PBS]), incubated with primary anti-PrP antibody 6D11 (Covance) (1:1,000 dilution in wash buffer), and placed at 4°C for 1 h. Following three 5-min washes with wash buffer, cells were treated with either cold 1% Triton X-100 (TX-100) in PBS at 4°C for 10 min or 2 mM methyl-beta cyclodextrin (MβCD) at 37°C for 1 h followed by 1% TX-100 or were left untreated. After three more 5-min washes with wash buffer (at 4°C for TX-100 samples), cells were fixed with 4% paraformaldehyde (PFA) (Electron Microscope Supplies)-0.02% glutaraldehyde-4% sucrose in PBS for 15 min, followed by incubation with 50 mM glycine in PBS for 10 min. Cells were then washed as before, blocked using Superblock for 30 min, and incubated with an Alexa Fluor 568-conjugated anti-mouse secondary antibody (Gibco) (1:1,000 dilution in wash buffer) for 1 h. Cells were finally washed with PBS and imaged as outlined below.
SDS-PAGE and Western blotting.
For detection of PrPC, cells from an approximately 90% confluent T25 flask were first washed twice with warm phosphate-buffered balanced saline (PBBS) and then lysed in 1.5 ml ice-cold lysis buffer (PBS [10 mM Na2HPO4-10 mM NaH2PO4-130 mM NaCl, pH 7.4], 0.5% TX-100, 0.5% sodium deoxycholate supplemented with a protease inhibitor cocktail tablet [Roche; 1 tablet in 10 ml lysis buffer]) for 2 min. The resulting lysate was kept on ice and centrifuged at 2,700 × g for 5 min at 4°C to pellet the nuclei, which were discarded. Total protein concentrations were determined using a bicinchoninic acid (BCA) assay (Pierce), and 20 μg of total protein was precipitated overnight at −20°C using 4 volumes of cold methanol. Following centrifugation at 22,000 × g for 20 min at 4°C, methanol was aspirated and the pellet was allowed to dry before resuspension in 1× sample buffer (0.125 M Tris-HCl, pH 6.8, 5% glycerol, 6 mM EDTA, 10% SDS, 0.04% bromophenol blue supplemented immediately prior to use with 0.05 M dithiothreitol [DTT]), heating at 95°C for 8 min, loading onto a 10% Bis-Tris NuPAGE gel (Gibco), and being run at 200 V with 2-(N-morpholino)ethanesulfonic acid (MES) running buffer (Gibco). Semidry transfers were carried out at 150 mA/gel onto Immobilon-P membranes (Millipore) that were subsequently blocked with 5% milk (Bio-Rad) prepared in Tris-buffered saline (TBS; 25 mM Tris-HCl, 150 mM NaCl, pH 7.5) plus 0.1% Tween 20. Blots were probed with anti-PrP antibody D13 (a gift from J. Striebel, Rocky Mountain Laboratories) (diluted 1:100 in blocking solution) and an anti-human alkaline phosphatase-conjugated secondary antibody (Jackson ImmunoResearch) diluted 1:2,500 in blocking solution. Membranes were incubated in Attophos substrate (Promega) and allowed to dry completely before imaging using a Typhoon scanner. Quantification was carried out using ImageQuant TL software.
For detection of PrPres, the procedure was carried out as described above with the adjustments outlined below. Protease inhibitors were omitted from the lysis buffer, and lysates containing 0.5 mg (for WT) or 1.0 mg (for NpL2 and TM) total protein were digested with 20 μg/ml PK for 1 h at 37°C in order to digest any PrPC. The PK digest was stopped by adding 1 mM Pefabloc and then incubating the mixture on ice for 5 min. To concentrate aggregated PrPres, prewarmed phosphotungstic acid (PTA) was added to an 0.3% final concentration and samples were incubated at 37°C with agitation for 1 h. Following centrifugation at 22,000 × g for 30 min at room temperature, the supernatant was aspirated and the pellet was resuspended in 2× sample buffer, boiled, and loaded onto a gel. Where alternative proteases to PK were used, these were at 20 μg/ml (thermolysin) or 100 μg/ml (pronase E and chymotrypsin). All enzymes were purchased from Sigma, and digestion was carried out according to the manufacturer's protocol.
For samples deglycosylated using peptide-N-glycosidase (PNGase) F (New England BioLabs), 10 μg of total protein was methanol precipitated, and then 10 μl of glycoprotein denaturing buffer was used to resuspend the pellet. Denaturation was achieved by heating at 100°C for 10 min, and then the mixture was made up to 20 μl with 2 μl G7 reaction buffer (supplied in kit), 2 μl NP-40, 2 μl Milli-Q water, and 4 μl PNGase F. The reaction mixture was incubated at 37°C for 1 h and precipitated using methanol-chloroform, and the pellet was loaded onto a 10% Bis-Tris NuPAGE gel run with 3-(N-morpholino)ethanesulfonic acid (MOPS) running buffer (Gibco).
Immunolabeling.
For labeling cell surface PrP, cells were fixed for 15 min with 3.7% PFA in PBS supplemented with 4% sucrose to prevent artifactual blebbing of the cells. After a wash with PBS, cells were incubated with 50 mM glycine in PBS for 5 min to block any unreacted aldehydes and then blocked using Superblock for 30 min. Primary anti-PrP antibody 6D11 (1:1,000 dilution in wash buffer) was incubated with cells at 4°C for 1 h. After washing with PBS, anti-mouse-Alexa Fluor 568 (1:1,000 dilution in wash buffer) secondary antibody was added for 1 h at room temperature. Cells were then washed and imaged in PBS within 24 h. For labeling intracellular PrP, a similar protocol was followed except that the concentration of PFA was lowered to 2%, and following treatment with glycine, cells were permeabilized with 0.3% TX-100 in PBS for 10 min.
Wide-field fluorescence microscopy images were taken using a 40× Plan Fluor numerical aperture 0.6 objective and Hamamatsu Orca ER II camera. Confocal images were acquired on a Nikon LiveScan confocal microscope described previously (48, 49). Huygens software (Scientific Volume Imaging) was used to deconvolve confocal images. Images were analyzed using NIS-Elements software.
RT-QuIC assay.
Real-time quaking-induced conversion was carried out as described elsewhere (26, 28). In order to concentrate the lysate, a confluent layer of cells from a T25 flask was scraped into PBS and centrifuged at 2,700 × g for 5 min. The pellet was washed with PBS and then resuspended into 50 to 100 μl of 0.05% TX-100 in PBS. This low level of detergent was required to break up the cell membranes and release any misfolded protein that might be able to seed conversion of the substrate. The pellet was then manually homogenized using a Kimble Chase Kontes pellet pestle, and the total protein concentration was measured using a BCA assay. Total protein was normalized so that equal amounts were loaded into the RT-QuIC reaction mixture diluted either 10-fold (cell lysates) or 1,000-fold (control brain homogenates) in PBS-0.1% SDS.
ACKNOWLEDGMENTS
We thank Louise Serpell for critical reading of the manuscript.
This research was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health (project numbers AI000982-06, AI000982-07, and AI000982-08).
REFERENCES
- 1.Prusiner SB. 1991. Molecular biology of prion diseases. Science 252:1515–1522. doi: 10.1126/science.1675487. [DOI] [PubMed] [Google Scholar]
- 2.Pan KM, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick RJ, Cohen FE, Prusiner SB. 1993. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. Proc Natl Acad Sci U S A 90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wuthrich K. 1996. NMR structure of the mouse prion protein domain PrP(121-231). Nature 382:180–182. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- 4.Riek R, Hornemann S, Wider G, Glockshuber R, Wuthrich K. 1997. NMR characterization of the full-length recombinant murine prion protein, mPrP(23-231). FEBS Lett 413:282–288. doi: 10.1016/S0014-5793(97)00920-4. [DOI] [PubMed] [Google Scholar]
- 5.Oesch B, Westaway D, Walchli M, McKinley MP, Kent SB, Aebersold R, Barry RA, Tempst P, Teplow DB, Hood LE, Prusiner SB, Weissmann C. 1985. A cellular gene encodes scrapie PrP 27-30 protein. Cell 40:735–746. doi: 10.1016/0092-8674(85)90333-2. [DOI] [PubMed] [Google Scholar]
- 6.Caughey B, Raymond GJ. 1991. The scrapie-associated form of PrP is made from a cell surface precursor that is both protease- and phospholipase-sensitive. J Biol Chem 266:18217–18223. [PubMed] [Google Scholar]
- 7.Baron GS, Wehrly K, Dorward DW, Chesebro B, Caughey B. 2002. Conversion of raft associated prion protein to the protease-resistant state requires insertion of PrP-res (PrPSc) into contiguous membranes. EMBO J 21:1031–1040. doi: 10.1093/emboj/21.5.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baron GS, Magalhaes AC, Prado MA, Caughey B. 2006. Mouse-adapted scrapie infection of SN56 cells: greater efficiency with microsome-associated versus purified PrP-res. J Virol 80:2106–2117. doi: 10.1128/JVI.80.5.2106-2117.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flechsig E, Hegyi I, Enari M, Schwarz P, Collinge J, Weissmann C. 2001. Transmission of scrapie by steel-surface-bound prions. Mol Med 7:679–684. [PMC free article] [PubMed] [Google Scholar]
- 10.Gilch S, Kehler C, Schatzl HM. 2006. The prion protein requires cholesterol for cell surface localization. Mol Cell Neurosci 31:346–353. doi: 10.1016/j.mcn.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 11.Goold R, Rabbanian S, Sutton L, Andre R, Arora P, Moonga J, Clarke AR, Schiavo G, Jat P, Collinge J, Tabrizi SJ. 2011. Rapid cell-surface prion protein conversion revealed using a novel cell system. Nat Commun 2:281. doi: 10.1038/ncomms1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeffrey M, McGovern G, Siso S, Gonzalez L. 2011. Cellular and sub-cellular pathology of animal prion diseases: relationship between morphological changes, accumulation of abnormal prion protein and clinical disease. Acta Neuropathol 121:113–134. doi: 10.1007/s00401-010-0700-3. [DOI] [PubMed] [Google Scholar]
- 13.Hope J, Morton LJ, Farquhar CF, Multhaup G, Beyreuther K, Kimberlin RH. 1986. The major polypeptide of scrapie-associated fibrils (SAF) has the same size, charge distribution and N-terminal protein sequence as predicted for the normal brain protein (PrP). EMBO J 5:2591–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barron RM, Campbell SL, King D, Bellon A, Chapman KE, Williamson RA, Manson JC. 2007. High titers of transmissible spongiform encephalopathy infectivity associated with extremely low levels of PrPSc in vivo. J Biol Chem 282:35878–35886. doi: 10.1074/jbc.M704329200. [DOI] [PubMed] [Google Scholar]
- 15.Gambetti P, Dong Z, Yuan J, Xiao X, Zheng M, Alshekhlee A, Castellani R, Cohen M, Barria MA, Gonzalez-Romero D, Belay ED, Schonberger LB, Marder K, Harris C, Burke JR, Montine T, Wisniewski T, Dickson DW, Soto C, Hulette CM, Mastrianni JA, Kong Q, Zou WQ. 2008. A novel human disease with abnormal prion protein sensitive to protease. Ann Neurol 63:697–708. doi: 10.1002/ana.21420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Head MW, Knight R, Zeidler M, Yull H, Barlow A, Ironside JW. 2009. A case of protease sensitive prionopathy in a patient in the UK. Neuropathol Appl Neurobiol 35:628–632. doi: 10.1111/j.1365-2990.2009.01040.x. [DOI] [PubMed] [Google Scholar]
- 17.Sajnani G, Silva CJ, Ramos A, Pastrana MA, Onisko BC, Erickson ML, Antaki EM, Dynin I, Vazquez-Fernandez E, Sigurdson CJ, Carter JM, Requena JR. 2012. PK-sensitive PrP is infectious and shares basic structural features with PK-resistant PrP. PLoS Pathog 8:e1002547. doi: 10.1371/journal.ppat.1002547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, Cohen FE, Prusiner SB. 1998. Eight prion strains have PrPSc molecules with different conformations. Nat Med 4:1157–1165. doi: 10.1038/2654. [DOI] [PubMed] [Google Scholar]
- 19.Tzaban S, Friedlander G, Schonberger O, Horonchik L, Yedidia Y, Shaked G, Gabizon R, Taraboulos A. 2002. Protease-sensitive scrapie prion protein in aggregates of heterogeneous sizes. Biochemistry 41:12868–12875. doi: 10.1021/bi025958g. [DOI] [PubMed] [Google Scholar]
- 20.Cronier S, Gros N, Tattum MH, Jackson GS, Clarke AR, Collinge J, Wadsworth JD. 2008. Detection and characterization of proteinase K-sensitive disease-related prion protein with thermolysin. Biochem J 416:297–305. doi: 10.1042/BJ20081235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D'Castro L, Wenborn A, Gros N, Joiner S, Cronier S, Collinge J, Wadsworth JD. 2010. Isolation of proteinase K-sensitive prions using pronase E and phosphotungstic acid. PLoS One 5:e15679. doi: 10.1371/journal.pone.0015679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silveira JR, Raymond GJ, Hughson AG, Race RE, Sim VL, Hayes SF, Caughey B. 2005. The most infectious prion protein particles. Nature 437:257–261. doi: 10.1038/nature03989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pastrana MA, Sajnani G, Onisko B, Castilla J, Morales R, Soto C, Requena JR. 2006. Isolation and characterization of a proteinase K-sensitive PrPSc fraction. Biochemistry 45:15710–15717. doi: 10.1021/bi0615442. [DOI] [PubMed] [Google Scholar]
- 24.Thackray AM, Hopkins L, Bujdoso R. 2007. Proteinase K-sensitive disease-associated ovine prion protein revealed by conformation-dependent immunoassay. Biochem J 401:475–483. doi: 10.1042/BJ20061264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colby DW, Wain R, Baskakov IV, Legname G, Palmer CG, Nguyen HO, Lemus A, Cohen FE, DeArmond SJ, Prusiner SB. 2010. Protease-sensitive synthetic prions. PLoS Pathog 6:e1000736. doi: 10.1371/journal.ppat.1000736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vascellari S, Orru CD, Hughson AG, King D, Barron R, Wilham JM, Baron GS, Race B, Pani A, Caughey B. 2012. Prion seeding activities of mouse scrapie strains with divergent PrPSc protease sensitivities and amyloid plaque content using RT-QuIC and eQuIC. PLoS One 7:e48969. doi: 10.1371/journal.pone.0048969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Atarashi R, Wilham JM, Christensen L, Hughson AG, Moore RA, Johnson LM, Onwubiko HA, Priola SA, Caughey B. 2008. Simplified ultrasensitive prion detection by recombinant PrP conversion with shaking. Nat Methods 5:211–212. doi: 10.1038/nmeth0308-211. [DOI] [PubMed] [Google Scholar]
- 28.Wilham JM, Orru CD, Bessen RA, Atarashi R, Sano K, Race B, Meade-White KD, Taubner LM, Timmes A, Caughey B. 2010. Rapid end-point quantitation of prion seeding activity with sensitivity comparable to bioassays. PLoS Pathog 6:e1001217. doi: 10.1371/journal.ppat.1001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stahl N, Borchelt DR, Hsiao K, Prusiner SB. 1987. Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell 51:229–240. doi: 10.1016/0092-8674(87)90150-4. [DOI] [PubMed] [Google Scholar]
- 30.Gorodinsky A, Harris DA. 1995. Glycolipid-anchored proteins in neuroblastoma cells form detergent-resistant complexes without caveolin. J Cell Biol 129:619–627. doi: 10.1083/jcb.129.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vey M, Pilkuhn S, Wille H, Nixon R, DeArmond SJ, Smart EJ, Anderson RG, Taraboulos A, Prusiner SB. 1996. Subcellular colocalization of the cellular and scrapie prion proteins in caveolae-like membranous domains. Proc Natl Acad Sci U S A 93:14945–14949. doi: 10.1073/pnas.93.25.14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naslavsky N, Stein R, Yanai A, Friedlander G, Taraboulos A. 1997. Characterization of detergent-insoluble complexes containing the cellular prion protein and its scrapie isoform. J Biol Chem 272:6324–6331. doi: 10.1074/jbc.272.10.6324. [DOI] [PubMed] [Google Scholar]
- 33.Madore N, Smith KL, Graham CH, Jen A, Brady K, Hall S, Morris R. 1999. Functionally different GPI proteins are organized in different domains on the neuronal surface. EMBO J 18:6917–6926. doi: 10.1093/emboj/18.24.6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pimpinelli F, Lehmann S, Maridonneau-Parini I. 2005. The scrapie prion protein is present in flotillin-1-positive vesicles in central- but not peripheral-derived neuronal cell lines. Eur J Neurosci 21:2063–2072. doi: 10.1111/j.1460-9568.2005.04049.x. [DOI] [PubMed] [Google Scholar]
- 35.Baron GS, Hughson AG, Raymond GJ, Offerdahl DK, Barton KA, Raymond LD, Dorward DW, Caughey B. 2011. Effect of glycans and the glycophosphatidylinositol anchor on strain dependent conformations of scrapie prion protein: improved purifications and infrared spectra. Biochemistry 50:4479–4490. doi: 10.1021/bi2003907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jen A, Parkyn CJ, Mootoosamy RC, Ford MJ, Warley A, Liu Q, Bu G, Baskakov IV, Moestrup S, McGuinness L, Emptage N, Morris RJ. 2010. Neuronal low-density lipoprotein receptor-related protein 1 binds and endocytoses prion fibrils via receptor cluster 4. J Cell Sci 123:246–255. doi: 10.1242/jcs.058099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor DR, Hooper NM. 2007. The low-density lipoprotein receptor-related protein 1 (LRP1) mediates the endocytosis of the cellular prion protein. Biochem J 402:17–23. doi: 10.1042/BJ20061736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pike LJ. 2006. Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function. J Lipid Res 47:1597–1598. doi: 10.1194/jlr.E600002-JLR200. [DOI] [PubMed] [Google Scholar]
- 39.Brown DA, Rose JK. 1992. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell 68:533–544. doi: 10.1016/0092-8674(92)90189-J. [DOI] [PubMed] [Google Scholar]
- 40.Brown DA. 2006. Lipid rafts, detergent-resistant membranes, and raft targeting signals. Physiology (Bethesda) 21:430–439. doi: 10.1152/physiol.00032.2006. [DOI] [PubMed] [Google Scholar]
- 41.Wilson EM, Tureckova J, Rotwein P. 2004. Permissive roles of phosphatidyl inositol 3-kinase and Akt in skeletal myocyte maturation. Mol Biol Cell 15:497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taraboulos A, Scott M, Semenov A, Avrahami D, Laszlo L, Prusiner SB. 1995. Cholesterol depletion and modification of COOH-terminal targeting sequence of the prion protein inhibit formation of the scrapie isoform. J Cell Biol 129:121–132. doi: 10.1083/jcb.129.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marella M, Lehmann S, Grassi J, Chabry J. 2002. Filipin prevents pathological prion protein accumulation by reducing endocytosis and inducing cellular PrP release. J Biol Chem 277:25457–25464. doi: 10.1074/jbc.M203248200. [DOI] [PubMed] [Google Scholar]
- 44.Chesebro B, Trifilo M, Race R, Meade-White K, Teng C, LaCasse R, Raymond L, Favara C, Baron G, Priola S, Caughey B, Masliah E, Oldstone M. 2005. Anchorless prion protein results in infectious amyloid disease without clinical scrapie. Science 308:1435–1439. doi: 10.1126/science.1110837. [DOI] [PubMed] [Google Scholar]
- 45.Chesebro B, Race B, Meade-White K, Lacasse R, Race R, Klingeborn M, Striebel J, Dorward D, McGovern G, Jeffrey M. 2010. Fatal transmissible amyloid encephalopathy: a new type of prion disease associated with lack of prion protein membrane anchoring. PLoS Pathog 6:e1000800. doi: 10.1371/journal.ppat.1000800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rogers M, Yehiely F, Scott M, Prusiner SB. 1993. Conversion of truncated and elongated prion proteins into the scrapie isoform in cultured cells. Proc Natl Acad Sci U S A 90:3182–3186. doi: 10.1073/pnas.90.8.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McNally KL, Ward AE, Priola SA. 2009. Cells expressing anchorless prion protein are resistant to scrapie infection. J Virol 83:4469–4475. doi: 10.1128/JVI.02412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Speare JO, Offerdahl DK, Hasenkrug A, Carmody AB, Baron GS. 2010. GPI anchoring facilitates propagation and spread of misfolded Sup35 aggregates in mammalian cells. EMBO J 29:782–794. doi: 10.1038/emboj.2009.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marshall KE, Offerdahl DK, Speare JO, Dorward DW, Hasenkrug A, Carmody AB, Baron GS. 2014. Glycosylphosphatidylinositol anchoring directs the assembly of Sup35NM protein into non-fibrillar, membrane-bound aggregates. J Biol Chem 289:12245–12263. doi: 10.1074/jbc.M114.556639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaneko K, Vey M, Scott M, Pilkuhn S, Cohen FE, Prusiner SB. 1997. COOH-terminal sequence of the cellular prion protein directs subcellular trafficking and controls conversion into the scrapie isoform. Proc Natl Acad Sci U S A 94:2333–2338. doi: 10.1073/pnas.94.6.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baron GS, Caughey B. 2003. Effect of glycosylphosphatidylinositol anchor-dependent and -independent prion protein association with model raft membranes on conversion to the protease-resistant isoform. J Biol Chem 278:14883–14892. doi: 10.1074/jbc.M210840200. [DOI] [PubMed] [Google Scholar]
- 52.Walmsley AR, Zeng F, Hooper NM. 2001. Membrane topology influences N-glycosylation of the prion protein. EMBO J 20:703–712. doi: 10.1093/emboj/20.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walmsley AR, Zeng F, Hooper NM. 2003. The N-terminal region of the prion protein ectodomain contains a lipid raft targeting determinant. J Biol Chem 278:37241–37248. doi: 10.1074/jbc.M302036200. [DOI] [PubMed] [Google Scholar]
- 54.Taylor DR, Watt NT, Perera WS, Hooper NM. 2005. Assigning functions to distinct regions of the N-terminus of the prion protein that are involved in its copper-stimulated, clathrin-dependent endocytosis. J Cell Sci 118:5141–5153. doi: 10.1242/jcs.02627. [DOI] [PubMed] [Google Scholar]
- 55.Rambold AS, Muller V, Ron U, Ben-Tal N, Winklhofer KF, Tatzelt J. 2008. Stress-protective signalling of prion protein is corrupted by scrapie prions. EMBO J 27:1974–1984. doi: 10.1038/emboj.2008.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Winklhofer KF, Heske J, Heller U, Reintjes A, Muranyi W, Moarefi I, Tatzelt J. 2003. Determinants of the in vivo folding of the prion protein. A bipartite function of helix 1 in folding and aggregation. J Biol Chem 278:14961–14970. [DOI] [PubMed] [Google Scholar]
- 57.Nishimura T, Sakudo A, Hashiyama Y, Yachi A, Saeki K, Matsumoto Y, Ogawa M, Sakaguchi S, Itohara S, Onodera T. 2007. Serum withdrawal-induced apoptosis in ZrchI prion protein (PrP) gene-deficient neuronal cell line is suppressed by PrP, independent of Doppel. Microbiol Immunol 51:457–466. doi: 10.1111/j.1348-0421.2007.tb03920.x. [DOI] [PubMed] [Google Scholar]
- 58.Bueler H, Fischer M, Lang Y, Bluethmann H, Lipp HP, DeArmond SJ, Prusiner SB, Aguet M, Weissmann C. 1992. Normal development and behaviour of mice lacking the neuronal cell-surface PrP protein. Nature 356:577–582. doi: 10.1038/356577a0. [DOI] [PubMed] [Google Scholar]
- 59.Hosokawa T, Tsuchiya K, Sato I, Takeyama N, Ueda S, Tagawa Y, Kimura KM, Nakamura I, Wu G, Sakudo A, Casalone C, Mazza M, Caramelli M, Takahashi H, Sata T, Sugiura K, Baj A, Toniolo A, Onodera T. 2008. A monoclonal antibody (1D12) defines novel distribution patterns of prion protein (PrP) as granules in nucleus. Biochem Biophys Res Commun 366:657–663. doi: 10.1016/j.bbrc.2007.11.163. [DOI] [PubMed] [Google Scholar]
- 60.Nishimura T, Sakudo A, Xue G, Ikuta K, Yukawa M, Sugiura K, Onodera T. 2008. Establishment of a new glial cell line from hippocampus of prion protein gene-deficient mice. Biochem Biophys Res Commun 377:1047–1050. doi: 10.1016/j.bbrc.2008.10.087. [DOI] [PubMed] [Google Scholar]
- 61.Oliveira-Martins JB, Yusa S, Calella AM, Bridel C, Baumann F, Dametto P, Aguzzi A. 2010. Unexpected tolerance of alpha-cleavage of the prion protein to sequence variations. PLoS One 5:e9107. doi: 10.1371/journal.pone.0009107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eberl H, Tittmann P, Glockshuber R. 2004. Characterization of recombinant, membrane-attached full-length prion protein. J Biol Chem 279:25058–25065. doi: 10.1074/jbc.M400952200. [DOI] [PubMed] [Google Scholar]
- 63.Hicks MR, Gill AC, Bath IK, Rullay AK, Sylvester ID, Crout DH, Pinheiro TJ. 2006. Synthesis and structural characterization of a mimetic membrane-anchored prion protein. FEBS J 273:1285–1299. doi: 10.1111/j.1742-4658.2006.05152.x. [DOI] [PubMed] [Google Scholar]
- 64.Zuegg J, Gready JE. 2000. Molecular dynamics simulation of human prion protein including both N-linked oligosaccharides and the GPI anchor. Glycobiology 10:959–974. doi: 10.1093/glycob/10.10.959. [DOI] [PubMed] [Google Scholar]
- 65.Zacharias DA, Violin JD, Newton AC, Tsien RY. 2002. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 66.Bosque PJ, Prusiner SB. 2000. Cultured cell sublines highly susceptible to prion infection. J Virol 74:4377–4386. doi: 10.1128/JVI.74.9.4377-4386.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Caughey B, Race RE, Ernst D, Buchmeier MJ, Chesebro B. 1989. Prion protein biosynthesis in scrapie-infected and uninfected neuroblastoma cells. J Virol 63:175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.DeArmond SJ, Sanchez H, Yehiely F, Qiu Y, Ninchak-Casey A, Daggett V, Camerino AP, Cayetano J, Rogers M, Groth D, Torchia M, Tremblay P, Scott MR, Cohen FE, Prusiner SB. 1997. Selective neuronal targeting in prion disease. Neuron 19:1337–1348. doi: 10.1016/S0896-6273(00)80424-9. [DOI] [PubMed] [Google Scholar]
- 69.Wiseman FK, Cancellotti E, Piccardo P, Iremonger K, Boyle A, Brown D, Ironside JW, Manson JC, Diack AB. 2015. The glycosylation status of PrPC is a key factor in determining transmissible spongiform encephalopathy transmission between species. J Virol 89:4738–4747. doi: 10.1128/JVI.02296-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cancellotti E, Mahal SP, Somerville R, Diack A, Brown D, Piccardo P, Weissmann C, Manson JC. 2013. Post-translational changes to PrP alter transmissible spongiform encephalopathy strain properties. EMBO J 32:756–769. doi: 10.1038/emboj.2013.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cancellotti E, Bradford BM, Tuzi NL, Hickey RD, Brown D, Brown KL, Barron RM, Kisielewski D, Piccardo P, Manson JC. 2010. Glycosylation of PrPC determines timing of neuroinvasion and targeting in the brain following transmissible spongiform encephalopathy infection by a peripheral route. J Virol 84:3464–3475. doi: 10.1128/JVI.02374-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tuzi NL, Cancellotti E, Baybutt H, Blackford L, Bradford B, Plinston C, Coghill A, Hart P, Piccardo P, Barron RM, Manson JC. 2008. Host PrP glycosylation: a major factor determining the outcome of prion infection. PLoS Biol 6:e100. doi: 10.1371/journal.pbio.0060100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Priola SA, Lawson VA. 2001. Glycosylation influences cross-species formation of protease-resistant prion protein. EMBO J 20:6692–6699. doi: 10.1093/emboj/20.23.6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nishina KA, Deleault NR, Mahal SP, Baskakov I, Luhrs T, Riek R, Supattapone S. 2006. The stoichiometry of host PrPC glycoforms modulates the efficiency of PrPSc formation in vitro. Biochemistry 45:14129–14139. doi: 10.1021/bi061526k. [DOI] [PubMed] [Google Scholar]
- 75.Lee KS, Magalhaes AC, Zanata SM, Brentani RR, Martins VR, Prado MA. 2001. Internalization of mammalian fluorescent cellular prion protein and N-terminal deletion mutants in living cells. J Neurochem 79:79–87. [DOI] [PubMed] [Google Scholar]
- 76.Santuccione A, Sytnyk V, Leshchyns'ka I, Schachner M. 2005. Prion protein recruits its neuronal receptor NCAM to lipid rafts to activate p59fyn and to enhance neurite outgrowth. J Cell Biol 169:341–354. doi: 10.1083/jcb.200409127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sunyach C, Jen A, Deng J, Fitzgerald KT, Frobert Y, Grassi J, McCaffrey MW, Morris R. 2003. The mechanism of internalization of glycosylphosphatidylinositol-anchored prion protein. EMBO J 22:3591–3601. doi: 10.1093/emboj/cdg344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gaus K, Gratton E, Kable EP, Jones AS, Gelissen I, Kritharides L, Jessup W. 2003. Visualizing lipid structure and raft domains in living cells with two-photon microscopy. Proc Natl Acad Sci U S A 100:15554–15559. doi: 10.1073/pnas.2534386100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Safar JG, DeArmond SJ, Kociuba K, Deering C, Didorenko S, Bouzamondo-Bernstein E, Prusiner SB, Tremblay P. 2005. Prion clearance in bigenic mice. J Gen Virol 86:2913–2923. doi: 10.1099/vir.0.80947-0. [DOI] [PubMed] [Google Scholar]
- 80.Lee IS, Long JR, Prusiner SB, Safar JG. 2005. Selective precipitation of prions by polyoxometalate complexes. J Am Chem Soc 127:13802–13803. doi: 10.1021/ja055219y. [DOI] [PubMed] [Google Scholar]
- 81.Caughey B, Raymond GJ, Ernst D, Race RE. 1991. N-terminal truncation of the scrapie-associated form of PrP by lysosomal protease(s): implications regarding the site of conversion of PrP to the protease-resistant state. J Virol 65:6597–6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yadavalli R, Guttmann RP, Seward T, Centers AP, Williamson RA, Telling GC. 2004. Calpain-dependent endoproteolytic cleavage of PrPSc modulates scrapie prion propagation. J Biol Chem 279:21948–21956. doi: 10.1074/jbc.M400793200. [DOI] [PubMed] [Google Scholar]
- 83.Tremblay P, Ball HL, Kaneko K, Groth D, Hegde RS, Cohen FE, DeArmond SJ, Prusiner SB, Safar JG. 2004. Mutant PrPSc conformers induced by a synthetic peptide and several prion strains. J Virol 78:2088–2099. doi: 10.1128/JVI.78.4.2088-2099.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chiesa R, Piccardo P, Biasini E, Ghetti B, Harris DA. 2008. Aggregated, wild-type prion protein causes neurological dysfunction and synaptic abnormalities. J Neurosci 28:13258–13267. doi: 10.1523/JNEUROSCI.3109-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Monaco S, Fiorini M, Farinazzo A, Ferrari S, Gelati M, Piccardo P, Zanusso G, Ghetti B. 2012. Allelic origin of protease-sensitive and protease-resistant prion protein isoforms in Gerstmann-Straussler-Scheinker disease with the P102L mutation. PLoS One 7:e32382. doi: 10.1371/journal.pone.0032382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hegde RS, Mastrianni JA, Scott MR, DeFea KA, Tremblay P, Torchia M, DeArmond SJ, Prusiner SB, Lingappa VR. 1998. A transmembrane form of the prion protein in neurodegenerative disease. Science 279:827–834. doi: 10.1126/science.279.5352.827. [DOI] [PubMed] [Google Scholar]
- 87.Taguchi Y, Shi ZD, Ruddy B, Dorward DW, Greene L, Baron GS. 2009. Specific biarsenical labeling of cell surface proteins allows fluorescent- and biotin-tagging of amyloid precursor protein and prion proteins. Mol Biol Cell 20:233–244. doi: 10.1091/mbc.E08-06-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Orru CD, Wilham JM, Raymond LD, Kuhn F, Schroeder B, Raeber AJ, Caughey B. 2011. Prion disease blood test using immunoprecipitation and improved quaking-induced conversion. mBio 2:e00078-11. doi: 10.1128/mBio.00078-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bian J, Nazor KE, Angers R, Jernigan M, Seward T, Centers A, Green M, Telling GC. 2006. GFP-tagged PrP supports compromised prion replication in transgenic mice. Biochem Biophys Res Commun 340:894–900. doi: 10.1016/j.bbrc.2005.12.085. [DOI] [PubMed] [Google Scholar]
- 90.Priola SA, Caughey B, Race RE, Chesebro B. 1994. Heterologous PrP molecules interfere with accumulation of protease-resistant PrP in scrapie-infected murine neuroblastoma cells. J Virol 68:4873–4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Supattapone S, Muramoto T, Legname G, Mehlhorn I, Cohen FE, DeArmond SJ, Prusiner SB, Scott MR. 2001. Identification of two prion protein regions that modify scrapie incubation time. J Virol 75:1408–1413. doi: 10.1128/JVI.75.3.1408-1413.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Marbiah MM, Harvey A, West BT, Louzolo A, Banerjee P, Alden J, Grigoriadis A, Hummerich H, Kan HM, Cai Y, Bloom GS, Jat P, Collinge J, Klohn PC. 2014. Identification of a gene regulatory network associated with prion replication. EMBO J 33:1527–1547. doi: 10.15252/embj.201387150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Goold R, McKinnon C, Rabbanian S, Collinge J, Schiavo G, Tabrizi SJ. 2013. Alternative fates of newly formed PrPSc upon prion conversion on the plasma membrane. J Cell Sci 126:3552–3562. doi: 10.1242/jcs.120477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jacobson K, Mouritsen OG, Anderson RG. 2007. Lipid rafts: at a crossroad between cell biology and physics. Nat Cell Biol 9:7–14. doi: 10.1038/ncb0107-7. [DOI] [PubMed] [Google Scholar]
- 95.Allen JA, Halverson-Tamboli RA, Rasenick MM. 2007. Lipid raft microdomains and neurotransmitter signalling. Nat Rev Neurosci 8:128–140. doi: 10.1038/nrn2059. [DOI] [PubMed] [Google Scholar]
- 96.Enari M, Flechsig E, Weissmann C. 2001. Scrapie prion protein accumulation by scrapie-infected neuroblastoma cells abrogated by exposure to a prion protein antibody. Proc Natl Acad Sci U S A 98:9295–9299. doi: 10.1073/pnas.151242598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vorberg I, Raines A, Story B, Priola SA. 2004. Susceptibility of common fibroblast cell lines to transmissible spongiform encephalopathy agents. J Infect Dis 189:431–439. doi: 10.1086/381166. [DOI] [PubMed] [Google Scholar]
- 98.Stahl N, Baldwin MA, Hecker R, Pan KM, Burlingame AL, Prusiner SB. 1992. Glycosylinositol phospholipid anchors of the scrapie and cellular prion proteins contain sialic acid. Biochemistry 31:5043–5053. doi: 10.1021/bi00136a600. [DOI] [PubMed] [Google Scholar]
- 99.Bate C, Williams A. 2012. Neurodegeneration induced by clustering of sialylated glycosylphosphatidylinositols of prion proteins. J Biol Chem 287:7935–7944. doi: 10.1074/jbc.M111.275743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bate C, Nolan W, Williams A. 2016. Sialic acid on the glycosylphosphatidylinositol anchor regulates PrP-mediated cell signaling and prion formation. J Biol Chem 291:160–170. doi: 10.1074/jbc.M115.672394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Katorcha E, Daus ML, Gonzalez-Montalban N, Makarava N, Lasch P, Beekes M, Baskakov IV. 2016. Reversible off and on switching of prion infectivity via removing and reinstalling prion sialylation. Sci Rep 6:33119. doi: 10.1038/srep33119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Katorcha E, Srivastava S, Klimova N, Baskakov IV. 2016. Sialylation of glycosylphosphatidylinositol (GPI) anchors of mammalian prions is regulated in a host-, tissue-, and cell-specific manner. J Biol Chem 291:17009–17019. doi: 10.1074/jbc.M116.732040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Castilla J, Saa P, Hetz C, Soto C. 2005. In vitro generation of infectious scrapie prions. Cell 121:195–206. doi: 10.1016/j.cell.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 104.Deleault NR, Harris BT, Rees JR, Supattapone S. 2007. Formation of native prions from minimal components in vitro. Proc Natl Acad Sci U S A 104:9741–9746. doi: 10.1073/pnas.0702662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shikiya RA, Bartz JC. 2011. In vitro generation of high-titer prions. J Virol 85:13439–13442. doi: 10.1128/JVI.06134-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Deleault NR, Geoghegan JC, Nishina K, Kascsak R, Williamson RA, Supattapone S. 2005. Protease-resistant prion protein amplification reconstituted with partially purified substrates and synthetic polyanions. J Biol Chem 280:26873–26879. doi: 10.1074/jbc.M503973200. [DOI] [PubMed] [Google Scholar]
- 107.Geoghegan JC, Valdes PA, Orem NR, Deleault NR, Williamson RA, Harris BT, Supattapone S. 2007. Selective incorporation of polyanionic molecules into hamster prions. J Biol Chem 282:36341–36353. doi: 10.1074/jbc.M704447200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Deleault NR, Piro JR, Walsh DJ, Wang F, Ma J, Geoghegan JC, Supattapone S. 2012. Isolation of phosphatidylethanolamine as a solitary cofactor for prion formation in the absence of nucleic acids. Proc Natl Acad Sci U S A 109:8546–8551. doi: 10.1073/pnas.1204498109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang F, Wang X, Yuan CG, Ma J. 2010. Generating a prion with bacterially expressed recombinant prion protein. Science 327:1132–1135. doi: 10.1126/science.1183748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Murayama Y, Yoshioka M, Masujin K, Okada H, Iwamaru Y, Imamura M, Matsuura Y, Fukuda S, Onoe S, Yokoyama T, Mohri S. 2010. Sulfated dextrans enhance in vitro amplification of bovine spongiform encephalopathy PrPSc and enable ultrasensitive detection of bovine PrP(Sc). PLoS One 5:e13152. doi: 10.1371/journal.pone.0013152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mays CE, Yeom J, Kang HE, Bian J, Khaychuk V, Kim Y, Bartz JC, Telling GC, Ryou C. 2011. In vitro amplification of misfolded prion protein using lysate of cultured cells. PLoS One 6:e18047. doi: 10.1371/journal.pone.0018047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pan T, Wong BS, Liu T, Li R, Petersen RB, Sy MS. 2002. Cell-surface prion protein interacts with glycosaminoglycans. Biochem J 368:81–90. doi: 10.1042/bj20020773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hundt C, Peyrin JM, Haik S, Gauczynski S, Leucht C, Rieger R, Riley ML, Deslys JP, Dormont D, Lasmezas CI, Weiss S. 2001. Identification of interaction domains of the prion protein with its 37-kDa/67-kDa laminin receptor. EMBO J 20:5876–5886. doi: 10.1093/emboj/20.21.5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Caughey B, Baron GS. 2006. Prions and their partners in crime. Nature 443:803–810. doi: 10.1038/nature05294. [DOI] [PubMed] [Google Scholar]
- 115.Taylor DR, Whitehouse IJ, Hooper NM. 2009. Glypican-1 mediates both prion protein lipid raft association and disease isoform formation. PLoS Pathog 5:e1000666. doi: 10.1371/journal.ppat.1000666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Horiuchi M, Caughey B. 1999. Specific binding of normal prion protein to the scrapie form via a localized domain initiates its conversion to the protease-resistant state. EMBO J 18:3193–3203. doi: 10.1093/emboj/18.12.3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Horiuchi M, Baron GS, Xiong LW, Caughey B. 2001. Inhibition of interactions and interconversions of prion protein isoforms by peptide fragments from the C-terminal folded domain. J Biol Chem 276:15489–15497. doi: 10.1074/jbc.M100288200. [DOI] [PubMed] [Google Scholar]
- 118.Prusiner SB, McKinley MP, Bowman KA, Bolton DC, Bendheim PE, Groth DF, Glenner GG. 1983. Scrapie prions aggregate to form amyloid-like birefringent rods. Cell 35:349–358. doi: 10.1016/0092-8674(83)90168-X. [DOI] [PubMed] [Google Scholar]
- 119.McKinley MP, Meyer RK, Kenaga L, Rahbar F, Cotter R, Serban A, Prusiner SB. 1991. Scrapie prion rod formation in vitro requires both detergent extraction and limited proteolysis. J Virol 65:1340–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jeffrey M, McGovern G, Goodsir CM, Siso S, Gonzalez L. 2009. Strain-associated variations in abnormal PrP trafficking of sheep scrapie. Brain Pathol 19:1–11. doi: 10.1111/j.1750-3639.2008.00150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D. 2005. Structure of the cross-beta spine of amyloid-like fibrils. Nature 435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Medof ME, Nagarajan S, Tykocinski ML. 1996. Cell-surface engineering with GPI-anchored proteins. FASEB J 10:574–586. [DOI] [PubMed] [Google Scholar]
- 123.Liu T, Li R, Pan T, Liu D, Petersen RB, Wong BS, Gambetti P, Sy MS. 2002. Intercellular transfer of the cellular prion protein. J Biol Chem 277:47671–47678. doi: 10.1074/jbc.M207458200. [DOI] [PubMed] [Google Scholar]
- 124.Premkumar DR, Fukuoka Y, Sevlever D, Brunschwig E, Rosenberry TL, Tykocinski ML, Medof ME. 2001. Properties of exogenously added GPI-anchored proteins following their incorporation into cells. J Cell Biochem 82:234–245. doi: 10.1002/jcb.1154. [DOI] [PubMed] [Google Scholar]
- 125.Legler DF, Doucey MA, Schneider P, Chapatte L, Bender FC, Bron C. 2005. Differential insertion of GPI-anchored GFPs into lipid rafts of live cells. FASEB J 19:73–75. [DOI] [PubMed] [Google Scholar]
- 126.Horonchik L, Tzaban S, Ben-Zaken O, Yedidia Y, Rouvinski A, Papy-Garcia D, Barritault D, Vlodavsky I, Taraboulos A. 2005. Heparan sulfate is a cellular receptor for purified infectious prions. J Biol Chem 280:17062–17067. doi: 10.1074/jbc.M500122200. [DOI] [PubMed] [Google Scholar]
- 127.Hijazi N, Kariv-Inbal Z, Gasset M, Gabizon R. 2005. PrPSc incorporation to cells requires endogenous glycosaminoglycan expression. J Biol Chem 280:17057–17061. doi: 10.1074/jbc.M411314200. [DOI] [PubMed] [Google Scholar]
- 128.Morillas M, Swietnicki W, Gambetti P, Surewicz WK. 1999. Membrane environment alters the conformational structure of the recombinant human prion protein. J Biol Chem 274:36859–36865. doi: 10.1074/jbc.274.52.36859. [DOI] [PubMed] [Google Scholar]
- 129.Sanghera N, Pinheiro TJ. 2002. Binding of prion protein to lipid membranes and implications for prion conversion. J Mol Biol 315:1241–1256. doi: 10.1006/jmbi.2001.5322. [DOI] [PubMed] [Google Scholar]
- 130.Wong C, Xiong LW, Horiuchi M, Raymond L, Wehrly K, Chesebro B, Caughey B. 2001. Sulfated glycans and elevated temperature stimulate PrPSc-dependent cell-free formation of protease-resistant prion protein. EMBO J 20:377–386. doi: 10.1093/emboj/20.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ben-Zaken O, Tzaban S, Tal Y, Horonchik L, Esko JD, Vlodavsky I, Taraboulos A. 2003. Cellular heparan sulfate participates in the metabolism of prions. J Biol Chem 278:40041–40049. doi: 10.1074/jbc.M301152200. [DOI] [PubMed] [Google Scholar]
- 132.Waneck GL, Stein ME, Flavell RA. 1988. Conversion of a PI-anchored protein to an integral membrane protein by a single amino acid mutation. Science 241:697–699. doi: 10.1126/science.3399901. [DOI] [PubMed] [Google Scholar]