Abstract
The pathogenesis of prion diseases, a class of transmissible fatal neurodegenerative diseases in humans and animals, is still unclear. The aim of this study was to identify the differentially regulated genes that correlate with the development of prion diseases for a better understanding of their pathological mechanisms. We employed Affymetrix Mouse Expression Arrays 430A containing >22,000 transcripts and compared the global gene expression profiles from brains of mice who were intracerebrally inoculated with scrapie strains ME7 and RML with those from brains of uninfected and mock-infected mice. The microarray data were analyzed by Significance Analysis of Microarrays, revealing 121 genes whose expression increased at least twofold in both ME7- and RML-infected mouse brains, with an estimated false discovery rate of ≤5%. These genes encode proteins involved in proteolysis, protease inhibition, cell growth and maintenance, the immune response, signal transduction, cell adhesion, and molecular metabolism. The time course of expression generally showed up-regulation of these genes from 120 days postinoculation (dpi) for ME7-inoculated mouse brains and from 90 dpi for RML-inoculated mouse brains. The onset of elevated expression correlated temporally with the onset of PrPSc accumulation and the activation of glia, which may have contributed to neuronal cell death. Among the differentially regulated genes reported in the present study, the emergence of genes for several cathepsins and S100 calcium binding proteins was conspicuous. These and other genes reported here may represent novel potential diagnostic and therapeutic targets for prion disease.
Prion diseases are a class of transmissible fatal neurodegenerative diseases which include Creutzfeldt-Jakob disease (CJD) in humans, bovine spongiform encephalopathy (BSE) in cattle, and scrapie in goats and sheep. A hallmark of prion diseases is the posttranslational conformational conversion of a host-encoded protein, the prion protein (PrP), to a disease-associated isoform, PrPSc. PrPSc can be distinguished from physiological PrP (PrPc) by its high beta-sheet content and its partial resistance to protease digestion. According to the prion hypothesis, PrPSc is the principal component of the infectious particle, which is called the prion (46).
The neuropathology of prion diseases is characterized by the appearance and accumulation of PrPSc in the brain, spongiform degeneration, neuronal loss, and the activation of glial cells (23, 31). Intensive research has been carried out to verify the correlation between the conversion of PrPc to PrPSc and the pathogenesis of prion disease. There is evidence indicating that the accumulation of PrPSc contributes to neuronal loss and gliosis. In addition, activated microglia and astrocytes release proinflammatory and neurotoxic factors, which may also contribute to neurodegeneration in prion diseases (21, 23, 27). However, the exact pathogenic mechanisms of neurodegeneration still remain unclear.
In addition to the involvement of the conformational change of PrP in the development of prion diseases, allelic variants of the prion protein gene (Prnp) affect the incubation time or susceptibility in humans, mice, and sheep (8, 26, 43, 54). In humans, a polymorphism in the coding region of the prion protein gene at codon 129 influences both the susceptibility of an individual to sporadic CJD and the clinical and pathological phenotype of the disease. In mice, two genotypes of Prnp, Prnpa and Prnpb, which are defined by the polymorphic codons 108 and 189, affect the incubation time in scrapie-infected mice. However, other lines of evidence indicate that the coding region of PrP is not the sole genetic influence. A comparison of several inbred mouse strains with the same Prnp genotype revealed notable differences in the incubation times of a defined prion strain, indicating that other factors contribute to the observed variation (34, 35).
The identification of genes which show differential expression during prion infection could help us to identify novel risk genes aside from the well-established Prnp gene as well as to find the abnormal intracellular or intercellular pathways that are responsible for the pathogenesis of prion diseases. The use of global expression analysis platforms such as oligonucleotide microarrays has become a robust technique for the identification of differentially expressed genes, and this technique has recently been used for the identification of novel cellular targets associated with a large variety of diseases. For this study, we applied this technique to identify differentially expressed genes in scrapie-infected mice by investigating the transcriptional expression of >22,000 transcripts with Affymetrix (Santa Clara, Calif.) high-density oligonucleotide probe arrays. We identified 121 genes that were up-regulated in the brains of mice who were infected with two distinct strains of scrapie, ME7 and RML (Rocky Mountain Laboratory). The majority of these genes have not been previously described for prion diseases and may serve as potential therapeutic targets and novel markers for these diseases.
MATERIALS AND METHODS
Inoculation of animals.
Four groups of 6-week-old C57BL/6 mice (Harlan Winkelmann, Borchen, Germany) were used for this study. The mice were treated as follows: for group 1, the mice were killed at the age of 6 weeks without any treatment (day 0); for group 2, the mice were inoculated intracerebrally with 30 μl of a 10% brain homogenate from healthy mice and were killed on day 30, 60, 90, 120, or 150 postinoculation (mock infection); for groups 3 and 4, the mice were inoculated intracerebrally with 30 μl of a 10% brain homogenate from mice infected with either ME7 or RML and were killed on day 30, 60, 90, or 120 postinoculation or when they showed clinical signs associated with terminal prion disease. Standard diagnostic criteria were used to identify animals exhibiting signs of scrapie (8, 47).
The mice were killed by cervical dislocation, and their brains were removed, frozen immediately in liquid nitrogen, and stored at −80°C until needed.
Microarray analysis.
Sample preparation and processing procedures were performed as described in the Affymetrix GeneChip Expression Analysis manual. Total RNAs were prepared with a phenol-guanidine isothiocyanate reagent (Trizol; Invitrogen, Karlsruhe, Germany) and cleaned by the use of RNeasy columns (Qiagen, Hilden, Germany). From 21-μg samples of total RNA, first-strand cDNAs were synthesized with SuperScript II (Invitrogen) and the T7-(dT24) primer (MWG, Ebersberg, Germany), followed by the generation of double-stranded cDNAs. The resultant cDNAs were purified by use of a GeneChip sample clean-up module (Affymetrix) and served as templates for the generation of biotinylated antisense RNAs (cRNAs) by use of an EnzoBioarray kit (Affymetrix). The biotinylated cRNAs were hybridized to Mouse Expression Arrays 430A (ME430A; Affymetrix), which contain 22,690 transcripts, at 45°C for 16 h with constant rotation at 60 rpm. The microarrays were then washed and stained with an Affymetrix fluidics station and scanned on Affymetrix scanners. The images were processed with Microarray Analysis Suite 5.0 (Affymetrix). All samples demonstrated characteristics of high-quality cRNA (3′/5′ ratio of probe sets for glyceraldehyde-3-phosphate dehydrogenase and beta-actin of <1.5) and were subjected to subsequent analysis.
The raw data for microarray results were scaled from each array to a target intensity value of 500 so that interarray comparisons could be performed and then were imported into the Significance Analysis of Microarrays (SAM) software (http://www-stat.stanford.edu/∼tibs/SAM/index.html; also see reference 53). SAM identifies genes with statistically significant changes in expression by assimilating a set of gene-specific t tests. Each gene, i, is assigned a score, d(i), representing the relative difference of this gene. Because the distribution of d values is independent of the level of gene expression, the use of the d value should help to correct for possible over- or underestimation by determinations of the fold change. The d(i) value is generated based on the ratio of the change in expression of gene i in different states (e.g., in treated and untreated animals) relative to the standard deviation of repeated measurements. In addition to the generation of a d(i) value and the fold change of each gene i, SAM provides an estimate of the false discovery rate (FDR) (the percentage of genes identified by chance alone) from randomly permuted data. Genes with scores higher than a threshold value or genes with FDR values lower than the threshold value were deemed potentially significant (53). For this study, SAM analysis was performed with the following settings: two-class response type, log transformation of data (on a base 2 scale), and 100 permutations. Genes were considered differentially expressed if they changed more than 1.5- or 2-fold (as defined in Results) with an estimated FDR of ≤5%.
Western blots.
Mouse brains were homogenized in lysis buffer containing 100 mM NaCl, 100 mM EDTA, 0.5% NP-40, 0.5% deoxycholate, and 10 mM Tris-HCl (pH 7.4). For the detection of protease-resistant PrPSc, equal volumes of brain homogenates were digested with proteinase K (PK; Roche, Mannheim, Germany) at a final concentration of 10 μg/ml for 30 min at 37°C. Digestion reactions were stopped by the addition of 2 mM phenylmethylsulfonyl fluoride. Equal volumes of brain homogenates that were treated or not treated with PK were boiled in 1× Laemmli buffer for 5 min, subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and subsequently transferred to polyvinylidene difluoride membranes (Millipore, Eschborn, Germany). The rabbit polyclonal antibody RA3153, derived against a peptide comprising amino acids 90 to 103 of murine PrP, was generated by Sigma Genosys (Cambridge, United Kingdom). RA3153 was used against murine PrP at a dilution of 1:3,000, followed by incubation of the membranes with an alkaline phosphatase-conjugated goat anti-rabbit antibody (1:3,000; Dako, Hamburg, Germany). The signals were visualized by chemiluminescence and autoradiography on X-ray films (Amersham Biosciences, Freiburg, Germany).
Quantitative RT-PCR.
Real-time quantitative reverse transcription-PCR (RT-PCR) was performed by using the Roche Light Cycler system. Briefly, 5-μg samples of total RNAs isolated from mouse brains as described above were used to make single-stranded cDNAs (Superscript II; Invitrogen) according to the manufacturer's instructions. Two microliters of diluted cDNA (1:10) was subjected to further PCR cycles, which were done with Faststart DNA Master SYBR Green I (Roche). The PCR conditions were 95°C for 10 min for a hot start, followed by denaturing at 95°C for 10 s, annealing at 57°C for 5 s, and extension at 72°C for 10 s for 45 cycles. The beta-actin gene was used as a general housekeeping gene to normalize target gene mRNA expression levels. Primer sets for beta-actin and target genes were chosen from published studies. Care was taken to select primers that bound to exon-intron boundaries or spanned exon-exon splice sites to avoid amplification of contaminating traces of genomic DNA.
The primer sets used for PCRs were as follows: for beta-actin (modified from reference 20), 5′ AAC CCT AAG GCC AAC CGT GAA AAG 3′ and 5′ CTA GGA GCC AGA GCA GTA ATC T 3′; for glial fibrillary acidic protein (GFAP) (20), 5′ AGT CCC TCC GCG GCA CGA ACG A 3′ and 5′ ACC ATC CCG CAT CTC CAC AGT CTT TAC CAC 3′; for S100 calcium binding protein A6 (S100A6) (17), 5′ CAG TGA TCA GTC ATG GCA TGC C 3′ and 5′ ACG GTC CCA TTT TAT TTC AGA GCT 3′; and for cathepsin D (14), 5′ AGG TGA AGG AGC TGC AGA AG 3′ and 5′ ATT CCC ATG AAG CCA CTC AG 3′.
Relative transcriptional expression levels of the target genes were generated by a relative quantification method as recommended by Roche (technical note LC 13/01).
RESULTS
Development of prion disease in mice inoculated intracerebrally with scrapie strains.
All ME7- and RML-infected mice that were not sacrificed during the time course experiment showed the clinical signs associated with terminal prion disease. The incubation time (the time between inoculation and terminal disease) for ME7-infected mice was longer (152 ± 4 days; n = 24) than that for RML-infected mice (144 ± 6 days; n = 24). The onset of terminal disease was more synchronous in ME7-infected mice than in RML-infected mice, as indicated by the standard deviation. No prion-associated clinical signs were observed in mock-infected and uninfected mice beyond day 250 postinoculation.
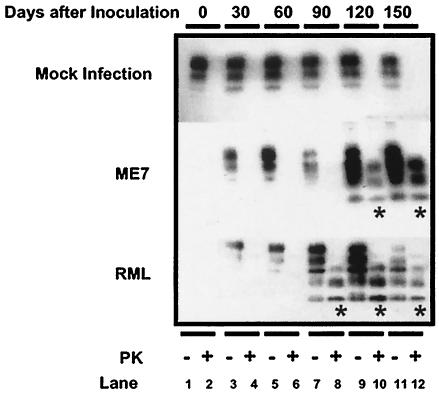
To assess the accumulation of PrPSc in mouse brains after inoculation with the scrapie agent, we detected PrPSc by treating the brain homogenates with PK and performing a subsequent immunoblot analysis. As shown in Fig. 1, the accumulation of PrPSc was readily detectable in RML-infected mouse brains at 90 days postinoculation (dpi), while the signal from PrPSc emerged at 120 dpi in ME7-infected mouse brains. This finding is in agreement with the different incubation times caused by RML and ME7 that were observed in this and previous studies (51).
FIG. 1.
Detection of PrPSc in mouse brains. Brains of mice inoculated with a healthy brain homogenate (mock infection) or an ME7 or RML inoculum were homogenized on days 30, 60, 90, 120, and 150 postinoculation and then treated (+) or not treated (−) with PK prior to Western blot analysis. Western blots of homogenates from uninfected mouse brains (day 0) are shown in the first two lanes of the top panel (mock infection). The accumulation of PK-resistant PrPSc is indicated with stars.
Up-regulated genes in mouse brains inoculated with ME7 and RML.
To identify differentially expressed genes in scrapie-infected mouse brains, we analyzed the expression levels of all transcripts on the ME430A array by using SAM to compare the expression levels in scrapie-infected brains from mice at the terminal stage with those in brains of control animals (i.e., uninoculated mouse brains and mock-infected mouse brains at 150 dpi). The inclusion of mock-infected mouse brains ensured the elimination of factors caused by the aging process and/or inoculation. Since SAM revealed only subtle differences in gene expression between uninoculated and mock-infected mouse brains (data not shown), these mice were combined as control animals and compared with ME7- or RML-infected mice by SAM analysis.
To achieve a high stringency for the identification of differentially expressed genes, we used the following strategy after ranking all genes by their relative difference d values, generated as described in Materials and Methods. Firstly, genes that had expression levels below the detection limits of the Affymetrix platform and that therefore generated an absent call based on a proprietary algorithm developed by Affymetrix in all experiments were eliminated from further analysis in order to avoid critical information caused by these genes; secondly, the threshold of the estimated FDR was set at 5%, indicating that the percentage of genes identified by chance was <5%; finally, only genes expressed at more-than-twofold higher levels in scrapie-infected mice than in control mice were considered. After the purging of redundant transcripts from the same gene, 190 genes in ME7-infected mouse brains and 179 genes in RML-infected mouse brains were identified. All of these genes demonstrated increased expression in scrapie-infected mouse brains compared to those of control animals (i.e., uninoculated and mock-infected mice). To recognize the genes that showed expression changes in both ME7- and RML-infected mouse brains, we compared the two data sets derived from ME7- and RML-infected mice and found 121 overlapping genes that were commonly up-regulated in both ME7- and RML-infected mouse brains.
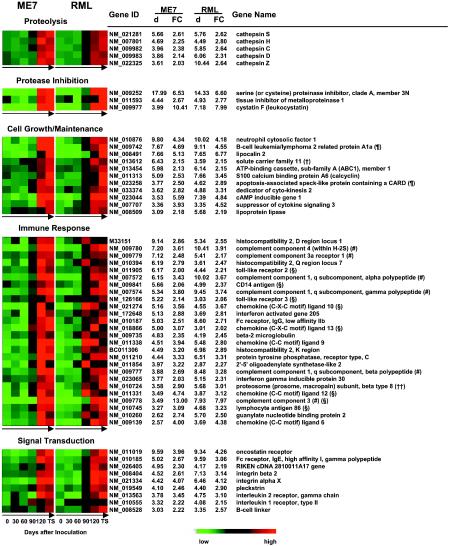
By applying the gene ontology mining tool of Affymetrix, we found that the 121 commonly up-regulated genes were involved in multiple biological processes. As highlighted in Fig. 2, several members of the cathepsin family (cathepsins S, H, C, D, and Z), which is involved in proteolysis, showed up-regulation in scrapie-infected mouse brains. Moreover, two additional cathepsins, cathepsins B and L, showed increases in expression of >1.5-fold in RML- and ME7-infected mice, respectively (data not shown). Since changes in gene expression between one- and twofold were not taken into account in this data analysis, cathepsins B and L were excluded from the most up-regulated genes shown in Fig. 2. Interestingly, several protease inhibitors, including cystatin F, a cysteine protease inhibitor (40), were concomitantly up-regulated in diseased brains.
FIG. 2.
Time course of expression of 121 genes identified as commonly up-regulated genes in both ME7 (left)- and RML (right)-infected brains. The expression of each gene in ME7- and RML-infected mouse brains on days 0, 30, 60, 90, and 120 postinoculation and at the terminal stage (TS) is shown. Each gene is represented by a single row, and each time point is represented by a single column. The genes are indicated by their GenBank accession numbers (gene ID) and gene descriptions (gene name). Gene expression is displayed colorimetrically. For each gene, red indicates an expression level higher than the average expression level of this gene in all samples, green indicates an expression level lower than the average, and black indicates that the expression level was close to the average. The data points on day 0 and at the terminal stage are averages of the expression levels in three mouse brains, and data points between 30 and 120 dpi were generated from the data for one mouse brain each. The d value and fold change (FC) in the expression of each gene in terminally diseased mouse brains (infected by ME7 and RML) compared to those in uninoculated and mock-infected mouse brains combined were generated by SAM. The genes are grouped according to function and are sorted in the order of d values of ME7-infected mouse brains. #, genes involved in the complement activation pathway; §, genes involved in the inflammatory response pathway; ¶, genes associated with apoptosis; †, the full gene name is solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1; ††, the full gene name is proteasome (prosome, macropain) subunit, beta type 8 (large multifunctional protease 7); †††, the full gene name is C-type (calcium dependent, carbohydrate recognition domain) lectin, superfamily member 12.
Previous studies have linked abnormalities in cell growth and maintenance, such as the induction of apoptosis, to neurodegeneration in prion disease (21, 22). In the present study, several novel genes from this category were identified, including S100A6, which is thought to be involved in the regulation of the cell cycle (5), as well as two apoptosis-associated genes, Bcl2a1a (B-cell leukemia/lymphoma 2-related protein A1a) (10) and Asc-pending (apoptosis-associated speck-like protein containing a CARD) (36).
A significant number of genes associated with the immune response, including genes involved in the complement activation and inflammatory response pathways, showed increased expression in scrapie-infected mouse brains. Additionally, genes involved in signal transduction and cell adhesion showed altered expression, as did genes involved in metabolism, including nucleic acid, protein, and lipid metabolism. Genes that are thought to be involved in other biological processes or whose functions are still not clarified were also identified as displaying increased transcription (Fig. 2, other). It is important to bear in mind that gene ontology analysis is a dynamic and flexible tool, as the state of biological knowledge of what genes and proteins do is very incomplete and is changing rapidly (1). In addition, the product of a particular gene may be multifunctional. Therefore, it cannot be ruled out that a gene which falls into a particular category may function in several other processes or that a gene which has unknown functions according to the current gene ontology data (e.g., many genes that are categorized as “other”) may be involved in one of the defined biological processes shown in Fig. 2.
An analysis of the time course of changes in expression demonstrated a generally earlier onset of up-regulation of most of the 121 genes in RML-infected mouse brains than that for genes in ME7-infected mice (Fig. 2). The onset of up-regulation of the 121 differentially regulated genes in ME7-infected mouse brains was generally rapid and readily detectable on day 120. In RML-infected mouse brains, up-regulation of the majority of genes became visible on day 90 and became obvious on day 120, showing a more gradual increase in expression. These different expression patterns correlating with different prion strains can be representatively observed in the genes involved in proteolysis (Fig. 2, proteolysis).
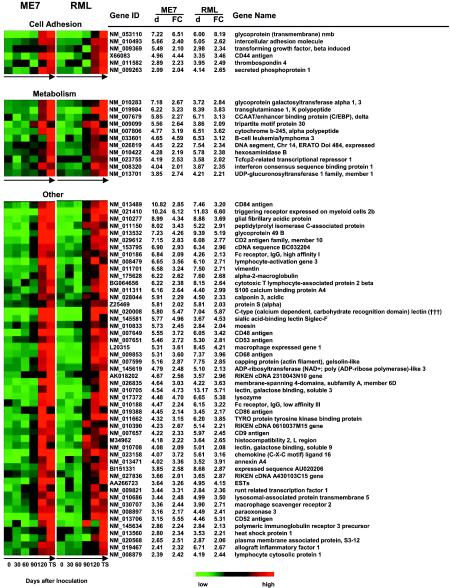
Up-regulated genes associated with particular scrapie strains.
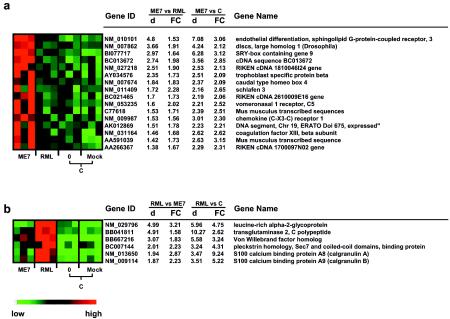
The ME7- and RML-infected mouse brains shared an increased expression of 121 genes, as described above, corresponding to 64% (121 of 190) and 68% (121 of 179) of the most differentially expressed genes in ME7- and RML-infected mouse brains, respectively. However, the existence of genes that did not overlap (67 and 56 genes in ME7- and RML-infected mice, respectively) indicated that different scrapie strains may cause various expression patterns in these genes. Several of the 67 genes that were predominantly up-regulated in ME7-infected mouse brains even showed suppressed expression in RML-infected mouse brains. The same held true for some of the 56 genes that were predominantly up-regulated in RML-infected mouse brains. Using SAM, we compared the expression data for the 67 ME7-associated genes and the 56 RML-associated genes derived from mice infected by one strain (e.g., ME7) with those derived from mice infected by the other strain (e.g., RML). With an estimated FDR of ≤5%, SAM identified 16 genes that were up-regulated at least 1.5-fold in ME7-infected mouse brains compared to RML-infected mouse brains (Fig. 3a). With the same criteria, six genes, including two S100 calcium binding proteins (S100A8 and S100A9), were identified as being highly correlated with RML (Fig. 3b). The unique expression pattern of these genes in brains of mice infected by a specific strain (ME7 or RML) indicated transcriptional distinctions which may contribute to the distinct characteristics caused by ME7 and RML.
FIG. 3.
Differentially expressed genes associated with ME7 (a) and RML (b). The genes are indicated by their GenBank accession numbers (gene ID) and gene descriptions (gene name). Gene expression in the brains of scrapie-infected mice at the terminal stage (for ME7, n = 3, and for RML, n = 3) and in the brains of control animals (C), including uninfected mice (0; n = 3) and mock-infected mice on day 150 after inoculation (mock; n = 3), is displayed colorimetrically. For each gene, red indicates an expression level higher than the average expression level of this gene in all samples, green indicates an expression level lower than the average, and black indicates that the expression level was close to the average. The d value and fold change (FC) in the expression of each gene in the brains of mice infected by one strain, e.g., ME7, compared to the mice infected by the other strain, e.g., RML, and to control animals were generated by SAM. The genes are sorted in the order of d values generated by the comparison between ME7- and RML-infected mouse brains.
Validation of gene expression patterns by RT-PCR.
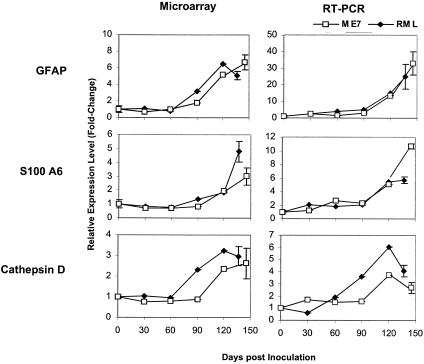
To validate the expression patterns detected by the microarray approach, we assessed the expression levels of three selected genes, GFAP, cathepsin D, and S100A6, by quantitative RT-PCR. These genes were identified as being commonly up-regulated in the brains of mice inoculated with ME7 and RML. Concordant trends in the expression levels yielded by microarrays and RT-PCR were observed for all three genes (Fig. 4). The changes in expression in scrapie-infected mouse brains versus uninoculated mouse brains (day 0) observed by microarray analysis were generally lower than those derived by RT-PCR. This could be explained by the different detection ranges of these two methods. Microarrays tend to have a low dynamic range, which could lead to under-representations of changes in gene expression, while RT-PCR has a high dynamic range (38).
FIG. 4.
Time course of expression of GFAP, S100A6, and cathepsin D in ME7- and RML-inoculated mouse brains. Analysis results for microarray assays and quantitative PCR are compared. For the microarray analysis, averages and standard deviations of three experiments on day 0 and at the terminal stage of ME7- or RML-inoculated mouse brains are shown. Data points between 30 and 120 dpi were generated from data for one mouse brain. For RT-PCR, averages and standard deviations of at least three experiments on day 0 and at the terminal stage of ME7- or RML-inoculated mouse brains are shown. Data points between 30 and 120 dpi are representative of average expression levels of at least two mouse brains. The relative expression levels of the genes are indicated by the fold changes in expression level in scrapie-infected mouse brains versus uninoculated mouse brains (day 0).
DISCUSSION
To identify the differentially expressed genes and the associated molecular pathological events in prion disease, we applied Affymetrix ME430A expression microarrays that allowed us to examine the expression levels of about 22,000 genes on a single platform. In order to identify the common expression changes in different scrapie models, we studied the gene expression levels in brains of mice infected with two scrapie strains, ME7 and RML. ME7 and RML were derived, respectively, from the spleen of a scrapie-infected sheep and from brains of experimentally infected goats that had previously been inoculated with the sheep scrapie brain homogenate SSBP/1 (6, 9, 51). RML and ME7 are known to have differences in incubation periods, clinical signs, biochemical patterns of PrPSc, and lesion profiles such as vacuolar degeneration (7, 32, 49, 51).
Global transcriptional expression analysis with SAM software, a robust program for the analysis of microarray expression data (53), revealed a total of 190 or 179 genes that changed at least twofold, with an FDR of ≤5%, in ME7- and RML-infected mouse brains, respectively. Among these genes, 121 genes were identified as being commonly up-regulated in both ME7- and RML-infected mouse brains. In addition, 16 and 6 genes showed a remarkable increase in expression only in ME7- or RML-infected mouse brains, respectively, suggesting a specific association of the expression changes of these genes with the two scrapie strains studied. The elevated expression of 121 genes observed in mouse brains infected with two distinct scrapie strains indicates that these genes may be common to different scrapie models and might therefore be especially important for the common molecular basis of prion disease.
Using quantitative RT-PCR, the standard validation procedure, we observed an overall concordance of trends between quantitative RT-PCR and microarray results for three genes, including the S100A6 gene, a novel differentially regulated gene identified in this study. This strengthens the power of our analysis based on the microarray technique.
The up-regulation of several genes which had been described in previous studies of scrapie-infected brains, such as cathepsin D, cathepsin S, complement C1qB, GFAP, lysosomal-associated protein transmembrane 5, β-2 microglobulin, and vimentin, was confirmed by our study (Table 1, group 1). In addition, the up-regulation of cathepsins S, L, and H, cystatin F, the CD48, CD84, and CD86 antigens, and cytochrome b-245-a, as well as several Fc receptors, which was observed in another experimental model, i.e., in microglia isolated from CJD-infected mouse brains (2), was also confirmed in this study. Several previously identified genes did not emerge in the list of 121 commonly dysregulated genes, which was generated with settings of an FDR of ≤5% and a more-than-twofold change, but they showed an enhanced expression of >1.5-fold in ME7- or in both ME7- and RML-infected mouse brains, with an FDR ≤5% (data not shown). These genes included apolipoproteins D and E, aquaporin 4, F4/80, metallothionein II, 2′,5′-oligoadenylate synthetase, and ScRG-1 (Table 1, group 2). The identification of up-regulated genes by independent techniques and in various models as described above confirms the validity of the present study based on the global gene expression technique.
TABLE 1.
Up-regulated genes identified by previous studies of brain tissue of scrapie-infected animals
| Groupa | Name of gene product | Model | Methodb | Reference |
|---|---|---|---|---|
| 1 | Cathepsin D | Mouse | NB, ISH | 13 |
| Cathepsin S | Mouse | DD, NB | 12 | |
| Complement ClqB | Mouse | DD, NB | 12 | |
| GFAP | Hamster, mouse | SH, NB | 12, 48, 15 | |
| LAPTtm5 | Hamster | SH, NB | 48 | |
| β-Microglobulin | Hamster, mouse | DD, NB, SH | 12, 15 | |
| Vimentin | Hamster | SH, NB | 48 | |
| 2 | Apolipoprotein D** | Mouse | DD, NB | 12 |
| Apolipoprotein E* | Mouse | NB, ISH | 13 | |
| AQP-4* | Hamster | SH, NB | 48 | |
| F4/80** | Mouse | DD | 12 | |
| Metallothionein II** | Mouse | DD, NB | 12 | |
| 2′,5′-oligoadenylate synthetase* | Hamster | SH, NB | 48 | |
| ScRG-1* | Mouse | DD, NB | 12 | |
| 3 | Complement ClqC | Hamster | SH, NB | 48 |
| HSP70 | Mouse | NB | 29 | |
| IL-1α | Mouse | RT-PCR | 30 | |
| IL-1β | Mouse | RT-PCR | 30 | |
| IL-6 | Mouse | RT-PCR | 30 | |
| iNOS | Mouse | RT-PCR | 30 | |
| Polyubiquitin | Mouse | NB | 29 | |
| Transferrin | Hamster | SH | 15 | |
| 4 | B-lymphocyte chemoattractant | Hamster | SH, NB | 48 |
| gp39 precursor | Hamster | SH, NB | 48 | |
| IIGP protein | Hamster | SH, NB | 48 | |
| IP-10 | Hamster | SH, NB | 48 | |
| Lhx7 | Hamster | SH, NB | 48 | |
| Mx protein (p78) | Hamster | SH, NB | 48 | |
| ScRG-2 | Mouse | DD, NB | 12 |
Group 1, the genes showed more than twofold increases in expression and an FDR of ≤5% in both ME7- and RML-infected mouse brains in this study; group 2, the genes demonstrated 1.5 to 2-fold increases in expression and an FDR of ≤5% in ME7 (*)-or both ME7- and RML-infected mouse brains (**) in this study; group 3, the genes were not identified by SAM with a setting of an FDR of ≤5% used in this study; group 4, the genes are not included in the Affymetrix ME430A microarray.
NB, Northern blots; DD, differential display; ISH, in situ hybridization; SH: subtractive hybridization.
Several genes which had previously been identified as showing up-regulation at the transcriptional level were not confirmed in the present study. Some of them were not represented on the Affymetrix ME430A microarray (Table 1, group 4). For the remaining genes (Table 1, group 3), the reason for our inability to confirm their increased expression in this study is unknown, but it was likely due to the use of different rodent and scrapie strains in the previous studies. Additionally, the different detection sensitivities of the techniques used may have caused the discrepancies between the findings of our study and those of previous studies.
Remarkably, a total of 26 genes involved in the immune response, including genes participating in the complement activation and inflammatory response pathways and astrocyte-associated genes such as GFAP and vimentin, showed increased expression in both ME7- and RML-infected brains. These findings support the hypothesis that the enhancement of complement proteins and inflammatory factors and the associated glial activation in the diseased brain are important pathogenic events in prion disease.
Previous studies have shown an association of microglial activation and neurodegeneration with prion disease. Apoptosis is known to be a mode of neuronal cell death in prion disease (21, 22). In the present work, we identified two apoptosis-associated genes, Bcl2ala and Asc-pending, as well as genes that influence cellular fate, such as S100A6. Since the differentially regulated genes reported here are classified according to current gene ontology data, a large number of these genes are or may be involved in multiple biological processes. Thus, genes from other functional categories may also contribute to apoptosis or neuronal cell death in scrapie-infected mouse brains. For example, CD14, a gene involved in the immune response, is known to bind to the endotoxin lipopolysaccharide, and this complex in turn binds to Toll-like receptor 4 and triggers neurodegeneration (33).
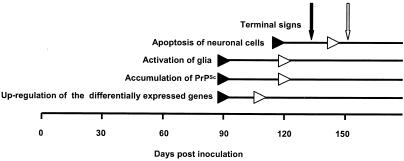
The time course of expression generally showed up-regulation of the 121 genes from 120 dpi in ME7-inoculated mouse brains and from 90 dpi in RML-inoculated mouse brains. This coincided temporally with the onset of PrPSc accumulation detected by Western blot analysis. Combining the time course experiments of this study with the results of a previous study (21), we additionally observed a temporal correlation of activated expression in the 121 genes with the glial activation which clearly preceded the detection of apoptotic neuronal cell death (21). These findings support a link between the 121 commonly up-regulated genes and the relevant pathological events in scrapie-infected brains, including the accumulation of PrPSc and the activation of glia, which may contribute to neurodegeneration (Fig. 5).
FIG. 5.
Onset of relevant pathological events in scrapie-infected mouse brains. The approximate onset of changes in ME7 (white arrows)- and RML (black arrows)-infected mouse brains are indicated. The approximate onset of terminal signs in scrapie-infected mice is highlighted by vertical arrows (black, ME7; white, RML). After the appearance of terminal signs, the mice were sacrificed for later analysis. The results of the present study (the onset of the most up-regulated genes, PrPSc accumulation, the activation of astrocytes, as confirmed by the up-regulation of GFAP, and the appearance of terminal signs) and previous work (the onset of PrPSc accumulation and the activation of glia as well as apoptosis induction) (21) are summarized.
For a large number of genes reported in this study, their functional relevance to the pathogenesis of prion disease is unclear at present. However, the altered expression of several genes with known functions and their associated biological processes are conspicuous. Several cathepsins, including cathepsins S, H, C, D, and Z, emerged in the list of commonly up-regulated genes in infected mouse brains. Additionally, cathepsins B and L demonstrated increased expression in RML- and ME7-infected mouse brains, respectively. An imbalance of cathepsins, a family of lysosomal proteases, has been associated with neurodegenerative diseases, including Alzheimer's disease and prion disease (2, 11, 12, 13, 56). Our observations of up-regulated cathepsin expression again confirmed these previous findings. In addition, genome-wide screening for differentially expressed genes enabled us to identify cathepsins, including cathepsins C and Z, that have not been described in the context of prion disease. The activation of cathepsins was found to be an upstream event in the apoptotic process (28). A recent study demonstrated that the β-amyloid protein, a pathological protein that undergoes fibrillogenesis during the development of Alzheimer's disease, caused an increased activation of cathepsin L in neuronal cells. This in turn accelerated the β-amyloid-mediated induction of the apoptotic cascade (4). Moreover, activated microglia are known to release some members of the cathepsin family which induce neuronal death by degrading extracellular matrix proteins (39). These previous findings suggest that cathepsins may have an impact on neuronal loss in different populations of brain cells.
Interestingly, the up-regulation of cathepsins after scrapie infection was accompanied by the up-regulation of several protease inhibitors, including cystatin F. An increased expression of cystatin F in microglia isolated from CJD-infected mouse brains was also reported by Baker and Manuelidis (2), indicating a correlation of cystatin F with microglial cells. Cystatin F is a cysteine protease inhibitor and binds cathepsin L with a high affinity (40). The up-regulation of cystatin F may be a secondary cellular response to the induction of lysosomal proteases such as cathepsins. There is also evidence that cystatin family members contribute to amyloid formation (50). Thus, cystatin F may also be involved in PrPSc replication.
In this study, we identified several S100 calcium binding proteins, including S100A6, S100A4, S100A8, and S100A9, which showed increased expression in scrapie-infected mouse brains. S100 proteins comprise a family of about 20 proteins and are characterized by EF-hand calcium binding motifs displaying different affinities for Ca2+, Zn2+, and Cu2+ ions (25). Recently, S100 proteins have received increased attention due to their close association with several human diseases, including neurodegenerative disorders such as Down syndrome, Alzheimer's disease, and Parkinson's disease (24, 37). For prion diseases, only S100β was reported to be up-regulated in the sera of CJD patients and prion-infected hamsters (3, 41, 42). The S100 proteins have been detected in both neurons and glial cells (18, 52, 55) and are thought to be involved in gliosis and neuronal apoptosis (16, 52). S100 proteins have numerous targets, including annexins and calponins (44). Both annexins and calponins were found to interact with cytoskeletal filaments such as F-actin. In addition, calponin was observed to colocalize with the two major intermediate filaments GFAP and vimentin in astrocytes (19, 44). A recent work demonstrated that the blockage of expression of GFAP and vimentin in astrocytes supports axonal regeneration in the central nervous system of mammals (45), indicating a correlation between the intermediate filaments in astrocytes and the maintenance of neuronal cells. Interestingly, annexin A4, calponin 3, and the calponin-associated proteins GFAP and vimentin were identified in our studies as up-regulated genes in both ME7- and RML-infected mouse brains. These results suggest that changes in the expression of genes for cytoarchitecture may contribute to the neurodegeneration associated with prion disease.
In summary, we identified the most differentially expressed genes in brains of scrapie-infected mice and uninfected animals by comparing their expression levels. We observed a more-than-twofold increase in expression in a total of 121 genes in both ME7- and RML-infected mouse brains. The increased expression of most of these genes was temporally well correlated with the accumulation of PrPSc and the activation of glial cells, the relevant pathological events which precede neuronal cell death. Since about 100 of the 121 differentially up-regulated genes have been described in the context of scrapie for the first time, the present work provides opportunities for the establishment of novel targets for the diagnosis for human or animal prion diseases as well as for the development of pharmacological agents that will be useful for halting or retarding the progression of human prion disease.
Acknowledgments
We thank the Laboratory for Leukemia Diagnostics, led by T. Haferlach (Department of Internal Medicine III, University Hospital Grosshadern, Ludwig-Maximilians-University), for the use of their Affymetrix workstation.
This work was supported by research grant 01GS0166 from the German National Genome Research Network (NGFN) of Bundesministerium für Bildung und Forschung of Germany.
REFERENCES
- 1.Ashburner, M., C. A. Ball, J. A. Blake, D. Botstein, H. Butler, J. M. Cherry, A. P. Davis, K. Dolinski, S. S. Dwight, J. T. Eppig, M. A. Harris, D. P. Hill, L. Issel-Tarver, A. Kasarskis, S. Lewis, J. C. Matese, J. E. Richardson, M. Ringwald, G. M. Rubin, and G. Sherlock. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25:25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, C. A., and L. Manuelidis. 2003. Unique inflammatory RNA profiles of microglia in Creutzfeldt-Jakob disease. Proc. Natl. Acad. Sci. USA 100:675-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beekes, M., M. Otto, J. Wiltfang, E. Bahn, S. Poser, and M. Baier. 1999. Late increase of serum S100 beta protein levels in hamsters after oral or intraperitoneal infection with scrapie. J. Infect. Dis. 180:518-520. [DOI] [PubMed] [Google Scholar]
- 4.Boland, B., and V. Campbell. 2004. A beta-mediated activation of the apoptotic cascade in cultured cortical neurones: a role for cathepsin-L. Neurobiol. Aging 25:83-91. [DOI] [PubMed] [Google Scholar]
- 5.Breen, E. C., and K. Tang. 2003. Calcyclin (S100A6) regulates pulmonary fibroblast proliferation, morphology, and cytoskeletal organization in vitro. J. Cell Biochem. 88:848-854. [DOI] [PubMed] [Google Scholar]
- 6.Bruce, M. E. 1976. Cerebral amyloidosis in scrapie in the mouse: effect of agent strain and mouse genotype. Neuropathol. Appl. Neurobiol. 2:471-478. [Google Scholar]
- 7.Bruce, M. E., I. McConnell, H. Fraser, and A. G. Dickinson. 1991. The disease characteristics of different strains of scrapie in Sinc congenic mouse lines: implications for the nature of the agent and host control of pathogenesis. J. Gen. Virol. 72:595-603. [DOI] [PubMed] [Google Scholar]
- 8.Carlson, G. A., D. T. Kingsbury, P. A. Goodman, S. Coleman, S. T. Marshall, S. DeArmond, D. Westaway, and S. B. Prusiner. 1986. Linkage of prion protein and scrapie incubation time genes. Cell 46:503-511. [DOI] [PubMed] [Google Scholar]
- 9.Chandler, R. L. 1961. Encephalopathy in mice produced by inoculation with scrapie brain material. Lancet i:1378-1379. [DOI] [PubMed] [Google Scholar]
- 10.Chuang, P. I., S. Morefield, C. Y. Liu, S. Chen, J. M. Harlan, and D. M. Willerford. 2002. Perturbation of B-cell development in mice overexpressing the Bcl-2 homolog A1. Blood 99:3350-3359. [DOI] [PubMed] [Google Scholar]
- 11.Combarros, O., A. Alvarez-Arcaya, M. Sanchez-Guerra, J. Infante, and J. Berciano. 2002. Candidate gene association studies in sporadic Alzheimer's disease. Dement. Geriatr. Cogn. Disord. 14:41-54. [DOI] [PubMed] [Google Scholar]
- 12.Dandoy-Dron, F., F. Guillo, L. Benboudjema, J. P. Deslys, C. Lasmezas, D. Dormont, M. G. Tovey, and M. Dron. 1998. Gene expression in scrapie. Cloning of a new scrapie-responsive gene and the identification of increased levels of seven other mRNA transcripts. J. Biol. Chem. 273:7691-7697. [DOI] [PubMed] [Google Scholar]
- 13.Diedrich, J. F., H. Minnigan, R. I. Carp, J. N. Whitaker, R. Race, W. Frey, and A. T. Haase. 1991. Neuropathological changes in scrapie and Alzheimer's disease are associated with increased expression of apolipoprotein E and cathepsin D in astrocytes. J. Virol. 65:4759-4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong, Z., M. Katar, B. E. Linebaugh, B. F. Sloane, and R. S. Berk. 2001. Expression of cathepsins B, D and L in mouse corneas infected with Pseudomonas aeruginosa. Eur. J. Biochem. 268:6408-6416. [DOI] [PubMed] [Google Scholar]
- 15.Duguid, J. R., and M. C. Dinauer. 1990. Library subtraction of in vitro cDNA libraries to identify differentially expressed genes in scrapie infection. Nucleic Acids Res. 18:2789-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fano, G., M. A. Mariggio, P. Angelella, I. Nicoletti, A. Antonica, S. Fulle, and P. Calissano. 1993. The S-100 protein causes an increase of intracellular calcium and death of PC12 cells. Neuroscience 53:919-925. [DOI] [PubMed] [Google Scholar]
- 17.Farnsworth, R. L., and F. Talamantes. 1998. Calcyclin in the mouse decidua: expression and effects on placental lactogen secretion. Biol. Reprod. 59:546-552. [DOI] [PubMed] [Google Scholar]
- 18.Filipek, A., M. Puzianowska, B. Cieslak, and J. Kuznicki. 1993. Calcyclin-Ca(2+)-binding protein homologous to glial S-100 beta is present in neurones. Neuroreport 4:383-386. [DOI] [PubMed] [Google Scholar]
- 19.Gerke, V., and S. E. Moss. 2002. Annexins: from structure to function. Physiol. Rev. 82:331-371. [DOI] [PubMed] [Google Scholar]
- 20.Ghazanfari, F. A., and R. R. Stewart. 2001. Characteristics of endothelial cells derived from the blood-brain barrier and of astrocytes in culture. Brain Res. 890:49-65. [DOI] [PubMed] [Google Scholar]
- 21.Giese, A., D. R. Brown, M. H. Groschup, C. Feldmann, I. Haist, and H. A. Kretzschmar. 1998. Role of microglia in neuronal cell death in prion disease. Brain Pathol. 8:449-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giese, A., M. H. Groschup, B. Hess, and H. A. Kretzschmar. 1995. Neuronal cell death in scrapie-infected mice is due to apoptosis. Brain Pathol. 5:213-221. [DOI] [PubMed] [Google Scholar]
- 23.Giese, A., and H. A. Kretzschmar. 2001. Prion-induced neuronal damage—the mechanisms of neuronal destruction in the subacute spongiform encephalopathies. Curr. Top. Microbiol. Immunol. 253:203-217. [DOI] [PubMed] [Google Scholar]
- 24.Griffin, W. S., L. C. Stanley, C. Ling, L. White, V. MacLeod, L. J. Perrot, C. L. White III, and C. Araoz. 1989. Brain interleukin 1 and S-100 immunoreactivity are elevated in Down syndrome and Alzheimer's disease. Proc. Natl. Acad. Sci. USA 86:7611-7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heizmann, C. W., and J. A. Cox. 1998. New perspectives on S100 proteins: a multi-functional Ca(2+)-, Zn(2+)- and Cu(2+)-binding protein family. Biometals 11:383-397. [DOI] [PubMed] [Google Scholar]
- 26.Hunter, N., W. Goldmann, G. Smith, and J. Hope. 1994. The association of a codon 136 PrP gene variant with the occurrence of natural scrapie. Arch. Virol. 137:171-177. [DOI] [PubMed] [Google Scholar]
- 27.Hur, K., J. I. Kim, S. I. Choi, E. K. Choi, R. I. Carp, and Y. S. Kim. 2002. The pathogenic mechanisms of prion diseases. Mech. Ageing Dev. 123:1637-1647. [DOI] [PubMed] [Google Scholar]
- 28.Ishisaka, R., T. Utsumi, T. Kanno, K. Arita, N. Katunuma, J. Akiyama, and K. Utsumi. 1999. Participation of a cathepsin L-type protease in the activation of caspase-3. Cell Struct. Funct. 24:465-470. [DOI] [PubMed] [Google Scholar]
- 29.Kenward, N., J. Hope, M. Landon, and R. J. Mayer. 1994. Expression of polyubiquitin and heat-shock protein 70 genes increases in the later stages of disease progression in scrapie-infected mouse brain. J. Neurochem. 62:1870-1877. [DOI] [PubMed] [Google Scholar]
- 30.Kim, J. I., W. K. Ju, J. H. Choi, E. Choi, R. I. Carp, H. M. Wisniewski, and Y. S. Kim. 1999. Expression of cytokine genes and increased nuclear factor-kappa B activity in the brains of scrapie-infected mice. Brain Res. Mol. Brain Res. 73:17-27. [DOI] [PubMed] [Google Scholar]
- 31.Kretzschmar, H. A., J. W. Ironside, S. J. DeArmond, and J. Tateishi. 1996. Diagnostic criteria for sporadic Creutzfeldt-Jakob disease. Arch. Neurol. 53:913-920. [DOI] [PubMed] [Google Scholar]
- 32.Kuczius, T., I. Haist, and M. H. Groschup. 1998. Molecular analysis of bovine spongiform encephalopathy and scrapie strain variation. J. Infect. Dis. 178:693-699. [DOI] [PubMed] [Google Scholar]
- 33.Lehnardt, S., L. Massillon, P. Follett, F. E. Jensen, R. Ratan, P. A. Rosenberg, J. J. Volpe, and T. Vartanian. 2003. Activation of innate immunity in the CNS triggers neurodegeneration through a Toll-like receptor 4-dependent pathway. Proc. Natl. Acad. Sci. USA 100:8514-8519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lloyd, S. E., O. N. Onwuazor, J. A. Beck, G. Mallinson, M. Farrall, P. Targonski, J. Collinge, and E. M. Fisher. 2001. Identification of multiple quantitative trait loci linked to prion disease incubation period in mice. Proc. Natl. Acad. Sci. USA 98:6279-6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manolakou, K., J. Beaton, I. McConnell, C. Farquar, J. Manson, N. D. Hastie, M. Bruce, and I. J. Jackson. 2001. Genetic and environmental factors modify bovine spongiform encephalopathy incubation period in mice. Proc. Natl. Acad. Sci. USA 98:7402-7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinon, F., K. Hofmann, and J. Tschopp. 2001. The pyrin domain: a possible member of the death domain-fold family implicated in apoptosis and inflammation. Curr. Biol. 11:R118-R120. [DOI] [PubMed] [Google Scholar]
- 37.Muramatsu, Y., R. Kurosaki, H. Watanabe, M. Michimata, M. Matsubara, Y. Imai, and T. Araki. 2003. Expression of S-100 protein is related to neuronal damage in MPTP-treated mice. Glia 42:307-313. [DOI] [PubMed] [Google Scholar]
- 38.Mutch, D. M., A. Berger, R. Mansourian, A. Rytz, and M. A. Roberts. 2001. Microarray data analysis: a practical approach for selecting differentially expressed genes. Genome Biol. 2:9. [DOI] [PubMed] [Google Scholar]
- 39.Nakanishi, H. 2003. Neuronal and microglial cathepsins in aging and age-related diseases. Ageing Res. Rev. 2:367-381. [DOI] [PubMed] [Google Scholar]
- 40.Ni, J., M. A. Fernandez, L. Danielsson, R. A. Chillakuru, J. Zhang, A. Grubb, J. Su, R. Gentz, and M. Abrahamson. 1998. Cystatin F is a glycosylated human low molecular weight cysteine proteinase inhibitor. J. Biol. Chem. 273:24797-24804. [DOI] [PubMed] [Google Scholar]
- 41.Otto, M., M. Beekes, J. Wiltfang, E. Bahn, S. Poser, and H. Diringer. 1998. Elevated levels of serum S100 beta protein in scrapie hamsters. J. Neurovirol. 4:572-573. [DOI] [PubMed] [Google Scholar]
- 42.Otto, M., J. Wiltfang, E. Schutz, I. Zerr, A. Otto, A. Pfahlberg, O. Gefeller, M. Uhr, A. Giese, T. Weber, H. A. Kretzschmar, and S. Poser. 1998. Diagnosis of Creutzfeldt-Jakob disease by measurement of S100 protein in serum: prospective case-control study. BMJ 316:577-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parchi, P., A. Giese, S. Capellari, P. Brown, W. Schulz-Schaeffer, O. Windl, I. Zerr, H. Budka, N. Kopp, P. Piccardo, S. Poser, A. Rojiani, N. Streichemberger, J. Julien, C. Vital, B. Ghetti, P. Gambetti, and H. Kretzschmar. 1999. Classification of sporadic Creutzfeldt-Jakob disease based on molecular and phenotypic analysis of 300 subjects. Ann. Neurol. 46:224-233. [PubMed] [Google Scholar]
- 44.Plantier, M., A. Fattoum, B. Menn, Y. Ben Ari, T. E. Der, and A. Represa. 1999. Acidic calponin immunoreactivity in postnatal rat brain and cultures: subcellular localization in growth cones, under the plasma membrane and along actin and glial filaments. Eur. J. Neurosci. 11:2801-2812. [DOI] [PubMed] [Google Scholar]
- 45.Privat, A. 2003. Astrocytes as support for axonal regeneration in the central nervous system of mammals. Glia 43:91-93. [DOI] [PubMed] [Google Scholar]
- 46.Prusiner, S. B. 1998. Prions. Proc. Natl. Acad. Sci. USA 95:13363-13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prusiner, S. B., S. P. Cochran, D. F. Groth, D. E. Downey, K. A. Bowman, and H. M. Martinez. 1982. Measurement of the scrapie agent using an incubation time interval assay. Ann. Neurol. 11:353-358. [DOI] [PubMed] [Google Scholar]
- 48.Riemer, C., I. Queck, D. Simon, R. Kurth, and M. Baier. 2000. Identification of upregulated genes in scrapie-infected brain tissue. J. Virol. 74:10245-10248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Somerville, R. A., A. Chong, O. U. Mulqueen, C. R. Birkett, S. C. Wood, and J. Hope. 1997. Biochemical typing of scrapie strains. Nature 386:564. [DOI] [PubMed] [Google Scholar]
- 50.Staniforth, R. A., S. Giannini, L. D. Higgins, M. J. Conroy, A. M. Hounslow, R. Jerala, C. J. Craven, and J. P. Waltho. 2001. Three-dimensional domain swapping in the folded and molten-globule states of cystatins, an amyloid-forming structural superfamily. EMBO J. 20:4774-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thackray, A. M., M. A. Klein, A. Aguzzi, and R. Bujdoso. 2002. Chronic subclinical prion disease induced by low-dose inoculum. J. Virol. 76:2510-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tiu, S. C., W. Y. Chan, C. W. Heizmann, B. W. Schafer, S. Y. Shu, and D. T. Yew. 2000. Differential expression of S100B and S100A6(1) in the human fetal and aged cerebral cortex. Brain Res. Dev. Brain Res. 119:159-168. [DOI] [PubMed] [Google Scholar]
- 53.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Windl, O., M. Dempster, J. P. Estibeiro, R. Lathe, R. de Silva, T. Esmonde, R. Will, A. Springbett, T. A. Campbell, K. C. Sidle, M. S. Palmer, and J. Collinge. 1996. Genetic basis of Creutzfeldt-Jakob disease in the United Kingdom: a systematic analysis of predisposing mutations and allelic variation in the PRNP gene. Hum. Genet. 98:259-264. [DOI] [PubMed] [Google Scholar]
- 55.Yamashita, N., K. Kosaka, E. C. Ilg, B. W. Schafer, C. W. Heizmann, and T. Kosaka. 1997. Selective association of S100A6 (calcyclin)-immunoreactive astrocytes with the tangential migration pathway of subventricular zone cells in the rat. Brain Res. 778:388-392. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, Y., E. Spiess, M. H. Groschup, and A. Burkle. 2003. Up-regulation of cathepsin B and cathepsin L activities in scrapie-infected mouse Neuro2a cells. J. Gen. Virol. 84:2279-2283. [DOI] [PubMed] [Google Scholar]