Abstract
Dendritic cells (DCs) link innate and adaptive immunity by sensing pathogens or vaccinogens and signaling a variety of defense responses. Since human papillomavirus type 16 L1 virus-like particles (HPV16 VLPs) induce a potent, protective immune response after vaccination, we examined their recognition by DCs. HPV16 VLPs cause phenotypic maturation of murine bone marrow-derived DCs (BMDCs), and immunization of mice with HPV16 VLP-loaded BMDCs or HPV16 VLPs alone induced T helper 1 (Th1)-biased immune responses. Analysis of transcriptional responses of murine BMDCs by microarray suggested that alpha/beta interferon (IFN-α/β) transcripts and numerous proinflammatory cytokines and chemokines are up regulated in response to HPV16 VLPs. Indeed, the induction of IFN-α, IFN-γ, and interleukin-12 (IL-12) production by BMDCs after stimulation with HPV16 VLPs was demonstrated by quantitative enzyme-linked immunosorbent assay. Many microbial products that induce proinflammatory responses are recognized via Toll-like receptor (TLR) signaling through the key adaptor protein MyD88 and activation of NF-κB, nuclear factor of activated T cells (NF-AT), and activating protein 1 (AP-1). Reporter assays indicated that HPV16 VLPs activated NF-κB-, NF-AT-, and AP-1-dependent transcription in the RAW264.7 macrophage cell line. Knockdown of MyD88 transcripts by small interfering RNA in the RAW264.7 macrophage cell line inhibited the activation of NF-κB-, NF-AT- and AP-1-dependent transcription by HPV16 VLP. Furthermore, MyD88−/− BMDCs failed to up regulate IL-12 and IFN-α and -γ in response to HPV16 VLPs. Finally, Th1-biased immune responses to HPV16 VLPs are dramatically impaired in MyD88 and IFN-α/β receptor-deficient mice. This implicates TLR recognition as central to immune recognition of HPV16 L1 VLPs.
Antigen-presenting cells, and especially dendritic cells (DCs), employ pattern recognition receptors including the Toll family to sense infections (41). In response to a pathogen-associated molecular pattern, Toll family members signal via an adaptor molecule, MyD88, activating NF-κB and activating protein 1 (AP-1) transcription factors and innate, proinflammatory responses (3). Importantly, these innate responses are largely responsible for shaping adaptive immune responses to a pathogen or vaccine antigen. The adaptive immune system contains a balanced repertoire of T helper 1 (Th1) and Th2 CD4+ cells. Th1 cells promote inflammatory responses through the secretion of cytokines such as gamma interferon (IFN-γ) that activate macrophages and provide help to CD8+ cytotoxic T cells. In contrast, Th2 cells provide help to antibody-producing B cells and secrete certain cytokines, such as interleukin 10 (IL-10), that antagonize the Th1 response.
Oncogenic human papillomavirus (HPV), notably the prototypic genotype 16 (HPV16), represents the primary causative agent of cervical cancer (44). The major papillomavirus capsid protein, L1, self assembles to form empty capsids termed virus-like particles (VLPs) that are morphologically and immunologically very similar to native virions but lack the oncogenic viral genome (26). Vaccination with L1 VLPs protects against papillomavirus infection (6, 28, 39).
HPV16 VLPs exhibit a highly ordered, closely packed foreign structure (4) and engender a potent Th1 immune response in humans, inducing both high-titer neutralizing antibody and cell-mediated immune responses (16, 21). A single L1 VLP vaccination without adjuvant affords mice CD8+ T-cell-dependent protection against tumor formation by the HPV16 genome-transformed cell line C3 (15). The immunogenicity of other peptide or whole-protein antigens is dramatically enhanced upon fusion with VLPs (19, 24). Indeed, vaccination with chimeric VLPs elicits a potent cytotoxic T lymphocyte response against a self antigen (33) or an otherwise weak antigen (37) and protects against tumor challenge in preclinical models (19). Notably, presentation of self antigens at high density on the surfaces of VLPs is sufficient to overcome B-cell tolerance (10-12).
Recent studies demonstrate that VLPs bind to DCs and stimulate their maturation, including the up regulation of major histocompatibility complex class I and II, CD80, CD86, and CD40 costimulatory molecules and cytokine production (30, 31). Here, we demonstrate that VLPs effectively activate bone marrow-derived DCs (BMDCs) to produce a range of responses, notably expression of IFN-α and -γ and IL-12. These immune mediators favor the differentiation of Th1 cells, consistent with the responses of mice immunized with VLP-loaded DCs. We show that production of IFN-α, IFN-γ, and IL-12 and Th1-biased immune responses to HPV16 VLPs are dependent upon MyD88 (3), consistent with a role for the Toll/IL-1 receptor family in immune recognition of HPV16 VLPs.
MATERIALS AND METHODS
Mice.
Six- to 8-week-old male 129/C57BL/6 Myd88+/+, 129/C57BL/6 Myd88−/−, 129-IFNα/βR−/−, 129-IFNα/βR+/+, and C57BL/6 mice (B&K Universal) were maintained in a pathogen-free animal facility at least 1 week before use. C57BL/6 mice were immunized intravenously with 5 × 105 murine DCs preloaded with HPV16 VLPs (10 μg/ml) or with HPV16 VLPs alone (10 μg/ml) at 0, 7, and 14 days. Ten days after the final inoculation, spleen cells and sera were harvested and stored at −80°C. Experiments were done in accordance with institutional guidelines.
Cytokines, antibodies, and reagents.
Murine recombinant granulocyte-macrophage colony-stimulating factor (mrGM-CSF) and lipopolysaccharide (LPS) (Escherichia coli serotype 026:B6) were purchased from R&D Systems (Minneapolis, Minn.) and Sigma-Aldrich, respectively. Poly(I:C) and mouse CpG (ODN1826) oligonucleotides were purchased from InvivoGen. Fluorescein isothiocyanate (FITC)- and phycoerythrin (PE)-labeled goat anti-mouse immunoglobulin G (IgG) were purchased from Jackson ImmunoResearch, and anti-IFN-γ antibody (clone XMG1.2) was purchased from PharMingen. CD4+ splenic T cells were purified with magnetic cell sorting (MACS) CD4 (L3T4) microbeads. A total of 106 CD4+ T cells per well of a six-well plate were stimulated with 105 phosphate-buffered saline (PBS)-treated or VLP-loaded BMDCs. The supernatants were harvested after 24 h and analyzed with the mouse Th1/Th2 cytokine cytometric bead array kit (Becton Dickinson).
Preparation of papillomavirus VLPs.
HPV16 L1 VLPs were generated by infection of Sf9 cells with recombinant baculoviruses and purified as previously reported (46). Briefly, recombinant virus was propagated in Sf9 insect cells (ATCC CRL 1711; American Type Culture Collection, Manassas, Va.). Sf9 cells were cultured in SF-900 II serum-free medium supplemented with 10% inactivated fetal bovine serum, 10 U of penicillin G/ml, and 10 μg of streptomycin/ml. For VLP preparation, Hi 5 cells were adjusted to 3 × 106 cells/ml and infected with recombinant baculovirus. After 72 h at 26.6°C, cells were harvested and washed once in ice-cold PBS. The pelleted cells were lysed by short-pulsed sonification on ice (Fisher Sonic Dismembrator). The lysates were loaded on top of 40% (wt/vol) sucrose (ICN Biomedicals, Aurora, Ohio)-PBS cushions and centrifuged at 25,000 rpm for 2.5 h (SW28 centrifuge; Beckman, Fullerton, Calif.). The resulting pellet was suspended in 2 ml of 27% (wt/wt) cesium chloride (CsCl; Life Technologies)-PBS and sonicated a second time. Cell lysates were subjected to two subsequent centrifugations to equilibrium in 27% (wt/wt) CsCl (20 h at 28,000 rpm and 15°C). After centrifugation, VLPs were purified again with an A-15 mGel (Bio-Rad, Richmond, Calif.) and a strong cation-exchange packing column (PerSeptive Biosystems, Framingham, Mass.). Fractions were harvested and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue and silver staining. VLPs were suspended in PBS, and the protein content was evaluated with a microBCA kit (Pierce). To investigate the possible effects of contaminating Hi 5 cell proteins in VLP preparations, a negative control was prepared by processing uninfected Hi 5 cells as for VLP purification. To determine the efficiency of VLP assembly, transmission electron microscopy was performed by absorbing particles onto carbon-coated grids and negative staining with 1% uranyl acetate. The grids were examined with a Philips CM120 transmission electron microscope operating at 80 kV.
Preparation of HPV16 L1 in E. coli.
Mutant L1 genes were inserted into pGEX-6P-2 E. coli strain BL21 cells transformed with a pGEX plasmid (pGEX16L1) were grown at room temperature in Luria-Bertani medium containing 100 μg of ampicillin/ml. At an optical density at 600 nm of 0.3, recombinant protein expression was induced by adding 0.25 mM isopropyl-β-d-thiogalactoside (IPTG) to the 0.5-liter 2YT culture at 30°C (13). Soluble glutathione S-transferase-L1 fusion proteins were purified on a 1-ml glutathione-Sepharose Fast Flow column according to the manufacturer's instructions. The column was incubated overnight at 4°C with PreScission protease to release L1 from the column. The protein content was evaluated using a microBCA kit (Pierce) and SDS-PAGE. Samples were absorbed onto carbon-coated grids and stained with 1% uranyl acetate. The grids were examined with a Philips CM120 transmission electron microscope operating at 80 kV.
Preparation and flow cytometric analysis of BMDCs.
BMDCs were prepared from bone marrow cells that were collected by removing the femur bones of mice, cutting off the ends of the bones, and flushing out the bone marrow with RPMI 1640 medium injected with a syringe. The pooled cells were harvested by centrifugation at 1,600 rpm for 10 min and resuspended in 2 ml of ammonium chloride lysing buffer (Biofluids, Biosource International) for 5 min at room temperature to lyse the red blood cells. These cells were washed in RPMI 1640 medium and cultured in RPMI 1640 medium containing 500 U of mrGM-CSF/ml for 6 days prior to analysis. BMDCs were collected in ice-cold PBS, and surface markers were immunolabeled with the following antibodies: FITC-conjugated anti-CD11C (N418), anti-mouse CD86 (GL1), anti-mouse CD80 (16-10A1), anti-mouse CD40 (3/23), PE-labeled anti-CD4 (L3T4), anti-mouse CD8α (53-6.6), anti-mouse CD45 R/B220 (RA36B2), anti-mouse CD11b (M1/70), purified anti-mouse I-Ab (25-9-17), and purified anti-mouse CD3 molecular complex (17A2). All of these antibodies were purchased from PharMingen. Single or double staining was performed with different monoclonal antibodies (MAbs). Cells were then washed twice before being resuspended in PBS containing 1% paraformadehyde and 1% fetal calf serum and kept at 4°C prior to flow cytometric analysis (FAScan; Becton Dickinson). For each analysis, isotype-matched control MAb was used a negative control.
Expression profiling with DNA microarrays.
Total RNA was isolated from DCs with TRIzol reagent (Invitrogen), followed by RNA cleanup with the RNeasy minikit (QIAGEN). The processing of the sample was done according to Affymetrix specifications. Briefly, 5 μg of total RNA was used to synthesize first-strand cDNA with oligonucleotide probes with 24 oligo(dT) plus T7 promoter as a primer (Proligo LLC) and the SuperScript Choice system (Invitrogen). Following the double-stranded cDNA synthesis, the product was purified by phenol-chloroform extraction, and biotinylated anti-sense cRNA was generated through in vitro transcription with the BioArray RNA High Yield transcript labeling kit (ENZO Life Sciences, Inc.). Fifteen micrograms of the biotinylated cRNA was fragmented at 94°C for 35 min (100 mM Tris-acetate [pH 8.2], 500 mM potassium acetate, 150 mM magnesium acetate), and 10 μg of total fragmented cRNA was hybridized to the Affymetrix murine genome U74Av2 GeneChip array, on different days, for 16 h at 45°C with constant rotation (60 rpm). The Affymetrix Fluidics Station 400 was then used to wash and stain the chips, removing the nonhybridized target and incubating with a streptavidin-PE conjugate to stain the biotinylated cRNA. The staining was then amplified with goat IgG as a blocking reagent and biotinylated goat anti-streptavidin antibody, followed by a second staining step with a streptavidin-PE conjugate. Fluorescence was detected with the Hewlett-Packard G2500 GeneArray scanner, and image analysis of each GeneChip sample was done with the Micro Array Suite 5.0 software from Affymetrix, using standard default settings. For comparison between different chips, global scaling was used, scaling all probe sets to a user-defined target intensity of 150. To ascertain the quality control of the total RNA from the samples, we used the Agilent Bioanalyzer Lab with GeneChip technology and confirmed that all the samples had optimal rRNA ratios (1:2 ratio of 18S to 28S) and clean run patterns. Likewise, this technology was used to confirm the quality of the RNA in the form of cRNA and fragmented cRNA. To assess the quality control of the hybridization, GeneChip image, and comparison between chips, we studied the following parameters: scaling factor, background, percentage of present calls, housekeeping genes (3′/5′ ratios of GAPDH), and presence or absence of internal spike controls. To assess quality control interreplicates, we observed the percentages of differential calls (up or down regulated) between pairwise comparisons. The initial expression results were based on pairwise comparisons between the different experimental conditions represented by the samples. Any transcript that showed at least a twofold change in expression level between experimental sample and control sample was considered significant. For duplicated samples, the results were filtered independently for significance on each of the four pairwise comparisons. Transcripts that were consistently significant in at least two of four iterative comparisons were selected for the final candidate list.
Transfection and reporter assays.
Mouse NF-κB, nuclear factor of activated T cells (NF-AT), and activating protein 1 (AP-1) promoter reporter plasmids were purchased from Clontech. RAW264.7 cells were transfected with 4 μg of plasmids pNF-κB SEAP, pNF-AT SEAP, pAP-1 SEAP, positive-control pSEAP2, and negative-control pTAL-SEAP with Lipofectamine 2000 (Invitrogen) in 24-well plates. The cells were stimulated 24 h later by adding 25 μg of VLPs/ml or buffer alone. The supernatants were collected at 12 h. Alkaline phosphatase assays were performed according to the manufacturer's instructions (Clontech). To generate the MyD88 knockdown in RAW264.7cells, small interfering RNA (siRNA) target sequences in MyD88 were designed according to a siRNA target finder and design tool program (Ambion) and previously reported methods (7). The MyD88 target sequences used were as follows: KD1, 5′ GATCCCCGTGGGAGTGAGGCGCCGCTTCAAGAGAGCGGCGCCTCACTCCCACGTTTTTGGAAA and 5′AGCTTTTCCAAAAACGTGGGAGTGAGGCGCCGCTCTCTTGAAGCGGCGCCTCACTCCCACGGGG; KD2, 5′GATCCCCGGAGCTGAAGTCGCGCATCTTCAAGAGAGATGCGCGACTTCAGCTCCTTTTTGGAAA and 5′AGCTTTTCCAAAAAGGAGCTGAAGTCGCGCATCTCTCTTGAAGATGCGCGACTTCAGCTCCGGG. The TLR9 target sequence was generated with the sequences 5′GATCCCCGGACTCTGCACCCCTTGTCTTCAAGAGAGACAAGGGGTGCAGAGTCCTTTTTGGAAA and 5′AGCTTTTCCAAAAAGGACTCTGCACCCCTTGTCTCTCTTGAAGACAAGGGGTGCAGAGTCCGGG. After hybridization, the oligonucleotides were inserted into the pSUPER vector (kindly provided by R. Agami, NKI, Amsterdam, The Netherlands, and modified to carry hygromycin B resistance) at the Bg1II and HindIII sites and then transfected into macrophage cell line RAW264. 7. After selection in hygromycin B, MyD88 mRNA expression in cell lines was tested by reverse transcription-PCR (RT-PCR). Total cellular RNA was prepared with TRIzol (GIBCO BRL) according to the manufacturer's instructions. To eliminate possible DNA contamination, total RNA preparations were incubated for 15 min at 37°C with 10 U of RNase-free DNase (Boehringer Mannheim). RT-PCR was performed by SuperScript one-step RT-PCR with Platinum Taq according to the protocol provided (Invitrogen). Amplification conditions were as follows: for cDNA synthesis and predenaturation, 1 cycle at 50°C for 15 min and 94°C for 2 min; for PCR amplification, 40 cycles (each cycle included a denaturing step at 94°C for 15 s, an annealing step at 55°C for 30 s, and an extension step at 72°C for 1 min/kb). A final extension consisted of 1 cycle at 72°C for 10 min. The primers used were as follows: for GAPDH, 5′ATGGTGAAGGTCGGTGTGAACGGATTTGGC and 5′CATCGAAGGTGGAAGAGTGGGAGTTGCTGT; for MyD88, 5′ATGTCTGCGGGAGACCCCCGCGTG and 5′ TCAGGGCAGGGACAAAGCCTTGG; for TLR9, 5′ATGGTTCTCCGTCGAAGGACTCTGCAC and 5′CTATTCTGCTGTAGGTCCCCGGCAG.
Analysis of IFN-α, IFN-γ, and IL-12 by ELISA.
The culture supernatants of different subsets of DCs were harvested at various times after stimulation by VLPs (25 μg/ml), buffer alone, LPS (0.1 μg/ml), CpG (5 μM), or Poly(I:C) (25 μg/ml). Commercial sandwich enzyme-linked immunosorbent assay (ELISA) kits were used for quantitation of IFN-γ (Pierce Endogen), IL-12p70 (R&D Systems), and IFN-α (PBL Biomedical Laboratories). The optical density at 450 nm of each sample was measured with a SpectraMax 190 ELISA plate reader. Cytokine levels were quantified from two to three titrations with standard curves and expressed as the number of picograms per milliliter.
Indirect immunofluorescence and confocal microscopy.
DCs were incubated with VLPs for 1 h at 4°C in Dulbecco's PBS and then washed with medium at 37°C. At the time points indicated, the cells were washed with PBS, fixed with 3.7% formaldehyde solution for 10 min, permeabilized with 0.1% (vol/vol) Triton X-100 in PBS for 5 min, and blocked with PBS containing 1% bovine serum albumin for 30 min. MAb H16.V5 was used at a 1:100 dilution for detection of HPV16 L1, and FITC-conjugated goat anti-mouse IgG (Sigma) was added sequentially at a concentration of 5 μg/ml for 20 min at 4°C. Actin was stained with rhodamine-phalloidin (Molecular Probes). Samples were examined by confocal fluorescence microscopy (UltraView confocal imaging system; Perkin-Elmer) (46). The cells were viewed on a Nikon Eclipse TE200 inverted microscope equipped with a ×40 plan fluor or ×60 or ×100 plan apochromatic objective lens with corresponding 1-, 0.8-, or 0.45-μm optical z-slices. Twelve-bit images were merged and analyzed with Ultraview acquisition software in RGB mode.
RESULTS
HPV16 VLPs activate NF-AT-, NF-κB-, and AP-1-dependent transcription.
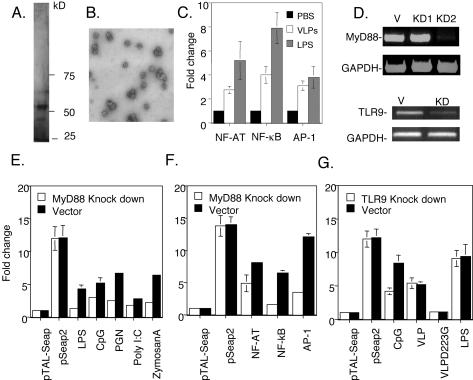
The mouse macrophage cell line RAW264.7 represents a useful model to study innate responses such as transcription factor activation and, unlike DCs, may be readily transduced with reporter plasmids (47). The NF-κB, NF-AT, and AP-1 transcription factors are involved in the transcriptional activation of many innate response genes by microbial products (41). Therefore, we employed the reporter constructs pNF-κBSEAP, pAT-1SEAP, and pNF-AT SEAP that express secreted alkaline phosphatase (SEAP) to determine whether the HPV16 VLPs activate NF-κB-, AP-1-, or NF-AT-dependent transcription, respectively (Fig. 1). The pSEAP2 and pTAL-SEAP plasmids were used as positive and negative controls, respectively. HPV16 VLPs were generated by infection of Hi 5 cells with recombinant baculovirus. The purity of HPV16 VLP preparations was assessed by SDS-PAGE, and the gels were developed with silver staining (Fig. 1A). The predominant band corresponds to HPV16 L1, which has a molecular weight of ∼50 kDa (27). Western blot analysis suggests that the minor bands represent glycosylated forms and degradation products of L1 (32, 49). To ensure that the HPV16 L1 had assembled into VLPs, the preparations were examined by transmission electron microscopy (Fig. 1B). As expected, the preparation contained empty particles ∼55 nm in diameter (27). RAW264.7 cells were transfected with each plasmid; after 18 h, the cells were stimulated by VLPs. The level of SEAP in the supernatant was assayed as a surrogate measure of promoter activity. The reporter constructs indicated that HPV16 VLP up regulated NF-κB, NF-AT, and AP-1 transcription factor-mediated activity (Fig. 1C). Additional steps were included in the purification scheme to ensure homogeneity of the HPV16 VLPs (Fig. 1A) and to ensure that the responses to the VLP preparations were not due to impurities, notably endotoxin. The endotoxin level in the VLP preparations was <0.058 endotoxin units/ml in our 1.05-mg/ml stock of HPV16 L1 VLPs, as determined by a Limulus assay (E-Toxate; Sigma). Thus, the data suggest that VLPs activate multiple key transcription factor pathways, although less efficiently than does LPS.
FIG. 1.
Activation of transcription factors in the RAW264.7 macrophage cell line by HPV16 VLPs. (A) SDS-PAGE analysis of HPV16 L1 VLP preparation from baculovirus-infected Sf9 cells visualized with silver stain. (B) A transmission electron micrograph of an HPV16 L1 VLP preparation. (C) pNF-κB-SEAP, pNF-AT SEAP, pAP-1 SEAP, negative-control pTAL-SEAP, and positive-control pSEAP2 DNAs were each transfected into macrophage cell line RAW264.7. After 24 h, the cells were stimulated with 25 μg of VLPs/ml. The supernatants were collected 12 h later and analyzed with a chemiluminescence-based SEAP assay. Fold change in chemiluminescent intensity compared to unstimulated cells is plotted. (D) RAW264.7 cells were stably transfected with pSUPER alone (V) or expressing siRNAs KD1 and KD2 that target MyD88 transcripts (top panels) or KD that targets TLR9 (bottom panels). Expression of MyD88, TLR9, and GAPDH mRNA was analyzed in these cell lines by RT-PCR. (E) Validation of the MyD88 knockdown phenotype. The pSUPER- and pSUPER-KD2-transfected RAW264.7 cell lines were transfected with pNF-κB-SEAP; after 24 h, the cells were stimulated with 0.1 μg of LPS (026:B6)/ml, 10 μg of PGN (peptidoglycan from S. aureus)/ml, 25 μg of poly(I:C)/ml, 5 μM CpG (ODN 1826), or 25 μg of zymosan A/ml. The supernatants were collected 12 h later and analyzed by a chemiluminescence-based SEAP assay. Fold change in chemiluminescent intensity compared to unstimulated cells is plotted. (F) VLP-induced activation of NF-κB-, NF-AT-, and AP-1-regulated transcription is reduced in the MyD88 knockdown macrophage cell line. The vector and MyD88 knockdown KD2-transfected RAW264.7 cells were transfected by pNF-κB-SEAP, pNF-AT SEAP, or pAP-1 SEAP. After 24 h, the cells were stimulated with 25 μg of VLPs/ml. The supernatants were collected 12 h later and analyzed as described above. (G) RAW264.7 cells were stably transfected with pSUPER alone or with expressing siRNA that targets TLR9 transcripts. The pSUPER- and pSUPER-TLR9 KD-transfected RAW264.7 cell lines were transfected with pTAL-SEAP (negative control) or pSEAP2 (positive control). The remaining samples were transfected with pNF-κB-SEAP and were then stimulated with 5 μM CpG (ODN 1826), 0.1 μg of LPS (026:B6)/ml, 25 μg of in vitro-assembled and polymyxin B-treated HPV16 VLPs/ml, or 25 μg of assembly-deficient HPV16 L1 D223G/ml (after polymyxin B treatment). The supernatants were collected 12 h later and analyzed by a chemiluminescence-based SEAP assay. Fold change in chemiluminescent intensity compared to unstimulated cells is plotted.
MyD88 knockdown in RAW264.7 cells reduces transcription factor activation by HPV16 VLPs.
Upon recognition of the characteristic components of pathogens, the Toll family of receptors employs the adaptor protein MyD88 to initiate innate responses via activation of NF-κB, AP-1, and other transcription factors (3). Therefore, we examined whether MyD88 mediates HPV16 VLP activation of NF-κB, NF-AT, and AP-1 in RAW264.7 cells. MyD88 transcripts in stably transfected RAW264.7 cell lines were targeted with two different siRNA constructs expressed from a polymerase III H1-RNA gene promoter in the pSUPER vector (7). The efficacy of MyD88 transcript knockdown by the stably transfected siRNA constructs was assessed by RT-PCR. While no change in MyD88 transcript levels in RAW264.7 cells transfected with the pSUPER vector alone or the KD1 construct was noted, the KD2 construct drastically reduced levels of MyD88, but not GAPDH, mRNA (Fig. 1D). To validate the KD2 knockdown, the cell line was stimulated with LPS (serotype 026:B6), peptidoglycan from Staphylococcus aureus, poly(I:C), CpG oligonucleotide, or zymosan A, all of which are dependent upon MyD88 to activate the NF-κB transcription factor (41). Activation of the NF-κB transcription factor pathway by each agent was significantly impaired in the MyD88 knockdown cell line KD2, providing functional evidence of a reduction in MyD88 expression (Fig. 1E). The ability of HPV16 VLP to activate NF-AT-, NF-κB-, and AP-1-dependent transcription was then tested with RAW264.7 cells transfected with vector alone or KD2. In the KD2 MyD88 knockdown line, NF-κB- and AP-1-dependent transcription was significantly impaired compared with that of the vector control, while the effect upon the NF-AT pathway was more modest (Fig. 1F).
It was possible that contaminating unmethylated insect cell DNA was responsible for the MyD88-dependent activation of NF-κB-dependent transcription by the HPV16 VLP preparation. To address this possibility, we knocked down transcripts of the Toll-like receptor (TLR) that recognizes unmethylated DNA, TLR9 (Fig. 1D). To further rule out activation mediated by contaminating baculovirus or endotoxin, we affinity purified HPV16 L1 from E. coli (13, 14). After in vitro assembly of the HPV16 L1 and verification by transmission electron microscopy, the preparations were depleted of endotoxin contamination with polymyxin B (34). When compared to vector-transfected RAW264.7 cells, RAW264.7 cells stably transfected with the TLR9 knockdown construct showed reduced NF-κB activation by CpG, but by in vitro-assembled and polymyxin B-treated HPV16 L1 VLPs or LPS that is recognized via TLR4 (Fig. 1G). This indicates that HPV16 VLPs are responsible for the MyD88-dependent activation of NF-κB, rather than unmethylated insect cell or baculoviral DNA. Furthermore, an assembly-deficient mutant of HPV16 L1, D223G, failed to activate NF-κB in control RAW264.7 cells, indicating that the correct capsid structure is required. Similar results with a different assembly-deficient HPV16 L1 mutant have been reported (30).
VLP-loaded BMDCs induce Th1-biased responses in naïve C57BL/6 mice.
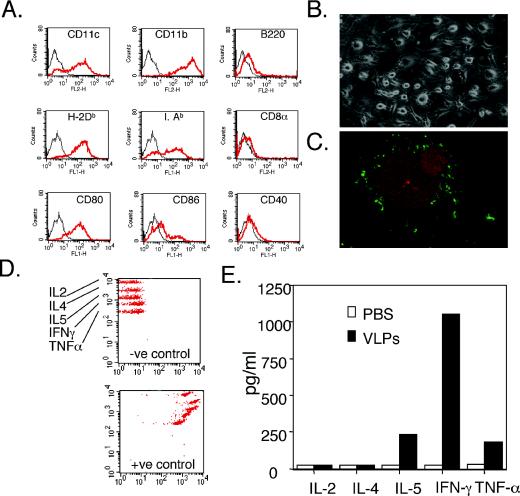
To further investigate the role of DCs in the adaptive immune response to HPV16 VLPs, mice were immunized with VLP-loaded BMDCs, and the nature of the immune response was determined. BMDCs were prepared by culture of bone marrow cells in RPMI 1640 medium containing 500 U of mrGM-CSF/ml for 6 days. Expression of surface markers characteristic of BMDCs was confirmed by flow cytometry (Fig. 2A), and the BMDCs exhibited typical morphology (Fig. 2B). BMDCs were loaded with HPV VLPs for 1 hour in PBS at 4°C. Binding of the HPV VLPs to BMDCs was visualized by immunofluorescent staining (Fig. 2C) (46). Mice were immunized intravenously with 5 × 105 murine DCs preloaded with HPV16 VLPs (10 μg/ml) at 0, 7, and 14 days. Ten days after the final inoculation, the splenic CD4+ T cells derived from these immunized mice were stimulated in vitro, and cytokines that were released into the supernatant were assayed. As shown in Fig. 2D and E, HPV16 VLPs clearly induced the generation of IFN-γ and tumor necrosis factor alpha (TNF-α) in spleen CD4+ T cells of mice immunized with HPV16 VLP-loaded BMDCs. While the Th2 cytokine IL-5 was also induced, the high level of IFN-γ and TNF-α responses, as well as an absence of IL-4 production (Fig. 2E), are consistent with Th1 bias and responses observed with VLP-vaccinated patients (14). The spleen cells from mice vaccinated with PBS-treated DCs did not induce similar Th1 responses.
FIG. 2.
Vaccination with VLP-loaded BMDCs induces Th1 responses. Surface marker (red line) and isotype (black line) control staining (A) and morphology (B) of murine BMDCs cultured in RPMI 1640 medium with 500 U of mrGM-CSF/ml for 6 days. (C) HPV16 VLPs bound to BMDCs viewed by confocal fluorescence microscopy. Green coloring represents H16.V5-stained VLPs; red coloring indicates tetramethyl rhodamine isocyanate-phalloidin-stained actin. (D and E) CD4+ T cells were isolated by positive selection with CD4+ microbeads (MACS; Mitenyi Biotec GmbH) from splenocytes of mice immunized three times with HPV16 VLP-loaded BMDCs. Levels of Th1 and Th2 cytokines, IL-2, IL-4, IL-5, IFN-γ, and TNF-α in supernatants of mouse splenic CD4+ T cells were assessed 24 h after stimulation with VLP-loaded BMDC (10:1 ratio) was detected with a cytokine bead array. Positive (+ve) and negative (−ve) controls for a cytokine bead array incubated with 5,000 pg/ml each of IL-2, IL-4, IL-5, IFN-γ, TNF-α, or buffer alone are shown in panel D.
Time course of VLP-induced transcriptional changes in mouse BMDCs.
To address the mechanism driving Th1-biased immune responses, we analyzed the transcriptional response of BMDCs to HPV16 VLPs in vitro. Lenz et al. described the delay in HPV16 VLP-mediated maturation of BMDCs compared to LPS-induced responses (30). Therefore, we initially performed a time course study to examine transcriptional changes in BMDCs at 6, 24, and 48 h after exposure to HPV16 VLPs or PBS (Fig. 3). RNA was prepared from cells at each time point for transcriptional profiling with Affymetrix U74A oligonucleotide microarrays. Six hours after exposure to HPV16 VLPs, transient up regulation of IFNα/β transcripts, notably those of IFN-α4, -α1-9, and -β, and delayed, persistent up regulation of the several IFN-induced transcripts were observed (e.g., IFN-induced protein 10 [IP-10/CXCL10], IP with tetratricopeptide repeats 1 and 2, and IFN-activated genes 203 and 204). A number of other cytokine and/or chemokine transcripts were also up regulated in BMDCs by HPV16 VLPs, including IL-1β, IL-6, IL-10, IL-18, IP-10, RANTES, macrophage inflammatory protein 1β (MIP-1β), monocyte chemotactic protein 3 (MCP-3), MCP-5, and granulocyte colony-stimulating factor (G-CSF). Using the Bio-Plex 18 cytokine assay (Bio-Rad), we demonstrated the production of IL-1β, IL-6, IL-10, IL-12, IFN-γ, RANTES, MIP-1β, and G-CSF in the supernatant of BMDC 14 h after stimulation with HPV16 VLP (data not shown), thus validating the microarray analysis. Furthermore, a cell lysate of insect cells processed by the methods applied during the purification of VLPs failed to up regulate CD40, CD80, and CD86 on BMDCs, as assessed by flow cytometric staining (data not shown), suggesting that an insect cell contaminant in the VLP preparations is unlikely to be the cause of DC activation (data not shown). By 6 h after exposure to HPV16 VLPs, therefore, BMDCs have up regulated the transcription of numerous defense response genes.
FIG. 3.
Time course of transcriptional changes in BMDCs upon stimulation with HPV16 VLPs. RNA was prepared from BMDCs 6, 24, and 48 h after exposure to HPV16 VLPs or PBS. RNA was analyzed by Affymetric microarray with the U74A chip. Data were curated to show genes exhibiting a greater-than-twofold transcriptional changes and sorted by ontogeny related to immune responses. Transcriptional changes of two- to fivefold or greater-than-fivefold up regulation are coded orange and red, respectively, and down regulations of two- to fivefold and greater-than-fivefold are shaded light green and dark green, respectively. NS, no significant change, i.e., less-than-twofold transcriptional change.
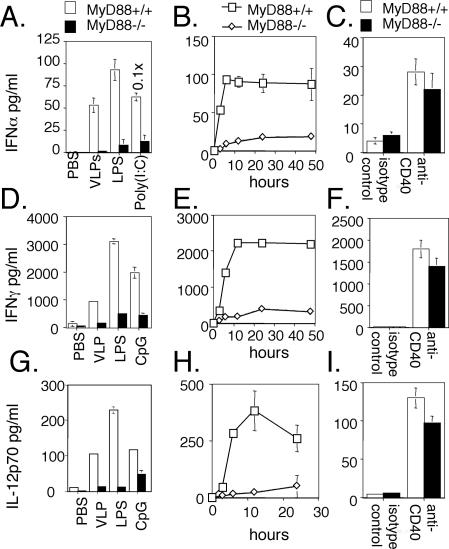
Induction of IFN-α, IFN-γ, and IL-12 by VLPs is dependent upon MyD88.
Knockdown studies implicated MyD88 in signaling innate responses to HPV16 VLPs (Fig. 1F). To further address the role of MyD88 in the development of an immune response to HPV16 VLPs in DCs, BMDCs from MyD88 knockout or wild-type control mice were exposed to HPV16 VLPs or (as positive controls) to LPS or CpG (1). Exposure to HPV16 VLPs, LPS, and CpG induced the production of IL-12, IFN-α, and IFN-γ by BMDCs generated from wild-type mice (Fig. 4). Levels of IL-12 and IFN-γ released by DCs peaked by about 12 h after stimulation with HPV16 VLPs, whereas IFN-α peaked earlier at 6 h. Interestingly, production of IL-12, IFN-α, and IFN-γ in response to HPV16 VLPs was dramatically lower in the absence of MyD88, consistent with the innate recognition of HPV16 VLPs via the TLR family.
FIG. 4.
MyD88-dependent up regulation of IFN-α, IFN-γ, and IL-12 by HPV16 VLPs. BMDCs generated from wild-type or MyD88 knockout mice were stimulated by VLPs (25 μg/ml), LPS (0.1 μg/ml), CpG (5 μM), poly(I:C) (25 μg/ml), or anti-CD40 or its isotype control (10 μg/ml). Supernatants were harvested at 12 h (A, C, D, F, G, and I) or at the times indicated (B, E, and H) after stimulation with HPV16 L1 VLPs. Levels of IFN-α (A to C), IFN-γ (D to F), and IL-12p70 (G to I) were quantified by ELISA.
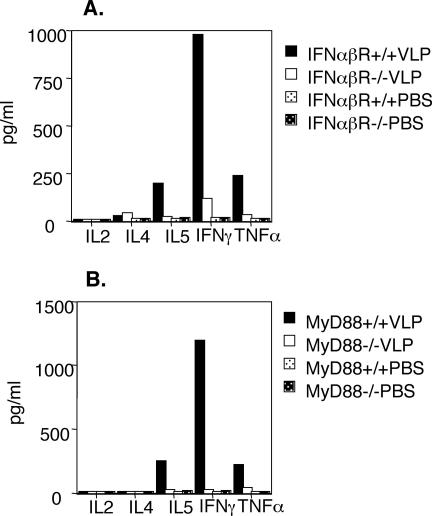
Impaired Th1-biased responses to HPV16 L1 VLPs in IFN-α/β receptor- and MyD88-deficient mice.
To investigate the contribution of IFN-α/β and MyD88 signaling to the adaptive immune response to VLPs, IFN-α/β receptor-deficient mice or MyD88-deficient mice were immunized with HPV16 VLPs. Splenic CD4+ T cells derived from these immunized mice were stimulated in vitro, and their supernatants were analyzed by Th1 and Th2 cytokine detection kits. HPV16 VLP restimulation clearly induced the generation of IFN-γ (and the generation of TNF-α and IL-5) by spleen CD4+ T cells of mice immunized with VLP-loaded DCs, consistent with a Th1 response. However, restimulation of CD4+ T cells of IFN-α/β receptor knockout mice severely impaired Th1 cytokine production (Fig. 5A). This effect was even more pronounced for restimulation of MyD88−/− CD4+ T cells (Fig. 5B).
FIG. 5.
IFN-α/β and MyD88 signaling play critical roles in the Th1-biased response to HPV16 VLPs. CD4+ T cells were isolated by positive selection with CD4+ microbeads (MACS; Mitenyi Biotec GmbH) from splenocytes of IFN-α/β receptor- or MyD88-deficient mice immunized three times with HPV16 VLPs. Levels of Th1 and Th2 cytokines, IL-2, IL-4, IL-5, IFN-γ, and TNF-α in supernatants of CD4+ T cells were assessed 24 h after stimulation with VLP-loaded BMDCs (10:1 ratio). Absolute values were calculated from a standard curve, and controls were as shown in Fig. 2D.
DISCUSSION
Rational vaccine development requires knowledge of the pathways that regulate innate and adaptive immune responses to clinically effective vaccine antigens such as HPV16 L1 VLPs. The murine model is useful for studies of the immune response to HPV16 L1 VLPs because of the availability of MyD88-deficient mice, but observations should be validated with humans. Both vaccination of patients with HPV16 VLPs (21) and treatment of naïve C57BL/6 mice with HPV16 VLP-loaded BMDCs induce Th1-biased responses (Fig. 2D), suggesting that the mouse is an appropriate model. Mice represent a heterologous host for HPV16, whereas humans are homologous. Yet human DCs and Langerhans cells respond to HPV16 VLPs in a manner similar to that of their murine counterparts. Notably, both murine BMDCs and human myeloid DCs produce IL-12 in response to VLP, and neither human nor murine Langerhans cells respond to HPV16 VLPs (17, 31). Like IFN-γ and IL-12, IFN-α/β plays a central role in the innate response against viral infections and is also involved in the regulation of adaptive immune responses (5). IFN-α/β can influence adaptive immune responses by modifying the maturation of DCs, e.g., by driving DC subpopulations to promote Th1 cells. Murine BMDCs also produce IFN-α/β in response to HPV16 VLPs. Herpes simplex virus and influenza virus infections, CpG, and poly(I:C) can activate DCs to produce high levels of IFN-α/β (8, 9, 38). Poly(I:C) induces ∼10-fold-higher levels of IFN-α by BMDCs (Fig. 3 and 4) than by HPV16 VLPs, although the latter are clearly sufficient to support a robust Th1 response.
The regulation of IL-12 production is complex (for a review, see reference 43). IFN-γ can enhance production of IL-12 (22), thereby forming a positive-feedback mechanism during inflammatory and Th1 responses. IFN-γ also plays a role by enhancing mRNA expression of the IL-12 receptor β1 and β2 chains (20, 40). Indeed, autocrine-paracrine stimulation of IL-12 production by IFN-γ and up regulation of both IL-12 receptor β chains was observed in BMDCs after VLP stimulation (Fig. 3 and data not shown). IFN production is also associated with expression of the IFN-regulated transcripts. For example, BMDCs strongly up regulate IP-10, IFN-induced 15-kDa protein, Mx1, glucocorticoid attenuated response gene (GARG)-16, GARG-39, GARG-49, and IFN-activated genes 203 and 204.
Chemokines cause recruitment and polarization of T cells (45). To respond to a chemotactic gradient of chemokines, T cells spatially reorganize components to form a leading edge (termed polarization) containing CCR2 and CCR5. This polarization facilitates the formation of the immunological synapse. Furthermore, certain chemokines such as RANTES are able to costimulate T cell proliferation. For example, RANTES-deficient mice exhibit impaired T-cell proliferation and IFN-γ and IL-2 production upon antigen stimulation. Chemokines can also influence T-cell fate (45). Receptors CCR5 and CXCR3 are markers for Th1 cells. RANTES, MIP-1α, and MIP-1β are produced by BMDCs upon HPV16 VLP stimulation and are ligands for CCR5. Similarly, IP-10 mRNA is up regulated by BMDCs in response to HPV16 VLP, and IP-10 is a ligand for CXCR3 (45). Thus, BMDCs up regulate numerous chemokines that ligand the Th1 markers CCR5 and CXCR3 and Th1-associated cytokines such as IL-12 and IFN, which likely drive the potent Th1-biased response of VLP-immunized mice. However, CCR2 is a marker for Th2 cells (45); its ligands MCP-1, MCP-3, and MCP-5 are up regulated in HPV16 VLP-stimulated BMDCs. The induction of these CCR2 ligands and the other Th2-associated cytokines IL-6, IL-10, and G-CSF may contribute to the weak Th2-type IL-5 production by stimulation of T helper cells derived from mice vaccinated with VLP-loaded BMDCs and by patients vaccinated with HPV16 VLPs (21). Further experimentation is necessary to clarify the contribution of these chemokines to VLP-induced immunity.
DCs promote innate and adaptive immunity by sensing pathogens. Recognition of HPV16 VLPs initiates signaling through the MyD88 adaptor protein and innate responses, including the activation of NF-κB, NF-AT, and AP-1 in RAW264.7 cells and production of IFN-α, IFN-γ, and IL-12 by BMDCs. This dependence on MyD88 signaling is suggestive of recognition of HPV16 VLPs by the Toll family of pattern recognition receptors (3). HPV L1 binds to heparan sulfate GAGs on the cell surface during infection (25). These interactions may be related to HPV16 L1-induced signaling via Toll receptors, as the oligosaccharides of hyaluronan can activate DCs via TLR4 (42). It is also notable that respiratory syncytial virus and mouse mammary tumor virus have been shown to bind heparan sulfate on the cell surface (18, 48) and are also recognized by TLR4-dependent pathways (29, 36). We are studying the TLRs that mediate HPV16 VLP recognition in TLR-deficient mice with reconstruction assays with human TLRs.
Several studies indicate that MyD88 signaling is involved in the initial commitment of naïve CD4+ T cells to differentiate into Th1 and Th2 lineage cells (2, 23). MyD88-deficient mice showed an inability to generate Th1-type immune responses to antigen in the presence of complete Freund's adjuvant containing heat-killed mycobacterium tuberculosis, components of which can activate TLRs such as TLR2 and TLR4 (23). Indeed, MyD88 is also critical to the induction of Th1-biased immune responses to HPV16 L1 VLPs.
Many attenuated recombinant microorganisms have been used as vaccine vectors (35). The immunogenicity of these vectors likely reflects recognition by TLRs expressed on antigen-presenting cells and consequent stimulation of DC function and immunity. The ability of HPV16 VLPs to induce such potent immune responses suggests that the highly ordered, closely packed structure (4) of HPV16 VLPs represents a pathogen-associated molecular pattern recognized by the TLR/MyD88 pathway (41).
Acknowledgments
We thank Mike Delannoy of the Microscopy Facility, Johns Hopkins School of Medicine, for expert confocal and electron microscopy.
This research was supported by grants to R.B.S.R. from the National Institutes of Health (PO1 AI 48203 and P50 CA098252); the Tobacco Restitution Fund, State of Maryland; and the American Cancer Society (RSG MBC-103111).
REFERENCES
- 1.Adachi, O., T. Kawai, K. Takeda, M. Matsumoto, H. Tsutsui, M. Sakagami, K. Nakanishi, and S. Akira. 1998. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity 9:143-150. [DOI] [PubMed] [Google Scholar]
- 2.Ahmad-Nejad, P., H. Hacker, M. Rutz, S. Bauer, R. M. Vabulas, and H. Wagner. 2002. Bacterial CpG-DNA and lipopolysaccharides activate Toll-like receptors at distinct cellular compartments. Eur. J. Immunol. 32:1958-1968. [DOI] [PubMed] [Google Scholar]
- 3.Akira, S., M. Yamamoto, and K. Takeda. 2003. Role of adapters in Toll-like receptor signalling. Biochem. Soc. Trans. 31:637-642. [DOI] [PubMed] [Google Scholar]
- 4.Bachmann, M. F., U. H. Rohrer, T. M. Kundig, K. Burki, H. Hengartner, and R. M. Zinkernagel. 1993. The influence of antigen organization on B cell responsiveness. Science 262:1448-1451. [DOI] [PubMed] [Google Scholar]
- 5.Biron, C. A. 2001. Interferons alpha and beta as immune regulators—a new look. Immunity 14:661-664. [DOI] [PubMed] [Google Scholar]
- 6.Breitburd, F., R. Kirnbauer, N. L. Hubbert, B. Nonnenmacher, C. Trin-Dinh-Desmarquet, G. Orth, J. T. Schiller, and D. R. Lowy. 1995. Immunization with viruslike particles from cottontail rabbit papillomavirus (CRPV) can protect against experimental CRPV infection. J. Virol. 69:3959-3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 8.Cella, M., D. Jarrossay, F. Facchetti, O. Alebardi, H. Nakajima, A. Lanzavecchia, and M. Colonna. 1999. Plasmacytoid monocytes migrate to inflamed lymph nodes and produce large amounts of type I interferon. Nat. Med. 5:919-923. [DOI] [PubMed] [Google Scholar]
- 9.Cella, M., M. Salio, Y. Sakakibara, H. Langen, I. Julkunen, and A. Lanzavecchia. 1999. Maturation, activation, and protection of dendritic cells induced by double-stranded RNA. J. Exp. Med. 189:821-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chackerian, B., P. Lenz, D. R. Lowy, and J. T. Schiller. 2002. Determinants of autoantibody induction by conjugated papillomavirus virus-like particles. J. Immunol. 169:6120-6126. [DOI] [PubMed] [Google Scholar]
- 11.Chackerian, B., D. R. Lowy, and J. T. Schiller. 2001. Conjugation of a self-antigen to papillomavirus-like particles allows for efficient induction of protective autoantibodies. J. Clin. Investig. 108:415-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chackerian, B., D. R. Lowy, and J. T. Schiller. 1999. Induction of autoantibodies to mouse CCR5 with recombinant papillomavirus particles. Proc. Natl. Acad. Sci. USA 96:2373-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, X. S., G. Casini, S. C. Harrison, and R. L. Garcea. 2001. Papillomavirus capsid protein expression in Escherichia coli: purification and assembly of HPV11 and HPV16 L1. J. Mol. Biol. 307:173-182. [DOI] [PubMed] [Google Scholar]
- 14.Chen, X. S., R. L. Garcea, I. Goldberg, G. Casini, and S. C. Harrison. 2000. Structure of small virus-like particles assembled from the L1 protein of human papillomavirus 16. Mol. Cell 5:557-567. [DOI] [PubMed] [Google Scholar]
- 15.De Bruijn, M. L., H. L. Greenstone, H. Vermeulen, C. J. Melief, D. R. Lowy, J. T. Schiller, and W. M. Kast. 1998. L1-specific protection from tumor challenge elicited by HPV16 virus-like particles. Virology 250:371-376. [DOI] [PubMed] [Google Scholar]
- 16.Evans, T. G., W. Bonnez, R. C. Rose, S. Koenig, L. Demeter, J. A. Suzich, D. O'Brien, M. Campbell, W. I. White, J. Balsley, and R. C. Reichman. 2001. A phase 1 study of a recombinant viruslike particle vaccine against human papillomavirus type 11 in healthy adult volunteers. J. Infect. Dis. 183:1485-1493. [DOI] [PubMed] [Google Scholar]
- 17.Fausch, S. C., D. M. Da Silva, M. P. Rudolf, and W. M. Kast. 2002. Human papillomavirus virus-like particles do not activate Langerhans cells: a possible immune escape mechanism used by human papillomaviruses. J. Immunol. 169:3242-3249. [DOI] [PubMed] [Google Scholar]
- 18.Feldman, S. A., R. M. Hendry, and J. A. Beeler. 1999. Identification of a linear heparin binding domain for human respiratory syncytial virus attachment glycoprotein G. J. Virol. 73:6610-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenstone, H. L., J. D. Nieland, K. E. de Visser, M. L. De Bruijn, R. Kirnbauer, R. B. Roden, D. R. Lowy, W. M. Kast, and J. T. Schiller. 1998. Chimeric papillomavirus virus-like particles elicit antitumor immunity against the E7 oncoprotein in an HPV16 tumor model. Proc. Natl. Acad. Sci. USA 95:1800-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grohmann, U., M. L. Belladonna, C. Vacca, R. Bianchi, F. Fallarino, C. Orabona, M. C. Fioretti, and P. Puccetti. 2001. Positive regulatory role of IL-12 in macrophages and modulation by IFN-gamma. J. Immunol. 167:221-227. [DOI] [PubMed] [Google Scholar]
- 21.Harro, C. D., Y. Y. Pang, R. B. Roden, A. Hildesheim, Z. Wang, M. J. Reynolds, T. C. Mast, R. Robinson, B. R. Murphy, R. A. Karron, J. Dillner, J. T. Schiller, and D. R. Lowy. 2001. Safety and immunogenicity trial in adult volunteers of a human papillomavirus 16 L1 virus-like particle vaccine. J. Natl. Cancer Inst. 93:284-292. [DOI] [PubMed] [Google Scholar]
- 22.Hayes, M. P., F. J. Murphy, and P. R. Burd. 1998. Interferon-gamma-dependent inducible expression of the human interleukin-12 p35 gene in monocytes initiates from a TATA-containing promoter distinct from the CpG-rich promoter active in Epstein-Barr virus-transformed lymphoblastoid cells. Blood 91:4645-4651. [PubMed] [Google Scholar]
- 23.Jankovic, D., M. C. Kullberg, S. Hieny, P. Caspar, C. M. Collazo, and A. Sher. 2002. In the absence of IL-12, CD4(+) T cell responses to intracellular pathogens fail to default to a Th2 pattern and are host protective in an IL-10(−/−) setting. Immunity 16:429-439. [DOI] [PubMed] [Google Scholar]
- 24.Jochmus, I., K. Schafer, S. Faath, M. Muller, and L. Gissmann. 1999. Chimeric virus-like particles of the human papillomavirus type 16 (HPV 16) as a prophylactic and therapeutic vaccine. Arch. Med. Res. 30:269-274. [DOI] [PubMed] [Google Scholar]
- 25.Joyce, J. G., J. S. Tung, C. T. Przysiecki, J. C. Cook, E. D. Lehman, J. A. Sands, K. U. Jansen, and P. M. Keller. 1999. The L1 major capsid protein of human papillomavirus type 11 recombinant virus-like particles interacts with heparin and cell-surface glycosaminoglycans on human keratinocytes. J. Biol. Chem. 274:5810-5822. [DOI] [PubMed] [Google Scholar]
- 26.Kirnbauer, R., F. Booy, N. Cheng, D. R. Lowy, and J. T. Schiller. 1992. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc. Natl. Acad. Sci. USA 89:12180-12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirnbauer, R., J. Taub, H. Greenstone, R. Roden, M. Durst, L. Gissmann, D. R. Lowy, and J. T. Schiller. 1993. Efficient self-assembly of human papillomavirus type 16 L1 and L1-L2 into virus-like particles. J. Virol. 67:6929-6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koutsky, L. A., K. A. Ault, C. M. Wheeler, D. R. Brown, E. Barr, F. B. Alvarez, L. M. Chiacchierini, and K. U. Jansen. 2002. A controlled trial of a human papillomavirus type 16 vaccine. N. Engl. J. Med. 347:1645-1651. [DOI] [PubMed] [Google Scholar]
- 29.Kurt-Jones, E. A., L. Popova, L. Kwinn, L. M. Haynes, L. P. Jones, R. A. Tripp, E. E. Walsh, M. W. Freeman, D. T. Golenbock, L. J. Anderson, and R. W. Finberg. 2000. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat. Immunol. 1:398-401. [DOI] [PubMed] [Google Scholar]
- 30.Lenz, P., P. M. Day, Y. Y. Pang, S. A. Frye, P. N. Jensen, D. R. Lowy, and J. T. Schiller. 2001. Papillomavirus-like particles induce acute activation of dendritic cells. J. Immunol. 166:5346-5355. [DOI] [PubMed] [Google Scholar]
- 31.Lenz, P., C. D. Thompson, P. M. Day, S. M. Bacot, D. R. Lowy, and J. T. Schiller. 2003. Interaction of papillomavirus virus-like particles with human myeloid antigen-presenting cells. Clin. Immunol. 106:231-237. [DOI] [PubMed] [Google Scholar]
- 32.Li, M., P. Beard, P. A. Estes, M. K. Lyon, and R. L. Garcea. 1998. Intercapsomeric disulfide bonds in papillomavirus assembly and disassembly. J. Virol. 72:2160-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nieland, J. D., D. M. Da Silva, M. P. Velders, K. E. de Visser, J. T. Schiller, M. Muller, and W. M. Kast. 1999. Chimeric papillomavirus virus-like particles induce a murine self-antigen-specific protective and therapeutic antitumor immune response. J. Cell Biochem. 73:145-152. [PubMed] [Google Scholar]
- 34.Ohlschlager, P., W. Osen, K. Dell, S. Faath, R. L. Garcea, I. Jochmus, M. Muller, M. Pawlita, K. Schafer, P. Sehr, C. Staib, G. Sutter, and L. Gissmann. 2003. Human papillomavirus type 16 L1 capsomeres induce L1-specific cytotoxic T lymphocytes and tumor regression in C57BL/6 mice. J. Virol. 77:4635-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pardoll, D. M. 2002. Spinning molecular immunology into successful immunotherapy. Nat. Rev. Immunol. 2:227-238. [DOI] [PubMed] [Google Scholar]
- 36.Rassa, J. C., J. L. Meyers, Y. Zhang, R. Kudaravalli, and S. R. Ross. 2002. Murine retroviruses activate B cells via interaction with toll-like receptor 4. Proc. Natl. Acad. Sci. USA 99:2281-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rudolf, M. P., J. D. Nieland, D. M. DaSilva, M. P. Velders, M. Muller, H. L. Greenstone, J. T. Schiller, and W. M. Kast. 1999. Induction of HPV16 capsid protein-specific human T cell responses by virus-like particles. Biol. Chem. 380:335-340. [DOI] [PubMed] [Google Scholar]
- 38.Siegal, F. P., N. Kadowaki, M. Shodell, P. A. Fitzgerald-Bocarsly, K. Shah, S. Ho, S. Antonenko, and Y. J. Liu. 1999. The nature of the principal type 1 interferon-producing cells in human blood. Science 284:1835-1837. [DOI] [PubMed] [Google Scholar]
- 39.Suzich, J. A., S. J. Ghim, F. J. Palmer-Hill, W. I. White, J. K. Tamura, J. A. Bell, J. A. Newsome, A. B. Jenson, and R. Schlegel. 1995. Systemic immunization with papillomavirus L1 protein completely prevents the development of viral mucosal papillomas. Proc. Natl. Acad. Sci. USA 92:11553-11557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szabo, S. J., A. S. Dighe, U. Gubler, and K. M. Murphy. 1997. Regulation of the interleukin (IL)-12R beta 2 subunit expression in developing T helper 1 (Th1) and Th2 cells. J. Exp. Med. 185:817-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takeda, K., T. Kaisho, and S. Akira. 2003. Toll-like receptors. Annu. Rev. Immunol. 21:335-376. [DOI] [PubMed] [Google Scholar]
- 42.Termeer, C., F. Benedix, J. Sleeman, C. Fieber, U. Voith, T. Ahrens, K. Miyake, M. Freudenberg, C. Galanos, and J. C. Simon. 2002. Oligosaccharides of hyaluronan activate dendritic cells via toll-like receptor 4. J. Exp. Med. 195:99-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trinchieri, G. 2003. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat. Rev. Immunol. 3:133-146. [DOI] [PubMed] [Google Scholar]
- 44.Walboomers, J. M., M. V. Jacobs, M. M. Manos, F. X. Bosch, J. A. Kummer, K. V. Shah, P. J. Snijders, J. Peto, C. J. Meijer, and N. Munoz. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189:12-19. [DOI] [PubMed] [Google Scholar]
- 45.Wong, M. M., and E. N. Fish. 2003. Chemokines: attractive mediators of the immune response. Semin. Immunol. 15:5-14. [DOI] [PubMed] [Google Scholar]
- 46.Yang, R., W. H. Yutzy IV, R. P. Viscidi, and R. B. S. Roden. 2003. Interaction of L2 with β-actin directs intracellular transport of papillomavirus and infection. J. Biol. Chem. 278:12546-12553. [DOI] [PubMed] [Google Scholar]
- 47.Yeo, S. J., J. G. Yoon, S. C. Hong, and A. K. Yi. 2003. CpG DNA induces self and cross-hyporesponsiveness of RAW264.7 cells in response to CpG DNA and lipopolysaccharide: alterations in IL-1 receptor-associated kinase expression. J. Immunol. 170:1052-1061. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, Y., J. C. Rassa, M. E. deObaldia, L. M. Albritton, and S. R. Ross. 2003. Identification of the receptor binding domain of the mouse mammary tumor virus envelope protein. J. Virol. 77:10468-10478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou, J., X. Y. Sun, and I. H. Frazer. 1993. Glycosylation of human papillomavirus type 16 L1 protein. Virology 194:210-218. [DOI] [PubMed] [Google Scholar]