Abstract
The genome sequence of the genetically tractable, mesophilic, hydrogenotrophic methanogen Methanococcus maripaludis contains 1,722 protein-coding genes in a single circular chromosome of 1,661,137 bp. Of the protein-coding genes (open reading frames [ORFs]), 44% were assigned a function, 48% were conserved but had unknown or uncertain functions, and 7.5% (129 ORFs) were unique to M. maripaludis. Of the unique ORFs, 27 were confirmed to encode proteins by the mass spectrometric identification of unique peptides. Genes for most known functions and pathways were identified. For example, a full complement of hydrogenases and methanogenesis enzymes was identified, including eight selenocysteine-containing proteins, with each being paralogous to a cysteine-containing counterpart. At least 59 proteins were predicted to contain iron-sulfur centers, including ferredoxins, polyferredoxins, and subunits of enzymes with various redox functions. Unusual features included the absence of a Cdc6 homolog, implying a variation in replication initiation, and the presence of a bacterial-like RNase HI as well as an RNase HII typical of the Archaea. The presence of alanine dehydrogenase and alanine racemase, which are uniquely present among the Archaea, explained the ability of the organism to use l- and d-alanine as nitrogen sources. Features that contrasted with the related organism Methanocaldococcus jannaschii included the absence of inteins, even though close homologs of most intein-containing proteins were encoded. Although two-thirds of the ORFs had their highest Blastp hits in Methanocaldococcus jannaschii, lateral gene transfer or gene loss has apparently resulted in genes, which are often clustered, with top Blastp hits in more distantly related groups.
The methanogenic Archaea (methanogens) occupy a unique metabolic niche, as they produce methane, which is a useful energy source and a powerful greenhouse gas. These organisms are found in diverse anaerobic habitats, ranging from aquatic and marine sediments to sewage digesters and the rumens and large intestines of herbivores and other mammals (127). In these habitats, the degradation of organic matter results in the production of H2 and other intermediates by fermentative organisms. By maintaining an extremely low partial pressure of H2, the methanogens keep fermentative pathways energetically favorable. In addition, some methanogens may occupy niches where hydrogen is produced predominately by geothermal reactions.
Metabolically, methanogens are divided into those that specialize in CO2 reduction and those that also use acetate and/or methyl compounds. The former group, the hydrogenotrophs, use H2 as an electron donor to reduce CO2 to methane. Many hydrogenotrophic species can substitute formate or certain low-molecular-weight alcohols and ketones for H2. Complete genome sequences have been published for three hydrogenotrophic methanogens, Methanocaldococcus jannaschii (13), Methanothermobacter thermautotrophicus (105), and Methanopyrus kandleri (104), all of which are thermophiles or hyperthermophiles. Of the methanogens that utilize acetate and methyl compounds, complete genome sequences have been published for two species, Methanosarcina acetivorans (26) and Methanosarcina mazei (19), both of which are mesophiles. In addition, partial sequences have been published for two psychrophiles, the hydrogenotroph Methanogenium frigidum and the methylotroph Methanolobus burtonii (97).
Genome sequences of methanogens have answered many questions, but they have inspired many others. More than half of the genes in Methanocaldococcus jannaschii lack a predicted function (13), and this proportion has not declined significantly as other methanogen sequences have been determined. The proportions of genes of unknown functions, which are either homologous to other genes of unknown function or have no known homologs at all, are 55% for the Methanothermobacter thermautotrophicus genome (105) and 51% for the Methanosarcina acetivorans genome (26).
These observations demonstrate a pressing need to identify the functions of genes in the methanogenic Archaea. Many of the most effective approaches involve genetic manipulation, determining the phenotypes of mutants, or affinity tagging proteins in vivo to facilitate their purification. Nevertheless, few genetic tools are available for sequenced species of the methanogenic Archaea. Genetics can be used for Methanosarcina acetivorans (88) but not for any previously sequenced hydrogenotrophic methanogenic species. Here we present the genome sequence of the genetically tractable species Methanococcus maripaludis.
M. maripaludis is a mesophilic hydrogenotrophic methanogen that was isolated from salt marshes (48). Like all methanogens, M. maripaludis belongs to the kingdom Euryarchaeota in the domain Archaea. M. maripaludis belongs to the family Methanococcaceae in the order Methanococcales (12). Although M. maripaludis is related to Methanocaldococcus jannaschii, it possesses many novel features, and approximately one-third of its genes lack orthologs in Methanocaldococcus jannaschii (see below). Extensive studies of physiology and regulation have already been performed with M. maripaludis, and many of them used genetic tools (53, 65, 113). The virtues of M. maripaludis as a model species are apparent. Dense liquid cultures are obtained overnight and colonies grow on agar medium in 2 days (49). Chemostat cultures can now be established reproducibly (36a). Many important genetic manipulations are routine, including transformation, complementation with shuttle vectors, gene deletions, and insertions of reporters (28). New approaches to genetic manipulation are being implemented (B. C. Moore and J. A. Leigh, submitted for publication), and the first comprehensive expression array and proteomic analyses have been completed (E. L. Hendrickson, M. Hackett, and J. A. Leigh, unpublished data).
MATERIALS AND METHODS
Strain.
M. maripaludis strain S2 (120) is a wild-type isolate that has also been designated strain LL.
Genome sequencing.
M. maripaludis strain S2 was sequenced by the use of standard DNA sequencing protocols and data collection tools. Initially, 43,950 small insert shotgun reads and 1,536 fosmid-end sequencing reads were collected by using Big Dye terminator sequencing chemistries. The sequences were assembled and viewed with phred/phrap/consed software. To facilitate opening and viewing of the genome assemblies in consed, we created a phd.ball file from each phd file. The creation of phd.ball files reduced the time to open the genome assembly in consed to <10 min. The initial assembly provided 8.14× Q20 sequence coverage (Q20, error rate of <1% [22]) and provided 99.46% coverage of the 1.66-Mb genome. The M. maripaludis genome was finished by using the autofinish tool of consed (30). In all, 770 finishing reads were attempted and four PCR templates were generated to finish the sequence. The fosmid-end sequence reads were tiled along the postshotgun sequence assembly with SeqTile software (W. Gillett, unpublished software tool), which identified two grossly misassembled regions. The misassemblies identified were all due to the presence of nearly identical ribosomal DNA repeats. Two unique fosmid clones that spanned the misassembled regions were selected and sequenced to 8× Q20 coverage. A third fosmid clone spanning difficult-to-finish regions was mutagenized by a transposon mutagenesis protocol suggested by the manufacturer (Epicenter Technologies). Random clones from mutagenesis experiments were picked, and DNAs were prepared and sequenced by using standard Big Dye terminator chemistry. The backbones from these independently assembled fosmid clones were imported into the genome assembly to resolve misassemblies and to improve the sequence quality of difficult-to-finish regions.
Sequence validation.
The final assembly contained 38,601 reads, including reads from autofinish and advanced finishing experiments as well as the backbones from the three independently sequenced fosmid clones. The final validation of the sequence assembly was performed by using SeqTile software and comparing the restriction fingerprint patterns of 417 fosmid clones with the virtual fingerprint pattern of the finished sequence assembly by using three enzymes, BglII, EcoRI, and HindIII. The 417 fosmid clones provided 10× clone coverage and uninterrupted 2× fingerprint coverage for the finished sequence assembly.
Genome analysis and annotation.
The genome sequence was analyzed, and annotations were entered at the Genome Channel facility at the Oak Ridge National Laboratories (http://genome.ornl.gov/microbial/mmar/). Automated annotations were accomplished for all open reading frames (ORFs) by Blastp comparisons to protein databases, Pfam, InterPro (incorporating Pfam, TIGRFams, SmartHMM, Prosite, Prints, and ProDom algorithms), and Clusters of Orthologous Groups (COGs). Most ORFs were also annotated by hand. In brief, preliminary identifications were first made by Blastp analysis, and high expectation values covering at least 80% of the ORF were sought. The list of Blast hits was then scanned for highly homologous proteins whose functions had been experimentally determined. The other analysis tools mentioned above, as well as the presence of a gene in an operon with functionally related genes, were then examined for supporting evidence. ORFs with clear homologies but uncertain functions were designated members of gene families, relatives of genes of known function, or conserved hypothetical proteins. Putative transporters were checked against M. Saier's transport protein classification web site (http://www.biology.ucsd.edu/∼msaier/transport). Genes were viewed graphically with Artemis (http://www.sanger.ac.uk/Software/Artemis/).
Proteomics.
During the course of our work, we analyzed 18 protein samples from a variety of M. maripaludis cultures. Protein mixtures were digested with trypsin and separated by multidimensional liquid chromatography as described previously (117, 118). “Bottom-up” proteomics was performed by tandem mass spectrometry using a Finnegan LCQ classic quadrupole ion trap mass spectrometer equipped with an electrospray ion source. Peptide sequences derived from proteolytic fragments were matched to M. maripaludis ORFs by computational reference to the genome sequence by using Sequest (21), DTASelect (109), and d2g (118) software and by manual interpretations of individual collision-induced dissociation mass spectra.
Nucleotide sequence accession number.
The M. maripaludis genome sequence is available at the EMBL/GenBank/DDBJ database under accession number BX950229 and at the Oak Ridge National Laboratories Genome Channel at http://genome.ornl.gov/microbial/mmar/.
RESULTS AND DISCUSSION
General features and organization.
The genome of M. maripaludis consists of a single circular chromosome of 1,661,137 bp (Table 1). Genome modeling predicts 1,722 protein-coding genes, with 52% carried on the forward strand and 48% carried on the complementary strand. The genome encodes four 5S rRNAs, three 23S rRNAs, three 16S rRNAs, 38 tRNAs, and RNase P. Since no distinct origin of replication can be discerned (see below), nucleotide numbering was begun at the end of an rRNA gene cluster. ORFs were numbered consecutively along the genome and given the prefix “Mmp.” Functional categories of protein-coding genes (ORFs) are listed in Table 2 and mapped in Fig. 1. A complete list of ORFs and their functional annotations is available at http://www.ncbi.nlm.nih.gov/genomes/altik.cgi?db=G&gi=394. The M. maripaludis sequence is included in the National Center for Biotechnology Information list of completed microbial genomes at http://www.ncbi.nlm.nih.gov/genomes/MICROBES/Complete.html.
TABLE 1.
General features of the M. maripaludis genome
| Parameter | Value |
|---|---|
| Total no. of bases | 1,661,137 |
| No. of protein-coding genes | 1,722 |
| No. of predicted transmembrane proteins | 350 |
| Gene density (genes/kb) | 1.036 |
| Average gene length (bp) | 857 |
| Protein-coding percentage | 88.9 |
| No. of tRNAs | 38 |
| No. of rRNA operons | 3 |
| %GC | 33.1 |
TABLE 2.
Functional categories of proteins encoded by M. maripaludis genome
| Functiona | No. of ORFs | % of ORFs |
|---|---|---|
| Amino acid biosynthesis | 82 | 4.8 |
| Biosynthesis of cofactors, prosthetic groups, and carriers | 68 | 3.9 |
| Cell envelope | 8 | 0.56 |
| Cellular processes (cell division, chemotaxis, and motility) | 30 | 1.7 |
| Central intermediary metabolism | 112 | 6.5 |
| DNA metabolism | 40 | 2.3 |
| Energy metabolism (methanogenesis, hydrogen metabolism, and ATPase) | 81 | 4.7 |
| Fatty acid and phospholipid metabolism | 6 | 0.35 |
| Hypothetical proteins | 102 | 5.9 |
| Unique proteins of unknown function | 27 | 1.6 |
| Conserved hypothetical proteins | 656 | 38 |
| Protein fate | 22 | 1.3 |
| Protein synthesis | 119 | 6.9 |
| Purines, pyrimidines, nucleosides, and nucleotides | 44 | 2.6 |
| Regulatory functions | 38 | 2.2 |
| Transcription | 22 | 1.3 |
| Transport and binding proteins | 86 | 5.0 |
| Unclassified and unknown function | 179 | 10.4 |
| Total | 1,722 | 100 |
Adapted from The Institute for Genome Research's functional categories.
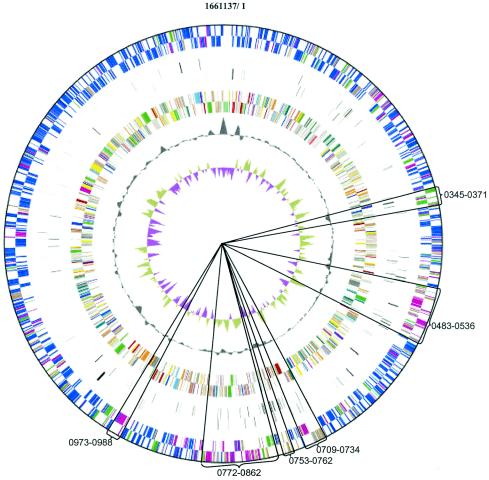
FIG. 1.
Circular map of M. maripaludis genome. First (outer) double ring, top Blastp hits; second double ring, ORFs unique to M. maripaludis; third double ring, functional categories; black single ring, deviation from average mol% G+C; inner ring, GC skew. Top Blast hits are coded as follows: blue, Methanocaldococcus jannaschii; magenta, other methanogens; green, other Archaea; brown, Bacteria and Eukarya. Sectors containing top Blast hits predominately to groups other than Methanocaldococcus jannaschii are shown, with ORF number intervals. Functional categories are coded as follows: red, replication and repair; green, energy metabolism; blue, carbohydrate metabolism; cyan, lipid metabolism; magenta, transcription; yellow, translation; sky blue, cellular processes; orange, amino acid metabolism; pink, metabolism of cofactors; light red, nucleotide metabolism; gray, conserved hypothetical proteins; white, hypothetical proteins; brown, unassigned proteins; black, other; pale green, RNAs.
M. maripaludis has a low G+C content, 33.1%, which is fairly homogeneous across the genome (Fig. 1). The only large deviations are in the regions of the rRNAs. Compared to the overall G+C content, the intergenic regions have a lower percentage, 25.7% G+C, while the ORFs contain 34% G+C.
Like those of all Bacteria and Archaea, most of the genes of M. maripaludis appear to be present in polycistronic operons. While some genes with common functionality are clustered into operons in M. maripaludis, many are not. Clustered genes include many of those encoding ribosomal components, methanogenic enzymes, conserved hypothetical proteins, and other multicomponent enzymes. However, compared to the case for the Bacteria, a striking feature of the M. maripaludis genome is the tendency for some functionally related genes to be unlinked. Genes for amino acid, purine, or pyrimidine biosynthesis are rarely linked, but instead are often present in operons with genes of unrelated or unknown function. Tryptophan biosynthetic genes, which are clustered in an operon, are a notable exception.
Although 19 inteins were found in the Methanocaldococcus jannaschii genome (84), none were found in M. maripaludis. Nevertheless, M. maripaludis encodes close homologs of all but two of the intein-containing ORFs.
Lateral gene transfer and gene loss.
Among the protein-coding genes of M. maripaludis, the highest frequency (64% of ORFs) of high-scoring Blastp hits occurred with genes of Methanocaldococcus jannaschii, the closest relative of M. maripaludis with a known genome sequence (13). The frequencies of top Blastp hits with other groups were as follows: other methanogens, 12%; Euryarchaeota, 18%; Crenarchaeota, 0.2%; Bacteria, 9.6%; and Eukarya, 0.6% (see the supplemental material). These figures suggest that lateral gene transfer into the M. maripaludis lineage from distant lineages has occurred but that it has not been as frequent as in the mesophilic methylotroph Methanosarcina mazei (19) or Methanosarcina acetivorans (26). The lack of any significant deviations from the average mol% G+C among the ORFs implies that any lateral transfers into M. maripaludis occurred long ago, allowing the G+C content to equilibrate over time, or were from organisms with similar G+C percentages.
For Fig. 1, the highest-scoring Blastp hits in Methanocaldococcus jannaschii, other methanogens, other Archaea, and Bacteria plus Eukarya were color coded around the genome. The distribution was nonrandom. Top Blastp hits to groups other than Methanocaldococcus jannaschii were noticeably less frequent in a wide sector centered around base 1,500,000 than in the opposite sector. Furthermore, discrete clusters of genes had top hits to predominantly one or more of the more distant groups at the expense of Methanocaldococcus jannaschii (Table 3 and Fig. 1). The most notable of these clusters (Mmp0483 to -0536) contains the genes encoding the molybdenum formylmethanofuran dehydrogenase (Fmd; top hits to other methanogens) as well as a gene for molybdopterin biosynthesis and three ABC transporters, two of which were for molybdate. Methanocaldococcus jannaschii lacks Fmd (see below), and the genes for Fmd and molybdenum-related functions could have been transferred laterally to the M. maripaludis lineage from outside of the methanococci or could have been present in an ancestor and lost from Methanocaldococcus jannaschii. The cluster from Mmp0973 to Mmp0988 contains carbon monoxide dehydrogenase/acetyl coenzyme A (CoA) synthase (Cdh); in this case, Methanocaldococcus jannaschii has the enzyme, yet all seven subunits yielded top hits to Methanothermobacter thermautotrophicus. The clustered nature of these genes is consistent with the idea that clustering can both facilitate and result from the lateral transfer of functionally related genes (61).
TABLE 3.
Phylogenetic distributions and functions of clustered non-Methanocaldococcus jannaschii top Blastp hit categories
| ORF intervala (Mmp no.) | Predominant group(s)b | Represented functions |
|---|---|---|
| 0345-0371 | Other Archaea | Conserved hypothetical proteins, carbohydrate metabolism |
| 0483-0536 | Other methanogens and Bacteria | Conserved hypothetical proteins, Fmd, Mo transport, molybdopterin biosynthesis, metal chelatase, probable cation transport |
| 0709-0734 | Bacteria and other methanogens | Divalent cation transport, UvrABC |
| 0753-0762 | Bacteria | Conserved hypothetical proteins |
| 0772-0862 | Other methanogens and Bacteria | Conserved hypothetical proteins, Vhc |
| 0973-0988 | Other methanogens | Cdh |
Intervals containing 70% or more top Blastp hits for other methanogens, other Archaea, and Bacteria combined.
Groups containing the top Blastp hits for at least 20% of the ORFs in the interval.
Interestingly, a family of putative ATPases known only in Methanocaldococcus jannaschii (Methanocaldococcus jannaschii ORFs MJ0625, MJECL26, MJ1076, and MJ1006, with more distant relatives in Methanocaldococcus jannaschii and Pyrococcus species) is entirely absent from M. maripaludis. Also, ribulose biosphosphate carbxylase, which is present in Methanocaldococcus jannaschii and other methanogens (25), is not encoded in the M. maripaludis genome.
Proteins of known and unknown function.
Of the 1,722 predicted proteins, a function was assigned to 758 (44%) of them. Another 835 (48%) ORFs were either homologous to genes of unknown function (conserved hypothetical proteins) or had uncertain affiliations with genes of known function. The remaining 129 (7.5%) were unique to M. maripaludis and had no known homologs.
For the 129 predicted proteins that were unique to M. maripaludis, the existence of 27 was confirmed by proteomics. Peptides that belong to these proteins were identified unequivocally in samples from M. maripaludis by mass spectrometry, and these proteins were therefore designated unique proteins of unknown function (Table 2; see the supplemental material). The remaining unique proteins were designated hypothetical proteins.
Information systems. (i) Replication.
Like most Archaea (8, 9), M. maripaludis contains a subset of the eukaryal replication proteins. However, the M. maripaludis replication apparatus has some distinctive features. At the replication initiation stage, both Methanocaldococcus jannaschii (31) and M. maripaludis lack a homolog of Cdc6, which forms part of the prereplication complex in Eukarya and other Archaea (75). Both species also lack a discrete transition in GC skew. Since the location of cdc6 and the GC skew transition typically provide the major evidence for the origin of replication, there is no clear indication for the origin in M. maripaludis. In fact, M. maripaludis has many GC skew transitions distributed around the chromosome (Fig. 1). Methanocaldococcus jannaschii is known to maintain 3 to 15 copies of the chromosome during its life cycle (72), and both species may employ multiple origins to achieve this end.
Despite the lack of Cdc6, M. maripaludis has four homologs of the minichromosome maintenance (MCM) proteins (Mmp0030, Mmp0470, Mmp0748, and Mmp1024) that are recruited to the initiation complex and provide helicase activity (62). In contrast, Methanothermobacter thermautotrophicus contains only one MCM protein, which forms a homomultimeric ring structure (100). In M. maripaludis, each protein may act independently to form a helicase, or all four may be required. Like the case for nearly all DNA processes, topoisomerases are also important for replication initiation (116), and a bacterial-type topoisomerase I (Mmp0956) is encoded in M. maripaludis, as in many Archaea. As expected, there is no gene for reverse gyrase, which is known only for hyperthermophiles, including Methanocaldococcus jannaschii.
M. maripaludis has all of the expected components for polymerization. Like other Euryarchaeota, M. maripaludis encodes a single family B DNA polymerase (Mmp0380), but it also contains an archaeon-specific two-subunit DNA polymerase (Mmp0008 and Mmp0026) (45). Processivity factors (Mmp1126 and Mmp1711), enabling long-range DNA polymerization, and the clamp-loading proteins (Mmp032 and Mmp0427) that bind the processivity factors are also encoded. Notable distinctions include the observation that M. maripaludis, like Methanocaldococcus jannaschii and Methanothermobacter thermautotrophicus, has only one subunit of the single-stranded DNA binding protein, Mmp1032 (56). Other Euryarchaeota have a protein with three different subunits (56). Like other Archaea (11), M. maripaludis has a single-subunit primase, p48 (Mmp0071), in contrast to Eukarya, which require p58 as well (68). Interestingly, M. maripaludis also has a homolog of DnaG (Mmp1286), the bacterial primase (86). Hence, M. maripaludis may have two separate primase systems.
Like other Archaea, M. maripaludis removes the primers from the lagging strand of DNA replication by using flap endonuclease (Fen1/Rad2 and Mmp1313) and RNase HII (Mmp1374) (58, 89). M. maripaludis is unique among Archaea in that it also encodes a homolog of RNase HI (Mmp0837), the main RNase in Bacteria (58). The M. maripaludis homolog is similar to RNase HI from Clostridium and may have been acquired by lateral gene transfer. Okazaki fragments are probably ligated together by a homolog of eukaryal ATP-dependent DNA ligase I (Mmp0970) (45).
M. maripaludis encodes a homolog of Smc (structural maintenance of chromosome; Mmp1397), which is believed by analogy with those of the Eukarya to play a part in archaeal chromosome segregation and condensation (7). The archaeal type II topoisomerase (Mmp0989 and Mmp1437), a two-subunit protein that decatenates chromosomes (7), is encoded, as are distant homologs of the Escherichia coli proteins XerC and XerD (Mmp0472 and Mmp0743) (24), suggesting the possibility of two separate systems for chromosome decatenation. M. maripaludis also contains two homologs of the plasmid partitioning gene parA (Mmp0704 and Mmp0593) and parB (Mmp0592) (29).
(ii) Cell division.
In bacterial cell division, a ring of proteins forms at the cell center and constricts as the septum grows (94). This system is shared by the Euryarchaeota, which often have multiple ftsZ homologs (8). Two homologs, Mmp1436 and Mmp1500, are found in M. maripaludis. M. maripaludis also carries a homolog of bacterial minD (Mmp1145), which is thought to encode an inhibitor of FtsZ ring formation (44). M. maripaludis lacks homologs of the two E. coli proteins that bind to the FtsZ ring, FtsA and ZipA (94). Surprisingly, M. maripaludis encodes a homolog of Cdc48 (Mmp0176), which in Saccharomyces cerevisiae plays a role in the membrane fusion of organelles (60). Since Archaea do not possess organelles, the role of the Cdc48 homolog is unknown.
(iii) Recombination and repair.
The M. maripaludis genome contains several genes that are predicted to code for recombination and repair systems. These include the Mre11-Rad50 double-stranded-break repair system (Mmp1340 to -1341), the archaeal RecA homolog RadA (Mmp1222), the related protein RadB (Mmp0617), which is thought to amplify RadA activity, a RecJ homolog (Mmp1682), and the unique archaeal Holliday junction resolvase, Mmp0336 (57). Several base excision repair proteins were found, including ExoA (Mmp1012) and an endonuclease III-related protein (Mmp0586), but no DNA photolyase was present. M. maripaludis, like Methanocaldococcus jannaschii, is missing a homolog of the mismatch repair gene mutS found in other archaeal species. However, unlike Methanocaldococcus jannaschii, M. maripaludis has an E. coli-like excinuclease, UvrABC (Mmp0727 to -0729), which functions as a wide-substrate-range nucleotide excision repair system (82). Weak homologs are also present for the MutT nucleotide diphosphate hydrolase (Mmp0339) and the O6-methylguanine-DNA methyltransferase (Mmp0069), which repair specific damage to nucleotides (101).
(iv) Transcription.
M. maripaludis contains a complete set of genes for the archaeal transcriptional machinery. Single homologs of the TATA box binding protein (Mmp0257) and transcription factors B (TFB; Mmp0041) and E (TFE; Mmp0036) are present (38). Homologs are also found for all 13 subunits of the archaeal RNA polymerase (71).
(v) Transcriptional regulators.
Like other sequenced Archaea, M. maripaludis encodes a few bacterial regulatory family members, including TetR (the most numerous, with four members), ArsR, LysR, and PadR (see supplemental material). M. maripaludis also encodes regulators that are found only in Archaea, including the known nitrogen repressor NrpR (Mmp0607) (65) and a member of an Archaea-specific COG that is predicted to be a transcriptional regulator (Mmp0907). Two-component regulators, which are numerous in Methanosarcina acetivorans (26) and Methanothermobacter thermautotrophicus (105), are absent from Methanocaldococcus jannaschii and Methanopyrus kandleri (104). Excluding those involved in chemotaxis (see below), only one two-component regulator is encoded by M. maripaludis (Mmp1303 and -1304). In total, 24 transcriptional regulators are predicted with confidence, which is about the number expected for a genome of this size (114).
(vi) Translation.
The factors governing translation in Archaea have been determined by homology to eukaryotic and prokaryotic systems (5, 27). M. maripaludis possesses homologs of the four archaeal translation elongation factors (Mmp1131, -1369, -1370, and -1401) and all but one of the archaeal translation initiation factors (Mmp0061, -0284, -0297, -0457, -0603, -0952, -1208, -1618, and -1707). While other Archaea, including Methanocaldococcus jannaschii, encode two subunits of initiation factor 2B (5), M. maripaludis apparently has only subunit 1 (Mmp1618).
Aminoacyl tRNA synthetases were identified for 18 amino acids. Two genes for the alpha subunit of the two-subunit phenylalanyl-tRNA synthetase were found (Mmp0688 and -1496), as is the case for Methanocaldococcus jannaschii and Methanothermobacter thermautotrophicus. No aminoacyl tRNA synthetases were found for asparagine or glutamine. Instead, asparaginyl-tRNA and glutaminyl-tRNA are made by tRNA-dependent amidotransferases, and all of the subunits for the enzyme that forms both asparaginyl-tRNA and glutaminyl-tRNA (GatABC; Mmp1510, -0946, and -0575) and the enzyme specific for glutaminyl-tRNA synthesis (GatDE; Mmp1266 and -1265) are encoded (108).
M. maripaludis makes use of selenocysteine, the 21st cotranslationally inserted amino acid. In Bacteria, selenocysteinyl-tRNA synthesis begins with the charging of tRNASec with serine, followed by dehydration and the addition of a selenide moiety from selenophosphate. A homolog of the Methanocaldococcus jannaschii selenophosphate synthetase was identified (SelD; Mmp0904) which, as in Methanocaldococcus jannaschii, appears itself to be a selenocysteine-containing protein. No selenocysteine synthase has been identified with confidence (93). Selenocysteine incorporation into proteins involves SelB (Mmp1336) (91).
tRNAs were identified for all 21 amino acids.
(vii) Protein folding.
The putative chaperoning systems of M. maripaludis consist of a chaperonin subunit (Mmp1515) of the group II, or thermosome, type (54) and two prefoldins (Mmp1470 and Mmp0245) similar to known archaeal prefoldin subunits alpha and beta, respectively (63). M. maripaludis does not have genes encoding the components of the molecular chaperone machine, namely Hsp70 (DnaK), Hsp40 (DnaJ), and GrpE, or genes encoding group I chaperonins GroEL and GroES. In this respect, M. maripaludis resembles many other Archaea (69), but it contrasts with Methanosarcina species (19, 26).
Metabolism. (i) Methanogenesis.
As a hydrogenotrophic methanogen, M. maripaludis obtains energy and carbon from H2 and CO2 by the methanogenic pathway (see Fig. 2 for the major pathways in M. maripaludis). The first step in CO2 reduction to methane is catalyzed by formylmethanofuran dehydrogenase, which is found in both tungsten (Fwd) (Mmp1244 to -1249 and -1691) and molybdenum (Fmd) (Mmp0200 and -0508 to -0512) forms in M. maripaludis. In contrast, Methanocaldococcus jannaschii possesses only the tungsten form (41), possibly due to its hyperthermophilicity (40). Unlike most methanogens, fwdB (Mmp1691) in M. maripaludis is not in an operon with the genes for the other Fwd subunits but is encoded adjacent to the Vhu hydrogenase (see below). Even more unusual, the fmd operon has two adjacent fmdB (Mmp0511 and -0512) genes, with one encoding a selenocysteine version of the protein.
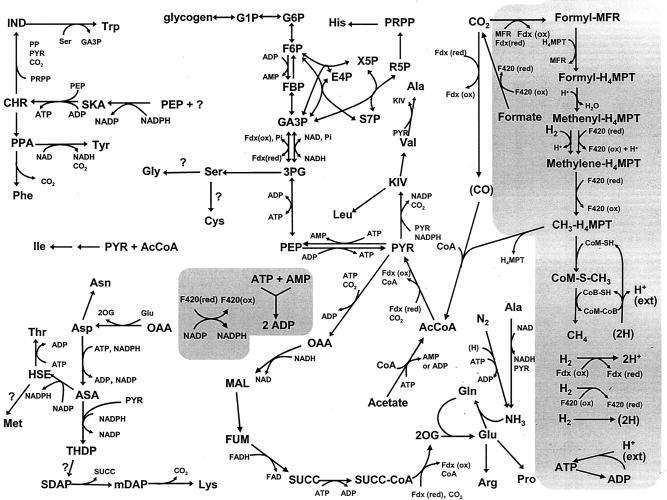
FIG. 2.
Map of major metabolic pathways in M. maripaludis. Shown are energy and redox-related pathways (shaded areas), CO2 fixation, the reductive branch of the TCA cycle, nitrogen assimilation, glycolysis and gluconeogenesis, the nonoxidative pentose phosphate pathway, and amino acid biosynthesis. Some reactions and minor substrates and products were omitted. Abbreviations: ASA, aspartate semialdehyde; CHR, chorismate; (CO), enzyme-bound carbon monoxide; CoA, coenzyme A; CoB, coenzyme B; CoM, coenzyme M; E4P, erythrose-4-phosphate; F6P, fructose-6-phosphate; FBP, fructose-bis-phosphate; Fdx, ferredoxin; FUM, fumarate; F420, coenzyme F420; GA3P, glyceraldehyde-3-phosphate; G1P, glucose-1-phosphate; G6P, glucose-6-phosphate; (2H), low-potential hydride on unknown carrier; H+ (ext), proton-motive force; H4MPT, tetrahydromethanopterin; HSE, homoserine; IND, indole-3-glycerol-phosphate; KIV, 2-ketoisovalerate; MAL, malate; mDAP, meso-diaminopimelate; MFR, methanofuran; OAA, oxaloacetate; 2OG, 2-oxoglutarate; PEP, phosphoenolpyruvate; 3PG, 3-phosphoglycerate; PP, pyrophosphate; PPA, prephenate; PRPP, phosphoribosylpyrophosphate; PYR, pyruvate; R5P, ribose-5-phosphate; SDAP, succinyldiaminopimelate; SKA, shikimate; SUCC, succinate; S7P, sedoheptulose-7-phosphate; THDP, tetrahydrodipicolinate; X5P, xylulose-5-phosphate; ?, incomplete knowledge of pathway.
M. maripaludis contains typical genes for the second (formyltransferase [Ftr]) (Mmp1609) and third (cyclohydrolase [Mch]) (Mmp1191) steps in methanogenesis. The fourth step can be catalyzed by two different methylene tetrahydromethanopterin dehydrogenases, one that is coenzyme F420 dependent (Mtd; Mmp0372) and one that is H2 dependent (Hmd; Mmp0127). M. maripaludis has genes for both of these enzymes. In addition, M. maripaludis has one Hmd paralog of unknown function, Mmp1716 (1).
A typical gene encoding the enzyme for the fifth step, methylene tetrahydromethanopterin reductase (Mer; Mmp0058), is present. The enzyme for the sixth step, methyltetrahydromethanopterin-coenzyme M methyltransferase (Mtr; Mmp1560 to -1567), is a multisubunit complex. While M. maripaludis contains all of the known Mtr subunits, mtrF (Mmp1565) encodes what appears to be a fusion between a duplicated N-terminal region of MtrA and the traditional MtrF protein.
Methanothermobacter thermautotrophicus and Methanocaldococcus jannaschii have two sets of enzymes that catalyze the final step in methanogenesis, namely methyl coenzyme M reductases I (Mcr) and II (Mrt) (31, 90). M. maripaludis encodes only one methylreductase complex (Mmp1555 to -1559), and due to the high levels of homology between Mcr and Mrt, sequence similarity was insufficient to distinguish which complex is present. However, the operon configuration and position next to the Mtr operon are characteristic of Mcr (85).
The reduction of methyl-coenzyme M produces a mixed disulfide from coenzymes M and B (37), and heterodisulfide reductase (Hdr) reduces this disulfide to the free coenzymes (18). Like other obligate hydrogenotrophs, M. maripaludis encodes an Hdr with three subunits, A, B, and C. Two hdrBC clusters are present (Mmp0642-Mmp0643 and Mmp1054-Mmp1053), as are two hdrA genes, one for a selenocysteine-type (Mmp1697) protein, adjacent to the Vhu hydrogenase genes, and the other a cysteine-type (Mmp0825) protein, adjacent to the Vhc hydrogenase cluster. In contrast, Methanocaldococcus jannaschii contains only the selenocysteine-type enzyme (31).
(ii) Hydrogenases.
M. maripaludis contains six nickel-iron hydrogenases. Like Methanococcus voltae, M. maripaludis contains complete gene clusters for two coenzyme F420-reducing hydrogenases, one of which is a selenocysteine-containing cluster (Fru) (Mmp1382 to -1385) and the other of which is a cysteine-containing cluster (Frc) (Mmp0817 to -0820) cluster, and for two non-F420-reducing hydrogenases, which also contain selenocysteine (Vhu) (Mmp1692 to -1696) and cysteine (Vhc) (Mmp0821 to -0824) (6). M. maripaludis also encodes two separate multisubunit energy-conserving hydrogenases, Eha (Mmp1448 to -1467) and Ehb (Mmp0400, -0940, -1049, -1073, -1074, -1153, -1469, and -1621 to -1629), which are homologous to those first identified in Methanothermobacter thermautotrophicus (111). These hydrogenases are thought to couple ion gradients to certain endergonic reduction steps in methanogenesis and biosynthesis. Some members of these gene clusters are predicted to encode polyferredoxins and integral membrane proteins. Like Methanocaldococcus jannaschii, M. maripaludis Eha is encoded by one cluster that is colinear with the cluster found in Methanothermobacter thermautotrophicus. In contrast, while some Ehb subunits are encoded by one small cluster, most of the genes appear to be scattered throughout the genome (31). EhbH and EhbI are fused into one ORF (Mmp1626).
(iii) Formate dehydrogenases.
M. maripaludis contains two formate dehydrogenases (Fdh) (Mmp0138-Mmp0139 and Mmp1297-Mmp1298), either one of which enables growth on formate as an alternative to hydrogen and CO2 (122). A formate transporter (Mmp1301) is encoded upstream of the latter formate dehydrogenase. Interestingly, while Methanococcus vannielii has selenium-dependent and -independent formate dehydrogenases (47), both M. maripaludis α subunits contain selenocysteine. As a result, M. maripaludis cannot grow on formate in the absence of selenocysteine incorporation (91).
(iv) Selenocysteine-containing proteins.
Nine selenocysteine-containing proteins are encoded by the genome. Of these, eight are subunits of methanogenic enzymes, hydrogenases, or formate dehydrogenases. These selenocysteine-containing proteins are the B subunits of the molybdenum (Mmp0511)- and tungsten (Mmp1691)-containing formylmethanofuran dehydrogenases, one of the two Hdr subunit A proteins (Mmp1697), subunit A of the Fru hydrogenase (Mmp1382), subunits D (Mmp1696) and U (Mmp1693) of the Vhu hydrogenase, and the A subunits of both formate dehydrogenases (Mmp0138 and -1300). The ninth selenocysteine-containing protein is selenophosphate synthetase (SelD; Mmp0904). These observations are in agreement with the experimental detection of selenocysteine-containing proteins in M. maripaludis (92).
(v) Acetyl-coenzyme A synthesis.
M. maripaludis is capable of autotrophic growth and uses carbon monoxide dehydrogenase/acetyl-coenzyme A synthase (CODH/ACS, or Cdh) to fix CO2 and form acetyl-CoA. Like many other methanogens (76), the CODH/ACS genes in M. maripaludis are found in a single cluster (Mmp0980 to -0985). In addition to genes for CODH/ACS itself, gene Mmp0979 encodes an iron-sulfur protein that may be involved in electron transfer to CODH/ACS (67). Mmp0977, carried in an adjacent operon, is related to the nickel insertion protein for the Rhodospirillum rubrum carbon monoxide dehydrogenase and may be involved in biosynthesis or maturation of the prosthetic group (46).
As an alternative to autotrophy, M. maripaludis can assimilate acetate. Acetyl-CoA is then synthesized by acetyl-CoA synthetase. M. maripaludis has both the ADP-forming enzyme characteristic of Archaea and Eukarya (Mmp0253) (81) and the AMP-forming type (Mmp0148) found commonly in Bacteria and Eukarya. Methanocaldococcus jannaschii has only the former.
(vi) Reductive branch of TCA cycle.
Once acetyl-CoA is produced, it is converted by the incorporation of another CO2 into pyruvate by a multisubunit pyruvate:ferredoxin oxidoreductase (Por; Mmp1502 to -1507) (67) and thence to oxaloacetate by pyruvate carboxylase (Pyc; Mmp0340 and -0341) (80, 102). Oxaloacetate enters the tricarboxylic acid (TCA) cycle, which proceeds in the reductive direction. All of the enzymes for the reductive arm of the TCA cycle are present, leading from oxaloacetate to 2-oxoglutarate. 2-Oxoglutarate oxidoreductase (Kor; Mmp0003, -1315, -1316, and -1687) belongs to a family of multisubunit ferredoxin oxidoreductases that also includes pyruvate oxidoreductase (67). Unlike the other family members, the genes for the subunits of 2-oxoglutarate oxidoreductase are not all linked: the beta and gamma subunit genes are adjacent, but the alpha and delta (ferredoxin) subunits are each encoded in a different location. The oxidative branch of the TCA cycle is absent.
(vii) Glycolysis and gluconeogenesis.
As in Methanocaldococcus jannaschii, most of the genes for glycolysis and gluconeogenesis are present in M. maripaludis (99). These genes include those for two noncanonical phosphoglycerate mutases, Mmp0112 and Mmp1439 (33), an unusual ADP-dependent enzyme (Mmp1296) with both glucokinase and phosphofructokinase activities (95), and an archaeal-type fructose bisphosphate aldolase (Mmp0686) (103). Genes for glycogen synthesis and degradation are also present, including glycogen synthase (Mmp1294).
(viii) Nitrogen metabolism.
M. maripaludis can meet its nitrogen needs from several sources, including ammonia assimilation, the fixing of diatomic nitrogen, and the assimilation of alanine, the last of which is unusual for an archaeon (119). Ammonia is assimilated by a Iα-type glutamine synthetase (Mmp1206) (15). Glutamate synthase provides glutamate. As in other Archaea, the glutamate synthase large chain is encoded in three separate subunits (Mmp0080 to -0082), which correspond to domains of a single protein in Bacteria. The glutamate synthase small chain seems to be absent. The presence of an alanine dehydrogenase (Mmp1513), an alanine racemase (Mmp1512), and an alanine permease (Mmp1511) account for the unusual ability of M. maripaludis to use l- and d-alanine (Moore and Leigh, submitted).
M. maripaludis has a nitrogenase operon (51) that contains nifH, -D, -K, -E, -N, and -X (Mmp0853 and -0856 to -0860), encoding the nitrogenase complex and proteins that participate in the synthesis of the nitrogenase cofactor, as well as nifI1 (Mmp0854) and nifI2 (Mmp0855), encoding proteins that regulate nitrogenase activity (52, 53). Homocitrate, a component of the nitrogenase cofactor, is synthesized by NifV in Bacteria. The NifV homolog in M. maripaludis that is responsible for homocitrate synthesis is probably AksA (Mmp0153), which is also involved in the synthesis of 2-oxosuberate, an intermediate in biotin and coenzyme B synthesis (42).
(ix) Ferredoxins and iron-sulfur proteins.
Several steps in the pathways mentioned above require low-potential electrons. To transport these electrons, M. maripaludis encodes numerous iron-sulfur proteins. A total of 59 proteins are predicted to have 4Fe-4S centers, characterized by the motif CXXCXXCXXXC. These proteins include ferredoxins whose sole predicted function is to carry low-potential electrons and subunits of enzymes that catalyze low-potential redox reactions. Of particular interest are ferredoxins associated with hydrogenases and oxidoreductases, since several of these enzymes require low-potential electrons to drive enzymatic reactions. Among the hydrogenases, three ferredoxins are found in the Eha cluster, namely Mmp1463 (a polyferredoxin, containing many iron-sulfur centers), Mmp1464, and Mmp1465. The Ehb cluster contains two ferredoxins, Mmp1623 and Mm1624 (a polyferredoxin). Vhc contains Mmp0824 (a polyferredoxin), Vhu contains Mmp1692 (a polyferredoxin), Fru contains Mmp1384, and Frc contains Mmp0818. Among the multisubunit oxidoreductases, one subunit for each enzyme contains 4Fe-4S motifs as follows: indolepyruvate oxidoreductases (Ior) 1 and 2 (Mmp0316 and Mmp0713), 2-oxoisovalarate oxidoreductase (Vor; Mmp1273), 2-oxoglutarate oxidoreductase (Kor; Mmp1687), and pyruvate oxidoreductase (Por; Mmp1506). In addition, the fifth and sixth Por subunits, PorE and PorF (Mmp1503 and Mmp1502) (66) contain iron-sulfur motifs, as does Mmp0979, a PorE homolog predicted to be the electron carrier associated with CODH/ACS. The CODH/ACS alpha subunit (Mmp0985) also contains a 4Fe-4S motif that is presumably involved in electron transfer from the carrier to the active site.
Several additional enzymes have subunits containing 4Fe-4S motifs, which is indicative of electron transfer via unknown ferredoxins. These include the A and C subunits of the heterodisulfide reductases (Mmp0825, Mmp1697, Mmp1154, and Mmp1054), subunits H, F, and G of the formylmethanofuran dehydrogenase (Mmp1244 to -1246), the β subunits of the formate dehydrogenases (Mmp0139 and Mmp1297), and the large subunit of glutamate synthase (Mmp0081). Additional enzymes that include 4Fe-4S motifs are succinate dehydrogenase/fumarate reductase (Mmp1067) and one of the two thymidylate synthases (Mmp0986). In addition, the functions of many iron-sulfur proteins found in the genome are not known.
Flagella and chemotaxis.
M. maripaludis has the same arrangement of archaeal flagellar genes as that found in Methanococcus voltae (Mmp1666 to -1676) (50). Also present is an almost complete set of bacterial chemotaxis homologs (Mmp0925 to -0933). As in the α-Proteobacteria (2), no cheZ gene is present. Four homologs of sensory methyl-accepting chemotaxis proteins are present (Mmp0413, -0487, -0788, and -0929), suggesting the ability to respond to many different chemoattractants.
Transporters.
The M. maripaludis genome encodes 86 predicted transporter and binding proteins comprising approximately 48 transporter systems (see the supplemental material). The majority fall into the ABC transporter class. Iron transporters are highly prevalent, with one ferric iron ABC transporter cluster (Mmp0108 to -0110) and two iron-chelating ABC transporter clusters (Mmp0196 to -0198 and Mmp1181 to -1183) as well as a cluster of three ORFs that are predicted to be iron binding periplasmic proteins (Mmp1176 to -1178; however, Mmp1176 and -1177 encode separate amino and carboxyl ends of an iron binding protein and may represent a nonfunctional frame shift). There is also a homolog of a non-ABC ferrous iron uptake protein (Mmp0630) that is believed to be powered by ATP or GTP hydrolysis. M. maripaludis employs both molybdenum and tungsten as metal cofactors, and besides iron transporters, putative molybdenum transporters make up the other large group of transporters. There are four clusters of molybdenum ABC transporters (Mmp0205 to -0207, Mmp0504 to -0506, Mmp0514 to -0516, and Mmp1650 to -1652) as well as a separately encoded periplasmic molybdenum binding protein (Mmp1111). Two ORFs homologous to ABC sulfate transporter proteins (Mmp1518 to -1519) may actually comprise a tungsten transporter. M. maripaludis also has two members (Mmp0711 and -1108) of the CorA family of aqueous pore transporters, which transport divalent metal ions.
M. maripaludis can assimilate both ammonia and alanine as nitrogen sources, and two ammonia transporters (Mmp0065 and Mmp0068) and an alanine-cation symporter (Mmp1511) are encoded. M. maripaludis strains have been reported to take up a variety of amino acids (121), and several ORFs may encode additional amino acid transporters. Scattered around the genome are homologs of an ABC polar amino acid transporter system. While only one homolog of an ATP binding protein (Mmp0229) and a permease (Mmp0551) are seen, there are six putative periplasmic amino acid binding proteins (Mmp0455, -0550, -0712, -0770, -1224, and -1225). There is also a proline-Na+ symporter (Mmp0221) as well as a member of the amino acid-polyamine symporter-antiporter family (Mmp0850). An ABC phosphate transporter system is also present (Mmp1095 to -1099).
A series of three ORFs (Mmp0165 to -0167) showed homology to genes for drug efflux transporters, encoding the two components of an ABC drug efflux system separated by a predicted Na+-drug antiporter gene. Finally, a predicted Na+-H+ antiporter (Mmp0587) may allow for the interconversion of a sodium-motive force (produced by the methyltransferase step of methanogenesis) with a proton-motive force.
S layer.
M. maripaludis encodes an S-layer precursor (Mmp0383) with high sequence similarity to that of Methanococcus vannielii (3).
Amino acid synthesis. (i) Glutamate family.
As mentioned above, glutamine and glutamate are synthesized by glutamine synthetase and an archaeal-type glutamate synthase. Arginine is synthesized from glutamate via the intermediate ornithine. Homologs to all of the enzymes of the pathway except the initial enzyme are present (Mmp0013, -0063, -0073, -0116, -0553, -0897, -1013, -1101, and -1589). A homolog of the argJ gene (Mmp0897) is also present, which is characteristic of the acetyl cycle version of the pathway of ornithine biosynthesis (73) that is known to occur in Methanococcus vannielii (78). Like Methanocaldococcus jannaschii, the enzyme that catalyzes the first step in ornithine biosynthesis is unknown for M. maripaludis (73).
Methanocaldococcus jannaschii generates proline by the cyclization of ornithine, but the enzyme is evidently not a homolog of any known ornithine cyclodeaminase and has not been characterized (34). As in Methanocaldococcus jannaschii, the genes for proline biosynthesis in M. maripaludis are unknown.
(ii) Pyruvate family.
Alanine is produced from pyruvate by a type I aminotransferase (77). Five type I aminotransferases (Mmp0096, -1072, -1216, -1396, and -1527; see below) are encoded, and experiments are needed to determine their specificities. Unlike other Archaea, M. maripaludis also has the potential to use the alanine dehydrogenase pathway since this enzyme is present; however, the gene is required only for alanine utilization, not alanine synthesis (Moore and Leigh, submitted).
As in other methanogens (20), isoleucine is synthesized by the citramalate pathway by use of the enzyme (R)-citramalate synthase (CimA; Mmp1018), which was first identified in Methanocaldococcus jannaschii (43).
Leucine and valine are synthesized by standard pathways. Recently, the leuA gene encoding isopropylmalate synthase (Mmp1063) was distinguished from its paralogs in the citramalate and α-ketosuberate pathways by mutagenesis in M. maripaludis (36a). The presence of 2-oxoisovalerate oxidoreductase (Mmp1271 to -1273) suggests the additional ability to produce the branched-chain amino acids from the corresponding branched-chain fatty acids.
(iii) Aspartate family.
Aspartate is evidently synthesized by an aspartate aminotransferase (AspC) orthologous to the one identified in Methanothermobacter thermautotrophicus (Mmp0391) (110). Asparagine is synthesized by glutamine-hydrolyzing asparagine synthase (Mmp0918) (59); no homolog of ammonia-utilizing asparagine synthase is present. Hence, as in certain other organisms (79), a tRNA-independent pathway for asparagine synthesis appears to exist, despite the lack of an asparaginyl-tRNA synthetase (see above).
Threonine, methionine, and lysine are synthesized largely by standard pathways. Threonine and methionine share a common intermediate, homoserine, whose biosynthesis (Mmp1017, -1391, and -1702) is distinguished by separate enzymes for aspartate kinase (Mmp1017) and homoserine dehydrogenase (Mmp1702), in contrast to the bifunctional enzymes typical of Bacteria. From homoserine, threonine is synthesized by standard enzymes (Mmp0135 and -0295). Like the case for many other archaeal genomes, only one ORF (MetE; Mmp0401) for the methionine biosynthesis pathway was found (39). Nevertheless, labeling studies with Methanocaldococcus jannaschii showed that methionine was formed from aspartate (107). Lysine is evidently synthesized by the diaminopimelic acid pathway (Mmp0576, -0917, -0923, -1200, and -1398) (4), despite the lack of known orthologs for certain steps (99, 105).
(iv) Serine family.
Three steps synthesize serine from 3-phosphoglycerate. Like Methanothermobacter thermautotrophicus and Methanocaldococcus jannaschii (99, 105), M. maripaludis has SerA (Mmp1588) and SerB (Mmp0541) but is missing a homolog for SerC. Glycine is normally formed from serine by glycine hydroxymethyltransferase (GlyA), and while homologs were found in Methanothermobacter thermautotrophicus and Methanocaldococcus jannaschii (99, 105), no homolog is present in the M. maripaludis genome.
As in Methanocaldococcus jannaschii (99) and Methanothermobacter thermautotrophicus (105), no genes involved in cysteine synthesis were identified.
(v) Aromatic amino acids.
Chorismate is the branch point for phenylalanine, tyrosine, and tryptophan synthesis. Five of the seven enzymes in the known pathway of chorismate synthesis (Mmp0320, -0936, -1205, -1333, and -1394) were found in M. maripaludis. Like the case for many Euryarchaeota, homologs for the initial steps are not apparent (17, 99). Because erythrose-4-phosphate is not a precursor for chorismate in M. maripaludis (112), it seems likely that the initial steps in this pathway are different from those found in Bacteria. Recently, the presence of a dehydroquinate dehydratase, which catalyzes the third step in the pathway, was confirmed by the construction of a deletion mutation of Mmp1394 (87). Phenylalanine and tyrosine biosynthesis are initiated by chorismate mutase (Mmp0578), followed by separate prephenate dehydratase (PheA; Mmp1528) and prephenate dehydrogenase (TyrA; Mmp1514) enzymes (70). In contrast, in many other organisms aroQ (encoding chorismate mutase) is fused with other aromatic amino acid biosynthetic genes or a regulatory domain (14). The entire standard pathway for tryptophan synthesis is also present (Mmp1002 to -1008). In addition to the de novo pathway, M. maripaludis can synthesize the aromatic amino acids by reductive carboxylation of the aryl acids phenylacetate, p-hydroxyphenylacetate, and indoleacetate (87). Indolepyruvate oxidoreductase catalyzes the key step in this pathway, and M. maripaludis contains two homologs of this enzyme system, Mmp0315-Mmp0316 and Mmp0713-Mmp0714.
(vi) Histidine.
All of the genes for the biosynthesis of histidine (Mmp0051, -0256, -0280, -0417, -0548, -0947, -0968, -1082, -1083, -1216, -1690, and -1722) were found except that for histidinol phosphate phosphatase (HisJ).
Nucleotide synthesis. (i) Purines.
Almost all of the genes for the biosynthesis of purines are present in M. maripaludis, although there are variations from the pathways of Bacteria and Eukarya. Ribose phosphate is synthesized by the nonoxidative pentose phosphate pathway, as in Methanocaldococcus jannaschii (99, 126), and phosphoribosylpyrophosphate is synthesized by phosphoribosylpyrophosphate synthase (Mmp0410). Like the genomes of several other Archaea (55), the genome of M. maripaludis is missing the purN homolog for phosphoribosylglycinamide formyltransferase, but it does have the alternative enzyme purT (Mmp0123) (74). As with certain other Archaea and Bacteria, M. maripaludis encodes a two-subunit phosphoribosylformyl glycinamidine synthase (PurL [Mmp0179] and PurQ [Mmp0178]) (98). M. maripaludis does not encode PurS even though it is encoded by Methanocaldococcus jannaschii and is required for phosphoribosylformyl glycinamidine synthase activity in Bacillus subtilis (98).
As in other methanogens, N5-carboxyaminoimidazole ribonucleotide synthetase (PurK) and N5-carboxyaminoimidazole ribonucleotide mutase (PurE) activities seem to be fused into a single PurE homolog constituting phosphoribosylaminoimidazole carboxylase (Mmp0282) (106). For the final two steps in the de novo synthesis of IMP, which are normally catalyzed in Bacteria and Eukarya by a single bifunctional enzyme, only the archaeal IMP cyclohydrolase (PurO; Mmp1310) (35) has been identified; no homolog of PurH is encoded, and the gene for aminoimidazole carboxamide ribonucleotide transformylase has yet to be identified. All of the genes necessary for converting IMP to AMP and GMP are present (PurA, Mmp1432; PurB, Mmp0971; GuaA, Mmp1445; and GuaB, Mmp0133).
Many of the genes involved in the biosynthesis of ATP, dATP, GTP, and dGTP from AMP and GMP are present (AdkA, Mmp1031; Ndk, Mmp0283; and NrdD, Mmp0227). However, neither bacterial nor archaeal (55) ribonucleotide diphosphate reductase is present, and M. maripaludis presumably generates any dADP from dATP. No guanylate kinase, which catalyzes the formation of GDP from GMP, has been found in any of the Archaea (55).
(ii) Pyrimidines.
All of the genes involved in the biosynthesis of UTP, the precursor of pyrimidines, are found in the genome of M. maripaludis. Like some other archaeal genomes, the M. maripaludis genome has two ORFs encoding orotate phosphoribosyltransferase (PyrE; Mmp0079 and Mmp1492) (10, 55). M. maripaludis contains all of the genes required for the conversion of UTP to CTP (PyrG; Mmp0893) and thence to CDP (Ndk; Mmp0283). In addition, CTP is converted to dCTP by ribonucleoside triphosphate reductase (NrdD; Mmp0227). dTTP is evidently made from dCTP by a pathway that was recently elucidated in Methanocaldococcus jannaschii that avoids the production of toxic dUTP as an intermediate (64) as follows. A bifunctional dCTP deaminase-dUTP diphosphatase (Mmp1426) converts dCTP to dUMP, which is then converted to dTMP by thymidylate synthase (ThyA; Mmp0986 and Mmp1379). dTMP is converted to dTDP by thymidylate kinase (Tmk; Mmp1034) and thence to dTTP by nucleoside diphosphate kinase (Ndk; Mmp0227). dUTP diphosphatase (Dut; Mmp1075) may be present merely to scavenge dUTP produced by the spontaneous deamination of dCTP.
For dCDP synthesis, neither the typical ribonucleotide-diphosphate reductase nor the alternative enzyme found in some Archaea is present. Thus, as with dADP, any dCDP presumably comes entirely from the triphosphate.
(iii) Salvage pathways.
As in Methanothermobacter thermautotrophicus, M. maripaludis has a hypoxanthine phosphoribosyltransferase (Hpt; Mmp0145) that may also serve as a guanine phosphoribosyltransferase (96). M. maripaludis also has homologs of adenine phosphoribosyltransferase (Mmp0660) and uracil phosphoribosyltransferase (Mmp0680).
Coenzymes.
Numerous genes for the known biosynthetic pathways of the conventional coenzymes were identified. Among the coenzymes of methanogenesis, all of the coenzyme M biosynthesis genes in Methanocaldococcus jannaschii (32) were found to have orthologs in M. maripaludis. Several genes encoding steps in the synthesis of coenzyme F420 have been identified in Methanocaldococcus jannaschii (32), and orthologs were found in M. maripaludis. Unlike Methanocaldococcus jannaschii, M. maripaludis has three homologs of F390 synthetase, encoded by Mmp0160, Mmp0314, and Mmp0715 (115). However, methanococci are not known to produce F390, which suggests that these genes must have some other purpose. Only a few genes in methanopterin biosynthesis have been identified. Although a dihydropteroate synthase (MptH) was identified in Methanocaldococcus jannaschii (125), no clear ortholog could be found in M. maripaludis. A series of condensation (AksA; Mmp0153), spontaneous hydration, and oxidative decarboxylation (AksF; Mmp0880) reactions are involved in the synthesis of 2-oxosuberate, an intermediate in coenzyme B as well as biotin synthesis (42).
Aminotransferases.
M. maripaludis contains 11 ORFs that have been identified as aminotransferases. In general, aminotransferases are divided into four subgroups based on structural similarity (77). M. maripaludis has five aminotransferases from subgroup I (Mmp0096, -1072, -1216, -1396, and -1527), which is generally the most common subgroup, comprising aspartate, alanine, tyrosine, phenylalanine, and histidinol phosphate aminotransferases (77). Because the substrate specificities of the subgroup I aminotransferases are highly variable, it is often difficult to assign a specific function based only on homology to a characterized enzyme. However, Mmp1216 could be assigned unambiguously as histidinol phosphate aminotransferase (HisC) based on its homology to the enzyme from Halobacterium volcanii (16), in which the function was demonstrated by genetic complementation of a histidine auxotroph.
M. maripaludis has three subgroup II aminotransferases, encoded by Mmp0224, -0865, and -1101. Mmp0224 was assigned as glutamate-1-semialdehyde aminotransferase (HemL) based on its homology to the enzyme from Sulfolobus solfataricus (83). One subgroup III aminotransferase (Mmp0132) was identified whose homology suggests that it could account for the branched-chain amino acid aminotransferase activity detected in Methanococcus spp. (123, 124). One subgroup IV aminotransferase, Mmp0391, was assigned as aspartate aminotransferase (AspC) based on its homology to the enzyme from Methanothermobacter thermautotrophicus (110). A final aminotransferase, Mmp1680, does not belong to any of the recognized subgroups and was assigned as glucosamine-fructose-6-phosphate aminotransferase (GlmS) based on its homology to the enzyme from Thermus thermophilus (23).
Conclusions.
The M. maripaludis genome reveals much about the organism, with nearly half of the genes having assignable functions. Many of these functions will no doubt be confirmed as experiments continue with this genetically manipulable species. However, the absence of assignable functions is at least equally striking, with nearly half of the proteins designated as conserved hypothetical proteins. Genes with unassignable functions include 129 predicted proteins with no homologs in any other species. For these cases, the availability of genetic tools will also be important. In addition, the small genome, which is typical of many Archaea, facilitates studies of global regulation, which should complement genetic analyses.
As expected, the genome size of M. maripaludis is in line with those of its hydrogenotrophic relatives Methanocaldococcus jannaschii, Methanothermobacter thermautotrophicus, and Methanopyrus kandleri, and it is much smaller than the genome sequence of the nutritionally versatile Methanosarcina species. The M. maripaludis genome provides ample opportunity for comparisons with its nearest relative with a sequenced genome, Methanocaldococcus jannaschii. Approximately two-thirds of the M. maripaludis ORFs had their highest-scoring Blastp hits in Methanocaldococcus jannaschii, and many pathways and functions are held in common. The two species also share some novel features, including an unusual mechanism of replication initiation implied by the absence of a Cdc6 protein and the lack of a discrete transition in GC skew. However, the contrasts between M. maripaludis and Methanocaldococcus jannaschii are interesting as well. Some differences can be attributed to the growth temperatures, with M. maripaludis being mesophilic and Methanocaldococcus jannaschii being hyperthermophilic. Reverse gyrase, which is present only in Methanocaldococcus jannaschii, is needed only in hyperthermophiles, and the presence in M. maripaludis only of a molybdenum formylmethanofuran dehydrogenase also agrees with the correlation of molybdenum- and tungsten-containing enzymes with the temperature (40). A systematic difference in amino acid preferences for homologs between the two species has already been reported (36). Other differences include the presence of only one methyl-coenzyme M reductase in M. maripaludis and the absence of inteins from M. maripaludis. Insights into some of the contrasts between M. maripaludis and Methanocaldococcus jannaschii came from our analysis of top Blast hit categories. ORFs with top Blastp hits to groups more distant than Methanocaldococcus jannaschii are often clustered, possibly due to the lateral transfer of functionally related genes (61) or the loss of clustered genes in the Methanocaldococcus jannaschii lineage.
Supplementary Material
Acknowledgments
This work was supported by grant GM60403 from the National Institutes of Health, grant NCC 2-1273 from the NASA Astrobiology Institute, and grant DE-FG03-01ER15252 from the Department of Energy's Microbial Cell Program.
We thank Alberto J. L. Macario and Mona Malz for their contributions to this work.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Afting, C., E. Kremmer, C. Brucker, A. Hochheimer, and R. K. Thauer. 2000. Regulation of the synthesis of H2-forming methylenetetrahydromethanopterin dehydrogenase (Hmd) and of HmdII and HmdIII in Methanothermobacter marburgensis. Arch. Microbiol. 174:225-232. [DOI] [PubMed] [Google Scholar]
- 2.Armitage, J. P., and R. Schmitt. 1997. Bacterial chemotaxis: Rhodobacter sphaeroides and Sinorhizobium meliloti—variations on a theme? Microbiology 143:3671-3682. [DOI] [PubMed] [Google Scholar]
- 3.Bahl, H., H. Scholz, N. Bayan, M. Chami, G. Leblon, T. Gulik-Krzywicki, E. Shechter, A. Fouet, S. Mesnage, E. Tosi-Couture, P. Gounon, M. Mock, E. Conway de Macario, A. J. L. Macario, L. A. Fernandez-Herrero, G. Olabarria, J. Berenguer, M. J. Blaser, B. Kuen, W. Lubitz, M. Sara, P. H. Pouwels, C. P. Kolen, H. J. Boot, and S. Resch. 1997. Molecular biology of S-layers. FEMS Microbiol. Rev. 20:47-98. [DOI] [PubMed] [Google Scholar]
- 4.Bakhiet, N., F. W. Forney, D. P. Stahly, and L. Daniels. 1984. Lysine biosynthesis in Methanobacterium thermoautotrophicum is by the diaminopimelic acid pathway. Curr. Microbiol. 10:195-198. [Google Scholar]
- 5.Bell, S. D., and S. P. Jackson. 1998. Transcription and translation in Archaea: a mosaic of eukaryal and bacterial features. Trends Microbiol. 6:222-228. [DOI] [PubMed] [Google Scholar]
- 6.Berghofer, Y., K. Agha-Amiri, and A. Klein. 1994. Selenium is involved in the negative regulation of the expression of selenium-free [NiFe] hydrogenases in Methanococcus voltae. Mol. Gen. Genet. 242:369-373. [DOI] [PubMed] [Google Scholar]
- 7.Bernander, R. 1998. Archaea and the cell cycle. Mol. Microbiol. 29:955-961. [DOI] [PubMed] [Google Scholar]
- 8.Bernander, R. 2003. The archaeal cell cycle: current issues. Mol. Microbiol. 48:599-604. [DOI] [PubMed] [Google Scholar]
- 9.Bernander, R. 2000. Chromosome replication, nucleoid segregation and cell division in archaea. Trends Microbiol. 8:278-283. [DOI] [PubMed] [Google Scholar]
- 10.Bitan-Banin, G., R. Ortenberg, and M. Mevarech. 2003. Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. J. Bacteriol. 185:772-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bocquier, A. A., L. Liu, I. K. Cann, K. Komori, D. Kohda, and Y. Ishino. 2001. Archaeal primase: bridging the gap between RNA and DNA polymerases. Curr. Biol. 11:452-456. [DOI] [PubMed] [Google Scholar]
- 12.Boone, D. R., and R. W. Castenholz (ed.). 2001. The Archaea and the deeply branching phototrophic bacteria, 2nd ed., vol. 1. Springer-Verlag, New York, N.Y.
- 13.Bult, C. J., O. White, G. J. Olsen, L. Zhou, R. D. Fleischmann, G. G. Sutton, J. A. Blake, L. M. Fitzgerald, R. A. Clayton, J. D. Gocayne, A. R. Kerlavage, B. A. Dougherty, J. F. Tomb, M. D. Adams, C. I. Reich, R. Overbeek, E. F. Kirkness, K. G. Weinstock, J. M. Merrick, A. Glodek, J. L. Scott, N. S. Geoghagen, and J. C. Venter. 1996. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 273:1058-1073. [DOI] [PubMed] [Google Scholar]
- 14.Calhoun, D. H., C. A. Bonner, W. Gu, G. Xie, and R. A. Jensen. 2001. The emerging periplasm-localized subclass of AroQ chorismate mutases, exemplified by those from Salmonella typhimurium and Pseudomonas aeruginosa. Genome Biol. 2:RESEARCH0030.1-30.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen-Kupiec, R., C. J. Marx, and J. A. Leigh. 1999. Function and regulation of glnA in the methanogenic archaeon Methanococcus maripaludis. J. Bacteriol. 181:256-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conover, R. K., and W. F. Doolittle. 1990. Characterization of a gene involved in histidine biosynthesis in Halobacterium (Haloferax) volcanii: isolation and rapid mapping by transformation of an auxotroph with cosmid DNA. J. Bacteriol. 172:3244-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daugherty, M., V. Vonstein, R. Overbeek, and A. Osterman. 2001. Archaeal shikimate kinase, a new member of the GHMP-kinase family. J. Bacteriol. 183:292-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deppenmeier, U. 2002. The unique biochemistry of methanogenesis. Prog. Nucleic Acids Res. Mol. Biol. 71:223-283. [DOI] [PubMed] [Google Scholar]
- 19.Deppenmeier, U., A. Johann, T. Hartsch, R. Merkl, R. A. Schmitz, R. Martinez-Arias, A. Henne, A. Wiezer, S. Baumer, C. Jacobi, H. Bruggemann, T. Lienard, A. Christmann, M. Bomeke, S. Steckel, A. Bhattacharyya, A. Lykidis, R. Overbeek, H. P. Klenk, R. P. Gunsalus, H. J. Fritz, and G. Gottschalk. 2002. The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J. Mol. Microbiol. Biotechnol. 4:453-461. [PubMed] [Google Scholar]
- 20.Ekiel, I., I. C. P. Smith, and G. D. Sprott. 1984. Biosynthesis of isoleucine in methanogenic bacteria: a 13C NMR study. Biochemistry 23:1683-1687. [Google Scholar]
- 21.Eng, J. K., A. L. McCormack, and J. R. Yates. 1994. An approach to correlate tandem mass spectra of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976-989. [DOI] [PubMed] [Google Scholar]
- 22.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 23.Fernandez-Herrero, L. A., M. A. Badet-Denisot, B. Badet, and J. Berenguer. 1995. glmS of Thermus thermophilus HB8: an essential gene for cell-wall synthesis identified immediately upstream of the S-layer gene. Mol. Microbiol. 17:1-12. [DOI] [PubMed] [Google Scholar]
- 24.Ferreira, H., B. Butler-Cole, A. Burgin, R. Baker, D. J. Sherratt, and L. K. Arciszewska. 2003. Functional analysis of the C-terminal domains of the site-specific recombinases XerC and XerD. J. Mol. Biol. 330:15-27. [DOI] [PubMed] [Google Scholar]
- 25.Finn, M. W., and F. R. Tabita. 2003. Synthesis of catalytically active form III ribulose 1,5-bisphosphate carboxylase/oxygenase in archaea. J. Bacteriol. 185:3049-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galagan, J. E., C. Nusbaum, A. Roy, M. G. Endrizzi, P. Macdonald, W. FitzHugh, S. Calvo, R. Engels, S. Smirnov, D. Atnoor, A. Brown, N. Allen, J. Naylor, N. Stange-Thomann, K. DeArellano, R. Johnson, L. Linton, P. McEwan, K. McKernan, J. Talamas, A. Tirrell, W. Ye, A. Zimmer, R. D. Barber, I. Cann, D. E. Graham, D. A. Grahame, A. M. Guss, R. Hedderich, C. Ingram-Smith, H. C. Kuettner, J. A. Krzycki, J. A. Leigh, W. Li, J. Liu, B. Mukhopadhyay, J. N. Reeve, K. Smith, T. A. Springer, L. A. Umayam, O. White, R. H. White, E. Conway de Macario, J. G. Ferry, K. F. Jarrell, H. Jing, A. J. Macario, I. Paulsen, M. Pritchett, K. R. Sowers, R. V. Swanson, S. H. Zinder, E. Lander, W. W. Metcalf, and B. Birren. 2002. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12:532-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ganoza, M. C., M. C. Kiel, and H. Aoki. 2002. Evolutionary conservation of reactions in translation. Microbiol. Mol. Biol. Rev. 66:460-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gardner, W. L., and W. B. Whitman. 1999. Expression vectors for Methanococcus maripaludis: overexpression of acetohydroxyacid synthase and beta-galactosidase. Genetics 152:1439-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Godfrin-Estevenon, A. M., F. Pasta, and D. Lane. 2002. The parAB gene products of Pseudomonas putida exhibit partition activity in both P. putida and Escherichia coli. Mol. Microbiol. 43:39-49. [DOI] [PubMed] [Google Scholar]
- 30.Gordon, D., C. Desmarais, and P. Green. 2001. Automated finishing with autofinish. Genome Res. 11:614-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graham, D. E., N. Kyrpides, I. J. Anderson, R. Overbeek, and W. B. Whitman. 2001. Genome of Methanocaldococcus (Methanococcus) jannaschii. Methods Enzymol. 330:40-123. [DOI] [PubMed] [Google Scholar]
- 32.Graham, D. E., and R. H. White. 2002. Elucidation of methanogenic coenzyme biosyntheses: from spectroscopy to genomics. Nat. Prod. Rep. 19:133-147. [DOI] [PubMed] [Google Scholar]
- 33.Graham, D. E., H. Xu, and R. H. White. 2002. A divergent archaeal member of the alkaline phosphatase binuclear metalloenzyme superfamily has phosphoglycerate mutase activity. FEBS Lett. 517:190-194. [DOI] [PubMed] [Google Scholar]
- 34.Graupner, M., and R. H. White. 2001. Methanococcus jannaschii generates l-proline by cyclization of l-ornithine. J. Bacteriol. 183:5203-5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Graupner, M., H. Xu, and R. H. White. 2002. New class of IMP cyclohydrolases in Methanococcus jannaschii. J. Bacteriol. 184:1471-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haney, P. J., J. H. Badger, G. L. Buldak, C. I. Reich, C. R. Woese, and G. J. Olsen. 1999. Thermal adaptation analyzed by comparison of protein sequences from mesophilic and extremely thermophilic Methanococcus species. Proc. Natl. Acad. Sci. USA 96:3578-3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36a.Haydock, A. K., I. Porat, W. B. Whitman, and J. A. Leigh. 2004. Continuous culture of Methanococcus maripaludis under defined nutrient conditions. FEMS Microbiol. Lett. 238:85-91. [DOI] [PubMed] [Google Scholar]
- 37.Hedderich, R., J. Koch, D. Linder, and R. K. Thauer. 1994. The heterodisulfide reductase from Methanobacterium thermoautotrophicum contains sequence motifs characteristic of pyridine-nucleotide-dependent thioredoxin reductases. Eur. J. Biochem. 225:253-261. [DOI] [PubMed] [Google Scholar]
- 38.Hickey, A. J., E. Conway de Macario, and A. J. L. Macario. 2002. Transcription in the archaea: basal factors, regulation, and stress-gene expression. Crit. Rev. Biochem. Mol. Biol. 37:537-599. [DOI] [PubMed] [Google Scholar]
- 39.Higuchi, S., T. Kawashima, and M. Suzuki. 1999. Comparison of pathways for amino acid biosynthesis in archaebacteria using their genomic DNA sequences. Proc. Jpn. Acad. 75:241-245. [Google Scholar]
- 40.Hochheimer, A., R. Hedderich, and R. K. Thauer. 1998. The formylmethanofuran dehydrogenase isoenzymes in Methanobacterium wolfei and Methanobacterium thermoautotrophicum: induction of the molybdenum isoenzyme by molybdate and constitutive synthesis of the tungsten isoenzyme. Arch. Microbiol. 170:389-393. [DOI] [PubMed] [Google Scholar]
- 41.Hochheimer, A., D. Linder, R. K. Thauer, and R. Hedderich. 1996. The molybdenum formylmethanofuran dehydrogenase operon and the tungsten formylmethanofuran dehydrogenase operon from Methanobacterium thermoautotrophicum. Structures and transcriptional regulation. Eur. J. Biochem. 242:156-162. [DOI] [PubMed] [Google Scholar]
- 42.Howell, D. M., K. Harich, H. Xu, and R. H. White. 1998. Alpha-keto acid chain elongation reactions involved in the biosynthesis of coenzyme B (7-mercaptoheptanoyl threonine phosphate) in methanogenic archaea. Biochemistry 37:10108-10117. [DOI] [PubMed] [Google Scholar]
- 43.Howell, D. M., H. Xu, and R. H. White. 1999. (R)-Citramalate synthase in methanogenic archaea. J. Bacteriol. 181:331-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu, Z., E. P. Gogol, and J. Lutkenhaus. 2002. Dynamic assembly of MinD on phospholipid vesicles regulated by ATP and MinE. Proc. Natl. Acad. Sci. USA 99:6761-6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishino, Y., and I. K. Cann. 1998. The euryarchaeotes, a subdomain of Archaea, survive on a single DNA polymerase: fact or farce? Genes Genet. Syst. 73:323-336. [DOI] [PubMed] [Google Scholar]
- 46.Jeon, W. B., J. Cheng, and P. W. Ludden. 2001. Purification and characterization of membrane-associated CooC protein and its functional role in the insertion of nickel into carbon monoxide dehydrogenase from Rhodospirillum rubrum. J. Biol. Chem. 276:38602-38609. [DOI] [PubMed] [Google Scholar]
- 47.Jones, J. B., and T. C. Stadtman. 1981. Selenium-dependent and selenium-independent formate dehydrogenases of Methanococcus vannielii. Separation of the two forms and characterization of the purified selenium-independent form. J. Biol. Chem. 256:656-663. [PubMed] [Google Scholar]
- 48.Jones, W. J., M. J. B. Paynter, and R. Gupta. 1983. Characterization of Methanococcus maripaludis sp. nov., a new methanogen isolated from salt marsh sediment. Arch. Microbiol. 135:91-97. [Google Scholar]
- 49.Jones, W. J., W. B. Whitman, R. D. Fields, and R. S. Wolfe. 1983. Growth and plating efficiency of methanococci on agar media. Appl. Environ. Microbiol. 46:220-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kalmokoff, M. L., and K. F. Jarrell. 1991. Cloning and sequencing of a multigene family encoding the flagellins of Methanococcus voltae. J. Bacteriol. 173:7113-7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kessler, P. S., C. Blank, and J. A. Leigh. 1998. The nif gene operon of the methanogenic archaeon Methanococcus maripaludis. J. Bacteriol. 180:1504-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kessler, P. S., C. Daniel, and J. A. Leigh. 2001. Ammonia switch-off of nitrogen fixation in the methanogenic archaeon Methanococcus maripaludis: mechanistic features and requirement for the novel GlnB homologues, NifI1 and NifI2. J. Bacteriol. 183:882-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kessler, P. S., and J. A. Leigh. 1999. Genetics of nitrogen regulation in Methanococcus maripaludis. Genetics 152:1343-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klumpp, M., and W. Baumeister. 1998. The thermosome: archetype of group II chaperonins. FEBS Lett. 430:73-77. [DOI] [PubMed] [Google Scholar]
- 55.Koike, H., T. Kawashima, and M. Suzuki. 1999. Enzymes identified using genomic DNA sequences suggest some typical characteristics of de novo biosynthesis of purines in archaebacteria. Proc. Jpn. Acad. 75:263-268. [Google Scholar]
- 56.Komori, K., and Y. Ishino. 2001. Replication protein A in Pyrococcus furiosus is involved in homologous DNA recombination. J. Biol. Chem. 276:25654-25660. [DOI] [PubMed] [Google Scholar]
- 57.Komori, K., S. Sakae, H. Shinagawa, K. Morikawa, and Y. Ishino. 1999. A Holliday junction resolvase from Pyrococcus furiosus: functional similarity to Escherichia coli RuvC provides evidence for conserved mechanism of homologous recombination in Bacteria, Eukarya, and Archaea. Proc. Natl. Acad. Sci. USA 96:8873-8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lai, L., H. Yokota, L. W. Hung, R. Kim, and S. H. Kim. 2000. Crystal structure of archaeal RNase HII: a homologue of human major RNase H. Struct. Fold Des. 8:897-904. [DOI] [PubMed] [Google Scholar]
- 59.Larsen, T. M., S. K. Boehlein, S. M. Schuster, N. G. Richards, J. B. Thoden, H. M. Holden, and I. Rayment. 1999. Three-dimensional structure of Escherichia coli asparagine synthetase B: a short journey from substrate to product. Biochemistry 38:16146-16157. [DOI] [PubMed] [Google Scholar]
- 60.Latterich, M., K. U. Frohlich, and R. Schekman. 1995. Membrane fusion and the cell cycle: Cdc48p participates in the fusion of ER membranes. Cell 82:885-893. [DOI] [PubMed] [Google Scholar]
- 61.Lawrence, J. G. 2003. Gene organization: selection, selfishness, and serendipity. Annu. Rev. Microbiol. 57:419-440. [DOI] [PubMed] [Google Scholar]
- 62.Lei, M., and B. K. Tye. 2001. Initiating DNA synthesis: from recruiting to activating the MCM complex. J. Cell Sci. 114:1447-1454. [DOI] [PubMed] [Google Scholar]
- 63.Leroux, M. R., M. Fandrich, D. Klunker, K. Siegers, A. N. Lupas, J. R. Brown, E. Schiebel, C. M. Dobson, and F. U. Hartl. 1999. MtGimC, a novel archaeal chaperone related to the eukaryotic chaperonin cofactor GimC/prefoldin. EMBO J. 18:6730-6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li, H., H. Xu, D. E. Graham, and R. H. White. 2003. The Methanococcus jannaschii dCTP deaminase is a bifunctional deaminase and diphosphatase. J. Biol. Chem. 278:11100-11106. [DOI] [PubMed] [Google Scholar]
- 65.Lie, T. J., and J. A. Leigh. 2003. A novel repressor of nif and glnA expression in the methanogenic archaeon Methanococcus maripaludis. Mol. Microbiol. 47:235-246. [DOI] [PubMed] [Google Scholar]
- 66.Lin, W., and W. B. Whitman. 2004. The importance of porE and porF in the anabolic pyruvate oxidoreductase of Methanococcus maripaludis. Arch. Microbiol. 181:68-73. [DOI] [PubMed] [Google Scholar]
- 67.Lin, W. C., Y. L. Yang, and W. B. Whitman. 2003. The anabolic pyruvate oxidoreductase from Methanococcus maripaludis. Arch. Microbiol. 179:444-456. [DOI] [PubMed] [Google Scholar]
- 68.Liu, L., K. Komori, S. Ishino, A. A. Bocquier, I. K. Cann, D. Kohda, and Y. Ishino. 2001. The archaeal DNA primase: biochemical characterization of the p41-p46 complex from Pyrococcus furiosus. J. Biol. Chem. 276:45484-45490. [DOI] [PubMed] [Google Scholar]
- 69.Macario, A. J., M. Malz, and E. Conway de Macario. 2004. Evolution of assisted protein folding: the distribution of the main chaperoning systems within the phylogenetic domain archaea. Front. Biosci. 9:1318-1332. [DOI] [PubMed] [Google Scholar]
- 70.MacBeath, G., P. Kast, and D. Hilvert. 1998. A small, thermostable, and monofunctional chorismate mutase from the archaeon Methanococcus jannaschii. Biochemistry 37:10062-10073. [DOI] [PubMed] [Google Scholar]
- 71.Mackereth, C. D., C. H. Arrowsmith, A. M. Edwards, and L. P. McIntosh. 2000. Zinc-bundle structure of the essential RNA polymerase subunit RPB10 from Methanobacterium thermoautotrophicum. Proc. Natl. Acad. Sci. USA 97:6316-6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Malandrin, L., H. Huber, and R. Bernander. 1999. Nucleoid structure and partition in Methanococcus jannaschii: an archaeon with multiple copies of the chromosome. Genetics 152:1315-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marc, F., P. Weigel, C. Legrain, Y. Almeras, M. Santrot, N. Glansdorff, and V. Sakanyan. 2000. Characterization and kinetic mechanism of mono- and bifunctional ornithine acetyltransferases from thermophilic microorganisms. Eur. J. Biochem. 267:5217-5226. [DOI] [PubMed] [Google Scholar]
- 74.Marolewski, A., J. M. Smith, and S. J. Benkovic. 1994. Cloning and characterization of a new purine biosynthetic enzyme: a non-folate glycinamide ribonucleotide transformylase from E. coli. Biochemistry 33:2531-2537. [DOI] [PubMed] [Google Scholar]
- 75.Matsunaga, F., P. Forterre, Y. Ishino, and H. Myllykallio. 2001. In vivo interactions of archaeal Cdc6/Orc1 and minichromosome maintenance proteins with the replication origin. Proc. Natl. Acad. Sci. USA 98:11152-11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maupin-Furlow, J. A., and J. G. Ferry. 1996. Analysis of the CO dehydrogenase/acetyl-coenzyme A synthase operon of Methanosarcina thermophila. J. Bacteriol. 178:6849-6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mehta, P. K., T. I. Hale, and P. Christen. 1993. Aminotransferases: demonstration of homology and division into evolutionary subgroups. Eur. J. Biochem. 214:549-561. [DOI] [PubMed] [Google Scholar]
- 78.Meile, L., and T. Leisinger. 1984. Enzymes of arginine biosynthesis in methanogenic bacteria. Experientia 40:899-900. [Google Scholar]
- 79.Min, B., J. T. Pelaschier, D. E. Graham, D. Tumbula-Hansen, and D. Soll. 2002. Transfer RNA-dependent amino acid biosynthesis: an essential route to asparagine formation. Proc. Natl. Acad. Sci. USA 99:2678-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mukhopadhyay, B., V. J. Patel, and R. S. Wolfe. 2000. A stable archaeal pyruvate carboxylase from the hyperthermophile Methanococcus jannaschii. Arch. Microbiol. 174:406-414. [DOI] [PubMed] [Google Scholar]
- 81.Musfeldt, M., and P. Schonheit. 2002. Novel type of ADP-forming acetyl coenzyme A synthetase in hyperthermophilic archaea: heterologous expression and characterization of isoenzymes from the sulfate reducer Archaeoglobus fulgidus and the methanogen Methanococcus jannaschii. J. Bacteriol. 184:636-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ogrunc, M., D. F. Becker, S. W. Ragsdale, and A. Sancar. 1998. Nucleotide excision repair in the third kingdom. J. Bacteriol. 180:5796-5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Palmieri, G., M. Di Palo, A. Scaloni, S. Orru, G. Marino, and G. Sannia. 1996. Glutamate-1-semialdehyde aminotransferase from Sulfolobus solfataricus. Biochem. J. 320:541-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Perler, F. B. 2002. InBase: the Intein Database. Nucleic Acids Res. 30:383-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pihl, T. D., S. Sharma, and J. N. Reeve. 1994. Growth-phase-dependent transcription of the genes that encode the two methyl coenzyme M reductase isoenzymes and N5-methyltetrahydromethanopterin:coenzyme M methyltransferase in Methanobacterium thermoautotrophicum delta H. J. Bacteriol. 176:6384-6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Podobnik, M., P. McInerney, M. O'Donnell, and J. Kuriyan. 2000. A TOPRIM domain in the crystal structure of the catalytic core of Escherichia coli primase confirms a structural link to DNA topoisomerases. J. Mol. Biol. 300:353-362. [DOI] [PubMed] [Google Scholar]
- 87.Porat, I., B. W. Waters, Q. Teng, and W. B. Whitman. 2004. Two biosynthetic pathways for the aromatic amino acids in the archaeon Methanooccus maripaludis. J. Bacteriol. 186:4940-4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pritchett, M. A., J. K. Zhang, and W. W. Metcalf. 2004. Development of a markerless genetic exchange method for Methanosarcina acetivorans C2A and its use in construction of new genetic tools for methanogenic archaea. Appl. Environ. Microbiol. 70:1425-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rao, H. G., A. Rosenfeld, and J. G. Wetmur. 1998. Methanococcus jannaschii flap endonuclease: expression, purification, and substrate requirements. J. Bacteriol. 180:5406-5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rospert, S., D. Linder, J. Ellermann, and R. K. Thauer. 1990. Two genetically distinct methyl-coenzyme M reductases in Methanobacterium thermoautotrophicum strain Marburg and delta H. Eur. J. Biochem. 194:871-877. [DOI] [PubMed] [Google Scholar]
- 91.Rother, M., I. Mathes, F. Lottspeich, and A. Bock. 2003. Inactivation of the selB gene in Methanococcus maripaludis: effect on synthesis of selenoproteins and their sulfur-containing homologs. J. Bacteriol. 185:107-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rother, M., A. Resch, W. L. Gardner, W. B. Whitman, and A. Bock. 2001. Heterologous expression of archaeal selenoprotein genes directed by the SECIS element located in the 3′ non-translated region. Mol. Microbiol. 40:900-908. [DOI] [PubMed] [Google Scholar]
- 93.Rother, M., A. Resch, R. Wilting, and A. Bock. 2001. Selenoprotein synthesis in archaea. Biofactors 14:75-83. [DOI] [PubMed] [Google Scholar]
- 94.Rueda, S., M. Vicente, and J. Mingorance. 2003. Concentration and assembly of the division ring proteins FtsZ, FtsA, and ZipA during the Escherichia coli cell cycle. J. Bacteriol. 185:3344-3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sakuraba, H., I. Yoshioka, S. Koga, M. Takahashi, Y. Kitahama, T. Satomura, R. Kawakami, and T. Ohshima. 2002. ADP-dependent glucokinase/phosphofructokinase, a novel bifunctional enzyme from the hyperthermophilic archaeon Methanococcus jannaschii. J. Biol. Chem. 277:12495-12498. [DOI] [PubMed] [Google Scholar]
- 96.Sauer, J., and P. Nygaard. 1999. Expression of the Methanobacterium thermoautotrophicum hpt gene, encoding hypoxanthine (guanine) phosphoribosyltransferase, in Escherichia coli. J. Bacteriol. 181:1958-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Saunders, N. F., T. Thomas, P. M. Curmi, J. S. Mattick, E. Kuczek, R. Slade, J. Davis, P. D. Franzmann, D. Boone, K. Rusterholtz, R. Feldman, C. Gates, S. Bench, K. Sowers, K. Kadner, A. Aerts, P. Dehal, C. Detter, T. Glavina, S. Lucas, P. Richardson, F. Larimer, L. Hauser, M. Land, and R. Cavicchioli. 2003. Mechanisms of thermal adaptation revealed from the genomes of the Antarctic archaea Methanogenium frigidum and Methanococcoides burtonii. Genome Res. 13:1580-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Saxild, H. H., and P. Nygaard. 2000. The yexA gene product is required for phosphoribosylformylglycinamidine synthetase activity in Bacillus subtilis. Microbiology 146:807-814. [DOI] [PubMed] [Google Scholar]
- 99.Selkov, E., N. Maltsev, G. J. Olsen, R. Overbeek, and W. B. Whitman. 1997. A reconstruction of the metabolism of Methanococcus jannaschii from sequence data. Gene 197:GC11-GC26. [DOI] [PubMed] [Google Scholar]
- 100.Shechter, D. F., C. Y. Ying, and J. Gautier. 2000. The intrinsic DNA helicase activity of Methanobacterium thermoautotrophicum delta H minichromosome maintenance protein. J. Biol. Chem. 275:15049-15059. [DOI] [PubMed] [Google Scholar]
- 101.Sheikh, S., S. F. O'Handley, C. A. Dunn, and M. J. Bessman. 1998. Identification and characterization of the Nudix hydrolase from the archaeon, Methanococcus jannaschii, as a highly specific ADP-ribose pyrophosphatase. J. Biol. Chem. 273:20924-20928. [DOI] [PubMed] [Google Scholar]
- 102.Shieh, J. S., and W. B. Whitman. 1987. Pathway of acetate assimilation in autotrophic and heterotrophic methanococci. J. Bacteriol. 169:5327-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Siebers, B., H. Brinkmann, C. Dorr, B. Tjaden, H. Lilie, J. van der Oost, and C. H. Verhees. 2001. Archaeal fructose-1,6-bisphosphate aldolases constitute a new family of archaeal type class I aldolase. J. Biol. Chem. 276:28710-28718. [DOI] [PubMed] [Google Scholar]
- 104.Slesarev, A. I., K. V. Mezhevaya, K. S. Makarova, N. N. Polushin, O. V. Shcherbinina, V. V. Shakhova, G. I. Belova, L. Aravind, D. A. Natale, I. B. Rogozin, R. L. Tatusov, Y. I. Wolf, K. O. Stetter, A. G. Malykh, E. V. Koonin, and S. A. Kozyavkin. 2002. The complete genome of hyperthermophile Methanopyrus kandleri AV19 and monophyly of archaeal methanogens. Proc. Natl. Acad. Sci. USA 99:4644-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Smith, D. R., L. A. Doucette-Stamm, C. Deloughery, H. Lee, J. Dubois, T. Aldredge, R. Bashirzadeh, D. Blakely, R. Cook, K. Gilbert, D. Harrison, L. Hoang, P. Keagle, W. Lumm, B. Pothier, D. Qiu, R. Spadafora, R. Vicaire, Y. Wang, J. Wierzbowski, R. Gibson, N. Jiwani, A. Caruso, D. Bush, and J. N. Reeve. 1997. Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: functional analysis and comparative genomics. J. Bacteriol. 179:7135-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sorensen, I. S., and G. Dandanell. 1997. Identification and sequence analysis of Sulfolobus solfataricus purE and purK genes. FEMS Microbiol. Lett. 154:173-180. [DOI] [PubMed] [Google Scholar]
- 107.Sprott, G. D., I. Ekiel, and G. B. Patel. 1993. Metabolic pathways in Methanococcus jannaschii and other methanogenic bacteria. Appl. Environ. Microbiol. 59:1092-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Stathopoulos, C., I. Ahel, K. Ali, A. Ambrogelly, H. Becker, S. Bunjun, L. Feng, S. Herring, C. Jacquin-Becker, H. Kobayashi, D. Korencic, B. Krett, N. Mejlhede, B. Min, H. Nakano, S. Namgoong, C. Polycarpo, G. Raczniak, J. Rinehart, G. Rosas-Sandoval, B. Ruan, J. Sabina, A. Sauerwald, H. Toogood, D. Tumbula-Hansen, M. Ibba, and D. Soll. 2001. Aminoacyl-tRNA synthesis: a postgenomic perspective. Cold Spring Harbor Symp. Quant. Biol. 66:175-183. [DOI] [PubMed] [Google Scholar]
- 109.Tabb, D. L., W. H. McDonald, and J. R. Yates III. 2002. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1:21-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tanaka, T., S. Yamamoto, T. Moriya, M. Taniguchi, H. Hayashi, H. Kagamiyama, and S. Oi. 1994. Aspartate aminotransferase from a thermophilic formate-utilizing methanogen, Methanobacterium thermoformicicum strain SF-4: relation to serine and phosphoserine aminotransferases, but not to the aspartate aminotransferase family. J. Biochem. (Tokyo) 115:309-317. [DOI] [PubMed] [Google Scholar]
- 111.Tersteegen, A., and R. Hedderich. 1999. Methanobacterium thermoautotrophicum encodes two multisubunit membrane-bound [NiFe] hydrogenases. Transcription of the operons and sequence analysis of the deduced proteins. Eur. J. Biochem. 264:930-943. [DOI] [PubMed] [Google Scholar]
- 112.Tumbula, D. L., Q. Teng, M. G. Bartlett, and W. B. Whitman. 1997. Ribose biosynthesis and evidence for an alternative first step in the common aromatic amino acid pathway in Methanococcus maripaludis. J. Bacteriol. 179:6010-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tumbula, D. L., and W. B. Whitman. 1999. Genetics of Methanococcus: possibilities for functional genomics in Archaea. Mol. Microbiol. 33:1-7. [DOI] [PubMed] [Google Scholar]
- 114.van Nimwegen, E. 2003. Scaling laws in the functional content of genomes. Trends Genet. 19:479-484. [DOI] [PubMed] [Google Scholar]
- 115.Vermeij, P., R. J. van der Steen, J. T. Keltjens, G. D. Vogels, and T. Leisinger. 1996. Coenzyme F390 synthetase from Methanobacterium thermoautotrophicum Marburg belongs to the superfamily of adenylate-forming enzymes. J. Bacteriol. 178:505-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang, J. C. 1996. DNA topoisomerases. Annu. Rev. Biochem. 65:635-692. [DOI] [PubMed] [Google Scholar]
- 117.Wang, T., Y. Zhang, W. Chen, Y. Park, R. J. Lamont, and M. Hackett. 2002. Reconstructed protein arrays from 3D HPLC/tandem mass spectrometry and 2D gels: complementary approaches to Porphyromonas gingivalis protein expression. Analyst 127:1450-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Washburn, M. P., D. Wolters, and J. R. Yates III. 2001. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19:242-247. [DOI] [PubMed] [Google Scholar]
- 119.Whitman, W. B., D. R. Boone, and Y. Koga. 2001. Methanococcaceae, p. 236-240. In D. R. Boone, R. W. Castenholz, and G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer-Verlag, New York, N.Y. [Google Scholar]
- 120.Whitman, W. B., J. Shieh, S. Sohn, D. S. Caras, and U. Premachandran. 1986. Isolation and characterization of 22 mesophilic methanococci. Syst. Appl. Microbiol. 7:235-240. [Google Scholar]
- 121.Whitman, W. B., S. Sohn, S. Kuk, and R. Xing. 1987. Role of amino acids and vitamins in nutrition of mesophilic Methanococcus spp. Appl. Environ. Microbiol. 53:2373-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wood, G. E., A. K. Haydock, and J. A. Leigh. 2003. Function and regulation of the formate dehydrogenase genes of the methanogenic archaeon Methanococcus maripaludis. J. Bacteriol. 185:2548-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xing, R. Y., and W. B. Whitman. 1992. Characterization of amino acid aminotransferases of Methanococcus aeolicus. J. Bacteriol. 174:541-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Xing, R. Y., and W. B. Whitman. 1991. Characterization of enzymes of the branched-chain amino acid biosynthetic pathway in Methanococcus spp. J. Bacteriol. 173:2086-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Xu, H., R. Aurora, G. D. Rose, and R. H. White. 1999. Identifying two ancient enzymes in Archaea using predicted secondary structure alignment. Nat. Struct. Biol. 6:750-754. [DOI] [PubMed] [Google Scholar]
- 126.Yu, J. P., J. Ladapo, and W. B. Whitman. 1994. Pathway of glycogen metabolism in Methanococcus maripaludis. J. Bacteriol. 176:325-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zinder, S. H. 1993. Physiological ecology of methanogens, p. 128-206. In J. G. Ferry (ed.), Methanogenesis. Chapman and Hall, London, United Kingdom.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.