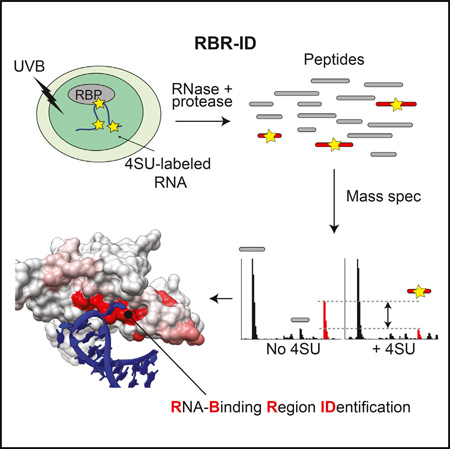
SUMMARY
Interactions between noncoding RNAs and chromatin proteins play important roles in gene regulation, but the molecular details of most of these interactions are unknown. Using protein-RNA photo-crosslinking and mass spectrometry on embryonic stem cell nuclei, we identified and mapped, at peptide resolution, the RNA-binding regions in ~800 known and previously unknown RNA-binding proteins, many of which are transcriptional regulators and chromatin modifiers. In addition to known RNA-binding motifs, we detected several protein domains previously unknown to function in RNA recognition, as well as non-annotated and/or disordered regions, suggesting that many functional protein-RNA contacts remain unexplored. We identified RNA-binding regions in several chromatin regulators, including TET2, and validated their ability to bind RNA. Thus, proteomic identification of RNA-binding regions (RBR-ID) is a powerful tool to map protein-RNA interactions and will allow rational design of mutants to dissect their function at a mechanistic level.
Graphical Abstract
INTRODUCTION
In addition to their central roles as messengers and translators of genetic information, RNA molecules have key roles in gene regulation (Bonasio and Shiekhattar, 2014; Holoch and Moazed, 2015). Nowhere is this more evident than in the nucleus of mammalian cells, where many classes of poorly understood noncoding RNAs (ncRNAs) continue to be discovered (Goff and Rinn, 2015; Quinn and Chang, 2016; Rinn and Chang, 2012; Wilusz et al., 2009).
Although some RNAs catalyze chemical reactions, they usually require association with proteins to function properly. Thus, it is reasonable to assume that the thousands of ncRNAs whose biochemical and biological roles are largely unknown exert their functions via protein-RNA interactions. Identifying proteins that interact with a given ncRNA has become a successful strategy to begin to dissect its biological roles; for example, the identification of Xist-associated proteins has provided important advances in understanding how this long ncRNA (lncRNA) controls X chromosome inactivation (Chu et al., 2015; McHugh et al., 2015; Minajigi et al., 2015).
To identify RNA-binding proteins (RBPs) in a more general, unbiased way, multiple groups have used polyA+ selection followed by mass spectrometry (MS). These studies identified hundreds of previously unknown RBPs bound to mRNAs in human cell lines (Baltz et al., 2012; Beckmann et al., 2015; Castello et al., 2012; Conrad et al., 2016) and mouse embryonic stem cells (ESCs) (Kwon et al., 2013). However, most small RNAs and many lncRNAs are not polyadenylated, including abundant nuclear RNAs like MALAT1 (Brown et al., 2012; Wilusz et al., 2012), enhancer-derived RNAs (eRNAs) (Lam et al., 2014), and circular RNAs (Wilusz, 2016). Proteins interacting with these and other polyA− ncRNAs have thus been missed by existing approaches.
The identification of a protein as being RNA-associated is only the first step toward understanding the role of RNA interactions in its biochemistry. Mapping RNA-binding residues allows for the rational design of mutants to study the functional relationship between the protein and its cognate RNAs (Bonasio et al., 2014; Kaneko et al., 2014a). Prediction of RNA-binding regions (RBRs) within RBPs is facilitated by the existence of well-characterized structural motifs that function as conserved RNA-binding domains (RBDs). The distinction between these two terms is important for this study: we refer to “RBRs” as minimal protein regions that make direct physical contacts with RNA (Bonasio et al., 2014), whereas “RBDs” are well-known, conserved domains that can be predicted from the primary sequence and typically function as RNA binders (Lunde et al., 2007). Examples of the latter category are the RNA-recognition motif (RRM) (Maris et al., 2005), the hnRNPK-homology domain (KH) (Grishin, 2001), and the double-stranded RNA-binding domain (dsRBD) (Chang and Ramos, 2005).
Until recently, it was widely believed that RBPs would contain one or more known RBDs and that the protein-RNA interaction could be assumed to take place within these domains. However, this simple concept has been challenged as the number of “non-canonical” RBPs (proteins that bind RNA without containing a classical RBD) continue to increase, especially for proteins that interface with chromatin and noncoding RNAs (G Hendrickson et al., 2016). Because the RBRs of these proteins cannot be predicted a priori, we and others have resorted to various biochemical methods to identify them with candidate-based, low-throughput methods (Bonasio et al., 2014; Kaneko et al., 2014a; Kaneko et al., 2010; Saldaña-Meyer et al., 2014).
Here, we report a high-throughput approach that exploits protein-RNA photocrosslinking and quantitative MS to identify proteins and protein regions interacting with RNA in vivo, regardless of the RNA polyadenylation status. As this approach not only identifies RNA-binding proteins, but also their respective RNA-binding regions, we named the technique RBR-ID. We applied RBR-ID to nuclei from mouse ESCs and identified RBRs within 803 proteins, more than half of which had not previously been reported as RBPs. We validated six RBRs, two in known RBPs whose mode of interaction with RNA was unknown and four in chromatin-associated proteins that had not been previously shown to bind RNA. Rational mutant design informed by RBR-ID nearly abolished RNA binding in vivo for these proteins, demonstrating the predictive power and practical utility of our technique for characterizing functional protein-RNA interactions.
RESULTS
Development and Optimization of RBR-ID
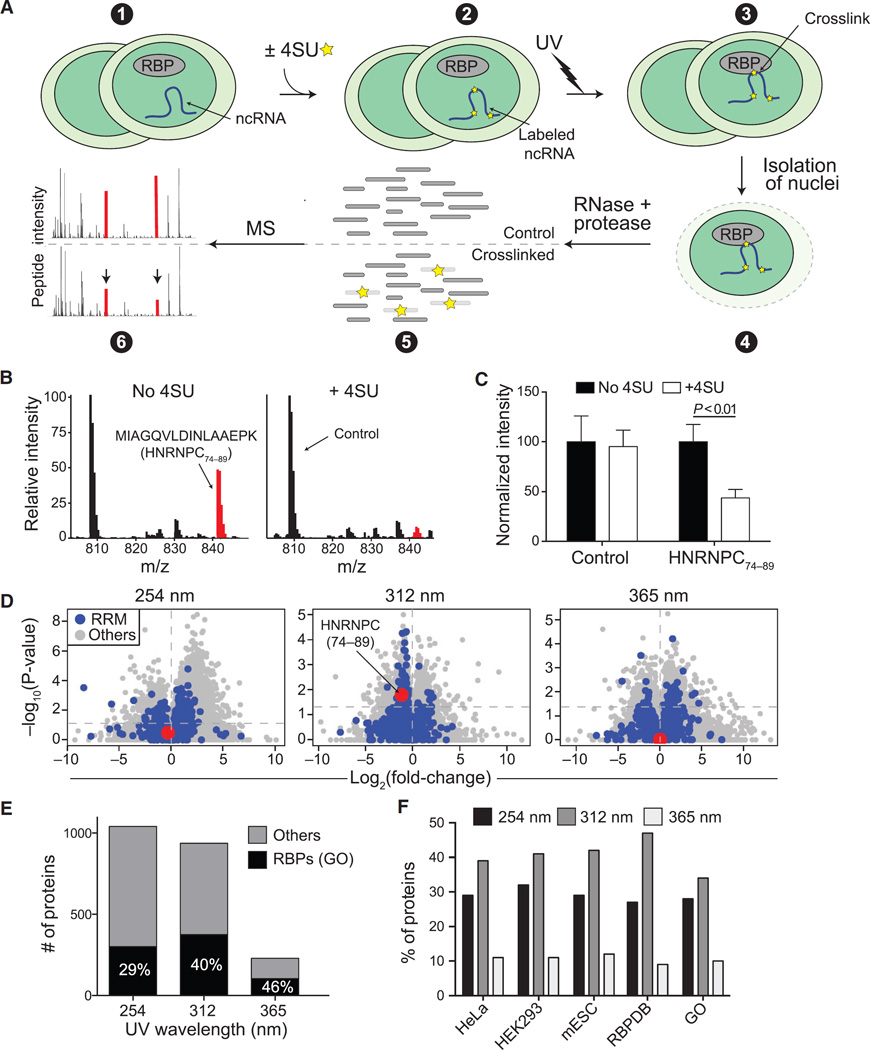
UV-mediated protein-RNA photocrosslinking generates adducts of RBPs with the covalent attachment localized at or near the site of physical interaction because of the short range of this type of crosslinking (Greenberg, 1979). Thus, it should be possible to detect the RBR of a protein by MS, as an RNA-crosslinked peptide would have a different mass, causing the intensity of the signal for the non-crosslinked peptide to be lower in the irradiated sample (Figure 1A).
Figure 1. Development and Optimization of RBR-ID.
(A) Mouse ESCs were pulsed with 4SU or not treated (1 and 2) and irradiated with different UV wavelengths (3). We isolated nuclei (4) and digested the crosslinked extracts with protease and RNase, producing a mixture of crosslinked and uncrosslinked peptides (5). Covalent crosslinks to RNA alters the peptide mass, and the mass spectrum of the corresponding uncrosslinked peptide decreases in intensity (6; red peaks).
(B) Example averaged spectra from comparable retention time windows from untreated (left) and 4SU-treated ESCs (right). UV (312 nm) crosslinking caused decreased intensity of the highlighted spectrum in the 4SU sample.
(C) Quantification of the extracted chromatogram for the control and HNRNPC peptides highlighted in (B). Bars indicate the average of the peak intensities normalized to the untreated sample (no 4SU) in six replicates + SEM.
(D) Volcano plots showing log-fold changes in peptide intensities on the x axis and p values on the y axis for ±UV (254 nm) and ±4SU (312 and 365 nm). Peptides overlapping annotated RRM domains are in blue. The RNA-binding peptide from HNRNPC is highlighted in red.
(E) Number of proteins with consistently (p < 0.05) depleted peptides and annotated as RBPs in the GO database (black) or not (gray).
(F) Percentage of known RBPs according to the indicated studies and databases that were identified using different UV wavelengths for the crosslink.
See also Figure S1.
Comparing mass spectra of UV-irradiated ESCs pulsed or not with 4-thiouridine (4SU), a uridine analog selectively activated by long-wavelength UV (Favre et al., 1986; Hafner et al., 2010), we observed that most peptides were unchanged in intensity (Figure 1B, black lines) but some were depleted in the 4SU-treated samples; for example, peptide 74–89 from HNRNPC (Figure 1B, red lines). We performed the experiment in three biological replicates, each acquired in duplicate MS runs, and noticed that the same HNRNPC peptide was consistently depleted by more than 50% (Figure 1C), suggesting that 4SU incorporation and UVB-mediated crosslinking had caused a fraction of these peptides to change mass and thus not be counted toward the peak intensity of the non-crosslinked peptide. As HNRNPC is a well-known RBP (Görlach et al., 1992) and the HRNPC74–89 peptide overlaps its RRM, we concluded that this analysis had the potential to reveal protein-RNA contacts in the entire proteome.
There are different ways to crosslink RNA to proteins. Conventional UV crosslinking exploits the excitation peak of natural nucleotides in the short-wavelength UVC range (254 nm) (Hockensmith et al., 1986; Stiege et al., 1988), whereas incorporation of 4SU allows for more selective and less damaging crosslinking, typically using 365 nm UV (UVA). We previously showed that some protein-RNA interactions can only be captured with 4SU-aided crosslinking when an intermediate wavelength of 312 nm (UVB) is used (Kaneko et al., 2013). We irradiated ESCs with the three different wavelengths and compared mass spectra obtained from isolated nuclei with those obtained from non-crosslinked samples (no 4SU treatment for 312 and 365 nm UV; no UV irradiation for 254 nm). Irradiation with 312 nm yielded the best compromise between sensitivity and specificity (Figures 1D and 1E); 254 nm UVC yielded a large number of peptides with decreased intensities but with no preference for peptides overlapping known RRMs (Figure 1D, blue dots); whereas, 365 nm UVA were too weak to consistently deplete a large number of peptides. For example, the RNA-binding peptide HNRNPC74–89 was significantly depleted only upon irradiation at 312 nm (Figure 1D, red dot), similar to the RNA-binding peptides from SNRNP70, SPEN, and HNRNPM (Figure S1A), three other well-known RBPs. Consistent with this, 254 nm UVC identified more candidates compared to 312 nm UVB (Figure 1E), but a smaller fraction of them were annotated as RBPs, suggesting that the increased sensitivity came at the cost of decreased specificity. Crosslinking with 365 nm UVA resulted in more accurate identification (46% versus 40% of proteins identified were RBPs) than 312 nm UVB but with a considerable loss in sensitivity (Figure 1E). Overall, 312 nm UVB crosslinking identified a larger fraction of all known RBPs (Figure 1F), whether from previous empirically determined lists from HeLa, HEK293, or mouse ESCs, or from digital annotations such as the GO and Toronto RBP databases (Cook et al., 2011). These proteome-wide observations were consistent with the higher efficiency of 4SU-dependent protein-RNA crosslinking, as measured by RNA pull-down followed by western blot for the U1-SNRNP70 complex (Figure S1B).
There was no correlation between the depletion of peptides by 4SU after UV crosslinking and depletion of peptides by 4SU alone (Figure S1C), suggesting that changes in protein isoform representation or post-translational modification in response to the 4SU treatment could not explain the bulk of depletion observed after UV. Furthermore, although some peptides showed an increase in apparent abundance upon 4SU crosslinking (Figure 1D), the majority of significant UV-induced changes were toward depletion in +4SU conditions (Figure S1D).
We conclude that RBR-ID can identify known and unknown RBPs and that comparison of 4SU-treated versus untreated samples after irradiation with 312 nm UVB is the best compromise between sensitivity and specificity.
Protein-Level Analyses
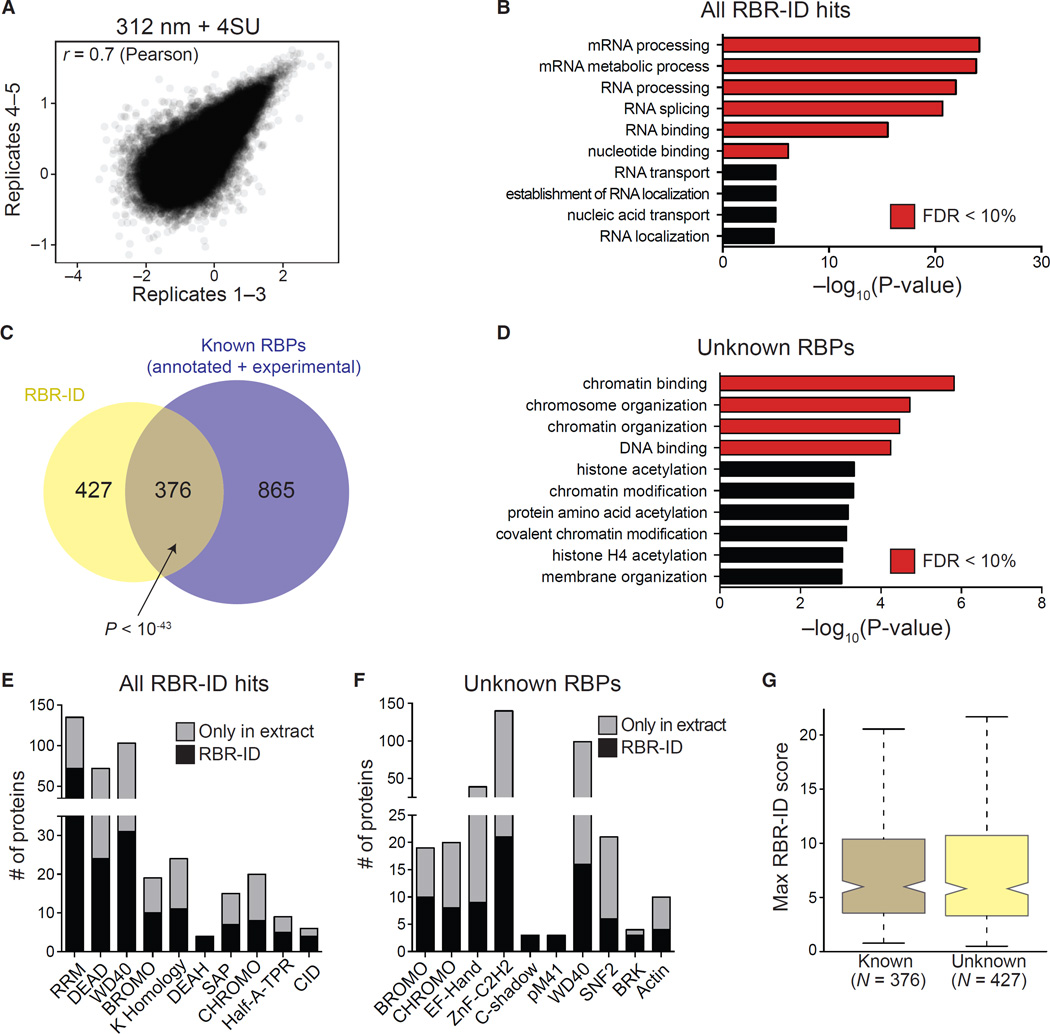
To increase the confidence in peptide quantification, we acquired two technical replicates each for two additional biological replicates of 312 nm UVB irradiation ± 4SU. Despite the noise in each individual run, once aggregated, the first set (three replicates) and second set (two replicates) of RBR-ID results were consistent (Figure 2A), suggesting that high replication could reduce artifactual identification of RBRs due to fluctuations in the MS signal.
Figure 2. Protein-Level Analyses of Proteins Identified by RBR-ID.
(A) Scatterplot showing log-converted and normalized average intensities for peptides from biological replicates 1–3 and the additional replicates 4 and 5.
(B) Top ten enriched GO terms (biological process and molecular function) for primary RBR-ID protein hits. p values are plotted on the x axis, and terms with false discovery rate (FDR) < 10% are shown in red.
(C) Overlap of RBR-ID protein hits and all known RBPs, both experimentally identified and annotated in databases. p value is from the hypergeometric distribution.
(D) Top ten enriched GO terms as in (B) but only for the RBR-ID protein hits not found in the set of already known RBPs.
(E and F) Top ten non-redundant protein domains enriched in the primary RBR-ID protein hits (E) or only in the unknown RBP set (F). The black section of the stacked bar plots indicates the number of proteins containing the domain and found in the primary RBR-ID candidate list; the gray section indicates the number of proteins not in the RBR-ID list but detected by MS in the ESC nuclear extract.
(G) Tukey boxplot for the distribution of maximum RBR-ID scores per protein comparing known RBPs and unknown putative RBPs from (C).
See also Tables S1, S2, S3, S4, S5, S6, and S7 and Figure S2.
In total, we detected 75,441 unique peptides from 4,929 proteins in mouse ESC nuclei; of these, 1,475 were consistently (p < 0.05) depleted by 4SU and UV, but not by 4SU alone (Table S1). These peptides belonged to 814 proteins (corresponding to 803 unique protein symbols), which we considered “primary hits” (Table S2). An additional set of 721 proteins identified with relaxed requirement (0.05 < p < 0.1) was used for some of the subsequent analyses and, along with the primary hits, constitutes our “extended” set (Table S2). GO annotations for the primary hits were enriched for functional terms related to RNA metabolism and function, including “RNA binding” (Figure 2B; Table S3). Primary hits also showed large overlaps with a variety of existing RBP lists (Figure S2A), either empirically determined (Baltz et al., 2012; Beckmann et al., 2015; Castello et al., 2012; Conrad et al., 2016; Kwon et al., 2013) or digitally annotated (Cook et al., 2011; UniProt Consortium, 2015). Based on these lists, 376 of the 803 primary candidates were previously known RBPs (Figure 2C), a significant overlap (p < 10−43, hypergeometric distribution). Among the previously known RBPs that were not recovered by RBR-ID, a large proportion (~40%, Figure S2A, compare bottom left with bottom right) could not be detected at all in ESC nuclei, likely because they were not expressed or were localized to the cytoplasm, as shown by their enrichment for ribosomal proteins and translation factors (Table S4, left). Nonetheless, 865 previously known RBPs were detectable in the ESC nuclear fraction and not recovered by RBR-ID (Figure 2C). This set of proteins was also enriched for ribosomal biogenesis and translation-related GO terms (Table S4, right), suggesting that some might be present in the nucleus but only bind RNA in the cytosol. It is also possible that a substantial number of true nuclear RBPs cannot be crosslinked efficiently to 4SU.
We then turned our attention to the 427 previously unknown RBPs that were identified by RBR-ID. Even when considering only the detectable nuclear proteome as background, these non-canonical RBPs were enriched for GO terms related to gene regulation and chromatin biology (Figure 2D; Table S5), consistent with the notion that many chromatin-associated proteins bind RNA (G Hendrickson et al., 2016; Khalil et al., 2009). Non-canonical RBPs identified by RBR-ID also contained different types of protein domains. The list of primary hits as a whole was enriched in known RBDs (RRM and KH) and RNA helicase domains, DEAD and DEAH (Figure 2E; Table S6), whereas the 427 unknown RBPs were enriched for chromatin-related domains, such as bromodomain and chromodomain, (Figure 2F; Table S7), which bind acetylated and methylated histones, respectively (Taverna et al., 2007), and the SNF2-related domain found in ATP-dependent chromatin remodelers (Eisen et al., 1995).
To estimate the confidence of identification for these proteins, we calculated an RBR-ID “score” for each peptide that captured both the extent of depletion (i.e., the log-converted fold-change between 4SU-treated and non-treated cells) and the consistency across replicates (i.e., the p value for the depletion; see Experimental Procedures). The previously unknown 427 RBPs had a distribution of RBR-ID scores comparable to that of the known 376 RBPs recovered RBR-ID (Figure 2G).
We validated the unknown RBPs by performing photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) using conventional 365 nm UVA (Hafner et al., 2010). We tested five candidate RBPs from the set of 427 previously unknown primary hits and four of them (RARG, CDKN2AIPNL, PCED1B, and PCGF2/MEL18) showed 4SU-dependent radioactive labeling (Figure S2B) indicative of RNA binding in vivo. A fifth one (CCDC115) was undetectable by PAR-CLIP, either because it was a false positive or because epitope tagging and overexpression interfered with its RNA binding activity. We also confirmed that NANOG, which was in the extended set, crosslinked to RNA in vivo (Figure S2C), indicating that these additional candidates might also comprise previously unknown RBPs.
Thus, RBR-ID identified a considerable portion of previously known nuclear RBPs and at least 427 unknown, non-canonical RBPs enriched for GO terms and protein domains related to chromatin function.
Known and Unknown RBPs with Chromatin-Related Function
Confirming the GO and domain enrichment analysis, visual inspection of the primary list of proteins identified by RBR-ID revealed many with chromatin-related functions whose moonlighting RNA-binding activities have been reported by candidate-based approaches (Table 1) but were missed by previous unbiased RBP identification endeavors. This included EZH2, the catalytic subunit of Polycomb repressive complex-2 (PRC2), which is responsible for formation of facultative heterochromatin and interacts with lncRNAs (Kaneko et al., 2010; Rinn et al., 2007; Tsai et al., 2010; Zhao et al., 2010) and nascent transcripts (Kaneko et al., 2013). We also recovered SUZ12, another subunit of PRC2 that binds RNA (Beltran et al., 2016; Kanhere et al., 2010), and HP1, a central component of constitutive heterochromatin and known RNA binder (Maison et al., 2011), as well as four chromatin factors whose binding to RNA was only recently reported: CTCF, ATRX, HDAC1, and DNMT3 (Castellanos-Rubio et al., 2016; Holz-Schietinger and Reich, 2012; Kung et al., 2015; Saldaña-Meyer et al., 2014; Sarma et al., 2014; Sun et al., 2013).
Table 1.
Examples of Chromatin Factors Identified as RBPs
| Name | Functions | References |
|---|---|---|
| ATRX | Chromatin remodelling | Sarma et al., 2014 |
| CBX1/3/5 (HP1α/β/γ) |
Heterochromatin binding |
Maison et al., 2011; Muchardt et al., 2002 |
| CTCF | Chromatin organization |
Kung et al., 2015; Saldaña-Meyer et al., 2014; Sun et al., 2013 |
| DNMT3A | DNA methylation | Holz-Schietinger and Reich, 2012 |
| EZH2 | Histone methylation |
Kaneko et al., 2010; Rinn et al., 2007; Tsai et al., 2010; Zhao et al., 2010 |
| HDAC1 | Histone deacetylation | Castellanos-Rubio et al., 2016 |
| SUZ12 | Histone methylation |
Beltran et al., 2016; Kanhere et al., 2010 |
| TET1 | DNA demethylation | – |
| TET2 | DNA demethylation | – |
Among the many candidate chromatin proteins identified by RBR-ID that have not previously been reported to bind RNA, we noted TET1 and TET2, two methylcytosine oxidases required for the epigenetic process of DNA demethylation (Pastor et al., 2013).
High-Resolution Mapping of RNA-Interacting Residues In Vitro
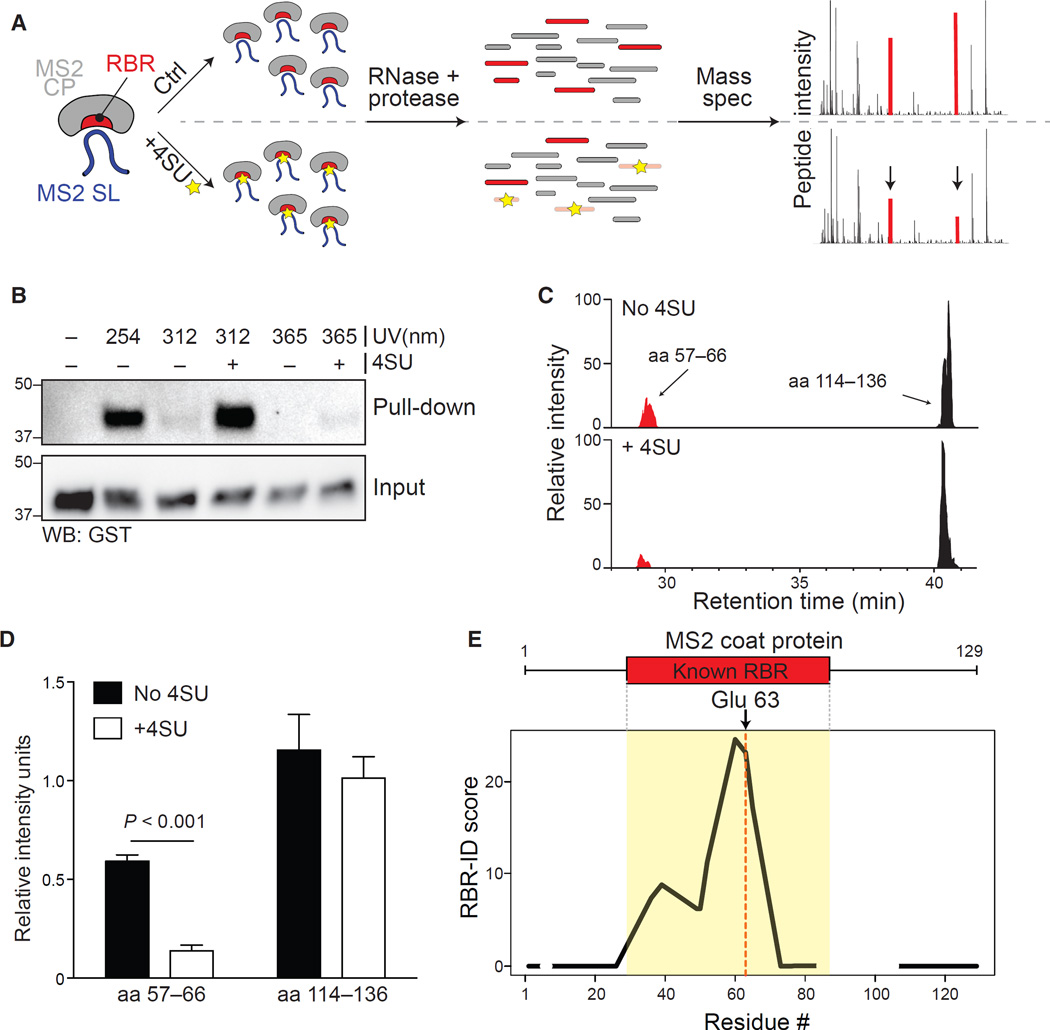
We reasoned that the real power of RBR-ID would rely in its ability to identify not only RNA-binding proteins, but also their RNA-binding regions. We first sought to characterize a well-defined protein-RNA interaction in a fully reconstituted system. We chose the phage MS2 coat protein (MS2-CP) and its cognate stem-loop RNA (MS2-SL), a well-known protein-RNA pair with several high-resolution crystal structures available (Grahn et al., 2001; Valegård et al., 1994; Valegârd et al., 1997).
We incubated recombinant MS2-CP and MS2-SL RNA transcribed in vitro in the presence or absence of 4SU, subjected the complexes to UVB irradiation, and analyzed the crosslinks using MS (Figure 3A). Incorporation of 4SU did not affect the ability of the coat protein to interact with the RNA (Figure S3A). Similar to what we had observed in vivo (Figure S1B), UVB were more efficient than UVA, although at the cost of some low-level background crosslinking even in absence of 4SU (Figure 3B; Figure S3B). Analysis of extracted ion chromatograms revealed a subset of peptides whose intensity was decreased in the 4SU sample (Figure 3C). We generated three biological replicates for this in vitro RBR-ID assay and acquired them in technical duplicates. The most consistently depleted peptide corresponded to the 57–66 region of MS2-CP (Figure 3D), which contains several residues known to form hydrogen bonds with RNA (Valegård et al., 1994; Valegârd et al., 1997).
Figure 3. Mapping of the RBR for MS2-CP with RBR-ID In Vitro.
(A) Recombinant MS2 coat protein and in vitro-transcribed stem-loop RNA with or without incorporated 4SU were allowed to form a complex, then crosslinked, digested, and analyzed by mass spectrometry.
(B) Pull-down of MS2-SL RNA with or without 4SU and crosslinked to MS2-CP with different UV wavelengths. MS2-CP was detected via its fusion tag, GST.
(C) Extracted ion chromatogram showing the elution profile of an RBR-overlapping peptide (red) and a peptide from an MS2-CP region that does not bind RNA (black) from MS2-CP crosslinked to natural (top) or 4SU-containing (bottom) MS2-SL RNA using 312 nm UV.
(D) Quantification of peak intensities for the two peptides shown in (C) for three biological replicates each acquired in duplicates. Bars show average intensity + SEM.
(E) Averaged and smoothed residue-level RBR-ID scores plotted along the primary sequence of MS2-CP. Regions with no peptide coverage are shown as gaps. Data are from three biological replicates each acquired in duplicates. Position of the known RBR and the uridine-interacting glu 63 residue are shown.
Next, we calculated RBR-ID scores (combining extent and consistency of depletion) for each residue and plotted them along the primary sequence of MS2-CP. The RBR-ID score was a good metric for protein-RNA crosslinking, as positive scores precisely mapped to the known RBR of the protein (Figure 3E), with the peak corresponding to glutamic acid 63, which forms hydrogen bonds with a uridine at position −5 in the stem loop (Valegârd et al., 1997).
The availability of crystal structures for the MS2-CP–MS2-SL complex allowed us to visualize the RBR-ID score in a more direct and powerful way. We converted the scores into a heat-map and used it to color the surface of the MS2 protein from the crystal structure (Valegârd et al., 1997), which revealed that the highest RBR-ID scores mapped to the pocket where most RNA contacts occur (Figure 4A).
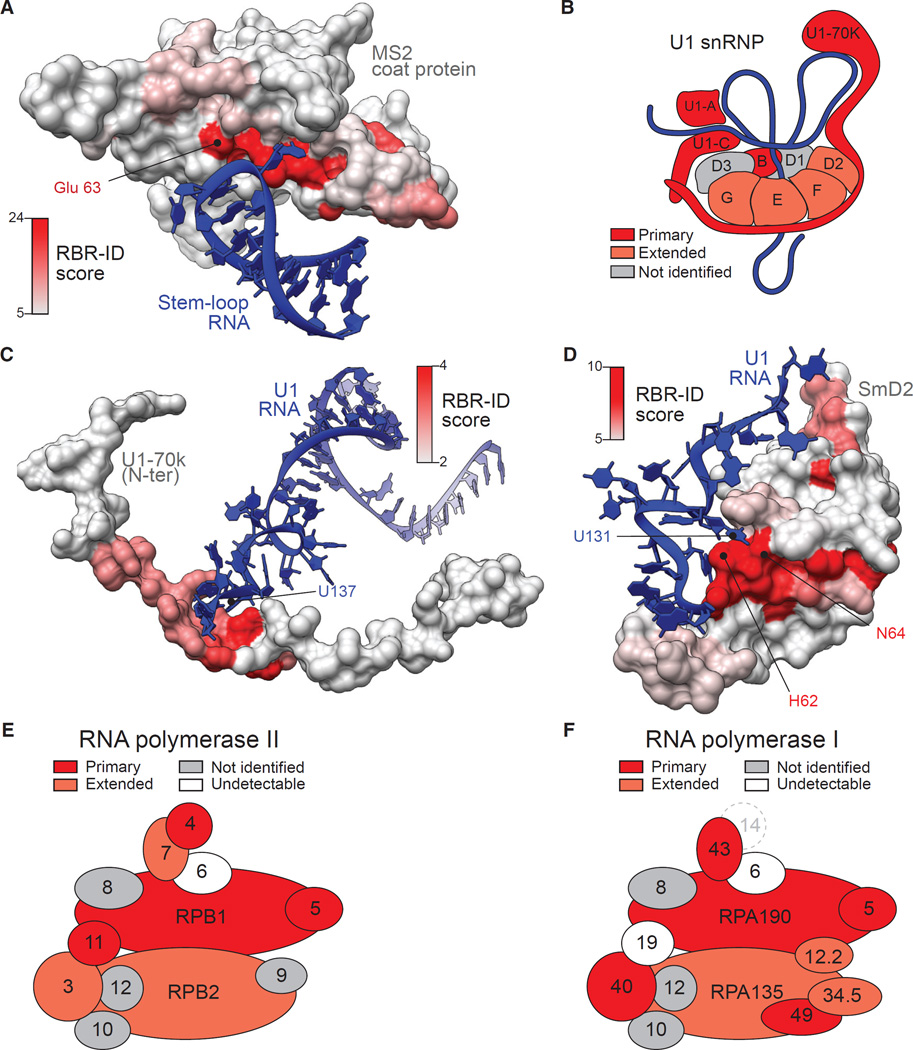
Figure 4. RBR-ID Maps the Sites of Protein-RNA Interactions In Vivo.
(A) The surface rendering of the MS2 coat protein in complex with its cognate RNA (PDB: 1ZDI; Valegârd et al., 1997) was color coded according to the residue-level RBR-ID score from the experiment shown in Figure 3.
(B) Schematic representation of the U1 snRNP particle (Kondo et al., 2015; Pomeranz Krummel et al., 2009). Subunits found in the primary list of RBR-ID candidates are in dark red; proteins in the extended list are in light red.
(C and D) Zoomed-in regions of the crystal structure of U1 snRNP (PDB: 4PJO; Kondo et al., 2015) showing protein surfaces color coded according to their RBR-ID score and interacting RNAs for two regions of U1-70K (C) and SmD2 (D).
(E and F) Schematic representation of the mammalian RNA pol II (E) and RNA pol I (F) complex according to Wild and Cramer (2012). Color coding is same as in (B). Subunits detected in the nuclear proteome, but not identified by RBR-ID, are in gray, undetectable subunits in white. The mammalian homolog for yeast RNA pol I subunit A14 (dashed circle) is unknown.
See also Figure S4.
Mapping of RBRs In Vivo
We returned to our in vivo RBR-ID dataset from ESCs and assessed its precision in mapping RNA-interacting residues in known protein-RNA complexes in vivo at a proteome-wide level. We analyzed the subunits of the U1 small nuclear ribonucleoprotein (snRNP), a component of the spliceosome for which a high-resolution crystal structure was obtained (Kondo et al., 2015). The mouse U1 snRNP is composed of a polyA− ncRNA, U1, and ten protein subunits; we recovered four in the primary RBR-ID candidate list and four more in the extended list (Figure 4B).
We calculated the single-residue RBR-ID scores for the identified subunits and used them to color the respective regions of the crystal structure. For the U1-70K subunit, our approach correctly identified two primary sites of RNA interactions, one within the conserved RRM that caps stem-loop I of the U1 RNA (Figure S4A) and another within the stretch of residues that wraps around the ring formed by the Sm subunits to reach U1-C (Figure 4C). Here, the highest RBR-ID scores were directly adjacent to uridine 137, which forms hydrogen bond contacts with the protein (Figure 4C).
To determine whether spatial RBR mapping was also accurate for proteins in the extended RBR-ID candidate list, we analyzed the SmD2 subunit of the U1 snRNP particle (Figure 4B), which contacts uridine 131 within U1 using H62 and N64. Even for this protein from the extended list, RBR-ID mapped the interacting region with great accuracy, with a peak in signal at the site of interactions as seen in the crystal structure (Figure 4D). Similarly, the highest scores for SmB were near histidine 37, which interacts with uridine 129 (Figure S4B).
To further validate the power of RBR-ID to identify known RBPs in vivo, we analyzed the subunits of RNA polymerase I and II (Figures 4E and 4F), protein complexes responsible for transcription of rRNA and mRNA, respectively (Wild and Cramer, 2012). The two large subunits that form opposite sides of the active center cleft were recovered in both cases, as well as several of the smaller subunits. Interestingly, we recovered both subunits forming the “stalk” structure of RNA pol II (Figure 4E), which were previously shown to crosslink to RNA in vitro (Hahn, 2004; Ujvári and Luse, 2006). We also recovered the corresponding protein subunit from RNA pol I, RPA43 (Figure 4F), suggesting that interactions of the polymerase stalk with nascent RNA might be a conserved feature in these related complexes.
Domain-Level Analyses on RBR-ID Candidates
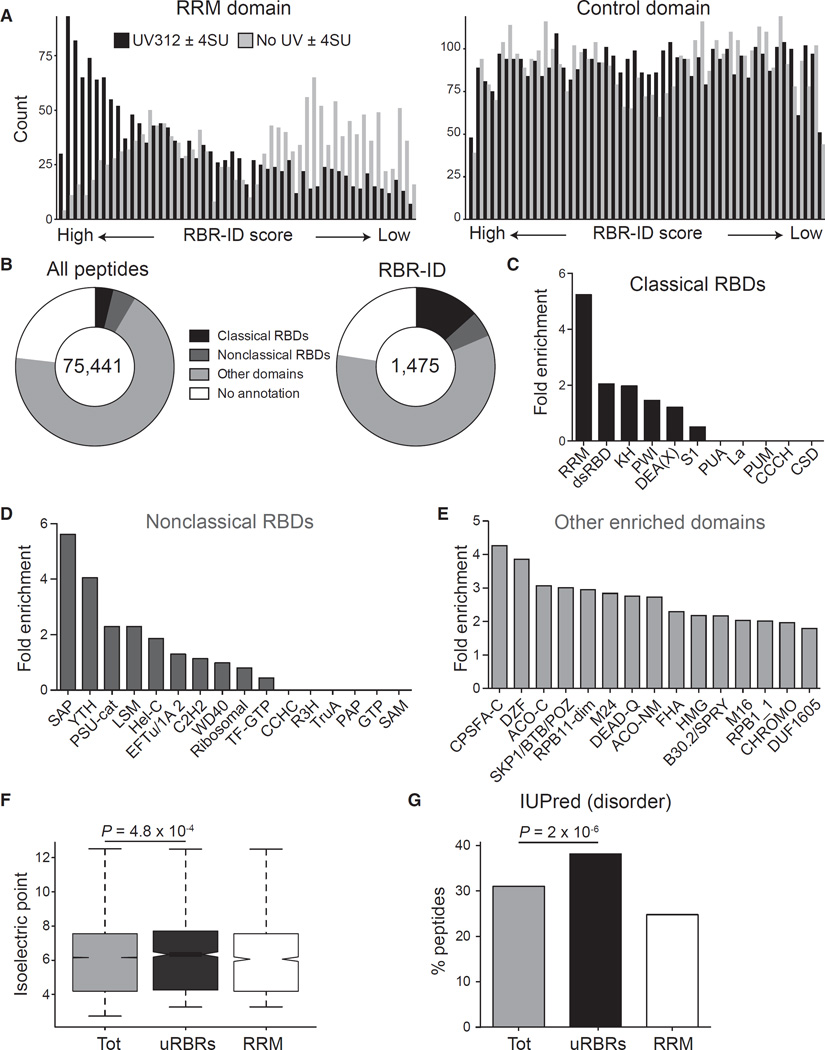
The mapping accuracy of RBR-ID was not restricted to the specific protein-RNA complexes discussed above, but extended to the entire proteome. Peptides overlapping RRM domains showed a strong bias for high RBR-ID scores compared to mock scores calculated from samples not irradiated with UV (Figure 5A, left). Because the RRM domain is relatively frequent in mouse proteins, we also analyzed the distribution of RBR-ID scores for a control domain with similar frequency, the Ploop containing nucleoside triphosphate hydrolase (Interpro: IPR027417), which did not show the same bias (Figure 5A, right). This demonstrated the selectivity of RBR-ID for a bona fide RBD in a proteome-wide manner.
Figure 5. Known and Unknown RNA-Binding Regions in the ESC Proteome.
(A) All detected peptides were sorted according to their RBR-ID score (UV312 ± 4SU) or a control score (no UV ± 4SU). The frequency of peptides overlapping the RRM domain (left) or a control, non-RNA binding domain (IPR027417, right) in these ranked lists is shown.
(B) Categories of Interpro annotations for all peptides detected (left) or peptides in the primary list from RBR-ID (right).
(C–E) Enrichment of selected domains in the top-tier RBR-ID peptides compared to the full list of detected peptides. Classical (C) and non-classical (D) RNA-binding domains are shown as well as enriched domains not previously reported to bind RNA (E).
(F) Tukey boxplot of the isoelectric point for the indicated sets of peptides. p value is from a Student’s t test. Tot, all detected peptides in the nuclear proteome; uRBRs, peptides in the primary candidate lists that did not overlap known RBDs; RRM, all detected peptides overlapping with the RRM domain.
(G) Percentage of peptides overlapping with disordered regions from IUPred (Dosztányi et al., 2005; Oates et al., 2013). Values are shown for all detected peptides (tot), all top-tier RBR-ID peptides not mapping to a known RNA-binding domain (uRBRs), and all peptides overlapping RRM domains. p value is from a chi-square test.
See also Figure S5.
Compared to all detected peptides, the primary list of RBR-ID peptides overlapped significantly with known RBDs but also contained a large proportion of peptides mapping to domains with no known RNA-related function (~59%) or no domain annotations at all (~23%, Figure 5B). RBR-ID hits were enriched in peptides overlapping the three best-known RBDs—RRM, dsRBD, and KH (Lunde et al., 2007)—as well as DEAD and DEAH RNA helicase domains (Figure 5C). We also analyzed a list of non-classical RBDs (Castello et al., 2012) and found many of them enriched (Figure 5D). In particular, the SAP domain, previously thought to mediate DNA binding (Aravind and Koonin, 2000), was enriched more than 5-fold compared to background. The reclassification of the SAP domain as a putative RBD was previously suggested based on its occurrence in empirically identified RBPs (Castello et al., 2012). The enrichment of peptides overlapping the SAP domain in our RBR-ID candidate list provides direct evidence that this domain participates in RNA binding in vivo.
Annotated domains enriched in the RBR-ID list, but not typically considered as possible RBDs (Figure 5E), contained a few domains typically associated with chromatin-related functions, such as the high mobility group domain (HMG) and chromodomain, as well as domains known to participate in nuclear processes but whose function remain nebulous, such as DZF (Doerks et al., 2002; Parker et al., 2001; Wolkowicz and Cook, 2012) and DUF1605 (Kim et al., 2010; Walbott et al., 2010).
Peptides that scored high in our RBR-ID screen, but could not be assigned to annotated domains, showed a slight tendency toward higher isoelectric points (Figure 5F; Figure S5A), consistent with a frequent role for positively charged amino acid in mediating direct interactions with RNA (Jones et al., 2001). However, we saw no global correlation between the isoelectric point of a peptide and its RBR-ID score, excluding the possibility that crosslinking to RNA strongly favored patches of positive amino-acids in a non-specific fashion (Figure S5B). Peptides identified by RBR-ID were also more likely to fall in protein regions predicted to be disordered (Figure 5G; Figure S5C), suggesting that in some cases the disordered regions might directly serve as RNA binding sites.
Validation of RBRs In Vivo
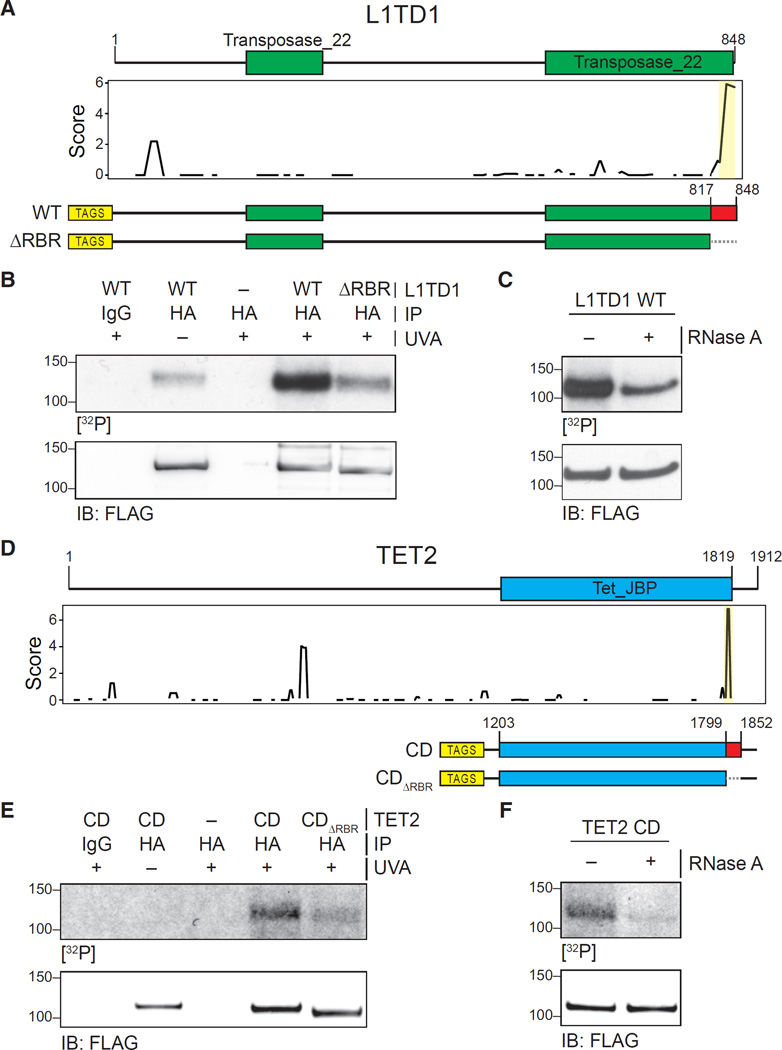
To validate the RBRs predicted by RBR-ID, we selected proteins for which the RBR was previously unknown. We started with L1TD1, a protein whose RNA-binding activity had been previously reported but not mapped (Kwon et al., 2013; Närvä et al., 2012). The RBR-ID score plot pointed to a small region at residues 833–848 in the C terminus as a likely site for RNA interaction (Figure 6A). We expressed epitope-tagged L1TD1 and a truncation mutant lacking the predicted RBR (ΔRBR) in HEK293 cells and performed PAR-CLIP using conventional 365 nm UVA (Hafner et al., 2010). We observed a radioactive signal that overlapped with the L1TD1 band (Figure 6B) and could be assigned to protein-RNA crosslinks because its intensity was reduced after treatment with RNase A (Figure 6C). Importantly, the mutant lacking the region predicted to interact with RNA by RBR-ID showed much lower PAR-CLIP signal despite equal expression levels and pull-down efficiencies for wild-type (WT) and mutant protein (Figure 6B), suggesting that this region is a primary site of RNA interactions.
Figure 6. Validation of RBRs in L1TD1 and TET2.
(A) Primary sequence and known domains for L1TD1 (top); smoothed residue-level RBR-ID score plotted along the primary sequence (middle); and scheme of epitope-tagged WT and RBR-deleted (ΔRBR) constructs used for validation (bottom).
(B) PAR-CLIP of transiently expressed WT and ΔRBR L1TD1 in HEK293 cells. Autoradiography for 32P-labeled RNA (top) and control western blot (bottom).
(C) PAR-CLIP for WT L1TD1 with and without treatment with RNase A (top) and control western blot (bottom).
(D) Primary sequence and known domains for TET2 (top); smoothed residue-level RBR-ID score plotted along the primary sequence (middle); and scheme of epitope-tagged catalytic domain fragment (CD) and RBR-deleted (CDΔRBR) constructs used for validation (bottom).
(E) PAR-CLIP of transiently expressed TET2-CD and TET2-CDΔRBR in HEK293 cells. Autoradiography for 32P-labeled RNA (top) and control western blot (bottom).
(F) PAR-CLIP for TET2 CD with and without treatment with RNase A (top) and control western blot (bottom).
Next, we sought to validate a predicted RBR within a protein previously not known to interact with RNA. RBR-ID identified a nine-residue peptide adjacent to the catalytic domain of TET2 as the most likely site of RNA interaction (Figure 6D). Indeed, a C-terminal fragment encompassing this predicted RBR was sufficient to bind to RNA in vitro (Figure S6A) and in vivo (Figures 6E and 6F), and the identified RBR was required for the interaction, as demonstrated by the drastically reduced PAR-CLIP signal in the ΔRBR mutant (Figure 6E). We made similar observations for MYCN and its predicted RBR (Figure S6B).
To validate additional candidate RBRs with a crosslinking-independent method, we switched to a native RNA immunoprecipitation assay (Bonasio et al., 2014). Although lack of crosslinking renders this technique more prone to non-specific interactions, we reasoned that differences in RNA immunoprecipitation efficiency between WT and ΔRBR versions of the same protein would strongly suggest that the predicted RBR mediated binding to RNA. Epitope-tagged versions of stem cell transcription factors POU5F1/OCT4 and NANOG as well as Polycomb protein MEL18 co-purified with RNA (Figures S6C–S6E), and deletion of their predicted RBR impaired RNA binding (Figures S6C–S6F), suggesting that the regions identified by RBR-ID were mainly responsible for RNA interactions.
Therefore, RBR-ID correctly identified six RBRs within two known (L1TD1 and OCT4) and four previously unknown (TET2, MYCN, MEL18, and NANOG) RBPs and guided the design of protein mutants that showed reduced RNA binding, demonstrating the validity of the predictions and the practical utility of RBR identification.
DISCUSSION
Interactions with RNA constitute an important regulatory layer for the protein machinery that controls chromatin structure and gene expression. To obtain a mechanistic understanding of the biological and biochemical roles of these protein-RNA interactions, comprehensive lists of proteins bound to various classes of RNAs are needed, as well as detailed mapping of the protein regions involved. In vivo photocrosslinking followed by MS allows for the identification of hundreds of protein-RNA interactions in an unbiased manner and with peptide-level resolution.
Rationale for the Development of RBR-ID
The identification of RBRs within non-canonical RBPs, such as Polycomb proteins SCML2 and JARID2, (Bonasio et al., 2014; Kaneko et al., 2014a), as well as CTCF (Saldaña-Meyer et al., 2014), were important steps toward defining the biochemical roles of their interactions with RNA. Using ΔRBR mutants is particularly advantageous when the RBR of a given protein interacts with many RNAs so that depleting individual RNAs generally does not cause overt phenotypes. For example, a subset of the protein-RNA interactions within the PRC2 complex lack sequence specificity despite high affinities (Davidovich et al., 2013, 2015), suggesting that the presence of any RNA, not a particular transcript, modulates the enzymatic activity of this complex (Kaneko et al., 2013, 2014b).
Mapping the RBRs of these non-canonical RBPs one at a time using recombinant protein fragments was a slow and labor-intensive strategy prone to in vitro artifacts. RBR-ID allowed us to identify the potential RBRs of 376 known and 427 unknown RBPs in ESC nuclei. These data will help focus future experiments on proteins and protein regions with the highest likelihood of forming protein-RNA contacts in vivo.
Advantages and Limitations of RBR-ID
Previous endeavors to identify RBPs have relied on enrichment of complexes containing polyadenylated RNA (Baltz et al., 2012; Castello et al., 2012; Kwon et al., 2013). Because of this experimental step, those approaches require up to 108–109 cells. RBR-ID can be performed with starting populations of 106 cells, making comparisons between cellular states (e.g., different differentiation trajectories) and studies in primary cells technically feasible.
Kramer et al. (2014) previously developed an MS pipeline capable, like RBR-ID, of assigning RNA binding sites within proteins based on UV crosslinking. They utilized their approach on human RBPs in a semi-artificial in vitro system, and even in those controlled conditions, crosslinks were identified in only 64 peptides from 49 proteins. This low number of RBPs was likely due to the difficulties in the positive identification of the complex mass spectra created by the heterogeneous products of protein-RNA crosslinking (Kramer et al., 2014).
While our manuscript was being revised, Hentze and colleagues used a different technique to map RBRs in HeLa cells (Castello et al., 2016). Their approach relies on two sequential oligo-dT pull-downs and therefore might have lower false positive rates than RBR-ID; however, it can only be used to identify RBPs that bind polyA+ RNA and requires 10–100 times more input material than RBR-ID.
The potential for false positives in RBR-ID should be curtailed by extensive replication, as was done for the experiments presented here. This is made possible by the low sample requirements, as only 2 µg of total nuclear protein were used per replicate. Even with replication, RBR-ID hits, as in any unbiased screen, will contain some false positives and therefore any candidate should be validated before pursuing the functional significance of its interactions with RNA. Identification by RBR-ID requires efficient protein-RNA crosslinks at a site of 4SU incorporation and therefore a substantial false negative rate is also to be expected, as shown by the missed identification of some known RBPs (Figure 2C). This limitation could be mitigated in the future by utilizing other nucleotide analogs (e.g., 6-thio-guanine; Hafner et al., 2010), different crosslink strategies, and/or more sensitive MS instruments.
Non-Canonical RNA Binding in Chromatin Proteins
Using RBR-ID, we identified 803 RBPs as well as their likely RBRs. Over 50% of these proteins were not present in previous lists from polyA+ RNA purifications or annotation databases. Among these are several chromatin proteins that have been identified by candidate-based approaches, such as EZH2, SUZ12, and CTCF (Kaneko et al., 2010; Kanhere et al., 2010; Rinn et al., 2007; Tsai et al., 2010), but were missed in previous unbiased screens, either because they bind to polyA− ncRNAs or because the stoichiometry of their interactions with RNA is too low for pull-down purification. These 427 unknown nuclear RBPs were enriched for GO annotations related to chromatin structure, chromosome organization, and transcriptional regulation. This observation lends further support to the idea that protein-RNA crosstalk plays a central role in epigenetic regulation (Bonasio and Shiekhattar, 2014; G Hendrickson et al., 2016; Holoch and Moazed, 2015; Rinn and Chang, 2012).
At the peptide level, several domains of interest were enriched, including the chromodomain, which was proposed as a potential RBD (Akhtar et al., 2000), before its role in recognizing lysine methylation was discovered (Lachner et al., 2001). Our RBR-ID data suggest that some chromodomains might indeed moonlight as RNA binders. A conspicuous number of putative RBRs map to protein regions that lack domain annotations. Although some of these might reflect incomplete annotation, the slight, but significant, enrichment of predicted disordered regions suggests that some of them might mediate RNA contacts, as in the case of FMRP and LAF-1 (Elbaum-Garfinkle et al., 2015; Phan et al., 2011). This is particularly relevant in light of the prominent role of RBPs with disordered regions in disease (Castello et al., 2013).
Use of RBR-ID Predictions
The validation of the RBR of TET2 provides an example of the utility of our RBR-ID dataset as a resource. In Drosophila, the TET2 homolog dTET is partially responsible for cytosine hydroxymethylation on RNA (Delatte et al., 2016). Although no RNA-binding evidence has been obtained for the Drosophila protein, the fact that mouse TETs can use RNA as a substrate in vitro (Fu et al., 2014) suggests that this function might be conserved in mammals. The presence of both TET1 and TET2 in the list of primary RBR-ID candidates strongly supports this hypothesis. Furthermore, the identification and validation of the TET2 RBR provides a useful starting point to study the biological role of this biochemical function for the TET family of epigenetic regulators.
Outlook and Conclusion
We applied RBR-ID to ESC nuclei and identified hundreds of RBRs within proteins previously unknown to bind RNA. Because the approach is easily implemented and versatile, we anticipate that variations on this theme will provide even more comprehensive and precise lists of RBRs than the one presented here. Improvements on MS instrumentation and quantification methods, such as “Tandem Mass Tagging” (Cheng et al., 2016), will increase sensitivity, and alternative photoactivatable nucleotides and protease treatments could expand the range of crosslinked peptides, improving resolution.
RBR mapping data are available at http://rbrid.bonasiolab.org. We anticipate that the community will find this a useful resource to design functional experiments aimed at decrypting the complex regulatory language of protein-RNA interactions on chromatin and elsewhere in cells.
EXPERIMENTAL PROCEDURES
Plasmids and Sequences
All oligonucleotide and synthetic DNA sequences used are in Table S8.
RNA Immunoprecipitation
Nuclear extracts were incubated with hemagglutinin (HA) antibody for 3 hr at 4°C and immunocomplexes recovered with protein G Dynabeads. Beads were washed in RIP-W buffer (20 mM Tris [pH 7.94° C], 1 mM MgCl2, 200 mM KCl, and 0.05% IGEPAL CA-630) twice and incubated with TURBO DNase to eliminate potential bridging effects of protein-DNA and DNA-RNA interactions. After two additional washes, RNA was eluted from the beads with TRIzol and purified. We quantified the RNA abundance after immunoprecipitations by measuring the intensity of the bands with ImageJ and normalizing to the IgG background.
PAR-CLIP
HEK293 cells were transiently transfected, pulsed with 100 µM 4-SU for 24 hr, crosslinked with 400 mJ/cm2 UVA (365 nm), and lysed in CLIP buffer (20 mM HEPES [pH 7.4], 5 mM EDTA, 150 mM NaCl, and 2% Empigen) with protease inhibitors, DNase, and RNase inhibitor. HA and StrepTag-fused proteins were first bound to StrepTactin beads in CLIP buffer for 3 hr at 4°C. Beads were washed five times using CLIP buffer and eluted with 2 mM biotin. Next, proteins were incubated with HA antibody overnight at 4°C and recovered with protein G Dynabeads. DNA was removed with DNase, and crosslinked RNA was dephosphorylated with Antarctic phosphatase and labeled with T4 PNK and [γ-32P] ATP. Labeled complexes were resolved on 4%–12% bis-tris gels, transferred to nitrocellulose membrane, and imaged. For NANOG PAR-CLIP, we used E14Tg2A (E14) ESCs pulsed with 500 µM 4-SU for 2 hr and crosslinked with 400 mJ/cm2 UVB (312 nm).
RBR-ID
Cells were pulsed with 500 µM 4SU for 2 hr and crosslinked with 1 J/cm2 UVA, 1 J/cm2 UVB, or 800 mJ/cm2 UVC. We verified that 2 hr was sufficient to incorporate 4SU in virtually all coding and noncoding transcripts by 4SU sequencing (data not shown). Cells were lysed in buffer A (10 mM Tris [pH 7.94° C], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, and 0.2 mM PMSF) with 0.2% IGEPAL CA-630 for 5 min on ice to isolate nuclei, which were lysed in 9 M urea and 100 mM Tris (pH 8RT). The lysate was diluted in 50 mM ammonium bicarbonate and reduced with 5 mM dithiothreitol for 45 min at 56°C. Cysteines were alkylated with 20 mM iodoacetamide for 30 min. Trypsinization was performed at an trypsin:sample ratio of 1:100 overnight at 37°C and blocked with 1% trifluoroacetic acid. Peptides were desalted, dried, and resuspended in 0.1% formic acid prior to MS analysis. Crosslinked RNA was removed with Benzonase.
For in vitro RBR-ID of MS2, preformed protein-RNA complexes were crosslinked with 1 J/cm2 UVB, treated with RNase A, and the protein digested with trypsin (as described above) or chymotrypsin at an enzyme:sample ratio of 1:20 overnight at 25°C.
LC-MS/MS
Nano-LC was performed with a 0%–30% A–B gradient (A, 0.1% formic acid; B, 95% acetonitrile, 0.1% formic acid) over 120 min for the nuclear proteome and over 45 min for MS2. The gradient proceeded from 30% to 85% solvent B in 5 min and 10 min isocratic at 85% B. MS was performed with an Orbitrap Fusion for the nuclear proteome or an Orbitrap Elite for MS2. MS/MS data for both experiment types were collected in centroid mode in the ion trap mass analyzer (normal scan rate). Only charge states 2–5 were included.
MS/MS spectra were processed through MaxQuant (Cox and Mann, 2008) using the Uniprot Mus musculus database. For each peptide, the maximum intensity of the corresponding extracted chromatogram was considered and inter-run variability was accounted for by normalizing for the sum of all peptide intensities in each MS run. Depletion was calculated as the log2-converted ratio of the mean intensity of each peptide in the +4SU samples divided by the mean intensity of the same peptide in −4SU samples. Peptides with log2(fold-change) < 0 and p value < 0.05 (Student’s t test) were considered primary hits. For the extended list, we relaxed the p value requirement to 0.1. For both the primary and extended lists, we filtered out peptides that passed the same cutoffs when comparing signals for +4SU and −4SU in absence of UV. RBR-ID scores were calculated by combining the extent of depletion and the p value according to the following formula:
score = −log2 (normalized + 4SU intensity/normalized − 4SU intensity) × (log10 (p value))2
GO and Interpro Enrichment
For protein list comparisons, all proteins identifiers were converted to official mouse symbols using the Biomart database (version 84). One-to-one human-mouse orthologs were mapped directly, whereas one-to-many and many-to-many homologs were reduced to one-to-one by considering the protein with highest percentage of homology, according to the Biomart database (version 84). For GO and Interpro annotation, tables were downloaded from the Uniprot and Interpro websites directly. Enrichment values and statistics were obtained using the DAVID web server (Huang et al., 2009) either using the unique Uniprot accession identifiers or the converted symbols, when needed.
Supplementary Material
In Brief.
Using 4SU-mediated photocrosslinking and quantitative mass spectrometry, He et al. map RNA-binding regions in hundreds of known and unknown RNA-binding proteins in the nuclei of embryonic stem cells, suggesting that RNA binding is a common feature of chromatin-associated proteins and transcriptional regulators.
Highlights.
RBR-ID identifies RNA-binding regions by 4SU photocrosslinking and mass spectrometry
RBRs were mapped in 803 nuclear RNA-binding proteins (RBPs) in embryonic stem cells
Many previously unknown RBPs regulate chromatin structure and transcription
RBRs were found in disordered regions and domains associated with chromatin function
Acknowledgments
The authors thank T. Christopher for technical support, R. Kohli for his generous gifts of TET2 reagents, and S. Berger and D. Reinberg for comments on the manuscript. R.B. was supported by the Searle Scholars Program, the W.W. Smith Foundation (C1404), the March of Dimes Foundation (1-FY-15-344). B.A.G acknowledges support from NIH grant R01GM110174 and DOD grant BC123187P1. J.E.W. is a Rita Allen Foundation Scholar and was supported by NIH grants R00-GM104166 and R35-GM119735. R.W.-T. was supported in part by NIH training grant T32GM008216.
Footnotes
ACCESSION NUMBERS
The accession number for the proteomics data reported in this paper is Chorus: 1128.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, six figures, and eight tables and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2016.09.034.
AUTHOR CONTRIBUTIONS
C.H. and R.B. conceived the project and designed the experiments with help from S.S. and B.A.G. C.H. carried out all experimental work with help from R.W.-T. S.S. analyzed samples by MS under the supervision of B.A.G. D.C.T. and J.E.W. provided critical reagents. C.H. and R.B. wrote the manuscript with input from all authors.
REFERENCES
- Akhtar A, Zink D, Becker PB. Chromodomains are protein-RNA interaction modules. Nature. 2000;407:405–409. doi: 10.1038/35030169. [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. SAP—a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 2000;25:112–114. doi: 10.1016/s0968-0004(99)01537-6. [DOI] [PubMed] [Google Scholar]
- Baltz AG, Munschauer M, Schwanhäusser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell. 2012;46:674–690. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- Beckmann BM, Horos R, Fischer B, Castello A, Eichelbaum K, Alleaume AM, Schwarzl T, Curk T, Foehr S, Huber W, et al. The RNA-binding proteomes from yeast to man harbour conserved enigmRBPs. Nat. Commun. 2015;6:10127. doi: 10.1038/ncomms10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran M, Yates CM, Skalska L, Dawson M, Reis FP, Viiri K, Fisher CL, Sibley CR, Foster BM, Bartke T, et al. The interaction of PRC2 with RNA or chromatin is mutually antagonistic. Genome Res. 2016;26:896–907. doi: 10.1101/gr.197632.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R, Shiekhattar R. Regulation of transcription by long noncoding RNAs. Annu. Rev. Genet. 2014;48:433–455. doi: 10.1146/annurev-genet-120213-092323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R, Lecona E, Narendra V, Voigt P, Parisi F, Kluger Y, Reinberg D. Interactions with RNA direct the Polycomb group protein SCML2 to chromatin where it represses target genes. eLife. 2014;3:e02637. doi: 10.7554/eLife.02637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JA, Valenstein ML, Yario TA, Tycowski KT, Steitz JA. Formation of triple-helical structures by the 3′-end sequences of MALAT1 and MENβ noncoding RNAs. Proc. Natl. Acad. Sci. USA. 2012;109:19202–19207. doi: 10.1073/pnas.1217338109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellanos-Rubio A, Fernandez-Jimenez N, Kratchmarov R, Luo X, Bhagat G, Green PH, Schneider R, Kiledjian M, Bilbao JR, Ghosh S. A long noncoding RNA associated with susceptibility to celiac disease. Science. 2016;352:91–95. doi: 10.1126/science.aad0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- Castello A, Fischer B, Hentze MW, Preiss T. RNA-binding proteins in Mendelian disease. Trends Genet. 2013;29:318–327. doi: 10.1016/j.tig.2013.01.004. [DOI] [PubMed] [Google Scholar]
- Castello A, Fischer B, Frese CK, Horos R, Alleaume AM, Foehr S, Curk T, Krijgsveld J, Hentze MW. Comprehensive identification of RNA-binding domains in human cells. Mol. Cell. 2016;63:696–710. doi: 10.1016/j.molcel.2016.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KY, Ramos A. The double-stranded RNA-binding motif, a versatile macromolecular docking platform. FEBS J. 2005;272:2109–2117. doi: 10.1111/j.1742-4658.2005.04652.x. [DOI] [PubMed] [Google Scholar]
- Cheng L, Pisitkun T, Knepper MA, Hoffert JD. Peptide labeling using isobaric tagging reagents for quantitative phosphoproteomics. Methods Mol. Biol. 2016;1355:53–70. doi: 10.1007/978-1-4939-3049-4_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161:404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad T, Albrecht AS, de Melo Costa VR, Sauer S, Meierhofer D, Ørom UA. Serial interactome capture of the human cell nucleus. Nat. Commun. 2016;7:11212. doi: 10.1038/ncomms11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook KB, Kazan H, Zuberi K, Morris Q, Hughes TR. RBPDB: a database of RNA-binding specificities. Nucleic Acids Res. 2011;39:D301–D308. doi: 10.1093/nar/gkq1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Davidovich C, Zheng L, Goodrich KJ, Cech TR. Promiscuous RNA binding by Polycomb repressive complex 2. Nat. Struct. Mol. Biol. 2013;20:1250–1257. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovich C, Wang X, Cifuentes-Rojas C, Goodrich KJ, Gooding AR, Lee JT, Cech TR. Toward a consensus on the binding specificity and promiscuity of PRC2 for RNA. Mol. Cell. 2015;57:552–558. doi: 10.1016/j.molcel.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delatte B, Wang F, Ngoc LV, Collignon E, Bonvin E, Deplus R, Calonne E, Hassabi B, Putmans P, Awe S, et al. RNA biochemistry. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science. 2016;351:282–285. doi: 10.1126/science.aac5253. [DOI] [PubMed] [Google Scholar]
- Doerks T, Copley RR, Schultz J, Ponting CP, Bork P. Systematic identification of novel protein domain families associated with nuclear functions. Genome Res. 2002;12:47–56. doi: 10.1101/gr.203201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosztányi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- Eisen JA, Sweder KS, Hanawalt PC. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CC, Eckmann CR, Myong S, Brangwynne CP. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. USA. 2015;112:7189–7194. doi: 10.1073/pnas.1504822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre A, Moreno G, Blondel MO, Kliber J, Vinzens F, Salet C. 4-Thiouridine photosensitized RNA-protein crosslinking in mammalian cells. Biochem. Biophys. Res. Commun. 1986;141:847–854. doi: 10.1016/s0006-291x(86)80250-9. [DOI] [PubMed] [Google Scholar]
- Fu L, Guerrero CR, Zhong N, Amato NJ, Liu Y, Liu S, Cai Q, Ji D, Jin SG, Niedernhofer LJ, et al. Tet-mediated formation of 5-hydroxymethylcytosine in RNA. J. Am. Chem. Soc. 2014;136:11582–11585. doi: 10.1021/ja505305z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- G Hendrickson D, Kelley DR, Tenen D, Bernstein B, Rinn JL. Widespread RNA binding by chromatin-associated proteins. Genome Biol. 2016;17:28. doi: 10.1186/s13059-016-0878-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff LA, Rinn JL. Linking RNA biology to lncRNAs. Genome Res. 2015;25:1456–1465. doi: 10.1101/gr.191122.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlach M, Wittekind M, Beckman RA, Mueller L, Dreyfuss G. Interaction of the RNA-binding domain of the hnRNP C proteins with RNA. EMBO J. 1992;11:3289–3295. doi: 10.1002/j.1460-2075.1992.tb05407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grahn E, Moss T, Helgstrand C, Fridborg K, Sundaram M, Tars K, Lago H, Stonehouse NJ, Davis DR, Stockley PG, Liljas L. Structural basis of pyrimidine specificity in the MS2 RNA hairpin-coat-protein complex. RNA. 2001;7:1616–1627. [PMC free article] [PubMed] [Google Scholar]
- Greenberg JR. Ultraviolet light-induced crosslinking of mRNA to proteins. Nucleic Acids Res. 1979;6:715–732. doi: 10.1093/nar/6.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishin NV. KH domain: one motif, two folds. Nucleic Acids Res. 2001;29:638–643. doi: 10.1093/nar/29.3.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S. Structure and mechanism of the RNA polymerase II transcription machinery. Nat. Struct. Mol. Biol. 2004;11:394–403. doi: 10.1038/nsmb763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockensmith JW, Kubasek WL, Vorachek WR, von Hippel PH. Laser cross-linking of nucleic acids to proteins. Methodology and first applications to the phage T4 DNA replication system. J. Biol. Chem. 1986;261:3512–3518. [PubMed] [Google Scholar]
- Holoch D, Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 2015;16:71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holz-Schietinger C, Reich NO. RNA modulation of the human DNA methyltransferase 3A. Nucleic Acids Res. 2012;40:8550–8557. doi: 10.1093/nar/gks537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Jones S, Daley DT, Luscombe NM, Berman HM, Thornton JM. Protein-RNA interactions: a structural analysis. Nucleic Acids Res. 2001;29:943–954. doi: 10.1093/nar/29.4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Li G, Son J, Xu CF, Margueron R, Neubert TA, Reinberg D. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 2010;24:2615–2620. doi: 10.1101/gad.1983810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Son J, Shen SS, Reinberg D, Bonasio R. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat. Struct. Mol. Biol. 2013;20:1258–1264. doi: 10.1038/nsmb.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Bonasio R, Saldaña-Meyer R, Yoshida T, Son J, Nishino K, Umezawa A, Reinberg D. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol. Cell. 2014a;53:290–300. doi: 10.1016/j.molcel.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Son J, Bonasio R, Shen SS, Reinberg D. Nascent RNA interaction keeps PRC2 activity poised and in check. Genes Dev. 2014b;28:1983–1988. doi: 10.1101/gad.247940.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanhere A, Viiri K, Araújo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol. Cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Pazhoor S, Bao M, Zhang Z, Hanabuchi S, Facchinetti V, Bover L, Plumas J, Chaperot L, Qin J, Liu YJ. Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proc. Natl. Acad. Sci. USA. 2010;107:15181–15186. doi: 10.1073/pnas.1006539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y, Oubridge C, van Roon AM, Nagai K. Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5′ splice site recognition. eLife. 2015;4:4. doi: 10.7554/eLife.04986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer K, Sachsenberg T, Beckmann BM, Qamar S, Boon KL, Hentze MW, Kohlbacher O, Urlaub H. Photo-cross-linking and high-resolution mass spectrometry for assignment of RNA-binding sites in RNA-binding proteins. Nat. Methods. 2014;11:1064–1070. doi: 10.1038/nmeth.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung JT, Kesner B, An JY, Ahn JY, Cifuentes-Rojas C, Colognori D, Jeon Y, Szanto A, del Rosario BC, Pinter SF, et al. Locus-specific targeting to the X chromosome revealed by the RNA interactome of CTCF. Mol. Cell. 2015;57:361–375. doi: 10.1016/j.molcel.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SC, Yi H, Eichelbaum K, Föhr S, Fischer B, You KT, Castello A, Krijgsveld J, Hentze MW, Kim VN. The RNA-binding protein repertoire of embryonic stem cells. Nat. Struct. Mol. Biol. 2013;20:1122–1130. doi: 10.1038/nsmb.2638. [DOI] [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Lam MT, Li W, Rosenfeld MG, Glass CK. Enhancer RNAs and regulated transcriptional programs. Trends Biochem. Sci. 2014;39:170–182. doi: 10.1016/j.tibs.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison C, Bailly D, Roche D, Montes de Oca R, Probst AV, Vassias I, Dingli F, Lombard B, Loew D, Quivy JP, Almouzni G. SUMOylation promotes de novo targeting of HP1α to pericentric heterochromatin. Nat. Genet. 2011;43:220–227. doi: 10.1038/ng.765. [DOI] [PubMed] [Google Scholar]
- Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521:232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minajigi A, Froberg JE, Wei C, Sunwoo H, Kesner B, Colognori D, Lessing D, Payer B, Boukhali M, Haas W, Lee JT. Chromosomes. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science. 2015;349:349. doi: 10.1126/science.aab2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C, Guilleme M, Seeler J-S, Trouche D, Dejean A, Yaniv M. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1alpha. EMBO Rep. 2002;3:975–981. doi: 10.1093/embo-reports/kvf194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näarväa E, Rahkonen N, Emani MR, Lund R, Pursiheimo JP, Nästi J, Autio R, Rasool O, Denessiouk K, Lähdesmäki H, et al. RNA-binding protein L1TD1 interacts with LIN28 via RNA and is required for human embryonic stem cell self-renewal and cancer cell proliferation. Stem Cells. 2012;30:452–460. doi: 10.1002/stem.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oates ME, Romero P, Ishida T, Ghalwash M, Mizianty MJ, Xue B, Dosztányi Z, Uversky VN, Obradovic Z, Kurgan L, et al. D2P2: database of disordered protein predictions. Nucleic Acids Res. 2013;41:D508–D516. doi: 10.1093/nar/gks1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker LM, Fierro-Monti I, Mathews MB. Nuclear factor 90 is a substrate and regulator of the eukaryotic initiation factor 2 kinase double-stranded RNA-activated protein kinase. J. Biol. Chem. 2001;276:32522–32530. doi: 10.1074/jbc.M104408200. [DOI] [PubMed] [Google Scholar]
- Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat. Rev. Mol. Cell Biol. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan AT, Kuryavyi V, Darnell JC, Serganov A, Majumdar A, Ilin S, Raslin T, Polonskaia A, Chen C, Clain D, et al. Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction. Nat. Struct. Mol. Biol. 2011;18:796–804. doi: 10.1038/nsmb.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeranz Krummel DA, Oubridge C, Leung AK, Li J, Nagai K. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature. 2009;458:475–480. doi: 10.1038/nature07851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldaña-Meyer R, González-Buendía E, Guerrero G, Narendra V, Bonasio R, Recillas-Targa F, Reinberg D. CTCF regulates the human p53 gene through direct interaction with its natural antisense transcript, Wrap53. Genes Dev. 2014;28:723–734. doi: 10.1101/gad.236869.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma K, Cifuentes-Rojas C, Ergun A, Del Rosario A, Jeon Y, White F, Sadreyev R, Lee JT. ATRX directs binding of PRC2 to Xist RNA and Polycomb targets. Cell. 2014;159:869–883. doi: 10.1016/j.cell.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiege W, Kosack M, Stade K, Brimacombe R. Intra-RNA cross-linking in Escherichia coli 30S ribosomal subunits: selective isolation of cross-linked products by hybridization to specific cDNA fragments. Nucleic Acids Res. 1988;16:4315–4329. doi: 10.1093/nar/16.10.4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Del Rosario BC, Szanto A, Ogawa Y, Jeon Y, Lee JT. Jpx RNA activates Xist by evicting CTCF. Cell. 2013;153:1537–1551. doi: 10.1016/j.cell.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujvári A, Luse DS. RNA emerging from the active site of RNA polymerase II interacts with the Rpb7 subunit. Nat. Struct. Mol. Biol. 2006;13:49–54. doi: 10.1038/nsmb1026. [DOI] [PubMed] [Google Scholar]
- UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valegård K, Murray JB, Stockley PG, Stonehouse NJ, Liljas L. Crystal structure of an RNA bacteriophage coat protein-operator complex. Nature. 1994;371:623–626. doi: 10.1038/371623a0. [DOI] [PubMed] [Google Scholar]
- Valegârd K, Murray JB, Stonehouse NJ, van den Worm S, Stockley PG, Liljas L. The three-dimensional structures of two complexes between recombinant MS2 capsids and RNA operator fragments reveal sequence-specific protein-RNA interactions. J. Mol. Biol. 1997;270:724–738. doi: 10.1006/jmbi.1997.1144. [DOI] [PubMed] [Google Scholar]
- Walbott H, Mouffok S, Capeyrou R, Lebaron S, Humbert O, van Tilbeurgh H, Henry Y, Leulliot N. Prp43p contains a processive helicase structural architecture with a specific regulatory domain. EMBO J. 2010;29:2194–2204. doi: 10.1038/emboj.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild T, Cramer P. Biogenesis of multisubunit RNA polymerases. Trends Biochem. Sci. 2012;37:99–105. doi: 10.1016/j.tibs.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Wilusz JE. Circular RNAs: unexpected outputs of many protein-coding genes. RNA Biol. 2016:1–11. doi: 10.1080/15476286.2016.1227905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, JnBaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, Sharp PA. A triple helix stabilizes the 30 ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012;26:2392–2407. doi: 10.1101/gad.204438.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolkowicz UM, Cook AG. NF45 dimerizes with NF90, Zfr and SPNR via a conserved domain that has a nucleotidyltransferase fold. Nucleic Acids Res. 2012;40:9356–9368. doi: 10.1093/nar/gks696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.