Abstract
Controlled oxidation reactions catalyzed by the large, proton-pumping complexes of the respiratory chain generate an electrochemical gradient across the mitochondrial inner membrane that is harnessed for ATP production. However, several alternative respiratory pathways in plants allow the maintenance of substrate oxidation while minimizing the production of ATP. We have investigated the role of light in the regulation of these energy-dissipating pathways by transcriptional profiling of the alternative oxidase, uncoupling protein, and type II NAD(P)H dehydrogenase gene families in etiolated Arabidopsis seedlings. Expression of the nda1 and ndc1 NAD(P)H dehydrogenase genes was rapidly up-regulated by a broad range of light intensities and qualities. For both genes, light induction appears to be a direct transcriptional effect that is independent of carbon status. Mutant analyses demonstrated the involvement of two separate photoreceptor families in nda1 and ndc1 light regulation: the phytochromes (phyA and phyB) and an undetermined blue light photoreceptor. In the case of the nda1 gene, the different photoreceptor systems generate distinct kinetic induction profiles that are integrated in white light response. Primary transcriptional control of light response was localized to a 99-bp region of the nda1 promoter, which contains an I-box flanked by two GT-1 elements, an arrangement prevalent in the promoters of photosynthesis-associated genes. Light induction was specific to nda1 and ndc1. The only other substantial light effect observed was a decrease in aox2 expression. Overall, these results suggest that light directly influences the respiratory electron transport chain via photoreceptor-mediated transcriptional control, likely for supporting photosynthetic metabolism.
The electron transport chain (ETC) and ATP synthase catalyze the final steps of aerobic respiration, whereby reduced organic compounds are converted into chemical energy in the form of ATP. The ETC is located in the inner mitochondrial membrane and is composed of four large, multiprotein complexes common to both plants and animals. Complexes I and II catalyze the oxidation of matrix NADH and succinate, respectively, transferring electrons to lipid-soluble ubiquinone. Reduced ubiquinone is then oxidized via complex III, which donates electrons to the cytochrome c protein. Complex IV then transfers electrons from cytochrome c to the terminal electron acceptor, O2, generating water. The oxidation reactions mediated by complexes I, III, and IV are coupled to the pumping of protons across the inner membrane, generating an electrochemical gradient. This membrane gradient is harnessed by the FoF1-ATP synthase for the production of ATP (Siedow and Day, 2000).
In addition to the basal ETC described above, plants possess several alternative respiratory pathways that bypass energy conservation by circumventing the formation or utilization of the electrochemical proton gradient. These energy-dissipating pathways are formed by several simple proteins: type II NAD(P)H dehydrogenases, which bypass proton-pumping complex I or allow oxidation of cytoplasmic NAD(P)H; alternative oxidases, which bypass proton-pumping complexes III and IV; and uncoupling proteins, which bypass the ATP synthase by directly dissipating the proton gradient (Vanlerberghe and McIntosh, 1997; Vercesi, 2001; Rasmusson et al., 2004). Direct experimental evidence demonstrating the physiological significance of the energy-dissipating respiratory proteins in plants is generally lacking, although it has been shown that alternative oxidase is required for thermogenesis during floral maturation in several species of lilies (Siedow and Day, 2000). In addition, these enzymes are likely involved in balancing cellular redox and energy status (van Lis and Atteia, 2004) and in minimizing the production of reactive oxygen species (ROS) generated by overreduction of basal respiratory chain components (Purvis and Shewfelt, 1993; Maxwell et al., 1999; Møller, 2001; Svensson et al., 2002; Brandalise et al., 2003). Interestingly, the expression of several genes encoding energy-dissipating respiratory proteins is tightly regulated by various environmental stimuli (Finnegan et al., 1997; Laloi et al., 1997; Svensson and Rasmusson, 2001; Svensson et al., 2002), which contrasts with the more constitutive expression of studied basal ETC components (with some exceptions; see e.g. Hilton and Owen, 1985).
Light is a key regulator of gene expression in plants, altering the transcription of thousands of genes through direct (photoreceptor-mediated) or indirect (photosynthetic product-mediated) pathways (Ma et al., 2001; Tepperman et al., 2001). However, the vast majority of detailed studies on light regulation have focused on photosynthesis-associated nuclear genes (Terzaghi and Cashmore, 1995; Argüello-Astorga and Herrera-Estrella, 1998). Apart from the light-induced photorespiratory enzymes Gly decarboxylase (e.g. Srinivasan and Oliver, 1995; Vauclare et al., 1998) and Ser hydroxymethyltransferase (McClung et al., 2000), little is known about the effects of light on mitochondria and the respiratory chain. Requirements for both ATP synthesis and NADH reoxidation substantially increase in the light with the up-regulation of Suc production, photorespiration, and Krebs cycle α-ketoglutarate synthesis for nitrogen assimilation (Krömer, 1995; Lancien et al., 1999; Ma et al., 2001). Thus, dynamic alterations in the ETC, most likely the structurally simple energy-dissipating proteins, may be necessary to accommodate increased electron flux in the light without substantially increasing ROS production. Indeed, it has recently been demonstrated that the type II NAD(P)H dehydrogenase gene nda1 is regulated by light in potato (Solanum tuberosum) and Arabidopsis (Svensson and Rasmusson, 2001; Michalecka et al., 2003) and that the alternative oxidase gene aox2a is light induced in soybean (Glycine max; Finnegan et al., 1997). The mode of signaling underlying light-regulated changes in mitochondria has not been investigated, but van Lis and Atteia (2004) have suggested a primary role for redox regulation.
Whole genome sequencing of Arabidopsis has allowed the identification of the complete gene families encoding the alternative oxidases (aox1a-d and aox2; Saisho et al., 1997; Thirkettle-Watts et al., 2003), type II NAD(P)H dehydrogenases (nda1-2, ndb1-4, and ndc1; Michalecka et al., 2003), and uncoupling proteins (ucp1-2; Watanabe et al., 1999). However, RNA hybridization-based studies (northern analysis and microarrays) have provided no insight into the regulation of these genes (Ma et al., 2001; Tepperman et al., 2001), many of which are expressed at extremely low levels under most conditions (Michalecka et al., 2003; Thirkettle-Watts et al., 2003). Indeed, a recent study utilizing the Affymetrix ATH1 Arabidopsis genome chip showed that the majority of genes-encoding energy-dissipating respiratory components are expressed at levels below or near the limit of reliable microarray detection (Wang et al., 2003). In contrast to hybridization-based methodologies, real time reverse transcription (RT)-PCR is an extremely sensitive and reliable technique for the quantitation of low-abundance transcripts (Czechowski et al., 2004), including type II NAD(P)H dehydrogenase mRNA (Svensson et al., 2002; Michalecka et al., 2003). In this study, we have utilized a real-time RT-PCR approach to perform a semi-global analysis of the role of light in the regulation of energy-dissipating respiratory gene families. Our results show that the respiratory chain of Arabidopsis seedlings is dynamically adjusted upon exposure to light through the action of several photoreceptor families which specifically mediate induction of the NAD(P)H dehydrogenase genes nda1 and ndc1.
RESULTS
Light Regulation of Genes Encoding Energy-Dissipating Components of the ETC
Light-regulated changes in gene expression were examined for the alternative oxidase, type II NAD(P)H dehydrogenase, and uncoupling protein gene families of Arabidopsis. A rapid real-time PCR assay was used to screen for relative alterations in transcript abundance in 5-d-old etiolated Arabidopsis seedlings maintained in continuous darkness or exposed to 10 μmol m−2 s−1 white light for 4 or 12 h (Table I). The non-light-responsive gene encoding the 76-kD subunit of ETC complex I was used as a negative internal control (Svensson and Rasmusson, 2001) and the light-induced gene encoding Gly decarboxylase subunit H (gdcH) was used as a positive control (Srinivasan and Oliver, 1995; Tepperman et al., 2004).
Table I.
A real-time RT-PCR screen for light-regulated respiratory genes
| 4-h Light Treatment
|
12-h Light Treatment
|
|||||
|---|---|---|---|---|---|---|
| Ct Lighta | Ct Dark | ΔCtb | Ct Light | Ct Dark | ΔCt | |
| aox1a | 21.93 ± 0.15 | 23.16 ± 0.03 | 1.23 | 23.75 ± 0.19 | 24.01 ± 0.16 | 0.26 |
| aox1b | 34.88 ± 0.09 | 35.52 ± 0.48 | 0.64 | 33.83 ± 0.05 | 34.26 ± 0.12 | 0.43 |
| aox1c | 31.72 ± 0.06 | 32.46 ± 0.25 | 0.74 | 31.44 ± 0.11 | 32.63 ± 0.05 | 1.19 |
| aox1d | NDc | ND | ND | ND | ND | ND |
| aox2 | 31.39 ± 0.32 | 29.34 ± 0.15 | −2.05 | 32.15 ± 0.01 | 29.84 ± 1.35 | −2.31 |
| nda1 | 26.78 ± 0.01 | 31.80 ± 0.42 | 5.02 | 25.89 ± 0.10 | 32.15 ± 0.23 | 6.26 |
| nda2 | 22.74 ± 0.62 | 22.80 ± 0.04 | 0.06 | 24.17 ± 0.01 | 23.84 ± 0.26 | −0.33 |
| ndb1 | 30.32 ± 0.72 | 29.02 ± 0.22 | −1.30 | 29.09 ± 0.27 | 28.29 ± 0.07 | −0.80 |
| ndb2 | 34.43 ± 0.56 | 34.91 ± 0.32 | 0.48 | 36.28 ± 0.37 | 35.50 ± 0.47 | −0.78 |
| ndb3 | ND | ND | ND | ND | ND | ND |
| ndb4 | 32.73 ± 0.09 | 33.55 ± 0.20 | 0.82 | 32.48 ± 0.33 | 32.64 ± 0.62 | 0.16 |
| ndc1 | 23.89 ± 0.33 | 25.48 ± 0.22 | 1.59 | 24.28 ± 0.30 | 26.31 ± 0.01 | 2.03 |
| ucp1 | 23.23 ± 0.47 | 23.14 ± 0.10 | −0.09 | 23.38 ± 0.03 | 23.02 ± 0.06 | −0.36 |
| ucp2 | 27.56 ± 0.55 | 27.53 ± 0.08 | −0.03 | 27.26 ± 0.03 | 27.78 ± 0.15 | 0.52 |
| 76 kD | 22.72 ± 0.28 | 22.43 ± 0.20 | −0.29 | 22.68 ± 0.31 | 22.74 ± 0.11 | 0.06 |
| GdcH | 21.07 ± 0.32 | 24.14 ± 0.43 | 3.07 | 20.01 ± 0.13 | 24.31 ± 0.17 | 4.30 |
Higher Ct values indicate lower transcript abundance; theoretically, a ΔCt of x corresponds to a change of 2x in template abundance at maximum amplification efficiency. Respiratory genes for which a substantial change in transcript abundance is seen (ΔCt > 1.6) are denoted in bold.
Ct values are ±sd from two separate RNA preparations (i.e. a total of least four independent PCR reactions) for each point.
ΔCt = Ct dark − Ct light.
ND, Not consistently detected.
Strong light responsiveness is not a general feature of energy-dissipating respiratory genes, as most of the investigated transcripts displayed less than a 3-fold (predicted maximum) change in abundance upon extended light exposure (ΔCt < 1.59). However, expression of the type II NAD(P)H dehydrogenase genes nda1 and ndc1, and the alternative oxidase gene aox2, was substantially altered after both 4 and 12 h of light exposure. nda1 and ndc1 are light induced, with nda1 transcript levels displaying a massive increase (ΔCt > 6) compared to the dark control. Although several independent experiments demonstrated that aox2 gene expression is consistently down-regulated by light, aox2 transcript levels in the dark were found to be highly variable between different experiments, precluding further quantitative assessment of this light effect (data not shown).
Kinetics of nda1 and ndc1 Light Induction
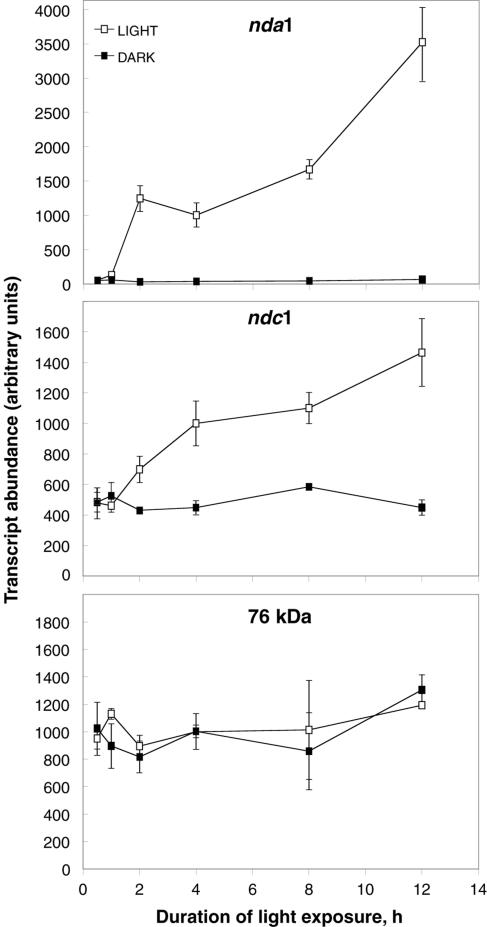
The baseline kinetics of nda1 and ndc1 light induction were established by performing quantitative real-time PCR on cDNA from seedlings exposed to 0.5, 1, 2, 4, 8, and 12 h of continuous white light (Fig. 1). As expected from the initial screening studies, induction of nda1 is rapid and substantial, with transcript abundance doubling in the 1st h of light exposure and increasing to approximately 70-fold over dark levels after 12 h. Interestingly, the nda1 induction curve displays two phases of increase in transcript abundance (from 1 to 2 h and 4 to 12 h) interrupted by a 2-h period in which there is no significant change in transcript levels. In comparison, ndc1 displays more linear induction kinetics and a modest overall transcript increase (approximately 3-fold).
Figure 1.
Induction kinetics of nda1 and ndc1 white light response. Five-day-old etiolated Arabidopsis seedlings were either maintained in darkness or exposed to continuous 10 μmol m−2 s−1 white light for the periods indicated. Transcript abundance was quantified by real-time PCR analysis of cDNA generated from total seedling RNA. The 76-kD subunit of respiratory complex I was utilized as a non-light-responsive control. Data are presented as means ± sd from three separate RNA preparations (i.e. a total of at least six independent PCR reactions) for each point.
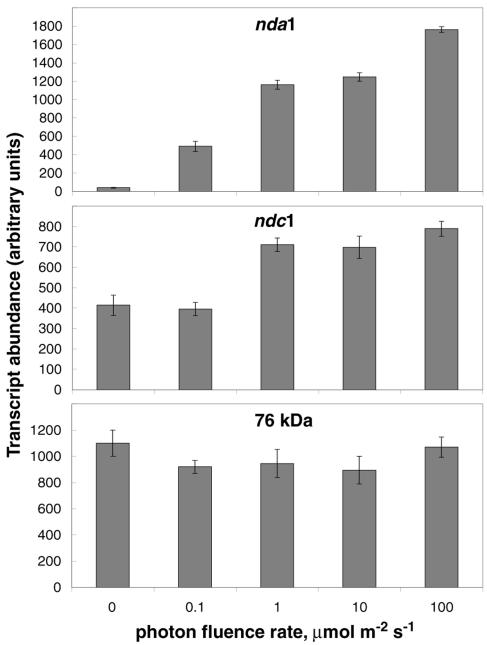
Effect of Photon Fluence Rate, Carbon Status, and Light Quality on nda1 and ndc1 Induction
Etiolated seedlings were exposed to various photon fluence rates (0.1, 1, 10, or 100 μmol m−2 s−1 continuous white light) for 2 h in order to determine the importance of light fluence rate in nda1 and ndc1 regulation (Fig. 2). All of the tested fluence rates induced substantial increases in nda1 transcript abundance, and there is a clear fluence rate dependence in the magnitude of the effect, with the 100 μmol m−2 s−1 treatment producing approximately 3-fold more nda1 transcript than the 0.1 μmol m−2 s−1 treatment. In contrast, only the 1, 10, and 100 μmol m−2s−1 light treatments effectively induced the ndc1 gene, with no significant differences in the magnitude of induction.
Figure 2.
Fluence-rate dependence of nda1 and ndc1 light induction. Etiolated Arabidopsis seedlings were exposed to various intensities of white light for 2 h, and transcript abundance was quantified by real-time RT-PCR. Data are presented as means ± sd from three separate RNA preparations for each point.
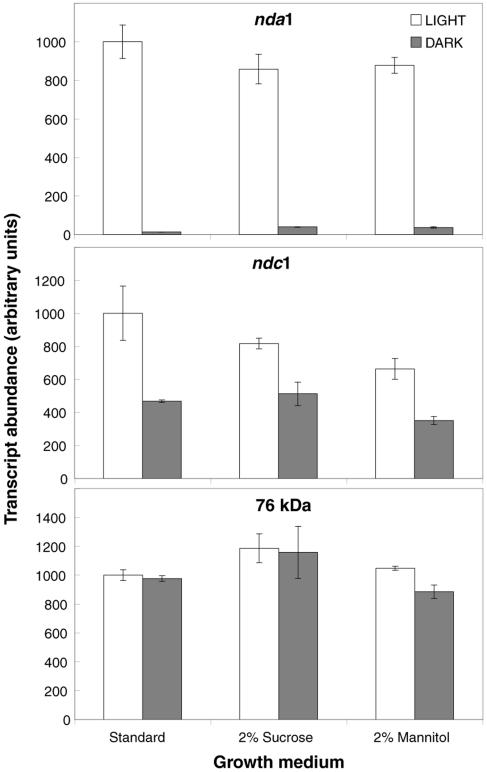
Light can alter gene expression both directly, through the action of photoreceptors, and indirectly, through the activation of photosynthetic metabolism. In particular, photosynthetic carbohydrate production appears to be the primary driver of a variety of light responses (Cheng et al., 1992; Hsieh et al., 1998). In order to investigate the effects of carbon status on nda1 and ndc1 light induction, seedlings grown on basal medium (lacking carbohydrate) were compared with seedlings grown on medium supplemented with 2% Suc or 2% mannitol (a nonmetabolizable osmotic control). As illustrated in Figure 3, Suc had no substantial inductive effect on nda1 or ndc1 gene expression in the light or dark, suggesting that carbon status plays little role in the regulation of the genes under these conditions.
Figure 3.
Effects of carbon status on nda1 and ndc1 expression. Etiolated Arabidopsis seedlings were grown on carbohydrate-free standard medium (Somerville and Ogren, 1982) or standard medium supplemented with 2% (w/v) Suc or 2% (w/v) mannitol (a nonmetabolizable osmotic control). Seedlings were maintained in darkness or exposed to 10 μmol m−2 s−1 white light for 4 h. Real-time RT-PCR measurements of transcript abundance are presented as means ± sd from two separate RNA preparations for each point.
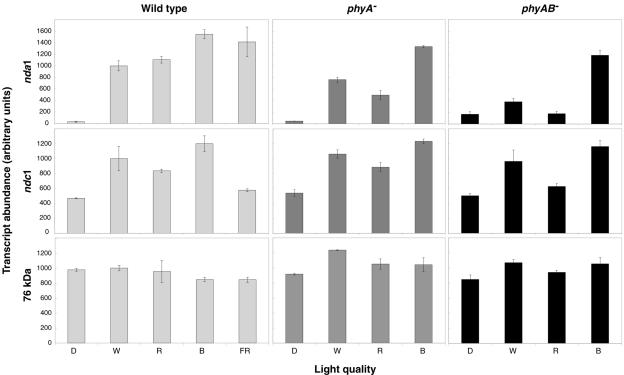
The rapidity, sensitivity to low light fluence rates (0.1 and/or 1 μmol m−2 s−1), and carbon independence of nda1 and ndc1 light response in etiolated seedlings suggested a direct (photoreceptor-mediated) light effect rather than an indirect effect of photosynthetic metabolism. In order to determine which photoreceptor families may mediate light induction, etiolated seedlings were exposed to 10 μmol m−2 s−1 continuous blue, red, far-red, or white light for 4 h (Fig. 4). Induction of nda1 was apparent under all tested light qualities, though at somewhat varying magnitudes, with blue-exposed plants displaying the highest response. ndc1 was up-regulated approximately 2- to 3-fold under red, blue, and white light, but exposure to far-red light had a minimal inductive effect. Induction by red and/or far-red light implicates the phytochrome photoreceptor family, encoded by the PHYA-E genes in Arabidopsis (Wang and Deng, 2002), while blue light response could be mediated by the cryptochromes (cry1-3), the phototropins (phot1-2), or phytochrome A (Whitelam et al., 1993; Liscum et al., 2003).
Figure 4.
The regulation of nda1 and ndc1 by different light qualities in wild-type, phyA−, and phyAB− genotypes. Five-day-old etiolated seedlings were exposed to 10 μmol m−2 s−1 white (W), red (R), or blue (B) light for 4 h. D indicates maintenance in complete darkness. Real-time RT-PCR measurements of transcript abundance are presented as means ± sd from two separate RNA preparations for each point.
Molecular Mechanisms of Light Response in nda1 and ndc1
phyA and phyB are the dominant phytochromes involved in Arabidopsis seedling germination and deetiolation and share several common signal transduction components (Gyula et al., 2003). It is well established that phyA is the only photoreceptor active under continuous far-red light (Tepperman et al., 2001; Sullivan and Deng, 2003). Thus, the strong induction of nda1 by far-red light clearly implicates phyA as a component of nda1 light response. In order to investigate which photoreceptor(s) regulate nda1 and ndc1 under other light qualities, we examined light response in phyA and phyAB mutant backgrounds (Fig. 4).
The magnitude of nda1 induction under red light was approximately halved in the phyA mutant and completely abolished in the phyAB double mutant, demonstrating that phyA and phyB are necessary and sufficient for nda1 red light response. ndc1 induction under red light illumination is generally weak, however, a clear decrease in transcript abundance is apparent in the phyAB double mutant (but not the phyA single mutant). This suggests that either phyA and phyB play redundant roles in this response or that phyB alone is the dominant red light receptor. Red light induction of ndc1 in a phyB monogenic mutant was comparable to the wild-type response (data not shown), demonstrating that phyA and phyB can act redundantly in this case.
Induction of ndc1 by blue light was not affected in the phyA and phyAB mutant backgrounds, and nda1 blue light induction showed only a small decrease in the phyA and phyAB mutants. These results demonstrate that induction by blue light is not primarily a phyA-mediated response, clearly implicating the involvement of a separate class of blue light photoreceptors. Thus, light regulation of nda1 and ndc1 is mediated by at least two photoreceptor families: the phytochromes and the phototropins and/or cryptochromes.
Distinct Temporal Profiles of Red and Blue Light Responses
Under white light conditions, both the phytochromes and a separate class of blue light photoreceptors appear to be active in nda1 and ndc1 induction (Fig. 4). Thus, multiple signal transduction pathways, each possessing unique response times, probably contribute to nda1 and ndc1 light response. Indeed, kinetic analyses of nda1 transcript accumulation under monochromatic light illumination demonstrate that red light induction of this gene is relatively transient, with an observed maximum at 4 h of exposure, while the magnitude of the blue light response steadily increases over the 12-h treatment (Fig. 5).
Figure 5.
nda1 and ndc1 induction kinetics under red and blue light illumination. Accumulation of the nda1 and ndc1 transcripts was measured by real-time PCR analysis of cDNA from wild-type (WT) and phyA− seedlings exposed to 10 μmol m−2 s−1 blue (B) or red (R) light for 0, 2, 4, and 12 h. Data are presented as means ± sd from two separate RNA preparations for each point.
phyA is relatively abundant in etiolated plants and is very rapidly transported from the cytosol into the nucleus upon exposure to red light (Nagy and Schäfer, 2002). However, phyA is degraded to nearly undetectable levels after 2 to 3 h of red light exposure (Hennig et al., 1999). In contrast, phyB is photostable but imported to the nucleus slowly, with a half-saturation time of 1 to 2 h (Nagy and Schäfer, 2002). Thus, the transient kinetics of nda1 red light response could be due to distinct temporal activity profiles of the photoreceptors controlling its expression. Indeed, the kinetics of red light response in the phyA mutant (Fig. 5) clearly demonstrate that phyA is absolutely required for the rapid, high-level induction of nda1 in the first 4 h of red light exposure, but it appears to play a minimal role in the maintenance of induction thereafter. Thus, distinct temporal inputs from at least three photoreceptors are likely integrated into nda1 induction under natural (white light) conditions, with blue light receptor(s) and phyA controlling response in the initial hours of light exposure and blue light effects (and to lesser extent phyB-mediated red light effects) dominating later.
Unlike nda1, the kinetics of ndc1 transcript accumulation under red and blue illumination regimes are similar, displaying steady increase throughout the experiment. As in previous experiments (Fig. 4), the loss of phyA has no effect on ndc1 red light induction (Fig. 5).
Fusion of the nda1 and ndc1 Promoter Regions to the β-Glucuronidase Transgene
To investigate the transcriptional control of nda1 and ndc1 gene expression, a series of nda1 and ndc1 promoter-β-glucuronidase (GUS) fusions was generated. The full-length nda1pro-GUS fusion (nda1Δ1043) contains the nda1 5′-untranslated region (UTR) and 1,043 bp of sequence upstream of the transcriptional start site, while the corresponding ndc1 construct (ndc1Δ1068) contains the ndc1 5′-UTR with 1,068 bp of upstream sequence. When expressed in transgenic Arabidopsis plants, both the nda1pro- and ndc1pro-GUS fusions display clear induction upon exposure to red, blue, or white light for 12 h (Fig. 6A), suggesting that nda1 and ndc1 light induction is likely controlled at the transcriptional level. (It should be noted that the presence of native 5′-UTR regions in the fusion constructs means that possible posttranscriptional effects cannot be absolutely excluded.) The nda1pro-GUS fusion may lack some cis-elements required for complete recapitulation of nda1-type light regulation, as the general magnitude of light induction is quantitatively reduced and the efficacy of white light (as compared to blue light) as an inducer of GUS expression is lower than expected. Transgenic lines expressing ndc1pro-GUS more precisely mirror native ndc1 light regulation characteristics, generally displaying modest (2- to 3-fold) induction upon exposure to white, red, or blue light.
Figure 6.
Light responsiveness of nda1pro-GUS and ndc1pro-GUS fusions. A, Seedlings of transgenic Arabidopsis lines expressing full length nda1 (Δ1043-1, -2, -9) or ndc1 (Δ1068-2, -5, -8) promoters fused to the GUS gene were exposed to 10 μmol m−2 s−1 light for 12 h, and GUS transcript accumulation was quantified by real-time RT-PCR analysis. The highest measured transcript abundance was arbitrarily assigned a value of 100. B, Five-day-old etiolated seedlings expressing a series of 5′ deletions of the nda1 promoter fused to GUS (Δ1043-1, 9; Δ426-6, 7; Δ281-8, 11; Δ182-2, 10) were exposed to 100 μmol m−2 s−1 white light for 24 h or maintained in darkness. GUS activity was quantified in seedling protein extracts by fluorometric MUG analysis. Data are presented as means ± sd from three separate protein extracts for each point.
Transgenic plants expressing GUS under the transcriptional control of a series of 5′ deletions of the nda1 promoter (1,043; 426; 281; or 182 bp upstream of the transcriptional start) were examined in order to localize key cis-regions of the nda1 promoter involved in light response. In order to simplify the detection and quantitation of GUS protein activity, light response was maximized by increasing the duration of light exposure to 24 h, increasing fluence rate to 100 μmol m−2 s−1, and utilizing a white metal halide bulb with strong emission in the blue region of the spectrum. As shown in Figure 6B, light-induced GUS activity is high in the Δ1,043 transgenic lines, is progressively reduced in the Δ426 and Δ281 lines, and is essentially lost in the Δ182 lines. These results suggest that separate positive quantitative elements related to light response exist in the −1,043 to −426 and −426 to −281 regions of the nda1 promoter, and that critical element(s) for the initiation of light induction are present in the 99-bp region from −281 to −182. This pattern of several upstream quantitative elements and a core response element near −250 is typical of many characterized light-responsive promoters from photosynthesis-associated nuclear genes (Terzaghi and Cashmore, 1995).
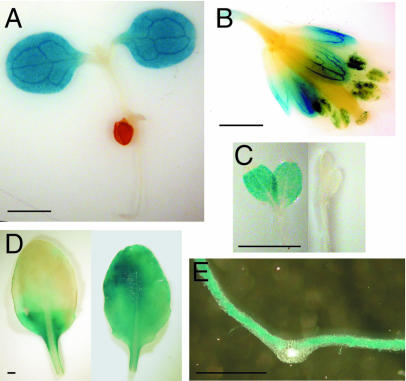
In addition to facilitating analysis of transcriptional control of nda1 and ndc1, transgenic plants expressing the nda1Δ1,043 and ndc1Δ1,068 promoter-GUS fusions were utilized for histochemical GUS analysis, allowing cell and tissue localization of GUS expression. Expression patterns of nda1pro-GUS, but not ndc1pro-GUS, were consistent with previous crude organ-level localization of the native transcripts (Michalecka et al., 2003), so data pertaining only to the former construct are reported here. In both light-grown seedlings and etiolated seedlings exposed to light for 24 h, the nda1pro-GUS construct is expressed solely in cotyledons (Fig. 7, A and C). In mature light-grown plants, GUS activity is detected in essentially all green tissue (leaves, stems, sepals) except siliques, and is also detected in anthers (Fig. 7, B and D). GUS expression in mature leaves is specifically localized in mesophyll cells of the lamina and is light dependent, as demonstrated by the loss of GUS activity in leaf segments maintained in the dark for 3 d (Fig. 7, D and E). High abundance of native nda1 transcript in the lamina compared to the midvein region of the leaf (Fig. 7E) was verified by real-time PCR analysis of dissected leaves (data not shown).
Figure 7.
Histochemical localization of GUS expression in transgenic Arabidopsis plants expressing nda1pro-GUS fusions. A, Ten-day-old light-grown seedling. B, Mature flower from light-grown plant. C, Five-day-old etiolated seedling exposed to 100 μmol m−2 s−1 white light for 24 h (left) or maintained in darkness (right). D, Leaf from light-grown plant whose distal half was partially covered (shielded from light) for 3 d (left) or left uncovered (right). E, Cross section of mature leaf from light-grown seedling. All photos are from plants expressing the Δ1043 (full-length) nda1 promoter-GUS construct. Scale bars = 1 mm.
DISCUSSION
A small number of studies have demonstrated that short- and/or long-term light exposure can be an important regulator of the transcript abundance, protein abundance, and/or protein activity of several energy-dissipating components of the ETC (Atkin et al., 1993; Finnegan et al., 1997; Ribas-Carbo et al., 2000; Svensson and Rasmusson, 2001; Michalecka et al., 2003). However, several global analyses of light regulation using microarrays have failed to identify respiratory chain genes regulated by light, although several other mitochondrial pathways (Krebs cycle and photorespiration) are light responsive (Ma et al., 2001; Tepperman et al., 2001). We have examined the light regulation of all putative energy-dissipating ETC proteins in Arabidopsis, a species in which the alternative oxidase, uncoupling protein, and type II NAD(P)H dehydrogenase gene families are well characterized at the genetic level. These studies have demonstrated that two NAD(P)H dehydrogenase genes, nda1 and ndc1, and one alternative oxidase gene, aox2, are substantially regulated by light in etiolated seedlings.
The down-regulation of aox2 by light was not studied in detail due to high variability in its dark transcript levels, but its general pattern of regulation provides an interesting contrast to previous studies of alternative oxidase light regulation in soybean and potato. Soybean has two aox2-type genes: aox2a, which is up-regulated at the mRNA and protein levels after 12 to 24 h of illumination in etiolated seedlings, and aox2b, which is down-regulated at the mRNA level under identical conditions (Finnegan et al., 1997; gene nomenclature according to Considine et al., 2002). Consistent with soybean aox2a, the abundance of alternative oxidase protein is decreased in potato after 4 d of dark treatment (Svensson and Rasmusson, 2001). Thus, it is perhaps less surprising that Arabidopsis aox2 transcript levels decrease in the light (analogous to soybean aox2b) than that none of the other members of the Arabidopsis gene family display strong light induction. It is possible, however, that the observed weak up-regulation of aox1a and/or aox1c is enhanced after longer periods of light exposure, potentially via a secondary (nonphotoreceptor-mediated) light effect associated with the activation of photosynthetic metabolism, which generally begins after approximately 12 h of light exposure in etiolated plants (Bradbeer, 1981).
There are three families of type II NAD(P)H dehydrogenase genes in Arabidopsis: nda (1-2), ndb (1-4), and ndc (1), all of which encode mitochondrially localized proteins (Michalecka et al., 2003). The precise function of each gene has not, however, been assigned. NDA1 in potato and Arabidopsis is an internal dehydrogenase most likely oxidizing matrix NADH (Rasmusson et al., 1999; Moore et al., 2003; Rasmusson et al., 2004), while NDB1 in potato faces the intermembrane space and oxidizes cytoplasmic NADPH (Michalecka et al., 2004). Potato NDC1 appears to be matrix facing, but its substrate specificity is unknown (Michalecka, 2004). In this study, the light regulation of the nda1 and ndc1 genes was characterized in detail, allowing a partial determination of the mechanisms of light response.
The Arabidopsis nda1 gene displayed an exceptionally strong light induction in seedlings (Fig. 1), comparable in rapidity and magnitude to the well-characterized light responses of the Rubisco small subunit (RBCS) and chlorophyll a/b binding protein genes involved in photosynthesis (Ma et al., 2001; Tepperman et al., 2001). nda1 transcript accumulation was induced by far-red, red, and blue light via the action of phytochrome A, phytochromes A and B, and an unknown blue light photoreceptor, respectively. Several recent studies suggest that many light-regulated genes in Arabidopsis are similarly controlled by multiple distinct photoreceptor inputs (Ma et al., 2001; Tepperman et al., 2004). phyA and phyB act sequentially in nda1 red light response, likely reflecting the differential photostability and nuclear localization kinetics of the photoreceptors (Nagy and Schäfer, 2002). A similar temporal pattern of phytochrome activity has been observed in red light regulation of the cessation of hypocotyl elongation, with phyA controlling early red light response (≤3 h light exposure) and phyB dominating subsequently (Parks et al., 2001). The identity of the nda1 blue light photoreceptor was not investigated; however, a recent study suggests that the vast majority of blue light-induced changes in the Arabidopsis transcriptome are mediated by the cryptochromes (Ohgishi et al., 2004).
The promoter regions of both nda1 and ndc1 drive light-inducible expression of a GUS transgene, and the genetic basis of light-regulated transcription was examined further through analysis of a series of 5′ deletions of the nda1 promoter. Light responsiveness is maintained (albeit at a quantitatively reduced level) in a 281-bp fragment of the nda1 promoter, but is lost by deletion of an additional 99 bp. Interestingly, this 99-bp core light-responsive region contains an I-box motif (GATAAG) at −218 flanked by two GT-1 consensus elements (GPu[A/T]AA[A/T]) at −234 and −202 (Higo et al., 1999). The I-box is a functionally important GATA-type light regulatory element located approximately 130 to 330 bp upstream of the transcriptional start of many RBCS genes (Terzaghi and Cashmore, 1995). GT-1 sites are relatively degenerate cis-elements that are commonly, but not exclusively, associated with light-regulated promoters (Zhou, 1999). The combination of these two elements can form a potent light-responsive core, as a GT-1 and I-box cluster in the pea RBCS-3A gene is required for high-level light regulation (Sarokin and Chua, 1992), and a synthetic tetramer of GT-1 and I-box elements can confer responsiveness to far-red, red, and blue light upon the otherwise non-light-regulated NOS101 promoter (Chattopadhyay et al., 1998). Combinations of GT-1 elements and I-box-like GATA elements are also present in the light-responsive promoters of the genes encoding the H and T subunits of Gly decarboxylase, which catalyzes the first step of photorespiration in the mitochondria (Srinivasan and Oliver, 1995; Vauclare et al., 1998). Thus, the −234 to −202 I-box/GT-1 cluster in the nda1 promoter is a strong candidate as the primary controller of light-regulated transcription in this gene.
The localization of nda1pro-GUS expression generally in green tissue and specifically in leaf lamina mesophyll cells is strikingly similar to expression patterns of several light-regulated photosynthetic genes, such as Rubisco activase (Liu et al., 1996) and the Rubisco small subunit family (Meier et al., 1995). Taken together, the nda1 gene's spatial-expression patterning, light-induction kinetics, and modular arrangement of cis-transcriptional elements suggest that the major physiological role of this respiratory NAD(P)H dehydrogenase is in photosynthetic metabolism.
Although quantitatively weaker than nda1 light response, the light regulation of ndc1 in Arabidopsis seedlings also appears to involve multiple photoreceptor families. Of the tested monochromatic light treatments, only red and blue light efficiently induce ndc1 expression. Red light response was redundantly regulated by either phyA or phyB, however the loss of both phyA and phyB has no effect on white (or blue) light induction. Because the kinetics of ndc1 light induction are not consistent with an effect arising from photosynthetic metabolism (Bradbeer, 1981), it thus appears that the primary driver of ndc1 induction in white light is a cryptochrome or phototropin.
Three action modes have been defined for the phytochromes: the red/far-red reversible low-fluence responses, the high-irradiance responses (HIRs) which require prolonged high light exposure, and the very low fluence responses, which are activated by very low light fluences (Wang and Deng, 2002). Little can be posited about the mode of ndc1 red light response, but phytochrome regulation of nda1 appears to involve a far-red HIR, as suggested by the inductive effect of far-red light, the fluence rate dependence of nda1 induction, and the previously characterized deficiency of the very low fluence response mode in the Columbia ecotype (Yanovsky et al., 1997). Far-red HIRs are a prominent response mode in several aspects of Arabidopsis seedling development, including seed germination and deetiolation (Wang and Deng, 2002).
Unfortunately, it is unclear whether NDA1 and NDC1 protein levels correlate with mRNA abundance, as attempts to detect these proteins in light-exposed etiolated seedlings by immunoblotting were unsuccessful. At least for nda1, this is likely due to the limited spatial domain (cotyledons) and relatively low expression levels in etiolated plants. Although NDA protein(s) have previously been detected at low levels in light-grown Arabidopsis plants, specific quantitation of NDA1 is complicated by the presence of the non-light-responsive nda2 gene, which is highly similar to nda1 throughout its length and of similar molecular mass (Michalecka et al., 2003). It should be noted, however, that expression of the nda1 homolog in potato is light regulated at the RNA, protein, and activity levels (Svensson and Rasmusson, 2001).
An etiolated plant that is exposed to light undergoes a massive burst of transcriptional and translational activity and initiates rapid assembly of the photosynthetic machinery (Bradbeer, 1981; Tepperman et al., 2001). Why would these energy-intensive processes be accompanied by an increase in NAD(P)H dehydrogenases (and in some cases alternative oxidases) whose primary function is to bypass energy conservation in the mitochondrial respiratory chain? The activation of photorespiration in the light leads to major NADH generation by Gly decarboxylase in the mitochondrial matrix. A corresponding increase in the redox capacity of the respiratory chain is presumably required to maintain photorespiration without causing overreduction of the basal ETC and matrix NADH and resultant ROS formation (Maxwell et al., 1999; Møller, 2001; Rasmusson et al., 2004). The structurally simple type II NAD(P)H dehydrogenases and alternative oxidases would seem to be ideally suited for such dynamic, short-term redox adjustments that could maintain multiprotein complexes I and III, and ubiquinone, in a relatively oxidized state (Rasmusson et al., 1998). Indeed, several studies have shown that Gly oxidation is generally insensitive to the inhibition of complex I, suggesting that type II NAD(P)H dehydrogenases can participate in reoxidation of photorespiratory NADH (Dry and Wiskich, 1985; Igamberdiev et al., 1997). Thus, it is possible that light regulation of nda1 and ndc1 reflects a critical role in the maintenance of photorespiratory metabolism.
MATERIALS AND METHODS
Plant Material
Arabidopsis (ecotype Columbia) seeds were sown on carbohydrate-free nutrient medium (Somerville and Ogren, 1982) supplemented with 0.8% agar and stratified at 4°C for 4 d, unless otherwise noted. Seeds were then exposed to 100 μmol m−2 s−1 white light for 1 h to induce germination and incubated in the dark at room temperature for 5 d prior to light treatment. The phytochrome loss of function genotypes phyA-211 (Reed et al., 1994), phyB-9 (Reed et al., 1993), and phyA-211× phyB-9 were utilized in mutant analyses.
Light Sources
White light treatments were performed using white fluorescent tubes (Asea Skandia 36W/83; Elektroskandia, Sollentuna, Sweden) except for the use of a metal halide lamp (Osram powerstar HQI-T; Osram, Munich) in the GUS fluorometric analyses (Fig. 6B). Far-red light was provided by light-emitting diodes with a λmax of 735 nm (Farnell, Leeds, UK). Blue light was provided by internally coated blue fluorescent tubes (TLD 36W/18; Philips, Eindhoven, Holland), filtered through two layers of blue cellulose acetate film (99.6% of total fluence between 320 and 550 nm), and red light was from internally coated red fluorescent tubes (TLD 36W/15; Philips; 95.7% of total fluence between 600 and 700 nm). Unless otherwise noted, seedlings were exposed to continuous light at a fluence rate of 10 μmol m−2 s−1. Light fluence rates were measured using a LI-COR LI-189 quantum meter (LI-COR, Lincoln, NE), and light spectra were determined using an Optronics 754-6S spectroradiometer (Optronics, Goleta, CA).
RNA Isolation and cDNA Synthesis
Total RNA was isolated from 100 mg of whole etiolated seedlings using the RNeasy Plant Mini kit (Qiagen USA, Valencia, CA). RNA was quantified on a Shimadzu UV-160A spectrophotometer (Shimadzu, Kyoto) and intactness was verified by visual inspection of rRNA banding in electrophoretically separated total RNA (Sambrook et al., 1989). For analysis of GUS transcript levels, RNA was DNAse treated using Ambion's DNA-free kit (Austin, TX). First strand cDNA synthesis was performed on 1 μg of total RNA per reaction of the Superscript II First Strand Synthesis system (Invitrogen, Carlsbad, CA). cDNA was diluted 5-fold with 10 mm Tris-HCl, pH 8.0, for use in real-time PCR analysis.
Real-Time PCR Analysis
PCR primers for the aox1a, aox1b, aox1c, aox1 d, aox2, nda1, nda2, ndb1, ndb2, ndb3, ndb4, ndc1, ucp1, ucp2, gdcH, gusA, and 76-kD genes were designed using the Primer3 software program (Rozen and Skaletsky, 2000) such that one primer in each pair spans an exon-exon border (except for the intronless gusA transgene). Primer sequences were, aox1a (locus At3g22370): 5′-CCGATTTGTTCTTCCAGAGG-3′, 5′-GCGCTCTCTCGTACCATTTC-3′; aox1b (locus At3g22360): 5′-CTTTTCTTCCAGAGGCGGTA-3′, 5′-TTAGGTTTCGCGACTTCCAT-3′; aox1c (locus At3g27620): 5′-CCGATCTTTTCTTCCAGAGG-3′, 5′-TGGGAGAGATTATGTATCCGATT-3′; aox1d (locus At1g32350): 5′-TGTTCGGCTATTGAGCTCTG-3′, 5′-ATCGCTCGTTCGTACCATTT-3′; aox2 (locus At5g64210): 5′-TGACGGTAAAGAAGGGTCAAA-3′, 5′-TGCATCCATATCGTCTCTGAA-3′; ucp1 (locus At3g54110): 5′-TCTGCTCTTGCTGGTGATGT-3′, 5′-TACCCAGTGCACCTGTTGTC-3′; ucp2 (locus At5g58970): 5′-GGATTTCAAACCAAGGATCG-3′, 5′-AGCGCACTAACTCCTTCCAG-3′; ndb4 (locus At2g20800): 5′-TTGTGGGAGTGACTGCTGAT-3′, 5′-TCGCAGCTATATCTTCCATGAC-3′; gdcH (locus At2g35370): 5′-CACAGAATCACCTGGCTTGA-3′, 5′-GCATGAGAATTGATAGAACTTGGA-3′; gusA (accession U12639): 5′-TGTGGAGTATTGCCAACGAA-3′, 5′-GGCACAGCACATCAAAGAGA-3′. The nda1 (At1g07180), nda2 (At2g29990), ndb1 (At4g28220), ndb2 (At4g05020), ndb3 (At4g21490), ndc1 (At5g08740), and 76-kD subunit (At5g37510) primer sequences have been reported previously (Michalecka et al., 2003). A small region of the reported genomic sequence of the ndc1 gene was found to be inconsistent with existing cDNA sequences and expressed sequence tags. We amplified this region using Advantage HF-2 polymerase (CLONTECH Laboratories, Palo Alto, CA) and found its sequence to be consistent with cDNA. A correction of this region of ndc1 genomic sequence has been submitted to GenBank with accession number AJ715502.
Real-time PCR reactions were run in a Rotor Gene 2072 Real-Time Cycler (Corbett Research, Sydney) as described previously (Svensson et al., 2002), except for the use of SYBR green at a 1:20,000 dilution, an extension temperature of 72°C, and an acquiring temperature of 81°C. For initial screening studies (Table I), real-time reactions were run without relative quantitation standards, so comparison of transcript abundance was based upon the number of amplification cycles required to meet a fluorescence threshold (Ct). This threshold was set manually to detect in the early exponential phase of amplification. In all later experiments, serial 10-fold dilutions of PCR-amplified DNA from the gene to be investigated were utilized as relative quantitation standards, and the fluorescence threshold was calculated based upon line of best fit. Control PCRs minus the RT step were run to verify lack of genomic DNA amplification. All presented PCR data were generated from a minimum of two independent reactions for each biological replicate.
In Figures 1 to 5, a value of 1,000 (arbitrary units) is defined as the abundance of the measured transcript in wild-type plants exposed to 10 μmol m−2 s−1 white light for 4 h. (Note that the 4-h time point is not shown in all figures.) All values are thus transcript specific, and not comparable between the different studied genes.
Vector Construction and Plant Transformation
A fragment of the nda1 promoter consisting of the 94-bp 5′-UTR and 1,043 bp of sequence upstream of the transcriptional start was PCR amplified from Arabidopsis DNA using the Advantage HF-2 polymerase (CLONTECH) and cloned into the pCR4-TOPO vector (Invitrogen). The ndc1 promoter, consisting of a 100-bp 5′-UTR and 1,068 bp of upstream sequence was similarly cloned. The promoter regions were sequenced to verify amplification fidelity and then cloned into the pBI101 binary vector (Jefferson et al., 1987), generating the GUS transcriptional fusion plasmids pA1Δ1043 (nda1) and pC1Δ1068 (ndc1). A series of 5′ deletions of the nda1 promoter were generated by digesting the cloned nda1 promoter (in pCR4-TOPO) with HindIII and BamHI, excising a 520-bp fragment (426-bp upstream sequence + 5′-UTR), or NsbI and BamHI, excising a 375-bp fragment (281-bp upstream sequence + 5′-UTR). These fragments, in addition to a sequence-verified 276-bp nda1 promoter deletion amplified by PCR (182-bp upstream sequence + 5′UTR), were subsequently cloned into pBI101, producing pA1Δ426, pA1Δ281, and pA1Δ182. The binary vector constructions were electroporated into Agrobacterium tumefaciens GV3101, and floral dip transformation of Arabidopsis was performed as described by Clough and Bent (1998). Transformed seedlings were selected on half-strength Murashige and Skoog medium containing 50 mg/L kanamycin. Transformants containing a single T-DNA insertion locus were identified by segregation analysis and utilized in all presented experiments.
GUS Analysis
Histochemical GUS staining was performed as described by Lindsey and Wei (2000) except that Triton X-100 was utilized at a concentration of 1% (v/v; Pretova et al., 2003). All samples were cleared of chlorophyll in 70% ethanol following staining. Quantitative fluorometric GUS analysis (MUG [4-methylumbelliferyl β-d-glucoronide analysis]) was performed as described by Jefferson (1987). Fluorescence was measured on a Shimadzu RF-5301PC fluorometer calibrated with standard solutions of 4-methyl umbelliferone. Fluorescence readings from protein extracts incubated with MUG substrate at 37°C for T = 60 min were subtracted from corresponding readings at T = 0 min to generate net fluorescence values. Protein isolations and MUG enzyme assays were performed in triplicate for each genotype/treatment.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AJ715502.
This work was supported by the Swedish Research Council for Environment, Agricultural Sciences, and Spatial Planning, by Carl Tesdorpfs Stiftelse, and by the Wenner-Gren Foundations (fellowship to M.A.E.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.046698.
References
- Argüello-Astorga G, Herrera-Estrella L (1998) Evolution of light-regulated plant promoters. Annu Rev Plant Physiol Plant Mol Biol 49: 525–555 [DOI] [PubMed] [Google Scholar]
- Atkin OK, Cummins WR, Collier DE (1993) Light induction of alternative pathway capacity in leaf slices of Belgium endive. Plant Cell Environ 16: 231–235 [Google Scholar]
- Bradbeer JW (1981) Development of photosynthetic function during chloroplast biogenesis. In PK Stumpf, EE Conn, eds, The Biochemistry of Plants, Vol 8. Academic Press, New York, pp 423–472
- Brandalise M, Maia IG, Borecky J, Vercesi AE, Arruda P (2003) Overexpression of plant uncoupling mitochondrial protein in transgenic tobacco increases tolerance to oxidative stress. J Bioenerg Biomembr 35: 203–209 [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S, Puente P, Deng X-W, Wei N (1998) Combinatorial interaction of light-responsive elements plays a critical role in determining the response characteristics of light-regulated promoters in Arabidopsis. Plant J 15: 69–77 [DOI] [PubMed] [Google Scholar]
- Cheng CL, Acedo GN, Cristinsin M, Conkling MA (1992) Sucrose mimics the light induction of Arabidopsis nitrate reductase gene transcription. Proc Natl Acad Sci USA 89: 1861–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Considine MJ, Holtzapffel RC, Day DA, Whelan J, Millar AH (2002) Molecular distinction between alternative oxidase from monocots and dicots. Plant Physiol 129: 949–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Bari RP, Stitt M, Scheible W-R, Udvardi MK (2004) Real time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecendented sensitivity reveals novel root- and shoot-specific genes. Plant J 38: 366–379 [DOI] [PubMed] [Google Scholar]
- Dry IB, Wiskich JT (1985) Characteristics of glycine and malate oxidation by pea leaf mitochondria: evidence of differential access to NAD and respiratory chains. Aust J Plant Physiol 12: 329–339 [Google Scholar]
- Finnegan PM, Whelan J, Millar H, Zhang Q, Smith MK, Wiskich JT, Day DA (1997) Differential expression of the multigene family encoding the soybean mitochondrial alternative oxidase. Plant Physiol 114: 455–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyula P, Schäfer E, Nagy F (2003) Light perception and signaling in higher plants. Curr Opin Plant Biol 6: 446–452 [DOI] [PubMed] [Google Scholar]
- Hennig L, Büche C, Eichenberg K, Schäfer E (1999) Dynamic properties of endogenous phytochrome A in Arabidopsis seedlings. Plant Physiol 121: 571–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton JR, Owen PD (1985) Phytochrome regulation of extractable cytochrome oxidase activity during early germination of Bromus sterilis L. and Lactuca sativa L. CV. Grand Rapids seeds. New Phytol 100: 163–171 [Google Scholar]
- Hsieh MH, Lam HM, Van De Loo FJ, Coruzzi G (1998) A PII-like protein in Arabidopsis: putative role in nitrogen sensing. Proc Natl Acad Sci USA 95: 13965–13970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igamberdiev AU, Bykova NV, Gardeström P (1997) Involvement of the cyanide-resistant and rotenone-insensitive pathways of mitochondrial electron transport during oxidation of glycine in higher plants. FEBS Lett 412: 256–269 [DOI] [PubMed] [Google Scholar]
- Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5: 387–405 [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krömer S (1995) Respiration during photosynthesis. Annu Rev Plant Physiol Plant Mol Biol 46: 45–70 [Google Scholar]
- Laloi M, Klein M, Riesmeier JW, Müller-Röber B, Fleury C, Bouillaud F, Ricquier D (1997) A plant cold-induced uncoupling protein. Nature 389: 135–136 [DOI] [PubMed] [Google Scholar]
- Lancien M, Ferrario-Mery S, Roux Y, Bismuth E, Masclaux C, Hirel B, Gadal P, Hodges M (1999) Simultaneous expression of NAD-dependent isocitrate dehydrogenase and other Krebs cycle genes after nitrate resupply to short-term nitrogen-starved tobacco. Plant Physiol 120: 717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey K, Wei W (2000) Tissue culture, transformation, and transient gene expression in Arabidopsis. In ZA Wilson, ed, Arabidopsis: A Practical Approach. Oxford University Press, Oxford, pp 125–143
- Liscum E, Hodgson DW, Campbell TJ (2003) Blue light signaling through the cryptochromes and phototropins. Plant Physiol 133: 1429–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Taub CC, McClung CR (1996) Identification of an Arabidopsis thaliana ribulose-1,5-bisphosphate carboxylase/oxygenase activase (RCA) minimal promoter regulated by light and the circadian clock. Plant Physiol 112: 43–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Li J, Qu L, Hager J, Chen Z, Zhao H, Deng XW (2001) Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13: 2589–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell DP, Wang Y, McIntosh L (1999) The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc Natl Acad Sci USA 96: 8271–8276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CR, Hsu M, Painter JE, Gagne JM, Karlsberg SD, Salomé PA (2000) Integrated temporal regulation of the photorespiratory pathway: circadian regulation of two Arabidopsis genes encoding serine hydroxymethyltransferase. Plant Physiol 123: 381–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier I, Callan KL, Fleming AJ, Gruissem W (1995) Organ-specific differential regulation of a promoter subfamily for the ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit genes in tobacco. Plant Physiol 107: 1105–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalecka AM (2004) Alternative NAD(P)H Dehydrogenases in Plant Mitochondria. PhD thesis. Lund University, Lund, Sweden
- Michalecka AM, Agius SC, Møller IM, Rasmusson AG (2004) Identification of a mitochondrial external NADPH dehydrogenase by overexpression in transgenic Nicotiana sylvestris. Plant J 37: 415–425 [DOI] [PubMed] [Google Scholar]
- Michalecka AM, Svensson ÅS, Johansson FI, Agius SC, Johanson U, Brennicke A, Rasmusson AG (2003) Arabidopsis genes encoding mitochondrial type II NAD(P)H dehydrogenases have different evolutionary origin and show distinct responses to light. Plant Physiol 133: 642–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller IM (2001) Plant mitochondria and oxidative stress: electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu Rev Plant Physiol Plant Mol Biol 52: 561–591 [DOI] [PubMed] [Google Scholar]
- Moore CS, Cook-Johnson RJ, Rudhe C, Whelan J, Day DA, Wiskich JT, Soole KL (2003) Identification of AtNDI1, an internal non-phosphorylating NAD(P)H dehydrogenase in Arabidopsis mitochondria. Plant Physiol 133: 1968–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy F, Schäfer E (2002) Phytochromes control photomorphogenesis by differentially regulated, interacting signaling pathways in higher plants. Annu Rev Plant Biol 53: 329–355 [DOI] [PubMed] [Google Scholar]
- Ohgishi M, Saji K, Okada K, Sakai T (2004) Functional analysis of each blue light receptor, cry1, cry2, phot1, and phot2, by using combinatorial multiple mutants in Arabidopsis. Proc Natl Acad Sci USA 101: 2223–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BM, Folta KM, Spalding EP (2001) Photocontrol of stem growth. Curr Opin Plant Biol 4: 436–440 [DOI] [PubMed] [Google Scholar]
- Pretova A, Obert B, Wetzstein HY (2003) Substrate infiltration and histological fixatives affect the in situ localization of β-glucuronidase activity in transgenic tissues. Acta Biotechnol 23: 383–388 [Google Scholar]
- Purvis AC, Shewfelt RL (1993) Does the alternative pathway ameliorate chilling injury in sensitive plant tissues? Physiol Plant 88: 712–718 [DOI] [PubMed] [Google Scholar]
- Rasmusson AG, Heiser VV, Zabaleta E, Brennicke A, Grohmann L (1998) Physiological, biochemical and molecular aspects of mitochondrial complex I in plants. Biochim Biophys Acta 1364: 101–111 [DOI] [PubMed] [Google Scholar]
- Rasmusson AG, Soole KL, Elthon TE (2004) Alternative NAD(P)H dehydrogenases of plant mitochondria. Annu Rev Plant Biol 55: 23–39 [DOI] [PubMed] [Google Scholar]
- Rasmusson AG, Svensson ÅS, Knoop V, Grohmann L, Brennicke A (1999) Homologues of yeast and bacterial rotenone-insensitive NADH dehydrogenases in higher eukaryotes: two enzymes are present in potato mitochondria. Plant J 20: 79–88 [DOI] [PubMed] [Google Scholar]
- Reed JW, Nagatani A, Elich TD, Fagan M, Chory J (1994) Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol 104: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J (1993) Mutations in the gene for red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5: 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas-Carbo M, Robinson SA, Gonzalez-Meler MA, Lennon AM, Giles L, Siedow JN, Berry JA (2000) Effects of light on respiration and oxygen isotope fractionation in soybean cotyledons. Plant Cell Environ 23: 983–989 [Google Scholar]
- Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In S Krawetz, S Misener, eds, Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press, Totowa, NJ, pp 365–386 [DOI] [PubMed]
- Saisho D, Nambara E, Naito S, Tsutsumi N, Hirai A, Nakazono M (1997) Characterization of the gene family for alternative oxidase from Arabidopsis thaliana. Plant Mol Biol 35: 585–596 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sarokin LP, Chua N-H (1992) Binding sites for two novel phosphoproteins, 3AF5 and 3AF3, are required for rbcS-3A expression. Plant Cell 4: 473–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siedow JN, Day DA (2000) Respiration and photorespiration. In BB Buchanan, W Gruissem, RL Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD, pp 676–728
- Somerville CR, Ogren WL (1982) Isolation of photorespiration mutants in Arabidopsis thaliana. In M Edelman, RB Hallick, NH Chua, eds, Methods in Chloroplast Biology. Elsevier Biomedical Press, New York, pp 129–138
- Srinivasan R, Oliver DJ (1995) Light-dependent and tissue specific expression of the H-protein of the glycine decarboxylase complex. Plant Physiol 109: 161–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JA, Deng XW (2003) From seed to seed: the role of photoreceptors in Arabidopsis development. Dev Biol 260: 289–297 [DOI] [PubMed] [Google Scholar]
- Svensson ÅS, Johansson FI, Møller IM, Rasmusson AG (2002) Cold stress decreases the capacity for respiratory NADH oxidation in potato leaves. FEBS Lett 517: 79–82 [DOI] [PubMed] [Google Scholar]
- Svensson ÅS, Rasmusson AG (2001) Light-dependent gene expression for proteins in the respiratory chain of potato leaves. Plant J 28: 73–82 [DOI] [PubMed] [Google Scholar]
- Tepperman JM, Hudson ME, Khanna R, Zhu T, Chang SH, Wang X, Quail PH (2004) Expression profiling of phyB mutant demonstrates substantial contribution of other phytochromes to red-light-regulated gene expression during seedling de-etiolation. Plant J 38: 725–739 [DOI] [PubMed] [Google Scholar]
- Tepperman JM, Zhu T, Chang H-S, Wang X, Quail PH (2001) Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc Natl Acad Sci USA 98: 9437–9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi WB, Cashmore AR (1995) Light-regulated transcription. Annu Rev Plant Physiol Plant Mol Biol 46: 445–474 [Google Scholar]
- Thirkettle-Watts D, McCabe TC, Clifton R, Moore C, Finnegan PM, Day DA, Whelan J (2003) Analysis of the alternative oxidase promoters from soybean. Plant Physiol 133: 1158–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lis R, Atteia A (2004) Control of mitochondrial function via photosynthetic redox signals. Photosynth Res 79: 133–148 [DOI] [PubMed] [Google Scholar]
- Vanlerberghe GC, McIntosh L (1997) Alternative oxidase: from gene to function. Annu Rev Plant Physiol Plant Mol Biol 48: 703–734 [DOI] [PubMed] [Google Scholar]
- Vauclare P, Macherel D, Douce R, Bourguignon J (1998) The gene encoding T protein of the glycine decarboxylase complex involved in the mitochondrial step of the photorespiratory pathway in plants exhibits features of light-induced genes. Plant Mol Biol 37: 309–318 [DOI] [PubMed] [Google Scholar]
- Vercesi AE (2001) The discovery of an uncoupling mitochondrial protein in plants. Biosci Rep 21: 195–200 [DOI] [PubMed] [Google Scholar]
- Wang H, Deng XW (2002) Phytochrome signaling mechanism. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/10.1199/tab.0074., http://www.aspb.org/publications/arabidopsis
- Wang R, Okamoto M, Xing X, Crawford NM (2003) Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol 132: 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe A, Nakazono M, Tsutsumi N, Hirai A (1999) AtUCP2: a novel isoform of the mitochondrial uncoupling protein of Arabidopsis thaliana. Plant Cell Physiol 40: 1160–1166 [DOI] [PubMed] [Google Scholar]
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Harberd NP (1993) Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5: 757–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky MJ, Casal JJ, Luppi JP (1997) The VLF loci, polymorphic between ecotypes Landsberg erecta and Columbia, dissect two branches of phytochrome A signal transduction that correspond to very-low-fluence and high-irradiance responses. Plant J 12: 659–667 [DOI] [PubMed] [Google Scholar]
- Zhou D-X (1999) Regulatory mechanism of plant gene transcription by GT-elements and GT-factors. Trends Plant Sci 4: 210–214 [DOI] [PubMed] [Google Scholar]