Abstract
Objectives
Current pancreatic cancer diagnostics cannot reliably detect early disease or distinguish it from chronic pancreatitis. We test the hypothesis that optical spectroscopy can accurately differentiate cancer from chronic pancreatitis and normal pancreas. We developed and tested clinically-compatible multimodal optical spectroscopy technology to measure reflectance and endogenous fluorescence from human pancreatic tissues.
Methods
Freshly-excised pancreatic tissue specimens (39 normal, 34 chronic pancreatitis, 32 adenocarcinoma) from 18 patients were optically interrogated, with site-specific histopathology representing the gold standard. A multinomial logistic model using principal component analysis and generalized estimating equations provided statistically rigorous tissue classification.
Results
Optical spectroscopy distinguished pancreatic cancer from normal pancreas and chronic pancreatitis (sensitivity 91%, specificity 82%, positive predictive value 69%, negative predictive value 95%, area under receiver operating characteristic curve = 0.89). Reflectance alone provided essentially the same classification accuracy as reflectance and fluorescence combined, suggesting that a rapid, low-cost, reduced-footprint, reflectance-based device could be deployed without notable loss of diagnostic power.
Conclusions
Our novel, clinically-compatible, label-free optical diagnostic technology accurately characterizes pancreatic tissues. These data provide the scientific foundation demonstrating that optical spectroscopy can potentially improve diagnosis of pancreatic cancer and chronic pancreatitis.
Keywords: Pancreatic Cancer Detection, Chronic Pancreatitis Detection, Reflectance Spectroscopy, Fluorescence Spectroscopy
INTRODUCTION
Pancreatic cancer is the third-leading cause of cancer death in the United States, with a five-year survival rate of only 7% (1), in part because no clinical method currently exists to reliably diagnose the disease in its early stages. The current method of choice to detect early-stage pancreatic adenocarcinoma is endoscopic ultrasound-guided fine-needle aspiration (EUS-FNA), which has shown the potential to detect small tumors (2) but is hindered by wide variations in negative predictive value (reported values vary from 16–96%) (2, 3). A recent meta-analysis of 28 clinical studies concluded that it was not advisable to use EUS-FNA to exclude the presence of cancer because “malignancy cannot be ruled out with adequate reliability (3).” Furthermore, EUS-FNA is unable to consistently distinguish pancreatic adenocarcinoma from chronic pancreatitis (4). This key limitation is driven by the failure of current methods to provide reliable needle-based tissue sampling suitable for histology, as tissue architecture is essential to accurately characterize and differentiate inflammatory from neoplastic pancreatic mass lesions. In a prospective clinical study, the sensitivity of EUS-FNA for detecting pancreatic adenocarcinoma in the setting of chronic pancreatitis was only 54% (4). Therefore, there is a significant clinical need to develop a method to more accurately detect and differentiate cancer and chronic pancreatitis.
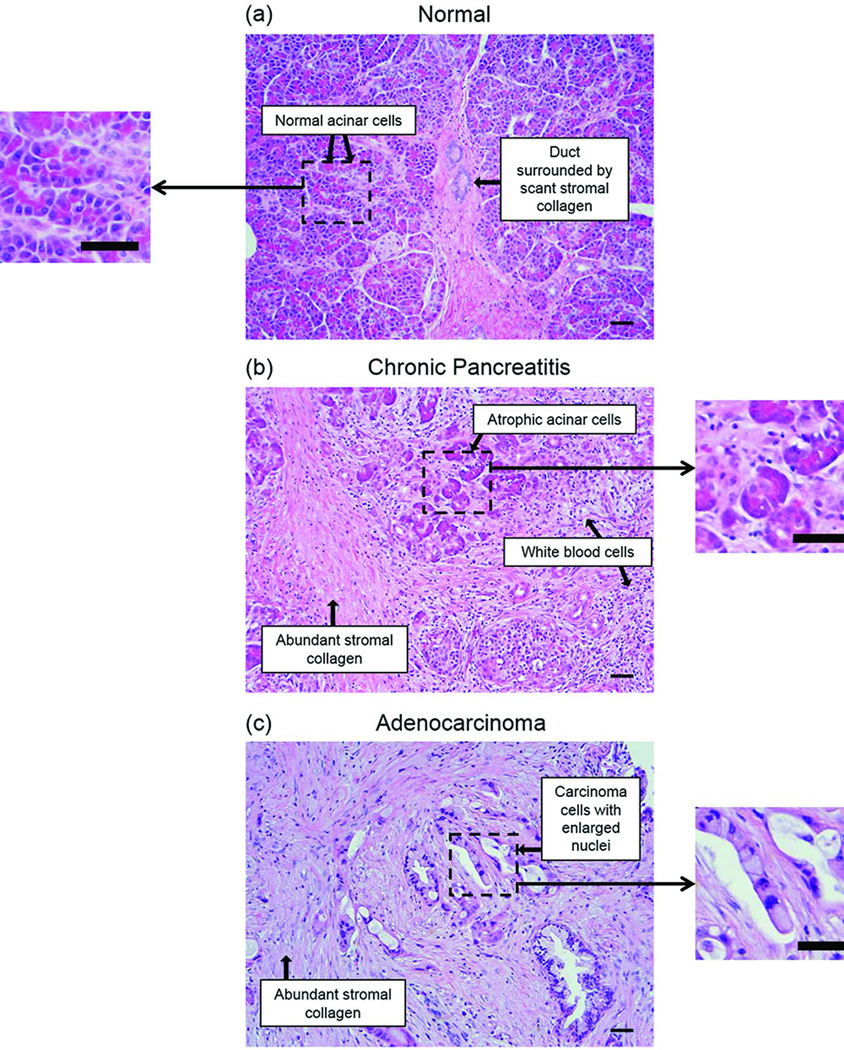
Optical spectroscopy has shown potential to assist with clinical (pre-) cancer diagnostics in many organs and tissues (5, 6), including needle-based in vivo diagnostics (7, 8). The feasibility of tissue optical spectroscopy for characterizing human pancreatic disease has been demonstrated in ex vivo pilot studies (9–13). Pancreatic tissue histology (Fig. 1) illustrates some of the key morphological and biochemical changes that are typically associated with pancreatic cancer and chronic pancreatitis, relative to normal pancreas. These changes are expected to affect the manner in which incident light propagates through the tissue before returning to the surface, influencing the detected optical reflectance and fluorescence spectra.
Fig. 1.
Representative histology images of (A) normal pancreatic tissue, (B) chronic pancreatitis (inflammation), and (C) adenocarcinoma. Histopathological analysis shows increased stromal collagen (light pink stain) in adenocarcinoma and chronic pancreatitis (relative to normal pancreas), as well as morphological changes in cell nuclei (purple stain) in adenocarcinoma (relative to both normal pancreas and chronic pancreatitis). These biochemical and morphological changes in diseased tissues provide a useful source of endogenous contrast for optical disease detection in the pancreas. Scale bar = 50 µm.
Here, we report a clinically-compatible optical spectroscopy technology and test the hypothesis that this technology can accurately distinguish between pancreatic cancer, chronic pancreatitis, and normal pancreatic tissue. The analysis of optical measurements from a medium as complex as human tissue, and the subsequent use of this optical data for tissue classification, is a challenging task due to the multifaceted information present in each measurement. Therefore, mathematical models (12, 13) play a crucial role in optical detection of disease by extracting quantitative diagnostic parameters from the measured optical spectra of human tissues. Furthermore, biostatistical algorithms are critical for rigorously assessing the potential of new diagnostic techniques, as they can quantitatively assess diagnostic accuracy while also correcting for effects of correlations in the data set (such as the measurement of multiple tissue sites from each patient) (14). With these considerations in mind, our technology includes instrumentation for measuring optical reflectance and fluorescence, a principal component analysis model to analyze these complicated optical signals, and a set of biostatistical algorithms to rigorously test the ability of the optical method to accurately classify pancreatic tissues. By performing this ex vivo study, we were able to compare the results of our technique against the corresponding “gold standard” histopathology readings obtained from the same tissue sites that we measured optically.
MATERIALS AND METHODS
Clinically-compatible instrumentation
To acquire optical spectra from human pancreatic tissues, prototype clinically compatible investigational instrumentation was developed (10, 11, 15) and employed. The instrumentation (Supplementary Figure 1) included two light sources: a continuous-wave tungsten-halogen lamp (HL 2000FHSA, Ocean Optics, Dunedin, FL) for reflectance and a 355 nm laser (PNV001525-140, JDS Uniphase, San Jose, CA) to excite tissue fluorescence. To deliver light to and collect light from the tissue surface, a custom fiber-optic probe (Ocean Optics) was employed. The probe contained three fibers, each with diameter 660 µm, arranged in a triangular configuration (Supplementary Figure 1). One fiber delivered light from the lamp to the tissue, the second fiber delivered laser light to the tissue, and the third fiber collected light that returned to the tissue surface. The collected light was sent to a spectrograph (MS 125, Oriel Instruments, Stratford, CT) and an intensified charge-coupled device (ICCD 2063, Andor Technology, Belfast, Northern Ireland) for wavelength-resolved detection. The reflectance and fluorescence measurements were performed sequentially, with data acquisition times less than 500 ms for reflectance and less than 1 s for fluorescence.
Acquisition of optical and histopathological data
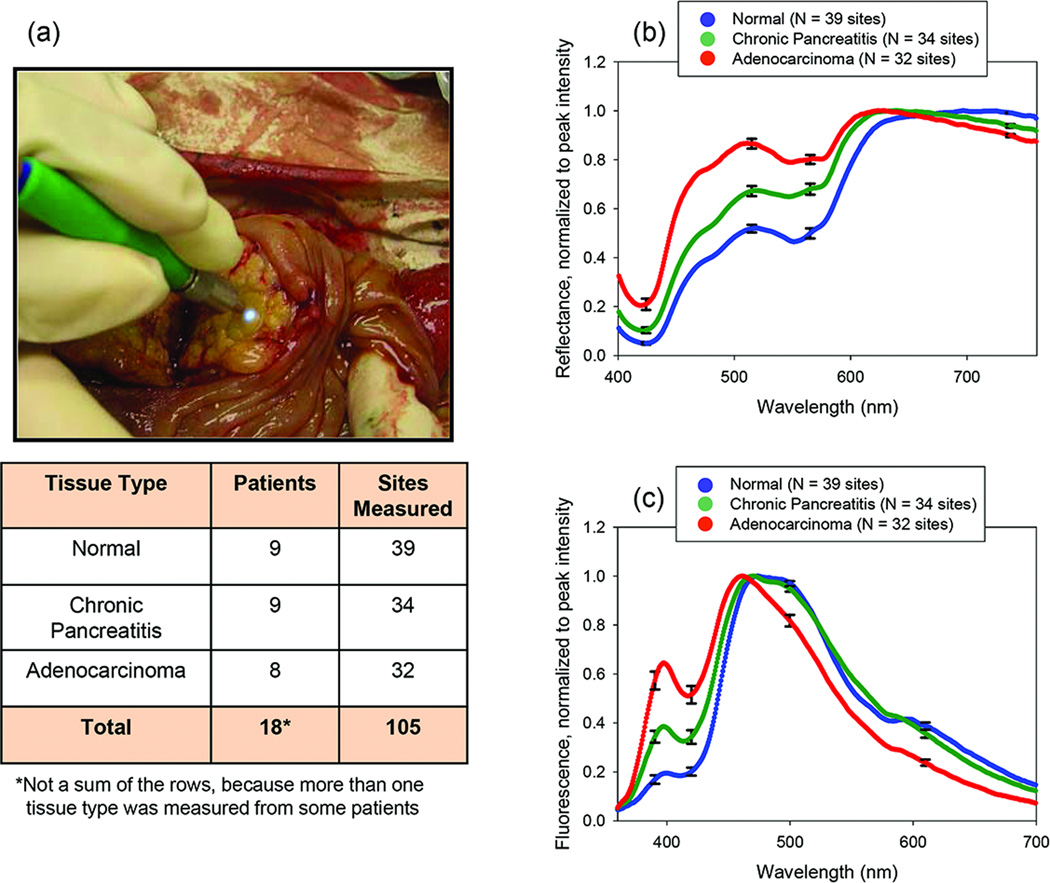
Reflectance and fluorescence spectra were acquired from freshly-excised human pancreatic tissues within 30 minutes of excision (Fig. 2). The Institutional Review Board of the University of Michigan approved the study (HUM00017352) and patient consent was obtained prior to all measurements. Multiple sites from each tissue specimen were measured, and two reflectance and two fluorescence measurements were acquired from each site (with the exception of one site, from which only one of each measurement was taken). For each measured site on each tissue specimen, biopsy and histopathology were performed to obtain the diagnostic “gold standard.” After the optical measurements were acquired at a given site, the fiber-optic probe was kept in position at that site to mark the location of the site until the pathologist was ready to perform a tissue biopsy of the site, and the biopsy was obtained by the pathologist immediately after the probe was removed. Biopsies were 3–5 mm in diameter, comparable to the ~1 mm-diameter region of tissue (16) interrogated by the fiber-optic probe. These tissue biopsies were subsequently fixed in formalin, paraffin-embedded, sectioned, stained with hematoxylin and eosin for histologic examination, and analyzed via histopathology. In all, 18 patients (39 normal sites, 34 pancreatitis sites, and 32 adenocarcinoma sites) were evaluated.
Fig. 2.
(A) Pancreatic tissue measurement (image from (10), used with permission) and patients and sites measured in this study. Mean reflectance spectra (B) and fluorescence spectra (C) from these measurements revealed significant differences between normal tissue, chronic pancreatitis, and adenocarcinoma. Error bars represent the standard error.
Data pre-processing
Reflectance spectra for which the detected signal at 550 nm divided by the detected signal at 650 nm (R550/R650) was less than 0.2 were removed from the data set because these spectra were dominated by absorption from blood. It is important to note that the presence of blood is not considered problematic in tissue-optics studies because blood can be avoided by repositioning the tip of the probe (8), removed by suctioning the region of interest (7), or accounted for mathematically (12, 13). Fluorescence spectra with a signal-to-noise ratio (SNR) of less than 30 were also removed, because these spectra were considered too noisy to properly analyze. Prior to data analysis, all raw spectra were background-corrected, corrected for instrument response, smoothed, and normalized to peak intensity. The resulting corrected spectra were used for data analysis.
Identification and removal of outlier sites from the data set
A residual-based method (17) was employed to detect and remove outlier sites for which the two measured fluorescence or reflectance spectra were extremely different from each other. This was done through a series of linear mixed models where the spectrum measurements at specific wavelengths were regressed on the tissue diagnosis with a random intercept to account for correlation among sites that originated from the same patient. Ten regressions were performed on the fluorescence spectra at wavelengths of 375.18, 417.19, 423.38, 427.52, 444.04, 452.99, 466.08, 479.16, 502.57, and 511.52 nm, and ten regressions were also performed on the reflectance spectra at wavelengths of 426.827, 440.598, 461.255, 481.913, 490.175, 541.130, 544.573, 612.053, 625.135, and 755.964 nm.
For each of the regressions, the difference between the residuals of the two duplicate measurements at each site were calculated and ranked in order of magnitude. The ranks for the ten regressions were then averaged to give an overall fluorescence or reflectance average rank for each site. Sites whose overall mean rank exceeded the 85th percentile of the total number of sites were flagged as potential outliers. Through this method five sites were identified as having a pair of very different reflectance spectra, and four additional sites were identified as having a pair of very different fluorescence spectra. Each site with two extremely different measurements was removed from the analysis (unless one of those two measurements had been removed from the data set previously due to low R550/R650 value or low fluorescence SNR), and for the remaining sites, the two measured spectra were averaged. Of the 117 sites from which measurements were acquired, a total of 12 sites were removed from the data set due to low R550/R650 value, low fluorescence SNR, or high degree of difference between the two reflectance or fluorescence measurements.
Principal Component Analysis model
Differences among the tissue types were observed in both the reflectance and the fluorescence spectra (Figs. 2(B, C)). To quantitatively characterize these differences, a standard Principal Component Analysis (PCA) model (18) was employed to extract the principal components from each reflectance and fluorescence spectrum. The inputs to the PCA code were the corrected reflectance and fluorescence spectra after pre-processing and outlier removal were performed (see data pre-processing and outlier identification/removal paragraphs above), without any additional correction of the spectra for attenuation artifacts. The PCA code was written in MATLAB (Mathworks, Natick, MA) using functions that were built into the MATLAB programming language.
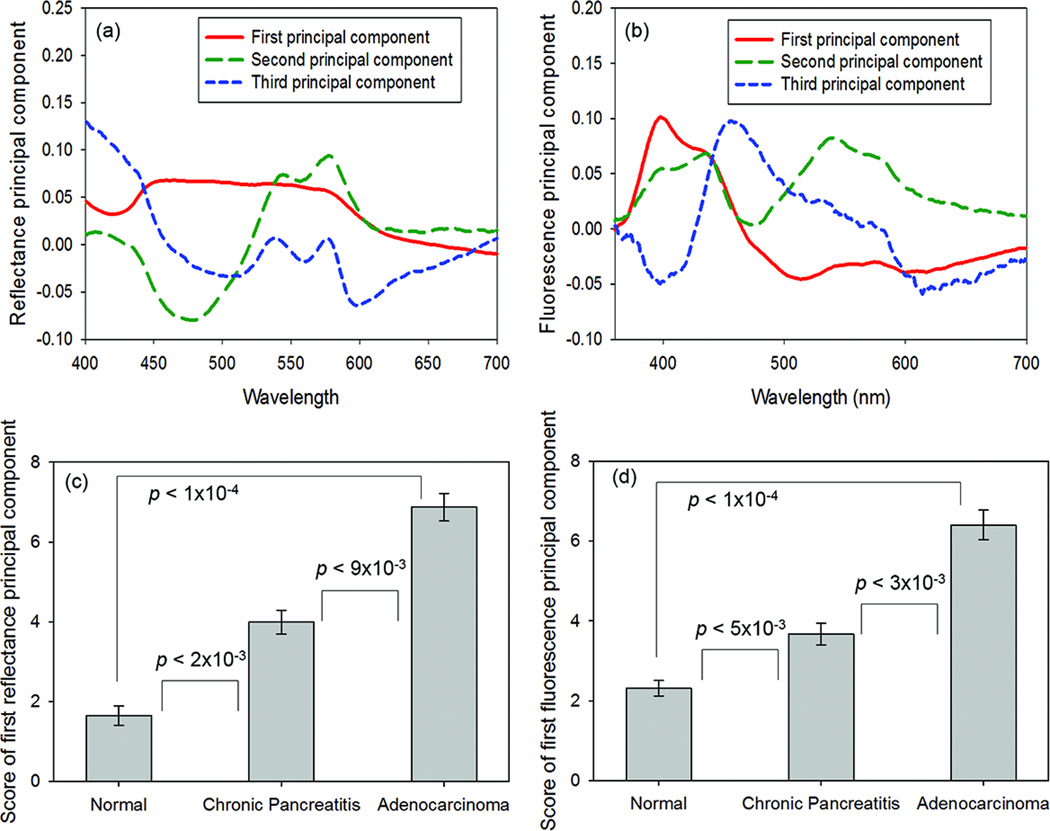
The first three reflectance and fluorescence principal components, their scores, and their statistical significance for distinguishing between the tissue types are displayed in Fig. 3 and Supplementary Table 1. The first reflectance principal component score (RPC1) and the first fluorescence principal component score (FPC1) were the most statistically significant (P < 0.01) for distinguishing adenocarcinoma from normal pancreatic tissue, distinguishing adenocarcinoma from chronic pancreatitis, and distinguishing chronic pancreatitis from normal pancreas.
Fig. 3.
(A, B) First three principal components (accounting for 95% of the spectral variation) of the reflectance (A) and fluorescence (B) data sets. (C, D) Parameters extracted from principal component analysis (PCA) of tissue optical spectra, shown with their statistical significance for tissue classification. (C) The first reflectance principal component score was statistically significant for distinguishing adenocarcinoma from normal pancreatic tissue (P < 1×10−4), distinguishing adenocarcinoma from chronic pancreatitis (P < 9×10−3), and distinguishing chronic pancreatitis from normal pancreatic tissue (P < 2×10−3). (D) The first fluorescence principal component score was also statistically significant for distinguishing adenocarcinoma from normal pancreatic tissue (P < 1×10−4), distinguishing adenocarcinoma from chronic pancreatitis (P < 3×10−3), and distinguishing chronic pancreatitis from normal pancreatic tissue (P < 5×10−3). To remove negative values for ease of display, an offset of 4 was added to all principal component scores before they were plotted on the bar graph.
Tissue classification algorithm – statistical methods
Selection of the variables used in the tissue classification algorithm was determined through multinomial logistic regression using Generalized Estimating Equations (GEE) (19–21) to statistically account for the intra-patient correlations in the data arising from the fact that the data set included multiple tissue sites from each patient. GEE has been previously employed in medical physics studies to correct for intra-patient correlations (14, 22–24), for instance, in ophthalmology, in which there are correlations between data taken from each of the two eyes of a single patient (14). However, optical methods for tissue classification typically do not account for statistical correlations between data obtained from multiple sites on a given patient. Here, we present an optics-based tissue classification algorithm that incorporates GEE to rigorously account for these intra-patient correlations, as we believe this to be a key improvement that should be widely adopted by the biomedical optics community. For tissue classification, a “ leave-two-patients-out” cross-validation (25) was employed (Supplementary Figure 2).
Tissue classification algorithm – selection of variables
For each variable extracted by the PCA model, a GEE model with an exchangeable correlation structure was employed to contrast the differences between the mean values of that variable for each pair of tissue types. For each contrast, a Wald test statistic using the robust covariance matrix was calculated to account for the repeatedly measured observations from the same patients (26). Following the identification of significant predictor variables (Supplementary Table 1), correlations between the selected variables were calculated and highly correlated variables were removed to minimize the multicollinearity.
The variables used in the algorithm that included reflectance and fluorescence were the scores of the first three reflectance principal components (RPC1, RPC2, RPC3), and the score of the first fluorescence principal component (FPC1). The score of the second fluorescence principal component (FPC2) was dropped due to extreme correlation with RPC2 (Pearson correlation = 0.76). The third fluorescence principal component FPC3 was dropped because it was not statistically significant for any of the tissue comparisons investigated in this study (Supplementary Table 1).
Tissue classification algorithm – “leave-two-patients-out” cross-validation
The classification algorithm (Supplementary Figure 2) was run nine times, and each time, the data set was split into a training set of 16 patients and a test set of two patients. The algorithm was executed nine times because there were 18 patients overall and each patient was put into the test set exactly once. In this way, the algorithm was always blinded to the patient data in the testing set. For each pair of training and test data sets a standard multinomial logistic regression was fit to the training dataset, which is then used to generate optical diagnosis probabilities of adenocarcinoma, chronic pancreatitis, and normal tissue types for the observations in the test data set. Thresholds were then applied to these probabilities to determine the sensitivity, specificity, positive predictive value, negative predictive value, and area under the receiver operating characteristic (ROC) curve for distinguishing cancer sites from non-cancer (normal and chronic pancreatitis) sites. Ternary plots of the diagnosis probabilities of adenocarcinoma, chronic pancreatitis, and normal pancreas are shown in Supplementary Figure 3 (using a combination of reflectance and fluorescence parameters) and Supplementary Figure 4 (using (a) only reflectance parameters and (b) only fluorescence parameters).
RESULTS
Variables from the PCA model were input into the tissue classification algorithm (Supplementary Figure 2) to distinguish between adenocarcinoma, chronic pancreatitis, and normal pancreas. The outputs of the classification algorithm were the optical diagnosis probabilities P(N), P(CP), and P(A) of each tissue site being normal, chronic pancreatitis, or adenocarcinoma. Ternary plots of the optical diagnosis probabilities P(N), P(CP), and P(A) were created for classification procedures that used both reflectance and fluorescence (Supplementary Figure 3), as well as reflectance only (Supplementary Figure 4(a)) and fluorescence only (Supplementary Figure 4(b)).
To determine the accuracy of the optical method for detecting pancreatic adenocarcinoma using reflectance and fluorescence PCA parameters, a threshold was applied to P(A). Specifically, a site was diagnosed as cancer if a manually-defined threshold P(A) > 0.3 was met. The resulting sensitivity, specificity, PPV, and NPV for distinguishing adenocarcinoma from non-cancerous tissues (Table 1) were calculated using cross-validation to be 91%, 82%, 69%, and 95%, respectively. The user-defined threshold on P(A) can be raised or lowered depending on whether the operator wishes to optimize the procedure for maximum sensitivity or maximum specificity. To demonstrate this concept, the sensitivity and specificity calculated for different thresholds on P(A) were employed to generate ROC curves for distinguishing adenocarcinoma from non-cancerous tissues (Fig. 4). The area under the ROC curve was 0.89 for the algorithm including both reflectance and fluorescence PCA parameters. These results show the high diagnostic accuracy of the optical method employing both reflectance and fluorescence, as described in this report.
Table 1.
Sensitivity, specificity, positive predictive value, negative predictive value, and area under the receiver operating characteristic curve of optical spectroscopy for distinguishing between pancreatic adenocarcinoma (A), chronic pancreatitis (CP), and normal pancreatic tissue (N). Results are shown for principal component analysis with reflectance and fluorescence together, reflectance only, and fluorescence only. The threshold P(A) > 0.3 was employed to achieve the shown classification accuracy.
| Method | Classification | Sensitivity | Specificity | Positive Predictive Value |
Negative Predictive Value |
Area Under Curve |
|---|---|---|---|---|---|---|
| Reflectance &Fluorescence |
A vs. (CP and N) | 91% | 82% | 69% | 95% | 0.89 |
| (A and CP) vs. N | 83% | 87% | 92% | 76% | 0.90 | |
| A vs. N | 97% | 90% | 89% | 97% | 0.96 | |
| A vs. CP | 91% | 71% | 74% | 89% | 0.81 | |
| CP vs. N | 77% | 82% | 79% | 80% | 0.84 | |
| Reflectance | A vs. (CP and N) | 91% | 84% | 71% | 95% | 0.89 |
| (A and CP) vs. N | 82% | 87% | 92% | 74% | 0.89 | |
| A vs. N | 94% | 95% | 94% | 95% | 0.97 | |
| A vs. CP | 91% | 68% | 73% | 89% | 0.81 | |
| CP vs. N | 71% | 87% | 83% | 77% | 0.83 | |
| Fluorescence | A vs. (CP and N) | 91% | 78% | 65% | 95% | 0.86 |
| (A and CP) vs. N | 76% | 87% | 91% | 68% | 0.81 | |
| A vs. N | 94% | 90% | 88% | 94% | 0.91 | |
| A vs. CP | 81% | 77% | 77% | 81% | 0.78 | |
| CP vs. N | 68% | 85% | 79% | 75% | 0.73 | |
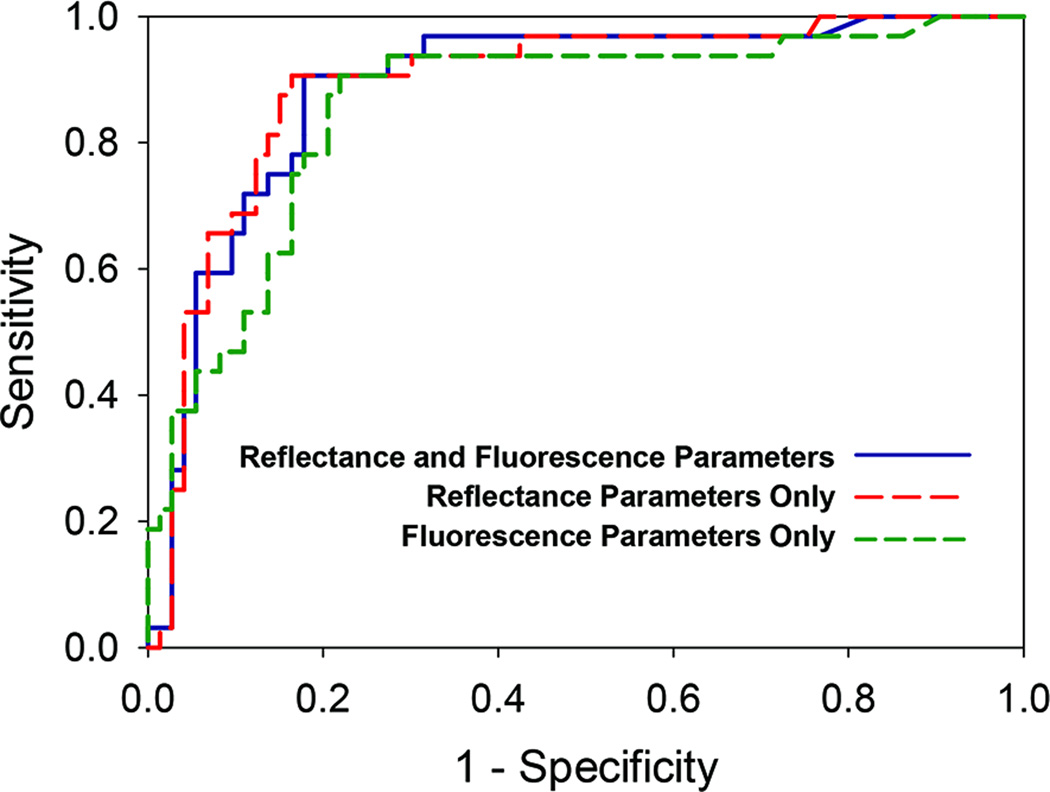
Fig. 4.
Receiver operating characteristic (ROC) curves for distinguishing malignant (adenocarcinoma) tissue sites from non-malignant (normal and chronic pancreatitis) tissue sites using PCA parameters from both reflectance and fluorescence data (solid blue line), PCA parameters from only reflectance data (dashed red line), and PCA parameters from only fluorescence data (dashed green line). The ROC curves were generated by applying a threshold to the optical diagnosis probability of adenocarcinoma. The area under the ROC curve was 0.89 for combined reflectance and fluorescence parameters, 0.89 for only reflectance parameters, and 0.86 for only fluorescence parameters.
Additional variations of the classification algorithm were run to compare the results obtained by using only reflectance variables (RPC1, RPC2, RPC3) and only fluorescence variables (FPC1, FPC2) with those obtained by using a combination of reflectance and fluorescence variables (RPC1, RPC2, RPC3, FPC1). For these comparisons, thresholds were manually selected such that the resulting sensitivity was the same (91%) for all three algorithms, and then the corresponding specificity, PPV, and NPV of the three algorithms were compared.
Using only reflectance variables, and choosing the threshold of P(A) > 0.20 for cancer diagnosis, the sensitivity, specificity, PPV, and NPV for distinguishing adenocarcinoma from non-cancerous tissues were 91%, 84%, 71%, and 95% (Table 1). The area under the ROC curve was 0.89 when only reflectance variables were employed (Fig. 4). Using only fluorescence variables, and choosing the threshold of P(A) > 0.22 for cancer diagnosis, the sensitivity, specificity, PPV, and NPV for distinguishing adenocarcinoma from non-cancerous tissues were 91%, 78%, 65%, and 95% (Table 1). The area under the ROC curve was 0.86 when only fluorescence variables were employed (Fig. 4).
These results show that optical spectroscopy provided accurate classification of pancreatic disease. Interestingly, for distinguishing between the tissue types measured in this study, using reflectance alone provided essentially the same diagnostic accuracy as using reflectance and fluorescence combined (Table 1). This is a significant finding, as an optical device that used only the reflectance modality would be less expensive, more compact, and faster when performing measurements.
DISCUSSION
In this study, we have developed and employed clinically-compatible optical spectroscopy technology to distinguish pancreatic cancer from non-cancerous pancreatic tissues (normal and chronic pancreatitis). The tissue optical data were analyzed with a novel mathematical algorithm that combined principal component analysis with a statistically rigorous tissue classification procedure. This technology was assessed, for the first time, on a set of 18 patients by using a “leave-two-out” cross-validation technique in which the classification results for each test set of two patients were compared with those of histopathology, as the diagnostic gold standard. The optical method accurately classified pancreatic cancer sites, with a sensitivity, specificity, positive predictive value, and negative predictive value of 91%, 82%, 69%, and 95%, respectively, with an area under the receiver operating characteristic curve of 0.89. These results suggest that optical spectroscopy has the potential to distinguish pancreatic cancer from non-malignant pancreatic tissues to provide immediate diagnostic feedback in a clinical setting, since the data collection and analysis can be performed in a few seconds. This novel approach has the potential to revolutionize needle-based pancreatic tissue characterization and improve clinical diagnostics in this most challenging patient population. The technology could also be employed surgically, using a hand-held optical probe for tumor margin detection.
In this study, tissue classification employing only reflectance provided nearly identical results to those obtained with a combination of reflectance and fluorescence. This finding can possibly be attributed to an overlap between the information encoded in the reflectance and fluorescence principal components. The reflectance data is known to contain notable information about tissue scattering, which is partially due to collagen in the tissue. However, collagen is also a prominent endogenous fluorophore in human pancreatic tissue, so the information about collagen is also found in the fluorescence spectra. Therefore, it is to be expected that the reflectance and fluorescence principal components would include overlapping information.
In Fig. 3(B), the first fluorescence principal component contains a spectral feature near the peak emission wavelength (~400 nm) of collagen fluorescence, which has been shown to distinguish between these tissue types in our previous work (13). Fig. 3(A) shows that the second and third reflectance principal components display spectral features similar to those of hemoglobin absorption, which has a distinctive Soret peak near 420 nm and secondary peaks near 545 nm and 575 nm. Since absorption (from hemoglobin) and scattering (from tissue components including collagen) play prominent roles in determining the measured reflectance spectrum, it makes sense that the first reflectance principal component could primarily include information about collagen scattering and the second and third reflectance principal components could primarily include information about hemoglobin absorption. Since the fluorescence spectrum also contains prominent information about collagen, as well as artifacts of hemoglobin absorption, it is quite possible that the fluorescence data is not providing a significant amount of diagnostic information that is different from that provided by the reflectance.
The diagnostic dilemma facing clinicians who care for patients with possible pancreatic disease is to accurately distinguish malignancy from chronic pancreatitis. Delay or failure to identify cancer contributes to the dismal survival rate associated with pancreatic adenocarcinoma. Using traditional methods of imaging, malignancy and inflammation share many histological features, and the dense fibrosis observed in both conditions renders aspirated cells (cytology via FNA) insensitive in many clinical situations. In addition, chronic pancreatitis has different diagnostic and treatment strategies, so there remains intrinsic value to confirming an enigmatic diagnosis in the patient with severe, unexplained abdominal pain, where pancreatic disease remains a possibility.
This work has demonstrated for the first time that quantitative tissue optical spectroscopy using visible light can be employed for classification of human pancreatic disease. Traditionally, the pancreas has been considered a challenging organ to study due to its location deep in the retroperitoneum and the lack of a non-invasive tissue gold standard for comparison. Our study, using freshly-excised surgical specimens containing the changes of both chronic pancreatitis and adenocarcinoma (as typically seen in this patient population) and the gold standard of tissue histopathology, provides the scientific proof of principle for the potential role of optical tissue characterization of the pancreas in vivo. Current pancreatic cancer detection methods, which employ ultrasound as the source of contrast, are unable to reliably detect the disease (27), especially in the setting of concurrent inflammation (pancreatitis) (4). Our technique has the potential to provide significantly increased contrast to reliably detect pancreatic cancer by optically characterizing disease-related changes in the tissue. The innovative optical technology developed offers rapid data acquisition and analysis to provide immediate diagnostic feedback to the clinician, using statistically rigorous algorithms that can be tailored to the specific clinical question of interest (cancer versus non-cancer, cancer versus pancreatitis, cancer versus normal, pancreatitis versus normal).
To establish the feasibility of clinical translation, we have addressed several limitations that must be overcome before the technology can be employed clinically, either as an intraoperative tool for tumor margin detection or as an endoscopic tool for needle-based tissue diagnostics. The data analyzed in this study were from freshly-excised human pancreatic tissues (ex vivo); therefore, the efficacy of the data analysis methods must be verified to be the same for in vivo human tissue. For minimally invasive applications, the technology must be incorporated into an endoscopic device that is compatible with the EUS procedure. Toward these goals, we have performed three important studies. First, we employed tissue-simulating “phantoms” to verify the accuracy of our method for both an ex vivo fiber-probe and an endoscope-compatible fiber-probe. Second, we verified that the method extracted consistent values of ex vivo human pancreatic tissue morphology properties despite large variations in tissue blood content over time. These two studies provided further evidence that the method is translatable to in vivo human pancreatic tissue. Third, we conducted a pilot study to optically interrogate human pancreatic tissues in vivo during surgery (28). Preliminary results from this in vivo study demonstrated that the optical method is able to extract the same diagnostically-relevant tissue parameters from in vivo data as were extracted from ex vivo data, and that these parameters can be employed to distinguish cancerous tissues from normal tissues in vivo in the same manner as they were employed for ex vivo data.
The results of these studies will be employed to appropriately inform the design of clinical optical diagnostic technology. We note that only data from normal, chronic pancreatitis, and adenocarcinoma sites were analyzed in the work reported here, although the methods could also be applicable to diagnosis of pre-malignant conditions and detection of cancer within a site containing several different tissue types. With this goal in mind, we conducted a pilot study that suggested the potential of the method to distinguish premalignant human pancreatic tissue sites from normal human pancreatic tissue sites (29).
In conclusion, we have presented an optical method for detection of pancreatic disease and employed a rigorous biostatistical method to demonstrate its accuracy for classifying pancreatic cancer, chronic pancreatitis, and normal pancreatic tissue. By showing the ability to reliably detect pancreatic cancer and distinguish cancer from chronic pancreatitis, our method provides a potentially valuable inroad toward addressing an important unmet clinical need. The technology developed does not require the administration of exogenous contrast agents to patients and the portable clinical device is constructed with components at a reasonable manufacturing cost. Furthermore, our study suggests that it may be possible to streamline the device even further (in terms of cost, footprint, and data acquisition and processing time) by employing only reflectance instrumentation, as using reflectance alone was shown to provide nearly identical diagnostic accuracy to using both reflectance and fluorescence. In addition to needle-based disease diagnosis, our technique is potentially applicable for rapid on-site detection of tumor margins during pancreatic surgery. Thus, our results provide the scientific “proof of principle” that the optical spectroscopic method described here has the potential to assist with the detection of pancreatic cancer, as well as chronic pancreatitis, in a clinical setting.
Supplementary Material
Acknowledgments
We thank Drs. C. Sonnenday, L. M. Colletti, R. Minter, and M. Mulholland for permitting us to collect data during the pancreatic surgeries they performed. We thank Dr. J. Purdy for histopathological analysis. We thank S. Korsnes for helping with patient recruitment for the study. This research was also made possible, in part, by the use of the University of Michigan Comprehensive Cancer Center Biostatistics Core.
Conflicts of Interest: The authors are co-inventors on issued patents related to tissue optical diagnostics using methods described in this report.
Source of Funding: This work was supported in part by the Wallace H. Coulter Foundation, the National Pancreas Foundation, the University of Michigan Comprehensive Cancer Center, a grant from the University of Michigan Medical School Translational Research Program, the National Institutes of Health (NIH-CA-114542, NIH T32 CA083654), and an ASGE Senior Mentoring Career Development Award (J. M. S.).
Abbreviations
- AUC
Area Under Curve
- EUS-FNA
Endoscopic Ultrasound-guided Fine-Needle Aspiration
- GEE
Generalized Estimating Equations
- NPV
Negative Predictive Value
- PCA
Principal Component Analysis
- PPV
Positive Predictive Value
- ROC
Receiver Operating Characteristic
- SE
Sensitivity
- SP
Specificity
- SNR
Signal-to-Noise Ratio
REFERENCES
- 1.Cancer Facts & Figures 2015. American Cancer Society; Available from: www.cancer.org. [Google Scholar]
- 2.Hewitt MJ, McPhail MJ, Possamai L, et al. EUS-guided FNA for diagnosis of solid pancreatic neoplasms: a meta-analysis. Gastrointest Endosc. 2012;75:319–331. doi: 10.1016/j.gie.2011.08.049. [DOI] [PubMed] [Google Scholar]
- 3.Hartwig W, Schneider L, Diener MK. Preoperative tissue diagnosis for tumours of the pancreas. Br J Surg. 2009;96:5–20. doi: 10.1002/bjs.6407. [DOI] [PubMed] [Google Scholar]
- 4.Fritscher-Ravens A, Brand L, Knöfel WT. Comparison of endoscopic ultrasound-guided fine needle aspiration for focal pancreatic lesions in patients with normal parenchyma and chronic pancreatitis. Am J Gastroenterol. 2002;97:2768–2775. doi: 10.1111/j.1572-0241.2002.07020.x. [DOI] [PubMed] [Google Scholar]
- 5.Brown JQ, Vishwanath K, Palmer GM, et al. Advances in quantitative UV-visible spectroscopy for clinical and pre-clinical application in cancer. Curr Opin Biotechnol. 2009;20:119–131. doi: 10.1016/j.copbio.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lloyd WR, Chen L-C, Wilson RH, et al. Biophotonics: Clinical Fluorescence Spectroscopy and Imaging. In: Maitland DJ, Moore JE Jr, editors. Biomedical Technology and Devices Handbook. 2nd. Boca Raton, FL: CRC Press - Taylor & Francis; 2013. pp. 335–358. [Google Scholar]
- 7.Zhu C, Burnside ES, Sisney GA. Fluorescence spectroscopy: an adjunct diagnostic tool to image-guided core needle biopsy of the breast. IEEE Trans Biomed Eng. 2009;56:2518–2528. doi: 10.1109/TBME.2009.2015936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanick SC, Leest Cvd, Djamin RS. Characterization of mediastinal lymph node physiology in vivo by optical spectroscopy during endoscopic ultrasound-guided fine needle aspiration. J Thorac Oncol. 2010;5:981–987. doi: 10.1097/jto.0b013e3181ddbc0e. [DOI] [PubMed] [Google Scholar]
- 9.Kondepati VR, Zimmermann J, Keese M, et al. Near-infrared fiber optic spectroscopy as a novel diagnostic tool for the detection of pancreatic cancer. J Biomed Opt. 2005;10:054016. doi: 10.1117/1.2060568. [DOI] [PubMed] [Google Scholar]
- 10.Chandra M, Scheiman J, Heidt D, et al. Probing pancreatic disease using tissue optical spectroscopy. J Biomed Opt. 2007;12:060501. doi: 10.1117/1.2818029. [DOI] [PubMed] [Google Scholar]
- 11.Chandra M, Scheiman J, Simeone D, et al. Spectral areas and ratios classifier algorithm for pancreatic tissue classification using optical spectroscopy. J Biomed Opt. 2010;15:010514. doi: 10.1117/1.3314900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson RH, Chandra M, Scheiman J, et al. Optical spectroscopy detects histological hallmarks of pancreatic cancer. Opt Express. 2009;17:17502–17516. doi: 10.1364/OE.17.017502. [DOI] [PubMed] [Google Scholar]
- 13.Wilson RH, Chandra M, Chen LC, et al. Photon-tissue interaction model enables quantitative optical analysis of human pancreatic tissues. Opt Express. 2010;18:21612–21621. doi: 10.1364/OE.18.021612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vizzeri G, Weinreb RN, Gonzalez-Garcia AO, et al. Agreement between spectral-domain and time-domain OCT for measuring RNFL thickness. Br J Ophthalmol. 2009;93(6):775–781. doi: 10.1136/bjo.2008.150698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chandra M, Vishwanath K, Fichter GD, et al. Quantitative molecular sensing in biological tissues: an approach to non-invasive optical characterization. Opt Express. 2006;14:6157–6171. doi: 10.1364/oe.14.006157. [DOI] [PubMed] [Google Scholar]
- 16.Kanick SC, Robinson DJ, Sterenborg HJCM. Monte Carlo analysis of single fiber reflectance spectroscopy: photon path length and sampling depth. Phys Med Biol. 2009;54:6991–7008. doi: 10.1088/0031-9155/54/22/016. [DOI] [PubMed] [Google Scholar]
- 17.Montgomery DC, Peck EA, Vining GG. Introduction to Linear Regression Analysis. Hoboken, NJ: John Wiley & Sons; 2012. [Google Scholar]
- 18.Jolliffe IT. Principal Component Analysis. New York: Springer-Verlag; 2002. [Google Scholar]
- 19.Hanley JA, Negassa A, deB Eduardes MD, et al. Statistical Analysis of Correlated Data Using Generalized Estimating Equations: An Orientation. Am J Epidemiol. 2003;157:364–375. doi: 10.1093/aje/kwf215. [DOI] [PubMed] [Google Scholar]
- 20.Burton P, Gurrin L, Sly P. Extending the simple linear regression model to account for correlated responses: An introduction to generalized estimating equations and multi-level mixed modelling. Stat Med. 1998;17:1261–1291. doi: 10.1002/(sici)1097-0258(19980615)17:11<1261::aid-sim846>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 21.Kuss O, McLerran D. A note on the estimation of the multinomial logistic model with correlated responses in SAS. Comput Methods Programs Biomed. 2007;87:262–269. doi: 10.1016/j.cmpb.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 22.Fortes DL, Allen MS, Lowe VJ, et al. The sensitivity of 18F-fluorodeoxyglucose positron emission tomography in the evaluation of metastatic pulmonary nodules. Eur J Cardiothorac Surg. 2008;34:1223–1227. doi: 10.1016/j.ejcts.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 23.Brem RF, Floerke AC, Rapelyea JA, et al. Breast-specific Gamma imaging as an adjunct imaging modality for the diagnosis of breast cancer. Radiology. 2008;247:651–657. doi: 10.1148/radiol.2473061678. [DOI] [PubMed] [Google Scholar]
- 24.Toi A, Neill MG, Lockwood GA, et al. The continuing importance of transrectal ultrasound identification of prostatic lesions. J Urol. 2007;177:516–520. doi: 10.1016/j.juro.2006.09.061. [DOI] [PubMed] [Google Scholar]
- 25.Volynskaya Z, Haka AS, Bechtel KL. Diagnosing breast cancer using diffuse reflectance spectroscopy and intrinsic fluorescence spectroscopy. J Biomed Opt. 2008;13:024012. doi: 10.1117/1.2909672. [DOI] [PubMed] [Google Scholar]
- 26.Harrell FE., Jr . Regression Modeling Strategies. New York: Springer-Verlag; 2001. [Google Scholar]
- 27.Eloubeidi M, Varadarajulu S, Desai S, et al. A Prospective Evaluation of an Algorithm Incorporating Routine Preoperative Endoscopic Ultrasound-Guided Fine Needle Aspiration in Suspected Pancreatic Cancer. J Gastrointest Surg. 2007;11:813–819. doi: 10.1007/s11605-007-0151-x. [DOI] [PubMed] [Google Scholar]
- 28.Lloyd WR, Wilson RH, Lee SY, et al. In vivo optical spectroscopy for improved detection of pancreatic adenocarcinoma: a feasibility study. Biomed Opt Express. 2014;5:9–15. doi: 10.1364/BOE.5.000009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee SY, Lloyd WR, Chandra M, et al. Characterizing human pancreatic cancer precursor using quantitative tissue optical spectroscopy. Biomed Opt Express. 2013;4:2828–2834. doi: 10.1364/BOE.4.002828. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.