Abstract
Amino acid replacements encoded by the prion protein gene (PRNP) have been associated with transmissible and hereditary spongiform encephalopathies in mammalian species. However, an association between bovine spongiform encephalopathy (BSE) and bovine PRNP exon 3 has not been detected. Moreover, little is currently known regarding the mechanisms of evolution influencing the bovine PRNP gene. Therefore, in this study we evaluated the patterns of nucleotide variation associated with PRNP exon 3 for 36 breeds of domestic cattle and representative samples for 10 additional species of Bovinae. The results of our study indicate that strong purifying selection has intensely constrained PRNP over the long-term evolutionary history of the subfamily Bovinae, especially in regions considered to be of functional, structural, and pathogenic importance in humans as well as other mammals. The driving force behind this intense level of purifying selection remains to be explained.
Transmissible spongiform encephalopathies, or prion diseases, are inevitably fatal neurodegenerative diseases that occur in humans as well as domestic and wild animals (1-3). Traditionally, human spongiform encephalopathies have been classified into Creutzfeldt-Jakob disease (CJD), Gerstmann-Sträusler-Scheinker disease, and kuru, with more recent classification into variant CJD (1). Animal transmissible spongiform encephalopathies include transmissible mink encephalopathy, scrapie of sheep and goats, chronic wasting disease of deer and elk, feline spongiform encephalopathy, and bovine spongiform encephalopathy (BSE) (1). Central to the development of these diseases is the accumulation of an infectious protease-resistant isoform (PrPSc) of the host-encoded cellular prion protein (PrPC) in tissues of the central nervous system (2-5).
The prototypical transmissible spongiform encephalopathy, scrapie, has been observed in European sheep for >200 years, whereas BSE in domestic cattle (Bos taurus and Bos indicus; hereafter, cattle) dates to 1986, presumably resulting from scrapie- and/or BSE-infected cattle feed (6-9). Thus, BSE seems to be a more recent phenomenon associated with modern agricultural practices. This idea is supported by several lines of evidence. First, at least 14 amino acid polymorphisms encoded by exon 3 of the ovine prion protein gene (PRNP) have been described (10-20), and those associated with codons 136 and 171 have been shown to influence expression of scrapie (8, 10). However, no amino acid polymorphisms associated with BSE have been identified in cattle, although an insertion-deletion (indel) polymorphism in the putative bovine PRNP promoter was shown to exhibit an association with BSE in a few German cattle breeds (21). Second, recorded patterns of variation within cattle PRNP exon 3 are markedly different from those observed in sheep, with variability in cattle primarily restricted to 11 synonymous nucleotide sites and two nonsynonymous sites where low-frequency variation has been observed (20-29). Notably, a substantially higher number of nonsynonymous polymorphic sites has been recorded for sheep (10-20). Additionally, indel polymorphism has not been observed within the octapeptide repeat region of ovine PRNP exon 3 (8, 10-20), whereas studies of cattle and other bovine species have yielded three indel isoforms possessing five to seven octapeptide repeats (20-31).
Despite the importance of cattle both to agricultural practices worldwide and to the global economy, surprisingly little is known about PRNP allelic diversity for cattle collectively and/or how this gene evolves in this lineage. In addition, although several nondomesticated species of Bovinae contracted transmissible spongiform encephalopathy-like diseases contemporaneous with the BSE epidemic (1), even less is known regarding how the PRNP gene evolves in these species. Herein we provide a detailed investigation of genetic variation within PRNP exon 3 for 36 breeds of cattle and 10 additional species of Bovinae. In particular, we examine patterns of variation across PRNP exon 3 and evaluate the dynamics of this variation in terms of evolutionary processes that may have brought about changes at the amino acid level observed among species, especially as they relate to structural and/or functional constraints on the prion protein.
Materials and Methods
To evaluate PRNP exon 3, we used a DNA panel of 119 artificial insemination (AI) sires from 36 cattle breeds. The source of DNA was spermatozoa. Breed names and sample sizes (in parentheses) were as follows: Angus (4), Beefalo (1), Beefmaster (4), Belgian Blue (4), Blonde D'Aquitaine (5), Braford (4), Brahman (2), Brahmousin (2), Brangus (2), Braunvieh (5), Brown Swiss (4), Charolais (5), Chianina-Chiangus (5), Corriente (1), Gelbvieh (4), Hereford (3), Holstein (4), Limousin (3), Maine Anjou (4), Murray Gray (2), Nelore (8), Normande (1), Piedmontese (2), Red Angus (4), Red Brangus (1), Red Poll (1), Romagnola (2), Salers (3), Santa Gertrudis (2), Scottish Highland (1), Senepol (2), Shorthorn (5), Simbrah (3), Simmental (8), Tarentaise (1), Texas Longhorn (4), Three-way-cross (2), and White Park (1). Six Nelore were not AI sires. Care was taken to select unrelated sires.
For comparison, we used a second DNA panel of 286 members of the subfamily Bovinae. Species included and sample sizes (in parentheses) were as follows: Tragelaphus strepsiceros, greater kudu (1); Tragelaphus imberbis, lesser kudu (1); Boselaphus tragocamelus, nilgai (1); Bubalus bubalis, Asian water buffalo (2); Bubalus depressicornis, lowland anoa (1); Syncerus caffer caffer, African buffalo (1); Syncerus caffer nanus, forest buffalo (1); Bos javanicus, banteng (2); Bos gaurus, gaur (2); and Bison bison, American bison (274). American bison were sampled from four U.S. federal herds and one private herd. For banteng, gaur, and bison, mtDNA analysis was used to exclude individuals showing evidence of interspecific hybridization with cattle (32). Likewise, Asian water buffalo that displayed karyotypic evidence of hybridization (river × swamp; ref. 33) were excluded from statistical analyses. One blackbuck, Antilope cervicapra, was sampled and used as the outgroup species to Bovinae.
Flanking primers SAF1 and SAF2 (34) were used to PCR amplify and sequence PRNP exon 3. For American bison, 96 were sequenced bidirectionally, and 178 were sequenced by using SAF1 only. Amplification, amplicon sequencing, and single-nucleotide polymorphism (SNP) detection followed previously published methods (19). Representative alleles from each genotypic class with more than one SNP were validated by a second PCR amplification, cloning, and bidirectional sequencing of multiple clones (19). This method also was used to assign indels to their respective alleles.
PRNP exon 3 sequences for all Bovinae taxa (all alleles) were aligned by using clustalx, Version. 1.81 (35). The alignment was corrected by using the published cattle octapeptide repeat units as a guide (25, 30). dnasp, Version 3.53 (36), was used to compute estimates of the number of segregating sites (S, ref. 37) and nucleotide diversity (π, ref. 38) from an intraspecific file of all cattle alleles. Estimates of Watterson's genetic diversity parameter (θ) were based on the number of segregating sites (37). The degree of nonrandom association between nucleotide variants (parsimony informative sites only, excluding gaps) for all cattle alleles was estimated by using linkage disequilibrium parameters implemented in dnasp (36), and the significance of the associations was evaluated by using Bonferroni-corrected two-tailed Fisher's exact tests (39). Estimates for the recombination parameter (R) per gene and between adjacent sites (40), as well as the minimum number of recombination events (RM, ref. 41) for all cattle alleles (excluding gaps), were computed in dnasp (36).
We used several approaches to evaluate the degree of selective constraint exerted upon bovine PRNP exon 3. Tajima's D (42) and Fu and Li's F* and D* (43) were calculated by using all cattle alleles as implemented in dnasp (36). To further assess the potential for selective constraint within cattle PRNP exon 3, these statistics were computed by using a sliding window approach (window size, 100 bp; step size, 25 bp). The McDonald and Kreitman test (44) was performed to assess intraspecific patterns of selective constraint in cattle and bison, using various wild bovine species as outgroup taxa. Estimates of the numbers of synonymous substitutions per synonymous site (dS) and nonsynonymous substitutions per nonsynonymous site (dN) were calculated by using mega, Version 2.1 (45), with the modified Nei-Gojobori method (46) with Jukes-Cantor correction and complete deletion of gaps. Standard errors for dS and dN were estimated from 1,000 bootstrap pseudoreplicates. A phylogenetic approach (47, 48) was used to examine the difference between dS and dN on both terminal and interior branches of the bovine phylogeny. In short, interior branch sequences were reconstructed by using a distance-based Bayesian method (49), allowing the observed numbers of synonymous (s) and nonsynonymous (n) substitutions to be plotted on each branch in the phylogeny and compared with the potential numbers of synonymous (S) and nonsynonymous (N) substitutions. The statistical significance of the difference between s/(S - s) and n/(N - n) was assessed by using Fisher's exact test.
Results
Assessment of PRNP Allelic Differences. PRNP exon 3 analysis of 812 chromosomes and >643,000 bp across all taxa investigated yielded 45 polymorphic sites, including polymorphisms within octapeptide repeats where indel variation also was detected. The distribution of polymorphic sites, nucleotide variants associated with each site, and predicted amino acid replacements are presented in Table 1. PRNP exon 3 alleles possessing four to seven octapeptide repeats were observed for cattle (Table 2). The unprecedented four-octapeptide repeat allele was detected in a single Brown Swiss sire, where repeats R3 and R4 (25) were predicted to be deleted by multiple sequence alignment. The seven-octapeptide repeat allele (30) was observed only for Brown Swiss cattle, whereas alleles possessing five and six repeats were noted for other domestic breeds and species of Bovinae (Table 2).
Table 1. Bovinae PRNP exon 3 polymorphic nucleotide sites.
| Bovinae species | Polymorphic nucleotide sites and single-nucleotide polymorphisms (IUPAC/IUB codes) | Total | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| B. taurus/B. indicus | 57(Y) | 69(Y) | 75(R) | 108(W) | 126(R) | 183(Y) | 189(Y) | 195(W) | 207(M) | 30 |
| 210(R) | 231(Y) | 234(R) | 237(Y) | 255(W) | 261(M) | 267(Y) | 270(Y) | 294(Y) | ||
| 315(Y) | 327(R) | 339(Y) | 378(Y) | 405(B) | 461(R) | 534(Y) | 555(Y) | 576(Y) | ||
| 630(Y) | 675(Y) | 678(Y) | ||||||||
| B. javanicus | 110(K) | 255(Y) | 267(W) | 554(R) | 4 | |||||
| Bison bison | 50(Y) | 1 | ||||||||
| Syncerus c. caffer | 231(Y) | 234(R) | 237(Y) | 255(W) | 261(M) | 267(W) | 270(W) | 554(R) | 585(M) | 11 |
| 693(R) | 700(S) | |||||||||
| Syncerus c. nanus | 679(W) | 751(R) | 2 | |||||||
| Bubalus depressicornis | 38(Y) | 351(R) | 548(R) | 3 | ||||||
| Bubalus bubalis | 126(R) | 127(R) | 234(R) | 285(R) | 322(R) | 678(Y) | 6 | |||
| Boselaphus tragocamelus | 423(R) | 1 | ||||||||
Bolded nucleotide sites are predicted to result in amino acid replacements. All others represent synonymous variation. Single-nucleotide polymorphisms and predicted amino acid replacements are as follows: 461A(S154N), 110T(G37V), 554G(N185S), 50C(M17T), 700G(Q234E), 679T(T227S), 751A(V251M), 38C(L13P), 548G(Q183R), 127A(G43R), and 322A(G108S). Nucleotide sites and variation underlined are shared among taxa. Numbering was derived from the six-octapeptide cattle allele (ref. 25; GenBank accession no. X55882). Novel cattle polymorphisms include: 183(Y), 189(Y), 195(W), 207(M), 231(Y), 237(Y), 255(W), 261(M), 267(Y), 270(Y), 294(Y), 315(Y), 327(R), 378(Y), 405(B), and 534(Y). IUPAC, International Union of Pure and Applied Chemistry; IUB, International Union of Biochemistry. The number of total unique polymorphic sites was 45.
Table 2. Observed octapeptide repeat genotypes.
| Species | Observed genotype | n | Observed frequency | Indel repeat unit* |
|---|---|---|---|---|
| B. taurus/B. indicus | 4:6 | 1 | 0.0084 | R3, R4 del |
| 5:5 | 1 | 0.0084 | R3 del | |
| 6:5 | 5 | 0.0420 | R3 del | |
| 6:6 | 109 | 0.9160 | ||
| 6:7 | 1 | 0.0084 | RN2 dup | |
| 7:7 | 2 | 0.0168 | RN2 dup | |
| B. javanicus | 6:5 | 2 | 1.0000 | R4 del |
Evaluation of the degree of nonrandom association among polymorphic sites for cattle PRNP exon 3 based on Fisher's exact test and the Bonferroni procedure revealed 12 significant (P < 0.001; Bonferroni significant for α′ = 0.05) comparisons among all parsimony informative sites. These nonrandom associations correspond to polymorphic sites in three cattle breeds: Nelore (n = 5) 69/555, 69/630, 555/630; and Brahman (n = 1) and Brahmousin (n = 1) 75/108, 75/126, 75/461, 108/126, 108/461, 108/678, 126/461, 126/678, and 461/678 (see Table 1). Significant associations result from several unique low-frequency PRNP exon 3 alleles detected in seven individuals of three cattle breeds, including three unique alleles possessing the G461A mutation (predicted amino acid replacement S154N).
PRNP exon 3 sequence analysis for other species of Bovinae yielded 19-amino acid replacements not found in cattle. The S154N amino acid replacement predicted for 3 of 238 (frequency, 0.0126) cattle PRNP exon 3 alleles was observed in all alleles for the lesser kudu, nilgai, Asian water buffalo, lowland anoa, African buffalo, and forest buffalo. Fixed amino acid replacements observed for nondomestic species when compared with cattle are as follows: lowland anoa, S4R, A16V, P54S, G108S, V123M, and F257L; Asian water buffalo, S4R, A16V, P54S, G108S, V123M, and F257L; greater kudu, G22A, P54S, H166Y, E197Q, and Q234E; lesser kudu, P54S, H166Y, E197Q, and Q234E; African buffalo, A16V, P54S, and I214V; forest buffalo, A16V, P54S, N185S, and I214V; and nilgai, G59S, G75S, S146N, H188R, R231T, I244V, and I252V.
Analyses of Intraspecific Polymorphism and Interspecific Tests of Selection. Overall, genetic variability in cattle was low. By excluding regions with gaps due to indels (25, 30), 22 unique PRNP exon 3 alleles were determined for our panel (238 total alleles) of cattle. Most of these alleles possessed one to three synonymous changes, and only one polymorphic site noted for cattle (G461A; Table 1) was associated with a predicted amino acid replacement (S154N). Thirteen of the 26 segregating sites observed, excluding gaps, were singletons. The number of total mutations was 27. Furthermore, estimates of θ (0.00578 ± 0.00163) and R (0.0000 per base pair and 0.0010 per gene) for cattle exon 3 alleles are low in comparison to what has been observed for other nuclear genes (38). Nucleotide diversity (π) was estimated at 0.00125 (± 0.00014). Thus, it is not surprising that Tajima's test yielded a significantly negative D value (-2.14, P < 0.01), as did Fu and Li's tests using both the D* and F* statistics (-4.12, P < 0.02 and -3.99, P < 0.02, respectively).
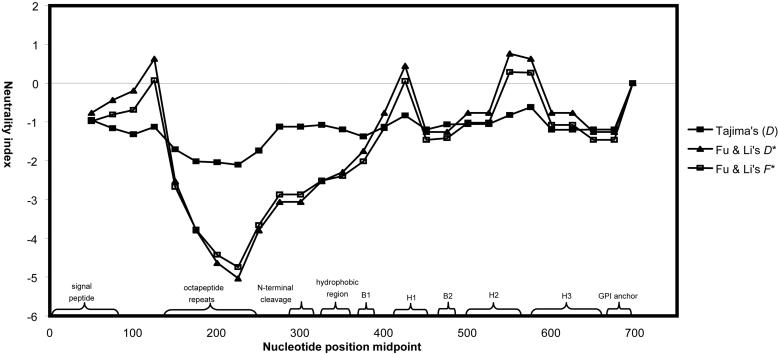
Application of a sliding-windows approach to Tajima's and Fu and Li's tests revealed several significantly negative 100-bp windows (Fig. 1) within cattle PRNP exon 3 (Tajima's test, P < 0.05; Fu and Li's tests, P < 0.05) that correspond to known structural features of the mammalian prion protein (50). Significantly negative windows by all tests correspond to the amino acid residues preceding the proline hydroxylation site, the octapeptide repeat region (repeat units R1-R6 and RN2; refs. 25 and 30), and the region C-terminal to R6 (R3, R4, and RN2 deleted by gap handling). In addition, Fu and Li's tests yielded significantly negative values for a window corresponding to the region of cattle PRNP exon 3 extending from R5 to the amino acid residues preceding the N-terminal cleavage site (Fu and Li's tests, P < 0.02; Tajima's D test, P < 0.10 for cleavage site). Fu and Li's tests also revealed several significantly negative windows (P < 0.05) corresponding to the hydrophobic region (transmembrane domain component) and β region 1 (B1) of cattle PRNP exon 3.
Fig. 1.
Sliding-window analyses of Tajima's (D) and Fu and Li's tests (D* and F*) for cattle PRNP exon 3 (repeats R3, R4, and RN2 deleted by gap handling). Significantly negative 100-bp windows (P < 0.05; all tests) extend from the amino acid residues preceding the proline hydroxylation site, through the octapeptide repeat region, and into the region C-terminal to R6. Fu and Li's tests also revealed significantly negative windows corresponding to the hydrophobic region and (B1)β region 1.
Examination of the pattern of synonymous and nonsynonymous substitutions was indicative of strong purifying selection, which is not surprising given the low levels of polymorphism observed and the results obtained from Tajima's and Fu and Li's tests. The average values of dS (d̄S) and dN (d̄N), respectively, were 0.0045 ± 0.0015 and 0.000046 ± 0.000045 for all cattle PRNP exon 3 alleles, yielding a d̄N/d̄S ratio of 0.01. All McDonald and Kreitman (MK) tests between cattle and other species of Bovinae were significant, except in cases when cattle were compared with closely related species (i.e., bison, gaur, and banteng; Table 3). These species diverged from cattle very recently, probably within the last 1-2 million years (51, 52), which explains the lack of fixed differences between them. MK tests performed between Bison bison and all other taxa were not significant.
Table 3. McDonald-Kreitman tests for PRNP exon 3 of domestic cattle and nondomestic taxa of Bovinae.
| Comparison species* | Fisher's† | G test‡ | Substitutions | Fixed | Polymorphic |
|---|---|---|---|---|---|
| Syncerus c. nanus | 0.0271 | 0.0266 | Synonymous | 4 | 26 |
| Nonsynonymous | 4 | 3 | |||
| Syncerus c. caffer | 0.0359 | 0.0392 | Synonymous | 3 | 30 |
| Nonsynonymous | 3 | 3 | |||
| Bubalus bubalis | 0.0006 | 0.0004 | Synonymous | 4 | 26 |
| Nonsynonymous | 6 | 1 | |||
| Bubalus depressicornis | 0.0033 | 0.0028 | Synonymous | 4 | 27 |
| Nonsynonymous | 6 | 3 | |||
| Boselaphus tragocamelus | 0.0009 | 0.0006 | Synonymous | 5 | 27 |
| Nonsynonymous | 6 | 1 | |||
| T. strepsiceros | 0.0127 | 0.0098 | Synonymous | 9 | 26 |
| Nonsynonymous | 5 | 1 | |||
| T. imberbis | 0.0136 | 0.0114 | Synonymous | 6 | 26 |
| Nonsynonymous | 4 | 1 | |||
| Bison bison | — | — | Synonymous | 0 | 26 |
| Nonsynonymous | 0 | 2 | |||
| B. javanicus | 1.0000 | — | Synonymous | 1 | 27 |
| Nonsynonymous | 0 | 3 | |||
| B. gaurus | — | — | Synonymous | 0 | 26 |
| Nonsynonymous | 0 | 1 |
Dash indicates that statistical tests could not be performed. No complex codons were encountered.
Between-species comparisons were made with domestic cattle with complete deletion of gaps.
P value for Fisher's exact test.
P value for G test with Williams correction.
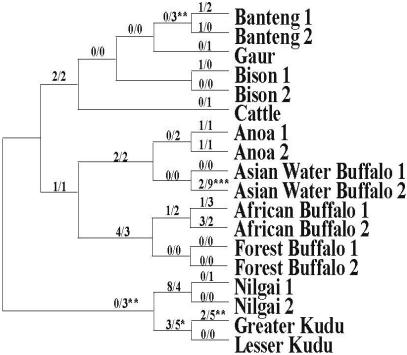
The observed numbers of synonymous and nonsynonymous changes were plotted for each branch in the bovine phylogeny (Fig. 2). Clearly, these numbers are small, indicating that an intense level of selection constrains the number of variable sites within PRNP exon 3. Thus, when the differences between levels of synonymous and nonsynonymous changes are compared, Fisher's exact test must be used (47). When we conducted this test, we found evidence for strong purifying selection (P ≪ 0.001) when levels of synonymous and nonsynonymous nucleotide substitution were compared over the entire phylogenetic tree. On the other hand, with few exceptions, levels of synonymous and nonsynonymous substitution were not different from each other along individual branches, particularly at shallow levels (i.e., terminal branches). There are two possible explanations for this. First, all variable sites observed are selectively neutral. This is consistent with the fact that no amino acid polymorphisms have been identified to date that either confer resistance to BSE (advantageous mutations) or augment expression of BSE (deleterious mutations). The second explanation for the observed pattern is that there is a lack of power to detect selection along individual branches due to only a very small number (e.g., one to three) of variable sites. This is the most likely explanation.
Fig. 2.
Phylogeny used for conducting tests of selection on bovine PRNP exon 3 sequences. The phylogeny is based on published molecular and morphological studies (51, 52). The numbers along branches represent observed nonsynomymous (n) and synonymous (s) substitutions, respectively. The average values of potential synonymous (S) and nonsynonymous (N) substitutions for all extant taxa were 226 and 563, respectively. The posterior probabilities for all inferred ancestral sequences were ≥99%. Note that the use of alternative alleles and/or tree topologies did not change the interpretation of the results. *, 0.02 < P ≤ 0.05; **, 0.005 < P ≤ 0.02; ***, P ≤ 0.004.
Discussion
Although rare, detection of the predicted S154N polymorphism encoded by cattle PRNP exon 3 is not unprecedented, because this polymorphism was previously detected at low frequencies (20, 23, 24). Given the low frequency of the S154N polymorphism and the overall pattern of nucleotide variation observed for cattle PRNP exon 3, additional functional, transgenic, and challenge experiments seem appropriate. Moreover, amino acid 154 (143, human numbering) has previously been implicated in the susceptibility of humans to cattle-derived prions (53). Significant nonrandom associations between polymorphic nucleotides of the three unique cattle PRNP alleles possessing the G461A mutation (S154N; Table 1) is unusual but not surprising, given the low estimates obtained for recombination (0.0000 per base pair and 0.0010 per gene). Also of interest is the commonality of the amino acid asparagine at position 154 (N154) for other species of Bovinae, as well as among other mammalian taxa (50). Notably, bovine amino acid position 154 (six-octapeptide repeat allele) corresponds to ovine position 146, which has not been described as polymorphic (N146) in sheep nor been implicated in the expression of scrapie. Therefore, additional studies are needed to evaluate what effects, if any, the bovine S154N polymorphism has on BSE expression.
The distribution of observed PRNP exon 3 octapeptide repeat genotypes for cattle was somewhat unexpected (Table 2), because we anticipated more individuals of the 6:5 genotype based on earlier studies (20, 21, 28, 29, 31). The underlying reason for this may be due to the fact that our cattle samples are almost exclusively from artificial insemination sires. However, the observed genotypic frequency of the 6:6 genotype for young bulls (0.894) in a study of Polish Black-and-White cattle (54) approached that observed in this study (0.916; Table 2). Furthermore, a study of full families of Polish Black-and-White cattle demonstrated abnormal segregation of octapeptide repeat alleles, as evidenced by nearly twice the expected number of 6:6 genotypes (55).
Frequency-distribution tests provided significant statistical support for an excess of rare alleles and/or singletons in our overall sample of cattle PRNP exon 3 alleles. Significantly negative values for Tajima's D and Fu and Li's tests (D* and F*) are often interpreted as purifying or directional selection but may also indicate violations of the mutation-drift equilibrium assumptions (42) and/or the random sample requirement (43) of these tests. For cattle PRNP exon 3, all of the singletons and most of the rare alleles (19/22; 86%) resulted from synonymous variation, which is unlikely to be subject to directional selection (56). Such an excess of rare synonymous variants, given a theoretically large random mating population, might indicate recent population expansion followed by insufficient time to establish a balance between the occurrence of new mutations and their loss via genetic drift (56). Nevertheless, the pattern of nucleotide variation observed for cattle PRNP exon 3 overall and within significantly negative 100-bp windows corresponding to regions preceding the N-terminal cleavage site, the cleavage site itself, the hydrophobic region, and β region 1 (B1) suggests that selection may be acting to preserve the amino acid sequence of cattle PrPC within regions of functional, structural, or potential pathogenic importance.
Interestingly, amino acid substitutions associated with human hereditary and sporadic spongiform encephalopathies form two clusters: (i) the region C-terminal to the octapeptide repeat region and N-terminal to the first α-helix and (ii) the second and third α-helices (refs. 1 and 57; Fig. 1). Several significantly negative windows obtained for cattle PRNP exon 3 correspond to regions where three human mutations (P102L, P105L, and A117V) associated with Gerstmann-Sträusler-Scheinker disease, a human hereditary spongiform encephalopathy, have been identified (1). Synonymous variation (C339T) was observed at the third position of the cattle equivalent to human codon 102 in our study and in previous studies (20, 26). Significantly negative windows also were obtained for the region of cattle PRNP exon 3 corresponding to human codon 129 (1). Also intriguing is the inclusion of β region 1 (B1) within significantly negative windows identified by sliding-window analysis, given that the conversion of PrPC to PrPSc is considered to bring about changes in secondary structure hallmarked by increased β-sheet formation (1).
Although the total number of fixed amino acid changes is relatively small in any given comparison, it is interesting to note the number of shared fixations among the taxa in our study. For example, our samples for lowland anoa and Asian water buffalo possessed the same fixed changes (S4R, A16V, P54S, G108S, V123M, and F257L) when compared with cattle. Likewise, both subspecies of Syncerus (African and forest buffalo) shared the A16V, P54S, and I214V fixed changes, and three fixed replacements also were observed for both species of Tragelaphus when compared with cattle. Previous phylogenetic studies suggested convergence between great apes and cattle at specific amino acid residues (53, 57). Although convergence remains possible, shared amino acid replacements between bovine species may represent shared primitive characters, given the short time frame since these species diverged from one another (51, 52).
Several fixed amino acid replacements noted in between-species comparisons with cattle warrant further investigation in light of previous studies on ovine scrapie and PrPC biogenesis (8, 10, 58, 59). The fixed replacement H188R in nilgai, equivalent to codon 180 in sheep, is proximal to the ovine Q171R polymorphism associated with scrapie resistance (8, 10). Furthermore, a study of amino acid replacements within the signal peptide of PrPC, proximally relevant to fixed replacements S4R and A16V, has demonstrated that signal peptide mutations influence the ratio of three topological forms in which PrPC is synthesized at the endoplasmic reticulum (ER) (58, 59). The predominant form (secPrP) is fully translocated into the ER lumen, whereas the other two forms (NtmPrP and CtmPrP) are single-spanning membrane proteins named to reflect the terminus inserted into the lumen (58-61). Signal sequence mutations may increase or decrease the ratio of CtmPrP relative to the other topological forms (58, 59). In addition, mutations that increase the generation of CtmPrP have been associated with neurodegenerative disease (58, 60, 61). Therefore, it is important to evaluate the amino acid replacements M17T and L13P, observed within the signal sequences of bison and lowland anoa, respectively (Table 1).
Several other nonsynonymous PRNP exon 3 polymorphisms observed for taxa of Bovinae warrant investigation based on ovine PRNP and scrapie (8, 10). For example, the amino acid replacement Q183R observed for lowland anoa corresponds to ovine position 175, which is near the ovine Q171R polymorphism. Likewise, the N185S replacement, polymorphic for banteng (24) but fixed for our sample of forest buffalo, is equivalent to ovine position 177, which is also proximal to ovine Q171R. Predicted amino acid replacements presented herein that are proximal to ovine amino acid 171 represent suitable candidates for future challenge experiments related to BSE resistance and/or susceptibility.
Conclusion
We have further documented and evaluated the emerging pattern of nucleotide variation for cattle PRNP exon 3, revealing evidence for highly intense purifying selection within regions previously suggested and/or demonstrated to be of functional, structural, or pathogenic importance in humans and other mammalian species (1, 8, 10, 50). In addition, several polymorphic sites and corresponding amino acid replacements for taxa not included in previous studies were identified (Table 1). These polymorphisms, as well as the fixed amino acid replacements identified in between-species comparisons with cattle, provide an opportunity to evaluate a new battery of amino acid residues with respect to prion disease in domestic and wild bovids.
Perhaps the most interesting outcome of this study concerns the nature of selection on PRNP exon 3. Knockout mice suffer either very subtle or no deleterious effects upon losing the PRNP gene (1, 62), suggesting that PRNP may be an evolutionary “appendix” not necessarily needed by the body. Yet, if this were true, why would purifying selection be so intense on this protein? Such strong levels of purifying selection are normally seen only among proteins such as histones that are essential to eukaryotic life (63). Thus, what is the driving force behind such intense purifying selection on PRNP? The answer to this question is currently unknown.
Acknowledgments
We thank J. W. Templeton (Texas A&M University) and Christopher Schutta (Texas A&M University) for seqscape; S. N. White (U.S. Department of Agriculture-Agriculture Research Station, Clay Center, NE), J. E. Womack (Texas A&M University), and L. C. Skow (Texas A&M University) for several DNA samples used in this project; and B. P. Chowdhary (Texas A&M University), T. Raudsepp (Texas A&M University), and E. Owens (Texas A&M University) for B. bubalis karyo-types. We also thank Texas A&M University and the Texas Agricultural Experiment Station for supporting this research.
Author contributions: C.M.S., R.L.H., and J.N.D. designed research; C.M.S. performed research; C.M.S., R.L.H., A.P.R., and N.D.H. analyzed data; and C.M.S., R.L.H., A.P.R., N.D.H., and J.N.D. wrote the paper.
Abbreviations: PRNP, prion protein gene; BSE, bovine spongiform encephalopathy; PrPC, cellular prion protein; indel, insertion-deletion.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY720445-AY720706).
References
- 1.Collinge, J. (2001) Annu. Rev. Neurosci. 24, 519-550. [DOI] [PubMed] [Google Scholar]
- 2.Prusiner, S. B. (1982) Science 216, 136-144. [DOI] [PubMed] [Google Scholar]
- 3.Prusiner, S. B. (1991) Science 252, 1515-1522. [DOI] [PubMed] [Google Scholar]
- 4.Bossers, A., de Vries, R. & Smits, M. A. (2000) J. Virol. 74, 1407-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griffith, J. S. (1967) Nature 215, 1043-1044. [DOI] [PubMed] [Google Scholar]
- 6.McGowan, J. P. (1922) Scot. J. Agric. 5, 365-375. [Google Scholar]
- 7.Wells, G. A. H., Scott, A. C., Johnson, C. T., Gunning R. F., Hancock, R. D., Jeffrey, M., Dawson, M. & Bradley, R. (1987) Vet. Rec. 31, 419-420. [DOI] [PubMed] [Google Scholar]
- 8.Hunter, N. (1997) in The Genetics of Sheep, ed. Ruvinsky, A. (CAB International, Wallingford, U.K.), pp. 225-240.
- 9.Wilesmith, J. W. (1991) Vet. Rec. 128, 310 (lett.). [DOI] [PubMed] [Google Scholar]
- 10.Belt, P. B., Muileman, I. H., Schreuder, B. E., Bos-de Ruijter, J., Gielkens, A. L. & Smits, M. A. (1995) J. Gen. Virol. 76, 509-517. [DOI] [PubMed] [Google Scholar]
- 11.Belt, P. B., Bossers, A., Schreuder, B. E. & Smits, M. A. (1996) in Bovine Spongiform Encephalopathy: The BSE Dilemma, ed. Gibbs, C. J., Jr. (Springer, New York), pp. 294-305.
- 12.Bossers, A., Schreuder, B. E., Muileman, I. H., Belt, P. B. & Smits, M. A. (1996) J. Gen. Virol. 77, 2669-2673. [DOI] [PubMed] [Google Scholar]
- 13.Goldmann, W., Hunter, N., Benson, G., Foster, J. D. & Hope, J. (1991) J. Gen. Virol. 72, 2411-2417. [DOI] [PubMed] [Google Scholar]
- 14.Goldmann, W., Hunter, N., Foster, J. D., Salbaum, J. M., Beyreuther, K. & Hope, J. (1990) Proc. Natl. Acad. Sci. USA 87, 2476-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunter, N., Foster, J. D., Goldmann, W., Stear, M. J., Hope, J. & Bostock, C. (1996) Arch. Virol. 141, 809-824. [DOI] [PubMed] [Google Scholar]
- 16.Laplanche, J. L., Chatelain, J., Westaway, D., Thomas, S., Dussaucy, M., Brugere-Picoux, J. & Launay, J. M. (1993) Genomics 15, 30-37. [DOI] [PubMed] [Google Scholar]
- 17.Tranulis, M. A., Osland, A., Bratberg, B. & Ulvund, M. J. (1999) J. Gen. Virol. 80, 1073-1077. [DOI] [PubMed] [Google Scholar]
- 18.Gombojav, A., Ishiguro, N., Horiuchi, M., Serjmyadag, D., Byambaa, B. & Shinagawa, M. (2003) J. Vet. Med. Sci. 65, 75-81. [DOI] [PubMed] [Google Scholar]
- 19.Seabury, C. M. & Derr, J. N. (2003) Cytogenet. Genome Res. 102, 85-88. [DOI] [PubMed] [Google Scholar]
- 20.Heaton, M., Leymaster, K., Freking, B., Hawk, D., Smith T., Keele, J., Snelling, W., Fox, J., Chitko-McKown, C. & Laegreid, W. (2003) Mamm. Genome 14, 765-777. [DOI] [PubMed] [Google Scholar]
- 21.Sander, P., Hamann, H., Pfeiffer, I., Wenheuer, W., Brenig, B., Groschup, M., Ziegler, U., Distl, O. & Leeb, T. (2004) Neurogenetics 5, 19-25. [DOI] [PubMed] [Google Scholar]
- 22.Casalone, C., Zanusso, G., Acutis, P., Ferrari, S., Capucci, L., Tagliavini, F., Monaco, S. & Caramelli, M. (2004) Proc. Natl. Acad. Sci. USA 101, 3065-3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wopfner, F., Weidenhöfer, G., Schneider, R., von Brunn, A., Gilch, S., Schwarz, T., Werner, T. & Schätzl, H. (1999) J. Mol. Biol. 289, 1163-1178. [DOI] [PubMed] [Google Scholar]
- 24.Takasuga, A., Abe, T., Ito, T., Watanabe, T., Kamatani, N. & Sugimoto, Y. (2003) Anim. Genet. 34, 396-397. [DOI] [PubMed] [Google Scholar]
- 25.Goldmann, W., Hunter, N., Martin, T., Dawson, M. & Hope, J. (1991) J. Gen. Virol. 72, 201-204. [DOI] [PubMed] [Google Scholar]
- 26.Humeny, A., Schiebel, K., Seeber, S. & Becker, C.-M. (2002) Neurogenetics 4, 59-60. [DOI] [PubMed] [Google Scholar]
- 27.Hills, D., Schlaepfer, J., Comincini, S., MacLean, I., Dolf, G., Ferretti, L., Olsaker, I. & Williams, J. L. (2003) Anim. Genet. 34, 183-190. [DOI] [PubMed] [Google Scholar]
- 28.Hunter, N., Goldmann, W., Smith, G. & Hope, J. (1994) Vet. Rec. 135, 400-403. [DOI] [PubMed] [Google Scholar]
- 29.Neibergs, H. L., Ryan, A. M., Womack, J. E., Spooner, R. L. & Williams, J. L. (1994) Anim. Genet. 25, 313-317. [DOI] [PubMed] [Google Scholar]
- 30.Schlapfer, J., Saitbekova, N., Gaillard, C. & Dolf, G. (1999) Anim. Genet. 30, 386-387. [DOI] [PubMed] [Google Scholar]
- 31.Premzl, M., Bozic, P. & Gamulin, V. (2000) Anim. Genet. 31, 408-409. [DOI] [PubMed] [Google Scholar]
- 32.Ward, T. J., Bielawski, J. P., Davis, S. K., Templeton, J. W. & Derr, J. N. (1999) Anim. Conserv. 2, 51-57. [Google Scholar]
- 33.Chowdhary, B.P., Gustavsson, I., Kunavongkrit, A., Lohachit, C. & Mäkinen, A. (1989) Buffalo J. 1, 41-49. [Google Scholar]
- 34.Prusiner, S. B., Füzi, M., Scott, M., Serban, D., Serban, H., Taraboulos, A., Gabriel, J.-M., Wells, G. A., Wilesmith, J. W., Bradley, R., et al. (1993) J. Infect. Dis. 167, 602-613. [DOI] [PubMed] [Google Scholar]
- 35.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997) Nucleic Acids Res. 25, 4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rozas, J. & Rozas, R. (1999) Bioinformatics 15, 174-175. [DOI] [PubMed] [Google Scholar]
- 37.Watterson, G. A. (1975) Theor. Popul. Biol. 7, 256-276. [DOI] [PubMed] [Google Scholar]
- 38.Nei, M. (1987) Molecular Evolutionary Genetics (Columbia Univ. Press, New York).
- 39.Sokal, R. R. & Rohlf, F. J. (1981) Biometry (Freeman, New York).
- 40.Hudson, R. R. (1987) Genet. Res. 50, 245-250. [DOI] [PubMed] [Google Scholar]
- 41.Hudson, R. R. & Kaplan, N. L. (1985) Genetics 111, 147-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tajima, F. (1989) Genetics 123, 585-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu, Y.-X. & Li, W.-H. (1993) Genetics 133, 693-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McDonald, J. H. & Kreitman, M. (1991) Nature 351, 652-654. [DOI] [PubMed] [Google Scholar]
- 45.Kumar, S., Tamura, K., Jakobsen, I. B. & Nei, M. (2001) Bioinformatics 17, 1244-1245. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, J., Rosenberg, H. & Nei, M. (1998) Proc. Natl. Acad. Sci. USA 95, 3708-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, J., Kumar, S. & Nei, M. (1997) Mol. Biol. Evol. 14, 1335-1338. [DOI] [PubMed] [Google Scholar]
- 48.Rooney, A. & Zhang, J. (1999) Mol. Biol. Evol. 16, 706-710. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, J. & Nei, M. (1997) J. Mol. Evol. 44, Suppl. 1, S139-S146. [DOI] [PubMed] [Google Scholar]
- 50.van Rheede, T., Smolenaars, M. W., Madsen, O. & de Jong, W. W. (2003) Mol. Biol. Evol. 20, 111-121. [DOI] [PubMed] [Google Scholar]
- 51.Ritz, L. R., Glowatzki-Mullis, M.-L., MacHugh, D. E. & Gaillard, C. (2000) Anim. Genet. 31, 178-185. [DOI] [PubMed] [Google Scholar]
- 52.Janecek, L. L., Honeycutt, R. L., Adkins, R. M. & Davis, S. K. (1996) Mol. Phylogenet. Evol. 6, 107-119. [DOI] [PubMed] [Google Scholar]
- 53.Krakauer, D., Pagel, M., Southwood, T. & Zanotto, P. M. (1996) Nature 380, 675 (lett.). [DOI] [PubMed] [Google Scholar]
- 54.Walawski, K. & Czarnik, U. (2003) J. Appl. Genet. 44, 191-195. [PubMed] [Google Scholar]
- 55.Walawski, K., Czarnik, U., Wojciechowski, R. & Pareek, C. S. (2003) J. Appl. Genet. 44, 375-378. [PubMed] [Google Scholar]
- 56.Glatt, C. E., DeYoung, J. A., Delgado, S., Service, S. K., Giacomini, K. M., Edwards, R. H., Risch, N. & Freimer, N. B. (2001) Nat. Genet. 27, 435-438. [DOI] [PubMed] [Google Scholar]
- 57.Krakauer, D., Zanotto, P. & Pagel, M. (1998) J. Mol. Evol. 47, 133-145. [DOI] [PubMed] [Google Scholar]
- 58.Kim, S. J., Rahbar, R. & Hegde, R. (2001) J. Biol. Chem. 276, 26132-26140. [DOI] [PubMed] [Google Scholar]
- 59.Kim, S. J. & Hegde, R. (2002) Mol. Biol. Cell. 13, 3775-3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hegde, R. S., Tremblay, P., Groth, D., DeArmond, S. J., Prusiner, S., B. & Lingappa, V. R. (1999) Nature 402, 822-826. [DOI] [PubMed] [Google Scholar]
- 61.Hegde, R. S., Mastrianni, J. A., Scott, M. R., Defea, K. A., Tremblay, P., Torchia, M., DeArmond, S. J., Prusiner, S. B. & Lingappa, V. R. (1998) Science 279, 827-834. [DOI] [PubMed] [Google Scholar]
- 62.Estibeiro, J. P. (1996) Trends Neurosci. 19, 257-259. [DOI] [PubMed] [Google Scholar]
- 63.Piontkivska, H., Rooney, A. P. & Nei, M. (2002) Mol. Biol. Evol. 19, 689-697. [DOI] [PubMed] [Google Scholar]