Abstract
The small GTPase Arf1p is involved in different cellular processes that require its accumulation at specific cellular locations. The recruitment of Arf1p to distinct points of action might be achieved by association of Arf1p with different proteins. To identify new interactors of Arf1p, we performed an affinity chromatography with GTP- or GDP-bound Arf1p proteins. A new interactor of Arf1p-GTP was identified as Pab1p, which binds to the polyA-tail of mRNAs. Pab1p was found to associate with purified COPI-coated vesicles generated from Golgi membranes in vitro. The stability of the Pab1p–Arf1p complex depends on the presence of mRNA. Both symmetrically distributed mRNAs as well as the asymmetrically localized ASH1 mRNA are found in association with Arf1p. Remarkably, Arf1p and Pab1p are both required to restrict ASH1 mRNA to the bud tip. Arf1p and coatomer play an unexpected role in localizing mRNA independent and downstream of the SHE machinery. Hereby acts the SHE machinery in long-range mRNA transport, whereas COPI vesicles could act as short-range and localization vehicles. The endoplasmic reticulum (ER)–Golgi shuttle might be involved in concentrating mRNA at the ER.
INTRODUCTION
Proteins destined for secretion are packaged into coated vesicles and transported through different cellular compartments. One major factor involved in this process is the small GTPase Arf1p. The protein is required for the formation of COPI-coated and subclasses of clathrin-coated vesicles (Kirchhausen, 2000). In addition, there is growing evidence that Arf1p plays a role in rearrangements of the cytoskeleton as well as in lipid metabolism (Fucini et al., 2000; Skippen et al., 2002). In higher eukaryotes, different members of the class I family of ARF proteins may play these distinct roles. However, in the yeast Saccharomyces cerevisiae only three ARF proteins have been identified. Arf1p and Arf2p are 96% identical, but Arf1p is ∼10 times more abundant than Arf2p (Stearns et al., 1990). Arf3p seems to fulfill similar functions to mammalian ARF6, a class III ARF, and plays a role in endocytosis (Lee et al., 1994; Givan and Sprague, 1997). Given the diverse requirements for Arf1p, its function needs to be tightly regulated in a temporally and spatially correlated manner and must therefore pivot on effector molecules that determine the specificity of the different Arf1p-dependent events. The guanine nucleotide-bound state of Arf1p is controlled by a guanine nucleotide exchange factor (ARFGEF) and a GTPase-activating protein (ARF-GAP). In yeast, three ARF-GEFs with established functions have been identified: Gea1p, Gea2p, and Sec7p. Gea1p and Gea2p have overlapping functions in retrograde transport from the Golgi to the endoplasmic reticulum (ER), whereas Sec7p has been implicated in vesicle formation from the trans-Golgi (Franzusoff et al., 1992; Spang et al., 2001). The cellular role of the fourth Sec7-domain containing protein, Syt1p, is still unclear, but it might act at the post-Golgi level (Jones et al., 1999). Which of the GEFs mediates the other Arf1p-dependent transport steps remains to be established. ARF-GAPs outnumber the ARF-GEFs in the cell. At least six putative ARF-GAPs are present in the yeast genome. Gcs1p and Glo3p have overlapping functions in retrograde transport from the Golgi to the ER (Poon et al., 1999). Yet, only Glo3p interacts directly with coatomer (Lewis et al., 2004). Age2p can be replaced by Gcs1p in the endocytic pathway (Poon et al., 2001). In addition, Gcs1p influences actin polymerization dynamics in yeast (Blader et al., 1999). The reuse of the same molecules for different tasks implies that these known regulators of Arf1p GTPase activity might be insufficient to target Arf1p to the different cellular locations required to perform its multiple functions. Furthermore, additional requirements for Arf1p in the cell might exist that are still veiled.
To identify additional interactors and effectors of Arf1p, we took a differential affinity chromatography approach with mutants restricted to the either GTP- or GDP-bound form of Arf1p. Unexpectedly, we found that Pab1p specifically interacts with the activated form of Arf1p (Arf1p-GTP), and formation of the Pab1p–Arf1p complex requires the presence of mRNA. Pab1p is the major polyA-binding protein in yeast and is required for translation and mRNA stability in the cell. Arf1p-Pab1p ribonucleotide particles contain both symmetrically and asymmetrically distributed mRNAs. These ribonucleotide particles are associated with COPI vesicles derived from the Golgi apparatus. Importantly, our results indicate that Pab1p and Arf1p cooperate to correctly localize ASH1 mRNA in the bud tip of growing yeast cells. ASH1 mRNA is one of at least two mRNAs that are asymmetrically localized to the bud of exponentially growing yeast cells (Long et al., 1997; Takizawa et al., 2000; Shepard et al., 2003). The mechanism of the asymmetric localization of the mRNA is dependent on the She proteins. These proteins include the motor protein Myo4p (She1p) as well as an RNA binding protein (She2p) (Bohl et al., 2000; Long et al., 2000; Takizawa and Vale, 2000). No role in mRNA localization for proteins involved in vesicular traffic had been evident. However, we now show that components of the early secretory transport machinery are required for keeping the asymmetrically distributed mRNA in the bud tip and additionally might bring symmetrically distributed mRNA to the ER. This process is independent of the SHE machinery.
MATERIALS AND METHODS
Yeast Methods and Protein Expression
Escherichia coli BL21(DE3)pLysS (Novagen, Madison, WI) was used for protein expression. Yeast strains used in this study are listed in Table 1. Standard genetic techniques were used throughout (Sherman, 1991). Chromosomal tagging and deletions were performed as described in Knop et al. (1999).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| GPY60 | MATα ura3-52 leu2,3-112 his4-579 pep4::ura3 prb1 trp4-579 | R. Schekman |
| YPH500 | MATα ade2-101 his3-200 leu2-1 lys2-801 trp-63 ura3-52 | P. Hieter |
| W303-1Aa | MATa ade2-1 ura3-1 trp1-1 his3-11,15 leu2,3-112 can1-100 | P. Poon |
| YAS321 | MATα ade2-101 his3-200 leu2-1 lys2-801 trp-63 ura3-52 arf1::HIS3MX6 | This study |
| YAS147 | MATα ade2-101 his3-200 leu2-1 lys2-801 trp-63 ura3-52 pG14-MS2-GFP (LEU2) YEP15-Lz-MS2-ASH1–3′UTR (URA3) | This study |
| YAS324 | MATα ade2-101 his3-200 leu2-1 lys2-801 trp-63 ura3-52 arf1::HIS3MX6 pG14-MS2-GFP (LEU2) YEP15-Lz-MS2-ASH1–3′UTR (URA3) | This study |
| YAS212 | MATα ade2-101 his3-200 leu2-1 lys2-801 trp-63 ura3-52 ARF1::ARF1-6HA-klTRP1 | This study |
| YAS225 | MATa ade2-101 his3-200 leu2-1 lys2-801 trp-63 ura3-52 PAB1::PAB1-9myc-klTRP1 | This study |
| YAS238 | ade2-101 his3-200 leu2-1 lys2-801 trp-63 ura3-52 ARF1::ARF1–6HA-klTRP1 PAB1::PAB1-9myc-klTRP1 | This study |
| YAS258-A | ade2-101 his3-200 leu2-1 lys2-801 trp-63 ura3-52 ARF1::ARF1-YFP-HIS3MX6 | This study |
| YAS258-B | ade2-101 his3-200 leu2-1 lys2-801 trp-63 ura3-52 ARF1::ARF1-YFP-HIS3MX6 PAB1::PAB1-CFP-KANMX6 | This study |
| YAS258-C | ade2-101 his3-200 leu2-1 lys2-801 trp-63 ura3-52 PAB1::PAB1-CFP-KANMX6 | This study |
| YAS258-D | ade2-101 his3-200 leu2-1 lys2-801 trp-63 ura3-52 | This study |
| YAS259-A | ade2-101 his3-200 leu2-1 lys2-801 trp-63 ura3-52 ARF1::ARF1-CFP-KANMX6 PAB1::PAB1-YFP-HIS3MX6 | This study |
| YAS259-B | ade2-101 his3-200 leu2-1 lys2-801 trp-63 ura3-52 ARF1::ARF1-CFP-KANMX6 | This study |
| YAS259-C | ade2-101 his3-200 leu2-1 lys2-801 trp-63 ura3-52 | This study |
| YAS259-D | ade2-101 his3-200 leu2-1 lys2-801 trp-63 ura3-52 PAB1::PAB1-YFP-HIS3MX6 | This study |
| NYY0-1 | MATa ade2::ARF1::ADE2 arf1::HIS3 arf2::HIS3 ura3 lys2 trp1 his3 leu2 | A. Nakano |
| NYY11-1 | MATa ade2::arf1-11::ADE2 arf1::HIS3 arf2::HIS3 ura3 lys2 trp1 his3 leu2 | A. Nakano |
| NYY17-1 | MATa ade2::arf1-17::ADE2 arf1::HIS3 arf2::HIS3 ura3 lys2 trp1 his3 leu2 | A. Nakano |
| NYY18-1 | MATa ade2::arf1-18::ADE2 arf1::HIS3 arf2::HIS3 ura3 lys2 trp1 his3 leu2 | A. Nakano |
| RSY248 | MATα his4-619 | R. Schekman |
| RSY271 | MATα his4-619 sec18-1 | R. Schekman |
| RSY1017 | MATα his3-200 ura3-52 pep4::URA3 sec21-1 | R. Schekman |
| RSY281 | MATα his4-619 ura3-52 sec23-1 | R. Schekman |
| RSY1312 | MATa leu2,3-112 trp1 ura3-52 sec27-1 | R. Schekman |
| sar1-D32G | MATα ura3 leu2 trp1 his3 ade2 lys2 sar1::HIS3 pep4::ADE2 pMYY3-1(YCp[sar1-D32G TRP1]) | A. Nakano |
| RJY585 | MATa ade2-1 his3-11 leu2-3 trp1-1 ura3 HO-ADE2 HO-CAN1 ASH1::HIS3 (S. pombe) | R. Jansen |
| YAS2314 | MATa ade2 his3 leu2 trp1 ura3 spb8::LEU2 pab1::HIS3 | A. Sachs |
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | EUROSCARF |
| Δarf1 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 arf1::KanMX4 | EUROSCARF |
| Δarf2 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 arf2::KanMX4 | EUROSCARF |
| Δbfr1 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 bfr1::KanMX4 | EUROSCARF |
| YAS318-D | his3Δ1 leu2Δ0 MET15 lys2Δ0 ura3Δ0 arf1::HIS3MX6 bfr1::KanMX4 | This study |
| Δscp160 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 scp160::KanMX4 | EUROSCARF |
Arf1p Affinity Chromatography
Recombinant ΔN17-Arf1Q71Lp and ΔN17-Arf1T31Np were expressed in E. coli and purified as described by Rein et al. (2002). Fifty milligrams of each protein was covalently coupled to 1.2 ml of N-hydroxysuccinimide (NHS)-activated Sepharose 4 fast flow (Amersham Biosciences, Freiburg, Germany). Yeast cytosol was prepared as described in Spang and Schekman (1998).
For the affinity chromatography, the columns were preequilibrated with 10 ml of NE buffer (25 mM HEPES, pH 7.7, 100 mM NaCl, 1 mM EDTA, 0.5 mM MgCl2, 0.1% Na-cholate). The nucleotide exchange was performed in 3 ml of NE buffer with 200 μM of the corresponding nucleotide for 1 h at 37°C (GTP for Arf1Q71Lp, GDP for Arf1T31Np). The control column without protein was treated in the same way as the Arf1Q71Lp-column. The columns were washed with 10 ml of NS buffer (20 mM HEPES, pH 7.4, 100 mM NaCl, 1 mM dithiothreitol, 1 mM EDTA, 2 mM MgCl2, 10 μM GXP). Yeast cytosol (125 mg) was allowed to bind for 1 h at 4°C. The columns were washed with 60 ml of NS buffer (+ 0.1% Triton X-100) and subsequently with 10 ml of prewarmed exchange buffer containing 10 μM GXP. Proteins were eluted using 4× 1 ml exchange buffer containing 500 μM GXP (GDP for Arf1Q71Lp, GTP for Arf1T31Np). For the first elution, the columns were incubated for 30 min at 37°C, whereas for the following elution steps 10 min was used. The samples were analyzed by immunoblot. The mass spectrometric analysis was performed as described by Sandmann et al. (2003).
Two-Hybrid Assay
The two-hybrid assay was performed with the LexA two-hybrid system (Golemis and Khazak, 1997). The yeast reporter strain EGY48 with the bait plasmid pEG202, the prey plasmid pJG4-5, and the reporter plasmid pSH18-34 were used. PAB1 and PUB1 were amplified from genomic yeast DNA by polymerase chain reaction (PCR) and cloned into pJG4–5. Constructs were verified by DNA sequencing.
Coimmunoprecipitation Experiments and Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
Yeast lysates were prepared by spheroplasting as described by Spang and Schekman (1998). Spheroplasts were sedimented and lysed in 20 mM HEPES, pH 6.8, 150 mM KAc, 5 mM Mg(Ac)2, 1% Tween 20. The lysates were cleared by centrifugation. Immunoprecitpitations were performed with 10 μl of α-Arf1p serum, 10 μl of α-COPI, or control serum per 1 ml of lysate for 1 h at 4°C. In other cases, 7 μl of α-HA.11 (Babco, Richmond, CA) or 5 μl of α-myc 9E10 (Sigma-Aldrich, St. Louis, MO) were added to the lysate.
For Bfr1p coimmunoprecipitation, the lysate was incubated with 10 μl of affinity-purified anti-Arf1p antibody cross-linked to 25 μl of protein A magnetic beads (New England Biolabs, Beverly, MA) by using dimethyl pimelimidate (Pierce Chemical, Rockford, IL). The antibody–protein interaction was severed by elution with 0.2 M glycine, pH 2.5.
For RNA digestion experiments, the lysates were treated for 35 min at 4°C with 200 μg of RNase A (Roche Diagnostics, Indianapolis, IN) or 500 U of RNase-free DNase I and centrifuged before precipitation.
For RT-PCR, the beads and the lysates were extracted as described by Gonsalvez et al. (2003). RT-PCR was performed using One-Step RT-PCR kit (QIAGEN, Hilden, Germany) for 25 or 30 cycles.
ASH1 mRNA Localization
For the visualization of ASH1 mRNA localization by green fluorescent protein (GFP) particles, the reporter system by Bertrand et al. (1998) was used.
In situ ASH1 mRNA hybridization with digoxigenin-labeled ASH1 antisense probe was performed as described previously (Takizawa et al., 1997). Where necessary, cells were shifted for 1 h to the nonpermissive temperature. At least 100 cells were counted for each condition, and a minimum of three independent experiments were performed per strain.
Northern Blot Analysis
Cells were grown overnight to logarithmic phase. When necessary, cells were shifted for 1 h to the nonpermissive temperature. Total RNA was extracted using the RNeasy kit (QIAGEN), and equal amounts of RNA were resolved on agarose gels containing formaldehyde. The RNA was transferred onto Hybond N+ (Amersham Biosciences) and subsequently hybridized to ASH1 and ADH1 probes, which were generated using an AlkPhos direct labeling kit (Amersham Biosciences). The probes were detected using the CDP-Star reagent (Amersham Biosciences) according to manufacturer's recommendations.
Golgi Budding Assay
The Golgi membranes were prepared and the Golgi budding assay was performed as described by Spang and Schekman (1998) with the following modifications. For the Golgi budding reactions, membranes were incubated with 0.1 mM GTP or guanosine 5′-O-(3-thio)triphosphate (GTP-γ-S), coatomer (125 μg/ml), and Arf1p (25 μg/ml) at 30°C for 30 min in a total volume of 400 μl. After chilling on ice, we loaded the samples on a Ficoll-sucrose gradient consisting of 0.3 ml of 60% (wt/vol) sucrose, 0.8 ml of 7.5%, 1 ml of 5, 4, and 3%, and 0.8 ml of 2% (wt/wt) Ficoll in 15% (wt/vol) sucrose in 20 mM HEPES, pH 6.8, 5 mM MgAc2, 150 mM KAc (B88*). The vesicles were separated from the Golgi apparatus by centrifugation for 2 h at 35,000 rpm (SW50.1 rotor; Beckman Instruments, Fullerton, CA). Fractions (400 μl) were collected from the top. Fractions 4–6 of the gradients were pooled, mixed with an equal volume of 80% Nycodenz in B88*, and overlaid with 600 μl of 35, 25, 20, and 15% and 400 μl of 10% Nycodenz in B88*. The gradients were centrifuged for 16 h at 40,000 rpm (SW50.1 rotor; Beckman Instruments). Fractions (300 μl) were collected from the top, trichloroacetic acid (TCA) precipitated, and analyzed by immunoblot.
RESULTS
Identification of Interactors of Arf1p
To identify new interacting proteins for Arf1p, we performed affinity chromatography with dominant active (ΔN17-Arf1Q71Lp) and dominant inactive (ΔN17-Arf1T31Np) forms of Arf1p (Kahn et al., 1995). We chose an N-terminal truncated form of Arf1p for our experiments because the N terminus contains a myristoylation site and forms an amphipathic helix. The replacement of this region of the proteins by a His6-tag should minimize nonspecific interactions on the affinity column, and it greatly facilitated the purification (Rein et al., 2002). Equal amounts of ΔN17-Arf1Q71Lp and ΔN17-Arf1T31Np were covalently coupled to NHS-agarose. Guanine nucleotide exchange reactions were performed on the immobilized proteins to ensure that Arf1Q71Lp and Arf1T31Np were bound to GTP and GDP, respectively. Cytosolic extracts from a wild-type yeast strain were passed over the columns. For the elution, the Arf1Q71Lp column was incubated with an excess of GDP and the Arf1T31Np column with an excess of GTP. Although the restricted mutants are predominantly in either the GTP- or the GDP-bound form, an excess of the other nucleotide was able to elicit a conformational change under conditions that favor spontaneous guanine nucleotide exchange. In this way, proteins were released that bound specifically to the active or inactive form of Arf1p (Figure 1A). An NHS-agarose column, which had been mock treated, served as a negative control.
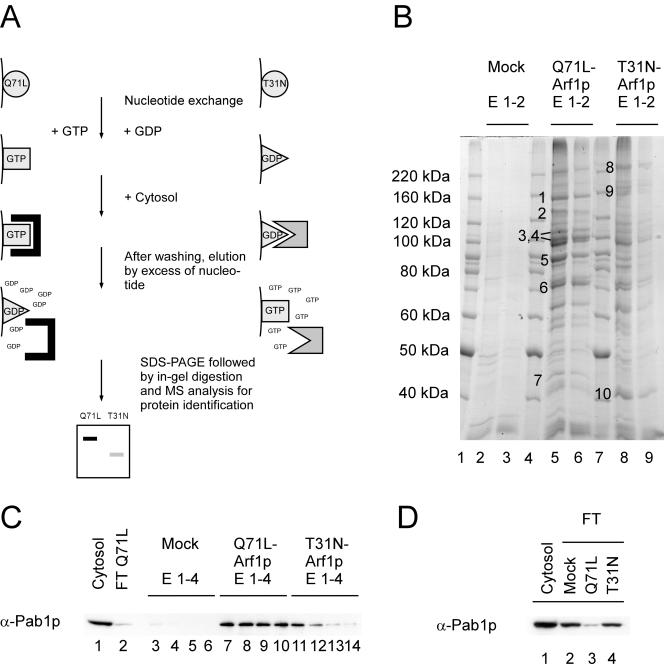
Figure 1.
Differential affinity chromatography. Yeast cytosol was incubated with either the Arf1-Q71Lp (Arf1p-GTP) or Arf1-T31Np (Arf1p-GDP) column. After washing, spontaneous nucleotide exchange was elicited resulting in a conformational change on Arf1p and the release of conformation-specific bound proteins. (A) Schematic drawing of the procedure. (B) The eluates from a differential affinity chromatography experiment were TCA precipitated and separated on an SDS gel. Bands that were enriched in eluates from either the Arf1p-GDP or Arf1p-GTP column were excised and subjected to mass spectrometric analysis. (C) Pab1p binds preferentially to ΔN17Arf1-Q71Lp. Fractions from the columns were blotted and incubated with antibodies against Pab1p. Pab1p is highly enriched in eluates from the Arf1p-Q71L column. (D) The flow-through from the Arf1-Q71Lp column is depleted of Pab1p. The starting cytosol as well as the flow-through from the three columns was analyzed for the presence of Pab1p by immunoblot. Pab1p was less abundant in the flow-through of the Arf1p-GTP column compared with the mock-treated and the Arf1p-GDP column.
We validated our approach with immunoblots of eluates from the different columns and antibodies directed against known interactors of Arf1p, e.g., coatomer, ARF-GEFs, and the ARF-GAP Glo3p. These proteins behaved as predicted by their biological function (our unpublished data). Coatomer is heptameric protein complex that forms together with Arf1p the COPI coat. This coat is involved in vesicular transport within the Golgi and from the Golgi to the ER.
Eluted proteins were precipitated with trichloracetic acid and then separated by SDS-PAGE and stained with Coomassie Blue (Figure 1B). Multiple proteins were eluted from both Arf1p columns. Whereas some proteins occurred in eluates from both Arf1p columns, a few proteins were at least predominantly present in fractions from only one of the columns. Bands present in the eluates from only one column were excised and analyzed by mass spectrometry. The results of the analysis are summarized in Table 2. Six of the analyzed bands corresponded to known Arf1p binding proteins: subunits of the coatomer complex, the ARF-GEFs Gea2p and Sec7p as well as the ARF-GAP Gcs1p. Similar results were obtained using columns with immobilized ΔN17-Arf1p bound to GTP or GDP. Surprisingly, Gcs1p was found in the eluate from the GDP column. However, ARF-GAPs have been shown to fulfill an additional function on recruitment of SNAREs and cargo to sites of vesicle emergence (Rein et al., 2002). This recruitment does not necessarily require the activated state of Arf1p.
TABLE 2.
Proteins identified by mass spectrometry
| Band | Protein | Description |
|---|---|---|
| 1 | Cop1p | Coatomer α-subunit |
| 3 | Sec26p | Coatomer β-subunit |
| 4 | Sec27p | Coatomer β′-subunit |
| 6 | Pab1p | Poly(A)-binding protein |
| 8 | Sec7p | ARF-GEF |
| 9 | Gea2p | ARF-GEF |
| 10 | Gcs1p | ARF-GAP |
Pab1p Binds to a ΔN17-Arf1Q71Lp Affinity Matrix
One of the new bands that we identified by mass spectrometry corresponded to Pab1p (Figures 1B, band 6, and Table 2). Pab1p is one of two major polyA-tail binding proteins in yeast. It is localized in the cytoplasm as well as in the nucleus, and it is part of the 3′ end RNA processing complex (cleavage factor I). Four RNA recognition domains mediate the interaction with the polyA-tail of the mRNA. To confirm the presence of Pab1p, we tested eluates from the affinity columns by immunoblot. Pab1p did not bind to the column material, but it was detected in fractions from the ΔN17-Arf1Q71Lp column (Figure 1C). A small amount of Pab1p also was present in eluates from the ΔN17-Arf1T31Np column. However, Pab1p showed a higher affinity toward the activated form of Arf1p, because the signal from the Arf1T31Np fractions trailed off quickly, whereas it persisted throughout the eluates from the Arf1Q71Lp matrix. This was consistent with the signal that we observed for coatomer. If the Pab1p–Arf1Q71Lp interaction represents a high-affinity contact, one would expect that the cytosol loaded on the column would be at least partially depleted of Pab1p. Therefore, the flow-throughs from different columns were analyzed by immunoblot. Although the signal from the control and the ΔN17-Arf1T31Np columns did not alter significantly, the cytosol that had passed through the ΔN17-Arf1Q71Lp matrix contained substantially less Pab1p than the starting material (Figure 1D).
Figure 6.
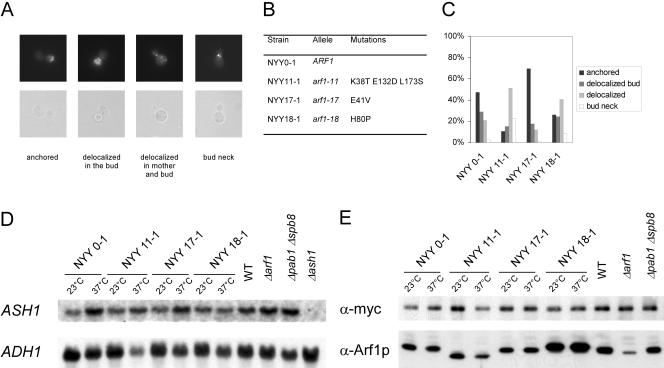
Arf1p and Pab1p are required for ASH1 mRNA localization to the bud tip. (A) Overview of scored phenotypes. (B) List of mutants used in the analysis in C. (C) The defect in mRNA localization is allele specific. Different mutants in ARF1 were grown to early log phase at 23°C and then shifted for 1 h to 37°C. ASH1 mRNA in the cells was visualized by FISH. At least 100 cells/strain were scored. (D) ASH1 mRNA levels are not sensitive to shift to the restrictive temperature. Different mutants in ARF1 were grown to early log phase at 23°C and then shifted for 1 h to 37°C; the deletion mutants in ARF1 and PAB1 SPB8 as well as a wild-type strain were grown at 30°C. Total RNA was extracted from the different strains and analyzed by Northern blot. The blot was incubated with probes for ASH1 mRNA and ADH1 mRNA. On temperature shift, no significant changes in mRNA levels were observed. (E) Ash1p levels are not altered in different mutants. The strains were transformed with a CEN plasmid encoding C-terminally myc9-tagged Ash1p and grown as described in D. Soluble extracts were prepared and analyzed by immunoblot for the presence of Ash1p-myc and Arf1p. The translation of ASH1 mRNA seemed to be unaffected by shift to restrictive temperature. The Arf-signal in the Δarf1 mutant corresponds to Arf2p, which is up-regulated in Δarf1 strains. Arf2p is 10 times less than abundant than Arf1p in wild-type strains.
ΔN17-Arf1Q71Lp Interacts with Pab1p but Not Pub1p
Pab1p is known to bind nonspecifically to many yeast proteins (Gavin et al., 2002). Therefore, to validate the interaction between Arf1p and Pab1p, we performed a two-hybrid assay. We used ΔN17-ARF1Q71L as the bait and PAB1, PUB1, and GLO3 as prey. Pub1p is the other major polyA binding protein in the cell, and like Pab1p, it localizes to the cytoplasm and the nucleus. We used growth on –LEU plates as the indicator for interactions. Whereas PAB1 and ΔN17-ARF1Q71L showed an interaction in the two-hybrid assay, no growth on –LEU plates could be detected in strains carrying PUB1 in conjunction with ΔN17-ARF1Q71L. GLO3 served as a positive and the empty vector as a negative control, respectively (Table 3). Eugster et al. (2000) have shown that GLO3 specifically interacts with the active form of ΔN17-ARF1 but neither with the wild-type nor the inactive mutant.
TABLE 3.
Two-hybrid assay
| Prey
|
||||
|---|---|---|---|---|
| Bait | PAB1 | PUB1 | GLO3 | pJG4-5 |
| ΔN17-ARF1-Q71L | + | - | + | - |
| pEG202 | - | n.d. | - | n.d. |
n.d., not done.
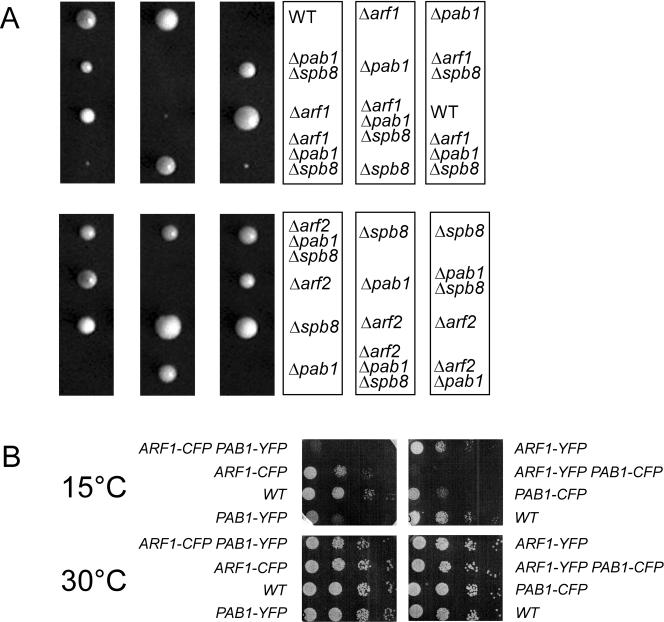
Pab1p is essential for cell viability. However, a suppressor mutant has been identified, Δspb8, that allows for survival in the absence of functional Pab1p (Boeck et al., 1998). ARF1 is not essential because its homolog ARF2, which is 96% identical, can at least partially substitute for ARF1. Yet Arf1p and Arf2p are not fully redundant (Spang et al., 2001). If Arf1p and Pab1p cooperate in an essential pathway, one would predict that a double mutant should show at least an enhanced phenotype compared with the single mutants. Thus, we crossed a Δarf1 strain with a Δpab1 Δspb8 strain (Figure 2). Although the Δpab1 Δspb8 mutant had a somewhat reduced growth rate compared with wild type, the Δarf1 Δpab1 Δspb8 spores were very sick and hardly grew at any temperature. In contrast, Δarf2 did not show the same phenotype with Δpab1Δspb8, most likely because Arf2p is much less abundant in the cell than Arf1p and Arf1p is still present in Δarf2 strains. In addition, we constructed a strain containing chromosomally tagged Arf1p-YFP and Pab1p-CFP, which was cold sensitive (Figure 2). The singly tagged strains do not show this phenotype. Therefore, the cold sensitivity provides additional evidence for the specific interaction between Arf1p and Pab1p in vivo. Together, these results are consistent with an interaction of Arf1p and Pab1p in vivo.
Figure 2.
ARF1 but not ARF2 interacts genetically with PAB1. (A) Δarf1 or Δarf2 haploid strains were crossed with Δpab1Δspb8 strains. The resulting diploid strains were sporulated and tetrads dissected. The spores were grown for 2 d at 30°C. (B) ARF1 and PAB1 were chromosomally appended with either yellow fluorescent protein (YFP) or cyan fluorescent protein (CFP). The resulting strains were grown to early log phase, and serial dilutions were spotted on YPD plates. The plates were incubated at the indicated temperatures.
Pab1p and Arf1p Form a Complex In Vivo
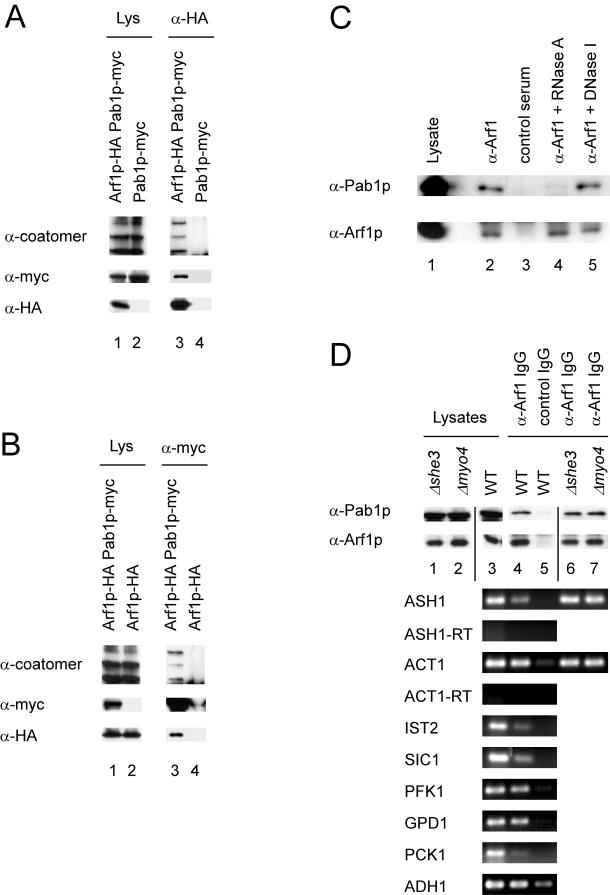
As a further test of binding specificity, we performed coimmunoprecipitation experiments. First, we appended chromosomally Arf1p and Pab1p with a hemagglutinin (HA)-tag and a myc-tag, respectively. The resulting strain, YAS238, did not show an altered phenotype compared with the isogenic wild-type (our unpublished data). Lysates of strain YAS238 were incubated with anti-HA or anti-myc antibodies and protein G-Sepharose. The immunoprecipitates were resolved by SDS-PAGE followed by immunoblot with antibodies against coatomer, and the HA- and the myc-tag (Figure 3, A and B). As expected, Pab1p-myc was coprecipitated with Arf1p-HA. This precipitation was dependent on the presence of Arf1p-HA. A similar result was obtained when the Pab1p-myc was precipitated: Arf1p-HA cosedimented only in the presence of Pab1p-myc (Figure 3B). Unexpectedly, coatomer also was precipitated with Pab1pmyc. Because coatomer also seemed to be part of an Arf1p–Pab1p complex, we repeated the immunoprecipitation with wild-type lysate and anti-coatomer antibodies. Indeed, Pab1p was found in the immunoprecipitate but not in the control (Figure 3D; our unpublished data). Moreover, Pab1p failed to coimmunoprecipitate when antibodies against Sar1p or Sec23p were used. Sec23p is the GTPase activating protein of Sar1p, and both are essential for the formation of COPII-coated vesicles from the ER. Thus, Pab1p does not associate nonspecifically with vesicle coat proteins. Together, these results provide further evidence for the existence of an in vivo Pab1p-Arf1p interaction and that coatomer is part of this complex.
Figure 3.
Arf1p and Pab1p are present in a ribonucleotide particle. (A and B) Pab1p and Arf1p coimmunoprecipitate. Pab1p and Arf1p were chromosomally appended with either a myc- or HA-tag. Yeast lysates were prepared from single- or double-tagged strains and subjected to immunoprecipitation with anti-myc or anti-HA antibodies. The precipitates were analyzed by immunoblot. Lanes 1 and 2 correspond to 1.7% of the lysate. (C) Pab1p–Arf1p interaction depends on mRNA. Yeast lysate from a wild-type strain was incubated with RNase A, DNase I, or mock treated. After the treatment an immunoprecipitation was performed with anti-Arf1p serum or a control serum and protein A-Sepharose. The precipitated proteins were detected by immunoblot. In lane 1, 1.7% of the lysate was loaded. (D) ASH1 mRNA is part of the Pab1p-Arf1p ribonucleotide particle even in the absence of the SHE machinery. A coimmunoprecipitation experiment was performed with affinity-purified anti-Arf1p antibodies. RNA was prepared from the precipitate and subjected to RT-PCR with primer specific for the indicated mRNAs. -RT indicates reactions in the absence of reverse transcriptase. In lanes 1 and 2, 1.7% of the lysate was loaded.
Pab1p Is Associated with Golgi-derived COPI Vesicles
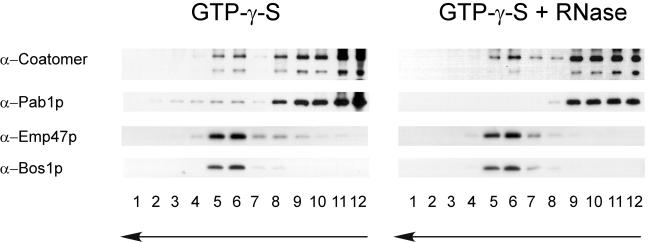
The results presented thus far support an in vivo interaction of coatomer, Arf1p, and Pab1p. Such an interaction could take place on the Golgi or on COPI vesicles. Pab1p is evenly distributed throughout the cytoplasm (Anderson et al., 1993), making it difficult to assess the place of interaction in vivo. Therefore, we used an in vitro Golgi budding assay, in which COPI-coated vesicles are formed from Golgi membranes (Spang and Schekman, 1998). Enriched Golgi membranes that were devoid of ER were incubated in the presence of coatomer, Arf1p, and guanine nucleotide. The generated COPI vesicles were separated from the Golgi membranes by velocity centrifugation. The vesicle peak was collected and refractionated based on coated membrane buoyant density. By using this purification scheme, vesicles are highly enriched, because contaminating particles would have to show the same behavior on a sedimentation gradient as well as sharing the same buoyant density, which is very unlikely. When GTP-γ-S was used to generate the COPI-coated vesicles, the vesicles peaked in fractions 5 and 6 as judged by the presence of the integral membrane cargo Emp47p and the v-SNARE Bos1p (Figure 4, GTP-γ-S). Pab1p peaked in the same fractions. A strong signal of Pab1p also was observed in the load. This was not surprising because we expected Pab1p to travel piggyback on the COPI vesicles. Thus, some protein if not most was lost during the flotation process. We did not detect any Sec61p, the translocon at the ER, or Pgk1p, a very abundant cytosolic protein in the vesicle fraction (our unpublished data). The peak changed in appearance when GTP was used instead of GTP-γ-S. In this case, the coat partially disassembled due to GTP hydrolysis, which also resulted in a partial loss of the Pab1p signal in the vesicle fraction. Pab1p was not recovered in the vesicle peak in the absence of coatomer and Arf1p (our unpublished data). Thus, Pab1p is associated with COPI vesicles generated from Golgi membranes, and this association is lost after the disassembly of the COPI coat.
Figure 4.
Pab1p association with COPI-coated vesicles is dependent on mRNA. COPI vesicles were generated from Golgi membranes in the presence of GTP-γ-S and COPI components. In one experiment, RNase (200 μg) was added to the budding reaction. The vesicles were purified over a velocity gradient and subsequently floated on a Nycodenz gradient. Fractions were collected from the top, separated by SDS-PAGE, and analyzed by immunoblot. The arrows indicate the direction of movement of lipid particles within the gradient. Nonmembrane-associated proteins remain in the load at the bottom of the gradient. The vesicles peak in fractions 5 and 6.
The Pab1p–Arf1p Complex Is Dependent on the Presence of mRNA
Pab1p is the major mRNA binding protein of the cell. Therefore, we wanted to test whether RNA is present in the Pab1p–Arf1p complex and whether RNA is required for the stability of the complex. To address these questions, we performed an immunoprecipitation with anti-Arf1p antibodies and digested the lysate with either RNase A or DNase I (Figure 3C). Pab1p was coimmunoprecipitated with anti-Arf1p antibodies, and it did not bind to nonrelated IgGs (Figure 3C). Interestingly, the interaction of Arf1p and Pab1p was severed upon addition of RNase A. DNA digestion did not have any influence on the stability of the Pab1p–Arf1p complex. Furthermore, the addition of an excess of purified RNA or DNA did not have any impact on the Pab1p–Arf1p interaction, indicating that binding is not mediated by a nonspecific association of protein and RNA. Thus, Arf1p seems to be part of a ribonucleotide particle. Next, we wondered whether Arf1p might be part of special 3′ end RNA processing complexes. Therefore, we performed RT-PCRs on anti-Arf1p immunoprecipitates (Figure 3D). We tested eight mRNAs that differ in their abundance and that are either symmetrically or asymmetrically localized. In S. cerevisiae, at least two asymmetrically localized mRNAs have been identified: ASH1 and IST2. They are transported into the bud along actin cables via the SHE machinery and anchored at the bud tip of a growing cell (Long et al., 1997; Munchow et al., 1999; Takizawa et al., 2000). The motor protein Myo4p forms a complex with the RNA binding protein She2p via She3p (Bohl et al., 2000; Long et al., 2000; Takizawa and Vale, 2000). This complex delivers the mRNA to the bud tip where it is anchored.
As shown before, Pab1p specifically precipitated with affinity-purified anti-Arf1p antibodies and not with control IgGs (Figure 3D). Using this precipitate as template for the RT-PCR for different mRNAs resulted in a positive signal for all tested mRNAs. These signals were specific because no product was detected in the lanes where a precipitate with control IgGs had been used at 25 cycles. Increasing the cycle number to 30 resulted in a signal also in the control for most of the mRNAs except for ASH1. This result indicates that the Arf1p-Pab1p particles contain a variety of different mRNAs.
Because we also detected the asymmetrically localized ASH1 and IST2 mRNAs in Arf1p-Pab1p ribonucleotide particles, we asked whether interfering with the SHE machinery also would affect the association of the asymmetrically localized ASH1 mRNA with the COPI vesicles. Therefore, we repeated the Arf1p immunoprecipitation in strains where either SHE1/MYO4 or SHE3 had been deleted. As observed in the wild type, Pab1p cosedimented with Arf1p (Figure 3D, compare lane 4 with lanes 6 and 7). Furthermore, the precipitated mRNA contained ASH1 mRNA. Therefore, the association of the asymmetrically localized mRNA ASH1 with the Arf1p–Pab1p complex is independent of its transport by the SHE machinery.
We have established that Pab1p is present on COPI vesicles. Furthermore, ASH1 mRNA was detected on the COPI vesicle fraction of a Golgi budding experiment (our unpublished data). Yet, does this interaction of Pab1p and Arf1p depend on the presence of mRNA? To address this question, we again performed a Golgi budding experiment (Figure 4). This time, however, we added RNase during COPI vesicle generation. Treatment of the Golgi membranes with RNase did not affect vesicle generation, but the association of Pab1p with the COPI vesicles was completely lost. Therefore, the attachment of Pab1p with Golgi-derived COPI vesicles requires the presence of mRNA.
Asymmetric mRNA Localization Is Disturbed in arf1 Mutants
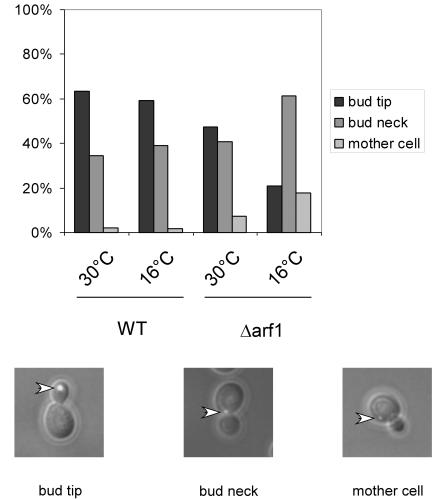
The Arf1p-Pab1p ribonucleotide particles contain asymmetrically and symmetrically localized mRNA. Thus, COPI vesicles are likely to contain a wide repertoire of mRNAs. These COPI vesicles are required for intra-Golgi as well as Golgi-to-ER retrograde transport. Because COPI vesicles are transport carrier, it seemed reasonable to look for a link to mRNA transport. One possible role for the mRNA association with the vesicles could be to bring the mRNA to the ER. The ER is a reticulate structure distributed throughout the cell, and we found it hard to study the transport of symmetrically localized mRNAs. Therefore, we investigated an asymmetrically distributed mRNA, ASH1, that also was found on COPI vesicles. First, we tested whether deletion of ARF1 interferes with the localization of ASH1 mRNA by using a GFP-based mRNA-localization assay (Bertrand et al., 1998). GFP is fused to sequences encoding the viral MS2 coat protein, whereas an ASH1 mRNA reporter construct contains MS2 binding sites. A nuclear localization signal on the GFP-containing plasmid restricts GFP to the nucleus. Only after interaction of the MS2 coat protein with MS2 binding sites can GFP exit the nucleus with ASH1 mRNA. We compared the ASH1 mRNA localization of wild-type yeast cells to that in a Δarf1 strain (Figure 5). We scored three different phenotypes: bud tip (correct localization), bud neck, and mother cell. In the wild-type strain, >60% of cells showed the correct localization of ASH1 mRNA in the bud tip at 30 and 16°C. In contrast, ASH1 mRNA seemed to be mislocalized in Δarf1 cells at the permissive temperature and even more severely at the restrictive temperature. Only in ∼20% of the Δarf1 cells incubated at the restrictive temperature was the GFP signal confined to the bud tip. The ASH1 mRNA never seemed to reach the bud tip in the remaining 80% of the cells. The GFP particle was either stuck in the bud neck or was randomly localized in the mother cell. This result indicates that Arf1p is required for the correct localization of ASH1 mRNA.
Figure 5.
ASH1 mRNA is mislocalized in Δarf1 mutant cells. An ASH1 mRNA-GFP reporter system was transformed in either wild-type or the isogenic Δarf1 strain. Cells were grown to early log phase at 30°C. One-half of the cultures were shifted to 16°C for 3 h. At least 100 cells per strain and temperature were analyzed for the localization of the GFP particle. The scored phenotypes are indicated. The arrowheads point toward the green fluorescent particle.
We extended our results by using temperature-sensitive arf1 alleles in a Δarf2 background generated by Yahara et al. (2001). In these strains, ARF function is only provided by the arf1 ts-allele. In addition, we switched our detection method to fluorescence in situ hybridization (FISH). The strains were grown overnight under permissive conditions, shifted for 1 h to the restrictive temperature, and then prepared for FISH analysis. Strain NYY 0-1 corresponds to the “wild type” in this experiment, because it contains the wild-type allele of ARF1 in a Δarf2 background, whereas NYY11-1, NYY17-1, and NYY18-1 represent different point mutants (Figure 6B). NYY11-1 carries three point mutations, one of which is in proximity to the switch 1 domain. The other two mutations are toward the C terminus, which is probably involved in protein–protein interaction. NYY11-1 has a strong defect in Golgi-to-ER transport. NYY17-1 (mutated in the switch 1 region) seems to be a weaker allele and is a member of a different intragenic complementation group. NYY18-1 is supposed to disturb transport at the Golgi and carries a mutation in the switch 2 region (Figure 6B). At the restrictive temperature, only ∼50% of the ARF1 wild type showed correct localization of the ASH1 mRNA (Figure 6C). However, the mutant NYY17-1 was not defective in mRNA localization and showed ∼70% correctly anchored ASH1 mRNA. In contrast, the mRNA localization in NYY11-1 and NYY18-1 was severely affected. The predominant defect was delocalized mRNA throughout the cell, indicating that these mutants were either deficient in transport or anchoring of the mRNA in the bud tip. These results indicate that the mRNA mislocalization phenotype displayed by the different mutants is due to a specific involvement of Arf1p in this process.
arf1 Mutants Show No Defects in Transcription or Translation of ASH1
Anchoring of ASH1 mRNA depends at least in part on Ash1 protein translation (Gonzalez et al., 1999). Therefore, the results presented above, although specific for Arf1p, also could be interpreted as a defect in transcription, mRNA instability, or down-regulation of translation of ASH1. In addition, some mutants of components along the secretory pathway led to a defect in ribosome biosynthesis (Mizuta and Warner, 1994). Moreover, Deloche et al. (2004) reported that translation initiation was attenuated very rapidly, and protein synthesis was reduced in secretion mutants upon shift to restrictive conditions. To test effects on ASH1 mRNA stability and Ash1p protein synthesis in arf1 and pab1 mutant strains before and after shift to the restrictive temperature, we performed Northern and Western blots (Figure 6, D and E). Yet, no significant variations in ASH1 mRNA levels were detected in arf1 ts-mutant strains upon shift to nonpermissive conditions or in strains where ARF1 or PAB1 had been deleted (Figure 6D). Detection of ADH1 mRNA served as internal control. A myc-tagged version of Ash1p is functional and has been used to assess the Ash1p content in cells (Cosma et al., 1999; McBride et al., 2001). Therefore, we used Ash1p-myc expressed from a CEN plasmid (single copy) to determine Ash1p levels in arf1 and pab1 mutants. Soluble yeast lysates were analyzed for the presence of Ash1p-myc and Arf1p (Figure 6E). As observed for the mRNA levels, no significant differences in the Ash1p levels were eminent. Thus, it is unlikely, that transcription or translation of ASH1 is disturbed in the arf1 mutants and that this would be the cause of the ASH1 mRNA localization defect.
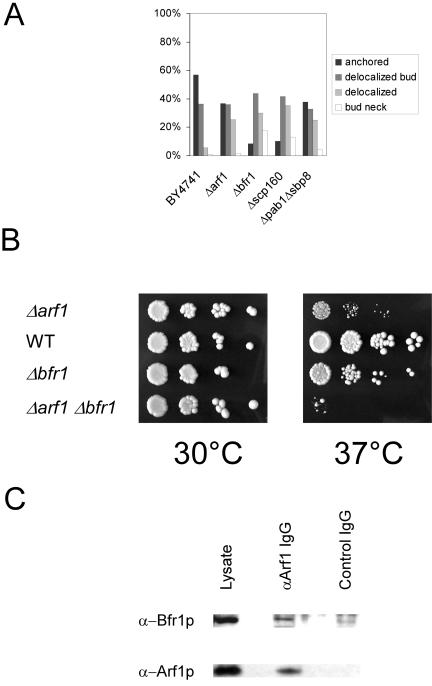
Δpab1Δsbp8, Δscp160, and Δbfr1 Are Defective in ASH1 mRNA Localization
Next, we wanted to check whether Pab1p also is required for asymmetric mRNA localization. Cells of a Δpab1Δsbp8 strain were grown at 30°C and prepared for FISH as described above. Indeed, a similar phenotype was observed as for a Δarf1 mutant (Figure 7A). In both cases, ∼60% of the ASH1 mRNA was mislocalized, whereas in the wild type 60% of the cells the signal was observed anchored in the bud tip. Pab1p is known to interact with two other proteins, Scp160p and Bfr1p (Lang and Fridovich-Keil, 2000). Moreover, Scp160p has been identified as a multicopy suppressor of a gea1-4 Δgea2 mutant and physically interacts with Gea1p (Peyroche and Jackson, 2001). Gea1p and Gea2p are ARFGEFs that have overlapping functions in retrograde transport from the Golgi to the ER (Peyroche et al., 1996; Spang et al., 2001). Scp160p is a polysome-associated protein that was shown to be defective in anchoring asymmetrically localized mRNA (Irie et al., 2002). Bfr1p is a protein of unknown function that was found in a screen for resistance against brefeldin A (Jackson and Kepes, 1994). Brefeldin A is a noncompetitive inhibitor of ARF-GEFs (Peyroche et al., 1999). Bfr1p is required for the association of Scp160p with polysomes but not with Pab1p (Lang et al., 2001). In addition, we found that Δbfr1 and Δarf1 are synthetically lethal at 37°C (Figure 7B). Furthermore, Bfr1p coimmunoprecipitates with Arf1p (Figure 7C). Therefore, at least a part of Arf1p function might be connected to Scp160p and Bfr1p, and it seemed likely that Δbfr1 also might be deficient in correct mRNA localization. Indeed, Δbfr1 showed the same phenotype as Δscp160, which was stronger than the one observed for Δarf1 and Δpab1 Δsbp8 (Figure 7A). This is consistent with the probable involvement of Scp160p and Bfr1p in the general translation machinery. In the absence of Arf1p, some mRNA could still get to the ER and interact there with Scp160p or Bfr1p. However, upon deletion of BFR1 or SCP160 the translation and thus restriction of mRNA at the ER could be more severely affected.
Figure 7.
Pab1p binding proteins display abnormal ASH1 mRNA localization. Cells were grown to early log phase at 30°C, and FISH for ASH1 mRNA was performed. BY4741 is the corresponding wild-type for the strains in A. At least 100 cells/strain were scored. (B) ARF1 and BFR1 interact genetically. Single and double mutants in ARF1 and BFR1 were grown at 30°C. Serial dilutions were dropped on plates and incubated at the indicated temperatures for 2 d. (C) Bfr1p interacts with Arf1p. An immunoprecipitation from yeast lysate was performed with affinity-purified antibodies against Arf1p or control IgGs. Bfr1p was coprecipitated with Arf1p.
Characterization of the Cytoskeleton in the arf1 Mutants
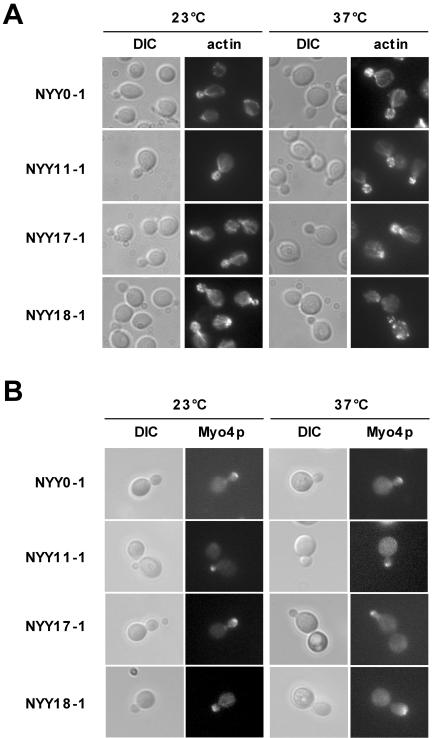
So far, we presented evidence that Arf1p is involved in mRNA transport and that the existence of an Arf1p-Pab1p-ASH1 ribonucleotide particle seemed to be independent of the SHE machinery. ASH1 mRNA was mislocalized in ARF mutants as determined by two different methods (Figures 5 and 6C). However, it is known that mutations in ARF also can affect the actin cytoskeleton and also might have some effect on microtubules. To rule out any general defects in cytoskeletal organization or polarity, we examined the microtubules and actin cytoskeleton in the NYY strains at the permissive and restrictive temperature. All strains showed wild-type microtubules at both temperatures as judged by indirect immunofluorescence (our unpublished data). In addition, none of the strains was sensitive toward benomyl, a microtubule depolymerizing drug. Next, we examined the actin localization by staining with rhodamine-phalloidin. NYY0-1, NYY11-1, and NYY17-1 did show correctly polarized actin cables and patches at the permissive and the restrictive temperature (Figure 8A). In contrast, whereas NYY18-1 behaved like wild type at 23°C, after the shift to the nonpermissive temperature the actin cables were missing and the actin patches were distributed randomly throughout the cell. Thus, the mRNA localization defect in NYY18-1 might be explained by the aberrant actin cytoskeleton. However, the polarity of the cell was not entirely lost, because bud site selection and budding still seemed to be normal as in the other strains tested (our unpublished data). The second arf1 mutant with an ASH1 mRNA localization defect, NYY11-1, behaved like wild type. Thus, the mRNA distribution phenotype in at least NYY11-1 is not caused by general defects in polarity and cytoskeletal organization.
Figure 8.
Localization of She1p/Myo4p and actin in arf1 point mutants. (A) The actin cytoskeleton is only disturbed in NYY18-1 at the restrictive temperature. Different mutants in ARF1 were grown to early log phase at 23°C and then shifted for 1 h to 37°C. Cells were fixed and the actin cytoskeleton was stained with rhodamine-phalloidin. (B) She1p/Myo4p localizes to the bud tip in arf1mutants. MYO4 was chromosomally appended with 2 × GFP. Different mutants in ARF1 were grown to early log phase at 23°C and then shifted for 1 h to 37°C. Cells were examined directly. For the restrictive temperature, a heated stage was mounted onto the microscope to keep the cells at 37°C during the observation.
She1p/Myo4p Localizes Correctly to the Bud Tip in arf1 Mutants
The actin cytoskeleton was visibly disturbed only in NYY18-1 at the restrictive temperature. Nonetheless, more subtle defects might conceivably prevent the SHE machinery from reaching the bud tip in the arf mutants. To test this possibility, we determined the localization of the motor protein She1p/Myo4p in the arf1 point mutants. The localization of She1p/Myo4p in the bud tip depends on the presence of She2p and She3p (Kruse et al., 2002). Thus, by checking She1p/Myo4p, we can examine the functionality of the entire SHE machinery. MYO4 was chromosomally appended by 2 × GFP in the different NYY strains as described by Kruse et al. (2002). At the permissive temperature as well as after a shift to the restrictive temperature for at least 1 h, Myo4p-GFP localized correctly to the bud tip in the arf1 point mutants irrespective to their defect in ASH1 mRNA localization (Figure 8B). Most importantly, Myo4p-GFP was localized correctly at the bud tip in NYY11-1 and NYY18-1 (Figure 8B). Our data indicate that the SHE machinery is functional in the arf1 mutants and that Arf1p is most likely involved in retaining ASH1 mRNA in the bud tip after the SHE proteins fulfilled their duty. These results demonstrate that the involvement of Arf1p in mRNA localization is independent of the SHE machinery. This is in agreement with our findings that the coatomer–Arf1p–Pab1p complex contains also symmetrically localized mRNAs that are not a substrate for the SHE machinery and that the deletion of members of the SHE machinery did not sever the ASH1 mRNA interaction with Pab1p and Arf1p.
Components of the Early Secretory Pathway Are Required for ASH1 mRNA Localization
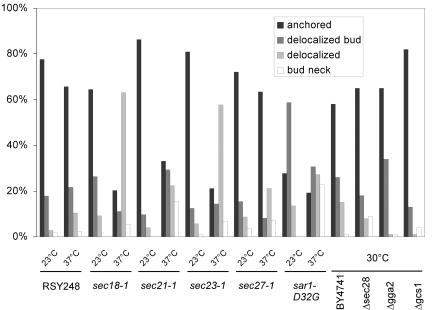
We found that mRNA is associated with COPI-coated vesicles that are destined to the ER. Therefore, we wondered whether vesicular traffic, especially through COPI transport carriers, might play a role in mRNA localization. To this end, we performed FISH with temperature-sensitive mutants in components of the early secretory pathway. Surprisingly, not only sec21-1, a mutant in the γ-subunit of coatomer was defective but also sec23-1 and sar1-D32G (Figure 9). Sar1p is the counterpart of Arf1p in the generation of COPII vesicles, and Sec23p is the GTPase activating protein for Sar1p (Barlowe, 2000). Furthermore, the N-ethylmaleimide-sensitive factor Sec18p, which is involved in homotypic and heterotypic membrane fusion (Novick et al., 1981; Graham and Emr, 1991), also seemed to be required for the anchoring of ASH1 mRNA because in ∼80% of the cells the FISH signal was mostly distributed all over the cell and could not be found concentrated in the bud tip. In contrast, two other mutants in the β-subunit (sec27) and ε-subunit (sec28) of coatomer behaved like wild type, indicating that the mRNA localization is a specific event involving coatomer. The phenotype that we detected was not due to a general secretion defect, because mutants in two Arf1p interacting proteins, Gga2p and Gcs1p, were not defective in ASH1 mRNA localization. Our results indicate that restricting ASH1 mRNA to the bud tip requires functional ER-Golgi transport.
Figure 9.
A functional early secretory pathway is necessary for ASH1 mRNA localization. Strains were grown to early log phase at 30°C or 23°C for ts-strains. In case of the ts-strains, one-half of the cultures were shifted to 37°C for 1 h. RSY248 corresponds to the wild-type strain for the tsmutants and is isogenic to sec18-1. BY4741 is the corresponding wild type for the deletion strains. At least 100 cells were scored per strain and condition.
The Actin Cytoskeleton Is Not Generally Disturbed in ERGolgi Transport Mutants
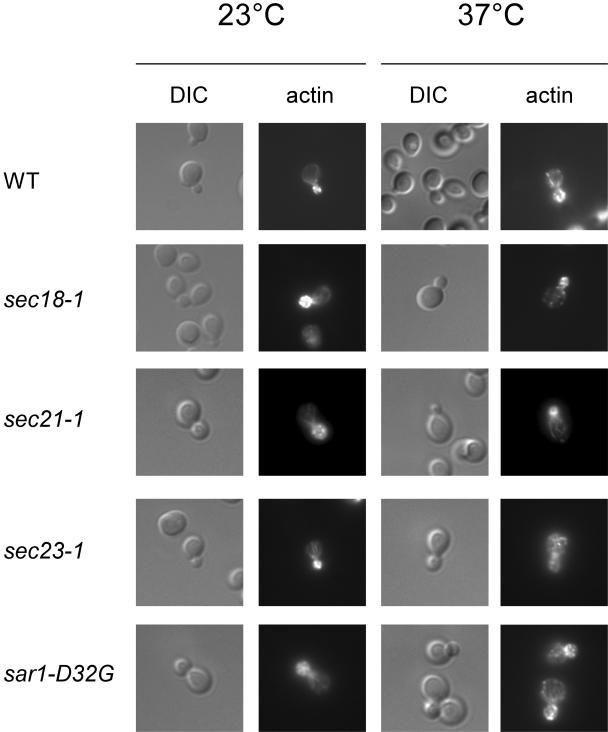
We repeated the actin-phalloidin staining with the temperature-sensitive secretion mutants that were defective in ASH1 mRNA localization (Figure 10). The mutant strains sec18-1, sec21-1, and sar1-D32G displayed a correctly polarized actin cytoskeleton at the permissive and at the restrictive temperature. In contrast, sec23-1 cells had a disorganized actin cytoskeleton after 1-h shift to the restrictive temperature. Thus, the defect in mRNA localization in these mutants (except for sec23-1) is not due to a lack of actin organization in the cell.
Figure 10.
The actin cytoskeleton is not disturbed as a result of a defect in secretion. Secretion mutants that displayed an ASH1 mRNA localization defect were examined for a functional actin cytoskeleton. The strains were grown to early log-phase at 23°C and then shifted for 1 h to 37°C. Cells were fixed and actin was stained with rhodamine-phalloidin.
Together, our results indicate a novel and unexpected role for components of the early secretory pathway in short-range mRNA localization most likely through interaction with Pab1p. This process is independent of the SHE machinery. We have only investigated the localization of the asymmetrically localized mRNA ASH1. However, these results also might reflect a mechanism by which symmetrically localized mRNA is concentrated at the ER.
DISCUSSION
We have used a differential affinity chromatography approach to identify new interactors of the small GTPase Arf1p. This approach is valid because known regulators were reidentified. We have identified Pab1p as a new interactor of the small GTPase Arf1p. Pab1p interacts predominantly with the GTP-bound form of Arf1p. In a high-throughput protein complex analysis in yeast, Pab1p was found to be one of the major contaminants (Gavin et al., 2002), indicating that Pab1p might interact nonspecifically with many other proteins. Therefore, we confirmed the specificity of interaction by two-hybrid analysis and genetic interaction as well as by mutual coimmunoprecipitation. Furthermore, the interaction between Pab1p and Arf1p is dependent on the presence of mRNA in the complex. Moreover, we found that Pab1p is associated peripherally with COPI vesicles generated from enriched Golgi membranes. Together, these results argue strongly that the Arf1p–Pab1p complex is specific. If and how Pab1p is incorporated into the vesicle coat remains elusive at present. Pab1p might not become an intrinsic part of the coat but might be peripherally associated with the vesicles through interactions with Arf1p and coatomer. COPI vesicles still form upon RNase treatment, which prohibits the Pab1p interaction with the coat.
We tested Arf1p immunoprecipitates for the presence of a number of different mRNAs that varied in their abundance and in their subcellular localization, by RT-PCR. After the immunoprecipitation, essentially all mRNAs were amplified. Thus, no specialized 3′ end processing complexes were associated with COPI vesicles. To further investigate the role of Arf1p, Pab1p, and the COPI coat in mRNA transport, we concentrated our efforts in studying the asymmetrically distributed ASH1 mRNA. In wild-type cells, ASH1 mRNA is asymmetrically localized to the bud tip of a growing yeast cell. However, when we investigated the role of mutants in ARF1 and PAB1 in asymmetric mRNA distribution, we found that they mislocalize ASH1 mRNA. Furthermore, by using enriched COPI vesicles from the Golgi budding assay, we could detect ASH1 mRNA by RT-PCR.
The fact that the ASH1 mRNA mislocalization phenotype in arf1 mutants was allele specific indicates that not the whole repertoire of ARF-dependent traffic is needed for mRNA localization. Moreover, this finding provides strong evidence that the ASH1 mRNA localization defect is not brought about by a secondary effect. This idea is corroborated by the data showing that some coatomer mutants did show a defect, whereas others did not. Furthermore, the extent of ASH1 mRNA mislocalization was variable in the different mutants of the early secretory pathway. However, other mutants of the early secretory pathway localized ASH1 mRNA correctly to the bud tip. Thus, the asymmetric mRNA distribution phenotype cannot be attributed to a general secretion defect. In addition to mutants in the early secretory pathway, we tested also mutants in late-acting components. They either had no effects (Δgga2) or the ASH1 mRNA mislocalization phenotype could be related to a defect in the actin cytoskeleton (sec1-1 and sec6-4; Trautwein and Spang, unpublished data). Moreover, the ASH1 mRNA localization defect was not due to defects in the actin cytoskeleton, the microtubules' organization, or cell polarity. Finally, we ruled out the possibility that ASH1 mRNA stability or translation efficiency was compromised in arf1 mutants. Ribosome biosynthesis and translation efficiency are affected in secretion mutants (Mizuta and Warner, 1994; Deloche et al., 2004). However, we did not observe this effect in arf1 and Δpab1Δspb8 mutants, which is most likely due to the use of different mutants and different strain backgrounds: the pool of mutants tested in the various studies is nonoverlapping. Yet, we cannot exclude that there are subtle effects on translation or on the cytoskeleton that we were not able to detect by our methods of choice. Nevertheless, we think it unlikely that the mRNA localization defect results entirely from such secondary effects.
Transport of asymmetrically distributed mRNA into the bud tip was shown to be dependent upon the SHE proteins: She1p/Myo4p is an unconventional myosin that transports She2p, an RNA binding protein. The interaction between these two proteins is mediated by She3p. The role of She4p and She5p/Bni1p in mRNA localization is less well understood, but they seem to be involved in stabilization and polarization of the actin cytoskeleton. They are thought to play a role in anchoring mRNA by a thus far unknown mechanism (Beach and Bloom, 2001). However, Bloom and Beach (1999) speculated that once mRNA reaches the vicinity of its final destination, cytoplasmic flow or passive diffusion may enable mRNA accumulation at that site (the bud tip in yeast). Thus, a mechanism for restricting ASH1 mRNA in the bud is required, which might be independent of the SHE machinery. It further indicates the transport by the SHE machinery is a prerequisite but not sufficient for anchoring ASH1 mRNA at the bud tip. This is in perfect agreement with our finding that the aberrant ASH1 mRNA localization observed could not be correlated with defects in the SHE machinery, because Myo4p-GFP was restricted to the bud tip in arf1 mutants. Furthermore, deletion of SHE1/MYO4 or SHE3 did not disrupt the Arf1p-Pab1p ribonucleotide particle. Therefore, the defects in ASH1 mRNA localization in the early secretion mutants are independent and most likely downstream of the SHE machinery. Our data are suggestive of COPI vesicles acting as short-range mRNA transport and localization vehicles: The asymmetric distribution of the ASH1 mRNA is brought about by the SHE machinery and does not depend on Arf1p and COPI vesicles; they may act only to restrict the mRNA in the bud tip when it is not efficiently anchored. The long-range mRNA transport is dependent on the SHE machinery, whereas the short-range localization might be reliant on COPI vesicles.
We concentrated our efforts on the study of the asymmetrically distributed ASH1 mRNA, because the pathway of ASH1 mRNA localization is well known and more amenable for experiments than symmetrically distributed mRNA. However, because the defects in mRNA localization are independent of the SHE machinery, our observations also could apply to the symmetrically distributed mRNAs. What might be the role then of the early secretory pathway in mRNA localization? We would like to suggest a model in which the mRNA is transported to the ER via the COPI machinery. This mRNA COPI interaction could take place at the Golgi. Pab1p was detected on purified Golgi membranes that we used to generate COPI vesicles in vitro. These vesicles contained proteins (e.g., the ER-Golgi v-SNARE Bos1p) that cycle between the Golgi and the ER. Thus, we believe that these COPI vesicles are destined to the ER. Alternatively, the COPI vesicles might be directly loaded on route. This would lead to a fast and efficient way to bring mRNA to the ER. If the mRNA cannot be anchored at the ER and diffuses away, the COPI vesicles destined to the ER might be an efficient transport system for relocation of the mRNA. Thus, COPI vesicles could act as molecular sieve to bring Pab1p containing ribonucleotide particles to the ER. Yet, one other possible explanation for the transport of mRNA on COPI vesicles might be the clearance of the ribosome-free Golgi region from mRNA. Although this might be a likely scenario for mammalian cells, the Golgi in S. cerevisiae is scattered throughout the cell, and the single different cisternae are surrounded by ribosomes in the cytoplasm. There is no ribosome-free Golgi region in S. cerevisiae. Still, we cannot exclude the possibility that there might be an ancient mechanism for mRNA clearance. Alternatively, symmetrically and asymmetrically mRNA transport to the ER might result in an enhancement of membrane-bound ribosome turnover at the ER. At least in mammalian cells, it has been shown that membrane-bound ribosomes do not distinguish between mRNA substrates and therefore can initiate the translation of any protein, regardless of whether it is cytosolic or destined for translocation/secretion (Potter and Nicchitta, 2000). However, when membrane-bound ribosomes were provided with mRNAs encoding model cytosolic proteins, subsequent translation yielded the release of the ribosomenascent chain complex from the ER to the cytosol (Potter and Nicchitta, 2000; Potter et al., 2001). Furthermore, mRNAs encoding cytosolic proteins are well represented in the ER membrane-bound polysome fraction (Diehn et al., 2000; Lerner et al., 2003). Nicchitta (2002) proposed a model suggesting that the exchange of ribosomes on the ER membrane is dependent on and driven by the translation of cytosolic proteins by membrane-bound ribosomes. Our data are in agreement with this hypothesis because COPI vesicles could act as carriers for mRNAs to the ER to allow for the ribosome exchange.
The mutant in the COPII component Sar1p could display such a strong defect for two reasons, which might lead to an additive effect: Because COPII vesicles are no longer formed in these mutants, the lipid and protein composition of the ER membrane is changed. As a result, the mRNA complex might “adhere” less efficiently at the ER membrane. The mRNA complex diffuses away and cannot be brought back because blocking anterograde COPII transport from the ER to the Golgi immediately abrogates retrograde COPI transport from the Golgi to the ER. Hence, one would observe a stronger phenotype than in the retrograde transport mutants. Although we cannot exclude a direct involvement of COPII components in mRNA localization, Pab1p did not interact with Sar1p or Sec23p. Therefore, we favor an indirect role of the COPII components in mRNA transport.
Together, our results provide the first evidence for a link between COPI vesicles and mRNA. Our data are suggestive of a role of Pab1p and COPI vesicles in concentrating mRNA at the ER. However, given the multiple roles of Arf1p in the cell and the interdependence of vesicular transport and metabolism, other interpretations for the in vivo role of RNA-loaded vesicles also are conceivable.
Supplementary Material
Acknowledgments
We are grateful to P.P. Poon, M. Swanson, R. Duden, A. Sachs, A. Nakano, R. Singer, K. Bloom, R.P. Jansen, and R. Schekman for strains and reagents. We thank S. Wahl for excellent technical assistance and A. Nordheim for allowing us to use the mass spectrometer. I.G. Macara and E. Hartmann are acknowledged for critical reading of the manuscript. We thank members of the Spang laboratory for stimulating discussions. This work was supported by the Max Planck Society. A.S. is an European Molecular Biology Organization Young Investigator.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–05–0411. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–05–0411.
The online version of this article contains supplementary material accessible through http://www.molbiolcell.org.
References
- Anderson, J.T., Paddy, M.R., and Swanson, M.S. (1993). PUB1 is a major nuclear and cytoplasmic polyadenylated RNA-binding protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 13, 6102-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe, C. (2000). Traffic COPs of the early secretory pathway. Traffic 1, 371-377. [DOI] [PubMed] [Google Scholar]
- Beach, D.L., and Bloom, K. (2001). ASH1 mRNA localization in three acts. Mol. Biol. Cell 12, 2567-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand, E., Chartrand, P., Schaefer, M., Shenoy, S.M., Singer, R.H., and Long, R.M. (1998). Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2, 437-445. [DOI] [PubMed] [Google Scholar]
- Blader, I.J., Cope, M.J., Jackson, T.R., Profit, A.A., Greenwood, A.F., Drubin, D.G., Prestwich, G.D., and Theibert, A.B. (1999). GCS1, an Arf guanosine triphosphatase-activating protein in Saccharomyces cerevisiae, is required for normal actin cytoskeletal organization in vivo and stimulates actin polymerization in vitro. Mol. Biol. Cell 10, 581-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom, K., and Beach, D.L. (1999). mRNA localization: motile RNA, asymmetric anchors. Curr. Opin. Microbiol. 2, 604-609. [DOI] [PubMed] [Google Scholar]
- Boeck, R., Lapeyre, B., Brown, C.E., and Sachs, A.B. (1998). Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant. Mol. Cell. Biol. 18, 5062-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohl, F., Kruse, C., Frank, A., Ferring, D., and Jansen, R.P. (2000). She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p. EMBO J. 19, 5514-5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma, M.P., Tanaka, T., and Nasmyth, K. (1999). Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97, 299-311. [DOI] [PubMed] [Google Scholar]
- Deloche, O., de la Cruz, J., Kressler, D., Doere, M., and Linder, P. (2004). A membrane transport defect leads to a rapid attenuation of translation initiation in Saccharomyces cerevisiae. Mol. Cell 13, 357-366. [DOI] [PubMed] [Google Scholar]
- Diehn, M., Eisen, M.B., Botstein, D., and Brown, P.O. (2000). Large-scale identification of secreted and membrane-associated gene products using DNA microarrays. Nat. Genet. 25, 58-62. [DOI] [PubMed] [Google Scholar]
- Eugster, A., Frigerio, G., Dale, M., and Duden, R. (2000). COP I domains required for coatomer integrity, and novel interactions with ARF and ARFGAP. EMBO J. 19, 3905-3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzusoff, A., Lauze, E., and Howell, K.E. (1992). Immuno-isolation of Sec7p-coated transport vesicles from the yeast secretory pathway. Nature 355, 173-175. [DOI] [PubMed] [Google Scholar]
- Fucini, R.V., Navarrete, A., Vadakkan, C., Lacomis, L., Erdjument-Bromage, H., Tempst, P., and Stamnes, M. (2000). Activated ADP-ribosylation factor assembles distinct pools of actin on Golgi membranes. J. Biol. Chem. 275, 18824-18829. [DOI] [PubMed] [Google Scholar]
- Gavin, A.C., et al. (2002). Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141-147. [DOI] [PubMed] [Google Scholar]
- Givan, S.A., and Sprague, G.F., Jr. (1997). The ankyrin repeat-containing protein Akr1p is required for the endocytosis of yeast pheromone receptors. Mol. Biol. Cell 8, 1317-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golemis, E.A., and Khazak, V. (1997). Alternative yeast two-hybrid systems. The interaction trap and interaction mating. Methods Mol. Biol. 63, 197-218. [DOI] [PubMed] [Google Scholar]
- Gonsalvez, G.B., Lehmann, K.A., Ho, D.K., Stanitsa, E.S., Williamson, J.R., and Long, R.M. (2003). RNA-protein interactions promote asymmetric sorting of the ASH1 mRNA ribonucleoprotein complex. RNA 9, 1383-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, I., Buonomo, S.B., Nasmyth, K., and von Ahsen, U. (1999). ASH1 mRNA localization in yeast involves multiple secondary structural elements and Ash1 protein translation. Curr. Biol. 9, 337-340. [DOI] [PubMed] [Google Scholar]
- Graham, T.R., and Emr, S.D. (1991). Compartmental organization of Golgi-specific protein modification and vacuolar protein sorting events defined in a yeast sec18 (NSF) mutant. J. Cell Biol. 114, 207-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie, K., Tadauchi, T., Takizawa, P.A., Vale, R.D., Matsumoto, K., and Herskowitz, I. (2002). The Khd1 protein, which has three KH RNA-binding motifs, is required for proper localization of ASH1 mRNA in yeast. EMBO J. 21, 1158-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, C.L., and Kepes, F. (1994). BFR1, a multicopy suppressor of brefeldin A-induced lethality, is implicated in secretion and nuclear segregation in Saccharomyces cerevisiae. Genetics 137, 423-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, S., Jedd, G., Kahn, R.A., Franzusoff, A., Bartolini, F., and Segev, N. (1999). Genetic interactions in yeast between Ypt GTPases and Arf guanine nucleotide exchangers. Genetics 152, 1543-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn, R.A., Clark, J., Rulka, C., Stearns, T., Zhang, C.J., Randazzo, P.A., Terui, T., and Cavenagh, M. (1995). Mutational analysis of Saccharomyces cerevisiae ARF1. J. Biol. Chem. 270, 143-150. [DOI] [PubMed] [Google Scholar]
- Kirchhausen, T. (2000). Three ways to make a vesicle. Nat. Rev. Mol. Cell. Biol. 1, 187-198. [DOI] [PubMed] [Google Scholar]
- Knop, M., Siegers, K., Pereira, G., Zachariae, W., Winsor, B., Nasmyth, K., and Schiebel, E. (1999). Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15, 963-972. [DOI] [PubMed] [Google Scholar]
- Kruse, C., Jaedicke, A., Beaudouin, J., Bohl, F., Ferring, D., Guttler, T., Ellenberg, J., and Jansen, R.P. (2002). Ribonucleoprotein-dependent localization of the yeast class V myosin Myo4p. J. Cell Biol. 159, 971-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, B.D., and Fridovich-Keil, J.L. (2000). Scp160p, a multiple KH-domain protein, is a component of mRNP complexes in yeast. Nucleic Acids Res. 28, 1576-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, B.D., Li, A., Black-Brewster, H.D., and Fridovich-Keil, J.L. (2001). The brefeldin A resistance protein Bfr1p is a component of polyribosome-associated mRNP complexes in yeast. Nucleic Acids Res. 29, 2567-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, F.J., Stevens, L.A., Kao, Y.L., Moss, J., and Vaughan, M. (1994). Characterization of a glucose-repressible ADP-ribosylation factor 3 (ARF3) from Saccharomyces cerevisiae. J. Biol. Chem. 269, 20931-20937. [PubMed] [Google Scholar]
- Lerner, R.S., Seiser, R.M., Zheng, T., Lager, P.J., Reedy, M.C., Keene, J.D., and Nicchitta, C.V. (2003). Partitioning and translation of mRNAs encoding soluble proteins on membrane-bound ribosomes. RNA 9, 1123-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, S.M., Poon, P.P., Singer, R.A., Johnston, G.C., and Spang, A. (2004). The ArfGAP Glo3 is required for the generation of COPI vesicles. Mol. Biol. Cell 14, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, R.M., Gu, W., Lorimer, E., Singer, R.H., and Chartrand, P. (2000). She2p is a novel RNA-binding protein that recruits the Myo4p-She3p complex to ASH1 mRNA. EMBO J. 19, 6592-6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, R.M., Singer, R.H., Meng, X., Gonzalez, I., Nasmyth, K., and Jansen, R.P. (1997). Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science 277, 383-387. [DOI] [PubMed] [Google Scholar]
- McBride, H.J., Sil, A., Measday, V., Yu, Y., Moffat, J., Maxon, M.E., Herskowitz, I., Andrews, B., and Stillman, D.J. (2001). The protein kinase Pho85 is required for asymmetric accumulation of the Ash1 protein in Saccharomyces cerevisiae. Mol. Microbiol. 42, 345-353. [DOI] [PubMed] [Google Scholar]
- Mizuta, K., and Warner, J.R. (1994). Continued functioning of the secretory pathway is essential for ribosome synthesis. Mol. Cell. Biol. 14, 2493-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munchow, S., Sauter, C., and Jansen, R.P. (1999). Association of the class V myosin Myo4p with a localised messenger RNA in budding yeast depends on She proteins. J. Cell Sci. 112, 1511-1518. [DOI] [PubMed] [Google Scholar]
- Nicchitta, C.V. (2002). A platform for compartmentalized protein synthesis: protein translation and translocation in the ER. Curr. Opin. Cell Biol. 14, 412-416. [DOI] [PubMed] [Google Scholar]
- Novick, P., Ferro, S., and Schekman, R. (1981). Order of events in the yeast secretory pathway. Cell 25, 461-469. [DOI] [PubMed] [Google Scholar]
- Peyroche, A., Antonny, B., Robineau, S., Acker, J., Cherfils, J., and Jackson, C.L. (1999). Brefeldin A acts to stabilize an abortive ARF-GDP-Sec7 domain protein complex: involvement of specific residues of the Sec7 domain. Mol. Cell 3, 275-285. [DOI] [PubMed] [Google Scholar]
- Peyroche, A., and Jackson, C.L. (2001). Functional analysis of ADP-ribosylation factor (ARF) guanine nucleotide exchange factors Gea1p and Gea2p in yeast. Methods Enzymol. 329, 290-300. [DOI] [PubMed] [Google Scholar]
- Peyroche, A., Paris, S., and Jackson, C.L. (1996). Nucleotide exchange on ARF mediated by yeast Gea1 protein. Nature 384, 479-481. [DOI] [PubMed] [Google Scholar]
- Poon, P.P., Cassel, D., Spang, A., Rotman, M., Pick, E., Singer, R.A., and Johnston, G.C. (1999). Retrograde transport from the yeast Golgi is mediated by two ARF GAP proteins with overlapping function. EMBO J. 18, 555-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon, P.P., Nothwehr, S.F., Singer, R.A., and Johnston, G.C. (2001). The Gcs1 and Age2 ArfGAP proteins provide overlapping essential function for transport from the yeast trans-Golgi network. J. Cell Biol. 155, 1239-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter, M.D., and Nicchitta, C.V. (2000). Regulation of ribosome detachment from the mammalian endoplasmic reticulum membrane. J. Biol. Chem. 275, 33828-33835. [DOI] [PubMed] [Google Scholar]
- Potter, M.D., Seiser, R.M., and Nicchitta, C.V. (2001). Ribosome exchange revisited: a mechanism for translation-coupled ribosome detachment from the ER membrane. Trends Cell Biol. 11, 112-115. [DOI] [PubMed] [Google Scholar]
- Rein, U., Andag, U., Duden, R., Schmitt, H.D., and Spang, A. (2002). ARFGAP-mediated interaction between the ER-Golgi v-SNAREs and the COPI coat. J. Cell Biol. 157, 395-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmann, T., Herrmann, J.M., Dengjel, J., Schwarz, H., and Spang, A. (2003). Suppression of coatomer mutants by a new protein family with COPI and COPII binding motifs in Saccharomyces cerevisiae. Mol. Biol. Cell 14, 3097-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard, K.A., Gerber, A.P., Jambhekar, A., Takizawa, P.A., Brown, P.O., Herschlag, D., DeRisi, J.L., and Vale, R.D. (2003). Widespread cytoplasmic mRNA transport in yeast: identification of 22 bud-localized transcripts using DNA microarray analysis. Proc. Natl. Acad. Sci. USA 100, 11429-11434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F. (1991). Getting started with yeast. Methods Enzymol. 194, 3-21. [DOI] [PubMed] [Google Scholar]
- Skippen, A., Jones, D.H., Morgan, C.P., Li, M., and Cockcroft, S. (2002). Mechanism of ADP ribosylation factor-stimulated phosphatidylinositol 4,5-bisphosphate synthesis in HL60 cells. J. Biol. Chem. 277, 5823-5831. [DOI] [PubMed] [Google Scholar]
- Spang, A., Herrmann, J.M., Hamamoto, S., and Schekman, R. (2001). The ADP ribosylation factor-nucleotide exchange factors Gea1p and Gea2p have overlapping, but not redundant functions in retrograde transport from the Golgi to the endoplasmic reticulum. Mol. Biol. Cell 12, 1035-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang, A., and Schekman, R. (1998). Reconstitution of retrograde transport from the Golgi to the ER in vitro. J. Cell Biol. 143, 589-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns, T., Willingham, M.C., Botstein, D., and Kahn, R.A. (1990). ADP-ribosylation factor is functionally and physically associated with the Golgi complex. Proc. Natl. Acad. Sci. USA 87, 1238-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa, P.A., DeRisi, J.L., Wilhelm, J.E., and Vale, R.D. (2000). Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science 290, 341-344. [DOI] [PubMed] [Google Scholar]
- Takizawa, P.A., Sil, A., Swedlow, J.R., Herskowitz, I., and Vale, R.D. (1997). Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature 389, 90-93. [DOI] [PubMed] [Google Scholar]
- Takizawa, P.A., and Vale, R.D. (2000). The myosin motor, Myo4p, binds Ash1 mRNA via the adapter protein, She3p. Proc. Natl. Acad. Sci. USA 97, 5273-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahara, N., Ueda, T., Sato, K., and Nakano, A. (2001). Multiple roles of Arf1 GTPase in the yeast exocytic and endocytic pathways. Mol. Biol. Cell 12, 221-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.