Abstract
Attack by the host powdery mildew Erysiphe cichoracearum usually results in successful penetration and rapid proliferation of the fungus on Arabidopsis. By contrast, the nonhost barley powdery mildew Blumeria graminis f. sp. hordei (Bgh) typically fails to penetrate Arabidopsis epidermal cells. In both instances the plant secretes cell wall appositions or papillae beneath the penetration peg of the fungus. Genetic screens for mutations that result in increased penetration of Bgh on Arabidopsis have recently identified the PEN1 syntaxin. Here we examine the role of PEN1 and of its closest homologue, SYP122, identified as a syntaxin whose expression is responsive to infection. pen1 syp122 double mutants are both dwarfed and necrotic, suggesting that the two syntaxins have overlapping functions. Although syp122-1 and the cell wall mur mutants have considerably more pronounced primary cell wall defects than pen1 mutants, these have relatively subtle or no effects on penetration resistance. Upon fungal attack, PEN1 appears to be actively recruited to papillae, and there is a 2-h delay in papillae formation in the pen1-1 mutant. We conclude that SYP122 may have a general function in secretion, including a role in cell wall deposition. By contrast, PEN1 appears to have a basal function in secretion and a specialized defense-related function, being required for the polarized secretion events that give rise to papilla formation.
INTRODUCTION
Fungi typically initiate an invasion of plant tissues by penetrating the plant cell wall. If the intruding pathogen can be intercepted at this early stage, cellular integrity and homeostasis are maintained, and damage to plant tissues is greatly reduced. In numerous studies of cereals attacked by powdery mildew, resistance to fungal penetration has been intimately associated with the papilla response (see Belanger and Bushnell, 2002). Papillae are dome-shaped appositions deposited by the epidermal cell in the apoplasm (between the cell wall and plasma membrane) directly subtending the appressorium and penetration peg of the fungus. Degenerate penetration pegs are often found trapped in the papilla matrix (Ebrahim-Nesbat et al., 1986; Hippe-Sanwald et al., 1992; Kunoh et al., 1996) and the phenolics, reactive oxygen species, and hydrolases found to accumulate in papillae are likely to constitute effective barriers to fungal penetration (Belanger and Bushnell, 2002). Although there has been extensive research on papillae in the grasses, very little is known about the genes that regulate papilla formation aside from the MLO locus of barley. The wild-type MLO gene product is a negative regulator of papilla formation (Buschges et al., 1997), and mutation at this locus confers resistance to barley powdery mildew infection.
Nonhost resistance, in which no genotype of a given plant species is susceptible to any genotype of a particular pathogen, is the most common form of disease resistance exhibited by plants (see Thordal-Christensen, 2003). Nonhost resistance is highly effective and durable and therefore exploitation of this form of resistance is potentially of great agronomic importance. However, the underlying mechanisms are poorly understood. Whereas Arabidopsis is a host for certain powdery mildew species such as Erysiphe cichoracearum, it does not host the barley powdery mildew, Blumeria graminis-f. sp. hordei (Bgh). Attack by the host powdery mildew E. cichoracearum usually results in successful penetration and rapid proliferation of the fungus on Arabidopsis. By contrast, the nonhost Bgh germinates at the surface of Arabidopsis tissues but only rarely succeeds in entering the cells. As a consequence, the nonhost/pathogen Arabidopsis/Bgh interaction provides an excellent model system for studies on penetration resistance, and the contrast between the host and nonhost interactions can be used to compare successful vs. failed penetration events.
Genetic screens for mutations that result in increased penetration by the nonhost Bgh on Arabidopsis have identified the PENETRATION1 (PEN1) locus. PEN1, also referred to as SYP121 (Sanderfoot et al., 2000), encodes a syntaxin that has been shown to reside on the plasma membrane (Collins et al., 2003). The pen1 increased penetration phenotype suggests that the nonhost fungus encounters effective barriers to penetration that are defective in the mutant. There are at least three potential barriers to fungal penetration. Although this is poorly documented, the first two barriers to fungal penetration are thought to be the preexisting cuticle and the primary cell wall (Howard and Valent, 1996). Papillae, whose formation is induced upon infection, are likely to constitute a third barrier to fungal penetration. As a plasma membrane syntaxin presumably involved in secretion, PEN1 could be implicated in the deposition of cuticle precursors, of the primary cell wall and of papillae. Its role in these processes remains to be determined.
In this study, we examined the role of the vesicle trafficking apparatus in defense. A survey of 13,000 cDNAs across 72 defense-related arrays identified the syntaxin SYP122 as being induced upon pathogen infection. SYP122 is the closest homologue of PEN1, and we examined its role in cell wall deposition and defense. Comparison of the roles of SYP122 and PEN1 highlights the unique features of PEN1 as a key player in nonhost resistance.
MATERIALS AND METHODS
Transcriptional Profiling
SYP121/PEN1 (At3g11820) corresponds to AFGC array element 188A19XP (Affymetrix, Santa Clara, CA; 258786_at) and SYP122 (At3g52400) to array element 86D5T7 (Affymetrix 252053_at). Microarray analysis was carried out using tools available on the Stanford microarray database (www.smd.stanford.edu). Channel intensity cutoffs were placed at 200. Statistical analysis was performed according to Tusher et al. (2001) for datasets with three or more biological replicates. For data sets with only two biological replicates, statistical analysis could not be performed. Instead, we compared the expression of the clone of interest to 20 baseline clones (see Supplementary Table 1), the 20 most stably expressed clones on the array. The specific data sets discussed in this article are available from the Stanford Microarray Database and are listed in Supplementary Table 2. The Affymetrix experiments were carried out as described (Hamann et al., 2005).
For real time PCR, two primers (GCTCCTTTATCAGAGGCGGA and TTTTCGCGTGTTCTTCTGGTAA for PEN1; AGTTGCACCAAGTGTTTCTTGATATG and TTGCCTTCAATGTCATCAAGCT for SYP122) and an internal, fluorescent-labeled TaqMan probe (CTGACCAGCTACAAACCGCTCGGG for PEN1; CTGTGTTGGTTGAGCATCAGGGTGCA for SYP122; 5′ end, reporter dye FAM [6-carboxyflourescein], 3′ end, quencher dye TAMRA [6-carboxytetramethylrhodamine]) were designed for each gene using Primer Express software (Applied Biosystems, Foster City, CA). As an endogenous control for the normalization of SYP122 signals, a predeveloped TaqMan PCR system from Applied Biosystems (Foster City, CA) was used to target Arabidopsis 18S rRNA sequences (part number 4310893). The calibrator for fungal infections was the uninoculated sample at the same time point as the inoculated sample. RNA extraction used the TRIzol (Invitrogen, Carlsbad, CA) method, as described (Chomczynski and Sacchi, 1987). RNA from three biological replicates was pooled for each time point, and three Taqman reactions run on each pool. RNA was digested for 60 min at 37°C with RNase-free DNase I (Roche Diagnostics, Indianapolis, IN), heated for 5 min at 95°C, and chilled on ice. Reverse transcriptase-reaction, real-time TaqMan PCR and the relative quantization of SYP gene transcription were essentially as described (Leutengger et al., 2000).
Plant Growth Conditions and Fungal Inoculations
The mur mutants were obtained from the ABRC stock center, Ohio. Plants were propagated under controlled greenhouse conditions and potted in Promix HP (Premier Horticulture, Red Hill, PA). For host and nonhost inoculations, plants were grown in growth chambers at 21°C, with an 8-h photoperiod and a light intensity of ∼100 μE m-2 s-1, for 2 to 3 wk. To assess penetration frequency in the double mutant, seedlings were grown in sterile tissue culture on half-strength MS medium at 100% relative humidity. Penetration was assessed 36 h after inoculation with Bgh. Host powdery mildew fungal (E. cichoracearum UCSC1 inoculum) maintenance and inoculations were as described (Vogel and Somerville, 2000). Nonhost powdery mildew fungal (Blumeria graminis f.sp. hordei race CR3 [Bgh], maintained on barley) inoculations were carried out as described (Collins et al., 2003). Inoculations were carried out at medium density for light microscopy and at high density for confocal and electron microscopy.
Characterization of Homozygous and Double Mutant Lines and Constructs
Homozygous mutants in the SALK insertion line, SALK_008617, were identified using two primer pairs (122nf TCAAAACCGCGTTTTAATCAG with 122nb GACCGAACAAGCGAGTTAGC, as well as 122f AGGTTTTAAACGAGCCGGAGAGAAG with 122b AAATTCTGGTCTGGTTTCCCTTCAG). For this purpose DNA was isolated using CTAB minipreparations, as described (Assaad et al., 2001). Flanking sequences were isolated using the T-DNA primers LBa1 TGGTTCACGTAGTGGGCCATCG (together with 122nb) for amplification and primer LBb1 (GCGTGGACCGCTTGCTGCAACT) for sequencing. Sequencing reactions were carried out with an Applied Biosystems prism semiautomated sequencer using Big Dye (Perkin Elmer Applied Biosystems, Foster City, CA).
For double mutant analysis, SALK_008617 or syp122-1, was crossed to pen1-1 and pen1-4 (Collins et al., 2003). Lines homozygous at the PEN1 locus and segregating the SYP122 T-DNA insertion were identified phenotypically by Bgh infection and callose staining (to select for pen1 mutants) and by PCR analysis with the LBa1 122nb primer pair described above (to establish the presence of the SYP122 T-DNA insertion SALK_008617). Among a total of 741 progeny of such lines, 184 plants (24.8%) were necrotic and severely dwarfed. Forty-seven necrotic plants and 48 “normal” siblings were genotyped using the SYP122 primer pairs described above. Although the 48 normal siblings were homozygous or heterozygous for the wild-type allele of SYP122, the 47 necrotic siblings were homozygous for the SALK_008617 T-DNA insertion. We conclude that the severely dwarfed and necrotic siblings were pen1 syp122-1 double mutants. Furthermore, the segregation analysis (24.8% double mutant progeny) suggests that there was no failure of transmission through the pollen.
For the P35S::CFP-SYP122 construct, the SYP122 coding sequence was amplified from the BAC F2206 using the following primer pair (f: ATCTCTTAAGCCGAATTCCATGAACGATCTTCTCT and rev: GATATGCTCATGGGATCCCATAGACAGAGG). The amplicon was digested with EcoRI and BamHI and inserted between the EcoRI and BamHI sites of the ECAD vector, a derivative of EGAD in which GFP was replaced by CFP (see www.deepgreen.stanford.edu/html/vectors.html; Cutler et al., 2000). Plants were transformed by agrobacterium infiltration as described (Cutler et al., 2000). Western analysis (with the SYP122 antibody, see below) of wild-type plants harboring the CFP-SYP122 fusion detected an additional band c. 30 kDa higher than the endogenous band, showing that the gene fusion was intact. The CFP-SYP122 gene fusion was transformed into pen1-1 mutants and crossed into pen1 sypp122-1 double mutants. Segregation analysis showed that the gene fusion was capable of rescuing the double mutant phenotype, i.e. that it is functional. The syp122-1 and pen1 syp122-1 single and double mutants will be made available to the public upon publication.
Immunoblot Analysis
Two-week-old seedlings were ground in cold extraction buffer (100 mM HEPESKOH, pH 7.5, 5% glycerol, 200 mM glucose, 50 mM sodium pyrophosphate, 15 mM EGTA, 5 mM EDTA, 0.5% polyvinylpyrrolidone, 3 mM DTT, 10 μM leupeptin, and 1 mM PMSF). After preclearing by centrifugation (10 min at 10,000 × g, 4°C), the supernatant was removed and centrifuged at 100,000 × g for 30 min at 4°C. The microsomal pellet was solubilized in SDS loading buffer, subjected to SDS-PAGE and transferred in the presence of 0.1% SDS for 2 h at 250 V onto nitrocellulose (Hybond-ECL, Amersham, Piscataway, NJ). The membrane was blocked using 5% skimmed milk powder in TBS containing 0.005% Tween-20 and probed with anti-SYP122 polyclonal antiserum (Nühse et al., 2003). Peroxidase-conjugated goat anti-rabbit IgG (Sigma, St. Louis, MO) was used as a secondary antibody, and the reaction was visualized by using the Supersignal Pico detection kit (Pierce, Rockford, IL).
Cell Wall Analysis
Seedlings were sterilized and grown on MS plates (Assaad et al., 2001) without sucrose for 2 to 3 wk. Seedlings were cleared in 70% ethanol and dried after an acetone rinse. For Fourier transform infrared analysis (FT-IR; but not for uronic acids or GC analysis), dried leaves were pooled and homogenized by ball-milling. Four to six biological replicates were used per sample for uronic acids and cell wall monosaccharide content, and 12 replicates for FT-IR analysis. FT-IR analysis was carried out using a Nexus 470 FT-IR spectrometer (ThermoElectric Corporation, Chicago, IL), and spectral data processed using OMNIC software (Thermo Nicolet, Madison, WI). Principal component analysis using a covariance-matrix separation was performed in the Win-DAS software (Kelmsley 1998). Figures were processed using Sigma Plot (Microsoft, Redmond, WA). Total uronic acids were measured spectrophotometrically, using authentic galacturonic acid as a standard (Blumenkrantz and Asboe-Hansen, 1973). Cell wall monosaccharide content was determined by gas chromatography essentially as described (Blakeney et al., 1983; Reiter et al., 1997), except that separations were performed on an Agilent 6890N gas chromatograph (Wilmington, DE).
Light, Electron, and Confocal Microscopy
Aniline blue stains for callose, and trypan blue stains for fungal hyphae or cell lesions were as described (Vogel and Somerville, 2000). A DMB fluorescence microscope (Leica Microsystems, Wetzlar, Germany) was used. Penetration frequencies were computed as the number of haustorium-containing cells (36-72 h postinoculation [hpi] in 16-21-d-old plants; haustoria visualized by staining for callose) divided by the total number of attacked cells. Cells attacked by more than one appressorium were discounted. Statistical analysis using the t test was carried out using Excel software (Microsoft), and for this purpose we used arcsine conversions of the percentage and computed the difference between this and 1, in order to better approximate a normal distribution.
To visualize cell wall halos, leaves were inoculated with Bgh on the lower epidermis. Twenty-four hours postinoculation, epidermal peels were taken, washed in 0.1% SDS for 4 h at 60°C, stained in Coomassie (0.1% in 40% ethanol, 10% acetic acid) at 90°C for 10 min, and destained in 40% ethanol, 10% acetic acid for 5 min.
Samples for electron microscopy were prepared as described (Krüger et al., 2000). The electron microscope was a Zeiss EM912 (Göttingen, Germany). Confocal imaging was performed using an MRC1024 laser scanning confocal head (Bio-Rad, Hercules, CA) mounted on a Diaphot 200 inverted microscope (Nikon, Tokyo, Japan), a Zeiss 510 laser scanning confocal microscope, and a Leica TCS SP2 AOBS. The objectives used were a 60× Nikon PlanApo water immersion (WI) 1.2 NA (numerical aperture; Technical Instruments, San Francisco), a 40× Nikon PlanApo WI 0.9 NA, and a HCX PL APO 63×/1.2 W Corr/0.17 Lbd. Bl. objective. Excitation was at 568 nm for FM4-64, 488 nm for GFP, and 442 nm for CFP and emission at 585 nm for FM4-64, 522/535 for GFP, and 465 nm for CFP. In addition, using a Leica 405-nm laser, CFP was excited at 405 nm and detected from 450 to 550 nm. Chlorophyll autofluorescence was detected from 610 to 700 nm. Samples were mounted on cover slips in 1.5% mannitol, 0.01% silvet, and fungal structures were stained with 1 μM FM4-64 (Molecular Probes, Eugene, Oregon). Three-dimensional reconstructions of image stacks and quantization of fluorescence (using free-hand drawing tool in Image J) were carried out with Volocity (Improvision, Lexington, MA) or Image J (http://rsb.info.nih.gov/ij/) software. The Mann-Whitney U test (nonparametric) was run with Statview (SAS, Cary, NC). All images were processed with Photoshop software (Adobe Systems, Mountain View, CA), and figures were assembled with QuarkXpress (Quark, Denver, CO) or Illustrator (Adobe Systems, San Jose, CA) software.
RESULTS
Expression of PEN1's Closest Homologue, SYP122, Is Responsive to Infection
To identify vesicle trafficking genes implicated in defense, we surveyed 13,000 cDNAs across 72 defense-related microarrays. The syntaxin SYP122 was found to be significantly induced upon infection with the host powdery mildew fungus E. cichoracearum (Figure 1A). SYP122 expression was also induced by bacterial (Xanthomonas campestris) and viral (tobacco mosaic virus [TMV]) pathogens (Figure 1A), consistent with the finding that SYP122 is phosphorylated in response to the bacterial elicitor flagellin (Nühse et al., 2003). Although the expression of PEN1 also appeared to be responsive to pathogen infection on some arrays, signal intensities were low and statistical analysis (performed where possible; see Materials and Methods) failed to show that this up-regulation was significant. We focused our attention on two powdery mildew fungi, E. cichoracearum, referred to as the host pathogen, and B. graminis f sp. hordei (Bgh), referred to as the nonhost pathogen (compare Figure 1B with 1C). Real-time PCR showed that PEN1 and SYP122 were up-regulated by both pathogens, whereby this was considerably more pronounced for SYP122. Thus, PEN1 was induced 5.2 ± 0.5-fold by the nonhost pathogen (at 18 hpi) and 4.7 ± 0.7-fold by the host pathogen (at 48 hpi). By contrast, SYP122 was induced 31.1 ± 2.4-fold by the nonhost pathogen (at 18 hpi) and 58.5 ± 10.9-fold by the host pathogen (at 48 hpi). SYP122 is the closest homologue of PEN1. PEN1 and SYP122 are in the KNOLLE family, in a subfamily with five members, SYP121 through SYP125 (Sanderfoot et al., 2000). Within this subfamily, SYP121 and SYP122 were the only two that were significantly expressed in vegetative tissues (Figure 1D; Table 1).
Figure 1.
Expression of SYP122 is responsive to infection. (A) Up-regulation of SYP122 by fungal (host powdery mildew), bacterial (virulent X. campestris), and viral (systemic infection with TMV) pathogens. The ratio of channel 2 (cy5, inoculated sample) to channel 1 (cy3, uninoculated control) intensities and a color-coded scale bar are shown. The data were compared with 20 baseline clones (the most stably expressed clones on the arrays; see Supplementary Table 1), averaged in the figure. For a full description of the array experiments used, see Supplementary Table 2. (B) Host fungus, E. cichoracearum, 4 d after inoculation. Note the extensive proliferation of the fungus. (C) Nonhost fungus, Bgh, 4 d after inoculation. A Bgh conidium germinates at the surface but fails to penetrate the plant cell. (D) Tissue-specific differences in the expression of the nine syntaxins in the SYP1/KNOLLE family on the full genome Affymetrix chip. Signal intensities (arbitrary units on the Y axis) are shown from left to right for roots (dark gray), leaves (black), flowers (white), and stems (light gray). In vegetative tissues, SYP121/PEN1 and SYP122 are the only two significantly expressed syntaxins in the SYP12 subfamily. Data based on four biological replicates. The baseline clone is 190EXP (SKIP5). Bar, 200 μm in B, 50 μm in C.
Table 1.
Overview of the differences between PEN1 and SYP122
| Feature | PEN1 or pen1 | SYP122 or syp122-1 |
|---|---|---|
| Tissue-specific expression | Upregulated in stem | Down-regulated in flower |
| AFGC array database | Up-regulated by fungal, bacterial, and viral pathogens | |
| Real-time PCR | About fivefold up-regulation by nonhost and host pathogens | 31-59-fold up-regulation by nonhost and host pathogens |
| Primary cell wall FT-IR phenotype | Subtle (1.5% of spectral variance) | Strong (98.2% of spectral variance) |
| Bgh penetration in mutant | Strong increase (50-70%) | No reproducible increase (10%) |
| Polarization at Bgh attack site | Strong (6.3-fold higher than at pm) | Slight (2-fold higher than at pm) |
| Conclusion | Polarized secretion during papilla formation | Suggestive of a general function in secretion, including a role in cell wall deposition |
Because expression of the SYP122 gene was responsive to infection, we tested whether a mutation in SYP122 would confer a defense-related phenotype. The SALK line SALK_008617, which we refer to as syp122-1, was sequenced and the T-DNA insertion found to be 35-40 base pairs upstream of the ATG, in the 5′ untranslated region. Immunoblot analysis with a specific antibody (Nühse et al., 2003) showed no detectable protein in syp122-1, consistent with the interpretation that this is a null allele (Figure 2). We failed to discern a qualitative difference in E. cichoracearum hyphal growth or conidiation between the wild-type and syp122-1 or pen1-1 mutants. Similarly, even though SYP122 was considerably more up-regulated by the nonhost pathogen Bgh than by PEN1, we failed to detect a reproducible increase in Bgh penetration frequency in the syp122-1 mutant (Table 1).
Figure 2.
syp122-1 is a null mutation with no detectable SYP122 protein. Left panel, Ponceau stain; right panel, Western blot probed with an anti-SYP122 antibody (Nühse et al., 2003). Col-0, wild-type; syp122, syp122 mutant.
The pen1 syp122-1 Double Mutant Is Both Severely Dwarfed and Necrotic
As single mutations, pen1 and syp122-1 mutants conferred no visible defects in growth rate or morphology. We crossed syp122-1 to the EMS-induced, null alleles of PEN1, pen1-1, and pen1-4 (Collins et al., 2003). For both allelic combinations, the double mutants (identified as described in Materials and Methods) grew on soil as severely dwarfed plants with necrotic rosettes (Figure 3, A-E). When plated on MS medium, double mutant seedlings grew well for the first 2 to 3 wk. Thereafter, necrotic lesions were seen on the rosette leaves, even though these were propagated in sterile tissue culture (Figure 3A). The lesions were more frequent under high light than under low light (compare Figure 3C with 3D), suggesting an enhanced susceptibility to oxidative stress. In spite of their lesions, the double mutants yielded a small amount of seed and could be propagated as true breeding lines. Processes requiring polarized secretion such as root hair tip growth, trichome morphogenesis (Figure 3A), and transmission through the pollen did not appear to be perturbed in the pen1 and syp122-1 single and double mutants; similarly, seed mucilage, which requires a specialized form of secretion at the seed coat, did not appear to be reduced (Materials and Methods and our unpublished results). Because the cuticle is thought to be a barrier to fungal penetration, we examined this layer by electron microscopy. In the double mutant, the cuticular layers were present (compare Figure 3F with 3G). The double mutant did not differ from pen1-1 with respect to penetration frequency with the nonhost pathogen Bgh (compare a penetration frequency of 26.6 ± 2.5% for pen1-1 with 26.1 ± 3% for pen1-1 syp122-1).
Figure 3.
The pen1-1 syp122-1 double mutant is severely dwarfed and necrotic. (A, C, D, E [left], and G) double mutant; (B, E [right], and F) wild-type. (A) Necrotic lesions in the double mutant grown under sterile conditions on plates. (B-D) Roughly 3-wk-old soil-grown seedlings. (B and D) High light (c. 400 μE m-2 s-1); (C) low light (c. 100 μE m-2 s-1). The seedling grown under low light in C has fewer lesions and is larger than the one in D, grown under high light. (E) Note the dwarfed phenotype of the double mutant (left) compared with the wild-type (right). (F and G) Electron micrographs of wild-type (F) and double mutant (G) seedlings show the presence of two cuticular layers, a dark layer of cutin interwoven with cell wall polymers (star) and a white layer of cutin. Above the cuticle at the leaf surface is an omiophillic layer that may correspond to epicuticular waxes, partially removed (see especially F) by solvent extraction during sample preparation.
The syp122-1 Mutant Has a Primary Cell Wall Defect
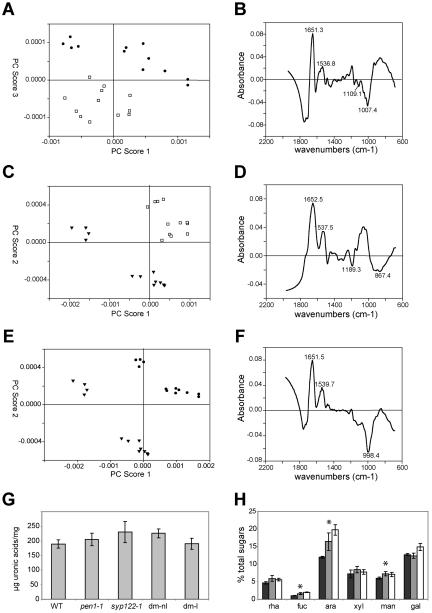
Because the components of the cell wall are largely secreted, we tested for alterations in the primary cell wall of pen1 and syp122-1 single and double mutants. FT-IR measurements of solvent-extracted cell walls revealed differences from the wild type in both the single and double mutants. For the pen1-1 mutant, we detected a separation along principal component 3 (for principal component [PC] analysis see Chen et al., 1997; Figure 4A), which accounts for only 1.5% of the spectral variance. By contrast, for syp122-1, we detected a separation along PC1 and PC2, which together account for 98.2% of the spectral variance (Figure 4C). The double mutant, harvested from plates before lesioning, showed a separation along PC1 and PC2 as well as PC1 and PC3, which together account for 99.9% of the spectral variance (our unpublished results). A direct comparison of pen1-1 and syp122-1 revealed differences in the spectra, with a good separation along PC1 and PC2 (Figure 4E). In all spectra, there appears to be a difference in the protein peaks (wavenumbers 1651-1653 and 1536-1540; Figure 4, B, D, and F). In comparisons to the wild-type for both pen1-1 and syp122, the troughs at wavenumbers 1109, 1007 (Figure 4B), and 998 (Figure 4F) are consistent with an altered pectin content or composition in the mutants (Coimbra et al., 1998; Wilson et al., 2000). However, a direct measure of the uronic acid content, a component of pectin, failed to reveal a difference from the wild-type in both the single and double mutants (Figure 4G). Although we failed to see a change in cell wall composition in syp122-1, the double mutants, harvested before or after lesioning, showed a complex change in cell wall monosaccharide content, with a slight but significant (p < 0.03) increase in fucose, arabinose, and mannose (Figure 4H).
Figure 4.
syp122-1 has a primary cell wall defect; 3-wk-old seedlings. (A-E) Principal component analysis of FT-IR spectra show a separation between the single mutants and the wild type along principal components (PC) 3 for pen1-1 (A, •) and along both PC1 and PC2 for syp122-1 (C, ▾), compared with the wild-type (□). (B) Wild-type vs. pen1-1, PC3. (D) Wild-type vs. syp122-1, PC1. (E) pen1-1 (•) and syp122-1 (▾) separate along PC1 and PC2. (F) Wild-type vs. syp122-1, PC2. (G) The single and double mutants, harvested before (dm-nl) and after (dm-l) lesioning, did not differ with respect to the wild type in terms of their uronic acid content, a measure of the pectin moiety of the cell wall. Y axis: uronic acids in μg per mg dry weight. (H) Cell wall composition showed a complex change in the double mutants, harvested before (gray columns) and after (white columns) lesioning. The wild type is depicted by black columns. From left to right: rhamnose, fucose, arabinose, xylose, mannose, and galactose. The increase in fucose, arabinose, and mannose in the double mutants is significant (denoted with an asterisk). Y axis: monosaccharide content represented as a percentage of total sugars.
Cell Wall and syp122-1 Mutants Have Very Subtle Nonhost Phenotypes Compared with pen1 Mutants
We compared pen1 with the mur mutants, isolated on the basis of their altered cell wall composition (Reiter et al., 1997). The mur1-1 mutant has <2% of the normal amounts of fucose in aerial parts of the plant (Reiter et al., 1997), and a tensile strength of elongating influorescences <50% that of the wild type (Bonin et al., 1997). MUR3 encodes a xyloglucan-specific galactosyl transferase (Madson et al., 2003) that considerably affects the mechanical properties of the cell wall (Ryden et al., 2003). The mur8-1 and mur10-1 mutants have complex changes in their cell wall composition (Reiter et al., 1997). In wild-type plants inoculated with the nonhost powdery mildew fungus, callose stains generally reveal a large, bright secondary cell wall apposition or papilla deposited at the site of attempted penetration (Figure 5C). In the pen1, syp122-1, and mur mutants, the plant's general ability to lay down papillae, measured 3 d after inoculation, did not appear to be impaired. In pen1 mutants, the most frequent plant response was to outline the attacked cell with callose (Figure 5B). The percentage of callose-outlined cells was roughly 70% in the pen1 mutants, compared with roughly 10% in the wild type, in syp122-1, and in the mur mutants. Bgh haustoria, arising as a result of penetration, were invariably encased in callose (Figure 5A) and failed to support an invasive growth of the fungus. Of the mur mutants, only mur1-1 showed a reproducible and significant (p = 0.002) decrease in its penetration resistance to Bgh (Figure 5D). The pen1-1 mutant had very subtle cell wall defects compared to the mur mutants or to syp122-1 mutants, but show a considerably greater increase in fungal penetration (Figure 5D and Table 1). The results show that not all cell wall defects cause an increase in fungal penetration. The Bgh phenotype of mur1-1 suggests that the composition of the primary cell wall may constitute a barrier to fungal penetration.
Figure 5.
The cell wall mutant mur1-1 has a relatively subtle nonhost phenotype compared with pen1 mutants. (A and B) pen1-1; (C) wild type. (A) pen1-1 mutants are characterized by an increased incidence of fungal penetration, as evidenced by the presence of haustoria (arrows). Note the callose encasement of both the haustoria and infected cells. (B) The most frequent response to nonhost infection in pen1-1 mutants is to outline the infected cell with callose. (C) On Bgh infection, wild-type plants generally deposit a cell wall apposition or papilla at the site of attempted penetration. (D) Percent Bgh penetration in mur1-1 and pen1-1 mutants. Bar in A is 10 μm; other panels are to scale.
PEN1 Defines of a Novel Cellular Compartment upon Fungal Infection
Double mutant analysis and the comparison between pen1 and syp122-1 mutants suggest that PEN1 may, in addition to a basal role in secretion, play a highly specific role in penetration resistance. A GFP-PEN1 fusion was shown to rescue the pen1 phenotype and was localized to the plasma membrane (Collins et al., 2003), but its localization upon fungal infection has not been reported. A CFP-SYP122 fusion protein was shown to be both intact and functional and, like GFP-PEN1, localized to the plasma membrane (Materials and Methods, Figure 6D, supplementary Figure 1). This is consistent with cell fractionation studies (Nühse et al. 2003). Upon infection with both the host and nonhost powdery mildews, GFP-PEN1 and CFP-SYP122 accumulate at papillae (Figure 6, A-E, and our unpublished results). Papillar localization could take several forms. GFP-PEN1 was not only seen in cup-shaped plasma membrane domains around the papillae (Figure 6, G and I), but surprisingly, in the apparent interior of the papillae as well (Figure 6B). Negative controls with nontransgenic leaves detected no signal at the papillae, showing that the signal seen with GFP-PEN1 and CFP-SYP122 at papillae was not due to autofluorescence. The penetration pore was clearly demarcated (Figure 6, B, E, and H) and concentric rings around the penetration pore with or without halos were also seen, forming “bull's eye” like shapes (Figure 6H). The halos were sometimes very large, spanning more than one cell without being affected by cellular boundaries. GFP-PEN1 was not only on the plasma membrane, but also on an endomembrane compartment ∼1 μm in diameter (Figure 6, E and H), sometimes seen to form a cloud apparently associated with papillae (Figure 6E).
Figure 6.
Accumulation of GFP-PEN1 at infection sites takes different forms. Three-dimensional reconstructions of image stacks. Fungal structures are in red, GFP-PEN1 or CFP-SYP122 (D) are shown in green. Images were taken 17-24 hpi with Bgh (A-D and F-I) or E. chiccoracearum (E). (A-C) Different projections of the same sample showing accumulation of GFP-PEN1 at papillae. B is the same as A, but without the red channel. (B) Accumulation of GFP-PEN1 inside papillae (p, compare to G and H where rings are seen), more clearly seen in serial sections (our unpublished results). Arrow points to the penetration hole, in which no GFP-PEN1 is seen. (C) Side view of A. (D) CFP-SYP122 localization to the plasma membrane (arrowheads) and to papillae (arrow). (E) Localization of GFP-PEN1 to papillae (p) upon infection with the host powdery mildew. GFPPEN1 also localizes to an endomembrane compartment, which forms clouds close to the infection site (arrowheads). (F) Accumulation of GFPPEN1 at the cell surface at the point of contact with the appressorium. The novel GFP-PEN1 domain is contiguous with the plasma membrane, too faint to be seen in the image. (G) A bright ring of GFP-PEN1 is seen. Note the relative intensity of the ring compared with the adjacent plasma membrane. In 3D reconstructions, this ring forms the rim of a cup-shaped structure (our unpublished results). (H) Concentric rings of GFP-PEN1 fluorescence form a bull's eye at the penetration hole. Arrow points to an endomembrane compartment, roughly 1 μm in diameter. (I) The second ring of the bull's eye (arrowhead in H and I) is the rim of a hollow cup-shaped structure. a, appressorium; p, papilla. Scale bar, 5 μm in F, 10 μm in all other panels.
The patterns of GFP-PEN-1 and CFP-SYP122 accumulation at the attack site show an interesting difference. Whereas the local intensity of CFP-SYP122 at the attack site was about twofold higher than that found at the plasma membrane (2.0 ± 0.94), the relative intensity of GFP-PEN1 at the attack site was significantly greater, being about sixfold higher (6.3 ± 3.05). This difference in relative intensity for GFP-PEN1 and CFP-SYP122 was highly significant (p = 1.7 e-6 with the t test; p < 0.0001 with the nonparametric Mann-Whitney U test). We compared the focal accumulation of GFP-PEN1 with that of two additional GFP-based plasma membrane markers and a number of GFP-based endomembrane markers (described in Cutler et al., 2000). Focal accumulation was not detected with the plasma membrane marker GFP-PIP2a. With the plasma membrane marker GFP-LTI6b a slight degree of focal accumulation was detected, but the ratio of the signal at the papillae vs. the signal at the plasma membrane was 3.9-fold less than that seen for GFP-PEN1 (p = 2 e-6). No focal accumulation was detected with a presumptive Golgi marker, with a vacuolar marker (Q5), and with freely soluble cytoplasmic GFP (EGAD; Cutler et al., 2000). A very weak focal accumulation of an ER marker (Q4; Cutler et al., 2004) was sometimes detected, and the ratio of the signal at the attack site vs. the signal at the endomembranes or plasma membrane was at least 5.8-fold less than seen with GFP-PEN1 (p = 0.005). These results suggest that GFP-PEN1 shows a greater degree of recruitment to the site of attempted penetration than seen with any other marker and that it serves as a specific marker for a cellular compartment that is associated with fungal attack.
A Delay in Papillae Formation in the pen1-1 Mutant
In light of the accumulation of PEN1 at papillae, we examined whether differences could be detected between wild-type and pen1 papillae by electron microscopy. Forty-eight hours after inoculation with the nonhost powdery mildew, pen1-1 mutant papillae were indistinguishable from the wild type (compare Figure 7, A and B). In the medial sections shown in Figure 7, A and B, a nonhomogeneous layering of the papillae can be seen, with osmiophillic substances especially concentrated in the center below the penetration peg. Three additional features of the micrographs are noteworthy. First, the papillae, like the rings or halos seen around papillae upon callose stain or with the GFP-PEN1 marker, do not respect cellular boundaries, but may form in two adjacent cells (Figure 7, A and E). Second, structures reminiscent of membranes can be detected within the papillae (Figure 7E). These may be an artifact of fixation, but could also possibly be due to an extrusion of membranes at the junction between the plasma membrane and the cell wall. Third, a thinning of the plant cell wall at the site of attempted penetration can be seen (Figure 7C), concomitant in some instances with a deformation of the cell wall (Figure 7D). An investigation of the cell wall at attack sites with Coomassie staining after washing in SDS revealed the presence of halos at the cell wall (Figure 7F), reminiscent of the halos we observed with GFP-SYP121 at the plasma membrane (Figure 6H). These had the appearance of ripples originating at the penetration pore (Figure 7F) and may reflect the presence of cross-linked protein (or other Coomassie-staining entity) intimately associated with the cell wall.
Figure 7.
Properties of nonhost papillae in the wild-type and in pen1 mutants. (A-E) Transmission electron micrographs of Bgh on Arabidopsis, 48 hpi. (A) Medial section of a wild-type papilla (p) that spans two adjacent cells. Note the presence of osmiophillic (darkly stained) bodies in the papilla and the layering of osmiophillic substances (arrowhead). (B) Medial section through a pen1-1 mutant papilla. Note the osmiophillic layering, as seen in A (arrowhead). Arrows point to a membrane contiguous with the plasma membrane (PL); note that papillae are apoplastic structures, residing between the plasma membrane and cell wall. (C) Enlargement of A (same infection site but different section). Note the thinning of the cell wall at the penetration pore (arrows), showing that the plant cell wall is digested. (D) Enlargement of B (same infection site but different section). Osmiophillic bodies (encircled) seen beneath the penetration peg. Note the deformation of the cell wall (arrows), suggesting that pressure is exerted by the Bgh appressorium. (E) Enlargement of A (different section) showing that the papilla spans two adjacent cells. Membrane-like entities are encircled. (F) Cell wall halos around papillae, visualized in a light micrograph of Coomassie-stained wild type 24 hpi with Bgh. CP, cytoplasm; CW, cell wall; M, mitochondria; P, papilla; PL, membrane contiguous with plasma membrane; V, vacuole.
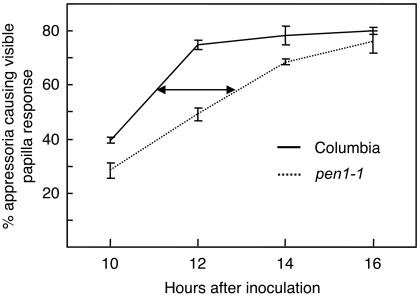
Having failed to detect a difference from the wild type in mature pen1-1 papillae, we looked at the onset of papillae formation. We found this to be significantly delayed in the pen1-1 mutant (Figure 8). Bgh has a very synchronous development, and penetration as well as GFP-PEN1 accumulation at papillae typically occurs in a narrow window of time around 12 hpi. At this time point, the delay in papilla formation is of ∼2 h (Figure 8), and we suggest that this delay causes the decreased penetration resistance observed in pen1 mutants.
Figure 8.
Papillae formation is delayed in pen1-1 inocculated with Bgh. Black line: wild-type; dotted line: pen1-1. Arrow highlights the delay in papilla formation.
DISCUSSION
The Arabidopsis genome encodes an unusually large number of SNAREs (soluble N-ethylmaleimide-sensitive factor adaptor protein receptors) relative to the number found in the human genome and other animal or fungal genomes (Sanderfoot et al., 2000; Assaad, 2001). However, phylogenetic analysis failed to detect Arabidopsis orthologues of plasma membrane SNAREs of yeast or mammals involved in secretion. This function may be supplied by a novel class of plant syntaxins including the plasma membrane SNAREs PEN1 and SYP122. Although the PEN1 and SYP122 single mutants have no visible phenotypes, pen1 syp122 double mutants are necrotic and dwarfed. The double mutant phenotype suggests that the two syntaxins have an overlapping, basal function in secretion. Tobacco expressing a dominant negative form of the PEN1 tobacco homologue, namely a truncated form of the NtSyr1 syntaxin, have a dwarfed phenotype, as observed in the pen1-1 syp122-1 double mutant, but necrotic lesions have not been reported in these lines; NtSyr1 has been shown to play a role in secretion and growth (Geelen et al., 2002). Analysis of the double mutant failed to reveal a role for PEN1 and SYP122 in specialized forms of secretion, polarized or otherwise, giving rise to root hair, trichome, or pollen tube morphogenesis or to seed mucilage. These functions may be supplied by the PEN1 and SYP122 paralogues, SYP123, SYP124, and SYP125. These three paralogues may also be able to compensate for PEN1 and SYP122 at the plasma membrane in the double mutant, which is viable. In light of the striking double mutant phenotype, it is interesting to note that the PEN1 and SYP122 syntaxins differ with respect to their transcriptional regulation and that the pen1-1 and syp122-1 single mutants differ with respect to the nature of both their cell wall defects and their nonhost phenotype (for overview see Table 1; compare to VTI SNAREs; Surpin et al., 2003). This suggests that although the two genes can partially complement each other, they play distinct, cooperative roles in cell wall biogenesis and defense. In light of its role in cell wall deposition, its up-regulation in response to bacterial and viral pathogens and its phosphorylation in response to the bacterial elicitor flagellin, SYP122 could play a role in diffuse secretion (see Table 1 and Nühse et al., 2003). By contrast, our results suggest that, in the context of defense, PEN1 is more specifically involved in the polarized secretion events that give rise to papilla formation.
Use of Array or Chip Data to Guide a Reverse Genetic Approach
A meta-analysis of 72 defense-related arrays pointed to a role for SYP122 in defense against a broad range of bacterial, viral, and fungal pathogens. However, we failed to uncover a defense-related phenotype in the syp122-1 mutant. By contrast, PEN1 showed a relatively weak up-regulation of gene expression, as detected by real-time PCR, and this correlated with a strong nonhost phenotype (this study, Collins et al., 2003). Our results echo those of Giaever et al. (2002) in yeast, who report that <7% of genes that exhibited a significant increase in mRNA expression are also required for optimal growth under four studied conditions. Functional overlap between closely related family members and the existence of compensation mechanisms may impact phenotypic analyses.
A Role for PEN1 in Papilla Formation
Our localization and genetic analyses suggest that PEN1 plays an active role in the polarized secretion events that give rise to the formation of papillae during fungal attack. The extent of accumulation of GFP-PEN1 at papillae is unique, suggesting that PEN1 is actively recruited to papillae. Papillae indistinguishable from the wild-type by electron microscopy were able to form in pen1 mutants, but their appearance was delayed by ∼2 h at the time of fungal penetration and of GFP-PEN1 accumulation at papillae. Thus, the pen1 phenotype is the converse of the mlo phenotype. Indeed, the papilla response occurs earlier in mlo mutants of barley (Aist and Bushnell 1991; Zeyen et al., 1993, 1995), but is delayed in pen1 mutants. Similarly, mlo mutants have an increase in penetration resistance, whereas pen1 mutants have a decreased penetration resistance (Collins et al., 2003). To this effect, it is intriguing that the barley orthologue of PEN1, ROR2, was identified as a locus “required for mlo resistance” in screens for suppressors of mlo (Freialdenhoven et al., 1996; Collins et al., 2003). In the race between the fungus and the plant, the timing of papilla formation is potentially critical and may suffice to affect the frequency of fungal penetration. We propose that delayed papillae formation is the primary defect in pen1 mutants and that the decreased penetration resistance observed in these mutants is a secondary consequence of this primary defect.
Surprisingly, GFP-PEN1 signal was found not only at the periphery of the papillae, but also apparently inside the papillae. Electron micrographs revealed the presence of membrane-like entities and membrane-bound bodies within the papillae. These may be a result of tissue fixation, but could possibly be due to an extrusion membranes at the junction between the plasma membrane and the cell wall. The latter possibility, though unexpected, is consistent with the observation of GFP-PEN1 fluorescence in the apparent interior of the papillar structure. Unusual trafficking and fusion events, involving the fusion of unusually large “vesicles” 1 μm in diameter at the papillae, have been reported during papilla formation (Zeyen and Bushnell, 1979), and such compartments have been shown to target reactive oxygen species toward infection sites (Hückelhoven et al., 1999). Collins et al. (2003) report a decrease in the number of large vesicles observed at sites of failed penetration by the fungus in pen1 mutants. A priori, this finding appears counterintuitive. Indeed, were PEN1 involved in docking and fusing vesicles at the plasma membrane, one would expect such vesicles to accumulate rather than to decrease in the pen1 mutant. In this study, we found that, in addition to its localization to the plasma membrane and to papillae, GFPPEN1 was found on an endomembrane compartment roughly 1 μm in diameter. This compartment was seen to accumulate in the vicinity of papillae. We speculate that this PEN1-related compartment might correspond to the “large vesicles” described previously in the defense literature (Zeyen and Bushnell, 1979; Hückelhoven et al., 1999) and that PEN1 might mediate its fusion at infection sites during papilla formation.
Electron micrographs of the nonhost interaction on Arabidopsis show both a deformation as well as a thinning of the plant cell wall at the site of attempted penetration. This suggests that the nonhost barley powdery mildew exerts a mechanical force on the plant cell wall and that it also has the ability to enzymatically digest the wall (compare to Howard and Valent, 1996 and Zeyen and Bushnell, 1979). Our genetic analyses suggest that the nonhost Bgh fungus encounters barriers to penetration at both the level of the primary cell wall and papillae, but the papillae may constitute a more effective barrier. Indeed, a significant decrease in penetration resistance was seen in mur1 mutants, isolated on the basis of their dramatically altered cell wall composition (Reiter et al., 1997), but this was considerably weaker than seen in pen1 mutants. Conversely, the pen1-1 mutant had very subtle primary cell wall defects compared with syp122-1 and the mur mutants but a considerable delay in papilla formation, coupled to a considerable decrease in penetration resistance.
The Plant Trafficking Apparatus in Host vs. Nonhost Responses
Localization of PEN1 to papillae was seen both when the plant acts as host and as nonhost. Similarly, cytoplasmic aggregation of ER and Golgi did not differ between compatible and incompatible host interactions or nonhost interactions in studies of Arabidopsis attacked by oomycetes (Takemoto et al., 2003). It appears that there is no difference in the way the secretory apparatus is mobilized in response to host and nonhost powdery mildew fungi. However, electron microscopy revealed differences in the properties of papillae associated with successful vs. failed penetration events. Indeed, a concentration of osmiophillic substances beneath the penetration peg, as observed here in nonhost papillae, has been correlated with penetration resistance in a number of studies on barley (Ebrahim-Nesbat et al., 1986; Hippe-Sanwald et al., 1992; Kunoh et al., 1996) and was not observed during compatible host interactions (Kunoh et al., 1996). Osmium has affinity for phospholipids, unsaturated fatty acids, tannins, and phenolic polymers. The degenerate penetration pegs often found trapped in the electron-dense center of layered papillae (Ebrahim-Nesbat et al., 1986; Hippe-Sanwald et al., 1992; Kunoh et al., 1996) are consistent with the interpretation that these osmiophillic substances correspond to phenolics and other antifungal agents.
Although the papilla response is often associated with resistance to fungal penetration, papilla may play distinct and/or additional roles in instances of successful penetration. Upon successful penetration, the penetration peg differentiates into a haustorium that enables the fungus to draw nutrients from the plant. In such instances, the papilla may seal the wound in the epidermal cell wall caused by the penetration peg, thereby potentially enabling the fungus to avoid recognition by the plant's surveillance mechanism. Consistent with this interpretation is the finding that the pmr4 callose synthase mutant of Arabidopsis has no callose in the papilla matrix and is resistant rather than hypersusceptible to the host powdery mildew (Jacobs et al., 2003; Nishimura et al., 2003). It appears that the host fungus evades recognition by the plant's surveillance mechanism and/or that it is capable of deflecting or detoxifying the plant's defenses. By contrast, recognition in the case of the nonhost may trigger changes in metabolism that give rise to effective chemical, physical, and enzymatic barriers to fungal establishment.
Our analysis of the pen1 phenotype suggests that there are multiple layers of resistance in the context of the nonhost interaction. Thus, in pen1 mutants, which fail to mount a rapid papilla response, the haustoria are encased and most often the attacked cells are outlined with callose. As a consequence of this or because of other or additional layers of protection, successful penetration does not result in successful growth and proliferation of the fungus in pen1 mutants. Even when the syntaxin that, to our knowledge, best defines the papillae as a novel cellular compartment is missing, papillae formation is delayed but not abolished. This points not only to redundancy at the level of the plasma membrane syntaxins, but also to the resilience of nonhost resistance.
GFP-PEN1 was occasionally seen to form concentric bull's-eye rings around the site of attempted penetration, showing a remodeling of the plasma membrane upon non-host powdery mildew infection. Halos were also visible at the cell wall and have also been reported in the literature (Belanger and Bushnell, 2002). Interestingly, the plasma membrane halos form perfect circles that, like the papillae, do not respect cellular boundaries but seem to depend solely on the site of attack by the fungus. This suggests that contact between the plant and the fungus at the level of the plant cell wall or cell surface initiates a signal transduction cascade that defines novel plasma membrane domains. Were the initial signal intracellular, one would expect it to respect cellular boundaries. Thus, we speculate that the cell wall halos are due to a signal, such as a wall degradation product, diffusing locally in the cell wall. Once established, the bull'seye-like plasma membrane domains could translate into a cortical cue that would reorganize the plant cytoskeleton and cytoplasmic contents, as seen during fungal attack.
Biological Function of Novel Plant SNAREs
To date, three biological functions have been attributed to the SYP1 syntaxins and/or to their interacting partners: cytokinesis, root hair morphogenesis, and resistance to pathogen attack (Lukowitz et al., 1996; Assaad et al., 2001; Collins et al., 2003; and this study). All three processes involve highly regulated and dynamic forms of membrane traffic. Cytokinesis takes on average 50 min for an Arabidopsis cell and root hair tip growth occurs at a rate of 100 μm/h. Similarly, upon pathogen attack a dramatic and rapid reorganization occurs as the plant cell confronts the pathogen with structural, chemical, and enzymatic weapons. Membrane traffic in these three instances is regulated by a variety of cues including cell cycle progression, nutrient and water availability, phytohormones, developmental stage, or pathogen attack. Thus, the unprecedented sophistication of the plant vesicle trafficking apparatus translates into a remarkably nimble cell, capable of fine-tuned and rapid growth or change in response to changes in the environment.
Supplementary Material
Acknowledgments
We thank Shauna Somerville and her lab members for providing resources and space for many of the defense-related experiments and for useful suggestions. Tze Yang Jeremy Lee helped with cell wall analysis and Ted Raab helped in interpreting the FT-IR data. F.A. is especially grateful to Monica Stein for an introduction to Bgh biology and histology and for sharing the pen1-4 allele before publication. We are indebted to Ralph Hückelhoven, Susanne Liebe, and Serry Koh for help with confocal analysis. We thank Jesse Coull for help with statistical analysis, Arthur Grossman and Klaus Lenzian for useful discussion, an anonymous reviewer for very thorough criticism as well as Christine Gietl and Erwin Grill for a critical evaluation of the manuscript. Sylvia Dobler prepared samples for electron microscopy, Erin Osborne ran the Affymetrix experiments, and Nick Mosenko at TAIR provided us with the baseline clones for microarray analysis. Finally, we thank Christian Leutengger at AppliedGenex for help with real time PCR. This research was made possible by a grant from the Danish Agricultural and Veterinary Research Council SJVF 53-00-0255 to H.T.-C., and U.S. Department of Energy grant DE-FG02-03ER20133 to C.S., Deutscheforschungsgemeinschaft grant AS110/2-1 to F.A. as well as by support from the Gatsby Foundation to S.C.P.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-02-0140. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-02-0140.
The online version of this article contains supplementary material accessible through www.molbiolcell.org.
References
- Aist, J.R., and Bushnell, W.R. (1991). Invasion of plants by powdery mildew fungi, and cellular mechanisms of resistance. In: The Spore and Disease Initiation in Plants and Animals, ed. G.T. Cole and H.C. Hoch, New York: Plenum Press, 321-345.
- Assaad, F.F. (2001). Of weeds and men: lessons on the genome for plant cell biology. Curr. Opin. Plant Biol. 4, 478-487. [DOI] [PubMed] [Google Scholar]
- Assaad, F.F., Huet, Y., Mayer, U., and Jürgens, G. (2001). The cytokinesis gene KEULE encodes a Sec1 protein that binds the syntaxin KNOLLE. J. Cell Biol. 152, 531-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belanger, R.R., and Bushnell, W.R. (2002). The Powdery Mildews: A Comprehensive Treatise, St. Paul, MN: APS Press.
- Blakeney, A.B., Harris, P.J., Henry, R.J., and Stone, B.A. (1983). A simple and rapid preparation of alditol acetates for monosaccharide analysis. Carbohydr. Res. 113, 291-299. [Google Scholar]
- Blumenkrantz, N., and Asboe-Hansen, G. (1973). New method for quantitative determination of uronic acids. Anal. Biochem. 54, 484-489. [DOI] [PubMed] [Google Scholar]
- Bonin, C.P., Potter, I., Vanzin, G.F., and Reiter, W.D. (1997). The MUR1 gene of Arabidopsis thaliana encodes an isoform of GDP-d-mannose-4,6-dehydratase, catalyzing the first step in the de novo synthesis of GDP-l-fucose. Proc. Natl. Acad. Sci. USA 94, 2085-2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschges, R. et al. (1997). The barley Mlo gene: a novel control element of plant pathogen resistance. Cell 88, 695-705. [DOI] [PubMed] [Google Scholar]
- Chen, L., Wilson, R.H., and McCann, M.C. (1997). Infra-red microspectroscopy of hydrated biological systems: design and construction of a new cell with atmospheric control for study of plant cell walls. J. Microsc. 188, 62-71. [Google Scholar]
- Chomczynski, P., and Sacchi, N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156-159. [DOI] [PubMed] [Google Scholar]
- Coimbra, M.A., Barros, A., Barros, M., Rutledge, D.N., and Delgadillo, I. (1998). Multivariate analysis of uronic acid and neutral sugars in whole pectic samples by FT-IR spectroscopy. Carbohydr. Polym. 37, 241-248. [Google Scholar]
- Collins, N.C. et al. (2003). SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425, 973-977. [DOI] [PubMed] [Google Scholar]
- Cutler, S.R., Ehrhardt, D.W., Grifitts, J.S., and Somerville, C.R. (2000). Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis thaliana at high frequency. Proc. Natl. Acad. Sci. USA 97, 3718-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahim-Nesbat, F., Heitefuss, R., and Rohringer, R. (1986). Ultrasctructural and histochemical studies on mildew of barley (Erysiphe graminis DC. f.sp. hordei Marchal) IV Characterization of papillae in fifth leaves exhibiting adult plant resistance. J. Phytopathol. 117, 289-300. [Google Scholar]
- Freialdenhoven, A., Peterhansel, C., Kurth, J., Kreuzaler, F., and Schulze-Lefert, P. (1996). Identification of genes required for the function of non-race-specific mlo resistance to powdery mildew in barley. Plant Cell 8, 5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geelen, D., Leyman, B., Batoko, H., Di Sansebastiano, G.P., Moore, I., Blatt, M.R., and Di Sansabastiano, G.P. (2002). The abscisic acid-related SNARE homolog NtSyr1 contributes to secretion and growth: evidence from competition with its cytosolic domain. Plant Cell 14, 387-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, et al. (2002). Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387-391. [DOI] [PubMed] [Google Scholar]
- Hamann, T., Osborne, E., Youngs, H.L., Misson, J., Nussaume, L., and Somerville, C.R. (2005). Tissue-specific expression of CESA and CSL genes in Arabidopsis. Cellulose (in press).
- Hippe-Sanwald, S., Hermanns, M., and Somerville, S.C. (1992). Ultrastructural comparison of incompatible and compatible interactions in the barley powdery mildew disease. Protoplasma 168, 27-40. [Google Scholar]
- Howard, R.J., and Valent, B. (1996). Breaking and entering: host penetration by the fungal rice blast pathogen Magnaporthe grisea. Annu. Rev. Microbiol. 50, 491-512. [DOI] [PubMed] [Google Scholar]
- Hückelhoven, R., Fodor, J., Preis, C., and Kogel, K.H. (1999). Hypersensitive cell death and papilla formation in barley attacked by the powdery mildew fungus are associated with hydrogen peroxide but not with salicylic acid accumulation. Plant Physiol. 119, 1251-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, A.K., Lipka, V., Burton, R.A., Panstruga, R., Strizhov, N., Schulze-Lefert, P., and Fincher, G.B. (2003). An Arabidopsis callose synthase, GSL5, is required for wound and papillary callose formation. Plant Cell 15, 2503-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelmsley, E.K. (1998). Discriminant Analysis and Class Modeling of Spectroscopic Data, New York: John Wiley and Sons.
- Kunoh, H., Matsuoka, K., and Kobayashi, I. (1996). Ultrastructure of papillae induced in barley coleoptile cells by a pathogen, Erysiphe graminis, and a non-pathogen, E. pisi. Fitopatol. Brasil 21, 418-425. [Google Scholar]
- Krüger, J., Loubradou, G., Wanner, G., Regenfelder, E., Feldbrügge, M., and Kahmann, R. (2000). Activation of the cAMP pathway in Ustilago maydis reduces fungal proliferation and teliospore formation in plant tumors. Mol. Plant Microbe Interactions 13, 1034-1040. [DOI] [PubMed] [Google Scholar]
- Leutenegger, C.M. et al. (2000). Immunization of cats against feline immuno-deficiency virus infection using minimalistic immunogenic defined gene expression vector vaccines expressing F.IV-gp140 alone or with feline IL-12, IL-16 or CpG. J. Virol. 74, 10447-10457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz, W., Mayer, U., and Jürgens, G. (1996). Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell 12, 61-71. [DOI] [PubMed] [Google Scholar]
- Madson, M., Dunand, C., Li, X., Verma, R., Vanzin, G.F., Caplan, J., Shoue, D.A., Carpita, N.C., and Reiter, W.D. (2003). The MUR3 gene of Arabidopsis thaliana encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell 15, 1662-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, M.T., Stein, M., Hou, B.H., Vogel, J.P., Edwards, H., and Somerville, S.C. (2003). Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301, 969-972. [DOI] [PubMed] [Google Scholar]
- Nühse, T.S., Boller, T., and Peck, S.C. (2003). A plasma membrane syntaxin is phosphorylated in response to the bacterial elicitor flagellin. J. Biol. Chem. 278, 45248-45254. [DOI] [PubMed] [Google Scholar]
- Reiter, W.D., Chapple, C., and Somerville, C.R. (1997). Mutants of Arabidopsis thaliana with altered cell wall polysaccharide composition. Plant J. 12, 335-345. [DOI] [PubMed] [Google Scholar]
- Ryden, P., Sugimoto-Shirasu, K., Smith, A.C., Findlay, K., Reiter, W.D., and McCann, M.C. (2003). Tensile properties of Arabidopsis cell walls depend on both a xyloglucan cross-linked microfibrillar network and rhamnogalacturonan II-borate complexes. Plant Physiol. 132, 1033-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot, A.A., Assaad, F.F., and Raikhel, N.V. (2000). The Arabidopsis genome: an abundance of soluble N-ethylmaleimide-sensitive factor adaptor protein receptors. Plant Physiol. 124, 1558-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surpin, M. et al. (2003). The VTI family of SNARE proteins is necessary for plant viability and mediates different protein transport pathways. Plant Cell 15, 2885-2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto, D., Jones, D.A., and Hardham, A.R. (2003). GFP-tagging of cell components reveals the dynamics of subcellular re-organization in response to infection of Arabidopsis by oomycete pathogens. Plant J. 33, 775-792. [DOI] [PubMed] [Google Scholar]
- Thordal-Christensen, H. (2003). Fresh insights into processes of non-host resistance. Curr. Opin. Plant Biol. 6, 351-357. [DOI] [PubMed] [Google Scholar]
- Tusher, V.G., Tibshirani, R., and Chu, G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98, 5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, J., and Somerville, S. (2000). Isolation and characterization of powdery mildew-resistant Arabidopsis mutants. Proc. Natl. Acad. Sci. USA 97, 1897-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, R.H., Smith, A.C., Kačuráková, M., Saunders, P.K., Wellner, N., and Waldron, K.W. (2000). The mechanical properties and molecular dynamics of plant cell wall polysaccharides studied by Fourier-Transform infrared spectroscopy. Plant Physiol. 124, 397-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeyen, R.J., Ahlstrand, G.G., and Carver, T.L.W. (1993). X-ray microanalysis of frozen-hydrated, freeze-dried, and critical point dried leaf specimens: determination of soluble and insoluble chemical elements of Erysiphe graminis epidermal cell papilla sites in barley isolines containing ML-o and ml-o alleles. Can. J. Bot. 71, 284-296. [Google Scholar]
- Zeyen, R.J., Bushnell, W.R., Carver, T.L.W., Vance, C.P., Robbins, M.P., and Clark, T.A. (1995). Inhibiting phenylalanine ammonia lyase and cinnamylalcohol dehydrogenase suppresses Mla1 (HR) but not mlo5 (non-HR) barley powdery mildew resistances. Physiol. Mol. Plant Pathol. 47, 119-140. [Google Scholar]
- Zeyen, R.J., and Bushnell, W.R. (1979). Papilla response of barley epidermal cells caused by Erysiphe graminis. Rate and method of deposition determined by microcinematography and transmission microscopy. Can. J. Bot. 57, 898-913. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.