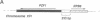Abstract
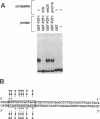
The gene encoding Saccharomyces cerevisiae transcription factor TFIIIA has been found adjacent to RPB6, a gene that specifies a subunit shared by nuclear RNA polymerases. Analysis of DNA upstream of the RPB6 gene revealed an open reading frame that predicts a protein, designated PZF1, with nine C2H2 zinc fingers. The presence of nine C2H2 zinc fingers in PZF1 protein, a hallmark of amphibian TFIIIA proteins, suggested that PZF1 might be a TFIIIA homologue. We found that purified recombinant PZF1 specifically binds the internal control region (ICR) of the 5S rRNA gene in S. cerevisiae. The presence of nine C2H2 zinc fingers, the specific binding to ICR DNA, and the similarity of the predicted molecular mass of PZF1 with that determined for purified yeast TFIIIA, together indicate that PZF1 is TFIIIA. The yeast and amphibian TFIIIA proteins share only a limited number of residues outside of those normally conserved in C2H2 zinc fingers; these conserved residues may provide clues to the sequence specificity of these proteins. The PZF1 gene was found to be single copy, transcribed into a 1.5-kilobase mRNA, and essential for yeast cell viability. Interestingly, the yeast RPB6 and TFIIIA coding sequences are divergently transcribed and are separated by only 233 base pairs, providing the potential for coregulated expression of components of RNA polymerases and the 5S rRNA component of ribosomes.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bieker J. J., Martin P. L., Roeder R. G. Formation of a rate-limiting intermediate in 5S RNA gene transcription. Cell. 1985 Jan;40(1):119–127. doi: 10.1016/0092-8674(85)90315-0. [DOI] [PubMed] [Google Scholar]
- Braun B. R., Riggs D. L., Kassavetis G. A., Geiduschek E. P. Multiple states of protein-DNA interaction in the assembly of transcription complexes on Saccharomyces cerevisiae 5S ribosomal RNA genes. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2530–2534. doi: 10.1073/pnas.86.8.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R. S., Sander C., Argos P. The primary structure of transcription factor TFIIIA has 12 consecutive repeats. FEBS Lett. 1985 Jul 8;186(2):271–274. doi: 10.1016/0014-5793(85)80723-7. [DOI] [PubMed] [Google Scholar]
- Carey M. F., Gerrard S. P., Cozzarelli N. R. Analysis of RNA polymerase III transcription complexes by gel filtration. J Biol Chem. 1986 Mar 25;261(9):4309–4317. [PubMed] [Google Scholar]
- Challice J. M., Segall J. Transcription of the 5 S rRNA gene of Saccharomyces cerevisiae requires a promoter element at +1 and a 14-base pair internal control region. J Biol Chem. 1989 Nov 25;264(33):20060–20067. [PubMed] [Google Scholar]
- Engelke D. R., Ng S. Y., Shastry B. S., Roeder R. G. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell. 1980 Mar;19(3):717–728. doi: 10.1016/s0092-8674(80)80048-1. [DOI] [PubMed] [Google Scholar]
- Evans R. M., Hollenberg S. M. Zinc fingers: gilt by association. Cell. 1988 Jan 15;52(1):1–3. doi: 10.1016/0092-8674(88)90522-3. [DOI] [PubMed] [Google Scholar]
- Fiser-Littell R. M., Duke A. L., Yanchick J. S., Hanas J. S. Deletion of the N-terminal region of Xenopus transcription factor IIIA inhibits specific binding to the 5 S RNA gene. J Biol Chem. 1988 Feb 5;263(4):1607–1610. [PubMed] [Google Scholar]
- Gaskins C. J., Hanas J. S. Sequence variation in transcription factor IIIA. Nucleic Acids Res. 1990 Apr 25;18(8):2117–2123. doi: 10.1093/nar/18.8.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiduschek E. P., Tocchini-Valentini G. P. Transcription by RNA polymerase III. Annu Rev Biochem. 1988;57:873–914. doi: 10.1146/annurev.bi.57.070188.004301. [DOI] [PubMed] [Google Scholar]
- Ginsberg A. M., King B. O., Roeder R. G. Xenopus 5S gene transcription factor, TFIIIA: characterization of a cDNA clone and measurement of RNA levels throughout development. Cell. 1984 Dec;39(3 Pt 2):479–489. doi: 10.1016/0092-8674(84)90455-0. [DOI] [PubMed] [Google Scholar]
- Hanas J. S., Littell R. M., Gaskins C. J., Zebrowski R. Internal deletion mutants of Xenopus transcription factor IIIA. Nucleic Acids Res. 1989 Dec 11;17(23):9861–9870. doi: 10.1093/nar/17.23.9861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartshorne T. A., Blumberg H., Young E. T. Sequence homology of the yeast regulatory protein ADR1 with Xenopus transcription factor TFIIIA. Nature. 1986 Mar 20;320(6059):283–287. doi: 10.1038/320283a0. [DOI] [PubMed] [Google Scholar]
- Honda B. M., Roeder R. G. Association of a 5S gene transcription factor with 5S RNA and altered levels of the factor during cell differentiation. Cell. 1980 Nov;22(1 Pt 1):119–126. doi: 10.1016/0092-8674(80)90160-9. [DOI] [PubMed] [Google Scholar]
- Johnston M., Davis R. W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Aug;4(8):1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga J. T., Carner K. R., Masiarz F. R., Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987 Dec 24;51(6):1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- Klevit R. E., Herriott J. R., Horvath S. J. Solution structure of a zinc finger domain of yeast ADR1. Proteins. 1990;7(3):215–226. doi: 10.1002/prot.340070303. [DOI] [PubMed] [Google Scholar]
- Klevit R. E. Recognition of DNA by Cys2,His2 zinc fingers. Science. 1991 Sep 20;253(5026):1367–1393. doi: 10.1126/science.1896847. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. S., Gippert G. P., Soman K. V., Case D. A., Wright P. E. Three-dimensional solution structure of a single zinc finger DNA-binding domain. Science. 1989 Aug 11;245(4918):635–637. doi: 10.1126/science.2503871. [DOI] [PubMed] [Google Scholar]
- Miller J., McLachlan A. D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardelli J., Gibson T. J., Vesque C., Charnay P. Base sequence discrimination by zinc-finger DNA-binding domains. Nature. 1991 Jan 10;349(6305):175–178. doi: 10.1038/349175a0. [DOI] [PubMed] [Google Scholar]
- Omichinski J. G., Clore G. M., Appella E., Sakaguchi K., Gronenborn A. M. High-resolution three-dimensional structure of a single zinc finger from a human enhancer binding protein in solution. Biochemistry. 1990 Oct 9;29(40):9324–9334. doi: 10.1021/bi00492a004. [DOI] [PubMed] [Google Scholar]
- Pavletich N. P., Pabo C. O. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991 May 10;252(5007):809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Pelham H. R., Brown D. D. A specific transcription factor that can bind either the 5S RNA gene or 5S RNA. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4170–4174. doi: 10.1073/pnas.77.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Párraga G., Horvath S. J., Eisen A., Taylor W. E., Hood L., Young E. T., Klevit R. E. Zinc-dependent structure of a single-finger domain of yeast ADR1. Science. 1988 Sep 16;241(4872):1489–1492. doi: 10.1126/science.3047872. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sakonju S., Brown D. D., Engelke D., Ng S. Y., Shastry B. S., Roeder R. G. The binding of a transcription factor to deletion mutants of a 5S ribosomal RNA gene. Cell. 1981 Mar;23(3):665–669. doi: 10.1016/0092-8674(81)90429-3. [DOI] [PubMed] [Google Scholar]
- Segall J., Matsui T., Roeder R. G. Multiple factors are required for the accurate transcription of purified genes by RNA polymerase III. J Biol Chem. 1980 Dec 25;255(24):11986–11991. [PubMed] [Google Scholar]
- Setzer D. R., Brown D. D. Formation and stability of the 5 S RNA transcription complex. J Biol Chem. 1985 Feb 25;260(4):2483–2492. [PubMed] [Google Scholar]
- Shastry B. S., Ng S. Y., Roeder R. G. Multiple factors involved in the transcription of class III genes in Xenopus laevis. J Biol Chem. 1982 Nov 10;257(21):12979–12986. [PubMed] [Google Scholar]
- Vrana K. E., Churchill M. E., Tullius T. D., Brown D. D. Mapping functional regions of transcription factor TFIIIA. Mol Cell Biol. 1988 Apr;8(4):1684–1696. doi: 10.1128/mcb.8.4.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. K., Weil P. A. Purification and characterization of Saccharomyces cerevisiae transcription factor IIIA. J Biol Chem. 1989 Jan 15;264(2):1092–1099. [PubMed] [Google Scholar]
- Woychik N. A., Liao S. M., Kolodziej P. A., Young R. A. Subunits shared by eukaryotic nuclear RNA polymerases. Genes Dev. 1990 Mar;4(3):313–323. doi: 10.1101/gad.4.3.313. [DOI] [PubMed] [Google Scholar]