ABSTRACT
Adaptation to alternating CO2 concentrations is crucial for all organisms. Carbonic anhydrases—metalloenzymes that have been found in all domains of life—enable fixation of scarce CO2 by accelerating its conversion to bicarbonate and ensure maintenance of cellular metabolism. In fungi and other eukaryotes, the carbonic anhydrase Nce103 has been shown to be essential for growth in air (~0.04% CO2). Expression of NCE103 is regulated in response to CO2 availability. In Saccharomyces cerevisiae, NCE103 is activated by the transcription factor ScCst6, and in Candida albicans and Candida glabrata, it is activated by its homologues CaRca1 and CgRca1, respectively. To identify the kinase controlling Cst6/Rca1, we screened an S. cerevisiae kinase/phosphatase mutant library for the ability to regulate NCE103 in a CO2-dependent manner. We identified ScSch9 as a potential ScCst6-specific kinase, as the sch9Δ mutant strain showed deregulated NCE103 expression on the RNA and protein levels. Immunoprecipitation revealed the binding capabilities of both proteins, and detection of ScCst6 phosphorylation by ScSch9 in vitro confirmed Sch9 as the Cst6 kinase. We could show that CO2-dependent activation of Sch9, which is part of a kinase cascade, is mediated by lipid/Pkh1/2 signaling but not TORC1. Finally, we tested conservation of the identified regulatory cascade in the pathogenic yeast species C. albicans and C. glabrata. Deletion of SCH9 homologues of both species impaired CO2-dependent regulation of NCE103 expression, which indicates a conservation of the CO2 adaptation mechanism among yeasts. Thus, Sch9 is a Cst6/Rca1 kinase that links CO2 adaptation to lipid signaling via Pkh1/2 in fungi.
IMPORTANCE
All living organisms have to cope with alternating CO2 concentrations as CO2 levels range from very low in the atmosphere (0.04%) to high (5% and more) in other niches, including the human body. In fungi, CO2 is sensed via two pathways. The first regulates virulence in pathogenic yeast by direct activation of adenylyl cyclase. The second pathway, although playing a fundamental role in fungal metabolism, is much less understood. Here the transcription factor Cst6/Rca1 controls carbon homeostasis by regulating carbonic anhydrase expression. Upstream signaling in this pathway remains elusive. We identify Sch9 as the kinase controlling Cst6/Rca1 activity in yeast and demonstrate that this pathway is conserved in pathogenic yeast species, which highlights identified key players as potential pharmacological targets. Furthermore, we provide a direct link between adaptation to changing CO2 conditions and lipid/Pkh1/2 signaling in yeast, thus establishing a new signaling cascade central to metabolic adaptation.
INTRODUCTION
Carbon dioxide is key to life on earth. Besides being RuBisCO’s substrate in the carbon-fixing component of the Calvin cycle, it is the final product of cellular respiration (1). Consequently CO2 plays a decisive role as a signaling molecule, and its levels are sensed by organisms as diverse as bacteria, plants, fungi, nematodes, insects, fish, and mammals (2, 3). In fungi, CO2 impacts fundamental biological characteristics, including growth, morphology, and virulence (4). For pathogenic fungi, the ability to adapt to changing CO2 is particularly relevant. When surviving as commensals on skin, fungi are exposed to atmospheric low CO2 concentrations, but when they invade their host, CO2 concentrations can rise to 150-fold higher levels (5% or more) (5–7).
In fungi, CO2 is sensed via two distinct signaling pathways. In the first, HCO3−, which is in equilibrium with membrane-permeable CO2 via action of the catalytic enzyme carbonic anhydrase (CA), directly activates adenylyl cyclases to signal via downstream protein kinase A (PKA) (8). This pathway was shown to trigger the yeast-to-hyphae switch of Candida albicans and capsule biosynthesis of Cryptococcus neoformans, representing well-characterized virulence factors of these major human pathogens (9, 10).
The second pathway is independent of adenylyl cyclase and much less characterized. This pathway directly affects cellular CO2 fixation and HCO3− homeostasis and thus central metabolism. One readout of this pathway is regulation of expression of the β-class fungal CA Nce103p in response to environmental CO2 (11, 12). CAs are ubiquitous zinc-containing metalloenzymes that accelerate the CO2 to HCO3− interconversion and thus impact carboxylation reactions and pH homeostasis (13–15).
Notably yeast CA expression is strongly induced under CO2-limiting atmospheric conditions and downregulated in elevated CO2 (11). The overarching significance of this second CO2-sensing pathway for fungal biology is emphasized by the fact that deletion of yeast CA abolishes growth in low CO2 (16).
CO2-dependent regulation of CA is conserved in Saccharomyces cerevisiae, C. albicans, and Candida glabrata. Some insight into the signaling events mediating CO2-dependent regulation of CA expression in yeast was recently gained by identification of the transcriptional activator regulator of carbonic anhydrase 1 (Rca1). Loss of Rca1 in the pathogenic yeasts C. albicans and C. glabrata and loss of the Rca1 orthologue Cst6 in S. cerevisiae led to a common phenotype displaying lack of CA induction in low CO2 (17, 18). Rca1 and Cst6 belong to the ATF/CREB transcription factor family and have a C-terminal basic leucine zipper domain (19). Comparison of the protein sequence of yeast Rca1/Cst6 revealed 3 conserved serine phosphorylation sites, and loss of serine 124 in the C. albicans orthologue abolished CO2-dependent regulation of downstream CA (17). This supports our hypothesis that Rca1/Cst6 phosphorylation is a critical step in yeast CO2 sensing.
To unravel this novel CO2-sensing pathway, we used S. cerevisiae as a model. Using high-throughput kinase/phosphatase mutant library screening, we identified Sch9 as the Cst6-specific kinase candidate. We confirmed direct interaction and phosphorylation of Cst6 by the Sch9 kinase and show that Cst6/Rca1 signaling is conserved in C. albicans and C. glabrata CO2 sensing. Finally, we provide evidence that Sch9 links CO2 adaptation to lipid signaling via Pkh1/2.
RESULTS
High-throughput screening identifies potential Cst6 kinases.
Previous work suggested that phosphorylation of the Cst6/Rca1 family of transcription factors is key to fungal CO2 sensing, carbon fixation, and metabolism (17). To test this hypothesis, we opted for a high-throughput approach to identify candidate kinases/phosphatases. CO2 regulation in the model organism S. cerevisiae was previously reported to be particularly pronounced in response to CO2 variations of up to 20-fold between atmospheric and high CO2 (11). Using quantitative reverse transcription-PCR (qRT-PCR), we confirmed this finding and found that CA mRNA levels are induced more than 23-fold when cultures are transferred from 5% CO2 (NCE103CO2) to low-CO2 conditions (NCE103air), reaching maximum induction levels at 60 min (23.3 ± 4.9-fold induction) (see Fig. S1A in the supplemental material). Furthermore, we established that induction of NCE103air was reduced in an S. cerevisiae cst6Δ mutant (Fig. 1) and that CST6 expression itself was not affected by CO2 levels (Fig. S1B). Since CA expression by CO2 is conserved in these ascomycete yeasts, we decided to screen an S. cerevisiae kinase and phosphatase deletion mutant library of 155 strains (see Table S1 in the supplemental material) and quantify CO2-dependent gene regulation (see Table S2 in the supplemental material for all data). Deletion of a Cst6 kinase should result in derepression of CA in high CO2 and consequently increased NCE103CO2 expression at levels comparable to growth in air (see Fig. S2 in the supplemental material) (17). Accordingly, mutants with mean NCE103CO2 expression levels twice as high (≥2.0-fold) as the wild type (WT) were considered putative kinase candidates. Of the 155 strains screened, 5 mutants met these criteria: the tpd3Δ, ptp1Δ, bud32Δ, sch9Δ, and ptk2Δ strains (Fig. 1; see Table S2 and Table S3 in the supplemental material). Among those, the sch9Δ mutant showed the highest upregulation of ScNCE103CO2 expression (3.55 ± 1.55-fold), whereas ScNCE103air expression (6.12 ± 2.98-fold) was similar to that of the WT. This deregulation pattern resulted in a low fold change between ScNCE103air and ScNCE103CO2 of 1.91 ± 0.7-fold (Fig. 1; Table S2). Furthermore, the sch9Δ strain did not display general growth defects, and SCH9 encodes a kinase known to be involved in stress response via nutritional sensing in S. cerevisiae and adaptation to hypoxia in C. albicans (20–24), making Sch9 the most probable candidate as Cst6 kinase. To confirm our hypothesis, we analyzed the Nce103 protein level and fluorescence levels in a CA promoter-green fluorescent protein (GFP) fusion, then carried out immunoprecipitation (IP) experiments, and finally showed that Sch9 phosphorylates Cst6.
FIG 1 .
Mutant library screening reveals putative Cst6 kinase. The S. cerevisiae wild type (WT) and mutants were grown in 5% CO2 to the exponential phase and were either maintained at 5% CO2 or transferred to air for 60 min. ScNCE103 expression was measured by qRT-PCR and normalized to WT expression under 5% CO2 (for all data, see Table S2). ScSch9 was the most promising Cst6 kinase candidate. The bars show means ± standard deviations from ≥5 independent experiments. Significance against WT expression under the corresponding condition was calculated with a two-sided t test for unpaired samples and defined as P ≤ 0.05 (*) and P ≤ 0.01 (**).
CO2-dependent time course expression of ScNCE103 and ScCST6. The S. cerevisiae wild type was grown in 5% CO2 to the exponential phase and was either maintained at 5% CO2 or transferred to air. Time course expression of ScNCE103 (A) and ScCST6 (B) was measured by qRT-PCR and normalized to expression at time point 0 in 5% CO2. ScNCE103 mRNA levels increase significantly in air; the highest expression was measured after 60 min. In contrast, ScCST6 expression is independent of CO2 levels. The graphs show means ± standard deviations. Significance against expression in 5% CO2 was defined as P ≤ 0.05 (*) and P ≤ 0.01 (**). n = 3. Download FIG S1, PDF file, 0.1 MB (128.1KB, pdf) .
Copyright © 2017 Pohlers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strains, plasmids, and primers used in the study. Download TABLE S1, PDF file, 0.2 MB (196.4KB, pdf) .
Copyright © 2017 Pohlers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mean ScNCE103 expression and standard deviation (SD) of S. cerevisiae kinase/phosphatase mutants in a changing CO2 environment. Download TABLE S2, PDF file, 0.2 MB (186.4KB, pdf) .
Copyright © 2017 Pohlers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Workflow of mutant library screening for Cst6 kinase identification. S. cerevisiae wild-type and deletion mutant strains were grown in 5% CO2 to exponential phase and were either maintained at 5% CO2 or transferred to air for 60 min. RNA was extracted, and ScNCE103 expression was analyzed by qRT-PCR. Data were normalized to WT expression in 5% CO2. Putative Cst6 kinase deletion mutants were identified according to elevated ScNCE103 expression in 5% CO2 compared to the WT. Download FIG S2, PDF file, 0.04 MB (45.6KB, pdf) .
Copyright © 2017 Pohlers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Kinase candidate genes identified in mutant library screening. Download TABLE S3, PDF file, 0.1 MB (146KB, pdf) .
Copyright © 2017 Pohlers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Loss of Sch9 deregulates CO2 sensing.
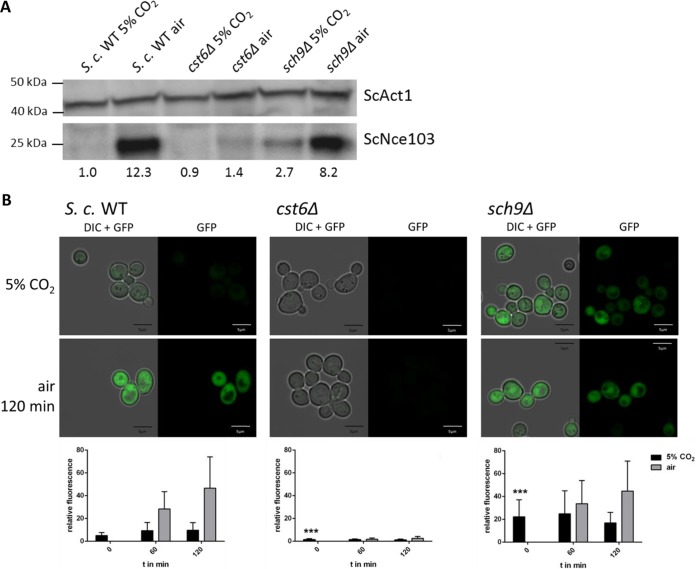
Using a specific antibody against S. cerevisiae CA, we measured protein levels in the sch9Δ mutant cultivated under different CO2 conditions. In contrast to the WT, Nce103 levels in the sch9Δ strain were elevated regardless of the strain being exposed to low or high CO2 (Fig. 2A). Similar to NCE103 gene expression, the Nce103 protein level of the sch9Δ strain was not strongly affected by environmental CO2. Next we transformed WT, cst6Δ and sch9Δ cells with a plasmid containing GFP under control of the ScNCE103 promoter and exposed to low and high concentrations of CO2. GFP expression was analyzed using confocal microscopy (Fig. 2B). GFP fluorescence of WT cells under 5% CO2 was constantly very low, but fluorescence increased after incubation in air (6-fold after 60 min and 9-fold after 120 min) (Fig. 2B). The persistent increase over time is likely due to the stability of GFP. In contrast, the cst6Δ strain showed nearly undetectable fluorescence levels under both conditions. The sch9Δ strain exhibited significantly higher fluorescence in 5% CO2 compared to the WT, but there were also higher differences in intensity between cells. Fluorescence in air was comparable to that of the WT. Noticeably, whereas NCE103 expression levels of the cst6Δ mutant in air and the sch9Δ mutant in 5% CO2 were not significantly different (Fig. 1), the GFP reporter signal for the cst6Δ mutant in air was lower than that for the sch9Δ mutant in high CO2. This could be due to the intrinsically high variation of NCE103 expression in air, which ranged from 3-fold to 10-fold when normalized to expression in 5% CO2. Furthermore, the experimental settings differ, with the GFP reporter being expressed from a plasmid rather than from its native locus. However, results for NCE103 levels in 5% CO2 are consistent in both assays and confirm that loss of Sch9 leads to a loss of CO2-dependent NCE103 repression.
FIG 2 .
Loss of ScSch9 enhances ScNce103 protein level and ScNCE103 promoter activation under 5% CO2. (A) Endogenous ScNce103 levels of the WT and mutants cultivated under 5% CO2 and air conditions were detected by Western blot analysis. The sch9Δ mutant showed elevated ScNce103 in 5% CO2 compared to the WT. Nce103 levels relative to the loading control and normalized to WT in 5% CO2 are indicated below the blot. (B) The S. cerevisiae WT and mutants were transformed with GFP under the control of the ScNCE103 promoter and grown under 5% CO2. Cells were either maintained under this condition or transferred to air for up to 120 min. Fluorescence was detected at the time point of the switch (0 min) and after 60 min and 120 min. Quantification was done by measuring the fluorescence intensity of at least 50 cells per time point. DIC, differential inference contrast. The significance of mutant against WT fluorescence at time point 0, calculated by two-sided t test for unpaired samples, was defined as P ≤ 0.001 (***).
Sch9 is the Cst6 kinase.
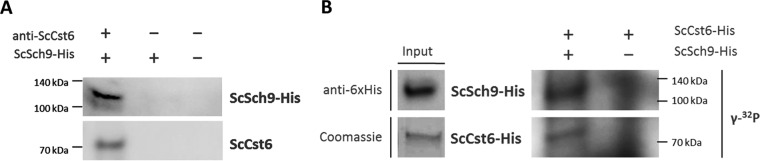
To demonstrate physical interaction between Cst6 and Sch9, immunoprecipitation experiments were performed. Endogenous ScCst6 was precipitated from WT lysate using a specific anti-Cst6 antibody. Immunocomplexes were bound to Sepharose beads and incubated with recombinantly expressed Sch9-His. After extensive washing, Western blot analysis of bound proteins was performed (Fig. 3A). Beads without addition of anti-Cst6 antibody did not show a signal for either Cst6 or Sch9. Addition of Sch9-His without prior binding of Cst6 by antibodies also did not exhibit any signal, which excluded the possibility of unspecific binding. Importantly, when Cst6 was previously bound by anti-Cst6 antibody, we could detect specific binding of Sch9, thus demonstrating the ability of Sch9 to bind its expected substrate, ScCst6.
FIG 3 .
ScSch9 interacts with and phosphorylates ScCst6. (A) ScCst6 was immunopurified from S. cerevisiae using specific antibodies and subsequently incubated with purified ScSch9-His. ScSch9 was only detectable when ScCst6 was previously bound. (B) For in vitro phosphorylation assays, ScSch9-His was immunopurified from S. cerevisiae grown under 5% CO2 and incubated with recombinant ScCst6-His in the presence of [γ-32P]ATP. Incorporation of γ-32P was detected by autoradiography. A clear band for radioactively labeled ScCst6 was visible, indicating phosphorylation of ScCst6 by ScSch9.
To provide further evidence that Sch9 is the Cst6 kinase, we carried out phosphorylation experiments. We overexpressed Sch9-His in S. cerevisiae under control of the inducible ScGAL1 promoter. Sch9 expression was induced by shifting the cells from glucose- to galactose-containing medium. S. cerevisiae-overexpressing cells were cultivated in 5% CO2, assuming that the kinase should be most active under this condition. ScSch9-His was purified from cell lysate via immunoprecipitation. Successful purification was followed by Western blot analysis resulting in a strong single band at ~120 kDa in the bead-bound fraction (Fig. 3B, left panel). Immunocomplexes with bound kinase were used for the radioactive kinase assay. The substrate Cst6-His was recombinantly expressed and purified using affinity chromatography. Purity was verified by Coomassie staining, represented by a single band at ~70 kDa (Fig. 3B, left panel). Sch9-His and Cst6-His were incubated in the presence of [γ-32P]ATP before separation by 4 to 20% SDS-PAGE. By means of autoradiography, incorporation of γ-32P was visualized. In control experiments, where the putative kinase ScSch9 was absent, no definite bands were visible, but a high background corresponding to nonincorporated radioactivity was seen (Fig. 3B, right panel). If Sch9 was present, two distinct bands of ~70 kDa and ~120 kDa were apparent, representing Cst6 and Sch9, respectively. These data demonstrate the phosphorylation of Cst6 by Sch9 in vitro and, furthermore, imply an extensive autophosphorylation of ScSch9, which was previously reported by Huber et al. (25).
Cst6 phosphorylation analysis demonstrates that CO2 signals via phosphorylation of S266.
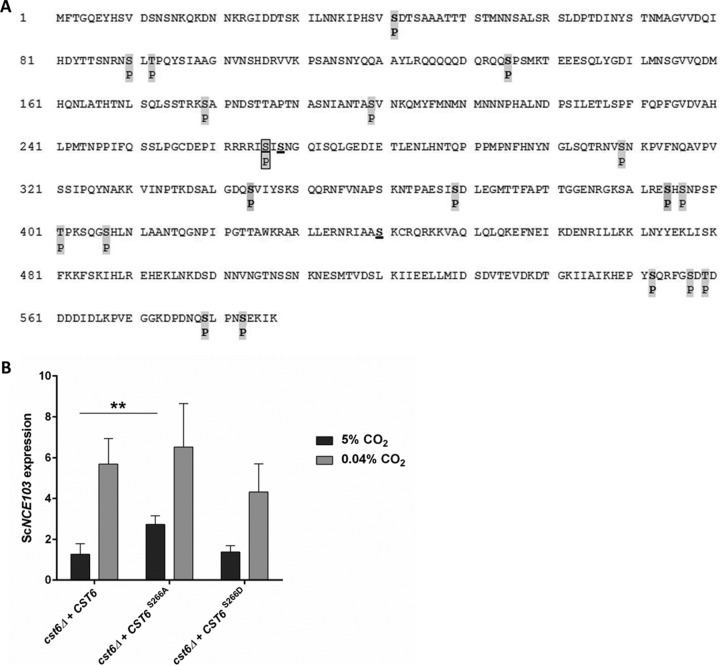
CO2 regulation of fungal CA expression is conserved, and S. cerevisiae Cst6 is an orthologue to Rca1 in C. albicans and C. glabrata. Accordingly we hypothesized fungal CO2-sensing signals via phosphorylation of defined Cst6/Rca1 amino acid residues. Comparison of the Cst6/Rca1 protein sequences identified three conserved serine residues at positions S124, S126, and S222 in C. albicans Rca1 corresponding to S266, S268, and S440 in S. cerevisiae Cst6. Previous work suggested that S124 but not S126 or S222 disrupts CA regulation in C. albicans (17). Hence, we assumed that phosphorylation of S124 regulates fungal CO2 sensing. In order to analyze the phosphorylation probability of the three serine residues, we overexpressed Cst6-His in the S. cerevisiae WT in 5% CO2, assuming that phosphorylation status is at maximum. Cst6-His was affinity purified via immobilized metal ion affinity chromatography (IMAC) and subjected to 4 to 20% SDS-PAGE. Total protein was Coomassie stained, corresponding bands were excised, and trypsin-LysC, AspN, LysargiNase, or GluC digestion was performed to achieve high sequence coverage. In addition, an in-solution digestion with trypsin-LysC and GluC was done to also detect peptides of extended length. Phosphopeptides were enriched using TiO2 and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Our results clearly showed that multiple sites of the Cst6 protein can be subject of phosphorylation. In the WT, we detected 19 different phosphorylation sites in at least two independent experiments (Fig. 4A). Thereof, 11 residues were previously identified by high-throughput approaches (26, 27). We identified another 8 sites, which were unknown to be phosphorylated, printed in boldface in Fig. 4A. In Cst6, the conserved serine residues correspond to S266, S268, and S440. Of those, only serine at position 266 was found to be phosphorylated in S. cerevisiae in 5% CO2, while S268 and S440 were found to be unphosphorylated. In addition, the number of detected phosphorylations suggests the possibility of additional kinases being involved in regulation of Cst6, although we cannot draw any conclusions about biological relevance from the phosphorylation site mapping.
FIG 4 .
ScCst6 phosphorylation at position S266 is crucial for ScNCE103 regulation. (A) ScCst6-His, overexpressed in the WT under 5% CO2, was affinity purified and subjected to SDS-PAGE. Corresponding bands were excised and digested with trypsin-LysC, AspN, GluC, or LysargiNase. Additionally, in-solution digestion for trypsin-LysC and GluC was performed. Phosphorylated peptides were enriched and analyzed by LC-MS/MS. Nineteen different phosphorylation sites, indicated by “P,” were identified in at least 2 of 10 independent experiments. Newly identified phosphorylation sites are shown in boldface, and previously known sites are not in boldface. Underlined are the 3 serine residues that were conserved in S. cerevisiae, C. glabrata, and C. albicans. Among them, only S266 was found to be phosphorylated in the WT, noted within a box. (B) Site-specific mutation of S266 was performed to investigate direct influence on ScNCE103 expression under changing CO2 conditions. Expression levels were normalized to WT ScNCE103 expression in 5% CO2. Changing S266 to alanine (cst6Δ + CST6S266A) showed a significant upregulation of ScNCE103CO2, while ScNCE103air levels were unaltered. This is comparable to expression changes of the sch9Δ mutant and proves the critical role of Cst6 S266 for CA regulation. The bars show means ± standard deviations from 5 independent experiments. Significance against the cst6Δ + CST6 strain, calculated by a two-sided t test for unpaired samples, was defined as P ≤ 0.01 (**).
S266 is the only residue within ScCST6 matching the Sch9 consensus motif R-(R/K)-X-S (28). Hence, we mutated serine at position 266 to alanine (S266A) or aspartic acid (S266D) and cloned these sequences or WT CST6 into the cst6Δ mutant to prove influence on ScNCE103 expression. The cst6Δ mutant reverted with WT CST6 showed ScNCE103 expression comparable to that of the WT (Fig. 4B). Phosphoablative mutation of S266 to A led to a significant upregulation of ScNCE103CO2 (2.73 ± 0.43-fold), while ScNCE103air (6.52 ± 2.12-fold) expression was unaltered. Interestingly, the pattern of expression of the cst6Δ + CST6S266A strain corresponds to the expression pattern of the sch9Δ mutant. This confirms our hypothesis and emphasizes S266 to be phosphorylated in 5% CO2 in order to inhibit NCE103 expression. Phosphomimetic mutation of CST6 (CST6S266D) led to slightly decreased ScNCE103air expression in comparison to WT CST6, but differences were not statistically significant. This might point to alternative regulation mechanisms regulating NCE103 expression, especially under the air condition.
CO2 sensing is conserved in yeast.
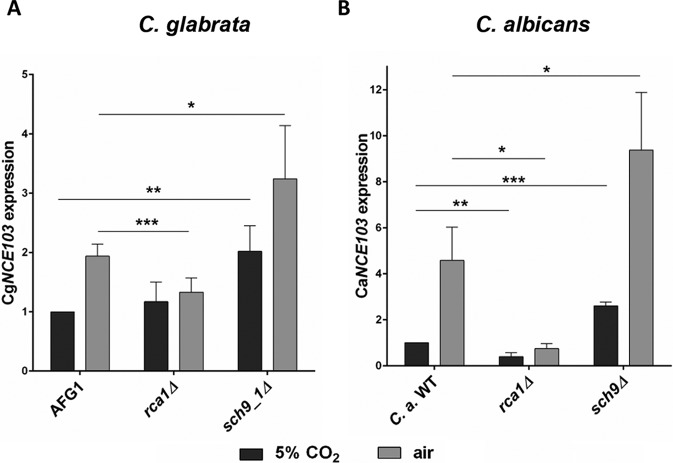
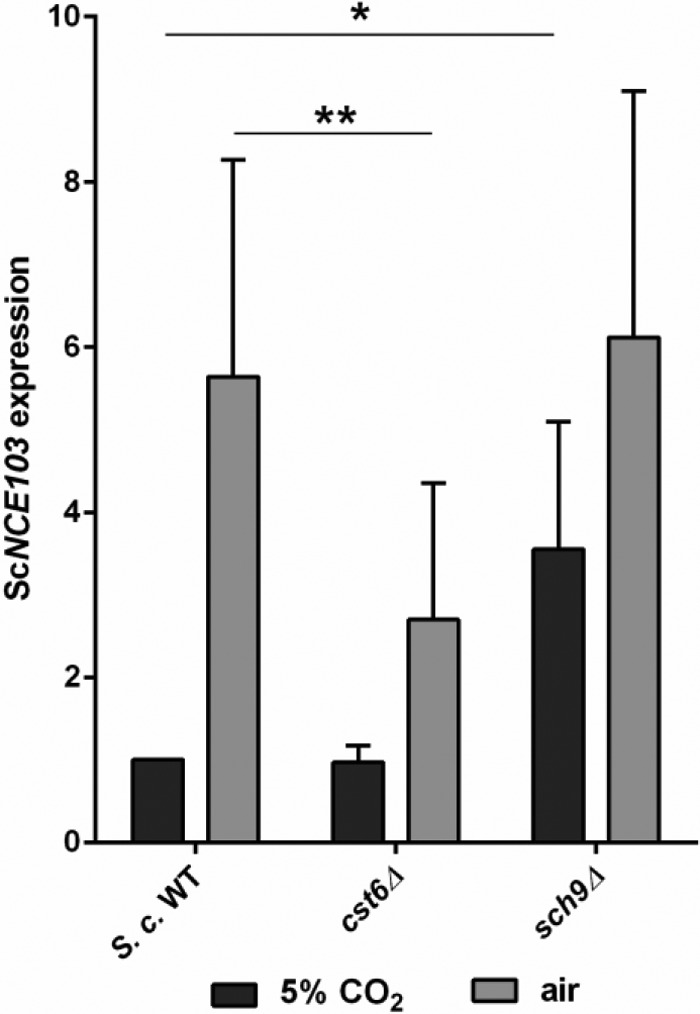
Our findings and previous work suggest that CO2-dependent regulation mediated via Cst6/Rca1 is conserved in some yeast species (17, 18). Due to the closer phylogenetic relationship between S. cerevisiae and C. glabrata, we first analyzed the influence of C. glabrata SCH9 (CgSCH9) on CgNCE103 expression under changing CO2 conditions. Expression of CgNCE103 in 5% CO2 and air after 60 min was quantified by qRT-PCR according to the previously described protocol for S. cerevisiae (Fig. 5A). The C. glabrata control strain, AFG1, revealed a 2-fold increase of CgNCE103air compared to CgNCE103CO2 expression. This increase was completely absent in the rca1Δ mutant. In fact, CgNCE103 expression of the rca1Δ mutant in air was as low as in 5% CO2. This is in contrast to the S. cerevisiae cst6Δ regulation pattern, where at least a small upregulation in air was observed. As expected, deletion of CgSch9 led to significantly elevated CgNCE103CO2 levels (2.02 ± 0.43-fold) compared to the control. Noteworthy and in contrast to the pattern found in S. cerevisiae, CgNCE103air expression was also increased. However, despite these differences, Sch9 clearly affects NCE103 regulation in C. glabrata.
FIG 5 .
SCH9 function in CO2 signaling is conserved in pathogenic yeasts. C. glabrata (A) and C. albicans (B) strains were grown under different CO2 conditions, and expression of NCE103 was measured by qRT-PCR. CgNCE103 expression was normalized to control expression (AFG1), and CaNCE103 expression was normalized to the C. albicans WT, cultivated in 5% CO2. Both species showed comparable regulation mechanisms. Deletion of RCA1 led to significantly decreased NCE103air expression. sch9Δ mutants of C. glabrata and C. albicans showed increased NCE103CO2 expression, consistent with findings for S. cerevisiae. This suggests a conserved role of SCH9 in CO2 signaling in all three yeast species. The bars show means ± standard deviations from ≥4 independent experiments. Significance against control expression, calculated by two-sided t test for unpaired samples, was defined as P ≤ 0.05 (*), P ≤ 0.01 (**), and P ≤ 0.001 (***).
We also verified the influence of SCH9 on C. albicans NCE103 (CaNCE103) using similar protocols (Fig. 5B). The C. albicans WT exhibited a 4.6-fold increase of CaNCE103 expression in cells transferred to air conditions. In comparison, deletion of CaRCA1 resulted in significantly lower CaNCE103air expression. A C. albicans sch9Δ mutant strain (24) showed significantly increased CaNCE103CO2 expression (2.61 ± 0.16-fold), similar to the effects of SCH9 deletion in S. cerevisiae and C. glabrata. Remarkably, also CaNCE103air levels were elevated, which coincides with what we found for C. glabrata, but not for S. cerevisiae. Taken together, these findings clearly indicate a conserved mechanism of CO2 adaptation in pathogenic and nonpathogenic yeast species and emphasize Sch9 is key to CO2 sensing in all three species.
Lipid signaling via Pkh1/2 but not TORC1 regulates fungal CO2 sensing.
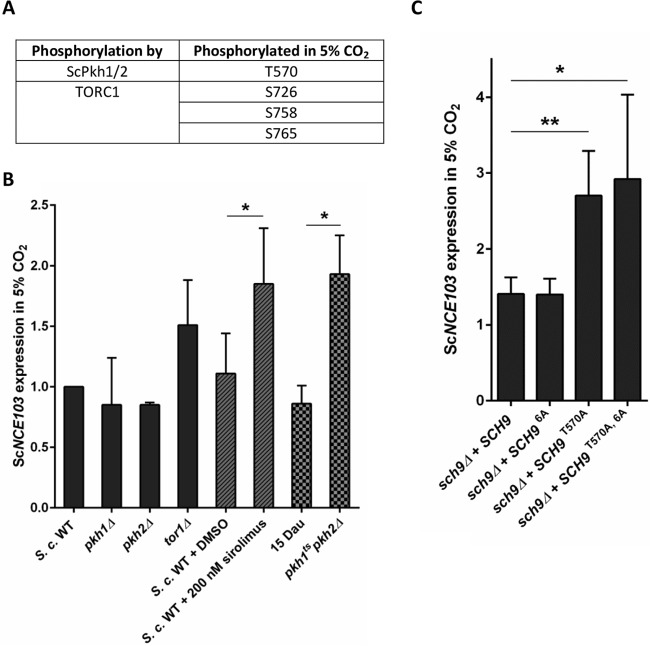
Previous work showed that in S. cerevisiae Sch9 is regulated by target-of-rapamycin (US approved name [USAN] sirolimus)-complex 1 (TORC1) and homologues of the mammalian 3-phosphoinositide-dependent protein kinase (mPDK1), Pkh1 and Pkh2, which phosphorylate Sch9 at specific sites (29, 30). TORC1 phosphorylates at least 6 residues of Sch9 (S711, T723, S726, T737, S758, and S765) (31), while ScPkh1/2 phosphorylates Sch9 at the PDK1 site T570 (32).
Analysis of LC-MS/MS data of overexpressed ScCST6-His allowed us to detect phosphorylated peptides specific for endogenous ScSch9. This enabled analysis of Sch9 phosphorylation in vivo to draw conclusions about putative activators. We detected the PDK1 site T570 and 3 of the phosphorylation sites addressed by TORC1 (S726, S758, and S765) to be phosphorylated in 5% CO2 (Fig. 6A). Therefore, we hypothesized that TOR and lipid signaling is involved in fungal CO2 sensing and that loss or inhibition of Sch9 upstream regulators mimics the sch9Δ mutation. Because activation of Sch9 is especially important under 5% CO2, we concentrated on changes of NCE103CO2 expression (see Table S2 and Table S4 in the supplemental material for NCE103air expression). Notably, the tor1Δ mutant had been included in our initial qRT-PCR screening and showed slightly but not significantly increased CA expression in high CO2 (Fig. 6B). Catalytic subunits of TORC1 are either ScTor1 or ScTor2, and TORC1 is inhibited by sirolimus (33). Since Tor2 is an essential gene in S. cerevisiae, we investigated the impact of TORC1 signaling on fungal CO2 sensing via Sch9 by using sirolimus. Inhibition of TORC1 by sirolimus aggravated the effect of tor1Δ on CA gene expression and led to a significant upregulation of NCE103CO2 (1.85 ± 0.46-fold) (Fig. 6B). However, it did clearly not reach the NCE103CO2 levels of the sch9Δ mutant. PKH1/PKH2 single mutants did not show altered NCE103CO2 expression (Fig. 6B), probably because of Pkh1/2 redundancy in S. cerevisiae (34). As a conventional Pkh1/2 double deletion mutant is known to be lethal in S. cerevisiae (35), we used a temperature-sensitive double mutant (35) to investigate the influence of Pkh1/2-mediated activation of Sch9 on NCE103CO2 expression. Indeed, expression levels of NCE103CO2 increased significantly to 2-fold in the pkh1ts pkh2Δ mutant compared to its control strain, 15 Dau. This implies that Sch9-mediated effects on CA expression could be dependent on both Sch9 regulators TORC1 and Pkh1/2.
FIG 6 .
Pkh1/2 but not TORC1 mediates activation of ScSch9-ScCst6 signaling in 5% CO2. (A) Phosphorylation of ScSch9 in 5% CO2 was detected using LC-MS/MS. Phosphorylation of residues known to be addressed by ScPkh1/2 and TORC1 was detected. (B) ScNCE103CO2 expression of the WT and ScSch9 activator mutants was measured by qRT-PCR and normalized to WT expression. ScTOR1, ScPKH1, or ScPKH2 single mutants did not change ScNCE103CO2 expression significantly. Furthermore, S. cerevisiae WT was treated with 200 nM sirolimus to inactivate TORC1 or DMSO as a solvent control. Sirolimus treatment led to upregulation of ScNCE103CO2 expression, but it does not reach the ScNCE103CO2 levels of the sch9Δ mutant. Because of ScPKH1/2 redundancy, a temperature-sensitive double mutant in the background of 15 Dau was tested and revealed significantly increased ScNCE103CO2 levels. The bars show means ± standard deviations from ≥3 independent experiments. Significance against expression of the WT plus DMSO or 15 Dau, calculated by two-sided t test for unpaired samples, was defined as P ≤ 0.05 (*). (C) ScNCE103 expression of the sch9Δ + SCH9 strain and revertants with SCH9 possessing S/T-to-A mutations of known activation sites was measured in changing CO2 and normalized to WT expression. Integration of SCH9 in the sch9Δ strain restored the WT regulation pattern of ScNCE103. Phosphoablative mutation of the 6 phosphorylation sites known to be addressed by TORC1 (S711A, T723A, S726A, T737A, S758A, and S765A), indicated as SCH96A, showed no expression changes. Mutation of the PKH1/2 site T570 increased ScNCE103CO2 expression significantly, indicating reduced activation of ScSch9. Combination of mutations of phosphorylation sites addressed by both activators, SCH9T570A, 6A, had no additional effect on ScNCE103CO2 expression, but ScNCE103air expression was significantly enhanced. The bars show means ± standard deviations from ≥4 independent experiments. Significance against the sch9Δ + SCH9 strain, calculated by two-sided t test for unpaired samples, was defined as P ≤ 0.05 (*) and P ≤ 0.01 (**).
Mean ScNCE103 expression and standard deviation (SD) of S. cerevisiae SCH9 regulator mutants and S. cerevisiae under rapamycin (USAN sirolimus) treatment. Download TABLE S4, PDF file, 0.1 MB (150KB, pdf) .
Copyright © 2017 Pohlers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To analyze the influence of Sch9 activation by TORC1 and Pkh1/2 more precisely, we investigated NCE103 expression of strains with site-specific mutations of known Sch9 phosphorylation sites addressed by TORC1 and Pkh1/2. As a control, the sch9Δ mutant reverted with WT SCH9 showed NCE103 expression levels in 5% CO2 and air comparable to those of the WT strain. Surprisingly, also phosphoablative mutation of all 6 TORC1 sites (S711A, T723A, S726A, T737A, S758A, and S765A), referred to as SCH96A, resulted in WT-like NCE103 expression levels (Fig. 6C). In contrast, T570A mutation of the PDK1 site resulted in significantly elevated ScNCE103CO2 expression (2.7 ± 0.59), while ScNCE103air expression was not changed. This is comparable with the deregulation pattern of the sch9Δ mutant. Additional S/T-to-A mutation of the 6 TORC1 sites within SCH9 (SCH9T570A, 6A) showed no additive effect on ScNCE103 expression (Fig. 6C).
Taken all together, we concluded that phosphorylation of Sch9 by Pkh1/2 at T570 is crucial for Sch9 activation in a 5% CO2 environment in order to decrease ScNCE103 expression. Although phosphorylation sites addressed by TORC1 were detected in Sch9, CO2-dependent activation of Sch9 and thus CO2 adaptation seems to be independent of TORC1.
DISCUSSION
Sensing and adapting to alterations in ambient CO2 are of paramount importance for all living organisms. Fungi adjust their metabolism to CO2 availability, and CAs are essential in this process. CAs catalyze the reversible conversion of mobile CO2 to HCO3−, and disruption of their function using CA inhibitors or deletion distorts fungal metabolism or abolishes growth altogether.
Expression of the fungal CA NCE103 is regulated by the transcription factors Cst6 and Rca1, which belong to the ATF/CREB family (17). To identify enzymes responsible for Cst6/Rca1 phosphorylation and unravel fungal CO2 sensing, we screened a kinase/phosphatase mutant library in the model organism S. cerevisiae. The library consisted of 111 kinase and 44 phosphatase mutants, and Sch9 was the most promising identified kinase candidate.
Sch9 regulates ribosome biogenesis, cell cycle progression, and life span and is involved in heat shock, osmotic and oxidative stress response by integrating nutrient signals (20–23, 36–38). Several lines of evidence support our hypothesis that Sch9 is indeed the Cst6 kinase and relays fungal CO2 sensing: immunoprecipitation using endogenous Cst6 showed a physical interaction between Cst6 and Sch9. Furthermore, in vitro phosphorylation showed that Sch9 mediates the incorporation of radioactively labeled phosphate groups into Cst6, clearly demonstrating the kinase activity. Finally, the preferred consensus motif for Sch9 phosphorylation events is R-(R/K)-X-S (28).
Our work provides evidence that regulation of NCE103 in S. cerevisiae, C. glabrata, and C. albicans is conserved. Deletion of ScSCH9, CgSCH9, and CaSCH9 affected NCE103 expression in comparable ways as they lead to a significant upregulation of NCE103CO2 (Fig. 1 and 5). Previous work provided some indirect evidence suggesting that S124 in C. albicans may be subject to phosphorylation impacting Rca1/Cst6 function (17). S124 is conserved in S. cerevisiae Cst6 (corresponding position S266) and C. glabrata Rca1 (S387). We now show proof that S266, but not other candidate residues, is phosphorylated in CST6 under elevated CO2 and thus provide a molecular explanation for the conservation of CO2 signaling in yeast (Fig. 4). Furthermore, we showed that phosphorylation of ScCST6 at S266 is crucial for repression of NCE103CO2, because the phosphoablative mutation S266A resulted in increased NCE103CO2 expression. This is in accordance with the results Cottier et al. found for S124 in C. albicans Rca1 (17).
Sch9 is a member of the AGC family (homologous to protein kinases A, G, and C) of proteins. It is conserved in S. cerevisiae, C. glabrata, and C. albicans but is also found in less-related fungal species like Fusarium graminearum (39). In mammalian cells, it has been hypothesized that PKB/Akt is the Sch9 homologue (40), although later studies suggest that Sch9 is more closely related to S6K1 (31), which acts downstream of TORC1. The best-characterized regulator of CA in human cells is HIF-1α, which induces human CA IX expression in response to hypoxia (41, 42). HIF-1α has been found to be regulated by mTOR, which suggested the assumption that Cst6/Rca1 regulation in yeast is also TOR dependent. Surprisingly, our experiments demonstrated that Sch9 phosphorylation by TORC1 does not contribute to CO2-dependent NCE103 regulation (Fig. 6C). Indeed, inhibition of TORC1 by sirolimus showed effects on NCE103CO2 (Fig. 6B), but these effects seem not to be dependent on Sch9 signaling. Furthermore, it was shown that addition of sirolimus is not always similar to inhibition of TORC1 (43). Instead of TORC1, phosphorylation by Pkh1/2 at T570 seems to be the driving force for Sch9 activation in order to repress NCE103 expression under 5% CO2 (Fig. 7). It should be considered that Pkh1/2 might not be the sole activator of Sch9. Deletion of SCH9 results in a 3.55-fold upregulation of ScNCE103CO2, whereas increase following deletion of PKH1/2 was only 1.93-fold. Also T570A mutation of Sch9 leading to unresponsiveness to Pkh1/2 activation did not reach the high NCE103CO2 levels. Thus, there is the possibility that Sch9 is directly influenced by CO2 levels and not only via Pkh1/2 (Fig. 7). An influence of oxygen levels on Sch9 activity should also be considered: it has been previously reported that Sch9 inhibits hyphae formation in the fungal pathogen C. albicans under high-CO2 conditions (>1%) and hypoxia (<10%) but not under either condition alone (24).
FIG 7 .
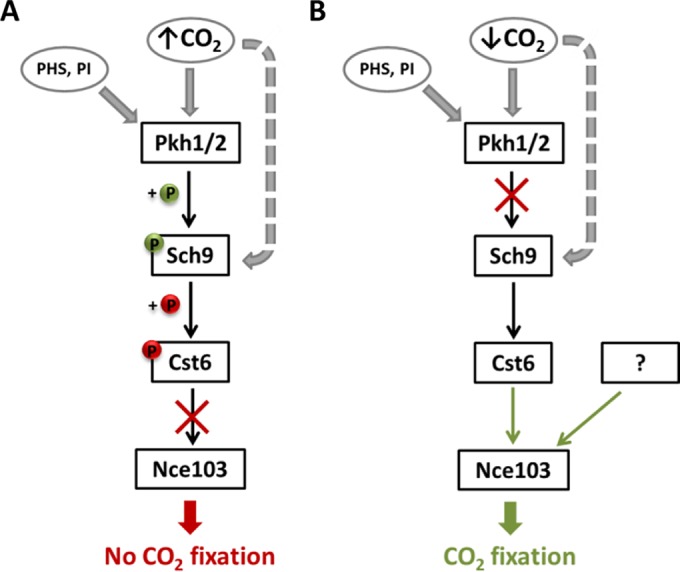
Model of Nce103 regulation under changing CO2 conditions. (A) In the 5% CO2 environment, Sch9 is activated by phosphorylation of T570 via Pkh1/2, whereby Pkh1/2 activity is influenced by phytosphingosine (PHS) and phosphatidylinositol (PI). Moreover, a direct influence of CO2 levels on Sch9 activation is possible. Activated Sch9 phosphorylates the transcription factor Cst6, which is subsequently not able to induce NCE103 expression. This results in very low Nce103 levels, which do not enhance CO2 fixation. (B) Under air conditions, Sch9 is not activated by Pkh1/2 and does not inhibit Cst6. Active Cst6 induces NCE103 expression and enhances the Nce103 protein level, leading to fixation of CO2. Furthermore, Nce103 seems to be induced by another, so far unknown mechanism.
It is noteworthy that another candidate identified in our screen was ScTpd3, which is a part of the protein phosphatase 2A complex. Although no kinase function has been reported, the protein phosphatase 2A complex is linked to nutrient signaling via Tor (44) and is suspected to dephosphorylate Sch9. Overexpression of ScTPD3 and deletion of SCH9 result in comparable phenotypes (45). Thus, ScTpd3 might indeed play a role in NCE103 regulation—maybe as a regulator of ScSch9.
The complexity of NCE103 regulation is further confirmed by our finding that despite the absence of ScCst6 as a transcriptional activator, we could observe upregulation of ScNCE103air resulting in elevated ScNce103p, although this was clearly less pronounced than in the WT (Fig. 2A). Therefore, besides the ScSch9-ScCst6 pathway, there seem to be other mechanisms regulating ScNCE103, maybe another—so far unknown—transcription factor (Fig. 7B). This could be an explanation for the viability of cst6Δ mutants even under air conditions, whereas nce103Δ mutants are only able to grow in elevated CO2 environment (16). Only a moderate induction of NCE103 expression seems to be sufficient for fungal survival, but it will result in growth delay and, probably, affect other cell properties.
In this study, we provide insight into fungal CO2 sensing and show that lipid signaling activating Pkh1/2 regulates metabolic adaptation via Sch9. More work is needed to identify the precise molecular events and complex feedback mechanisms involved in CO2 signal reception and transduction to Sch9. However, the Sch9-Cst6/Rca1 cascade is conserved in yeasts and central to fungal metabolism, and Rca1 has previously been reported to play a role in biofilm formation, which is instrumental in fungal pathogenesis (46). Thus, targeting fungus-specific components in organisms CO2 sensing, such as CA or Rca1/Cst6, constitutes a valid approach to new treatment option.
MATERIALS AND METHODS
Strains, plasmids, and primers.
For the strains, plasmids, and primers used in this study, see Table S1.
ScNCE103 expression analysis.
Exponential cultures of S. cerevisiae WT (BY4741) were cultivated at 37°C as duplicates in 5% CO2 and transferred to air for 10, 30, 60, 90, 120, or 180 min to induce ScNCE103 expression. Pellets were harvested, and RNA was extracted using a hot phenol-chloroform method (47). The quantity of RNA was measured with NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA), and 100 ng/µl RNA was utilized as the template for qRT-PCR using a Brilliant III Ultra Fast SYBR green qRT-PCR kit on a Stratagene Mx3005P (both Agilent Technologies, Santa Clara, CA). Gene expression was calculated by the ΔΔCT method (48) using ScACT1 as the housekeeping gene. The same procedure was done for the screening of S. cerevisiae kinase/phosphatase mutant library (Euroscarf, Frankfurt, Germany). Exponential cultures were transferred to air for 60 min to induce the highest ScNCE103 expression. ScNCE103 expression of mutants was normalized to WT expression in 5% CO2 (Fig. S2). The mutants were tested at least twice, those that showed dysregulation were run 3 to 6 times. Expression of the WT strain was measured in every qRT-PCR as an internal control. For sirolimus experiments, 200 nM sirolimus of a 1 mM stock solution in dimethyl sulfoxide (DMSO) was added. DMSO served as a solvent control. ScNCE103 expression of the 15 Dau and pkh1ts pkh2Δ strains was measured with slight differences in the protocol due to temperature sensitivity of the pkh1ts pkh2Δ mutant. Strains were preincubated overnight in 30°C and 5% CO2 and transferred to 37°C for 3 h to inhibit Pkh1 synthesis. Then samples were transferred to air for another hour to induce ScNCE103.
Expression and purification of His-tagged proteins from E. coli.
S. cerevisiae CST6, NCE103, and SCH9 were synthesized with a C-terminal 6×His tag (GeneArt Gene Synthesis, Thermo Fisher Scientific) and cloned into E. coli expression vector p41. NCE103 and SCH9 were expressed in E. coli strain BL21(DE3) (New England Biolabs, Ipswich, MA). Bacteria were grown in LB medium (10 g/liter NaCl, 5 g/liter yeast extract, 10 g/liter tryptone [pH 7]) with 50 µg/ml ampicillin at 37°C. ScNCE103 expression was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at an optical density at 600 nm (OD600) of 0.2 and cultivated overnight until harvesting. Expression of ScSCH9 was induced at an OD600 of 0.8 for 4 h at room temperature. ScCST6 expression was performed using E. coli Rosetta. Cells were cultivated in LB medium with 50 µg/ml ampicillin and 34 µg/ml chloramphenicol at 37°C until they reached an OD600 of 0.4. Expression was induced with 1 mM IPTG for 4 h at 37°C. Bacteria were harvested, resuspended in purification buffer (25 mM sodium phosphate [pH 7.4], 250 mM NaCl, 10 mM imidazole, 1 mM phenylmethylsulfonyl fluoride [PMSF]), and disrupted by freezing and thawing for a total of 3 rounds. Recombinant proteins were purified by IMAC using Ni-nitrilotriacetic acid (NTA) Sepharose (Biozol, Eching, Germany). His-tagged proteins were eluted with purification buffer containing 250 mM imidazole. For further purification of ScCst6-His, size exclusion chromatography using an Äkta with a Sephadex 200 GL column (both GE Healthcare, Little Chalfont, United Kingdom) was performed. Protein purity was determined by silver staining, and quantity was measured with a bicinchoninic acid (BCA) assay. At least 1 mg purified protein was used to produce specific rabbit antibodies against ScCst6-His and ScNce103-His (Thermo Fisher Scientific).
Western blot analysis.
For detection of ScNce103 levels, 10 ml of S. cerevisiae exponential cultures was cultivated in 5% CO2 or transferred to air for 90 min. Cells were harvested, resuspended in 500 µl yeast lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 0.1% Triton X-100, 1 mM dithiothreitol [DTT], 10% glycerol, PhosStop, Complete EDTA-free protease inhibitor, PMSF), and lysed with glass beads (diameter, 425 to 600 µm) five times for 1 min each with incubation on ice between lyses. Lysates were centrifuged (16,000 × g, 10 min, 4°C), and total protein was measured using a BCA protein assay kit (Thermo Fisher Scientific). After addition of 5× SDS loading dye and heating at 95°C for 5 min, samples (1 mg total protein) were subjected to 4 to 20% SDS-PAGE. Transfer to polyvinylidene difluoride (PVDF) membrane was performed using the Bio-Rad Wet/Tank blotting system for 1 h. Three percent nonfat dry milk in Tris-buffered saline (TBS) was used for membrane blocking and antibody binding. Membranes were incubated with custom-made rabbit anti-ScNce103 (1:1,000 [Thermo Fisher Scientific]) and mouse anti-β-actin (AB8224; 1:1,000 [Abcam, Inc., Cambridge, United Kingdom]) overnight at 4°C with gentle rotation. After incubation with fluorophore-labeled secondary antibodies (Jackson ImmunoResearch, West Grove, PA) for 2 h at 4°C, signals were detected with the FluorChem Q Imager (Alpha Innotec, Kasendorf, Germany). Quantitative analysis was done using the AlphaView software (ProteinSimple).
NCE103 promoter-driven GFP expression.
The S. cerevisiae WT, cst6Δ, and sch9Δ strains were transformed with pRS316 plasmid, containing GFP under control of the ScNCE103 promoter using the lithium-acetate (LiAc) method according to Gietz et al. (49). In brief, yeast cells of the early lag phase were harvested, washed, and resuspended in Tris-EDTA (TE) buffer with 100 mM LiAc. Fifty-microliter cell aliquots were incubated with heat-denatured salmon sperm DNA and 240 µl 50% polyethylene glycol (PEG) 3640, and then DNA was added. Samples were incubated at 30°C for 30 min. After addition of 45 µl DMSO, heat shock at 42°C for 15 min was performed. Centrifuged and resuspended cells were incubated for 2 h at 30°C and washed again before plating. Picked colonies were cultivated at 30°C overnight in 5% CO2 in minimal medium supplemented with histidine, methionine, and leucine. After inoculation of duplicates, one culture was transferred to air, while the second one was further incubated under 5% CO2. Fluorescence was detected with LSM 780 (Zeiss, Jena, Germany) at the time point of the switch from CO2 to air and after 60 and 120 min of incubation. GFP fluorescence of 50 cells per time point was measured using ZEN software (Zeiss), and mean fluorescence was calculated.
Protein expression in S. cerevisiae.
Overexpression of ScCST6-His and ScSCH9-His in S. cerevisiae was achieved using the ScGAL1 promoter. Yeast cells were cultivated in minimal medium, supplemented with histidine, methionine, and leucine at 30°C overnight. Overexpression of ScCST6 and ScSCH9 was induced by a shift from minimal medium to YEP medium (1% yeast extract, 2% peptone) containing 2% raffinose and after overnight culture to YEP with 2% galactose for an additional 6 h. Cells were harvested, resuspended in yeast lysis buffer, and lysed with glass beads as described. Cell extracts were tested by Western blotting for overexpression.
Immunoprecipitation.
Exponential S. cerevisiae WT or ScSCH9-His-overexpressing cultures (50 ml) were harvested, resuspended in 500 µl immunoprecipitation (IP) buffer (20 mM sodium phosphate [pH 7], PhosStop, Complete protease inhibitor EDTA-free, PMSF), and lysed with glass beads as described. Extracts were incubated with either custom-made rabbit anti-Cst6, saturated with 5 µg 6×His tag peptide (Gentaur, Aachen, Germany), or anti-6×His antibody (GTX115045 [GeneTex, Hsinchu City, Taiwan]) overnight at 4°C. Fifty microliters of protein-G Sepharose beads (GE Healthcare) was washed with IP wash buffer (IP buffer plus 500 mM NaCl) and incubated with immunocomplexes for 1 h at 4°C. Beads were centrifuged at 2,500 rpm for 2 min, supernatant was taken off with a syringe and a 25-gauge needle, and beads were resuspended in wash buffer and incubated 1 min with shaking. This washing procedure was repeated four times. For coimmunoprecipitation (co-IP), beads were furthermore incubated with 20 µg recombinant ScSch9-His lysate for 3 h at 4°C. After extensive washing, beads were boiled in SDS sample buffer, subjected to 4 to 20% SDS-PAGE, and analyzed by Western blotting. Rabbit anti-ScCst6 (1:1,000) was used for detection of endogenous ScCst6 and fluorescein isothiocyanate (FITC)-labeled mouse anti-6×His (MA1-81891; 1:250 [Thermo Fisher Scientific]) for detection of 6×His-tagged ScSch9.
Radioactive kinase assay.
ScSch9-His was immunoprecipitated from S. cerevisiae ScSCH9-His-overexpressing culture. After washing of bead-bound immunocomplexes with IP wash buffer, an additional washing step with 1× kinase buffer (phosphate-buffered saline [PBS; pH 7.4], 4 mM MgCl, 10 mM DTT, 20% glycerol, 1 mM PMSF) was performed. One microgram of bacterially expressed and purified ScCst6-His was added to immunocomplexes in 50 µl kinase buffer. After addition of 100 µM ATP and 25 µCi [γ-32P]ATP (BLU002250UC; specific activity, 10 Ci/mmol [PerkinElmer, Waltham, MA]), reaction mixtures were incubated at 30°C for 30 min. The kinase reaction was stopped by heating at 95°C for 5 min in 4× SDS loading dye, and samples were subjected to 4 to 20% SDS-PAGE. Gels were dried, and radioactive incorporation was detected by autoradiography.
LC-MS/MS analysis and protein database search.
To detect CO2-dependent phosphorylation in ScCst6, ScCST6-overexpressing S. cerevisiae cultures were harvested, and ScCst6-His was purified using IMAC. For mass spectrometry analysis, in-gel digestion with trypsin-LysC, AspN, and GluC (all Promega, Madison, WI) or LysargiNase (50) and in-solution digestion with trypsin-LysC and GluC was performed. Peptides were extracted with trifluoroacetic acid and increasing concentrations of acetonitrile (50 to 90%) after in-gel digestion. Phosphopeptide enrichment was carried out using the TiO2 method (TiO2 Spin Tip sample prep kit, Protea Biosciences, Morgantown, WV), and the remaining peptides were purified with C18 minicolumns (C18 Spin Tip sample prep kit [Protea Biosciences, Morgantown, WV]). Dried peptides were solubilized in MS buffer (0.05% trifluoroacetic acid in 2% acetonitrile–98% H2O) and applied to LC-MS/MS analysis, carried out on an Ultimate 3000 nano-rapid-separation liquid chromatography (nano-RSLC) system coupled to a QExactive Plus mass spectrometer (both from Thermo Fisher Scientific). Thermo raw files were processed via the Proteome Discoverer (PD) v1.4.0.288. Tandem mass spectra were searched against the NCBI protein database of S. cerevisiae using the algorithms of Mascot v2.4.1 (Matrix Science, Inc., London, United Kingdom), Sequest HT, and MS Amanda. For further information, see Text S1 in the supplemental material.
Supplemental methods. Download TEXT S1, PDF file, 0.1 MB (13.8KB, pdf) .
Copyright © 2017 Pohlers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Site-specific mutagenesis of ScCST6 and ScSCH9.
Phosphoablative (S266A) and phosphomimetic (S266D) sequences of ScCST6 (GeneArt Gene Synthesis) were inserted in the pRS316 plasmid using PacI/AscI restriction sites. The ScCST6 promoter was cloned in pRS316 using SacI/PacI. After transformation of the S. cerevisiae cst6Δ mutant, ScNCE103 expression in 5% and 0.04% CO2 was measured as described and normalized to WT expression in 5% CO2. For construction of ScSCH9 with S/T-to-A mutation of all 6 TORC1 sites (SCH96A), the sequence of SCH9T570A, 6A was used as the template. A570 was remutated to T570 using QuikChange II site-directed mutagenesis kit (Agilent Technologies) with mutation-specific primers. The plasmid with the corrected sequence was checked via sequencing and used for transformation of the sch9Δ mutant.
Construction of the C. glabrata sch9Δ mutant.
Deletion of CgSCH9 was achieved by replacement of the CgSCH9 open reading frame (ORF) with a NAT1 cassette, which mediates resistance against nourseothricin. NAT1 was amplified by PCR with EcoRI and SacI restriction sites and cloned into pBluescript. A homologous region upstream of the CgSCH9 ORF with a length of 1,000 bp was amplified by colony PCR and cloned into pBluescript using KpnI/XhoI. The same was done for a 1,000-bp fragment located downstream of CgSCH9 ORF using restriction sites SacII and SacI. The whole construct, the NAT1 cassette flanked by homologous regions, was restricted with KpnI/SacI and purified in order to transform C. glabrata. AFG1, a strain lacking LIG4, was used for transformation because of its increased efficiency of homologous recombination (51). Transformation was performed using the lithium-acetate method, with cells plated on YPD with 100 µg/ml ClonNAT, and positive clones were selected. Integration of the NAT1 cassette and deletion of CgSCH9 were checked by colony PCR with specific internal primers. Furthermore, deletion mutants were verified using external primers (see Fig. S3 in the supplemental material).
Verification of C. glabrata sch9 deletion. (A) Insertion of NAT1 and deletion of CgSCH9 were analyzed by colony PCR of the untransformed parental strain (AFG1) and sch9 mutant. For amplification of NAT1, internal primers G1-CgSCH9 and X2-NAT1 were used for amplification of CgSCH9 G1-CgSCH9 and I2-CgSCH9. The strain AFG1 showed no signal for NAT1 but did show signal for SCH9. In contrast, the sch9 deletion mutant exhibited a PCR product for NAT1 but not for SCH9. As a marker, the GeneRuler 1-kb DNA ladder (Thermo Fisher Scientific) was used in lane 1. (B) For verification of the correct position of NAT1 cassette insertion, PCR with external primers G1-CgSCH9 and G4-CgSCH9 was used; these primers bind outside the inserted cassette. If the SCH9 ORF is present, a PCR product of 4,385 bp occurs. If the SCH9 ORF is replaced by NAT1, a shift to 3,463 bp is visible. As a marker, the GeneRuler 1-kb DNA ladder (Thermo Fisher Scientific) was used in lane 1. Download FIG S3, PDF file, 0.2 MB (233.6KB, pdf) .
Copyright © 2017 Pohlers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental references. Download TEXT S2, PDF file, 0.1 MB (130.9KB, pdf) .
Copyright © 2017 Pohlers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
NCE103 expression measurement of C. glabrata and C. albicans.
Measurement of CgNCE103 and CaNCE103 expression was performed according to the protocol for ScNCE103 expression described in the first section. As housekeeping genes, CgACT1 for measurement of CgNCE103 and CaACT1 for measurement of CaNCE103 were used.
Statistical analysis.
Data are presented as arithmetic means ± standard deviation, and statistical significance (P < 0.05) was calculated using a two-sided t test for unpaired samples.
ACKNOWLEDGMENTS
We thank Robbie Loewith (University of Geneva, Switzerland) for providing ScSCH9 plasmids pRS416-promScSCH9-ScSCH9-3HA, pRS416-promScSCH9-ScSCH9-3HA (T570A), pGEX-6P-1 ScSCH9-3HA (S711A, T723A, S726A, T737A, S758A, S765A), and pRS416-promScSCH9-ScSCH9-3HA (T570A, S711A, T723A, S726A, T737A, S758A, S765A). Furthermore, we are grateful to Cindy Reichmann for expert technical assistance and Ilaria Granata for help with the library screening.
This work was funded by the Deutsche Forschungsgemeinschaft DFG (KU1540/3-1 to O. Kurzai). Work in the lab of A.B., O. Kniemeyer, and T.K. was funded by the Deutsche Forschungsgemeinschaft within the Collaborative Research Center TR124 (project A1 and Z2).
S.P., R.M., D.H., T.K., and F.H. performed experiments and analyzed data. O. Kniemeyer, H.P.S., A.B., P.V.D., J.F.E., F.M., and O. Kurzai contributed strains and reagents and interpreted data. F.M. and O. Kurzai designed experiments. S.P., F.M., and O. Kurzai wrote the manuscript.
Footnotes
Citation Pohlers S, Martin R, Krüger T, Hellwig D, Hänel F, Kniemeyer O, Saluz HP, Van Dijck P, Ernst JF, Brakhage A, Mühlschlegel FA, Kurzai O. 2017. Lipid signaling via Pkh1/2 regulates fungal CO2 sensing through the kinase Sch9. mBio 8:e02211-16. https://doi.org/10.1128/mBio.02211-16.
REFERENCES
- 1.Hetherington AM, Raven JA. 2005. The biology of carbon dioxide. Curr Biol 15:R406–R410. doi: 10.1016/j.cub.2005.05.042. [DOI] [PubMed] [Google Scholar]
- 2.Cummins EP, Selfridge AC, Sporn PH, Sznajder JI, Taylor CT. 2014. Carbon dioxide-sensing in organisms and its implications for human disease. Cell Mol Life Sci 71:831–845. doi: 10.1007/s00018-013-1470-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharabi K, Lecuona E, Helenius IT, Beitel GJ, Sznajder JI, Gruenbaum Y. 2009. Sensing, physiological effects and molecular response to elevated CO2 levels in eukaryotes. J Cell Mol Med 13:4304–4318. doi: 10.1111/j.1582-4934.2009.00952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bahn YS, Mühlschlegel FA. 2006. CO2 sensing in fungi and beyond. Curr Opin Microbiol 9:572–578. doi: 10.1016/j.mib.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Frame GW, Strauss WG, Maibach HI. 1972. Carbon dioxide emission of the human arm and hand. J Invest Dermatol 59:155–159. doi: 10.1111/1523-1747.ep12625939. [DOI] [PubMed] [Google Scholar]
- 6.Polke M, Hube B, Jacobsen ID. 2015. Candida survival strategies. Adv Appl Microbiol 91:139–235. doi: 10.1016/bs.aambs.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 7.Brunke S, Hube B. 2013. Two unlike cousins: Candida albicans and C. glabrata infection strategies. Cell Microbiol 15:701–708. doi: 10.1111/cmi.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klengel T, Liang WJ, Chaloupka J, Ruoff C, Schröppel K, Naglik JR, Eckert SE, Mogensen EG, Haynes K, Tuite MF, Levin LR, Buck J, Mühlschlegel FA. 2005. Fungal adenylyl cyclase integrates CO2 sensing with cAMP signaling and virulence. Curr Biol 15:2021–2026. doi: 10.1016/j.cub.2005.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rocha CR, Schröppel K, Harcus D, Marcil A, Dignard D, Taylor BN, Thomas DY, Whiteway M, Leberer E. 2001. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol Biol Cell 12:3631–3643. doi: 10.1091/mbc.12.11.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alspaugh JA, Pukkila-Worley R, Harashima T, Cavallo LM, Funnell D, Cox GM, Perfect JR, Kronstad JW, Heitman J. 2002. Adenylyl cyclase functions downstream of the Galpha protein Gpa1 and controls mating and pathogenicity of Cryptococcus neoformans. Eukaryot Cell 1:75–84. doi: 10.1128/EC.1.1.75-84.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amoroso G, Morell-Avrahov L, Müller D, Klug K, Sültemeyer D. 2005. The gene NCE103 (YNL036w) from Saccharomyces cerevisiae encodes a functional carbonic anhydrase and its transcription is regulated by the concentration of inorganic carbon in the medium. Mol Microbiol 56:549–558. doi: 10.1111/j.1365-2958.2005.04560.x. [DOI] [PubMed] [Google Scholar]
- 12.Aguilera J, Petit T, de Winde JH, Pronk JT. 2005. Physiological and genome-wide transcriptional responses of Saccharomyces cerevisiae to high carbon dioxide concentrations. FEMS Yeast Res 5:579–593. doi: 10.1016/j.femsyr.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 13.Supuran CT. 2008. Carbonic anhydrases—an overview. Curr Pharm Des 14:603–614. doi: 10.2174/138161208783877884. [DOI] [PubMed] [Google Scholar]
- 14.Aguilera J, Van Dijken JP, De Winde JH, Pronk JT. 2005. Carbonic anhydrase (Nce103p): an essential biosynthetic enzyme for growth of Saccharomyces cerevisiae at atmospheric carbon dioxide pressure. Biochem J 391:311–316. doi: 10.1042/BJ20050556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elleuche S, Pöggeler S. 2010. Carbonic anhydrases in fungi. Microbiology 156:23–29. doi: 10.1099/mic.0.032581-0. [DOI] [PubMed] [Google Scholar]
- 16.Götz R, Gnann A, Zimmermann FK. 1999. Deletion of the carbonic anhydrase-like gene NCE103 of the yeast Saccharomyces cerevisiae causes an oxygen-sensitive growth defect. Yeast 15:855–864. doi:. [DOI] [PubMed] [Google Scholar]
- 17.Cottier F, Raymond M, Kurzai O, Bolstad M, Leewattanapasuk W, Jiménez-López C, Lorenz MC, Sanglard D, Váchová L, Pavelka N, Palková Z, Mühlschlegel FA. 2012. The bZIP transcription factor Rca1p is a central regulator of a novel CO(2) sensing pathway in yeast. PLoS Pathog 8:e1002485. doi: 10.1371/journal.ppat.1002485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cottier F, Leewattanapasuk W, Kemp LR, Murphy M, Supuran CT, Kurzai O, Mühlschlegel FA. 2013. Carbonic anhydrase regulation and CO(2) sensing in the fungal pathogen Candida glabrata involves a novel Rca1p ortholog. Bioorg Med Chem 21:1549–1554. doi: 10.1016/j.bmc.2012.05.053. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Gimeno MA, Struhl K. 2000. Aca1 and Aca2, ATF/CREB activators in Saccharomyces cerevisiae, are important for carbon source utilization but not the response to stress. Mol Cell Biol 20:4340–4349. doi: 10.1128/MCB.20.12.4340-4349.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morano KA, Thiele DJ. 1999. The Sch9 protein kinase regulates Hsp90 chaperone complex signal transduction activity in vivo. EMBO J 18:5953–5962. doi: 10.1093/emboj/18.21.5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pascual-Ahuir A, Proft M. 2007. The Sch9 kinase is a chromatin-associated transcriptional activator of osmostress-responsive genes. EMBO J 26:3098–3108. doi: 10.1038/sj.emboj.7601756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weinberger M, Mesquita A, Caroll T, Marks L, Yang H, Zhang Z, Ludovico P, Burhans WC. 2010. Growth signaling promotes chronological aging in budding yeast by inducing superoxide anions that inhibit quiescence. Aging (Albany, NY) 2:709–726. doi: 10.18632/aging.100215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qie B, Lyu Z, Lyu L, Liu J, Gao X, Liu Y, Duan W, Zhang N, Du L, Liu K. 2015. Sch9 regulates intracellular protein ubiquitination by controlling stress responses. Redox Biol 5:290–300. doi: 10.1016/j.redox.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stichternoth C, Fraund A, Setiadi E, Giasson L, Vecchiarelli A, Ernst JF. 2011. Sch9 kinase integrates hypoxia and CO2 sensing to suppress hyphal morphogenesis in Candida albicans. Eukaryot Cell 10:502–511. doi: 10.1128/EC.00289-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huber A, Bodenmiller B, Uotila A, Stahl M, Wanka S, Gerrits B, Aebersold R, Loewith R. 2009. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev 23:1929–1943. doi: 10.1101/gad.532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swaney DL, Beltrao P, Starita L, Guo A, Rush J, Fields S, Krogan NJ, Villén J. 2013. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat Methods 10:676–682. doi: 10.1038/nmeth.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Albuquerque CP, Smolka MB, Payne SH, Bafna V, Eng J, Zhou H. 2008. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteomics 7:1389–1396. doi: 10.1074/mcp.M700468-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kennelly PJ, Krebs EG. 1991. Consensus sequences as substrate specificity determinants for protein kinases and protein phosphatases. J Biol Chem 266:15555–15558. [PubMed] [Google Scholar]
- 29.Jacinto E, Lorberg A. 2008. TOR regulation of AGC kinases in yeast and mammals. Biochem J 410:19–37. doi: 10.1042/BJ20071518. [DOI] [PubMed] [Google Scholar]
- 30.Liu K, Zhang X, Lester RL, Dickson RC. 2005. The sphingoid long chain base phytosphingosine activates AGC-type protein kinases in Saccharomyces cerevisiae including Ypk1, Ypk2, and Sch9. J Biol Chem 280:22679–22687. doi: 10.1074/jbc.M502972200. [DOI] [PubMed] [Google Scholar]
- 31.Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, Wanke V, Anrather D, Ammerer G, Riezman H, Broach JR, De Virgilio C, Hall MN, Loewith R. 2007. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell 26:663–674. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 32.Voordeckers K, Kimpe M, Haesendonckx S, Louwet W, Versele M, Thevelein JM. 2011. Yeast 3-phosphoinositide-dependent protein kinase-1 (PDK1) orthologs Pkh1-3 differentially regulate phosphorylation of protein kinase A (PKA) and the protein kinase B (PKB)/S6K ortholog Sch9. J Biol Chem 286:22017–22027. doi: 10.1074/jbc.M110.200071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN. 2002. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell 10:457–468. doi: 10.1016/S1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 34.Casamayor A, Torrance PD, Kobayashi T, Thorner J, Alessi DR. 1999. Functional counterparts of mammalian protein kinases PDK1 and SGK in budding yeast. Curr Biol 9:186–197. doi: 10.1016/S0960-9822(99)80088-8. [DOI] [PubMed] [Google Scholar]
- 35.Inagaki M, Schmelzle T, Yamaguchi K, Irie K, Hall MN, Matsumoto K. 1999. PDK1 homologs activate the Pkc1-mitogen-activated protein kinase pathway in yeast. Mol Cell Biol 19:8344–8352. doi: 10.1128/MCB.19.12.8344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huber A, French SL, Tekotte H, Yerlikaya S, Stahl M, Perepelkina MP, Tyers M, Rougemont J, Beyer AL, Loewith R. 2011. Sch9 regulates ribosome biogenesis via Stb3, Dot6 and Tod6 and the histone deacetylase complex RPD3L. EMBO J 30:3052–3064. doi: 10.1038/emboj.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang X, Liu J, Dickson RC. 2012. Down-regulating sphingolipid synthesis increases yeast lifespan. PLoS Genet 8:e1002493. doi: 10.1371/journal.pgen.1002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M. 2004. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev 18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen D, Wang Y, Zhou X, Wang Y, Xu JR. 2014. The Sch9 kinase regulates conidium size, stress responses, and pathogenesis in Fusarium graminearum. PLoS One 9:e105811. doi: 10.1371/journal.pone.0105811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sobko A. 2006. Systems biology of AGC kinases in fungi. Sci STKE 2006:re9. doi: 10.1126/stke.3522006re9. [DOI] [PubMed] [Google Scholar]
- 41.Wykoff CC, Beasley NJ, Watson PH, Turner KJ, Pastorek J, Sibtain A, Wilson GD, Turley H, Talks KL, Maxwell PH, Pugh CW, Ratcliffe PJ, Harris AL. 2000. Hypoxia-inducible expression of tumor-associated carbonic anhydrases. Cancer Res 60:7075–7083. [PubMed] [Google Scholar]
- 42.Schofield CJ, Ratcliffe PJ. 2005. Signalling hypoxia by HIF hydroxylases. Biochem Biophys Res Commun 338:617–626. doi: 10.1016/j.bbrc.2005.08.111. [DOI] [PubMed] [Google Scholar]
- 43.Thoreen CC, Sabatini DM. 2009. Rapamycin inhibits mTORC1, but not completely. Autophagy 5:725–726. doi: 10.4161/auto.5.5.8504. [DOI] [PubMed] [Google Scholar]
- 44.Zabrocki P, Van Hoof C, Goris J, Thevelein JM, Winderickx J, Wera S. 2002. Protein phosphatase 2A on track for nutrient-induced signalling in yeast. Mol Microbiol 43:835–842. doi: 10.1046/j.1365-2958.2002.02786.x. [DOI] [PubMed] [Google Scholar]
- 45.Sugajska E, Swiatek W, Zabrocki P, Geyskens I, Thevelein JM, Zolnierowicz S, Wera S. 2001. Multiple effects of protein phosphatase 2A on nutrient-induced signalling in the yeast Saccharomyces cerevisiae. Mol Microbiol 40:1020–1026. doi: 10.1046/j.1365-2958.2001.02449.x. [DOI] [PubMed] [Google Scholar]
- 46.Riera M, Mogensen E, d’Enfert C, Janbon G. 2012. New regulators of biofilm development in Candida glabrata. Res Microbiol 163:297–307. doi: 10.1016/j.resmic.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 47.Martin R, Moran GP, Jacobsen ID, Heyken A, Domey J, Sullivan DJ, Kurzai O, Hube B. 2011. The Candida albicans-specific gene EED1 encodes a key regulator of hyphal extension. PLoS One 6:e18394. doi: 10.1371/journal.pone.0018394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gietz D, St Jean A, Woods RA, Schiestl RH. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huesgen PF, Lange PF, Rogers LD, Solis N, Eckhard U, Kleifeld O, Goulas T, Gomis-Rüth FX, Overall CM. 2015. LysargiNase mirrors trypsin for protein C-terminal and methylation-site identification. Nat Methods 12:55–58. doi: 10.1038/nmeth.3177. [DOI] [PubMed] [Google Scholar]
- 51.Cen Y, Fiori A, Van Dijck P. 2015. Deletion of the DNA ligase IV gene in Candida glabrata significantly increases gene-targeting efficiency. Eukaryot Cell 14:783–791. doi: 10.1128/EC.00281-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CO2-dependent time course expression of ScNCE103 and ScCST6. The S. cerevisiae wild type was grown in 5% CO2 to the exponential phase and was either maintained at 5% CO2 or transferred to air. Time course expression of ScNCE103 (A) and ScCST6 (B) was measured by qRT-PCR and normalized to expression at time point 0 in 5% CO2. ScNCE103 mRNA levels increase significantly in air; the highest expression was measured after 60 min. In contrast, ScCST6 expression is independent of CO2 levels. The graphs show means ± standard deviations. Significance against expression in 5% CO2 was defined as P ≤ 0.05 (*) and P ≤ 0.01 (**). n = 3. Download FIG S1, PDF file, 0.1 MB (128.1KB, pdf) .
Copyright © 2017 Pohlers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strains, plasmids, and primers used in the study. Download TABLE S1, PDF file, 0.2 MB (196.4KB, pdf) .
Copyright © 2017 Pohlers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mean ScNCE103 expression and standard deviation (SD) of S. cerevisiae kinase/phosphatase mutants in a changing CO2 environment. Download TABLE S2, PDF file, 0.2 MB (186.4KB, pdf) .
Copyright © 2017 Pohlers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Workflow of mutant library screening for Cst6 kinase identification. S. cerevisiae wild-type and deletion mutant strains were grown in 5% CO2 to exponential phase and were either maintained at 5% CO2 or transferred to air for 60 min. RNA was extracted, and ScNCE103 expression was analyzed by qRT-PCR. Data were normalized to WT expression in 5% CO2. Putative Cst6 kinase deletion mutants were identified according to elevated ScNCE103 expression in 5% CO2 compared to the WT. Download FIG S2, PDF file, 0.04 MB (45.6KB, pdf) .
Copyright © 2017 Pohlers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Kinase candidate genes identified in mutant library screening. Download TABLE S3, PDF file, 0.1 MB (146KB, pdf) .
Copyright © 2017 Pohlers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mean ScNCE103 expression and standard deviation (SD) of S. cerevisiae SCH9 regulator mutants and S. cerevisiae under rapamycin (USAN sirolimus) treatment. Download TABLE S4, PDF file, 0.1 MB (150KB, pdf) .
Copyright © 2017 Pohlers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental methods. Download TEXT S1, PDF file, 0.1 MB (13.8KB, pdf) .
Copyright © 2017 Pohlers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Verification of C. glabrata sch9 deletion. (A) Insertion of NAT1 and deletion of CgSCH9 were analyzed by colony PCR of the untransformed parental strain (AFG1) and sch9 mutant. For amplification of NAT1, internal primers G1-CgSCH9 and X2-NAT1 were used for amplification of CgSCH9 G1-CgSCH9 and I2-CgSCH9. The strain AFG1 showed no signal for NAT1 but did show signal for SCH9. In contrast, the sch9 deletion mutant exhibited a PCR product for NAT1 but not for SCH9. As a marker, the GeneRuler 1-kb DNA ladder (Thermo Fisher Scientific) was used in lane 1. (B) For verification of the correct position of NAT1 cassette insertion, PCR with external primers G1-CgSCH9 and G4-CgSCH9 was used; these primers bind outside the inserted cassette. If the SCH9 ORF is present, a PCR product of 4,385 bp occurs. If the SCH9 ORF is replaced by NAT1, a shift to 3,463 bp is visible. As a marker, the GeneRuler 1-kb DNA ladder (Thermo Fisher Scientific) was used in lane 1. Download FIG S3, PDF file, 0.2 MB (233.6KB, pdf) .
Copyright © 2017 Pohlers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental references. Download TEXT S2, PDF file, 0.1 MB (130.9KB, pdf) .
Copyright © 2017 Pohlers et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.