Abstract
Autophagy is an intracellular process for vacuolar degradation of cytoplasmic components. Thus far, plant autophagy has been studied primarily using morphological analyses. A recent genome-wide search revealed significant conservation among autophagy genes (ATGs) in yeast and plants. It has not been proved, however, that Arabidopsis thaliana ATG genes are required for plant autophagy. To evaluate this requirement, we examined the ubiquitination-like Atg8 lipidation system, whose component genes are all found in the Arabidopsis genome. In Arabidopsis, all nine ATG8 genes and two ATG4 genes were expressed ubiquitously and were induced further by nitrogen starvation. To establish a system monitoring autophagy in whole plants, we generated transgenic Arabidopsis expressing each green fluorescent protein–ATG8 fusion (GFP-ATG8). In wild-type plants, GFP-ATG8s were observed as ring shapes in the cytoplasm and were delivered to vacuolar lumens under nitrogen-starved conditions. By contrast, in a T-DNA insertion double mutant of the ATG4s (atg4a4b-1), autophagosomes were not observed, and the GFP-ATG8s were not delivered to the vacuole under nitrogen-starved conditions. In addition, we detected autophagic bodies in the vacuoles of wild-type roots but not in those of atg4a4b-1 in the presence of concanamycin A, a V-ATPase inhibitor. Biochemical analyses also provided evidence that autophagy in higher plants requires ATG proteins. The phenotypic analysis of atg4a4b-1 indicated that plant autophagy contributes to the development of a root system under conditions of nutrient limitation.
INTRODUCTION
For plants, which are nonmotile multicellular organisms, protein degradation is important to adapt to various severe environmental conditions, such as nutrient deprivation (Vierstra, 1996; Marty, 1999). Autophagy is the major system responsible for bulk protein degradation in the vacuole/lysosome. There are two types of autophagy, called microautophagy and macroautophagy (Klionsky and Ohsumi, 1999). In microautophagy, cytoplasmic components are engulfed by an invaginated vacuolar membrane. During macroautophagy, bulk cytosolic constituents and organelles are sequestered into a double-membrane structure called an autophagosome. The outer membrane of the autophagosome then fuses to the vacuolar membrane, thus delivering the inner membrane structure, the autophagic body, into the vacuolar lumen for degradation. So far, in plants, both microautophagy and macroautophagy have been reported (Matile, 1975; Van Der Wilden et al., 1980; Herman et al., 1981; Aubert et al., 1996; Moriyasu and Ohsumi, 1996; Robinson et al., 1998; Swanson et al., 1998; Rojo et al., 2001; Toyooka et al., 2001).
Although most studies of autophagy have been based on morphological observations, genetic screens in the yeast Saccaromyces cerevisiae have expanded the molecular dissection of autophagy. We, together with Thumm's group, have previously isolated autophagy-defective mutants and identified 16 autophagy genes (ATGs) essential for autophagosome formation (Tsukada and Ohsumi, 1993; Thumm et al., 1994; Barth et al., 2001). Upon recent comprehensive genome sequencing, significant conservation of autophagy genes among yeast, slime mold, nematode, fruit fly, mammals, and plants was revealed, indicating that the molecular basis of autophagy is well conserved in eukaryotes (Mizushima et al., 1998; Thumm and Kadowaki, 2001; Doelling et al., 2002; Hanaoka et al., 2002; Juhász et al., 2003; Meléndez et al., 2003; Otto et al., 2003). In Arabidopsis thaliana, 25 ATG genes that are homologous to 12 of the yeast ATG genes were found (Doelling et al., 2002; Hanaoka et al., 2002). Little is known, however, about the molecular mechanisms underlying autophagy in higher plants. Phenotypic analysis of Arabidopsis mutants with a T-DNA insert in either the ATG7 or ATG9 gene suggested that autophagy is required for proper senescence and efficient nutrient recycling (Doelling et al., 2002; Hanaoka et al., 2002). Up until now, however, no direct evidence has been obtained showing that the mutant phenotypes are actually caused by a defect in autophagy because of the absence of a system that allows monitoring of plant autophagy. To establish such a monitoring system, we focused on the Atg8 molecule, which contains a ubiquitin fold like GATE-16 and LC3 (Paz et al., 2000; Sugawara et al., 2004).
In yeast and mammalian cells, Atg8 and LC3 provide useful molecular markers for monitoring the autophagic process (Kirisako et al., 1999; Kabeya et al., 2000). Atg8, which is induced by starvation, is modified with the lipid molecule phosphatidylethanolamine (PE) by ubiquitination-like reactions that occur after the cleavage of a C-terminal extension by the Cys protease Atg4 (Ohsumi, 2001). The C-terminal Arg of newly synthesized Atg8 is cleaved by Atg4, resulting in an Atg8 with an exposed Gly residue at its C terminus (hereafter Atg8-G). The Atg8 is then activated by the E1-like enzyme, Atg7, through a thioester bond between the Gly residue of Atg8 and the Cys residue of Atg7, and is subsequently donated to the Cys residue of the E2-like enzyme Atg3. Finally, the Atg8 is conjugated to PE via an amide bond between the Gly residue of Atg8 and the amino group of PE (Ichimura et al., 2000). After these modification reactions, the Atg8 conjugated to PE (Atg8-PE) is localized to a preautophagosomal structure where it is thought to play a role in autophagosome formation. In addition to Atg8 lipidation, the deconjugation of Atg8 by Atg4 is also essential for autophagosome formation (Kirisako et al., 2000). Recently, several reports have shown that the Atg8 modification system is conserved in mammalian cells (Tanida et al., 2002; Hemelaar et al., 2003; Mariño et al., 2003).
In this report, we have obtained an Arabidopsis mutant with T-DNA inserts in the ATG4 genes, and we have examined whether the Atg8 lipidation system in Arabidopsis functions in a similar manner to that in yeast and mammals. Moreover, we have established a system to monitor the autophagic process in an intact plant. This is the first report directly demonstrating the ATG-dependent autophagic process in a higher plant.
RESULTS
ATG8s and ATG4s Were Ubiquitously Expressed in All Organs and Were Further Induced by Nitrogen Starvation
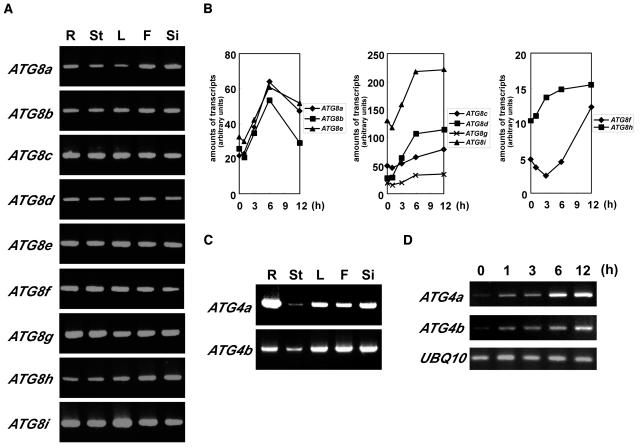
In our previous report, we searched for homologs of yeast ATG genes in the Arabidopsis genome and identified nine candidate ATG8s (Hanaoka et al., 2002). All the ATG8 proteins display a high degree of identity (∼70%) with the yeast Atg8 protein, and the Gly residues at their C-terminal regions are conserved. Interestingly, two of the nine homologs, ATG8h and ATG8i, had no C-terminal extension after the Gly residue.
To examine where the ATG8s function, we performed expression analysis of ATG8a through ATG8i by RT-PCR. Because the nucleotide sequences of the coding regions of ATG8s are highly conserved, gene-specific primers corresponding to each ATG8 from the 5′ or 3′ untranslated regions were designed based on sequences found in the Arabidopsis EST database (see Methods). Total cellular RNAs were isolated from roots, stems, leaves, flowers, and siliques of wild-type Arabidopsis grown in hydroponic culture for 4 to 5 weeks and subjected to RT-PCR. As shown in Figure 1A, all ATG8s were ubiquitously expressed in every organ tested. Next, in intact plants, the expression of ATG8a-i mRNA under nitrogen starvation was examined by quantitative RT-PCR because the abundance of both the ATG8 and ATG7 mRNAs increased when leaves were artificially senescent under nutrient-depleted conditions in the dark (Doelling et al., 2002). For nitrogen starvation, 5-week-old wild-type Arabidopsis plants grown in nutrient medium (7 mM nitrate) were transferred to nitrogen-depleted medium (0 mM nitrate). After 1, 3, 6, and 12 h of nitrogen starvation, total cellular RNA isolated from leaves were subjected to quantitative RT-PCR analysis. The expression of all ATG8s was moderately induced by nitrogen starvation, however, each ATG8 showed a different pattern of induction (Figure 1B).
Figure 1.
RT-PCR Analysis of ATG8 and ATG4 Expression in Arabidopsis.
(A) and (C) Total cellular RNA from roots (R), stems (St), leaves (L), flowers (F), and siliques (Si) of wild-type Arabidopsis grown hydroponically for 4 to 5 weeks was isolated and subjected to RT-PCR using gene-specific primers. The RT-PCR was terminated after 35 cycles for all the reactions, and the products were electrophoresed and detected.
(B) and (D) Wild-type Arabidopsis were grown for 5 weeks in nutrient-rich hydroponic medium (7 mM nitrate) and then transferred to nitrogen-depleted hydroponic medium (0 mM nitrate) for 1 d. Total cellular RNA from leaves after 1, 3, 6, and 12 h of nitrogen starvation was isolated and subjected to quantitative RT-PCR (B) or semiquantitative RT-PCR (D). The amounts of transcripts were presented as arbitrary units. Semiquantitative RT-PCR was terminated after 22 cycles for ATG4a and ATG4b and 20 cycles for UBQ10. The products were electrophoresed and detected. The ubiquitin gene, UBQ10, which is constitutively expressed under all conditions, was used as an internal control for RNA level.
Next, in addition to ATG8s, we also focused on Atg4 homologs, which may function in the C-terminal cleavage of ATG8s required for their modification and in the deconjugation of their modified forms. Two homologs of the ATG4 gene, ATG4a and 4b, were identified in the Arabidopsis genome (Hanaoka et al., 2002). ATG4 proteins had a significant but low level of identity with the yeast Atg4 protein (ATG4a-yeast Atg4, 26%; ATG4b-yeast Atg4, 29%). However, the two ATG4s displayed 71% overall amino acid identity. The Cys, Asp, and His residues, which comprise the catalytic site, were well conserved in ATG4s. In addition, both ATG4s complemented the autophagic defect caused by the yeast atg4 mutation (Hanaoka et al., 2002). These results suggest that ATG4a and 4b are yeast Atg4 orthologs.
We also performed expression analysis of ATG4a and 4b by RT-PCR using gene-specific primers (see Methods). As shown in Figure 1C, the two ATG4s were ubiquitously expressed like ATG8s. The induction of ATG4 transcription under nitrogen starvation was examined by semiquantitative RT-PCR. With 1 h of starvation, both ATG4s were immediately and drastically induced (Figure 1D).
ATG8s Are Cleaved at the C Terminus by an ATG4 Protease
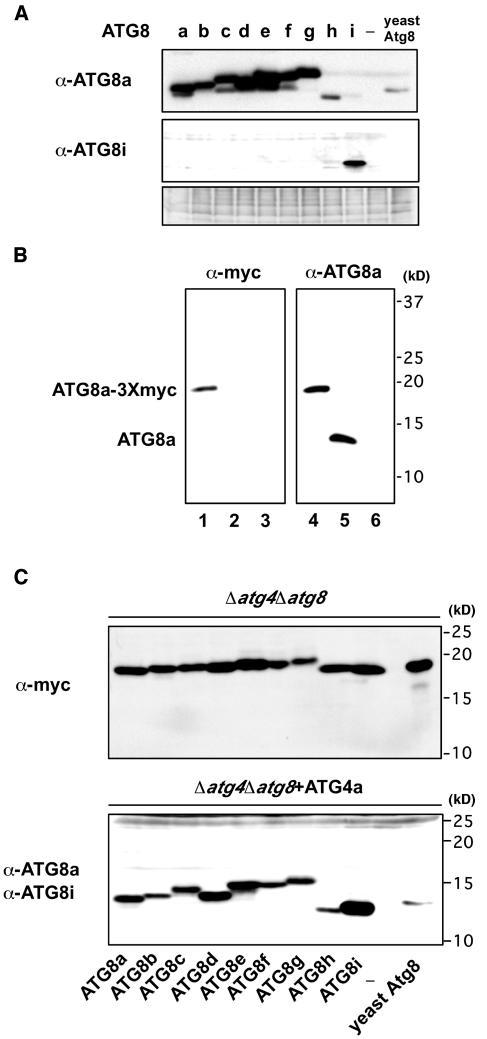
First of all, to detect ATG8 proteins, antibodies against ATG8a and 8i (hereafter anti-ATG8a and anti-ATG8i) were made. Because of their high conservation of the primary sequences, the cross-reactivity of these antibodies was tested in lysates of yeast cells expressing each ATG8. The anti-ATG8a antibodies recognized all ATG8 proteins, with varying degrees of reactivity, whereas the anti-ATG8i antibodies recognized mostly ATG8i but faintly ATG8h (Figure 2A).
Figure 2.
ATG8s Are Cleaved at Their C Termini by ATG4 Proteases.
(A) Cross-reactivity of anti-ATG8a and anti-ATG8i antibodies. Protein extracts from yeast Δatg4Δatg8 mutant cells that expressed Arabidopsis ATG4a in addition to Arabidopsis ATG8s-3Xmyc proteins or yeast Atg8-3Xmyc were analyzed by immunoblots using anti-ATG8a (top panel) and anti-ATG8i antibodies (middle panel), respectively. Nonspecific bands detected by anti-ATG8i antibodies are shown in the bottom panel as control for equivalent amounts of protein loaded into lanes.
(B) Representative data of cleavage assay. Protein extracts from yeast Δatg4Δatg8 mutant cells that expressed Arabidopsis ATG8a-3Xmyc proteins (lanes 1 and 4) or those that expressed Arabidopsis ATG4a in addition to Arabidopsis ATG8a-3Xmyc proteins (lanes 2 and 5) were analyzed by immunoblots using an anti-myc antibody (left) or anti-ATG8a antibodies (right), respectively. Protein extracts from only yeast Δatg4Δatg8 mutant cells were applied as negative control (lanes 3 and 6).
(C) Cleavage assay for all ATG8s. Protein extracts from yeast Δatg4Δatg8 mutant cells that expressed Arabidopsis ATG8s-3Xmyc proteins or yeast Atg8-3Xmyc (top panel) or those that expressed Arabidopsis ATG4a in addition to Arabidopsis ATG8s-3Xmyc proteins or yeast Atg8-3Xmyc (bottom panel) were analyzed by immunoblots using an anti-myc antibody (top panel) or anti-ATG8a and anti-ATG8i antibodies (bottom panel), respectively.
To investigate whether ATG8s are also processed at the C terminus by ATG4s, each ATG8 was tagged with three consecutive myc epitopes at the C terminus and was expressed in the yeast atg4atg8 mutant. Total cell lysates were then subjected to immunoblot analysis with an anti-myc antibody. In every cell, a single band of ∼18 kD was detected, corresponding to the molecular sizes of the ATG8-3Xmyc fusion protein (Figures 2B, lane 1, and 2C, top panel). Next, ATG4a or 4b was coexpressed in these cells followed by an immunoblot analysis with anti-ATG8a and 8i antibodies (Figures 2B, lane 5, and 2C, bottom panel; data not shown). A single band of ∼13 kD appeared, which was not detectable with the anti-myc antibody (Figure 2B, lane 2; data not shown). These data indicate that the C-terminal extensions of ATG8a-g had been cleaved by the ATG4s. ATG8h and 8i are novel types of ATG8, which did not possess additional amino acids after Gly, and it is not supposed to be subjected to the cleavage by ATG4 as in the other ATG8s. However, it is noteworthy that ectopically fused C termini (3Xmyc) of ATG8h and 8i were also susceptible to the cleavage in the ATG4s dependent manner.
Most ATG8s Are Tightly Membrane-Associated Proteins in Arabidopsis
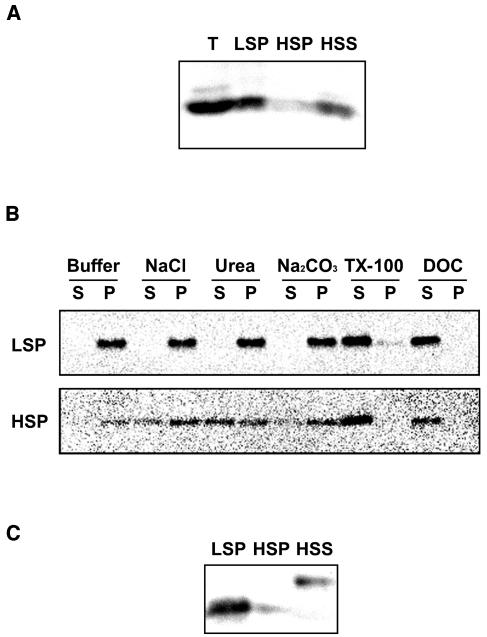
Next, we investigated the intracellular distribution of ATG8s by subcellular fractionation. Leaves of 4-week-old wild-type Arabidopsis were homogenized, and the post-nuclear supernatant was fractionated by successive centrifugation to generate a 13,000g pellet fraction (low-speed pellet [LSP]), a 100,000g pellet fraction (high-speed pellet [HSP]), and a 100,000g supernatant fraction (high-speed supernatant [HSS]). Bands of ∼13 kD were detected by the anti-ATG8a antibodies, corresponding to the predicted size of ATG8s. The ATG8s were predominantly detected in the pellet fraction, mostly in the LSP. Small amounts were recovered from the HSP and HSS (Figure 3A). Next, we tried to solubilize the ATG8s from the pellet fractions by resuspension in a buffer containing 1 M NaCl, 2 M urea, 0.1 M Na2CO3, 2% Triton X-100, and 1% deoxycholate (DOC). Samples were centrifuged again, and the supernatant and the pellet were analyzed by immunoblotting with the anti-ATG8a antibodies. ATG8s in the LSP fraction could not be extracted by salt, urea, or alkali but were almost completely solubilized by Triton X-100 or DOC (Figure 3B, top panel). These results indicate that ATG8s in the LSP fraction are tightly associated with the membrane. On the other hand, ATG8s in the HSP fraction were partially solubilized by salt, urea, or alkali and almost completely by detergent (Figure 3B, bottom panel), suggesting that some of the protein in the HSP fraction was peripherally membrane associated.
Figure 3.
Association of ATG8s with Membranes.
(A) Subcellular distrubution of ATG8s. The total lysates (T) prepared from 4-week-old wild-type leaves were centrifuged at 13,000g for 15 min to generate supernatants and pellets (LSP). The resulting supernatants were then centrifuged at 100,000g for 1 h to generate further supernatants (HSS) and pellets (HSP). These samples were subjected to immunoblot analysis with purified anti-ATG8a antibodies.
(B) Solubilization of ATG8s recovered in the LSP (top panel) and HSP (bottom panel). The LSP and HSP of wild-type leaves were incubated on ice in a buffer containing salt, urea, sodium carbonate (Na2CO3), Triton X-100 (TX-100), or DOC. Samples were centrifuged again, and the resulting supernatants (S) and pellets (P) were analyzed by immunoblotting with purified anti-ATG8a antibodies.
(C) Pellet fractions were putative modified forms. Cell fractions (LSP, HSP, and HSS) from 4-week-old wild-type leaves were subjected to SDS-PAGE with 6 M urea and immunoblot analysis with purified anti-ATG8a antibodies.
Previous reports have shown that the modified form of yeast Atg8, which is conjugated with PE, migrates faster than the unmodified form of yeast Atg8 on a 6 M urea SDS gel (Kirisako et al., 2000). Taking advantage of this gel system, we attempted to detect the modified forms of ATG8s. On a urea SDS gel, most of the ATG8s in the LSP fraction migrated faster, but not those from the HSS fraction (Figure 3C). In addition, recently it was reported that LC3 also conjugated with PE in animal cells (Kabeya et al., 2004). Thus, we postulated these faster-migrating forms to be putative PE-conjugated forms (hereafter ATG8*). Most of the ATG8s in the LSP fraction were the ATG8* but those in the HSS fractions were the unmodified form. These experiments suggest that most ATG8s exist as the ATG8* and are associated with membranes. These results are in principle consistent with the biochemical properties of yeast Atg8 (Kirisako et al., 2000).
ATG8 Proteins in Various Organs at Different Developmental Stages
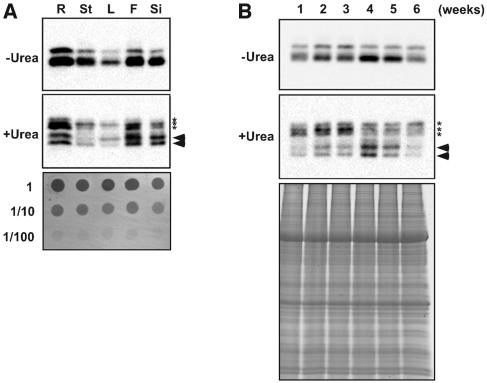
We examined the expression of ATG8 proteins in different organs and different growth stages by immunoblot analysis using anti-ATG8a antibodies (Figure 4). As shown in Figure 4A (top panel), ATG8s were ubiquitously detected in all organs examined, and the protein levels were higher in root, flower, and silique. In addition to unmodified forms (Figure 4A, middle panel, asterisks), two faster-migrating forms, the ATG8*, were detected (Figure 4A, middle panel, arrowheads). The banding patterns differed slightly among all organs (Figure 4A, middle panel), suggesting that different ATG8 may be functionally assigned to each organ. Furthermore, ATG8s were detected throughout all growth stages; the level of protein expression increased until the plant reached 4 weeks, followed by a gradual decrease (Figure 4B, top panel). We also investigated the ATG8* in leaves of different growth stages. Interestingly, the level of the ATG8* also increased as growth progressed, peaking in 4-week-old plants, which is the stage just before bolting (Figure 4B, middle panel).
Figure 4.
Expression of ATG8 Proteins in Various Organs at Developmental Stages.
Total protein samples prepared from roots (R), stems (St), leaves (L), flowers (F), and siliques (Si) (A) or total protein samples prepared from a series of wild-type plants grown for 1 to 6 weeks (B) were subjected to SDS-PAGE without (top panel) or with (middle panel) 6 M urea and immunoblot analysis with purified anti-ATG8a antibodies. Twenty micrograms of total protein was applied to each column. The asterisks indicate the unmodified forms of ATG8s, and the arrowheads indicate putative PE conjugated forms. Dot-blotted polyvinylidene difluoride membrane and SDS-PAGE stained by Coomassie Brilliant Blue are shown in the bottom panels as control for equivalent amounts of protein loaded into lanes.
Subcellular Localization of ATG8s
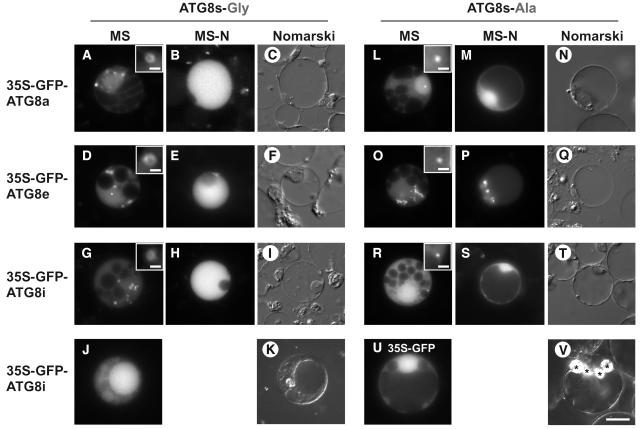
Next, the subcellular localization of green fluorescent protein–ATG8s (GFP-ATG8s) was examined by polyethylene glycol–mediated transient expression assays using Arabidopsis cultured cells. We made nine constructs in which each ATG8 open reading frame (ORF) was fused to the C terminus of GFP and placed under the control of the constitutive 35S promoter of Cauliflower mosaic virus. GFP alone was expressed as a negative control. GFP-ATG8s exhibited rings and punctate structures, which are thought to be autophagosomes and their precursors, in addition to the evenly stained cytosol (Figures 5A, 5D, and 5G; data not shown). Interestingly, in some cases, GFP-ATG8h and GFP-ATG8i localized also to the inside of the vacuole (Figure 5J; data not shown). Because ATG8h and ATG8i do not require processing of the C-terminal extension by ATG4s, they may be delivered to vacuole more quickly than other GFP-ATG8s. In the case of ATG8a-g, the level of ATG4 expression may be limiting because only GFP-ATG8s were transiently overexpressed after strong promoter. We also constructed GFP-ATG8a-A, e-A, and i-A, in which each C-terminal Gly was substituted by an Ala residue. The resultant fusion proteins were found in dot structures smaller than the rings that were observed upon expression of the wild-type GFP-ATG8s (Figures 5L, 5O, and 5R; data not shown). These experiments indicate that the substitution of the C-terminal Gly results in defective localization to preautophagosomal structures.
Figure 5.
Localization of GFP-ATG8 Fusion Proteins in Protoplasts of a Suspension Culture of Arabidopsis Cells.
(A) to (K) Arabidopsis cells expressing GFP-ATG8a, 8e, and 8i grown in MS ([A], [D], [G], and [J]) and MS-N ([B], [E], and [H]) medium. Nomarski images of (B), (E), (H), and (J) are shown in (C), (F), (I), and (K), respectively.
(L) to (T) Arabidopsis cells expressing GFP-ATG8a-A, 8e-A, and 8i-A,which harbor Gly-to-Ala substitutions at their C termini, grown in MS ([L], [O], and [R]) and MS-N ([M], [P], and [S]) medium. Nomarski images of (M), (P), and (S) are shown in (N), (Q), and (T), respectively.
(U) and (V) Arabidopsis cells expressing GFP alone as a control (U). Nomarski image of (U) is shown in (V). Asterisks indicate starch granules.
All scales are same as in (V); bar = 10 μm. Insets in (A), (D), (G), (L), (O), and (R) show ring and dot structures at high magnification, respectively (bar = 1.25 μm).
The behavior of GFP-ATG8s was also investigated under nutrient starvation conditions. As shown in Figures 5B, 5E and 5H, vacuoles swelled after 48 h of incubation in nitrogen-free medium, and GFP-ATG8a, e, and i were delivered to the vacuole. GFP-ATG8a-A, e-A, and i-A, however, were not delivered to the vacuole (Figures 5M, 5P, and 5S). The most probable explanation is that the ATG8-As could not be cleaved by the ATG4s or modified at their C termini because of the substitution of the C-terminal Gly to an Ala residue; consequently, the GFP-ATG8-As were not able to localize to the putative preautophagosomal structures, preventing the effective delivery of the GFP-ATG8-As to the vacuole. These results suggest that the C-terminal Gly of all ATG8s is essential for autophagy, namely, the ATG8 system functions in a similar manner in plant autophagy as in yeast. From these results, we have also shown that GFP-ATG8s are functional and good molecular markers for monitoring plant autophagy.
Isolation of the T-DNA Insertion Double Mutant atg4a4b-1
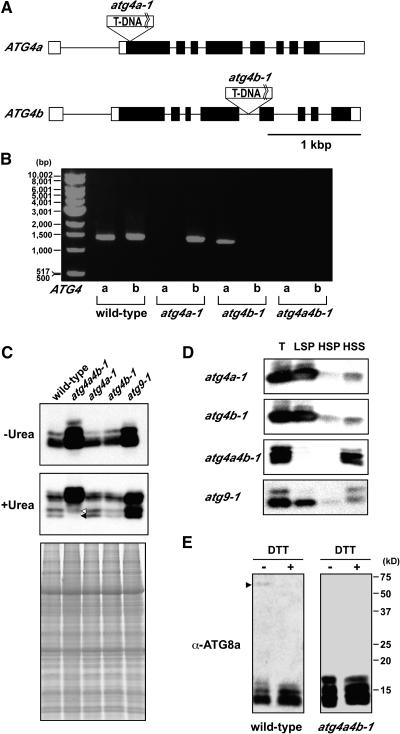
To elucidate the role of ATG4s in whole plants, we generated the T-DNA insertion double mutant atg4a4b-1. The Arabidopsis ATG4 proteins are encoded by two genes located on the bottom arm of chromosomes 2 and 3. We screened a population of T-DNA insertional mutants obtained from the University of Wisconsin Arabidopsis Knockout Facility and Kazusa DNA Research Institute and isolated two T-DNA insertion single mutants, which we designated atg4a-1 and atg4b-1. atg4a-1 and atg4b-1 plants contain T-DNA insertions in the second exon, 32 bp downstream of the translational initiation codon, and in the fifth intron, 1280 bp downstream of the translational initiation codon, respectively (Figure 6A). The segregation pattern of antibiotic resistance revealed a single T-DNA insertion in atg4b-1, but not atg4a-1 plants. We performed a backcross and obtained an atg4a-1 mutant with only a single T-DNA insertion, in which the antibiotic resistance gene was deleted. RT-PCR analysis using gene-specific primers that span the T-DNA insertion confirmed the absence of full-length ATG4a and 4b mRNA in both of the respective mutants (Figure 6B). We conclude that these alleles represent null mutations. To obtain double mutant plants, plants homozygous for atg4a-1 and for atg4b-1 were crossed. Homozygous double mutant plants (atg4a4b) were identified by PCR in F2 progeny after F1 self-pollination. In the atg4a4b-1 mutant, neither of the full-length mRNAs encoding ATG4a and 4b were detected (Figure 6B), indicating that this mutant is devoid of functional ATG4 proteins. After three backcrosses to the wild type, homozygous atg4a4b-1 plants were used for further study.
Figure 6.
Characterization of the atg4a-1, atg4b-1, and atg4a4b-1 Mutants.
(A) Schematic representation of the T-DNA insertion sites in the two ATG4 genes. Lines indicate introns, and boxes show exons (open boxes, untranslated regions; closed boxes, translated regions).
(B) RT-PCR analysis of the expression of ATG4 genes in wild-type and T-DNA insertional mutants. Total cellular RNA was isolated from wild-type, atg4a-1, atg4b-1, and atg4a4b-1 plants and subjected to RT-PCR. RT-PCR was performed for 35 cycles using gene-specific primers to amplify full-length ORFs.
(C) Expression of ATG8s in atg disruption mutants. Total protein samples from 6-week-old wild-type, atg4a4b-1, atg4a-1, atg4b-1, and atg9-1 leaves were subjected to SDS-PAGE without (top panel) and with (middle panel) 6 M urea and immunoblot analysis with purified anti-ATG8a antibodies. Forty micrograms of total protein was applied to each column. SDS-PAGE stained by Coomassie Brilliant Blue is shown in the bottom panel as control for equivalent amounts of protein loaded into lanes.
(D) Subcellular distribution of ATG8 proteins in atg disruption mutants. The total lysates (T) prepared from 4-week-old atg4a-1, atg4b-1, atg4a4b-1, and atg9-1 leaves were centrifuged at 13,000g for 15 min to generate supernatants and pellets (LSP). The resulting supernatants were then centrifuged at 100,000g for 1 h to generate further supernatants (HSS) and pellets (HSP). These samples were subjected to immunoblot analysis with purified anti-ATG8a antibodies.
(E) Putative ATG8-ATG3 conjugates. Total lysates (40 μg) from wild-type and atg4a4b-1 leaves with and without DTT were subjected to immunoblot analysis with purified anti-ATG8a antibodies. The arrowhead indicates putative ATG8-ATG3 conjugates.
Behavior of ATG8 Proteins in atg4a4b-1 Mutants Is Different from That in Wild-Type Plants
To investigate the behavior of ATG8s in atg4a4b-1 mutant plants, total proteins prepared from leaves of wild-type, atg4a-1, atg4b-1, atg4a4b-1, and atg9-1 plants were subjected to urea SDS-PAGE and immunoblot analysis with anti-ATG8a antibodies (Figure 6C). As shown in Figure 6C (bottom panel), faster-migrating bands were detected in wild-type, atg4a-1, and atg4b-1 plants. Like in yeast, the faster-migrating ATG8 bands were detected also in atg9-1 plants. By contrast, the fastest-migrating band was completely absent in atg4a4b-1 plants (Figure 6C, closed arrowhead). These results support the conclusion that the fastest-migrating band, which is observed in wild-type plants, represents the ATG8*. The other faster-migrating band was faintly detected even in atg4a4b-1 (Figure 6C, opened arrowhead). The band was not consistent with the band detected with anti-ATG8i antibodies (data not shown). The level of ATG8s was drastically increased in atg4a4b-1 and atg9-1 plants (Figure 6C, top panel), but in two single mutants, atg4a-1 and atg4b-1, the ATG8 levels were similar to those in the wild type. Therefore, it is likely that, in atg4a4b-1, the band of unmodified form that was not detected in the wild type appeared in the same position as ATG8*. Why did the level of ATG8s increase in atg disruption mutants? One possibility is that each ATG8 transcript increases in disruption mutants to compensate for the defect in autophagy. It is more likely, however, that ATG8 proteins are not delivered to the vacuole because of the defect in autophagy, and consequently are not degraded.
Next, we examined the intracellular distribution of ATG8s in atg mutants by subcellular fractionation. In atg4a-1, atg4b-1, and atg9-1 plants, ATG8s were mainly recovered in the LSP fraction, as they are in the wild-type plants (Figure 6D). By contrast, as expected, in atg4a4b-1 plants, ATG8s were recovered primarily in the HSS and only at low levels in the LSP and HSP fractions (Figure 6D), indicating that ATG8s could not be converted into ATG8* in atg4a4b-1.
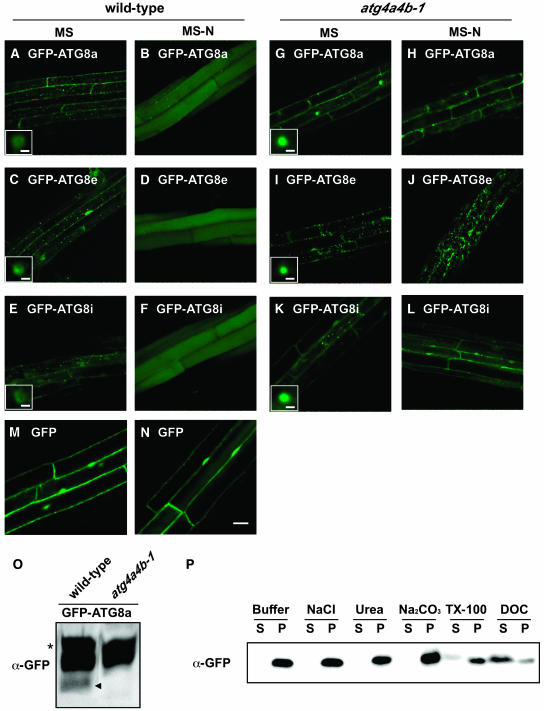
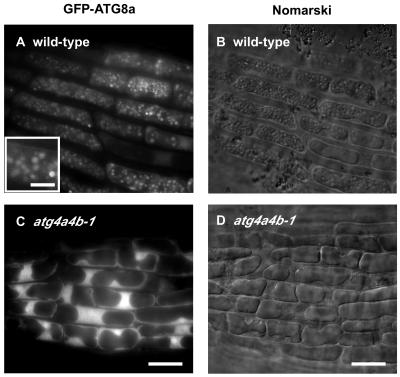
Wild-type and atg4a4b-1 plants expressing GFP-ATG8a, 8e, 8i, and GFP alone were generated to confirm whether the disruption of ATG4s affect intracellular localization of ATG8s in whole plants. As shown in Figure 7, in roots of wild-type plants grown under nutrient-rich conditions, every fusion protein was observed in many ring-shaped and punctate structures, which are interpreted to be autophagosomes and their intermediates (Figures 7A, 7C, and 7E). By contrast, these structures were not observed in roots of atg4a4b-1 plants, although some small dot structures were observed (Figures 7G, 7I, and 7K). The putative PE-conjugated form was detected only in the wild-type plants expressing GFP-ATG8a but not in the atg4a4b-1 plants expressing GFP-ATG8a (Figure 7O), supporting that GFP-ATG8 fusion proteins are functional. In the solubilization experiment, the GFP-ATG8a fusion protein was effectively solubilized by ionic detergent, DOC, but faintly solubilized by nonionic detergent, Triton X-100, suggesting that the small dot structures observed in atg4a4b-1 cells could be aggregated proteins (Figure 7P). These results indicate that ATG4s are required for formation of autophagosomes by permitting the association of ATG8s to membranes.
Figure 7.
Effect of atg4a4b-1 Mutations on Behavior of GFP-ATG8 Fusion Proteins in Stably Transformed Arabidopsis Roots.
(A) to (F) Wild-type Arabidopsis roots stably expressing GFP-ATG8a ([A] and [B]), 8e ([C] and [D]), and 8i ([E] and [F]).
(G) to (L) atg4a4b-1 mutant roots stably expressing GFP-ATG8a ([G] and [H]), 8e ([I] and [J]), and 8i ([K] and [L]).
(M) and (N) Wild-type Arabidopsis roots stably expressing GFP as control.
The roots of one-week-old transgenic seedlings grown on MS medium were observed using confocal laser scanning microscopy ([A], [C], [E], [G], [I], [K], and [M]). For nitrogen starvation, seedlings grown on MS medium were transferred to MS-N medium for an additional 7 d and then observed ([B], [D], [F], [H], [J], [L], and [N]). Bars = 20 μm. Insets in (A), (C), (E), (G), (I), and (K) show ring and dot structures at high magnification, respectively (bar = 1 μm).
(O) Putative PE-conjugated form of GFP-ATG8a. Total protein samples (20 μg) from wild-type and atg4a4b-1 expressing GFP-ATG8a fusion protein were subjected to 8% (w/v) SDS-PAGE with 6 M urea and analyzed by immunoblots using anti-GFP antibodies (Molecular Probes, Eugene, OR). The arrowhead indicates putative PE-conjugated form of GFP-ATG8a. The asterisk indicates a cross-reacting band that is not related to GFP-ATG8a.
(P) Solubilization of GFP-ATG8a in the atg4a4b-1 mutant. Subcellular fractionation was performed as described in Methods. The LSP fraction from the atg4a4b-1 mutant expressing GFP-ATG8a was incubated on ice in a buffer containing salt, urea, sodium carbonate (Na2CO3), Triton X-100 (TX-100), or DOC. The samples were centrifuged again, and the resulting supernatants (S) and pellets (P) were analyzed by immunoblotting with anti-GFP antibodies (Molecular Probes).
Monitoring of Autophagy in Whole Plants
To monitor autophagy in a whole plant, we examined the behavior of GFP-ATG8s under nitrogen-starved conditions in wild-type plants. For nitrogen starvation, 7-d-old seedlings grown in nutrient-rich medium (MS medium [21 mM NH4NO3, 18 mM KNO3, and 88 mM sucrose]) were transferred to nitrogen-depleted medium (MS-N [0 mM nitrogen and 88 mM sucrose]). After 6 d of nitrogen starvation, the roots were observed by fluorescence laser scanning confocal microscopy. Under these conditions, every fusion protein was delivered to vacuolar lumens in wild-type roots (Figures 7B, 7D, and 7F). On the other hand, in atg4a4b-1, none of these fusion proteins, even GFP-ATG8i, which does not have the C-terminal extension after the Gly residue, were delivered to the vacuoles, suggesting that ATG4s are essential for plant autophagy via deconjugation of ATG8* (Figures 7H, 7J, and 7L).
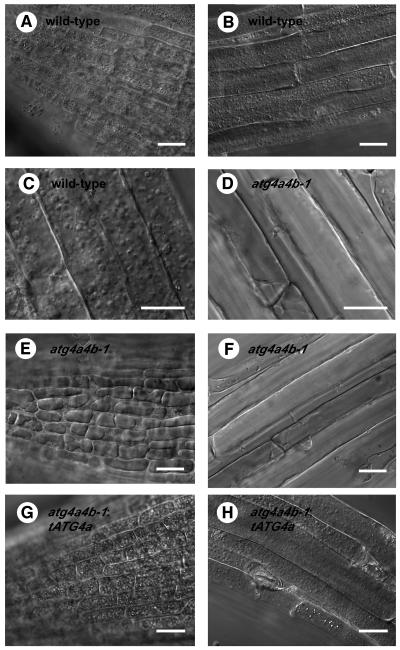
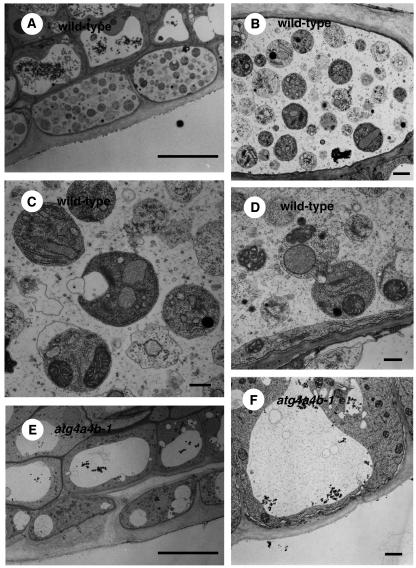
Concanamycin A, a V-ATPase inhibitor, is known to raise the pH in vacuolar lumens when exogenously added to plant cells (Drose et al., 1993; Matsuoka et al., 1997). Consequently, it is expected that, under such high pH conditions, vacuolar hydrolases cannot act, leading to the accumulation of autophagic bodies in the vacuole (Y. Moriyasu, personal communication). Roots of 1-week-old wild-type and atg4a4b-1 seedlings expressing GFP-ATG8s were treated with 1 μM concanamycin A for 6 h under nitrogen-starved conditions and then observed by microscopy. Nomarski images are shown in Figures 8, 9B, and 9D. In wild-type roots treated with concanamycin A, there were many spherical structures in well-developed vacuoles (Figures 8A to 8C). These structures were stained by GFP-ATG8s (Figure 9A), suggesting that they were plant autophagic bodies. By contrast, vacuoles in atg4a4b-1 were small and did not contain spherical structures (Figures 8D to 8F). Next, electron microscopy was performed. In wild-type roots, single-membrane-bound vesicles ∼1.5 μm in diameter were observed. These vesicles were electron dense and contained cytoplasmic structures, such as mitochondria, endoplasmic reticulum, and Golgi bodies (Figures 10A to 10D), demonstrating that they were indeed plant autophagic bodies. By contrast, there were no autophagic bodies in the vacuoles of atg4a4b-1 plants. Furthermore, the vacuoles of atg4a4b-1 mutants were smaller than those of wild-type plants in the cells around the root cap or meristem (Figures 10E and 10F; data not shown). In yeast, swollen vacuoles are typically observed as autophagy progresses under nitrogen-starved conditions (Takeshige et al., 1992). Our results suggest that autophagy might contribute to the swelling of vacuoles relatively soon after exposure to nutrient-depleted conditions.
Figure 8.
Roots of Wild-Type and atg4a4b-1 Mutants Treated with Concanamycin A under Nitrogen-Starved Conditions.
(A) to (C) Wild-type roots.
(D) to (F) atg4a4b-1 mutant roots.
(G) and (H) Complemented atg4a4b-1 mutant (atg4a4b-1;tATG4a) roots.
Roots of 1-week-old wild-type ([A] to [C]), atg4a4b-1 ([D] to [F]), and atg4a4b-1 transformed with the ATG4a gene ([G] and [H]) seedlings grown on MS medium were incubated in MS-N liquid medium containing concanamycin A for 12 h and then observed by conventional transmission light microscopy. Bars = 20 μm for (A), (B), and (E) to (H) and 10 μm for (C) and (D).
Figure 9.
Many Vesicles Accumulated in the Vacuoles upon Concanamycin A Treatment Were Stained with GFP-ATG8 Proteins.
(A) and (C) Fluorescence micrographs of roots from wild-type (A) and atg4a4b-1 (C) seedlings expressing GFP-ATG8a.
(B) and (D) Nomarski images of (A) and (C), respectively.
Roots of transgenic seedlings expressing GFP-ATG8 proteins were treated with concanamycin A (1 μM) for 6 to 12 h and observed using light and fluorescence microscopy. Representative pictures are shown in this figure. Bars = 20 μm. Inset in (A) shows enlargements of spherical structures (bar = 5 μm).
Figure 10.
Electron Micrographs of Plant Autophagic Bodies.
(A) to (D) Electron micrographs of wild-type roots treated with concanamycin A.
(E) and (F) Electron micrographs of atg4a4b-1 mutant roots treated with concanamycin A.
Roots of 1-week-old wild-type ([A] to [D]) and atg4a4b-1 ([E] and [F]) seedlings were treated with concanamycin A (1 μM) under nitrogen-starved conditions for 6 h. The same regions of the root tip were compared. Bars = 10 μm for (A) and (E), 500 nm for (C) and (D), and 1 μm for (B) and (F).
Complementation of the atg4a4b-1 Mutant
To verify that the defect in accumulation of autophagic bodies, that is, autophagy, in the atg4a4b-1 mutant was caused by the T-DNA insertion in each of the two ATG4 genes, we performed a functional complementation experiment. A genomic ATG4a fragment encompassing its 5′ flanking area and coding region (tATG4a) was transformed into the atg4a4b-1 mutant. Five independent transformants harboring the tATG4a transgene were selected by antibiotic resistance and confirmed by PCR for the presence of the tATG4a transgene and for the absence of the wild-type ATG4a and ATG4b alleles (data not shown). The expression of the tATG4a transgene rescued the defective accumulation of autophagic bodies (Figures 8G and 8H), indicating that the defect in autophagy was caused by the combination of the atg4a and atg4b mutations.
Defective Autophagy Results in the Arrested Development of the Root System under Nitrogen-Starved Conditions
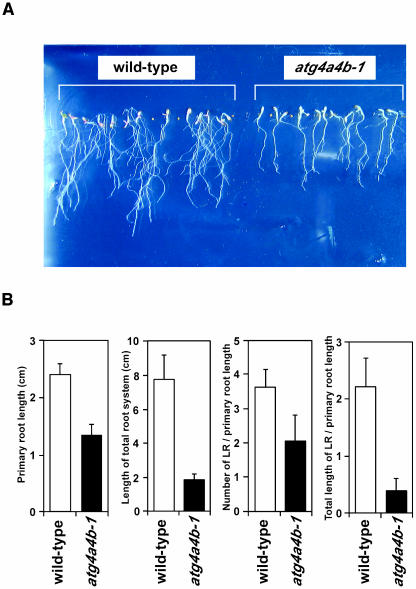
The above results led us to conclude that atg4a4b-1 plants are defective in autophagy. This is the first direct evidence that ATG genes are essential for plant autophagy. Previous studies by our group and Vierstra's group demonstrated that disruption of ATG7 and ATG9— atg7-1 and atg9-1, respectively—accelerated the senescence of cotyledon and rosette leaves, and, moreover, that both of the mutant plants develop fewer flowers and siliques, resulting in fewer seeds (Doelling et al., 2002; Hanaoka et al., 2002). These phenotypes were also observed in the atg4a4b-1 mutant (data not shown). In addition, when 10-d-old seedlings grown in nutrient-rich medium were transferred to nitrogen-depleted medium, it appeared that lateral-root elongation was severely inhibited in atg4a4b-1 plants (data not shown). To confirm the inhibition of lateral-root elongation in atg4a4b-1 plants under nitrogen-starved conditions, seeds of wild-type and atg4a4b-1 plants were sowed on nitrogen-starvation medium (MS-N), and primary root length, number of lateral roots, and total length of lateral roots were scored in 21-d-old seedlings (Figure 11). The rate of primary root elongation of atg4a4b-1 mutants was lower than that of wild-type plants. The total number of lateral roots per primary root in atg4a4b-1 seedlings was also reduced compared with wild-type seedlings, and the total length of lateral roots per primary root was even more severely decreased, indicating that the length of the total root system was reduced in this autophagy-defective mutant (Figure 11B). These results indicate that autophagy contributes to root elongation under nutrient-depleted conditions.
Figure 11.
Phenotype of atg4a4b-1 Mutants Caused by Defects in Autophagy under Nitrogen-Depleted Conditions.
(A) Roots of wild-type (left) and atg4a4b-1 (right) plants grown on nitrogen-free medium for 21 d.
(B) Statistical evaluation of primary root length, length of the entire root system, number of lateral roots (LR) per primary root length, and total length of lateral roots per primary root length. Seeds of wild-type and atg4a4b-1 plants were sowed on nitrogen-free medium, and after 21 d, primary root length, number of lateral roots, and total length of lateral roots were counted using NIH Image. Error bars represent sd. All measurements were made on at least 10 individual plants.
DISCUSSION
The Arabidopsis genome contains many ATG homologs (ATG) that encode proteins displaying all the characteristics of yeast Atg proteins, including the functional domains and amino acid residues essential for yeast autophagy, suggesting that autophagy depending on Atg proteins exists in higher plants. Until now, however, there has been no direct experimental evidence for this process in higher plants. Here, we have developed a monitoring system that assesses the autophagic process in a whole plant and have clearly demonstrated the existence of Atg protein–dependent autophagy in higher plants. Therefore, we are now able to discuss the physiological roles of autophagy in plants using mutants in which the ATG genes have been disrupted.
When and Where Does Autophagy Occur in Higher Plants?
When and where autophagy occurs in whole plants is an interesting and important issue for understanding the physiological roles of autophagy in plants. Expression analysis revealed that ATG8s and ATG4s are ubiquitously expressed and that ATG8 proteins are present in most organs. The presence of the mRNA and proteins in many different organs implies that autophagy may occur in all organs as a fundamental cell process. We also found that ATG8 proteins are expressed throughout all developmental stages, suggesting that autophagy is always occurring at basal level in addition to occurring under nutrient-limiting conditions. However, there is also a possibility that the nine ATG8s may have variable tissue specificity, and, accordingly, autophagy may occur in a tissue-specific manner. Furthermore, we found that the level of ATG8 proteins and the rate of conversion to ATG8* are highest in 4-week-old plants, which is the stage just before bolting. It is thought that plant autophagy is especially active when plants convert from the vegetative stage to the reproductive stage. However, it is still unknown whether the induction of Atg8 protein is necessary for the occurrence of autophagy and what actual roles their PE-conjugated form plays, even in yeast. In addition, there remains a possibility that ATG8s with a C-terminal extension may have other functions that are unrelated to autophagy; in this study, however, we have shown that all nine ATG8s do function in plant autophagy, at least after cleavage by ATG4s.
Under nutrient-rich conditions, autophagic bodies accumulated, and GFP-ATG8s were delivered to vacuoles in wild-type roots (data not shown), suggesting that basal autophagy occurs constitutively irrespective of nutrient conditions.
Membrane Binding and Subcellular Localization of ATG8s
Yeast Atg8, a key molecule for autophagy, displays several interesting behaviors during this process, and details of its molecular functions are continuously accumulating. Subcellular fractionation experiments have demonstrated that yeast Atg8 exists mainly in the pellet fraction, specifically in the HSP. According to the results of the solubilization experiment, the LSP contained exclusively the Atg8-PE, whereas the HSP contained both the unmodified and the PE-conjugated forms. Based on the evidence described above, in yeast it is thought that Atg8 undergoes conjugation to PE on membrane structures in the HSP before the formation of autophagosomes, whereas the Atg8-PE in the LSP was associated with the autophagosomes and its precursor structures (Kirisako et al., 2000). Consistent with these results, in plants, most ATG8s were also found in the pellet fraction. In addition, although ATG8s in the LSP were all tightly bound to membranes, there were two forms of ATG8s in the HSP, loosely membrane-bound and tightly membrane-bound, suggesting that a portion of ATG8-Gs undergo modification of their C termini on membrane structures found in the HSP. However, a discrepancy was observed between plants and yeast in the distribution of ATG8s within the pellet fraction. In plants, the majority of ATG8s in the pellet fraction were contained in the LSP, not in the HSP. These results suggest that in plants autophagosomes and preautophagosomal structures may exist stably in significant amounts. In fact, many rings and punctate structures, which are thought to be autophagosome or preautophagosomal structures, were constitutively observed in the roots of wild-type plants that expressed GFP-ATG8s. Unlike yeasts and animals, plant autophagy may be occurring constantly, but slowly, in significant amounts.
The Atg System Functions in a Similar Manner in Plants as in Yeast
The presence of Arabidopsis Atg8 homologs that display a high degree of identity with yeast Atg8, as well as the conservation of important catalytic domains in Arabidopsis, ATG4, ATG7, and ATG3, strongly suggest that Arabidopsis has similar mechanisms of autophagy to that seen in yeast. This prediction has been further supported by the demonstration that wild-type ATG7 can complement the phenotype seen in the Arabidopsis T-DNA mutant of ATG7, but ATG7C/S, which contains a mutation in a catalytic amino acid and is predicted to be enzymatically inactive, cannot (Doelling et al., 2002). In this study, we have clearly shown that in plant autophagy a ubiquitination-like Atg8 lipidation system functions in a similar fashion to that in yeast. This conclusion is based on the observation that the C terminus of ATG8s was cleaved in an ATG4-dependent fashion and that GFP-ATG8-As were not transported to the vacuoles under nitrogen-starved conditions, but instead accumulated in the cytosol with a diffuse or small dot-staining pattern. It is also supported by the observations that GFP-ATG8s, in wild-type plants, were localized to putative autophagosomes under nutrient-rich conditions and transported to the vacuoles under nitrogen starvation, but not in the ATG4-disrupted mutant, atg4a4b-1. In addition, protein blot analysis showed that ATG8* was detected in wild-type plants but not in atg4a4b-1. Furthermore, we could detect the putative ATG8-ATG3 conjugates in wild-type plants but not in atg4a4b-1 by immunoblotting using sample buffer without reducing agent (Figure 6E). However, it remains to be conclusively shown that ATG8s are modified with PE.
It is likely that ATG8i is able to be modified at its C terminus without processing because the Gly residue is exposed in a newly synthesized molecule. Therefore, GFP-ATG8i was expected to be transported to the vacuoles under nitrogen-starved conditions even in the atg4a4b-1 mutant. We observed, however, that in atg4a4b-1 under nitrogen-starved conditions, GFP-ATG8i was not transported to the vacuoles, but behaved in a similar manner to ATG8a and 8e. ATG8i would not have been able to deconjugate from the putative PE-conjugated form even though it could be modified at its C terminus in the absence ATG4s. We therefore concluded that the deconjugation reaction is also essential for plant autophagy.
Physiological Roles of Plant Autophagy
Recent reports of the phenotypes of Arabidopsis atg mutants, atg7-1 and atg9-1, in which the ATG genes had been disrupted by T-DNA, have discussed the physiological roles of autophagy in plants (Doelling et al., 2002; Hanaoka et al., 2002). In both cases, the mutants exhibited early senescence of rosette leaves even under nutrient-rich conditions and fewer seeds under nutrient-limiting conditions; however, these plants were nevertheless able to undergo a complete life cycle. Surpin et al. also reported that the T-DNA insertion mutant of VTI12, whose yeast homolog has been shown to be involved in autophagosome docking and fusion, showed an accelerated senescence phenotype under nutrient-limiting conditions (Surpin et al., 2003). The phenotype of atg4a4b-1 was similar to that of atg7-1, atg9-1, and vti12. This work provides further support for the hypothesis that autophagy is required for the maintenance of cellular viability. In addition, atg7-1 displayed a reduction in the growth rate of roots under nutrient-limiting conditions (Doelling et al., 2002). In this study, we also focused on the growth rate of roots under nutrient-limiting conditions. We were able to examine this aspect in detail because we had developed a way to monitor the autophagic process in the Arabidopsis roots and to observe the autophagic defect in the roots of atg4a4b-1. Our detailed examination indicated that autophagic defect in atg4a4b-1 led not only to a reduced growth rate in the primary root, but also to that of lateral roots under nutrient-limiting conditions. It appeared that this reduction was caused by a reduction in cell number because there was almost no difference in the cell size within the roots in the same regions (data not shown). When plants are exposed to more extreme nitrogen-deficient conditions, a general inhibition of growth is observed, but root growth is decreased by a smaller amount than shoot growth (Bloom et al., 1985; Schulze and Chapin, 1987). Plant autophagy might play a role in such a situation; it will contribute to the improvement of nutrient acquisition by developing a root system when nitrogen source is limiting. We are currently investigating how autophagy contributes to root elongation, and furthermore, why atg mutants exhibit an early senescence phenotype.
The careful phenotypic analysis of atg4a4b-1 also provides new knowledge. The phenotypes seen in atg4a4b-1 were more severe to those of atg9-1 (data not shown). In atg9-1 roots, autophagic bodies slowly accumulated in the presence of concanamycin A, despite the presence of a null mutation. This was also confirmed by another mutant allele, atg9-2 (data not shown), indicating that the autophagic defect in atg9 is leaky. However, in the yeast atg9 mutant, autophagy is completely blocked. These results suggest that higher plants have a complicated, plant-specific autophagy pathway. Further analyses of atg mutants using the monitoring systems established in this study will further elucidate the molecular mechanisms and physiological roles of plant autophagy.
METHODS
Plant Material and Growth Conditions
The Arabidopsis thaliana ecotype Wassilewskija was used in this study. The plants were grown on rockwool using vermiculite as soil or hydroponically at 22°C with 16-h-light/8-h-dark cycles. The hydroponic culture was performed as described by Hanaoka et al. (2002). For nitrogen starvation, 5-week-old plants hydroponically grown in nutrient medium were transferred to nitrogen-depleted medium that had been prepared by replacing KNO3 and Ca(NO3)2 with KCl and CaCl2. For plate-grown plants, seeds were surface sterilized, chilled at 4°C for 4 d, and then sown and grown on MS medium. For nitrogen-depleted MS medium (MS-N), the medium was prepared by depleting the NH4NO3 and replacing the KNO3 with KCl.
Identification of ATG Genes
Genes homologous to yeast (Saccharomyces cerevisiae) ATG were searched for in the Arabidopsis whole genome database using the Arabidopsis Information Resource BLAST 2.2.8 program (Altschul et al., 1997), and the amino acid sequence alignment was performed using the program Megalign (DNASTAR, Madison, WI).
RT-PCR Analysis of ATG8 and ATG4 Expression
To examine the tissue-specific expression of the ATG8a-i, ATG4a, and 4b genes, total cellular RNA was prepared, using the guanidium isothiocyanate/CsCl method, from roots, stems, leaves, flowers, and siliques of wild-type Arabidopsis that had been grown in hydroponic culture for 4 to 5 weeks (Cox and Goldberg, 1988). cDNA was generated using the ProSTAR RT-PCR kit (Stratagene, La Jolla, CA) following the manufacturer's instructions. cDNA derived from 0.2 μg of total RNA was used as template for each PCR reaction. Gene-specific primers used are as follows: for ATG8a (At4g21980), 5′-TCGGAGACTAATCGAATCGC-3′ and 5′-CATCAAAGTCATCCAAAGATCG-3′; for ATG8b (At4g04620.1 and At4g04620.2), 5′-CATCGTAGATACTAACCGAATCATC-3′ and 5′-GATCAGACGTAGAAGCTGAGG-3′; for ATG8c (At1g62040), 5′-AATCTTTGATTCTTTAATCGCC-3′ and 5′-CAAAGCAACATTTACATTAATAGTAG-3′; for ATG8d (At2g05630), 5′-TCTCTTCTCTGTTTCTCTCTCG-3′ and 5′-TCGATCCACATATTCCAAGC-3′; for ATG8e (At2g45170.1), 5′-TTCCATCAAATTCTCTCTCTAAG-3′ and 5′-CGGATTCTCAGAGGTCAGAG-3′; for ATG8f (At4g16520.1 and At4g16520.2), 5′-GTAGTCTACAGGCGTGGAAGG-3′ and 5′-TTATGGAGATCCAAATCCAAATG-3′; for ATG8g (At3g60640), 5′-CGCATAATCCAGAGAGGACC-3′ and 5′-CATTCAATTAAAAGCAAGAACACC-3′; for ATG8h (At3g06420), 5′-TCATTCGTCGTGAAATCTG-3′ and 5′-AAATCTTTTGTTAGCCGAAAG-3′; for ATG8i (At3g15580), 5′-CCGGGCGGTCGAAGAAG-3′ and 5′-ACACAGACACTAACATCATTATTGG-3′; for ATG4a (At2g44140.1), 5′-ATGAAGGCTTTATGTGATAGATTTGTTC-3′ and 5′-TCAGAGCATTTGCCAGTCATCTTCAC-3′; for ATG4b (At3g59950.1), 5′-ATGAAGGCTTTATGTGATAGATTTGTTC-3′ and 5′-GTCACACAATGAAAAGAATGGCTAGGAG-3′; for UBQ10 (At4g05320), 5′-TTCACTTGGTCCTGCGTCTTCGTGGTGGTTTC-3′ and 5′-CATCAGGGATTATACAAGGCCCC-3′. To examine the effect of nitrogen starvation on the expression of ATG8a-i, ATG4a, and 4b genes, 0.2 μg of total cellular RNA from leaves was isolated after 1, 3, 6, and 12 h of nitrogen starvation and subjected to quantitative or semiquantitative RT-PCR. Quantitative RT-PCR was performed by the method described previously (Kamada et al., 2003). Gene-specific primers used for quantitative RT-PCR are as follows: for ATG8a, 5′-TCGATTCTTCTTCTCCAGTTTCAATCA-3′ and 5′-CCATTGCGATTCGATTAGTCTCCGAAG-3′; for ATG8b (At4g04620.1 and At4g04620.2), 5′-TTACCTCAGCTTCTACGTCTGATCCTC-3′ and 5′-AGAGAGTCAAATTAGAATCTTGTTTCA-3′; for ATG8c, 5′-CCTAATTCTGTTTTCTACACGAAACCC-3′ and 5′-GGCGATTAAAGAATCAAAGATTAGGGC-3′; for ATG8d, 5′-TGCGCTTGGAATATGTGGATGCATCAG-3′ and 5′-CACCATGAACGGAAGACCTTATCAT-3′; for ATG8e (At2g45170.1 and At2g45170.2), 5′-ATTAGTCTATGTACATTTTAACAAGTG-3′ and 5′-TAATCTCTGACCTCTGAGAATCCGCCA-3′; for ATG8f (At4g16520.1), 5′-GCTTATTTGCTTTTTCTTTCATATTTTTTCG-3′ and 5′-CATTCCTTCCACGCCTGTAGACTACAAAG-3′; for ATG8g, 5′- CCAACACTTTGTTTGCAAACCGATTGG-3′ and 5′-GGTGTTCTTGCTTTTAATTGAATGTAA-3′; for ATG8 h, 5′-GAACAATCAAAAACGAACTAACCAAAC-3′ and 5′-ATCCCCATTACAGATTTCACGACGAATGA-3′; for ATG8i, 5′-GCAGTGAGAAAACCTTTGGTTGATCC-3′ and 5′-GCTTAACAAGTGTTTGATGTCTTTTG-3′. The gene specificity of each primer was confirmed by PCR using plasmids subcloned each full-length cDNA (data not shown).
Antibody Production and Immunoblot Analysis
The ATG8a and 8i coding regions, from nucleotides 1 to 351 of ATG8a and 1 to 345 of ATG8i, were amplified by PCR from the full-length cDNA using the following primers: for ATG8a, 5′-CGCggatccCAATGGCTAAGAGTTCCTTC-3′ and 5′-CGCggatccTCCAAAAGTGTTCTCTCCACTGT-3′; and for ATG8i, 5′-CGCggatccAGATGAAATCGTTCAAGGAACAATAC-3′ and 5′-CGCggatccACCAAAGGTTTTCTCACTGCT-3¢, designed to contain a BamHI site (lowercased) at both ends. The resulting products were digested with BamHI and inserted into pGEX-3X (Amersham Biosciences, Piscataway, NJ) for expression in Escherichia coli. After 5 h of induction with 0.05 mM isopropyl-β-d-thiogalactopyranoside, the ATG8a and 8i recombinant proteins were purified from the lysates by affinity chromatography on a glutathione-Sepharose 4B column using Factor Xa (Amersham Biosciences) and used to raise polyclonal antibodies in rabbits. Antisera against ATG8a and 8i were subsequently affinity purified by passing the sera over a column of ATG8a or 8i recombinant proteins coupled to cyanogen bromide–activated Sepharose 4B (Amersham Biosciences).
Plant protein samples were prepared as follows: 300 mg of each plant were homogenized in 300 μL of SDS sample buffer without a reducing agent and bromophenol blue (0.1 M Tris-HCl, pH 6.8, 4% [w/v] SDS, and 20% [v/v] glycerol) and boiled for 2 to 3 min, then centrifuged at 500g for 1 min. The resultant supernatant was used as total protein. Total proteins were quantified by the BCA protein assay reagent kit (Pierce, Rockford, IL).
For immunoblot analyses, total protein extracts that had been subjected to SDS-PAGE with or without 6-M urea were transferred to polyvinylidene difluoride membranes (Millipore, Bedford, MA), and the membranes were blocked using 5% milk powder. Primary and secondary antibodies were diluted 1:200 and 1:5000 in TBS containing 0.1% Tween 20, respectively. The secondary antibodies used were peroxidase-conjugated goat anti-rabbit immunoglobulins (Jackson ImmunoResearch Laboratories, West Grove, PA). Signals were detected using the Western Lightning Chemiluminescence Reagent Plus kit (Perkin-Elmer Life Sciences, Boston, MA).
Cleavage Assay for Arabidopsis ATG8s in Yeast atg Mutants
To express Arabidopsis ATG4 proteins in yeast, Arabidopsis ATG4 genes were inserted into the yeast expression vector pKT10, which contains the URA3 marker, under the control of the GAP promoter (Hanaoka et al., 2002). To express 3xmyc-tagged ATG8s proteins in yeast, ATG8 genes were amplified by PCR from the full-length cDNA using the following primers: for ATG8a, 5′-CGCggatccCAATGGCTAAGAGTTCCTTC-3′ and 5′-CGCggatccTCCAAAAGTGTTCTCTCCACTGT-3′; for ATG8b, 5′-CGCggatccTCATGGAGAAGAACTCCTTCAAGC-3′ and 5′-CGCggatccACCAAATGTGTTCTCTCCACTGT-3′; for ATG8c, 5′-CGCggatccCCATGGCTAATAGCTCTTTCAAG-3′ and 5′-CGCggatccACCAAAGGTGTTCTCTCCACTG-3′; for ATG8d, 5′-CGCggatccTCATGGCGATTAGCTCCTTCAA-3′ and 5′-CGCggatccCCCGAACGTGTTCTCACCACTG-3′; for ATG8e, 5′-CGCggatccAGATGAATAAAGGAAGCATCTTTAAGATGG-3′ and 5′-CGCggatccACCGAATGTGTTCTCGCCACTG-3′; for ATG8f, 5′-CGCggatccGAATGGCAAAAAGCTCGTTCAAG-3′ and 5′-CGCggatccTCCAAATGTGTTTTCTCCGCTG-3′; for ATG8g, 5′-CGCggatccAGATGAGTAACGTCAGCTTCAGG-3′ and 5′-CGCggatccTCCAAAAGTGTTTTCCCCACTG-3′; for ATG8h, 5′-CGCggatccTAATGGGGATTGTTGTCAAGTCTTTC-3′ and 5′-CGCggatccGCCGAAAGTTTTCTCGGTGCTG-3′; and for ATG8i, 5′-CGCggatccAGATGAAATCGTTCAAGGAACAATAC-3′ and 5′-CGCggatccACCAAAGGTTTTCTCACTGCT-3′, designed to contain a BamHI site (lowercased) at both ends. The fragments digested with BamHI were substituted for the BamHI-BamHI ATG8 ORF fragment of pRS315:ATG8, which contains an ATG8 fragment that includes the promoter region and the inserted 3xmyc sequence. pRS315:ATG8 was generated by cloning 1.3 kb of the SpeI-EcoRI fragment from an ATG8 plasmid, which was derived from the pRS316 vector (Kirisako et al., 2000), into the yeast expression vector pRS315 containing a LEU2 marker. Cells of Δatg4Δatg8 (MATα leu2 ura3 his3 trp1 lys2 suc2-Δ9 Δatg8::HIS3 Δatg4::TRP1 Δpho8::PHO8Δ60) were transformed with these plasmids. The transformants were cultured in SC-without-leucine medium to express ATG8s or SC-minus-leucine-and-uracil medium to express ATG8s and ATG4s. All cultures were then transferred to SD-N medium (Burke et al., 2000).
Yeast whole-cell lysates were prepared as previously described (Hanaoka et al., 2002) and subjected to 13.5% (w/v) polyacrylamide SDS-PAGE and analyzed by immunoblotting using an anti-myc antibody (9E10) (Berkeley Antibody, Richmond, CA) or anti-ATG8a and anti-ATG8i antibodies.
Subcellular Fractionation and Solubilization of ATG8s
Leaves were chopped in chilled extraction buffer (100 mM Tris-HCl, pH 7.5, 400 mM sucrose, 1 mM EDTA, 0.1 mM phenylmethylsulfonyl fluoride, 10 μg/mL of leupeptin, 10 μg/mL of pepstatin A, and 4% [v/v] protease inhibitor cocktail; Roche, Penzberg, Germany) using a razor and were then filtered through four layers of cheesecloth and centrifuged at 500g for 5 min to remove cell debris. The supernatant (T) was centrifuged at 13,000g for 15 min to generate the 13,000g membrane fraction (LSP). The resulting 13,000g supernatant was then centrifuged at 100,000g for 1 h to generate the 100,000g membrane fraction (HSP) and the soluble fraction (HSS). Pellets were resuspended in extraction buffer, and fractions were subjected to SDS-PAGE and immunoblot analysis using purified anti-ATG8a antibodies.
For solubilization of ATG8s, the LSP and HSP fractions were resuspended in extraction buffer plus 1 M NaCl, 2 M urea, 0.1 M Na2CO3, 2% Triton X-100, and 1% DOC and incubated on ice for 1 h. Samples were then centrifuged at 100,000g for 1 h to separate soluble from insoluble fractions. The pellets were resuspended in SDS sample buffer. The supernatants were precipitated using TCA, and the protein pellets were washed in acetone and resuspended in SDS sample buffer. Samples were analyzed by SDS-PAGE and immunoblotting with purified anti-ATG8a antibodies.
Transient Expression of GFP-ATG8 Fusion Proteins in Arabidopsis Suspension Culture Cells
The full-length ORF of ATG8a, 8e, and 8i with BamHI sites at both ends were amplified by PCR and fused to the C terminus of the ORF of EGFP in pEZS-CL plasmids. Primers used are as follows, each designed to introduce a BamHI site (indicated by lowercase letters): for ATG8a, 5′-CGCggatccCAATGGCTAAGAGTTCCTTC-3′ and 5′-CGCggatccTCAAGCAACGGTAAGAGATCCAA-3′; for ATG8e, 5′-CGCggatccAGATGAATAAAGGAAGCATCTTTAAGATGG-3′ and 5′-CGCggatccTTAGATTGAAGAAGCACCGAATGTG-3′; for ATG8i, 5′-CGCggatccAGATGAAATCGTTCAAGGAACAATAC-3′ and 5′-CGCggatccTCAACCAAAGGTTTTCTCACTGC-3′. GFP-ATG8-A plasmids were produced by site-directed mutagenesis using the above wild-type GFP-ATG8 plasmids as templates. Primers used are as follows: for ATG8a-A, 5′-GAGAGAACACTTTTGcATCTCTTACCGTTGC-3′ and 5′-GCAACGGTAAGAGATgCAAAAGTGTTCTCTC-3′; for ATG8e-A, 5′-GGCGAGAACACATTCGcTGCTTCTTCAATCTA-3′ and 5′-TAGATTGAAGAAGCAgCGAATGTGTTCTCGCC-3′; for ATG8i-A, 5′- GTGAGAAAACCTTTGcTTGAGGATCCACCG-3′ and 5′-CGGTGGATCCTCAAgCAAAGGTTTTCTCAC-3′ (lowercase letters represent point mutation sites). A pEZS-CL plasmid containing GFP alone was used as a control.
Transient expression of GFP-fused ATG8 proteins in a suspension of Arabidopsis cultured cells was performed by a method described previously (Ueda et al., 2001). For nitrogen starvation, transformed cells were transferred to MS-N medium supplemented with 0.4 M mannitol and incubated under gentle agitation at 23°C for 48 h in the dark. Transformed cells were observed with a fluorescence microscope (Olympus IX81; Tokyo, Japan).
Identification of T-DNA Mutants
The atg4a-1 plant was isolated from an Arabidopsis T-DNA–mutagenized population created at the University of Wisconsin Arabidopsis Knockout Facility using a PCR-based reverse genetic screen (Krysan et al., 1996, 1999). The atg4b-1 plant was isolated using the T-DNA insertion line screening system engineered at the Kazusa DNA Research Institute as described previously (Hanaoka et al., 2002). The primers used to detect T-DNA insertions into the ATG4a and ATG4b genes are as follows: for atg4a-1, ATG4a-specific primers were 5′-ATGAAGGCTTTATGTGATAGATTTGTTC-3′ and 5′-TCAGAGCATTTGCCAGTCATCTTCAC-3′, and the T-DNA–specific primers were 5′-CATTTTATAATAACGCTGCGGACATCTAC-3′ and 5′-TTTCTCCATATTGACCATCATACTCATTG-3′. For atg4b-1, ATG4b-specific primers were 5′-GTCACACAATGAAAAGAATGGCTAGGAG-3′ and 5′-CGGAATTCGTTGTATTTGGTGCTTAATGA-3′, and the T-DNA–specific primers were 5′-TAGATCCGAAACTATCAGTG-3′ and 5′-ATAACGCTGCGGACATCTAC-3′. DNA sequencing of PCR-amplified fragments carrying the T-DNA genome junctions determined the position of the T-DNA insertion in each gene.
Plasmid Constructions and Arabidopsis Transformation
To construct plasmids for generating stable transformants constitutively expressing GFP-ATG8 fusion proteins, plasmids that transiently expressed each GFP-ATG8 protein under control of a 35S promoter of Cauliflower mosaic virus and octopine synthase terminator were digested with NotI and blunt-end ligated into the SmaI site of pCAMBIA 2300 (CAMBIA, Black Mountain, Australia). A pEGAD plasmid containing GFP alone (Cutler et al., 2000) was used as control. To construct the plasmid for complementation of the atg4a4b-1 mutant, a 5357-bp genomic fragment containing a 3-kb sequence upstream from the start codon and the ATG4a coding region was amplified by PCR using a primer set, 5′-CGGggtaccTCTGCAACCTACAGCCCCACAATGGCAGTC-3′ and 5′-CGGggtaccCTCTACTCTTCCATTTTCGCATGCTTC-3¢, designed to introduce a KpnI site (lowercased) into both ends. The fragment was digested with KpnI and then cloned into the KpnI site of the pCAMBIA2300 vector (CAMBIA), resulting in the tATG4a construct. These constructs were verified by sequencing and introduced into wild-type and/or atg4a4b-1 Arabidopsis plants by the floral-dip method of in planta Agrobacterium tumefaciens–mediated transformation (Clough and Bent, 1998). Transgenic plants expressing GFP-ATG8s or atg4a4b-1 mutants expressing the tATG4a transgene were selected on MS medium without sucrose containing 50 μg/mL of kanamycin. At least five independent transgenic plants were obtained, and representative data are shown in Figures 7 and 8.
Analysis of the Cellular Localization of GFP Fusion Proteins in Arabidopsis Roots
Primary roots of transgenic plants expressing GFP-ATG8s grown on MS medium for 1 week were cut and mounted in water for microscopic observation. For nitrogen-starved conditions, transgenic plants grown on MS medium for 1 week were transferred to MS-N medium for an additional 7 d in the dark and then observed. The subcellular localization of GFP fusion proteins was analyzed with a Zeiss LSM 510 confocal laser scanning microscope (Zeiss, Jena, Germany) using a 488/568-nm ArKr laser in combination with a 505- to 550-nm band-pass filter set. Image acquisition and processing were performed using a Zeiss laser scanning microscope (LSM 510, version 2.01) and Adobe Photoshop 5.5 (Adobe Systems, San Jose, CA).
Concanamycin A Treatment
Primary roots of wild-type, atg4a4b-1, or atg4a4b-1;tATG4a plants vertically grown on MS medium for 1 week were cut and then incubated in MS-N liquid medium containing 1 μM concanamycin A (C-9705; Sigma-Aldrich, St. Louis, MO) under gentle agitation at 23°C for 6 to 12 h in the dark. The roots were mounted in water and observed by conventional transmission light microscopy (Olympus IX81). Primary roots of transgenic plants expressing GFP-ATG8s were also treated with concanamycin A as described above and observed by epifluorescence microscopy (Olympus IX81).
Electron Microscopy
The root tips of wild-type and atg4a4b-1 primary roots treated with concanamycin A were fixed in a 0.1 M cacodylate buffer containing 2% glutaraldehyde for 3 h in an ice bath. The specimens were rinsed in the same buffer and postfixed in 2% osmium tetroxide for 3 h in an ice bath. Dehydrated specimens were embedded in epoxy resin (Epon 812). Ultrathin sections (70 to 80 nm) were made with an LKB-8800 ultramicrotome (Amersham Biosciences) using a diamond knife and mounted on 200-mesh copper grids. Sections were stained with 2% uranyl acetate and lead citrate. The sections were observed with a JEM-2000EX transmission electron microscope (JEOL, Tokyo, Japan) operating at 100 kV.
Phenotypic Analysis of atg4a4b-1 Mutants
Phenotypic analyses were performed as described previously (Hanaoka et al., 2002), except for examination of root elongation under nitrogen-starved conditions. For measurement of root elongation under nitrogen-starved conditions, wild-type and atg4a4b-1 mutant seeds were sowed on MS-N medium and then, after 21 d, primary root length, number of lateral roots, and total length of lateral roots were counted using NIH Image 1.44 (Bethesda, MD).
Acknowledgments
We thank the Wisconsin Arabidopsis Knockout Facility team for the T-DNA insertion pools. We also thank Hirokazu Tsukaya of the National Institute for Basic Biology and Yuji Moriyasu of the University of Shizuoka for their helpful comments, Masami Miwa for technical assistance, and Chieko Nanba and Keiko Suzuki of the Plant Culture Laboratory for expert care of our plants. The pEZS-CL plasmid was a kind gift from David Ehrhardt of the Carnegie Institution of Washington. This study was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Yoshinori Ohsumi (yohsumi@nibb.ac.jp).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.025395.
References
- Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert, S., Gout, E., Bligny, R., Marty-Mazars, D., Barrieu, F., Alabouvette, J., Marty, F., and Douce, R. (1996). Ultrastructural and biochemical characterization of autophagy in higher plant cells subjected to carbon deprivation: Control by the supply of mitochondria with respiratory substrates. J. Cell Biol. 133, 1251–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth, H., Meiling-Wesse, K., Epple, U.D., and Thumm, M. (2001). Autophagy and cytoplasm to vacuole targeting pathway both require Aut10p. FEBS Lett. 508, 23–28. [DOI] [PubMed] [Google Scholar]
- Bloom, A.J., Chapin, F.S., and Mooney, H.A. (1985). Resource limitation in plants—An economic analogy. Annu. Rev. Ecol. Syst. 16, 363–392. [Google Scholar]
- Burke, D., Dawson, D., and Stearns, T. (2000). Methods in Yeast Genetics: Appendix A, P. Barker and M. Mazzullo, eds (New York: CSHI Press), pp. 171–181.
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cox, H., and Goldberg, R.B. (1988). Analysis of plant gene expression. In Plant Molecular Biology, H.C. Shaw, ed (Oxford: IRL Press), pp. 1–35.
- Cutler, S.R., Ehrhardt, D.W., Griffitts, J.S., and Somerville, C.R. (2000). Randon GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. USA 97, 3718–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling, J.H., Walker, J.M., Friedman, E.M., Thompson, A.R., and Vierstra, R.D. (2002). The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 277, 33105–33114. [DOI] [PubMed] [Google Scholar]
- Drose, S., Bindseil, K.U., Bowmana, E.J., Siebers, A., Zeeck, A., and Altendorf, K. (1993). Inhibitory effect of modified bafilomycins and concanamycins on P- and V-type adenosinetriphosphatases. Biochemistry 32, 3902–3906. [DOI] [PubMed] [Google Scholar]
- Hanaoka, H., Noda, T., Shirano, Y., Kato, T., Hayashi, H., Shibata, D., Tabata, S., and Ohsumi, Y. (2002). Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 129, 1181–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman, E.M., Baumgarther, B., and Chrispeels, M.J. (1981). Uptake and apparent digestion of cytoplasmic organelles by protein bodies (protein storage vacuoles) in mung bean cotyledons. Eur. J. Cell Biol. 24, 226–235. [PubMed] [Google Scholar]
- Hemelaar, J., Lelyveld, V.S., Kessler, B.M., and Ploegh, H.L. (2003). A single protease, Apg4B, is specific for the autophagy-related ubiquitin-like proteins GATE-16, MAP-LC3, GABARAP, and Apg8L. J. Biol. Chem. 278, 51841–51850. [DOI] [PubMed] [Google Scholar]
- Ichimura, Y., Kirisako, T., Takao, T., Satomi, Y., Shimonishi, Y., Ishihara, N., Mizushima, N., Tanida, I., Kominami, E., Ohsumi, M., Noda, T., and Ohsummi, Y. (2000). A ubiquitin-like system mediates protein lipidation. Nature 408, 488–492. [DOI] [PubMed] [Google Scholar]
- Juhász, G., Csikós, G., Sinka, R., Erdélyi, M., and Sass, M. (2003). The Drosophila homolog of Aut1 is essential for autophagy and development. FEBS Lett. 543, 154–158. [DOI] [PubMed] [Google Scholar]
- Kabeya, Y., Mizushima, N., Ueno, T., Yamamoto, A., Kirisako, T., Noda, T., Kominami, E., Ohsumi, Y., and Yoshimori, T. (2000). LC3, a mammalian homologue of yeast Apg8p is processed and localized in autophagosome membranes. EMBO J. 19, 5720–5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya, Y., Mizushima, N., Yamamoto, A., Oshitani-Okamoto, S., Ohsumi, Y., and Yoshimori, T. (2004). LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 117, 2805–2812. [DOI] [PubMed] [Google Scholar]
- Kamada, T., Nito, K., Hayashi, H., Mano, S., Hayashi, M., and Nishimura, M. (2003). Functional differentiation of peroxisomes revealed by expression profiles of peroxisomal genes in Arabidopsis thaliana. Plant Cell Physiol. 44, 1275–1289. [DOI] [PubMed] [Google Scholar]
- Kirisako, T., Baba, M., Ishihara, N., Miyazawa, K., Ohsumi, M., Yoshimori, T., Noda, T., and Ohsumi, Y. (1999). Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147, 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako, T., Ichimura, Y., Okada, H., Kabeya, Y., Mizushima, N., Yoshimori, T., Ohsumi, M., Noda, T., and Ohsumi, Y. (2000). Reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 151, 263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky, D.J., and Ohsumi, Y. (1999). Vacuolar import of proteins and organelles from the cytoplasm. Annu. Rev. Cell Dev. Biol. 15, 1–32. [DOI] [PubMed] [Google Scholar]
- Krysan, P.J., Young, J.C., and Sussman, M.R. (1999). T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11, 2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan, P.J., Young, J.C., Tax, F., and Sussman, M.R. (1996). Identification of transferred DNA insertions within Arabidopsis genes involved in signal transduction and ion transport. Proc. Natl. Acad. Sci. USA 23, 8145–8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariño, G., Uría, J.A., Puente, X.S., Quesada, V., Bordallo, J., and López-Otín, C. (2003). Human autophagins, a family of cysteine proteinases potentially implicated in cell degradation by autophagy. J. Biol. Chem. 278, 3671–3678. [DOI] [PubMed] [Google Scholar]
- Marty, F. (1999). Plant vacuoles. Plant Cell 11, 587–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matile, P. (1975). The lytic compartment of plant cells. In Cell Biology Monographs, Vol. 1, M. Alfert, W. Beermann, G. Rudkin, W. Sandritter, and P. Sitte, eds (Berlin: Springer-Verlag), pp. 1–175.
- Matsuoka, K., Higuchi, T., Maeshima, M., and Nakamura, K. (1997). A vacuolar-type H+-ATPase in a nonvacuolar organelle is required for the sorting of soluble vacuolar protein precursors in tobacco cells. Plant Cell 9, 533–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meléndez, A., Tallóczy, Z., Seaman, M., Eskelinen, E.L., Hall, D.H., and Levine, B. (2003). Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 301, 1387–1391. [DOI] [PubMed] [Google Scholar]
- Mizushima, N., Sugita, H., Yoshimori, T., and Ohsumi, Y. (1998). A new protein conjugation system in human: The counterpart of the yeast Apg12p conjugation system essential for autophagy. J. Biol. Chem. 273, 33889–33892. [DOI] [PubMed] [Google Scholar]
- Moriyasu, Y., and Ohsumi, Y. (1996). Autophagy in tobacco suspension-cultured cells in response to sucrose starvation. Plant Physiol. 111, 1233–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi, Y. (2001). Molecular dissection of autophagy: Two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2, 211–216. [DOI] [PubMed] [Google Scholar]
- Otto, G.P., Wu, M.Y., Kazgan, N., Anderson, O.R., and Kessin, R.H. (2003). Macroautophagy is required for multicellular development of the social amoeba Dictyostelium discoideum. J. Biol. Chem. 278, 17636–17645. [DOI] [PubMed] [Google Scholar]
- Paz, Y., Elazar, Z., and Fass, D. (2000). Structure of GATE-16, membrane transport modulator and mammalian ortholog of autophagocytosis factor Aut7p. J. Biol. Chem. 275, 25445–25450. [DOI] [PubMed] [Google Scholar]
- Robinson, D.G., Galili, G., Herman, E., and Hillmer, S. (1998). Topical aspects of vacuolar protein transport: Autophagy and prevacuolar compartments. J. Exp. Bot. 49, 1263–1270. [Google Scholar]
- Rojo, E., Gillmor, C.S., Kovaleva, V., Somerville, C.R., and Raikhel, N.V. (2001). VACUOLELESS1 is an essential gene required for vacuole formation and morphogenesis in Arabidopsis. Dev. Cell 1, 303–310. [DOI] [PubMed] [Google Scholar]
- Schulze, E.D., and Chapin, F.S. (1987). Plant specialization to environments of different resource availability. In Potentials and Limitations of Ecosystem Analysis: Ecological Studies, Vol. 61, E.D. Shulze and H. Zwölfer, eds (Berlin: Springer-Verlag), pp. 120–148.
- Sugawara, K., Suzuki, N.N., Fujioka, Y., Mizushima, N., Ohsumi, Y., and Inagaki, F. (2004). The crystal structure of microtubule-associated protein light chain 3, a mammalian homologue of Saccharomyces cerevisiae Atg8. Genes Cells 9, 611–618. [DOI] [PubMed] [Google Scholar]
- Surpin, M., Zheng, H., Morita, M.T., Saito, C., Avila, E., Blakeslee, J.J., Bandyopadhyay, A., Kovaleva, V., Carter, D., Murphy, A., Tasaka, M., and Raikhel, N. (2003). The VTI family of SNARE protein is necessary for plant viability and mediates different protein transport pathways. Plant Cell 15, 2885–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, S.J., Bethke, P.C., and Jones, R.L. (1998). Barley aleurone cells contain two types of vacuoles: Characterization of lytic organelles by use of fluorescent probes. Plant Cell 10, 685–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshige, K., Baba, M., Tsuboi, S., Noda, T., and Ohsumi, Y. (1992). Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 119, 301–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida, I., Tanida-Miyake, E., Komatsu, M., Ueno, T., and Kominami, E. (2002). Human Apg3p/Aut1 homologue is an authentic E2 enzyme for multiple substrates, GATE-16, GABARAP, and MAP-LC3, and facilitates the conjugation of hApg12p to hApg5p. J. Biol. Chem. 277, 13739–13744. [DOI] [PubMed] [Google Scholar]
- Thumm, M., Egner, R., Kock, B., Schlumpberger, M., Straub, M., Veenhuis, M., and Wolf, D.H. (1994). Isolation of autophagocytosis mutants of Saccaharomyces cerevisiae. FEBS Lett. 349, 275–280. [DOI] [PubMed] [Google Scholar]
- Thumm, M., and Kadowaki, T. (2001). The loss of Drosophila APG4/AUT2 function modifies the phenotypes of cut and Notch signaling pathway mutants. Mol. Genet. Genomics 266, 657–663. [DOI] [PubMed] [Google Scholar]
- Toyooka, K., Okamoto, T., and Minamikawa, T. (2001). Cotyledon cells of Vigna mungo seedlings use at least two distinct autophagic machineries for degradation of starch granules and cellular components. J. Cell Biol. 154, 973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada, M., and Ohsumi, Y. (1993). Isolation and characterization of autophagy-defective mutants of Saccaharomyces cerevisiae. FEBS Lett. 333, 169–174. [DOI] [PubMed] [Google Scholar]
- Ueda, T., Yamaguchi, M., Uchimiya, H., and Nakano, A. (2001). Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J. 20, 4730–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Wilden, W., Herman, E.M., and Chrispeels, M.J. (1980). Protein bodies of mung bean cotyledons as autophagic organelles. Proc. Natl. Acad. Sci. USA 77, 428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra, R.D. (1996). Proteolysis in plants: Mechanisms and functions. Plant Mol. Biol. 32, 275–302. [DOI] [PubMed] [Google Scholar]