Abstract
Development is a complex and well-defined process characterized by rapid cell proliferation and apoptosis. At this stage in life, a developmentally young organism is more sensitive to toxicants as compared to an adult. In response to pro-oxidant exposure, members of the Cap’n’Collar (CNC) basic leucine zipper (b-ZIP) transcription factor family (including Nfe2 and Nfe2-related factors, Nrfs) activate the expression of genes whose protein products contribute to reduced toxicity. Here, we studied the role of the CNC protein, Nfe2, in the developmental response to pro-oxidant exposure in the zebrafish (Danio rerio). Following acute waterborne exposures to diquat or tert-buytlhydroperoxide (tBOOH) at one of three developmental stages, wildtype (WT) and nfe2 knockout (KO) embryos and larvae were morphologically scored and their transcriptomes sequenced. Early in development, KO animals suffered from hypochromia that was made more severe through exposure to pro-oxidants; this phenotype in the KO may be linked to decreased expression of alas2, a gene involved in heme synthesis. WT and KO eleutheroembryos and larvae were phenotypically equally affected by exposure to pro-oxidants, where tBOOH caused more pronounced phenotypes as compared to diquat. Comparing diquat and tBOOH exposed embryos relative to the WT untreated control, a greater number of genes were up-regulated in the tBOOH condition as compared to diquat (tBOOH: 304 vs diquat: 148), including those commonly found to be differentially regulated in the vertebrate oxidative stress response (OSR) (e.g. hsp70.2, txn1, and gsr). When comparing WT and KO across all treatments and times, there were 1170 genes that were differentially expressed, of which 33 are known targets of the Nrf proteins Nrf1 and Nrf2. More specifically, in animals exposed to pro-oxidants a total of 968 genes were differentially expressed between WT and KO across developmental time, representing pathways involved in coagulation, embryonic organ development, body fluid level regulation, erythrocyte differentiation, and oxidation-reduction, amongst others. The greatest number of genes that changed in expression between WT and KO occurred in animals exposed to diquat at 2 h post fertilization (hpf). Across time and treatment, there were six genes (dhx40, cfap70, dnajb9b, slc35f4, spi-c, and gpr19) that were significantly up-regulated in KO compared to WT and four genes (fhad1, cyp4v7, nlrp12, and slc16a6a) that were significantly down-regulated. None of these genes have been previously identified as targets of Nfe2 or the Nrf family. These results demonstrate that the zebrafish Nfe2 may be a regulator of both primitive erythropoiesis and the OSR during development.
Keywords: Nfe2, Nrf, oxidative stress, development, zebrafish, RNA-Seq
1. Introduction
Development is a multifaceted process that depends on the delicate balance and timing of cellular proliferation, differentiation, and apoptosis. Reactive Oxygen Species (ROS), produced endogenously via respiration and oxygenating enzymes, play an important role in normal development by functioning as messengers in cell signal transduction and differentiation (Hitchler and Domann, 2007). In excess, ROS create a disruption of redox signaling and control (Jones, 2006), and consequently may damage lipids as well as proteins (Livingstone, 2001; Valavanidis et al., 2006), leading to premature cell cycle arrest or differentiation (Li et al., 2007; Smith et al., 2000).
To combat oxidative stress, organisms have both basal antioxidant molecules (Mandal et al., 2009) and inducible antioxidant proteins (Hu et al., 2006; Mathers et al., 2004; McMahon et al., 2001; Nair et al., 2007; Rangasamy et al., 2004; Timme-Laragy et al., 2013). Despite the fact that embryos create and need ROS for cellular signaling, they have a reduced although ontogenetically dynamic antioxidant capacity as compared to adults (Juchau, 2003; Timme-Laragy et al., 2013; Wells and Winn, 1996). This leaves embryos susceptible to toxicity associated with increases in oxidative stress, whether that arises from endogenous or exogenous sources. The essential mediators of the basal and inducible antioxidant response are known as the oxidative stress response (OSR). The OSR has been identified in adult vertebrates and cells in culture, including the NRF (Nfe2-related factor) signaling pathway (Kensler et al., 2007; Lee et al., 2005; Motohashi and Yamamoto, 2004; Osburn et al., 2006b), but only recently has embryonic sensitivity to chemically-induced oxidative stress and the role of NRF signaling become known.
The inducible response to ROS, electrophiles and pro-oxidant chemicals proceeds through a family of Cap’n’collar (CNC) basic leucine zipper (bZIP) transcription factors (Andrews et al., 1993). In mammals, there are four NFE2-related CNC-bZIP proteins: Nuclear Factor Erythroid, 2 (NFE2), Nuclear Factor Erythroid, 2 like 1 (NRF1), Nuclear Factor Erythroid, 2 like 2 (NRF2), and Nuclear Factor Erythroid, 2 like 3 (NRF3) (Motohashi et al., 2002) and two distantly related proteins BACH1 and BACH2 (Oyake et al., 1996). All vertebrate CNC members regulate transcription by binding to MAF recognition elements, also known as Antioxidant Response Elements (ARE), upon heterodimerizing with one of three small MAF proteins. (Motohashi et al., 2002, 1997). NRF2 is the most well-characterized of the family, especially as it relates to the OSR (Kensler et al., 2007; Nguyen et al., 2003). The OSR is not solely regulated by NRF2; distinct and non-overlapping functions of the various NRFs during the response have been identified (Motohashi et al., 2010; Ohtsuji et al., 2008), despite sharing binding affinity to the same cis-acting ARE motifs.
Antioxidant defenses of fish (Di Giulio et al., 1989; Hahn et al., 2014; Kelly et al., 1998; Timme-Laragy et al., 2013, 2012; Valavanidis et al., 2006; Williams et al., 2013; Winston and Di Giulio, 1991) include enzyme systems and low molecular weight antioxidants are similar to those found in mammals (George, 1994; Richard and Joel, 2008; Stegeman et al., 1992; Timme-Laragy et al., 2013). However, as a result of fish-specific whole genome duplication (Amores et al., 1998; Postlethwait et al., 2004; Taylor et al., 2001), there are several OSR genes in zebrafish that are co-orthologous to mammalian genes that exist as paralogs in fish. Thus, there are six nrf family genes: nfe2, nrf1a, nrf1b, nrf2a, nrf2b, and nrf3, all of which all share strong relationships with NRF orthologs in vertebrates (Timme-Laragy et al., 2012). Unlike nrf1 and nrf2 which are found as paralogs in zebrafish, nfe2 is found as a single ortholog, with little known about its function, although its expression has been documented (Pratt et al., 2002). nfe2 is expressed throughout development, with the highest concentration of transcript found in the unfertilized egg (Williams et al., 2013). Spatially, it is concentrated in erythroid cells from 10 somites (~12hpf) to 36 hpf and in the developing ear at 48 hpf (Pratt et al., 2002). Given its sequence similarity to human NFE2, spatial expression, and lack of expression in cloche mutants, it has been hypothesized that Nfe2 function is similar to its mammalian ortholog and is involved in hematopoiesis (Pratt et al., 2002). Phenotypic outcomes of transient Nfe2 knockdown in zebrafish and knockout in mice have provided some insight into the potential molecular targets of Nfe2.
In the mouse model, Nfe2 null mice lack circulating platelets due to a late block in megakaryocyte maturation, and most die of hemorrhage in the neonatal period (Shivdasani et al., 1995). Further examination of megakaryocytes from null embryonic mice indicate the novelty of NFE2 in regulating ROS signaling, a crucial step in the maturation of these cells. NFE2 competes with NRF2 to regulate cytoprotective genes such as heme oxygenase 1(Ho-1) and NADP(H):quinine oxidoreductase (Nqo1) (Motohashi et al., 2010). However, since the Nfe2 knockout is neonatally lethal in most mice, the role of NFE2 could be examined only in the few surviving adults from these litters. In mice that survive the knockout, it has been found that NFE2 is involved in the production of proplatelets (Lecine et al., 1998). Using the zebrafish model and transient morpholino knockdown of Nfe2, additional biological roles of Nfe2 have been elucidated including roles in swimbladder inflation and otic vesicle formation (Williams et al., 2013). However, the role of Nfe2 in responding to and regulating the OSR during development has not been explored in zebrafish.
In this study, we used a zebrafish nfe2 knockout model, which is not developmentally lethal, to examine the role of Nfe2 in regulating the response to oxidative stress. Zebrafish at three distinct developmental periods (blastula/gastrula, hatching, and larval) were acutely exposed to two model pro-oxidants: diquat (Sandy et al., 1987; Stancliffe and Pirie, 1971) and tert-butylhydroperoxide (Ahmed-Choudhury et al., 1998). Following the waterborne exposure, phenotypic outcomes were compared between wildtype and nfe2 knockout fish. In addition, transcriptome analyses were completed to identify differential expression between treatment groups and strains to ascertain the transcriptional regulatory role of Nfe2.
2. Methods
2.1. Chemicals
Diquat dibromide monohydrate was purchased from Sigma-Alrich (St. Louis, MO, USA), and freshly dissolved in 0.3X Danieau’s. Luperox® TBH70X tert-butylhydroperoxide (tBOOH) solution was purchased from Sigma-Aldrich (St. Louis, MO, USA) and freshly added to 0.3X Danieau’s. o-dianisidine was purchased from Sigma-Aldrich (St. Louis, MO, USA) and freshly dissolved in nuclease-free water.
2.2. Fish husbandry and strains
Adults were maintained and embryos were collected as previously described (Jonsson et al., 2007). Wildtype animals were on the AB background (Zebrafish International Resource Center, Eugene, OR). To create a germline nfe2 knockout, a pair of vectors containing TAL effector nucleases (TALENs) targeting exon 3 of nfe2 were generated using the REAL (Restriction Enzyme And Ligation) assembly method. Component plasmids were obtained from Addgene (www.addgene.org/talengineering/talenkit/). Briefly, target sites were selected and TALENs designed using Zifit (http://zifit.partners.org/ZiFiT/), followed by assembly. mRNA was synthesized from the vectors and injected into single cell zebrafish embryos on an AB/TL hybrid background (Rost et al., in preparation). The TALEN target sequences are: 5′-TCACCCACCTCTTATGAG-3′ and 5′-CATGACTACACGTGGTCA-3′. A subsequent founder deletion of eight base pairs (GCACATGA) was found via sequencing in exon 3 starting at nucleotide position 468 from the translational start site; this deletion resulted in a frame shift, causing a change in protein sequence starting at amino acid 111 (M → D) and the introduction of a premature stop codon 13 amino acids later (Rost et al., in preparation). The frameshift was introduced 161 amino acids prior to the Cap’n’collar (CNC) family basic leucine zipper domain that is responsible for DNA binding (Pratt et al., 2002).
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Bates College Institutional Animal Care and Use Committee (Animal Welfare AssuranceNumber A3320-01) and the University of Michigan Institutional Animal Care and Use Committee (Animal Welfare AssuranceNumber A3114-01).
2.3. Chemical exposure of embryos to diquat and tBOOH for phenotypic and RNA-Seq analyses
Waterborne exposures were carried out at three different developmental stages: 2 h post fertilization (hpf) (experiment 1), 48hpf (experiment 2), and 96hpf (experiment 3) in 3 pools of 30 embryos per condition (Fig. 1). Animals were exposed to no treatment, 20 μM diquat or 1 mM tBOOH for a 4-h dosing period in glass scintillation vials in a 20 mL volume of 0.3 × Danieau’s. Chemicals were chosen for their aqueous solubility and ability to induce oxidative stress (Higuchi et al., 2011; Osburn et al., 2006a; Timme-Laragy et al., 2009). For phenotypic analysis, animals were moved to 0.3 × Danieau’s for 48 h post-exposure (hpe) and imaged with a Leica M165 FC stereoscope and DFC310FX camera. For RNA-Seq experiments, animals were moved to 0.3 × Danieau’s for 4 hpe before being flash frozen using liquid nitrogen and stored at −80 °C.
Fig. 1.
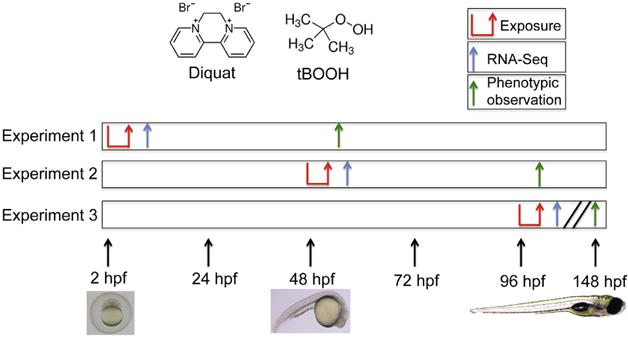
Experimental overview for phenotypic and RNA-Seq experiments. Early developmental exposure to water, tBOOH and diquat is shown in experiment one, mid-developmental exposure is shown in experiment two, and late-developmental exposure is shown in experiment three. For each experiment, the exposure duration was 4 h. RNA was collected for RNA-Seq 4 h post-exposure (hpe) and phenotypic observations were made 48 hpe.
2.4. Phenotypic assessment
Forty-five developing animals in each of the three treatment pools (water, tBOOH, diquat) were randomly selected and blindly scored in WT and KO at 48 hpe using a stage-specific 6 point scale (0.5 = extreme morphological deformities to 5 = normal morphology) as previously described (Brannen et al., 2010; Panzica-Kelly et al., 2010) at 100 hpf (dosed at 48hpf) and 148 hpf (dosed at 96hpf). For 54hpf larvae (dosed at 2 hpf), the same scale and morphological endpoints were used as described above, but normal developmental morphology was modified for the pec fin stage (Kimmel et al., 1995). Statistical differences between treatment groups and strains were assessed using a two-way ANOVA and Tukey’s multiple comparisons test in Prism v.6.
2.5. Hemoglobin staining
To detect hemoglobin in hatched zebrafish, an o-dianisidine staining method was used (Iuchi and Yamamoto, 1983; Paffett-Lugassy and Zon, 2005), in which 20 animals per treatment and strain were incubated in 0.6 mg/mL of o-dianisidine, 0.1 M sodium acetate (pH 4.5), and 0.65% hydrogen peroxide for 15–30 min. Following washes with phosphate-buffered saline (PBS), embryos were fixed at room temperature for 1 h in freshly prepared 4% paraformaldehyde, and bleached in a solution containing 0.8% KOH, 0.9% hydrogen peroxide, and 0.1% Tween-20. Images then were obtained with a Leica M165 FC stereoscope and DFC310FX camera. Animals were blindly scored, with a 3 given to an animal with normally stained erythrocytes, a 2 given to an animal with mild hypochromia, and a 1 given to an animal with severe hypochromia. Statistical differences between treatment groups and strains were assessed using a two-way ANOVA and Tukey’s multiple comparisons test in Prism v.6.
2.6. RNA extraction & sequencing
Total RNA was isolated from pooled animals using the Aurum Total RNA Mini Kit (BioRad, Hercules, California, USA) following the manufacturer’s protocol. At HudsonAlpha Institute for Biotechnology (Huntsville, AL, USA), the quality of the RNA was checked using Caliper Instrumentation (PerkinElmer, Waltham, MA, USA) and RiboGreen reagents (Invitrogen, Carlsbad, CA, USA). Directional mRNA libraries with poly(A) selection were made using New England Biolabs (Ipswich, MA, USA) reagents: NEBNext Poly (A) mRNA Isolation Magnetic Module, NEBNext First Strand Synthesis Module, NEBNext Second Strand Synthesis Module (with dUTP), NEBNext End Repair Module, NEBNext dA Tailing Module, and the NEBNext Quick Ligation Module. Quality analysis of the libraries was carried out using PicoGreen (Thermo Fisher, Waltham, MA, USA) and Caliper instrumentation (PerkinElmer, Waltham, MA, USA). 50 base pair (bp), paired end, libraries were sequenced using the Illumina HiSeq 200 platform with v4 chemistry platform at HudsonAlpha Institute for Biotechnology (Huntsville, AL, USA).
2.7. RNA-Seq data processing
Raw RNA-Sequencing (RNA-Seq) reads were obtained from HudsonAlpha as 50 bp paired-end reads in FASTQ format with approximately 25 million reads per sample. The data for each sample was concatenated into a single FASTQ file and uploaded to the McMaster instance of the online Galaxy analysis platform (galaxylab.mcmaster.ca) (Afgan et al., 2016). Reads were analyzed for various quality control parameters including: per base sequence quality, per sequence GC content, sequence length distribution and sequence duplication levels and Kmer content using the FastQC tool (Version 0.11.4) (Andrews, 2015). All files passed the quality control thresholds such that no adjustments were made on the data. Reads were then aligned to the Ensembl GRCz10 zebrafish reference genome using Tophat2 (Version 2.0.14), which functions based on Bowtie2 alignment software (Version 2.2.5) (Kim et al., 2013) using a mean inner distance of 300 bp, a maximum intron length of 380,000 bp and all other parameters set to default values. Read counts for each gene were then obtained for the aligned reads using the Htseq-counts tool (Version 0.6.1) (Anders et al., 2015). The Tophat accepted hits BAM file was analyzed against the Ensembl GTF zebrafish gene annotation reference, with pre-sorting by name through Samtools Sort (Samtools version 0.1.19) within the Htseq-count software.
2.8. Statistical analysis of RNA-Seq counts data
Raw counts data obtained from Htseq-counts were normalized using DESeq2 (Version 1.10.1) (Love et al., 2014) in R (version 3.2.3, 64bit platform) using size factors obtained from the total dataset and virtual reference sample based on the data, then filtered to keep only those genes for which at least one sample had normalized counts greater than 10. Remaining zero-counts post-filtering were assigned a value of 1 to facilitate downstream log2 transformation of the data. The filtered and normalized dataset was imported into MeV (version 4.9.0) (Saeed et al., 2003) where the data was log2 transformed and median-centered for statistical analysis. A two-factor ANOVA was performed for time, overall treatment (including both strain and compound), and their interaction with p value based on 1000 permutations of the data and α=0.01. The genes found to be significantly affected in the two-factor ANOVA were subsequently analyzed using Rank Product testing (Breitling et al., 2004) to identify the genes that were up- and down-regulated in response to compound treatment and nfe2 knockout. Rank Product tests were performed as a two-class unpaired analysis using 5000 permutations of the data and a false discovery rate of ≤10%. The unique genes found significant among all Rank Product tests were annotated using the Ensembl BioMart data-mining tool (Yates et al., 2016), then input into Gene Cluster 3.0 (C Clustering library version 1.52) and Java TreeView (version 1.1.6r4) (Saldanha, 2004) for heat map generation. Heat maps were generated from fold change data in average normalized signal for each treatment relative to average normalized signal for untreated wildtype zebrafish at 2 hpf, and the data was log2 transformed with genes and/or treatments clustered using Pearson’s correlation (i.e. centered) and average linkage clustering. Gene clusters identified visually within the heat map were analyzed using the DAVID Bioinformatics Database Functional Annotation Tool (Version 6.7) (Huang da et al., 2009a,b) for gene ontology and pathway enrichment, using an EASE value of 0.05. KEGG Pathways and Gene Ontology (GO) terms (cellular component, molecular function, biological process) were used for the DAVID analysis. If no enriched terms were detected, the top terms, regardless of significance, were identified through the Functional Annotation Clustering results. GO terms identified through the Functional Annotation analysis at an EASE value cutoff of <0.1 were visualized using Revigo (Supek et al., 2011) to obtain a scatter-plot of the enriched ontology terms. The significant genes from the Rank Product testing were additionally analyzed using the R version of the software UpSet (Lex et al., 2014) to visualize overlapping gene sets among treatments.
2.9. ARE motif search
To determine if Nfe2 is capable of directly regulating genes that were differentially expressed in the KO, a search for cis-AREs in the 10,000 base pairs upstream of the start site plus the entire length of zebrafish genes was completed as previously described (Williams et al., 2013).
2.10. Quantitative real-time PCR validation of gene expression
To validate the ten genes that were up-regulated or down-regulated in the wildtype versus nfe2 knockout model across all treatments and times, quantitative real-time PCR was conducted. Total RNA (1 μg) isolated for the RNA-Seq experiment was used to synthesize cDNA using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). Using the Agilent Mx3000 qPCR system (Agilent, Santa Clara, CA), qPCR was conducted with the Brillant II SYBR Master-mix on ten candidate genes and one housekeeping gene. For each sample, triplicate technical reactions in separate wells were run containing 5 ng of cDNA. The PCR conditions were 95 °C for 10 min followed by 35 cycles of 95 °C for 30 s, 55 °C for 60 s, and 72 °C for 60 s. Following each run, a melt curve was generated to ensure the amplification of single product. Gene-specific primers are listed in Table 1. All primers were tested for amplification efficiency using a calibration dilution curve and slope calculation approach (Rutledge and Cote, 2003). β2-Microglobulin (b2m) was chosen as a housekeeping gene due to its limited variation in expression with embryonic development and chemical exposure (McCurley and Callard, 2008). Its stability was confirmed with a One-Way ANOVA with the samples used in this study. Expression of genes was analyzed using the comparative ΔΔCT method (Livak and Schmittgen, 2001).
Table 1.
qRT-PCR Primers used for gene amplification of commonly up- and down-regulated genes across treatment and time in nfe2 knockout. Primer sequences are written 5′–3′ and amplicon size is shown.
| Gene | Primer Sequence (5′ – 3′) | Amplicon Size |
|---|---|---|
| b2m | F: CTGAAGAACGGACAGGTTATGT | 125 |
| R: ACGCTGCAGGTATATTCATCTC | ||
| cfap70 | F:CACGGAGACCTTACTGACATAGAG | 187 |
| R: GTACCGTCTGAGTGACTTTCATG | ||
| cyp4v7 | F: AGCAATTCCAGACACCTTGAC | 135 |
| R: TGGAGAAATGGAAGGTTGGAG | ||
| dhx40 | F: CACGCAGCTACCTCAGTACTTG | 85 |
| R: GACACTCTCTGGGCTACAGTG | ||
| dnajb9b | F: CTTTCACAAACTAGCCATGCG | 138 |
| R: GCTTCTCGTCTGATCGTATTCC | ||
| fhad1 | F: CTACCTCTGTTTCACCCCATC | 129 |
| R: CATCCTCTGTTAGCACTCCAG | ||
| gpr19 | F: TCTACGTGCTGCTGTCCATC | 243 |
| R: AGGAACCCAAACAGCAAATG | ||
| nlrp12 | F: CTTCAGGCCTGTTTCTCCAG | 197 |
| R: AGGAACAGTGTGGGGAGATG | ||
| slc16a6a | F: TCAACTGCATTCACCAGCTC | 184 |
| R: GGAGCGAAAGCAAATGTAGC | ||
| slc35f4 | F: TTTATGGGTGTGGCCTTAGC | 214 |
| R: CCCAGGGTAAAGAGGAGAGG | ||
| spi-c | F: TGGCTCTGTGGACACTTCTG | 202 |
| R: TACTGCTGGGCAGTGACTTG |
3. Results
3.1. Phenotypic assessments
During early development, nfe2 knockout (KO) embryos had slight hypochromia as compared to WT (Fig. 2A; Fig. 3A; Suppl Fig. 1A, B; Suppl Fig. 2A, A′, B, B′). Upon an acute pro-oxidant exposure at 2hpf and subsequent scoring at 48hpe, WT animals had slight hypochromia, but KO had an almost total loss of hemoglobin (Suppl Fig. 2C, C′ D, D′, E, E′, F, F′). Additionally, treated KOs also had slower heart rates (85–87% of untreated WT) and minor yolk sac and pericardial edema (Suppl Fig. 1D). KOs scored at 100hpf were significantly different from WT (Fig. 2B), in that they displayed delayed inflation of the swimbladder, minor delays in otolith migration, and had hypochromia (Suppl Fig. 1 E, F; Suppl Fig. 3 A, A′, B, B′). Upon acute exposure to either pro-oxidant, both WT and KO larvae were affected as compared to their respective untreated control (Fig. 2B). Exposure to either pro-oxidant caused pericardial edema and elevated heart rate (105–112% of untreated WT), hypochromia, and lack of swimbladder inflation (Suppl Fig. 1 G, H; SSuppl Fig. 2C, C′, D, D′, E, E′, F, F′). Additionally, KO fish had minor otolith migration delays (Suppl Fig. 1H) and significantly more severe hypochromia as compared to WT exposed to the pro-oxidants (Fig. 3B; Suppl Fig. 2 D, D′, F, F′). WT and KO animals dosed with tBOOH displayed similar, but more pronounced phenotypes (e.g. more severe pericardial edema) compared to animals exposed to diquat. Under control conditions, there were no differences between WT and KO at 148hpf (Fig. 2C) except for slight, but significant, hypochromia (Fig. 3C; Suppl Fig. 1 I, J; Suppl Fig. 4 Fig. 4A, A′, B, B′). When acutely exposed to either pro-oxidant at 96hpf, WT and KO were affected as compared to their respective untreated controls, but there were no significant differences between the two strains (Fig. 2C, Suppl Fig. 1K,L) except for a greater amount of hypochromia in the KO (Fig. 3C; Suppl Fig. 4C, C′, D, D′, E, E′, F, F′). Like treatment at 48hpf, tBOOH was more toxic at 96hpf as compared to diquat due to more pronounced phenotypes (Fig. 2C). Shared phenotypic outcomes upon pro-oxidant exposure between WT and KO at 96hpf included a slower heart rate (53–62% of untreated WT control) and lack of swimbladder inflation in most (88%) animals (Suppl Fig. 1K,L). In pro-oxidant exposed WT, blood pooled near the caudal fin in 32 out of 45 animals and there was slight curvature of the dorsal/ventral body axis in all animals (Suppl Fig. 1K). KO treated with pro-oxidants had a slightly more exaggerated curvature of the dorsal/ventral body axis, almost total loss of circulation, a reduced number of red blood cells which were hypochromic (Suppl Fig. 4 D, D′, F, F′), and 65% of larvae had pericardial edema (Suppl Fig. 1L).
Fig. 2.
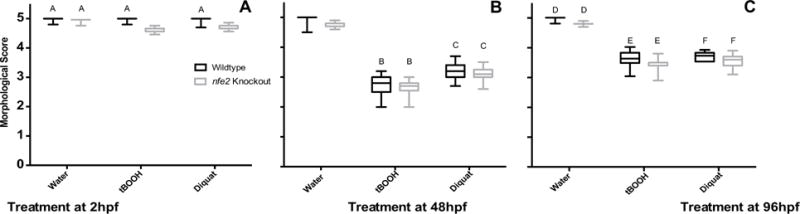
Phenotypic effects of pro-oxidant exposure. Animals treated at either 2hpf (Panel A), 48hpf (Panel B), or 96hpf (Panel C) were imaged and morphologically scored 48 hpe on a six-point scale where morphological scores ranged from 0.5 (anatomical features missing) to 5 (normal anatomical features). Boxes represent upper and lower quartiles of scores for the group, with median score marked by the internal line. Whiskers represent the range of scores outside of the middle 50% (excluding outliers). Shared letters indicate no statistical difference within a time point (two-way ANOVA, Tukey’s multiple comparisons test, p ≤ 0.05). No outliers were present.
Fig. 3.
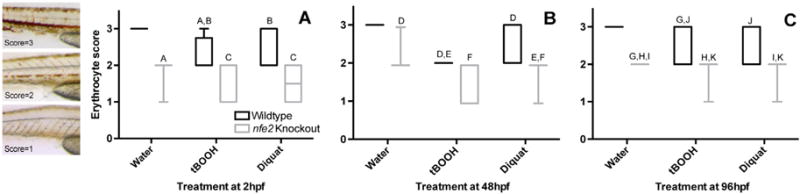
Staining and scoring of erythrocytes in developing zebrafish. Animals treated at either 2hpf (Panel A), 48hpf (Panel B), or 96hpf (Panel C) were stained with o-dianisidine, imaged and morphologically scored 48 hpe where morphological scores ranged from 1 (severe hypochromia), to 2 (mild hypochromia), to 3 (normal). Boxes represent upper and lower quartiles of scores for the group, with median score marked by the internal line. Whiskers represent the range of scores outside of the middle 50% (excluding outliers). Shared letters indicate no statistical difference within a time point (two-way ANOVA, Tukey’s multiple comparisons test, p ≤ 0.05). No outliers were present.
3.2. Transcriptomics
HiSeq read alignment to the Ensembl GRCz10 zebrafish reference genome averaged 91.6% overall read mapping with 84.0% concordant pair alignment. A total of 27,919 out of 31,953 genes in the zebrafish genome had reads subsequently assigned through Tophat alignment and a total of 14,969 genes were retained for statistical analysis based on read count filtering, with 6223 genes found significant by the two-factor ANOVA. The DESeq2 normalized Htseq-counts for all genes with RNA-Seq data, significant or otherwise, have been deposited into the Gene Expression Omnibus (GEO) database with NCBI GEO accession number GSE83466.
After ANOVA, there were 1534 unique genes found significant among all Rank Product tests that were subsequently hierarchically clustered based on fold change in average normalized signal for each treatment relative to average normalized signal for wild-type zebrafish in 0.3 × Danieau’s at 2 hpf (Fig. 4). In general, there were clusters of genes that were up-regulated (represented in red) or down-regulated (represented in green) as compared to the 2hpf WT untreated control (Fig. 4). Of particular note was the transcriptomic signature of tBOOH at later stages (48 and 96hpf), with 224 genes up-regulated and 167 genes down-regulated (Fig. 5A). In comparison to diquat exposure, tBOOH caused more genes to be up-regulated than diquat (tBOOH: 304 vs diquat: 148) and approximately the same number (tBOOH: 167 vs diquat: 175) of down-regulated genes. The greatest induction of genes by tBOOH occurred at 96hpf where 88 genes were up-regulated in WT as compared to 32 at 48hpf and 27 at 2hpf, respectively (Fig. 5A). Like WT, the largest numbers of genes were up-regulated in the KO upon exposure to tBOOH at 96hpf (135), compared to 20 at 48hpf and 2 at 2hpf, respectively. As shown in red, the thioredoxin 1 (txn1) gene was up-regulated in both strains at 48hpf and 96hpf when comparing water and tBOOH treated animals (Fig. 5A). Across developmental time within WT, three genes (mmp9 and hsp70.3, and sla) were up-regulated.
Fig. 4.
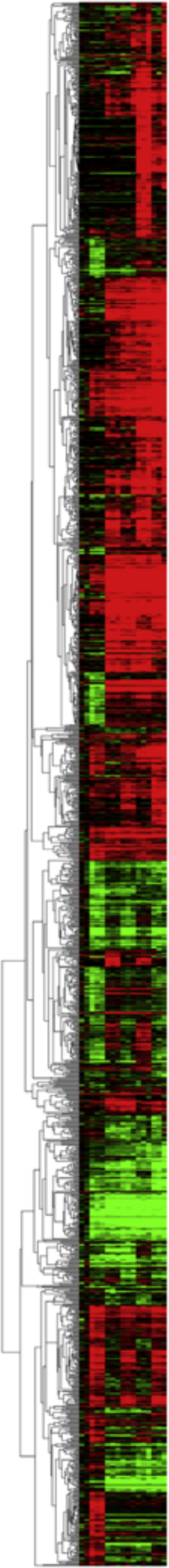
Differentially expressed genes due to treatment and strain effects. A total of 1534 genes were identified as differentially expressed in at least one Rank Product test. The heatmap of these genes shows fold change data relative to the untreated 2hpf wildtype (WT) strain, where red indicates up-regulation and green indicates down-regulation relative to this control(For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
Fig. 5.
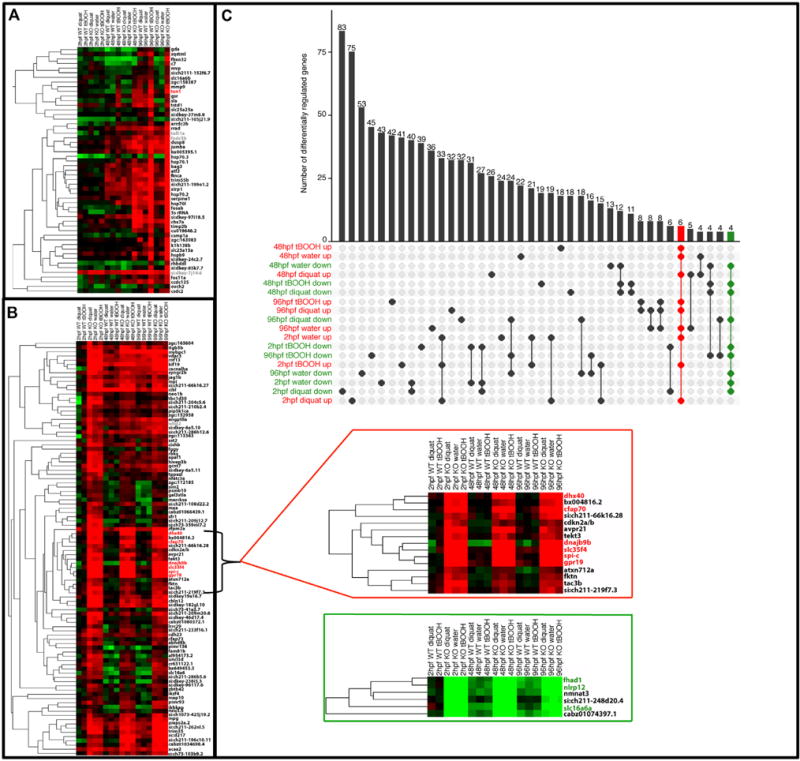
Gene regulation with tBOOH treatment, diquat treatment, and nfe2 knockout. All heatmaps show fold change data relative to the untreated 2hpf wildtype (WT), with red indicating up-regulation and green indicating down-regulation. Black gene names indicate genes significant in at least one Rank Product test, red indicate up-regulation across all relevant conditions, green indicate down-regulation across all relevant conditions, and grey indicates non-significance for the conditions of interest. A) Effect of tBOOH treatment at 48 and 96 hpf. Black genes represent those significant in at least one strain (WT or KO) or time point (48 or 96hpf) and grey represent those that were not significant for tBOOH up-regulation. txn1, in red, was found to be up-regulated across both time points and strains. B) Effect of nfe2 knockout. Up-regulation of this gene set was observed across all time points in the nfe2 knockout strain. The red-highlighted subset of genes were significantly up-regulated across all times and treatments as compared to the untreated WT strain. C) Up and down-regulated gene commonalities across wildtype versus knockout fish. The upper plot indicates gene overlaps between the Rank Product gene lists for WT versus KO tests. The dot matrix of the plot shows the intersections and the bars represent the size of the overlaps. A set of 6 genes were found to be common across all tests for up-regulation (red), and 4 were found to be common across all tests for down-regulation (green). All 6 up-regulated genes and 3 of the 4 down-regulated genes were also found to be clustered closely within the fold change heat maps with the relevant overlapping genes for up and down-regulation highlighted red and green, respectively (for interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
A strain effect was present within the heatmap, with a large number of genes being up-regulated across the various treatments upon loss of Nfe2 (Fig. 5B). In total, there were 1170 genes that were differentially regulated (up or down) across all experimental conditions when comparing WT and KO, a subset of which are shown in Fig. 5B. The largest shared gene set comprising 83 down-regulated genes is between WT and KO dosed at 2hpf with diquat (Fig. 5C). The next largest shared gene set is for 75 up-regulated genes in WT versus KO dosed at 2hpf with diquat (Fig. 5C). There is also a conserved set of 33 up-regulated genes within 2hpf samples across treatment (Fig. 5C). Across treatment and time, there were six genes commonly up-regulated (dhx40, cfap70, dnajb9b, slc35f4, spi-c, and gpr19) and four down-regulated (fhad1, cyp4v7, nlrp12, and slc16a6a) upon nfe2 knockout (Fig. 5C). These gene expression patterns were validated with qRT-PCR (Table 1, Fig. 6). Of these commonly regulated genes, all had multiple putative Nfe2 binding sites (Suppl Table 1). Given that Nrfs share a cis-ARE binding motif (Motohashi et al., 2002, 1997), all genes that were differentially regulated by the loss of Nfe2 were compared against known Nrf1 and Nrf2 targets from the literature. In total, 33 of the 1170 genes were shared targets of the Nrf family of proteins (Suppl Table 2 Table 2).
Fig. 6.
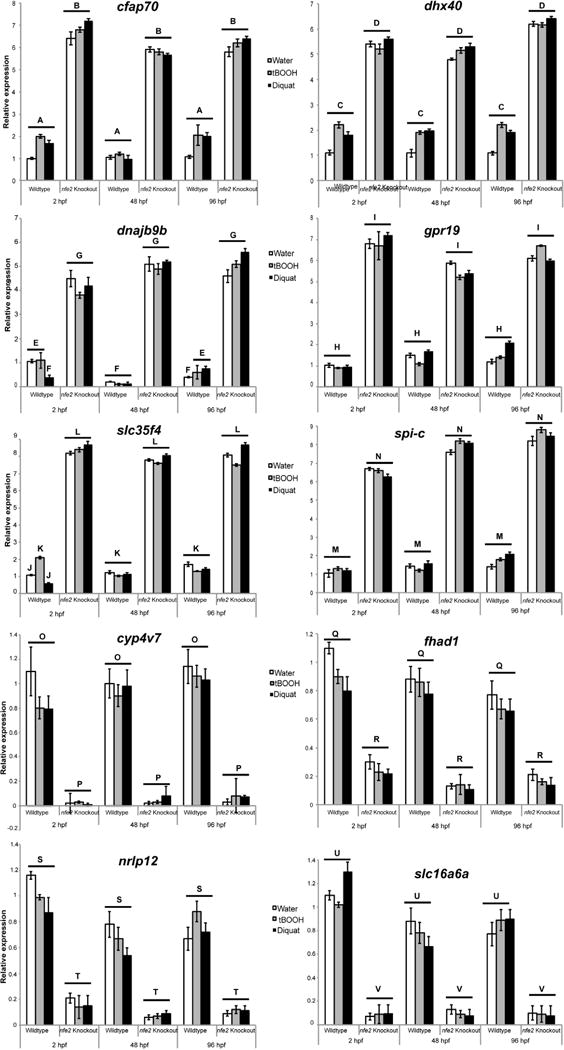
Relative expression of commonly up- and down-regulated genes across treatment and time in nfe2 knockout as measured by qRT-PCR. Relative expression values are shown for each gene, where b2 m was used as a housekeeping gene and values normalized to 2hpf WT water control. Data are presented as the mean ± S.E.M. (error bars), and N = 3 pools of 30 embryos. Within each gene, differences in expression were assessed using a two-way ANOVA and shared letters indicate no statistical difference.
To determine the overall role of Nfe2 in regulating the cellular processes of the developing animal, enriched biological themes were identified with DAVID and plotted in Revigo from a gene set containing those that were differentially regulated in wild-type versus knockout across all treatments and times. The most significantly enriched terms were related to coagulation, regulation of body fluid levels, and embryonic development (Fig. 7). Less significantly enriched terms included the development of glial cells, differentiation of erythrocytes, homeostasis, response to wounding, transport of ions and oxygen, oxidation-reduction, and adhesion of cells (Fig. 7). The genes encompassed in these GO terms are listed in Supplementary Table 3.
Fig. 7.
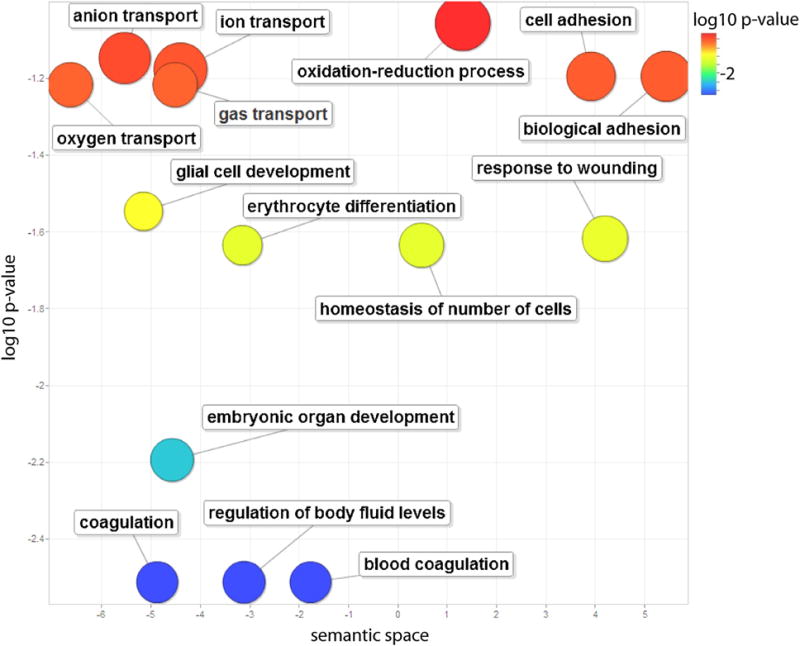
Enriched GO terms for genes differentially regulated in wildtype versus nfe2 knockout animals. GO terms meeting an EASE cutoff value of 0.1 from the DAVID Functional Annotation analysis are plotted based on enrichment p-value and ontology semantic space for genes that were differentially regulated in WT versus KO across developmental time and pro-oxidant treatment. Colors closer to the blue end of the spectrum indicate higher levels of significance, with a log10 p-value of −1.3 corresponding to a p-value of 0.05. Gene names within each term are reported in supplemental Table 3(for interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
4. Discussion
The focus of research on NFE2 has centered on its regulatory role in hematopoietic cell development in mammals and mammalian cell culture (Chan et al., 2001; Chen et al., 2007; Fujita et al., 2013; Gasiorek et al., 2012; Kacena et al., 2005; Lecine et al., 1998; Levin et al., 1999; Motohashi et al., 2010; Shivdasani and Orkin, 1995; Shivdasani et al., 1995). The mechanisms by which NFE2 regulates these processes have become clearer, including its role in regulating the response to ROS (Chan et al., 2001; Motohashi et al., 2010). Like other NRF proteins, NFE2 can bind antioxidant response elements (Motohashi et al., 2000), through which it can regulate cytoprotective genes, amongst others. Due to viviparity, the ease at which the role of NFE2 can be studied in the context of mammalian development is limited. In zebrafish, the presence of a single nfe2 ortholog (Pratt et al., 2002; Timme-Laragy et al., 2012; Williams et al., 2013) and oviparous reproduction allows for a greater observation of the role of Nfe2 throughout development and in the context of oxidative stress. Combining phenotypic assessment and transcriptomics, our study is the first to describe the role of Nfe2 throughout zebrafish development. We accomplished this by using a loss of function model, determined early developmental sensitivity to pro-oxidants, and ascertained the role of Nfe2 in regulating that response.
Phenotypic assessments in the zebrafish model point to the importance of Nfe2 in regulating primitive erythropoiesis. Hematopoiesis is well conserved across vertebrates (Galloway and Zon, 2003), and Nfe2 has been implicated as a critical regulator of globin gene expression (Blank et al., 1997; Kotkow and Orkin, 1995; Lu et al., 1994; Woon Kim et al., 2011). Both the mouse model (Shivdasani and Orkin, 1995) and our KO early larval zebrafish have hypochromia (Suppl Fig. 2 B, B′). Treated with pro-oxidants, KO and WT also have hypochromia, although it was more severe in the KO (Fig. 3, Suppl Figs. 2–4). In the KO fish under control and pro-oxidant conditions at 2 and 48hpf an erythroid-specific gene, alas2, was down-regulated upon loss of nfe2. ALAS2, or erythroid-specific δ-aminolevulinate synthase, is the first enzyme in the heme biosynthetic pathway (Riddle et al., 1989). Like our zebrafish, Alas2-null mice have erythroblast cell pellets that were deficient of heme (Harigae et al., 2003). Interestingly, mammalian NFE2 has not been directly implicated in the regulation of Alas2, but rather genes downstream of the protein such as Pgbd, Uros, Urod, Cpox, Ppox, and Fech (Rheinemann et al., 2014). In our model, Nfe2 could be playing a role in the direct regulation of alas2 and our motif analysis supports that hypothesis (Suppl Table 1). One of these motifs was within 250 bp of the transcriptional start site (TSS) on the plus strand (Suppl Table 1); given that mammalian NFE2 has been shown to bind relatively near the transcriptional start site (TSS) of genes that it directly regulates (Fujita et al., 2013), this is a likely target worthy of further investigation with chromatin immunoprecipitation (ChIP). Rescue experiments using capped mRNA would also contribute to our understanding of the role of alas2 in this phenotype. Our data also suggest that Nfe2 plays a part in regulating the development of the swimbladder and the otolith (Suppl Fig. 1).
Nfe2 morphants have similar, but not identical, morphological deformities (Williams et al., 2013), such as defects in swimbladder inflation and otolith formation, to those shown in our knockout model. To explain the morphant phenotypes it was suggested that embryos have deficient hedgehog signaling that is directly regulated by Nfe2 through cis-ARE motifs (Williams et al., 2013). In support of this suggestion, KO fish had decreased indian hedgehog a (ihha) expression compared to the untreated WT at 2hpf and 96hpf, as well as in tBOOH exposed animals at 96hpf. Zebrafish that are mutant in ihha (Korzh et al., 2011) have a similar swimbladder phenotype to our KO, likely due to aberrant swimbladder bud formation. Otolith formation and migration has also been linked to hedgehog signaling (Sapede and Pujades, 2010). In our KO, there are slight delays in otolith migration up to 100hpf; however, they are overcome by 148hpf. This phenotype is not as pronounced as was observed in the Nfe2 morphant (Williams et al., 2013), perhaps due to genetic compensation (Kok et al., 2015; Rossi et al., 2015). Our data also reveal up-regulation of cfap70, a cilia and flagella associated protein of unknown function (McClintock et al., 2008), also known as ttc18, across all time points and treatment conditions in the KO versus WT fish (Fig. 4C). This finding reveals a novel target for Nfe2 and a potential role in regulating the development of cilia, which are important to the development of the inner ear (Stooke-Vaughan et al., 2012). Further functional studies and in situ analysis may reveal the importance of cfap70 in early zebrafish development and a regulatory role for Nfe2 in that process.
While Nfe2 is likely regulating some early developmental basal functions, the intensification of the KO phenotypes upon pro-oxidant exposure during development is consistent with a role of Nfe2 in regulating the inducible OSR. It has been suggested that during early developmental zebrafish cannot mount a defense against oxidative stress (Kobayashi et al., 2002, 2009), although Hahn et al. (Hahn et al., 2014) found induction of genes known to respond to oxidative stress (gstp1, gclc, and nrf2a) in 24hpf zebrafish treated with a model pro-oxidant tert-butylhydroquinone (tBHQ). However, the response to pro-oxidant exposure has not been shown in younger embryos before this study. While prototypic OSR genes were not up-regulated at 2hpf, there are several genes that were up-regulated upon exposure to tBOOH and diquat. The mechanisms of tBOOH and diquat differ slightly, with tBOOH creating peroxyl (tBOO.) and alkoxyl (tBO.) radicals (Ahmed-Choudhury et al., 1998), and diquat creating superoxide anion radicals (Sandy et al., 1987). Several genes (strbp, trit1, cyp2p10, and asf1bb) were up-regulated in WT at 2hpf by both chemicals, and one gene (ulk2) was down-regulated by both chemicals at 2hpf compared to their untreated controls. Of these genes, trit1, or RNA-isopentenyltransferase (tRNA-IPT) is responsible for the formation of isopentenyladenosine (i6A), a modification that is found at position 37 of several tRNAs, including Selenocysteine (tRNASec) (Gu et al., 2014). The resulting tRNA (i6A-tRNA) has been linked to changes in expression levels of genes involved in the NRF2-mediated oxidative stress response (Dassano et al., 2014). However, the other genes listed above have not been ascribed roles in the OSR. Later in development (>48 h) an OSR response, like that mounted in other vertebrate models, was shown (Fig. 5A). Six (hsp70.3, hsp70l, hsp90a, dnajb1b, txn1, and sqstm1) of 20 candidate mammalian OSR genes (Hahn et al., 2014) were up-regulated in at least one pro-oxidant condition in our 48 and/or 96hpf animals compared to their respective untreated controls. Additionally, six others (atf3, dnajb1, gstpo2, igfbp1, junb, and mafb) were also up-regulated in either 48hpf or 96hpf pro-oxidant treated fish and found to be effective in developing zebrafish treated with tBHQ (Hahn et al., 2014; Timme-Laragy et al., 2012). Thus, pro-oxidants with differing modes of action can induce a similar molecular level OSR later in development. At the phenotypic level, exposure to pro-oxidants elicited changes to heart rate that were stage specific. Early-developmental (2hpf) and late-developmental (96hpf) exposure to tBOOH and diquat caused a lower heart rate compared to the untreated control whereas exposure during the hatching stage (48hpf) caused higher heart rates. This complex phenotype, combined with anemia, may be a result of compensation and decompensation processes. Early developmental exposure to excess ROS may disrupt cardiac neural crest cell migration (Tsatmali et al., 2006), leading to slower heart rates. During mid-development, increases in ROS due to chemical exposure has been shown to increase heart rate (Johnson et al., 2007), potentially as a compensatory mechanism. Later in development, decompensation as measured via a slower heart rate has been linked to chemically-induced disruptions of hematopoiesis (Li et al., 2016). Given limited studies on the topic of oxidative stress and heart development in zebrafish, future studies should focus on the cell signaling pathways that lead to these phenotypes and the role of Nfe2 and other Nrf proteins in modulating that response.
When comparing the genes that were up- and down-regulated in previous studies (Chan et al., 2001; Motohashi et al., 2010) with our own, none were found to overlap. This lack of overlap could be due to differences in species (mouse vs. zebrafish) or the biological material used (current study: whole embryo/larvae; Chan: red blood cells; Motohashi: cultured fetal liver cells). With respect to differences due to species, there is variation of tissue expression of Nfe2 (mouse: hematopoietic tissues and trophoblast cells (Gasiorek and Blank, 2015); zebrafish: intermediate cell mass, circulating blood, and inner ear (Pratt et al., 2002)), and in nfe2 nucleotide conservation outside of the DNA binding domain (Pratt et al., 2002; Timme-Laragy et al., 2012). However, a battery of other genes were dysregulated in the KO including 158 under diquat conditions, 80 under tBOOH conditions, and an additional 21 shared between the two conditions (Fig. 4C). For example, the gene for Erythrocyte membrane protein 4.1b (epb4.1b) is down-regulated with exposure to both tBOOH and diquat at 48hpf. This protein anchors the spectrin-actin based cytoskeleton to the plasma membrane of the cell (Conboy, 1993; Yawata et al., 1997) and is a known Nrf2 target (Chorley et al., 2012). In the absence of Epb4.1b, red blood cells are abnormal in their morphology and have fragile membranes (Conboy et al., 1990; Tchernia et al., 1981). The severe hypochromia in KO animals as compared to WT treated with pro-oxidants may be due to a combination of decreased levels of alas2 and epb4.1b. In WT treated with pro-oxidants, however, it is not surprising to see hypochromia as ROS accumulation and erythrocyte sensitivity has been previously linked (Ghaffari, 2008). The increased severity of the hypochromia phenotype may be due to additional changes in gene expression upon loss of Nfe2 such as the reduction of glutathione s-transferase pi-2 (gstp2) that is down-regulated in KO as compared to WT at 96hpf treated with diquat. gstp2 is a cytosolic glutathione S-transferase that is a known target of Nrf1 and Nrf2 (Chanas et al., 2002; Miyazaki et al., 2014; Suzuki et al., 2005; Xu et al., 2005), and up-regulated due to exposure to pro-oxidants like tBOOH and polycyclic aromatic hydrocarbons in developing zebrafish (Timme-Laragy et al., 2009). Other genes that were down-regulated in the KO fish were identified as genes involved in oxidation-reduction processes, oxidoreductase activity, protein tyrosine kinase activity, phosphatidylinositol signaling, and ATP binding (Fig. 5). While many genes in the OSR are up-regulated due to pro-oxidant exposure, there are likely some that are down-regulated and also regulated via Nfe2.
Half of the genes dysregulated upon pro-oxidant exposure in nfe2 knockouts, as compared to the untreated WT control, were up-regulated suggesting that Nfe2 may have a role as a repressor under certain contexts. Of note is rad17, a component of a PCNA-like complex that is involved in double-strand break repair (Nunes et al., 2008) and has several cis-AREs (Suppl Table 1). Interestingly, Nfe2 seems to be down-regulating genes involved in DNA damage repair, a response that was induced in tBHQ treated zebrafish (Hahn et al., 2014). Another gene that is up-regulated in the KO is the CCAAT/enhancer binding protein-β, cebp-b, a basic leucine zipper transcription factor that has been linked to endoplasmic reticulum mediated cell death (Meir et al., 2010) and is up-regulated in response to tBHQ (Hahn et al., 2014). A known Nfe2 target, human immunodeficiency virus type I enhancer binding protein 3b (Hivep3b) (Fujita et al., 2013), which is also a known regulator of endoplasmic stress (Imamura et al., 2014), was up-regulated in the KO at 2hpf upon pro-oxidant exposure. Thus, Nfe2 may be playing a role in suppressing factors that induce ER stress, perhaps in conjunction with Nrf2 (Cullinan et al., 2003).
In our study we were able to identify 10 novel targets for zebrafish Nfe2. Genes up-regulated across all times and treatments in the KO include dhx40, cfap70, dnajb9b, slc35f4, spi-c, and gpr19. dhx40 is a RNA helicase involved in RNA metabolism (Xu et al., 2002), cfap70 as previously mentioned is a cilia and flagella associated protein (McClintock et al., 2008), dnajb9 is an inhibitor of the pro-apoptotic function of p53 (Lee et al., 2015), and slc35f4 is a nucleotide sugar transporter (Nishimura and Naito, 2008). s-pic controls the development of red pulp macrophages (Kohyama et al., 2009) and gpr19 is an orphan G protein-coupled receptor that binds adropin (Stein et al., 2015). Genes down-regulated across all times and treatment in the KO include fhad1, nlrp12, slc16a6a, and cyp4v7. fhad1is a Forkhead-Associated (FHA) Phosphopeptide Binding Domain 1, nlrp12 is a gene that encodes a member of the CATERPILLER family of cytoplasmic proteins (Pinheiro et al., 2011), slc16a6a is a monocarboxylate transporter (Hugo et al., 2012), and cyp4v7 is a common hepatotoxicity marker (Driessen et al., 2014). While no obvious features exist between them except for their cis-ARE binding motifs (Suppl Table 1), the regulation of these genes by Nfe2 in the zebrafish model shows that its cellular and molecular role may reach beyond platelets and cytoprotection.
Nfe2 is one of the least studied of the Nrf proteins and has been hypothesized to have limited regulatory functions (Gasiorek and Blank, 2015; Hahn et al., 2015). Through the use of a knockout model in the developing zebrafish, this study highlighted the importance of Nfe2 in primitive erythropoiesis and in the potential direct regulation of genes of various functions. The exposure of early developing embryos to two different pro-oxidants allowed us to show that an OSR was mounted in blastula/gastrula embryos which was distinct from the OSR response mounted in hatched larvae. The response to pro-oxidants on the phenotypic and transcriptomic level in the KO model also identified the importance of Nfe2 through several stages of development. Several genes up- and down-regulated in the KO were also shown to be Nrf1 and Nrf2 targets, implicating Nfe2 as a co-regulator of genes with cis-ARE motifs. This study has thus identified a greater role for Nfe2 during development and in response to pro-oxidants through the use of the tractable zebrafish model that is worthy of future investigation.
Supplementary Material
Acknowledgments
We would like to thank Mary Hughes and her staff, as well as Katie Richter, for excellent fish husbandry. Dr. John Stegeman provided helpful feedback on an earlier version of this manuscript. Dr. Michaela Spitzer kindly provided help with application of the R suite of software for RNA-Seq statistical analysis. Dr. Brandon Aubie and Arjun Sharma assisted with in silico regulatory protein binding site prediction.
Funding: An Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM103423 supported LMW, NP, NS, SS, KP, ARR, BAL, and RSM. The Center for Regenerative Biology and Medicine at MDI Biological Laboratory (grant USAMRMC W81XWH-11-1-0425), and the Bodi Schmidt-Nielsen Fund supported LMW, NS, SS, KP, and RSM. AGM holds a Cisco Research Chair in Bioinformatics, supported by Cisco Systems Canada, Inc. BAL is supported by the McMaster University Faculty of Science Interdisciplinary Research Fund and a M.G. DeGroote Institute for Infectious Disease Research Summer Student Fellowship. Computer resources were supplied by the McMaster Service Lab and Repository (MSLR) computing cluster, funded in part by the Canadian Foundation for Innovation. JS, YL, AV were supported by a research project grant from the National Heart, Lung, and Blood Institute of the National Institutes of Health under grant number R01HL124232.
Abbreviations
- ARE
antioxidant response element
- bp
base pair
- bZIP
basic leucine zipper
- ChIP
chromatin immunoprecipitation
- CNC
Cap’n’Collar (CNC)
- hpe
hours post exposure
- hpf
hours post fertilization
- KO
knockout
- NFE2
Nuclear Factor (Erythroid-Derived 2)
- NRF1
Nuclear Factor (Erythroid-Derived 2)-like 1
- NRF2
Nuclear Factor (Erythroid-Derived 2)-like 2
- NRF3
Nuclear Factor (Erythroid-Derived 2)-like 3
- OSR
oxidative stress response
- PBS
phosphate buffered saline
- ROS
reactive oxygen species
- tBOOH
tert-butylhydroperoxide
- tBHQ
tert-butylhydroquinone
- tRNA-IPT
RNA-isopentenyltransferase
- TSS
transcriptional start site
- WT
wildtype
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.aquatox.2016.09.019.
Footnotes
Author contributions
LMW designed and carried out phenotypic and RNA-Seq experiments, analyzed the results, and drafted and revised the manuscript.
BAL, AGM, and ARR analyzed the RNA-Seq data and helped draft and revise the manuscript. ARR additionally designed the computational infrastructure for RNA-Seq analysis.
NS, SS, KP, RSM conceived of and refined the stage-specific morphological defect scale, carried out initial phenotypic experiments with tert-buytlhydroperoxide and diquat, and helped revise the manuscript.
NP carried out initial gene expression experiments with diquat and helped revise the manuscript
JAS, YL, and AHV developed the nfe2 knockout model and helped revise the manuscript.
All authors have read and approved the final manuscript.
References
- Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, ÄOEech M, Chilton J, Clements D, Coraor N, Eberhard C, Grüning BR, Guerler A, Hillman-Jackson J, Von Kuster G, Rasche E, Soranzo N, Turaga N, Taylor J, Nekrutenko A, Goecks J. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses. Nucl Acids Res. 2016 doi: 10.1093/nar/gkw343. http://nar.oxfordjournals.org/content/early/2016/05/02/nar.gkw343.full. [DOI] [PMC free article] [PubMed]
- Ahmed-Choudhury J, Orsler DJ, Coleman R. Hepatobiliary effects of tertiary-butylhydroperoxide (tBOOH) in isolated rat hepatocyte couplets. Toxicol Appl Pharmacol. 1998;152:270–275. doi: 10.1006/taap.1998.8495. [DOI] [PubMed] [Google Scholar]
- Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL, Westerfield M, Ekker M, Postlethwait JH. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq? a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews NC, Erdjumentbromage H, Davidson MB, Tempst P, Orkin SH. Erythroid transcription factor nf-E2 is a hematopoietic-Specific basic leucine zipper protein. Nature. 1993;362:722–728. doi: 10.1038/362722a0. [DOI] [PubMed] [Google Scholar]
- Andrews S. FastQC: a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- Blank V, Kim MJ, Andrews NC. Human MafG is a functional partner for p45 NF-E2 in activating globin gene expression. Blood. 1997;89:3925–3935. [PubMed] [Google Scholar]
- Brannen KC, Panzica-Kelly JM, Danberry TL, Augustine-Rauch KA. Development of a zebrafish embryo teratogenicity assay and quantitative prediction model. Birth Defects Res B: Dev Reprod Toxicol. 2010;89:66–77. doi: 10.1002/bdrb.20223. [DOI] [PubMed] [Google Scholar]
- Breitling R, Amtmann A, Herzyk P. Iterative Group Analysis (iGA): a simple tool to enhance sensitivity and facilitate interpretation of microarray experiments. BMC Bioinform. 2004;5:1–8. doi: 10.1186/1471-2105-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JY, Kwong M, Lo M, Emerson R, Kuypers FA. Reduced oxidative-stress response in red blood cells from p45NFE2-deficient mice. Blood. 2001;97:2151–2158. doi: 10.1182/blood.v97.7.2151. [DOI] [PubMed] [Google Scholar]
- Chanas SA, Jiang Q, McMahon M, McWalter GK, McLellan LI, Elcombe CR, Henderson CJ, Wolf CR, Moffat GJ, Itoh K, Yamamoto M, Hayes JD. Loss of the Nrf2 transcription factor causes a marked reduction in constitutive and inducible expression of the glutathione S-transferase Gsta1, Gsta2, Gstm1, Gstm2, Gstm3 and Gstm4 genes in the livers of male and female mice. Biochem J. 2002;365:405–416. doi: 10.1042/BJ20020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Hu M, Shivdasani RA. Expression analysis of primary mouse megakaryocyte differentiation and its application in identifying stage-specific molecular markers and a novel transcriptional target of NF-E2. Blood. 2007;109:1451–1459. doi: 10.1182/blood-2006-08-038901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorley BN, Campbell MR, Wang X, Karaca M, Sambandan D, Bangura F, Xue P, Pi J, Kleeberger SR, Bell DA. Identification of novel NRF2-regulated genes by ChIP-Seq: influence on retinoid X receptor alpha. Nucleic Acids Res. 2012;40:7416–7429. doi: 10.1093/nar/gks409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy J, Marchesi S, Kim R, Agre P, Kan YW, Mohandas N. Molecular analysis of insertion/deletion mutations in protein 4.1 in elliptocytosis: II. Determination of molecular genetic origins of rearrangements. J Clin Invest. 1990;86:524–530. doi: 10.1172/JCI114739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy JG. Structure function, and molecular genetics of erythroid membrane skeletal protein 4.1 in normal and abnormal red blood cells. Semin Hematol. 1993;30:58–73. [PubMed] [Google Scholar]
- Cullinan SB, Zhang D, Hannink M, Arvisais E, Kaufman RJ, Diehl JA. Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol Cell Biol. 2003;23:7198–7209. doi: 10.1128/MCB.23.20.7198-7209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassano A, Mancuso M, Giardullo P, Cecco LD, Ciuffreda P, Santaniello E, Saran A, Dragani TA, Colombo F. N(6)-isopentenyladenosine and analogs activate the NRF2-mediated antioxidant response. Redox Biol. 2014;2:580–589. doi: 10.1016/j.redox.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giulio RT, Washburn PC, Wenning RJ, Winston GW, Jewell CS. Biochemical responses in aquatic animals: a review of determinants of oxidative stress. Environ Toxicol Chem. 1989;8:1103–1123. [Google Scholar]
- Driessen M, Kienhuis AS, Vitins AP, Pennings JL, Pronk TE, van den Brandhof EJ, Roodbergen M, van de Water B, van der Ven LT. Gene expression markers in the zebrafish embryo reflect a hepatotoxic response in animal models and humans. Toxicol Lett. 2014;230:48–56. doi: 10.1016/j.toxlet.2014.06.844. [DOI] [PubMed] [Google Scholar]
- Fujita R, Takayama-Tsujimoto M, Satoh H, Gutierrez L, Aburatani H, Fujii S, Sarai A, Bresnick EH, Yamamoto M, Motohashi H. NF-E2 p45 is important for establishing normal function of platelets. Mol Cell Biol. 2013;33:2659–2670. doi: 10.1128/MCB.01274-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway JL, Zon LI. Ontogeny of hematopoiesis: examining the emergence of hematopoietic cells in the vertebrate embryo. Curr Top Dev Biol. 2003;53:139–158. doi: 10.1016/s0070-2153(03)53004-6. [DOI] [PubMed] [Google Scholar]
- Gasiorek JJ, Blank V. Regulation and function of the NFE2 transcription factor in hematopoietic and non-hematopoietic cells. Cell Mol Life Sci. 2015;72:2323–2335. doi: 10.1007/s00018-015-1866-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiorek JJ, Nouhi Z, Blank V. Abnormal differentiation of erythroid precursors in p45 NF-E2(−/−) mice. Exp Hematol. 2012;40:393–400. doi: 10.1016/j.exphem.2012.01.007. [DOI] [PubMed] [Google Scholar]
- George SG. Enzymology and molecular biology of phase II xenobiotic-conjugating enzymes in fish. In: Malins DC, Ostrander GK, editors. Aquatic Toxicology: Molecular, Biochemical, and Cellular Perspectives. CRC/Lewis; Boca Raton: 1994. [Google Scholar]
- Ghaffari S. Oxidative stress in the regulation of normal and neoplastic hematopoiesis. Antioxid Redox Signal. 2008;10:1923–1940. doi: 10.1089/ars.2008.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C, Begley TJ, Dedon PC. tRNA modifications regulate translation during cellular stress. FEBS Lett. 2014;588:4287–4296. doi: 10.1016/j.febslet.2014.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn ME, McArthur AG, Karchner SI, Franks DG, Jenny MJ, Timme-Laragy AR, Stegeman JJ, Woodin BR, Cipriano MJ, Linney E. The transcriptional response to oxidative stress during vertebrate development: effects of tert-butylhydroquinone and 2,3,7,8-tetrachlorodibenzo-p-dioxin. PLoS One. 2014;9:e113158. doi: 10.1371/journal.pone.0113158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn ME, Timme-Laragy AR, Karchner SI, Stegeman JJ. Nrf2 and Nrf2-related proteins in development and developmental toxicity: insights from studies in zebrafish (Danio rerio) Free Radic Biol Med. 2015;88:275–289. doi: 10.1016/j.freeradbiomed.2015.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harigae H, Nakajima O, Suwabe N, Yokoyama H, Furuyama K, Sasaki T, Kaku M, Yamamoto M, Sassa S. Aberrant iron accumulation and oxidized status of erythroid-specific delta-aminolevulinate synthase (ALAS2)-deficient definitive erythroblasts. Blood. 2003;101:1188–1193. doi: 10.1182/blood-2002-01-0309. [DOI] [PubMed] [Google Scholar]
- Higuchi M, Yoshikawa Y, Orino K, Watanabe K. Effect of diquat-induced oxidative stress on iron metabolism in male Fischer-344 rats. Biometals. 2011;24:1123–1131. doi: 10.1007/s10534-011-9471-0. [DOI] [PubMed] [Google Scholar]
- Hitchler MJ, Domann FE. An epigenetic perspective on the free radical theory of development. Free Radic Biol Med. 2007;43:1023–1036. doi: 10.1016/j.freeradbiomed.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R, Xu C, Shen G, Jain MR, Khor TO, Gopalkrishnan A, Lin W, Reddy B, Chan JY, Kong AN. Identification of Nrf2-regulated genes induced by chemopreventive isothiocyanate PEITC by oligonucleotide microarray. Life Sci. 2006;79:1944–1955. doi: 10.1016/j.lfs.2006.06.019. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hugo SE, Cruz-Garcia L, Karanth S, Anderson RM, Stainier DY, Schlegel A. A monocarboxylate transporter required for hepatocyte secretion of ketone bodies during fasting. Genes Dev. 2012;26:282–293. doi: 10.1101/gad.180968.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura K, Maeda S, Kawamura I, Matsuyama K, Shinohara N, Yahiro Y, Nagano S, Setoguchi T, Yokouchi M, Ishidou Y, Komiya S. Human immunodeficiency virus type 1 enhancer-binding protein 3 is essential for the expression of asparagine-linked glycosylation 2 in the regulation of osteoblast and chondrocyte differentiation. J Biol Chem. 2014;289:9865–9879. doi: 10.1074/jbc.M113.520585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi I, Yamamoto M. Erythropoiesis in the developing rainbow trout, Salmo gairdneri irideus: histochemical and immunochemical detection of erythropoietic organs. J Exp Zool. 1983;226:409–417. doi: 10.1002/jez.1402260311. [DOI] [PubMed] [Google Scholar]
- Johnson A, Carew E, Sloman KA. The effects of copper on the morphological and functional development of zebrafish embryos. Aquat Toxicol. 2007;84:431–438. doi: 10.1016/j.aquatox.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Jones DP. Redefining oxidative stress. Antioxid Redox Signal. 2006;8:1865–1879. doi: 10.1089/ars.2006.8.1865. [DOI] [PubMed] [Google Scholar]
- Jonsson ME, Jenny MJ, Woodin BR, Hahn ME, Stegeman JJ. Role of AHR2 in the expression of novel cytochrome p450 1 family genes, cell cycle genes, and morphological defects in developing zebra fish exposed to 3,3′,4,4′,5-pentachlorobiphenyl or 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol Sci. 2007;100:180–193. doi: 10.1093/toxsci/kfm207. [DOI] [PubMed] [Google Scholar]
- Juchau MR. Bioactivation in chemical teratogenesis. Annu Rev Pharmacool Toxicol. 2003;29:165–187. doi: 10.1146/annurev.pa.29.040189.001121. [DOI] [PubMed] [Google Scholar]
- Kacena MA, Gundberg CM, Nelson T, Horowitz MC. Loss of the transcription factor p45 NF-E2 results in a developmental arrest of megakaryocyte differentiation and the onset of a high bone mass phenotype. Bone. 2005;36:215–223. doi: 10.1016/j.bone.2004.09.024. [DOI] [PubMed] [Google Scholar]
- Kelly KA, Havrilla CM, Brady TC, Abramo KH, Levin ED. Oxidative stress in toxicology: established mammalian and emerging piscine model systems. Environ Health Perspect. 1998;106:375–384. doi: 10.1289/ehp.98106375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensler TW, Wakabayash N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacool Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic-vevelopment of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Itoh K, Suzuki T, Osanai H, Nishikawa K, Katoh Y, Takagi Y, Yamamoto M. Identification of the interactive interface and phylogenic conservation of the Nrf2-Keap1 system. Genes Cells. 2002;7:807–820. doi: 10.1046/j.1365-2443.2002.00561.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Li L, Iwamoto N, Nakajima-Takagi Y, Kaneko H, Nakayama Y, Eguchi M, Wada Y, Kumagai Y, Yamamoto M. The antioxidant defense system keap1-Nrf2 comprises a multiple sensing mechanism for responding to a wide range of chemical compounds. Mol Cell Biol. 2009;29:493–502. doi: 10.1128/MCB.01080-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohyama M, Ise W, Edelson BT, Wilker PR, Hildner K, Mejia C, Frazier WA, Murphy TL, Murphy KM. Role for Spi-C in the development of red pulp macrophages and splenic iron homeostasis. Nature. 2009;457:318–321. doi: 10.1038/nature07472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok O FatmaÂ, Shin M, Ni CW, Gupta A, Grosse Ann S, van Impel A, Kirchmaier Bettina C, Peterson-Maduro J, Kourkoulis G, Male I, DeSantis Dana F, Sheppard-Tindell S, Ebarasi L, Betsholtz C, Schulte-Merker S, Wolfe Scot A, Lawson Nathan D. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev Cell. 2015;32:97–108. doi: 10.1016/j.devcel.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzh S, Winata CL, Zheng W, Yang S, Yin A, Ingham P, Korzh V, Gong Z. The interaction of epithelial Ihha and mesenchymal Fgf10 in zebrafish esophageal and swimbladder development. Dev Biol. 2011;359:262–276. doi: 10.1016/j.ydbio.2011.08.024. [DOI] [PubMed] [Google Scholar]
- Kotkow KJ, Orkin SH. Dependence of globin gene expression in mouse erythroleukemia cells on the NF-E2 heterodimer. Mol Cell Biol. 1995;15:4640–4647. doi: 10.1128/mcb.15.8.4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecine P, Villeval JL, Vyas P, Swencki B, Xu Y, Shivdasani RA. Mice lacking transcription factor NF-E2 provide in vivo validation of the proplatelet model of thrombocytopoiesis and show a platelet production defect that is intrinsic to megakaryocytes. Blood. 1998;92:1608–1616. [PubMed] [Google Scholar]
- Lee JM, Li J, Johnson DA, Stein TD, Kraft AD, Calkins MJ, Jakel RJ, Johnson JA. Nrf2, a multi-organ protector? FASEB J. 2005;19:1061–1066. doi: 10.1096/fj.04-2591hyp. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Kim JM, Kim KH, Heo JI, Kwak SJ, Han JA. Genotoxic stress/p53-induced DNAJB9 inhibits the pro-apoptotic function of p53. Cell Death Differ. 2015;22:86–95. doi: 10.1038/cdd.2014.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin J, Peng JP, Baker GR, Villeval JL, Lecine P, Burstein SA, Shivdasani RA. Pathophysiology of thrombocytopenia and anemia in mice lacking transcription factor NF-E2. Blood. 1999;94:3037–3047. [PubMed] [Google Scholar]
- Lex A, Gahlenborg N, Strobelt H, Vuillemot R, Pfister H. UpSet: visualization of intersecting sets. IEEE Trans Vis Comput Graph. 2014;20:1983–1992. doi: 10.1109/TVCG.2014.2346248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Dong T, Proschel C, Noble M. Chemically diverse toxicants converge on Fyn and c-Cbl to disrupt precursor cell function. PLoS Biol. 2007;5:e35. doi: 10.1371/journal.pbio.0050035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Bonneton F, Tohme M, Bernard L, Chen XY, Laudet V. In vivo screening using transgenic zebrafish embryos reveals new effects of HDAC inhibitors trichostatin a and valproic acid on organogenesis. PLoS One. 2016;11:e0149497. doi: 10.1371/journal.pone.0149497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Livingstone DR. Contaminant-stimulated reactive oxygen species production and oxidative damage in aquatic organisms. Mar Pollut Bull. 2001;42:656–666. doi: 10.1016/s0025-326x(01)00060-1. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SJ, Rowan S, Bani MR, Ben-David Y. Retroviral integration within the Fli-2 locus results in inactivation of the erythroid transcription factor NF-E2 in Friend erythroleukemias: evidence that NF-E2 is essential for globin expression. Proc Natl Acad Sci U S A. 1994;91:8398–8402. doi: 10.1073/pnas.91.18.8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal S, Yadav S, Yadav S, Nema RK. Antioxidants: a review. J Chem Pharm Res. 2009;1:102–104. [Google Scholar]
- Mathers J, Fraser JA, McMahon M, Saunders RD, Hayes JD, McLellan LI. Antioxidant and cytoprotective responses to redox stress. Biochem Soc Symp. 2004:157–176. doi: 10.1042/bss0710157. [DOI] [PubMed] [Google Scholar]
- McClintock TS, Glasser CE, Bose SC, Bergman DA. Tissue expression patterns identify mouse cilia genes. Physiol Genomics. 2008;32:198–206. doi: 10.1152/physiolgenomics.00128.2007. [DOI] [PubMed] [Google Scholar]
- McCurley AT, Callard GV. Characterization of housekeeping genes in zebrafish: male-female differences and effects of tissue typedevelopmental stage and chemical treatment. BMC Mol Biol. 2008;9:1–12. doi: 10.1186/1471-2199-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon M, Itoh K, Yamamoto M, Chanas SA, Henderson CJ, McLellan LI, Wolf CR, Cavin C, Hayes JD. The cap ‘n’ collar basic leucine zipper transcription factor Nrf2 (NF-E2 p45-related factor 2) controls both constitutive and inducible expression of intestinal detoxification and glutathione biosynthetic enzymes. Cancer Res. 2001;61:3299–3307. [PubMed] [Google Scholar]
- Meir O, Dvash E, Werman A, Rubinstein M. C/EBP-beta regulates endoplasmic reticulum stress-triggered cell death in mouse and human models. PLoS One. 2010;5:e9516. doi: 10.1371/journal.pone.0009516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki Y, Shimizu A, Pastan I, Taguchi K, Naganuma E, Suzuki T, Hosoya T, Yokoo T, Saito A, Miyata T, Yamamoto M, Matsusaka T. Keap1 inhibition attenuates glomerulosclerosis. Nephrol Dial Transplant. 2014;29:783–791. doi: 10.1093/ndt/gfu002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi H, Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol Med. 2004;10:549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Motohashi H, Shavit JA, Igarashi K, Yamamoto M, Engel JD. The world according to Maf. Nucleic Acids Res. 1997;25:2953–2959. doi: 10.1093/nar/25.15.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi H, Katsuoka F, Shavit JA, Engel JD, Yamamoto M. Positive or negative MARE-dependent transcriptional regulation is determined by the abundance of small Maf proteins. Cell. 2000;103:865–875. doi: 10.1016/s0092-8674(00)00190-2. [DOI] [PubMed] [Google Scholar]
- Motohashi H, O’Connor T, Katsuoka F, Engel JD, Yamamoto M. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene. 2002;294:1–12. doi: 10.1016/s0378-1119(02)00788-6. [DOI] [PubMed] [Google Scholar]
- Motohashi H, Kimura M, Fujita R, Inoue A, Pan XQ, Takayama M, Katsuoka F, Aburatani H, Bresnick EH, Yamamoto M. NF-E2 domination over Nrf2 promotes ROS accumulation and megakaryocytic maturation. Blood. 2010;115:677–686. doi: 10.1182/blood-2009-05-223107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S, Xu C, Shen G, Hebbar V, Gopalakrishnan A, Hu R, Jain MR, Liew C, Chan JY, Kong AN. Toxicogenomics of endoplasmic reticulum stress inducer tunicamycin in the small intestine and liver of Nrf2 knockout and C57BL/6J mice. Toxicol Lett. 2007;168:21–39. doi: 10.1016/j.toxlet.2006.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T, Sherratt PJ, Pickett CB. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu Rev Pharmacol Toxicol. 2003;43:233–260. doi: 10.1146/annurev.pharmtox.43.100901.140229. [DOI] [PubMed] [Google Scholar]
- Nishimura M, Naito S. Tissue-specific mRNA expression profiles of human solute carrier transporter superfamilies. Drug Metab Pharmacokinet. 2008;23:22–44. doi: 10.2133/dmpk.23.22. [DOI] [PubMed] [Google Scholar]
- Nunes E, Candreva E, Bracesco N, Sanchez A, Dell M. HDF1 and RAD17 genes are involved in DNA double-strand break repair in stationary phase Saccharomyces cerevisiae. J Biol Phys. 2008;34:63–71. doi: 10.1007/s10867-008-9105-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuji M, Katsuoka F, Kobayashi A, Aburatani H, Hayes JD, Yamamoto M. Nrf1 and nrf2 play distinct roles in activation of antioxidant response element-dependent genes. J Biol Chem. 2008;283:33554–33562. doi: 10.1074/jbc.M804597200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osburn WO, Wakabayashi N, Misra V, Nilles T, Biswal S, Trush MA, Kensler TW. Nrf2 regulates an adaptive response protecting against oxidative damage following diquat-Mediated formation of superoxide anion. Arch Biochem Biophys. 2006a;454:7–15. doi: 10.1016/j.abb.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osburn WO, Wakabayashi N, Misra V, Nilles T, Biswal S, Trush MA, Kensler TW. Nrf2 regulates an adaptive response protecting against oxidative damage following diquat-mediated formation of superoxide anion. Arch Biochem Biophys. 2006b;454:7–15. doi: 10.1016/j.abb.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyake T, Itoh K, Motohashi H, Hayashi N, Hoshino H, Nishizawa M, Yamamoto M, Igarashi K. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol Cell Biol. 1996;16:6083–6095. doi: 10.1128/mcb.16.11.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paffett-Lugassy NlN, Zon LI. Analysis of hematopoietic development in the zebrafish. In: Baron MH, editor. Developmental Hematopoiesis: Methods and Protocols. Humana Press; Totowa, NJ: 2005. pp. 171–198. [DOI] [PubMed] [Google Scholar]
- Panzica-Kelly JM, Zhang CX, Danberry TL, Flood A, DeLan JW, Brannen KC, Augustine-Rauch KA. Morphological score assignment guidelines for the dechorionated zebrafish teratogenicity assay. Birth Defects Res B Dev Reprod Toxicol. 2010;89:382–395. doi: 10.1002/bdrb.20260. [DOI] [PubMed] [Google Scholar]
- Pinheiro AS, Eibl C, Ekman-Vural Z, Schwarzenbacher R, Peti W. The NLRP12 pyrin domain †structure, dynamics and functional insights. J Mol Biol. 2011;413:790–803. doi: 10.1016/j.jmb.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postlethwait J, Amores A, Cresko W, Singer A, Yan YL. Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet. 2004;20:481–490. doi: 10.1016/j.tig.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Pratt SJ, Drejer A, Foott H, Barut B, Brownlie A, Postlethwait J, Kato Y, Yamamoto M, Zon LI. Isolation and characterization of zebrafish NFE2. Physiol Genomics. 2002;11:91–98. doi: 10.1152/physiolgenomics.00112.2001. [DOI] [PubMed] [Google Scholar]
- Rangasamy T, Cho CY, Thimmulappa RK, Zhen L, Srisuma SS, Kensler TW, Yamamoto M, Petrache I, Tuder RM, Biswal S. Genetic ablation of Nrf2 enhances susceptibility to cigarette smoke-induced emphysema in mice. J Clin Invest. 2004;114:1248–1259. doi: 10.1172/JCI21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheinemann L, Seeger TS, Wehrle J, Pahl HL. NFE2 regulates transcription of multiple enzymes in the heme biosynthesis pathway. Haematologica. 2014;99:e208–210. doi: 10.3324/haematol.2014.106393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard TDG, Joel NM. Reactive Oxygen Species and Oxidative Stress The Toxicology of Fishes. CRC Press; 2008. pp. 273–324. [Google Scholar]
- Riddle RD, Yamamoto M, Engel JD. Expression of delta-aminolevulinate synthase in avian cells: separate genes encode erythroid-specific and nonspecific isozymes. Proc Natl Acad Sci U S A. 1989;86:792–796. doi: 10.1073/pnas.86.3.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A, Kontarakis Z, Gerri C, Nolte H, Holper S, Kruger M, Stainier DYR. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524:230–233. doi: 10.1038/nature14580. [DOI] [PubMed] [Google Scholar]
- Rost MS, Shestopalov I, Liu Y, Vo A, Barrett FG, Stachura D, Zon L, Shavit J. Nfe2 is Dispensable for Early, but Required for Adult, Thrombocyte Formation and Function in Zebrafish. In preparation. doi: 10.1182/bloodadvances.2018021865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge RG, Cote C. Mathematics of quantitative kinetic PCR and the application of standard curves. Nucleic Acids Res. 2003;31:e93. doi: 10.1093/nar/gng093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview–extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Sandy MS, Moldeus P, Ross D, Smith MT. Cytotoxicity of the redox cycling compound diquat in isolated hepatocytes: involvement of hydrogen peroxide and transition metals. Arch Biochem Biophys. 1987;259:29–37. doi: 10.1016/0003-9861(87)90466-8. [DOI] [PubMed] [Google Scholar]
- Sapede D, Pujades C. Hedgehog signaling governs the development of otic sensory epithelium and its associated innervation in zebrafish. J Neurosci. 2010;30:3612–3623. doi: 10.1523/JNEUROSCI.5109-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivdasani RA, Orkin SH. Erythropoiesis and globin gene expression in mice lacking the transcription factor NF-E2. Proc Natl Acad Sci U S A. 1995;92:8690–8694. doi: 10.1073/pnas.92.19.8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivdasani RA, Rosenblatt MF, Zuckerfranklin D, Jackson CW, Hunt P, Saris CJM, Orkin SH. Transcription factor nf-E2 is required for platelet formation independent of the actions of Thrombopoietin/Mgdf in megakaryocyte development. Cell. 1995;81:695–704. doi: 10.1016/0092-8674(95)90531-6. [DOI] [PubMed] [Google Scholar]
- Smith J, Ladi E, Mayer-Proschel M, Noble M. Redox state is a central modulator of the balance between self-renewal and differentiation in a dividing glial precursor cell. Proc Natl Acad Sci U S A. 2000;97:10032–10037. doi: 10.1073/pnas.170209797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancliffe TC, Pirie A. The production of superoxide radicals in reactions of the herbicide diquat. FEBS Lett. 1971;17:297–299. doi: 10.1016/0014-5793(71)80168-0. [DOI] [PubMed] [Google Scholar]
- Stegeman JJ, Brouwer M, Di Giulio R, Forlin L, Fowler BM, Sanders BM, Van Veld P. Molecular responses to environmental contamination: enzyme and protein systems as indicators of contaminant exposure and effect. In: Huggett RJ, editor. Biomarkers for Chemical Contaminants. CRC; Boca Raton: 1992. [Google Scholar]
- Stein LM, Yosten GL, Samson WK. Adropin acts in brain to inhibit water drinking: potential interaction with the orphan G protein-coupled receptor, GPR19. Am J Physiol Regul Integr Comp Physiol. 2015;310:R476–480. doi: 10.1152/ajpregu.00511.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stooke-Vaughan GA, Huang P, Hammond KL, Schier AF, Whitfield TT. The role of hair cells, cilia and ciliary motility in otolith formation in the zebrafish otic vesicle. Development. 2012;139:1777–1787. doi: 10.1242/dev.079947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bosnjak M, Skunca N, Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Takagi Y, Osanai H, Li L, Takeuchi M, Katoh Y, Kobayashi M, Yamamoto M. Pi class glutathione S-transferase genes are regulated by Nrf2 through an evolutionarily conserved regulatory element in zebrafish. Biochem J. 2005;388:65–73. doi: 10.1042/BJ20041860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JS, Van de Peer Y, Braasch I, Meyer A. Comparative genomics provides evidence for an ancient genome duplication event in fish. Philos Trans R Soc Lond B Biol Sci. 2001;356:1661–1679. doi: 10.1098/rstb.2001.0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchernia G, Mohandas N, Shohet SB. Deficiency of skeletal membrane protein band 4.1 in homozygous hereditary elliptocytosis. Implications for erythrocyte membrane stability. J Clin Invest. 1981;68:454–460. doi: 10.1172/JCI110275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme-Laragy AR, Van Tiem LA, Linney EA, Di Giulio RT. Antioxidant responses and NRF2 in synergistic developmental toxicity of PAHs in zebrafish. Toxicol Sci. 2009;109:217–227. doi: 10.1093/toxsci/kfp038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme-Laragy AR, Karchner SI, Franks DG, Jenny MJ, Harbeitner RC, Goldstone JV, McArthur AG, Hahn ME. Nrf2b, novel zebrafish paralog of oxidant-responsive transcription factor NF-E2-related factor 2 (NRF2) J Biol Chem. 2012;287:4609–4627. doi: 10.1074/jbc.M111.260125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme-Laragy AR, Goldstone JV, Imhoff BR, Stegeman JJ, Hahn ME, Hansen JM. Glutathione redox dynamics and expression of glutathione-related genes in the developing embryo. Free Radic Biol Med. 2013;65:89–101. doi: 10.1016/j.freeradbiomed.2013.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsatmali M, Walcott EC, Makarenkova H, Crossin KL. Reactive oxygen species modulate the differentiation of neurons in clonal cortical cultures. Mol Cell Neurosci. 2006;33:345–357. doi: 10.1016/j.mcn.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valavanidis A, Vlahogianni T, Dassenakis M, Scoullos M. Molecular biomarkers of oxidative stress in aquatic organisms in relation to toxic environmental pollutants. Ecotoxicol Environ Saf. 2006;64:178–189. doi: 10.1016/j.ecoenv.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Wells PG, Winn LM. Biochemical toxicology of chemical teratogenesis. Crit Rev Biochem Mol Biol. 1996;31:1–40. doi: 10.3109/10409239609110574. [DOI] [PubMed] [Google Scholar]
- Williams LM, Timme-Laragy AR, Goldstone JV, McArthur AG, Stegeman JJ, Smolowitz RM, Hahn ME. Developmental expression of the nfe2-related factor (Nrf) transcription factor family in the zebrafish, Danio rerio. PLoS One. 2013;8:e79574. doi: 10.1371/journal.pone.0079574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston GW, Di Giulio RT. Prooxidant and antioxidant mechanisms in aquatic organisms. Aquat Toxicol. 1991;19:137–161. [Google Scholar]
- Woon Kim Y, Kim S, Geun Kim C, Kim A. The distinctive roles of erythroid specific activator GATA-1 and NF-E2 in transcription of the human fetal gamma-globin genes. Nucleic Acids Res. 2011;39:6944–6955. doi: 10.1093/nar/gkr253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Wu H, Zhang C, Cao Y, Wang L, Zeng L, Ye X, Wu Q, Dai J, Xie Y, Mao Y. Identification of a novel human DDX40gene, a new member of the DEAH-box protein family. J Hum Genet. 2002;47:681–683. doi: 10.1007/s100380200104. [DOI] [PubMed] [Google Scholar]
- Xu Z, Chen L, Leung L, Yen TS, Lee C, Chan JY. Liver-specific inactivation of the Nrf1 gene in adult mouse leads to nonalcoholic steatohepatitis and hepatic neoplasia. Proc Natl Acad Sci U S A. 2005;102:4120–4125. doi: 10.1073/pnas.0500660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates A, Akanni W, Amode MR, Barrell D, Billis K, Carvalho-Silva D, Cummins C, Clapham P, Fitzgerald S, Gil L, GirÃ3n CG, Gordon L, Hourlier T, Hunt SE, Janacek SH, Johnson N, Juettemann T, Keenan S, Lavidas I, Martin FJ, Maurel T, McLaren W, Murphy DN, Nag R, Nuhn M, Parker A, Patricio M, Pignatelli M, Rahtz M, Riat HS, Sheppard D, Taylor K, Thormann A, Vullo A, Wilder SP, Zadissa A, Birney E, Harrow J, Muffato M, Perry E, Ruffier M, Spudich G, Trevanion SJ, Cunningham F, Aken BL, Zerbino DR, Flicek P. Ensembl 2016 Nucl Acids Res. 2016 doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yawata A, Kanzaki A, Gilsanz F, Delaunay J, Yawata Y. A markedly disrupted skeletal network with abnormally distributed intramembrane particles in complete protein 4.1-deficient red blood cells (allele 4.1 Madrid): implications regarding a critical role of protein 4.1 in maintenance of the integrity of the red blood cell membrane. Blood. 1997;90:2471–2481. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


