Abstract
Background
Dengue control and prevention rely heavily on insecticide-based interventions. However, insecticide resistance in the dengue vector Aedes aegypti, threatens the continued effectiveness of these tools. The molecular basis of the resistance remains uncharacterised in many endemic countries including Malaysia, preventing the design of evidence-based resistance management. Here, we investigated the underlying molecular basis of multiple insecticide resistance in Ae. aegypti populations across Malaysia detecting the major genes driving the metabolic resistance.
Methodology/Principal Findings
Genome-wide microarray-based transcription analysis was carried out to detect the genes associated with metabolic resistance in these populations. Comparisons of the susceptible New Orleans strain to three non-exposed multiple insecticide resistant field strains; Penang, Kuala Lumpur and Kota Bharu detected 2605, 1480 and 425 differentially expressed transcripts respectively (fold-change>2 and p-value ≤ 0.05). 204 genes were commonly over-expressed with monooxygenase P450 genes (CYP9J27, CYP6CB1, CYP9J26 and CYP9M4) consistently the most up-regulated detoxification genes in all populations, indicating that they possibly play an important role in the resistance. In addition, glutathione S-transferases, carboxylesterases and other gene families commonly associated with insecticide resistance were also over-expressed. Gene Ontology (GO) enrichment analysis indicated an over-representation of GO terms linked to resistance such as monooxygenases, carboxylesterases, glutathione S-transferases and heme-binding. Polymorphism analysis of CYP9J27 sequences revealed a high level of polymorphism (except in Joho Bharu), suggesting a limited directional selection on this gene. In silico analysis of CYP9J27 activity through modelling and docking simulations suggested that this gene is involved in the multiple resistance in Malaysian populations as it is predicted to metabolise pyrethroids, DDT and bendiocarb.
Conclusion/significance
The predominant over-expression of cytochrome P450s suggests that synergist-based (PBO) control tools could be utilised to improve control of this major dengue vector across Malaysia.
Author Summary
Aedes aegypti is the major vector of emerging or re-emerging arboviruses such as dengue, chikungunya and Zika viruses worldwide. As there is still no vaccine or specific treatment for these diseases, their prevention relies mainly on insecticide-based interventions. Unfortunately, vector control programs are facing operational challenges with mosquitoes becoming resistant to commonly used insecticides in several regions across the world. To facilitate the design of suitable resistance management strategies for control of this vector, we investigated the molecular basis of insecticide resistance across four Malaysian Ae. aegypti populations. Transcriptional analysis revealed that 204 genes were commonly over-expressed with cytochrome P450 genes (CYP9J27, CYP6CB1, CYP9J26 and CYP9M4) consistently found as the most up-regulated detoxification genes in all populations indicating that they likely play an important role in the observed resistance. Quantitative real-time polymerase chain reaction (qRT-PCR) validated the over-expression of all cytochrome P450s except for CYP6CB1. Polymorphism analysis of CYP9J27 sequences revealed a high polymorphism (except in Joho Bharu), suggesting a lack of directional selection on this gene. The predominant expression of cytochrome P450s suggests that synergist-based control tools could be used to improve control of this major dengue vector across Malaysia.
Introduction
The mosquito Aedes aegypti Linnaeus, 1762 (Diptera: Culicidae) is the most important vector of dengue, Zika, yellow fever [1, 2] and chikungunya [3, 4] viruses to human throughout the tropical world. This domestic mosquito usually bites during daylight, feeding predominantly on humans, mating and resting indoors and breeding in man-made containers in and around human dwellings [5].
Control of Ae. aegypti relies on reducing breeding sites and insecticide-based interventions such as treatment of breeding sites, Ultra Low Volume (ULV) space sprays, fogging and thermal spraying [6]. Unfortunately, many vector control programs are threatened by the development of insecticide resistance in Ae. aegypti [7, 8]. Resistance to multiple insecticides such as pyrethroids, dichlorodiphenyltrichloroethane (DDT), bendiocarb and organophosphates has been reported in Ae. aegypti [9–12]. Resistance to pyrethroid insecticides, the primary insecticide class used against adult mosquitoes is particularly worrying in the context of the re-emergence of dengue and other arboviruses worldwide, including Zika virus [13, 14]. The elucidation of the mechanisms of insecticide resistance may aid in the in the design of suitable resistance management strategies to prolong the effectiveness of the existing insecticide-based control tools.
The two main causes of insecticide resistance are alterations in the target sites and increase in the rate of insecticide metabolism [15]. Target site resistance is caused by mutations in target genes such as the voltage gated sodium channel (VGSC) which causes knockdown (kdr) resistance, mutations in the acetylcholinesterase (Ace-1) gene and GABA receptors [15, 16]. The most important target site resistance for mosquitoes is kdr as it confers resistance to both pyrethroids and DDT. kdr occurs as a result of a change in the affinity of the insecticides to their binding sites, because of mutations in the sodium channel [17]. Several kdr mutations have been identified in Ae. aegypti, and the association between the V1016G/I and the F1534C mutations and pyrethroid resistance has been established [18–20]. In Malaysia, a recent study revealed that the frequency of the 1534C resistant mutation ranges from 40 to 80% whereas the 1016G mutation is found at around 20 to 39% [10]. A significant correlation was also established between F1534C genotypes and pyrethroid resistance [10] in Malaysian mosquitoes, whereas no significant correlation was found for the V1016G mutation. However, an additive effect to pyrethroid resistance was observed when both 1534C and 1016G were present [10].
Another key resistance mechanism is metabolic resistance through up-regulation of detoxification genes. The three main enzymes families responsible for insecticide resistance in mosquitoes are the monooxygenases (cytochrome P450s), glutathione S-transferases (GSTs) and carboxylesterases (COEs) [21, 22]. Metabolic resistance can occur as a result of point-mutations affecting protein activity (e.g. change in binding affinity or an altered substrate specificity) [23–25] or via mutations in cis/trans regulatory loci of these three enzyme families [15]. In the case of cytochrome P450s elevated expression of several genes from this family has previously been shown to be primarily responsible for resistance towards pyrethroids, carbamates and organophosphates [7, 15, 26]. Multiple and widespread resistance to insecticides was recently reported in Ae. aegypti populations across Malaysia [10]. Although the F1534C kdr mutation was shown to play some role in the case of pyrethroids and DDT resistance, PBO synergist assays suggested that metabolic resistance mechanisms play an important role in the resistance patterns. Furthermore, it remains unknown whether differences observed in resistance profiles notably in Kuala Lumpur (KL), where high and multiple resistance was observed, is supported by differences in underlying molecular basis of the resistance.
Therefore, to aid the design of suitable resistance management strategies for control of Ae. aegypti in Malaysia, we investigated the molecular basis of multiple insecticide resistance using a microarray-based genome-wide transcriptional analysis of various populations of this species across Malaysia. This study detected key resistance genes and revealed that metabolic resistance is primarily driven by the cytochrome P450 gene family.
Materials and Methods
Ethical clearance
An institutional clearance for this study, including the sampling of mosquito, was granted by the Ministry of Health, Malaysia.
Mosquito samples
Ae. aegypti mosquitoes were collected using ovitraps in July and August 2010 in four states (Fig 1A), namely: Penang (Northwest), Kota Bharu (Northeast), Kuala Lumpur (Central) and Johor Bharu (South) in Malaysia as previously described [10]. Old tyres, flower pots, tree holes and containers that held water were also inspected for larvae. The larvae were reared at the Vector Control Research Unit (VCRU), Universiti Sains Malaysia, in Penang (Malaysia) as recently described [10]. Adult Ae. aegypti were induced to lay eggs on filter papers that were later dried and shipped to the Liverpool School of Tropical Medicine (LSTM) under the LSTM import license from DEFRA. The egg batches were then allowed to hatch in the insectary in water supplemented with hay infusion solution. Larvae were reared as above and the adults fed with 10% sucrose, and kept at a room temperature of 27 ± 2°C with relative humidity of 70 ± 10%. The bioassays were performed in the same conditions with F1 generation or subsequent F2 generations. Analysis revealed that all four populations were resistant to permethrin and to deltamethrin. The highest resistance levels to both insecticides were observed in Kuala Lumpur with nearly all mosquitoes surviving the 1 h exposure. However, in Kota Bharu, the high permethrin resistance (10% mortality) contrasted with only a moderate resistance to deltamethrin (82% mortality). These populations were also resistant to DDT with the highest resistance level recorded again in Kuala Lumpur with no mortality after 1 h exposure. Widespread resistance is also observed against the carbamate bendiocarb except in Kota Bharu where 91% mortality was observed in females. Full susceptibility was observed for the organophosphate malathion, except for Kuala Lumpur where a probable resistance is observed with 91% mortality. Similarly, a full susceptibility was observed against dieldrin except in Johru Bharu where a moderate resistance is observed with 88% mortality in females [10]. These samples have been used for the transcriptional analysis.
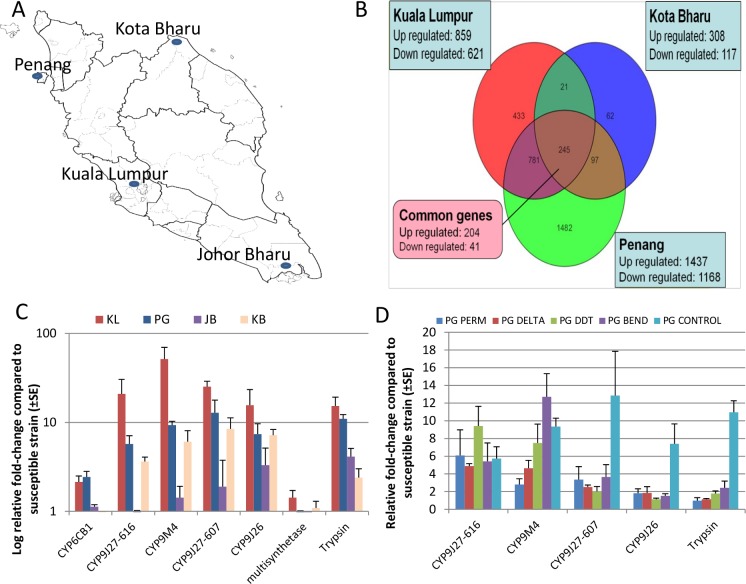
Fig 1. Transcription profiling of Ae. aegypti in Malaysia.
A) Map of the Malaysian peninsular showing the four collection sites of Penang (PG), Kota Bharu (KB), Kuala Lumpur (KL) and Johor Bharu (JB). B) Venn diagram of differentially transcribed genes from microarray data (p = 0.01); C) Relative fold-change of candidate genes from qRT-PCR analysis; D) Relative fold-change of candidate genes from qRT-PCR analysis for resistant samples from Penang. Perm stands for Permethrin, Delta for deltamethrin, Bend for bendiocarb.
Investigating metabolic resistance using genome-wide transcriptional analysis
A series of microarray experiments were conducted to identify the genes potentially associated with the metabolic resistance in the Ae. aegypti populations across Malaysia. The genome-wide transcription profiling was carried out in comparison with a susceptible strain (New Orleans) to investigate the differential expression profiles of the populations. Microarray hybridizations were done using the 8 x 15k Agilent Ae. aegypti chip containing eight replicated arrays of 60-mers oligo-probes representing more than 14,320 different Ae. aegypti transcripts from AaegL1.2 Vectorbase annotation and several control (ArrayExpress accession number A-MEXP-1966). This 8 x 15k microarray enables a high coverage across the whole genome [27] and at the same time reduces cost. It also increases throughput compared to the previous Aedes Detox chip which includes only 204 detoxification genes probes [26].Total RNA was extracted from 3 replicates of pools of 10 adult unexposed mosquitoes from all four strains. Only samples from Penang (PG), Kuala Lumpur (KL) and Kota Bharu (KB) were used in the microarray. Samples from Johor Bharu (JB) were omitted from the microarray experiment due to the population having similar resistance profile to PG (but at a lower level) and therefore possibly sharing similar resistance mechanisms [10]. However, samples from JB were used in the validation of the candidate genes through qRT-PCR. RNA from the susceptible New Orleans (NO) strain was also used. Quality and quantity of RNA were assessed using the Nanodrop ND-1000 (Thermo Scientific, Delaware, USA) and the Agilent 2100 Bioanalyzer (Agilent Technologies, California, USA). RNA (100ng) sample from each sample were amplified and labelled using the two-colour low input Quick Amp Labelling kit (Agilent Technologies). Labelled cRNAs were hybridized to the arrays for 17h at 65°C according to the manufacturer’s protocol. Three different comparisons were made: NO susceptible lab strain vs. PG, NO vs. KL and NO vs. KB (non-exposed vs susceptible) (S1 Fig). A total of five replicates (including three biological replicates and two dye swaps) were performed for each comparison as previously successfully done in other species to detect the resistance genes [28, 29].
Microarray data analysis and gene ontology (GO) enrichment analysis
Microarray data were analysed using the Genespring GX 12.0 software (Agilent Technologies, US). Mean expression ratios were assessed using a t-test against zero with a multiple testing correction (Benjamini-Hochberg false discovery rate). Genes showing both t-test p-values less than 0.05 and a fold change (FC) value greater than 2 were considered significantly differentially transcribed between the two samples compared.
Genes or entities that were considered as significantly differentially expressed were used for Gene Ontology (GO) enrichment analysis Blast2GO software (BioBam Bioinformatics S.L., Valencia, Spain). Descriptions and GO-terms of transcript-IDs were generated from Blast2GO extracted from VectorBase. GO term enrichment analysis was performed on the significantly up-regulated genes (72% of transcripts present on microarray have GO-terms) using Blast2GO software with Fisher’s exact test and false discovery rate (FDR) < 0.05 [30].
Validation of candidate genes using quantitative real time-polymerase chain reaction (qRT-PCR)
Some of the most significantly differentially expressed genes from the microarray analysis were selected for qRT-PCR validation. Firstly, control samples not exposed to insecticides from all four locations (including the JB sample that was not used in the microarray to also confirm their potential role in this location) were compared to the susceptible NO strain to assess the geographical variation of these key genes. Secondly, samples alive after exposure to various insecticides were also compared to the control non exposed mosquitoes and to the NO strain to detect possible induction of these genes or whether their over-expression was more associated to a specific insecticide than others. This was done only in PG as this population is moderately resistant to all insecticides allowing a comparison to the control sample. Total RNA from 3 replicates of each samples were used for the qRT-PCR. Primers used are listed in S1 Table. Standard curves for each gene were generated using a serial dilution of cDNA to assess PCR efficiency and quantitative differences between samples. qRT-PCR amplification was performed as described previously [28, 29]. The relative expression level and fold change (FC) of each target gene in field samples relative to the susceptible NO (S) were calculated according to the 2-ΔΔCT method incorporating the PCR efficiency [31] after normalization with the housekeeping genes ribosomal protein S7 (RSP7; AAEL009496-RA) and Tubulin (AAEL009496-RA).
Polymorphism analysis of the candidate resistance P450 gene CYP9J27
Patterns of polymorphism for CYP9J27 were explored across Malaysian Ae. aegypti populations to detect possible correlation with resistance profile using the permethrin susceptible (NO) and unexposed Ae. aegypti mosquitoes from the four sites in Malaysia: JB, KL, KB and PG. Full-length coding region of CYP9J27 was amplified from cDNA using the same cDNA synthesized for qRT-PCR with the Phusion High-Fidelity DNA Polymerase (Thermo Scientific), cloned into the pJET1.2/blunt cloning vector (Thermo Scientific), and sequenced as described previously [24]. Primers used are listed in S2 Table. Polymorphic positions were detected through manual analysis of sequence traces using BioEdit version 7.2.1 and as sequence differences in multiple alignments using ClustalW [32]. Basic sequence statistics, including the number of haplotypes (h), the number of polymorphism sites (S), haplotype diversity (Hd) and nucleotide diversity (π), were computed with DnaSP 5.10 [33]. The statistical tests of Tajima [34], Fu and Li [35] was used with DnaSP to test non-neutral evolution and deviation from mutation-drift equilibrium. Different haplotypes were compared by constructing a maximum likelihood phylogenetic tree and polymorphic positions of amino acid sequences were generated using MEGA 6.0 [36].
Homology modelling of CYP9J27 and docking simulations with various insecticides
To predict the ability of the CYP9J27 to bind the various insecticides homology models of the P450 were created using query amino acid sequences from the various study locations (KL, PG, JB, KB), as well as the sequence from susceptible strain, NO. The 3D models of the P450s were created using the standalone tool EasyModeller [37]. CYP3A4 (PDB: 1TQN) [38] was used as a template with sequence identity of 31% for all the five CYP9J27 amino acid sequences. Virtual datasets of ligand insecticides: 1R-cis permethrin (ZINC01850374), deltamethrin (ZINC01997854), DDT (ZINC01530011) and bendiocarb (ZINC02015426) were retrieved from the library in ZINC12 database (https://zinc.docking.org/) [39]. Docking simulations were carried out using the Blind Docking Web Server (http://bio-hpc.ucam.edu/webBD/index.php/entry). For each ligand, 30 binding poses were generated and sorted according to the binding energy and conformation in the protein’s active site. Figures were prepared using the PyMOL 1.7 [40].
Results
1-Genome-wide microarray-based transcriptional profiling
The microarrays were used to perform a genome-wide transcription analysis between the susceptible strain (New Orleans, NO) and non-exposed field populations from Penang (PG), Kuala Lumpur (KL) and Kota Bharu (KB). The number of differentially transcribed genes after analysis is presented in the Venn diagram (Fig 1B). Penang had the most number of differentially transcribed probes with 2605 probes, followed by Kuala Lumpur with 1480 gene probes, and Kota Bharu with 425 gene probes. The number of commonly up-regulated probes in all population is 204 while 41 probes were down-regulated.
Genes commonly up-regulated in the three locations
To detect the genes associated with insecticide resistance across Malaysian Ae. aegypti, priority was given to the genes commonly up-regulated in the three locations since their consistent over-expression increases the likelihood of them playing a role in the observed resistance. The 204 up-regulated gene probes comprise various gene families such as those involved in protein synthesis, ion transport, detoxification, etc (S3 Table). The most commonly over-expressed gene was the anionic-trypsin with very high fold change notably in KL (FC = 759) and PG (FC = 581) but with moderate over-expression in KB populations (FC = 43.8). This gene is known to be expressed abundantly in mosquito’s midgut where it hydrolyses proteins after blood meals [41]. Over-expression of midgut proteases has been commonly associated with insecticide resistance in various mosquito species [28, 29, 41, 42]. The list of the top 50 up-regulated probes include several probes associated with regulation of gene expression including the transcription elongation regulator 1 (FC = 448 in KL), five finger zinc proteins and two probes from the GATA transcription factor. It is possible that these genes are involved in the transcriptional regulation of the major insecticide detoxification genes in these populations of Ae. aegypti. Among the commonly up-regulated detoxification genes, cytochrome P450s were the most predominant with nine genes over-expressed, while only a single carboxylesterase was over-expressed. Of the P450s, CYP6CB1 was the most over-expressed gene with the highest FC value in PG (212.00) followed by Kuala Lumpur and Kota Bharu with FC values of 124.70 and 36.40, respectively. Another detoxification gene that was over-expressed was the CYP9J26 gene (AAEL014609-RA) with a similar expression level in PG and KL with 7.20 and 7.70 FC values. The expression was lower for this gene in KB with FC value of 4.40. CYP9M4 and CYP9J27-616 had the similar expression level in the three locations. In KL the FC value for CYP9M4 and CYP9J27-616 (AAEL014616) was 13.20 and 13.80, followed by PG with 7.20 and 7.40 and KB with 4.10 and 3.70. Another cytochrome P450 which was over-expressed in all the locations was the transcript AAEL014614-RA which had the closest hit to CYP9J4 in An. gambiae after NCBI BLAST. The FC value for this gene was highest in KL with 13.80 followed by PG and KB with 2.90 and 2.10 FC values respectively. The unique carboxylesterase gene (AAEL004724-RA) commonly up-regulated in the three locations had similar low FC values for all the locations ranging from 2.00 to 2.80 (Table 1).
Table 1. Probes from detoxification genes & genes linked with resistance up-regulated in Malaysian populations in comparison with susceptible New Orleans strain.
FC = fold change (p = 0.01).
| Probe Name | Systematic Name | Blast2GO Annotation | Kota Bharu vs NO | Kuala Lumpur vs NO | Penang vs NO | |||
|---|---|---|---|---|---|---|---|---|
| Absolute FC | Corrected p-value | Absolute FC | Corrected p-value | Absolute FC | Corrected p-value | |||
| Common to all three locations | ||||||||
| CUST_2845_PI424980000 | AAEL009018-RA | cytochrome p450 (CYP6CB1) | 36.40 | 0.04881 | 124.70 | 0.00128 | 212.00 | 0.00076 |
| CUST_9292_PI424980000 | AAEL003349-RA | nadph-cytochrome p450 reductase | 6.60 | 0.0442 | 24.20 | 0.00421 | 8.90 | 0.00012 |
| CUST_12457_PI424980000 | AAEL014689-RA | nadph cytochrome p450 | 5.30 | 0.0303 | 15.10 | 0.00163 | 11.00 | 0.00022 |
| CUST_105_PI424980000 | AAEL014609-RA | cytochrome p450 (CYP9J26) | 4.40 | 0.01914 | 7.70 | 0.00435 | 7.20 | 0.00033 |
| CUST_157_PI424980000 | AAEL001320-RA | cytochrome p450 (CYP9M4) | 4.10 | 0.01679 | 13.20 | 0.00317 | 7.20 | 0.00174 |
| CUST_106_PI424980000 | AAEL014616-RA | cytochrome p450 (CYP9J27) | 3.70 | 0.01236 | 13.80 | 0.00178 | 7.80 | 0.00022 |
| CUST_140_PI424980000 | AAEL014614-RA | cytochrome p450(as CYP9J4 in An. gambiae) | 2.10 | 0.00352 | 13.80 | 0.00178 | 2.90 | 0.00104 |
| CUST_228_PI424980000 | AAEL004724-RA | carboxylesterase | 2.00 | 0.00243 | 2.10 | 0.00808 | 2.80 | 0.00046 |
| Common to KB and KL but not PG | ||||||||
| CUST_151_PI424980000 | AAEL001288-RA | cytochrome p450(as CYP9M1 in An. gambiae) | 2.50 | 0.00908 | 2.40 | 0.00835 | ||
| Common to KB and PG but not KL | ||||||||
| CUST_10688_PI424980000 | AAEL012836-RA | cytochrome b561 | 4.40 | 0.00986 | 9.50 | 0.00022 | ||
| CUST_67_PI424980000 | AAEL014893-RA | cytochrome p450 (CYP6BB2) | 2.00 | 0.00104 | 5.20 | 0.00019 | ||
| Common to KL and PG but not KB | ||||||||
| CUST_162_PI424980000 | AAEL014607-RA | cytochrome p450 (CYP9J27) | 356.70 | 0.00214 | 395.70 | 0.00022 | ||
| CUST_145_PI424980000 | AAEL014606-RA | cytochrome p450 (CYP9J7) | 16.90 | 0.00349 | 12.20 | 0.00069 | ||
| CUST_131_PI424980000 | AAEL008023-RA | cytochrome p450 (CYP4C52) | 12.70 | 0.00512 | 3.50 | 0.00376 | ||
| CUST_148_PI424980000 | AAEL014891-RA | cytochrome p450 (as CYP6P4 in An. gambiae) | 8.50 | 0.00978 | 9.70 | 0.00117 | ||
| CUST_64_PI424980000 | AAEL007473-RA | cytochrome p450 (CYP6AH1) | 5.90 | 0.00752 | 2.50 | 0.0024 | ||
| CUST_176_PI424980000 | AAEL007951-RA | glutathione-s-transferase gst (GSTE2) | 4.50 | 0.00397 | 2.40 | 0.00291 | ||
| CUST_256_PI424980000 | AAEL004118-RA | aldo-keto reductase | 3.20 | 0.00252 | 5.00 | 0.0006 | ||
| CUST_352_PI424980000 | AAEL008672-RA | abc transporter | 2.50 | 0.00711 | 3.40 | 0.00056 | ||
| KB only | ||||||||
| CUST_8318_PI424980000 | AAEL014246-RA | glucosyl glucuronosyl transferases | 3.60 | 0.04007 | ||||
| CUST_348_PI424980000 | AAEL004331-RA | abc transporter | 3.20 | 0.00636 | ||||
| CUST_112_PI424980000 | AAEL001292-RA | cytochrome p450 (CYP9M7) | 3.00 | 0.03684 | ||||
| KL only | ||||||||
| CUST_143_PI424980000 | AAEL014617-RA | cytochrome p450 (CYP9J28) | 10.80 | 0.00209 | ||||
| CUST_6_PI424980000 | AAEL004870-RA | cytochrome p450 (CYP18A1) | 3.20 | 0.00725 | ||||
| CUST_7415_PI424980000 | AAEL010157-RA | microsomal glutathione s-transferase | 2.80 | 0.00212 | ||||
| CUST_111_PI424980000 | AAEL001312-RA | cytochrome p450 (CYP9M6) | 2.40 | 0.00402 | ||||
| PG only | ||||||||
| CUST_88_PI424980000 | AAEL012491-RA | cytochrome p450 (CYP6P12) | 6.50 | 0.00091 | ||||
| CUST_67_PI424980000 | AAEL014893-RA | cytochrome p450 (CYP6BB2) | 5.20 | 0.00019 | ||||
| CUST_86_PI424980000 | AAEL009126-RA | cytochrome p450 (CYP6N6) | 4.90 | 0.00041 | ||||
| CUST_129_PI424980000 | AAEL005695-RA | cytochrome p450 (CYP325X1) | 4.60 | 0.00542 | ||||
| CUST_184_PI424980000 | AAEL011741-RB | glutathione s-transferase (GSTS1) | 4.30 | 0.00456 | ||||
| CUST_233_PI424980000 | AAEL000905-RA | carboxylesterase | 4.00 | 0.00082 | ||||
| CUST_3471_PI424980000 | AAEL005937-RA | atp-binding cassette transporter | 3.60 | 0.00039 | ||||
| CUST_44_PI424980000 | AAEL007795-RA | cytochrome p450 (CYP4D37) | 3.40 | 0.00936 | ||||
| CUST_338_PI424980000 | AAEL008624-RA | abc transporter | 3.40 | 0.00029 | ||||
| CUST_87_PI424980000 | AAEL009121-RA | cytochrome p450 (CYP6N9) | 3.50 | 0.00045 | ||||
| CUST_44_PI424980000 | AAEL007795-RA | cytochrome p450 (CYP4D37) | 3.40 | 0.00936 | ||||
| CUST_31_PI424980000 | AAEL017136-RA | cytochrome p450 (CYP325V1) | 3.10 | 0.00046 | ||||
| CUST_95_PI424980000 | AAEL006798-RA | cytochrome p450 (CYP9J10) | 3.00 | 0.00064 | ||||
| CUST_11833_PI424980000 | AAEL014612-RA | cytochrome p450 (as CYP9J5 in An. gambiae) | 2.90 | 0.00109 | ||||
| CUST_93_PI424980000 | AAEL009131-RA | cytochrome p450 (CYP6Z8) | 2.50 | 0.00639 | ||||
| CUST_6046_PI424980000 | AAEL009119-RA | cytochrome p450 (as CYP6M2 in An. gambiae) | 2.40 | 0.00168 | ||||
| CUST_10102_PI424980000 | AAEL012838-RA | cytochrome b561 as CYP325K1 in An. gambiae) | 2.30 | 0.00055 | ||||
| CUST_130_PI424980000 | AAEL008017-RA | cytochrome p450 (CYP4C50) | 2.30 | 0.00943 | ||||
| CUST_102_PI424980000 | AAEL017217-RA | cytochrome p450 (as CYP9J5 in An. gambiae) | 2.30 | 0.00039 | ||||
| CUST_7589_PI424980000 | AAEL015641-RA | cytochrome p450 (as CYP6AH1 in An. gambiae) | 2.20 | 0.00047 | ||||
| CUST_108_PI424980000 | AAEL002638-RA | cytochrome p450 (CYP9J6) | 2.20 | 0.00046 | ||||
| CUST_4_PI424980000 | AAEL002031-RA | cytochrome p450 (CYP12F7) | 2.10 | 0.00123 | ||||
| CUST_346_PI424980000 | AAEL006717-RA | abc transporter | 2.10 | 0.00138 | ||||
| CUST_194_PI424980000 | AAEL002367-RA | Carboxylesterase (CCEAE1B) | 2.10 | 0.00228 | ||||
| CUST_357_PI424980000 | AAEL005249-RA | abc transporter | 2.00 | 0.00778 | ||||
Genes commonly up-regulated only in two locations
Commonly up-regulated only in KL and PG but not KB
From the list of probes over-expressed only between KL and PG but not in KB, eight genes linked to detoxification and resistance were observed. Another transcript of CYP9J27-607 (AAEL014607-RA) was the most over-expressed cytochrome P450 with FC values more than 350 in both locations which is by far more than the FC values for the same gene but with a different transcript (AAEL014616-RA) in the commonly over-expressed genes for all three locations. Similarly, a different transcript of the CYP9J7 (AAEL014606-RA) is also up-regulated in KL (FC value 16.90) and PG (FC value 12.20) only. Other cytochrome P450s included CYP4C52, CYP6AH1 and a transcript with the closest hit to CYP6P4 in An. gambiae, a cytochrome P450 associated with permethrin resistance in An. gambiae [43] and recently showed to metabolise pyrethroids in An. arabiensis [25, 44]. Also, the glutathione S-transferase gene GSTe2, a known DDT metaboliser [45] [23] was commonly up-regulated in KL and PG, however with low FC values of 4.50 and 2.40 respectively (Table 1).
Genes commonly up-regulated only in KB and PG but not KL
Only two genes were established as commonly over-expressed in KB and PG. One cytochrome P450 CYP6BB2 with FC value of 2.00 in KB and 5.20 in PG was recorded. Another gene up-regulated was the cytochrome b561 which has been previously established to play a role in insecticide resistance in Ae. aegypti [23, 30] (Table 1).
Commonly up-regulated only in KL and KB but not PG
Only one cytochrome P450 transcript with the closest hit to CYP9M1 in An. gambiae was commonly up-regulated in KL and KB. The FC values were low with 2.50 in KB and 2.40 in KL (Table 1).
Genes up-regulated only in a single location
From the list of the genes up-regulated in a single location only, PG had the most number of up-regulated detoxification genes exclusively expressed in this location with a predominance of cytochrome P450 among which is the P450 CYP6P12 (FC value 6.50). The ortholog of this gene in Ae. albopictus was recently shown to be the main pyrethroid resistance gene in this species in KL [42]. CYP6P12 is also the ortholog of CYP6P4 in the malaria vectors An. gambiae, An. arabiensis and An. funestus. Other cytochrome P450 genes are the CYP6BB2 (a different probe than in the commonly up-regulated in PG and KB), CYP6N6, CYP325X1, CYP4D37, CYP9J10, CYP6Z8 and others. Other genes belong to GSTs (GSTS1), carboxylesterase (transcript AAEL000905-RA and CCEAE1B) and the ABC transporters (Table 1).
The set of probes up-regulated only in KL is mainly made of cytochrome P450s among which the CYP9J28 gene exhibited the highest FC value of 10.80. This gene has previously been shown to confer pyrethroid resistance in Ae. aegypti [46]. Other cytochrome P450s over-expressed only in KL are CYP18A1 and CYP9M6 (Table 1).
The genes up-regulated in KB only are a cytochrome P450 (CYP9M7), an ABC transporter and a UDP glucuronosyl transferase (Table 1).
Genes commonly down-regulated in the three locations
Among the top genes commonly down-regulated in PG, KL and KB the highest down-regulated was the domain-containing protein cg6693. Glutamine synthetase seems to be consistently down-regulated with two probes being consistently at the top. One detoxification gene CYP325M2 was found to be the commonly down-regulated in all three locations (S4 Table and S5 Table). Among the detoxification genes down-regulated only in one or two locations, were several cytochrome P450s notably from CYP325 (CYP325F2, CYP325G2, CYP325C2, CYP325T2 and more), CYP4J (CYP4J10, CYP4J13, CYP4J14, CYP4J15) and some CYP6 (CYP6N13, CYP6N14, CYP6N17, CYP6AG8, CYP6BY1).
2-Validation of candidate genes through qRT-PCR
Seven candidate genes commonly up-regulated in all three locations were chosen to validate the microarray expression pattern. These genes were trypsin (AAEL013623-RA), multisynthetase complex, CYP6CB1, CYP9J26 (AAEL014609-RA), CYP9J27-607 (AAEL014607-RA), CYP9M4 and CYP9J27-616 (AAEL014616-RA). The most over-expressed P450 for microarray in all 3 locations, CYP6CB1 was not significantly over-expressed from the qRT-PCR results in all four locations including JB (Fig 1C). However, the over-expression of other five P450 genes was confirmed whereas the over-expression of the multisynthetase gene was not confirmed. The JB sample consistently exhibited a lower expression level for all these genes in comparison to the other 3 locations. This difference could be associated with the lower resistance level to pyrethroids and DDT in JB. Overall, for most of the genes, the highest over-expression was observed for the KL population which correlates with the high resistance level to pyrethroids and DDT observed in KL from bioassays [10]. The expression profile of the five significantly over-expressed genes from qRT-PCR was further assessed for various samples from mosquitoes alive after exposure to different insecticides. From this analysis, CYP9J27-616 was more over-expressed, although not significantly, in the DDT resistant mosquitoes from PG than the other insecticides (Fig 1D). CYP9M4 was significantly more over-expressed in the PG bendiocarb resistant sample than other mosquito samples. No significant difference was observed between samples for CYP9J26, CYP9J27-607 and for the trypsin gene.
3. Gene ontology (GO) enrichment analysis
GO enrichment analysis was used to identify particular Gene Ontology (GO) terms that were over represented in the data set of transcripts up-regulated in all three resistant populations and in single populations. When comparing the commonly up-regulated genes in all three populations at p = 0.01, a few GO terms that relates to detoxification was observed. These included NADPH-hemoprotein reductase activity, ATP binding and others (S2 Fig). When observing the GO terms of single locations either in PG (S3 Fig), KB (S4 Fig) or KL (S5 Fig), interesting terms such as ATP binding, heme-binding and monooxygenase activity were over-represented possibly associated with the multiple resistance to insecticides detected in these locations.
4-Genetic variability of CYP9J27 in relation to pyrethroid resistance
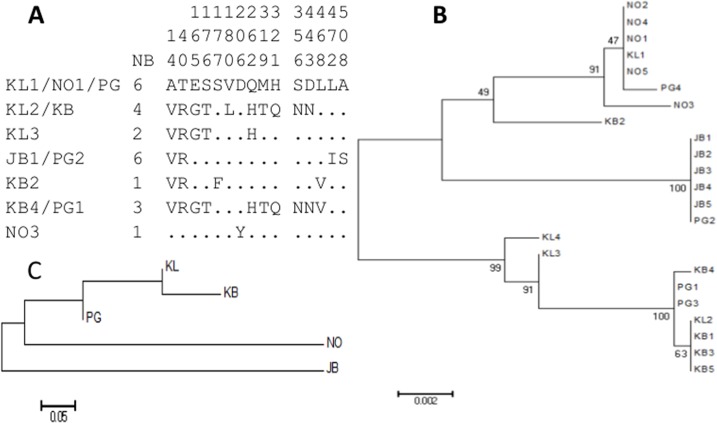
In order to functionally investigate the potential involvement of the commonly over-expressed CYP9J27 gene in conferring resistance to insecticides in Ae. aegypti, further attention was paid on this gene since particularly as the role of other P450s has already been assessed [46]. Full-length cDNA sequences (1611 bp) of CYP9J27-616 (hereafter CYP9J27), one of the genes commonly over-expressed all the three populations from microarray were successfully generated for 23 clones from four locations across Malaysia and the susceptible NO strain allowing to assess their polymorphism (GenBank Accesion Number: KX394421-KX394443). Overall, analysis of the genetic variability revealed 54 polymorphic sites (s), 10 haplotypes (h), and 15 amino acid changes in total, with the lowest polymorphism observed in JB (Table 2, Fig 2A). Phylogenetic tree constructed using maximum likelihood confirm the relatively high genetic variability of this gene in each location except in JB (Fig 2B). Haplotype diversity is high (Hd>0.73) in most samples except JB suggesting that little directional pressure is acting in this population. However, the lack of diversity in JB should be further investigated. The difference between JB and other populations is further highlighted on the neighbour-joining tree of the genetic distances between various populations, with JB exhibiting high level of genetic differentiation based on KST estimates whereas KL and KB cluster together. When all the sequences were analysed as a unique sample, the Tajima D, was positive (D = 2.0675) and statistically significant (P<0.05) (Table 2). In the other hand, when the sequences were analysed according to the origin of each sample this statistics was negative but not statistically significant (Table 2). Negative values for these indexes may indicate an excess of rare polymorphisms in a population and suggest either population expansion or background selection [35]. However, further analysis with more sequences is needed to test for a signature of selection on this gene.
Table 2. Summary statistics for polymorphism of CYP9J27 haplotypes from unexposed samples and susceptible NO strains, across Malaysia.
| Samples | N | S | h | Hd | Syn | Nonsyn | π (κ) | D | D* |
|---|---|---|---|---|---|---|---|---|---|
| Johor Bharu | 5 | 0 | 1 | 0.00 | 0 | 0 | 0.0000 (NA) | NA | NA |
| Kuala Lumpur | 4 | 36 | 4 | 1.00 | 26 | 10 | 0.1159 (18.66) | -0.5119 | 0.5119 |
| Kota Bharu | 5 | 30 | 3 | 0.70 | 20 | 10 | 0.0077 (12.40) | -1.0384 | -1.0384 |
| Penang | 4 | 45 | 3 | 0.83 | 34 | 11 | 0.0163 (26.33) | 0.7572 | 0.7572 |
| New Orleans | 5 | 5 | 2 | 0.40 | 4 | 1 | 0.0012 (2.00) | -1.1239 | -1.1239 |
| All | 23 | 54 | 10 | 0.87 | 39 | 15 | 0.0138 (22.33) | 2.0675* | 1.3035 |
N = number of sequences (n); S, number of polymorphic sites; h, haplotype; Hd, haplotype diversity; Syn, Synonymous mutations; Nonsyn, Non-synonymous mutations; π, nucleotide diversity (k = mean number of nucleotide differences); Tajima’s D and Fu and Li’s D statistics, ns, not significant.
* Statistically significant at p<0.05; NA, not applicable.
Fig 2. Genetic diversity of CYP9J27 across Malaysian populations of Aedes aegypti.
A) Schematic representation of CYP9J27 haplotypes from across Malaysia and in the NO susceptible strain. The polymorphic positions for CYP9J27 amino acid sequences are highlighted. A number is given to each haplotype preceded by location origin (KL, KB, JB, PG and NO, for Kuala Lumpur, Kota Bharu, Johor Bharu, Penang and New Orleans, respectively). The column Nb stands for the number of individual mosquitoes sharing a haplotype. B) Neighbor joining tree of CYP9J27 haplotypes. C) Genetic distance tree between the different samples based on the Nst genetic distances.
5. Prediction of CYP9J27 activity through modelling and docking simulations with various insecticides
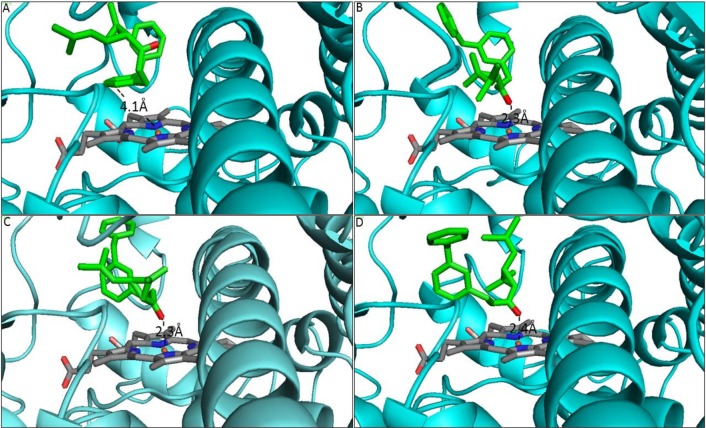
For each of the four locations as well the NO, CYP9J27 model with the highest ERRAT score (which is based on the patterns of non-bonded interaction) [47] was chosen for molecular docking. For the type I pyrethroid permethrin, the KL model exhibited the lowest binding energy (10.1 kcal/mol) which was comparable to values obtained from JB and KB (-9.9 kcal/mol respectively); but lower than observed with PG (-9.6 kcal/mol) (S6 Table). NO model exhibited the highest binding energy of -9.1 kcal/mol, however this binding energy predicts good binding of the permethrin to the model from the susceptible strain, even though lower than obtained from the other models. However, with the exception of KL model in which permethrin binds with the benzyl ring perpendicular above the heme plane (2 spot of benzyl ring located 4.1Å from the heme iron (Fig 3A), predicting ring hydroxylation to generate 2-hydroxypermethrin), the insecticide docked in all the other models with the ester oxygen projecting toward the heme iron, susceptible to ester hydrolysis (Fig 3B–3D; S9A Fig).
Fig 3.
Binding conformation of permethrin in (A) KL, (B) JB, (C) PG and (D) NO CYP9J27 models. Distances between the probable sites of attack on permethrin and the heme iron are annotated.
As with permethrin, docking of type II pyrethroid deltamethrin to the active site of KL CYP9J27 model produced the lowest binding energy (-10.3 kcal/mol), lower than observed with the other models, with NO model producing the highest binding free energy (-9.3 kcal/mol) (S6 Table). Also, as with permethrin, deltamethrin docked in the active site of KL model with the phenoxy ring above the heme iron and the 4´ spot located within 4.2Å of the heme iron (S6A Fig). In this posture ring hydroxylation to generate 4´-hydroxydeltamethrin is predicted. For JB and the NO models the α-cyano group docked above the heme iron within 2.4Å of the heme iron (S6B and S6D Fig), while for PG it is the ester oxygen which projects toward the heme iron at a distance of 2.3Å (S6C Fig). For KB, it’s the benzyl ring which docked within 2.9Å from the heme iron with possibility of ring hydroxylation (S9B Fig).
With the exception of NO model, DDT docked in the models of KL, JB, PG and KB with low binding energy (S6 Table) and productively with carbon atom of the trichloromethyl group positioned within a distance of 5.4Å, 4.3Å and 4.8Å respectively for KL, JB and PG (S7A–S7C Fig). The same pose was obtained with KB model with the carbon atom of the trichloromethyl group at a distance of 5.6Å from the heme iron (S9C Fig). In this posture reductive dechlorination to generate DDE is predicted. In contrast, DDT docked unproductively in the active site of NO with the chlorine atoms projecting toward the heme iron at a distance of 3.6Å (S7D Fig). Generally, the free energy of DDT binding in the models from the resistant populations is higher than obtained with the pyrethroids but low enough to warrant good binding, with the exception of NO model to which the binding energy of DDT is very high (-5.3 kcal/mol).
In KL, JB, PG and KB models, bendiocarb bind productively with the carbamic acid ester group oriented toward heme at a reasonable distance of 3.4Å, 3.9Å, 3.7Å and 3.9Å, respectively (S8A–S8C and S9D Figs). In this posture ester hydrolysis to generate benzodioxol-4-ol and carbamic acid is predicted. Free binding energy for all the models is lower than obtained from docking with pyrethroids and DDT; however, NO has the highest binding energy of all (-5.1 kcal/mol) indicative of low affinity compared with the other models. Not surprisingly, bendiocarb docked in the active site of NO model away (13.2Å) from heme catalysis centre for any meaningful interaction to take place.
Discussion
Elucidating the molecular basis of insecticide resistance in the dengue vector Ae. aegypti is an important step for the design of suitable resistance management strategies to control this vector. In this study genome-wide transcriptional analyses carried out using microarray showed that metabolic resistance plays an important role in conferring resistance to insecticides in Ae. aegypti across Malaysia. This further supports the synergist assay result with PBO showing a recovery of susceptibility notably for pyrethroids [10]. The role of metabolic resistance was supported by the over-expression of many genes belonging to detoxification families in PG, KL and KB when comparing them to the susceptible NO strain. The most prominent detoxification gene family established in this Aedes populations were the cytochrome P450 genes which were the only detoxification family discovered except for one unique carboxylesterase commonly over-expressed in the three locations.
The most over-expressed cytochrome P450 is the CYP6CB1 which has also been reported in a strain from Isla Mujeres in Mexico [46]. Unfortunately, the microarray over-expression of CYP6CB1 was not supported by the qRT-PCR validation for all four populations tested. This discrepancy between the two methods could be caused by differences in the sequences of the microarray probes and the qRT-PCR primers. Recent functional analysis has shown that this CYP6CB1 could not metabolise pyrethroids [46] although it could metabolise other insecticides. CYP9J26 (AAEL014609) gene was among the top up-regulated detoxification gene which has also been reported in Cuba, Thailand and Grand Cayman [30] and has also been functionally validated to confer pyrethroid resistance [46].
Overall, several P450 genes belonging to the CYP9 family were over-expressed in the control- versus susceptible across Malaysia including two transcripts of CYP9J27, CYP9J26, CYP9J28, CYP9M6 while only few cytochrome P450s from the CYP6 family (CYP6P12, CYP6BB2) were over-expressed and usually at lower fold change. This is further supported by previous studies worldwide showing that contrary to Anopheles mosquitoes, genes from the CYP9 family play a more important role than those from the CYP6 family in insecticide resistance in Ae. aegypti [7, 26, 30, 48, 49]. This was different than the Aedes albopictus transcription analyses performed with samples collected from same locations at the same time as more P450 genes belonging to the CYP6 family were over-expressed in the C-S comparison of Ae. albopictus samples in Malaysia including CYP6N3, CYP6P12, CYP6Z6, CYP6AG6, while only few cytochrome P450s from the CYP9 family were over-expressed [42].
Interestingly, the top most commonly over-expressed gene was the anionic-trypsin which is found in the midgut of mosquitoes and shown to hydrolyse proteins after blood meals. This serine proteinase is found to be over-expressed in deltamethrin resistant strain of Culex pipiens pallens from China [50].
Few glutathione S-transferases were detected compared to cytochrome P450s despite the very high DDT resistance notably in KL. The PBO synergist assay previously indicated a recovery of susceptibility from 0 to 35% in KL for DDT [10]. The low expression of GSTs notably that of the known DDT metaboliser GSTe2 (FC = 4.5) [42, 45] suggests that knockdown resistance could be responsible for most of the remaining 65% loss of DDT susceptibility. Similar assessment of pyrethroids shows a recovery of susceptibility after PBO assay from 1% to 26% for permethrin and from 0% to 71% for deltamethrin. This suggests that metabolic resistance through P450 up-regulation is more important for deltamethrin than permethrin resistance and kdr playing a more important role for permethrin than deltamethrin. This will be in line with the higher correlation previously observed between permethrin and F1534C genotypes than with deltamethrin [10]. However, the molecular docking predicted CYP9J27 to bind and metabolise pyrethroids and DDT, especially KL model, compared to NO model to which DDT binds unproductively indicating lack of affinity and activity towards this organochlorine insecticide by NO strain.
Because PBO assays with bendiocarb also revealed a nearly full recovery of the susceptibility to this insecticide [10] it is likely that some of the cytochrome P450 genes detected in this study are responsible for this resistance although future functional characterization will identify the specific genes. Of course, with the exception of NO model, docking analyses with CYP9J27-616 models predicted productive binding and good affinity towards bendiocarb suggesting the ability to metabolise this carbamate insecticide. The possible role of P450s in carbamate resistance will explain the low expression of carboxylesterase genes observed in this study and suggests an absence of any Ace-1 mutations as previously reported [49].
The higher polymorphism level of CYP9J27-616 gene across Malaysian samples (apart from JB) (Table 2) suggests of little directional selection pressure favouring a specific SNP or amino acid change is acting on this gene in Malaysia despite it consistent over-expression. This suggests that CYP9J27 potential role in the resistance if confirmed would be through a mechanism involving genetic variation in the regulatory regions such as promoter beside potential variation in the coding sequence. This is similar to cases of some P450 genes in other mosquito species for which despite a high over-expression in resistant individuals, no significant association has been observed between polymorphism level and resistance phenotype, such as CYP6M7 in the malaria vector An. funestus [24]. Further analysis of these regulatory regions could help detect the specific genomic changes driving resistance through CYP9J27. Nevertheless, the lowest binding affinity from docking exhibited by the CYP9J27 allele from the susceptible NO strain compared to those from Malaysian could suggest that allelic variation could also be playing a role in the resistance observed as recently demonstrated in the African malaria vector An. funestus [25]. Interestingly, negative values of neutrality tests Tajima’s, and Fu and Li’s observed for CYP9J27 suggest either recent demographic expansion or background selection [35] on this gene across Ae. aegypti Malaysian population.
Conclusion
This study revealed that the cytochrome P450 genes are associated in insecticide resistance in Ae. aegypti populations across Malaysia. However, further functional characterization work using either transgenic expression in Drosophila flies or recombinant enzymes expression in Escherichia coli coupled with metabolic assay has to be done to confirm the exact contribution of these candidate genes in the resistance profile. An alternative to pyrethroid is the organophosphate, malathion since all of the populations are susceptible to this insecticide [10].This and other proper resistance management strategies should be adopted to reverse the spread and evolution of this resistance problem in Malaysia before it compromises control programmes.
Supporting Information
Green arrows refer to Cy-3 dye and red arrows refer to Cy-5 dye.
(PPTX)
(PPTX)
(PPTX)
(PPTX)
(PPTX)
Binding conformation of deltamethrin in (A) KL, (B) JB, (C) PG and (D) NO CYP9J27 models. Distances between the probable sites of attack on deltamethrin and heme iron are annotated.
(PPTX)
Binding conformation of DDT in (A) KL, (B) JB, (C) PG and (D) NO CYP9J27 models. Distances between the probable sites of attack on DDT and the heme iron are annotated.
(PPTX)
Binding conformation of bendiocarb in (A) KL, (B) JB, (C) PG and (D) NO CYP9J27 models. Distances between the probable sites of attack on bendiocarb and the heme iron are annotated.
(PPTX)
Binding conformation of (A) permethrin, (B) deltamethrin, (C) DDT and (D) bendiocarb in KB CYP9J27 model. Distances between the probable sites of attack on various insecticides and the heme iron are annotated.
(PPTX)
(DOCX)
(DOCX)
FC = fold change. (p = 0.01).
(DOCX)
FC = fold change. (p = 0.01).
(DOCX)
FC = fold change. (p = 0.01).
(DOCX)
(DOCX)
Data Availability
GenBank Accesion Number: KX394421-KX394443
Funding Statement
This work was supported by a PhD Studentship to IHI by the Malaysian Ministry of Higher Education and Universiti Sains Malaysia and a Wellcome Trust Research Career Development Fellowship to CSW (083515/Z/07/Z). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Gubler DJ. Epidemic dengue/dengue hemorrhagic fever as a public health, social and economic problem in the 21st century. Trends Microbiol. 2002;10(2):100–3. Epub 2002/02/06. [DOI] [PubMed] [Google Scholar]
- 2.Reiter P. Yellow fever and dengue: a threat to Europe? Euro Surveill. 2010;15(10):19509 Epub 2010/04/21. [PubMed] [Google Scholar]
- 3.Paupy C, Ollomo B, Kamgang B, Moutailler S, Rousset D, Demanou M, et al. Comparative role of Aedes albopictus and Aedes aegypti in the emergence of Dengue and Chikungunya in central Africa. Vector Borne Zoonotic Dis. 2010;10(3):259–66. Epub 2009/09/04. 10.1089/vbz.2009.0005 [DOI] [PubMed] [Google Scholar]
- 4.Charrel RN, de Lamballerie X, Raoult D. Chikungunya outbreaks—the globalization of vectorborne diseases. N Engl J Med. 2007;356(8):769–71. Epub 2007/02/23. 10.1056/NEJMp078013 [DOI] [PubMed] [Google Scholar]
- 5.Morrison AC, Zielinski-Gutierrez E, Scott TW, Rosenberg R. Defining challenges and proposing solutions for control of the virus vector Aedes aegypti. PLoS Med. 2008;5(3):e68 Epub 2008/03/21. 07-PLME-PF-2051 [pii]. PubMed Central PMCID: PMC2267811. 10.1371/journal.pmed.0050068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO. Dengue: Guidelines for diagnosis, treatment, prevention and control: New edition. Geneva, World Health Organization; 2009:147. Epub 2013/06/14. [PubMed] [Google Scholar]
- 7.Marcombe S, Poupardin R, Darriet F, Reynaud S, Bonnet J, Strode C, et al. Exploring the molecular basis of insecticide resistance in the dengue vector Aedes aegypti: a case study in Martinique Island (French West Indies). BMC Genomics. 2009;10:494 Epub 2009/10/28. PubMed Central PMCID: PMC2770535. 10.1186/1471-2164-10-494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ranson H, Burhani J, Lumjuan N, Black W,. Insecticide resistance in dengue vectors. TropIKA net Journal. 2010;1(1). [Google Scholar]
- 9.Jirakanjanakit N, Rongnoparut P, Saengtharatip S, Chareonviriyaphap T, Duchon S, Bellec C, et al. Insecticide susceptible/resistance status in Aedes (Stegomyia) aegypti and Aedes (Stegomyia) albopictus (Diptera: Culicidae) in Thailand during 2003–2005. J Econ Entomol. 2007;100(2):545–50. Epub 2007/04/28. [DOI] [PubMed] [Google Scholar]
- 10.Ishak IH, Jaal Z, Ranson H, Wondji CS. Contrasting patterns of insecticide resistance and knockdown resistance (kdr) in the dengue vectors Aedes aegypti and Aedes albopictus from Malaysia. Parasit Vectors. 2015;8:181 PubMed Central PMCID: PMC4377062. 10.1186/s13071-015-0797-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan HH, Mustafa FF, Zairi J. Assessing the susceptibility status of Aedes albopictus on Penang Island using two different assays. Trop Biomed. 2011;28(2):464–70. Epub 2011/11/02. [PubMed] [Google Scholar]
- 12.Chen CD, Nazni WA, Lee HL, Sofian-Azirun M. Susceptibility of Aedes aegypti and Aedes albopictus to temephos in four study sites in Kuala Lumpur City Center and Selangor State, Malaysia. Trop Biomed. 2005;22(2):207–16. Epub 2006/08/03. [PubMed] [Google Scholar]
- 13.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, et al. The global distribution and burden of dengue. Nature. 2013;496(7446):504–7. PubMed Central PMCID: PMC3651993. 10.1038/nature12060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hennessey M, Fischer M, Staples JE. Zika Virus Spreads to New Areas—Region of the Americas, May 2015-January 2016. Am J Transplant. 2016;16(3):1031–4. [DOI] [PubMed] [Google Scholar]
- 15.Hemingway J, Ranson H. Insecticide resistance in insect vectors of human disease. Annual Review of Entomology. 2000;45:371–91. 10.1146/annurev.ento.45.1.371 [DOI] [PubMed] [Google Scholar]
- 16.Sathantriphop S, Paeporn P, Supaphathom K. Detection of insecticides resistance status in Culex quinquefasciatus and Aedes aegypti to four major groups of insecticides. Trop Biomed. 2006;23(1):97–101. Epub 2006/10/17. [PubMed] [Google Scholar]
- 17.Davies TGE, Field L, M., Usherwood PNR, Williamson MS. A comparative study of voltage-gated sodium channels in the Insecta: implications for pyrethroid resistance in Anopheline and other Neopteran species. Insect Molecular Biology. 2007;16(3):361–75. 10.1111/j.1365-2583.2007.00733.x [DOI] [PubMed] [Google Scholar]
- 18.Brengues C, Hawkes NJ, Chandre F, McCarroll L, Duchon S, Guillet P, et al. Pyrethroid and DDT cross-resistance in Aedes aegypti is correlated with novel mutations in the voltage-gated sodium channel gene. Med Vet Entomol. 2003;17(1):87–94. Epub 2003/04/12. [DOI] [PubMed] [Google Scholar]
- 19.Yanola J, Somboon P, Walton C, Nachaiwieng W, Somwang P, Prapanthadara LA. High-throughput assays for detection of the F1534C mutation in the voltage-gated sodium channel gene in permethrin-resistant Aedes aegypti and the distribution of this mutation throughout Thailand. Trop Med Int Health. 2011;16(4):501–9. Epub 2011/02/24. 10.1111/j.1365-3156.2011.02725.x [DOI] [PubMed] [Google Scholar]
- 20.Saavedra-Rodriguez K, Urdaneta-Marquez L, Rajatileka S, Moulton M, Flores AE, Fernandez-Salas I, et al. A mutation in the voltage-gated sodium channel gene associated with pyrethroid resistance in Latin American Aedes aegypti. Insect Mol Biol. 2007;16(6):785–98. Epub 2007/12/21. 10.1111/j.1365-2583.2007.00774.x [DOI] [PubMed] [Google Scholar]
- 21.Li X, Schuler MA, Berenbaum MR. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu Rev Entomol. 2007;52:231–53. Epub 2006/08/24. 10.1146/annurev.ento.51.110104.151104 [DOI] [PubMed] [Google Scholar]
- 22.Perry T, Batterham P, Daborn PJ. The biology of insecticidal activity and resistance. Insect Biochem Molec. 2011;41(7):411–22. [DOI] [PubMed] [Google Scholar]
- 23.Riveron JM, Yunta C, Ibrahim SS, Djouaka R, Irving H, Menze BD, et al. A single mutation in the GSTe2 gene allows tracking of metabolically-based insecticide resistance in a major malaria vector. Genome Biol. 2014;15(2):R27 10.1186/gb-2014-15-2-r27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riveron JM, Ibrahim SS, Chanda E, Mzilahowa T, Cuamba N, Irving H, et al. The highly polymorphic CYP6M7 cytochrome P450 gene partners with the directionally selected CYP6P9a and CYP6P9b genes to expand the pyrethroid resistance front in the malaria vector Anopheles funestus in Africa. BMC Genomics. 2014;15:817 Epub 2014/09/28. PubMed Central PMCID: PMC4192331. 10.1186/1471-2164-15-817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ibrahim SS, Riveron JM, Bibby J, Irving H, Yunta C, Paine MJ, et al. Allelic Variation of Cytochrome P450s Drives Resistance to Bednet Insecticides in a Major Malaria Vector. PLoS Genet. 2015;11(10):e1005618 PubMed Central PMCID: PMCPMC4627800. 10.1371/journal.pgen.1005618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strode C, Wondji CS, David JP, Hawkes NJ, Lumjuan N, Nelson DR, et al. Genomic analysis of detoxification genes in the mosquito Aedes aegypti. Insect Biochem Mol Biol. 2008;38(1):113–23. Epub 2007/12/12. 10.1016/j.ibmb.2007.09.007 [DOI] [PubMed] [Google Scholar]
- 27.Poupardin R, Riaz MA, Jones CM, Chandor-Proust A, Reynaud S, David JP. Do pollutants affect insecticide-driven gene selection in mosquitoes? Experimental evidence from transcriptomics. Aquat Toxicol. 2012;114–115:49–57. Epub 2012/03/13. 10.1016/j.aquatox.2012.02.001 [DOI] [PubMed] [Google Scholar]
- 28.Kwiatkowska RM, Platt N, Poupardin R, Irving H, Dabire RK, Mitchell S, et al. Dissecting the mechanisms responsible for the multiple insecticide resistance phenotype in Anopheles gambiae s.s., M form, from Vallee du Kou, Burkina Faso. Gene. 2013;519(1):98–106. Epub 2013/02/06. PubMed Central PMCID: PMC3611593. 10.1016/j.gene.2013.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riveron JM, Irving H, Ndula M, Barnes KG, Ibrahim SS, Paine MJ, et al. Directionally selected cytochrome P450 alleles are driving the spread of pyrethroid resistance in the major malaria vector Anopheles funestus. Proc Natl Acad Sci U S A. 2013;110(1):252–7. Epub 2012/12/19. 10.1073/pnas.1216705110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bariami V, Jones CM, Poupardin R, Vontas J, Ranson H. Gene Amplification, ABC Transporters and Cytochrome P450s: Unraveling the Molecular Basis of Pyrethroid Resistance in the Dengue Vector, Aedes aegypti. PLoS Negl Trop Dis. 2012;6(6):e1692 10.1371/journal.pntd.0001692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nature protocols. 2008;3(6):1101–8. Epub 2008/06/13. [DOI] [PubMed] [Google Scholar]
- 32.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–2. Epub 2009/04/07. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- 34.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123(3):585–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133(3):693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–9. PubMed Central PMCID: PMC3840312. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuntal BK, Aparoy P, Reddanna P. EasyModeller: A graphical interface to MODELLER. BMC research notes. 2010;3:226 PubMed Central PMCID: PMC2936912. 10.1186/1756-0500-3-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yano JK, Wester MR, Schoch GA, Griffin KJ, Stout CD, Johnson EF. The structure of human microsomal cytochrome P450 3A4 determined by X-ray crystallography to 2.05-A resolution. J Biol Chem. 2004;279(37):38091–4. 10.1074/jbc.C400293200 [DOI] [PubMed] [Google Scholar]
- 39.Irwin JJ, Shoichet BK. ZINC—a free database of commercially available compounds for virtual screening. Journal of chemical information and modeling. 2005;45(1):177–82. PubMed Central PMCID: PMC1360656. 10.1021/ci049714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.PyMOL User's Guide [Internet]. The PyMOL Molecular Graphics System 2004 [Google Scholar]
- 41.Vontas J, Blass C, Koutsos AC, David JP, Kafatos FC, Louis C, et al. Gene expression in insecticide resistant and susceptible Anopheles gambiae strains constitutively or after insecticide exposure. Insect Mol Biol. 2005;14(5):509–21. 10.1111/j.1365-2583.2005.00582.x [DOI] [PubMed] [Google Scholar]
- 42.Ishak IH, Riveron JM, Ibrahim SS, Stott R, Longbottom J, Irving H, et al. The Cytochrome P450 gene CYP6P12 confers pyrethroid resistance in kdr-free Malaysian populations of the dengue vector Aedes albopictus. Sci Rep. 2016;6:24707 PubMed Central PMCID: PMCPMC4837359. 10.1038/srep24707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tene BF, Poupardin R, Costantini C, Awono-Ambene HP, Wondji C, Ranson H, et al. Resistance to DDT in an Urban Setting: Common Mechanisms Implicated in Both M and S Forms of Anopheles gambiae in the City of Yaoundé Cameroon. Plos One. 2013;8(4):e61408 10.1371/journal.pone.0061408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Witzig C, Parry M, Morgan JC, Irving H, Steven A, Cuamba N, et al. Genetic mapping identifies a major locus spanning P450 clusters associated with pyrethroid resistance in kdr-free Anopheles arabiensis from Chad. Heredity (Edinb). 2013. Epub 2013/01/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lumjuan N, Rajatileka S, Changsom D, Wicheer J, Leelapat P, Prapanthadara LA, et al. The role of the Aedes aegypti Epsilon glutathione transferases in conferring resistance to DDT and pyrethroid insecticides. Insect Biochem Mol Biol. 2011;41(3):203–9. Epub 2011/01/05. 10.1016/j.ibmb.2010.12.005 [DOI] [PubMed] [Google Scholar]
- 46.Stevenson BJ, Pignatelli P, Nikou D, Paine MJI. Pinpointing P450s Associated with Pyrethroid Metabolism in the Dengue Vector, Aedes aegypti: Developing New Tools to Combat Insecticide Resistance. PLoS Negl Trop Dis. 2012;6(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Colovos C, Yeates TO. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2(9):1511–9. 10.1002/pro.5560020916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saavedra-Rodriguez K, Suarez AF, Salas IF, Strode C, Ranson H, Hemingway J, et al. Transcription of detoxification genes after permethrin selection in the mosquito Aedes aegypti. Insect Mol Biol. 2012;21(1):61–77. PubMed Central PMCID: PMC3540788. 10.1111/j.1365-2583.2011.01113.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vontas J, Kioulos E, Pavlidi N, Morou E, della Torre A, Ranson H. Insecticide resistance in the major dengue vectors Aedes albopictus and Aedes aegypti. Pesticide Biochemistry and Physiology. 2012;104(2):126–31. [Google Scholar]
- 50.Yang Q, Zhou D, Sun L, Zhang D, Qian J, Xiong C, et al. Expression and characterization of two pesticide resistance-associated serine protease genes (NYD-tr and NYD-ch) from Culex pipiens pallens for metabolism of deltamethrin. Parasitol Res. 2008;103(3):507–16. 10.1007/s00436-008-0997-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Green arrows refer to Cy-3 dye and red arrows refer to Cy-5 dye.
(PPTX)
(PPTX)
(PPTX)
(PPTX)
(PPTX)
Binding conformation of deltamethrin in (A) KL, (B) JB, (C) PG and (D) NO CYP9J27 models. Distances between the probable sites of attack on deltamethrin and heme iron are annotated.
(PPTX)
Binding conformation of DDT in (A) KL, (B) JB, (C) PG and (D) NO CYP9J27 models. Distances between the probable sites of attack on DDT and the heme iron are annotated.
(PPTX)
Binding conformation of bendiocarb in (A) KL, (B) JB, (C) PG and (D) NO CYP9J27 models. Distances between the probable sites of attack on bendiocarb and the heme iron are annotated.
(PPTX)
Binding conformation of (A) permethrin, (B) deltamethrin, (C) DDT and (D) bendiocarb in KB CYP9J27 model. Distances between the probable sites of attack on various insecticides and the heme iron are annotated.
(PPTX)
(DOCX)
(DOCX)
FC = fold change. (p = 0.01).
(DOCX)
FC = fold change. (p = 0.01).
(DOCX)
FC = fold change. (p = 0.01).
(DOCX)
(DOCX)
Data Availability Statement
GenBank Accesion Number: KX394421-KX394443