Abstract
Common diseases often show sex differences in prevalence, onset, symptomology, treatment, or prognosis. Although studies have been performed to evaluate sex differences at specific SNP associations, this work aims to comprehensively survey a number of complex heritable diseases and anthropometric traits. Potential genetically encoded sex differences we investigated include differential genetic liability thresholds or distributions, gene–sex interaction at autosomal loci, major contribution of the X-chromosome, or gene–environment interactions reflected in genes responsive to androgens or estrogens. Finally, we tested the overlap between sex-differential association with anthropometric traits and disease risk. We utilized complementary approaches of assessing GWAS association enrichment and SNP-based heritability estimation to explore explicit sex differences, as well as enrichment in sex-implicated functional categories. We do not find consistent increased genetic load in the lower-prevalence sex, or a disproportionate role for the X-chromosome in disease risk, despite sex-heterogeneity on the X for several traits. We find that all anthropometric traits show less than complete correlation between the genetic contribution to males and females, and find a convincing example of autosome-wide genome-sex interaction in multiple sclerosis (P = 1 × 10−9). We also find some evidence for hormone-responsive gene enrichment, and striking evidence of the contribution of sex-differential anthropometric associations to common disease risk, implying that general mechanisms of sexual dimorphism determining secondary sex characteristics have shared effects on disease risk.
Keywords: sex differences, gene–sex interactions, heritability, sex heterogeneity, hormone-responsive genes
SEX differences are a major predictor in many common diseases, used in diagnosis, prognosis, and treatment recommendations. We know much about the biological basis of sex determination (She and Yang 2014), and research in model organisms allows us to separate the effects of sex chromosomes and hormonal differences (Arnold and Chen 2009; Cox et al. 2014). However, we do not fully understand how the biology of sex shapes disease risk and outcomes in humans (Ober et al. 2008; Ngo et al. 2014; Austad and Bartke 2015). While some studies in model organisms suggest major roles for gene–sex interaction in complex traits (Mackay 2009; Lehtovaara et al. 2013; Bearoff et al. 2015; Parks et al. 2015), a recent study using mouse models found few true sex interaction effects (Krohn et al. 2014). Human studies of disease-relevant quantitative traits in founder populations suggested major sex differences in heritability and identifiable genetic loci (Weiss et al. 2006), as well as a major role for the X-chromosome (Pan et al. 2007). Twin studies have been used to investigate gene–sex interaction in a variety of complex diseases and traits, with a range of findings from little to substantial sex difference (Vink et al. 2012; Mitchem et al. 2014; Richmond-Rakerd et al. 2014). Additionally, several studies have examined loci identified in combined-sex samples to identify gene–sex interactions in these candidate regions (Avery et al. 2006; Silander et al. 2008; Loisel et al. 2011; Gilks et al. 2014; Yao et al. 2014; de Castro-Catala et al. 2015; Mersha et al. 2015). However, few studies have applied more sophisticated genome-wide methodologies for assessing association and determining additive SNP-based heritability to comprehensively assess sex differences (Zillikens et al. 2008; Chiu et al. 2010; Luo et al. 2010; Myers et al. 2014).
In this work, we selected nine common diseases, and nine heritable traits, with rich genetic datasets available and a variety of sex biases, to investigate several genetic hypotheses about the drivers of sexual dimorphism. For discrete traits, we examine consistent adherence to liability threshold (LT) models (Hayeck et al. 2015; Weissbrod et al. 2015), which are commonly used in contemporary heritability analyses (Cross-Disorder Group of the Psychaitric Genomics Consortium 2013; Lee et al. 2011). Under an LT model of disease, individuals have an underlying normally distributed phenotype, φ, called the liability. When an individual’s liability is greater than a threshold, t, they are a case, and are a control otherwise. In order to induce a sex-biased disease prevalence, the liability distributions, and/or thresholds, must differ between males and females. When males and females have identical distributions of liability, but a sex-biased prevalence exists due to a difference in sex-specific thresholds, the lower prevalence sex will have an enrichment of genetic associations due to the increased genetic load required to exceed the higher threshold. This is mathematically equivalent to one sex having an environmental risk factor, (e.g., if androgens affect the mean liability, the disease prevalence will differ between males and females), and again the lower prevalence sex will have a relatively increased genetic load among cases.
To evaluate this LT model (hypothesis 1, Figure 1), we consider autosomal genetic load in both sexes to determine whether differences in polygenic burden can account for sex differences in prevalence. We use enrichment of autosome-wide association signal in male-specific and female-specific datasets (1a) and polygenic additive SNP-based heritability estimates (h2g) to determine whether males and females have different genetic loads for common disease and anthropometric traits (1b), and whether the genetic patterns follow those expected under the liability threshold model based on prevalence differences, or imply the existence of nongenetic sex differences in the mean or variance of liability or trait distributions.
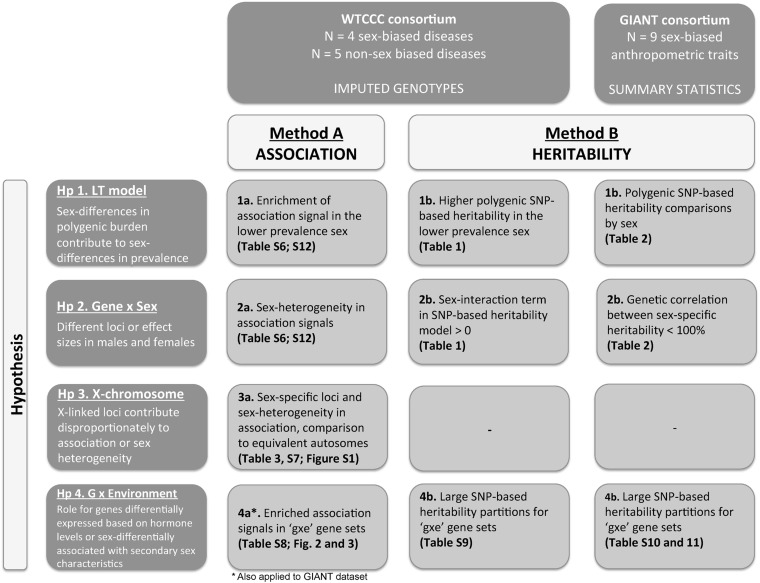
Figure 1.
Analyses flow chart. The scheme reports four main hypothesis tested with two approaches (method A: association; method B: heritability). Method A was applied to the imputed genotypes available for WTCCC dataset (when possible also to summary statistics available for GIANT dataset), whereas method B was applied to both WTCCC and GIANT datasets. For each hypothesis and method, the results are reported in the Results section, and summarized in figures and tables: results for hypothesis 1 tested in the WTCCC dataset are reported in Table 1 and in the GIANT dataset in Table 2. Table 1 and Table 2 also report results for hypothesis 2 tested in the WTCCC dataset and GIANT dataset, respectively. Results for hypothesis 3 are shown in Table 3. Finally, hypothesis 4 results are shown in Figure 2 and Figure 3.
Second, we globally test for evidence of gene–sex interaction (hypothesis 2, Figure 1) to determine whether similar or different autosomal loci might contribute to disease risk across the sexes. Much of the gene–sex interaction literature is focused on specific genetic loci that might differ in their effects by sex. We assess more globally whether evidence exists for sex-heterogeneity in association signal (2a), significant sex-interaction terms in h2g models, or a genetic correlation <1 for the same trait across the sexes (2b). We also use simulation to examine the effects of liability variance differences between sexes on disease prevalence that can occur in the presence of gene–sex interactions.
Third, we dissect the role of the X-chromosome (hypothesis 3, Figure 1), the major genomic sex difference (Ross et al. 2005). The X-chromosome is gene-rich, contrasting with the small gene-poor Y chromosome (Ellis and Affara 2006; Mulugeta et al. 2016), and, despite dosage-compensation mechanisms, shows gene expression differences across the sexes (Jansen et al. 2014). The X-chromosome has been proposed to contribute to autoimmune disease; metabolic and cardiovascular traits such as fasting insulin, blood pressure, and cholesterol levels; and anthropometric traits such as height (Pan et al. 2007; Chen et al. 2012; Gao et al. 2014; Tukiainen et al. 2014). We examine sex differences in association signal on the X-chromosome between males and females (3a).
Finally, we postulate that gene-environment interactions might generate sexual dimorphism (hypothesis 4, Figure 1), and, at the cellular level, steroid hormones could be a significant contributor. Thus, we specifically consider genes whose expression is known to be responsive to androgens or estrogens, and assess whether SNPs in these genes contribute disproportionately to association signal (4a), or heritability (4b), in complex disease or anthropometric traits. Similarly, we ask whether the same underlying biology is responsible for secondary sex characteristics like height, weight, and body proportions, and we assess whether SNPs showing differential association to anthropometric traits by sex contribute markedly to common, complex heritable disease (Roach et al. 2015).
Materials and Methods
Samples
WTCCC1 and WTCCC2 data were gathered from the Wellcome Trust Case Control Consortium (WTCCC; http://www.wtccc.org.uk/). WTCCC1 includes BD (prevalence 0.005), CAD (prevalence 0.06), CD (prevalence 0.001), HT (prevalence 0.26), RA (prevalence 0.005), T1D (prevalence 0.005), and T2D (prevalence 0.08); WTCCC2 includes AS (prevalence 0.003) and MS (prevalence 0.001) (Table S1).
Genetic Investigation of Anthropometric Traits (GIANT) genome-wide meta-analyzed data were gathered from the Broad Institute (http://portals.broadinstitute.org/collaboration/giant/index.php/GIANT_consortium_data_files). The 2015 datasets were used for all anthropometric traits, except for when unavailable [such as for Height and Weight; 2012 uploads were used instead (Yang et al. 2012)] (Table S1).
Software
PLINK v 1.90b2n and R v 3.0.1, and METASOFT v2.0.0 were used in association enrichment and heterogeneity analyses. GCTA v 1.24.2 was used for REML estimates of heritability. PCGC regression was used for Haseman-Elston regression estimates heritability. HAPI-UR v 1.01 was used for prephasing, and IMPUTE v 2.3.0 was used for imputation.
Creation of the hormone-responsive gene sets
An androgen-responsive gene list was gathered from ARGDB (Jiang et al. 2009) (Table S2), with duplicates in name removed for a total of 2616 genes. Of these 2616 genes, 2508 are autosomal. CNTNAP2 was removed from androgen-responsive analyses due to difficulty of matching on gene-length in gene-permutations, as detailed below.
An estrogen-responsive gene list was gathered from ERGDB (Tang et al. 2004) (Table S3), with duplicates in name removed for a total of 1431 genes. Of these genes, 1150 are autosomal. GRID2 was removed from estrogen-responsive analyses due to difficulty of matching on gene-length in gene-permutations, as detailed below.
Creation of top heterogeneous anthropometric hits
METASOFT was used on GIANT consortium data (further elaborated on below) to ascertain the most significantly heterogeneous SNPs between males and females for each anthropometric trait separately (P ≤ 0.001). These markers were combined for a total of 8423 SNPs. This list contained no duplicates, and was LD-pruned (r2 = 0.5) for a final total of 8162 SNPs.
Determination of sex
PLINK –sex-check flags were used to determine the sexes of all subjects within the WTCCC1/2 datasets. Ambiguously identifiable individuals per PLINK’s –sex-check flag were excluded from sex-specific analyses.
Imputation
WTCCC data were prephased using three runs of HAPI-UR with default parameters, and merged by consensus vote. Phased genomes were then imputed to the 1000 Genomes (phase 1 integrated v3) reference panels in 1-MB windows using default parameters. For chrX imputation, only the nonpseudoautosomal reference was used. The X chromosome contained ∼10,000 and ∼14,000 genotyped SNPs in the WTCCC1 and WTCCC2 data, respectively. After imputation, 64 and 69% of all reference SNPs with MAF >1% were imputed with high accuracy (INFO score >0.9) in the respective cohorts; indicative of sufficient SNP density to impute a majority of the reference variants with high quality. After imputation, variants with INFO score >0.5 were retained, and dosages were rounded to hard calls. An additional round of stringent QC was performed, removing any variants with: MAF <0.01; missingness >5%; Hardy-Weinberg equilibrium P < 0.01; or case-control missingness P < 0.05.
HLA removal in autoimmune diseases
For all autoimmune diseases—rheumatoid arthritis, Crohn’s disease, type-1 diabetes, multiple sclerosis, and ankylosing spondylitis—the HLA region was removed due to the known significant enrichment associated within that region (of extended linkage disequilibrium) that violates the polygenic assumption of many modest effects across the genome. The HLA region was defined as chr6:26,000,000:34,000,000 (de Bakker et al. 2006).
Method A—association analyses
We applied two different approaches, i.e., association enrichment (method A) and SNP-based heritability (method B), to test our four main hypotheses. A complete analysis flowchart is reported in Figure 1.
Association analyses and false discovery rate plots:
In each complex trait, we analyzed the sexes combined and separately using the appropriate –filter-males and –filter-females PLINK flags where needed. Through PLINK, we ran logistic regressions, while using as covariates the first 10 principal components of each phenotype to account for differences in genetic ancestry. Sex-specific association signal enrichment was also tested separately for the nonpseudoautosomal X chromosome (and chromosome 7 and 17 for comparison). The X chromosome was coded in standard PLINK format, where male genotypes are A = 0 and B = 1, and female genotypes are AA = 0, AB = 1, and BB = 2. In order to compare across permutations, we set a false discovery rate (FDR) threshold, and assessed the percent of SNPs with q-value exceeding this threshold. A threshold of q = 0.7 was chosen to maximize the power of this comparison in small datasets. This was determined after calculating the percentage of autosomal SNPs above a range of FDR thresholds (0.1, 0.3, 0.5, 0.7, and 0.9) in the male, female, and combined-sex datasets for WTCCC diseases. For each FDR threshold, we calculated the number of datasets with SNP proportion below 0.0001%, and above 1%. An FDR threshold 0.7 minimized datasets for which our comparisons would lack power, or be based on implausibly high proportions of SNPs (Table S4). Note that we do not use this FDR threshold to assess significance, only as a means of comparison across permuted datasets, as recommended previously (Liu et al. 2012a). The proportion of SNPs with q-values that meet or fall below the FDR threshold is referred to here as the original proportion.
This procedure was implemented for genome-wide, chromosome-specific, hormone-responsive, and sex-heterogeneous SNP set analyses.
Sex permutations:
To assess whether association signals are truly female- or male-specific, we ran a series of sex permutations. The null hypothesis we assess is that differences observed in male-specific vs. female-specific datasets are random sampling differences; our alternative hypothesis is that sex is driving a difference. For each complex trait, we randomly permuted sex labels within case and control strata. We produced a complete list of permuted individuals, combining sex-permuted controls with the sex-permuted cases. We repeated this 100 times for a total collection of 100 permuted male lists and 100 permuted female lists. R was used to generate these lists in the format of phenotype files for consequent association analyses in PLINK, and then multiple-test corrections via FDR in R, as described above. As a result of this step, each permutation results in a data point—a proportion of SNPs that meet or fall below the FDR threshold. Empirical P-values were calculated by tallying the number of permuted sets with a proportion of significant SNPs that exceeded the original proportion. For analysis with nominally significant and/or borderline significant empirical P-values, P ≤ 0.1, we repeated the random selection 1000 times, and we obtained 1000 permuted male lists, and 1000 permuted female lists, and we estimated empirical P-values as described earlier. We replaced initial P-values estimated with 100 permutations with empirical P-values obtained with 1000 permutations.
Test for heterogeneity:
METASOFT was used to assess extent of significant heterogeneity between the sexes by means of Cochran’s Q test for heterogeneity. To discern whether a phenotype was particularly (read: significantly) heterogeneous, we used the above FDR test and sex-permutation approach. Empirical P-values were used to determine significance. This analysis was done on the Wellcome-Trust cohorts, and in genome-wide and X-linked data. For significantly heterogeneous phenotypes, we applied a binomial sign test to assess the top 0.1% of hits in sex-specific datasets for effect directions matching across the sexes. Empirical P-values were calculated with the sex-permutation approach. For analysis with nominally significant empirical P-values P ≤ 0.1, we repeated the sex-permutation approach 1000 times, and we estimated empirical P-values as described earlier. We replaced initial P-values estimated with 100 permutations with empirical P-values obtained with 1000 permutations.
Gene/SNP permutations:
Similar to the purpose of the sex permutations, gene permutations are meant to assess whether elevated enrichment when investigating a subset of genes is due to those genes, and not to other factors like gene size. In gene permutations, we sample genes that met the following criteria: (1) the sampled gene locus includes at least one SNP that is represented within our dataset, and (2) the sampled gene is matched on gene length, selected within a window of 100 genes closest in length to the original gene of interest. SNPs for these genes (all SNPs within 5 kb of longest transcript) were then compiled into lists. Because association analyses were previously run, the SNPs in each permuted list were extracted with their corresponding P-value and quantified for the proportion of SNPs that met or fell below the FDR threshold. In other words, the same FDR analysis was conducted on each permuted gene list as it had been for the original gene set of interest. We repeated this 100 times for a total collection of 100 permuted estrogen-responsive gene lists, and 100 permuted androgen-responsive gene lists.
For sex-heterogeneous SNPs, SNP permutations were performed using a similar procedure, but matching individual SNPs for test statistic in the combined-sex dataset of origin (i.e., a SNP with sex-heterogeneity for height will be matched on its test statistic for combined-sex association for height, but tested for association enrichment in each anthropometric trait).
Empirical P-values were determined as described earlier. For analysis with nominally significant empirical P-values P ≤ 0.1, we repeated the random selection 1000 times, and we obtained 1000 permuted estrogen-responsive gene lists, 1000 permuted androgen-responsive gene lists, and 1000 permuted sex-heterogeneous SNPs. We estimated empirical P-values as described earlier. We replaced initial P-values estimated with 100 permutations with empirical P-values obtained with 1000 permutations.
Reference gene annotations for sampling were downloaded from UCSC’s genome annotation database. For WTCCC1/2 analyses, we used the hg19 refGene.txt.gz annotation file. The file was truncated to contain: (1) only autosomes, (2) the longest version of a gene when duplicates were found, and (3) removal of CNTNAP2 and GRID2 due to their inability to be properly matched.
Method B—heritability analyses
Variance-component estimation:
SNP-heritability (h2g) was estimated using variance-components and restricted maximum-likelihood (REML) (Yang et al. 2011) for studies with individual-level data. Briefly, the variance-component model assumes the phenotype is drawn from a multivariate normal distribution, with variance modeled by a linear combination of components computed from the SNPs and a normal residual. For each annotation (e.g., AR genes) a genetic relatedness matrix (GRM) from SNPs in that annotation was jointly evaluated with a GRM from all remaining SNPs and the identity matrix to estimate the corresponding variance parameters and The heritability proportion was then computed as and the corresponding SE estimated using the delta method. Twenty principal components were always included as fixed-effects in the estimation procedure to account for population structure. We observed a significant association between sex and the top 20 PCs for 2/9 cohorts: AS (P = 0.003, R2 = 0.008) and MS (P = 4 × 10−21, R2 = 0.011). However, because the R2 is low (explaining ∼1% of the sex label), and principal components were always included as fixed effects, we do not expect this to impact the results. We separately evaluated the potential confounding effect of case-control ascertainment by estimating genome-wide SNP-heritability using Haseman-Elston regression (Golan et al. 2014), where the product of individual phenotypes is regressed on the corresponding off-diagonal GRM entries (Table S5). The same GRMs, individuals, and principal components were used for both estimation procedures.
A similar procedure was used for estimating autosomal heritability of SNPs-by-sex (). The sex GRM () was computed by setting entries in the standard GRM to 0 for individuals of different sex. The phenotype was then modeled as and significance of assessed by likelihood ratio test against the one-component model.
We did not use a bivariate variance-components model to evaluate genetic correlation between traits due to small sample size. Power calculations showed that, for the average trait evaluated here (1300 cases and 1700 controls), the SE on the genetic correlation is expected to be 0.21, yielding little power to detect deviations.
Heritability permutations:
To evaluate significance of the difference in h2g between males and females, sex label permutation was again used. For each trait, sex labels were randomly shuffled and sex-specific partitioned heritability re-estimated to get the estimates from a single permutation. The permutation procedure was performed 1000 times, and the fraction of instances where the absolute permuted difference was higher than the absolute observed difference used to compute the P-value. The same variance-components and fixed effects were used in the permutations as in the real data.
Summary statistic-based estimation:
SNP-heritability was estimated using LD-score regression (LDSC) for studies with only summary-level data. For a single trait and functional annotation, LDSC regresses the χ2 (or Z2) association statistic from each SNP onto the “LD-score” of that SNP: computed as the sum of LD across all neighboring SNPs from a reference panel. Under assumptions of independent causal effect sizes, the slope of this regression is then proportional to the SNP-heritability, and the intercept is proportional to the effects of population stratification. For multiple functional annotations, the model naturally extends to include annotation-specific LD-scores computed only to the neighboring SNPs that belong to the given annotation. The coefficients from this multiple regression are then proportional to the partitioned SNP-heritability for each annotation. For multiple traits, replacing the χ2 statistic with the product of association βs from each trait in either the single or multiple LD-score regression yields an estimate of genetic correlation between the traits.
LDSC was run on sex-specific summary statistics to estimate total h2g and genetic correlation using default parameters and default LD-scores (computed in the 1000 Genomes EUR samples). The GIANT GWAS data were imputed (by the original study) to ∼2 M HapMap3 variants. This is the recommended SNP set to use for LD-score regression, and has been shown to yield comparable results to high-quality 1000G imputation (Finucane et al. 2015). SE were estimated using the weighted block-jackknife. For the novel annotations of AR genes, ER genes, and GIANT heterogeneous SNPs, additional LD-scores were computed using the 1000 Genomes EUR samples, and used to partition the heritability and genetic correlation. Heritability partitioning was evaluated with and without the “baseline” annotations from Finucane et al. (2015) to account for potential background enrichment from overlapping functional categories. When baseline annotations were included, the P-value of the coefficient is reported, which corresponds to the significance of the given annotation beyond all other annotations in the model. For the AR/ER annotations this will yield a conservative estimate because multiple “genic” annotations are already in the baseline model. In contrast to the previous studies with raw data, we are not aware of any method to evaluate G × Sex heritability from summary data, and could not assess this effect for the anthropometric traits.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article and are publicly available.
Results
Autosomal genetic load
We studied nine common, complex diseases from the WTCCC studies (WTCCC 2007; Evans et al. 2011; Sawcer et al. 2011), and nine anthropometric traits from the GIANT studies (Randall et al. 2013; Shungin et al. 2015), to capture a variety of sex biases (or lack thereof). Male biased diseases included ankylosing spondylitis (AS, with M:F prevalence of 2:1) (Chen et al. 2011; Haroon et al. 2014), and type 1 diabetes (T1D, 3:2) (Gale and Gillespie 2001; Liu et al. 2012a; Orozco et al. 2012). Female biased diseases included multiple sclerosis (MS, 1:3) (Pakpoor and Ramagopalan 2014) and rheumatoid arthritis (RA, 1:2) (Emery et al. 2014). Disorders estimated to have similar lifetime prevalence by sex included bipolar disorder (BD) (Almeida-Filho et al. 1997; Negash et al. 2005; Diflorio and Jones 2010), coronary artery disease (CAD) (Sharma and Gulati 2013), Crohn’s disease (CD) (Liu et al. 2012b; Law and Li 2014), hypertension (HT) (Nwankwo et al. 2013), and type 2 diabetes (T2D) (Orozco et al. 2012; Hilawe et al. 2013), although sex differences often exist in age of onset, subtype, and comorbidities. Anthropometric traits with different means by sex were body mass index (BMI), hip circumference (hip), hip adjusted for BMI (hip-a), waist circumference (WC), WC adjusted for BMI (WC-a), waist-hip-ratio (WHR), WHR adjusted for BMI (WHR-a), height, and weight.
Under a threshold difference LT model of sex bias, we would anticipate that diseases with increased prevalence in one sex would have more association enrichment due to increased genetic load in the opposite sex. This pattern did not occur for any of the disorders with sex-biased prevalence, when sex-specific datasets were compared with sex-permuted datasets [Figure 1 (Method 1a) and Table S6]. We excluded the HLA region for autosomal analysis of autoimmune diseases, as its large effect could bias interpretation. However, when we tested the HLA region separately, we observed greater association enrichment in the HLA region in AS males (P < 0.001). As is male-biased in prevalence, neither finding supports the threshold difference LT model (Table S6).
In order to confirm our observations using an independent approach, we evaluated polygenic variance components for differences in total heritability, gene set specific heritability, and genetic correlation [Figure 1 (Method 1b)]. These analyses consider both “quantitative” differences between sexes due to different genetic variance, and “qualitative” differences due to different regions contributing to heritability. First, we estimated SNP-based additive heritability (h2g) on the liability scale, utilizing imputed datasets for each disease trait, and utilizing summary statistics for each anthropometric trait. This estimate is a ratio of the SNP-genetic and environmental terms, and resulting h2g differences between sexes can be indicative of quantitative differences in either component. The HLA region was excluded from autosomal heritability estimates for MHC-linked autoimmune disorders as in our association enrichment results, as its major effect (with extensive LD) violates the polygenic model, and could thus impact interpretation.
As in our association enrichment results, we did not find evidence for a threshold difference LT model based on heritability estimates. Of the two disorders with higher male prevalence (AS, T1D), neither showed increased heritability in the lower-prevalence sex (Table 1), with similar heritability estimates for each sex in AS, and higher estimate in males for T1D, which was not significantly different from female estimates. Of the disorders with higher female prevalence (MS, RA), we observed similar estimates across the sexes for MS, and a higher estimate of heritability for males with RA, which was not significant.
Table 1. Autosomal SNP-based heritability and G × Sex interaction in WTCCC diseases.
| Disease | Heritabilitya | G × Sex Interactiona | |
|---|---|---|---|
| M h2g (SE) | F h2g (SE) | h2gxs (SE) | |
| ASb | 0.11 (0.04) | 0.15 (0.07) | 0.01 (0.05) |
| BD | 0.18 (0.08) | 0.22 (0.06) | −0.01 (0.06) |
| CAD | 0.25 (0.09) | 0.50 (0.22) | 0.12 (0.11) |
| CDb | 0.14 (0.06) | 0.17 (0.04) | 0.03 (0.05) |
| HT | 0.18 (0.21) | 0.73 (0.17)*** | 0.19 (0.17) |
| MSb | 0.17 (0.02) | 0.17 (0.02) | 0.06 (0.01)**** |
| RAb | 0.20 (0.11) | 0.08 (0.05) | 0.03 (0.06) |
| T1Db | 0.14 (0.06) | 0.08 (0.06) | 0.04 (0.06) |
| T2D | 0.31 (0.12) | 0.37 (0.15) | −0.06 (0.12) |
h2g, heritability; SE, standard error. **** P < 0.0005, *** P < 0.001.
Empirical P-value for difference between M and F h2g: estimation based on 1000 permutations.
HLA locus was tested separately from the rest of the autosomes.
One disease thought to have similar lifetime prevalence showed evidence for sexually dimorphic heritability, with HT showing higher heritability in females in the WTCCC dataset (P < 0.001). We re-evaluated the dichotomous traits using a regression-based method that is not biased by case-control ascertainment, and observed no substantial change in sex differences (see Materials and Methods, Table S5). Several anthropometric traits showed higher heritability in males compared with females, including WC-a, hip-a, WHR (P < 0.05) (Table 2).
Table 2. Autosomal SNP-based heritability and genetic correlation in GIANT anthropometric traits.
| Phenotype | Heritability | Genetic Correlation | ||||
|---|---|---|---|---|---|---|
| M h2g (SE) | F h2g (SE) | P-valuea | covg (SE) | rg (SE) | P-valuea | |
| BMI | 0.18 (0.01) | 0.17 (0.01) | NS | 0.15 (0.01) | 0.89 (0.02) | 5.6 × 10−10 |
| HIP | 0.19 (0.02) | 0.18 (0.01) | NS | 0.17 (0.01) | 0.90 (0.03) | 6.2 × 10−4 |
| HIP-a | 0.21 (0.01) | 0.15 (0.01) | 5.5 × 10−3 | 0.17 (0.01) | 0.92 (0.03) | 2.6 × 10−3 |
| WC | 0.18 (0.02) | 0.16 (0.01) | NS | 0.16 (0.01) | 0.91 (0.03) | 1.6 × 10−3 |
| WC-a | 0.18 (0.01) | 0.11 (0.01) | 2.1 × 10−5 | 0.11 (0.01) | 0.75 (0.05) | 2.0 × 10−8 |
| WHR | 0.14 (0.01) | 0.11 (0.01) | 4.0 × 10−2 | 0.09 (0.01) | 0.74 (0.05) | 9.2 × 10−7 |
| WHR-a | 0.13 (0.01) | 0.11 (0.01) | NS | 0.08 (0.01) | 0.67 (0.06) | 2.3 × 10−9 |
| Height | 0.26 (0.02) | 0.25 (0.02) | NS | 0.25 (0.02) | 0.95 (0.02) | 2.0 × 10−2 |
| Weight | 0.19 (0.01) | 0.19 (0.01) | NS | 0.18 (0.01) | 0.93 (0.03) | 1.0 × 10−2 |
h2g, heritability; SE, standard error; covg, genetic covariance; rg, genetic correlation; NS, not significant.
Empirical P-value for difference between M and F h2g: estimation based on block jackknife.
Gene × sex
We did not detect autosome-wide heterogeneity by comparing the distribution of Cochran’s Q [Figure 1 (Method 2a)] to the distribution of sex-permuted datasets. However, when we separately assessed the HLA region, we observed significant sex-heterogeneity for association signal on the HLA region for MS (P < 0.001) (Table S6). For traits with, or without, polygenic heritability differences, gene–sex interaction could occur at specific loci [Figure 1 (Method 2b)]. For example, a disease could have equivalent overall heritability but with different loci contributing in males vs. females; a difference in environmental contribution could result in different heritability, with the same loci contributing, or different loci could result in disparate heritability estimates. For traits with individual-level data, we were able to include an overall G × Sex interaction term in the heritability estimates. We observed highly significant heritability of G × Sex for MS (0.06 SE 0.01; P = 1 × 10−9); this was striking, given the remaining additive SNP-heritability was 0.13 (SE 0.01) (Table 1). Although we could not specify an interaction term due to lack of genotype data for the anthropometric traits, we assessed traits for which genetic correlation between male-specific and female-specific datasets was significantly <1, which occurred for every anthropometric trait (P < 0.05) (Table 2). This estimate of genetic correlation is not affected by quantitative differences in total genetic variance, and only reflects differences in individual effect-sizes.
X-chromosome
Genetic sex differences not contained on the autosomes could be attributable to the sex chromosomes. Since the X-chromosome is relatively gene-rich, contrasting with the small gene-poor Y chromosome (Ellis and Affara 2006; Mulugeta et al. 2016), and our disease datasets had available data, we assessed the contribution of the X-chromosome. We detect a relatively modest contribution from the X-chromosome to this group of common, complex diseases [Figure 1 (Method 3a)]. Association enrichment signal appears similar to autosomes matched by physical length (chr7), and SNP content (chr17) (Table S7).
We next assessed sex differences on the X-chromosome. We note that, although females have twice the number of alleles, and thus increased power compared to males, one might expect greater impact of nonpseudoautosomal loci in males, who are hemizygous, and thus express an associated allele in every cell as the sole copy. AS and CAD showed increased male signal (P = 0.050 and P = 0.037, respectively) (Table 3).
Table 3. Sex-differences in X-chromosome association signal and heterogeneity in WTCCC diseases.
| Prev | Association | Heterogeneity | Sign Testa | |||
|---|---|---|---|---|---|---|
| Disease | M:F | M Pb | F Pb | Pb | M%c | F%d |
| AS | 2:1 | 0.0500e | NS | 0.0600e | 77.3 | 9.3** |
| BD | 1:1 | NS | NS | NS | — | — |
| CAD | 1:1 | 0.0370e | NS | 0.0040e | 71.3* | 8.0* |
| CD | 1:1 | NS | NS | NS | — | — |
| HT | 1:1 | NS | NS | NS | — | — |
| MS | 1:3 | NS | NS | 0.0016e | 82.0 | 75.7 |
| RA | 1:2 | NS | 0.1000e | NS | — | — |
| T1D | 3:2 | NS | NS | NS | — | — |
| T2D | 3:2 | NS | NS | NS | — | — |
Prev, ratio in prevalence.
Sign test based on 0.1% of male and female top hits applied only to significant heterogeneity results.
Empirical P-value estimation based on 100 permutations.
Empirical P-value estimation based on 100 permutations showing increase.
Empirical P-value estimation based on 100 permutations showing decrease.
Empirical P ≤ 0.1 were replaced with empirical P-values estimated with 1000 permutations. ** P < 0.01 ; * P < 0.05
Of all the equivalent sets of permutations performed on both chromosomes 7 and 17, only one showed a significant female increase (T2D, chr7 P = 0.046) (Table S7). To support these data suggesting sex differences specific to the X-chromosome, we assessed the chromosome-wide heterogeneity via Cochran’s Q, and compared the distribution to sex-permuted datasets. AS, CAD, and MS showed suggestive or significant sex-heterogeneity for association signal on the X-chromosome (P = 0.059, P = 0.004, and P = 0.016, respectively) (Table 3 and Figure S1). It is difficult to consider equivalent male and female statistical models considering X-inactivation (or escape), because the biology of mosaicism is challenging to predict and compare with the typical biallelic models (or hemizygosity in males). We thus performed the binomial test to compare the direction of association, as direction should not be impacted by model specification, regardless of effect size estimates. While MS did not show differences in direction of association in the top 0.1% of X-chromosome SNPs, male and female top associations show significant differences in AS (P = 0.01 females) and in CAD (P = 0.04 males; P = 0.02 females). For AS and CAD, while male top X-chromosome results showed an excess of top SNPs in the same direction in females, female top X results showed a significantly decreased proportion of top SNPs in the same direction in males compared with permutations.
Gene set “environmental” contribution analyses
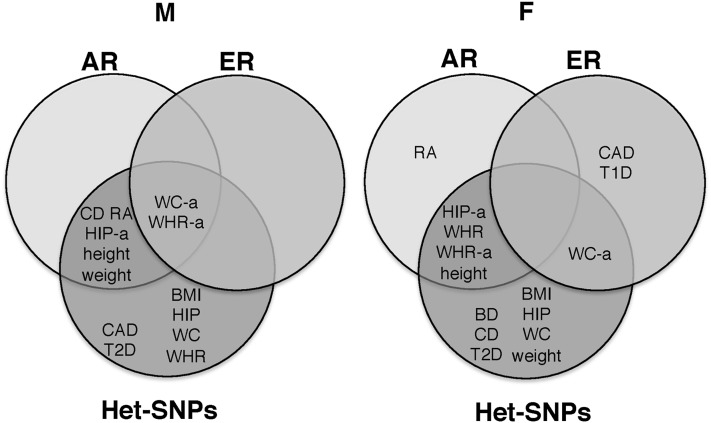
At the biological level, a major “environmental” influence on the genome might be differences in levels of steroid hormones, beginning early in development, and influencing the expression of many genes. The first hypothesis we wanted to assess with regard to steroid-responsive genes (Tang et al. 2004; Jiang et al. 2009) (Tables S2 and S3) was whether these genes contribute disproportionately to association signals compared to matched gene sets [Figure 1 (Method 4a)]. In fact, compared with permuted gene sets matched for gene length (which resulted in matching for SNP number), androgen-responsive genes contributed disproportionately to association signal in CD, RA, hip-a, WC-a, WHR, WHR-a, height, and weight, in at least one sex (Figure 2 and Table S8). Although the set of estrogen-responsive genes is smaller, we identified significant contribution to association signal in CAD, T1D, WC-a, and WHR-a (Figure 2 and Table S8).
Figure 2.
Venn diagram of association enrichment of androgen-responsive (AR) and estrogen-responsive (ER) genes and heterogeneous SNPs (Het-SNPs) in WTCCC diseases and GIANT anthropometric traits. Diseases/traits with significant association enrichment in males (M) are represented in the left diagram, diseases/traits with significant association enrichment in females (F) are represented in the right diagram.
The second putative mechanism would be gene–sex interaction with respect to steroid-responsive genes, such that these sets of genes would show evidence of sex difference in their trait association [Figure 1 (Method 4a)]. We observed sex differences significant by sex permutation for androgen-responsive genes in WC-a (males), WHR (females), WHR-a (females), height (both sexes), and weight (males), in association signal (Table S8). We observed sex differences for estrogen-responsive genes in CAD (females) and T1D (females), and WHR-a (males) in association signal (Table S8).
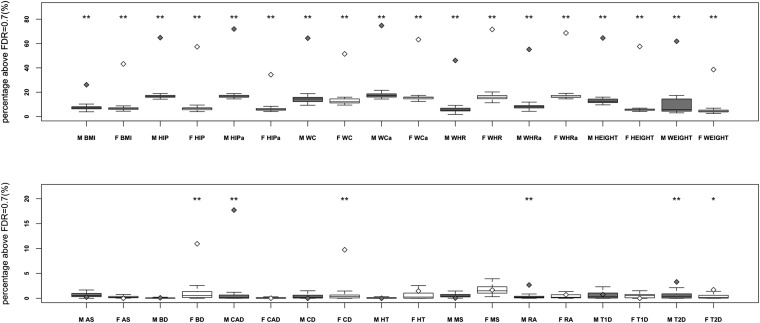
Next, we wondered whether the same SNPs showing sexual dimorphism in anthropometric traits (P < 10−3, Randall et al. 2013) would show excess contribution to common disease and trait genetic signal (compared with SNPs matched on test statistic in the combined sample) [Figure 1 (Method 4a)]. With the exception of AS, MS, HT, and T1D, we observed a disproportionate enrichment of SNPs with sex-heterogeneous effects in the remaining diseases and every trait (Figure 2, Figure 3, and Table S8).
Figure 3.
True enrichment and permutation-based enrichment for heterogeneous markers in WTCCC diseases and GIANT anthropometric traits. Enrichment distributions for 100 permuted males and 100 permuted females are represented with boxplots. Diamonds show the true significant enrichment for males and females. Males are represented in light grey, females in white. ** P < 0.01, *P < 0.05.
We partitioned the heritability to quantify the contribution of all SNPs in AR/ER gene regions (Tables S9 and S10) [Figure 1 (Method 4b)]. In contrast to sex differences in total h2g, partitioned heritability is normalized by total heritability, and therefore will be unaffected by differences in the overall environmental component. An additional 2 kb flank was added to include nearby regulatory elements in the promoter, and these heritability estimates should be interpreted as corresponding to SNPs in or near the genes (Table S11). For the anthropometric traits that are well-powered to model overlapping annotations, we included “baseline” annotations (Finucane et al. 2015) to account for potential background enrichment from overlapping functional categories, such as our selection of genic regions. In this joint model, AR genes account for a significantly increased proportion of heritability in both sexes for height, and for females in BMI. ER genes account for increased proportion of heritability in males in WHR-a and weight.
We observed significant sex differences in heritability for androgen-responsive genes in CAD (females) and RA (females) (Table S9). Genetic correlation in AR genes was significantly <1 between males and females for BMI, hip, WC-a, WHR, WHR-a, height, and weight, suggesting heterogeneity in this functional category (Table S10). Sex differences in heritability for estrogen-responsive genes were significant for WHR-a (females), weight (males), and CAD (females) (Tables S9 and S10). Genetic correlation in ER genes was significantly <1 between males and females in WHR-a but the analysis could not be performed in hip or weight as the estimate for one sex was nonsignificant (Table S10). Despite the tiny size of the sex-heterogeneous SNP set, heritability was significantly different between the sexes after permutation in MS, RA, and T2D (P < 0.05) (Table S9).
We separately analyzed each of the “baseline” annotations—which include major regulatory and evolutionary functional groups—for deviations in genetic correlation. After correcting for the 51 annotations tested, conserved regions (Lindblad-Toh et al. 2011) were the only annotation that remained significant, with genetic correlation <1 for BMI, hip, WC, and WHR. Conserved regions have previously been identified as enriched for heritability across many traits (Finucane et al. 2015), and this depletion in genetic correlation implicates conserved regions in harboring sex-specific effects. The depletion is striking given that these regions harbor 43% of the anthropometric trait h2g on average.
Interpretation of results under the LT model
To better understand the results of the analyses presented above, we consider several LT models that can lead to sex biased prevalence. As we observed little difference in genetic load by sex (Table 1), we evaluated the effects of variance instead of threshold differences. The variance of the liability can have dramatic effects on disease prevalence, which will be larger in the higher variance sex if the mean liabilities and thresholds are equal. To compute the disease prevalence for a given threshold and variance, we computed the area under the normal curve with mean 0, and specified variance that falls beyond the specified threshold. This was achieved using the pnorm function in the R statistical package (Table S12). For rare diseases, even a small increase in the variance of the liability can have a dramatic effect on disease prevalence. Consider a disease similar to MS with prevalence 0.1%. If the liability variance is increased to 1.2 in females vs. 1.0 in males, the disease prevalence will be 2.4 times higher in females.
Gene–sex interactions arising from the autosomes as we observed for MS (or dominance effects on the sex chromosomes, not evaluated here) can result in differences in liability between sexes without having large-scale effects on heritability estimates. We simulated a disease under an LT model with population prevalence 2%. (We used a prevalence of 2% instead of 0.1% for computational efficiency, but the same principles will hold at any prevalence.) We sampled 1000 individuals genotyped at 10 SNPs, which were drawn from a binomial distribution with allele frequencies drawn uniformly between 0.05 and 0.95. Males and females were sampled randomly, and an unobserved environmental factor was drawn for each individual from a normal distribution, with mean 1.0 in males and 2.0 in females, and variance 1.0 in both sexes. The total additive heritability was 20%, and the G × Sex interaction heritability was 30%. In each of 100 simulations, we estimated the heritability in the entire population as well as males and females separately. We also estimated the prevalence of the disease in each sex. As expected (Hill et al. 2008), we observed similar average heritabilities on the liability scale of 35.3, 34.9, and 36.1% in all individuals, females, and males, respectively. Our observed G × Sex heritability in MS was only 6%, suggesting that the absorption of this effect into the sex specific estimates is even more likely to occur.
Discussion
In this study, we set out to perform a survey of the influence of sex on the genetic risk for common, complex heritable disease, and anthropometric traits. Utilizing SNP GWAS results (method A), we assessed sex-specific association enrichment (hypothesis 1), global gene–sex interaction (hypothesis 2), sex differences on the X-chromosome (hypothesis 3), and the influence of steroid-responsive genes and sex-heterogeneous loci associated with secondary sex characteristics (hypothesis 4). We examined polygenic heritability (method B) to substantiate our observations. We found that sex-specific mechanisms were not limited to diseases or traits with notable prevalence or mean differences. In general, we sought to perform a broad survey of potential mechanisms for sexual dimorphism, and to examine patterns of genetic evidence supporting these mechanisms across traits. Because many tests were performed, individual results with respect to a given disease/trait should be interpreted with caution in the absence of further follow-up. However, general principles of sexual dimorphism are substantiated by observations replicated in several complex traits.
When assessing sex differences in genetic load, we did not find global evidence for the predicted threshold difference LT model (hypothesis 1), e.g., the lower-prevalence sex showing higher genetic load. We found an example of association enrichment in the HLA locus in males for male-biased AS. We also found evidence that hypertension shows increased heritability in females despite similar prevalence to males (Biino et al. 2013). Sex specific heritability was previously evaluated in much smaller sample for the anthropometric traits, with inconsistent effects, and the large studies used here resolves this difference (Shungin et al. 2015). We found evidence for sex differences in heritability in several anthropometric traits, although, interestingly, for traits with potentially greater selective impact for females (hip-a, WC-a, and WHR), males have significantly higher heritability, in contrast to some previous findings for fitness traits (Pettay et al. 2005). While the LT is a popular model, alternatives exist, and these may better explain the observed data. Further research should include delineating the impact of sex differences in genetically or environmentally mediated trait or liability variance on heritability estimates, and assessment of the influence of ascertainment, which may differ by sex for case-control traits (Zaitlen et al. 2012a, 2012b; Yang et al. 2014). The extent to which data do, or do not, reflect given sex bias models constrains the set of possible biological mechanisms inducing sex-biased prevalence. Further, clinical interpretations of sex-specific risk should consider these results. For example, counseling relatives of a proband of the lower-prevalence sex that they are at increased risk compared to relatives of a proband of the higher-prevalence sex is based on the threshold difference LT model. In the context of GWAS, alternative analysis strategies could both provide biological insights as well as improve power. Random effects meta-analysis (Han and Eskin 2011) can reveal variants with different effect sizes between sexes, and retrospective likelihood models can reduce power-loss when sex is included as a covariate in ascertained studies (Zaitlen et al. 2012b).
We estimated a substantial G × Sex interaction term in heritability for MS. MS is by far the largest and best-powered study evaluated, and, based on estimated SE, we still cannot rule out the presence of interaction for other diseases. For every anthropometric trait, genetic correlation appeared significantly <1. Although the genetic correlation between sexes was still relatively high for many of these traits, the large anthropometric datasets allowed for powerful tests, confirming that less than complete correlation in genetic contribution to males and females is pervasive (hypothesis 2). Similarly, a study (Rawlik et al. 2016) published while this work was under review found that the genetic correlation between the traits measured in men and women was significantly below 1 (complete correlation) across several quantitative traits including height, BMI, WC, HIP, and WHR, supporting our evidence for G × Sex interaction. There is precedent in both model organism (Nuzhdin et al. 1997; Dilda and Mackay 2002; Leips and Mackay 2002; Mackay and Anholt 2006) and gene expression (Trabzuni et al. 2013; Yao et al. 2014; Kukurba et al. 2016) data for autosomal G × Sex interaction, supporting the plausibility of our results. In addition, consistent with Rawlik et al. (2016), our well-powered study highlighted no significant differences for male and female heritability for BMI and height. However, we observed significant male enrichment for WHR heritability, in contrast with the female enrichment showed by Rawlik et al. (2016). Heritability estimates can differ due to a different amount of genetic variance present in a population, or different amounts of nongenetic variance in a trait. The study designs for GIANT (meta-analysis of 34 studies from different populations) and Rawlik et al. (2016) (UK Biobank) likely lead to the observed differences in heritability estimates. Population-specific and sex-specific properties of environment, such as lifestyle, diet, and smoking status, are important for anthropometric measures, and could contribute to differences between studies, as could populations with different genetic ancestry leading to differing genetic variation.
In order to better understand our lack of evidence for threshold differences and strong evidence for G × Sex interaction, we performed simulations. Our simulated models showed a small difference in estimated heritability, but, in the presence of G × Sex interaction, led to an increase in variance in one sex. We thus demonstrated how a 66.6% increase in disease prevalence can exist between sexes with only a 1.1% difference in additive heritability estimates. Given that the phenotypic variance of many traits is different between the sexes in the GIANT data (Randall et al. 2013; Shungin et al. 2015), difference in variance of disease-related risk factors, as well as differences in mean values of risk factors, may contribute to observed differences in prevalence. In the context of GWAS, a difference in variance between sexes, as opposed to difference in mean, is not captured by a standard fixed effect regression term for sex. Performing sex-stratified analyses, or using a double generalized linear model to account for the difference in variance between sexes, will improve power.
When assessing the most obvious difference in the genome, the X-chromosome, in contrast to previous studies (Pan et al. 2007), we did not find strikingly disproportionate contribution from the X chromosome, although we did observe sex differences in X-chromosome signal, including evidence for female-limited X associations and effect size heterogeneity (hypothesis 3). A previous linkage study concluded there is no role for the X-chromosome in AS (Hoyle et al. 2000), in contrast to our results suggesting association enrichment on the X specific to males. Although blood pressure and cholesterol levels have been associated with X-linked loci, similar evidence for X-chromosome sex differences have not previously been associated with cardiovascular disease (Chang et al. 2014), and a study of stroke specifically suggested a role for hormones, but not sex chromosomes (Manwani et al. 2014; Winham et al. 2015). Additionally, a recent well-powered X-WAS meta-analysis showed no evidence of genome-wide X-chromosome loci contributing to CAD (Loley et al. 2016). Consistently, we did not find any genome-wide associated locus on X chromosome (Figure S1). However, our study was designed to assess global rather than locus-specific genetic contributions to disease, thus we were able to identify significant chromosome-wide heterogeneity across sexes and male association enrichment for CAD on the X chromosome. Likewise, MS has been suggested to have X-linked risk via mouse models and association with X aneuploidy (D’Alessandro et al. 1990; Smith-Bouvier et al. 2008; Seminog et al. 2015), but, to our knowledge, not genome-wide studies before ours (Chang et al. 2014). Additionally, a recent study (Chang et al. 2014) hypothesized sexually dimorphic effect sizes for X-linked genes in autoimmune diseases. Although the latter authors showed individual X-linked genes with sexually dimorphic association, our study did not detect significant chromosome-wide sex heterogeneity for the same disorders, i.e., CD, RA, and T2D. Our power for single-chromosome analyses was extremely limited, and these data were not available for the anthropometric data—our most powerful datasets. For example, we observe a large estimated difference in RA, but it does not reach significance. Thus larger data sets are needed to robustly estimate the contribution of the X-chromosome to prevalence differences through changes in mean and variance of the liability. The X-chromosome may have a different role in traits subject to strong natural selection, such as those directly related to reproduction, in comparison to common diseases with onset primarily after historic reproductive ages (Kosova et al. 2010).
In order to assess potential consequences to genetic risk of sex differences in steroid hormone levels, we assessed the contribution of steroid-responsive genes to each trait. We found a strong contribution of hormone-responsive genes to several diseases and anthropometric traits, with examples of sex-interaction in this contribution (hypothesis 4). Recently, a gene involved in androgen synthesis was associated with RA (Stark et al. 2015), and we show that androgen-responsive genes across the genome are enriched for association signal and sex differences. Although estrogen biology has been associated with comorbidities of T1D (Ryba et al. 2011; Ryba-Stanisławowska et al. 2014; Słomiński et al. 2015), our results are the first to suggest a global impact on risk. Interestingly, we observed several examples of an increased proportion of heritability in AR genes in females, and for ER genes in males. These results could imply that increased levels or variance in androgens in males and estrogens in females reduce the relative impact of genetic variation in genes responsive to these hormones.
Finally, and strikingly, we observed that SNPs showing heterogeneity in association with anthropometric traits make an exceptional contribution to common, complex traits, including those without major prevalence differences. Although there has been a previously noted relationship between hypertension and height, our findings suggest that many observed disease differences by sex are generated by the same mechanisms determining secondary sex characteristics. Our observation is one of association enrichment for anthropometric sex-heterogeneous SNPs, and thus does not directly correspond with sex-heterogeneity in the target disease, but does highlight the pleiotropy of the biology involved in sexual dimorphism. This implies that sex differences in disease prevalence, symptoms, and outcomes may be governed by universal biological pathways, rather than disease-specific pathophysiology or environmental/behavioral risk factors. Further research may clarify whether identification of these putative factors could improve sex-specific diagnosis, treatment, or prevention.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.193623/-/DC1.
Acknowledgments
We thank Bogdan Pasaniuc, Kathryn Tsang, Ileena Mitra, and Jonathan Bravier for helpful discussion and assistance. A full list of the investigators who contributed to the generation of the WTCCC data is available from www.wtccc.org.uk. This study makes use of data generated by the WTCCC. We gratefully acknowledge the datasets made available by the GIANT consortium. This work was supported by a Staglin Family/International Mental Health Research Organization Assistant Professorship (L.A.W.), by K25HL121295 (N.Z., D.S.P.), by National Institutes of Health (NIH) F32 GM106584 (A.G.), and by NIH grant CA08816 (J.A.M.). Funding for the WTCCC project was provided by the Wellcome Trust under award 076113.
Footnotes
Communicating editor: E. R. Hauser
Literature Cited
- Almeida-Filho N., Mari Jde J., Coutinho E., Franca J. F., Fernandes J., et al. , 1997. Brazilian multicentric study of psychiatric morbidity. Methodological features and prevalence estimates. Br. J. Psychiatry 171: 524–529. [DOI] [PubMed] [Google Scholar]
- Arnold A. P., Chen X., 2009. What does the “four core genotypes” mouse model tell us about sex differences in the brain and other tissues? Front. Neuroendocrinol. 30: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austad S. N., Bartke A., 2015. Sex differences in longevity and in responses to anti-aging interventions: a mini-review. Gerontology 62: 40–46. [DOI] [PubMed] [Google Scholar]
- Avery C. L., Freedman B. I., Kraja A. T., Borecki I. B., Miller M. B., et al. , 2006. Genotype-by-sex interaction in the aetiology of type 2 diabetes mellitus: support for sex-specific quantitative trait loci in hypertension genetic epidemiology network participants. Diabetologia 49: 2329–2336. [DOI] [PubMed] [Google Scholar]
- Bearoff F., Case L. K., Krementsov D. N., Wall E. H., Saligrama N., et al. , 2015. Identification of genetic determinants of the sexual dimorphism in CNS autoimmunity. PLoS One 10: e0117993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biino G., Parati G., Concas M. P., Adamo M., Angius A., et al. , 2013. Environmental and genetic contribution to hypertension prevalence: data from an epidemiological survey on Sardinian genetic isolates. PLoS One 8: e59612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D., Gao F., Slavney A., Ma L., Waldman Y. Y., et al. , 2014. Accounting for eXentricities: analysis of the X chromosome in GWAS reveals X-linked genes implicated in autoimmune diseases. PLoS One 9: e113684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.-H., Chen T.-J., Chen Y.-M., Ying-Ming C., Chen D.-Y., 2011. Gender differences in ankylosing spondylitis-associated cumulative healthcare utilization: a population-based cohort study. Clinics (Sao Paulo) 66: 251–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., McClusky R., Chen J., Beaven S. W., Tontonoz P., et al. , 2012. The number of X chromosomes causes sex differences in adiposity in mice. PLoS Genet. 8: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu Y.-F., Chuang L.-M., Kao H.-Y., Shih K.-C., Lin M.-W., et al. 2010. Sex-specific genetic architecture of human fatness in Chinese: the SAPPHIRe Study. Hum. Genet. 128: 501–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox K. H., Bonthuis P. J., Rissman E. F., 2014. Mouse model systems to study sex chromosome genes and behavior: relevance to humans. Front. Neuroendocrinol. 35: 405–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium. Lee S. H., Ripke S., Neale B. M., Faraone S. V., 2013. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat. Genet. 45: 984–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alessandro E., Di Cola M., Lo Re M. L., Ligas C., Vaccarella C., et al. , 1990. Nonrandom chromosome changes in multiple sclerosis. Am. J. Med. Genet. 37: 406–411. [DOI] [PubMed] [Google Scholar]
- de Bakker P. I. W., McVean G., Sabeti P. C., Miretti M. M., Green T., et al. , 2006. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat. Genet. 38: 1166–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro-Catala M., Barrantes-vidal N., Sheinbaum T., Moreno-Fortuny A., Kwapil T. R., et al. , 2015. COMT-by-sex interaction effect on psychosis proneness. BioMed Res. Int. 2015: 829237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diflorio A., Jones I., 2010. Is sex important? Gender differences in bipolar disorder. Int. Rev. Psychiatry 22: 437–452. [DOI] [PubMed] [Google Scholar]
- Dilda C. L., Mackay T. F. C., 2002. The genetic architecture of Drosophila sensory bristle number. Genetics 162: 1655–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis P. J., Affara N. A., 2006. Spermatogenesis and sex chromosome gene content: an evolutionary perspective. Hum. Fertil. 9: 1–7. [DOI] [PubMed] [Google Scholar]
- Emery P., Kavanaugh A., Bao Y., Ganguli A., Mulani P., 2014. Comprehensive disease control (CDC): what does achieving CDC mean for patients with rheumatoid arthritis? Ann. Rheum. Dis. 74: 2165–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D. M., Spencer C. C. A., Pointon J. J., Su Z., Harvey D., et al. 2011. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat. Genet. 43: 761–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finucane H. K., Bulik-Sullivan B., Gusev A., Trynka G., Reshef Y., et al. , 2015. Partitioning heritability by functional category using genome-wide association summary statistics. Nat. Genet. 47: 1228–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale E. A., Gillespie K. M., 2001. Diabetes and gender. Diabetologia 44: 3–15. [DOI] [PubMed] [Google Scholar]
- Gao F., Chang D., Biddanda A., Ma L., Guo Y., et al. , 2014. XWAS: a software toolset for genetic data analysis and association studies of the X chromosome. J. Hered. 106: 666–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks W. P., Abbott J. K., Morrow E. H., 2014. Sex differences in disease genetics: evidence, evolution, and detection. Trends Genet. 30: 453–463. [DOI] [PubMed] [Google Scholar]
- Golan D., Lander E. S., Rosset S., 2014. Measuring missing heritability: inferring the contribution of common variants. Proc. Natl. Acad. Sci. USA 111: E5272–E5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B., Eskin E., 2011. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am. J. Hum. Genet. 88: 586–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haroon N. N., Paterson J. M., Li P., Haroon N., 2014. Increasing proportion of female patients with ankylosing spondylitis: a population-based study of trends in the incidence and prevalence of AS. BMJ Open 4: e006634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayeck T. J., Zaitlen N. A., Loh P. R., Vilhjalmsson B., Pollack S., et al. , 2015. Mixed model with correction for case-control ascertainment increases association power. Am. J. Hum. Genet. 96: 720–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilawe E. H., Yatsuya H., Kawaguchi L., Aoyama A., 2013. Differences by sex in the prevalence of diabetes mellitus, impaired fasting glycaemia and impaired glucose tolerance in sub-Saharan Africa: a systematic review and meta-analysis. Bull. World Health Organ. 91: 671–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W. G., Goddard M. E., Visscher P. M., 2008. Data and theory point to mainly additive genetic variance for complex traits. PLoS Genet. 4: e1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyle E., Laval S. H., Calin A., Wordsworth B. P., Brown M. A., 2000. The X-chromosome and susceptibility to ankylosing spondylitis. Arthritis Rheum. 43: 1353–1355. [DOI] [PubMed] [Google Scholar]
- Jansen R., Batista S., Brooks A. I., Tischfield J. A., Willemsen G., et al. , 2014. Sex differences in the human peripheral blood transcriptome. BMC Genomics 15: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M., Ma Y., Chen C., Fu X., Yang S., et al. , 2009. Androgen-responsive gene database: integrated knowledge on androgen-responsive genes. Mol. Endocrinol. 23: 1927–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosova G., Abney M., Ober C., 2010. Colloquium papers: heritability of reproductive fitness traits in a human population. Proc. Natl. Acad. Sci. USA 107(Suppl. 1): 1772–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krohn J., Speed D., Palme R., Touma C., Mott R., et al. , 2014. Genetic interactions with sex make a relatively small contribution to the heritability of complex traits in mice. PLoS One 9: e96450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukurba K. R., Parsana P., Balliu B., Smith K. S., Zappala Z., et al. , 2016. Impact of the X chromosome and sex on regulatory variation. Genome Res. 26: 768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law S., Li K. K., 2014. Gender-related differences in clinical course of Crohn’s disease in an Asian population: a retrospective cohort review. Arq. Gastroenterol. 51: 90–96. [DOI] [PubMed] [Google Scholar]
- Lee S. H., Wray N. R., Goddard M. E., Visscher P. M., 2011. Estimating missing heritability for disease from genome-wide association studies. Am. J. Hum. Genet. 88: 294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtovaara A., Schielzeth H., Flis I., Friberg U., 2013. Heritability of life span is largely sex limited in Drosophila. Am. Nat. 182: 653–665. [DOI] [PubMed] [Google Scholar]
- Leips J., Mackay T. F. C., 2002. The complex genetic architecture of Drosophila life span. Exp. Aging Res. 28: 361–390. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K., Garber M., Zuk O., Lin M. F., Parker B. J., et al. , 2011. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478: 476–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. Y., Schaub M. A., Sirota M., Butte A. J., 2012a Sex differences in disease risk from reported genome-wide association study findings. Hum. Genet. 131: 353–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. Y., Schaub M. A., Sirota M., Butte A. J., 2012b Transmission distortion in Crohn’s disease risk gene ATG16L1 leads to sex difference in disease association. Inflamm. Bowel Dis. 18: 312–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loisel D. A., Tan Z., Tisler C. J., Evans M. D., Gangnon R. E., et al. , 2011. IFNG genotype and sex interact to influence the risk of childhood asthma. J. Allergy Clin. Immunol. 128: 524–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loley C., Alver M., Assimes T. L., Bjonnes A., Goel A., et al. , 2016. No association of coronary artery disease with X-chromosomal variants in comprehensive international meta-analysis. Sci. Rep. 6: 35278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B. F., Du L., Li J. X., Pan B. Y., Xu J. M., et al. , 2010. Heritability of metabolic syndrome traits among healthy younger adults: a population based study in China. J. Med. Genet. 47: 415–420. [DOI] [PubMed] [Google Scholar]
- Mackay T. F. C., 2009. The genetic architecture of complex behaviors: lessons from Drosophila. Genetica 136: 295–302. [DOI] [PubMed] [Google Scholar]
- Mackay T. F. C., Anholt R. H., 2006. Of flies and man: Drosophila as a model for human complex traits. Annu. Rev. Genomics Hum. Genet. 7: 339–367. [DOI] [PubMed] [Google Scholar]
- Manwani B., Bentivegna K., Benashski S. E., Venna V. R., Xu Y., et al. , 2014. Sex differences in ischemic stroke sensitivity are influenced by gonadal hormones, not by sex chromosome complement. J. Cereb. Blood Flow Metab. 35: 221–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersha T. B., Martin L. J., Biagini Myers J. M., Kovacic M. B., He H., et al. , 2015. Genomic architecture of asthma differs by sex. Genomics 106: 15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchem D. G., Purkey A. M., Grebe N. M., Carey G., Garver-Apgar C. E., et al. , 2014. Estimating the sex-specific effects of genes on facial attractiveness and sexual dimorphism. Behav. Genet. 44: 270–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulugeta E., Wassenaar E., Sleddens-linkels E., Van IJcken W. F., Heard E., et al. , 2016. Genomes of Ellobius species provide insight into the evolutionary dynamics of mammalian sex chromosomes. Genome Res. 26: 1202–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers R. A., Scott N. M., Gauderman W. J., Qiu W., Mathias R. A., et al. , 2014. Genome-wide interaction studies reveal sex-specific asthma risk alleles. Hum. Mol. Genet. 23: 5251–5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negash A., Alem A., Kebede D., Deyessa N., Shibre T., et al. , 2005. Prevalence and clinical characteristics of bipolar I disorder in Butajira, Ethiopia: a community-based study. J. Affect. Disord. 87: 193–201. [DOI] [PubMed] [Google Scholar]
- Ngo S. T., Steyn F. J., McCombe P. A., 2014. Gender differences in autoimmune disease. Front. Neuroendocrinol. 35: 347–369. [DOI] [PubMed] [Google Scholar]
- Nuzhdin S. V., Pasyukova E. G., Dilda C. L., Zeng Z. B., Mackay T. F. C., 1997. Sex-specific quantitative trait loci affecting longevity in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 94: 9734–9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwankwo T., Yoon S. S., Burt V., Gu Q., 2013. Hypertension among adults in the United States: National Health and Nutrition Examination Survey, 2011–2012. NCHS Data Brief (133): 1–8. [PubMed] [Google Scholar]
- Ober C., Loisel D. A., Gilad Y., 2008. Sex-specific genetic architecture of human disease. Nat. Rev. Genet. 9: 911–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco G., Ioannidis J. P. A., Morris A., Zeggini E., 2012. Sex-specific differences in effect size estimates at established complex trait loci. Int. J. Epidemiol. 41: 1376–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakpoor J., Ramagopalan S. V., 2014. Russell W Brain and the aetiology of multiple sclerosis—a historical perspective. QJM 107: 423–427. [DOI] [PubMed] [Google Scholar]
- Pan L., Ober C., Abney M., 2007. Heritability estimation of sex-specific effects on human quantitative traits. Genet. Epidemiol. 31: 338–347. [DOI] [PubMed] [Google Scholar]
- Parks B. W., Sallam T., Mehrabian M., Psychogios N., Hui S. T., et al. , 2015. Genetic architecture of insulin resistance in the mouse. Cell Metab. 21: 334–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettay J. E., Kruuk L. E. B., Jokela J., Lummaa V., 2005. Heritability and genetic constraints of life-history trait evolution in preindustrial humans. Proc. Natl. Acad. Sci. USA 102: 2838–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall J. C., Winkler T. W., Kutalik Z., Berndt S. I., Jackson A. U., et al. , 2013. Sex-stratified genome-wide association studies including 270,000 individuals show sexual dimorphism in genetic loci for anthropometric traits. PLoS Genet. 9: e1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlik K., Canela-xandri O., Tenesa A., 2016. Evidence for sex-specific genetic architectures across a spectrum of human complex traits. Genome Biol. 17: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond-Rakerd L. S., Slutske W. S., Heath A. C., Martin N. G., 2014. Genetic and environmental influences on the ages of drinking and gambling initiation: evidence for distinct aetiologies and sex differences. Addiction 109: 323–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach R. E. J., Venemans A., Cannegieter S. C., Lijfering W. M., 2015. Differential risks in men and women for first and recurrent venous thrombosis: the role of genes and environment: reply. J. Thromb. Haemost. 13: 886–887. [DOI] [PubMed] [Google Scholar]
- Ross M. T., Grafham D. V., Coffey A. J., Scherer S., McLay K., et al. , 2005. The DNA sequence of the human X chromosome. Nature 434: 325–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba M., Malinowska E., Rybarczyk-Kapturska K., Brandt A., Myśliwiec M., et al. , 2011. The association of the IVS1-397T>C estrogen receptor α polymorphism with the regulatory conditions in longstanding type 1 diabetic girls. Mol. Immunol. 49: 324–328. [DOI] [PubMed] [Google Scholar]
- Ryba-Stanisławowska M., Rybarczyk-Kapturska K., Brandt A., Myśliwiec M., Myśliwska J., 2014. IVS1–397T>C estrogen receptor α polymorphism is associated with low-grade systemic inflammatory response in type 1 diabetic girls. Mediators Inflamm. 2014: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawcer S., Hellenthal G., Pirinen M., Spencer C. C. A., Patsopoulos N. A., et al. , 2011. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 476: 214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seminog O. O., Seminog A. B., Yeates D., Goldacre M. J., 2015. Associations between Klinefelter’s syndrome and autoimmune diseases: English national record linkage studies. Autoimmunity 48: 125–128. [DOI] [PubMed] [Google Scholar]
- Sharma K., Gulati M., 2013. Coronary artery disease in women: a 2013 update. Glob. Heart 8: 105–112. [DOI] [PubMed] [Google Scholar]
- She Z.-Y., Yang W.-X., 2014. Molecular mechanisms involved in mammalian primary sex determination. J. Mol. Endocrinol. 53: R21–R37. [DOI] [PubMed] [Google Scholar]
- Shungin D., Winkler T. W., Croteau-Chonka D. C., Ferreira T., Locke A. E., et al. , 2015. New genetic loci link adipose and insulin biology to body fat distribution. Nature 518: 187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silander K., Alanne M., Kristiansson K., Saarela O., Ripatti S., et al. , 2008. Gender differences in genetic risk profiles for cardiovascular disease. PLoS One 3: e3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Słomiński B., Myśliwska J., Brandt A., 2015. Grade of inflammation in boys with type 1 diabetes depends on the IVS1–397T>C estrogen receptor α polymorphism. J. Diabetes Complications 29: 801–807. [DOI] [PubMed] [Google Scholar]
- Smith-Bouvier D. L., Divekar A. A., Sasidhar M., Du S., Tiwari-Woodruff S. K., et al. , 2008. A role for sex chromosome complement in the female bias in autoimmune disease. J. Exp. Med. 205: 1099–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark K., Straub R. H., Rovenský J., Blažičková S., Eiselt G., et al. , 2015. CYB5A polymorphism increases androgens and reduces risk of rheumatoid arthritis in women. Arthritis Res. Ther. 17: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S., Han H., Bajic V. B., 2004. ERGDB: estrogen responsive genes database. Nucleic Acids Res. 32: D533–D536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabzuni D., Ramasamy A., Imran S., Walker R., Smith C., et al. , 2013. Widespread sex differences in gene expression and splicing in the adult human brain. Nat. Commun. 4: 2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukiainen T., Pirinen M., Sarin A.-P., Ladenvall C., Kettunen J., et al. , 2014. Chromosome X-wide association study identifies loci for fasting insulin and height and evidence for incomplete dosage compensation. PLoS Genet. 10: e1004127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink J. M., Bartels M., van Beijsterveldt T. C. E. M., van Dongen J., van Beek J. H. D. A., et al. , 2012. Sex differences in genetic architecture of complex phenotypes? PLoS One 7: e47371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss L. A., Pan L., Abney M., Ober C., 2006. The sex-specific genetic architecture of quantitative traits in humans. Nat. Genet. 38: 218–222. [DOI] [PubMed] [Google Scholar]
- Weissbrod O., Lippert C., Geiger D., Heckerman D., 2015. Accurate liability estimation improves power in ascertained case-control studies. Nat. Methods 12: 332–334. [DOI] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium , 2007. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447: 661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winham S. J., de Andrade M., Miller V. M., 2015. Genetics of cardiovascular disease: importance of sex and ethnicity. Atherosclerosis 241: 219–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Lee S. H., Goddard M. E., Visscher P. M., 2011. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88: 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Loos R. J. F., Powell J. E., Medland S. E., Speliotes E. K., et al. , 2012. FTO genotype is associated with phenotypic variability of body mass index. Nature 490: 267–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Zaitlen N. A., Goddard M. E., Visscher P. M., Price A. L., 2014. Advantages and pitfalls in the application of mixed-model association methods. Nat. Genet. 46: 100–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C., Joehanes R., Johnson A. D., Huan T., Esko T., et al. , 2014. Sex- and age-interacting eQTLs in human complex diseases. Hum. Mol. Genet. 23: 1947–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaitlen N., Pasaniuc B., Patterson N., Pollack S., Voight B., et al. , 2012a Analysis of case-control association studies with known risk variants. Bioinformatics 28: 1729–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaitlen N., Lindström S., Pasaniuc B., Cornelis M., Genovese G., et al. , 2012b Informed conditioning on clinical covariates increases power in case-control association studies. PLoS Genet. 8: e1003032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillikens M. C., Yazdanpanah M., Pardo L. M., Rivadeneira F., Aulchenko Y. S., et al. , 2008. Sex-specific genetic effects influence variation in body composition. Diabetologia 51: 2233–2241. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article and are publicly available.