Abstract
The rickettsial pathogen Anaplasma marginale assembles an actin filament bundle during intracellular infection. Unlike other bacterial pathogens that generate actin filament tails, A. marginale infects mature erythrocytes, and the F-actin appendages are assembled on the cytoplasmic surface of a vacuole containing several organisms. To identify A. marginale molecules associated with these filaments, two complementary approaches were used: matrix-assisted laser desorption ionization-time-of-flight mass spectrometry and tandem mass spectrometry of A. marginale proteins identified with an appendage-specific monoclonal antibody and expression screening of an A. marginale phage library. Amino acid and nucleotide sequences were mapped to a full-length gene in the genome of the St. Maries strain of A. marginale; the correct identification was confirmed by expression of full-length recombinant protein and its reactivity with appendage-specific antibodies. Interestingly, there is marked variation in the abilities of diverse A. marginale strains to assemble the F-actin appendages. Comparison of four strains, the Florida, Illinois, St. Maries, and Virginia strains, revealed substantial polymorphism in the gene encoding the appendage-associated protein, with amino acid sequence identity of as low as 34% among strains. However, this variation does not underlie the differences in expression, as there is no specific polymorphism associated with loss of ability to assemble actin appendages. In contrast, the ability to assemble an actin filament bundle reflected dramatic strain-specific differences in the expression level of the appendage-associated protein. Understanding how this protein influences the cycle of invasion, replication, and egress in the host cell may provide new insights into pathogen-host interactions.
Actin-based motility is important in the invasion and replication of intracellular bacterial pathogens, including Listeria monocytogenes, Rickettsia rickettsii, Rickettsia conorii, and Shigella flexneri (8, 18, 20). Uniquely among these bacterial pathogens, the rickettsia Anaplasma marginale parasitizes mature erythrocytes (21). Within the erythrocyte, A. marginale replicates within a parasitophorous vacuole formed from the invaginated erythrocyte membrane (15). During replication within this vacuole, a structure initially described as a tail and better described as an inclusion appendage forms on the erythrocyte cytoplasmic face of the vacuole membrane (23, 25, 38). Several studies indicated that inclusion appendages contain a parasite-derived component (19, 24, 28). More recently, this inclusion appendage was also shown to contain host actin filaments (F-actin) (41). Thus, unlike the classic pattern in which actin is assembled on the bacterial surface, the A. marginale-associated appendage assembles on the external vacuolar surface. Any parasite-encoded protein associated with the inclusion appendage would have to be secreted across the bacterial membrane and the membrane surrounding the parasitophorous vacuole.
Ultrastructurally, the A. marginale-associated inclusion appendage is composed of highly ordered F-actin bundles that are similar to 1,0 and 1,1 views of hexagonally packed actin filaments present in stable structures such as Limulus sp. sperm and sterocilia of the inner ear (11, 12). This high degree of order reflects regular cross-linking of F-actin into bundles. Thus, the extremely dynamic behavior of ActA and the Arp2/3 complex in actin tail polymerization associated with L. monocytogenes may not be applicable to the A. marginale inclusion appendage (9, 39, 41, 42, 44). However, this highly ordered bundle structure strongly suggests that host F-actin is not the only molecule involved and that additional molecules must be present for cross-linking. While these molecules could be derived from either the host or the pathogen, the presence of the appendage in A. marginale-infected erythrocytes but not in erythrocytes parasitized with other microbial agents suggests an active and specific role of the pathogen rather than a nonspecific cellular response to parasitism.
The idea that a specific A. marginale molecule associates with the cross-linked F-actin bundles was also supported by marked variation in formation of the appendage among A. marginale strains. While most strains examined assemble intraerythrocytic appendages clearly identifiable by light microscopy, strains that do not assemble the F-actin-laden appendage have been isolated (26, 32). Notably, the Florida strain, which does not form appendages, was observed to be unreactive by immunofluorescence microscopy with two monoclonal antibodies (MAbs), AnaO23A5 and AnaO24D5, that bound all strains assembling appendages and to the appendage structure itself (28). As F-actin itself should be available to all pathogen strains, we hypothesized that the difference among strains in F-actin appendage formation is due to the presence or absence of a unique A. marginale appendage-associated protein. In this paper, we report the testing of this hypothesis by identification of the appendage-associated protein and its encoding gene and examination of whether strain-specific appendage formation is attributable to gene loss, polymorphism in the encoded protein, or variation in level of expression.
MATERIALS AND METHODS
Colocalization of anti-A. marginale antibodies to the F-actin appendage.
Thin blood smears from calves infected with the Florida, Illinois, or Virginia strain of A. marginale were prepared as previously described (41). All vertebrate animals were cared for in accordance with a protocol approved by and on file with the Ohio State University Institutional Laboratory Animal Care and Use Committee. F-actin was labeled with phalloidin conjugated to rhodamine (Molecular Probes, Eugene, Oreg.). A. marginale DNA was labeled with 4′,6-diamidino-2-phenylindole (DAPI) (Molecular Probes). The unknown A. marginale appendage-associated protein was bound by using either MAb AnaO23A5 or MAb AnaO24D5 followed by goat anti-mouse immunoglobulin G (IgG) labeled with Alexa 488 (Molecular Probes). AnaO23A5 and AnaO24D5 have been previously reported to bind to inclusion appendages of the North Texas, South Idaho, Virginia, Washington-O, and Washington-C strains of A. marginale (28). Blood smears were incubated at 37°C for 45 min with AnaO23A5 or AnaO24D5 (2 μg/ml) in phosphate-buffered saline (PBS), rinsed twice with PBS, and incubated for 45 min at 37°C with 10 μg of Alexa 488 per ml conjugated to goat anti-mouse IgG. Phalloidin, conjugated to rhodamine, and DAPI were added to the secondary antibody solution for colocalization of F-actin and DNA, respectively. Slides were rinsed three times with PBS and mounted with the Prolong antifade kit (Molecular Probes) as recommended by the manufacturer. Fluorescence from DAPI, Alexa 488, and rhodamine was observed with a Zeiss Aksioskop microscope with filter cubes CZ 902, 41001, and 41002b (Chroma Technology, Brattleboro, Vt.) at the Ohio State University Campus Microscopy and Imaging Facility or with an Olympus BX51 microscope with filter cubes 31000, 11001v2, and 41004 (Chroma Technology). Micrographs were digitally recorded from both microscopes, and fluorescence images were merged by using PhotoShop version 5.0 software.
Identification of the A. marginale appendage-associated protein.
Two approaches were used to identify and verify the identification of the appendage-associated protein. In the first approach, two-dimensional electrophoretic separation of A. marginale proteins was followed by immunoblotting with MAbs AnaO23A5 and AnaO24D5. The identified spot was excised, trypsinized, and subjected to matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS) analysis and capillary liquid chromatography-nanospray tandem MS (MS-MS) on a quadrupole time-of-flight (Q-TOF) mass spectrometer to derive a protein map and specific peptide sequences. In the second approach, the two MAbs were used to screen an A. marginale expression library and identify a clone from which a partial gene sequence was obtained. Detailed methods for both approaches are provided in the following section. In both approaches, the full-length sequence was obtained by a BLAST search using the partial amino acid sequence (approach 1) or the partial nucleotide sequence (approach 2) to search the complete A. marginale St. Maries strain genome sequence (http://www.vetmed.wsu.edu/research_vmp/anagenome/).
Two-dimensional electrophoresis.
Erythrocytes infected with the Illinois strain of A. marginale were lysed, and hemoglobin was removed by washing as previously described (5, 40), with inclusion of protease inhibitors (7.5 mM sodium phosphate, 1 mM sodium EDTA, 20 μg of phenylmethylsulfonyl fluoride per ml, 2 μg of pepstatin A per ml) at all wash steps. For electrophoresis, 250 μg of the sample preparation was lyophilized and redissolved in 185 μl of immobilized pH gradient (IPG) rehydration-sample buffer {8 M urea, 2% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS], 40 mM dithiothreitol [DTT], 0.2% Bio-Lyte 3/10 ampholyte, and 0.0002% bromophenol blue). The mixture was subsequently centrifuged, and the supernatant was applied to either pH 3 to 10 or pH 4 to 7 11-cm IPG gel strips (Bio-Rad, Hercules, Calif.). The IPG strips were overlaid with mineral oil and rehydrated at 20°C for 12 h. The isoelectric focusing step was performed with a Protean isoelectric focusing cell (Bio-Rad) and consisted of electrophoresis at 250 V (linear gradient over 20 min), 8,000 V (linear gradient over 2.5 h), and 8,000 V (fixed for 50,000 V · h), all at 20°C. Afterwards, the IPG strips were reduced at 20°C for 15 min with DTT equilibration buffer (6 M urea, 2% sodium dodecyl sulfate [SDS], 50 mM Tris-HCl, 20% glycerol, 2% DTT) and alkylated at 20°C for 15 min with iodoacetamide equilibration buffer (6 M urea, 2% SDS, 50 mM Tris-HCl, 20% glycerol, 2.5% iodoacetoamide). The second dimension SDS-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out on 8 to 16% gradient polyacrylamide gels at 200 V for 70 min. Gels were either placed in transfer buffer (20 mM Tris, 192 mM glycine, 20% methanol) and stained with SYPRO Orange (Bio-Rad) for protein spot viewing or fixed for 12 h in 50% ethanol and 2% phosphoric acid. Fixed gels were stained with Coomassie blue for 48 h and destained in nanopure water for protein spot visualization before further processing by mass spectrometry (see below). For immunoblotting, SYPRO Orange-stained gels were transferred to 0.45 μm-pore-size porous Hybond-P polyvinylidene difluoride transfer membranes (Amersham Pharmacia) at a fixed current of 100 mA for 12 h. The membranes were rinsed with transfer buffer and then blocked in PBS containing 1% casein and 0.04% Tween 20 for 30 min. Membranes were rinsed twice with PBS-Tween 20 (0.05%) for 5 min each and incubated for 1 h with MAb AnaO23A5 or AnaO24D5 (80 ng/ml) in blocking solution, followed by two more wash steps and incubation for 1 h with sheep anti-mouse IgG (heavy and light chains) conjugated to horseradish peroxidase (1:4,000) (Amersham Pharmacia). The blots were then washed three times, rinsed in water, and detected by using the 3-amino-9-ethylcarbazole substrate as previously described (16).
Mass spectrometry.
Proteins of interest were separated by two-dimensional electrophoresis, stained with Coomassie blue, excised from the gel, and washed in 50% methanol-5% acetic acid for several hours. The gel bands were dried with acetonitrile and reconstituted with DTT solution to reduce the cysteines. Iodoacetamide was added to alkylate the cysteines, and the gels were washed again with acetonitrile and ammonium bicarbonate prior to digestion overnight at room temperature with sequencing-grade trypsin (Promega, Madison, Wis.), using the Montage in-gel digestion kit (Millipore, Billerica, Mass.). The peptides were extracted from the polyacrylamide with 50% acetonitrile and 5% formic acid several times, pooled, and concentrated to 25 μl. MALDI-TOF was performed on a Reflex III (Bruker, Breman, Germany) mass spectrometer operated in the linear, positive-ion mode with an N2 laser. Laser power was used at the threshold level required to generate a signal, and the accelerating voltage was set to 28 kV. The instrument was calibrated with protein standards bracketing the molecular weights of the protein samples (typically mixtures of bradykinin fragment 1 to 5 and adrenocorticotropic hormone fragment 18 to 39 as appropriate). For the matrix, α-cyano-4-hydroxy-cinamic acid was prepared as a saturated solution in 50% acetonitrile-0.1% trifluoroacetic acid in water. Aliquots of 1 μl of matrix and 1 μl of sample were mixed together; 0.5 μl of this was spotted on the target plate and allowed to dry. Resulting values for monoisotopic peaks were used for database searches with the computer program Mascot (33).
Peptide sequences were determined with MS-MS. Capillary liquid chromatography-nanospray tandem mass spectrometry was performed on a hybrid Q-TOF II (Micromass, Wythenshawe, United Kingdom) mass spectrometer equipped with an orthogonal nanospray source from New Objective, Inc. (Woburn, Mass.) operated in the positive-ion mode. The liquid chromatography system was an Alliance 2690 separation module (Waters, Milford, Mass.). Ten microliters of each sample was first injected onto the trapping column, which was washed with 50 mM acetic acid, and the peptides were eluted with acetonitrile onto a BioBasic C18 column (New Objectives) for chromatographic separations. Peptides were eluted directly off the column into the Q-TOF system by using a gradient of 3 to 80% B over 30 min, with a flow rate of 280 μl/min. Mass spectra were recorded by using MassLynx 4.0 with automatic switching functions and were acquired from 300 to 2,000 Da every s with a resolution of 8,000 (full-width half maximum). When the desired peak was detected at a minimum of eight ion counts, the mass spectrometer automatically switched to acquire the collision-induced dissociation MS-MS spectrum of the individual peptide. Collision energy was set depending on charge state recognition properties. Sequence information from the MS-MS data was processed by using the MassLynx 4.0 Biolynx software. Database searches were performed with Mascot and Genomic Solutions, and de novo sequences were analyzed by hand and with Biolynx from Micromass.
Expression library screening.
The library was generated by partial Sau3A1 digestion of A. marginale St. Maries strain genomic DNA, ligation into λZap Express (Stratagene, La Jolla, Calif.), and packaged as previously described (7). The library was screened for positive plaques with a pool of MAbs AnaO23A5 and AnaO24D5 by using the picoBlue immunoscreening kit (Stratagene). Positive plaques were isolated and plaque purified three times. The insert was recovered in the pBK-CMV phagemid by using EXAssist helper phage with Escherichia coli strain XLOLR (Stratagene). The phagemid was rescued in E. coli XL1-Blue MRF′ plated on Luria-Bertani-kanamycin agar plates. Single colonies of E. coli XL1-Blue containing the phagemid were grown in 5 ml of Luria-Bertani medium overnight, and minipreparations were prepared by using the Promega miniprep kit. Inserts were sequenced in both directions with the Big Dye kit and an ABI PRISM automated sequencer (PE-Applied Biosystems), using T7 and T3 primers (Stratagene).
Cloning and characterization of the full-length A. marginale appendage-associated protein.
The sequences obtained from the mass spectrometry and expression library screening approaches were used to identify the full-length open reading frame by search and alignment against the complete A. marginale St. Maries strain genome. To sequence the full-length genes from the Florida, Illinois, and Virginia strains, a forward primer at the 5′ end incorporating the ATG start codon (primer 1, 5′ ATGATTGTGACATATGGCACTGTGG 3′) and a reverse primer at the 3′ end (primer 2, 5′ GGACCCCAAGCATCCAAGAAA 3′) were synthesized and used to prime PCR amplification. PCR cycling conditions were 40 cycles of melting at 95°C for 15 s, annealing at 58°C for 15 s, and extension at 72°C for 1 min, with a final extension at 72°C for 7 min. Amplification products were detected by electrophoresis in 1% agarose gels containing ethidium bromide and sequenced as described above. Sequences were compiled and analyzed by using the VECTOR NTI software package (InforMAX). Protein sequence analyses were performed with the Wisconsin package version 10.3 (Accelrys Inc., San Diego, Calif.) through the Ohio State University College of Biological Sciences Computational Biology Facility. A consensus for sequences derived from the Florida, Illinois, Virginia, and St. Maries strains was estimated with Pileup and Pretty software. The consensus sequence was analyzed with SPScan, Hmmerpfam, Peptidestructure, and Profilescan software, and consensus and individual strain sequences were analyzed with the Coilscan and Transmem programs.
Expression of A. marginale appendage-associated protein.
The level of expression was determined by using quantitative Western blotting (37). For expression of the full-length appendage-associated genes of the Florida, Illinois, and Virginia strains, the amplicons were ligated into pTrcHis TOPO TA (Invitrogen) and used to transform E. coli TOP 10 cells (Invitrogen). Correct orientation of the inserts and the reading frames was confirmed by sequencing. A single clone of each was selected and subsequently used for protein expression. Freshly grown cultures of transformed bacteria were induced with 1 mM isopropyl-β-d-thiogalactoside (IPTG) and harvested after 3 h by centrifugation. Equal organism numbers of the Florida, Illinois, and Virginia strains of A. marginale and equal protein quantities of the recombinant appendage-associated protein of each strain were added to individual wells and separated by SDS-PAGE. Western blotting was done as described above.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the appendage associated protein sequences are as follows: St. Maries strain, AY514450; Florida strain, AY514451; Illinois strain, AY514452; and Virginia strain, AY514453.
RESULTS
Monoclonal antibodies AnaO23A5 and AnaO24D5 colocalize with F-actin to the A. marginale appendage.
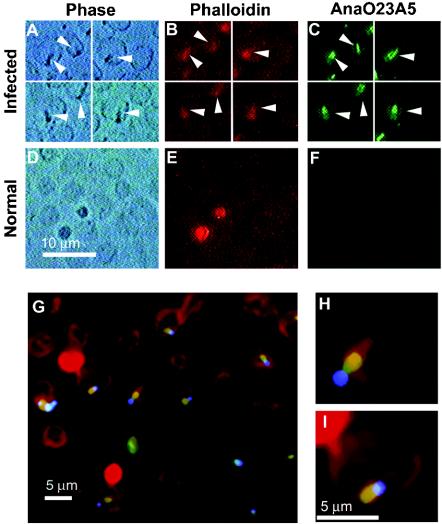
Binding of each MAb colocalized with binding of phalloidin to F-actin and, as a positive control, with DAPI-bound A. marginale DNA within infected erythrocytes (Fig. 1). Phalloidin bound only to the cortical cytoskeleton, and there was no binding of DAPI or either MAb AnaO23A5 or MAb AnaO24D5 within identically treated uninfected erythrocytes. Goat anti-mouse secondary antibody alone did not colocalize with phalloidin or DAPI within infected erythrocytes. The appearance of the appendages resembled the “comets, rings and dumbbells” previously described (14). There was no difference in this appearance when the MAbs and phalloidin were used separately or in combination (data not shown) compared to the triple-labeling experiments (Fig. 1).
FIG. 1.
Colocalization of the A. marginale protein bound by MAb AnaO23A5 with F-actin in the intraerythrocytic appendage. (A) Phase-contrast microscopy was used to locate intraerythrocytic A. marginale and the associated inclusion appendage; (B) rhodamine-phalloidin was used to label F-actin; and (C) goat-anti-mouse IgG conjugated to Alexa 488 was used to detect MAb AnaO23A5 bound to the A. marginale protein. Micrographs in the same row are images of the same field, and arrowheads designate the appendages. Detection using MAb AnaO24D5 gave the same result. (D to F) Uninfected erythrocytes were tested identically to negative controls. The bright circular entities external to erythrocytes in panel E are platelets. (G to I) MAb AnaO23A5 binding (detected with Alexa 488, green) colocalizes (yellow-orange) with F-actin (labeled with rhodamine-phalloidin, red) to the appendage adjacent to the intraerythrocytic vacuole containing A. marginale (DNA labeled with DAPI, blue).
Identification of the A. marginale appendage-associated protein.
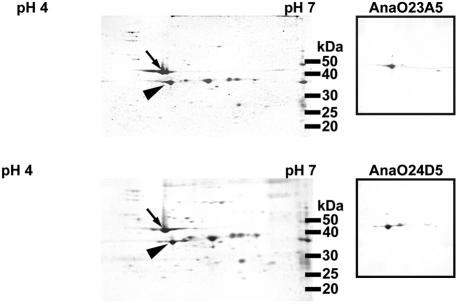
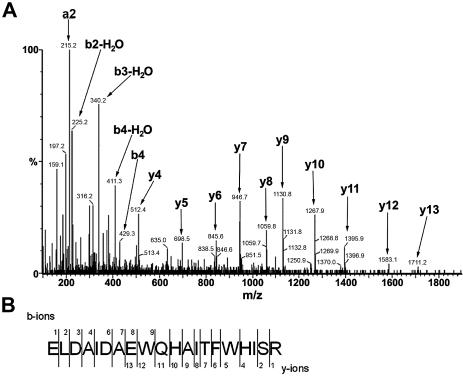
Two-dimensional gel electrophoresis followed by immunoblotting with MAbs AnaO23A5 and AnaO24D5 was used to target a protein for analysis by mass spectrometry. The separation of the Illinois strain proteins by two-dimensional electrophoresis (Fig. 2) resembled that previously reported for the 35S-labeled proteins of the Florida and Washington-O strains (3). Both MAb AnaO23A5 and MAb AnaO24D5 bound to a protein with an approximate molecular size of 39 kDa (Fig. 2, right panels). There was no binding of either MAb AnaO23A5 or MAb AnaO24D5 when uninfected erythrocytes were analyzed under identical conditions (data not shown). Protein spots were excised, digested with trypsin, and subjected to MALDI-TOF and MS-MS analysis. The de novo amino acid sequence of precursor ion 813.43+, ELDAIDAEWQHAITFWHISR, was determined by MS-MS (Fig. 3; Table 1) and used to identify a putative open reading frame in the A. marginalegenome sequence (http://www.vetmed.wsu.edu/research_vmp/anagenome/). The observed m/z values of four additional peptides matched the same open reading frame (Fig. 4) with one peptide representing a repeated sequence. To confirm that the correct open reading frame was identified, a phagemid insert identified by MAb AnaO23A5-AnaO24D5 screening of a Sau3A1 A. marginale expression library was sequenced. The 483-bp insert carried a partial open reading frame of 167 amino acids. This partial open reading frame included three peptides identified by MALDI-TOF, including the peptide repeat described below (Fig. 4), and was contained within the complete open reading frame identified in the genome. The complete gene as identified in the genome sequence of the St. Maries strain of A. marginale is 1,275 bp and encodes a protein of 419 amino acids with a predicted molecular size of 46,106 Da.
FIG. 2.
Two-dimensional gel electrophoresis and detection of the A. marginale appendage-associated protein with MAbs AnaO23A5 and AnaO24D5. Sypro Orange-stained two-dimensional gels of two different preparations of A. marginale infected erythrocytes are shown. The arrows and arrowheads designate the positions of actin and the A. marginale appendage-associated protein, respectively. Proteins from each gel were transferred to a polyvinylidene difluoride membrane, and the position of the protein was confirmed by using MAbs AnaO23A5 and AnaO24D5. This protein was excised for analysis by mass spectrometry.
FIG. 3.
MS-MS spectrum of a tryptic peptide from AAAP. (A) Collision-induced dissociation product ion mass spectrum of a triply charged tryptic peptide ion from the protein spot bound by MAbs AnaO23A5 and AnaO24D5. The arrows indicate the commonly observed theoretical daughter ions for different residues (Table 1). (B) The nomenclature for the peptide ion fragments is as proposed by Roepstorff and Fohlman (35), where N- and C-terminal peptide fragment ions are classed as a, b, and c or x, y, and z, respectively, based on where the peptide bonds are cleaved.
TABLE 1.
Commonly observed theoretical daughter ions for a de novo peptide sequence mapped to the A. marginale appendage-associated protein
| Residue | Daughter ion (m/z)a
|
|||||
|---|---|---|---|---|---|---|
| N terminalb
|
C terminalc
|
|||||
| No.d | a | b | No.d | y | y − H2Oe | |
| E | 1 | 102.6 | 130.1 | 20 | NAf | NA |
| L | 2 | 215.1 | 243.1 | 19 | 2309.1 | 2291.1 |
| D | 3 | 330.2 | 358.2 | 18 | 2196.1 | 2178.0 |
| A | 4 | 401.2 | 429.2 | 17 | 2081.0 | 2063.0 |
| I | 5 | 514.3 | 542.3 | 16 | 2010.0 | 1992.0 |
| D | 6 | 629.3 | 657.3 | 15 | 1896.9 | 1878.9 |
| A | 7 | 700.4 | 728.4 | 14 | 1781.9 | 1763.9 |
| E | 8 | 829.4 | 857.4 | 13 | 1710.9 | 1692.8 |
| W | 9 | 1015.5 | 1043.5 | 12 | 1581.8 | 1563.8 |
| Q | 10 | 1143.5 | 1171.5 | 11 | 1395.7 | 1377.2 |
| H | 11 | 1280.6 | 1308.6 | 10 | 1267.7 | 1249.7 |
| A | 12 | 1351.6 | 1379.6 | 9 | 1130.6 | 1112.6 |
| I | 13 | 1464.7 | 1492.7 | 8 | 1059.6 | 1041.6 |
| T | 14 | 1565.8 | 1593.8 | 7 | 946.5 | 928.5 |
| F | 15 | 1712.8 | 1740.8 | 6 | 845.4 | 827.4 |
| W | 16 | 1899.0 | 1926.9 | 5 | 698.4 | 680.4 |
| H | 17 | 2054.0 | 2064.0 | 4 | 512.3 | 494.3 |
| I | 18 | 2149.0 | 2177.0 | 3 | 357.2 | 357.2 |
| S | 19 | 2236.1 | 2264.0 | 2 | 262.2 | 244.1 |
| R | 20 | NA | NA | 1 | 175.1 | 157.1 |
A peptide carrying a positively charged fragment in its backbone primarily generates a, b, and y ions. Boldface indicates observed ions with good intensity (see Fig. 3).
Ions where the charge is retained on the N-terminal fragment of the peptide.
Ions where the charge is retained on the C-terminal fragment.
Numbering indicates which peptide bond is cleaved, counting from the N or C terminus, respectively.
Peaks seen for ions which have lost water (−18 Da).
NA, not applicable.
FIG. 4.
Open reading frame encoding the A. marginale appendage-associated protein in the St. Maries strain. The underlined boldface sequences were derived from the de novo sequence determined by MS-MS, and the boldface sequences indicate peptides mapped by MALDI-TOF following protein identification. The shaded sequences were derived from a clone identified by MAbs AnaO23A5 and AnaO24D5 by screening a Sau3A1 expression library. The complete open reading frame was identified by using the partial sequences (amino acid and nucleotide) to search the complete A. marginale genome.
Strain-specific differential expression of the appendage-associated protein.
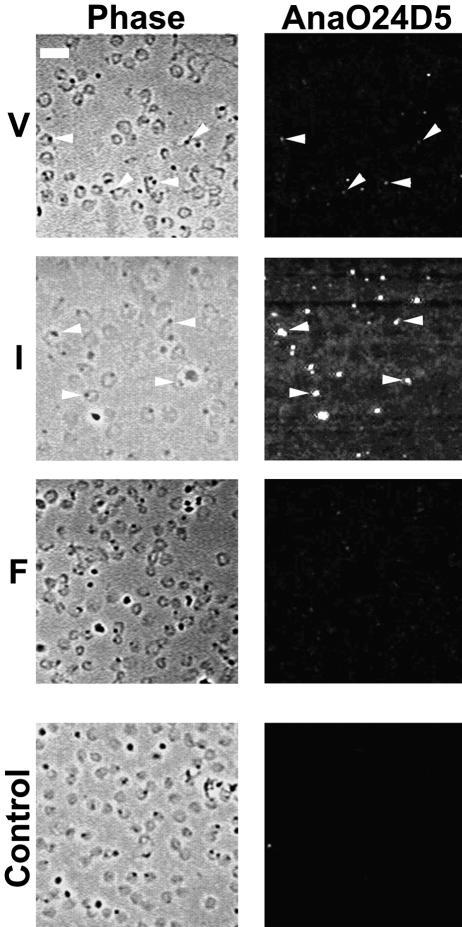
MAbs AnaO23A5 and AnaO24D5 were previously reported to bind the North Texas, South Idaho, St. Maries, Virginia, Washington-C, and Washington-O strains of A. marginale but not the Florida strain (28, 45). Importantly, the Florida strain does not assemble an appendage within infected erythrocytes (1, 13, 28, 31, 34, 45). To confirm this previous observation and to test whether there are differences in expression among appendage-bearing strains, binding was assessed by fluorescence microscopy. Infected erythrocytes were demonstrated by phase-contrast microscopy, and the appendage-associated protein was localized in situ in the same field (Fig. 5). Both the Illinois and Virginia strains bound each MAb, while binding of either MAb to the Florida strain was very weak (Fig. 5). Comparison of the Illinois and Virginia strains revealed consistently more intense immunofluorescence with the Illinois strain when either MAb was used. This apparently greater binding occurred despite a lower percentage of infected erythrocytes in the Illinois strain blood smears (19%) than in the Virginia strain smears (24%).
FIG. 5.
Genetically distinct A. marginale strains differ in reactivity with a MAb specific for the appendage-associated protein. The Virginia (V), Illinois (I), and Florida (F) strains were examined by phase-contrast (Phase) and fluorescence (AnaO24D5) microscopy of the same field labeled with MAb AnaO24D5. Replicates with MAb AnaO23A5 gave the same result (not shown). Omission of the MAb (control) or use of an unrelated MAb was used as a negative control for nonspecific immunofluorescence. The white bar in the upper left corner of panel V represents 10 μm.
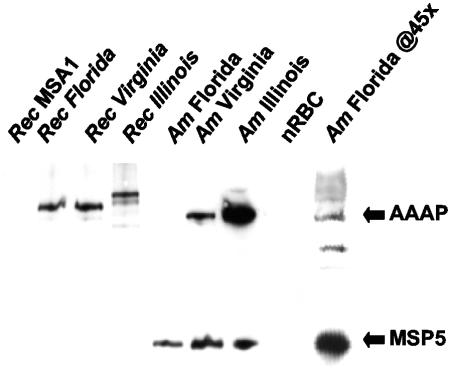
The apparent low level of expression detected by immunofluorescence in the Florida strain could have been influenced by the lack of a tail appendage, because antigen concentrated in a local region is much more readily detected by using this approach. To directly assess the level of expression of this polypeptide in the Illinois, Virginia, and Florida strains, equal numbers of organisms were separated by SDS-PAGE and immunoblotted with MAb AnaO23A5. The equivalent loading of each strain was confirmed by reactivity with MAb ANAF16C1, which binds a constitutively expressed protein (MSP5) that is highly conserved in all A. marginale strains examined (37, 43). Using equal numbers of organisms, clearly the greatest level of expression was detected in the Illinois strain (Fig. 6), which was consistent with the immunofluorescence results. No expression could be detected in the Florida strain until the number of organisms was increased 10-fold (not shown), and clearly detectable binding required a 45-fold increase (Fig. 6). There was no binding to uninfected erythrocytes. To control for the differences in MAb binding reflecting strain-specific sequence polymorphism rather than strain-specific expression, the open reading frame encoding the appendage-associated protein in each strain was cloned and expressed as a recombinant His-tagged fusion protein, and equal amounts were tested for MAb binding. The recombinant proteins from the Florida, Virginia, and Illinois strains were bound by MAb AnaO24D5 with no apparent differences (Fig. 6). There was no binding to recombinant Babesia bovis MSA1, an unrelated protein expressed in the same vector system. The SDS-PAGE and immunoblotting were repeated three times with identical results.
FIG. 6.
Genetically distinct A. marginale strains differ in level of expression of the appendage-associated protein. Equal organism numbers of the Florida (Am Florida), Virginia (Am Virginia), and Illinois (Am Illinois) strains and equal protein amounts of the recombinant fusion protein of each strain (Rec Florida, Rec Virginia, and Rec Illinois) were separated by SDS-PAGE and reacted in Western blots with MAb AnaO23A5 (to detect the A. marginale appendage-associated protein [AAAP]) and ANAF16C1 (to confirm equal numbers of organisms among strains by detection of major surface protein 5 [MSP5]). Uninfected erythrocytes (nRBC) were used as a negative antigen control for the A. marginale strains, and recombinant B. bovis MSA1 (Rec MSA1) was used as a negative control for the recombinant A. marginale appendage-associated proteins. A 45-fold increase in the number of A. marginale organisms loaded was used to show expression by the Florida strain (Am Florida @45×).
Primary sequence polymorphism and conserved secondary structure in the appendage-associated protein among strains.
The genes encoding the full-length appendage-associated proteins in the Florida, Illinois, and Virginia strains were sequenced and aligned along with the St. Maries strain sequences derived from the genome (Fig. 7). The encoded proteins differed in length (Florida, 324 amino acids [34,874 Da]; Illinois, 364 amino acids [39,798 Da]; St. Maries, 419 amino acids [46,106 Da]; and Virginia, 320 amino acids [34,576 Da]). Alignment of the amino acid sequences revealed the presence of multiple imperfect peptide repeats centered around the sequence ELKAIDA; seven repeats occur in the Florida, Illinois, and Virginia strains, while nine repeats are present in the St. Maries strain. Aside from the two extra repeats in the St. Maries strain, size polymorphism among strains reflected insertions and deletions in regions between the imperfect repeats (Fig. 7). Together with numerous individual amino acid substitutions, the insertions and deletions resulted in identities of 34 to 98% between strain pairs (Florida-Illinois, 63%; Florida-Virginia, 98%; Florida-St. Maries, 49%; Illinois-Virginia, 62%; Illinois-St. Maries, 34%; and Virginia-St. Maries 48%).
FIG. 7.
The appendage-associated protein is polymorphic among A. marginale strains. The amino acid sequences for the full-length protein in the Florida (FL), Illinois (IL), St. Maries (StM), and Virginia (VA) strains are aligned. The numbers designate amino acid residue numbers. Block shading indicates amino acids that are identical in all four strains, and hyphens indicate deletions in a strain relative to one or more other strains.
Sequence analyses were performed to identify predicted motifs and secondary structures conserved among the protein sequences from the different A. marginale strains. Conserved secretory signal peptide sequences were not found, and none of the motifs listed in the Prosite dictionary were found on the consensus sequence. Predicted secondary structures of the consensus sequence indicated that the N- and C-terminal ends consist of numerous turns and that the internal region has five alpha helices. Each ELKAIDA-like repeat was associated with a helix. A coiled-coil segment was predicted for each strain sequence, at residues 221 to 256, 141 to 174, 228 to 267, and 218 to 253 for the Florida, Illinois, St. Maries, and Virginia strains, respectively. A transmembrane helix was predicted downstream of the coiled coil segment, at residues 265 to 283, 324 to 342, 335 to 357, and 262 to 280 for the Florida, Illinois, St. Maries, and Virginia strain sequences, respectively. This software also predicted internal N and external C termini for each sequence except that of the St. Maries strain, where the N and C termini were external and internal, respectively. Biochemical and structural studies of the purified protein are required to provide data to support or refute these theoretical predictions.
DISCUSSION
The colocalization of both MAbs, AnaO23A5 and AnaO24D5, with phalloidin binding to the A. marginale-associated appendage confirmed two previously separate observations: specific recognition of the appendage by these MAbs (28, 32) and phalloidin binding to F-actin in the appendage (41). The lack of reactivity of MAbs AnaO23A5 and AnaO24D5 with the Florida strain of A. marginale (28) (Fig. 5), which also fails to assemble the intraerythrocytic appendage, suggested an important role for the protein recognized by these MAbs. Several observations indicate that we have identified this protein, now designated the A. marginale appendage-associated protein (AAAP): (i) mass spectrometric analysis of proteins excised from four different two-dimensional gels identified the same protein, based on MS-MS determination of a peptide sequence and MALDI-TOF determination of mass/charge ratios for individual tryptic peptides; (ii) the DNA sequence of a clone expressing a recombinant protein bound by both MAbs encoded a 167-amino-acid fragment that overlapped the peptides identified by mass spectrometry and mapped to the same open reading frame in the complete genome sequence; and (iii) the full-length open reading frame that was identified expressed a recombinant fusion protein that was bound by both MAbs. Interestingly, neither the protein nor its encoding gene was found to have closely related homologs in other bacteria or in eukaryotic cells (no BLASTN or BLASTP scores of <10−5).
Previous findings suggested that expression of the appendage-associated protein was all or none, a pattern typified by the lack of MAb binding to the Florida strain and strong reactivity with multiple genetically distinct strains (28, 32). However, the immunofluorescence microscopy of the Illinois strain along with the Florida and Virginia strains (Fig. 5) revealed a gradient of expression from high to minimal for the Illinois, Virginia, and Florida strains, respectively. This microscopically observed pattern within infected erythrocytes was confirmed by quantitative immunoblotting. Unexpectedly, the data showed that, in addition to differences in the level of expression between the Illinois and Virginia strains, the Florida strain also expresses the appendage-associated protein at a low level. Equal reactivity of the MAbs with the individual recombinant proteins of each strain indicated that this observation was due to actual differences in the level of protein expression rather than diminished reactivity of the Florida strain protein with the MAbs (Fig. 6).
We had hypothesized that the difference in appendage formation among strains is due to the presence or absence of a unique A. marginale appendage-associated protein. This hypothesis, as stated, is rejected, as the encoding gene is present and protein is expressed in the Florida strain. The dramatically lower level of protein expression in the Florida strain is an obvious alternative explanation. A second possible alternative is polymorphism in the appendage-associated protein among strains. The high degree of polymorphism was unexpected, as most A. marginale proteins, with the exception of outer membrane protein families under immune selective pressure, are highly conserved among strains (2, 4, 6, 7, 10, 17, 27, 29, 30, 36, 37, 43). Notably, the greatest differences in the amino acid sequence are between the Illinois and St. Maries strains (34% identity), both of which assemble F-actin-containing appendages during intraerythrocytic infection. Thus, secretion of the appendage-associated protein into the cytoplasmic compartment and its association with the F-actin filaments are maintained despite relatively marked polymorphism. In contrast, there is 98% identity between the Florida and Virginia strains. Only one of the five amino acid differences between the Florida and Virginia strains was unique to the Florida strain (G at position 258 in Florida, D in Illinois and Virginia, and S in St. Maries). Based on the marked polymorphism in the protein among appendage-bearing strains, it seems unlikely that this single unique amino acid change is responsible for the inability of the Florida strain to assemble an appendage. The smaller amount of expressed protein in the Florida strain appears to provide an explanation for the lack of the tail appendage. Yet, the need for additional, currently unidentified A. marginale proteins in the assembly or stabilization of the appendage cannot be excluded.
In conclusion, we report the identification of a novel pathogen-derived protein associated with the F-actin appendages that are assembled during A. marginale infection of the host erythrocyte. Further, strain-specific differences in protein expression correlate with appendage assembly. The lack of homologs in other well-characterized bacteria that develop actin tails during intracellular infection is consistent with structural observations that the actin filament organization in A. marginale-infected erythrocytes differs from those in L. monocytogenes-, R. rickettsii-, and S. flexneri-infected cells. It is also noteworthy that no significant evidence of a homolog to R. conorii RickA, a rickettsial protein recently reported to nucleate actin polymerization (20), was found within the A. marginale St. Maries strain genome (no BLASTN or BLASTP scores of <10−5). Understanding how A. marginale assembles the actin appendages and how AAAP might influence the cycle of invasion, replication, and egress in the host cell may provide new insights into pathogen-erythrocyte interactions. Finally, because appendages are associated with A. marginale in the tick blood meal and after movement to the tick midgut epithelium (22), investigation into a possible role for AAAP in infection of the tick host is warranted.
Acknowledgments
This work was supported by NIH grants AI44005 and AI47932, USDA-ARS Cooperative Agreement 58-5348-3-212, The Ohio State University Office of Research Seed Grant Program, and The Ohio State University College of Veterinary Medicine Office of Research.
We thank Larry Capitini and David Anderson for splenectomy of experimental calves and Brian Kemmenoe of the Ohio State University Campus Microscopy and Imaging Facility. The excellent technical assistance of Jacqueline Farst, Debra Grover, Peter Hetrick, and Beverly Hunter is greatly appreciated.
Editor: J. F. Urban, Jr.
REFERENCES
- 1.Adams, J. H., R. D. Smith, and M. S. Kuhlenschmidt. 1986. Identification of antigens of two isolates of Anaplasma marginale, using a Western blot technique. Am. J. Vet. Res. 47:501-506. [PubMed] [Google Scholar]
- 2.Allred, D. R., T. C. McGuire, G. H. Palmer, S. R. Leib, T. M. Harkins, T. F. McElwain, and A. F. Barbet. 1990. Molecular basis for surface antigen size polymorphisms and conservation of a neutralization-sensitive epitope in Anaplasma marginale. Proc. Natl. Acad. Sci. USA 87:3220-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbet, A. F., L. W. Anderson, G. H. Palmer, and T. C. McGuire. 1983. Comparison of proteins synthesized by two different isolates of Anaplasma marginale. Infect. Immun. 40:1068-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbet, A. F., A. Lundgren, J. Yi, F. R. Rurangirwa, and G. H. Palmer. 2000. Antigenic variation of Anaplasma marginale by expression of MSP2 mosaics. Infect. Immun. 68:6133-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennet, V. 1983. Proteins involved in membrane-cytoskeleton association in human erythrocytes: spectrin, ankyrin and band 3. Methods Enzymol. 96:313-324. [DOI] [PubMed] [Google Scholar]
- 6.Brayton, K. A., D. P. Knowles, T. C. McGuire, and G. H. Palmer. 2001. Efficient use of a small genome to generate antigenic diversity in tick-borne ehrlichial pathogens. Proc. Natl. Acad. Sci. USA 98:4130-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brayton, K. A., G. H. Palmer, A. Lundgren, J. Yi, and A. F. Barbet. 2002. Antigenic variation of Anaplasma marginale msp2 occurs by combinatorial gene conversion. Mol. Microbiol. 43:1151-1159. [DOI] [PubMed] [Google Scholar]
- 8.Cossart, P. 1995. Actin-based bacterial motility. Curr. Opin. Cell Biol. 7:94-101. [DOI] [PubMed] [Google Scholar]
- 9.Cossart, P. 2000. Actin-based motility of pathogens: the Arp2/3 complex is a central player. Cell. Microbiol. 2:195-205. [DOI] [PubMed] [Google Scholar]
- 10.de la Fuente, J., R. A. Van Den Bussche, J. C. Garcia-Garcia, S. D. Rodriguez, M. A. Garcia, A. A. Guglielmone, A. J. Mangold, L. M. Friche Passos, M. F. Barbosa Ribeiro, E. F. Blouin, and K. M. Kocan. 2002. Phylogeography of New World isolates of Anaplasma marginale based on major surface protein sequences. Vet. Microbiol. 88:275-285. [DOI] [PubMed] [Google Scholar]
- 11.DeRosier, D. J., and L. G. Tilney. 1982. How actin filaments pack into bundles. Cold Spring Harbor Symp. Quant. Biol. 46:525-540. [DOI] [PubMed] [Google Scholar]
- 12.DeRosier, D. J., L. G. Tilney, E. M. Bonder, and P. Frankl. 1982. A change in twist of actin provides the force for the extension of the acrosomal process in Limulus sperm: the false-discharge reaction. J. Cell Biol. 93:324-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eriks, I. S., D. Stiller, W. L. Goff, M. Panton, S. M. Parish, T. F. McElwain, and G. H. Palmer. 1994. Molecular and biological characterization of a newly isolated Anaplasma marginale strain. J. Vet. Diagn. Investig. 6:435-441. [DOI] [PubMed] [Google Scholar]
- 14.Espana, C., E. M. Espana, and D. Gonzalez. 1959. Anaplasma marginale. I. Studies with phase contrast and electron microscopy. Am. J. Vet. Res. 20:795-805. [PubMed] [Google Scholar]
- 15.Francis, D. H., D. A. Kinden, and G. M. Buening. 1979. Characterization of the inclusion limiting membrane of Anaplasma marginale by immunoferritin labeling. Am. J. Vet. Res. 40:777-782. [PubMed] [Google Scholar]
- 16.French, D. M., W. C. Brown, and G. H. Palmer. 1999. Emergence of Anaplasma marginale antigenic variants during persistent rickettsemia. Infect. Immun. 67:5834-5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.French, D. M., T. F. McElwain, T. C. McGuire, and G. H. Palmer. 1998. Expression of Anaplasma marginale major surface protein 2 variants during persistent cyclic rickettsemia. Infect. Immun. 66:1200-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldberg, M. B. 2001. Actin-based motility of intracellular microbial pathogens. Microbiol. Mol. Biol. Rev. 65:595-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goncalves Ruiz, P. M., L. M. Passos, M. S. Martins, J. H. Patarroyo, and M. F. Ribeiro. 2002. Antigenic characterization of morphologically distinct Anaplasma marginale isolates using a panel of monoclonal antibodies. Vet. Parasitol. 107:169-177. [DOI] [PubMed] [Google Scholar]
- 20.Gouin, E., C. Egile, P. Dehoux, V. Villiers, J. Adams, F. Gertler, R. Li, and P. Cossart. 2004. The RickA protein of Rickettsia conorii activates the Arp2/3 complex. Nature 427:457-461. [DOI] [PubMed] [Google Scholar]
- 21.Kessler, R. H., and M. Ristic. 1979. In vitro cultivation of Anaplasma marginale: invasion of and development in noninfected erythrocytes. Am. J. Vet. Res. 40:1774-1776. [PubMed] [Google Scholar]
- 22.Kocan, K. M., S. A. Ewing, J. A. Hair, and S. J. Barron. 1984. Demonstration of the inclusion appendage of Anaplasma marginale in nymphal Dermacentor andersoni. Am. J. Vet. Res. 45:1800-1807. [PubMed] [Google Scholar]
- 23.Kocan, K. M., J. H. Venable, and W. E. Brock. 1978. Ultrastructure of anaplasmal inclusions (Pawhuska isolate) and their appendages in intact and hemolyzed erythrocytes and in complement-fixation antigen. Am. J. Vet. Res. 39:1123-1130. [PubMed] [Google Scholar]
- 24.Kocan, K. M., J. H. Venable, K. C. Hsu, and W. E. Brock. 1978. Ultrastructural localization of anaplasmal antigens (Pawhuska isolate) with ferritin-conjugated antibody. Am. J. Vet. Res. 39:1131-1135. [PubMed] [Google Scholar]
- 25.Koidzumi, M. 1912. On the nature of “marginal points” occurring in the blood corpuscles of cattle. Zentbl. Bakteriol. Mikrobiol. Hyg. 65:337-340. [Google Scholar]
- 26.Kreier, J. P., and M. Ristic. 1963. Anaplasmosis. X. Morphologic characteristics of the parasites present in the blood of calves infected with the Oregon strain of Anaplasma marginale. Am. J. Vet. Res. 24:676-687. [PubMed] [Google Scholar]
- 27.Lohr, C. V., K. A. Brayton, V. Shkap, T. Molad, A. F. Barbet, W. C. Brown, and G. H. Palmer. 2002. Expression of Anaplasma marginale major surface protein 2 operon-associated proteins during mammalian and arthropod infection. Infect. Immun. 70:6005-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGuire, T. C., G. H. Palmer, W. L. Goff, M. I. Johnson, and W. C. Davis. 1984. Common and isolate-restricted antigens of Anaplasma marginale detected with monoclonal antibodies. Infect. Immun. 45:697-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meeus, P. F., K. A. Brayton, G. H. Palmer, and A. F. Barbet. 2003. Conservation of a gene conversion mechanism in two distantly related paralogues of Anaplasma marginale. Mol. Microbiol. 47:633-643. [DOI] [PubMed] [Google Scholar]
- 30.Oberle, S. M., G. H. Palmer, and A. F. Barbet. 1993. Expression and immune recognition of the conserved MSP4 outer membrane protein of Anaplasma marginale. Infect. Immun. 61:5245-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palmer, G. H., A. F. Barbet, A. J. Musoke, J. M. Katende, F. Rurangirwa, V. Shkap, E. Pipano, W. C. Davis, and T. C. McGuire. 1988. Recognition of conserved surface protein epitopes on Anaplasma centrale and Anaplasma marginale isolates from Israel, Kenya and the United States. Int. J. Parasitol. 18:33-38. [DOI] [PubMed] [Google Scholar]
- 32.Palmer, G. H., S. D. Waghela, A. F. Barbet, W. C. Davis, and T. C. McGuire. 1987. Characterization of a neutralization-sensitive epitope on the Am 105 surface protein of Anaplasma marginale. Int. J. Parasitol. 17:1279-1285. [DOI] [PubMed] [Google Scholar]
- 33.Perkins, D. N., D. J. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551-3567. [DOI] [PubMed] [Google Scholar]
- 34.Ristic, M. 1960. Anaplasmosis. Adv. Vet. Sci. 6:111-192. [Google Scholar]
- 35.Roepstorff, P., and J. Fohlman. 1984. Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomed. Mass Spectrom. 11:601. [DOI] [PubMed] [Google Scholar]
- 36.Rurangirwa, F. R., K. A. Brayton, T. C. McGuire, D. P. Knowles, and G. H. Palmer. 2002. Conservation of the unique rickettsial rRNA gene arrangement in Anaplasma. Int. J. Syst. Evol. Microbiol. 52:1405-1409. [DOI] [PubMed] [Google Scholar]
- 37.Rurangirwa, F. R., D. Stiller, D. M. French, and G. H. Palmer. 1999. Restriction of major surface protein 2 (MSP2) variants during tick transmission of the ehrlichia Anaplasma marginale. Proc. Natl. Acad. Sci. USA 96:3171-3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simpson, C. F., J. M. Kling, and F. C. Neal. 1965. The nature of bands in parasitized bovine erythrocytes. J. Cell Biol. 27:225-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Southwick, F. S., and D. L. Purich. 1996. Intracellular pathogenesis of listeriosis. N. Engl. J. Med. 334:770-776. [DOI] [PubMed] [Google Scholar]
- 40.Steck, T. L., and J. A. Kant. 1974. Preparation of impermeable ghosts and inside-out vesicles from human erythrocyte membranes. Methods Enzymol. 31:172-180. [DOI] [PubMed] [Google Scholar]
- 41.Stich, R. W., Kocan, K. M., Damian, R. T., and M. Fechheimer. 1997. Inclusion appendages associated with the intraerythrocytic rickettsial parasite Anaplasma marginale are composed of bundled actin filaments. Protoplasma 199:93-98. [Google Scholar]
- 42.Theriot, J. A., J. Rosenblatt, D. A. Portnoy, P. J. Goldschmidt-Clermont, and T. J. Mitchison. 1994. Involvement of profilin in the actin-based motility of L. monocytogenes in cells and in cell-free extracts. Cell 76:505-517. [DOI] [PubMed] [Google Scholar]
- 43.Visser, E. S., T. C. McGuire, G. H. Palmer, W. C. Davis, V. Shkap, E. Pipano, and D. P. Knowles, Jr. 1992. The Anaplasma marginale msp5 gene encodes a 19-kilodalton protein conserved in all recognized Anaplasma species. Infect. Immun. 60:5139-5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Welch, M. D., A. Iwamatsu, and T. J. Mitchison. 1997. Actin polymerization is induced by Arp2/3 protein complex at the surface of Listeria monocytogenes. Nature 385:265-269. [DOI] [PubMed] [Google Scholar]
- 45.Wickwire, K. B., K. M. Kocan, S. J. Barron, S. A. Ewing, R. D. Smith, and J. A. Hair. 1987. Infectivity of three Anaplasma marginale isolates for Dermacentor andersoni. Am. J. Vet. Res. 48:96-99. [PubMed] [Google Scholar]