An Arabidopsis transcription factor negatively regulates the accumulation of seed oil by targeting WRI1 and several other genes involved in the seed oil biosynthetic pathway.
Abstract
In many higher plants, seed oil accumulation is precisely controlled by intricate multilevel regulatory networks, among which transcriptional regulation mainly influences oil biosynthesis. In Arabidopsis (Arabidopsis thaliana), the master positive transcription factors, WRINKLED1 (WRI1) and LEAFY COTYLEDON1-LIKE (L1L), are important for seed oil accumulation. We found that an R2R3-MYB transcription factor, MYB89, was expressed predominantly in developing seeds during maturation. Oil and major fatty acid biosynthesis in seeds was significantly promoted by myb89-1 mutation and MYB89 knockdown; thus, MYB89 was an important repressor during seed oil accumulation. RNA sequencing revealed remarkable up-regulation of numerous genes involved in seed oil accumulation in myb89 seeds at 12 d after pollination. Posttranslational activation of a MYB89-glucocorticoid receptor fusion protein and chromatin immunoprecipitation assays demonstrated that MYB89 inhibited seed oil accumulation by directly repressing WRI1 and five key genes and by indirectly suppressing L1L and 11 key genes involved in oil biosynthesis during seed maturation. These results help us to understand the novel function of MYB89 and provide new insights into the regulatory network of transcriptional factors controlling seed oil accumulation in Arabidopsis.
Seed storage reserves in many higher plants usually constitute starch, oil stored as triacylglycerols (TAGs), and storage proteins. Seed TAGs not only serve as the main source of nutrients for humans and livestock but also facilitate postgerminative growth and subsequent seedling establishment (Li et al., 2006; Graham, 2008). They are also widely utilized by various industries (Lu et al., 2011) and serve as feedstock for the production of biofuels (Durrett et al., 2008). Thus, understanding the mechanisms underlying seed oil accumulation is of great interest, particularly in terms of its social and economic significance.
Arabidopsis (Arabidopsis thaliana) is considered an excellent model system for investigating oil biosynthesis in seeds (Baud and Lepiniec, 2009). Several previous studies have shown that a complex regulatory network of master positive transcription factors (TFs), including WRINKLED1 (WRI1), LEAFY COTYLEDON1 (LEC1), LEAFY COTYLEDON1-LIKE (L1L), LEC2, FUSCA3 (FUS3), and ABSCISIC ACID INSENSITIVE3 (ABI3), regulate the accumulation of storage reserves during seed maturation (Santos-Mendoza et al., 2008). WRI1, which is the TF of the APETALA2-ethylene-responsive element-binding protein family, directly or indirectly targets some enzymes involved in the late glycolysis and plastidial fatty acid (FA) biosynthetic network (Cernac and Benning, 2004; Baud et al., 2007; Maeo et al., 2009). Mutations in wri1 cause an 80% reduction in Arabidopsis seed oil content (Focks and Benning, 1998), and overexpression of WRI1 leads to a significant increase in the oil content (Cernac and Benning, 2004; Baud et al., 2009; Maeo et al., 2009; Sanjaya et al., 2011; Grimberg et al., 2015; Adhikari et al., 2016). LEC1 encodes a protein that is homologous to the Saccharomyces cerevisiae HEME ACTIVATOR PROTEIN3 or mammalian NUCLEAR FACTOR YB subunit of the heterotrimeric CCAAT box-binding factor (Lotan et al., 1998; Lee et al., 2003). LEC2, FUS3, and ABI3 encode closely related plant-specific TFs of the conserved B3 DNA-binding domain family (Giraudat et al., 1992; Luerssen et al., 1998; Stone et al., 2001). Activation of LEC1 or LEC2 induces ectopic embryogenesis in the vegetative tissues of Arabidopsis (Lotan et al., 1998; Santos Mendoza et al., 2005; Mu et al., 2008). Induced expression of LEC1 causes a global increase in the expression of genes involved in FA biosynthesis, thereby substantially promoting the accumulation of oil and major FA species (Mu et al., 2008). L1L is closely related to LEC1 and genetically suppresses the lec1 mutation when driven by the LEC1 promoter (Kwong et al., 2003). L1L also plays an important positive role in seed FA biosynthesis (Mu et al., 2008; Tan et al., 2011) and has been implicated in storage oil biosynthesis (Baud and Lepiniec, 2010). The ectopic expression of LEC2 activates the expression of genes encoding seed storage proteins, enzymes required for oil biosynthesis, and oil body-associated proteins, which, in turn, trigger oil deposition in rosette leaves (Santos Mendoza et al., 2005; Braybrook et al., 2006). FUS3 promotes oil deposition by positively regulating the expression of genes involved in photosynthesis and FA biosynthesis (Wang et al., 2007; Yamamoto et al., 2010; Zhang et al., 2016). Both LEC1 and LEC2 function as positive regulators upstream of WRI1, ABI3, and FUS3, which jointly control the expression of genes that contribute to the accumulation of seed oil and storage proteins (Kroj et al., 2003; Kagaya et al., 2005; Mu et al., 2008; Pan et al., 2010). In addition, ABI3 and FUS3 positively regulate themselves and each other, thereby forming feedback loops (To et al., 2006). L1L induction also is dependent on both LEC1 and FUS3 (Mu et al., 2008).
Recently, important progress regarding TFs in the regulation of seed oil accumulation has been made in Arabidopsis. Loss of GL2 activity in the seed coat, rather than the embryo, produces the high seed oil phenotype, partially by influencing MUCILAGE MODIFIED4 expression, which promotes mucilage biosynthesis in the seed coat (Shi et al., 2012). bZIP67 regulates seed α-linolenic acid content by binding G-boxes in the FATTY ACID DESATURASE3 (FAD3) promoter (Mendes et al., 2013). TRANSPARENT TESTA8 (TT8) inhibits seed FA accumulation by targeting LEC1, LEC2, and FUS3 in the seeds (Chen et al., 2014). The negative feedback effect of TTG1 on LEC2 and ABI3 plays a role in mediating FUS3 expression in seeds, thereby regulating the rate of seed storage reserve accumulation (Chen et al., 2015). An R2R3-MYB protein, MYB123 (TT2), was found to be a key regulator of not only proanthocyanidin deposition but also oil accumulation in seeds (Nesi et al., 2001; Chen et al., 2012; Wang et al., 2014). Knockout of MYB118, which also is a member of the R2R3-MYB TFs, caused the derepression of maturation-related genes, thus stimulating storage processes in endosperm (Barthole et al., 2014). Another R2R3-MYB TF, MYB96, directly regulates FATTY ACID ELONGATION1 (FAE1) to stimulate the accumulation of very-long-chain fatty acids (VLCFAs; C ≥ 20) during seed maturation (Lee et al., 2015). However, TFs and the regulatory network controlling oil biosynthesis in plant seeds remain largely unknown and, thus, require further investigation.
In this study, we found that an R2R3-MYB TF, MYB89, is highly expressed in developing seeds during seed maturation in Arabidopsis. We found that MYB89 directly and indirectly targets some key genes that contribute to oil accumulation during seed maturation. Our results suggest that MYB89 functions as a negative regulator of seed oil accumulation during maturation in Arabidopsis seeds.
RESULTS
MYB89 Is Expressed Predominantly in Developing Seeds
MYB89 encodes an unknown MYB protein that belongs to the R2R3-MYB family TFs (Dubos et al., 2010). According to the Arabidopsis eFP Browser, MYB89 is localized predominantly in mature pollen and developing seeds, particularly at the green cotyledon stages, suggesting that it regulates seed-related traits. Therefore, MYB89 was selected as the potential target in investigating the role of MYB TFs in the regulation of seed oil accumulation.
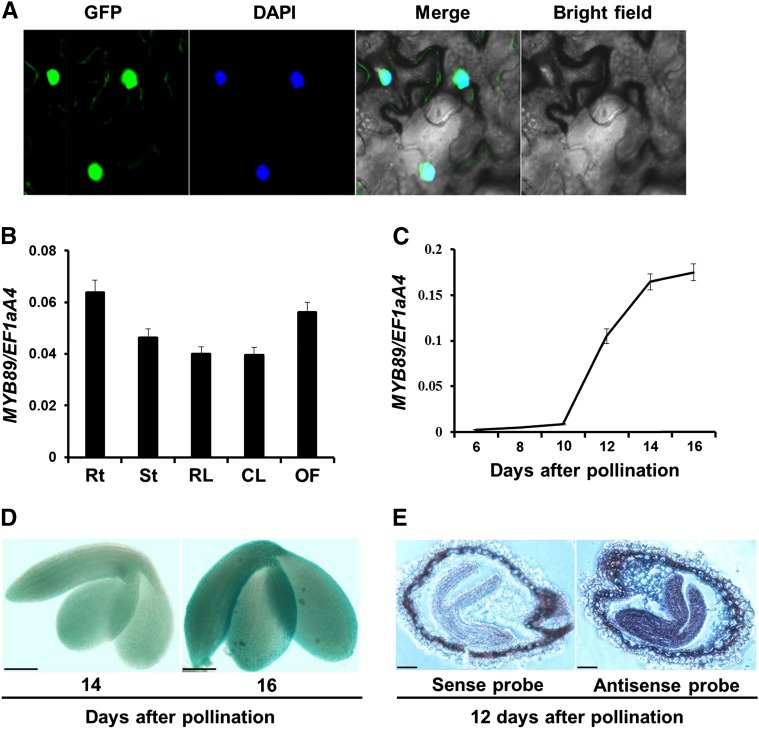
We investigated the subcellular localization of MYB89 in Nicotiana benthamiana leaves by using the GFP fusion construct, 35S:MYB89-GFP. MYB89-GFP was specifically localized in the nucleus (Fig. 1A), indicating that MYB89 functions as a TF.
Figure 1.
Analysis of MYB89 expression pattern. A, Subcellular localization of the MYB89 protein fused with GFP (35S:MYB89-GFP) in N. benthamiana leaves. DAPI, Fluorescence of 4′,6-diamino-2-phenylindole; Merge, merge of GFP, DAPI, and bright-field images. B, qRT-PCR analysis of MYB89 expression in various tissues of the wild type Columbia-0 (Col-0). Rt, Roots; St, stems; RL, rosette leaves; CL, cauline leaves; OF, open flowers. Results were normalized against the expression of EF1aA4 as an internal control. Values are means ± sd (n = 3). C, qRT-PCR analysis of MYB89 expression in developing seeds of the wild type (Col-0). Results were normalized against the expression of EF1aA4 as an internal control. Values are means ± sd (n = 3). D, Representative GUS staining of pMYB89:GUS transgenic plants shows MYB89 expression in developing embryos at 14 and 16 DAP in the wild type (Col-0). Bars = 100 µm. E, mRNA in situ localization of MYB89 in wild-type (Col-0) developing seeds at 12 DAP. These seeds were hybridized with the antisense or sense MYB89 probe as indicated. Bars = 100 µm.
We further investigated MYB89 expression in various tissues of wild-type plants by using quantitative real-time (qRT)-PCR and found the highest expression in developing seeds (Fig. 1, B and C). During seed development, MYB89 expression remained relatively low at the early seed maturation stage, from 6 to 10 d after pollination (DAP), but increased progressively afterward at the mid seed maturation stage, until it reached the maximal level at 16 DAP (Fig. 1C). To monitor the detailed expression pattern of MYB89, we generated a pMYB89:GUS reporter construct, in which a 1.199-kb MYB89 5ʹ regulatory region upstream of the ATG start codon was fused to the GUS reporter gene. No GUS staining signal was observed in the nontransgenic embryo (data not shown). Among the 17 pMYB89:GUS independent lines in the wild-type background, most lines showed similar GUS staining patterns in developing seeds (Fig. 1D), which is highly consistent with the results of qRT-PCR analysis (Fig. 1C). Positive GUS staining also was observed in other tissues, including root tips and true leaves of young seedlings (Supplemental Fig. S1). Furthermore, in situ hybridization revealed high MYB89 expression in developing seeds at 12 DAP (Fig. 1E). This expression pattern indicated that the dynamic regulation of MYB89 was relevant to the accumulation of seed oil, which occurs mainly at the seed maturation stage (Fait et al., 2006; Baud et al., 2008).
MYB89 Represses Seed Oil Accumulation in Arabidopsis
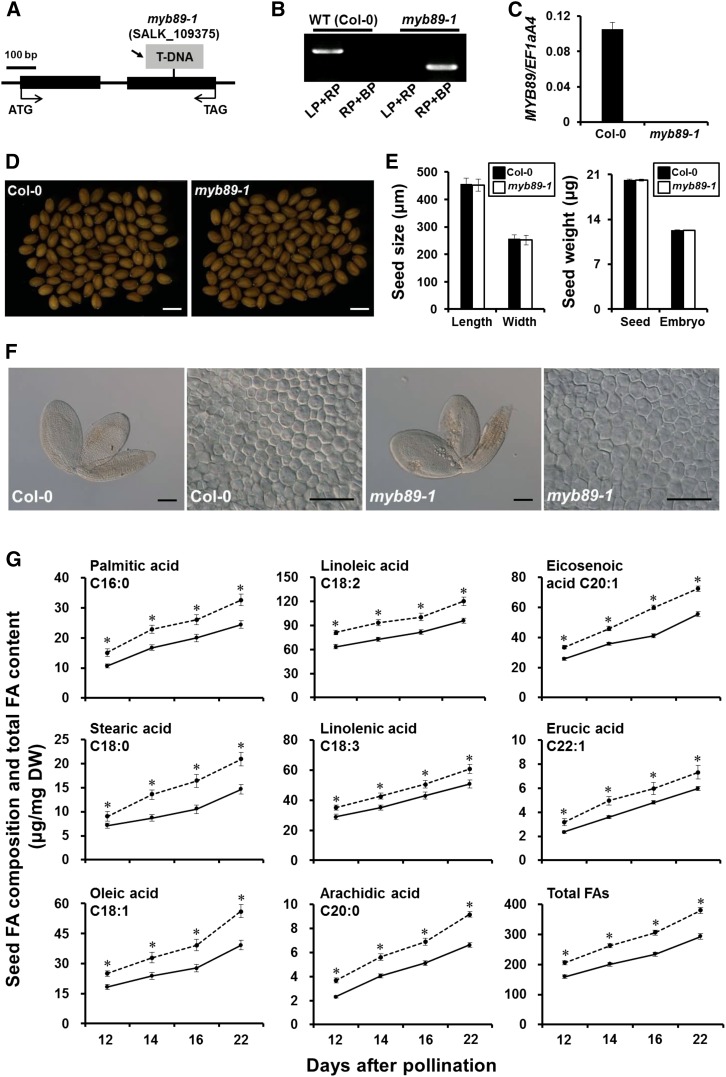
To investigate its biological function on seed oil accumulation, we isolated a corresponding T-DNA insertion mutant (SALK_109375) in the Col-0 background in the MYB89 exon from the Arabidopsis Biological Resource Center (Fig. 2A). The SALK mutant was designated myb89-1, which was backcrossed three times with Col-0, in case other mutations were present in this mutant. qRT-PCR showed that MYB89 was hardly expressed in developing seeds of myb89-1 homozygous plants but was highly expressed in the wild-type plants (Fig. 2, B and C).
Figure 2.
Comprehensive characterization of myb89-1 seeds. A, Molecular identification of the myb89-1 mutation. The structure of the MYB89 gene shows the position of the T-DNA insertion in the myb89-1 (SALK_109375) mutant. Black boxes represent exons, whereas black lines stand for the intron and other genomic regions. The arrow indicates the left border of the T-DNA. B, PCR-based genotyping of the wild type (WT; Col-0) and the homozygous myb89-1 mutant. LP and RP refer to the MYB89 gene-specific primers, and BP refers to the T-DNA right-border primer. C, qRT-PCR analysis of MYB89 expression in the wild type (Col-0) and the myb89-1 mutant. RNA samples were extracted from developing seeds at 12 DAP. Results were normalized against the expression of EF1aA4 as an internal control. Values are means ± sd (n = 3). D, Microscopic observation of mature seeds randomly selected from wild-type (Col-0) and myb89-1 plants. Bars = 500 µm. E, Quantitative comparisons of seed size (length and width) and dry weight of seeds and embryos between wild-type (Col-0) and myb89-1 plants. Values are means ± sd (n = 5), and each of the three assays for each biological replicate contained 200 seeds from 12 individual plants grown in different pots arranged randomly within one of three blocks. F, Microscopic observations of mature embryos and epidermal cell layers of the central region of cotyledons from wild-type (Col-0) and myb89-1 plants. Bars = 100 µm (embryos) and 20 µm (epidermal cell layers). G, Dynamic accumulation of major FA species and total FAs in developing seeds (12, 14, and 16 DAP) and mature seeds (22 DAP) of wild-type (Col-0) and myb89-1 plants. Solid and dotted lines indicate the content of FAs in wild-type (Col-0) and myb89-1 seeds, respectively. Values are means ± sd (n = 5). Asterisks indicate significant differences in the contents of FA composition and total FAs between wild-type and myb89-1 plants (two-tailed paired Student’s t test, P ≤ 0.05). DW, Dry weight.
Arabidopsis seed oil contains six major FA species, palmitic acid (C16:0), stearic acid (C18:0), oleic acid (C18:1n9), α-linoleic acid (C18:2n6), α-linolenic acid (C18:3n3), and eicosenoic acid (C20:1n11), and many minor species (less than 3 mol % each). To test whether MYB89 affects the accumulation of seed oil, we measured the contents of these major FAs in developing seeds at 12, 14, and 16 DAP and in mature seeds (22 DAP) of myb89-1 and wild-type plants. The contents of all FA species detected in wild-type and myb89-1 seeds increased, with the highest levels in mature seeds at 22 DAP (Fig. 2G; Supplemental Table S2). Notably, the levels of total FAs and each FA composition were uniformly considerably higher in myb89-1 than in the wild-type seeds at all stages of seed development investigated. We also found no obvious differences in several seed morphological traits, such as seed coat color (Fig. 2D), seed size and dry weight (Fig. 2E), embryo weight and size (Fig. 2, E and F), and embryo cell size and number (Fig. 2F; Supplemental Table S1), between mature seeds of myb89-1 and wild-type plants. Furthermore, the loss of MYB89 activity did not alter plant growth size (Supplemental Fig. S2) and the responses to the abiotic stresses caused by high concentrations of NaCl and Glc during seedling establishment (Supplemental Fig. S3). These results indicated that MYB89 negatively regulates seed oil accumulation without affecting other aspects of plant growth and development that influence seed oil deposition.
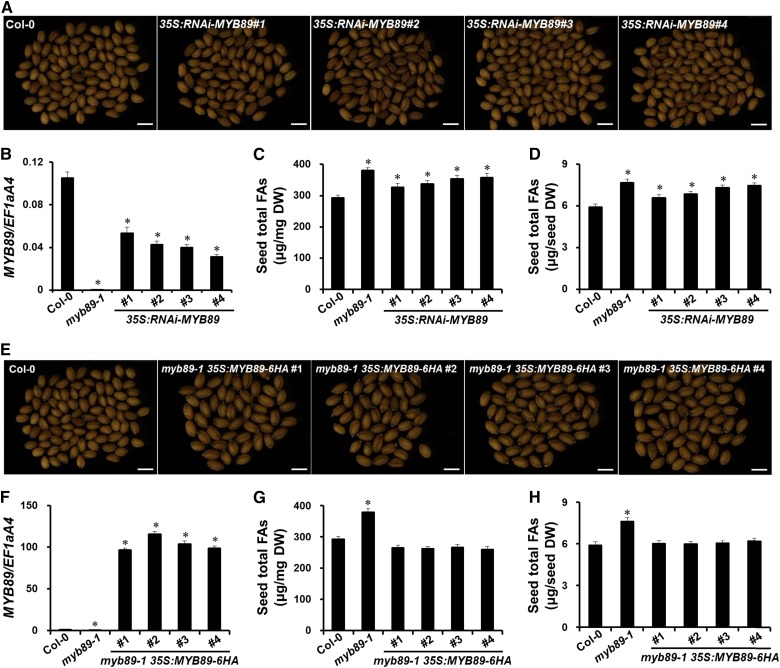
To verify that the loss of MYB89 function is responsible for the high oil phenotype, we introduced the MYB89 double-stranded RNA interference construct (35S:MYB89-RNAi) into wild-type Arabidopsis using Agrobacterium tumefaciens-mediated transformation. We obtained a total of 19 transformants, of which four independent 35S:MYB89-RNAi T3 homozygous transgenic lines with the strongest effect on MYB89 expression were selected (Fig. 3B). No obvious differences in seed coat color (Fig. 3A), seed size (Fig. 3A; Supplemental Fig. S4A), and dry weight (Supplemental Fig. S4B) were observed between mature seeds of the 35S:MYB89-RNAi and wild-type plants. This result showed that the contents of total FAs and each FA composition per seed or milligram of seed were uniformly considerably higher in 35S:MYB89-RNAi plants than in wild-type plants (Fig. 3, C and D; Supplemental Table S2). Furthermore, a MYB89-6 HEMAGGLUTININ (6HA) fusion gene driven by the 35S promoter was transformed into myb89-1 plants. Four selected independent myb89-1 35S:MYB89-6HA T3 homozygous transgenic lines with the strongest effect on MYB89 expression (Fig. 3F) exhibited comparable total FA quantity per seed or milligram of seed to that of the wild-type plants (Fig. 3, G and H; Supplemental Table S2). In addition, the content of each FA detected also was almost restored to the wild-type level (Supplemental Table S2). It is worth mentioning that myb89-1 35S:MYB89-6HA plants produced significantly larger and heavier seeds (Fig. 3E; Supplemental Fig. S4, C and D). Lower seed production often is correlated with larger seeds. Consistently, the total number of seeds per myb89-1 35S:MYB89-6HA plant was reduced markedly, whereas that of the myb89-1 and 35S:MYB89-RNAi plants did not change relative to the wild-type plants (Supplemental Fig. S5A). We also found that the seed yield per plant was not changed, but the oil yield per plant was increased significantly in myb89-1 and 35S:MYB89-RNAi lines compared with the wild-type control (Supplemental Fig. S5, B and C). However, the seed yield and oil yield per plant were decreased significantly in myb89-1 35S:MYB89-6HA lines (Supplemental Fig. S5, B and C). Taken together, our findings suggest that MYB89 represses seed oil accumulation during seed maturation.
Figure 3.
Effects of MYB89 knockdown in the wild-type (Col-0) background and MYB89 overexpression in the myb89-1 mutant background on seed traits. For each construct (35S:RNAi-MYB89 and 35S:MYB89-6HA), four independent homozygous T3 transgenic lines with the strongest effect on MYB89 expression were selected and analyzed. A, Microscopic observation of mature seeds randomly selected from the wild type (Col-0) and 35S:RNAi-MYB89 transgenic plants. Bars = 500 µm. B, qRT-PCR analysis of MYB89 expression in wild-type (Col-0), myb89-1, and four selected independent 35S:RNAi-MYB89 transgenic plants. RNA samples were extracted from developing seeds at 12 DAP. Results were normalized against the expression of EF1aA4 as an internal control. Values are means ± sd (n = 3). C, Comparisons of seed total FA content (μg mg−1) among wild-type (Col-0), myb89-1, and 35S:RNAi-MYB89 transgenic plants. Values are means ± sd (n = 5). Asterisks indicate significant differences in the seed total FA content compared with that in the wild type (two-tailed paired Student’s t test, P ≤ 0.05). DW, Dry weight. D, Comparisons of seed total FA content (μg seed−1) among wild-type (Col-0), myb89-1, and myb89-1 35S:MYB89-6HA transgenic plants. Values are means ± sd (n = 5). Asterisks indicate significant differences in the seed total FA content compared with that in the wild type (two-tailed paired Student’s t test, P ≤ 0.05). E, Microscopic observation of mature seeds randomly selected from wild-type (Col-0) and myb89-1 35S:MYB89-6HA transgenic plants. Bars = 500 µm. F, qRT-PCR analysis of MYB89 expression in wild-type (Col-0), myb89-1, and four selected independent myb89-1 35S:MYB89-6HA transgenic plants. RNA samples were extracted from developing seeds at 12 DAP. Results were normalized against the expression of EF1aA4 as an internal control. Values are means ± sd (n = 3). G, Comparisons of seed total FA content (μg mg−1) among wild-type (Col-0), myb89-1, and myb89-1 35S:MYB89-6HA transgenic plants. Values are means ± sd (n = 5). Asterisks indicate significant differences in the seed total FA content compared with that in the wild type (two-tailed paired Student’s t test, P ≤ 0.05). H, Comparisons of seed total FA content (μg seed−1) among wild-type (Col-0), myb89-1, and myb89-1 35S:MYB89-6HA transgenic plants. Values are means ± sd (n = 5). Asterisks indicate significant differences in the seed total FA content compared with that in the wild type (two-tailed paired Student’s t test, P ≤ 0.05).
Differentially Expressed Genes in Developing Seeds at 12 DAP between Wild-Type and myb89-1 Plants
We carefully selected one critical stage of seed oil accumulation to compare the expression profiles between myb89-1 and the wild-type control. At 12 DAP, large differences in the contents of total FAs and major FA composition in seeds between the mutant and wild-type plants were identified (Fig. 2G; Supplemental Table S2). At this stage, the total FA content in the myb89-1 seeds was 205.94 μg mg−1, which was significantly higher than that in the wild-type seeds (159.15 μg mg−1), and the FA content of developing seeds accounted for about 54% of the final FA content in mature seeds in both genotypes (Supplemental Table S2). The accumulation of seed oil increased markedly during the late embryonic maturation stages from 12 to 16 DAP and reached the final seed FA content at the end of embryo maturation (Fig. 2G), thereby indicating an active involvement of the genes that contribute to seed oil accumulation. Therefore, transcriptome analysis of developing seeds at 12 DAP could provide useful information on the downstream targets of MYB89 that contribute to oil biosynthesis as well as facilitate a better understanding of the regulatory networks underlying MYB89-mediated seed oil accumulation.
RNA sequencing (RNA-seq) analysis identified 1,171 differentially expressed genes (DEGs), among which 649 were up-regulated (Supplemental Table S3) and 522 were down-regulated (Supplemental Table S4) in myb89-1 developing seeds at 12 DAP. Functional analysis indicated that 71 (10.9%) and 45 (6.9%) of the up-regulated genes were related to carbohydrate metabolism and general protein biosynthesis, respectively (Table I). Notably, numerous genes (34) involved in oil metabolism, accounting for 5.2% of the total up-regulated genes, were significantly up-regulated in myb89-1 seeds (Table I; Supplemental Table S3). However, the expression of some genes that play major roles in oil accumulation did not change in myb89-1 seeds relative to the wild-type seeds (Supplemental Table S5). Obviously, the number of up-regulated genes involved in the metabolism of carbohydrates, proteins, and oil was significantly higher than that of the down-regulated genes in myb89-1 seeds (Table I).
Table I. Functional classification of DEGs in developing seeds of myb89-1 plants at 12 DAP.
Functional classification of DEGs was performed using the biological process category of Arabidopsis Gene Ontology (http://www.geneontology.com). Percentage refers to the ratio of genes of each functional category relative to total up-regulated or down-regulated DEGs identified in the RNA-seq experiment. The DEGs with log2 ratios greater than 1 or less than −1 (only GO Slim identifiers with P ≤ 0.05 and false discovery rate ≤ 0.05) are listed.
| Category |
Up-Regulated DEGs |
Down-Regulated DEGs |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| ≥2 |
1 to 2 |
Total |
Percentage |
≤−2 |
−2 to −1 |
Total |
Percentage |
||
| log2 ratio | log2 ratio | ||||||||
| Metabolism | |||||||||
| Photosynthesis | 3 | 14 | 17 | 2.6 | 3 | 2 | 5 | 1.0 | |
| Cell wall | 4 | 11 | 15 | 2.3 | 1 | 3 | 4 | 0.8 | |
| Oil metabolism | 4 | 30 | 34 | 5.2 | 4 | 17 | 21 | 4.0 | |
| Carbohydrate metabolism | 4 | 67 | 71 | 10.9 | 16 | 38 | 54 | 10.3 | |
| Nucleic acid | 3 | 16 | 19 | 2.9 | 3 | 24 | 27 | 5.2 | |
| Amino acid and protein | 8 | 37 | 45 | 6.9 | 5 | 11 | 16 | 3.1 | |
| Growth and development | |||||||||
| Leaf and root development | 0 | 12 | 12 | 1.8 | 1 | 9 | 10 | 1.9 | |
| Shoot development | 2 | 2 | 4 | 0.6 | 0 | 0 | 0 | 0 | |
| Embryo/seed development | 1 | 11 | 12 | 1.8 | 1 | 9 | 10 | 1.9 | |
| Flower development | 5 | 7 | 12 | 1.8 | 2 | 3 | 5 | 1.0 | |
| Cell growth | 5 | 42 | 47 | 7.2 | 5 | 6 | 11 | 2.1 | |
| Hormone | 5 | 15 | 20 | 3.1 | 13 | 12 | 25 | 4.8 | |
| Stress/defense response | 9 | 40 | 49 | 7.6 | 27 | 76 | 103 | 19.7 | |
| Cell regulation | |||||||||
| Transcriptional regulation | 0 | 15 | 15 | 2.3 | 1 | 4 | 5 | 1.0 | |
| Signaling transduction | 6 | 34 | 40 | 6.2 | 10 | 23 | 33 | 6.3 | |
| Transport facilitation | 3 | 28 | 31 | 4.8 | 8 | 27 | 35 | 6.7 | |
| Others | 30 | 176 | 206 | 31.7 | 45 | 113 | 158 | 30.3 | |
In all, 20 genes involved in transcriptional regulation were regulated by MYB89, of which 15 genes were up-regulated and five genes were down-regulated in myb89-1 seeds (Table I). Interestingly, WRI1 and L1L, the master regulators of oil accumulation, were significantly up-regulated in myb89-1 seeds, whereas no significant differences in the expression of the other TFs, including LEC1, LEC2, ABI3, FUS3, bZIP67, TT2, and TTG1, were observed between wild-type and myb89-1 developing seeds at 12 DAP (Supplemental Tables S3–S5). Therefore, MYB89 inhibits seed oil accumulation by repressing two master regulators, WRI1 and L1L, and a range of genes in the oil biosynthetic pathway during seed development.
Verification of Regulated Master Regulators and Genes Contributing to Oil Biosynthesis at Different Developmental Stages in myb89-1 Seeds
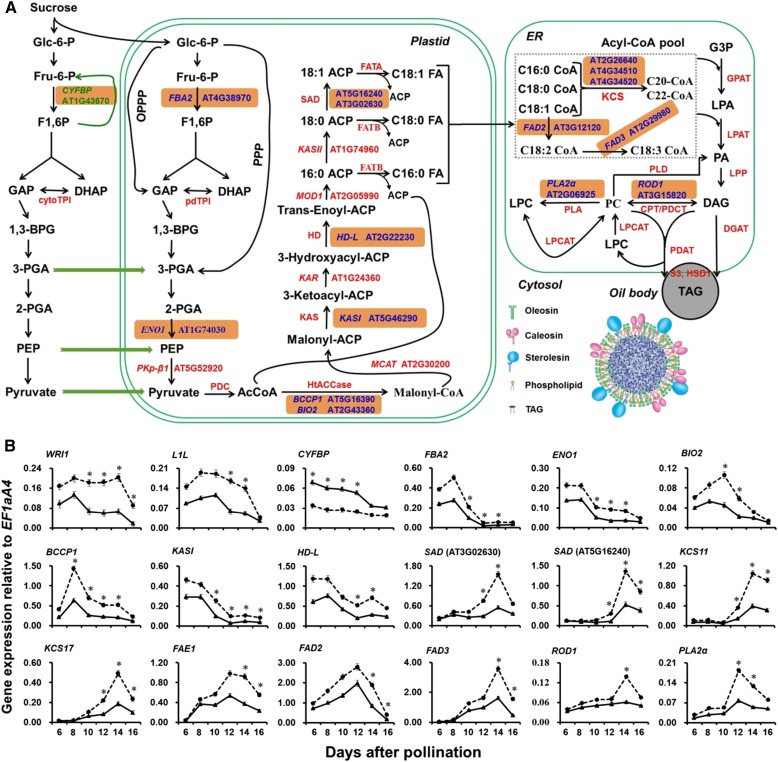
Seed oil content is a complex quantitative trait controlled by multiple genes. TAG biosynthesis could be briefly classified into three phases in plant cells (Baud and Lepiniec, 2009; Itabe, 2010; Chapman and Ohlrogge, 2012). First, pyruvate and other substances produced during glycolysis are catabolized into acetyl-CoA, which is the precursor for FA biosynthesis, and then C16-18 FAs are synthesized in the plastid. Subsequently, most of these are exported to the cytoplasm, and, after chain elongation and desaturation, they form various FA derivatives at the acyl chains. Finally, oil is formed and stored in the oil body in the form of TAGs (Fig. 4A).
Figure 4.
Dynamic expression analysis of genes involved in the processes of glycolysis, FA biosynthesis and modification, and TAG accumulation in developing seeds of wild-type (Col-0) and myb89-1 plants. A, Simplified scheme showing the altered expression levels of genes involved in the oil biosynthetic pathway in developing seeds between wild-type (Col-0) and myb89-1 plants. The seed oil body generated from the endoplasmic reticulum (ER) accumulates TAG inside and is surrounded by a layer of phospholipids and proteins, including oleosin, caleosin, and steroleosin. The genes marked by blue color with an orange box were significantly up-regulated, and the gene CYFBP indicated by green color with an orange box was down-regulated in myb89-1 developing seeds. F1,6P, Fru-1,6-diphosphate; GAP, glyceraldehyde-3-phosphate; DHAP, dihydroxy-acetone-phosphate; cytoTPI, cytosolic triose phosphate isomerase; pdTPI, plastid TPI; 1,3-BPG, 1,3-bisphosphoglycerate; 3PGA, glycerate-3-phosphate; 2PGA, glycerate-2-phosphate; PEP, phosphoenolpyruvate; PDC, pyruvate dehydrogenase complex; AcCoA, acetyl-CoA; HtACCase, heteromeric acetyl-CoA carboxylase; ACP, acyl carrier protein; KAS, 3-ketoacyl-ACP synthase; HD, hydroxyacyl-ACP dehydratase; SAD, stearoyl-ACP desaturase; FAT, fatty acyl-ACP thioesterase; KCS, 3-ketoacyl-CoA synthase; G3P, glycerol 3-phosphate; GPAT, G3P acyltransferase; LPA, lysophosphatidic acid; LPAT, LPA acyltransferase; PA, phosphatidic acid; LPP, lipid phosphate phosphatase; DAG, diacylglycerol; PC, phosphatidylcholine; PDCT, PC:DAG phosphocholine transferase; CPT, choline-phosphotransferase; PLA, phospholipase A; PLD, phospholipase D; LPCAT, lyso-PC acyltransferase; LPC, lyso-PC; DGAT, DAG acyltransferase; PDAT, phospholipid:DAG acyltransferase; TAG, triacylglycerol; HSD, hydroxysteroid dehydrogenase; CYFBP, CYTOSOL FRU-1,6-BISPHOSPHATASE; FBA2, FRU-1,6-BISPHOSPHATE ALDOLASE2; ENO1, PLASTID-LOCALIZED PHOSPHOENOLPYRUVATE ENOLASE1; PKp-β1, PLASTIDIC PYRUVATE KINASE β-SUBUNIT1; BCCP1, BIOTIN CARBOXYL-CARRIER PROTEIN1; BIO2, BIOTIN AUXOTROPH2; MCAT, MALONYL-COA:ACP TRANSACYLASE; KAR, 3-KETOACYL-ACP REDUCTASE; MOD1, MOSAIC DEATH1; FAD2, FATTY ACID DESATURASE2; ROD1, REDUCED OLEATE DESATURATION1; PLA2α, PHOSPHOLIPASE A2α; OPPP, oxidative pentose phosphate pathway; PPP, nonoxidative pentose phosphate pathway. B, qRT-PCR analysis of the expression of genes involved in the oil biosynthetic pathway in developing seeds between wild-type (Col-0) and myb89-1 plants. Solid and dotted lines indicate the dynamic expression of genes in wild-type and myb89-1 plants, respectively. Results were normalized against the expression of EF1aA4 as an internal control. Values are means ± sd (n = 3). Asterisks indicate significant differences in gene expression levels in myb89-1 plants compared with those in wild-type plants (two-tailed paired Student’s t test, P ≤ 0.05).
To confirm the regulation of the master regulators, WRI1 and L1L, and genes involved in oil biosynthesis in myb89-1 developing seeds at 12 DAP, and to extensively explore potential genes involved in the oil biosynthetic pathway that is regulated by MYB89, we performed qRT-PCR to compare the expression patterns at the seed maturation stages (6–16 DAP) between the wild-type and myb89-1 plants. First, we selected 13 highly up-regulated TFs or genes, WRI1, L1L, FBA2, ENO1, BIO2, BCCP1, KASI, HD-L, SAD (AT3G02630 and AT5G16240), KCS11, KCS17, and PLA2α, and one significantly down-regulated gene, CYFBP, in myb89-1 developing seeds at 12 DAP (Fig. 4B). The expression of WRI1 was significantly higher at the midmaturation stage from 10 DAP in myb89-1 developing seeds than in the wild-type seeds. Relative to that of the wild type, myb89 mutation resulted in at least a 2-fold higher level of L1L transcripts in developing seeds at 12 and 14 DAP. The expression of the other genes was significantly higher at the seed maturation stage between wild-type and myb89-1 plants at 12 DAP and other examined stages, except for CYFBP expression, which was decreased significantly from 6 to 12 DAP and was always lower during seed maturation in myb89-1 than in the wild type. The expression of nearly all of these up-regulated genes was higher than in the wild-type seeds from 6 to 16 DAP (Fig. 4).
Furthermore, another 28 genes (Fig. 4B; Supplemental Fig. S6), the expression of which was not altered significantly in myb89-1 developing seeds at 12 DAP (Supplemental Table S5), were selected to investigate whether these are regulated by MYB89 during seed maturation. No significant differences in the expression of 24 genes were noted at the seed maturation stage between wild-type and myb89-1 plants (Supplemental Fig. S6); however, the expression of FAD2, FAD3, FAE1, and ROD1 was significantly up-regulated in myb89-1 developing seeds at 14 and/or 16 DAP (Fig. 4B).
These significantly up-regulated genes, except for the two master regulators, WRI1 and L1L, are marked in blue with an orange box, and the down-regulated gene, CYFBP, is indicated in green with an orange box, in the oil biosynthetic pathway (Fig. 4A). Taken together, these findings suggest that MYB89 regulates the expression of WRI1 and L1L, the TFs critical for oil accumulation, and a class of genes in the oil biosynthetic pathway, which is concomitant with a steady increase in FA levels in seeds at the midmaturation stage (Fig. 2G; Supplemental Table S2), indicating that MYB89 mediates seed oil accumulation by affecting the expression of these genes.
MYB89 Directly Targets WRI1, ENO1, BCCP1, KASI, KCS11, and PLA2α in Arabidopsis Developing Seeds
To determine how MYB89 regulates the mRNA expression of downstream targeted genes, we generated a steroid-inducible version of MYB89 in the background of myb89-1, in which the MYB89 gene was fused to the rat glucocorticoid receptor (GR) and driven by the 35S promoter. We isolated a myb89-1 35S:MYB89-GR transgenic line, which rescued the higher oil content phenotype of myb89-1 seeds after dexamethasone treatment every alternate day after germination, whereas the mock-treated myb89-1 35S:MYB89-GR accumulated higher oil like myb89-1 (Supplemental Fig. S7). This showed that the MYB89-GR fusion protein has a biological function like that of the wild-type MYB89 upon steroid induction.
By using this established steroid-inducible activation of MYB89, we further determined whether the expression of these genes is repressed by MYB89 activity (Fig. 4). Dexamethasone treatment of myb89-1 35S:MYB89-GR siliques at 12 or 14 DAP for 2 or 4 h caused a reduction in the expression of all these genes, except for CYFBP, relative to that of the mock-treated controls (Fig. 5; Supplemental Fig. S8). However, the combined treatment of dexamethasone and cycloheximide for 2 or 4 h only significantly reduced the expression of WRI1, ENO1, BCCP1, KASI, KCS11, and PLA2α (Fig. 5; Supplemental Fig. S8), suggesting that these six genes are immediate targets of transcriptional repression by MYB89 in developing seeds, whereas the induction of CYFBP and repression of the other 12 genes by MYB89 are dependent on other intermediate proteins.
Figure 5.
Induced MYB89 activity transcriptionally represses the expression of WRI1, ENO1, BCCP1, KASI, KCS11, and PLA2α during seed development. The myb89-1 35S:MYB89-GR siliques at 12 DAP were mock treated (Mock) or treated with 10 μm dexamethasone (DEX), 10 μm cycloheximide (CYC), or 10 μm CYC plus 10 μm DEX (CYC+DEX). The expression of these genes was examined after 2 or 4 h of treatment using qRT-PCR analyses. Results were normalized against the expression of EF1aA4 as an internal control. Values are means ± sd (n = 3). Asterisks indicate significant differences in gene expression in dexamethasone-treated samples compared with their respective controls (two-tailed paired Student’s t test, P ≤ 0.05).
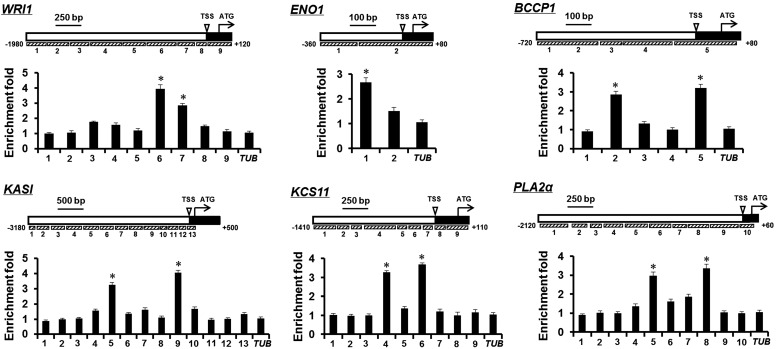
To investigate whether MYB89 binds directly to the promoter regions of WRI1, ENO1, BCCP1, KASI, KCS11, and PLA2α to control its expression, we performed chromatin immunoprecipitation (ChIP) assays using a functional transgenic line that expressed a MYB89-6HA fusion gene driven by the 35S promoter. 35S:MYB89-6HA completely rescued the higher oil content phenotype of myb89-1 in the myb89-1 35S:MYB89-6HA lines (Fig. 3, G and H; Supplemental Table S2), indicating that the fusion protein of MYB89-6HA at least retains the same biological function as MYB89 in developing seeds. We designed sufficient pairs of primers to cover all the possible cis-elements bound by MYB89 in the promoter regions of these six genes (Fig. 6). ChIP assays performed using a representative line, myb89-1 35S:MYB89-6HA#1, showed that MYB89-6HA was associated with the promoter regions near fragments 6 and 7 of WRI1, fragment 1 of ENO1, fragments 2 and 5 of BCCP1, fragments 5 and 9 of KASI, fragments 4 and 6 of KCS11, and fragments 5 and 8 of PLA2α (Fig. 6). These results collectively suggested that MYB89 binds directly to the loci of ENO1, BCCP1, KASI, KCS11, and PLA2α to suppress their expression.
Figure 6.
Schematic diagrams show the promoter regions of WRI1, ENO1, BCCP1, KASI, KCS11, and PLA2α, and ChIP analysis shows MYB89-6HA binding to their promoter regions in developing siliques at 12 DAP. The transcriptional start site (TSS) and exon are represented by black boxes, whereas promoter regions are represented by white boxes. Gray boxes represent the DNA fragments amplified in ChIP analysis for each gene. The enrichment fold of each fragment was calculated first by normalizing the amount of a target DNA fragment against a genomic fragment of EF1aA4 as an internal control and then by normalizing the value for myb89-1 35S:MYB89-6HA against that for myb89-1. A TUB2 (TUB) fragment was amplified as a negative control. Values are means ± sd (n = 3). Significant differences in comparison with the enrichment of the TUB fragment are indicated with asterisks (two-tailed paired Student’s t test, P ≤ 0.05).
DISCUSSION
Seed oil accumulation in many higher plants is precisely controlled by intricate regulatory networks that coordinate various environmental and developmental signals and by multilevel regulation, among which transcriptional regulation is a major factor influencing FA supply for TAG biosynthesis. However, the mechanism underlying how TFs and the regulatory network control the overall amount of oil stored in plant seeds is not completely understood to date. The MYB family includes numerous proteins and is functionally involved in the regulatory networks that control the development, metabolism, and responses to biotic and abiotic stresses (Dubos et al., 2010). We used the reverse genetic approach and found that myb89 mutation causes a significant increase in the levels of total FAs and each major FA composition during seed maturation (Fig. 2G; Supplemental Table S2), which leads to the remarkable accumulation of seed FAs (Baud et al., 2008). In contrast, knockdown of MYB89 resulted in a significant increase of total FAs and each FA composition per seed or milligram of seed (Fig. 3, B–D; Supplemental Table S2). Furthermore, the ectopic expression of MYB89 in the myb89-1 background could completely restore the higher seed oil content of myb89-1 plants to the wild-type level (Fig. 3, F–H; Supplemental Table S2). These results, together with the observation of the increased expression of MYB89 in developing embryos at the mid seed maturation stage (Fig. 1, B–E), suggest that MYB89 plays an important role in the regulatory network that suppresses the accumulation of seed oil in Arabidopsis.
The coordinated expression of genes involved in the oil biosynthetic pathway is essential for seed oil accumulation (Ohlrogge and Jaworski, 1997; Ruuska et al., 2002; Graham, 2008). Loss of function of MYB89 regulated the expression of many genes involved in various important metabolic processes, including glycolysis, FA biosynthesis and modification, and TAG deposition in the plastids or endoplasmic reticulum, which, in turn, contribute to oil accumulation (Fig. 4; Supplemental Tables S3 and S4). Of these enzymes, FBA2, which is localized in the plastids, is an essential enzyme that generates metabolites for starch biosynthesis, and fba2 seedlings accumulate less starch (Sonnewald et al., 1994; Lu et al., 2012). CYFBP, which is expressed in the cytoplasm and nucleus, catalyzes the formation of Fru-6-P for Suc biosynthesis, whereas its disruption elevates the levels of starch and glycerate-3-phosphate in the leaves during the day (Fig. 4A; Rojas-González et al., 2015). ENO1 encodes the plastid-localized phosphoenolpyruvate enolase that functions metabolically in the conversion of glycerate-2-phosphate into phosphoenolpyruvate (Fig. 4A; Prabhakar et al., 2009), which mediates the production of acetyl-CoA, the precursor for FA biosynthesis. Previous studies have proposed that starch serves as a carbon source for the accumulation of seed storage reserves such as oil and storage proteins (Norton and Harris, 1975; daSilva et al., 1997; Periappuram et al., 2000; Andriotis et al., 2010). The master regulator, WRI1, plays an essential role in the developmental regulation of carbohydrate metabolism, particularly in the incorporation of Suc and Glc into TAGs during seed filling (Focks and Benning, 1998). Notably, WRI1 is significantly up-regulated in myb89-1 developing seeds from 10 to 16 DAP (Fig. 4; Supplemental Table S3-2). Thus, the decreased expression of CYFBP (Fig. 4; Supplemental Table S4-2) and the subsequent elevated expression of WRI1, FBA2, and ENO1 (Fig. 4; Supplemental Table S3-2) might assist in the partitioning of carbons for oil accumulation in myb89-1 developing seeds (Fig. 2G).
The ATP-dependent carboxylation of acetyl-CoA to yield malonyl-CoA is a primary reaction that occurs during de novo FA biosynthesis (Baud and Lepiniec, 2009). This process is mediated by HtACCase, which also acts as a sensor or gating system that controls the overall flux of FA biosynthesis (Reverdatto et al., 1999; Mu et al., 2008). Our results showed that BCCP1, which encodes an HtACCase subunit and BIO2, was significantly up-regulated in myb89-1 developing seeds (Fig. 4; Supplemental Table S3-2). Reduced BCCP1 activity severely affects normal vegetative growth and also markedly decreases seed FA accumulation (Li et al., 2011). BIO2 converts dethiobiotin to biotin in the biotin biosynthetic pathway (Picciocchi et al., 2001), and the loss of BIO2 reduces the content of seed oil (Pommerrenig et al., 2013). Thus, more active HtACCase and more biotin might promote oil accumulation in myb89-1 developing seeds at the very early stage of the biosynthetic pathway (Figs. 2G and 4A; Supplemental Table S2). Three separate condensing enzymes, or 3-ketoacyl-ACP synthases (KASI–KASIII), are necessary for the production of C18 FAs. Among these, KASI is responsible for producing six- to 16-carbon chains (Fig. 4A; Pidkowich et al., 2007), and its deficiency markedly reduces seed FA accumulation (Wu and Xue, 2010). Furthermore, 3-hydroxyacyl-ACP dehydratase (HD) is required after each condensation step to obtain a saturated and two-carbon-longer FA (Fig. 4A; Mou et al., 2000), and some 16:0-ACP molecules are elongated to C18:0-ACP, which is efficiently desaturated by a stromal SAD (Fig. 4A; Browse and Somerville, 1991). Hence, the suppression of KASI, HD-L, and SADs, including AT5G16240 and AT3G02630, by MYB89 (Fig. 4; Supplemental Table S3-2) influences the accumulation of FA composition such as C16:0, C18:0, and C18:1 in seeds at the middle stage of the biosynthetic pathway (Figs. 2G and 4; Supplemental Table S2). The biosynthesis of long-chain FAs in the plastids is terminated when the acyl group is removed from ACP by fatty acyl-ACP thioesterase (Fig. 4A; Salas and Ohlrogge, 2002; Bonaventure et al., 2003). Once activated to CoA esters and transported to the endoplasmic reticulum, these FA species can be modified (i.e. desaturated and elongated). FAD2 is essential for polyunsaturated lipid synthesis (Fig. 4A; Okuley et al., 1994), and FAD3 is responsible for the biosynthesis of C18:3 from phospholipids using cytochrome b5 as an electron donor (Fig. 4A; Shah et al., 1997). Thus, the highly up-regulated expression of the genes FAD2 and FAD3 would accelerate the accumulation of C18:2 and C18:3, respectively, in myb89-1 developing seeds (Fig. 2G; Supplemental Table S2). The KCS genes are essential for the biosynthesis of VLCFAs and facilitate the extension of the chain length of FAs from C18 to C20 and C22 (Fig. 4A). Notably, MYB89 largely suppresses the expression of genes involved in the elongation of FAs, such as KCS11, KCS17, and KCS18 (FAE1; Fig. 4; Supplemental Table S3-2). This could explain why myb89-1 seeds accumulated considerably more VLCFAs than wild-type seeds (Fig. 2G; Supplemental Table S2). Furthermore, the expression of PLA2α and ROD1, which participate in TAG deposition at the late stage of the biosynthetic pathway, was increased significantly in the myb89 developing seeds (Fig. 4; Supplemental Table S3-2). PLA2α, a secretory phospholipase A2 enzyme, specifically hydrolyzes the sn-2 position of phospholipids and has a preference toward the linoleoyl acyl chain (Ryu, 2004; Ryu et al., 2005). ROD1 encodes a phosphatidylcholine:diacylglycerol cholinephosphotransferase that is involved in a major reaction for the transfer of C18:1 to phosphatidylcholine for desaturation as well as for the reverse transfer of C18:2 and C18:3 to the TAG synthesis pathway (Fig. 4A; Lu et al., 2009). Therefore, MYB89 participates in multiple regulatory events at the transcriptional level that mediate oil accumulation in seeds (Fig. 4A).
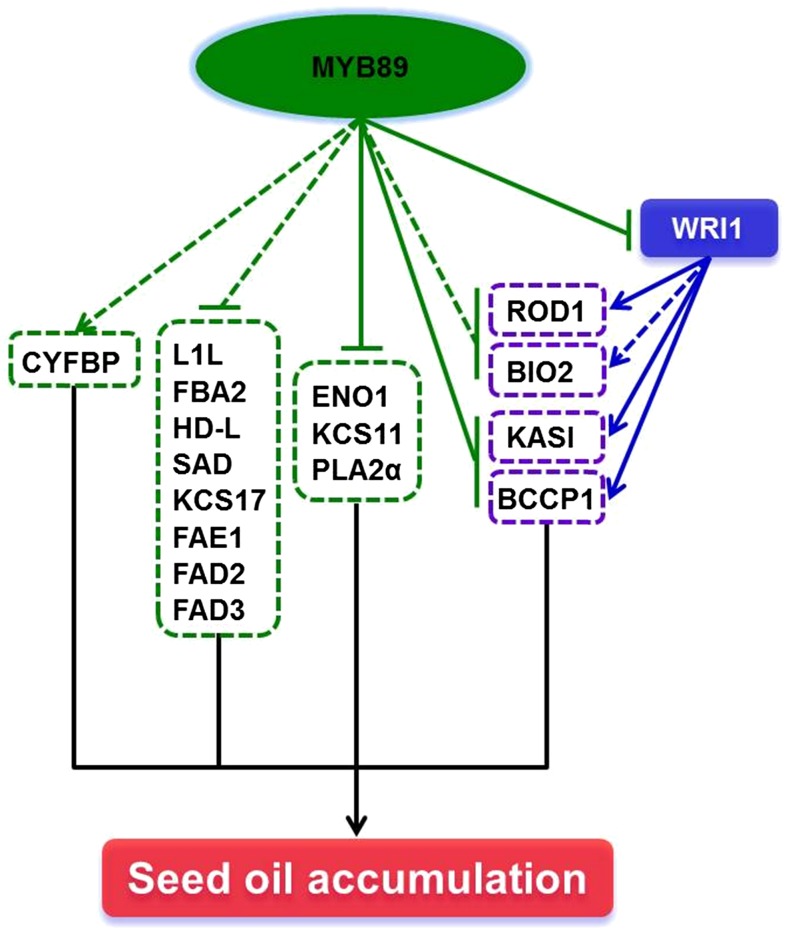
RNA-seq and qRT-PCR analyses showed that LEC2 expression was not altered (Supplemental Fig. S6; Supplemental Table S5), whereas WRI1 expression was increased significantly, in myb89-1 developing seeds (Fig. 4; Supplemental Table S3-2). By using the GR-inducible system and the ChIP assay, we found the novel result that WRI1 is repressed directly by MYB89 in developing seeds (Figs. 5 and 6). A previous study showed that the genes encoding enzymes required for FA biosynthesis are not direct targets of LEC2 in leaves (Santos Mendoza et al., 2005). Further genetic analyses indicated that WRI1 is a direct target of LEC2 and functions as a global regulator, specifying the regulatory action of LEC2 toward FA biosynthesis, and fine-tunes the expression of lipogenic genes during seed maturation in Arabidopsis (Baud et al., 2007). We reasoned that MYB89 represses WRI1 expression directly, not via LEC2 at the transcriptional level. These findings indicated that MYB89 serves as a novel negative regulator in directly suppressing the expression of WRI1, the master regulator of oil accumulation, in maturing Arabidopsis seeds (Fig. 7).
Figure 7.
A simplified model shows that MYB89 directly and indirectly regulates the expression of key regulators and genes that control the accumulation of seed oil in Arabidopsis. Arrows and T bars, respectively, indicate promoting and inhibitory effects, whereas solid and dotted lines indicate direct and indirect transcriptional regulation, respectively.
Molecular analyses have revealed that ENO1, BCCP1, KASI, KCS11, and PLA2α are repressed directly (Figs. 5 and 6), whereas L1L and the remaining 11 key genes, CYFBP, FBA2, BIO2, HD-L, SAD (AT3G02630 and AT5G16240), KCS17, FAE1, FAD2, FAD3, and ROD1, are regulated indirectly (Supplemental Fig. S8), by MYB89 during seed maturation. Previous studies indicated that WRI1 acts as the master regulator of FA biosynthesis by directly promoting the expression of SUS2, PI-PKβ1, BCCP1, BCCP2, KASI, GLB1, and ROD1 and indirectly inducing ACP1, CAC2, CAC3, BIO2, PDH E1α, KASIII, MOD1, and LAS during Arabidopsis seed development (Focks and Benning, 1998; Cernac and Benning, 2004; Masaki et al., 2005; Baud et al., 2007; Maeo et al., 2009; To et al., 2012). ROD1 also was demonstrated to be up-regulated by overexpressing Arabidopsis WRI1 in N. benthamiana leaves (Grimberg et al., 2015) and castor (Ricinus communis) seeds (Adhikari et al., 2016). Thus, alterations in the FA profile of myb89 seeds become complicated and cannot be attributed to specific genes. The loss of MYB89 and the elevated expression of WRI1 together might promote the expression of KASI and BCCP1, and the other two genes, ROD1 and BIO2, might be induced via the up-regulation of WRI1 in myb89-1 developing seeds, thereby accelerating seed oil accumulation (Fig. 7). These four genes might be regulated by multiple forms during seed development. Theoretically, except for KASI, BCCP1, ROD1, and BIO2, the up-regulation of WRI1 induces the expression of all the other genes that are positively regulated by WRI in myb89-1 developing seeds. However, no changes in transcriptional levels of the genes, including SUS2, PI-PKβ1, BCCP2, GLB1, ACP1, CAC2, CAC3, PDH E1α, KASIII, MOD1, and LAS, in myb89-1 seeds were noted compared with those in the wild-type seeds (Supplemental Fig. S6; Supplemental Table S5). These genes up-regulated by WRI1 (Fig. 7) are likely promoted by MYB89 or regulated by unknown TFs, and other unknown TFs might independently act or interact with MYB89 and/or master regulators described here to regulate those genes that were targeted by MYB89. A previous study showed that the loss of BTB/POZ-MATH (BPM) proteins widely affects plant development and causes altered FA contents in Arabidopsis mutant seeds, and BPM proteins function as negative regulators of WRI1 activities by mediating assembly with the CULLIN3-based REALLY INTERESTING NEW GENE E3 ligases core and ultimately causing its degradation via the 26S proteasome (Chen et al., 2013). However, the expression of the BPM genes is not changed in myb89-1 developing seeds compared with wild-type seeds (Supplemental Fig. S9; Supplemental Table S5), suggesting that MYB89 controls the expression of WRI1 not via BPM genes at the transcriptional level to affect seed oil accumulation in Arabidopsis but possibly via a distinct regulatory pathway. These interesting questions need to be investigated further.
In summary, to our knowledge, this study is the first to show that an R2R3-MYB TF, MYB89, inhibits Arabidopsis seed oil accumulation by directly repressing the expression of the master regulator, WRI1, and that of five key genes, ENO1, BCCP1, KASI, KCS11, and PLA2α, and indirectly suppressing another master regulator, L1L, and various other key genes in the oil biosynthetic pathway during seed maturation (Fig. 7). In addition, the deletion of MYB89 does not lead to the formation of impaired phenotypes in plants and alter the responses to abiotic stresses during seedling establishment (Fig. 2, D–F; Supplemental Figs. S2 and S3). These factors ensure the appropriate manipulation of seed oil production in oil-producing crops and plants by antagonizing MYB89 activity at the late stages of seed development to promote the expression of WRI1 and other various key genes that contribute to oil accumulation, thereby improving both the quantity and the quality of seed oil.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Col-0 was used as the wild-type control. The mutant myb89-1 (SALK_109375) was in the Col-0 background, and its genotyping primers are listed in Supplemental Table S6. All transgenic plants were generated in the Col-0 background through Agrobacterium tumefaciens-mediated transformation and selected by Basta on soil. Since the growth conditions can markedly affect the total oil accumulation in seeds (Li et al., 2006), all plants for FA measurements were grown at the same time in the same chamber under long-day conditions (16 h of light/8 h of dark) at 22°C. The overhead light intensity was 160 µmol m−2 s−1, as detected at the middle region of the plant.
Light Microscopy
Embryo observation was performed as described previously (Ohto et al., 2005; Chen et al., 2014). Isolated embryos were incubated in a buffer (50 mm sodium phosphate [pH 7], 10 mm EDTA, 1% [v/v] Triton X-100, and 1% [v/v] dimethyl sulfoxide) at 37°C overnight, fixed in a formaldehyde-acetic acid solution (10% [v/v] formalin, 5% [v/v] acetic acid, 45% [v/v] ethanol, and 0.01% [v/v] Triton X-100) for 45 min, and then rehydrated using a graded ethanol series. Embryos were then cleared in Hoyer’s solution (chloral hydrate:water:glycerol, 3:0.8:0.4) and observed with a light microscope (Olympus SZ 61). The cell size of embryos was measured using NIH Image software (http://rsb.info.nih.gov/nih-image/index.html).
Measurement of Seed FAs
The seeds for FA measurement were harvested from the lower part of the main stem of 16 individual plants grown in different pots arranged randomly within one of three blocks. Seed FAs were extracted and analyzed as reported previously in detail (Poirier et al., 1999; Chen et al., 2012). Briefly, total FAs were converted to FA methyl esters in methanol solution containing 1 m HCl for 2 h at 80°C. FAs in seeds were subsequently measured using a gas chromatograph (GC-2014; Shimadzu).
Plasmid Construction
The plasmids 35S:MYB89-6HA and 35S:MYB89-GR were constructed by amplifying the full-length coding region of MYB89 and cloning it into pGreen-35S-6HA (Liu et al., 2008) and pGreen-35S-GR (Yu et al., 2004), respectively. To construct 35S:RNAi-MYB89, the amplified antisense and sense fragments were digested with restriction enzymes into the pGreen-HY104 vector (Yu et al., 2004) before and after the GUS fragment, respectively. The GUS reporter construct pMYB89:GUS was constructed by amplifying its 5ʹ regulatory region upstream of the ATG start codon and cloning it into pHY107 (Liu et al., 2007). The primers used for plasmid construction are listed in Supplemental Table S6.
RNA-Seq and Data Analyses
The flowers of wild-type (Col-0) and myb89-1 plants were tagged with different colored threads to indicate DAP. Only developing seeds from the siliques on the primary shoots of 80 individual plants for each genotype in one biological replicate, which were grown in different pots arranged randomly, were used for the RNA-seq experiment. Two independent biological replicates from two different plantings were conducted for the wild type and myb89-1 in the RNA-seq experiment. The following analysis was performed using the services of Gene Denovo (http://www.genedenovo.com/) following the standard protocol (http://www.genedenovo.com/product/41.html). The Excel add-in for significance analysis of RNA-seq was used to identify DEGs between the wild type and myb89-1. The DEGs were functionally classified using the biological process category of Arabidopsis Gene Ontology (http://www.geneontology.com). The DEGs with log2 ratios greater than 1 or less than −1 (only GO Slim identifiers with P ≤ 0.05 and false discovery rate ≤ 0.05) are listed in Supplemental Tables S3 and S4.
Expression Analysis
Total RNA from siliques or developing seeds was extracted using the MiniBEST Plant RNA Extraction Kit (TaKaRa) and reverse transcribed using PrimerScript RT (TaKaRa). qRT-PCR was performed for three biological replicates using SYBR Green Master Mix (TaKaRa). The siliques or developing seeds were from at least 12 individual plants grown in different pots arranged randomly, and three independent biological replicates from three different plantings were used for the expression analysis. qRT-PCR is the preferred method for targeted gene expression measurements because of its sensitivity and reproducibility. However, normalization using the internal control, which is necessary to correct for sample input and reverse transcriptase efficiency, is a crucial step to obtain reliable qRT-PCR results. EF1aA4 was regarded as the internal control in this study. However, the ubiquitously expressed genes AP2M (AT5G46630) and MON1 (AT2G28390) during seed development (Czechowski et al., 2005; Dekkers et al., 2012) might be more appropriate internal controls for gene expression measurements in Arabidopsis developing seeds. GUS staining were performed as described previously (Jefferson et al., 1987). In situ hybridization experiments were performed as described elsewhere (Yu et al., 2002). GR induction and RNA analysis were performed as reported previously (Xu et al., 2014). Primers used for in situ hybridization and qRT-PCR analyses are listed in Supplemental Tables S6 and S7, respectively.
ChIP Assay
The ChIP assay was performed as reported previously (Chen et al., 2015). Siliques were fixed on ice in a buffer (10 mm potassium phosphate [pH 7], 50 mm NaCl, and 0.1 m Suc) with 1% (v/v) formaldehyde under vacuum for 1 h. Chromatin was extracted and sonicated to produce DNA fragments of sizes about 250 bp. MYB89-6HA was immunoprecipitated using an anti-HA agarose conjugate (Sigma-Aldrich). The relative enrichment of each fragment was determined by qRT-PCR, and ChIP assays were repeated using three biological replicates. The siliques for the three independent biological replicates were from at least 50 individual plants grown in different pots arranged randomly and not a resample from the same 50 plants. Primer pairs used in the ChIP assays are listed in Supplemental Table S8.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: MYB89 (AT5G39700), WRI1 (AT3G54320), LEC1 (AT1G21970), LEC2 (AT1G28300), L1L (AT5G47670), FUS3 (AT3G26790), ABI3 (AT3G24650), CYFBP (AT1G43670), SUS2 (AT5G49190), FBA2 (AT4G38970), ENO1 (AT1G74030), PI-PKβ1 (AT5G52920), BCCP1 (AT5G16390), BCCP2 (AT5G15530), CAC2 (AT5G35360), CAC3 (AT2G38040), PDH E1α (AT1G01090), ACP1 (AT3G05020), KASI (AT5G46290), KASII (AT1G74960), KASIII (AT1G62640), KAR (AT1G24360), HD-L (AT2G22230), MOD1 (AT2G05990), SAD (AT3G02630), SAD (AT5G16240), LAS (AT5G08415), KCS11 (AT2G26640), KCS17 (AT4G34510), FAD2 (AT3G12120), FAD3 (AT2G29980), FAE1 (AT4G34520), SSI2 (AT2G43710), CDS2 (AT4G22340), PIS2 (AT4G38570), BIO2 (AT2G43360), LPAT1 (AT4G30580), LPAT2 (AT3G57650), LPAT3 (AT1G51260), LPAT4 (AT1G75020), LPAT5 (AT3G18850), LPP1 (AT2G01180), LPP2 (AT1G15080), LPP3 (AT3G02600), ROD1 (AT3G15820), PLA2α (AT2G06925), DGAT1 (AT2G19450), DGAT2 (AT3G51520), PDAT1 (AT5G13640), PDAT2 (AT3G44830), CCT1 (AT2G32260), and CCT2 (AT4G15130).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Representative GUS staining of pMYB89:GUS transgenic plants shows MYB89 expression in true leaves and a root tip of a 15-d-old seedling.
Supplemental Figure S2. Comparison of plant size between wild-type (Col-0) and myb89-1 plants.
Supplemental Figure S3. Comparison of wild-type (Col-0) and myb89-1 young seedlings under stressed environments.
Supplemental Figure S4. Effect of MYB89 knockdown in the wild-type (Col-0) background and MYB89 overexpression in the myb89-1 mutant background on seed size and seed weight.
Supplemental Figure S5. Effect of altering MYB89 expression on seed number, seed yield, and oil yield.
Supplemental Figure S6. qRT-PCR analysis of the expression of genes involved in FA biosynthesis and TAG deposition in developing seeds of wild-type (Col-0) and myb89-1 plants.
Supplemental Figure S7. Confirmation of the biologically active MYB89-GR fusion.
Supplemental Figure S8. The genes involved in glycolysis, FA biosynthesis and modification, and TAG deposition in developing seeds are not immediate targets of transcriptional regulation by MYB89 in developing seeds.
Supplemental Figure S9. qRT-PCR analysis of the expression of six BPM genes in developing seeds of wild-type (Col-0) and myb89-1 plants.
Supplemental Table S1. Quantitative comparison of the cell size of epidermal cell layers of the cotyledons from wild-type (Col-0) and myb89-1 mature embryos.
Supplemental Table S2. Seed major FA compositions of the mutants and transgenic plants in this study.
Supplemental Table S3. List of up-regulated genes in developing seeds of myb89-1 plants at 12 DAP.
Supplemental Table S4. List of down-regulated genes in developing seeds of myb89-1 plants at 12 DAP.
Supplemental Table S5. List of TFs and genes encoding key enzymes for oil biosynthesis whose expression was not altered in developing seeds of myb89-1 plants at 12 DAP.
Supplemental Table S6. Primers used for various constructs, in situ hybridization, and genotyping of SALK mutants in this study.
Supplemental Table S7. Primers used for qRT-PCR analysis in this study.
Supplemental Table S8. Primers used for ChIP analysis in this study.
Supplementary Material
Acknowledgments
We thank Dr. X. Du and Dr. W. Tong for assistance in operating equipment and H. Yu for critical reading of the article.
Glossary
- TAG
triacylglycerol
- TF
transcription factor
- FA
fatty acid
- VLCFAs
very-long-chain fatty acids
- qRT
quantitative real-time
- DAP
days after pollination
- Col-0
Columbia-0
- RNA-seq
RNA sequencing
- DEGs
differentially expressed genes
- ChIP
chromatin immunoprecipitation
Footnotes
This work was supported by the Natural Science Foundation of China (grant no. 31501336), the Science Fund for the Cultivation of Excellent Youth Scholars (grant no. Z109021517), and the Startup Fund for Talents of Northwest A&F University (grant no. Z111021402).
Articles can be viewed without a subscription.
References
- Adhikari ND, Bates PD, Browse J (2016) WRINKLED1 rescues feedback inhibition of fatty acid synthesis in hydroxylase-expressing seeds. Plant Physiol 171: 179–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andriotis VME, Pike MJ, Kular B, Rawsthorne S, Smith AM (2010) Starch turnover in developing oilseed embryos. New Phytol 187: 791–804 [DOI] [PubMed] [Google Scholar]
- Barthole G, To A, Marchive C, Brunaud V, Soubigou-Taconnat L, Berger N, Dubreucq B, Lepiniec L, Baud S (2014) MYB118 represses endosperm maturation in seeds of Arabidopsis. Plant Cell 26: 3519–3537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Dubreucq B, Miquel M, Rochat C, Lepiniec L (2008) Storage reserve accumulation in Arabidopsis: metabolic and developmental control of seed filling. The Arabidopsis Book 6: e0113, doi/10.1199/tab.0113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Lepiniec L (2009) Regulation of de novo fatty acid synthesis in maturing oilseeds of Arabidopsis. Plant Physiol Biochem 47: 448–455 [DOI] [PubMed] [Google Scholar]
- Baud S, Lepiniec L (2010) Physiological and developmental regulation of seed oil production. Prog Lipid Res 49: 235–249 [DOI] [PubMed] [Google Scholar]
- Baud S, Mendoza MS, To A, Harscoët E, Lepiniec L, Dubreucq B (2007) WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J 50: 825–838 [DOI] [PubMed] [Google Scholar]
- Baud S, Wuillème S, To A, Rochat C, Lepiniec L (2009) Role of WRINKLED1 in the transcriptional regulation of glycolytic and fatty acid biosynthetic genes in Arabidopsis. Plant J 60: 933–947 [DOI] [PubMed] [Google Scholar]
- Bonaventure G, Salas JJ, Pollard MR, Ohlrogge JB (2003) Disruption of the FATB gene in Arabidopsis demonstrates an essential role of saturated fatty acids in plant growth. Plant Cell 15: 1020–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braybrook SA, Stone SL, Park S, Bui AQ, Le BH, Fischer RL, Goldberg RB, Harada JJ (2006) Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc Natl Acad Sci USA 103: 3468–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J, Somerville C (1991) Glycerolipid synthesis: biochemistry and regulation. Ann Rev Plant Physiol Mol Bio 42: 467–506 [Google Scholar]
- Cernac A, Benning C (2004) WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J 40: 575–585 [DOI] [PubMed] [Google Scholar]
- Chapman KD, Ohlrogge JB (2012) Compartmentation of triacylglycerol accumulation in plants. J Biol Chem 287: 2288–2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Lee JH, Weber H, Tohge T, Witt S, Roje S, Fernie AR, Hellmann H (2013) Arabidopsis BPM proteins function as substrate adaptors to a cullin3-based E3 ligase to affect fatty acid metabolism in plants. Plant Cell 25: 2253–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Wang Z, Zhu Y, Li Z, Hussain N, Xuan L, Guo W, Zhang G, Jiang L (2012) The effect of TRANSPARENT TESTA2 on seed fatty acid biosynthesis and tolerance to environmental stresses during young seedling establishment in Arabidopsis. Plant Physiol 160: 1023–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Xuan L, Wang Z, Zhou L, Li Z, Du X, Ali E, Zhang G, Jiang L (2014) TRANSPARENT TESTA8 inhibits seed fatty acid accumulation by targeting several seed development regulators in Arabidopsis. Plant Physiol 165: 905–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Zhang B, Li C, Kulaveerasingam H, Chew FT, Yu H (2015) TRANSPARENT TESTA GLABRA1 regulates the accumulation of seed storage reserves in Arabidopsis. Plant Physiol 169: 391–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- daSilva PMFR, Eastmond PJ, Hill LM, Smith AM, Rawsthorne S (1997) Starch metabolism in developing embryos of oilseed rape. Planta 203: 480–487 [Google Scholar]
- Dekkers BJW, Willems L, Bassel GW, van Bolderen-Veldkamp RP, Ligterink W, Hilhorst HW, Bentsink L (2012) Identification of reference genes for RT-qPCR expression analysis in Arabidopsis and tomato seeds. Plant Cell Physiol 53: 28–37 [DOI] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15: 573–581 [DOI] [PubMed] [Google Scholar]
- Durrett TP, Benning C, Ohlrogge J (2008) Plant triacylglycerols as feedstocks for the production of biofuels. Plant J 54: 593–607 [DOI] [PubMed] [Google Scholar]
- Fait A, Angelovici R, Less H, Ohad I, Urbanczyk-Wochniak E, Fernie AR, Galili G (2006) Arabidopsis seed development and germination is associated with temporally distinct metabolic switches. Plant Physiol 142: 839–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focks N, Benning C (1998) wrinkled1: a novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol 118: 91–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM (1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4: 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham IA. (2008) Seed storage oil mobilization. Annu Rev Plant Biol 59: 115–142 [DOI] [PubMed] [Google Scholar]
- Grimberg Å, Carlsson AS, Marttila S, Bhalerao R, Hofvander P (2015) Transcriptional transitions in Nicotiana benthamiana leaves upon induction of oil synthesis by WRINKLED1 homologs from diverse species and tissues. BMC Plant Biol 15: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itabe H. (2010) Intracellular lipid droplet-associated proteins: unique members and their biological functions. Biol Pharm Bull 33: 341. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya Y, Toyoshima R, Okuda R, Usui H, Yamamoto A, Hattori T (2005) LEAFY COTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3. Plant Cell Physiol 46: 399–406 [DOI] [PubMed] [Google Scholar]
- Kroj T, Savino G, Valon C, Giraudat J, Parcy F (2003) Regulation of storage protein gene expression in Arabidopsis. Development 130: 6065–6073 [DOI] [PubMed] [Google Scholar]
- Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, Goldberg RB, Harada JJ (2003) LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15: 5–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Fischer RL, Goldberg RB, Harada JJ (2003) Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc Natl Acad Sci USA 100: 2152–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HG, Park BY, Kim HU, Seo PJ (2015) MYB96 stimulates C18 fatty acid elongation in Arabidopsis seeds. Plant Biotechnol Rep 9: 161–166 [Google Scholar]
- Li X, Ilarslan H, Brachova L, Qian HR, Li L, Che P, Wurtele ES, Nikolau BJ (2011) Reverse-genetic analysis of the two biotin-containing subunit genes of the heteromeric acetyl-coenzyme A carboxylase in Arabidopsis indicates a unidirectional functional redundancy. Plant Physiol 155: 293–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Beisson F, Pollard M, Ohlrogge J (2006) Oil content of Arabidopsis seeds: the influence of seed anatomy, light and plant-to-plant variation. Phytochemistry 67: 904–915 [DOI] [PubMed] [Google Scholar]
- Liu C, Chen H, Er HL, Soo HM, Kumar PP, Han JH, Liou YC, Yu H (2008) Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 135: 1481–1491 [DOI] [PubMed] [Google Scholar]
- Liu C, Zhou J, Bracha-Drori K, Yalovsky S, Ito T, Yu H (2007) Specification of Arabidopsis floral meristem identity by repression of flowering time genes. Development 134: 1901–1910 [DOI] [PubMed] [Google Scholar]
- Lotan T, Ohto M, Yee KM, West MAL, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93: 1195–1205 [DOI] [PubMed] [Google Scholar]
- Lu C, Napier JA, Clemente TE, Cahoon EB (2011) New frontiers in oilseed biotechnology: meeting the global demand for vegetable oils for food, feed, biofuel, and industrial applications. Curr Opin Biotechnol 22: 252–259 [DOI] [PubMed] [Google Scholar]
- Lu C, Xin Z, Ren Z, Miquel M, Browse J (2009) An enzyme regulating triacylglycerol composition is encoded by the ROD1 gene of Arabidopsis. Proc Natl Acad Sci USA 106: 18837–18842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Tang X, Huo Y, Xu R, Qi S, Huang J, Zheng C, Wu CA (2012) Identification and characterization of fructose 1,6-bisphosphate aldolase genes in Arabidopsis reveal a gene family with diverse responses to abiotic stresses. Gene 503: 65–74 [DOI] [PubMed] [Google Scholar]
- Luerssen H, Kirik V, Herrmann P, Miséra S (1998) FUSCA3 encodes a protein with a conserved VP1/AB13-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J 15: 755–764 [DOI] [PubMed] [Google Scholar]
- Maeo K, Tokuda T, Ayame A, Mitsui N, Kawai T, Tsukagoshi H, Ishiguro S, Nakamura K (2009) An AP2-type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW-box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. Plant J 60: 476–487 [DOI] [PubMed] [Google Scholar]
- Masaki T, Mitsui N, Tsukagoshi H, Nishii T, Morikami A, Nakamura K (2005) ACTIVATOR of Spomin:LUC1/WRINKLED1 of Arabidopsis thaliana transactivates sugar-inducible promoters. Plant Cell Physiol 46: 547–556 [DOI] [PubMed] [Google Scholar]
- Mendes A, Kelly AA, van Erp H, Shaw E, Powers SJ, Kurup S, Eastmond PJ (2013) bZIP67 regulates the omega-3 fatty acid content of Arabidopsis seed oil by activating fatty acid desaturase3. Plant Cell 25: 3104–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Z, He Y, Dai Y, Liu X, Li J (2000) Deficiency in fatty acid synthase leads to premature cell death and dramatic alterations in plant morphology. Plant Cell 12: 405–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J, Tan H, Zheng Q, Fu F, Liang Y, Zhang J, Yang X, Wang T, Chong K, Wang XJ, et al. (2008) LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol 148: 1042–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L (2001) The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13: 2099–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton G, Harris JF (1975) Compositional changes in developing rape seed (Brassica napus L.). Planta 123: 163–174 [DOI] [PubMed] [Google Scholar]
- Ohlrogge JB, Jaworski JG (1997) Regulation of fatty acid synthesis. Annu Rev Plant Physiol Plant Mol Biol 48: 109–136 [DOI] [PubMed] [Google Scholar]
- Ohto MA, Fischer RL, Goldberg RB, Nakamura K, Harada JJ (2005) Control of seed mass by APETALA2. Proc Natl Acad Sci USA 102: 3123–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuley J, Lightner J, Feldmann K, Yadav N, Lark E, Browse J (1994) Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell 6: 147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan YL, Tchagangl A, Berube H, Phan S, Shearer H, Liu ZY, Fobert P, Famili F (2010) Integrative data mining in functional genomics of Brassica napus and Arabidopsis thaliana. Lect Notes Artif Int 6098: 92–101 [Google Scholar]
- Periappuram C, Steinhauer L, Barton DL, Taylor DC, Chatson B, Zou J (2000) The plastidic phosphoglucomutase from Arabidopsis: a reversible enzyme reaction with an important role in metabolic control. Plant Physiol 122: 1193–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picciocchi A, Douce R, Alban C (2001) Biochemical characterization of the Arabidopsis biotin synthase reaction: the importance of mitochondria in biotin synthesis. Plant Physiol 127: 1224–1233 [PMC free article] [PubMed] [Google Scholar]
- Pidkowich MS, Nguyen HT, Heilmann I, Ischebeck T, Shanklin J (2007) Modulating seed beta-ketoacyl-acyl carrier protein synthase II level converts the composition of a temperate seed oil to that of a palm-like tropical oil. Proc Natl Acad Sci USA 104: 4742–4747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier Y, Ventre G, Caldelari D (1999) Increased flow of fatty acids toward beta-oxidation in developing seeds of Arabidopsis deficient in diacylglycerol acyltransferase activity or synthesizing medium-chain-length fatty acids. Plant Physiol 121: 1359–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommerrenig B, Popko J, Heilmann M, Schulmeister S, Dietel K, Schmitt B, Stadler R, Feussner I, Sauer N (2013) SUCROSE TRANSPORTER 5 supplies Arabidopsis embryos with biotin and affects triacylglycerol accumulation. Plant J 73: 392–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhakar V, Löttgert T, Gigolashvili T, Bell K, Flügge UI, Häusler RE (2009) Molecular and functional characterization of the plastid-localized phosphoenolpyruvate enolase (ENO1) from Arabidopsis thaliana. FEBS Lett 583: 983–991 [DOI] [PubMed] [Google Scholar]
- Reverdatto S, Beilinson V, Nielsen NC (1999) A multisubunit acetyl coenzyme A carboxylase from soybean. Plant Physiol 119: 961–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-González JA, Soto-Súarez M, García-Díaz Á, Romero-Puertas MC, Sandalio LM, Mérida Á, Thormählen I, Geigenberger P, Serrato AJ, Sahrawy M (2015) Disruption of both chloroplastic and cytosolic FBPase genes results in a dwarf phenotype and important starch and metabolite changes in Arabidopsis thaliana. J Exp Bot 66: 2673–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruuska SA, Girke T, Benning C, Ohlrogge JB (2002) Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14: 1191–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu SB. (2004) Phospholipid-derived signaling mediated by phospholipase A in plants. Trends Plant Sci 9: 229–235 [DOI] [PubMed] [Google Scholar]
- Ryu SB, Lee HY, Doelling JH, Palta JP (2005) Characterization of a cDNA encoding Arabidopsis secretory phospholipase A2-alpha, an enzyme that generates bioactive lysophospholipids and free fatty acids. Biochim Biophys Acta 1736: 144–151 [DOI] [PubMed] [Google Scholar]
- Salas JJ, Ohlrogge JB (2002) Characterization of substrate specificity of plant FatA and FatB acyl-ACP thioesterases. Arch Biochem Biophys 403: 25–34 [DOI] [PubMed] [Google Scholar]
- Sanjaya, Durrett TP, Weise SE, Benning C (2011) Increasing the energy density of vegetative tissues by diverting carbon from starch to oil biosynthesis in transgenic Arabidopsis. Plant Biotechnol J 9: 874–883 [DOI] [PubMed] [Google Scholar]
- Santos-Mendoza M, Dubreucq B, Baud S, Parcy F, Caboche M, Lepiniec L (2008) Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J 54: 608–620 [DOI] [PubMed] [Google Scholar]
- Santos Mendoza M, Dubreucq B, Miquel M, Caboche M, Lepiniec L (2005) LEAFY COTYLEDON 2 activation is sufficient to trigger the accumulation of oil and seed specific mRNAs in Arabidopsis leaves. FEBS Lett 579: 4666–4670 [DOI] [PubMed] [Google Scholar]
- Shah S, Xin Z, Browse J (1997) Overexpression of the FAD3 desaturase gene in a mutant of Arabidopsis. Plant Physiol 114: 1533–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Katavic V, Yu Y, Kunst L, Haughn G (2012) Arabidopsis glabra2 mutant seeds deficient in mucilage biosynthesis produce more oil. Plant J 69: 37–46 [DOI] [PubMed] [Google Scholar]
- Sonnewald U, Lerchl J, Zrenner R, Frommer W (1994) Manipulation of sink-source relations in transgenic plants. Plant Cell Environ 17: 649–658 [Google Scholar]
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ (2001) LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA 98: 11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H, Yang X, Zhang F, Zheng X, Qu C, Mu J, Fu F, Li J, Guan R, Zhang H, et al. (2011) Enhanced seed oil production in canola by conditional expression of Brassica napus LEAFY COTYLEDON1 and LEC1-LIKE in developing seeds. Plant Physiol 156: 1577–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To A, Joubès J, Barthole G, Lécureuil A, Scagnelli A, Jasinski S, Lepiniec L, Baud S (2012) WRINKLED transcription factors orchestrate tissue-specific regulation of fatty acid biosynthesis in Arabidopsis. Plant Cell 24: 5007–5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To A, Valon C, Savino G, Guilleminot J, Devic M, Giraudat J, Parcy F (2006) A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell 18: 1642–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Guo J, Lambert KN, Lin Y (2007) Developmental control of Arabidopsis seed oil biosynthesis. Planta 226: 773–783 [DOI] [PubMed] [Google Scholar]
- Wang Z, Chen M, Chen T, Xuan L, Li Z, Du X, Zhou L, Zhang G, Jiang L (2014) TRANSPARENT TESTA2 regulates embryonic fatty acid biosynthesis by targeting FUSCA3 during the early developmental stage of Arabidopsis seeds. Plant J 77: 757–769 [DOI] [PubMed] [Google Scholar]
- Wu GZ, Xue HW (2010) Arabidopsis β-ketoacyl-[acyl carrier protein] synthase I is crucial for fatty acid synthesis and plays a role in chloroplast division and embryo development. Plant Cell 22: 3726–3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Grain D, Bobet S, Le Gourrierec J, Thévenin J, Kelemen Z, Lepiniec L, Dubos C (2014) Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. New Phytol 202: 132–144 [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Kagaya Y, Usui H, Hobo T, Takeda S, Hattori T (2010) Diverse roles and mechanisms of gene regulation by the Arabidopsis seed maturation master regulator FUS3 revealed by microarray analysis. Plant Cell Physiol 51: 2031–2046 [DOI] [PubMed] [Google Scholar]
- Yu H, Ito T, Wellmer F, Meyerowitz EM (2004) Repression of AGAMOUS-LIKE 24 is a crucial step in promoting flower development. Nat Genet 36: 157–161 [DOI] [PubMed] [Google Scholar]
- Yu H, Xu Y, Tan EL, Kumar PP (2002) AGAMOUS-LIKE 24, a dosage-dependent mediator of the flowering signals. Proc Natl Acad Sci USA 99: 16336–16341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Cao X, Jia Q, Ohlrogge J (2016) FUSCA3 activates triacylglycerol accumulation in Arabidopsis seedlings and tobacco BY2 cells. Plant J 88: 95–107 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.