Significance
Despite long-standing interest in the contribution of cis-regulatory changes to adaptation, we still have a limited understanding of the selective importance and genomic determinants of cis-regulatory variation in natural populations. We use a combination of analyses of allele-specific expression and population genomic analyses to investigate the selective forces and genomic determinants of cis-regulatory variation in the outcrossing plant species Capsella grandiflora. We conclude that gene-specific functional constraints shape cis-regulatory variation and that genes with cis-regulatory variation are under relaxed purifying selection compared with other genes. Finally, we identify a link between gene body methylation and the extent of cis-regulatory constraint in natural populations.
Keywords: allele-specific expression, fitness effects, purifying selection, positive selection, gene body methylation
Abstract
Understanding the causes of cis-regulatory variation is a long-standing aim in evolutionary biology. Although cis-regulatory variation has long been considered important for adaptation, we still have a limited understanding of the selective importance and genomic determinants of standing cis-regulatory variation. To address these questions, we studied the prevalence, genomic determinants, and selective forces shaping cis-regulatory variation in the outcrossing plant Capsella grandiflora. We first identified a set of 1,010 genes with common cis-regulatory variation using analyses of allele-specific expression (ASE). Population genomic analyses of whole-genome sequences from 32 individuals showed that genes with common cis-regulatory variation (i) are under weaker purifying selection and (ii) undergo less frequent positive selection than other genes. We further identified genomic determinants of cis-regulatory variation. Gene body methylation (gbM) was a major factor constraining cis-regulatory variation, whereas presence of nearby transposable elements (TEs) and tissue specificity of expression increased the odds of ASE. Our results suggest that most common cis-regulatory variation in C. grandiflora is under weak purifying selection, and that gene-specific functional constraints are more important for the maintenance of cis-regulatory variation than genome-scale variation in the intensity of selection. Our results agree with previous findings that suggest TE silencing affects nearby gene expression, and provide evidence for a link between gbM and cis-regulatory constraint, possibly reflecting greater dosage sensitivity of body-methylated genes. Given the extensive conservation of gbM in flowering plants, this suggests that gbM could be an important predictor of cis-regulatory variation in a wide range of plant species.
Understanding the causes of regulatory variation is of major importance for many areas of biology and medicine (1). Much interest has centered on cis-regulatory variation, which has long been thought to be particularly important for adaptation (2–5). Like other quantitative traits, cis-regulatory variation is expected to be shaped by the interplay of mutation, selection, and drift. However, the relative importance of these forces remains unclear in most species.
Recently, prospects for quantifying cis-regulatory variation have greatly improved, and, as a result, ample heritable cis-regulatory variation has been identified in many species (6); this is resulting in a growing consensus that a large amount of standing cis-regulatory variation is under weak purifying selection (7–9). Clarifying why the impact of purifying selection varies across the genome is therefore important to understand the maintenance of cis-regulatory variation.
Variation in the intensity of purifying selection across the genome can result from differences in selective constraint that are due to the specific functions of the genes involved. For example, according to the dosage balance hypothesis, genes that encode interacting proteins are expected to experience stronger constraint than other genes (10). In yeast, there is evidence that purifying selection on expression noise constrains regulatory evolution of dosage-sensitive genes (11–13), and, in plants, dosage sensitivity affects the retention of duplicate genes following whole-genome duplication (14). However, many other genomic features, including expression level, tissue specificity and gene body methylation (gbM), are also known to be associated with constraint (15–18) and could affect cis-regulatory variation.
Variation in purifying selection can also result from broad, genome-scale forces that affect genes mainly as a result of their genomic environment, and not due to their specific function. For instance, in the self-fertilizing species Caenorhabditis elegans, variation in the impact of background selection across the genome had a major effect on the distribution of cis-regulatory variation across the genome (8). If background selection is important, then one might generally expect levels of cis-regulatory variation to be associated with recombination rate and/or gene density (19). At present, however, the relative importance of gene-level constraint vs. genome-scale evolutionary forces for the distribution of cis-regulatory variation remains unclear in most species.
In this study, we have investigated the selective importance and genomic correlates of common cis-regulatory variation in the outcrossing crucifer species Capsella grandiflora. This species is particularly well suited for studying differences in the impact of selection across the genome, as it has relatively low population structure (20) and a large, stable effective population size (21, 22). Indeed, selection on both protein-coding (23) and regulatory regions (18) is highly efficient in C. grandiflora, and high levels of polymorphism enhance the power to detect cis-regulatory variation and quantify selection. Genomic studies are facilitated by the close relationship between C. grandiflora and the selfing species Capsella rubella, for which a genome sequence is available (22).
Here, we identified genes with common cis-regulatory variation in C. grandiflora based on analyses of allele-specific expression (ASE) in deep transcriptome sequencing data. To quantify the impact of positive and purifying selection on genes with cis-regulatory variation, we conducted population genomic analyses of high-coverage whole-genome resequencing data from 32 C. grandiflora individuals. Finally, we identified genomic predictors of cis-regulatory variation. Our results show that there is pervasive cis-regulatory variation in C. grandiflora, and genes that harbor cis-regulatory variation are under weaker purifying selection and undergo less frequent positive selection than other genes. We find no evidence for a role of recombination rate or gene density in shaping cis-regulatory variation, suggesting that gene-specific variation in functional constraint is more important in this species. We further identify gbM as a major factor constraining cis-regulatory variation, whereas presence of nearby transposable elements (TEs) and tissue specificity of expression increase the odds of ASE. Our results provide evidence for a link between gbM and cis-regulatory constraint, possibly reflecting greater dosage sensitivity of body-methylated genes.
Results
Widespread Cis-Regulatory Variation in C. grandiflora.
To identify genes with cis-regulatory variation, we quantified ASE based on deep whole transcriptome sequencing data (total 95.2 Gbp with Q ≥ 30) from flower buds and leaves of three C. grandiflora F1s (SI Appendix, Table S1). Each F1 harbored an average of about 235,700 high-confidence heterozygous coding SNPs, which were phased before analyses of ASE. After filtering, ∼14,000 genes per F1 were amenable to ASE analyses (Table 1).
Table 1.
Genes amenable to analysis of ASE in flower buds (F) and leaves (L), and ASE results
| F1 | Analyzed* | ASE genes† | ASE prop.‡ | FDR |
| 6.3 F | 13,521 | 3,065 | 0.33 | 0.0013 |
| 7.2 F | 14,390 | 3,829 | 0.36 | 0.0024 |
| 8.2 F | 14,232 | 3,601 | 0.35 | 0.0020 |
| 6.3 L | 12,390 | 3,425 | 0.34 | 0.0018 |
| 7.2 L | 13,074 | 3,749 | 0.39 | 0.0024 |
| 8.2 L | 12,796 | 3,550 | 0.34 | 0.0020 |
Number of genes with expression data for at least one replicate, and with a phased fragment containing at least three transcribed SNPs after filtering.
Number of genes with posterior probability of ASE ≥ 0.95.
Estimated proportion of genes with ASE.
We assessed ASE using a Bayesian method (24), accounting for technical variation in allelic counts using high-coverage whole-genome resequencing data for each F1 (mean coverage of 40×, total 26.6 Gbp with Q ≥ 30; SI Appendix, Table S2). We estimated that a mean of 35% (range 33 to 39%) of analyzed genes show ASE in individual C. grandiflora F1s (Table 1). Similar proportions of genes had ASE in both leaves and flower buds (Table 1), and allelic expression biases were moderate for most genes with ASE, with strong allelic expression biases (0.2 ≤ ASE ratio ≥ 0.8) shown by an average of 5.1% of genes (Fig. 1 and SI Appendix, Figs. S1 and S2).
Fig. 1.
The magnitude of ASE (deviation from equal expression of both alleles) in (A and B) leaves and (C and D) flower buds of one representative C. grandiflora F1. A and C show the deviation from equal expression for all assayed genes. B and D show the magnitude of ASE for genes with strong evidence for ASE (posterior probability of ASE ≥ 0.95).
Out of a total of 11,532 genes that were amenable to analysis of ASE in all F1s, there were 1,010 genes that showed ASE in either leaves or flower buds, 313 genes showed that ASE in flower buds but not leaves, 404 genes that showed ASE in leaf samples but not flower buds, and 293 genes that had ASE in both flower buds and leaves of all F1s (SI Appendix, Fig. S3). Among the 1,010 genes with ASE leaves or flower buds of all F1s, one Gene Ontology (GO) category, GO:0006952, “defense response,” was significantly enriched at false discovery rate (FDR) ≤ 0.01; this was likely driven by genes with ASE in leaves, as there was no significant enrichment of GO terms among genes with ASE in flower buds, whereas six biological process GO terms associated with photosynthesis and defense responses were significantly enriched (FDR ≤ 0.01) among genes with ASE in leaves (SI Appendix, Table S3). Among genes without ASE, there was a nominally significant enrichment of genes in only two GO terms, protein binding (GO:0005515) and zinc ion binding (GO:0008270) (Weighted Fisher P ≤ 0.01), but this was not significant at FDR ≤ 0.01.
Lower Intensity of Purifying Selection on Genes with Cis-Regulatory Variation.
To assess the impact of selection on genes showing cis-regulatory variation in C. grandiflora, we sequenced the genomes of 21 individuals from one population in the Zagory region of Greece (the “population sample”) as well as 12 individuals from separate populations across the species range (the “range-wide sample”) using 233.2 Gbp of high-quality (Q ≥ 30) paired-end 100-bp Illumina reads and a mean coverage of 25× per individual (SI Appendix, Table S2).
We compared levels of polymorphism at genes that show ASE in all of our F1s (1,010 genes; “ASE genes”), using as a control set the 10,552 genes that were amenable to ASE analyses in all F1s but did not show significant ASE in leaves or flower buds (termed “control genes”) (SI Appendix, Fig. S3). To reduce bias resulting from the requirement of expressed polymorphisms for analyses of ASE, all population genetic analyses were conducted only on these paired gene sets, and genes that were not amenable to analysis of ASE were not included. ASE genes had elevated polymorphism levels compared with the control at all investigated site classes, as well as an elevated ratio of nonsynonymous to synonymous polymorphism (Table 2 and SI Appendix, Table S4), suggesting that the impact of purifying selection might differ between ASE and control genes (Table 2 and SI Appendix, Table S4).
Table 2.
Population genetic summary statistics and divergence estimates for the different site classes, separately for ASE and control genes
| Sites* | Genes | Mean θW | πsiteclass/π4-f† | d |
| Fourfold | ASE | 0.029 | NA | 0.16 |
| Control | 0.024 | NA | 0.15 | |
| Zerofold | ASE | 0.011 | 0.32 | 0.04 |
| Control | 0.007 | 0.23 | 0.03 | |
| 3′-UTR | ASE | 0.021 | 0.62 | 0.13 |
| Control | 0.018 | 0.62 | 0.12 | |
| 5′-UTR | ASE | 0.016 | 0.55 | 0.12 |
| Control | 0.012 | 0.54 | 0.12 | |
| 500 bp up | ASE | 0.020 | 0.6 | 0.16 |
| Control | 0.019 | 0.68 | 0.15 | |
| Intron | ASE | 0.022 | 0.69 | 0.15 |
| Control | 0.020 | 0.79 | 0.14 |
d, divergence between Capsella and Arabidopsis.
Class of sites investigated, including fourfold degenerate sites (fourfold), zerofold degenerate sites (zerofold), 5′-UTRs, 3′-UTRs, 500 bp upstream of the TSS (500 bp up), and introns.
Ratio of nucleotide diversity at focal site class to nucleotide diversity at fourfold synonymous sites.
To quantify the impact of purifying selection on ASE genes and control genes, we used the DFE-alpha method (25, 26), which allows estimation of a gamma distribution of negative fitness effects (DFE) based on site frequency spectra at putatively neutral and selected sites. We found that ASE genes have a significantly higher proportion of nearly neutral nonsynonymous mutations than control genes, as well as a significantly reduced proportion of nonsynonymous mutations under strong purifying selection (strength of purifying selection Nes > 10) (Fig. 2). This result applies broadly, both for the population and the range-wide samples, and when assuming a constant population size as well as after correcting for population size change (SI Appendix, Fig. S4). The result also holds after controlling for differences in the expression level among genes with and without ASE (SI Appendix, Figs. S5 and S6), when controlling for differences in coding polymorphism level (SI Appendix, Figs. S5 and S7), and when classifying genes based on a single F1 individual (SI Appendix, Fig. S8), suggesting that the results hold broadly for common cis-regulatory variation. Our results further remain unchanged after removing defense response genes (GO:0006952) with ASE (SI Appendix, Fig. S9) before DFE-alpha analyses, and thus strong balancing selection on these genes does not drive the patterns we observe.
Fig. 2.
The impact of purifying selection differs between genes with and without ASE in C. grandiflora. The estimated proportion of mutations in each bin of the distribution of negative fitness effects (DFE) is shown, with whiskers corresponding to 95% confidence intervals. The strength of purifying selection is given in units of the effective population size times the selection coefficient (Nes). Shown are the DFE for (A) nonsynonymous sites (zerofold degenerate sites), (B) introns, (C) promoter regions 500 bp upstream of the transcription start site, and (D) 3′-UTRs. Significance levels for comparisons of ASE and control genes are indicated by asterisks (*P ≤ 0.05; **P ≤ 0.01). These results are based on the population sample and the 1-epoch model.
In contrast to the clear evidence for weaker purifying selection on nonsynonymous sites for genes with ASE, there were no significant differences in the DFE depending on ASE status at 5′-UTRs (SI Appendix, Fig. S10). For introns, results were inconsistent, with some but not all analyses pointing to weaker purifying selection on control genes (Fig. 2 and SI Appendix, Fig. S10 and Table S5). This finding could suggest that patterns of selection differ among coding and noncoding regions. However, at noncoding regions other than introns, such as promoter regions 500 bp upstream of the transcription start site (TSS) and at 3′-UTRs, there was some evidence for relaxed purifying selection at ASE genes (Fig. 2 and SI Appendix, Fig. S10 and Table S5). These results held only under the 1-epoch model, which could in part be due to a lack of power, as regulatory motifs are expected to make up a small fraction of the analyzed sites. Consistent with this, we infer weaker purifying selection on upstream regions and UTRs than on nonsynonymous mutations (SI Appendix, Fig. S10 and Table S5).
Genes with Cis-Regulatory Variation Undergo Less Frequent Adaptive Evolution.
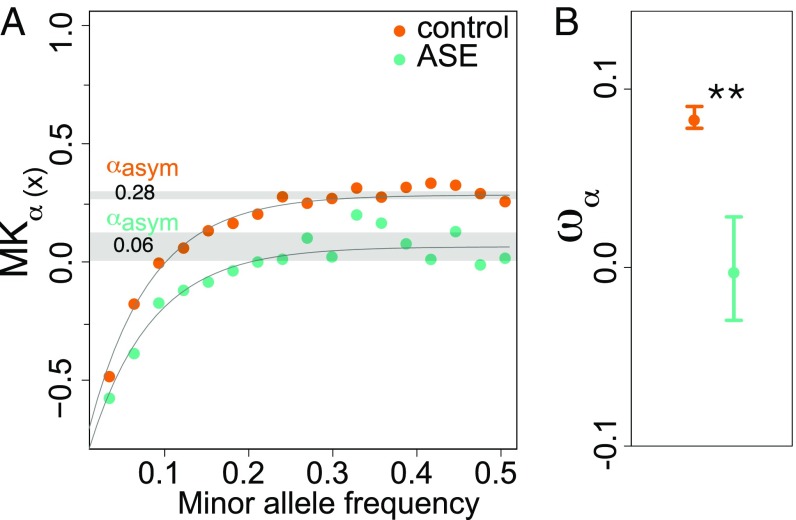
To investigate the impact of positive selection on genes with and without ASE, we obtained estimates of ωα, the rate of adaptive substitutions relative to neutral divergence (27) in DFE-alpha. For this purpose, we relied on genome-wide divergence between Capsella and Arabidopsis, with fourfold synonymous sites considered to be evolving mainly neutrally (Materials and Methods). Using this method, we find that ASE genes show a significantly lower proportion of adaptive nonsynonymous substitutions than do control genes (Fig. 3). In contrast, we found no significant differences in ωα among ASE genes or control genes for UTRs or regions 500 bp upstream of the TSS (SI Appendix, Table S5). Second, we estimated α, the proportion of adaptive fixations in the selected site class, based on the approximate method of ref. 28, designed to yield accurate estimates in the presence of linked selection. Results generated with this method were consistent with DFE-alpha, with a significantly lower estimate of the proportion of adaptive nonsynonymous substitutions at ASE genes than at control genes (Fig. 3).
Fig. 3.
A lower proportion of adaptive nonsynonymous fixations (α) at genes with ASE. (A) Estimation of α using an asymptotic method that fits an exponential function to estimates of α based on polymorphisms at different frequencies. Orange dots show values for control genes, and green dots show values for genes with ASE. The gray shaded area indicates 95% confidence intervals. The point estimate for genes with and without ASE is 0.06 and 0.28, respectively. (B) The estimated proportion of adaptive fixations relative to fourfold synonymous substitutions (ωα) for genes with and without ASE. Whiskers correspond to 95% confidence intervals, and significance levels for comparisons of ASE and control genes are indicated by asterisks (*P ≤ 0.05; **P ≤ 0.01).
Determinants of cis-Regulatory Variation in C. grandiflora.
To identify genomic factors and potential drivers of cis-regulatory variation, we conducted logistic regression analyses with presence/absence of ASE as the response variable. We included a total of 12 predictor variables, chosen to include proxies for variation in mutation rate, recombination rate, gene density, expression level, and degree of constraint, which could be expected to affect levels of cis-regulatory variation (Materials and Methods). The best-fit model based on the Akaike Information Criterion (AIC) retained eight of these predictor variables (Table 3). In this model, gbM had the greatest effect on cis-regulatory variation, resulting in a reduction of 49% in the odds of observing ASE (Table 3), whereas the presence of polymorphic TEs within 1 kb of the gene also had a substantial effect, increasing the odds of ASE by 38%, followed, in turn, by tissue specificity of expression, promoter diversity, expression level, gene length, and nonsynonymous/synonymous polymorphism, all of which increased the odds of ASE (Table 3). Including network connectivity improved model fit, although the effect was not individually significant (Table 3). Notably, gene density and recombination rate, which affect the intensity of linked selection, were not included in the best-fit model based on AIC (Table 3) or the Bayesian Information Criterion (BIC) (SI Appendix, Table S6) and had low importance based on model averaging (SI Appendix, Table S7). Similar results were obtained in an analysis that followed the approach of ref. 29 to ensure orthogonality of predictors by using principal components of all continuous predictors in logistic regression analyses (SI Appendix, Tables S8−S10). These analyses suggest that variation in gene-specific constraint is important for the distribution of cis-regulatory variation across the C. grandiflora genome, and that gbM and presence of nearby TEs are strong predictors of cis-regulatory constraint.
Table 3.
Predictor importance for the best-fit logistic regression model predicting ASE from genomic features, selected using AIC (AIC = 3,086.9)
| Model parameter | Coeff. (SE) | z value | P value | OR |
| gbM | −0.67 (0.20) | −3.41 | <10−3 | 0.51 |
| πN/πS | 0.08 (0.04) | 2.25 | 0.024 | 1.09 |
| Expression level | 0.20 (0.06) | 3.31 | <0.001 | 1.22 |
| Promoter polymorphism | 0.21 (0.05) | 4.45 | <10−3 | 1.23 |
| Tissue specificity | 0.30 (0.06) | 5.03 | <10−3 | 1.35 |
| TE within 1 kb | 0.32 (0.13) | 2.50 | 0.013 | 1.38 |
| Coexpression module size | −0.08 (0.05) | −1.59 | NS | 0.92 |
| Gene length | 0.08 (0.06) | 1.49 | NS | 1.09 |
| Intercept | −2.60 (0.06) | −42.91 | <10−3 | 0.07 |
Regression coefficients (Coeff.) and their SE, z statistics and associated P values, and odds ratios (OR) are shown.
Discussion
Our results show that genes that harbor common cis-regulatory variation in C. grandiflora are under weaker purifying selection and experience less frequent positive selection than other genes. We further find that gene-specific features that likely reflect the degree of functional constraint and mutational input are better predictors of cis-regulatory variation than those that are expected to shape the broad impact of linked selection across the genome. These functional constraints do not appear to limit the potential for adaptation at coding sequences, as positive selection had a greater impact on coding divergence at genes that did not exhibit common cis-regulatory variation in C. grandiflora.
Our findings support the view that most standing cis-regulatory variation in natural populations is weakly deleterious (7), and our robust inference of relaxed purifying selection on genes with common cis-regulatory variation agrees well with those of a recent expression quantitative trait locus mapping study in C. grandiflora (9). Our results are also complementary to previous findings in Arabidopsis, where genes with elevated divergence at upstream putative regulatory regions also show elevated rates of nonsynonymous divergence (30). Our inference of relaxed purifying selection on genes with common cis-regulatory variation does not appear to be driven by balancing selection or conditional neutrality affecting a subset of defense-related genes that show ASE, as our results remain unchanged after removing such genes.
The major association between gbM and cis-regulatory constraint that we detected is particularly interesting, because the function of gbM is currently unclear (31, 32). The conservation of gbM of orthologs in very distantly related plant species suggests that gbM has functional importance, but, intriguingly, some plants lack gbM (31–33). Body-methylated genes tend to be longer than other genes, are expressed at intermediate levels, evolve slowly at the sequence level (17, 34, 35), and are stably expressed under different conditions (36). A recent study found that Arapidopsis thaliana from northern Sweden show elevated gbM, mainly due to trans-acting loci (36), but, as far as we are aware, no study has directly linked gbM to cis-regulatory variation in natural plant populations.
It is possible that these associations between genomic features and cis-regulatory variation are caused by underlying drivers that were not directly measured. One natural candidate is gene essentiality. However, although gbM is significantly associated with predicted gene essentiality (37) (Fisher exact test P < 0.001), our results do not appear to be driven by essentiality, which was not retained in our best-fit logistic regression model for cis-regulatory variation. Instead, we hypothesize that selection for increased stability of expression of dosage-sensitive genes could underlie several of the associations we observe. Dosage-sensitive genes exhibit less expression noise (12, 38), show less variation in expression among tissues, and are expected to be part of larger regulatory network modules (10, 12). In our study, reduced tissue specificity of expression and increased network connectivity were associated with a reduced likelihood of ASE (Table 3). Furthermore, expression variation among three biological replicates of a C. rubella genotype (39) that likely represents mainly noise is significantly lower for genes with no ASE than for those with ASE [median coefficient of variation of fragments per kilobase of exon per million fragments mapped (FPKM) = 0.28 for genes with ASE, 0.18 for control genes, Wilcoxon rank sum test, P value < 10−5]. Finally, defense-related genes, which are thought to be dosage-insensitive in plants (40), were significantly enriched among genes with cis-regulatory variation in our study, whereas protein-binding genes were nominally enriched among control genes without ASE. Both promoter polymorphism and TE insertions, which can impact expression in several ways (41), might be more likely to be tolerated near dosage-insensitive genes. Our results are therefore consistent with dosage sensitivity causing strong constraint on cis-regulatory variation and shaping the impact of positive and purifying selection on coding variation. Thus, similar functional constraints that shape duplicate gene retention after whole-genome duplication (14) may also be key for the genomic distribution of cis-regulatory variation in natural plant populations. Future studies should explore the connection between dosage sensitivity, gbM, and cis-regulatory variation in greater detail across a wider range of plant species.
Materials and Methods
Plant Material.
For analyses of ASE, we generated three intraspecific C. grandiflora F1s by crossing six individuals sampled across the range of C. grandiflora (SI Appendix, Table S11). For population genomic analyses, we grew a single offspring from field-collected seeds of each of 32 plants (“the population genomic sample”; SI Appendix, Table S12), representing 21 plants from one population from Greece (the population sample), and 11 additional plants from 11 separate Greek populations covering the species’ range. Combined with an individual from the population sample, these represent a 12-plant range-wide sample. We grew plants at standard long-day conditions and collected leaves and mixed stage flower buds for RNA sequencing, and collected leaves for whole-genome sequencing as previously described (39).
Sample Preparation and Sequencing.
We extracted total RNA from the three intraspecific F1s using a Qiagen RNEasy Plant Mini Kit (Qiagen). RNAseq libraries were constructed using the TruSeq RNA v2 kit. For genomic resequencing, we extracted genomic DNA using a modified cetyl trimethyl ammonium bromide (CTAB) extraction method. Whole-genome sequencing libraries with an insert size of 300 to 400 bp were prepared using the TruSeq DNA v2 protocol. Sequencing of 100-bp paired-end reads was done on an Illumina HiSeq 2000 instrument. All sequence data have been submitted to the European Bioinformatics Institute (www.ebi.ac.uk), with study accession numbers PRJEB12070 and PRJEB12072.
Sequence Quality and Trimming.
RNA and DNA reads from the three F1s were trimmed as previously described (39). Adapters and low quality sequence were trimmed using CutAdapt 1.3. We analyzed genome coverage using BEDTools v.2.17.0 (42) and removed potential PCR duplicates using Picard v.1.92 (picard.sourceforge.net).
Read Mapping, Variant Calling, and Filtering.
We mapped RNAseq reads from the F1s to the v1.0 reference C. rubella assembly (22) using STAR software v.2.3.0.1 (43) with default parameters. For genomic reads from F1s, we mapped reads with STAR as in ref. 25. Genomic reads from the population genomic sample were mapped using BWA-MEM software v.0.7.12 (44) using default parameters and the –M flag.
Variant calling was done using GATK (The Genome Analysis Toolkit) UnifiedGenotyper (45–47). We conducted duplicate marking, local realignment around indels, and recalibrated base quality scores using a set of 1,538,085 SNPs identified in C. grandiflora (18) as known variants, and retained only SNPs considered high quality by GATK. An example script with all program versions and flags used for read mapping and variant calling is found on Figshare.
We removed centromeric and pericentromeric regions where we have low confidence in variant calls, and, before ASE analysis, we conducted additional filtering to remove SNPs that showed strongly biased allelic ratios in the genomic data and that were located in regions with overlapping genes, as in ref. 39. Using this procedure, we identified an average of 235,719 heterozygous coding SNPs in 17,973 genes in each F1. For population genomic analyses, we further filtered all genomic regions annotated as repeats using RepeatMasker 4.0.1, and removed sites with extreme coverage (depth of coverage < 15 or > 200) and too many missing individuals (≥20%) using VCFtools (48). Indels and nonbiallelic SNP were also pruned before analysis. Filtered vcf files are available on Figshare.
Expression Levels.
We mapped RNA seq reads of the three F1s to the C. rubella v.1.0 reference genome using TopHat v.2.0.4 (49) using standard settings. FPKM values were generated using Cufflinks v.2.0.2 (50) and standard settings. We estimated overall expression by taking the maximum of the FPKM measurements from leaves and flower buds in each F1, following ref. 16, and averaging these FPKM values over all F1s.
Phasing.
Before ASE analysis, we conducted read-backed phasing of genomic variants in F1s using GATK v. 2.5-2 ReadBackPhasing (-phaseQualityThresh 10). RNAseq data from all F1s were then phased by reference to the phased genomic variants. Read counts for all phased fragments were obtained using Samtools mpileup. This resulted in a mean number of 31,313 contiguous phased fragments per F1 (Table 1).
To validate our phasing procedure, we compared the phased fragments, based on reads, with the phased chromosomes, based on heritage, in three interspecific C. grandiflora × C. rubella F1s from ref. 39. For most genes, over 95% of SNPs were correctly phased in the interspecific F1s, demonstrating that our phasing procedure is reliable (SI Appendix, Figs. S11 and S12). Example scripts and phased vcf files are available on Figshare.
Analyses of Allele-Specific Expression.
We analyzed ASE using a hierarchical Bayesian method that requires phased data, in the form of read counts at heterozygous SNPs for genomic and transcriptomic data (24). Genomic read counts are used to obtain an empirical estimate of technical variation, which is then used in analyses of the RNAseq data. We used this method to estimate the posterior probability and degree of ASE, for the longest phased fragment per gene with at least three transcribed SNPs. We excluded genes with no read counts at the phased SNPs and analyzed ∼14,000 genes for ASE in flower buds, and ∼13,400 genes in leaves (Table 1). We ran three independent chains per sample with 200,000 iterations sampled every 1,000 generations, resulting in a final posterior of 2,000 samples per chain. We checked that the three chains converged to the same stationary distribution, with sufficient mixing, by inspecting the trace plot for each parameter and estimating the effective sample size. We used Gelman−Rubin−Brooks plots (51) to estimate a shrink factor among chains (SI Appendix, Fig. S13) as implemented in the R package “coda” (52). Runs were completed on a high-performance computing cluster at Uppsala University (UPPMAX) using the pqR version of R (www.pqr-project.org). The first 10% of each run was discarded as burn-in, and parameter estimates were then obtained as in ref. 24.
Population Genomic Analyses.
To assess whether patterns of polymorphism differ among ASE and control genes, we tested for a difference in median levels of polymorphism and Tajima’s D in the C. grandiflora population sample, using Mann−Whitney u tests, with Benjamini−Hochberg correction (53) for multiple comparisons. Estimates of nucleotide diversity (π), Watterson's theta (θW), and Tajima's D (DT) were obtained using custom R scripts. Separate estimates were obtained for six classes of sites: fourfold degenerate sites, zerofold degenerate sites, 3′- and 5′-UTRs, introns, and intergenic regions 500 bp upstream of the TSS.
Selection on Genes with ASE.
To test whether there was evidence for a difference in the strength and direction of natural selection on ASE and control genes, we first estimated the distribution of fitness effects (DFE) as in ref. 25, and the proportion of adaptive substitutions relative to the total number of synonymous substitutions (ωα) (27). The DFE was estimated under a constant population size model and under a model with stepwise population size change. We obtained confidence intervals for our estimates of three bins of the DFE (0 < Nes < 1; 1 < Nes < 10; 10 < Nes) and for α and ωα by resampling genes in 200 bootstrap replicates and tested for a difference in the DFE, and ωα among sets of genes with ASE and control genes, as in ref. 26. Separate estimates were obtained for zerofold degenerate sites, 3′- and 5′-UTRs, introns, and promoter regions 500 bp upstream of the TSS likely enriched for regulatory elements, using fourfold degenerate sites as neutral standard. For estimates of α and ωα, we relied on divergence to Arabidopsis; specifically, we generated a whole-genome alignment using lastz v. 1.03.54, with chaining of C. rubella, A. thaliana, and Arabidopsis lyrata as described in ref. 54, and counted divergence differences and sites as in ref. 18. DFE-alpha analyses were run using Method I (26).
To assess the effect of expression level on our DFE-alpha inference, we selected genes among the control set of genes to match the distribution of expression levels of ASE genes (SI Appendix, Fig. S5). We first assigned genes to 10 equal-sized bins with respect to overall expression level (average FPKM over all F1s). We calculated the proportion of genes in each bin for the ASE gene set, and then subsampled control genes to achieve the same proportion of genes in each bin as in the ASE gene set. Finally, we excluded extreme bins (first and last) for both ASE and control genes. Purifying and positive selection were then reestimated in DFE-alpha for the subsampled control gene set and ASE set.
To assess whether the differences in purifying selection we observed could be an artifact of higher power to detect ASE for high-polymorphism genes, we used a similar strategy of subsampling control gene sets to match the distribution of π in control and ASE gene sets (SI Appendix, Fig. S5). We then reran DFE-alpha on the matched gene sets. To assess whether our results were robust to the sampling strategy for ASE, we based our classification of ASE and control genes based on a single F1 individual and repeated the DFE analyses. To test whether our results could be driven by the inclusion of defense-related genes, we removed genes annotated as defense response genes (GO:0006952), and repeated the DFE-alpha analyses.
Genomic Determinants of Cis-Regulatory Variation.
We assessed the relative importance of a number of genomic features for presence/absence of ASE using logistic regression on a set of genes that was restricted to those for which we could assess ASE. We included the following genomic features that may affect linked selection: recombination rate and gene density (in 50-kb windows). Gene density was based on the annotation of C. rubella v1.0 reference genome (22). By fitting a smooth spline, we obtained recombination rates per 50-kb windows based on 878 markers from ref. 55. We further included gene length, tissue specificity (τ; ref. 16), expression level (log FPKM values), and as a proxy for mutation rate variation, we included fourfold synonymous divergence to Arabidopsis (dS). Because promoter polymorphism may cause cis-regulatory variation, we included nucleotide diversity (π) for the region 500 bp upstream of the TSS. We included nonsynonymous/synonymous nucleotide diversity (πN/πS) to reflect the level of constraint at the coding sequence level. According to the dosage balance hypothesis, genes in smaller coexpression modules may be under reduced regulatory constraint. We therefore included information on A. thaliana coexpression module size (37) in our analyses. We further included information on the presence of retained paralogs from the Brassicaceae α whole-genome duplication or the β and γ whole-genome duplication (37). We identified a set of genes with gbM in both C. rubella (33) and A. thaliana (17), which are highly likely to also harbor gbM in C. grandiflora. Finally, we included information on polymorphic TEs within 1 kb of genes in the range-wide sample. We identified TE insertions in our range-wide sample as in ref. 39, except that we required a minimum of five reads to call a TE insertion. A collated data set with all of these variables is available on Figshare. All continuous variables were centered and scaled before analysis. We conducted a logistic regression with ASE as the response variable and genomic features as predictors. We conducted model selection using a stepwise procedure with backward and forward selection of variables to find the best-fit model, using AIC and BIC as selection criteria. We also evaluated all possible models, ranked them using BIC and AIC, and calculated average coefficient and variable importance based on the relative weight of each model. We conducted an additional analysis using a strategy that is superior to partial correlation and robust in the presence of noisy genomic data and multicollinearity of predictor variables (29). We used a set of orthogonal predictor variables obtained by identifying principal components for a data set including all of the continuous variables using the “pls” package in R, as well as gbM and presence of heterozygous TEs as binary factors, and conducted AIC model selection as described above. Code for these analyses is found on Figshare.
Supplementary Material
Acknowledgments
We thank Lauren McIntyre and Stephen Wright for valuable discussion, Daniel Halligan for sharing scripts for DFE-alpha analyses, and Daniel Skelly for advice on ASE analyses. We thank Veronika Scholz and Michael Nowak for bioinformatic assistance and Julia Dankanich and Cindy Canton for experimental assistance. Sequencing was performed by the SNP&SEQ Technology Platform in Uppsala. Computations were performed on resources provided by Science for Life Laboratory and the Swedish National Infrastructure for Computing (SNIC) through UPPMAX under Projects b2012122 and b2012190. This study was funded by grants from the Swedish Research Council, the Nilsson-Ehle Foundation, the Magnus Bergvall Foundation, and the Erik Philip-Sörensen Foundation (to T.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All sequence data have been submitted to the European Bioinformatics Institute (www.ebi.ac.uk; accession nos. PRJEB12070 and PRJEB12072). An example script with all program versions and flags used for bioinformatic analyses, filtered vcf files, and a collated dataset and scripts used to identify genomic predictors of ASE are available on Figshare: https://doi.org/10.17045/sthlmuni.c.3654650.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612561114/-/DCSupplemental.
References
- 1.Albert FW, Kruglyak L. The role of regulatory variation in complex traits and disease. Nat Rev Genet. 2015;16(4):197–212. doi: 10.1038/nrg3891. [DOI] [PubMed] [Google Scholar]
- 2.King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188(4184):107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 3.Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8(3):206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- 4.Carroll SB. Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell. 2008;134(1):25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 5.Wittkopp PJ, Kalay G. Cis-regulatory elements: Molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet. 2011;13(1):59–69. doi: 10.1038/nrg3095. [DOI] [PubMed] [Google Scholar]
- 6.Fraser HB. Genome-wide approaches to the study of adaptive gene expression evolution: Systematic studies of evolutionary adaptations involving gene expression will allow many fundamental questions in evolutionary biology to be addressed. BioEssays. 2011;33(6):469–477. doi: 10.1002/bies.201000094. [DOI] [PubMed] [Google Scholar]
- 7.Fay JC, Wittkopp PJ. Evaluating the role of natural selection in the evolution of gene regulation. Heredity (Edinb) 2008;100(2):191–199. doi: 10.1038/sj.hdy.6801000. [DOI] [PubMed] [Google Scholar]
- 8.Rockman MV, Skrovanek SS, Kruglyak L. Selection at linked sites shapes heritable phenotypic variation in C. elegans. Science. 2010;330(6002):372–376. doi: 10.1126/science.1194208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Josephs EB, Lee YW, Stinchcombe JR, Wright SI. Association mapping reveals the role of purifying selection in the maintenance of genomic variation in gene expression. Proc Natl Acad Sci USA. 2015;112(50):15390–15395. doi: 10.1073/pnas.1503027112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Birchler JA, Veitia RA. Gene balance hypothesis: Connecting issues of dosage sensitivity across biological disciplines. Proc Natl Acad Sci USA. 2012;109(37):14746–14753. doi: 10.1073/pnas.1207726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lemos B, Meiklejohn CD, Hartl DL. Regulatory evolution across the protein interaction network. Nat Genet. 2004;36(10):1059–1060. doi: 10.1038/ng1427. [DOI] [PubMed] [Google Scholar]
- 12.Lehner B. Selection to minimise noise in living systems and its implications for the evolution of gene expression. Mol Syst Biol. 2008;4(1):170. doi: 10.1038/msb.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Metzger BPH, Yuan DC, Gruber JD, Duveau F, Wittkopp PJ. Selection on noise constrains variation in a eukaryotic promoter. Nature. 2015;521(7552):344–347. doi: 10.1038/nature14244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, et al. Gene duplicability of core genes is highly consistent across all angiosperms. Plant Cell. 2016;28(2):326–344. doi: 10.1105/tpc.15.00877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rocha EPC. The quest for the universals of protein evolution. Trends Genet. 2006;22(8):412–416. doi: 10.1016/j.tig.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 16.Slotte T, et al. Genomic determinants of protein evolution and polymorphism in Arabidopsis. Genome Biol Evol. 2011;3:1210–1219. doi: 10.1093/gbe/evr094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takuno S, Gaut BS. Body-methylated genes in Arabidopsis thaliana are functionally important and evolve slowly. Mol Biol Evol. 2012;29(1):219–227. doi: 10.1093/molbev/msr188. [DOI] [PubMed] [Google Scholar]
- 18.Williamson RJ, et al. Evidence for widespread positive and negative selection in coding and conserved noncoding regions of Capsella grandiflora. PLoS Genet. 2014;10(9):e1004622. doi: 10.1371/journal.pgen.1004622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slotte T. The impact of linked selection on plant genomic variation. Brief Funct Genomics. 2014;13(4):268–275. doi: 10.1093/bfgp/elu009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.St Onge KR, Källman T, Slotte T, Lascoux M, Palmé AE. Contrasting demographic history and population structure in Capsella rubella and Capsella grandiflora, two closely related species with different mating systems. Mol Ecol. 2011;20(16):3306–3320. doi: 10.1111/j.1365-294X.2011.05189.x. [DOI] [PubMed] [Google Scholar]
- 21.Foxe JP, et al. Recent speciation associated with the evolution of selfing in Capsella. Proc Natl Acad Sci USA. 2009;106(13):5241–5245. doi: 10.1073/pnas.0807679106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slotte T, et al. The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat Genet. 2013;45(7):831–835. doi: 10.1038/ng.2669. [DOI] [PubMed] [Google Scholar]
- 23.Slotte T, Foxe JP, Hazzouri KM, Wright SI. Genome-wide evidence for efficient positive and purifying selection in Capsella grandiflora, a plant species with a large effective population size. Mol Biol Evol. 2010;27(8):1813–1821. doi: 10.1093/molbev/msq062. [DOI] [PubMed] [Google Scholar]
- 24.Skelly DA, Johansson M, Madeoy J, Wakefield J, Akey JM. A powerful and flexible statistical framework for testing hypotheses of allele-specific gene expression from RNA-seq data. Genome Res. 2011;21(10):1728–1737. doi: 10.1101/gr.119784.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keightley PD, Eyre-Walker A. Joint inference of the distribution of fitness effects of deleterious mutations and population demography based on nucleotide polymorphism frequencies. Genetics. 2007;177(4):2251–2261. doi: 10.1534/genetics.107.080663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eyre-Walker A, Keightley PD. Estimating the rate of adaptive molecular evolution in the presence of slightly deleterious mutations and population size change. Mol Biol Evol. 2009;26(9):2097–2108. doi: 10.1093/molbev/msp119. [DOI] [PubMed] [Google Scholar]
- 27.Gossmann TI, et al. Genome wide analyses reveal little evidence for adaptive evolution in many plant species. Mol Biol Evol. 2010;27(8):1822–1832. doi: 10.1093/molbev/msq079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Messer PW, Petrov DA. Frequent adaptation and the McDonald−Kreitman test. Proc Natl Acad Sci USA. 2013;110(21):8615–8620. doi: 10.1073/pnas.1220835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drummond DA, Raval A, Wilke CO. A single determinant dominates the rate of yeast protein evolution. Mol Biol Evol. 2006;23(2):327–337. doi: 10.1093/molbev/msj038. [DOI] [PubMed] [Google Scholar]
- 30.Yang L, Takuno S, Waters ER, Gaut BS. Lowly expressed genes in Arabidopsis thaliana bear the signature of possible pseudogenization by promoter degradation. Mol Biol Evol. 2011;28(3):1193–1203. doi: 10.1093/molbev/msq298. [DOI] [PubMed] [Google Scholar]
- 31.Takuno S, Ran J-H, Gaut BS. Evolutionary patterns of genic DNA methylation vary across land plants. Nat Plants. 2016;2(2):15222. doi: 10.1038/nplants.2015.222. [DOI] [PubMed] [Google Scholar]
- 32.Bewick AJ, et al. On the origin and evolutionary consequences of gene body DNA methylation. Proc Natl Acad Sci USA. 2016;113(32):9111–9116. doi: 10.1073/pnas.1604666113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niederhuth CE, et al. Widespread natural variation of DNA methylation within angiosperms. Genome Biol. 2016;17(1):194. doi: 10.1186/s13059-016-1059-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takuno S, Gaut BS. Gene body methylation is conserved between plant orthologs and is of evolutionary consequence. Proc Natl Acad Sci USA. 2013;110(5):1797–1802. doi: 10.1073/pnas.1215380110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seymour DK, Koenig D, Hagmann J, Becker C, Weigel D. Evolution of DNA methylation patterns in the Brassicaceae is driven by differences in genome organization. PLoS Genet. 2014;10(11):e1004785. doi: 10.1371/journal.pgen.1004785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dubin MJ, et al. DNA methylation in Arabidopsis has a genetic basis and shows evidence of local adaptation. eLife. 2015;4:e05255. doi: 10.7554/eLife.05255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lloyd JP, Seddon AE, Moghe GD, Simenc MC, Shiu S-H. Characteristics of plant essential genes allow for within- and between-species prediction of lethal mutant phenotypes. Plant Cell. 2015;27(8):2133–2147. doi: 10.1105/tpc.15.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fraser HB, Hirsh AE, Giaever G, Kumm J, Eisen MB. Noise minimization in eukaryotic gene expression. PLoS Biol. 2004;2(6):e137. doi: 10.1371/journal.pbio.0020137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steige KA, Reimegård J, Koenig D, Scofield DG, Slotte T. Cis-regulatory changes associated with a recent mating system shift and floral adaptation in Capsella. Mol Biol Evol. 2015;32(10):2501–2514. doi: 10.1093/molbev/msv169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coate JE, Song MJ, Bombarely A, Doyle JJ. Expression-level support for gene dosage sensitivity in three Glycine subgenus Glycine polyploids and their diploid progenitors. New Phytol. 2016;212(4):1083–1093. doi: 10.1111/nph.14090. [DOI] [PubMed] [Google Scholar]
- 41.Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9(5):397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v2.
- 45.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van der Auwera GA, et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Danecek P, et al. 1000 Genomes Project Analysis Group The variant call format and VCFtools. Bioinformatics. 2011;27(15):2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brooks SP, Gelman A. General methods for monitoring convergence of iterative simulations. J Comput Graph Stat. 1998;7(4):434–455. [Google Scholar]
- 52.Plummer M, Best N, Cowles K, Vines K. CODA: Convergence diagnosis and output analysis for MCMC. R News. 2006;6(1):7–11. [Google Scholar]
- 53.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57(1):289–300. [Google Scholar]
- 54.Haudry A, et al. An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nat Genet. 2013;45(8):891–898. doi: 10.1038/ng.2684. [DOI] [PubMed] [Google Scholar]
- 55.Slotte T, Hazzouri KM, Stern D, Andolfatto P, Wright SI. Genetic architecture and adaptive significance of the selfing syndrome in Capsella. Evolution. 2012;66(5):1360–1374. doi: 10.1111/j.1558-5646.2011.01540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.