Abstract
Whole-genome sequencing (WGS) allows for a comprehensive view of the sequence of the human genome. We present and apply integrated methodologic steps for interrogating WGS data to characterize the genetic architecture of 10 heart- and blood-related traits in a sample of 1,860 African Americans. In order to evaluate the contribution of regulatory and non-protein coding regions of the genome, we conducted aggregate tests of rare variation across the entire genomic landscape using a sliding window, complemented by an annotation-based assessment of the genome using predefined regulatory elements and within the first intron of all genes. These tests were performed treating all variants equally as well as with individual variants weighted by a measure of predicted functional consequence. Significant findings were assessed in 1,705 individuals of European ancestry. After these steps, we identified and replicated components of the genomic landscape significantly associated with heart- and blood-related traits. For two traits, lipoprotein(a) levels and neutrophil count, aggregate tests of low-frequency and rare variation were significantly associated across multiple motifs. For a third trait, cardiac troponin T, investigation of regulatory domains identified a locus on chromosome 9. These practical approaches for WGS analysis led to the identification of informative genomic regions and also showed that defined non-coding regions, such as first introns of genes and regulatory domains, are associated with important risk factor phenotypes. This study illustrates the tractable nature of WGS data and outlines an approach for characterizing the genetic architecture of complex traits.
Keywords: whole-genome sequence, quantitative traits, genomic analysis
Introduction
Common complex traits, such as blood glucose and cholesterol levels, underlie some of the most common diseases burdening human health. Genetic analysis of these complex traits has followed the development of the fields of genetics and genomics, beginning with familial aggregation and linkage, transitioning through candidate genes and genome-wide association studies (GWASs), and arriving at the emerging promise of whole-genome sequencing (WGS). Declining costs have catalyzed accelerated adoption of WGS in large-scale genetics studies. However, few studies have utilized WGS to assess the contribution of low-frequency and rare genetic variation to complex traits.
Morrison et al.1 conducted WGS analysis of high-density lipoprotein cholesterol and described initial steps for an unbiased and coordinated approach to evaluating WGS data in a population-based sample of European Americans (EA). The UK10K Consortium explored association testing of common, low-frequency, and rare variants for quantitative traits using WGS data among European individuals.2 These initial studies, along with the results of numerous GWASs, support more comprehensive evaluation of non-coding regions in relation to complex quantitative traits, and also suggest that tests of association involving WGS would benefit from variant selection strategies that incorporate annotation of functional genomic elements. In fact, WGS analyses conducted in deeply phenotyped sample sets may be an efficient strategy for fine-mapping established GWAS signals. However, association studies involving WGS are challenged by the large number of very rare variants, especially singletons,3 and tests that aggregate the cumulative effects of rare variants have been proposed and implemented.4 These aggregate tests require an a priori defined region of the genome within which the combined effect of rare variants are assessed, and by far the most common units are the protein-encoding genes. WGS data offer the opportunity to aggregate variants over the full spectrum of annotated motifs, from specifically defined regulatory domains to an agnostic sliding window. In this study, we offer practical approaches to WGS analysis of complex traits using aggregate tests across a variety of annotated functional motifs. We also show how WGS may be informative for fine-mapping loci associated with traits of interest and identification of presumed single-nucleotide variants (SNVs) responsible for the observed associations. In addition, we consider weighted analyses using nucleotide-specific information and provide guidance on p values for defining thresholds of statistical significance. Because previous applications have focused on samples from populations of European descent, we provide an example application in a sample of African Americans (AA) measured for multiple cardiovascular risk factors.
Subjects and Methods
Study Population and Phenotype Measurements
The Atherosclerosis Risk in Communities (ARIC) study has been described in detail previously.5 In brief, participants aged 45 to 64 years at baseline were recruited from four communities: Forsyth County, North Carolina; Jackson, Mississippi; Minneapolis, Minnesota; and Washington County, Maryland. A total of 15,792 individuals, predominantly of European and African ancestry, participated in the baseline examination in 1987–1989, with four follow-up examinations. The example application presented here focuses on 1,860 AA study participants with WGS data and measurements for 10 heart- and blood-related factors, including circulating neutrophil count, platelet count, and levels of hemoglobin, lipoprotein(a) (Lp(a)), magnesium (Mg), and phosphorus (P) that were measured at the baseline exam. Small dense low-density lipoprotein cholesterol (sdLDL-C), C-reactive protein (CRP), cardiac troponin T (cTnT), and N-terminal pro-B-type natriuretic peptide (NT-proBNP) were measured at the fourth examination between 1996 and 1998. Detailed descriptions of the methods for each phenotype measurement are summarized in the Supplemental Note. A sample of 1,705 EA individuals with WGS and measures for the 10 heart- and blood-related factors were available for replication analyses. The ARIC study has been approved by Institutional Review Boards (IRBs) at all participating institutions: University of North Carolina at Chapel Hill IRB, Johns Hopkins University IRB, University of Minnesota IRB, and the University of Mississippi Medical Center IRB. Study participants provided written informed consent at all study visits.
Whole-Genome Sequencing, Variant Calling, and Quality Control
WGS data were generated by the Baylor College of Medicine Human Genome Sequencing Center. DNA samples were constructed into Illumina paired-end libraries according to the manufacturer’s protocol (Illumina Multiplexing_SamplePrep_Guide_1005361_D) and sequenced on the Hiseq 2000 (Illumina) in a pooled format to generate a minimum of 18 unique aligned Gbp per sample. Methods for WGS of ARIC study participants have been described in detail in Morrison et al.1 Individuals of African and European ancestry were sequenced at 7.4-fold average depth on Illumina HiSeq instruments and variant calling was completed using goSNAP, which employed GATK, SNPTools, and GotCloud as callers, each in joint calling mode, and took an ensemble consensus approach to generate a high-quality variant call set. Per-sample genotyping and reference-panel-independent imputation and phasing were done using SNPTools. The majority (59.7%) of the SNVs were novel compared to dbSNP v142. Compared to a subset of the samples with whole-exome sequencing, the sensitivity and specificity of the WGS call set is 63.6% and 99.9%, respectively, and compared to an overlapping set of single-nucleotide polymorphism (SNP) array data, the false discovery rate (FDR) is 1.6%. Variant-level quality assurance was achieved by excluding variants with a site level inbreeding coefficient < −0.9. Variants not meeting Hardy-Weinberg equilibrium exact test expectations in ancestry-specific groups (p value < 1 × 10−14) were also excluded. Sample-level quality control and quality assurance checks included principal-component analysis (PCA) to identify possible population substructure and sample abnormalities. The set of variants for PCA was restricted to variants with minor allele frequency (MAF) >5% and linkage disequilibrium between variants of r2 < 0.30. A total of 40 ARIC AA individuals identified as outliers by PCA were removed from further analyses. Higher-order principal components showed minor levels of population structure. After sample-level quality control, a total of 1,860 AA and 1,705 EA from the ARIC study were available for the genotype-phenotype analyses reported here.
Statistical Analyses
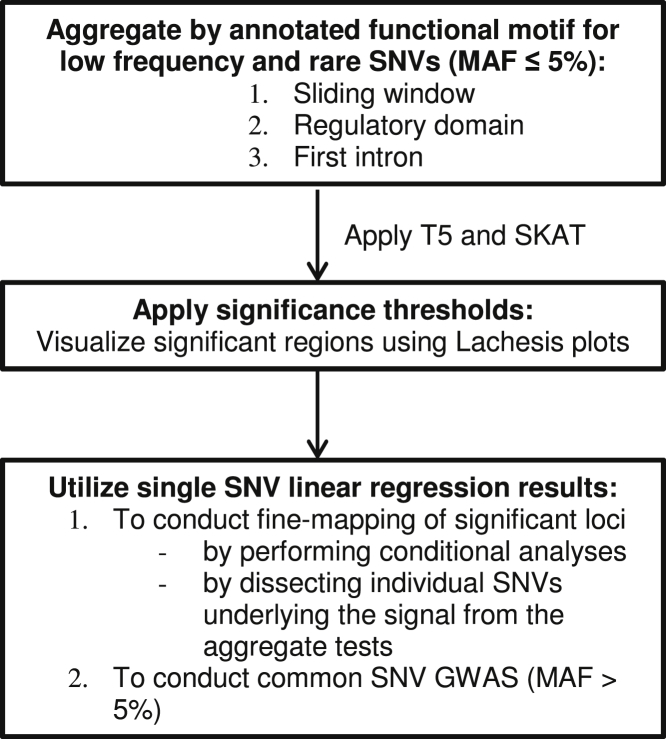
Each of the ten cardiovascular risk factors were analyzed separately. Figure 1 shows each step for the overall analytic approach. Because our primary focus was on rare variant sequence analysis, analyses within annotated functional motifs considered only low-frequency and rare variants (MAF ≤ 5%), and we required that the aggregate set of SNVs had an overall minor allele count (MAC) of ≥3. Within each annotated functional motif, a burden test (T56) and the Sequence Kernel Association Test (SKAT7) were used adjusting for age, sex, and the first three principal components (PCs), with additional adjustment of body mass index (BMI) for CRP and current smoking status (yes or no) for neutrophil counts. The T5 test collapses variants with MAF ≤ 5% into a single genetic score, while SKAT takes into account the possibility that the effects of the SNVs are in both directions. As a default, SKAT weights the variants according to their MAF using beta density weights with parameters 1 and 25. For completeness, we also conducted an additional survey of the genome investigating all individual variants using an additive genetic model with the same adjustments, and provide a focused look specifically at those with MAF > 5%. All analyses were carried out using the R seqMeta package. The results from the single SNV analyses were used to conduct focused fine-mapping at significant loci, including conditional analyses as well as to aid in the dissection of the SNVs underlying the signal from the aggregate tests.
Figure 1.
Overall Analytic Approach
We evaluated the WGS data using the Combined Annotation Dependent Depletion (CADD) scores8 as variant weights. The CADD algorithm integrates multiple functional annotations including conservation scores, functional prediction scores for missense SNVs, epigenomic markers, and others. CADD scores are available for both coding and non-coding variants. The weights were defined as the difference between raw CADD scores and the minimum CADD score scaled by the range of the raw CADD scores and were introduced into both the T5 test and SKAT. Previous studies show that using the quartic form of prediction scores as a variant weight can improve the discriminative power,9 so the quartic form of the CADD score was also used in the aggregate tests. The analytical models were the same as described above.
We also evaluated the WGS data using Eigen scores10 as variant weights. Only Eigen scores in non-coding regions were used for analysis. The weights were defined as the difference between raw Eigen scores and the minimum Eigen score scaled by the range of the raw Eigen scores and were introduced into both the T5 and SKAT. The analytical models were the same as described above.
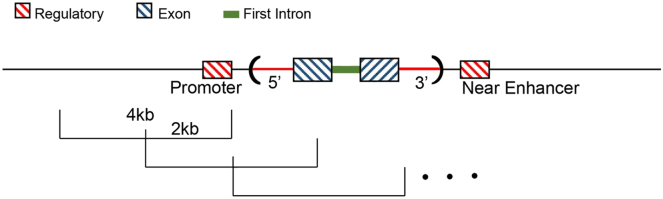
An initial step for analyzing the association between WGS data and phenotypes involves defining tractable analytical units for the proposed aggregate tests. On the one extreme, these units could simply be annotated protein-encoding genes, which would approximately recapitulate whole-exome sequence data without the vagaries of exon capture. On the other extreme, an agnostic sliding window would consider the whole genome, regardless of annotation or presumed functionality. Studies such as ENCODE11 are defining candidate regulatory elements and helping formulate a better understanding of non-translated RNA-encoding genes. There is ample and still emerging evidence of the critical role of the first intron in regulating gene expression.12, 13 Taken together, Figure 2 shows a schematic diagram of the annotated functional motifs used in the analyses presented here. We did not conduct analyses focusing only on exons because they are well known in the literature and have been demonstrated with much larger sample sizes. Based on our previous experience,1 physical windows were defined as 4 kb in length and begin at position 0 bp for each chromosome, with a skip length of 2 kb. WGS variation was annotated across the genome and functional domains using the Whole Genome Sequencing Annotation (WGSA) pipeline.14 The first intron of a gene was determined using SnpEff15 annotation based on the RefSeq16 gene model. The 3′ and 5′ untranslated region (UTR) of a gene was determined using ANNOVAR17 annotations based on the RefSeq gene model. The promoter of a gene was defined based on the overlap between the permissive set of CAGE peaks reported by the FANTOM5 project18 and the 5 kb upstream region determined by the ANNOVAR annotation based on the RefSeq gene model. The enhancers and the target genes of the enhancers were defined based on the permissive set of enhancers and enhancer-promoter pairs reported by the FANTOM5 project. In the case of an undesignated enhancer-gene pair, we assigned an enhancer to the nearest gene. Therefore, the regulatory domain motif utilized for aggregate variant tests includes enhancers, the 3′ and 5′ UTRs, and promoter of a gene. Variants could be included in multiple groupings in the case of overlapping genes. For example, we determined that 770,137 SNVs (MAF ≤ 5% and with MAC ≥ 3) are annotated as belonging to the annotated regulatory domains (i.e., in an enhancer, promoter, 3′ or 5′ UTR), and 11.4% (n = 87,718 SNVs) of the time they also belong to a regulatory domain for a neighboring gene. In order to visualize the contribution from each of these annotated functional motifs, we utilized the online tool Lachesis to view any region of interest.
Figure 2.
Annotated Functional Motifs
We defined a priori thresholds of statistical significance for each annotated motif. For the sliding window approach, we considered 668,836 contiguous and non-overlapping windows and 10 traits and therefore set a significance threshold at p < 7.5 × 10−9 (equal to 0.05 / 668,836 / 10). We next applied the T5 test and SKAT to the set of low-frequency and rare variants (MAF ≤ 5%) among the annotated regulatory domains and also for the first intron of each gene. Associations were considered significant with p < 2.3 × 10−7 accounting for 21,414 regulatory domains and 10 traits for regulatory domain analyses, and p < 3.5 × 10−7 accounting for 14,202 first introns and 10 traits for first intron analyses. We restricted our analyses in sliding windows, regulatory domains, and first introns to MAC ≥ 3 within a motif based on our prior work.19 Finally, for the common variants with MAF > 5% evaluated individually, a threshold of p < 5 × 10−8 was used for genome-wide significance accounting for ∼1 million independent common variants.20
Results
WGS was completed for 1,860 AA and 1,705 EA individuals from the ARIC study. For these analyses, we selected heart and blood traits related to cardiovascular outcomes that were measured across the entire cohort to maximize sample size. Descriptive characteristics for these ten traits are provided in Table S1.
Among the AA individuals sequenced at 7.8-fold depth of coverage, there were 51,350,433 total SNVs. Figure S1 shows the proportion of variants within frequency bins characterized as very rare (43.6%, MAF ≤ 0.1%), rare (25.7%, 0.1% ≤ MAF ≤ 1%), low-frequency (13.4%, 1% ≤ MAF ≤ 5%), and common (17.3%, MAF > 5%). This study primarily focuses on low-frequency, rare, and very rare variants aggregated by various motifs such as a sliding window across the genome, in annotated regulatory domains, or residing in the first intron of coding genes. A total of 1,337,673 4-kb overlapping windows in AA have a distribution of 1 to 694 SNVs per window (Figure S2) with median MAC of 1,131. Among the 21,414 annotated regulatory domains in AA, we observed a distribution of 1 to 750 SNVs per domain (Figure S2) and a median MAC of 500. In comparison, an assessment of the first intron of all 14,202 coding genes in AA showed a range of 1 to 15,552 SNVs (Figure S2) with a median MAC of 956.
Test Results for Low-Frequency and Rare Variation in Annotated Functional Motifs
We applied tests aggregating low-frequency and rare variation within annotated functional motifs: sliding windows, regulatory domains, and first introns. Index windows are shown representing the most significant window from the T5 test (Table 1) or SKAT (Table 2) within a set of contiguous sliding windows. We report results for all underlying significant overlapping windows for the T5 test (Table S2) and SKAT (Table S3). Significant T5 and SKAT results are shown for the regulatory domains in Tables 3 and 4, respectively. Significant SKAT results are detailed for the first intron in Table 5. There were no significant T5 test results for the first intron motif. Figure S12 shows the quantile-quantile (QQ) plots related to the results in Tables 1, 2, 3, 4, and 5.
Table 1.
Index Sliding Windows Demonstrating a Significant Association for the T5 Test in African Americans
| Trait | Chr. | Start Position (bp) | Stop Position (bp) | cMAF | # SNVs | p Value |
|---|---|---|---|---|---|---|
| Lp(a) | 6 | 160,928,009 | 160,932,008 | 0.2353 | 40 | 7.73 × 10−11 |
| Lp(a) | 6 | 160,990,009 | 160,994,008 | 0.4608 | 84 | 5.01 × 10−11 |
| Lp(a) | 6 | 161,006,009 | 161,010,008 | 0.5767 | 79 | 2.17 × 10−9 |
| Lp(a) | 6 | 161,068,009 | 161,072,008 | 0.4991 | 92 | 3.00 × 10−11 |
| Neutrophil count | 1 | 159,174,150 | 159,178,149 | 0.2706 | 56 | 8.39 × 10−19 |
| Neutrophil count | 1 | 159,290,150 | 159,294,149 | 0.1818 | 38 | 2.49 × 10−11 |
| Neutrophil count | 1 | 159,316,150 | 159,320,149 | 0.2667 | 74 | 1.80 × 10−10 |
| Neutrophil count | 1 | 159,410,150 | 159,414,149 | 0.4092 | 54 | 1.50 × 10−10 |
| Neutrophil count | 1 | 159,446,150 | 159,450,149 | 0.2261 | 62 | 4.14 × 10−13 |
| Neutrophil count | 1 | 159,478,150 | 159,482,149 | 0.1459 | 26 | 2.61 × 10−15 |
| Neutrophil count | 1 | 159,514,150 | 159,518,149 | 0.2364 | 44 | 1.26 × 10−12 |
| Neutrophil count | 1 | 159,540,150 | 159,544,149 | 0.4025 | 82 | 4.99 × 10−9 |
| Neutrophil count | 1 | 159,546,150 | 159,550,149 | 0.1751 | 45 | 3.00 × 10−14 |
| Neutrophil count | 1 | 161,664,150 | 161,668,149 | 0.2557 | 48 | 2.69 × 10−10 |
Base pair (bp) position based on hg19. Significant: p < 7.5 × 10−9. Abbreviations are as follows: Chr, chromosome; cMAF, cumulative minor allele frequency.
Table 2.
Index Sliding Windows Demonstrating a Significant Association for the SKAT Test in African Americans
| Trait | Chr. | Start Position (bp) | Stop Position (bp) | cMAF | # SNVs | p Value |
|---|---|---|---|---|---|---|
| Lp(a) | 6 | 160,660,009 | 160,664,008 | 0.396 | 74 | 4.26 × 10−9 |
| Lp(a) | 6 | 160,710,009 | 160,714,008 | 0.740 | 87 | 1.58 × 10−10 |
| Lp(a) | 6 | 160,750,009 | 160,754,008 | 0.415 | 62 | 4.03 × 10−14 |
| Lp(a) | 6 | 160,772,009 | 160,776,008 | 0.358 | 63 | 3.45 × 10−11 |
| Lp(a) | 6 | 160,794,009 | 160,798,008 | 0.323 | 49 | 5.08 × 10−9 |
| Lp(a) | 6 | 160,800,009 | 160,804,008 | 0.365 | 58 | 5.62 × 10−9 |
| Lp(a) | 6 | 160,810,009 | 160,814,008 | 0.322 | 56 | 4.17 × 10−12 |
| Lp(a) | 6 | 160,824,009 | 160,828,008 | 0.392 | 48 | 2.38 × 10−10 |
| Lp(a) | 6 | 160,832,009 | 160,836,008 | 0.191 | 49 | 3.08 × 10−10 |
| Lp(a) | 6 | 160,842,009 | 160,846,008 | 0.348 | 57 | 9.56 × 10−12 |
| Lp(a) | 6 | 160,852,009 | 160,856,008 | 0.349 | 60 | 1.55 × 10−14 |
| Lp(a) | 6 | 160,880,009 | 160,884,008 | 0.139 | 40 | 3.92 × 10−12 |
| Lp(a) | 6 | 160,888,009 | 160,892,008 | 0.530 | 69 | 8.18 × 10−12 |
| Lp(a) | 6 | 160,900,009 | 160,904,008 | 0.416 | 75 | 6.23 × 10−15 |
| Lp(a) | 6 | 160,928,009 | 160,932,008 | 0.235 | 40 | 1.70 × 10−13 |
| Lp(a) | 6 | 160,944,009 | 160,948,008 | 0.214 | 42 | 4.11 × 10−28 |
| Lp(a) | 6 | 161,008,009 | 161,012,008 | 0.457 | 77 | 6.18 × 10−34 |
| Lp(a) | 6 | 161,052,009 | 161,056,008 | 0.352 | 49 | 2.57 × 10−14 |
| Lp(a) | 6 | 161,090,009 | 161,094,008 | 0.300 | 61 | 4.67 × 10−30 |
| Lp(a) | 6 | 161,100,009 | 161,104,008 | 0.338 | 56 | 5.25 × 10−11 |
| Lp(a) | 6 | 161,120,009 | 161,124,008 | 0.504 | 71 | 2.94 × 10−11 |
| Lp(a) | 6 | 161,134,009 | 161,138,008 | 0.256 | 61 | 2.66 × 10−12 |
| Lp(a) | 6 | 161,178,009 | 161,182,008 | 0.417 | 64 | 1.43 × 10−14 |
| Lp(a) | 6 | 161,290,009 | 161,294,008 | 0.364 | 65 | 2.94 × 10−9 |
| Lp(a) | 6 | 161,302,009 | 161,306,008 | 0.352 | 53 | 2.32 × 10−9 |
| Neutrophil count | 1 | 156,746,150 | 156,750,149 | 0.308 | 60 | 4.79 × 10−9 |
| Neutrophil count | 1 | 158,764,150 | 158,768,149 | 0.234 | 39 | 7.64 × 10−12 |
| Neutrophil count | 1 | 159,168,150 | 159,172,149 | 0.411 | 62 | 3.68 × 10−11 |
| Neutrophil count | 1 | 159,290,150 | 159,294,149 | 0.182 | 38 | 6.75 × 10−14 |
| Neutrophil count | 1 | 159,370,150 | 159,374,149 | 0.424 | 60 | 3.77 × 10−11 |
| Neutrophil count | 1 | 159,402,150 | 159,406,149 | 0.259 | 58 | 1.11 × 10−10 |
| Neutrophil count | 1 | 159,408,150 | 159,412,149 | 0.455 | 59 | 2.20 × 10−10 |
| Neutrophil count | 1 | 159,416,150 | 159,420,149 | 0.731 | 102 | 1.48 × 10−9 |
| Neutrophil count | 1 | 159,446,150 | 159,450,149 | 0.226 | 62 | 3.91 × 10−16 |
| Neutrophil count | 1 | 159,470,150 | 159,474,149 | 0.290 | 46 | 1.12 × 10−10 |
| Neutrophil count | 1 | 159,488,150 | 159,492,149 | 0.164 | 54 | 1.82 × 10−13 |
| Neutrophil count | 1 | 159,502,150 | 159,506,149 | 0.240 | 54 | 2.06 × 10−15 |
| Neutrophil count | 1 | 159,514,150 | 159,518,149 | 0.236 | 44 | 2.08 × 10−12 |
| Neutrophil count | 1 | 159,520,150 | 159,524,149 | 0.289 | 39 | 3.76 × 10−11 |
| Neutrophil count | 1 | 159,536,150 | 159,540,149 | 0.459 | 65 | 5.97 × 10−11 |
| Neutrophil count | 1 | 159,548,150 | 159,552,149 | 0.195 | 45 | 1.42 × 10−15 |
| Neutrophil count | 1 | 159,556,150 | 159,560,149 | 0.221 | 57 | 3.27 × 10−13 |
| Neutrophil count | 1 | 159,580,150 | 159,584,149 | 0.299 | 73 | 4.95 × 10−10 |
| Neutrophil count | 1 | 159,798,150 | 159,802,149 | 0.350 | 83 | 1.23 × 10−12 |
| Neutrophil count | 1 | 161,508,150 | 161,512,149 | 0.455 | 92 | 1.88 × 10−9 |
Significant: p < 7.5 × 10−9.
Table 3.
Regulatory Domains Demonstrating a Significant Association for the T5 Test in African Americans
| Trait | Gene | cMAF | # SNVs | p Value | Beta | SE |
|---|---|---|---|---|---|---|
| Lp(a) | PLG | 0.179 | 33 | 5.55 × 10−8 | −0.17 | 0.03 |
| Neutrophil count | DARC | 0.121 | 21 | 3.91 × 10−8 | 2.71 | 0.49 |
Significant: p < 2.3 × 10−7. Abbreviation is as follows: cMAF, cumulative minor allele frequency.
Table 4.
Regulatory Domains Demonstrating a Significant Association for SKAT in African Americans
| Trait | Gene | cMAF | # SNVs | p Value |
|---|---|---|---|---|
| cTnT | CA9 | 0.001 | 2 | 9.16 × 10−9 |
| Lp(a) | MFAP5 | 0.001 | 4 | 5.09 × 10−8 |
| Lp(a) | SLC22A3 | 0.231 | 35 | 2.29 × 10−7 |
| Neutrophil count | MNDA | 0.324 | 38 | 1.10 × 10−9 |
| Neutrophil count | IFI16 | 0.412 | 54 | 1.85 × 10−9 |
| Neutrophil count | HSPA6 | 0.460 | 65 | 1.69 × 10−8 |
| Neutrophil count | DARC | 0.121 | 21 | 1.89 × 10−8 |
| Neutrophil count | CADM3 | 0.204 | 54 | 2.08 × 10−8 |
| Neutrophil count | PYHIN1 | 0.300 | 34 | 5.34 × 10−8 |
Significant: p < 2.3 × 10−7. Abbreviation is as follows: cMAF, cumulative minor allele frequency.
Table 5.
First Introns Demonstrating a Significant Association for SKAT in African Americans
| Trait | Gene | cMAF | # SNVs | p Value |
|---|---|---|---|---|
| Lp(a) | SLC22A3 | 4.194 | 757 | 3.24 × 10−11 |
| Neutrophil count | EFNA3 | 0.250 | 64 | 2.59 × 10−7 |
Significant: p < 3.5 × 10−7. Abbreviation is as follows: cMAF, cumulative minor allele frequency.
Significant findings in AAs are investigated by the T5 test and SKAT in EAs for the sliding window (Tables S2 and S3), regulatory domains (Tables S4 and S5), and first intron (Table S6). The key to understanding the genome-phenotype relationship for complex traits is to assess the joint contribution from each annotated functional motif for each trait. Lachesis plots aid in this visualization and we review the results in AAs in the following vignettes for the three heart- and blood-related traits (Lp(a), neutrophil count, and cTnT) that demonstrated significant findings across various motifs in AAs.
Test Results for Common Variants
For completeness, we conducted a survey of the genome investigating all common variants with MAF > 5%. Common variants in five genomic regions reached our pre-defined significance threshold for five traits, including neutrophil count, CRP, Lp(a), P, and sdLDL-c (Figure S3). The sentinel SNV with the lowest p value for each trait is shown in Table S7 and results for all significant associations (p < 5 × 10−8) are shown in Table S8. Four loci—DARC, CRP, LPA, and APOE—with their corresponding traits have been reported by previous GWASs.21, 22, 23, 24, 25, 26, 27 We identified a signal at 9p21, a well-known cardiovascular disease locus, associated with serum phosphorus levels. However, the index SNV, rs60456827 (MAF = 15% in AA), was not significantly associated with P levels in ARIC EAs (MAF = 2%, beta = 0.02, p = 0.77).
Lp(a)
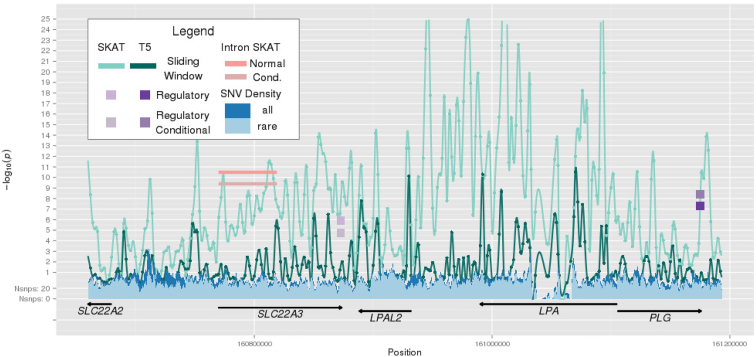
A 646-kb region (from 160,660,009 to 161,306,008 bp on chromosome 6) consisting of 107 windows showed a significant association with Lp(a) (lowest p = 3.0 × 10−11 for the T5 test and lowest p = 6.18 × 10−34 for SKAT; Tables S2 and S3) among AAs. The windows reside in 6q25.3-6q26, covering 218 kb upstream and 292 kb downstream of LPA, and include four other genes (PLG, SLC22A2, SLC22A3, and LPAL2). Investigation of annotated regulatory domain motifs showed that there are two regulatory domains significantly associated with Lp(a) levels in this region. The first regulatory domain involves SLC22A3 (2.29 × 10−7, SKAT; Table 4) and the signal is driven by three SNVs in the 3′ UTR and one intronic SNV that resides in a defined enhancer, all with p < 0.01 (Table S9). The target for the FANTOM5 enhancer involving the intronic SNV is unknown and therefore was assigned to SLC22A3. The first intron of SLC22A3 (p = 3.24 × 10−11, SKAT; Table 5) was also significantly associated with Lp(a) levels. The second regulatory domain involves PLG (p = 5.55 × 10−8, T5 test; Table 3) and the aggregate test result is largely due to four SNVs in the 3′ UTR with p < 0.01 (Table S10). Figure 3 shows the entire genomic landscape of this region, incorporating all of these test results. An additional regulatory domain on chromosome 12 was identified near MFAP5 (5.09 × 10−8, SKAT; Table 4) and all four intergenic SNVs included in the aggregate test reside in an enhancer (Table S11).
Figure 3.
Survey of the Genomic Landscape on Chromosome 6q25.3-6q26 for Lp(a) Levels via Lachesis
The sliding window trace shows the results from tests (SKAT and T5) aggregating low-frequency and rare variation within overlapping physical windows of 4 kb. Significant results are shown for regulatory domains in SLC22A3 (SKAT) and PLG (T5) and the first intron of SLC22A3 (SKAT). Results after conditioning on rs115848955 in LPA are also shown.
Lp(a) is encoded by LPA, and an intronic SNV of LPA, rs115848955 (MAF = 5%), showed the strongest signal with Lp(a) in a recent AA study.21 The Lp(a) sentinel common SNV, rs41271018 (MAF = 5%), identified in our study of AA is in linkage disequilibrium with rs115848955 (r2 = 0.93). We re-examined our region of interest located 6q25.3-6q26 after adjusting for rs115848955, and several windows in the region remained significant (lowest p = 2.72 × 10−12 for T5 test and lowest p = 7.70 × 10−25 for SKAT; Table S12). Interestingly, the identified regulatory domain at PLG increased in significance after conditioning on rs115848955 and the SLC22A3 regulatory domain and first intron decreased in significance. Figure 3 depicts these conditional results in the context of the unconditional results. We further investigated replication in EA individuals for each significant motif and showed that many of the sliding windows were also strongly associated with Lp(a) levels (Tables S2 and S3). No association was seen in EAs for the regulatory domains of PLG (p = 0.11, Table S4) or MFAP5 (p = 0.40, Table S5). Replication in EA was observed for the regulatory domain of SLC22A3 (p = 0.004, Table S5) and the first intron of SLC22A3 (p = 0.0002, Table S6).
We characterized the overall range of risk across this locus around LPA by utilizing the most significant driving SNV from each identified motif, resulting in a total of seven SNVs: one from each of the four sliding windows (Table 2), one from the first intron of SLC22A3 (Table 5), one from the regulatory domain of SLC22A3 (Table 4), and one from the regulatory domain of PLG (Table 3). Of the seven SNVs, three variants had an effect of increasing Lp(a) levels and they were from the regulatory domain of SLC22A3, the first intron of SLC22A3, and one of the sliding windows. These three SNVs constituted a risk score and individuals with two or more risk alleles were contrasted with the individuals with no risk alleles (Figure S4). Overall, the magnitude of effects considered together shows that individuals with risk variants related to increased Lp(a) levels across this region on chromosome 6 have higher Lp(a) levels. Of the seven SNVs across this region, the other four variants had an effect of decreasing Lp(a) levels and they were from the regulatory domain of PLG and the remaining three sliding windows. These four SNVs constituted a risk score and individuals with two or more risk alleles were contrasted with the individuals with no risk alleles (Figure S5). Overall, the magnitude of effects considered together shows that individuals with risk variants related to decreased Lp(a) levels across this region on chromosome 6 generally have lower Lp(a) levels.
Neutrophil Count
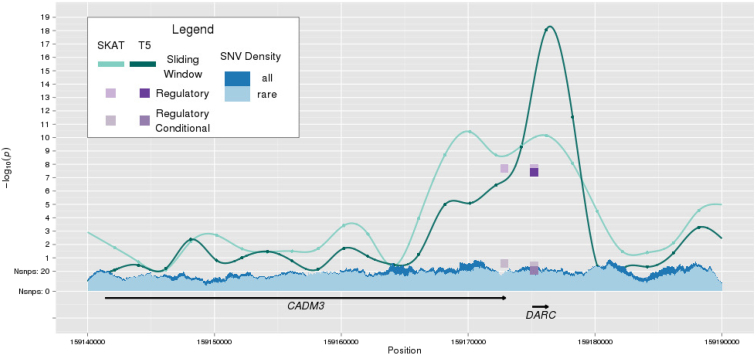
For neutrophil count, we observed a 4.92-Mb region spanning 1q23 that showed significant association for the sliding windows (lowest p = 8.39 × 10−19 for the T5 test and lowest p = 3.91 × 10−16 for SKAT) among AA individuals. Within this region, as shown in Tables 3 and 4, the regulatory domain motif for DARC was significantly associated with neutrophil count in both the T5 test and SKAT and is driven by three SNVs (p < 0.01) in the 5′ UTR of the gene (Table S13). The regulatory domain of a neighboring gene, CADM3, also was significantly associated with neutrophil count and can be explained by four SNVs in the 3′ UTR (Table S14). It is notable that these CADM3 3′ UTR SNVs are also upstream of DARC, and two are suggested to be in promoter of DARC by funseq.28 Figure 4 shows the genomic landscape encompassing DARC and CADM3. DARC is a well-studied locus for neutrophil count26, 27 and a common SNV (rs2814778, MAF = 0.17, Table S7) was the sentinel SNV identified in AA from this study. After accounting for rs2814778, we observed decreased significance for the nearby windows, regulatory domains, and first introns (lowest p = 0.02; Tables S4–S6). None of the sliding windows on 1q23 that were significantly associated with neutrophil count in AA demonstrated a significant association in EAs (lowest p = 0.007; Tables S2 and S3). The findings for DARC and CADM3 regulatory domains did not replicate in EAs (Tables S4 and S5).
Figure 4.
Survey of the Genomic Landscape on Chromosome 1q23 for Neutrophil Count via Lachesis
The sliding window trace shows the results from tests (SKAT and T5) aggregating low-frequency and rare variation within overlapping physical windows of 4 kb. Significant results are shown for regulatory domains in DARC (T5 and SKAT) and CADM3 (SKAT). Results after conditioning on rs2814778 in DARC are also shown.
Additional significant signal in the 1q23 region comes from regulatory domains for three overlapping genes (MNDA, PYHIN1, and IFI16) and is driven by three SNVs residing in an enhancer for all three genes (Tables S15–17). Similarly, the significant regulatory domain for HSPA6 contains two SNVs (p < 0.01) in the 5′ UTR and two in an enhancer targeting HSPA6 and other genes (Table S18). The first intron of EFNA3 (p = 2.59 × 10−7, SKAT; Table 5) was also significantly associated with neutrophil count. None of these additional motifs identified on 1q23 replicated in EAs (Tables S4–S6).
cTnT
A single regulatory motif on chromosome 9 involving the gene carbonic anhydrase IX (CA9) was significantly associated with cTnT in AA (p = 9.16 × 10−9; Table 4) and showed modest association in EA (p = 0.03; Table S5). This motif contained only two SNVs in the 5′ UTR of CA9 with a total MAC = 3 in AA, and only one of the SNV in the 5′ UTR with a total MAC = 6 was observed in EA. Based on single SNV results, the primary signal is driven by the SNV located at 35,673,953 bp (Table S19).
Sliding Window Approach Incorporating CADD or Eigen Score as a Variant Weight
Tests aggregate low-frequency and rare SNVs within a motif to increase statistical power, but noise also increases given the equal consideration for functional and non-functional SNVs, in particular for non-coding regions. We evaluated the impact of signal to noise on aggregate tests of association by introducing CADD score or Eigen score predictions of nucleotide function as weights. Overall, we do not observe a clear enhancement for weighted tests versus tests that do not use functional predictions as a weight. As an example, Figure S6 shows the quantile-quantile (QQ) plots for incorporating CADD score in the sliding window motif analyses for Lp(a) levels and neutrophil counts, the two traits for which we saw genome-wide significant windows. Figure S7 shows similar plots for Eigen scores. The T5 test does not show appreciable difference between tests that do not use functional predictions as a weight and those weighted by CADD or Eigen score. The SKAT analysis shows some differences between CADD or Eigen weighted tests and tests that do not use functional predictions as a weight. These observations hold true for analyses including the quartic form of the CADD score (Figure S8).
We next investigated the average CADD score for all significant windows for these two traits (from Tables S2 and S3). The average CADD score for each significant window is shown as a vertical red line in Figures S9A and S9B for the T5 test and SKAT, respectively, compared to the distribution of average CADD for all windows. The average CADD score for these significant windows is significantly larger than those of a random sample of windows (p value = 0.03 for windows in Figure S9A, Kolmogorov-Smirnov test, one-tail; p value = 0.02 for windows in Figure S9B, Kolmogorov-Smirnov test, one-tail). The average quartic CADD scores for the sliding windows are plotted in Figure S10. Similarly, we investigated the average Eigen score for all significant windows covering non-coding regions for Lp(a) levels and neutrophil count from Tables S2 and S3. The average Eigen score for each significant non-coding window is shown as a vertical red line in Figures S11A and S11B for the T5 test and SKAT, respectively, compared to the distribution of average Eigen scores in non-coding windows. In Figure S11A, the average non-coding Eigen scores for those significant non-coding windows in Table S2 is larger than those of a random sample of non-coding windows, but not significantly different (p value = 0.067, Kolmogorov-Smirnov test, one-tail). In Figure S11B, we observe that the average non-coding Eigen scores for those significant non-coding windows in Table S3 are significantly larger than those of a random sample of windows (p value = 1.41 × 10−5, Kolmogorov-Smirnov test, one-tail).
Discussion
This study provides a practical approach to WGS analysis of complex traits using aggregate tests across a variety of annotated functional motifs: sliding window, regulatory domains, and first introns. Inclusion of annotated regulatory domains, such as those from FANTOM5, as a focus of aggregate tests and use of predicted functional scores (e.g., CADD and EIGEN) as variant weights in the tests represent important additions to the series of analysis steps outlined in this practical approach to WGS analysis. Considering the current results, the relationship between regulatory domains neighboring LPA and Lp(a) level, the regulatory domains near DARC and neutrophil count, or the regulatory domain of CA9 and cTnT level would not have been discovered without conducting motif-based association tests of rare variants. Our investigation revealed that components of the genomic landscape were significantly associated with six out of the ten heart and blood traits related to cardiovascular outcomes. For two traits, Lp(a) and neutrophil count, aggregate tests of low-frequency and rare variation were significantly associated across multiple motifs. For a third trait, cTnT, investigation of regulatory domains may have identified a locus on chromosome 9.
The results presented here outline a series of practical steps for both analyzing WGS data and synthesizing the results (Figure 1) with the goal of utilizing as much information as possible to identify loci contributing to a complex trait. The results also demonstrate how WGS analyses can be used to fine-map significantly associated loci and to identify driver SNVs that may be responsible for the underlying observed associations. One key component to this approach is the ability to visualize the contribution from multiple sources of information. As shown in the Lachesis plot for Lp(a), the sliding window trace provides a background context for interpreting the signals observed from annotated regulatory domains. For Lp(a) levels, the dominant signal is upstream of LPA, the coding gene for Lp(a). In the region of interest at 6q25.3-6q26 encompassing LPA, there were distinct signals coming from three distinct regulatory elements. Our analytic strategy allowed for inspection of SNVs included in the aggregate tests that appear to be driving the regulatory domain signals. In this way, we could determine that the four most significant SNVs (p < 0.01) contributing to the significant T5 test results for Lp(a) are located in the 3′ UTR of PLG (upstream of LPA) and range in MAF from 0.05% to 4%. This defined regulatory domain increased in significance after conditioning on the most significant common variant, unlike the other two regulatory domain signals in the region near SLC22A3 that decreased in significance. These association results for Lp(a) also highlight that an existing knowledge gap in the field is the need for additional refinement of enhancer-gene target pairing. We identified that the regulatory domain motif for SLC22A3 is in part significantly driven by an intronic SNV residing in an enhancer with unknown target, and therefore was assigned as an enhancer of SLC22A3. It is plausible that the reason this regulatory element is identified in our analysis of Lp(a) is because it may indeed be an enhancer for LPA.
Using the outlined analytic strategy, we were also able to identify and interpret genomic regions contributing to neutrophil count in AA. For neutrophil count, the Lachesis plot clearly shows how the regulatory motif signal is also picked up by the sliding window and involves the known gene DARC, emphasizing again that the sliding window approach provides an informative background context for overall elucidation of the genomic contribution to complex traits.
The results for cTnT identified a locus downstream of the 9p21 region associated with cardiovascular disease.29 CA9 is induced by hypoxia in humans and is a significant serologic predictor of right ventricular dysfunction in patients with pulmonary embolism along with cTnT.30 This is a promising finding, but also an example of how results from WGS must be interpreted with caution as the primary signal comes from a single SNV (at 35,673,953 bp) with a total MAC of 2 (Table S19). This SNV was monomorphic in EA. To further evaluate the validity of these findings for CA9, we conducted a permutation test whereby cTnT levels were permutated 1 million times and the SKAT test was repeated, resulting in the ranking of the original p value 165th out of the million tests for a permutation test p value 165 / 1,000,001 = 1.65 × 10−4. For WGS analyses employing aggregate tests of association involving rare variants, we recommend that investigators set a lower bound on the MAC that takes into account sample size for their study.
More than GWASs or exome sequencing, careful annotation is a key feature of WGS analysis. In the analyses presented here, annotation provided different sources of information. First, annotation provided the boundaries of units for aggregate testing, such as the regulatory domain motif. In this context, linked databases (e.g., RefSeq) and national efforts to define functional genome elements (e.g., ENCODE) are invaluable. In this analysis, aggregate tests of annotated regulatory elements yielded few novel significant results. This result may be specific to the traits analyzed in this study. The full value of aggregate tests of annotated functional motifs remains to be seen and may rely on improved annotation and statistical methods or increased sample sizes. The second type of information gained from annotation was whether or not a variant was predicted to have a functional impact on protein or genome function. The most obvious examples include nonsynonymous substitutions and nonsense mutations, although more subtle examples exist, such as splice variants. Related to predicted function, but more nuanced, was our use of predicted deleteriousness (i.e., CADD score) as weights for the genotype-phenotype analyses. Such weights take into account the fact that all amino acid substitutions or all promotor variants are not equal, and one can predict the impact based on knowledge of the location and type of substitution. Studies validating these predictions for protein-encoding genes have been carried out with mixed success,19, 31, 32 and studies validating such predictions for regulatory elements are almost nonexistent. Databases of weights such as MetaLR33 and CADD score8 have been linked to popular annotation tools. In the analyses presented here, we did not see marked benefit of weighted analyses across the full range of weights, nor did we observe improved benefit of weights scaled to accentuate predicted highly impactful deleterious variants. Others have enthusiastically argued that predicted functional annotation must take into account population genetic principles and the effects of natural selection34 and some progress has been made in this area.35, 36 Clearly, more work is necessary in the area of whole-genome annotation and the success of whole-genome sequencing for understanding the genetic architecture of complex disease may indeed depend on it. In converse, the results of whole-genome sequence analysis may become the fodder of future annotation tools.
This study provides an example application of WGS analysis in a sample of AA measured for multiple cardiovascular-related traits. As such, we included EA individuals with WGS only as a source of replication for the top signals from AA. Future studies may wish to consider WGS analyses that pool data from multiple ethnicities under the assumption of similar effect sizes on the traits of interest for causal rare variants in each ethnicity. This is in contrast with common-variant GWASs, where variants were not expected to be causal but rather in linkage disequilibrium with causal variants, and therefore the effects in each ethnic group were expected to be different because of differences underlying linkage disequilibrium across populations. For many traits, the majority of GWASs have detected significant loci located in non-coding regions, with a much smaller percentage of significant loci lying in coding sequences. Therefore, the practical approach for evaluating WGS outlined here focuses on motifs involving primarily non-coding functional domains. However, our strategy can easily extend to analysis of only exonic regions, thereby recapitulating previous exome studies, or incorporate exonic information into regulatory domain motifs. Where large effect sizes are present for coding elements, the sliding window is likely to capture this signal as well. Additionally, as studies continue to accrue WGS, sample sizes will increase such that the sliding window motif will begin to characterize novel loci.
In conclusion, we demonstrate a guideline for analyzing and interpreting WGS for complex traits and demonstrate the tractable nature of WGS for characterizing the architecture of complex traits.
Acknowledgments
The Atherosclerosis Risk in Communities (ARIC) study is carried out as a collaborative study supported by the National Heart, Lung, and Blood Institute (NHLBI) contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C). The authors thank the staff and participants of the ARIC study for their important contributions. Funding support for “Building on GWAS for NHLBI-diseases: the U.S. CHARGE consortium” was provided by the NIH through the American Recovery and Reinvestment Act of 2009 (ARRA) (5RC2HL102419). Sequencing was carried out at the Baylor College of Medicine Human Genome Sequencing Center, also supported by the National Human Genome Research Institute grants U54 HG003273 and UM1 HG008898 to R.G.
Published: January 12, 2017
Footnotes
Supplemental Data include Supplemental Methods, 12 figures, and 19 tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.12.009.
Contributor Information
Alanna C. Morrison, Email: alanna.c.morrison@uth.tmc.edu.
Eric Boerwinkle, Email: eric.boerwinkle@uth.tmc.edu.
Web Resources
Supplemental Data
References
- 1.Morrison A.C., Voorman A., Johnson A.D., Liu X., Yu J., Li A., Muzny D., Yu F., Rice K., Zhu C., Cohorts for Heart and Aging Research in Genetic Epidemiology (CHARGE) Consortium Whole-genome sequence-based analysis of high-density lipoprotein cholesterol. Nat. Genet. 2013;45:899–901. doi: 10.1038/ng.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walter K., Min J.L., Huang J., Crooks L., Memari Y., McCarthy S., Perry J.R., Xu C., Futema M., Lawson D., UK10K Consortium The UK10K project identifies rare variants in health and disease. Nature. 2015;526:82–90. doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu F., Lu J., Liu X., Gazave E., Chang D., Raj S., Hunter-Zinck H., Blekhman R., Arbiza L., Van Hout C. Population genomic analysis of 962 whole genome sequences of humans reveals natural selection in non-coding regions. PLoS ONE. 2015;10:e0121644. doi: 10.1371/journal.pone.0121644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee S., Abecasis G.R., Boehnke M., Lin X. Rare-variant association analysis: study designs and statistical tests. Am. J. Hum. Genet. 2014;95:5–23. doi: 10.1016/j.ajhg.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The ARIC investigators The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. Am. J. Epidemiol. 1989;129:687–702. [PubMed] [Google Scholar]
- 6.Li B., Leal S.M. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am. J. Hum. Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu M.C., Lee S., Cai T., Li Y., Boehnke M., Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim T., Wei P. Incorporating ENCODE information into association analysis of whole genome sequencing data. BMC Proc. 2016;10(Suppl 7):257–261. doi: 10.1186/s12919-016-0040-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ionita-Laza I., McCallum K., Xu B., Buxbaum J.D. A spectral approach integrating functional genomic annotations for coding and noncoding variants. Nat. Genet. 2016;48:214–220. doi: 10.1038/ng.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.ENCODE Project Consortium The ENCODE (ENCyclopedia Of DNA Elements) project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 12.Majewski J., Ott J. Distribution and characterization of regulatory elements in the human genome. Genome Res. 2002;12:1827–1836. doi: 10.1101/gr.606402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park S.G., Hannenhalli S., Choi S.S. Conservation in first introns is positively associated with the number of exons within genes and the presence of regulatory epigenetic signals. BMC Genomics. 2014;15:526. doi: 10.1186/1471-2164-15-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X., White S., Peng B., Johnson A.D., Brody J.A., Li A.H., Huang Z., Carroll A., Wei P., Gibbs R. WGSA: an annotation pipeline for human genome sequencing studies. J. Med. Genet. 2016;53:111–112. doi: 10.1136/jmedgenet-2015-103423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cingolani P., Platts A., Wang L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forrest A.R., Kawaji H., Rehli M., Baillie J.K., de Hoon M.J., Haberle V., Lassmann T., Kulakovskiy I.V., Lizio M., Itoh M., FANTOM Consortium and the RIKEN PMI and CLST (DGT) A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li A.H., Morrison A.C., Kovar C., Cupples L.A., Brody J.A., Polfus L.M., Yu B., Metcalf G., Muzny D., Veeraraghavan N. Analysis of loss-of-function variants and 20 risk factor phenotypes in 8,554 individuals identifies loci influencing chronic disease. Nat. Genet. 2015;47:640–642. doi: 10.1038/ng.3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pe’er I., Yelensky R., Altshuler D., Daly M.J. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet. Epidemiol. 2008;32:381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 21.Wu Y., Marvelle A.F., Li J., Croteau-Chonka D.C., Feranil A.B., Kuzawa C.W., Li Y., Adair L.S., Mohlke K.L. Genetic association with lipids in Filipinos: waist circumference modifies an APOA5 effect on triglyceride levels. J. Lipid Res. 2013;54:3198–3205. doi: 10.1194/jlr.P042077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qi Q., Workalemahu T., Zhang C., Hu F.B., Qi L. Genetic variants, plasma lipoprotein(a) levels, and risk of cardiovascular morbidity and mortality among two prospective cohorts of type 2 diabetes. Eur. Heart J. 2012;33:325–334. doi: 10.1093/eurheartj/ehr350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okada Y., Takahashi A., Ohmiya H., Kumasaka N., Kamatani Y., Hosono N., Tsunoda T., Matsuda K., Tanaka T., Kubo M. Genome-wide association study for C-reactive protein levels identified pleiotropic associations in the IL6 locus. Hum. Mol. Genet. 2011;20:1224–1231. doi: 10.1093/hmg/ddq551. [DOI] [PubMed] [Google Scholar]
- 24.Ober C., Nord A.S., Thompson E.E., Pan L., Tan Z., Cusanovich D., Sun Y., Nicolae R., Edelstein C., Schneider D.H. Genome-wide association study of plasma lipoprotein(a) levels identifies multiple genes on chromosome 6q. J. Lipid Res. 2009;50:798–806. doi: 10.1194/jlr.M800515-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ridker P.M., Pare G., Parker A., Zee R.Y., Danik J.S., Buring J.E., Kwiatkowski D., Cook N.R., Miletich J.P., Chasman D.I. Loci related to metabolic-syndrome pathways including LEPR,HNF1A, IL6R, and GCKR associate with plasma C-reactive protein: the Women’s Genome Health Study. Am. J. Hum. Genet. 2008;82:1185–1192. doi: 10.1016/j.ajhg.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crosslin D.R., McDavid A., Weston N., Nelson S.C., Zheng X., Hart E., de Andrade M., Kullo I.J., McCarty C.A., Doheny K.F., Electronic Medical Records and Genomics (eMERGE) Network Genetic variants associated with the white blood cell count in 13,923 subjects in the eMERGE Network. Hum. Genet. 2012;131:639–652. doi: 10.1007/s00439-011-1103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reiner A.P., Lettre G., Nalls M.A., Ganesh S.K., Mathias R., Austin M.A., Dean E., Arepalli S., Britton A., Chen Z. Genome-wide association study of white blood cell count in 16,388 African Americans: the continental origins and genetic epidemiology network (COGENT) PLoS Genet. 2011;7:e1002108. doi: 10.1371/journal.pgen.1002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khurana E., Fu Y., Colonna V., Mu X.J., Kang H.M., Lappalainen T., Sboner A., Lochovsky L., Chen J., Harmanci A., 1000 Genomes Project Consortium Integrative annotation of variants from 1092 humans: application to cancer genomics. Science. 2013;342:1235587. doi: 10.1126/science.1235587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harismendy O., Notani D., Song X., Rahim N.G., Tanasa B., Heintzman N., Ren B., Fu X.D., Topol E.J., Rosenfeld M.G., Frazer K.A. 9p21 DNA variants associated with coronary artery disease impair interferon-γ signalling response. Nature. 2011;470:264–268. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abul Y., Ozsu S., Mentese A., Durmus I., Bektas H., Pehlivanlar M., Turan O.E., Sumer A., Orem A., Ozlu T. Carbonic anhydrase IX in the prediction of right ventricular dysfunction in patients with hemodynamically stable acute pulmonary embolism. Clin. Appl. Thromb. Hemost. 2014;20:838–843. doi: 10.1177/1076029613486540. [DOI] [PubMed] [Google Scholar]
- 31.Schick U.M., Auer P.L., Bis J.C., Lin H., Wei P., Pankratz N., Lange L.A., Brody J., Stitziel N.O., Kim D.S., Cohorts for Heart and Aging Research in Genomic Epidemiology. National Heart, Lung, and Blood Institute GO Exome Sequencing Project Association of exome sequences with plasma C-reactive protein levels in >9000 participants. Hum. Mol. Genet. 2015;24:559–571. doi: 10.1093/hmg/ddu450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu B., Pulit S.L., Hwang S.J., Brody J.A., Amin N., Auer P.L., Bis J.C., Boerwinkle E., Burke G.L., Chakravarti A., CHARGE Consortium and the National Heart, Lung, and Blood Institute GO ESP∗ Rare exome sequence variants in CLCN6 reduce blood pressure levels and hypertension risk. Circ Cardiovasc Genet. 2016;9:64–70. doi: 10.1161/CIRCGENETICS.115.001215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong C., Wei P., Jian X., Gibbs R., Boerwinkle E., Wang K., Liu X. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet. 2015;24:2125–2137. doi: 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Graur D., Zheng Y., Price N., Azevedo R.B., Zufall R.A., Elhaik E. On the immortality of television sets: “function” in the human genome according to the evolution-free gospel of ENCODE. Genome Biol. Evol. 2013;5:578–590. doi: 10.1093/gbe/evt028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kellis M., Wold B., Snyder M.P., Bernstein B.E., Kundaje A., Marinov G.K., Ward L.D., Birney E., Crawford G.E., Dekker J. Defining functional DNA elements in the human genome. Proc. Natl. Acad. Sci. USA. 2014;111:6131–6138. doi: 10.1073/pnas.1318948111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corbett-Detig R.B., Hartl D.L., Sackton T.B. Natural selection constrains neutral diversity across a wide range of species. PLoS Biol. 2015;13:e1002112. doi: 10.1371/journal.pbio.1002112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.