Abstract
Background
We have previously described the association between rheumatoid arthritis (RA) prevalence and the two mutY Homolog (E. coli) (MUTYH) SNPs (rs3219463 and rs3219476) among the Taiwanese population. This present study will aim to elucidate whether the SNPs can alter the expression of EGFR in the progression of RA.
Methods
The cohort study included 368 Taiwan’s Han Chinese RA patients and 364 healthy controls. Blood samples collected from the participants were analyzed to determine their serum MUTYH levels and to identify rs3219463 SNP of MUTYH from their genomic DNA.
Results
Our data resulted in a statistically significant difference in genotype frequency distributions at rs3219463 for RA patients and controls (p < 0.0002). Also, the patients with G carrier at rs3219463 were less likely to suffer from painful joints (p < 0.006) and DAS28 scores (p < 0.003). Furthermore, the increase in serum level of MUTYH was also observed in RA patients (p < 0.005).
Conclusions
Our study showed that RA is associated with rs3219463 SNP in EGFR gene and an increased serum level of the MUTYH protein. These findings suggest MUTYH is worthy of further investigation as a therapeutic target for RA.
Keywords: RA, MUTYH, SNPs, CNVs, Oxidative DNA damage. DAS28
Background
Rheumatoid arthritis (RA) is a chronic systemic inflammatory disease characterized by persistent leukocytes infiltration, suppressed synovial fluid leukocyte apoptosis and sustained synovial hyperplasia [1–5]. If not properly treated, chronic joint inflammation can lead to permanent joint damage, and thus, lead to deformity. Although the pathophysiological causes of RA are not fully known, oxidative stress-induced cellular deoxyribonucleic acid (DNA) damage has been implicated in its pathogenesis [6–9].
In human cells, oxidative DNA attacks are happening thousands and millions of times per cell per day [10]. We are concerned that these attacks may lead to unfavorable genetic alternations. Normal cellular metabolism and environmental factors such as radiation and UV light appear to be the endogenous and exogenous instigators of DNA damages [10]. For this, the evolutionary process has equipped us with sophisticated DNA repair systems to preserve genetic stability. The major mechanism that cells use to repair oxidative DNA lesions, such as 8-oxo-7,8-dihydroguanine (8-oxo-G) and numerous types of oxidative DNA base damage products, is known as the base excision repair (BER) system [11, 12]. BER is initiated by DNA glycosylases, which recognize and remove specific damaged or mispaired bases, forming AP sites. These AP sites are then cleaved by AP endonucleases to yield a 3' hydroxyl adjacent to a 5' deoxyribosephosphate (dRP). The resulting single-strand break can subsequently be repaired by either short-patch or long-patch BER [10, 13]. The association of base excision repair (BER) of oxidative DNA damage and oxidative stress with RA were described previously [6–9]. Here, we are interested in the enzymes involved in the BER pathway and its association with RA.
mutY Homolog (E. coli) (MUTYH) is a unique DNA glycosylase that proficiently recognizes and catalyzes the removal of a mispaired adenine (A) from A:8-oxo-G, a frequent DNA lesion estimated to emerge around 1000–7000 times per cell per day [14–16]. Thus MUTYH is a key factor for giving a way to the supreme BER that eventually restores the undamaged guanine (G) [17, 18]. Most DNA is packaged in chromosomes within the nucleus (nDNA), but it can also be found in the mitochondria (mtDNA). Both nDNA and mtDNA are essential for proper cellular functioning; they are needed to flawlessly replicate from one cell cycle to the next. This may be the reason why MUTYH, as a genome caretaker, is localized to both the nucleus and the mitochondria [19, 20]. Gene polymorphism and mutations that lead to MUTYH inactivation have been associated with many cancers and cancer-associated inflammatory responses [21–24]. A number of studies have demonstrated that oxidative DNA strand break, and oxidative stress are significantly increased in the mononuclear leukocytes, serum and synovial fluid from RA patients than healthy controls [6–9]. The presence of oxidized DNA in joints has also been linked with arthritis in both mice [25] and humans [26]. We have previously described the association of rheumatoid arthritis (RA) prevalence and two mutY Homolog (E. coli) (MUTYH) SNPs (rs3219463 and rs3219476) among the Taiwanese population [27]. In this present study, we increased the sample size of the cohort and aimed to find out whether the SNPs can alter the expression of MUTYH in the progression of RA.
Methods
Patient selection
The study subjects included 368 RA patients and 364 healthy subjects, which were recruited from China Medical University Hospital in Taiwan. Patients with RA, according to the revised America College of Rheumatology criteria [28, 29], were enrolled. Nephelometry detected the rheumatoid factor (RF), and values ≧ 30 IU/ml were defined as positive. The gender-age-matched healthy controls from the general population were selected by health examination. Blood samples were collected by venipuncture for genomic DNA isolation. Informed consent was obtained from all participants, and it was according to the guidelines approved by the local ethics committee.
Genomic DNA extraction, genotyping and quantitation of MUTYH copy number
Genomic DNA (gDNA) was prepared from peripheral blood using the genomic DNA kit from Roche. Polymerase chain reaction was used to identify the MUYTH rs3219463 polymorphism (Fig. 1). A pre-designed copy number assay (assay ID: Hs01177408_cn) from Applied Biosystems was used to quantify MUTYH copy number by TaqMan®Real-Time PCR. Primer sequences, reaction components, thermal cycling profiles and identification of various MUTYH genotypes by restriction enzymatic digestion and gel electrophoresis are all provided in the supplementary section.
Fig. 1.
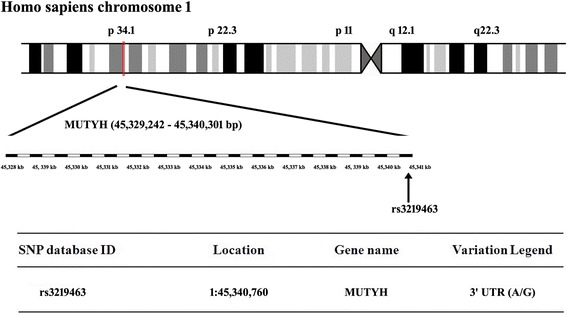
Map of MUTYH (rs3219463) located within Chromosome 1p34.1 region (45,329,242-45,340,301 bp)
Quantitative determination of MUTYH in serum
Serum samples were diluted 1/200 in dilution buffer to be in the range of standard curve and then directly quantified by enzyme linked immunosorbent assay (Cat No. 30–7110, ALPCO Diagnostics, USA). The human MUTYH in the serums samples were bound to monoclonal mouse antibodies against human MUTYH, which were immobilized on the surface of the microtiter plates. After a washing step, the quantification of bound human MUTYH was carried out by adding a rabbit anti-human MUTYH antibody. Detection of the bound rabbit antibody was performed by a peroxidase labeled goat anti-rabbit antibody. The amount of converted substrate was directly proportional to the amount of bound human MUTYH and was photo metrically determined at 450 nm.
Statistical analysis
The genotypic and allelic frequencies of MUTYH rs3219463 SNP for the RA patients and controls were compared using the chi-square test. When one cell had an expected count of <1 or >20% of the cells had an expected count of < 5, Fisher’s exact test was used. Results were considered statistically significant when p values less than 0.05. The odds ratios (OR) were calculated from the genotypic frequency and allelic frequency with a 95% confidence interval (95% CI) for the MUTYH rs3219463 SNP. The statistical analysis was performed by using SPSS version 11. The Mann–Whitney U test and Kruskal-Wallis method were used for non-parametric comparisons, and Student’s t test was used for parametric comparisons [30].
Results
Genotypic and allelic frequency distributions of MUTYH rs3219463 SNP among Taiwan’s Han Chinese population
The genotypic and allelic frequency distributions of rs3219463 SNP in the MUTYH gene were summarized in Table 1. The Hardy-Weinberg model was used to describe and predict genotype and allele frequencies in our study. We observed G allele to be the major allele in the population, regardless of whether they were in the patient group or the control group – at rs3219463 SNP site, the G allele frequencies were 57.6% (424 out of 736) for patients and 67.2% (489 out of 728) for the controls. By comparing the genotypic distributions between RA patients and healthy controls, our data indicated that individuals who carried GG or GA (G carrier) at rs3219463 SNP site are statistically significantly at a lower risk for developing RA (p < 0.0002).
Table 1.
Genotypic and allelic frequency of MUTYH-437 (rs3219463) genetic polymorphism in the RA patients and controls
| RA patient | Control | OR (95% CI) | p value | |
|---|---|---|---|---|
| MUTYH-437 (rs3219463) | n = 368 (%) | n = 364 (%) | ||
| AA | 54 (14.7) | 47 (12.9) | 1.80 (1.14-2.84) | 6E-06 * |
| AG | 204 (55.4) | 145 (39.8) | 2.20 (1.60-3.03) | |
| GG | 110 (29.9) | 172 (47.3) | Ref | |
| Allelic frequency | ||||
| Allele A | 312 (42.4) | 239 (32.8) | 1.51 (1.22-1.86) | 0.0002 * |
| Allele G | 424 (57.6) | 489 (67.2) | Ref | |
CI confidence interval, OR odds ratio
* Statistically significant
Normal MUTYH CNV in RA patients
Blood leukocytes gDNA samples were available from 227 RA patients and 223 healthy controls. For the remaining patients and controls, insufficient gDNA was collected to quantify MUTYH CNV. RA patients were not associated with abnormal MUTYH CNVs (Table 2). All patients and controls had 2 copies of the MUTYH gene present in their genome.
Table 2.
Distribution of MUTYH CNV in the RA patients and controls
| RA patient | Control | OR (95% CI) | Chi-Square | Fisher’s Exact test | |
|---|---|---|---|---|---|
| n = 227 (%) | n = 223 (%) | p value | p value | ||
| MUTYH CNV | |||||
| 0, 1 or 3 | 0 | 0 | --- | --- | --- |
| 2 | 227 (100%) | 223 (100%) | |||
CI confidence interval, OR odds ratio
Increased serum level of MUTYH in RA patients
Serum levels of MUTYH were available from 40 RA patients and 38 healthy controls. For the remaining patients and controls, insufficient serum was collected to quantify MUTYH. There was a statistically significant increase in serum MUTYH concentration among RA patients (Fig. 2, p < 0.005). The mean MUTYH concentrations per milliliter of serum samples from RA patients and healthy controls are as follows: 21.62 ± 16.53 ng/mL for RA patients and 19.87 ± 27.47 for healthy controls. RA patients had 8.8% higher serum MUTYH levels than their age-, gender- and race-matched healthy controls.
Fig. 2.
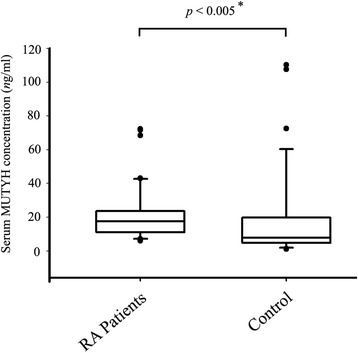
The expression level of MUTYH in the serum of RA patients and controls. p value was calculated by T-Test and Mann–Whitney U test. * means data is statistically significant
Classification of functional capacity and measurement of joint changes in RA patients carry risk- and non-risk-associated MUTYH SNP at rs3219463 site
RA patients can be categorized into four groups based on their functional capacity assessment. Class 1 patients can do all the usual activities without help. Class 2 patients can also do most of the normal activities despite feeling uncomfortable and thus limiting their mobility of one or more joints. Class 3 patients can do only a few tasks, and they are unable to work or they may not take care of themselves very well. Class 4 patients rely solely on others to take care them. There were no statistically significant difference between the RA-risk-associated group (AA at rs3219463) and the RA-non-risk-associated group (G carriers at rs3219463) in regards to the various RA diagnosis classes (Table 3). For the assessment of joint pain and joint stiffness, those RA patients who are G carriers at rs3219463 are less likely to have painful joints (Table 3, p < 0.01). We also performed quantitative joint examinations for our RA patients: the golden standard 66 swollen/68 tender joint counts (SJC66/TJC68), the disease activity score in 28 joints (DAS28), patient global assessment of disease activity (PtGA) and provider global assessment of disease activity (PrGA) were investigated among our RA patients. In our cohort, RA patients who are G carriers at rs3219463 showed statistically significant lower DAS 28 value (Table 3, p < 0.01) than the ones who carried AA at rs3219463. In addition, although the data was not statistical significance, those RA patients who were G carriers at rs3219463 tend to have lower SJC 66, TJC66, PtGA and PrGA values than those who carry AA at rs3219463 (Table 3).
Table 3.
Classification of functional capacity and measurement of joint changes in RA patients
| RA patients | Chi-Square a
p value |
Fisher’s Exact test a
p value |
||
| Genotype at rs3219463 | ||||
| G carrier - n (%) | AA - n (%) | |||
| RA Diagnosis class | ||||
| I | 88 (75.2) | 13 (56.5) | 0.315 | 0.199 |
| II | 19 (16.2) | 6 (26.1) | ||
| III | 8 (6.8) | 3 (13.0) | ||
| IV | 2 (1.7) | 1 (4.3) | ||
| RA patients | T-Test a
p value |
Mann–Whitney U test a
p value |
||
| Genotype at rs3219463 | ||||
| G carrier - n (mean ± SD) | AA - n (mean ± SD) | |||
| MUTYH serum level b | 27 (20.98 ± 16.51) | 13 (23.07 ± 17.16) | 0.714 | 0.593 |
| Joint pain | 135 (31.93 ± 24.33) | 28 (45.89 ± 25.46) | 0.007 * | 0.006 * |
| Joint stiffness | 102 (0.62 ± 0.97) | 19 (0.58 ± 0.72) | 0.871 | 0.540 |
| SJC 66 b | 142 (1.96 ± 2.69) | 26 (2.81 ± 3.18) | 0.156 | 0.250 |
| TJC 68 b | 142 (3.83 ± 4.07) | 26 (6.69 ± 7.06) | 0.055 | 0.075 |
| DAS 28 | 133 (3.88 ± 1.2) | 25 (4.63 ± 1.23) | 0.005 * | 0.003 * |
| PtGA b | 135 (39.65 ± 26.72) | 27 (50.48 ± 27.82) | 0.058 | 0.052 |
| PrGA b | 142 (23.73 ± 17.73) | 27 (32.78 ± 23.38) | 0.065 | 0.078 |
a Nonparametric test
b Mean value is lower in the G carrier / RA-non-risk-associated but data does not reach statistical significance
* Statistically significant
Discussion
Rheumatoid arthritis (RA) can affect any joint in the body; the severity varies from person to person. Although the exact causes of RA are unknown, oxidative stress-induced DNA damage plays a big part. We performed a candidate gene study to investigate the association of MUTYH genetic variants with their expression and disease severity in RA patients. MUTYH was chosen as a candidate gene because MUTYH polymorphisms and mutations have been associated with various cancers and cancer-linked inflammatory responses [21–25]. RA is characterized by proliferative and invasive synovial fibroblasts in the synovium, a cell population with properties similar to cancer cells [1–5]. We initially tested the association of four SNPs (rs3219463, rs3219476, rs3219489 and rs3219493; data not shown) tagging MUTYH with the incidence of RA in a Taiwan Han Chinese cohort study. The MUTYH G to A polymorphism at rs3219463 SNP site was identified as a susceptible allele in relation to RA prevalence. Then, we carried out several follow-up studies that gave us a more detailed picture of MUTYH rs3219463 polymorphism and its association with RA severity.
The present study extends our earlier publication in 2015 on SNP rs3219463 of MUTYH in Taiwan-Chinese RA [27]. The sample sizes of RA increased from 192 to 368, and healthy control subjects from 192 to 364. As a follow-up study, the sample sizes for RA patients and controls were still relatively small. It would have been more impactful if the sample size for each group was greater than 500 samples. Currently, compared with other studies using Asian individual groups, we have the biggest sample size RA research. Here, we not only reporting the characterization of a SNP and potential copy number variation of MUTYH in rheumatoid arthritis patients, but also the MUTYH protein levels in RA patient’s serum. Our results indicate a significantly higher level of MUTYH in patients than in controls. To our knowledge, this is the first report on MUTYH serum protein levels in RA patients. And we think that is a very important information in RA development.
MUTYH is a DNA glycosylase that recognizes and excises mispaired adenine (A) bases from the DNA backbone. Post-translational phosphorylation of MUTYH is particularly important to grant itself the unique glycosylase activity on A:8-oxo-G and A:G mispairs [18, 31]. This enzyme, found in either nucleus or mitochondria, predominantly removes A from A:8-oxo-G mispairs under physiological salt concentrations [17]. As a genome caretaker that defends genome integrity, MUTYH is ubiquitously expressed and its activity is directed to newly synthesized DNA strands [32]. When MUTYH function is compromised, A:8-oxo-G mispairs on the newly synthesized DNA are not fixed, CG → AT transversion mutations can be generated in the next round of replication. Accumulations of deleterious mutations in normal cells ultimately transform them into cancer cells [33]. MUTYH-associated polyposis (MAP) and colorectal cancer (CRC) are the well-characterized hereditary conditions related to MUTYH mutations [23]. Indeed, the BER deficiency associated with MUTYH inactivation can happen to any cell type and affect any gene. The MAP tumor spectrum has been expanded to extra-colonic organs [22, 24, 34–37]. Synovial hyperplasia in RA is similar to a hyperplastic tumor, together with the fact that increased oxidative DNA damage and oxidative stress have been demonstrated in RA patients [6–9], we believed that MUTYH might have a role to play in RA pathology.
In this study, we examined the association of rheumatoid arthritis prevalence and the MUTYH rs3219463 polymorphism among Taiwan’s Han Chinese population. MUTYH rs3219463 SNP is located in 3’non-coding regions. This SNP has been linked with higher treatment-related mortality (TRM) risk and disease relapse in patients who have undergone allogeneic hematopoietic cell transplantation (HCT) [38]. Thyagarajan et al. proposed that lower BER activity can increase damage to normal tissues, resulting in higher cell death and thus increasing the risk of TRM [38]. MUTYH composes of localization sequence, a DNA binding domain, an adenine bind motif and several interaction domains for APE1, PCNA, RPA and MSH6 [39]. Various MUTYH isoforms differ in their 5’ sequence or first exon and they can be grouped into three categories, namely α, β and γ [40]. Studies have described MUTYH isoforms to differ in their 5’ sequence which will put them into different cellular compartments, either nucleus or mitochondria [19, 20, 40–42]. However, the distribution and functional statuses of different MUTYH isoforms to the different subcellular locations in different cell and tissue types are still not known. Indeed, examples of inter-isoformal regulation of their functional counterparts are held almost everywhere [43, 44]. Therefore, we propose to proceed further with the study of MUTYH isoform profiling and distribution in association with MUTYH SNPs.
Our pilot study has attempted to demonstrate whether MUTYH rs3219463 is associated with RA susceptibility and disease severity. There was a statistically significant increase in serum MUTYH concentration among RA patients. Unfortunately, we lacked samples to monitor the A/8oxoG base repair. Increased MUTYH expression may compensate for the loss of functional MUTYH isoform but further experiments are needed to validate this hypothesis. Also, a detailed questionnaire evaluated the joint damage (using joint pain, joint stiffness, SJC66/TJC68 and DAS28) and the global assessment of disease activity (using PtGA and PrGA) for our RA patients [45]. Although there were no statistically significant differences between various genotypes regarding RA function classes, indeed, RA patients who carried a potentially protective genotype (i.e. G carrier at rs3219463) had less severe disease symptoms – They were statistically significant in lower joint pain values and DAS scores. In addition, although the data did not reach statistical significance, RA patients who are G carriers at rs3219463 tend to have lower SJC 66, TJC66, PtGA and PrGA values than those who carried AA at rs3219463. Actually, the evaluation was somehow based on subjective analysis rather than on rigorous criteria. Pain level, stiffness level and tenderness level are subjective experiences of individuals. RA is a chronic illness and many patients with RA will suffer from depression. We cannot rule out that some patients may report physical inability and uncomfortableness were caused by psychological factors. In order to get statistically significant findings, a larger study group will be required to further verify the relationship between disease phenotypes and genotypes.
Both RF and anti-CCP are specific markers for RA because they are produced as part of the process that leads to joint inflammation in rheumatoid arthritis [46]. CRP is usually ordered along with ESR, and they measure how much inflammation is in the body [47]. Although ESR and CRP are not specific tests, flaring up of these values do indicate that you have inflammation somewhere in your body. The RA patients who are G carriers at rs3219463 tend to have lower mean RF and ESR values than the group who carried AA, but the difference did not reach statistical significance, possibly because of the small sample size of the subgroup analysis. The sample size is closely tied to statistical power. We have to admit that we have a small sample size in some subgroups, and sometimes we cannot get enough blood for both routine and RA-specific blood tests. Therefore, additional studies are needed to validate these results by using a larger cohort of RA patients.
Medications that can reduce joint inflammation, relieve pain and slow down joint damage are prescribed to treat or reduce the symptoms of RA. However, some of these medications can cause serious side effects [48, 49]. In addition, RA itself can increase the risk of developing certain cancers and organ dysfunctions in patients. These complications may affect serum MUTYH levels. We, thus, carefully documented the drug use patterns of each RA patient and no serious side effects were observed.
Conclusions
In conclusion, our pilot study showed that RA is associated with rs3219463 SNP in MUTYH gene and increased serum level of the MUTYH protein. These findings suggest EGFR is a valuable therapeutic target in the treatment of RA, and thus, is worthy of further investigation.
Acknowledgements
We are grateful to the patients for donating their samples to our research studies, staff within the research and clinical teams at Genetic Center, China Medical University Hospital for help in obtaining and processing samples for funding this research.
Funding
This work was supported by China Medical University Hospital in Taiwan (DMR-102-110 and DMR-103-102), Taiwan Ministry of Health and Welfare Clinical Trial and Research Center of Excellence (MOHW105-TDU-B-212-133019) and National Science Council in Taiwan (NSC99-2314-B-039-005-MY3).
Availability of data and materials
The data cannot be publicly shared because permission was not obtained from the participants but are available from the corresponding author on reasonable request.
Authors’ contributions
According to the definition given by the International Committee of Medical Journal Editors (ICMJE), the authors listed above qualify for authorship based on making one or more of the substantial contributions to the intellectual content of: (i) Conception and design [SYC, CMH, FJT]; and/or, (ii) Analysis and interpretation of data [SYC, CMH, FJT, YCH, SPL, YJL, YYC, HWL]; and/or (iii) Participated in drafting of the manuscript [SYC, CMH, FJT, HHC]; and/or (iv) Critical revision of the manuscript for important intellectual content [SYC, CMH, FJT, HHC, SFL]. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
This study was approved by the institutional review board (IRB) of china medical university hospital (Taichung, Taiwan) prior to patient enrollment. All participants provided written informed consent.
Abbreviations
- 95% CI
95% Confidence interval
- CNVs
Copy number variations
- DAS
Disease activity score
- MUTYH
mutY Homolog (E. coli)
- RA
Rheumatoid arthritis
- SJC66/TJC68
Swollen joint count (SJC66 joints) and tender joint count (TJC68 joints)
- SNPs
Single nucleotide polymorphisms
Contributor Information
Chung-Ming Huang, Email: hcm.jeffrey@msa.hinet.net.
Fuu-Jen Tsai, Email: d0704@mail.cnuh.org.tw.
References
- 1.Müller-Ladner U, Ospelt C, Gay S, et al. Cells of the synovium in rheumatoid arthritis. Synovial fibroblasts. Arthritis Res Ther. 2007;9:223. doi: 10.1186/ar2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palmer G, Gabay C, Imhof BA. Leukocyte migration to rheumatoid joints: Enzymes take over. Arthritis Rheum. 2006;54:2707–10. doi: 10.1002/art.22062. [DOI] [PubMed] [Google Scholar]
- 3.Silverman GJ, Carson DA. Roles of B cells in rheumatoid arthritis. Arthritis Res Ther. 2003;5:S1–6. doi: 10.1186/ar1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weyand CM, Bryl E, Goronzy JJ. The role of T cells in rheumatoid arthritis. Arch Immunol Ther Exp (Warsz) 2000;48:429–35. [PubMed] [Google Scholar]
- 5.Raza K, Scheel-Toellner D, Lee CY, et al. Synovial fluid leukocyte apoptosis is inhibited in patients with very early rheumatoid arthritis. Arthritis Res Ther. 2006;8:R120. doi: 10.1186/ar2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hajizadeh S, DeGroot J, TeKoppele JM, et al. Extracellular mitochondrial DNA and oxidatively damaged DNA in synovial fluid of patients with rheumatoid arthritis. Arthritis Res Ther. 2003;5:R234–40. doi: 10.1186/ar787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altindag O, Karakoc M, Kocyigit A, et al. Increased DNA damage and oxidative stress in patients with rheumatoid arthritis. Clin Biochem. 2007;40:167–71. doi: 10.1016/j.clinbiochem.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 8.Bhusate LL, Herbert KE, Scott DL, Perrett D. Increased DNA strand breaks in mononuclear cells from patients with rheumatoid arthritis. Ann Rheum Dis. 1992;51:8–12. doi: 10.1136/ard.51.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee SH, Chang DK, Goel A, et al. Microsatellite instability and suppressed DNA repair enzyme expression in rheumatoid arthritis. J Immunol. 2003;170:2214–20. doi: 10.4049/jimmunol.170.4.2214. [DOI] [PubMed] [Google Scholar]
- 10.Lodish H, Berk A, Kaiser CA, et al. Molecular Cell Biology, 7th ed. New York: W.H.Freeman & Company; 2012.
- 11.David SS, O’Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–50. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003;17:1195–214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 13.Robertson AB, Klungland A, Rognes T, Leiros I. Base excision repair: the long and short of it. Cell Mol Life Sci. 2009;66:981–93. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.(ESCODD) ESC on ODD Measurement of DNA oxidation in human cells by chromatographic and enzymic methods. Free Radic Biol Med. 2003;34:1089–99. doi: 10.1016/S0891-5849(03)00041-8. [DOI] [PubMed] [Google Scholar]
- 15.Gedik CM, Collins A, ESCODD (European Standards Committee on Oxidative DNA Damage) Establishing the background level of base oxidation in human lymphocyte DNA: results of an interlaboratory validation study. FASEB J. 2005;19:82–4. doi: 10.1096/fj.04-1767fje. [DOI] [PubMed] [Google Scholar]
- 16.Friedberg EC, Walker GC, Siede W, et al. DNA Repair and Mutagenesis: Second Edition. Washington, DC: American Society for Microbiology; 2006. [Google Scholar]
- 17.Shinmura K, Yamaguchi S, Saitoh T, et al. Adenine excisional repair function of MYH protein on the adenine:8-hydroxyguanine base pair in double-stranded DNA. Nucleic Acids Res. 2000;28:4912–8. doi: 10.1093/nar/28.24.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu Y, Lu AL. Differential DNA recognition and glycosylase activity of the native human MutY homolog (hMYH) and recombinant hMYH expressed in bacteria. Nucleic Acids Res. 2001;29:2666–74. doi: 10.1093/nar/29.12.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsai-Wu JJ, Su HT, Wu YL, et al. Nuclear localization of the human mutY homologue hMYH. J Cell Biochem. 2000;77:666–77. doi: 10.1002/(SICI)1097-4644(20000615)77:4<666::AID-JCB14>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 20.Boldogh I, Milligan D, Lee M. hMYH cell cycle-dependent expression, subcellular localization and association with replication foci: evidence suggesting replication-coupled repair of adenine: 8-oxoguanine. Nucleic Acids Res. 2001;29:2802–9. doi: 10.1093/nar/29.13.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oka SNY. DNA glycosylase encoded by MUTYH functions as a molecular switch for programmed cell death under oxidative stress to suppress tumorigenesis. Cancer Sci. 2011;102:677–82. doi: 10.1111/j.1349-7006.2011.01869.x. [DOI] [PubMed] [Google Scholar]
- 22.Goto M, Shinmura K, Yamada H, et al. OGG1, MYH and MTH1 gene variants identified in gastric cancer patients exhibiting both 8-hydroxy-2’-deoxyguanosine accumulation and low inflammatory cell infiltration in their gastric mucosa. J Genet. 2008;87:181–6. doi: 10.1007/s12041-008-0028-0. [DOI] [PubMed] [Google Scholar]
- 23.Yamaguchi S, Ogata H, Katsumata D, et al. MUTYH-associated colorectal cancer and adenomatous polyposis. Surg Today. 2014;44(4):593–600. doi: 10.1007/s00595-013-0592-7. [DOI] [PubMed] [Google Scholar]
- 24.Mazzei F, Viel A, Bignami M. Role of MUTYH in human cancer. Mutat Res. 2013;743–744:33–43. doi: 10.1016/j.mrfmmm.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 25.Deng GM, Nilsson IM, Verdrengh M, et al. Intra-articularly localized bacterial DNA containing CpG motifs induces arthritis. Nat Med. 1999;5:702–5. doi: 10.1038/9554. [DOI] [PubMed] [Google Scholar]
- 26.van der Heijden IM, Wilbrink B, Tchetverikov I, et al. Presence of bacterial DNA and bacterial peptidoglycans in joints of patients with rheumatoid arthritis and other arthritides. Arthritis Rheum. 2000;43:593–8. doi: 10.1002/1529-0131(200003)43:3<593::AID-ANR16>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 27.Kung YJ, Tsai KS, Huang CM, et al. MUTYH gene polymorphisms as risk factors for rheumatoid arthritis. Biomed Res Int. 2015;2015:893796. doi: 10.1155/2015/893796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arnett FC, Edworthy SM, Bloch DA, The American Rheumatism Association et al. revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1987;1988:315–24. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 29.Aletaha D, Neogi T, Silman AJ, et al. 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum. 2010;62:2569–81. doi: 10.1002/art.27584. [DOI] [PubMed] [Google Scholar]
- 30.Lin YJ, Liu X, Chang JS, et al. Coronary artery aneurysms occurrence risk analysis between Kawasaki disease and LRP1B gene in Taiwanese children. Biomedicine (Taipei) 2014;4:10. doi: 10.7603/s40681-014-0010-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parker AR, O’Meally RN, Sahin F, et al. Defective human MutY phosphorylation exists in colorectal cancer cell lines with wild-type MutY alleles. J Biol Chem. 2003;278:47937–45. doi: 10.1074/jbc.M306598200. [DOI] [PubMed] [Google Scholar]
- 32.Hayashi H, Tominaga Y, Hirano S. Replication-associated repair of adenine: 8-oxoguanine mispairs by MYH. Curr Biol. 2002;12:335–9. doi: 10.1016/S0960-9822(02)00686-3. [DOI] [PubMed] [Google Scholar]
- 33.Greenman C, Stephens P, Smith R, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vogt S, Jones N, Christian D, et al. Expanded extracolonic tumor spectrum in MUTYH-associated polyposis. Gastroenterology. 2009;137:1976–85. doi: 10.1053/j.gastro.2009.08.052. [DOI] [PubMed] [Google Scholar]
- 35.Win AK, Cleary SP, Dowty JG, et al. Cancer risks for monoallelic MUTYH mutation carriers with family history of colorectal cancer. Int J Cancer. 2011;129:2256–2262. doi: 10.1002/ijc.25870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ponti G, Ponz de Leon M, Maffei S, et al. Attenuated familial adenomatous polyposis and Muir-Torre syndrome linked to compound biallelic constitutional MYH gene mutations. Clin Genet. 2005;68:442–7. doi: 10.1111/j.1399-0004.2005.00519.x. [DOI] [PubMed] [Google Scholar]
- 37.Miyaishi A, Osawa K, Osawa Y, et al. MUTYH Gln324His gene polymorphism and genetic susceptibility for lung cancer in a Japanese population. J Exp Clin Cancer Res. 2009;28:10. doi: 10.1186/1756-9966-28-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thyagarajan B, Lindgren B, Basu S, et al. Association between genetic variants in the base excision repair pathway and outcomes after hematopoietic cell transplantations. Biol Blood Marrow Transplant. 2010;16:1084–9. doi: 10.1016/j.bbmt.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Half E, Bercovich D, Rozen P. Familial adenomatous polyposis. Orphanet J Rare Dis. 2009;4:22. doi: 10.1186/1750-1172-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohtsubo T, Nishioka K, Imaiso Y, et al. Identification of human MutY homolog (hMYH) as a repair enzyme for 2-hydroxyadenine in DNA and detection of multiple forms of hMYH located in nuclei and mitochondria. Nucleic Acids Res. 2000;28:1355–64. doi: 10.1093/nar/28.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takao M, Aburatani H, Kobayashi K, Yasui A. Mitochondrial targeting of human DNA glycosylases for repair of oxidative DNA damage. Nucleic Acids Res. 1998;26:2917–22. doi: 10.1093/nar/26.12.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takao M, Zhang QM, Yonei S, Yasui A. Differential subcellular localization of human MutY homolog (hMYH) and the functional activity of adenine:8-oxoguanine DNA glycosylase. Nucleic Acids Res. 1999;27:3638–44. doi: 10.1093/nar/27.18.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang ET, Sandberg R, Luo S, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–6. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pelechano V, Wei W, Steinmetz LM. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature. 2013;497:127–31. doi: 10.1038/nature12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anderson J, Caplan L, Yazdany J, Robbins ML, Neogi T, Michaud K, Saag KG, O'Dell JR, Kazi S. Rheumatoid arthritis disease activity measures: American College of Rheumatology recommendations for use in clinical practice. Arthritis Care Res (Hoboken) 2012;64:640–7. doi: 10.1002/acr.21649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lal P, Su Z, Holweg CT, et al. Inflammation and autoantibody markers identify rheumatoid arthritis patients with enhanced clinical benefit following rituximab treatment. Arthritis Rheum. 2011;63:3681–91. doi: 10.1002/art.30596. [DOI] [PubMed] [Google Scholar]
- 47.Crowson CS, Rahman MU, Matteson EL. Which measure of inflammation to use? A comparison of erythrocyte sedimentation rate and C-reactive protein measurements from randomized clinical trials of golimumab in rheumatoid arthritis. J Rheumatol. 2009;36:1606–10. doi: 10.3899/jrheum.081188. [DOI] [PubMed] [Google Scholar]
- 48.Klinge SA, Sawyer GA. Effectiveness and safety of topical versus oral nonsteroidal anti-inflammatory drugs: a comprehensive review. Phys Sportsmed. 2013;41:64–74. doi: 10.3810/psm.2013.05.2016. [DOI] [PubMed] [Google Scholar]
- 49.Armstrong AW, Brezinski EA, Follansbee MR, Armstrong EJ. Effects of biologic agents and other disease-modifying antirheumatic drugs on cardiovascular outcomes in psoriasis and psoriatic arthritis: a systematic review. Curr Pharm Des. 2014;20:500–12. doi: 10.2174/138161282004140213123505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data cannot be publicly shared because permission was not obtained from the participants but are available from the corresponding author on reasonable request.


