Abstract
Chromosome replication in Escherichia coli is initiated by DnaA. DnaA binds ATP which is essential for formation of a DnaA-oriC nucleoprotein complex that promotes strand opening, helicase loading and replisome assembly. Following initiation, DnaAATP is converted to DnaAADP primarily by the Regulatory Inactivation of DnaA process (RIDA). In RIDA deficient cells, DnaAATP accumulates leading to uncontrolled initiation of replication and cell death by accumulation of DNA strand breaks. Mutations that suppress RIDA deficiency either dampen overinitiation or permit growth despite overinitiation. We characterize mutations of the last group that have in common that distinct metabolic routes are rewired resulting in the redirection of electron flow towards the cytochrome bd-1. We propose a model where cytochrome bd-1 lowers the formation of reactive oxygen species and hence oxidative damage to the DNA in general. This increases the processivity of replication forks generated by overinitiation to a level that sustains viability.
Author Summary
In most bacteria chromosome replication is initiated by the DnaA protein. In Escherichia coli, DnaA binds ATP and ADP with similar affinity but only the ATP bound form is active. An increased level of DnaAATP causes overinitiation and cell death by accumulation of DNA strand breaks. These strand breaks often result from forks encountering gapped DNA formed during repair of oxidative damage. We provide evidence that cell death in overinitiating cells can be prevented by rewiring the metabolism to favor the micro-aerobic respiratory chain with the cytochrome bd-1 as terminal oxidase. Cytochrome bd-1 is found in aerobic as well as anaerobic bacteria. Its role is to reduce O2 in micro-aerobic conditions and work as an electron sink to prevent the formation of reactive oxygen species. Our results suggest that bacteria can cope with replication stress by increasing respiration through cytochrome bd-1 to reduce the formation of reactive oxygen species, and hence oxidative damage to a level that does not interfere with replication fork progression.
Introduction
Initiation of chromosome replication from the unique replication origin of E. coli oriC, is tightly controlled and happens once and only once per cell cycle [1, 2]. Chromosome replication is initiated by the DnaA initiator protein. DnaA is an AAA+ ATPase that exists in an ATP bound form and an ADP bound form [3]. DnaA associated with either ATP or ADP binds a set of strong recognition sites in oriC throughout the cell cycle [4] to form the origin recognition complex (ORC;[5]). Upon initiation the DnaA protein associated with ATP forms the orisome by binding to numerous additional sites in oriC. This displaces Fis (Factor for Inversion Stimulation), a protein that binds oriC for most of the cell cycle. With Fis gone, the IHF (Integration Host Factor) protein can bind oriC, which ultimately leads to duplex opening [6, 7], helicase loading and assembly of two replisomes [8].
The level of DnaAATP fluctuates during the cell cycle and is high at the time of initiation [9]. Following initiation, DnaAATP is converted to DnaAADP by the RIDA (Regulatory Inactivation of DnaA) and DDAH (datA-dependent DnaAATP hydrolysis) processes. During RIDA, the Hda protein complexed with the DNA loaded β-clamp stimulates the intrinsic ATPase activity of DnaA thereby converting DnaAATP to the non-active DnaAADP [10, 11]. DDAH is less efficient and takes place at the datA locus where a complex of datA and IHF promotes DnaAATP hydrolysis [12]. If extra initiation events are triggered by loss of RIDA or by conditional mutations in DnaA [13], DNA strand breaks progressively accumulate, eventually resulting in cell death. It was shown that the lethal accumulation of strand breaks in such cells resulted from replication forks encountering DNA damage repair intermediates, particularly resulting from oxidative damage to the DNA during normal aerobic growth. Therefore, growth could be restored in the absence of oxygen or by removing the predominant glycosylase of oxidized bases [14].
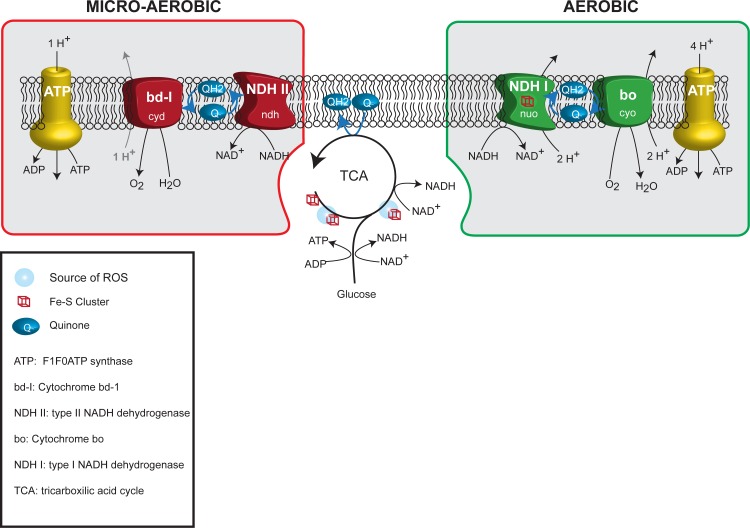
During aerobic growth, a proton gradient is generated by a respiratory chain made of the type I dehydrogenases containing iron-sulfur proteins and the cytochrome bo that is efficient and has low affinity for oxygen (Fig 1)[15]. It is controversial how Reactive Oxygen Species (ROS) are produced in E. coli [16]. Respiration per se is not generating ROS [17, 18]. In contrast, respiration is thought to limit ROS formation by pulling away electrons from potential ROS-sources [19]. For example, mutants lacking NAD dehydrogenases I and II or cytochrome oxidases bo and bd-1 produces more H2O2. The main cellular sources of ROS are thought to be free iron, flavins and iron sulfur cluster proteins with the dehydratase enzymes of the TCA cycle as the main culprits [20].
Fig 1. Simplistic representation of oxidative phosphorylation pathways in E. coli.
The aerobic (green frame) and micro-aerobic (red frame) respiratory chain are represented together with the tricarboxylic acid cycle and glycolysis. Iron sulfur clusters and known sources of ROS associated with these pathways are also shown.
During micro-aerobic growth, another set of proteins which are less efficient in generating a proton gradient dominates the respiratory chain. These consist of a copper containing dehydrogenase (NDHII) and the cytochrome bd-I terminal oxidase that has a high affinity for oxygen (Fig 1). This micro-aerobic chain is also predominant when iron is scarce or during oxidative stress [21] (for review [15, 22]). During these stress conditions the cytochrome bd-I is thought to act as an electron sink to reduce the ROS level [19].
We previously identified two mutations in the iscU and fre genes (iscUC63F and freΔ68) that suppress RIDA deficiency [23]. These genes encode an Iron-Sulphur cluster scaffold protein and Flavin reductase, respectively. Here, we provide evidence that the mechanism of suppression is not linked to DnaA or replication initiation activity. Global transcription analysis of iscUC63F and freΔ68 cells showed that genes encoding enzymes of the TCA cycle were down regulated in both mutants while respiration was altered to favor the use of the micro-aerobic respiratory chain. Therefore, these two mutants may tolerate overinitiation in a manner similar to cells growing in the absence of oxygen. For the freΔ68 mutant, we show that the ArcA regulon plays a crucial role for suppression in part by upregulating cyd transcription to overproduce cytochrome bd-1 [24, 25].
Results
The iscUC63F and freΔ68 mutations suppress Hda deficiency without reducing initiations from oriC
Hda mutant cells accumulate strand breaks under aerobic conditions resulting in progressive growth inhibition, and loss of colony forming ability, unless a suppressor mutation is acquired [23, 26, 27]. The nature of several suppressor mutations was previously identified [23]. One suppressor is a missense mutation in iscU resulting in cysteine being replaced with phenylalanine at position 63 of the scaffold protein for assembly of iron sulfur clusters, IscU (IscUC63F). Iron sulfur clusters are used in a variety of cellular activities such as respiration, amino acid synthesis and DNA repair. A second suppressor is a 380 bp deletion between two imperfect repeats starting at position 497bp after the start codon of the fre gene and ending in the intergenic fre-fadA region. This results in a premature stop codon and loss of the 68 C-terminal amino acids of the flavin reductase (FreΔ68). The Flavin reductase catalyzes the reduction of free flavins by NAD(P)H. It is thought that Fre accounts for more than 80 percent of the free Flavin reduction in E. coli [28, 29] and may serve as a general cytosolic source of electrons [30].
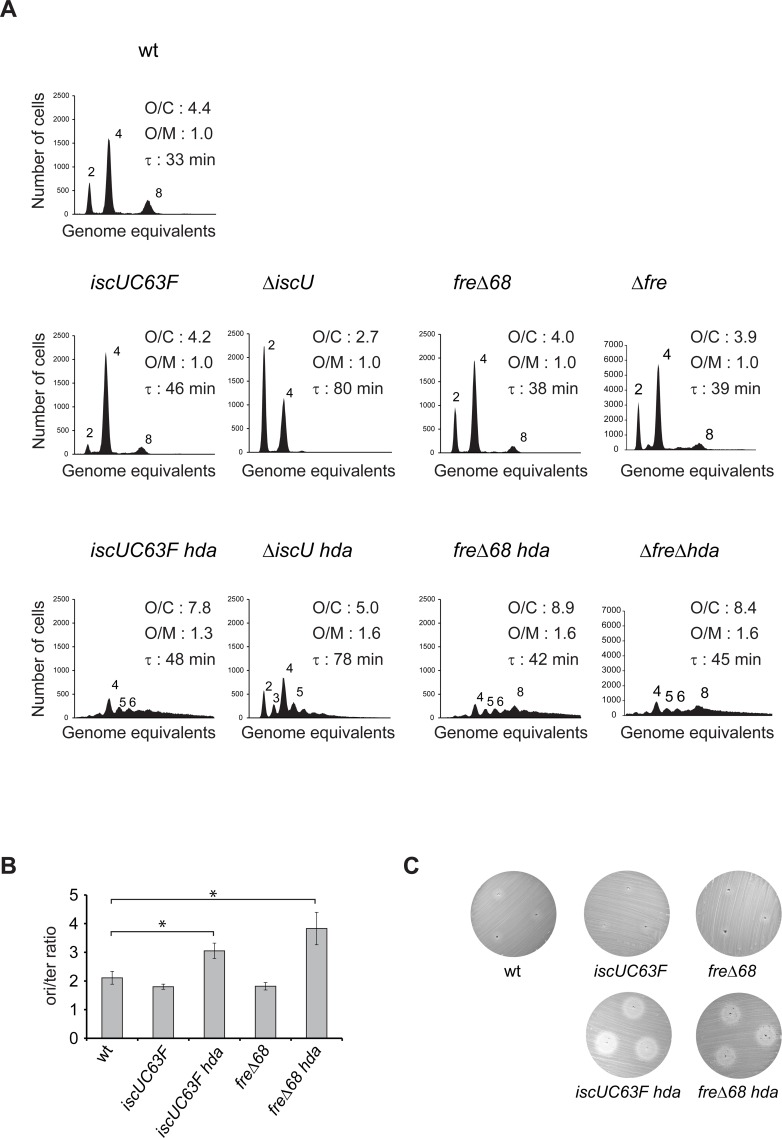
Cells carrying iscUC63F and freΔ68 mutations grew somewhat slower than the wild-type (Fig 2A). Analysis of chromosome replication by flow cytometry revealed that both mutants initiated in synchrony with a small reduction in numbers of origins per cell corresponding to the slower growth rate but the origin concentration remained unchanged (Fig 2A). Loss of RIDA activity by deletion of hda (referred to as loss of RIDA throughout this work), resulted in initiation asynchrony, increased the average number of origins per cell from around 4 to 7.8 and 8.9 for iscUC63F and freΔ68, respectively, and increased the origin concentration (Fig 2A). Note that these are minimum estimates for numbers of origins per cell as runout was too poor to allow for an exact enumeration. This shows that the iscUC63F and freΔ68 mutations restore growth of Hda deficient cells despite of continued overinitiation.
Fig 2. Cell cycle profile of iscU and fre mutants.
A) Cells were grown exponentially in AB medium supplemented with 0.2% glucose and 0.5% casamino acids and treated with rifampicin and cephalexin prior to flow cytometric analysis. Each panel represents a minimum of 30000 cells. The average ori/cell (O/C), ori/mass (O/M) relative to wild-type and mass doubling time (τ) are inserted in the histograms. B) ori/ter ratio determined by qPCR analysis. Shown is the mean ± s.d. (n = 3),* = p<0.05. C) IscUC63F and freΔ68 mutations do not alter the sensitivity to hydroxyurea. Sensitivity to hydroxyurea is tested by a diffusion assay on cells plated on LB agar medium. Cells are plated evenly on LB plates, 3μl of 500mM HU was introduced into 3 separated holes and plates incubated at 37°C for 16h.
In order to determine whether the iscUC63F and freΔ68 mutations resulted in change or loss of function of their respective proteins we proceeded to delete iscU and fre from otherwise wild-type cells. Complete loss of Flavin reductase resulted in a cell cycle profile similar to that observed for the freΔ68 mutant cells suggesting that the FreΔ68 protein is not functional (Fig 2A). Loss of iscU on the other hand resulted in cells that grew slower than iscUC63F cells and that had a reduced number of cellular origins (Fig 2A). Loss of iscU restored growth of Hda deficient cells while these continued to overinitiate in an asynchronous manner (Fig 2A). The relative increase in origin concentration caused by the deletion of hda in ΔiscU relative to iscUC63F may be a simple consequence of a better runout in ΔiscU cells. This suggests that ΔiscU is a better suppressor than iscUC63F but altogether the data indicate that the IscUC63F protein is partly functional which is in agreement with an earlier report showing that an IscUC63A mutant protein is able to form and transfer [2Fe-2S] clusters with reduced activity compared to the wild-type enzyme [31]. The ori/ter ratio of iscUC63F and freΔ68 also increased from a level of 1.8 similar to wild-type cells, to about 3 and 3.8 upon loss of Hda (Fig 2B). Taken together, the origin concentration and ori/ter ratios indicates that replication fork processivity was uncompromised in Hda deficient cells also carrying iscUC63F and freΔ68 mutations, despite of overinitiation. A Pulse Field Gel Electrophoresis analysis confirmed this, as the levels of fragmented chromosomes in hda iscUC63F and hda freΔ68 was greatly reduced compared to Hda deficient cells (S1 Fig).
iscUC63F and freΔ68 mutations suppress the loss of Hda in a growth rate independent manner
We previously suggested that formation of lethal double strand breaks in an hda mutant is intimately linked to the number of replicative forks moving on the chromosome [14]. The number of ongoing replication forks in a cell can be reduced simply by reducing growth rate. We therefore streaked anaerobically generated hda cells on minimal medium plates supplemented with glycerol (S2A Fig) and with glucose and casamino acids (S2B Fig) and incubated aerobically at 37°C. Hda cells failed to form colonies on minimal plates with glucose and casamino acids, but formed colonies as well as wild-type cells on minimal medium plates supplemented with glycerol. To ensure that the cells had not accumulated suppressor mutations, glycerol grown hda cells were restreaked on LB agar in aerobic condition (S2C Fig). These cells failed to form colonies, implying that the glycerol selected hda clones had not acquired suppressor mutations. The growth rate of hda cells in minimal medium supplemented with glycerol was similar to that of the wild-type but initiation of replication was asynchronous and origin concentration was increased (S2D Fig). Hda cells were then shifted from growth in glycerol to glucose and casamino acids (S2E Fig) or LB medium (S2F Fig) for three mass doubling time. As expected, this resulted in an increased origin concentration (relative origin per mass equal to 1.7 or 1.8) reminiscent of the shift observed between anaerobic to aerobic conditions [14]. Overall, these observations indicate that hda is dispensable in slow growing cells, consistent with what has been observed for other overinitiation mutants [32].
Cells carrying iscUC63F and freΔ68 mutations had a reduced growth rate in minimal medium supplemented with glucose and casamino acids relative to wild-type cells (46 and 38 versus 33 minutes, respectively; Fig 2A) and contained fewer origins and hence fewer ongoing replication forks per cell. A possibility was therefore that the ability of these mutants to suppress Hda deficiency resulted from slow growth. The iscUC63F and freΔ68 mutations were originally isolated as hda suppressors from cultures grown in LB [23, 26]. In this medium, the mutants grew with doubling times of 30 and 21 minutes, respectively, and contained a high number of origins and ongoing replication forks per cell, yet loss of Hda was tolerated (S3 Fig). A reduced growth rate is therefore not the main mechanism of suppression in these cells.
Hda deficient iscUC63F and freΔ68 cells are sensitive to hydroxyurea
Since overexpression of Ribonucleotide Reductase Ia (RNRIa) encoded by nrdAB genes or RNRIb encoded by nrdEF genes were previously shown to suppress the loss of hda [27, 33], we tested whether an increase in amount or activity of these enzymes is present in iscUC63F and freΔ68 cells. To assess the Ribonucleotide Reductase activity we made use of the RNR inhibitor hydroxyurea (HU). The sensitivities to hydroxyurea of iscUC63F and freΔ68 mutant cells were similar to wild-type in our assay (Fig 2C) although a fre mutant has been previously shown to have a 10 to 50% growth reduction in the presence of 10 to 40 mM HU. In the absence of Hda, both the iscUC63F and freΔ68 mutants became hypersensitive to HU, indicating that the RNR activity is indeed critical for survival and that the dNTP pool is limiting in such cells.
Global transcription profile of iscUC63F and freΔ68 cells
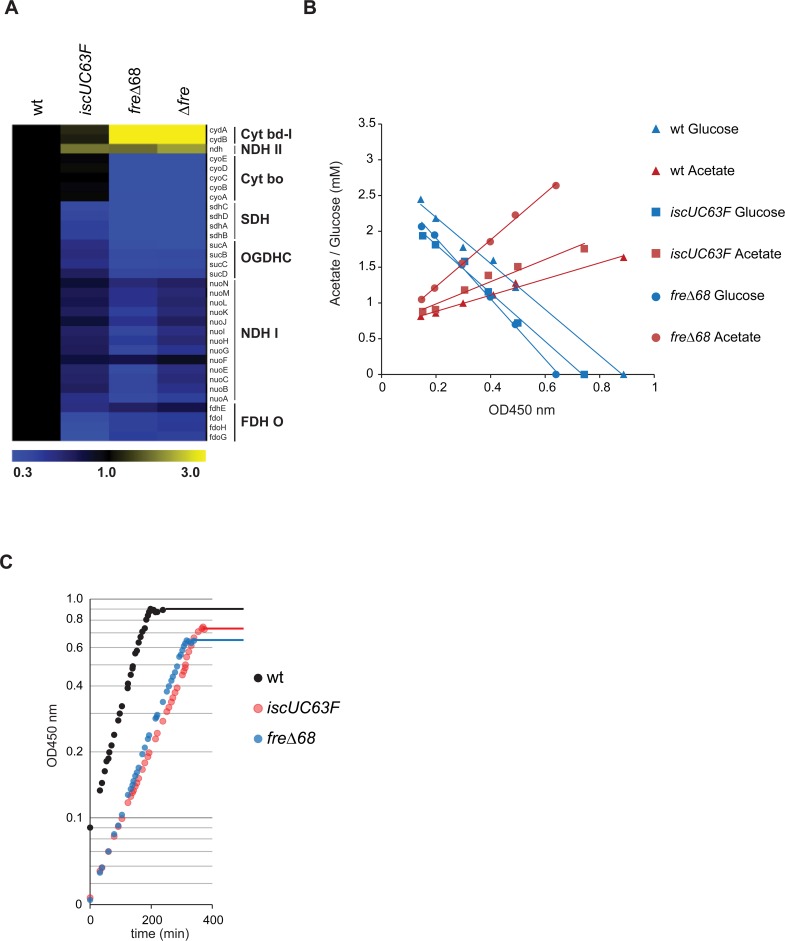
To gain insight into the mechanism of hda suppression by the iscUC63F and freΔ68 mutations we performed a global transcription analysis of cells grown in minimal medium supplemented with glucose and casamino acids using microarrays. The two mutants had both changes in gene expression that were specific to each mutant and changes that were in common. Genes that were specifically upregulated in the iscUC63F mutant included those encoded by the iscRSUA hsbB and sufABCDSE operons (S1 Table) which is in agreement with a poorly functioning IscUC63F protein [34–36]. Note that we also observe an increased expression of nrdHEIF (see discussion). In the freΔ68 strain, specifically the cyd operon encoding cytochrome bd-1 was upregulated and the majority of dehydrogenases were downregulated (S2 Table). Overproduction of Fre has little consequences on the transcription profile (S2 Table). Similarities between iscUC63F, freΔ68 and Δfre cells included expression of genes whose products are involved in cellular respiration (Fig 3A, S1 and S2 Tables). The sdh operon encoding succinate dehydrogenase (succinate-coenzyme Q reductase; SDH), the suc operon encoding 2-oxoglutarate dehydrogenase (OGDHC), the nuo operon encoding NADH dehydrogenase I (NDH-I)and the fdo operon encoding formate dehydrogenase (FDH O), most of which contain [Fe-S] clusters, are all part of the respiratory chain and are down-regulated. In addition the cyo genes encoding cytochrome o oxidase (Cyt bo) (S2 Table), are downregulated in freΔ68 and fre deleted cells (Fig 3A, S1 and S2 Tables). On the other hand ndh encoding the type II NADH dehydrogenase (NDH-II), which is a metalo enzyme dependent on copper, is overexpressed in iscUC63F, freΔ68 and Δfre cells. This is in accordance with a report showing that in iscU mutants, NDH-I activity is reduced to background level while NDH-II activity is increased [37]. Finally, the cydAB genes encoding cytochrome bd-1 (Cyt bdI) (S2 Table) and to some extent the genes encoding appBC cytochrome bd-2 (Cyt bdII) are overproduced in freΔ68 and fre deleted cells. The microarray results are coming from single experiments, therefore, the expression patterns of cydA, sdhD and cyoA genes were confirmed by RT-qPCR (S3 Table).
Fig 3. The respiro-fermentative metabolism of IscUC63F and freΔ68 mutant cells.
A) Expression profiles of genes involved in the respiratory chain as obtained by whole genome microarray. Expression intensities relative to wild-type are displayed by a two color gradient from blue (low) to yellow (high). Shown, the expression profile of operons encoding succinate dehydrogenase (SDH), 2-oxoglutarate dehydrogenase (OGDHC), NADH dehydrogenase I (NDH-I) and II (NDH-II), formate dehydrogenase (FDH O), cytochrome o oxidase (Cyt bo) and bd-1 (Cyt bd-1). B) Measurement of glucose consumption and acetate production during growth in glucose limited medium (AB minimal medium supplemented with 0.04% glucose). The plot shows the concentration of glucose (blue symbols), and acetate (red symbols) over optical density (no lactate, formate, citrate, ethanol and succinate were detected). Samples for final optical density for wild-type, IscUC63F and freΔ68 were measured at growth arrest (OD450 nm 0.88, 0.75, 0.64 respectively). C) Growth of wild-type, IscUC63F and freΔ68 cultures in AB minimal medium supplemented with 0.04% glucose.
Overall these expression data points towards IscUC63F and freΔ68 cells being shifted from a normal aerobic respiration pattern towards what would be characteristic of cells growing under micro-aerobic or iron limited conditions (Fig 1).
A metabolic shift in IscUC63F and freΔ68 cells
We proceeded to determine whether IscUC63F and freΔ68 cells had their metabolism shifted from the normal aerobic respiration towards the less efficient Cytochrome bd-1 dependent respiratory pathway or even fermentation, despite of growing in aerobic conditions. In order to obtain precise measurements of metabolites, cells were grown in minimal medium supplemented with 0.04% glucose. Surprisingly, we found that the ATP/ADP ratio was increased by 50–60% in both IscUC63F and freΔ68 cells (Table 1). This rules out that these mutations suppress overinitiation by lowering the ATP/ADP ratio which in turn could lower the DnaAATP/DnaAADP ratio, as both nucleotides bind DnaA with similar affinities [3]. The NADH/NAD+ ratio was similar in wild-type and IscUC63F cells but was increased by more than two-fold in freΔ68 cells. The production of acetate per mole of glucose was also increased by 100% and 40% in freΔ68 and IscUC63F cells, respectively (Table 1; Fig 3B). Formate, ethanol, succinate, citrate and lactate were not produced in any of the strains. The IscUC63F and freΔ68 strains also reached lower optical density when glucose consumption ended, i.e. had a lower yield of biomass per mole of glucose (Fig 3C).
Table 1. Metabolic parameters of IscUC63F and freΔ68 mutant cells.
| Wild-type | IscUC63F | freΔ68 | |
|---|---|---|---|
| Doubling time (min)1 | 60 | 89 | 81 |
| Growth rate (ln2/td)/h1 | 0.70 | 0.47 | 0.51 |
| Specific rate of Glucose consumption 1 (mmol/gDW/h) | 11.80 | 8.28 | 11.44 |
| Specific rate of Acetate production 1 (mmol/gDW/h) | 4.15 | 3.80 | 8.82 |
| Acetate yield on Glucose (mol/mol) 1 | 0.35 | 0.46 | 0.77 |
| Biomass yield on Glucose (gDW/mmol) 1 | 0.059 | 0.056 | 0.045 |
| Relative Respiration rate (%) 2 † (% oxygen/gDW/min) | 100 (7.6) | 77.2 (5.5) | 63.8 (4.0) |
| ATP/ADP ratio1 | 7.5 (0.7) | 12.1 (2.0) | 11.4 (1.9) |
| NADH/NAD+ ratio1 | 0.2* | 0.2* | 0.5* |
*Average of two samples: wild-type = (0.24 + 0.18), IscUC63F = (0.21 + 0.21), freΔ68 = (0.55 + 0.48)
1Cells were grown in AB medium supplemented with 0.04% glucose at 37°C.
2Cells were grown in AB medium supplemented with 0.2% glucose at 37°C.
† wild-type respiration rate = 885% oxygen/gDW/min.
These results agree with the microarray data (Fig 3A) and show that the metabolism of iscUC63F and freΔ68 cells is partly re-routed toward acetate production as would be expected when the TCA cycle is downregulated. The increase in acetate production in freΔ68 cells is consistent with a high NADH/NAD+ ratio [38, 39], which in turn agrees with the suggestion that flavin reductase accounts for a significant part of the NADH cellular oxidation [28]. The high ATP/ADP ratio in iscUC63F and freΔ68 suggests that anabolic reactions are reduced in the strains. This increased ratio could also decrease the flux in the TCA cycle by allosteric inhibition [40]. Our microarray analysis indicates that respiration in iscUC63F and freΔ68 cells could be shifted towards the less efficient Cytochrome bd-1, therefore it is expected that ATP production through oxidative phosphorylation is reduced in these strains. The oxygen consumption of iscUC63F and freΔ68 cells was decreased somewhat as expected (Table 1).
Role of the Arc regulon in hda suppression
The transcription profiles of freΔ68 and Δfre cells are by and large consistent with the activation of the ArcA regulon [25, 41]. The ArcBA (anoxic redox control) two-component system, senses the redox state of the cell and reprograms the metabolism to increase the availability of NAD+ required for glycolysis. ArcA activity is coupled to the NADH/NAD+ balance and conditions that artificially increase the ratio NADH/NAD+ have been shown to activate the ArcA regulon [38].
When oxygen becomes limiting, ArcA represses genes involved in the generation of NADH, including those specifying enzymes of the TCA cycle, while genes encoding enzymes involved in fermentative regeneration of NADH into NAD+ are activated [25].
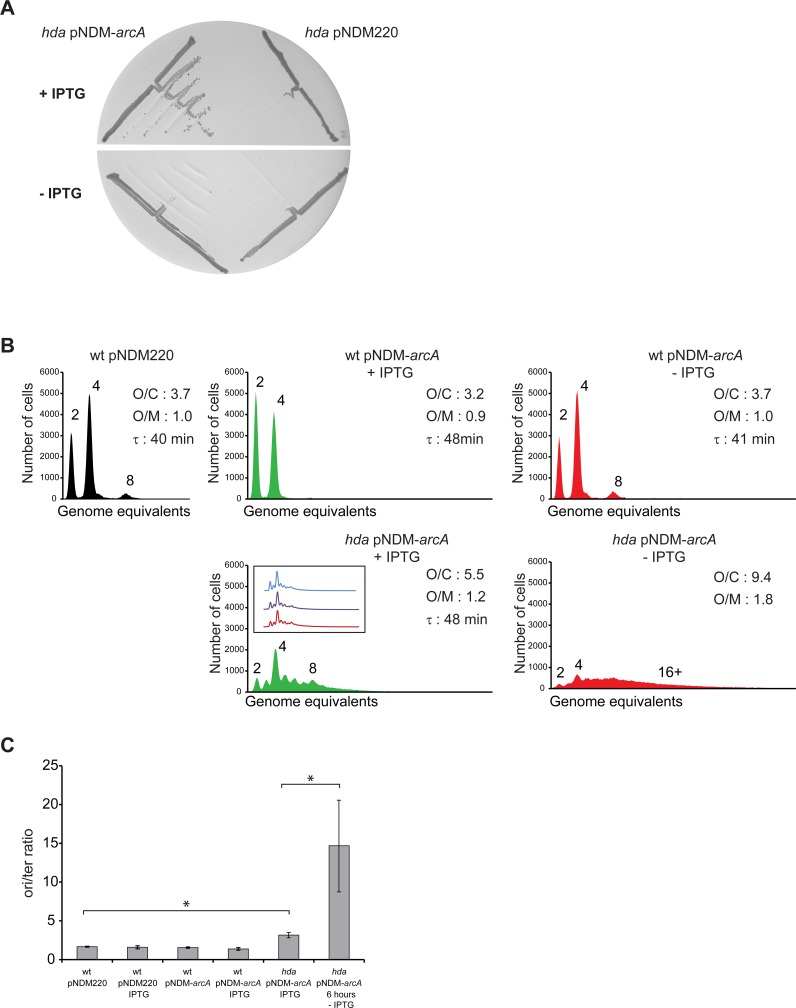
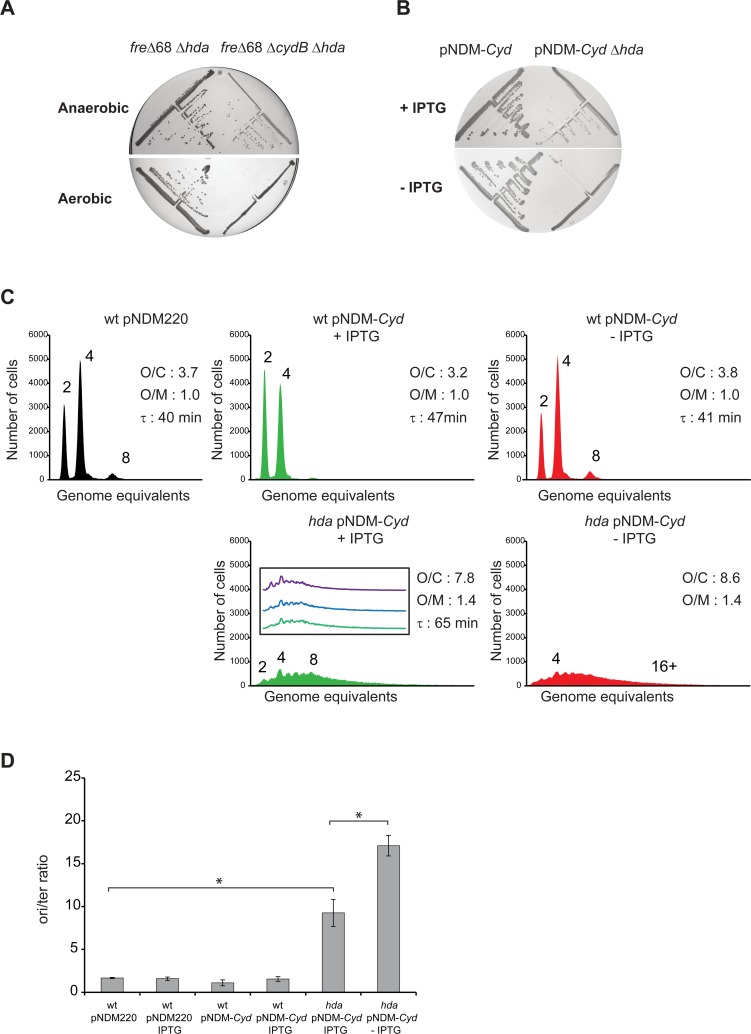
We cloned arcA under control of the IPTG regulated promoter pA1/O4/O3 promoter [42] in the R1 based plasmid pNDM220 [43] and proceeded to delete hda in the presence of IPTG. Restreaking of the resultant colonies indicated that colony formation depended on IPTG (Fig 4A) and suggests that ArcA is necessary for freΔ68 dependent suppression of Hda deficiency. Flow cytometry analysis revealed that initiation of replication in independent clones of hda cells overexpressing arcA was more frequent and asynchronous than in wild-type cells (Fig 4B). Similar to what was observed for freΔ68Δhda cells (Fig 2), the ori/ter ratio was somewhat increased (Fig 4C). When ArcA was depleted by shifting the cells to a medium lacking IPTG for six hours, the number of origins per cell increased dramatically as did the ori/ter ratio (Fig 4B and 4C). The relative origin concentration was also increased. This indicates that cells were not able to complete chromosome replication, probably due to DNA damage associated arrest or collapse of replication forks, while initiation of replication remained unperturbed [14].
Fig 4. Hda deficiency is suppressed by ArcA overexpression.
A) The hda::cat mutation was introduced into wild-type cells containing plasmid pNDM220 or pNDM-arcA under anaerobic conditions. Cells were restreaked on LB plates with or without the IPTG and incubated aerobically at 32°C for 24 hours. B) Cells were grown exponentially at 32°C in AB medium supplemented with 0.2% glucose and 0.5% casamino acids and treated with rifampicin and cephalexin prior to flow cytometric analysis. When indicated cells were grown with 1mM IPTG. The flow diagram of “hda/pNDM-arcA -IPTG” was obtained from cells grown in presence of 1mM IPTG then washed and regrown in absence of IPTG for 6 hours. Each panel represents a minimum of 30000 cells. The average ori/cell (O/C), ori/mass (O/M) relative to wt/pNDM220 and mass doubling time (τ) are inserted in the histograms. C) The cellular ori/ter ratio determined by qPCR analysis. Shown is the mean ± s.d (n = 3),* = p<0.05.
Altogether these results indicate that the freΔ68 mutation results in a high NADH/NAD+ ratio which activates the ArcA regulon to shift cells from the normal aerobic respiration towards the less efficient Cytochrome bd-1 dependent respiratory pathway, despite of growing in aerobic conditions. In the absence of ArcA overproduction, initiation of replication continued but cells were not able to complete chromosome replication.
Loss of ATPase activity suppress RIDA (Hda) Deficiency
We proceeded to mimic the metabolism of fermenting cells while growing aerobically by deleting the atpA or atpB genes encoding the F1F0 ATP synthase. In such cells the respiration rate increases but is uncoupled, i.e. does not result in ATP production end energy is solely produced by fermentation. Consequently the cellular ATP/ADP ratio is reduced to about one third of the wild-type level [44]. In ATPase deficient cells, cytochrome bo remains unchanged, cytochrome bd-1 and NDHII are overproduced while the TCA cycle is repressed leading to a redirection of the glycolytic flux towards acetate production [45]. The composition of the respiratory chain used by ATPase deficient cells thus resembles the one used under micro-aerobic conditions and the one used by freΔ68 cells (Fig 1), which is thought to favor NAD+ regenerations [45]. We tested whether cells lacking ATPase could tolerate the loss of Hda similar to freΔ68 cells. Hda was deleted in ΔatpA or ΔatpB cells under anaerobic conditions. Restreaking in the presence of oxygen demonstrated that Hda is indeed dispensable for growth of ATPase deficient cells (Fig 5A). Cells deleted for atpA or atpB had doubling times and cell cycle parameters similar to wild-type cells (Fig 5B). A further loss of hda in the atpA and atpB mutant cells resulted in an elevated number of chromosomes per cell, an increased origin concentration and an increased ori/ter ratio (Fig 5C) relative to wild-type. Note that although increased, the ori/ter ratio in hda atpA and hda atpB is still lower than that of hda cells shifted from anaerobic to aerobic growth for 4 hours (Fig 5C), indicating that replication fork progression is affected in hda atpA and hda atpB but to a tolerable level. Again this demonstrates that the cellular ATP/ADP ratio has little influence on replication initiation, and that loss of ATPase function permits cells to survive despite of overinitiation similar to what we observed for iscUC63F and freΔ68 cells.
Fig 5. Loss of ATP synthase restores growth of Hda deficient cells.
A) The hda::cat mutation was introduced into wild-type,ΔatpA and ΔatpB cells under anaerobic conditions, restreaked on LB plates and incubated under aerobic and anaerobic conditions. B) Wild-type, atpA, atpB, atpA hda and atpB hda mutants cells were grown exponentially in AB medium supplemented with 0.2% glucose and 0.5% casamino acids and treated with rifampicin and cephalexin prior to flow cytometric analysis. Each panel represents a minimum of 30000 cells. The average ori/cell (O/C), ori/mass (O/M) relative to wild-type and mass doubling time (τ) are inserted in the histograms. C) The cellular ori/ter ratio were determined by qPCR analysis. Shown is the mean ± s.d (n = 3),* = p<0.05.
Cytochrome bd-1 function is critical for survival in absence of hdA
The cytochrome bd-1 complex is upregulated in fre (S2 and S3 Tables) and ATPase mutants [45] and may provide protection against ROS because of its high affinity to O2 and potential peroxidase activity [21, 46–48].
We deleted the cydB gene, essential for the cytochrome bd-1 activity, in wild-type, freΔ68 and freΔ68 hda, mutant at 32°C under anaerobic conditions. The low temperature was chosen as cydB mutants grow poorly at 37°C [49]. Colonies were restreaked anaerobically and aerobically and revealed that loss of cydB in the freΔ68 hda mutant led to severe growth inhibition under aerobic conditions (Fig 6A). Therefore cytochrome bd-1 is instrumental in the freΔ68 mechanism of hda suppression.
Fig 6. Cytochrome bd-1 function is required for freΔ68 survival in absence of Hda.
A) The hda::cat mutation was introduced into freΔ68 and freΔ68ΔcydB cells under anaerobic conditions, restreaked on LB plates under aerobic and anaerobic conditions. Plates were incubated at 32°C for 24 hours. B) The hda::cat mutation was introduced into wild-type cells containing plasmid pNDM220 or pNDM220-cyd under anaerobic conditions. Cells were restreaked on LB plates with or without the IPTG and incubated aerobically at 32°C for 24 hours. C) Cells were grown exponentially at 32°C in AB medium supplemented with 0.2% glucose and 0.5% casamino acids and treated with rifampicin and cephalexin prior to flow cytometric analysis. When indicated cells were grown with 1mM IPTG. The flow diagram of “hda/pNDM-cyd -IPTG” was obtained from cells grown in presence of 1mM IPTG that was washed and regrown in absence of IPTG for 16 hours. The average ori/cell (O/C), ori/mass (O/M) relative to wild-type pNDM220 and mass doubling time (τ) are inserted in the histograms. D) The cellular ori/ter ratios were determined by qPCR analysis. Shown is the mean ± s.d (n = 3),* = p<0.05.
We also deleted cydB in atpA hda and atpB hda mutants and similar to freΔ68 hda, the cells were unable to grow aerobically showing that cytochrome bd-1 is also essential for hda suppression through loss of ATPase activity (S4 Fig). Although not overproduced in iscU mutants the cytochrome bd-1 was also found essential when hda iscUC63F cells were grown aerobically, indicating its function is critical despite the presence of the cytochrome bo (S4 Fig).
We proceeded to overproduce cytochrome bd-1 (pNDM-Cyd) and found that hda could be deleted in such cells without loss of ability to form colonies (Fig 6B) although the small colony size suggested that pNDM-Cyd was a relatively poor hda suppressor. In agreement with this, independent clones of hda cells overexpressing cytochrome bd-1 grow with a relative high cellular origin concentration but were all similar when analyzed by flow cytometry (Fig 6C). When depleted for cytochrome bd-1, the origin concentration increased overtime although this was difficult to assess due to incomplete runout after rifampicin and cephalexin treatment. The ori/ter ratio was increased from about 10 to about 20 following 16 hours of cytochrome bd-1 depletion (Fig 6D).
ROS levels in iscUC63F and freΔ68 cells
In E. coli, a series of dedicated enzymes detoxify endogenously generated reactive oxygen species. Among the different systems that sense ROS, the OxyR system that perceives and reacts to the threat of hydrogen peroxide accumulation is the best understood. To preserve the cellular homeostasis in case of an H2O2 assault, OxyR activates the transcription of genes involved in protective or detoxifying processes [50]. Although OxyR activation is mostly considered a sensor of extracellular elevated H2O2 levels [50], down regulation of the OxyR regulon in iscUC63F, freΔ68 cells could imply that these cells generate less H2O2 endogenously. The microarray data did however not reveal down regulation of the OxyR regulon in iscUC63F, freΔ68 cells (S5 Fig). As microarray data may not reveal minor differences in gene expression we decided determine expression of katG, encoding a catalase strongly induced by OxyR, by using a katG::lacZ transcriptional fusion carried on the chromosome [51]. We did not observe major changes in katG expression in iscUC63F and freΔ68 (S6 Fig) which could indicate that the cytoplasmic level of H2O2 is not reduced in the mutants. We did not find the Sox regulon is affected in iscUC63F and freΔ68 cells either (S5 Fig; S1 Appendix). Although this may indicate that superoxide levels are not changed in the mutants, the SoxS/R system is now believed to react to redox cycling molecules rather than superoxide directly [52].
We also assessed the impact of superoxide dismutase sodA and sodB mutations on survival of overinitiating cells. Deletion of sodA had little effect on the growth of wild-type, hda freΔ68 cells while deletion of sodB appeared deleterious (S7 Fig). This implies that superoxide accumulation is toxic in overinitiating cells but tolerated in wild-type cells. SodA and SodB differ especially by the nature of the metal cofactor Mn and Fe respectively [53]. MnSodA is involved in oxidative stress response, while FeSodB normally provides the generic scavenging activity. The sensitivity of hda freΔ68 to the loss of sodB only may reflect the fact that sodA is repressed by ArcA directly at the transcriptional level and potentially also indirectly at the translational level through the small RNA FnrS (not present in our microarray) [54]. Altogether, this indicates that sodB hda freΔ68 may be low in cytoplasmic superoxide dismutase activity, a stressing situation already for wild-type cells.
Discussion
In E. coli excessive initiations from oriC result in progressive growth inhibition due to the accumulation of DNA strand breaks [13, 14]. The isolation and characterization of second site suppressor mutations that overcome this inviability has revealed that they fall into two categories. The first category includes mutations that reduce oriC activity and are found in oriC itself [55], in dnaA or in genes that affect DnaA function or activity [23, 27, 33, 56–59]. The second category of suppressor mutations have in common that they do not reduce initiation frequency but allow cells to survive in spite of overinitiation. These mutations facilitate replication fork progression along the chromosome either by increasing the size of the dNTP pool (overexpression of Ribonucleotide Reductase; [27, 33]), by altering the DNA topology (ex: mutation in hns; [60]), or by limiting the repair of DNA damages (i.e. mutM;[14]). Here we characterize three hda suppressor mutations in iscU, fre and atpAB that seemingly promote viability by limiting oxidative damage to DNA.
An altered energy charge does not affect replication initiation from oriC
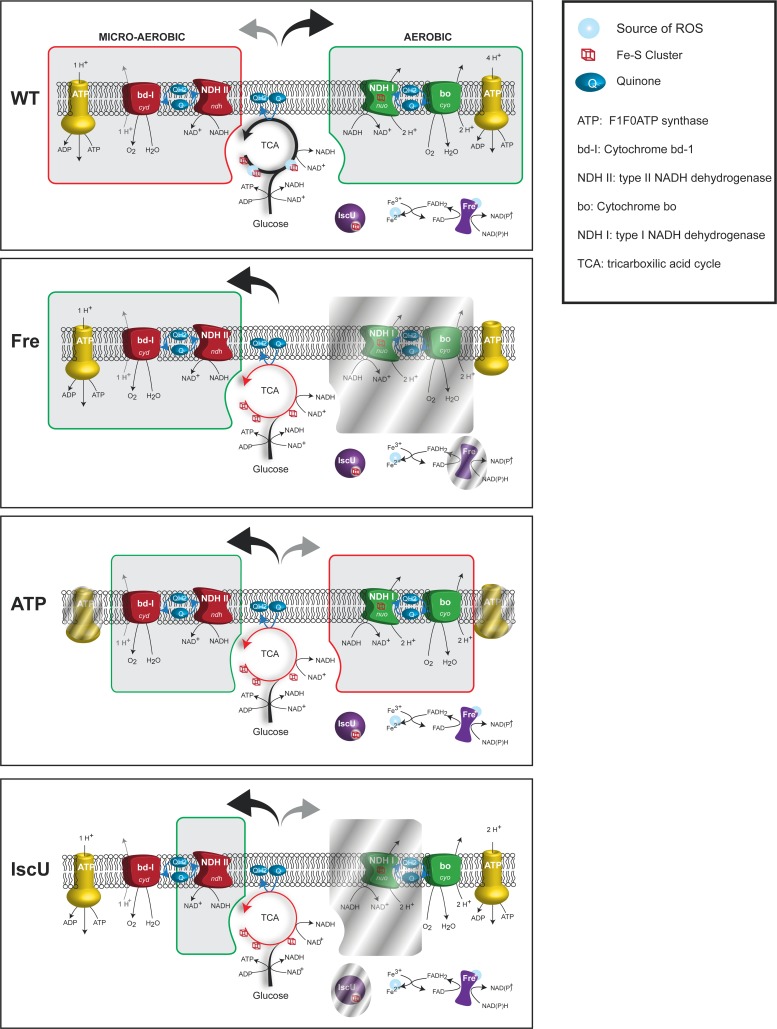
The iscUC63F, freΔ68 and atpAB mutants all have in common that the TCA cycle is downregulated and all, or parts of the micro-aerobic respiratory chain is upregulated (Fig 7). While the micro-aerobic respiratory chain is relatively inefficient in generating a proton gradient (Fig 7), the ATPase mutants cannot even utilize the proton gradient for ATP production and one might therefore suspect that the ATP/ADP ratio is lowered in all of these cell types. Because DnaA has the same affinity for ATP and ADP [3] a lowered ATP/ADP ratio would result in generation of less DnaAATP upon rejuvenation or de novo synthesis, which could contribute to a lowered overall DnaAATP/DnaAADP ratio. In turn, this could explain the ability of the iscUC63F mutation to suppress RIDA deficiency by lowering initiations from oriC. However, two lines of evidence argue against this. First, we found that initiation of replication was not significantly affected in any of the mutants with respect to origin concentration and initiation synchrony. Second, the ATP/ADP ratio is actually increased in iscUC63F and freΔ68, and only reduced in atpAB mutants [44].This argues that the changes in ATP/ADP ratio observed here has little influence on initiation frequency. This also implies that either the ATP/ADP ratio must be changed more dramatically to affect initiation of replication or that other mechanisms dedicated to maintain the balance DnaAATP/DnaAADP such as RIDA, DDAH, and DARS mediated rejuvenation counteract any gross variations in the cellular ATP/ADP ratio to maintain an initiation frequency that is tightly coupled to cell mass increase.
Fig 7. Respiratory pathways in cells carrying hda suppressor mutations.
Representation of the TCA cycle and the respiratory chain components of cells grown aerobically in presence of glucose. For each strain, the predominant respiratory path is framed in green while secondary or repressed paths are framed in red or shaded respectively.
It should also be noted that the energy charge of E. coli is normally relatively invariable [61–63] at different growth rates and whether cells are grown aerobically or anaerobically. This is thought to be controlled by changes in the glycolysis flux in response to the demands in ATP [44]. In wild-type cells, the cellular energy charge is only affected during adaptation to environmental changes or stress [62, 63].
Increased dNTP synthesis is not the primary suppression mechanism in iscUC63F and freΔ68
Overexpression of the Ribonucleotide Reductase is known to suppress the loss of hda [27, 33]. In iscUC63F cells we observed that transcription of nrdHEIF genes encoding RNR1b was increased about two-fold (S1 Table). RNRIa and RNRIb differ mostly in the dinuclear metal cluster required for their activity: a 2Fe-tyrosyl radical (RNRIa) or a 2Mn-tyrosyl radical (RNRIb). However, the activation of nrdHEIF transcription is assumed to palliate a deficiency of the RNRIa only during iron limitation or oxidative stress [64, 65] and requires a concomitant overexpression of the manganese transporter MntH [64] which is not upregulated in the iscUC63F mutant (S1 Table). The RNRIa activity is likely to be reduced in iscUC63F mutant cells because RNR1a function is normally helped by the [Fe-S] cluster protein YfaE. The net result of the iscUC63F mutation is therefore unlikely to be cells with an increased dNTP pool. This is corroborated by a similar sensitivity of IscUC63F cells to the RNR inhibitor hydroxyurea as wild-type cells. Importantly, Hda deficient cells suppressed by overproduction of RNRIa or RNRIb no longer overinitiate replication, i.e. the origin concentration is similar to wild-type. RNR overproduction therefore reduces oriC activity in hda cells [27]. This is quite different from the continued overinitiation of hda iscUC63F and hda freΔ68 cells, which suggest that suppression is not mediated through an increased dNTP level in these mutants. The efficient HU-mediated killing of Hda deficient iscUC63F cells also shows that RNR activity is critical for survival of these cells and therefore that the dNTP pool is still limiting. The situation is similar in the freΔ68 mutant. Fre is a well-known in vitro activator of RNRIa [66]. The freΔ68 mutant sensitivity to HU was similar to wild-type in our assay but hda freΔ68 cells were effectively killed by HU treatment. This implies that in absence of Hda, the dNTP pool is limiting in iscUC63F and freΔ68, although we cannot exclude that the dNTP pool is larger in comparison to what would be found in Fre+ hda and IscU+ hda cells had they been viable.
Survival through a decrease in cellular ROS production?
The enzymes of the TCA cycle are downregulated in iscUC63F, freΔ68 and atpAB mutants (Fig 7) resulting in reduced ROS production arising from the dehydratases [16]. ROS production is expected to be further reduced in iscUC63F cells as this mutant is partly defective in synthesis of the [Fe-S] clusters required for function of many of the dehydratases (Fig 7). ROS production is also likely to be further reduced in the freΔ68 mutant because the generation of FADH2 is affected in the flavin reductase mutant [28]. FADH2 reduces Fe+++ to Fe++ which participates in the Fenton reaction to generate ROS [17] (Fig 7).
The aerobic respiratory chain is also altered in the mutants (Fig 7). For the iscUC63F mutant, this is due to the lack of Fe-S cluster required for several key enzymes in the pathway along with a transcriptional reprograming towards expression of enzymes devoid of Fe-S (S1 Table). For freΔ68 and atpAB mutants, an increased NADH/NAD ratio triggers a transcriptional repression of major aerobic chain components while the micro-aerobic chain is induced, notably by overproduction of cytochrome bd-1 (S2 Table; [38]), an enzyme known to scavenge or even process ROS species (Fig 7) [48, 67–69]. Cytochrome bd-1 was, although not upregulated, also necessary for suppression of overinitiation in IscUC63F cells, indicating that cytochrome bd-1 plays an essential role for growth even when the aerobic terminal oxidase cytochrome bo is also expressed. The importance of cytochrome bd-1 was demonstrated by showing that overproduction of cytochrome bd-1 directly or indirectly through ArcA overproduction could suppress RIDA deficiency and by showing that cytochrome bd-1 was absolutely required for freΔ68 mediated suppression of RIDA deficiency. There are in principle two known ways that cytochrome bd-1 can act as ROS scavenger. First, through its peroxidase activity, where the cellular localization of cytochrome bd-1 is found would suggest that it sanitizes the periplasm or exogenously created H2O2. Second, and probably most relevant in this context, by oxidizing quinone’s, that are also a source of ROS [70], cytochrome bd-1 could maintain a flux of electrons, thereby preventing flavoproteins (and quinones) to retain electrons and adventitiously passed them on to O2. Because ArcA overproduction did not significantly reduce the origin concentration of wild-type cells it is unlikely that ArcA binding to oriC [71] contribute to its ability to suppress overinitiation.
We previously proposed that aerobic inviability of Hda deficient cells results from an increased number of replication forks of which some may encounter an intermediary in the repair of primarily 8-oxodG to create double stranded breaks [14]. As 8-oxodG arises by oxidation of guanine residues in the DNA, primarily by hydroxyl radicals [72], growth can be restored in anaerobic conditions or by removal the GO-repair system, that is responsible for repairing 8-oxodG lesions.
During aerobic growth hydrogen peroxide (or superoxide) is generated as a consequence of flavin auto oxidation in dehydratases such as aconitase and fumarase of the TCA cycle, a process that could be reduced in freΔ68 and iscUC63F cells, due to down regulation of the flavoproteins in freΔ68 cells (S2 Table) and or inactivation of these enzymes due to the absence of iron sulfur clusters in iscUC63F cells. As ROS levels are notoriously difficult to determine directly [16], we turned to the OxyR regulon that is induced by H2O2. We did not observe major changes in expression of the OxyR regulated katG gene in iscUC63F and freΔ68 cells (S6 Fig) which could indicate that the cytoplasmic level of H202 is not reduced in the mutants. However, the observation that katG expression is not reduced during anaerobic growth either [73], suggest that katG is not a good reporter for H2O2 levels close to or especially below wild-type level. Our observation that katG expression was reduced when ArcA was overproduced could be independent of H2O2 and result in repression of rpoS which in turn resulted in production of less KatG [74]
Although we have not been able to measure a reduction in the cellular ROS level, we favor a model in which iscUC63F, freΔ68 and atpAB mutants sustain growth of overinitiating cells by reducing ROS and hence oxidative damage to the DNA. This is supported by the observation that deletion of sodB which presumably result in accumulation of superoxide, is deleterious for overinitiating cells, but tolerated by wild-type cells. Overall we suggest that iscUC63F, freΔ68 and atpAB mutations are just a few of many putative RIDA suppressors that affect the cellular redox balance and ultimately chromosome stability. We do however recognize that this is a model only, and that iscUC63F, freΔ68 and atpAB mutations have pleiotropic effects on cellular physiology and that mechanisms not involving ROS reduction may at least in part be involved.
RIDA deficient cells carrying the iscUC63F, freΔ68 and atpAB suppressor mutations grow relatively poorly compared to other suppressors that downregulate initiation of replication (i.e. affecting DnaA, SeqA; [23]). This most likely reflects the partial suppression of DNA damages, but a too high DNA concentration could per se be a challenge for the cells in relation to processes such as supercoiling, repair and segregation. This may explain the slow growth of this type of suppressors relative to those that affect the initiation of replication. This also stresses the fact that E. coli has evolved a multitude of mechanisms that ensure that the origin concentration remains relatively invariant regardless of the growth conditions [2].
Materials and Methods
Growth conditions
Cells were grown in Luria–Bertani (LB) medium or AB minimal medium [75] supplemented with 0.2% or 0.04% glucose, 0.5% casamino acids and 10 μg/ml thiamine. LB with 0.2% glucose medium was used for anaerobic growth. Unless specified, all cells were cultured at 37°C. When necessary, antibiotic selection was maintained at the following concentrations: kanamycin 50 μg/ml; chloramphenicol, 20 μg/ml; ampicillin, 150 μg/ml. Cell growth was monitored by measuring optical density at 450 nm for AB minimal medium. Anaerobic growth was performed in an anaerobic jar using anaerobic atmosphere generation bags (BD).
Bacterial strains and plasmids
All strains used are derivatives of MG1655 (F-λ-rph-1) [76]. The hda deletion described previously [26] was moved by P1 mediated generalized transduction [77].
Mutations from the KEIO collection [78] were moved by P1 transduction into MG1655 using lysates of: JW4364 (ΔarcA), JW0723-2 (ΔcydB), JW3712-1 (ΔatpA), JW3716-1 (ΔatpB), JW2513 (ΔiscU) JW3879 (ΔsodA), JW1648 (ΔsodB).
The katG::lacZ gene fusion from strain AL441 [51] was moved into MG1655, iscUC63F and freΔ68 by P1 transduction.
The cyd operon and arcA gene were amplified from strain MG1655 using primers pairs 5’-CTCTAGATTAAGGAGGCCATATGTTAGATATAGTCGAACT-3’/ 5’-AGAGAATTCTGATTTAAAAGAA-3’ and 5’-CTCTAGATTAAGGAGGCCATATGCAGACCCCGCACATTCT-3’/ 5’-CGAATTCTTAATCTTCCAGATCACCGC-3’ respectively. The PCR products were digested with XbaI / EcoRI and cloned into plasmid pFH2102 digested with the same enzymes resulting in plasmids pFH2102CYDOP and pFH2102ARCA. The cyd operon and the arcA gene were then amplified from pFH2102CYDOP and pFH2102ARCA using a primer annealing upstream of the Shine Dalgarno GTTGACTTGTGAGCGGATAA and 5’-AGAGAATTCTGATTTAAAAGAA-3’ (for cyd) or 5’-CGAATTCTTAATCTTCCAGATCACCGC-3’ (for arcA). The PCR products were digested with XhoI/EcoRI and cloned into plasmid pNDM220 digested with the same enzymes, resulting in plasmids pNDM-cyd and pNDM-arcA
Microarray analysis
Strains were grown exponentially in ABTG supplemented with 0.5% casamino acids. At an optical density OD450 = 0.3, 35 ml of culture was transferred to a cold tube containing 5 ml frozen water and centrifugated at 4°C at 10000g for 5min. Pellets were resuspended in 0.5ml ice cold TE-Buffer, transferred to an Eppendorph tube containing 250μl lysis buffer (2% SDS, 16mM EDTA and 200mM NaCl) and 750μl Phenol, whirly mixed and placed at 65°C for 10min with whirly mixing every 3–4 min. Following phenol extraction, each sample was treated with Dnase I for one hour at 4°C and an RNA clean up was made using the RNeasy Mini Spin Column kit from QIAGEN.
cDNA was synthesized using the Revert Aid H Minus first strand synthesis Kit from Thermo Scientific. The cDNA was fragmented with DNAse I in One-Phor-All buffer (Amersham Biosciences) and end-labeled with Biotin using the Enzo BioArray Terminal Labeling Kit. The fluorescent labeled cDNA hybrizized to GeneChip® E. coli Genome 2.0 Array was scanned as described in the Affymetrix UserGuide (www.affymetrix.com) and analyzed using GeneChip Analysis Suite software. Following Robust Multi-array Average (RMA) normalization, a cut off for low expression was applied and genes whose expression varied in the wt strains excluded from the analysis. Raw RMA normalization data can be found in S1 Appendix.
Determination of ori/ter ratio
Was done by quantitative PCR was performed as previously described [14] with modifications. One milliliter of exponentially growing cells (OD450 ~ 0.2) is harvested and put on ice, centrifuged 5 minute 8000 g, the supernatant discarded and cells resuspended in 100 μl of cold 10 mM Tris pH7.5 and fixed by adding 1 ml of 77% ethanol. The samples were stored at 4°C. For qPCR analysis, 100 μl of ethanol fixed cells were centrifuged 7 minutes at 17000 g, the supernatant discarded and the samples were centrifuged for another 30 seconds at 17000 g and the remaining ethanol removed. The cell pellet was resuspended in 1ml cold water and 2 μl of the cell suspension was used as template for qPCR analysis. The Quantitative-PCR was performed using Takara SYBR Premix Ex Taq II (RR820A) in a BioRAD CFX96. All ori/ter ratios were normalized to the ori/ter ratio of MG1655 treated with rifampicin for 2h. The origin and terminus were quantified using primers 5′-TTCGATCACCCCTGCGTACA-3′ and 5′-CGCAACAGCATGGCGATAAC-3′ for the origin and 5′-TTGAGCTGCGCCTCATCAAG-3′ and 5′-TCAACGTGCGAGCGATGAAT-3′ for terminus as previously reported [26].
Flow cytometry
Flow cytometry was performed as described previously [79] using an Apogee A10 Bryte instrument. For each sample, 30 000 to 200 000 cells were analyzed. Numbers of origins per cell and relative cell mass were determined as described previously [79].
Extraction and quantification of intracellular cofactors
Intracellular ATP and ADP were extracted as previously described [80]. ATP/ADP ratios were measured using a luciferin-luciferase ATP kit (Microbial ATP Kit HS, BioThema AB, Sweden). Intracellular NADH and NAD+ was extracted as previously described [81]. The NADH/NAD+ ratio was quantified by a luciferase assay provided by the kit NAD+/NADH-Glo Assay (Promega). The luminescence from each assay was measured using the Infinite® M1000 PRO microplate reader (TECAN) with the 96-well microplates from Greiner Bio-one (Cat. No. 655901).
Quantification of glucose and fermentation products
High-performance liquid chromatography (HPLC) was used to measure the concentration of glucose, acetate, formate, ethanol, succinate, citrate and lactate as previously described [81]
Quantification of oxygen consumption was made in a Bioreactor (Sartorius Biostat Q, 500 mL working volume) equipped with an O2 electrode (Mettler Toledo, Switzerland).
Pulsed-field gel electrophoresis
Was done as described previously [14]
RT-qPCR
RT qPCR was performed on phenol extracted total RNA using QuantiNova SYBR Green RT-PCR Kit using supplier recommendations and specific primers for cydA (5’-TGCGGCCTGTATACCCTGTTCC-3’ and 5’-CGTGCCGGCTGAGTAGTCGTG-3’), cyoA (5’-CCGCTGGCACACGACGAGA-3’ and 5’-AAGCGATTTCATTCACGGTAGCA-3’) and sdhD(5’-GATCGGTTTCTTCGCCTCTG-3’ and 5’-CGGTCAACACCTGCCACAT-3’) [82] and normalized to rpoA (5’-TTGATATCGAGCAAGTGAGTTCG-3’ and 5’- GCATCGATGAGAGCAGAATACG-3’) [27].
Supporting Information
Gene expression is represented as ratio relative to MG1655. Fold changes for genes that are repressed are expressed in negative values. All genes in a given operon where a relevantly expressed gene is located are shown; the gene whose expression is most affected in the operon is highlighted in red (repressed) or green (overexpressed).
(DOCX)
Gene expression is represented as ratio relative to MG1655. Fold changes for genes that are repressed are expressed in negative values. All genes in a given operon where a relevantly expressed gene is located are shown; the gene whose expression is most affected in the operon is highlighted in red (repressed) or green (overexpressed). Genes coding for dehydrogenases are highlighted in yellow Genes coding for cytochrome terminal oxidase are highlighted in blue. Gene products inhibited by NADH are followed by an asterisk. (1) Operons or genes that are differentially expressed in a ΔarcA strain compared to wild-type [25]. (2) Operons or genes directly regulated by ArcA [25]. (3) Operons or genes that are differentially expressed in a ΔarcA strain compared to wild-type [24].
(DOCX)
The indicated cells were grown in AB minimal medium supplemented with 0.2% glucose and 0.5% casamino acids. Relative abundance of cydA, sdhD and cyoA mRNA relative to rpoA mRNA was measured by quantitative RT-PCR. Shown is the mean ± s.d. (n = 3). For comparison, values for the same genes derived from the microarray analysis are included.
(DOCX)
Cells were grown aerobically in AB minimal medium supplemented with 0.2% glucose and 0.5% casamino acids except hda cells that were grown anaerobically and shifted for 4 hours to aerobic conditions. Wild-type cells treated for 30 minutes with 2μg/ml ciprofloxacin are used as strand break control. Top: PFGE gel. Bottom: line scan analysis of relevant lanes.
(PDF)
A) The hda::cat mutation was introduced into wild-type cells under anaerobic conditions, restreaked on AB minimal medium plates supplemented with 0.2% glycerol along with wild-type and hda iscUC63F cells and incubated aerobically. B) The hda::cat mutation was introduced into wild-type cells under anaerobic conditions, restreaked on AB minimal medium plates supplemented with 0.2% glucose and 0.5% casamino acids along with wild-type and hda iscUC63F cells and incubated aerobically. C) Hda clones obtained on AB minimal medium supplemented with 0.2% glycerol were restreaked on LB plates along with wild-type and hda iscUC63F cells and incubated aerobically. D) Cells were grown exponentially in AB minimal medium supplemented with 0.2% glycerol and treated with rifampicin and cephalexin prior to flow cytometric analysis. E) Cells were grown exponentially in AB minimal medium then shifted to AB minimal medium supplemented with 0.2% glucose and 0.5% casamino acids medium for 3 mass doubling time or F) LB medium for 3 mass doubling time and treated with rifampicin and cephalexin prior to flow cytometric analysis. Each panel represents a minimum of 30000 cells. The average ori/cell (O/C), ori/mass (O/M) relative to wild-type and mass doubling time (τ) are inserted in the histograms.
(PDF)
Cells were grown exponentially in LB medium and treated with rifampicin and cephalexin prior to flow cytometric analysis. Each panel represents a minimum of 30000 cells. The average ori/cell (O/C), ori/mass (O/M) relative to wild-type and mass doubling time (τ) are inserted in the histograms.
(PDF)
The hda::cat mutation was introduced into the indicated strains under anaerobic conditions, restreaked on LB agar and incubated aerobically.
(PDF)
The expression level of selected OxyR and SoxRS controlled genes relative to wild-type extracted from the microarray experiment.
(PDF)
Expression of katG::lacZ in cells growing in LB medium was measured by β-galactosidase assay. The results are expressed relative to wild-type.
(PDF)
The sodA::kan and sodB::kan mutations were introduced into hda freΔ68 cells under anaerobic conditions and restreaked under aerobic conditions on LB agar.
(PDF)
(XLSX)
Acknowledgments
We thank Elsa Germain and James Imlay for bacterial strains, Jianming Liu for help with fermenter set up. Jakob Frimodt-Møller for helpful discussions.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was part of the Center for Bacterial Stress Response and Persistence (BASP) funded by a grant from the Danish National Research Foundation (DNRF120) and grants from the Lundbeck Foundation and the NovoNordisk Foundation and the Brazilian Scientific Mobility Program “Science without Borders” supported by CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior; grant number 12179/13-2). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Leonard AC, Mechali M. DNA replication origins. Cold Spring Harb Perspect Biol. 2013;5(10):a010116 PubMed Central PMCID: PMCPMC3783049. 10.1101/cshperspect.a010116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Skarstad K, Katayama T. Regulating DNA replication in bacteria. Cold Spring Harb Perspect Biol. 2013;5(4):a012922 PubMed Central PMCID: PMCPMC3683904. 10.1101/cshperspect.a012922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sekimizu K, Bramhill D, Kornberg A. ATP activates dnaA protein in initiating replication of plasmids bearing the origin of the E. coli chromosome. Cell. 1987;50(2):259–65. [DOI] [PubMed] [Google Scholar]
- 4.Samitt CE, Hansen FG, Miller JF, Schaechter M. In vivo studies of DnaA binding to the origin of replication of Escherichia coli. EMBO J. 1989;8(3):989–93. PubMed Central PMCID: PMCPMC400901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leonard AC, Grimwade JE. The orisome: structure and function. Front Microbiol. 2015;6:545 PubMed Central PMCID: PMCPMC4451416. 10.3389/fmicb.2015.00545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leonard AC, Grimwade JE. Regulation of DnaA assembly and activity: taking directions from the genome. Annu Rev Microbiol. 2011;65:19–35. PubMed Central PMCID: PMCPMC4075013. 10.1146/annurev-micro-090110-102934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ozaki S, Katayama T. Highly organized DnaA-oriC complexes recruit the single-stranded DNA for replication initiation. Nucleic Acids Res. 2012;40(4):1648–65. PubMed Central PMCID: PMCPMC3287180. 10.1093/nar/gkr832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chodavarapu S, Kaguni JM. Replication Initiation in Bacteria. Enzymes. 2016;39:1–30. 10.1016/bs.enz.2016.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurokawa K, Nishida S, Emoto A, Sekimizu K, Katayama T. Replication cycle-coordinated change of the adenine nucleotide-bound forms of DnaA protein in Escherichia coli. EMBO J. 1999;18(23):6642–52. PubMed Central PMCID: PMCPMC1171727. 10.1093/emboj/18.23.6642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katayama T, Kubota T, Kurokawa K, Crooke E, Sekimizu K. The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E. coli chromosomal replicase. Cell. 1998;94(1):61–71. [DOI] [PubMed] [Google Scholar]
- 11.Kato J, Katayama T. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 2001;20(15):4253–62. PubMed Central PMCID: PMCPMC149159. 10.1093/emboj/20.15.4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kasho K, Katayama T. DnaA binding locus datA promotes DnaA-ATP hydrolysis to enable cell cycle-coordinated replication initiation. Proc Natl Acad Sci U S A. 2013;110(3):936–41. PubMed Central PMCID: PMCPMC3549119. 10.1073/pnas.1212070110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simmons LA, Breier AM, Cozzarelli NR, Kaguni JM. Hyperinitiation of DNA replication in Escherichia coli leads to replication fork collapse and inviability. Mol Microbiol. 2004;51(2):349–58. 10.1046/j.1365-2958.2003.03842.x [DOI] [PubMed] [Google Scholar]
- 14.Charbon G, Bjorn L, Mendoza-Chamizo B, Frimodt-Moller J, Lobner-Olesen A. Oxidative DNA damage is instrumental in hyperreplication stress-induced inviability of Escherichia coli. Nucleic Acids Res. 2014;42(21):13228–41. PubMed Central PMCID: PMCPMC4245963. 10.1093/nar/gku1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poole RK, Cook GM. Redundancy of aerobic respiratory chains in bacteria? Routes, reasons and regulation. Adv Microb Physiol. 2000;43:165–224. [DOI] [PubMed] [Google Scholar]
- 16.Imlay JA. Diagnosing oxidative stress in bacteria: not as easy as you might think. Curr Opin Microbiol. 2015;24:124–31. PubMed Central PMCID: PMCPMC4380616. 10.1016/j.mib.2015.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woodmansee AN, Imlay JA. Reduced flavins promote oxidative DNA damage in non-respiring Escherichia coli by delivering electrons to intracellular free iron. J Biol Chem. 2002;277(37):34055–66. 10.1074/jbc.M203977200 [DOI] [PubMed] [Google Scholar]
- 18.Seaver LC, Imlay JA. Are respiratory enzymes the primary sources of intracellular hydrogen peroxide? J Biol Chem. 2004;279(47):48742–50. 10.1074/jbc.M408754200 [DOI] [PubMed] [Google Scholar]
- 19.Korshunov S, Imlay JA. Two sources of endogenous hydrogen peroxide in Escherichia coli. Mol Microbiol. 2010;75(6):1389–401. PubMed Central PMCID: PMCPMC3049997. 10.1111/j.1365-2958.2010.07059.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imlay JA. Pathways of oxidative damage. Annu Rev Microbiol. 2003;57:395–418. 10.1146/annurev.micro.57.030502.090938 [DOI] [PubMed] [Google Scholar]
- 21.Mason MG, Shepherd M, Nicholls P, Dobbin PS, Dodsworth KS, Poole RK, et al. Cytochrome bd confers nitric oxide resistance to Escherichia coli. Nat Chem Biol. 2009;5(2):94–6. 10.1038/nchembio.135 [DOI] [PubMed] [Google Scholar]
- 22.Giuffre A, Borisov VB, Arese M, Sarti P, Forte E. Cytochrome bd oxidase and bacterial tolerance to oxidative and nitrosative stress. Biochim Biophys Acta. 2014;1837(7):1178–87. 10.1016/j.bbabio.2014.01.016 [DOI] [PubMed] [Google Scholar]
- 23.Charbon G, Riber L, Cohen M, Skovgaard O, Fujimitsu K, Katayama T, et al. Suppressors of DnaA(ATP) imposed overinitiation in Escherichia coli. Mol Microbiol. 2011;79(4):914–28. 10.1111/j.1365-2958.2010.07493.x [DOI] [PubMed] [Google Scholar]
- 24.Liu X, De Wulf P. Probing the ArcA-P modulon of Escherichia coli by whole genome transcriptional analysis and sequence recognition profiling. J Biol Chem. 2004;279(13):12588–97. 10.1074/jbc.M313454200 [DOI] [PubMed] [Google Scholar]
- 25.Park DM, Akhtar MS, Ansari AZ, Landick R, Kiley PJ. The bacterial response regulator ArcA uses a diverse binding site architecture to regulate carbon oxidation globally. PLoS Genet. 2013;9(10):e1003839 PubMed Central PMCID: PMCPMC3798270. 10.1371/journal.pgen.1003839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riber L, Olsson JA, Jensen RB, Skovgaard O, Dasgupta S, Marinus MG, et al. Hda-mediated inactivation of the DnaA protein and dnaA gene autoregulation act in concert to ensure homeostatic maintenance of the Escherichia coli chromosome. Genes Dev. 2006;20(15):2121–34. PubMed Central PMCID: PMCPMC1536062. 10.1101/gad.379506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujimitsu K, Su'etsugu M, Yamaguchi Y, Mazda K, Fu N, Kawakami H, et al. Modes of overinitiation, dnaA gene expression, and inhibition of cell division in a novel cold-sensitive hda mutant of Escherichia coli. J Bacteriol. 2008;190(15):5368–81. PubMed Central PMCID: PMCPMC2493284. 10.1128/JB.00044-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaudu P, Touati D, Niviere V, Fontecave M. The NAD(P)H:flavin oxidoreductase from Escherichia coli as a source of superoxide radicals. J Biol Chem. 1994;269(11):8182–8. [PubMed] [Google Scholar]
- 29.Campbell ZT, Baldwin TO. Fre Is the Major Flavin Reductase Supporting Bioluminescence from Vibrio harveyi Luciferase in Escherichia coli. J Biol Chem. 2009;284(13):8322–8. PubMed Central PMCID: PMCPMC2659190. 10.1074/jbc.M808977200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niviere V, Fieschi F, Decout JL, Fontecave M. Is the NAD(P)H:flavin oxidoreductase from Escherichia coli a member of the ferredoxin-NADP+ reductase family?. Evidence for the catalytic role of serine 49 residue. J Biol Chem. 1996;271(28):16656–61. [DOI] [PubMed] [Google Scholar]
- 31.Bonomi F, Iametti S, Morleo A, Ta D, Vickery LE. Facilitated transfer of IscU-[2Fe2S] clusters by chaperone-mediated ligand exchange. Biochemistry. 2011;50(44):9641–50. 10.1021/bi201123z [DOI] [PubMed] [Google Scholar]
- 32.Sutera VA Jr., Lovett ST. The role of replication initiation control in promoting survival of replication fork damage. Mol Microbiol. 2006;60(1):229–39. 10.1111/j.1365-2958.2006.05093.x [DOI] [PubMed] [Google Scholar]
- 33.Gon S, Camara JE, Klungsoyr HK, Crooke E, Skarstad K, Beckwith J. A novel regulatory mechanism couples deoxyribonucleotide synthesis and DNA replication in Escherichia coli. EMBO J. 2006;25(5):1137–47. PubMed Central PMCID: PMCPMC1409723. 10.1038/sj.emboj.7600990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai Y, Outten FW. The E. coli SufS-SufE sulfur transfer system is more resistant to oxidative stress than IscS-IscU. FEBS Lett. 2012;586(22):4016–22. PubMed Central PMCID: PMCPMC3511050. 10.1016/j.febslet.2012.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Outten FW, Djaman O, Storz G. A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol Microbiol. 2004;52(3):861–72. 10.1111/j.1365-2958.2004.04025.x [DOI] [PubMed] [Google Scholar]
- 36.Nachin L, Loiseau L, Expert D, Barras F. SufC: an unorthodox cytoplasmic ABC/ATPase required for [Fe-S] biogenesis under oxidative stress. EMBO J. 2003;22(3):427–37. PubMed Central PMCID: PMCPMC140745. 10.1093/emboj/cdg061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ezraty B, Vergnes A, Banzhaf M, Duverger Y, Huguenot A, Brochado AR, et al. Fe-S cluster biosynthesis controls uptake of aminoglycosides in a ROS-less death pathway. Science. 2013;340(6140):1583–7. 10.1126/science.1238328 [DOI] [PubMed] [Google Scholar]
- 38.Vemuri GN, Altman E, Sangurdekar DP, Khodursky AB, Eiteman MA. Overflow metabolism in Escherichia coli during steady-state growth: transcriptional regulation and effect of the redox ratio. Appl Environ Microbiol. 2006;72(5):3653–61. PubMed Central PMCID: PMCPMC1472329. 10.1128/AEM.72.5.3653-3661.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andersen KB, von Meyenburg K. Are growth rates of Escherichia coli in batch cultures limited by respiration? J Bacteriol. 1980;144(1):114–23. PubMed Central PMCID: PMCPMC294601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holm AK, Blank LM, Oldiges M, Schmid A, Solem C, Jensen PR, et al. Metabolic and transcriptional response to cofactor perturbations in Escherichia coli. J Biol Chem. 2010;285(23):17498–506. PubMed Central PMCID: PMCPMC2878514. 10.1074/jbc.M109.095570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Federowicz S, Kim D, Ebrahim A, Lerman J, Nagarajan H, Cho BK, et al. Determining the control circuitry of redox metabolism at the genome-scale. PLoS Genet. 2014;10(4):e1004264 PubMed Central PMCID: PMCPMC3974632. 10.1371/journal.pgen.1004264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lanzer M, Bujard H. Promoters largely determine the efficiency of repressor action. Proc Natl Acad Sci U S A. 1988;85(23):8973–7. PubMed Central PMCID: PMCPMC282639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gotfredsen M, Gerdes K. The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol Microbiol. 1998;29(4):1065–76. [DOI] [PubMed] [Google Scholar]
- 44.Jensen PR, Michelsen O. Carbon and energy metabolism of atp mutants of Escherichia coli. J Bacteriol. 1992;174(23):7635–41. PubMed Central PMCID: PMCPMC207475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noda S, Takezawa Y, Mizutani T, Asakura T, Nishiumi E, Onoe K, et al. Alterations of cellular physiology in Escherichia coli in response to oxidative phosphorylation impaired by defective F1-ATPase. J Bacteriol. 2006;188(19):6869–76. PubMed Central PMCID: PMCPMC1595526. 10.1128/JB.00452-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Green GN, Fang H, Lin RJ, Newton G, Mather M, Georgiou CD, et al. The nucleotide sequence of the cyd locus encoding the two subunits of the cytochrome d terminal oxidase complex of Escherichia coli. J Biol Chem. 1988;263(26):13138–43. [PubMed] [Google Scholar]
- 47.VanOrsdel CE, Bhatt S, Allen RJ, Brenner EP, Hobson JJ, Jamil A, et al. The Escherichia coli CydX protein is a member of the CydAB cytochrome bd oxidase complex and is required for cytochrome bd oxidase activity. J Bacteriol. 2013;195(16):3640–50. PubMed Central PMCID: PMCPMC3754557. 10.1128/JB.00324-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Al-Attar S, Yu Y, Pinkse M, Hoeser J, Friedrich T, Bald D, et al. Cytochrome bd Displays Significant Quinol Peroxidase Activity. Sci Rep. 2016;6:27631 PubMed Central PMCID: PMCPMC4899803. 10.1038/srep27631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wall D, Delaney JM, Fayet O, Lipinska B, Yamamoto T, Georgopoulos C. arc-dependent thermal regulation and extragenic suppression of the Escherichia coli cytochrome d operon. J Bacteriol. 1992;174(20):6554–62. PubMed Central PMCID: PMCPMC207623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Imlay JA. Transcription Factors That Defend Bacteria Against Reactive Oxygen Species. Annu Rev Microbiol. 2015;69:93–108. PubMed Central PMCID: PMCPMC4618077. 10.1146/annurev-micro-091014-104322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Y, Imlay JA. Cell death from antibiotics without the involvement of reactive oxygen species. Science. 2013;339(6124):1210–3. PubMed Central PMCID: PMCPMC3731989. 10.1126/science.1232751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gu M, Imlay JA. The SoxRS response of Escherichia coli is directly activated by redox-cycling drugs rather than by superoxide. Mol Microbiol. 2011;79(5):1136–50. PubMed Central PMCID: PMCPMC3071027. 10.1111/j.1365-2958.2010.07520.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hopkin KA, Papazian MA, Steinman HM. Functional Differences between Manganese and Iron Superoxide Dismutases in Escherichia-Coli K-12. J Biol Chem. 1992;267(34):24253–8. [PubMed] [Google Scholar]
- 54.Durand S, Storz G. Reprogramming of anaerobic metabolism by the FnrS small RNA. Mol Microbiol. 2010;75(5):1215–31. 10.1111/j.1365-2958.2010.07044.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Riber L, Fujimitsu K, Katayama T, Lobner-Olesen A. Loss of Hda activity stimulates replication initiation from I-box, but not R4 mutant origins in Escherichia coli. Mol Microbiol. 2009;71(1):107–22. 10.1111/j.1365-2958.2008.06516.x [DOI] [PubMed] [Google Scholar]
- 56.Fujimitsu K, Senriuchi T, Katayama T. Specific genomic sequences of E. coli promote replicational initiation by directly reactivating ADP-DnaA. Genes Dev. 2009;23(10):1221–33. PubMed Central PMCID: PMCPMC2685538. 10.1101/gad.1775809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Katayama T. The mutant DnaAcos protein which overinitiates replication of the Escherichia coli chromosome is inert to negative regulation for initiation. J Biol Chem. 1994;269(35):22075–9. [PubMed] [Google Scholar]
- 58.Chodavarapu S, Felczak MM, Simmons LA, Murillo A, Kaguni JM. Mutant DnaAs of Escherichia coli that are refractory to negative control. Nucleic Acids Res. 2013;41(22):10254–67. PubMed Central PMCID: PMCPMC3905854. 10.1093/nar/gkt774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ishida T, Akimitsu N, Kashioka T, Hatano M, Kubota T, Ogata Y, et al. DiaA, a novel DnaA-binding protein, ensures the timely initiation of Escherichia coli chromosome replication. J Biol Chem. 2004;279(44):45546–55. 10.1074/jbc.M402762200 [DOI] [PubMed] [Google Scholar]
- 60.Katayama T, Takata M, Sekimizu K. The nucleoid protein H-NS facilitates chromosome DNA replication in Escherichia coli dnaA mutants. J Bacteriol. 1996;178(19):5790–2. PubMed Central PMCID: PMCPMC178422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schneider DA, Gourse RL. Relationship between growth rate and ATP concentration in Escherichia coli: a bioassay for available cellular ATP. J Biol Chem. 2004;279(9):8262–8. 10.1074/jbc.M311996200 [DOI] [PubMed] [Google Scholar]
- 62.Chapman AG, Fall L, Atkinson DE. Adenylate energy charge in Escherichia coli during growth and starvation. J Bacteriol. 1971;108(3):1072–86. PubMed Central PMCID: PMCPMC247190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Petersen C, Moller LB. Invariance of the nucleoside triphosphate pools of Escherichia coli with growth rate. J Biol Chem. 2000;275(6):3931–5. [DOI] [PubMed] [Google Scholar]
- 64.Martin JE, Imlay JA. The alternative aerobic ribonucleotide reductase of Escherichia coli, NrdEF, is a manganese-dependent enzyme that enables cell replication during periods of iron starvation. Mol Microbiol. 2011;80(2):319–34. PubMed Central PMCID: PMCPMC3097424. 10.1111/j.1365-2958.2011.07593.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang M, Parker MJ, Stubbe J. Choosing the right metal: case studies of class I ribonucleotide reductases. J Biol Chem. 2014;289(41):28104–11. PubMed Central PMCID: PMCPMC4192465. 10.1074/jbc.R114.596684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Coves J, Niviere V, Eschenbrenner M, Fontecave M. NADPH-sulfite reductase from Escherichia coli. A flavin reductase participating in the generation of the free radical of ribonucleotide reductase. J Biol Chem. 1993;268(25):18604–9. [PubMed] [Google Scholar]
- 67.Lindqvist A, Membrillo-Hernandez J, Poole RK, Cook GM. Roles of respiratory oxidases in protecting Escherichia coli K12 from oxidative stress. Antonie Van Leeuwenhoek. 2000;78(1):23–31. [DOI] [PubMed] [Google Scholar]
- 68.Goldman BS, Gabbert KK, Kranz RG. The temperature-sensitive growth and survival phenotypes of Escherichia coli cydDC and cydAB strains are due to deficiencies in cytochrome bd and are corrected by exogenous catalase and reducing agents. J Bacteriol. 1996;178(21):6348–51. PubMed Central PMCID: PMCPMC178510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Borisov VB, Forte E, Davletshin A, Mastronicola D, Sarti P, Giuffre A. Cytochrome bd oxidase from Escherichia coli displays high catalase activity: an additional defense against oxidative stress. FEBS Lett. 2013;587(14):2214–8. 10.1016/j.febslet.2013.05.047 [DOI] [PubMed] [Google Scholar]
- 70.Korshunov S, Imlay JA. Detection and quantification of superoxide formed within the periplasm of Escherichia coli. J Bacteriol. 2006;188(17):6326–34. PubMed Central PMCID: PMCPMC1595388. 10.1128/JB.00554-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee YS, Han JS, Jeon Y, Hwang DS. The arc two-component signal transduction system inhibits in vitro Escherichia coli chromosomal initiation. J Biol Chem. 2001;276(13):9917–23. 10.1074/jbc.M008629200 [DOI] [PubMed] [Google Scholar]
- 72.Keyer K, Gort AS, Imlay JA. Superoxide and the production of oxidative DNA damage. J Bacteriol. 1995;177(23):6782–90. PubMed Central PMCID: PMCPMC177543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Seaver LC, Imlay JA. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J Bacteriol. 2001;183(24):7173–81. PubMed Central PMCID: PMCPMC95566. 10.1128/JB.183.24.7173-7181.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ivanova A, Miller C, Glinsky G, Eisenstark A. Role of Rpos (Katf) in Oxyr-Independent Regulation of Hydroperoxidase-I in Escherichia-Coli. Mol Microbiol. 1994;12(4):571–8. [DOI] [PubMed] [Google Scholar]
- 75.Clark DJ, Maaløe O. DNA replication and the division cycle in Escherichia coli. JMolBiol. 1967;23:99–112. [Google Scholar]
- 76.Guyer MS, Reed RR, Steitz JA, Low KB. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring HarbSympQuantBiol. 1981;45:135–40. [DOI] [PubMed] [Google Scholar]
- 77.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972 1972. [Google Scholar]
- 78.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006 0008. PubMed Central PMCID: PMCPMC1681482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lobner-Olesen A, Skarstad K, Hansen FG, von Meyenburg K, Boye E. The DnaA protein determines the initiation mass of Escherichia coli K-12. Cell. 1989;57(5):881–9. [DOI] [PubMed] [Google Scholar]
- 80.Koebmann BJ, Westerhoff HV, Snoep JL, Nilsson D, Jensen PR. The glycolytic flux in Escherichia coli is controlled by the demand for ATP. J Bacteriol. 2002;184(14):3909–16. PubMed Central PMCID: PMCPMC135175. 10.1128/JB.184.14.3909-3916.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu J, Chan SH, Brock-Nannestad T, Chen J, Lee SY, Solem C, et al. Combining metabolic engineering and biocompatible chemistry for high-yield production of homo-diacetyl and homo-(S,S)-2,3-butanediol. Metab Eng. 2016;36:57–67. 10.1016/j.ymben.2016.02.008 [DOI] [PubMed] [Google Scholar]
- 82.Steinsiek S, Stagge S, Bettenbrock K. Analysis of Escherichia coli Mutants with a Linear Respiratory Chain. Plos One. 2014;9(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene expression is represented as ratio relative to MG1655. Fold changes for genes that are repressed are expressed in negative values. All genes in a given operon where a relevantly expressed gene is located are shown; the gene whose expression is most affected in the operon is highlighted in red (repressed) or green (overexpressed).
(DOCX)
Gene expression is represented as ratio relative to MG1655. Fold changes for genes that are repressed are expressed in negative values. All genes in a given operon where a relevantly expressed gene is located are shown; the gene whose expression is most affected in the operon is highlighted in red (repressed) or green (overexpressed). Genes coding for dehydrogenases are highlighted in yellow Genes coding for cytochrome terminal oxidase are highlighted in blue. Gene products inhibited by NADH are followed by an asterisk. (1) Operons or genes that are differentially expressed in a ΔarcA strain compared to wild-type [25]. (2) Operons or genes directly regulated by ArcA [25]. (3) Operons or genes that are differentially expressed in a ΔarcA strain compared to wild-type [24].
(DOCX)
The indicated cells were grown in AB minimal medium supplemented with 0.2% glucose and 0.5% casamino acids. Relative abundance of cydA, sdhD and cyoA mRNA relative to rpoA mRNA was measured by quantitative RT-PCR. Shown is the mean ± s.d. (n = 3). For comparison, values for the same genes derived from the microarray analysis are included.
(DOCX)
Cells were grown aerobically in AB minimal medium supplemented with 0.2% glucose and 0.5% casamino acids except hda cells that were grown anaerobically and shifted for 4 hours to aerobic conditions. Wild-type cells treated for 30 minutes with 2μg/ml ciprofloxacin are used as strand break control. Top: PFGE gel. Bottom: line scan analysis of relevant lanes.
(PDF)
A) The hda::cat mutation was introduced into wild-type cells under anaerobic conditions, restreaked on AB minimal medium plates supplemented with 0.2% glycerol along with wild-type and hda iscUC63F cells and incubated aerobically. B) The hda::cat mutation was introduced into wild-type cells under anaerobic conditions, restreaked on AB minimal medium plates supplemented with 0.2% glucose and 0.5% casamino acids along with wild-type and hda iscUC63F cells and incubated aerobically. C) Hda clones obtained on AB minimal medium supplemented with 0.2% glycerol were restreaked on LB plates along with wild-type and hda iscUC63F cells and incubated aerobically. D) Cells were grown exponentially in AB minimal medium supplemented with 0.2% glycerol and treated with rifampicin and cephalexin prior to flow cytometric analysis. E) Cells were grown exponentially in AB minimal medium then shifted to AB minimal medium supplemented with 0.2% glucose and 0.5% casamino acids medium for 3 mass doubling time or F) LB medium for 3 mass doubling time and treated with rifampicin and cephalexin prior to flow cytometric analysis. Each panel represents a minimum of 30000 cells. The average ori/cell (O/C), ori/mass (O/M) relative to wild-type and mass doubling time (τ) are inserted in the histograms.
(PDF)
Cells were grown exponentially in LB medium and treated with rifampicin and cephalexin prior to flow cytometric analysis. Each panel represents a minimum of 30000 cells. The average ori/cell (O/C), ori/mass (O/M) relative to wild-type and mass doubling time (τ) are inserted in the histograms.
(PDF)
The hda::cat mutation was introduced into the indicated strains under anaerobic conditions, restreaked on LB agar and incubated aerobically.
(PDF)
The expression level of selected OxyR and SoxRS controlled genes relative to wild-type extracted from the microarray experiment.
(PDF)
Expression of katG::lacZ in cells growing in LB medium was measured by β-galactosidase assay. The results are expressed relative to wild-type.
(PDF)
The sodA::kan and sodB::kan mutations were introduced into hda freΔ68 cells under anaerobic conditions and restreaked under aerobic conditions on LB agar.
(PDF)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.