Abstract
Background
Prenatal alcohol exposure (PAE) can result in an array of morphological, behavioural and neurobiological deficits that can range in their severity. Despite extensive research in the field and a significant progress made, especially in understanding the range of possible malformations and neurobehavioral abnormalities, the molecular mechanisms of alcohol responses in development are still not well understood. There have been multiple transcriptomic studies looking at the changes in gene expression after PAE in animal models, however there is a limited apparent consensus among the reported findings. In an effort to address this issue, we performed a comprehensive re-analysis and meta-analysis of all suitable, publically available expression data sets.
Methods
We assembled ten microarray data sets of gene expression after PAE in mouse and rat models consisting of samples from a total of 63 ethanol-exposed and 80 control animals. We re-analyzed each data set for differential expression and then used the results to perform meta-analyses considering all data sets together or grouping them by time or duration of exposure (pre- and post-natal, acute and chronic, respectively). We performed network and Gene Ontology enrichment analysis to further characterize the identified signatures.
Results
For each sub-analysis we identified signatures of differential expressed genes that show support from multiple studies. Overall, the changes in gene expression were more extensive after acute ethanol treatment during prenatal development than in other models. Considering the analysis of all the data together, we identified a robust core signature of 104 genes down-regulated after PAE, with no up-regulated genes. Functional analysis reveals over-representation of genes involved in protein synthesis, mRNA splicing and chromatin organization.
Conclusions
Our meta-analysis shows that existing studies, despite superficial dissimilarity in findings, share features that allow us to identify a common core signature set of transcriptome changes in PAE. This is an important step to identifying the biological processes that underlie the etiology of FASD.
Keywords: meta-analysis, gene expression, fetal alcohol spectrum disorder, prenatal alcohol exposure
Introduction
Fetal Alcohol Spectrum Disorders (FASD) is an umbrella term that describes a wide variety of deficits caused by maternal alcohol consumption during pregnancy. These developmental deficits can be morphological, behavioural and neurobiological and can range in their severity. The prevalence of FASD is currently estimated to range between 2–5% (May et al. 2009).
Since the birth anomalies resulting from the prenatal alcohol exposure (PAE) were first described in the modern scientific literature (Lemoine et al. 1968, Jones and Smith 1973), thousand of papers have been published on the topic of FASD. It is well known that the outcome of PAE can vary immensely and that it can be influenced by dose, duration, frequency and timing of PAE relative to the developmental stage of the fetus. The genetic backgrounds of the mother and fetus, as well as various environmental effects, such as socioeconomic status, maternal nutrition and exposure to other harmful substances, also play important roles in susceptibility to adverse effects of PAE (Guerri et al. 2009, Jones 2011). This array of complicating factors, as well as ethical considerations limiting access to tissue, makes FASD research in humans challenging. Thus animal models, which allow for a direct control over alcohol exposure parameters, genetic background and environment variables, with the ability to assay cells and tissue at any time during or after development, are extremely important in investigating the adverse effects of PAE. Experimental research in animal models has allowed studies to focus on the underlying cellular and molecular mechanisms of alcohol-induced teratogenesis and has contributed significantly to our current knowledge of FASD.
While significant progress has been made in understanding the spectrum of possible malformations and neurobehavioral abnormalities after PAE, the molecular mechanisms of alcohol responses in development still remain unclear. One approach has been to apply genome-wide assays of gene expression, leading to elucidation of several candidate mechanisms and gene/pathway targets (for review see Ramsay 2010). However, the resulting gene lists identified as altered and the associated molecular pathways are rarely concordant across studies. Integration and harmonization of these disparate results is a challenge facing FASD research (Haycock 2009).
Focusing only on the rodent animal models, which are among the most common, the discrepancy of results of transcriptome studies after PAE can be attributed to several sources. Besides differences in the genetic background of animals used and potential effect of diverse environments, the studies can widely vary in choice of treatment paradigm, including timing, dosage, duration and means of ethanol administration, as well as sample collection details (timing and organs/tissues collected). In addition, there are methodological differences in sample preparation, choice of gene-expression quantification platform and data analysis. Finally, many of the older studies focused on a small subset of genes or particular molecular pathways, offering a very limited view of the transcriptome in PAE. Whole transcriptome studies, on the other hand, often have small sample sizes, resulting in limited power to detect more subtle differences in expression.
One approach to resolve some of these issues is to re-analyze and meta-analyze available gene expression data sets. Combining information across multiple existing data sets can increase the reliability and generalizability of findings and allow for detection of relatively small but consistent perturbations in gene expression that can be undetectable in a single, underpowered study. While meta-analytical approaches for combining high-throughput gene expression profiling data have been successfully applied in other areas of neuroscience and alcohol research (Mulligan et al. 2006, 2011, Hu et al. 2008, Tabakoff et al. 2008, Rogic and Pavlidis 2009, Roder et al. 2012, Mistry et al. 2013a, Uddin and Singh 2013, Raddatz et al. 2014, Ch’ng et al. 2015), such a synthesis is lacking in FASD research.
In this study, we sought to identify commonalities in gene expression changes across multiple publically available data sets of prenatal alcohol exposure in rodent models. We re-analyzed and meta-analyzed ten publicly available gene expression data sets, considering them all together or grouping them by time or duration of exposure. We identified a number of genes that are consistently differentially expressed in PAE animals across multiple studies, which otherwise seemed to be largely discordant. Our results indicate that PAE acts predominantly as a net inhibitor of gene expression and that genes involved in protein synthesis, mRNA splicing and chromatin organization are primary targets of PAE.
Materials and Methods
Data set selection
We searched public data repositories (GEO, ArrayExpress) as well as published literature for gene expression profiling studies relating to prenatal alcohol exposure in murine animal models. We selected only data sets with case-control experimental design and excluded the data sets or samples using maternal tissue. The following initially selected GEO data sets were excluded: GSE19436 (ethanol-treated and control samples were treated with additional mitogenic agents), GSE23579 (small data set with a strong batch effect not deemed correctable), GSE5186 (the GEO record was ambiguous about the source of extracted RNA; no publication available), GSE43324 and GSE1997 (excluded due to the shape of the p-value distribution for differential expression, indicating violation of assumptions of our statistical approach (Barton et al. 2013); see Figure S1 and further explanation in Supplemental Materials).
After additional screening based on QC parameters described below we retained 8 mouse and one rat data set, with of a total of 63 alcohol-exposed and 83 control samples (Table 1). One of the mouse data sets, GSE9545, contained samples processed on two different microarray platforms and for the purposes of further analyses was split in two separate data sets (GSE9545.1 and GSE9545.2; see Supplemental Materials).
Table 1.
Data sets used in the study. The last column indicates the number of ethanol-treated and control samples in the study. The numbers in parentheses represent the number of samples remaining after excluding samples due to QC or batch effect problems. For GSE1996 the reported rat developmental stages is shown.
| Data set | Platform | Reference | Species (Strain) | Dev Stage | Tissue | PAE:Control |
|---|---|---|---|---|---|---|
| Downing | GPL1261 | (Downing et al. 2012) | mouse (C57BL/6J and DBA/2J) | E9 | whole embryo | 20:40 (20:38) |
| GSE1074 | GPL1261 | (Green et al. 2007) | mouse (C57BL/6J and C57BL/6N) | E8 | headfolds | 4:4 (4:3) |
| GSE9545.1 | GPL1261 | (Zhou et al. 2011) | mouse (C57BL/6) | E10 | whole embryo | 7:4 |
| GSE9545.2 | GPL339 | (Zhou et al. 2011) | mouse (C57BL/6) | E10 | whole embryo | 4:4 |
| GSE34469 | GPL6246 | (Laufer et al. 2013) | mouse (C57BL/6J) | P70 | whole brain | 4:4 |
| GSE34549 | GPL6246 | (Laufer et al. 2013) | mouse (C57BL/6J) | P60 | whole brain | 2:2 |
| GSE34305 | GPL6246 | (Kleiber et al. 2012) | mouse (C57BL/6J) | P70 | whole brain | 5:5 |
| GSE23105 | GPL6887 | (Kaminen-Ahola et al. 2010) | mouse (C57BL/6J) | P28 | kidney | 6:6 |
| GSE23106 | GPL6887 | (Kaminen-Ahola et al. 2010) | mouse (C57BL/6J) | P28 | liver | 3:4 |
| GSE1996 | GPL341 | unpublished | rat (Sprague-Dawley) | P100 | hippocampus | 8:10 |
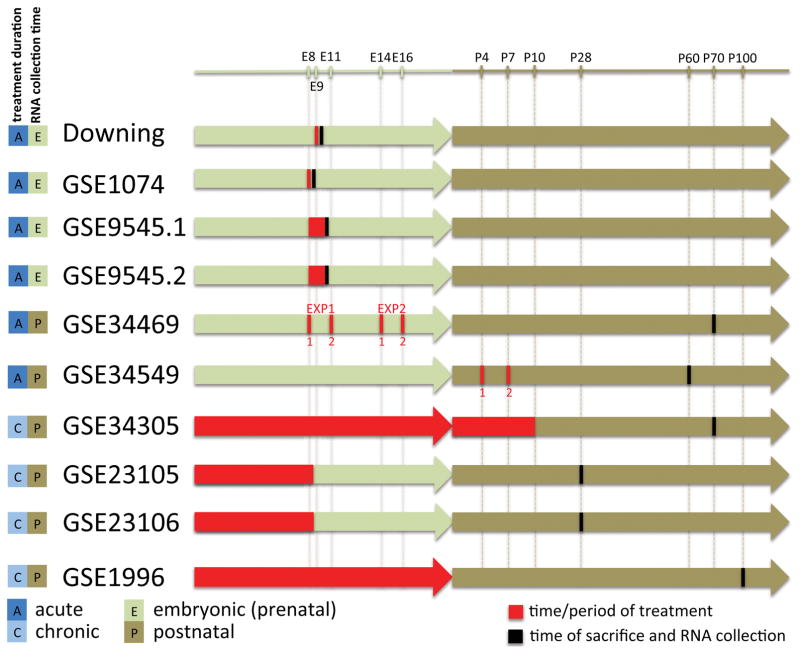
The selected data sets were generated on five different microarray platforms, including four different Affymetrix platforms and one Illumina platform (details in Table S1). In addition to differences in microarray platforms used, the collected data sets were heterogeneous in a number of other ways; they differed with respect to organ or tissue used for RNA extraction, developmental stage at the time of treatment and at the time of RNA extraction, and alcohol exposure method and duration. The timeline depicting time and duration of ethanol treatment and time of RNA extraction is shown in Figure 1.
Figure 1.
Timeline of treatment and sample collection across the data sets. Numbered red vertical lines indicate timing of the first and second injection. For GSE1996 the plotted time periods are based on reported rat developmental stages.
Data Acquisition and Quality Control
For all Affymetrix data sets we retrieved raw data (CEL files) from GEO. Since the only two Illumina data sets were pre-processed and normalized the same way, we downloaded normalized data from GEO. All data sets were subjected to comprehensive quality control. Based on this we excluded one sample from the Downing data set and one sample from the GSE1074 data set (see Supplemental Materials for details).
Differential Expression Analysis of Individual Data sets
The raw expression data were pre-processed using Robust Multi-array Average (RMA) implemented in the affy (Gautier et al. 2004) or oligo (Carvalho and Irizarry 2010) R packages where appropriate. For two Illumina data sets, GSE23105 and GSE23106, the normalized data were downloaded from GEO and further processed to exclude probes that were not expressed in more than half of the samples and then log2-transformed. Probeset annotations were obtained from Gemma (Zoubarev et al. 2012), which performs sequence analysis and gene assignment based on the current genome annotations. Probesets that map to multiple genes or do not map to a gene were excluded from the analysis. Three of our data sets, Downing, GSE34305, and GSE34469, were generated in multiple batches. We used ComBat (Johnson et al. 2007) to correct for batch effects (see Supplemental Materials for more details).
Differential expression analysis based on the case-control model was performed using analysis of variance (ANOVA) implemented in the R package limma (Smyth 2004). For some data sets additional factors were used to model gene expression values: for Downing and GSE1074, the additional factor was the strain of mice and for GSE1996, the additional factor was the differential training. In the case of two-factor design, the additive linear model was used and the interactions between factors were ignored. The resulting p-values were adjusted for multiple testing using Benjamini-Hochberg method (Benjamini and Hochberg 1995).
For the purposes of meta-analysis, in order to take into account the direction of expression change, we computed one-sided p-values based on the two-sided p-values derived from ANOVA.
Meta-analysis
We conducted five separate meta-analyses: “all”, including all 10 data sets, “prenatal”, including Downing, GSE1074, GSE9545.1, GSE9545.2, “postnatal”, including GSE34469, GSE34549, GSE34305, GSE23105, GSE23106, GSE1996, “acute”, including Downing, GSE1074, GSE9545.1, GSE9545.2, GSE34469, GSE34549, and “chronic”, including GSE34305, GSE23105, GSE23106, GSE1996 data sets (Figure 1). We used Fisher’s combined probability test (Fisher 1928) that combines p-values resulting from the individual differential expression analyses, an effective method for expression data meta-analysis (Chang et al. 2013).
We conducted separate meta-analyses for up-regulated and down-regulated genes. For each individual data set, we computed one-sided p-values corresponding to two alternative hypotheses (gene expression does not increase after PAE and gene expression does not decrease after PAE) and used them to compute F statistics for each direction separately. This approach allowed us to consider all the genes in all the data sets. Since data sets were generated on different platforms we used gene-level data to allow for cross-platform integration (see Supplemental Materials for more details). The p-values obtained from Fisher’s test were adjusted for multiple testing using the Benjamini-Hochberg method. The genes that meet the threshold of FDR<0.05 were considered to be meta-signature genes. For the purposes of integration, rat genes from data set GSE1996 were mapped to their mouse homologs using NCBI’s resource HomoloGene (NCBI Resource Coordinators 2014; http://www.ncbi.nlm.nih.gov/homologene).
To obtain core signature genes we employed a jackknife procedure, which performs n sub-meta-analyses, where n is the number of data sets considered, removing sequentially one data set at a time and then finally combining the results of all n runs. We combined the results by intersecting n resulting meta-signatures at FDR<0.1.
Functional enrichment analysis
We conducted functional enrichment analysis using ermineJ version 3.0 (Gillis et al. 2010, http://erminej.chibi.ubc.ca; see more details in Supplemental Materials). For summarizing and visualizing statistically significant GO terms based on their semantic similarity we used REVIGO (Supek et al. 2011).
Network analysis
We examined local network properties of our meta- and core signatures using protein-protein interaction networks (PPIN). Both mouse and human PPINs were used to assemble a more complete, integrated network consisting of 14,325 unique genes and 160,663 unique interactions. We computed shortest path length, node degree and clustering coefficients for the sub-networks composed of our genes of interest and evaluated their statistical significance using the permutation test (more details in Supplemental Materials).
Results
Data set selection and pre-processing
After an exhaustive search of the literature and public data repositories, followed by a careful selection process and quality control, we identified ten suitable microarray data sets relating to prenatal alcohol exposure in rodent models (see Materials and Methods). The data sets were heterogeneous with respect to species, organ or tissue collected and treatment paradigm but could be clustered into groups on the basis of experimental design (Figure 1 and Table 1). For each data set we performed a comprehensive quality analysis of raw data using an array of quality measures (see Materials and Methods). The filtered final data set consisted of a total of 63 ethanol-exposed and 80 control samples across 10 data sets (Table 1).
Differential expression analysis of individual data sets and comparison with published results
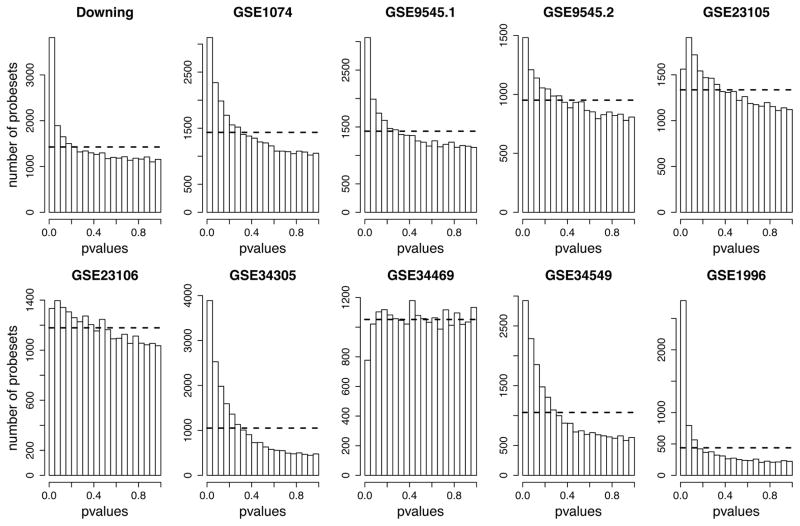
Our initial analysis considered each data set independently. We analyzed each data set for differential expression using a standard linear modeling approach. Most of the data sets showed evidence of differential expression based on the distribution of p-values observed (Figure 2). However, applying a standard multiple test correction that controls the false discovery rate (FDR), only two data sets, Downing and GSE1996, yielded a relatively large amount of significant results (FDR<0.05; Table S2). We compared these results to the original reported findings of the study authors using two methods. First, we computed the overlap between the hits we obtained at FDR<0.05 and the genes reported. Overall, the direct overlap is poor, except for the Downing data set, which we believe is due to the lower stringency used for the selection of significant genes in the original studies. In our analysis, at an FDR threshold of 0.05, many of the studies yielded no significant results. Because this thresholding might hide similarities to our gene rankings, we examined the rankings of the originally reported hits in our meta-analysis, quantified using the area under the receiver operating characteristic curve (AUC) (Table 2). The AUC is equivalent to the Wilcoxon rank-sum test and provides a measure for the extent genes selected by the authors rank highly in our own analysis, regardless of p-value. These analyses reveal reasonably high agreement between our re-analysis and the previously published results, especially for down-regulated genes, despite differences in data pre-processing and analytic approach. The data set yielding the lowest AUC, GSE34469, had been analyzed in two batches in the published study and the provided lists of significant probesets are not as directly comparable to our results. We conclude that our re-analyses are reasonable starting points for meta-analysis.
Figure 2.
Distributions of p-values resulting from the individual DE analyses of data sets included in the study. For each data set the raw p-values were computed using a standard linear modeling approach. The dashed line represents the uniform distribution expected if there was no differential expression in the data set. Most data sets have a peak of p-values near zero indicating differential expression.
Table 2.
Overlap between published gene lists and results of our re-analysis of individual data sets. The Overlap column shows the fraction of significant probesets/genes reported in the original paper that overlap with the re-analysis hit lists (see Supplemental Materials for more details). For GSE34469 there are separate probeset lists available for early (top) and late exposure, and we compared both to our results obtained on the unified data. ‘?’ indicates that the number of genes or probe sets was not reported by the authors.
| Data set | # of significant probesets/genes reported in paper | Gene/probeset list available | # of significant probesets (FDR<0.05) in our analysis | Overlap | AUC | ||
|---|---|---|---|---|---|---|---|
| up | down | up | down | ||||
| Downing | 283/50 | yes | 622 | 6/7 | 28/43 | 0.99 | 0.92 |
| GSE1074 | 2340/? | no | 0 | - | - | - | - |
| GSE9545.1 | 2519/? | partial (87) | 2 | 0/38 | 0/49 | 0.76 | 0.87 |
| GSE9545.2 | 850/? | partial (87) | 1 | 0/38 | 1/49 | 0.74 | 0.90 |
| GSE34469 | 195/195 | yes | 0 | 0/61 | 0/134 | 0.61 | 0.73 |
| 231/231 | yes | 0/54 | 0/177 | 0.48 | 0.79 | ||
| GSE34549 | 376/374 | yes | 0 | 0/15 | 0/361 | 0.79 | 0.73 |
| GSE34305 | ?/161 | yes | 0 | 0/73 | 0/90 | 0.73 | 0.91 |
| GSE23105 | 148/138 | yes | 0 | 0/46 | 0/102 | 0.52 | 0.90 |
| GSE23106 | ?/11 | yes | 14 | 0/2 | 0/9 | 0.997 | 0.81 |
| GSE1996 | - | no | 1286 | - | - | - | - |
Meta-analysis of differential expression
We began our data integration by simply comparing the results of our differential expression re-analyses across the data sets (Supplemental Figure S2), expressed as an overlap of hits. These overlaps, although limited, were encouraging and we hypothesized that a comprehensive statistical meta-analysis would identify further concordances.
We chose to employ a p-value combination approach, using Fisher’s combined probability test (Fisher 1928), which is one of the most widely used meta-analytical methods. In the recent review by Chang et al., 2013, which compares 12 meta-analytical approaches for combining multiple gene expression profiles, it was shown to have a high detection power and robustness and to yield biologically plausible results, especially if the goal is to detect genes that are differentially expressed in one or more studies. Due to the heterogeneity of the data sets, we conducted five separate meta-analyses: one including all 10 data sets (referred to as “all”), and four analyses on overlapping subsets of the data, grouped together by their experimental designs, first considering tissue sampling time (“prenatal” or “postnatal”) and second by duration of alcohol exposure (“acute” or “chronic”) (Figure 1). In order to take into account direction of expression change when performing a meta-analysis, we conducted separate analyses for up-regulated and down-regulated genes, using gene-level one-sided p-values. Altogether, this analysis yielded 10 ranked gene lists (“meta-signatures”): for each of the five analyses, and for “up” and “down” direction of expression change.
The results, in terms of the total number of genes found to be significant in a meta-analysis at FDR<0.05, are shown in Table S3 (the full results are given in the Supplemental Material and at our supplemental website: http://www.chibi.ubc.ca/faculty/paul-pavlidis/pavlidis-lab/data-and-supplementary-information/pae/). Overall, there are more down-regulated than up-regulated genes, with the exception of “prenatal” and “chronic” meta-signatures. The agreement between meta-results and individual differential expression analysis results are shown in Figure S3. Meta-results for each of the analyses show a positive and reasonably high correlation with the results of data sets included in that analysis. We also examined the different meta-signatures for overlap with each other. The “prenatal” group of data sets represents a subset of “acute” data sets; as expected, the meta-signatures computed for these two groups have a large overlap (Figure S4). The agreement between the results is further confirmed using Spearman’s correlation between the full lists of Fisher’s p-values from each meta-analysis (0.92 for up-regulated and 0.79 for down-regulated) (for complete results see Table S4). The situation is similar for the “postnatal” and “chronic” groups. The “all” meta-signature overlaps with every other meta-signature and the full meta-results correlate well with the results of other meta-analyses. This agreement suggests that the “all” meta-analysis captures commonalities across the ten data sets, especially for down-regulated genes.
Identification of robust core signatures
Closer examination of the top genes from the “all” meta-analysis suggested that, while there were genes that had concordant changes of expression across multiple data sets (e.g. down-regulated Crebzf; Figure S6), inclusion of some genes seemed to be solely due to the influence of one data set (e.g. up-regulated Rbm33; Figure S5). Due to its relatively large sample size, the Downing data set yielded genes with very small p-values (down to 10−12), which were sufficient to generate low Fisher’s p-values, regardless of p-values from other data sets (the other data set with many differentially expressed genes, GSE1996, had a smaller sample size and the p-values were more moderate (down to 10−7), so its contribution to the meta-analysis results was less). This behavior of Fisher’s method with respect to the influence of individual studies was expected. Therefore, as in our previous meta-analyses (Mistry et al. 2013a, 2013b, Ch’ng et al. 2015), to control for the susceptibility of Fisher’s method to outliers, we employed a jackknife procedure, described in the Materials and Methods. The procedure provides a core signature that is not due to the influence of any single data set and would therefore be considered a more robust set of findings. The results, in terms of the total number of genes in the core signatures, are shown in Table S5. The core signatures are subsets of their corresponding meta-signatures, but contain fewer genes. Importantly, the core signatures necessarily show higher concordance across data sets. For example, down-regulated gene Srfs7, which was among the top six genes in the “all” meta-signature, but was heavily influenced by the Downing data set, is not included in the core signature (Figure S6). Similarly, none of the up-regulated genes shown in Supplemental Figure S5 survived the jackknife procedure.
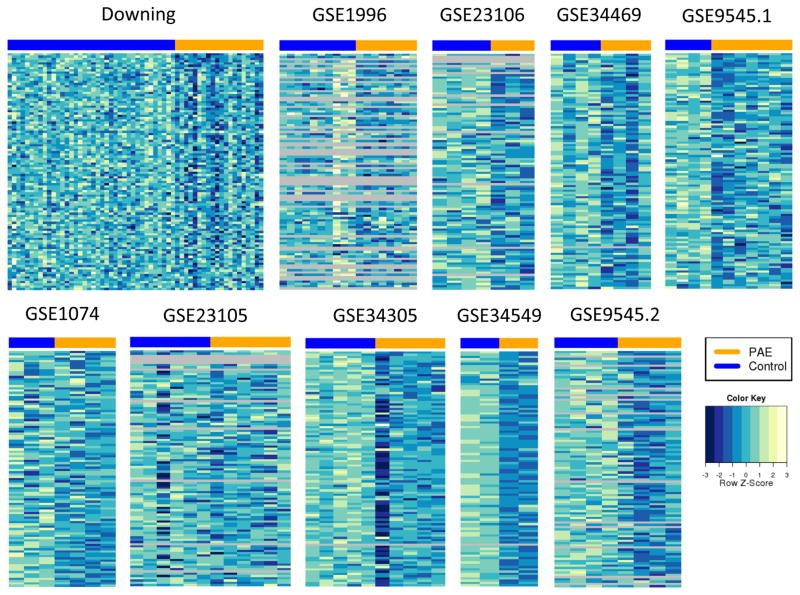
In general, there are very few genes in the up-regulated core signatures, which is in agreement with observed lower numbers of up-regulated genes in meta-signatures, smaller overlap between meta-signatures (Figure S4) and an indication of an overall lower concordance among up-regulated genes across studies. The “prenatal”, “postnatal” and “acute” down-regulated core signatures are mostly contained within the “all” core signature (Supplemental Figure S7). We also looked at the expression patterns of core signature genes. The heatmaps for down-regulated “all” core signature genes are shown in Figure 3. For most of the data sets there is a small, but clear difference between expression levels of core signature genes between PAE and control samples. Closer inspection of expression levels of top core signature genes (ranking based on their Fisher’s p-value from the initial meta-analysis) revealed that the direction of expression change is usually consistent across all data sets. An example is given for the top-ranked core signature gene, Crebzf, in Supplemental Figure S8. Down-regulation of this gene can be observed in the majority of data sets, not only the ones that yielded low p-values (this gene achieved statistical significance at FDR<0.05 only for the Downing data set).
Figure 3.
Visualization of expression differences between PAE and control samples for 104 genes in “all” down-regulated core signature. The heatmaps are based on expression data that was normalized and batch corrected within each data set. For display purposes, the expression values for each gene were normalized to have mean 0 and variance 1 to facilitate comparison between genes in the same heatmap. The heatmap colors were binned to allow for comparison across data sets. Grey rows represent genes that were not present in a particular data set (microarray platform).
Network analysis
To characterize our meta- and core signatures we first performed a gene network analysis. The goal was to assess whether our signature genes have unusual network properties when compared to carefully selected groups of background genes. To this end, we focused on local network properties of our meta- and core signature genes within a large aggregated mouse and human protein-protein interaction network (PPIN) and computed their shortest path length, node degree and clustering coefficients. The results are given in Supplemental Table S11 and Figure S9. Our main finding was that our down-regulated core signatures and several meta-signatures are unusually close (p-value for shortest path length <0.05) in the examined PPINs, suggesting a higher likelihood of functional relatedness.
Functional characterization of core and meta-signatures
Next, we compared our results with known FASD candidate genes downloaded from Phenocarta (Portales-Casamar et al. 2013). Two of the genes in our core signatures, Gpx1 and Ntf3, have been previously associated with FASD. An additional 9 genes are associated with “developmental disorder of mental health”. Across all meta-signatures there were 8 known FASD candidate genes and this enrichment was statistically significant for “prenatal” and “chronic” down-regulated meta-signatures (Table S6). The associations of these genes with FASD phenotype were not based on microarray gene expression measurements and thus are independent from the data sets used in our meta-analysis study. The full lists of known FASD candidate genes that are present in the meta- and core signatures are given in Supplemental Tables S6 and S7.
We computed enrichment of Gene Ontology (GO) terms associated with our signatures, using a FDR of 0.1 across the terms tested to identify significant enrichment. For the “all” down-regulated core signature, the most significant GO term was “peptidyl-proline modification” (Supplemental Table S8), with 5 out of 29 genes with this annotation present in the core signature (Egln3, Fkbp1b, Fkbp7, Fkbp14, and Ppig). The p-value support of these genes across ten data sets is shown in Figure S10. The three genes from the FKBP family along with Ppig have prolyl isomerase activity and function as protein folding chaperones for proteins containing proline residues (“protein folding” was also one of the enriched GO categories). Egln3 is involved in peptidyl-proline hydroxylation, which is a vital component of the hypoxia response. Some earlier studies have shown that prenatal alcohol exposure attenuates cerebrovascular responses to hypoxia (Gleason et al. 1997, Mayock et al. 2007), which might be caused by down-regulation of Egln3, which was consistently observed across the data sets in our study. Egln1, another peptidyl-proline hydroxylase, showed similar down-regulation after PAE (significant at a FDR of 0.09 in the “all” meta-analysis).
Two significant GO terms were related to bisphosphate biosynthetic/metabolic process and came up as significant due to three genes in the core signature, Pank3, Ppcdc and Papss1. Finally, genes annotated with “RNA splicing” and “protein folding”, two of the fundamental steps in gene expression, were also found to be significantly enriched in the “all” down-regulated core signature. The other core signatures did not have any GO functional categories that pass the significance threshold.
We found significant GO enrichment for “all”, “acute”, “prenatal” and “postnatal” down-regulated meta- signatures. Based on semantic similarity and GO hierarchy, enriched GO terms can be summarized by two groups: RNA metabolism and macromolecular complex biogenesis (Table S9 and Figure S11). Additionally, “acute” and “prenatal” meta-signature functional enrichment yielded GO categories that relate to nucleosome organization and chromatin assembly.
The concentration of histone coding genes at the top of our “all” down-regulated meta-signature (Figure S5) as well as the functional and pathway (see Supplemental Materials) enrichment results led us to believe that our meta- and core signatures might be enriched for this group of genes. We assembled a list of all 83 histone-coding genes in the mouse genome (see Supplemental Materials for details) and used it to assess the presence and enrichment of these genes in our signatures. We found that histone genes are significantly enriched in “all”, “acute”, and “prenatal” down-regulated meta-signatures as well as in “all” and “acute” down-regulated core signatures (Table S10, Figure S12).
Discussion
Using meta-analytical approaches we were able to identify a number of genes that are consistently differentially expressed in PAE animals across multiple studies, which otherwise seemed to be discordant. The majority of data sets that were included in our study had a small sample size and thus limited statistical power to detect differentially expressed genes at reasonable false discovery rates, even though p-value distribution plots suggested the presence of differential expression (Figure 2). Integrated analysis of data sets overcame this limitation by identifying consistent signals across the data sets.
Although the fact that selected data sets were heterogeneous in a number of ways represented a challenge, it gave us an opportunity to compare molecular effects of different treatment paradigms and, at the same time, look for unifying PAE gene expression signature. We did not consider different tissues separately primarily due to the limited availability of data, but also because three of the data sets included in our study already used a mixture of tissues for RNA extraction (Downing, GSE9545.1, GSE9545.2 used whole embryos). It is important to point out that, while this approach allowed us to look for commonalities across the tissues, it would not be able to identify tissue-specific expression changes.
While four of our meta-analyses subgrouped the data sets based on their experimental designs, considering the overlap between these subgroups (e.g. “prenatal” with “acute” and “chronic” with “postnatal”) and the overlap between computed meta-signatures (Figure S4), we can simplify the discussion by summarizing the findings for overlapping subgroups together. Overall, the changes in gene expression were more extensive after acute ethanol treatment during prenatal development. This is somewhat expected considering that for the majority of experiments in this group the RNA was extracted soon after the treatment, so the observed changes would include acute and transient effects of alcohol exposure, such as cell death (Kleiber et al. 2014).
Altogether, our results suggest that PAE acts as a net inhibitor of gene expression. Most of our down- regulated meta-signatures were larger than their up-regulated counterparts; they also showed higher concordance across different sub-analyses. In addition, we found very few up-regulated genes in our core signatures. This is in agreement with previous interpretations of gene expression studies in the field (Hard et al. 2005, Zhou et al. 2011, Downing et al. 2012, Kleiber et al. 2013). In addition, down-regulated genes in our core and meta-signatures seemed to be more functionally related based on our network and functional enrichment analyses. Finally, all but one of the previously known FASD candidate genes differentially expressed in our data showed a pattern of down-regulation (Table S6).
Using GO functional enrichment analysis, we identified a number of basic biological processes that are consistently affected after PAE, and which can be summarized under the umbrella of RNA processing, RNA metabolism, macromolecular complex biogenesis and chromatin organization. The genes involved in these processes were all found to be down-regulated across the studies. A caveat to our discussion of the functional significance of our findings is that changes in RNA levels do not automatically indicate downstream functional changes e.g. to protein levels (Gygi et al. 1999, Wilhelm et al. 2014).
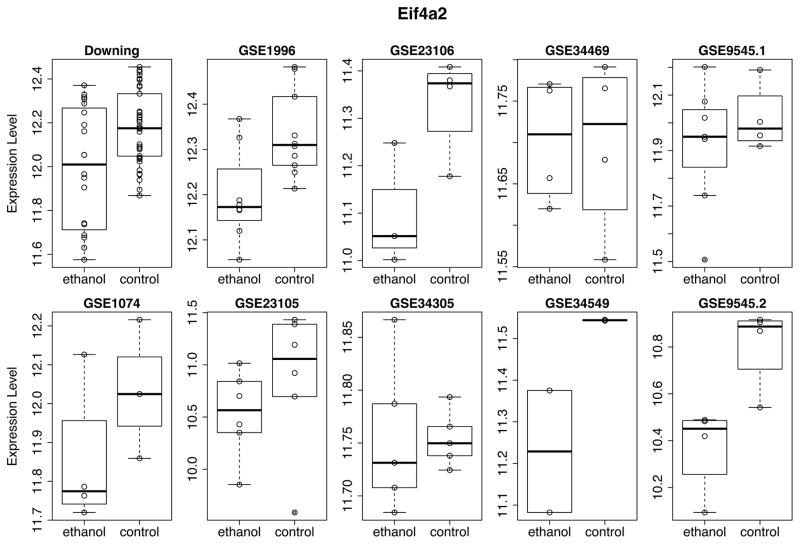
One of the earliest hypotheses to explain the widespread and heterogeneous effects of prenatal alcohol exposure proposed protein synthesis inhibition as the main mechanism. Impairment in protein synthesis following PAE results in cellular growth restriction at critical periods of development and is not typically restored postnatally. This integrating hypothesis brought forward by Kennedy in 1984 relied on earlier experimental studies that reported reduced protein synthesis in fetal tissue after alcohol exposure. One study that appeared after Kennedy’s review demonstrated that protein synthesis rates were significantly reduced in the brains of ethanol-exposed rat embryos (Rawat 1985). This was further confirmed in more recent studies, which established that the rate of protein synthesis reduction in the brain can be as much as 40% (Bonner et al. 2003, 2003, Narasimhan et al. 2013). The results of our meta-analyses are consistent with this hypothesis: we found enrichment of genes involved in ribosome biogenesis (or more generally ribonucleoprotein complex biogenesis) in our down-regulated “all”, “acute” and “prenatal” meta-signatures. In addition, other important cellular processes that can result in reduced numbers of functional proteins, such as mRNA processing and protein folding are found to be affected (core and meta-signatures). Furthermore, we found that one of the key initiators of translation, RNA helicase Eif4a2, which is required for unwinding of double stranded mRNA and its binding to the 40S ribosomal subunit, was consistently down-regulated across all the data sets in our study (Figure 4) and was found in down-regulated “all” core signature and “all”, “acute” and “prenatal” meta-signatures. Previously, this gene was reported to be significantly down-regulated in rat-derived ethanol-treated primary cortical neurons and it was suggested that ethanol-induced blocking of eukaryotic initiation factor-4A (eIF4A) complex is the main mechanism of de novo protein synthesis inhibition (Narasimhan et al. 2013). A second member of the eIF4A family, Eif4a3, was also in “all” down-regulated meta-signature.
Figure 4.
Expression pattern of RNA helicase and translation initiator Eif4a2. Normalized batch-corrected expression values are plotted for each data set. Note the differences in the ordinate scales of each plot, which reflect expression levels as measured in each data set.
We found that genes involved in RNA splicing are consistently down-regulated across our data sets. The RNA splicing GO category was found to be significantly enriched in down-regulated “all” core signature and down-regulated “all”, “prenatal” and “acute” meta-signatures. Congruently, pathway enrichment analysis revealed that these meta-signatures are enriched in spliceosome pathway genes (see Supplemental Materials). Similar functional enrichment was noted in (Downing et al. 2012), which discusses one of the data sets included in our study. However, the down-regulation of these genes goes beyond the influence of this one data set and is supported by the expression patterns in multiple data sets (Figure S13). We are not aware of any other studies identifying the impairment of spliceosome machinery as one of the consequences of PAE, but based on our results this seems like a viable target of investigation.
Lastly, we observed consistent dysregulation of genes involved in chromatin organization, indicated by significant functional enrichment of multiple GO categories related to this biological process. In particular, we found that a subgroup of these genes, histone genes, were significantly enriched in “all”, “acute”, and “prenatal” down-regulated meta-signatures as well as in “all” and “acute” down-regulated core signatures. Down-regulation of epigenetic-related genes was also reported in some of the gene-expression studies that produced data sets included in our meta-analyses (Liu et al. 2009, Zhou et al. 2011, Downing et al. 2012, Kleiber et al. 2013). The hypothesis that ethanol-induced abnormalities could arise through epigenetic reprograming has gained much interest lately. This area of FASD research is still in its infancy but there is emerging evidence that ethanol can cause changes in both DNA methylation (Garro et al. 1991, Liu et al. 2009, Kaminen-Ahola et al. 2010) and histone modifications (evidence mostly from studies using chronic alcohol exposure, reviewed in Shukla et al. 2008 and Haycock 2009). Our results provide further evidence for epigenetic dysregulation following PAE and suggest an important role of histone genes themselves.
Supplementary Material
Acknowledgments
We are grateful to all the investigators and institutions who made their data publically available. We thank Dr. Joanne Weinberg and Dr. Elodie Portales-Casamar for valuable discussions and feedback on the manuscript. This work was supported by NeuroDevNet Network of Centres of Excellence and the NIH (grant GM076990 to P.P.).
References
- Barton SJ, Crozier SR, Lillycrop KA, Godfrey KM, Inskip HM. Correction of unexpected distributions of P values from analysis of whole genome arrays by rectifying violation of statistical assumptions. BMC Genomics. 2013;14(1):161. doi: 10.1186/1471-2164-14-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57(1):289–300. [Google Scholar]
- Bonner AB, Dalwai S, Marway JS, Preedy VR. Acute exposure to the nutritional toxin alcohol reduces brain protein synthesis in vivo. Metabolism. 2003;52(4):389–396. doi: 10.1053/meta.2003.50009. [DOI] [PubMed] [Google Scholar]
- Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics (Oxford, England) 2010;26(19):2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang LC, Lin HM, Sibille E, Tseng GC. Meta-analysis methods for combining multiple expression profiles: comparisons, statistical characterization and an application guideline. BMC Bioinformatics. 2013;14(1):368. doi: 10.1186/1471-2105-14-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ch’ng C, Kwok W, Rogic S, Pavlidis P. Meta-Analysis of Gene Expression in Autism Spectrum Disorder. Autism Research: Official Journal of the International Society for Autism Research. 2015 doi: 10.1002/aur.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing C, Flink S, Florez-McClure ML, Johnson TE, Tabakoff B, Kechris KJ. Gene Expression Changes in C57BL/6J and DBA/2J Mice Following Prenatal Alcohol Exposure. Alcoholism, clinical and experimental research. 2012;36(9):1519–1529. doi: 10.1111/j.1530-0277.2012.01757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher SRA. Statistical Methods for Research Workers. Oliver and Boyd; 1928. [Google Scholar]
- Garro AJ, McBeth DL, Lima V, Lieber CS. Ethanol consumption inhibits fetal DNA methylation in mice: implications for the fetal alcohol syndrome. Alcoholism, clinical and experimental research. 1991;15(3):395–398. doi: 10.1111/j.1530-0277.1991.tb00536.x. [DOI] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics (Oxford, England) 2004;20(3):307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- Gillis J, Mistry M, Pavlidis P. Gene function analysis in complex data sets using ErmineJ. Nature protocols. 2010;5(6):1148–1159. doi: 10.1038/nprot.2010.78. [DOI] [PubMed] [Google Scholar]
- Gleason CA, Iida H, Hotchkiss KJ, Northington FJ, Traystman RJ. Newborn Cerebrovascular Responses after First Trimester Moderate Maternal Ethanol Exposure in Sheep. Pediatric Research. 1997;42(1):39–45. doi: 10.1203/00006450-199707000-00007. [DOI] [PubMed] [Google Scholar]
- Green ML, Singh AV, Zhang Y, Nemeth KA, Sulik KK, Knudsen TB. Reprogramming of genetic networks during initiation of the Fetal Alcohol Syndrome. Developmental Dynamics. 2007;236(2):613–631. doi: 10.1002/dvdy.21048. [DOI] [PubMed] [Google Scholar]
- Guerri C, Bazinet A, Riley EP. Foetal Alcohol Spectrum Disorders and Alterations in Brain and Behaviour. Alcohol and Alcoholism (Oxford, Oxfordshire) 2009;44(2):108–114. doi: 10.1093/alcalc/agn105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Molecular and Cellular Biology. 1999;19(3):1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hard ML, Abdolell M, Robinson BH, Koren G. Gene-expression analysis after alcohol exposure in the developing mouse. The Journal of laboratory and clinical medicine. 2005;145(1):47–54. doi: 10.1016/j.lab.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Haycock PC. Fetal alcohol spectrum disorders: the epigenetic perspective. Biology of reproduction. 2009;81(4):607–617. doi: 10.1095/biolreprod.108.074690. [DOI] [PubMed] [Google Scholar]
- Hu W, Saba L, Kechris K, Bhave SV, Hoffman PL, Tabakoff B. Genomic Insights into Acute Alcohol Tolerance. The Journal of pharmacology and experimental therapeutics. 2008;326(3):792–800. doi: 10.1124/jpet.108.137521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Jones KL. The effects of alcohol on fetal development. Birth defects research. Part C, Embryo today: reviews. 2011;93(1):3–11. doi: 10.1002/bdrc.20200. [DOI] [PubMed] [Google Scholar]
- Jones KL, Smith DW. Recognition of the fetal alcohol syndrome in early infancy. Lancet. 1973;302(7836):999–1001. doi: 10.1016/s0140-6736(73)91092-1. [DOI] [PubMed] [Google Scholar]
- Kaminen-Ahola N, Ahola A, Flatscher-Bader T, Wilkins SJ, Anderson GJ, Whitelaw E, Chong S. Postnatal growth restriction and gene expression changes in a mouse model of fetal alcohol syndrome. Birth Defects Research Part A: Clinical and Molecular Teratology. 2010;88(10):818–826. doi: 10.1002/bdra.20729. [DOI] [PubMed] [Google Scholar]
- Kaminen-Ahola N, Ahola A, Maga M, Mallitt KA, Fahey P, Cox TC, Whitelaw E, Chong S. Maternal Ethanol Consumption Alters the Epigenotype and the Phenotype of Offspring in a Mouse Model. PLoS Genet. 2010;6(1):e1000811. doi: 10.1371/journal.pgen.1000811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy LA. The pathogenesis of brain abnormalities in the fetal alcohol syndrome: an integrating hypothesis. Teratology. 1984;29(3):363–368. doi: 10.1002/tera.1420290306. [DOI] [PubMed] [Google Scholar]
- Kleiber ML, Diehl EJ, Laufer BI, Mantha K, Chokroborty-Hoque A, Alberry B, Singh SM. Long-term genomic and epigenomic dysregulation as a consequence of prenatal alcohol exposure: a model for fetal alcohol spectrum disorders. [Accessed 18 Jul 2014];Frontiers in Genetics [online] 2014 :5. doi: 10.3389/fgene.2014.00161. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4040446/ [DOI] [PMC free article] [PubMed]
- Kleiber ML, Laufer BI, Wright E, Diehl EJ, Singh SM. Long-term alterations to the brain transcriptome in a maternal voluntary consumption model of fetal alcohol spectrum disorders. Brain Research. 2012;1458:18–33. doi: 10.1016/j.brainres.2012.04.016. [DOI] [PubMed] [Google Scholar]
- Kleiber ML, Mantha K, Stringer RL, Singh SM. Neurodevelopmental alcohol exposure elicits long-term changes to gene expression that alter distinct molecular pathways dependent on timing of exposure. Journal of Neurodevelopmental Disorders. 2013;5(1):6. doi: 10.1186/1866-1955-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufer BI, Mantha K, Kleiber ML, Diehl EJ, Addison SMF, Singh SM. Long-lasting alterations to DNA methylation and ncRNAs could underlie the effects of fetal alcohol exposure in mice. Disease models & mechanisms. 2013;6(4):977–992. doi: 10.1242/dmm.010975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine P, Harousseau H, Borteyru JP, Menuet JC. Children of alcoholic parents--observed anomalies: discussion of 127 cases. Ouest Med. 1968;8:476–482. doi: 10.1097/00007691-200304000-00002. [DOI] [PubMed] [Google Scholar]
- Liu Y, Balaraman Y, Wang G, Nephew KP, Zhou FC. Alcohol exposure alters DNA methylation profiles in mouse embryos at early neurulation. Epigenetics3: official journal of the DNA Methylation Society. 2009;4(7):500–511. doi: 10.4161/epi.4.7.9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayock DE, Ness D, Mondares RL, Gleason CA. Binge alcohol exposure in the second trimester attenuates fetal cerebral blood flow response to hypoxia. Journal of Applied Physiology. 2007;102(3):972–977. doi: 10.1152/japplphysiol.00956.2006. [DOI] [PubMed] [Google Scholar]
- May PA, Gossage JP, Kalberg WO, Robinson LK, Buckley D, Manning M, Hoyme HE. Prevalence and epidemiologic characteristics of FASD from various research methods with an emphasis on recent in-school studies. Developmental disabilities research reviews. 2009;15(3):176–192. doi: 10.1002/ddrr.68. [DOI] [PubMed] [Google Scholar]
- Mistry M, Gillis J, Pavlidis P. Genome-wide expression profiling of schizophrenia using a large combined cohort. Molecular psychiatry. 2013a;18(2):215–225. doi: 10.1038/mp.2011.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry M, Gillis J, Pavlidis P. Meta-analysis of gene coexpression networks in the post-mortem prefrontal cortex of patients with schizophrenia and unaffected controls. BMC Neuroscience. 2013b;14(1):105. doi: 10.1186/1471-2202-14-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan MK, Ponomarev I, Hitzemann RJ, Belknap JK, Tabakoff B, Harris RA, Crabbe JC, Blednov YA, Grahame NJ, Phillips TJ, Finn DA, Hoffman PL, Iyer VR, Koob GF, Bergeson SE. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proceedings of the National Academy of Sciences. 2006;103(16):6368–6373. doi: 10.1073/pnas.0510188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan MK, Rhodes JS, Crabbe JC, Mayfield RD, Harris RA, Ponomarev I. Molecular Profiles of Drinking Alcohol to Intoxication in C57BL/6J Mice. Alcoholism, Clinical and Experimental Research. 2011;35(4):659–670. doi: 10.1111/j.1530-0277.2010.01384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan M, Rathinam M, Riar A, Patel D, Mummidi S, Yang HS, Colburn NH, Henderson GI, Mahimainathan L. Programmed cell death 4 (PDCD4): a novel player in ethanol-mediated suppression of protein translation in primary cortical neurons and developing cerebral cortex. Alcoholism, clinical and experimental research. 2013;37(1):96–109. doi: 10.1111/j.1530-0277.2012.01850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic acids research. 2014;42(Database issue):D7–17. doi: 10.1093/nar/gkt1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portales-Casamar E, Ch’ng C, Lui F, St-Georges N, Zoubarev A, Lai AY, Lee M, Kwok C, Kwok W, Tseng L, Pavlidis P. Neurocarta: aggregating and sharing disease-gene relations for the neurosciences. BMC Genomics. 2013;14(1):129. doi: 10.1186/1471-2164-14-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raddatz BBR, Hansmann F, Spitzbarth I, Kalkuhl A, Deschl U, Baumgärtner W, Ulrich R. Transcriptomic Meta-Analysis of Multiple Sclerosis and Its Experimental Models. PLoS ONE. 2014;9(1):e86643. doi: 10.1371/journal.pone.0086643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay M. Genetic and epigenetic insights into fetal alcohol spectrum disorders. Genome Medicine. 2010;2(4):1–8. doi: 10.1186/gm148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat AK. Nucleic acid and protein synthesis inhibition in developing brain by ethanol in the absence of hypothermia. Neurobehavioral toxicology and teratology. 1985;7(2):161–166. [PubMed] [Google Scholar]
- Roder C, Kasuya H, Harati A, Tatagiba M, Inoue I, Krischek B. Meta-analysis of microarray gene expression studies on intracranial aneurysms. Neuroscience. 2012;201:105–113. doi: 10.1016/j.neuroscience.2011.10.033. [DOI] [PubMed] [Google Scholar]
- Rogic S, Pavlidis P. Meta-Analysis of Kindling-Induced Gene Expression Changes in the Rat Hippocampus. [Accessed 21 May 2013];Frontiers in Neuroscience [online] 2009 :3. doi: 10.3389/neuro.15.001.2009. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2858611/ [DOI] [PMC free article] [PubMed]
- Shukla SD, Velazquez J, French SW, Lu SC, Ticku MK, Zakhari S. Emerging Role of Epigenetics in the Actions of Alcohol. Alcoholism: Clinical and Experimental Research. 2008;32(9):1525–1534. doi: 10.1111/j.1530-0277.2008.00729.x. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical applications in genetics and molecular biology. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE. 2011;6(7):e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabakoff B, Saba L, Kechris K, Hu W, Bhave SV, Finn DA, Grahame NJ, Hoffman PL. The genomic determinants of alcohol preference in mice. Mammalian genome3: official journal of the International Mammalian Genome Society. 2008;19(5):352–365. doi: 10.1007/s00335-008-9115-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin RK, Singh SM. Hippocampal Gene Expression Meta-Analysis Identifies Aging and Age-Associated Spatial Learning Impairment (ASLI) Genes and Pathways. PLoS ONE. 2013;8(7):e69768. doi: 10.1371/journal.pone.0069768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M, Schlegl J, Hahne H, Moghaddas Gholami A, Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx H, Mathieson T, Lemeer S, Schnatbaum K, Reimer U, Wenschuh H, Mollenhauer M, Slotta-Huspenina J, Boese JH, Bantscheff M, Gerstmair A, Faerber F, Kuster B. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509(7502):582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- Zhou FC, Zhao Q, Liu Y, Goodlett CR, Liang T, McClintick JN, Edenberg HJ, Li L. Alteration of gene expression by alcohol exposure at early neurulation. BMC Genomics. 2011;12(1):124. doi: 10.1186/1471-2164-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoubarev A, Hamer KM, Keshav KD, McCarthy EL, Santos JRC, Van Rossum T, McDonald C, Hall A, Wan X, Lim R, Gillis J, Pavlidis P. Gemma: a resource for the reuse, sharing and meta-analysis of expression profiling data. Bioinformatics (Oxford, England) 2012;28(17):2272–2273. doi: 10.1093/bioinformatics/bts430. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.