SUMMARY
The advent of heme during evolution allowed organisms possessing this compound to safely and efficiently carry out a variety of chemical reactions that otherwise were difficult or impossible. While it was long assumed that a single heme biosynthetic pathway existed in nature, over the past decade, it has become clear that there are three distinct pathways among prokaryotes, although all three pathways utilize a common initial core of three enzymes to produce the intermediate uroporphyrinogen III. The most ancient pathway and the only one found in the Archaea converts siroheme to protoheme via an oxygen-independent four-enzyme-step process. Bacteria utilize the initial core pathway but then add one additional common step to produce coproporphyrinogen III. Following this step, Gram-positive organisms oxidize coproporphyrinogen III to coproporphyrin III, insert iron to make coproheme, and finally decarboxylate coproheme to protoheme, whereas Gram-negative bacteria first decarboxylate coproporphyrinogen III to protoporphyrinogen IX and then oxidize this to protoporphyrin IX prior to metal insertion to make protoheme. In order to adapt to oxygen-deficient conditions, two steps in the bacterial pathways have multiple forms to accommodate oxidative reactions in an anaerobic environment. The regulation of these pathways reflects the diversity of bacterial metabolism. This diversity, along with the late recognition that three pathways exist, has significantly slowed advances in this field such that no single organism's heme synthesis pathway regulation is currently completely characterized.
KEYWORDS: heme, biosynthetic pathways, metabolic regulation, pathway evolution, tetrapyrroles
INTRODUCTION
With the exception of a few organisms, tetrapyrroles are ubiquitous in their distribution in nature. While most commonly found as metallated macrocycles, biologically significant linear forms such as bilins also exist, which play important roles as both photosynthetic accessory pigments and photopigment photoreceptor proteins. The metallated modified tetrapyrroles are involved in nearly all the major metabolic and respiratory processes found in biological systems, from photosynthesis to methanogenesis. From a prokaryotic perspective, the ability to make modified tetrapyrroles enriches the metabolic capacity, providing ecological advantages and allowing the host to rapidly switch between environmental conditions. It is no coincidence, therefore, that the richest diversity of tetrapyrrole compounds is found among microorganisms that generally possess all the necessary enzymatic machinery to synthesize their own complement of modified tetrapyrroles.
The metallated modified tetrapyrrole fraternity includes among its members the hemes, the chlorophylls and bacteriochlorophylls, the corrins (vitamin B12), siroheme, coenzyme F430, and heme d1. Most of these molecules are associated with just one key process. For instance, the chlorophylls and bacteriochlorophylls are the light-absorbing pigments that are the essential components of oxygenic and anoxygenic photosynthesis, respectively (1). Similarly, siroheme, an Fe-containing isobacteriochlorin, acts as a prosthetic group in the six-electron reduction of either sulfite or nitrite in assimilatory sulfite and nitrite reductase. Coenzyme F430, a yellow-colored nickel-containing modified tetrapyrrole, plays an essential role in the final step in methanogenesis in the enzyme methyl coenzyme M (CoM) reductase (2). Heme d1, which is technically not a heme but an Fe-containing dioxoisobacteriochlorin, is a prosthetic group in the cytochrome cd1 nitrite reductase (3). In contrast, the corrins and hemes are functionally much more diverse. Biological corrins, which are sometimes referred to as cobamides, cobalamins, and vitamin B12, possess a cobalt-containing corrin macrocycle. These molecules play a role in at least four major classes of enzymes, including B12-dependent isomerases, methyltransferases, reductive dehalogenases, and radical S-adenosyl-l-methionine (SAM) enzymes (4). Moreover, adenosylcobalamin has recently been shown to play a role as a light sensor in the control of transcription (5). However, the most versatile and ubiquitous metalloprosthetic group is heme. It is known to play roles in gas sensing and transport, catalysis, signaling, electron transport, and transcription (6). This review explores how heme, an Fe-containing porphyrin, is made in bacteria and archaea but in so doing also highlights how the other Fe-containing modified tetrapyrroles, siroheme and heme d1, are constructed.
In the Bacteria, de novo heme biosynthesis appears to have been developed evolutionarily very early, by the time that deep-branching phyla such as the Planctomycetes or Aquificae had diverged. The absence of the pathway (or incomplete pathways) in various bacterial pathogens and symbionts appears to result from secondary gene losses. In free-living organisms, the absence of the protoheme biosynthetic branch is not uncommon; however, the total absence of the de novo synthesis of all tetrapyrroles is almost nonexistent. Exceptions are extremely few and include only genera at the root of the bacterial phylogeny: Dehalococcoides and Thermotoga. The loss of tetrapyrrole biosynthesis in the Thermotogaceae appears secondary (in spite of the fact that it is missing in all Thermotoga genomes available to date), since the genome of Thermosipho melanesiensis contains the full complement of genes for the biosynthesis of uroporphyrinogen (URO) III (the last common precursor of various tetrapyrrolic cofactors) and several porphinoids.
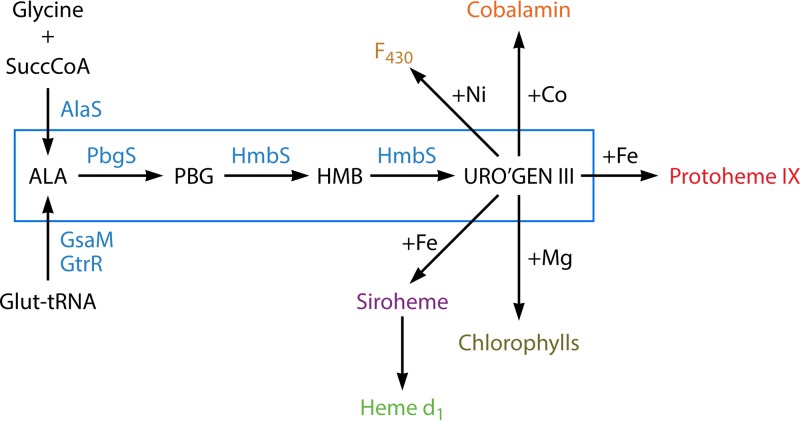
The core pathway for the biosynthesis of tetrapyrroles is an ancient one (Fig. 1). Two distinct mechanisms to synthesize the first committed pathway intermediate, 5-aminolevulinic acid (ALA), have evolved, but the next three enzymes, porphobilinogen (PBG) synthase (PBGS), hydroxymethylbilane (HMB) synthase (HMBS), and uroporphyrinogen synthase, are highly conserved, being found in archaea to eukaryotes. Indeed, these three enzymes can be considered the trunk of the biological tetrapyrrole synthesis tree. The end product of this core pathway, uroporphyrinogen III, serves as the precursor to the synthetic branches for the Ni-containing F430 that is utilized in archaeal methanogenesis (7, 8): the Co-containing corrin that is an important nutrient in the form of vitamin B12 but synthesized by a limited number of archaea and eubacteria (9, 10), the Fe-containing siroheme (11, 12), heme d1 and protoheme (13, 14), and the Mg-containing chlorophylls and bacteriochlorophylls found in photosynthetic organisms (15). A casual examination of these pathways and the branches of life in which they are found makes clear that the order of appearance in evolution must be F430 and cobalamin first, followed by siroheme, then protoheme, and finally chlorophyll. Indeed, while it is generally stated that the advent of chlorophyll-based photosynthesis was an early and key evolutionary event, it is obvious that highly varied life existed well before nature happened upon protoporphyrin, the key building block for this compound. Additionally, the presence of oxygen respiration in Gram-positive bacteria, which predated Gram-negative bacterial chlorophyll synthesis, suggests that the generation of oxygen by other means, such as by the hemoprotein enzyme chlorite dismutase, may have played more significant roles than generally credited (16).
FIG 1.
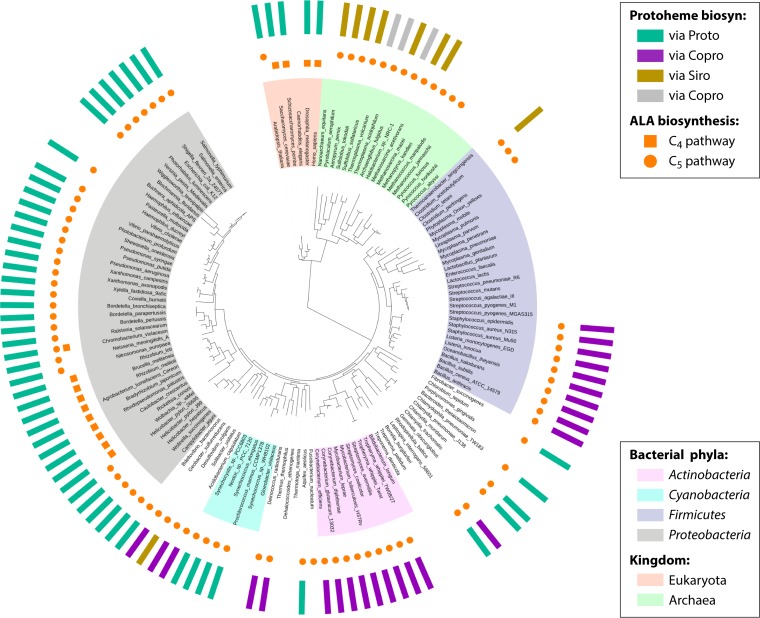
Tetrapyrrole biosynthetic pathways in prokaryotes. An outline of prokaryotic tetrapyrrole biosynthetic pathways is shown with the conserved three-enzymatic-step core path from ALA (5-aminolevulinate) to uroporphyrinogen III boxed in blue. Among prokaryotes, one finds one of two generally nonoverlapping paths to ALA. The synthesis of ALA from glutamyl-tRNA (5-carbon pathway) is most common, with a limited number of bacteria possessing the enzyme AlaS to form ALA from glycine and succinyl-CoA (4-carbon pathway). The 4-carbon pathway to ALA is found only in bacteria that possess the protoporphyrin-dependent pathway branch. Uroporphyrinogen III is the precursor to Ni-containing F430 in methanogens, Co-containing cobalamin, and Fe-containing siroheme and heme d1 as well as coproporphyrinogen, which is the common precursor of Fe-containing protoheme and Mg-containing chlorophylls. Abbreviations: PBG, porphobilinogen; HMB, hydroxymethylbilane; uro'gen III, uroporphyrinogen III.
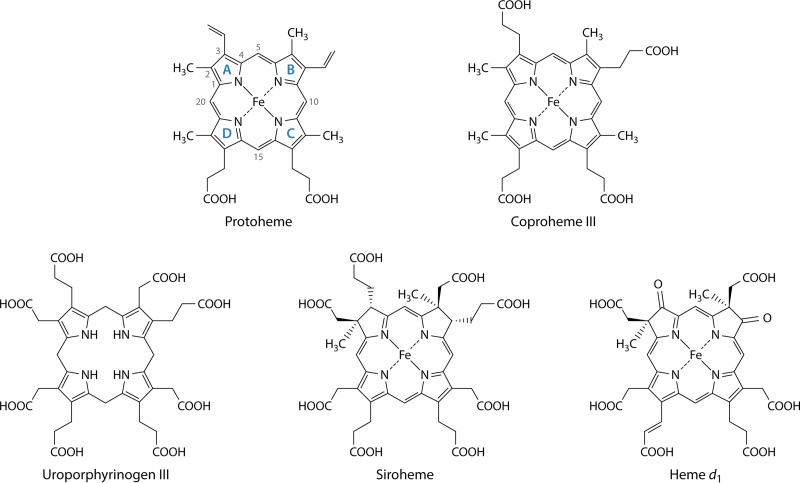
Iron-containing porphyrins (hemes) (Fig. 2) are widely distributed, being found in eukaryotes, most characterized bacteria, and some archaea. The functions served by hemes are diverse and dependent upon the nature of the heme moiety and the protein to which it is bound. Many bacteria contain a diversity of environmentally regulated heme-containing respiratory cytochromes in which the heme has a redox function as a one-electron carrier (17). There are other non-respiratory-chain cytochromes, such as cytochrome P450s, which utilize protoheme as a cofactor to metabolize a broad range of substrates, most commonly serving as a monooxygenase (18). There are also some noncytochrome hemoproteins, such as the enzyme chlorite dismutase, which converts chlorite to chloride and molecular oxygen (19), and may serve as individual respiratory proteins. Another class of hemoproteins in bacteria serves as gas sensors. Unlike the respiratory proteins, the heme of gas-binding hemoproteins does not undergo a redox reaction. Examples of these are the well-characterized CO-sensing CooA transcription factor (20), nitric oxide/oxygen-sensing (H-NOX) proteins (21), globin-coupled oxygen sensors (22), and the DosS and DosT gas sensors of Mycobacterium tuberculosis (23). Additionally, many eubacteria possess heme-containing peroxidases and catalases. In some photosynthetic bacteria, endogenously synthesized protoheme serves as the substrate for heme oxygenase in the synthesis of the linear tetrapyrrole biliverdin as a precursor to the photosynthesis accessory pigments named phycobilins (24).
FIG 2.
Structures of protoheme IX, coproheme III, uroporphyrinogen III, siroheme, and heme d1. The structure of protoheme IX is shown with the four pyrrole rings labeled by convention as rings A, B, C, and D. The numbering of side chains is also shown. Of note is that the D ring is inverted to form the IX isomer of protoporphyrin, which corresponds to the III isomer of uroporphyrinogen and coproporphyrinogen. Siroheme is not a true porphyrin but rather is an isobacteriochlorin, since its B and C rings have methyl groups substituted on the A and B rings, which prevents full macrocycle desaturation. Likewise, heme d1 is also not a porphyrin but is a dioxoisobacteriochlorin.
Most bacteria that contain heme possess the necessary machinery to synthesize it. However, this is not universal, since a few bacteria, such as Enterococcus faecalis and lactic acid bacteria (25), possess genes encoding apohemoproteins but cannot synthesize their own heme (26, 27). They are instead equipped to acquire and utilize exogenously supplied heme for incorporation into these apoproteins to make the mature hemoprotein. Many bacteria have heme acquisition systems whose main function is to procure heme for subsequent degradation and release of iron (28, 29). The fields of heme trafficking and degradation are diverse, rapidly expanding, and outside the scope of this review.
Research on heme biosynthesis dates back over 50 years and is summarized in the first comprehensive overview of tetrapyrrole synthesis in the 1964 monograph by Lascelles in the Microbial and Molecular Biology Series edited by Bernard Davis (30). Initially, investigators interchangeably employed eukaryotic and prokaryotic sources of material in their studies, believing that the pathway was conserved among all organisms. Pathway intermediates were guessed, in part, from the excreted porphyrin and porphyrin precursors found in human clinical samples and in microbial cultures. The assumption of a universally conserved pathway, as described below, was inaccurate and over the years has occasionally led to considerable confusion. Interestingly, data that suggested alternative pathways to heme were reported even in the 1950s (31), but they were routinely ignored or explained as experimental artifacts. Nevertheless, by the early 1960s, most of the steps in the classical heme biosynthesis pathway were known, although there were significant voids, since none of the enzymes had been purified and characterized and a few were yet to be identified. It was apparent that microorganisms possessing the ability to synthesize hemoproteins had multiple and diverse mechanisms to regulate the synthesis of their tetrapyrrole compounds (30). In particular, the fact that oxygen played a significant role in regulation was known and had been studied in a variety of organisms, with Saccharomyces cerevisiae and Rhodobacter sphaeroides being the most extensively examined. What was not appreciated at that time was the diversity in pathway enzymes that had evolved to cope with the need for oxygen-dependent and oxygen-independent mechanisms for two steps in the pathway.
Multiple reviews of eukaryotic heme synthesis have been reported in the past decade (13, 14, 32–34), but there is no comprehensive and current review of prokaryotic heme synthesis. Short reviews within otherwise focused manuscripts can be found (35–39), and there is an excellent review of tetrapyrrole biosynthesis in Rhodobacter capsulatus (40). The last broad-scope genome-based review of prokaryotic heme synthesis was reported over a dozen years ago by O'Brian and Thony-Meyer (41). This, along with a parallel review on whole-microbial-genome analysis (39), began to challenge the dogmatic view of a conserved pathway model, when it was found that even among a modest number of prokaryotic genomes (69 genomes), there were missing genes for essential pathway steps in most bacteria, and no heme-synthesizing archaea possessed an intact pathway.
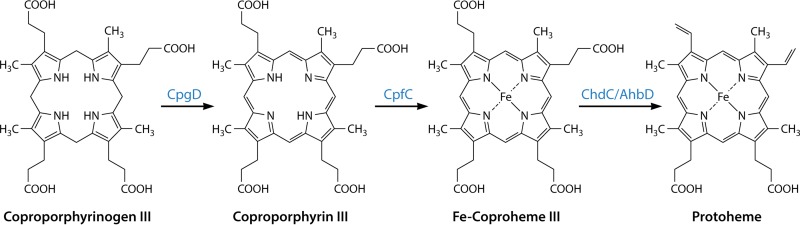
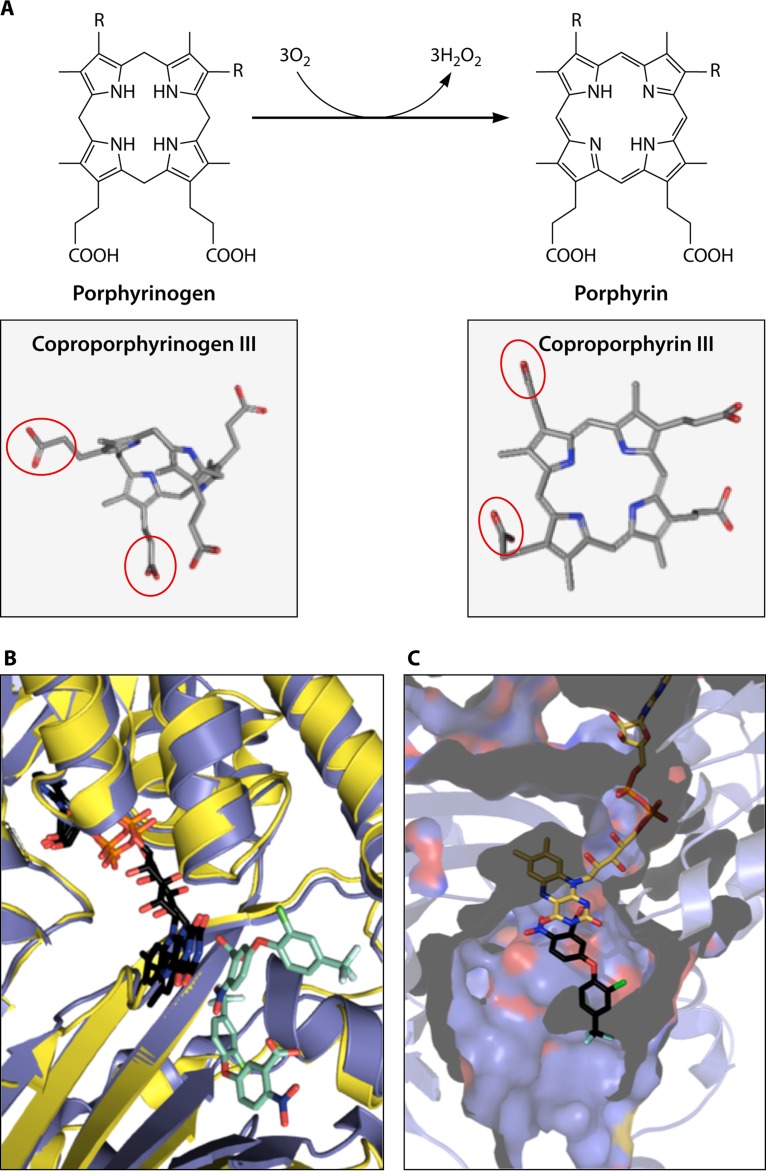
More recent studies have revealed that there are multiple pathways for heme synthesis in prokaryotes (11, 35, 42, 43), and in only limited instances does one find a set of enzymes in bacteria that closely resembles those of the so-called classic pathway found in metazoans. It is now known that some archaea and sulfate-reducing bacteria synthesize siroheme and then convert it to protoheme (11, 42), Gram-positive heme-synthesizing bacteria go through a set of enzymes that utilize a coproporphyrin intermediate (35, 43–45), and Gram-negative bacteria have a set of enzymes that go through a protoporphyrin intermediate (35). These various pathways to heme in prokaryotes have acquired a variety of common names, such as primitive (46), alternate (11), transitional (43), and classic. While this nomenclature may find favor among those closely involved in the study of these pathways, it is not particularly obvious or transparent to the general audience. Here we have chosen to name the pathway branches based upon compounds unique to each one. Thus, the primitive, or alternative, pathway becomes the siroheme-dependent branch, the transitional pathway is named the coproporphyrin-dependent (CPD) branch, and the classic pathway is named the protoporphyrin-dependent (PPD) branch.
This review of protoheme biosynthesis is presented loosely in an evolutionarily based manner rather than a historical fashion. The core tetrapyrrole synthesis “trunk” of the tree is first, followed by the siroheme branch, then the coproporphyrin-based branch, and finally the protoporphyrin-based branch. With the discovery of the coproporphyrin- and protoporphyrin-dependent branches, we have employed new nomenclature for the annotation of the pathway genes/proteins to eliminate the confusion created by overlapping nomenclature. Our newly proposed nomenclature, along with the old nomenclature, is given in Table 1 and is based upon enzyme activity rather than the pathway name and number/letter.
TABLE 1.
Names and abbreviations for bacterial heme synthesis enzymes
| Enzyme | Old abbreviation(s) | New abbreviation |
|---|---|---|
| ALA synthase | HemA | AlaS |
| Glu-tRNA reductase | HemA/GtrA | GtrR |
| GSA mutase | HemL/GSAM | GsaM |
| PBG synthase | HemB/ALAD/PBGS | PbgS |
| HMB synthase | HemC/HMBS/PBGD | HmbS |
| URO synthase | HemD/UROS | UroS |
| URO decarboxylase | HemE/UROD | UroD |
| Coproporphyrinogen decarboxylase | HemF/CPOX | CgdC |
| Coproporphyrinogen dehydrogenase | HemN/HemZ/CPDH | CgdH |
| Protoporphyrinogen dehydrogenase | HemG | PgdH1 |
| Protoporphyrinogen dehydrogenase | HemJ | PgdH2 |
| Protoporphyrinogen oxidase | HemY | PgoX |
| Protoporphyrin ferrochelatase | HemH | PpfC |
| Coproporphyrin ferrochelatase | HemH/HemZ | CpfC |
| Coproporphyrinogen oxidase | HemY | CgoX |
| Coproheme decarboxylase | HemQ | ChdC |
THE TETRAPYRROLE BIOSYNTHESIS PATHWAY COMMON CORE
5-Aminolevulinate Synthesis: Two Pathways to ALA
In nature, there are two independent, unrelated, biosynthetic routes for the formation of ALA. The first of these routes to be discovered was the so-called “Shemin” or “C4” pathway, which involves the condensation of succinyl coenzyme A (CoA) and glycine by 5-aminolevulinic acid synthase (AlaS) (Fig. 3). This reaction is used by metazoans, fungi, and the alphaproteobacteria, with few exceptions, and for many years, this was thought to be the sole mechanism for the production of ALA, even though AlaS activity in plants and some bacteria had never been detected. It was 2 decades before an alternate route to ALA was discovered. This route, which is likely the evolutionarily older process, termed the “C5 pathway,” utilizes the C5 skeleton of a tRNA-bound glutamate to generate ALA in a two-step reaction. This route is found in plants, archaea, and most bacteria. The initial substrate, glutamyl-tRNA, is synthesized by glutamyl-tRNA synthetase (GluRS), is utilized in both protein and tetrapyrrole biosynthesis, and is one of the few examples of a charged tRNA species being used for something other than translation. Glutamyl-RNA is converted into the labile glutamate-1-semialdehyde (GSA) intermediate by glutamyl-tRNA reductase (GtrR) and is then converted into ALA by glutamate-1-semialdehyde-2,1-aminomutase (GsaM) (Fig. 4 and 5). Only in very rare instances have both pathways for ALA formation been found in a single organism, such as Euglena gracilis (14, 33, 47–49) and Chromobacterium violaceum (http://www.theseed.org/SubsystemStories/Porphyrin,_Heme,_and_Siroheme_Biosynthesis/story.pdf).
FIG 3.
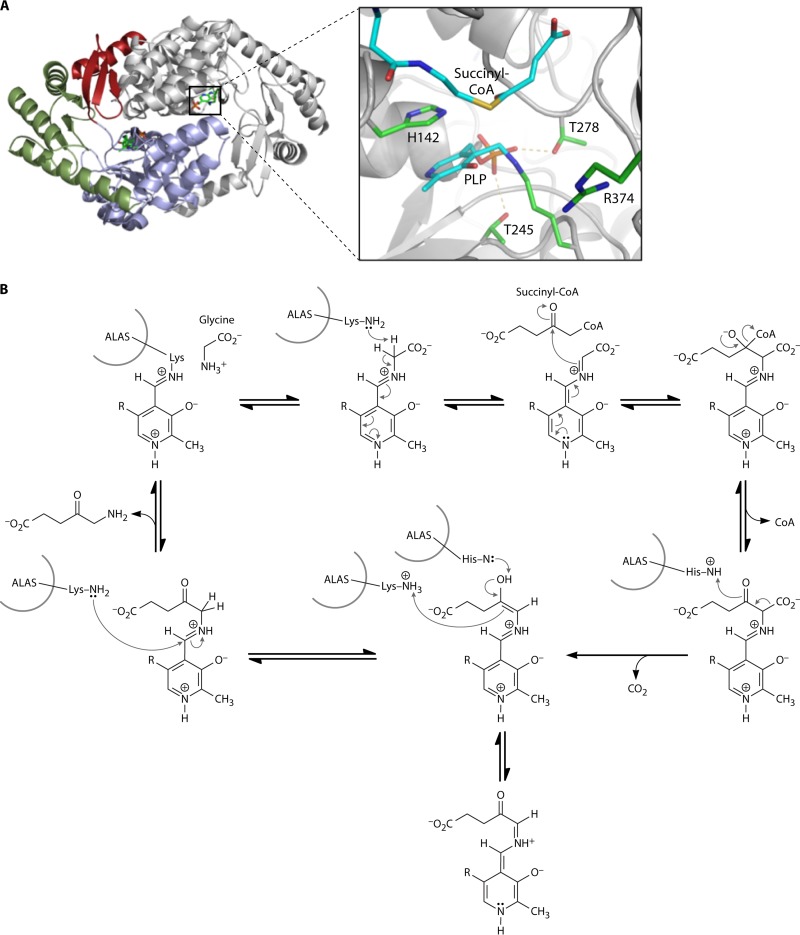
5-Aminolevulinic acid synthase (AlaS). (A) Crystal structure of AlaS from R. capsulatus. (Left) AlaS is a homodimeric protein in which each monomer (shown in color or gray) consists of three domains (red, green, and blue). (Right) In the active site of the enzyme, the PLP cofactor is covalently attached as a Schiff base to a conserved lysine residue. (Adapted from reference 14.) (B) Proposed reaction mechanism by which AlaS catalyzes the PLP-dependent condensation of glycine and succinyl-CoA to form ALA (see the text for a detailed explanation). (Adapted from reference 61.)
FIG 4.
Glutamyl-tRNA reductase (GtrR). (A, left) Crystal structure of M. kandleri GtrR showing the unusual V-shaped form of the dimeric enzyme. (Right) In the active site of GluTR, C-48 is ideally positioned to attack the α-carboxyl group of the substrate. (Adapted from reference 14.) (B) Proposed reaction mechanism by which GtrR catalyzes the NADPH-dependent reduction of glutamyl-tRNA to glutamate-1-semialdehyde (see the text for a detailed explanation).
FIG 5.
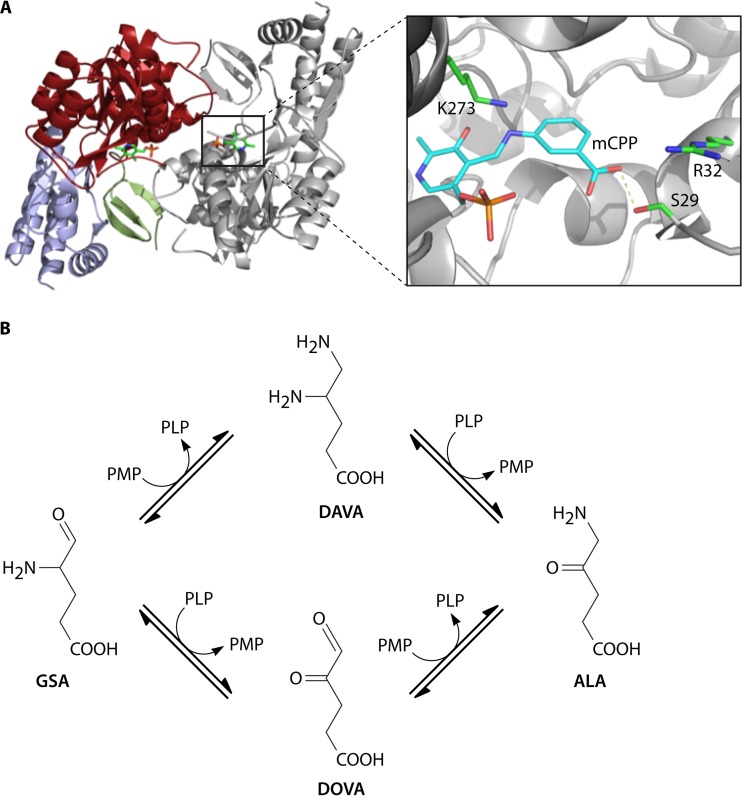
Glutamate-1-semialdehyde-2,1-aminomutase (GsaM). (A, left) Crystal structure of T. elongatus GsaM. (Right) In the active site of Synechococcus GsaM, the inhibitor gabaculine was observed to bind covalently to the PLP cofactor (m-carboxyphenylpyridoxalamine [mCPP]). (Adapted from reference 14.) (B) Depending on the initial form of the pyridoxal phosphate cofactor, GsaM is able to convert GSA into ALA via either DAVA or DOVA (4,5-dioxovalerate) (see the text for a detailed explanation). Kinetic and other biochemical data suggested that formation via DAVA is used by the enzyme. (Adapted from reference 77.)
The Shemin pathway for ALA biosynthesis: 5-aminolevulinic acid synthase.
AlaS catalyzes the condensation of succinyl-CoA and glycine to ALA with the release of CO2 and coenzyme A. In 1945, Shemin and Rittenberg reported the incorporation of nitrogen atoms from glycine into heme (50). Subsequently, in the 1950s and 1960s, the groups of Shemin and Neuberger identified succinyl-CoA as the other source of carbon atoms in heme, with ALA being identified as the first committed precursor molecule for heme formation and AlaS being identified as the first enzyme of heme biosynthesis (51–56). AlaS is a homodimeric protein and belongs to the α-oxoamine synthase class of pyridoxal-5′-phosphate (PLP)-dependent enzymes, which catalyze the condensation of an acid-CoA thioester and small amino acids with the concomitant decarboxylation of the amino acid (57, 58) (Fig. 3). However, the cleavage of two α-carbon bonds during the AlaS reaction is unusual for PLP-dependent enzymes. The reaction mechanism has been elucidated through the employment of region-specific labeled substrates coupled with detailed kinetic studies using high-resolution stopped-flow techniques with the knowledge of the only currently solved AlaS crystal structure, that of the R. capsulatus enzyme (49, 59–63) (Fig. 3A). Initially, an active-site lysine covalently binds the PLP cofactor to form the internal aldimine (Fig. 3B). The external aldimine is formed via a Schiff base to the first substrate, glycine, and the PLP cofactor. Stereospecific abstraction of the pro-R-hydrogen as a proton from the PLP-bound glycine generates quinonoid intermediate I. Subsequently, the electrophilic carbonyl group of succinyl-CoA binds to the α-carbon of glycine with the formation of the 2-amino-3-ketoadipate as an intermediate. The quinonoid intermediate II is formed after the release of coenzyme A from the tetrahedral intermediate and the decarboxylation of the resultant α-amino-β-ketoadipate aldimine. The AlaS-ALA-aldimine is formed via the protonation of the quinonoid, and ALA is finally released from the enzyme as the product after the completion of this cycle (49, 59–62).
The C5 pathway of ALA biosynthesis: glutamyl-tRNA reductase and glutamate-1-semialdhyde-2,1-aminomutase.
In 1973, a second pathway for ALA formation from the C5 skeleton of glutamate was discovered by Beale et al. in cucumber cotyledons (64, 65). Subsequently, the conversion of glutamate via GSA to ALA was demonstrated, and the relevant enzymes from barley were isolated and biochemically characterized (66, 67). As a result of these efforts, glutamyl-tRNA was identified as the starting molecule of the pathway (68). The C5 pathway enzymes GtrR and GsaM were subsequently purified from multiple bacterial, archaeal, and plant sources (48).
For many years, the crystal structure of Methanopyrus kandleri GtrR was the only one available for this enzyme (69) (Fig. 4A). Recently, a second structure representing Arabidopsis thaliana GluTR has become available (70). GtrR is a highly asymmetric, V-shaped, dimeric enzyme composed of catalytic (domain I), nucleotide-binding fold-containing (domain II), and dimerization (domain III) domains. These three domains are arranged along a curved spinal helix (69). This structure allows the necessary movement of NADPH-binding domain II toward active-site domain I during catalysis and the formation of a complex with GsaM to permit the metabolic channeling of solvent-sensitive GSA. During GtrR catalysis, an active-site cysteine residue acts as a nucleophile to attack the tRNA-activated α-carboxyl group of glutamate with the formation of an enzyme-bound thioester and the release of tRNAGlu (71, 72) (Fig. 4B). Hydride transfer from NADPH to the thioester leads to GSA formation and its release (73). Glutamyl-tRNA is recognized by GtrR from the overall shape of the tRNA molecule rather than by specific single nucleotides and the attached glutamate (74).
Crystal structures of various GsaM enzymes (i.e., Synechococcus, Thermosynechococcus elongatus, Bacillus subtilis, Bacillus anthracis, Thermus thermophilus, Aeropyrum pernix, and Yersinia pestis) have been solved (75–77). GsaM is a functional dimer, with large central domains harboring the active sites close to the dimer surface (75–77) (Fig. 5A). The subunits are asymmetric and act cooperatively during catalysis through a process involving structural signaling between them (78, 79). GsaM is part of the α-family of PLP-dependent enzymes and has interesting structural similarity to AlaS. This had led to speculation that GsaM is an evolutionary precursor of AlaS (77). In contrast to typical aminotransferases, GsaM is an aminomutase that uses GSA as the sole substrate, transferring the amino group intramolecularly. During catalysis, a complex is first formed between the pyridoxamine-5′-phosphate (PMP) form of GsaM and GSA, with the subsequent generation of the ketamine-5 form (Fig. 5B). Double-bond shifts lead to the formation of the external aldimine between PLP and 5′-diaminovalerate (DAVA). Next, the internal aldimine between an active-site lysine and PLP is formed with the release and reorientation of DAVA (80, 81). An active-site gating loop of GsaM prevents the escape of DAVA (82). A second external aldimine between DAVA and PMP is formed, followed by the formation of the ketamine-4 form between PMP and ALA. Finally, the PMP of GsaM is regenerated with the release of ALA (83–86). Structures revealed that the GsaM and GtrR enzymes form a stable complex for channeling of the labile GSA intermediate (87, 88). In this complex, GsaM sits in the middle of the V-shaped GtrR dimer within close proximity of the GtrR active-site exit and the entrance to the GsaM active site (69, 70).
Synthesis of a Monopyrrole, Porphobilinogen Synthase
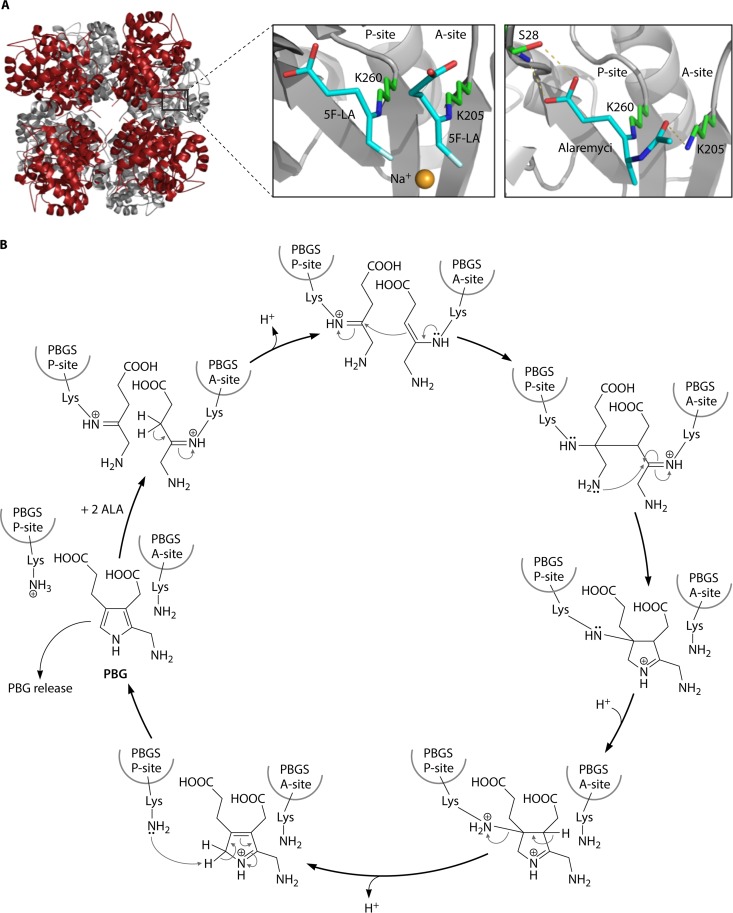
Porphobilinogen synthase (PbgS) catalyzes the asymmetric condensation of two ALA molecules to the monopyrrole porphobilinogen. There are two ALA-binding sites, the so-called A and P sites, corresponding to the ALA that contributes to the acetate and propionate portions of PBG, respectively (Fig. 6). The P-site ALA forms a Schiff base with a conserved lysine residue (K247 in Escherichia coli). Features of the A site may differ among PbgS enzymes from different organisms depending on the metal requirement. The enzyme is a homooctomer in both eukaryotes and prokaryotes (89–93) and requires metal for activity (94). Three binding sites for divalent metals have been described, but not all porphobilinogen synthase proteins have all three sites. Animals and yeast have two Zn2+-binding sites (ZnA and ZnB), but the plant enzyme binds Mg2+ in an MgA/MgB/MgC configuration. Prokaryotes also have an MgC site, which is allosteric. Most bacterial PbgSs contain either Zn2+ or Mg2+ in the active site, but the R. capsulatus enzyme is metal independent (95). The introduction of cysteines in positions of Mg2+ PbgS normally found in the Zn2+ enzymes is sufficient to alter the metal specificity from Mg2+ to Zn2+ (96, 97). The modified enzyme also acquires sensitivity to lead and has a pH optimum that is 3.5 units lower than that of the wild type, as is observed for the naturally occurring Zn2+-containing enzyme. For Pseudomonas aeruginosa PbgS, a series of mutant combinations intermediate between the two types of enzymes was extensively characterized (97). These variants were similar to naturally occurring PbgS enzymes from other organisms that vary in their dependence on Zn2+ or Mg2+. Of the nine variants constructed, five have no known natural equivalent, and four of these are inactive. Thus, a plausible evolutionary path of metal dependence could be deduced.
FIG 6.
Porphobilinogen synthase (PbgS). (A, left) Crystal structure of P. aeruginosa PbgS showing the octameric assembly of the protein representing a tetramer of homodimers. (Top right) In the active site of P. aeruginosa PbgS, two molecules of the substrate analog 5-fluorolevulinic acid (5F-LA) were observed to bind covalently to the enzyme through Schiff bases, with the catalytically essential lysine residues supporting a double-Schiff-base mechanism. (Bottom right) The antibiotic alaremycin was also observed to bind covalently to the enzyme through a Schiff base with the P-site lysine. (Adapted from reference 14.) (B) Proposed reaction mechanism by which PbgS catalyzes the asymmetric condensation of two ALA molecules to the pyrrole porphobilinogen (see the text for a detailed explanation). (Adapted from reference 97.)
Binding and kinetic studies indicate that Bradyrhizobium japonicum PbgS contains four active-site Mg2+ atoms per octamer and eight allosteric Mg2+ ions (98). This enzyme is also stimulated by the monovalent cation K+ (98, 99). The Mg2+ enzyme from P. aeruginosa contains only four allosteric ions per octamer, but activity is not strictly metal dependent (93). The crystal structure of the P. aeruginosa enzyme shows that it is composed of 4 asymmetric dimers. The monomers in each dimer differ by one having a closed active-site pocket shielded from the solvent by a “lid” that is disordered in the open active site (93). Mg2+ is 14 Å away from the Schiff base lysine, which is too far away to play a direct catalytic role. However, it has been suggested that the open and closed forms of each monomer are governed by Mg2+ binding (93). Unlike the P. aeruginosa enzyme, E. coli PbgS is symmetric. Interestingly, crystal structures of P. aeruginosa variants with cysteine substitutions show decreasing asymmetry with increasing numbers of cysteines (97).
Although structural information on porphobilinogen synthases from many sources shows that the predominant active form of the enzyme is a homooctamer, alternate oligomeric states have been identified (89, 100–103). Jaffe (100) introduced the term morpheein to describe this phenomenon. A morpheein is a homooligomeric protein that exists in an equilibrium of functionally distinct, quaternary structural isoforms. The morpheein isoforms of human PBGS are an octamer, a hexamer, and two different dimers. The interconversion of the octamer to the hexamer involves dissociation to a dimer. The hexamer isoform was initially recognized in a rare human PBGS allele, whereby rearrangement of the N-terminal arm caused by an F12L substitution was responsible for the oligomeric switch leading to a low-activity enzyme (89). The variant N-terminal arm structures vary among the morpheeins, and the relationship between the arm and the β-barrel domain dictates the isoforms. An R240A mutation in PBGS that affects this relationship stabilizes the hexamer, which can be converted to the octamer in the presence of the substrate (103). The physiological relevance of these morpheeins is apparent from a study of PBGS variants found in patients with the disease aminolevulinate dehydratase (ALAD) (now known as PBGS) porphyria (101). All eight known porphyria-associated variants shifted the morpheein isoform from the octamer toward the hexamer, which can explain the deficiency in these patients. Morpheein structures have not been described for any bacterial PbgS enzyme, but interestingly, PbgS of R. capsulatus is normally active as a hexamer and does not require any metal (95).
From Porphobilinogen to Tetrapyrrole
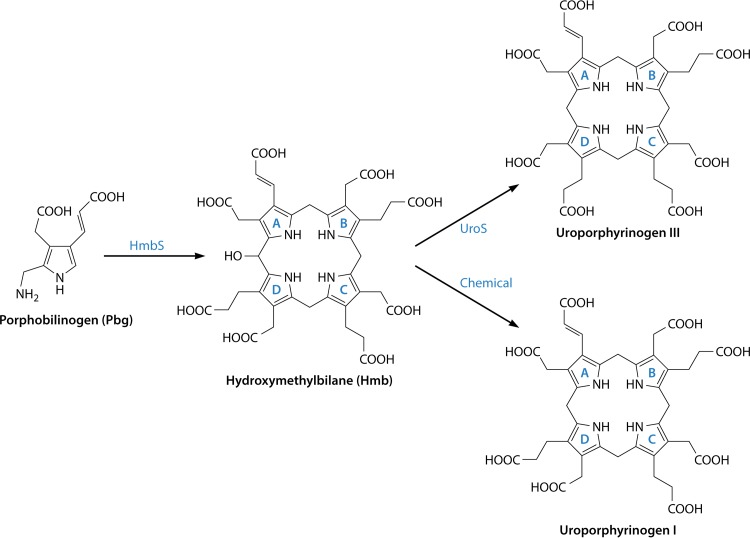
The transformation of the pyrrole building block, PBG, into the first macrocyclic intermediate, uroporphyrinogen III, requires the actions of two enzymes, hydroxymethylbilane synthase (HmbS) and uroporphyrinogen synthase (UroS). In bacteria, these enzymes are encoded by hemC and hemD. The first of these enzymes, HmbS, was initially termed porphobilinogen deaminase (PBGD), as the enzyme deaminates the aminomethyl moiety of PBG and releases this as ammonia (104). The deaminated pyrroles are then strung together to generate a linear tetrapyrrole, or bilane, which is subsequently cyclized in a very specific manner by the proceeding enzyme, UroS, to generate the type III isomer of uroporphyrinogen (104) (Fig. 7). In early studies investigating the transformation of PBG into uroporphyrinogen III, researchers found that the two enzymes tended to purify together, and hence, the latter enzyme was referred to as the cosynthase. However, this enzyme is now more commonly referred to as uroporphyrinogen III synthase (UroS). The two enzymes HmbS and UroS were initially differentiated on the basis of their heat stability: HmbS is normally heat stable at temperatures of up to ∼70°C, whereas UroS is heat labile at temperatures much above 50°C. Thus, an extract containing the two enzymes would transform PBG into uroporphyrinogen III, whereas a heat-treated extract would convert PBG into HMB, which would then spontaneously cyclize into the type I isomer (Fig. 7). This established that the two enzymes required for uroporphyrinogen III production acted separately. As with the earlier enzymes in the pathway, much has been learned about HmbS and UroS from human genetic disorders that cause porphyria, in these cases acute intermittent porphyria and congenital erythropoietic porphyria, respectively (105).
FIG 7.
Transformation of porphobilinogen into uroporphyrinogen III. Four molecules of porphobilinogen are deaminated and polymerized in an ordered sequential fashion (rings A to D) into a linear tetrapyrrole called hydroxymethylbilane by the action of hydroxymethylbilane synthase. The unstable bilane is acted upon by the enzyme uroporphyrinogen synthase, which inverts ring D and cyclizes the macrocycle to give the type III isomer of uroporphyrinogen. In the absence of the enzyme, hydroxymethylbilane spontaneously cyclizes to give the type I isomer.
Early work on these two enzymes was hampered by a lack of knowledge of the reactions actually catalyzed by the two enzymes. In this respect, the product of HmbS was unknown, and hence, the substrate for UroS was likewise a mystery. The problem was solved by the application of nuclear magnetic resonance (NMR). By using purified HmbS from R. sphaeroides, it was demonstrated that incubation of [11-13C]PBG gave rise to a transient intermediate, with a half-life of about 5 min, which spontaneously cyclized to give uroporphyrinogen I (106). However, in the presence of UroS, this transient intermediate was rapidly converted into uroporphyrinogen III. The intermediate was initially called preuroporphyrinogen (106), and it did not take long before it was shown to be HMB. This was unambiguously demonstrated by total chemical synthesis of the compound, which was revealed to be the preferred substrate for UroS (107). Aminomethyl bilane also acted as a substrate for the enzyme but was converted by HmbS at a much lower rate (108–110). HmbS is evidently able to deaminate the aminomethyl bilane and convert it to HMB (111). The identification of HMB as the product of HmbS and the substrate for UroS then permitted more detailed analyses of the mechanism for the two enzymes.
Hydroxymethylbilane synthase.
Considerable evidence suggested that HmbS must form a covalent complex with its substrate during the pyrrole polymerization process, but the amino acid residue involved in this attachment process was unknown. The formation and identification of enzyme-substrate complexes were shown by native polyacrylamide gel electrophoresis of enzyme samples that had been incubated with increasing stoichiometric quantities of PBG (112, 113). This revealed that the enzyme ran as either a free enzyme or an enzyme with one, two, three, or four substrates attached. These enzyme-substrate complexes were referred to as ES1, ES2, ES3, and ES4 (Fig. 8A). Around the same time, through the stoichiometric addition of either radiolabeled ([3,5-14C]PBG) or heavy ([11-13C]PBG) isotopes, it was demonstrated that HMB was synthesized in an ordered and sequential fashion, with the first PBG ending up as ring A, the second as ring B, the third as ring C, and the final PBG as ring D within the final macrocycle (114, 115). Together, all of this evidence indicated that the enzyme forms HMB by forming a growing polypyrrole chain attached presumably to the active site (Fig. 8A).
FIG 8.
Mechanism and structure of hydroxymethylbilane synthase (HmbS). (A) Mechanism of action of HmbS. HmbS contains a dipyrromethane cofactor, which is attached to an active-site cysteine residue and constitutes the holoenzyme form. The two rings of the cofactor are termed C1 and C2. During the polymerization process, the first substrate PBG molecule, ring A of the final product, undergoes deamination to generate an azafulvene species. This is then attached to the C2 ring of the cofactor to generate the ES1 substrate complex. This process is then repeated three more times to generate the ES2, ES3, and ES4 substrate complexes. The ES4 complex then undergoes hydrolysis between the C2 cofactor ring and ring A of the final product to generate the holoenzyme and the hydroxymethylbilane product. The acetate and propionate side chains are designated A and P, respectively. (B) Structure of E. coli HmbS shown in cartoon format, colored according to secondary structure. The 312 amino acid residues of the enzyme are folded into three α/β-domains with a large active-site cavity formed in the space between the three domains. The dipyrromethane cofactor is seen attached to C242 with a key catalytic residue, D84, located just below the two N atoms of the dipyrromethane.
The advent of recombinant-DNA technology played a key role in the identification of the key active-site residues involved in forming the growing bilane chain attached to the enzyme. The discovery, isolation, and sequencing of the E. coli hemC gene allowed the enzyme to be overproduced, making the protein much more readily available (116, 117). Most HmbSs have a molecular mass of around 35 kDa and are monomeric. Two independent pieces of research established that the key active-site residue to which the first PBG unit attaches was, in fact, a dipyrromethane cofactor (118–120) (Fig. 8A). This was deduced from several pieces of evidence. An elegant NMR approach, whereby [11-13C]PBG was added to a large amount of the enzyme, resulted in an NMR spectrum that suggested that the first PBG unit had formed a methylene bridge with the enzyme. Moreover, it was also observed that the free enzyme, to which no PBG had been added, formed significant quantities of porphyrin when treated with a strong acid. Both of these pieces of evidence suggested that some pyrrole species was already attached to the enzyme. Finally, treatment of the free enzyme with Ehrlich's reagent gave a reaction consistent with the presence of a dipyrrole. The ES1 substrate complex reacted with Ehrlich's reagent to give a spectrum typical of a tripyrrole, while the ES2 substrate complex gave a reaction typical of a linear tetrapyrrole.
Further evidence of the presence of a dipyrromethane cofactor came from the radioactive incorporation of ALA into the enzyme during the production of the recombinant enzyme in E. coli (121). In this case, a hemA strain of E. coli was used to overproduce the enzyme and was grown in the presence of added exogenous [5-14C]ALA. This resulted in the incorporation of the label into the enzyme, a label that was not turned over when the purified enzyme was incubated with the unlabeled substrate. This indicated that the ALA was converted into PBG, which was subsequently incorporated into the active site of the enzyme. It had also been noted previously that a hemB (PbgS)-deficient strain of E. coli not only was deficient in PbgS activity but also lacked HmbS activity, presumably because the strain was unable to provide the PBG for cofactor assembly (122).
NMR was again employed to determine how the dipyrromethane cofactor was attached to the enzyme. By the incorporation of either [5-13C]ALA or [11-13C]PBG into the enzyme, the resulting spectra confirmed not only the presence of the dipyrromethane cofactor but also that the cofactor was attached to the sulfur atom of a cysteine residue (123, 124). At that time, there were only comparatively few sequences of HmbS available, and when aligned, these sequences highlighted the presence of two conserved cysteine groups within the protein. Site-directed mutagenesis then identified that cysteine 242 of the E. coli enzyme was the residue responsible for the binding of the dipyrromethane cofactor. The presence of the cofactor has since been confirmed in a broad range of HMBSs that have been studied.
The next question to be addressed was related to how the cofactor was assembled and inserted into the enzyme. In fact, it soon became apparent that the enzyme was able to generate its own cofactor. This was demonstrated by showing that the apoenzyme, an enzyme without any cofactor, generated either by isolating the enzyme from an E. coli gtrR or pbgS strain or by treating the holoenzyme with 1 M HCl, could have its activity restored by incubating it with PBG on ice (121, 123). The apoenzyme is able to deaminate PBG to allow the attachment of the first pyrrole ring of the cofactor to the enzyme, a process that is repeated for the incorporation of the second PBG-derived pyrrole unit for the completion of the cofactor. Subsequently, it was shown that hydroxymethylbilane was incorporated more quickly than PBG to give an active enzyme, presumably allowing the direct formation of an ES2 complex (125, 126).
The recombinant production of E. coli HmbS allowed the enzyme to be crystallized, and it was the first enzyme of tetrapyrrole biosynthesis to have its structure determined by X-ray crystallography (116, 127). This structure revealed that the protein was composed of three domains, with the dipyrromethane cofactor being attached to domain 3 and with the majority of the active site being formed within a cleft between domains 1 and 2 (128, 129) (Fig. 8B). The cofactor was observed to be positioned just above an aspartic acid (D84) residue, which appears to be the main catalytic group, although the active site is lined with arginine residues not only to help hold the cofactor but also to accommodate the growing polypyrrole chain via the negatively charged carboxylates of the pyrrole units (130, 131). The role of these residues in catalysis and substrate binding was confirmed by site-directed mutagenesis. Since then, the structures of a number of HmbSs have been determined, including those of Arabidopsis, human, and Bacillus megaterium, revealing similar overall topologies (132–135). Interestingly, no one has been able to grow crystals of the enzyme in the presence of a substrate, so it is not known how the growing polypyrrole product is held within the active site of the enzyme or how the domains of the enzyme move during the catalytic process. Conformational change is known to take place during the reaction, and indeed, computation molecular analysis suggests that the structure of the enzyme becomes progressively less compact during the catalytic reaction (121, 136). Moreover, those studies also suggest that the cofactor moves toward the second domain to generate more space for the growing product (137).
Collectively, all of these data have allowed an overarching mechanism for the enzyme to be developed (121) (Fig. 8A). The holoenzyme binds the first substrate PBG unit in the active site, in close proximity to the dipyrromethane cofactor with the C-11 position of PBG above the catalytic aspartate acid residue, which participates in acid-base catalysis (Fig. 8B). This promotes the deamination of the PBG to generate an azafulvene on C-11 (Fig. 8A), which is then able to react with the free alpha position of the cofactor. In doing so, the cofactor is extended to a tripyrrole, generating the ES1 substrate complex. This process is then repeated three more times, with incoming PBG substrates undergoing deamination and polymerization onto the free alpha position of the growing polypyrrole. These processes generate the ES2, ES3, and ES4 complexes, which represent tetra-, penta-, and hexapyrrole species at the active site of the enzyme. Once the ES4 complex has formed, the bond between ring A of the product and the dipyrromethane cofactor is hydrolyzed to generate the hydroxymethylbilane (Fig. 8A). It is likely that the same aspartate residue (D84 in the E. coli enzyme) is involved in the hydrolysis process, although there is also evidence to suggest that another conserved residue, D46, may also play a role in the disassembly of the complex (138).
Uroporphyrinogen synthase.
In contrast to the relatively stable and abundant HmbS enzyme, UroS has proven to be a difficult enzyme to study because it is very labile to proteolysis and temperature. UroS is a much more active enzyme than its predecessor in the pathway, and hence, the quantity of UroS within the cell is significantly smaller than that of HmbS. Moreover, the assay employed at that time for UroS was not straightforward, making the detection of the enzyme rather difficult. However, several things came together to help permit more detailed studies of this enzyme. The advent of recombinant-DNA technology allowed the enzyme to be produced in much larger quantities (139, 140), and the development of a fluorescence-based assay (141) coupled with more accurate high-performance liquid chromatography (HPLC) analysis to separate the type I and III isomers made the assays much more rapid and accurate (142). Consequently, after the initial laborious methods to isolate the first purified UroS enzyme from human blood (143), quite a few UroSs from a variety of organisms were subsequently purified as a consequence of recombinant-DNA approaches (144–147). UroSs are generally monomeric species with molecular masses of around 30 kDa.
UroS undertakes quite a remarkable reaction. Not only does it cyclize its bilane substrate, it also inverts the D ring to generate the type III isomer (Fig. 7). A simple cyclization alone would generate the type I isomer (Fig. 7). However, nature has opted for the type III isomer as the template upon which all the major biologically modified tetrapyrroles are based. Of the four possible isomers, the type III isomer is the only unsymmetrical variant, and it could be that nature has selected this lack of symmetry as a molecular handle or reference point for further modification of the molecule (148). However, the type III isomer is also the most abundant isomer formed when PBG is left under acidic (nonenzymatic) conditions, suggesting that this variant would have been more prevalent within a prebiotic soup and therefore more available for biological selection (149).
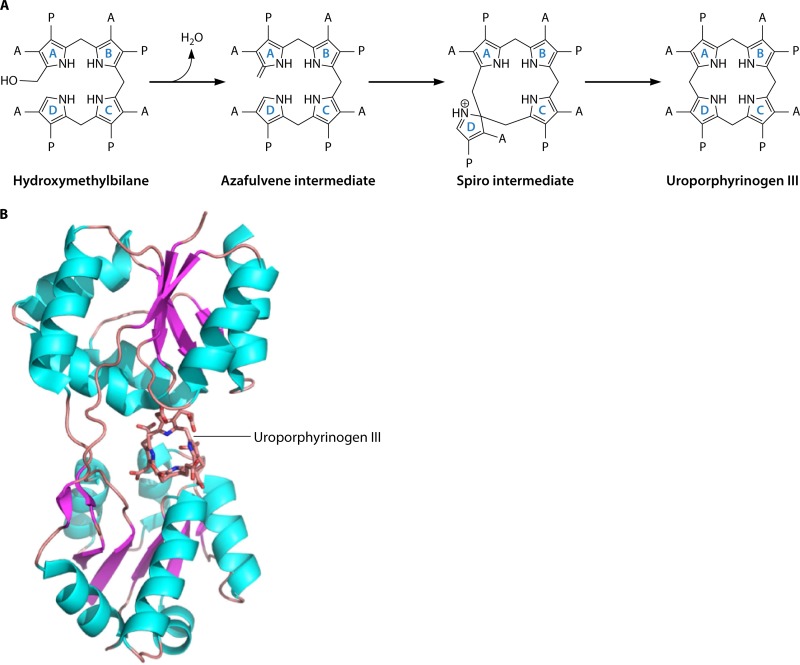
The type III isomer means that the order of the acetate and propionate side chains of ring D is inverted with respect to rings A, B, and C. This ability to invert ring D of the bilane and enforce ring closure obviously makes the reaction mechanistically interesting. The favored mechanism for UroS was proposed by Mathewson and Corwin over 50 years ago and involves a cyclic spiro intermediate (150). Overall, the reaction involves the loss of the hydroxyl group from the substrate HMB to allow it to form the spiro intermediate by generating a bond between the C-1 and C-16 positions of the substrate (Fig. 9A). This involves breaking the bond between the C-15 methylene and ring D to generate an azafulvene in ring C, which can then react with the free alpha position on ring D to generate the type III isomer of uroporphyrinogen. Evidence for this mechanism comes from studies using a spirolactam analog, which acts as a strong competitive inhibitor, suggesting that the molecule resembles the intermediate (151–153). The chemical synthesis of this spirolactam has actually generated two enantiomers, and X-ray analysis revealed that it is the R configuration that acts as the inhibitor (154, 155). Previous work with HMB analogs had shown that the enzyme inverts the terminal D ring (108–110). Thus, given a bilane with the D ring already inverted, the enzyme forms the type I isomer.
FIG 9.
Mechanism and structure of uroporphyrinogen III synthase (UroS). (A) Mechanism of action of UroS. The transformation of hydroxymethylbilane into uroporphyrinogen III is thought to proceed via a spiro intermediate, which is itself formed from an azafulvene intermediate generated from the loss of the hydroxyl group. Rearrangement of the spiro intermediate is then able to produce the type III isomer of uroporphyrinogen. The acetate and propionate side chains are designated A and P, respectively. (B) Structure of UroS. The structure of uroporphyrinogen III synthase from T. thermophilus, together with its product uroporphyrinogen III, is shown in cartoon format. The 252 amino acid residues of the enzyme are folded into two α/β-domains with a large active site formed at their juncture. There are a few highly conserved amino acid residues found in the active site, suggesting that the binding of the substrate in the correct orientation promotes the chemistry outlined in panel A.
Although uroS sequences from a broad range of sources have been determined, a comparison of the encoded proteins reveals only a relatively low level of conservation when their sequences are aligned (156). Attempts to identify key catalytic groups associated with the activity of the enzyme identified only a tyrosine residue whose mutagenesis reduced, but did not abolish, activity (157). All this suggested that there was no key amino acid residue involved in the reaction mechanism. Significant progress in the understanding of the function of UroS was next to come from the crystal structure of the human enzyme (158). The human enzyme was the first to have its structure solved, and this structure revealed that the protein is bilobed, composed of two domains that are separated by a two-stranded β-sheet. This arrangement generates a large active site that is easily able to accommodate the substrate (Fig. 9B). Again, mutagenesis of potential catalytic residues within the active site did not identify any essential amino acid. A follow-up structural study, this time with Thermus thermophilus UroS crystallized in the presence of uroporphyrinogen III, revealed that the protein is able to adopt a range of different conformations (159). Significantly, however, the product was observed to bind between the two domains and is held in place through an intricate series of hydrogen bonds (Fig. 9B). In this product complex, rings A and B are held more tightly than are rings C and D. In contrast, a density functional analysis of the reaction suggested that the D ring of HMB binds to the enzyme in a conformation that protects it from reacting with ring A and thereby prevents the formation of the type I isomer as a product (160). Interestingly, however, the crystal structure of the T. thermophilus enzyme with uroporphyrinogen III identified a conserved tyrosine residue that may help in the elimination of the hydroxyl group from HMB (159). A crystal structure of the Pseudomonas syringae enzyme has also been determined and shows a similar overall topology but a conformation different from those of both the human and T. thermophilus enzymes (161). Collectively, all this information confirms a mechanism involving a spiro intermediate, as outlined in Fig. 9A.
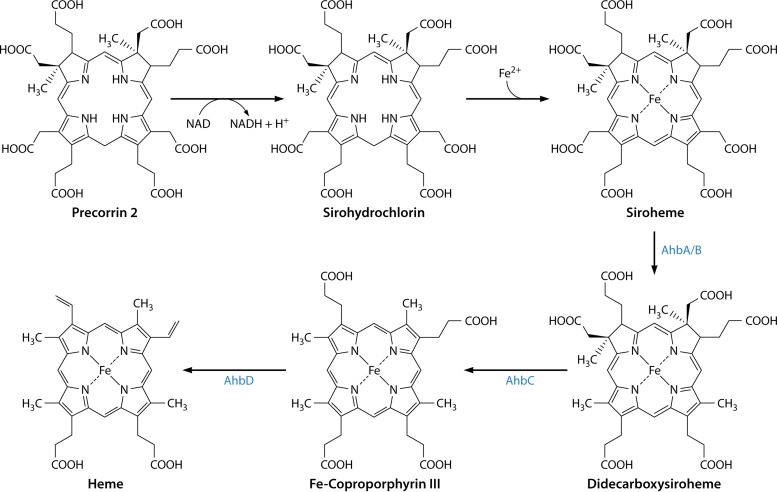
SIROHEME TO PROTOHEME/HEME d1 BRANCH
Discovery of a Novel Alternative Pathway
Serendipity often plays a role in scientific discovery, and so it was with the finding of an alternative route for the biosynthesis of heme. As part of a comprehensive study of Desulfovibrio vulgaris cytochrome c3, a small tetraheme protein that acts as an electron carrier for the hydrogenase, Akutsu and colleagues were trying to correlate a number of microscopic redox potentials with the hemes present within the protein (162). To achieve this, they labeled the protein with a number of deuterated amino acids, including phenylalanine, tyrosine, histidine, and methionine. To their surprise, they found that when the protein was isolated from a strain that had been fed methionine, deuterated in the methyl group, the deuterium was incorporated not only into the protein but also into two of the methyl groups of the hemes. More specifically, by NMR, those researchers observed that the hemes had a reduced signal from the methyl groups attached to C-2 and C-7 and concluded that these methyl groups likely had been derived from SAM rather from the decarboxylation of the acetic acid side chains of uroporphyrinogen III (162).
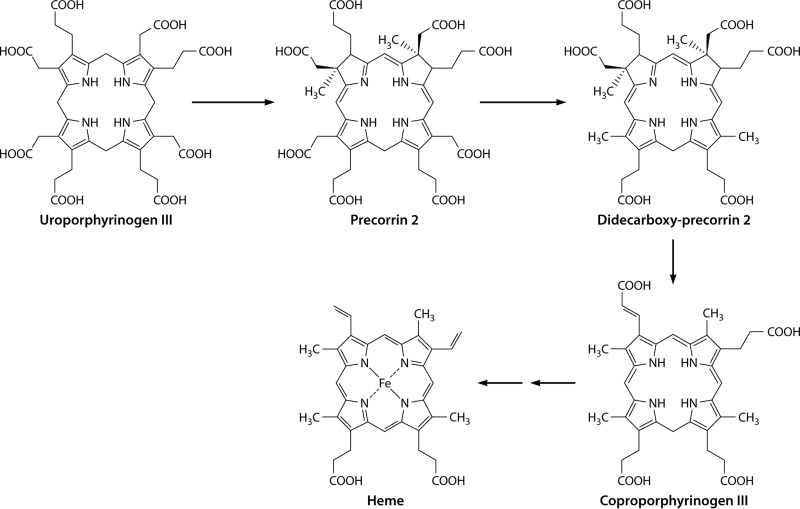
The classic heme biosynthetic pathway does not involve the methylation of the tetrapyrrole framework, as the four methyl groups within the final (heme) product result from the decarboxylation of the acetic acid side chains of uroporphyrinogen III. In contrast, the biosyntheses of siroheme (163, 164) and vitamin B12 (165–167) involve the SAM-dependent methylation of the tetrapyrrole framework at positions C-2 and C-7. This is achieved by the action of an enzyme called S-adenosyl-l-methionine uroporphyrinogen III methyltransferase (SUMT), which specifically methylates uroporphyrinogen III at positions C-2 and C-7 in a SAM-dependent fashion to generate precorrin-2, or dihydrosirohydrochlorin (166, 168) (Fig. 10). Precorrin-2 is a known intermediate in the biosynthesis of siroheme (163, 169, 170), cobalamin (165, 171), coenzyme F430 (172), and heme d1 (173, 174) (Fig. 10). As many of these prosthetic groups are made under anaerobic conditions and are thought to be more evolutionarily ancient than heme, it has been postulated that this alternative heme biosynthetic pathway may represent a “primitive” pathway for porphyrin metabolism.
FIG 10.
Transformation of uroporphyrinogen III (Uro'gen III) into precorrin-2, a key precursor in the biogenesis of heme d1, vitamin B12, coenzyme F430, siroheme, and heme. Precorrin-2 is synthesized from uroporphyrinogen III by the action of the enzyme uroporphyrinogen III methyltransferase, which adds two S-adenosylmethionine-derived methyl groups to C-2 and C-7 of the macrocycle.
This radically different idea broke all dogma concerning the biosynthesis of heme. It was thought at the time that heme was made via a specific set of pathway intermediates, and although there was some variation in the nature of the enzymes, anaerobic or aerobic, that catalyzed the synthesis of the intermediate, the intermediates remained constant between anaerobic and aerobic biosynthesis of heme.
Identification of New Intermediates?
Having provided evidence that heme in D. vulgaris was likely made from precorrin-2, Ishida and colleagues then attempted to dissect the pathway (46). Initially, that research group supplemented the growth medium of D. vulgaris cultures with 5-ALA and methionine-methyl-d3 and analyzed any deuterated compound-containing modified tetrapyrroles that they found within the strain. The porphinoid compounds were isolated from cells after lysis and esterification to allow their extraction into organic solvents in their methyl ester form. This approach resulted in the identification of deuterated coproporphyrin III, protoporphyrin IX, and a previously unknown hexacarboxylic acid. The latter was subsequently structurally identified as 12,18-didecarboxysirohydrochlorin (oxidized 12,18-didecarboxyprecorrin-2). From this, those authors predicted an alternative pathway for the biosynthesis of heme from uroporphyrinogen III (Fig. 11) (46). For this pathway, they made the assumption that the compounds that they had isolated from D. vulgaris were likely oxidized as a result of the extraction-and-esterification process. Thus, they predicted that uroporphyrinogen III undergoes bis-methylation by a SUMT-dependent uroporphyrinogen III methyltransferase at positions C-2 and C-7 to generate precorrin-2, followed by the decarboxylation of the acetic acid side chains attached to C-12 and C-18, to give 12,18-didecarboxy-precorrin-2, which then undergoes elimination of the acetic acid side chains attached to C-2 and C-7, yielding coproporphyrinogen III. It was suggested that coproporphyrinogen III is converted to heme via the more familiar intermediates of the classic pathway; i.e., coproporphyrinogen III is transformed into protoporphyrinogen IX, then protoporphyrin, and finally heme (46). However, aspects of this proposed pathway remained intellectually unsatisfying, as the route seemed rather laborious, effectively having three enzymes to cover the role played by the uroporphyrinogen decarboxylase within the classic pathway.
FIG 11.
Proposed alternative heme biosynthetic pathway. Initially, it was thought that the alternative heme pathway involved the decarboxylation of precorrin-2 to give a didecarboxy compound, which then underwent a loss of the acetic acid side chains on rings A and B to give coproporphyrinogen III. The latter could then be converted into heme via the classic pathway.
Further evidence for the novel transformation of uroporphyrinogen III into coproporphyrinogen III was provided by enzymatic assays using partially purified D. vulgaris cell lysates (46). Incubation of uroporphyrinogen III with SAM and a D. vulgaris DEAE low-salt elution fraction yielded precorrin-2, which was isolated and characterized as its oxidized octamethyl ester. Incubation of precorrin-2, generated as described above, with a DEAE-eluted medium-salt cell fraction then yielded a mixture of 12/18-monodecarboxysirohydrochlorin and 12,18-didecarboxysirohydrochlorin (46). Finally, when uroporphyrinogen III and SAM were incubated with the DEAE-eluted low-salt, medium-salt, and high-salt fractions, coproporphyrin III was identified as its tetramethyl ester (46). Although these enzyme activities were observed, the actual proteins were not purified to homogeneity and therefore were not formally identified or characterized. Similarly, as the products of the reactions were identified only after isolation as their oxidized esters, the exact nature of the enzymatic transformation was not determined with absolute certainty.
Siroheme as a Precursor to Heme d1 and Protoheme
With the discovery of this novel alternative pathway for the biosynthesis of heme, it was suggested that perhaps this represented a more widespread and general, but previously undetected, route for porphyrin formation. However, this idea was quickly dispelled when it was shown that no radioactivity from 14C-labeled methyl-l-methionine was incorporated into heme when added to cultures of Chlorobium vibrioforme (175). In a similar vein, heme was found to still be produced when a Salmonella enterica ΔcysG strain, a strain that is deficient in the ability to make precorrin-2, was grown anaerobically (175). CysG, in fact, is a multifunctional protein that not only houses the SAM-dependent methyltransferase activity required to make precorrin-2 but also contains precorrin-2 dehydrogenase and sirohydrochlorin ferrochelatase activities and is therefore able to transform uroporphyrinogen III into siroheme (163, 164, 169, 170).
The alternative heme pathway was therefore viewed very much as an evolutionary remnant from an anaerobic world, representing a primitive pathway that would likely be found in only a limited group of modern-day bacteria (46). In this respect, the alternative pathway was envisaged as an ancient route that had been superseded by the classic pathway, which became universally adopted in the eukaryotic world. However, increasing amounts of genome sequencing data started to reveal some interesting insights into heme biosynthesis. In particular, most archaeal genomes were found to be missing orthologs of hemE, hemF or hemN, hemG or hemY, and hemH even though these genomes also encoded a number of hemoproteins, highlighting the fact that they must be able to make heme through some alternative process (176). A similar observation was also made for the genomes of sulfate-reducing bacteria, including that of D. vulgaris (177). It was therefore proposed that heme-requiring archaea may make heme by the alternative heme pathway first put forward by Ishida and colleagues (176). This idea proved to be correct, as when non-covalently bound heme was extracted from cell lysates of Methanosarcina barkeri that had been grown on medium containing l-(methyl-d3)methionine and analyzed by matrix-assisted laser desorption ionization (MALDI) mass spectrometry, the data clearly showed that a significant proportion of the deuterated label had been incorporated into heme (176). This finding is consistent with the presence of the alternative heme biosynthetic pathway that had been observed by Ishida et al. (46).
Moreover, this result demonstrated that the alternative pathway was likely to be much more widespread than previously thought, representing the major pathway for heme synthesis within the archaeal kingdom of life (176). This prompted a more detailed bioinformatic approach in order to help identify the proteins and enzymes associated with the transformation of uroporphyrinogen III into heme within the Archaea. This analysis involved searching 59 completely sequenced archaeal genomes and looking for the clustering of genes associated with the known early genes of tetrapyrrole synthesis (hemA, hemL, hemB, hemC, and hemD) (178). Interestingly, out of the 59 genomes sequenced, 47 were found to have the early hemA, hemL, hemB, hemC, and hemD genes required for the transformation of glutamyl-tRNA into uroporphyrinogen III. These genomes were also found to encode both a SUMT enzyme as well as a precorrin-2 dehydrogenase, which together convert uroporphyrinogen III into sirohydrochlorin via precorrin-2 (178).
In 32 of the genomes studied, genes associated with the biosynthesis of heme d1 were also found (178). Heme d1 is a prosthetic group required by the cytochrome cd1 nitrite reductase, NirS, an enzyme that mediates the reduction of nitrite to nitric oxide (179). NirS houses both a c-type cytochrome as well as heme d1. Technically, heme d1 is not actually a heme in that the macrocycle is not a porphyrin but rather is a dioxoisobacteriochlorin. In this respect, the molecule is more similar to siroheme (also an isobacteriochlorin) but differs in that the two propionate side chains attached to C-3 and C-8 are replaced by oxygen atoms, generating carbonyls at these positions. The biosynthesis of heme d1 had not been elucidated, but gene clusters associated with the construction of the prosthetic group had been identified (180, 181). The genes involved in heme d1 synthesis were thought to include nirDL, nirE, nirF, nirG, nirH, nirJ, and nirN. Furthermore, labeling studies established that the biosynthesis of heme d1 proceeded via precorrin-2 (182), and consistent with this view, NirE was found to have a high degree of similarity to SUMTs. Indeed, the activity of NirE was confirmed by enzymological studies (173, 174), and a structure of the enzyme (178) revealed that it had an overall topology similar to that of the SUMT associated with cobalamin biosynthesis (168).
Within the archaeal genomes, however, homologs of nirD, nirH, and nirJ were found (178). Significantly, the presence of these genes did not coincide with the presence of NirS, the nitrite reductase, suggesting that these genes must have another function. Often, these nir genes were found clustered with the hem genes (178). All of these data were consistent with the nir-like genes being involved in the alternative heme biosynthetic pathway. Interestingly, the nir-like genes were initially given the prefix ahb (for archaeal heme biosynthesis), but subsequently, the acronym was changed to stand for alternative heme biosynthesis, as these same nir-like genes were also found in the sulfate-reducing bacteria and were also presumed to be associated with the alternative heme pathway. A consensus of the genome data suggested that the following genes were required for the transformation of uroporphyrinogen III into heme: the SUMT, precorrin-2 dehydrogenase, nirD (ahbA), nirH (ahbB), nirJ1 (ahbC), and nirJ2 (ahbD) genes (178). The next step was to try to assign a function to these gene products and thus to elucidate the pathway.
Color Changes Highlight the Pathway
The major breakthrough in our understanding of the alternative heme biosynthesis pathway and heme d1 synthesis came from experiments conducted with the nir cluster of genes associated with heme d1 construction (11). When E. coli extracts containing recombinant Paracoccus denitrificans NirDL, NirG, and NirH were incubated with sirohydrochlorin, a significant color change was observed. No such color change was observed when the purified proteins were incubated with sirohydrochlorin. The reaction product was extracted as its free acid, no esterification was involved, and the compound was analyzed initially by liquid chromatography-mass spectrometry (LCMS). What was remarkable was that the product had a mass consistent with that of didecarboxysiroheme rather than didecarboxysirohydrochlorin (11). What had happened was that adventitious iron, present in the crude cell lysate, was chelated nonenzymatically into sirohydrochlorin to give siroheme. It was siroheme that then acted as a substrate for NirDL, NirG, and NirH, which catalyzed the decarboxylation of the acetic acid side chains on C-12 and C-18 of siroheme to give 12,18-didecarboxysiroheme (Fig. 12). This was confirmed when siroheme was incubated with purified NirDL, NirG, and NirH and was converted into the didecarboxy form. Incubation of purified NirDL, NirG, and NirH with either sirohydrochlorin or precorrin-2 did not give a product. The structure of 12,18-didecarboxysiroheme was confirmed by NMR (11).
FIG 12.
The alternative heme biosynthetic pathway via siroheme as an intermediate. Precorrin-2 is converted into siroheme via sirohydrochlorin by utilizing the siroheme biosynthetic enzyme system. Siroheme then undergoes decarboxylation of the acetic acid side chains attached to C-12 and C-18 to generate didecarboxysiroheme. The loss of the acetic acid side chains attached to C-2 and C-7 is mediated by a radical SAM enzyme, AhbC, to give Fe-coproporphyrin (coproheme). The final step in biosynthesis is mediated by another radical SAM enzyme, AhbD, which promotes the loss of the carboxylic acid groups on the propionate side chains attached to C-3 and C-8 to generate heme.
NirDL, NirG, and NirH are proteins that share a degree of similarity with each other and also display sequence similarity with the Lrp/AsnC family of transcriptional regulators. NirD and NirL proteins are found as separate proteins in some organisms such as P. aeruginosa but are fused together in bacteria such as P. denitrificans. There is also similarity between NirDL, NirG, and NirH and the AhbA/B proteins found in the Archaea and sulfate-reducing bacteria. Indeed, incubation of siroheme with AhbA/B also resulted in the complete conversion of siroheme into didecarboxysiroheme (11). The relationship between the Nir proteins associated with heme d1 synthesis and the Ahb proteins of heme synthesis is shown in Fig. 12. The identification of siroheme as an intermediate for the biosynthesis of heme d1 and the alternative heme biosynthetic pathway made it immediately clear how the two pathways must work and explained the commonality of the genes required for their biosynthesis.
For heme d1 synthesis, siroheme is produced through a combination of a SUMT (NirE), a precorrin-2 dehydrogenase, and a sirohydrochlorin ferrochelatase (183). In P. denitrificans, this ferrochelation is mediated by a CbiX-like protein (12), which is a member of the type II chelatase family (184) and displays significant structural similarity to the cobaltochelatases (185, 186). Chromosomal inactivation of this cbiX gene in P. denitrificans prevents respiration on nitrate, thereby confirming the role of siroheme in heme d1 synthesis (12). Siroheme is next converted into 12,18-didecarboxysiroheme by a siroheme decarboxylase that is formed from NirDL, NirG, and NirH. NirDL also appears to perform decarboxylation by itself but is more efficient in the presence of NirG and NirH (11). The reaction is both oxygen and cofactor independent and is analogous to the decarboxylation reactions mediated by the uroporphyrinogen III decarboxylase UroD. Following the decarboxylation reaction, didecarboxysiroheme undergoes a loss of the propionate acid side chains in a reaction catalyzed by NirJ (11), which is a member of the radical SAM family (187). Such a reaction would generate a dihydro-heme d1 molecule, which then has to undergo dehydrogenation of the propionate acid side chain attached to C-17 to give the acrylate functionality and generate heme d1. In fact, although the NirJ reaction has not yet been demonstrated, the formation of the double bond on the C-17 propionate has been shown to be mediated by NirN within the periplasm (188, 189). This was demonstrated when a NirN knockout gave rise to a NirS enzyme containing dihydro-heme d1. Moreover, NirN was then shown to convert dihydro-heme d1 into heme d1, most likely by employing an electron bifurcation mechanism for this 2-electron oxidation, utilizing both the cytochrome c prosthetic group and the heme d1 product (188). The role of NirF in this biosynthetic process is still to be elucidated, but interestingly, it too is located within the periplasm (189, 190). How dihydro-heme d1 gets across the membrane is unknown.
The Siroheme Branch to Protoheme
The alternative heme biosynthesis pathway can also be explained in relatively simple terms with the knowledge that siroheme is a key primogenitor (Fig. 12). Archaeal genome data mining revealed that organisms that require heme also have the genes that encode the enzymes for siroheme synthesis. After siroheme is made, it is converted to 12,18-didecarboxysiroheme through the action of AhbA and AhbB, in a reaction identical to that mediated by NirDL, NirG, and NirH (11). The elimination of the two acetic acid residues attached to C-2 and C-7 of didecarboxysiroheme generates Fe-coproporphyrin (coproheme). The oxidative decarboxylation of the two propionates on C-3 and C-8 then yields heme. This series of reactions was proven to be correct when it was demonstrated that incubation of 12,18-didecarboxysiroheme with AhbC, a radical SAM enzyme, together with SAM was enough to transform the substrate into Fe-coproporphyrin III (11). Finally, AhbD, another radical SAM enzyme, was able to mediate the transformation of Fe-coproporphyrin III into heme in the presence of SAM by promoting the decarboxylation of the two propionate side chains to give vinyl groups (11). This final step is analogous to the reaction on the classic pathway catalyzed by CgdH, which is also a radical SAM enzyme.
The alternative heme biosynthesis pathway also explains the observations made by Ishida and colleagues when they tried to dissect the pathway (46). In essence, when they isolated intermediates from either whole cells or cell extracts, they then esterified the intermediates with methanol and sulfuric acid to generate the more solvent-soluble methyl ester derivatives. However, this process also removed the iron from the tetrapyrrole, and hence, when those researchers isolated the extracted compounds by chromatography and mass spectrometry, they observed only sirohydrochlorin, 12,18-didecarboxysirohydrochlorin, coproporphyrin III, and protoporphyrin IX rather than the Fe-containing intermediates.
The role of siroheme in the alternative heme pathway was key to its elucidation. Siroheme was not expected to act as an intermediate, as it was assumed, wrongly, that iron chelation would be the final step as it is in the classic pathway. The role of siroheme also explains why Ishida and colleagues were unsuccessful in the accurate description of the pathway. From this research, it was concluded that the alternative heme biosynthesis pathway progresses by using siroheme as a key biosynthetic intermediate, as is outlined in Fig. 12. There are many unanswered questions concerning the individual enzymes in the biosynthesis of both heme and heme d1. However, some insights into the decarboxylation of siroheme have been deduced from the structure determinations of AhbA/B of D. desulfuricans and Hydrogenobacter thermophilus NirDL (191, 192). In the latter case, unlike most other heme d1 synthesis systems, NirD/L works in the absence of a NirG or NirH. The structures of both enzymes reveal, as expected, similarity to the Lrp/AsnC family of transcription factors (191, 192). A large active site is formed between the two subunits, and crystallization with either a substrate analog (Fe-uroporphyrin III) (191) or product (12,18-didecarboxysiroheme) (192) has allowed models for the decarboxylation of the kinetically stable carboxyl groups on the substrate to be proposed. These mechanisms involve a number a conserved residues involving histidine, tyrosine, and arginine residues.
TWO BRANCHES TO SYNTHESIZE PROTOHEME
Uroporphyrinogen Decarboxylase
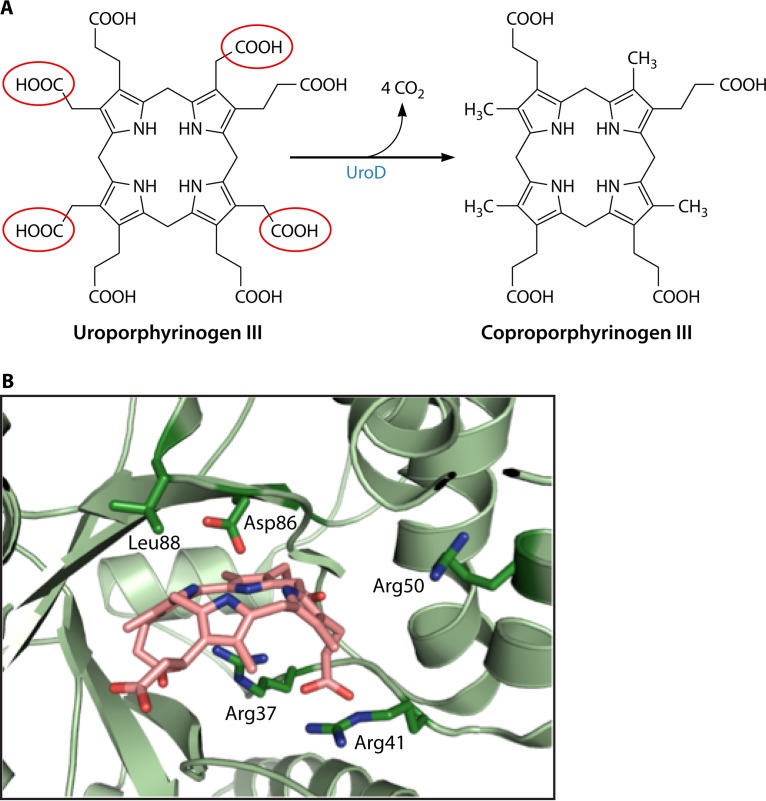
Protoheme is synthesized by archaea and denitrifying and sulfate-reducing bacteria via the siroheme-dependent pathway detailed above. For all other bacteria, a distinct protoheme synthesis branch exists. In all of these organisms, the first step of the pathway toward protoheme is the conversion of uroporphyrinogen III into coproporphyrinogen III. The enzyme responsible for this is named uroporphyrinogen III decarboxylase (UroD) (EC 4.1.1.37) (14, 193).
UroD catalyzes the stepwise decarboxylation of each of the four pyrrole ring acetic acid side chains, yielding four methyl groups at the C-2, C-7, C-12, and C-18 positions (also numbered ring side chains 1, 3, 5, and 8, respectively) (Fig. 13A). Early studies on the uroporphyrinogen decarboxylase enzyme mechanism focused on the order of decarboxylation and were conducted largely on the metazoan form of the enzyme. The rationale for this choice was the known linkage between the disease porphyria cutanea tarda (PCT) and diminished UROD activity levels (194). An excellent review of early chemically based studies by Jackson's group (195) is available, which outlines the experiments that helped to determine the stepwise decarboxylations that start with the D ring and proceed clockwise to the A, B, and, finally, C rings (196). UROD utilizes both the I and III isomers of uroporphyrinogen as well as all 14 possible intermediates between uroporphyrinogen and coproporphyrinogen. Interestingly, the enzyme catalyzes this reaction without any associated cofactor.
FIG 13.
Uroporphyrinogen decarboxylase (UroD). (A) Overall reaction of four sequential decarboxylations. The acetic acid side chains that are removed are highlighted in red circles. Details of the reaction are outlined in the text. (B) Active site of human UROD with a bound product (coproporphyrinogen III). While structures of bacterial UroDs are available, there are none with a bound substrate or product. However, given the homology between the bacterial and human enzymes, one would assume that the reaction mechanism is the same. The positions of the essential Asp residue and four Arg residues are shown. Numbering is according to that of the human enzyme.
To date, all major studies on the enzyme's mechanism have been carried out with the human enzyme. The enzyme from R. capsulatus was identified in 1995, but no biochemical studies have been reported (197). An X-ray crystallographic structure at 2.3 Å of B. subtilis UroD was reported in 2007 (198), but there have been no follow-up studies. The Protein Data Bank (PDB) contains a 1.95-Å structure (accession number 4WSH) of a “probable” UroD enzyme from P. aeruginosa as well as a 1.9-Å structure (PDB accession number 2EJA) from Aquifex aeolicus, a 2.8-Å structure (accession number 3CYV) from Shigella flexneri, and a 1.6-Å structure (accession number 4EXQ) from Burkholderia thailandensis. Of these structures deposited in the PDB, only the one reported under PDB accession number 4EXQ has an associated publication, which is the report of a structural genomic study of 406 putative essential genes of B. thailandensis (199). No biochemical studies were carried out on any of these UroD proteins.
The X-ray crystal structures reveal that UROD from human (200, 201) and Nicotiana tabacum (202) and the listed structures for bacterial UroDs are homodimers of ∼40 kDa per subunit, with the two active-site clefts being adjacent to the dimer interface and with no associated cofactor or prosthetic group. The latter observation validated the proposal that the enzyme is unique in that it catalyzes decarboxylations without any cofactor. Nevertheless, the bacterial UroD enzymes are structurally highly similar to the human and tobacco enzymes, even though there is only about 37% amino acid sequence identity. The main difference between the structures of the human and B. subtilis enzymes is a subtle movement of two small surface loops (L1 and L2). The spatial orientation of conserved active-site residues of the UroD enzymes is highly similar to that of the human structure, suggesting that the catalytic mechanisms of these proteins are identical.
A structure of human UROD with the product coproporphyrinogen III bound revealed that the substrate/product binds as a dome-shaped molecule with the four pyrrole NH groups facing inward within hydrogen-bonding distance of a conserved Asp residue (Fig. 13B). The tilt of the four pyrrole rings relative to the plane that passes through the four N atoms ranges from −12° for the C ring to 35°, 45°, and 58° for the B, D, and A rings, respectively. The active-site pocket is relatively hydrophobic but is ringed with charged residues that include the three conserved Arg residues, one conserved His residue, and one conserved Tyr residue (203). These polar residues have been suggested to interact with the propionate side chains of the tetrapyrrole to spatially orient the substrate, although it has been strongly suggested that one of the Arg residues participates in the catalytic cycle. One model proposes that the conserved Asp residue donates a proton to the substrate pyrrole ring adjacent to the acetate side chain (204). This results in decarboxylation, creating an intermediate and requiring that the conserved Arg residue contribute a proton to the methylene group as the ring is deprotonated by the Asp side chain. Regardless of the exact mechanism, it has been suggested that UROD is a “benchmark” for catalytic proficiency among enzymes without cofactors, with an estimated value for enzyme enhancement of the decarboxylation reactions of ∼1017 (204).
As mentioned above, UROD is a homodimer with juxtaposed and facing active sites. Given that four decarboxylations of a single uroporphyrinogen molecule are required for the production of coproporphyrinogen, there are at least three distinct models that may be considered. In one model, the homodimer shuttles, or channels, a single porphyrinogen back and forth between active sites without equilibration with a bulk solvent. The second model is a variation of the first model where transfer between active sites could occur, but there is not obligatory channeling and equilibration with bulk medium. In the third model, the substrate remains in a single active site and would rotate 90° following each decarboxylation until all four acetyl side chains are converted to methyl groups. To address this question, experiments in which the two subunits of the homodimeric UROD were tethered together were conducted (205). In this construct, one monomer's active site was left unaltered, and the second monomer's active site was mutated to result in a catalytically impaired monomer. This engineered enzyme was active, yielding the expected four decarboxylated products, demonstrating that shuttling between subunits is not necessary for activity. Taking advantage of the fact that UROD utilizes both the I and III isomers of uroporphyrinogen and that the I and III isomers and resulting decarboxylation products can be independently identified via HPLC, a series of experiments demonstrated that porphyrinogen release and equilibration with bulk medium occurred following each decarboxylation in the in vitro assay system. Thus, the experimental data are inconsistent with obligatory channeling and/or substrate rotation without intermediate release. However, given the reactive nature of the porphyrinogen substrates, one may anticipate that generalized release to and equilibration with bulk medium would be inefficient in vivo.
The Coproporphyrin-Dependent Branch
Among bacteria, with the exception of sulfate-reducing and denitrifying bacteria (11), protoheme synthesis necessarily involves the intermediate coproporphyrinogen III. This is true for all eukaryotes as well. It has long been known that in eukaryotes, the pathway from uroporphyrinogen III involves the decarboxylation of uroporphyrinogen III to coproporphyrinogen III, followed by the oxidative decarboxylation of coproporphyrinogen III to protoporphyrinogen IX, the oxidation of this to protoporphyrin IX, and, finally, the insertion of iron to create protoheme (32). This has frequently been referred to as the classic pathway, and until recently, it was believed to be universal in protoheme-synthesizing bacteria. This generic model was demonstrated to be incorrect with the discovery that Gram-positive bacteria utilize an alternate route, which can be viewed as an evolutionary transition between the siroheme-dependent pathway found first in archaea and described above and the classical pathway found in Gram-negative bacteria and eukaryotes (35, 43, 44). We refer to this pathway as the coproporphyrin-dependent (CPD) branch since it uniquely contains coproporphyrin III as an intermediate (Fig. 14). The coproporphyrin-dependent pathway is found in Gram-positive bacteria and a few bacteria that are transitional between Gram-positive and Gram-negative bacteria, such as Deinococcus (35, 44), but has not been found in archaea, eukaryotes, or Gram-negative bacteria. In the CPD pathway, coproporphyrinogen III is first oxidized to coproporphyrin III by coproporphyrinogen oxidase (CgoX), iron is inserted to make coproheme III by coproporphyrin ferrochelatase (CpfC), and, finally, coproheme is decarboxylated to protoheme by coproheme decarboxylase (ChdC). It is of note that neither the coproporphyrin-dependent nor the siroheme-dependent pathways have protoporphyrinogen or protoporphyrin as intermediates. The evolutionary significance of this is not immediately apparent, but since chlorophyll synthesis requires protoporphyrin, the advent of chlorophyll-based photosynthesis could not have occurred prior to the evolution of the protoporphyrin-dependent (PPD) pathway discussed later in this review.
FIG 14.
The coproporphyrin-dependent pathway to protoheme. The pathway from coproporphyrinogen III to protoheme is shown along with the structures of the intermediates and enzymes responsible for each reaction. Details for each individual step are outlined in the text and in figures below.
Porphyrin oxidation by coproporphyrinogen oxidase (CgoX).
The first committed step in the CPD pathway is the oxidation of coproporphyrinogen III to coproporphyrin III. The enzyme responsible for this reaction was named HemY when it was first cloned, expressed, and characterized in B. subtilis and was believed to be a protoporphyrinogen oxidase (206–208). Interestingly, those groups reported that B. subtilis HemY catalyzed the oxidation of coproporphyrinogen to coproporphyrin at a rate higher than that at which it oxidized protoporphyrinogen to protoporphyrin. With the recent elucidation of the CPD pathway and to prevent confusion with protoporphyrinogen oxidase (also previously named HemY), we now name this enzyme coproporphyrinogen oxidase (CgoX).
Most of what we currently know about this enzyme comes from studies on the B. subtilis and Staphylococcus aureus enzymes (43, 206–208). The reaction catalyzed is the six-electron oxidation of coproporphyrinogen III to coproporphyrin III. In the in vitro reaction, three molecules of molecular oxygen accept the six protons from the porphyrinogen, forming three molecules of H2O2 and the oxidized porphyrin (Fig. 15). This reaction converts the flexible, cyclic tetrapyrrole porphyrinogen into the fully conjugated, planar macrocyclic porphyrin. CgoX is a flavin adenine dinucleotide (FAD)-containing, soluble, monomeric protein with a molecular mass of ∼50 kDa. Since there is only a single FAD, it is clear that the full six-electron reaction must proceed via three two-electron steps, yielding the tetrahydro and dihydro intermediates, although this has yet to be demonstrated experimentally. S. aureus CgoX has been reported to utilize coproporphyrinogen III, but not protoporphyrinogen IX, as a substrate, with a Km of 0.3 μM and a kcat of 1.3 min−1 (43). CgoX of B. subtilis can oxidize both coproporphyrinogen III and protoporphyrinogen IX but at significantly different rates (206–208). This CgoX enzyme is reported to have a Km of 0.6 μM and a Vmax of 7.0 min−1 with coproporphyrinogen III, compared with a Km of 1.3 μM and a Vmax of 0.8 min−1 with protoporphyrinogen IX. As discussed below, the diphenyl ether herbicide acifluorfen, which effectively inhibits eukaryotic and Gram-negative protoporphyrinogen oxidases, is a poor inhibitor of CgoX, although other compounds in this class, such as oxyfluorfen, inhibit the enzyme at low-micromolar concentrations.
FIG 15.
(A) Conversion of porphyrinogen to porphyrin. The conversion of coproporphyrinogen to coproporphyrin is catalyzed by CgoX, while the conversion of protoporphyrinogen to protoporphyrin is catalyzed by the homologous protein PgoX. The only difference between the two reactions is that CgoX acts upon coproporphyrinogen, while PgoX acts upon protoporphyrinogen. For coproporphyrinogen, “R” groups on the diagram are propionates, while for protoporphyrinogen, R groups are vinyls. The reaction in vitro utilizes three molecules of molecular oxygen and produces three molecules of hydrogen peroxide. Below the structure drawings are three-dimensional representations of coproporphyrinogen and coproporphyrin. The A- and B-ring propionates of coproporphyrinogen III, which are vinyl groups in protoporphyrinogen IX, are outlined in red. The diagram illustrates the flexibility of the porphyrinogen allowed by the presence of the four saturated methyl mesobridges in the tetrapyrrole compared to the planarity of the aromatic porphyrin macrocycle. (B) Cartoon structure of the active site of B. subtilis CgoX (blue) on M. xanthus PgoX (yellow) (PDB accession numbers 3I6D and 2IVD, respectively) showing the close similarity of the structures of the two proteins. The overlaid FAD molecules are shown in a black stick representation, and the positions of the two inhibitor acifluorfen molecules are shown in turquoise. Previously, others presented the B. subtilis CgoX structure with a molecule of protoporphyrinogen modeled into the active site based upon the positions of the acifluorfen molecules (211). However, in the absence of clearly identified catalytic residues along with a lack of knowledge about potential molecular rearrangements that may occur upon substrate binding, we did not reproduce that model. (C) Cutaway view through the active site of B. subtilis CgoX. The position of the acifluorfen is shown as black sticks, and FAD is in yellow. This view illustrates the spacious active-site pocket that can easily accommodate the tetrapyrrole molecule. Both panels B and C show the close proximity of the flavin and inhibitor ring structures.
Two X-ray crystallographic structures for CgoX enzymes of Gram-positive bacteria have been deposited in the PDB. One of these, at 2.06 Å, for the Exiguobacterium sp. strain SP255-15 enzyme (PDB accession number 3LOV), was determined by the Joint Center for Structural Genomics effort and has no associated publication. The second structure, at 2.9 Å, is for the B. subtilis protein (PDB accession number 3I6D) (Fig. 15) and is associated with a previous report by Qin et al. (209) that contains discussion about the structure as well as some enzyme kinetics of the wild type and several variants. These two structures are highly similar to the reported structures of protoporphyrinogen oxidase of the Gram-negative bacterium Myxococcus xanthus (210) and the human (211) and tobacco (212) proteins. The root mean square deviation (RMSD) between C-α atoms of B. subtilis CgoX and the eukaryotic enzymes is ∼1.5 to 1.8 Å, which compares favorably with the RMSD between the two Gram-positive CgoX structures (1.7 Å).
There are two distinctions between the Gram-positive CgoXs and the protoporphyrinogen oxidases described below. The first of these distinctions is the size of the putative active-site pocket. In the CgoXs, this region is significantly larger than that found in protoporphyrinogen oxidase. B. subtilis CgoX has a pocket calculated to be 1,173 Å3, compared with 627 Å3 and 440 Å3 for the M. xanthus and human enzymes (209), respectively. The second difference is that the CgoX active-site pocket has more positively charged surface areas than those found in the protoporphyrinogen oxidase structures. Both of these observations are consistent with the binding by CgoX of coproporphyrinogen III as the substrate, which has larger and negatively charged propionates at the 3 and 8 positions on the A and B pyrrole rings compared with the smaller, uncharged vinyl groups present at these two positions in protoporphyrinogen.
Porphyrin metalation by coproporphyrin ferrochelatase (CpfC).
Following the oxidation of coproporphyrinogen to coproporphyrin, a metal chelatase, coproporphyrin ferrochelatase (CpfC), catalyzes the insertion of ferrous iron to form coproheme III. Metal chelatases identified to date fall mainly into two broad classes (213). One group, class I, is composed of ATP-dependent heteromeric complexes, and the second group, class II, is composed of ATP-independent monomeric or homodimeric proteins. Enzymes of class I include Mg chelatases for both chlorophyll and bacteriochlorophyll (BchD, BchH, and BchI), aerobic cobalamin biosynthetic cobalt chelatases (CobN), and probably an uncharacterized Ni chelatase for the synthesis of coenzyme F430. Members of class II are the sirohydrochlorin ferrochelatase (SirB), the anaerobic cobalamin biosynthetic cobalt chelatases (CbiK and CbiX), coproporphyrin III ferrochelatase (CpfC), and protoporphyrin IX ferrochelatase (PpfC). A third smaller class of chelatases, class III, is a group of enzymes possessing both dehydrogenase and iron chelation abilities. To date, class III chelatases are known only for siroheme biosynthesis (CysG and Met8p) (169). The class II chelatases, of which coproporphyrin and protoporphyrin ferrochelatases are members (Fig. 16), are generally deemed as being more simple enzymes given their size and lack of heteromeric tertiary structure (184, 214–217). However, even these simple proteins appear to utilize relatively complex methods to achieve their goal (217–221). From an evolutionary and gene size viewpoint, the most simple of the class II chelatases is CbiXs of Archaeoglobus fulgidus and other archaea (185). There is minimal amino acid sequence identity among all ferrochelatases, but there is clear structural homology among members of this group, and the few conserved residues are generally found in the active-site pocket.
FIG 16.
Iron insertion by ferrochelatase. Two ferrochelatases are described in the text. One, CpfC, catalyzes the insertion of ferrous iron into coproporphyrin III, and the other, PpfC, inserts iron into protoporphyrin IX. Both ferrochelatases are structurally homologous and probably possess the same catalytic features. (A) The PpfC reaction. This reaction differs from the CpfC reaction in the presence of the vinyl groups on the A and B rings of protoporphyrin, which are propionates for coproporphyrin for the CpfC reaction. (B) Cartoon structures of B. subtilis CpfC, in wheat color on the left (PDB accession number 1C1H), and human ferrochelatase (the best-characterized PpfC enzyme), in yellow on the right (PDB accession number 2QD1). The red arrows point to the position of the bound porphyrin in the active sites. It should be noted that no structures of a CpfC enzyme with the bound substrate porphyrin or a product are available in the PDB, so the structure with the tight-binding, competitive inhibitor N-methylmesoporphyrin is shown. PpfC enzymes possess a larger lip on the active site that closes over the “mouth” when porphyrin is bound. This enclosure of the active site precludes the binding of coproporphyrin with its propionate groups on the A and B rings. (C) Cartoon representation of the active site of human ferrochelatase illustrating the positions of conserved active-site residues in relation to the bound porphyrin substrate. Current models for enzyme function propose that the essential His residue is the acceptor for the two pyrrole nitrogen protons, Lys and Tyr help align the porphyrin macrocycle in the active site, and Met (Tyr in some CpfC enzymes) is the site of iron donation. Details and alternative models are presented in the text. (D) Ribbon cartoon representation demonstrating the three structural conformations of human ferrochelatase. The substrate-free resting state is wheat colored. Upon binding of the porphyrin substrate, the upper lip (shown in green) closes the active-site pocket. (E) Following metalation, the lower lip/π-helix extends (shown in red) to facilitate product release. Upon metalation, the 14-propionate (C ring) of heme flips conformation. This is accompanied by a reorientation of the essential His residue, and this movement is proposed to cause pocket opening and π-helix extension.
Coproporphyrin ferrochelatase is found only in bacteria that possess the CPD pathway (35). Archaea do not have either coproporphyrin or protoporphyrin ferrochelatases and form protoheme via a coproheme intermediate that is derived from the already metallated siroheme and not coproporphyrin (11, 42). Eukaryotes and Gram-negative bacteria all possess a protoporphyrin-specific ferrochelatase. The true identity of CpfC as a chelatase with specificity for coproporphyrin, rather than protoporphyrin, as is found for PpfC, was revealed with the discovery of the CPD pathway (35, 43). Both coproporphyrin and protoporphyrin ferrochelatases catalyze the insertion of divalent iron, cobalt, nickel, or zinc into coproporphyrin (43, 215).
The best-characterized CpfC enzyme is that from B. subtilis (222–227). In this organism, as with all firmicutes, the enzyme is a water-soluble, monomeric protein with a molecular mass of about 35 kDa and no bound cofactors or metals. CpfCs from actinobacteria have also been shown to preferentially utilize coproporphyrin II as the substrate (35). These enzymes are water soluble, monomeric proteins with molecular masses of 41 kDa and possess a [2Fe-2S] cluster (228). As with the well-characterized human ferrochelatase that has a [2Fe-2S] cluster, the clusters of the actinobacterial CpfCs coordinate the irons of the [2Fe-2S] cluster via one internal and three carboxyl-terminal cysteine residues (215, 228). At present, the role played by the [2Fe-2S] cluster has not been identified.
A number of X-ray crystallographic structures at about a 2-Å resolution have been determined for wild-type and variant forms of B. subtilis CpfC (222–227). Additionally, structures with the inhibitor N-methylmesoporphyrin (NMMP) and 2,4-disulfonic acid deuteroporphyrin IX (dSDP) (similar to coproporphyrin III but with sulfonate groups replacing the propionate groups of the A and B rings) have been determined. In neither of these structures is the porphyrin macrocycle in the same spatial position and orientation within the active-site pocket as those found with the substrate- and/or product-bound human ferrochelatase. Additionally, NMMP and dSDP do not bind in the same position or orientation (relative to each other) within the CpfC active site. Unfortunately, no structures of any bacterial CpfC enzyme with the substrate or product bound have been reported. Thus, at present, it is not possible to unequivocally determine a catalytically relevant binding orientation for coproporphyrin III within the CpfC active-site pocket or assign roles served by specific residue side chains.
The structures that are available reveal an active-site pocket of a sufficient size to engulf the four pyrrole rings of the porphyrin macrocycle (Fig. 16B). Within the pocket is an invariant and essential histidine residue found in all ferrochelatases that has been proposed to be involved in binding the substrate iron and/or serving as an acceptor for the pyrrole nitrogen protons that are removed to allow metal insertion (215, 218, 229). There is also a highly conserved π-helix with a patch of conserved acidic residues that has been shown for human ferrochelatase to be mobile and essential for the catalytic cycle (217, 220). Although the primary sequences of the human and B. subtilis enzymes are <15% identical, their structures are highly similar, with an RMSD of 2.4 Å for C-α atoms, and the active-site pockets contain most of the conserved residues. Given the structural and enzymological similarities, it is quite likely that both enzymes utilize the same catalytic mechanism. Why it is that the Gram-positive CpfCs utilize coproporphyrin as the substrate while protoporphyrin ferrochelatases do not is most likely attributable to the absence of a dozen residues in CpfCs that form a “lip” on one side of the active site on protoporphyrin ferrochelatases (35, 224). In the ferrochelatases that possess this lip, the lip closes down over the active-site pocket during the catalytic cycle (219, 220). In this closed position, there is insufficient space within the pocket for the 3,8-propionate side chains of the A and B rings of coproporphyrin (35). The coproporphyrin ferrochelatases lack this lip, so even with the expected molecular movement during catalysis, the pocket would remain open on one side to allow for the two propionate side chains of the A and B rings to project into the surrounding milieu (Fig. 16B).
Decarboxylation of coproheme III to protoheme.
The last step in the coproporphyrin-dependent pathway is the decarboxylation of coproheme III to protoheme. In the siroheme-dependent branch discussed above, the conversion of coproheme to protoheme was shown to be catalyzed anaerobically by the enzyme named AhbD (11). AhbD is a putative radical SAM enzyme that was initially reported to be found only in denitrifying and sulfate-reducing bacteria and in protoheme-synthesizing archaea (11, 42) but has recently been identified in Gram-positive bacteria that possess the CPD pathway. In CDP pathway-using organisms, coproheme is an intermediate product synthesized by CgoX/CpfC and used as a substrate by the enzyme coproheme decarboxylase (ChdC), which catalyzes the oxidative decarboxylation of the A- and B-ring propionates into vinyl groups to form protoheme (Fig. 17) (206–208). Thus, the complete coproporphyrin-dependent pathway in Gram-positive bacteria requires the action of CgoX plus CpfC plus ChdC. Protoporphyrin is not an intermediate as is found for heme-synthesizing Gram-negative bacteria and eukaryotes. This explains why the complementation of E. coli lacking either protoporphyrinogen oxidase or protoporphyrin ferrochelatase requires the presence of CgoX, CpfC, and ChdC acting in unison (35). Interestingly, the lack of protoporphyrin in all examined Gram-positive bacteria and the early observation that cell extracts of the Gram-positive bacterium Micrococcus lysodeikticus (31) could produce coproheme hinted long ago at what was recently demonstrated (35, 43, 44).
FIG 17.
Decarboxylation of coproheme III to form protoheme IX. (A) Reaction catalyzed by the terminal enzyme in the CPD pathway, ChdC. The two propionates that are converted to vinyl groups are circled in red. The reaction goes through a monovinyl monopropionate deuteroheme intermediate. (B) Structure of ChdC from Geobacillus stearothermophilus (PDB accession number 1T0T) with the putative active-site pocket shown in red. On the left is a space-filling model, and on the right is the same protein rotated 90° and shown in a ribbon format to illustrate the hole through the middle of the pentamer doughnut. (C) Structure of L. monocytogenes CpdC with bound coproheme (PDB accession number 5LOQ). The residues shown are the conserved His174 residue (which is ligated to the iron of the coproheme) as well as Arg133 and Lys151. The latter two residues are hydrogen bonded to the propionates of rings C and D (230). This pocket tightly binds heme, so there must be some structural difference for ChdC to allow the substrate to bind and the product to be released.
While they share the same substrate and product, ChdC has no similarity to AhbD. In fact, ChdC is a member of the chlorite dismutase family (Clds) and was originally annotated as a possible chlorite dismutase, even though most organisms possessing this gene lacked the ability to metabolize chlorite (19, 35, 44). Crystal structures are available for a number of chlorite dismutases and several ChdCs (PDB accession numbers 1T0T, 3DZT, 1VDH, 4WWS, and 5LOQ). Both enzymes are soluble homopentamers with subunit molecular masses of ∼30,000 Da, and both enzymes assemble into a structure that resembles a tall donut with heme-binding pockets located on the outside rim at one end (Fig. 17B). The crystal structures for all chlorite dismutases have bound protoheme, but only ChdC of Listeria monocytogenes has a bound heme, and this is the substrate coproheme (230) (Fig. 17C). ChdC is found in both heme-synthesizing firmicutes and actinobacteria. While the proteins are clearly similar and share conserved active-site residues, firmicute ChdC differs from actinobacterial ChdC in that it possesses an additional 8 to 12 residues that form a lip adjacent to the putative active site (35, 44). This feature, which has been suggested to be flexible and to possibly serve a gatekeeper function for the active site (231), is visible in only two subunits (subunits A and D) of the crystal structure of coproheme-bound L. monocytogenes ChdC. The fact that actinobacterial ChdCs do not possess this loop feature suggests that if it serves a function, it is not essential for enzyme activity. Interestingly, the active-site residues are relatively conserved between chlorite dismutase and ChdC, which has allowed models of heme binding within the ChdC active site to be proposed (231). The L. monocytogenes crystal structure clearly demonstrates the orientation of coproheme within the active site and reveals that coproheme is anchored by a conserved proximal histidine residue (230). Unfortunately, the heterogeneity in the orientations of the proposed essential side chains as well as the fact that few active-site residues are conserved between firmicute and actinobacterial ChdC enzymes leave a number of questions about catalysis unresolved.
The reaction occurs in a stepwise fashion, much like what is seen with UroD. The A-ring propionate is first decarboxylated, yielding the intermediate monovinyl monopropionate deuteroheme IX (231) (Fig. 17A). This is then decarboxylated in a reaction that is significantly slower than the first reaction to yield the final product protoheme. It has been shown that both the III and IV isomers of the monovinyl monopropionate deuteroheme can be decarboxylated, although the IV isomer (propionate on the A ring) reacts more rapidly, but even the “fast” reaction is relatively slow, with a half-life (t1/2) of 1 to 2 s. Lacking structural data for the monovinyl monopropionate intermediate-bound ChdC enzyme, it is not possible to say if the macrocycle rotates to move the B ring into the previous A-ring site, thus replacing the B ring with the C ring as it goes from the decarboxylation of the A ring to the B ring, or if the macrocycle is flipped, essentially replacing the A ring with the B ring and vice versa. The second model would be favored if the alignment of the substrate heme within the active site involves the propionates of the C and D rings, since inversion of the macrocycle of the coproheme III/protoheme IX isomers would leave the 13,17-position propionates of the C and D rings unaltered. If, however, the absolute position of the propionate being decarboxylated with respect to the overall macrocycle in the active site is the essential feature, then the rotation model would be favored. Also, the fate of the released carboxylate is yet to be demonstrated. As is the case with the CgdC reaction, the expectation is that this is released as CO2, but data conclusively showing this have yet to be presented. Possible reaction pathways have been proposed, and they all involve the participation of the heme iron (19, 43, 231). This is consistent with the observation that cobalt-coproporphyrin III is not decarboxylated (35, 44).
ChdC decarboxylation reactions require the presence of a proton acceptor. When a variety of possible common acceptors (e.g., NADP, NAD, FAD, and flavin mononucleotide [FMN]) were tried in in vitro assays, only FMN supported turnover and product formation (35, 44). Since the previous reaction of CgoX results in the in vitro production of three molecules of H2O2 for each coproporphyrinogen molecule oxidized, there is a possibility that these peroxides could serve as acceptors for the ChdC reaction. This proposition was additionally supported by the observation that in the presence of purified recombinant CgoX plus CpfC plus ChdC, coproporphyrinogen III plus ferrous iron are stoichiometrically converted into protoheme IX (35, 43). However, incubation of CpfC plus ChdC (no CgoX) with coproporphyrin III plus ferrous iron yielded only coproheme and not protoheme. Finally, it was shown that in the presence of H2O2, ChdC alone catalyzes the conversion of coproheme to protoheme. Interestingly the first decarboxylation occurs in the presence of one equivalent of H2O2, as anticipated, but the second decarboxylation is reported to require 5 to 10 equivalents of H2O2 in ChdC of S. aureus (43, 231) but only 1 equivalent for ChdC of L. monocytogenes (230).
Bioinformatic analysis revealed that some Gram-positive bacteria possess both chdC and ahbD (35). In fact, in Gram-positive bacteria, ahbD is found more frequently with chdC and cpfC than it is found with ahbABC, and some chdC- and cpfC-containing Gram-positive organisms possess only ahbD and not chdC.
The Protoporphyrin-Dependent Branch
The protoporphyrin-dependent branch is commonly called the classic pathway, since it was the first one to be discovered and was long believed to be the sole biosynthetic route to protoheme (Fig. 18). It is now recognized that while this is the single pathway utilized in eukaryotes, among prokaryotes, it is found only within Gram-negative bacteria. The first committed step, the conversion of coproporphyrinogen III into protoporphyrinogen IX, requires the oxidative decarboxylation of the propionate side chains on rings A and B of the substrate to the corresponding vinyl groups (232–234) (Fig. 19). Currently, two different, structurally and mechanistically unrelated enzymes are known to catalyze this reaction (234, 235). Coproporphyrinogen decarboxylase (CgdC) requires molecular oxygen for this reaction. Most eukaryotes and a small group of Gram-negative bacteria utilize this enzyme for aerobic heme and chlorophyll biosynthesis (236). The second enzyme, coproporphyrinogen dehydrogenase (CgdH), is oxygen independent and utilizes a radical SAM mechanism (14, 33). Both enzymes catalyze the decarboxylation of ring A prior to that of ring B with the formation of a monovinyl monopropionate deuteroporphyrin intermediate (237–239).
FIG 18.
Protoporphyrin-dependent pathway to protoheme IX synthesis. An overview of the three-step reaction and intermediates from coproporphyrinogen to protoheme, along with the enzymes responsible for the reaction, is shown. Coproporphyrinogen III is oxidatively decarboxylated at two propionate groups to yield protoporphyrinogen IX. This intermediate is oxidized to protoporphyrin, which is converted into heme upon iron insertion.
FIG 19.
Coproporphyrinogen III oxidase (CgdC). (A, left) Crystal structure of yeast CgdC (245). (Right) In the structure of human CgdC, a citrate molecule was observed to bind, indicating the localization of the active site. (Adapted from reference 14.) (B) Proposed reaction mechanism by which CgdC catalyzes the two oxidative decarboxylation reactions of coproporphyrinogen III to protoporphyrinogen IX with oxygen as the terminal electron acceptor (see the text for a detailed explanation). (Adapted from reference 246.)
Conversion of coproporphyrinogen III into protoporphyrinogen IX.
(i) Oxygen-dependent coproporphyrinogen III oxidase (CgdC).
In 1961, Sano and Granick reported the first partial purification and initial characterization of coproporphyrinogen decarboxylase (oxidase) from bovine liver (234). Characterizations of the enzymes from rat liver, yeast, and, later, various recombinant sources followed (232–234, 240–244). The usually dimeric protein has been proposed to initially catalyze base-mediated deprotonation of the pyrrole NH group, yielding an azacyclopentadienyl anion. The reaction of this anion with molecular oxygen at the α-position leads to the formation of a pyrrole peroxide anion. The exocyclic double bond is formed by proton abstraction at the β-position of the substrate propionate side chain by peroxide and the resulting six-membered-ring transition. The reaction concludes with the elimination of CO2 and H2O2 followed by bond rearrangements with the formation of the product vinyl group (160, 245–248). Currently, the structures of CgdC from yeast (PDB accession number 1TLB), human (accession number 2AEX), Leishmania major (accession number 3DWR), Leishmania donovani (accession number 3EJO), Leishmania naiffi (accession number 3E8J), and Acinetobacter baumannii (accession number 5EO6) have been solved, with all of them revealing an unprecedented fold for the monomer of large seven-stranded antiparallel β-sheets covered on both sides by α-helices (Fig. 19). The L. major structure was determined with the inhibitor 5-fluoroindole-2-carboxylic acid bound in the active site. A conserved aspartate residue in the active site was identified and suggested to be the initial base for catalysis. In addition, two conserved arginine residues were proposed to form hydrogen bonds with propionate side chains of the pyrrole rings so as to properly orient the substrate in the active-site pocket (249–251).
(ii) Oxygen-independent coproporphyrinogen III dehydrogenase (CgdH).
The first description of CgdH activity was given by Tait in 1969 and 1972 for a cell extract prepared from R. sphaeroides (252, 253). Around 10 years later, the stereospecific loss of the pro-S-hydrogen atom at the β-carbon of the substrate propionate side chain, identical to that shown for the CgdC reaction, was demonstrated (254). However, genetic approaches were necessary to identify the corresponding cgdH gene and allow recombinant production of the E. coli enzyme, its detailed biochemical characterization, and determination of its crystal structure (255–260) (PDB accession number 1OLT). This enzyme is a member of the radical SAM protein family and thus contains an oxygen-labile [4Fe-4S] cluster, ligated by 3 conserved cysteine residues of the CX3CX2C motif and a SAM molecule. Catalysis starts by the reduction of the iron sulfur cluster by an as-yet-unknown electron donor. Transfer of the electron from the iron sulfur cluster to the bound SAM leads to the homolytic cleavage of SAM into methionine and the formation of a 5′-deoxyadenosyl radical. Stereospecific abstraction of the hydrogen atom from the substrate propionate side chain by the radical yields 5′-deoxyadenosine and an allylic substrate radical. Transfer of the remaining electron to an unidentified electron acceptor with the elimination of CO2 concludes the reaction (255, 257, 258). Interestingly, the crystal structure of the E. coli enzyme revealed an N-terminal domain of a curved, 12-stranded, largely parallel β-sheet decorated by α-helices at its outer surface (Fig. 20). This (β/α)6 repeat is structurally similar to known TIM barrel domains. Two SAM molecules were found bound to the enzyme (256).
FIG 20.
Coproporphyrinogen III dehydrogenase (CgdH). (A, left) Crystal structure of E. coli CgdC. (Right) In the active site of the enzyme, the [4Fe-4S] cluster is coordinated by the three cysteine residues of the conserved CX3CX2C sequence motif and by one of the two bound SAM molecules. (Adapted from reference 14.) (B) Proposed reaction mechanism by which the radical SAM enzyme CgdH catalyzes two oxidative decarboxylation reactions of the coproporphyrinogen III propionate side chains on rings A and B to the corresponding vinyl groups of protoporphyrinogen without oxygen as the terminal electron acceptor (see the text for a detailed explanation).
Oxidation of protoporphyrinogen IX.
The penultimate step in the protoporphyrin-dependent branch is the oxidation of protoporphyrinogen IX to protoporphyrin IX. This is a six-electron oxidation, and as with the previous step, there are oxygen-dependent and oxygen-independent enzymes. The oxygen-dependent form is found in all heme-synthesizing eukaryotes but few Gram-negative bacteria (35, 38). The protein, named PgoX (previously HemY) for bacteria, is a FAD-containing homodimer and utilizes molecular oxygen as the terminal electron acceptor (261, 262). Bacterial PgoX is homologous to the eukaryotic oxygen-dependent protoporphyrinogen oxidase enzymes. This enzyme is part of a FAD-containing amine oxidase superfamily (262). It is of interest to note that there is as much sequence similarity between bacterial PgoX and other amine oxidases, such as monoamine oxidases or phytoene desaturases, as there is with eukaryotic protoporphyrinogen oxidase. As with AlaS and CgdH, PgoX is an example of the evolutionary diversity of function found in some protein families. There are structures available for protoporphyrinogen oxidases of tobacco (212) and human (211) and PgoX of the bacterium M. xanthus (210). All of these enzymes are membrane-associated homodimers with noncovalently bound FAD in each subunit and a subunit molecular mass of ∼50 kDa. The enzyme of the hyperthermophile A. aeolicus (263), however, has been reported to be membrane associated but monomeric. Unlike CgoX of Gram-positive bacteria, these enzymes use protoporphyrinogen IX, but not coproporphyrinogen III, as the substrate and are inhibited by the diphenyl ether herbicide acifluorfen at a 1 μM concentration (261, 263).
The reaction mechanism of this enzyme appears identical to that of eukaryotic Ppox and Gram-positive CgoX enzymes (Fig. 13). In vitro, the six-electron oxidation utilizes three O2 molecules and generates three H2O2 molecules. Given that the enzyme has only a single FAD, the reaction is assumed to proceed via three two-electron steps from the porphyrinogen to the tetrahydro and then dihydro intermediates and finally to fully oxidized porphyrin. The possibility that, in vivo, the enzyme may utilize an electron acceptor other than O2 has not been examined, although it is of note that all PgoX-possessing Gram-negative bacteria are aerobes. All mechanistic studies of this enzyme have been done with the eukaryotic enzyme, but it is likely that the reaction mechanism is conserved among all porphyrinogen oxidase-type enzymes. Unfortunately, there are no crystal structures with a bound substrate or product that would help to identify essential interactions between the tetrapyrrole and the enzyme. However, there are structures with a bound inhibitor, which allowed in silico modeling of substrate binding (210–212, 264). Based upon kinetic studies with a tritium-labeled substrate, it was proposed that three meso-carbon hydride ions are removed from the same surface of the tetrapyrrole in a sequential fashion with the concomitant removal of the NH proton (265). More recently, it was suggested that all hydride abstractions occur from the C-20 meso-carbon with total ring hydrogen rearrangements via enamine-imine tautomerizations (212). Whether the tetrapyrrole remains bound throughout the process or is released and rebound has not been determined. However, if all hydride abstractions occur from the single C-20 meso-carbon, it may be anticipated that there is not intermediate release and rebinding. Overall, the reaction is relatively slow, with a kcat of <10 s−1, although selected mutations to active-site residues of tobacco Ppox resulted in a 100-fold increase in the kcat, which led to the suggestion that the enzyme evolved to maximize substrate recognition and specificity but not catalytic turnover (264). Given that all heme synthesis enzymes exhibit similarly low turnover rates, there appears to be no need to maximize catalytic efficiency.
As mentioned above, PgoX is found in relatively few Gram-negative bacteria, and given its homology to Gram-positive CgoX, it is clearly derived from this family of proteins. Structural comparison of the eukaryotic and Gram-negative protoporphyrinogen oxidases (PgoX) with the Gram-positive coproporphyrinogen oxidase (CgoX) reveals a less constricted active-site pocket for the Gram-positive enzyme, which allows the binding of the larger and more charged coproporphyrinogen (209) (see above). Unfortunately, and unlike bacterial ferrochelatases, there are no characteristic, defining motifs that allow one to differentiate PgoX from CgoX by amino acid sequence alignments. However, realizing that PgoX is associated with PPD pathway-containing bacteria and CgoX is associated with CPD pathway-containing bacteria, it is possible to estimate that 65% of the members of this class of enzymes in bacteria whose genome sequences are currently known are CgoX enzymes, and only 35% are PgoX enzymes (Table 2). Overall, PgoX is found in only about 16% of heme-synthesizing Gram-negative bacteria.
TABLE 2.
Taxonomic distribution of enzymes specific for the siroheme-, coproporphyrin-, and protoporphyrin-dependent heme synthesis pathways
| Major taxon analyzed | No. of organisms capable of heme biosynthesis | No. of UroD+ genomes per taxon that utilize: |
No. of UroD+ genomes per taxon that utilize: |
|||||
|---|---|---|---|---|---|---|---|---|
| ChdC | AhbD | CgdH | CgdC | HemYa | PgdH2 | PgdH1 | ||
| Total | 645/651 | 153 | 61 | 390 | 387 | 231 | 326 | 99 |
| Actinobacteria | 81 | 83 | 16 | 2 | 79 | 1 | ||
| Firmicutes | 38 | 37 | 12 | 39 | ||||
| Chloroflexi (green nonsulfur bacteria) | 7 | 6 | 3 | 2 | 1 | 7 | 2 | |
| Deinococcus-Thermus | 9 | 9 | 2 | 2 | 1 | 9 | 1 | |
| Proteobacteria | ||||||||
| Alphaproteobacteria | 106 | 70 | 101 | 1 | 105 | 6 | ||
| Betaproteobacteria | 61 | 52 | 55 | 2 | 58 | 3 | ||
| Gammaproteobacteria | 163 | 2 | 3 | 136 | 144 | 11 | 70 | 82 |
| Epsilonproteobacteria | 27 | 27 | 25 | |||||
| Zetaproteobacteria | 1 | 1 | 1 | 1 | ||||
| Deltaproteobacteria | 19 | 13 | 4 | 4 | 17 | 1 | 2 | |
| Acidobacteria | 3 | 3 | 1 | 1 | 1 | 3 | ||
| Aquificae (class) | 9 | 2 | 7 | 6 | 3 | |||
| Armatimonadetes | 1 | |||||||
| Bacteroidetes-Chlorobi; Bacteroidetes | 52 | 49 | 44 | 12 | 44 | |||
| Bacteroidetes-Chlorobi; Chlorobi (green sulfur) | 9 | 9 | 9 | 1 | ||||
| Chlamydiae-Verrucomicrobia group | 10 | 2 | 7 | 6 | 10 | |||
| Cyanobacteria | 25 | 14 | 25 | 8 | 18 | |||
| Deferribacteres | 2 | 2 | 2 | |||||
| Gemmatimonadetes | 1 | 1 | 1 | 1 | ||||
| Nitrospira | 3 | 1 | 2 | 1 | 3 | |||
| Planctomycetes | 7 | 7 | 1 | 7 | ||||
| Spirochaetes | 5 | 5 | 1 | 4 | 1 | |||
| Synergistetes | 1 | |||||||
| Thermodesulfobacteria | 2 | |||||||
| Chrysiogenetes | 1 | 1 | 1 | |||||
| Caldithrix | 1 | 1 | 1 | |||||
| Thermobaculum | 1 | 1 | ||||||
“HemY” refers to the combined number of CgoX and PgoX enzymes.
Our knowledge of the oxygen-independent form of protoporphyrinogen oxidase comes from early work done by the Jacobs laboratory (266–272). Those researchers found that isolated membrane fractions of E. coli were capable of oxidizing protoporphyrinogen to protoporphyrin in the absence of oxygen so long as an electron acceptor that could be utilized by the organism was present. A mutant of E. coli that lacked the ability to oxidize protoporphyrinogen was isolated and characterized by Sasarman et al. (273), and the gene was named hemG (here renamed pgdH1). This gene clearly is involved in the activity documented by the Jacobs group and predated the identification and characterization of the oxygen-dependent PgoX enzyme described above. PgdH1 has been found mainly in gammaproteobacteria such as E. coli and PPD-possessing bacteria examined to date. From this analysis, it is clear that PgdH1 is found in only about 20% of Gram-negative bacteria.
PgdH1 is a protein with similarity to long-chain flavodoxins. It is distinct and unrelated to PgoX (274). By convention, PgdH1 is a member of what is referred to as the long-chain flavodoxin family. Flavodoxins are a structurally and biophysically well-characterized class of FMN-containing small-protein electron carriers (275, 276). Thus, PgdH1, like AlaS, CgdH, and PgoX, represents an instance of patchwork evolution. In comparison to characterized flavodoxins, PgdH1 contains a 7-amino-acid residue insertion between E. coli flavodoxin A helix 3 and sheet 4. There is minimal primary sequence identity between PgdH1 and other known flavodoxins, and the ability of this protein to oxidize protoporphyrinogen appears to reside in the so-called long-chain loop region. Transferring a 22-residue-long segment from this region of PgdH1 into E. coli flavodoxin A confers PgdH1 activity to the flavodoxin in vivo (274). PgdH1 has specificity for protoporphyrinogen IX and does not oxidize uro- or coproporphyrinogen. PgdH1 does not directly utilize molecular oxygen but has been shown to interact with the cell's respiratory chain, hence the name protoporphyrinogen dehydrogenase. This reaction not only catalyzes the formation of protoporphyrin but also may generate cellular ATP, as the six electrons generated in the reaction can be funneled down the respiratory chain (277). In vitro, this enzyme has been reported to utilize menadione, triphenyltetrazolium chloride, ubiquinone, and menaquinone as well as to interact via quinones with purified terminal oxidoreductases in the presence of their respective substrates, although in vivo studies on menaquinone-deficient E. coli mutants found no evidence for the utilization of ubiquinone by PgdH1 (269).
A third protoporphyrinogen oxidase/dehydrogenase, PgdH2, was identified and characterized in Synechocystis (278) and Acinetobacter (279). Genome-wide analyses revealed that all heme-synthesizing Gram-negative bacteria that lack pgdH1 and pgoX possess pgdH2 (38, 279). Thus, pgdH2 is the most commonly occurring form of protoporphyrinogen oxidase, occurring in almost two-thirds of heme-synthesizing Gram-negative bacteria. Unfortunately, relatively little is known about PgdH2. It is a membrane-bound protein but may require a soluble factor for activity (278, 279). This protein has not been purified to homogeneity, and there are no data to support the presence or absence of a cofactor. Based upon the variety of heme-synthesizing Gram-negative bacteria in which pgdH2 is found, it seems likely that it, like PgdH1, may interact with cellular respiratory chains and not directly with an electron acceptor such as O2. Interestingly, it was reported that pgdH2 does not complement an E. coli ΔpgdH1 mutant, suggesting that the mechanism of electron transfer from PgdH2 to the respiratory chain is distinct from that for PgdH1 (279).
Metalation of protoporphyrin IX by protoporphyrin ferrochelatase (PpfC).
The terminal step in the protoporphyrin-dependent branch is the insertion of ferrous iron into protoporphyrin IX. This is catalyzed by the enzyme protoporphyrin ferrochelatase (PpfC). Although initial studies of this enzyme were done with metazoan sources (see references 13 and 215), publications from as early as the 1960s described ferrochelatase activity in the denitrifying bacterium P. denitrificans and the facultative photosynthetic bacterium R. sphaeroides (280–283) as well as the yeasts Candida utilis and S. cerevisiae. Later, characterization of the enzyme from R. sphaeroides demonstrated that it was membrane associated and could utilize Co2+, Zn2+, Ni2+, Mn2+, and Fe2+, but not Fe3+, as metal substrates and meso-, deutero-, and protoporphyrin IX as the porphyrin substrates (284, 285). The fact that metal insertion was enzyme dependent rather than non-enzyme dependent, as was suggested in the early 1960s (286, 287), was conclusively demonstrated by the isolation and characterization of a heme auxotroph of Aquaspirillum itersonii that lacked ferrochelatase activity (288).
Diverse but limited biochemical studies have been performed on the Gram-negative protoporphyrin ferrochelatases from R. sphaeroides (289, 290), A. itersonii (291), B. japonicum (292), Caulobacter crescentus (293), M. xanthus, Pseudomonas putida, Bdellovibrio bacteriovorus (228), and A. aeolicus (263). In general, these enzymes all utilize Co2+, Zn2+, Ni2+, and Fe2+ as metal substrates and deutero-, meso-, hemato-, and protoporphyrin IX as the porphyrin substrates. While reported kinetic parameters vary due to differences in assay conditions utilized among research groups, the Kms for both the Fe2+ and protoporphyrin IX substrates are in the 1 to 10 μM range. Unfortunately, at present, there are no crystal structures for any Gram-negative bacterial PpfC enzyme.
Earlier reviews of ferrochelatase stated that with the exception of the enzyme in actinobacteria, most bacterial ferrochelatases do not possess a [2Fe-2S] cluster (215). This was based upon a relatively small sample size of sequenced bacterial genomes available at the time. Among these, a small number of ferrochelatases, as exemplified by those from M. xanthus, Bd. bacteriovorus, Azotobacter vinelandii, and P. syringae, have a [2Fe-2S] cluster that is coordinated by 4 cysteine residues that are located within a 20-residue internal segment (228). Interestingly, it appears from in silico modeling that the cluster of these Gram-negative PpfCs may occupy a spatially similar site as that found for the cluster of the human enzyme. The coordination by internal cysteines varies from what exists for actinobacterial, metazoan, as well as a limited number of Gram-negative bacterial (such as C. crescentus) ferrochelatases that have [2Fe-2S] clusters that are coordinated by 3 cysteine residues located within the carboxyl-terminal 30 residues and 1 internal cysteine residue. An examination of bacterial genomes available in 2016 reveals that the carboxyl-terminal or internal cluster-coordinating motifs are scattered and relatively rare among ferrochelatases of the Proteobacteria. However, based upon sequence alignment models, the internal coordination motif is found in ferrochelatases of the Bacteroidetes, Planctomycetes-Chlamydia-Verrucomicrobia, Cytophaga, and Flavobacteria. Additionally, there is anecdotal evidence suggesting that there may be a third cluster-coordinating motif among some proteobacterial ferrochelatases. Thus, rather than being a rare feature among bacteria, the [2Fe-2S] cluster may be more common than not. As mentioned above, the specific role played by the [2Fe-2S] cluster in the ferrochelatases that possess it is not currently known.
The mechanisms of action of all ferrochelatases would seem to be similar, although only the human enzyme has a model for catalysis that is supported by kinetic and structural data (217–221, 294) (Fig. 16). This model, which is based upon multiple structures of variant and wild-type enzymes with and without bound porphyrin or heme, projects that the enzyme active site exists in an “open” conformation to which the porphyrin molecule binds (Fig. 16C and D). Binding of this substrate triggers a rearrangement of a hydrogen bond network among conserved active-site residues (possibly due to an abstraction of one of the pyrrole hydrogens by the enzyme), with the result that the lips of the pocket close (“closed conformation”). This closure engulfs the macrocycle and causes an ∼15° distortion of the bound porphyrin. This distortion facilitates the chelation of the metal by the macrocycle to form the metallated heme with the simultaneous displacement of the second pyrrole hydrogen to the conserved histidine residue. The last act causes the movement of the imidazole side chain of the conserved histidine that results in the unwinding of the conserved π-helix, thereby reopening the active site with an extension of the π-helix (“release conformation”). This allows/promotes product release.
As detailed above, it appears that one significant difference between the coproporphyrin and protoporphyrin ferrochelatases is that one of the active-site lips that is present in the protoporphyrin ferrochelatases is truncated in the coproporphyrin ferrochelatases (Fig. 16B). The absence of the one lip would be anticipated to leave open the active-site pocket even in the closed conformation, thereby allowing the propionate side chains on the A and B rings of coproporphyrin to extend out. This would explain why protoporphyrin ferrochelatases do not utilize coproporphyrin as a substrate, but it does not explain why coproporphyrin ferrochelatases have such a poor affinity for protoporphyrin. Resolution of this issue will require additional crystal structures of ferrochelatases with bound coproporphyrin.
The source and supply of ferrous iron for the metalation of tetrapyrrole by ferrochelatase are frequently overlooked in the discussion of heme synthesis. This is true whether one is considering siroheme, coproheme, or protoheme synthesis. Microbial iron metabolism is an active field, including the acquisition and trafficking of free iron or heme-derived iron, yet few studies touch upon direct iron supply for ferrochelatases. Given that ferrochelatases utilize ferrous, and not ferric, iron, both the transport and reduction of iron for delivery to ferrochelatase must be considered. One of the few examinations of this topic is also the earliest study that experimentally linked iron reduction to heme synthesis in A. itersonii, R. sphaeroides, P. denitrificans, and E. coli (295). By using the ferrous iron chelator ferrozine as an iron trap, it was found that ferric iron reduction was linked to the cell's respiratory chain in the organisms examined but that there was not a stoichiometric link between iron reduction and heme synthesis (296, 297). The identity of the enzyme(s) specifically involved in ferric iron reduction for heme synthesis in bacteria has not been elucidated, although a variety of ferric iron reductases have been identified in metazoans.
Putative multienzyme complexes involving enzymes of heme synthesis in bacteria are also poorly documented. For eukaryotes, there is an ever-increasing amount of data to support the presence of multienzyme complexes for at least some heme synthesis enzymes (298–301), but for bacteria, the only data available are those supporting a complex involving PgoX and PpfC in the thermophilic cyanobacterium Thermosynechococcus elongatus (302).
DIVERSITY OF HEME SYNTHESIS PATHWAYS AMONG PROKARYOTES
With the recent identification of additional pathways to synthesize protoheme, along with an explosion in the numbers of microbial genomes sequenced, it is now possible to get a more comprehensive view of the diversity that exists among prokaryotes for the biosynthesis of heme. Among microorganisms, the capacity to synthesize uroporphyrinogen III from ALA is extremely common but not ubiquitous. Among the analyzed set of 982 representative organisms, 9 archaea (17% of 54 genomes studied) and 120 eubacteria (13%) lack recognizable homologs of any tetrapyrrole biosynthesis genes (see Tables S1 to S3 in the supplemental material) (see http://pubseed.theseed.org//SubsysEditor.cgi?page=ShowFunctionalRoles&subsystem=Heme_Biosynthesis%3A_protoporphyrin-%2C_coproporphyrin-_and_siroheme-dependent_pathways). The complete absence of a tetrapyrrole biosynthetic capacity (or incomplete pathways) appears to result from secondary gene losses. This is not uncommon in various bacterial pathogens and symbionts (e.g., in Mollicutes, Bacteroidetes, Bifidobacterium, or uncultured termite group 1 bacterium phylotype Rs-D17) for which heme or other essential tetrapyrrole compounds can be available from the host or other colonizing microorganisms in the same niche. Other organisms lacking a tetrapyrrole biosynthetic capability are domesticated microorganisms living in rich and stable environments such as milk (e.g., Lactobacillus, Leuconostoc, Lactococcus, and Streptococcus thermophilus), fruit juice (Oenococcus), or sausage (Weissella) and selected for their fermentation capacity when respiration, and, hence, the availability of heme, is nonessential. In free-living organisms, however, the complete absence of tetrapyrrole biosynthesis is extremely rare and occurs in only a few genera at the root of the eubacterial phylogeny (Dehalococcoides, Thermotoga, and Thermoanaerobacterales) and in several classes of Archaea (Table S3). The majority of these organisms are extreme thermophiles.
In sharp contrast to the nearly invariant tetrapyrrole pathway from ALA to URO III, the conversion of URO III to heme has long been known to differ significantly among species. This multiplicity was puzzling until recent studies revealed that there are multiple pathways of heme synthesis in prokaryotes. As detailed above, there are three distinct pathways used to synthesize protoheme from uroporphyrinogen. With this knowledge, a more informed analysis of the global phylogenetic distribution of the known enzymes of heme biosynthesis becomes possible. By identifying unique enzyme participants in each pathway, signature gene/protein patterns diagnostic for the presence of each of the 3 pathways are identifiable (see Tables S1 and S2 in the supplemental material). The selected proteins are (i) AhbA, AhbB, and AhbC for the siroheme-dependent pathway; (ii) the coproporphyrinogen decarboxylases CgdC and/or CgdH and the protoporphyrinogen oxidase PgdH1 or PgdH2 for the PPD pathway; and (iii) ChdC for the CPD pathway. It should be noted that while only ChdC can be used as a reliable diagnostic maker for the CPD route, some CPD pathway-possessing bacteria may utilize AhbD in place of ChdC. Of the 806 (82% of the total) representative prokaryotic genomes for which de novo URO III biosynthesis can be asserted, the majority (713; 88.5%) also encode one of the 3 known routes for the production of protoheme (Fig. 21), as deduced from the presence of the corresponding signature genes (Tables S1 and S2). Only 8.8% of these 806 organisms lack any known heme biosynthesis genes (71 out of 806), while a small fraction (22; 2.7%) contain unusual combinations of such genes. The PPD route seems to be the most abundant, occurring in over half of the analyzed genomes (471 out of 806; 58.4%), while the CPD and siroheme-dependent pathways are encoded in 166 (20.6%) and 76 (9.4%) genomes, respectively. Thus, the 3 routes accurately describe heme biosynthesis for the vast majority of the prokaryotic world with sequenced genomes available. However, the observed relative occurrence ratios for the 3 pathways across the Prokaryota likely reflect the current uneven sampling of various microbial taxa for genome sequencing. There are a few oddities, only 22 in the set of nearly a thousand, that do not clearly fit into the siroheme, CPD, or PPD pathways. In some cases, these deviations may be due to errors in sequencing, assembly, or annotation. However, if the same unusual pattern occurs in multiple genomes, it is likely a valid biological phenomenon and not an artifact.
FIG 21.
Phylogenic distribution of the three currently characterized pathways for protoheme synthesis mapped onto the Tree of Life. The outside ring shows (as vertical rectangles) the presence or absence of the siroheme-dependent branch (via Siro), the coproporphyrin-dependent branch (via Copro), or the classic protoporphyrin-dependent branch (via Proto). Gray rectangles mark the organisms that contain unusual combinations of genes normally involved in different pathways for protoheme synthesis (hybrid paths). The distribution of the two routes used to synthesize 5-aminolevulinic acid (ALA) are also presented: the Shemin or C4 pathway and the C5 pathway. The absence of a circle or square shows the inability of an organism to produce tetrapyrroles of any kind. Likewise, the absence of a rectangle in the outside ring indicates the absence of any known route for protoheme synthesis in an organism. This illustration covers only 133 representative organisms (17 archaea, 110 eubacteria, and 6 eukaryotes) included in the Tree of Life (391, 392). A full analysis of the 978 representative microorganisms performed in this work is available in the SEED subsystem “Heme Biosynthesis: Protoporphyrin-, Coproporphyrin-, and Siroheme-Dependent Pathways” (see http://pubseed.theseed.org//SubsysEditor.cgi?page=ShowFunctionalRoles&subsystem=Heme_Biosynthesis%3A_protoporphyrin-%2C_coproporphyrin-_and_siroheme-dependent_pathways). Mapped onto this tree, the siroheme-dependent pathway occurs largely in the Archaea, in the Thermodesulfobacteria, and, rarely, in several other taxa (see Table S3 in the supplemental material). The CPD route (teal) is found primarily in slow-evolving monoderm Firmicutes and Actinobacteria and in evolutionarily early-branching (Acidobacteria, Planctomyces, and Aquificae) and transitional (Deinococcus-Thermus group) diderm phyla, while the PPD pathway has a wider distribution, occurring in Proteobacteria, cyanobacteria, the Bacteroidetes-Chlorobi and Chlamydiae-Verrucomicrobia groups, Aquificae, Gemmatimonadetes, Caldithrix, and several other taxa. This is the main route for protoheme production in the evolutionarily younger Proteobacteria, with an illuminating exception of the Deltaproteobacteria. Deltaproteobacteria and epsilonproteobacteria are believed to be the oldest phyla within the Proteobacteria, with the other main proteobacterial groups being derived from them linearly in a directional rather than in a tree-like manner in the order delta/epsilonproteobacteria → alphaproteobacteria → betaproteobacteria → gammaproteobacteria (393). Notably, the phylum Deltaproteobacteria is the only proteobacterial phylum in which all 3 routes leading to protoheme are represented in different species (Table S3). A complete reconstruction of the heme biosynthetic pathways in the 38 representative deltaproteobacterial genomes is available online in the SEED subsystem (limit view to “deltaproteobacteria”). As a few examples, genomes of Stigmatella aurantiaca and Myxococcus xanthus encode the PPD pathway, and genomes of Geobacter metallireducens and Desulfuromonas acetoxidans harbor the CPD pathway, while genomes of Desulfovibrio vulgaris, Desulfatibacillum alkenivorans, and Desulfobacula toluolica harbor the siroheme-dependent route.
From an analysis of the bacterial genomes available in the SEED heme subsystem (Fig. 21) (see Tables S1 to S3 in the supplemental material), one finds that among representative examples, ∼82% of Actinobacteria possess the CPD pathway, 3% possess the siroheme-dependent (ahbABCD) pathway, and 13% lack a functional heme biosynthetic pathway (Table 2). Among the available genome sequences of representative members of the Firmicutes, 62% lack a heme biosynthetic pathway, 15% have the siroheme-dependent pathway, and 21% have the CPD pathway. Among members of the Proteobacteria that possess the ability to synthesize heme, the vast majority utilize the PPD pathway, with members of the Deltaproteobacteria being the outliers.
Organisms that possess multiple complete heme synthesis pathways are quite rare in the currently available genomes. Methylomirabilis oxyfera is an example of an organism with gene sets for both the siroheme-dependent and CPD pathways. There are organisms in the Rhodobacteraceae (deltaproteobacteria) that possess genes for both the CPD and PPD pathways. In these organisms, the CPD pathway utilizes ChdH, and not AhbD, to convert coproheme to protoheme, while in the PPD pathway, both CgdH and PpdH2 are present, suggesting that the PPD pathway may be favored by these organisms under low-oxygen conditions. It should be noted, however, that there are currently no biochemical data to support the presence of multiple, functional pathways in any bacterium.
While the present review focuses largely on bacterial systems, it is of value to observe what is known to exist in the Archaea. The CPD or PPD pathways have not been found in archaea. In an analysis of representative members of the Archaea, 12 of 17 Crenarchaeota possess ahbABCD, and only 8 of 35 Euryarchaeota have ahbABCD (Table 3). As noted above, some ChdC-like proteins are present in genomes, but none are associated with a complete set of heme-synthesizing enzymes (44). In addition, several of them are fusion proteins with other unidentified proteins whose function is probably not associated with heme synthesis.
TABLE 3.
Taxonomic distribution of three pathways used to synthesize protoheme
| Major taxon analyzed | No. of genomes analyzed (in the 981-taxon set) | No. of organisms per taxon encoding main pathway |
|||
|---|---|---|---|---|---|
| None | Siroheme dependent | Copro dependent | Proto dependent | ||
| Archaea | |||||
| Crenarchaeota | 17 | 3 | 12 | ||
| Euryarchaeota | 35 | 19 | 8 | ||
| Actinobacteria | 99 | 13 | 3 | 81 | |
| Firmicutes | 179 | 112 | 27 | 38 | |
| Chloroflexi (green nonsulfur bacteria) | 11 | 2 | 1 | 6 | 2 |
| Thermotogae | 10 | 10 | |||
| Deinococcus-Thermus | 10 | 8 | 1 | ||
| Proteobacteria | |||||
| Alphaproteobacteria | 109 | 4 | 106 | ||
| Betaproteobacteria | 62 | 1 | 61 | ||
| Gammaproteobacteria | 174 | 11 | 4 | 159 | |
| Epsilonproteobacteria | 27 | 27 | |||
| Zetaproteobacteria | 1 | 1 | |||
| Deltaproteobacteria | 38 | 19 | 13 | 5 | |
| Acidobacteria | 4 | 1 | 3 | ||
| Aquificae (class) | 9 | 2 | 7 | ||
| Armatimonadetes | 1 | ||||
| Bacteroidetes-Chlorobi; Bacteroidetes | 70 | 15 | 51 | ||
| Bacteroidetes-Chlorobi; Chlorobi (green sulfur) | 9 | 9 | |||
| Chlamydiae-Verrucomicrobia group | 12 | 1 | 9 | ||
| Cyanobacteria | 25 | 25 | |||
| Deferribacteres | 2 | 2 | |||
| Fusobacteria | 6 | 6 | |||
| Gemmatimonadetes | 1 | 1 | |||
| Nitrospira | 3 | 2 | 1 | ||
| Planctomycetes | 7 | 7 | |||
| Spirochaetes | 18 | 13 | 5 | ||
| Synergistetes | 9 | 7 | 2 | ||
| Thermodesulfobacteria | 2 | 2 | |||
| Chrysiogenetes | 1 | 1 | |||
| Tenericutes; Mollicutes | 17 | 17 | |||
| Caldithrix | 1 | 1 | |||
| Thermobaculum | 1 | ||||
REGULATION OF HEME BIOSYNTHESIS
There are considerable obstacles for any comprehensive review of the regulation of heme biosynthesis in bacteria. Foremost among these are the existence of at least three distinct pathways used to synthesize heme and wide metabolic diversity among bacteria. Compounding this is that historically, studies of pathway regulation have been based upon the assumption of a single classical pathway. Thus, data obtained in diverse bacterial systems have sometimes been interpreted via an inaccurate pathway model. Below, we present data that currently exist, realizing full well that it is an overly simplistic approach that will necessarily change as future studies focus on distinctions among the three protoheme-synthesizing systems.
The fact that prokaryotic heme synthesis is highly sensitive to environmental factors is obvious to anyone who has noted the color of cell pellets and/or colonies on a plate of facultative bacteria grown under conditions of high versus low aeration. Perhaps one of the more profound visual examples is that of B. japonicum grown in liquid culture, where modest levels of hemes are produced versus those found in leguminous nodules that are bright red from the accumulation of leghemoglobin. Early studies on porphyrin metabolism in microorganisms, as outlined previously (30), were limited to relatively few organisms, with a major focus on the facultative photosynthetic bacterium R. sphaeroides. This selection was based upon the fact that whereas other subject bacteria such as M. lysodeikticus and P. denitrificans modulate their porphyrin content about 10-fold, the tetrapyrrole content in R. sphaeroides can vary over 100-fold. Depending upon one's view, the selection of R. sphaeroides as a model organism was either fortuitous or unfortunate. This is because R. sphaeroides, as an organism that possesses AlaS (the C4, or Shemin, pathway) as the major rate-limiting step in tetrapyrrole synthesis, is not representative of the majority of bacteria and archaea that were later shown to utilize the C5 pathway to synthesize ALA. Also, as a model for the regulation of heme synthesis, R. sphaeroides introduces the confounding issue that it utilizes tetrapyrrole synthesis for both chlorophyll and heme. Following the discovery of the C5 pathway for ALA synthesis that employs the enzymes GtrR and GsaM, it was shown that the overproduction of GtrR in E. coli results in the accumulation of ALA and porphyrins (303). This suggested that GtrR serves a regulatory role in heme synthesis in organisms that possess the C5 pathway. Thus, whatever the organism, the clear consensus was that the biosynthesis of ALA, the first committed compound of the pathway, is the key rate-limiting step in tetrapyrrole synthesis.
Regulation of Heme Biosynthesis by Iron
Iron can be a limiting nutrient, and protoporphyrin and other heme precursors can be toxic. These facts make a strong prima facie argument for the regulation of heme biosynthesis by iron to prevent porphyrin synthesis from exceeding iron availability. Starting with Pappenheimer in 1947 (304), there have been numerous studies demonstrating the impact of iron limitation on heme synthesis. Interestingly, many of those studies reported that under iron limitation, coproporphyrin, and not protoporphyrin, accumulates in the growth medium (30). This is not surprising for the Actinobacteria and Firmicutes, which lack the ability to synthesize protoporphyrin (35). However, the Gram-negative bacterium R. sphaeroides also accumulates large amounts of coproporphyrin (30). It was found that additions of low levels of iron, considerably lower than what would be required to form a stoichiometric amount of heme from the excreted coproporphyrin, largely eliminated the accumulation of coproporphyrin (305). It was suggested that the heme formed from the added iron served to inhibit AlaS and reduce the overproduction of porphyrin (306). Why this was coproporphyrin rather than protoporphyrin in this organism has never been completely explained. The nonphotosynthetic Gram-negative bacteria E. coli and P. denitrificans were noted to accumulate coproporphyrin and protoporphyrin in amounts that are responsive to the nature of the available terminal electron acceptor, although in all cases, the amount of coproporphyrin found in the medium significantly exceeded the amount of protoporphyrin (267, 272).
Surprisingly, iron-based transcriptome and proteome analyses of numerous bacterial species failed to turn up heme biosynthesis genes and proteins (142, 307–315). In these cases, either gene expression is not regulated by iron or one or more other factors are limiting or in excess under the growth conditions used, which could mask the effects of iron. Iron-dependent control of heme biosynthesis genes has been described for the rhizobiales and R. capsulatus.
In B. japonicum, Rhizobium leguminosarum, and Brucella abortus, the iron response regulator (Irr) protein coordinates the heme biosynthetic pathway with iron availability to prevent the accumulation of toxic porphyrin precursors under iron limitation (316–318). A loss of function of the irr gene is sufficient to uncouple the pathway from iron-dependent control, as discerned by the accumulation of protoporphyrin under iron limitation (316, 317). In B. japonicum, Irr accumulates to a high level in iron-limited cells to negatively regulate heme biosynthesis at pbgS and alaS, the genes encoding the heme biosynthetic enzymes porphobilinogen synthase (PbgS) and ALA synthase, respectively (316, 319). Predicted Irr-binding sites are found upstream of the alaS gene in many species of the Rhizobiales (318), the taxonomic order that includes the rhizobia, Brucella, and Agrobacterium.
Irr does not sense iron directly; rather, it perceives and responds to an iron-dependent catalytic reaction, namely, the synthesis of heme. Protoporphyrin ferrochelatase catalyzes the insertion of iron into protoporphyrin to form heme in the final step of the heme biosynthetic pathway in Gram-negative bacteria. Irr interacts directly with ferrochelatase and responds to iron via the status of heme and protoporphyrin localized at the site of heme synthesis (320). Under iron-replete conditions, Irr forms a complex with ferrochelatase. Heme synthesized by ferrochelatase binds to specific sites on Irr, leading to the degradation of Irr (320, 321). The understanding of the mechanism by which heme triggers Irr degradation is incomplete, but evidence supports the conclusion that heme catalyzes Irr oxidation, leading to turnover (319, 321). When iron is limiting, ferrochelatase will be bound by protoporphyrin only, and it cannot form a complex with Irr, leaving Irr free to regulate genes under its control (Fig. 22). Thus, Irr senses heme directly through binding to heme and to protoporphyrin and iron indirectly through its interaction with ferrochelatase. This mechanism bypasses the need to invoke a free-heme pool, a concept not readily compatible with the cytotoxicity of heme, and it may represent the simplest type of heme signaling because there is no obvious need for a factor to chaperone heme from the site of synthesis to its target. Heme exerts an effect on the activation function of B. japonicum Irr in vivo in addition to its role in degradation, but it does not impair Irr occupancy of target genes; hence, DNA binding must remain intact (322). The Irr protein from R. leguminosarum binds heme, but the cellular Irr level is not diminished in response to iron (323). In vitro studies show that binding of heme to Irr inhibits its DNA-binding activity.
FIG 22.
Irr senses iron indirectly through the status of heme biosynthesis. Under high-iron conditions, Irr forms a complex with protoporphyrin ferrochelatase (FC), which inactivates Irr, followed by heme-dependent degradation. Heme is represented in red, and protoporphyrin is shown in blue. Under low-iron conditions, ferrochelatase is bound to protoporphyrin but not iron, and ferrochelatase cannot form a complex with Irr. In Rhizobium leguminosarum, Irr is not degraded in response to iron, but heme inhibits DNA binding.
Irr was initially identified in a genetic screen for mutants defective in the control of the heme biosynthetic pathway, but it is now known to be a global regulator of iron-responsive genes (318, 319, 324). This means that iron homeostasis and metabolism are regulated by an iron-intensive process rather than by iron directly. Transcriptome analyses show that, at least in B. japonicum, Irr has a large regulon and that most genes strongly controlled by iron at the mRNA level are also regulated by Irr (319). Irr is present and active under iron limitation, and genes under positive control are involved in iron acquisition and other functions that allow adaptation to this environment. Irr represses genes encoding proteins that function optimally under iron-replete conditions, including those that contain iron, synthesize heme and iron-sulfur clusters, and respond to high-iron stress (210, 274, 318, 324–332).
Global transcriptional control by iron is mediated by the Fur protein in E. coli and in many other bacteria, and Irr-mediated regulation appears to be confined to the Alphaproteobacteria (333). Fur binds iron directly, which confers DNA-binding activity on the protein. The discovery that Irr is a global iron regulator raises the question as to why the rhizobia and related alphaproteobacteria employ a complex iron-sensing mechanism when, from a naive perspective, a simple one should suffice. The rhizobia have coopted a Fur homolog to respond to manganese instead of iron, and it has been renamed Mur (manganese uptake regulator) (334–338). This is consistent with other findings suggesting that B. japonicum and its relatives are more reliant on manganese than are other bacteria (311, 336, 339, 340). Interestingly, B. japonicum Mur behaves as an iron-responsive regulator when expressed in E. coli cells (341), suggesting that the cellular environment confers metal specificity. Consistent with this, E. coli Fur responds to manganese and not iron in B. japonicum cells (341). Thus, adaptation to manganese likely results in a cellular milieu that prohibits Mur (or Fur) from responding to iron, thereby requiring a different sensing strategy, as observed for Irr.
B. japonicum cells grown under low-iron conditions respire and synthesize heme proteins, and therefore, heme biosynthesis genes cannot be completely repressed by Irr. However, the Irr-repressible gene encoding the iron exporter protein MbfA is needed only under high-iron stress and appears not to have a housekeeping role (330). This finding suggests that genes within the Irr regulon must be differentially regulated. This problem appears to be solved, at least in part, with promoters of Irr regulon genes with a wide range of affinities for the regulator (Fig. 23) (322). pbgS and other housekeeping genes have promoters with a weak affinity for Irr to ensure some level of expression even under the repressing conditions of iron limitation. In contrast, bacterioferritin and MbfA are important in managing high-iron stress (330, 342); hence, strong repression of genes that encode them is appropriate under conditions of low or moderate iron levels, with derepression occurring only under conditions of high iron levels. This control is achieved with bfr and mbfA gene promoters that bind Irr with high affinity (322).
FIG 23.
Differential control of Irr-repressed genes by the variable affinity of Irr for target promoters. Low-affinity promoters such as those found in the housekeeping gene leuC or pbgS allow some expression (derepression) even under low-iron conditions, where Irr levels are highest. High-affinity promoters such as those found for mbfA and bfr repress expression strongly at low and intermediate levels of iron and are derepressed only at high iron levels.
Regulation of Heme Biosynthesis by Heme
Biosynthetic pathways are often regulated by their end product, and heme controls its own synthesis at the levels of transcription, protein stability, and enzyme activity. However, there does not appear to be a universal model or mechanism that is widely distributed throughout the bacterial kingdom. Interestingly, most sites of regulation characterized to date occur with early pathway enzymes.
B. japonicum Irr responds to heme as described above to mediate iron control of heme biosynthesis. Expression levels of pbgS and other genes normally repressed by Irr are constitutively low in a heme biosynthesis mutant because Irr is not degraded (319, 322). Thus, heme does not serve as a feedback mechanism to downregulate its own synthesis. Rather, Irr responds indirectly to the substrates of heme synthesis, protoporphyrin and iron, by interacting with ferrochelatase. Thus, it responds to the status of the synthesis of heme and not merely the product.
The gtrR gene of the firmicute Corynebacterium diphtheriae is negatively regulated by heme through the actions of the two-component regulatory systems ChrA-ChrS and HrrA-HrrS (343). These regulatory systems were originally discovered in studies to elucidate the mechanism of positive control of the heme oxygenase (hmuO) gene by heme and hemoglobin (344, 345). Heme oxygenase catalyzes the degradation of heme for use as an iron source, and the positive control of hmuO by heme makes physiological sense. ChrA is a response regulator, and ChrS is the sensor histidine kinase that responds to the heme status. Mutations in genes encoding these proteins diminish but do not abolish activity, which led to the search for and discovery of HrrA and HrrS. A loss of function in both systems abolishes the control of hmuO and gtrR. In the related organism Corynebacterium glutamicum, HrrA/HrrS controls numerous steps in heme biosynthesis as well as heme acquisition from the environment (346). Collectively, the findings show that the biosynthesis of heme and its subsequent metabolism are coordinately controlled in Corynebacterium species.
The expression levels of several heme biosynthesis genes in R. capsulatus that are upregulated by the transcriptional regulator are downregulated by heme (347). Moreover, HbrL binds heme directly (347), suggesting that heme attenuates the activation function of HbrL. Heme has also been shown to directly inhibit the activity of R. sphaeroides AlaS in vivo (306).
The glutamyl-tRNA reductase (GtrR) protein of Salmonella enterica serovar Typhimurium is destabilized by heme, resulting in a 25-fold increase in protein levels in heme-starved cells, with only a 2-fold change in gtrR promoter activity (348, 349). Heme-dependent degradation of GtrR is lost in mutants defective in genes encoding the Lon and ClpAP proteases. Cysteine 170 of GtrR was shown to be necessary for heme binding, and a C170A mutant protein is constitutively expressed in vivo (350).
Heme inhibits GtrR activity in cell extracts from several bacterial species by an allosteric feedback mechanism. Recombinant GtrR from Chlorobium vibrioforme is a homodimer purified as a recombinant protein from E. coli. It contains one tightly bound heme molecule per monomer, and heme inhibits activity in vitro (351). GtrR levels are diminished by ALA supplementation in Acidithiobacillus ferrooxidans (352). Synechocystis sp. strain PCC6803 with diminished ferrochelatase activity accumulates protoporphyrin and has elevated ALA synthesis activity (353). This mutant grows normally and synthesizes phycobilins, which are derived from heme. It is likely that the diminution in heme synthesis in the mutant is not severe enough to limit heme for most cell functions, but it is below a level needed to allosterically inhibit ALA synthesis, presumably at GtrR (353).
ALA synthesis by the C5 pathway is initiated by the charging of tRNAGlu with glutamate by glutamyl-tRNA synthetase (GluRS), which is not a committed step in tetrapyrrole formation because it also is used in protein synthesis. Therefore, it is surprising that GluRS is regulated by heme status in A. ferrooxidans, an organism that can accumulate 9-fold more heme when grown with ferrous iron as an electron donor than can E. coli cells (352). In this organism, heme affects both the protein levels and enzyme activity of GluRS. Interestingly, and perhaps coincidentally, heme also binds to human arginyl-tRNA synthetase (ArgRS), even though this enzyme is not involved in heme biosynthesis (209). ArgRS is the source of arginine in arginyltransferase activity for targeting proteins for degradation via the N-end rule of protein degradation. Heme promotes ArgRS oligomerization, which may contribute to the inhibition of enzyme activity.
Regulation of Heme Biosynthesis by Oxygen
AlaS of R. sphaeroides was shown to be sensitive to negative-feedback inhibition by micromolar concentrations of heme (306), and its activity in whole cells was sensitive to oxygen tension (354). The second observation was expanded by Neuberger's group in a series of papers (355–357). That group demonstrated that AlaS from cell extracts could be resolved into two fractions, high-activity and low-activity enzyme fractions. The ratio of these two forms was oxygen sensitive, with a preponderance of the low-activity form occurring in cells that had been grown under high aeration and the high-activity one being the major form in cells grown under low aeration. It was further demonstrated that in vitro, the low-activity form is converted to the high-activity form by cystine trisulfide or the mixed trisulfide of cystine and glutathione. The model proposed for this was that the trisulfide compound reacted with the enzyme to produce a protein-bound persulfide or trisulfide, which somehow activated the enzyme. The modulation of this activation is attributable to the regulation (decrease) of the trisulfide concentration by (increased) oxygen tension. Unfortunately, in the ensuing 4 decades, no additional studies have further tested this model by identifying the site and nature of the AlaS posttranslational modification (PTM) or to determine how the trisulfide level is regulated.
In animals and fungi, hypoxia or anaerobiosis is associated with fermentative metabolism, since oxidative phosphorylation is strictly dependent on O2. Thus, oxygen and heme are positively correlated. Furthermore, three heme biosynthetic enzymes, coproporphyrinogen III decarboxylase (CgdC), coproporphyrinogen oxidase (CgoX), and protoporphyrinogen IX oxidase (PgoX), use O2 as a substrate. In fact, O2 control of genes involved in aerobic metabolism occurs indirectly through heme in S. cerevisiae (358). For many prokaryotes, however, this correlation is not observed, because they can use electron acceptors other than O2 to support oxidative respiration. In addition, some bacteria have cytochrome oxidases with a very high affinity for O2, conferring to them the ability to grow under oxygen limitation. Thus, hypoxia or anaerobiosis does not necessarily trigger a switch to fermentative metabolism and may actually require an increase in tetrapyrrole synthesis. Moreover, most heme-synthesizing Gram-negative bacteria have an O2-independent coproporphyrinogen III dehydrogenase (CgdH). Finally, low O2 levels are a cue to induce photosynthesis-related genes in bacteria, which requires enzymes common to heme and chlorophyll synthesis. Photosynthesis requires steps in tetrapyrrole synthesis for chlorophyll synthesis and also for electron transfer reactions.
The regulation of tetrapyrrole synthesis genes by oxygen in R. sphaeroides and R. capsulatus is under the control of numerous regulatory circuits for photosynthetic and anaerobic metabolism (40). The fnrL locus was identified in a mutant screen for elevated alaS gene expression levels under aerobic conditions and is required for photosynthetic growth and anaerobic growth with dimethyl sulfoxide (359, 360). FnrL binds to both promoters of alaS for the anaerobic induction of this gene (361). Moreover, FnrL affects numerous other genes in heme biosynthesis in both R. sphaeroides and R. capsulatus (216, 362). In addition to FnrL, the two-component response regulator PrrA (RegA) has been shown to control heme synthesis gene expression in Rhodobacter (216, 362, 363). PrrA/RegA responds to the cellular redox state to control anaerobic metabolism in numerous bacterial species. PrrA binds to both promoters of R. sphaeroides alaS under aerobic conditions, but anaerobic activation requires binding to the P2 promoter (363). PpsR/CrtJ is a repressor of photosystem development under aerobic conditions, and this transcriptional regulator also controls numerous heme synthesis genes (362, 364).
R. sphaeroides contains two putative hemN (cgdH) gene homologs encoding O2-independent coproporphyrinogen dehydrogenases, designated cgdH1 (hemN1) and cgdH2 (hemN2 or hemZ) (359). hemN1 is negatively regulated by the outer membrane protein TspO, apparently by a posttranscriptional control mechanism (365). TspO also negatively regulates carotenoid and bacteriochlorophyll production aerobically, and so cgdH1 may be primarily involved in chlorophyll synthesis.
Rhizobia can establish symbiosis with leguminous plants, which culminates with the development of a specialized plant organ called a root nodule. The bacteria convert atmospheric nitrogen to ammonia to fulfill the nutritional nitrogen requirement of the plant host. The oxygen tension within a nodule is very low, and this serves as a signal for the induction of genes necessary for nitrogen fixation and microaerobic metabolism. De novo heme biosynthesis for the synthesis of cytochrome cbb3 oxidase and other cytochromes is required for microaerobic metabolism, hence the need to control heme biosynthesis. The RegRS system in B. japonicum is homologous to the RegAB/PrrAB proteins in Rhodobacter. Although heme synthesis is regulated by O2 in B. japonicum and is phylogenetically related to Rhodobacter, the RegR regulon does not appear to include heme synthesis genes, as judged from transcriptional profiling data (366). Rather, O2 control of heme synthesis and cytochrome production occurs by a regulatory cascade initiated by another two-component regulatory system, FixLJ. FixL senses O2 directly by binding directly to the protein via a heme moiety (367). Deoxygenated FixL phosphorylates FixJ to activate target genes, including fixK2, encoding a Crp/Fnr family regulator. FixK2 positively controls alaS, pbgS, and cgdH and is also needed to downregulate cytochrome aa3 levels, which are not needed in microaerobic nodules (366, 368–371). B. japonicum also has two putative hemN (cgdH) genes, designated hemN1 and hemN2 (369). Neither gene is expressed aerobically, but both genes are strongly induced under microaerobic or anaerobic conditions. A hemN1 mutant does not have a discernible phenotype, but a hemN2 mutant is unable to grow anaerobically on nitrate and does not elicit functional nodules on its plant host (369). The presence of multiple hemN homologs and control by Fnr-like proteins are not confined to the alphaproteobacteria Rhodobacter and Rhizobium. A hemN gene of P. aeruginosa is under the control of two Fnr-type regulators, Dnr and Anr, both of which are needed for anaerobic induction (372). The hemN gene in this organism is also expressed aerobically, which is dependent only on Anr. Surprisingly, the cgoX gene encoding the O2-dependent oxidase is also strongly induced by anaerobiosis in an Anr- and Dnr-dependent manner. Unfortunately, hemN has frequently been inappropriately assigned the annotation of being an “oxygen-independent coproporphyrinogen oxidase” without appropriate biochemical support (35). Thus, many proposed HemNs are, in fact, not bona fide CgdH enzymes. Some annotated HemN proteins are known to be HemW, a heme chaperone (reviewed in reference 35), and ChuW, an oxygen-independent heme-degrading enzyme (373). This makes the interpretation of reported putative heme regulation data even more challenging.
Regulation of Heme Biosynthesis by Reactive Oxygen Species
The generation of reactive oxygen species (ROS) is a consequence of aerobic respiration as a result of the partial reduction of molecular oxygen and the subsequent reactions of these products with transition metals and other compounds (374). The environment can also be a source of ROS given the permeability of bacterial membranes to hydrogen peroxide (375). Superoxide (O2−) and hydrogen peroxide (H2O2), the initial partial reduction products of oxygen, can damage cell components but to only a limited extent. However, O2− is capable of destroying exposed iron-sulfur clusters with the release of free iron, and the released iron subsequently reacts with H2O2 via the Fenton reaction, generating extremely reactive hydroxyl radicals (HO˙). Hydroxyl radicals can directly attack most macromolecules such as DNA, lipids, and proteins, which is the basis of oxygen toxicity (reviewed in reference 376).
Bacteria have multiple defense strategies against oxidative stress, including the direct detoxification of ROS by catalase, peroxidases, and superoxide dismutase. Catalase and peroxidases are heme proteins, and their synthesis must be maintained or perhaps elevated under oxidative stress. Oxidative stress responses require the activation of regulatory proteins and the induction of genes under their control. In many bacteria, the transcriptional regulator OxyR senses H2O2 through disulfide bond formation that locks the protein in an active conformation (377). In E. coli, the ppfC and cgdC genes, encoding ferrochelatase and the O2-dependent coproporphyrinogen oxidase, respectively, are induced in response to H2O2 stress by OxyR-dependent activation (378, 379). H2O2 stress induces iron limitation by sequestration by the DNA-protective, iron storage protein Dps (DNA-binding protein under starvation) in order to minimize Fenton chemistry that creates hydroxyl radicals from H2O2 and Fe2+. Elevated ferrochelatase levels may allow the enzyme to compete with Dps for iron for heme synthesis (378). Additionally, CgdH is an iron-sulfur cluster-containing protein whose clusters are susceptible to H2O2 damage (380), and its synthesis may be compromised under iron limitation induced by H2O2 stress. These cellular responses underscore the roles that iron plays as both a problem and a solution in managing H2O2 toxicity.
In B. subtilis, PerR is the major peroxide regulator and represses a large PerR regulon (381). The OhrR family of antioxidant regulators is responsible for organic hydroperoxide resistance (382). B. subtilis PerR is an H2O2-responsive transcriptional repressor belonging to the Fur family of metalloregulators (383). The oxidation of PerR by H2O2 inactivates it, resulting in the derepression of its regulon. These genes include the hemAXCDBL operon, encoding enzymes for the early steps of heme synthesis (384, 385). Heme synthesis in response to H2O2 provides the prosthetic group for the catalase KatA. A perR mutation has a growth phenotype that is contributed by heme sequestration due to derepression of the katA gene (386). This finding suggests that a substantial quantity of heme produced is committed to KatA synthesis. B. japonicum Irr, a Fur family protein, is also affected by H2O2, by promoting its degradation (319, 322). A loss of Irr derepresses alaS, pbgS, and the major catalase gene katG (319).
Unexplored Territory
Over the past decade, studies have revealed that a multitude of metabolic pathways are subject to regulation by PTM. For bacteria in particular, a growing body of data has implicated reversible lysine acetylation as an essentially universal modulator of pathways (387), whereas in eukaryotes, one also frequently finds lysine succinylation (388). In addition, for pathways that are sensitive to oxygen and or redox potential, cysteine glutathionylation is not infrequently encountered (389). PTM regulatory mechanisms have yet to be studied for bacterial heme biosynthesis, but one would anticipate that these mechanisms exist and play a significant role in heme homeostasis in bacteria.
ONE MODEL FOR THE EVOLUTION OF TETRAPYRROLE BIOSYNTHESIS AND SOME PIECES THAT ARE STILL MISSING
The manner in which the various pathways for heme synthesis evolved has led to a number of models that each tried to resolve the question based upon currently available data. This topic has most recently been reviewed by Ducluzeau and Nitschke (36). Unfortunately, that overview lacked information about the CPD pathway and therefore fell short in its final proposal. Assuming that there are no major revelations about the diversity of heme synthesis, we believe that some simple observations can be made.
The most ancient of the tetrapyrrole biosynthetic pathways exists in the Archaea. Here one finds the ability to synthesize ALA from glutamyl-tRNA and the presence of the core pathway enzymes to make uroporphyrinogen III. This core set of enzymes is highly conserved from the Archaea through bacteria to eukaryotes, suggesting that all tetrapyrrole-synthesizing organisms descended from a common pathway. There was a primitive chelatase (similar to CbiXs) that evolved to function with different tetrapyrroles and a variety of metals (184, 185, 213). Among the Archaea, pathways for the synthesis of cobalamin (Co), F430 (Ni), and siroheme (Fe) evolved. Siroheme appears to be the most ancient surviving iron-containing tetrapyrrole, although the possibility that a less decorated iron-containing tetrapyrrole structure first evolved cannot be ruled out. The ability to modify siroheme into heme d1 and protoheme resulted in the first appearance of protoheme in nature (11). Interestingly, this pathway led through coproheme III, which was not adopted as a cofactor and, thus, was clearly less ideal for the biochemical processes involving heme than was protoheme.
Among eubacteria, UroD is first found as an extension of the common pathway from uroporphyrinogen III to protoheme (35, 43). CgoX, CpfC, and ChdC appear in the firmicutes and actinobacteria to form protoheme via coproheme, and this pathway exists in some ancient Gram-negative bacteria. The siroheme-dependent pathway to protoheme (AhbABCD) persists for a limited number of eubacteria, but AhbD of this pathway is more widely adopted by the CPD pathway as an apparent anaerobic alternative to ChdC. Nevertheless, no Gram-positive organism that has the ability to synthesize protoporphyrin has currently been identified. The switch from the CPD pathway to the PPD pathway is an interesting one without explanation at present, but a recent report that ChdC stimulates CgoX to oxidize protoporphyrinogen to protoporphyrin may provide an interesting first clue (390).
In Gram-negative bacteria, after a coproporphyrinogen “oxidative decarboxylase” (CgdH/CgdC) became available to synthesize protoporphyrinogen IX, CdoX evolved a specificity for protoporphyrinogen IX over coproporphyrinogen III to become PgoX, and a select set of bacteria evolved PgdH1 from a flavodoxin to replace PgoX, while the majority of Gram-negative bacteria evolved PgdH2 for this purpose. CpfC evolved to PpfC with a specificity for protoporphyrin. It is difficult to see what drove these various evolutionary alterations, but the major result was the availability of protoporphyrin, a necessity for chlorophyll biosynthesis.
Finally, in a limited number of bacteria, AlaS replaces GtrR plus GsaM. The last change may represent the evolutionary conversion of GsaM to AlaS (77). The ultimate outcome of such an event was the elimination of one enzyme (GtrR) and the switch of the ALA precursor from glutamyl-tRNA to succinyl-CoA. Among the 806 organisms contributing to the SEED representative genome set (see Table S3 in the supplemental material), 102 (12.6%) possess the C4 route catalyzed by AlaS, 680 (84.4%) contain the C5 route (encoded by GtrR and GsaM), 5 (0.6%) encode both pathways, and 19 (1.9%) have neither. The one-step pathway to ALA that utilizes succinyl-CoA plus glycine rather than depleting the cellular pool of glutamyl-tRNA apparently proved to be beneficial and spread by horizontal gene transfer (HGT) to some organisms already possessing the C5 route. Only a few genomes in the SEED collection of 983 genomes encode both functional pathways: Chromobacterium violaceum ATCC 12472, Serratia proteamaculans 568, Pseudomonas fulva 12-X, Streptosporangium roseum DSM 43021, and Streptomyces griseus NBRC 13350. These organisms belong to four different classes (Actinobacteria, Betaproteobacteria, Gammaproteobacteria, and Firmicutes) and represent rare events in their respective taxonomic groups, where the majority of members encode exclusively the C5 route (from 0.4% to 1.5% of genomes analyzed outside the representative set). There are no clear examples of HGT events in the opposite direction (i.e., that of an organism in the AlaS-possessing class of Alphaproteobacteria acquiring the 2-step C5 pathway).
From a distance, it may appear that the jigsaw puzzle of bacterial heme synthesis is largely complete, but there are still a few crucial missing pieces. The most obvious of these is in the CPD pathway, where only an oxygen-dependent enzyme (CpoX) has been identified to convert coproporphyrinogen into coproporphyrin. Given that a number of CPD-containing organisms are capable of heme synthesis under anaerobic conditions, it is clear that an oxygen-independent form of CpoX must exist, just as there are oxygen-dependent and oxygen-independent forms of protoporphyrinogen oxidase for the PPD pathway. Whole-genome studies hint that a gene encoding such a protein may exist in the DUF1444 superfamily of the Firmicutes, but no biochemical studies have verified this possibility, and interestingly, no such protein appears in the Actinobacteria.
There also are major deficiencies in our knowledge of the catalytic mechanism for many pathway enzymes. While X-ray crystal structures exist for a majority of pathway enzymes, there are only a few structures of enzymes with the substrate or product bound. These data are essential since, as clearly outlined above for ferrochelatase and as suggested for HmbS, molecular rearrangements are a requirement for catalysis by some enzymes. For the enzymes AhbD, PgdH1, and PgdH2, there are currently no structures and little catalytic information. Additionally, little is known about possible multiprotein complexes among the enzymes of the core or any of the three branches (87, 302), although limited data suggesting that “metabolons” exist for heme synthesis proteins in eukaryotic mitochondria are now available (300).
Without a doubt, the largest “black box” for all three biosynthetic pathways to protoheme is an understanding of regulatory mechanisms. For instance, the half-century-old observation that coproporphyrin is commonly excreted by many bacteria under certain growth conditions is still not understood or explained. Is this a common random event, or is the coproporphyrinogen purposefully overproduced to serve a specific biological function, perhaps as an emergency electron sink? Given the diversity of cellular metabolism among heme-synthesizing prokaryotes, it is obvious that the regulatory mechanisms employed to modulate heme homeostasis are quite diverse, and lessons learned from an individual group of organisms may not be applicable to others. This, then, is the major challenge going forward and should occupy researchers for decades to come.
Supplementary Material
ACKNOWLEDGMENTS
The research laboratory of H.A.D. is supported by NIH grant DK096051. D.J. and M.J. are supported by the Deutsche Forschungsgemeinschaft. M.R.O. is funded by NIH grant GM099667. M.J.W. is supported by grants from the Biotechnology and Biological Sciences Research Council. We acknowledge that we have no conflict of interest regarding the contents of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MMBR.00048-16.
REFERENCES
- 1.Johnson MP. 2016. Photosynthesis. Essays Biochem 60:255–273. doi: 10.1042/EBC20160016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ragsdale SW. 2014. Biochemistry of methyl-coenzyme M reductase: the nickel metalloenzyme that catalyzes the final step in synthesis and the first step in anaerobic oxidation of the greenhouse gas methane. Met Ions Life Sci 14:125–145. doi: 10.1007/978-94-017-9269-1_6. [DOI] [PubMed] [Google Scholar]
- 3.Rinaldo S, Sam KA, Castiglione N, Stelitano V, Arcovito A, Brunori M, Allen JW, Ferguson SJ, Cutruzzola F. 2011. Observation of fast release of NO from ferrous d(1) haem allows formulation of a unified reaction mechanism for cytochrome cd(1) nitrite reductases. Biochem J 435:217–225. doi: 10.1042/BJ20101615. [DOI] [PubMed] [Google Scholar]
- 4.Banerjee R, Ragsdale SW. 2003. The many faces of vitamin B12: catalysis by cobalamin-dependent enzymes. Annu Rev Biochem 72:209–247. doi: 10.1146/annurev.biochem.72.121801.161828. [DOI] [PubMed] [Google Scholar]
- 5.Jost M, Fernandez-Zapata J, Polanco MC, Ortiz-Guerrero JM, Chen PY, Kang G, Padmanabhan S, Elias-Arnanz M, Drennan CL. 2015. Structural basis for gene regulation by a B12-dependent photoreceptor. Nature 526:536–541. doi: 10.1038/nature14950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang L. 2011. Heme biology: the secret life of heme in regulating diverse biological processes. World Scientific Publishing Company, Singapore, Singapore. [Google Scholar]
- 7.Thauer RK, Bonacker LG. 1994. Biosynthesis of coenzyme F430, a nickel porphinoid involved in methanogenesis. Ciba Found Symp 180:210–222, discussion 222–227. [DOI] [PubMed] [Google Scholar]
- 8.Warren MJ, Deery E, Rose R-S. 2009. Biosynthesis of siroheme and coenzyme F430, p 343–351. In Warren MJ, Smith AG (ed), Tetrapyrroles: birth, life and death. Landes Bioscience, Austin, TX. [Google Scholar]
- 9.Escalante-Semerena JC, Woodson JD, Buan NR, Zayas CL. 2009. Conversion of cobinamide into coenzyme B12, p 300–316. In Warren MJ, Smith AG (ed), Tetrapyrroles: birth, life and death. Landes Bioscience, Austin, TX. [Google Scholar]
- 10.Moore SJ, Lawrence AD, Biedendieck R, Deery E, Frank S, Howard MJ, Rigby SE, Warren MJ. 2013. Elucidation of the anaerobic pathway for the corrin component of cobalamin (vitamin B12). Proc Natl Acad Sci U S A 110:14906–14911. doi: 10.1073/pnas.1308098110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bali S, Lawrence AD, Lobo SA, Saraiva LM, Golding BT, Palmer DJ, Howard MJ, Ferguson SJ, Warren MJ. 2011. Molecular hijacking of siroheme for the synthesis of heme and d1 heme. Proc Natl Acad Sci U S A 108:18260–18265. doi: 10.1073/pnas.1108228108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bali S, Rollauer S, Roversi P, Raux-Deery E, Lea SM, Warren MJ, Ferguson SJ. 2014. Identification and characterization of the ‘missing’ terminal enzyme for siroheme biosynthesis in alpha-proteobacteria. Mol Microbiol 92:153–163. doi: 10.1111/mmi.12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dailey HA. 1990. Biosynthesis of heme and chlorophylls. McGraw-Hill, New York, NY. [Google Scholar]
- 14.Layer G, Reichelt J, Jahn D, Heinz DW. 2010. Structure and function of enzymes in heme biosynthesis. Protein Sci 19:1137–1161. doi: 10.1002/pro.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heyes DJ, Hunter CN. 2009. Biosynthesis of chlorophyll and bacteriochlorophyll, p 235–249. In Warren MJ, Smith AG (ed), Tetrapyrroles: birth, life and death. Landes Bioscience, Austin, TX. [Google Scholar]
- 16.Coates JD, Achenbach LA. 2004. Microbial perchlorate reduction: rocket-fueled metabolism. Nat Rev Microbiol 2:569–580. doi: 10.1038/nrmicro926. [DOI] [PubMed] [Google Scholar]
- 17.Mayfield JA, Dehner CA, DuBois JL. 2011. Recent advances in bacterial heme protein biochemistry. Curr Opin Chem Biol 15:260–266. doi: 10.1016/j.cbpa.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moody SC, Loveridge EJ. 2014. CYP105—diverse structures, functions and roles in an intriguing family of enzymes in Streptomyces. J Appl Microbiol 117:1549–1563. doi: 10.1111/jam.12662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Celis AI, DuBois JL. 2015. Substrate, product, and cofactor: the extraordinarily flexible relationship between the CDE superfamily and heme. Arch Biochem Biophys 574:3–17. doi: 10.1016/j.abb.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts GP, Kerby RL, Youn H, Conrad M. 2005. CooA, a paradigm for gas sensing regulatory proteins. J Inorg Biochem 99:280–292. doi: 10.1016/j.jinorgbio.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 21.Plate L, Marletta MA. 2013. Nitric oxide-sensing H-NOX proteins govern bacterial communal behavior. Trends Biochem Sci 38:566–575. doi: 10.1016/j.tibs.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wan X, Tuckerman JR, Saito JA, Freitas TA, Newhouse JS, Denery JR, Galperin MY, Gonzalez G, Gilles-Gonzalez MA, Alam M. 2009. Globins synthesize the second messenger bis-(3′-5′)-cyclic diguanosine monophosphate in bacteria. J Mol Biol 388:262–270. doi: 10.1016/j.jmb.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar A, Toledo JC, Patel RP, Lancaster JR Jr, Steyn AJ. 2007. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc Natl Acad Sci U S A 104:11568–11573. doi: 10.1073/pnas.0705054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frankenberg-Dinkel N, Terry MJ. 2009. Synthesis and role of bilins in photosynthetic organisms, p 208–220. In Warren MJ, Smith AG (ed), Tetrapyrroles: birth, life and death. Landes Bioscience, Austin, TX. [Google Scholar]
- 25.Baureder M, Hederstedt L. 2013. Heme proteins in lactic acid bacteria. Adv Microb Physiol 62:1–43. doi: 10.1016/B978-0-12-410515-7.00001-9. [DOI] [PubMed] [Google Scholar]
- 26.Baureder M, Hederstedt L. 2012. Genes important for catalase activity in Enterococcus faecalis. PLoS One 7:e36725. doi: 10.1371/journal.pone.0036725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Torres VJ, Stauff DL, Pishchany G, Bezbradica JS, Gordy LE, Iturregui J, Anderson KL, Dunman PM, Joyce S, Skaar EP. 2007. A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host Microbe 1:109–119. doi: 10.1016/j.chom.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammer ND, Skaar EP. 2011. Molecular mechanisms of Staphylococcus aureus iron acquisition. Annu Rev Microbiol 65:129–147. doi: 10.1146/annurev-micro-090110-102851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Runyen-Janecky LJ. 2013. Role and regulation of heme iron acquisition in Gram-negative pathogens. Front Cell Infect Microbiol 3:55. doi: 10.3389/fcimb.2013.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lascelles J. 1964. Tetrapyrrole biosynthesis and its regulation. WA Benjamin, Inc, New York, NY. [Google Scholar]
- 31.Townsley PM, Neilands JB. 1957. The iron and porphyrin metabolism of Micrococcus lysodeikticus. J Biol Chem 224:695–705. [PubMed] [Google Scholar]
- 32.Hamza I, Dailey HA. 2012. One ring to rule them all: trafficking of heme and heme synthesis intermediates in the metazoans. Biochim Biophys Acta 1823:1617–1632. doi: 10.1016/j.bbamcr.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heinemann IU, Jahn M, Jahn D. 2008. The biochemistry of heme biosynthesis. Arch Biochem Biophys 474:238–251. doi: 10.1016/j.abb.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 34.Warren MJ, Smith AG (ed). 2009. Tetrapyrroles: birth, life and death. Landes Bioscience, Austin, TX. [Google Scholar]
- 35.Dailey HA, Gerdes S, Dailey TA, Burch JS, Phillips JD. 2015. Noncanonical coproporphyrin-dependent bacterial heme biosynthesis pathway that does not use protoporphyrin. Proc Natl Acad Sci U S A 112:2210–2215. doi: 10.1073/pnas.1416285112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ducluzeau A-L, Nitschke W. 2016. When did hemes enter the scene of life? On the natural history of heme cofactors and heme-containing enzymes, p 13–24. In Cramer WA, Kallas T (ed), Cytochrome complexes: evolution, structures, energy transduction, and signaling. Advances in photosynthesis and respiration, vol 41 Springer Science, Dordrecht, Netherlands. [Google Scholar]
- 37.Frankenberg N, Moser J, Jahn D. 2003. Bacterial heme biosynthesis and its biotechnological application. Appl Microbiol Biotechnol 63:115–127. doi: 10.1007/s00253-003-1432-2. [DOI] [PubMed] [Google Scholar]
- 38.Kobayashi K, Masuda T, Tajima N, Wada H, Sato N. 2014. Molecular phylogeny and intricate evolutionary history of the three isofunctional enzymes involved in the oxidation of protoporphyrinogen IX. Genome Biol Evol 6:2141–2155. doi: 10.1093/gbe/evu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Panek H, O'Brian MR. 2002. A whole genome view of prokaryotic haem biosynthesis. Microbiology 148:2273–2282. doi: 10.1099/00221287-148-8-2273. [DOI] [PubMed] [Google Scholar]
- 40.Zappa S, Li K, Bauer CE. 2010. The tetrapyrrole biosynthetic pathway and its regulation in Rhodobacter capsulatus. Adv Exp Med Biol 675:229–250. doi: 10.1007/978-1-4419-1528-3_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Brian MR, Thony-Meyer L. 2002. Biochemistry, regulation and genomics of haem biosynthesis in prokaryotes. Adv Microb Physiol 46:257–318. doi: 10.1016/S0065-2911(02)46006-7. [DOI] [PubMed] [Google Scholar]
- 42.Kuhner M, Haufschildt K, Neumann A, Storbeck S, Streif J, Layer G. 2014. The alternative route to heme in the methanogenic archaeon Methanosarcina barkeri. Archaea 2014:327637. doi: 10.1155/2014/327637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lobo SA, Scott A, Videira MA, Winpenny D, Gardner M, Palmer MJ, Schroeder S, Lawrence AD, Parkinson T, Warren MJ, Saraiva LM. 2015. Staphylococcus aureus haem biosynthesis: characterisation of the enzymes involved in final steps of the pathway. Mol Microbiol 97:472–487. doi: 10.1111/mmi.13041. [DOI] [PubMed] [Google Scholar]
- 44.Dailey HA, Gerdes S. 2015. HemQ: an iron-coproporphyrin oxidative decarboxylase for protoheme synthesis in Firmicutes and Actinobacteria. Arch Biochem Biophys 574:27–35. doi: 10.1016/j.abb.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dailey TA, Boynton TO, Albetel AN, Gerdes S, Johnson MK, Dailey HA. 2010. Discovery and characterization of HemQ: an essential heme biosynthetic pathway component. J Biol Chem 285:25978–25986. doi: 10.1074/jbc.M110.142604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ishida T, Yu L, Akutsu H, Ozawa K, Kawanishi S, Seto A, Inubushi T, Sano S. 1998. A primitive pathway of porphyrin biosynthesis and enzymology in Desulfovibrio vulgaris. Proc Natl Acad Sci U S A 95:4853–4858. doi: 10.1073/pnas.95.9.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jahn D, Heinz DW. 2009. Biosynthesis of 5-aminolevulinic acid, p 29–42. In Warren MJ, Smith AG (ed), Tetrapyrroles: birth, life and death. Landes Bioscience, Austin, TX. [Google Scholar]
- 48.Jahn D, Verkamp E, Soll D. 1992. Glutamyl-transfer RNA: a precursor of heme and chlorophyll biosynthesis. Trends Biochem Sci 17:215–218. doi: 10.1016/0968-0004(92)90380-R. [DOI] [PubMed] [Google Scholar]
- 49.Kaufholz AL, Hunter GA, Ferreira GC, Lendrihas T, Hering V, Layer G, Jahn M, Jahn D. 2013. Aminolaevulinic acid synthase of Rhodobacter capsulatus: high-resolution kinetic investigation of the structural basis for substrate binding and catalysis. Biochem J 451:205–216. doi: 10.1042/BJ20121041. [DOI] [PubMed] [Google Scholar]
- 50.Shemin D, Rittenberg D. 1945. The utilization of glycine for the synthesis of a porphyrin. J Biol Chem 159:567–568. [Google Scholar]
- 51.Gibson KD, Laver WG, Neuberger A. 1958. Initial stages in the biosynthesis of porphyrins. 2. The formation of delta-aminolaevulic acid from glycine and succinyl-coenzyme A by particles from chicken erythrocytes. Biochem J 70:71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kikuchi G, Kumar A, Talmage P, Shemin D. 1958. The enzymatic synthesis of delta-aminolevulinic acid. J Biol Chem 233:1214–1219. [PubMed] [Google Scholar]
- 53.Muir HM, Neuberger A. 1950. The biogenesis of porphyrins. 2. The origin of the methyne carbon atoms. Biochem J 47:97–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Radin NS, Rittenberg D, Shemin D. 1950. The role of acetic acid in the biosynthesis of heme. J Biol Chem 184:755–767. [PubMed] [Google Scholar]
- 55.Shemin D, Kumin S. 1952. The mechanism of porphyrin formation; the formation of a succinyl intermediate from succinate. J Biol Chem 198:827–837. [PubMed] [Google Scholar]
- 56.Shemin D, Russell CS. 1953. Aminolevulinic acid, its role in the biosynthesis of porphyrins and purines. J Am Chem Soc 75:4873–4874. doi: 10.1021/ja01115a546. [DOI] [Google Scholar]
- 57.Alexander FW, Sandmeier E, Mehta PK, Christen P. 1994. Evolutionary relationships among pyridoxal-5′-phosphate-dependent enzymes. Regio-specific alpha, beta and gamma families. Eur J Biochem 219:953–960. doi: 10.1111/j.1432-1033.1994.tb18577.x. [DOI] [PubMed] [Google Scholar]
- 58.Nandi DL. 1978. Delta-aminolevulinic acid synthase of Rhodopseudomonas spheroides. Binding of pyridoxal phosphate to the enzyme. Arch Biochem Biophys 188:266–271. doi: 10.1016/S0003-9861(78)80008-3. [DOI] [PubMed] [Google Scholar]
- 59.Astner I, Schulze JO, van den Heuvel J, Jahn D, Schubert WD, Heinz DW. 2005. Crystal structure of 5-aminolevulinate synthase, the first enzyme of heme biosynthesis, and its link to XLSA in humans. EMBO J 24:3166–3177. doi: 10.1038/sj.emboj.7600792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hunter GA, Ferreira GC. 1999. Pre-steady-state reaction of 5-aminolevulinate synthase. Evidence for a rate-determining product release. J Biol Chem 274:12222–12228. doi: 10.1074/jbc.274.18.12222. [DOI] [PubMed] [Google Scholar]
- 61.Hunter GA, Ferreira GC. 2009. 5-Aminolevulinate synthase: catalysis of the first step of heme biosynthesis. Cell Mol Biol (Noisy-le-grand) 55(1):102–110. [PMC free article] [PubMed] [Google Scholar]
- 62.Stojanovski BM, Hunter GA, Jahn M, Jahn D, Ferreira GC. 2014. Unstable reaction intermediates and hysteresis during the catalytic cycle of 5-aminolevulinate synthase: implications from using pseudo and alternate substrates and a promiscuous enzyme variant. J Biol Chem 289:22915–22925. doi: 10.1074/jbc.M114.574731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zaman Z, Jordan PM, Akhtar M. 1973. Mechanism and stereochemistry of the 5-aminolaevulinate synthetase reaction. Biochem J 135:257–263. doi: 10.1042/bj1350257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beale SI, Castelfranco PA. 1973. 14C incorporation from exogenous compounds into delta-aminolevulinic acid by greening cucumber cotyledons. Biochem Biophys Res Commun 52:143–149. doi: 10.1016/0006-291X(73)90966-2. [DOI] [PubMed] [Google Scholar]
- 65.Beale SI, Gough SP, Granick S. 1975. Biosynthesis of delta-aminolevulinic acid from the intact carbon skeleton of glutamic acid in greening barley. Proc Natl Acad Sci U S A 72:2719–2723. doi: 10.1073/pnas.72.7.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang DD, Wang WY, Gough SP, Kannangara CG. 1984. Delta-aminolevulinic acid synthesizing enzymes need an RNA moiety for activity. Science 225:1482–1484. doi: 10.1126/science.6206568. [DOI] [PubMed] [Google Scholar]
- 67.Kannangara CG, Gough SP. 1978. Biosynthesis of delta-aminolevulinate in greening barley leaves: glutamate-1-semialdehyde aminotransferase. Carlsberg Res Commun 43:185–194. doi: 10.1007/BF02914241. [DOI] [PubMed] [Google Scholar]
- 68.Schon A, Krupp G, Gough S, Berry-Lowe S, Kannangara CG, Soll D. 1986. The RNA required in the first step of chlorophyll biosynthesis is a chloroplast glutamate tRNA. Nature 322:281–284. doi: 10.1038/322281a0. [DOI] [PubMed] [Google Scholar]
- 69.Moser J, Schubert WD, Beier V, Bringemeier I, Jahn D, Heinz DW. 2001. V-shaped structure of glutamyl-tRNA reductase, the first enzyme of tRNA-dependent tetrapyrrole biosynthesis. EMBO J 20:6583–6590. doi: 10.1093/emboj/20.23.6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao A, Fang Y, Chen X, Zhao S, Dong W, Lin Y, Gong W, Liu L. 2014. Crystal structure of Arabidopsis glutamyl-tRNA reductase in complex with its stimulator protein. Proc Natl Acad Sci U S A 111:6630–6635. doi: 10.1073/pnas.1400166111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moser J, Lorenz S, Hubschwerlen C, Rompf A, Jahn D. 1999. Methanopyrus kandleri glutamyl-tRNA reductase. J Biol Chem 274:30679–30685. doi: 10.1074/jbc.274.43.30679. [DOI] [PubMed] [Google Scholar]
- 72.Schauer S, Chaturvedi S, Randau L, Moser J, Kitabatake M, Lorenz S, Verkamp E, Schubert WD, Nakayashiki T, Murai M, Wall K, Thomann HU, Heinz DW, Inokuchi H, Soll D, Jahn D. 2002. Escherichia coli glutamyl-tRNA reductase. Trapping the thioester intermediate. J Biol Chem 277:48657–48663. doi: 10.1074/jbc.M206924200. [DOI] [PubMed] [Google Scholar]
- 73.Luer C, Schauer S, Virus S, Schubert WD, Heinz DW, Moser J, Jahn D. 2007. Glutamate recognition and hydride transfer by Escherichia coli glutamyl-tRNA reductase. FEBS J 274:4609–4614. doi: 10.1111/j.1742-4658.2007.05989.x. [DOI] [PubMed] [Google Scholar]
- 74.Randau L, Schauer S, Ambrogelly A, Salazar JC, Moser J, Sekine S, Yokoyama S, Soll D, Jahn D. 2004. tRNA recognition by glutamyl-tRNA reductase. J Biol Chem 279:34931–34937. doi: 10.1074/jbc.M401529200. [DOI] [PubMed] [Google Scholar]
- 75.Ge H, Lv X, Fan J, Gao Y, Teng M, Niu L. 2010. Crystal structure of glutamate-1-semialdehyde aminotransferase from Bacillus subtilis with bound pyridoxamine-5′-phosphate. Biochem Biophys Res Commun 402:356–360. doi: 10.1016/j.bbrc.2010.10.033. [DOI] [PubMed] [Google Scholar]
- 76.Hennig M, Grimm B, Contestabile R, John RA, Jansonius JN. 1997. Crystal structure of glutamate-1-semialdehyde aminomutase: an alpha2-dimeric vitamin B6-dependent enzyme with asymmetry in structure and active site reactivity. Proc Natl Acad Sci U S A 94:4866–4871. doi: 10.1073/pnas.94.10.4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schulze JO, Schubert WD, Moser J, Jahn D, Heinz DW. 2006. Evolutionary relationship between initial enzymes of tetrapyrrole biosynthesis. J Mol Biol 358:1212–1220. doi: 10.1016/j.jmb.2006.02.064. [DOI] [PubMed] [Google Scholar]
- 78.Campanini B, Bettati S, di Salvo ML, Mozzarelli A, Contestabile R. 2013. Asymmetry of the active site loop conformation between subunits of glutamate-1-semialdehyde aminomutase in solution. Biomed Res Int 2013:353270. doi: 10.1155/2013/353270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stetefeld J, Ruegg MA. 2005. Structural and functional diversity generated by alternative mRNA splicing. Trends Biochem Sci 30:515–521. doi: 10.1016/j.tibs.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 80.Grimm B, Smith MA, von Wettstein D. 1992. The role of Lys272 in the pyridoxal 5-phosphate active site of Synechococcus glutamate-1-semialdehyde aminotransferase. Eur J Biochem 206:579–585. doi: 10.1111/j.1432-1033.1992.tb16962.x. [DOI] [PubMed] [Google Scholar]
- 81.Ilag LL, Jahn D. 1992. Activity and spectroscopic properties of the Escherichia coli glutamate 1-semialdehyde aminotransferase and the putative active site mutant K265R. Biochemistry 31:7143–7151. doi: 10.1021/bi00146a016. [DOI] [PubMed] [Google Scholar]
- 82.Sorensen JL, Stetefeld J. 2011. Kinemage of action—proposed reaction mechanism of glutamate-1-semialdehyde aminomutase at an atomic level. Biochem Biophys Res Commun 413:572–576. doi: 10.1016/j.bbrc.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 83.Brody S, Andersen JS, Kannangara CG, Meldgaard M, Roepstorff P, von Wettstein D. 1995. Characterization of the different spectral forms of glutamate 1-semialdehyde aminotransferase by mass spectrometry. Biochemistry 34:15918–15924. doi: 10.1021/bi00049a006. [DOI] [PubMed] [Google Scholar]
- 84.Pugh CE, Harwood JL, John RA. 1992. Mechanism of glutamate semialdehyde aminotransferase. Roles of diamino- and dioxo-intermediates in the synthesis of aminolevulinate. J Biol Chem 267:1584–1588. [PubMed] [Google Scholar]
- 85.Smith MA, King PJ, Grimm B. 1998. Transient-state kinetic analysis of Synechococcus glutamate 1-semialdehyde aminotransferase. Biochemistry 37:319–329. doi: 10.1021/bi9717587. [DOI] [PubMed] [Google Scholar]
- 86.Tyacke RJ, Harwood JL, John RA. 1993. Properties of the pyridoxaldimine form of glutamate semialdehyde aminotransferase (glutamate-1-semialdehyde 2,1-aminomutase) and analysis of its role as an intermediate in the formation of aminolaevulinate. Biochem J 293(Part 3):697–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Luer C, Schauer S, Mobius K, Schulze J, Schubert WD, Heinz DW, Jahn D, Moser J. 2005. Complex formation between glutamyl-tRNA reductase and glutamate-1-semialdehyde 2,1-aminomutase in Escherichia coli during the initial reactions of porphyrin biosynthesis. J Biol Chem 280:18568–18572. doi: 10.1074/jbc.M500440200. [DOI] [PubMed] [Google Scholar]
- 88.Nogaj LA, Beale SI. 2005. Physical and kinetic interactions between glutamyl-tRNA reductase and glutamate-1-semialdehyde aminotransferase of Chlamydomonas reinhardtii. J Biol Chem 280:24301–24307. doi: 10.1074/jbc.M502483200. [DOI] [PubMed] [Google Scholar]
- 89.Breinig S, Kervinen J, Stith L, Wasson AS, Fairman R, Wlodawer A, Zdanov A, Jaffe EK. 2003. Control of tetrapyrrole biosynthesis by alternate quaternary forms of porphobilinogen synthase. Nat Struct Biol 10:757–763. doi: 10.1038/nsb963. [DOI] [PubMed] [Google Scholar]
- 90.Coates L, Beaven G, Erskine PT, Beale SI, Wood SP, Shoolingin-Jordan PM, Cooper JB. 2005. Structure of Chlorobium vibrioforme 5-aminolaevulinic acid dehydratase complexed with a diacid inhibitor. Acta Crystallogr D Biol Crystallogr 61:1594–1598. doi: 10.1107/S0907444905030350. [DOI] [PubMed] [Google Scholar]
- 91.Erskine PT, Newbold R, Roper J, Coker A, Warren MJ, Shoolingin-Jordan PM, Wood SP, Cooper JB. 1999. The Schiff base complex of yeast 5-aminolaevulinic acid dehydratase with laevulinic acid. Protein Sci 8:1250–1256. doi: 10.1110/ps.8.6.1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Erskine PT, Norton E, Cooper JB, Lambert R, Coker A, Lewis G, Spencer P, Sarwar M, Wood SP, Warren MJ, Shoolingin-Jordan PM. 1999. X-ray structure of 5-aminolevulinic acid dehydratase from Escherichia coli complexed with the inhibitor levulinic acid at 2.0 A resolution. Biochemistry 38:4266–4276. doi: 10.1021/bi982137w. [DOI] [PubMed] [Google Scholar]
- 93.Frankenberg N, Erskine PT, Cooper JB, Shoolingin-Jordan PM, Jahn D, Heinz DW. 1999. High resolution crystal structure of a Mg2+-dependent porphobilinogen synthase. J Mol Biol 289:591–602. doi: 10.1006/jmbi.1999.2808. [DOI] [PubMed] [Google Scholar]
- 94.Jaffe EK. 2000. The porphobilinogen synthase family of metalloenzymes. Acta Crystallogr D Biol Crystallogr 56:115–128. doi: 10.1107/S0907444999014894. [DOI] [PubMed] [Google Scholar]
- 95.Bollivar DW, Clauson C, Lighthall R, Forbes S, Kokona B, Fairman R, Kundrat L, Jaffe EK. 2004. Rhodobacter capsulatus porphobilinogen synthase, a high activity metal ion independent hexamer. BMC Biochem 5:17. doi: 10.1186/1471-2091-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chauhan S, O'Brian MR. 1995. A mutant Bradyrhizobium japonicum δ-aminolevulinic acid dehydratase with an altered metal requirement functions in situ for tetrapyrrole synthesis in soybean root nodules. J Biol Chem 270:19823–19827. doi: 10.1074/jbc.270.34.19823. [DOI] [PubMed] [Google Scholar]
- 97.Frere F, Reents H, Schubert WD, Heinz DW, Jahn D. 2005. Tracking the evolution of porphobilinogen synthase metal dependence in vitro. J Mol Biol 345:1059–1070. doi: 10.1016/j.jmb.2004.10.053. [DOI] [PubMed] [Google Scholar]
- 98.Petrovich RM, Jaffe EK. 1997. Magnetic resonance studies on the active site and metal centers of Bradyrhizobium japonicum porphobilinogen synthase. Biochemistry 36:13421–13427. doi: 10.1021/bi971642a. [DOI] [PubMed] [Google Scholar]
- 99.Petrovich RM, Litwin S, Jaffe EK. 1996. Bradyrhizobium japonicum porphobilinogen synthase uses two Mg(II) and monovalent cations. J Biol Chem 271:8692–8699. doi: 10.1074/jbc.271.15.8692. [DOI] [PubMed] [Google Scholar]
- 100.Jaffe EK. 2005. Morpheeins—a new structural paradigm for allosteric regulation. Trends Biochem Sci 30:490–497. doi: 10.1016/j.tibs.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 101.Jaffe EK, Stith L. 2007. ALAD porphyria is a conformational disease. Am J Hum Genet 80:329–337. doi: 10.1086/511444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Selwood T, Tang L, Lawrence SH, Anokhina Y, Jaffe EK. 2008. Kinetics and thermodynamics of the interchange of the morpheein forms of human porphobilinogen synthase. Biochemistry 47:3245–3257. doi: 10.1021/bi702113z. [DOI] [PubMed] [Google Scholar]
- 103.Tang L, Breinig S, Stith L, Mischel A, Tannir J, Kokona B, Fairman R, Jaffe EK. 2006. Single amino acid mutations alter the distribution of human porphobilinogen synthase quaternary structure isoforms (morpheeins). J Biol Chem 281:6682–6690. doi: 10.1074/jbc.M511134200. [DOI] [PubMed] [Google Scholar]
- 104.Jordan PM. 1991. The biosynthesis of 5-aminolaevulinic acid and its transformation into uroporphyrinogen III, p 1–66. In Neurberger A, van Deenen LLM (ed), Biosynthesis of tetrapyrroles: new comprehensive biochemistry. Elsevier, Amsterdam, Netherlands. [Google Scholar]
- 105.Bonkovsky HL, Guo JT, Hou W, Li T, Narang T, Thapar M. 2013. Porphyrin and heme metabolism and the porphyrias. Compr Physiol 3:365–401. doi: 10.1002/cphy.c120006. [DOI] [PubMed] [Google Scholar]
- 106.Burton G, Fagerness PE, Hosozawa S, Jordan PM, Scott AI. 1979. 13C NMR evidence for a new intermediate, pre-uroporphyrinogen, in the enzymic transformation of porphobilinogen into uroporphyrinogens I and III. J Chem Soc Chem Commun 1979:202–204. [Google Scholar]
- 107.Lewis NG, Neier R, Matcham GWJ, Mcdonald E, Battersby AR. 1979. Biosynthesis of vitamin-B12—experiments on loss of C-20 from the precursor macrocycle. J Chem Soc Chem Commun 1979:541–542. [Google Scholar]
- 108.Battersby AR, Fookes CJR, Pandey PS. 1983. Biosynthesis of porphyrins and related macrocycles. 19. Linear tetrapyrrolic intermediates for biosynthesis of the natural porphyrins—experiments with modified substrates. Tetrahedron 39:1919–1926. [Google Scholar]
- 109.Battersby AR, Fookes CJR, Matcham GWJ, Pandey PS. 1981. Biosynthesis of natural porphyrins—studies with isomeric hydroxymethylbilanes on the specificity and action of cosynthetase. Angew Chem Int 20:293–295. doi: 10.1002/anie.198102931. [DOI] [Google Scholar]
- 110.Battersby AR, Fookes CJR, Hart G, Matcham GWJ, Pandey PS. 1983. Biosynthesis of porphyrins and related macrocycles. 21. The interaction of deaminase and its product (hydroxymethylbilane) and the relationship between deaminase and co-synthetase. J Chem Soc Perkin Trans 1:3041–3047. [Google Scholar]
- 111.Battersby AR, Fookes CJR, Gustafson-Potter KE, McDonald E, Matcham GWJ. 1979. Biosynthesis of the natural porphyrins: experiments on the ring-closure steps and with the hydroxy-analogue of porphobilinogen. J Chem Soc Chem Commun 1979:316–319. [Google Scholar]
- 112.Anderson PM, Desnick RJ. 1980. Purification and properties of uroporphyrinogen-I synthase from human-erythrocytes—identification of stable enzyme-substrate intermediates. J Biol Chem 255:1993–1999. [PubMed] [Google Scholar]
- 113.Jordan PM, Berry A. 1981. Mechanism of action of porphobilinogen deaminase—the participation of stable enzyme substrate covalent intermediates between porphobilinogen and the porphobilinogen deaminase from Rhodopseudomonas-Spheroides. Biochem J 195:177–181. doi: 10.1042/bj1950177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Battersby AR, Fookes CJR, Matcham GWJ, McDonald E. 1979. Order of assembly of the four pyrrole rings during biosynthesis of the natural porphyrins. J Chem Soc Chem Commun 1979:539–541. [Google Scholar]
- 115.Jordan PM, Seehra JS. 1979. Biosynthesis of uroporphyrinogen-III—order of assembly of the 4 porphobilinogen molecules in the formation of the tetrapyrrole ring. FEBS Lett 104:364–366. doi: 10.1016/0014-5793(79)80853-4. [DOI] [PubMed] [Google Scholar]
- 116.Jordan PM, Thomas SD, Warren MJ. 1988. Purification, crystallization and properties of porphobilinogen deaminase from a recombinant strain of Escherichia coli K12. Biochem J 254:427–435. doi: 10.1042/bj2540427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Thomas SD, Jordan PM. 1986. Nucleotide sequence of the hemC locus encoding porphobilinogen deaminase of Escherichia coli K12. Nucleic Acids Res 14:6215–6226. doi: 10.1093/nar/14.15.6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hart GJ, Miller AD, Beifuss U, Leeper FJ, Battersby AR. 1990. Biosynthesis of porphyrins and related macrocycles. 35. Discovery of a novel dipyrrolic cofactor essential for the catalytic action of hydroxymethylbilane synthase (porphobilinogen deaminase). J Chem Soc Perkin Trans 1:1979–1993. [Google Scholar]
- 119.Hart GJ, Miller AD, Leeper FJ, Battersby AR. 1987. Biosynthesis of the natural porphyrins: proof that hydroxymethylbilane synthase (porphobilinogen deaminase) uses a novel binding group in its catalytic action. J Chem Soc Chem Commun 1987:1762–1765. [Google Scholar]
- 120.Jordan PM, Warren MJ. 1987. Evidence for a dipyrromethane cofactor at the catalytic site of E. coli porphobilinogen deaminase. FEBS Lett 225:87–92. doi: 10.1016/0014-5793(87)81136-5. [DOI] [PubMed] [Google Scholar]
- 121.Warren MJ, Jordan PM. 1988. Investigation into the nature of substrate binding to the dipyrromethane cofactor of Escherichia coli porphobilinogen deaminase. Biochemistry 27:9020–9030. doi: 10.1021/bi00425a021. [DOI] [PubMed] [Google Scholar]
- 122.Umanoff H, Russell CS, Cosloy SD. 1988. Availability of porphobilinogen controls appearance of porphobilinogen deaminase activity in Escherichia coli K-12. J Bacteriol 170:4969–4971. doi: 10.1128/jb.170.10.4969-4971.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hart GJ, Miller AD, Battersby AR. 1988. Evidence that the pyrromethane cofactor of hydroxymethylbilane synthase (porphobilinogen deaminase) is bound through the sulfur atom of a cysteine residue. Biochem J 252:909–912. doi: 10.1042/bj2520909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jordan PM, Warren MJ, Williams HJ, Stolowich NJ, Roessner CA, Grant SK, Scott AI. 1988. Identification of a cysteine residue as the binding-site for the dipyrromethane cofactor at the active-site of Escherichia coli porphobilinogen deaminase. FEBS Lett 235:189–193. doi: 10.1016/0014-5793(88)81260-2. [DOI] [PubMed] [Google Scholar]
- 125.Awan SJ, Siligardi G, Shoolingin-Jordan PM, Warren MJ. 1997. Reconstitution of the holoenzyme form of Escherichia coli porphobilinogen deaminase from apoenzyme with porphobilinogen and preuroporphyrinogen: a study using circular dichroism spectroscopy. Biochemistry 36:9273–9282. doi: 10.1021/bi9702602. [DOI] [PubMed] [Google Scholar]
- 126.Shoolingin-Jordan PM, Warren MJ, Awan SJ. 1996. Discovery that the assembly of the dipyrromethane cofactor of porphobilinogen deaminase holoenzyme proceeds initially by the reaction of preuroporphyrinogen with the apoenzyme. Biochem J 316:373–376. doi: 10.1042/bj3160373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jordan PM, Warren MJ, Mgbeje BIA, Wood SP, Cooper JB, Louie G, Brownlie P, Lambert R, Blundell TL. 1992. Crystallization and preliminary X-ray investigation of Escherichia coli porphobilinogen deaminase. J Mol Biol 224:269–271. doi: 10.1016/0022-2836(92)90590-G. [DOI] [PubMed] [Google Scholar]
- 128.Louie GV, Brownlie PD, Lambert R, Cooper JB, Blundell TL, Wood SP, Malashkevich VN, Hadener A, Warren MJ, Shoolingin-Jordan PM. 1996. The three-dimensional structure of Escherichia coli porphobilinogen deaminase at 1.76-angstrom resolution. Proteins 25:48–78. doi:. [DOI] [PubMed] [Google Scholar]
- 129.Louie GV, Brownlie PD, Lambert R, Cooper JB, Blundell TL, Wood SP, Warren MJ, Woodcock SC, Jordan PM. 1992. Structure of porphobilinogen deaminase reveals a flexible multidomain polymerase with a single catalytic site. Nature 359:33–39. doi: 10.1038/359033a0. [DOI] [PubMed] [Google Scholar]
- 130.Brownlie PD, Lambert R, Louie GV, Jordan PM, Blundell TL, Warren MJ, Cooper JB, Wood SP. 1994. The 3-dimensional structures of mutants of porphobilinogen deaminase—toward an understanding of the structural basis of acute intermittent porphyria. Prot Sci 3:1644–1650. doi: 10.1002/pro.5560031004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Woodcock SC, Jordan PM. 1994. Evidence for participation of aspartate-84 as a catalytic group at the active site of porphobilinogen deaminase obtained by site-directed mutagenesis of the hemC gene from Escherichia coli. Biochemistry 33:2688–2695. doi: 10.1021/bi00175a043. [DOI] [PubMed] [Google Scholar]
- 132.Azim N, Deery E, Warren MJ, Wolfenden BA, Erskine P, Cooper JB, Coker A, Wood SP, Akhtar M. 2014. Structural evidence for the partially oxidized dipyrromethene and dipyrromethanone forms of the cofactor of porphobilinogen deaminase: structures of the Bacillus megaterium enzyme at near-atomic resolution. Acta Crystallogr D Biol Crystallogr 70:744–751. doi: 10.1107/S139900471303294X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gill R, Kolstoe SE, Mohammed F, Al d-Bass A, Mosely JE, Sarwar M, Cooper JB, Wood SP, Shoolingin-Jordan PM. 2009. Structure of human porphobilinogen deaminase at 2.8 A: the molecular basis of acute intermittent porphyria. Biochem J 420:17–25. doi: 10.1042/BJ20082077. [DOI] [PubMed] [Google Scholar]
- 134.Roberts A, Gill R, Hussey RJ, Mikolajek H, Erskine PT, Cooper JB, Wood SP, Chrystal EJ, Shoolingin-Jordan PM. 2013. Insights into the mechanism of pyrrole polymerization catalysed by porphobilinogen deaminase: high-resolution X-ray studies of the Arabidopsis thaliana enzyme. Acta Crystallogr D Biol Crystallogr 69:471–485. doi: 10.1107/S0907444912052134. [DOI] [PubMed] [Google Scholar]
- 135.Song G, Li Y, Cheng C, Zhao Y, Gao A, Zhang R, Joachimiak A, Shaw N, Liu ZJ. 2009. Structural insight into acute intermittent porphyria. FASEB J 23:396–404. doi: 10.1096/fj.08-115469. [DOI] [PubMed] [Google Scholar]
- 136.Warren MJ, Gul S, Aplin RT, Scott AI, Roessner CA, O'Grady P, Shoolingin-Jordan PM. 1995. Evidence for conformational changes in Escherichia coli porphobilinogen deaminase during stepwise pyrrole chain elongation monitored by increased reactivity of cysteine-134 to alkylation by N-ethylmaleimide. Biochemistry 34:11288–11295. doi: 10.1021/bi00035a038. [DOI] [PubMed] [Google Scholar]
- 137.Bung N, Pradhan M, Srinivasan H, Bulusu G. 2014. Structural insights into E. coli porphobilinogen deaminase during synthesis and exit of 1-hydroxymethylbilane. PLoS Comput Biol 10:e1003484. doi: 10.1371/journal.pcbi.1003484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Li N, Chu X, Wu L, Liu X, Li D. 2008. Functional studies of rat hydroxymethylbilane synthase. Bioorg Chem 36:241–251. doi: 10.1016/j.bioorg.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 139.Jordan PM, Mgbeje BI, Alwan AF, Thomas SD. 1987. Nucleotide sequence of hemD, the second gene in the hem operon of Escherichia coli K-12. Nucleic Acids Res 15:10583. doi: 10.1093/nar/15.24.10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sasarman A, Nepveu A, Echelard Y, Dymetryszyn J, Drolet M, Goyer C. 1987. Molecular cloning and sequencing of the hemD gene of Escherichia coli K-12 and preliminary data on the Uro operon. J Bacteriol 169:4257–4262. doi: 10.1128/jb.169.9.4257-4262.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jordan PM. 1982. Uroporphyrinogen III cosynthetase: a direct assay method. Enzyme 28:158–169. [DOI] [PubMed] [Google Scholar]
- 142.Lim CK, Li F, Peters TJ. 1987. High-performance liquid chromatography of type-III heptocarboxylic porphyrinogen isomers. Biochem J 247:229–232. doi: 10.1042/bj2470229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tsai SF, Bishop DF, Desnick RJ. 1987. Purification and properties of uroporphyrinogen III synthase from human erythrocytes. J Biol Chem 262:1268–1273. [PubMed] [Google Scholar]
- 144.Alwan AF, Mgbeje BI, Jordan PM. 1989. Purification and properties of uroporphyrinogen III synthase (co-synthase) from an overproducing recombinant strain of Escherichia coli K-12. Biochem J 264:397–402. doi: 10.1042/bj2640397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Amillet JM, Labbe-Bois R. 1995. Isolation of the gene HEM4 encoding uroporphyrinogen III synthase in Saccharomyces cerevisiae. Yeast 11:419–424. doi: 10.1002/yea.320110504. [DOI] [PubMed] [Google Scholar]
- 146.Mohr CD, Sonsteby SK, Deretic V. 1994. The Pseudomonas aeruginosa homologs of hemC and hemD are linked to the gene encoding the regulator of mucoidy AlgR. Mol Gen Genet 242:177–184. doi: 10.1007/BF00391011. [DOI] [PubMed] [Google Scholar]
- 147.Stamford NP, Capretta A, Battersby AR. 1995. Expression, purification and characterisation of the product from the Bacillus subtilis hemD gene, uroporphyrinogen III synthase. Eur J Biochem 231:236–241. doi: 10.1111/j.1432-1033.1995.0236f.x. [DOI] [PubMed] [Google Scholar]
- 148.Warren MJ, Scott AI. 1990. Tetrapyrrole assembly and modification into the ligands of biologically functional cofactors. Trends Biochem Sci 15:486–491. doi: 10.1016/0968-0004(90)90304-T. [DOI] [PubMed] [Google Scholar]
- 149.Lindsey JS, Ptaszek M, Taniguchi M. 2009. Simple formation of an abiotic porphyrinogen in aqueous solution. Orig Life Evol Biosph 39:495–515. doi: 10.1007/s11084-009-9168-3. [DOI] [PubMed] [Google Scholar]
- 150.Mathewson JH, Corwin AH. 1961. Biosynthesis of pyrrole pigments: a mechanism for porphobilinogen polymerization. J Am Chem Soc 83:135–137. doi: 10.1021/ja01462a026. [DOI] [Google Scholar]
- 151.Stark WM, Baker MG, Leeper FJ, Raithby PR, Battersby AR. 1988. Biosynthesis of porphyrins and related macrocycles. 30. Synthesis of the macrocycle of the spiro system proposed as an intermediate generated by cosynthetase. J Chem Soc Perkin Trans 1:1187–1201. [Google Scholar]
- 152.Stark WM, Baker MG, Raithby PR, Leeper FJ, Battersby AR. 1985. The spiro intermediate proposed for biosynthesis of the natural porphyrins—synthesis and properties of its macrocycle. J Chem Soc Chem Commun 1985:1294–1296. [Google Scholar]
- 153.Stark WM, Hawker CJ, Hart GJ, Philippides A, Petersen PM, Lewis JD, Leeper FJ, Battersby AR. 1993. Biosynthesis of porphyrins and related macrocycles. 40. Synthesis of a spiro-lactam related to the proposed spiro-intermediate for porphyrin biosynthesis—inhibition of cosynthetase. J Chem Soc Perkin Trans 1:2875–2892. [Google Scholar]
- 154.Spivey AC, Capretta A, Frampton CS, Leeper FJ, Battersby AR. 1995. Stereochemical studies on the proposed spiro intermediate for the biosynthesis of the natural porphyrins—determination by a novel X-ray method of the absolute configuration of the spirolactam which inhibits cosynthetase. J Chem Soc Chem Commun 1995:1789–1790. [Google Scholar]
- 155.Spivey AC, Capretta A, Frampton CS, Leeper FJ, Battersby AR. 1996. Biosynthesis of porphyrins and related macrocycles. Part 45. Determination by a novel X-ray method of the absolute configuration of the spiro lactam which inhibits uroporphyrinogen III synthase (cosynthetase). J Chem Soc Perkin Trans 1:2091–2102. [Google Scholar]
- 156.Tan FC, Cheng Q, Saha K, Heinemann IU, Jahn M, Jahn D, Smith AG. 2008. Identification and characterization of the Arabidopsis gene encoding the tetrapyrrole biosynthesis enzyme uroporphyrinogen III synthase. Biochem J 410:291–299. doi: 10.1042/BJ20070770. [DOI] [PubMed] [Google Scholar]
- 157.Roessner CA, Ponnamperuma K, Scott AI. 2002. Mutagenesis identifies a conserved tyrosine residue important for the activity of uroporphyrinogen III synthase from Anacystis nidulans. FEBS Lett 525:25–28. doi: 10.1016/S0014-5793(02)03056-9. [DOI] [PubMed] [Google Scholar]
- 158.Mathews MA, Schubert HL, Whitby FG, Alexander KJ, Schadick K, Bergonia HA, Phillips JD, Hill CP. 2001. Crystal structure of human uroporphyrinogen III synthase. EMBO J 20:5832–5839. doi: 10.1093/emboj/20.21.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Schubert HL, Phillips JD, Heroux A, Hill CP. 2008. Structure and mechanistic implications of a uroporphyrinogen III synthase-product complex. Biochemistry 47:8648–8655. doi: 10.1021/bi800635y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Silva PJ, Ramos MJ. 2008. Comparative density functional study of models for the reaction mechanism of uroporphyrinogen III synthase. J Phys Chem B 112:3144–3148. doi: 10.1021/jp076235f. [DOI] [PubMed] [Google Scholar]
- 161.Peng S, Zhang H, Gao Y, Pan X, Cao P, Li M, Chang W. 2011. Crystal structure of uroporphyrinogen III synthase from Pseudomonas syringae pv. tomato DC3000. Biochem Biophys Res Commun 408:576–581. doi: 10.1016/j.bbrc.2011.04.064. [DOI] [PubMed] [Google Scholar]
- 162.Akutsu H, Park JS, Sano S. 1993. l-Methionine methyl is specifically incorporated into the C-2 and C-7 positions of the porphyrin of cytochrome c(3) in a strictly anaerobic bacterium, Desulfovibrio vulgaris. J Am Chem Soc 115:12185–12186. doi: 10.1021/ja00078a075. [DOI] [Google Scholar]
- 163.Spencer JB, Stolowich NJ, Roessner CA, Scott AI. 1993. The Escherichia coli cysG gene encodes the multifunctional protein, siroheme synthase. FEBS Lett 335:57–60. doi: 10.1016/0014-5793(93)80438-Z. [DOI] [PubMed] [Google Scholar]
- 164.Warren MJ, Bolt EL, Roessner CA, Scott AI, Spencer JB, Woodcock SC. 1994. Gene dissection demonstrates that the Escherichia coli cysG gene encodes a multifunctional protein. Biochem J 302(Part 3):837–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Battersby AR, McDonald E, Thompson M, Bykhovsky VY. 1978. Biosynthesis of vitamin-B12—proof of a B structure for sirohydrochlorin by its specific incorporation into cobyrinic acid. J Chem Soc Chem Commun 1978:150–151. [Google Scholar]
- 166.Blanche F, Debussche L, Thibaut D, Crouzet J, Cameron B. 1989. Purification and characterization of S-adenosyl-l-methionine:uroporphyrinogen III methyltransferase from Pseudomonas denitrificans. J Bacteriol 171:4222–4231. doi: 10.1128/jb.171.8.4222-4231.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Bergmann KH, Deeg R, Gneuss KD, Kriemler HP, Muller G. 1977. Preparation of intermediates in cobyrinic acid biosynthesis by suspensions of Propionibacterium shermanii. Hoppe Seylers Z Physiol Chem 358:1315–1323. doi: 10.1515/bchm2.1977.358.2.1315. [DOI] [PubMed] [Google Scholar]
- 168.Vevodova J, Graham RM, Raux E, Schubert HL, Roper DI, Brindley AA, Scott AI, Roessner CA, Stamford NP, Stroupe ME, Getzoff ED, Warren MJ, Wilson KS. 2004. Structure/function studies on a S-adenosyl-l-methionine-dependent uroporphyrinogen III C methyltransferase (SUMT), a key regulatory enzyme of tetrapyrrole biosynthesis. J Mol Biol 344:419–433. doi: 10.1016/j.jmb.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 169.Stroupe ME, Leech HK, Daniels DS, Warren MJ, Getzoff ED. 2003. CysG structure reveals tetrapyrrole-binding features and novel regulation of siroheme biosynthesis. Nat Struct Biol 10:1064–1073. doi: 10.1038/nsb1007. [DOI] [PubMed] [Google Scholar]
- 170.Warren MJ, Roessner CA, Santander PJ, Scott AI. 1990. The Escherichia coli cysG gene encodes S-adenosylmethionine-dependent uroporphyrinogen III methylase. Biochem J 265:725–729. doi: 10.1042/bj2650725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Deeg R, Kriemler HP, Bergmann KH, Muller G. 1977. Cobyrinic acid biosynthesis—novel, methylated hydroporphyrins and their role in cobyrinic acid formation. Hoppe Seylers Z Physiol Chem 358:339–352. doi: 10.1515/bchm2.1977.358.1.339. [DOI] [PubMed] [Google Scholar]
- 172.Mucha H, Keller E, Weber H, Lingens F, Trosch W. 1985. Sirohydrochlorin, a precursor of factor F430 biosynthesis in Methanobacterium thermoautotrophicum. FEBS Lett 190:169–171. doi: 10.1016/0014-5793(85)80451-8. [DOI] [Google Scholar]
- 173.Storbeck S, Walther J, Muller J, Parmar V, Schiebel HM, Kemken D, Dulcks T, Warren MJ, Layer G. 2009. The Pseudomonas aeruginosa nirE gene encodes the S-adenosyl-l-methionine-dependent uroporphyrinogen III methyltransferase required for heme d(1) biosynthesis. FEBS J 276:5973–5982. doi: 10.1111/j.1742-4658.2009.07306.x. [DOI] [PubMed] [Google Scholar]
- 174.Zajicek RS, Bali S, Arnold S, Brindley AA, Warren MJ, Ferguson SJ. 2009. d(1) haem biogenesis—assessing the roles of three nir gene products. FEBS J 276:6399–6411. doi: 10.1111/j.1742-4658.2009.07354.x. [DOI] [PubMed] [Google Scholar]
- 175.Bollivar DW, Elliott T, Beale SI. 1995. Anaerobic protoporphyrin biosynthesis does not require incorporation of methyl groups from methionine. J Bacteriol 177:5778–5783. doi: 10.1128/jb.177.20.5778-5783.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Buchenau B, Kahnt J, Heinemann IU, Jahn D, Thauer RK. 2006. Heme biosynthesis in Methanosarcina barkeri via a pathway involving two methylation reactions. J Bacteriol 188:8666–8668. doi: 10.1128/JB.01349-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lobo SA, Brindley A, Warren MJ, Saraiva LM. 2009. Functional characterization of the early steps of tetrapyrrole biosynthesis and modification in Desulfovibrio vulgaris Hildenborough. Biochem J 420:317–325. doi: 10.1042/BJ20090151. [DOI] [PubMed] [Google Scholar]
- 178.Storbeck S, Rolfes S, Raux-Deery E, Warren MJ, Jahn D, Layer G. 2010. A novel pathway for the biosynthesis of heme in Archaea: genome-based bioinformatic predictions and experimental evidence. Archaea 2010:175050. doi: 10.1155/2010/175050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ferguson SJ, Fulop V. 2000. Cytochrome cd1 nitrite reductase structure raises interesting mechanistic questions. Subcell Biochem 35:519–540. [DOI] [PubMed] [Google Scholar]
- 180.Kawasaki S, Arai H, Kodama T, Igarashi Y. 1997. Gene cluster for dissimilatory nitrite reductase (nir) from Pseudomonas aeruginosa: sequencing and identification of a locus for heme d1 biosynthesis. J Bacteriol 179:235–242. doi: 10.1128/jb.179.1.235-242.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Palmedo G, Seither P, Korner H, Matthews JC, Burkhalter RS, Timkovich R, Zumft WG. 1995. Resolution of the nirD locus for heme d1 synthesis of cytochrome cd1 (respiratory nitrite reductase) from Pseudomonas stutzeri. Eur J Biochem 232:737–746. doi: 10.1111/j.1432-1033.1995.tb20868.x. [DOI] [PubMed] [Google Scholar]
- 182.Yap-Bondoc F, Bondoc LL, Timkovich R, Baker DC, Hebbler A. 1990. C-methylation occurs during the biosynthesis of heme d1. J Biol Chem 265:13498–13500. [PubMed] [Google Scholar]
- 183.Raux E, Leech HK, Beck R, Schubert HL, Santander PJ, Roessner CA, Scott AI, Martens JH, Jahn D, Thermes C, Rambach A, Warren MJ. 2003. Identification and functional analysis of enzymes required for precorrin-2 dehydrogenation and metal ion insertion in the biosynthesis of sirohaem and cobalamin in Bacillus megaterium. Biochem J 370:505–516. doi: 10.1042/bj20021443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Brindley AA, Raux E, Leech HK, Schubert HL, Warren MJ. 2003. A story of chelatase evolution: identification and characterization of a small 13-15-kDa “ancestral” cobaltochelatase (CbiXS) in the Archaea. J Biol Chem 278:22388–22395. doi: 10.1074/jbc.M302468200. [DOI] [PubMed] [Google Scholar]
- 185.Romao CV, Ladakis D, Lobo SA, Carrondo MA, Brindley AA, Deery E, Matias PM, Pickersgill RW, Saraiva LM, Warren MJ. 2011. Evolution in a family of chelatases facilitated by the introduction of active site asymmetry and protein oligomerization. Proc Natl Acad Sci U S A 108:97–102. doi: 10.1073/pnas.1014298108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Yin J, Xu LX, Cherney MM, Raux-Deery E, Bindley AA, Savchenko A, Walker JR, Cuff ME, Warren MJ, James MN. 2006. Crystal structure of the vitamin B12 biosynthetic cobaltochelatase, CbiXS, from Archaeoglobus fulgidus. J Struct Funct Genomics 7:37–50. doi: 10.1007/s10969-006-9008-x. [DOI] [PubMed] [Google Scholar]
- 187.Brindley AA, Zajicek R, Warren MJ, Ferguson SJ, Rigby SE. 2010. NirJ, a radical SAM family member of the d1 heme biogenesis cluster. FEBS Lett 584:2461–2466. doi: 10.1016/j.febslet.2010.04.053. [DOI] [PubMed] [Google Scholar]
- 188.Adamczack J, Hoffmann M, Papke U, Haufschildt K, Nicke T, Broring M, Sezer M, Weimar R, Kuhlmann U, Hildebrandt P, Layer G. 2014. NirN protein from Pseudomonas aeruginosa is a novel electron-bifurcating dehydrogenase catalyzing the last step of heme d1 biosynthesis. J Biol Chem 289:30753–30762. doi: 10.1074/jbc.M114.603886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Nicke T, Schnitzer T, Munch K, Adamczack J, Haufschildt K, Buchmeier S, Kucklick M, Felgentrager U, Jansch L, Riedel K, Layer G. 2013. Maturation of the cytochrome cd1 nitrite reductase NirS from Pseudomonas aeruginosa requires transient interactions between the three proteins NirS, NirN and NirF. Biosci Rep 33:e00048. doi: 10.1042/BSR20130043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Bali S, Warren MJ, Ferguson SJ. 2010. NirF is a periplasmic protein that binds d(1) heme as part of its essential role in d(1) heme biogenesis. FEBS J 277:4944–4955. doi: 10.1111/j.1742-4658.2010.07899.x. [DOI] [PubMed] [Google Scholar]
- 191.Haufschildt K, Schmelz S, Kriegler TM, Neumann A, Streif J, Arai H, Heinz DW, Layer G. 2014. The crystal structure of siroheme decarboxylase in complex with iron-uroporphyrin III reveals two essential histidine residues. J Mol Biol 426:3272–3286. doi: 10.1016/j.jmb.2014.07.021. [DOI] [PubMed] [Google Scholar]
- 192.Palmer DJ, Schroeder S, Lawrence AD, Deery E, Lobo SA, Saraiva LM, McLean KJ, Munro AW, Ferguson SJ, Pickersgill RW, Brown DG, Warren MJ. 2014. The structure, function and properties of sirohaem decarboxylase—an enzyme with structural homology to a transcription factor family that is part of the alternative haem biosynthesis pathway. Mol Microbiol 93:247–261. doi: 10.1111/mmi.12656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Jordan PM. 1990. The biosynthesis of 5-aminolevulic acid and its transformation into coproporphyrinogen in animals and bacteria, p 55–122. In Dailey HA. (ed), Biosynthesis of heme and chlorophylls. McGraw-Hill, New York, NY. [Google Scholar]
- 194.Karim Z, Lyoumi S, Nicolas G, Deybach JC, Gouya L, Puy H. 2015. Porphyrias: a 2015 update. Clin Res Hepatol Gastroenterol 39:412–425. doi: 10.1016/j.clinre.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 195.Jackson AH, Sancovich HA, Ferramola AM, Evans N, Games DE, Matlin SA, Elder GH, Smith SG. 1976. Macrocyclic intermediates in the biosynthesis of porphyrins. Philos Trans R Soc Lond B Biol Sci 273:191–206. doi: 10.1098/rstb.1976.0009. [DOI] [PubMed] [Google Scholar]
- 196.Akhtar M. 1991. Mechanism and stereochemistry of the enzymes involved in the conversion of uroporphyrinogen III into haem, p 67–100. In Jordan PM. (ed), Biosynthesis of tetrapyrroles, vol 19 Elsevier, Amsterdam, Netherlands. [Google Scholar]
- 197.Ineichen G, Biel AJ. 1995. Nucleotide sequence of the Rhodobacter capsulatus hemE gene. Plant Physiol 108:423. doi: 10.1104/pp.108.1.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Fan J, Liu Q, Hao Q, Teng M, Niu L. 2007. Crystal structure of uroporphyrinogen decarboxylase from Bacillus subtilis. J Bacteriol 189:3573–3580. doi: 10.1128/JB.01083-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Baugh L, Gallagher LA, Patrapuvich R, Clifton MC, Gardberg AS, Edwards TE, Armour B, Begley DW, Dieterich SH, Dranow DM, Abendroth J, Fairman JW, Fox D III, Staker BL, Phan I, Gillespie A, Choi R, Nakazawa-Hewitt S, Nguyen MT, Napuli A, Barrett L, Buchko GW, Stacy R, Myler PJ, Stewart LJ, Manoil C, Van Voorhis WC. 2013. Combining functional and structural genomics to sample the essential Burkholderia structome. PLoS One 8:e53851. doi: 10.1371/journal.pone.0053851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Phillips JD, Whitby FG, Kushner JP, Hill CP. 1997. Characterization and crystallization of human uroporphyrinogen decarboxylase. Protein Sci 6:1343–1346. doi: 10.1002/pro.5560060624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Whitby FG, Phillips JD, Kushner JP, Hill CP. 1998. Crystal structure of human uroporphyrinogen decarboxylase. EMBO J 17:2463–2471. doi: 10.1093/emboj/17.9.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Martins BM, Grimm B, Mock HP, Huber R, Messerschmidt A. 2001. Crystal structure and substrate binding modeling of the uroporphyrinogen-III decarboxylase from Nicotiana tabacum. Implications for the catalytic mechanism. J Biol Chem 276:44108–44116. doi: 10.1074/jbc.M104759200. [DOI] [PubMed] [Google Scholar]
- 203.Phillips JD, Whitby FG, Kushner JP, Hill CP. 2003. Structural basis for tetrapyrrole coordination by uroporphyrinogen decarboxylase. EMBO J 22:6225–6233. doi: 10.1093/emboj/cdg606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Lewis CA Jr, Wolfenden R. 2008. Uroporphyrinogen decarboxylation as a benchmark for the catalytic proficiency of enzymes. Proc Natl Acad Sci U S A 105:17328–17333. doi: 10.1073/pnas.0809838105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Phillips JD, Warby CA, Whitby FG, Kushner JP, Hill CP. 2009. Substrate shuttling between active sites of uroporphyrinogen decarboxylase is not required to generate coproporphyrinogen. J Mol Biol 389:306–314. doi: 10.1016/j.jmb.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Corrigall AV, Siziba KB, Maneli MH, Shephard EG, Ziman M, Dailey TA, Dailey HA, Kirsch RE, Meissner PN. 1998. Purification of and kinetic studies on a cloned protoporphyrinogen oxidase from the aerobic bacterium Bacillus subtilis. Arch Biochem Biophys 358:251–256. doi: 10.1006/abbi.1998.0834. [DOI] [PubMed] [Google Scholar]
- 207.Dailey TA, Meissner P, Dailey HA. 1994. Expression of a cloned protoporphyrinogen oxidase. J Biol Chem 269:813–815. [PubMed] [Google Scholar]
- 208.Hansson M, Hederstedt L. 1994. Bacillus subtilis HemY is a peripheral membrane protein essential for protoheme IX synthesis which can oxidize coproporphyrinogen III and protoporphyrinogen IX. J Bacteriol 176:5962–5970. doi: 10.1128/jb.176.19.5962-5970.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Qin X, Sun L, Wen X, Yang X, Tan Y, Jin H, Cao Q, Zhou W, Xi Z, Shen Y. 2010. Structural insight into unique properties of protoporphyrinogen oxidase from Bacillus subtilis. J Struct Biol 170:76–82. doi: 10.1016/j.jsb.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 210.Corradi HR, Corrigall AV, Boix E, Mohan CG, Sturrock ED, Meissner PN, Acharya KR. 2006. Crystal structure of protoporphyrinogen oxidase from Myxococcus xanthus and its complex with the inhibitor acifluorfen. J Biol Chem 281:38625–38633. doi: 10.1074/jbc.M606640200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Wang B, Wen X, Qin X, Wang Z, Tan Y, Shen Y, Xi Z. 2013. Quantitative structural insight into human variegate porphyria disease. J Biol Chem 288:11731–11740. doi: 10.1074/jbc.M113.459768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Koch M, Breithaupt C, Kiefersauer R, Freigang J, Huber R, Messerschmidt A. 2004. Crystal structure of protoporphyrinogen IX oxidase: a key enzyme in haem and chlorophyll biosynthesis. EMBO J 23:1720–1728. doi: 10.1038/sj.emboj.7600189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Schubert HL, Raux E, Wilson KS, Warren MJ. 1999. Common chelatase design in the branched tetrapyrrole pathways of heme and anaerobic cobalamin synthesis. Biochemistry 38:10660–10669. doi: 10.1021/bi9906773. [DOI] [PubMed] [Google Scholar]
- 214.Al-Karadaghi S, Franco R, Hansson M, Shelnutt JA, Isaya G, Ferreira GC. 2006. Chelatases: distort to select? Trends Biochem Sci 31:135–142. doi: 10.1016/j.tibs.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Dailey HA, Dailey TA. 2003. Ferrochelatase, p 93–122. In Kadish KM, Smith KM, Guilard R (ed), The porphyrin handbook, vol 12 Academic Press, San Diego, CA. [Google Scholar]
- 216.Dailey HA, Dailey TA, Wu CK, Medlock AE, Wang KF, Rose JP, Wang BC. 2000. Ferrochelatase at the millennium: structures, mechanisms and [2Fe-2S] clusters. Cell Mol Life Sci 57:1909–1926. doi: 10.1007/PL00000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Lanzilotta WN, Dailey HA. 2011. Human ferrochelatase, p 138–146. In Messerschmidt A. (ed), Handbook of metalloproteins, vol 5 and 6 John Wiley & Sons, Chichester, United Kingdom. [Google Scholar]
- 218.Dailey HA, Wu CK, Horanyi P, Medlock AE, Najahi-Missaoui W, Burden AE, Dailey TA, Rose J. 2007. Altered orientation of active site residues in variants of human ferrochelatase. Evidence for a hydrogen bond network involved in catalysis. Biochemistry 46:7973–7979. doi: 10.1021/bi700151f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Medlock A, Swartz L, Dailey TA, Dailey HA, Lanzilotta WN. 2007. Substrate interactions with human ferrochelatase. Proc Natl Acad Sci U S A 104:1789–1793. doi: 10.1073/pnas.0606144104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Medlock AE, Carter M, Dailey TA, Dailey HA, Lanzilotta WN. 2009. Product release rather than chelation determines metal specificity for ferrochelatase. J Mol Biol 393:308–319. doi: 10.1016/j.jmb.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Medlock AE, Dailey TA, Ross TA, Dailey HA, Lanzilotta WN. 2007. A pi-helix switch selective for porphyrin deprotonation and product release in human ferrochelatase. J Mol Biol 373:1006–1016. doi: 10.1016/j.jmb.2007.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Al-Karadaghi S, Hansson M, Nikonov S, Jonsson B, Hederstedt L. 1997. Crystal structure of ferrochelatase: the terminal enzyme in heme biosynthesis. Structure 5:1501–1510. doi: 10.1016/S0969-2126(97)00299-2. [DOI] [PubMed] [Google Scholar]
- 223.Hansson MD, Karlberg T, Rahardja MA, Al-Karadaghi S, Hansson M. 2007. Amino acid residues His183 and Glu264 in Bacillus subtilis ferrochelatase direct and facilitate the insertion of metal ion into protoporphyrin IX. Biochemistry 46:87–94. doi: 10.1021/bi061760a. [DOI] [PubMed] [Google Scholar]
- 224.Karlberg T, Hansson MD, Yengo RK, Johansson R, Thorvaldsen HO, Ferreira GC, Hansson M, Al-Karadaghi S. 2008. Porphyrin binding and distortion and substrate specificity in the ferrochelatase reaction: the role of active site residues. J Mol Biol 378:1074–1083. doi: 10.1016/j.jmb.2008.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Lecerof D, Fodje M, Hansson A, Hansson M, Al-Karadaghi S. 2000. Structural and mechanistic basis of porphyrin metallation by ferrochelatase. J Mol Biol 297:221–232. doi: 10.1006/jmbi.2000.3569. [DOI] [PubMed] [Google Scholar]
- 226.Olsson U, Billberg A, Sjovall S, Al-Karadaghi S, Hansson M. 2002. In vivo and in vitro studies of Bacillus subtilis ferrochelatase mutants suggest substrate channeling in the heme biosynthesis pathway. J Bacteriol 184:4018–4024. doi: 10.1128/JB.184.14.4018-4024.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Shipovskov S, Karlberg T, Fodje M, Hansson MD, Ferreira GC, Hansson M, Reimann CT, Al-Karadaghi S. 2005. Metallation of the transition-state inhibitor N-methyl mesoporphyrin by ferrochelatase: implications for the catalytic reaction mechanism. J Mol Biol 352:1081–1090. doi: 10.1016/j.jmb.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 228.Shepherd M, Dailey TA, Dailey HA. 2006. A new class of [2Fe-2S]-cluster-containing protoporphyrin (IX) ferrochelatases. Biochem J 397:47–52. doi: 10.1042/BJ20051967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Lecerof D, Fodje MN, Alvarez Leon R, Olsson U, Hansson A, Sigfridsson E, Ryde U, Hansson M, Al-Karadaghi S. 2003. Metal binding to Bacillus subtilis ferrochelatase and interaction between metal sites. J Biol Inorg Chem 8:452–458. [DOI] [PubMed] [Google Scholar]
- 230.Hofbauer S, Mlynek G, Milazzo L, Puhringer D, Maresch D, Schaffner I, Furtmuller PG, Smulevich G, Djinovic-Carugo K, Obinger C. 14 November 2016. Hydrogen peroxide-mediated conversion of coproheme to heme b by HemQ—lessons from the first crystal structure and kinetic studies. FEBS J doi: 10.1111/febs.13930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Celis AI, Streit BR, Moraski GC, Kant R, Lash TD, Lukat-Rodgers GS, Rodgers KR, DuBois JL. 2015. Unusual peroxide-dependent, heme-transforming reaction catalyzed by HemQ. Biochemistry 54:4022–4032. doi: 10.1021/acs.biochem.5b00492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.del Batlle AM, Benson A, Rimington C. 1965. Purification and properties of coproporphyrinogenase. Biochem J 97:731–740. doi: 10.1042/bj0970731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Porra RJ, Falk JE. 1964. The enzymic conversion of coproporphyrinogen 3 into protoporphyrin 9. Biochem J 90:69–75. doi: 10.1042/bj0900069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Sano S, Granick S. 1961. Mitochondrial coproporphyrinogen oxidase and protoporphyrin formation. J Biol Chem 236:1173–1180. [PubMed] [Google Scholar]
- 235.Ehteshamuddin AF. 1968. Anaerobic formation of protoporphyrin IX from coproporphyrinogen III by bacterial preparations. Biochem J 107:446–447. doi: 10.1042/bj1070446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Cavallaro G, Decaria L, Rosato A. 2008. Genome-based analysis of heme biosynthesis and uptake in prokaryotic systems. J Proteome Res 7:4946–4954. doi: 10.1021/pr8004309. [DOI] [PubMed] [Google Scholar]
- 237.Cavaleiro JAS, Kenner GW, Smith KM. 1974. Pyrroles and related compounds. Part XXXII. Biosynthesis of protoporphyrin-IX from coproporphyrinogen-III. J Chem Soc Perkin Trans 1:1188–1194. [DOI] [PubMed] [Google Scholar]
- 238.Elder GH, Evans JO, Jackson JR, Jackson AH. 1978. Factors determining the sequence of oxidative decarboxylation of the 2- and 4-propionate substituents of coproporphyrinogen III by coproporphyrinogen oxidase in rat liver. Biochem J 169:215–223. doi: 10.1042/bj1690215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Rand K, Noll C, Schiebel HM, Kemken D, Dulcks T, Kalesse M, Heinz DW, Layer G. 2010. The oxygen-independent coproporphyrinogen III oxidase HemN utilizes harderoporphyrinogen as a reaction intermediate during conversion of coproporphyrinogen III to protoporphyrinogen IX. Biol Chem 391:55–63. doi: 10.1515/BC.2010.006. [DOI] [PubMed] [Google Scholar]
- 240.Breckau D, Mahlitz E, Sauerwald A, Layer G, Jahn D. 2003. Oxygen-dependent coproporphyrinogen III oxidase (HemF) from Escherichia coli is stimulated by manganese. J Biol Chem 278:46625–46631. doi: 10.1074/jbc.M308553200. [DOI] [PubMed] [Google Scholar]
- 241.Kohno H, Furukawa T, Tokunaga R, Taketani S, Yoshinaga T. 1996. Mouse coproporphyrinogen oxidase is a copper-containing enzyme: expression in Escherichia coli and site-directed mutagenesis. Biochim Biophys Acta 1292:156–162. doi: 10.1016/0167-4838(95)00188-3. [DOI] [PubMed] [Google Scholar]
- 242.Labbe P. 1997. Purification and properties of coproporphyrinogen III oxidase from yeast. Methods Enzymol 281:367–378. doi: 10.1016/S0076-6879(97)81044-0. [DOI] [PubMed] [Google Scholar]
- 243.Poulson R, Polglase WJ. 1974. Aerobic and anaerobic coproporphyrinogenase activities in extracts from Saccharomyces cerevisiae. J Biol Chem 249:6367–6371. [PubMed] [Google Scholar]
- 244.Yoshinaga T, Sano S. 1980. Coproporphyrinogen oxidase. I. Purification, properties, and activation by phospholipids. J Biol Chem 255:4722–4726. [PubMed] [Google Scholar]
- 245.Battersby AR, McDonald E, Wurziger HKW, James KJ. 1975. Stereochemistry of biosynthesis of the vinyl groups of protoporphyrin-IX: a short synthesis of porphobilinogen. J Chem Soc Chem Commun 1975:493–494. [Google Scholar]
- 246.Lash TD. 2005. The enigma of coproporphyrinogen oxidase: how does this unusual enzyme carry out oxidative decarboxylations to afford vinyl groups? Bioorg Med Chem Lett 15:4506–4509. doi: 10.1016/j.bmcl.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 247.Seehra JS, Jordan PM, Akhtar M. 1983. Anaerobic and aerobic coproporphyrinogen III oxidases of Rhodopseudomonas spheroides. Mechanism and stereochemistry of vinyl group formation. Biochem J 209:709–718. doi: 10.1042/bj2090709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Zaman Z, Akhtar M. 1976. Mechanism and stereochemistry of vinyl-group formation in haem biosynthesis. Eur J Biochem 61:215–223. doi: 10.1111/j.1432-1033.1976.tb10014.x. [DOI] [PubMed] [Google Scholar]
- 249.Lee DS, Flachsova E, Bodnarova M, Demeler B, Martasek P, Raman CS. 2005. Structural basis of hereditary coproporphyria. Proc Natl Acad Sci U S A 102:14232–14237. doi: 10.1073/pnas.0506557102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Phillips JD, Whitby FG, Warby CA, Labbe P, Yang C, Pflugrath JW, Ferrara JD, Robinson H, Kushner JP, Hill CP. 2004. Crystal structure of the oxygen-dependent coproporphyrinogen oxidase (Hem13p) of Saccharomyces cerevisiae. J Biol Chem 279:38960–38968. doi: 10.1074/jbc.M406050200. [DOI] [PubMed] [Google Scholar]
- 251.Stephenson JR, Stacey JA, Morgenthaler JB, Friesen JA, Lash TD, Jones MA. 2007. Role of aspartate 400, arginine 262, and arginine 401 in the catalytic mechanism of human coproporphyrinogen oxidase. Protein Sci 16:401–410. doi: 10.1110/ps.062636907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Tait GH. 1969. Coproporphyrinogenase activity in extracts from Rhodopseudomonas spheroides. Biochem Biophys Res Commun 37:116–122. doi: 10.1016/0006-291X(69)90888-2. [DOI] [PubMed] [Google Scholar]
- 253.Tait GH. 1972. Coproporphyrinogenase activities in extracts of Rhodopseudomonas spheroides and Chromatium strain D. Biochem J 128:1159–1169. doi: 10.1042/bj1281159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Coomber SA, Jones RM, Jordan PM, Hunter CN. 1992. A putative anaerobic coproporphyrinogen III oxidase in Rhodobacter sphaeroides. I. Molecular cloning, transposon mutagenesis and sequence analysis of the gene. Mol Microbiol 6:3159–3169. doi: 10.1111/j.1365-2958.1992.tb01772.x. [DOI] [PubMed] [Google Scholar]
- 255.Layer G, Grage K, Teschner T, Schunemann V, Breckau D, Masoumi A, Jahn M, Heathcote P, Trautwein AX, Jahn D. 2005. Radical S-adenosylmethionine enzyme coproporphyrinogen III oxidase HemN: functional features of the [4Fe-4S] cluster and the two bound S-adenosyl-l-methionines. J Biol Chem 280:29038–29046. doi: 10.1074/jbc.M501275200. [DOI] [PubMed] [Google Scholar]
- 256.Layer G, Moser J, Heinz DW, Jahn D, Schubert WD. 2003. Crystal structure of coproporphyrinogen III oxidase reveals cofactor geometry of radical SAM enzymes. EMBO J 22:6214–6224. doi: 10.1093/emboj/cdg598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Layer G, Pierik AJ, Trost M, Rigby SE, Leech HK, Grage K, Breckau D, Astner I, Jansch L, Heathcote P, Warren MJ, Heinz DW, Jahn D. 2006. The substrate radical of Escherichia coli oxygen-independent coproporphyrinogen III oxidase HemN. J Biol Chem 281:15727–15734. doi: 10.1074/jbc.M512628200. [DOI] [PubMed] [Google Scholar]
- 258.Layer G, Verfurth K, Mahlitz E, Jahn D. 2002. Oxygen-independent coproporphyrinogen-III oxidase HemN from Escherichia coli. J Biol Chem 277:34136–34142. doi: 10.1074/jbc.M205247200. [DOI] [PubMed] [Google Scholar]
- 259.Troup B, Hungerer C, Jahn D. 1995. Cloning and characterization of the Escherichia coli hemN gene encoding the oxygen-independent coproporphyrinogen III oxidase. J Bacteriol 177:3326–3331. doi: 10.1128/jb.177.11.3326-3331.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Xu K, Elliott T. 1994. Cloning, DNA sequence, and complementation analysis of the Salmonella typhimurium hemN gene encoding a putative oxygen-independent coproporphyrinogen III oxidase. J Bacteriol 176:3196–3203. doi: 10.1128/jb.176.11.3196-3203.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Dailey HA, Dailey TA. 1996. Protoporphyrinogen oxidase of Myxococcus xanthus. Expression, purification, and characterization of the cloned enzyme. J Biol Chem 271:8714–8718. doi: 10.1074/jbc.271.15.8714. [DOI] [PubMed] [Google Scholar]
- 262.Dailey TA, Dailey HA. 1998. Identification of an FAD superfamily containing protoporphyrinogen oxidases, monoamine oxidases, and phytoene desaturase. Expression and characterization of phytoene desaturase of Myxococcus xanthus. J Biol Chem 273:13658–13662. doi: 10.1074/jbc.273.22.13658. [DOI] [PubMed] [Google Scholar]
- 263.Wang KF, Dailey TA, Dailey HA. 2001. Expression and characterization of the terminal heme synthetic enzymes from the hyperthermophile Aquifex aeolicus. FEMS Microbiol Lett 202:115–119. doi: 10.1111/j.1574-6968.2001.tb10789.x. [DOI] [PubMed] [Google Scholar]
- 264.Heinemann IU, Diekmann N, Masoumi A, Koch M, Messerschmidt A, Jahn M, Jahn D. 2007. Functional definition of the tobacco protoporphyrinogen IX oxidase substrate-binding site. Biochem J 402:575–580. doi: 10.1042/BJ20061321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Akhtar M. 2003. Coproporphyrinogen III and protoporphyrinogen IX oxidase, p 75–92. In Kadish KM, Smith KM, Guilard R (ed), The porphyrin handbook, vol 12 Academic Press, San Diego, CA. [Google Scholar]
- 266.Jacobs JM, Jacobs NJ. 1977. The late steps of anaerobic heme biosynthesis in E. coli: role for quinones in protoporphyrinogen oxidation. Biochem Biophys Res Commun 78:429–433. doi: 10.1016/0006-291X(77)91272-4. [DOI] [PubMed] [Google Scholar]
- 267.Jacobs NJ, Jacobs JM. 1975. Fumarate as alternate electron acceptor for the late steps of anaerobic heme synthesis in Escherichia coli. Biochem Biophys Res Commun 65:435–441. doi: 10.1016/S0006-291X(75)80112-4. [DOI] [PubMed] [Google Scholar]
- 268.Jacobs NJ, Jacobs JM. 1977. Evidence for involvement of the electron transport system at a late step of anaerobic microbial heme synthesis. Biochim Biophys Acta 459:141–144. doi: 10.1016/0005-2728(77)90017-2. [DOI] [PubMed] [Google Scholar]
- 269.Jacobs NJ, Jacobs JM. 1978. Quinones as hydrogen carriers for a late step in anaerobic heme biosynthesis in Escherichia coli. Biochim Biophys Acta 544:540–546. doi: 10.1016/0304-4165(78)90328-8. [DOI] [PubMed] [Google Scholar]
- 270.Jacobs NJ, Jacobs JM, Brent P. 1970. Formation of protoporphyrin from coproporphyrinogen in extracts of various bacteria. J Bacteriol 102:398–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Jacobs NJ, Jacobs JM, Brent P. 1971. Characterization of the late steps of microbial heme synthesis: conversion of coproporphyrinogen to protoporphyrin. J Bacteriol 107:203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Jacobs NJ, Jacobs JM, Morgan HE Jr. 1972. Comparative effect of oxygen and nitrate on protoporphyrin and heme synthesis from delta-amino levulinic acid in bacterial cultures. J Bacteriol 112:1444–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Sasarman A, Letowski J, Czaika G, Ramirez V, Nead MA, Jacobs JM, Morais R. 1993. Nucleotide sequence of the hemG gene involved in the protoporphyrinogen oxidase activity of Escherichia coli K12. Can J Microbiol 39:1155–1161. doi: 10.1139/m93-174. [DOI] [PubMed] [Google Scholar]
- 274.Boynton TO, Daugherty LE, Dailey TA, Dailey HA. 2009. Identification of Escherichia coli HemG as a novel, menadione-dependent flavodoxin with protoporphyrinogen oxidase activity. Biochemistry 48:6705–6711. doi: 10.1021/bi900850y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Goni G, Serrano A, Frago S, Hervas M, Peregrina JR, De la Rosa MA, Gomez-Moreno C, Navarro JA, Medina M. 2008. Flavodoxin-mediated electron transfer from photosystem I to ferredoxin-NADP+ reductase in Anabaena: role of flavodoxin hydrophobic residues in protein-protein interactions. Biochemistry 47:1207–1217. doi: 10.1021/bi7017392. [DOI] [PubMed] [Google Scholar]
- 276.Hoover DM, Drennan CL, Metzger AL, Osborne C, Weber CH, Pattridge KA, Ludwig ML. 1999. Comparisons of wild-type and mutant flavodoxins from Anacystis nidulans. Structural determinants of the redox potentials. J Mol Biol 294:725–743. doi: 10.1006/jmbi.1999.3152. [DOI] [PubMed] [Google Scholar]
- 277.Mobius K, Arias-Cartin R, Breckau D, Hannig AL, Riedmann K, Biedendieck R, Schroder S, Becher D, Magalon A, Moser J, Jahn M, Jahn D. 2010. Heme biosynthesis is coupled to electron transport chains for energy generation. Proc Natl Acad Sci U S A 107:10436–10441. doi: 10.1073/pnas.1000956107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Kato K, Tanaka R, Sano S, Tanaka A, Hosaka H. 2010. Identification of a gene essential for protoporphyrinogen IX oxidase activity in the cyanobacterium Synechocystis sp. PCC6803. Proc Natl Acad Sci U S A 107:16649–16654. doi: 10.1073/pnas.1000771107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Boynton TO, Gerdes S, Craven SH, Neidle EL, Phillips JD, Dailey HA. 2011. Discovery of a gene involved in a third bacterial protoporphyrinogen oxidase activity through comparative genomic analysis and functional complementation. Appl Environ Microbiol 77:4795–4801. doi: 10.1128/AEM.00171-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Jones MS, Jones OT. 1969. The structural organization of haem synthesis in rat liver mitochondria. Biochem J 113:507–514. doi: 10.1042/bj1130507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Neuberger A, Tait GH. 1964. Studies on the biosynthesis of porphyrin and bacteriochlorophyll by Rhodopseudomonas spheroides. 5. Zinc-protoporphyrin chelatase. Biochem J 90:607–616. doi: 10.1042/bj0900607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Porra RJ, Lascelles J. 1965. Haemoproteins and haem synthesis in facultative photosynthetic and denitrifying bacteria. Biochem J 94:120–126. doi: 10.1042/bj0940120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Porra RJ, Ross BD. 1965. Haem synthase and cobalt porphyrin synthase in various micro-organisms. Biochem J 94:557–562. doi: 10.1042/bj0940557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Barrett J, Jones OT. 1978. Localization of ferrochelatase and of newly synthesized haem in membrane fractions from Rhodopseudomonas spheroides. Biochem J 174:277–281. doi: 10.1042/bj1740277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Jones MS, Jones OT. 1970. Ferrochelatase of Rhodopseudomonas spheroides. Biochem J 119:453–462. doi: 10.1042/bj1190453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Kassner RJ, Walchak H. 1973. Heme formation from Fe(II) and porphyrin in the absence of ferrochelatase activity. Biochim Biophys Acta 304:294–303. doi: 10.1016/0304-4165(73)90247-X. [DOI] [PubMed] [Google Scholar]
- 287.Tokunaga R, Sano S. 1972. Comparative studies on nonenzymic and enzymic protoheme formation. Biochim Biophys Acta 264:263–271. doi: 10.1016/0304-4165(72)90290-5. [DOI] [PubMed] [Google Scholar]
- 288.Dailey HA Jr, Lascelles J. 1974. Ferrochelatase activity in wild-type and mutant strains of Spirillum itersonii. Solubilization with chaotropic reagents. Arch Biochem Biophys 160:523–529. doi: 10.1016/0003-9861(74)90429-9. [DOI] [PubMed] [Google Scholar]
- 289.Dailey HA. 1982. Purification and characterization of membrane-bound ferrochelatase from Rhodopseudomonas sphaeroides. J Biol Chem 257:14714–14718. [PubMed] [Google Scholar]
- 290.Dailey HA, Fleming JE, Harbin BM. 1986. Ferrochelatase from Rhodopseudomonas sphaeroides: substrate specificity and role of sulfhydryl and arginyl residues. J Bacteriol 165:1–5. doi: 10.1128/jb.165.1.1-5.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Dailey HA., Jr 1977. Purification and characterization of the membrane-bound ferrochelatase from Spirillum itersonii. J Bacteriol 132:302–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Frustaci JM, O'Brian MR. 1992. Characterization of a Bradyrhizobium japonicum ferrochelatase mutant and isolation of the hemH gene. J Bacteriol 174:4223–4229. doi: 10.1128/jb.174.13.4223-4229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Dailey TA, Dailey HA. 2002. Identification of [2Fe-2S] clusters in microbial ferrochelatases. J Bacteriol 184:2460–2464. doi: 10.1128/JB.184.9.2460-2464.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Hoggins M, Dailey HA, Hunter CN, Reid JD. 2007. Direct measurement of metal ion chelation in the active site of human ferrochelatase. Biochemistry 46:8121–8127. doi: 10.1021/bi602418e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Dailey HA Jr, Lascelles J. 1977. Reduction of iron and synthesis of protoheme by Spirillum itersonii and other organisms. J Bacteriol 129:815–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Moody MD, Dailey HA. 1984. Siderophore utilization and iron uptake by Rhodopseudomonas sphaeroides. Arch Biochem Biophys 234:178–186. doi: 10.1016/0003-9861(84)90339-4. [DOI] [PubMed] [Google Scholar]
- 297.Moody MD, Dailey HA. 1985. Ferric iron reductase of Rhodopseudomonas sphaeroides. J Bacteriol 163:1120–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Chen W, Dailey HA, Paw BH. 2010. Ferrochelatase forms an oligomeric complex with mitoferrin-1 and Abcb10 for erythroid heme biosynthesis. Blood 116:628–630. doi: 10.1182/blood-2009-12-259614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Ferreira GC, Andrew TL, Karr SW, Dailey HA. 1988. Organization of the terminal two enzymes of the heme biosynthetic pathway. Orientation of protoporphyrinogen oxidase and evidence for a membrane complex. J Biol Chem 263:3835–3839. [PubMed] [Google Scholar]
- 300.Medlock AE, Shiferaw MT, Marcero JR, Vashisht AA, Wohlschlegel JA, Phillips JD, Dailey HA. 2015. Identification of the mitochondrial heme metabolism complex. PLoS One 10:e0135896. doi: 10.1371/journal.pone.0135896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Proulx KL, Woodard SI, Dailey HA. 1993. In situ conversion of coproporphyrinogen to heme by murine mitochondria: terminal steps of the heme biosynthetic pathway. Protein Sci 2:1092–1098. doi: 10.1002/pro.5560020703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Masoumi A, Heinemann IU, Rohde M, Koch M, Jahn M, Jahn D. 2008. Complex formation between protoporphyrinogen IX oxidase and ferrochelatase during haem biosynthesis in Thermosynechococcus elongatus. Microbiology 154:3707–3714. doi: 10.1099/mic.0.2008/018705-0. [DOI] [PubMed] [Google Scholar]
- 303.Chen W, Wright L, Li S, Cosloy SD, Russell CS. 1996. Expression of glutamyl-tRNA reductase in Escherichia coli. Biochim Biophys Acta 1309:109–121. doi: 10.1016/S0167-4781(96)00117-0. [DOI] [PubMed] [Google Scholar]
- 304.Pappenheimer AM., Jr 1947. Diphtheria toxin; a reinvestigation of the effect of iron on toxin and porphyrin production. J Biol Chem 167:251–259. [PubMed] [Google Scholar]
- 305.Lascelles J. 1956. The synthesis of porphyrins and bacteriochlorophyll by cell suspensions of Rhodopseudomonas spheroides. Biochem J 62:78–93. doi: 10.1042/bj0620078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Burnham BF, Lascelles J. 1963. Control of porphyrin biosynthesis through a negative-feedback mechanism. Biochem J 87:462–472. doi: 10.1042/bj0870462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Alvarez Hayes J, Lamberti Y, Surmann K, Schmidt F, Völker U, Rodriguez ME. 2015. Shotgun proteome analysis of Bordetella pertussis reveals a distinct influence of iron availability on the bacterial metabolism, virulence, and defense response. Proteomics 15:2258–2266. doi: 10.1002/pmic.201400512. [DOI] [PubMed] [Google Scholar]
- 308.Basler M, Linhartová I, Halada P, Novotná J, Bezoušková S, Osička R, Weiser J, Vohradský J, Šebo P. 2006. The iron-regulated transcriptome and proteome of Neisseria meningitidis serogroup C. Proteomics 6:6194–6206. doi: 10.1002/pmic.200600312. [DOI] [PubMed] [Google Scholar]
- 309.Beltrán NC, Horváthová L, Jedelský PL, Šedinová M, Rada P, Marcinčiková M, Hrdý I, Tachezy J. 2013. Iron-induced changes in the proteome of Trichomonas vaginalis hydrogenosomes. PLoS One 8:e65148. doi: 10.1371/journal.pone.0065148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Ducey TF, Carson MB, Orvis J, Stintzi AP, Dyer DW. 2005. Identification of the iron-responsive genes of Neisseria gonorrhoeae by microarray analysis in defined medium. J Bacteriol 187:4865–4874. doi: 10.1128/JB.187.14.4865-4874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Hohle TH, O'Brian MR. 2012. Manganese is required for oxidative metabolism in unstressed Bradyrhizobium japonicum cells. Mol Microbiol 84:766–777. doi: 10.1111/j.1365-2958.2012.08057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Kopf M, Klähn S, Scholz I, Matthiessen JKF, Hess WR, Voß B. 2014. Comparative analysis of the primary transcriptome of Synechocystis sp. PCC 6803. DNA Res 21:527–539. doi: 10.1093/dnares/dsu018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.McHugh JP, Rodriguez-Quinones F, Abdul-Tehrani H, Svistunenko DA, Poole RK, Cooper CE, Andrews SC. 2003. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J Biol Chem 278:29478–29486. doi: 10.1074/jbc.M303381200. [DOI] [PubMed] [Google Scholar]
- 314.Singh AK, McIntyre LM, Sherman LA. 2003. Microarray analysis of the genome-wide response to iron deficiency and iron reconstitution in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol 132:1825–1839. doi: 10.1104/pp.103.024018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Tuanyok A, Kim HS, Nierman WC, Yu Y, Dunbar J, Moore RA, Baker P, Tom M, Ling JML, Woods DE. 2005. Genome-wide expression analysis of iron regulation in Burkholderia pseudomallei and Burkholderia mallei using DNA microarrays. FEMS Microbiol Lett 252:327–335. doi: 10.1016/j.femsle.2005.09.043. [DOI] [PubMed] [Google Scholar]
- 316.Hamza I, Chauhan S, Hassett R, O'Brian MR. 1998. The bacterial Irr protein is required for coordination of heme biosynthesis with iron availability. J Biol Chem 273:21669–21674. doi: 10.1074/jbc.273.34.21669. [DOI] [PubMed] [Google Scholar]
- 317.Martinez M, Ugalde RA, Almiron M. 2005. Dimeric Brucella abortus Irr protein controls its own expression and binds haem. Microbiology 151:3427–3433. doi: 10.1099/mic.0.28213-0. [DOI] [PubMed] [Google Scholar]
- 318.Rodionov DA, Gelfand MS, Todd JD, Curson AR, Johnston AW. 2006. Computational reconstruction of iron- and manganese-responsive transcriptional networks in alpha-Proteobacteria. PLoS Comput Biol 2:e163. doi: 10.1371/journal.pcbi.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Yang J, Panek HR, O'Brian MR. 2006. Oxidative stress promotes degradation of the Irr protein to regulate haem biosynthesis in Bradyrhizobium japonicum. Mol Microbiol 60:209–218. doi: 10.1111/j.1365-2958.2006.05087.x. [DOI] [PubMed] [Google Scholar]
- 320.Qi Z, O'Brian MR. 2002. Interaction between the bacterial iron response regulator and ferrochelatase mediates genetic control of heme biosynthesis. Mol Cell 9:155–162. doi: 10.1016/S1097-2765(01)00431-2. [DOI] [PubMed] [Google Scholar]
- 321.Yang J, Ishimori K, O'Brian MR. 2005. Two heme binding sites are involved in the regulated degradation of the bacterial iron response regulator (Irr) protein. J Biol Chem 280:7671–7676. doi: 10.1074/jbc.M411664200. [DOI] [PubMed] [Google Scholar]
- 322.Jaggavarapu S, O'Brian MR. 2014. Differential control of Bradyrhizobium japonicum iron stimulon genes through variable affinity of the iron response regulator (Irr) for target gene promoters and selective loss of activator function. Mol Microbiol 92:609–624. doi: 10.1111/mmi.12584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Singleton C, White GF, Todd JD, Marritt SJ, Cheesman MR, Johnston AW, Le Brun NE. 2010. Heme-responsive DNA binding by the global iron regulator Irr from Rhizobium leguminosarum. J Biol Chem 285:16023–16031. doi: 10.1074/jbc.M109.067215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Rudolph G, Semini G, Hauser F, Lindemann A, Friberg M, Hennecke H, Fischer HM. 2006. The iron control element, acting in positive and negative control of iron-regulated Bradyrhizobium japonicum genes, is a target for the Irr protein. J Bacteriol 188:733–744. doi: 10.1128/JB.188.2.733-744.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Anderson ES, Paulley JT, Martinson DA, Gaines JM, Steele H, Roop RM II. 2011. The iron-responsive regulator Irr is required for wild-type expression of the gene encoding the heme transporter BhuA in Brucella abortus 2308. J Bacteriol 193:5359–5364. doi: 10.1128/JB.00372-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Battisti JM, Smitherman LS, Sappington KN, Parrow NL, Raghavan R, Minnick MF. 2007. Transcriptional regulation of the heme binding protein gene family of Bartonella quintana is accomplished by a novel promoter element and iron response regulator. Infect Immun 75:4373–4385. doi: 10.1128/IAI.00497-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Martinez M, Ugalde RA, Almiron M. 2006. Irr regulates brucebactin and 2,3-dihydroxybenzoic acid biosynthesis, and is implicated in the oxidative stress resistance and intracellular survival of Brucella abortus. Microbiology 152:2591–2598. doi: 10.1099/mic.0.28782-0. [DOI] [PubMed] [Google Scholar]
- 328.Nienaber A, Hennecke H, Fischer HM. 2001. Discovery of a haem uptake system in the soil bacterium Bradyrhizobium japonicum. Mol Microbiol 41:787–800. [DOI] [PubMed] [Google Scholar]
- 329.Ojeda JF, Martinson DA, Menscher EA, Roop RM II. 2012. The bhuQ gene encodes a heme oxygenase that contributes to the ability of Brucella abortus 2308 to use heme as an iron source and is regulated by Irr. J Bacteriol 194:4052–4058. doi: 10.1128/JB.00367-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Sankari S, O'Brian MR. 2014. A bacterial iron exporter for maintenance of iron homeostasis. J Biol Chem 289:16498–16507. doi: 10.1074/jbc.M114.571562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Small SK, O'Brian MR. 2011. The Bradyrhizobium japonicum frcB gene encodes a diheme ferric reductase. J Bacteriol 193:4088–4094. doi: 10.1128/JB.05064-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Wexler M, Yeoman KH, Stevens JB, de Luca NG, Sawers G, Johnston AW. 2001. The Rhizobium leguminosarum tonB gene is required for the uptake of siderophore and haem as sources of iron. Mol Microbiol 41:801–816. [DOI] [PubMed] [Google Scholar]
- 333.O'Brian MR. 2015. Perception and homeostatic control of iron in the Rhizobia and related bacteria. Annu Rev Microbiol 69:229–245. doi: 10.1146/annurev-micro-091014-104432. [DOI] [PubMed] [Google Scholar]
- 334.Chao TC, Becker A, Buhrmester J, Puhler A, Weidner S. 2004. The Sinorhizobium meliloti fur gene regulates, with dependence on Mn(II), transcription of the sitABCD operon, encoding a metal-type transporter. J Bacteriol 186:3609–3620. doi: 10.1128/JB.186.11.3609-3620.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Diaz-Mireles E, Wexler M, Sawers G, Bellini D, Todd JD, Johnston AW. 2004. The Fur-like protein Mur of Rhizobium leguminosarum is a Mn(2+)-responsive transcriptional regulator. Microbiology 150:1447–1456. doi: 10.1099/mic.0.26961-0. [DOI] [PubMed] [Google Scholar]
- 336.Hohle TH, O'Brian MR. 2009. The mntH gene encodes the major Mn(2+) transporter in Bradyrhizobium japonicum and is regulated by manganese via the Fur protein. Mol Microbiol 72:399–409. doi: 10.1111/j.1365-2958.2009.06650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Menscher EA, Caswell CC, Anderson ES, Roop RM II. 2012. Mur regulates the gene encoding the manganese transporter MntH in Brucella abortus 2308. J Bacteriol 194:561–566. doi: 10.1128/JB.05296-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Platero R, Peixoto L, O'Brian MR, Fabiano E. 2004. Fur is involved in manganese-dependent regulation of mntA (sitA) expression in Sinorhizobium meliloti. Appl Environ Microbiol 70:4349–4355. doi: 10.1128/AEM.70.7.4349-4355.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Anderson ES, Paulley JT, Gaines JM, Valderas MW, Martin DW, Menscher E, Brown TD, Burns CS, Roop RM II. 2009. The manganese transporter MntH is a critical virulence determinant for Brucella abortus 2308 in experimentally infected mice. Infect Immun 77:3466–3474. doi: 10.1128/IAI.00444-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Hohle TH, Franck WL, Stacey G, O'Brian MR. 2011. Bacterial outer membrane channel for divalent metal ion acquisition. Proc Natl Acad Sci U S A 108:15390–15395. doi: 10.1073/pnas.1110137108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Hohle TH, O'Brian MR. 2016. Metal-specific control of gene expression mediated by Bradyrhizobium japonicum Mur and Escherichia coli Fur is determined by the cellular context. Mol Microbiol 101:152–166. doi: 10.1111/mmi.13381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Sankari S, O'Brian MR. 2016. The Bradyrhizobium japonicum ferrous iron transporter FeoAB is required for ferric iron utilization in free living aerobic cells and for symbiosis. J Biol Chem 291:15653–15662. doi: 10.1074/jbc.M116.734129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Bibb LA, Kunkle CA, Schmitt MP. 2007. The ChrA-ChrS and HrrA-HrrS signal transduction systems are required for activation of the hmuO promoter and repression of the hemA promoter in Corynebacterium diphtheriae. Infect Immun 75:2421–2431. doi: 10.1128/IAI.01821-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Bibb LA, King ND, Kunkle CA, Schmitt MP. 2005. Analysis of a heme-dependent signal transduction system in Corynebacterium diphtheriae: deletion of the chrAS genes results in heme sensitivity and diminished heme-dependent activation of the hmuO promoter. Infect Immun 73:7406–7412. doi: 10.1128/IAI.73.11.7406-7412.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Schmitt MP. 1999. Identification of a two-component signal transduction system from Corynebacterium diphtheriae that activates gene expression in response to the presence of heme and hemoglobin. J Bacteriol 181:5330–5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Frunzke J, Gätgens C, Brocker M, Bott M. 2011. Control of heme homeostasis in Corynebacterium glutamicum by the two-component system HrrSA. J Bacteriol 193:1212–1221. doi: 10.1128/JB.01130-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Smart JL, Bauer CE. 2006. Tetrapyrrole biosynthesis in Rhodobacter capsulatus is transcriptionally regulated by the heme-binding regulatory protein, HbrL. J Bacteriol 188:1567–1576. doi: 10.1128/JB.188.4.1567-1576.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Wang L, Elliott M, Elliott T. 1999. Conditional stability of the HemA protein (glutamyl-tRNA reductase) regulates heme biosynthesis in Salmonella typhimurium. J Bacteriol 181:1211–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Wang LY, Brown L, Elliott M, Elliott T. 1997. Regulation of heme biosynthesis in Salmonella typhimurium: activity of glutamyl-tRNA reductase (HemA) is greatly elevated during heme limitation by a mechanism which increases abundance of the protein. J Bacteriol 179:2907–2914. doi: 10.1128/jb.179.9.2907-2914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Jones AM, Elliott T. 2010. A purified mutant HemA protein from Salmonella enterica serovar Typhimurium lacks bound heme and is defective for heme-mediated regulation in vivo. FEMS Microbiol Lett 307:41–47. doi: 10.1111/j.1574-6968.2010.01967.x. [DOI] [PubMed] [Google Scholar]
- 351.Srivastava A, Beale SI. 2005. Glutamyl-tRNA reductase of Chlorobium vibrioforme is a dissociable homodimer that contains one tightly bound heme per subunit. J Bacteriol 187:4444–4450. doi: 10.1128/JB.187.13.4444-4450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Levican G, Katz A, de Armas M, Nunez H, Orellana O. 2007. Regulation of a glutamyl-tRNA synthetase by the heme status. Proc Natl Acad Sci U S A 104:3135–3140. doi: 10.1073/pnas.0611611104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Sobotka R, McLean S, Zuberova M, Hunter CN, Tichy M. 2008. The C-terminal extension of ferrochelatase is critical for enzyme activity and for functioning of the tetrapyrrole pathway in Synechocystis strain PCC 6803. J Bacteriol 190:2086–2095. doi: 10.1128/JB.01678-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Lascelles J. 1960. The synthesis of enzymes concerned in bacteriochlorophyll formation in growing cultures of Rhodopseudomonas spheroides. J Gen Microbiol 23:487–498. doi: 10.1099/00221287-23-3-487. [DOI] [PubMed] [Google Scholar]
- 355.Neuberger A, Sandy JD, Tait GH. 1973. Control of 5-aminolaevulinate synthetase activity in Rhodopseudomonas spheroides. The purification and properties of an endogenous activator of the enzyme. Biochem J 136:491–499. doi: 10.1042/bj1360491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Neuberger A, Sandy JD, Tait GH. 1973. Control of 5-aminolaevulinate synthetase activity in Rhodopseudomonas spheroides. The involvement of sulphur metabolism. Biochem J 136:477–490. doi: 10.1042/bj1360477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Sandy JD, Davies RC, Neuberger A. 1975. Control of 5-aminolaevulinate synthetase activity in Rhodopseudomonas spheroides a role for trisulphides. Biochem J 150:245–257. doi: 10.1042/bj1500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Mense SM, Zhang L. 2006. Heme: a versatile signaling molecule controlling the activities of diverse regulators ranging from transcription factors to MAP kinases. Cell Res 16:681–692. doi: 10.1038/sj.cr.7310086. [DOI] [PubMed] [Google Scholar]
- 359.Zeilstra-Ryalls JH, Kaplan S. 1995. Aerobic and anaerobic regulation in Rhodobacter sphaeroides 2.4.1: the role of the fnrL gene. J Bacteriol 177:6422–6431. doi: 10.1128/jb.177.22.6422-6431.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Zeilstra-Ryalls JH, Kaplan S. 1998. Role of the fnrL gene in photosystem gene expression and photosynthetic growth of Rhodobacter sphaeroides 2.4.1. J Bacteriol 180:1496–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Fales L, Kryszak L, Zeilstra-Ryalls J. 2001. Control of hemA expression in Rhodobacter sphaeroides 2.4.1: effect of a transposon insertion in the hbdA gene. J Bacteriol 183:1568–1576. doi: 10.1128/JB.183.5.1568-1576.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Smart JL, Willett JW, Bauer CE. 2004. Regulation of hem gene expression in Rhodobacter capsulatus by redox and photosystem regulators RegA, CrtJ, FnrL, and AerR. J Mol Biol 342:1171–1186. doi: 10.1016/j.jmb.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 363.Ranson-Olson B, Jones DF, Donohue TJ, Zeilstra-Ryalls JH. 2006. In vitro and in vivo analysis of the role of PrrA in Rhodobacter sphaeroides 2.4.1 hemA gene expression. J Bacteriol 188:3208–3218. doi: 10.1128/JB.188.9.3208-3218.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Moskvin OV, Gomelsky L, Gomelsky M. 2005. Transcriptome analysis of the Rhodobacter sphaeroides PpsR regulon: PpsR as a master regulator of photosystem development. J Bacteriol 187:2148–2156. doi: 10.1128/JB.187.6.2148-2156.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Yeliseev AA, Kaplan S. 1999. A novel mechanism for the regulation of photosynthesis gene expression by the TspO outer membrane protein of Rhodobacter sphaeroides 2.4.1. J Biol Chem 274:21234–21243. doi: 10.1074/jbc.274.30.21234. [DOI] [PubMed] [Google Scholar]
- 366.Lindemann A, Moser A, Pessi G, Hauser F, Friberg M, Hennecke H, Fischer HM. 2007. New target genes controlled by the Bradyrhizobium japonicum two-component regulatory system RegSR. J Bacteriol 189:8928–8943. doi: 10.1128/JB.01088-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Gilles-Gonzalez MA, Gonzalez G. 2005. Heme-based sensors: defining characteristics, recent developments, and regulatory hypotheses. J Inorg Biochem 99:1–22. doi: 10.1016/j.jinorgbio.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 368.Chauhan S, O'Brian MR. 1997. Transcriptional regulation of δ-aminolevulinic acid dehydratase synthesis by oxygen in Bradyrhizobium japonicum and evidence for developmental control of the hemB gene. J Bacteriol 179:3706–3710. doi: 10.1128/jb.179.11.3706-3710.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Fischer HM, Velasco L, Delgado MJ, Bedmar EJ, Scharen S, Zingg D, Gottfert M, Hennecke H. 2001. One of two hemN genes in Bradyrhizobium japonicum is functional during anaerobic growth and in symbiosis. J Bacteriol 183:1300–1311. doi: 10.1128/JB.183.4.1300-1311.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Nellen-Anthamatten D, Rossi P, Preisig O, Kullik I, Babst M, Fischer HM, Hennecke H. 1998. Bradyrhizobium japonicum FixK2, a crucial distributor in the FixLJ-dependent regulatory cascade for control of genes inducible by low oxygen levels. J Bacteriol 180:5251–5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Page KM, Guerinot ML. 1995. Oxygen control of the Bradyrhizobium japonicum hemA gene. J Bacteriol 177:3979–3984. doi: 10.1128/jb.177.14.3979-3984.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Rompf A, Hungerer C, Hoffmann T, Lindenmeyer M, Romling U, Gross U, Doss MO, Arai H, Igarashi Y, Jahn D. 1998. Regulation of Pseudomonas aeruginosa hemF and hemN by the dual action of the redox response regulators Anr and Dnr. Mol Microbiol 29:985–997. doi: 10.1046/j.1365-2958.1998.00980.x. [DOI] [PubMed] [Google Scholar]
- 373.LaMattina JW, Nix DB, Lanzilotta WN. 2016. Radical new paradigm for heme degradation in Escherichia coli O157:H7. Proc Natl Acad Sci U S A 113:12138–12143. doi: 10.1073/pnas.1603209113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Park S, Imlay JA. 2003. High levels of intracellular cysteine promote oxidative DNA damage by driving the Fenton reaction. J Bacteriol 185:1942–1950. doi: 10.1128/JB.185.6.1942-1950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Seaver LC, Imlay JA. 2001. Hydrogen peroxide fluxes and compartmentalization inside growing Escherichia coli. J Bacteriol 183:7182–7189. doi: 10.1128/JB.183.24.7182-7189.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Imlay JA. 2008. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem 77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Lee C, Lee SM, Mukhopadhyay P, Kim SJ, Lee SC, Ahn WS, Yu MH, Storz G, Ryu SE. 2004. Redox regulation of OxyR requires specific disulfide bond formation involving a rapid kinetic reaction path. Nat Struct Mol Biol 11:1179–1185. doi: 10.1038/nsmb856. [DOI] [PubMed] [Google Scholar]
- 378.Mancini S, Imlay JA. 2015. The induction of two biosynthetic enzymes helps Escherichia coli sustain heme synthesis and activate catalase during hydrogen peroxide stress. Mol Microbiol 96:744–763. doi: 10.1111/mmi.12967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Zheng M, Wang X, Templeton LJ, Smulski DR, LaRossa RA, Storz G. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J Bacteriol 183:4562–4570. doi: 10.1128/JB.183.15.4562-4570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Jang S, Imlay JA. 2007. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J Biol Chem 282:929–937. doi: 10.1074/jbc.M607646200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Herbig AF, Helmann JD. 2001. Roles of metal ions and hydrogen peroxide in modulating the interaction of the Bacillus subtilis PerR peroxide regulon repressor with operator DNA. Mol Microbiol 41:849–859. [DOI] [PubMed] [Google Scholar]
- 382.Mongkolsuk S, Praituan W, Loprasert S, Fuangthong M, Chamnongpol S. 1998. Identification and characterization of a new organic hydroperoxide resistance (ohr) gene with a novel pattern of oxidative stress regulation from Xanthomonas campestris pv. phaseoli. J Bacteriol 180:2636–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Moore CM, Helmann JD. 2005. Metal ion homeostasis in Bacillus subtilis. Curr Opin Microbiol 8:188–195. doi: 10.1016/j.mib.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 384.Chen L, Keramati L, Helmann JD. 1995. Coordinate regulation of Bacillus subtilis peroxide stress genes by hydrogen peroxide and metal ions. Proc Natl Acad Sci U S A 92:8190–8194. doi: 10.1073/pnas.92.18.8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Mongkolsuk S, Helmann JD. 2002. Regulation of inducible peroxide stress responses. Mol Microbiol 45:9–15. doi: 10.1046/j.1365-2958.2002.03015.x. [DOI] [PubMed] [Google Scholar]
- 386.Faulkner M, Ma Z, Fuangthong M, Helmann JD. 2012. Derepression of the Bacillus subtilis PerR peroxide stress response leads to iron deficiency. J Bacteriol 194:1226–1235. doi: 10.1128/JB.06566-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Hentchel KL, Escalante-Semerena JC. 2015. Acylation of biomolecules in prokaryotes: a widespread strategy for the control of biological function and metabolic stress. Microbiol Mol Biol Rev 79:321–346. doi: 10.1128/MMBR.00020-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Weinert BT, Scholz C, Wagner SA, Iesmantavicius V, Su D, Daniel JA, Choudhary C. 2013. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep 4:842–851. doi: 10.1016/j.celrep.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 389.Go YM, Jones DP. 2013. The redox proteome. J Biol Chem 288:26512–26520. doi: 10.1074/jbc.R113.464131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Hobbs C, Dailey HA, Shepherd M. 2016. The HemQ coprohaem decarboxylase generates reactive oxygen species: implications for the evolution of classical haem biosynthesis. Biochem J 473:3997–4009. doi: 10.1042/BCJ20160696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. 2006. Toward automatic reconstruction of a highly resolved tree of life. Science 311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 392.Letunic I, Bork P. 2016. Interactive Tree of Life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Gupta RS. 2000. The phylogeny of proteobacteria: relationships to other eubacterial phyla and eukaryotes. FEMS Microbiol Rev 24:367–402. doi: 10.1111/j.1574-6976.2000.tb00547.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.