Abstract
Aldehyde/alcohol dehydrogenases (ADHEs) are bifunctional enzymes that commonly produce ethanol from acetyl-CoA with acetaldehyde as intermediate and play a key role in anaerobic redox balance in many fermenting bacteria. ADHEs are also present in photosynthetic unicellular eukaryotes, where their physiological role and regulation are, however, largely unknown. Herein we provide the first molecular and enzymatic characterization of the ADHE from the photosynthetic microalga Chlamydomonas reinhardtii. Purified recombinant ADHE catalyzed the reversible NADH-mediated interconversions of acetyl-CoA, acetaldehyde, and ethanol but seemed to be poised toward the production of ethanol from acetaldehyde. Phylogenetic analysis of the algal fermentative enzyme supports a vertical inheritance from a cyanobacterial-related ancestor. ADHE was located in the chloroplast, where it associated in dimers and higher order oligomers. Electron microscopy analysis of ADHE-enriched stromal fractions revealed fine spiral structures, similar to bacterial ADHE spirosomes. Protein blots showed that ADHE is regulated under oxic conditions. Up-regulation is observed in cells exposed to diverse physiological stresses, including zinc deficiency, nitrogen starvation, and inhibition of carbon concentration/fixation capacity. Analyses of the overall proteome and fermentation profiles revealed that cells with increased ADHE abundance exhibit better survival under dark anoxia. This likely relates to the fact that greater ADHE abundance appeared to coincide with enhanced starch accumulation, which might reflect ADHE-mediated anticipation of anaerobic survival.
Keywords: algae, carbohydrate metabolism, Chlamydomonas, chloroplast, electron microscopy (EM), metabolic regulation, metalloprotein, western blot, zinc
Introduction
Oxygenic photosynthetic microorganisms are typically associated with illuminated oxic environments, and so, little attention is paid to energy generation in conditions of dark anoxia. The study of the anaerobic heterotrophic metabolism of both microalgae and cyanobacteria is, however, highly relevant because these organisms regularly face conditions of dark anoxia in their natural habitats, especially in eutrophized and/or polluted waters. To meet the energy demand for life or cell maintenance in these specific conditions, photosynthetic cells carry out fermentation at the expense of endogenous carbohydrates (glycogen or starch). Anaerobic respiration has also been reported, i.e. sulfur respiration in cyanobacterial species such as Oscillatoria limnetica and Microcoleus chthonoplastes (1, 2), and fumarate respiration in Euglena gracilis (3, 4).
In cyanobacteria, the fermentation routes are diverse and include homofermentation (lactate or acetate), heterofermentation, and mixed acid fermentation (5). Among microalgae, fermentative metabolism has been investigated mainly in chlorophytes. It is known for some time that in absence of oxygen, green algae such as Chlamydomonas and Chlorella have the ability to produce organic acids (acetate, formate, lactate, and succinate), alcohols (ethanol), and gases (CO2 and H2) (6–9). The genome sequences of Chlamydomonas reinhardtii (10) and Chlorella variabilis NC64 (11) have greatly helped to get a good grasp of the anaerobic pathways in the two algae (12–14). Since then, in C. reinhardtii, these pathways have been actively investigated through molecular and biochemical studies of the fermentative enzymes (15–19), as well as physiological studies of mutant strains (20–25). As we understand it, the “core” anaerobic network in the chlorophyte combines enzymes commonly found in eukaryotes, i.e. pyruvate decarboxylase (PDC),2 alcohol dehydrogenases (ADHs), and lactate dehydrogenase, with enzymes that are typical for bacteria (referred to as bacterial-type enzymes), i.e. pyruvate formate lyase (PFL; EC 2.3.1.54), pyruvate:ferredoxin oxidoreductase (PFO or PFR; EC 1.2.7.1), iron-only hydrogenases, and aldehyde/alcohol dehydrogenase (ADHE or ADH1) (Fig. 1). The presence of these bacterial-type enzymes raises questions about their evolutionary origin and their integration in a mitochondriate eukaryote that we and others have tackled since their discovery. PFL and PFO were both shown to be functional in C. reinhardtii (16, 18, 19) and can, under anaerobic conditions, catalyze the production of acetyl-CoA from pyruvate. PFL accumulates under aerobic conditions and is dually targeted to chloroplast and mitochondria (12). PFO is expressed under anaerobic conditions and localizes to the chloroplast where it is coupled to H2 production by iron-only hydrogenases via FDX1 (PetF) (18, 19). ADHE is a mono-iron enzyme, which was found to be the major ethanol-producing enzyme in conditions of dark anoxia (22). Although it is now clear that the bacterial-type enzymes fully participate in the anaerobic life of C. reinhardtii, their physiological significance in the intricate anaerobic network (Fig. 1), as well as their regulation, remains largely enigmatic.
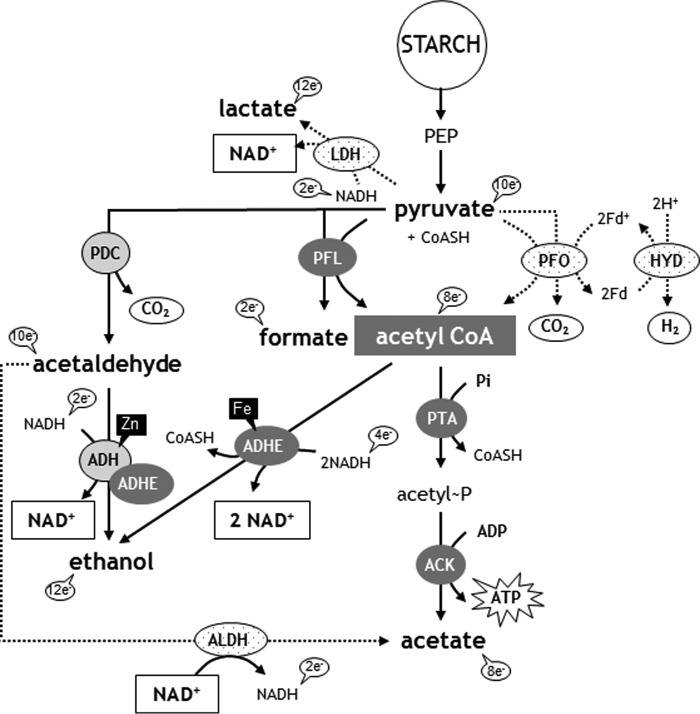
FIGURE 1.
Anerobic metabolic routes in C. reinhardtii. Under dark anoxia, starch is metabolized to pyruvate via glycolysis, thereby generating ATP and reducing power (NADH). The network shows a pyruvate branch point with four enzymes. Another branch point is acetyl-CoA that can be used to produce ethanol via ADHE or acetate via the PTA-ACK pathway. Network is simplified by the omission of the subcellular localization of the different enzymes. Fe and Zn indicate the metal in the active site of the alcohol dehydrogenases. To better understand the metabolic responses, the number of oxidizable electrons is indicated for each metabolite. The main fermentation route used by the alga incubated in AIB under dark anoxia is the “PFL-gated pathway,” which consists of PFL, ADHE, and PTA-ACK.
To better understand the integration and the regulation of the ADHE in the context of a photosynthetic eukaryotic cell, we explored different aspects of C. reinhardtii ADHE, in particular its evolutionary origin, subcellular localization, and enzymatic properties. We also followed ADHE abundance in different mutant strains and relevant physiological conditions. This is the first report on the factors that influence the ADHE abundance in C. reinhardtii and on the consequences of enhanced protein levels in fermentation behavior.
Results
Phylogenetic Analyses of C. reinhardtii ADHE
ADHE genes are present in the five eukaryotic supergroups described by Adl et al. (26), i.e. the Archaeplastida, Amoebozoa, Excavates, Opisthokonta, and the Stramenopiles-Alveolata-Rhizaria (SAR) (Table 1). However, the gene remains poorly represented among eukaryotes, where it appears to be restricted to unicellular species, most of which are pathogenic anaerobes. The only free-living eukaryotes in which an ADHE gene has been found so far are microalgae. In addition to the chlorophytes C. reinhardtii, Chlorella, and Polytomella (Archaeplastida), an ADHE gene is also present in two photosynthetic species of the SAR supergroup: the cryptophyte Guillardia theta and the chlorarachniophyte Bigelowiella natans (14) (Table 1).
TABLE 1.
Distribution of ADHE among the five eukaryotic supergroups defined by Adl et al. (26)
| Supergroup | Species name | Lifestyle | Identitya | Accession number |
|---|---|---|---|---|
| Archaeplastida | ||||
| Chlorophytes | C. reinhardtii | Free-living photosythetic alga | gi 92084840 | |
| Chlorophytes | Volvox carteri | Free-living photosythetic alga | 84 | gi 302853679 |
| Chlorophytes | C. variabilis | Free-living photosythetic alga | 68 | gi 552841275 |
| Amoebozoa | ||||
| Entamoebids | Entamoeba histolytica | Human pathogen (intestine) | 47 | gi 2492737 |
| Pelobionts | Mastigamoeba | Free-living species | 55 | gi 21435953 |
| Excavata | ||||
| Diplomonads | Giardia intestinalis | Human pathogen (intestine) | 52 | gi 2052472 |
| Diplomonads | Spironucleus barkhanus | Fish pathogen | 53 | gi 27983190 |
| Opisthokonta | ||||
| Fungi (chytrid) | Neocallimastix frontalis | Anaerobic rumen fungus | 52 | gi 387233067 |
| Fungi (chytrid) | Piromyces sp. E2 | Anaerobic rumen fungus | 52 | gi 33578055 |
| Fungi | Togninia minima UCRPA7 | Grapevine pathogen | 47 | gi 631237462 |
| SAR | ||||
| Cryptophytes | G. theta CCMP2712 | Free-living photosythetic alga | 47 | gi 551672365 |
| Cercozoa | B. natans CCMP2755 | Free-living photosythetic alga | 43 | jgi Bigna1 85335 |
| Apicomplexa | Cryptosporidium hominis TU502 | Anaerobic pathogen | 48 | gi 67623585 |
a Amino acid identity with C. reinhardtii ADHE.
Earlier phylogenies have shown most eukaryotic ADHEs wedged in between two clusters of bacterial enzymes (27). The ADHE phylogenetic analysis that we present here includes a significantly larger set of bacterial sequences, as well as all eukaryotic sequences publicly available (supplemental Data Sets S1 and S2). The current analysis further substantiates the dichotomy of the bacterial ADHEs (clusters I and II) and the unclear rooting of most eukaryote proteins (Fig. 2).
FIGURE 2.
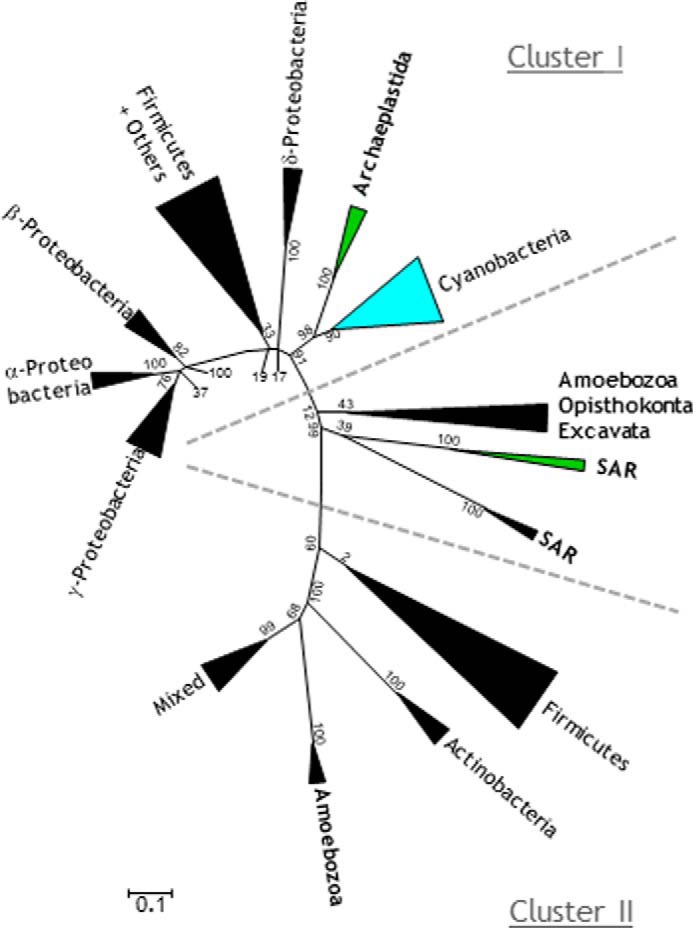
Schematic phylogenetic tree of aldehyde/alcohol dehydrogenases from bacterial and eukaryotic sources. The tree was constructed using the maximum likelihood algorithm. Eukaryotic supergroups are indicated in bold type. Photosynthetic eukaryotes are indicated in green and cyanobacteria in blue. The list of species used in this analysis can be found in supplemental Data Set S1. Multiple sequence alignment is shown in supplemental Data Set S2.
The ADHEs from the marine algae B. natans and G. theta are found in the undefined “eukaryotic” region of the tree, whereas the enzymes of C. reinhardtii and other chlorophytes are found in cluster I, where they branch with cyanobacterial ADHEs (Fig. 2). Our phylogenetic analysis thus suggests that (i) the origin of the chlorophyte enzymes is distinct from that of the other eukaryotic enzymes and (ii) the ADHE gene in green algae might have been inherited vertically from the cyanobacterial ancestor. The unique phylogenetic position of C.reinhardtii ADHE compared with that of the eukaryotic enzymes studied so far, i.e. Entamoeba (Amoebozoa) (28, 29) and Giardia (Excavata) (30), was a motive to study in detail its biochemical characteristics and its regulation.
Subcellular Localization and Oligomerization of C. reinhardtii ADHE
Relative to bacterial ADHEs, the C. reinhardtii 954-amino acid protein exhibits an extended N terminus (∼60 residues) (Fig. 3A), which could serve as an intracellular targeting signal to the chloroplast and/or the mitochondrion. The targeting of ADHE to the bioenergetic organelles was evaluated by protein blots using antibodies raised against a truncated form of the algal ADHE (tADHE, Val354–Pro703) (12). These antibodies recognize native ADHE with an apparent molecular mass of ∼100 kDa (Fig. 4A). To isolate chloroplasts and mitochondria, we used mutant strain 10-6C, in which ADHE abundance was found to be 3–4-fold higher than in wild-type strain CC-124 (supplemental Fig. S1). Strain 10-6C is a photosynthetic mutant impaired in CO2 assimilation because of a point mutation in the RBCL gene (31, 32) (supplemental Fig. S2). The relative purity of the organelle fractions was evaluated with antibodies against the light harvesting complex proteins (LHCs) and subunit β of mitochondrial F0F1-ATPase (ATP2). Immunoblot analysis indicated that ADHE is present in the chloroplast fraction but not in the fraction enriched in mitochondria (Fig. 3A). Our result, obtained with cells grown under oxic conditions, thus corroborates proteomics data on organelles isolated from anaerobic cells (33). C. reinhardtii is the first organism for which an ADHE chloroplast location is found. Indeed, ADHE was so far described in the cytosol of parasites such as Piromyces and Giardia (4, 34) and in the mitochondria of the non-photosynthetic chlorophyte Polytomella (27).
FIGURE 3.

Structural and enzymatic characteristics of ADHE. A, the C. reinhardtii enzyme consists of an N-terminal ALDH domain (cd07077) followed by a C-terminal ADH domain (cd08178). The catalytic Cys residue in the ALDH domain (Cys323) is indicated. Two signatures for iron-binding were identified in the ADH domain: ADH_iron_1 (residues 706–734) (Prosite PS00913) and ADH_iron_2 (residues 794–814) (Prosite PS00060). The position of the residues potentially involved in iron coordination (Asp727, His731, His797, and His811) was inferred from the structure of the E. coli iron-dependent alcohol dehydrogenase FucO (69). Black boxes indicate NADH binding sites (Gly270–Gly291 and Glu599–Met622) based on (70). Compared with bacterial enzymes, the algal ADHE exhibits at its N terminus an extension (gray box), which likely serves for intracellular targeting. B, enzymatic activities catalyzed by ADHE.
FIGURE 4.
C. reinhardtii ADHE is located in the chloroplast stroma. A, organelle fractionation from strain 10-6C. Ce, cell extract; Cp, chloroplast fraction; Mt, mitochondrial fraction. Proteins were separated on SDS-PAGE (5–12% acrylamide gradient). Immunoblots show the distribution in isolated organelles of LHCs, subunit β of the mitochondrial F0F1-ATPase (ATP2), and ADHE. B, two-dimensional resolution of proteins in a stromal fraction. Proteins were subjected to BN (3–12%)/SDS-PAGE (10%) and further transferred to nitrocellulose for immunodetection. The blot was first probed for ADHE and then for PFL. ★, position of the ADHE dimer; ▾, position of the PFL dimer. C, EM images of a stromal protein fraction using uranyl acetate as negative stain.
To gain insights into the intraorganellar localization of the ADHE in C. reinhardtii, chloroplasts were broken by two freeze-thaw cycles and fractionated by low speed centrifugation. In the resulting supernatant, which contains primarily stromal proteins (not shown), the ADHE was identified by mass spectrometry-based proteomic analysis with a coverage of more than 50% over the entire sequence (supplemental Fig. S3).
To study the oligomerization state of the native ADHE, the enzyme was further purified by anion exchange chromatography and affinity chromatography on Blue Sepharose resin. Assessment of the protein oligomerization was done by two-dimensional Blue Native (BN)/SDS-PAGE and immunoblotting. Protein blots showed that ADHE occurred in various complexes of similar abundance (Fig. 4B); the smallest complex of ∼180 kDa is likely a dimer, and the complex in the range of ∼400 kDa might represent a tetramer. PFL, present in the same stromal fraction, was found mainly as a monomer at ∼80-kDa and a dimer at ∼160 kDa (Fig. 4B). The various ADHE forms observed on BN-PAGE might be indicative of the protein tendency to aggregate. Alternatively, these different forms might indicate the progressive dissociation of large assemblies during purification and/or migration on the native gel. ADHE that was partially purified by anion exchange chromatography could be pelleted after ultracentrifugation, thus supporting the possibility that this protein associates in large molecular assemblies (not shown). EM images of the resulting ADHE-containing pellet revealed the presence of spiral-like filaments of an average length of 100 nm (Fig. 4C), morphologically indistinguishable from bacterial ADHE oligomers, also known as spirosomes (35–37). Attempts to obtain samples enriched in these filaments by either centrifugation on concentrating devices or ammonium sulfate precipitation have failed, possibly because these structures are relatively fragile.
Molecular Characterization and Kinetic Analysis of Recombinant ADHE
Considering the low amounts of ADHE present in C. reinhardtii, even in strain 10-6C, we chose to study the recombinant enzyme. The ADHE gene was cloned into the pET24a vector, and the protein was expressed in Escherichia coli. A hexahistidine tag added to the C terminus of the ADHE allowed fast purification via affinity chromatography on a nickel column (Fig. 5A).
FIGURE 5.
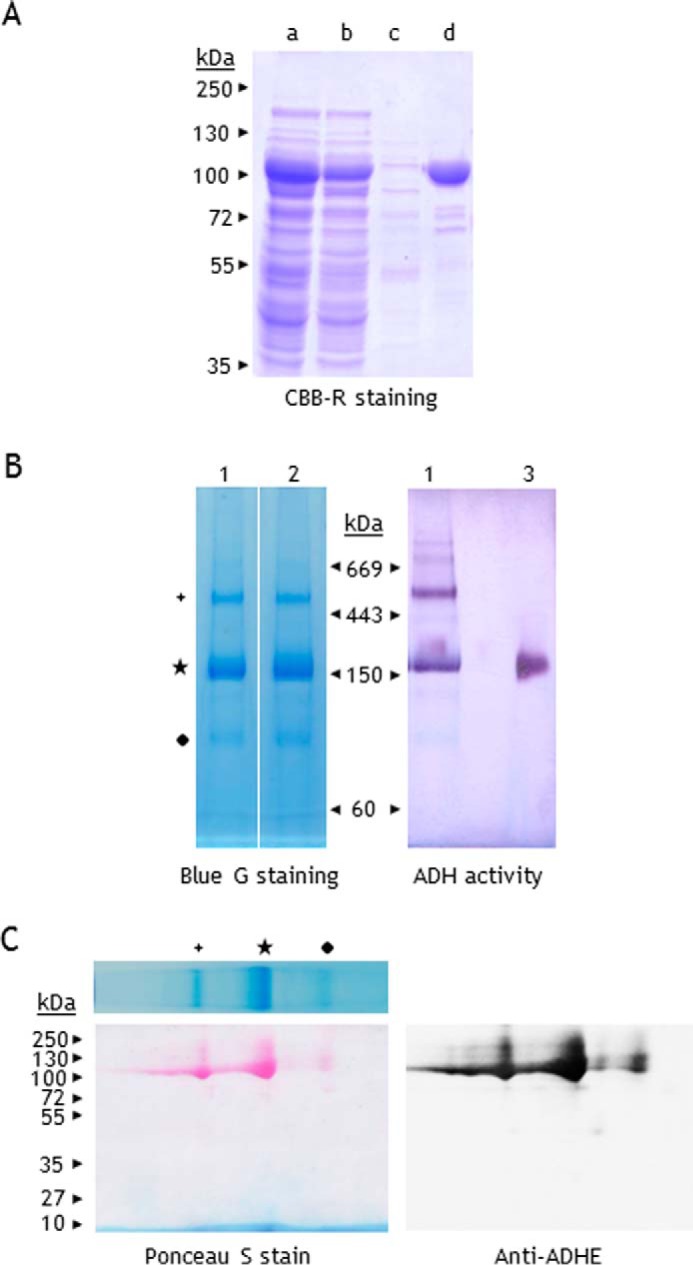
PAGE analysis of C. reinhardtii recombinant ADHE. A, purification of His-tagged C. reinhardtii ADHE (rADHE). Coomassie Brilliant Blue R stain of proteins separated on SDS-PAGE (10%). Lane a, E. coli soluble fraction; lane b, flow-through; lane c, 10 mm imidazole wash; lane d, rADHE eluted with 100 mm imidazole (∼10 μg). B, freshly purified rADHE (25 μg) was subjected to BN-PAGE (3–12%). Lane 1, rADHE purified under anaerobic conditions; lane 2, rADHE purified under atmospheric conditions; lane 3, S. cerevisiae ADH1 (0.5 μg). C, detection of rADHE by immunoblotting after two-dimensional BN/SDS-PAGE. One BN gel lane was excised, subjected to denaturation in 1% SDS and 1% β-mercaptoethanol, and further loaded on SDS-PAGE (10%). Proteins were then transferred to a nitrocellulose membrane and stained with Ponceau S. The membrane was probed for ADHE.
The oligomerization of freshly purified rADHE was first assessed by gel filtration. The elution profile in 50 mm potassium phosphate, pH 7.0, supplemented with 150 mm NaCl revealed the heterogeneity of the sample, with different complexes of molecular masses ranging between 200 and 600 kDa (not shown). On a BN gel, rADHE was resolved as several bands (Fig. 5B): one major complex of ∼180 kDa, a complex of ∼500 kDa, and several minor bands at lower (<120 kDa) and higher molecular mass ranges (>669 kDa). This BN profile was reproducible, also when enzyme purification was carried out under atmospheric conditions (Fig. 5B, second lane). All protein bands, except the lowest at ∼90-kDa, showed a NAD+-dependent alcohol dehydrogenase activity (Fig. 5B). The enzymatically active complex of ∼180 kDa that runs in the same region as Saccharomyces cerevisiae tetrameric alcohol dehydrogenase ADH1 (Mr = ∼160 kDa) is likely an ADHE dimer. Two-dimensional BN/SDS-PAGE confirmed that rADHE occurs in multiple oligomeric forms, like the native ADHE (Fig. 4B). However, in contrast to the native enzyme, the rADHE associates most predominantly into dimers (Fig. 5C).
Bifunctional aldehyde/alcohol dehydrogenases consist of a coenzyme A-dependent aldehyde dehydrogenase (ALDH) (EC 1.2.1.10) followed by an iron-containing ADH (EC 1.1.1.1) (Fig. 3A). ADHEs catalyze the two distinct activities that are typically reversible (30) (Fig. 3B). Here we first assessed the ability of the purified rADHE enzyme to catalyze each of the four different partial reactions. For the NADH-dependent acetyl-CoA and acetaldehyde reduction activities, the enzyme exhibited Vmax values of 1 and 4 units/mg, respectively (Table 2, reactions 1 and 2). Both reductive reactions show highest activities in the pH range of 6.0–7.0 (Table 2). Reductions of acetyl-CoA and acetaldehyde appear specific for NADH, because the Vmax for NADPH was only 1–2% of that for NADH (not shown). The specific activities of NAD+-dependent ethanol and acetaldehyde oxidations were both found to be in the range of 1–1.5 units/mg (reactions 3 and 4) (Table 2). Overall, reaction 2 showed the highest activity, which may represent an adaptation to quickly remove acetaldehyde, a compound toxic for the cell. Also, from the fact that the Vmax of reaction 4 is much lower than that of reaction 2, it could be hypothesized that the enzyme is poised to function toward the production of ethanol and not vice versa, at least under the saturating conditions used for the measurements. Iron addition to rADHE exhibited a (moderate) stimulating effect on all activities (Table 2), whereas an incubation in 100 mm EDTA had no clear inhibitory effect.
TABLE 2.
Enzymatic data on the recombinant ADHE
Standard reaction medium contains 50 mm potassium phosphate (pH 7.0). The substrates were used at the following concentrations: NAD (1.5 mm), NADH (0.4 mm), acetyl-CoA (100 μm), coenzyme A (100 μm), ethanol (200 mm), acetaldehyde (1 mm). “Fe” indicates incubation with 0.3 mm FeCl3.
| Reaction no. | Vmax | pHmax |
|---|---|---|
| μmol·min−1·mg−1 | ||
| 1 | 0.88 ± 0.18 | 6.5–7.0 |
| 1 (+Fe) | 1.07 ± 0.09 | |
| 2 | 4.00 ± 0.63 | 6.5–7.0 |
| 2 (+Fe) | 4.99 ± 1.21 | |
| 3 | 0.85 ± 0.06 | |
| 3 (+Fe) | 1.20 ± 0.40 | |
| 4 | 0.69 ± 0.29 | |
| 4 (+Fe) | 0.83 ± 0.33 |
The recombinant enzyme rADHE was further characterized for the substrate affinities of its forward reactions (reactions 1 and 2), which are metabolically relevant. Acetyl-CoA reduction (reaction 1) obeyed Michaelis-Menten kinetics with an apparent Km for acetyl-CoA of 12.7 ± 2.9 μm (Table 3). The kinetics of NADH is less clear and likely confounded by the fact that NADH is also used in reaction 2, using the reaction product of reaction 1. A typical Michaelis-Menten curve was obtained for NADH in acetaldehyde reduction (reaction 2) with a Km of 20.9 ± 3.5 μm (Table 3).
TABLE 3.
Km values of rADHE for the activities directed towards its physiological role, the production of ethanol
| Reaction no. | Substrate | Km |
|---|---|---|
| 1 | NADH | 120.9 ± 48.0 μm |
| 1 | Acetyl-CoA | 12.7 ± 2.9 μm |
| 2 | NADH | 20.9 ± 3.5 μm |
| 2 | Acetaldehyde | 35.0 ± 17.6 mm |
Analysis of Fermentation Products by ADHE Overexpression Strain
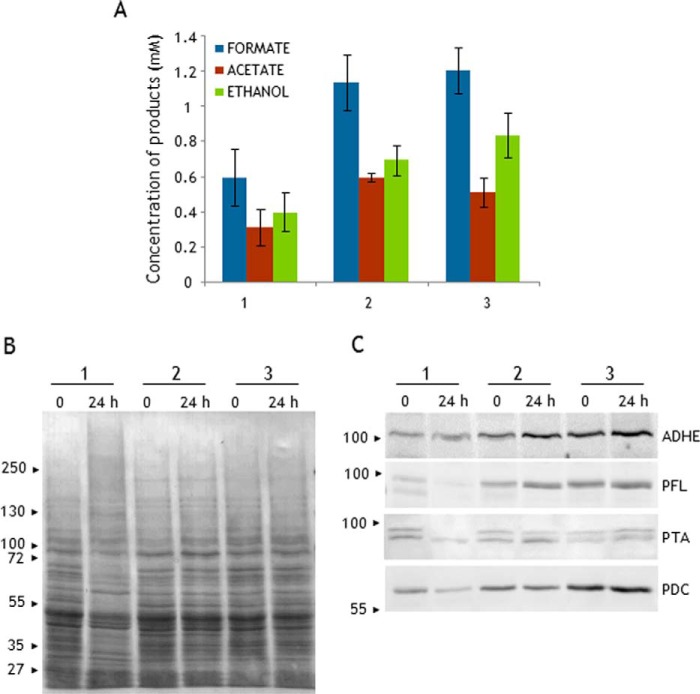
To gain insights into the regulation of the carbon fluxes in the green alga, we investigated the fermentative capabilities of strain 10-6C, which accumulates ∼3.5 times more ADHE than the wild-type strain CC-124 (supplemental Fig. S1 and Table 4). Algal cells were incubated in dark conditions in AIB medium, a standard medium used to study algal fermentation (13). The products excreted by the cells were analyzed by HPLC at 0–8 h and at 24 h. 45 min after the shift to anoxia, formate, ethanol, and acetate were already detected in the extracellular medium, and their production continued steadily up to 8 h (Fig. 6). After 8 h of dark anoxia, the formate production by strains CC-124 and 10-6C was comparable at ∼0.6 mm (1 × 107 cells ml−1), indicating that the ADHE-overaccumulating strain catabolizes endogenous carbon reserves in a similar manner (products and kinetics) as the wild type, i.e. pyruvate stemming from glycolysis is directed mostly to pyruvate formate lyase (Fig. 1). When acetyl-CoA produced by PFL is equally distributed between ADHE and the phosphotransacetylase-acetate kinase (PTA-ACK) route, ethanol and acetate are expected to be produced in equimolar amounts. A balanced production of acetate and ethanol was indeed observed for the WT strain, with 0.34 mm of each product after an incubation of 8 h. In the case of strain 10-6C, the production of acetate appeared somewhat lower than that of ethanol with 0.28 and 0.43 mm, respectively (Fig. 6), which suggests a slight shift in the distribution of acetyl-CoA toward ADHE. This imbalance could be explained by the increased ADHE levels in the mutant strain, although the increase in ethanol production (<1.4) is not in proportion with the increase in ADHE abundance (∼3.5; Table 4). After 24 h of dark anoxia, the fermentation profiles of the WT strain were comparable with those at 8 h, indicating that no further fermentation occurred after 8 h. In contrast, the levels of each product excreted by the mutant strain after 24 h had approximately doubled relative to 8 h, reaching 1.2 mm formate, 0.5 mm acetate, and 0.83 mm ethanol (Fig. 6).
TABLE 4.
Relative abundance of ADHE and PFL in C. reinhardtii cells exposed to various physiological conditions
| Strain | Phenotype | Medium | Light intensity | ADHE relative | PFL relative |
|---|---|---|---|---|---|
| μE m−2 s−1 | |||||
| CC-124 | WT | TAP | 10–15 | 1 | 1 |
| CC-124 | WT | TAP | 40–50 | 1 | 1 |
| CC-124 | WT | TAP | 200 | 1.6–1.8 | 1.1–1.2 |
| CC-124 | WT | HSM + 2% CO2 | 40–50 | 0.3 | NDa |
| CC-124 | WT | TAP + DCMU | 40–50 | 0.9–1.2 | 1.3–1.6 |
| CC-124 | WT | TAP (no ZnSO4) | 40–50 | 3.3–3.5 | 1.5–1.7 |
| CC-124 | WT | N-free TAP (24 h) | 40–50 | 2.5 | 0.75 |
| 10-6C | Lacks RubisCO carboxylase activity | TAP | 40–50 | 3.4–3.6 | 1.4–1.6 |
| 10-6C | Lacks RubisCO carboxylase activity | TAP (no ZnSO4) | 40–50 | 3.3–3.5 | ND |
| ΔRBCL | Lacks RubisCO | TAP | 10–15 | 0.2–0.3 | 1.1–1.4 |
| CIA3 | Lacks carbonic anhydrase 3 | TAP | 40–50 | 1.4–1.7 | 1.1–1.4 |
| CIA5 | Lacks CCM1 | TAP | 40–50 | 1.5–1.7 | 1.1–1.3 |
| sta6 | Impaired in starch synthesis | TAP (no ZnSO4) | 40–50 | 1.0–1.1 | ND |
| sta6 | Impaired in starch synthesis | TAP (no ZnSO4) | 40–50 | 3.0–3.2 | ND |
a ND, not determined.
FIGURE 6.
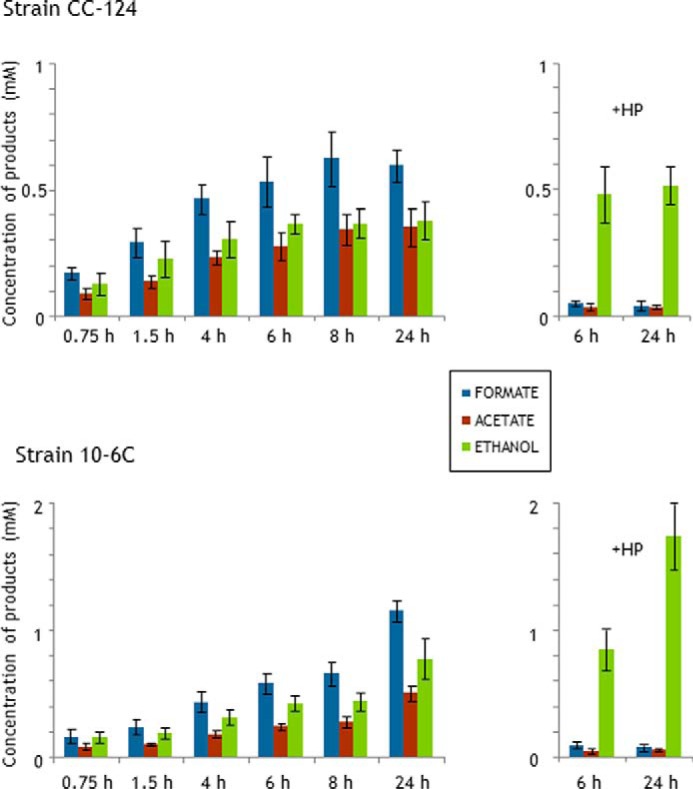
Fermentative products excreted by C. reinhardtii incubated in dark anoxia. Shown are the kinetics of acetate, formate, and ethanol production by the wild-type strain (upper graph) and the mutant strain (lower graph) incubated in AIB medium. Metabolites in the extracellular medium were identified by HPLC at the indicated times. HP indicates that 10 mm sodium hypophosphite was added to the incubation medium to inhibit PFL. Each value is the mean of at least four independent experiments carried out with cell suspensions at 107 cells ml−1. The error bars indicate standard deviations. Note the different scales for the two strains.
To further investigate the fermentative abilities of strain 10-6C in view of its elevated ADHE levels, we blocked the PFL-gated pathway by adding to the cells the PFL inhibitor sodium (HP) (16). As shown in Fig. 6, strains CC-124 and 10-6C displayed the same overall response to HP: a production of formate and acetate nearly abolished, and the yield of ethanol increased by 1.5–2-fold. The HPLC profiles did not reveal any other metabolites, not even lactate. Thus, irrespective of the strain, ethanol homofermentation is the main (only) metabolic route when PFL is blocked, which is likely to proceed via PDC and ADHE (Fig. 1).
The HP-inhibited cells showed the same fermentation trend during extended anoxia as compared with non-inhibited cells, i.e. fermentation by strain CC-124 stopped after 6 h but continued in strain 10-6C. The ethanol production by the latter strain almost doubled between 6 and 24 h, reaching concentrations of 1.75 mm (Fig. 6). Altogether, our data indicated that the ADHE-overaccumulating strain exhibits an extended fermentative capacity compared with the wild-type strain, whether PFL is inactivated or not.
Protein Levels in Dark Anoxia
Protein extracts from CC-124 and 10-6C cells exposed to dark anoxia were analyzed by SDS-PAGE. As observed in Fig. 7A, a drastic change in the overall protein profile of wild-type cells was observed between 6 and 24 h, after fermentation stopped. In contrast, protein profiles of mutant strain 10-6C appeared stable through the 24-h period. To gain insights into the respective acclimation of the two strains to dark anoxia, selected proteins were followed by immunoblotting experiments (Fig. 7B). For both strains, a slight increase in ADHE levels could be observed after 6 h of incubation. After 24 h of incubation, the ADHE signal was unchanged in strain 10-6C but appeared fuzzy in strain CC-124 (Fig. 7B). We also followed PFL and PFO, the two enzymes that under anoxia can potentially produce acetyl-CoA, the substrate of ADHE (Fig. 1). During the first hours of dark incubation (up to 6 h), no variation in the PFL levels were noticed. After 24 h, the PFL abundance was unchanged in the mutant strain, whereas the protein could no longer be detected in the wild-type strain. For PFO, the antibodies failed to detect the protein under both oxic and anoxic conditions (not shown), in agreement with earlier studies (18). The RBCL abundance appeared stable over the incubation period in the mutant strain; in contrast, in the wild-type strain, RBCL was no longer detected after 24 h of incubation (Fig. 7B). Notably, the levels of subunit β of the mitochondrial ATPase (ATP2) in both strains were stable over the whole period of dark anoxia (Fig. 7B). The increased abundance of subunit γ of chloroplast ATPase (ATPC) observed in WT strain may be due to a relative over-representation of stable proteins when the total number and amount of proteins is dwindling. Of note, neither the overall protein profiles nor the ADHE abundance were affected by the HP treatment (supplemental Fig. S4). Thus, rather than inducing a protein remodeling in the WT strain, it seems that prolonged dark anoxia has a deleterious effect on its overall proteome, likely representing a degradation of cell integrity. In contrast, the proteome of strain 10-6C, which exhibits higher ADHE levels, appears quite stable despite the prolonged anoxia. Strain 10-6C has been poorly characterized so far, and the lack of knowledge on its genetic background hampered drawing firm conclusions as to the regulation of ADHE levels during aerobic growth and its importance in prolonged dark anoxia.
FIGURE 7.
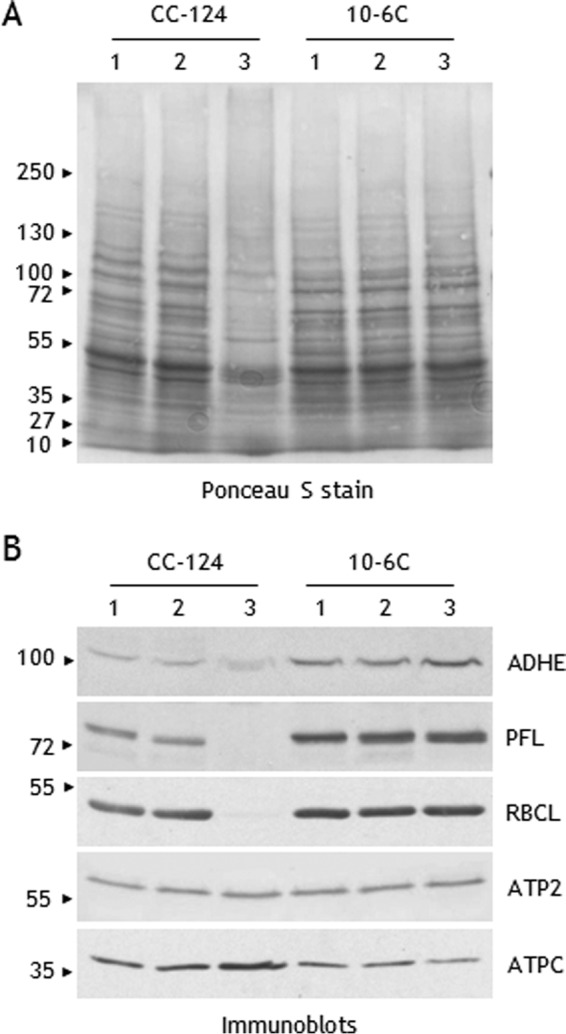
Protein abundance in response to dark anoxia in strains CC-124 (wild type) and 10-6C (lacks Rubisco carboxylase activity). Cells were kept under dark anoxia in AIB medium at a cell concentration of 107 cells ml−1. Proteins in cell extracts (40 μg) were separated by urea/SDS-PAGE (6 m urea, 5–12% acrylamide gel) and transferred to nitrocellulose membrane. Lane 1, exponentially grown cells; lane 2, 6 h of incubation; lane 3, 24 h of incubation. A, nitrocellulose membrane was stained with Ponceau Red S. B, select proteins were detected by immunoblot analyses with antisera against ADHE, PFL, the large subunit of Rubisco (RBCL), subunit β of mitochondrial ATPase (ATP2), and subunit γ of chloroplast ATPase (ATPC). Note that strain 10-6C exhibits higher levels of ADHE and PFL.
Zinc Deficiency Increases ADHE Accumulation and Fermentative Abilities
In a study based on quantitative proteomics, it was reported that aerobic growth in conditions of zinc deficiency increased the ADHE intracellular levels by 3–4-fold in strain CC-4532 (38). The increased ADHE abundance in zinc-deficient cells could be interpreted as a way to maintain a basal ADH capacity in the cell. Indeed, the replacement of the four predicted zinc-dependent ADHs (Cre09.g392134, Cre14.g623650, Cre03.g207800, and Cre03.g207550) by functional homologs that rely on other metals, iron in particular, could ensure optimal (fermentative) metabolism. We first checked the response of our reference strain CC-124 to zinc deficiency. The alga was inoculated in Tris-acetate-Phosphate (TAP) medium without ZnSO4 and later transferred twice into the same medium to reduce the intracellular zinc content. The establishment of metal deficiency was confirmed by the increased abundance of chaperone Zcp2 (supplemental Fig. S5) (38). Protein blots showed that the ADHE levels in zinc-deficient CC-124 were ∼3-fold higher than in zinc-replete cells (supplemental Fig. S5 and Table 4), thus making these cells of value for our fermentation studies.
The response of zinc-deficient CC-124 cells to 24 h of dark anoxia was examined through the analysis of proteins and fermentation products (Fig. 8). HPLC analysis of excreted metabolites indicated the presence of formate, acetate, and ethanol in a molar ratio of 2:1:1, being thus comparable with the ratio obtained with cells grown on standard TAP medium. The respective amounts of these products were, however, found to be 2-fold higher in zinc-deficient cells as compared with zinc-replete cells (Fig. 8A), revealing the higher fermentative capacities of the metal-deficient cells. In contrast to zinc-replete CC-124 cells, the overall protein profile of zinc-deficient cells hardly changed after prolonged anoxia, as observed in strain 10-6C (Fig. 8B). Notably, the levels of ADHE, PFL, and the PTAs, which compete with ADHE for acetyl-CoA (Fig. 1), remained rather stable in zinc-deficient cells (Fig. 8B). On the whole, our data had several implications: (i) strain CC-124 is not intrinsically unfit to withstand prolonged anoxia, (ii) zinc deficiency enhances the cell capacity to survive anoxia, and (iii) in C. reinhardtii, the PFL-gated pathway is the preferred fermentative route irrespective of the zinc status of the cell.
FIGURE 8.
Fermentation by zinc-deficient cells. Cells were incubated for 24 h under dark anoxia in AIB medium, at a concentration of 107 cells ml−1. Sample 1, strain CC-124 grown on TAP medium; sample 2, strain CC-124 grown on zinc-deficient TAP medium; sample 3, strain 10-6C grown on TAP medium. A, fermentation products excreted after 24 h in dark anoxia. Metabolite concentrations are given for 107 cells ml−1. B and C, proteins in extracts from cells before (t = 0) and after a prolonged incubation in anoxia (t = 24 h) were analyzed on urea/SDS-PAGE (6 m urea, 5–12% acrylamide) and transferred to nitrocellulose membrane. B, Ponceau S-stained nitrocellulose membrane. C, immunoblots showing the levels of specific fermentative enzymes. The two bands detected by the anti-PTA serum likely correspond to the chloroplast and mitochondrial isoforms of phosphotransacetylase (supplemental Fig. S6 and S7).
Evaluating ADHE Accumulation in Relation to Starch Levels
In the conditions of fermentation applied here, i.e. dark incubation in a potassium phosphate medium, starch content is believed to be key for the maintenance of cell integrity and survival. Indeed, starch is the source of glucose, directly fueling glycolysis to produce ATP under anoxia (Fig. 1). We therefore examined the starch content in the cells analyzed above. As shown in Fig. 9A, the amounts of starch in strain CC-124 increased 3–4-fold when zinc was removed from medium, reaching up to 5–6 μg starch/106 cells. The analysis of cell sections by electron microscopy confirmed the higher starch content in zinc-deficient cells relative to zinc-replete cells (Fig. 9B). Of note, starch content in TAP-grown 10-6C cells was also found to be higher than in the reference CC-124 cells, with ∼6–7 μg/106 cells (not shown). These data thus suggest that the higher fermentation capacity of zinc-deficient CC-124 cells and strain 10-6C, relative to standard CC-124 cells, is a direct result of a higher content in C-reserves. The associated increase in ADHE levels may be then projected to facilitate the ethanol fermentation of starch over longer periods.
FIGURE 9.
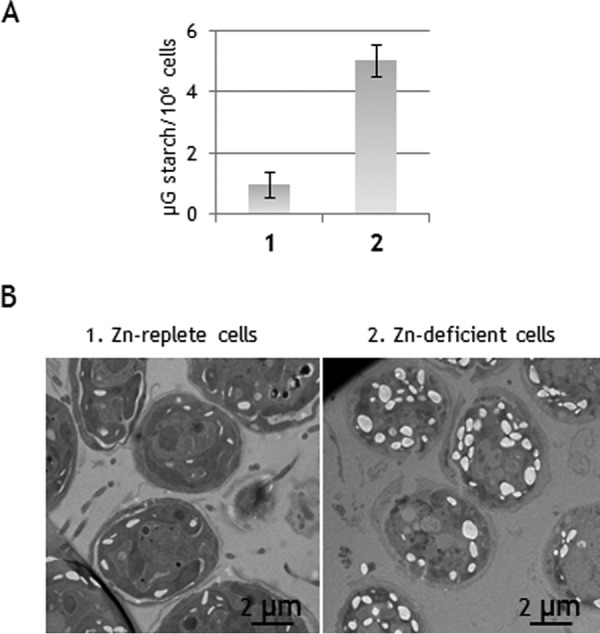
Impact of zinc on starch accumulation. A, starch content in exponentially growing C. reinhardtii cells (strain CC-124). The data are the means of at least three independent replicates. B, electron micrographs of cell sections showing higher starch content in cells deficient in zinc compared with cells grown in presence of zinc.
To further evaluate the link between ADHE up-regulation and starch accumulation, the protein abundance was followed after transferring the cells to TAP medium without ammonium (N-free medium). Such a medium is known to trigger the accumulation of large amounts of starch in the green alga C. reinhardtii (39). Time course experiments with long term sampling (0, 17, 24, 40, 49, and 72 h) were carried out with strain CC-124. In our conditions, cellular starch content increased steadily during the first 2 days after transfer to N-free medium (Fig. 10). Followed by protein blots, the ADHE steady-state levels were found to increase progressively during the first 24 h (∼2.5-fold) (Table 4), thus correlating with the increase in starch content (Fig. 10). Later a slow decrease in the protein abundance was observed (Fig. 10). PFL levels appeared rather stable throughout the whole period, although a slight decrease was observed in the first hours of N-deprivation (Fig. 10). The behavior of ADHE and PFL in response to N-starvation is interesting because it contrasts with that of other chloroplast proteins, including enzymes of the Calvin-Benson-Bassham (CBB) cycle and components of the photosynthetic chain, whose levels decreased shortly after the transfer to N-free medium (40, 41). Here we observed within the first 24 h of N-starvation an important decrease (more than 50%) in the abundance of RBCL and phosphoribulokinase, two key enzymes of the CBB cycle, and of ADP-glucose pyrophosphorylase (ADPG; EC 2.7.7.27), the chloroplast enzyme that catalyzes the first committed step in starch synthesis. The decrease in ADPG while starch content increases is puzzling (Fig. 10) but could indicate that ADPG is present at overcapacity in TAP-grown cells and is tuned down to a minimum level to sustain starch production in N-free medium. Altogether, our observations indicate that ADHE, which is synthesized in conditions where N is limiting, occupies a position of priority among chloroplast proteins.
FIGURE 10.
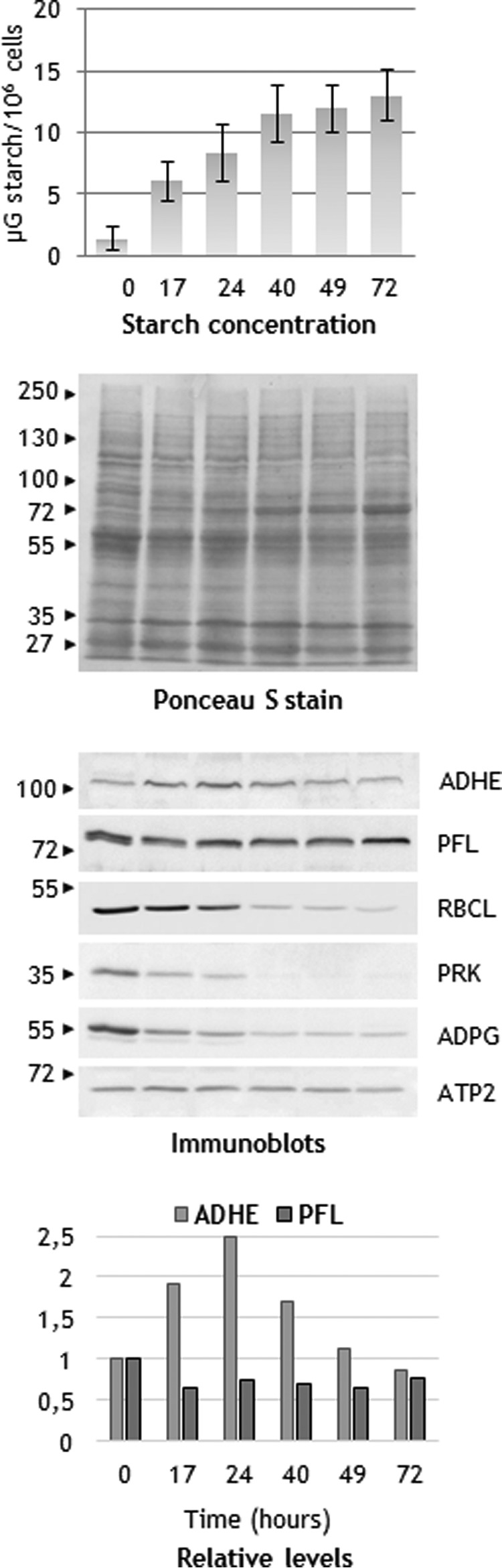
ADHE levels increase upon exposure to nitrogen starvation conditions. Starch content and select proteins were analyzed after transfer of C. reinhardtii strain CC-124 to N-free TAP medium. Proteins in cell extracts were separated on SDS-PAGE (10%) and transferred to nitrocellulose membrane.
Finally, we addressed the potential link between ADHE accumulation and starch synthesis. For that, we used mutant strain Sta6, which is unable to synthesize starch because it lacks the ADPG small subunit (42). In this strain growing on standard TAP medium, ADHE is present at low levels, comparable with those in wild-type strain CC-124 (Table 4). The transfer of Sta6 cells to zinc-deficient TAP medium led to a ∼3-fold increase in ADHE abundance (Table 4), similar to that observed with wild-type strain CC-124. From the study of starch-less strain Sta6, it can be inferred that ADHE accumulation is not directly controlled by starch synthesis. This does, however, not exclude a common origin of the regulation of ADHE and starch.
Evaluating ADHE Accumulation in Relation to CO2 Requirements
A well documented effect of zinc deficiency in C. reinhardtii is the impairment of carbon-concentrating mechanism (CCM) under atmospheric conditions (38, 43). Therefore we considered the possibility that the aforementioned rise in ADHE levels in zinc-deprived cells CC-124 and Sta6 might be a consequence of reduced CO2 availability.
All the experiments described in this work were carried out with cells grown on TAP-derived medium under atmospheric conditions (0.04% CO2) at a light intensity of 40 μE m−2 s−1. In these conditions, the cells rely on CCM to concentrate CO2 at the active site of the Rubisco (44). The ADHE abundance was first assessed in two CCM mutant strains: strain CIA5, which lacks Ccm1, a master gene regulator that controls the induction of CCM by sensing CO2 availability in C. reinhardtii (45, 46), and strain CIA3, which lacks CAH3, the thylakoid lumen carbonic anhydrase, which provides CO2 to the Rubisco (47). The ADHE abundance in these two strains grown under atmospheric conditions was similar, being 1.5–1.8-fold higher than that in wild-type strain CC-124 (Table 4).
Additional observations provided further support for a link between the ADHE levels and the CO2 availability: (i) ADHE was hardly detected in wild-type strain CC-124 grown on TAP medium supplemented with 2% CO2 (in these conditions CCM is not required) (Table 4); (ii) ADHE was only faintly observed in strain ΔRBCL (Table 4), which lacks Rubisco because of a large deletion in the chloroplast RBCL gene (48). This strain, which is light-sensitive, relies mostly on mitochondrial metabolism for growth (49); and (iii) at the 5′-UTR of the ADHE gene a typical low CO2 cis-acting enhancer element was identified (GAGTTGC; position −299 to −293 from the ATG) (50), which argues for the up-regulation of the ADHE expression under low CO2.
Light was also found to influence the ADHE abundance in C. reinhardtii. Wild-type CC-124 cells were grown to early exponential-phase on TAP medium at a light intensity of 40 μmol photons m−2 s−1 and later exposed to different light intensities for 20 h. Cells grown at 15 and 40 μE m−2 s−1 exhibited similar amounts of ADHE, whereas cells grown at 200 μE m−2 s−1 contained slightly more ADHE (Table 4). At higher light intensity, the cells require more CO2, which may not be fulfilled under the atmospheric conditions and thus constitute a signal for ADHE accumulation.
Discussion
Bifunctional aldehyde/alcohol dehydrogenases are present in a variety of facultative and strict anaerobic bacteria but remain so far undetected in Archaea. In the eukaryotic world, ADHEs have long been thought to be restricted to anaerobic species (pathogens), but over the last decade, ADHE genes have been uncovered in species of varied lifestyles. In particular, ADHEs have been found in photosynthetic microalgae thriving in fresh or marine waters (14). Despite the significant set of bacterial and eukaryotic ADHE sequences used in our phylogenetic analysis (supplemental Data Set S1), the evolutionary history of eukaryote enzymes remains confusing. Only in the case of photosynthetic microalgae, the ADHE is found in two distinct positions in the tree (Fig. 2): the enzymes in B. natans and G. theta are found among most of their counterparts in anaerobic eukaryotes, between the two bacterial protein clusters, whereas the chlorophyte ADHEs are found in cluster I, in close proximity to the cyanobacterial counterparts (Fig. 2). Hence, at least two different ADHE gene donors have to be considered in the evolution of unicellular algae.
Microalgae with primary plastids, i.e. chlorophytes, rhodophytes, glaucophytes, are believed to have emerged from the ancient endosymbiosis of a cyanobacterium in a heterotrophic eukaryote (51). As shown here, a vertical inheritance of ADHE from cyanobacteria appears sound for the chlorophytes, especially because the enzyme is present in the species of the orders Nostocales and Stigonematales, proposed to be at the origin of all primary plastids (52). Up to now, only few genomes of glaucophytes and rhodophytes have been sequenced, none of them containing any ADHE genes. The evolution of the ADHE gene (retention versus loss) among species with primary plastids cannot be understood without more genome sequences from these glaucophyte and rhodophyte algal lineages.
C. reinhardtii is the first eukaryote known so far in which the ADHE localizes to the chloroplast; such a compartmentalization is likely reminiscent of the cyanobacterial ancestor. Despite the chloroplast confinement, the algal ADHE exhibits molecular and enzymatic traits comparable with those of its counterparts in bacteria and amitochondriate eukaryotes. This is significant because ADHE is known as a major target of metal-catalyzed oxidation (53). In this process, Fe2+ ions in the protein active site (Fig. 1) react with ROS and generate hydroxyl radicals. In turn, these radicals can selectively oxidize the nearby His residue, leading to the irreversible inactivation of the enzyme. Also interesting is the fact that spirosome-like structures were found in an ADHE-enriched stromal fraction. Assuming that these assemblies are indeed ADHE oligomers, this would indicate that the formation of spirosomes can also take place in the oxygen-rich atmosphere of the chloroplast.
Previous studies on the fermentative capacities of C. reinhardtii indicated that both the chloroplast and the mitochondria contained a complete PFL-gated pathway, composed of a PFL, a PTA-ACK, and an ADHE (8). Although the presence of a PFL and PTA-ACK in mitochondria has been confirmed by biochemical and molecular approaches (12, 24), the attempts to detect an ADHE have failed as yet (12, 33) (Fig. 4). Further studies will determine whether PFL and PTA-ACK form a pathway leading to formate and acetate or whether they act independently in the mitochondrion.
Our study adds further weight to the notion that decreased oxygen availability is not a signal for ADHE accumulation in C. reinhardtii (Fig. 6). In this respect, the microalga differs from the facultative anaerobic bacteria studied to date. In E. coli, for example, ADHE abundance increases up to 10-fold upon exposure to anoxia (54). In Staphylococcus aureus, ADHE is a target of the redox sensing transcriptional regulator Rex, which regulates most genes encoding fermentative enzymes (55). The few data on photosynthetic bacteria (cyanobacteria) also point at a regulation by anoxia: the AdhE transcripts in Synechococcus species in microbial mats (Octopus Spring, Yellowstone National Park) were shown to increase during the evening (56). The rationale for the different enzyme regulation between facultative anaerobic bacteria and C. reinhardtii is likely to be found in differences in lifestyle and cell complexity. The constitutive accumulation in the alga might specify the need to respond quickly to dark anoxia. It could make good sense for C. reinhardtii to produce ADHE, a large protein, under aerobic conditions when energy is not usually limiting rather than under anoxia, when ATP is in short supply. This also holds for PFL, which accumulates under aerobic conditions (12) (Figs. 7 and 8). Alternatively, the accumulation of ADHE under oxic conditions could indicate that the enzyme is also required in aerated environments. Production of ethanol under atmospheric conditions, as it happens in S. cerevisiae, seems unlikely because ethanol has not been detected in the culture medium of aerobically growing algae (not shown). The ADHE in the alga could also have a function not directly linked to ethanol metabolism, as reported recently in E. coli (57, 58).
Throughout this work we have identified situations (mutant strains and physiological conditions) that induce fluctuations in ADHE abundance in C. reinhardtii. Increased ADHE abundance was observed in cells growing on TAP medium lacking zinc or a nitrogen source (Figs. 8 and 10), in mutant strains impaired in the carbon concentrating mechanism (CIA5 and CIA3), in cells lacking Rubisco carboxylase activity (strain 10-6C), or even in conditions of high light (Table 4). Inversely, ADHE was hardly detected in cells that do not rely on CCM for growth (Table 4). A possible rationale for an increased ADHE abundance in conditions where carbon concentration is impaired may relate to an over-reduction state of the chloroplast: low CO2 or the absence of CBB cycle activity will strongly decrease CO2 reduction, which is a major electron sink in the light. Interestingly, our data showed that enhanced ADHE levels coincide with higher starch content in the cells; that is the case for cells growing on TAP medium lacking zinc or a nitrogen source and in cells lacking Rubisco carboxylase activity (strain 10-6C) (Figs. 8 and 10). As described earlier, we have identified a cis-acting element in the promotor region of the ADHE gene that could provide a rationale for the observed regulatory influence of CO2. Importantly, the same type of element was found in the AGDP gene, the rate-limiting enzyme involved in starch synthesis. A concerted induction by low CO2 could constitute a direct link between ADHE and starch synthesis. Still, a probable factor in the concerted accumulation of ADHE and starch is an increase in reducing equivalent levels, as is known for bacterial ADHEs. Indeed, all conditions where ADHE is up-regulated imply a diminished use of reducing equivalents, for example a defunct Rubisco. The reason why this is not sufficient in C. reinhardtii is that, unlike bacteria, the alga can only ferment internal starch reserves, which thus have to be synthesized first. For this, an increase in ATP status is also required, and therefore we theorize that regulation in C. reinhardtii requires increases in both redox equivalents and adenylate status.
In our experimental conditions, the alga ferments its carbon stores mainly into formate, acetate, and ethanol in a molar ratio of 2:1:1. This mixed acid fermentation indicates the involvement of a metabolic pathway gated through PFL. The use of this route has a clear advantage over other more typical routes in eukaryotes (PDC/ADH or lactate dehydrogenase) (Fig. 1) as it balances NAD+ regeneration with the need for energy production in conditions of dark anoxia. Here we showed that cells with elevated ADHE levels (strain 10-6C and zinc-deprived strain CC-124) exhibit the same mixed acid fermentation rate but have extended fermentation abilities. This means that ADHE is not rate-limiting in this fermentative pathway. From the product yields at 24 h (Figs. 6 and 8), it is inferred that the fermentation span is at least 16 h. Furthermore, our results revealed the higher stability of the overall proteome in cells with enhanced ADHE levels. After 24 h of dark anoxia, the protein profiles of the cells with higher ADHE content were shown to be unaltered, contrasting with the situation in the reference cells where protein degradation, in particular degradation of fermentative enzymes, takes place. Cells with higher ADHE are better suited to survive under dark anoxia, which appears most immediately linked to the content in endogenous carbohydrate content. If we assume that ADHE is only required for anaerobic metabolism, it could be hypothesized that the high ADHE content, which seems intricately linked to high starch levels, anticipates the loss of protein because of inactivation and/or degradation that inevitably occurs over the extended fermentation periods that higher starch stores will allow. The question of whether an increased ADHE level is indeed crucial for prolonged anoxic survival remains to be answered because we have not identified a condition or strain with high starch and low amounts of ADHE. The amazing ability of C. reinhardtii to survive over long periods of dark anoxia was in some way unexpected for a photosynthetic alga that typically lives in mid latitudes where day (light) length varies only moderately. It would thus appear that other conditions than light also determine to what extent the algal cell is in fact exposed to conditions of dark anoxia.
We believe that the present work forms a solid basis to disclose the molecular details of the ADHE regulation in C. reinhardtii. The predominant role of ADHE in the fermentation metabolism of the green alga is confirmed here, but our studies cannot rule out the possibility that this enzyme has other functions (under oxic conditions). To elucidate the mechanism underlying the aldehyde/alcohol dehydrogenase up-regulation as well as to identify potential alternative functions for ADHE in the green alga, the examination of counterparts from diverse biochemical context and thriving in different environments is expected to provide valuable insights. Among the ADHEs that would deserve scrutiny are those from cyanobacteria and from non-chlorophyte photosynthetic algae, such as the members of the SAR supergroup, in which the repertoire of fermentative enzymes is distinct from that in chlorophytes (14).
Experimental Procedures
C. reinhardtii Strains, Media, and Culture Conditions
From the Chlamydomonas Resource Center of Minnesota University, the following strains were obtained: CC-124 (137c mt− nit1− nit 2−), CC-4348 (Sta6-1 mt+), and CC-2702 (CIA5). From the ChlamyStation Collection (CNRS UMR7141, Paris, France) were obtained the Rubisco mutants 10-6C.arg2.mt+ and ΔRbcL 1.7.5. The presence of the point mutation G171D in RBCL, which confers impaired CO2 fixation in strain 10-6C (31, 32), was confirmed in this work by protein-based mass spectrometry analysis (supplemental Fig. S2). Strain CIA3 was a gift from Göran Samuelsson (Umeå University, Umeå, Sweden). All strains were maintained at 24 °C on TAP medium (59) containing 1.5% agar. For strain 10-6C.arg2.mt+, TAP medium was supplemented with arginine (100 mg/ml). All media were prepared with MilliQ water.
The cells were grown in flasks on TAP medium unless otherwise stated. Flasks were placed in an Innova incubator (New Brunswick Scientific) at 125 rpm under continuous light at 24 °C. The cells were routinely grown at a light intensity of 40–50 μE m−2 s−1, except strain ΔRbcL 1.7.5 at 10–15 μE m−2 s−1. Wild-type strain CC-124 was also grown at 15 and 200 μE m−2 s−1. For growth in absence of zinc, the culture medium was prepared by omitting ZnSO4 from the standard TAP medium. Cells from TAP plates were inoculated into 100-ml of “zinc-free” medium and grown to their late exponential phase. The cells were then transferred into a fresh medium without zinc at a cell concentration of ∼ 1 × 105 cells ml−1. Zinc deficiency was reached after a third round of transfer into “zinc-free” medium. The analyses of zinc-deficient cells were carried out on cells harvested in their mid-exponential phase. N-free medium was prepared by omitting ammonium chloride from the TAP medium. The cells subjected to N-deprivation were grown to 3–4 × 106 cells ml−1 and collected by centrifugation at 1,500 × g for 5 min at room temperature. The cells were washed in N-free TAP medium and finally resuspended in N-free TAP to ∼1 × 106 cells ml−1. For experiments involving CO2 supplementation, the cultures were grown in an incubator bubbled with an atmosphere of 2% CO2 in air. The cell densities were determined using a Malassez hemocytometer after addition of Lugol's solution (Sigma-Aldrich) to the cell suspension to immobilize the cells.
Organelle Isolation and Partial Purification of Chloroplast ADHE
Isolation of chloroplasts and mitochondria from mutant strain 10-6C was essentially as described in (60). For the ADHE partial purification, the chloroplasts were resuspended in 50 mm HEPES (pH 7.0) in presence of protease inhibitors (0.5 mm benzamidine and 1 mm aminocaproic acid) to a final protein concentration of 5 mG/ml, frozen and thawed twice, and centrifuged at 11,000 × g for 15 min at 4 °C. The ADHE-containing supernatant was applied to an anion exchange Ceramic HyperD® resin (GE Healthcare), equilibrated, and eluted with 50 mm HEPES (pH 7.0). In these conditions, ADHE was not retained on the column. ADHE in the flow-through was either pelleted by ultracentrifugation at 110,000 × g for 1 h (4 °C) or applied on a Blue Sepharose 6 (GE Healthcare). Elution from the Blue resin was achieved with 1 m NaCl in 50 mm HEPES (pH 7.0).
Mass Spectrometry Analysis
For mass spectrometry analysis, different samples were analyzed. Whole chloroplasts were loaded on 6 m urea/SDS-PAGE (5–12% acrylamide), and gel bands containing PTAs (80 kDa), RBCL (45 kDa), and ADHE (100-kDa) were cut out. ADHE was also identified in whole cell lysate and in a chloroplast soluble fraction. For the two latter samples, 3–4 μg of proteins were stacked at the top of a 10% acrylamide gel, and gel bands containing the concentrated proteins were cut out. All gel bands were cut in pieces before being washed by six successive incubations of 15 min in 25 mm NH4HCO3 and in 25 mm NH4HCO3 containing 50% (v/v) acetonitrile. Gel pieces were then dehydrated with 100% acetonitrile and incubated for 45 min at 53 °C with 10 mm DTT in 25 mm NH4HCO3 and for 35 min in the dark with 55 mm iodoacetamide in 25 mm NH4HCO3. Alkylation was stopped by adding 10 mm DTT in 25 mm NH4HCO3 and mixing for 10 min. Gel pieces were then washed again by incubation in 25 mm NH4HCO3 before dehydration with 100% acetonitrile. Modified trypsin (Promega, sequencing grade) in 25 mm NH4HCO3 was added to the dehydrated gel pieces for an overnight incubation at 37 °C. Peptides were then extracted in three sequential extraction steps (15 min each) in 30 μl of 50% acetonitrile, 30 μl of 5% formic acid, and finally 30 μl of 100% acetonitrile. The pooled supernatants were then dried under vacuum.
The dried extracted peptides were resuspended in 5% acetonitrile and 0.1% trifluoroacetic acid and analyzed by online nanoLC-MS/MS (Ultimate 3000, Dionex, and LTQ-Orbitrap Velos pro; Thermo Scientific). Peptides were sampled on a 300 μm × 5 mm PepMap C18 precolumn and separated on a 75 μm × 250 mm C18 column (PepMap, Dionex). The nanoLC methods consisted of 25- and 120-min acetronitrile gradients in 0.1% formic acid at a flow rate of 300 nl/min for, respectively, gel band and stacking analyses. MS and MS/MS (Top20) data were acquired using Xcalibur (Thermo Scientific) and processed automatically using Mascot Daemon software (version 2.5; Matrix Science). Searches against the Phytozome database (version 9.0, C. reinhardtii taxonomy) (61), a homemade classical contaminants database, and the corresponding reversed databases were performed using Mascot (version 2.5.1). ESI-TRAP was chosen as the instrument, trypsin/P was chosen as the enzyme, and two missed cleavages allowed. Precursor and fragment mass error tolerances were set, respectively, at 10 ppm and at 0.6 Da. Peptide modifications allowed during the search were carbamidomethyl (C, fixed) acetyl (N-terminal, variable), and oxidation (M, variable). The IRMa software (62) (version 1.31.1) was used to filter the results: conservation of rank 1 peptides, peptide identification false discovery rate < 1% (as calculated on peptide scores by employing the reverse database strategy), and minimum of 1 specific peptide/identified protein group.
Dark Fermentation
500–700 ml of cultures in their mid-exponential phase were harvested, washed in 10–20 ml of filtered-sterilized anaerobic induction buffer (AIB) medium, and finally resuspended in sterile AIB medium to a cell concentration of ∼1 × 107 cells ml−1. 30 ml of cell suspension was transferred to a 60-ml glass serum bottle. Each bottle was closed with a butyl rubber stopper, wrapped in aluminum foil, degassed for 10 min, and then purged with argon for 15 min. The bottles were placed in an incubator at 24 °C and agitated at 125 rpm. Where indicated, sodium hypophosphite (at a final concentration of 10 mm) was added to the cell suspension. Anerobic samples were taken regularly and processed for the analysis of protein, fermentation products, and starch content (see below). After each sampling, the bottles were flushed with argon.
Analysis of Fermentation Products
Cells acclimated to dark anoxia were harvested by centrifugation (7 min at 14,000 × g). The resulting supernatants were filtered through a nylon filter (0.2 μm) and frozen until use. Prior to the analysis by HPLC (Agilent 1260), the samples were diluted twice in 10 mm H2SO4 and centrifuged briefly. The fermentative products were then separated using a HiPlex-H column for carbohydrates, organic acids, and alcohols (300 × 7.7 mm, 8 μm; Agilent) at 50 °C; the mobile phase was 10 mm sulfuric acid, and the flow rate was 0.6 ml/min. Detection was done with a refractive index detector (50 °C) and a photodiode array detector operating at 210 and 280 nm. The predominant products were identified as formate (14 min), acetate (15.3 min), and ethanol (21.7 min). Signals were integrated, and metabolites were quantified using a standard curve generated for each metabolite detected.
Starch Quantification
Cells (2–5 × 106 cells) from cultures or anaerobic incubations were collected by centrifugation (5 min at 14,000 × g) and frozen in liquid nitrogen until use. To disrupt cell integrity and extract pigments, 1 ml of 80% acetone is added to each cell pellet. The cell residues obtained after centrifugation were rinsed with 0.5 ml of 100 mm sodium acetate buffer (pH 4.5) and then resuspended in 300 μl of the same buffer and heat-treated for 20 min at 120 °C for starch solubilization. Total starch was quantified using an enzymatic starch assay kit including amyloglucosidase, hexokinase, and glucose-6-phosphate dehydrogenase (R-Biopharm, Darmstadt, Germany). Alternatively, solubilized starch was stained using a commercial iodine solution (Sigma) as follows: 100 μl of solubilized starch were mixed to 800 μl of water and 100 μl of Lugol's solution. Absorbance was measured spectrophotometrically at 580 nm. Quantification was done using commercial starch (R-Biopharm). The results from both methods were comparable.
Determination of Protein Concentration, SDS-PAGE, and Immunodetection
Protein concentrations were routinely determined using the bicinchoninic acid-based Thermo Pierce® protein assay kit, and bovine serum albumin as standard. When protein concentrations of cell extracts were to be determined, proteins were first precipitated by CHCl3/MeOH (63) and then resuspended in 2% SDS (w/v). Proteins were analyzed either on 10% acrylamide SDS-PAGE, 7.5–12% acrylamide SDS-PAGE, or 5–12% acrylamide gels containing 6 m urea (urea/SDS-PAGE). Protein samples (40 μg) were resuspended in 2% SDS (w/v) and 2% β-mercaptoethanol and boiled for 90 s. Insoluble material was then removed via a 2-min centrifugation at 13,000 × g. Apparent molecular masses were estimated using prestained protein ladder Plus (Euromedex). After electrophoresis, proteins were either stained with Coomassie Blue R250 or with Ponceau S after transfer onto nitrocellulose membranes.
Immunoblotting experiments were carried out following a standard protocol at room temperature. Nitrocellulose membranes were first blocked for 1 h in 20 mm Tris, 300 mm NaCl, pH 7.5, supplemented with 0.1% Tween 20 before incubation with primary antibodies for 1 h. The primary antibodies were from various sources. From Agrisera AB (Vännäs, Sweden) were purchased antibodies against ADGP (AS11 1739), ATPC (AS08 312), CAH3 (AS05073), HydA (AS09514), PDC (AS10691), and RbcL (AS03037). Antibodies against ADHE, PFL, and ATP2 were described by (12). Antibodies to PTAs were generated by immunization using heterologous protein (supplemental Figs. S6 and S7). Anti-LHC was from O. Vallon (Institut de Biologie Physico-Chimique (IBPC), Paris, France), and anti-phosphoribulokinase was from Mike Salvucci (U.S. Department of Agriculture, Washington, D. C.). The Zcp2 antibodies were a gift from S. Merchant (UCLA, Los Angeles, CA). Anti-rabbit IgG peroxidase conjugate (Sigma-Aldrich) was used as secondary antibody. Immunochemical detection was carried out using the homemade enhanced chemiluminescence system (64) and Image Quant LAS 4000 mini (GE).
BN-PAGE and Two-dimensional SDS-PAGE
BN-PAGE was carried out using a 3–12% linear polyacrylamide gradient (27). Freshly purified rADHE (25 μg) was supplemented with loading buffer that contains 30 mG/ml Blue G in 50 mm Tricine (pH 8.0). Electrophoresis was carried out at 4 °C at constant current (10 mA) for ∼1 h. The molecular mass of the rADHE oligomers separated on BN-PAGE were evaluated using commercial molecular mass markers (bovine serum albumin (66 kDa), S. cerevisiae alcohol dehydrogenase (150 kDa), horse spleen apoferritin (443 kDa), and bovine thyroglobulin (669 kDa) (Sigma-Aldrich)). In-gel alcohol dehydrogenase activity assay was performed at room temperature as follows. A BN gel lane was washed twice in 100 mm Tris (pH 8.8) and then incubated in 25 ml of 100 mm Tris-Cl (pH 8.8) containing 20 mg of NAD, 5 mg of nitroblue tetrazolium, 1 mg of phenazine methosulfate, and 25 μl of absolute ethanol. In the presence of phenazine methosulfate, nitroblue tetrazolium reacts with NADH produced by dehydrogenases to produce an insoluble blue-purple formazan precipitate. For two-dimensional BN/SDS-PAGE, a BN gel lane was cut out after the run, washed three times for 5 min in 1% SDS and 1% 2-mercaptoethanol, and then applied on top of a 10% SDS-PAGE gel. After electrophoresis, the proteins were transferred onto nitrocellulose membrane.
Expression and Purification of C. reinhardtii ADHE
The coding region of C. reinhardtii ADHE was amplified using the following pair of primers: CrADHE_NdeI gaccatatggccgctgccccggccaccc (61AAAPAT66) and CrADHE_XhoI gtcctcgagGTTGATCTTGGAGAAGAACTC (948EFFSKIN954) using as a template the full-length cDNA (CAF04128). The amplified fragment was cloned into the pET-24a expression vector (Novagen) using the XhoI and NdeI restriction sites within the multiple cloning site to incorporate a C-terminal His6 tag. The sequence of the clone pET24a-CrADHE was confirmed at GATC Biotech. E. coli BL21 (DE3) cells were transformed with this construct. Transformed cells from plates were inoculated in the autoinduction medium ZYM2052 (65) and grown overnight at 37 °C. The cells were harvested, washed once in buffer 1 (300 mm NaCl, 50 mm sodium phosphate, pH 8.0) and frozen. Purification of ADHE was performed the same day at 4 °C under argon. The cells were resuspended in buffer 1 supplemented with 0.5 mg/ml lysozyme, 0.5 mg/ml DNase, and 2 mm MgCl2 and protease inhibitors (PMSF, amino caproic acid, and benzamidine) and then broken by three cycles of freezing (liquid nitrogen) and thawing. The broken cells were centrifuged at 10,000 × g for 15 min at 4 °C. The supernatant was collected and gently mixed with HIS-Select nickel affinity gel (Sigma-Aldrich). The mixture was then poured into an empty column. The gel was extensively washed with 10 mm imidazole in buffer 1. The His-tagged ADHE was eluted with 100 mm imidazole in buffer 1, quickly desalted on a PD10 column pre-equilibrated with 50 mm Tricine, pH 8.0, and finally concentrated on a Vivaspin 30 (cut off 30 kDa) (GE Healthcare) to reach a final concentration of 5–7 mg protein/ml.
Enzyme Assays
Freshly prepared ADHE was tested for the different enzymatic activities, at 25 °C. Standard assay medium for NADH-dependent reactions consisted of 50 mm potassium phosphate (pH 7.0) and 0.4 2 mm NADH. Acetyl-CoA reductase activity was determined in the presence of 0.21 mm acetyl-CoA; acetaldehyde reductase was determined with 1 mm acetaldehyde and 0.21 mm CoASH. The rate of both reductase reactions were monitored spectrophotometrically following the disappearance of NADH at 340 nm. Standard assay medium for NAD+-dependent reactions contained 1.5 mm NAD+ in 50 mm glycine/NaOH buffer (pH 9.0). Ethanol dehydrogenase activity was assayed in 170 mm ethanol (1% v/v); acetaldehyde dehydrogenase activity was assayed in 1 mm acetaldehyde and 0.2 mm CoASH. The rates of both dehydrogenases were monitored by the formation of NADH at 340 nm. All reactions were started by the addition of 25–40 μg of purified enzyme. In all the assays, one unit of enzyme activity was defined as 1 μmol of NADH converted per min.
Electron Microscopy
For negative staining: 5-μl drops of the protein suspension were placed directly on glow discharged carbon coated grids (EMS) for 3 min. The grids were then washed with two drops of 2% uranyl acetate in water and stained with a third drop for 1 min. The grids were dried on filter paper and the samples were analyzed using a Tecnai 200KV (operated at 120 kV) electron microscope (FEI). Digital acquisitions were made with a numeric camera (Eagle, FEI). For morphology, in preparation for high-pressure freezing, aliquots of the liquid cultures were transferred to 50-ml conical centrifuge tubes, spun at 500 × g for 5 min, and then gently resuspended in the growth medium containing 150 mm mannitol (66). Subsequently the cells were pelleted, high pressure frozen, freeze substituted, and embedded in LR White resin (hard grade), according to Ref. 67. Ultrathin sections were prepared with an ultracryomicrotome (EM UC7 Leica).
Bioinformatic and Phylogenetic Analyses
Sequence alignments were done with the Muscle module as part of the MEGA 6 program (68) and manually adjusted to optimize alignments. The maximum likelihood phylogenetic tree was also produced using the MEGA 6 program. Conserved domains in CrADHE were identified using the NCBI-CDD database. Iron-binding signatures were identified using Prosite, a database of protein domains, families, and functional sites. Protein quantification was done on Western blots using ImageJ. Km values were calculated with the help of the program Hyper32 for hyperbolic regression analysis.
Author Contributions
A. A., R. V. L., and D. D. designed the research. A. A., R. V. L., and M. P. performed the research. R. V. L. and W. N. phylogenetic analyses. Y. C. carried out the proteomic analyses. A. K. performed electron microscopy. A. A. and R. V. L. wrote the article with contributions from all the authors.
Supplementary Material
Acknowledgments
We thank are due to Dr. Göran Samuelsson (Umeå University) for providing strain cia3, Dr. Joanna Porankiewicz-Asplund (Agrisera) for several useful insights regarding the antibodies, Sabeeha Merchant (UCLA) for the antibodies to ZCP2 and discussions on zinc deficiency at the early stage of this work, Sandrine Bujaldon (IBPC) for various discussions around the strains and for fluorescence measurements, and Jean-François Sassi (Cité des Energies, CEA-Cadarache) for the cultures in 2% CO2.
This work was supported by the Centre National de la Recherche Scientifique and partly supported by Agence Nationale pour la Recherche Grant ANR-10-BIOE-0004 (to A. A., R. V. L., and M. P.), and ProFi Infrastructure Grant ANR-10-INBS-08-01 (to Y. C.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Figs. S1–S7 and Data Sets S1 and S2.
- PDC
- pyruvate decarboxylase
- ADHE
- aldehyde/alcohol dehydrogenase
- ADH
- alcohol dehydrogenase
- PFL
- pyruvate formate lyase
- PFO
- pyruvate:ferredoxin oxidoreductase
- RBCL
- large subunit of the Rubisco
- SAR
- Stramenopiles-Alveolata-Rhizaria
- LHC
- light-harvesting complex protein
- BN
- Blue Native
- ALDH
- aldehyde dehydrogenase
- PTA
- phosphotransacetylase
- ACK
- acetate kinase
- HP
- hypophosphite
- CBB
- Calvin-Benson-Bassham
- ADPG
- ADP-glucose pyrophosphorylase
- CCM
- carbon-concentrating mechanism
- Rubisco
- ribulose-bisphosphate carboxylase/oxygenase
- TAP
- Tris-acetate-phosphate
- AIB
- anaerobic induction buffer.
References
- 1. Oren A., and Shilo M. (1979) Anaerobic heterotrophic dark metabolism in the cyanobacterium Oscillatoria limnetica: sulfur respiration and lactate fermentation. Arch. Microbiol. 122, 77–84 [Google Scholar]
- 2. Moezelaar R., Bijvank S. M., and Stal L. J. (1996) Fermentation and sulfur reduction in the Mat-building cyanobacterium Microcoleus chthonoplastes. Appl. Environ. Microbiol. 62, 1752–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hoffmeister M., van der Klei A., Rotte C., van Grinsven K. W., van Hellemond J. J., Henze K., Tielens A. G., and Martin W. (2004) Euglena gracilis rhodoquinone:ubiquinone ratio and mitochondrial proteome differ under aerobic and anaerobic conditions. J. Biol. Chem. 279, 22422–22429 [DOI] [PubMed] [Google Scholar]
- 4. Müller M., Mentel M., van Hellemond J. J., Henze K., Woehle C., Gould S. B., Yu R. Y., van der Giezen M., Tielens A. G., and Martin W. F. (2012) Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol. Mol. Biol. Rev. 76, 444–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stal L. J., and Moezelaar R. (1997) Fermentation in cyanobacteria. FEMS Microbiol. Rev. 21, 179–211 [Google Scholar]
- 6. Kreuzberg K. (1984) Starch fermentation via a formate producing pathway in Chlamydomonas reinhardtii, Chlorogonium elongatum and Chlorella fusca. Physiol. Plant 61, 87–94 [Google Scholar]
- 7. Gfeller R. P., and Gibbs M. (1984) Fermentative metabolism of Chlamydomonas reinhardtii: I. Analysis of fermentative products from starch in dark and light. Plant Physiol. 75, 212–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kreuzberg K., Klock G., and Grobheiser D. (1987) Subcellular distribution of pyruvate-degrading enzymes in Chlamydomonas reinhardtii studied by an improved protoplast fractionation procedure. Physiol. Plant 69, 481–488 [Google Scholar]
- 9. Maione T. E., and Gibbs M. (1986) Hydrogenase-mediated activities in isolated chloroplasts of Chlamydomonas reinhardii. Plant Physiol. 80, 360–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Merchant S. S., Prochnik S. E., Vallon O., Harris E. H., Karpowicz S. J., Witman G. B., Terry A., Salamov A., Fritz-Laylin L. K., Maréchal-Drouard L., Marshall W. F., Qu L. H., Nelson D. R., Sanderfoot A. A., Spalding M. H., et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Blanc G., Duncan G., Agarkova I., Borodovsky M., Gurnon J., Kuo A., Lindquist E., Lucas S., Pangilinan J., Polle J., Salamov A., Terry A., Yamada T., Dunigan D. D., Grigoriev I. V., et al. (2010) The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell 22, 2943–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Atteia A., van Lis R., Gelius-Dietrich G., Adrait A., Garin J., Joyard J., Rolland N., and Martin W. (2006) Pyruvate formate-lyase and a novel route of eukaryotic ATP-synthesis in Chlamydomonas mitochondria. J. Biol. Chem. 281, 9909–9918 [DOI] [PubMed] [Google Scholar]
- 13. Mus F., Dubini A., Seibert M., Posewitz M. C., and Grossman A. R. (2007) Anaerobic acclimation in Chlamydomonas reinhardtii: anoxic gene expression, hydrogenase induction, and metabolic pathways. J. Biol. Chem. 282, 25475–25486 [DOI] [PubMed] [Google Scholar]
- 14. Atteia A., van Lis R., Tielens A. G., and Martin W. F. (2013) Anaerobic energy metabolism in unicellular photosynthetic eukaryotes. Biochim. Biophys. Acta 1827, 210–223 [DOI] [PubMed] [Google Scholar]
- 15. Happe T., and Naber J. D. (1993) Isolation, characterization and N-terminal amino acid sequence of hydrogenase from the green alga Chlamydomonas reinhardtii. Eur. J. Biochem. 214, 475–481 [DOI] [PubMed] [Google Scholar]
- 16. Hemschemeier A., Jacobs J., and Happe T. (2008) Biochemical and physiological characterization of the pyruvate formate-lyase Pfl1 of Chlamydomonas reinhardtii, a typically bacterial enzyme in a eukaryotic alga. Eukaryot. Cell 7, 518–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Silakov A., Kamp C., Reijerse E., Happe T., and Lubitz W. (2009) Spectroelectrochemical characterization of the active site of the [FeFe] hydrogenase HydA1 from Chlamydomonas reinhardtii. Biochemistry 48, 7780–7786 [DOI] [PubMed] [Google Scholar]
- 18. van Lis R., Baffert C., Couté Y., Nitschke W., and Atteia A. (2013) Chlamydomonas reinhardtii chloroplasts contain a homodimeric pyruvate:ferredoxin oxidoreductase that functions with FDX1. Plant Physiol. 161, 57–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Noth J., Krawietz D., Hemschemeier A., and Happe T. (2013) Pyruvate:ferredoxin oxidoreductase is coupled to light-independent hydrogen production in Chlamydomonas reinhardtii. J. Biol. Chem. 288, 4368–4377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dubini A., Mus F., Seibert M., Grossman A. R., and Posewitz M. C. (2009) Flexibility in anaerobic metabolism as revealed in a mutant of Chlamydomonas reinhardtii lacking hydrogenase activity. J. Biol. Chem. 284, 7201–7213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Philipps G., Krawietz D., Hemschemeier A., and Happe T. (2011) A pyruvate formate lyase-deficient Chlamydomonas reinhardtii strain provides evidence for a link between fermentation and hydrogen production in green algae. Plant J. 66, 330–340 [DOI] [PubMed] [Google Scholar]
- 22. Magneschi L., Catalanotti C., Subramanian V., Dubini A., Yang W., Mus F., Posewitz M. C., Seibert M., Perata P., and Grossman A. R. (2012) A mutant in the ADH1 gene of Chlamydomonas reinhardtii elicits metabolic restructuring during anaerobiosis. Plant Physiol. 158, 1293–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Catalanotti C., Dubini A., Subramanian V., Yang W., Magneschi L., Mus F., Seibert M., Posewitz M. C., and Grossman A. R. (2012) Altered fermentative metabolism in Chlamydomonas reinhardtii mutants lacking pyruvate formate lyase and both pyruvate formate lyase and alcohol dehydrogenase. Plant Cell 24, 692–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang W., Catalanotti C., D'Adamo S., Wittkopp T. M., Ingram-Smith C. J., Mackinder L., Miller T. E., Heuberger A. L., Peers G., Smith K. S., Jonikas M. C., Grossman A. R., and Posewitz M. C. (2014) Alternative acetate production pathways in Chlamydomonas reinhardtii during dark anoxia and the dominant role of chloroplasts in fermentative acetate production. Plant Cell 26, 4499–4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Burgess S. J., Taha H., Yeoman J. A., Iamshanova O., Chan K. X., Boehm M., Behrends V., Bundy J. G., Bialek W., Murray J. W., and Nixon P. J. (2016) Identification of the elusive pyruvate reductase of Chlamydomonas reinhardtii chloroplasts. Plant Cell Physiol. 57, 82–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Adl S. M., Simpson A. G., Lane C. E., Lukeš J., Bass D., Bowser S. S., Brown M. W., Burki F., Dunthorn M., Hampl V., Heiss A., Hoppenrath M., Lara E., Le Gall L., Lynn D. H., et al. (2012) The revised classification of eukaryotes. J. Eukaryot. Microbiol. 59, 429–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Atteia A., van Lis R., Mendoza-Hernández G., Henze K., Martin W., Riveros-Rosas H., and González-Halphen D. (2003) Bifunctional aldehyde/alcohol dehydrogenase (ADHE) in chlorophyte algal mitochondria. Plant Mol. Biol. 53, 175–188 [DOI] [PubMed] [Google Scholar]
- 28. Bruchhaus I., and Tannich E. (1994) Purification and molecular characterization of the NAD+-dependent acetaldehyde/alcohol dehydrogenase from Entamoeba histolytica. Biochem. J. 303, 743–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang W., Li E., Kairong T., and Stanley S. L. Jr. (1994) Entamoeba histolytica has an alcohol dehydrogenase homologous to the multifunctional adhE gene product of Escherichia coli. Mol. Biochem. Parasitol. 64, 253–260 [DOI] [PubMed] [Google Scholar]
- 30. Sánchez L. B. (1998) Aldehyde dehydrogenase (CoA-acetylating) and the mechanism of ethanol formation in the amitochondriate protist, Giardia lamblia. Arch. Biochem. Biophys. 354, 57–64 [DOI] [PubMed] [Google Scholar]
- 31. Spreitzer R. J., and Mets L. J. (1980) Non-Mendelian mutation affecting ribulose-1,5-biphosphate carboxylase structure and activity. Nature 285, 114–115 [Google Scholar]
- 32. Dron M., Rahire M., Rochaix J. D., and Mets L. (1983) First DNA sequence of a chloroplast mutation: a missense alteration in the ribulose biphosphate carboxylase large subunit gene. Plasmid 9, 321–324 [DOI] [PubMed] [Google Scholar]
- 33. Terashima M., Specht M., Naumann B., and Hippler M. (2010) Characterizing the anaerobic response of Chlamydomonas reinhardtii by quantitative proteomics. Mol. Cell. Proteomics 9, 1514–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Boxma B., Voncken F., Jannink S., van Alen T., Akhmanova A., van Weelden S. W., van Hellemond J. J., Ricard G., Huynen M., Tielens A. G., and Hackstein J. H. (2004) The anaerobic chytridiomycete fungus Piromyces sp. E2 produces ethanol via pyruvate:formate lyase and an alcohol dehydrogenase E. Mol. Microbiol. 51, 1389–1399 [DOI] [PubMed] [Google Scholar]
- 35. Kawata T., Masuda K., and Yoshino K. (1975) Presence of fine spirals (spirosomes) in Lactobacillus fermenti and Lactobacillus casei. Jpn. J. Microbiol. 19, 225–227 [DOI] [PubMed] [Google Scholar]
- 36. Kessler D., Leibrecht I., and Knappe J. (1991) Pyruvate-formate-lyase-deactivase and acetyl-CoA reductase activities of Escherichia coli reside on a polymeric protein particle encoded by adhE. FEBS Lett. 281, 59–63 [DOI] [PubMed] [Google Scholar]
- 37. Laurenceau R., Krasteva P. V., Diallo A., Ouarti S., Duchateau M., Malosse C., Chamot-Rooke J., and Fronzes R. (2015) Conserved Streptococcus pneumoniae spirosomes suggest a single type of transformation pilus in competence. PLoS Pathog. 11, e1004835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hsieh S. I., Castruita M., Malasarn D., Urzica E., Erde J., Page M. D., Yamasaki H., Casero D., Pellegrini M., Merchant S. S., and Loo J. A. (2013) The proteome of copper, iron, zinc, and manganese micronutrient deficiency in Chlamydomonas reinhardtii. Mol. Cell. Proteomics 12, 65–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ball S. G., Dirick L., Decq A., Martiat J. C., and Matagne R. F. (1990) Physiology of starch storage in the monocellular alga Chlamydomonas reinhardtii. Plant Sci. 66, 1–9 [Google Scholar]
- 40. Schmöllinger S., Mühlhaus T., Boyle N. R., Blaby I. K., Casero D., Mettler T., Moseley J. L., Kropat J., Sommer F., Strenkert D., Hemme D., Pellegrini M., Grossman A. R., Stitt M., Schroda M., et al. (2014) Nitrogen-sparing mechanisms in Chlamydomonas affect the transcriptome, the proteome, and photosynthetic metabolism. Plant Cell 26, 1410–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Valledor L., Furuhashi T., Recuenco-Muñoz L., Wienkoop S., and Weckwerth W. (2014) System-level network analysis of nitrogen starvation and recovery in Chlamydomonas reinhardtii reveals potential new targets for increased lipid accumulation. Biotechnol. Biofuels 7, 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zabawinski C., Van Den Koornhuyse N., D'Hulst C., Schlichting R., Giersch C., Delrue B., Lacroix J. M., Preiss J., and Ball S. (2001) Starchless mutants of Chlamydomonas reinhardtii lack the small subunit of a heterotetrameric ADP-glucose pyrophosphorylase. J. Bacteriol. 183, 1069–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Malasarn D., Kropat J., Hsieh S. I., Finazzi G., Casero D., Loo J. A., Pellegrini M., Wollman F. A., and Merchant S. S. (2013) Zinc deficiency impacts CO2 assimilation and disrupts copper homeostasis in Chlamydomonas reinhardtii. J. Biol. Chem. 288, 10672–10683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang Y., Duanmu D., and Spalding M. H. (2011) Carbon dioxide concentrating mechanism in Chlamydomonas reinhardtii: inorganic carbon transport and CO2 recapture. Photosynth. Res. 109, 115–122 [DOI] [PubMed] [Google Scholar]
- 45. Moroney J. V., Husic H. D., Tolbert N. E., Kitayama M., Manuel L. J., and Togasaki R. K. (1989) Isolation and characterization of a mutant of Chlamydomonas reinhardtii deficient in the CO2 concentrating mechanism. Plant Physiol. 89, 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fukuzawa H., Miura K., Ishizaki K., Kucho K. I., Saito T., Kohinata T., and Ohyama K. (2001) Ccm1, a regulatory gene controlling the induction of a carbon-concentrating mechanism in Chlamydomonas reinhardtii by sensing CO2 availability. Proc. Natl. Acad. Sci. U.S.A. 98, 5347–5352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Karlsson J., Clarke A. K., Chen Z. Y., Hugghins S. Y., Park Y. I., Husic H. D., Moroney J. V., and Samuelsson G. (1998) A novel α-type carbonic anhydrase associated with the thylakoid membrane in Chlamydomonas reinhardtii is required for growth at ambient CO2. EMBO J. 17, 1208–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Johnson X., Wostrikoff K., Finazzi G., Kuras R., Schwarz C., Bujaldon S., Nickelsen J., Stern D. B., Wollman F. A., and Vallon O. (2010) MRL1, a conserved pentatricopeptide repeat protein, is required for stabilization of rbcL mRNA in Chlamydomonas and Arabidopsis. Plant Cell 22, 234–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Johnson X., Steinbeck J., Dent R. M., Takahashi H., Richaud P., Ozawa S., Houille-Vernes L., Petroutsos D., Rappaport F., Grossman A. R., Niyogi K. K., Hippler M., and Alric J. (2014) Proton gradient regulation 5-mediated cyclic electron flow under ATP- or redox-limited conditions: a study of ΔATpase pgr5 and ΔrbcL pgr5 mutants in the green alga Chlamydomonas reinhardtii. Plant Physiol. 165, 438–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kucho K., Yoshioka S., Taniguchi F., Ohyama K., and Fukuzawa H. (2003) cis-Acting elements and DNA-binding proteins involved in CO2-responsive transcriptional activation of Cah1 encoding a periplasmic carbonic anhydrase in Chlamydomonas reinhardtii. Plant Physiol. 133, 783–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Keeling P. J. (2010) The endosymbiotic origin, diversification and fate of plastids. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 729–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dagan T., Roettger M., Stucken K., Landan G., Koch R., Major P., Gould S. B., Goremykin V. V., Rippka R., Tandeau de Marsac N., Gugger M., Lockhart P. J., Allen J. F., Brune I., Maus I., et al. (2013) Genomes of stigonematalean cyanobacteria (subsection V) and the evolution of oxygenic photosynthesis from prokaryotes to plastids. Genome Biol. Evol. 5, 31–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cabiscol E., Aguilar J., and Ros J. (1994) Metal-catalyzed oxidation of Fe2+ dehydrogenases. Consensus target sequence between propanediol oxidoreductase of Escherichia coli and alcohol dehydrogenase II of Zymomonas mobilis. J. Biol. Chem. 269, 6592–6597 [PubMed] [Google Scholar]
- 54. Leonardo M. R., Cunningham P. R., and Clark D. P. (1993) Anaerobic regulation of the adhE gene, encoding the fermentative alcohol dehydrogenase of Escherichia coli. J. Bacteriol. 175, 870–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pagels M., Fuchs S., Pané-Farré J., Kohler C., Menschner L., Hecker M., McNamarra P. J., Bauer M. C., von Wachenfeldt C., Liebeke M., Lalk M., Sander G., von Eiff C., Proctor R. A., and Engelmann S. (2010) Redox sensing by a Rex-family repressor is involved in the regulation of anaerobic gene expression in Staphylococcus aureus. Mol. Microbiol. 76, 1142–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Steunou A. S., Bhaya D., Bateson M. M., Melendrez M. C., Ward D. M., Brecht E., Peters J. W., Kühl M., and Grossman A. R. (2006) In situ analysis of nitrogen fixation and metabolic switching in unicellular thermophilic cyanobacteria inhabiting hot spring microbial mats. Proc. Natl. Acad. Sci. U.S.A. 103, 2398–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Beckham K. S., Connolly J. P., Ritchie J. M., Wang D., Gawthorne J. A., Tahoun A., Gally D. L., Burgess K., Burchmore R. J., Smith B. O., Beatson S. A., Byron O., Wolfe A. J., Douce G. R., and Roe A. J. (2014) The metabolic enzyme AdhE controls the virulence of Escherichia coli O157:H7. Mol. Microbiol. 93, 199–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shasmal M., Dey S., Shaikh T. R., Bhakta S., and Sengupta J. (2016) E. coli metabolic protein aldehyde-alcohol dehydrogenase-E binds to the ribosome: a unique moonlighting action revealed. Sci. Rep. 6, 19936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Harris E. (1989) The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use, Academic Press, San Diego, CA: [DOI] [PubMed] [Google Scholar]
- 60. van Lis R., Atteia A., Nogaj L. A., and Beale S. I. (2005) Subcellular localization and light-regulated expression of protoporphyrinogen IX oxidase and ferrochelatase in Chlamydomonas reinhardtii. Plant Physiol. 139, 1946–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Goodstein D. M., Shu S., Howson R., Neupane R., Hayes R. D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N., and Rokhsar D. S. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dupierris V., Masselon C., Court M., Kieffer-Jaquinod S., and Bruley C. (2009) A toolbox for validation of mass spectrometry peptides identification and generation of database: IRMa. Bioinformatics. 25, 1980–1981 [DOI] [PubMed] [Google Scholar]
- 63. Wessel D., and Flügge U. I. (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141–143 [DOI] [PubMed] [Google Scholar]
- 64. Durrant I. (1990) Light-based detection of biomolecules. Nature 346, 297–298 [DOI] [PubMed] [Google Scholar]
- 65. Studier F. W. (2005) Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 66. O'Toole E. T. (2010) Chlamydomonas cryopreparation methods for the 3-D analysis of cellular organelles. Methods Cell Biol. 96, 71–91 [DOI] [PubMed] [Google Scholar]
- 67. McDonald K. L. (2014) Rapid embedding methods into epoxy and LR White resins for morphological and immunological analysis of cryofixed biological specimens. Microsc. Microanal. 20, 152–163 [DOI] [PubMed] [Google Scholar]
- 68. Tamura K., Stecher G., Peterson D., Filipski A., and Kumar S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Montella C., Bellsolell L., Pérez-Luque R., Badía J., Baldoma L., Coll M., and Aguilar J. (2005) Crystal structure of an iron-dependent group III dehydrogenase that interconverts l-lactaldehyde and l-1,2-propanediol in Escherichia coli. J. Bacteriol. 187, 4957–4966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zheng T., Olson D. G., Tian L., Bomble Y. J., Himmel M. E., Lo J., Hon S., Shaw A. J., van Dijken J. P., and Lynd L. R. (2015) Cofactor specificity of the bifunctional alcohol and aldehyde dehydrogenase (AdhE) in wild-type and mutant Clostridium thermocellum and Thermoanaerobacterium saccharolyticum. J. Bacteriol. 197, 2610–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.