Abstract
Variation in phenotypic traits that contribute to fitness influences a population's evolutionary response and its impact on ecosystem function following environmental change, yet its amount and nature are rarely known. Here, we investigated variation in nitrogen (N) fixation activity and its genetic basis for a random sample of laboratory strains of the cyanobacterium Mastigocladus laminosus from a N-limited, geothermally influenced stream in Yellowstone National Park. In a linear mixed-effects model, temperature and genetic differences among strains were the most important factors explaining variation in activity. Genome-wide analyses of genetic divergence between groups of strains that varied in N fixation activity revealed that few loci were strongly associated with these phenotypic differences. Notably, a single nonsynonymous polymorphism in the sulfate assimilation gene apsK explained >25% of the variation in activity at high temperature. We further identified a role for allelic variation of multiple terminal cytochrome oxidases for different aspects of N fixation. In addition, genomes of strains that fixed the most N overall contained a nonsense mutation in a histidine kinase gene that is expected to disrupt normal protein function and may result in transcriptional rewiring. This study illustrates how taking complementary approaches to link phenotype and genotype can inform our understanding of microbial population diversity.
Introduction
Microbial populations often exhibit extensive genetic diversity (Cadillo-Quiroz et al., 2012; Shapiro et al., 2012), and patterns of genome variation are consistent with the action of past or ongoing selection resulting from spatial and/or temporal heterogeneity in the environment (Rosen et al., 2015). Recently, it has been proposed that much of this diversity may be maintained by negative frequency-dependent selection on gene content variation underlying ecological and social interactions, such as phage resistance and the sharing of public goods, respectively (Rodriguez-Valera et al., 2009; Cordero and Polz, 2014). However, the nature and genetic basis of the functional variation within microbial populations, as well as the evolutionary mechanisms that maintain it, remain poorly understood in general.
Developing a better understanding of the extent of variation for fitness-related traits within populations is important for several reasons. As first pointed out by Darwin, heritable variation within a population represents the raw material of evolution by natural selection. The amount of this variation can impact how populations respond to environmental change, because the rate at which beneficial mutations arise and subsequently attain high frequencies is slow compared with the rate at which populations can potentially adapt from pre-existing genetic variation (Barrett and Schluter, 2008). Genetic diversity within populations also has potential implications for ecosystem resilience in a changing environment (Hughes et al., 1997; Luck et al., 2003; Reusch et al., 2005; Ehlers et al., 2008).
The ability to obtain sufficient nitrogen (N) is an important fitness component for all cells, and many bacteria and archaea can fix N in the absence of a preferred source of this nutrient. At White Creek, an N-limited, geothermally influenced stream in the Lower Geyser Basin of Yellowstone National Park, a population of the multicellular cyanobacterium Mastigocladus (Fischerella) laminosus fixes N by developing heterocysts (Miller et al., 2006), which are terminally differentiated cells that spatially separate the oxygen-sensitive N fixation process from photosynthetic oxygen produced by surrounding vegetative cells. N fixation has a complex genetic basis in heterocystous cyanobacteria (Kumar et al., 2010), requiring genes that regulate and specify the morphological and physiological changes that occur during the differentiation process, in addition to N fixation (nif) genes that are common to all diazotrophs. Estimates of the number of genes that are differentially regulated during heterocyst development range from ~500 in Nostoc punctiforme (Campbell et al., 2007) to over 1000 in Anabaena PCC 7120 (Ehira et al., 2003). Among these are 100–140 'Fox' genes that are required for N fixation in the presence of oxygen (Wolk, 2000), including those involved in the remodeling of the cell surface to form an envelope of glycolipid and polysaccharide layers that provides a passive gas diffusion barrier to limit the entry of oxygen (Nicolaisen et al., 2009).
While much is known from model cyanobacteria regarding the genes required to develop a heterocyst and perform N fixation, we are ignorant of both the amount and the genetic basis of phenotypic variation for this important biogeochemical process in strains from natural bacterial populations. Here, we assay variation in N fixation activity for 22 M. laminosus strains isolated from along the White Creek thermal gradient of 39–54 °C mean annual temperature (Miller et al., 2009). We next used existing genome sequence data available for most of these strains (Sano et al., submitted) to identify genes associated with phenotypic differences among strains. Genetic exchange along White Creek is generally high throughout much of the M. laminosus genome (Wall et al., 2014), and, consequently, we could take advantage of historic recombination events in the population to identify regions of the genome that exhibit unusually high levels of genetic differentiation between phenotypically divergent strains. These complementary approaches provide insights into the connection between genotype and phenotype for a complex fitness-related trait.
Materials and methods
Nitrogen and carbon fixation assays
Axenic M. laminosus strains were grown in 75 ml of nitrate-containing D mineral salts medium (Castenholz, 1988) at a maintenance temperature of 50 °C and a light intensity of 105±5 μmol m−2 s−1 with cool white fluorescent bulbs. The first digit in a strain's name indicates the White Creek sampling site from which it was isolated (with ‘1' representing the most downstream and ‘5' the most upstream sites; Miller et al., 2009). Subsamples of growing cultures were transferred to flasks containing ND medium (D medium without the addition of combined N; Castenholz, 1988) to establish steady-state growth in the absence of combined N, as in Miller et al. (2006, 2009). After 2 weeks, cultures were split into six sublines, with three each moved to 37 °C and 55 °C growth chambers, respectively. Sublines were maintained in ND medium with a 12/12 h light/dark cycle. Growing cells from each subline were assayed for N fixation, as described below. For each strain, assays were performed two times using independent starting culture inocula.
Subsamples from each subline were homogenized with a tissue grinder and normalized to an OD750 of 0.05±0.003 with a Beckman Coulter DU 530 spectrophotometer (Brea, CA, USA). For dilution series of growing cultures from two White Creek strains, we determined that optical density has the expected linear relationship with dry weight for 3 ml of M. laminosus cell suspensions that had been filtered onto a 0.45 μm pore size Millipore filter and dried for 48 h in a desiccation chamber (Pearson's correlation coefficient R=0.98, P<0.001; Supplementary Figure 1). Cultures were harvested at the beginning of the light cycle and then homogenized such that clumps were broken up but long chains containing vegetative cells and heterocysts remained intact. N fixation rates were estimated by ethylene production in acetylene reduction assays (Stewart et al., 1967). Assays were carried out in 10 ml of ND medium in 20 ml crimp-sealed vials at a light intensity of 105±5 μmol m−2 s−1. Samples were incubated for 4 h following the addition of 5 ml of acetylene gas (generated by adding 5 g of calcium carbide to 100 ml of deionized H2O) and terminated by aspirating as much sample headspace as possible (~15 ml) and injecting it into a pre-evacuated 5 ml crimp vial. Ethylene production was measured by flame-ionization detection gas chromatography with a Shimadzu GC-2014 (Kyoto, Japan). Ethylene production measurements were estimated using a standard curve, blank corrected against parallel incubation vials that contained only ND growth medium and normalized by sample optical density. Microscopic counts of heterocyst frequency were performed for a randomly selected subline at each temperature treatment.
Carbon fixation was concurrently estimated by 14C-bicarbonate incorporation (Miller et al., 1998). Briefly, incubations were initiated with the addition of 0.2 μCi of 14C-bicarbonate to 3 ml aliquots of each subline, carried out for 1 h under the same conditions as for the acetylene reduction assays and then terminated by the addition of 200 μl of formalin. Samples with formalin added at the start of the incubation were included to correct for non-biological uptake of radioisotope. Each sample was filtered onto a 0.45 μm GN-6 membrane filter (PALL Life Sciences, Port Washington, NY, USA), rinsed first with 3% HCl to remove unincorporated radioisotope and then rinsed with deionized water. Filters were placed into 20 ml scintillation vials and ventilated in a fume hood for 1 h before adding 1.5 ml of EcoLite scintillation fluid (MP Biomedicals, Santa Ana, CA, USA). Samples were read by a Beckman LS6000SE scintillation counter (Brea, CA, USA) and normalized to sample OD as above.
Statistical analysis
Because of the crossed experimental design and the heteroskedastic nature of the data, a linear mixed-effects model was used using the R 'lme4' package (Bates et al., 2014) to analyze the factors that contribute to variation in N fixation. Fixed factors of the model included normalized carbon fixation rate, temperature, heterocyst frequency and all possible interactions. The random-effects structure was designed such that the model accounted for variation within incubations and among strains across the two temperature treatments. Other variables in the model were removed via backwards stepwise nested hypothesis testing using the F-test until the lowest Akaike information criterion score was obtained. A post hoc pseudo-R2 for linear mixed models (Nakagawa and Schielzeth, 2013) was used to approximate the model fit and estimate the amount of variation explained by fixed factors and individual random effects.
Identification of candidate genes associated with phenotypic variation
M. laminosus strains were grouped into classes based on N fixation activity at each temperature treatment and for overall pooled performance. For each data set, strains in the upper quartile of mean normalized ethylene production were treated as the 'upper' class, and those below this benchmark were categorized as the 'lower' class. Genome data for M. laminosus strains used in the analysis were obtained previously (Sano et al., submitted). Briefly, paired-end Illumina sequence data were acquired to >100 × coverage for 20 White Creek strains randomly selected from the laboratory strain collection. The genome sequence data have been deposited in the NCBI Sequence Read Archive (GenBank accession no. SRP075623). Draft genomes were assembled de novo using Velvet (Zerbino and Birney, 2008), and genome contigs were autoannotated with the RAST server (http://rast.nmpdr.org/; Aziz et al., 2008).
For each genome, protein-coding genes (CDS) were extracted from GenBank files with extract_feature_seq.PLS (bioperl.org) to create individual FASTA-formatted files for each CDS. Orthologous CDS from individual genomes were combined into single files using sequential local blastn queries of a non-redundant database of CDS from the population. This procedure was used to create separate FASTA-formatted files of each CDS for the total sample and for the individual phenotypic classes described above.
Custom Perl scripts (Supplementary Information) were next used to estimate nucleotide diversity π (i.e., the probability that two randomly selected sequences in a sample differ in nucleotide identity at a site) within phenotypic classes and for the total sample. Only full-length CDS were included, and loci with fewer than 10 sequences were excluded from the analysis. From these π estimates, both the relative amount of genetic differentiation (ΦST, the fraction of total nucleotide diversity between classes; Nei, 1982) and absolute genetic differentiation between phenotypic classes (DXY; i.e., the numerator of ΦST) were estimated for each polymorphic CDS. In addition, we used PERL scripts to identify individual alleles at each locus and build contingency tables of allele counts for both phenotypic classes. These tables were used to test for a statistical association between allele frequencies at a locus and phenotype with Fisher's exact tests. This test was chosen owing to the small sample sizes involved and to avoid the assumption that the data conform to a χ distribution. Although necessitated by the labor-intensive nature of assaying N fixation, a consequence of the small sample sizes in the study is that we did not have the statistical power to accurately discriminate false positives for Fisher's tests. For select candidate genes that exhibited extreme genetic differentiation by one or more of the above approaches, we therefore used generalized linear models both to test the significance (with Bonferroni correction) of the association between allelic and phenotypic variation and to estimate the amount of observed phenotypic variation explained by the locus. In addition, to assess whether any flanking non-coding variation was linked to a candidate gene, sliding windows with a window length of 100 nt and a step size of 25 nt were estimated with DNASP version 5.10 (Librado and Rozas, 2009) for aligned genome contigs containing the candidate gene.
Expression of candidate genes
Transcription during growth under different N conditions was assessed by reverse transcription-PCR for five strains representing different alleles for candidate genes 167-28586 (histidine kinase (HK)) and 28-39736 (apsK). Strains were grown in semicontinuous batch cultures in D medium until ~5 ml of cell biomass had accumulated and then washed two times in ND medium before transfer to triplicate flasks containing 250 ml of either ND or D medium. Cultures were maintained at 37 °C with a 12/12 h light/dark cycle. Approximately 0.5 ml of cell mass was collected at 0.5 (T0), 6, 12, 18, 24, 36 and 48 h after transfer for ND cultures and once for mid-growth phase cultures in D medium. All samples were immediately snap frozen in liquid nitrogen and stored at −80 °C. A Qiagen RNeasy Mini Extraction Kit (Hilden, Germany) was used to isolate RNA according to the manufacturer's instructions. RNA quantity and quality was checked on a NanoDrop spectrometer (Wilmington, DE, USA). We used reverse transcriptase-negative controls to confirm the absence of genomic DNA contamination of the isolated RNAs using the PCR cycling conditions described below. If needed, an additional DNase digestion was performed before cDNA synthesis. A Thermo Scientific Maxima First-Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA) for reverse transcription-PCR was used to construct first-strand cDNA according to the manufacturer's instructions. First-strand synthesis was accompanied by a template-negative control. For HK gene 167-28586, an ~1 kbp cDNA was amplified by touchdown PCR on an MJR PTC-100 thermal cycler (MJ Research, Waltham, MA, USA) with primers 5′-GGAATCCACCAACTATGG-3′ and 5′-CCAGGTGTAGAGTAGCAC-3′. An initial 3 min denaturation step at 94 °C was followed by 30 cycles of 1 min at 94 °C, 30 s at variable annealing temperatures, and 1 min at 72 °C. The initial annealing temperature was 54 °C and decreased every 10 cycles, reaching a final annealing temperature of 50 °C. For adenylsulfate kinase gene 28-39736, a ~300 bp cDNA product was amplified with primers 5′-GAAAACATCCGTCGCATTGG-3′ and 5′-TGATGTAACCGAGTTCTGCC-3′. An initial denaturation step at 94 °C for 3 min was followed by 30 cycles of 1 min at 94 °C, 30 s at 52 °C and 30 s at 72 °C. The presence of cDNA amplicons was assessed by agarose gel electrophoresis.
Evolutionary history of the apsK candidate
A neighbor-net splits network was inferred with Splittstree (Huson and Bryant, 2006) using default settings for sequence data from White Creek and for orthologs of the APS kinase candidate 28-39736 from representative M. laminosus strains from throughout its range (GenBank accession nos KX267709–KX267733), includingMastigocladus (=Fischerella) strain PCC 7521 (Dagan et al., 2013).
Results and discussion
Nitrogen fixation activity of White Creek M. laminosus strains
We assayed N fixation by the acetylene reduction method (Stewart et al., 1967) for 22 laboratory strains randomly selected from our culture collection for the White Creek M. laminosus population to estimate the extent of phenotypic variation for this trait. Strains were assayed at 37 °C and 55 °C, temperatures that delimit the approximate bounds of the population range. N fixation is energetically expensive, with high reductant requirements, and fixed carbon provisioned by adjacent vegetative cells contributes both electrons and a portion of the ATP budget for the process (Kumar et al., 2010). We therefore simultaneously estimated carbon fixation to test whether rates of N fixation and carbon fixation are correlated. We might expect a positive correlation, as has previously been reported for Anabaena sp. (Oh et al., 1991). On the other hand, because enhanced carbon fixation implies increased photosynthetic oxygen generation, which can potentially inhibit nitrogenase, the relationship between the two might be more complicated. Finally, we estimated heterocyst frequencies to test whether strains with more heterocysts fix more N.
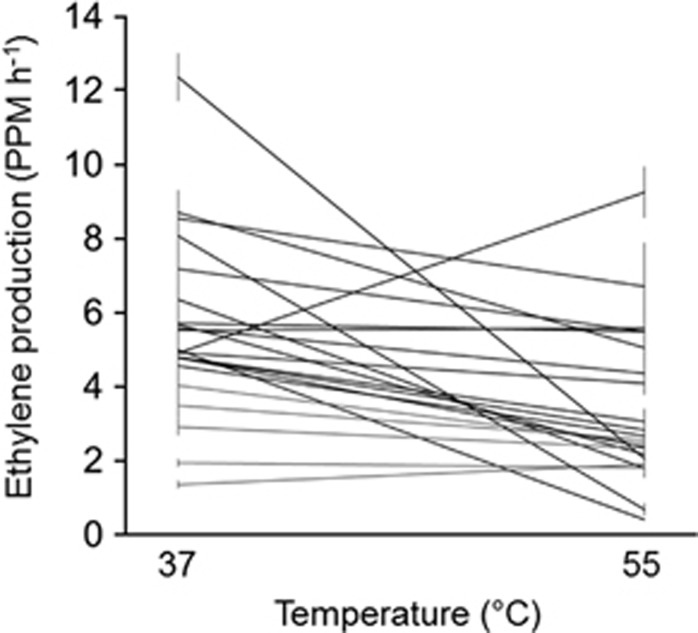
We observed considerable variation among strains in N fixation activity (i.e., ethylene production normalized to cell density; Figure 1). The final linear mixed model included two fixed factors (heterocyst frequency and a carbon fixation × temperature interaction) and two random effects (inoculum, i.e., assays of a strain derived from independent culture inocula, and a strain × temperature interaction). The pseudo-R2 for the model was 0.74, with random effects explaining 53% of the total variation. Of the random effects, strain identity and temperature explained comparable amounts of variation (19% and 20%, respectively), emphasizing the importance of both genetic differences among strains and the environment in determining activity. By contrast, the effect of inoculum was small (3%), indicating that a strain's activity was generally consistent between independent cultures.
Figure 1.
Ethylene production normalized by cell optical density among M. laminosus strains at 37 °C and 55 °C. Bars are s.e. Each trace represents a strain.
With respect to the effects of temperature on N fixation, individual strains generally fixed more N at 37 °C than at 55 °C (Figure 1). This finding is corroborated by both 15N2-uptake experiments (Stewart, 1970) and acetylene reduction assays performed with M. laminosus-containing microbial mats from White Creek (Sano et al., submitted). N fixation activity was significantly higher at the lower temperature for eight strains (t-tests, P<0.05), and estimated activities for most other strains also tended to be higher at 37 °C. In only one case (strain WC344) was N fixation significantly greater at 55 °C.
Fixed factors explained roughly half as much variation as the random effects (21%). Average heterocyst frequency was 2.4±0.3% (mean±95% confidence interval). This is lower than the ~5–10% frequency typically reported for model heterocystous cyanobacteria (Kumar et al., 2010) but comparable to previous observations for M. laminosus (unpublished data). A significant correlation between heterocyst frequency and N fixation was only observed at 55 °C (Pearson's correlation coefficient R=0.50, P<0.01) but was positive in sign for all data sets (R=0.17 and 0.30 for the 37 °C and pooled data sets, respectively). Mean strain-specific normalized carbon fixation rates for the pooled data set ranged between 33 and 133 μg C l−1 h−1. In general, C and N fixation activities are positively associated. The correlation between C and N fixation was significantly positive in the 55 °C (R=0.73, P<0.01) and pooled data sets (R=0.59, P<0.01), and positive, although not significant, at 37 °C (R=0.50, P=0.25).
Overview of genome-wide analyses of genetic variation associated with differences in N fixation activity
Differences among strains in N fixation activity accounted for more than one-quarter of the total variation explained by the model. We used annotated genome data that were available for 18 of the strains (Sano et al., submitted) to identify the genes most strongly associated with these differences for the 37 °C, 55 °C and pooled data sets, respectively. To do so, we first binned strains into 'upper' (upper quartile of mean N fixation activity) and 'lower' phenotypic classes for each data set. Four of the six strains (WC114, WC119, WC1110 and WC245) in the upper quartile at 37 °C were isolated from lower temperature, downstream sites at White Creek, whereas four of the six strains (WC344, WC438, WC439 and WC542) in the upper quartile at 55 °C were isolated from higher temperature, upstream sites (Figure 1 and Supplementary Table 1). This observation is in accord with a general interaction between the temperature dependence of N fixation and the location along White Creek from which the strain was isolated (F(1,24.4)=4.1; P<0.05). Specifically, although upstream (sites WC3–5) and downstream (sites WC1–2) strains did not significantly differ in N fixation at 37 °C (mean (s.e.) of 5.8 (0.45) versus 5.7 (0.32) p.p.m. ethylene produced per h for downstream and upstream strains, respectively), strains from the downstream sites exhibited lower rates of N fixation at 55 °C than did strains from upstream sites (3.5 (0.44) versus 5.0 (0.42) p.p.m. ethylene produced per h). This pattern is in agreement with our previous finding that upstream strains have much higher fitness at 55 °C than do downstream strains (Miller et al., 2009). Still, there was considerable variation among strains with respect to the temperature dependence of N fixation and location along White Creek, which is of potential importance for ecosystem function given seasonal heterogeneity in temperature at different sites (Miller et al., 2009). Among the strains in the upper quartile for the pooled data set, strains WC119, WC245 and WC439 were also in the upper quartiles for both temperature-specific data sets, whereas the other strains were in the upper quartile for one of the temperature treatments (Figure 1 and Supplementary Table 1).
We next estimated three measures of genetic differentiation between classes for each of the 2072 polymorphic protein-coding genes in the sample. We first used Fisher's exact test to evaluate whether allele frequencies at a locus were statistically different between classes. The second and third measures were both based on the amount of genetic variation (nucleotide diversity, π) within and between phenotypic classes: ΦST is the fraction of the total variation that is between phenotypic classes, whereas DXY is the average number of nucleotide differences per site between randomly selected sequences from different phenotypic classes. These metrics capture different aspects of divergence. For example, recently diverged alleles that contribute to phenotypic differentiation but differ by only one single-nucleotide polymorphism would be expected to be a candidate for the ΦST data set, whereas more divergent alleles would be expected to only be a DXY candidate if they did not also exhibit an extreme difference in allele frequency between phenotypic classes.
For all three data sets, genetic differentiation between phenotypic classes was very low for the majority of the M. laminosus genome (i.e., Fisher's exact tests for allele frequency contingency tables were not significant and values of ΦST and DXY were low; Supplementary Figures 2 and 3). Genes in the right tails of the respective frequency distributions represent the best candidates for loci that contribute to the observed genetic differences in N fixation activity among strains. Consequently, we focused on the loci exhibiting the most extreme differentiation between classes (Table 1). These included genes at a significance level of P<0.01 for a Fisher's exact test, a ΦST value >0.5 and a DXY value >0.005. For all three measures, these cutoffs represented <0.3% of polymorphic genes in any of the analyses.
Table 1. Genes associated with differences in N fixation activity between phenotypic classes of M. laminosus strains.
| Analysis | Annotation (gene ID) | PFET | ΦST | DXY |
|---|---|---|---|---|
| 55 °C | Adenylsulfate kinase (28-39736) | 0.0034 | 0.73 | |
| Hypothetical protein (26-13568) | 0.0079 | |||
| Glycosyltransferase (1-33964) | 0.57 | |||
| ATPase (49-34361) | 0.53 | |||
| CoxCI; Cyt oxidase subunit 3(65-42545) | 0.52 | |||
| Histidine kinase (93-42545) | 0.52 | |||
| ABC transporter (77-48944) | 0.0081 | |||
| UreA urease (10-32834) | 0.0053 | |||
| 37 °C | Riboflavin reductase (43-56937) | 0.0082 | ||
| Hypothetical protein (33-9029) | 0.0057 | |||
| Pooled | Histidine kinase (167-28586) | 0.54 | 0.0161 | |
| Glycine cleavage system (27-2489) | 0.0057 | |||
| CoxAII; Cyt oxidase subunit 1 (21-20552) | 0.0070 | |||
| ArsA ATPase (7-34642) | 0.0070 | |||
| Hypothetical protein (48-37089) | 0.0083 | |||
| Pleiotropic regulatory protein (84-29888) | 0.0087 | |||
| Cytochrome P450 (20-24813) | 0.53 | |||
| ABC transporter (77-48944) | 0.0088 | |||
| Hypothetical protein (33-9029) | 0.0057 | |||
| UreA urease (10-32834) | 0.0056 | |||
| Hypothetical protein (93-17867) | 0.0054 | |||
| Chlorophyll-binding protein (62-37089) | 0.0052 |
Abbreviation: PFET, Fisher's exact test P-value.
There was little overlap among candidate loci for the three data sets (Table 1), which suggests that the genetic factors underlying variation between phenotypic classes largely differ for general N fixation performance (i.e., reflected in the pooled data set) as well as specific activities at the different temperatures. These apparent differences in genetic architecture make sense given that mean strain activities at 37 °C and 55 °C were not correlated overall (P=0.43). Similarly, only a minority of genes within each data set were candidates for multiple measures of genetic differentiation (Table 1). Below, we highlight a few particularly noteworthy candidates.
APS kinase at 55 °C
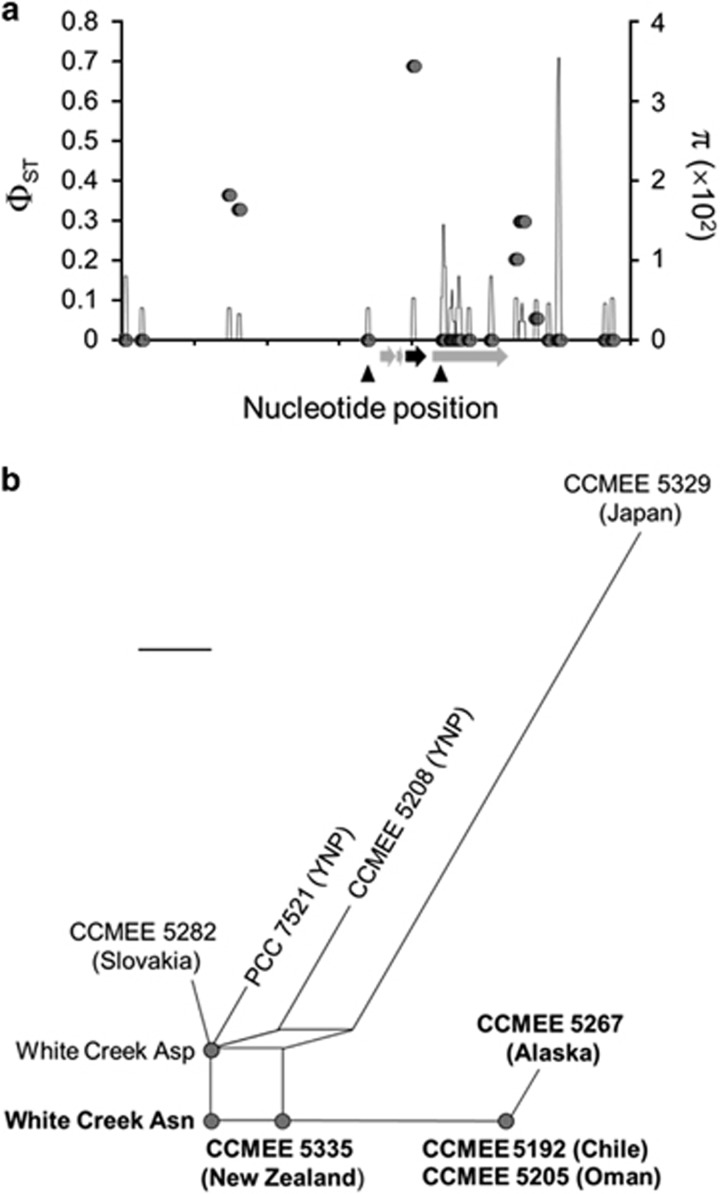
A 55 °C candidate with the greatest ΦST value and the most extreme differences in allele frequency between phenotypic classes of any analysis encodes the sulfate assimilation enzyme adenylylsulfate (APS) kinase (gene ID 28-39736; Table 1). The two alternative alleles at this locus differed by only a single nonsynonymous polymorphism, resulting in either an asparagine or an aspartate residue at codon 77, with the latter fixed (i.e., 100% frequency) in the upper class and the former at 83% frequency in the lower class. A sliding window analysis of the variation surrounding apsK, including flanking regulatory DNA, which was invariant in the sample, confirmed that only this single-nucleotide polymorphism (SNP) is tightly associated with the differences between phenotypic classes (Figure 2a). The two SNP variants are not restricted to White Creek but, rather, are geographically widespread (Figure 2b), which suggests that the SNP is old and has been maintained by selection during M. laminosus diversification. This illustrates that functional variation observed within a population may pre-date earlier phylogenetic divergence. Such trans-specific polymorphisms are generally thought to be rare but may occur when spatiotemporal variation or other mechanisms of balancing selection actively maintain ancestral variation in the descendent lineages of a common ancestor over long evolutionary time scales (Hedrick, 2006). Both alleles are expressed during growth with combined N, during the heterocyst differentiation process, and during steady-state growth under N-fixing conditions (Supplementary Figure 4).
Figure 2.
(a) Sliding window of genetic variation (nucleotide diversity π, line) and differentiation between phenotypic classes (ΦST, closed circles) for the 55 °C analysis for a ca. 14 kb region (axis ticks are 2 kb increments) surrounding the apsK candidate. The window length was 100 nt with a step size of 25 nt. SNPs flanking apsK (black arrow) are not associated with the phenotype (low ΦST) as a result of historic recombination events (closed triangles) that have broken up linkage between aspK and surrounding DNA. (b) Neighbor-net splits network of the evolutionary relationships among apsK alleles for M. laminosus strains from White Creek and throughout its global range. Because this history includes recombination, these relationships are better described by a network, which connects alleles related by recombination with cycles, than by a bifurcating tree. Nucleotide identity at position 229 is indicated, with A229 conferring the asparagine allele (bold) and G229 conferring the aspartate allele (normal font), respectively. Scale bar is 0.002 expected nucleotide substitutions per nucleotide site.
A longstanding question in evolutionary biology is whether functional differences within populations are primarily due to selection on variation for a few genes of major effect (e.g., >10% of the variation explained) or, alternatively, for many genes of small effect (Phillips, 2005). More than 25% of the variation in N fixation activity at 55 °C could be explained by which apsK allele a strain possessed (R2=0.28; P<10−6, significant after Bonferroni correction), indicating that this gene makes a major contribution to phenotypic variation. This contrasts with the 37 °C analysis, for which no strong candidate emerged (Table 1); in this case, a much larger sample size will be required to dissect the genetic basis of variation in N fixation. Although the mechanism by which the different apsK alleles impact N fixation at 55 °C remains to be resolved, we note that the sulfur requirements for N fixation are high due to the demand for iron-sulfur clusters by nitrogenase (Rubio and Ludden, 2008).
Terminal cytochrome oxidases
Enhanced respiratory activity within a mature heterocyst by cytochrome oxidases both reduces oxygen concentration in the vicinity of nitrogenase and contributes ATP to N fixation. Anabaena PCC 7120 has three copies of the coxBAC operon (coxBACI, coxBACII and coxBACIII), and all are strongly upregulated following heterocyst maturation, with coxBACII and coxBACIII exhibiting particularly large expression changes compared with nitrate-grown cells (Flaherty et al., 2011). The M. laminosus genome contains orthologs of all three operons, and components of each appear to be genetically differentiated between phenotypic classes for one or more of the analyses. For the allele frequency analyses, coxAII (gene ID 21-20552; Cyt c oxidase subunit 1) was the second-most differentiated gene for overall N fixation (Table 1), whereas coxAIII (gene ID 135-35450) was near our significance cutoff (P=0.011) for both the pooled and 37 °C analyses. At 55 °C, coxCI (gene ID 65-42545; Cyt c oxidase subunit 3) was a ΦST candidate. Given the importance of oxygen consumption for both ATP production and the maintenance of a microoxic environment in heterocysts, these results raise the possibility that this genetic variation may contribute to N fixation differences among strains by impacting aerobic respiration activity, and these loci represent future targets for functional validation.
HK gene 167-28586
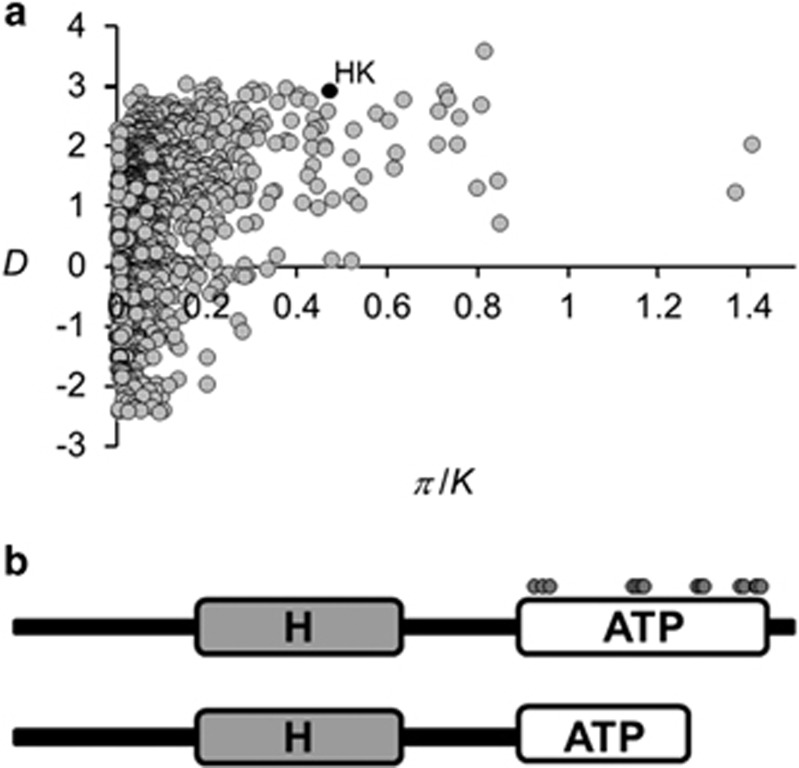
The most extreme outlier in both the ΦST and DXY distributions for the pooled data set was an annotated HK gene (gene ID 167-28586; Table 1). Four percent of nucleotide sites were variable at this locus, 40 times greater than the genome-wide average (Sano et al., submitted). Three alleles were identified: one with a nonsense mutation 150 nucleotides upstream of the 3′ end that was fixed in the upper class and at 36% frequency in the lower class; a full-length allele; and an apparently rare (N=1 in our sample) recombinant allele that is identical to the latter at the 5′ end and to the former at the 3′ end and therefore contains the nonsense mutation.
The comparatively high sequence divergence at this locus suggests either an idiosyncratically high evolutionary rate or that genetic variation has been maintained by some mechanism of balancing selection. The latter alternative is strongly supported by two molecular evolutionary signatures of long-term balancing selection observed for a random sample of 20 White Creek M. laminosus genomes (Figure 3a; Sano et al., submitted). One is an extremely positively skewed value of Tajima's D (among the top 0.5% most positive values; Tajima, 1989), which indicates that the common ancestor of the alleles is older than expected under selective neutrality. The second is an excess of nucleotide polymorphism in the White Creek population compared with divergence from the sister taxon Fischerella PCC 7414, among the 2% highest values observed. This evidence for long-term balancing selection maintaining variation at this locus suggests that different alleles may be favored in different environments.
Figure 3.
(a) HK candidate gene 167-28586 (HK, black circle) exhibits extreme values of both Tajima's D and the ratio of polymorphism (π) to divergence (K) from Fischerella strain PCC 7414 for 1623 orthologs that are polymorphic at White Creek. (b) Both the full-length (top) and nonsense (bottom) alleles of HK 167-28586 possess an intact H domain, but the nonsense mutation in the latter results in the loss of more than one-half of the ATP-binding residues (gray circles) in the binding pocket of the ATP domain.
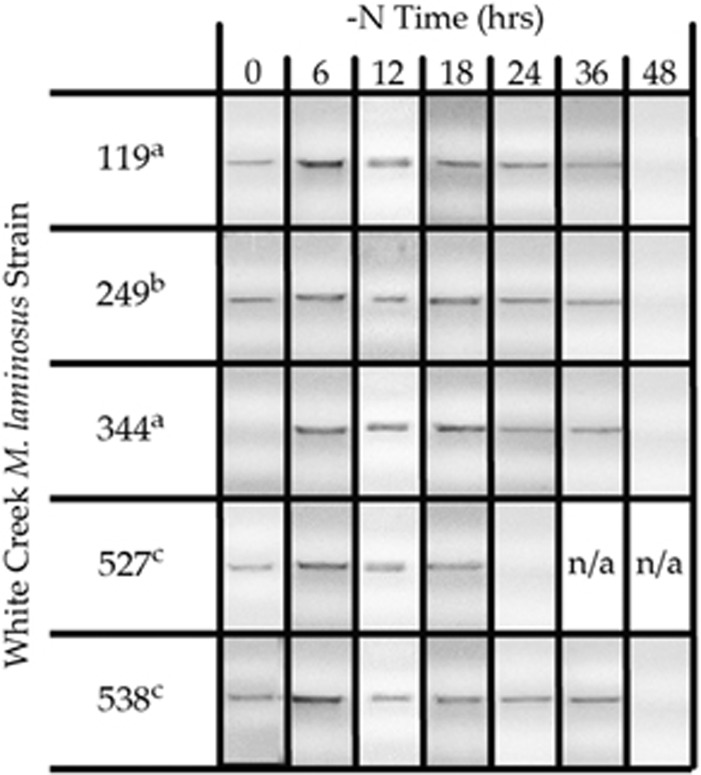
HKs participate in two-component signal-transduction systems, the principal means by which bacteria sense and respond to environmental changes (Gao and Stock, 2009). Following an environmental stimulus, the ATP-binding domain at the C-terminal end of an HK catalyzes the autophosphorylation of a conserved histidine residue. The HK then transfers the phosphoryl group to a cognate response regulator to effect a change in gene expression or protein activity (Galperin, 2010). The nonsense mutation in HK 167-28586 results in the loss of more than one-half of the ATP-binding pocket and of key ATP-binding residues (Figure 3b and Supplementary Figure 5), including the highly conserved G2 and G3 boxes required for autophosphorylation and kinase activities (Dutta and Inouye, 2000). This is expected to render this allele nonfunctional for these activities, with potential implications for the downstream regulation of gene expression. All three observed HK 167-28586 alleles were expressed during growth with nitrate (Supplementary Figure 6), immediately following N deprivation as well as during heterocyst development (Figure 4). However, because mature heterocysts were clearly visible at 36 h, the last time point for which the transcript was detected (Figure 4), we conclude that the gene is not expressed during steady-state growth under N-fixing conditions, following heterocyst differentiation. This suggests that any contribution of this locus to differences among strains in N fixation activity is a consequence of expression during the heterocyst development process itself, or, potentially, before the onset of N limitation.
Figure 4.
Presence or absence of an HK gene 167-28586 transcript after nitrogen step-down in five M. laminosus strains from White Creek. Subscripts indicate strain genotype ('a' is the nonsense allele, 'b' is the recombinant allele and 'c' is the full-length allele). With the exception of strain WC527, which had insufficient biomass after 24 h and was therefore not available for further analysis (N/A), the transcript could no longer be detected 48 h after the onset of N limitation.
Loss-of-function mutations that alter regulatory networks may be a common mechanism of bacterial adaptation to environmental change (Hottes et al., 2013), and we speculate that the nonsense mutation in HK 167-28586 may result in transcriptional 'rewiring' that is somehow favorable with respect to N fixation. Resolution of this issue, however, will require characterization of its function, including the identification of its cognate response regulator(s) and the signal-transduction network in which it participates. In addition, addressing whether there are fitness trade-offs in alternative environments (beyond N availability) among strains with different HK 167-28586 alleles will help to clarify the precise role that balancing selection has in the maintenance of variation at this locus.
Conclusion
Tests of genetic differentiation between phenotypic classes are a powerful way to identify the genetic variation that specifically contributes to functional differences among members of a population. Our results have revealed several candidate genes associated with differences in N fixation activity, some with clear links to heterocyst metabolism. In the case of apsK, we have resolved a single, geographically widespread SNP with a major effect on this ecologically important trait. Future efforts will be aimed at developing a conjugal transfer system for M. laminosus analogous to what has been recently accomplished for its close relative, Fischerella muscicola PCC 7414 (Stucken et al., 2012). This will enable us to investigate the phenotypic effects of individual genetic variants (e.g., apsK and HK 167-28586 alleles) in an otherwise identical genetic background and thereby begin to develop a mechanistic understanding of their consequences for organism function.
Acknowledgments
We thank the Cory Cleveland lab group for training and use of their gas chromatograph. Elizabeth Crone for statistical advice regarding the mixed-effects model and Kayli Anderson for assistance with the expression studies and DNA sequencing. We also thank two reviewers for their helpful comments and suggestions for improving the paper. This work was supported by US National Science Foundation award IOS-1110819 to SRM.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett RD, Schluter D. (2008). Adaptation from standing genetic variation. Trends Ecol Evol 23: 38–44. [DOI] [PubMed] [Google Scholar]
- Bates D, Maechler M, Bolker B, Walker S. (2014). lme4: Linear mixed-effects models using Eigen and S4. R package version 1.0-6. Available at: http://CRAN.R-project.org/package=lme4 (accessed 1 March 2014).
- Cadillo-Quiroz H, Didelot X, Held NL, Herrera A, Darling A, Reno ML et al. (2012). Patterns of gene flow define species of thermophilic Archaea. PLoS Biol 10: e1001265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell EL, Summers ML, Christman H, Martin ME, Meeks JC. (2007). Global gene expression patterns of Nostoc punctiforme in steady-state dinitrogen-grown heterocyst-containing cultures and at single time points during the differentiation of akinetes and hormogonia. J Bacteriol 189: 5247–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castenholz RW. (1988). Culturing methods for cyanobacteria. Methods Enzymol 167: 68–93. [Google Scholar]
- Cordero OX, Polz MF. (2014). Explaining microbial genomic diversity in light of evolutionary ecology. Nat Rev Microbiol 12: 263–273. [DOI] [PubMed] [Google Scholar]
- Dagan T, Roettger M, Stucken K, Landan G, Koch R, Major P et al. (2013). Genomes of Stigonematalean cyanobacteria (Subsection V) and the evolution of oxygenic photosynthesis from prokaryotes to plastids. Genome Biol Evol 5: 31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta R, Inouye M. (2000). GHKL, an emergent ATPase/kinase superfamily. Trends Biochem Sci 25: 24–28. [DOI] [PubMed] [Google Scholar]
- Ehira S, Ohmori M, Sato N. (2003). Genome-wide expression analysis of the responses to nitrogen deprivation in the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. DNA Res 10: 97–113. [DOI] [PubMed] [Google Scholar]
- Ehlers A, Worm B, Reusch T. (2008). Importance of genetic diversity in eelgrass Zostera marina for its resilience to global warming. Mar Ecol Prog Ser 355: 1–7. [Google Scholar]
- Flaherty BL, van Nieuwerburgh FV, Head SR, Golden JW, van Nieuwerburgh F. (2011). Directional RNA deep sequencing sheds new light on the transcriptional response of Anabaena sp. strain PCC 7120 to combined-nitrogen deprivation. BMC Genomics 12: 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY. (2010). Diversity of structure and function of response regulator output domains. Curr Opin Microbiol 13: 150–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Stock AM. (2009). Biological insights from structures of two-component proteins. Annu Rev Microbiol 63: 133–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick PW. (2006). Genetic polymorphism in heterogeneous environments: the age of genomics. Annu Rev Ecol Evol. Syst. 37: 67–93. [Google Scholar]
- Hottes AK, Freddolino PL, Khare A, Donnell ZN, Liu JC, Tavazoie S. (2013). Bacterial adaptation through loss of function. PLoS Genet 9: e1003617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JB, Daily GC, Ehrlich PR. (1997). Population diversity: its extent and extinction. Science 278: 689–692. [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D. (2006). Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23: 254–267. [DOI] [PubMed] [Google Scholar]
- Kumar K, Mella-Herrera RA, Golden JW. (2010). Cyanobacterial heterocysts. Cold Spring Harb Perspect Biol 2: a000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- Luck GW, Daily GC, Ehrlich PR. (2003). Population diversity and ecosystem services. Trends Ecol Evol 18: 331–336. [Google Scholar]
- Miller SR, Wingard CE, Castenholz RW. (1998). Effects of visible light and UV radiation on photosynthesis in a population of a hot spring cyanobacterium, a Synechococcus sp., subjected to high-temperature stress. Appl Environ Microbiol 64: 3893–3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SR, Purugganan MD, Curtis SE. (2006). Molecular population genetics and phenotypic diversification of two populations of the thermophilic cyanobacterium Mastigocladus laminosus. Appl Environ Microbiol 72: 2793–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SR, Williams C, Strong AL, Carvey D. (2009). Ecological specialization in a spatially structured population of the thermophilic cyanobacterium Mastigocladus laminosus. Appl Environ Microbiol 75: 729–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Schielzeth H. (2013). A general and simple method for obtaining from generalized linear mixed-effects models. Methods Ecol Evol 4: 133–142. [Google Scholar]
- Nei M. (1982) Evolution of human races at gene level. In: Bonne-Tamir B (ed). Human Genetics, Part A: The Unfolding Genome. Alan R Liss Inc.: New York, NY, USA, pp 167–181. [Google Scholar]
- Nicolaisen K, Hahn A, Schleiff E. (2009). The cell wall in heterocyst formation by Anabaena sp. PCC 7120. J Basic Microbiol 49: 5–24. [DOI] [PubMed] [Google Scholar]
- Oh H-M, Maeng J, Rhee G-Y. (1991). Nitrogen and carbon fixation by Anabaena sp. isolated from a rice paddy and grown under P and light limitations. J Appl Phycol 3: 335–343. [Google Scholar]
- Phillips P. (2005). Testing hypotheses regarding the genetics of adaptation. Genetica 123: 15–24. [DOI] [PubMed] [Google Scholar]
- Reusch TBH, Ehlers A, Hammerli A, Worm B. (2005). Ecosystem recovery after climatic extremes enhanced by genotypic diversity. Proc Natl Acad Sci USA 102: 2826–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Valera F, Martin-Cuadrado A, Rodriguez-Brito B, Pašić L, Thingstad TF, Rohwer F et al. (2009). Explaining microbial population genomics through phage predation. Nat Rev Microbiol 7: 828–836. [DOI] [PubMed] [Google Scholar]
- Rosen MJ, Davison M, Bhaya D, Fisher DS. (2015). Fine-scale diversity and extensive recombination in a quasisexual bacterial population occupying a broad niche. Science 348: 1019–1023. [DOI] [PubMed] [Google Scholar]
- Rubio LM, Ludden PW. (2008). Biosynthesis of the iron-molybdenum cofactor of nitrogenase. Annu Rev Microbiol 62: 93–111. [DOI] [PubMed] [Google Scholar]
- Shapiro BJ, Friedman J, Cordero OX, Preheim SP, Timberlake SC, Szabo G et al. (2012). Population genomics of early events in the ecological differentiation of bacteria. Science 336: 48–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart WD. (1970). Nitrogen fixation by blue-green algae in Yellowstone thermal areas. Phycologia 9: 261–268. [Google Scholar]
- Stewart WD, Fitzgerald GP, Burns RH. (1967). In situ studies on N2 fixation using the acetylene reduction technique. Proc Natl Acad Sci USA 58: 2071–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucken K, Ilhan J, Roettger M, Dagan T, Martin WF. (2012). Transformation and conjugal transfer of foreign genes into the filamentous multicellular cyanobacteria (Subsection V) Fischerella and Chlorogloeopsis. Curr Microbiol 65: 552–560. [DOI] [PubMed] [Google Scholar]
- Tajima F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall CA, Koniges GJ, Miller SR. (2014). Divergence with gene flow in a population of thermophilic bacteria: a potential role for spatially varying selection. Mol Ecol 23: 3371–3383. [DOI] [PubMed] [Google Scholar]
- Wolk CP. (2000). Heterocyst formation in Anabaena. In: Brun Y, Shimkets LJ (eds). Prokaryotic Development. ASM Press: Washington, DC, USA, pp 83–104.
- Zerbino DR, Birney E. (2008). Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.