Abstract
Low-grade, chronic inflammation has been associated with many diseases of aging, but the mechanisms responsible for producing this inflammation remain unclear. Inflammasomes can drive chronic inflammation in the context of an infectious disease or cellular stress, and they trigger the maturation of interleukin-1β (IL-1β). Here we find that the expression of specific inflammasome gene modules stratifies older individuals into two extremes: those with constitutive expression of IL-1β, nucleotide metabolism dysfunction, elevated oxidative stress, high rates of hypertension and arterial stiffness; and those without constitutive expression of IL-1β, who lack these characteristics. Adenine and N4-acetylcytidine, nucleotide-derived metabolites that are detectable in the blood of the former group, prime and activate the NLRC4 inflammasome, induce the production of IL-1β, activate platelets and neutrophils and elevate blood pressure in mice. In individuals over 85 years of age, the elevated expression of inflammasome gene modules was associated with all-cause mortality. Thus, targeting inflammasome components may ameliorate chronic inflammation and various other age-associated conditions.
Low-grade chronic inflammation has been associated with many of the diseases associated with aging1–7, but the mechanisms that produce this inflammation are poorly understood. IL-1β is a potent inflammatory cytokine that is elevated in the blood of older people8; this elevated IL-1β is linked to an increased risk of cardiovascular disease9, cancer10, functional decline11 and various degenerative diseases3,12. Inflammasomes, which are intracellular structures composed of NOD-like receptors (NLRs) or the ‘Absent in Melanoma 2’ (AIM2) protein that are triggered by the presence of pathogens or cellular stress13, are one source of IL-1β. In a recent study of aging in rats, the expression of a number of molecules that are associated with inflammasome activation was elevated, as was that of IL-1β14. However, it is unknown whether inflammasomes are activated in human aging and whether they contribute to the onset of age-associated disease. Therefore, we set out to investigate this question by making use of the large, immunology-focused data sets that we have accumulated on the Stanford–Ellison longitudinal cohort8,15–17. Starting with the gene expression data, we found that the levels of two gene modules that contained some inflammasome genes were consistently elevated over a 5-year period in individuals who were hypertensive and who also exhibited other comorbidities. Metabolomic data identified two novel DAMP molecules that were also elevated in hypertensive individuals and that could trigger inflammasome activity in vitro. In combination, these molecules could also induce hypertension and lymphocyte infiltration of the kidneys in mice. These data provide another direct link between an innate immune response against pathogens with chronic inflammation and cardiovascular disease.
RESULTS
Higher expression of inflammasome gene modules in older adults
To investigate the changes in the expression of genes from immune cells during human aging, we first analyzed age-related gene expression in the Stanford–Ellison longitudinal cohort8,15–17 using a modular approach18. An important feature of this approach is that the genes are organized into modules based on the coordinated expression of their components; such modules may contain genes that are previously known to be involved in a function along with those whose function has yet to be discovered. Using this approach, we found that of a total 109 gene modules, 41 were correlated with age (false discovery rate (FDR) Q ≤ 0.05, by Benjamini–Hochberg method)19, and among these, only two (modules 62 and 78, composed of 82 and 17 genes, respectively) (Supplementary Fig. 1) participate in cytokine production, as indicated by functional analysis20 (P < 0.01). To confirm this result, we conducted hypergeometric tests and found significant enrichment for these same modules (FDR Q < 0.01) (Supplementary Fig. 2a). Module 78 contained NLRC4, and module 62 contained NLRC5 and IL1B among other genes that are related to inflammasome activity, such as IL1RN, TLR6 and TLR8 (module 62), as well as IFAR1 and TLR5 (module 78) (Supplementary Fig. 1).
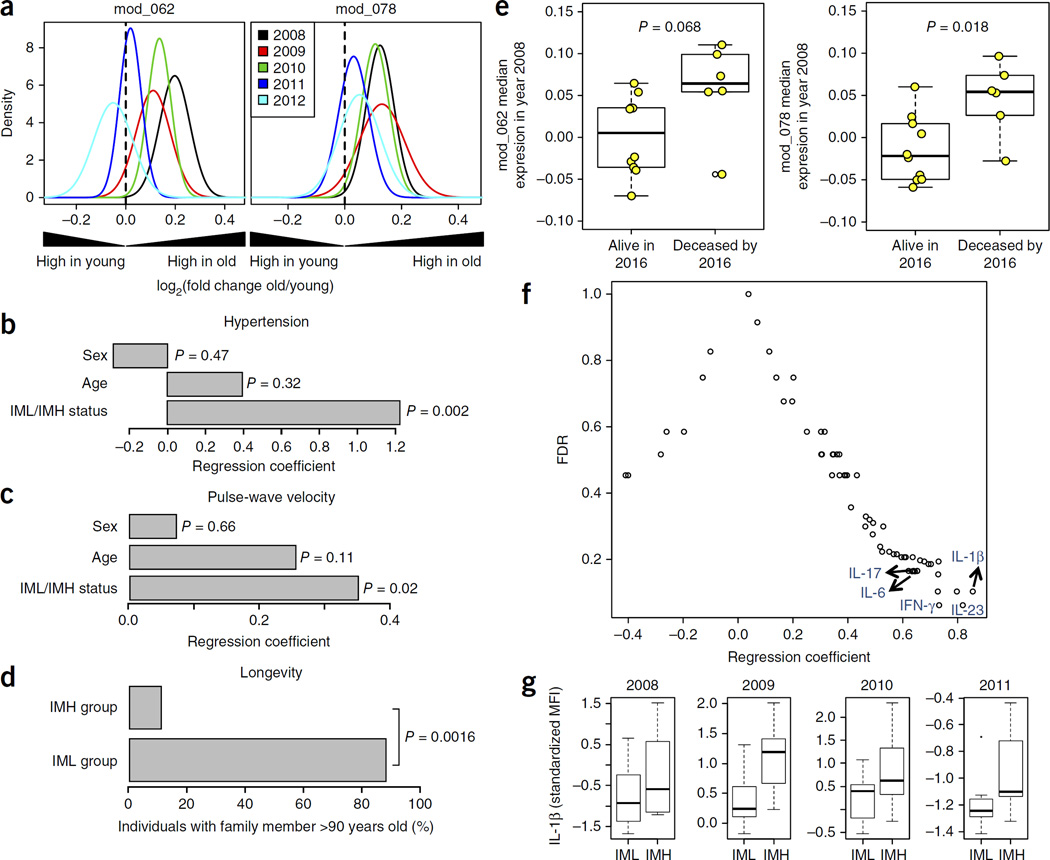
To determine the stability of the age associations for modules 62 and 78, we analyzed data from blood samples collected from the cohort over five consecutive years (2008–2012)8,15,16. Each year’s data consisted of data from both new subjects and subjects from previous years who were able to return (Supplementary Table 1), and the expression of these two gene modules in young (ages 20–30 years) versus older (ages 60 to >89 years) adults was compared using the QuSAGE method21. For this analysis, samples from each individual’s first appearance in the study (n = 114) were used to analyze how module expression is associated with age. When they are considered together, these data sets show a significant age-dependent increase for both gene modules (Fig. 1a; P < 10−3; see Online Methods).
Figure 1.
Expression of inflammasome gene modules in older adults and its association with human health and longevity. (a) Gene expression data from the Stanford–Ellison longitudinal cohort4,12,13 (n = 114) were used to find age-associated gene modules that participate in cytokine production and were enriched for inflammasome genes (see Supplementary Figs. 1 and 2). For the determination of significant differences in the expression of inflammasome gene modules 62 and 78, the QuSAGE gene set analysis method19 was used. Positive fold change values (x-axis) indicate higher expression in aged individuals in samples taken from 2008–2012. P-value for age on combined data for each gene module, 0.001. (b) A logistic regression analysis was conducted on IML (n = 11) or IMH (n = 12) group status and hypertension (shown are regression coefficients for age, sex and IML/IMH status). (c) Seventeen individuals from the year 2011 cohort (the same 8 IML and 9 IMH individuals as in b) were studied to measure the association of IML versus IMH status with the degree of arterial stiffness, as measured by pulse-wave velocity. Multiple regression analysis was performed on the pulse-wave velocity of each individual against their age, sex and IML/IMH status (shown are regression coefficients for each variable). P-values in b,c for each regression coefficient were calculated based on permutation methods (see Online Methods). (d) In the same 17 individuals from the year 2011 cohort, familial longevity was determined on the basis of membership in a family with at least one member over 90 years of age. The P-value was obtained by chi-square test. (e) Association between the expression of inflammasome gene modules 62 and 78 with all-cause mortality. Each point is representative of one individual. The P-value was obtained by Student’s t-test. (f) Serum levels of 62 different cytokines, chemokines and growth factors were compared between IML and IMH subjects using data from year 2013 (IML n = 8, IMH n = 8). Multiple regression analysis on each analyte’s MFI against their age, sex and IML/IMH status was conducted and significance (y-axis) was obtained via permutation tests. (g) IL-1β serum abundance, as shown by longitudinal analysis of data collected during the years 2008–2011 (IML n for 2008, 2009, 2010 and 2011 = 10, 10, 8 and 7, respectively; IMH n 2008, 2009, 2010 and 2011 = 12, 11, 12 and 8, respectively). Whisker bars represent maximum and minimum values.
Inflammasome modules correlate with health and longevity
Next, we investigated whether the expression of both modules 62 and 78 was associated with any clinical phenotype in our aging cohorts. To do so, we first defined the extreme phenotypes of older adults on the basis of both the magnitude and the chronicity in the expression levels of modules 62 and 78. Subjects were assigned into inflammasome module high (IMH) or inflammasome module low (IML) groups if they were in the upper or lower quartiles, respectively, for each gene module in 3 or more of the 5 years that were analyzed (see Online Methods). This sorting yielded 19 individuals with extreme phenotypes for module 62, 9 categorized as IMH and 10 as IML, and 16 individuals with extreme phenotypes for module 78, 9 IMH and 7 IML. We noted a significant degree of overlap for modules 62 and 78 in each category (6 IMH and 6 IML, P-value for enrichment < 0.001). Furthermore, the expression levels of these two modules across all examined individuals were highly correlated (R2 = 0.76, P < 10−5) (Supplementary Fig. 2b). Thus, to improve statistical power, IMH (age range 66–86) or IML (age range 62–90) individuals from modules 62 and 78 were combined (n = 23) for further analysis.
We next performed regression analysis to compare IMH and IML phenotype with respect to the clinical history of diabetes, hypertension and psychiatric disorders of an individual. The scoring of all these clinical outcomes was binary and based on clinical history. No significant associations were found for diabetes or psychiatric disorders. However, 75% (9/12) of IMH subjects were hypertensive (known as ‘essential’ hypertension), as compared to very few (1/11 or 9%) in the IML group. The hypertension rate for all the individuals in the older cohort was 52%, which is equivalent to the rate in the general US population of that age22. The rate of hypertension in unclassified older adults was 51% (Supplementary Fig. 3). Because the age range of our older cohorts was relatively large, age and sex were included the logistic regression models. In addition, we adjusted for other confounding factors such as medication history (Supplementary Table 2) and body mass index (BMI) (see Online Methods), yet we still found a significant association between hypertension and IMH/IML status (P = 0.002) (Fig. 1b).
On the basis of our observation that the IMH and IML groups differed in their history of hypertension and in the potential contribution of other confounders, we conducted a follow-up study to determine arterial stiffness, which is a stable risk factor for cardiovascular complications, using carotid-femoral pulse wave velocity (PWV) testing. The PWV was significantly lower in the IML group (7.9 ± 2.4 m/s) than in the IMH group (10.7 ± 2.1 m/s) (P = 0.02) (Fig. 1c) and was not significantly different from that in unclassified individuals (Supplementary Fig. 3). Interestingly, self-reported familial longevity, as determined by membership in a family with at least one family member over 90 years of age, was significantly higher in IML subjects (88%) compared with IMH subjects (11%) (P < 10−4) (Fig. 1d). Moreover, when we classified individuals over 85 years of age according to whether or not they died between the years 2009–2016, we noted that expression of module 78 at the beginning of the study (year 2008) was higher in those who were deceased when compared with those still living as of 2016 (P = 0.018). This was also true, to a lesser extent, for module 62 (P = 0.068) (Fig. 1e).
Stable expression of circulating IL-1β in IMH subjects
We measured the abundance of 62 cytokines and chemokines in serum samples collected in all the individuals recruited in 2013 (n = 92), and we conducted a regression analysis to search for associations between circulating cytokine levels and IML versus IMH status. For this analysis, we used samples from the subset of IML and IMH classified groups after adjusting for the age and sex of those included. To control for multiple comparisons, we estimated the statistical significance of each regression coefficient by permutation analysis using 500 resamplings, which enabled us to calculate a corrected P-value for each regression coefficient. At a FDR of Q < 0.2, we found that the levels of 17 cytokines were increased in the serum of individuals in the IMH group (Supplementary Table 3), with IL-1β increasing the most (Fig. 1f). The differences in IL-1β were stable during the years 2008–2011 (Fig. 1g). The levels of IL-1β in unclassified older adults were intermediate for the years 2008, 2009 and 2011 and were not significantly different for the year 2010 when compared with the IMH and IML groups (Supplementary Fig. 3).
Metabolic dysfunction and oxidative stress in the IMH group
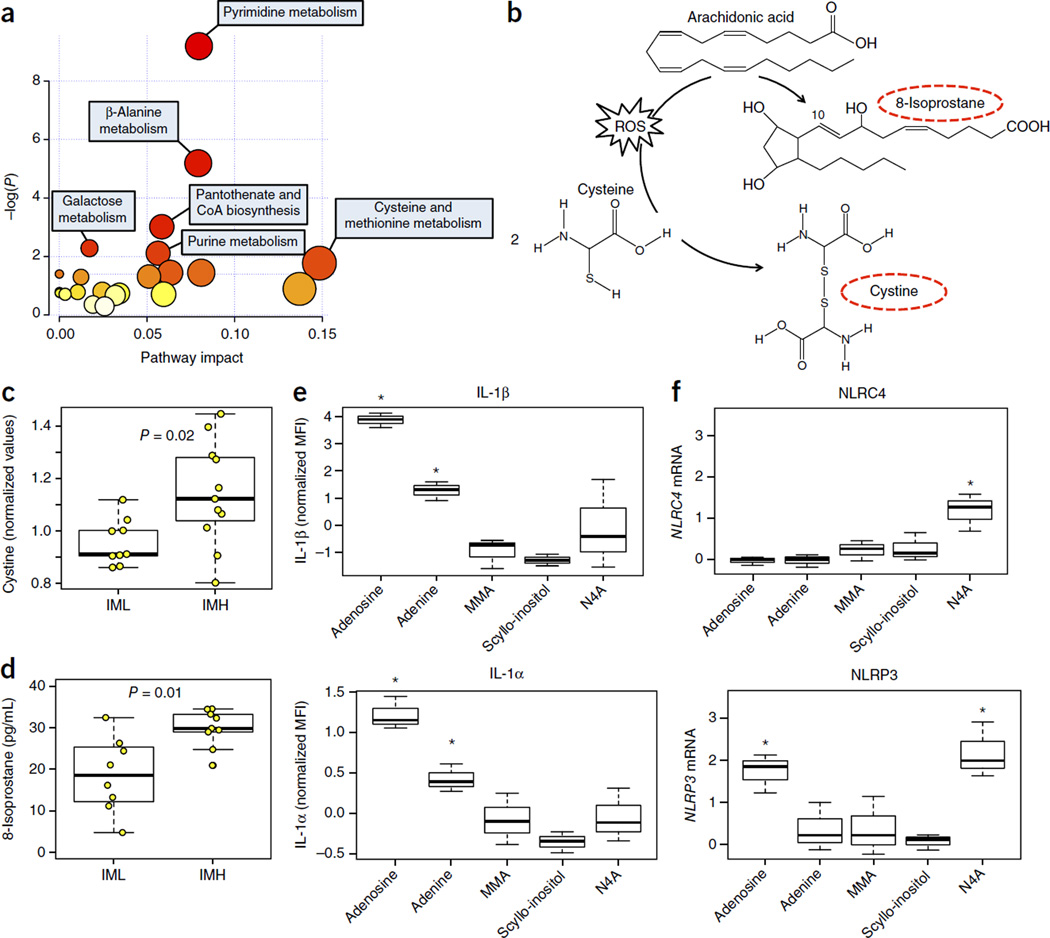
The main inflammasome machinery requires both priming, to initiate transcription, and a subsequent activation step in order to function. To identify molecules that could influence either process, we analyzed 692 metabolites from the sera of 11 IML and 9 IMH individuals by mass spectrometry. Using the significance analysis of micro-array (SAM)23 analysis (see Online Methods), 67 metabolites were found to be significantly different between the IML and IMH groups (FDR Q < 0.2); all were more abundant in the IMH group than in the IML group (Supplementary Table 4). Pathway enrichment analysis24 revealed that the metabolites that were upregulated in the IMH individuals were associated with the metabolism of pyrimidine, β-alanine, galactose, purine, and the biosynthesis of pantothenate and CoA (P ≤ 0.01) (Fig. 2a). We next analyzed the differences in the expression levels of the genes involved in pyrimidine metabolism, β-alanine metabolism, pantothenate and CoA biosynthesis, and purine metabolism (cut-off P-value < 0.01 and pathway impact > 0.05) (see Online Methods) (Fig. 2a). Our analysis revealed statistically significant differences between the two groups in the expression of genes involved in the pyrimidine and purine pathways (Supplementary Fig. 4a,b), but not of genes involved in the β-alanine metabolism or pantothenate and CoA biosynthesis pathways (P < 0.05). Genes that encode proteins that degrade nucleotide triphosphates (UTP, CTP) to generate uracil and thymine-derived species, such as CMPK1 (UMP-CMP kinase), NT5E (5′-nucleotidase), UPRT (uracil phosphoribosyltransferase homolog), ENTPD1 (ectonucleoside triphosphate diphosphohydrolase 1) and others, were consistent with the metabolite species found in the IMH group (Supplementary Fig. 4a). Therefore, integration of metabolomics and gene expression data demonstrate that IMH individuals exhibit signs of nucleotide metabolism dysfunction.
Figure 2.
Metabolites present in IMH individuals induce IL-1β and upregulate the expression of inflammasome genes. (a) Broad-coverage metabolomics profiling was conducted on available serum samples from year 2011 (n = 9 IML, n = 11 IMH). From a total of 692 metabolites analyzed, 67 were differentially expressed (all upregulated) in IMH versus IML at an FDR of Q < 0.2 (by SAM analysis; see Online Methods). Functional annotation and pathway analysis were conducted using MetPA33. A significant enrichment for several metabolic pathways was identified for these metabolites (P < 0.05). Darker color indicates higher level of significance. (b) The conversions of cysteine to cystine and of arachidonic acid to 8-isoprostane n the presence of ROS. Circulating levels of (c) cystine and (d) 8-isoprostane are greater in IMH as compared to I ML individuals. (e) Adenine, dl-4-hydroxy-3-methoxymandelic acid (vanillylmandelate) (MMA), scyllo-inositol and N4-acetylcytidine (N4A) were selected for further study on the basis of their levels of significance (Q < 0.001; see Supplementary Table 4) and their representation of different metabolic pathways. We assessed each compound’s ability to alter IL-1α and IL-1β levels and the expression of NLRC4 in primary monocytes from four healthy young adults. Results show one representative experiment. Adenosine was used as a positive control. A significant induction of IL-1α and IL-1β was observed when using adenosine and adenine but not the other compounds. (f) The highest dose of each compound (100 µM) was used to determine expression of NLRC4 and NLRP3 by qPCR on the same samples used for cytokine determination. A significant increase in NLRC4 and NLRP3 is shown only for N4-acetylcytidine (P < 0.05, by one-sided Student’s t-test). Adenosine treatment upregulated NLRP3 gene expression (P < 0.01 by one-sided Student’s t-test). Expression of GAPDH was used to standardize the samples, and the results are expressed as the normalized ratio in relation to the control. Error bars reflect experimental variability.
We also investigated the presence of markers of oxidative stress in IMH versus IML subjects (Fig. 2b). Using the metabolomics data, we determined whether the levels of circulating cystine (an oxidized product of cysteine) differed between the IMH and IML groups. This compound is generated from a direct reaction between cysteine and a reactive oxygen species (ROS) (Fig. 2b), and thus its presence is an important marker of oxidative stress. Significantly higher levels of circulating cystine were detected in IMH as compared with IML individuals(Fig. 2c). In addition, we measured the circulating levels of 8-isoprostane from available sera (n = 17) and found that these were significantly higher in IMH as compared with IML individuals (Fig. 2d). Therefore, in addition to the defects in nucleotide metabolism, metabolic reprogramming of mitochondrial bioenergetics in IMH individuals may lead to constitutive high levels of ROS and subsequent chronic oxidative stress.
Metabolites in IMH subjects elicit inflammation
To determine whether the circulating metabolites that are found in higher levels in the sera from IMH as compared to IML subjects upregulate NLRC4 gene expression and/or cytokine production, we selected four candidate compounds identified from our analysis that each represent distinct metabolic pathways. These compounds included adenine (purine metabolism), dl-4-hydroxy-3-methoxymandelic acid (vanillylmandelate; phenylalanine and tyrosine metabolism), scyllo-inositol (inositol metabolism) and N4-acetylcytidine (N4A; pyrimidine metabolism) (Supplementary Table 4). Adenosine was included as a positive control for IL-1β production25. Primary monocytes from four young, healthy donors were isolated from fresh blood and incubated with increasing concentrations (0, 3, 10, 30 and 100 µM) of either adenosine or adenine for 6 h. The highest concentration was chosen on the basis of previous reports showing that adenosine (used at 100 µM) can regulate inflammasome activity that is initiated by a wide range of PAMPs and DAMPs25. The other compounds were also used at the same concentration (100 µM), which for N4A corresponds to approximately one-half of the concentration observed in the blood of healthy individuals26. A significant dose-dependent increase in IL-1β was observed only for the adenosine and adenine treatments (Supplementary Fig. 5), and no induction was observed for the other compounds tested (Fig. 2e). We also investigated whether these stimuli upregulate the expression of NLRC4 and NLRP3. As expected25, adenosine treatment increased the expression of NLRP3 (Fig. 2f). However, no increase in NLRC4 expression was observed. In contrast, treatment with N4A induced the expression of both NLRP3 and NLRC4 (P < 0.01). No significant effect of adenine, dl-4-hydroxy-3-methoxymandelic acid or scyllo-inositol was observed (Fig. 2f). These results indicate that N4A, an endogenous nucleoside product in the degradation tRNA (a marker of oxidative stress), may prime NLRC4 expression; in contrast, adenosine and adenine may generate a second signal for NLRC4 activation that results in the induction and secretion of IL-1β. More biochemical studies are necessary to validate this hypothesis. That adenine treatment does not upregulate NLRP3 or NLRC4 but induces IL-1β production from primary monocytes suggests that there is at least a degree of in vivo priming causing background expression of inflammasome genes.
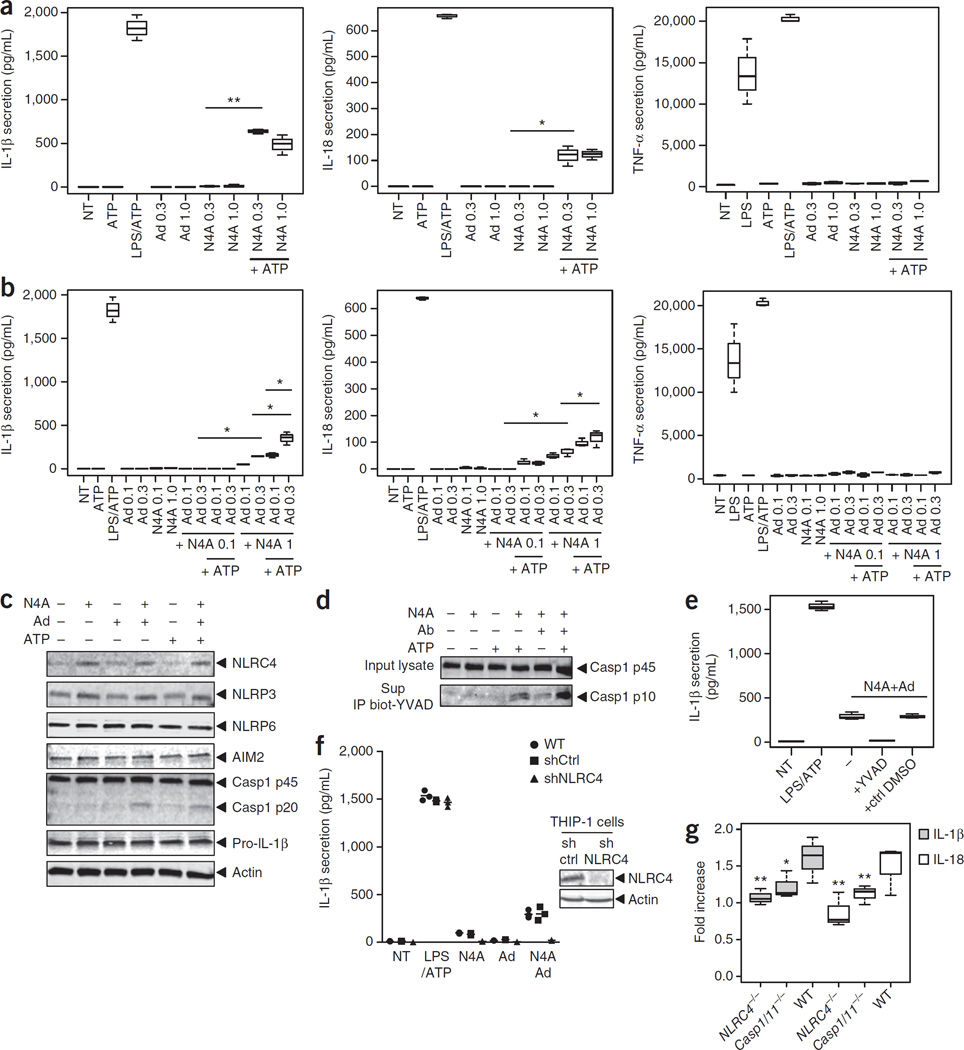
In parallel experiments, we treated differentiated THP-1 cells, a human monocytic cell line, with N4A and adenine either alone or in combination. Neither adenine nor N4A alone significantly influenced production of IL-1β, IL-18 (another member of the IL-1 family of cytokines) or TNF-α (Fig. 3a). However, the addition of ATP in the presence of N4A induced the secretion of IL-1β and IL-18 but not of TNF-α (Fig. 3a). Moreover, treating cells with adenine and N4A together induced a significant increase in IL-1β and IL-18 levels (Fig. 3b), which was further augmented by pulsing cells with ATP. This combinatorial effect of N4A and adenine on the production of IL-1β and IL-18 was not observed for TNF-α, which indicates that the observed effect is likely dependent on inflammasome activation. In accordance with the N4A-dependent increase of NLRC4 and NLRP3 mRNAs, we also observed an N4A-induced increase in the abundance of NLRC4 and NLRP3 protein by western-blot analysis (Fig. 3c and Supplementary Fig. 6). Consistently with the observed increase in IL-1β secretion that occurs upon stimulation of THP-1 cells with N4A and adenine, procaspase-1 cleavage into the active p10 fragment was confirmed by specifically immunoprecipitating the active p10 fragment with biotinyl-YVAD-fmk from THP-1 cells stimulated with these two compounds (Fig. 3c,d and Supplementary Fig. 6). Caspase-1 dependency was further demonstrated by showing the complete abrogation of compound-mediated IL-1β secretion by the caspase-1 inhibitor YVAD-fmk (Fig. 3e).
Figure 3.
Metabolites in IMH individuals activate the NLRC4 inflammasome. (a,b) Differentiated THP-1 cells were treated with ATP (5 mM, 30 min), or they were primed with LPS (1 µg/ml, 4 h) and then pulsed with ATP, or they were treated with the indicated concentrations (mM) of either adenine (Ad) or N4-acetylcytidine (N4A) alone (a) or in combination (b) for 6 h. NT, nontreated. They were then either pulsed with ATP or not. Secretion of cytokines IL-1β, IL-18 and TNF-α was measured by ELISA from cell culture supernatants. (c) Differentiated THP-1 cells were treated with compounds as indicated (1 mM N4A; 300 µM adenine (Ad)) for 6 h or with ATP 5 mM for 30 min. Then cells were lysed, and the lysates were immunoblotted with various antibodies to monitor cellular expression of NLRs, caspase-1 (casp1) and pro-IL-1β. (d) Differentiated THP-1 cells were treated with compounds using the same process outlined earlier, and the cell lysates were submitted to immunoprecipitation using biotinyl-YVAD-fmk peptide. Complexes were then recovered by using streptavidin-Sepharose beads and were immunoblotted with anti-caspase-1 p10 antibody. (e) Differentiated THP-1 cells were treated with compounds as before in the presence or absence of Ac-YVAD-fmk or of control DMSO, and then IL-1β secretion was measured. (f) Differentiated wild-type (WT) or stable shTHP-1 cell lines were treated with compounds as before (1 mM N4A; 300 µM Ad) and IL-1β secretion was measured. Right, western blots showing protein expression of NLRC4 in stable shTHP-1 cell lines. Ctrl, control. (g) Bone-marrow-derived macrophages from WT or NLRC4-KO mice were obtained as previously described46. Cells were plated in triplicates, treated with combination of compounds (1 mM N4A; 300 µM adenine) and IL-1β secretion was measured. Data in panels a,b,e–g are expressed as the concentration of cytokines (pg/mL) or as the fold increase with respect to baseline as indicated (mean ± s.d.; n = 3 for a,b,e,f; n = 9 for g). P-values were determined by Student’s t-test. *P < 0.05, **P < 0.01.
We then investigated the role of NLRC4 in IL-1β secretion by using THP-1 cells that stably express shNLRC4 and bone marrow-derived macrophages from NLRC4 knockout mice. Knockdown or knockout of NLRC4 significantly reduced the IL-1β secretion induced by N4A + adenine (Fig. 3f,g). shNLRC4 had no effect on the LPS/ATP-induced IL-1β secretion, which is consistent with the dependence of these stimuli on NLRP3 inflammasome activation.
N4A and adenine activate platelets and neutrophils in vitro
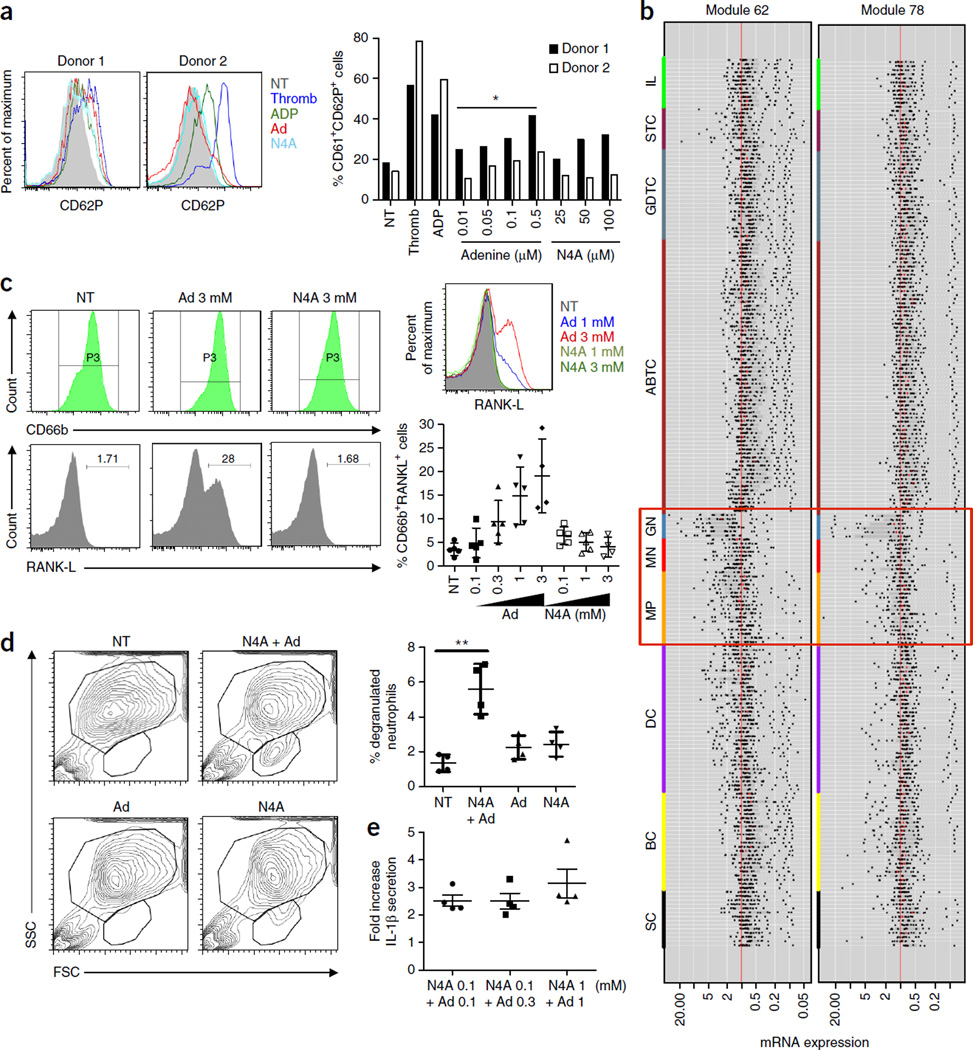
Platelet activation occurs in various inflammatory conditions of both infectious and non-infectious origins, and accumulating evidence indicates that patients with hypertension exhibit high levels of activated platelets when compared with healthy, disease-free controls.
Putative mechanisms that contribute to platelet activation in hypertension include endothelial dysfunction, neurohumoral (sympathetic and renin–angiotensin systems) overactivity, decreased platelet nitric oxide (NO) biosynthesis and platelet degranulation that is secondary to increased shear27–30. Since viral infection can activate platelets through the inflammasome (NLRP3) pathway28, we sought to determine whether N4A and adenine were able to activate human platelets in vitro using platelets that were isolated from the blood of two healthy young adults. The tested concentrations of adenine were chosen based on standard activating concentrations used for ADP (5 µM). N4A-activated platelets were more sensitive than those activated by THP-1 cells or primary monocytes, and thus we used lower concentrations of N4A than those previously used in these cells to reach maximal effects. Activation was monitored by measuring the concentration of CD61+ and CD62P+ cells by flow cytometry. Only adenine resulted in a statistically significant dose-response effect (P < 0.05 for each donor) (Fig. 4a) with higher doses of adenine inducing increasing numbers of activated granulocytes.
Figure 4.
Metabolites in IMH individuals activate human primary platelets and neutrophils. (a) Primary platelets were isolated from blood of healthy donors and were incubated for 6 h with thrombin, ADP (3 µM) or various concentrations of adenine or N4A. Platelet activation was monitored by measuring the membrane expression of CD61 and CD62P in healthy cells by flow cytometry. Histograms show overlay of CD62P staining within the CD61 positive population for Adenine (0.5 µM) and N4A (100 µM) for each donor. Bar graph summarizes the results obtained from each donor. Student’s t-test with *P < 0.05 applies to each donor. (b) Analysis of immune cell type populations (ImmGen database) show that modules 62 and 78 are predominantly expressed in macrophages, monocytes and granulocytes (orange, red and blue respectively) (P < 10−10). SC, stem cells; BC, B cells; DC, dendritic cells; MP, macrophages; MN, monocytes; GN, granulocytes; ABTC, αβ T cells; GDTC, γδ T cells; STC, stromal cells; IL, innate lymphocytes. (c) Primary neutrophils were isolated from blood of healthy donors and then incubated for 24 h with various concentrations of adenine or N4A either separately or in combination. the concentration of RANK-L+ cells present within the CD66b+ population was determined. Histograms show overlay of RANK-L staining within the CD66b+ population. Graph below summarizes the results of each donor. (d) Primary neutrophils were treated with N4A (1 mM) and/or adenine (1 mM), and the percentage of degranulated population was measured as shown in the scatter plot. Graph at right summarizes results obtained from each donor. (e) Primary neutrophils were treated as described previously with the compounds as indicated in the figure, and IL-1β secretion was measured from the cell culture supernatants obtained from each donor. Data are expressed either as the concentration of cytokines (pg/mL) or as the fold increase with respect to the non-treated (NT) condition as indicated. P-values in a,d,e were determined by Student’s t-test. *P < 0.05, **P < 0.01.
Computational analysis revealed that modules 62 and 78 were preferentially expressed in monocytes, macrophages and neutrophils (P < 10−10; see Online Methods (Fig. 4b and Supplementary Fig. 7). Thus, we also investigated the effects of N4A + adenine on primary human neutrophil activation and IL-1β secretion. Adenine, in contrast to N4A, induced a potent increase in RANKL+ cells within the CD66b cell population (Fig. 4c). In addition to promoting expression of RANKL, the combination of N4A + adenine induced an increase in a degranulated population of neutrophils (Fig. 4d). Finally, N4A + adenine-treated neutrophils were able to secrete relatively low concentrations of IL-1β (a 2- to 3-fold increase compared to untreated cells) (Fig. 4e).
IMH metabolites induce hypertension and inflammation in mice
To study whether N4A and adenine had a direct effect on blood pressure in vivo, we injected mice with these compounds daily and monitored changes in blood pressure using a tail cuff method for 34 d (time where the increase in blood pressure in treated mice reached plateau; see below). We chose to use a final in vivo concentration of these compounds of 1 mM, which is 10 times the concentration used for the in vitro studies in primary monocytes (see Online Methods).
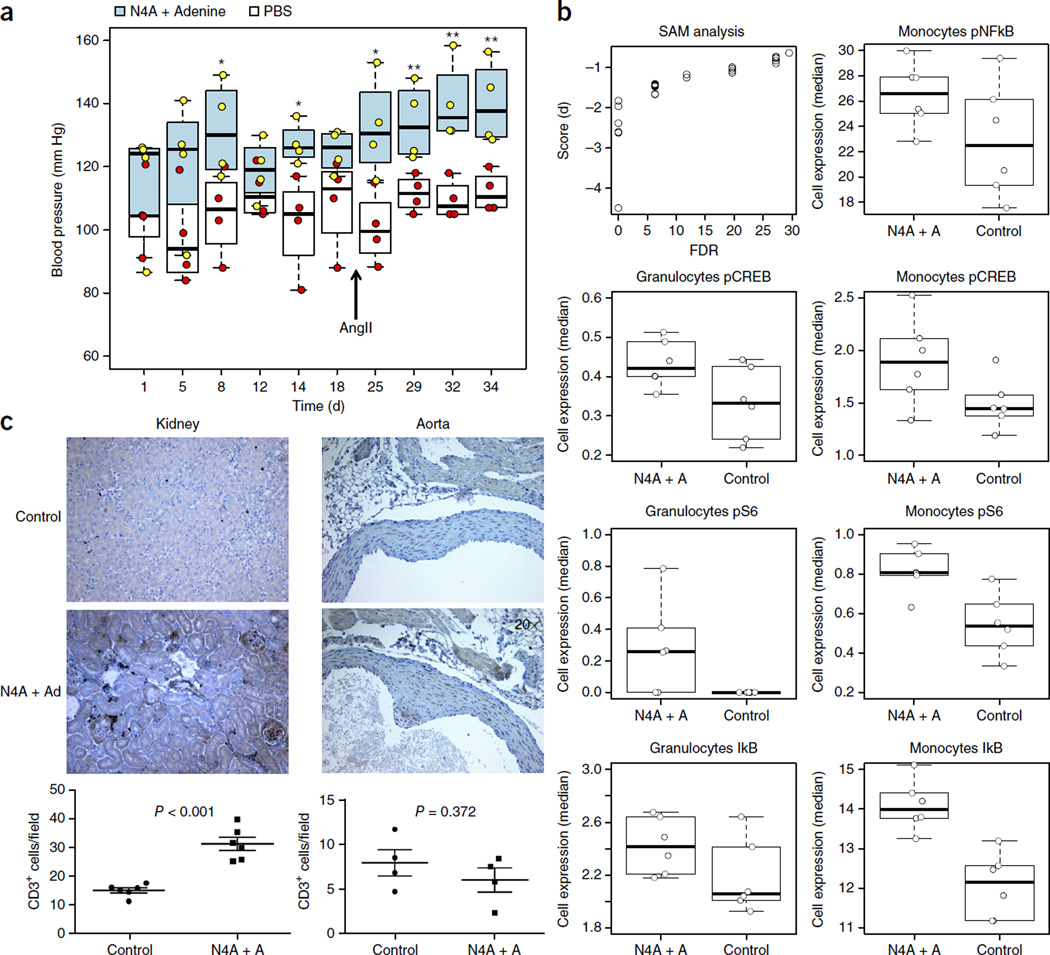
Treatment with N4A and adenine had a mild effect with borderline-significant increases in blood pressure (pre-hypertension) as early as 8 d after the first injection (P = 0.04 for group comparison) (Fig. 5a). On the basis of the previous observation that pre-hypertensive stimuli, such as angiotensin II (angII), and an oxidative stress-dependent inflammatory response act jointly during sustained hypertension31, we implanted angII pumps (at 140 ng/kg/min) and administered angII in combination with the compounds (in the treated mice) or with PBS (in the control mice) at day 20. This resulted in a significant increase in average systolic blood pressure in the treated mice when compared to the control group (140 (±7) versus 112 (±3) mmHg; P = 0.016) (Fig. 5a). Therefore, N4A and adenine can elevate blood pressure in mice, but pre-hypertensive stimuli may be needed in addition to the effect of these compounds to induce sustained hypertension.
Figure 5.
Metabolites in IMH individuals induce high blood pressure in mice. (a) Mice were randomly put into either the control (n = 4) or the treatment (n = 4) group. The treatment group mice were injected with N4A + adenine at 20 mM stock solution of 100 µL per 25 g body weight once daily. Treatment with N4A + adenine had a mild effect with pre-hypertension states as early as 8 d after the first injection (P = 0.02, by one-tailed Student’s t-test). Significant increases were observed after day 20 in the treated mice, with an average systolic blood pressure of 140 (±7) mmHg in that group, as compared to 112 (±3) mmHg in the control group (P = 0.008). *P < 0.05, **P < 0.01 by one-tailed t-test. These experiments were repeated using 10 mice per group with similar results. Filled and empty horizontal lines represent the mean values for the treated and control groups, respectively. Whisker bars represent maximum and minimum values. (b) Increased phosphorylation levels of signaling proteins (FDR Q < 0.05) in blood cells from N4A + adenine-angII treated mice versus control mice (treated with angII alone). Shown are the results of SAM analysis comparing the two groups of mice (see Online Methods); the x-axes represent the FDR or significance (cutoff value of 5%) as a function of score (d) parameter (y-axis), which is equivalent to the t-statistic value of a t-test when comparing two samples. P-values were adjusted for multiple comparisons using SAM, which provides an estimate of FDR. pNF-κB, phosphorylated form of NF-κB (p65 Ser529); pCREB, phosphorylated form of CREB (Ser133); pS6, phosphorylated form of 40S ribosomal protein S6. (c) Higher levels of infiltrating T cells in the kidney, but not the aorta, are observed in the treatment group versus the control group.
To gain more insight into this effect, we repeated this experiment using 10 mice per group and collected peripheral blood, kidney and aorta samples, from 6 out of the 10 mice per group at the end of study. We used mass cytometry (CyTOF) to compare the expression of the NF-κB inhibitor IκB and the activation marker CD62L. We also compared the levels of expression of a series of phosphorylated intracellular signaling proteins, including CREB, STAT1, STAT3, STAT5, p38, S6, NF-κB, ERK and MAPKAPK2, in blood cell subsets including granulocytes, monocytes, NK cells, CD4+ and CD8+ T cells, T regulatory CD4+ T dcells and B cells from both compound-treated and control mice (see Supplementary Table 5 for antibody panel composition). We observed a state of immune activation in compound-treated mice, as evidenced by higher levels of pS6 and pCREB in monocytes and in granulocytes and increased pNF-κB in monocytes in these mice as compared to untreated controls (Fig. 5b). IκB levels were also elevated in treated mice as compared to controls. This apparent disagreement can be explained by the fact that it is only under acute conditions that the reduction of total IκB tracks with increased pNF-κB levels; under chronic conditions, the initial reduced IκB signaling following stimulation returns to baseline at a time when pNF-κB levels are still elevated32.
We also observed significant T cell infiltration in the kidneys (cortex) (P < 0.001) but not the aorta (P = 0.372) in N4A + adenine-treated mice when compared to the controls (Fig. 5c). These differences did not reach significance in the kidney medulla, but we observed a similar trend in the cortex with higher levels of T cell infiltration present in treated mice than those that were found in control mice (P = 0.09) (Supplementary Fig. 8).
Caffeine inhibits metabolite-induced IL-1β production
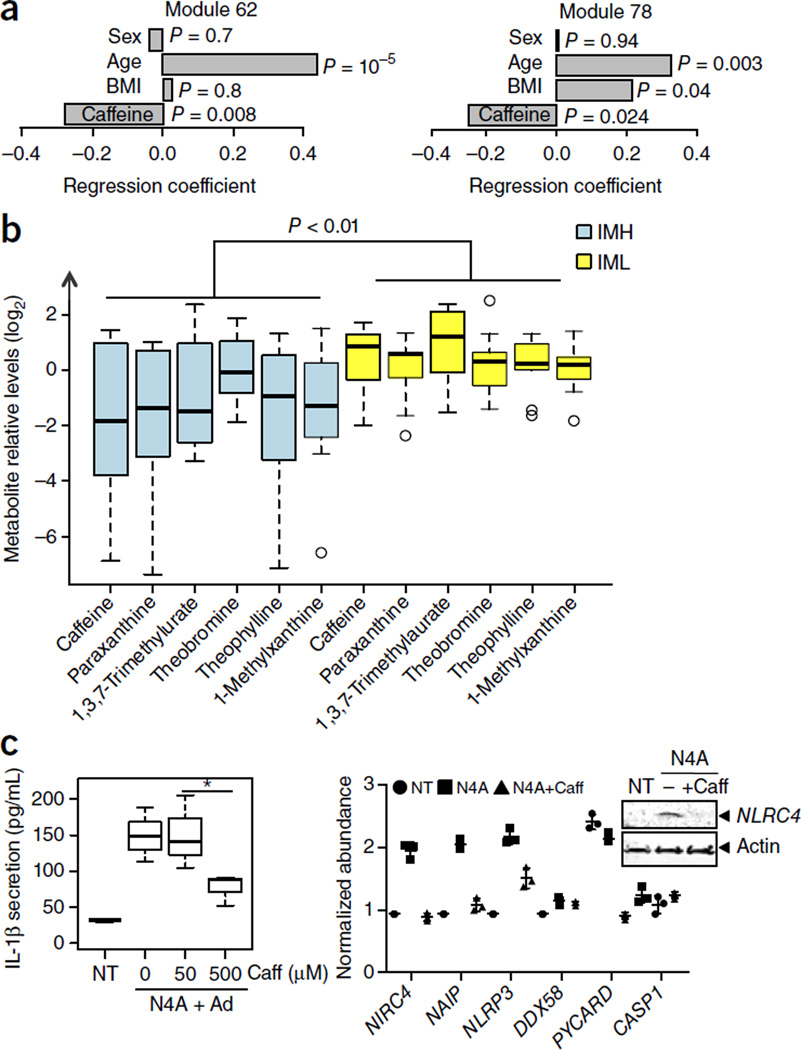
Since caffeine antagonizes adenosine due to their structural similarity, we next asked whether caffeine levels were associated with inflammasome-module gene expression. To do this, we used a questionnaire consisting of a 15-item survey of dietary and pharmaceutical sources of caffeine (see Online Methods), derived from 120 of the most commonly consumed caffeinated products in the United States in 2007 (Center for Science in the Public Interest; see www.cspinet.org/new/cafchart.htm). The questionnaire was administered to 91 subjects in the year 2008. We performed multiple regression analysis using the data from all individuals in the year 2008 (N = 89) on the expression of modules 62 and 78 and their caffeine intake (in mg/week). We also adjusted for BMI, a known confounding factor that is associated with caffeine intake. We found a significant age-, sex- and BMI-adjusted association between caffeine intake and the expression of modules 62 (P < 0.01) and 78 (P = 0.024) (Fig. 6a).
Figure 6.
Caffeine negatively regulates the NLRC4 inflammasome. Multiple regression analysis was conducted on expression levels of module 62 and 78 (from data collected in year 2008, n = 89) and caffeine intake for each individual in mg/week (adjusted for age, sex and BMI). (a) A significant association was found between caffeine intake and the expression of modules 62 (P < 0.01) and 78 (P = 0.024). (b) Differences in the circulating levels of coffee and coffee-derived metabolites between the IML and IMH groups were computed using a one-tail Student’s t-test, and P-values were combined using a modified generalized Fisher method for combining probabilities from dependent tests33 (P < 0.01). Whisker bars represent maximum and minimum values. (c) Left: Differentiated THP-1 cells were incubated with various concentrations of caffeine (caff) and were then treated with N4A (at 1 mM) + adenine (at 300 µM) for 6 h. IL-1β secretion was measured from cell culture supernatants. Data are expressed as concentration of cytokines (pg/mL) (mean ± s.d.; n = 3). P-values were determined by Student’s t-test. *P < 0.05. (c) Right: Differentiated THP-1 cells were treated with N4A (at 1 mM) in the presence or absence of caffeine (at 0.5 mM) for 6 h, and expression of various genes as indicated was assessed by qPCR. Cell lysates were submitted to immunoblotting with indicated antibodies for protein expression.
We then compared the levels of caffeine and caffeine-derived metabolites in the sera from IMH and IML subjects. To do so, we used the metabolomics data that was previously generated and compared serum levels of caffeine and its metabolites paraxanthine, 1,3,7-trimethyluric acid, theophylline, theobromine and 1-methylxanthine, without adjusting for multiple comparisons. We found that, when considered jointly33, the IML group exhibited significantly elevated levels of these metabolites when compared to the IMH group (P < 0.01) (Fig. 6b). We also tested the effect of caffeine in vitro at concentrations that matched the estimated concentrations found in the plasma of coffee drinkers (10–50 µM)34. We observed a significant inhibition of IL-1β secretion (Fig. 6c) that is due to a decrease in NLRC4 gene expression, as indicated by both qPCR and protein analysis (Fig. 6d). Therefore, caffeine seems to impair the priming of the NLRC4 inflammasome.
DISCUSSION
Here we show that the elevated and persistent expression of particular inflammasome gene modules in older individuals correlates with the occurrence of elevated blood pressure, arterial stiffness, chronic levels of inflammatory cytokines, metabolic dysfunction, oxidative stress and a lack of familial longevity. The inflammasome genes in these modules (62 and 78 here) are known to be a part of the response to infectious pathogens, but their role in age-related inflammation has not been studied extensively. For example, among the 23 NLR genes in humans, two were identified in modules 62 and 78. These correspond to NLRC4 (module 78) and NLRC5 (module 62). As cited previously, a role for NLRC4 expression in aging was suggested by a recent study in rats14. NLRC5, which is associated with a viral response, can also interact with NLRP3, which triggers immunosenescence and functional decline in mice11. Although the expression of the NLRP3 gene was not strongly associated with age in our human cohorts (Q = 0.202, R = 1.91; see http://cs.unc.edu/~vjojic/fluy2-upd/mod63), NLRC5 expression may promote the NLRP3 inflammasome activation at the protein level. The regulation of NLRC5 expression depends on type-I IFN signaling35, and consistently, we found type-I-IFN-dependent transcription factor genes, including IRF7/9 (see http://cs.unc.edu/~vjojic/fluy2-upd/mod62), in the regulatory program of module 62.
In addition to the reported elevation of IL-1β in patients with essential hypertension36–38, a role for inflammasome components has been suggested in two other studies: first, in a mouse model of kidney damage39, and second, in humans, where non-coding NLRP3 mutations were linked to susceptibility to hypertension40. Our data suggest that other inflammasome genes, such as NLRC4, are involved in hypertension and vascular dysfunction. Two recent independent studies identifying the existence of single-nucleotide polymorphisms (rs385076, rs17229943, rs1842893, rs2290414, rs34649619 and rs149451729) that affect both NLRC4 expression and function and that simultaneously occur with increased IL-18 (another member of the IL-1 family of cytokines) serum levels41,42 in patients with coronary artery disease corroborate the suggestions drawn from our data.
Our results also show that the elevation of the levels of the metabolites adenine and N4A correlates with the occurrence of hypertension in older adults and actively causes hypertension in mice. The ribosylation of adenine forms adenosine, which is known to sustain inflammasome activity and IL-1β secretion25. Consistently, we also found differences in adenosine and adenosine-derivatives, including N1-methyladenosine, N6-methyladenosine, N6-carbamoylthreonyladenosine, N6-succinyladenosine and 5-methylthioadenosine (all being increased in IMH compared with IML) (Supplementary Fig. 9), suggesting that these compounds may also contribute to inflammasome activity and IL-1β production in the IMH group.
We find that N4A and adenine, when applied in combination, were able to trigger inflammasome activation in vitro, inducing the THP-1 cells to secrete IL-1β and IL-18. Since N4A induces cytokine secretion in the presence of ATP, and N4A (but not adenine) increases NLRC4 gene and protein expression, it appears that N4A primes monocytes to enable NLRC4 expression (signal 1) and that adenine then triggers inflammasome activation (signal 2) and cytokine maturation. Therefore, these two metabolites can be considered danger-associated molecular pattern molecules (DAMPs) that act together to induce IL-1β expression in monocytes and neutrophils, along with enhancing platelet and neutrophil activation. That caffeine negatively correlates with NLRC4 gene and protein expression, and its metabolites are found at higher levels in the IML group, indicates that caffeine has an antagonistic effect on N4A-mediated upregulation of NLRC4 in addition to its role in antagonizing adenosine and adenine. Thus, moderate coffee consumption may suppress systemic inflammation that is caused by inflammasome activation, which may account for its correlation with decreased mortality43.
With respect to the link between angII and the higher levels of oxidative stress in IMH subjects, our results support recent findings showing that a state of oxidative stress triggers an inflammatory response to isoketal-modified self-proteins. These proteins involve the production of IL-1β, IL-6 and IL-23 by dendritic cells and IFN-γ and IL-17 by T cells, which, in turn, infiltrate the kidneys and vessels, causing sustained hypertension31. Pre-hypertensive stimuli, such as elevated angII, track with ROS production, and the role of ROS in inflammation and aging has been extensively studied, with recent data showing that mitochondrial and other sources of ROS may converge in the activation of the inflammasome NLRP3 (ref. 44). However, so far, no activation of NLRC4 has been reported in response to production of ROS. Thus, our data suggest the existence of a different mechanism that may also be ROS-dependent. Since tRNA degradation is a marker for oxidative stress (both acute and chronic)45, and the IMH subjects show clear signs of oxidative stress, we speculate that a rise in oxidative stress in some elderly individuals may result in an increase in tRNA degradation, which results in the generation of N4A and the subsequent upregulation of NLRC4 gene expression. Thus, several pathways may be involved in the inflammatory response to these nucleotide metabolites, resulting in the activation of immune cells, the occurrence of chronic inflammation, the development of hypertension, and the potential onset of other aging-associated diseases.
ONLINE METHODS
Study design, subjects and sample collection
One hundred and fourteen donors (ages 20 to >89) were enrolled in an influenza vaccine study at the Stanford–LPCH Vaccine Program during the years 2008 to 20138,15,16 (ClinicalTrials.gov registration NCT#01827462). Since baseline samples were obtained from all the individuals before vaccination with the influenza vaccine, no randomization or blinding was done for this study. The protocol for this study was approved by the Institutional Review Board of the Research Compliance Office at Stanford University. Informed consent was obtained from all subjects. All individuals were ambulatory and were generally healthy, as determined by a clinical assessment. At the time of initial enrollment, the volunteers had no acute systemic or serious concurrent illness, no history of immunodeficiency nor any known or suspected impairment of immunologic function. This definition includes the absence of clinically observed liver disease, diabetes mellitus treated with insulin, moderate to severe renal disease, blood pressure >150/95 at screening, chronic hepatitis B or C and recent or current use of immunosuppressive medication. In addition, on each annual vaccination day, none of the volunteers had been recipients or donors of blood or blood products within the past 6 months and 6 weeks respectively, and none showed any signs of febrile illness on day of baseline blood draw. Peripheral blood samples were obtained from venipuncture, and whole blood was used for gene expression analysis (below). Serum was separated by centrifugation of clotted blood, and it was stored at −80 °C before cytokine and chemokine determination. Gene expression analysis can be found in the Supplementary Methods, gene expression analysis section. Estimation of inflammasome gene module expression levels in myeloid cells and granulocytes versus other cell populations. To estimate differences in the expression levels of modules 62 and 78 across different populations, we used the geneset enrichment algorithm (ImmGen)47. Available data were analyzed across genes in each module. We performed an unpaired t-test on gene expression between the cell lines/populations. We treated all of the genes in a module as though they were drawn from the same distribution.
Determination of cytokines, chemokines and growth factors
I. Polystyrene bead kits
This assay was performed in the Human Immune Monitoring Center at Stanford University. Human 50-plex (for year 2008) or 51-plex (for yeas 2009–2011) kits were purchased from Affymetrix and used according to the manufacturer’s recommendations with the usage modifications as described below. Briefly, samples were mixed with antibody-linked polystyrene beads on 96-well filter-bottom plates and were incubated at room temperature for 2 h, which was followed by overnight incubation at 4 °C. Room temperature incubation steps were performed on an orbital shaker at 500–600 r.p.m. Plates were vacuum filtered and washed twice with a wash buffer, and then were incubated with a biotinylated detection antibody for 2 h at room temperature. Samples were then filtered and washed twice as above, and then were re-suspended in streptavidin-PE. After incubation for 40 min at room temperature, two additional vacuum washes were performed, and then the samples were re-suspended in reading buffer. Each sample was measured in duplicate. Plates were read using a Luminex 200 instrument with a lower bound of 100 beads per sample per cytokine. Custom assay control beads manufactured by Radix Biosolutions were added to all wells.
II. Magnetic bead kits
Serum specimens were collected from the human blood samples as previously described8 and were frozen in aliquots at −80 °C. Human 63-plex (for year 2013) kits were purchased from eBiosciences/Affymetrix, of which 62 analytes passed QC. They were used according to the manufacturer’s recommendations with the modifications as described below. Beads were added to a 96-well plate and washed in a Biotek ELx405 washer. Samples were added to the plate containing the mixed antibody-linked beads and were incubated at room temperature for 1 h, followed by overnight incubation at 4 °C with shaking. Both cold and room temperature incubation steps were performed on an orbital shaker at 500–600 r.p.m. Following the overnight incubation, plates were washed in a Biotek ELx405 washer and then a biotinylated detection antibody was added to the plates for 75 min at room temperature with shaking. The plate was washed as above, and streptavidin-PE was added to it. After samples were incubated for 30 min at room temperature, a wash was performed as above and the reading buffer was added to the wells. Each sample was measured in duplicate. Plates were read using a Luminex 200 instrument, with a lower bound of 50 beads per sample per cytokine. Custom assay control beads by Radix Biosolutions were added to all of the wells. Mean fluorescence intensities (MFIs) were recorded and then used for further analysis.
To identify differences between IH and IL individuals in an unbiased fashion, we first analyzed data from year 2013 since this was the year with the largest number of measured analytes (N = 62). We conducted multiple regression analysis on each analyte’s MFI against the IH/IL status, age and sex and obtained the significance for each regression coefficient via permutation tests48 over 500 resamplings. To study whether the differences in IL-1α and IL-1β observed in IH subjects when compared to IL subjects were longitudinally stable, we directly compared the levels of IL-1α and IL-1β from the data generated in the years 2008–2011 between the two groups, using a regression model with IL-1β or IL-1α MFI against IH/IL status, age and sex without multiple hypothesis correction. Combined data showed homoscedasticity by using Bartlett’s test. Cytokine data from 2012 was not included in this analysis because data from only 14 extreme phenotype individuals was available from that year. P-values for years 2008 through 2011 were combined using a modified generalized Fisher method for combining P-values obtained from dependent tests33.
Cardiovascular phenotyping
A subgroup of subjects (N = 39; 17 extreme phenotypes + 22 unclassified) from the Stanford–Ellison longitudinal cohort underwent comprehensive cardiovascular assessment at Stanford Cardiovascular Institute Biomarker and Phenotypic Core Laboratory. Vascular studies included the measurement of both carotid intima-media thickness (cIMT) and central aortic pulse wave velocity (PWV). We used a 9.0 MHz Philips linear array probe for carotid and femoral measurements. The cIMT was the average of the anterior, lateral and posterior measurements and was averaged for both the right and left carotid artery. Aortic PWV was calculated as the path length traveled divided by the transit time of the aortic pulse wave; path length (D) was measured as the distance from the sternal notch to the femoral artery minus the echocardiographic distance from the sternal notch to proximal descending aorta. This measurement reflects arterial stiffness49.
Metabolomics data generation and analysis
Metabolomic data analyses were conducted at Metabolon as described previously using non-targeted metabolomic profiling50. Serum samples from IMH (N = 11) and IML (N = 9) were subjected to methanol extraction and then were split into aliquots for analysis by ultrahigh performance liquid chromatography/mass spectrometry (UHPLC/MS) in the positive, negative or polar ion mode and by gas chromatography/mass spectrometry (GC/MS). Metabolites were identified by automated comparison of ion features to a reference library of chemical standards followed by visual inspection for quality control. For statistical analyses and data display, any missing values were assumed to be below the limits of detection; these values were imputed with the compound minimum (minimum value imputation). To determine statistical significance, significance analysis of microarrays (SAM)23 was conducted on the residuals from a multiple regression model, which included the age and sex as covariates. A Q-value <0.05 was used as an indication of high confidence in a result. A total of 67 differentially regulated metabolites were observed in IML versus IMH individuals. Pathway analysis was conducted using MetPA24, which combines several advanced pathway enrichment analyses along with the analysis of pathway topological characteristics across more than 874 metabolic pathways. For over-representation and pathway topology analyses, a hypergeometric test and the relative-betweeness centrality were used, respectively.
Determination of circulating isoketals
Sera available from a total of 17 extreme phenotypes (8 IML and 9 IMH) were used to quantify 8-isoprostane by using the Enzyme Immunoassay EIA Kit (Cayman Chemical Company) according to the manufacturer’s instructions.
Differential expression of purine and pyrimidine metabolism genes
A total of 104 pyrimidine metabolism genes (PYR) and 54 genes participating in purine metabolism (PUR) were obtained from KEGG51. Regression analysis was conducted on each gene’s expression using microarray data from year 20088,15 against IML/IMH status after adjusting for age and sex. Significance for each regression coefficient was obtained via permutation tests48. Genes that were differentially expressed were subjected to enrichment analysis by hypergeometric test. A P-value < 0.05 was used as an indication of high confidence in a result.
Compound treatment, cytokine secretion and qPCR assays
Adenosine, adenine, dl-4-hydroxy-3-methoxymandelic acid and scyllo-inositol were all purchased form Sigma (Sigma-Aldrich, St. Louis, MO) and N4-acetylcytidine was purchased from Santa Cruz Biotechnology (Dallas, TX).
Whole blood was obtained from venipuncture (30 mL), and monocytes were enriched using the RosetteSep Human Monocytes Enrichment Cocktail (cat #15068, Stemcell Technologies, Vancouver, BC, Canada) according to the manufacturer’s recommendations. Cells were plated on 96-well plates at a density of 3 × 10−5 cells in 200 µL LGM-3 serum-free media (Lonza), and plates were incubated for 6 h at 37 °C. Supernatants were collected, frozen immediately and stored at −80 °C. Samples were then transferred to the Human Immune Monitoring Core at Stanford for quantification of cytokines, chemokines and growth factors using the 63-plex Luminex system, as described above. Compounds were tested at increasing concentrations (0, 3, 10, 30 and 100 µM) and displayed significant dose-responses (P < 0.01) in the concentration of cytokines found in the supernatants (Supplementary Fig. 5). To assess significance for the dose-response experiments, we used short time-series expression miner (STEM), which uses clustering methods for time-series or dose-response experiments and allows for the identification of significant dose-dependent profiles52. The highest concentration was chosen on the basis of previous reports showing that adenosine (when used at 100 µM) can regulate inflammasome activity that is initiated by a wide range of PAMPs and DAMPs. The highest concentration (100 µM) was also used for all other compounds and stimulation assays. For the studies of caffeine, we chose to test several concentrations (0, 50 and 500 µM) that match the estimated concentrations that are found in the plasma of coffee drinkers (10–50 µM).
RNA was extracted from cell pellets using the RNeasy micro kit (Qiagen) following the manufacturer’s recommendations. cDNA was prepared using the SuperScript® VILO™ cDNA Synthesis Kit (Life Technologies). NLRC4 and NLRP3 expression was measured by quantitative PCR using pre-design TaqMan gene expression assays (Life Technologies), and plates were run on a StepOne real time PCR system (Applied Biosciences). The expression of GAPDH was used to standardize the samples, and the results are expressed as the normalized ratio of this expression relative to the control.
Hypertension studprifeies in mice
Hypertension studies in mice. Adult male C57BL/6 WT mice (aged 12–18 weeks) were randomly placed into two groups: PBS or compound treated-mice (N4-acetylcytidine + adenine dissolved in PBS). The investigator was not blinded to treatment or blood pressure measurement. For an expected value of >140 mmHg and sample average of 120 mmHg (s.d. = 10) (pre-hypertension due to short-term angII treatment in control mice), we estimated a sample size of 4 mice (statistical power = 92%, α = 0.01). Compound-treated mice were injected with N4-acetylcytidine and adenine (stock solution 20 mM each, 100 µL/25 g body weight, retro-orbital injection, once daily) or PBS. This concentration was chosen so that the in vivo concentration reaches 1 mM of circulating compound. After 3 weeks of treatment, while they continued receiving daily injections of PBS or N4A + Adenine, mice from both groups were administered an infusion of human angiotensin II (angII, #A9525, Sigma-Aldrich, St. Louis, MO) for another 2 weeks. AngII (140 ng per kg body weight per min) was dissolved in 100 µL 20 mM (N4A + Adenine) or in 100 µL PBS and loaded into a small osmotic pump (Durect Corporation, Cupertino, CA, USA). The osmotic pump was then implanted subcutaneously on the dorsal side around the neck of mice under anesthesia (2% oxygen, 2.5% isoflurane). The systolic blood pressures of the mice were measured every other day in conscious mice by use of tail-cuff plethysmography (Vistech System BP-2000, Apex, NC, USA). The experiment was repeated once with 10 mice per group. In the second experiment, we collected peripheral blood from 6/10 mice per group (treated versus control) by cardiac puncture. Aortic tissue and kidneys were obtained and fixed in 4% paraformaldehyde. Aortic and kidney sectioning and anti-CD3 staining procedures were conducted by Histo-Tec Laboratory (Hayward, CA). Infiltrating cells were investigated by counting the number of positive cells per 10× microscope field. Multiple fields were evaluated, and representative pictures were taken from 5–6 fields per condition. Mouse peripheral blood was kept in heparinized collection tubes for 2–4 h at room temperature before it was fixed using SmartTubes (Smart Tube Inc. cat # MTS1P) in a 5-tube base station (Smart Tube Inc. cat # PBS05). Cytometry by time-of-flight was used to characterize the activation status of blood cells as described below. This study was conducted in compliance with the requirements of ethical regulations for animal studies of Stanford University.
Mass cytometry experiments are described in the Supplementary Methods, mass cytometry (CyTOF) section.
Reagents and antibodies
Anti-human NLRP3 was purchased from Enzo Life Sciences (ALX-804-819); anti-human caspase-1 p10 antibody from Santa Cruz Biotechnology (sc-515); anti-human caspase-1 p20 (D7F10) antibody from Cell Signaling (#3866); anti-human IL-1β (3A6) antibody from Cell Signaling (#12242); anti-human NLRC4 from Abcam (AB115537); anti-human NLRP6 from Merck Millipore (ABF29); and anti-human AIM2 from Clinisciences (OAEB01074). Caspase-1 inhibitor Ac-YVAD-fmk was purchased from Calbiochem (caspase-1 inhibitor VI; 218746), and biotinyl-VAD-fmk from Enzo Life Sciences (ALX-260-098). LPS from E. Coli serotype 055:B5 (L6529) and ATP were purchased from SIGMA. Fluorescent dye-conjugated secondary antibodies were purchased from LI-COR Biosciences. ELISA kits to measure IFN-γ and TNF-α secretion were purchased from Mabtech (3420-1H-20 and 3510-1H-20; respectively). Validation for antibody reactivity against the relevant species and applications is provided on the manufacturer’s website.
Immunoblotting and recovery of active caspase-1
Immunoblotting was performed as described previously53. THP-1 cell lines in suspension were treated with 50 ng/mL PMA (phorbol 12-myristate 13-acetate) for 12 h at 37 °C. For recovery of active caspase-1, cell culture supernatants of adherent THP-1 cells that were treated with one of the four compounds of interest were incubated with biotinyl-VAD-fmk (30 µM) overnight at 4 °C in order to precipitate the active, cleaved fragment p10 of caspase-1. The biotinyl-VAD-fmk/caspase-1 p10 complex was recovered by using streptavidin-Sepharose beads (Sigma), adding 30 µL of 1:1 streptavidin-Sepharose suspension per 250 mL of cell supernatant overnight at 4 °C. Beads were pelleted by centrifugation (3000 rpm/10 min/4 °C) and were washed 3 times in cold IP buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 50 mM NaF, 0.3% NP-40, 0.1 mM Na3VO4) before adding SDS loading buffer on top of the beads. Beads were then heated for 5 min and pelleted. Supernatant was analyzed by SDS-PAGE/immunoblotting using an antibody that detects the p10 small subunit of processed human caspase-1 (Santa Cruz Biotechnology, sc515).
Isolation and growth of bone marrow-derived macrophages
Bone marrow cells were obtained from bones from wild type, NLRC4 and Casp1/11 knockout mice, by flushing pre-chilled Bone Marrow Media (BMM) (RPMI, 10% heat-inactivated fetal calf serum, 1% glutamine, 5% 3T3-CSF-cell conditioned media and antibiotics) through femur and tibia and by disrupting bone marrow mechanically. Cells were then gently centrifuged for 1 minute at 4 °C at 500 r.p.m. to remove debris. Supernatant was then centrifuged for 10 minutes at 4 °C at 1,000 r.p.m. and cells were resuspended in 30 ml BMM media and grown in 150 × 15 mm petri dishes. At day 4, 10 ml BMM was added to the cell cultures. At day 8, bone marrow-derived macrophages were used for stimulation assays with N4A + adenine.
Isolation of primary human neutrophils
Neutrophils were isolated from the blood samples of healthy donors by using a Ficoll gradient. The pellet was retained and suspended in 50 mL pH7 hypotonic lysis buffer (155 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA), and the lysis of red blood cells was finished on ice between 5–15 min later. Neutrophils were then isolated by centrifugation (460g, 7 min at 4 °C), were washed once in lysis buffer, were washed a second time in PBS and finally were washed in complete RPMI medium at a final concentration of 5.105 cells/ml. Then, cells were seeded in a 12-well plate and were incubated overnight at 37 °C.
Lentivirus infections
Lentiviral plasmid constructs expressing shRNAs that silence the human NLRC4 gene were purchased from Sigma (MISSION shRNAs). Recombinant lentiviruses were then generated by the “Plateforme de Vectorologie” at Bordeaux University, and THP-1 cells were transduced in a suspension with each virus at various MOIs (1, 10 and 30) in a 6-well plate. At 2 d post-infection, the infected cells were selected on 1 µg/mL puromycin and were amplified to test the silencing of gene expression by western-blots by using anti-human NLRC4 antibody.
Experiments with human THP-1 cells and primary blood platelets
Human THP-1 monocytic cell lines (ATCC, TIB202) were cultured in 6-well plates in RPMI media (supplemented with 10% Fetal Bovine Serum) and were differentiated overnight with phorbol 12-myristate 13-acetate (PMA; Sigma, cat # P1585) (10 ng/ml). The day after, adherent cells were washed with fresh media and were treated with agonists LPS (Sigma, cat # L6529; 1 ng/ml, 4 h) and ATP (Sigma, cat # 1852; 5 mM, 30 min) or the compounds Adenine (Sigma, cat # A2786)/N4-acetylcytidine (Santa Cruz, cat # sc-222023) at the indicated concentrations. After 6 h, the incubation cells were pulsed either with or without ATP. The secretion of cytokines IL-1β, IL-18, and TNF-α was then measured by ELISA from cell culture supernatants by using IL-1β and TNF-α kits (Mabtech, cat # 3415-1H-20, and cat # 3510-1H-20 respectively). IL-18 was measured using a custom-made ELISA combining anti-IL18 coating (MBL, cat # D044-3) and anti-IL18-biotin capture antibodies (MBL, cat # D045-6) and then adding streptavidin-horseradish peroxidase (Mabtech, cat # 3310-9). Cell lines were obtained from and identified by ATCC(R). Cells were tested for mycoplasma contamination.
Human primary platelets were obtained from the whole blood of healthy donors by venipuncture in EDTA tubes. After a 20 min centrifugation at 1000 r.p.m. without a break, the supernatant was harvested and 1µM prostaglandin E1 (Sigma, cat # P8908) was added to the plate. After gentle centrifugation (10 min at 2000 r.p.m. without a break), the supernatant was removed and Tyrode buffer (134 mM NaCl, 12 mM NaHCO3, 2.9 mM KCl, 0.34 mM Na2HPO4, 1 mM MgCl2, 10 mM HEPES, pH 7.4) was added to the plate. Platelets were then stimulated with thrombin (Sigma, cat # T9326; at 0.5 U/ml), ADP (Sigma, cat # A2754) or with the indicated concentrations of N4A or adenine, and platelet activation was monitored by flow cytometry using immunostaining of membrane markers with anti-CD61-PC7 (Beckman Coulter, cat # IM3716) (marker of platelet population) and anti-CD62-PE (Beckman Coulter, cat # 1759U) (marker of activation involved in aggregation).
Supplementary Material
Acknowledgments
We thank the Stanford–Ellison longitudinal cohort volunteers for their participation; Project/Regulatory/Data Manager S. Mackey; Research Nurses S. Swope, C. Walsh, S. French, B. Sullivan, S. Cathey, T. Trela and N. Mastman; Clinical Research Associates A. Goel, T. Quan, K. Span, R. Fleischmann, B. Tse, I. Chang and S. Batra. We also are grateful to The Ellison Medical Foundation for initial support and to the NIH (U19 AI090019) and the Howard Hughes Medical Institute for the remainder (M.M.D.). We also thank H. Maecker and Y. Rosenberg-Hasson (Human Immune Monitoring Core) at Stanford, and R.E. Vance and I. Rauch at the University of California, Berkeley, for kindly providing us with material from NLRC4 and caspase-1 knockout mice. B.F., J.D-M. and J.F.M. were funded by Fondation pour la Recherche Médicale (DEQ20110421287), INCa-Cancéropôle GSO, Ligue contre le Cancer de la Dordogne, and the Conseil Régional d’Aquitaine. We thank the Metabolon Inc. for the metabolite analysis.
Footnotes
Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
Gene expression data were generated at the Human Immune Monitoring Core (Stanford University). D.F. and B.F. conducted stimulation, inhibition and qPCR assays; D.F., C.R.B. and V.J. analyzed data; D.F., B.F., S.D., L.L., I.D. and P.B, designed or conducted in vitro studies of monocyte and platelet activation; J.C. and C.J.K. helped with the design and conducted the hypertension studies in mice; D.F., B.G., E.A.G., G.P.N., G.K.F. and M.H.S. designed or conducted the mass cytometry studies F.H. supervised cardiovascular phenotyping data generation; J.-F.M. and J.D.-M. helped with clinical insights and discussion; C.L.D. coordinated, organized and conducted the clinical studies and contributed to study design; G.P.N. and M.M.D. provided support, contributed with the planning of the immunological studies and contributed to study design; D.F., M.M.D. and B.F. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Howcroft TK, et al. The role of inflammation in age-related disease. Aging. 2013;5:84–93. doi: 10.18632/aging.100531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Okin D, Medzhitov R. Evolution of inflammatory diseases. Curr. Biol. 2012;22:R733–R740. doi: 10.1016/j.cub.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scrivo R, Vasile M, Bartosiewicz I, Valesini G. inflammation as “common soil” of the multifactorial diseases. Autoimmun. Rev. 2011;10:369–374. doi: 10.1016/j.autrev.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Kotas ME, Medzhitov R. Homeostasis, inflammation, and disease susceptibility. Cell. 2015;160:816–827. doi: 10.1016/j.cell.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Proctor MJ, et al. Systemic inflammation predicts all-cause mortality: a Glasgow inflammation outcome study. PLoS One. 2015;10:e0116206. doi: 10.1371/journal.pone.0116206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arai Y, et al. inflammation, but not telomere length, predicts successful ageing at extreme old age: a longitudinal study of semi-supercentenarians. EBioMedicine. 2015;2:1549–1558. doi: 10.1016/j.ebiom.2015.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen-Orr SS, et al. Defective signaling in the JAK-STAT pathway tracks with chronic inflammation and cardiovascular risk in aging humans. Cell Syst. 2016;3:374–384. doi: 10.1016/j.cels.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furman D, et al. Apoptosis and other immune biomarkers predict infuenza vaccine responsiveness. Mol. Syst. Biol. 2013;9:659. doi: 10.1038/msb.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duewell P, et al. NLRP3 infammasomes are required for atherogenesis and activated by cholesterol crystals. Nature. 2010;464:1357–1361. doi: 10.1038/nature08938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zitvogel L, Kepp O, Galluzzi L, Kroemer G. Infammasomes in carcinogenesis and anticancer immune responses. Nat. Immunol. 2012;13:343–351. doi: 10.1038/ni.2224. [DOI] [PubMed] [Google Scholar]
- 11.Youm YH, et al. Canonical Nlrp3 infammasome links systemic low-grade inflammation to functional decline in aging. Cell Metab. 2013;18:519–532. doi: 10.1016/j.cmet.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sardi F, et al. Alzheimer’s disease, autoimmunity and inflammation. The good, the bad and the ugly. Autoimmun. Rev. 2011;11:149–153. doi: 10.1016/j.autrev.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Martinon F, Burns K, Tschopp J. The infammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 14.Song F, Ma Y, Bai XY, Chen X. The expression changes of inflammasomes in the aging rat kidneys. J. Gerontol. A Biol. Sci. Med. Sci. 2016;71:747–756. doi: 10.1093/gerona/glv078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furman D, et al. Systems analysis of sex differences reveals an immunosuppressive role for testosterone in the response to influenza vaccination. Proc. Natl. Acad. Sci. USA. 2014;111:869–874. doi: 10.1073/pnas.1321060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang C, et al. Effects of aging, cytomegalovirus infection, and EBV infection on human B cell repertoires. J. Immunol. 2014;192:603–611. doi: 10.4049/jimmunol.1301384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furman D, et al. Cytomegalovirus infection enhances the immune response to influenza. Sci. Transl. Med. 2015;7:281ra43. doi: 10.1126/scitranslmed.aaa2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Segal E, et al. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat. Genet. 2003;34:166–176. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- 19.Benjamini YHY. Controlling the false Discovery rate: a practical and powerful approach to multiple testing. J Royal Statistical Society. 1995;57:289–300. [Google Scholar]
- 20.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 21.Yaari G, Bolen CR, Thakar J, Kleinstein SH. Quantitative set analysis for gene expression: a method to quantify gene set differential expression including gene-gene correlations. Nucleic Acids Res. 2013;41:e170. doi: 10.1093/nar/gkt660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nwankwo T, Yoon SS, Burt V, Gu Q. Hypertension among adults in the United States: National Health and Nutrition Examination Survey, 2011–2012. NCHS Data Brief. 2013;133:1–8. [PubMed] [Google Scholar]
- 23.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia J, Wishart DS. MetPA: a web-based metabolomics tool for pathway analysis and visualization. Bioinformatics. 2010;26:2342–2344. doi: 10.1093/bioinformatics/btq418. [DOI] [PubMed] [Google Scholar]
- 25.Ouyang X, et al. Adenosine is required for sustained infammasome activation via the AA receptor and the HIF-1α pathway. Nat. Commun. 2013;4:2909. doi: 10.1038/ncomms3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niwa T, Takeda N, Yoshizumi H. RNA metabolism in uremic patients: accumulation of modified ribonucleosides in uremic serum. Technical note. Kidney Int. 1998;53:1801–1806. doi: 10.1046/j.1523-1755.1998.00944.x. [DOI] [PubMed] [Google Scholar]
- 27.Gkaliagkousi E, Passacquale G, Douma S, Zamboulis C, Ferro A. Platelet activation in essential hypertension: implications for antiplatelet treatment. Am. J. Hypertens. 2010;23:229–236. doi: 10.1038/ajh.2009.247. [DOI] [PubMed] [Google Scholar]
- 28.Hottz ED, et al. Platelets mediate increased endothelium permeability in dengue through NLRP3-infammasome activation. Blood. 2013;122:3405–3414. doi: 10.1182/blood-2013-05-504449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minuz P, et al. Determinants of platelet activation in human essential hypertension. Hypertension. 2004;43:64–70. doi: 10.1161/01.HYP.0000105109.44620.1B. [DOI] [PubMed] [Google Scholar]
- 30.Preston RA, et al. Effects of severe hypertension on endothelial and platelet microparticles. Hypertension. 2003;41:211–217. doi: 10.1161/01.hyp.0000049760.15764.2d. [DOI] [PubMed] [Google Scholar]
- 31.Kirabo A, et al. DC isoketal-modified proteins activate T cells and promote hypertension. J. Clin. Invest. 2014;124:4642–4656. doi: 10.1172/JCI74084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nelson DE, et al. Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science. 2004;306:704–708. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- 33.Dai H, Leeder JS, Cui Y. A modified generalized Fisher method for combining probabilities from dependent tests. Front. Genet. 2014;5:32. doi: 10.3389/fgene.2014.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lelo A, Miners JO, Robson R, Birkett DJ. Assessment of caffeine exposure: caffeine content of beverages, caffeine intake, and plasma concentrations of methylxanthines. Clin. Pharmacol. Ther. 1986;39:54–59. doi: 10.1038/clpt.1986.10. [DOI] [PubMed] [Google Scholar]
- 35.Staehli F, et al. NLRC5 deficiency selectively impairs MHC class I- dependent lymphocyte killing by cytotoxic T cells. J. Immunol. 2012;188:3820–3828. doi: 10.4049/jimmunol.1102671. [DOI] [PubMed] [Google Scholar]
- 36.Dörffel Y, et al. Preactivated peripheral blood monocytes in patients with essential hypertension. Hypertension. 1999;34:113–117. doi: 10.1161/01.hyp.34.1.113. [DOI] [PubMed] [Google Scholar]
- 37.Fearon WF, Fearon DT. inflammation and cardiovascular disease: role of the interleukin-1 receptor antagonist. Circulation. 2008;117:2577–2579. doi: 10.1161/CIRCULATIONAHA.108.772491. [DOI] [PubMed] [Google Scholar]
- 38.Lamkanf M, Dixit VM. inflammasomes and their roles in health and disease. Annu. Rev. Cell Dev. Biol. 2012;28:137–161. doi: 10.1146/annurev-cellbio-101011-155745. [DOI] [PubMed] [Google Scholar]
- 39.Zhang C, et al. Activation of Nod-like receptor protein 3 inflammasomes turns on podocyte injury and glomerular sclerosis in hyperhomocysteinemia. Hypertension. 2012;60:154–162. doi: 10.1161/HYPERTENSIONAHA.111.189688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Omi T, et al. An intronic variable number of tandem repeat polymorphisms of the cold-induced autoinflammatory syndrome 1 (CIAS1) gene modifies gene expression and is associated with essential hypertension. Eur. J. Hum. Genet. 2006;14:1295–1305. doi: 10.1038/sj.ejhg.5201698. [DOI] [PubMed] [Google Scholar]
- 41.Johansson Å, et al. NLRC4 infammasome is an important regulator of Interleukin-18 levels in patients with acute coronary syndromes: genome-wide association study in the PLATelet inhibition and patient outcomes trial (PLATO) Circ Cardiovasc Genet. 2015;8:498–506. doi: 10.1161/CIRCGENETICS.114.000724. [DOI] [PubMed] [Google Scholar]
- 42.Zeller T, et al. Molecular characterization of the NLRC4 expression in relation to Interleukin-18 levels. Circ Cardiovasc Genet. 2015;8:717–726. doi: 10.1161/CIRCGENETICS.115.001079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crippa A, Discacciati A, Larsson SC, Wolk A, Orsini N. Coffee consumption and mortality from all causes, cardiovascular disease, and cancer: a dose-response meta-analysis. Am. J. Epidemiol. 2014;180:763–775. doi: 10.1093/aje/kwu194. [DOI] [PubMed] [Google Scholar]
- 44.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 infammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 45.Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:2095–2103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rauch I, et al. NAIP proteins are required for cytosolic detection of specific bacterial ligands in vivo. J. Exp. Med. 2016;213:657–665. doi: 10.1084/jem.20151809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heng TS, Painter MW. The immunological genome project: networks of gene expression in immune cells. Nat. Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 48.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Statistical Society, Series B. 1995;B-57:289–300. [Google Scholar]
- 49.Reference Values for Arterial Stiffness’ Collaboration. Determinants of pulse wave velocity in healthy people and in the presence of cardiovascular risk factors: ‘establishing normal and reference values’. Eur. Heart J. 2010;31:2338–2350. doi: 10.1093/eurheartj/ehq165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shin SY, et al. An atlas of genetic influences on human blood metabolites. Nat. Genet. 2014;46:543–550. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kanehisa M, et al. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ernst J, Bar-Joseph Z. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics. 2006;7:191. doi: 10.1186/1471-2105-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Faustin B, et al. Reconstituted NALP1 infammasome reveals two-step mechanism of caspase-1 activation. Mol. Cell. 2007;25:713–724. doi: 10.1016/j.molcel.2007.01.032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.