Summary
Joint models for longitudinal and survival data are routinely used in clinical trials or other studies to assess a treatment effect while accounting for longitudinal measures such as patient-reported outcomes (PROs). In the Bayesian framework, the deviance information criterion (DIC) and the logarithm of the pseudo marginal likelihood (LPML) are two well-known Bayesian criteria for comparing joint models. However, these criteria do not provide separate assessments of each component of the joint model. In this paper, we develop a novel decomposition of DIC and LPML to assess the fit of the longitudinal and survival components of the joint model, separately. Based on this decomposition, we then propose new Bayesian model assessment criteria, namely, ΔDIC and ΔLPML, to determine the importance and contribution of the longitudinal (survival) data to the model fit of the survival (longitudinal) data. Moreover, we develop an efficient Monte Carlo method for computing the Conditional Predictive Ordinate (CPO) statistics in the joint modeling setting. A simulation study is conducted to examine the empirical performance of the proposed criteria and the proposed methodology is further applied to a case study in mesothelioma.
Keywords: CPO, DIC, LPML, Monte Carlo method, Patient-reported outcome (PRO)
1 Introduction
Recently, joint modeling of longitudinal and time-to-event outcomes has become more popular in the analysis of patient-reported outcomes (PROs) for the purpose of evaluating the efficacy and tolerability of cancer treatment. In oncology applications, information from the patients’ perspectives can be useful in evaluating actual patients’ experiences on dimensions known to be important to them and also associated with treatment outcomes. The field of PROs has evolved and reached a common understanding about good clinical practices for the use of PROs (Rothman et al., 2009). In addition, the U.S. and European regulators have published guidance on the use of these measures to support PRO-based claims in pharmaceutical product labeling (European Medicines Agency, 2005; US Food and Drug Administration Guidance for Industry, 2009) (DeMuro et al., 2013). Siddiqui et al. (2014) reviewed and addressed issues regarding the “why, how, and what” of PROs as well as cancer survivorship because it closely relates to PROs. Building on previous joint modeling work in a highly symptomatic and particularly fatal cancer (Wang et al., 2012; Hatfield et al., 2011, 2012; and Zhang et al., 2014, 2015a), we develop new Bayesian methodology on how to evaluate the distinct effects of longitudinal and time-to-event outcomes on the fit of a joint model.
A popular approach in joint modeling of longitudinal and survival data is based on shared random effects, where the longitudinal component and the survival component of the joint model share common random effects and these random effects then induce correlation between the longitudinal and survival data. There are two basic formulations of the joint model. The first is the “trajectory model” (TM), where one substitutes the time trajectory function from the longitudinal component into the hazard function of the survival component, and in this case, the trajectory function acts like a time-varying covariate in the survival component. The second formulation is the shared parameter model (SPM), which directly includes the random effects as covariates in the survival component. One of the main advantages of the TM is that it leads to a straightforward interpretation of the association between the longitudinal measure and survival time through the direct inclusion of the trajectory function in the hazard. For the SPM, the characterization of the association is much more complex and can only be analytically determined once the random effects have been integrated out, as the two components of the model are independent conditional on these random effects. However, the TM is computationally more expensive compared to the SPM. In addition, the TM requires extrapolation beyond the last time at which the longitudinal measure is observed in the survival component. The SPM typically fits the survival component of the joint model better as it directly includes the random effects as covariates in the survival component. There is a very rich literature concerning these two basic approaches. The TM has been considered in Schluchter (1992), Hogan and Laird (1997), Law et al. (2002), Brown and Ibrahim (2003), Chen et al. (2004) Ibrahim et al. (2004), Brown et al. (2005), Chi and Ibrahim (2006), Chi and Ibrahim (2007), and Ibrahim et al. (2010) for joint modeling with biomedical applications. There has also been much work on using the SPM, including Pawitan and Self (1993), DeGruttola and Tu (1994), Lavalley and DeGruttola (1996), Henderson et al. (2000), Xu and Zeger (2001a, 2001b), and Song et al. (2002) for univariate or multivariate longitudinal data. An excellent review on joint modeling of longitudinal and survival data is given in Tsiatis and Davidian (2004) and an overview of joint models for longitudinal and time-to-event data can be found in Ibrahim, Chen, and Sinha (2001, Chapter 7) and Rizopoulos (2012a). There are several R packages available in fitting joint models, including JM (Rizopoulos, 2012b), JMbayes (Rizopoulos, 2014), and joineR (Philipson et al., 2012). There is also a Stata module stjm (Crowther, 2012; Crowther et al., 2013), which fits shared random effects models. In addition, another R package, lcmm (Proust-Lima et al., 2014), fits joint models based on shared latent classes.
One important issue in the joint modeling of longitudinal and survival data concerns the separate contribution of the model components to the overall goodness-of-fit of the joint model. Zhang et al. (2014) developed a decomposition of AIC and BIC to assess the fit of each component of the joint model. A SAS macro, called JMFit, (Zhang et al., 2015b) implements a variety of popular joint models and provides several model assessment measures including the decomposition of AIC and BIC as well as ΔAIC and ΔBIC. Within the Bayesian framework, Hanson et al. (2011) proposed to use LPML to predict survival times conditional on the longitudinal component of the model. In this paper, we derive a novel decomposition of the DIC and LPML criteria into additive components that will allow us to assess the goodness of fit for each component of the joint model. Such a development is extremely important since it not only allows us to quantify the contribution of the longitudinal data to the fit of the survival data or the contribution of the survival data to the fit of the longitudinal data, but it also allows us to identify which PROs are most highly associated with survival outcomes, a finding with significant clinical implications. In addition, we also develop a new Monte Carlo (MC) method for computing the CPO statistics which may involve intractable high-dimensional integrals. The proposed MC approach for computing the CPO has a potential to lead to a gain in computing time compared to a numerical approximation approach, particularly in the joint modeling setting. To illustrate our proposed method, we only consider (i) polynomial trajectories and independent and identically distributed Gaussian noise for longitudinal measures and (ii) the Cox model with a piecewise constant baseline hazard function for survival data in our simulation study and real data analysis. However, the proposed method can be applied to other types of longitudinal trajectories and other types of survival models such as those considered in Hanson et al. (2011).
The rest of the paper is organized as follows. Section 2 presents the joint models and the likelihood and posterior. The first decomposition (Decomposition I) of DIC and LPML (i.e., DIC= DICLong + DICSurv|Long and LPML= LPMLLong+LPMLSurv|Long), the corresponding two new criteria (i.e., ΔDICSurv and ΔLPMLSurv), as well as a new Monte Carlo method for computing CPO are also developed in Section 2. A simulation study is conducted in Section 3, and a comprehensive analysis of the longitudinal and survival data from a cancer clinical trial is carried out in Section 4. We conclude the paper with a brief discussion in Section 5. Prior specification and posterior computation are discussed in Appendix A of the supplementary material. In addition, we develop the second decomposition (Decomposition II) of DIC and LPML (i.e., DIC= DICSurv + DICLong|Surv and LPML= LPMLSurv + LPMLLong|Surv) and the corresponding ΔDICLong and ΔLPMLLong criteria to assess the fit of the longitudinal data using the information from the survival data in Appendix B of the supplementary material.
2 Bayesian Assessment of Model Fit in the Joint Model
2.1 The Joint Models
Suppose that there are n subjects. For the ith subject, let yi(t) denote the longitudinal measure, which is observed at time t ∈ {ai1, ai2, . . . , aimi}, where 0 ≤ ai1 < ai2 < · · · < aimi and mi > 1. Note that yi(0) corresponds to the baseline value. Also let xi and zi denote two vectors of covariates, which may include the treatment indicator. We assume a mixed effects regression model for yi(t) given by
| (2.1) |
where g(aij) denotes a (q+1)-dimensional vector of functions of aij for j = 1, . . . , mi, θi denotes a (q+1)-dimensional vector of random effects, and γ denotes a vector of regression coefficients. In (2.1), we further assume θi ~ N(θ, Ω), where θ is a (q+1)-dimensional vector of overall effects, Ω is a (q+1) × (q+1) positive definite covariance matrix, εi(aij) is the measurement error term, which is assumed to follow a N(0, σ2) distribution and is independent of θi. We note that in (2.1), if q = 1, g(aij) = (1, aij)′ and (g(aij))′ θi represents a linear trajectory; if q = 2, and (g(aij))′ θi leads to a quadratic trajectory; and if g(aij) = (1, B1(aij), . . . , Bq(aij))′, where {Bk(·), k = 1, 2, . . . , q} is a q-dimensional basis for spline functions over a finite interval, (g(aij))′θi represents a spline trajectory considered in Brown et al. (2005).
Let ti and δi denote the failure time and the censoring indicator, respectively, where δi = 1 if ti is a failure time and 0 if ti is right-censored for the ith subject. The hazard function for failure time ti is assumed to have the form
| (2.2) |
where λ0(t) is the baseline hazard function, h(·) is a linear function of g(t) and θi with α being a vector of regression coefficients. Note that λ0, α, and β are the fixed effects parameters pertaining to the survival component of the joint model. When h(α, θi, g(t)) = {g(t)′θi}α, where α is a scalar, the hazard function (2.2) leads to the TM. When h does not depend on g(t), that is, , where α is a (q + 1)-dimensional vector, the hazard function specified by (2.2) defines the SPM. Under the TM, g(t)′θi acts a time-varying covariate in the survival component while under the SPM, the random effects θi are included as q + 1 covariates in the survival component.
2.2 The Likelihood and Posterior
We first introduce some notation. We rewrite (2.1) as
where yi = (yi(ai1), . . . , yi(aimi))′, , and εi = (εi(ai1), . . . , εi(aimi))′ ~ N(0, σ2Imi). Then, the probability density function (pdf) of yi conditional on θi is given by
and the pdf of θi is given by
for i = 1, . . . , n. Letting λ be a vector of parameters for the baseline hazard function λ0(t), we write
where λ(t|λ0, α,β,θi, g(t), zi) is defined in (2.2). We note that when δi = 1, f(ti|λ, α, β, θi, δi = 1, zi) reduces to the density of ti, and when δi = 0, f(ti|λ, α, β, θi,δi = 0, zi) is the survival function evaluated at ti.
Let φ = (θ, γ, σ2, Ω, λ, α, β). The joint distribution of (yi, ti, θi) is written as
| (2.3) |
and the marginal joint distribution of (yi, ti) is given by
| (2.4) |
for i = 1, . . . , n. Letting Dobs = {(yi, ti, θi, xi, zi), i = 1, . . . , n} denote the observed data, the observed-data likelihood is given by
| (2.5) |
Using (2.5), the joint posterior of φ takes the form
| (2.6) |
where π(φ) is the joint prior, which is specified in Appendix A, and the normalizing constant is given by
| (2.7) |
We write , which is the vector of all the random effects. Then, the augmented posterior distribution of (φ, θR) is given by
| (2.8) |
where f(yi, ti, θi|φ, δi, xi, zi) is defined in (2.3). It is easy to see that ∫ π(φ, θR|Dobs)dθR = π(φ|Dobs). The implementation details of the Gibbs sampling algorithm to sample (φ, θR) from (2.8) are given in Appendix A.
2.3 Deviance Information Criterion
The Deviance Information Criterion (DIC) (Spiegelhalter et al., 2002) for the joint model is defined as
| (2.9) |
where Dev(φ) is the deviance function, is the effective number of model parameters, and and are the posterior means of φ and Dev(φ), respectively, with respect to the posterior distribution in (2.6). To assess the overall fit of the joint model, we specify the deviance function as
where L(φ|Dobs) is given by (2.5). From (2.5), we see that Dev(φ) involves the computation of n integrals as shown in (2.4).
The integration over the random effects specified in (2.4) always poses a major challenge in computing the observed-data likelihood of the joint model. One possible approach is to use a Monte Carlo (MC) approach, but this may be computationally intensive. Adaptive Gaussian quadrature (AGQ) (Pinheiro and Bates, 1995) is another approach to approximate (2.4), and is implemented here to calculate DIC when the dimension of θi is low.
2.3.1 DIC Decomposition
To assess the contribution of the longitudinal data to the fit of the survival data, we develop a novel decomposition of DIC in (2.9). Specifically, we decompose DIC into two parts: one part for the longitudinal data and the other part for the survival data conditional on the longitudinal data. Write φ1 = (θ, γ, σ2,Ω) and φ2 = (λ, α, β). Let f(θi|φ1, yi, xi) be the conditional density of the random effects θi given yi, and also let f(yi|φ1, xi) = ∫ f(yi| γ, σ2, θi, xi)f(θi|θ, Ω)dθi, which is the marginal density of yi. Let and denote the posterior means of φ1 and φ2. Define , , , and . We are led to the following result.
Result 1
DIC and pD in (2.9) have the following decomposition:
| (2.10) |
where , and .
In (2.10), DICLong measures the contribution of the longitudinal data to the total DIC while DICSurv|Long quantifies the contribution to the total DIC due to the survival data given the additional information from the longitudinal data.
The marginal distribution of yi follows
and the conditional distribution of the random effects θi given the longitudinal data takes the form
where . These are the quantities needed to apply Result 1.
2.3.2 ΔDICSurv
When we fit the survival data alone, i.e., α = 0 in (2.2), the hazard function reduces to and the density for ti becomes Write DSurv, obs = {(ti, δi, zi), i = 1,...,n} and let
where , and . We now propose the following model assessment criterion:
| (2.11) |
In (2.11), ΔDICSurv measures the gain of the fit in the survival component due to the longitudinal data with a penalty for the additional parameters in the survival component of the joint model. A model with a large value of ΔDICSurv is more preferred. When , then ΔDICSurv < 0. That is, when the penalty for the additional parameters in the survival component outweighs the gain of the fit in the survival component, ΔDICSurv can be negative.
2.4 Conditional Predictive Ordinate
2.4.1 CPO Computation
Let denote the observed data with the ith subject deleted. The Conditional Predictive Ordinate (CPO) (e.g., Geisser and Eddy, 1979; Gelfand et al., 1992; and Gelfand and Dey, 1994) for the ith subject is defined as
| (2.12) |
where
| (2.13) |
and is the normalizing constant, i.e., . Following Chen et al. (2000), we obtain the first CPO identity.
CPO Identity I
CPOi in (2.12) can be rewritten as
| (2.14) |
The proof of this identity directly follows from Chapter 10 of Chen et al. (2000). CPO Identity I leads to the development of a popular Monte Carlo estimate of CPO using Gibbs samples from the posterior distribution given Dobs instead of . Letting {φb, b = 1, . . . , B} denote a Gibbs sample of φ from π(φ|Dobs) and using (2.14), a Monte Carlo estimate of is given by
| (2.15) |
The numerical approximation of in (2.15) involves the integral over the random effects and can be calculated using AGQ to approximate (2.4). However, this method would likely be computationally intensive when the dimension of the random effects is high. To circumvent this numerical integration issue in (2.15), we develop a second CPO identity and then propose a new efficient MC method which directly uses the Gibbs samples generated from the augmented posterior distribution π(φ, θR|Dobs) in (2.8) to calculate .
CPO Identity II
Let wi(θi) be a normalized weight function such that ∫ wi(θi)dθi = 1. Then, CPOi in (2.12) can be expressed as
| (2.16) |
Now, let denote a Gibbs sample of (φ, θR) from π(φ, θR|Dobs). Using the CPO Identity II in (2.16), a Monte Carlo estimate of is given by
Under certain ergodic conditions, is unbiased and consistent for any normalized weight function wi. However, the Monte Carlo error of depends on the choice of wi. The following theorem characterizes the optimal choice of wi in minimizing the variance of the Monte Carlo estimator when is a sample from π(φ, θR|Dobs).
Theorem 1
Let
Then, for any normalized weight function wi, we have
where the variance is taken with respect to the posterior distribution π(φ, θR|Dobs).
Remark 1
The result established in Theorem 1 provides the best choice of wi. However, this optimal weight function is expensive to compute. Since the optimal weight function wi,opt is analogous to the optimal weight function in the importance-weighted marginal density estimation (IWMDE) of the marginal posterior density proposed by Chen (1994), we may follow the guidelines discussed in Geweke (1989) and Chen (1994) to construct a good weight function wi which is similar to wi,opt. One possible choice of wi is a multivariate normal density, which is constructed via the Laplace approximation to the joint density f(yi, ti, θi|φ, δi, xi, zi) in (2.3). Another possible choice of wi is wi,cond(θi) = f(θi|φ1, yi, xi), which is the conditional density of the random effects θi given yi. Note that when yi and ti are independent, wi,cond(θi) = wi,opt. Therefore, wi,cond(θi) may be a reasonable choice for computing the CPOi.
2.4.2 CPO Decomposition
In this subsection, we first establish the third CPO identity which will lead to the decomposition of CPO.
CPO Identity III
The CPO in (2.12) can also be expressed as
| (2.17) |
which is true for all φ.
Since plugging in any numerical value for φ in (2.17) results in the CPO, we have
| (2.18) |
where φ* is a fixed value of φ, which may be chosen as the posterior mean. We note that (2.18) is similar to the identity of Chib (1995). Let and denote the posterior means of φ1 and φ2. From (2.4) and (C.1), we have
where . We also observe that
| (2.19) |
Using (2.18) and the facts of the joint densities stated above, we propose the CPO decomposition:
| (2.20) |
where
| (2.21) |
and
| (2.22) |
Remark 2
Let DLong,obs = {(yi, xi), i = 1, . . . , n} denote the observed longitudinal data and DSurv,obs = {(ti, δi, zi), i = 1, . . . , n} denote the survival data, respectively. Also let and denote the observed longitudinal and survival data with the ith subject deleted, respectively. Assume that DLong,obs and DSurv,obs are independent and π(φ1, φ2) = π(φ1)π(φ2). Under these assumptions, we have
| (2.23) |
| (2.24) |
and
where
and f0(tj|φ2, δj, zj) is defined in Section 2.3.2 with φ2 = (λ α = 0, β). Therefore, CPOi,Long and CPOi,Surv|Long reduce to the usual CPOs for the longitudinal data and the survival data separately, and the CPO decomposition (2.20) holds under the usual definition of CPO.
Next, we develop useful in the following theorem for , CPOi,Long, and CPOi,Surv|Long, which facilitate the computation and further understanding of these quantities.
Theorem 2
For CPOi, CPOi,Long, and CPOi,Surv|Long, we have the following identities:
| (2.25) |
and
| (2.26) |
Remark 3
The identity in (2.26) is quite attractive as it has a similar form as the usual CPOi in (2.14). We also see from (2.26) that CPOi,Surv|Long is free of . In addition, CPOi,Surv|Long can be directly calculated from (2.26). Thus, if only CPOi,Surv|Long is of interest, it is not necessary to compute the overall CPOi. However, it does not appear possible that CPOi,Long can be computed directly without knowing CPOi and CPOi,Surv|Long.
Remark 4
To avoid the calculation of , we use the same idea as in (2.16) and obtain
where the optimal choice of wi(θi) is . Similarly,
where the optimal choice of wi(θi) is .
2.4.3 LPML and LPML Decomposition
The logarithm of the Pseudo marginal likelihood (LPML) (Ibrahim et al., 2001) is defined as
We note that there is a relationship between the DIC and the LPML in large samples (see Draper and Krnjajić (2005, Section 4)). Using the decomposition of CPO in (2.20), we are led to the following result.
Result 2
LPML can be decomposed as
where , and CPOi,Long and CPOi,Surv|Long are given by (2.21) and (2.22), respectively.
2.4.4 ΔLPMLSurv
Define , where CPOi,Surv0 is given by (2.24). We propose the model assessment criterion
ΔLPMLSurv quantifies the gain of the fit in the survival component due to the longitudinal data with a penalty for the additional parameters in the survival component of the joint model. A model with a large value of ΔLPMLSurv is more preferred. From Remark 3, it is easy to see that if our interest is on ΔLPMLSurv only, we do not need to compute the overall LPML for the joint model. Similar to ΔDICSurv, it is not guaranteed that ΔLPMLSurv is non-negative.
3 A Simulation Study
We conduct a simulation study to evaluate the empirical performance of ΔDICSurv and ΔLPMLSurv in selecting the true model or identifying the true longitudinal data. We generate longitudinal and survival data under the SPM with the simple exponential baseline. Specifically, we first simulate θi = (θ0i, θ1i)′ N(θ, Ω), where θ = (θ0, θ1)′ = (0.1, 0.5)′ and . We then simulate the longitudinal data from a N(μi(aij), σ2) distribution with a linear trajectory μi(aij) = θ0i + aij θ1i + xiγ. For the survival data, we set zi = xi and generate t* from an exponential regression model, i.e., , where U ~ U(0, 1), and draw the censoring times Ci from an exponential distribution with mean 10. Then, we compute and δi = 1 if and 0 otherwise. The treatment indicator xi is generated from a Bernoulli(0.5) distribution. For each subject, 6 or 7 time points (aij, j = 1, . . . , 6 or 7) for the longitudinal measures are chosen to be (0 + ζi1, 21 + ζi2, 42 + ζi3, 63 + ζi4, 84 + ζi5, 105 + ζi6)/30.4375 if ζi7 > 0 and (0 + ζi1, 21 + ζi2, 42 + ζi3, 63 + ζi4, 84 + ζi5, 105 + ζi6, 126 + ζi7)/30.4375 if ζi7 ≤ 0, where ζij ~ U(−3, 3) for j = 1, . . . , 7, and 30.4375 = 365.25/12. The design values of the parameters are given as Ω00 = 0.7, Ω10 = Ω01 = −0.1, Ω11 = 0.06, δ2 = 0.3, θ0 = 0.1, θ1 = 0.5, γ = −0.2, α1 = 0.3, α2 = 1.6, β= −0.4, and λ = 0.08. 500 datasets are simulated independently with n = 400 subjects in each simulated dataset. The resulting censoring percentage is about 40%.
Let DT denote the dataset generated from the true SPM model. One additional set of longitudinal data is generated by adding noise to the true longitudinal measures. More specifically, it is simulated from a N(μℓi(aij), δ2) distribution with linear trajectories μℓi(aij) = (θ0i + τℓ0i) + aij(θ1i + τℓ1i) + xiγ, where (τℓ0i, τℓ1i)′ ~ N(0, 0.22I2), and the values of the other parameters remain the same as before. By combining this longitudinal dataset with the same survival data in DT, we obtain the additional dataset and denote it as DW.
We consider the following scenarios to fit different joint models to the datasets DT and DW:
Long: Fit the joint model with (3.1) and (3.2) to DW. In this case, DW is fit by the joint model with misspecified longitudinal submodel.
SurvI: Fit the joint model with (3.1) and misspecified survival submodel to DT. In this joint model, (3.2) reduces to λexp{θ0i α1 + zi β}.
SurvII: Fit the joint model with (3.1) and misspecified survival submodel to DT. In this joint model, (3.2) reduces to λexp{θ1i α2 + zi β}.
TM: Fit the joint model with (3.1) and misspecified survival submodel to DT. In this joint model, (3.2) becomes λexp{(θ0i + θ1it)α + zi β}.
Long&Surv: Fit the joint model with misspecified longitudinal and survival submodels to DT. In this joint model, (3.1) becomes yi(aij) = θ0i + xiγ + εi(aij), and (3.2) reduces to γexp{θ0i α1 + zi β}.
In all the six scenarios, the exponential regression model, namely, λexp{zi β}, fits the true survival data DT in computing DICSurv, 0 and LPMLSurv, 0. Thus, the values of DICSurv, 0 and LPMLSurv, 0 are the same for all of the six scenarios. Since ΔDICSurv = DICSurv, 0 − DICSurv|Long and ΔLPMLSurv = LPMLSurv|Long − LPMLSurv, 0, ΔDICSurv and ΔLPMLSurv can be used to assess the fit of the survival component of the joint model for all of the six scenarios. We also note that in scenario (ii), both components of the joint model are correctly specified but fit to the longitudinal data, which are less correlated to the survival data; in scenarios (iii), (iv), and (v), the longitudinal component is correctly specified, the survival component is misspecified, and both components fit the true longitudinal and survival data; and in scenario (vi), both components of the joint model are misspecified but fit the true data.
For each simulated dataset, we take 5000 Gibbs samples with 100 burn-in iterations. The means of ΔDICSurv and ΔLPMLSurv as well as the frequencies of ranking each model as best based on ΔDICSurv and ΔLPMLSurv are reported in Table 1. From this table, we see that True has the largest means of ΔDICSurv and ΔLPMLSurv, which are 18.72 and 9.37, and gets ranked as the best with 423 times out of 500 by both criteria, while SurvI has the smallest means of ΔDICSurv and ΔLPMLSurv and never gets ranked as the best by these two criteria in these 500 simulated datasets. These results show that both ΔDICSurv and ΔLPMLSurv can correctly identify the true model or the true data.
Table 1.
Means of ΔDICSurv and ΔLPMLSurv and frequencies of ranking each model as best based on ΔDICSurv and ΔLPMLSurv
| ΔDICSurv |
ΔLPMLSurv |
|||
|---|---|---|---|---|
| Data | Mean | Frequency | Mean | Frequency |
| True | 18.72 | 423 | 9.37 | 423 |
| Long | 10.67 | 27 | 5.35 | 28 |
| SurvI | 1.31 | 0 | 0.63 | 0 |
| SurvII | 10.54 | 19 | 5.23 | 22 |
| TM | 4.09 | 8 | 1.92 | 6 |
| Long&Surv | 10.59 | 23 | 5.30 | 21 |
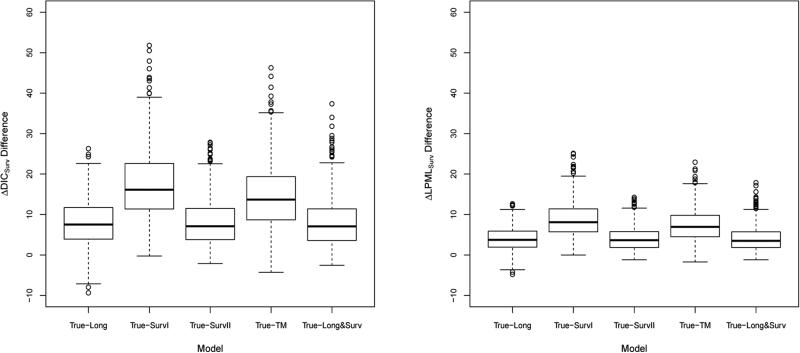
Figure 1 shows boxplots of the ΔDICSurv and ΔLPMLSurv differences between True and each of Long, SurvI, SurvII, TM, and Long&Surv. We see from this figure that boxplots for ΔDICSurv and ΔLPMLSurv differences are almost above zero, indicating that the true model does fit the true data much better than other models based on either ΔDICSurv or ΔLPMLSurv. These results are consistent with those based on the means of ΔDICSurv and ΔLPMLSurv and the frequencies of ranking each model as best as shown in Table 1.
Figure 1.
Boxplots of the ΔDICSurv differences and the ΔLPMLSurv differences between True and each of Long, SurvI, SurvII, TM, and Long&Surv.
4 Analysis of the EMPHACIS Data
We consider a dataset from a multicenter, randomized, single-blind, EMPHACIS lung cancer clinical trial (Evaluation of MTA in Mesothelioma in a Phase III Study with Cisplatin) (Vogelzang et al., 2003). The study drug was multi-targeted antifolate (MTA) pemetrexed given in combination with cisplatin (the PEM/Cis arm), and the active-treatment comparator was cisplatin alone (the Cis arm). The treatment for both arms was structured as six 21-day cycles of therapy; patients receiving treatment benefit could receive additional cycles based on investigator discretion. Malignant pleural mesothelioma is characterized by rapid disease progression, high symptom burden, and a relatively short median survival of 12 months after diagnosis (Thompson et al., 2014). Accordingly, patient-reported assessments are important for evaluation of disease progression and patients’ response to therapy. In oncology, the patients’ importance ratings on the magnitude of progression-free survival improvement has been shown to depend on the severity of disease-related symptoms (Bridges et al., 2012). We analyzed the disease-specific patient-reported Lung Cancer Symptom Scales (LCSS) (Patricia et al., 2006) to evaluate the patient-level association of five of the six instrument items (i.e., anorexia, cough, dyspnea, fatigue, and pain) with progression-free survival using the EMPHACIS trial data. Progression free survival time (PFS) is defined as the time from randomization to the time until documented progression or death from any cause. We are interested in the association between post-baseline LCSS measurements and PFS. The main goal of applying joint models in this study is to assess the association of each longitudinal LCSS symptom with PFS and the treatment effects on each LCSS item and PFS simultaneously. More importantly, with the decomposition of DIC and LPML, the longitudinal LCSS symptoms can be compared in terms of their contribution to the fit of the PFS data as well as the gain in the fit of the longitudinal data for each LCSS symptom using the information from the PFS data can be determined.
Our study cohort consists of 425 patients with at least one post-baseline value of each longitudinal measure and seven binary covariates, including race (xi1 = 1 if white), gender (xi2 = 1 if male), age (xi3 = 1 if age ≥ 65), Karnofsky status (xi4 = 1 if Karnofsky status is high), baseline stage of disease (xi5 = 1 if stage I/II), vitamin supplementation (xi6 = 1 if full vitamin supplementation), and treatment assignment (xi7 = 1 if the ith patient is in the pemetrexed/cisplatin arm). Among the 425 patients, 394 patients experienced disease progression. Among these 394 patients, there were only 129 distinct disease progression times. In all the computations, we used zi = xi and standardized these five LCSS measures, where the means and standard deviations were 30.79 and 27.19, 11.48 and 17.93, 31.41 and 26.33, 39.38 and 27.06, and 24.64 and 24.90 for anorexia, cough, dyspnea, fatigue, and pain, respectively. The total numbers of longitudinal measures (i.e., ) including the baseline measures were 5504, 5544, 5553, 5530, and 5546 for anorexia, cough, dyspnea, fatigue, and pain.
In (2.2), we assume a piecewise constant hazard function for λ0(t) defined as
| (4.1) |
where 0 = s0 < s1 < s2 < . . . < sK−1 < sK = ∞ is a finite partition of the time axis. The sk's in (4.1) were constructed based on the percentiles such as the first (Q1), second (Q2), and third (Q3) quartiles of the PFS times. Let Danorexia, Dcough, Ddyspnea, Dfatigue, and Dpain denote the five observed LCSS longitudinal datasets and also let DSurv denote the observed PFS data. We fit the shared parameter model and the trajectory model with a linear trajectory, denoted by SPML and TML, respectively, to each pair of the PFS data and one of the five LCSS longitudinal outcomes corresponding to anorexia, cough, dyspnea, fatigue, and pain, namely, Danorexia + DSurv, Dcough + DSurv, Ddyspnea + DSurv, Dfatigue + DSurv, and Dpain + DSurv. The prior π(g=f) in (2.6) is specified in Appendix A of the supplementary material. For TML, we specify a N(0, 10000) prior distribution for α.
To construct the partition {sk, k = 0, 1, . . . , K}, we adopt the left bi-sectional quantile partition (LBSQP) method proposed in Zhang et al. (2015b). We use DICSurv, 0 and LPMLSurv, 0 to determine the number of intervals (K) in (4.1). We start with a large value of K, which is close to the number of distinct PFS times, and work down to a smaller value of K. For the EMPHACIS data, K = 100 should be sufficiently large given that there were only 129 distinct PFS times. We determine an “optimal” value of K according to DICSurv, 0 and LPMLSurv, 0 by fitting the PFS data alone. Table S1 of the supplementary material shows the results for various values of K. From Table S1, we see that the respective values of DICSurv, 0 and LPMLSurv, 0 were 2070.61 and −1070.94 for K = 100; 2022.56 and −1012.62 for K = 35; 2018.49 and −1010.07 for K = 30; 2026.85 and −1014.27 for K = 25; and 2206.05 and −1103.10 for K = 2. Thus, the piecewise constant baseline hazard function with K = 30 fit the PFS data alone best according to both DICSurv, 0 and LPMLSurv, 0. We then fit each of the LCSS longitudinal and PFS data, Danorexia + DSurv, Dcough + DSurv, Ddyspnea + DSurv, Dfatigue + DSurv, and Dpain + DSurv, with the “best” value of K = 30 in fitting the PFS data alone along with K = 25 and K = 35. We used the Laplace approximation to construct a multivariate normal density for wi in computing LPML (MC), LPMLSurv|Long (MC), and ΔLPMLSurv (MC). Table 2 shows DIC, DICSurv|Long, ΔDICSurv, LPML, LPMLSurv|Long, and ΔLPMLSurv using the proposed MC method for each of the five PROs for K = 25, 30, and 35 under SPML and TML, respectively. The values of pD and pD[Surv|Long] associated with DIC and DICSurv|Long are given in Table S2 of the supplemental material. Table S3 of the supplemental material shows LPML, LPMLSurv|Long, and ΔLPMLSurv using the AGQ approach. We see from Table 2 and Table S3 that LPML (MC), LPMLSurv|Long (MC), and ΔLPMLSurv (MC) are very close to LPML (GQ), LPMLSurv|Long (GQ), and ΔLPMLSurv (GQ). We also see from Table 2 that (a) according to DICSurv|Long and LPMLSurv|Long, the joint model with K = 30 fit the longitudinal and survival data better than those models with K = 25 and K = 35 under both SPML and TML; and (b) according to DIC and LPML, SPML fit Danorexia + DSurv, Ddyspnea + DSurv, Dfatigue + DSurv, and Dpain + DSurv better than TML except for Dcough + DSurv. Among the five PROs, pain had the largest values of ΔDICSurv and ΔLPMLSurv while cough had the smallest values of ΔDICSurv and ΔLPMLSurv under both SPML and TML. These results indicate that pain led to the most gain in fitting the PFS data while cough had the least contribution to the fit of the PFS data. We mention here that the overall DIC and LPML were not able to determine the contribution of the longitudinal data in fitting the survival data for these five LCSS longitudinal measures under the joint modeling framework. From Table 2, we observe that the smallest DICLong (or largest LPMLLong) value was the main reason for dyspnea having the smallest DIC (largest LPML) value, which had no implication on the contribution of the LCSS data to the fit of the PFS data. In addition, DIC and LPML were not directly comparable among these five PROs since the total numbers of longitudinal measures were different.
Table 2.
The Decompositions of DIC and LPML for five PROs under SPML and TML with different K
| K | Model | Anorexia | Cough | Dyspnea | Fatigue | Pain | |
|---|---|---|---|---|---|---|---|
| 25 | SPML | DIC | 14022.37 | 14271.39 | 11920.73 | 13001.30 | 12843.70 |
| DICSurv|Long | 2004.52 | 2022.77 | 2007.75 | 1995.93 | 1975.15 | ||
| ΔDICSurv | 22.33 | 4.08 | 19.10 | 30.91 | 51.70 | ||
| LPML | −7015.02 | −7145.62 | −5965.58 | −6504.33 | −6428.93 | ||
| LPMLSurv|Long | −1003.19 | −1012.15 | −1004.82 | −998.63 | −988.28 | ||
| ΔLPMLSurv | 11.08 | 2.12 | 9.45 | 15.63 | 25.99 | ||
| TML | DIC | 14024.40 | 14269.30 | 11927.25 | 13007.29 | 12858.02 | |
| DICSurv|Long | 2006.64 | 2020.66 | 2015.58 | 2001.91 | 1990.13 | ||
| ΔDICSurv | 20.20 | 6.19 | 11.27 | 24.93 | 36.71 | ||
| LPML | −7016.13 | −7144.96 | −5968.82 | −6507.33 | −6436.08 | ||
| LPMLSurv|Long | −1004.31 | −1011.09 | −1008.75 | −1001.89 | −995.99 | ||
| ΔLPMLSurv | 9.96 | 3.17 | 5.51 | 12.38 | 18.27 | ||
| 30 | SPML | DIC | 14014.67 | 14262.97 | 11911.96 | 12993.00 | 12835.64 |
| DICSurv|Long | 1996.62 | 2014.65 | 1999.37 | 1987.86 | 1967.00 | ||
| ΔDICSurv | 21.86 | 3.84 | 19.11 | 30.62 | 51.49 | ||
| LPML | −7011.19 | −7141.36 | −5960.94 | −6500.14 | −6424.85 | ||
| LPMLSurv|Long | −999.00 | −1008.05 | −1000.58 | −994.65 | −984.21 | ||
| ΔLPMLSurv | 11.07 | 2.02 | 9.48 | 15.41 | 25.86 | ||
| TML | DIC | 14016.65 | 14260.43 | 11919.21 | 12999.26 | 12849.74 | |
| DICSurv|Long | 1998.91 | 2012.10 | 2007.31 | 1994.24 | 1982.24 | ||
| ΔDICSurv | 19.58 | 6.39 | 11.18 | 24.25 | 36.24 | ||
| LPML | −7012.14 | −7140.22 | −5964.61 | −6503.19 | −6431.90 | ||
| LPMLSurv|Long | −1000.25 | −1006.79 | −1004.63 | −997.96 | −991.98 | ||
| ΔLPMLSurv | 9.82 | 3.28 | 5.44 | 12.11 | 18.09 | ||
| 35 | SPML | DIC | 14018.84 | 14267.09 | 11914.40 | 12997.21 | 12839.63 |
| DICSurv|Long | 2000.43 | 2018.43 | 2002.76 | 1991.57 | 1970.54 | ||
| ΔDICSurv | 22.13 | 4.13 | 19.81 | 31.00 | 52.02 | ||
| LPML | −7013.76 | −7143.93 | −5962.76 | −6502.81 | −6427.37 | ||
| LPMLSurv|Long | −1001.53 | −1010.42 | −1002.91 | −997.12 | −986.54 | ||
| ΔLPMLSurv | 11.09 | 2.20 | 9.71 | 15.50 | 26.07 | ||
| TML | DIC | 14019.89 | 14264.71 | 11923.25 | 13002.92 | 12853.22 | |
| DICSurv|Long | 2002.30 | 2015.98 | 2011.28 | 1998.05 | 1986.04 | ||
| ΔDICSurv | 20.27 | 6.58 | 11.28 | 24.52 | 36.52 | ||
| LPML | −7014.31 | −7143.10 | −5967.13 | −6505.60 | −6434.11 | ||
| LPMLSurv|Long | −1002.67 | −1009.37 | −1007.04 | −1000.34 | −994.39 | ||
| ΔLPMLSurv | 9.95 | 3.25 | 5.58 | 12.28 | 18.23 | ||
Tables 3 and 4 show the posterior estimates and 95% highest posterior density (HPD) intervals of the hazard ratio (HR) of the overall treatment effect on PFS (β1) and the estimates (Est) of the regression coefficients α associated with the random effects under SPML and TML with K = 30, respectively. We observe that except for dyspnea under SPML, the HRs under the joint model (ranging from 0.614 to 0.634 under SPML and ranging from 0.608 to 0.636 under TML) were smaller than or close to the HR of 0.638 when fitting the PFS data alone.
Table 3.
Parameter estimates under SPML with K = 30
|
β
1
|
α
1
|
α
2
|
||||
|---|---|---|---|---|---|---|
| PRO | HR | 95% HPD Int. | Est | 95% HPD Int. | Est | 95% HPD Int. |
| Anorexia | 0.614 | (0.495, 0.756) | 0.365 | (0.202, 0.530) | 1.178 | (0.449, 1.893) |
| Cough | 0.634 | (0.516, 0.777) | 0.200 | (0.060, 0.343) | 0.608 | (−0.060, 1.230) |
| Dyspnea | 0.641 | (0.522, 0.790) | 0.203 | (0.068, 0.343) | 1.412 | (0.770, 2.069) |
| Fatigue | 0.620 | (0.498, 0.765) | 0.367 | (0.205, 0.534) | 1.437 | (0.706, 2.176) |
| Pain | 0.622 | (0.502, 0.776) | 0.349 | (0.206, 0.489) | 1.938 | (1.354, 2.537) |
When fitting the PFS data alone, the estimate and 95% HPD interval of exp(β1) are 0.638 and (0.526, 0.785).
Table 4.
Parameter estimates under TML with K = 30
|
β
1
|
α
|
|||
|---|---|---|---|---|
| PRO | HR | 95% HPD Int. | Est | 95% HPD Int. |
| Anorexia | 0.620 | (0.501, 0.760) | 0.320 | (0.186, 0.455) |
| Cough | 0.636 | (0.520, 0.782) | 0.192 | (0.064, 0.318) |
| Dyspnea | 0.631 | (0.518, 0.776) | 0.223 | (0.098, 0.340) |
| Fatigue | 0.620 | (0.501, 0.759) | 0.343 | (0.215, 0.478) |
| Pain | 0.608 | (0.491, 0.751) | 0.391 | (0.273, 0.515) |
We used the overlapping batch statistics approach with a batch size of 2000 (Meketon and Schmeiser, 1984; and Chen et al., 2000, Section 3.3) to compute the Monte Carlo (MC) standard errors of DICSurv|Long, ΔDICSurv, LPMLSurv|Long, and ΔLPMLSurv under SPML and TML. The results are reported in Table 5. From this table, we see that (i) the MC standard errors ranged from 0.074 to 0.620 for all of DICSurv|Long, ΔDICSurv, LPMLSurv|Long, and ΔLPMLSurv, which were reasonably small compared to the magnitudes of their estimated values; and (ii) the MC standard errors of LPMLSurv|Long (GQ) and LPMLSurv|Long (MC), and ΔLPMLSurv (GQ) and ΔLPMLSurv (MC) were very close, which empirically confirmed that the proposed MC approach for estimating LPMLSurv|Long and ΔLPMLSurv were as accurate as the numerical approximation approach for computing these quantities. Table 6 shows the running times in minutes on an Intel i686 processor machine with 16 GB of RAM memory using a GNU/Linux operating system for computing ΔDICSurv, ΔLPMLSurv (GQ), and ΔLPMLSurv (MC) under SPML and TML with K = 30 based on an Markov chain Monte Carlo (MCMC) sample size of 20,000. From Table 6, we see that (i) the running times for computing ΔLPMLSurv (MC) were similar to those for computing ΔDICSurv under SPML though ΔLPMLSurv (MC) required two MCMC samples; (ii) SPML required much less running time than TML; and (iii) ΔLPMLSurv (GQ) required the most running time.
Table 5.
MC Standard Errors of DICSurv|Long, ADICSurv, LPMLSurv|Long, and ΔLPMLSurv under SPML and TML with K = 30 based on an MC sample size of 20,000
| Model | Anorexia | Cough | Dyspnea | Fatigue | Pain | |
|---|---|---|---|---|---|---|
| SPML | DICSurv|Long | 0.391 | 0.263 | 0.397 | 0.396 | 0.458 |
| ΔDICSurv | 0.530 | 0.444 | 0.535 | 0.534 | 0.581 | |
| LPMLSurv|Long (GQ) | 0.248 | 0.246 | 0.200 | 0.196 | 0.225 | |
| LPMLSurv|Long (MC) | 0.248 | 0.246 | 0.201 | 0.197 | 0.222 | |
| ΔLPMLSurv (GQ) | 0.314 | 0.313 | 0.278 | 0.275 | 0.297 | |
| ΔLPMLSurv (MC) | 0.314 | 0.313 | 0.279 | 0.275 | 0.294 | |
| TML | DICSurv|Long | 0.288 | 0.346 | 0.451 | 0.512 | 0.370 |
| ΔDICSurv | 0.460 | 0.498 | 0.576 | 0.624 | 0.515 | |
| LPMLSurv|Long (GQ) | 0.104 | 0.125 | 0.114 | 0.073 | 0.074 | |
| LPMLSurv|Long (MC) | 0.104 | 0.125 | 0.114 | 0.073 | 0.074 | |
| ΔLPMLSurv (GQ) | 0.219 | 0.230 | 0.224 | 0.206 | 0.207 | |
| ΔLPMLSurv (MC) | 0.219 | 0.230 | 0.224 | 0.206 | 0.207 | |
Table 6.
Running Times in Minutes for Computing ΔDICSurv, ΔLPMLSurv (GQ), and ΔLPMLSurv (MC) under SPML and TML with K = 30 based on an MC sample size of 20,000
| Model | Anorexia | Cough | Dyspnea | Fatigue | Pain | |
|---|---|---|---|---|---|---|
| SPML | MCMC Sampling | 6.0 | 5.9 | 5.8 | 6.3 | 5.9 |
| ΔDICSurv | 16.0 | 14.9 | 18.3 | 17.7 | 16.9 | |
| ΔLPMLSurv (GQ) | 18.2 | 17.6 | 21.0 | 20.3 | 21.4 | |
| ΔLPMLSurv (MC) | 17.6 | 16.0 | 18.5 | 17.9 | 18.5 | |
| TML | MCMC Sampling | 523.9 | 526.2 | 525.7 | 521.9 | 516.6 |
| ΔDICSurv | 1142.7 | 1070.9 | 1195.1 | 1153.4 | 1245.6 | |
| ΔLPMLSurv (GQ) | 1681.2 | 1711.8 | 1704.2 | 1718.3 | 1665.4 | |
| ΔLPMLSurv (MC) | 1488.4 | 1494.2 | 1397.0 | 1486.3 | 1373.8 | |
Finally, we computed relevant quantities under the second decomposition of DIC and LPML given in Appendix B of the supplementary material to quantify the contribution of the PFS data to the fit of the longitudinal data. The results are shown in Table 7. As mentioned earlier, the total numbers of observations for these five symptoms were different, implying that ΔDICLong and ΔLPMLLong were not directly comparable for the EMPHACIS data. Therefore, we consider the relative ΔDICLong and ΔLPMLLong defined by
and
From Table 7, we see that pain had the largest relative improvement in terms of both RΔDICLong and RΔLPMLLong (MC), which were 5.00 and 6.18 under SPML and 3.52 and 4.83 under TML, and cough had the smallest relative improvement with RΔDICLong = 0.35 and RΔLPMLLong = 1.46 (MC) under SPML and RΔDICLong = 0.57 and RΔLPMLLong = 1.90 (MC) under TML. The values of RΔDICLong and RΔLPMLLong (MC) for anorexia, dyspnea, and fatigue, were 1.89 and 2.77, 1.89 and 3.04, and 2.91 and 3.89, respectively, under SPML; and 1.68 and 2.76, 1.09 and 2.33, and 2.26 and 3.43, respectively, under TML.
Table 7.
Decomposition II of DICs and LPMLs under SPML and TML with K = 30
| Model | Anorexia | Cough | Dyspnea | Fatigue | Pain | |
|---|---|---|---|---|---|---|
| Long alone | DICLong,alone | 12017.52 | 12248.60 | 9911.66 | 11005.02 | 10867.34 |
| LPMLLong,alone | −6011.72 | −6133.68 | −4959.88 | −5505.25 | −5439.62 | |
| SPML | DICSurv | 2019.87 | 2018.65 | 2019.26 | 2019.99 | 2022.59 |
| (pD[Surv]) | 37.28 | 37.32 | 36.94 | 37.06 | 36.42 | |
| DICLong|Surv | 11994.80 | 12244.31 | 9892.94 | 10973.01 | 10813.05 | |
| (pD[Long|Surv]) | 15.17 | 14.95 | 15.70 | 15.10 | 15.94 | |
| ΔDICLong | 22.72 | 4.28 | 18.73 | 32.00 | 54.29 | |
| RΔDICLong | 1.89 | 0.35 | 1.89 | 2.91 | 5.00 | |
| LPMLLong|Surv (GQ) | −5995.08 | −6124.71 | −4944.80 | −5483.84 | −5405.97 | |
| LPMLLong|Surv (MC) | −5995.08 | −6124.70 | −4944.78 | −5483.83 | −5406.03 | |
| ΔLPMLLong (GQ) | 16.64 | 8.97 | 15.08 | 21.41 | 33.65 | |
| ΔLPMLLong (MC) | 16.64 | 8.98 | 15.09 | 21.42 | 33.59 | |
| RΔLPMLLong (GQ) | 2.77 | 1.46 | 3.04 | 3.89 | 6.19 | |
| RΔLPMLLong (MC) | 2.77 | 1.46 | 3.04 | 3.89 | 6.18 | |
| TML | DICSurv | 2019.31 | 2018.87 | 2018.34 | 2019.08 | 2020.69 |
| (pD[Surv]) | 37.29 | 37.20 | 37.28 | 37.26 | 36.89 | |
| DICLong|Surv | 11997.34 | 12241.56 | 9900.87 | 10980.18 | 10829.05 | |
| (pD[Long|Surv]) | 14.15 | 14.12 | 14.03 | 14.12 | 14.53 | |
| ΔDICLong | 20.18 | 7.04 | 10.79 | 24.84 | 38.29 | |
| RΔDICLong | 1.68 | 0.57 | 1.09 | 2.26 | 3.52 | |
| LPMLLong|Surv (GQ) | −5995.20 | −6122.07 | −4948.57 | −5486.61 | −5413.59 | |
| LPMLLong|Surv (MC) | −5995.11 | −6122.04 | −4948.30 | −5486.38 | −5413.33 | |
| ΔLPMLLong (GQ) | 16.51 | 11.61 | 11.30 | 18.64 | 26.03 | |
| ΔLPMLLong (MC) | 16.60 | 11.64 | 11.58 | 18.87 | 26.29 | |
| RΔLPMLLong (GQ) | 2.75 | 1.89 | 2.28 | 3.39 | 4.78 | |
| RΔLPMLLong (MC) | 2.76 | 1.90 | 2.33 | 3.43 | 4.83 | |
5 Discussion
In this paper, we have developed two versions of the DIC and CPO decomposition as well as two sets of new criteria in Section 2 (ΔDICSurv, ΔLPMLSurv) and in Appendix B (ΔDICLong, ΔLPMLLong). The decompositions, DIC = DICLong + DICSurv|Long and LPML = LPMLLong + LPMLSurv|Long (Decomposition I), are most useful when our primary goal is to make inferences about the parameters in the survival component of the joint model while using the information from longitudinal data through the joint model. In practice, DICSurv|Long and LPMLSurv|Long can be used to select the survival component of the joint model and the main utility of ΔDICSurv and ΔLPMLSurv is to determine which longitudinal marker leads to the most gain in the fit of the survival data or which longitudinal marker is most highly associated with the survival outcome. The simulation study in Section 3 and the real data analysis in Section 4 empirically demonstrated that DICSurv|Long, LPMLSurv|Long, ΔDICSurv, and ΔLPMLSurv are quite effective and promising in selecting the survival component of the joint model and identifying the importance of longitudinal biomarkers in fitting the survival data. Decomposition II and the corresponding RΔDICLong and RΔLPMLLong criteria are useful when the main focus of a clinical trial is on the longitudinal markers and the primary goal is to make inferences about the parameters in the longitudinal component of the joint model while using the information from the survival data through the joint model. Similar to Decomposition I, DICLong|Surv and LPMLLong|Surv can be used to choose the longitudinal component of the joint model and RΔDICLong and RΔLPMLLong are useful to determine the gain in the fit of the longitudinal data while using the information from the survival data through the joint model.
In the AIC decomposition developed in Zhang et al. (2014), dim(φ1) and dim(φ2) were manually allocated to AICLong and AICSurv|Long, respectively, as the dimensions of the parameters. However, the parameters φ1 are also involved in computing AICSurv|Long. Thus, the appropriateness of these dimension allocations needs to be further validated. The DIC decomposition developed in this paper automatically calculates the dimensions of the parameters, pD[Long] and pD[Surv|Long], in DICLong and DICSurv|Long. The real data analysis in Section 4 and the results shown in Table S2 of the supplementary material empirically demonstrated that pD[Long] ≈ dim(φ1) and pD[Surv|Long] ≈ dim(φ2). Since the DIC approximates the AIC as discussed in Spiegelhalter et al. (2002) for Gaussian posteriors (or very large samples), our empirical results based on the DIC decomposition confirm that the dimension allocations of the model parameters in the AIC decomposition are quite appropriate. Both the AIC decomposition and the DIC decomposition require the numerical approximation of an intractable integral ∫ f(yi, ti, θi|φ, δi, xi, zi)dθi in (2.4) for computing the joint distribution of yi and ti. The proposed LPML decomposition avoids the calculation of this integral. As demonstrated in both the simulation study and the real data analysis, LPMLSurv|Long and ΔLPMLSurv performed equally well as DICSurv|Long and ΔDICSurv in selecting the survival model and identifying the important longitudinal markers. In addition, as shown in Table 6, LPMLSurv|Long (MC) and ΔLPMLSurv (MC) require less computing time than LPMLSurv|Long (GQ) and ΔLPMLSurv (GQ). Thus, the LPML decomposition may be potentially more useful in practice.
In Section 2, we proposed two approaches (AGQ and MC) for computing CPO related criteria. As shown in Section 4, both approaches yielded almost identical results. However, the proposed MC method requires less computing time and is more applicable to models involving high-dimensional random effects than the AGQ approach. In Section 4, the LPMLSurv|Long's were calculated based on the CPO decomposition in (2.18) by taking φ* as the posterior mean of φ. We also calculated the LPMLSurv|Long's by taking φ* as the posterior median of φ. For the EMPHACIS Data, under SPML with K = 30, the LPMLSurv|Long's calculated based on the posterior medians were −998.95, −1008.13, −1000.64, −994.54, and −984.13 for anorexia, cough, dyspnea, fatigue, pain, respectively. These values are very close to those given in Table 2. Thus, LPMLSurv|Long is relatively robust to the choice of φ*.
Hanson et al. (2011) introduced the conditional CPO and LPML. Using our notation, the conditional CPO is defined as
where is the joint posterior of (φ, θR) given DLong,oba and with the survival data deleted for the ith subject. The conditional LPML in Hanson et al. (2011) is thus defined by
For the purpose of assessing the fit of the survival data, and do correspond to CPOi,Surv|Long and LPMLSurv|Long. However, they are not the same unless the longitudinal data are independent of the survival data. Although and cannot be used to assess the overall fit of the joint model or to determine the gain in the fit of the longitudinal data while using the information from the survival data through the joint model, they are quite attractive due to computational simplicity if the primary goal is to make inferences about the parameters in the survival component. We defer to a future project for further investigation of theoretical and empirical comparisons between and LPMLSurv|Long.
Although the proposed Bayesian criteria are developed under the joint model in Section 2, they can be easily extended to models for other types of data such as longitudinal binary/ordinal response or count data as well as other types of survival models such as cure rate models, nonproportional hazards models, and competing risks models discussed in Klein et al. (2013). Furthermore, the proposed MC method for computing CPO is applicable for a variety of Bayesian models involving random effects or latent variables. The potential applications of the proposed methodology to other types of longitudinal data such as multi-dimensional longitudinal data and more complex survival data, such as survival data in the presence of competing risks and/or semi-competing risks, are currently under investigation.
In Sections 3 and 4, we carried out all computations using the FORTRAN 95 software with double precision and IMSL subroutines. The FORTRAN 95 code is available upon request. We are currently working on a user-friendly R interface of the FORTRAN code that has been developed for this paper so that it would be available to practitioners.
Supplementary Material
Acknowledgments
We would like to thank the Editor, the Associate Editor, and the two anonymous reviewers for their very helpful comments and suggestions, which have led to a much improved version of the paper. Dr. M.-H. Chen and Dr. J. G. Ibrahim's research was partially supported by NIH grants #GM70335 and #P01CA142538.
Footnotes
Supplementary Materials
In the supplementary material, we provide the details of prior specification and posterior computation (Appendix A); the development of the second decomposition (Decomposition II) of DIC and LPML (Appendix B); the proofs of identities, results, and theorems (Appendix C); and additional tables (Appendix D) for DIC, pD, and LPML for fitting survival alone with different K, pD's and pD[Surv|Long]'s for five PROs under SPML and TML with different K associated with Table 2, and the decomposition of LPML for five PROs under SPML and TML with different K using Gaussian quadrature associated with Table 2 for the EMPHACIS data in Section 4.
References
- Bridges JFP, Mohamed AF, Finnern HW, Woehl A, Hauber AB. Patients’ preferences for treatment outcomes for advanced non-small cell lung cancer: A conjoint analysis. Lung Cancer. 2012;77:224–231. doi: 10.1016/j.lungcan.2012.01.016. [DOI] [PubMed] [Google Scholar]
- Brown ER, Ibrahim JG, DeGruttola V. A flexible B-spline model for multiple longitudinal biomarkers and survival. Biometrics. 2005;61:64–73. doi: 10.1111/j.0006-341X.2005.030929.x. [DOI] [PubMed] [Google Scholar]
- Brown ER, Ibrahim JG. Bayesian approaches to joint cure rate and longitudinal models with applications to cancer vaccine trials. Biometrics. 2003;59:686–693. doi: 10.1111/1541-0420.00079. [DOI] [PubMed] [Google Scholar]
- Chen M-H. Importance-weighted marginal Bayesian posterior density estimation. Journal of the American Statistical Association. 1994;89:818–824. [Google Scholar]
- Chen M-H, Ibrahim JG, Sinha D. A new joint model for longitudinal and survival data with a cure fraction. Journal of Multivariate Analysis. 2004;91:18–34. [Google Scholar]
- Chen M-H, Shao Q-M, Ibrahim JG. Monte Carlo Methods in Bayesian Computation. Springer-Verlag; New York: 2000. [Google Scholar]
- Chi Y-Y, Ibrahim JG. Bayesian approaches to joint longitudinal and survival models accommodating both zero and nonzero cure fractions. Statistica Sinica. 2007;17:445–462. [Google Scholar]
- Chi Y-Y, Ibrahim JG. Joint models for multivariate longitudinal and multivariate survival data. Biometrics. 2006;62:432–445. doi: 10.1111/j.1541-0420.2005.00448.x. [DOI] [PubMed] [Google Scholar]
- Chib S. Marginal likelihood from the Gibbs output. Journal of the American Statistical Association. 1995;90:1313–1321. [Google Scholar]
- Crowther MJ. STJM: Stata module to fit shared parameter joint models of longitudinal and survival data. 2012 http://econpapers.repec.org/software/bocbocode/s457502.htm/
- Crowther MJ, Abrams KR, Lambert PC. Joint modeling of longitudinal and survival data. The Stata Journal. 2013;13:165–184. [Google Scholar]
- Draper D, Krnjajić M. Technical Report. Department of Applied Mathematics and Statistics, University of California; Santa Cruz: 2005. Bayesian model specification. [Google Scholar]
- DeGruttola V, Tu XM. Modeling progression of CD4-lymphocyte count and its relationship to survival time. Biometrics. 1994;50:1003–1014. [PubMed] [Google Scholar]
- DeMuro C, Clark M, Doward L, Mordin M, Gnanasakthy A. Assessment of PRO label claims granted by the FDA as compared to the EMA (2006-2010) Value in Health. 2013;16:1150–1155. doi: 10.1016/j.jval.2013.08.2293. [DOI] [PubMed] [Google Scholar]
- Geisser S, Eddy WF. A predictive approach to model selection. Journal of the American Statistical Association. 1979;74:153–160. [Google Scholar]
- Gelfand AE, Dey DK. Bayesian model choice: Asymptotics and exact calculations. Journal of the Royal Statistical Society, Series B. 1994;56:501–514. [Google Scholar]
- Gelfand AE, Dey DK, Chang H. Model Determinating Using Predictive Distributions with Implementation via Sampling-based Methods (with Discussion) In: Bernado JM, Berger JO, Dawid AP, Smith AFM, editors. Bayesian Statistics. Vol. 4. Oxford University Press; Oxford: 1992. pp. 147–167. [Google Scholar]
- Geweke J. Bayesian inference in econometrics models using Monte Carlo integration. Econometrica. 1989;57:1317–1340. [Google Scholar]
- Hanson TE, Branscum AJ, Johnson WO. Predictive comparison of joint longitudinal-survival modeling: a case study illustrating competing approaches (with Discussion) Lifetime Data Analysis. 2011;17:3–42. doi: 10.1007/s10985-010-9162-0. [DOI] [PubMed] [Google Scholar]
- Hatfield LA, Boye ME, Carlin BP. Joint modeling of multiple longitudinal patient-reported outcomes and survival. Journal of Biopharmaceutical Statistics. 2011;21:971–991. doi: 10.1080/10543406.2011.590922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield LA, Boye ME, Hackshaw MD, Carlin BP. Multilevel bayesian models for survival times and longitudinal patient-reported outcomes with many zeros. Journal of the American Statistical Association. 2012;107:875–885. [Google Scholar]
- Henderson R, Diggle PJ, Dobson A. Joint modelling of longitudinal measurements and event time data. Biostatistics. 2000;1:465–480. doi: 10.1093/biostatistics/1.4.465. [DOI] [PubMed] [Google Scholar]
- Hogan JW, Laird NM. Mixture models for the joint distribution or repeated measures and event times. Statistics in Medicine. 1997;16:239–257. doi: 10.1002/(sici)1097-0258(19970215)16:3<239::aid-sim483>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Ibrahim JG, Chen M-H, Sinha D. Bayesian methods for joint modeling of longitudinal and survival data with applications to cancer vaccine studies. Statistica Sinica. 2004;14:863–883. [Google Scholar]
- Ibrahim JG, Chen M-H, Sinha D. Bayesian Survival Analysis. Springer-Verlag; New York: 2001. [Google Scholar]
- Ibrahim JG, Chu H, Chen LM. Basic concepts and methods for joint models of longitudinal and survival data. Journal of Clinical Oncology. 2010;28:2796–2801. doi: 10.1200/JCO.2009.25.0654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein JP, van Houwelingen HC, Ibrahim JG, Scheike TH, editors. Handbook of Survival Analysis. Chapman & Hall; Boca Raton, FL: 2013. [Google Scholar]
- Lavalley MP, DeGruttola V. Model for empirical Bayes estimators of longitudinal CD4 counts. Statistics in Medicine. 1996;15:2289–2305. doi: 10.1002/(SICI)1097-0258(19961115)15:21<2289::AID-SIM449>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Law NJ, Taylor JMG, Sandler H. The joint modeling of a longitudinal disease progression marker and the failure time process in the presence of cure. Biostatistics. 2002;3:547–563. doi: 10.1093/biostatistics/3.4.547. [DOI] [PubMed] [Google Scholar]
- Meketon MS, Schmeiser BW. Overlapping batch means: Something for nothing? Proceedings of the Winter Simulation Conference. 1984:227–230. [Google Scholar]
- Patricia HJ, Gralla RJ, Liepa AM, Symanowski JT, Rusthoven JJ. Measuring quality of life in patients with pleural mesothelioma using a modified version of the Lung Cancer Symptom Scale (LCSS): psychometric properties of the LCSS-Meso. Supportive Care in Cancer. 2006;14:11–21. doi: 10.1007/s00520-005-0837-0. [DOI] [PubMed] [Google Scholar]
- Pawitan Y, Self S. Modeling disease marker processes in AIDS. Journal of the American Statistical Association. 1993;88:719–726. [Google Scholar]
- Philipson P, Sousa I, Diggle P, Williamson P, Kolamunnage-Dona R, Henderson R. joineR: Joint modelling of repeated measurements and time-to-event data. R package version 1.0-3. 2012 http://cran.r-project.org/web/packages/joineR/index.html.
- Pinheiro JC, Bates DM. Approximations to the log-likelihood function in the nonlinear mixed-effects model. Journal of Computational and Graphical Statistics. 1995;4:12–35. [Google Scholar]
- Proust-Lima C, Philipps V, Diakite A, Liquet B. lcmm: Estimation of extended mixed models using latent classes and latent processes. R package version 1.6-4. 2014 http://cran.r-project.org/web/packages/lcmm/index.html.
- Rizopoulos D. Joint Models for Longitudinal and Time-to-Event Data: With Applications in R. CRC Press/Chapman & Hall; Boca Raton, FL: 2012a. [Google Scholar]
- Rizopoulos D. JM: Joint modeling of longitudinal and survival data. R package version 1.1-0. 2012b http://rwiki.sciviews.org/doku.php?id=packages:cran:jm.
- Rizopoulos D. JMbayes: Joint modeling of longitudinal and time-to-event data under a Bayesian approach. R package version 0.5-3. 2014 http://cran.r-project.org/web/packages/JMbayes/index.html.
- Rothman M, Burke L, Erickson P, Leidy NK, Patrick DL, Petrie CD. Use of existing patient-reported outcome (PRO) instruments and their modification: the ISPOR good research practices for evaluating and documenting content validity for the use of existing instruments and their modification PRO task force report. Value in Health. 2009;12:1075–1083. doi: 10.1111/j.1524-4733.2009.00603.x. [DOI] [PubMed] [Google Scholar]
- Schluchter MD. Methods for the analysis of informatively censored longitudinal data. Statistics in Medicine. 1992;11:1861–1870. doi: 10.1002/sim.4780111408. [DOI] [PubMed] [Google Scholar]
- Siddiqui F, Liu AK, Watkins-Bruner D, Movsas B. Patient-reported outcomes and survivorship in radiation oncology: overcoming the cons. Journal of Clinical Oncology. 2014;32:2920–2927. doi: 10.1200/JCO.2014.55.0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Davidian M, Tsiatis AA. An estimator for the proportional hazards model with multiple longitudinal covariates measured with error. Biostatistics. 2002;3:511–528. doi: 10.1093/biostatistics/3.4.511. [DOI] [PubMed] [Google Scholar]
- Spiegelhalter DJ, Best NG, Carlin BP, van der Linde A. Bayesian measures of model complexity and fit. Journal of the Royal Statistical Society: Series B. 2002;64:583–639. [Google Scholar]
- Thompson JK, Westbom CM, Shukla A. Malignant mesothelioma: development to therapy. Journal of Cellular Biochemistry. 2014;115:1–7. doi: 10.1002/jcb.24642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiatis AA, Davidian M. Joint modeling of longitudinal and time-to-event data: An overview. Statistica Sinica. 2004;14:809–834. [Google Scholar]
- Vogelzang NJ, Rusthoven JJ, Symanowski J, Denham C, Kaukel E, Ruffie P, Gatzemeier U, Boyer M, Emri S, Manegold C, Niyikiza C, Paoletti P. Phase III study of pemetrexed in combination with cisplatin versus cisplatin alone in patients with malignant pleural mesothelioma. Journal of Clinical Oncology. 2003;21:2636–2644. doi: 10.1200/JCO.2003.11.136. [DOI] [PubMed] [Google Scholar]
- Wang P, Shen W, Boye ME. Joint modeling of longitudinal outcomes and survival using latent growth modeling approach in a mesothelioma trial. Health Services and Outcomes Research Methodology. 2012;12:182–199. doi: 10.1007/s10742-012-0092-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Zeger SL. Joint analysis of longitudinal data comprising repeated measures and times to events. Applied Statistics. 2001a;50:375–387. [Google Scholar]
- Xu J, Zeger SL. The evaluation of multiple surrogate endpoints. Biometrics. 2001b;57:81–87. doi: 10.1111/j.0006-341x.2001.00081.x. [DOI] [PubMed] [Google Scholar]
- Zhang D, Chen M-H, Ibrahim JG, Boye ME, Wang P, Shen W. Assessing model fit in joint models of longitudinal and survival data with applications to cancer clinical trials. Statistics in Medicine. 2014;33:4715–4733. doi: 10.1002/sim.6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Chen M-H, Ibrahim JG, Boye ME, Shen W. Assessment of fit in longitudinal data for joint models with applications to cancer clinical trials. In: Chen Z, Liu A, Qu Y, Tang L, Ting N, Tsong Y, editors. Applied Statistics in Biomedicine and Clinical Trials Design - Selected Papers from 2013 ICSA/ISBS Joint Statistical Meetings. Springer; New York: 2015a. pp. 347–365. New York: Springer. In press. [Google Scholar]
- Zhang D, Chen M-H, Ibrahim JG, Boye ME, Shen W. JMFit: A SAS Macro for Joint Models of Longitudinal and Survival Data. Journal of Statistical Software. 2015b doi: 10.18637/jss.v071.i03. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.